- 1State Key Laboratory of Cotton Biology, Institute of Cotton Research of Chinese Academy of Agricultural Sciences, Anyang, China
- 2Forest Department, Forestry College, Hebei Agricultural University, Baoding, China
- 3College of Life Sciences, Xinyang Normal University, Xinyang, China
The membrane attack complex/perforin (MACPF) domain-containing proteins are involved in the various developmental processes and in responding to diverse abiotic stress. The function and regulatory network of the MACPF genes are rarely reported in Gossypium spp. We study the detailed identification and partial functional verification of the members of the MACPF family. Totally, 100 putative MACPF proteins containing complete MACPF domain were identified from the four cotton species. They were classified into three phylogenetic groups and underwent multifold pressure indicating that selection produced new functional differentiation. Cotton MACPF gene family members expanded mainly through the whole-genome duplication (WGD)/segmental followed by the dispersed. Expression and cis-acting elements analysis revealed that MACPFs play a role in resistance to abiotic stresses, and some selected GhMACPFs were able to respond to the PEG and cold stresses. Co-expression analysis showed that GhMACPFs might interact with valine-glutamine (VQ), WRKY, and Apetala 2 (AP2)/ethylene responsive factor (ERF) domain-containing genes under cold stress. In addition, silencing endogenous GhMACPF26 in cotton by the virus-induced gene silencing (VIGS) method indicated that GhMACPF26 negatively regulates cold tolerance. Our data provided a comprehensive phylogenetic evolutionary view of Gossypium MACPFs. The MACPFs may work together with multiple transcriptional factors and play roles in acclimation to abiotic stress, especially cold stress in cotton.
Introduction
The membrane attack complex/perforin (MACPF) proteins are pore-forming proteins across the cellular membrane in plants and other organisms. They have a signature motif, Y/S-G-G/S-H-X7-G-G, and play important roles in mammalian immunity and bacterial pathogenesis (Hadders et al., 2007; Wade and Tweten, 2015; Johnson et al., 2017; Moreno-Hagelsieb et al., 2017; Ni and Gilbert, 2017; Yu et al., 2020). In Arabidopsis, the necrotic spotted lesion 1 (NSL1) protein, containing a putative MACPF domain, was involved in the negative regulation of programmed cell death (PCD) and defense response (Noutoshi et al., 2006). Moreover, loss function of the MACPF domain could trigger cell death through activating salicylic acid (SA) signaling in Arabidopsis (Morita-Yamamuro et al., 2005; Fukunaga et al., 2017).
Environmental stress is a major challenge in agricultural production, affecting plant photosynthesis efficiency, cell membrane homeostasis, and plant growth and development (Hasegawa et al., 2000; Odukoya et al., 2019). Plants have developed sophisticated physiological and biochemical adaptation that enables them to survive through the regulation of multiple hormones including salicylic acid (SA), ethylene, and abscisic acid (ABA) (Zhu, 2002; Vlot et al., 2009; Hasanuzzaman et al., 2017; Emamverdian et al., 2020). SA exerts key effects in disease resistance by modulating host cell death and defense gene expression (Vlot et al., 2009). An SA-responsive protein is known as constitutively activated cell death 1 (CAD1) has been found to have a conservative MACPF domain. The CAD 1 mutant altered endophytic phyllosphere microbiota and displays leaf tissue damage in Arabidopsis. CAD1 is important for plant defense and likely negatively regulates plant immunity (Morita-Yamamuro et al., 2005; Tsutsui et al., 2006; Chen T. et al., 2020).
In recent years, different transcriptomic data showed that the MACPF genes were involved in growth, development, and response to abiotic stresses in sorghum, rice, and maize (Yu et al., 2020). However, the evolution and function of the MACPF genes in Gossypium spp. were still unknown. The underlying interaction among the MACPFs and other genes was also unclear. The weighted gene co-expression network analysis (WGCNA) was a useful method to identify the potential network through the transcriptomic analysis, which had been used to study development and abiotic stress in many core traits (Chen X. et al., 2020; Cheng et al., 2021).
As a pioneer crop in the barren land, cotton has a natural stress resistance through the complex genome structure (Li et al., 2015). Understanding the potential function of the MACPFs in cotton will be helpful to the cotton breeding and functional analysis. In this study, we performed a genome-wide identification of the MACPF family members in tetraploid species Gossypium hirsutum (AD1) and Gossypium barbadense (AD2) and their ancestor diploid Gossypium arboreum (A2) and Gossypium raimondii (D5) (Paterson et al., 2012; Du et al., 2018; Hu et al., 2019). The evolutionary relationships of the MACPFs were investigated and the putative interactions of the MACPFs with other genes were analyzed by the WGCNA. In total, 38 GhMACPFs, 33 GbMACPFs, 14 GaMACPFs, and 15 GrMACPFs were identified from the four Gossypium spp. genomes. Phylogenetic, conserved structural motif, the whole-genome duplication (WGD), non-synonymous substitution (Ka), and synonymous substitution (Ks) showed that the MACPFs were subjected to functional gene selection. Co-expression indicated that GhMACPFs played an important role in responding to abiotic stress. and Apetala 2 (AP2)/ethylene responsive factor (ERF), valine-glutamine (VQ), and WRKY transcription factors (TFs) were the co-expression genes of GhMACPFs in the regulation of cold stress response. In addition, silencing of GhMACPF26 in cotton enhanced the tolerance to cold stress. This study will lay a solid foundation for further exploration of the functions of GhMACPFs in cotton.
Materials and Methods
Identification and Molecular Characterization of the MACPF Domain Family in Multiple Plants
A total of 15 genome databases, which contained Arabidopsis thaliana (L.) Heynh., Oryza sativa Linn., Solanum lycopersicum Linn., Vitis vinifera Linn., Ananas comosus (L.) Merr., Amborella trichopoda Baill., Carica papaya Linn., Theobroma cacao Linn., G. hirsutum, G. barbadense, Gossypium arboreum, G. raimondii, Brassica napus Linn., Brassica oleracea Linnaeus, and Brassica rapa Linn., were obtained from the CottonGen1 (Yu et al., 2014), the Joint Genome Institute (JGI) database2 (Goodstein et al., 2012), and the National Center for Biotechnology Information (NCBI) database3 (Sayers et al., 2010), respectively. Four AtMACPF genes, including AtMACPF01 (AT1G14780), AtMACPF02 (AT1G28380), AtMACPF03 (AT1G29690), and AtMACPF04 (AT4G24290), were used as query sequences to blast the selected proteins database via the protein-protein BLAST (BLASTP) method (E-value = 1 × 10–3). The hidden Markov model (HMM) profile of the MACPF domain (Pfam01823) was downloaded from the Pfam web4 (El-Gebali et al., 2019) and the Hmmsearch (version 3.2.1) software (Eddy, 2011) was used to scan the 14 genome proteins. In addition, the web of the SMART database5 (Ivica et al., 2009) and the NCBI conserved domain database6 (Marchlerbauer et al., 2015) were used to confirm the conservative MACPF domain. Finally, we identified the molecular characterization through the Softberry web7 and the ExPASy website8.
Phylogenetic and Collinearity Analysis of the MACPF Genes
The Multiple Alignment using Fast Fourier Transform (MAFFT) (version 7.4.0.7) (Katoh and Standley, 2014), the Gblocks (version 0.91), and the PhyML9 (Guindon et al., 2005) were used to align 184 MACPF domain-containing proteins, analyze the conserved site sequences, and construct the maximum likelihood (ML) tree, respectively. R/ggtree (Yu et al., 2016) was applied to color the final tree file.
The MCScanX software (Wang et al., 2012) was used to analyze gene collinearity with default parameter among in Gossypium spp., which contained G. hirsutum and G. barbadense; G. arboreum, G. raimondii, and G. hirsutum; and G. arboreum, G. raimondii, and G. barbadense. The duplication event was implemented through the commands of the MCScanX/duplicate_gene_classifier code. To determine selection pressure, the BLASTP (Mark et al., 2008), the ParaAT (Zhang et al., 2012), and the KaKs_Calculator (version 2.0) (Wang et al., 2010) were used to calculate the Ka and Ks values.
Gene Location, Cis Element, and Conserved Motif Analysis
The MACPF domain-containing gene locations were obtained from the general feature format (GTF), which contained gene id, source, feature, start, end, and score via the python script. The intron/exon structure of the MACPF genes was extracted from the GTF. The conserved motifs in the MACPF proteins were obtained by submitting the protein sequences to the MEME web10 (Bailey et al., 2009) with the default of 30 amino acids in length. Gene structure view of the TBtools software (Chen C. et al., 2020) was used to integrate the motif and gene structure. Finally, the cis-elements of the promoter (2 kb) in the Gossypium MACPF genes were analyzed through the PlantCARE web11 (Lescot et al., 2002).
