
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Plant Sci. , 17 June 2021
Sec. Plant Development and EvoDevo
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.687416
This article is part of the Research Topic Epigenetics in Plant Development View all 20 articles
Trimethylation of histone H3 lysine 27 (H3K27me3) is a highly conserved repressive histone modification that signifies transcriptional repression in plants and animals. In Arabidopsis thaliana, the demethylation of H3K27 is regulated by a group of JUMONJI DOMAIN-CONTANING PROTEIN (JMJ) genes. Transcription of JMJ genes is spatiotemporally regulated during plant development and in response to the environment. Once JMJ genes are transcribed, recruitment of JMJs to target genes, followed by demethylation of H3K27, is critically important for the precise control of gene expression. JMJs function synergistically and antagonistically with transcription factors and/or other epigenetic regulators on chromatin. This review summarizes the latest advances in our understanding of Arabidopsis H3K27me3 demethylases that provide robust and flexible epigenetic regulation of gene expression to direct appropriate development and environmental responses in plants.
Chromatin is critically important for gene expression during plant development and in response to the environment (Eccleston et al., 2013; Bruneau et al., 2019). Chromatin is composed of genomic DNA, histones, and accessory proteins, with approximately 150 base pairs of DNA wrapped around each octameric histone protein complex (Vergara and Gutierrez, 2017; van Steensel and Furlong, 2019). Each histone protein consists of a structural core at the C terminus and an unstructured tail domain at the N terminus. The N-terminal flexible histone tails often possess extensive posttranslational modifications, such as acetylation, methylation, and ubiquitination on lysine residues, methylation and citrullination on arginine residues, and phosphorylation of serine, threonine, and tyrosine residues. These modifications cause epigenetic changes in chromatin and lead to changes in gene expression.
One chromatin modification, trimethylation of histone H3 lysine 27 (H3K27me3), mediates epigenetic silencing of gene expression (Xiao and Wagner, 2015; Xiao et al., 2016). In general, H3K27me3 marks occur within facultative heterochromatin, in which gene expression is repressed but can be activated in response to developmental or environmental cues. In animals and plants, H3K27me3 deposition and removal are mediated by specific enzymes termed “writers” and “erasers”, respectively. Polycomb repressive complex 2 (PRC2), a multisubunit epigenetic repressor complex, writes H3K27me3 marks associated with gene repression. By contrast, histone demethylases, such as the Jumonji C (JmjC)-containing eraser demethylases, can demethylate H3K27me3 and thereby counteract the action of writer methylases (Crevillén, 2020). Understanding the role of JmjC-containing demethylases is crucial to understanding the effects of H3K27me3 in plant development and environmental responses. Although PRC2 and its actions have been reasonably well characterized through decades of research, existing knowledge about H3K27me3 demethylases and demethylation is relatively limited. In the last decade, however, research on H3K27me3 removal in Arabidopsis thaliana (Arabidopsis) has made great progress. To date, five JMJ proteins have been identified as H3K27me3 demethylases: EARLY FLOWERING 6 (ELF6)/JUMONJI DOMAIN-CONTAINING PROTEIN11 (JMJ11), RELATIVE OF ELF6 (REF6)/JMJ12, JMJ13, JMJ30, and JMJ32 (Lu F. et al., 2011; Crevillén et al., 2014; Gan et al., 2014; Cui et al., 2016; Yan et al., 2018). Here, we summarize current understanding of a group of JmjC-containing demethylases of H3K27me3, with emphasis on the most recent advances in knowledge.
Upon sensing developmental or environmental cues, JMJ proteins make genomic regions accessible by removing repressive H3K27me3 marks to generate a legible genome that is specific to a particular cell type, developmental stage, or environmental condition. Functional analysis of loss-of-function jmj mutants in Arabidopsis has indicated that JMJ proteins make major contributions to developmentally or environmentally triggered transcriptional reprogramming events. REF6, ELF6, and JMJ13 make a broader contribution to plant growth and development than JMJ30 and JMJ32, which play more specific and redundant roles in environmental responses.
The divergence in the biological roles of H3K27me3 demethylases might be due to their different spatial and temporal expression patterns. The spatial distribution of REF6, ELF6, JMJ13, JMJ30, and JMJ32 proteins was examined by introducing constructs harboring their upstream and coding sequences fused with sequences encoding the β-glucuronidase (GUS) reporter into wild-type plants (Noh et al., 2004; Gan et al., 2014; Zheng et al., 2019). Among these five GUS reporters, JMJ30-GUS highly accumulated in various plant organs, such as leaves, roots, and flowers (Gan et al., 2014). REF6-GUS, JMJ13-GUS, and JMJ32-GUS show moderate accumulation in young leaves near the shoot apical meristem and in root tips but lower accumulation in the leaf vasculature (Noh et al., 2004; Gan et al., 2014; Zheng et al., 2019). By contrast, ELF6-GUS accumulates only in the distal part of young leaves. Current spatial expression data were obtained mainly by whole-mount GUS staining. Our understanding of JMJ accumulation is still limited largely to the organ level. Expression analysis derived from GUS staining may not be precise, due to diffusion of the enzyme outside the tissue, as compared with fluorescent protein–based experiments.
Transcriptome data from publicly available databases increase the understanding of demethylase function in Arabidopsis and allow functions to be inferred. Shoot apex–specific RNA sequencing (RNA-seq) and cell type–specific single-cell (sc) RNA-seq data revealed different expression patterns for the six Arabidopsis H3K27me3 demethylase genes (Winter et al., 2007; Ryu et al., 2019; Tian et al., 2019). In the shoot apical meristem, JMJ30 is highly expressed, whereas the other genes are weakly expressed (Tian et al., 2019). Among eight different domains within the shoot apical meristem, JMJ30 expression is higher in the CLAVATA3 (CLV3) and ARABIDOPSIS THALIANA MERISTEM LAYER 1 (ATML1) expression domains (Lu et al., 1996; Brand et al., 2002). Because CLV3 and ATML1 are specifically expressed in the central zone and layer 1, respectively, these observations suggest that JMJ30 is also highly expressed in the center and/or epidermis of the shoot apical meristem. In the root, JMJ13, ELF6, JMJ30, and RFF6 are expressed in a cell type–specific manner (Ryu et al., 2019). High JMJ13 expression in the protoxylem suggests that it has a specific function in this tissue. These high-resolution differential expression patterns suggest that histone demethylation is tissue or cell type specific. Expression specificity at the cell-type or cellular levels needs to be characterized in detail to further our understanding of when and where JMJ proteins work.
