
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 18 June 2021
Sec. Plant Metabolism and Chemodiversity
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.685654
This article is part of the Research Topic Phenylpropanoid Systems Biology and Biotechnology View all 34 articles
Anthocyanins and proanthocyanidins (PAs) are important flavonoids in Chinese bayberry (Morella rubra), which functions in fruit color and exhibits multiple health promoting and disease-preventing effects. To investigate the regulation of their biosynthesis in Chinese bayberries, we isolated and identified a subgroup 4 MYB transcription factor (TF), MrMYB6, and found MrMYB6 shared similar repressor domains with other MYB co-repressors of anthocyanin and PA biosynthesis after sequence analysis. Gene expression results revealed the transcripts of MrMYB6 were negatively correlated with the anthocyanin and insoluble PA contents and also with the gene expressions involved in anthocyanin biosynthesis and PA specific genes such as MrLAR and MrANR during the late ripening stages of bayberries. In addition, overexpression of MrMYB6 in tobacco inhibited the transcript levels of NtCHI, NtLAR, and NtANR2, resulting into a decline in the levels of anthocyanins and PAs in tobacco flowers. We further found that MrMYB6 interacted with MrbHLH1 and MrWD40-1 to form functional complexes that acted to directly repress the promoter activities of the PA-specific gene MrLAR and MrANR and the anthocyanin-specific gene MrANS and MrUFGT. Taken together, our results suggested that MrMYB6 might negatively regulate anthocyanin and PA accumulation in Chinese bayberry.
Flavonoids, widespread secondary metabolites, play many important roles in the development of plants (Winkel-Shirley, 2001). Anthocyanins and proanthocyanidins (PAs) are two major classes of flavonoids in fruits. The former is associated with the wide range of colors including bright orange, pink, red, violet, and blue in fruits, and the latter contribute to the astringency and flavor of fruits. Furthermore, both anthocyanins and PAs are considered as dietary antioxidants that are beneficial to human health in reducing free radical mediated injury and cardiovascular disease (Middleton et al., 2000). Therefore, they play important roles in influencing fruit quality.
The biosynthesis of anthocyanins and PAs share many enzymes in the upstream pathway, including chalcone synthase (CHS), chalcone isomerase (CHI), flavanone 3-hydroxylase (F3H), flavonoid 3′-hydroxylase (F3′H), and dihydroflavonol 4-reductase (DFR). Leucoanthocyanidins produced by DFR are the first branch point between the anthocyanin and PA biosynthesis pathways. Then leucoanthocyanidin reductase (LAR) and anthocyanidin synthase (ANS) can use leucoanthocyanidins to produce trans-flavan-3-ols and anthocyanidins, respectively. After that, anthocyanidins will be converted either to cis-flavan-3-ols by anthocyanidin reductase (ANR) or to anthocyanins by UDP-glucose flavonoid 3-O-glucosyltransferase (UFGT).
Increasing evidences have shown that the synthesis of anthocyanins and PAs is transcriptionally regulated by a conserved MBW complex that consists of R2R3-MYB, bHLH, and WD40 transcription factors (TFs) (Ramsay and Glover, 2005). Of these TFs, R2R3-MYB protein displays the functional specificity of the complex and determines which pathways are regulated. To date, many R2R3-MYB activators responsible for anthocyanin and PA accumulation have been characterized in various plants. For instance, PpMYB10.1 was able to activate anthocyanin biosynthesis and PpMYB7 regulated PA biosynthesis in peach (Rahim et al., 2014; Zhou et al., 2015). In apple, MdMYB1, MdMYB10, and MdMYB110a were activators responsible for anthocyanin accumulation (Takos et al., 2006; Espley et al., 2007; Chagne et al., 2013), while MdMYB9 and MdMYB11 played a positive role in regulating PA biosynthesis (Gesell et al., 2014; An et al., 2015). The accumulation of anthocyanins in grapevine was enhanced by VvMYBA1 and VvMYBA2 (Walker et al., 2007), whereas VvMYBPA1, VvMYBPA2, and VvMYBPAR promoted PA biosynthesis (Bogs et al., 2007; Terrier et al., 2009; Koyama et al., 2014).
Besides MYB activators, R2R3-MYB inhibitors are also important contributors to flavonoid regulation. Many MYB repressors, such as strawberry FaMYB1 (Aharoni et al., 2001), petunia PhMYB27 (Albert et al., 2014), peach PpMYB17-20 (Zhou et al., 2016), grapevine VvMYB4-like (Ricardo Perez-Diaz et al., 2016), apple MdMYB16 (Xu et al., 2017), and MdMYB15L (Xu et al., 2018), have been shown to play key roles in inhibiting anthocyanin biosynthesis. Grapevine VvMYBC2-L1 was identified as the first PA pathway repressor (Huang et al., 2014). Subsequently, some other R2R3-MYB repressors have been identified. For example, FtMYB8 from Tartary buckwheat inhibited both anthocyanin and PA biosynthesis, and interacted with AtTT8/FtTT8/FtGL3 in yeast (Huang et al., 2019). Overexpression of poplar MYB182 reduced anthocyanin and PA accumulation, which depended on the interaction between MYB182 and bHLH131 (Yoshida et al., 2015). In Medicago truncatula, MYB2 was identified as an inhibitor of both anthocyanin and PA biosynthesis, binding to bHLH factors TT8 (Jun et al., 2015). Interestingly, all these repressors typically belonged to the subgroup 4 R2R3-MYB TFs (Kranz et al., 1998), and shared the C1 motif (LlsrGIDPxT/SHRxI/L) and C2 motif (pdLNLD/ELxiG/S) with a core consensus sequence of either LxLxL or DLNxxP in the C-terminal region, which were identified as repression domains (Dubos et al., 2010; Zhou et al., 2019).
Chinese bayberry (Morella rubra Sieb. et Zucc.) contains large amounts of anthocyanins and PAs, and is an important source of natural antioxidants (Bao et al., 2005; Shi et al., 2018a). To date, only one MBW transcription complex, MrMYB1-MrbHLH1-MrWD40-1, has been observed to promote fruit anthocyanin formation Liu, X. et al., 2013; Liu, X. F. et al., 2013), however, there is no available information on TFs negatively regulating anthocyanin biosynthesis. Furthermore, the MYB TFs that specifically regulate PA biosynthesis in Chinese bayberry have not been reported yet. From the transcriptome data, our group previously isolated three putative subgroup 4 MYB repressors (c24596_g1, c28754_g2, and c48297_g1) from Chinese bayberry (Shi et al., 2018b). Here, we cloned and characterized MrMYB6 (c24596_g1) as the ortholog of other MYB TFs that have previously revealed as a negative controller of flavonoid biosynthesis in fruit species (Albert et al., 2014; Cavallini et al., 2015; Yoshida et al., 2015). Functional analysis indicated that MrMYB6 could regulate the transcripts of flavonoid biosynthesis genes, and its overexpression in tobacco repressed both anthocyanin and PA accumulation, suggesting that MrMYB6 might be an inhibitor of anthocyanin and PA synthesis in Chinese bayberry.