Expression Profiles and Construction of Co-expression Network
The transcripts of the MACPF genes in different tissues and multiabiotic stresses were collected from previous research (PRJNA490626) (Hu et al., 2019). Transcriptomic analysis was performed according to the previously described protocols (Chen P. et al., 2020). Differentially expressed genes (DEGs) were analyzed by using the R/edgeR package (Robinson et al., 2010). The R/WGCNA package (Langfelder and Horvath, 2008) was used to identify the co-expression module and construct the co-expression networks. In addition, hub genes were identified via the cytoHubba/Cytoscape software (Shannon et al., 2003; Chin et al., 2014) and the final network was showed by the Cytoscape.
Plant Cultivation, RNA Isolation, and Quantitative Real-Time-PCR Analysis
The Texas Marker-1 (TM-1) cotton seeds were provided by the Institute of Cotton Research of the Chinese Academy of Agricultural Sciences and planted in artificial growth conditions under a photoperiod of 16 h light/8 h darkness at 28/22°C. In order to verify the results of the transcriptome, the cotton plants grown at the three-leaf stage were subjected to stress treatment. For the drought treatment, the seedlings of TM-1 were irrigated with 400 mM polyethylene glycol 600 (PEG 600) for 0, 1, 6, 12, and 24 h. For the cold stress, the seedlings were irrigated with continuous cold stress (4°C) in a plant incubator/illumination incubator for 0, 2, 4, 6, 8, 12, and 24 h. Leaf samples were collected from five uniform plants, then quickly frozen in liquid nitrogen and stored at −80°C. All the experiments were repeated three times.
Total RNA was isolated by using the RNAprep Pure Plant Kit (Polysaccharides and Polyphenolics-rich, DP441) (TIANGEN, Beijing, China). The Mir-XTM miRNA First-Strand Synthesis Kit (Takara Biotechnology Corporation Ltd., Dalian, China), the SYBR Green PCR Supermix Kit (Bio-Rad Laboratories), and the 7500 Real-Time PCR System (Applied Biosystems) were used to synthesize complementary DNA (cDNA) and analyze gene expression levels, respectively. GhUBQ7 (NCBI accession: DQ116441) (Tu et al., 2007) was used as an endogenous control. All the primers designed by Primer 6 were listed in Supplementary Table 1.
Virus-Induced Gene Silencing and Cold Stress
To silence GhMACPF26 in cotton, a 300-bp fragment of the gene was PCR amplified and cloned into a pTRV2 vector; the primers used for implication were listed in Supplementary Table 1. A specific fragment of GhMACPF26 was amplified from the upland cotton accession of the TM-1 cDNA library. The constructs were transformed into Agrobacterium tumefaciens strain LBA4404. The virus-induced gene silencing (VIGS) method and the growth condition of the TM-1 cotton seedlings were described previously by Gu et al. (2018). The VIGS and control plants were treated at 4°C for 24 h and the leaves were sampled for biochemical experimental analysis. Malondialdehyde (MDA) was measured as the manual of the MDA assay kit [thiobarbituric acid (TBA) method]. Each experiment was performed in triplicate.
Results
Membrane Attack Complex/Perforin Domain Proteins in the Allotetraploid and Diploid Cotton
All the protein sequences annotated as the MACPF in four Gossypium and other 11 species were performed. A total of 184 MACPF domain-containing sequences were retained for this study (Supplementary Table 2). The number of the MACPFs is approximately similar to the genome size of the evolution biodiversity (Qiao et al., 2019). Similarly, the number of the MACPF genes in each allotetraploid is nearly equal to the sum of those in the two diploids. With the ML method and the conservative MACPF domain, the phylogenetic tree was constructed and classified into three branches (clades I–III) (Figure 1). However, the majority of the MACPF genes in group III (35 MACPFs) was specific and belonging to the Malvaceae, indicating that those genes likely experienced a sequence divergence event during polyploidization.
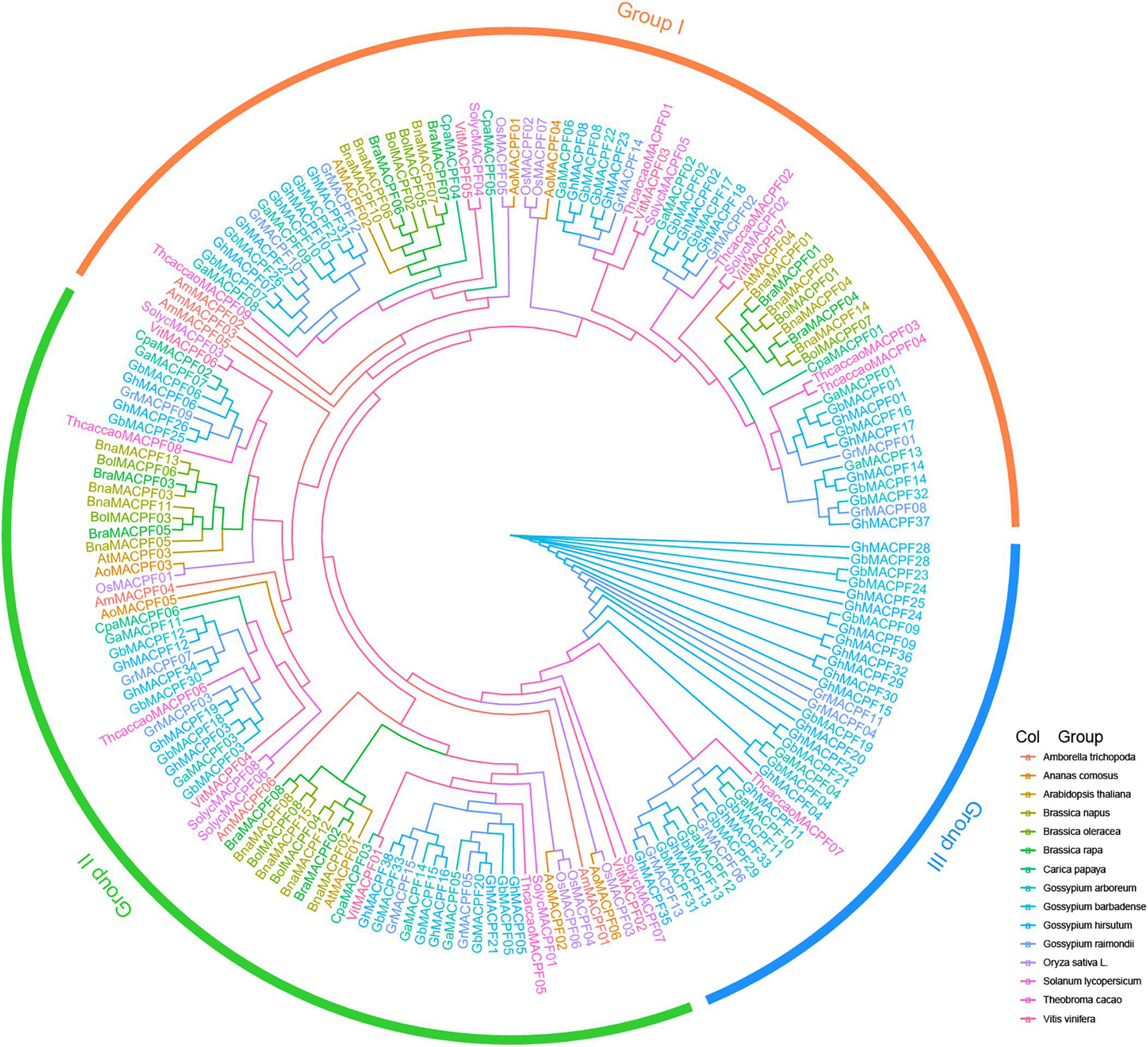
Figure 1. A phylogenetic tree of the membrane attack complex/perforin (MACPF) gene family. All the sequences of the MACPF domain gene in four Gossypium spp. and 11 other plants were carried out by using the maximum likelihood (ML) method in PhyML and members in the same species belong to the same color.
Basic information of the MACPF genes in Gossypium was listed in Supplementary Table 3 including genomic length, transcript length, GC count (%), exon number, molecular weight (MW), charge, and isoelectric point (PI). The genomic length of the MACPF genes was ranged from 790 (GhMACPF22) to 9,893 bp (GrMACPF14), the coding sequence (CDS) length of those range from 348 (GhMACPF22 and GbMACPF21) to 1,872 bp (GbMACPF19), and the average GC content of the transcript was 43.13. Moreover, the exon numbers varied from 2 (GhMACPF15, GbMACPF21, and GhMACPF22) to 9 (GbMACPF33), and more than half of the MACPFs containing seven exons. The MW value ranged from 12.884 (GbMACPF21) to 69.677 kDa (GbMACPF19) and the PI values varied from 6.228 (GhMACPF28) to 9.889 (GbMACPF15) (Supplementary Table 3).
