- 1College of Forestry, Henan Agricultural University, Zhengzhou, China
- 2Department of Biological Sciences, University of Wisconsin-Milwaukee, Milwaukee, WI, United States
MicroRNAs (miRNAs) are small (∼21-nucleotides) non-coding RNAs found in plant and animals. MiRNAs function as critical post-transcriptional regulators of gene expression by binding to complementary sequences in their target mRNAs, leading to mRNA destabilization and translational inhibition. Plant miRNAs have some distinct characteristics compared to their animal counterparts, including greater evolutionary conservation and unique miRNA processing methods. The lifecycle of a plant begins with embryogenesis and progresses through seed germination, vegetative growth, reproductive growth, flowering and fruiting, and finally senescence and death. MiRNAs participate in the transformation of plant growth and development and directly monitor progression of these processes and the expression of certain morphological characteristics by regulating transcription factor genes involved in cell growth and differentiation. In woody plants, a large and rapidly increasing number of miRNAs have been identified, but their biological functions are largely unknown. In this review, we summarize the progress of miRNA research in woody plants to date. In particular, we discuss the potential roles of these miRNAs in growth, development, and biotic and abiotic stresses responses in woody plants.
Introduction
MiRNAs are small (∼21-nucleotides) non-coding RNAs that have emerged as key post-transcriptional regulators of gene expression in both plants and animals (Jones-Rhoades et al., 2006; Chen, 2008; Filipowicz et al., 2008; Ramachandran and Chen, 2008). In plants, a primary miRNA (pri-miRNA) is transcribed from a MIRNA (MIR) gene, forming a stem-loop structure. RNase III-type endonucleases (also known as Dicer proteins) release the stem segment to produce a paired precursor miRNA (pre-miRNA) of approximately 21 nucleotides and 2-nucleotide 3′overhangs. One strand of miRNA in the miRNA duplex becomes the mature miRNA, which is loaded into an RNA-induced silencing complex (RISC). The mature miRNA recognizes its target mRNA(s) by base pairing, resulting in its cleavage or translational attenuation. Some miRNAs also function in chromatin modification by mediating DNA methylation (Wu et al., 2010; Jia et al., 2011). A study of target transcripts of plant miRNAs found that most encode transcription factors involved in plant development patterns or cell differentiation (Rhoades et al., 2002). To date, a vast number of miRNAs have been identified from a variety of plants, including woody plants.
MiRNAs play remarkably widespread roles in controlling almost all aspects of plant growth and development, ranging from determining organ polarity to meristem function, vascular and root development, flower formation, seed and fruit development, and biotic and abiotic stress responses. In general, the functions of miRNAs are characterized by analyzing mutants with impaired miRNA biogenesis (Griffiths-Jones, 2004; Griffiths-Jones et al., 2006; Griffiths-Jones et al., 2008; Liu et al., 2010; Kozomara and Griffiths-Jones, 2011, 2014; Zhang, 2015; Kozomara et al., 2019). However, only a small fraction of known miRNAs have so far been studied. Overexpressing certain miRNAs or expressing of the resistant version of a target mRNA allows the roles of miRNAs to be investigated by analyzing the resulting phenotypes. The characterization of loss-of-function mutants of MIR genes and the strategy of interfering with mature miRNAs have also been successfully employed to identify miRNA functions (Baker et al., 2005; Allen et al., 2007; Franco-Zorrilla et al., 2007; Sieber et al., 2007; Voinnet, 2009; Liu et al., 2010; Todesco et al., 2010; Yan et al., 2012).
Thus far, the general and specific functions of miRNAs have mainly been studied in Arabidopsis (Arabidopsis thaliana) and Populus (poplar; Populus trichocarpa). The miRNAs from these species share similar and yet different functions. For example, miR397 is a conserved miRNA in flowering plants (Kozomara and Griffiths-Jones, 2011). In the woody model plant P. trichocarpa, miR397 is the main regulator of lignin biosynthesis (Lu et al., 2013). In addition, miR397b affects lignin biosynthesis by regulating its target gene LACCASE4 (LAC4) in Arabidopsis (Wang C. Y. et al., 2014). These findings indicate that miR397 has a conserved role in the regulatory network of lignin biosynthesis in herbaceous and woody plants. However, upregulating miR397 expression increased grain size and panicle branching in rice (Oryza sativa) by regulating the expression of its target gene LAC and thereby significantly improving grain yield (Zhang Y. C. et al., 2013), indicating that conserved miRNAs might exhibit different functions across species. Therefore, identifying and characterizing miRNAs in woody plants would greatly facilitate further research on the evolution and functions of miRNAs.
According to the miRNA database (1, Release 22.1; Kozomara et al., 2019), among the 10,414 mature miRNAs identified in 82 plant species, 2,656 were from 26 tree species. Information about plant miRNAs is also hosted in other databases, such as PmiREN (2, Guo et al., 2020), a comprehensive functional plant miRNA database that shares 44 species with miRBase. PmiREN contains 20,388 miRNA loci (MIR) belonging to 5,757 families in 88 species ranging phylogenetically from chlorophytes to angiosperms. (The data of the two databases are listed in Table 1).
Based on the identified miRNA families in plants and their sequence homology, cross-species comparisons have been performed to determine the conservation of certain miRNA families in different plant lineages (Jones-Rhoades, 2012). In this review, we compare the miRNAs of 25 tree species with those of the model plants A. thaliana, O. sativa, and P. trichocarpa. MiRNAs that are found in model plants and at least three woody tree species with high sequence homology are defined as conservative miRNAs. Based on these criteria, 22 highly conserved miRNAs families have been identified in plants, including miR156, miR159/miR319, miR160, miR162, miR164, miR166, miR167, miR168, miR169, miR170/171, miR172, miR319, miR390, miR393, miR394, miR395, miR396, miR397, miR398, miR399, miR408, and miR827 families. These findings are consistent with the results of a previous analysis of flowering plants (Cuperus et al., 2011). Conserved miRNAs families also include miR403, miR828, miR2111, miR477, miR482, and miR3627 (Solofoharivelo et al., 2015). Here, we focus on the potential functions of miRNAs in growth, development, and responses to abiotic and biotic stresses in woody plants.
Growth and Development
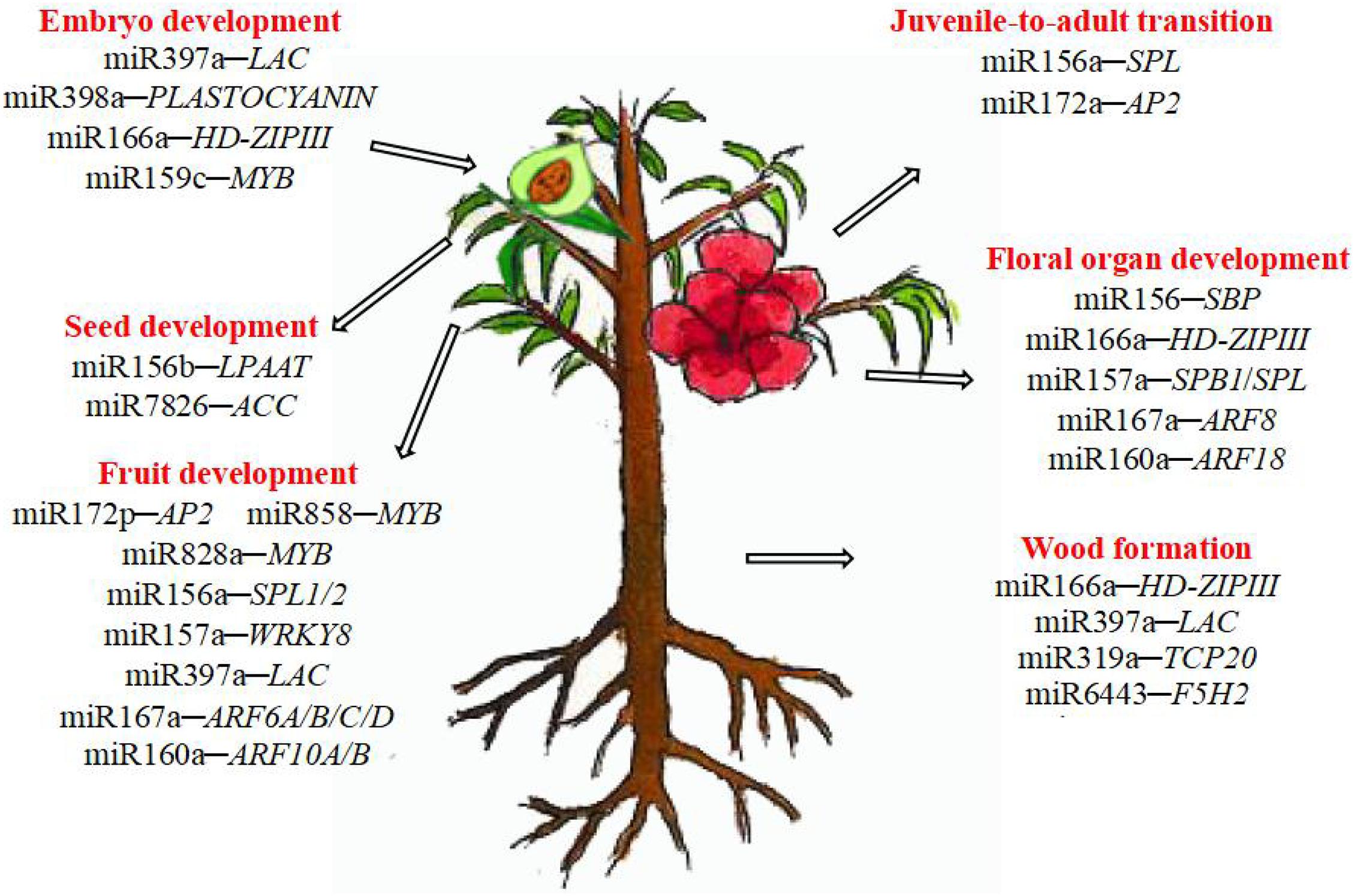
Wood Formation
Lignin, an important component of the secondary cell wall, forms support tissues in vascular plants. In Chinese white poplar (Populus tomentosa), 36 miRNAs were shown to be involved in wood formation by targeting lignin biosynthesis genes (Lu et al., 2005; Quan et al., 2016, 2019). Overexpression of miR6443 in P. tomentosa specifically downregulated the expression of the miR6443 target gene, FERULATE 5-YDROXYLASE 2 (F5H2), resulting in a significant decrease in S lignin content (Fan et al., 2019). During vascular development in P. tomentosa, miR319a and PtoTCP20 are differentially expressed; overexpression of miR319a in seedlings caused a delay in secondary growth and a decreased xylem yield. A dual-luciferase assay confirmed the targeted cleavage of PtoTCP20 by miR319a. Yeast two-hybrid and yeast one-hybrid screens demonstrated that PtoTCP20 interacts with PtoWOX4a and activates the expression of PtoWND6A/B, thereby promoting the differentiation of cambium into secondary xylem cells and regulating the secondary vascular tissue development (Hou et al., 2020).
In hybrid aspen (Populus tremula × Populus alba), Pta-miR166 targets the Class III HD-Zip transcription factor-encoding gene Pta-HB1 (the ortholog of Arabidopsis REVOLUTA). The expression patterns of Pta-HB1 are associated with the wood formation in aspen and vary upon seasonal changes (Ko et al., 2006). Moreover, knocking out the Class III HD ZIP transcription factor gene POPCORONA led to aberrant lignification in pith cells; conversely, expression of the miR165/166-resistant gene POPCORONA delayed the lignification of xylem and phloem fibers during secondary growth (Du et al., 2011). HD-Zip III subfamily genes, the targets of miR165/166, were also highly expressed in the cambium and xylem of plum blossom (Prunus mume) and Acacia mangium, indicating that miR165/166 is indispensable for vascular differentiation (Ong and Wickneswari, 2012; Li et al., 2019). Interestingly, miR166 was not detected in the xylem of apple (Malus domestica “Royal Gala”) (Varkonyi-Gasic et al., 2010).
MiR397a negatively regulates the lignin-biosynthesis-related LAC genes in poplar: P. tomentosa overexpressing Ptr-MIR397a showed decreased expression levels of 17 Ptr-LAC genes as well as reduced lignin content (Lu et al., 2013; Chen J. et al., 2015). Similarly, in alpine ash (Eucalyptus grandis), 17 LAC genes were predicted to be targets of miR397, of which 9 were mainly expressed in vascular tissues involved in the synthesis of xylem lignin (Arcuri et al., 2020). Additionally, degradation sequencing revealed that MYB, the target of miR159, is involved in the wood formation of E. grandis (Pappas et al., 2015). In cork oak (Quercus variabilis), miR167, miR165/166, miR396, and miR159 are highly expressed in phellogen, pointing to roles in cork formation (Chaves et al., 2014). A recent study comparing the miRNA transcriptomes of different tissues of P. alba × P. glandulosa and Larix kaempferi showed that miRNAs have different roles in regulating wood formation in these two tree species, providing a basis for understanding the wood formation mechanisms of gymnosperms and angiosperms (Li et al., 2020).