Chinese bayberry (Myrica rubra Sieb. and Zucc. cv. Biqi) trees were planted in an orchard in Cixi, Zhejiang Province. Fruits were picked at 57, 71, 85, 99, and 113 days after full bloom (DAFB) from four trees. The mixed fruits from the four trees were divided into three biological replicates for each developmental stage. All samples were frozen rapidly in liquid nitrogen and kept at −80°C for further use. Tobacco plants (Nicotiana tabacum cv. Samsunand Nicotiana benthamiana) were planted in a greenhouse with a 16 h light/8 h dark photoperiod at 25°C.
Total anthocyanin content was determined using the method reported by our group (Shi et al., 2014). Total soluble PA content was measured by reaction with dimethylaminocinnamaldehyde (DMACA), and total insoluble PA content was evaluated according to the butanol-HCl method reported in our previous study (Shi et al., 2018a).
Total RNA was obtained using a Plant RNA Kit (Omega, Norcross, GA) followed by treatment with RNase-free DNase I. First-strand cDNA synthesis was performed using the SuperRT First Strand cDNA Synthesis Kit (CWBIO, Beijing, China) following to the manufacturer's recommendations. qRT-PCR analysis was conducted using LightCycler 480 SYBR Green Master (Roche, Shanghai, China) on a Bio-Rad CFX96 Real-Time PCR System (BioRad, Hercules, CA, USA). Each reaction was conducted with a 12.5 μl reaction volume containing 6.25 μl of SYBR Green PCR Master Mix, 4.75 μl of RNase-free water, 0.5 μl of cDNA, and 0.5 μl of each primer (10 mM). The transcript levels were calculated using the 2−ΔΔCt method and normalized using the housekeeping gene MrACT (GQ340770) for Chinese bayberry and NtACTIN (GQ339768) for tobacco. All analyses were determined using three biological replicates with 40 fruits per replicate. Primers used for qRT-PCR are shown in Supplementary Table 1.
The 3′-end sequence of MrMYB6 gene was amplified using the SMART RACE cDNA amplification kit (Clontech, Mountain View, CA, USA). Nested primers were designed based on the partial MrMYB6 sequence found in the transcriptome data, and are shown in Supplementary Table 1 (Shi et al., 2018b). The product of PCR was recombined into pMD18-T cloning vector (TaKaRa, China). The deduced amino acid sequences of MrMYB6 and other R2R3-MYB repressors were aligned using the ClustalX2 program. Phylogenetic analysis was performed by the MEGA 7 program with the neighbor-joining (NJ) method and 1,000 bootstrap replicates.
The full-length MrMYB6 coding region without the stop codon was amplified and ligated into the pCAMBIA1301-GFP vector using primers listed in Supplementary Table 1. After sequencing, the recombinant plasmid and the GFP control vector were electroporated into Agrobacterium tumefaciens strain EHA105, and the cultures were adjusted to an OD 600 of 0.5 with infiltration buffer (10 mM MES, 10 mM MgCl2, 150 mM acetosyringone, pH 5.6) and then infiltrated into tobacco (N. benthamiana) leaves using needleless syringes. The GFP fluorescence of the tobacco leaves was detected 3 days after infiltration using the A1+ confocal laser scanning microscope (Nikon, Tokyo, Japan).
The full-length MrMYB6 coding region was inserted into the pCAMBIA-1300 binary vector using primers shown in Supplementary Table 1. The resulting binary vector was then transformed into A. tumefaciens strain EHA105 by electroporation. Transgenic tobacco plants overexpressing of MrMYB6 were obtained using the leaf disc infection technology reported by our group (Horsch et al., 1985). The putative transformed plants were selected on MS medium containing 250 mg/L carbenicillin and 25 mg/L hygromycin and finally identified by qRT-PCR analysis.
The possible interactions between proteins were tested in yeast using the Matchmaker® Gold Yeast Two-Hybrid System (Clontech, Japan). Full-length coding regions of Arabidopsis AtTT8, AtTTG, and bayberry MrHLH1 and MrWD40-1 were cloned into pGBKT7 (bait) vector, and the full-length MrMYB6 was ligated into pGADT7 (prey) vector (primers are listed in Supplementary Table 1). Then, each pair of recombinant plasmids was co-transferred into Y2H yeast by the PEG/LiAC method. Yeast transformants were selected on SD/-Leu/-Trp medium and the interactions were detected on SD/-Leu/-Trp/His/-Ade medium in the presence of X-α-Gal. pGADT7-T co-transformed with pGBKT7-p53 or pGBKT7-Lam were used as positive or negative controls.
The promoter regions of MrANR and MrLAR were isolated using the GenomeWalkerTM Universal Kit (Clontech, USA) according to the manufacturer's protocol, and the promoters of MrANS and MrUFGT were directly amplified based on the sequences reported (Liu, X. F. et al., 2013). These promoter regions were then ligated into the pGreenII 0800-LUC reporter vector. The full-length coding sequences of bayberry MrMYB6, MrbHLH1, MrWD40-1, and Arabidopsis AtTT8, AtTTG were amplified and cloned into the pGreenII 62-SK effector vector. Primer information is shown in Supplementary Table 1. These recombinant vectors were then electroporated into Agrobacterium strain EHA105. After cultivation, cells were resuspended in infiltration buffer (10 mM MgCl2, 10 mM MES, 150 mM acetosyringone, pH 5.6), adjusted to an OD 600 of 0.2, and incubated for 2 h at room temperature without shaking before infiltration. Four-week-old leaves of N. benthamiana were infected with mixed agrobacteria and collected for dual luciferase test according to the method described previously (Hellens et al., 2005).
In our previous study, three putative subgroup 4 MYB repressors (c24596_g1, c28754_g2, and c48297_g1) of Chinese bayberry were obtained from the transcriptome data (Shi et al., 2018b). Here, we cloned and identified MrMYB6 (c24596_g1), which was named based upon sequence homology to the genes found through the BLAST in the NCBI database, and the characterization of the other two MYB repressors will be carried out in future work. The 3′-end of MrMYB6 open reading frame (ORF) was amplified via 3′RACE-PCR and an 879 bp fragment was obtained. Sequence analysis indicated MrMYB6 contained an ORF of 684 bp encoding 228 amino acid resides with a predicted molecular mass of 25.72 kD and a calculated PI of 8.38.
