- 1Institute of Plant Biology and Department of Life Science, National Taiwan University, Taipei, Taiwan
- 2Institute of Plant Biology, Center of Evolutionary Biology, School of Life Sciences, Fudan University, Shanghai, China
The manganese (Mn) tracking factor for mitochondrial Mn superoxide dismutase (MnSOD) has been annotated as yMTM1 in yeast, which belongs to the mitochondrial carrier family. We confirmed that Arabidopsis AtMTM1 and AtMTM2 are functional homologs of yMTM1 as they can revive yeast MnSOD activity in yMTM1-mutant cells. Transient expression of AtMnSOD-3xFLAG in the AtMTM1 and AtMTM2-double mutant protoplasts confirmed that AtMTM1 and AtMTM2 are required for AtMnSOD activation. Our study revealed that AtMnSOD interacts with AtMTM1 and AtMTM2 in the mitochondria. The expression levels of AtMTM1, AtMTM2, and AtMnSOD respond positively to methyl viologen (MV) and metal stress. AtMTM1 and AtMTM2 are involved in Mn and Fe homeostasis, root length, and flowering time. Transient expression of chloroplast-destined AtMnSOD revealed that an evolutionarily conserved activation mechanism, like the chloroplastic-localized MnSOD in some algae, still exists in Arabidopsis chloroplasts. This study strengthens the proposition that AtMTM1 and AtMTM2 participate in the AtMnSOD activation and ion homeostasis.
Introduction
Cellular reactive oxygen species (ROS) contain superoxide anion radicals, hydroxyl radicals, singlet oxygen, and hydrogen peroxide, whose generation is induced by high light intensity, heat, drought, and salt stress. Superoxide anion radicals are mainly generated from the respiratory and photosynthetic electron transport chains in the mitochondria and chloroplasts and can rapidly damage nearby cell components. The superoxide dismutases (SODs) catalyze the conversion of toxic superoxide anion radicals to oxygen and hydrogen peroxide; the corresponding cofactors in SODs are transition metal ions that accept or donate an electron during the dismutation process (Fridovich, 1975; Halliwell, 1994; Apel and Hirt, 2004).
Superoxide dismutases are classified as CuZnSOD, FeSOD, Mn superoxide dismutase (MnSOD), or NiSOD based on their metal cofactors. These metalloenzymes are important for cell survival under oxidative stress (Bowler et al., 1994). Most eukaryotes contain CuZnSOD and MnSOD, while plants also contain FeSOD (Alscher et al., 2002; Fink and Scandalios, 2002). NiSOD is present in Streptomyces and cyanobacteria (Choudhury et al., 1999; Barondeau et al., 2004; Priya et al., 2007). In Arabidopsis (Arabidopsis thaliana), seven SOD isoforms are distributed among various organelles, including cytosolic CuZnSOD1, chloroplastic CuZnSOD2, and peroxisomal CnZnSOD3, as well as three chloroplastic FeSODs and one mitochondrial MnSOD (Kliebenstein et al., 1998; Kuo et al., 2013a,b). MnSOD is also found in the thylakoid membrane of some species of green and blue–green algae.
The SOD enzyme activation requires a metallochaperone or transporter that captures and loads the metal cofactor into the SOD apoprotein. The pathway using copper chaperone of SOD1 (CCS) for CuZnSOD activation is referred to as the CCS-dependent pathway (Casareno et al., 1998; Rae et al., 2001; Chu et al., 2005; Culotta et al., 2006). The chloroplast chaperonin Cpn20 functions as an Fe chaperone for FeSOD activation (Kuo et al., 2013a,b). The manganese (Mn) tracking factor for mitochondrial MnSOD activation was annotated as yMTM1 in yeast; its homolog AtMTM1 can complement yeast MnSOD (ySOD2) activity in yMTM1 mutant cells (Luk et al., 2003; Su et al., 2007). Notably, Mn supply increased the ySOD2 activity in yeast cells, and the ySOD2 protein could not be activated in the cytosol, suggesting that Mn insertion is linked to the ySOD2 importing process via yMTM1 in mitochondria (Luk et al., 2005).
There are 35 members of the mitochondrial carrier family in yeast, and more than 50 members in plants and humans, which are evolutionarily conserved for transporting cofactors and specific substrates (Haferkamp and Schmitz-Esser, 2012). Arabidopsis AtMTM1 and AtMTM2 are classified as mitochondrial carriers with conserved transmembrane sequences, and their amino acid sequences are highly homologous in the phylogeny (Picault et al., 2004; Palmieri et al., 2011). To date, the physiological roles of AtMTM1 and AtMTM2 in MnSOD activation have not been fully elucidated.
This study revealed that the effects of AtMTM1 and AtMTM2 on MnSOD activation in yMTM1 mutant cells and AtMTM1 and AtMTM2-double mutant plants are similar. We confirmed that AtMTM1 and AtMTM2 are associated with root length and flowering time. We also found that AtMTM1 and AtMTM2 have different responses under stress, and both play a role in Mn homeostasis. Finally, our study revealed that the activation mechanism of the chloroplastic-localized MnSOD in algae is retained in Arabidopsis chloroplasts.
Materials and Methods
Plants and Growth Condition
Arabidopsis (A. thaliana) accession Columbia-0 (Col) was used as the wild-type (WT). T-DNA insertion mutants of mtm1-1 (SALK 023286), mtm1-2 (SALK 054287C), mtm2-1 (SALK 005166), mtm2-2 (SALK 103984), and mtm2-3 (SALK 025071) were obtained from the Arabidopsis Biological Resource Center (ABRC, Ohio State University, Columbus, United States). Plants were grown in growth chambers at 22–24°C with 8 h dark/16 h light at 80–100 μmol m–2 s–1. For root length measurements, sterile seeds were placed on solid half-strength Murashige and Skoog (1962) basal medium (1/2 MS; Sigma M5519) containing 1% sucrose and 0.8% (w/v) phytagel (Sigma, Ronkonkoma, NY, USA). For gene regulation, metal stress, and metal ion homeostasis, 14-day-old complete seedlings were transferred to Milli-Q water containing methyl viologen (MV; paraquat), metals, or MnCl2. Transgenic plants were generated by Agrobacterium tumefaciens GV3101-mediated transformation and the floral dip method (Clough and Bent, 1998). The flowering time was scored when the primary inflorescence reached 5 cm in length.
Yeast Strains and Growth Condition
Yeast (Saccharomyces cerevisiae) used in this study contained WT BY4741 (Y00000; MATa, his3Δ1, leu2Δ0, met15Δ0, and ura3Δ0) and BY4742 (Y10000; MATα, his3Δ1, leu2Δ0, lys2Δ0, and ura3Δ0). The mutant strains of ysod2Δ (Y06605; sod2:kanMX4) and ymtm1Δ (Y07288; ygr257c:kanMX4) were obtained from the Saccharomyces Genome Deletion Project1. Enriched yeast extract–peptone–dextrose (YPD) medium was supplemented with 2% glucose to culture yeast at 30°C under aerobic conditions (Brachmann et al., 1998). For yeast expression, all cDNAs were cloned into the yeast expression vector pADNS (Colicelli et al., 1989). Yeast transformation was performed according to the lithium acetate procedure (Gietz and Schiestl, 1991), and transformants were selected on minimal synthetic dextrose (SD) media.
Western Blotting and SOD Activity Analysis
Yeast lysate was extracted by using the glass bead lysis method (Culotta et al., 1997). Total protein from Arabidopsis seedling was isolated by grinding 100 mg of frozen tissue in 300 μl of ice-cold buffer of 50 mM potassium phosphate (pH 7.8), 0.1% BSA, 0.1% ascorbate, and 0.05% 2-mercaptoethanol (Van Camp et al., 1994), as well as in 150 mM Tris–HCl buffer (pH 7.2) (Pan and Yau, 1992; Chen and Pan, 1996; Chu et al., 2005; Kuo et al., 2013c). Supernatants were collected by centrifuging twice for 10 min at 16,000 × g at 4°C in Eppendorf tube, and protein concentration in the supernatants was determined using the Bradford protein assay method (Bradford, 1976). An equal amount of proteins were separated immediately on a 10% non-denaturing polyacrylamide gel for in-gel SOD activity assay (Beauchamp and Fridovich, 1971; Kliebenstein et al., 1998; Kuo et al., 2013c). A 12.5% denaturing polyacrylamide gel electrophoresis was used for western blotting with antibodies of α-ADH1 (Sigma-Aldrich, St. Louis, MO, USA), α-FLAG (Sigma-Aldrich), α-Actin (Agrisera), and α-AtMnSOD (Agrisera, Västerbäck, Vännäs, Sweden). The SOD activity was quantified by the UVP ChemStudio PLUS imaging system (Analytik Jena US LLC, Upland, CA, United States).
Genomic DNA and RNA Preparations, cDNA Synthesis, and Quantitative Real-Time PCR
Genomic DNA was extracted for genotyping using PCR, as previously reported (Edwards et al., 1991). Total RNA was prepared with TRIZOL reagent (Invitrogen, Carlsbad, CA, USA) and TURBO DNA-free Kit (Applied Biosystems, Foster City, CA, USA). cDNA synthesis was performed with Ready-To-Go Kit (GE Healthcare Life Sciences, Stockholm, Sweden) and analyzed by reverse transcription PCR (RT-PCR). PCR primers were designed by Primer32. Quantitative real-time PCRs (qPCRs) with iQ SYBR Green Supermix (Bio-Rad, Techview, Singapore) were performed by using MyiQ thermocycler and iQ5 optical system (Bio-Rad). qPCR data were normalized to the internal control of PP2AA3 (PP2A) (Czechowski et al., 2005).
Subcellular Localization and Bimolecular Fluorescence Complementation
The coding regions of AtMTM1, AtMTM2, and AtMnSOD genes were amplified by RT-PCR, and cloned into pCR8/GW/TOPO vector (Invitrogen) for sequencing. Constructs were further recombined into pEarleyGate101 vector that contains yellow fluorescent protein (YFP) for subcellular localization or into pEarleyGate201-YN and pEarleyGate202-YC for bimolecular fluorescence complementation (BiFC) assay (Lu et al., 2010). Four-week-old plants were used for protoplast preparation and transfection (Yoo et al., 2007). Transient expression of 15 μg plasmid DNA in 2 × 104 protoplasts was performed in BiFC assay, and YFP signals were observed by using a TCS SP5 confocal microscope (Leica, Wetzlar, German). Mitochondria were stained with 200 nM MitoTracker CMTMRos (Thermo Fisher Scientific, Fremont, CA, USA). The fluorescence signals of YFP, MitoTracker, and chlorophyll were all excited at 514 nm and recorded at 530, 580, and 660 nm, respectively.
Generation of AtMTM1-miRNA Mutants, Complementation Lines With AtMTM2, and AtMTM2 Histochemical Analysis
The 21mer artificial miRNA of AtMTM1 was cloned into the pRS300 vector which contains the miR319a precursor (Schwab et al., 2006). The artificial miRNA3 was designed and cloned into pPZP200GB vector (Chu et al., 2005) for constructing miRNA-mediated AtMTM1 suppression (mtm1-i) lines. The construct of 35S:AtMTM2 with pCAMBIA1300 vector (CAMBIA) was transformed into mtm1-i, mtm2, and mtm1-i mtm2-double mutants background. These complementation lines with AtMTM2 transgene were selected by hygromycin and kanamycin. In addition, a 2-kb AtMTM2 promoter region was subcloned into pCAMBIA3300 vector (CAMBIA) with a GUS reporter for analyzing the AtMTM2 promoter activity. The GUS staining method was performed as described (Weigel and Glazebrook, 2002).
Inductively Coupled Plasma Optical Emission Spectrometry
Inductively coupled plasma optical emission spectrometry (ICP-OES) (Perkin Elmer Optima 5300 DV) was conducted to evaluate the metal ion content (Lahner et al., 2003; Yang et al., 2008; Lin et al., 2009); 14-day-old complete seedlings were transferred to the 1/2 MS medium containing 100 μM MnCl2 for 24 h with agitation. Aerial parts (shoots) and roots were separated and dried at 60°C for 3 days. An amount of 0.1 g dried tissue sample was analyzed by using the ICP-OES. Spinach and tomato leaves (SRM-1570a and SRM-1573a; US National Institute of Standards and Technology) were the standard references to validate the measurement.
Constructs of Different Organelle-Destined AtMnSOD
The cytosol-destined and chloroplast-destined AtMnSOD constructs were generated (Supplementary Figure 1A). The deletion of the mitochondrial transit peptide of AtMnSOD resulted in cytosol-localized AtMnSOD (Δ-TP-AtMnSOD). The mitochondrial transit peptide of AtMnSOD replaced by the chloroplast transit peptide of Arabidopsis CuZnSOD2 (Huang et al., 2012) became chloroplast-destined AtMnSOD (Chl-TP-AtMnSOD). They were further recombined into the pEarleyGate101 vector for confirmation of subcellular localization. Besides, AtMnSOD-Tag (a tag of 15 amino acids derived from the vector) and AtMnSOD-3xFLAG were generated (Supplementary Figures 1B,C) in pEarleyGate101 vector for transient expression assay in protoplasts.