The functions of miRNAs are affected by allelic variation; one single-nucleotide polymorphism (SNP) in the pre-miRNA region of Pto-MIR160a altered the number of loops in its secondary structure and was significantly associated with tree growth and wood properties (Tian et al., 2016). In addition, variation of an SNP at the binding site of a target gene affected its binding affinity to miRNA (Chen J. et al., 2016). Target gene prediction and transcriptome sequencing suggested that miR257 and its 12 potential target genes control wood formation and tree growth. SNP association analysis to explore the functions of Pto-miR257 and its 12 target genes revealed a significant epistatic interaction between Pto-miR257 and these genes, expanding our understanding of gene regulation and the mechanisms underlying how miRNA-target interactions affect phenotypic variation (Chen B. et al., 2016).
Single-nucleotide polymorphisms in miRNA biogenesis genes (miRBGs) associated with lignin can alter the expression correlation of miRBG with miRNA and miRNA with target genes, indicating that miRBG mutations regulate the expression of downstream target genes by affecting miRNA expression abundance. Thus, a miRBG-miRNA-target epistasis interaction network that affects forest growth and wood quality variation was constructed, providing a new theory and strategy for the genetic improvement of forests (Chen B. et al., 2018). The allelic variation and interaction between the Pto-miR319 family and its 12 target genes related to wood formation were recently analyzed based on their association, representing a step forward in the use of association genetics to explain the multigene interactions of trees (Si et al., 2020).
Flowering Time and Flower Development
miRNAs such as miR156 and miR172 play important roles in the reproductive development of trees, which are similar to their functions in the model plant Arabidopsis. In small Philippine acacia (Acacia confusa), Cole’s wattle (Acacia colei), blue gum (Eucalyptus globulus), ivy (Hedera helix), German oak (Quercus acutissima), and Canadian poplar (Populus × Canadensis), miR156 expression levels remain high at the seedlings stage and decrease during the juvenile-to-adult transition. However, the expression pattern of miR172 is different (Wang et al., 2011). An integrated analysis of mRNA and miRNA found that the MsSVP/miR172/mssap2-like module controls the expression of MsLFY by responding to gibberellin signals and stimulates the transition from vegetative growth to reproductive growth in Magnolia × soulangeana (Sun et al., 2020). Furthermore, overexpression of miR156 in transgenic Canadian poplar plants reduced the expression of miR156-targeted SPL genes and miR172, leading to the prolonged juvenile phase (Wang et al., 2011). Overexpression of miR172a caused early flowering, aberrant flowers formation, and abnormal leaf development in physic nut (Jatropha curcas) (Tang et al., 2018). Similarly, the expression level of miR156 was high, whereas the expression levels of miR172 was low, in the leaves of Chinese crabapple (Malus hupehensis) at the juvenile phase (Xing et al., 2014). The finding that miR156 and miR172 have similar expression patterns in other woody plants, such as trifoliate orange (Poncirus trifoliata) (Zhu and Helliwell, 2011; Zhang X. N. et al., 2014), kiwifruit (Actinidia deliciosa) (Erika et al., 2012), Chinese plum (Prunus mume) (Wang T. et al., 2014), apple (M. domestica) varieties “Golden Delicious” (Guo et al., 2017), Fuji, Fuji/M9, and M9 (An et al., 2018), and Rhododendrons (Rhododendron arboreum), implies that these two miRNAs have conserved functions in controlling the juvenile-to-adult transition (Choudhary et al., 2018).
In Chinese hickory (Carya cathayensis), miR156, miR160, miR167, miR172, and miR394 were upregulated during flower development (Wang et al., 2012). Likewise, miR156, miR166, miR167, miR169, and miR171 together with their target genes were differentially expressed, providing further support for critical roles of miRNAs in male and female flower development in C. cathayensis (Sun et al., 2017). The target ARF4 genes of miR167, miR529, and miR2950 affect flower development in Chinese jujube (Ziziphus jujuba) infected by phytoplasma, causing floral organs to develop into leaf-like structures (Ma et al., 2020). Nine miRNAs (miR157a/b, miR161, miR164b, miR166a/b, miR172, miR399, and miR776) are expressed in mature or germinated pollen of loblolly pine (pinus taeda), pointing to their potential functions in pollen germination and pollen tube growth (Quinn et al., 2015). In addition, miR157, miR160, miR167, and miR171 might control the induction of female flowers formation in walnut (Juglans regia), since they were mainly detected in female flowers (Zhou et al., 2018). The target gene of miR156 was mapped to the gender locus in dustpan willow (Salix suchowensis), pointing to the function of miR156 in controlling flower gender (Wei et al., 2017). Furthermore, the methylation of miR172b might play an important role in flower gender determination, as miR172b is hypermethylated in female P. tomentosa flowers (Song et al., 2013, 2015).
It is likely that miR156, miR164, miR166, and miR5139 control rose petal development by regulating the ethylene signaling (Kim et al., 2012; Pei et al., 2013). An analysis of differentially expressed miRNAs in simple flowers (wild-type) and double flowers (mutant) of shiny-leaved yellowhorn (Xanthoceras sorbifolia) suggested that miR159, miR164, miR166, and miR319 are involved in floral organ development (Ao et al., 2012). In M. domestica, miR159 might control male organ and embryo development by targeting MYB genes (Xia et al., 2012). Moreover, expressing the precursor of miR396c of P. trichocarpa in tobacco altered the identity of stamens and carpels, suggesting that miR396 is essential for floral organ specification (Baucher et al., 2013).
Seed Development
MiRNAs play important roles in seed development, dormancy, and germination. MiR156b, miR414, miR2673b, and miR7826 control seed development by regulating the expression of genes required for fatty acid biosynthesis and lipid metabolism in tree peony (Paeonia suffruticosa) (Yin et al., 2018). The seed oil of J. curcas is a promising substitute for petro-diesel. A variety of miRNAs, including miR156, miR157, miR159, miR166, miR167, miR168, and miR396, target genes associated with cellular structure, nuclear function, translation, transport, hormone synthesis, and lipid metabolism during seed development in J. curcas (Galli et al., 2014). Similarly, deep sequencing of the woody oil seed plant of Pongamia (Pongamia pinnata) at three stages of development (embryogenesis phase, seed-filling phase, and desiccation phase) revealed that miR482a-3p regulates fatty acid biosynthesis during seed development (Jin et al., 2019). In coconut (Cocos nucifera), miR166a, miR397a, and miR398a are highly expressed in the immature endosperm (Li D. et al., 2009). Additionally, during somatic embryogenesis in longan (Dimocarpus longan), many miRNAs were preferentially expressed at different stages: miR156 and miR166c∗ are expressed in the early embryonic cultures; miR160, miR159, miR390, and miR398 are expressed during the heart-shaped and torpedo-shaped stages; miR167, miR168a∗, miR397, miR398, miR808, and miR5077 are expressed during the cotyledon stage; while miR167, miR808, and miR5077 are expressed during the mature embryo stage (Lin and Lai, 2013). In loblolly pine (P. taeda), miR166 is mainly expressed in zygotic embryos and female gametophytes. However, miR167 was only detected in zygotic embryos, representing the first report of miRNAs in gametophytic seed tissues (Oh et al., 2008). In Japanese larch (Larix kaempferi), miR-13, miR1313b, miR-14, miR319a, miR894g, miR159c, miR-12, miR397d, and miR162a are upregulated in dormant embryos, whereas miR168b, miR169b, miR4414b, and miR529a are upregulated in germinated embryos (Zhang J. et al., 2013). Similarly, overexpression of miR166 suppressed the formation of cotyledons during somatic embryogenesis, leading to abnormal development of the apical embryo (Li et al., 2016). MiR159 maintains somatic embryo maturation by negatively regulating its target LaMYB33 (Li W. F. et al., 2013). Finally, miR397 is involved in the process of converting dormant embryos into germinating embryos by regulating the thickness of cell walls. During this process, abscisic acid alters composition of miRNAs by regulating the expression levels of miRNA biogenesis genes (Zhang J. et al., 2013).
Fruit Development
MiRNAs, particularly miR156, miR160, miR167, miR172, miR390, miR393, miR397, miR828, and miR858, are important for fruit development (Damodharan et al., 2016; Chen C. et al., 2018). In M. domestica, the silencing of miR172 via a transposon insertion led to an increase in fruit size; conversely, overexpressing miR172 significantly reduced fruit size and altered flower structure (Yao et al., 2015). MiR408 regulates the biomass of Arabidopsis and rice: overexpressing MIR408 in these plant significantly enhanced photosynthesis by improving light utilization efficiency and carbon dioxide fixation capacity, thereby increasing biomass and seed yield (Pan et al., 2018; Song et al., 2018). In rice, miR408 can increase yield by regulating its target UCL8, encoding a member of the phytocyanin family, to increase the number of panicles and grains (Zhang et al., 2017). In addition, in P. trichocarpa, ptr-miR408 effectively targets plastocyanin-like protein genes (PCLs) (Lu et al., 2005, 2008). Although miR408 regulates different target genes in herbaceous and woody plants, all of these targets encode copper-containing proteins that are essential for photosynthetic electron transport. Therefore, miR408 is thought to plays an important role in the photosynthesis and fruit yield of woody plants.
In tea-oil camellia (Camellia oleifera), miR156, miR390, and miR395 are involved in regulating carbohydrate accumulation during fruit growth and development, and miR477 is a key factor in fatty acid synthesis (Liu et al., 2019). In litchi (Litchi chinensis), the miR156-targeted gene Lc-SPL1/2 encodes a transcription factor that interacts with Lc-MYB1 to control fruit coloration by promoting anthocyanin biosynthesis (Liu et al., 2016). Similarly, miR156, miR159, miR828, miR858, and miR5072 regulate the accumulation of anthocyanin in apple peel (Xia et al., 2012; Qu et al., 2016). Furthermore, restricting root cultivation in grapevine (Vitis Vinifera) inhibits the expression of miR828, which in turn enhances anthocyanin metabolism by increasing the expression of its potential target MYB genes (Chen et al., 2019). In barbary wolfberry (Lycium barbarum), miR156/157 and their target genes Lb-CNR and Lb-WRKY8, together with miR171 and its target gene Lb-GRAS, are thought to be involved in controlling the fruit ripening process. Finally, miR156 negatively regulates the expression of fructokinase and 1-deoxy-D-xylulose-5-phosphatesynthase genes in L. barbarum, and miR164 targets β-fructofuranosidase genes to control the quality and nutritional value of fruits (Zeng et al., 2015).
The secondary thickening of the parenchyma cell wall and the deposition of lignin result in the formation of stone cells during fruit development (Cai et al., 2010; Yan et al., 2014). Profiling of miRNAs expression in pear fruits at different developmental stages found that many miRNAs play essential roles in fruit development by regulating the expression of genes involved in lignin synthesis, glycometabolism, acid metabolism, and hormone signaling (Wu et al., 2014). Specifically, miR3711, miR419, and miR5260 downregulate the hydroxycinnamoyl transferase (HCT) gene, which promotes lignin formation (Wu et al., 2014). Examination of miRNAs in pear fruits with different amounts of stone cells revealed that miR1809 and miR144-3p negatively regulate genes encoding laccases (LAC), which are key lignin biosynthesis enzymes (Cheng et al., 2017). In addition, miR397 targets LAC genes to control fruit cell lignification in Chinese pear (Pyrus bretschneideri) (Xue et al., 2018). Finally, many ARF genes, which are potential targets of miR160 and miR167, act as repressors of fruit abscission in L. chinensis (Zhang et al., 2019).
MiRNAs are involved in various stages of woody plant growth (Figure 1). Studies using various approaches suggested that miRNAs largely target genes encoding transcription factors that regulate plant growth, development, and hormone signaling. The same miRNAs are sometimes involved in different stages of woody plant only limited studied of this topic development, and different miRNAs sometimes regulate the same type of targets, highlighting the complexity of post-transcriptional regulation via miRNAs. MiRNAs involved in growth and development of trees are summarized in Table 2.