Although epigenetic regulation is thought to be important in responses to environmental stimuli, few reports have described the relationship between environmental stress and the induction of JMJ genes. JMJ30 expression is further enhanced by the stress hormone abscisic acid (ABA) and by salt stress, drought stress, and heat stress compared to control conditions (Qian et al., 2015; Wu et al., 2019a; Yamaguchi et al., 2020). ABA treatment triggers a rapid increase in JMJ30 protein levels but does not change the area of JMJ30 expression, based on whole-mount GUS staining (Wu et al., 2019a). The expression of JMJ13 is affected by light and temperature conditions, according to GUS expression data (Zheng et al., 2019). REF6 expression is induced by long-term heat exposure (Liu et al., 2019). To date, no effects of environmental stress on the regulation of ELF6 and JMJ32 expression have been reported. Bulk transcriptome datasets also largely support these results (Qian et al., 2015). Expression specificity and subcellar localization of JMJ proteins in response to environmental stimuli should also be addressed with higher resolution in the future.
The seeds of the ref6 mutant germinate later than wild type (Li et al., 2016; Chen et al., 2020). REF6 induces two key genes for ABA catabolism, CYP707A1 and CYP707A3, through removal of H3K27me3. CYP707A1 and CYP707A2 encode ABA 8′-hydeoxylases and play key roles in reducing ABA levels (Okamoto et al., 2006). Overexpression of CYP707A1 by cauliflower mosaic virus (CaMV) 35S promoter rescues the dormancy phenotype of the ref6 mutant (Chen et al., 2020). The jmj30 jmj32 double mutant, by contrast, shows no difference in seed dormancy phenotype from wild type (Wu et al., 2019a).
Under normal growth conditions, ref6 and elf6 mutants have similar leaf phenotypes that include reduced petiole length, which is characteristic of brassinosteroid (BR)-defective mutants (Yu et al., 2008). A shorter leaf blade is seen in ref6 but not elf6 plants, suggesting that the REF6 and ELF6 proteins have tissue-specific roles. The ref6 mutation further enhances the phenotype of a BR-deficient mutant. In the ref6 elf6 double mutant, expression of BR-regulated genes, such as TOUCH 4 (TCH4), is reduced. Later in leaf development, ref6 delays chlorophyll degradation (Wang et al., 2019) and REF6 promotes general leaf senescence by directly activating senescence-related genes, including ETHYLENE INSENSITIVE 2 (EIN2), OLEOSIN 1 (ORE1), and NONYELLOWING genes (NYEs). The jmj13 single mutant does not display detectable abnormalities in leaf phenotype (Zheng et al., 2019), but the ref6 elf6 jmj13 triple mutant has shorter petioles than ref6 elf6, suggesting that REF6, ELF6, and JMJ13 are essential developmental regulators (Yan et al., 2018). The jmj30 jmj32 double mutant, by contrast, shows no difference in leaf phenotype from wild type (Yamaguchi et al., 2020).
All five Arabidopsis H3K27me3 demethylases regulate flowering time, but in distinct fashions. REF6 and ELF6 were originally identified on the basis of their influence on flowering-time phenotypes: under long-day conditions, ref6 mutants are late flowering and elf6 and jmj13 mutants are early flowering (Noh et al., 2004; Zheng et al., 2019). REF6 directly induces floral activator genes, such as SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1) and FRUITFULL (FUL) (Hou et al., 2014; Hyun et al., 2016). ELF6 binds to the regulatory region of the floral repressor gene FLOWERING LOCUS C (FLC) for transcriptional activation (Yang et al., 2016). In jmj13 mutants, the floral repressor gene SHORT VEGETATIVE PHASE (SVP) is downregulated. By contrast, flowering-time defects in jmj30 jmj32 are observed only at high ambient temperatures but not under long-day conditions (Gan et al., 2014; Yan et al., 2014). Thus, the H3K27me3 demethylases REF6, ELF6, JMJ13, JMJ30, and JMJ32 show differences as well as similarities in how they influence flowering time.
Differences are also observed between the phenotypes of elf6 and jmj13 during flower development (Keyzor et al., 2021). In wild type and the ref6 mutant, the initial one or two flowers do not undergo self-pollination and form very short fruits without seeds; elf6 plants display increased self-fertility and consistent fruit production. Conversely, the jmj13 mutant shows reduced fertility and gives rise to aborted fruits up to the eighth flower on the primary inflorescence. JASMONATE-ZIM-DOMAIN PROTEIN 7 (JAZ7), SMALL AUXIN UP RNA 26 (SAUR26) and ARABINOGALACTAN PROTEINs (AGPs) are downregulated in jmj13 buds. No defects in floral developmental have been reported for jmj30 and jmj32 mutants.
The functions of JMJ30 and JMJ32 appear to be relatively distinct from those of REF6, ELF6, and JMJ13. jmj30 mutants show a circadian phenotype (Jones et al., 2010), and the JMJ30 gene was originally identified due to its co-expression with TIMING OF CAB1 EXPRESSION 1 (TOC1). Consistent with the circadian oscillation in JMJ30 expression, circadian rhythms in reporter-gene activity in jmj30 mutants are significantly shorter than those in wild type. JMJ30 and TOC1 interact genetically to promote the expression of CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY). By contrast, no circadian oscillation in JMJ32 expression is observed (Lu S. X. et al., 2011), suggesting that JMJ30 and JMJ32 are regulated by distinct mechanisms.
jmj30 mutants also feature phenotypes that are dependent on environmental conditions. Callus formation induced by incubating leaf explants on callus-inducing medium is reduced in jmj30 mutants. JMJ30 promotes the expression of LATERAL ORGAN BOUNDARIES DOMAIN 16 (LBD16) and LBD29 to establish root primordium–like unorganized cell masses (Lee et al., 2018). Furthermore, stress hormone–induced growth arrest is compromised in jmj30 jmj32 double mutants (Wu et al., 2019a, b, 2020), and JMJ30 directly activates SNF1-RELATED PROTEIN KINASE 2.8 (SnRK2.8) and BRASSINAZOLE RESISTANT1 (BZR1) to maintain a balance between stress responses and growth. Acquired thermotolerance is also reduced in jmj30 jmj32 ref6 elf6 quadruple mutants (Yamaguchi et al., 2020). JMJ30 binds to HEAT SHOCK PROTEIN 17.6C (HSP17.6C) and HSP22 and activates their transcription in response to heat.
