- 1Institute of Biosciences and Bioresources (IBBR), Italian National Research Council (CNR), Napoli, Italy
- 2Department of Molecular Biology and Genetics, Aarhus University, Aarhus, Denmark
Nitrogen-fixing nodules are new organs formed on legume roots as a result of the beneficial interaction with the soil bacteria, rhizobia. Proteins of the nitrate transporter 1/peptide transporter family (NPF) are largely represented in the subcategory of nodule-induced transporters identified in mature nodules. The role of nitrate as a signal/nutrient regulating nodule functioning has been recently highlighted in the literature, and NPFs may play a central role in both the permissive and inhibitory pathways controlling N2-fixation efficiency. In this study, we present the characterization of the Lotus japonicus LjNPF3.1 gene. LjNPF3.1 is upregulated in mature nodules. Promoter studies show transcriptional activation confined to the cortical region of both roots and nodules. Under symbiotic conditions, Ljnpf3.1-knockout mutant’s display reduced shoot development and anthocyanin accumulation as a result of nutrient deprivation. Altogether, LjNPF3.1 plays a role in maximizing the beneficial outcome of the root nodule symbiosis.
Introduction
As sessile organisms, land plants developed mechanisms that enable them to cope with the dynamically changing availability of nutrients in the soil (Williams and Miller, 2001; Rogato et al., 2010). Nitrogen in the form of nitrates is often a limiting resource for supporting plant growth in temperate climates (Miller and Cramer, 2005). A primary role in the network governing nitrate uptake, assimilation, storage, and distribution among different plant tissues and organs is played by two protein families, the low-affinity nitrate transporter peptide family (NPF; LATS > 0.5 mM) and the high-affinity nitrate transporter system (NRT2; HATS < 0.5 mM). NPF is a large plant family consisting of 53, 93, and 86 members in Arabidopsis thaliana, Oryza sativa, and Lotus japonicus, respectively (Tsay et al., 2007; Léran et al., 2014; Sol et al., 2019). To date, nitrate transport activity has been reported for 17 A. thaliana NPF proteins (Corratgé-Faillie and Lacombe, 2017), with AtNPF6.3 being the only exception as this displays a switching dual nitrate affinity, controlled via the phosphorylation of the threonine residue at position 101, in response to fluctuating external concentrations (Liu et al., 1999; Liu and Tsay, 2003; Parker and Newstead, 2014; Sun et al., 2014). The extended functional characterization carried out in A. thaliana is allowing the unraveling of the network of the AtNPF distinct functional roles. A variegated pattern of spatiotemporal NPF gene expression through the whole plant body is crucial to provide nitrate uptake from the soil, upward and downward long-distance transport, distribution from source to sink tissues, intercellular flux, and cellular nitrate storage redistribution (Wang et al., 2018). NPF members are also able to transport substrates other than nitrate, including di-/tripeptides, amino acids, glucosinolates, malate, auxin, abscisic acid (ABA), gibberellic acid, and jasmonic acid (Frommer et al., 1994; Jeong et al., 2004; Waterworth and Bray, 2006; Krouk et al., 2010; Kanno et al., 2012; Nour-Eldin et al., 2012; Saito et al., 2015; Tal et al., 2016). The different transport capacities are distributed among the eight NPF subclades identified in plants, and the prediction of the transported substrate cannot be determined from the sequences data alone (Léran et al., 2014). In a few cases, the capability of the NPF members to transport both nitrate and hormones with different affinities has been reported, and this multispecificity could suggest intriguing roles of NPF proteins for the integration of environmental and physiological information linked to the relative availability of different nutrients (Krouk et al., 2010; Kanno et al., 2012; Saito et al., 2015; Corratgé-Faillie and Lacombe, 2017). So far, this role has been demonstrated only for the AtNPF6.3 gene, which also functions as an auxin transport facilitator to modulate lateral root elongation in response to nitrate (Krouk et al., 2010).
The nodule organogenesis competence evolved in legume plants as a result of the symbiotic interaction with rhizobia partner where an ideal microenvironmental niche enables the conversion of atmospheric N2 into the plant assimilable NH3. The invaded cells of the N2-fixing nodules are filled with symbiosomes, the organelles originated by an endocytosis-like process enclosing invading bacteria in a plant-derived membrane whose formation involves an exocytotic process [peri-bacteroidal membrane (PBM)]. Inside the symbiosome, bacteria stop dividing and differentiate into the N2-fixing bacteroids. For the correct function of the nodules, it is crucial to maintain the balance between the microaerophilic condition, which protects nitrogenase from inactivation, and the high rates of respiration taking place in the cytosol and bacteroid compartments of the invaded cells (Bergensen, 1996; Witty and Minchin, 1998). These conflicting demands are met via the oxygen barrier constituted by parenchyma cell layers and mainly by the action of the high-affinity O2− binding protein leghemoglobin (Lb), present at millimolar concentrations in the N2-fixing cells (Appleby, 1984). The nodulation process is an excellent example of adaptation to the changing environment as all the very demanding steps of nodule formation, development, and functioning are quickly inhibited when sufficient amounts of fixed N are readily available in the soil (Carroll and Gresshoff, 1983; Fujikake et al., 2003; Barbulova et al., 2007; Omrane and Chiurazzi, 2009; Naudin et al., 2011; Cabeza et al., 2014). In the case of nodule functioning, the responsiveness to nitrate has been mainly associated with the inhibitory pathway triggering a dramatic decrease in functional leghemoglobin and nitrogenase activity by the exposure of nodulated roots to high nitrate concentrations (5–10 mM; Arrese-Igor et al., 1998; Cabeza et al., 2014). However, a positive action played by low concentrations of nitrate to maintain the energy status required for efficient N2-fixation, ensuring the correct nodule functioning, has been also reported (Horchani et al., 2011; Hicri et al., 2015). More recently, a positive role played by both NPF and NRT2 nitrate transporters for satisfying the required nitrate allocation and distribution into the N2-fixing zone of the nodules has been proposed (Valkov et al., 2017, 2020; Wang et al., 2020). In particular, LjNRT2.4 and MtNPF7.6 seem to be involved in the nitrate-mediated regulation of nodule function by controlling the quick and fine-tuning of nitric oxide (NO) concentration in the nodule-invaded cells in response to rapid changes in O2 availability (Valkov et al., 2020; Wang et al., 2020).
In this study, we report the characterization of another member of the L. japonicus NPF family that may contribute to the completion of the route of nitrate toward the nodule N2-fixing zone required for a correct nodule functioning in the presence of low-permissive external concentrations.
Materials and Methods
Plant Material and Growth Conditions
All experiments were carried out with L. japonicus ecotype B-129 F14 GIFU (Handberg and Stougaard, 1992; Jiang and Gresshoff, 1997). Plants were cultivated in Petri dishes, in a growth chamber with a light intensity of 200 μmol m−2 s−1 at 23°C with a 16 h/8 h day/night cycle. Solid growth media had the same composition as B5 medium (Gamborg, 1970), except that (NH4)2SO4 and KNO3 were omitted and/or substituted by different concentrations of KNO3. KCl was added to the medium to replace the potassium source. The media containing vitamins (Duchefa catalogue G0415) were buffered with 2.5 mM 2-(N-morpholino) ethanesulfonic acid (MES; Duchefa, M1503.0250), and pH was adjusted to 5.7 with KOH.
For the phenotypic characterization at 6 weeks after inoculation, synchronized lotus seedlings were transferred after germination in pots containing lightweight expanded clay aggregate (LECA, Saint-Gobain Weber A/S,1 2–4 mm diameter) and vermiculite size M (Damolin A/S)2 in a 4:1 mixture. Plants were supplemented with 60 ml 0.25 × B&D medium supplemented with the required amount of KNO3. Plants were incubated at 23°C under a 16-h light/8-h dark cycle.
Mesorhizobium loti inoculation was performed as described in Barbulova et al. (2005). For phenotypic comparisons, after germination, unsynchronized seedlings were discarded. The strain R7A was used for the inoculation experiments and was grown in a liquid TYR medium supplemented with rifampicin (20 mg/L).
Lotus japonicus Hairy Root Transformation Procedures
Binary vectors were conjugated into the Agrobacterium rhizogenes 15,834 strain (Stougaard et al., 1987). A. rhizogenes-mediated L. japonicus transformations have been performed as described in Bastianelli et al. (2009). Inoculation of composite plants was described in Santi et al. (2003).
Protoplast Transformation
Leaf protoplasts were prepared and transformed according to Pedrazzini et al. (1997), using 3-week-old Nicotiana tabacum plants. DNA (40 μg of each construct) was introduced into 1 × 106 protoplasts by PEG-mediated transfection. After 16-h incubation in the dark at 25°C, yellow fluorescent protein (YFP) fluorescence in protoplast cells was detected by confocal microscopy.
LORE1 Lines Isolation
LORE1 lines 300121103 and 30082596 were obtained from the LORE1 collection (Fukai et al., 2012; Urbanski et al., 2012; Malolepszy et al., 2016). The plants in the segregating populations were genotyped, and the expression of homozygous plants tested with primers is listed in Supplementary Table S1.
Determination of Acetylene Reduction Activity
Detached roots with a comparable number of nodules were placed in glass vials. The vials were filled with an acetylene – air mixture (C2H2; air = 1:9 v/v). After 30 min of incubation at 25°C, the amount of ethylene in the gas phase was determined using a gas chromatograph (PerkinElmer Clarus 580).
The analysis of acetylene reduction activity (ARA) after a shift in high KNO3 conditions has been performed as described in the study by D’Apuzzo et al. (2015); nodulated plants were transferred at 4 wpi on slanted Petri dishes where roots are placed in sandwich position between two filter papers wet with a Gamborg B5 liquid media containing no KNO3 or 10 mM KNO3. Plants are maintained for 48 h in these conditions with filter papers wet with 20 ml liquid media. ARA was then tested as described above.
Estimation of Anthocyanin
Stem tissue from three plants per assay was weighed and then extracted with 99:1 methanol: HCl (v/v) at 4°C. The OD530 and OD657 for each sample were measured, and relative anthocyanin levels were determined with the equation OD530 – (0.25 × OD657) × extraction volume (ml) × 1/weight of tissue sample (g) = relative units of anthocyanin/g fresh weight of tissue.
