
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci., 26 July 2021
Sec. Plant Cell Biology
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.684626
This article is part of the Research TopicMolecular Organization, Evolution, and Function of Ribosomal DNAView all 15 articles
Eukaryotic ribosome assembly starts in the nucleolus, where the ribosomal DNA (rDNA) is transcribed into the 35S pre-ribosomal RNA (pre-rRNA). More than two-hundred ribosome biogenesis factors (RBFs) and more than two-hundred small nucleolar RNAs (snoRNA) catalyze the processing, folding and modification of the rRNA in Arabidopsis thaliana. The initial pre-ribosomal 90S complex is formed already during transcription by association of ribosomal proteins (RPs) and RBFs. In addition, small nucleolar ribonucleoprotein particles (snoRNPs) composed of snoRNAs and RBFs catalyze the two major rRNA modification types, 2′-O-ribose-methylation and pseudouridylation. Besides these two modifications, rRNAs can also undergo base methylations and acetylation. However, the latter two modifications have not yet been systematically explored in plants. The snoRNAs of these snoRNPs serve as targeting factors to direct modifications to specific rRNA regions by antisense elements. Today, hundreds of different sites of modifications in the rRNA have been described for eukaryotic ribosomes in general. While our understanding of the general process of ribosome biogenesis has advanced rapidly, the diversities appearing during plant ribosome biogenesis is beginning to emerge. Today, more than two-hundred RBFs were identified by bioinformatics or biochemical approaches, including several plant specific factors. Similarly, more than two hundred snoRNA were predicted based on RNA sequencing experiments. Here, we discuss the predicted and verified rRNA modification sites and the corresponding identified snoRNAs on the example of the model plant Arabidopsis thaliana. Our summary uncovers the plant modification sites in comparison to the human and yeast modification sites.
Ribosome biogenesis is an essential biochemical process in all existing organisms. The formation of functional ribosomes involves a huge number of different RNAs and proteins, which have to act in a defined order. These factors catalyze various steps during the maturation of ribosomal RNA (rRNA) from the initial precursor including, their folding, modifications of the rRNA and the assembly of ribosomal proteins. For model systems like yeast, the understanding of molecular events during ribosome biogenesis are already well described. For example, comprehensive number of ribosome assembly factors and their functions, in addition to availability of high resolution ribosome structure, paved the way for in-depth analysis of ribosome maturation in yeast (Woolford and Baserga, 2013; Klinge and Woolford, 2019). While for the same processes in plant systems, many aspects are yet to be given a detailed account. For the analysis of the processes in plants, Arabidopsis thaliana has become the model plant for the examination of ribosome biogenesis next to crop plants like wheat and rice (Armache et al., 2010b; Hang et al., 2018).
The maturation of 80S ribosomes is coordinated between three different compartments of the cell. It begins with the transcription of the 35S pre-rRNA by RNA-polymerase I in the nucleolus (Tsang et al., 2003; Henras et al., 2008; Missbach et al., 2013; Hellmann, 2020). The 35S pre-rRNA consists of the three rRNAs 18S, 5.8S, and 25S. The 18S and 5.8S rRNA are separated by the internal transcribed spacer 1 (ITS1), 5.8S and 25S rRNA by the internal transcribed spacer 2 (ITS2) and the three maturing rRNAs are additionally flanked by the 5′- and 3′-external transcribed spacers (ETSs); (Tollervey and Kiss, 1997; Lafontaine, 2015; Weis et al., 2015b; Figure 1). This precursor is subsequently processed and modified. The maturation of the rRNA is assisted by ribosome biogenesis factors (RBFs) and small nucleolar RNAs (snoRNAs) (Figure 2A). Initially, the 90S particle formation is followed by subsequent splitting into pre-40S and pre-60S particles. The maturation of these particles occurs in the nucleolus, nucleoplasm and in the cytosol (Figure 1). During the maturation of ribosomal subunits, the precursors of 18S, 5.8S, and 25S rRNA are processed, folded and modified, and the final steps occur in the cytoplasm before final assembly of the 80S ribosomes (Henras et al., 2008; Palm et al., 2019; Sáez-Vásquez and Delseny, 2019). In addition to the rRNAs transcribed on the 35S transcript, the large ribosomal subunit (LSU) contains a 5S rRNA. This rRNA is transcribed independently in the nucleus by RNA-polymerase III (Henras et al., 2008; Missbach et al., 2013; Bassham and MacIntosh, 2017). The 5S rRNA forms the 5S RNP together with the ribosomal proteins L5 and L18, which associates with the 60S pre-ribosomal particle in the nucleoplasm (Leidig et al., 2014; Lafontaine, 2015).
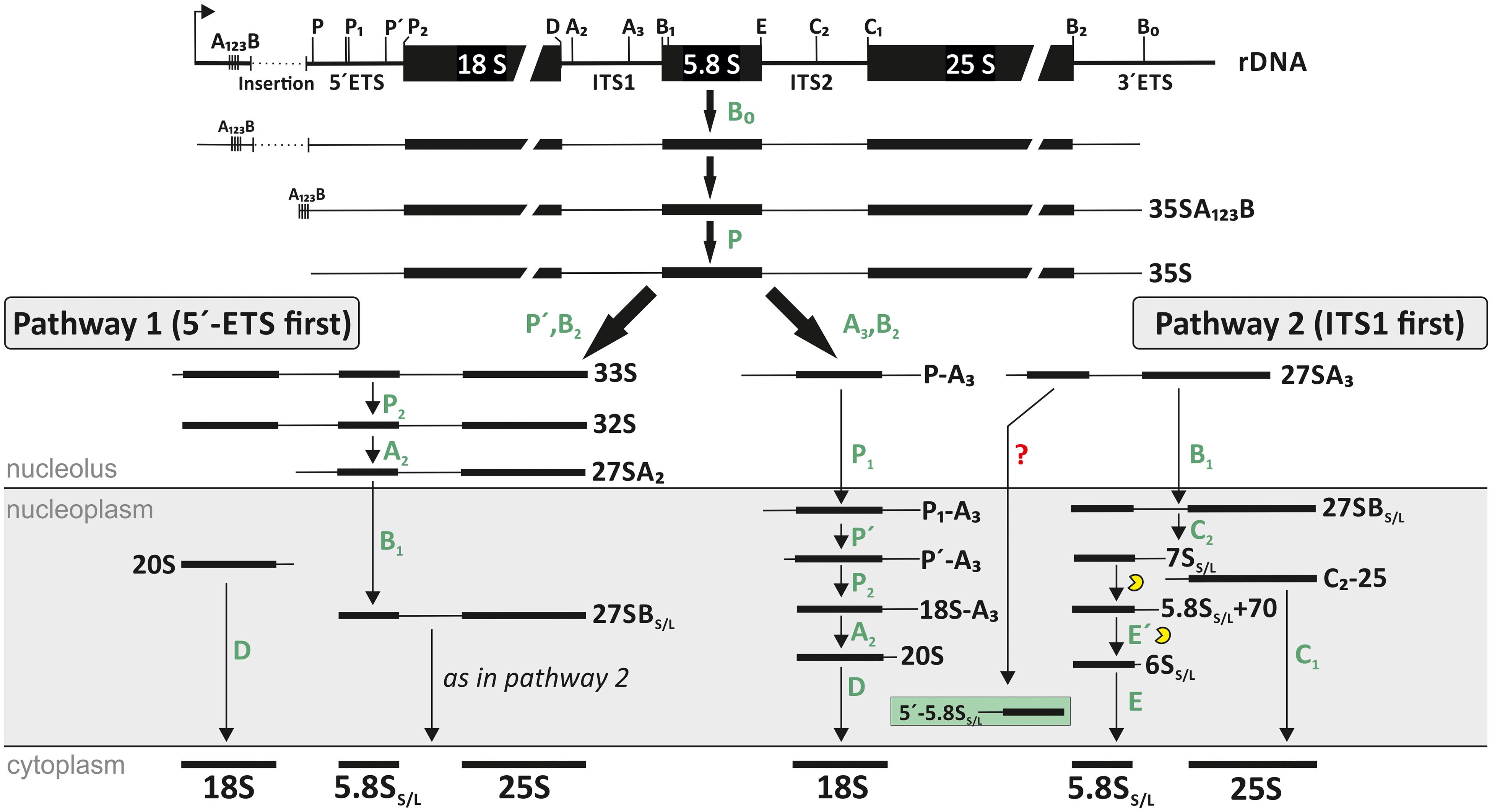
Figure 1. Ribosomal RNA (rRNA) processing pathway in A. thaliana. After transcription the 35S pre-rRNA is formed. After cleavage at site P, the further processing depends on the next cleavage site. While for pathway 1 the next cleavage occurs in the 5′-ETS (similar to the main pathway in yeast), the second and majorly used pathway is characterized by a first cleavage in the ITS1 region (similar to the human pathway). The location of each existing precursor of Arabidopsis is shown. However, the exact localization of 27S and 18S precursors are not fully analyzed yet. The figure was modified from Weis et al. (2015a).
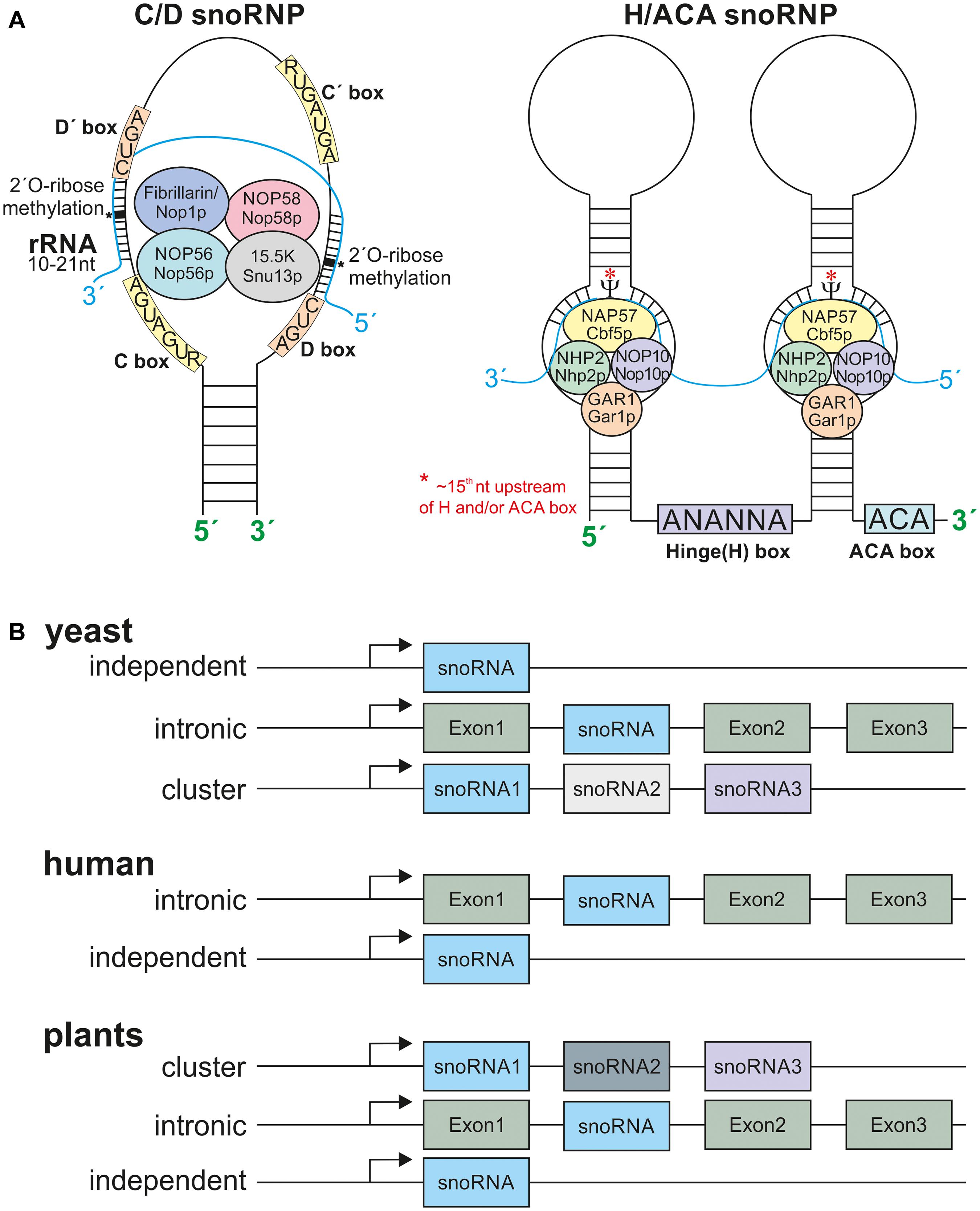
Figure 2. The snoRNPs and snoRNA localization. (A) Depicted are the C/D box snoRNPs and H/ACA snoRNPs. The C/D box snoRNPs contain the conserved C (RUGAUGA) and D (CUGA) boxes and the two less conserved C’ and D’-boxes. The C/D box snoRNPs are composed of the methyltransferase fibrillarin/Nop1p, NOP56/Nop56p, Nop58/Nop58p, and 15.5K/Snu13p. The rRNA has a 10–21 nt long complementary site to the according snoRNA. The 2′-O-ribose-methylation takes place 5 nt downstream of the D or D’ box (asterisks). The H/ACA box snoRNPs are composed of at least one stem loop. The Hinge Box (ANANNA) is located between two stem loops and the ACA box at the 3′ end of the snoRNA. The pseudouridine synthase NAP57/Cbf5p is modifying approx. the 15th nucleotide (asterisks) of the rRNA upstream of the H and or ACA box, further the proteins NHP2/Nhp2p, Nop10/Nop10p, and GAR1/Gar1p are required. (B) The snoRNA gene organization in different eukaryotes. Yeast snoRNAs are mainly localized in independent regions and lesser in intronic and polycistronic (cluster) regions. The human snoRNA gene organization is mostly intronic with few independent genes and plant snoRNAs are mostly located within clusters of many snoRNAs. Only very few examples for intronic and independent gene organization are known. For references see main text.
To date, more than 200 RBFs and more than 200 snoRNAs were described to regulate ribosome maturation. In plants, the inventory for both has been established by a combination of experimental evidence and bioinformatics prediction (Barneche et al., 2001; Brown et al., 2001; Qu et al., 2001; Chen C. L. et al., 2003; Chen and Wu, 2009; Kim et al., 2010; Liu et al., 2013; Palm et al., 2016; Azevedo-Favory et al., 2020). Considering the importance of rRNA modifications for proper processing and maturation of rRNA but also for the function of ribosomes, we discuss in the following the current knowledge on the rRNA modifications and snoRNAs in Arabidopsis and compare these with the human and yeast modifications sites.
