- Department of Plant and Microbial Biology, University of California, Berkeley, Berkeley, CA, United States
Nitrogen (N), phosphorus (P), and potassium (K) are three major macronutrients essential for plant life. These nutrients are acquired and transported by several large families of transporters expressed in plant roots. However, it remains largely unknown how these transporters are distributed in different cell-types that work together to transfer the nutrients from the soil to different layers of root cells and eventually reach vasculature for massive flow. Using the single cell transcriptomics data from Arabidopsis roots, we profiled the transcriptional patterns of putative nutrient transporters in different root cell-types. Such analyses identified a number of uncharacterized NPK transporters expressed in the root epidermis to mediate NPK uptake and distribution to the adjacent cells. Some transport genes showed cortex- and endodermis-specific expression to direct the nutrient flow toward the vasculature. For long-distance transport, a variety of transporters were shown to express and potentially function in the xylem and phloem. In the context of subcellular distribution of mineral nutrients, the NPK transporters at subcellular compartments were often found to show ubiquitous expression patterns, which suggests function in house-keeping processes. Overall, these single cell transcriptomic analyses provide working models of nutrient transport from the epidermis across the cortex to the vasculature, which can be further tested experimentally in the future.
Introduction
Plant growth and development depend on the constant supply of mineral nutrients through the root system. Therefore, a deficiency in macronutrients such as NPK (Nitrogen, phosphorus and potassium) can have a profound impact on root growth and architecture (Figure 1), which in turn can alter the efficiency of nutrient acquisition and transport (López-Bucio et al., 2003; Gruber et al., 2013; Giehl and von Wiren, 2014; Luan et al., 2017). Plants possess large sets of nutrient transporters to perform these functions, among which many remain uncharacterized. Because the levels of these macronutrients in natural soils are often limiting plant growth, crop production often relies on heavy use of chemical fertilizers, which is not sustainable and pollutes the environment (Poirier and Bucher, 2002; Luan et al., 2017). To support sustainable agriculture, there is an urgent need to enhance their nutrient use efficiency (NUE), which will require decoding the complex genetic and biochemical components of nutrient transport.
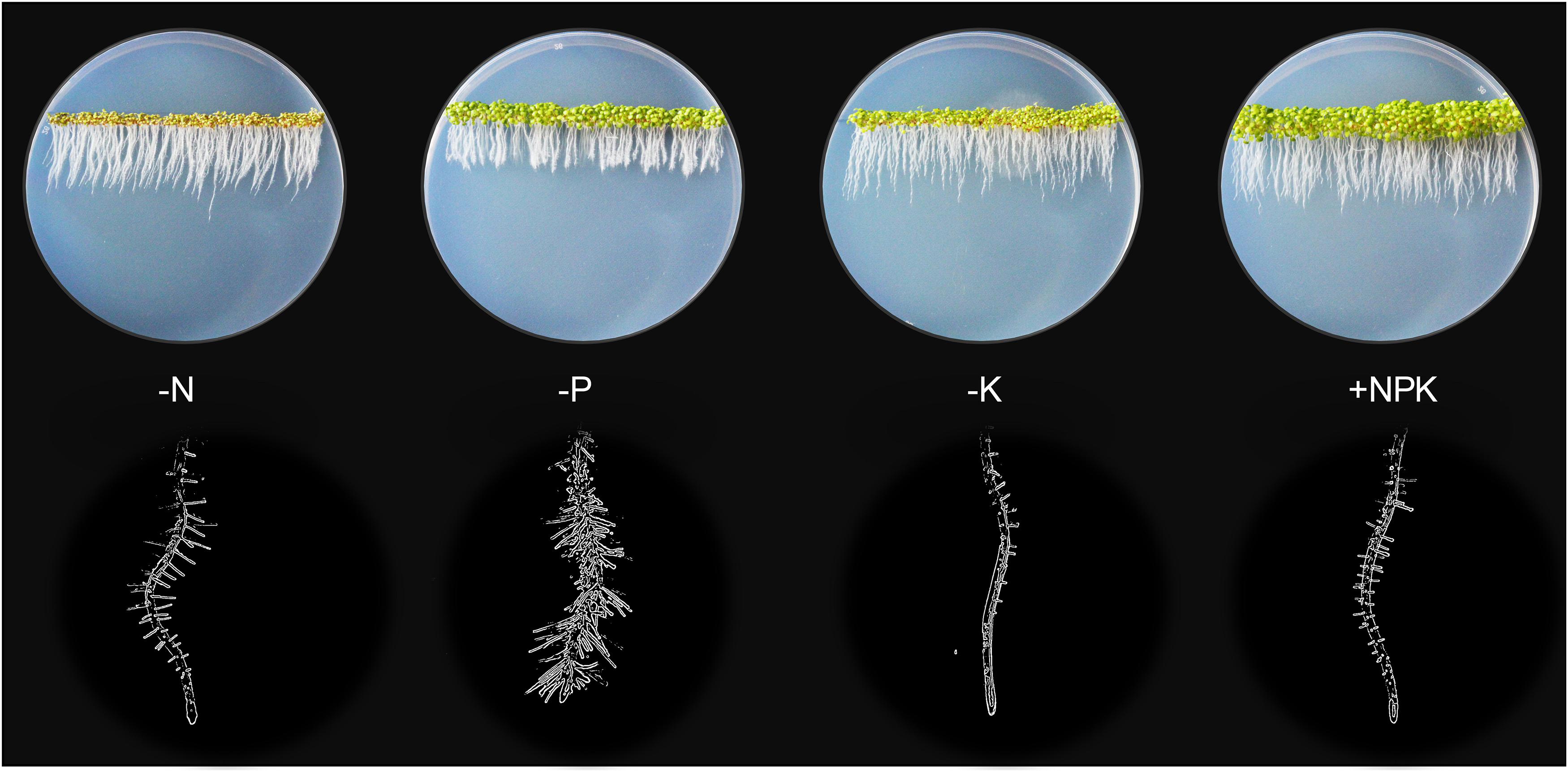
Figure 1. Plant growth highly depends on the continual nutrient supplies. Without N, P, or K (0 mM, 1/6 MS), 5-days old Arabidopsis seedlings displayed stunted shoot growth (top rows) and modified root architectures including primary root and root hair growth (bottom rows) compared to the sufficient-nutrient (1/6 MS) conditions. MS for Murashige and Skoog media.
Decades of research have advanced our understanding on the physiological roles of many nutrient transporters. One important aspect of transporter function is its site of activity. By combining genetic approaches with expression pattern analysis using promoter-reporter transgenic plant, studies can link the phenotype of a mutant to the tissue or cell-type where a particular transporter is expressed to make a functional prediction. This approach has identified a number of nutrient transporters that function in nutrient uptake in roots, root-to-shoot translocation, stomatal movement, pollen tube elongation and reproductive development (Wang and Wu, 2013; Gu et al., 2016; Wang et al., 2018). Although highly effective, this approach is time-consuming, labor-intensive, and limited by the number of genes and cells it can assess at a time. Recent breakthrough in single cell RNA-sequencing (scRNA-seq) provides a high-throughput platform to assess global gene expression at a single cell resolution, with the capacity to measure thousands of cells of different types and developmental stages simultaneously in a single experiment (Macosko et al., 2015; Ziegenhain et al., 2017). In the context of nutrient transport, scRNAseq can map the cell-type profiles of a large number of transporters so that we can predict where they may function and how they may be connected in a systematic process for nutrient handling at whole root or whole plant level.
The scRNAseq technology has been successfully applied to Arabidopsis thaliana roots for spatiotemporal characterization of distinct root cell-types, their developmental trajectories, and their transcriptional regulatory pathways (Denyer et al., 2019; Jean-Baptiste et al., 2019; Ryu et al., 2019; Shulse et al., 2019; Zhang et al., 2019). An Arabidopsis root atlas is generated by integrating two of the previously published datasets (Denyer et al., 2019; Ryu et al., 2019), and newly produced datasets from total of 110,427 single cells to profile gene expression dynamics in 11 different root cell-types (Shahan et al., 2020). Here, we leveraged this enormous dataset to map the cell-type expression pattern of NPK transporters in the Arabidopsis roots. Such analyses identified the putative transport map that allow nutrient transport longitudinally from the root cap (lateral root cap/LRC and columella) to the quiescent center (QC), and radially from the root epidermis (trichoblast and atrichoblast) to cortex and endodermis to reach stellar cells (pericycle, xylem, procambium and phloem) for root-to-shoot translocation. We first examined the expression patterns of known nutrient transporters in each family. This allowed us to gain more insights into the known NPK transporters at the root single cell resolution, which has not been fully achieved before. Then, we profile the expression patterns of uncharacterized nutrient transporter genes to predict their functional roles, considering their membrane localizations and phylogenetic relationship to the functionally characterized NPK transporters. However, this single cell profiling is currently limited to roots of young seedlings (5–7 days-old) under the normal growth conditions, which may not remain the same throughout the plant life cycle and under the changing nutrient status. For instance, 5-days-old seedlings respond to low-nutrient (N, P or K) stresses differently, with a different degree of modification to the root architectures and shoot growth (Figure 1). The transcriptional profiles of nutrient transporters under these stresses may be different compared to the controls and between the treatments. Therefore, low-nutrient-induced transporters may not be fully captured under the normal growth conditions presented in this study. However, these provide future opportunities to generate more detailed Arabidopsis root atlas that cover dynamic transcriptional changes of nutrient transporters over the course of plant development under changing nutrient status. This coupled with the physiological characterization of nutrient transporters will decipher the complexity of nutrient transport processes in plants. Our study identifies nutrient transporters that may function in collaborative fashion to distribute nutrients throughout the root system in concentric manners under non-stress conditions to support normal plant growth. In addition, it aims to lay the foundation for future research to dissect the complex networks of nutrient transport systems at a single cell resolution under varieties of biotic and abiotic stresses of different plant tissues and organs.
Results and Discussion
Nitrate Transport
Nitrate (NO3–) is the predominant form of N available in the soil and absorbed by plants for the synthesis of amino acids, nucleic acids, and many secondary metabolites (Noguero and Lacombe, 2016; Bellegarde et al., 2017; Fan et al., 2017). NO3– not only serves as a major macronutrient but also acts as a signaling molecule that regulates plant developmental processes including seed germination, lateral root development, and flowering time (Bellegarde et al., 2017; Fan et al., 2017; Fredes et al., 2019). However, NO3– is often limited in natural soils (<1 mM) and crop production relies on heavy fertilizer application that can reach up to 70 mM in agricultural lands, leading to runoff and pollution of aquatic ecological systems (Reisenauer, 1966; Krapp et al., 2014). To combat the constantly changing NO3– status in the environment, plants have evolved large sets of NO3– transporters for efficient uptake and distribution throughout plant cells and tissues (Krapp et al., 2014). These processes are mediated by four major NO3– transporter families, which include NPF (Nitrate Transporter 1 (NRT1)/Peptide Transporter (PTR) Family), NRT2 (Nitrate Transporter 2), CLC (Chloride Channel), and SLAC/SLAH (Slow Anion Channel/SLAC1 homolog) (Krapp et al., 2014; Fan et al., 2017; Wang et al., 2018). While some of these NO3– transporters in each family have been functionally characterized, the majority of them remains to be evaluated, especially those in the large NPF family. We examined the expression of these NO3– transporters at a single cell resolution to understand how plants spatially distribute these transporters in the root system to charge the uptake, and transport of NO3– from the external environment to the xylem vessels for long-distance translocation. The potential network we presented here of NO3– channels and transporters is mainly applicable to young seedlings under the sufficient nutrient supply. It is solely based on the transcriptional profiles, which needs to be backed up by sufficient physiological studies in the future.
NPF Transporters
The Arabidopsis NPF family includes 53 members that display high sequence homology with the SLC15/PepT/PTR/POT family of peptide transporters in animals (Léran et al., 2014; O’Brien et al., 2016; Wang et al., 2018). NPF transporters are mostly located at the plasma membrane (PM) and serve as low-affinity transporters that transport NO3– and other diverse metabolites and hormones (Krapp et al., 2014; Léran et al., 2014; Wang et al., 2018). NPFs are divided into eight subfamilies based on their phylogenetic relationships (Léran et al., 2014). NPF1 contains three members, among which the PM-localized NPF1.1 (NRT1.12) and NPF1.2 (NRT1.11) were involved in transferring root-derived NO3– into phloem of mature leaves for redistribution to young leaves (Hsu and Tsay, 2013). Root single cell transcriptomes indicated these NPF1s were broadly expressed in stellar cells, with the highest expression in the procambium (Figure 2A), the meristematic tissue that give rise to xylem and phloem (Jouannet et al., 2015; De Rybel et al., 2016). This expression pattern suggests NPF1.1/1.2 may function in NO3– loading to both meristematic and mature phloem and xylem cells in developing roots under the normal growth conditions. The function of NPF1.3 has not been previously characterized. Based on its single cell profile, NPF1.3 was expressed at a much lower level as compared to NPF1.1/1.2, mainly in endodermal and xylem cells (Figure 2A), possibly to translocate NO3– to shoots. Its expression may amplify during the later stage of development or under a specific nutrient stress condition. The broad expression of NPF1.1 in other cell-types except root cap suggests its potential contribution to the radial transport and distribution of NO3– to or from stellar cells.
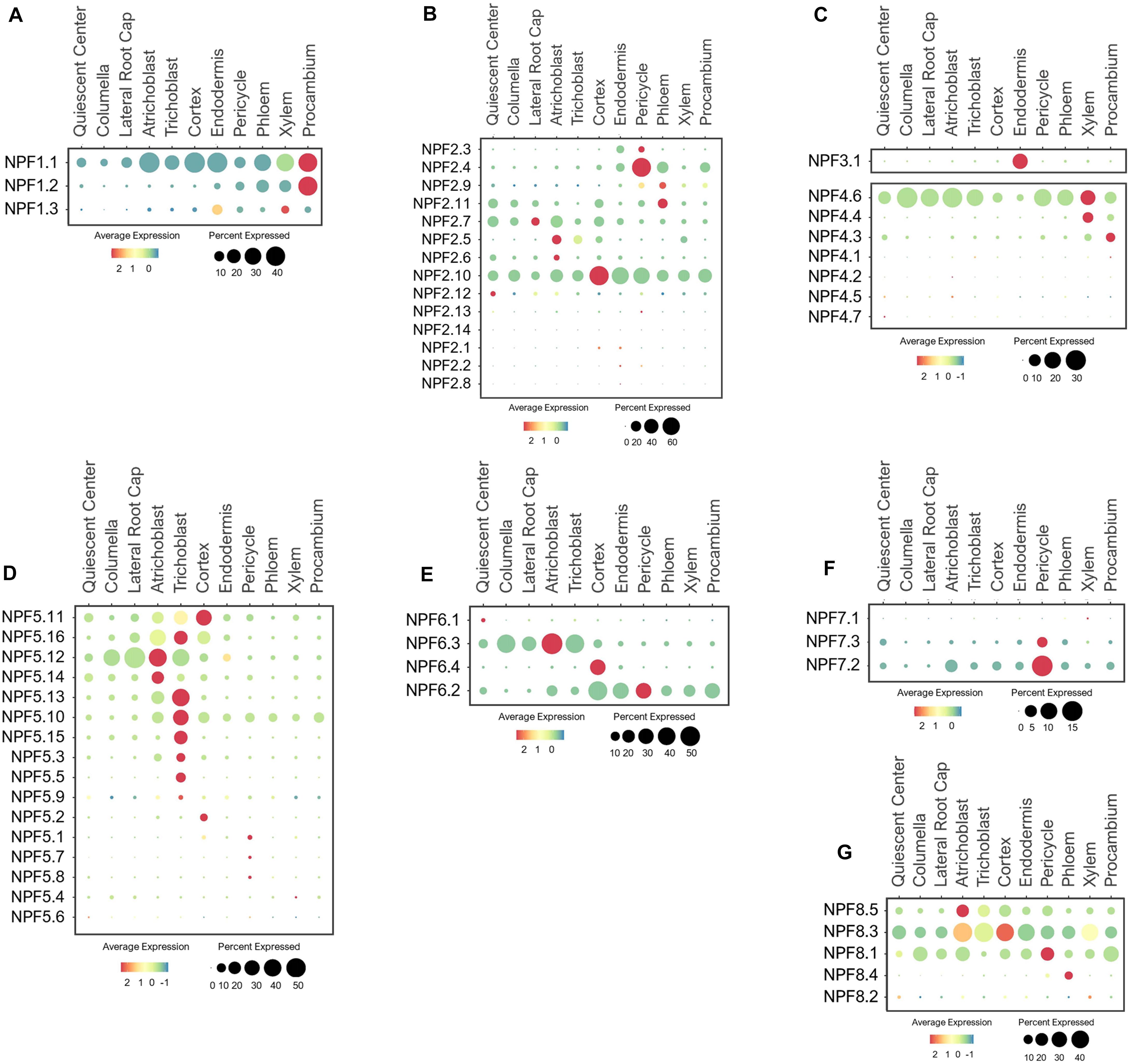
Figure 2. Expression of the low-affinity NPF transporters in Arabidopsis root single cells, (A,B) NPF1-2, (C) NPF3/4, and (D–G) NPF5-8. In dot plots, the circle size represents the proportion (%) of cells expressing a given gene, and the color of scale bar represents the mean expression (natural log +1 pseudocount).
The NPF2 subfamily contains 14 members, several of which have been functionally characterized. For example, the PM-localized NPF2.3 (NAXT2) mediates NO3– efflux from pericycle to xylem for root-to-shoot transport in response to salt stress (Taochy et al., 2015). As expected, NPF2.3 was mainly expressed in the pericycle, but at a low level under the normal conditions (Figure 2B), suggesting it may be more stress-responsive. NPF2.4 was reported to specifically mediate chloride (Cl–) loading to xylem in response to salt stress (Li et al., 2016). Consistent with this function, NPF2.4 was expressed in the pericycle (Figure 2B), but its high expression under the normal growth conditions suggests it may also mediate Cl– transport under the non-stress conditions. NPF2.9 (NRT1.9) was shown to recirculate NO3– from aerial tissues to roots via phloem loading (Wang and Tsay, 2011). As expected, NPF2.9 was expressed in phloem at a low level (Figure 2B), implying NO3– recirculation may not be a high priority under the nutrient sufficiency. NPF2.11 (GTR2) can transport NO3–/gibberellin/glucosinolate, and mediated source-to-sink transport of glucosinolate via phloem loading (Nour-Eldin et al., 2012; Andersen et al., 2013; Saito et al., 2015; Wang et al., 2018). Consistent with these studies, NPF2.11 was expressed in phloem (Figure 2B). Its function may overlap with NPF2.9 in NO3– loading to phloem based on their similar expression patterns. NPF2.7 (NAXT1) extrudes NO3– from root cells under the acidic environments (Segonzac et al., 2007). In compliance with its physiology, NPF2.7 was expressed in the cortex, QC, epidermis, and root cap (Figure 2B). NPF2.5/2.6 are closely related to NPF2.7, and may share similar functions in NO3– secretion from root epidermis to maintain ionic balance at the root-soil interface. A previous study reported the involvement of NPF2.5 in Cl– extrusion from roots under salt stress (Li et al., 2017a), however, its NO3– transport activity needs further evaluation. Among all the NPF2s, NPF2.10 displayed a ubiquitous expression, signifying its potential function in NO3– distribution across different root cell layers. Studies have shown that NPF2.13 (NRT1.7) recycles NO3– from old to developing leaves by directly loading NO3– to the phloem (Fan et al., 2009), and NPF2.12 (NRT1.6) delivers NO3– from source leaves to developing seeds (Almagro et al., 2008). As a result of specific expression and function in leaf and seed developments, these two transporters were barely expressed in roots (Figure 2B).
As the only member in the NPF3 clade, NPF3.1 was reported to be a PM-localized transporter involved in nitrite and NO3– transport in leaves, and GA transport to root endodermis (Sugiura et al., 2007; Pike et al., 2014; Tal et al., 2016). Consistently, it exhibited endodermis-specific expression in the developing roots under the normal conditions (Figure 2C). This suggests that NPF3.1 may enable NO3–/nitrite/GA transport across the casparian strip barrier.
Seven NPF4 members are present in Arabidopsis, among which, only NPF4;6 (NRT1.2/AIT1) has been characterized to function as a low-affinity NO3– uptake transporter (Huang et al., 1999). As anticipated, NPF4.6 was expressed in epidermis and root tip (QC, columella and LRC) to initiate NO3– acquisition (Figure 2C). In addition, its high expression in stellar cells, especially xylem, suggests a potential role in NO3– loading to xylem. We hypothesize that NPF4.4 may also be involved in xylem loading because its expression was exclusively associated with xylem. NPF4.3 was preferentially expressed in procambium, suggesting its potential role in xylem and/or phloem loading. Their potential functions in long-distance transport needs to be evaluated experimentally.
While the majority of NPFs are found at the PM, NPF5.11/5.12/5.16, among the sixteen NPF5 members, display the tonoplast localization to remobilize NO3– from the vacuoles of stellar cells, especially pericycle (He et al., 2017). However, these transporters were mainly expressed in root surface and cortical cells based on the single cell profiles of young seedlings under the normal growth conditions (Figure 2D), necessitating a more detailed examination of their function in the future. Intriguingly, the other NPF5s were also largely expressed in the epidermis, especially trichoblast, suggesting a role in soil-root exchange. Because the subcellular locations of these NPF5s remain to be determined, there is a gap of knowledge regarding their physiological functions. The single cell expression patterns here provide a reference point for functional analysis using genetic procedures. Among the NPF5 subfamily, NPF5.10 was expressed more broadly in stellar cells, and NPF5.1/5.7/5.8 specifically in a few pericycle cells. Depending on their subcellular locations and transport activities, these transporters could serve as candidates for NO3– transfer or remobilization from stellar cells to xylem vessels for long-distance transport. In addition, some of their expressions may be more responsive to nutrient stresses or during later-stages of the plant development.
NPF6 has four members, among which, NPF6.3 (NRT1.1/CHL1) functions as a dual-affinity transporter that mediate NO3– uptake and nitrate-dependent auxin transport (Huang et al., 1996; Wang et al., 1998; Liu et al., 1999; Krouk et al., 2010). As the key transporter in NO3– acquisition, NPF6.3 was highly expressed in epidermis and root cap (Figure 2E). Root cap cells, located at the root tip are essential for nutrient sensing and uptake, root penetration in soils, root growth, gravitropic and hydrotropic responses, which are mostly controlled by polar auxin transport (Svistoonoff et al., 2007; Arnaud et al., 2010; Ruiz Herrera et al., 2015). Studies have shown NPF6.3 at the root tip is able to sense the changes in soil NO3– status, and transduces NO3– signaling to regulate nitrate uptake and auxin distribution for lateral root development (Liu and Tsay, 2003; Ho et al., 2009; Krouk et al., 2010; Gojon et al., 2011; Mounier et al., 2014; O’Brien et al., 2016). Other members in the same subfamily displayed divergent expression patterns. For example, NPF6.4 (NRT1.3) showed cortex-specific expression while NPF6.2 (NRT1.4) was broadly expressed in cortical and stellar cells. The more distant member, NPF6.1 expression was not detected in roots under the conditions provided. These NPF6s may have evolved to perform distinct functions in transporting NO3– radially from root surfaces to inner vasculature.
Among the three NPF7 transporters, the closely related NPF7.3 (NRT1.5) and NPF7.2 (NRT1.8) mediate opposite functions in long-distance transport. NPF7.3 has been shown to export NO3– and K+ from pericycle to xylem for shoot translocation, whereas NPF7.2 retrieves NO3– from xylem and retain NO3– in root cells (Lin et al., 2008; Li et al., 2010, 2017b; Drechsler et al., 2015). Interestingly, these two homologs shared high expression in the pericycle (Figure 2F), suggesting that their transport mode may be opposite, one mediating influx and one efflux in the same cell-type.