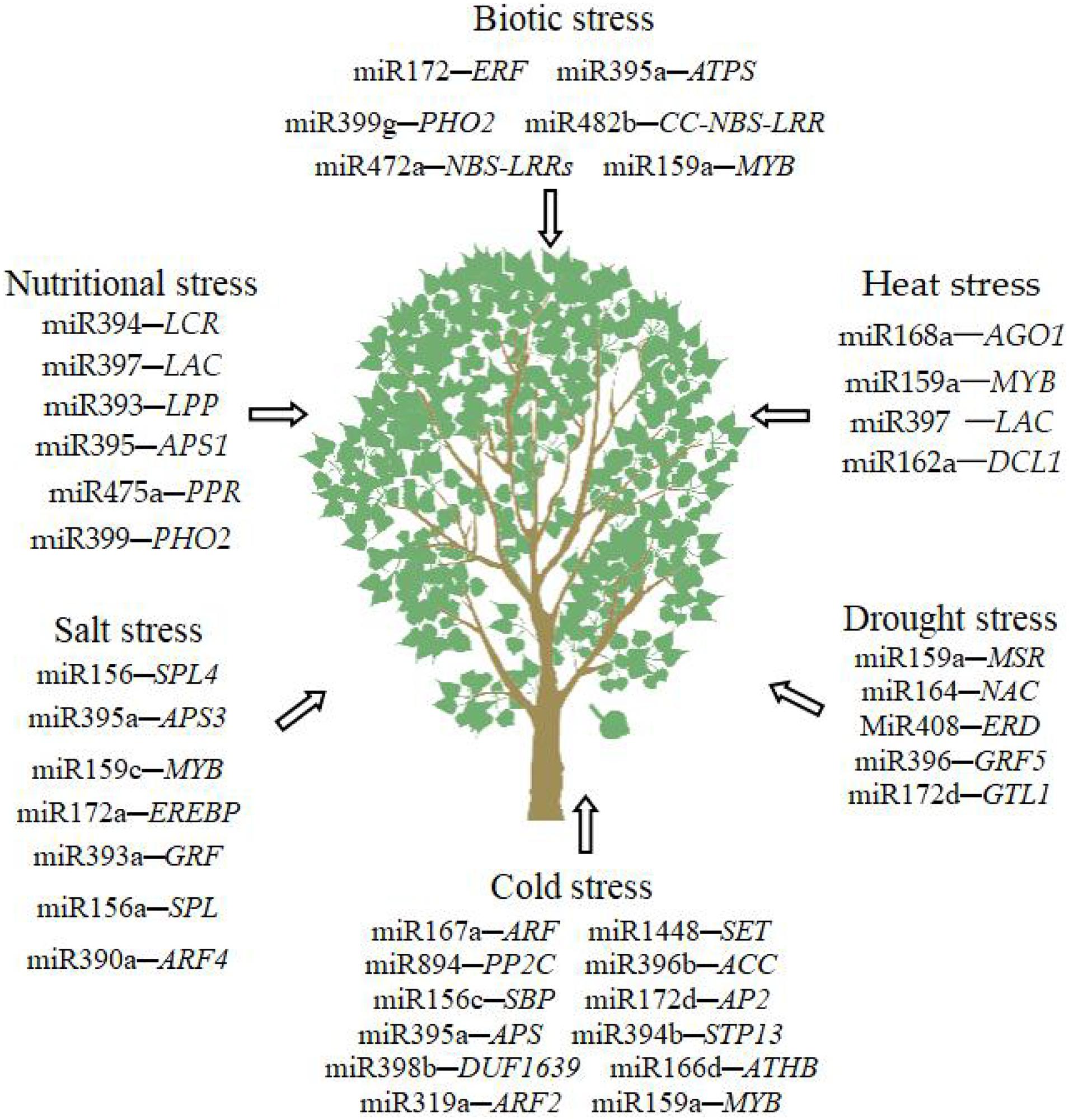
Abiotic Stress
Heat Stress
Heat stress affects the growth and development of trees. Both conserved and non-conserved heat-responsive miRNAs have been identified in various plant species. However, only limited studies of this topic have been conducted in just a few woody plants. In P. trichocarpa, miR156, miR159, miR164, miR171, and miR408 are responsive to stress in a tissue-specific manner (Lu et al., 2005). In addition, 52 miRNAs in P. tomentosa were found to respond to heat stress, most of which are downregulated (Chen et al., 2012a). In Betula luminifera, downregulation of the miR162-targeted gene DCL1 and the miR403-targeted gene AGO2 might modulate miRNA biosynthesis, thereby altering the abundance of miRNAs and their responses to heat stress (Pan et al., 2017). Furthermore, in cassava (Manihot esculenta), miR156a, miR159, miR160, miR397, and miR408 are downregulated by heat stress, indicating that they play important roles in heat stress responses by negatively regulating their target genes MYB and LAC (Ballén-Taborda et al., 2013).
Cold Stress
Cold stress negatively affects the growth and development of trees and restricts their spatial distribution. In P. trichocarpa, 156 genes were identified as potential targets of 14 cold-responsive miRNAs (Lu et al., 2008). The role of miR475b in the freezing response has been reported in sweet poplar (Populus suaveolens) (Niu et al., 2016). Cold stress induces the expression of similar miRNAs between P. suaveolens and P. trichocarpa with few exceptions, such as miR168a, miR168b, and miR475, suggesting that miRNAs play partially conserved roles in the responses of these trees to cold stress (Sun, 2011). MiR319a-c and miR395b-k are downregulated by cold stress, in P. tomentosa, resulting in enhanced resistance to this stress by regulating their targets MYBs and APS (Chen et al., 2012c). In addition, 21 upregulated and 43 downregulated miRNAs and 46 upregulated and 45 downregulated miRNAs were predicted to respond to cold stress in the tea plant (Camellia sinensis) varieties in “Yingshuang” and “Baiye 1,” respectively (Zhang Y. et al., 2014). In almond (Prunus dulcis), cold stress increases the expression of miR159, miR164a-c, miR171a/c/g/f, miR394a/b, miR482f, and miR7122a-5p, but decreases the expression of miR156a/b, miR162, miR166a-e, miR396b, miR398a-3p, miR6262, and miR8123-5p (Karimi et al., 2016). Furthermore, miR7122-3p might target the cold-tolerance gene Pd-HOS1 (Alisoltani et al., 2015). MiR169 is expressed at low levels in a cold-tolerant clone of rubber tree (Hevea brasiliensis), suggesting that miR169 might affect the expression of its target gene NF-YA to enhance cold resistance (Kuruvilla et al., 2017). In grapevine (V. vinifera), cold-induced miRNAs might negatively regulate the expression of various transcriptional factor genes, including AP2, SBP, MYB, bHLH, GRAS, and bZIP (Sun et al., 2015). Moreover, 36 conserved and 5 novel miRNAs are either up or downregulated in response to cold stress in trifoliate orange (P. trifoliata) (Zhang X. N. et al., 2014). Further studies showed that miR396b enhances cold tolerance in P. trifoliata by negatively regulating the expression of the 1-aminocyclopropane-1-carboxylic acid oxidase (ACO) gene, which consequently affects ethylene and polyamine synthesis (Zhang X.N. et al., 2016). In M. esculenta, miRNAs participate in the response and acclimation to low temperatures by regulating stress-related pathways, such as auxin signaling (Xia J. et al., 2014). By integrating miRNA and transcriptome analysis of lesser black poplar (Populus simonii × P. nigra), miR319, miR159, miR167, miR395, miR390, and miR172 were found to be differentially expressed under cold treatment and to negatively regulate their target genes, including ARF, MYB, SBP, LHW, and bZIP. These target genes played important roles in cold resistance (Zhou et al., 2019).
Drought Stress
Drought stress can severely affect the growth and productivity of trees. Many miRNAs are involved in regulating the drought response in trees. In P. trichocarpa, miR164, miR394, miR408, miR473, miR123, miR1444a, miR93a, miR93b, and their potential target genes are differentially expressed under drought stress, pointing to their essential roles in the drought response (Lu et al., 2008; Kreuzwieser et al., 2009; Li B. et al., 2009; Wilkins et al., 2009; David et al., 2010; Peng et al., 2013). In V. vinifera, various miRNAs, such as miR166, miR319, miR396, miR3631, mi3633, and miR3639, are downregulated in response to severe water stress (Pantaleo et al., 2016). In addition, the accumulation of miRNAs including miR156, miR159, miR164, miR166, miR393, miR394, miR396, miR3624, and miR3639 in grapevine varies under drought stress depending on the grape variety and rootstocks (Pagliarani et al., 2017). Moreover, in the stems and leaves of Mongolian Ammopiptanthus (Ammopiptanthus mongolicus), miR2118 and miR858 upregulate the expression of the OZF1 and various MYB genes that control flavonol production under drought stress (Gao et al., 2016). In H. brasiliensis, miR168 and miR160 are associated with drought tolerance (Kuruvilla et al., 2019). In mulberry (Morus alba), miR156, miR172, and miR396 as well as their target genes might be involved in the response to drought stress (Li et al., 2017). Additionally, in peach (Prunus persica), the expression levels of 262 miRNAs in leaves and 368 miRNAs in roots were significantly altered upon drought stress (Eldem et al., 2012). In citrus (Citrus junos Siebold cv. “Ziyang”), 19 known miRNAs and 15 novel miRNAs were shown to respond to dehydration treatment. Among these, 16 miRNAs were downregulated whereas 18 miRNAs were upregulated under these conditions (Xie et al., 2017).
In P. tomentosa, several miRNAs, such as pto-miR394, pto-miR160f, pto-miR472, and pto-miR1448, and their target genes showed opposite expression patterns under drought and flooding stress; their target genes encode transcription factors and signal transduction elements that participate in stress responses. These findings further emphasize the importance of miRNAs in plants under water stress (Ren et al., 2012). A recent study showed that Pu-miRNA172d reduces stomatal density, net photosynthetic rate, and transpiration by inhibiting the expression of its target gene PuGTL1 in Cathay poplar (Populus ussuriensis) (Liu et al., 2020). Similar findings have been reported in princess tree (Paulownia tomentosa) and caragana (Caragana korshinskii), supporting the notion that many miRNAs play important roles in plant responses to drought stress (Deng et al., 2017; Ning et al., 2017).
Salt Stress
The expression levels of many miRNAs vary in responsive to salt stress (Zhang Q. et al., 2013, Zhang, 2015; Sun et al., 2019). In P. tomentosa, salt stress alters the expression of 21 conserved miRNAs (19 downregulated and 2 upregulated) and 7 non-conserved miRNAs (5 downregulated and 2 upregulated) (Ren et al., 2013). In desert poplar (Populus euphratica), the expression levels of 15 miRNAs and their putative target genes are regulated by salt stress (Li B. et al., 2013). In addition, the expression level of miR156 significantly increased after salt treatment, and this was followed by the enhanced expression of miR166, miR167, miR169, miR172, miR827, miR2119, and miR5020. By contrast, the expression of miR393, miR645, miR860, and miR1444 decreased following salt treatment (Si et al., 2014). Furthermore, several similar studies have identified many conserved and non-conserved miRNAs that are responsive to salt stress in date palm (Phoenix dactylifera) (Yaish et al., 2015), mulberry (Morus alba) (Wu et al., 2015), and citrus (C. junos) (Xie et al., 2017). The Salicaceae family member Cathay poplar (Populus cathayana) is salt-sensitive, but the dryland willow (Salix matsudana) is highly salt-tolerant. Saline treatment decreased the expression of miR474c and miR398b in P. cathayana but increased their expression in S. matsudana, suggesting that miRNAs play key roles in the differential responses of different plant ecotypes to salt stress, which occurs via different regulatory mechanisms to enhance plant adaption to saline-alkali environments (Zhou et al., 2012). Moreover, in Caragana intermedia, miR157a, miR165a, miR172b, and miR396a were upregulated under salt treatment, while their potential target genes were downregulated, suggesting that miRNAs play important post-transcriptional roles in the adaptation of this plant to salt stress (Zhu et al., 2013). In P. tomentosa under salt stress, the increased expression of miR390 promotes an increase in the levels of TAS3-derived trans-acting short-interfering RNAs (tasiARFs) and the degradation of ARF4 transcripts. The decreased expression of ARF4 releases auxin signals to promote lateral root growth and to improve salt tolerance (He et al., 2018).
Other Abiotic Stresses
In sweet orange trees (Citrus sinensis), boron deficiency alters the expression of many miRNAs, such as miR159, miR160, miR164, miR393, miR394, miR408, miR472, miR474, miR782 miR821, miR830, miR2118, miR3465, miR5023, and miR5266, pointing to important roles for these miRNAs in boron uptake (Lu et al., 2014). Another study showed that miR397a functions in boron tolerance by targeting LAC17 and LAC4, which control secondary cell walls synthesis (Huang J. H. et al., 2016). Copper is an essential micronutrient for plant photosynthesis. In Arabidopsis, when the supply of copper was limited, miR398, miR397, miR408, and miR857 accumulated and downregulated their target genes, encoding a copper/zinc superoxide dismutase, a copper protein phytocyanin, and members of the copper protein family of laccases. This mechanism allows plants to preserve copper for some important functions (Abdel-Ghany and Pilon, 2008). Similar to the pattern in Arabidopsis, the target genes of miR397, miR398, and miR408 were also downregulated in P. trichocarpa under copper deficiency. In addition, miR1444, a Populus-specific miRNA, participates in the copper regulatory network by regulating the expression of its target polyphenol oxidase gene (Lu et al., 2011).
miR167 was significantly upregulated in roots of coffee (Coffea arabica) at the beginning of a period of nitrogen starvation. The target gene of miR167 encodes the auxin response factor ARF, which is involved in lateral roots development. The expression of miR159 increased after 10 days of nitrogen starvation; its target MYB and TCP genes play important roles in regulating root activity. These findings indicate that this miRNA responds to nitrogen starvation by regulating different biological pathways at different time points (Zhao M. et al., 2012; dos Santos et al., 2019). Similarly, nitrogen deficiency increases the expression of miR159, miR160, miR166, miR167, miR169, miR171, miR172, miR390, and miR475 but decreases the expression of miR393 and miR395 in P. tomentosa (Ren et al., 2015). A recent study showed that low-nitrogen-responsive miRNAs, including miR167 and miR171, also participate in the response to low phosphorus in P. tomentosa (Bao et al., 2019).