Conserved Characteristics Analysis in the Diploid and Allotetraploid Cotton
The 2,000-bp upstream from the initiation codon in the diploid and allotetraploid cotton was used for the cis-element analysis of the MACPF genes. Through the PlantCARE web analysis, 15 cis elements were identified from the allotetraploid cotton, 14 cis elements in G. arboreum, and 13 cis elements in G. raimondii, respectively. Among those, 13 cis elements were common. Five kinds of hormone-response cis-elements, including ABA-responsive element (ABRE), auxin-responsive element (TGA-element), gibberellin (GARE-motif, P-box, and TATC-box), MeJA-responsive element (CGTCA-motif, and TGACG-motif), and SA-responsive element (SARE and TCA-element) were identified in each gene. Totally, four regulatory elements, which contained the low temperature responsive element (LTR), MYB-binding site (MBS), MYB-binding site I (MBSI), and TC-rich repeats, were found to have a certain function in the cold, drought, and defense response in plants (Supplementary Figure 1). Furthermore, the RY elements (seed-specific regulation) and circadian elements (cis-acting regulatory element involved in circadian control) were also found in the promoters, indicated that the MACPF gene might be played a role in plant development. The diversity analysis of elements determined the response of the MACPFs in Gossypium spp. to endogenous hormones, abiotic stresses, and development.
Combining the phylogenetic, motif composition, and gene structure analysis in Figure 2, the MACPF domain-contained proteins were divided into three groups. In this study, with the 30 distinct conserved motifs set through the MEME program, motif 1 was shared in GhMACPFs and GbMACPFs with the conserved “(F/Y)GTH(F/Y)-X6-GG” structure (Supplementary Figure 2). Moreover, motif 20 and motif 28 were specific in group I and group II and motif dispersion is variable in group III (Figure 2). Multiple motif distribution suggested that those genes were conserved and might have similar functions in the allotetraploid cotton. Moreover, the gene structure and intronic phase were exhibited similar to the MEME dispersion in the diploid, especially in group I and group II (Supplementary Figure 3). As described, the MACPF members were more likely to have gene selection and expansion between the diploid and allotetraploid cotton.
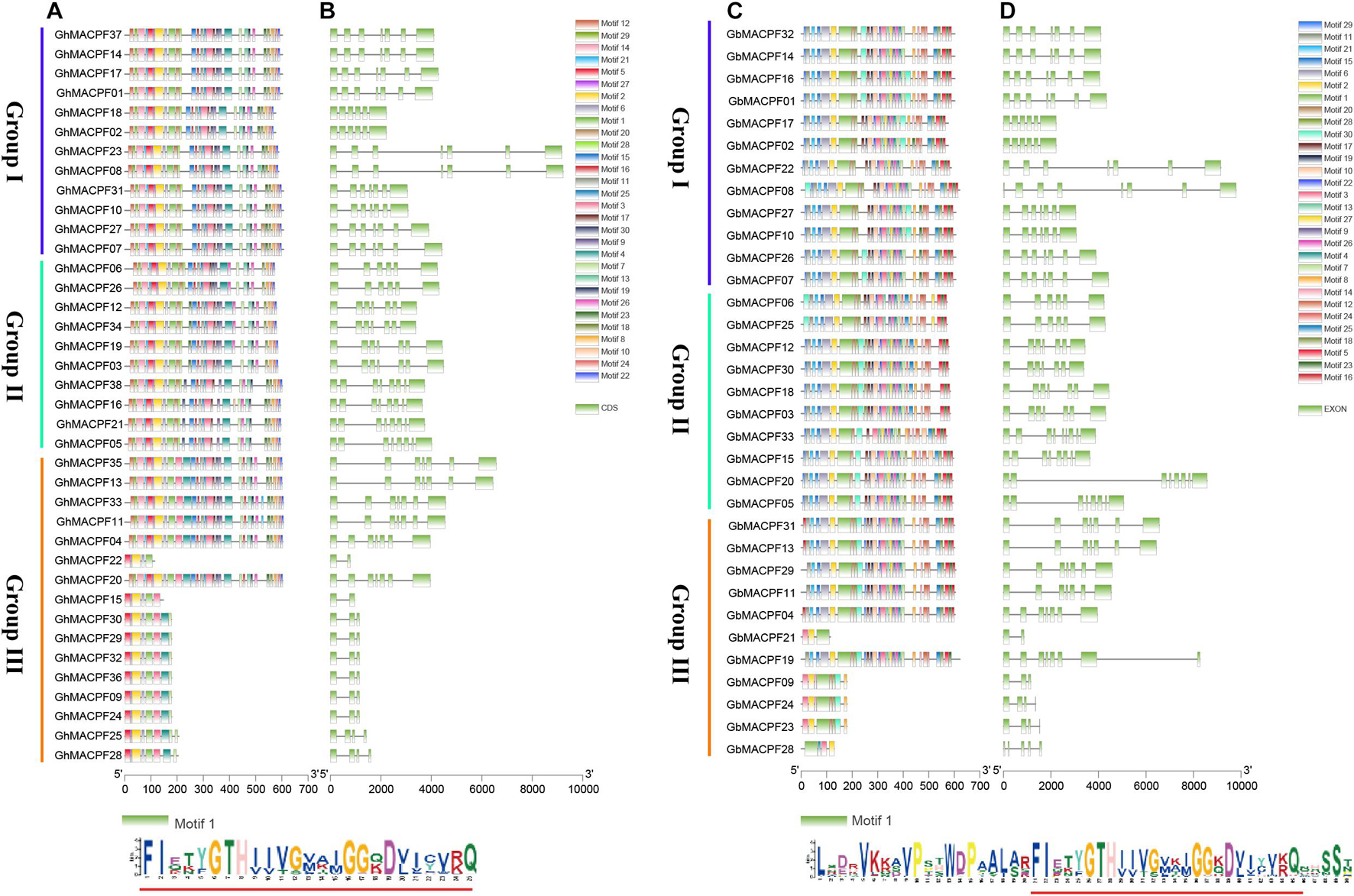
Figure 2. Motif and gene structure of the MACPF member in the allotetraploid cotton. (A,C) Are the converted motif and (B,D) are the gene structure of the MACPF member.
Orthologous Genes Analyses Between the Diploid and Allotetraploid Cotton
The chromosomal distribution of the MACPF genes in diploid cotton showed that 15 GrMACPFs were mapped to nine chromosomes of the G. raimondii. 14 GaMACPFs were also mapped to nine chromosomes of the G. arboreum (Supplementary Figure 4). A total of 38 GhMACPFs and 33 GbMACPFs were mapped to 19 chromosomes in the allotetraploid cotton, respectively. There was no MACPF gene located on chromosomes GhA04, GhA07, GhA08, GhA11, GhD03, GhD08, GhD11, GbA04, GbA07, GbA08, GbA11, GbD03, GbD08, and GbD11 (Figure 3). Numerous genes were varied from 1 to 4 on the mapped chromosomes (Figure 3, Supplementary Figure 4, and Supplementary Table 3). Duplication event analysis in the MACPF family indicated that there were 12 WGD/segmental events in Gh_At (75%), Gh_Dt (54.5%), Gb_At (80%), and Gb_Dt (66.7%); 9 WGD/segmental events (64.3%) in G. arboreum; and 11 events (73.3%) in G. raimondii, respectively. However, the singleton and proximal event were few in the MACPF family. It was suggested that the WGD/segmental was the main reason for the expansion of the MACPF gene family in Gossypium spp. (Supplementary Table 4).