Although the interactions between JMJ proteins and downstream targets is regulated in a spatiotemporal manner, it is not yet known how exactly JMJ proteins lead to H3K27me3 removal. Most phenotyping has been conducted in knock-out or knock-down mutants, while mutant rescue by expressing downstream targets has used CaMV35S-based overexpression lines (Wu et al., 2019a; Chen et al., 2020; Yamaguchi et al., 2020). To assess the precise roles of JMJ during plant development and environmental responses, conditional jmj mutants should be employed. Furthermore, organ-, tissue-, or cell type–specific phenotypic rescues using appropriate promoters are required to understand when and where JMJ proteins function.
Phylogenetic analysis of JmjC-containing demethylases defined 14 subfamilies and identified more than 10 members in land plants. Green algae such as Chlamydomonas and Volvox include only two members of this family, implying that the functions of JMJ proteins may have been important for plant adaptation to land (Qian et al., 2015). Arabidopsis contains 21 JMJ proteins (Lu et al., 2008). Although not all family members have been fully characterized, they include putative H3K9me3-, H3K36me3-, H3K4me3-, and H3K27me3-specific demethylases. Additional H3K27me3 demethylases may also exist. ELF6 and REF6 show highest sequence similarities to the H3K9me3- and H3K36me3-specific KMD4 demethylases. The precise functions of the remaining JMJ proteins need to be carefully examined in a manner that is unbiased by sequence similarity.
The ELF6, REF6, and JMJ13 proteins belong to the plant-specific KMD4 subfamily, which is present in land plants but not in green algae (Lu et al., 2008; Qian et al., 2015). REF6 contains JmjN, JmjC, and C2H2-type zinc-finger (ZnF) domains (Figure 1A). The REF6 protein characteristically possesses four tandem repeats of the ZnF domain. These domains are essential for REF6 function, as complementation of the ref6 mutant through the introduction of REF6 without a ZnF domain fails to rescue the ref6 mutant phenotype. Some histone demethylases, such as KDM2, interact with chromatin via direct binding to DNA through ZnF domains. ZnF is one of the largest class of DNA-binding domains. Consistent with this, REF6 functions as a DNA sequence–specific H3K27me3 demethylase. Genome-wide REF6 binding studies and crystal structure analysis revealed that the ZnF domains of REF6 recognize the CTCTGYTY DNA motif for H3K27me3 removal (Figures 1B, 2A) (Cui et al., 2016; Li et al., 2016; Tian et al., 2020). The ZnF domains of REF6 complex with NAC004 double-stranded (ds) DNA by forming a half-cross-braced structure (Figure 1B). Interactions at the interface between REF6 and dsDNA, such as hydrogen bonds, electrostatic interactions, and hydrophobic interactions, strengthen their binding. dsDNA binding induces profound conformation changes (Tian et al., 2020). Conformational plasticity of DNA allows REF6 to recognize diverse target genes.
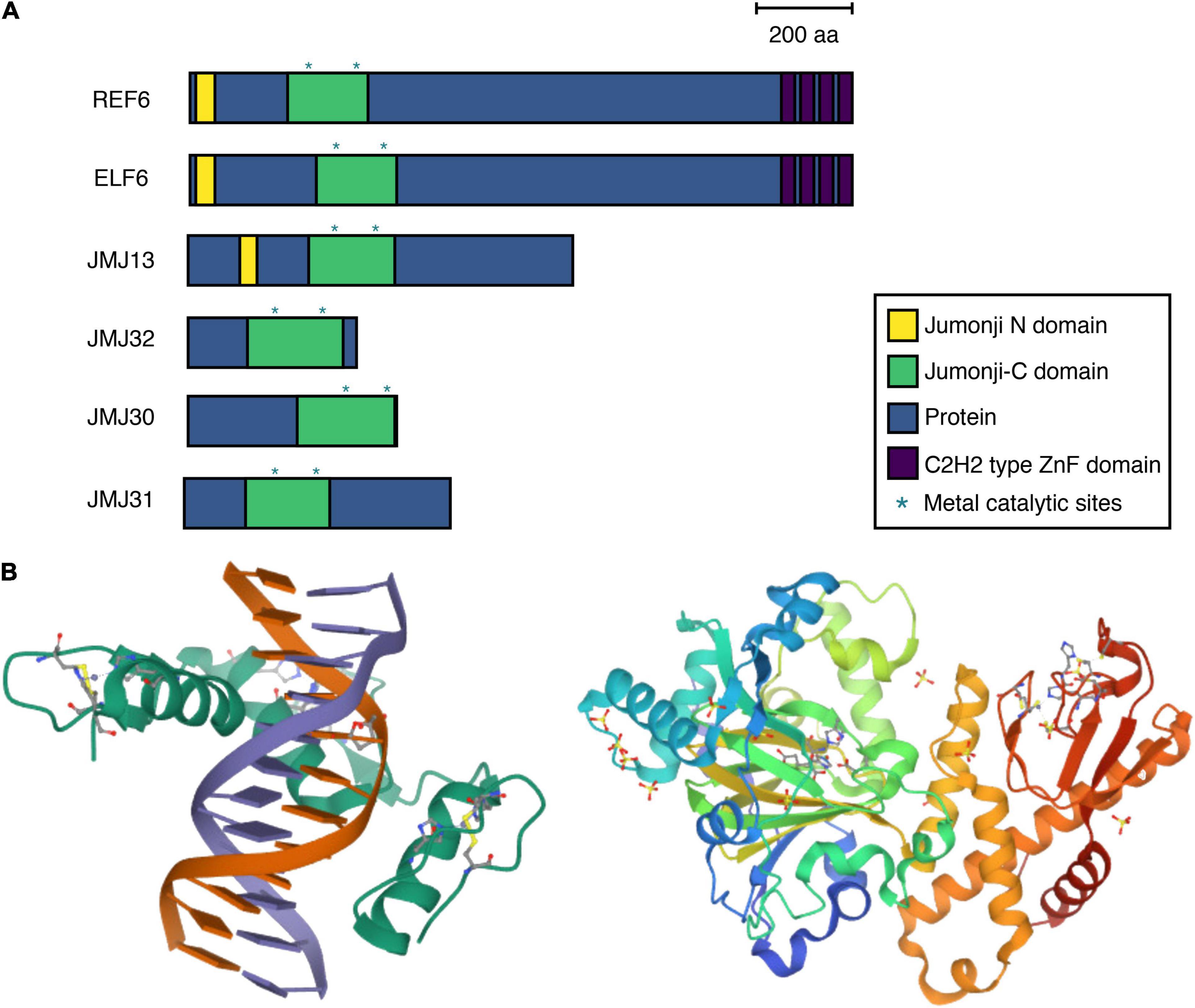
Figure 1. Conserved motifs, and three-dimensional structure of H3K27me3 demethylases in Arabidopsis thaliana. (A) The domain structures of Jumonji domain–containing proteins (JMJs) in A. thaliana. The positions of the Jumonji N domain, Jumonji C domain, protein, and C2H2-type zinc-finger (ZnF) domains are indicated in yellow, green, blue, and purple, respectively; metal catalytic sites are indicated with asterisks. Scale bar = 200 amino acids. (B) Crystal structure of JMJ proteins. Left REF6 ZnF and NAC004-mC3 double-stranded (ds) DNA. Ribbon representation of REF6-DNA structure. REF6 protein is shown in green, while dsDNA is shown in orange and purple. α-helices and β-sheets are represented by spiral ribbons and green arrows, respectively. A few residues engage Zn2+ ion. Right The JMJ13 catalytic domain in complex with AKG. Ribbon representation of JMJ13 structure. Blue and green ribbons represent JMJ domains. Orange and red ribbons show helical and ZnF domains, respectively. The data were obtained from the Protein Data Bank (https://biorender.com).
