- Department of Biological Sciences, Life Sciences I, Virginia Tech, Blacksburg, VA, United States
The development of host-microbe interactions between legumes and their cognate rhizobia requires localization of the bacteria to productive sites of initiation on the plant roots. This end is achieved by the motility apparatus that propels the bacterium and the chemotaxis system that guides it. Motility and chemotaxis aid rhizobia in their competitiveness for space, resources, and nodulation opportunities. Here, we examine studies on chemotaxis of three major model rhizobia, namely Sinorhizobium meliloti, Rhizobium leguminosarum, and Bradyrhizobium japonicum, cataloging their range of attractant molecules and correlating this in the context of root and seed exudate compositions. Current research areas will be summarized, gaps in knowledge discussed, and future directions described.
Introduction
The endosymbiosis between leguminous plants and rhizobia benefits both parties whereby the bacteria, sheltered and supplied nutrients from the plant, fix nitrogen into ammonia for their host. Several rhizobium-legume combinations have stood out as the model systems for this symbiosis, namely Sinorhizobium (Ensifer) meliloti – Medicago truncatula, Bradyrhizobium japonicum – Glycine max, and Rhizobium leguminosarum, the latter able to nodulate clovers, pea, common bean, or others depending on the biovar. These organisms have been used to build our knowledge on the genetics, biochemistry, development, and ecology of the many facets of this interaction. The initiation step of the symbiosis occurs at the tips of young root hairs, which curl and pinch in on a population of rhizobia, allowing access inside the plant cells. Prior to this, the rhizobia must localize themselves to the root hairs and outcomplete other bacteria for this niche (Ames et al., 1980; Ames and Bergman, 1981; Pinochet et al., 1993). This is achieved with chemotaxis and motility, the phenomenon by which bacteria move up a gradient of attractant or down a gradient of repellent. Attractant and repellent signals are many and diverse, as bacteria can respond to carbon sources, heavy metals, osmolytes, pH, light, and temperature (Tso and Adler, 1974; Hazelbauer, 1975; Croxen et al., 2006; Jekely, 2009; Paulick et al., 2017; Webb et al., 2017a). The typical mechanism of chemotactic sensing starts with transmembrane sensor proteins called methyl-accepting chemotaxis proteins (MCPs or receptors), which sense multiple and highly different signals such as single molecules, chemical classes, and physical stimuli (Paulick et al., 2017). MCP-ligand binding modulates the autokinase activity of the internal chemotaxis protein CheA. Phosphorylated CheA transfers its phosphoryl group to the response regulator CheY, which interacts with the flagellar motor to affect a change in its rotation. This two-component system thus controls the movement of the bacterium toward an attractant or away from a repellent by sensing increasing or decreasing ligand binding (Parkinson et al., 2015; Salah Ud-Din and Roujeinikova, 2017).
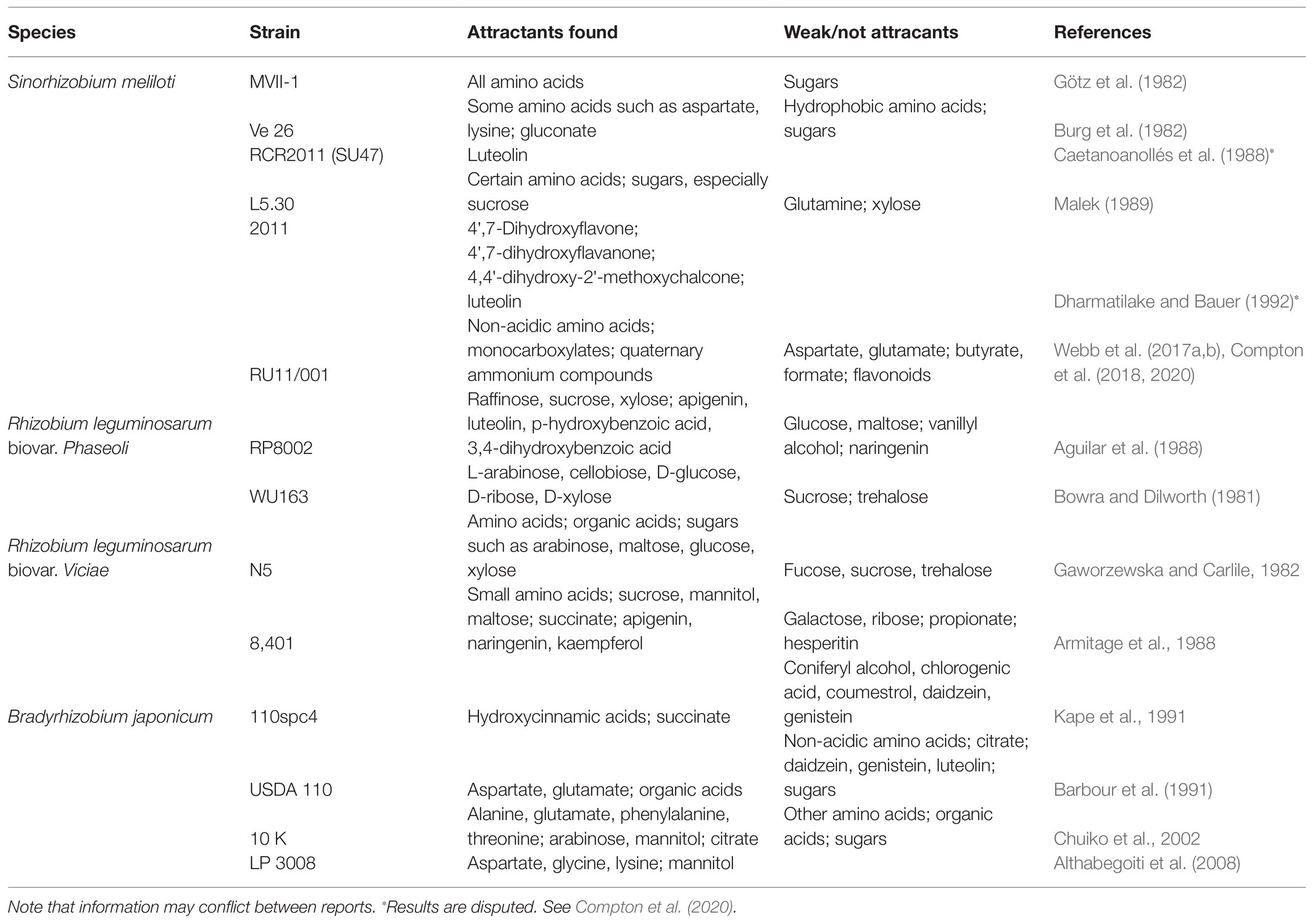
To study chemotaxis, numerous assays have been used to quantify bacterial behavior, but Adler’s capillary assay remains the gold standard (Adler, 1966, 1973). In short, the method involves filling glass capillaries with a putative attractant solution and placing the capillary into a suspension of bacteria. During incubation, the attractant solution forms a gradient which the bacteria follow inside the capillary. The result is typically measured with colony counts of the capillary contents. A reference capillary containing only buffer is included to account for diffusion of cells, acting as an internal negative control. Chemotaxis values are either reported by subtracting reference counts from test counts or as a coefficient with test counts being divided by the reference. Unfortunately, variations in growth conditions, technical procedures, and bacterial strains make comparing studies between lab groups difficult. For example, one study might define a few thousand cells above background as significant, while another might require >105 cells as a significant response (Aguilar et al., 1988; Barbour et al., 1991; Kape et al., 1991; Meier et al., 2007; Webb et al., 2014).
