- 1Graduate School of Science and Engineering, Teikyo University, Utsunomiya, Japan
- 2Department of Biosciences, Faculty of Science and Engineering, Teikyo University, Utsunomiya, Japan
- 3Advanced Instrumental Analysis Center, Teikyo University, Utsunomiya, Japan
- 4Agro-Biotechnology Research Center, Graduate School of Agricultural and Life Sciences, The University of Tokyo, Tokyo, Japan
- 5Graduate School of Science, Tohoku University, Sendai, Japan
- 6Institute of Plant Science and Resources, Okayama University, Kurashiki, Japan
- 7Proteo-Science Center, Ehime University, Matsuyama, Japan
- 8Collaborative Research Institute for Innovative Microbiology, The University of Tokyo, Tokyo, Japan
- 9Graduate School of Life Sciences, Tohoku University, Sendai, Japan
Jasmonic acid (JA) and its biologically active form jasmonoyl-L-isoleucine (JA-Ile) regulate defense responses to various environmental stresses and developmental processes in plants. JA and JA-Ile are synthesized from α-linolenic acids derived from membrane lipids via 12-oxo-phytodienoic acid (OPDA). In the presence of JA-Ile, the COI1 receptor physically interacts with JAZ repressors, leading to their degradation, resulting in the transcription of JA-responsive genes by MYC transcription factors. Although the biosynthesis of JA-Ile is conserved in vascular plants, it is not recognized by COI1 in bryophytes and is not biologically active. In the liverwort Marchantia polymorpha, dinor-OPDA (dn-OPDA), a homolog of OPDA with two fewer carbons, and its isomer dn-iso-OPDA accumulate after wounding and are recognized by COI1 to activate downstream signaling. The moss Calohypnum plumiforme produces the antimicrobial-specialized metabolites, momilactones. It has been reported that JA and JA-Ile are not detected in C. plumiforme and that OPDA, but not JA, can induce momilactone accumulation and the expression of these biosynthetic genes, suggesting that OPDA or its derivative is a biologically active molecule in C. plumiforme that induces chemical defense. In the present study, we investigated the biological functions of OPDA and its derivatives in C. plumiforme. Searching for the components potentially involving oxylipin signaling from transcriptomic and genomic data revealed that two COI1, three JAZ, and two MYC genes were present. Quantification analyses revealed that OPDA and its isomer iso-OPDA accumulated in larger amounts than dn-OPDA and dn-iso-OPDA after wounding. Moreover, exogenously applied OPDA, dn-OPDA, or dn-iso-OPDA induced the transcription of JAZ genes. These results imply that OPDA, dn-OPDA, and/or their isomers potentially act as biologically active molecules to induce the signaling downstream of COI1-JAZ. Furthermore, co-immunoprecipitation analysis showed the physical interaction between JAZs and MYCs, indicating the functional conservation of JAZs in C. plumiforme with other plants. These results suggest that COI1-JAZ-MYC mediated signaling is conserved and functional in C. plumiforme.
Introduction
The phytohormone jasmonic acid (JA; 1) and its biologically active form jasmonoyl-L-isoleucine (JA-Ile) regulate defense responses to various environmental stresses such as wounding, and plant growth and development (Wasternack and Hause, 2013; Wasternack, 2015; Guo et al., 2018; Ballaré and Austin, 2019). JA and JA-Ile are synthesized from fatty acids, α-linolenic acid or hexadecatrienoic acid, derived from membrane lipids. Oxygenation by lipoxygenase and dehydration-cyclization by allene oxide synthase and allene oxide cyclase (AOC) converts α-linolenic acid to 12-oxo-phytodienoic acid (OPDA; 2). The double bond hydrogenation by OPDA reductase (OPR3) and three sequential β-oxidations convert OPDA to JA (Wasternack and Hause, 2013). Finally, the GH3 family protein JAR1 conjugates JA to isoleucine to produce the biologically active form (+)-7-iso-JA-Ile (Staswick and Tiryaki, 2004; Fonseca et al., 2009).
JA-Ile is recognized by the F-box protein COI1 (Xie et al., 1998; Yan et al., 2009; Sheard et al., 2010). JAZ proteins repress transcription factors such as MYC, which activate the transcription of JA-responsive genes, when JA-Ile is at low levels. In the presence of high JA-Ile levels, COI1 physically interacts with JAZ repressors, leading to their degradation, and the transcription of JA-responsive genes by MYC and other transcription factors (Chini et al., 2007; Thines et al., 2007).
Although its biosynthesis is conserved in vascular plants (Wasternack and Hause, 2013; Wasternack, 2015; Pratiwi et al., 2017), JA-Ile is not a biologically active form recognized by COI1 in bryophytes. In the liverwort Marchantia polymorpha, 2,3-dinor-OPDA (dn-OPDA; 3) and its isomer 2,3-dinor-12-oxo-9(13),15(Z)-phytodienoic acid (dn-iso-OPDA; 4) accumulate after wounding, whereas JA-Ile is not found. dn-OPDA and dn-iso-OPDA are recognized by MpCOI1 to activate downstream signaling (Monte et al., 2018). The model moss Physcomitrium patens, formerly known as Physcomitrella patens, also produces dn-OPDA and dn-iso-OPDA (Stumpe et al., 2010; Mukhtarova et al., 2020). Additionally, 12-oxo-9(13),15(Z)-phytodienoic acid (iso-OPDA; 5) has been found in P. patens, but its biological activity has not been revealed (Mukhtarova et al., 2020). Comparative genomics and metabolomics suggest that dn-OPDA and dn-iso-OPDA might act as COI1 ligands in mosses (Monte et al., 2018; Mukhtarova et al., 2020).
The moss Calohypnum plumiforme like rice and barnyard grass produces specialized metabolites, momilactones, which have antimicrobial and allelochemical activities (Nozaki et al., 2007; Yamane, 2013; Miyamoto et al., 2014; Guo et al., 2017). These momilactone-producing plants possess the momilactone biosynthetic gene cluster, which is formed by the concentration of biosynthetic genes in a narrow region of the chromosome (Shimura et al., 2007; Guo et al., 2017; Mao et al., 2020). Therefore, the investigation of the regulatory mechanism(s) of momilactone production in C. plumiforme will contribute to a better understanding of the diversity and evolution of plant chemical defenses. Previous studies have revealed that JA and JA-Ile are not detected in C. plumiforme and that OPDA, but not JA, induces momilactone accumulation and the expression of these biosynthetic genes (Okada et al., 2016; Mao et al., 2020). These results suggest that OPDA or its derivative is a biologically active molecule in C. plumiforme that induces the production of momilactones.
In the present study, we focused on the function of OPDA and its derivatives and explored the potential involvement of protein components in oxylipin signaling in the moss C. plumiforme. We found two COI1, three JAZ, and two MYC genes from the transcriptome and genome data. The quantification of endogenous oxylipins revealed that OPDA and iso-OPDA accumulated in larger amounts than dn-OPDA and dn-iso-OPDA. We also found that exogenously applied OPDA, dn-OPDA, or dn-iso-OPDA induced the transcription of JAZ genes. Moreover, co-immunoprecipitation analysis revealed the physical interaction between JAZs and MYCs. These results imply that OPDA, dn-OPDA, and/or their isomers act as biologically active molecules to induce downstream signaling and that COI1-JAZ-MYC-mediated signaling is conserved in C. plumiforme.
Materials and Methods
Plant Materials and Growth Conditions
Calohypnum plumiforme kindly provided by Prof. Kenichiro Hayashi (Okayama University of Science) was the same line as that used in a previous report (Okada et al., 2016). Gametophores were cultured on BCDATG agar medium under continuous white light at 23°C (Nishiyama et al., 2000), transferred to a new BCDATG agar medium every 2 months, and used for chemical and wounding treatments 1 month after each transfer.
Chemicals
Racemic JA was purchased from Tokyo Chemical Industry Co. (Tokyo, Japan) and used as a standard for quantification analysis. (–)-JA was prepared as previously described (Miyamoto et al., 2019) and used for the treatment of C. plumiforme. [2H5]-JA was obtained from Nacalai Tesque, Inc. (Kyoto, Japan). (–)-JA-Ile and [13C6]-JA-Ile were synthesized as previously described (Jikumaru et al., 2004).
OPDA and [2H5]-OPDA were purchased from Olchemim (Olomouc, Czechia Republic) and used as standards for quantification analysis. For the treatment of C. plumiforme, OPDA and [2H5]-OPDA were synthesized from α-linolenic acid (FUJIFILM Wako Pure Chemical Co., Osaka, Japan) and [2H5]-α-linolenic acid (Cayman Chemical Company, MI, United States), respectively, as previously described with slight modifications (Zimmerman and Feng, 1978; Kajiwara et al., 2012). Briefly, the extracted protein fraction of flaxseed acetone powder supplemented with recombinant Oryza sativa AOC (OsAOC) was used for enzymatic conversion. Glutathione-S-transferase (GST)-fused OsAOC was expressed in Escherichia coli Rosetta2 (DE3) (Merck, Darmstadt, Germany) according to the manufacturer’s instructions (Riemann et al., 2013), precipitated with ammonium sulfate and suspended in 50 mM potassium phosphate (pH 7.0). α-Linolenic acid and [2H5]-α-linolenic acid were suspended in 200 mM borate-NaOH buffer (pH 9.0) containing 5% (v/v) Tween 20, mixed with the extract of flaxseed acetone powder supplemented with recombinant GST-OsAOC, and incubated at 25°C for 150 min. The reaction was stopped by adding 1 N HCl to adjust the pH to 3.0–4.0, and the mixture was flushed with N2 gas for 30 min. The products were extracted with ethyl acetate, dried using an evaporator, and dissolved in methanol.
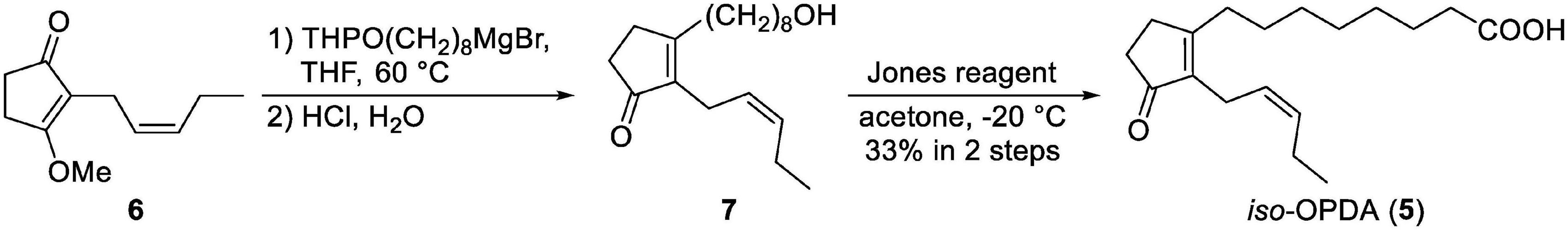
iso-OPDA was synthesized as shown in Scheme 1. All chemical reagents and solvents for chemical synthesis of iso-OPDA were obtained from Kanto Chemical Co., FUJIFILM Wako Pure Chemical Co., Nacalai Tesque Co., Tokyo Chemical Industry Co. (Tokyo, Japan), Sigma-Aldrich Co. (MO, United States), and GE Healthcare (IL, United States) and used without further purification. All anhydrous solvents were either dried by standard techniques and freshly distilled before use or purchased in anhydrous form and used as supplied.