The five NPF8 proteins are not well understood concerning their NO3– transport properties. NPF8.1/8.2/8.3 (PTR1/5/2) are shown to transport dipeptide, hormones and/or amino acid (Corratgé-Faillie and Lacombe, 2017). NPF8.2 is preferentially expressed in flowers, where it transported dipeptide across the PM of germinating pollen (Komarova et al., 2008; Hammes et al., 2010). Thus, its expression was barely detected in roots (Figure 2G). The tissue- and cell-specific expression for other NPF8 transporters have not been previously reported. In this study, we found that these NPF8.1/8.3/8.5 were broadly expressed in multiple root cell-types. If these transporters are localized to the PM and capable of transporting NO3–, they may function with NPF6.3 and NPF4.6 in NO3– uptake at the root surfaces. In addition, NPF8.1/8.3 were also expressed in stellar cells, suggesting their involvement in long-distance transport. NPF8.4 may have more specific function in NO3– loading to phloem based on its phloem-specific expression. While these expression patterns provide a valuable information regarding their potential functions, analyses of their subcellular localizations, transport properties and physiological roles are necessary for validations.
NRT2 Transporters
Unlike NPF, the NRT2 family comprises seven proteins that are considered high-affinity NO3– transporters (Orsel et al., 2002; Krapp et al., 2014). These proteins were identified by sequence homology to NRT2.1, which was isolated by a differential display approach due to highly NO3–inducible expression (Filleur and Daniel-Vedele, 1999; Orsel et al., 2002). NRT2.1/2.2/2.4/2.5 were shown to be PM-localized transporters, involved in NO3– uptake especially under low-NO3– stress (Cerezo et al., 2001; Filleur et al., 2001; Kiba et al., 2012; Lezhneva et al., 2014). Because their expressions are highly induced by low-NO3–, the single cell profiling detected only low levels of expression in roots under the normal conditions. Nevertheless, they were expressed in root epidermis (Figure 3), consistent with their uptake functions. NRT2.7 is the only transporter located at the tonoplast, where it is shown to import NO3– into vacuoles of seeds (Chopin et al., 2007). However, it was also broadly expressed in various cell-types in young roots compared to the other NRT2s (Figure 3). This suggests that NRT2.7 may serve as a housekeeping function to sequester excess NO3– into the vacuole in multiple tissues.
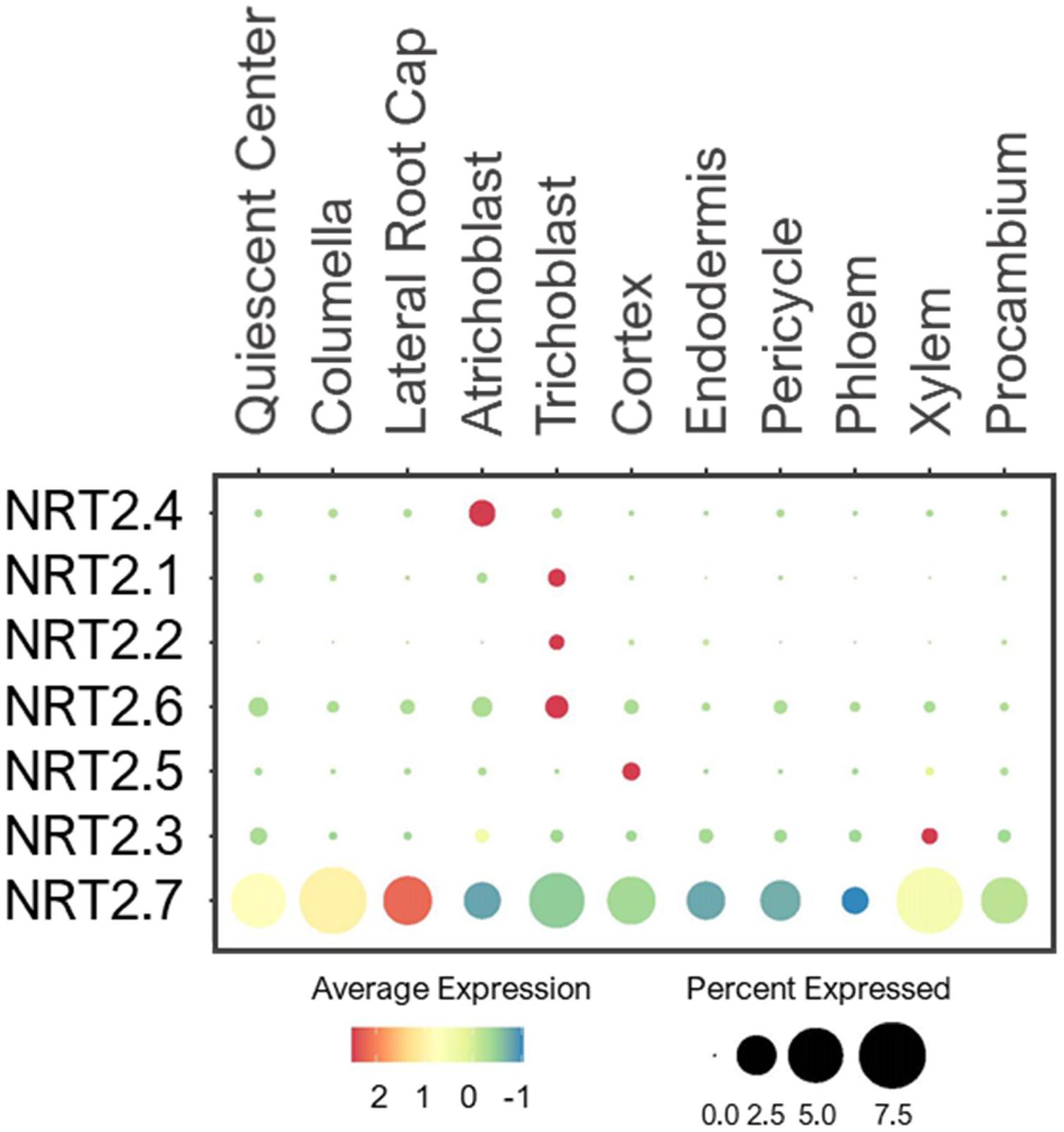
Figure 3. Expression of the high-affinity NRT transporters in Arabidopsis root single cells. In dot plot, the circle size represents the proportion (%) of cells expressing a given gene, and the color of scale bar represents the mean expression (natural log +1 pseudocount).
CLC Antiporters
Arabidopsis CLCs were identified by the sequence homology to voltage-gated chloride channels in animals (Hechenberger et al., 1996; Lv et al., 2009). The plant CLC family contains both channels and antiporters that transport anions including NO3– (Fan et al., 2017). The Arabidopsis genome encodes 7 CLC members. CLCa-c, and CLCg were localized to tonoplast, CLCd/f to Golgi, and CLCe to chloroplast (Fecht-Bartenbach et al., 2007; Marmagne et al., 2007; Lv et al., 2009; Barbier-Brygoo et al., 2011). Because CLCe is shown to be specifically expressed in green tissues (Monachello et al., 2009), its expression in roots was undetected (Figure 4A). CLCa is a major component that drives NO3– accumulation into vacuoles (Geelen et al., 2000; De Angeli et al., 2006). Its functional significance was evident from the ubiquitous expression in all cell-types in developing roots under the normal conditions (Figure 4A). Likewise, CLCc was widely expressed, thus may share similar tasks as CLCa in vacuolar NO3– sequestration. The expression of CLCb was specifically detected in the cortex, suggesting a specific function of this CLC in storing NO3– in the vacuole of cortical cells. Additionally, two Golgi-localized transporters, CLCd and -f showed ubiquitous expression in all cell-types, implicating them in the housekeeping function of anion distribution in Golgi apparatus of all cell-types.
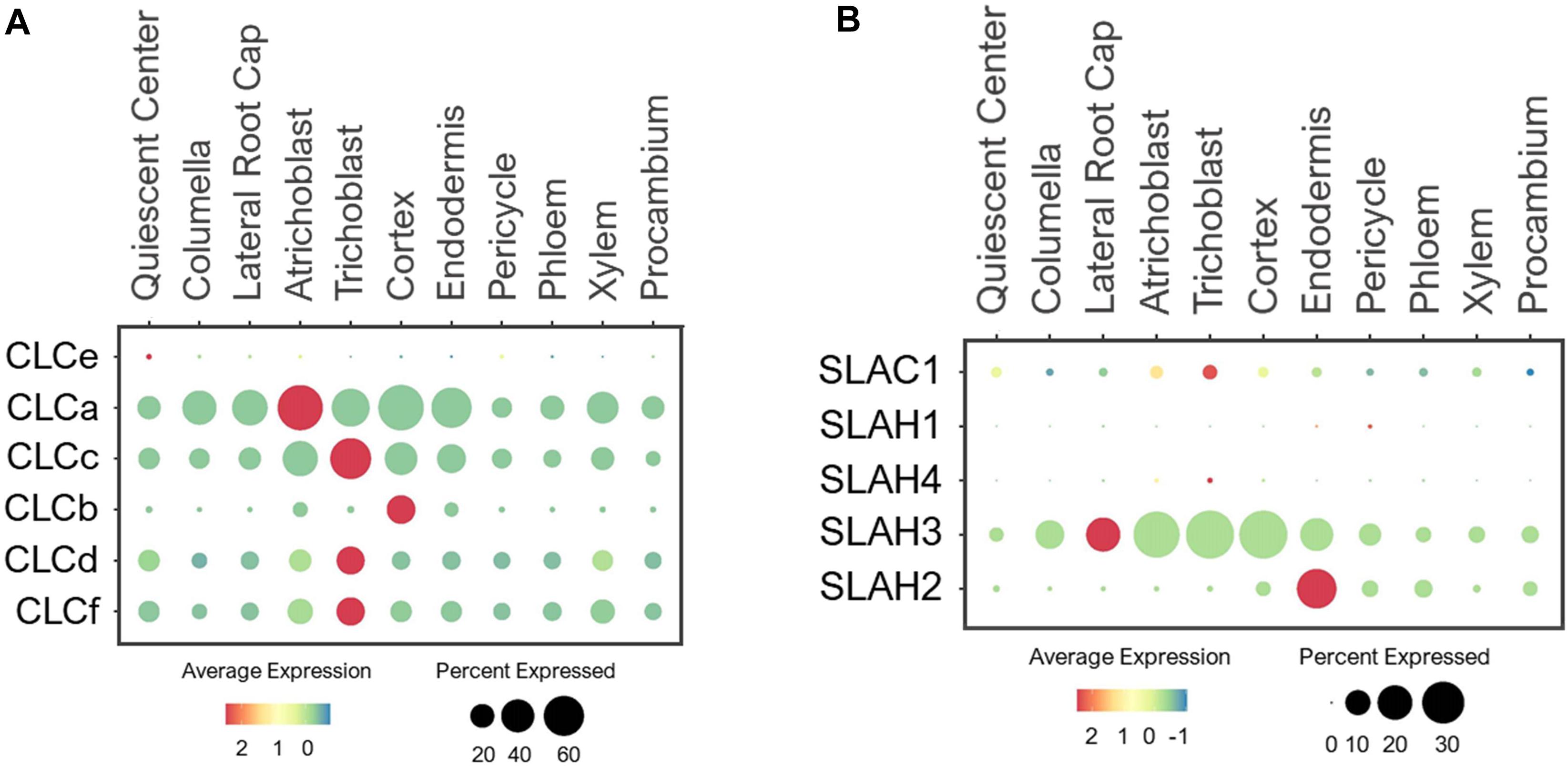
Figure 4. Expression of the (A) CLC antiporters and (B) SLAC1 channels in Arabidopsis root single cells. In dot plots, the circle size represents the proportion (%) of cells expressing a given gene, and the color of scale bar represents the mean expression (natural log +1 pseudocount). The expression of CLCg was not detected in any single cells.
SLAC Channels
SLAC1 shares homology to TehA transporters in bacteria, archaea and fungi (Chen et al., 2010; Dreyer et al., 2012). SLAC1 and its four homologs, SLAH1-4, are anion channels that conduct NO3– and Cl– transport across the PM (Negi et al., 2008; Barbier-Brygoo et al., 2011; Fan et al., 2017). SLAC1 and SLAH3 were shown to mediate NO3–/Cl– efflux and regulate stomatal closure (Schmidt and Schroeder, 1994; Vahisalu et al., 2008; Geiger et al., 2009b, 2011; Chen et al., 2010; Zheng et al., 2015). Because SLAC1 is preferentially expressed in guard cells (Negi et al., 2008), it was not detected in roots (Figure 4B). SLAH1/3 are earlier reported to be expressed in root pericycle to mediate Cl– efflux to xylem (Cubero-Font et al., 2016). SLAH2 is shown to be a NO3–-specific channel that may facilitate root-to-shoot NO3– transport (Maierhofer et al., 2014). Based on their expressions in stellar cells, especially pericycle, SLAH1-3 have been speculated to drive NO3– loading to xylem (Hedrich and Geiger, 2017). We examined their expressions based on single cell profiles and found that the closely related SLAH1/4 were barely detected in the developing roots under the normal conditions (Figure 4B). These genes may require specific growth conditions or plant developmental stages for induction. On the other hand, SLAH2/3 showed distinct expression patterns. While SLAH3 was broadly expressed in epidermis, root cap, cortical and endodermal cells, SLAH2 displayed predominant expression in endodermis (Figure 4B), suggesting an endodermis-specific function for SLAH2. The broad expression pattern of SLAH3 is consistent with its functions in diverse processes (Gutermuth et al., 2013; Zheng et al., 2015; Cubero-Font et al., 2016; Zhang et al., 2016; Hedrich and Geiger, 2017).
Potential Network of Nitrate Channels and Transporters in Roots
In Arabidopsis, 73 NO3– transporters belonging to the NPF, NRT2, CLC, and SLAC1/SLAH families are identified, among which, only several have been characterized to function as NO3– transporters in roots (Krapp et al., 2014; Wang et al., 2018). Through single cell transcriptomic analysis, we were able to expand this list, and map the potential network of NO3– channels and transporters in the root system of young seedlings under the normal growth conditions (Figure 5). The previous studies reported the roles of NPF4.6/6.3 and NRT2.1/2.2/2.4/2.5 in NO3– uptake from root surface cells (Huang et al., 1996, 1999; Wang et al., 1998; Liu et al., 1999; Cerezo et al., 2001; Filleur et al., 2001; Kiba et al., 2012). We additionally found several NPF2/5/8, CLCs, NRT2.7 and SLAH3 in the root tip and epidermis, where they might participate in NO3– uptake, efflux or storage/distribution to adjacent cells (Figure 5). Many of these transporters were also expressed in the cortex and endodermis to distribute NO3– toward stellar cells. Among these families, the putative PM-localized NPF2/4/6/8 members are likely involved in NO3– uptake, transfer and long-distance transport; SLAH3 in NO3– distribution; CLCs in NO3– sequestration into vacuoles (also NRT2.7) and Golgi; and the putative vacuolar NPF5s in NO3– remobilization.
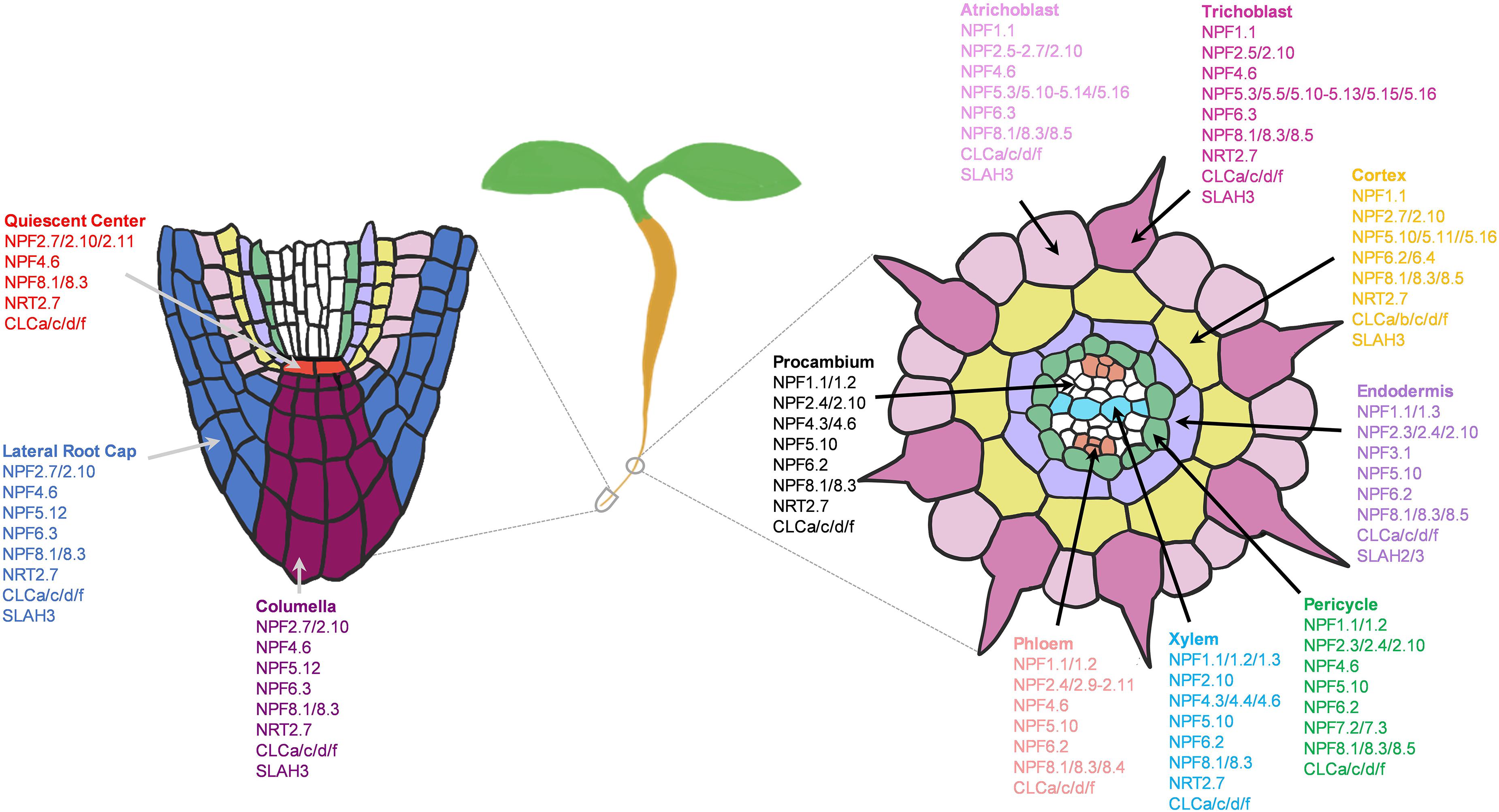
Figure 5. Potential network of nitrate (NO3–) channels and transporters in the root system of 5–7 days old Arabidopsis seedlings grown under the normal growth conditions. Expressed NO3– transporters involved in longitudinal transport (left) at the meristematic zone and radial transport (right) at the elongation/maturation zones were shown. Nitrate transporter genes not expressed under the normal growth conditions were excluded.
In addition, we also found few transporters with cell-type specific expressions such as NPF6.4 and CLCb in cortex, and NPF3.1 and SLAH2 in endodermis. These cortex-specific transporters may be the major contributors to the higher NO3– concentrations observed in the cortical cells compared to the epidermal cells, as reported in the previous study using the NO3–-selective microelectrodes (Zhen et al., 1991). Such single cell profiling of nitrate concentrations in NO3– transporter mutants compared to the wild type may determine the function of a nutrient transporter in a specific cell type.
NPF2.3/7.3 was previously reported to export NO3– from pericycle to xylem (Lin et al., 2008; Taochy et al., 2015; Li et al., 2017b), NPF7.2 in NO3– retrieval from xylem (Lin et al., 2008; Li et al., 2010, 2017b; Drechsler et al., 2015), and NPF2.9 in NO3– loading to phloem (Wang and Tsay, 2011). We additionally found several NPF1/2/4/5/6/8, NRT2.7 and CLC transporters expressed in stellar cells that might participate in long-distance NO3– transport and distribution (Figure 5). Among these, several showed cell-type specific expressions, such as NPF5.1/5.7/5.8 in pericycle, NPF4.3 in procambium, NPF4.4 xylem, and NPF8.4 in phloem. These transcriptional profiles provide functional predictions of these NO3– transporters in specific cell-types of small root tissues, grown under the normal conditions. More in-depth analysis of these genes under diverse nutritional and developmental conditions are required to reflect their main sites of action. Also, it is critical to determine the subcellular localizations and transport activities of these uncharacterized transporters to understand their mode of transport for NO3– distribution. So far, the majority of the functionally known NO3– transporters were PM-localized, with few found at the tonoplast and Golgi membranes. There still exists numerous novel NO3– transporters that might localize to different subcellular compartments such as plastids, mitochondria, and ER in addition to vacuole and Golgi.
Phosphate Transport
P is absorbed by plants in the form of inorganic phosphate (Pi) that serves as an integral component of nucleic acids, phospholipids and ATP, and participates in energy metabolism and signaling transduction (Poirier and Bucher, 2002; Chiou and Lin, 2011). Plants require 60–80 μM of cytosolic Pi to sustain growth, but the typical Pi concentration in the natural soil is extremely low (<10 μM) (Pratt et al., 2009; Plaxton and Tran, 2011; Luan et al., 2017). This is largely compensated by the intensive use of P-rock fertilizers, compromising the sustainability of natural resources and negatively impacting the environment (Chiou and Lin, 2011; López-Arredondo et al., 2014; Luan et al., 2017). In response to the changing Pi status in soils, plants have developed large families of Pi transporters for efficient Pi acquisition, translocation, storage and distribution throughout cells and tissues (Gu et al., 2016; Luan et al., 2017). These include five families of Phosphate Transporters (PHT1-5), Glycerol 3-phosphate permease (G3Pp) and Phosphate1 (PHO1) (Stefanovic et al., 2007; Gu et al., 2016; Xu et al., 2019; Lhamo et al., 2020). The membrane localizations of many of these Pi transporters have been elucidated, allowing us to better predict their functions in the root system using single cell transcriptomic datasets. This in silico approach has provided us the opportunity to create a potential network of Pi transporters that together direct the Pi flow from the epidermis to the vasculature. However, this is purely based on transcriptional profiling of Pi transporters at a single time-point (5–7 days old seedlings) and one condition (normal). While the present study serves as a starting point, the responses of Pi transporters during different plant life stages and stress conditions need to be evaluated in more details, backed up by sufficient genetic and physiological studies to support the putative model.
PHT1 Transporters
The PHT1 family contains nine members that share sequence homology to the yeast high-affinity H+/Pi symporter, PHO84 (Poirier and Bucher, 2002; Nussaume, 2011). PHT1s are located at the PM of root surface cells to mediate Pi uptake and translocation (Mudge et al., 2002; Nussaume, 2011). PHT1;1 and PHT1;4 are the dominant players in Pi absorption under a wide-range of Pi concentrations (Misson et al., 2004; Shin et al., 2004). Consistently, single cell profiles showed the high expression of these two transporters in root epidermis under the normal conditions (Figure 6). PHT1;4 was additionally expressed in LRC. This cell-type is shown to be important for sensing low-Pi stress to promote root growth arrest (Svistoonoff et al., 2007), as observed in Figure 1. In addition, a study using high-resolution real-time 33P imaging indicated that the root cap cells contribute 20% of the total seedling Pi uptake (Kanno et al., 2016). These suggest the role of PHT1;4 in Pi sensing and uptake at the root tip. This together with PHT1;1 at the epidermis makeup the largest portion of absorbed Pi in Arabidopsis seedlings (Shin et al., 2004).