The sequence alignment of MrMYB6 with MYB repressors of other species suggested that their N-terminus contained highly conserved R2R3 domain with a bHLH-binding domain (Grotewold et al., 2000) and their C-terminus included two conserved motifs of MYB subgroup 4 TFs, the C1 and C2 motif, of which the C2 motif associated with EAR repressor domain (Jin et al., 2000; Kagale and Rozwadowski, 2011; Shen et al., 2012) (Supplementary Figure 1A). MrMYB6 shared the LxLxL-type EAR motif with other subgroup 4 MYBs that inhibit both PA and anthocyanin biosynthesis, such as PtrMYB182 (Yoshida et al., 2015), VvMYBC2-L3 (Cavallini et al., 2015), FtMYB8 (Huang et al., 2019), and FaMYB1 (Aharoni et al., 2001; Paolocci et al., 2011). In addition, MrMYB6 contained a TLLLFR repressor motif, which has been identified in FaMYB1-like proteins such as PtrMYB182 (Yoshida et al., 2015) and VvMYBC2 (Cavallini et al., 2015), but not in AtMYB4-like repressors (Jin et al., 2000). Phylogenetic analysis showed that R2R3 MYB inhibitors were separated into two clades, AtMYB4-like and FaMYB1-like (Supplementary Figure 1B). MrMYB6 was clustered in FaMYB1-like clade, which contained co-repressors in anthocyanin and PA biosynthesis including VvMYBC2 from grapevine (Cavallini et al., 2015), PtrMYB182 from Poplar (Yoshida et al., 2015), PhMYB27 from petunia (Albert et al., 2014), FtMYB8 from buckwheat (Huang et al., 2019), MtMYB2 from Medicago (Jun et al., 2015), and FaMYB1 (Aharoni et al., 2001; Paolocci et al., 2011) from strawberry. Thus, MrMYB6 might act as a negative controller of flavonoid biosynthesis in Chinese bayberry.
During fruit development, the red color of bayberry fruit gradually deepened and reached the deepest point at 113 day after full bloom (DAFB) (Figure 1A). MYB6 gene exhibited low expression level at 57 DAFB and 113 DAFB, but was highly expressed from 71 DAFB to 99 DAFB (Figure 1B).
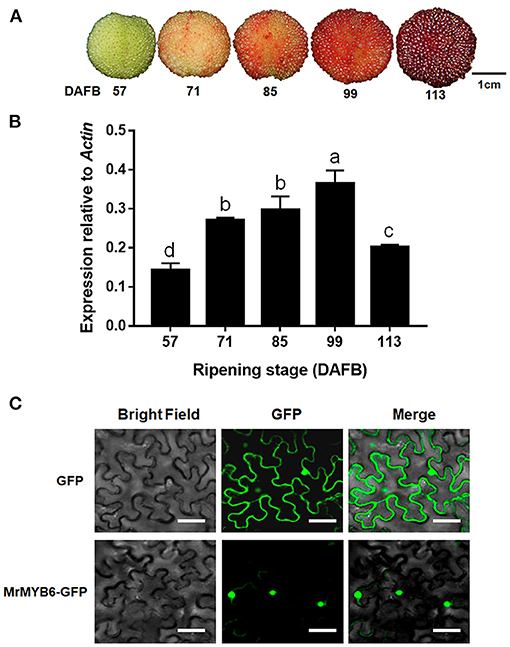
Figure 1. MrMYB6 expression pattern during bayberry fruit development. (A) The bayberry fruits at 57, 71, 85, 99, and 113 days after full bloom (DAFB) used in this study. Scale bar represents 1 cm. (B) Expression levels of MrMYB6 at different stages. The data were normalized to the MrACT expression level. Error bars indicate the mean ± SE of three replicate reactions. Letters (a, b, c, and d) represent significant difference between wild-type and transgenic plants according to the Duncan's multiple range test (P < 0.05). (C) Subcellular localization of MrMYB6 in tobacco leaf epidermal cells. Scale bar represents 50 μm.
In order to investigate the subcellular location of MrMYB6, the 35S:MrMYB6-GFP fusion protein and the 35S:GFP control protein were transiently expressed in tobacco leaves. By scanning the GFP signal, it was found that the 35S:MrMYB6-GFP fusion protein was exclusively localized in the nucleus, whereas the control 35S:GFP was distributed in both the cytoplasm and the nucleus (Figure 1C).
To characterize the function of this TF, transgenic tobacco plants overexpressing MrMYB6 were generated. From twelve independent T1 transgenic lines, nine plants showed weaker flower color than wild-type (WT) (date not shown). Among them, three transgenic lines (L1, L2, and L3) with obvious white colored flowers were selected for further study after confirmation of high MrMYB6 transcript levels using qRT-PCR analysis (Figures 2A,B). The result of anthocyanin analysis showed that the anthocyanin content in flowers of all transgenic lines was significantly reduced compared with WT (Figure 2C). The soluble and insoluble PA levels in transgenic tobacco petals were also significantly declined (Figures 2D,E). In addition, all three MrMYB6 overexpression lines exhibited decreased NtLAR, NtANR2, and NtCHI expression and increased NtCHS, NtF3H, NtF3′H, NtDFR, NtANS, NtUFGT, and NtFLS expression in transgenic flowers (Figure 2F).
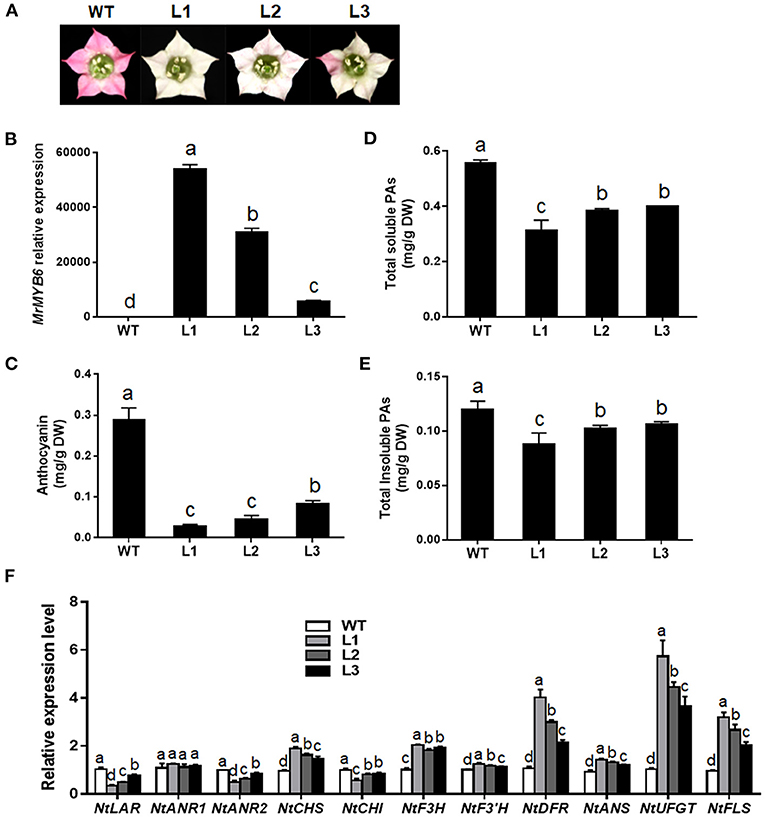
Figure 2. Ectopic expression of MrMYB6 gene in tobacco. (A) Comparison of flower color in wild type (WT) and three MrMYB6 transgenic (L1, L2, and L3) tobacco lines. (B) Relative expression levels of MrMYB6 in transgenic flowers. The data were normalized using the WT sample and the NtACTIN (GQ339768) gene as the internal controls and calculated using the 2−ΔΔCt method. (C) Anthocyanin content in flowers of WT and three MrMYB6 tobacco lines. (D) Total soluble PAs levels of WT and transgenic flowers. (E) Total insoluble PAs levels of WT and transgenic flowers. (F) Expression analysis of flavonoid biosynthetic genes in flowers of MrMYB6 transgenic tobacco. Error bars indicate the mean ± SE of three replicate reactions. Letters (a, b, c, and d) represent significant difference between wild-type and transgenic plants according to the Duncan's multiple range test (P < 0.05).