Here, we review studies that used the capillary assay to derive information about the attractants of B. japonicum, R. leguminosarum, and S. meliloti (Table 1). Other methods such as swim plates are available, but do not offer comparable resolution and so will be excluded (Sampedro et al., 2015). A discussion will focus on chemical classes that have been tested for chemotaxis and their prevalence in plant exudates.
Attractants Classes of Model Rhizobia
When comparing results from different behavioral chemotaxis assays, one should bear in mind that attractant profiles of different species and strains will vary. Growth conditions (such as variations in media, temperature, aeration, and growth phase) can cause variances in receptor expression, altering the sensory capability of the bacterium (Lopez-Farfan et al., 2017). In addition, the literature is biased because some compounds have been tested more frequently than others and not to the same extent in different rhizobial species. However, it is naive to expect every source to standardize the array of compounds tested since most studies have a focus on a particular compound or component of plant exudates. As it stands, comparative analyses are restricted.
Amino Acids
One of the most frequently tested compounds is amino acids because they are ubiquitous in exudates (Moe, 2013). Rhizobium leguminosarum biovar viciae is similarly attracted to all amino acids; only glutamate and proline stand out as stronger attractants (Gaworzewska and Carlile, 1982; Armitage et al., 1988). In contrast, B. japonicum was weakly attracted to non-acidic amino acids, while aspartate and glutamate were potent attractants (Barbour et al., 1991; Chuiko et al., 2002; Althabegoiti et al., 2008). S. meliloti senses all amino acids as attractants. Two groups presented aspartate or leucine as the strongest (Burg et al., 1982; Götz et al., 1982, respectively). However, these findings are in contrast with more recent reports that described the molecular mechanism for amino acid sensing, showing that arginine, phenylalanine, proline, and tryptophan are the strongest attractants of this class (Webb et al., 2017b). The chemoreceptor McpU directly binds all amino acids except for glutamate and aspartate, which do not serve as chemoattractants for S. meliloti RU11/001 (Webb et al., 2014, 2017b). The chemical nature of the R-group appears to be important for the bias of different bacterial species toward acidic or non-acidic amino acids.
Carboxylates
Carboxylates, also referred to as organic acids, are carbon sources commonly found in plant exudates, the rhizosphere, and bulk soil. Citrate, malate, and succinate are attractants for R. leguminosarum, but are generally weaker than the amino acids (Gaworzewska and Carlile, 1982; Armitage et al., 1988). In B. japonicum, malonate and succinate elicit a strong attractant response, equivalent to aspartate and glutamate. In addition, other 4-carbon carboxylates are also attractants, although citrate is not (Barbour et al., 1991; Kape et al., 1991). Sinorhizobium meliloti has a dedicated 2–3 carbon monocarboxylate sensor, McpV, and a sensor for small dicarboxylates, McpT (Baaziz et al., in review; Compton et al., 2018). The mono- and di-carboxylates are weak attractants compared to the amino acids (Webb et al., 2014).
Flavonoids
Flavonoids belong to a plant-borne compound group that are of interest because they induce the expression of symbiotic genes in their cognate rhizobia (Abdel-Lateif et al., 2012). Although apigenin and luteolin do not induce nod gene expression in R. leguminosarum, they are attractants in this organism. In contrast, naringenin is a nod gene inducer but not an attractant (Aguilar et al., 1988). Two reports on B. japonicum chemotaxis showed that all flavonoids tested have no attractant function (Barbour et al., 1991; Kape et al., 1991). Studies of S. meliloti chemotaxis reported that the nod gene inducers dihydroxyflavone and luteolin are attractants, but that responses were very low. The studies did not test any other compounds for comparison and, therefore, lack context (Caetanoanollés et al., 1988; Dharmatilake and Bauer, 1992). The conclusion that flavonoids are attractants was not replicated and recently disputed (Compton et al., 2020). Taken together, the evidence for flavonoid chemotaxis as a general phenomenon in rhizobia is debatable.
Phenolics
The phenolics comprise a class of compounds that include phenylpropanoid derivatives of aromatic acids, which are common plant metabolites and precursors to flavonoids. Bradyrhizobium japonicum senses several phenylpropanoid acids and coniferyl alcohol as attractants, but not chlorogenic acid (Kape et al., 1991). Benzoic alcohols are attractants for R. leguminosarum with acetosyringone being one of the strongest attractants reported, while umbelliferone and vanillyl alcohol are weak to moderate attractants (Aguilar et al., 1988).