Statistical Analysis
All experiments were repeated independently at least three times. Statistical analysis involved Student’s t-test and ANOVA with Tukey’s HSD post hoc test. P < 0.05 was considered statistically significant.
Primers and Accession Numbers
Primers and accession numbers are listed in Supplementary Table 1.
Results
Protein Structure of AtMTM1, AtMTM2, and yMTM1
We characterized AtMTM1 and AtMTM2 by sequence alignment with reported yMTM1 (Luk et al., 2003; Su et al., 2007), and the results showed that they have high sequence similarity (Figure 1). Each protein sequence consists of three tandem homologous domains, and each domain contains two transmembrane α-helices. Three copies of a 10-amino acid sequence motif are the mitochondrial energy transfer signatures of the mitochondrial carrier family (Millar and Heazlewood, 2003; Robinson and Kunji, 2006). AtMTM1 and AtMTM2 share a conserved Mn-binding site with yMTM1. Protein BLAST4 revealed that AtMTM2 has 59% protein identity with AtMTM1 and 34% protein identity with yMTM1. Sequence alignment and motif structure indicated that AtMTM1 and AtMTM2 could be homologs of yMTM1.
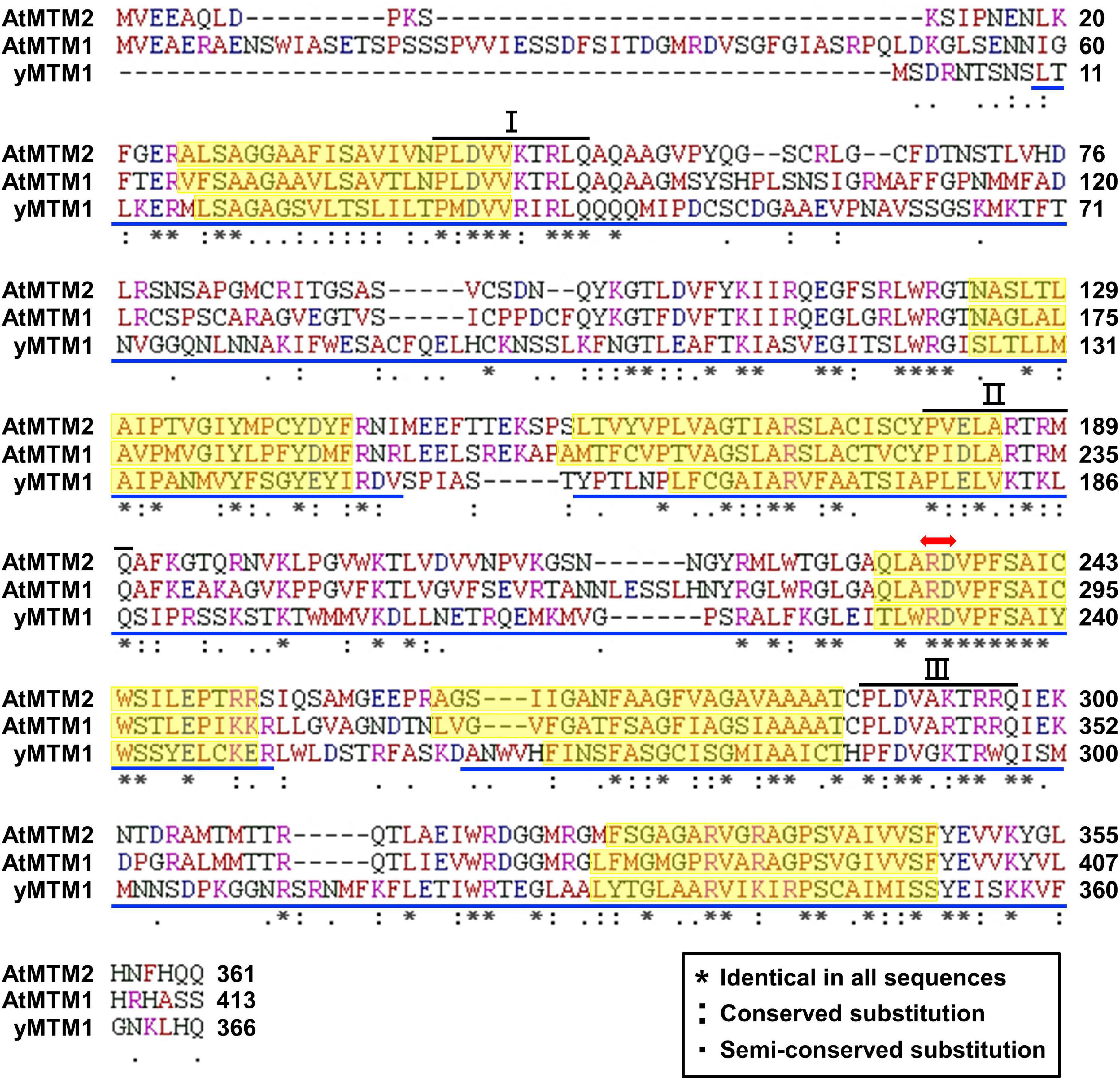
Figure 1. Sequence alignment of AtMTM1, AtMTM2, and yMTM1. Tandemly repeated domains (blue underline), transmembrane α-helices (yellow shading), mitochondrial energy transfer signature (I, II, and III), and substrate Mn-binding site (red double-headed arrow) are indicated. Consensus symbols of identical, conserved, and semi-conserved residues are shown in inset.
Characterization of AtMTM1, AtMTM2, and AtMTM1 AtMTM2-Double Mutant Lines
To elucidate the physiological functions of AtMTM1 and AtMTM2, we characterized T-DNA insertion mutants of mtm1-1 and mtm1-2 and established microRNA-mediated AtMTM1 mutant (mtm1-i) lines. Homozygous T-DNA insertion mutants of mtm2-1, mtm2-2, and mtm2-3 were also analyzed.
In mtm1-1 and mtm1-2 mutants, T-DNA was inserted in the 3′-UTR and 5′-UTR of the AtMTM1 gene, respectively (Supplementary Figure 2). Homozygous lines were confirmed by PCR-based genotyping. RT-PCR and qPCR revealed that mtm1-1 and mtm1-2 are knockdown mutants. Moreover, five independent T4 lines of the mtm1-i mutant were screened (Supplementary Figure 3). The lowest expression line retained approximately 20% of the AtMTM1 expression and was referred to as mtm1-i; however, AtMnSOD activity in mtm1-i remained the same as in the WT.
In the mtm2-1 mutant, T-DNA was inserted in the last exon of the AtMTM2 coding region (Supplementary Figure 4). Genotyping, RT-PCR, and qPCR confirmed that mtm2-1 is a null mutant. AtMnSOD activity in mtm2-1 was also similar to that in the WT. In mtm2-2 and mtm2-3 mutants, T-DNA was inserted in the intron, but AtMTM2 expression was not significantly affected. In the following part of the study, mtm2-1 refers to the null mutant mtm2.
To determine the synergistic effect of AtMTM1 and AtMTM2, we crossed mtm1-i and mtm2 to generate mtm1-i mtm2-double mutants and screened the lowest gene expression line (Supplementary Figure 5). Three individual mtm1-i mtm2-double mutants exhibited 70% AtMnSOD activity and early flowering phenotype.
Functional Homologs of AtMTM1, AtMTM2, and yMTM1 in MnSOD Activation
To analyze the role of AtMTM1 and AtMTM2 in yeast MnSOD (ySOD2) activation, we transformed AtMTM1 and AtMTM2 transgenes into ymtm1Δ cells (Figure 2). WT yeast cells contained two activity bands of ySOD2 and one band of CuZnSOD (ySOD1), with the residual ySOD2 activity retained in ymtm1Δ cells (Figure 2, lanes 1 and 2). The activity of ySOD2 was restored by the transformation of AtMTM1 and AtMTM2 transgenes in ymtm1Δ cells (Figure 2, lanes 3 and 4). Co-transformation of AtMTM1 and AtMTM2 restored the ySOD2 activity similar to that of the WT (Figure 2, lane 5). Thus, AtMTM1 and AtMTM2 are functional homologs of yMTM1.
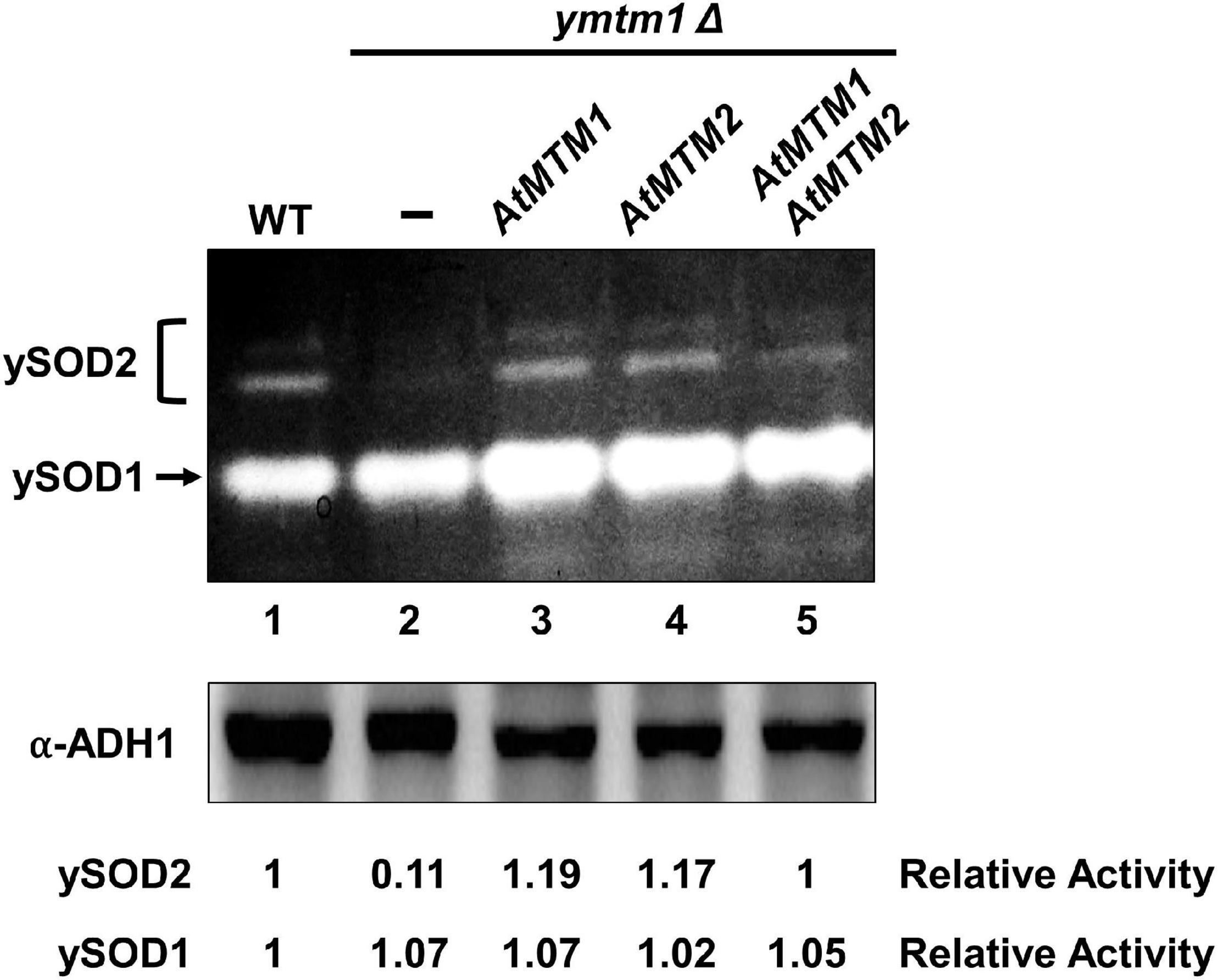
Figure 2. Restored yeast MnSOD activities by AtMTM1 and ATMTM2 in ymtm1Δ cells. ymtm1Δ cells were transformed without (–) and with AtMTM1, AtMTM2, and both. Yeast MnSOD (ySOD2) and CuZnSOD (ySOD1) activities are indicated by bracket and arrow, respectively (top). Western blotting with α-alcohol dehydrogenase1 (ADH1) antibody was a loading control (bottom). Relative activity represents ratio of ySOD2 or ySOD1 activity to that in WT (lane 1).