1H and 13C NMR spectra were recorded on a JNM–ECS–400 spectrometer (JEOL, Tokyo, Japan) in deuterated chloroform using TMS as an internal standard. Fourier transform infrared (FT/IR) spectra were recorded on an FT/IR–4100 (JASCO, Tokyo, Japan). High–resolution (HR) electrospray ionization (ESI)–mass spectrometry (MS) analyses were conducted using a microTOF II (Bruker Daltonics Inc., MA, United States).
To a solution of THPO(CH2)8MgBr (0.53 M in THF, 5.3 mL, 2.81 mmol) was added a solution of 6 (Wang et al., 2021) (127 mg, 702 μmol) in THF (6.5 mL) at reflux temperature under argon atmosphere. After being stirred at 60°C for 2 h, the reaction mixture was allowed to cool to rt and 2 M HCl aq. (10 mL) was added. After 12 h of stirring, H2O was added and the water layer was extracted with EtOAc. The combined organic layers were washed with saturated aqueous NaCl, dried over Na2SO4 and concentrated under reduced pressure. After evaporation, the residue was purified by medium-pressure chromatography (Isolera, eluent: 40:60 n-hexane/EtOAc to EtOAc) to give the mixture contained 7 (73.2 mg). The mixture was carried on to the next step.
To a solution of the mixture (73.2 mg) in acetone (20 mL) was added Jones reagent (4.0 M solution, 2.5 mL, 10 mmol) at −20°C. After 1 h of stirring at −20°C, i-PrOH was added to quench the remaining reagent. Then, H2O (200 mL) was added and the water layer was extracted with EtOAc. The combined organic layers were washed with saturated aqueous NaCl, dried over Na2SO4 and concentrated under reduced pressure. The residue was purified by medium-pressure chromatography (Isolera, eluent: 0.5:50:50 AcOH/n-hexane/EtOAc to 0.5:99.5 AcOH/EtOAc) to give iso-OPDA (68.3 mg, 33% in two steps) as a colorless oil. 1H NMR (400 MHz, CDCl3); δH 5.37 (dtt, J = 10.6, 7.4, 1.6 Hz, 1H), 5.21 (dtt, J = 10.6, 7.0, 1.5 Hz, 1H), 2.93 (brd, J = 7.0 Hz, 2H), 2.52–2.45 (m, 2H), 2.42 (t, J = 7.4 Hz, 2 H), 2.39–2.32 (m, 4H), 2.15 (quintet, J = 7.4 Hz, 2H), 1.64 (brquintet, J = 7.5 Hz, 2H), 1.52 (brquintet, J = 7.5 Hz, 2H), 1.34 (brs. 6H), 0.99 (t, J = 7.4 Hz, 3H); 13C NMR (100 MHz, CDCl3); δC 209.49, 178.88, 174.35, 139.16, 132.19, 125.33, 34.18, 33.81, 31.27, 29.52, 29.16, 29.04, 28.89, 27.35, 24.59, 21.23, 20.60, 14.16; IR (film) cm–1: 3205, 2931, 2858, 1732, 1701; HRMS (ESI, positive) m/z [M + Na]+ Calcd. for C18H28O3: 2315.1936, found: 315.1927. The 1H and 13C NMR spectra of iso-OPDA are shown in Supplementary Figures 1, 2, respectively.
dn-OPDA and dn-iso-OPDA were prepared as previously described (Wang et al., 2021). [2H5]-dn-OPDA was purchased from Cayman Chemical Company.
Chemical and Wounding Treatments
12-Oxo-phytodienoic acid, dn-OPDA, dn-iso-OPDA, JA, and JA-Ile were dissolved in dimethyl sulfoxide at a concentration of 10 mM and added to BCDATG liquid media to a final concentration of 50 μM, whereas [2H5]-OPDA, dissolved in 20% (v/v) aqueous dimethyl sulfoxide solution at a concentration of 1 mM, was added to BCDATG liquid media to 25 μM. C. plumiforme gametophores were incubated with media containing the chemicals for 2 h under continuous white light at 23°C.
For wounding treatment, C. plumiforme gametophores were cut by scissors into 5-mm long pieces, incubated under continuous white light at 23°C, and collected after 0, 0.5, 1, 2, and 4 h.
5′- and 3′-Rapid Amplification of cDNA Ends
To determine the nucleotide sequence of full-length CpJAZ2 cDNA, total RNA was extracted from C. plumiforme gametophores 1 h after wounding using an RNeasy Plant Mini Kit (QIAGEN, Venlo, Netherlands). 5′- and 3′-RACE analyses of CpJAZ2 were performed using the SMARTer RACE cDNA Amplification Kit (Clontech, CA, United States) and KOD FX (TOYOBO Co., Osaka, Japan) according to the manufacturer’s protocol. Amplified RACE products were cloned into pRACE using the In-Fusion HD Cloning Kit (Clontech) and sequenced to determine the full-length cDNA of CpJAZ2, which was amplified by end-to-end RT-PCR using KOD FX and a cDNA template prepared for 5′-RACE analysis. The full-length cDNA was cloned into the pCR-II-Blunt-TOPO vector (Thermo Fisher Scientific, MA, United States) to generate pCR-CpJAZ2, and the inserted fragment was sequenced. Sequences of primers used for RACE and cloning are provided in Supplementary Table 1.
Cloning of COI1, JAZ, and MYC2 Genes
According to information from the genome database, coding sequences of COI1, JAZ, and MYC2 genes of C. plumiforme were amplified by RT-PCR using KOD Plus Neo (TOYOBO Co.) and a cDNA template that was prepared for the 5′-RACE analysis. pCR-CpJAZ2 was used as a template for the amplification of CpJAZ2. Sequences of primers used for cloning are provided in Supplementary Table 1. Amplified fragments were cloned into pEU vectors (CellFree Sciences Co., Ehime, Japan), which are the expression vectors for a wheat germ cell-free protein expression system, using the In-Fusion HD Cloning Kit (Clontech) or SLiCE (Motohashi, 2015). COI1 and MYC2 genes were cloned into pEU-E01-GST-PS-MCS-N1, and JAZ genes were cloned into pEU-E01-DYKDDDDK-MCS-N1. The inserted fragments were sequenced.
Sequence Identification and Phylogenetic Analyses
Transcriptome data of C. plumiforme treated with CuCl2, which induces oxidative stress and defense responses in plants, were available based on our previous RNA-seq analyses, the results of which are deposited in the DDBJ Sequence Read Archive under accession no. DRA010138 (Okada et al., 2016). The C. plumiforme genome sequence and annotation for estimated coding proteins are deposited in the Genome Sequence Archive under accession no. PRJCA001833 (Mao et al., 2020). C. plumiforme genes homologous to COI1, JAZ, and MYC were obtained using the corresponding genes in M. polymorpha as a query for a BLAST search using transcriptome data and the genome sequence.
Phylogenetic and molecular evolutionary analyses were conducted using MEGA version X (Kumar et al., 2018) with the neighbor-joining method (Saitou and Nei, 1987). Sequence alignments were constructed in MEGA using Muscle (Edgar, 2004). Neighbor-joining trees were also constructed. Bootstrap values were evaluated with 1,000 replications. Sequence alignments were performed using the BioEdit software version 7.2.5.
Statistical Analysis
One-way ANOVA with Tukey’s post hoc test was performed using Excel (Microsoft Co., WA, United States) and Statcel 4 (OMS Publishing Inc., Saitama, Japan) software.
Quantification of OPDAs, JA, and JA-Ile
Wounded C. plumiforme gametophores (approximately 50 mg fresh weight) were homogenized and suspended in 2 mL of 80% (v/v) aqueous methanol, and [2H5]-OPDA (2.5 ng), [2H5]-JA (10 ng), [13C6]-JA-Ile (10 ng), and [2H5]-dn-OPDA (10 ng) were added as internal standards. The extracts were purified and concentrated as previously described (Miyamoto et al., 2016). [2H5]-OPDA-fed gametophores (approximately 7 mg fresh weight) were homogenized and purified using the same method without internal standards. The resultant samples were subjected to liquid chromatography with electrospray ionization tandem mass spectrometry (LC-ESI-MS/MS) composed of a quadrupole tandem mass spectrometer (Agilent 6460 Triple Quadrupole mass spectrometer) with an electrospray ion source and an Agilent 1200 separation module. OPDAs, JA, and JA-Ile were quantified as previously described, with slight modifications (Tamiru et al., 2016). The mobile phase consisted of 0.05% acetic acid in water (solvent A) and 0.05% acetic acid in acetonitrile (solvent B). Elution was conducted using a linear gradient of solvent B from 3 to 70% over 15 min and 70 to 98% over 5 min at a flow rate of 0.2 mL/min. The retention times of OPDA, iso-OPDA, dn-OPDA, dn-iso-OPDA, JA, and JA-Ile were 17.1, 16.8, 15.9, 15.5, 13.4, and 15.0 min, respectively. The precursor-to-product transitions monitored were m/z 291/165 for OPDA and iso-OPDA, m/z 296/170 for [2H5]-OPDA and [2H5]-iso-OPDA, m/z 263/165 for dn-OPDA and dn-iso-OPDA, m/z 268/170 for [2H5]-dn-OPDA and [2H5]-dn-iso-OPDA, m/z 209/59 for JA; m/z 214/62 for [2H5]-JA, m/z 322/130 for JA-Ile, and m/z 328/136 for [13C6]-JA-Ile. The concentrations of OPDAs and JA in the wounded samples were calculated based on the relative proportions of the peak area relative to the internal standards, whereas those of OPDAs in the [2H5]-OPDA-fed samples were determined based on the concentration of the authentic standard run under the same analysis conditions.