Saccharides
Saccharides or sugars and their subclasses such as sugar alcohols and sugar acids are some of the most readily available carbon sources (Geddes and Oresnik, 2014). Common energy sources like gluconate and glucose as well as structural components of pectins such as arabinose and xylose are logical candidates to serve as attractants for rhizobia. Multiple reports investigated the taxis of R. leguminosarum to sugars, but these gave conflicting results. Glucose, maltose, ribose, and sucrose were reported as attractants in certain studies, but the evidence in other references suggests that they are not (Bowra and Dilworth, 1981; Gaworzewska and Carlile, 1982; Aguilar et al., 1988; Armitage et al., 1988). However, arabinose and xylose were identified as attractants in all studies, although they were not necessarily the strongest chemoattractants (Bowra and Dilworth, 1981; Gaworzewska and Carlile, 1982; Aguilar et al., 1988). Information on B. japonicum and S. meliloti taxis to sugars is sparse. Mannitol, a sugar alcohol, is the only member of this class that was presented as an attractant for B. japonicum, while common sugars such as arabinose and glucose do not appear to serve as attractants (Barbour et al., 1991; Chuiko et al., 2002). In S. meliloti, gluconate and sucrose were reported to be the best attractants among the sugars over arabinose, fructose, and glucose (Malek, 1989). Currently, it is difficult to make conclusions on the prevalence of sugars as chemoattractants in rhizobia. Most compounds were only tested in a single study and at one concentration (Table 1). Sugars are a large, complex class because of the numerous structural and stereoisomers, which further complicates analysis. Currently, there is too little information to make a clear statement about the role of sugars as attractants.
Preferred Attractants for Rhizobia
A prominent aim in chemotaxis research is identifying dominant or preferred attractants for a given organism because it is indicative of its role in the ecosystem. The clearest body of information available for this conclusion comes from work done on S. meliloti strain RU11/001. All studies examining different classes of chemoattractants included comparative experiments with the chemoattractant proline and used similar techniques – thus creating a standard for comparison. This body of work allows the conclusion that amino acids are of similar attractant strength to quaternary ammonium compounds (QACs) such as betaines or choline (another class of attractants that was only recently recognized), and 5- to 10-fold stronger attractants than carboxylates (Webb et al., 2014, 2017a,b; Compton et al., 2018).
Work on the chemosensing of B. japonicum, albeit from a single study, revealed that glutamate, malonate, and succinate are the most potent attractants. It is noteworthy that glutamate was used as a nitrogen source in the growth medium, raising the possibility that chemotaxis responses are inducible in this organism (Barbour et al., 1991).
Acetosyringone is the strongest attractant reported for R. leguminosarum, followed by the flavonoids apigenin and luteolin. The evidence for rhizobial chemotaxis to flavonoids is strongest in R. leguminosarum, even though the bacterium only appears to be attracted to non-nod gene inducing flavonoid species (Aguilar et al., 1988). Arabinose and xylose are not the strongest attractants but have been consistently reported to attract multiple strains of R. leguminosarum (Bowra and Dilworth, 1981; Gaworzewska and Carlile, 1982; Aguilar et al., 1988).
Where are Attractants Found?
Seed Exudates
The compounds released from germinating seeds are a great source of attractants for rhizobia, readily available for study, and consistent between samples. When seeds are formed, the seed coat is impregnated with numerous chemicals from the mother plant (Radchuk and Borisjuk, 2014). Upon imbibition, these chemicals are leeched out as an exudate into the surrounding medium forming what is called the spermosphere (Nelson, 2004). Amino acids and carboxylates are some of the most sampled constituents of seed exudates and these compounds have the clearest link to known bacterial chemosensory systems (Götz et al., 1982; Rozan et al., 2001; Kuo et al., 2004; Kamilova et al., 2006; Lambers et al., 2013; Webb et al., 2016). However, there is great chemical diversity in exudates that include QACs such as betaines, saccharides, fatty acids, phenolics, and alcohols (Silva et al., 2013; Schiltz et al., 2015; Webb et al., 2017a; Mildaziene et al., 2020; Zuluaga et al., 2020). The gradients these chemicals form serve to recruit microorganisms to the surface of the nascent plant.
Root Exudates
Exudates from roots share the chemical diversity of seed exudates, but their specific composition will vary according to biotic and abiotic factors and the state of the plant. Changes in exudate profiles are strongly driven by environmental factors (Herz et al., 2018; Dietz et al., 2019). Conditions lacking mineral nutrients induce the release of organic acids to solubilize phosphorus and chelate iron (Shen et al., 2005; Preece and Penuelas, 2020; Vives-Peris et al., 2020). Young or immature plants tend to release more sugars, while the exudates of older plants are biased toward amino acids and phenolics (Chaparro et al., 2013). In another example, a biocontrol organism induces the release of a plant metabolite that is toxic to a pathogen, but not to the biocontrol agent (Wang et al., 2019). In fact, and at the risk of overgeneralizing, it appears that nearly any environmental or physiological perturbation influences the composition or quantity of root exudates (Canarini et al., 2019; Vives-Peris et al., 2020). It would follow that chemotactic recruitment of soil bacteria is, in many cases, a byproduct of plant activity. In support of this, the saprotrophic soil bacterium Pseudomonas putida possesses a chemoreceptor that preferentially senses citrate-magnesium complex compared to free citrate, possibly sourced from efforts of a plant to acquire magnesium or other chelated cations (Martin-Mora et al., 2016). However, plants are not indifferent to their rhizosphere neighbors. Recruitment of soil symbionts is a definite priority for plants because of the benefits their microbiota provide. Furthermore, stressed plants release additional carbon compounds to recruit symbionts and other beneficial bacteria in times of need (Ames and Bergman, 1981; Gulash et al., 1984; Badri and Vivanco, 2009; Canarini et al., 2019; O’Neal et al., 2020; Vives-Peris et al., 2020).
Other Attractant Sources
The vast majority of research focuses on rhizobial activities in the rhizosphere and nodule (Poole et al., 2018). Few studies examine rhizobial survival in the bulk soil. This is nonetheless an important topic, because effective inoculants and symbionts must persist in the soil to infect the next generation of hosts (Pinochet et al., 1993; Hirsch and Spokes, 1994; Da and Deng, 2003). In the absence of hosts, rhizobia can subsist on soil organic carbon, an amalgam containing energy and nutrient sources from detritus and decaying matter, which take the form of aromatics, alkyl compounds, carboxylates, and nitrogenous molecules such as proteins (Beyer et al., 1995; Chiu et al., 2002; Spielvogel et al., 2004). Rhizobia are adapted to saprophytic lifestyles, and chemotaxis to the carbon and nitrogen sources available is critical to survival in the bulk soil (Kennedy and Lawless, 1985; Turnbull et al., 2001; diCenzo et al., 2016; Poole et al., 2018).