Previous studies demonstrated that Arabidopsis AtTT2 formed the MBW complex with AtTT8 and AtTTG1 to promote PA accumulation (Baudry et al., 2004), and MrMYB1-MrbHLH1-MrWD40-1 complex was essential for anthocyanin biosynthesis in Chinese bayberry (Liu, X. F. et al., 2013; Liu, X. et al., 2013). Therefore, we carried out yeast two-hybrid assays to determine whether MrMYB6 interacts with AtTT8, AtTTG1, MrbHLH1, and MrWD40-1 to regulate the biosynthesis of PAs and anthocyanins.
As shown in Figure 3A, yeast transformants harboring the pGBKT7-MrMYB6/AtTT8/AtTTG1/MrbHLH1/MrWD40-1 constructs could not grow on SD/-Trp-His-Ade selection plates, indicating that these proteins had no self-activating activities. Nevertheless, the yeast cells possessing both pGADT7-MrMYB6 and pGBKT7-AtTT8/AtTTG1/MrbHLH1/MrWD40-1 grew well and showed α-galactosidase activity on the SD/-Trp/-Leu/-His/-Ade medium (Figure 3B). These results showed that MrMYB6 interacted physically with AtTT8, AtTTG1, MrbHLH1, and MrWD40-1 and implied that MrMYB6 might form a MBW complex to control PA and anthocyanin biosynthesis.
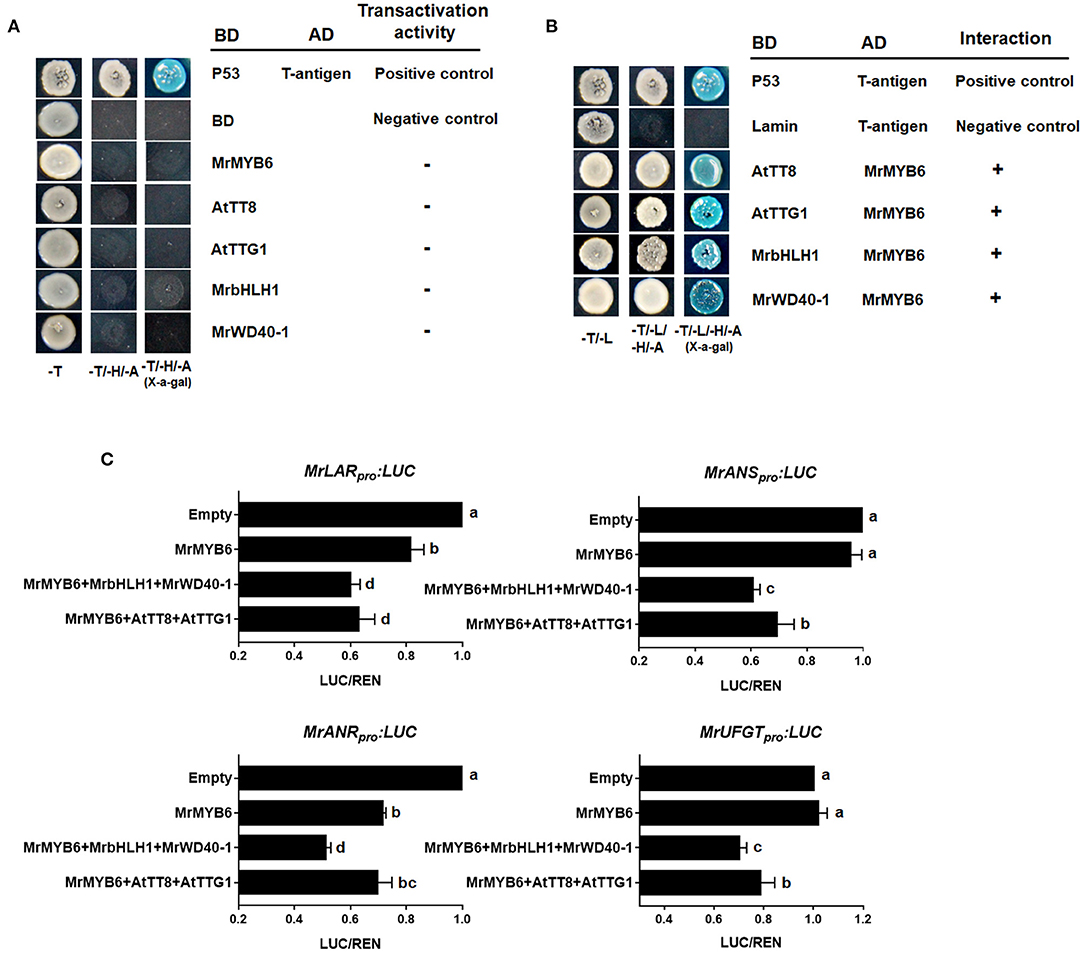
Figure 3. The role of MrMYB1-MrbHLH1-MrWD40-1 complex. (A) Autoactivation tests in yeast cells. (B) MrMYB6 interacts with AtTT8/AtTTG1/MrbHLH1/MrWD40-1 in yeast. -T, SD/-Trp medium; -T/-H/-A, SD/-Trp-His-Ade medium; -T/-L, SD/-Trp-Leu medium; -T/-L/-H/-A, SD/-Trp-Leu-His-Ade medium; BD, DNA-binding domain; AD, activation domain. (C) Transcriptional repression ability of MrMYB6 to the promoters of PA-specific gene MrLAR and MrANR and anthocyanin-specific gene MrANS and MrUFGT. The LUC/REN ratio of the empty vector plus promoter was set as 1. Different letters above the columns indicate significant differences at p < 0.05. Error bars indicate SE from four biological replicates.