Low sulfur levels induce the expression of miR395 and repress the expression of the miR395 target gene ATPS3 in poplar, thus inhibiting sulfur assimilation; high sulfur levels inhibits the expression of miR399, whose target gene PHO2 enhances sulfur assimilation (Shi et al., 2020). In several herbaceous plants, miR395 and miR399 accumulate in the phloem under sulfur and phosphorus deficiency and are transported from the stem to the root, where they regulate the uptake of sulfur and phosphorus by inhibiting the expression of their target genes PHO2 and APS4. In addition, these miRNAs are transported through the xylem to aboveground tissue, thereby regulating the homeostasis of the plant nutrition system (Pant et al., 2008; Buhtz et al., 2010; Chiou and Lin, 2011; Kehr, 2013). A similar phenomenon has been observed in B. luminifera: miR399 and miR395 were significantly upregulated under low -phosphorus conditions. Moreover, PLDD was identified as the cleavage target of miR399c (Zhang et al., 2021). These findings indicate that herbaceous species and woody perennials adapt to low-phosphorus environments by regulating different targets.
In sycamores (Platanus acerifolia), pla-miR482a-3p and pla-mir39 were differentially expressed under lead stress. Their targets are RPM1 and NBS, respectively, which encode disease resistance proteins, suggesting that lead stress induces signal transduction pathways related to plant defense against pathogens (Wang et al., 2015). In P. trichocarpa, 12 PtrDof genes appear to be targeted by 15 miRNAs, seven of which were upregulated in leaves and roots under osmotic stress at all time points, indicating that PtrDof transcription factors play an important role in the resistance of this tree to osmotic stress (Wang H. et al., 2017).
Biotic Stress
The fact that diseases affect the expression of many miRNAs and their target genes supports the idea that miRNAs play diverse roles in biotic stress responses. In P. trichocarpa, the potential target genes of miR472, miR482, miR1447, and miR1448 are disease-resistance genes (Lu et al., 2008). Twelve miRNAs (miR156, miR159, miR160, miR164, miR166, miR168, miR172, miR319, miR398, miR408, miR1448, and miR1450) were upregulated in poplar after infection with the stem canker pathogen Botryosphaeria dothidea (Zhao J. P. et al., 2012). Similar miRNAs that respond to the canker pathogen Valsa mali were identified in P. beijingensis (P. cathayana × P. nigra var. italica) (Chen et al., 2012b). Moreover, miR482b plays an important role in the response of M. domestica to the infection by V. mali by regulating its target genes, encoding CC-NBS-LRR class resistance proteins (Feng et al., 2017). In Mexican lime (Citrus aurantifolia), miR157 might be involved in altering tree structure and leaf morphology in response to phytoplasma infection (Ehya et al., 2013).
Following infection of loblolly pine (P. taeda) with fusiform rust gall, miRNAs from 10 families (including miR156, miR159, and miR319) were significantly downregulated in the stems. Although the expression of these miRNAs in the roots and upper stems of the infected parts of the tree did not change significantly, the expression of their target genes significantly increased. These findings suggest that the host produced an immune response in the uninfected parts of the tree following pathogen infection that led to the host’s defense response (Lu et al., 2007). In wax gourd poplar (Populus purdomii), 7 conserved and 20 novel miRNAs that are responsive to foliar rust fungus infection are predicted to regulate genes encoding disease-resistance proteins, serine/threonine protein kinases, transcription factors, and other related proteins (Chen and Cao, 2015). Furthermore, in V. vinifera, phytoplasma infection induced the differential expression of various conserved and novel miRNAs that might target genes involved in morphology, hormone signaling, nutrient homeostasis, and stress (Snyman et al., 2017). The number of differentially expressed miRNAs reflects the degree of symbiosis between fungi and roots (Mewalal et al., 2019). When P. tomentosa was inoculated with endophytic Streptomyces sp. SSD49, miR160 and miR156 were upregulated, and miR319 was downregulated. Their targets primarily encode auxin response factors, disease-resistant proteins, plant hormone oxidases, and so on, which could promote plant growth and improve disease resistance (Tian et al., 2019). NBS-LRRs, the targets of miR472, launch effective immune responses in the fungus Cytospora chrysosperma (Su et al., 2018).
Some miRNAs in the phloem function as signaling molecules for long-distance transport (Notaguchi and Okamoto, 2015). For example, after mulberry was infected by a phytoplasma, the expression of mul-miR482a-5p in the phloem sap increased. Although the expression levels of the target genes of this miRNA were not significantly altered in the phloem, these genes were downregulated in roots. Grafting experiment showed that mul-miR482a-5p could be transported from the scion to rootstock. Therefore, mul-miR482a-5p in the phloem sap may regulate its target genes in roots (Gai et al., 2018).
Many microRNAs are involved in abiotic and biotic stresses (Figure 2). Same miRNAs play vital roles in multiple abiotic and biotic stresses. The miRNAs that are important to the abiotic and biotic stresses of trees are summarized in Table 3.
Conclusion and Future Perspectives
Over the past decade, numerous conserved and species-specific miRNAs have been identified in many woody plants due to the advent of next-generation sequencing technologies. Several studies have revealed that a conserved regulatory mechanism between miRNAs and mRNAs could control various biological processes in different plant tissues. For example, miR166 participates in wood formation (Du et al., 2011), floral organ development (Sun et al., 2017), and embryo development (Li et al., 2016) by regulating the same target HD-Zip III gene. On the other hand, miRNAs can sometimes regulate different biological processes by targeting different genes. CKB3, a target of MiR397b, participates in circadian rhythms and affects flowering time in plants (Feng et al., 2020). LCS9, the target of miR397, participates in fatty acid metabolism in oil palm (Elaeis guineensis) (Zheng et al., 2019). In addition, miR397 is involved in embryo development (Lin and Lai, 2013; Zhang J. et al., 2013) and juice sac granulation (Zhang C. et al., 2016). Furthermore, the targets of miR159, miR828, and miR858 are homologous MYB genes with different biological functions. Different miRNAs can bind to the same target but regulate different downstream genes, thus performing different functions. These observations highlight the complexity of miRNA-mediated post-transcriptional regulation. The diversity of woody plants provides a great opportunity to study the evolution of miRNAs.
The bioinformatics tools currently available for miRNA target prediction based on sequence information are PsRobot (Wu et al., 2012), psRNATarget (Dai and Zhao, 2011), and Targetfinder (Kiełbasa et al., 2010). To a certain extent, these approaches allow the target genes of miRNAs to be mined. However, considering that one miRNA can target multiple mRNAs and that one mRNA can also be targeted by multiple miRNAs, the amount of information generated by bioinformatics will be huge, with numerous false-positive candidates. Therefore, it is necessary to identify key genes engaged in a large number of target relationships for experimental verification. At present, some of the functions of miRNAs identified in woody plants can be verified using model plants. However, due to the long generation cycle of perennial woody plants, the establishment of genetic systems for these plants has been restricted. It is difficult to investigate specific aspects of trees in model plants, such as wood formation, seasonal changes, and perennial growth characteristics. Virus-induced gene silencing (VIGS) technology could be utilized for functional verification of miRNAs in woody plants lacking a stable genetic system. CRISPR/Cas9 could be useful for generating a large number of site-directed mutants and to enrich experimental resources. Finally, the combined use of VIGS and CRISPR/Cas9 technology holds great promise for functional analysis of woody plants in the future.
Author Contributions
YW and LF wrote the manuscript. Both authors contributed to the article and approved the submitted version.
Funding
This research in the Laboratory of YW (a previous visiting scholar at UWM) was funded by Forestry Science and Technology Development Project, Science and Technology Development Center of State Forestry and Grassland Administration (KJZXSA2019041).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors would like to thank Xiaochen Yuan from Michigan State University and Robert Johnson from Concordia University Wisconsin for assisting with proofreading the manuscript.
Footnotes
References
Abdel-Ghany, S. E., and Pilon, M. (2008). MicroRNA-mediated systemic down-regulation of copper protein expression in response to low copper availability in Aarbidopsis. J. Biol. Chem. 283, 15932–15945. doi: 10.1074/jbc.M801406200
Alisoltani, A., Shiran, B., Fallahi, H., and Ebrahimie, E. (2015). Gene regulatory network in almond (Prunus dulcis Mill.) in response to frost stress. Tree Genet. Genome 11:100. doi: 10.1007/s11295-015-0929-z
Allen, R. S., Li, J., Stahle, M. I., Aurélie, D., and Millar, A. A. (2007). Genetic analysis reveals functional redundancy and the major target genes of the Arabidopsis miR159 family. Proc. Natl. Acad. Sci. U.S.A. 104, 16371–16376. doi: 10.1073/pnas.0707653104
Amborella Genome Project, Albert, V. A., Barbazuk, W. B., dePamphilis, C. W., Der, J. P., Leebens-Mack, J., et al. (2013). The Amborella genome and the evolution of flowering plants. Science 342, 1467–1467. doi: 10.1126/science.1241089
An, N., Fan, S., Yang, Y., Chen, X., Dong, F., Wang, Y., et al. (2018). Identification and characterization of miRNAs in self-rooted and grafted Malus reveals critical networks associated with flowering. Int. J. Mol. Sci. 19:2384. doi: 10.3390/ijms19082384
Ao, Y., Wang, Y., Chen, L., Wang, T., Yu, H., and Zhang, Z. (2012). Identification and comparative profiling of microRNAs in wild-type Xanthoceras sorbifolia and its double flower mutant. Genes Genomics 34, 561–568. doi: 10.1007/s13258-012-0065-1
Arcuri, M., Fialho, L. C., Nunes-Laitz, A. V., Fuchs-Ferraz, M., and Maia, I. G. (2020). Genome-wide identification of multifunctional laccase gene family in Eucalyptus grandis: potential targets for lignin engineering and stress tolerance. Trees 34, 745–758. doi: 10.1007/s00468-020-01954-3
Argout, X., Salse, J., Aury, J., Guiltinan, M. J., Droc, G., Gouzy, J., et al. (2011). The genome of Theobroma cacao. Nat. Genet. 43, 101–108. doi: 10.1038/ng.736
Aryal, R., Yang, X., Yu, Q., Sunkar, R., and Ming, R. (2012). Asymmetric purine-pyrimidine distribution in cellular small RNA population of papaya. BMC Genomics 13:682. doi: 10.1186/1471-2164-13-682
Baker, C. C., Sieber, P., Wellmer, F., and Meyerowitz, E. M. (2005). The early extra petals1 mutant uncovers a role for microRNA miR164c in regulating petal number in Arabidopsis. Curr. Biol. 15, 303–315. doi: 10.1016/j.cub.2005.02.017
Ballén-Taborda, C., Plata, G., Ayling, S., Rodríguez-Zapata, F., Becerra Lopez-Lavalle, L. A., Duitama, J., et al. (2013). Identification of cassava microRNAs under abiotic stress. Int. J. Genomics 2013:857986. doi: 10.1155/2013/857986