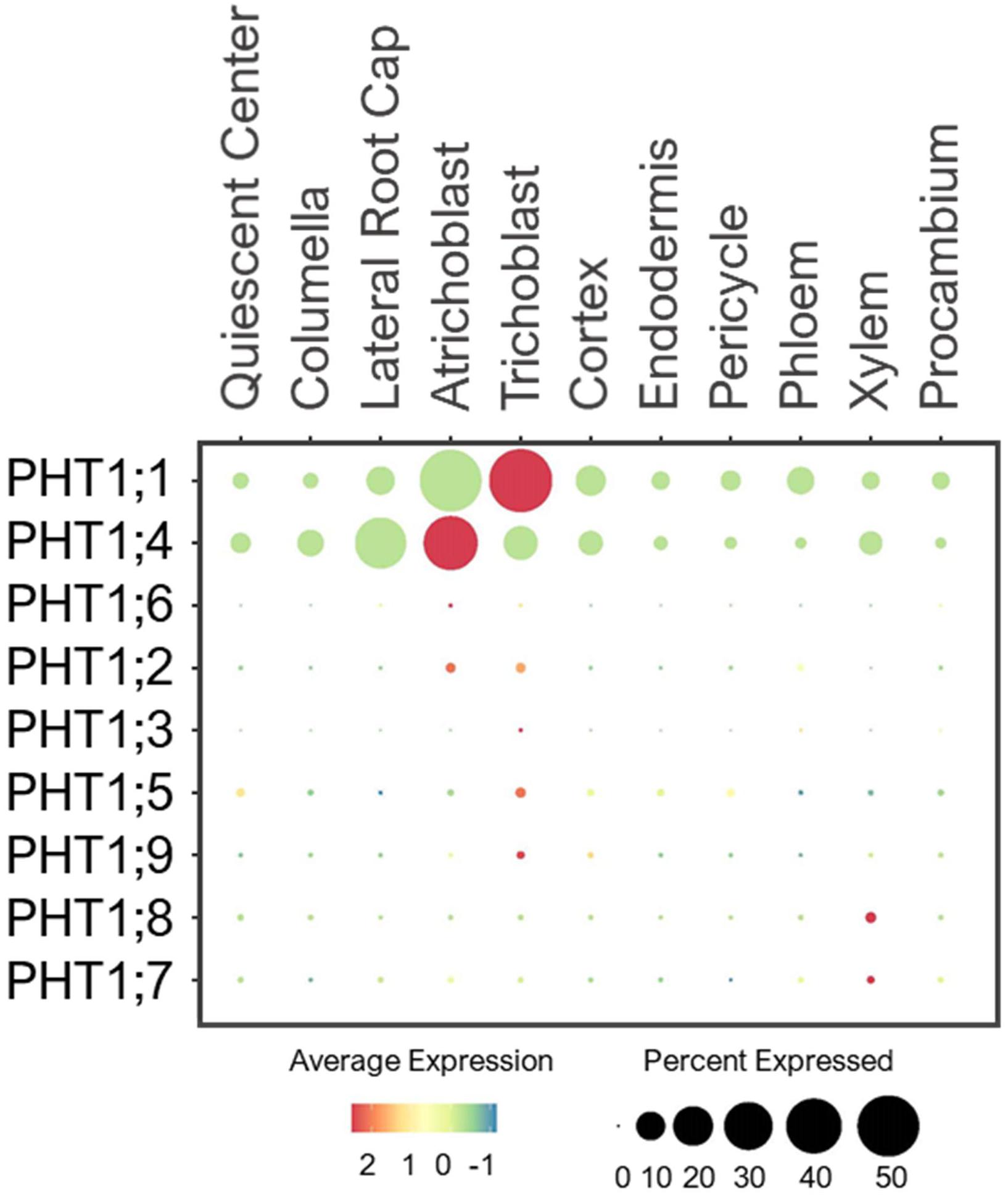
Figure 6. Expression of the high-affinity PHT1 transporters in Arabidopsis root single cells. In dot plot, the circle size represents the proportion (%) of cells expressing a given gene, and the color of scale bar represents the mean expression (natural log +1 pseudocount).
PHT1;8 and PHT1;9 were shown to mediate Pi uptake from roots and translocation to shoots especially under low-Pi (Remy et al., 2012; Lapis-Gaza et al., 2014). In line with these studies, the single cell profiles revealed a low expression of PHT1;9 in epidermis and PHT1;8 in xylem of developing roots under the normal conditions (Figure 6). PHT1;7 was also lowly detected in xylem, and may share overlapping functions with PHT1;8 in translocating Pi from root to shoot. Because no definitive Pi transporters at the PM of stellar cells have been reported, we speculate that some of the PHT1 members such as PHT1;7/1;8 may be present to promote xylem loading, and their expression may be more responsive to low-Pi than the normal conditions provided in this study.
Other PHT1 members were lowly expressed in epidermis of young seedlings under the normal conditions (Figure 6), and might be more responsive to Pi starvation to initiate Pi uptake (Mudge et al., 2002; Nussaume, 2011). In addition to the condition requirement, some may function in later stages of plant development. For instance, PHT1;5 was shown to recycle Pi from source-to-sink (senescing leaves to young leaves and roots) in response to low-Pi (Mudge et al., 2002; Nagarajan et al., 2011; Nussaume, 2011). These indicate plants may not need to activate the transcription of all PHT1 members during the early growth stage when the Pi supply is sufficient.
PHT2/3/4 Transporters
PHT2/3/4/5 family transporters are located at different subcellular compartments where they participate in Pi storage and distribution throughout the plant (Gu et al., 2016; Luan et al., 2017). Several of these transporters have been characterized and shown to have function in green tissues. However, their expression and physiology in the root system are not well understood. For example, PHT2;1, a single member of the PHT2 family was shown to transport Pi into chloroplasts in leaves (Daram et al., 1999; Versaw and Harrison, 2002; Rausch et al., 2004). We detected PHT2;1 expression exclusively in root endodermis (Figure 7A), raising a question about its intriguing function in root Pi transport. We hypothesize that PHT2;1 in endodermis may contribute to Pi translocation to shoots, where its function is more prominent.
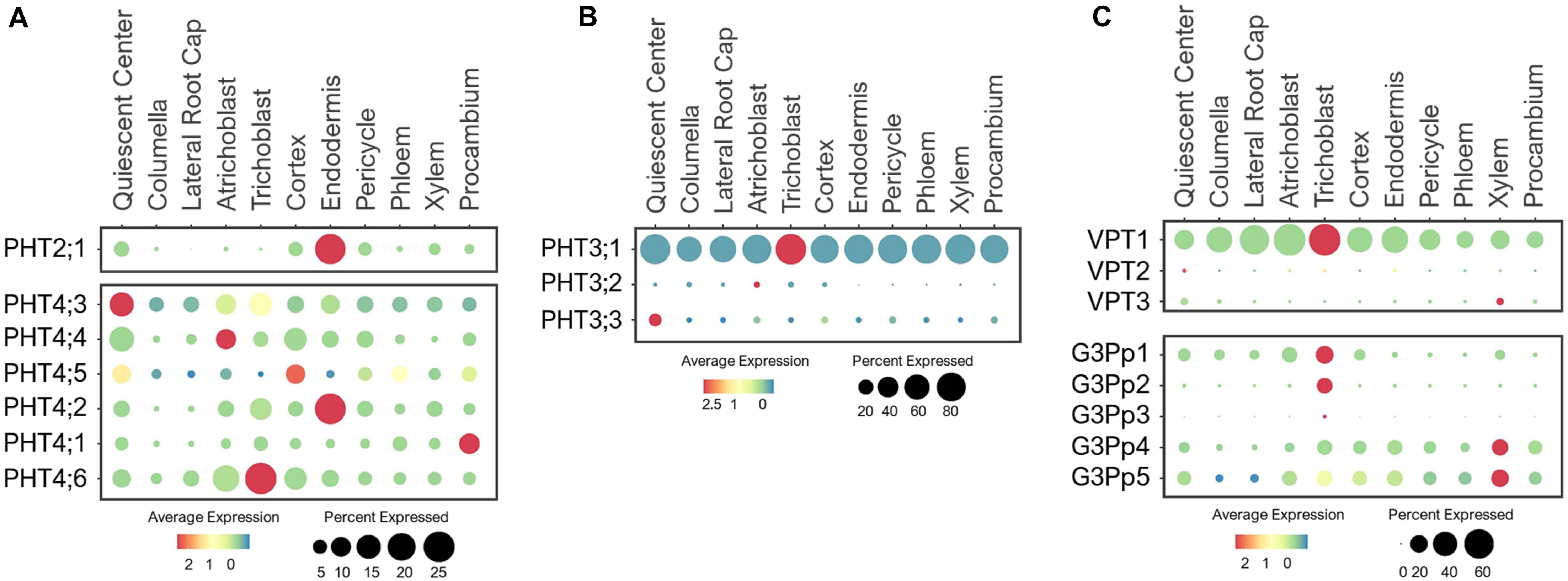
Figure 7. Expression of the (A) PHT2/4, (B) PHT3 and (C) VPT and G3Pp transporters in Arabidopsis root single cells. In dot plots, the circle size represents the proportion (%) of cells expressing a given gene, and the color of scale bar represents the mean expression (natural log +1 pseudocount).
Similar to PHT2;1, five PHT4 members (PHT4;1–4;5) are targeted to plastids, and one (PHT4;6) in Golgi (Guo et al., 2008a, b). These PHT4s were shown to mediate Pi transport when expressed in yeast Pi transport mutants (Guo et al., 2008b). PHT4;1, PHT4;4, and PHT4;5 were reported to be expressed in leaves, PHT4;2 exclusively in roots, and PHT4;3 and PHT4;6 in both tissues (Guo et al., 2008a, b). Among these, PHT4;1 was shown to compartmentalize Pi in chloroplasts for ATP synthesis and photosynthesis (Guo et al., 2008a; Karlsson et al., 2015). PHT4;4 was reported to transport ascorbate into chloroplasts, and capable of transporting both Pi and ascorbate in liposomes (Miyaji et al., 2015). PHT4;2 was shown to perform reversible Na+-dependent Pi transport in root plastids (Irigoyen et al., 2011). Expression analysis of these PHT4 members in root single cells revealed broad expression in multiple cell-types of young roots under the normal growth conditions (Figure 7A), likely to support their housekeeping functions in plastids. However, these transporters had distinct preferential expressions. For example, PHT4;3 and PHT4;5 were highly expressed in QC and PHT4;1 in procambium, suggesting their roles in balancing plastid Pi in meristematic cells of the root tip and the vasculature, respectively. Also, PHT4;3/4;4/4;6 were highly expressed in epidermis, PHT4;5 in cortex, and PHT4;2 in endodermis. These PHT4s may coordinate with each other to control Pi fluxes in plastids throughout the root system, which is essential for the synthesis and distribution of amino acids, fatty acids and secondary compounds (Flügge et al., 2003). PHT4;6 is the only PHT4 member localized to the Golgi, where it was shown to mediate Pi efflux from Golgi to cytosol (Cubero et al., 2009; Hassler et al., 2012, 2016). In a single cell profile of developing roots, PHT4;6 was largely expressed in epidermis and cortex (Figure 7A). This suggests a potential function of PHT4;6 in maintaining Pi homeostasis in Golgi of these transit cell-types. Currently, the function of many PHT4 members in Pi transport and distribution in the root system and whether these may participate in long-distance transport are vaguely understood. We provided assumptions based on the known subcellular localization and expression profiling of these Pi transporters at one time point and growth condition. Their functions in the root system need to be verified experimentally.
The PHT3 family contains three members in Arabidopsis that show sequence homology to the first mitochondrial Pi translocator (MPT1) cloned from birch plants and yeast MIR1 (Kiiskinen et al., 1997; Takabatake et al., 1999; Hamel et al., 2004). These PHT3s transport Pi from cytosol into the mitochondrial matrix to support ATP synthesis and redox homeostasis (Zhu et al., 2012; Jia et al., 2015). PHT3;1 (MPT3) was reported to be ubiquitously expressed in multiple tissues, while PHT3;2 (MPT2), and PHT3;3 (MPT1) were only expressed in the green and/or reproductive tissues (Zhu et al., 2012). Among these, PHT3;1 had the largest contribution to the mitochondrial Pi homeostasis, which was shown to be indispensable for plant development (Jia et al., 2015). Consistent with these studies, PHT3;1 displayed ubiquitous expression in all root cell-types, while the other two were barely detected in developing roots under the normal growth conditions (Figure 7B). This emphasizes the essential role of PHT3;1 in supporting mitochondrial functions throughout the root system. Further studies of these mitochondrial transporters have been compromised by the lethality of their homozygous mutants (Jia et al., 2015).
VPT and G3Pp Transporters
In Arabidopsis, three members of the PHT5 family are responsible for Pi storage into vacuoles (Liu et al., 2015, 2016). VPT1 (PHT5;1) is the major player in vacuolar Pi sequestration (Liu et al., 2015, 2016; Luan et al., 2019). Single cell profiling revealed a ubiquitous expression of VPT1 in all root cell-types, while VPT2 (PHT5;2) and VPT3 (PHT5;3) were expressed at low levels in roots of young seedlings under the normal growth conditions (Figure 7C). This is consistent with the earlier functional analysis (Liu et al., 2015, 2016). However, the expressions of VPT2/3 may become more noticeable in roots of later stages or in response to a specific stress, such as Pi toxicity. In addition, these VPTs may function in other tissues or organs. For example, VPT3 along with VPT1 are shown to play role in the later stage of plant development for systemic allocation of Pi between leaves and flowers (Luan et al., 2019).
The stored Pi in vacuoles is remobilized by the G3Pp family transporters (Xu et al., 2019). This family contains five G3Pp members in Arabidopsis, among which, G3Pp1-3 (VPE1-3) are localized to tonoplast (Ramaiah et al., 2011; Luan and Lan, 2019; Xu et al., 2019). Only G3Pp2 has been confirmed to be a functional vacuolar Pi exporter (Xu et al., 2019). G3Pp4 is localized to the plastid envelope and mediated lipid accumulation in seeds (Kawai et al., 2014). The membrane localization of G3Pp5 remains uncharacterized, so are the functions of the majority of G3Pps. Single cell profiling of young seedlings under the nutrient sufficiency displayed G3Pp1 expression in root tip, epidermis and cortex, and G3Pp2 exclusively in trichoblast, whereas G3Pp3 was barely detected (Figure 7C). Considering these G3Pps as efflux transporters, G3Pp1/2 may work concurrently with VPT1 in controlling Pi fluxes between vacuoles and cytosol of epidermal cells to maintain stable cytosolic Pi concentration, and might aid in Pi distribution to the adjacent cells under the normal conditions. Their roles in vacuolar Pi remobilization are critical for low-Pi adaptation, and as such, these genes were shown to be highly induced by low-Pi stress (Ramaiah et al., 2011). On the other hand, the G3Pp3 expression was highly upregulated under various low-nutrient (Pi, K+, NO3–, and Iron) stresses (Ramaiah et al., 2011), suggesting the specific role of this transporter in stress-response. Based on their putative functions, we anticipate a wider expression of these G3Pps in different root cell-types upon nutrient-stress conditions. G3Pp4 and its close homolog, G3Pp5, shared broad expression in multiple cell-types including xylem during normal growth conditions (Figure 7C). These G3Pps may contribute to Pi distribution via Pi efflux from plastids, however, their Pi transport mechanisms and physiological functions need to be confirmed.
PHO1 Transporters
Plants transfer Pi from root-to-shoot via xylem loading. This process requires Pi efflux from root pericycle to xylem, which is mediated by the Golgi-localized Pi exporter, PHO1 (Poirier et al., 1991; Hamburger et al., 2002; Stefanovic et al., 2011; Arpat et al., 2012). It contains 10 additional homologs in the Arabidopsis genome, referred to as PHO1;H1–H10 (Wang et al., 2004). Studies have shown their tissue- and organ-specific expressions (Hamburger et al., 2002; Wang et al., 2004; Stefanovic et al., 2007; Khan et al., 2014), but their cell-type-specific expression in the root system remains unknown. PHO1;H1 is closely related to PHO1, and shares a redundant function in xylem loading (Stefanovic et al., 2007). Consistent with their functions, single cell profiling revealed the overlapping expression of PHO1 and PHO1;H1 in pericycle (Figure 8). However, PHO1;H1 was expressed at a low level under the normal conditions because its function is more prominent in response to low-Pi (Stefanovic et al., 2007).
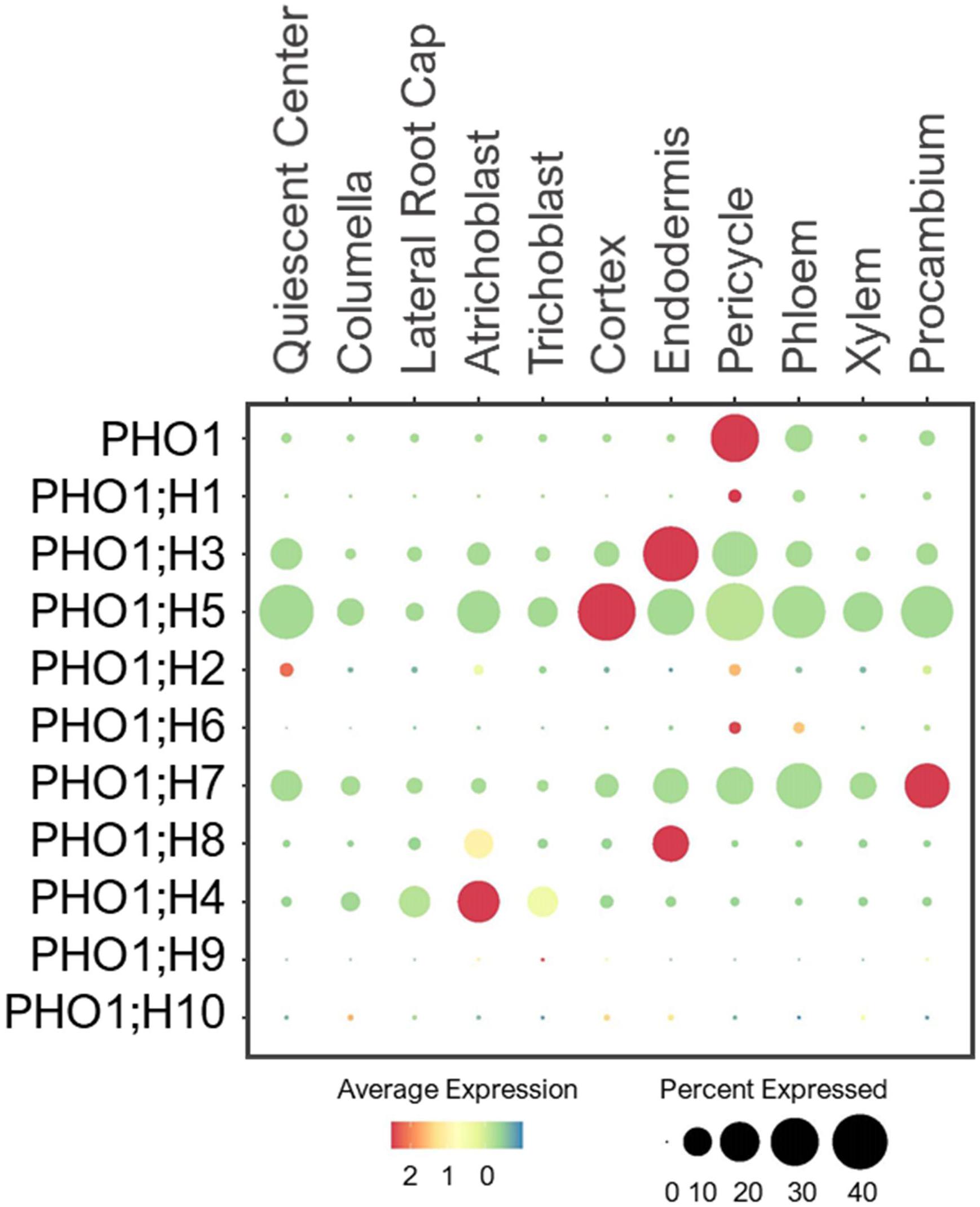
Figure 8. Expression of the PHO1 transporters in Arabidopsis root single cells. In dot plot, the circle size represents the proportion (%) of cells expressing a given gene, and the color of scale bar represents the mean expression (natural log +1 pseudocount).
PHO1;H3 shared a similar expression pattern and subcellular localization to PHO1, but it had the opposite function in long-distance transport (Hamburger et al., 2002; Khan et al., 2014). It was shown to restrict root-to-shoot Pi transport under zinc deficiency (Khan et al., 2014). In root single cell profile, PHO1;H3 was largely expressed in endodermis and pericycle (Figure 8). This implies PHO1;H3 may retrieve Pi from the xylem back to pericycle and endodermis. PHO1;H2, PHO1;H5 and PHO1;H6 are close homologs of PHO1;H3 (Wang et al., 2004), and shared overlapping expressions in pericycle (Figure 8). However, PHO1;H2 and PHO1;H6 were expressed at low level and may require specific environmental conditions to induce their expression and function in long-distance transport. In contrast, PHO1;H5 was broadly expressed in all cell-types except root cap, suggesting its potential role in radial Pi distribution from root epidermis to stellar cells or vice versa. PHO1;H4, PHO1;H7, and PHO1;H8 are closely related (Wang et al., 2004), but they showed distinct expression patterns. For example, PHO1;H4 was largely expressed in epidermis and lateral root cap (Figure 8), and may mediate root-soil Pi exchange. PHO1;H8 was highly expressed in endodermis, and likely transport Pi to pericycle for xylem loading. PHO1;H7 was more broadly expressed in endodermis and stellar cells, suggesting its role in xylem and/or phloem loading. However, these are putative functions based on their expression patterns, and need to be validated experimentally. Whether all these PHO1 homologs localize to Golgi, and how Golgi might engage in long-distance transport remain unknown.
Potential Network of Phosphate Transporters in Roots
Pi transporters are found at the PM and membranes of different subcellular compartments, and most of their tissue expressions have been elucidated (Wang et al., 2004; Ramaiah et al., 2011; Gu et al., 2016; Luan et al., 2017; Xu et al., 2019). However, many of their cell-type specific expressions, physiology and Pi transport network in the root system are not well understood. By examining the expression patterns of Pi transporters, we generated a potential network of Pi transporters involved in longitudinal and radial Pi transport in the root system of young Arabidopsis seedlings under the normal growth conditions (Figure 9). In general, PHT1s (esp. PHT1;1/1;4) mainly absorbed Pi from the root tip and epidermis, while other subcellular-localized PHT, G3Pp, and PHO1;H transporters might distribute Pi further to the adjacent cells. Many of these subcellular Pi transporters that are expressed under the normal conditions showed widespread expression to distribute Pi throughout the root system to support housekeeping functions. These include mitochondrial PHT3;1, vacuolar PHT5;1, most plastid/Golgi PHT4s, putative plastid G3Pp4/5, and putative Golgi PHO1;H5.