We also performed a transient expression assay to reveal the role of AtMTM1 and AtMTM2 in AtMnSOD activation in plant cells. We transfected the AtMnSOD-3xFLAG transgene in Arabidopsis WT and mtm1-i mtm2-double mutant protoplasts and monitored them for 16 h (Figure 3). Endogenous AtMnSOD and exogenous AtMnSOD-3xFLAG activities can be distinguished using the in-gel SOD activity assay (Figure 3, top). We compared the relative activity ratio (1:0.09) and the relative protein ratio (1:0.87) of transient-expressed AtMnSOD-3xFLAG between WT and double mutant, and the double mutant exhibited about 10-fold reduction in AtMnSOD-3xFLAG activity with the transfection of 10 μg plasmid DNA (Figure 3, bottom). In addition, endogenous AtMnSOD activity was lower in the double mutant than that in the WT. Overall, it was evident that AtMTM1 and AtMTM2 are crucial for the AtMnSOD activation.
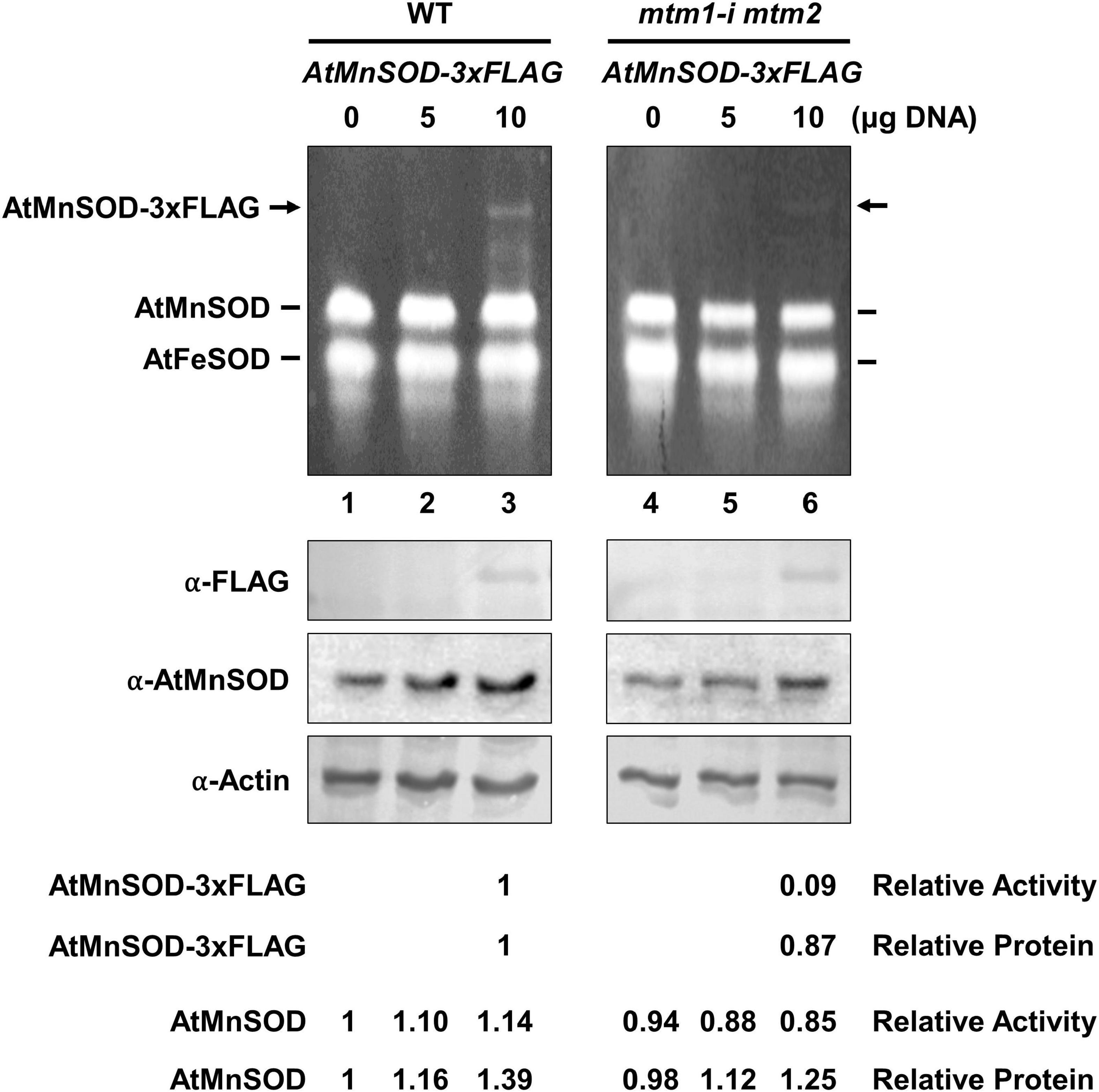
Figure 3. Transient expression of AtMnSOD-3xFLAG transgene in WT and mtm1-i mtm2-double mutant protoplasts; 5 and 10 μg plasmid DNA containing AtMnSOD-3xFLAG transgene were transfected in 106 protoplasts and monitored for 16 h. In-gel SOD activity assay (top) and western blotting with α-FLAG, α-AtMnSOD, and α-actin antibodies (bottom) were conducted. Actin was a loading control. AtMnSOD-3xFLAG activities and proteins in double mutant cells were measured relative to those in WT (lane 3). AtMnSOD activities and proteins were measured relative to those without transfection (lane 1). Data represent one of three independent repeats.
Subcellular Localization of AtMTM1 and AtMTM2 and Interaction With AtMnSOD
To examine the subcellular localization of AtMTM1 and AtMTM2, we transfected AtMTM1-YFP and AtMTM2-YFP transgenes in Arabidopsis protoplasts (Figure 4A). The subcellular localization of AtMTM1-YFP and AtMTM2-YFP was examined by using the confocal microscopy analysis. Using the MitoTracker dye, we showed that they merged well with the mitochondria rather than the chloroplasts. This result is in agreement with the proteomic evidence in mitochondrial AtMTM1 and AtMTM2 (Senkler et al., 2017).
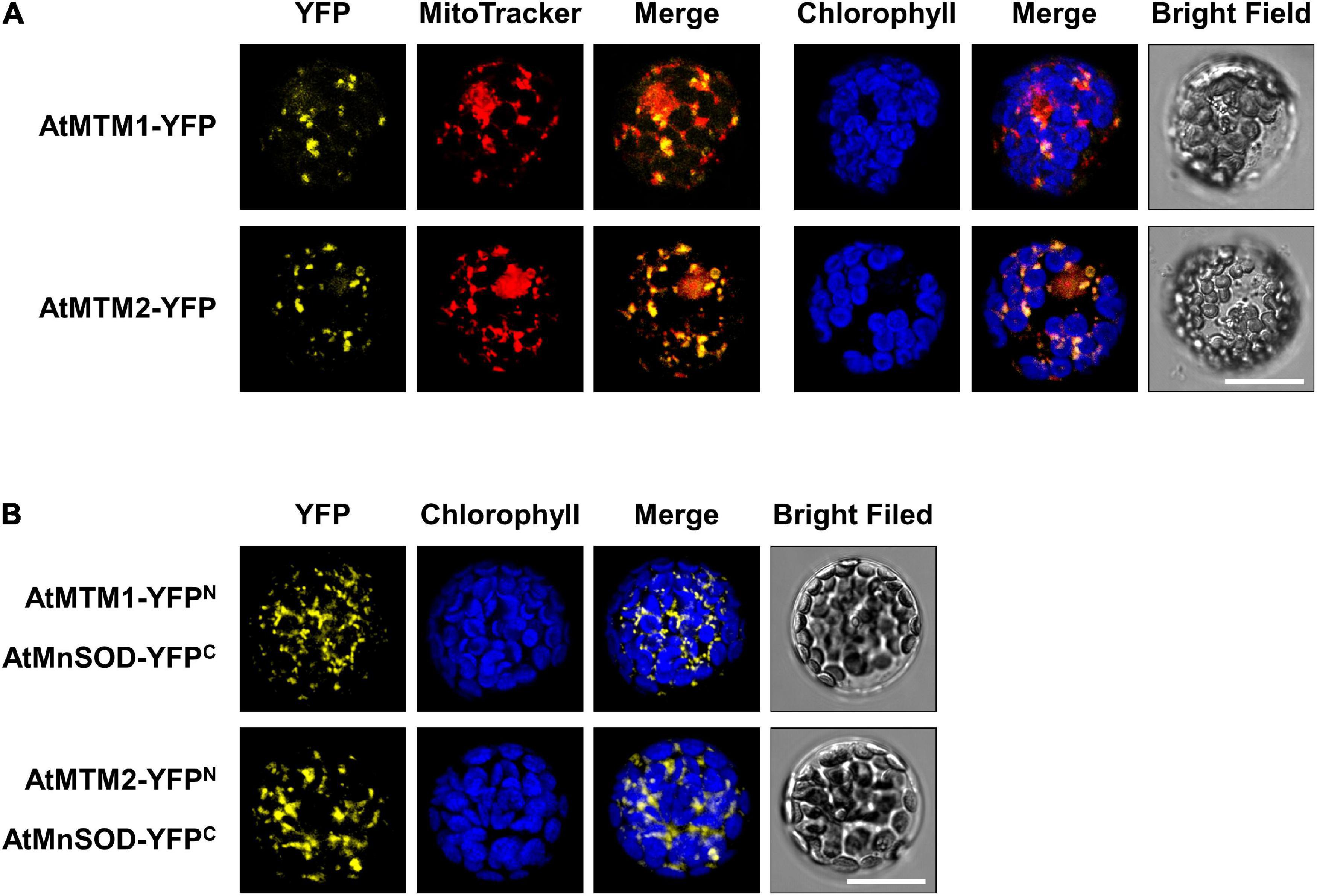
Figure 4. Localization of AtMTM1 and AtMTM2 to mitochondria and their interaction with AtMnSOD. (A) AtMTM1-YFP and AtMTM2-YFP were transfected in Arabidopsis protoplasts, and YFP signals were observed by confocal microscopy. (B) BiFC assay revealed that AtMnSOD interacted with AtMTM1 and AtMTM2. Reconstituted YFP signals were observed in mitochondria. MitoTracker staining and chlorophyll autofluorescence were used to identify mitochondria and chloroplasts, respectively. Bars = 20 μm.
A BiFC assay was carried out to examine the protein–protein interactions of AtMnSOD–AtMTM1 and AtMnSOD–AtMTM2 (Figure 4B). The reconstituted YFP signals revealed that AtMnSOD interacts with AtMTM1 and AtMTM2 in the mitochondria. There were no YFP fluorescent signals in the negative controls of the BiFC assay in healthy protoplasts (Supplementary Figure 6).
Ubiquitous Expressions of AtMTM1 and AtMTM2 During Development
The tissue-specific expression of AtMTM1 was ubiquitous, except in the siliques (Su et al., 2007). The transgenic plants of AtMTM2-promoter:GUS were established and stained for GUS activity (Supplementary Figure 7). AtMTM2 expression was clear in the hypocotyls, roots, and cotyledons at an early stage. It was also expressed in the trichomes, vascular bundles, stems, rosettes, and cauline leaves of seedlings. In particular, AtMTM2 was detected in the floral parts, namely, sepals, petals, anthers, pollen, and stigma, as well as in the young siliques. Overall, AtMTM2 was ubiquitously expressed during development.
Expression Levels of AtMTM1, AtMTM2, and AtMnSOD During Development and in Response to Metal Stress
The expression levels of AtMTM1, AtMTM2, and AtMnSOD genes in roots, rosette leaves, cauline leaves, stems, flowers, and siliques were examined by qPCR (Figure 5A). AtMTM1 was higher than AtMTM2 in the roots, stems, and siliques, while AtMTM2 was dominant in leaves. Notably, the expression levels of both AtMTM1 and AtMTM2 were substantial in flowers. In addition, AtMnSOD was consistently higher during development, but was lower in flowers and siliques.