Attractant signals need not be chemical compounds. Chemoreceptors can sense oxygen gradients or the cellular redox state as an indicator of the surrounding milieu, a strategy termed energy taxis. In the nitrogen fixing Azospirilum brasilense, metabolism mediates and is necessary for chemotaxis to attractants. This behavior is also critical to the colonization of host plants (Alexandre et al., 2000; Greer-Phillips et al., 2004). Utilizing a generalized strategy such as energy taxis allows a bacterium to seek areas suitable to its needs, regardless of the identity of the attractant source.
Conclusion and Future Directions
Chemotaxis is a major survival strategy for bacteria and greatly aids in seeking hosts (Raina et al., 2019). The rhizobia sense a wide array of chemical cues that are concomitantly found in the exudates of respective host plants but differ in their physiological role (Figure 1). Many of these attractants are primary metabolites and do not signify the identity of the source plant. A single chemical that is specific to a host would be difficult to pinpoint. Among semi polar metabolites, very few compounds are exclusively found in a particular plant species (Dietz et al., 2019). At best, specific metabolites might be characteristic of a genus or family (Rosenthal and Nkomo, 2000; Kidd et al., 2018). Rhizobia, therefore, have numerous chemoreceptors that may detect any of the thousands of compounds found in plant exudates, although in some cases, only a few chemoreceptors have any impact on host colonization (Feng et al., 2019). As each rhizobial species or strain analyzed seems to have different preferred attractants, the question remains how these preferences are formed. Since chemotactic preferences must be shaped by selection, perhaps each rhizobium specializes in obtaining particular types of nutrients. In effect, this is akin to a broad survival strategy that expands upon the phenomenon of catabolite repression, where a specific carbon source takes priority in the organism’s metabolism (Georgi and Ettinger, 1941; Bruckner and Titgemeyer, 2002; Iyer et al., 2016).
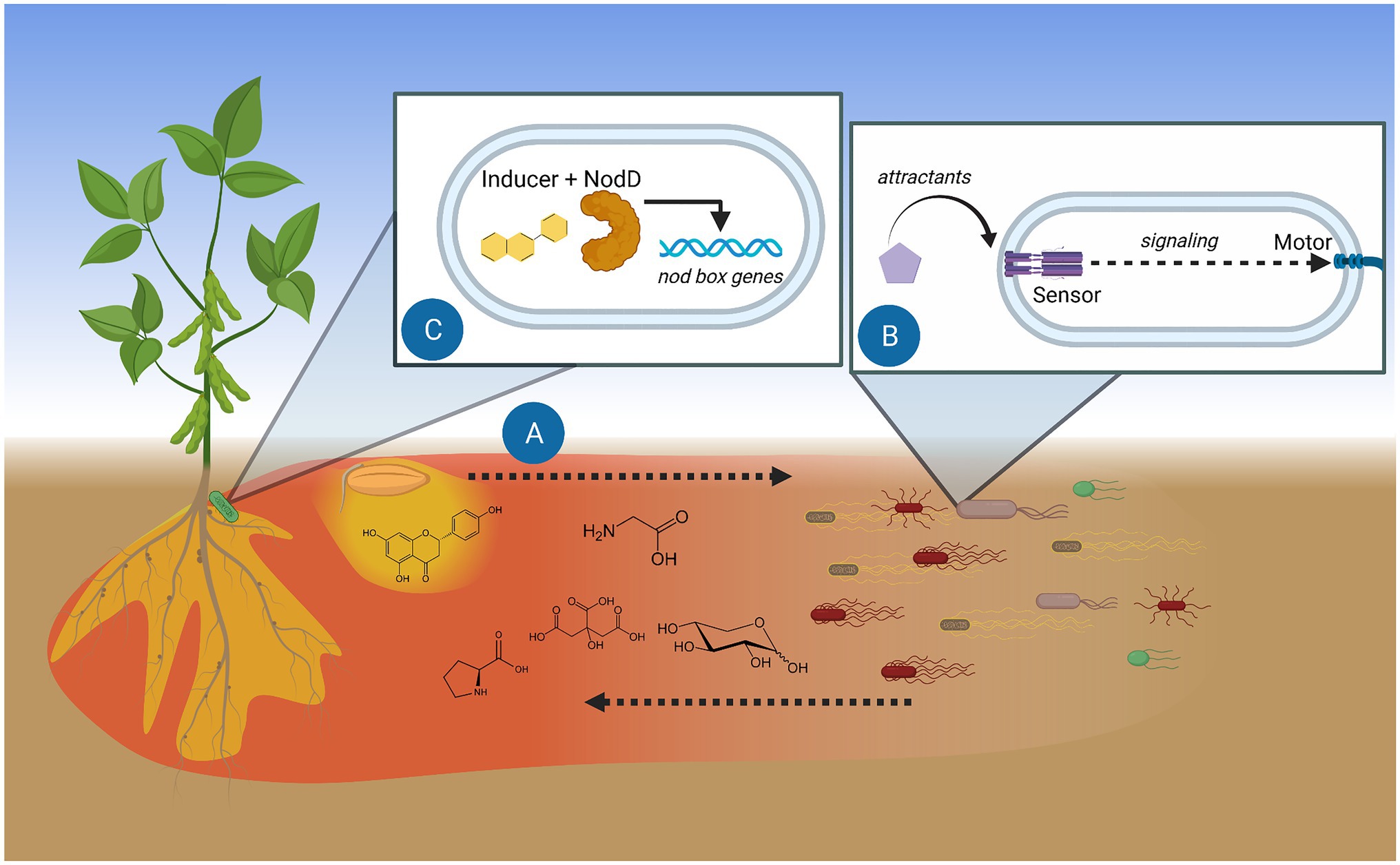
Figure 1. Model of chemotaxis function in the initiation of rhizobium-legume symbiosis. (A) Root and seed exudates diffuse and create chemical trails. Gradients of soluble exudates (red) travel furthest and act as chemoattractants. Hydrophobic exudate components (yellow) stay closer to the source. (B) Specific exuded compounds are detected by bacterial methyl-accepting chemotaxis proteins (MCPs), the dedicated sensors of attractants. Signaling between the chemosensory system and the motor guides the bacterium toward the attractant source. (C) Perception of plant flavonoids by rhizobial NodD initiates transcription of genes involved in infecting the proper host. Figure created with BioRender.