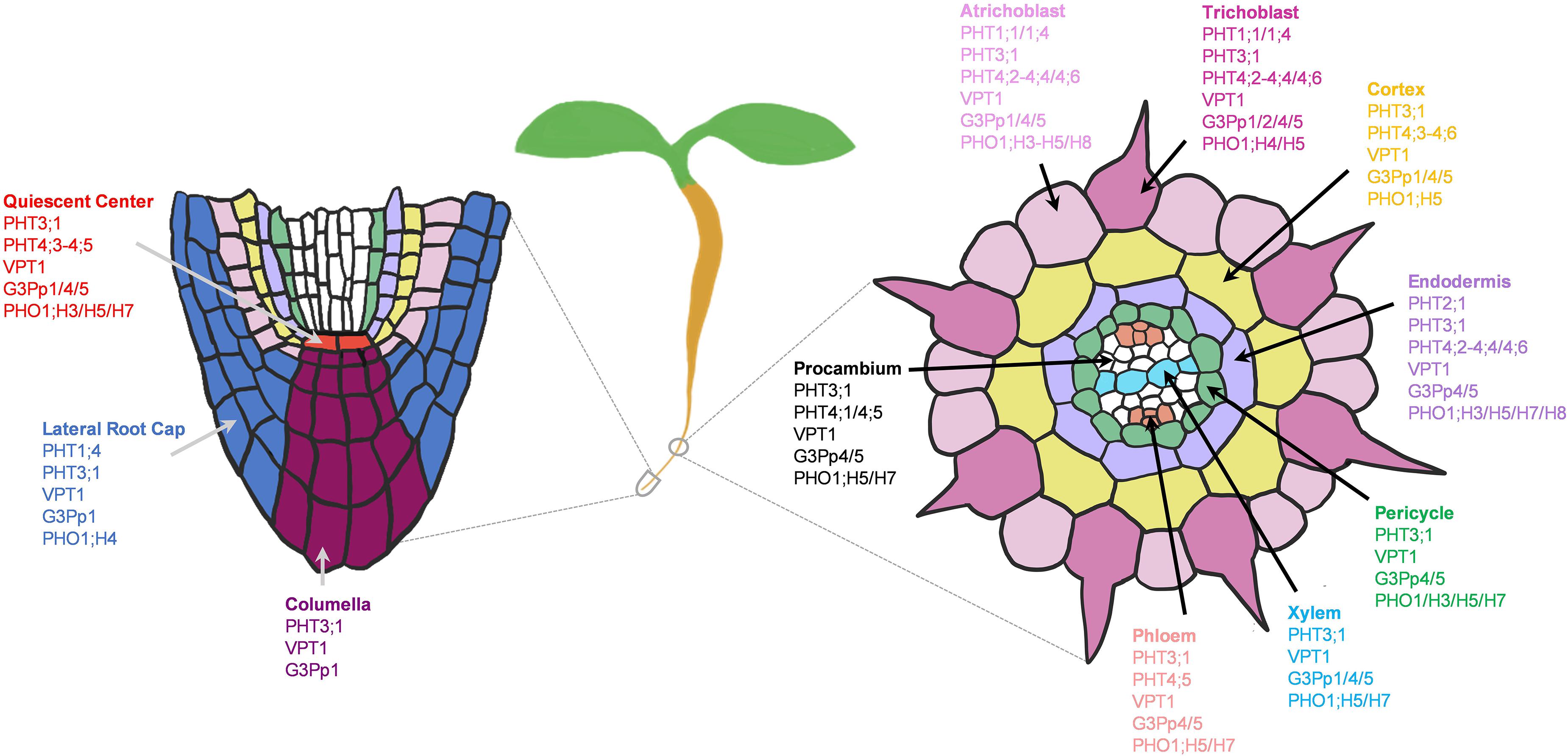
Figure 9. Potential network of phosphate (Pi) transporters in the root system of 5–7 days old Arabidopsis seedlings grown under the normal growth conditions. Expressed Pi transporters involved in longitudinal transport (left) at the meristematic zone and radial transport (right) at the elongation/maturation zones were shown. Pi transporter genes not expressed under the normal growth conditions were excluded.
Several Pi transporters showed cell-type specific expressions, including G3Pp2 and PHO1;H4 in epidermis, PHT2;1 in endodermis, and PHO1 in pericycle. Among these, G3Pp2 is shown to be involved in vacuolar Pi remobilization (Xu et al., 2019), PHT2;1 in Pi transport into plastids (Daram et al., 1999; Versaw and Harrison, 2002; Rausch et al., 2004), and PHO1 in root-to-shoot Pi translocation (Poirier et al., 1991; Hamburger et al., 2002). Except for PHO1, their contributions in specific root cell-types remain obscure, and may be subjected to modifications upon the changing environmental stresses and developmental stages. We additionally found several PHO1 homologs especially PHO1;H5/H7 that are most likely involved in long-distance transport during plant development (Figure 9). These are potential candidates for both xylem and/or phloem loading or unloading, which need to be confirmed genetically and physiologically.
To understand how these Pi transporters may contribute to the low-Pi adaptation, we also profiled their expression patterns from scRNA-seq analyzed under low-Pi (Wendrich et al., 2020). PHT1;1/1;4, PHT2;1, VPT1, G3Pp2, PHO1, and PHO1;H3/H5 were differentially expressed in response to low-Pi (Supplementary Figure 1), with similar expression patterns to our analysis. These Pi transporters may contribute to low-Pi adaptation, probably by enhancing Pi uptake, fine-tuning Pi fluxes from subcellular compartments, and redistributing Pi between shoots and roots. Therefore, these Pi transporters are worth further investigations especially PHT2;1, G3Pp2, and PHO1;H5, whose roles in the root system have not been studied yet. In addition, we need to identify novel Pi transporters that transfer Pi across the PM of different root cell-types besides PHT1s.
Potassium Transport
K+ is the most abundant cation in plants, required for the membrane potential maintenance, protein synthesis, enzyme activation, stomatal movement and photosynthesis (Wang et al., 2013; Luan et al., 2017; Wang and Wu, 2017; Hasanuzzaman et al., 2018). Plants need 100–200 mM of cytosolic K+ to sustain cell metabolism (Walker et al., 1996; Tester and Leigh, 2001; Sharma et al., 2013; Luan et al., 2017). However, K+ is often limited in the natural environment (Maathuis, 2009; White, 2013), leading to heavy fertilizer application that raises sustainability issues. To combat the large fluctuations in the soil K+, plants have evolved elaborate transport systems to maximize K+ absorption and distribution. These include Shaker-type and TPK (Tandem Pore K+) channels, KUP/HAK/KT (K+ Uptake/High-Affinity K+ Transporter) transporters, and four families of Cation/Proton Antiporters (CPA), including CHX (Cation/H+ Exchangers), KEA (K+ Efflux Antiporters) and NHX (Na+/H+ Exchangers) (Mäser et al., 2001; Gomez-Porras et al., 2012; Wang and Wu, 2013; Santa-María et al., 2018; Lhamo et al., 2021). Decades of research have elucidated the importance of these K+ channels and transporters in different plant tissues and organs. However, how these diverse K+ channels and transporters coordinate in the root system to translocate K+ to aerial tissues is unclear. We utilize the single cell profiles to provide a potential network of K+ channels and transporters that transport K+ from the root epidermis to the xylem vessels using young Arabidopsis seedlings grown under the normal conditions.
Shaker-Type Channels
The Arabidopsis Shaker-type family is a well-studied group of voltage-gated K+ channels, with nine members that are classified into three categories based on their voltage dependence (Pilot et al., 2003; Lebaudy et al., 2007; Wang and Wu, 2013). These include K+ inward-rectifying channels such as AKT1/5, KAT1/2, and SPIK (Hirsch et al., 1998; Pilot et al., 2001; Mouline et al., 2002), and weak-rectifying channel, AKT2 (Lacombe et al., 2000; Michard et al., 2005) that are activated by membrane hyperpolarization; and outward-rectifying channels such as SKOR and GORK that are activated by membrane depolarization (Gaymard et al., 1998; Ache et al., 2000). These Shaker-type channels are mainly localized to PM and participate in different physiological processes (Wang and Wu, 2013; Véry et al., 2014; Luan et al., 2017; Lhamo et al., 2021). For example, AKT1 channel is known to mediate K+ uptake from the soil (Lagarde et al., 1996; Hirsch et al., 1998; Gierth et al., 2005), and its activity is fine-tuned by KC1 channel via heteromerization (Reintanz et al., 2002; Duby et al., 2008; Geiger et al., 2009a). As such, these two K+ channels shared overlapping expression patterns in the root tip and epidermis (Figure 10A), where changes in K+ level can be sensed to initiate or prevent K+ uptake. AKT1 was additionally expressed in large portions of xylem and procambium, suggesting its additional role in K+ translocation. This is in line with a previous study reporting AKT1 involvement in K+ retrieval from the xylem sap under the sufficient-K+ condition (Nieves-Cordones et al., 2019).
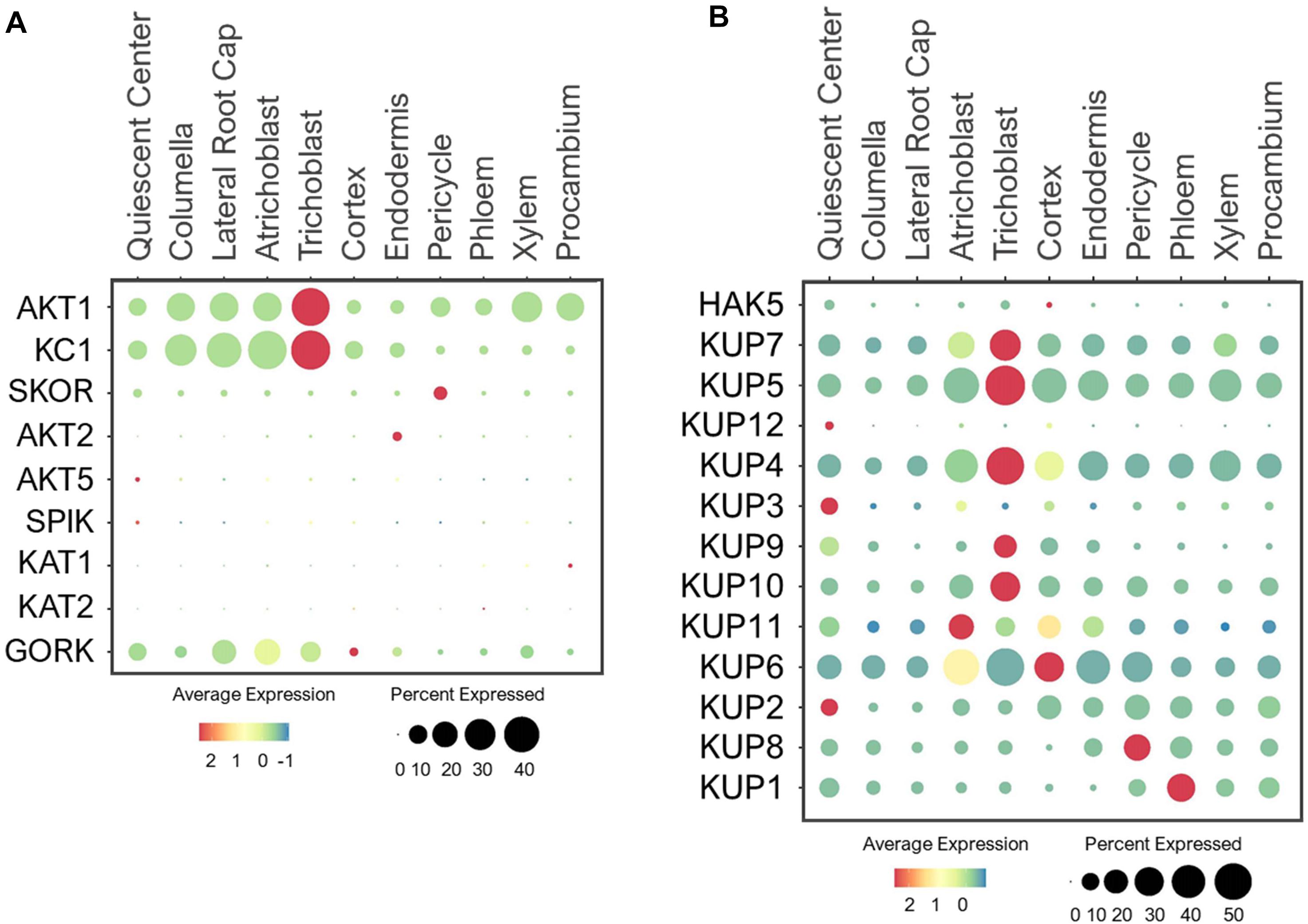
Figure 10. Expression of the (A) Shaker-type channels (B) KUP transporters in Arabidopsis root single cells. In dot plots, the circle size represents the proportion (%) of cells expressing a given gene, and the color of scale bar represents the mean expression (natural log +1 pseudocount).
For shoot translocation, K+ efflux channel, SKOR mediates K+ release from pericycle to xylem (Gaymard et al., 1998; Liu et al., 2006). As expected, SKOR was expressed specifically in pericycle (Figure 10A). However, its low expression level implies the existence of other transporters that may also function in K+ root-to-shoot translocation under the normal conditions since the K+ concentrations in shoots were previously reported to be twice the roots in different plant species using the K+-selective microelectrodes (Tester and Leigh, 2001). Once K+ is present in leaves, it can be recirculated back to roots by AKT2 via phloem loading (Marten et al., 1999; Lacombe et al., 2000). However, AKT2 was lowly detected in root single cells (Figure 10A), suggesting a minimal requirement for K+ recycling from shoot-to-root under the normal conditions. This recycling process may be activated during low-K+ stress to support root metabolic processes.
Several Shaker-type channels were shown to be specifically expressed and function in green tissues and floral organs. For example, AKT5 and SPIK displayed flower-specific expression, where SPIK channel was reported to conduct K+ influx into pollen for pollen tube growth (Lacombe et al., 2000; Mouline et al., 2002; Zhao et al., 2013). KAT1/2 are expressed in leaves and flowers, where they mediate K+ influx to control stomatal opening (Nakamura et al., 1995; Kwak et al., 2001; Pilot et al., 2001). Since these four Shaker-type channels are specifically expressed in aerial tissues, their expressions in root single cells were barely detected (Figure 10A). GORK is also expressed in leaf stomata and mediates K+ efflux to induce stomatal closure (Ache et al., 2000; Hosy et al., 2003). Studies have also revealed GORK expression in root epidermis, where it mediates K+ efflux from root cells to maintain osmotic balance (Ivashikina et al., 2001; Osakabe et al., 2013; Demidchik, 2014). We also found GORK to be expressed in large portions of epidermal, LRC and QC cells (Figure 10A), suggesting GORK may export K+ radially and longitudinally from the root surfaces. Because AKT1 and GORK expressions overlap, their coordination at the root surface might balance K+ influx and efflux, which may prevent epidermal and LRC cells from bursting under the nutrient-rich conditions.
KUP Transporters
The KUP family encodes 13 KUP/HAK/KT members in Arabidopsis that show high homology to the bacterial KUP and fungal TRK transporters (Schleyer and Bakker, 1993; Banuelos et al., 1995). Similar to Shaker-type channels, these KUP transporters also have diverse roles in transporting K+ throughout different cells and tissues. In addition, KUPs are localized to the PM and different subcellular compartments, and some are capable of transporting hormones (Li et al., 2018; Lhamo et al., 2021). Among these, the PM-localized HAK5 and KUP7 mediate K+ uptake in response to low-K+ stress, with an additional role of KUP7 in K+ translocation to shoots (Gierth et al., 2005; Qi et al., 2008; Pyo et al., 2010; Han et al., 2016). Since HAK5 functions specifically under low-K+ (10–100 μM) stress (Rubio et al., 2008; Nieves-Cordones et al., 2014), its expression was hardly detectable in young roots under the normal conditions (Figure 10B). However, KUP7 exhibited ubiquitous expression, suggesting its ability to transport K+ from epidermal to stellar cells for shoot translocation during the normal growth. Although an earlier study indicated probable role of HAK5 in K+ translocation under the sufficient-K+ condition (Nieves-Cordones et al., 2019), our single cell profiles indicated more probable role of KUP7 than HAK5 in this process during early plant development. HAK5 may be involved in K+ translocation during the later stages of development or specially under low-K+ stress, requiring more detailed analysis. Like KUP7, its close homolog, KUP5, was also ubiquitously expressed (Figure 10B), suggesting these two KUPs may functionally overlap with AKT1 in K+ uptake and SKOR in K+ translocation during normal growth conditions.
In addition to K+ transport, several KUPs function in root development. For instance, KUP4 (TRH1) located in the endomembrane promotes root hair growth and gravitropic responses by modulating auxin transport in the root apex (Rigas et al., 2001, 2013; Vicente-Agullo et al., 2004). The single cell profile indicated that KUP4 was ubiquitously expressed in young roots, suggesting it’s important role in root development (Figure 10B). The high expression of KUP4 in trichoblast and cortex may suggest its involvement in generating auxin maxima needed for root hair elongation (Jones et al., 2009; Vissenberg et al., 2020). This is supported by genetic analyses of the KUP4 null mutants that display no root hair growth, and reduced auxin accumulation in epidermal and cortical cells compared to the wild type upon exogenous auxin treatment (Rigas et al., 2013). In addition, the expression of KUP4 in stellar cells suggests it may be involved in both acropetal and basipetal auxin distribution (Sustr et al., 2019). Its close homolog, KUP3, was predominantly expressed in QC (Figure 10B), therefore, may fine-tune K+ and/or hormone distribution to support meristematic activity as observed for KUP9. Recently, KUP9 is shown to mediate auxin and K+ export from ER (Endoplasmic reticulum) to cytosol in response to low-K+, which is essential to maintain meristematic activity and primary root growth (Zhang et al., 2020). As expected, KUP9 was expressed in QC, but also in trichoblast and cortex (Figure 10B). This suggests KUP9 may also contribute to polar auxin transport and K+ distribution during plant growth. The closely related KUP10 and KUP11 displayed broad expressions, however, their single mutants did not show low-K+ sensitive phenotype observed for kup9 mutants (Zhang et al., 2020). Generation of higher-order mutants may dissect functional redundancies between these close homologs, with KUP9 as the dominant player.
KUP2/6/8 are K+ efflux transporters that are suggested to function redundantly in exporting K+ from stellar cells to epidermis (Osakabe et al., 2013). Consistent with this study, our single cell profiling revealed overlapping expressions of these KUPs in stellar cells, with the broader expression of KUP6 in other cell types (Figure 10B). The coordination among KUP2/6/8 might allow efficient K+ transport from stellar cells to root epidermis, where KUP6 along with GORK likely extrude K+ from roots back to the soil. It remains a question whether young plants use such a mechanism to remove K+ buildup under the sufficient-nutrient availability for osmotic balance. In addition, analysis of kup2/6/8 triple mutants revealed the importance of these KUPs in hormone responses that affect cell expansion and lateral root development (Osakabe et al., 2013), however, the mechanism is unclear. Similar to KUP4/9, we suspect that KUP2/6/8 may be capable of transporting hormones in addition to K+. KUP1, the founding member of the Arabidopsis KUP family, was previously shown to mediate both high- and low-affinity K+ transport (Fu and Luan, 1998; Kim et al., 1998). However, its tissue expression and physiological role have not been examined. Our study indicated KUP1 expression in stellar cells, especially the phloem in young seedlings (Figure 10B). This implicated the possible role of KUP1 in phloem loading, and may work in parallel or substitute AKT2 function under the normal growth conditions. Genetic and physiological analyses are needed to confirm its putative function.
CHX Antiporters
CHX is the largest family belonging to CPA superfamily, which in general plays important role in regulating pH and ion homeostasis of the subcellular organelles that affect protein processing, vesicular cargo composition, vesicle movement and protein trafficking (Bassil et al., 2012; Sze and Chanroj, 2018). The CHX family consists of 28 members in Arabidopsis, which are localized to the PM and endosomes where they perform K+/H+ exchange (Chanroj et al., 2011; Sze and Chanroj, 2018). The physiological functions of many of these CHXs remain uncharacterized. Earlier studies showed that a large number of CHXs transporters are expressed in pollen, among which, some are functionally characterized to be involved in male fertility, pollen tube guidance, pollen wall construction and seed development (Sze et al., 2004; Lu et al., 2011; Chanroj et al., 2013; Sze and Chanroj, 2018). Because our single cell profiling is focused on roots of developing seedlings, the expression of many of these CHXs were detected at low levels (Figure 11A).
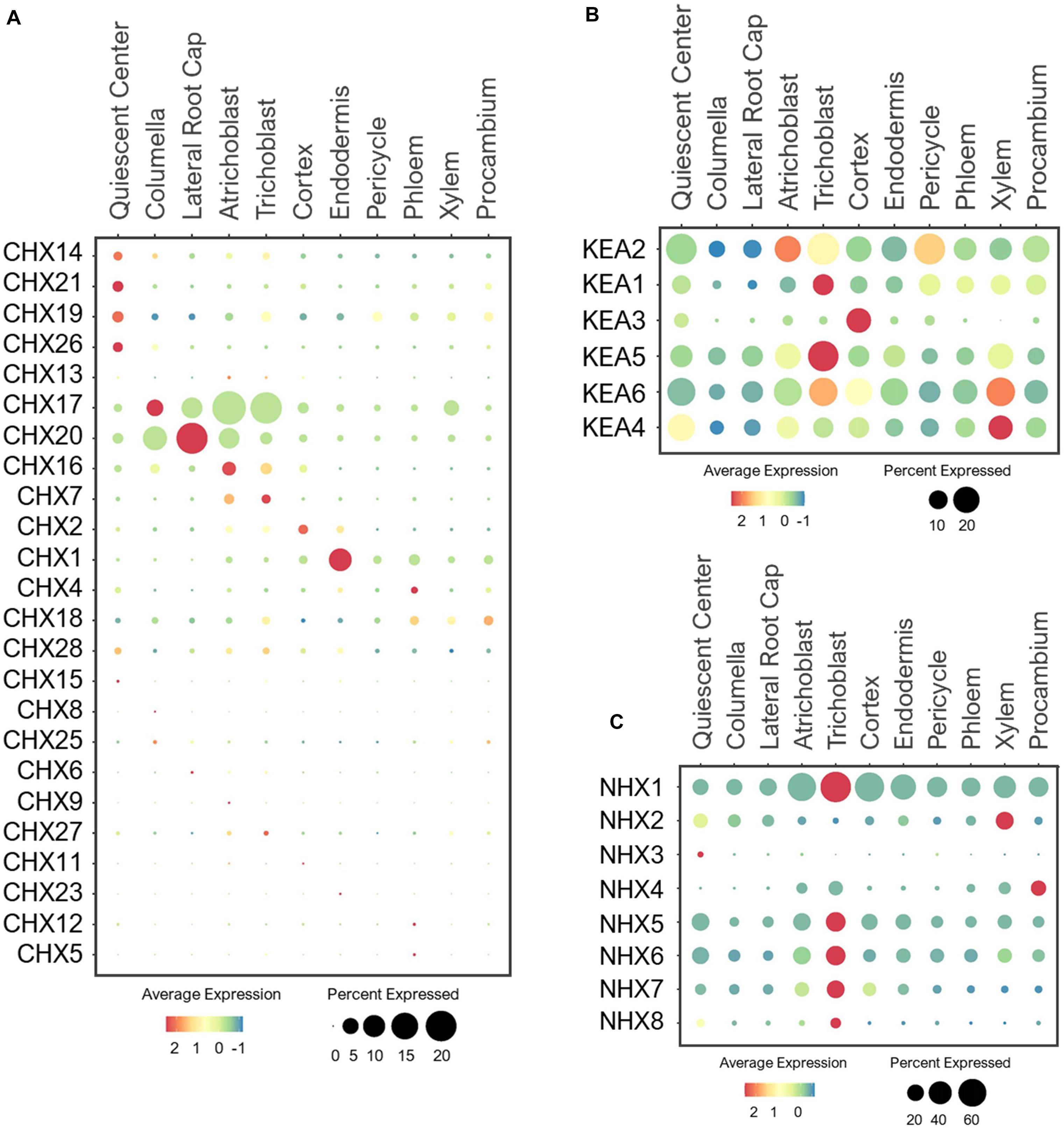
Figure 11. Expression of the (A) CHX, (B) KEA and (C) NHX antiporters in Arabidopsis root single cells. In dot plots, the circle size represents the proportion (%) of cells expressing a given gene, and the color of scale bar represents the mean expression (natural log +1 pseudocount).
However, several CHX members are known to be expressed and function in the root system. These include the PM-localized CHX13 and CHX17 that mediate high-affinity K+ uptake under low-K+ (Cellier et al., 2004; Zhao et al., 2008). While expression of CHX13 was not detected in the root under the normal conditions, CHX17 was highly expressed in the epidermis and root cap (Figure 11A). This implies CHX17, but not CHX13, may mediate K+ uptake in developing seedlings under the normal conditions. CHX13 is probably more responsive to low-K+ stress, similar to HAK5. CHX20 was previously shown to mediate K+/H+ exchanges in ER to control the stomatal movement (Padmanaban et al., 2007), but it’s role in roots have not been elucidated. We found CHX20 to be highly expressed in the root cap and atrichoblast, and may control K+ fluxes in ER of these cells, likely to regulate protein processing and trafficking as reported (Chanroj et al., 2011). CHX7 and CHX16 were expressed at low levels in epidermis (Figure 11A), and may also participate in K+ uptake or distribution depending on their membrane localizations. Between the two, CHX16 was shown to be located at the PM and ER, and implicated to function in pH homeostasis (Chanroj et al., 2011, 2013). In maintaining pH, CHX16 at the PM of root epidermis is likely to initiate K+ acquisition in exchange.