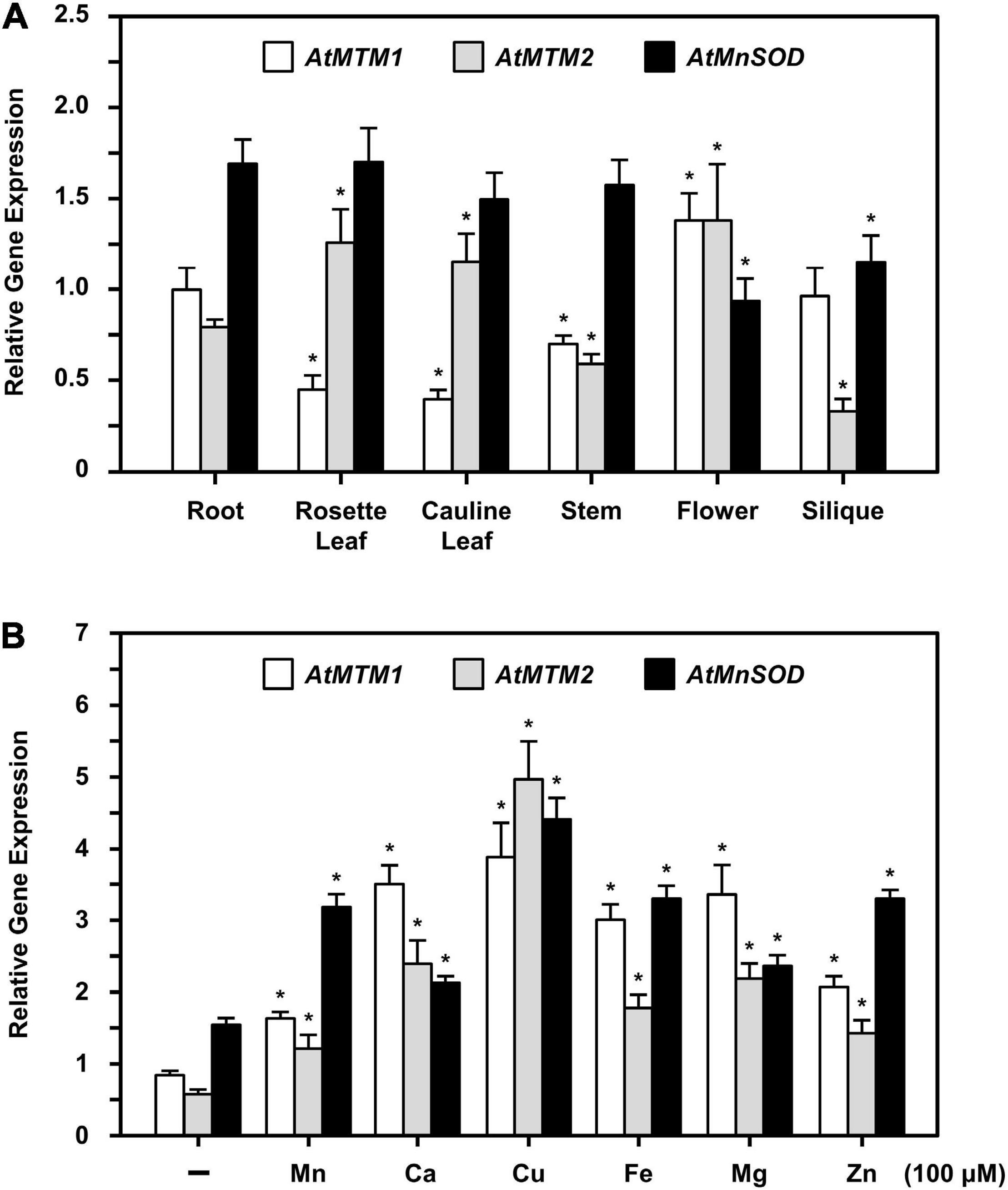
Figure 5. Quantitative PCR analysis of expressions of AtMTM1, AtMTM2, and AtMnSOD in different tissues and under metal stresses. (A) Expression levels of AtMTM1, AtMTM2, and AtMnSOD genes were measured relative to AtMTM1 level in root in WT. (B) 14-day-old complete WT seedlings were treated without (–; control) and with metal ions for 24 h. Gene expression levels in response to 100 μM MnCl2, CaCl2, CuSO4, Fe citrate, MgCl2, or ZnSO4 were measured relative to AtMTM1 level in control. Data are mean ± SE of three independent repeats. *Significant at P < 0.05 (Student’s t-test). PP2A was an internal control.
To elucidate the effect of metal stress on AtMTM1, AtMTM2, and AtMnSOD gene regulation, we transferred 14-day-old complete seedlings to Milli-Q water containing 100 μM of Mn (MnCl2), Ca (CaCl2), Cu (CuSO4), Fe (Fe citrate), Mg (MgSO4), or Zn (ZnSO4) with gentle agitation for 24 h, after which the gene expression corresponding to each metal stress was examined by qPCR (Figure 5B). All three genes were upregulated under metal stress as compared to those under the unstressed condition. In particular, the expression levels of AtMTM1 were higher than those of AtMTM2 under all metal stress conditions except Cu stress, implying that AtMTM1 and AtMTM2 have different expression sensitivities in response to different metal stresses.
Divergent Effects of AtMTM1 and AtMTM2 in Response to Oxidative Stress
Altered root elongation is an indication of oxidative stress during plant growth (Tsukagoshi, 2012); we used MV as an oxidative stress inducer. We monitored the root lengths of mtm1-i and mtm2 and the complementation lines with the AtMTM2 transgene in mtm1-i and mtm2 backgrounds (i.e., 35S:AtMTM2/mtm1-i and 35S:AtMTM2/mtm2) on MV-containing plates from day 8 to day 10 (Figure 6). The treatment period and MV concentration varied depending on the mutant, and thus, we used 8- to 10-day-old seedlings to compare the complementary effect of AtMTM2 on mutants with different backgrounds.
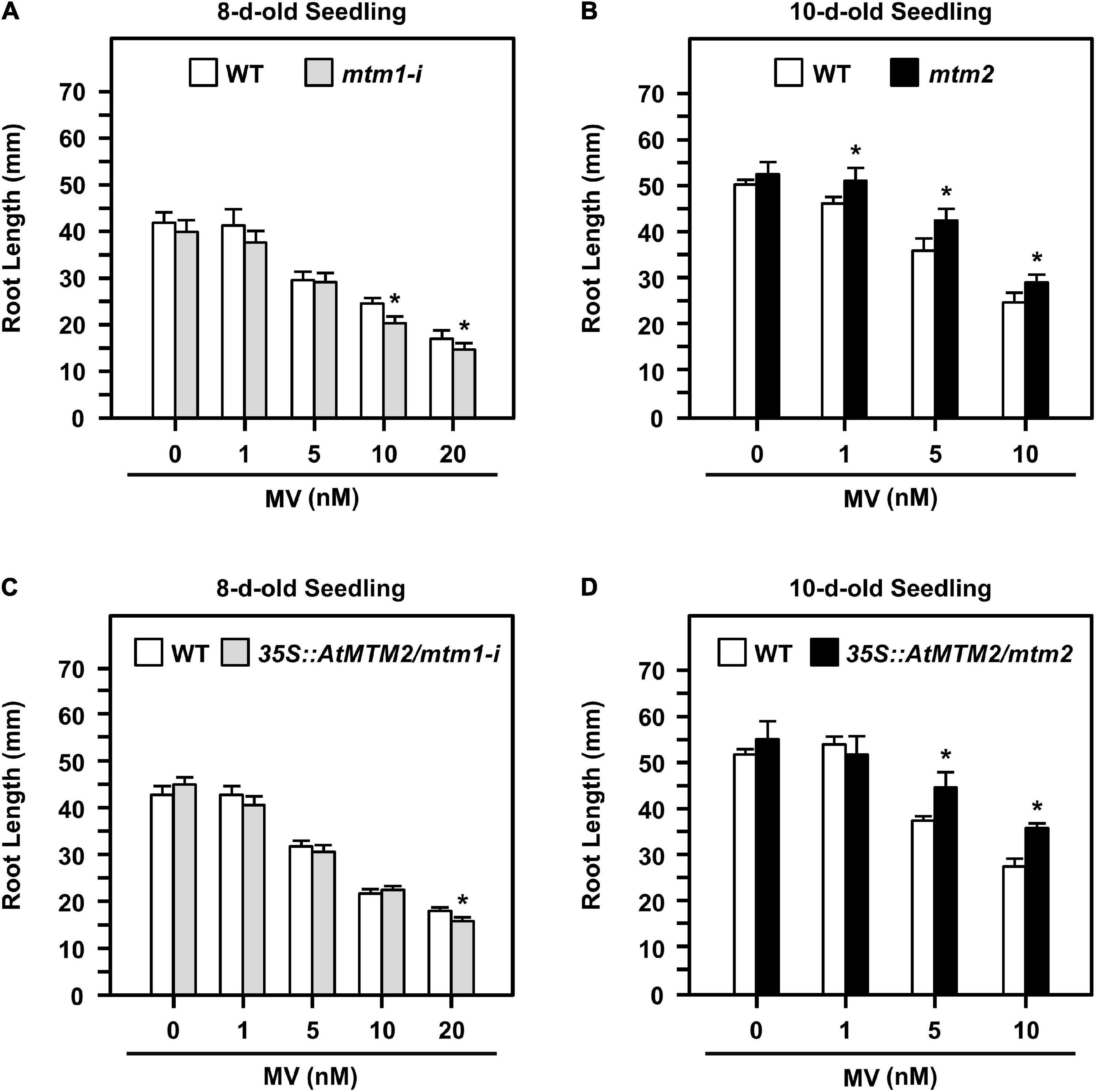
Figure 6. Root lengths of mtm1-i, mtm2, and complementation lines with AtMTM2 transgene in mtm1-i and mtm2 backgrounds in response to MV stress. (A,B) Root lengths of mtm1-i grown on 1–20 nM MV plates on day 8 and mtm2 grown on 1–10 nM MV plates on day 10, respectively. (C,D) Root lengths of complementation lines with AtMTM2 transgene in mtm1-i and mtm2 backgrounds (35S:AtMTM2/mtm1-i and 35S:AtMTM2/mtm2) on day 8 and day 10, respectively. Data are mean ± SE of three independent repeats (n = 50). *Significant at P < 0.05 (Student’s t-test).
The 8-day-old mtm1-i seedlings indicated a shorter root length phenotype compared with the WT in response to 10 and 20 nM MV treatments (Figure 6A); however, the 10-day-old mtm2 seedlings showed longer root length in response to 1, 5, and 10 nM MV treatments (Figure 6B). The complementation lines of 35S:AtMTM2/mtm1-i recovered root length under 10 nM MV on day 8 (Figure 6C), and the 35S:AtMTM2/mtm2 recovered root length under 1 nM MV on day 10 (Figure 6D). Overall, AtMTM1 and AtMTM2 are associated with primary root length control.
Quick and Positive Response of AtMnSOD, AtMTM1, and AtMTM2 to Oxidative Stress
To access the timing regulation of AtMTM1 and AtMTM2 for AtMnSOD activation, we treated seedlings with MV at different time periods (Figure 7). We examined AtMnSOD activity and protein levels in 14-day-old complete seedlings after 0.1, 1, 5, and 10 μM MV treatments for 24 h (Figure 7A). The AtMnSOD activity increased gradually with an increased dose of MV (Figure 7A, top), but the AtMnSOD protein level remained the same (Figure 7A, bottom). This result indicates the posttranscriptional regulation of AtMnSOD activity under MV stress.
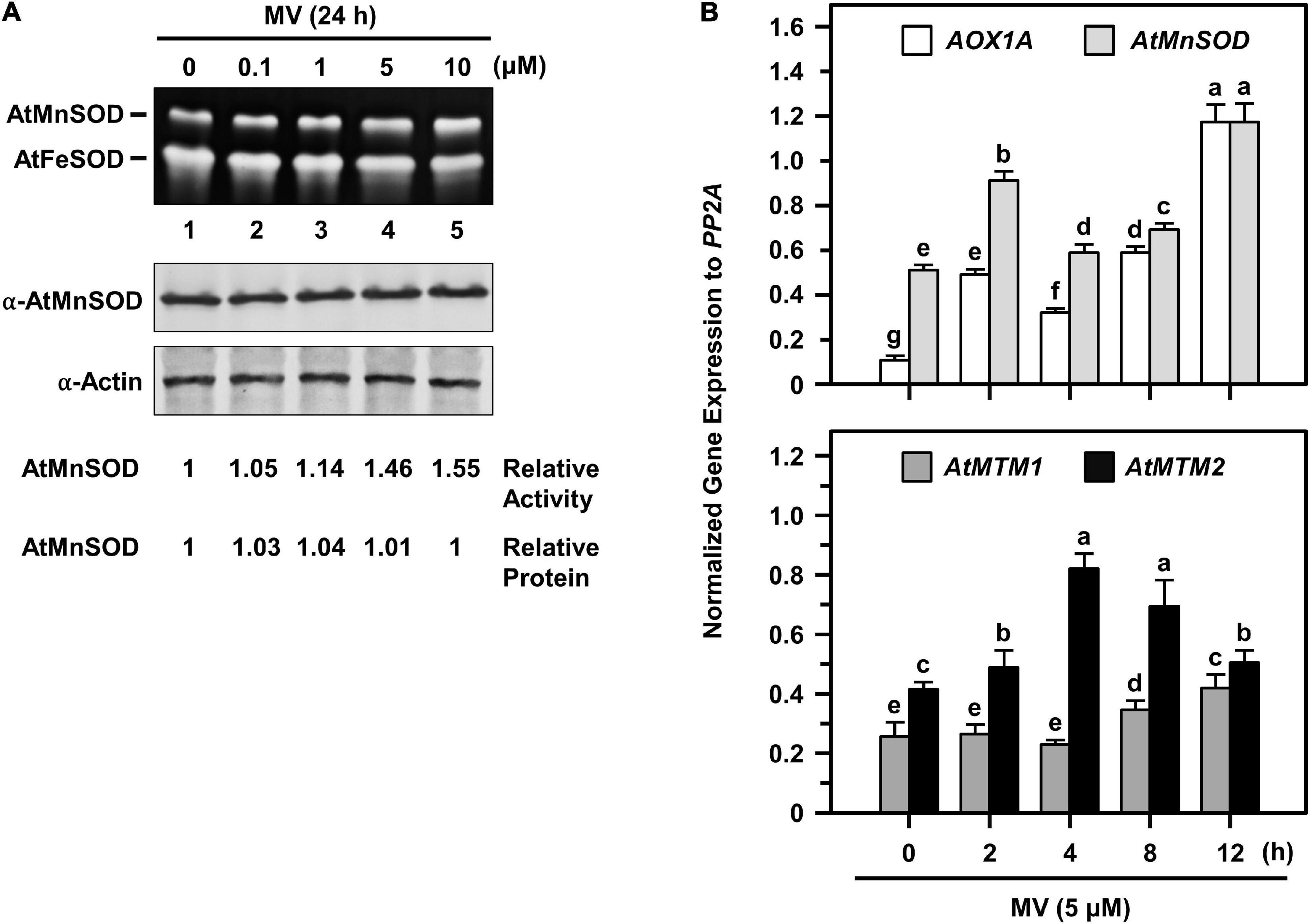
Figure 7. AtMnSOD regulation and gene expressions of AtMnSOD, AtMTM1, and AtMTM2 under MV stress. (A) 14-day-old complete WT seedlings were treated with 0.1–10 μM MV for 24 h. In-gel SOD activity assay (top) and western blotting with α-AtMnSOD and α-actin antibodies (bottom) were conducted. Actin was a loading control. AtMnSOD activities and proteins were measured relative to those in control without treatment (lane 1). (B) Complete WT seedlings were treated with 5 μM MV for 2–12 h. Expression levels of AOX1A, AtMnSOD, AtMTM1, and AtMTM2 were analyzed by qPCR. Mitochondrial oxidative-responsive gene AOX1A was used as a reference. Gene expression was normalized to PP2A. Data are mean ± SE of three independent repeats. Statistical significances (P < 0.05) among groups are indicated using different letters (Tukey’s HSD test).