The promoters of several flavonoid pathway genes, including MrLAR, MrANR, MrANS, and MrUFGT were cloned into reporter constructs. Among them, the MrLAR and MrANR promoters were selected to represent the PA-specific pathway, MrANS and MrUFGT for anthocyanin-specific pathway. Effector constructs expressing MrMYB6 were assayed with combinations of AtTT8, AtTTG1, MrbHLH1, and MrWD40-1. As shown in Figure 3C, MrMYB6 significantly reduced the activities of MrLAR and MrANR promoters. This suppression appeared stronger when MrMYB6 was co-transformed with AtTT8 and AtTTG or with MrbHLH1 and MrWD40-1. Similarly, the co-transformation of MrMYB6 with AtTT8 and AtTTG or with MrbHLH1 and MrWD40-1 inhibited the promoters of MrANS and MrUFGT. However, MrMYB6 alone showed no suppression on the promoters of either MrANS or MrUFGT.
In higher plants, R2R3-MYB subfamily, including activators and repressors, are major TFs coordinating flavonoid biosynthesis. However, no MYB repressors that regulate flavonoid synthesis have been identified in Chinese bayberry. In this research, we isolated a novel bayberry R2R3-MYB TF, named MrMYB6. Protein sequence analysis revealed that MrMYB6 contained two conserved motifs, C1 and C2 (Supplementary Figure 1A), which have been demonstrated to function as repression motifs of subgroup 4 MYB TFs (Jin et al., 2000; Shen et al., 2012). In addition, MrMYB6 shared the LxLxL-type EAR motif and TLLLFR repressor motif with FaMYB1-like proteins which inhibit both anthocyanin and PA biosynthesis (Cavallini et al., 2015; Yoshida et al., 2015) (Supplementary Figure 1A). The phylogenetic analysis clearly placed MrMYB6 in FaMYB1-like clade with co-repressors in anthocyanin and PA biosynthesis, including VvMYBC2 (Cavallini et al., 2015), PtrMYB182 (Yoshida et al., 2015), PhMYB27 (Albert et al., 2014), FtMYB8 (Huang et al., 2019), MtMYB2 (Jun et al., 2015), and FaMYB1 (Aharoni et al., 2001; Paolocci et al., 2011) (Supplementary Figure 1B). Thus, MrMYB6 displayed all structural characteristics of known inhibitors of flavonoid biosynthesis, and might exhibit similar regulatory functions. Moreover, the nuclear localization of MrMYB6 protein supported its potential function as a TF (Figure 1C).
It has been reported that many R2R3-MYB repressors were negatively correlated with the levels of anthocyanins and PAs and the transcripts of related structural genes. For instance, FtMYB18 was functionally characterized as an inhibitor of anthocyanin and PA synthesis in Tartary buckwheat, and its expression was negatively related to anthocyanin and PA contents (Dong et al., 2020). The elevated expression of VvMYBC2-L1 during berry development was negatively correlated with the anthocyanin and PA synthesis profiles and also with the expression of VvDFR, VvLDOX, VvLAR1, and VvANR (Huang et al., 2014; Cavallini et al., 2015). For Chinese bayberry, the soluble PA content gradually decreased, while the anthocyanin content greatly increased during fruit development (Shi et al., 2018a,b). Interestingly, in our present study, MrMYB6 experienced low expression level exactly at the early developmental stage (57 DAFB) and the harvest stage (113 DAFB), when the soluble PA content and anthocyanin levels were contradictingly the highest (Figure 1B), paralleling with the high expression levels of PA specific MrANR gene and many anthocyanin structural genes (Shi et al., 2018a,b). These results indicated that MrMYB6 expression also exhibited an opposite trend with anthocyanins and PAs accumulation during fruit development in bayberry.
Overexpression of MrMYB6 in tobacco in our present study resulted in a remarkable loss of red color in flowers due to a significant decrease in anthocyanin levels (Figures 2A,C). However, a recent study suggested that overexpression of gerbera GhMYB1a in tobacco plants reduced anthocyanin content but increased the expression of NtCHS, NtF3H, NtDFR, NtANS, and NtUFGT (Zhong et al., 2020). Ectopic expression of tea CsMYB5a in tobacco resulted into downregulated anthocyanin contents but elevated transcripts of NtCHS, NtCHI, NtF3H, NtF3′H, and NtANS in transgenic tobacco flowers (Jiang et al., 2018). Similarly, our results revealed the transcripts of all anthocyanin structural genes were significantly increased in MrMYB6-overexpression tobacco flowers except for NtCHI (Figure 2F). CHI has been identified as a key gene for anthocyanin biosynthesis. Silencing of CHI effectively inhibited anthocyanin accumulation in tobacco (Nishihara et al., 2005). Together with our findings, these results indicated that the inhibition of expression of NtCHI played the most important role in declining anthocyanin content in tobacco flowers when overexpressing MrMYB6 in tobacco. In addition, MrMYB6 was observed to down-regulate the expression of NtLAR and NtANR2, consistent with the observation that the PA content was reduced in transgenic tobacco flowers (Figures 2D–F). The transcription activity assay further showed that MrMYB6 could significantly inhibit the promoters of MrLAR and MrANR (Figure 3C). Taken together, these results suggested that MrMYB6 might be a negative regulator for anthocyanin and PA biosynthesis by inhibiting the transcription of related structural genes.