Alike messenger RNA (mRNA) and transfer RNA (tRNA), rRNA is highly post-transcriptional modified (Sloan et al., 2017). For yeast, it could be shown that the loss of individual modifications within the rRNA is non-essential, while the lack of more than one modification site, especially in important regions of the ribosome has led to alterations in ribosomal processing but also rRNA processing can be affected (Liang et al., 2009; Demirci et al., 2010; Polikanov et al., 2015). Furthermore, different distributions of modification sites can be related to different cell type as reported for human ribosomes, where cancer cells carry a different subset of modifications (Natchiar et al., 2017). Nonetheless, in contrast it could be demonstrated that the loss of a single modification site in zebrafish can have harmful effects during the early development (Higa-Nakamine et al., 2012). However, only a small number of rRNA modification types are known. The two major modifications are 2′-O-ribose-methylation (2′-O-ribose-me), where a methyl group is attached to the 2′ hydroxyl-group of the ribose within nucleosides, and pseudouridylation involving the conversion of uridine to pseudouridine (Zhao and Yu, 2004; Ito et al., 2014). Recent studies in yeast showed, that the acetyltransferase Kre33 acetylates the sites ac4C1773 and ac4C1280 of the 18S rRNAs, which are guided by the two orphan C/D box snoRNAs snR4 and snR45 (Ito et al., 2014; Sharma et al., 2017b). Some snoRNAs like the abundant C/D box snoRNAs U3 and U14 are rather involved in pre-rRNA cleavage at the 5′-ETS site and are therefore involved in 18S rRNA production (Brown and Shaw, 1998; Venema and Tollervey, 1999; Brown et al., 2003).
The 2′-O-ribose-me is the most frequently occurring modification within RNA and can be important for RNA degradation. For example, it was observed that miRNA and siRNAs lacking 2′-O-ribose-me on the 3′ terminal ribose are exposed to degradation (Zhao et al., 2012). In addition, 2′-O-ribose-me defines local secondary structures (Filippova et al., 2017). Likewise, the isomerization of uridine to pseudouridine confers stability of hairpins by base stacking (Desaulniers et al., 2008; Filippova et al., 2017). Modifications often occur in functionally relevant areas of the ribosomes such as A, P, and E sites, the peptidyl transferase center (PTC) and the intersubunit bridge (Decatur and Fournier, 2002; Watkins and Bohnsack, 2012). Remarkably, RNA and especially rRNA modifications appear to be altered during development in addition to environmental changes, which could indicate ribosome heterogeneity (Sloan et al., 2017).
Small nucleolar RNAs are small RNA molecules essential for the regulation and guidance of the post-transcriptional modifications of rRNA, tRNA, and snRNAs (Kiss, 2001; Kruszka et al., 2003; Chow et al., 2007). SnoRNAs exist in eukaryotes and archaea, but not in bacteria (Terns and Terns, 2002; Bhattacharya et al., 2016). Because of their importance for rRNA folding and modification, they are often localized in the nucleus where processing and modification of rRNA takes place (Kiss-László et al., 1998). They are re-localized to the cytoplasm in response to stress, which has so far only been described in yeast (Holley et al., 2015). Whether this holds true for eukaryotes in general needs to be elucidated.
The sizes of snoRNAs vary between 60 and 300 nucleotides (nt; Falaleeva et al., 2017) and they are mostly transcribed by RNA-polymerase II (Qu et al., 2015). Nevertheless, in cases of the U3 gene in plants and in dicistronic tRNA-snoRNA genes, RNA-Polymerase III is responsible for the transcription (Dieci et al., 2009). The snoRNA gene organization varies between organisms. Most snoRNAs in yeast are independently encoded, and only a minority is localized in intronic regions or in cluster with other snoRNA genes (Brown et al., 2003; Figure 2B). The majority of snoRNAs in humans are organized in intronic regions, and only few are encoded as independent genes (Figure 2B). In plant genomes snoRNAs are encoded either independently, in intronic regions or in intronic gene clusters as shown in rice (Figure 2B; Leader et al., 1997; Brown et al., 2003). In addition, snoRNAs are also organized in dicistronic tRNA-snoRNAs or snoRNA-miRNA clusters as described in A. thaliana and rice (Kruszka et al., 2003; Qu et al., 2015).
The snoRNAs are classified by the existence of conserved sequence motifs (Bachellerie and Cavaillé, 1997; Weinstein and Steitz, 1999). The so called C/D box and H/ACA box snoRNAs form the two major classes, while some minor classes have been identified as well. The C/D box snoRNAs are characterized by a “C box” with a consensus sequence RUGAUGA (R stands for any purine) and a “D box” (consensus sequence: CUGA) (Figure 2A). Frequently, these snoRNAs contain additional, less conserved boxes annotated as C’ and D’. The conserved C and D boxes fulfill a multitude of functions and are amongst necessary for the snoRNA import into the nucleolus (Samarsky et al., 1998; Newman et al., 2000; Bertrand and Fournier, 2013). In contrast, binding to the rRNA target region is accomplished by one or two antisense elements of about 10–21 nt positioned upstream of the D or D’-boxes. In most cases, the complement to the fifth nucleotide of this element is modified in the rRNA (Barneche et al., 2001; Kiss, 2001; Kruszka et al., 2003). The secondary structure of C/D box snoRNA is characterized by a K-turn motif that brings the C and D box (C’ and D’) in proximity through stem loop formation, and by guide elements carrying the antisense sequences (Matera et al., 2007). The C/D box snoRNAs are components of small nucleolar ribonucleoprotein particles (snoRNPs) that in addition consists of described four core proteins fibrillarin (methyltransferase; Nop1p in yeast), NOP58 (Nop58p in yeast), NOP56 (Nop56p in yeast), and 15.5K (Snu13p in yeast) (Rodor et al., 2011; Huang et al., 2016; Figure 2A).
The hinge box (H-box: sequence ANANNA, N stands for any nucleotide) and the 3′ terminal located ACA box characterize the H/ACA box snoRNA family (Brown and Shaw, 1998; Kiss, 2001). The H and ACA boxes are required for nucleolar import, for example (Bertrand and Fournier, 2013). H/ACA snoRNPs form a hairpin-hinge-hairpin-tail structure with the tail and the hinge region being single stranded (Dragon et al., 2006). The nucleotide to be modified is positioned about 15 nt upstream of the ACA or hinge motif, respectively (Lindsay et al., 2013; Figure 2A). Alike the C/D box snoRNAs, H/ACA box snoRNAs are components of snoRNPs. However, the known snoRNPs containing an H/ACA box snoRNA consist of the proteins dyskerin/NAP57 (pseudouridine synthase; Cbf5p in yeast), NHP2 (Nhp2p), NOP10 (Nop10p), and GAR1 (Gar1p) (Rodor et al., 2011; Figure 2A).
Another minor class of snoRNAs unifies the mitochondrial RNA processing (MRP)-RNAs, a snoRNA family which lacks conserved boxes, but harbors rRNA processing activity (Bertrand and Fournier, 2013). Additional snoRNAs without typical motifs are deposited in plant snoRNA databases as well (Yoshihama et al., 2013).
Since the first discoveries of snoRNAs in 1970’s (Reddy et al., 1974, 1979) different studies targeted the identification of plant snoRNAs by experimental approaches. Early on, the snoRNAs U3, U14, and U49 were identified in plants based on similarity to the snoRNAs of yeast and vertebrates (Kiss et al., 1991; Leader et al., 1994, 1997). Remarkably, U3 in plants is transcribed by the RNA polymerase III and possess a different capping than found for U3 in yeast or human (Kiss et al., 1991). By dot-matrix analysis of Fib1 and Fib2, the plant-specific snoRNAs U60.1f and U60.2f were discovered (Barneche et al., 2000).
After the release of the A. thaliana genome (Kaul et al., 2000) snoRNAs were identified by computational strategies searching for C/D box characteristics, rRNA complementarities or other structural attributes (Zhou et al., 2000; Barneche et al., 2001; Brown et al., 2001; Qu et al., 2001). The next boost for the discovery of plant snoRNAs came by RNomics on either total RNA from different tissues or from the nucleolar RNA of A. thaliana (Marker et al., 2002; Kim et al., 2010; Streit et al., 2020) and by re-analysis of existing small RNA datasets of different A. thaliana tissues and growth stages (Chen and Wu, 2009). This analysis was initially focused on Arabidopsis and was then extended to Oryza sativa (Chen C. L. et al., 2003; Liu et al., 2013). Today, 10,654 different H/ACA box snoRNA genes and 6064 different C/D box snoRNA genes are deposited in the database snOPY (Yoshihama et al., 2013).
In contrast to globally discovered snoRNAs, only a single plant snoRNA is functionally characterized. The C/D box type snoRNA HIDDEN TREASURE 2 (HID2) associates with 45S pre-rRNA but is not relevant for 2′-ribose methylation at position G2620 as this modification was not altered in an according mutant (Zhu et al., 2016). It was speculated that other snoRNAs might complement for HID2 function (Zhu et al., 2016), which needs to be verified. Thus, the analysis of the snoRNA function and the complementarity of the different snoRNA genes of the different families will be a major target of future research.
There are two major types of rRNA modifications, namely 2′-O-ribose-methylation and pseudouridylation. However, for yeast and human rRNAs, base methylations were additionally described (e.g., for yeast dimethylase Dim1p) (Lafontaine et al., 1995). Furthermore, yeast rRNA was also found to be acetylated (Ito et al., 2014; Sharma et al., 2017b). However, the latter two modification types have not been described so far in plants (Piekna-Przybylska et al., 2007).
Initially, the analysis of the individual snoRNAs was accompanied by the analysis of the rRNA modification sites, e.g., by primer extension analysis (Barneche et al., 2000). Recently, genome-wide pseudouridine sequencing verified predicted pseudouridine modifications in cytosolic and plastidic ribosomes (Sun et al., 2019). At the same time, this approach led to the discovery of yet unknown modification sites as well (Sun et al., 2019). Remarkably, the ITS1 separating the 18S rRNA from 5.8S is modified as well (Sun et al., 2019). However, future studies are required to explore whether this is a unique modification or whether ITS1, ITS2 and the ETS regions are generally modified, and to understand the role of modifications of the pre-rRNA. A complementary analysis using RiboMethSeq for detection of 2′-O-ribose-me modifications yielded novel modification sites as well (Azevedo-Favory et al., 2020).
Different approaches exploring the modifications of the rRNA in Arabidopsis yielded a total of 321 rRNA modification sites (Barneche et al., 2001; Qu et al., 2001; Chen and Wu, 2009; Kim et al., 2010; Sun et al., 2019; Azevedo-Favory et al., 2020; Streit et al., 2020). A total of 79 2′-O-ribose-me and 43 pseudouridylation sites were assigned for the 18S rRNA, of which 44 2′-O-ribose-me and 28 pseudouridylation sites were experimentally confirmed. For 25S rRNA, 132 2′-O-ribose-me and 64 pseudouridylation sites are proposed, of which 86 and 51, respectively, are experimentally confirmed. For 5.8S rRNA, three sites carrying 2′-O-ribose-me were predicted due to antisense elements found in three snoRNAs of which two are experimentally confirmed. Accordingly, a recent study confirmed the predicted U22 pseudouridylation and mapped a new site at U78 (Sun et al., 2019).
It has to be considered that the existing discrepancy between detected and predicted modification sites might result from a variability of the modification pattern in ribosomes of one cell, in different tissues, at different developmental stages or in response to environmental changes as discovered for other species named ribosome heterogeneity (Sloan et al., 2017). However, as the ribosome turn-over is comparatively slow, alterations in rRNA modifications are considered to be more meaningful for long-term changes (Ferretti and Karbstein, 2019). Although the final annotation and confirmation of predicted sites requires further research, in here the predicted sites are discussed as well. In the following sections, the positioning of the modifications in the rRNA, for selected modifications the function and the required snoRNAs are discussed.
For the 5.8S rRNA, three 2′-O-ribose-me sites were predicted (Figure 3 and Supplementary Table 1), of which two sites were mapped by primer extension (Barneche et al., 2001; Brown et al., 2001; Qu et al., 2001). Additionally, two pseudouridylation sites in 5.8S could be mapped as well (Sun et al., 2019). Thus, the modification of the 5.8S rRNA in A. thaliana is more similar to human with four modifications (two 2′-O-ribose-me and two pseudouridylation sites) than to yeast, where only a single Ψ-site at U73 is known so far (Piekna-Przybylska et al., 2007). However, the Ψ-site in yeast exists in A. thaliana at position Ψ78, although this Ψ-site was found at the adjacent uracil in Arabidopsis (Figure 3). The H/ACA box snoRNA snR43 targeting this site in yeast (Piekna-Przybylska et al., 2007) could not be identified in A. thaliana.