Every species or strain of rhizobia harbors numerous chemoreceptors that bestow perception of a particular signal. The number of MCPs per rhizobium varies greatly from less than 10 in Sinorhizobium and Ensifer spp. to over 30 in R. leguminosarum and B. japonicum isolates (for more information on chemoreceptor distributions, see Scharf et al., 2016). Currently, only four MCPs in S. meliloti have been assigned functions (Baaziz et al., in review; Webb et al., 2014, 2017b; Compton et al., 2018). The remainder of our knowledge on rhizobial chemotaxis is limited to dated behavioral studies. A complete understanding of rhizobium chemotaxis is an important goal for the following reasons: (1) total knowledge of a rhizobium’s chemotaxis system permits the modeling of its behavior in the environment and the prediction of its performance in host nodulation based on its exudate composition (Edgington and Tindall, 2018); (2) inoculant strains can be optimized to more efficiently nodulate crops, outcompete symbiotically inefficient native strains, and better survive in the soil, which increases the longevity of their application; and (3) chemoreceptors are often conserved among plant and animal pathogens, making information on MCPs translational to numerous fields (Brewster et al., 2016; Compton et al., 2018).
The way forward is, therefore, to increase the output of studies on MCP function that address the following goals: (1) identification of attractant classes and their relative strengths; (2) determination of sensors responsible for detecting attractants and the molecular mechanism of sensing; and (3) evaluation of the purpose of attractants in the survival or symbiosis of rhizobia. These aims can be easily initiated by revisiting known attractants and identifying their respective sensors. Two approaches are effective in characterizing MCP-ligand relationships. The first approach involves defining an attractant class and identifying a corresponding sensor, as exemplified by McpX in S. meliloti and the QACs (Webb et al., 2017a). The second approach identifies the ligand profile of an MCP using high-throughput screens followed by behavioral assays (McKellar et al., 2015). These pursuits will eventually illuminate each rhizobium’s attractome, the total range of compounds sensed through its chemotaxis systems. These findings will be a great boon to the economical, technical, and ecological challenges facing modern agriculture.
Author Contributions
KC and BS contributed to conception of the study. KC wrote the first draft of the manuscript. Both the authors contributed to manuscript revision, read, and approved the submitted version.
Funding
This work was supported by NSF grants MCB-1253234 and MCB-1817652 to BS.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Abdel-Lateif, K., Bogusz, D., and Hocher, V. (2012). The role of flavonoids in the establishment of plant roots endosymbioses with arbuscular mycorrhiza fungi, rhizobia and Frankia bacteria. Plant Signal. Behav. 7, 636–641. doi: 10.4161/psb.20039
Adler, J. (1973). A method for measuring chemotaxis and use of the method to determine optimum conditions for chemotaxis by Escherichia coli. J. Gen. Microbiol. 74, 77–91. doi: 10.1099/00221287-74-1-77
Aguilar, J. M. M., Ashby, A. M., Richards, A. J. M., Loake, G. J., Watson, M. D., and Shaw, C. H. (1988). Chemotaxis of Rhizobium leguminosarum biovar phaseoli towards flavonoid inducers of the symbiotic nodulation genes. J. Gen. Microbiol. 134, 2741–2746. doi: 10.1099/00221287-134-10-2741
Alexandre, G., Greer, S. E., and Zhulin, I. B. (2000). Energy taxis is the dominant behavior in Azospirillum brasilense. J. Bacteriol. 182, 6042–6048. doi: 10.1128/JB.182.21.6042-6048.2000
Althabegoiti, M. J., Lopez-Garcia, S. L., Piccinetti, C., Mongiardini, E. J., Perez-Gimenez, J., Quelas, J. I., et al. (2008). Strain selection for improvement of Bradyrhizobium japonicum competitiveness for nodulation of soybean. FEMS Microbiol. Lett. 282, 115–123. doi: 10.1111/j.1574-6968.2008.01114.x
Ames, P., and Bergman, K. (1981). Competitive advantage provided by bacterial motility in the formation of nodules by Rhizobium meliloti. J. Bacteriol. 148, 728–729. doi: 10.1128/JB.148.2.728-729.1981
Ames, P., Schluederberg, S. A., and Bergman, K. (1980). Behavioral mutants of Rhizobium meliloti. J. Bacteriol. 141, 722–727. doi: 10.1128/JB.141.2.722-727.1980
Armitage, J. P., Gallagher, A., and Johnston, A. W. (1988). Comparison of the chemotactic behaviour of Rhizobium leguminosarum with and without the nodulation plasmid. Mol. Microbiol. 2, 743–748. doi: 10.1111/j.1365-2958.1988.tb00085.x
Badri, D. V., and Vivanco, J. M. (2009). Regulation and function of root exudates. Plant Cell Environ. 32, 666–681. doi: 10.1111/j.1365-3040.2009.01926.x
Barbour, W. M., Hattermann, D. R., and Stacey, G. (1991). Chemotaxis of Bradyrhizobium japonicum to soybean exudates. Appl. Environ. Microbiol. 57, 2635–2639. doi: 10.1128/AEM.57.9.2635-2639.1991
Beyer, L., Blume, H. P., Elsner, D. C., and Willnow, A. (1995). Soil organic-matter composition and microbial activity in urban soils. Sci. Total Environ. 168, 267–278. doi: 10.1016/0048-9697(95)04704-5