Several CHXs have been reported to function in response to osmotic stresses. For example, the PM-localized CHX14 is suggested to mediate K+ efflux from vascular stellar cells under excess-K+ condition (Zhao et al., 2015). On the other hand, the PM-localized CHX21 in root endodermis is implicated in loading Na+ to stelar cells under salinity stress for sequestration into leaf vacuoles (Hall et al., 2006). These stress-responsive transporters, CHX14/21, were lowly detected in QC under the normal conditions, similar to CHX19/26 (Figure 11A). Based on their known functions, the QC-specific expression might be an artifact. It is more likely that CHX14/21 expression and function may be more pronounced in stellar cells under osmotic stresses as reported (Hall et al., 2006; Zhao et al., 2015). CHX1 was the only member in the CHX family to be highly expressed in the endodermis (Figure 11A), raising the interesting possibility for this CHX to play a role in long-distance transport.
KEA Antiporters
The KEA family is also part of the CPA superfamily, with six members that share high homology to the bacterial K+/H+ antiporters KefB and KefC (Mäser et al., 2001; Chanroj et al., 2012; Han et al., 2015; Sze and Chanroj, 2018). Previous studies reported their tissue-specific expressions (Han et al., 2015; Zhu et al., 2018), however, their expressions at a cellular level are unclear. At the subcellular level, the closely related KEA1/2 are located at the inner envelope membrane and KEA3 at the thylakoid membrane of chloroplasts (Kunz et al., 2014). These KEAs participate in the K+/H+ exchange within and across chloroplasts to maintain pH and osmotic balance for chloroplast development and photosynthesis (Mäser et al., 2001; Kunz et al., 2014). Interestingly, we detected a broad expression of KEA1/2 in multiple cell-types in developing roots under the normal growth conditions (Figure 11B). This suggests the possible roles of KEA1/2 in K+ and pH homeostasis of plastids in different cell-types of roots in addition to shoots. In contrast, the more distant member, KEA3, was exclusively expressed in the cortex, and may mediate K+/H+ exchange specifically in the cortical plastids, which require further analysis to understand its functional significance in this specific cell-type.
The closely related KEA4/5/6 mediate K+/H+ exchange across trans-Golgi networks for proper vesicle trafficking that allow better adaptation to various osmotic stresses (Zhu et al., 2018; Wang et al., 2019). These KEAs shared overlapping and ubiquitous expressions in young roots under the normal growth conditions (Figure 11B), emphasizing their importance in pH/ionic homeostasis to support the endomembrane function in all root cell-types. Although broadly expressed, most KEAs displayed the highest expression in root epidermis, mainly trichoblast. It would be interesting to examine if these KEAs have any specific roles in root hair development. KEAs were also widely expressed in stellar cells, with the high expression of KEA2 detected in pericycle and KEA4/5/6 in xylem. This is in agreement with the previous findings (Han et al., 2015), suggesting these KEAs may further contribute to K+ distribution across stellar cells for shoot translocation. Currently, the physiological function of KEA members in the root system, and their possible involvement in K+ partitioning between roots and shoots have not been elucidated. In addition, how these KEAs may allow adaptation to low-K+ and excess-K+ stresses at the tissue- and cell-specific levels need to be further evaluated since their higher-order mutants (e.g., kea4/5/6) were highly sensitive to these K+ stresses (Zhu et al., 2018; Wang et al., 2019).
NHX Antiporters
The NHX family is another group of the CPA superfamily. In Arabidopsis, there are eight members, originally described as Na+/H+ exchangers (thus NHX) (Blumwald and Poole, 1985). Later studies indicated that different NHX members can transport Na+, K+, or both in exchange of H+ (Venema et al., 2002; Liu et al., 2010; Bassil et al., 2011b; Barragán et al., 2012). These transporters are found at the PM and different subcellular compartments (Bassil et al., 2012). NHX1/2 are located at the tonoplast to mediate K+ sequestration into the vacuoles (Shi and Zhu, 2002; Bassil et al., 2011b; Barragán et al., 2012). In aerial parts, they regulate stomatal movement and flower development (Bassil et al., 2011b; Barragán et al., 2012). We found that NHX1 was ubiquitously expressed in various cell-types of roots, whereas NHX2 showed narrower expression in xylem and QC (Figure 11C). This is consistent with the previous reports of tissue-specific expressions (Shi and Zhu, 2002; Barragán et al., 2012). The expression dominance of NHX1 compared to NHX2 coincides with its functional dominance in maintaining turgor for cell expansion to support overall growth and development (Bassil et al., 2011b). Because NHX2 was highly expressed in xylem, it may be involved in maintaining high K+ pools necessary for shoot translocation when plants are challenged by K+ limitations. Their roles in the root system need to be further evaluated because the earlier studies mainly focus on aerial tissues, where NHX1/2 were also expressed. The K+-selective microelectrode analysis revealed the maintenance of constant cytosolic K+ concentration between different root cell-types under the sufficient-K+ condition, and suggested the involvement of vacuolar transporters in this process (Walker et al., 1996; Tester and Leigh, 2001). The vacuolar NHXs are the prime candidates to sequester the accumulating K+ under the nutrient sufficiency.
NHX3 and NHX4 are also localized to tonoplast, with the NHX3 expression detected in pollen grains and siliques, and NHX4 in roots and shoots (Li et al., 2009; Liu et al., 2010; Bassil et al., 2012). Consistently, NHX3 was barely detected in roots (Figure 11C), suggesting it may have developed a specific function in reproductive organs. In contrast, the NHX4 was highly expressed in procambium, where it may provide the K+ supply needed for procambium differentiation into mature xylem and phloem. Similar to NHX2, NHX4 may support a long-distance K+ transport to shoots. Its contribution to K+ nutrition in the root system needs to be further explored. One important reason is because a previous study reported the highest K+ concentration in vacuoles of xylem compared to all other cell types, followed by pericycle, endodermis and cortex by using the cryo-scanning microscopy with energy dispersive X-ray microanalysis (McCully, 1994). NHX1/2/4 may be the key players in maintaining the high K+ level in xylem.
NHX5 and NHX6 are closely related transporters with high homology to the yeast NHX1 and the human NHE6 and NHE7 (Khan et al., 2018). These NHXs maintain pH and K+/Na+ homeostasis in endosomes to mediate cell expansion, proliferation and vesicle trafficking by influencing auxin transport and distribution (Bassil et al., 2011a; Dragwidge et al., 2018, 2019). NHX5/6 shared broad expression in the root single cell profiles (Figure 11C), supporting their essential and redundant functions in the housekeeping processes. The PM-localized NHX7 (SOS1) and NHX8 are the most divergent members of the NHX family, which closely resemble the prokaryotic NhaP antiporter (Bassil et al., 2012). These NHXs maintain cellular ion homeostasis by extruding toxic Na+ and Li+ out of the root cells (Shi et al., 2002; An et al., 2007). Consistent with their functions, NHX7/8 were highly expressed in epidermis, especially trichoblast, to remove the toxic cations out of the cell. In return, K+ may serve as a counter ion of Na+ to be absorbed by NHX7 at the root surface (Wu et al., 1996).
TPK Channels
The TPK family is composed of five Tandem Pore K+ channels (TPKs) and one Kir-like K+ channel. All of them are localized to the tonoplast except TPK4, which is found at the PM (Voelker et al., 2006, 2010; Lhamo et al., 2021). Electrophysiological studies in heterologous systems revealed TPK1/4 to be voltage-independent, K+ selective channels, and their channel activities are regulated by calcium and pH (Becker et al., 2004; Bihler et al., 2005; Gobert et al., 2007; Tang et al., 2020). TPK4 is expressed predominantly in pollen, where it conducts K+ across the PM of pollen tubes (Becker et al., 2004). As reported, the expression of TPK4 was barely detected in root cells of young seedlings under the normal conditions (Figure 12). On the other hand, the tonoplast-localized TPK1 is ubiquitously expressed, and plays important role in low-K+ adaptation by releasing K+ from vacuoles (Czempinski et al., 2002; Bihler et al., 2005; Gobert et al., 2007; Tang et al., 2020). The physiological characterizations of other tonoplast-localized TPKs remain unelucidated. In root single cells, TPK1/3/5 were widely expressed in all cell-types although TPK5 was expressed at a lower level (Figure 12). This suggests these TPKs may share similar housekeeping functions in vacuolar K+ remobilization throughout the root system. While their functions are likely to dominate under low-K+ stress, TPK1/3/5 may modulate the vacuolar K+ fluxes in coordination with NHX1/2 to maintain the constant cytosolic K+ concentration under the normal growth conditions, similar to AKT1 and GORK at the PM. The tonoplast-localized TPK2 and KCO3 were lowly detected in trichoblast under the normal conditions. Their expressions may amplify in multiple root cell-types under low-K+, to optimize K+ remobilization along with TPK1/3/5. However, the presence of all vacuolar TPKs at the root epidermis might explain the previous findings that cytosolic K+ concentration declines faster in epidermal cells than cortical cells upon continual low-K+ treatment (Walker et al., 1996; Tester and Leigh, 2001).
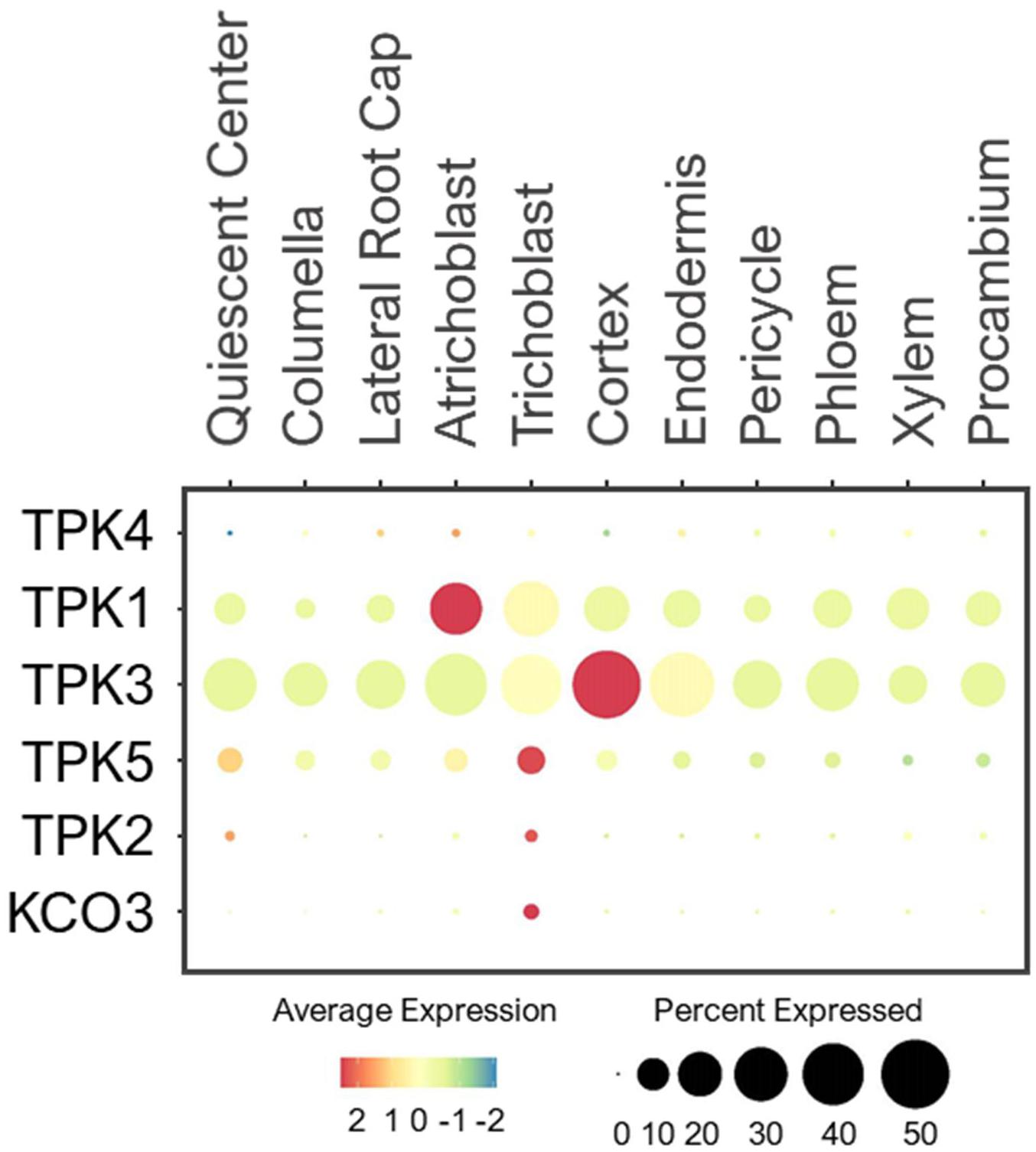
Figure 12. Expression of the TPK channels in Arabidopsis root single cells. In dot plot, the circle size represents the proportion (%) of cells expressing a given gene, and the color of scale bar represents the mean expression (natural log +1 pseudocount).
Potential Network of Potassium Channels and Transporters in Roots
Single cell profiling has allowed us to generate a working model of how diverse K+ channels and transporters might coordinate to transport K+ across different root cell-types of developing seedlings under the normal growth conditions (Figure 13). However, these are purely based on transcriptional profiles, which is subjected to change upon different developmental stages and K+ status. In general, K+ uptake and long-distance transport are largely mediated by Shaker-type channels, KUP and CHX transporters. The subcellular K+ distribution is controlled by TPK channels, KUP, KEA, NHX, and CHX transporters. Although many of their membrane localizations were elucidated, the functional studies of these K+ channels and transporters in the root system await further investigations. At the root surfaces, AKT1, HAK5, KUP7, and CHX13/17 are known to participate in K+ uptake especially in response to low-K+ stress (Hirsch et al., 1998; Cellier et al., 2004; Gierth et al., 2005; Qi et al., 2008; Zhao et al., 2008; Han et al., 2016), whereas KC1 channel inhibited AKT1 activity (Reintanz et al., 2002; Duby et al., 2008; Geiger et al., 2009a). Our expression analysis indicated AKT1, KUP7, and CHX17 to also mediate K+ acquisition under the normal growth conditions in young roots. We additionally found a number of KUP, CHX, KEA, NHX transporters, and TPK channels expressed at the root tip and epidermis that might be involved in K+ influx and efflux into and out of the root cells and subcellular compartments (Figure 13). Among these, several showed cell-type specific expressions at low level, including KUP2/3 in QC and TPK2/KCO3 in trichoblast. Several KUP, KEA, NHX, and TPK members were expressed in cortex and endodermis to distribute K+ within and toward stellar cells (Figure 13). KEA3 was predominantly expressed in the cortex and CHX1 in the endodermis, suggesting their unique functions in these cell-types, which might be worth further investigations.
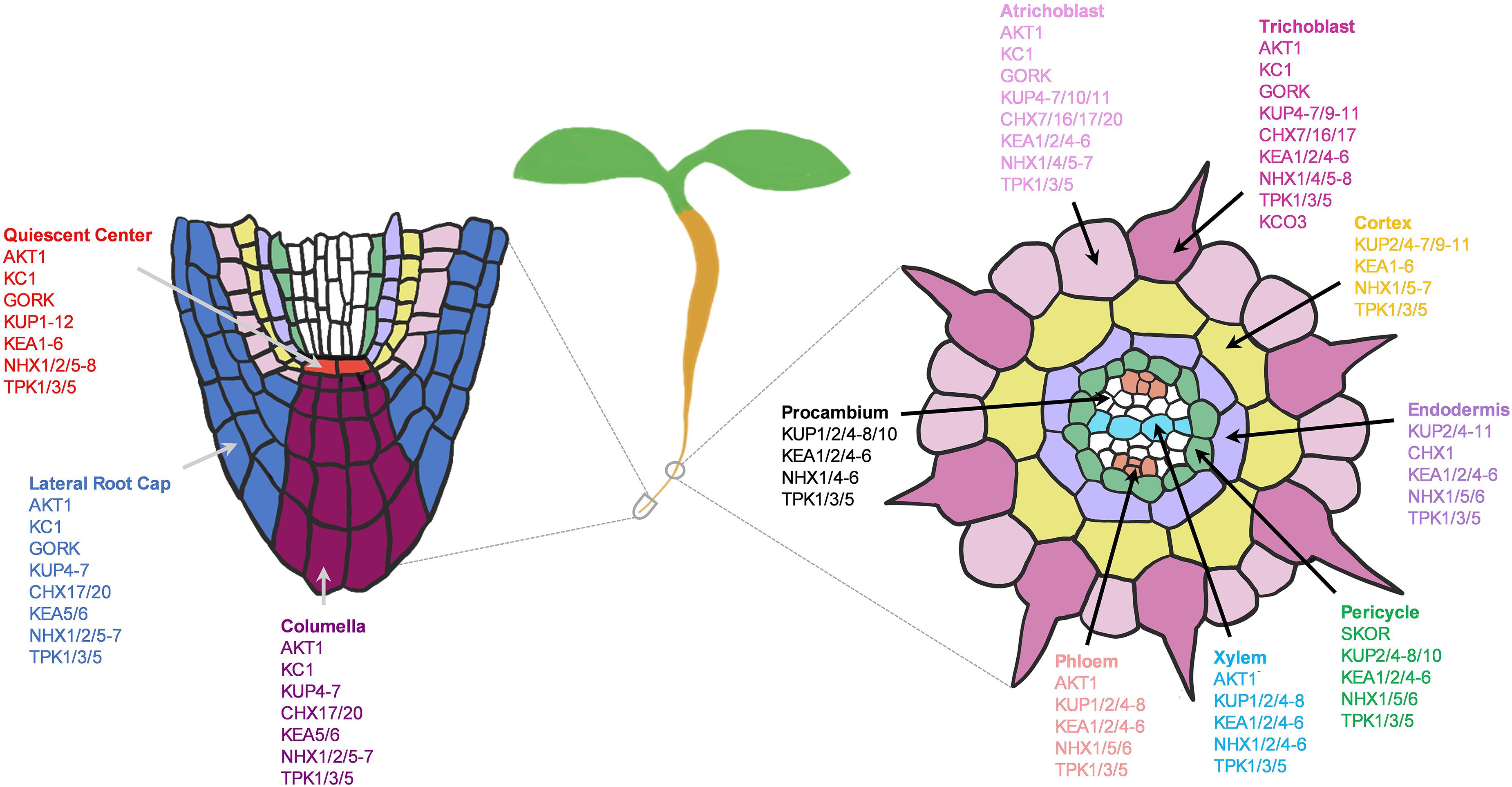
Figure 13. Potential network of potassium (K+) channels and transporters in the root system of 5–7 days old Arabidopsis seedlings grown under the normal growth conditions. Expressed K+ transporters involved in longitudinal transport (left) at the meristematic zone and radial transport (right) at the elongation/maturation zones were shown. K+ transporter genes not expressed under the normal growth conditions were excluded.
In stellar cells, SKOR and KUP7 were known to mediate xylem loading (Gaymard et al., 1998; Liu et al., 2006; Han et al., 2016) AKT2 in phloem loading (Marten et al., 1999; Lacombe et al., 2000), and AKT1 in xylem retrieval (Nieves-Cordones et al., 2019). Surprisingly, SKOR and AKT2 channels were lowly expressed in developing roots under the normal conditions; therefore, we anticipated the involvement of additional nutrient transporters in these processes. For instance, NPF7.3 (NRT1.5) in pericycle is shown to mediate K+ loading to xylem under low-K+ and the normal conditions, although its function is more pronounced under low-nutrient (K+ or NO3–) stresses (Drechsler et al., 2015; Li et al., 2017b). In addition to this, we found number of KUPs, KEAs, NHXs, and TPKs that might contribute to long-distance K+ transport (Figure 13). Among these, NHX2/4 might be more specifically involved in xylem loading and KUP1 in phloem loading. KUP2/6/8 and GORK might coordinate with each other to extrude K+ from stellar cells to the soil (Osakabe et al., 2013).
Overall, many KUPs were widely expressed, indicating their essential role in K+ transport in developing roots under the sufficient-K+ supply, and worth further studies to dissect the function of uncharacterized KUPs. Because there are large number of KUPs, functional redundancies among the KUP transporters may restrict further studies, which could be potentially resolved by generating higher-order mutants. Our analysis provides KUP genes that share overlapping expressions that may be used to generate higher-order mutants. KEAs are essential for K+/H+ balance in plastids and endomembranes, but their significance in K+ transport and distribution in the root system remain unelucidated despite their broad expressions. On the other hand, the large portions of Shaker-type channels and CHX transporters were not much expressed in young roots, suggesting their roles in other tissues and organs or in response to specific biotic and abiotic stresses. Several NHX members were ubiquitously expressed for housekeeping functions in tonoplast and endosomes. TPK1/3/5 were broadly expressed to remobilize K+ from vacuoles, and may function concurrently with the vacuolar influx transporter, NHX1 in maintaining cellular K+ homoeostasis during plant development. However, TPK3/5 are not physiologically characterized yet, and more studies of NHX members in roots are needed.
While we focus on the major families of K+ channels and transporters, there exist other voltage-independent, non-selective cation channels such as CNGCs (cyclic nucleotide gated channels) that are capable of transporting K+ in different heterologous systems, which include CNGC1/2/3/4/10/17 (Köhler et al., 1999; Leng et al., 1999; Balagué et al., 2003; Hua et al., 2003; Li et al., 2005; Gobert et al., 2006; Ladwig et al., 2015). This shows the complexity of K+ transport in plants, in which various channels and transporters are able to control K+ fluxes during different developmental stages and in response to various environmental stresses. Nevertheless, they share a similar goal of providing K+ supply needed to support growth, which involves the coordination among different nutrient transporters and channels to maintain cellular ion homeostasis.
Conclusion
In summary, single cell profiling has allowed us to predict the functions of many unknown NPK channels and transporters and generate their potential networks in the root system of young Arabidopsis seedlings. However, further molecular, electrophysiological and genetic studies are required to confirm their functions in the specific root cell-types, and determine functional redundancies among the closely-related transporters. In addition, generation of root atlas using a large population of cells under different developmental and low-nutrient stresses may unravel the putative channel and transporter networks that are crucial for plant development and low-nutrient adaptation. Moreover, the spatial profiling of cytosolic and subcellular nutrient concentrations in root single cells using technologies such as microelectrodes (Zhen et al., 1991; Radcliffe et al., 2005), nuclear magnetic resonance (NMR) (Lee and Ratcliffe, 1993; Radcliffe et al., 2005; Gout et al., 2011; Liu et al., 2016) and fluorescence resonance energy transfer (FRET)-based systems (Looger et al., 2005; Mukherjee et al., 2015; Sahu et al., 2020) are much needed to dissect the complexity of nutrient transport within and between different root cell-types.
Materials and Methods
Plant Growth Conditions for Low-Nutrient Stresses
Arabidopsis seeds were sterilized in 10% bleach, rinsed with distilled water four times, and plated directly on 1/6 strength MS (Murashige and Skoog) with (NPK) and without N, P, or K (0 mM) nutrients, supplemented with 1% sucrose and 1% phytoblend (Caisson labs), pH 5.7. The MS media with and without N or P were ordered from Caisson labs (North Logan, Utah). Low-K medium (1/6 MS) were prepared in-house using 0.5 mM Ca(NO3)2, 0.25 mM MgSO4, 0.21 mM NH4H2PO4, and 1/6 micronutrients (Caisson labs). Plated seeds were stratified at 4°C for 2 days and transferred to the growth chamber in vertical position for 5 days, with a 12-h light/12-h dark cycle (100 μmol m–2 s–1). Two sets of plates were prepared for each condition, one with plants clustered together, and the other with seedlings separated with spaces in-between for root hair phenotype, and the most representative one is presented.