Gene Expression Analysis
Total RNA was extracted from C. plumiforme gametophores using an RNeasy Plant Mini Kit (QIAGEN) and subjected to cDNA synthesis using a PrimeScript RT Reagent Kit with gDNA Eraser (Takara Bio, Shiga, Japan). Quantitative RT-PCR (qRT-PCR) was performed using a THUNDERBIRD SYBR qPCR Mix (TOYOBO Co.) on an ABI 7500 Fast Real-Time PCR System (Applied Biosystems, CA, United States). The ddCt method was used to calculate transcript levels. CpACT3 (DDBJ ID: LC129863) was used as an internal control to normalize the amount of mRNA. For each sample, the mean value from biological triplicate samples was used to calculate the transcript abundance. Sequences of primers used for qRT-PCR analysis are provided in Supplementary Table 1.
In vitro Cell-Free Protein Expression
GST- or FLAG tag-fused CpMYC2 and CpJAZ proteins were expressed using a WEPRO7240 Expression Kit (CellFree Sciences Co.), which is a wheat germ cell-free protein expression system, according to the manufacturer’s protocol. Expressed samples were centrifuged at 17,800 × g for 5 min at 4°C, and the supernatants were used for subsequent analyses. FLAG-CpJAZ2 was diluted five times before use because its expression was higher than that of FLAG-CpJAZ1 and FLAG-CpJAZ3. A portion of the supernatant was mixed with Laemmli SDS sample buffer and boiled at 96°C for 10 min; the boiled proteins were subjected to protein gel blot analysis. The remaining supernatants were subjected to co-immunoprecipitation analysis.
Co-immunoprecipitation Analysis
For each sample, 10 μL of the supernatant was used. GST-CpMYC2a, GST-CpMYC2b, and GST were mixed with FLAG-CpJAZ1, FLAG-CpJAZ2, or FLAG-CpJAZ3 in IP buffer [25 mM Tris pH 7.8, 50 mM NaCl, 5% (v/v) glycerol, 20 mM 2-mercaptoethanol, and 0.05% (v/v) Tween 20] in the presence of a cOmplete EDTA-free protease inhibitor cocktail (Merck). After incubation at 4°C for 60 min, 0.2 μg of anti-FLAG M2 monoclonal antibody (Merck) was added to each sample. The samples were further incubated at 4°C for 60 min with rotation, and SureBeads Protein G Magnetic Beads (Bio-Rad, CA, United States) were added. Finally, after incubation at 4°C for 60 min with rotation, the beads were washed three times with PBS containing 0.1% (v/v) Tween 20. The resultant beads were boiled in double-diluted sample buffer solution (2ME +) (× 2) (Fujifilm Co.) containing 100 mM dithiothreitol at 60°C for 10 min. The boiled proteins were subjected to protein gel blot analysis.
Protein Gel Blot Analysis
The boiled samples were subjected to SDS-PAGE on 10% (w/v) polyacrylamide gels and transferred to a nitrocellulose membrane (Bio-Rad). GST-CpMYC2a, GST-CpMYC2b, and GST were detected using an anti-GST HRP conjugate [dilution 1:5,000 (v/v)] (Cytiva, Tokyo, Japan) and iBind Flex Western Device (Thermo Fisher Scientific), whereas FLAG-CpJAZ1, FLAG-CpJAZ2, and FLAG-CpJAZ3 were detected using anti-FLAG M2 monoclonal antibody (Merck) [dilution 1:1,000 (v/v)] as the primary antibody and anti-mouse IgG, HRP-linked whole antibody sheep [dilution 1:10,000 (v/v)] (Cytiva) as the secondary antibody with an iBind Flex Western Device. Chemiluminescent detection was carried out using Immobilon Western Chemiluminescent HRP Substrate (Merck) and a ChemiDoc imaging system (Bio-Rad).
Results
Genomic Mining of COI1, JAZ, and MYC2 Gene Homologs in Calohypnum plumiforme
In the process of elucidating the molecular mechanism that induces momilactone production in C. plumiforme, we previously reported that OPDA induces momilactone accumulation, suggesting that OPDA or its derivatives act as signaling molecules (Okada et al., 2016). The OPDA/dn-OPDA signaling pathway has been identified in the bryophyte M. polymorpha. Therefore, we first searched for genes homologous to COI1, JAZ, and MYC using the corresponding genes in M. polymorpha as a query using transcriptome data and the genome sequence of C. plumiforme.
COI1 in Calohypnum plumiforme
We found two genes similar to MpCOI1 (Monte et al., 2018) from transcriptome data, and these genes were also found in the C. plumiforme genome database. These genes were designated as CpCOI1a and CpCOI1b (Table 1). We cloned the coding sequences of CpCOI1a and CpCOI1b based on the information from the genome annotation, which were identical to annotated sequences in the genome database and deposited in GenBank under accession no. MW775560 (CpCOI1a) and MW775561 (CpCOI1b).
Phylogenetic analysis was conducted using the amino acid sequences of COI1 in C. plumiforme, M. polymorpha, and P. patens with Arabidopsis thaliana COI1 as an outgroup. CpCOI1a and three sequences from P. patens, were grouped in the same clade as MpCOI1 (Figure 1A), suggesting that these COI1s are conserved in both mosses and liverworts. However, CpCOI1b and COI1 of P. patens formed another clade (Figure 1A), suggesting that these two COI1s are moss-specific. The alignment of these sequences revealed that the residues involved in the recognition of ligands or the interaction with JAZ proteins reported by Sheard et al. (2010) were conserved in COI1 of C. plumiforme (Supplementary Figure 3). Monte et al. (2018) reported that valine at position 377 in MpCOI1 is essential for the recognition of dn-OPDA and dn-iso-OPDA and that this residue is substituted with alanine on COI1s recognizing a JA-Ile such as AtCOI1. COIs in the mosses predominantly show isoleucine in this position, which is a hydrophobic residue similar to valine (Monte et al., 2018). Consistent with this, the isoleucine residue is conserved in CpCOI1a, although the corresponding residue in CpCOI1b is valine (Figure 1B).
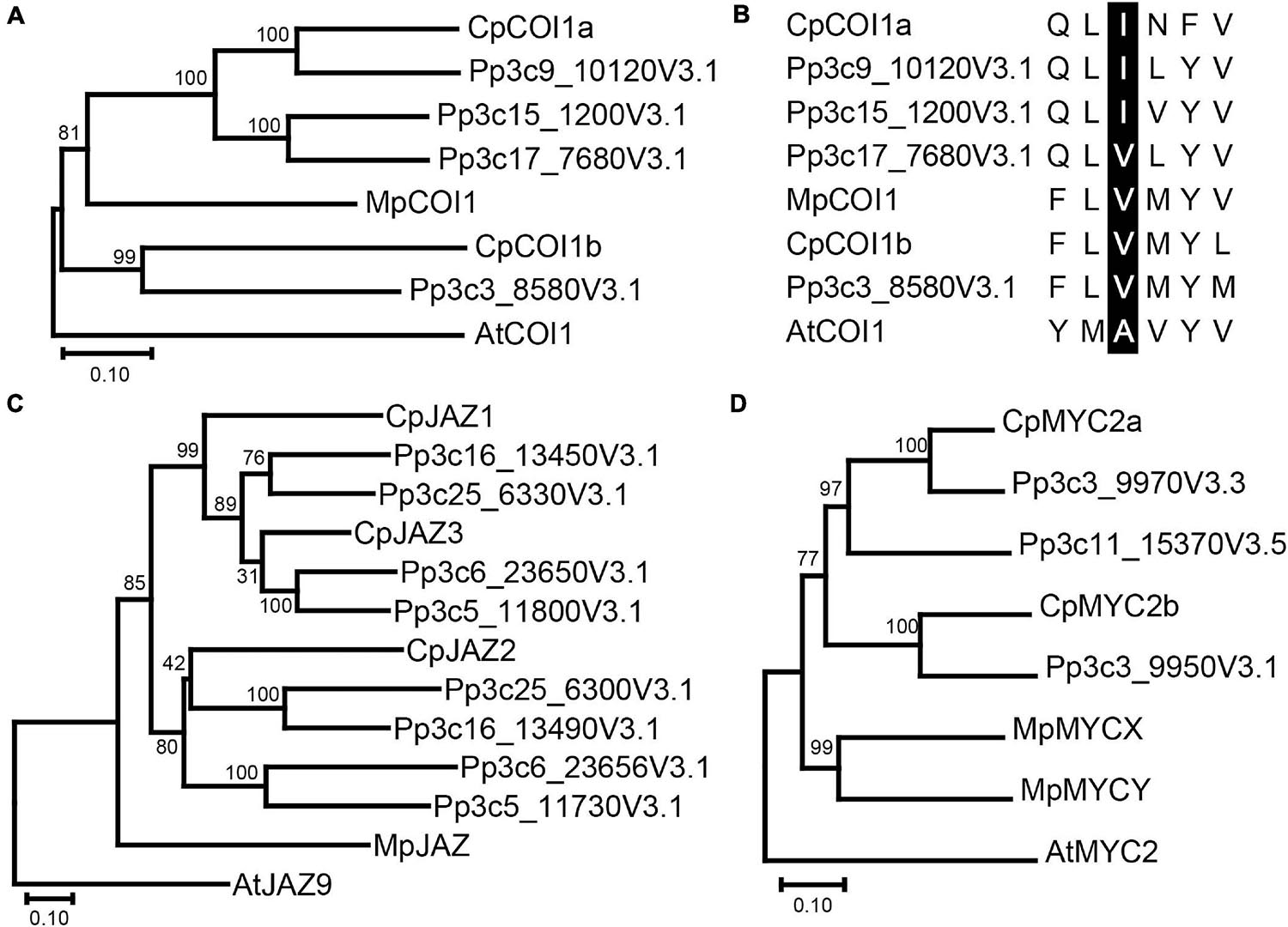
Figure 1. Phylogenetic analysis of COI1 (A), JAZ (C), MYC2 (D) homologs, and the residue essential for dn-OPDA and dn-iso-OPDA recognition (B). (A) Phylogenetic trees (amino acid sequences) were obtained using the neighbor-joining method as described in the “Materials and Methods.” The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches. COI1 homologs in M. polymorpha and P. patens are also included. The phylogenetic tree was rooted in AtCOI1. Gene IDs are as follows: MpCOI1: Mapoly0025s0025 and AtCOI1: AT2G39940. (B) The amino acid sequence of the valine residue at position 377 (V377) of MpCOI1 and its surroundings is shown with the corresponding sequences of other COI1 homologs. V377 in MpCOI1 and its corresponding residues in other COI1 homologs are highlighted by black. (C) Phylogenetic trees (amino acid sequences) were obtained using the neighbor-joining method as described in the “Materials and Methods.” The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches. JAZ homologs in M. polymorpha and P. patens are also included. The phylogenetic tree was rooted in AtJAZ9. Gene IDs are as follows: MpJAZ: Mapoly0097s0021 and AtJAZ9: AT1G70700. (D) Phylogenetic trees (amino acid sequences) were obtained using the neighbor-joining method as described in the “Materials and Methods.” The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) is shown next to the branches. MYC2 homologs in M. polymorpha and P. patens are also included. The phylogenetic tree was rooted in AtMYC2. Gene IDs are as follows: MpMYCX: Mapoly0018s0019; MpMYCY: MapolyY_B0018; and AtMYC2: AT1G32640.