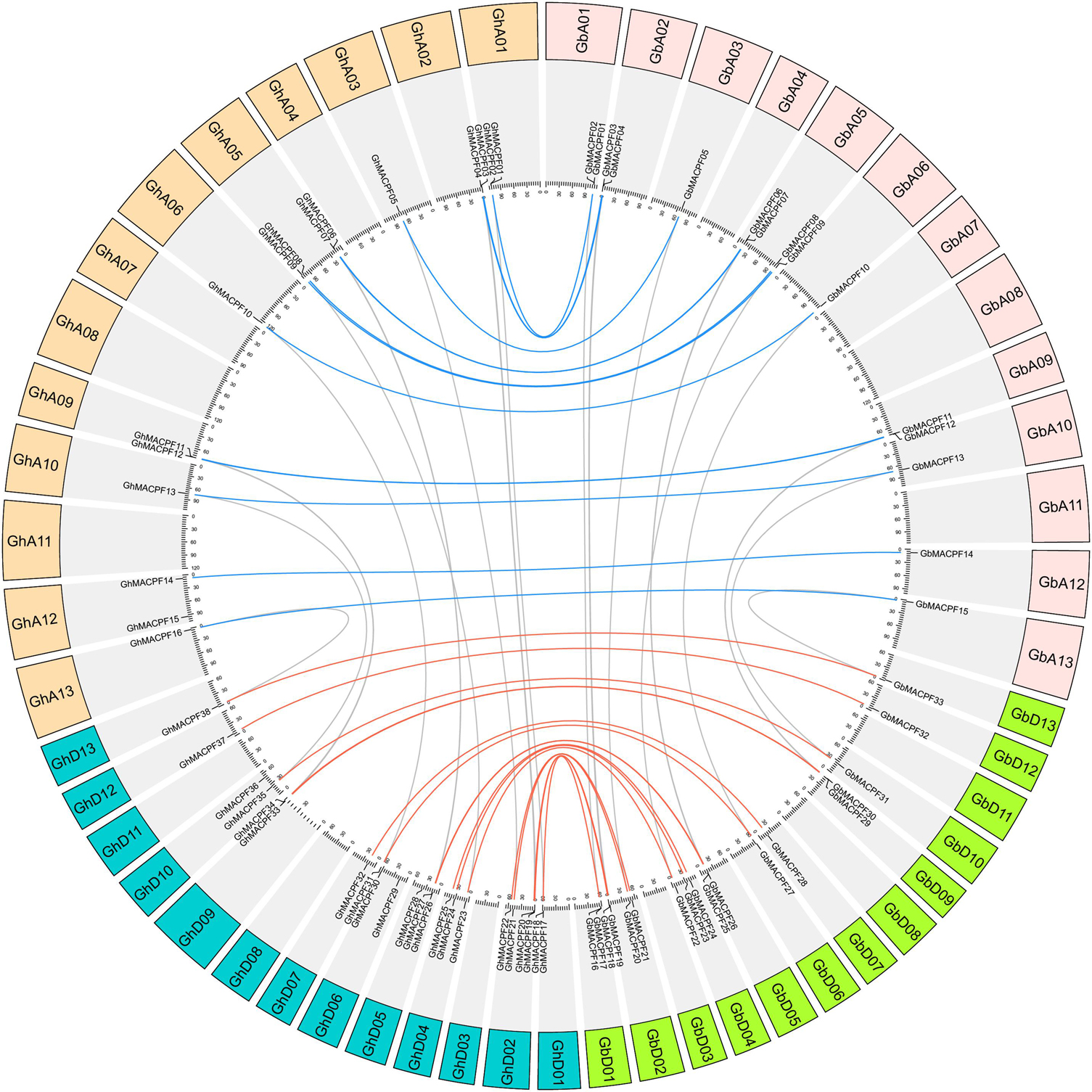
Figure 3. Duplication event among the MACPF members between G. hirsutum and G. barbadense. The Blue line indicates the homologous genes between the Gh_At and Gb_At subgenomes and the red line indicates the Gh_Dt and Gb_Dt subgenomes.
Orthologous analysis showed 31 MACPF gene pairs in the two allotetraploid pieces of cotton including 14 gene pairs in Gh_At and Gb_At subgenomes and 17 pairs in Gh_Dt and Gb_Dt subgenomes. Meanwhile, 12 paralogous gene pairs were found in G. hirsutum and 14 paralogous gene pairs were found in G. barbadense, respectively (Figure 3 and Supplementary Table 5). Subsequently, orthologous genes were also identified between the two allotetraploid pieces of cotton and two diploids. A total of 26 GhMACPFs and 26 GbMACPFs were orthologous genes in the two diploid cottons of which 13 gene pairs showed in G. arboreum, while 13 gene pairs showed in G. raimondii (Supplementary Figure 4 and Supplementary Table 5). Furthermore, the Ka/Ks ratios for the multi-MACPF pairs in Gossypium spp. were determined in Gb_Dt-Gr, Gh_Dt-Gr, Gb_At-Ga, Gh_At-Ga, Ga-Gr, Gb_At-Gb-Dt, Gb_At-Gh-At, Gh_At-Gh-Dt, and Gb_Dt-Gh-Dt. Among the 121 gene pairs, the Ka/Ks ratio of GbMACPF31-GhMACPF35, GbMACPF12-GaMACPF11, and GhMACPF12-GaMACPF11 were >1, the other 109 gene pairs were range from 0 to 0.94, indicating that these genes have undergone purifying selection (Supplementary Table 6).
Expression Patterns and Co-expression of the Homologous MACPF Genes in the Allotetraploid Cotton
To understand the potential function of GhMACPFs and GbMACPFs, we analyzed the expression patterns of the MACPFs in various tissues via the published RNA-seq data (Supplementary Figure 5 and Supplementary Table 7). Further, 27 GhMACPFs and 23 GbMACPFs were exhibited distinctive expression patterns in the different tissues. Most of the MACPFs were highly expressed in the anther, bract, filament, petal, pistil, and sepal (Supplementary Figures 5A,B). GhMACPFs and GbMACPFs were also highly expressed in the 5 DPA (day post anthesis), 10 DPA, and 15 DPA of the ovule, and low expression in fiber development stages (Supplementary Figures 5C,D). Different expression profiles of the MACPFs suggested that they widely participated in the various development stages of cotton. Furthermore, the expression pattern of the MACPFs in the allotetraploid cotton was regulated by cold, heat, salt, and drought stresses (Figure 4). Most of the selected GhMACPFs were predominantly expressed at 24 h under cold treatment, especially GhMACPF04, GhMACPF13, GhMACPF26, and GhMACPF35. We further carried out qRT-PCR to confirm the RNA-seq data after the PEG and cold treatments (Figure 5). The results suggested that GhMACPFs were involved in response to the cold and PEG stresses in cotton.
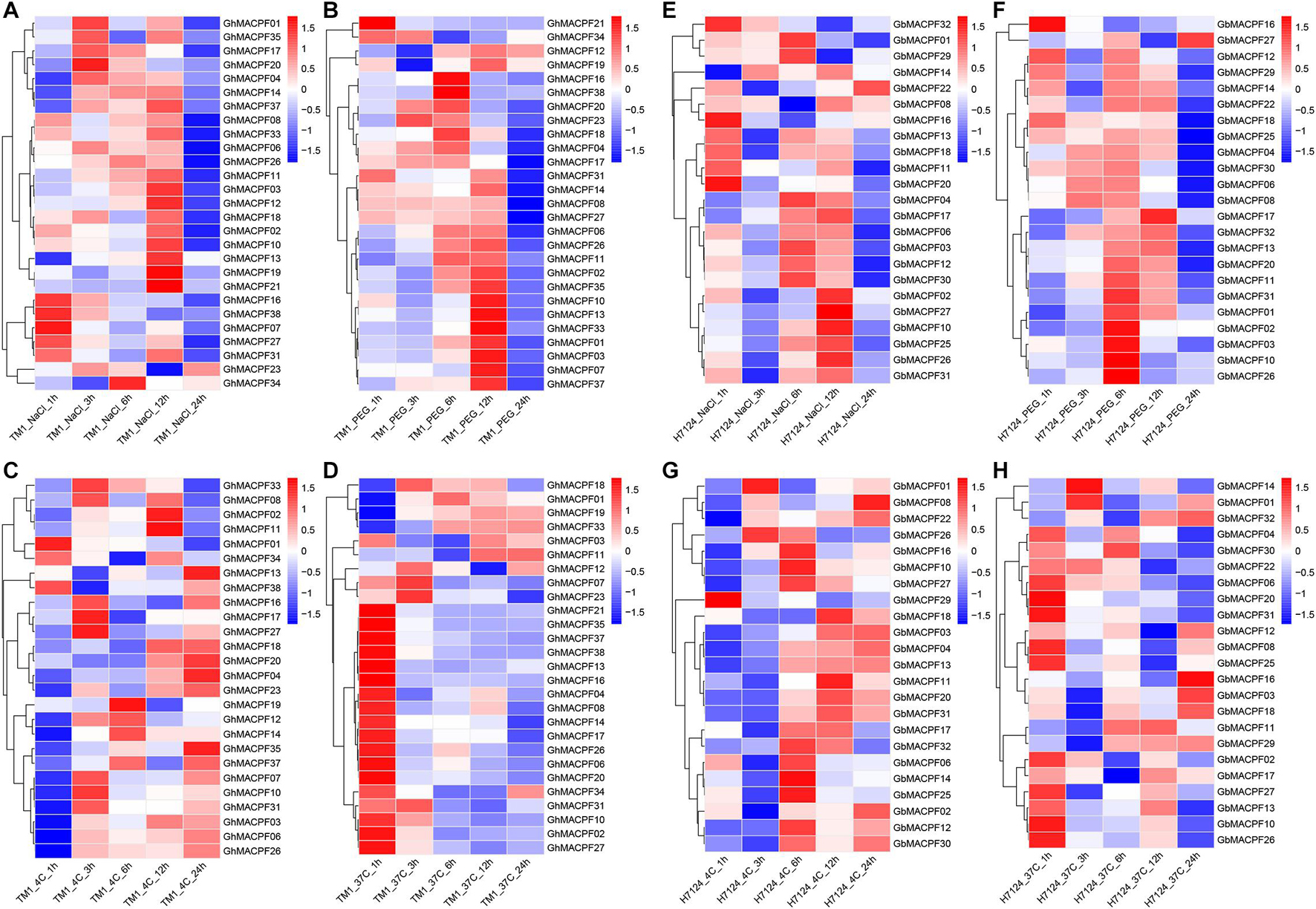
Figure 4. Expression profiles of the MACPF member in response to the different abiotic stresses. The expression levels of GhMACPFs and GbMACPFs under salt stresses (A,E), polyethylene glycol (PEG) stresses (B,F), cold stresses (C,G), and hot stresses (D,H), respectively.