Quantitative Real-Time PCR
RNA was extracted as described in Omrane et al. (2009). Real-time PCR was performed with a DNA Engine Opticon 2 System, MJ Research (MA, United States) using SYBR to monitor dsDNA synthesis. The procedure was described in the study by Moscatiello et al. (2018). The ubiquitin (UBI) gene (AW719589) was used as an internal standard. The concentration of primers was optimized for every PCR, and amplifications were carried out in triplicate. The PCR program used was as follows: 95°C for 3 min and 39 cycles of 94°C for 15 s, 60°C for 15 s, and 72°C for 15 s. Data were analyzed using Opticon Monitor Analysis Software version 2.01 (MJ Research). The quantitative real-time PCR (qRT-PCR) data were analyzed using the comparative Ct method. The relative level of expression was calculated with the following formula: The relative expression ratio of the gene of interest is 2−∆CT with ∆←←←CT = CtGENE minus CTUBI. The efficiency of the LjNPF3.1 primers was assumed to be two. Analysis of the melting curve of PCR product at the end of the PCR run revealed a single narrow peak for each amplification product, and fragments amplified from total complementary DNA (cDNA) were gel-purified and sequenced to assure accuracy and specificity. The oligonucleotides used for the qRT-PCR are listed in Supplementary Table S1.
Plasmid Preparation
The various constructs used for L. japonicus transformation were assembled using Golden Gate Cloning (Weber et al., 2011). The LjNPF3.1 sequences have been retrieved in the Lotus database (Gifu assembly; Mun et al., 2016)3 and synthesized by ThermoFisher Scientific. Overhangs extremities with the BsaI or BpiI restriction sites have been added to the sequences. For promoter activity studies, the putative promoter region of 2,503 bp upstream of the translation start of LjNPF3.1 was used to drive a triple YFP reporter carrying a nuclear localization signal on the C terminus (Reid et al., 2016). The construct was assembled in a pIV10-L2 vector backbone (ampicillin and spectinomycin/streptomycin resistant). For localization studies in tobacco protoplasts, the LjNPF3.1 CDS sequence was cloned downstream of the 35S promoter and upstream of the sequence encoding the eYFP directly into pICH binary vector backbone (Weber et al., 2011). Restriction ligase reactions were made using the BpiI restriction enzyme and T4 ligase of ThermoFisher Scientific.
Statistical Analysis
Statistical analyses were performed using the VassarStats analysis of variance program.
Confocal Imaging
For promoter activity using tYFPnls, transformed roots were fixed with paraformaldehyde and cleared as described previously (Warner et al., 2014). The samples were analyzed on a ZEISS confocal microscope LSM780. The whole root images were obtained using Z-stack and tile scan tools with 514/515–530 nm excitation/emission settings. Final images were generated by Maximum Intensity Projection in ZEN software (ZEISS) or ImageJ. For the 35S driven LjNPF3.1-YFP fusion in tobacco protoplast, confocal analyses were performed using a LeicaDMi8 (Leica Biosystems, Wetzlar, Germany) laser scanning confocal imaging system as described in Rogato et al. (2008). For YFP detection, excitation was set at 488 nm, and detection between 515 and 530 nm.
Results
LjNPF3.1 Expression Is Strongly Induced in N2-Fixing Nodules
We have recently reported the identification of the 86 members of the L. japonicus NPF family (accession MG20; Criscuolo et al., 2012; Sol et al., 2019) whose nomenclature was assigned based on the two-letter code established in Léran et al. (2014). The name LjNPF3.1 is assigned to the MG20 gene Lj2g3v1155500.1 (Sol et al., 2019) and the identical copy LotjaGi2g1v0278100.1, identified in the L. japonicus accession Gifu (Supplementary Table S2; Kamal et al., 2020).4 LjNPF3.1 is one of the three members of the clade 3 (Supplementary Table S2) coding for a 580-amino acid protein with a molecular mass of 64.4 kDa. LjNPF3.1 has 12 TM predicted domains (Tusnàdy and Simon, 2001) and shares the highest level of amino acid identity among the Arabidopsis NPF members, with the AtNPF3.1 protein (74%; AT1G68570.1). LjNPF3.1 was previously included in the subclass of eight LjNPF nodule-induced genes identified by in silico analysis (Valkov and Chiurazzi, 2014).5 We have now confirmed this specific profile of expression in a time-course experiment where L. japonicus seedlings grown in a derived Gamborg-B5 medium without N sources were inoculated with M. loti 1 week after sowing. The LjNPF3.1 transcript was barely detectable in uninoculated as well as roots tested up to 14 days after inoculation, whereas a strong induction was observed in mature nodules (tenfold induction; Figure 1A). To further characterize the profile of expression of LjNPF3.1, we have also analyzed the distribution of the LjNPF3.1 transcript in different organs of L. japonicus. The LjNPF3.1 gene showed a regulated transcriptional profile with a peak of expression in mature nodules and a significant level of transcript in leaves and mature flowers (Figure 1B).
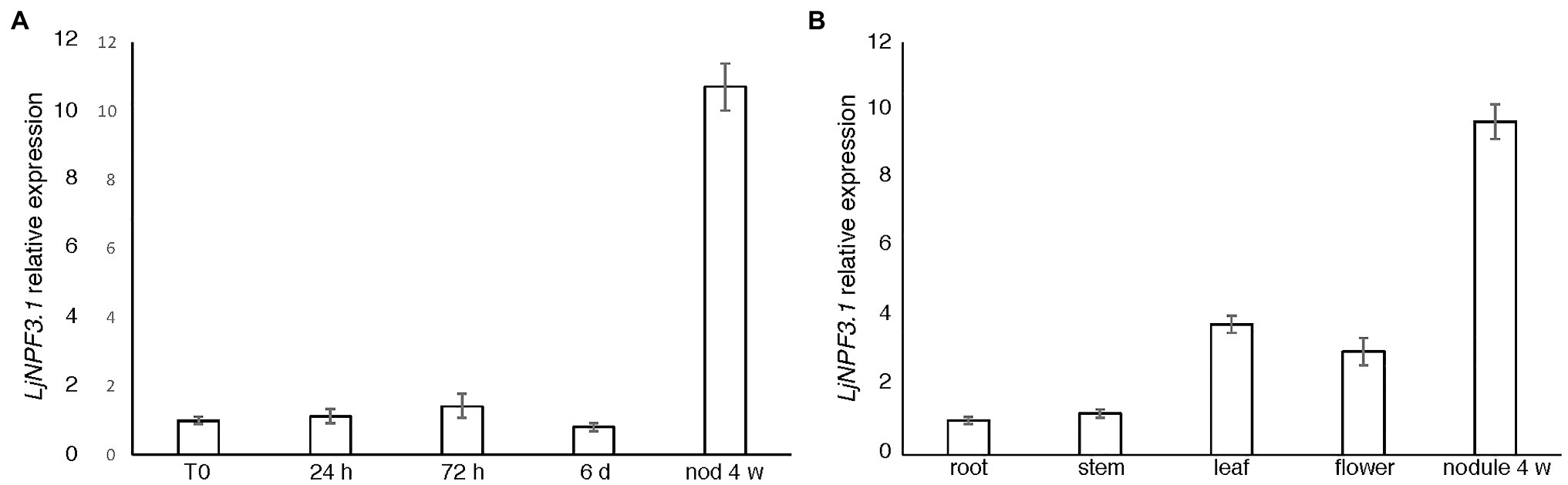
Figure 1. LjNPF3.1 transcriptional regulation. (A) Time-course analysis in wild-type Lotus japonicus root and nodule tissues after Mesorhizobium loti inoculation. RNAs were extracted from roots of seedlings grown in N-starvation conditions at different times after inoculation (R0, 24 h, 72 h, 6 days, and 14 days) and from mature nodules (28 dpi). (B) Expression in different organs. RNAs were extracted 4 weeks after inoculation. Mature flowers were obtained from lotus plants propagated in the growth chamber. Expression levels obtained by quantitative real-time PCR (qRT-PCR) were normalized with respect to the internal control ubiquitin (UBI) gene and plotted as relative to the expression of T0 (A) and root (B). Data bars represent the mean and SDs of data obtained with RNA extracted from three different sets of plants and three real-time PCR experiments.
LjNPF3.1 Is Expressed in Root and Nodule Cortical Regions
To gain information about the spatial distribution of the LjNPF3.1 transcript in root and nodule tissues, the putative promoter region of the LjNPF3.1 gene was obtained by amplifying a PCR fragment extending up to 2.5 kb upstream of the ATG. A proNPF3.1:tYFP-NLS reporter constructs localizing triple YFP in the nucleus of transgenic roots was used to visualize the LjNPF3.1 spatial gene expression at the cellular level. Confocal microscopy analyses indicated that the LjNPF3.1 promoter was active in the cortical cells of the inoculated hairy roots (Figure 2A), and the fluorescence level increased at the base of the nodules (Figure 2B), consistent with the increased expression shown in Figure 1. The section in Figure 2C shows a strong YFP signal also in the peripheral parenchymatic nodule region, most likely the outer cortex cell layers.