We further elucidated AtMnSOD, AtMTM1, and AtMTM2 gene expression profiles in 14-day-old complete seedlings after 5 μM MV treatment for 2, 4, 8, and 12 h by qPCR (Figure 7B). The rapidly upregulated AtMnSOD showed a trend similar to that of AOX1A in response to MV stress (Figure 7B, top), where AOX1A, a mitochondrial oxidative stress-responsive marker gene, was used as a reference. In particular, AtMTM2 had an earlier and higher expression compared to AtMTM1 in response to MV (Figure 7B, bottom). Overall, the AtMnSOD, AtMTM1, and AtMTM2 genes are immediately upregulated in response to MV stress.
Participation of AtMTM1 and AtMTM2 in Flowering-Time Control
Since the mtm1-i mtm2-double mutant indicated an early flowering phenotype (Supplementary Figure 5), we performed the flowering time analysis to elucidate the role of AtMTM1 and AtMTM2. Both mtm1-i and mtm2 single mutants displayed milder early flowering phenotypes compared with the WT, and the double mutant maintained the highest percentage of flowering plants (Figure 8A), indicating the synergistic effect of defective AtMTM1 and AtMTM2 at the flowering stage. The complementation lines of 35S:AtMTM2/mtm1-i, 35S:AtMTM2/mtm2, and 35S:AtMTM2/mtm1-i mtm2 showed flowering times similar to that of the WT (Figure 8B). These results indicate that AtMTM1 and AtMTM2 participate in flowering-time control.
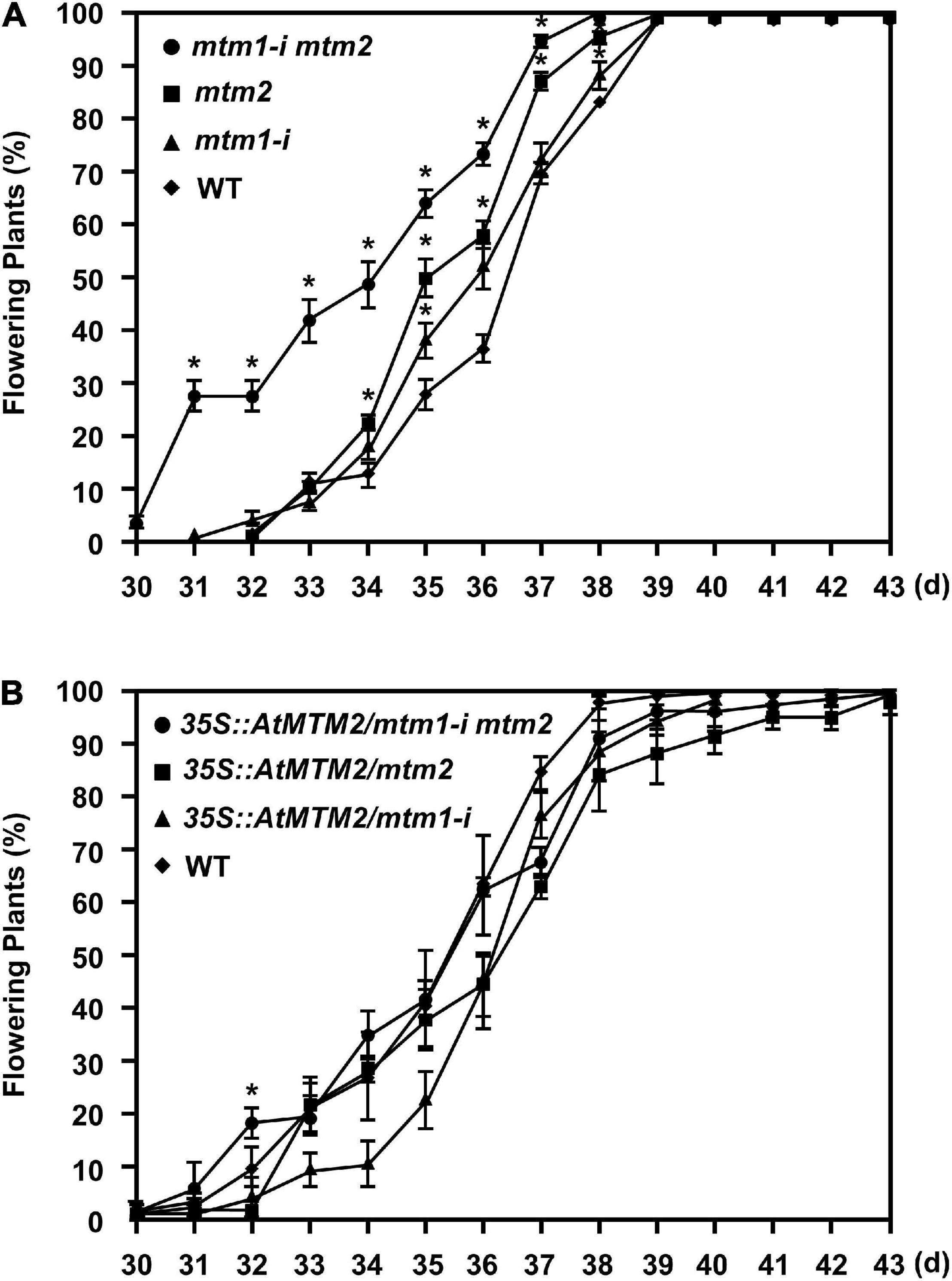
Figure 8. Flowering time analysis. (A,B) Percentages of flowering in WT, mtm1-i, mtm2, and mtm1-i mtm2-double mutants, as well as complementation lines with AtMTM2 transgene in mutant background. Flowering time was scored when primary inflorescence reached 5 cm in length. Data are mean ± SE of three independent repeats (n = 60). *Significant at P < 0.05 (Student’s t-test).
Phenotypic Complementation of Root Length in Mutants Through Mn Supply
To delineate the defective metal regulation in transporter mutants, researchers have applied metal supplementation and examined phenotypic complementation (Schneider et al., 2016; Eisenhuta et al., 2018). We monitored the root lengths of mtm1-i, mtm2, and mtm1-i mtm2-double mutants on 1/2 MS plates containing 10, 50, and 500 μM MnCl2 (Figure 9). The 6-day-old mtm1-i seedlings showed a shorter root length compared with the WT without extra Mn supply, but mtm2 and double mutants had longer root lengths. The 10, 50, and 500 μM Mn treatments restored the root length of mtm1-i on day 6, since mtm1-i and WT had similar root lengths through Mn supply. Moreover, the 500 μM Mn supply restored the root lengths of mtm2 and the double mutant, since mtm2 and mtm1-i mtm2-double mutants reached similar lengths compared with those in mtm1-i and WT. In summary, it is inferred that AtMTM1 and AtMTM2 are involved in root growth.
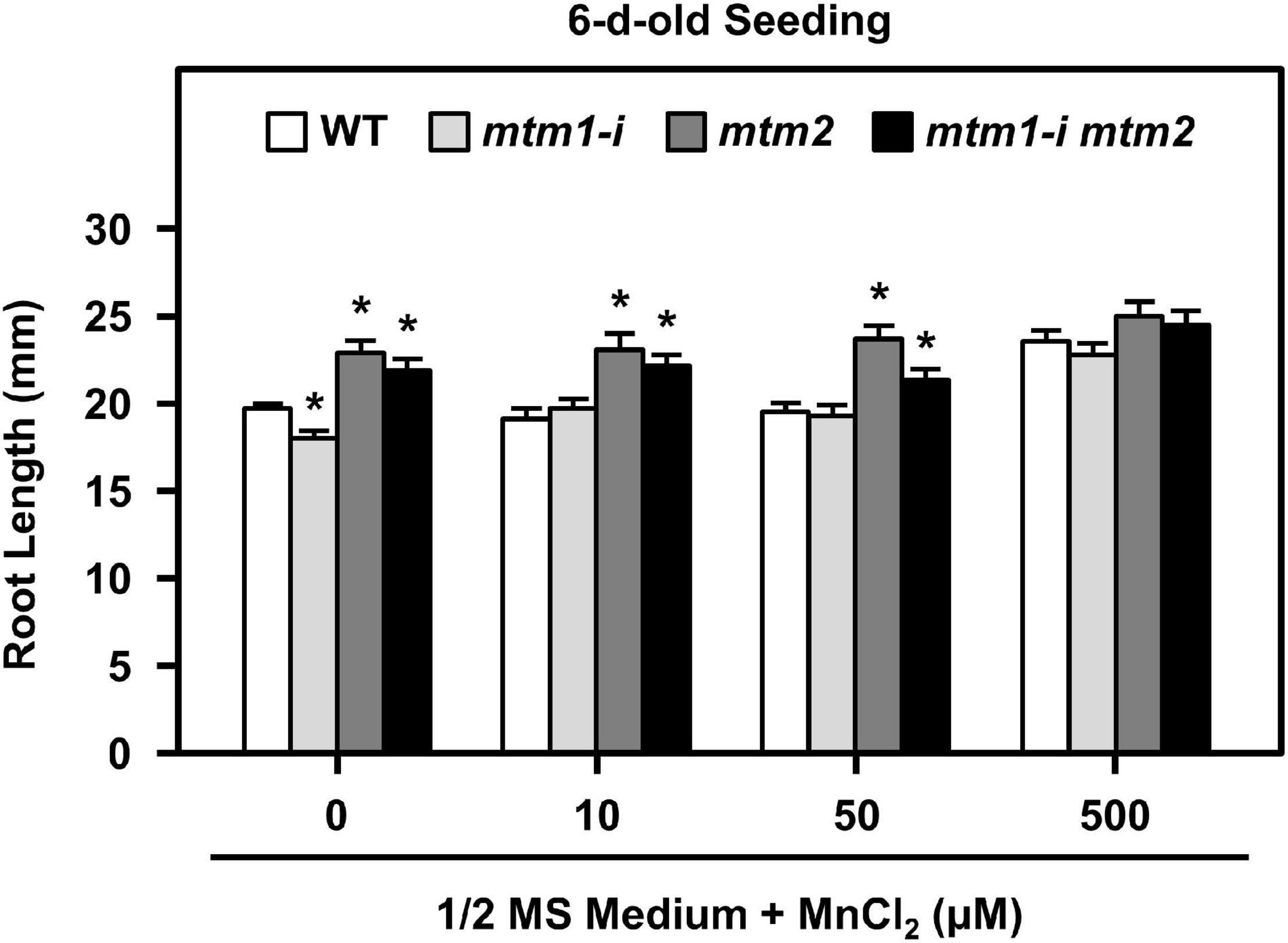
Figure 9. Root lengths of mtm1-i, mtm2, and mtm1-i mtm2-double mutants through Mn supply. Wild-type and three mutants were grown on 1/2 MS medium supplied without and with 10, 50, and 500 μM MnCl2 for 6 days. Data are mean ± SE of three independent repeats (n = 50). *Significant at P < 0.05 (Student’s t-test).