Single-Cell Root Altas Data Source and Information
The scRNA-seq data and Seurat objects for Arabidopsis root altas were obtained from Shahan et al. (2020) (GEO: GSE152766). Briefly, the primary roots of 5–7 days old Arabidopsis seedlings under normal growth conditions were used for root protoplast extractions. Shahan et al. (2020) generated the root atlas from 110,427 single cells, by integrating their datasets with the previously published datasets (Denyer et al., 2019; Ryu et al., 2019). Details about the data integrations can be found in Shahan et al. (2020). The large datasets cover all the major root cell-types including QC (1834 cells), columella (11,757), LRC (14,420), atrichoblast (13,972), trichoblast (11,946), cortex (10,318), endodermis (10,541), pericycle (11,603), phloem (6758), xylem (4984), and procambium (12,294). The identity for these cell-types were assigned based on the expression of cell-type-specific marker genes that are well-studied (Shahan et al., 2020).
Data Analyses and Visualizations
To understand the potential networks of NPK transport systems in Arabidopsis roots, the expression patterns of all the major NPK family channels and transporters were analyzed. Extensive literature searches were performed to identify all the major NPK transporter families in Arabidopsis. The Arabidopsis gene IDs for the NPK transporters were retrieved from The Arabidopsis Information Resource (TAIR1). All data were analyzed in R (version 3.5.2) using the batch-corrected (“RNA” assay in Seurat object) expression value. The expression profiles of specific N, P, or K transporter families were plotted using Seurat’s DotPlot function. Unlike the typically used heatmap, the dot plot visualization provides more detailed expression profiling that shows the percentage of cells expressing a specific nutrient channel or transporter gene, and the average expression of cells expressing the gene. This allows identification of the major nutrient transporters that contribute to NPK transport in specific cell type(s) under the normal growth conditions. The differentially expressed Pi transporters in response to low-Pi were extracted from root single cells analyzed by Wendrich et al. (2020) (Supplementary Figure 1). The artworks for potential networks of NPK channels and transporters in the root system were drawn using Adobe illustrator draw application. Nutrient channel and transporter genes expressed under the normal growth conditions were included in the networks.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/geo/, GSE152766.
Author Contributions
DL performed the literature searches, bioinformatic analysis, and wrote the manuscript. SL reviewed and revised the manuscript. Both the authors contributed to the article and approved the submitted version.
Funding
This project was supported by a grant from the National Science Foundation (MCB-1714795), the California Agricultural Experimental Station and the Innovative Genomics Institute of University of California, Berkeley.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are thankful to Drs. Benjamin J. Cole and Ronan C. O’Malley for providing us with the raw single cell transcriptomic datasets and suggestions.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.689545/full#supplementary-material
Supplementary Figure 1 | Differentially expressed Pi transporters in specific root cell-types under Pi starvation. The data were retrieved from Wendrich et al. (2020).
Footnotes
References
Ache, P., Becker, D., Ivashikina, N., Dietrich, P., Roelfsema, M. R., and Hedrich, R. (2000). GORK, a delayed outward rectifier expressed in guard cells of Arabidopsis thaliana, is a K(+)-selective, K(+)-sensing ion channel. FEBS Lett. 486, 93–98. doi: 10.1016/s0014-5793(00)02248-1
Almagro, A., Lin, S. H., and Tsay, Y. F. (2008). Characterization of the Arabidopsis nitrate transporter NRT1.6 reveals a role of nitrate in early embryo development. Plant Cell 20, 3289–3299. doi: 10.1105/tpc.107.056788
An, R., Chen, Q.-J., Chai, M.-F., Lu, P.-L., Su, Z., Qin, Z.-X., et al. (2007). AtNHX8, a member of the monovalent cation: proton antiporter-1 family in Arabidopsis thaliana, encodes a putative Li/H antiporter: AtNHX8 encodes an Li+/H+ antiporter. Plant J. 49, 718–728. doi: 10.1111/j.1365-313X.2006.02990.x
Andersen, T. G., Nour-Eldin, H. H., Fuller, V. L., Olsen, C. E., Burow, M., and Halkier, B. A. (2013). Integration of biosynthesis and long-distance transport establish organ-specific glucosinolate profiles in vegetative Arabidopsis. Plant Cell 25, 3133–3145. doi: 10.1105/tpc.113.110890
Arnaud, C., Bonnot, C., Desnos, T., and Nussaume, L. (2010). The root cap at the forefront. C. R. Biol. 333, 335–343. doi: 10.1016/j.crvi.2010.01.011
Arpat, A. B., Magliano, P., Wege, S., Rouached, H., Stefanovic, A., and Poirier, Y. (2012). Functional expression of PHO1 to the golgi and trans-golgi network and its role in export of inorganic phosphate: PHO1 localization to the Golgi/trans-Golgi network. Plant J. 71, 479–491. doi: 10.1111/j.1365-313X.2012.05004.x
Balagué, C., Lin, B., Alcon, C., Flottes, G., Malmström, S., Köhler, C., et al. (2003). HLM1, an essential signaling component in the hypersensitive response, is a member of the cyclic nucleotide-gated channel ion channel family. Plant Cell 15, 365–379. doi: 10.1105/tpc.006999
Banuelos, M. A., Klein, R. D., Alexander-Bowman, S. J., and Rodríguez-Navarro, A. (1995). A potassium transporter of the yeast Schwanniomyces occidentalis homologous to the Kup system of Escherichia coli has a high concentrative capacity. EMBO J. 14, 3021–3027. doi: 10.1002/j.1460-2075.1995.tb07304.x
Barbier-Brygoo, H., De Angeli, A., Filleur, S., Frachisse, J.-M., Gambale, F., Thomine, S., et al. (2011). Anion channels/transporters in plants: from molecular bases to regulatory networks. Ann. Rev. Plant Biol. 62, 25–51. doi: 10.1146/annurev-arplant-042110-103741
Barragán, V., Leidi, E. O., Andrés, Z., Rubio, L., De Luca, A., Fernández, J. A., et al. (2012). Ion exchangers NHX1 and NHX2 mediate active potassium uptake into vacuoles to regulate cell turgor and stomatal function in Arabidopsis. Plant Cell 24, 1127–1142. doi: 10.1105/tpc.111.095273
Bassil, E., Coku, A., and Blumwald, E. (2012). Cellular ion homeostasis: emerging roles of intracellular NHX Na+/H+ antiporters in plant growth and development. J. Exp. Bot. 63, 5727–5740. doi: 10.1093/jxb/ers250
Bassil, E., Ohto, M.-A., Esumi, T., Tajima, H., Zhu, Z., Cagnac, O., et al. (2011a). The Arabidopsis intracellular Na+/H+ antiporters NHX5 and NHX6 are endosome associated and necessary for plant growth and development. Plant Cell 23, 224–239. doi: 10.1105/tpc.110.079426
Bassil, E., Tajima, H., Liang, Y.-C., Ohto, M.-A., Ushijima, K., Nakano, R., et al. (2011b). The Arabidopsis Na+/H+ antiporters NHX1 and NHX2 control vacuolar pH and K+ homeostasis to regulate growth, flower development, and reproduction. Plant Cell 23, 3482–3497. doi: 10.1105/tpc.111.089581
Becker, D., Geiger, D., Dunkel, M., Roller, A., Bertl, A., Latz, A., et al. (2004). AtTPK4, an Arabidopsis tandem-pore K+ channel, poised to control the pollen membrane voltage in a pH- and Ca2+-dependent manner. Proc. Natl. Acad. Sci. U.S.A. 101, 15621–15626. doi: 10.1073/pnas.0401502101
Bellegarde, F., Gojon, A., and Martin, A. (2017). Signals and players in the transcriptional regulation of root responses by local and systemic N signaling in Arabidopsis thaliana. J. Exp. Bot. 68, 2553–2565. doi: 10.1093/jxb/erx062
Bihler, H., Eing, C., Hebeisen, S., Roller, A., Czempinski, K., and Bertl, A. (2005). TPK1 is a vacuolar ion channel different from the slow-vacuolar cation channel. Plant Physiol. 139, 417–424. doi: 10.1104/pp.105.065599
Blumwald, E., and Poole, R. J. (1985). Na+/H+ antiport in isolated tonoplast vesicles from storage tissue of Beta vulgaris. Plant Physiol. 78, 163–167.
Cellier, F., Conéjéro, G., Ricaud, L., Luu, D. T., Lepetit, M., Gosti, F., et al. (2004). Characterization of AtCHX17, a member of the cation/H+ exchangers, CHX family, from Arabidopsis thaliana suggests a role in K+ homeostasis. Plant J. 39, 834–846. doi: 10.1111/j.1365-313X.2004.02177.x
Cerezo, M., Tillard, P., Filleur, S., Muños, S., Daniel-Vedele, F., and Gojon, A. (2001). Major alterations of the regulation of root NO(3)(-) uptake are associated with the mutation of Nrt2.1 and Nrt2.2 genes in Arabidopsis. Plant Physiol. 127, 262–271. doi: 10.1104/pp.127.1.262
Chanroj, S., Lu, Y., Padmanaban, S., Nanatani, K., Uozumi, N., Rao, R., et al. (2011). Plant-specific cation/H+ exchanger 17 and its homologs are endomembrane K+ transporters with roles in protein sorting. J. Biol. Chem. 286, 33931–33941. doi: 10.1074/jbc.M111.252650
Chanroj, S., Padmanaban, S., Czerny, D. D., Jauh, G.-Y., and Sze, H. (2013). K+ transporter AtCHX17 with its hydrophilic C tail localizes to membranes of the secretory/endocytic system: role in reproduction and seed set. Mol. Plant 6, 1226–1246. doi: 10.1093/mp/sst032
Chanroj, S., Wang, G., Venema, K., Zhang, M., Delwiche, C., and Sze, H. (2012). Conserved and diversified gene families of monovalent cation/H+ antiporters from algae to flowering plants. Front. Plant Sci. 3:25. doi: 10.3389/fpls.2012.00025
Chen, Y.-H., Hu, L., Punta, M., Bruni, R., Hillerich, B., Kloss, B., et al. (2010). Homologue structure of the SLAC1 anion channel for closing stomata in leaves. Nature 467, 1074–1080. doi: 10.1038/nature09487
Chiou, T.-J., and Lin, S.-I. (2011). Signaling network in sensing phosphate availability in plants. Annu. Rev. Plant Biol. 62, 185–206. doi: 10.1146/annurev-arplant-042110-103849
Chopin, F., Orsel, M., Dorbe, M.-F., Chardon, F., Truong, H.-N., Miller, A. J., et al. (2007). The Arabidopsis ATNRT2.7 nitrate transporter controls nitrate content in seeds. Plant Cell 19, 1590–1602. doi: 10.1105/tpc.107.050542
Corratgé-Faillie, C., and Lacombe, B. (2017). Substrate (un)specificity of Arabidopsis NRT1/PTR FAMILY (NPF) proteins. J. Exp. Bot. 68, 3107–3113. doi: 10.1093/jxb/erw499
Cubero, B., Nakagawa, Y., Jiang, X.-Y., Miura, K.-J., Li, F., Raghothama, K. G., et al. (2009). The phosphate transporter PHT4;6 is a determinant of salt tolerance that is localized to the Golgi apparatus of Arabidopsis. Mol. Plant 2, 535–552. doi: 10.1093/mp/ssp013
Cubero-Font, P., Maierhofer, T., Jaslan, J., Rosales, M. A., Espartero, J., Díaz-Rueda, P., et al. (2016). Silent S-Type anion channel subunit SLAH1 gates SLAH3 open for chloride root-to-shoot translocation. Curr. Biol. 26, 2213–2220. doi: 10.1016/j.cub.2016.06.045
Czempinski, K., Frachisse, J.-M., Maurel, C., Barbier-Brygoo, H., and Mueller-Roeber, B. (2002). Vacuolar membrane localization of the Arabidopsis “two-pore”K+ channel KCO1. Plant J. 29, 809–820. doi: 10.1046/j.1365-313X.2002.01260.x
Daram, P., Brunner, S., Rausch, C., Steiner, C., Amrhein, N., and Bucher, M. (1999). Pht2;1 encodes a low-affinity phosphate transporter from arabidopsis. Plant Cell 11:2153. doi: 10.2307/3871016
De Angeli, A., Monachello, D., Ephritikhine, G., Frachisse, J. M., Thomine, S., Gambale, F., et al. (2006). The nitrate/proton antiporter AtCLCa mediates nitrate accumulation in plant vacuoles. Nature 442, 939–942. doi: 10.1038/nature05013
De Rybel, B., Mähönen, A. P., Helariutta, Y., and Weijers, D. (2016). Plant vascular development: from early specification to differentiation. Nat. Rev. Mol. Cell Biol. 17, 30–40. doi: 10.1038/nrm.2015.6
Demidchik, V. (2014). Mechanisms and physiological roles of K+ efflux from root cells. J. Plant Physiol. 171, 696–707. doi: 10.1016/j.jplph.2014.01.015
Denyer, T., Ma, X., Klesen, S., Scacchi, E., Nieselt, K., and Timmermans, M. C. P. (2019). Spatiotemporal developmental trajectories in the arabidopsis root revealed using high-throughput single-cell RNA sequencing. Dev. Cell 48, 840–852.e5. doi: 10.1016/j.devcel.2019.02.022
Dragwidge, J. M., Ford, B. A., Ashnest, J. R., Das, P., and Gendall, A. R. (2018). Two endosomal NHX-type Na+/H+ antiporters are involved in auxin-mediated development in Arabidopsis thaliana. Plant Cell Physiol. 59, 1660–1669. doi: 10.1093/pcp/pcy090
Dragwidge, J. M., Scholl, S., and Schumacher, K. (2019). NHX-type Na+ (K+)/H+ antiporters are required for TGN/EE trafficking and endosomal ion homeostasis in Arabidopsis thaliana. J. Cell 132:jcs226472.
Drechsler, N., Zheng, Y., Bohner, A., Nobmann, B., von Wirén, N., Kunze, R., et al. (2015). Nitrate-dependent control of shoot K homeostasis by the nitrate transporter1/peptide transporter family member NPF7.3/NRT1.5 and the stelar K+ outward rectifier SKOR in Arabidopsis. Plant Physiol. 169, 2832–2847. doi: 10.1104/pp.15.01152
Dreyer, I., Gomez-Porras, J. L., Riaño-Pachón, D. M., Hedrich, R., and Geiger, D. (2012). Molecular evolution of slow and quick anion channels (SLACs and QUACs/ALMTs). Front. Plant Sci. 3:263. doi: 10.3389/fpls.2012.00263
Duby, G., Hosy, E., Fizames, C., Alcon, C., Costa, A., Sentenac, H., et al. (2008). AtKC1, a conditionally targeted Shaker-type subunit, regulates the activity of plant K+ channels. Plant J. 53, 115–123. doi: 10.1111/j.1365-313X.2007.03324.x
Fan, S.-C., Lin, C.-S., Hsu, P.-K., Lin, S.-H., and Tsay, Y.-F. (2009). The Arabidopsis nitrate transporter NRT1.7, expressed in phloem, is responsible for source-to-sink remobilization of nitrate. Plant Cell 21, 2750–2761. doi: 10.1105/tpc.109.067603
Fan, X., Naz, M., Fan, X., Xuan, W., Miller, A. J., and Xu, G. (2017). Plant nitrate transporters: from gene function to application. J. Exp. Bot. 68, 2463–2475. doi: 10.1093/jxb/erx011
Fecht-Bartenbach, J., von der Fecht-Bartenbach, J., Bogner, M., Krebs, M., Stierhof, Y.-D., Schumacher, K., et al. (2007). Function of the anion transporter AtCLC-d in the trans-Golgi network. Plant J. 50, 466–474. doi: 10.1111/j.1365-313x.2007.03061.x
Filleur, S., and Daniel-Vedele, F. (1999). Expression analysis of a high-affinity nitrate transporter isolated from Arabidopsis thaliana by differential display. Planta 207, 461–469. doi: 10.1007/s004250050505
Filleur, S., Dorbe, M.-F., Cerezo, M., Orsel, M., Granier, F., Gojon, A., et al. (2001). An Arabidopsis T-DNA mutant affected in Nrt2 genes is impaired in nitrate uptake. FEBS Lett. 489, 220–224. doi: 10.1016/S0014-5793(01)02096-8
Flügge, U.-I., Häusler, R. E., Ludewig, F., and Fischer, K. (2003). Functional genomics of phosphate antiport systems of plastids. Physiol. Plant. 118, 475–482. doi: 10.1034/j.1399-3054.2003.00137.x
Fredes, I., Moreno, S., Díaz, F. P., and Gutiérrez, R. A. (2019). Nitrate signaling and the control of Arabidopsis growth and development. Curr. Opin. Plant Biol. 47, 112–118. doi: 10.1016/j.pbi.2018.10.004
Fu, H. H., and Luan, S. (1998). AtKuP1: a dual-affinity K+ transporter from Arabidopsis. Plant Cell 10, 63–73. doi: 10.1105/tpc.10.1.63
Gaymard, F., Pilot, G., Lacombe, B., Bouchez, D., Bruneau, D., Boucherez, J., et al. (1998). Identification and disruption of a plant shaker-like outward channel involved in K+ release into the xylem sap. Cell 94, 647–655. doi: 10.1016/s0092-8674(00)81606-2
Geelen, D., Lurin, C., Bouchez, D., Frachisse, J.-M., Lelièvre, F., Courtial, B., et al. (2000). Disruption of putative anion channel gene AtCLC-a in Arabidopsis suggests a role in the regulation of nitrate content. Plant J. 21, 259–267. doi: 10.1046/j.1365-313x.2000.00680.x
Geiger, D., Becker, D., Vosloh, D., Gambale, F., Palme, K., Rehers, M., et al. (2009a). Heteromeric AtKC1\textperiodcentered AKT1 channels in Arabidopsis roots facilitate growth under K+-limiting conditions. J. Biol. Chem. 284, 21288–21295.
Geiger, D., Maierhofer, T., Al-Rasheid, K. A. S., Scherzer, S., Mumm, P., Liese, A., et al. (2011). Stomatal closure by fast abscisic acid signaling is mediated by the guard cell anion channel SLAH3 and the receptor RCAR1. Sci. Signal 4:ra32. doi: 10.1126/scisignal.2001346
Geiger, D., Scherzer, S., Mumm, P., Stange, A., Marten, I., Bauer, H., et al. (2009b). Activity of guard cell anion channel SLAC1 is controlled by drought-stress signaling kinase-phosphatase pair. Proc. Natl. Acad. Sci. U.S A. 106, 21425–21430. doi: 10.1073/pnas.0912021106
Giehl, R. F. H., and von Wiren, N. (2014). Root Nutrient Foraging. Plant Physiol. 166, 509–517. doi: 10.1104/pp.114.245225
Gierth, M., Mäser, P., and Schroeder, J. I. (2005). The potassium transporter AtHAK5 functions in K(+) deprivation-induced high-affinity K(+) uptake and AKT1 K(+) channel contribution to K(+) uptake kinetics in Arabidopsis roots. Plant Physiol. 137, 1105–1114. doi: 10.1104/pp.104.057216
Gobert, A., Isayenkov, S., Voelker, C., Czempinski, K., and Maathuis, F. J. M. (2007). The two-pore channel TPK1 gene encodes the vacuolar K+ conductance and plays a role in K+ homeostasis. Proc. Natl. Acad. Sci. U.S.A. 104, 10726–10731. doi: 10.1073/pnas.0702595104
Gobert, A., Park, G., Amtmann, A., Sanders, D., and Maathuis, F. J. M. (2006). Arabidopsis thaliana cyclic nucleotide gated channel 3 forms a non-selective ion transporter involved in germination and cation transport. J. Exp. Bot. 57, 791–800. doi: 10.1093/jxb/erj064
Gojon, A., Krouk, G., Perrine-Walker, F., and Laugier, E. (2011). Nitrate transceptor(s) in plants. J. Exp. Bot. 62, 2299–2308. doi: 10.1093/jxb/erq419
Gomez-Porras, J., Riaño Pachón, D. M., Benito, B., Haro, R., Sklodowski, K., Rodríguez-Navarro, A., et al. (2012). Phylogenetic analysis of K+ transporters in bryophytes, lycophytes, and flowering plants indicates a specialization of vascular plants. Front. Plant Sci. 3:167. doi: 10.3389/fpls.2012.00167
Gout, E., Bligny, R., Douce, R., Boisson, A.-M., and Rivasseau, C. (2011). Early response of plant cell to carbon deprivation: in vivo31P-NMR spectroscopy shows a quasi-instantaneous disruption on cytosolic sugars, phosphorylated intermediates of energy metabolism, phosphate partitioning, and intracellular pHs. New Phytol. 189, 135–147. doi: 10.1111/j.1469-8137.2010.03449.x
Gruber, B. D., Giehl, R. F. H., Friedel, S., and von Wirén, N. (2013). Plasticity of the Arabidopsis root system under nutrient deficiencies. Plant Physiol. 163, 161–179. doi: 10.1104/pp.113.218453
Gu, M., Chen, A., Sun, S., and Xu, G. (2016). Complex regulation of plant phosphate transporters and the gap between molecular mechanisms and practical application: what is missing? Mol. Plant 9, 396–416. doi: 10.1016/j.molp.2015.12.012
Guo, B., Irigoyen, S., Fowler, T. B., and Versaw, W. K. (2008a). Differential expression and phylogenetic analysis suggest specialization of plastid-localized members of the PHT4 phosphate transporter family for photosynthetic and heterotrophic tissues. Plant Signal Behav. 3, 784–790. doi: 10.4161/psb.3.10.6666
Guo, B., Jin, Y., Wussler, C., Blancaflor, E. B., Motes, C. M., and Versaw, W. K. (2008b). Functional analysis of the Arabidopsis PHT4 family of intracellular phosphate transporters. New Phytol. 177, 889–898. doi: 10.1111/j.1469-8137.2007.02331.x
Gutermuth, T., Lassig, R., Portes, M.-T., Maierhofer, T., Romeis, T., Borst, J.-W., et al. (2013). Pollen tube growth regulation by free anions depends on the interaction between the anion channel SLAH3 and calcium-dependent protein kinases CPK2 and CPK20. Plant Cell 25, 4525–4543. doi: 10.1105/tpc.113.118463
Hall, D., Evans, A. R., Newbury, H. J., and Pritchard, J. (2006). Functional analysis of CHX21: a putative sodium transporter in Arabidopsis. J. Exp. Bot. 57, 1201–1210. doi: 10.1093/jxb/erj092
Hamburger, D., Rezzonico, E., MacDonald-Comber Petétot, J., Somerville, C., and Poirier, Y. (2002). Identification and characterization of the Arabidopsis PHO1 gene involved in phosphate loading to the xylem. Plant Cell 14, 889–902. doi: 10.1105/tpc.000745
Hamel, P., Saint-Georges, Y., de Pinto, B., Lachacinski, N., Altamura, N., and Dujardin, G. (2004). Redundancy in the function of mitochondrial phosphate transport in Saccharomyces cerevisiae and Arabidopsis thaliana. Mol. Microbiol. 51, 307–317. doi: 10.1046/j.1365-2958.2003.03810.x
Hammes, U. Z., Meier, S., Dietrich, D., Ward, J. M., and Rentsch, D. (2010). Functional properties of the Arabidopsis peptide transporters AtPTR1 and AtPTR5. J. Biol. Chem. 285, 39710–39717. doi: 10.1074/jbc.M110.141457
Han, L., Li, J. L., Wang, L., Shi, W. M., and Su, Y. H. (2015). Identification and localized expression of putative K+/H+ antiporter genes in Arabidopsis. Acta Physiol. Plant 37:101. doi: 10.1007/s11738-015-1845-4
Han, M., Wu, W., Wu, W.-H., and Wang, Y. (2016). Potassium transporter KUP7 Is involved in K(+) acquisition and translocation in arabidopsis root under K(+)-limited conditions. Mol. Plant 9, 437–446. doi: 10.1016/j.molp.2016.01.012
Hasanuzzaman, M., Bhuyan, M. H. M. B., Nahar, K., Hossain, M. S., Mahmud, J. A., Hossen, M. S., et al. (2018). Potassium: a vital regulator of plant responses and tolerance to abiotic stresses. Agronomy 8:31. doi: 10.3390/agronomy8030031
Hassler, S., Jung, B., Lemke, L., Novák, O., Strnad, M., Martinoia, E., et al. (2016). Function of the golgi-located phosphate transporter PHT4;6 is critical for senescence-associated processes in Arabidopsis. J. Exp. Bot. 67, 4671–4684. doi: 10.1093/jxb/erw249
Hassler, S., Lemke, L., Jung, B., Möhlmann, T., Krüger, F., Schumacher, K., et al. (2012). Lack of the Golgi phosphate transporter PHT4;6 causes strong developmental defects, constitutively activated disease resistance mechanisms and altered intracellular phosphate compartmentation in Arabidopsis. Plant J. 72, 732–744. doi: 10.1111/j.1365-313x.2012.05106.x
He, Y.-N., Peng, J.-S., Cai, Y., Liu, D.-F., Guan, Y., Yi, H.-Y., et al. (2017). Tonoplast-localized nitrate uptake transporters involved in vacuolar nitrate efflux and reallocation in Arabidopsis. Sci. Rep. 7:6417. doi: 10.1038/s41598-017-06744-5
Hechenberger, M., Schwappach, B., Fischer, W. N., Frommer, W. B., Jentsch, T. J., and Steinmeyer, K. (1996). A family of putative chloride channels from arabidopsis and functional complementation of a yeast strain with a CLC gene disruption. J. Biol. Chem. 271, 33632–33638. doi: 10.1074/jbc.271.52.33632
Hedrich, R., and Geiger, D. (2017). Biology of SLAC1-type anion channels - from nutrient uptake to stomatal closure. New Phytol. 216, 46–61. doi: 10.1111/nph.14685
Hirsch, R. E., Lewis, B. D., Spalding, E. P., and Sussman, M. R. (1998). A role for the AKT1 potassium channel in plant nutrition. Science 280, 918–921. doi: 10.1126/science.280.5365.918
Ho, C.-H., Lin, S.-H., Hu, H.-C., and Tsay, Y.-F. (2009). CHL1 functions as a nitrate sensor in plants. Cell 138, 1184–1194. doi: 10.1016/j.cell.2009.07.004
Hosy, E., Vavasseur, A., Mouline, K., Dreyer, I., Gaymard, F., Porée, F., et al. (2003). The arabidopsis outward K+ channel GORK is involved in regulation of stomatal movements and plant transpiration. Proc. Natl. Acad. Sci. U.S.A. 100, 5549–5554. doi: 10.1073/pnas.0733970100
Hsu, P.-K., and Tsay, Y.-F. (2013). Two phloem nitrate transporters, NRT1.11 and NRT1.12, are important for redistributing xylem-borne nitrate to enhance plant growth. Plant Physiol. 163, 844–856. doi: 10.1104/pp.113.226563
Hua, B.-G., Mercier, R. W., Leng, Q., and Berkowitz, G. A. (2003). Plants do it differently. a new basis for potassium/sodium selectivity in the pore of an ion channel. Plant Physiol. 132, 1353–1361. doi: 10.1104/pp.103.020560
Huang, N. C., Chiang, C. S., Crawford, N. M., and Tsay, Y. F. (1996). CHL1 encodes a component of the low-affinity nitrate uptake system in arabidopsis and shows cell type-specific expression in roots. Plant Cell 8, 2183–2191. doi: 10.1105/tpc.8.12.2183
Huang, N. C., Liu, K. H., Lo, H. J., and Tsay, Y. F. (1999). Cloning and functional characterization of an Arabidopsis nitrate transporter gene that encodes a constitutive component of low-affinity uptake. Plant Cell 11, 1381–1392. doi: 10.1105/tpc.11.8.1381
Irigoyen, S., Karlsson, P. M., Kuruvilla, J., Spetea, C., and Versaw, W. K. (2011). The sink-specific plastidic phosphate transporter PHT4;2 influences starch accumulation and leaf size in Arabidopsis. Plant Physiol. 157, 1765–1777. doi: 10.1104/pp.111.181925
Ivashikina, N., Becker, D., Ache, P., Meyerhoff, O., Felle, H. H., and Hedrich, R. (2001). K channel profile and electrical properties of Arabidopsis root hairs. FEBS Lett. 508, 463–469. doi: 10.1016/s0014-5793(01)03114-3
Jean-Baptiste, K., McFaline-Figueroa, J. L., Alexandre, C. M., Dorrity, M. W., Saunders, L., Bubb, K. L., et al. (2019). Dynamics of gene expression in single root cells of A. thaliana. Plant Cell 31, 993–1011. doi: 10.1101/448514
Jia, F., Wan, X., Zhu, W., Sun, D., Zheng, C., Liu, P., et al. (2015). Overexpression of mitochondrial phosphate transporter 3 severely hampers plant development through regulating mitochondrial function in arabidopsis. PLoS One 10:e0129717. doi: 10.1371/journal.pone.0129717
Jones, A. R., Kramer, E. M., Knox, K., Swarup, R., Bennett, M. J., Lazarus, C. M., et al. (2009). Auxin transport through non-hair cells sustains root-hair development. Nat. Cell Biol. 11, 78–84. doi: 10.1038/ncb1815
Jouannet, V., Brackmann, K., and Greb, T. (2015). (Pro) cambium formation and proliferation: two sides of the same coin? Curr. Opin. Plant Biol. 23, 54–60. doi: 10.1016/j.pbi.2014.10.010
Kanno, S., Arrighi, J.-F., Chiarenza, S., Bayle, V., Berthomé, R., Péret, B., et al. (2016). A novel role for the root cap in phosphate uptake and homeostasis. Elife 5:e14577. doi: 10.7554/eLife.14577
Karlsson, P. M., Herdean, A., Adolfsson, L., Beebo, A., Nziengui, H., Irigoyen, S., et al. (2015). The arabidopsis thylakoid transporter PHT4;1 influences phosphate availability for ATP synthesis and plant growth. Plant J. 84, 99–110. doi: 10.1111/tpj.12962
Kawai, H., Ishikawa, T., Mitsui, T., Kore-eda, S., Yamada-Kawai, M., and Ohnishi, J.-I. (2014). Arabidopsis glycerol-3-phosphate permease 4 is localized in the plastids and involved in the accumulation of seed oil. Plant Biotechnol. 31, 159–165.