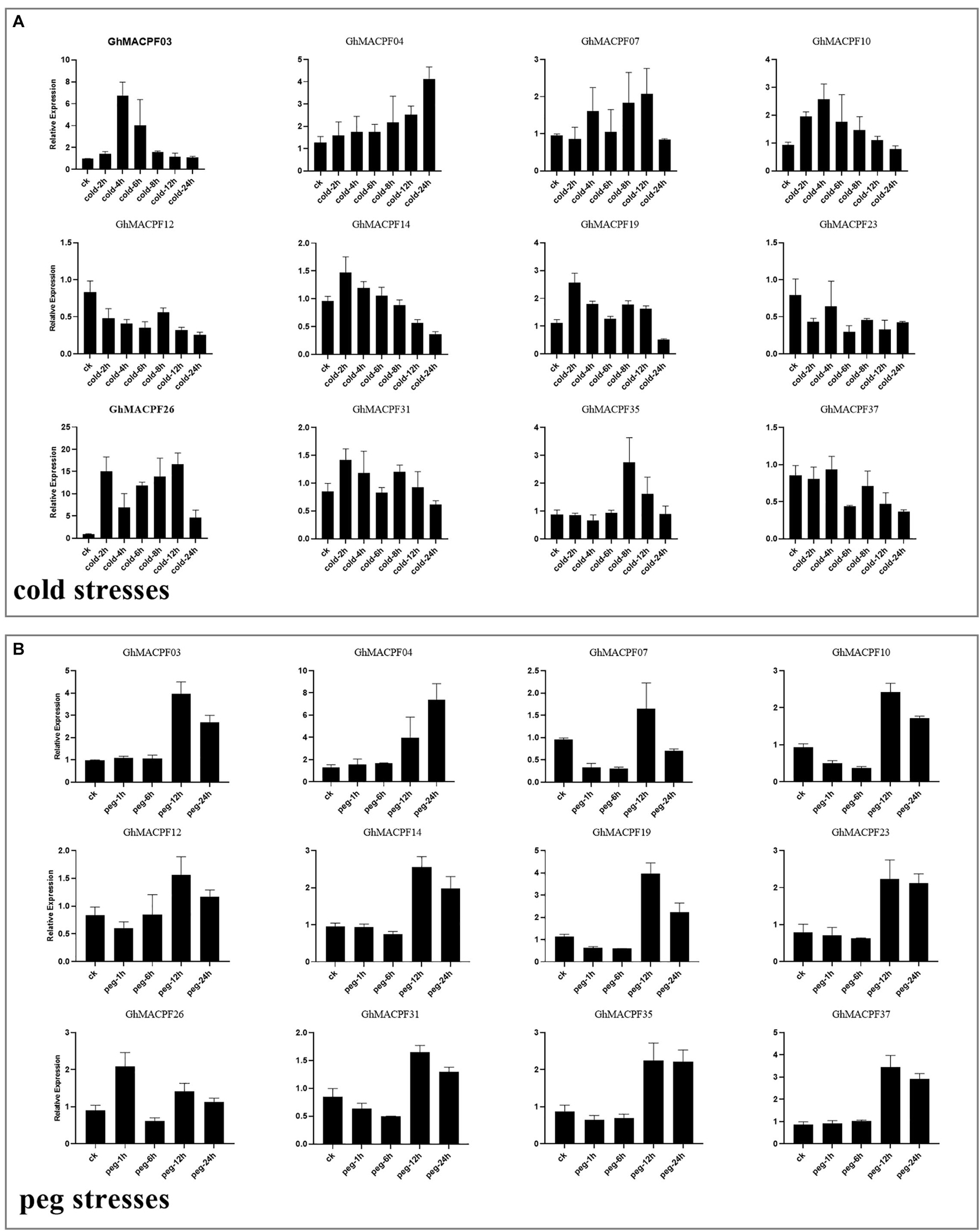
Figure 5. Expression levels of 12 GhMACPF genes in response to the (A) cold and (B) PEG treatment. Gene expression was analyzed by quantitative real-time-PCR (qRT-PCR). Error bars represent the SD of the three biological replicates.
Co-expression analysis was constructed to uncover the potential interaction of functional genes with the MACPFs in cotton under cold treatment. Figure 6A displayed nine GhMACPFs, including GhMACPF06, GhMACPF07, GhMACPF10, GhMACPF19, GhMACPF26, GhMACPF27, GhMACPF31, GhMACPF34, and GhMACPF39, which might be co-expressed with the AP2, GRAS, VQ, WRKY, and C2H2 TFs, which are widely involved in cold stress response (Supplementary Table 8). These results were confirmed with qRT-PCR analysis (Supplementary Figure 6). Moreover, Figure 6B (GhMACPF01, GhMACPF03, GhMACPF04, GhMACPF23, and GhMACPF35), Figure 6C (GhMACPF02, GhMACPF08, and GhMACPF11), and Figure 6D (GhMACPF18 and GhMACPF33) showed the other 10 GhMACPFs that were also participated in the cold responding interaction networks of TFs including AP2, p450, C2H2, basic helix-loop-helix protein (bHLH), and VQ domain-containing genes (Supplementary Table 8). GhMACPFs might interact with AP2, VQ, and WRKY TFs to enhance the cold resistance regulation of cotton.
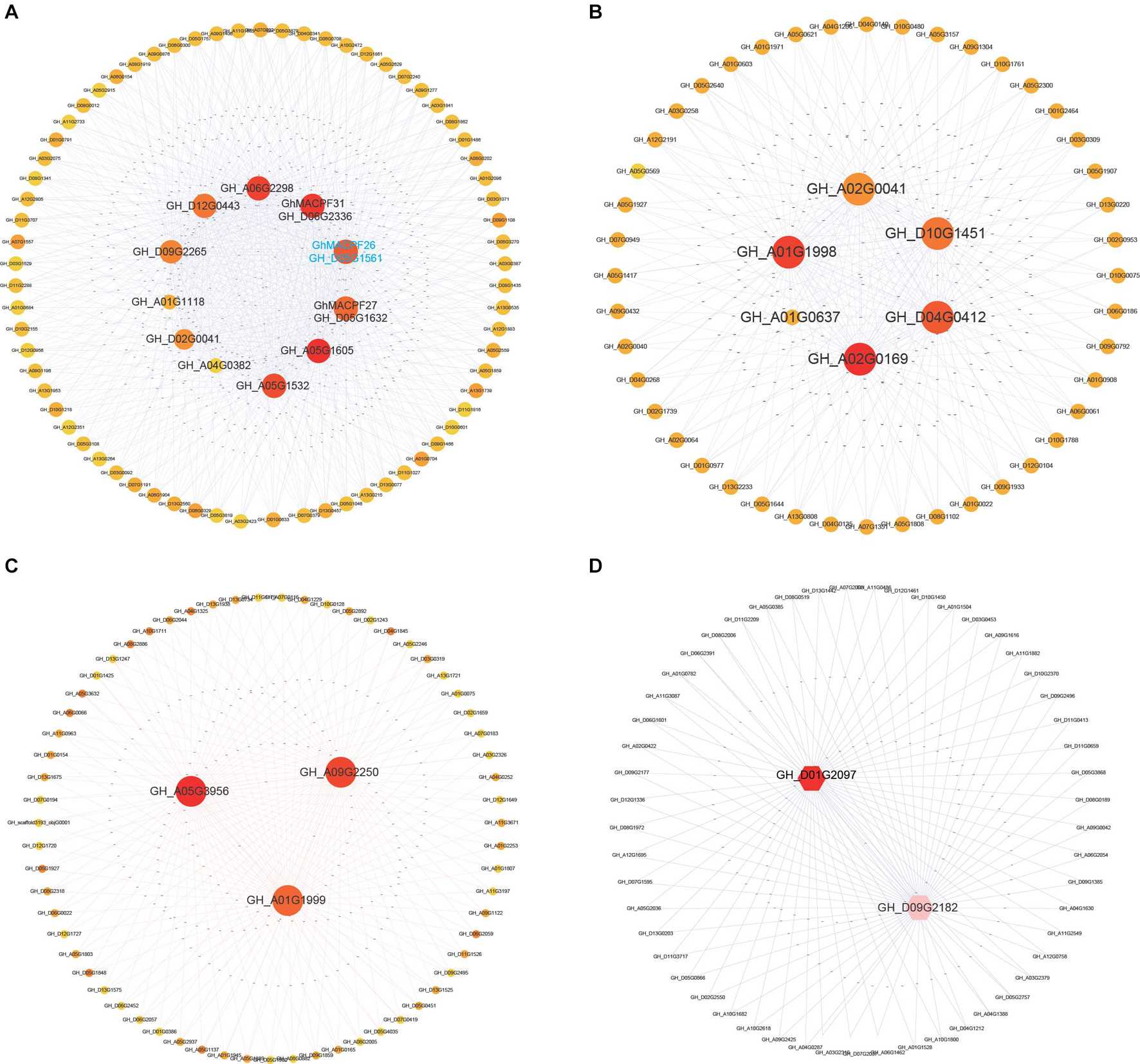
Figure 6. The correlation network of 19 GhMACPF members in cold stresses. All the gene networks are constructed by the weighted gene co-expression network analysis (WGCNA) in which each node represents a gene. (A) (nine GhMACPFs), (B) (five GhMACPFs), (C) (three GhMACPFs), and (D) (two GhMACPFs) indicate GhMACPFs in the different modules.