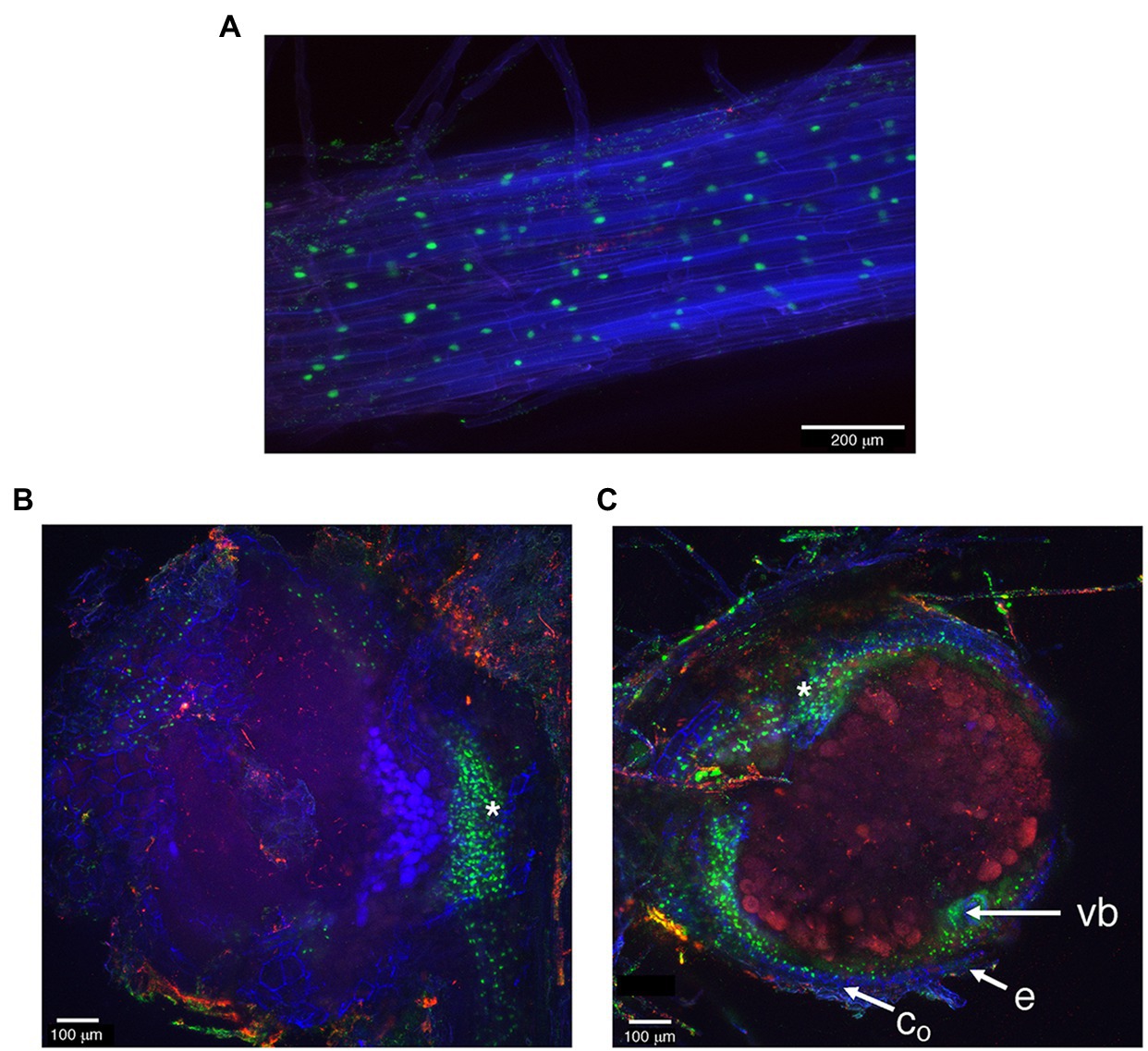
Figure 2. The activity of LjNPF3.1 2.5-kb promoter fused to the tYFP-NLS reporter (nuclear-localized, green) in inoculated hairy roots. (A) Expression in the root cortical region. (B) Increased YFP fluorescence was detected at the base of mature nodules. (C) YFP fluorescence at the periphery of the N2-fixing nodules in the outer cortex region. *, nodule base; vb, vascular bundle; co, outer cortex; e, epidermis.
To determine the LjNPF3.1 subcellular localization, we generated a construct that fused YFP to the C-terminus of LjNPF3.1 under the control of the 35S promoter and 35S terminator and introduced this construct in tobacco protoplasts. Although we did not provide evidence that the 35S-LjNPF3.1-YFF fusion maintains the correct biological function, confocal microscopy analysis indicated that this localizes at the plasma membrane of transformed protoplasts, mirroring the localization of the AtPIP2A-mCherry plasma membrane marker (Figures 3A–F).
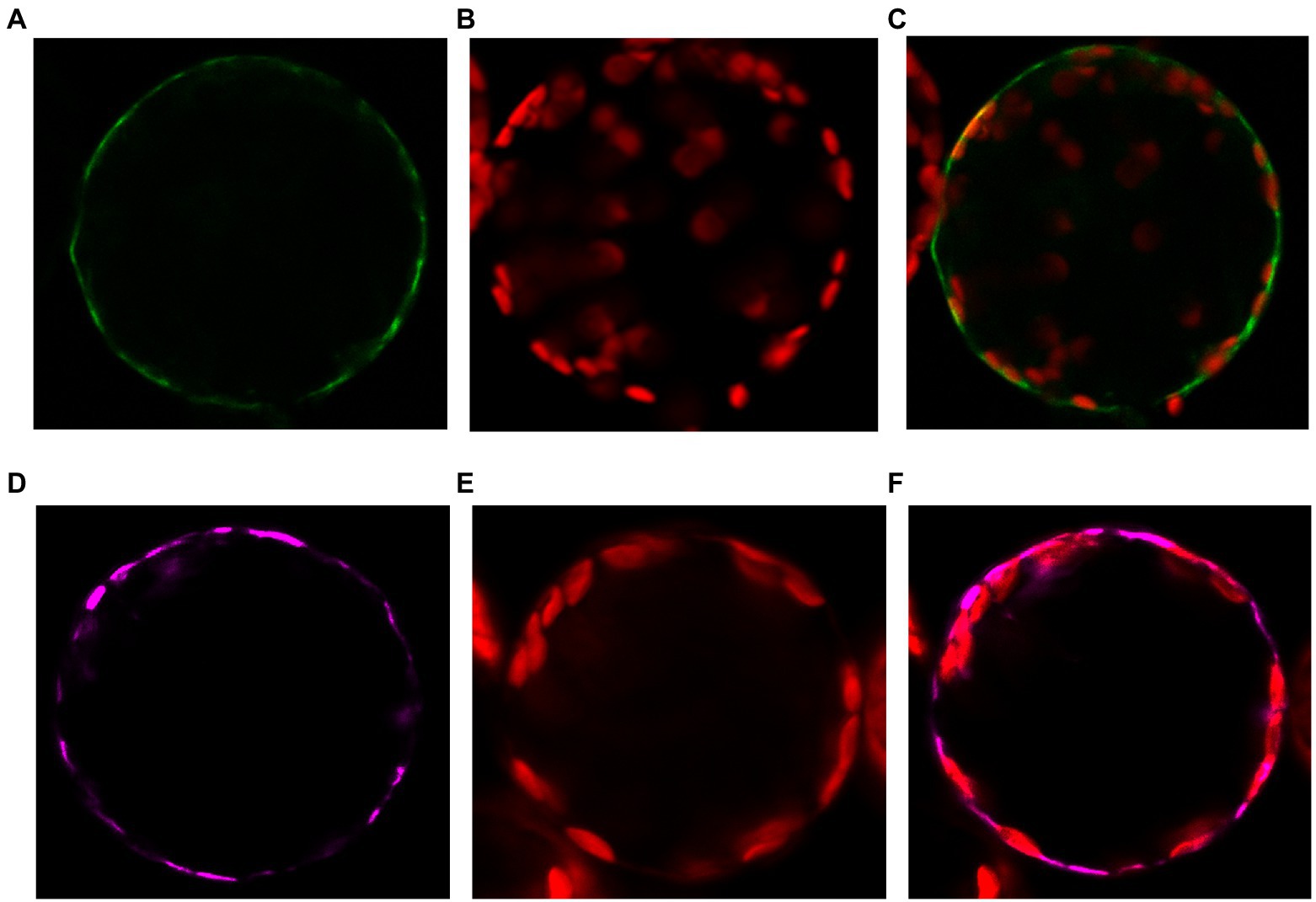
Figure 3. Plasma membrane localization of the p35S-LjNPF3.1-eYFP fusion transiently expressed in protoplasts of tobacco mesophyll cells. (A) Plasma membrane localization of the 35S-LjNPF3.1-eYFP. (B) Chlorophyll autofluorescence of the transformed protoplast. (C) Merged image of (A,B). (D) Plasma membrane localization of the pUBI-AtPIP2A-mCherry marker (purple). (E) Chlorophyll autofluorescence of the transformed protoplast. (F) Merged image of (C,D).
Isolation of LORE1-Insertion Null Mutants and Phenotypic Characterization
To determine the in vivo function of LjNPF3.1, two independent LORE1 insertion mutants have been isolated from the LORE1 lines collection established in the L. japonicus Gifu accession (Fukai et al., 2012; Urbanski et al., 2012; Malolepszy et al., 2016). Lines 300121103 and 30082596, bearing retrotransposon insertions in the third and fourth exon (Figure 4A), have been genotyped by PCR, and plants homozygous for those insertion events were selected and transferred to the plant chamber for the production of seeds. Endpoint RT-PCR analyses of homozygous plants from lines 300121103 and 30082596 revealed no detectable LjNPF3.1 mRNA in mature nodules and hence, considered null mutants and hereafter named Ljnpf3.1-1 and Ljnpf3.1-2, respectively (Figure 4B). Two individual homozygous mutant plants from each insertion line have been selected for analyses, and because their growth phenotypes did not significantly differ, the data obtained with the selected individual mutants have been pooled. As transcript abundance and promoter activity were related to mature nodules, we analyzed the symbiotic performances of the Ljnpf3.1 plants in terms of nodule formation capacity and shoot growth parameters exhibited at 4 weeks after inoculation with M. loti. Wild-type and Ljnpf3.1 synchronized seedlings with a 0.5 cm long root were transferred 5 days after sowing, in Petri dishes containing B5-derived medium without N sources or with different concentrations of KNO3 and inoculated with M. loti. Wild-type and mutant plants display the same nodule formation capacity, whereas a clear-cut reduction of the shoot length (21–32%) and fresh weight (16–26%) values were exhibited by Ljnpf3.1-1 and -2 when compared to the ones of the wild-type plants (Figures 5A–C; Supplementary Figure S1). Importantly, when the growth medium was supplemented with 5 mM KNO3, a concentration that is known to inhibit nodule initiation (Barbulova et al., 2007) and sufficient to support optimal plant growth, the mutants did not display any shoot growth-defective phenotype (Figures 5A–C; Supplementary Figure S1). The specificity of the symbiotic deficient phenotype was further confirmed by another set of experiments where wild-type and mutant genotypes were grown in pots to allow the evaluation of symbiotic phenotypes up to 6 weeks after inoculation. The scoring of nodulation, as well as shoot growth parameters, confirmed the deficient phenotype of both Ljnpf3.1 inoculated plants, whereas no differences were observed in uninoculated plants grown in the presence of 1 mM KNO3 (Figures 5D–F; Supplementary Figure S2). Furthermore, as a correlation between nitrate uptake/reallocation/assimilation and water stress responses has been well-characterized in plants (Chen et al., 2012; Gloser et al., 2020), we have also analyzed the dry weight of detached wild-type and mutant leaves to rule out the hypothesis that the reduced weight was due to different water contents. Data shown in Supplementary Figure S3 indicate that the dry weight of leaves was also significantly reduced in mutants as compared to wild-type and that the wt/Ljnpf3.1 fresh and dry weight ratios remain constant. The specific symbiotic deficient phenotype displayed by the Ljnpf3.1 plants was also confirmed by the substantially increased accumulation of anthocyanin exhibited in the stems of the inoculated mutants as compared to wild-type plants, with the exception of plants grown in the presence of sufficient N conditions (5 mM KNO3; Supplementary Figure S1). The anthocyanin accumulation was quantified, and the data shown in Figure 6 indicate a 3–5-fold increase in the mutants as compared to wild-type, whereas such a difference was not scored in the uninoculated plants (Figure 6A).