Bao, H., Chen, H., Chen, M., Xu, H. M., Huo, X. W., and Xu, Q. H. (2019). Transcriptome-wide identification and characterization of microRNAs responsive to phosphate starvation in Populus tomentosa. Funct. Integr. Genomics 15, 892–898. doi: 10.1007/s10142-019-00692-1
Baucher, M., Moussawi, J., Vandeputte, O. M., Monteyne, D., Mol, A., Perezmorga, D., et al. (2013). A role for the miR396/GRF network in specification of organ type during flower development, as supported by ectopic expression of Populus trichocarpa miR396c in transgenic tobacco. Plant Biol. 15, 892–898. doi: 10.1111/j.1438-8677.2012.00696.x
Buhtz, A., Pieritz, J., Springer, F., and Kehr, J. (2010). Phloem small RNAs, nutrient stress responses, and systemic mobility. BMC Plant Biol. 10:64. doi: 10.1186/1471-2229-10-64
Cai, Y., Li, G., Nie, J., Lin, Y., Nie, F., Zhang, J., et al. (2010). Study of the structure and biosynthetic pathway of lignin in stone cells of pear. Sci. Hortic. 125, 374–379. doi: 10.1016/j.scienta.2010.04.029
Chaves, I., Lin, Y. C., Pinto-Ricardo, C., Yves, V. P., and Célia, M. (2014). miRNA profiling in leaf and cork tissues of Quercus suber reveals novel miRNAs and tissue-specific expression patterns. Tree Gene Genomes 10, 721–731. doi: 10.1007/s11295-014-0717-1
Chen, B., Chen, J., Du, Q., Zhou, D., Wang, L., Xie, J., et al. (2018). Genetic variants in microRNA biogenesis genes as novel indicators for secondary growth in Populus. New Phytol. 219, 1263–1282. doi: 10.1111/nph.15262
Chen, B., Du, Q., Chen, J., Yang, X., Tian, J., Li, B., et al. (2016). Dissection of allelic interactions among Pto-miR257 and its targets and their effects on growth and wood properties in Populus. Heredity 117, 73–83. doi: 10.1038/hdy.2016.26
Chen, C., Zeng, Z., Liu, Z., and Xia, R. (2018). Small RNAs, emerging regulators critical for the development of horticultural traits. Hortic. Res. 5, 1–14. doi: 10.1038/s41438-018-0072-8
Chen, J., Chen, B., Yang, X., Tian, J., Du, Q., and Zhang, D. (2015). Association genetics in Populus reveals the interactions between Pto-miR397a and its target genes. Sci. Rep. 5:11672. doi: 10.1038/srep11672
Chen, J., Xie J., Chen, B., Quan, M., Li, Y., Li, B., et al. (2016). Genetic variations and miRNA-target interactions contribute to natural phenotypic variations in Populus. New Phytol. 212, 150–160. doi: 10.1111/nph.14040
Chen, L., Ren, Y., Zhang, Y., Xu, J., Sun, F., and Zhang, Z. (2012a). Genome-wide identification and expression analysis of heat-responsive and novel microRNAs in Populus tomentosa. Gene 504, 160–165. doi: 10.1016/j.gene.2012.05.034
Chen, L., Ren, Y., Zhang, Y., Xu, J., Zhang, Z., Wang, Y., et al. (2012b). Genome-wide profiling of novel and conserved Populus microRNAs involved in pathogen stress response by deep sequencing. Planta 235, 873–883. doi: 10.1007/s00425-011-1548-z
Chen, L., Zhang, Y., Ren, Y., Xu, J., Zhang, Z., and Wang, Y. (2012c). Genome-wide identification of cold-responsive and new microRNAs in Populus tomentosa by high-throughput sequencing. Biochem. Biophys. Res. Commun. 417, 892–896. doi: 10.1016/j.bbrc.2011.12.070
Chen, M., and Cao, Z. (2015). Genome-wide expression profiling of microRNAs in poplarupon infection with the foliar rust fungus melampsora larici-populina. BMC Genomics 16:696. doi: 10.1186/s12864-015-1891-8
Chen, Q. J., Deng, B. H., Gao, J., Zhao, Z. Y., Chen, Z. L., Song, S. R., et al. (2019). Comparative analysis of miRNA abundance revealed the function of Vvi-miR828 in fruit coloring in root restriction cultivation grapevine (Vitis vinifera L.). Int. J. Mol. Sci. 20:40582019. doi: 10.3390/ijms20164058
Chen, X. (2008). MicroRNA metabolism in plants. Curr. Top. Microbiol. Immunol. 320, 117–136. doi: 10.1007/978-3-540-75157-16
Chen, X., Xia, J., Xia, Z., Zhang, H., Zeng, C., and Lu, C. (2015). Potential functions of microRNAs in starch metabolism and development revealed by miRNA transcriptome profiling of cassava cultivars and their wild progenitor. BMC Plant Biol. 15:33. doi: 10.1186/s12870-014-0355-7
Cheng, X., Yan, C., Zhang, J., Ma, C., Li, S., Jin, Q., et al. (2017). The effect of different pollination on the expression of Dangshan Su pear microRNA. Biomed Res. Int. 2017:2794040. doi: 10.1155/2017/2794040
Chiou, T. J, and Lin, S. I. (2011). Signaling network in sensing phosphate availability in Plants. Annu. Rev. Plant Biol. 62, 185–206. doi: 10.1146/annurev-arplant-042110-103849
Choudhary, S., Thakur, S., Majeed, A., and Bhardwaj, P. (2018). Exploring microRNA profiles for circadian clock and flowering development regulation in Himalayan Rhododendron. Genomics 111, 1456–1463. doi: 10.1016/j.ygeno.2018.09.019
Cuperus, J. T., Fahlgren, N., and Carrington, J. C. (2011). Evolution and functional diversification of miRNA genes. Plant Cell 23, 431–442. doi: 10.1105/tpc.110.082784
Dai, X. B., and Zhao, P. X. (2011). PsRNATarget: a plant small RNA target analysis server. Nucleic Acids Res. 39, 155–159. doi: 10.1093/nar/gkr319
Damodharan, S., Zhao, D., and Arazi, T. (2016). A common miRNA160-based mechanism regulates ovary patterning, floral organ abscission and lamina outgrowth in tomato. Plant J. 86, 458–471. doi: 10.1111/tpj.13127
David, C., Marie, T., Emilie, T., Sandrine, B., Gaëlle, L., Nathalie, N., et al. (2010). Comparative transcriptomics of drought responses in Populus: a meta-analysis of genome-wide expression profiling in mature leaves and root apices across two genotypes. BMC Genomics 11:630. doi: 10.1186/1471-2164-11-630
Deng, M., Cao, Y., Zhao, Z., Yang, L., Zhang, Y., and Dong, Y. (2017). Discovery of microRNAs and their target genes related to drought in paulownia, “yuza 1” by high-throughput sequencing. Int. J. Genomics 2017:3674682. doi: 10.1155/2017/3674682
dos Santos, T. B. D., João, D. M., Lima, J. E., Silva, J. C., Ivamoto, S. T., Baba, V. Y., et al. (2019). An integrated analysis of mRNA and sRNA transcriptional profilesin Coffea arabica L. roots: insights on nitrogen starvation responses. Funct. Integr. Genomic 19, 151–169. doi: 10.1007/s10142-018-0634-8
Du, J., Miura, E., Robischon, M., Martinez, C., and Groover, A. (2011). The Populus class III HD ZIP transcription factor popcorona affects cell differentiation during secondary growth of woody stems. PLoS One 6:e17458. doi: 10.1371/journal.pone.0017458
Ehya, F., Monavarfeshani, A., Fard, E. M., Farsad, L. K., Nekouei, M. K., Mardi M, et al. (2013). Phytoplasmares-ponsive microRNAs modulate hormonal, nutritional, and stress signaling pathways in mexican lime trees. PLoS One 8:e66372. doi: 10.1371/journal.pone.0066372
Eldem, V., Akcay, U. C., Ozhuner, E., Bakir, Y., Uranbey, S., Unver, T., et al. (2012). Genome-wide identification of miRNAs responsive to drought in peach (Prunus persica) by high-throughput deep sequencing. PLoS One 7:e50298. doi: 10.1371/journal.pone.0050298
Erika, V. G., Lough, R. H., Moss, S. M. A., Wu, R., and Hellens, R. P. (2012). Kiwifruit floral gene APETALA2 is alternatively spliced and accumulates in aberrant indeterminate flowers in the absence of miR172. Plant Mol. Biol. 78, 417–429. doi: 10.1007/s11103-012-9877-2
Fan, D., Li, C., Fan, C., Hu, J., Li, J., Yao, S., et al. (2019). MicroRNA6443-mediated regulation of FERULATE 5-HYDROXYLASE gene alters lignin composition and enhances saccharification in Populus tomentosa. New Phytol. 226, 410–425. doi: 10.1111/nph.16379
Farooq, M., Mansoor, S., Guo, H., Amin, I., Chee, P. W., Azim, M. K., et al. (2017). Identification and characterization of miRNA transcriptome in Asiatic cotton (Gossypium arboreum) using high throughput sequencing. Front. Plant Sci. 8:969. doi: 10.3389/fpls.2017.00969
Feng, H., Xu, M., Zheng, X., Zhu, T., Gao, X., and Huang, L. (2017). MicroRNAs and their targets in apple (Malus domestica cv.”Fuji”) involved in response to infection of pathogen Valsa mali. Front. Plant Sci. 8:2081. doi: 10.3389/fpls.2017.02081
Feng, Y., Yu, Y., Zhou, Y. F., Yang, Y. W., Lei, M. Q., Lian, J. P., et al. (2020). A natural variant of miR397 mediates a feedback loop in circadian rhythm. Plant Physiol. 182, 204–214. doi: 10.1104/pp.19.00710
Filipowicz, W., Bhattacharyya, S. N., and Sonenberg, N. (2008). Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight. Nat. Rev. Genet. 9, 102–114. doi: 10.1038/nrg2290
Franco-Zorrilla, J. M., Valli, A., and Todesco, M. (2007). Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 39, 1033–1037. doi: 10.1038/ng2079
Gai, Y. P., Zhao, H. N., Zhao, Y. N., Zhu, B S., Yuan, S. S., Li S, et al. (2018). MiRNA-seq-based profles of miRNAs in mulberry phloem sap provide insight into the pathogenic mechanisms of mulberry yellow dwarf disease. Sci. Rep. 8:812. doi: 10.1038/s41598-018-19210-7
Galli, V., Guzman, F., de Oliveira, L. F., Loss-Morais, G., Korbes, A. P., Silva SD, et al. (2014). Identifying microRNAs and transcript targets in Jatropha seeds. PLoS One 9:e83727. doi: 10.1371/journal.pone.008372
Gao, F., Wang, N., Li, H., Liu, J., Fu, C., and Xiao, Z. (2016). Identification of drought-responsive microRNAs and their targets in Ammopiptanthus mongolicus by using high-throughput sequencing. Sci. Rep. 6:34601. doi: 10.1038/srep34601
Gébelin, V., Julie, L., Songnian, H., Chaorong, T., and Pascal, M. (2013). Regulation of MIR genes in response to abiotic stress in Hevea brasiliensis. Int. J. Mol. Sci. 14, 19587–19604. doi: 10.3390/ijms141019587
Griffiths-Jones, S. (2004). The microRNA Registry. Nucleic Acids Res. 32, D109–D111. doi: 10.1093/nar/gkh023
Griffiths-Jones, S., Grocock, R. J., Dongen, S., Bateman, A., Enright A. J., et al. (2006). miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 34, D140–D144. doi: 10.1093/nar/gkj112
Griffiths-Jones, S., Saini, H. K., Van, D. S., and Enright, A. J. (2008). miRBase: tools for microRNA genomics. Nucleic Acids Res. 36, D154–D158. doi: 10.1093/nar/gkm952
Guan, R., Zhao, Y., Zhang, H., Fan, G., Liu, X., Zhou, W., et al. (2016). Draft genome of the living fossil Ginkgo biloba. Giga Sci. 5:49. doi: 10.1186/s13742-016-0154-1
Guo, X., Ma, Z., Zhang, Z., Cheng, L., Zhang, X., Li, T., et al. (2017). Small RNA-sequencing links physiological changes and RdDM process to vegetative-to-floral transition in Apple. Front. Plant Sci. 8:873. doi: 10.3389/fpls.2017.00873
Guo, Z. L., Kuang, Z., Wang, Y., Zhao, Y. X., Tao, Y. H., Cheng, C., et al. (2020). PmiREN: a comprehensive encyclopedia of plant mirnas. Nucleic Acids Res. 48, D1114–D1121. doi: 10.1093/nar/gkz894
Guzman, F., Mauricio, P. A., Ana, P. K., Loss-Morais, G., and Margis, R. (2012). Identification of microRNAs from Eugenia uniflora by high-throughput sequencing and bioinformatics analysis. PLoS One 7:e0049811. doi: 10.1371/journal.pone.0049811
Hajieghrari, B., Farrokhi, N., Goliaei, B., and Kavousi, K. (2015). Computational identification, characterization and analysis of conserved miRNAs and their targets in Amborella Trichopoda. J. Data Min. Genomics Proteomics 6:168. doi: 10.4172/2153-0602.1000168
He, F., Xu, C. Z., Shen, X. K., and Guo, Y. (2018). The microRNA390/TRANS-ACTING SHORT INTERFERING RNA3 module mediates lateral root growth under salt stress via the auxin pathway. Plant Physiol. 177, 775–791. doi: 10.1104/pp.17.01559
Hou, J., Xu, H., Fan, D., Ran, L., Li, J., Wu, S., et al. (2020). MiR319a-targeted PtoTCP20 regulates secondary growth via interactions with PtoWOX4 and PtoWND6 in Populus tomentosa. New Phytol. 228, 1354–1368. doi: 10.1111/nph.16782