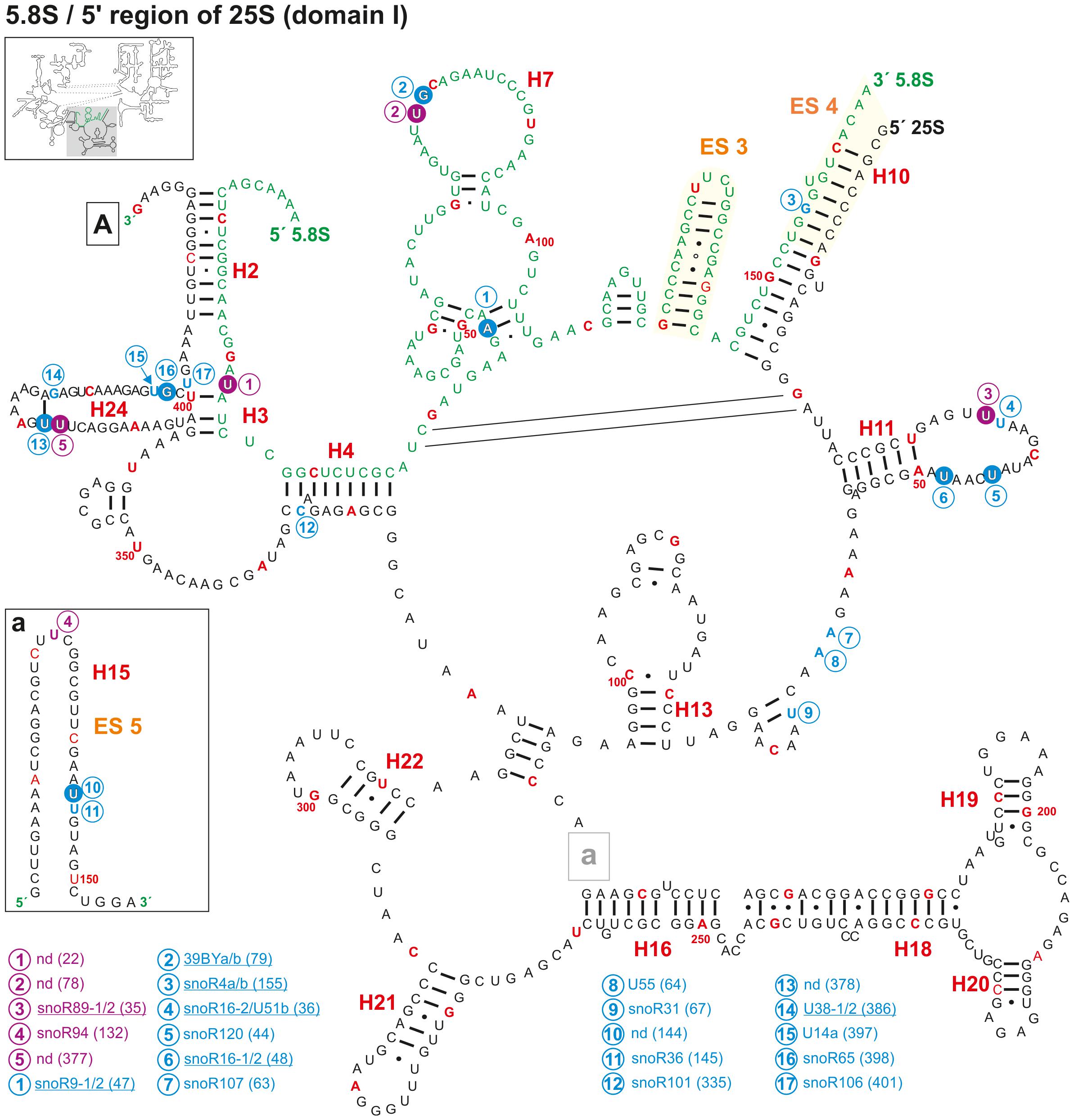
Figure 3. Secondary structure diagram of the 5.8S rRNA and domain I of 25S rRNA of A. thaliana. There is no experimental determined secondary structure map of the rRNA of A. thaliana. Hence, the secondary map of 5.8S (letters in green) and 25S (letters in black) was created based on the RNAcentral database (The RNAcentral Consortium, 2019) and http://www.rna.icmb.utexas.edu. The positions for predicted (blue letter) and verified 2′-O-ribose-me (white letter in blue circle) sites and the predicted (violet letter) and verified positions for pseudouridylation (white letter in violet circle) are shown. If several snoRNAs are annotated to target the same site, the name is underlined. Analyses were conducted by using the snoRNA databases snOPY (Yoshihama et al., 2013) and the plant snoRNA DB (Brown et al., 2003). Predicted and verified positions for 2′-O-ribose-methylations and pseudouridylations were obtained from Barneche et al. (2001), Brown et al. (2001), Qu et al. (2001), Sun et al. (2019), Azevedo-Favory et al. (2020). Every tenth nucleotide is marked in red and every 50th nucleotide is labeled with the according number. Framed small letters indicate the position of the structures shown separately and framed large letters indicate the position of the connections in subsequent images (for A see Figure 4). The sequence used for the secondary structure map refers to the sequence of 5.8S and 25S of chromosome 2. The number in brackets correspond to the modified nucleotide position. A small illustration of the whole 60S rRNA secondary structure highlighting the according region is enclosed.
The sites Am47, Gm79, and Ψ78 are localized in the 5.8S secondary structure, which is formed by three bulges between the helices 5, 6, and 7 (Figure 3 and Supplementary Table 1). Worth mentioning, the 2′-O-ribose-me at Gm79 in A. thaliana (Figure 3) represents the Gm75 modification site in humans (Piekna-Przybylska et al., 2007). Remarkably, in human ribosomes the region which includes helices 5, 6, and 7 is sandwiched between uL26, L35/uL29, L37, and eL39 (Khatter et al., 2015) and the structure changes between the mRNA free and the mRNA bound state (Graifer et al., 2005). A similar structure was obtained in plant ribosomes, where the helices 5, 6, and 7 are sandwiched by L24, L29, L37e, and L39e (Armache et al., 2010a). Moreover, L29 was identified as one of the ribosomal proteins with diurnal alteration of the phosphorylation state in Arabidopsis (Turkina et al., 2011). This suggest that the modification in this region of the 5.8S rRNA might be important for the ribosomal activity in translation (Gulay et al., 2017) or for the ribosomal translation elongation (translocation), which was found in cell-free extracts to be under the regulation of the 5.8S rRNA as well (Elela and Nazar, 1997).
In addition, one modification is found in the bulge between helix 2 and helix 3, and one in helix 10. All three helices are formed by base pairing between 5.8S and 25S rRNA (Figure 3) and are deeply buried in the ribosomal structure in the human ribosomes (Khatter et al., 2015). Thus, it is likely that the modifications are required for stabilizing the structure of the ribosomes. Interestingly, the predicted 2′-ribose-O-me site at position Gm155, which hypothetically is targeted by snoR4a/4b, could not be confirmed by radiographic labeling of modified nucleotides in wheat-embryo (Lau et al., 1974), suggesting that this snoRNA is probably not involved in the modification but rather in rRNA processing in the ITS2 region (Brown et al., 2001). However, it is known that certain modifications of the eukaryotic 5.8S are tissue specific (Nazar et al., 1975). Hence, it remains possible that modifications like Gm155 are only present in selected tissues or in developmental manner.
Furthermore, modifications such as 2′-O-ribose-methylations at Um14 in rat liver appeared to be present in a higher degree in the cytoplasmic fraction than in nuclear fractions (Nazar et al., 1980). However, the cellular distribution of the rRNA modifications in plant cells was not experimentally approached so far.
The 25S rRNA is the largest RNA within ribosomes and thus it contains numerous modifications. For better discussion, the 25S rRNA is dissected in here into five domains along the rRNA sequence. The 5′ region (bp 1–660) is assigned as domain I (Figures 3, 4), bp 660–1440 as domain II (Figure 5), bp 1440–1870 as domain III (Figure 4), bp 1870–2370 as domain IV (Figure 6) and the 3′ region (bp 2370–3375) is assigned as domain V/VI (Figure 7; Paci and Fox, 2015). The helical domains (H), the expansion segments (ES), and pivoting regions (PR) are in part numbered according to previous annotations (Taylor et al., 2009; Paci and Fox, 2015). Expansion segments are additional rRNA parts in eukaryotic rRNA compared to the prokaryotic rRNA. Though expansion segments can vary in their sequence and length but are rather conserved in their overall secondary structure (Ramesh and Woolford, 2016).
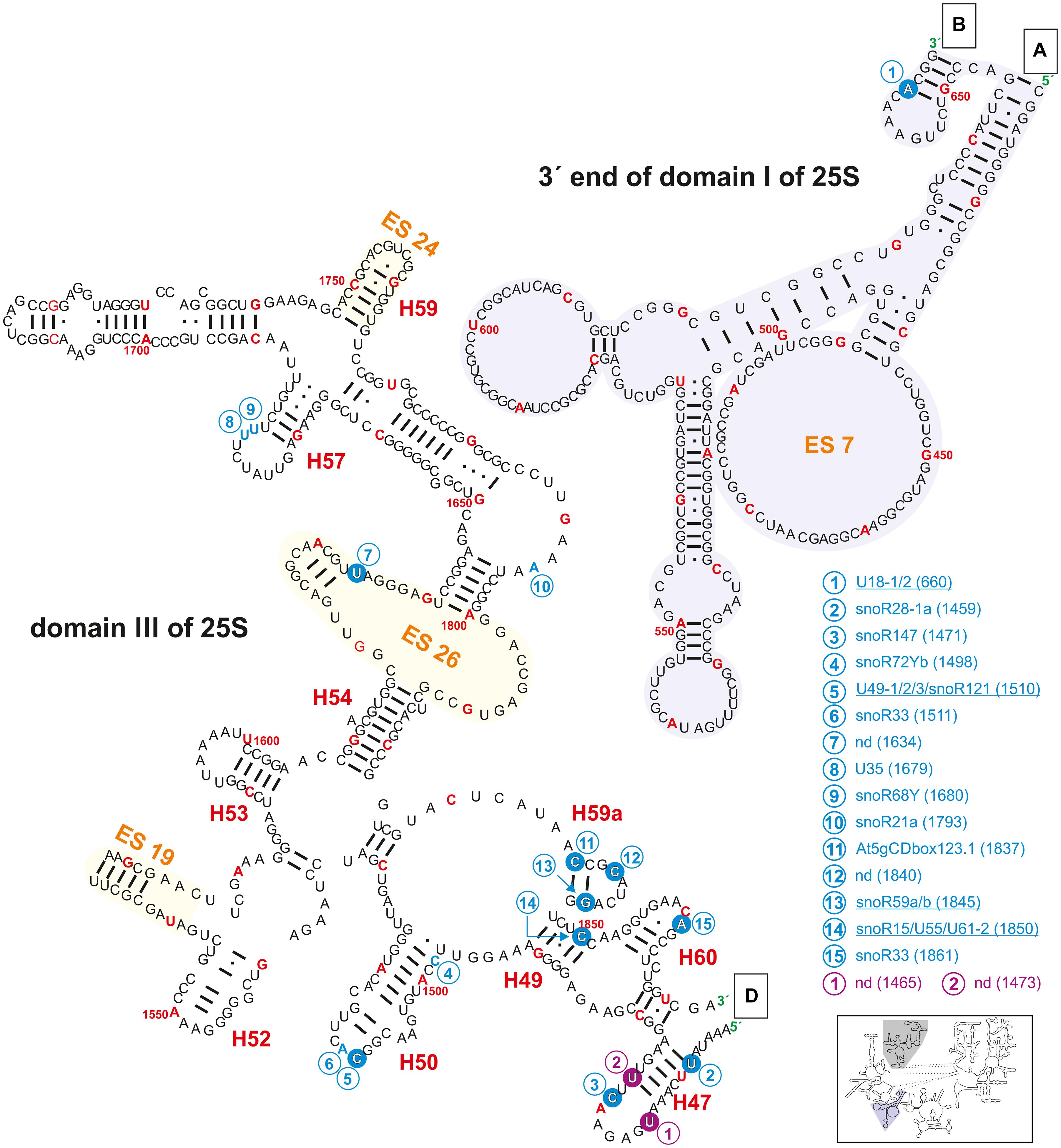
Figure 4. Secondary structure diagram of the 3′ end of domain I (gray background) as well as of domain III of 25S rRNA of A. thaliana. The image is shown according to the legend for Figure 3. Framed large letters indicate the position of the connections in subsequent images (for A see Figure 3; for B see Figure 6; for D see Figure 5). The number in brackets correspond to the modified nucleotide position. A small illustration of the whole 60S rRNA secondary structure highlighting the according region is enclosed.
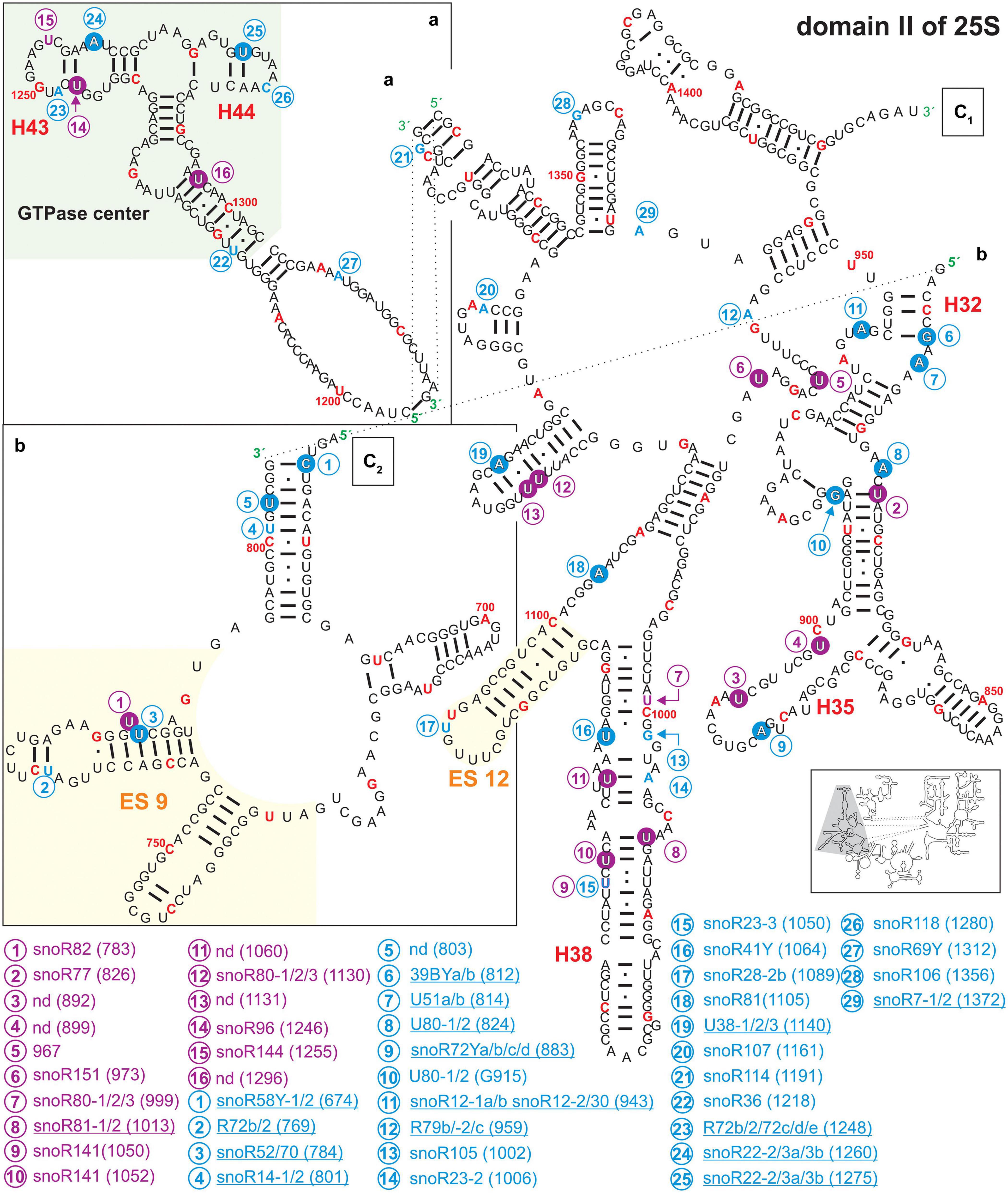
Figure 5. Secondary structure diagram of domain III of 25S rRNA of A. thaliana. The image is shown according to the legend for Figure 3. Framed large letters indicate the position of the connections in subsequent images (for C see Figure 6). PE annotates a pivoting element previously identified (Paci and Fox, 2015). The number in brackets correspond to the modified nucleotide position. A small illustration of the whole 60S rRNA secondary structure highlighting the according region is enclosed.