JAZ in Calohypnum plumiforme
We found three genes similar to MpJAZ (Monte et al., 2019) from transcriptome data designated as CpJAZ1, CpJAZ2, and CpJAZ3 (Table 1). CpJAZ1 and CpJAZ3 were also annotated in the C. plumiforme genome database. We cloned the coding sequences of CpJAZ1 and CpJAZ3 according to the information from the genome database, which were identical to annotated sequences in the genome database and deposited in GenBank under accession no. MW775562 (CpJAZ1) and MW775564 (CpJAZ3). CpJAZ2 was not annotated in the genome database, and only a partial sequence of CpJAZ2 was obtained from the RNA-seq data. Therefore, we performed 5′- and 3′- RACE to obtain the full-length sequence of CpJAZ2, which was deposited in GenBank under accession no. MW775563. Phylogenetic analysis was conducted using the amino acid sequences of JAZ in C. plumiforme, M. polymorpha, and P. patens, with A. thaliana JAZ9 as an outgroup. CpJAZ1 and CpJAZ3, along with four sequences from P. patens, were grouped in the same clade, whereas CpJAZ2 and four sequences of P. patens formed another clade (Figure 1C), suggesting that JAZ proteins evolved into two groups in mosses. The alignment of these sequences revealed that the Jas motif and TIFY domain were conserved in CpJAZ1, CpJAZ2, and CpJAZ3 (Figure 2). N-terminal sequences were conserved among several JAZs in C. plumiforme and P. patens, including CpJAZ1 and CpJAZ3. This conserved sequence was not found in MpJAZ or AtJAZ9 (Figure 2).
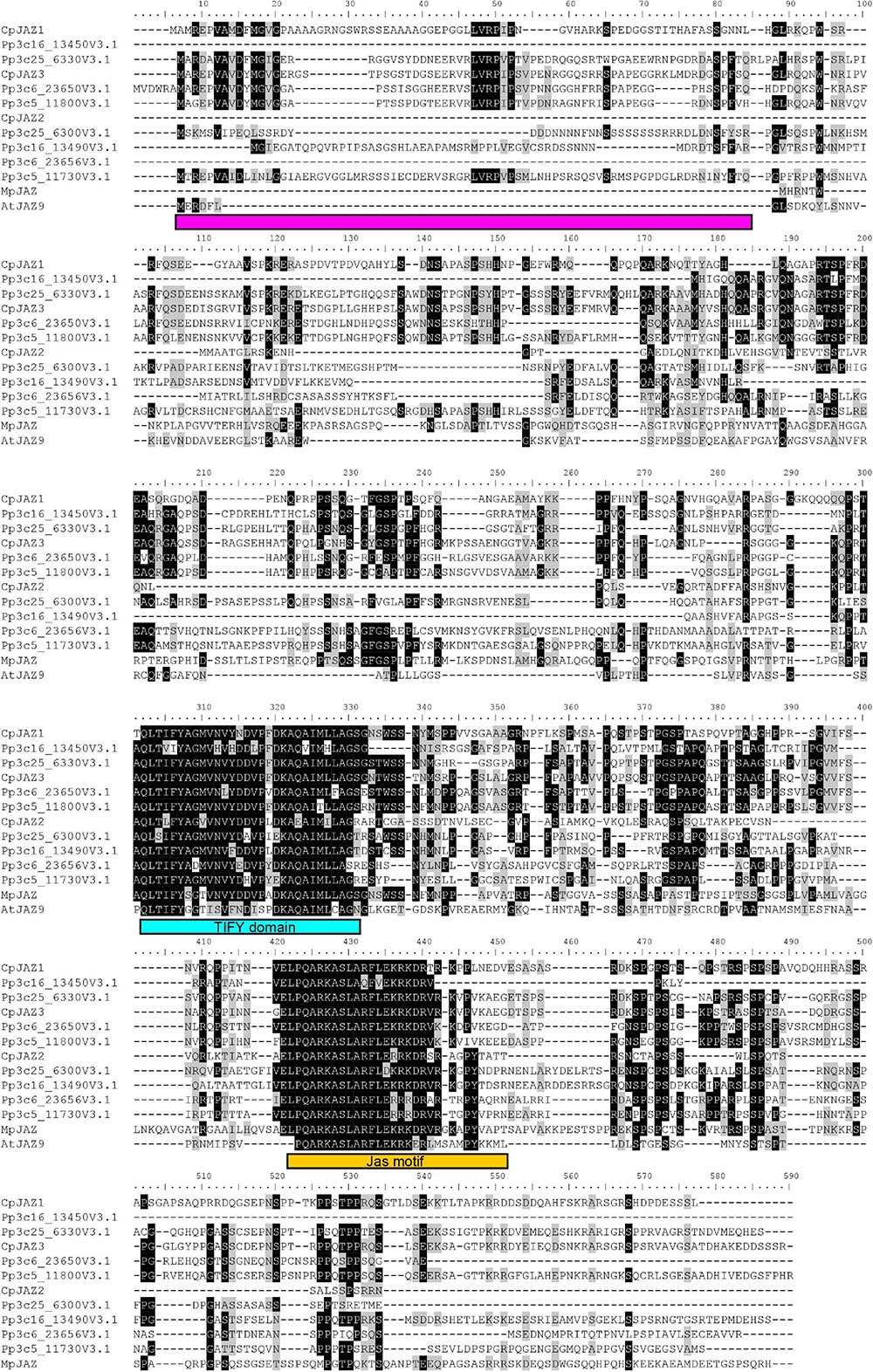
Figure 2. Sequence alignment of JAZ proteins. Sequences of JAZ homologs were aligned using MEGA version X using Muscle. Sequence alignments were drawn using the BioEdit software version 7.2.5. The shading thresholds for identical or similar residues were 40%. Identical and similar residues are highlighted by black and gray, respectively. The TIFY domain and Jas motifs are indicated by blue and orange squares, respectively. The conserved sequence in the N-terminus of the moss JAZs is indicated by magenta squares.
MYC2 in Calohypnum plumiforme
We found two genes similar to the M. polymorpha MYC2 ortholog gene, MpMYCY (Peñuelas et al., 2019), from transcriptome data, and these genes were also found in the C. plumiforme genome database. These two genes were designated as CpMYC2a, and CpMYC2b (Table 1). We cloned the coding sequences of CpMYC2a and CpMYC2b according to the information from the genome database; that of CpMYC2a had a 12 bp substitution with the annotation of the genome database, possibly due to misassembly of the genome sequence or polymorphism among the C. plumiforme lines used in genome sequencing and our experiments (Supplementary Figure 4A), whereas that of CpMYC2b lacked a 99 bp sequence that was annotated as exons in the genome database. This sequence had a putative 3′-splice site (5′-AG-3′) and branchpoint sequence (5′-CTNAN-3′; Brown et al., 1996), suggesting that the longer region was removed in the C. plumiforme gametophores used in the present study (Supplementary Figure 4B). The cloned coding sequences were deposited in GenBank under accession no. MW775565 (CpMYC2a) and MW775566 (CpMYC2b) and used for subsequent analyses.
Phylogenetic analysis was conducted using the amino acid sequences of MYC2 in C. plumiforme, M. polymorpha, and P. patens with A. thaliana MYC2 as an outgroup. CpMYC2a and two sequences from P. patens were grouped in the same clade, whereas CpMYC2b and one sequence from P. patens formed another clade (Figure 1D), suggesting that MYC2 proteins evolved into two groups in mosses. The alignment of these sequences revealed that the JID and bHLH domains were conserved in CpMYC2a and CpMYC2b (Supplementary Figure 5).
Quantification of Endogenous Levels of OPDA, dn-OPDA, and Their Isomers
We found putative OPDA/dn-OPDA signaling components in C. plumiforme; however, the endogenous levels of OPDA, dn-OPDA, and their isomers, which accumulate in gametophores after wounding, have not yet been investigated. Therefore, we measured these. The levels of OPDA and dn-OPDA increased and peaked at 30 min after wounding and then decreased (Figure 3). iso-OPDA and dn-iso-OPDA also accumulated after 30 min but remained at a high level for 4 h after wounding. After wounding, the level of OPDA was highest, followed by that of iso-OPDA, which was approximately one-tenth of that of OPDA. The amounts of dn-OPDA and dn-iso-OPDA were less than one-hundredth that of OPDA. We detected a small amount of JA, although this was below the detection limit of a previous study (Okada et al., 2016). Additionally, the level of JA 4 h after wounding was significantly higher than that in unwounded C. plumiforme gametophores (Figure 3). The amount of JA was about 1/1000 of that of OPDA, and it was almost consistent with a steady-state level of JA in angiosperms, such as A. thaliana and rice (Park et al., 2002; Riemann et al., 2013). JA-Ile, which is a biologically active form in seed plants, was not detected with or without wounding.