Huang, J. H., Qi, Y. P., Wen, S. X., Guo, P., Chen, X. M., and Chen, L. S. (2016). Illumina microrRNAs profiles reveal the involvement of miR397a in citrus adaptation to long-term boron toxicity via modulating secondary cell-wall biosynthesis. Sci. Rep. 6, 2978–2995. doi: 10.1038/srep22900
Huang, S., Ding, J., Deng, D., Tang, W., Sun, H., Liu, D., et al. (2013). Draft genome of the kiwifruit Actinidia chinensis. Nat. Commun. 4:2640. doi: 10.1038/ncomms3640
Ibrahim, R. I., Azuma, J., and Sakamoto, M. (2006). Complete nucleotide sequence of the Cotton (Gossypium barbadense L.) chloroplast genome with a comparative analysis of sequences among 9 dicot plants. Nat. Genet. 81, 311–321. doi: 10.1266/ggs.81.311
Jaillon, O., Aury, J., Noel, B., Policriti, A., Clepet, C., Casagrande, A., et al. (2007). The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449, 463–467. doi: 10.1038/nature06148
Jia, X., Yan, J., and Tang, G. (2011). MicroRNA-mediated DNA methylation in plants. Front. Biol. 6, 133–139. doi: 10.1007/s11515-011-1136-4
Jin, Y., Liu, L., Hao, X., Harry, D. E., Zheng, Y., Huang, T., et al. (2019). Unravelling the MicroRNA-Mediated Gene Regulation in Developing Pongamia Seeds by High-Throughput Small RNA Profiling. Int. J. Mol. Sci. 20:3509. doi: 10.3390/ijms20143509
Jones-Rhoades, M. W. (2012). Conservation and divergence in plant microRNAs. Plant Mol. Biol. 80, 3–16. doi: 10.1007/s11103-011-9829-2
Jones-Rhoades, M. W., Bartel, D. P., and Bartel, B. (2006). MicroRNAs and their regulatory roles in plants. Annu. Rev. Plant Biol. 57, 19–53. doi: 10.1146/annurev.arplant.57.033905.105218
Karimi, M., Ghazanfari, F., Fadaei, A., Ahmadi, L., Shiran, B., Rabei, M., et al. (2016). The small-RNA profiles of almond (Prunus dulcis mill.) reproductive tissues in response to cold stress. PLoS One 11:e56519. doi: 10.1371/journal.pone.0156519
Kehr, J. (2013). Systemic regulation of mineral homeostasis by micro RNAs. Front. Plant Sci. 4:145. doi: 10.3389/fpls.2013.00145
Khraiwesh, B., Pugalenthi, G., and Fedoroff, N. V. (2013). Identification and analysis of red sea mangrove (Avicennia marina) microRNAs by high-throughput sequencing and their association with stress responses. PLoS One 8:e60774. doi: 10.1371/journal.pone.0060774
Kiełbasa, S. M., Blïthgen, N., Fähling, M., and Mrowka, R. (2010). Targetfinder. org: a resource for systematic discovery of transcription factor target genes. Nucleic Acids Res. 38, 233–238. doi: 10.1093/nar/gkq374
Kim, J., Park, J., Lim, C., Lim, J., Ryu, J. Y., and Lee, B. W. (2012). Small RNA and transcriptome deep sequencing proffers insight into floral gene regulation in Rosa cultivars. BMC Genomics 13:657. doi: 10.1186/1471-2164-13-657
Kim, T., Park, J. H., Lee, S. G., Kim, S., and Shin, C. (2017). Small RNA transcriptome of Hibiscus Syriacus provides insights into the potential influence of MicroRNAs in flower development and terpene synthesis. Mol. Cells 40, 587–597. doi: 10.14348/molcells.2017.0086
Ko, J. H., Prassinos, C., and Han, K. H. (2006). Developmental and seasonal expression of PtaHB1, a Populus gene encoding a class III HD-Zip protein, is closely associated with secondary growth and inversely correlated with the level of microRNA (miR166). New Phytol. 169, 469–478. doi: 10.1111/j.1469-8137.2005.01623.x
Kozomara, A., Birgaoanu, M., and Griffiths-Jones, S. (2019). miRBase: from microRNA sequences to function. Nucleic Acids Res. 47, D155–D162. doi: 10.1093/nar/gky1141
Kozomara, A., and Griffiths-Jones, S. (2011). miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 39, D152–D157. doi: 10.1093/nar/gkq1027
Kozomara, A., and Griffiths-Jones, S. (2014). miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 42, D68–D73. doi: 10.1093/nar/gkt1181
Kreuzwieser, J., Hauberg, J., Howell, K. A., Carroll, A., Rennenberg, H., Millar, A. H., et al. (2009). Differential response of gray poplar leaves and roots underpins stress adaptation during hypoxia. Plant Physiol. 149, 461–473. doi: 10.1104/pp.108.125989
Kuruvilla, L., Sathik, M., Luke, L. P., and Thomas, M. (2019). Identification and validation of drought-responsive microRNAs from Hevea brasiliensis. Acta Physiol. Plant 41:14. doi: 10.1007/s11738-018-2803-8
Kuruvilla, L., Sathik, M. M., Thomas, M., Luke, L. P., and Sumesh, K. V. (2017). Identification and validation of cold responsive microRNAs of Hevea brasiliensis using high throughput sequencing. J. Crop Sci. Biotechnol. 20, 369–377. doi: 10.1007/s12892-017-0062-0
Lee, W. S., Leong, P. L., Ho, C. L., and Harikrishna, J. A. (2011). Identification of microRNA precursors in Bruguiera spp. Bot. Mar. 54, 313–324. doi: 10.1515/bot.2011.031
Li, B., Duan, H., Li, J., Deng, X. W., Yin, W., and Xia, X. (2013). Global identification of miRNAs and targets in Populus euphratica under salt stress. Plant Mol. Biol. 81, 525–539. doi: 10.1007/s11103-013-0010-y
Li, B., Yin, W., and Xia, X. (2009). Identification of microRNAs and their targets from Populus euphratica. Biochem. Biophys. Res. Commun. 388, 272–277. doi: 10.1016/j.bbrc.2009.07.161
Li, B. S., Qin, Y. R., Duan, H., Yin, W. L., and Xia, X. L. (2011). Genome-wide characterization of new and drought stress responsive microRNAs in Populus euphratica. J. Exp. Bot. 62, 3765–3779. doi: 10.1093/jxb/err051
Li, D., Zheng, Y., Wan, L., Zhu, X., and Wang, Z. (2009). Differentially expressed microRNAs during solid endosperm development in coconut (Cocos nucifera L.). Sci. Hortic. 122, 666–669. doi: 10.1016/j.scienta.2009.07.002
Li, H., Huang, X., Li, W., Lu, Y., Dai, X., Zhou, Z., et al. (2020). MicroRNA comparison between poplar and larch provides insight into the different mechanism of wood formation. Plant Cell Rep. 39, 1199–1217. doi: 10.1007/s00299-020-02559-3
Li, L., Zheng, T., Zhuo, X., Li, S., Qiu, L., Li, S., et al. (2019). Genome-wide identification, characterization and expression analysis of the HD-Zip gene family in the stem development of the woody plant Prunus mume. Peer J. 7:e7499. doi: 10.7717/peerj.7499
Li, R., Chen, D., Wang, T., Wan, Y., and Zhao, W. (2017). High throughput deep degradome sequencing reveals microRNAs and their targets in response to drought stress in mulberry (Morus alba). PLoS One 12:e0172883. doi: 10.1371/journal.pone.0172883
Li, W. F., Zhang, S. G., Han, S. Y., Wu, T., and Zhang, J. H. (2013). Regulation of LaMYB33 by miR159 during maintenance of embryogenic potential and somatic embryo maturation in Larix kaempferi (Lamb.) Carr. Plant Cell Tissue Organ Cult. 113, 131–136. doi: 10.1007/s11240-012-0233-7
Li, Z. X., Li, S. G., Zhang, L. F., Han, S. Y., Li, W. F., Xu, H. Y., et al. (2016). Over-expression of miR166a inhibits cotyledon formation in somatic embryos and promotes lateral root development in seedlings of Larix leptolepis. Plant Cell Tissue Organ Cult. 127, 461–473. doi: 10.1007/s11240-012-0233-7
Lin, Y., and Lai, Z. (2013). Comparative analysis reveals dynamic changes in miRNAs and their targets and expression during somatic embryogenesis in longan (Dimocarpus longan lour.). PLoS One 8:e60337. doi: 10.1371/journal.pone.0060337
Liu, Q., Wang, Z., Yu, S., Li, W., and Li, C. (2020). Pu-miRNA172d regulates stomatal density and water use efficiency via targeting pugtl1 in poplar. J. Exp. Bot. 72, 1370–1383. doi: 10.1093/jxb/eraa493
Liu, R., Lai, B., Hu, B., Qin, Y., Hu, G., and Zhao, J. (2016). Identification of microRNAs and their target genes related to the accumulation of anthocyanins in Litchi chinensis by high-throughput sequencing and degradome analysis. Front. Plant Sci. 7:2059. doi: 10.3389/fpls.2016.02059
Liu, X., Huang, J., Wang, Y., Khanna, K., Xie, Z., Owen, H. A., et al. (2010). The role of floral organs in carpels, an Arabidopsis los-of-function mutation in MicroRNA160a, in organogenesis and the mechanism regulating its expression. Plant J. 62, 416–428. doi: 10.1111/j.1365-313X.2010.04164.x
Liu, X. X., Luo, X. F., Luo, K. X., Liu, Y. L., Pan, T., Li, Z.-Z., et al. (2019). Small RNA sequencing reveals dynamic microrna expression of important nutrient metabolism during development of camellia oleifera fruit. Int. J. Biol. Sci. 15, 416–429. doi: 10.7150/ijbs.26884
Liu, Y., Wang, L., Chen, D., Wu, X., Ding, H., Chen, L., et al. (2014). Genome-wide comparison of microRNAs and their targeted transcripts among leaf, flower and fruit of sweet orange. BMC Genomics 15:695. doi: 10.1186/1471-2164-15-695
Lu, S., Li, Q., Wei, H., Chang, M. J., and Chiang, V. L. (2013). Ptr-miR397a is a negative regulator of laccase genes affecting lignin content in Populus trichocarpa. Proc. Natl. Acad. Sci. U.S.A. 110, 10848–10853. doi: 10.1073/pnas.1308936110
Lu, S., Sun, Y. H., Amerson, H., and Chiang, V. L. (2007). MicroRNAs in loblolly pine (Pinus taeda L.) and their association with fusiform rust gall development. Plant J. 51, 1077–1098. doi: 10.1111/j.1365-313X.2007.03208.x
Lu, S., Sun, Y. H., and Chiang, V. L. (2008). Stress-responsive microRNAs in Populus. Plant J. 55, 131–151. doi: 10.1111/j.1365-313X.2008.03497.x
Lu, S., Sun, Y. H., Shi, R., Clark, C., Li, L., and Chiang, V. L. (2005). Novel and mechanical stress-responsive MicroRNAs in Populus trichocarpa that are absent from Arabidopsis. Plant Cell 17, 2186–2203. doi: 10.1105/tpc.105.033456
Lu, S. F., Yang, C. M., and Chiang, V. L. (2011). Conservation and diversity of microRNA-associated copper-regulatory networks in Populus trichocarpa. J Integr. Plant Biol. 53, 879–891. doi: 10.1111/j.1744-7909.2011.01080.x
Lu, X., Dun, H., Lian, C., Zhang, X., Yin, W., Xia, X., et al. (2017). The role of Peu-miR164 and its target PeNAC genes in response to abiotic stress in, Populus euphratica. Plant Physiol. Biochem. 115, 418–438. doi: 10.1016/j.plaphy.2017.04.009
Lu, Y. B., Yang, L. T., Qi, Y. P., Li, Y., Li, Z., Chen, Y. B., et al. (2014). Identification of boron-deficiency-responsive microRNAs in Citrus sinensisroots by illumina sequencing. BMC Plant Biol. 14:123. doi: 10.1186/1471-2229-14-123
Ma, F., Huang, J., Yang, J., Zhou, J., and Sun, J. (2020). Identification, expression and miRNA targeting of auxin response factor genes related to phyllody in the witches’ broom disease of jujube. Gene 746, 144656. doi: 10.1016/j.gene.2020.144656
Mehrpooyan, F., Othman, R. Y., and Harikrishna, J. A. (2012). Tissue and temporal expression of miR172 paralogs and the AP2-like target in oil palm (Elaeis guineensis Jacq.). Tree Genet. Genome 8, 1331–1343. doi: 10.1007/s11295-012-0519-2
Mewalal, R., Yin, H., Hu, R., Jawdy, S., Vion, P., Tuskan, G. A., et al. (2019). Identification of populus small rnas responsive to mutualistic interactions with mycorrhizal fungi, laccaria bicolor and rhizophagus irregularis. Front. Microbiol. 10:151. doi: 10.3389/fmicb.2019.00515
Ming, R., Hou, S., Feng, Y., and Yu, Q. (2008). The draft genome of the transgenic tropical fruit tree papaya (Carica papaya Linnaeus). Nature 452, 991–996. doi: 10.1038/nature06856
Niu, J., Wang, J., Hu, H., Chen, Y., An, J., Cai, J., et al. (2016). Cross-talk between freezing response and signaling for regulatory transcriptions of MIR475b and its targets by miR475b promoter in Populus suaveolens. Sci. Rep. 6:20648. doi: 10.1038/srep20648
Notaguchi, M., and Okamoto, S. (2015). Dynamics of long-distance signaling via plant vascular tissues. Front. Plant Sci. 6:161. doi: 10.3389/fpls.2015.00161
Oh, T. J., Wartell, R. M., Cairney, J., and Pullman, G. S. (2008). Evidence for stage-specific modulation of specific microRNAs (miRNAs) and miRNA processing components in zygotic embryo and female gametophyte of loblolly pine (Pinus taeda). New Phytol. 179, 67–80. doi: 10.1111/j.1469-8137.2008.02448.x