Functional Role of AtMTM1 and AtMTM2 in Mn and Fe Homeostasis
To confirm the influence of defective AtMTM1 and AtMTM2 on Mn homeostasis, we maintained 14-day-old mtm1-i, mtm2, and mtm1-i mtm2-double mutants in 1/2 MS medium containing 100 μM MnCl2 for 24 h. The ICP-OES was performed to measure Mn and Fe ion contents in roots or shoots (Figure 10). The index of metal retention capability represents the ratio of the ion content with Mn supply to that without Mn supply.
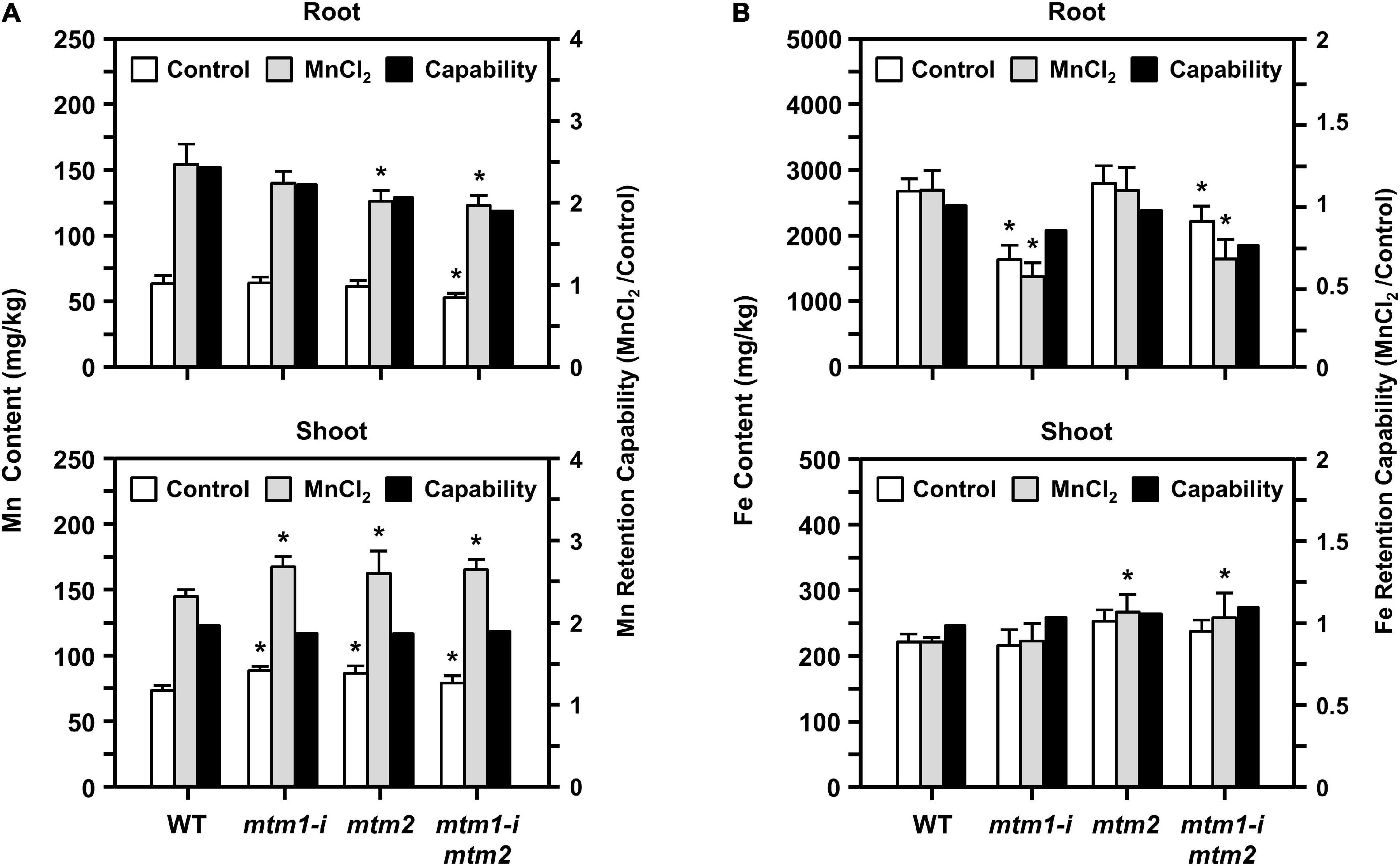
Figure 10. Manganese and Fe content in mtm1-i, mtm2, and mtm1-i mtm2-double mutants through Mn supply. (A,B) Metal contents (Mn and Fe) in root and shoot were measured by the ICP-OES; 14-day-old complete WT and mutant seedlings were incubated without (control) and with 100 μM MnCl2 for 24 h. Data are mean ± SE of three independent repeats. Metal retention capability represents the ratio of ion content with Mn supply (MnCl2) to that without Mn supply (control). *Significant at P < 0.05 (Student’s t-test).
All single and double mutants showed decreased Mn content and Mn retention capabilities in roots, especially in the mtm2 and mtm1-i mtm2-double mutants (Figure 10A, top). This result indicated that the absorbance of Mn ions was restricted to the roots of these mutants. All mutants showed markedly increased Mn content in shoots under Mn treatment and displayed slightly decreased Mn retention capabilities in shoots (Figure 10A, bottom). These results imply that both AtMTM1 and AtMTM2 are involved in Mn homeostasis and that Mn ions may accumulate in the shoot.
The mtm1 and double mutants showed decreased Fe content and Fe retention capabilities in the roots (Figure 10B, top). However, the mtm2 and double mutants showed increased Fe content and retention capabilities in shoots under Mn treatment (Figure 10B, bottom). Fe metal retention capabilities were not consistent between roots and shoots, indicating that AtMTM1 and AtMTM2 exhibit divergence in Fe homeostasis.
Activation of Chloroplast-Destined AtMnSOD
It has been shown that the Mn insertion is linked to the ySOD2 importing process via yMTM1 in mitochondria (Luk et al., 2005), and the chloroplastic-localized MnSOD exists in some algae. To analyze whether AtMnSOD can be activated in the cytosol and chloroplasts, we constructed cytosol-destined AtMnSOD (Δ-TP-AtMnSOD) and chloroplast-destined AtMnSOD (Chl-TP-AtMnSOD) (Supplementary Figure 1A), which were then fused with YFP to confirm their localization by transient expression in protoplasts (Figure 11A). Confocal microscopy revealed that the localization of AtMnSOD-YFP, Δ-TP-AtMnSOD-YFP, and Chl-TP-AtMnSOD-YFP was consistent with their destinations in the mitochondria, cytosol, and chloroplasts, respectively.
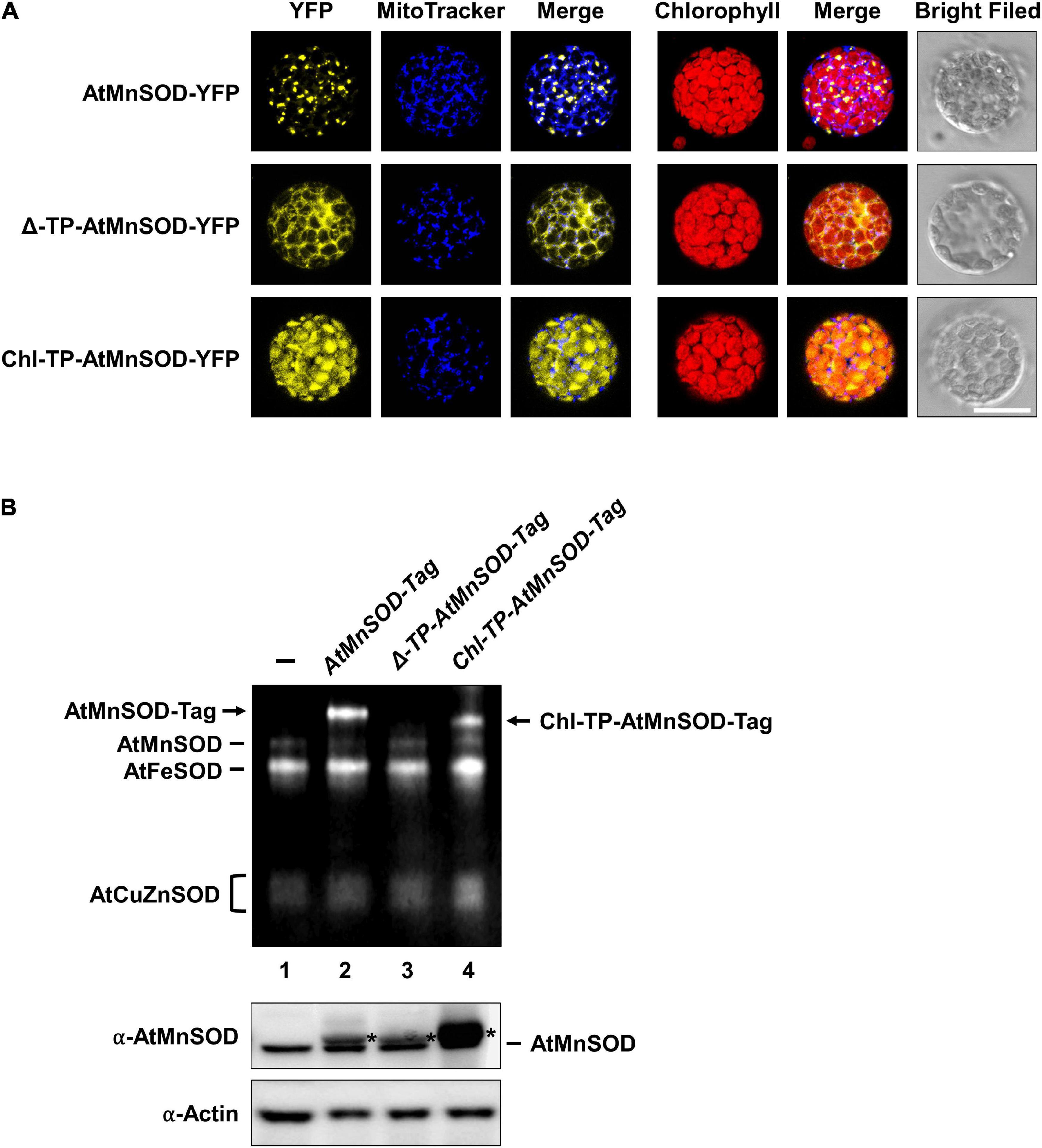
Figure 11. Subcellular localization and SOD activity assay of cytosol-destined and chloroplast-destined AtMnSOD. (A) Transfection of AtMnSOD-YFP, Δ-TP-AtMnSOD-YFP, and Chl-TP-AtMnSOD-YFP transgenes in WT protoplasts and YFP signals corresponded to the localization of mitochondrial, cytosol, and chloroplast, respectively. Bar = 20 μm. (B) Transient expressions without (–) and with AtMnSOD-Tag, Δ-TP-AtMnSOD-Tag, and Chl-TP-AtMnSOD-Tag transgenes in WT protoplasts were analyzed using the in-gel SOD activity assay (top) and western blotting with α-AtMnSOD and α-actin antibodies (bottom). Actin was a loading control. *Represents the exogenous-expressed AtMnSOD.
AtMnSOD and the modified AtMnSOD were also fused with a tag (Supplementary Figure 1B), and their activities could be distinguished from the endogenous AtMnSOD by using the in-gel SOD activity assay (Figure 11B). Exogenously expressed AtMnSOD-Tag and Chl-TP-AtMnSOD-Tag were activated (Figure 11B, lanes 2 and 4), but not the Δ-TP-AtMnSOD-Tag (Figure 11B, lane 3). This result agrees with the importance of the mitochondrial MTM for MnSOD activation (Luk et al., 2005), and an evolutionarily conserved activation mechanism of MnSOD still exists in Arabidopsis chloroplasts.
Discussion
Mitochondrial carrier family proteins are localized in the mitochondrial inner membrane, and they mediate the transport of inorganic ions, cofactors, metabolites, and nucleotides from the cytosol into the mitochondrial matrix (Haferkamp and Schmitz-Esser, 2012). The yMTM1 transgene complements ySOD2 activity in ymtm1Δ cells (Luk and Culotta, 2001; Luk et al., 2003, 2005). This study revealed that the mitochondrial carrier family proteins AtMTM1 and AtMTM2 are homologs of yMTM1 by sequence comparison and transformation assay (Figures 1, 2). The transformation of AtMTM1- or AtMTM2-enhanced ySOD2 activity reflected the maximum capacity in yeast mutant cells; however, co-transformation of AtMTM1 and AtMTM2 resulted in similar ySOD2 activity as the WT. There may be a competition effect of AtMTM1 and AtMTM2, or unknown cellular factors were involved during the co-transformation. We demonstrated that AtMTM1 and AtMTM2 are necessary for AtMnSOD activation by using transient expression assay (Figure 3). We confirmed that AtMTM1 and AtMTM2 interact with AtMnSOD in mitochondria by using the BiFC assay (Figure 4), which agrees with the proteomic evidence of mitochondrial AtMTM1 and AtMTM2 (Senkler et al., 2017).