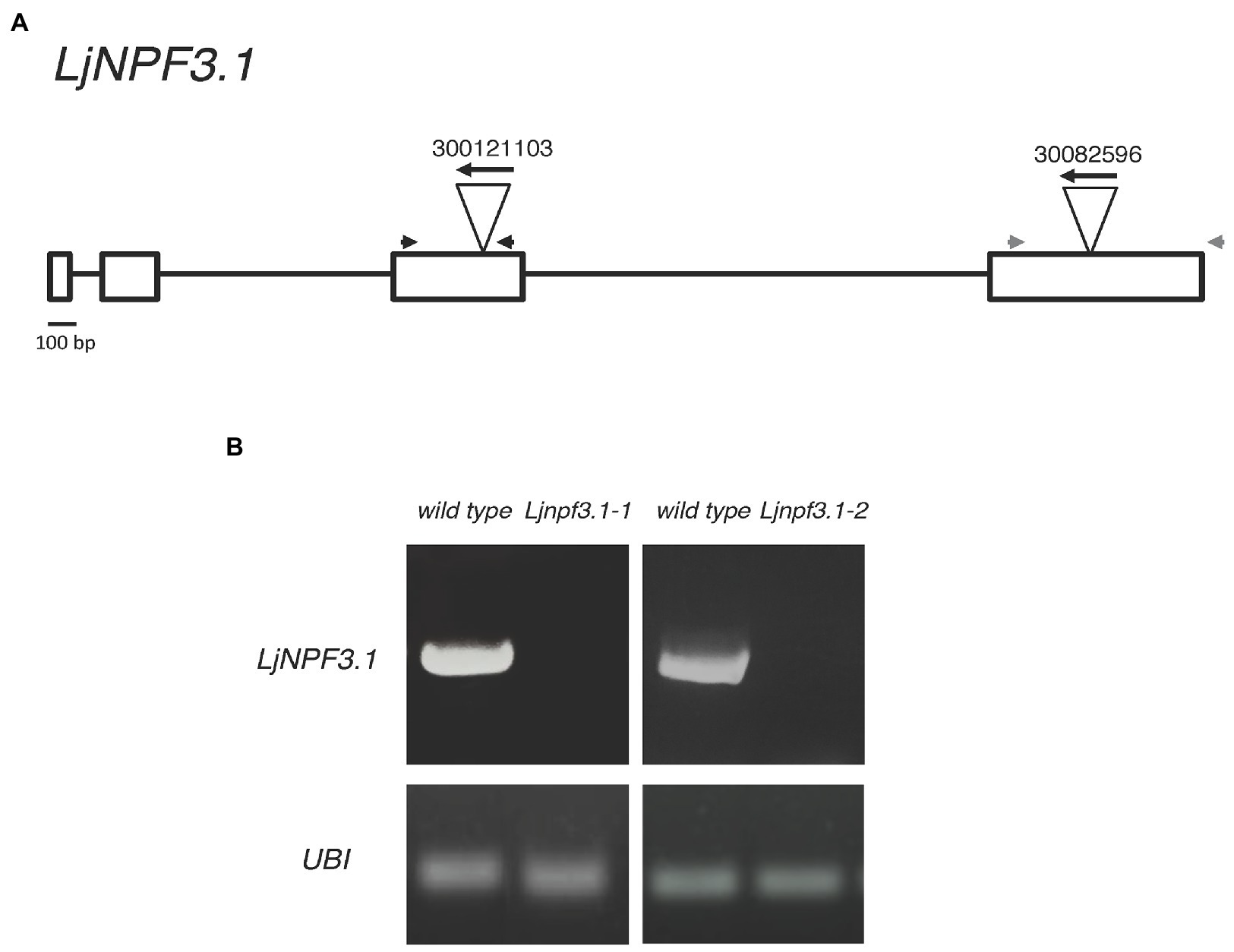
Figure 4. (A) Exon/intron organization of the LjNPF3.1 gene. Insertion sites, relative orientations of the LORE1 retrotransposon element in the 300121103 and 30082596 lines, and positions of a couple of primers (gray and black arrowheads) used for genotyping are indicated. (B) LjNPF3.1 is not expressed in LORE1 homozygous mutant lines.
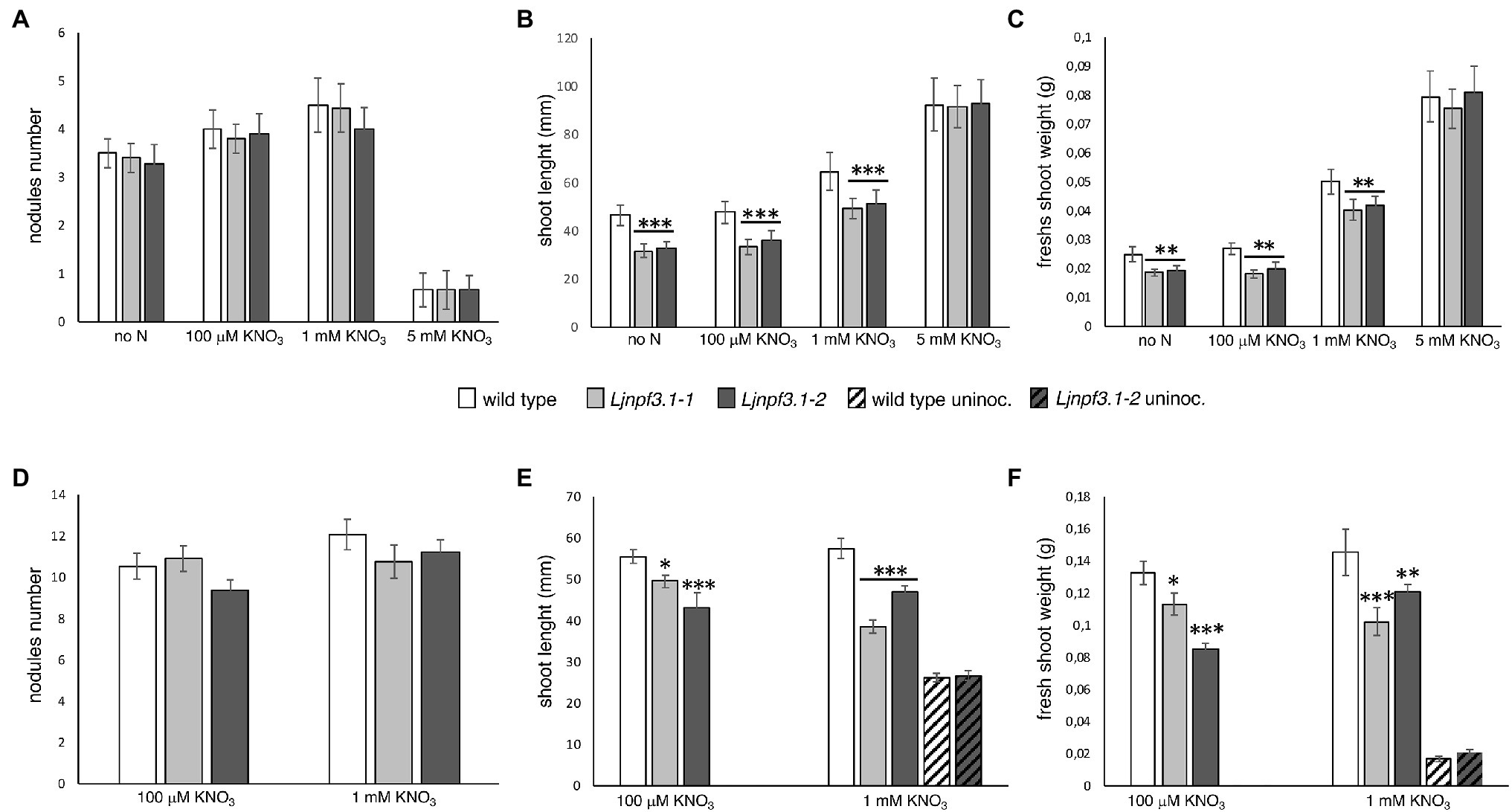
Figure 5. Phenotypic characterization of Ljnpf3.1-1 and Ljnpf3.1-2 mutants. Wild-type and Ljnpf3-1 plants, inoculated with M. loti and grown in the presence of different concentrations of KNO3. (A–C) Plants grown in Petri dishes, axenic conditions, scored 4 weeks after inoculation. (D–F) Plants were grown in pots scored at 6 weeks after inoculation. (A,D) Nodules number per plant. (B,D) Shoots length per plant. (C,F) Shoots fresh weight per plant. Different KNO3 regimes and plant genotypes are indicated [noninoculated plants in panels (E,F) are indicated by oblique-filled bars]. Bars represent the means and SE of measures from three experiments (at least 20 plants per experiment per condition). Asterisks indicate significant differences with wild-type levels. ∗p < 0.05; ∗∗p < 0.002; and ∗∗∗p < 0.0001.