Silencing GhMACPF26 Increased Tolerance to Cold Stress
To investigate the role of GhMACPF26 in response to cold stress, we performed a VIGS assay to decrease the GhMACPF26 expression in TM-1 plants. The albino phenotype ensured the success of the tobacco rattle virus (TRV)::CLA1 in cotton (Figure 7A) and the comparison of the expression level of TRV::00 and TRV::GhMACPF26 in cotton indicated that the gene expression had been successfully silenced (Figures 7B–D). TRV::GhMACPF26 and TRV::00 plants were subjected to cold stress for 24 h. The empty control plants (Figure 7B) and TRV::GhMACPF26 (Figure 7C) were similar before the cold treatment, but they contained MDA content that was significantly different based on the t-test analysis (Figure 7E). The phenotypic difference in the degree of leaf damage was clear between the TRV::00 (Figure 7F) and TRV::GhMACPF26 (Figure 7G) after cold treatments for 24 h. The MDA content in TRV::00 plants was about 1.6 times more than in TRV::GhMACPF26 content (Figure 7H). Additionally, the MDA content was examined to the degree of cell damage (Jouve et al., 1993). Our results preliminarily proved that silencing of the GhMACPF26 gene improves cold tolerance in cotton.

Figure 7. Silencing GhMACPF26 via virus-induced gene silencing (VIGS) enhances the cold resistance regulation of cotton. (A) Plant albino phenotypes of TRV::CLA. (B,C) Phenotypes of TRV::00 and TRV::GhMACPF26 before cold stress. (D) GhMACPF26 expression levels in leaves of TRV::00 and TRV::GhMACPF26 plants. (E) The malondialdehyde (MDA) content of TRV::00 and TRV::GhMACPF26 before cold stress. (F,G) Phenotypes of TRV::00 and TRV::GhMACPF26 after cold treatment for 24 h. (H) The MDA content of TRV::00 and TRV::GhMACPF26 after cold stress. *p < 0.05, **p < 0.01, and ***p < 0.001 (Student’s one-tailed t test).
Discussion
Many MACPF domain-containing proteins were involved in the innate and adaptive immunity against pathogens through multiple pathways (Esser, 1994; McCormack et al., 2013; Spicer et al., 2017; Chen T. et al., 2020) and more and more pieces of evidence showed that the MACPF genes might participate in plant immune system, development, and abiotic stresses (Yu et al., 2020). In this study, 184 MACPFs were identified through a comprehensive analysis among the 15 genomes. Transcriptomic and co-expression analysis revealed that the MACPF genes were involved in the cold stresses, while silenced GhMACPF26 enhanced cotton plant tolerance to cold stress. These results indicated that the MACPF genes might play an important role in cotton adaptation to abiotic stress.
Phylogenetic, Duplication, and Structural Characteristics of the MACPFs in Gossypium spp.
The WGD or polyploidization is important for genome evolution due to the neofunctionalization and subfunctionalization of redundant genes (Blanc and Wolfe, 2004; Jiao et al., 2011; Qiao et al., 2019). Polyploidy on selection and domestication among the Gossypium spp. drives parallel gene expression in the development and abiotic stresses (Hu et al., 2019; Chen Z.J. et al., 2020). In this study, we found that the MACPF genes were shared with conserved gene numbers and similar duplication events by comparing the allotetraploid and diploid cotton. Totally, 38 GhMACPFs, 33 GbMACPFs, 14 GaMACPFs, and 15 GrMACPFs were identified from the four Gossypium spp. through a comprehensive gene family analysis, respectively. The number of the MACPFs in the allotetraploid cotton was almost two times the number in diploid cotton. The duplication event analysis showed that the WGD event was likely leading to the expansion of the MACPF genes in the allotetraploid cotton. Meanwhile, 184 MACPF genes were divided into three groups, of which 4 AtMACPFs and 7 OsMACPFs were evenly distributed in the Group I and Group II. The results showed that group I was conserved and belonged to the mallow (Figure 1). The conserved motif analysis in Gossypium spp. indicated that the “(F/Y)GTH(F/Y)-X6-GG” motif was widely distributed in GhMACPFs, GbMACPFs, GrMACPFs, and GaMACPFs (Figure 2 and Supplementary Figure 3). Orthologous analysis suggested that the evolution of the MACPF genes was not balanced between the allotetraploid and diploid genomes and there were more genes in the Dt subgenomes (Figure 3 and Supplementary Figure 4). The imbalance transcript indicated that the donors of the Dt subgenomes and G. raimondii were important to the abiotic stresses in the allotetraploid cotton and in the Brassica napus allotetraploids (Wang et al., 2019; Meng et al., 2020; Zhang et al., 2020). In addition, Ka/Ks values showed that most gene pairs have undergone purification selection during evolution (Supplementary Table 6) and the purification selection in WRKY and plant homeodomain (PHD) gene family (Hurst, 2002; Gu et al., 2018; Wu et al., 2021). These results suggested that the MACPF genes have been replicated in the ancient genome duplication events.
Membrane Attack Complex/Perforins Play Important Roles in Abiotic Stress
Cis-regulatory elements within the promoter analysis are important to understand transcriptional regulation, which contained the development regulation and environmental responses (Chen et al., 2018; Yan et al., 2019). Cis-acting elements on the promoter were recruited and bound by TFs and the expression of the gene was regulated (Liu et al., 2013). As the previous results, the ABREs (ABA responsive elements) induced the expression of genes involved in the ABA signaling pathway (Hossain et al., 2010), while the GARE motif functions as a gibberellin-response element, TGA element acts as an auxin-response element, TGACG motif is the MeJA-response element (Chen and Qiu, 2020), and LTR is the low-temperature responsiveness element (Yamaguchi-Shinozaki and Shinozaki, 1994). ABRE, TGA, GARE- motif, CGTCA motif, TGACG motif, LTR, MBS, MBSI, and TC-rich repeats were also found in the MACPF promoters. These cis elements indicated that they participated in the different regulatory mechanisms of environmental adaptation including abiotic stress response (Supplementary Figure 1). Interestingly, the expression of the GhMACPFs and GbMACPFs widely responded to various abiotic stresses including the salt, PEG, cold, and heat treatments (Figure 4). In addition, most GhMACPFs and GbMACPFs were highly expressed in various tissues, and the ovule or fiber development, suggesting that the MACPFs also play an important role in the differentiation and fiber development of cotton tissue (Supplementary Figure 5). As reported in rice, maize, and Arabidopsis (Yu et al., 2020), MACPF genes were widely involved in abiotic stress response via the complex potential mechanism in Gossypium spp.
The Co-expression network was constructed to explore the potential connectivity of the genes involved in the plant development and abiotic stress response (Zhang and Horvath, 2005; van Dam et al., 2018; Chen P. et al., 2020; Cheng et al., 2020; Hu et al., 2020; Wang et al., 2020). TFs, including AP2/ERF, GRAS, VQ, and WRKY families, have recently been subjected to an intensive investigation because of increasing evidence of their response to abiotic stress (Mizoi et al., 2012; Grimplet et al., 2016; Gu et al., 2018; Chen P. et al., 2020). Constructing expression regulatory networks related to TFs is important for mining the candidate functional genes. Several GhMACPFs, including GhMACPF6, GhMACPF7, GhMACPF10, GhMACPF19, and GhMACPF26, were predicted to interact with AP2/ERF, WRKY, and VQ TFs (Figure 6 and Supplementary Table 8). The results indicated that the MACPF genes might act together with these TFs under cold stress in cotton.
Previous studies have shown that cold stress not only led to a decrease in growth and development but also affected plant metabolism and phytohormones (Thakur et al., 2010). Silencing GhMACPF26 indicated that the VIGS plant enhances the cotton cold resistance via the measure of the MDA content (Figure 7). The MDA content in TRV::00 was significantly higher than the TRV::GhMACPF26 after 24 h cold treatment, indicating that the plasma membrane damage is more serious in the TRV::00 plant (Figure 7). The MDA content is usually used to measure the degree of damage to plant cells and it also acts as the product of lipid oxidation (Leshem, 1987; Jouve et al., 1993). Transcriptomic results suggested that GhMACPF was also participating in the regulation of cotton tissue, ovule, fiber growth, and development. Moreover, we hypothesized that the MACPF members related to cotton resistance, especially resistance to cold stress. However, the underlying molecular mechanism requires further elucidation.