Analysis of the expression of Arabidopsis mitochondrial carrier family proteins has revealed that carriers usually have distinct specificities during plant growth and abiotic stress (Maia et al., 1998; Watanabe et al., 1999; Catoni et al., 2003; Hoyos et al., 2003). In this study, the gene expression levels of AtMTM1, AtMTM2, and AtMnSOD were measured relative to AtMTM1 level in the root or in the control, and thus, we can compare the relative ratio in tissues and in metal stresses. AtMTM1 and AtMTM2 were ubiquitously expressed in different tissues and were responsible for metal stress in the 14-day-old complete seedlings (Figure 5). However, the substrate specificity or the metal ion affinity of AtMTM1 and AtMTM2 remains to be clarified. We used MV as a superoxide generator and emphasized the complementary effect of AtMTM2 transgene in the mutants. The complementation lines of 35S:AtMTM2/mtm1-i and 35S:AtMTM2/mtm2 partially restored the root lengths to the WT in response to MV stress in 8- to 10-day-old seedlings (Figure 6). The MV sensitivities of the root lengths in these complementation lines may be restricted to different degrees based on their background or the germination process, and it is worthwhile to use several independent complementation lines with different AtMTM2 alleles or monitor the root length from germination to rule out these possibilities. The study further revealed an increase in AtMnSOD activity under MV stress, with AtMTM2 expression being dominant compared with AtMTM1 in the 14-day-old complete WT seedlings (Figure 7); thus, it is worthwhile to track the root lengths for a longer time in these complementation lines. WT seedling remained bright green rosette leaves after MV stress, and it is worthwhile to detect AtMTM1, AtMTM2, and AtMnSOD gene expressions at 24 h and monitor the AtMnSOD activity in mtm1-i, mtm2, and double mutants under MV stress to support the posttranslational regulation of AtMnSOD. Taken together, we assume that AtMTM1 and AtMTM2 have different sensitivities in response to metal and MV stress and that both AtMTM1 and AtMTM2 carriers are involved in the posttranscriptional regulation of AtMnSOD. Notably, the complementation lines of 35S:AtMTM2/mtm1-i, 35S:AtMTM2/mtm2, and 35S:AtMTM2/mtm1-i mtm2 reverted from the early flowering phenotype to the WT (Figure 8), which reflects the necessity of AtMTM1 and AtMTM2 for flowering-time control.
Yeast yMTM1 facilitates the insertion of the Mn cofactor into ySOD2, but ymtm1Δ retains normal Mn levels in the mitochondria (Luk et al., 2003). The loss of the Mn transporter SMF2 decreases ySOD2 activity and Mn levels in yeast (Luk and Culotta, 2001; Luk et al., 2005). The disruption of vacuole-localized Mn transporters of NRAMP3 and NRAMP4 decreases the amount of photosystem II, but does not affect MnSOD activity in Arabidopsis (Allen et al., 2007; Lanquar et al., 2010). Mn-deficient Arabidopsis has abnormal root lengths and altered Fe homeostasis (Yang et al., 2008; Rodríguez-Celma et al., 2016). In our study, the defective Mn carriers of AtMTM1 and AtMTM2 decreased AtMnSOD activity in the mtm1-i mtm2-double mutant. The Mn supply can restore the abnormal root lengths of mtm1-i, mtm2, and mtm1-i mtm2-double mutants to the WT (Figure 9); the strengthened uptake of Mn via AtMTM1 and AtMTM2 can compensate the altered root length phenotype on day 6. It is worthwhile to monitor the root length from the germination, since mtm1-i and mtm2 showed divergent root lengths on day 6 and reached similar lengths as that in WT on day 8 to day 10 under control conditions. We used the same seedling stage and the same condition in the ICP-OES assay and the gene expression assay under Mn stress, which showed upregulated gene expressions in WT. The decreased Mn content in the 14-day-old mtm1-i and mtm2 roots also indicates that AtMTM1 and AtMTM2 are associated with Mn homeostasis. The decreased Mn retention capability was more obvious in the mtm1-i mtm2-double mutant, implying that both AtMTM1 and AtMTM2 have a higher affinity for Mn (Figure 10). In contrast, ymtm1Δ mutant had decreased ySOD2 activity, with increased ISU protein and higher Fe content. ySOD2 inactivation has also been observed in the Fe–S cluster biogenesis mutants of atm1Δ, grx5Δ, and ssq1Δ (Luk et al., 2003; Naranuntarat et al., 2009). The decreased Fe content in the roots of mtm1-i and the double mutant in our study implies that AtMTM1 has a higher affinity for Fe.
In addition to land plants, MnSOD is present in the thylakoids of some prokaryotic and eukaryotic algae. MnSOD participates in photosynthetic water oxidation and scavenges superoxide in the chloroplasts of algae, which is influenced by the environment with fluctuating light and temperature (Kanematsu and Asada, 1979; Okada et al., 1979; Regelsberger et al., 2002). However, this function in the chloroplasts was taken over by FeSOD in Arabidopsis. In our study, we generated modified AtMnSOD constructs and noticed that chloroplast-destined AtMnSOD can be activated in Arabidopsis protoplasts (Figure 11). The factors involved in chloroplast-destined AtMnSOD activation may include envelop-localized or thylakoid-localized Mn transporters of NRAMP3, NRAMP4, CMT1, PAM71/CCHA1, Mn cluster-related factors, or released Mn from photosystem II (Schneider et al., 2016; Wang et al., 2016). Any chloroplast Mn transporter that has a structure similar to that of AtMTM1 or AtMTM2 may insert Mn into modified AtMnSOD polypeptides. Likewise, modified AtMnSOD may have a structure similar to that of certain chloroplast proteins and accept Mn via its Mn transporter. Thus, the activity of chloroplast-destined AtMnSOD highlighted an evolutionarily conserved mechanism that remains in Arabidopsis chloroplasts, as the chloroplastic-localized MnSOD in algae.
Conclusion
This study demonstrates that AtMTM1 and AtMTM2 interact with AtMnSOD, respond to oxidative stress, regulate the root growth, participate in flowering-time control, and involve in Mn homeostasis. We elucidated the gene expression profiles of AtMnSOD, AtMTM1, and AtMTM2 during development. The indispensability of mitochondrial carrier proteins AtMTM1 and AtMTM2 for AtMnSOD enzyme activation was well established by this study. We also modified AtMnSOD and found that chloroplastic factors for chloroplast-destined AtMnSOD activation still exist. Overall, this study reveals the physiological roles of AtMTM1 and AtMTM2 in MnSOD activation and links the Mn uptake via AtMTM1 and AtMTM2 to the MnSOD importing process in mitochondria.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Funding
This work was supported by the Ministry of Science and Technology, Taiwan (Grant Nos. 105-2311-B-002-033-MY3, 107-2923-B-002-003-MY3, and 109-2311-B-002-022-MY3) and the National Taiwan University (Grant No. 110L893602) to T-LJ.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are grateful to Fang-Jen Lee for the gift of yeast BY4741, ysod2Δ, and ymtm1Δ lines. We appreciate Kuo-Chen Yeh for helping with ICP-OES analysis, Lynne Stracovsky for editing, Hui-Chen Wu for commenting on the manuscript, and Pei-Chun Hsieh for technical support. We also appreciate the NTU Confocal Microscope Laboratory for performing fluorescence imaging.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.690064/full#supplementary-material
Supplementary Table 1 | Primers for genotyping, cloning, RT−PCR, and qPCR, as well as accession numbers of genes.
Supplementary Figure 1 | Constructs of modified AtMnSOD.
Supplementary Figure 2 | Characterization of mtm1-1 and mtm1-2 T-DNA insertion mutants.
Supplementary Figure 3 | Characterization of mtm1-i mutant.
Supplementary Figure 4 | Characterization of mtm2-1, mtm1-2, and mtm1-3 T-DNA insertion mutants.
Supplementary Figure 5 | Characterization of mtm1-i mtm2-double mutant.
Supplementary Figure 6 | Control experiments in BiFC.
Supplementary Figure 7 | Histochemical analysis of AtMTM2-promoter:GUS plants.
Footnotes
- ^ http://www-sequence.stanford.edu/group/yeast_deletion_project
- ^ http://primer3.ut.ee
- ^ http://wmd3.weigelworld.org/cgi-bin/webapp.cgi
- ^ http://blast.ncbi.nlm.nih.gov/Blast.cgi
References
Allen, M. D., Kropat, J., Tottey, S., Del Campo, J. A., and Merchant, S. S. (2007). Manganese deficiency in Chlamydomonas results in loss of photosystem II and MnSOD function, sensitivity to peroxides, and secondary phosphorus and iron deficiency. Plant Physiol. 143, 263–277. doi: 10.1104/pp.106.088609
Alscher, R. G., Erturk, N., and Heath, L. S. (2002). Role of superoxide dismutases (SODs) in controlling oxidative stress in plants. J. Exp. Bot. 53, 1331–1341. doi: 10.1093/jexbot/53.372.1331
Apel, K., and Hirt, H. (2004). Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55, 373–399. doi: 10.1146/annurev.arplant.55.031903.141701
Barondeau, D. P., Kassmann, C. J., Bruns, C. K., Tainer, J. A., and Getzoff, E. D. (2004). Nickel superoxide dismutase structure and mechanism. Biochemistry 43, 8038–8047.
Beauchamp, C., and Fridovich, I. (1971). Superoxide dismutase: improved assays and an assay applicable to acrylamide gels. Anal. Biochem. 44, 276–287. doi: 10.1016/0003-2697(71)90370-8
Bowler, C., Camp, W. V., Montagu, M. V., and Inzé, D. (1994). Superoxide dismutase in plants. Crit. Rev. Plant Sci. 13, 199–218.
Brachmann, C. B., Davies, A., Cost, G. J., Caputo, E., Li, J., Hieter, P., et al. (1998). Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14, 115–132. doi: 10.1002/(sici)1097-0061(19980130)14:2<115::aid-yea204>3.0.co;2-2
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. doi: 10.1016/0003-2697(76)90527-3
Casareno, L. B., Waggoner, D., and Gitlin, J. D. (1998). The copper chaperone CCS directly interacts with copper/zinc superoxide dismutase. J. Biol. Chem. 273, 23625–23628. doi: 10.1074/jbc.273.37.23625
Catoni, E., Schwab, R., Hilpert, M., Desimone, M., Schwacke, R., Flügge, U. I., et al. (2003). Identification of an Arabidopsis mitochondrial succinate-fumarate translocator. FEBS Lett. 534, 87–92. doi: 10.1016/s0014-5793(02)03782-1
Chen, C. N., and Pan, S. M. (1996). Assay of superoxide dismutase activity by combining electrophoresis and densitometry. Bot. Bull. Acad. Sin. 7, 107–111.
Choudhury, S. B., Lee, J. W., Davidson, G., Yim, Y. I., Bose, K., Sharma, M. L., et al. (1999). Examination of the nickel site structure and reaction mechanism in Streptomyces seoulensis superoxide dismutase. Biochemistry 38, 3744–3752.
Chu, C. C., Lee, W. C., Guo, W. Y., Pan, S. M., Chen, L. J., Li, H. M., et al. (2005). A copper chaperone for superoxide dismutase that confers three types of copper/zinc superoxide dismutase activity in Arabidopsis. Plant Physiol. 139, 425–436. doi: 10.1104/pp.105.065284
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Colicelli, J., Birchmeier, C., Michaeli, T., O’neill, K., Riggs, M., and Wigler, M. (1989). Isolation and characterization of a mammalian gene encoding a high-affinity cAMP phosphodiesterase. Proc. Natl. Acad. Sci. U. S. A. 86, 3599–3603. doi: 10.1073/pnas.86.10.3599
Culotta, V. C., Klomp, L. W., Strain, J., Casareno, R. L., Krems, B., and Gitlin, J. D. (1997). The copper chaperone for superoxide dismutase. J. Biol. Chem. 272, 23469–23472.
Culotta, V. C., Yang, M., and O’halloran, T. V. (2006). Activation of superoxide dismutases: putting the metal to the pedal. Biochim. Biophys. Acta 1763, 747–758. doi: 10.1016/j.bbamcr.2006.05.003
Czechowski, T., Stitt, M., Altmann, T., Udvardi, M. K., and Scheible, W. R. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139, 5–17. doi: 10.1104/pp.105.063743
Edwards, K., and Johnstone, C., and Thompson, C. (1991). A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res. 19:1349.