Bowra, B. J., and Dilworth, M. J. (1981). Motility and chemotaxis towards sugars in Rhizobium-leguminosarum. J. Gen. Microbiol. 126, 231–235. doi: 10.1099/00221287-126-1-231
Brewster, J. L., McKellar, J. L., Finn, T. J., Newman, J., Peat, T. S., and Gerth, M. L. (2016). Structural basis for ligand recognition by a cache chemosensory domain that mediates carboxylate sensing in Pseudomonas syringae. Sci. Rep. 6:35198. doi: 10.1038/srep35198
Bruckner, R., and Titgemeyer, F. (2002). Carbon catabolite repression in bacteria: choice of the carbon source and autoregulatory limitation of sugar utilization. FEMS Microbiol. Lett. 209, 141–148. doi: 10.1016/S0378-1097(02)00559-1
Burg, D., Guillaume, J., and Tailliez, R. (1982). Chemotaxis by Rhizobium meliloti. Arch. Microbiol. 133, 162–163. doi: 10.1007/BF00413532
Caetanoanollés, G., Cristestes, D. K., and Bauer, W. D. (1988). Chemotaxis of Rhizobium meliloti to the plant flavone luteolin requires functional nodulation genes. J. Bacteriol. 170, 3164–3169. doi: 10.1128/JB.170.7.3164-3169.1988
Canarini, A., Kaiser, C., Merchant, A., Richter, A., and Wanek, W. (2019). Root exudation of primary metabolites: mechanisms and their roles in plant responses to environmental stimuli. Front. Plant Sci. 10:157. doi: 10.3389/fpls.2019.00157
Chaparro, J. M., Badri, D. V., Bakker, M. G., Sugiyama, A., Manter, D. K., and Vivanco, J. M. (2013). Root exudation of phytochemicals in Arabidopsis follows specific patterns that are developmentally programmed and correlate with soil microbial functions. PLoS One 8:e55731. doi: 10.1371/journal.pone.0055731
Chiu, C. Y., Wang, M. K., Hwong, J. L., and King, H. B. (2002). Physical and chemical properties in rhizosphere and bulk soils of Tsuga and Yushania in a temperate rain forest. Commun. Soil Sci. Plant Anal. 33, 1723–1735. doi: 10.1081/Css-120004818
Chuiko, N. V., Antonyuk, T. S., and Kurdish, I. K. (2002). The chemotactic response of Bradyrhizobium japonicum to various organic compounds. Microbiology 71, 391–396. doi: 10.1023/A:1019881124077
Compton, K. K., Hildreth, S. B., Helm, R. F., and Scharf, B. E. (2018). Sinorhizobium meliloti chemoreceptor McpV senses short-chain carboxylates via direct binding. J. Bacteriol. 200:e00519–18. doi: 10.1128/JB.00519-18
Compton, K. K., Hildreth, S. B., Helm, R. F., and Scharf, B. E. (2020). An updated perspective on Sinorhizobium meliloti chemotaxis to alfalfa flavonoids. Front. Microbiol. 11:581482. doi: 10.3389/fmicb.2020.581482
Croxen, M. A., Sisson, G., Melano, R., and Hoffman, P. S. (2006). The Helicobacter pylori chemotaxis receptor TlpB (HP0103) is required for pH taxis and for colonization of the gastric mucosa. J. Bacteriol. 188, 2656–2665. doi: 10.1128/JB.188.7.2656-2665.2006
Da, H. N., and Deng, S. P. (2003). Survival and persistence of genetically modified Sinorhizobium meliloti in soil. Appl. Soil Ecol. 22, 1–14. doi: 10.1016/S0929-1393(02)00127-0
Dharmatilake, A. J., and Bauer, W. D. (1992). Chemotaxis of Rhizobium-meliloti towards todulation gene-inducing bompounds from alfalfa roots. Appl. Environ. Microbiol. 58, 1153–1158. doi: 10.1128/AEM.58.4.1153-1158.1992
diCenzo, G. C., Checcucci, A., Bazzicalupo, M., Mengoni, A., Viti, C., Dziewit, L., et al. (2016). Metabolic modelling reveals the specialization of secondary replicons for niche adaptation in Sinorhizobium meliloti. Nat. Commun. 7:12219. doi: 10.1038/ncomms12219
Dietz, S., Herz, K., Doll, S., Haider, S., Jandt, U., Bruelheide, H., et al. (2019). Semi-polar root exudates in natural grassland communities. Ecol. Evol. 9, 5526–5541. doi: 10.1002/ece3.5043
Edgington, M. P., and Tindall, M. J. (2018). Mathematical analysis of the Escherichia coli chemotaxis signalling pathway. Bull. Math. Biol. 80, 758–787. doi: 10.1007/s11538-018-0400-z
Feng, H., Zhang, N., Fu, R., Liu, Y., Krell, T., Du, W., et al. (2019). Recognition of dominant attractants by key chemoreceptors mediates recruitment of plant growth-promoting rhizobacteria. Environ. Microbiol. 21, 402–415. doi: 10.1111/1462-2920.14472
Gaworzewska, E. T., and Carlile, M. J. (1982). Positive chemotaxis of Rhizobium leguminosarum and other bacteria towards root exudates from legumes and other plants. Microbiology 128, 1179–1188. doi: 10.1099/00221287-128-6-1179
Geddes, B. A., and Oresnik, I. J. (2014). Physiology, genetics, and biochemistry of carbon metabolism in the alphaproteobacterium Sinorhizobium meliloti. Can. J. Microbiol. 60, 491–507. doi: 10.1139/cjm-2014-0306
Georgi, C. E., and Ettinger, J. M. (1941). Utilization of carbohydrates and sugar acids by the rhizobia. J. Bacteriol. 41, 323–340. doi: 10.1128/JB.41.3.323-340.1941
Götz, R., Limmer, N., Ober, K., and Schmitt, R. (1982). Motility and chemotaxis in 2 strains of rhizobium with complex flagella. J. Gen. Microbiol. 128, 789–798. doi: 10.1099/00221287-128-4-789
Greer-Phillips, S. E., Stephens, B. B., and Alexandre, G. (2004). An energy taxis transducer promotes root colonization by Azospirillum brasilense. J. Bacteriol. 186, 6595–6604. doi: 10.1128/JB.186.19.6595-6604.2004
Gulash, M., Ames, P., Larosiliere, R. C., and Bergman, K. (1984). Rhizobia are attracted to localized sites on legume roots. Appl. Environ. Microbiol. 48, 149–152. doi: 10.1128/AEM.48.1.149-152.1984