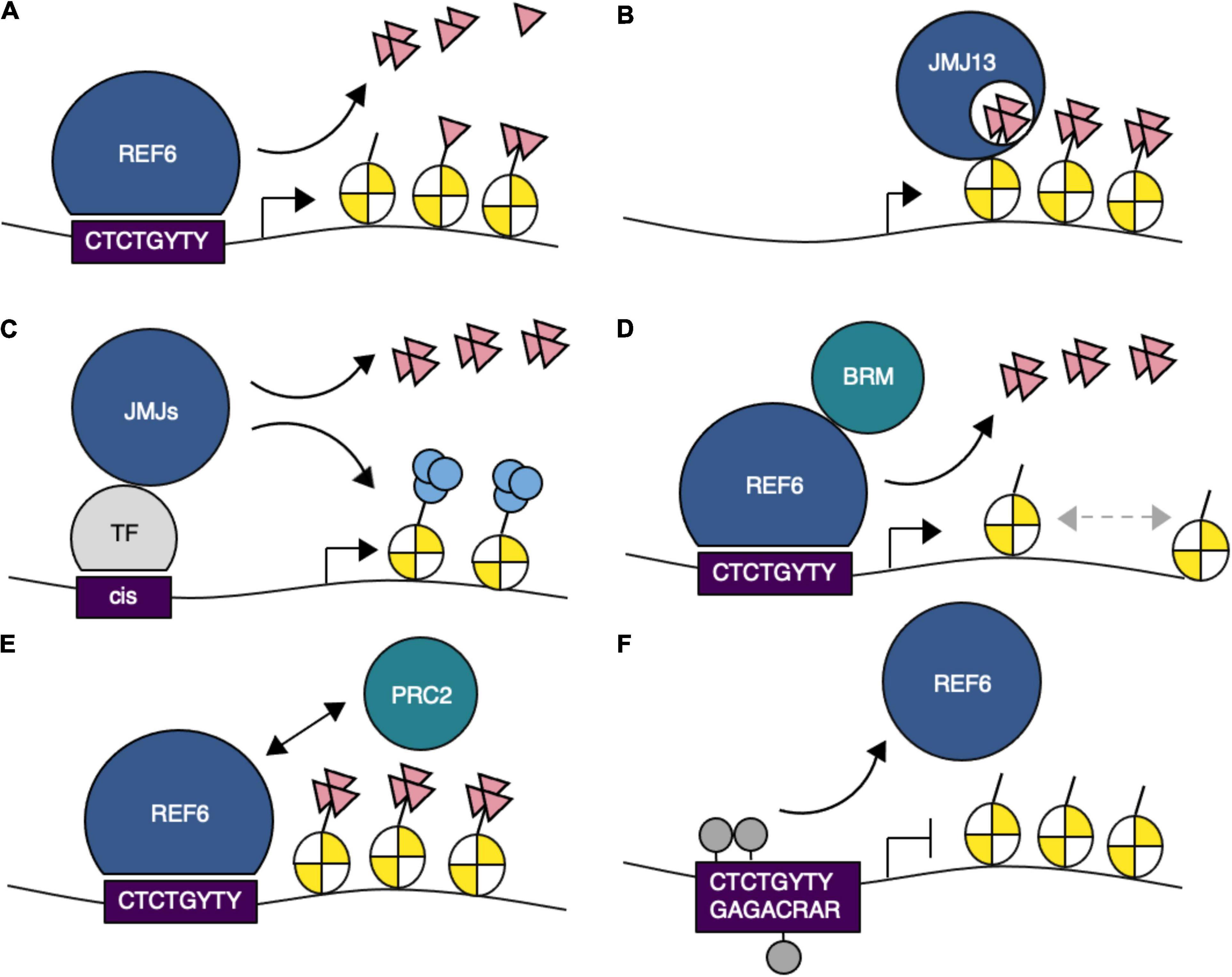
Figure 2. Schematic representation of the function of H3K27me3 demethylases in gene regulation. (A) REF6 demethylates H3K27me1/2/3 via recognition of CTCTGYTY (where Y is C or T) DNA motifs. (B) JMJ13 recognizes H3K27me3 marks. (C) JMJ proteins interact with transcription factors and are recruited to their target sites. (D) REF6 interacts with BRAHMA (BRM) and mediates nucleosome positioning. (E) REF6 prevents the uncontrolled spreading of PRC2-mediated chromatin silencing. (F) DNA methylation at CTCTGYTY motifs prevents REF6 targeting. H3K27me3, pink triangles; H3K9ac, light blue circles; DNA methylation, gray circles.