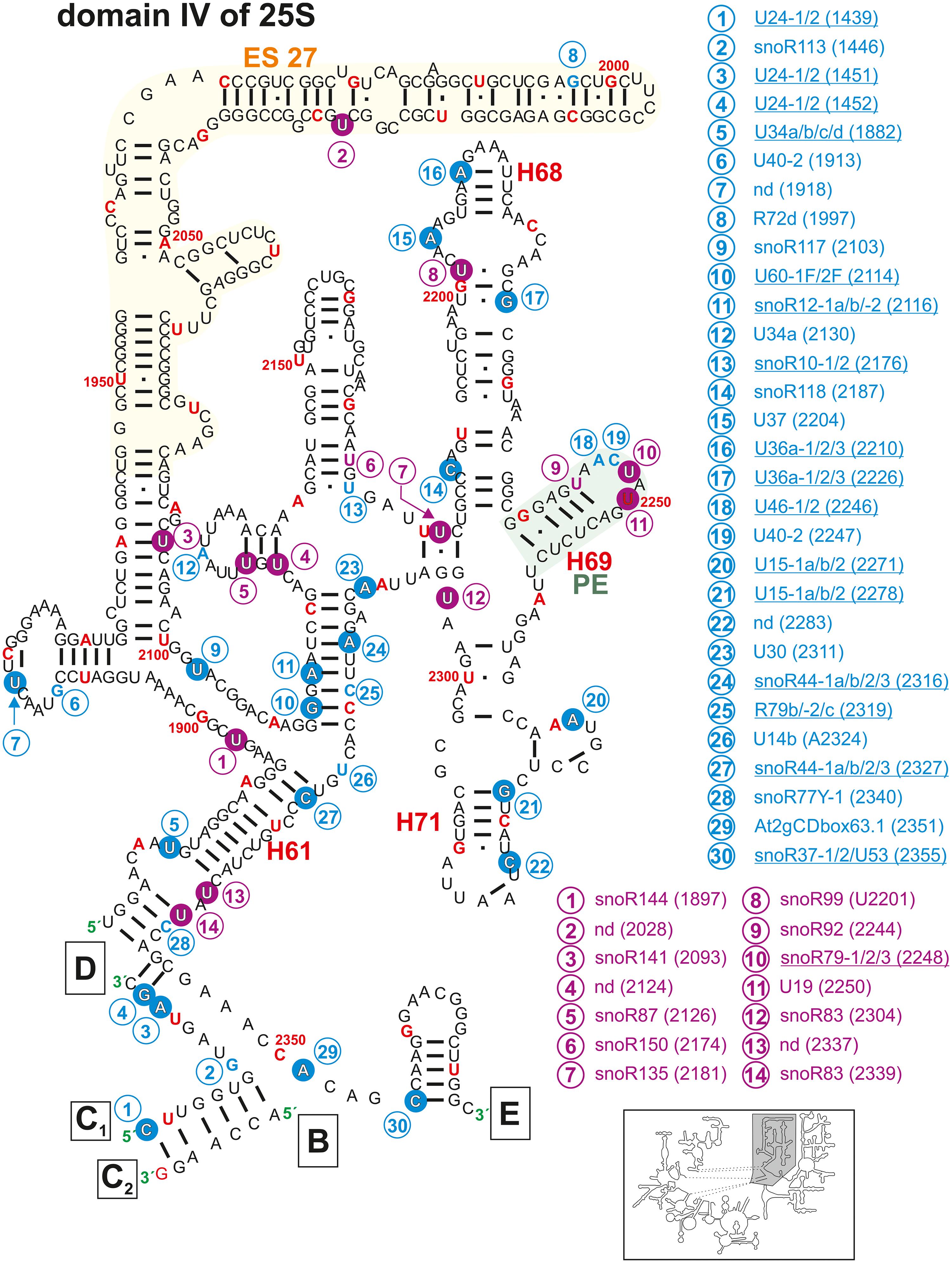
Figure 6. Secondary structure diagram of domain IV of 25S rRNA of A. thaliana. The image is shown according to the legend for Figure 3. Framed large letters indicate the position of the connections in subsequent images (for B see Figure 4; for C see Figure 5; for D see Figure 4; for E see Figure 7). The number in brackets correspond to the modified nucleotide position. A small illustration of the whole 60S rRNA secondary structure highlighting the according region is enclosed.
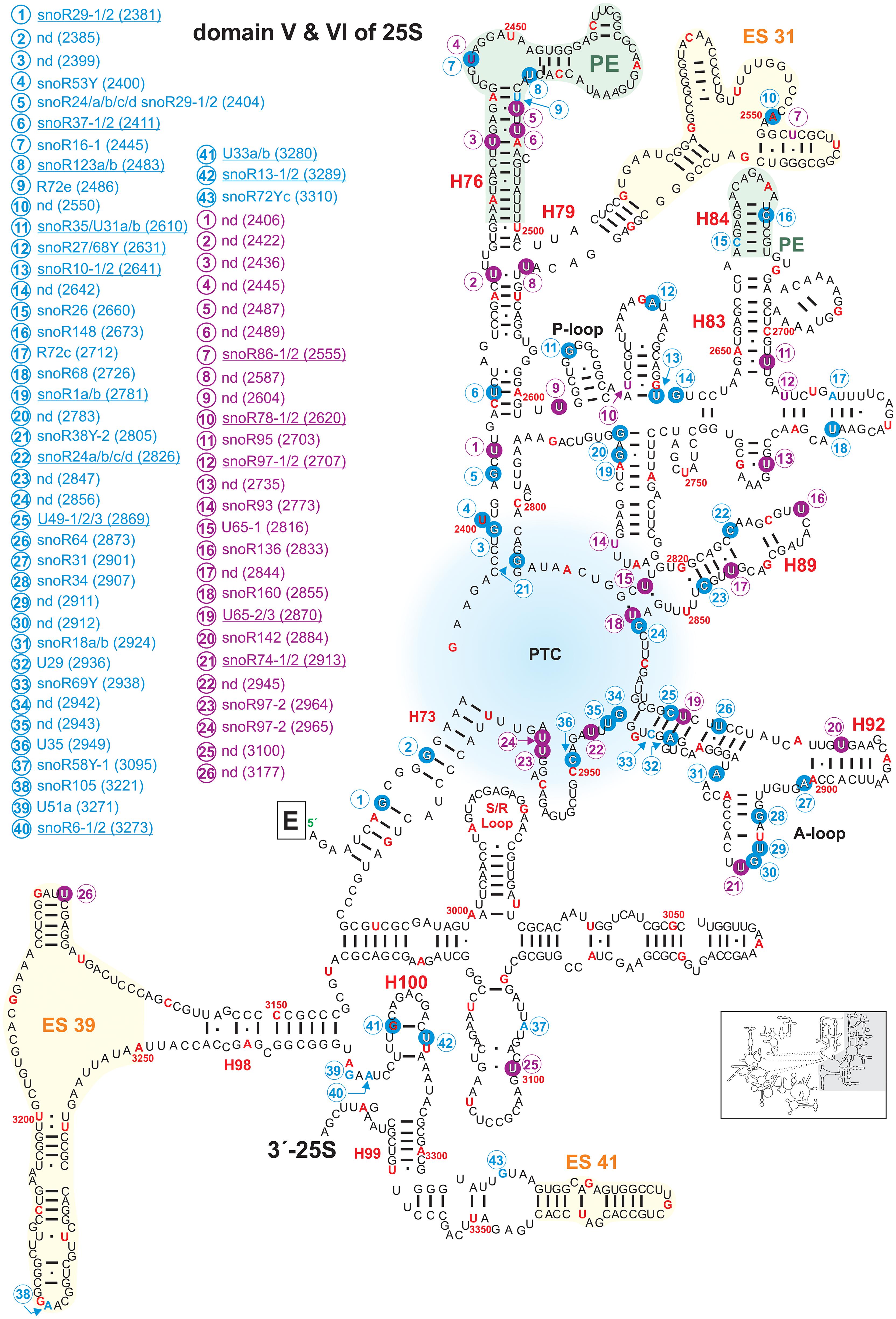
Figure 7. Secondary structure diagram of domain V and VI of 25S rRNA of A. thaliana. The image is shown according to the legend for Figure 3. Framed large letters indicate the position of the connections in subsequent images (for E see Figure 6). The region of the PTC is indicated in blue. The number in brackets correspond to the modified nucleotide position. A small illustration of the whole 60S rRNA secondary structure highlighting the according region is enclosed.
Domain I of the 25S rRNA contains five mapped and additionally nine predicted 2′-O-ribose-me sites (Figures 3, 4 and Supplementary Table 2), as well as two mapped and one predicted pseudouridylation site(s). The methylations can be found in helix 11, 15, and 24. Among others, helices 7, 18, 19, 20, and 24 surround the exit tunnel for the nascent polypeptide chain (Spahn et al., 2001). However, only in helix 24 two 2′-O-ribose-me sites and one pseudouridylation site were mapped (Figure 3). Accordingly, in human 28S rRNA, helix 24 carries a modification at site Am389 and Am391 (Sharma et al., 2017a), while no modification was found in domain I in yeast (Yang et al., 2016). In fact, signal recognition particles (SRPs) (Halic et al., 2004), which recognize specific sequences of nascent polypeptide chains from the translating ribosomes interact with the tip of helix 24 of the 25/28S rRNA (Beckmann et al., 2001). In turn one could conclude that human and plant ribosomes have evolved a similar mode of binding to such molecular mechanisms. More intriguingly, the presence or absence of modified nucleotides in this region could be used as a complex regulator for the interaction of such particles with the ribosomes of humans and plants. Furthermore, snoRNAs for helix 24 modification were identified for humans, but not for Arabidopsis (Supplementary Table 2). Hence, a stand-alone enzyme might be responsible for these modifications that could act in the cytoplasm of Arabidopsis.
Helix 11 of the 25S rRNA in Arabidopsis contains three positions with modifications (two 2′-O-ribose-me and one pseudouridylation site; Figure 3). However, this helix does not carry modifications in yeast or human (Piekna-Przybylska et al., 2007). Thus, it is highly likely that this helix has in plants or at least in Arabidopsis a special function within the 60S ribosomal subunits or even in the 80S ribosomes, which requires such modification.
The region annotated as expansion segment 7 (ES7) contains only one described modification site at Am660 which is targeted by the snoRNAs U18-1 and 2 (Figure 4 and Supplementary Table 2). The ES7 is known be localized at the ribosome surface and belongs to the largest expansion segments with the highest variability in eukaryotes (Ramos et al., 2016). It was found that proteins binding to ES7 were relevant for regulations upon environmental changes, 60S subunit biogenesis and transcription elongation (Ramos et al., 2016). In contrast, yeast and human ES7 of the 25/28S is substantially greater than in plants (Parker et al., 2018). Together with the fact that Arabidopsis ES7 carries a 2′-O-ribose-me it can be concluded that plants evolved a special way of regulating those important features during stress conditions as well as in other regulatory functions.
Domain II of Arabidopsis 25S rRNA carries 29 putative 2′-O-ribose-me sites of which 14 could be successfully mapped. For pseudouridylation, 16 sites were predicted of which 13 were mapped (Figure 5 and Supplementary Table 2). Domain II contains the GTPase center mainly composed of helix 43 and helix 44 (Figure 5), which is highly conserved in all ribosomes (Ryan and Draper, 1991). In Escherichia coli, this region including the ribosomal proteins L10 and L11 is involved in the regulation of GTP hydrolysis by the elongation factor G and TU (Egebjerg et al., 1990; Briones et al., 1998). The rRNA of the GTPase center in A. thaliana contains three mapped and two predicted modification sites, for which associated snoRNAs are assigned (Figure 5 and Supplementary Table 2). Interestingly the modifications seem to be unique for A. thaliana since this segment is not modified in human or yeast (Piekna-Przybylska et al., 2007).
In general, many of the mapped modification sites in domain II are localized in stem structures (Figure 3). The helices 27, 31, 32, 35, and 38 carry many modifications. In yeast and human, helix 38 is exceedingly pseudouridylated but does not contain any 2′-O-ribose-me modification (Piekna-Przybylska et al., 2007). In Arabidopsis, pseudouridylations and 2′-O-ribose-me modifications were predicted in this particular helix based on the detection of according snoRNAs. Moreover, three pseudouridylation sites and one 2′-O-ribose-me were experimentally confirmed in helix 38 of the rRNA of Arabidopsis. Nevertheless, for the pseudouridylation site at ψ1060 an according snoRNA could not be identified so far. Intriguingly, helix 38 is involved in the formation of the intersubunit bridge between the 60S and 40S subunit by interacting with S19p of the 40S particle, and it is contacting the A-site bound tRNA in yeast (Spahn et al., 2001). However, in comparison to the yeast and human ribosomes, it can be proposed that the modifications in this helix are involved in the structural stabilization of this important subunit-subunit interaction site in Arabidopsis (Karijolich et al., 2010; Sloan et al., 2017).
Further, helix 35 carries one mapped 2′-O-ribose-me site and two pseudouridylation sites at U892 and U899. For the latter two sites guiding snoRNAs were not discovered (Figure 4 and Supplementary Table 2), leading to the assumption that stand-alone enzymes may be responsible. In yeast, helix 35 carries two 2′-O-ribose-me sites and in humans one pseudouridylation site (Piekna-Przybylska et al., 2007). A second intersubunit bridge is formed by helix 34 with the 40S subunit. Thus, also in this case the modifications of helix 35 in plants are likely involved in the stabilization of the neighboring structural element.
In domain III, a high density of modifications is present in the region of helix 47, 50, 59a, and 60 (Figure 4). In yeast, it is assumed that Nop4 is binding to helices 47, 32, 26, 33 but also to helix 60 bringing domain II and III in proximity (Granneman et al., 2011). However, helix 47 in yeast carries no modifications, while Arabidopsis helix 47 is highly modified. It can be speculated that these modifications are required for proper processing of 25S precursors like 27SB or 27S-A2/27S-A3.
Domains IV (Figure 6) and V (Figure 7) of the 25S rRNA contain the highest degree of modifications. In both domains a total of 66 sites for 2′-O-ribose-me sites are annotated from which 53 could be successfully confirmed. However, for 13 sites no snoRNA could be identified (Figures 6, 7 and Supplementary Table 2). In contrast, 31 pseudouridylation sites were mapped, while seven sites could not be verified yet. For 13 of the mapped sites, the associated snoRNA is not known (Figures 6, 7 and Supplementary Table 2).
The secondary structure map of the core region of 25S rRNA of A. thaliana points to a high density of modifications surrounding the PTC (Figure 7), which parallels findings for other organisms (Decatur and Fournier, 2002). The PTC is required for the peptide bond formation and peptide release (Lilley, 2001; Polacek and Mankin, 2005; Torres de Farias et al., 2017). In yeast, defective rRNA modifications in this region lead to increased sensitivity to translational inhibitors or changes in translational fidelity (Baxter-Roshek et al., 2007). In Arabidopsis, especially the helices H73, H74, H75, H88, H89, H90, H91, H92, and H93 contain the highest density of modifications (Figure 7). Interestingly, Arabidopsis contains the highest number of modification sites (34) in these particular helices in contrast to yeast (16) and human (22). This leads to the conclusion that these modifications are of prime importance for the stability of the PTC structure. Nevertheless, Arabidopsis 60S subunit seems to be closer related to the human 60S regarding the high density of modifications. In human ribosomes, helix 74 is important for the accurate structure of the nascent polypeptide exit tunnel (NPET) (Wilson et al., 2020) and helix 93 is a contact site for hydroxylated uL2, which induces structural rearrangements in the PTC of the mature ribosomes (Yanshina et al., 2015). The same could hold true for plants as well. The tip of helix 89 interacts with the GTPase-associated center which might depend on the modifications (Figure 5; Sergiev et al., 2005; Baxter-Roshek et al., 2007) and the modifications in helix 92 were found to be necessary for the correct folding of helix 90–92 in yeast (Baxter-Roshek et al., 2007).