Khan, G. A., Bouraine, S., Wege, S., Li, Y., de Carbonnel, M., Berthomieu, P., et al. (2014). Coordination between zinc and phosphate homeostasis involves the transcription factor PHR1, the phosphate exporter PHO1, and its homologue PHO1;H3 in Arabidopsis. J. Exp. Bot. 65, 871–884. doi: 10.1093/jxb/ert444
Khan, I. U., Ali, A., and Yun, D.-J. (2018). Arabidopsis NHX transporters: sodium and potassium antiport mythology and sequestration during ionic stress. J. Plant Biol. 61, 292–300. doi: 10.1007/s12374-018-0244-y
Kiba, T., Feria-Bourrellier, A.-B., Lafouge, F., Lezhneva, L., Boutet-Mercey, S., Orsel, M., et al. (2012). The arabidopsis nitrate transporter NRT2.4 plays a double role in roots and shoots of nitrogen-starved plants. Plant Cell 24, 245–258. doi: 10.1105/tpc.111.092221
Kiiskinen, M., Korhonen, M., and Kangasjärvi, J. (1997). Isolation and characterization of cDNA for a plant mitochondrial phosphate translocator (Mpt1): ozone stress induces Mpt1 mRNA accumulation in birch (Betula pendula Roth). Plant Mol. Biol. 35, 271–279. doi: 10.1023/a:1005868715571
Kim, E. J., Kwak, J. M., Uozumi, N., and Schroeder, J. I. (1998). AtKUP1: an Arabidopsis gene encoding high-affinity potassium transport activity. Plant Cell 10, 51–62. doi: 10.1105/tpc.10.1.51
Köhler, C., Merkle, T., and Neuhaus, G. (1999). Characterisation of a novel gene family of putative cyclic nucleotide- and calmodulin-regulated ion channels in Arabidopsis thaliana. Plant J. 18, 97–104. doi: 10.1046/j.1365-313x.1999.00422.x
Komarova, N. Y., Thor, K., Gubler, A., Meier, S., Dietrich, D., Weichert, A., et al. (2008). AtPTR1 and AtPTR5 transport dipeptides in planta. Plant Physiol. 148, 856–869. doi: 10.1104/pp.108.123844
Krapp, A., David, L. C., Chardin, C., Girin, T., Marmagne, A., Leprince, A.-S., et al. (2014). Nitrate transport and signalling in Arabidopsis. J. Exp. Bot. 65, 789–798. doi: 10.1093/jxb/eru001
Krouk, G., Lacombe, B., Bielach, A., Perrine-Walker, F., Malinska, K., Mounier, E., et al. (2010). Nitrate-regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Dev. Cell 18, 927–937. doi: 10.1016/j.devcel.2010.05.008
Kunz, H.-H., Gierth, M., Herdean, A., Satoh-Cruz, M., Kramer, D. M., Spetea, C., et al. (2014). Plastidial transporters KEA1, -2, and -3 are essential for chloroplast osmoregulation, integrity, and pH regulation in Arabidopsis. Proc. Natl. Acad. Sci. 111, 7480–7485. doi: 10.1073/pnas.1323899111
Kwak, J. M., Murata, Y., Baizabal-Aguirre, V. M., Merrill, J., Wang, M., Kemper, A., et al. (2001). Dominant negative guard cell K+ channel mutants reduce inward-rectifying K+ currents and light-induced stomatal opening in arabidopsis. Plant Physiol. 127, 473–485.
Lacombe, B., Pilot, G., Michard, E., Gaymard, F., Sentenac, H., and Thibaud, J.-B. (2000). A shaker-like K+ channel with weak rectification is expressed in both source and sink phloem tissues of arabidopsis. Plant Cell 12, 837–851. doi: 10.1105/tpc.12.6.837
Ladwig, F., Dahlke, R. I., Stührwohldt, N., Hartmann, J., Harter, K., and Sauter, M. (2015). Phytosulfokine regulates growth in Arabidopsis through a response module at the plasma membrane that includes cyclic nucleotide-Gated channel17, H+-ATPase, and BAK1. Plant Cell 27, 1718–1729.
Lagarde, D., Basset, M., Lepetit, M., Conejero, G., Gaymard, F., Astruc, S., et al. (1996). Tissue-specific expression of Arabidopsis AKT1 gene is consistent with a role in K+ nutrition. Plant J. 9, 195–203. doi: 10.1046/j.1365-313x.1996.09020195.x
Lapis-Gaza, H. R., Jost, R., and Finnegan, P. M. (2014). Arabidopsis phosphate transporter1 genes PHT1;8 and PHT1;9 are involved in root-to-shoot translocation of orthophosphate. BMC Plant Biol. 14:334. doi: 10.1186/s12870-014-0334-z
Lebaudy, A., Véry, A.-A., and Sentenac, H. (2007). K+ channel activity in plants: genes, regulations and functions. FEBS Lett. 581, 2357–2366. doi: 10.1016/j.febslet.2007.03.058
Lee, R. B., and Ratcliffe, R. G. (1993). Subcellular distribution of inorganic phosphate, and levels of nucleoside triphosphate, in mature maize roots at low external phosphate concentrations: measurements with 31P-NMR. J. Exp. Bot. 44, 587–598. doi: 10.1093/jxb/44.3.587
Leng, Q., Mercier, R. W., Yao, W., and Berkowitz, G. A. (1999). Cloning and first functional characterization of a plant cyclic nucleotide-gated cation channel. Plant Physiol. 121, 753–761. doi: 10.1104/pp.121.3.753
Léran, S., Varala, K., Boyer, J.-C., Chiurazzi, M., Crawford, N., Daniel-Vedele, F., et al. (2014). A unified nomenclature of nitrate transporter 1/peptide transporter family members in plants. Trends Plant Sci. 19, 5–9. doi: 10.1016/j.tplants.2013.08.008
Lezhneva, L., Kiba, T., Feria-Bourrellier, A.-B., Lafouge, F., Boutet-Mercey, S., Zoufan, P., et al. (2014). The Arabidopsis nitrate transporter NRT2.5 plays a role in nitrate acquisition and remobilization in nitrogen-starved plants. Plant J. 80, 230–241. doi: 10.1111/tpj.12626
Lhamo, D., Shao, Q., Tang, R., and Luan, S. (2020). Genome-wide analysis of the five phosphate transporter families in camelina sativa and their expressions in response to low-P. Int. J. Mol. Sci. 21:8365. doi: 10.3390/ijms21218365
Lhamo, D., Wang, C., Gao, Q., and Luan, S. (2021). Recent advances in genome-wide analyses of plant potassium transporter families. Curr. Geno. 22, 1875–5488. doi: 10.2174/1389202922666210225083634
Li, B., Byrt, C., Qiu, J., Baumann, U., Hrmova, M., Evrard, A., et al. (2016). Identification of a stelar-localized transport protein that facilitates root-to-shoot transfer of chloride in arabidopsis. Plant Physiol. 170, 1014–1029. doi: 10.1104/pp.15.01163
Li, B., Qiu, J., Jayakannan, M., Xu, B., Li, Y., Mayo, G. M., et al. (2017a). AtNPF2.5 modulates chloride (Cl-) efflux from roots of Arabidopsis thaliana. Front. Plant Sci. 7:2013. doi: 10.3389/fpls.2016.02013
Li, H., Yu, M., Du, X.-Q., Wang, Z.-F., Wu, W.-H., Quintero, F. J., et al. (2017b). NRT1.5/NPF7.3 functions as a proton-coupled H /K antiporter for K loading into the Xylem in Arabidopsis. Plant Cell 29, 2016–2026. doi: 10.1105/tpc.16.00972
Li, H.-T., Liu, H., Gao, X.-S., and Zhang, H. (2009). Knock-out of arabidopsis AtNHX4 gene enhances tolerance to salt stress. Biochem. Biophys. Res. Commun. 382, 637–641. doi: 10.1016/j.bbrc.2009.03.091
Li, J.-Y., Fu, Y.-L., Pike, S. M., Bao, J., Tian, W., Zhang, Y., et al. (2010). The Arabidopsis nitrate transporter NRT1.8 functions in nitrate removal from the xylem sap and mediates cadmium tolerance. Plant Cell 22, 1633–1646. doi: 10.1105/tpc.110.075242
Li, W., Xu, G., Alli, A., and Yu, L. (2018). Plant HAK/KUP/KT K+ transporters: Function and regulation. Semin. Cell Dev. Biol. 74, 133–141. doi: 10.1016/j.semcdb.2017.07.009
Li, X., Borsics, T., Harrington, H. M., and Christopher, D. A. (2005). Arabidopsis AtCNGC10 rescues potassium channel mutants of E. coli, yeast and Arabidopsis and is regulated by calcium/calmodulin and cyclic GMP in E. coli. Funct. Plant Biol. 32, 643–653. doi: 10.1071/FP04233
Lin, S.-H., Kuo, H.-F., Canivenc, G., Lin, C.-S., Lepetit, M., Hsu, P.-K., et al. (2008). Mutation of the Arabidopsis NRT1.5 nitrate transporter causes defective root-to-shoot nitrate transport. Plant Cell 20, 2514–2528. doi: 10.1105/tpc.108.060244
Liu, H., Tang, R., Zhang, Y. U. E., Wang, C., Lv, Q., Gao, X., et al. (2010). AtNHX3 is a vacuolar K+/H+ antiporter required for low-potassium tolerance in Arabidopsis thaliana. Plant Cell Environ. 33, 1989–1999. doi: 10.1111/j.1365-3040.2010.02200.x
Liu, J., Yang, L., Luan, M., Wang, Y., Zhang, C., Zhang, B., et al. (2015). A vacuolar phosphate transporter essential for phosphate homeostasis in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 112, E6571–E6578. doi: 10.1073/pnas.1514598112
Liu, K., Li, L., and Luan, S. (2006). Intracellular K+ sensing of SKOR, a Shaker-type K+ channel from Arabidopsis. Plant J. 46, 260–268. doi: 10.1111/j.1365-313X.2006.02689.x
Liu, K. H., Huang, C. Y., and Tsay, Y. F. (1999). CHL1 is a dual-affinity nitrate transporter of Arabidopsis involved in multiple phases of nitrate uptake. Plant Cell 11, 865–874. doi: 10.1105/tpc.11.5.865
Liu, K.-H., and Tsay, Y.-F. (2003). Switching between the two action modes of the dual-affinity nitrate transporter CHL1 by phosphorylation. EMBO J. 22, 1005–1013. doi: 10.1093/emboj/cdg118
Liu, T.-Y., Huang, T.-K., Yang, S.-Y., Hong, Y.-T., Huang, S.-M., Wang, F.-N., et al. (2016). Identification of plant vacuolar transporters mediating phosphate storage. Nat. Commun. 7:11095. doi: 10.1038/ncomms11095
Looger, L. L., Lalonde, S., and Frommer, W. B. (2005). Genetically encoded FRET sensors for visualizing metabolites with subcellular resolution in living cells. Plant Physiol. 138, 555–557. doi: 10.1104/pp.104.900151
López-Arredondo, D. L., Leyva-González, M. A., González-Morales, S. I., López-Bucio, J., and Herrera-Estrella, L. (2014). Phosphate nutrition: improving low-phosphate tolerance in crops. Annu. Rev. Plant Biol. 65, 95–123. doi: 10.1146/annurev-arplant-050213-035949
López-Bucio, J., Cruz-Ramírez, A., and Herrera-Estrella, L. (2003). The role of nutrient availability in regulating root architecture. Curr. Opin. Plant Biol. 6, 280–287. doi: 10.1016/s1369-5266(03)00035-9
Lu, Y., Chanroj, S., Zulkifli, L., Johnson, M. A., Uozumi, N., Cheung, A., et al. (2011). Pollen tubes lacking a pair of K+ transporters fail to target ovules in Arabidopsis. Plant Cell 23, 81–93. doi: 10.1105/tpc.110.080499
Luan, M., and Lan, W. (2019). Escape routes for vacuolar phosphate. Nat. Plants 5, 9–10. doi: 10.1038/s41477-018-0343-2
Luan, M., Tang, R.-J., Tang, Y., Tian, W., Hou, C., Zhao, F., et al. (2017). Transport and homeostasis of potassium and phosphate: limiting factors for sustainable crop production. J. Exp. Bot. 68, 3091–3105. doi: 10.1093/jxb/erw444
Luan, M., Zhao, F., Han, X., Sun, G., Yang, Y., Liu, J., et al. (2019). Vacuolar phosphate transporters contribute to systemic phosphate homeostasis vital for reproductive development in arabidopsis. Plant Physiol. 179, 640–655. doi: 10.1104/pp.18.01424
Lv, Q.-D., Tang, R.-J., Liu, H., Gao, X.-S., Li, Y.-Z., Zheng, H.-Q., et al. (2009). Cloning and molecular analyses of the Arabidopsis thaliana chloride channel gene family. Plant Sci. 176, 650–661. doi: 10.1016/j.plantsci.2009.02.006
Maathuis, F. J. M. (2009). Physiological functions of mineral macronutrients. Curr. Opin. Plant Biol. 12, 250–258. doi: 10.1016/j.pbi.2009.04.003
Macosko, E. Z., Basu, A., Satija, R., Nemesh, J., Shekhar, K., Goldman, M., et al. (2015). Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214. doi: 10.1016/j.cell.2015.05.002
Maierhofer, T., Lind, C., Hüttl, S., Scherzer, S., Papenfuß, M., Simon, J., et al. (2014). A single-pore residue renders the arabidopsis root anion channel SLAH2 highly nitrate selective. Plant Cell 26, 2554–2567. doi: 10.1105/tpc.114.125849
Marmagne, A., Vinauger-Douard, M., Monachello, D., de Longevialle, A. F., Charon, C., Allot, M., et al. (2007). Two members of the Arabidopsis CLC (chloride channel) family, AtCLCe and AtCLCf, are associated with thylakoid and Golgi membranes, respectively. J. Exp. Bot. 58, 3385–3393. doi: 10.1093/jxb/erm187
Marten, I., Hoth, S., Deeken, R., Ache, P., Ketchum, K. A., Hoshi, T., et al. (1999). AKT3, a phloem-localized K+ channel, is blocked by protons. Proc. Natl. Acad. Sci. U.S.A. 96, 7581–7586. doi: 10.1073/pnas.96.13.7581
Mäser, P., Thomine, S., Schroeder, J. I., Ward, J. M., Hirschi, K., Sze, H., et al. (2001). Phylogenetic relationships within cation transporter families of Arabidopsis. Plant Physiol. 126, 1646–1667. doi: 10.1104/pp.126.4.1646
McCully, M. E. (1994). Accumulation of high levels of potassium in the developing xylem elements in roots of soybean and some other dicotyledons. Protoplasma 183, 116–125. doi: 10.1007/bf01276819
Michard, E., Dreyer, I., Lacombe, B., Sentenac, H., and Thibaud, J.-B. (2005). Inward rectification of the AKT2 channel abolished by voltage-dependent phosphorylation. Plant J. 44, 783–797. doi: 10.1111/j.1365-313X.2005.02566.x
Misson, J., Thibaud, M.-C., Bechtold, N., Raghothama, K., and Nussaume, L. (2004). Transcriptional regulation and functional properties of Arabidopsis Pht1;4, a high affinity transporter contributing greatly to phosphate uptake in phosphate deprived plants. Plant Mol. Biol. 55, 727–741. doi: 10.1007/s11103-004-1965-5
Miyaji, T., Kuromori, T., Takeuchi, Y., Yamaji, N., Yokosho, K., Shimazawa, A., et al. (2015). AtPHT4;4 is a chloroplast-localized ascorbate transporter in Arabidopsis. Nat. Commun. 6:5928. doi: 10.1038/ncomms6928
Monachello, D., Allot, M., Oliva, S., Krapp, A., Daniel-Vedele, F., Barbier-Brygoo, H., et al. (2009). Two anion transporters AtClCa and AtClCe fulfil interconnecting but not redundant roles in nitrate assimilation pathways. New Phytol. 183, 88–94. doi: 10.1111/j.1469-8137.2009.02837.x
Mouline, K., Véry, A. A., and Gaymard, F. (2002). Pollen tube development and competitive ability are impaired by disruption of a Shaker K+ channel in Arabidopsis. Genes Dev. 16, 339–350.