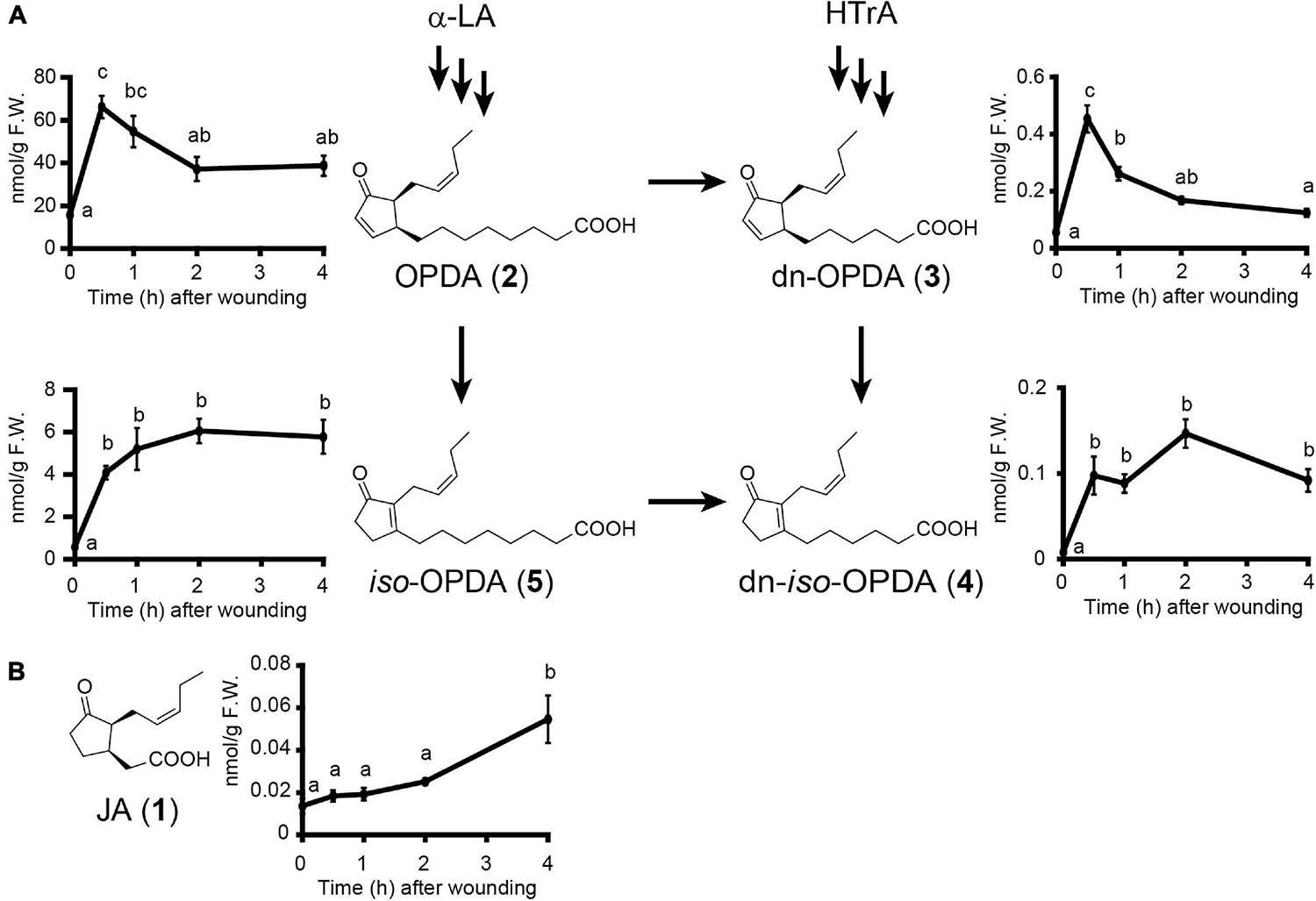
Figure 3. Effects of wounding on accumulation levels of endogenous OPDA, iso-OPDA, dn-OPDA, dn-iso-OPDA (A), and JA (B). (A) Values indicate the amount of OPDA, iso-OPDA, dn-OPDA, and dn-iso-OPDA in C. plumiforme gametophores after wounding (n = 3). (B) Values indicate the amount of JA in C. plumiforme gametophores after wounding (n = 3). Arrows indicate the proposed biosynthetic pathways in M. polymorpha. Bars indicate standard errors of the means. Statistically different data groups are indicated with different letters (p < 0.05, one-way ANOVA with Tukey’s post hoc test). LA, linolenic acid; HTrA, hexadecatrienoic acid.
In planta Conversion of Labeled-OPDA to iso-OPDA, dn-OPDA, and dn-iso-OPDA
Two synthetic pathways for dn-OPDA and dn-iso-OPDA are shown in M. polymorpha. One is the conversion of OPDA to dn-OPDA and dn-iso-OPDA; the other is the direct synthesis of dn-OPDA and dn-iso-OPDA from hexadecatrienoic acid, but not via OPDA (Monte et al., 2018). To investigate the conversion of OPDA to dn-OPDA and dn-iso-OPDA, we fed 25 μM deuterium-labeled OPDA [(2H5)-OPDA] to C. plumiforme gametophores and then measured the level of deuterium-labeled derivatives by LC-MS/MS. Both [2H5]-dn-OPDA and [2H5]-dn-iso-OPDA accumulated after [2H5]-OPDA treatment, indicating that OPDA can be efficiently converted to dn-OPDA in C. plumiforme (Figure 4). Isomerization of OPDA to iso-OPDA was also observed because [2H5]-iso-OPDA also accumulated after [2H5]-OPDA treatment (Figure 4). The ratio of the amounts of [2H5]-OPDA, [2H5]-iso-OPDA, [2H5]-dn-OPDA, and [2H5]-dn-iso-OPDA in [2H5]-OPDA-treated C. plumiforme was similar to that of the unlabeled compounds in wounded plants.
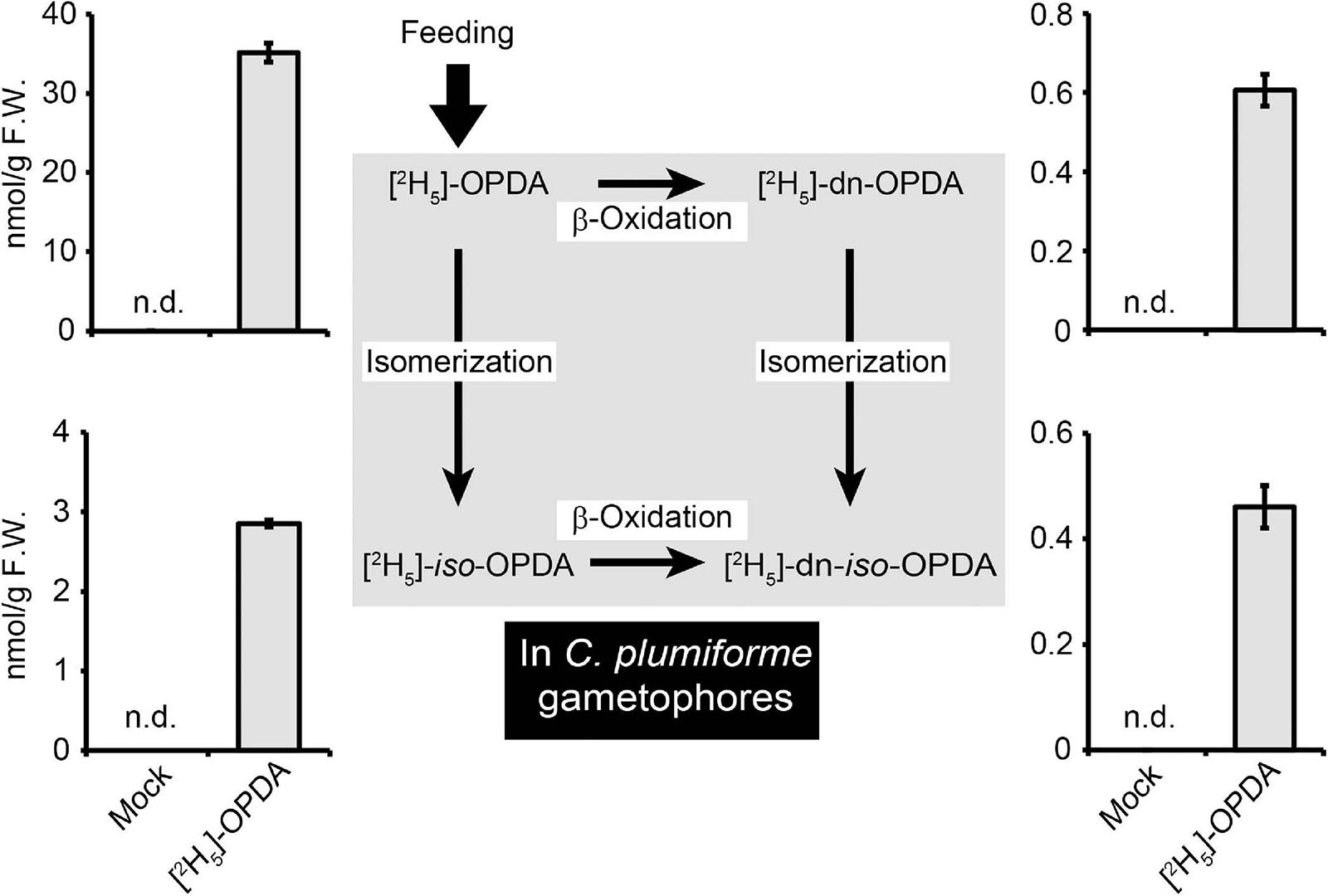
Figure 4. Conversion of deuterium-labeled OPDA [(2H5)-OPDA] in C. plumiforme gametophores. The values indicate the amount of [2H5]-OPDA, iso-OPDA, dn-OPDA, and dn-iso-OPDA in C. plumiforme gametophores after [2H5]-OPDA treatment at 25 μM (n = 3). Mock samples were treated with BCDATG liquid media containing 0.5% (v/v) dimethyl sulfoxide as a negative control. The arrows indicate the proposed biosynthetic pathways in M. polymorpha.
OPDA, dn-OPDA, and dn-iso-OPDA Induce the Expression of CpJAZ Genes
The expression of JAZ genes encoding repressors is induced by the ligand for COI1 to repress the signal (Chini et al., 2007; Thines et al., 2007; Monte et al., 2019). We examined the expression of JAZ genes after OPDA, dn-OPDA, or dn-iso-OPDA treatment in C. plumiforme. As shown in Figure 5, the expression of CpJAZ1, CpJAZ2, and CpJAZ3 was induced by treatment with OPDA, dn-OPDA, or dn-iso-OPDA. JA and JA-Ile did not affect the expression of these genes. These results suggest that OPDA and/or both isomers of dn-OPDA are biologically active. dn-OPDA and dn-iso-OPDA might be recognized by CpCOI1 as in M. polymorpha, although there remains the possibility that another metabolite from OPDA and dn-OPDA acts as a ligand of CpCOI1.