EARLY FLOWERING 6 is the closest homolog of REF6. Those two proteins share a high sequence similarity. How ELF6 recognizes DNA is currently unknown. Because the mutant phenotypes of elf6 and ref6 differ, the recognition motifs or mechanisms might also differ between REF6 and ELF; a recent study showed that REF6 and ELF6 play distinct roles in H3K27me3 and H3K27me1 homeostasis (Antunez-Sanchez et al., 2020). ELF6 regulates a small subset of genes compared to REF6. This could be due to less protein structural plasticity of ELF6 and/or a difference in DNA-binding affinity. Further analysis, such as determination of crystal structures of ELF6-DNA complexes, is required to reveal the structural basis for the epigenetic modification recognition.
Although JMJ13, like REF6, belongs to the KMD4 subfamily and functions to remove H3K27me3 in vitro and in vivo, it possesses a different DNA-recognition mechanism. JMJ13 does not contain ZnF domains at the C terminus (Figure 1A); instead, its catalytic domain (JMJ13CD) contains Jmj and helical domains, as well as a unique C4HCHC-type ZnF domain. Crystal structure analysis using JMJ13CD revealed that JMJ13 recognizes the H3K27me3 peptide and functions as a reader of the histone modification state (Figures 1B, 2B) (Zheng et al., 2019). The interactions between JMJ13 and the H3K27me3 peptide are restricted to the region between H3R26 and H3P30. Because other JMJ proteins are predicted to possess different putative ZnF domains, such as the C5HC2-type, the recognition of histone modifications by ZnF domains located within the JmjC domain might also occur for other JMJ proteins.
JMJ30 and JMJ32, as well as their close homolog JMJ31, belong to the JmjC-domain-only group. Consistent with their domain structure, two clades of protein homology are identified by phylogenetic analysis: one contains ELF6, REF6, and JMJ13 and the other contains JMJ30, JMJ31, and JMJ32 (Lu et al., 2008; Qian et al., 2015). Although the function and regulation of JMJ30 and JMJ32 are relatively well characterized, nothing is known about their three-dimensional protein structure or DNA–histone recognition mechanisms. As is often observed for REF6 and ELF6 recruitment (Figure 2C), JMJ30 physically interacts with tissue-specific transcription factors, such as EARLY FLOWERING MYB PROTEIN (EFM) and AUXIN RESPONSE FACTORs (ARFs) (Yan et al., 2014; Lee et al., 2018). Furthermore, JMJ30 activity affects H3K9me3 and H3K36me3 in addition to H3K27me3. Further experiments, such as JMJ30 chromatin immunoprecipitation and deep sequencing (ChIP-seq), are required to precisely understand their biochemical functions.
Genome-wide JMJ protein binding and histone modification data were obtained by ChIP-seq. Since ChIP-seq assays require large numbers of input cells/tissues, whole plants are often used for the assays. Hence, spatial information is completely lost. Recently, low-input binding tests in plants, such as CUT&Tag, CUT&RUN, nCUT&Tag, and ChIL, have been developed to study interactions between DNA and proteins using low-input samples or single live cells (Zheng and Gehring, 2019; Sakamoto et al., 2020; Tao et al., 2020; Ouyang et al., 2021). By combining these with cell sorting or laser microdissection techniques, cell type–specific JMJ protein binding and histone modification data can be obtained in the future. These analyses may contribute to our understanding of the precise spatiotemporal regulation of H3K27me3 demethylation.
Genome-wide binding analysis coupled with immunoprecipitation and mass spectrometry (IP-MS) identified an interaction between REF6 and the SWI/SNF-type chromatin remodeling ATPase BRAHMA (BRM) on chromatin in vivo (Li et al., 2016). REF6 and BRM bind to many common genomic loci that contain CTCTGYTY motifs. Recruitment of BRM to target loci is dependent on REF6 function, but REF6 does not require BRM activity for its own targeting. Thus, REF6 directly binds to chromatin containing the CTCTGYTY motifs and subsequently recruits BRM to activate targets, potentially through changes in nucleosome position (Figure 2D).
An antagonistic role between REF6 or BRM and PRC2 at their target loci has been demonstrated (Bezhani et al., 2007; Lu F. et al., 2011; Wu et al., 2012; Li et al., 2015). Antagonism is often mediated by competitive binding at the same sites on chromatin (Zhu et al., 2020). However, PRC2 preferentially binds to different motifs, such as the telobox and GAGA motifs (Hecker et al., 2015; Xiao et al., 2017; Zhou et al., 2018), suggesting that competitive antagonism is unlikely to occur between REF6/BRM and PRC2; moreover, the binding patterns of PRC2 and REF6 do not overlap. REF6 is localized to the boundaries of H3K27me3 regions, which are covered by PRC2 (Yan et al., 2018) (Figure 2E). The spreading of H3K27me3 observed in ref6 elf6 jmj13 triple mutants indicates that the function of REF6 binding inhibits the spreading of H3K27me3, but how ELF6 and JMJ13 contribute to preventing this spreading remains unclear.
Recognition of dsDNA by REF6 not only relies on DNA sequence but also is affected by DNA methylation and sequence-dependent conformations of DNA (Qiu et al., 2019). REF6 preferentially binds to hypomethylated CTCTGYTY motifs. Methylation of CHG within the motif attenuates REF6-binding affinity (Figure 2F), and the minor groove width of each nucleotide in the structure of the complex differs considerably. This difference affects recognition of the CTCTGYTY motifs by REF6 and its binding affinity in CUP-SHAPED COTYLEDON 1 (CUC1) and CUC2 (Tian et al., 2020). The possibility that factors other than DNA sequence may contribute to REF6 binding affinity is supported by the fact that REF6 recognizes only 15% of the CTCTGYTY motifs in the Arabidopsis genome.
Protein–protein interaction between JMJ proteins and other transcription/chromatin factors are critical for H3K27me3 removal. However, conclusive in vivo evidence of when and where exactly those factors interact each other is lacking. Innovative in vivo imaging techniques are used to understand plant development and environmental responses through spatiotemporal regulation of gene expression (Abe et al., 2019; Hirakawa et al., 2019). Application of these techniques in H3K27me3 demethylase research to reveal the distribution of JMJ protein complexes will provide new insights into the spatiotemporal regulation of H3K27me3 removal.