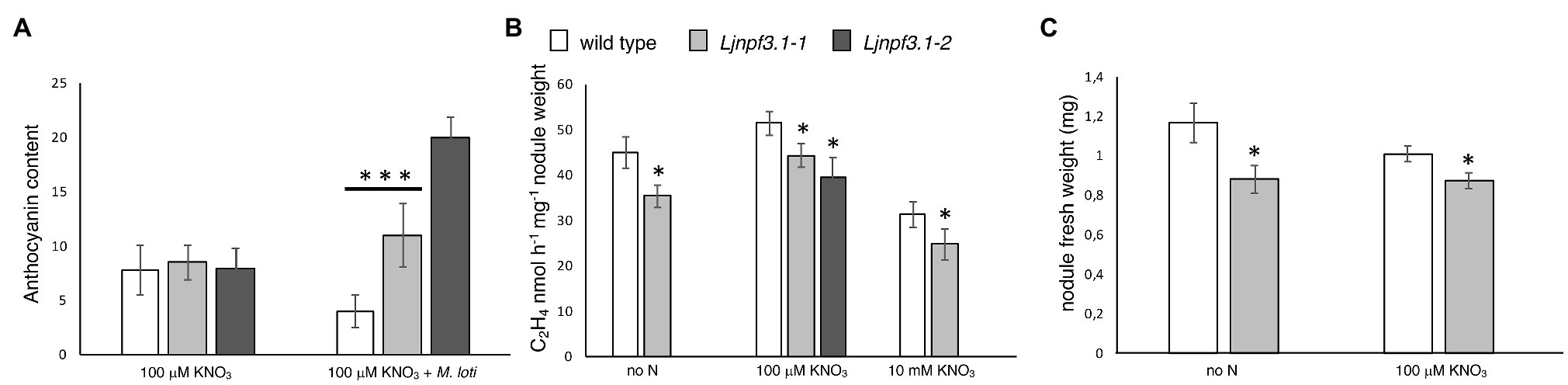
Figure 6. Phenotypic characterization of Ljnpf3.1-1 and Ljnpf3.1-2 mutants. (A) Analysis of anthocyanin content in the stems of wild-type and Ljnpf3.1 plants grown in Petri dishes with/without M. loti inoculation. Relative anthocyanin levels scored in the Y-axis were determined with the equation OD530 – (0.25 × OD657) × extraction volume (ml) × 1/weight of tissue sample (g) = relative units of anthocyanin/g fresh weight of tissue. Anthocyanin was extracted 4 weeks after sowing. (B) Acetylene reduction activity (ARA) per nodule weight at 4 weeks postinoculation. (C) Nodules fresh weight. Plants were grown in Petri dishes as in Figures 5A–C. N regimes and plant genotypes are indicated. Data bars represent the means and SE of measures from six experiments in (B; eight samples of five nodules per experiment per condition). Asterisks indicate significant differences with wild-type levels. ∗p < 0.05 and ∗∗∗p < 0.001.
To test whether the symbiotic phenotypes could be associated with altered nodule functionality, we have analyzed the N2-fixing performances of wild-type and Ljnpf3.1 mutant nodules. The ARA was measured in detached nodule samples from five independent experiments in plants grown in axenic conditions at 4 weeks postinoculation. A significant 22% reduction of activity in plants grown without N supply was scored, and this deficiency was reduced to still a significant 14% (Ljnpf3.1-1) and 24% (Ljnpf3.1-2) in nodules of mutants grown in the presence of 100 μM KNO3 (Figure 6B). Furthermore, the comparison of nodulation phenotypes in nitrate-free and 100 μM KNO3 revealed a significant increase in nodule biomass (fresh weight) in the wild-type as compared to Ljnpf3.1-1 (Figure 6C; 25 and 15%, respectively). To test whether LjNPF3.1 is involved in the pathway responsible for the rapid drop of the N2-fixation activity observed in nodules after the transfer in the presence of high external nitrate concentrations (Arrese-Igor et al., 1998; Cabeza et al., 2014), we compared the values of acetylene reduction in wild-type and mutant nodules after transfer in the presence of 10 mM KNO3 for 48 h. As shown in Figure 6B, the N2-fixation capacity is reduced to more than 30% in both wild-type and mutant nodules after the transfer in high nitrate conditions (Figure 6B).
The identical phenotypes displayed by the Ljnpf3.1-1 and -2 null mutants confirmed that the LORE1 insertions in the LjNPF3.1 gene are the causal mutations of the deficient phenotypes observed exclusively in symbiotic conditions. In addition, heterozygous plants for the LORE1 insertion, the LjNPF3.1 gene isolated in the two analyzed lines, displayed neither the shoot biomass deficient phenotypes nor the anthocyanin accumulation (data not shown).
Discussion
The important contribution of transporters of the NPF family to the functioning of N2-fixing nodules is suggested by the large numbers of NPF genes found to be upregulated in mature nodules (Takanashi et al., 2012; Valkov and Chiurazzi, 2014; Clarke et al., 2015; Wang et al., 2016). In this study, we report the characterization of one of the eight upregulated LjNPF genes identified in mature N2-fixing Lotus nodules, LjNPF3.1 (Valkov and Chiurazzi, 2014). Analyses of the expression profile revealed an increase in the LjNPF3.1 transcript in nodules compared to roots (more than 10-fold; Figure 1). The clade 3 of the L. japonicus NPF family consists of three members as in other legume plants (Medicago truncatula and Phaseolus vulgaris; Léran et al., 2014), and LjNPF3.1 is the only one upregulated in mature nodules.6 Interestingly, MtNPF3.1 that shares 84% of amino acid identity with LjNPF3.1 also exhibits the induced profile of expression in mature nodules (He et al., 2009).7
The phenotypic characterization of the two independent null mutants provides evidence for a positive role played by LjNPF3.1 for an efficient nodule functioning (Figures 5, 6). The reduction of shoot height and weight scored in the mutants in the whole range of permissive low N conditions tested (Figures 5B,C,E,F) might be explained, in the presence of an equal number of nodules (Figures 5A,D), by the partial, but significant, impairment of the nitrogenase activity reported in Figure 6B. Consistently, the shoot phenotypes displayed by the Ljnpf3.1 mutants (Figure 5; Supplementary Figure S1) are not as severe as those reported for the fix− mutants, rather resembling the ones of the mutants described as Fix+/Fix−, which show an impaired N2-fixation activity (Garcìa-Calderon et al., 2012; Pislariu et al., 2012). The relatively slight reduction of nitrogenase activity could also be due to the functional redundancy of LjNPF3.1 with other NPF genes upregulated in the mature nodule (Valkov and Chiurazzi, 2014). The relationship between the reduced shoot biomass and the N-starvation condition due to the partial efficiency of the N2-fixation activity is confirmed by the rescue of the stressed phenotype observed in the presence of 5 mM KNO3 (Figures 5B,C). Furthermore, the anthocyanin accumulation scored only in the stems of the inoculated Ljnpf3.1 plants represents another indication of an impaired N2-fixation activity in the mutant genetic background as anthocyanin accumulation in the stems has been reported as a phenotype associated with N-starvation condition associated with a deficiency of the nodule functioning or lack of nodulation (Krussell et al., 2005; Ott et al., 2005; Bourcy et al., 2013; Pal’ove-Balang et al., 2015). Although we cannot exclude the possibility that the spatial expression of LjNPF3.1 is also controlled by sequences located outside the 2.5 kb of the 5'-UTR region used in the promoter-fusion construct, the spatial expression profile (Figure 2) provides some clues about the action played by LjNPF3.1 for the control of nodule efficiency. The preferential expression in the root cortex shown in Figure 2A has been only reported for two Arabidopsis nitrate excretion transporters (NAXT) of the subgroup 2, AtNPF2.5 and AtNPF2.7, which mediate chloride and nitrate efflux from the root, respectively (Segonzac et al., 2007; Li et al., 2017). Recently, in M. truncatula, the flux of nitrate from the root tissue and from the external environment to the nodules has been traced (Wang et al., 2020). The MtNPF7.6 high-affinity nitrate transporter, specifically expressed in nodule vascular tissues, functions in nitrate uptake and transport through the nodule transfer cells (NTC) to fine-tune the nodule development and functioning in response to fluctuating environmental nitrate status (Wang et al., 2020). The spatial expression profile of LjNPF3.1 in the root cortical region at the base of the nodule (Figure 2B) and in the layers of cortex adjacent to the infected cells inside the nodule (Figure 2C) is consistent with possible involvement in the flux of nitrate from the root and from outside to inside the nodule toward the N2-fixation zone (Figure 7) where nitrate might contribute to regulate NO homeostasis and oxidative stress (Horchani et al., 2011; Hicri et al., 2015; Valkov et al., 2017, 2020; Signorelli et al., 2020; Villar et al., 2020; Wang et al., 2020). However, the involvement of LjNPF3.1 should be limited to conditions of low external concentration that are permissive for nodule functioning, as the inhibitory pathway occurring at high concentrations of nitrate is not altered in the Ljnpf3.1 mutants (Figure 6B). So far, it is noteworthy that the transporters reported being involved in the nodular transport of nitrate (Figure 7) exhibit capacity to transport this ion at low concentrations (0.5 mM; Valkov et al., 2017), with saturable kinetics in the micromolar range (Wang et al., 2020) or as members of the high-affinity NRT2 family (Valkov et al., 2020).