In domain IV, Helix 68, 69, and 71 (Figure 6), are involved in the inter-subunit bridge formation between the 40S and 60S (Spahn et al., 2001; Gigova et al., 2014). Helix 68 contains three mapped methylation and one pseudouridylation sites (Figure 6). One 2′-O-ribose-me site (Am 2210) is conserved in yeast (Am2220) and human (Am3703; Piekna-Przybylska et al., 2007; Supplementary Table 2). The yeast helix 68 contains two E-sites (exit sites), which most probably exist in A. thaliana as well (Xie et al., 2012).
Helix 69 is highly modified with two mapped pseudouridylation sites and one mapped 2′-O-ribose-me site in A. thaliana (Figure 6). This helix interacts with the tRNAs located in the A and P-site, respectively (Ge and Yu, 2013). Similarly, a cluster of modifications is localized in helix 69 in the yeast rRNA, and their deletions led to e.g., severe growth phenotypes and a lower translational rate (Liang et al., 2007). Helix 71 contains two mapped 2′-O-ribose-me sites at Cm2283 and Gm2278 (Figure 6 and Supplementary Table 2). The site Cm2283 was newly identified but the snoRNA targeting this region was not found (Azevedo-Favory et al., 2020). However, while the human rRNA is lacking this modification, it is conserved between yeast and plants (Piekna-Przybylska et al., 2007).
Interestingly, the enigmatic exceptionally large expansion segment 27 (ES27, Figure 6) was recently unveiled as essential for translational fidelity, in which it seems to regulate amino acid incorporation and by that prevents frameshift errors (Fujii et al., 2018). Furthermore, it was found that this very flexible region of the eukaryotic ribosomes serves as a scaffold for the conserved enzyme methionine amino peptidase (MetAP), which is required to remove co-translationally the first methionine from the nascent polypeptide chain (Fujii et al., 2018; Knorr et al., 2019). Just recently one pseudouridylation site (Ψ2028) without known snoRNA was found in ES27 of Arabidopsis (Sun et al., 2019).
The P-loop in helix 80 and the A-loop in helix 92 are direct pairing sites for A- and P-site tRNA (Kim and Green, 1999). While the P-loop contains one confirmed 2′-O-ribose-methylation site, the A-loop contains two mapped 2′-O-ribose-me sites with the site Gm2912 having a known snoRNA targeting this region. Moreover, the A-loop contains a pseudouridylation site (Figure 7).
Domain VI containing the 3′-end of 25S rRNA from nucleotide 2986 to 3375 contains the conserved sarcin/ricin loop (S/R-Loop; Figure 7). This loop is the site of attack of the two toxins α-sarcin, which is a ribonuclease produced by a fungus and ricin, which is an RNA N-glycosylase synthesized by plants (Endo et al., 1988; Macbeth and Wool, 1999). The attack inhibits proper binding of the elongation factors, and thus, translation is blocked (Szewczak and Moore, 1995). In human 60S subunits the S/L-Loop shows a high degree of modifications in comparison to plants or yeast (Figure 7; Piekna-Przybylska et al., 2007).
Domain VI harbors the lowest degree of modifications with two mapped 2′-O-ribose-me sites with associated snoRNAs and two mapped pseudouridylations sites in Arabidopsis. The two 2′-O-me sites are within helix 100, one pseudouridylation sites in H97 and one in H98 of ES39 (Figure 7). ES39 is exposed to the ribosome surface, the exact function remains elusive, however due to its presence in all eukaryotes it is obvious that eukaryotic ribosomes require this segment (Nygård et al., 2006). For the two pseudouridylation sites U3177 and U3100 a snoRNA is not known so far. Interestingly, the Arabidopsis U3100 is conserved in the human 28S rRNA (U4659), while yeast has not even one modification regarding this specific region (Piekna-Przybylska et al., 2007; Supplementary Table 2).
The A. thaliana genome encodes for two different 18S rRNA variants. While the 18S gene on chromosome 3 has a size of 1808 nt, the copies on chromosomes 2 and 4 contain 1804 nt. The secondary structure model of 18S in here refers to the gene in chromosomes 2 and 4, respectively (Figures 8, 9). The SSU binds the mRNA to decode the genetic information in the “decoding center‘’ (Schluenzen et al., 2000). For the 18S rRNA in total 79 sites are predicted to be 2′-O-ribose methylated, of which 44 are experimentally verified (Supplementary Table 3). Similarly, from 64 predicted pseudouridylation sites 28 were experimentally confirmed.
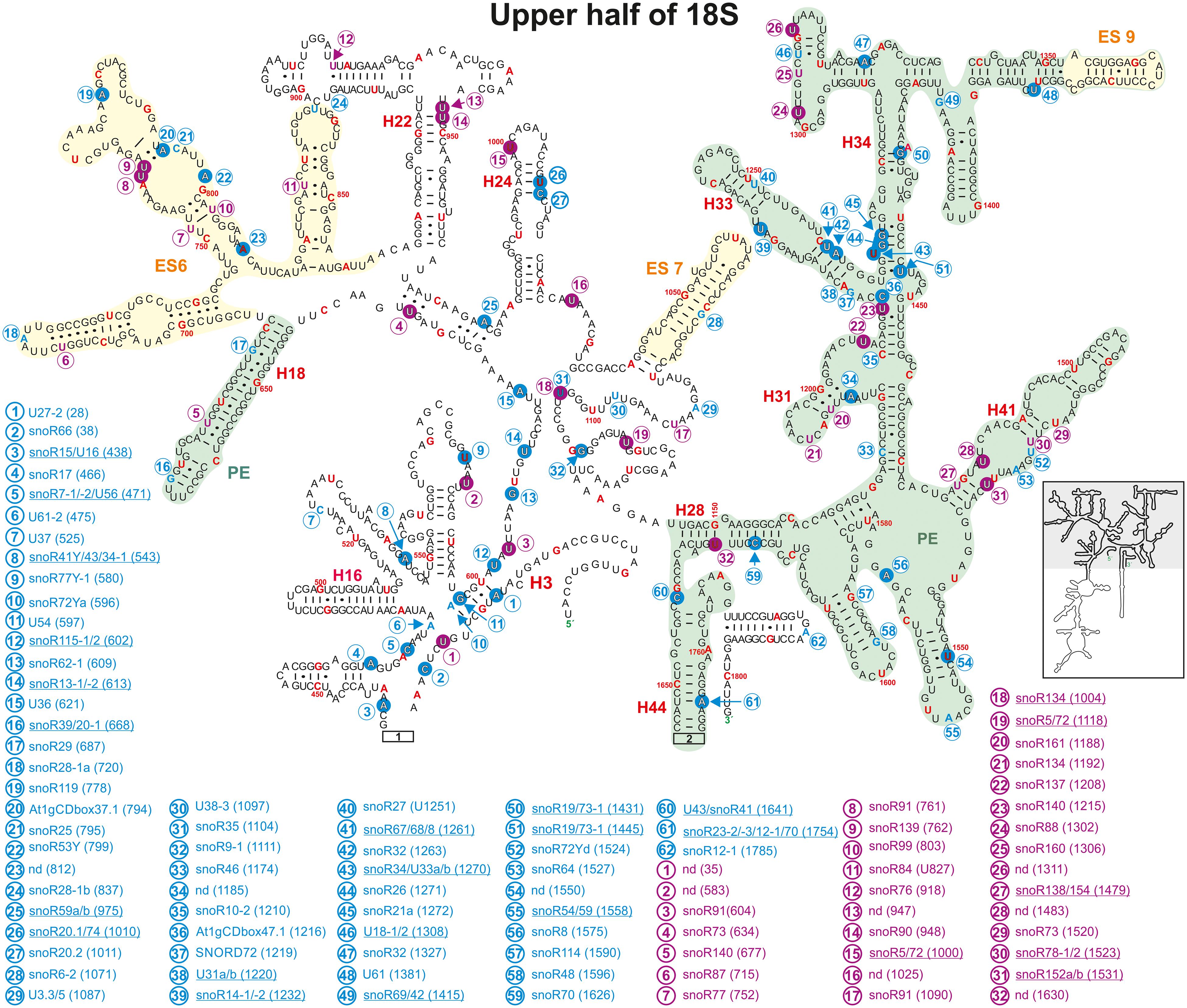
Figure 8. Secondary structure diagram of the 18S rRNA (At2g01010) of A thaliana according to The RNAcentral Consortium (2019). Depicted are the positions from 1 to 41, 436 to 1655, and 1750 to 1804. Predicted and verified positions for 2′-O-ribose-methylations and pseudouridylations, as well as of snoRNAs were obtained from Brown et al. (2001, 2003), Qu et al. (2001), Yoshihama et al. (2013), Sun et al. (2019), Streit et al. (2020). The coloring is according to Figure 3. Framed large numbers indicate the position of the connections in Figure 9. PE annotates a pivoting element and ES expansion elements previously identified (Paci and Fox, 2015). The number in brackets correspond to the modified nucleotide position. A small illustration of the whole 40S rRNA secondary structure highlighting the according region is enclosed.
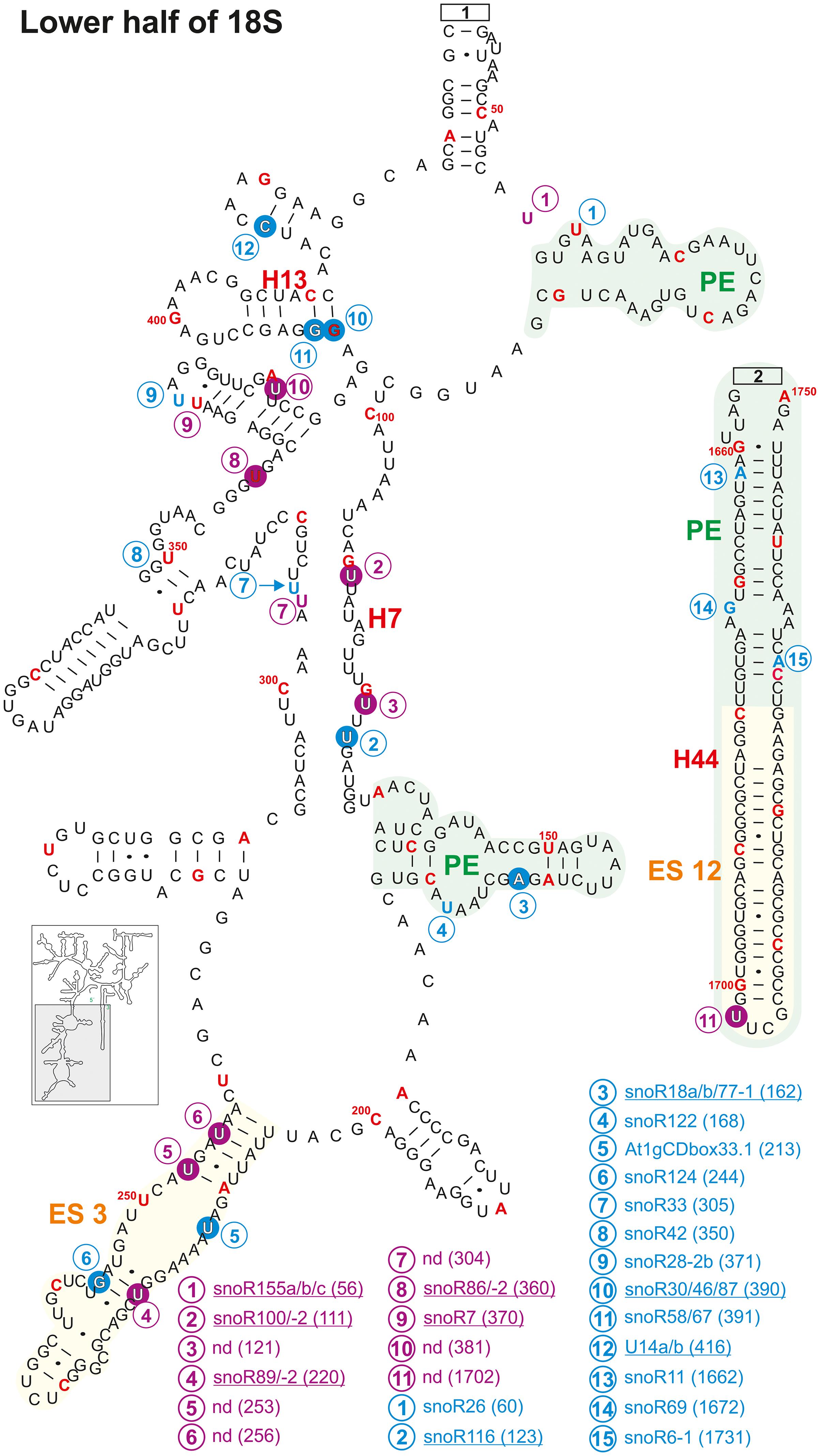
Figure 9. Continuation of the secondary structure diagram of the 18S rRNA (At2g01010) of A. thaliana according to The RNAcentral Consortium (2019). All informations are listed in the legend of Figure 8. The number in brackets correspond to the modified nucleotide position. A small illustration of the whole 40S rRNA secondary structure highlighting the according region is enclosed.
It is proposed that the decoding center within the SSU consists of the helices 18, 24, 31, 34, and 44 (Liang et al., 2009), which harbor many modifications in yeast. Helix 24 carries the same Ψ-modification in yeast, human and A. thaliana. The modifications in helix 18 vary between two 2′-O-ribose-me and one Ψ-site in human, one 2′-O-ribose-me site in yeast and just one Ψ-site in A. thaliana (Figure 8), while no methylation site exists in A. thaliana.
Helix 34 contains four mapped 2′-O-ribose-me sites in A. thaliana, while human and yeast contain three 2′-O-ribose-me sites. Both yeast and human 18S rRNA contain three pseudouridine sites in helix 31 with one site having the hypermodification m1acp3Ψ (N1-methyl-N3-aminocarboxypropyl-pseudouridine) at U1191 and U1248, respectively. A. thaliana contains two predicted Ψ-sites in this helix, while one of these sites U1192 is the yeast and human equivalent of m1acp3Ψ (Piekna-Przybylska et al., 2007). However, it needs to be analyzed whether the plant rRNA also carries hypermodifications as their vertebrate and yeast counterparts.