Ong, S. S., and Wickneswari, R. (2012). Characterization of microRNAs expressed during secondary wall biosynthesis in Acacia mangium. PLoS One 7:e49662. doi: 10.1371/journal.pone.0049662
Pagliarani, C., Vitali, M., Ferrero, M., Vitulo, N., Incarbone, M., Lovisolo, C., et al. (2017). The accumulation of miRNAs differentially modulated by drought stress is affected by grafting in grapevine. Plant Physiol. 173, 2180–2195. doi: 10.1104/pp.16.01119
Pan, J. W., Huang, D. H., Guo, Z. L., Kuang, Z., Zhang, H., Xie, X. Y., et al. (2018). Overexpression of microRNA408 enhances photosynthesis, growth, and seed yield in diverse plants. J. Integr. Plant Biol. 60, 323–340. doi: 10.1111/jipb.12634
Pan, Y., Niu, M. Y., Liang, J. S., Lin, E., Tong, Z. K., Zhang, J. H., et al. (2017). Identification of heat-responsive miRNAs to reveal the miRNA-mediated regulatory network of heat stress response in Betula luminifera. Trees 31, 1635–1652. doi: 10.1007/s00468-017-1575-x
Pant, B. D., Buhtz, A., Kehr, J., and Scheible, W. R. (2008). MicroRNA399 is a long-distance signal for the regulation of plant phosphate homeostasis. Plant J. 53, 731–738. doi: 10.1111/j.1365-313X.2007.03363.x
Pantaleo, V., Vitali, M., Boccacci, P., Miozzi, L., Cuozzo, D., Chitarra, W., et al. (2016). Novel functional microRNAs from virus-free and infected Vitis vinifera plants under water stress. Sci. Rep. 6:20167. doi: 10.1038/srep20167
Pappas, M. C. R., Pappas, G. J., and Grattapaglia, D. (2015). Genome-wide discovery and validation of Eucalyptus small RNAs reveals variable patterns of conservation and diversity across species of Myrtaceae. BMC Genomics 16:1113. doi: 10.1186/s12864-015-2322-6
Patanun, O., Lertpanyasampatha, M., and Punchapat, S. (2013). Computational identification of microRNAs and their targets in cassava (Manihot esculenta Crantz.). Mol. Biotechnol. 53, 257–269. doi: 10.1007/s12033-012-9521-z
Pei, H., Ma, N., Chen, J., Zheng, Y., Tian, J., Li, J., et al. (2013). Integrative analysis of miRNA and miRNA profiles in response to ethylene in rose petals during flower opening. PLoS One 8:e64290. doi: 10.1371/journal.pone.0064290
Peng, S., Dan, L., Zhang, Z. J., Yin, W., and Xia, X. L. (2013). Identification of drought-responsive and novel Populus trichocarpa microRNAs by high-throughput sequencing and their targets using degradome analysis. BMC Genomics 14:233. doi: 10.1186/1471-2164-14-233
Qu, D., Yan, F., Meng, R., Jiang, X., Yang, H., Gao, Z., et al. (2016). Identification of microRNAs and their targets associated with fruit-bagging and subsequent sunlight re-exposure in the”granny smith” apple exocarp using high-throughput sequencing. Front. Plant Sci. 7:27. doi: 10.3389/fpls.2016.00027
Quan, M., Du, Q., Xiao, L., Lu, W., Wang, L., Xie, J., et al. (2019). Genetic architecture underlying the lignin biosynthesis pathway involves noncoding RNAs and transcription factors for growth and wood properties in Populus. Plant Biotechnol. J. 17, 302–315. doi: 10.1111/pbi.12978
Quan, M., Wang, Q., Phangthavong, S., Yang, X., Song, Y., Du, Q., et al. (2016). Association studies in Populus tomentosa reveal the genetic interactions of Pto-MIR156c and its targets in wood formation. Front. Plant Sci. 7:1159. doi: 10.3389/fpls.2016.01159
Quinn, C. R., Iriyama, R., and Fernando, D. D. (2015). Computational predictions and expression patterns of conserved microRNAs in loblolly pine (Pinus taeda). Tree Genet. Genome 11:806. doi: 10.1007/s11295-014-0806-1
Ramachandran, V., and Chen, X. (2008). Degradation of microRNAs by a family of exoribonucleases in Arabidopsis. Science 321, 1490–1492. doi: 10.1126/science.1163728
Ramos, A., Usié, A., Barbosa, P., Barros, P. M., Capote, T., Chaves, I., et al. (2018). The draft genome sequence of cork oak. Sci. Data 5:180069. doi: 10.1038/sdata.2018.69
Ren, Y., Chen, L., Zhang, Y., Kang, X., Zhang, Z., and Wang, Y. (2012). Identification of novel and conserved Populus tomentosa microRNA as components of a response to water stress. Funct. Integr. Genomic 12, 327–339. doi: 10.1007/s10142-012-0271-6
Ren, Y., Chen, L., Zhang, Y., Kang, X., Zhang, Z., and Wang, Y. (2013). Identification and characterization of salt-responsive microRNAs in Populus tomentosa by high-throughput sequencing. Biochimie 95, 743–750. doi: 10.1016/j.biochi.2012.10.025
Ren, Y. Y., Sun, F. S., Hou, J., Chen, L., Zhang, Y. Y., Kang, X. Y., et al. (2015). Differential profiling analysis of miRNAs reveals a regulatory role in low N stress response of Populus. Funct. Integr. Genomics 15, 1–13. doi: 10.1007/s10142-014-0408-x
Rendón-Anaya, M., Ibarra-Laclette, E., Méndez-Bravo, A., Lan, T., Zheng, C., Carretero-Paulet, L., et al. (2019). The Avocado genome informs deep angiosperm phylogeny, highlights introgressive hybridization, and reveals pathogen-influenced gene space adaptation. Proc. Natl. Acad. Sci. U.S.A. 116, 17081–17089. doi: 10.1073/pnas.1822129116
Rhoades, M. W., Reinhart, B. J., Lim, L. P., Burge, C. B., Bartel, B., and Bartel, D. P. (2002). Prediction of plant microrna targets. Cell 110, 513–520. doi: 10.1016/S0092-8674(02)00863-2
Saminathan, T., Bodunrin, A., Singh, N. V., Devarajan, R., Nimmakayala, P., Jeff, M., et al. (2016). Genome-wide identification of MicroRNAs in Pomegranate (Punica granatum L.) by high-throughput sequencing. BMC Plant Biol. 16:122. doi: 10.1186/s12870-016-0807-3
Shi, W., Liu, W., Ma, C., Zhang, Y., Ding, S., Yu, W., et al. (2020). Dissecting microRNA–mRNA regulatory networks underlying sulfur assimilation and cadmium accumulation in poplar leaves. Plant Cell Physiol. 61, 1614–1630. doi: 10.1093/pcp/pcaa084
Si, J., Quan, M., Xiao, L., Xie, J., Du, Q., Zhang, D., et al. (2020). Genetic interactions among Pto-miR319 family members and their targets influence growth and wood properties in Populus tomentosa. Mol. Genet. Genomics 295, 855–870. doi: 10.1007/s00438-020-01667-9
Si, J. N., Zhou, T., Bo, W., Xu, F., and Wu, R. (2014). Genome-wide analysis of salt-responsive and novel microRNAs in Populus euphratica by deep sequencing. BMC Genet. 15:S6. doi: 10.1186/1471-2156-15-S1-S6
Sieber, P., Wellmer, F., Gheyselinck, J., Riechmann, J. L., and Meyerowitz, E. M. (2007). Redundancy and specialization among plant microRNAs: role of the MIR164 family in developmental robustness. Development 134, 1051–1060. doi: 10.1242/dev.02817
Snyman, M. C., Solofoharivelo, M. C., Souza-Richards, R., Stephan, D., Murray, S., Burger, J. T., et al. (2017). The use of high-throughput small RNA sequencing reveals differentially expressed microRNAs in response to aster yellows phytoplasma-infection in Vitis vinifera cv.’chardonnay’. PLoS One 12:e0182629. doi: 10.1371/journal.pone.0182629
Sollars, E. S., Harper, A. L., Kelly, L. J., Sambles, C. M., Ramirez-Gonzalez, R. H., Swarbreck, D., et al. (2017). Genome sequence and genetic diversity of European ash trees. Nature 12, 212–216. doi: 10.1038/nature20786
Solofoharivelo, M. C., Van, D., Stephan, D., Burger, J. T., Murray, S. L., Weber, A., et al. (2015). MicroRNAs in fruit trees: discovery, diversity and future research directions. Plant Biol. 16, 856–865. doi: 10.1111/plb.12153
Song, C., Fang, J., Li, X., Liu, H., and Thomas, C. (2009). Identification and characterization of 27 conserved microRNAs in citrus. Planta 230, 671–685. doi: 10.1007/s00425-009-0971-x
Song, Y., Ma, K., Ci, D., Zhang, Z., and Zhang, D. (2013). Sexual dimorphism floral microRNA profiling and target gene expression in Andromonoecious Poplar (Populus tomentosa). PLoS One 8:e062681. doi: 10.1371/journal.pone.0062681
Song, Y., Tian, M., Ci, D., and Zhang, D. (2015). Methylation of microRNA genes regulates gene expression in bisexual flower development in andromonoecious poplar. J. Exp. Bot. 66, 1891–1905. doi: 10.1093/jxb/eru531
Song, Z., Zhang, L., Wang, Y., Li, H., and Zhang, H. (2018). Constitutive expression of mir408 improves biomass and seed yield in Aarabidopsis. Front. Plant Sci. 8:2114. doi: 10.3389/fpls.2017.02114
Su, Y., Li, H. G., Wang, Y., Li, S., Wang, H. L., Yu, L., et al. (2018). Poplar mir472a targeting NBS-LRRs is involved in effective defense response to necrotrophic fungus cytospora chrysosperma. J. Exp. Bot. 69, 5519–5530. doi: 10.1093/jxb/ery304
Sun, L., Jiang, Z., Ju, Y., Zou, X., Wan, X., and Chen, Y. (2020). A potential endogenous gibberellin-mediated signaling cascade regulated floral transition in Magnolia × soulangeana ‘Changchun’. Mol. Genet. Genomics 296, 207–222. doi: 10.1007/s00438-020-01740-3
Sun, R. Z. (2011). Cloning and analysis of the low temperature stress-responsive microRNAs from Populus suaveolens. Genomics Appl. Biol. 30, 204–211. doi: 10.3969/gab.030.000204
Sun, X., Fan, G., Su, L., Wang, W., Liang, Z., Li, S., et al. (2015). Identification of cold-inducible microRNAs in grapevine. Front. Plant Sci. 6:595. doi: 10.3389/fpls.2015.00595
Sun, X., Lin, L., and Sui, N. (2019). Regulation mechanism of microRNA in plant response to abiotic stress and breeding. Mol. Biol. Rep. 46, 1447–1457. doi: 10.1007/s11033-018-4511-2
Sun, Z. C., Zhang, L. S., and Wang, Z. J. (2017). Genome-wide analysis of miRNAs in Carya cathayensis. BMC Plant Biol. 17:228. doi: 10.1186/s12870-017-1180-6
Tang, M., Bai, X., Niu, L., Chai, X., Chen, M., and Xu, Z. (2018). MiR172 regulates both vegetative and reproductive development in the perennial woody plant Jatropha curcas. Plant Cell Physiol. 59, 2549–2563. doi: 10.1093/pcp/pcy175
Taylor, R. S., Tarver, J. E., Foroozani, A., and Donoghue, P. C. (2017). MicroRNA annotation of plant genomes - Do it right or not at all. Bioessays 39:1600113. doi: 10.1002/bies.201600113
Tian, J., Chen, J., Li, B., and Zhang, D. (2016). Association genetics in Populus reveals the interactions between Pto-miR160a and its target Pto-ARF16. Mol. Genet. Genomics 291, 1069–1082. doi: 10.1007/s00438-015-1165-9
Tian, W., Ge, Y., Liu, X., Dou, G., and Ma, Y. (2019). Identification and characterization of populus micrornas in response to plant growth-promoting endophytic Streptomyces sp. SSD49. World J. Microbiol. Biotechnol. 35:97. doi: 10.1007/s11274-019-2671-4
Todesco, M., Rubiosomoza, I., Pazares, J., and Weigel, D. (2010). A collection of target mimics for comprehensive analysis of microRNA function in Arabidopsis thaliana. PLoS Genet. 6:e1001031. doi: 10.1371/journal.pgen.1001031
Tuskan, G. A., Difazio, S., Jansson, S., Bohlmann, J., Grigoriev, I., Hellsten, U., et al. (2006). The genome of black cottonwood, Populus trichocarpa (torr.& gray). Science 313, 1596–1604. doi: 10.1126/science.1128691
Vanessa, G., Frank, G., de Oliveira, L. F. V., Guilherme, L. M., Rbes, A., Silva, S., et al. (2014). Identifying MicroRNAs and transcript targets in Jatropha seeds. PLoS One 9:e83727. doi: 10.1371/journal.pone.0083727
Varkonyi-Gasic, E., Gould, N., Sandanayaka, M., Sutherland, P., and MacDiarmid, R. M. (2010). Characterisation of microRNAs from apple (Malus domestica ‘Royal Gala’) vascular tissue and phloem sap. BMC Plant Biol. 10:159. doi: 10.1186/1471-2229-10-159
Voinnet, O. (2009). Origin, biogenesis, and activity of plant microRNAs. Cell 136, 669–687. doi: 10.1016/j.cell.2009.01.046
Wan, L. C., Wang, F., Guo, X., Lu, S., Qiu, Z., and Zhao, Y. (2012). Identification and characterization of small non-coding RNAs from chinese fir by high throughput sequencing. BMC Plant Biol. 12:146–146. doi: 10.1186/1471-2229-12-146
Wang, C. Y., Zhang, S., Yu, Y., Luo, Y. C., Liu, Q., Ju, C., et al. (2014). MiR397b regulates both lignin content and seed number in Arabidopsis via modulating a laccase involved in lignin biosynthesis. Plant Biotechnol. J. 12, 1132–1142. doi: 10.1111/pbi.12222