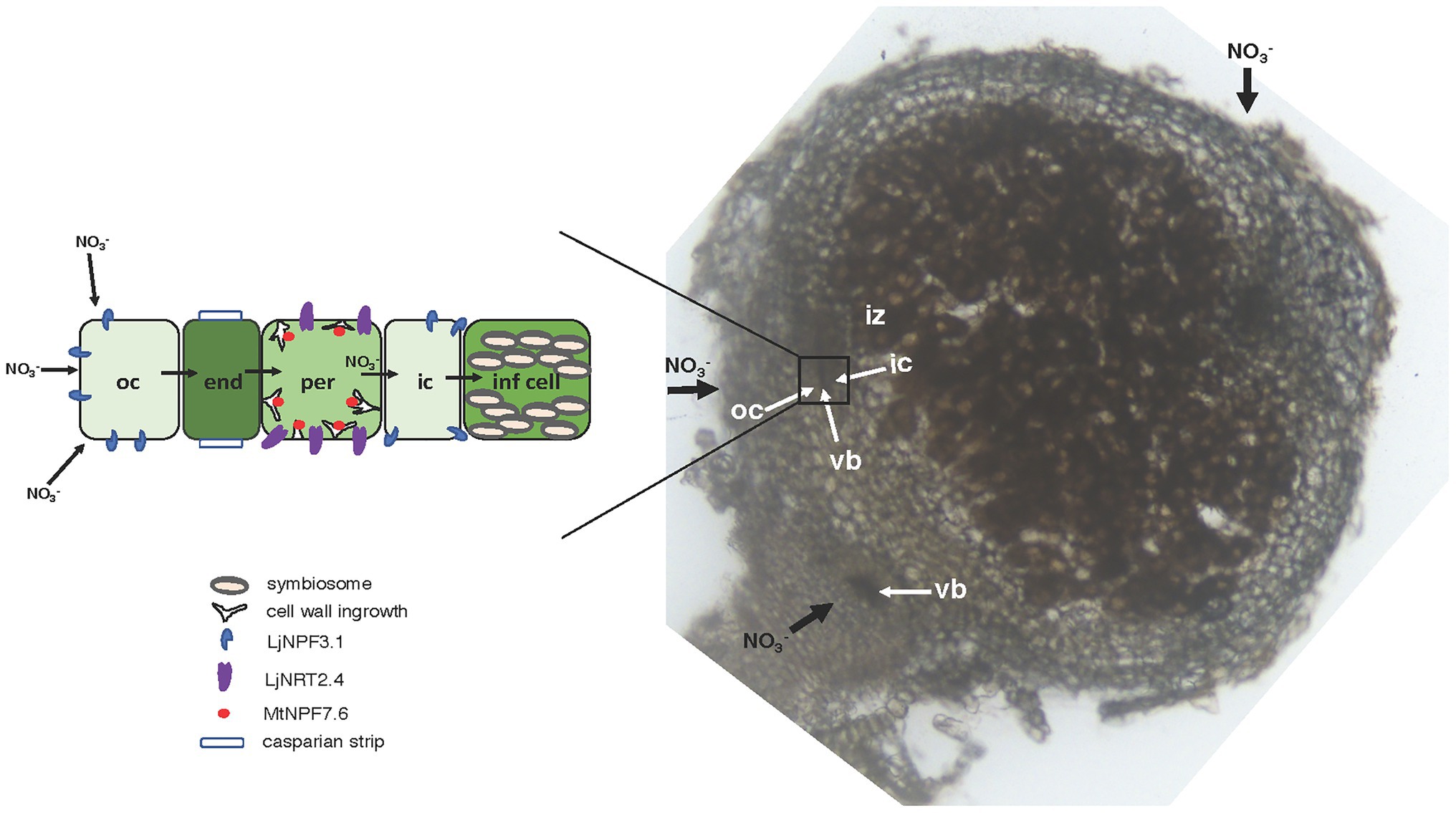
Figure 7. Model of nitrate flux in the N2-fixing nodule. Right side: 70 μm vibratome longitudinal section of a 4-week old L. japonicus nodule with an illustration of the flux of nitrate (thick black arrows) toward the infected cells of the N2-fixation zone either from the root or from outside to inside the nodule. The rectangle indicates the vascular bundle structure bracketed by the outer and inner cortex. Left side: scheme of the flux of nitrate through different cell layers with an illustration of the NPFs and NRT2 involved. The NPFs and NRT2 transporters involved in the transport of nitrate, subcellular structures, and organelles are indicated. oc, outer cortex; ic, inner cortex; vb, vascular bundle; iz, infection zone; end, endodermis; per, pericycle; inf cell, infected cell.
In A. thaliana, the clade 3 of the NPF family consists of a unique member, AtNPF3.1. AtNPF3.1 has also been reported to act as a low-affinity nitrate/nitrite transporter in Xenopus laevis oocytes (Pike et al., 2014) although the nitrate transport capacity could not be confirmed by the characterization of Atnpf3.1 mutants (David et al., 2016). However, as already mentioned, it has been well-documented that NPFs transport various substrates, including phytohormones, and hence, the involvement of LjNPF3.1 in other pathways such as the one governing the distribution of gibberellin (GA), recently reported as a positive regulator of nodule functioning (Serova et al., 2019), cannot be excluded.
In conclusion, although a further characterization of the LjNPF3.1 gene will certainly pass through the identification of the preferentially transported substrate(s), the data reported here clearly indicate its crucial role in the correct N2-fixation process, further highlighting the roles played by NPF transporters for an efficient nodule functioning.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
YV, MN, and AR designed, performed, and analyzed the experiments. SR designed and analyzed the experiments. VV designed, performed, and analyzed the experiments and contributed to the writing of this manuscript. MC designed the experiments and wrote the article. YV, MN, AR, SR, VV, and MC contributed to the experimental design and manuscript preparation. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by CNR Project (no.FOE-2019 DBA.AD003.139) and the European Community, Horizon 2020, Partnership for Research and Innovation in the Mediterranean Area, PRIMA, Section II, Legumes in biodiversity-based farming systems in Mediterranean Basin, LEGU-MED. YV was granted an Erasmus+ traineeship for graduate students from the University Federico II of Naples.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Marco Petruzziello and Giuseppina Zampi for technical assistance and Finn Pedersen for professional plant care.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.688187/full#supplementary-material
Supplementary Figure S1 | Representative images of the wild-type and Ljnpf3.1 plants at 4 weeks after inoculation. Nitrate conditions and plant genotypes are indicated. The white arrows indicate the zones displaying the evident accumulation of anthocyanin in the stems of the mutant plants as compared to the wild-type.
Supplementary Figure S2 | Representative images of the wild-type and Ljnpf3.1 plants at 6 weeks after inoculation. Nitrate condition and plant genotypes are indicated. The white arrows are pointed on stems of mutants displaying evident accumulation of anthocyanin.
Supplementary Figure S3 | Phenotypic characterization of Ljnpf3.1-1 and Ljnpf3.1-2 mutants. (A) Fresh weight of detached leaves. (B) The dry weight of detached leaves. Plants were grown in Petri dishes, in the presence of 1 mM KNO3. Leaves from single plants were detached and immediately weighted at 4 weeks after inoculation. Bars represent the means and SE of measures from three experiments (12 plants per experiment). Asterisks indicate significant differences with wild-type levels. *p < 0.05.
Supplementary Table S1 | Oligonucleotides are used in the present work.
Supplementary Table S2 | Description of the Lotus japonicus NPF clade 3.
Footnotes
References
Appleby, C. A. (1984). Leghemoglobin and Rhizobium respiration. Annu. Rev. Plant Physiol. Plant Mol. Biol. 35, 443–478. doi: 10.1146/annurev.pp.35.060184.002303
Arrese-Igor, C., Gordon, A. J., Minchin, F. R., and Denison, R. F. (1998). Nitrate entry and nitrite formation in the infected region of soybean nodules. J. Exp. Bot. 49, 41–48. doi: 10.1093/jxb/49.318.41
Barbulova, A., D’Apuzzo, E., Rogato, A., and Chiurazzi, M. (2005). Improved procedures for in vitro regeneration and for phenotypical analysis in the model legume Lotus japonicus. Funct. Plant Biol. 32, 529–536. doi: 10.1071/FP05015
Barbulova, A., Rogato, A., D’Apuzzo, E., Omrane, S., and Chiurazzi, M. (2007). Differential effects of combined N sources on early steps of the Nod factor-dependent transduction pathway in Lotus japonicus. Mol. Plant-Microbe Interact. 20, 994–1003. doi: 10.1094/MPMI-20-8-0994
Bastianelli, F., Costa, A., Vescovi, M., D’Apuzzo, E., Zottini, M., Chiurazzi, M., et al. (2009). Salicylic acid differentially affects suspension cell cultures of Lotus japonicus and one of its non-symbiotic mutants. Plant Mol. Biol. 72, 469–483. doi: 10.1007/s11103-009-9585-8
Bergensen, F. J. (1996). Delivery of O2 to bacteroids in soybean nodule cells: considerations of gradients of concentrations of free, dissolved O2 in and near symbiosomes and beneath intercellular spaces. Protoplasma 191, 9–20. doi: 10.1007/BF01280821
Bourcy, M., Brocard, L., Pislariu, C. I., Cosson, V., Mergaert, P., Tadege, M., et al. (2013). Medicago truncatula DNF2 is a PI-PLC-XD-containing protein required for bacteroid persistence and prevention of nodule early senescence and defence-like reactions. New Phytol. 197, 1250–1261. doi: 10.1111/nph.12091
Cabeza, R., Koester, B., Liese, R., Lingner, A., Baumgarten, V., Dirks, J., et al. (2014). An RNA sequencing transcriptome analysis reveals novel insights into molecular aspects of the nitrate impact on the nodule activity of Medicago truncatula. Plant Physiol. 164, 400–411. doi: 10.1104/pp.113.228312
Carroll, B., and Gresshoff, P. M. (1983). Nitrate inhibition of nodulation and nitrogen fixation in white clover. Z. Pflanzenphysiol. 110, 77–88. doi: 10.1016/S0044-328X(83)80218-9
Chen, C. Z., Lv, X. F., Li, J. Y., Yi, H. Y., and Gong, J. M. (2012). Arabidopsis NRT1.5 is another essential component in the regulation of nitrate reallocation and stress tolerance. Plant Physiol. 159, 1582–1590. doi: 10.1104/pp.112.199257
Clarke, V. C., Loughlin, C., Gavrin, A., Chen, C., Brear, E. M., Day, D. A., et al. (2015). Proteomic analysis of the soybean symbiosome identifies new symbiotic proteins. Mol. Cell. Proteomics 14, 1301–1322. doi: 10.1074/mcp.M114.043166
Corratgé-Faillie, C., and Lacombe, B. (2017). Substrate (un)specificity of Arabidopsis NRT1/PTR family (NPF) proteins. J. Exp. Bot. 68, 3107–3113. doi: 10.1093/jxb/erw499
Criscuolo, G., Valkov, V. T., Parlati, A., Martin-Alves, L., and Chiurazzi, M. (2012). Molecular characterization of the Lotus japonicus NRT1 (PTR) and NRT2 families. Plant Cell Environ. 35, 1567–1581. doi: 10.1111/j.1365-3040.2012.02510.x
D’Apuzzo, E., Valkov, V. T., Parlati, A., Omrane, S., Barbulova, A., Sainz, M. M., et al. (2015). PII overexpression in Lotus japonicus affects nodule activity in permissive low nitrogen conditions and increases nodule numbers in high nitrogen treated plants. Mol. Plant-Microbe Interact. 28, 432–442. doi: 10.1094/MPMI-09-14-0285-R