Flexible and robust gene expression during plant development and in response to the environment is primarily controlled by epigenetic regulation. In the past 5 years, plant epigenetic research on demethylases using transcriptome, epigenome, and crystal structure analyses has revealed the importance of H3K27me3. In Arabidopsis thaliana, the demethylation of H3K27 is regulated by a group of JUMONJI DOMAIN-CONTANING PROTEIN (JMJ) genes. JMJ30 expression is high in various organs and is further boosted in response to environmental cues. On the other hand, the expression levels of REF6, ELF6, JMJ13, and JMJ32 is moderate. REF6 and JMJ13 expression is also affected by environmental cues. These H3K27me3 demethylases bind to chromatin through generic or sequence-specific targeting mechanisms: direct binding to DNA via a ZnF domain, direct recognition of H3K27me3, or indirect binding through interactions with transcription factors. DNA methylation and minor groove width also fine-tune the binding affinity of these H3K27me3 demethylases. The targeting and occupancy of the histone demethylases on chromatin antagonize PRC2-mediated H3K27me3 deposition, and the removal of histone demethylases and prevention of their uncontrolled spread determine the shape of the H3K27me3 peak. Subsequently, H3K27me3 demethylases recruit a chromatin remodeler to activate gene transcription. One major limitation in current epigenome research is the scarcity of spatial information concerning the binding of epigenetic regulators and the nature of epigenetic modifications and co-factors. Furthermore, when and where target expression by H3K27me3 demethylases is mediated are poorly understood. Both binding patterns and DNA–protein structures and/or co-factors might vary among cells, tissues, and organs. In addition, growth conditions affect the epigenomic dynamics among individual plants. Therefore, specific genomic profiles obtained from plants grown under different conditions are needed to understand the specific roles of H3K27me3 demethylases during plant development and responses to the environment.
NY conceptualization and writing the manuscript.
This work was supported by a grant from the Japan Science and Technology Agency “PREST” (JPMJPR15QA), a JSPS KAKENHI Grant-in-Aid for Scientific Research on Innovative Areas (No. 18H04782), a JSPS KAKENHI Grant-in-Aid for Scientific Research B (No. 18H02465), a Grant-in-Aid for challenging Exploratory Research (No. 19K22431), and a grant from the SECOM Science and Technology Foundation to NY.
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author thank Sachi Ando for critical comments on this manuscript.
Abe, M., Kosaka, S., Shibuta, M., Nagata, K., Uemura, T., Nakano, A., et al. (2019). Transient activity of the florigen complex during the floral transition in Arabidopsis thaliana. Development 146:dev171504. doi: 10.1242/dev.171504
Antunez-Sanchez, J., Naish, M., Ramirez-Prado, J. S., Ohno, S., Huang, Y., Dawson, A., et al. (2020). A new role for histone demethylases in the maintenance of plant genome integrity. Elife 27:e58533. doi: 10.7554/eLife.58533
Bezhani, S., Winter, C., Hershman, S., Wanger, J. D., Kennedy, J. F., Kwan, C. S., et al. (2007). Unique, shared, and redundant roles for the Arabidopsis SWI/SNF chromatin remodeling ATPases BRHAMA and SPLAYED. Plant Cell 19, 403–416. doi: 10.1105/tpc.106.048272
Brand, U., Grunewald, M., Hobe, M., and Simon, R. (2002). Regulation of CLV3 expression by two homeobox genes in Arabidopsis. Plant Physiol. 129, 565–575. doi: 10.1104/pp.001867
Bruneau, B. G., Koseki, H., Strome, S., and Torres-Padilla, M.-E. (2019). Chromatin and epigenetics in development: a Special Issue. Development 146:5025. doi: 10.1242/dev.185025
Chen, H., Tong, J., Fu, W., Liang, Z., Ruan, J., Yu, Y., et al. (2020). The H3K27me3 Demethylase RELATIVE OF EARLY FLOWERING6 Suppresses Seed Dormancy by Inducing Abscisic Acid Catabolism. Plant Physiol. 184, 1969–1978. doi: 10.1104/pp.20.01255
Crevillén, P. (2020). Histone demethylases as counterbalance to H3K27me3 silencing in plants. iScience 23:101715. doi: 10.1016/j.isci.2020.101715
Crevillén, P., Yang, H., Cui, X., Greeff, C., Trick, M., Qiu, Q., et al. (2014). Epigenetic reprogramming that prevents transgenerational inheritance of the vernalized state. Nature 515, 587–590. doi: 10.1038/nature13722
Cui, X., Lu, F., Qiu, Q., Zhou, B., Gu, L., Zhang, S., et al. (2016). REF6 recognizes a specific DNA sequence to demethylate H3K27me3 and regulate organ boundary formation in Arabidopsis. Nat. Genet. 48, 694–699. doi: 10.1038/ng.3556
Eccleston, A., Cesari, F., and Skipper, M. (2013). Transcription and epigenetics. Nature 502:461. doi: 10.1038/502461a
Gan, E. S., Xu, Y., Wong, J. Y., Goh, J. G., Sun, B., Wee, W. Y., et al. (2014). Jumonji demethylases moderate precocious flowering at elevated temperature via regulation of FLC in Arabidopsis. Nat. Commun. 5:5098. doi: 10.1038/ncomms6098
Hecker, A., Brand, L. H., Peter, S., Simoncello, N., Kilian, J., Harter, K., et al. (2015). The Arabidopsis GAGA-Binding Factor BASIC PENTACYSTEINE6 Recruits the POLYCOMB-REPRESSIVE COMPLEX1 Component LIKE HETEROCHROMATIN PROTEIN1 to GAGA DNA Motifs. Plant Physiol. 168, 1013–1024. doi: 10.1104/pp.15.00409
Hirakawa, T., Kuwata, K., Gallego, M. E., White, C. I., Nomoto, M., Tada, Y., et al. (2019). LSD1-LIKE1-Mediated H3K4me2 Demethylation Is Required for Homologous Recombination Repair. Plant Physiol. 181, 499–509. doi: 10.1104/pp.19.00530
Hou, X., Zhou, J., Liu, C., Liu, L., Shen, L., and Yu, H. (2014). Nuclear factor Y-mediated H3K27me3 demethylation of the SOC1 locus orchestrates flowering responses of Arabidopsis. Nat. Commun. 5:4601. doi: 10.1038/ncomms5601