In helix 44 of the A. thaliana, two 2′-O-ribose-me are mapped and additional three are predicted, while only a single Ψ-site is annotated (Figures 8, 9). In contrast, human 18S rRNA contain two and yeast only one 2′-O-ribose-me sites in H44 (Piekna-Przybylska et al., 2007). Interestingly, A. thaliana harbors a novel and unique Ψ-site at U1702 in the helix 44 region of ES12 (Figure 9). In yeast, H44 just close to the 3′ end is one of the binding sites for the helicase Prp43, which is required for final maturation of 20S and 27S pre-rRNA, respectively (Bohnsack et al., 2009; Pertschy et al., 2009). For example, in the case of 20S maturation in yeast, Prp43 is assumed to be involved in unwinding of the pre-rRNA enabling endonuclease Nob1 for cleavage at site D (Figure 1; Bohnsack et al., 2009). It is likely, the modification at Am1754 could be necessary for binding of the Prp43 helicase. Yeast, of all things lacks this modification in H44 (Piekna-Przybylska et al., 2007).
Expansion segment 6 (ES6) is located at the surface of the small subunit and highly conserved in plants, vertebrates, and yeast (Alkemar and Nygård, 2003, 2006). As longest expansion segment it contains two mapped Ψ-sites, two mapped 2′-O-ribose-me sites and five predicted sites in A. thaliana (Figure 8). In contrast, the rRNA of human ES6 has six Ψ-sites and two 2′-O-ribose-me sites, while the same element in yeast contains two Ψ-sites and one 2′-O-ribose-me site (Piekna-Przybylska et al., 2007). Remarkably, Arabidopsis ES6 shows similarities to both human and yeast. The two Ψ-sites found in A. thaliana (Ψ761 and Ψ762) are conserved with the human sites (Ψ814 and Ψ815) and that one of the Arabidopsis 2′-O-ribose-me site (Am799) is conserved to the yeast site (Am796; Figure 8, Piekna-Przybylska et al., 2007; Supplementary Table 3). However, the function of this segment and thus, their modifications remain elusive. In vivo crosslinking in yeast suggested that snR30/U17 snoRNAs can bind to two conserved sites in the ES6 to permit proper 18S rRNA processing (Fayet-Lebaron et al., 2009). As snR30 and U17 could not be identified so far in A. thaliana, independent pathway for modification might have evolved for A. thaliana.
Ribosomal proteins and RBFs bind to specific regions within the rRNA. It could be shown that Enp1 (essential nuclear protein 1) binds to an AUU sequence in helix 33 in yeast, where it is required for pre-rRNA processing of 18S (Chen W. et al., 2003; Granneman et al., 2010). Helix 33 in Arabidopsis contains two 2′-O-ribose-methylations (Um1261 and Am1263) in the downstream adjacent region of the AUU site (Figure 8 and Supplementary Table 3). This seems to be unique for plants as yeast and human 18S rRNA lack these modifications (Piekna-Przybylska et al., 2007). Besides, a new modification site at Cm1219 in helix 33 was predicted based on the identification of the snoRNA SNORD72 (Streit et al., 2020).
Helix 41 of the 18S rRNA in A. thaliana contains two mapped Ψ-sites at U1483 and U1531 located within a region, which is a binding site for rpS5 in yeast (Figure 8; Supplementary Table 3; Granneman et al., 2010). However, human and yeast helix 41 of the 18S rRNA is not modified (Piekna-Przybylska et al., 2007).
Helix 28 of A. thaliana 18S rRNA contains a single mapped 2′-O-ribose-me site at Cm1626 targeted by snoR70 and a single novel Ψ-site at U1630 targeted from an unknown snoRNA (Figure 8). Human 18S rRNA contains a Ψ-site at the same position (U1692) in helix 28, whereas yeast lacks any modification is this region (Piekna-Przybylska et al., 2007; Supplementary Table 3). The helices 36 and 37 of A. thaliana contain each one mapped Ψ-site at U1302 (H36, snoR88) and U1311 (H37, unknown snoRNA; Figure 8). Although the human helix 37 contains at least the counterpart of U1311 of Arabidopsis at U1367, yeast helices are absent of these modification sites (Piekna-Przybylska et al., 2007; Supplementary Table 3).
Prediction and experimental verification suggest that the rRNA of A. thaliana is extensively decorated with different varieties of modifications, where pseudouridylations and 2′-O-ribose-methylations represent most of the modifications (Charette and Gray, 2000; Dimitrova et al., 2019). For Arabidopsis 18S rRNA almost 55% of all predicted 2′-O-ribose-methylation sites and 65% of all predicted pseudouridylation sites were successfully experimentally verified (Supplementary Table 3). For the 25S rRNA even 65% of all predicted 2′-O-ribose-methylation sites and almost 58% of predicted pseudouridylation sites were experimentally confirmed (Supplementary Table 2). In contrast, the 5.8S rRNA carries only a low number of modifications (Figure 3 and Supplementary Table 1). Two new modification sites at Ψ22, which is plant specific and Ψ78 (yeast Ψ73) were found in the past (Sun et al., 2019; Supplementary Table 1). In contrast, based on the antisense element of the snoRNA snoR4a/4b (Brown et al., 2001) the modification at the 3′-end of 5.8S rRNA (Gm155) was predicted, but could not be experimentally confirmed (Supplementary Table 1).
The absence of experimental confirmation of predicted sites can have three different reasons. (i) Although nowadays a huge repertoire of techniques is used for mapping of modification sites, a certain limitation in detection sensitivity still exists. An interesting technique would be the use of mung bean nuclease protection assay coupled to RP-HPLC (Yang et al., 2016). (ii) It is discussed those modifications of the rRNA can be tissue or development specific (Chen and Wu, 2009; Sloan et al., 2017; Streit et al., 2020). Thus, the absence of detection can be the result of the analysis of a specific type of ribosomal systems. (iii) It is known that snoRNAs are also involved in the folding of rRNA elements (Bertrand and Fournier, 2013). Hence, it cannot be excluded at stage that some of the modification sites predicted by the detection of snoRNAs might not exist, as the snoRNA is required for guiding a snoRNP involved in rRNA processing or folding.
The modifications in 5.8S varies between the three model species. While yeast 5.8S rRNA contains only one modification site in H7, human and plant 5.8S rRNAs carry two 2′-O-ribose-me and two pseudouridylations (Figure 3; Supplementary Table 1; Piekna-Przybylska et al., 2007). Modifications like pseudouridylations in rRNA are required for the stability of the RNA structure in the ribosomes and 2′-O-ribose-me are necessary for translational accuracy and efficiency (Wu et al., 2015; Erales et al., 2017). Consistent with this finding, the 5.8S rRNA plays a crucial role in translation elongation (translocation; Elela and Nazar, 1997), for which the modifications of 5.8S might be important. In turn, it appears that the 5.8S rRNA of human and plants share high similarities while yeast seems to have evolved a unique way for keeping the structural and translational balance.
Remarkably, human and A. thaliana rRNAs share many conserved sites, which are not present in yeast. This elucidates that plant and human rRNA, despite the different sizes, are closer related than plant to yeast rRNA. Moreover, a subset of modifications is clearly unique to Arabidopsis like modifications in the GTPase center (Figure 5), in ES27 (Figure 6) or ES3 in the 18S rRNA (Figure 9). All these regions are targets of a subset of RBFs and RPs. Hence, the plant specific modification pattern stands in relation to the observed plant specificities of the rRNA processing (Weis et al., 2015b; Sáez-Vásquez and Delseny, 2019; Palm et al., 2019) and the modifications might be required for stabilizing the binding of the rRNA to proteins.
Furthermore, alternative functions of snoRNAs were proposed for the plant system. It was suggested that snoRNAs may regulate the modification level of rRNAs and snRNAs under stress as found for drought stress (Zheng et al., 2019). Thus, snoRNAs might have additional functions in plants, which must be discovered.
In future, it will be important to identify the snoRNAs responsible for certain newly discovered modifications sites, and in turn to map rRNA modifications in ribosomes isolated form different tissues, from plants at different developmental stages and after various stress treatments to complete the picture of the Arabidopsis rRNA modification landscape. The latter would perhaps show whether the high number of predicted modifications sites argues for a high ribosome heterogeneity. This concept might even be valid for ribosomes within a single cell but required for the translation of different mRNA pools, which begs the analysis of the rRNA modification profile associated with different mRNAs. Moreover, it will be important to establish a complete profile of rRNA modifications of other plants to allow conclusions on globally conserved and species-specific modifications. The latter is of particular importance to transfer the knowledge based on model systems into agricultural applications.
DS and ES: conceptualization, data curation, writing – original draft preparation, and visualization. ES: supervision and funding acquisition. Both authors have read and agreed to the published version of the manuscript.
This research was funded by the Deutsche Forschungsgemein- schaft, SFB 902, B9 to ES.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to thank Sotirios Fragkostefanakis and Thiruvenkadam Shanmugam for reading the manuscript and for the helpful and critical discussion.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.684626/full#supplementary-material
Supplementary Table 1 | List of all mapped and predicted modification sites within 5.8S rRNA in Arabidopsis thaliana and corresponding human and yeast sites. Not determined (nd).
Supplementary Table 2 | List of all mapped or predicted modification sites within 25S rRNA in Arabidopsis thaliana and corresponding human and yeast sites. Not determined (nd).
Supplementary Table 3 | List of all predicted and mapped modification sites within 18S rRNA in Arabidopsis thaliana and corresponding human and yeast sites. Not determined (nd).
Alkemar, G., and Nygård, O. (2006). Probing the secondary structure of expansion segment ES6 in 18S ribosomal RNA. Biochemistry 45, 8067–8078. doi: 10.1021/bi052149z
Alkemar, G., and Nygård, O. D. D. (2003). A possible tertiary rRNA interaction between expansion segments ES3 and ES6 in eukaryotic 40S ribosomal subunits. RNA 9, 20–24. doi: 10.1261/rna.2108203
Armache, J. P., Jarasch, A., Anger, A. M., Villa, E., Becker, T., Bhushan, S., et al. (2010a). Cryo-EM structure and rRNA model of a translating eukaryotic 80S ribosome at 5.5-Å resolution. PNAS 107, 19748–19753. doi: 10.1073/pnas.1009999107
Armache, J. P., Jarasch, A., Anger, A. M., Villa, E., Becker, T., Bhushan, S., et al. (2010b). Localization of eukaryote-specific ribosomal proteins in a 5.5-Å cryo-EM map of the 80S eukaryotic ribosome. PNAS 107, 19754–19759. doi: 10.1073/pnas.1010005107
Azevedo-Favory, J., Gaspin, C., Ayadi, L., Montacié, C., Marchand, V., Jobet, E., et al. (2020). Mapping rRNA 2′-O-methylations and identification of C/D snoRNAs in Arabidopsis thaliana plants. RNA Biol. 1–18. doi: 10.1080/15476286.2020.1869892
Bachellerie, J. P., and Cavaillé, J. (1997). Guiding ribose methylation of rRNA. Trends Biochem. Sci. 22, 257–261. doi: 10.1016/s0968-0004(97)01057-8
Barneche, F., Gaspin, C., Guyot, R., and Echeverrìa, M. (2001). Identification of 66 box C/D snoRNAs in Arabidopsis thaliana: extensive gene duplications generated multiple isoforms predicting new ribosomal RNA 2′-O-methylation sites. J. Mol. Biol. 311, 57–73. doi: 10.1006/jmbi.2001.4851
Barneche, F., Steinmetz, F., and Echeverrìa, M. (2000). Fibrillarin genes encode both a conserved nucleolar protein and a novel small nucleolar RNA involved in ribosomal RNA methylation in arabidopsis thaliana. J. Biol. Chem. 275, 27212–27220. doi: 10.1016/s0021-9258(19)61499-7
Bassham, D. C., and MacIntosh, G. C. (2017). Degradation of cytosolic ribosomes by autophagy-related pathways. Plant Sci. 262, 169–174. doi: 10.1016/j.plantsci.2017.05.008
Baxter-Roshek, J. L., Petrov, A. N., and Dinman, J. D. (2007). Optimization of ribosome structure and function by rRNA base modification. PLoS One 2:e174. doi: 10.1371/journal.pone.0000174
Beckmann, R., Spahn, C. M., Eswar, N., Helmers, J., Penczek, P. A., Sali, A., et al. (2001). Architecture of the protein-conducting channel associated with the translating 80S ribosome. Cell 107, 361–372. doi: 10.1016/s0092-8674(01)00541-4
Bertrand, E., and Fournier, M. J. (2013). The snoRNPs and related machines: ancient devices that mediate maturation of rRNA and other RNAs. In Madame Curie Bioscience Database [Internet]. Austin, TX: Landes Bioscience.
Bhattacharya, D. P., Canzler, S., Kehr, S., Hertel, J., Grosse, I., and Stadler, P. F. (2016). Phylogenetic distribution of plant snoRNA families. BMC Genom. 17:969. doi: 10.1186/s12864-016-3301-2
Bohnsack, M. T., Martin, R., Granneman, S., Ruprecht, M., Schleiff, E., and Tollervey, D. (2009). Prp43 bound at different sites on the pre-rRNA performs distinct functions in ribosome synthesis. Mol. Cell 36, 583–592. doi: 10.1016/j.molcel.2009.09.039
Briones, E., Briones, C., Remacha, M., and Ballesta, J. P. (1998). The GTPase Center Protein L12 is required for correct ribosomal stalk assembly but Not for Saccharomyces cerevisiae Viability. J. Biol. Chem. 273, 31956–31961. doi: 10.1074/jbc.273.48.31956
Brown, J. W., Clark, G. P., Leader, D. J., Simpson, C. G., and Lowe, T. O. D. D. (2001). Multiple snoRNA gene clusters from Arabidopsis. RNA 7, 1817–1832.