David, L. C., Berquin, P., Kanno, Y., Seo, M., Daniel-Vedele, A., and Ferrario-Mery, S. (2016). N availability modulates the role of NPF3.1, a gibberellin transporter, in GA-mediated phenotypes in Arabidopsis. Planta 244, 1315–1328. doi: 10.1007/s00425-016-2588-1
Frommer, W. B., Hummel, S., and Rentsch, D. (1994). Cloning of an Arabidopsis histidine transporting protein related to nitrate and peptide transporters. FEBS Lett. 347, 185–189. doi: 10.1016/0014-5793(94)00533-8
Fujikake, H., Yamazaki, A., Ohtake, N., Sueyoshi, K., Matsuhashi, S., Ito, T., et al. (2003). Quick and reversible inhibition of soybean nodule growth by nitrate involves a decrease in sucrose supply to nodules. J. Exp. Bot. 54, 1379–1388. doi: 10.1093/jxb/erg147
Fukai, E., Soyano, T., Umehara, Y., Nakayama, S., Hirakawa, H., Tabata, S., et al. (2012). Establishment of a Lotus japonicus gene tagging population using the exon-targeting endogenous retrotransposon LORE1. Plant J. 69, 720–730. doi: 10.1111/j.1365-313X.2011.04826.x
Gamborg, O. L. (1970). The effects of amino acids and ammonium on the growth of plant cells in suspension culture. Plant Physiol. 45, 372–375. doi: 10.1104/pp.45.4.372
Garcìa-Calderon, M., Chiurazzi, M., Espuny, M. R., and Màrquez, A. J. (2012). Photorespiratory metabolism and nodule function. Behavior of Lotus japonicus mutants deficient in plastid glutamine synthetase. Mol. Plant-Microbe Interact. 25, 211–219. doi: 10.1094/MPMI-07-11-0200
Gloser, V., Dvoraskova, M., Mota, D. H., Petrovic, B., Gonzalez, P., and Geilfus, C. M. (2020). Early changes in nitrate uptake and assimilation under drought in relation to transpiration. Front. Plant Sci. 11:602065. doi: 10.3389/fpls.2020.602065
Handberg, K., and Stougaard, J. (1992). Lotus japonicus, an autogamous, diploid legume species for classical and molecular genetics. Plant J. 2, 487–496. doi: 10.1111/j.1365-313X.1992.00487.x
He, J., Benedito, V. A., Wang, M., Murray, J. D., Zhao, P. X., Tang, Y., et al. (2009). The Medicago truncatula gene expression atlas web server. BMC Bioinformatics 10:441. doi: 10.1186/1471-2105-10-441
Hicri, I., Boscari, A., Castella, C., Rovere, M., Puppo, A., and Brouquisse, R. (2015). Nitric oxide: a multifaceted regulator of the nitrogen-fixing symbiosis. J. Exp. Bot. 66, 2877–2887. doi: 10.1093/jxb/erv051
Horchani, F., Prévot, M., Boscari, A., Evangelisti, E., Meilhoc, E., Bruand, C., et al. (2011). Both plant and bacterial nitrate reductases contribute to nitric oxide production in Medicago truncatula nitrogen-fixing nodules. Plant Physiol. 155, 1023–1036. doi: 10.1104/pp.110.166140
Jeong, J., Suh, S., Guan, C., Tsay, Y. F., Moran, N., Oh, C. J., et al. (2004). A nodule-specific dicarboxylate transporter from alder is a member of the peptide transporter family. Plant Physiol. 134, 969–978. doi: 10.1104/pp.103.032102
Jiang, Q., and Gresshoff, P. M. (1997). Classical and molecular genetics of the model legume Lotus japonicus. Mol. Plant-Microbe Interact. 10, 59–68. doi: 10.1094/MPMI.1997.10.1.59
Kamal, N., Man, T., Reid, D., Lin, J. S., Akyol, T. Y., Sandal, N., et al. (2020). Insights into the evolution of symbiosis gene copy number and distribution from a chromosome-scale Lotus japonicus Gifu genome sequence. DNA Res. 1:27. doi: 10.1093/dnares/dsaa015
Kanno, Y., Hanada, A., Chiba, Y., Ichikawa, T., Nakazawa, M., Matsui, M., et al. (2012). Identification of an abscisic acid transporter by functional screening using the receptor complex as a sensor. Proc. Natl. Acad. Sci. U. S. A. 109, 9653–9658. doi: 10.1073/pnas.1203567109
Krouk, G., Lacombe, B., Bielach, A., Perrine-Walker, F., Malinska, K., Mounier, E., et al. (2010). Nitrate-regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Dev. Cell 18, 927–397. doi: 10.1016/j.devcel.2010.05.008
Krussell, L., Krause, K., Ott, T., Desbrosses, G., Kramer, U., Sato, S., et al. (2005). The sulfate transporter SST1 is crucial for symbiotic nitrogen fixation in Lotus japonicus root nodules. Plant Cell 17, 1625–1636. doi: 10.1105/tpc.104.030106
Léran, S., Varala, K., Boyer, J. C., Chiurazzi, M., Crawford, N., Daniel-Vedele, F., et al. (2014). A unified nomenclature of nitrate transporter 1/peptide transporter family members in plants. Trends Plant Sci. 19, 5–9. doi: 10.1016/j.tplants.2013.08.008
Li, B., Qiu, J., Jayakannan, M., Xu, B., Li, Y., Mayo, G. M., et al. (2017). AtNPF2.5 modulates chloride (Cl−) efflux from roots of Arabidopsis thaliana. Front. Plant Sci. 7:2013. doi: 10.3389/fpls.2016.02013
Liu, K. H., Huang, C. Y., and Tsay, Y. (1999). CHL1 is a dual-affinity nitrate transporter of Arabidopsis involved in multiple phases of nitrate uptake. Plant Cell 11, 865–874. doi: 10.1105/tpc.11.5.865
Liu, K. H., and Tsay, Y. F. (2003). Switching between the two action modes of the dual-affinity nitrate transporter CHL1 by phosphorylation. EMBO J. 22, 1005–1013. doi: 10.1093/emboj/cdg118
Malolepszy, A., Mun, T., Sandal, N., Gupta, V., Dubin, M., Urbański, D. F., et al. (2016). The LORE1 insertion mutant resource. Plant J. 88, 306–317. doi: 10.1111/tpj.13243
Miller, A. J., and Cramer, M. D. (2005). Root nitrogen acquisition and assimilation. Plant Soil 274, 1–36. doi: 10.1007/s11104-004-0965-1
Moscatiello, R., Sello, S., Ruocco, M., Barbulova, A., Cortese, E., Nigris, S., et al. (2018). The hydrophobin HYTLO1 secreted by the biocontrol fungus Trichoderma longibrachiatum triggers a NAADP-mediated calcium signalling pathway in Lotus japonicus. Int. J. Mol. Sci. 19, 2596–2612. doi: 10.3390/ijms19092596
Mun, T., Bachmann, A., Gupta, V., Stougaard, J., and Andersen, S. U. (2016). Lotus base: an integrated information portal for the model legume Lotus japonicus. Sci. Rep. 6:39447. doi: 10.1038/srep39447
Naudin, C., Corre-Hellou, G., Voisin, A. S., Oury, V., Salon, C., Crozat, Y., et al. (2011). Inhibition and recovery of symbiotic N2 fixation by peas (Pisum sativum L.) in response to short-term nitrate exposure. Plant Soil 346, 275–287. doi: 10.1007/s11104-011-0817-8
Nour-Eldin, H. H., Andersen, T. G., Burow, M., Madsen, S. R., Jørgensen, M. E., Olsen, C. E., et al. (2012). NRT/PTR transporters are essential for translocation of glucosinolate defence compounds to seeds. Nature 488, 531–534. doi: 10.1038/nature11285
Omrane, S., and Chiurazzi, M. (2009). A variety of regulatory mechanisms are involved in the nitrogen-dependent modulation of the nodule organogenesis program in legume roots. Plant Signal. Behav. 4, 1066–1068. doi: 10.4161/psb.4.11.9735
Omrane, S., Ferrarini, A., D’Apuzzo, E., Rogato, A., Delledonne, M., and Chiurazzi, M. (2009). Symbiotic competence in Lotus japonicus is affected by plant nitrogen status: transcriptomic identification of genes affected by a new signalling pathway. New Phytol. 183, 380–394. doi: 10.1111/j.1469-8137.2009.02873.x
Ott, T., van Dongen, J. T., Gunther, C., Krussell, L., Desbrosses, G., Vigeolas, H., et al. (2005). Symbiotic leghemoglobins are crucial for nitrogen fixation in legume root nodules but not for general plant growth and development. Curr. Biol. 15, 531–535. doi: 10.1016/j.cub.2005.01.042
Pal’ove-Balang, P., Garcia-Calderòn, M., Perez-Delgado, C. M., Pavlokin, J., and Màrquez, A. J. (2015). A Lotus japonicus mutant defective in nitrate uptake is also affected in the nitrate response to nodulation. Plant Biol. 17, 16–25. doi: 10.1111/plb.12169
Parker, J. L., and Newstead, S. (2014). Molecular basis of nitrate uptake by the plant nitrate transporter NRT1.1. Nature 507, 68–72. doi: 10.1038/nature13116
Pedrazzini, E., Giovinazzo, G., Bielli, A., de Virgilio, M., Frigerio, L., Pesca, M., et al. (1997). Protein quality control along the route to the plant vacuole. Plant Cell 9, 1869–1880. doi: 10.1105/tpc.9.10.1869