Hyun, Y., Richter, R., Vincent, C., Martinez-Gallegos, R., Porri, A., and Coupland, G. (2016). Multi-layered regulation of SPL15 and cooperation with SOC1 integrate endogenous flowering pathways at the Arabidopsis shoot meristem. Dev. Cell 37, 1–13. doi: 10.1016/j.devcel.2016.04.001
Jones, M. A., Covington, M. F., DiTacchio, L., Vollmers, C., Panda, S., and Harmer, S. L. (2010). Jumonji domain protein JMJD5 functions in both the plant and human circadian systems. Proc. Natl. Acad. Sci. U. S. A 107, 21623–21628. doi: 10.1073/pnas.1014204108
Keyzor, C., Mermaz, B., Trigazis, E., Jo, S., and Song, J. (2021). Histone demethylases ELF6 and JMJ13 antagonistically regulate self-fertility in Arabidopsis. Front. Plant Sci. 12:640135. doi: 10.3389/fpls.2021.640135
Lee, K., Park, O., and Seo, P. J. (2018). JMJ30-mediated H3K9me3 demethylation drives tissue identity changes to promote callus formation in Arabidopsis. Plant J. 95, 961–975. doi: 10.1111/tpj.14002
Li, C., Chen, C., Gao, L., Yang, S., Nguyen, V., Shi, X., et al. (2015). The Arabidopsis SWI2/SNF2 chromatin remodeler BRAHMA regulates Polycomb function during vegetative development and directly activates the flowering repressor gene SVP. PLoS Genet 11:e1004944. doi: 10.1371/journal.pgen.1004944
Li, C., Gu, L., Gao, L., Chen, C., Wei, C.-Q., Qiu, Q., et al. (2016). Concerted genomic targeting of H3K27 demethylase REF6 and chromatin remodeling ATPase BRM in Arabidopsis. Nat. Genet. 48, 687–693. doi: 10.1038/ng.3555
Liu, J., Feng, L., Gu, X., Deng, X., Qiu, Q., Li, Q., et al. (2019). An H3K27me3 demethylase-HSFA2 regulatory loop orchestrates transgenerational thermomemory in Arabidopsis. Cell Res. 29, 379–390. doi: 10.1038/s41422-019-0145-8
Lu, F., Cui, X., Zhang, S., Jenuwein, T., and Cao, X. (2011). Arabidopsis REF6 is a histone H3 lysine 27 demethylase. Nat. Genet. 43, 715–719. doi: 10.1038/ng.854
Lu, S. X., Knowles, S. M., Webb, C. J., Celaya, R. B., Cha, C., Siu, J. P., et al. (2011). The Jumonji C domain-containing protein JMJ30 regulates period length in the Arabidopsis circadian clock. Plant Physiol. 155, 906–915. doi: 10.1104/pp.110.167015
Lu, F., Li, G., Cui, X., Liu, C., Wang, X. J., and Cao, X. (2008). Comparative analysis of JmjC domain-containing proteins reveals the potential histone demethylases in Arabidopsis and rice. J. Integr. Plant Biol. 50, 886–896. doi: 10.1111/j.1744-7909.2008.00692.x
Lu, P., Porat, R., Nadeau, J. A., and O’Neill, S. D. (1996). Identification of a meristem L1 layer-specific gene in Arabidopsis that is expressed during embryonic pattern formation and defines a new class of homeobox genes. Plant Cell 8, 2155–2168. doi: 10.1105/tpc.8.12.2155
Noh, B., Lee, S., Kim, H., Yi, G., Shin, E., Lee, M., et al. (2004). Divergent roles of a pair of homologous jumonji/zinc-finger-class transcription factor proteins in the regulation of Arabidopsis flowering time. Plant Cell 16, 2601–2613. doi: 10.1105/tpc.104.025353
Okamoto, M., Kuwahara, A., Seo, M., Kushiro, T., Asami, T., Hirai, N., et al. (2006). CYP707A1 and CYP707A2, which encode abscisic acid 8’-hydroxylases, are indispensable for proper control of seed dormancy and germination in Arabidopsis. Plant Physiol. 141, 97–107. doi: 10.1104/pp.106.079475
Ouyang, W., Zhang, X., Peng, Y., Zhang, Q., Cao, Z., Li, G., et al. (2021). Rapid and Low-Input Profiling of Histone Marks in Plants Using Nucleus CUT&Tag. Front Plant Sci. 12, 634679. doi: 10.3389/fpls.2021.634679
Qian, S., Wang, Y., Ma, H., and Zhang, L. (2015). Expansion and Functional Divergence of Jumonji C-Containing Histone Demethylases: Significance of Duplications in Ancestral Angiosperms and Vertebrates. Plant Physiol. 168, 1321–1337. doi: 10.1104/pp.15.00520
Qiu, Q., Mei, H., Deng, X., He, K., Wu, B., Yao, Q., et al. (2019). DNA methylation repels targeting of Arabidopsis REF6. Nat. Commun. 10:2063. doi: 10.1038/s41467-019-10026-1
Ryu, L. H., Huang, L., Kang, H. M., and Schiefelbein, J. (2019). Single-cell RNA sequencing resolves molecular relationships among individual plant cells. Plant Physiol. 179, 1444–1456. doi: 10.1104/pp.18.01482
Sakamoto, Y., Sato, M., Sato, Y., Harada, A., Suzuki, T., Goto, C., et al. (2020). Subnuclear gene positioning through lamina association affects copper tolerance. Nat. Commun. 11:5914. doi: 10.1038/s41467-020-19621-z
Tao, X., Feng, S., Zhao, T., and Guan, X. (2020). Efficient chromatin profiling of H3K4me3 modification in cotton using CUT&Tag. Plant Methods 16:120. doi: 10.1186/s13007-020-00664-8
Tian, C., Wang, Y., Yu, H., He, J., Wang, J., Shi, B., et al. (2019). A gene expression map of shoot domains reveals regulatory mechanisms. Nat. Commun. 10:141. doi: 10.1038/s41467-018-08083-z
Tian, Z., Li, X., Li, M., Wu, W., Zhang, M., Tang, C., et al. (2020). Crystal structures of REF6 and its complex with DNA reveal diverse recognition mechanisms. Cell Discov. 6:17. doi: 10.1038/s41421-020-0150-6
van Steensel, B., and Furlong, E. E. M. (2019). The role of transcription in shaping the spatial organization of the genome. Nat. Rev. Mol. Cell Biol. 20, 327–337. doi: 10.1038/s41580-019-0114-6
Vergara, Z., and Gutierrez, C. (2017). Emerging roles of chromatin in the maintenance of genome organization and function in plants. Genome Biol. 18:96. doi: 10.1186/s13059-017-1236-9
Wang, X., Gao, J., Gao, S., Song, Y., Yang, Z., and Kuai, B. (2019). The H3K27me3 demethylase REF6 promotes leaf senescence through directly activating major senescence regulatory and functional genes in Arabidopsis. PLoS Genet. 15:1–24.