Conclusion
In this study, a comprehensive analysis of the MACPF family was performed in the diploid and allotetraploid cotton via the phylogenetic, structural, orthologous, and transcriptomic analysis. Our data revealed that the WGD events might be the MACPF genes expansion in the allotetraploid cotton and genes on the Dt subgenome were related to stress resistance. Cis element, expression, and VIGS results indicated that the MACPF genes were activated in cotton response to cold stress. Co-expression analysis predicted that the GhMACPF genes might interact with AP2/ERF, WRKY, and VQ TFs to enhance the cold resistance of cotton. Silenced GhMACPF26 increased the cotton tolerance to cold treatment. This study could provide new insights into the MACPF gene function in cotton and their potential interactions with other TFs.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author Contributions
HW and SY designed the experiments. PC, HJ, and FW collected the sequences and analyzed the transcriptome analysis. AW, TH, XL, XG, JL, and LM participated in cotton culture processing and RNA extraction. LG, FW, and HW revised the language. PC performed the experiments and wrote the manuscript. All authors read and approved the final manuscript.
Funding
This research was funded by the National Key Research and Development Program (2018YFD0100402) and Agricultural Science and Technology Innovation Program of the Chinese Academy of Agricultural Sciences.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We extend our thanks to the reviewers and editor for their careful reading and helpful comments on this manuscript. We would like to thank Fushun Hao from Henan University and Pei Li from the Huazhong Agricultural University for their advice and help in writing this article.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.684227/full#supplementary-material
Footnotes
- ^ https://www.cottongen.org/
- ^ https://www.phytozome.net
- ^ https://www.ncbi.nlm.nih.gov/
- ^ https://pfam.xfam.org/
- ^ https://smart.embl-heidelberg.de/
- ^ https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi
- ^ https://linux1.softberry.com/berry.phtml
- ^ https://web.expasy.org/translate/
- ^ http://phylogeny.lirmm.fr/phylo_cgi/index.cgi
- ^ http://meme-suite.org/
- ^ http://bioinformatics.psb.ugent.be/webtools/plantcare/html/
References
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. doi: 10.1093/nar/gkp335
Blanc, G., and Wolfe, K. H. (2004). Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell. 16, 1667–1678. doi: 10.1105/tpc.021345
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, P., Cheng, S., Ma, L., Wang, H., Zhang, M., Mao, G., et al. (2020). A comprehensive analysis of cotton VQ gene superfamily reveals their potential and extensive roles in regulating cotton abiotic stress. BMC Genom. 21:795. doi: 10.1186/s12864-020-07171-z
Chen, T., Nomura, K., Wang, X., Sohrabi, R., Xu, J., Yao, L., et al. (2020). A plant genetic network for preventing dysbiosis in the phyllosphere. Nature 580, 653–657. doi: 10.1038/s41586-020-2185-0
Chen, X., Mei, Q., Liang, W., Sun, J., Wang, X., Zhou, J., et al. (2020). Gene mapping, genome-wide transcriptome analysis, and WGCNA reveals the molecular mechanism for triggering programmed cell death in rice mutant pir1. Plants (Basel) 9:1607. doi: 10.3390/plants9111607
Chen, Z. J., Sreedasyam, A., Ando, A., Song, Q., De Santiago, L. M., Hulse-Kemp, A. M., et al. (2020). Genomic diversifications of five Gossypium allopolyploid species and their impact on cotton improvement. Nat. Genet. 52, 525–533. doi: 10.1038/s41588-020-0614-5
Chen, D., Yan, W., Fu, L. Y., and Kaufmann, K. (2018). Architecture of gene regulatory networks controlling flower development in Arabidopsis thaliana. Nat. Commun. 9:4534. doi: 10.1038/s41467-018-06772-3
Chen, S., and Qiu, G. (2020). Cloning and activity analysis of the promoter of nucleotide exchange factor gene ZjFes1 from the seagrasses Zostera japonica. Sci. Rep. 10:17291. doi: 10.1038/s41598-020-74381-6
Cheng, G., Zhang, L., Wang, H., Lu, J., Wei, H., and Yu, S. (2020). Transcriptomic profiling of young cotyledons response to chilling stress in two contrasting cotton (Gossypium hirsutum L.) genotypes at the seedling stage. Int. J. Mol. Sci. 21:5095. doi: 10.3390/ijms21145095
Cheng, S., Chen, P., Su, Z., Ma, L., Hao, P., Zhang, J., et al. (2021). High-resolution temporal dynamic transcriptome landscape reveals a GhCAL-mediated flowering regulatory pathway in cotton (Gossypium hirsutum L.). Plant Biotechnol. J. 19, 153–166. doi: 10.1111/pbi.13449
Chin, C. H., Chen, S. H., Wu, H. H., Ho, C. W., Ko, M. T., and Lin, C. Y. (2014). cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 8:S11. doi: 10.1186/1752-0509-8-S4-S11
Du, X., Huang, G., He, S., Yang, Z., Sun, G., Ma, X., et al. (2018). Resequencing of 243 diploid cotton accessions based on an updated A genome identifies the genetic basis of key agronomic traits. Nat. Genet. 50, 796–802. doi: 10.1038/s41588-018-0116-x
Eddy, S. R. (2011). Accelerated profile HMM searches. PLoS Comput. Biol. 7:e1002195. doi: 10.1371/journal.pcbi.1002195
El-Gebali, S., Mistry, J., Bateman, A., Eddy, S. R., Luciani, A., Potter, S. C., et al. (2019). The Pfam protein families database in 2019. Nucleic Acids Res. 47, D427–D432. doi: 10.1093/nar/gky995
Emamverdian, A., Ding, Y., and Mokhberdoran, F. (2020). The role of salicylic acid and gibberellin signaling in plant responses to abiotic stress with an emphasis on heavy metals. Plant Signal. Behav. 15:1777372. doi: 10.1080/15592324.2020.1777372
Esser, A. F. (1994). The membrane attack complex of complement. Assembly, structure and cytotoxic activity. Toxicology 87, 229–247. doi: 10.1016/0300-483x(94)90253-4
Fukunaga, S., Sogame, M., Hata, M., Singkaravanit-Ogawa, S., Pislewska-Bednarek, M., Onozawa-Komori, M., et al. (2017). Dysfunction of Arabidopsis MACPF domain protein activates programmed cell death via tryptophan metabolism in MAMP-triggered immunity. Plant J. 89, 381–393. doi: 10.1111/tpj.13391
Goodstein, D. M., Shu, S., Howson, R., Neupane, R., Hayes, R. D., Fazo, J., et al. (2012). Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40, D1178–D1186. doi: 10.1093/nar/gkr944
Grimplet, J., Agudelo-Romero, P., Teixeira, R. T., Martinez-Zapater, J. M., and Fortes, A. M. (2016). Structural and functional analysis of the GRAS gene family in grapevine indicates a Role of GRAS proteins in the control of development and stress responses. Front. Plant Sci. 7:353. doi: 10.3389/fpls.2016.00353
Gu, L., Wang, H., Wei, H., Sun, H., Li, L., Chen, P., et al. (2018). Identification, expression, and functional analysis of the group IId WRKY subfamily in upland cotton (Gossypium hirsutum L.). Front. Plant Sci. 9:1684. doi: 10.3389/fpls.2018.01684
Guindon, S., Lethiec, F., Duroux, P., and Gascuel, O. (2005). PHYML Online—a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res. 33, W557–W559. doi: 10.1093/nar/gki352
Hadders, M. A., Beringer, D. X., and Gros, P. (2007). Structure of C8alpha-MACPF reveals mechanism of membrane attack in complement immune defense. Science 317, 1552–1554. doi: 10.1126/science.1147103
Hasanuzzaman, M., Nahar, K., Bhuiyan, T. F., Anee, T. I., Inafuku, M., Oku, H., et al. (2017). “Salicylic acid: an all-rounder in regulating abiotic stress responses in plants,” in Phytohormones-Signaling Mechanisms and crosstalk in plant development and stress responses, ed. M. A. El-Esawi (London: IntechOpen), 31–75.