It is well known that flavonoid biosynthesis is transcriptionally regulated by MYB-bHLH-WD40 (MBW) complexes (Ramsay and Glover, 2005). The transcriptional activity of some MYBs require physical interaction with their bHLH and WD40 partners. In some plants, MYB activators and repressors can interact with the same bHLH and WD40 cofactors during regulation of flavonoid biosynthesis. As reported in petunia, repression of anthocyanin biosynthesis by MYB27 required the presence of AN1 (a bHLH protein), which was a necessary composition of the MBW activation complex for pigmentation (Albert et al., 2014). In poplar, MYB115 interacted with bHLH131 and TTG1 (WD40) to promote PA biosynthesis, while PtrMYB57 depended on the same bHLH131 and TTG1 cofactors for negatively regulating anthocyanin and PA biosynthesis (Wan et al., 2017; Wang et al., 2017). Consistent with these reports, our study found that MrMYB6 could interact with MrbHLH1 and MrWD40-1 (Figure 3B), which have been identified as members of the MrMYB1-MrbHLH1-MrWD40-1 complex that positively regulated anthocyanin biosynthesis in Chinese bayberry (Liu, X. et al., 2013; Liu, X. F. et al., 2013). In addition, MrMYB6 could also interact with AtTT8 and AtTTG that are essential for PA accumulation in Arabidopsis (Baudry et al., 2004). Meanwhile, our dual luciferase assay data confirmed that the interaction of MrMYB6 with these MBW components increased the inhibitory activity of MrMYB6 on the PA-specific genes such as MrLAR and MrANR and the anthocyanin-specific genes such as MrANS and MrUFGT (Figure 3C). Notably, the combination of MrMYB6 with MrbHLH1 and MrWD40-1 exhibited the strongest repression effect on the flavonoid biosynthesis genes compared with other factors. Therefore, these results indicated that MrMYB6 negatively regulated anthocyanin and PA synthesis through directly inhibiting the transcription of flavonoid pathway genes by forming a MrMYB6-MrbHLH1-MrWD40-1 complex. It must be pointed out that without any partners, MrMYB6 displayed no suppression on the promoters of either MrANS or MrUFGT in anthocyanin-specific pathway (Figure 3C). On the contrary, researches have revealed that several R2R3-MYB repressors such as VcMYBC2, PbMYB120, MaMYB4, and FaMYB1 on their own were sufficient to inhibit anthocyanin accumulation in many fruits such as blueberry, pear, banana, and strawberry (Aharoni et al., 2001; Song et al., 2020; Deng et al., 2021; Li et al., 2021). Therefore, we supposed that there might be other MYB TFs independently down-regulating anthocyanin accumulation in bayberries, which deserved further studies.
In conclusion, a R2R3-MYB transcriptional factor, MrMYB6, which functions as one of the negative regulators for anthocyanin and PA biosynthesis, was isolated from Chinese bayberry. The transcripts of MrMYB6 displayed a negative correlation with the anthocyanin and insoluble PA accumulation during the late ripening stages of bayberries. Ectopic overexpression of MrMYB6 caused reduced anthocyanin and PA contents in tobacco flowers because of the decline in the expression of NtCHI, NtLAR, and NtANR2 genes. In addition, MrMYB6 could directly inhibit anthocyanin and PA pathway gene expression by forming MrMYB6-MrbHLH1-MrWD40-1 complex.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
LS, ZY, and SC were involved in experimental design and data analysis. LS wrote the manuscript. XC, KW, and MY performed most of experiments. WC and SC revised the manuscript. All authors contributed to the article and approved the submitted version.
This study was supported by the National Natural Science Foundation of China (31671898) to ZY and the Natural Science Foundation of Zhejiang Province (LQ20C200006) to LS.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.685654/full#supplementary-material
Supplementary Table 1. Primers used in this study.
Supplementary Figure 1. Sequence analysis of MrMYB6. (A) Multiple sequence alignment of the R2R3-type motifs in MYB TFs from different plant species. (B) Phylogenetic analysis of MrMYB6 and other R2R3-MYB proteins. The phylogenetic tree was constructed using the NJ method with MEGA7 software. The numbers near the branches represents bootstrap value from 1,000 replicates. AmMYB308 (Antirrhinum majus MYB308, P81393), SmMYB39 (Salvia miltiorrhiza MYB39, AGS55356), GhMYB9 (Gossypium hirsutum MYB9, AAK19619), CmMYB1 (Chrysanthemum morifolium MYB1, AEO27497), EgMYB1 (Eucalyptus gunnii, MYB1 CAE09058), HvMYB1 (Hordeum vulgare MYB1, P20026), ZmMYB42 (Zea mays MYB42, NP_001106009), ZmMYB31 (Zea mays MYB31, NP_001105949.2), PhMYB4 (Petunia hybrida MYB4, ADX33331), AtMYB4 (Arabidopsis thaliana MYB4, AAC83582.1), AtMYB32 (Arabidopsis thaliana MYB32, NP_195225), AtMYB7 (Arabidopsis thaliana MYB7, NP_179263), NtMYB2 (Narcissus tazetta MYB2, ATO58377), FaMYB1 (Fragaria × ananassa MYB1, AAK84064.1), PhMYB27 (Petunia hybrida MYB27, AHX24372.1), VvMYBC2-L1 (Vitis vinifera MYBC2-L1, ABW34393), VvMYBC2-L3 (Vitis vinifera MYBC2-L3, AIP98385), FtMYB8 (Fagopyrum tataricum MYB8, MK128409), PtrMYB182 (Populus trichocarpa MYB182, XP_002305872), MtMYB2 (Medicago truncatula MYB2, XP_003616388).
Aharoni, A., De Vos, C. H. R., Wein, M., Sun, Z. K., Greco, R., Kroon, A., et al. (2001). The strawberry FaMYB1 transcription factor suppresses anthocyanin and flavonol accumulation in transgenic tobacco. Plant J. 28, 319–332. doi: 10.1046/j.1365-313X.2001.01154.x
Albert, N. W., Davies, K. M., Lewis, D. H., Zhang, H., Montefiori, M., Brendolise, C., et al. (2014). A conserved network of transcriptional activators and repressors regulates anthocyanin pigmentation in eudicots. Plant Cell 26, 962–980. doi: 10.1105/tpc.113.122069
An, X. H., Tian, Y., Chen, K. Q., Liu, X. J., Liu, D. D., Xie, X. B., et al. (2015). MdMYB9 and MdMYB11 are involved in the regulation of the JA-induced biosynthesis of anthocyanin and proanthocyanidin in apples. Plant Cell Physiol. 56, 650–662. doi: 10.1093/pcp/pcu205