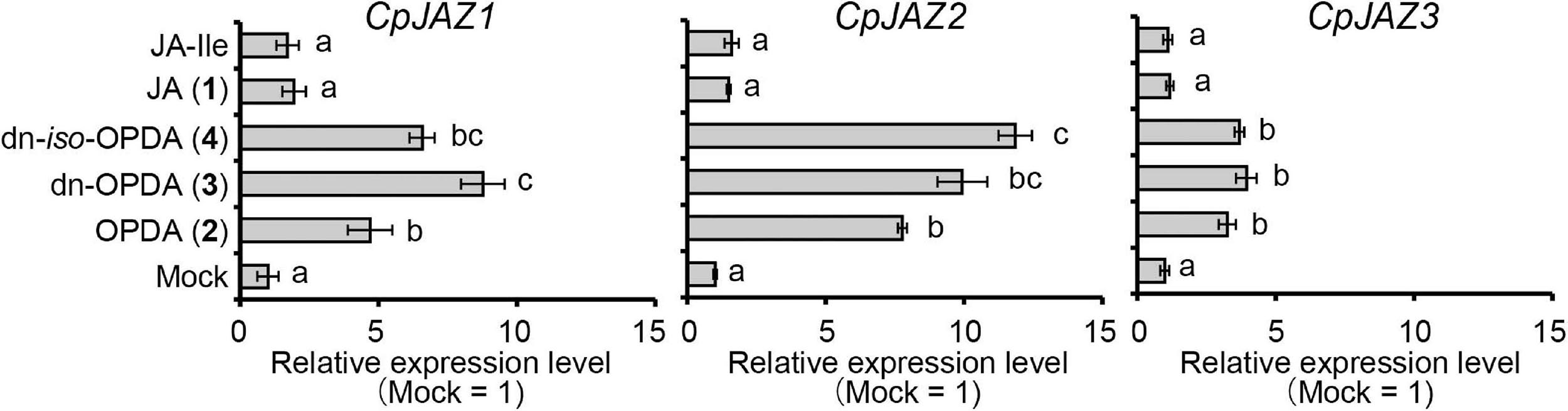
Figure 5. Effect of wounding on the expression of CpJAZ1, CpJAZ2, and CpJAZ3 as determined by qRT-PCR. Values indicate relative expression levels after treatment with OPDA, dn-OPDA, dn-iso-OPDA, JA, and JA-Ile at 50 μM (n = 3). Mock samples were treated with BCDATG liquid media containing 0.5% (v/v) dimethyl sulfoxide as a negative control. Expression levels were normalized to the expression of CpACT3, and the bars indicate the standard errors of the means. Statistically different data groups are indicated with different letters (p < 0.05, one-way ANOVA with Tukey’s post hoc test).
Physical Interaction Between CpJAZs and CpMYC2s
JAZ proteins physically interact with MYC2, MYC3 and MYC4 transcription factors to repress downstream signaling (Chini et al., 2007; Thines et al., 2007; Fernández-Calvo et al., 2011; Peñuelas et al., 2019). To investigate the physical interaction between JAZ and MYC2 proteins from C. plumiforme, we performed co-immunoprecipitation analyses. GST- or FLAG tag-fused CpMYC2 and CpJAZ proteins were expressed using a wheat germ cell-free protein expression system and detected by protein gel blot analysis at a position consistent with the predicted molecular weight; GST: 29.3 kDa, GST-CpMYC2a: 105.7 kDa, GST-CpMYC2b: 111.1 kDa, FLAG-CpJAZ1: 53.6 kDa, FLAG-CpJAZ2: 25.5 kDa, and FLAG-CpJAZ3: 54.8 kDa (Supplementary Figure 6). Expressed MYC2 and JAZ proteins were mixed and then immunoprecipitated using an anti-FLAG antibody. Immunoprecipitated proteins were analyzed by protein gel blot analysis using anti-GST or anti-FLAG antibodies. GST-CpMYC2a and CpMYC2b, but not GST, were detected after immunoprecipitation with CpJAZ1, CpJAZ2, or CpJAZ3 (Figure 6A). Additionally, GST-CpMYC2a and CpMYC2b in immunoprecipitated proteins with CpJAZs were more strongly detected than those without FLAG-tagged proteins. Immunoprecipitated FLAG-CpJAZs were detected using an anti-FLAG antibody (Figure 6B). These results indicate that CpJAZ1, CpJAZ2, and CpJAZ3 directly interact with CpMYC2a or CpMYC2b in vitro.
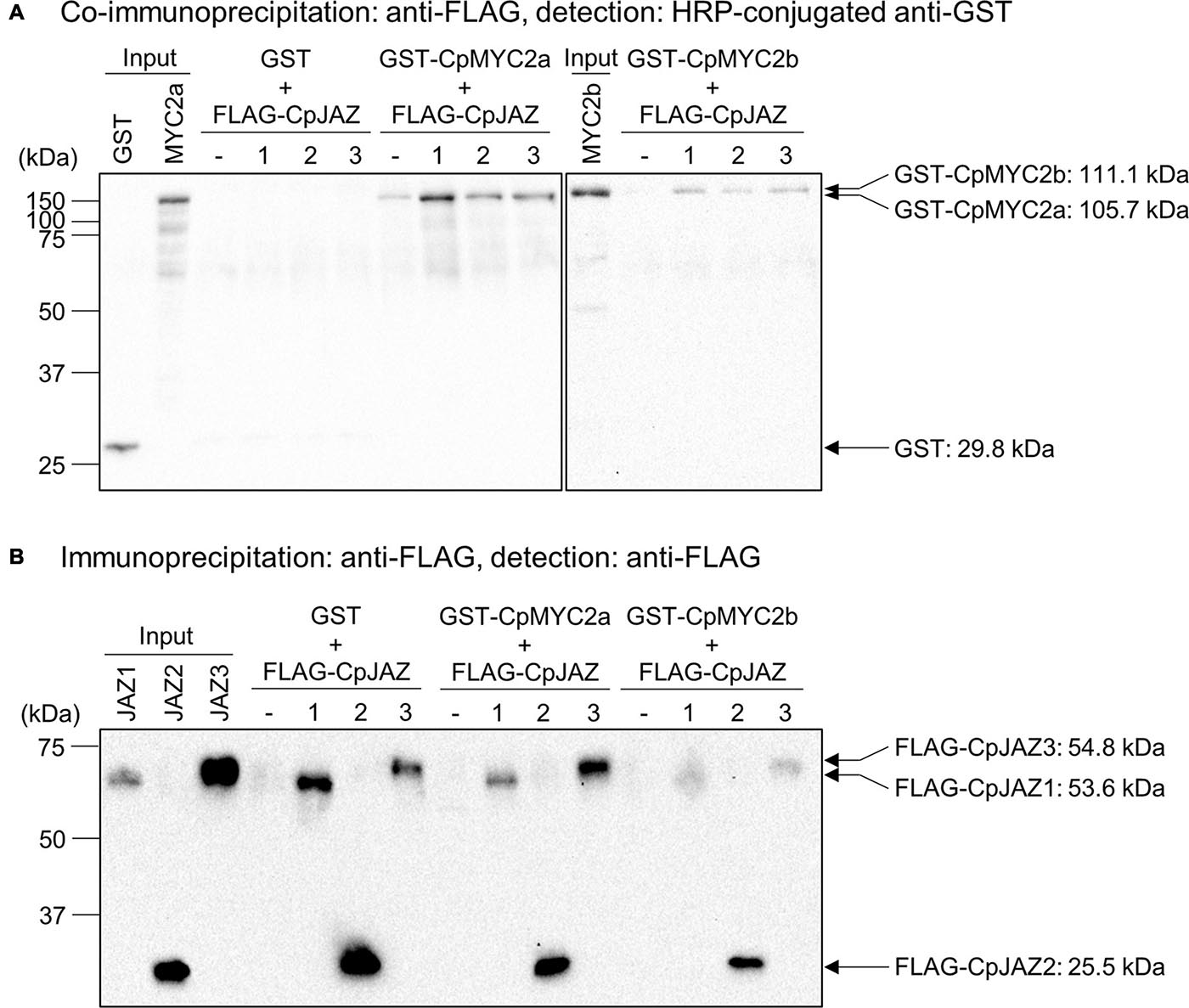
Figure 6. Co-immunoprecipitation analysis investigating the interaction between CpJAZs and CpMYC2s. (A) Proteins co-immunoprecipitated by anti-FLAG antibodies were analyzed. GST and GST-fused proteins were detected by protein gel blot analysis using a horseradish peroxidase-conjugated anti-GST antibody. (B) Proteins immunoprecipitated by anti-FLAG antibody were analyzed. FLAG-tagged proteins were detected by protein gel blot analysis using an anti-FLAG antibody.
Discussion
COI1, JAZ, and MYC2 Genes Were Conserved in Calohypnum plumiforme
In this study, we investigated signaling mechanisms promoted by OPDA and its derivatives in C. plumiforme. We found that COI, JAZ, and MYC2 sequences, including domains important for their functions, were conserved (Figures 1, 2 and Supplementary Figures 3, 5). Moreover, in CpCOI1a and CpCOI1b, a hydrophobic residue that is important for the recognition of dn-OPDA and dn-iso-OPDA was conserved (Figure 1B), suggesting that CpCOI1a and CpCOI1b recognize dn-OPDA and its related derivatives.
CpJAZ1, CpJAZ2, and CpJAZ3 conserved the Jas motif and TIFY domain as MpJAZ and AtJAZ9, implying that JAZs of C. plumiforme have similar functions to repress the downstream signaling of COI1 and MYCs. Several JAZs from mosses had conserved N-terminal sequences, which were not found in MpJAZ and AtJAZ9 (Figure 2). These sequences might be involved in the specific function of JAZs in mosses, although we could not find a similar sequence with identifiable functions by the BLAST search. Although the liverwort M. polymorpha has a single JAZ gene, mosses C. plumiforme and P. patens have at least three and eight JAZ genes, respectively. The number of JAZs in seed plants is even higher; for example, there are 13 and 15 JAZs in Arabidopsis and rice, respectively (Pauwels and Goossens, 2011; Chini et al., 2016; Uji et al., 2016). Genome duplication has been reported in mosses (Rensing et al., 2007; Gao et al., 2021), which may explain why there are more JAZs in mosses than in liverworts.
As C. plumiforme possessed fewer COI1, JAZ, and MYC2 genes than P. patens (Figure 1), less redundancy is expected. Unfortunately, we could not use reverse genetics approaches because of the absence of a transformation system in C. plumiforme. Because of the small number of COI1, JAZ, and MYC2 homologs and their importance in the research on the evolution of plant chemical defenses, the development of a transformation system for C. plumiforme is needed.
OPDA, dn-OPDA, and Their Isomers Accumulated After Wounding Treatment in Calohypnum plumiforme
To determine a biologically active molecule in OPDA signaling in C. plumiforme, we performed quantification analyses of OPDA, dn-OPDA, and their isomers. We observed that OPDA, iso-OPDA, dn-OPDA, and dn-iso-OPDA accumulated after wounding (Figure 3). A small amount of JA was detected and increased by wounding, although JA-Ile was not detected (Figure 3). Bryophytes do not have OPR3 orthologs, which catalyze the reduction of OPDA, but homologous genes to AtOPR2, which catalyzes 4,5-didehydro-JA reduction to produce JA (Han, 2016; Chini et al., 2018; Wasternack and Hause, 2018). Consistent with these reports, genes homologous to AtOPR2 were found in the C. plumiforme genome. These translation products possibly catalyze the reduction of OPDA homologs, such as dn-OPDA. However, the contribution of OPR should be small in C. plumiforme, since the level of JA is very low.