Winter, D., Venegar, B., Nahai, H., Ammar, R., Wilson, G. V., and Provart, N. J. (2007). An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One 2:e718. doi: 10.1371/journal.pone.0000718
Wu, J., Ichihashi, Y., Suzuki, T., Shibata, A., Shirasu, K., Yamaguchi, N., et al. (2019a). Abscisic acid-dependent histone demethylation during post-germination growth arrest in Arabidopsis. Plant Cell Environ. 42, 2198–2214. doi: 10.1371/journal.pgen.1008068
Wu, J., Yamaguchi, N., and Ito, T. (2019b). Histone demethylases control root elongation in response to stress-signaling hormone abscisic acid. Plant Signal. Behav. 14:1604019. doi: 10.1080/15592324.2019.1604019
Wu, J., Yan, M., Zhang, D., Zhou, D., Yamaguchi, N., and Ito, T. (2020). Histone demethylases coordinate the antagonistic interaction between abscisic acid and brassinosteroid signaling in Arabidopsis. Front. Plant Sci. 11:596835. doi: 10.3389/fpls.2020.596835
Wu, M. F., Sang, Y., Bezhani, S., Yamaguchi, N., Han, S. K., Li, Z., et al. (2012). SWI2/SNF2 chromatin remodeling ATPases overcome polycomb repression and control floral organ identity with the LEAFY and SEPALLATA3 transcription factors. Proc. Natl. Acad. Sci. U. S. A. 109, 3576–3581. doi: 10.1073/pnas.1113409109
Xiao, J., Jin, R., Yu, X., Shen, M., Wagner, J. D., Pai, A., et al. (2017). Cis and trans determinants of epigenetic silencing by Polycomb repressive complex 2 in Arabidopsis. Nat. Genet. 49, 1546–1552. doi: 10.1038/ng.3937
Xiao, J., Lee, U. S., and Wagner, D. (2016). Tug of war: adding and removing histone lysine methylation in Arabidopsis. Curr. Opin. Plant Biol. 34, 41–53. doi: 10.1016/j.pbi.2016.08.002
Xiao, J., and Wagner, D. (2015). Polycomb repression in the regulation of growth and development in Arabidopsis. Curr. Opin. Plant Biol. 23, 15–24. doi: 10.1016/j.pbi.2014.10.003
Yamaguchi, N., Matsubara, S., Yoshimizu, K., Seki, M., Hamada, K., Kamitani, M., et al. (2020) H3K27me3 demethylases alter HSP22 and HSP17.6C expression in response to recurring heat in Arabidopsis. Nat. Commun. (in press). doi: 10.1038/s41467-021-23766-w
Yan, W., Chen, D., Smaczniak, C., Engelhorn, J., Liu, H., Yang, W., et al. (2018). Dynamic and spatial restriction of Polycomb activity by plant histone demethylases. Nat. Plants 4, 681–689. doi: 10.1038/s41477-018-0219-5
Yan, Y., Shen, L., Chen, Y., Bao, S., Thong, Z., and Yu, H. (2014). A MYB-domain protein EFM mediates flowering responses to environmental cues in Arabidopsis. Dev. Cell 30, 437–448. doi: 10.1016/j.devcel.2014.07.004
Yang, H., Howard, M., and Dean, C. (2016). Physical coupling of activation and derepression activities to maintain an active transcriptional state at FLC. Proc. Natl. Acad. Sci. U. S. A. 113, 9369–9374. doi: 10.1073/pnas.1605733113
Yu, X., Li, L., Li, L., Guo, M., Chory, J., and Yin, Y. (2008). Modulation of brassinosteroid-regulated gene expression by Jumonji domain-containing proteins ELF6 and REF6 in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 105, 7618–7623. doi: 10.1073/pnas.0802254105
Zheng, S., Hu, H., Ren, H., Yang, Z., Qiu, Q., Qi, W., et al. (2019). The Arabidopsis H3K27me3 demethylase JUMONJI 13 is a temperature and photoperiod dependent flowering repressor. Nat. Commun. 10:1303. doi: 10.1038/s41467-019-09310-x
Zheng, X. Y., and Gehring, M. (2019). Low-input chromatin profiling in Arabidopsis endosperm using CUT&RUN. Plant Reprod. 32, 63–75. doi: 10.1007/s00497-018-00358-1
Zhou, Y., Wang, Y., Krause, K., Yang, T., Dongus, J. A., Zhang, Y., et al. (2018). Telobox motifs recruit CLF/SWN–PRC2 for H3K27me3 deposition via TRB factors in Arabidopsis. Nat. Genet. 50:638. doi: 10.1038/s41588-018-0109-9
Keywords: Arabidopsis, development, demethylases, epigenetics, environmental response, JUMONJI, histone modification, H3K27me3
Citation: Yamaguchi N (2021) Removal of H3K27me3 by JMJ Proteins Controls Plant Development and Environmental Responses in Arabidopsis. Front. Plant Sci. 12:687416. doi: 10.3389/fpls.2021.687416
Received: 29 March 2021; Accepted: 26 May 2021;
Published: 17 June 2021.
Edited by:
Stewart Gillmor, National Laboratory of Genomics for Biodiversity, Center for Research and Advanced Studies, National Polytechnic Institute of Mexico (CINVESTAV), MexicoReviewed by:
Gerardo del Toro, Swedish University of Agricultural Sciences, SwedenCopyright © 2021 Yamaguchi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nobutoshi Yamaguchi, bm9idXlAYnMubmFpc3QuanA=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.