Mounier, E., Pervent, M., Ljung, K., Gojon, A., and Nacry, P. (2014). Auxin-mediated nitrate signalling by NRT1.1 participates in the adaptive response ofArabidopsisroot architecture to the spatial heterogeneity of nitrate availability. Plant Cell Environ. 37, 162–174. doi: 10.1111/pce.12143
Mudge, S. R., Rae, A. L., Diatloff, E., and Smith, F. W. (2002). Expression analysis suggests novel roles for members of the Pht1 family of phosphate transporters in Arabidopsis. Plant J. 31, 341–353. doi: 10.1046/j.1365-313x.2002.01356.x
Mukherjee, P., Banerjee, S., Wheeler, A., Ratliff, L. A., Irigoyen, S., Garcia, L. R., et al. (2015). Live imaging of inorganic phosphate in plants with cellular and subcellular resolution. Plant Physiol. 167, 628–638. doi: 10.1104/pp.114.254003
Nagarajan, V. K., Jain, A., Poling, M. D., Lewis, A. J., Raghothama, K. G., and Smith, A. P. (2011). Arabidopsis Pht1;5 mobilizes phosphate between source and sink organs and influences the interaction between phosphate homeostasis and ethylene signaling. Plant Physiol. 156, 1149–1163. doi: 10.1104/pp.111.174805
Nakamura, R. L., McKendree, W. L. Jr., Hirsch, R. E., Sedbrook, J. C., Gaber, R. F., and Sussman, M. R. (1995). Expression of an Arabidopsis potassium channel gene in guard cells. Plant Physiol. 109, 371–374. doi: 10.1104/pp.109.2.371
Negi, J., Matsuda, O., Nagasawa, T., Oba, Y., Takahashi, H., Kawai-Yamada, M., et al. (2008). CO2 regulator SLAC1 and its homologues are essential for anion homeostasis in plant cells. Nature 452, 483–486. doi: 10.1038/nature06720
Nieves-Cordones, M., Alemán, F., Martínez, V., and Rubio, F. (2014). K+ uptake in plant roots. the systems involved, their regulation and parallels in other organisms. J. Plant Physiol. 171, 688–695. doi: 10.1016/j.jplph.2013.09.021
Nieves-Cordones, M., Lara, A., Ródenas, R., Amo, J., Rivero, R. M., Martínez, V., et al. (2019). Modulation of K+ translocation by AKT1 and AtHAK5 in Arabidopsis plants. Plant Cell Environ. 42, 2357–2371. doi: 10.1111/pce.13573
Noguero, M., and Lacombe, B. (2016). Transporters involved in root nitrate uptake and sensing by arabidopsis. Front. Plant Sci. 7:1391. doi: 10.3389/fpls.2016.01391
Nour-Eldin, H. H., Andersen, T. G., Burow, M., Madsen, S. R., Jørgensen, M. E., Olsen, C. E., et al. (2012). NRT/PTR transporters are essential for translocation of glucosinolate defence compounds to seeds. Nature 488, 531–534. doi: 10.1038/nature11285
Nussaume, L. (2011). Phosphate import in plants: focus on the PHT1 transporters. Front. Plant Sci. 2:83. doi: 10.3389/fpls.2011.00083
O’Brien, J. A., Vega, A., Bouguyon, E., Krouk, G., Gojon, A., Coruzzi, G., et al. (2016). Nitrate transport, sensing, and responses in plants. Mol. Plant 9, 837–856. doi: 10.1016/j.molp.2016.05.004
Orsel, M., Krapp, A., and Daniel-Vedele, F. (2002). Analysis of the NRT2 nitrate transporter family in Arabidopsis. structure and gene expression. Plant Physiol. 129, 886–896. doi: 10.1104/pp.005280
Osakabe, Y., Arinaga, N., Umezawa, T., Katsura, S., Nagamachi, K., Tanaka, H., et al. (2013). Osmotic stress responses and plant growth controlled by potassium transporters in Arabidopsis. Plant Cell 25, 609–624. doi: 10.1105/tpc.112.105700
Padmanaban, S., Chanroj, S., Kwak, J. M., Li, X., Ward, J. M., and Sze, H. (2007). Participation of endomembrane cation/H+ exchanger AtCHX20 in osmoregulation of guard cells. Plant Physiol. 144, 82–93. doi: 10.1104/pp.106.092155
Pike, S., Gao, F., Kim, M. J., Kim, S. H., Schachtman, D. P., and Gassmann, W. (2014). Members of the NPF3 transporter subfamily encode pathogen-inducible nitrate/nitrite transporters in grapevine and Arabidopsis. Plant Cell Physiol. 55, 162–170. doi: 10.1093/pcp/pct167
Pilot, G., Lacombe, B., Gaymard, F., Chérel, I., Boucherez, J., Thibaud, J.-B., et al. (2001). Guard cell inward K+ channel activity inarabidopsis involves expression of the twin channel subunits KAT1 and KAT2. J. Biol. Chem. 276, 3215–3221. doi: 10.1074/jbc.M007303200
Pilot, G., Pratelli, R., Gaymard, F., Meyer, Y., and Sentenac, H. (2003). Five-group distribution of the shaker-like K+ channel family in higher plants. J. Mol. Evol. 56, 418–434. doi: 10.1007/s00239-002-2413-2
Plaxton, W. C., and Tran, H. T. (2011). Metabolic adaptations of phosphate-starved plants. Plant Physiol. 156, 1006–1015. doi: 10.1104/pp.111.175281
Poirier, Y., and Bucher, M. (2002). Phosphate transport and homeostasis in Arabidopsis. Arabidopsis Book 1:e0024. doi: 10.1199/tab.0024
Poirier, Y., Thoma, S., Somerville, C., and Schiefelbein, J. (1991). Mutant of Arabidopsis deficient in xylem loading of phosphate. Plant Physiol. 97, 1087–1093. doi: 10.1104/pp.97.3.1087
Pratt, J., Boisson, A.-M., Gout, E., Bligny, R., Douce, R., and Aubert, S. (2009). Phosphate (Pi) starvation effect on the cytosolic Pi concentration and Pi exchanges across the tonoplast in plant cells: an in vivo 31P-nuclear magnetic resonance study using methylphosphonate as a Pi analog. Plant Physiol. 151, 1646–1657. doi: 10.1104/pp.109.144626
Pyo, Y. J., Gierth, M., Schroeder, J. I., and Cho, M. H. (2010). High-affinity K(+) transport in Arabidopsis: AtHAK5 and AKT1 are vital for seedling establishment and postgermination growth under low-potassium conditions. Plant Physiol. 153, 863–875. doi: 10.1104/pp.110.154369
Qi, Z., Hampton, C. R., Shin, R., Barkla, B. J., White, P. J., and Schachtman, D. P. (2008). The high affinity K+ transporter AtHAK5 plays a physiological role in planta at very low K+ concentrations and provides a caesium uptake pathway in Arabidopsis. J. Exp. Bot. 59, 595–607. doi: 10.1093/jxb/erm330
Radcliffe, S. A., Miller, A. J., and Ratcliffe, R. G. (2005). Microelectrode and 133Cs nuclear magnetic resonance evidence for variable cytosolic and cytoplasmic nitrate pools in maize root tips. Plant Cell Environ. 28, 1379–1387. doi: 10.1111/j.1365-3040.2005.01370.x
Ramaiah, M., Jain, A., Baldwin, J. C., Karthikeyan, A. S., and Raghothama, K. G. (2011). Characterization of the phosphate starvation-induced glycerol-3-phosphate permease gene family in Arabidopsis. Plant Physiol. 157, 279–291. doi: 10.1104/pp.111.178541
Rausch, C., Zimmermann, P., Amrhein, N., and Bucher, M. (2004). Expression analysis suggests novel roles for the plastidic phosphate transporter Pht2; 1 in auto-and heterotrophic tissues in potato and Arabidopsis. Plant J. 39, 13–28. doi: 10.1111/j.1365-313X.2004.02106.x
Reintanz, B., Szyroki, A., Ivashikina, N., Ache, P., Godde, M., Becker, D., et al. (2002). AtKC1, a silent Arabidopsis potassium channel α-subunit modulates root hair K+ influx. Proc. Natl. Acad. Sci. U.S.A. 99, 4079–4084. doi: 10.1073/pnas.052677799
Remy, E., Cabrito, T. R., Batista, R. A., Teixeira, M. C., Sá-Correia, I., and Duque, P. (2012). The Pht1;9 and Pht1;8 transporters mediate inorganic phosphate acquisition by the Arabidopsis thaliana root during phosphorus starvation. New Phytol. 195, 356–371. doi: 10.1111/j.1469-8137.2012.04167.x
Rigas, S., Debrosses, G., Haralampidis, K., Vicente-Agullo, F., Feldmann, K. A., Grabov, A., et al. (2001). TRH1 encodes a potassium transporter required for tip growth in Arabidopsis root hairs. Plant Cell 13, 139–151. doi: 10.1105/tpc.13.1.139
Rigas, S., Ditengou, F. A., Ljung, K., Daras, G., Tietz, O., Palme, K., et al. (2013). Root gravitropism and root hair development constitute coupled developmental responses regulated by auxin homeostasis in theArabidopsisroot apex. New Phytol. 197, 1130–1141. doi: 10.1111/nph.12092
Rubio, F., Nieves-Cordones, M., Alemán, F., and Martínez, V. (2008). Relative contribution of AtHAK5 and AtAKT1 to K uptake in the high-affinity range of concentrations. Physiol. Plantar. 134, 598–608. doi: 10.1111/j.1399-3054.2008.01168.x
Ruiz Herrera, L. F., Shane, M. W., and López-Bucio, J. (2015). Nutritional regulation of root development. wiley interdiscip. Rev. Dev. Biol. 4, 431–443. doi: 10.1002/wdev.183
Ryu, K. H., Huang, L., Kang, H. M., and Schiefelbein, J. (2019). Single-cell RNA sequencing resolves molecular relationships among individual plant cells. Plant Physiol. 179, 1444–1456. doi: 10.1104/pp.18.01482
Sahu, A., Banerjee, S., Raju, A. S., Chiou, T.-J., Garcia, L. R., and Versaw, W. K. (2020). Spatial profiles of phosphate in roots indicate developmental control of uptake, recycling, and sequestration. Plant Physiol. 184, 2064–2077. doi: 10.1104/pp.20.01008
Saito, H., Oikawa, T., Hamamoto, S., Ishimaru, Y., Kanamori-Sato, M., Sasaki-Sekimoto, Y., et al. (2015). The jasmonate-responsive GTR1 transporter is required for gibberellin-mediated stamen development in Arabidopsis. Nat. Commun. 6:6095. doi: 10.1038/ncomms7095
Santa-María, G. E., Oliferuk, S., and Moriconi, J. I. (2018). KT-HAK-KUP transporters in major terrestrial photosynthetic organisms: a twenty years tale. J. Plant Physiol. 226, 77–90. doi: 10.1016/j.jplph.2018.04.008
Schleyer, M., and Bakker, E. P. (1993). Nucleotide sequence and 3’-end deletion studies indicate that the K (+)-uptake protein kup from Escherichia coli is composed of a hydrophobic core linked to a large. J. Bacteriol. 175, 6925–6931.
Schmidt, C., and Schroeder, J. I. (1994). Anion selectivity of slow anion channels in the plasma membrane of guard cells (large nitrate permeability). Plant Physiol. 106, 383–391. doi: 10.1104/pp.106.1.383
Segonzac, C., Boyer, J.-C., Ipotesi, E., Szponarski, W., Tillard, P., Touraine, B., et al. (2007). Nitrate efflux at the root plasma membrane: identification of an Arabidopsis excretion transporter. Plant Cell 19, 3760–3777. doi: 10.1105/tpc.106.048173
Shahan, R., Hsu, C. W., Nolan, T. M., Cole, B. J., Taylor, I. W., Vlot, A. H. C., et al. (2020). A single cell Arabidopsis root atlas reveals developmental trajectories in wild type and cell identity mutants. bioRxiv [preprint] doi: 10.1101/2020.06.29.178863
Sharma, T., Dreyer, I., and Riedelsberger, J. (2013). The role of K(+) channels in uptake and redistribution of potassium in the model plant Arabidopsis thaliana. Front. Plant Sci. 4:224. doi: 10.3389/fpls.2013.00224
Shi, H., Quintero, F. J., Pardo, J. M., and Zhu, J.-K. (2002). The putative plasma membrane Na(+)/H(+) antiporter SOS1 controls long-distance Na(+) transport in plants. Plant Cell 14, 465–477. doi: 10.1105/tpc.010371
Shi, H., and Zhu, J.-K. (2002). Regulation of expression of the vacuolar Na+/H+ antiporter gene AtNHX1 by salt stress and abscisic acid. Plant Mol. Biol. 50, 543–550. doi: 10.1023/a:1019859319617
Shin, H., Shin, H.-S., Dewbre, G. R., and Harrison, M. J. (2004). Phosphate transport in Arabidopsis: Pht1; 1 and Pht1; 4 play a major role in phosphate acquisition from both low-and high-phosphate environments. Plant J. 39, 629–642. doi: 10.1111/j.1365-313X.2004.02161.x
Shulse, C. N., Cole, B. J., Ciobanu, D., Lin, J., Yoshinaga, Y., Gouran, M., et al. (2019). High-throughput single-cell transcriptome profiling of plant cell types. Cell Rep. 27, 2241–2247.e4. doi: 10.1016/j.celrep.2019.04.054
Stefanovic, A., Arpat, A. B., Bligny, R., Gout, E., Vidoudez, C., Bensimon, M., et al. (2011). Over-expression of PHO1 in Arabidopsis leaves reveals its role in mediating phosphate efflux. Plant J. 66, 689–699. doi: 10.1111/j.1365-313X.2011.04532.x
Stefanovic, A., Ribot, C., Rouached, H., Wang, Y., Chong, J., Belbahri, L., et al. (2007). Members of the PHO1 gene family show limited functional redundancy in phosphate transfer to the shoot, and are regulated by phosphate deficiency via distinct pathways. Plant J. 50, 982–994. doi: 10.1111/j.1365-313X.2007.03108.x
Sugiura, M., Georgescu, M. N., and Takahashi, M. (2007). A nitrite transporter associated with nitrite uptake by higher plant chloroplasts. Plant Cell Physiol. 48, 1022–1035. doi: 10.1093/pcp/pcm073
Sustr, M., Soukup, A., and Tylova, E. (2019). Potassium in root growth and development. Plants 8:435. doi: 10.3390/plants8100435
Svistoonoff, S., Creff, A., Reymond, M., Sigoillot-Claude, C., Ricaud, L., Blanchet, A., et al. (2007). Root tip contact with low-phosphate media reprograms plant root architecture. Nat. Genet. 39, 792–796. doi: 10.1038/ng2041
Sze, H., and Chanroj, S. (2018). Plant endomembrane dynamics: studies of K /H antiporters provide insights on the effects of PH and ion homeostasis. Plant Physiol. 177, 875–895. doi: 10.1104/pp.18.00142
Sze, H., Padmanaban, S., Cellier, F., Honys, D., Cheng, N.-H., Bock, K. W., et al. (2004). Expression patterns of a novel AtCHX gene family highlight potential roles in osmotic adjustment and K+ homeostasis in pollen development. Plant Physiol. 136, 2532–2547. doi: 10.1104/pp.104.046003
Takabatake, R., Hata, S., Taniguchi, M., Kouchi, H., Sugiyama, T., and Izui, K. (1999). Isolation and characterization of cDNAs encoding mitochondrial phosphate transporters in soybean, maize, rice, and Arabidopsis. Plant Mol. Biol. 40, 479–486. doi: 10.1023/A:1006285009435
Tal, I., Zhang, Y., Jørgensen, M. E., Pisanty, O., Barbosa, I. C. R., Zourelidou, M., et al. (2016). The Arabidopsis NPF3 protein is a GA transporter. Nat. Commun. 7:11486. doi: 10.1038/ncomms11486
Tang, R.-J., Zhao, F.-G., Yang, Y., Wang, C., Li, K., Kleist, T. J., et al. (2020). A calcium signalling network activates vacuolar K+ remobilization to enable plant adaptation to low-K environments. Nat. Plants 6, 384–393. doi: 10.1038/s41477-020-0621-7
Taochy, C., Gaillard, I., Ipotesi, E., Oomen, R., Leonhardt, N., Zimmermann, S., et al. (2015). The Arabidopsis root stele transporter NPF2.3 contributes to nitrate translocation to shoots under salt stress. Plant J. 83, 466–479. doi: 10.1111/tpj.12901
Tester, M., and Leigh, R. A. (2001). Partitioning of nutrient transport processes in roots. J. Exp. Bot. 52, 445–457. doi: 10.1093/jexbot/52.suppl_1.445
Vahisalu, T., Kollist, H., Wang, Y.-F., Nishimura, N., Chan, W.-Y., Valerio, G., et al. (2008). SLAC1 is required for plant guard cell S-type anion channel function in stomatal signalling. Nature 452, 487–491. doi: 10.1038/nature06608
Venema, K., Quintero, F. J., Pardo, J. M., and Donaire, J. P. (2002). The arabidopsis Na+/H+ exchanger AtNHX1 catalyzes low affinity Na+ and K+ transport in reconstituted liposomes. J. Biol. Chem. 277, 2413–2418. doi: 10.1074/jbc.M105043200
Versaw, W. K., and Harrison, M. J. (2002). A chloroplast phosphate transporter, PHT2;1, influences allocation of phosphate within the plant and phosphate-starvation responses. Plant Cell 14, 1751–1766. doi: 10.1105/tpc.002220
Véry, A.-A., Nieves-Cordones, M., Daly, M., Khan, I., Fizames, C., and Sentenac, H. (2014). Molecular biology of K+ transport across the plant cell membrane: what do we learn from comparison between plant species? J. Plant Physiol. 171, 748–769. doi: 10.1016/j.jplph.2014.01.011
Vicente-Agullo, F., Rigas, S., Desbrosses, G., Dolan, L., Hatzopoulos, P., and Grabov, A. (2004). Potassium carrier TRH1 is required for auxin transport in Arabidopsis roots. Plant J. 40, 523–535. doi: 10.1111/j.1365-313X.2004.02230.x
Vissenberg, K., Claeijs, N., Balcerowicz, D., and Schoenaers, S. (2020). Hormonal regulation of root hair growth and responses to the environment in Arabidopsis. J. Exp. Bot. 71, 2412–2427. doi: 10.1093/jxb/eraa048
Voelker, C., Gomez-Porras, J. L., Becker, D., Hamamoto, S., Uozumi, N., Gambale, F., et al. (2010). Roles of tandem-pore K+ channels in plants - a puzzle still to be solved. Plant Biol. 12(Suppl. 1), 56–63. doi: 10.1111/j.1438-8677.2010.00353.x
Voelker, C., Schmidt, D., Mueller-Roeber, B., and Czempinski, K. (2006). Members of the Arabidopsis AtTPK/KCO family form homomeric vacuolar channels in planta. Plant J. 48, 296–306. doi: 10.1111/j.1365-313X.2006.02868.x
Walker, D. J., Leigh, R. A., and Miller, A. J. (1996). Potassium homeostasis in vacuolate plant cells. Proc. Natl. Acad. Sci. U.S.A. 93, 10510–10514. doi: 10.1073/pnas.93.19.10510
Wang, M., Zheng, Q., Shen, Q., and Guo, S. (2013). The critical role of potassium in plant stress response. Int. J. Mol. Sci. 14, 7370–7390. doi: 10.3390/ijms14047370
Wang, R., Liu, D., and Crawford, N. M. (1998). The Arabidopsis CHL1 protein plays a major role in high-affinity nitrate uptake. Proc. Natl. Acad. Sci. U.S.A. 95, 15134–15139. doi: 10.1073/pnas.95.25.15134
Wang, Y., Ribot, C., Rezzonico, E., and Poirier, Y. (2004). Structure and expression profile of the Arabidopsis PHO1 gene family indicates a broad role in inorganic phosphate homeostasis. Plant Physiol. 135, 400–411. doi: 10.1104/pp.103.037945
Wang, Y., Tang, R.-J., Yang, X., Zheng, X., Shao, Q., Tang, Q.-L., et al. (2019). Golgi-localized cation/proton exchangers regulate ionic homeostasis and skotomorphogenesis in Arabidopsis. Plant Cell Environ. 42, 673–687. doi: 10.1111/pce.13452
Wang, Y., and Wu, W.-H. (2013). Potassium transport and signaling in higher plants. Ann. Rev. Plant Biol. 64, 451–476. doi: 10.1146/annurev-arplant-050312-120153
Wang, Y., and Wu, W.-H. (2017). Regulation of potassium transport and signaling in plants. Curr. Opin. Plant Biol. 39, 123–128. doi: 10.1016/j.pbi.2017.06.006
Wang, Y.-Y., Cheng, Y.-H., Chen, K.-E., and Tsay, Y.-F. (2018). Nitrate transport, signaling, and use efficiency. Annu. Rev. Plant Biol. 69, 85–122. doi: 10.1146/annurev-arplant-042817-040056
Wang, Y.-Y., and Tsay, Y.-F. (2011). Arabidopsis nitrate transporter NRT1.9 Is important in phloem nitrate transport. Plant Cell 23, 1945–1957. doi: 10.1105/tpc.111.083618
Wendrich, J. R., Yang, B., Vandamme, N., Verstaen, K., Smet, W., Van de Velde, C., et al. (2020). Vascular transcription factors guide plant epidermal responses to limiting phosphate conditions. Science 370:eaay4970. doi: 10.1126/science.aay4970
White, P. J. (2013). Improving potassium acquisition and utilisation by crop plants. J. Plant Nutr. Soil Sci. 176, 305–316. doi: 10.1002/jpln.201200121
Wu, S. J., Ding, L., and Zhu, J. K. (1996). SOS1, a genetic locus essential for salt tolerance and potassium acquisition. Plant Cell 8, 617–627. doi: 10.1105/tpc.8.4.617
Xu, L., Zhao, H., Wan, R., Liu, Y., Xu, Z., Tian, W., et al. (2019). Identification of vacuolar phosphate efflux transporters in land plants. Nat Plants 5, 84–94. doi: 10.1038/s41477-018-0334-3
Zhang, A., Ren, H.-M., Tan, Y.-Q., Qi, G.-N., Yao, F.-Y., Wu, G.-L., et al. (2016). S-type anion channels SLAC1 and SLAH3 function as essential negative regulators of inward K+ channels and stomatal opening in arabidopsis. Plant Cell 28, 949–955. doi: 10.1105/tpc.16.01050
Zhang, M.-L., Huang, P.-P., Ji, Y., Wang, S., Wang, S.-S., Li, Z., et al. (2020). KUP9 maintains root meristem activity by regulating K+ and auxin homeostasis in response to low K. EMBO Rep. 21:e50164. doi: 10.15252/embr.202050164
Zhang, T.-Q., Xu, Z.-G., Shang, G.-D., and Wang, J.-W. (2019). A single-cell RNA sequencing profiles the developmental landscape of arabidopsis root. Mol. Plant 12, 648–660. doi: 10.1016/j.molp.2019.04.004
Zhao, J., Cheng, N.-H., Motes, C. M., Blancaflor, E. B., Moore, M., Gonzales, N., et al. (2008). AtCHX13 is a plasma membrane K+ transporter. Plant Physiol. 148, 796–807. doi: 10.1104/pp.108.124248
Zhao, J., Li, P., Motes, C. M., Park, S., and Hirschi, K. D. (2015). CHX 14 is a plasma membrane K-efflux transporter that regulates K+ redistribution in A rabidopsis thaliana. Plant Cell Environ. 38, 2223–2238. doi: 10.1111/pce.12524
Zhao, L.-N., Shen, L.-K., Zhang, W.-Z., Zhang, W., Wang, Y., and Wu, W.-H. (2013). Ca2 -dependent protein kinase11 and 24 modulate the activity of the inward rectifying K channels in arabidopsis pollen tubes. Plant Cell 25, 649–661. doi: 10.1105/tpc.112.103184
Zhen, R. G., Koyro, H. W., Leigh, R. A., Tomos, A. D., and Miller, A. J. (1991). Compartmental nitrate concentrations in barley root cells measured with nitrate-selective microelectrodes and by single-cell sap sampling. Planta 185, 356–361. doi: 10.1007/BF00201056
Zheng, X., He, K., Kleist, T., Chen, F., and Luan, S. (2015). Anion channel SLAH 3 functions in nitrate-dependent alleviation of ammonium toxicity in A rabidopsis. Plant Cell Environ. 38, 474–486. doi: 10.1111/pce.12389
Zhu, W., Miao, Q., Sun, D., Yang, G., Wu, C., Huang, J., et al. (2012). The mitochondrial phosphate transporters modulate plant responses to salt stress via affecting ATP and gibberellin metabolism in Arabidopsis thaliana. PLoS One 7:e43530. doi: 10.1371/journal.pone.0043530
Zhu, X., Pan, T., Zhang, X., Fan, L., Quintero, F. J., Zhao, H., et al. (2018). K+ efflux antiporters 4, 5, and 6 mediate pH and K+ homeostasis in endomembrane compartments. Plant Physiol. 178, 1657–1678.
Keywords: root single cell transcriptomics, expression analysis, nutrient transporters, nitrate transport, phosphate transport, potassium transport, NPK transport networks
Citation: Lhamo D and Luan S (2021) Potential Networks of Nitrogen-Phosphorus-Potassium Channels and Transporters in Arabidopsis Roots at a Single Cell Resolution. Front. Plant Sci. 12:689545. doi: 10.3389/fpls.2021.689545
Received: 01 April 2021; Accepted: 24 May 2021;
Published: 16 June 2021.
Edited by:
Lourdes Rubio, University of Malaga, SpainReviewed by:
Francisco Rubio, Center for Edaphology and Applied Biology of Segura, Spanish National Research Council, SpainVadim Volkov, London Metropolitan University, United Kingdom
Copyright © 2021 Lhamo and Luan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dhondup Lhamo, ZGxoYW1vQGJlcmtlbGV5LmVkdQ==
 Dhondup Lhamo
Dhondup Lhamo Sheng Luan
Sheng Luan