We conducted a feeding experiment using [2H5]-OPDA to investigate whether iso-OPDA, dn-OPDA, and dn-iso-OPDA were synthesized from OPDA. [2H5]-dn-OPDA was detected after the feeding of [2H5]-OPDA (Figure 4), indicating that OPDA was converted to dn-OPDA by β-oxidation in C. plumiforme as reported in M. polymorpha (Monte et al., 2018). The detection of [2H5]-iso-OPDA in [2H5]-OPDA-fed plants revealed that the isomerization of OPDA occurred (Figure 4). [2H5]-dn-iso-OPDA was observed after feeding [2H5]-OPDA (Figure 4). dn-iso-OPDA may be synthesized from OPDA by the β-oxidation of iso-OPDA and/or isomerization of dn-OPDA as proposed in M. polymorpha and P. patens (Monte et al., 2018; Mukhtarova et al., 2020). In M. polymorpha, dn-OPDA is synthesized from hexadecatrienoic acid, not via OPDA, in addition to the pathway from OPDA. It is unknown whether a biosynthetic pathway from hexadecatrienoic acid exists in C. plumiforme. The ratio of [2H5]-OPDA, [2H5]-iso-OPDA, [2H5]-dn-OPDA, and [2H5]-dn-iso-OPDA in [2H5]-OPDA-treated plants was approximately 60:5:1:1, which is similar to that of the corresponding endogenous compounds after wounding. Therefore, the pathway for the synthesis of dn-OPDA from OPDA may also be functional in C. plumiforme after wounding. However, the metabolic flow of the β-oxidation of OPDA and/or iso-OPDA seems to be moderate compared with that in M. polymorpha.
OPDA, dn-OPDA, and dn-iso-OPDA Induce CpJAZ Gene Expression
We investigated the inductive activity of OPDA, dn-OPDA, and dn-iso-OPDA on the expression of JAZ genes. Although they induced the expression of CpJAZ1, CpJAZ2, and CpJAZ3, JA and JA-Ile did not (Figure 5). We attempted to reveal the ligand for CpCOI1a and CpCOI1b by analyzing the physical interaction between COI1-JAZ using co-immunoprecipitation and yeast two-hybrid assays in the presence of OPDA, dn-OPDA, or dn-iso-OPDA, but we could not prove this interaction. This may be because the expressed COI1 protein did not retain its activity or another molecule is the ligand for COI1 of C. plumiforme.
Difference in OPDA Profiles of Bryophytes
We showed that OPDA was the most abundant in wounded C. plumiforme, followed by iso-OPDA, dn-OPDA, and dn-iso-OPDA (Figure 3). In the model liverwort M. polymorpha, dn-iso-OPDA is most abundant after wounding, followed by OPDA and dn-OPDA, the levels of which are almost comparable (Monte et al., 2018). Additionally, in the model moss P. patens, iso-OPDA is predominant, and smaller amounts of OPDA and dn-iso-OPDA exist with or without the feeding of α-linolenic acid (Mukhtarova et al., 2020). The differences in the profiles of OPDA and its derivatives among the three species indicate that the metabolic rates of isomerization and β-oxidation are different. The biological significance of the different profiles of OPDAs requires further study.
CpJAZs Interact With CpMYC2s
JAZ proteins interact with MYC2, MYC3 and MYC4 and repress downstream genes in various plants (Chini et al., 2007; Thines et al., 2007; Fernández-Calvo et al., 2011; Wasternack and Hause, 2013; Peñuelas et al., 2019). Co-immunoprecipitation analyses revealed that CpJAZ1, CpJAZ2, and CpJAZ3 physically interacted with CpMYC2a or CpMYC2b (Figure 6). Although the target genes of CpMYC2s are still unknown, CpJAZs might repress the transcription of these target genes at a steady state. Because a histidine residue in the bHLH domain that determines which DNA sequence to bind was conserved in CpMYC2s (Supplementary Figure 5), we assumed that CpMYC2s might bind to a G-box motif (CACGTG), similar to AtMYC2 and MpMYCs (Chini et al., 2007; Peñuelas et al., 2019). We found a G-box motif approximately 60 and 100 bp upstream of the transcription start site of CpJAZ1 and CpJAZ2, respectively. There is no G-box motif in the 2,000 bp upstream region from the transcription start site of CpJAZ3, but a G-box motif exists in the 5′- untranslated region of CpJAZ3. CpMYC2a and CpMYC2b possibly bind to these G-box motifs and regulate the inductive expression of CpJAZ genes in response to OPDA or its derivatives.
Conclusion
We found that OPDA, dn-OPDA, and their isomers accumulated in C. plumiforme after wounding at different abundance ratios than in M. polymorpha and P. patens, and that OPDA, dn-OPDA, and dn-iso-OPDA induced CpJAZ gene expression, indicating that they all have biological activity in C. plumiforme. COI1, JAZ, and MYC2 genes were conserved, as were the physical interactions between JAZ and MYC2 proteins. These results indicate that COI1-JAZ-MYC mediated signaling is conserved. At this time, the ligand(s) of CpCOI1s remain unknown. Future research using genetic approaches and/or other biochemical methods will reveal the interaction between CpCOI1s and CpJAZs and the ligand(s) of CpCOI1.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) are as follows: https://www.ncbi.nlm.nih.gov/genbank/, MW775560; MW775561; MW775562; MW775563; MW775564; MW775565; MW775566.
Author Contributions
KMi and KO designed the study. HI, NA, KMu, KS, SM, and MT performed the biological experiments. EY carried out LC/MS/MS analyses. KU synthesized (–)-JA and JA-Ile. NK and MU synthesized dn-OPDA, dn-iso-OPDA, and iso-OPDA. YH, TSh, and IG synthesized OPDA and [2H5]-OPDA. TK, YT, and MU performed the protein-protein interaction analyses. AN and TSa expressed JAZ proteins. HI and KMi performed all the computational analyses. KMi, HN, and KO supervised the experiments. KMi and KO wrote the manuscript. HI, NK, TSh, and IG revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by JSPS KAKENHI (Grant Numbers JP18K14399 to KMi; JP20H02922 to KO; JP17H06407, JP18KK0162, and JP20H00402 to MU; JP18H02101 and JP19H05283 to YT), JSPS A3 Foresight Program to MU, Japan-Austria Research Cooperative Program between JSPS and FWF (Grant Number JPJSBP120202002) to KO, and the Joint Usage/Research Center, Institute of Plant Science and Resources, Okayama University.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Hisakazu Yamane (The University of Tokyo) for his advice; Peter Nick and Michael Riemann for providing an expression vector for OsAOC; Ryosuke Sato (Teikyo University) for providing the SLiCE; and Kenichiro Hayashi (Okayama University of Science) for providing C. plumiforme plants. We would also like to thank Izumi C. Mori and Takakazu Matsuura (Okayama University) for their insights and technical assistance in [2H5]-dn-OPDA synthesis.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.688565/full#supplementary-material
Supplementary Figure 1 | 1H NMR spectrum of iso-OPDA.
Supplementary Figure 2 | 13C NMR spectrum of iso-OPDA.
Supplementary Figure 3 | Sequence alignment of COIs.
Supplementary Figure 4 | Differences between cloned CpMYC2s sequences and annotated sequences in the genome database.
Supplementary Figure 5 | Sequence alignment of MYC2.
Supplementary Figure 6 | Expression of GST-CpMYC2s and FLAG-JAZs.
Supplementary Table 1 | Primers used in this study.
References
Ballaré, C. L., and Austin, A. T. (2019). Recalculating growth and defense strategies under competition: key roles of photoreceptors and jasmonates. J. Exp. Bot. 70, 3425–3434. doi: 10.1093/jxb/erz237
Brown, J. W. S., Smith, P., and Simpson, C. G. (1996). Arabidopsis consensus intron sequences. Plant Mol. Biol. 32, 531–535. doi: 10.1007/bf00019105
Chini, A., Fonseca, S., Fernández, G., Adie, B., Chico, J. M., Lorenzo, O., et al. (2007). The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448, 666–671. doi: 10.1038/nature06006
Chini, A., Gimenez-Ibanez, S., Goossens, A., and Solano, R. (2016). Redundancy and specificity in jasmonate signalling. Curr. Opin. Plant Biol. 33, 147–156. doi: 10.1016/j.pbi.2016.07.005
Chini, A., Monte, I., Zamarreño, A. M., Hamberg, M., Lassueur, S., Reymond, P., et al. (2018). An OPR3-independent pathway uses 4,5-didehydrojasmonate for jasmonate synthesis. Nat. Chem. Biol. 14, 171–178. doi: 10.1038/nchembio.2540
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Fernández-Calvo, P., Chini, A., Fernández-Barbero, G., Chico, J.-M., Gimenez-Ibanez, S., Geerinck, J., et al. (2011). The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23, 701–715. doi: 10.1105/tpc.110.080788
Fonseca, S., Chini, A., Hamberg, M., Adie, B., Porzel, A., Kramell, R., et al. (2009). (+)-7-iso-Jasmonoyl-l-isoleucine is the endogenous bioactive jasmonate. Nat. Chem. Biol. 5, 344–350. doi: 10.1038/nchembio.161
Gao, B., Chen, M.-X., Li, X.-S., Liang, Y.-Q., Zhang, D.-Y., Wood, A. J., et al. (2021). Ancestral gene duplications in mosses characterized by integrated phylogenomic analyses. J. Syst. Evol. (in press). doi: 10.1111/jse.12683
Guo, L., Qiu, J., Ye, C., Jin, G., Mao, L., Zhang, H., et al. (2017). Echinochloa crus-galli genome analysis provides insight into its adaptation and invasiveness as a weed. Nat. Commun. 8:1031. doi: 10.1038/s41467-017-01067-5
Guo, Q., Yoshida, Y., Major, I. T., Wang, K., Sugimoto, K., Kapali, G., et al. (2018). JAZ repressors of metabolic defense promote growth and reproductive fitness in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 115, E10768–E10777. doi: 10.1073/pnas.1811828115
Han, G.-Z. (2016). Evolution of jasmonate biosynthesis and signaling mechanisms. J. Exp. Bot. 68, 1323–1331. doi: 10.1093/jxb/erw470