Hazelbauer, G. L. (1975). Maltose chemoreceptor of Escherichia coli. J. Bacteriol. 122, 206–214. doi: 10.1128/JB.122.1.206-214.1975
Herz, K., Dietz, S., Gorzolka, K., Haider, S., Jandt, U., Scheel, D., et al. (2018). Linking root exudates to functional plant traits. PLoS One 13:e0204128. doi: 10.1371/journal.pone.0204128
Hirsch, P. R., and Spokes, J. D. (1994). Survival and dispersion of genetically modified rhizobia in the field and genetic interactions with native strains. FEMS Microbiol. Ecol. 15, 147–159. doi: 10.1111/j.1574-6941.1994.tb00239.x
Iyer, B., Rajput, M. S., Jog, R., Joshi, E., Bharwad, K., and Rajkumar, S. (2016). Organic acid mediated repression of sugar utilization in rhizobia. Microbiol. Res. 192, 211–220. doi: 10.1016/j.micres.2016.07.006
Jekely, G. (2009). Evolution of phototaxis. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 364, 2795–2808. doi: 10.1098/rstb.2009.0072
Kamilova, F., Kravchenko, L. V., Shaposhnikov, A. I., Azarova, T., Makarova, N., and Lugtenberg, B. (2006). Organic acids, sugars, and L-tryptophane in exudates of vegetables growing on stonewool and their effects on activities of rhizosphere bacteria. Mol. Plant-Microbe Interact. 19, 250–256. doi: 10.1094/MPMI-19-0250
Kape, R., Parniske, M., and Werner, D. (1991). Chemotaxis and nod gene activity of Bradyrhizobium japonicum in response to hydroxycinnamic acids and isoflavonoids. Appl. Environ. Microbiol. 57, 316–319. doi: 10.1128/AEM.57.1.316-319.1991
Kennedy, M. J., and Lawless, J. G. (1985). Role of chemotaxis in the ecology of denitrifiers. Appl. Environ. Microbiol. 49, 109–114. doi: 10.1128/AEM.49.1.109-114.1985
Kidd, D. R., Ryan, M. H., Hahne, D., Haling, R. E., Lambers, H., Sandral, G. A., et al. (2018). The carboxylate composition of rhizosheath and root exudates from twelve species of grassland and crop legumes with special reference to the occurrence of citramalate. Plant Soil 424, 389–403. doi: 10.1007/s11104-017-3534-0
Kuo, Y. H., Rozan, P., Lambein, F., Frias, J., and Vidal-Valverde, C. (2004). Effects of different germination conditions on the contents of free protein and non-protein amino acids of commercial legumes. Food Chem. 86, 537–545. doi: 10.1016/j.foodchem.2003.09.042
Lambers, H., Clements, J. C., and Nelson, M. N. (2013). How a phosphorus-acquisition strategy based on carboxylate exudation powers the success and agronomic potential of lupines (Lupinus, Fabaceae). Am. J. Bot. 100, 263–288. doi: 10.3732/ajb.1200474
Lopez-Farfan, D., Reyes-Darias, J. A., and Krell, T. (2017). The expression of many chemoreceptor genes depends on the cognate chemoeffector as well as on the growth medium and phase. Curr. Genet. 63, 457–470. doi: 10.1007/s00294-016-0646-7
Malek, W. (1989). Chemotaxis in Rhizobium meliloti strain L5.30. Arch. Microbiol. 152, 611–612. doi: 10.1007/BF00425496
Martin-Mora, D., Reyes-Darias, J. A., Ortega, A., Corral-Lugo, A., Matilla, M. A., and Krell, T. (2016). McpQ is a specific citrate chemoreceptor that responds preferentially to citrate/metal ion complexes. Environ. Microbiol. 18, 3284–3295. doi: 10.1111/1462-2920.13030
McKellar, J. L. O., Minnell, J. J., and Gerth, M. L. (2015). A high-throughput screen for ligand binding reveals the specificities of three amino acid chemoreceptors from Pseudomonas syringae pv. actinidiae. Mol. Microbiol. 96, 694–707. doi: 10.1111/mmi.12964
Meier, V. M., Muschler, P., and Scharf, B. E. (2007). Functional analysis of nine putative chemoreceptor proteins in Sinorhizobium meliloti. J. Bacteriol. 189, 1816–1826. doi: 10.1128/JB.00883-06
Mildaziene, V., Ivankov, A., Pauzaite, G., Nauciene, Z., Zukiene, R., Degutyte-Fomins, L., et al. (2020). Seed treatment with cold plasma and electromagnetic field induces changes in red clover root growth dynamics, flavonoid exudation, and activates nodulation. Plasma Process. Polym. 18:2000160. doi: 10.1002/ppap.202000160
Moe, L. A. (2013). Amino acids in the rhizosphere: from plants to microbes. Am. J. Bot. 100, 1692–1705. doi: 10.3732/ajb.1300033
Nelson, E. B. (2004). Microbial dynamics and interactions in the spermosphere. Annu. Rev. Phytopathol. 42, 271–309. doi: 10.1146/annurev.phyto.42.121603.131041
O’Neal, L., Vo, L., and Alexandre, G. (2020). Specific root exudate compounds sensed by dedicated chemoreceptors shape Azospirillum brasilense chemotaxis in the rhizosphere. Appl. Environ. Microbiol. 86:e01026–20. doi: 10.1128/AEM.01026-20
Parkinson, J. S., Hazelbauer, G. L., and Falke, J. J. (2015). Signaling and sensory adaptation in Escherichia coli chemoreceptors: 2015 update. Trends Microbiol. 23, 257–266. doi: 10.1016/j.tim.2015.03.003
Paulick, A., Jakovljevic, V., Zhang, S., Erickstad, M., Groisman, A., Meir, Y., et al. (2017). Mechanism of bidirectional thermotaxis in Escherichia coli. elife 6:e26607. doi: 10.7554/eLife.26607
Pinochet, X., Arnaud, F., and Cleyetmarel, J. C. (1993). Competition for nodule occupancy of introduced Bradyrhizobium japonicum strain Smgs1 in french soils already containing Bradyrhizobium japonicum strain G49. Can. J. Microbiol. 39, 1022–1028. doi: 10.1139/m93-155
Poole, P., Ramachandran, V., and Terpolilli, J. (2018). Rhizobia: from saprophytes to endosymbionts. Nat. Rev. Microbiol. 16, 291–303. doi: 10.1038/nrmicro.2017.171