Eisenhuta, M., Hoecker, N., Schmidt, S. B., Basgaran, R. M., Flachbart, S., Jahns, P., et al. (2018). The plastid envelope CHLOROPLAST MANGANESE TRANSPORTER1 is essential for manganese homeostasis in Arabidopsis. Mol. Plant 11, 955–969. doi: 10.1016/j.molp.2018.04.008
Fink, R. C., and Scandalios, J. G. (2002). Molecular evolution and structure-function relationships of the superoxide dismutase gene families in angiosperms and their relationship to other eukaryotic and prokaryotic superoxide dismutases. Arch. Biochem. Biophys. 399, 19–36. doi: 10.1006/abbi.2001.2739
Gietz, R. D., and Schiestl, R. H. (1991). Applications of high efficiency lithium acetate transformation of intact yeast cells using single-stranded nucleic acids as carrier. Yeast 7, 253–263. doi: 10.1002/yea.320070307
Haferkamp, I., and Schmitz-Esser, S. (2012). The plant mitochondrial carrier family: functional and evolutionary aspects. Front. Plant Sci. 3:2. doi: 10.3389/fpls.2012.00002
Halliwell, B. (1994). Free radicals, antioxidants, and human disease: curiosity, cause, or consequence? Lancet 344, 721–724. doi: 10.1016/s0140-6736(94)92211-x
Hoyos, M. E., Palmieri, L., Wertin, T., Arrigoni, R., Polacco, J. C., and Palmieri, F. (2003). Identification of a mitochondrial transporter for basic amino acids in Arabidopsis thaliana by functional reconstitution into liposomes and complementation in yeast. Plant J. 33, 1027–1035. doi: 10.1046/j.1365-313x.2003.01685.x
Huang, C. H., Kuo, W. Y., and Jinn, T. L. (2012). Models for the mechanism for activating copper-zinc superoxide dismutase in the absence of the CCS Cu chaperone in Arabidopsis. Plant Signal. Behav. 7, 429–431.
Kanematsu, S., and Asada, K. (1979). Ferric and manganese superoxide dismutases in Euglena gracilis. Arch. Biochem. Biophys. 195, 535–545. doi: 10.1016/0003-9861(79)90380-1
Kliebenstein, D. J., Monde, R. A., and Last, R. L. (1998). Superoxide dismutase in Arabidopsis: an eclectic enzyme family with disparate regulation and protein localization. Plant Physiol. 118, 637–650. doi: 10.1104/pp.118.2.637
Kuo, W. Y., Huang, C. H., and Jinn, T. L. (2013a). Chaperonin 20 might be an iron chaperone for superoxide dismutase in activating iron superoxide dismutase (FeSOD). Plant Signal. Behav. 8:e23074. doi: 10.4161/psb.23074
Kuo, W. Y., Huang, C. H., Liu, A. C., Cheng, C. P., Li, S. H., Chang, W. C., et al. (2013b). Chaperonin 20 mediates iron superoxide dismutase (FeSOD) activity independent of its co-chaperonin role in Arabidopsis chloroplasts. New Phytol. 197, 99–110. doi: 10.1111/j.1469-8137.2012.04369.x
Kuo, W. Y., Huang, C. H., Shih, C., and Jinn, T. L. (2013c). Cellular extract preparation for superoxide dismutase (SOD) activity assay. Bio Protoc. 3:e811.
Lahner, B., Gong, J., Mahmoudian, M., Smith, E. L., Abid, K. B., Rogers, E. E., et al. (2003). Genomic scale profiling of nutrient and trace elements in Arabidopsis thaliana. Nat. Biotech. 21, 1215–1221. doi: 10.1038/nbt865
Lanquar, V., Ramos, M. S., Lelievre, F., Barbier-Brygoo, H., Krieger-Liszkay, A., Kramer, U., et al. (2010). Export of vacuolar manganese by AtNRAMP3 and AtNRAMP4 is required for optimal photosynthesis and growth under manganese deficiency. Plant Physiol. 152, 1986–1999. doi: 10.1104/pp.109.150946
Lin, Y. F., Liang, H. M., Yang, S. Y., Boch, A., Clemens, S., Chen, C. C., et al. (2009). Arabidopsis IRT3 is a zinc-regulated and plasma membrane localized zinc/iron transporter. New Phytol. 182, 392–404. doi: 10.1111/j.1469-8137.2009.02766.x
Lu, Q., Tang, X., Tian, G., Wang, F., Liu, K., Nguyen, V., et al. (2010). Arabidopsis homolog of the yeast TREX-2 mRNA export complex: components and anchoring nucleoporin. Plant J. 61, 259–270. doi: 10.1111/j.1365-313x.2009.04048.x
Luk, E., Carroll, M., Baker, M., and Culotta, V. C. (2003). Manganese activation of superoxide dismutase 2 in Saccharomyces cerevisiae requires MTM1, a membrane of the mitochondrial carrier family. Proc. Natl. Acad. Sci. U. S. A. 100, 10353–10357. doi: 10.1073/pnas.1632471100
Luk, E., and Culotta, V. C. (2001). Manganese superoxide dismutase in Saccharomyces cerevisiae acquires its metal co-factor through a pathway involving the Nramp metal transporter, Smf2p. J. Biol. Chem. 276, 47556–47562. doi: 10.1074/jbc.m108923200
Luk, E., Yang, M., Jensen, L. T., Bourbonnais, Y., and Culotta, V. C. (2005). Manganese activation of superoxide dismutase 2 in the mitochondria of Saccharomyces cerevisiae. J. Biol. Chem. 280, 22715–22720. doi: 10.1074/jbc.m504257200
Maia, I. G., Benedetti, C. E., Leite, A., Turcinelli, S. R., Vercesi, A. E., and Arruda, P. (1998). AtPUMP: an Arabidopsis gene encoding a plant uncoupling mitochondrial protein. FEBS Lett. 429, 403–406. doi: 10.1016/s0014-5793(98)00634-6
Millar, A. H., and Heazlewood, J. L. (2003). Genomic and proteomic analysis of mitochondrial carrier proteins in Arabidopsis. Plant Physiol. 131, 443–453. doi: 10.1104/pp.009985
Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 15, 473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x
Naranuntarat, A., Jensen, L. T., Panicni, S., Penner-Hahn, J. E., and Culotta, V. C. (2009). The interaction of mitochondrial iron with manganese superoxide dismutase. J. Biol. Chem. 284, 22633–22640. doi: 10.1074/jbc.m109.026773
Okada, S., Kanematsu, S., and Asada, K. (1979). Intracellular distribution of manganese and ferric superoxide dismutases in blue-green algae. FEBS Lett. 103, 106–110. doi: 10.1016/0014-5793(79)81260-0
Palmieri, F., Pierri, C. L., Grassi, A. D., Nunes-Nesi, A., and Fernie, A. R. (2011). Evolution, structure and function of mitochondrial carriers: a review with new sights. Plant J. 66, 161–181. doi: 10.1111/j.1365-313x.2011.04516.x
Pan, S. M., and Yau, Y. Y. (1992). Characterization of superoxide dismutase in Arabidopsis. Taiwania 37, 58–66.
Picault, N., Hodges, M., Palmieri, L., and Palmieri, F. (2004). The growing family of mitochondrial carriers in Arabidopsis. Trends Plant Sci. 9, 138–146. doi: 10.1016/j.tplants.2004.01.007
Priya, B., Premanandh, J., Dhanalakshmi, R. T., Seethalakshmi, T., Uma, L., Prabaharan, D., et al. (2007). Comparative analysis of cyanobacterial superoxide dismutases to discriminate canonical forms. BMC Genomics 8, 435–444. doi: 10.1186/1471-2164-8-435
Rae, T. D., Torres, A. S., Pufahl, R. A., and O’halloran, T. V. (2001). Mechanism of Cu, Zn-superoxide dismutase activation by the human metallochaperone hCCS. J. Biol. Chem. 276, 5166–5176. doi: 10.1074/jbc.m008005200
Regelsberger, G., Atzenhofer, W., Ruker, F., Peschek, G. A., Jakopitsch, C., Paumann, M., et al. (2002). Biochemical characterization of a membrane-bound manganese-containing superoxide dismutase from the cyanobacterium Anabaena PCC 7120. J. Biol. Chem. 277, 43615–43622. doi: 10.1074/jbc.m207691200
Robinson, A. J., and Kunji, E. R. S. (2006). Mitochondrial carriers in the cytoplasmic state have a common substrate binding site. Proc. Natl. Acad. Sci. U. S. A. 103, 2617–2622. doi: 10.1073/pnas.0509994103
Rodríguez-Celma, J., Tsai, Y. H., Wen, T. N., Wu, Y. C., Curie, C., and Schmidt, W. (2016). Systems-wide analysis of manganese deficiency-induced changes in gene activity of Arabidopsis roots. Sci. Rep. 6:35846.
Schneider, A., Steinberger, I., Herdean, A., Gandini, C., Eisenhut, M., Kurz, S., et al. (2016). The evolutionarily conserved protein PHOTOSYNTHESIS AFFECTED MUTANT71 is required for efficient manganese uptake at the thylakoid membrane in Arabidopsis. Plant Cell 28, 892–910.
Schwab, R., Ossowski, S., Riester, M., Warthmann, N., and Weigel, D. (2006). Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18, 1121–1133. doi: 10.1105/tpc.105.039834
Senkler, J., Senkler, M., Eubel, H., Hildebrandt, T., Lengwenus, C., Schertl, P., et al. (2017). The mitochondrial complexome of Arabidopsis thaliana. Plant J. 89, 1079–1092. doi: 10.1111/tpj.13448
Su, Z., Chai, M. F., Lu, P. L., An, R., Chen, J., and Wang, X. C. (2007). AtMTM1, a novel mitochondrial protein, may be involved in activation of the manganese-containing superoxide dismutase in Arabidopsis. Planta 226, 1031–1039. doi: 10.1007/s00425-007-0547-6
Tsukagoshi, H. (2012). Defective root growth triggered by oxidative stress is controlled through the expression of cell cycle-related genes. Plant Sci. 197, 30–39. doi: 10.1016/j.plantsci.2012.08.011
Van Camp, W., Willekens, H., Bowler, C., Van Montagu, M., Inze, D., Reupold-Popp, P., et al. (1994). Elevated levels of superoxide dismutase protect transgenic plants against ozone damage. Biotechniques 12, 165–168. doi: 10.1038/nbt0294-165
Wang, C., Xu, W., Jin, H., Zhang, T., Lai, J., Zhou, X., et al. (2016). A putative chloroplast-localized Ca(2+)/H(+) antiporter CCHA1 is involved in calcium and pH homeostasis and required for PSII function in Arabidopsis. Mol. Plant 9, 1183–1196. doi: 10.1016/j.molp.2016.05.015
Watanabe, A., Nakazono, M., Tsutsumi, N., and Hirai, A. (1999). AtUCP2: a novel isoform of the mitochondrial uncoupling protein of Arabidopsis thaliana. Plant Cell Physiol. 40, 1160–1166. doi: 10.1093/oxfordjournals.pcp.a029501
Weigel, D., and Glazebrook, J. (2002). Arabidopsis: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
Yang, T. J., Perry, P. J., Ciani, S., Pandian, S., and Schmidt, W. (2008). Manganese deficiency alters the patterning and development of root hairs in Arabidopsis. J. Exp. Bot. 59, 3453–3464. doi: 10.1093/jxb/ern195
Keywords: metalloenzyme, mitochondrial carrier family, MnSOD, Mn transporter, reactive oxygen species, superoxide dismutase
Citation: Hu S-H, Lin S-F, Huang Y-C, Huang C-H, Kuo W-Y and Jinn T-L (2021) Significance of AtMTM1 and AtMTM2 for Mitochondrial MnSOD Activation in Arabidopsis. Front. Plant Sci. 12:690064. doi: 10.3389/fpls.2021.690064
Received: 02 April 2021; Accepted: 13 July 2021;
Published: 06 August 2021.
Edited by:
Luisa M. Sandalio, Departamento de Bioquímica, Biología Celular y Molecular de Plantas, Estación Experimental del Zaidín, SpainReviewed by:
Francisca Sevilla, Center for Edaphology and Applied Biology of Segura, Spanish National Research Council, SpainVivek Dogra, Institute of Himalayan Bioresource Technology (CSIR), India
Copyright © 2021 Hu, Lin, Huang, Huang, Kuo and Jinn. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tsung-Luo Jinn, amlubnRAbnR1LmVkdS50dw==
†These authors have contributed equally to this work
 Shu-Hsuan Hu
Shu-Hsuan Hu Shu-Fan Lin
Shu-Fan Lin Ya-Chen Huang
Ya-Chen Huang Chien-Hsun Huang
Chien-Hsun Huang Wen-Yu Kuo1
Wen-Yu Kuo1 Tsung-Luo Jinn
Tsung-Luo Jinn