Jikumaru, Y., Asami, T., Seto, H., Yoshida, S., Yokoyama, T., Obara, N., et al. (2004). Preparation and biological activity of molecular probes to identify and analyze jasmonic acid-binding proteins. Biosci. Biotechnol. Biochem. 68, 1461–1466. doi: 10.1271/bbb.68.1461
Kajiwara, A., Abe, T., Hashimoto, T., Matsuura, H., and Takahashi, K. (2012). Efficient synthesis of (+)-cis-12-oxo-phytodienoic acid by an in vitro enzymatic reaction. Biosci. Biotechnol. Biochem. 76, 2325–2328. doi: 10.1271/bbb.120506
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Mao, L., Kawaide, H., Higuchi, T., Chen, M., Miyamoto, K., Hirata, Y., et al. (2020). Genomic evidence for convergent evolution of gene clusters for momilactone biosynthesis in land plants. Proc. Natl. Acad. Sci. U.S.A. 117, 12472–12480. doi: 10.1073/pnas.1914373117
Miyamoto, K., Enda, I., Okada, T., Sato, Y., Watanabe, K., Sakazawa, T., et al. (2016). Jasmonoyl-l-isoleucine is required for the production of a flavonoid phytoalexin but not diterpenoid phytoalexins in ultraviolet-irradiated rice leaves. Biosci. Biotechnol. Biochem. 80, 1934–1938. doi: 10.1080/09168451.2016.1189319
Miyamoto, K., Matsumoto, T., Yumoto, E., Sakazawa, T., Yokota, T., Yamane, H., et al. (2019). Facile preparation of optically active jasmonates and their biological activities in rice. Biosci. Biotechnol. Biochem. 83, 876–881. doi: 10.1080/09168451.2019.1569500
Miyamoto, K., Shimizu, T., and Okada, K. (2014). Transcriptional regulation of the biosynthesis of phytoalexin: a lesson from specialized metabolites in rice. Plant Biotechnol. 31, 377–388. doi: 10.5511/plantbiotechnology.14.0730a
Monte, I., Franco-Zorrilla, J. M., García-Casado, G., Zamarreño, A. M., García-Mina, J. M., Nishihama, R., et al. (2019). A single JAZ repressor controls the jasmonate pathway in Marchantia polymorpha. Mol. Plant 12, 185–198. doi: 10.1016/j.molp.2018.12.017
Monte, I., Ishida, S., Zamarreño, A. M., Hamberg, M., Franco-Zorrilla, J. M., García-Casado, G., et al. (2018). Ligand-receptor co-evolution shaped the jasmonate pathway in land plants. Nat. Chem. Biol. 14, 480–488. doi: 10.1038/s41589-018-0033-4
Motohashi, K. (2015). A simple and efficient seamless DNA cloning method using SLiCE from Escherichia coli laboratory strains and its application to SLiP site-directed mutagenesis. BMC Biotechnol. 15:47. doi: 10.1186/s12896-015-0162-8
Mukhtarova, L. S., Lantsova, N. V., Khairutdinov, B. I., and Grechkin, A. N. (2020). Lipoxygenase pathway in model bryophytes: 12-Oxo-9(13),15-phytodienoic acid is a predominant oxylipin in Physcomitrella patens. Phytochemistry 180:112533. doi: 10.1016/j.phytochem.2020.112533
Nishiyama, T., Hiwatashi, Y., Sakakibara, K., Kato, M., and Hasebe, M. (2000). Tagged mutagenesis and gene-trap in the moss, Physcomitrella patens by shuttle mutagenesis. DNA Res. 7, 9–17. doi: 10.1093/dnares/7.1.9
Nozaki, H., Hayashi, K., Nishimura, N., Kawaide, H., Matsuo, A., and Takaoka, D. (2007). Momilactone A and B as allelochemicals from moss Hypnum plumaeforme: first occurrence in bryophytes. Biosci. Biotechnol. Biochem. 71, 3127–3130. doi: 10.1271/bbb.70625
Okada, K., Kawaide, H., Miyamoto, K., Miyazaki, S., Kainuma, R., Kimura, H., et al. (2016). HpDTC1, a stress-inducible bifunctional diterpene cyclase involved in momilactone biosynthesis, functions in chemical defence in the moss Hypnum plumaeforme. Sci. Rep. 6:25316. doi: 10.1038/srep25316
Park, J.-H., Halitschke, R., Kim, H. B., Baldwin, I. T., Feldmann, K. A., and Feyereisen, R. (2002). A knock-out mutation in allene oxide synthase results in male sterility and defective wound signal transduction in Arabidopsis due to a block in jasmonic acid biosynthesis. Plant J. 31, 1–12. doi: 10.1046/j.1365-313X.2002.01328.x
Pauwels, L., and Goossens, A. (2011). The JAZ proteins: a crucial interface in the jasmonate signaling cascade. Plant Cell 23, 3089–3100. doi: 10.1105/tpc.111.089300
Peñuelas, M., Monte, I., Schweizer, F., Vallat, A., Reymond, P., García-Casado, G., et al. (2019). Jasmonate-related MYC transcription factors are functionally conserved in Marchantia polymorpha. Plant Cell 31, 2491–2509. doi: 10.1105/tpc.18.00974
Pratiwi, P., Tanaka, G., Takahashi, T., Xie, X., Yoneyama, K., Matsuura, H., et al. (2017). Identification of jasmonic acid and jasmonoyl-isoleucine, and characterization of AOS, AOC, OPR and JAR1 in the model lycophyte Selaginella moellendorffii. Plant Cell Physiol. 58, 789–801. doi: 10.1093/pcp/pcx031
Rensing, S. A., Ick, J., Fawcett, J. A., Lang, D., Zimmer, A., Van de Peer, Y., et al. (2007). An ancient genome duplication contributed to the abundance of metabolic genes in the moss Physcomitrella patens. BMC Evol. Biol. 7:130. doi: 10.1186/1471-2148-7-130
Riemann, M., Haga, K., Shimizu, T., Okada, K., Ando, S., Mochizuki, S., et al. (2013). Identification of rice Allene Oxide Cyclase mutants and the function of jasmonate for defence against Magnaporthe oryzae. Plant J. 74, 226–238. doi: 10.1111/tpj.12115
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. doi: 10.1093/oxfordjournals.molbev.a040454
Sheard, L. B., Tan, X., Mao, H., Withers, J., Ben-Nissan, G., Hinds, T. R., et al. (2010). Jasmonate perception by inositol-phosphate-potentiated COI1–JAZ co-receptor. Nature 468, 400–405. doi: 10.1038/nature09430
Shimura, K., Okada, A., Okada, K., Jikumaru, Y., Ko, K.-W., Toyomasu, T., et al. (2007). Identification of a biosynthetic gene cluster in rice for momilactones. J. Biol. Chem. 282, 34013–34018. doi: 10.1074/jbc.M703344200
Staswick, P. E., and Tiryaki, I. (2004). The oxylipin signal jasmonic acid Is activated by an enzyme that conjugates it to isoleucine in Arabidopsis. Plant Cell 16, 2117–2127. doi: 10.1105/tpc.104.023549
Stumpe, M., Göbel, C., Faltin, B., Beike, A. K., Hause, B., Himmelsbach, K., et al. (2010). The moss Physcomitrella patens contains cyclopentenones but no jasmonates: mutations in allene oxide cyclase lead to reduced fertility and altered sporophyte morphology. New Phytol. 188, 740–749. doi: 10.1111/j.1469-8137.2010.03406.x
Tamiru, M., Takagi, H., Abe, A., Yokota, T., Kanzaki, H., Okamoto, H., et al. (2016). A chloroplast-localized protein LESION AND LAMINA BENDING affects defence and growth responses in rice. New Phytol. 210, 1282–1297. doi: 10.1111/nph.13864
Thines, B., Katsir, L., Melotto, M., Niu, Y., Mandaokar, A., Liu, G., et al. (2007). JAZ repressor proteins are targets of the SCFCOI1 complex during jasmonate signalling. Nature 448, 661–665. doi: 10.1038/nature05960
Uji, Y., Taniguchi, S., Tamaoki, D., Shishido, H., Akimitsu, K., and Gomi, K. (2016). Overexpression of OsMYC2 results in the up-regulation of early JA-responsive genes and bacterial blight resistance in rice. Plant Cell Physiol. 57, 1814–1827. doi: 10.1093/pcp/pcw101
Wang, J., Sakurai, H., Kato, N., Kaji, T., and Ueda, M. (2021). Syntheses of dinor-cis/iso-12-oxo-phytodienoic acid (dn-cis/iso-OPDAs), ancestral jasmonate phytohormones of the bryophyte Marchantia polymorpha L., and their catabolites. Sci. Rep. 11:2033. doi: 10.1038/s41598-021-81575-z
Wasternack, C. (2015). How jasmonates earned their laurels: past and present. J. Plant Growth Regul. 34, 761–794. doi: 10.1007/s00344-015-9526-5
Wasternack, C., and Hause, B. (2013). Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 111, 1021–1058. doi: 10.1093/aob/mct067
Wasternack, C., and Hause, B. (2018). A bypass in jasmonate biosynthesis – the OPR3-independent formation. Trends Plant Sci. 23, 276–279. doi: 10.1016/j.tplants.2018.02.011
Xie, D.-X., Feys, B. F., James, S., Nieto-Rostro, M., and Turner, J. G. (1998). COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280, 1091–1094. doi: 10.1126/science.280.5366.1091
Yamane, H. (2013). Biosynthesis of phytoalexins and regulatory mechanisms of it in rice. Biosci. Biotechnol. Biochem. 77, 1141–1148. doi: 10.1271/bbb.130109
Yan, J., Zhang, C., Gu, M., Bai, Z., Zhang, W., Qi, T., et al. (2009). The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor. Plant Cell 21, 2220–2236. doi: 10.1105/tpc.109.065730
Keywords: oxylipin, plant hormone, 12-oxo-phytodienoic acid, moss, Calohypnum plumiforme
Citation: Inagaki H, Miyamoto K, Ando N, Murakami K, Sugisawa K, Morita S, Yumoto E, Teruya M, Uchida K, Kato N, Kaji T, Takaoka Y, Hojo Y, Shinya T, Galis I, Nozawa A, Sawasaki T, Nojiri H, Ueda M and Okada K (2021) Deciphering OPDA Signaling Components in the Momilactone-Producing Moss Calohypnum plumiforme. Front. Plant Sci. 12:688565. doi: 10.3389/fpls.2021.688565
Received: 31 March 2021; Accepted: 03 May 2021;
Published: 31 May 2021.
Edited by:
Koichi Sugimoto, University of Tsukuba, JapanReviewed by:
Isabel Monte, University of Zurich, SwitzerlandMarcelo Lattarulo Campos, Federal University of Mato Grosso, Brazil
Copyright © 2021 Inagaki, Miyamoto, Ando, Murakami, Sugisawa, Morita, Yumoto, Teruya, Uchida, Kato, Kaji, Takaoka, Hojo, Shinya, Galis, Nozawa, Sawasaki, Nojiri, Ueda and Okada. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Koji Miyamoto, bWl5YW1vdG9AbmFzdS5iaW8udGVpa3lvLXUuYWMuanA=
 Hideo Inagaki1
Hideo Inagaki1 Koji Miyamoto
Koji Miyamoto Yousuke Takaoka
Yousuke Takaoka Ivan Galis
Ivan Galis Akira Nozawa
Akira Nozawa Hideaki Nojiri
Hideaki Nojiri Minoru Ueda
Minoru Ueda