Bao, J. S., Cai, Y. Z., Sun, M., Wang, G. Y., and Corke, H. (2005). Anthocyanins, flavonols, and free radical scavenging activity of Chinese bayberry (Myrica rubra) extracts and their color properties and stability. J. Agric. Food Chem. 53, 2327–2332. doi: 10.1021/jf048312z
Baudry, A., Heim, M. A., Dubreucq, B., Caboche, M., Weisshaar, B., and Lepiniec, L. (2004). TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J. 39, 366–380. doi: 10.1111/j.1365-313X.2004.02138.x
Bogs, J., Jaffe, F. W., Takos, A. M., Walker, A. R., and Robinson, S. P. (2007). The grapevine transcription factor VvMYBPA1 regulates proanthocyanidin synthesis during fruit development. Plant Physiol. 143, 1347–1361. doi: 10.1104/pp.106.093203
Cavallini, E., Matus, J. T., Finezzo, L., Zenoni, S., Loyola, R., Guzzo, F., et al. (2015). The phenylpropanoid pathway is controlled at different branches by a set of R2R3-MYB C2 repressors in grapevine. Plant Physiol. 167, 1448–1470. doi: 10.1104/pp.114.256172
Chagne, D., Kui, L. W., Espley, R. V., Volz, R. K., How, N. M., Rouse, S., et al. (2013). An ancient duplication of apple MYB transcription factors is responsible for novel red fruit-flesh phenotypes. Plant Physiol. 161, 225–239. doi: 10.1104/pp.112.206771
Deng, G. M., Zhang, S., Yang, Q. S., Gao, H. J., Sheng, O., Bi, F. C., et al. (2021). MaMYB4, an R2R3-MYB repressor transcription factor, negatively regulates the biosynthesis of anthocyanin in banana. Front. Plant Sci. 11:2023. doi: 10.3389/fpls.2020.600704
Dong, Q., Zhao, H., Huang, Y., Chen, Y., Wan, M., Zeng, Z., et al. (2020). FtMYB18 acts as a negative regulator of anthocyanin/proanthocyanidin biosynthesis in Tartary buckwheat. Plant Mol. Biol. 104, 309–325. doi: 10.1007/s11103-020-01044-5
Dubos, C., Stracke, R., Grotewold, E., Weisshaar, B., Martin, C., and Lepiniec, L. (2010). MYB transcription factors in Arabidopsis. Trends Plant Sci. 15, 573–581. doi: 10.1016/j.tplants.2010.06.005
Espley, R. V., Hellens, R. P., Putterill, J., Stevenson, D. E., Kutty-Amma, S., and Allan, A. C. (2007). Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J. 49, 414–427. doi: 10.1111/j.1365-313X.2006.02964.x
Gesell, A., Yoshida, K., Tran, L. T., and Constabel, C. P. (2014). Characterization of an apple TT2-type R2R3 MYB transcription factor functionally similar to the poplar proanthocyanidin regulator PtMYB134. Planta 240, 497–511. doi: 10.1007/s00425-014-2098-y
Grotewold, E., Sainz, M. B., Tagliani, L., Hernandez, J. M., Bowen, B., and Chandler, V. L. (2000). Identification of the residues in the Myb domain of maize C1 that specify the interaction with the bHLH cofactor R. Proc. Natl. Acad. Sci. U.S.A. 97, 13579–13584. doi: 10.1073/pnas.250379897
Hellens, R. P., Allan, A. C., Friel, E. N., Bolitho, K., Grafton, K., Templeton, M. D., et al. (2005). Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1, 1–14. doi: 10.1186/1746-4811-1-13
Horsch, R., Fry, J., Hoffman, N., Eichholtz, D., Rogers, S. A., and Fraley, R. (1985). A simple and general method for transferring genes into plants. Science 227, 1229–1231. doi: 10.1126/science.227.4691.1229
Huang, Y., Wu, Q., Wang, S., Shi, J., Dong, Q., Yao, P., et al. (2019). FtMYB8 from tartary buckwheat inhibits both anthocyanin/proanthocyanidin accumulation and marginal trichome initiation. BMC Plant Biol. 19:263. doi: 10.1186/s12870-019-1876-x
Huang, Y. F., Vialet, S., Guiraud, J. L., Torregrosa, L., Bertrand, Y., Cheynier, V., et al. (2014). A negative MYB regulator of proanthocyanidin accumulation, identified through expression quantitative locus mapping in the grape berry. New Phytol. 201, 795–809. doi: 10.1111/nph.12557
Jiang, X., Huang, K., Zheng, G., Hou, H., Wang, P., Jiang, H., et al. (2018). CsMYB5a and CsMYB5e from Camellia sinensis differentially regulate anthocyanin and proanthocyanidin biosynthesis. Plant Sci. 270, 209–220. doi: 10.1016/j.plantsci.2018.02.009
Jin, H. L., Cominelli, E., Bailey, P., Parr, A., Mehrtens, F., Jones, J., et al. (2000). Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. Embo J. 19, 6150–6161. doi: 10.1093/emboj/19.22.6150
Jun, J. H., Liu, C., Xiao, X., and Dixon, R. A. (2015). The transcriptional repressor MYB2 regulates both spatial and temporal patterns of proanthocyandin and anthocyanin pigmentation in Medicago truncatula. Plant Cell 27, 2860–2879. doi: 10.1105/tpc.15.00476
Kagale, S., and Rozwadowski, K. (2011). EAR motif-mediated transcriptional repression in plants: an underlying mechanism for epigenetic regulation of gene expression. Epigenetics 6, 141–146. doi: 10.4161/epi.6.2.13627
Koyama, K., Numata, M., Nakajima, I., Goto-Yamamoto, N., Matsumura, H., and Tanaka, N. (2014). Functional characterization of a new grapevine MYB transcription factor and regulation of proanthocyanidin biosynthesis in grapes. J. Exp. Bot. 65, 4433–4449. doi: 10.1093/jxb/eru213
Kranz, H. D., Denekamp, M., Greco, R., Jin, H., Leyva, A., Meissner, R. C., et al. (1998). Towards functional characterisation of the members of the R2R3-MYB gene family from Arabidopsis thaliana. Plant J. 16, 263–276. doi: 10.1046/j.1365-313x.1998.00278.x
Li, T., Yamane, H., and Tao, R. (2021). Preharvest long-term exposure to UV-B radiation promotes fruit ripening and modifies stage-specific anthocyanin metabolism in highbush blueberry. Hortic. Res. 8, 1–12. doi: 10.1038/s41438-021-00503-4
Liu, X., Feng, C., Zhang, M., Yin, X., Xu, C., and Chen, K. (2013). The MrWD40-1 Gene of Chinese bayberry (Myrica rubra) Interacts with MYB and bHLH to enhance anthocyanin accumulation. Plant Mol. Biol. Rep. 31, 1474–1484. doi: 10.1007/s11105-013-0621-0
Liu, X. F., Yin, X. R., Allan, A. C., Lin-Wang, K., Shi, Y. N., Huang, Y. J., et al. (2013). The role of MrbHLH1 and MrMYB1 in regulating anthocyanin biosynthetic genes in tobacco and Chinese bayberry (Myrica rubra) during anthocyanin biosynthesis. Plant Cell Tiss. Org. 115, 285–298. doi: 10.1007/s11240-013-0361-8
Middleton, E., Kandaswami, C., and Theoharides, T. C. (2000). The effects of plant flavonoids on mammalian cells: implications for inflammation, heart disease, and cancer. Pharmacol. Rev. 52, 673–751.