Preece, C., and Penuelas, J. (2020). A return to the wild: root exudates and food security. Trends Plant Sci. 25, 14–21. doi: 10.1016/j.tplants.2019.09.010
Radchuk, V., and Borisjuk, L. (2014). Physical, metabolic and developmental functions of the seed coat. Front. Plant Sci. 5:510. doi: 10.3389/fpls.2014.00510
Raina, J. B., Fernandez, V., Lambert, B., Stocker, R., and Seymour, J. R. (2019). The role of microbial motility and chemotaxis in symbiosis. Nat. Rev. Microbiol. 17, 284–294. doi: 10.1038/s41579-019-0182-9
Rosenthal, G. A., and Nkomo, P. (2000). The natural abundance of L-canavanine, an active anticancer agent, in alfalfa, Medicago sativa (L.). Pharm. Biol. 38, 1–6. doi: 10.1076/1388-0209(200001)3811-BFT001
Rozan, P., Kuo, Y. H., and Lambein, F. (2001). Amino acids in seeds and seedlings of the genus Lens. Phytochemistry 58, 281–289. doi: 10.1016/S0031-9422(01)00200-X
Salah Ud-Din, A. I. M., and Roujeinikova, A. (2017). Methyl-accepting chemotaxis proteins: a core sensing element in prokaryotes and archaea. Cell. Mol. Life Sci. 74, 3293–3303. doi: 10.1007/s00018-017-2514-0
Sampedro, I., Parales, R. E., Krell, T., and Hill, J. E. (2015). Pseudomonas chemotaxis. FEMS Microbiol. Rev. 39, 17–46. doi: 10.1111/1574-6976.12081
Scharf, B. E., Hynes, M. F., and Alexandre, G. M. (2016). Chemotaxis signaling systems in model beneficial plant-bacteria associations. Plant Mol. Biol. 90, 549–559. doi: 10.1007/s11103-016-0432-4
Schiltz, S., Gaillard, I., Pawlicki-Jullian, N., Thiombiano, B., Mesnard, F., and Gontier, E. (2015). A review: what is the spermosphere and how can it be studied? J. Appl. Microbiol. 119, 1467–1481. doi: 10.1111/jam.12946
Shen, H., He, L. F., Sasaki, T., Yamamoto, Y., Zheng, S. J., Ligaba, A., et al. (2005). Citrate secretion coupled with the modulation of soybean root tip under aluminum stress. Up-regulation of transcription, translation, and threonine-oriented phosphorylation of plasma membrane H+-ATPase. Plant Physiol. 138, 287–296. doi: 10.1104/pp.104.058065
Silva, L. R., Pereira, M. J., Azevedo, J., Mulas, R., Velazquez, E., Gonzalez-Andres, F., et al. (2013). Inoculation with Bradyrhizobium japonicum enhances the organic and fatty acids content of soybean (Glycine max (L.) Merrill) seeds. Food Chem. 141, 3636–3648. doi: 10.1016/j.foodchem.2013.06.045
Spielvogel, S., Knicker, H., and Kogel-Knabner, I. (2004). Soil organic matter composition and soil lightness. J. Plant Nutr. Soil Sci. 167, 545–555. doi: 10.1002/jpln.200421424
Tso, W. W., and Adler, J. (1974). Negative chemotaxis in Escherichia coli. J. Bacteriol. 118, 560–576. doi: 10.1128/JB.118.2.560-576.1974
Turnbull, G. A., Morgan, J. A. W., Whipps, J. M., and Saunders, J. R. (2001). The role of bacterial motility in the survival and spread of Pseudomonas fluorescens in soil and in the attachment and colonisation of wheat roots. FEMS Microbiol. Ecol. 36, 21–31. doi: 10.1111/j.1574-6941.2001.tb00822.x
Vives-Peris, V., de Ollas, C., Gomez-Cadenas, A., and Perez-Clemente, R. M. (2020). Root exudates: from plant to rhizosphere and beyond. Plant Cell Rep. 39, 3–17. doi: 10.1007/s00299-019-02447-5
Wang, N., Wang, L. Y., Zhu, K., Hou, S. S., Chen, L., Mi, D. D., et al. (2019). Plant root exudates are involved in Bacillus cereus AR156 mediated biocontrol against Ralstonia solanacearum. Front. Microbiol. 10:98. doi: 10.3389/fmicb.2019.00098
Webb, B. A., Compton, K. K., Castañeda Saldaña, R., Arapov, T., Ray, W. K., Helm, R. F., et al. (2017a). Sinorhizobium meliloti chemotaxis to quaternary ammonium compounds is mediated by the chemoreceptor McpX. Mol. Microbiol. 103, 333–346. doi: 10.1111/mmi.13561
Webb, B. A., Compton, K. K., del Campo, J. S. M., Taylor, D., Sobrado, P., and Scharf, B. E. (2017b). Sinorhizobium meliloti chemotaxis to multiple amino acids is mediated by the chemoreceptor McpU. Mol. Plant-Microbe Interact. 30, 770–777. doi: 10.1094/MPMI-04-17-0096-R
Webb, B. A., Helm, R. F., and Scharf, B. E. (2016). Contribution of individual chemoreceptors to Sinorhizobium meliloti chemotaxis towards amino acids of host and nonhost seed exudates. Mol. Plant-Microbe Interact. 29, 231–239. doi: 10.1094/MPMI-12-15-0264-R
Webb, B. A., Hildreth, S., Helm, R. F., and Scharf, B. E. (2014). Sinorhizobium meliloti chemoreceptor McpU mediates chemotaxis toward host plant exudates through direct proline sensing. Appl. Environ. Microbiol. 80, 3404–3415. doi: 10.1128/AEM.00115-14
Keywords: bacterial survival, flagellar motility, plant-host exudate, plant-microbe signaling, rhizosphere, symbiosis
Citation: Compton KK and Scharf BE (2021) Rhizobial Chemoattractants, the Taste and Preferences of Legume Symbionts. Front. Plant Sci. 12:686465. doi: 10.3389/fpls.2021.686465
Edited by:
Maria Jose Soto, Consejo Superior de Investigaciones Científicas (CSIC), SpainReviewed by:
Gladys Alexandre, The University of Tennessee, Knoxville, United StatesTino Krell, Consejo Superior de Investigaciones Científicas (CSIC), Spain
Copyright © 2021 Compton and Scharf. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Birgit E. Scharf, YnNjaGFyZkB2dC5lZHU=
 K. Karl Compton
K. Karl Compton Birgit E. Scharf
Birgit E. Scharf