Wang, H., Zhao, S. C., Gao, Y. C., and Yang, J. L. (2017). Characterization of Dof transcription factors and their responses to osmotic stress in Poplar (Populus trichocarpa). PLoS One 12:e0170210. doi: 10.1371/journal.pone.0170210
Wang, J., Park, M. Y., Wang, L., Koo, Y., Chen, X., Weigel, D., et al. (2011). MiRNA control of vegetative phase change in trees. PLoS Genet. 7:e1002012. doi: 10.1371/journal.pgen.1002012
Wang, T., Pan, H., Wang, J., Yang, W., Cheng, T., and Zhang, Q. (2014). Identification and profiling of novel and conserved microRNAs during the flower opening process in Prunus mume via deep sequencing. Mol. Genet. Genomics 289, 169–183. doi: 10.1007/s00438-013-0800-6
Wang, X., Xu, Y., Zhang, S., Cao, L., Huang, Y., Cheng, J., et al. (2017). Genomic analyses of primitive, wild and cultivated Citrus provide insights into asexual reproduction. Nat. Genet. 49, 765–772. doi: 10.1038/ng.3839
Wang, Y., Zhao, Z., Deng, M., Liu, R., Niu, S., and Fan, G. (2015). Identification and functional analysis of microRNAs and their targets in Platanus acerifolia under lead (pb) stress. Int. J. Mol. Sci. 16, 7098–7111. doi: 10.3390/ijms16047098
Wang, Z. J., Huang, J. Q., Huang, Y. J., Li, Z., and Zheng, B. S. (2012). Discovery and profiling of novel and conserved microRNAs during flower development in Carya cathayensis via deep sequencing. Planta 236, 613–621. doi: 10.1007/s00425-012-1634-x
Wei, S., Ye, N., and Yin, T. (2017). Detecting the candidate gender determinants by bioinformatic prediction of miRNAs and their targets from transcriptome sequences of the male and female flowers in Salix suchowensis. Biomed Res. Int. 2017:9614596. doi: 10.1155/2017/9614596
Wilkins, O., Waldron, L., Nahal, H., Provart, N. J., and Campbell, M. M. (2009). Genotype and time of day shape the Populus drought response. Plant J. 60, 703–715. doi: 10.1111/j.1365-313X.2009.03993.x
Wong, M. M., Cannon, C. H., and Wickneswari, R. (2011). Identification of lignin genes and regulatory sequences involved in secondary cell wall formation in Acacia auriculiformis and Acacia mangium via de novo transcriptome sequencing. BMC Genomics 12:342. doi: 10.1186/1471-2164-12-342
Wu, H. J., Ma, Y. K., Tong, C., Wang, M., and Wang, X. J. (2012). PsRobot: a web-based plant small RNA meta-analysis toolbox. Nucleic Acids Res. 40, 22–28. doi: 10.1093/nar/gks554
Wu, J., Wang, D., Liu, Y., Wang, L., Qiao, X., and Zhang, S. (2014). Identification of miRNAs involved in pear fruit development and quality. BMC Genomics 15:953. doi: 10.1186/1471-2164-15-953
Wu, L., Zhou, H., Zhang, Q., Zhang, J., and Qi, Y. (2010). DNA methylation mediated by a microRNA pathway. Mol. Cell 38, 465–475. doi: 10.1016/j.molcel.2010.03.008
Wu, P., Han, S., Zhao, W., Chen, T., Zhou, J., and Li, L. (2015). Genome-wide identification of abiotic stress-regulated and novel microRNAs in mulberry leaf. Plant Physiol. Biochem. 95, 75–82. doi: 10.1016/j.plaphy.2015.07.007
Xia, J., Zeng, C., Chen, Z., Zhang, K., Chen, X., Zhou, Y. F., et al. (2014). Endogenous small-noncoding RNAs and their roles in chilling response and stress acclimation in cassava. BMC Genomics 15:634. doi: 10.1186/1471-2164-15-634
Xia, R., Xu, J., Arikit, S., and Meyers, B. C. (2015). Extensive families of miRNAs and PHAS loci in Norway spruce demonstrate the origins of complex phasiRNA networks in seed plants. Mol. Biol. Evol. 32, 2905–2918. doi: 10.1093/molbev/msv164
Xia, R., Zhu, H., An, Y., Beers, E. P., and Liu, Z. (2012). Apple miRNAs and tasiRNAs with novel regulatory networks. Genome Biol. 13, 1–18. doi: 10.1186/gb-2012-13-6-r47
Xia, W., Li, J., and Zhou, L. X. (2014). Prediction and characterization of conserved microRNA in Cocos nucifera. Guangdong Agric. Sci. 14, 130–135. doi: 10.16768/j.issn.1004-874x.2014.14.030
Xie, R., Zhang, J., Ma, Y., Pan, X., Dong, C., Pang, S., et al. (2017). Combined analysis of mRNA and miRNA identifies dehydration and salinity responsive key molecular players in citrus roots. Sci. Rep. 7:42094. doi: 10.1038/srep42094
Xing, L., Zhang, D., Li, Y., Zhao, C., Zhang, S., Shen, Y., et al. (2014). Genome-wide identification of vegetative phase transition-associated microRNAs and target predictions using degradome sequencing in Malus hupehensis. BMC Genomics 15:1125. doi: 10.1186/1471-2164-15-1125
Xue, C., Yao, J., Qin, M., Zhang, M., Allan, A. C., Wang, D., et al. (2018). PbrmiR397a regulates lignification during stone cell development in pear fruit. Plant Biotechnol. J. 17, 103–117. doi: 10.1111/pbi.12950
Yaish, M. W., Ramanjulu, S., Yun, Z., Bo, J., Rashid, A. Y., Farooq, S. A., et al. (2015). A genome-wide identification of the miRNAome in response to salinity stress in date palm (Phoenix dactylifera L.). Front. Plant Sci. 6:946. doi: 10.3389/fpls.2015.00946
Yan, C., Yin, M., Zhang, N., Jin, Q., Fang, Z., Lin, Y., et al. (2014). Stone cell distribution and lignin structure in various pear varieties. Sci. Hortic. 174, 142–150. doi: 10.1016/j.scienta.2014.05.018
Yan, J., Gu, Y., Jia, X., Kang, W., Pan, S., and Tang, X. (2012). Effective small RNA destruction by the expression of a short tandem target mimic in Arabidopsis. Plant Cell 24, 415–427. doi: 10.1105/tpc.111.094144
Yanik, H., Turktas, M., Dundar, E., Hernandez, P., Dorado, G., and Unver, T. (2013). Genome-wide identification of alternate bearing-associated microRNAs (miRNAs) in olive (Olea europaea L.). BMC Plant Biol. 13:10. doi: 10.1186/1471-2229-13-10
Yao, J. L., Xu, J., Cornille, A., Tomes, S., Karunairetnam, S., Luo, Z., et al. (2015). A microRNA allele that emerged prior to apple domestication may underlie fruit size evolution. Plant J. 84, 417–427. doi: 10.1111/tpj.13021
Yin, D. D., Li, S. S., Shu, Q. Y., Gu, Z. Y., Wu, Q., Feng, C. Y., et al. (2018). Identification of microRNAs and long non-coding rnas involved in fatty acid biosynthesis in tree peonyseeds. Gene 666, 72–82. doi: 10.1016/j.gene.2018.05.011
Zeng, S., Liu, Y., Pan, L., Hayward, A., and Wang, Y. (2015). Identification and characterization of miRNAs in ripening fruit of Lycium barbarum using high-throughput sequencing. Front. Plant Sci. 6:778. doi: 10.3389/fpls.2015.00778
Zhang, B. H. (2015). MicroRNA: a new target for improving plant tolerance to abiotic stress. J. Exp. Bot. 66, 1749–1761. doi: 10.1093/jxb/erv013
Zhang, C., Zhang, B., Ma, R., Yu, M., Guo, S., Guo, L., et al. (2016). Identification of known and novel microRNAs and their targets in peach (prunus persica) fruit by high-throughput sequencing. PLoS One 11:e0159253. doi: 10.1371/journal.pone.0159253
Zhang, J., Lin, Y., Wu, F., Zhang, Y., Cheng, L., Huang, M., et al. (2021). Profiling of microRNAs and their targets in roots and shoots reveals a potential miRNA-mediated interaction network in response to phosphate deficiency in the forestry tree Betula luminifera. Front. Genet. 12:552454. doi: 10.3389/fgene.2021.552454
Zhang, J., Zhang, S., Han, S., Li, X., Tong, Z., and Qi, L. (2013). Deciphering small noncoding RNAs during the transition from dormant embryo to germinated embryo in larches (Larix leptolepis). PLoS One 8:e0081452. doi: 10.1371/journal.pone.0081452
Zhang, J. P., Yu, Y., Feng, Y. Z., Zhou, Y. F., Zhang, F., Yang, Y. W., et al. (2017). MiR408 regulates grain yield and photosynthesis via a phytocyanin protein. Plant Physiol. 175, 1175–1185. doi: 10.1104/pp.17.01169
Zhang, Q., Zhao, C., Li, M., Sun, W., and Zhao, Y. (2013). Genome-wide identification of Thellungiella salsuginea microRNAs with putative roles in the salt stress response. BMC Plant Biol. 13:180. doi: 10.1186/1471-2229-13-180
Zhang, X. N., Li, X., and Liu, J. H. (2014). Identification of conserved and novel cold-responsive microRNAs in trifoliate orange (Poncirus trifoliata (L.) raf.) using high-throughput sequencing. Plant Mol. Biol. Rep. 32, 328–341. doi: 10.1007/s11105-013-0649-1
Zhang, X. N., Wang, W., Wang, M., Zhang, H. Y., and Liu, J. H. (2016). The miR396b of Poncirus trifoliata functions in cold tolerance by regulating ACC oxidase gene expression and modulating ethylene-polyamine homeostasis. Plant Cell Physiol. 57, 1865–1878. doi: 10.1093/pcp/pcw108
Zhang, Y., Zhu, X., Chen, X., Song, C., Zou, Z., and Wang, Y. (2014). Identification and characterization of cold-responsive microRNAs in tea plant (Camellia sinensis) and their targets using high-throughput sequencing and degradome analysis. BMC Plant Biol. 14:271. doi: 10.1186/s12870-014-0271-x
Zhang, Y. C., Yu, Y., Wang, C. Y., Li, Z. Y., Liu, Q., Xu, J., et al. (2013). Overexpression of microRNA OsmiR397 improves rice yield by increasing grain size and promoting panicle branching. Nat. Biotechnol. 31, 848–852. doi: 10.1038/nbt.2646
Zhang, Y. Q., Zeng, Z. H., Chen, C. J., Li, C. Q., Xia, R., Li, J. G., et al. (2019). Genome-wide characterization of the auxin response factor (ARF) gene family of litchi (Litchi chinensis Sonn.): phylogenetic analysis, miRNA regulation and expression changes during fruit abscission. Peer J. 7:e6677. doi: 10.7717/peerj.6677
Zhao, J. P., Jiang, X. L., Zhang, B. Y., and Su, X. H. (2012). Involvement of microRNA-mediated gene expression regulation in the pathological development of stem canker disease in Populus trichocarpa. PLoS One 7:e44968. doi: 10.1371/journal.pone.0044968
Zhao, M., Tai, H., Sun, S., Zhang, F., Xu, Y., and Li, W. X. (2012). Cloning and characterization of maize miRNAs involved in responses to nitrogen deficiency. PLoS One 7:e29669. doi: 10.1371/journal.pone.0029669
Zheng, Y., Chen, C., Liang, Y., Sun, R., Gao, L., Liu, T., et al. (2019). Genomewide association analysis of the lipid and fatty acid metabolism regulatory network in the mesocarp of oil palm (Elaeis guineensis Jacq.) based on small noncoding RNA sequencing. Tree Physiol. 39, 356–371. doi: 10.1093/treephys/tpy091
Zhou, B., Kang, Y., Leng, J., and Xu, Q. (2019). Genome-wide analysis of the mirna-mrnas network involved in cold tolerance in Populus simonii × p. nigra. Genes 10:430. doi: 10.3390/genes10060430
Zhou, J., Liu, M., Jiang, J., Qiao, G., and Zhuo, R. (2012). Expression profile of miRNAs in Populus cathayana L. and Salix matsudana Koidz under salt stress. Mol. Biol. Rep. 39, 8645–8654. doi: 10.1007/s11033-012-1719-4
Zhou, L., Quan, S., Xu, H., Ma, L., and Niu, J. (2018). Identification and expression of miRNAs related to female flower induction in walnut (Juglans regia). Molecules 23:1202. doi: 10.3390/molecules23051202
Zhu, H., Xia, R., Zhao, B., An, Y. Q., Dardick, C. D., Callahan, A. M., et al. (2012). Unique expression, processing regulation, and regulatory network of peach (Prunus persica) miRNAs. BMC Plant Biol. 12:149. doi: 10.1186/1471-2229-12-149
Zhu, J., Li, W., Yang, W., Qi, L., and Han, S. (2013). Identification of microRNAs in Caragana intermediaby high-throughput sequencing and expression analysis of 12 microRNAs and their targets under salt stress. Plant Cell Rep. 32, 1339–1349. doi: 10.1007/s00299-013-1446-x
Keywords: MicroRNAs, functions, woody plants, growth and development, abiotic and biotic stresses
Citation: Fang L and Wang Y (2021) MicroRNAs in Woody Plants. Front. Plant Sci. 12:686831. doi: 10.3389/fpls.2021.686831
Received: 28 March 2021; Accepted: 03 August 2021;
Published: 31 August 2021.
Edited by:
Anna N. Stepanova, North Carolina State University, United StatesReviewed by:
Lee Jeong Hwan, Jeonbuk National University, South KoreaRonald Ross Sederoff, North Carolina State University, United States
Copyright © 2021 Fang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yanmei Wang, d3ltMzU1NDcxMEAxNjMuY29t
 Lisha Fang
Lisha Fang Yanmei Wang1,2*
Yanmei Wang1,2*