Brown, J. W., Echeverria, M., and Qu, L. H. (2003). Plant snoRNAs: functional evolution and new modes of gene expression. Trends Plant Sci. 8, 42–49. doi: 10.1016/s1360-1385(02)00007-9
Brown, J. W., and Shaw, P. J. (1998). Small nucleolar RNAs and pre-rRNA processing in plants. Plant Cell 10, 649–657. doi: 10.2307/3870654
Charette, M., and Gray, M. W. (2000). Pseudouridine in RNA: what, where, how, and why. IUBMB Life 49, 341–351. doi: 10.1080/152165400410182
Chen, C. L., Liang, D., Zhou, H., Zhuo, M., Chen, Y. Q., and Qu, L. H. (2003). The high diversity of snoRNAs in plants: identification and comparative study of 120 snoRNA genes from Oryza sativa. Nucleic Acids Res. 31, 2601–2613. doi: 10.1093/nar/gkg373
Chen, H. M., and Wu, S. H. (2009). Mining small RNA sequencing data: a new approach to identify small nucleolar RNAs in Arabidopsis. Nucleic Acids Res. 37:e69. doi: 10.1093/nar/gkp225
Chen, W., Bucaria, J., Band, D. A., Sutton, A., and Sternglanz, R. (2003). Enp1, a yeast protein associated with U3 and U14 snoRNAs, is required for pre-rRNA processing and 40S subunit synthesis. Nucleic Acids Res. 31, 690–699. doi: 10.1093/nar/gkg145
Chow, C. S., Lamichhane, T. N., and Mahto, S. K. (2007). Expanding the nucleotide repertoire of the ribosome with post-transcriptional modifications. ACS Chem. Biol. 2, 610–619. doi: 10.1021/cb7001494
Decatur, W. A., and Fournier, M. J. (2002). rRNA modifications and ribosome function. Trends Biochem. Sci. 27, 344–351. doi: 10.1016/s0968-0004(02)02109-6
Demirci, H., Murphy, F., Belardinelli, R., Kelley, A. C., Ramakrishnan, V., Gregory, S. T., et al. (2010). Modification of 16S ribosomal RNA by the KsgA methyltransferase restructures the 30S subunit to optimize ribosome function. RNA 16, 2319–2324. doi: 10.1261/rna.2357210
Desaulniers, J. P., Chang, Y. C., Aduri, R., Abeysirigunawardena, S. C., SantaLucia, J. Jr., and Chow, C. S. (2008). Pseudouridines in rRNA helix 69 play a role in loop stacking interactions. Org. Biomol. Chem. 6, 3892–3895. doi: 10.1039/b812731j
Dieci, G., Preti, M., and Montanini, B. (2009). Eukaryotic snoRNAs: a paradigm for gene expression flexibility. Genomics 94, 83–88. doi: 10.1016/j.ygeno.2009.05.002
Dimitrova, D. G., Teysset, L., and Carré, C. (2019). RNA 2′-O-methylation (Nm) modification in human diseases. Genes 10:117. doi: 10.3390/genes10020117
Dragon, F., Lemay, V., and Trahan, C. (2006). snoRNAs: biogenesis, structure and function. Encycl. Life Sci. 2006, 1–7. doi: 10.1002/9780470015902.a0001377
Egebjerg, J., Douthwaite, S. R., Liljas, A., and Garrett, R. A. (1990). Characterization of the binding sites of protein L11 and the L10.(L12) 4 pentameric complex in the GTPase domain of 23 S ribosomal RNA from Escherichia coli. J. Mol. Biol. 213, 275–288. doi: 10.1016/s0022-2836(05)80190-1
Elela, S. A., and Nazar, R. N. (1997). Role of the 5.8 S rRNA in ribosome translocation. Nucleic Acids Res. 25, 1788–1794. doi: 10.1093/nar/25.9.1788
Endo, Y., Chan, Y. L., Lin, A., Tsurugi, K., and Wool, I. G. (1988). The cytotoxins alpha-sarcin and ricin retain their specificity when tested on a synthetic oligoribonucleotide (35-mer) that mimics a region of 28 S ribosomal ribonucleic acid. J. Biol. Chem. 263, 7917–7920. doi: 10.1016/s0021-9258(18)68418-2
Erales, J., Marchand, V., Panthu, B., Gillot, S., Belin, S., Ghayad, S. E., et al. (2017). Evidence for rRNA 2′-O-methylation plasticity: Control of intrinsic translational capabilities of human ribosomes. PNAS 114, 12934–12939. doi: 10.1073/pnas.1707674114
Falaleeva, M., Welden, J. R., Duncan, M. J., and Stamm, S. (2017). C/D-box snoRNAs form methylating and non-methylating ribonucleoprotein complexes: Old dogs show new tricks. Bioessays 39:1600264. doi: 10.1002/bies.201600264
Fayet-Lebaron, E., Atzorn, V., Henry, Y., and Kiss, T. (2009). 18S rRNA processing requires base pairings of snR30 H/ACA snoRNA to eukaryote-specific 18S sequences. EMBO J. 28, 1260–1270. doi: 10.1038/emboj.2009.79
Ferretti, M. B., and Karbstein, K. (2019). Does functional specialization of ribosomes really exist? RNA 25, 521–538. doi: 10.1261/rna.069823.118
Filippova, J. A., Semenov, D. V., Juravlev, E. S., Komissarov, A. B., Richter, V. A., and Stepanov, G. A. (2017). Modern approaches for identification of modified nucleotides in RNA. Biochem Mosc. 82, 1217–1233. doi: 10.1134/s0006297917110013
Fujii, K., Susanto, T. T., Saurabh, S., and Barna, M. (2018). Decoding the function of expansion segments in ribosomes. Mol. Cell 72, 1013–1020. doi: 10.1016/j.molcel.2018.11.023
Ge, J., and Yu, Y. T. (2013). RNA pseudouridylation: new insights into an old modification. Trends Biochem. Sci. 38, 210–218. doi: 10.1016/j.tibs.2013.01.002
Gigova, A., Duggimpudi, S., Pollex, T. I. M., Schaefer, M., and Koš, M. (2014). A cluster of methylations in the domain IV of 25S rRNA is required for ribosome stability. RNA 20, 1632–1644. doi: 10.1261/rna.043398.113
Graifer, D., Molotkov, M., Eremina, A., Ven’Yaminova, A., Repkova, M., and Karpova, G. (2005). The central part of the 5.8 S rRNA is differently arranged in programmed and free human ribosomes. Biochem. J. 387, 139–145. doi: 10.1042/bj20041450
Granneman, S., Petfalski, E., Swiatkowska, A., and Tollervey, D. (2010). Cracking pre-40S ribosomal subunit structure by systematic analyses of RNA–protein cross-linking. EMBO J. 29, 2026–2036. doi: 10.1038/emboj.2010.86
Granneman, S., Petfalski, E., and Tollervey, D. (2011). A cluster of ribosome synthesis factors regulate pre-rRNA folding and 5.8 S rRNA maturation by the Rat1 exonuclease. EMBO J. 30, 4006–4019. doi: 10.1038/emboj.2011.256
Gulay, S. P., Bista, S., Varshney, A., Kirmizialtin, S., Sanbonmatsu, K. Y., and Dinman, J. D. (2017). Tracking fluctuation hotspots on the yeast ribosome through the elongation cycle. Nucleic Acids Res. 45, 4958–4971. doi: 10.1093/nar/gkx112
Halic, M., Becker, T., Pool, M. R., Spahn, C. M., Grassucci, R. A., Frank, J., et al. (2004). Structure of the signal recognition particle interacting with the elongation-arrested ribosome. Nature 427, 808–814. doi: 10.1038/nature02342
Hang, R., Wang, Z., Deng, X., Liu, C., Yan, B., Yang, C., et al. (2018). Ribosomal RNA biogenesis and its response to chilling stress in Oryza sativa. Plant physiol. 177, 381–397. doi: 10.1104/pp.17.01714
Hellmann, E. (2020). How to make an extraordinary machine: SMALL ORGAN4 regulates ribosome biogenesis in plants. Plant Physiol. 184, 1627–1629. doi: 10.1104/pp.20.01456
Henras, A. K., Soudet, J., Gerus, M., Lebaron, S., Caizergues-Ferrer, M., Mougin, A., et al. (2008). The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell. Mol. Life Sci. 65, 2334–2359. doi: 10.1007/s00018-008-8027-0
Higa-Nakamine, S., Suzuki, T., Uechi, T., Chakraborty, A., Nakajima, Y., Nakamura, M., et al. (2012). Loss of ribosomal RNA modification causes developmental defects in zebrafish. Nucleic Acids Res. 40, 391–398. doi: 10.1093/nar/gkr700
Holley, C. L., Li, M. W., Scruggs, B. S., Matkovich, S. J., Ory, D. S., and Schaffer, J. E. (2015). Cytosolic accumulation of small nucleolar RNAs (snoRNAs) is dynamically regulated by NADPH oxidase. J. Biol. Chem. 290, 11741–11748. doi: 10.1074/jbc.m115.637413
Huang, C., Karijolich, J., and Yu, Y. T. (2016). Detection and quantification of RNA 2′-O-methylation and pseudouridylation. Methods 103, 68–76. doi: 10.1016/j.ymeth.2016.02.003
Ito, S., Akamatsu, Y., Noma, A., Kimura, S., Miyauchi, K., Ikeuchi, Y., et al. (2014). A single acetylation of 18 S rRNA is essential for biogenesis of the small ribosomal subunit in Saccharomyces cerevisiae. J. Biol. Chem. 289, 26201–26212. doi: 10.1074/jbc.m114.593996
Karijolich, J., Kantartzis, A., and Yu, Y. T. (2010). RNA modifications: a mechanism that modulates gene expression. Methods Mol. Biol. 629, 1–19. doi: 10.1007/978-1-60761-657-3_1
Kaul, S., Koo, H. L., Jenkins, J., Rizzo, M., Rooney, T., Tallon, L. J., et al. (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. doi: 10.1038/35048692
Khatter, H., Myasnikov, A. G., Natchiar, S. K., and Klaholz, B. P. (2015). Structure of the human 80S ribosome. Nature 520, 640–645.
Kim, D. F., and Green, R. (1999). Base-pairing between 23S rRNA and tRNA in the ribosomal A site. Mol. Cell. 4, 859–864. doi: 10.1016/s1097-2765(00)80395-0
Kim, S. H., Spensley, M., Choi, S. K., Calixto, C. P., Pendle, A. F., Koroleva, O., et al. (2010). Plant U13 orthologues and orphan snoRNAs identified by RNomics of RNA from Arabidopsis nucleoli. Nucleic Acids Res. 38, 3054–3067. doi: 10.1093/nar/gkp1241
Kiss, T. (2001). Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs. EMBO J. 20, 3617–3622. doi: 10.1093/emboj/20.14.3617
Kiss, T., Marshallsay, C., and Filipowicz, W. (1991). Alteration of the RNA polymerase specificity of U3 snRNA genes during evolution and in vitro. Cell 65, 517–526. doi: 10.1016/0092-8674(91)90469-f
Kiss-László, Z., Henry, Y., and Kiss, T. (1998). Sequence and structural elements of methylation guide snoRNAs essential for site-specific ribose methylation of pre-rRNA. EMBO J. 17, 797–807. doi: 10.1093/emboj/17.3.797
Klinge, S., and Woolford, J. L. (2019). Ribosome assembly coming into focus. Nat. Rev. Mol. Cell Biol. 20, 116–131. doi: 10.1038/s41580-018-0078-y
Knorr, A. G., Schmidt, C., Tesina, P., Berninghausen, O., Becker, T., Beatrix, B., et al. (2019). Ribosome–NatA architecture reveals that rRNA expansion segments coordinate N-terminal acetylation. Nat. Struct. Mol. Biol 26, 35–39. doi: 10.1038/s41594-018-0165-y
Kruszka, K., Barneche, F., Guyot, R., Ailhas, J., Meneau, I., Schiffer, S., et al. (2003). Plant dicistronic tRNA–snoRNA genes: a new mode of expression of the small nucleolar RNAs processed by RNase Z. EMBO J. 22, 621–632. doi: 10.1093/emboj/cdg040
Lafontaine, D., Vandenhaute, J., and Tollervey, D. (1995). The 18S rRNA dimethylase Dim1pis required for pre-ribosomal RNA process-ing in yeast. Genes Dev. 9, 2470–2481. doi: 10.1101/gad.9.20.2470
Lafontaine, D. L. (2015). Noncoding RNAs in eukaryotic ribosome biogenesis and function. Nat. Struct. Mol. Biol. 22, 11–19. doi: 10.1038/nsmb.2939
Lau, R. Y., Kennedy, T. D., and Lane, B. G. (1974). Wheat-embryo ribonucleates. III. Modified nucleotide constituents in each of the 5.8 S, 18S and 26S ribonucleates. Can. J. Biochem. 52, 1110–1123. doi: 10.1139/o74-155
Leader, D. J., Clark, G. P., Watters, J., Beven, A. F., Shaw, P. J., and Brown, J. W. (1997). Clusters of multiple different small nucleolar RNA genes in plants are expressed as and processed from polycistronic pre-snoRNAs. EMBO J. 16, 5742–5751. doi: 10.1093/emboj/16.18.5742
Leader, D. J., Sanders, J. F., Waugh, R., Shaw, P., and Brown, J. W. (1994). Molecular characterisation of plant U14 small nucleolar RNA genes: closely linked genes are transcribed as polycistronic U14 transcripts. Nucleic Acids Res. 22, 5196–5203. doi: 10.1093/nar/22.24.5196
Leidig, C., Thoms, M., Holdermann, I., Bradatsch, B., Berninghausen, O., Bange, G., et al. (2014). 60S ribosome biogenesis requires rotation of the 5S ribonucleoprotein particle. Nat. Commun. 5, 1–8.
Liang, X. H., Liu, Q., and Fournier, M. J. (2007). rRNA modifications in an intersubunit bridge of the ribosome strongly affect both ribosome biogenesis and activity. Mol. Cell. 28, 965–977. doi: 10.1016/j.molcel.2007.10.012
Liang, X. H., Liu, Q., and Fournier, M. J. (2009). Loss of rRNA modifications in the decoding center of the ribosome impairs translation and strongly delays pre-rRNA processing. RNA 15, 1716–1728. doi: 10.1261/rna.1724409
Lilley, D. M. (2001). The ribosome functions as a ribozyme. Chembiochem. 2, 31–35. doi: 10.1002/1439-7633(20010105)2:1<31::aid-cbic31>3.0.co;2-p
Lindsay, M. A., Griffiths-Jones, S., Lui, L., and Lowe, T. (2013). Small nucleolar RNAs and RNA-guided post-transcriptional modification. Essays Biochem. 54, 53–77. doi: 10.1042/bse0540053
Liu, T. T., Zhu, D., Chen, W., Deng, W., He, H., He, G., et al. (2013). A global identification and analysis of small nucleolar RNAs and possible intermediate-sized non-coding RNAs in Oryza sativa. Mol. Plant. 6, 830–846. doi: 10.1093/mp/sss087
Macbeth, M. R., and Wool, I. G. (1999). The phenotype of mutations of G2655 in the sarcin/ricin domain of 23 S ribosomal RNA. J. Mol. Bio. 285, 965–975. doi: 10.1006/jmbi.1998.2388
Marker, C., Zemann, A., Terhörst, T., Kiefmann, M., Kastenmayer, J. P., Green, P., et al. (2002). Experimental RNomics: identification of 140 candidates for small non-messenger RNAs in the plant Arabidopsis thaliana. Curr. Biol. 12, 2002–2013.