Nishihara, M., Nakatsuka, T., and Yamamura, S. (2005). Flavonoid components and flower color change in transgenic tobacco plants by suppression of chalcone isomerase gene. Febs Lett. 579, 6074–6078. doi: 10.1016/j.febslet.2005.09.073
Paolocci, F., Robbins, M. P., Passeri, V., Hauck, B., Morris, P., Rubini, A., et al. (2011). The strawberry transcription factor FaMYB1 inhibits the biosynthesis of proanthocyanidins in Lotus corniculatus leaves. J. Exp. Bot. 62, 1189–1200. doi: 10.1093/jxb/erq344
Rahim, M. A., Busatto, N., and Trainotti, L. (2014). Regulation of anthocyanin biosynthesis in peach fruits. Planta 240, 913–929. doi: 10.1007/s00425-014-2078-2
Ramsay, N. A., and Glover, B. J. (2005). MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. 10, 63–70. doi: 10.1016/j.tplants.2004.12.011
Ricardo Perez-Diaz, J., Perez-Diaz, J., Madrid-Espinoza, J., Gonzalez-Villanueva, E., Moreno, Y., and Ruiz-Lara, S. (2016). New member of the R2R3-MYB transcription factors family in grapevine suppresses the anthocyanin accumulation in the flowers of transgenic tobacco. Plant Mol. Biol. 90, 63–76. doi: 10.1007/s11103-015-0394-y
Shen, H., He, X., Poovaiah, C. R., Wuddineh, W. A., Ma, J., Mann, D. G. J., et al. (2012). Functional characterization of the switchgrass (Panicum virgatum) R2R3-MYB transcription factor PvMYB4 for improvement of lignocellulosic feedstocks. New Phytol. 193, 121–136. doi: 10.1111/j.1469-8137.2011.03922.x
Shi, L., Cao, S., Chen, X., Chen, W., Zheng, Y., and Yang, Z. (2018a). Proanthocyanidin synthesis in Chinese bayberry (Myrica rubra Sieb. et Zucc.) fruits. Front. Plant Sci. 9:212. doi: 10.3389/fpls.2018.00212
Shi, L., Cao, S., Shao, J., Chen, W., Zheng, Y., Jiang, Y., et al. (2014). Relationship between sucrose metabolism and anthocyanin biosynthesis during ripening in Chinese bayberry fruit. J. Agric. Food Chem. 62, 10522–10528. doi: 10.1021/jf503317k
Shi, L., Chen, X., Chen, W., Zheng, Y., and Yang, Z. (2018b). Comparative transcriptomic analysis of white and red Chinese bayberry (Myrica rubra) fruits reveals flavonoid biosynthesis regulation. Sci. Hortic. 235, 9–20. doi: 10.1016/j.scienta.2018.02.076
Song, L., Wang, X., Han, W., Qu, Y., Wang, Z., Zhai, R., et al. (2020). PbMYB120 negatively regulates anthocyanin accumulation in pear. Int. J Mol. Sci. 21:1528. doi: 10.3390/ijms21041528
Takos, A. M., Jaffe, F. W., Jacob, S. R., Bogs, J., Robinson, S. P., and Walker, A. R. (2006). Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol. 142, 1216–1232. doi: 10.1104/pp.106.088104
Terrier, N., Torregrosa, L., Ageorges, A., Vialet, S., Verries, C., Cheynier, V., et al. (2009). Ectopic expression of VvMybPA2 promotes proanthocyanidin biosynthesis in grapevine and suggests additional targets in the pathway. Plant Physiol. 149, 1028–1041. doi: 10.1104/pp.108.131862
Walker, A. R., Lee, E., Bogs, J., McDavid, D. A. J., Thomas, M. R., and Robinson, S. P. (2007). White grapes arose through the mutation of two similar and adjacent regulatory genes. Plant J. 49, 772–785. doi: 10.1111/j.1365-313X.2006.02997.x
Wan, S., Li, C., Ma, X., and Luo, K. (2017). PtrMYB57 contributes to the negative regulation of anthocyanin and proanthocyanidin biosynthesis in poplar. Plant Cell Rep. 36, 1263–1276. doi: 10.1007/s00299-017-2151-y
Wang, L., Ran, L., Hou, Y., Tian, Q., Li, C., Liu, R., et al. (2017). The transcription factor MYB115 contributes to the regulation of proanthocyanidin biosynthesis and enhances fungal resistance in poplar. New Phytol. 215, 351–367. doi: 10.1111/nph.14569
Winkel-Shirley, B. (2001). Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 126, 485–493. doi: 10.1104/pp.126.2.485
Xu, H., Wang, N., Liu, J., Qu, C., Wang, Y., Jiang, S., et al. (2017). The molecular mechanism underlying anthocyanin metabolism in apple using the MdMYB16 and MdbHLH33 genes. Plant Mol. Biol. 94, 149–165. doi: 10.1007/s11103-017-0601-0
Xu, H., Yang, G., Zhang, J., Wang, Y., Zhang, T., Wang, N., et al. (2018). Overexpression of a repressor MdMYB15L negatively regulates anthocyanin and cold tolerance in red-fleshed callus. Biochem. Biophys. Res. Commun. 500, 405–410. doi: 10.1016/j.bbrc.2018.04.088
Yoshida, K., Ma, D., and Constabel, C. P. (2015). The MYB182 protein down-regulates proanthocyanidin and anthocyanin biosynthesis in poplar by repressing both structural and regulatory flavonoid genes. Plant Physiol. 167, 693–710. doi: 10.1104/pp.114.253674
Zhong, C., Tang, Y., Pang, B., Li, X., Yang, Y., Deng, J., et al. (2020). The R2R3-MYB transcription factor GhMYB1a regulates flavonol and anthocyanin accumulation in Gerbera hybrida. Hortic. Res. 7:78. doi: 10.1038/s41438-020-0296-2
Zhou, H., Kui, L. W., Liao, L., Chao, G., Lu, Z., Allan, A. C., et al. (2015). Peach MYB7 activates transcription of the proanthocyanidin pathway gene encoding leucoanthocyanidin reductase, but not anthocyanidin reductase. Front. Plant Sci. 6:908. doi: 10.3389/fpls.2015.00908
Zhou, H., Lin-Wang, K., Wang, F., Espley, R. V., Ren, F., Zhao, J., et al. (2019). Activator-type R2R3-MYB genes induce a repressor-type R2R3-MYB gene to balance anthocyanin and proanthocyanidin accumulation. New Phytol. 221, 1919–1934. doi: 10.1111/nph.15486
Keywords: Chinese bayberry, anthocyanins, proanthocyanidins, repressor, MYB transcription factor
Citation: Shi L, Chen X, Wang K, Yang M, Chen W, Yang Z and Cao S (2021) MrMYB6 From Chinese Bayberry (Myrica rubra) Negatively Regulates Anthocyanin and Proanthocyanidin Accumulation. Front. Plant Sci. 12:685654. doi: 10.3389/fpls.2021.685654
Received: 25 March 2021; Accepted: 27 May 2021;
Published: 18 June 2021.
Edited by:
Igor Cesarino, University of São Paulo, BrazilReviewed by:
Tina Agarwal, Purdue University, United StatesCopyright © 2021 Shi, Chen, Wang, Yang, Chen, Yang and Cao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shifeng Cao, c2hpZmVuZ2NhbzFAZ21haWwuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.