Pike, S., Gao, F., Kim, S. H., Schachtman, D. P., and Gassmann, W. (2014). Members of the NPF3 transporter subfamily encode pathogen-inducible nitrate/nitrite transporters in gravepine and Arabidopsis. Plant Cell Physiol. 55, 162–170. doi: 10.1093/pcp/pct167
Pislariu, C. I., Murray, J. D., Wen, J. Q., Cosson, V., Muni, R. R. D., Wang, M., et al. (2012). A Medicago truncatula tobacco retrotransposon insertion mutant collection with defects in nodule development and symbiotic nitrogen fixation. Plant Physiol. 159, 1686–1699. doi: 10.1104/pp.112.197061
Reid, D. E., Heckmann, A. B., Novák, O., and Stougaard, J. (2016). Cytokinin oxidase/dehydrogenase 3 maintains cytokinin homeostasis during root and nodule development in Lotus japonicus. Plant Physiol. 170, 1060–1074. doi: 10.1104/pp.15.00650
Rogato, A., D’Apuzzo, E., Barbulova, A., Omrane, S., Stedel, C., Simon-Rosin, U., et al. (2008). Tissue-specific down-regulation of LjAMT1;1 compromises nodule function and enhances nodulation in Lotus japonicus. Plant Mol. Biol. 68, 585–595. doi: 10.1007/s11103-008-9394-5
Rogato, A., D’Apuzzo, E., and Chiurazzi, M. (2010). The multiple plant response to high ammonium conditions. The Lotus japonicus AMT1;3 protein acts as a putative transceptor. Plant Signal. Behav. 5, 1584–1586. doi: 10.4161/psb.5.12.13856
Saito, H., Oiwawa, T., Hamamoto, S., Ishimaru, Y., Kanamori-Sato, M., Sasaki-Sekimoto, Y., et al. (2015). The jasmonate-responsive GTR1 transporter is required for gibberellin-mediated stamen development in Arabidopsis. Nat. Commun. 6, 6095–7006. doi: 10.1038/ncomms7095
Santi, C., von Groll, U., Ribeiro, A., Chiurazzi, M., Auguy, F., Bogusz, D., et al. (2003). Comparison of nodule induction in legume and actinorhizal symbioses: the induction of actinorhizal nodules does not involve ENOD40. Mol. Plant-Microbe Interact. 16, 808–816. doi: 10.1094/MPMI.2003.16.9.808
Segonzac, C., Boyer, J. C., Ipotesi, E., Szponarski, W., Tillard, P., Touraine, B., et al. (2007). Nitrate efflux at the root plasma membrane: identification of an Arabidopsis excretion transporter. Plant Cell 19, 3760–3777. doi: 10.1105/tpc.106.048173
Serova, T. A., Tsyganova, A. V., Tikhonovich, I. A., and Tsyganov, V. E. (2019). Gibberellins inhibit nodule senescence and stimulate nodule meristem bifurcation in pea (Pisum sativum L.). Front. Plant Sci. 10:285. doi: 10.3389/fpls.2019.00285
Signorelli, S., Sainz, M., Tabares-da Rosa, S., and Monza, J. (2020). The role of nitric oxide in nitrogen fixation by legumes. Front. Plant Sci. 11:521. doi: 10.3389/fpls.2020.00521
Sol, S., Valkov, V. T., Rogato, A., Noguero, M., Gargiulo, L., Mele, G., et al. (2019). Disruption of the Lotus japonicus transporter LjNPF2.9 increases shoot biomass and nitrate content without affecting symbiotic performances. BMC Plant Biol. 19:380. doi: 10.1186/s12870-019-1978-5
Stougaard, J., Abildsten, D., and Marcker, K. A. (1987). The Agrobacterium rhizogenes pRi TL-DNA segment as a gene vector system for transformation of plants. Mol. Gen. Genet. 207, 251–255. doi: 10.1007/BF00331586
Sun, J., Bankston, J. R., Payandeh, J., Hinds, T. R., Zagotta, W. N., and Zheng, N. (2014). Crystal structure of the plant dual-affinity nitrate transporter NRT1.1. Nature 507, 73–77. doi: 10.1038/nature13074
Takanashi, K., Takahashi, H., Sakurai, N., Sugiyama, A., Suzuki, H., Shibata, D., et al. (2012). Tissue-specific transcriptome analysis in nodules of Lotus japonicus. Mol. Plant-Microbe Interact. 25, 869–876. doi: 10.1094/MPMI-01-12-0011-R
Tal, I., Zhang, Y., Jorgensen, M. E., Pisanty, O., Barbosa, I. C., Zourelidou, M., et al. (2016). The Arabidopsis NPF3 protein is a GA transporter. Nat. Comm. 7:11486. doi: 10.1038/ncomms11486
Tsay, Y. F., Chiu, C. C., Tsai, C. B., Ho, C. H., and Hsu, P. K. (2007). Nitrate transporters and peptide transporters. FEBS Lett. 581, 2290–2300. doi: 10.1016/j.febslet.2007.04.047
Tusnàdy, G. E., and Simon, I. (2001). The HMMTOP transmembrane topology prediction server. Bioinformatics 17, 849–850. doi: 10.1093/bioinformatics/17.9.849
Urbanski, D. F., Małolepszy, A., Stougaard, J., and Andersen, S. U. (2012). Genome-wide LORE1 retrotransposon mutagenesis and high-throughput insertion detection in Lotus japonicus. Plant J. 69, 731–741. doi: 10.1111/j.1365-313X.2011.04827.x
Valkov, V. T., and Chiurazzi, M. (2014). “Nitrate transport and signaling,” in The Lotus Japonicus Genome. eds. S. Tabata and J. Stougaard (Berlin, Heidelberg: Compendium of Plant GenomesSpringer-Verlag), 125–136.
Valkov, V. T., Rogato, A., Alves, M. L., Sol, S., Noguero, M., Léran, S., et al. (2017). LjNPF8.6 controls the N-fixing nodule activity in Lotus japonicus. Plant Physiol. 175, 1269–1282. doi: 10.1104/pp.17.01187
Valkov, V. T., Sol, S., Rogato, A., and Chiurazzi, M. (2020). The functional characterization of LjNRT2.4 indicates a novel, positive role of nitrate for an efficient nodule N2-fixation activity. New Phytol. 228, 682–696. doi: 10.1111/nph.16728
Villar, I., Larrainzar, E., Milazzo, L., Pérez-Rontomé, C., Rubio, M. C., Smulevich, G., et al. (2020). A plant gene encoding one-heme and two-heme hemoglobins with extreme reactivities towards diatomic gases and nitrite. Front. Plant Sci. 11:600336. doi: 10.3389/fpls.2020.600336
Wang, Y. Y., Cheng, Y. H., Chen, K. E., and Tsay, Y. F. (2018). Nitrate transport, signaling, and use efficiency. Annu. Rev. Plant Biol. 69, 85–122. doi: 10.1146/annurev-arplant-042817-040056
Wang, Q., Huang, Y., Ren, Z., Zhang, X., Ren, J., Su, J., et al. (2020). Transfer cells mediate nitrate uptake to control root nodule symbiosis. Nat. Plants 6, 800–808. doi: 10.1038/s41477-020-0683-6
Wang, C., Yu, H., Luo, L., Duan, L., Cai, L., He, X., et al. (2016). Nodules with activated defense 1 is required for maintenance of rhizobial endosymbiosis in Medicago truncatula. New Phytol. 212, 176–191. doi: 10.1111/nph.14017
Warner, C. A., Biedrzycki, M. L., Jacobs, S. S., Wisser, R. J., Caplan, J. L., and Sherrier, D. J. (2014). An optical clearing technique for plant tissues allowing deep imaging and compatible with fluorescence microscopy. Plant Physiol. 166, 1684–1687. doi: 10.1104/pp.114.244673
Waterworth, W. M., and Bray, C. M. (2006). Enigma variations for peptides and their transporters in higher plants. Ann. Bot. 98, 1–8. doi: 10.1093/aob/mcl099
Weber, E., Engler, C., Gruetzner, R., Werner, S., and Marillonnet, S. (2011). A modular cloning system for standardized assembly of multigene constructs. PLoS One 6:e16765. doi: 10.1371/journal.pone.0016765
Williams, L., and Miller, A. (2001). Transporters responsible for the uptake and partitioning of nitrogenous solutes. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 659–688. doi: 10.1146/annurev.arplant.52.1.659
Keywords: nitrate transport, N2-fixation, nodule, insertion mutants, Lotus japonicus
Citation: Vittozzi Y, Nadzieja M, Rogato A, Radutoiu S, Valkov VT and Chiurazzi M (2021) The Lotus japonicus NPF3.1 Is a Nodule-Induced Gene That Plays a Positive Role in Nodule Functioning. Front. Plant Sci. 12:688187. doi: 10.3389/fpls.2021.688187
Edited by:
Katharina Pawlowski, Stockholm University, SwedenReviewed by:
Anis M. Limami, Université d’Angers, FranceRebecca Dickstein, University of North Texas, United States
Antonio J. Márquez, Sevilla University, Spain
Copyright © 2021 Vittozzi, Nadzieja, Rogato, Radutoiu, Valkov and Chiurazzi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maurizio Chiurazzi, bWF1cml6aW8uY2hpdXJhenppQGliYnIuY25yLml0; Vladimir Totev Valkov, dmxhZGltaXIudmFsa292QGliYnIuY25yLml0
 Ylenia Vittozzi
Ylenia Vittozzi Marcin Nadzieja
Marcin Nadzieja Alessandra Rogato
Alessandra Rogato Simona Radutoiu
Simona Radutoiu Vladimir Totev Valkov
Vladimir Totev Valkov Maurizio Chiurazzi
Maurizio Chiurazzi