Matera, A. G., Terns, R. M., and Terns, M. P. (2007). Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat. Rev. Mol. Cell Biol. 8, 209–220. doi: 10.1038/nrm2124
Missbach, S., Weis, B. L., Martin, R., Simm, S., Bohnsack, M. T., and Schleiff, E. (2013). 40S ribosome biogenesis co-factors are essential for gametophyte and embryo development. PloS One 8:e54084. doi: 10.1371/journal.pone.0054084
Natchiar, S. K., Myasnikov, A. G., Kratzat, H., Hazemann, I., and Klaholz, B. P. (2017). Visualization of chemical modifications in the human 80S ribosome structure. Nature 551, 472–477. doi: 10.1038/nature24482
Nazar, R. N., Sitz, T. O., and Busch, H. (1975). Tissue specific differences in the 2′-O-methylation of eukaryotic 5.8 S ribosomal RNA. FEBS Lett. 59, 83–87. doi: 10.1016/0014-5793(75)80346-2
Nazar, R. N., Sitz, T. O., and Somers, K. D. (1980). Cytoplasmic methylation of mature 5.8 S ribosomal RNA. J. Mol. Biol. 142, 117–121. doi: 10.1016/0022-2836(80)90209-0
Newman, D. R., Kuhn, J. F., Shanab, G. M., and Maxwell, E. S. (2000). Box C/D snoRNA-associated proteins: two pairs of evolutionarily ancient proteins and possible links to replication and transcription. RNA 6, 861–879. doi: 10.1017/s1355838200992446
Nygård, O., Alkemar, G., and Larsson, S. L. (2006). Analysis of the secondary structure of expansion segment 39 in ribosomes from fungi, plants and mammals. J. Mol. Biol. 357, 904–916. doi: 10.1016/j.jmb.2006.01.043
Paci, M., and Fox, G. E. (2015). Major centers of motion in the large ribosomal RNAs. Nucleic Acids Res. 43, 4640–4649. doi: 10.1093/nar/gkv289
Palm, D., Simm, S., Darm, K., Weis, B. L., Ruprecht, M., Schleiff, E., et al. (2016). Proteome distribution between nucleoplasm and nucleolus and its relation to ribosome biogenesis in Arabidopsis thaliana. RNA Biol. 13, 441–454. doi: 10.1080/15476286.2016.1154252
Palm, D., Streit, D., Shanmugam, T., Weis, B. L., Ruprecht, M., Simm, S., et al. (2019). Plant-specific ribosome biogenesis factors in Arabidopsis thaliana with essential function in rRNA processing. Nucleic Acids Res. 47, 1880–1895. doi: 10.1093/nar/gky1261
Parker, M. S., Balasubramaniam, A., Sallee, F. R., and Parker, S. L. (2018). The expansion segments of 28S Ribosomal RNA extensively match human messenger RNAs. Front. Genet. 9:66. doi: 10.3389/fgene.2018.00066
Pertschy, B., Schneider, C., Gnädig, M., Schäfer, T., Tollervey, D., and Hurt, E. (2009). RNA helicase Prp43 and its co-factor Pfa1 promote 20 to 18 S rRNA processing catalyzed by the endonuclease Nob1. J. Biol. Chem. 284, 35079–35091. doi: 10.1074/jbc.m109.040774
Piekna-Przybylska, D., Decatur, W. A., and Fournier, M. J. (2007). The 3D rRNA modification maps database: with interactive tools for ribosome analysis. Nucleic Acids Res. 36, D178–D183.
Polacek, N., and Mankin, A. S. (2005). The ribosomal peptidyl transferase center: structure, function, evolution, inhibition. Crit. Rev. Biochem. Mol. Biol. 40, 285–311. doi: 10.1080/10409230500326334
Polikanov, Y. S., Melnikov, S. V., Söll, D., and Steitz, T. A. (2015). Structural insights into the role of rRNA modifications in protein synthesis and ribosome assembly. Nat. Struct. Mol. Biol. 22, 342–344. doi: 10.1038/nsmb.2992
Qu, G., Kruszka, K., Plewka, P., Yang, S. Y., Chiou, T. J., Jarmolowski, A., et al. (2015). Promoter-based identification of novel non-coding RNAs reveals the presence of dicistronic snoRNA-miRNA genes in Arabidopsis thaliana. BMC Genom. 16:1009. doi: 10.1186/s12864-015-2221-x
Qu, L. H., Meng, Q., Zhou, H., and Chen, Y. Q. (2001). Identification of 10 novel snoRNA gene clusters from Arabidopsis thaliana. Nucleic Acids Res. 29, 1623–1630. doi: 10.1093/nar/29.7.1623
Ramesh, M., and Woolford, J. L. (2016). Eukaryote-specific rRNA expansion segments function in ribosome biogenesis. RNA 22, 1153–1162. doi: 10.1261/rna.056705.116
Ramos, L. M. G., Smeekens, J. M., Kovacs, N. A., Bowman, J. C., Wartell, R. M., Wu, R., et al. (2016). Yeast rRNA expansion segments: folding and function. J. Mol. Biol. 428, 4048–4059. doi: 10.1016/j.jmb.2016.08.008
Reddy, R., Henning, D., and Busch, H. (1979). Nucleotide sequence of nucleolar U3B RNA. J. Biol. Chem. 254, 11097–11105. doi: 10.1016/s0021-9258(19)86635-8
Reddy, R., Sitz, T. O., Ro-Choi, T. S., and Busch, H. (1974). Two-dimensional polyacrylamide gel electrophoresis separation of low molecular weight nuclear RNA. Biochem. Biophys. Res. Commun. 56, 1017–1022. doi: 10.1016/s0006-291x(74)80290-1
Rodor, J., Jobet, E., Bizarro, J., Vignols, F., Carles, C., Suzuki, T., et al. (2011). AtNUFIP, an essential protein for plant development, reveals the impact of snoRNA gene organisation on the assembly of snoRNPs and rRNA methylation in Arabidopsis thaliana. Plant J. 65, 807–819. doi: 10.1111/j.1365-313x.2010.04468.x
Ryan, P. C., and Draper, D. E. (1991). Detection of a key tertiary interaction in the highly conserved GTPase center of large subunit ribosomal RNA. PNAS 88, 6308–6312. doi: 10.1073/pnas.88.14.6308
Sáez-Vásquez, J., and Delseny, M. (2019). Ribosome biogenesis in plants: from functional 45S ribosomal DNA organization to ribosome assembly factors. Plant Cell. 31, 1945–1967. doi: 10.1105/tpc.18.00874
Samarsky, D. A., Fournier, M. J., Singer, R. H., and Bertrand, E. (1998). The snoRNA box C/D motif directs nucleolar targeting and also couples snoRNA synthesis and localization. EMBO J. 17, 3747–3757. doi: 10.1093/emboj/17.13.3747
Schluenzen, F., Tocilj, A., Zarivach, R., Harms, J., Gluehmann, M., Janell, D., et al. (2000). Structure of functionally activated small ribosomal subunit at 3.3 Å resolution. Cell 102, 615–623. doi: 10.1016/s0092-8674(00)00084-2
Sergiev, P. V., Lesnyak, D. V., Burakovsky, D. E., Kiparisov, S. V., Leonov, A. A., Bogdanov, A. A., et al. (2005). Alteration in location of a conserved GTPase-associated center of the ribosome induced by mutagenesis influences the structure of peptidyltransferase center and activity of elongation factor G. J. Biol. Chem. 280, 31882–31889. doi: 10.1074/jbc.m505670200
Sharma, S., Marchand, V., Motorin, Y., and Lafontaine, D. L. (2017a). Identification of sites of 2′-O-methylation vulnerability in human ribosomal RNAs by systematic mapping. Sci. Rep. 7, 1–15.
Sharma, S., Yang, J., van Nues, R., Watzinger, P., Kötter, P., Lafontaine, D. L., et al. (2017b). Specialized box C/D snoRNPs act as antisense guides to target RNA base acetylation. PLoS Genet. 13:e1006804. doi: 10.1371/journal.pgen.1006804
Sloan, K. E., Warda, A. S., Sharma, S., Entian, K. D., Lafontaine, D. L., and Bohnsack, M. T. (2017). Tuning the ribosome: The influence of rRNA modification on eukaryotic ribosome biogenesis and function. RNA Biol. 14, 1138–1152. doi: 10.1080/15476286.2016.1259781
Spahn, C. M., Beckmann, R., Eswar, N., Penczek, P. A., Sali, A., Blobel, G., et al. (2001). Structure of the 80S ribosome from Saccharomyces cerevisiae—tRNA-ribosome and subunit-subunit interactions. Cell 107, 373–386. doi: 10.1016/s0092-8674(01)00539-6
Streit, D., Shanmugam, T., Garbelyanski, A., Simm, S., and Schleiff, E. (2020). The existence and localization of nuclear snoRNAs in Arabidopsis thaliana revisited. Plants 9:1016. doi: 10.3390/plants9081016
Sun, L., Xu, Y., Bai, S., Bai, X., Zhu, H., Dong, H., et al. (2019). Transcriptome-wide analysis of pseudouridylation of mRNA and non-coding RNAs in Arabidopsis. J. Exp. Bot. 70, 5089–5600. doi: 10.1093/jxb/erz273
Szewczak, A. A., and Moore, P. B. (1995). The sarcin/ricin loop, a modular RNA. J. Mol. Biol. 247, 81–98. doi: 10.1006/jmbi.1994.0124
Taylor, D. J., Devkota, B., Huang, A. D., Topf, M., Narayanan, E., Sali, A., et al. (2009). Comprehensive molecular structure of the eukaryotic ribosome. Structure 17, 1591–1604. doi: 10.1016/j.str.2009.09.015
Terns, M. P., and Terns, R. M. (2002). Small nucleolar RNAs: versatile trans-acting molecules of ancient evolutionary origin. Gene Expr. 10, 17–39.
The RNAcentral Consortium (2019). RNAcentral: a hub of information for non-coding RNA sequences. Nucleic Acids Res. 47, D221–D229.
Tollervey, D., and Kiss, T. (1997). Function and synthesis of small nucleolar RNAs. Curr. Opin. Cell Biol. 9, 337–342. doi: 10.1016/s0955-0674(97)80005-1
Torres de Farias, S., Gaudêncio Rêgo, T., and José, M. V. (2017). Peptidyl transferase center and the emergence of the translation system. Life 7:21. doi: 10.3390/life7020021
Tsang, C. K., Bertram, P. G., Ai, W., Drenan, R., and Zheng, X. S. (2003). Chromatin-mediated regulation of nucleolar structure and RNA Pol I localization by TOR. EMBO J. 22, 6045–6056. doi: 10.1093/emboj/cdg578
Turkina, M. V., Årstrand, H. K., and Vener, A. V. (2011). Differential phosphorylation of ribosomal proteins in Arabidopsis thaliana plants during day and night. PloS One 6:e29307. doi: 10.1371/journal.pone.0029307
Venema, J., and Tollervey, D. (1999). Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet. 33, 261–311.
Watkins, N. J., and Bohnsack, M. T. (2012). The box C/D and H/ACA snoRNPs: key players in the modification, processing and the dynamic folding of ribosomal RNA. Wiley Interdiscip. Rev. RNA. 3, 397–414. doi: 10.1002/wrna.117
Weinstein, L. B., and Steitz, J. A. (1999). Guided tours: from precursor snoRNA to functional snoRNP. Curr. Opin. Cell Biol. 11, 378–384. doi: 10.1016/S0955-0674(99)80053-2
Weis, B. L., Kovacevic, J., Missbach, S., and Schleiff, E. (2015b). Plant-specific features of ribosome biogenesis. Trends Plant Sci. 20, 729–740. doi: 10.1016/j.tplants.2015.07.003
Weis, B. L., Palm, D., Missbach, S., Bohnsack, M. T., and Schleiff, E. (2015a). atBRX1-1 and atBRX1-2 are involved in an alternative rRNA processing pathway in Arabidopsis thaliana. RNA 21, 415–425. doi: 10.1261/rna.047563.114
Wilson, D. M., Li, Y., LaPeruta, A., Gamalinda, M., Gao, N., and Woolford, J. L. (2020). Structural insights into assembly of the ribosomal nascent polypeptide exit tunnel. Nat. commun. 11, 1–15.
Woolford, J. L. Jr., and Baserga, S. J. (2013). Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics 195, 643–681. doi: 10.1534/genetics.113.153197
Wu, G., Huang, C., and Yu, Y. T. (2015). Pseudouridine in mRNA: incorporation, detection, and recoding. Meth. Enzymol. 560, 187–217.
Xie, Q., Wang, Y., Lin, J., Qin, Y., Wang, Y., and Bu, W. (2012). Potential key bases of ribosomal RNA to kingdom-specific spectra of antibiotic susceptibility and the possible archaeal origin of eukaryotes. PloS One 7:e29468. doi: 10.1371/journal.pone.0029468
Yang, J., Sharma, S., Watzinger, P., Hartmann, J. D., Kötter, P., and Entian, K. D. (2016). Mapping of complete set of ribose and base modifications of yeast rRNA by RP-HPLC and mung bean nuclease assay. PloS One 11:e0168873. doi: 10.1371/journal.pone.0168873
Yanshina, D. D., Bulygin, K. N., Malygin, A. A., and Karpova, G. G. (2015). Hydroxylated histidine of human ribosomal protein uL2 is involved in maintaining the local structure of 28S rRNA in the ribosomal peptidyl transferase center. FEBS J. 282, 1554–1566. doi: 10.1111/febs.13241
Yoshihama, M., Nakao, A., and Kenmochi, N. (2013). snOPY: a small nucleolar RNA orthological gene database. BMC Res. Notes. 6:426. doi: 10.1186/1756-0500-6-426
Zhao, X., and Yu, Y. T. (2004). Detection and quantitation of RNA base modifications. RNA 10, 996–1002. doi: 10.1261/rna.7110804
Zhao, Y., Yu, Y., Zhai, J., Ramachandran, V., Dinh, T. T., Meyers, B. C., et al. (2012). The Arabidopsis nucleotidyl transferase HESO1 uridylates unmethylated small RNAs to trigger their degradation. Curr. Biol. 22, 689–694. doi: 10.1016/j.cub.2012.02.051
Zheng, J., Zeng, E., Du, Y., He, C., Hu, Y., Jiao, Z., et al. (2019). Temporal small RNA expression profiling under drought reveals a potential regulatory role of small nucleolar RNAs in the drought responses of Maize. Plant Genome 12, 1–15.
Zhou, H., Meng, Q., and Qu, L. (2000). Identification and structural analysis of a novel snoRNA gene cluster from Arabidopsis thaliana. Sci. China C. Life Sci. 43, 449–453.
Keywords: snoRNA, A. thaliana, rRNA, ribosome, 2cpsdummy′-O-ribose-methylation, pseudouridylation
Citation: Streit D and Schleiff E (2021) The Arabidopsis 2′-O-Ribose-Methylation and Pseudouridylation Landscape of rRNA in Comparison to Human and Yeast. Front. Plant Sci. 12:684626. doi: 10.3389/fpls.2021.684626
Received: 23 March 2021; Accepted: 16 June 2021;
Published: 26 July 2021.
Edited by:
Roman A. Volkov, Chernivtsi National University, UkraineReviewed by:
Jonas Barandun, Umeå University, SwedenCopyright © 2021 Streit and Schleiff. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Enrico Schleiff, c2NobGVpZmZAYmlvLnVuaS1mcmFua2Z1cnQuZGU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.