Hasegawa, P. M., Bressan, R. A., Zhu, J. K., and Bohnert, H. J. (2000). Plant cellular and molecular responses to high salinity. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51, 463–499. doi: 10.1146/annurev.arplant.51.1.463
Hossain, M. A., Cho, J., Han, M., Ahn, C.-H., Jeon, J.-S., An, G., et al. (2010). The ABRE-binding bZIP transcription factor OsABF2 is a positive regulator of abiotic stress and ABA signaling in rice. J. Plant Physiol. 167, 1512–1520. doi: 10.1016/j.jplph.2010.05.008
Hu, G., Grover, C. E., Arick, M. A., Liu, M., Peterson, D. G., and Wendel, J. F. (2020). Homoeologous gene expression and co-expression network analyses and evolutionary inference in allopolyploids. Brief Bioinform. 22, 1819–1835. doi: 10.1093/bib/bbaa035
Hu, Y., Chen, J., Fang, L., Zhang, Z., Ma, W., Niu, Y., et al. (2019). Gossypium barbadense and Gossypium hirsutum genomes provide insights into the origin and evolution of allotetraploid cotton. Nat. Genet. 51, 739–748. doi: 10.1038/s41588-019-0371-5
Hurst, L. D. (2002). The Ka/Ks ratio: diagnosing the form of sequence evolution. Trends Genet. 18:486. doi: 10.1016/s0168-9525(02)02722-1
Ivica, L., Tobias, D., and Peer, B. (2009). SMART 6: recent updates and new developments. Nucleic Acids Res. 37, 229–232. doi: 10.1093/nar
Jiao, Y., Wickett, N. J., Ayyampalayam, S., Chanderbali, A. S., Landherr, L., Ralph, P. E., et al. (2011). Ancestral polyploidy in seed plants and angiosperms. Nature 473, 97–100. doi: 10.1038/nature09916
Johnson, T. K., Henstridge, M. A., and Warr, C. G. (2017). MACPF/CDC proteins in development: Insights from Drosophila torso-like. Semin. Cell. Dev. Biol. 72, 163–170. doi: 10.1016/j.semcdb.2017.05.003
Jouve, L., Engelmann, F., Noirot, M., and Charrier, A. (1993). Evaluation of biochemical markers (sugar, proline, malonedialdehyde and ethylene) for cold sensitivity in microcuttingsof two coffee species. Plant Sci. 91, 109–116. doi: 10.1016/0168-9452(93)90194-5
Katoh, K., and Standley, D. M. (2014). MAFFT: iterative refinement and additional methods. Methods Mol. Biol. 1079, 131–146. doi: 10.1007/978-1-62703-646-7_8
Langfelder, P., and Horvath, S. (2008). WGCNA: an R package for weighted correlation network analysis. BMC Bioinform. 9:559. doi: 10.1186/1471-2105-9-559
Lescot, M., Dehais, P., Thijs, G., Marchal, K., Moreau, Y., Van De Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Leshem, Y. (1987). Membrane phospholipid catabolism and Ca2+ activity in control of senescence. Physiol. Plantarum 69, 551–559. doi: 10.1111/j.1399-3054.1987.tb09239.x
Li, F., Fan, G., Lu, C., Xiao, G., Zou, C., Kohel, R. J., et al. (2015). Genome sequence of cultivated Upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nat. Biotechnol. 33, 524–530. doi: 10.1038/nbt.3208
Liu, J.-H., Peng, T., and Dai, W. (2013). Critical cis-Acting elements and interacting transcription factors: key players associated with abiotic stress responses in plants. Plant Mol. Biol. Rep. 32, 303–317. doi: 10.1007/s11105-013-0667-z
Marchlerbauer, A., Derbyshire, M. K., Gonzales, N. R., Lu, S., Chitsaz, F., Geer, L. Y., et al. (2015). CDD: NCBI’s conserved domain database. Nucleic Acids Res. 43, 222–226. doi: 10.1093/nar/gku1221
Mark, J., Irena, Z., Yan, R., Yuri, M., Scott, M., and Madden, T. L. (2008). NCBI BLAST: a better web interface. Nucleic Acids Res. 36, W5–W9. doi: 10.1093/nar/gkn201
McCormack, R., De Armas, L., Shiratsuchi, M., and Podack, E. R. (2013). Killing machines: three pore-forming proteins of the immune system. Immunol. Res. 57, 268–278. doi: 10.1007/s12026-013-8469-9
Meng, F., Pan, Y., Wang, J., Yu, J., Liu, C., Zhang, Z., et al. (2020). Cotton duplicated genes produced by polyploidy show significantly elevated and unbalanced evolutionary rates, overwhelmingly perturbing gene tree topology. Front. Genet. 11:239. doi: 10.3389/fgene.2020.00239
Mizoi, J., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2012). AP2/ERF family transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta 1819, 86–96. doi: 10.1016/j.bbagrm.2011.08.004
Moreno-Hagelsieb, G., Vitug, B., Medrano-Soto, A., and Saier, M. H. Jr. (2017). The membrane attack complex/perforin superfamily. J. Mol. Microbiol. Biotechnol. 27, 252–267. doi: 10.1159/000481286
Morita-Yamamuro, C., Tsutsui, T., Sato, M., Yoshioka, H., Tamaoki, M., Ogawa, D., et al. (2005). The Arabidopsis gene CAD1 controls programmed cell death in the plant immune system and encodes a protein containing a MACPF domain. Plant Cell Physiol. 46, 902–912. doi: 10.1093/pcp/pci095
Ni, T., and Gilbert, R. J. C. (2017). Repurposing a pore: highly conserved perforin-like proteins with alternative mechanisms. Philos. Trans. R. Soc. Lond. B Biol. Sci. 372:20160212. doi: 10.1098/rstb.2016.0212
Noutoshi, Y., Kuromori, T., Wada, T., Hirayama, T., Kamiya, A., Imura, Y., et al. (2006). Loss of Necrotic Spotted Lesions 1 associates with cell death and defense responses in Arabidopsis thaliana. Plant Mol. Biol. 62, 29–42. doi: 10.1007/s11103-006-9001-6
Odukoya, J., Lambert, R., and Sakrabani, R. (2019). Understanding the impacts of crude oil and its induced abiotic stresses on agrifood production: A review. Horticulturae 5:47. doi: 10.3390/horticulturae5020047
Paterson, A., Wendel, J., Gundlach, H., Guo, H., Jenkins, J., Jin, D., et al. (2012). Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature 492, 423–427. doi: 10.1038/nature11798
Qiao, X., Li, Q., Yin, H., Qi, K., Li, L., Wang, R., et al. (2019). Gene duplication and evolution in recurring polyploidization-diploidization cycles in plants. Genome Biol. 20:38. doi: 10.1186/s13059-019-1650-2
Robinson, M. D., Mccarthy, D. J., and Smyth, G. K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Sayers, E. W., Barrett, T., Benson, D. A., Bolton, E., Bryant, S. H., Canese, K., et al. (2010). Database resources of the national center for biotechnology information. Nucleic Acids Res. 39, D38–D51. doi: 10.1093/nar/gkt1146
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi: 10.1101/gr.1239303
Spicer, B. A., Conroy, P. J., Law, R. H., Voskoboinik, I., and Whisstock, J. C. (2017). Perforin-A key (shaped) weapon in the immunological arsenal. Semin. Cell. Dev. Biol. 72, 117–123. doi: 10.1016/j.semcdb.2017.07.033
Thakur, P., Kumar, S., Malik, J. A., Berger, J., and Nayyar, H. (2010). Cold stress effects on reproductive development in grain crops: An overview. Environ. Exper. Bot. 67, 429–443. doi: 10.1016/j.envexpbot.2009.09.004
Tsutsui, T., Morita-Yamamuro, C., Asada, Y., Minami, E., Shibuya, N., Ikeda, A., et al. (2006). Salicylic acid and a chitin elicitor both control expression of the CAD1 gene involved in the plant immunity of Arabidopsis. Biosci. Biotechnol. Biochem. 70, 2042–2048. doi: 10.1271/bbb.50700
Tu, L., Zhang, X., Liu, D., Jin, S., Cao, J., Zhu, L., et al. (2007). Suitable internal control genes for qRT-PCR normalization in cotton fiber development and somatic embryogenesis. Chinese Sci. Bull. 52, 3110–3117. doi: 10.1007/s11434-007-0461-0
van Dam, S., Vosa, U., Van Der Graaf, A., Franke, L., and De Magalhaes, J. P. (2018). Gene co-expression analysis for functional classification and gene-disease predictions. Brief Bioinform. 19, 575–592. doi: 10.1093/bib/bbw139
Vlot, A. C., Dempsey, D. A., and Klessig, D. F. (2009). Salicylic Acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 47, 177–206. doi: 10.1146/annurev.phyto.050908.135202
Wade, K. R., and Tweten, R. K. (2015). The Apicomplexan CDC/MACPF-like pore-forming proteins. Curr. Opin. Microbiol. 26, 48–52. doi: 10.1016/j.mib.2015.05.001
Wang, D., Zhang, Y., Zhang, Z., Zhu, J., and Yu, J. (2010). KaKs_Calculator 2.0: a toolkit incorporating gamma-series methods and sliding window strategies. Genom Proteomics Bioinform. 8, 77–80. doi: 10.1016/S1672-0229(10)60008-3
Wang, M., Tu, L., Yuan, D., Zhu, D., Shen, C., Li, J., et al. (2019). Reference genome sequences of two cultivated allotetraploid cottons, Gossypium hirsutum and Gossypium barbadense. Nat. Genet. 51, 224–229. doi: 10.1038/s41588-018-0282-x
Wang, Y., Liu, J., Zhao, G., Geng, Z., Qi, H., Dou, H., et al. (2020). Dynamic transcriptome and co-expression network analysis of the cotton (Gossypium hirsutum) root response to salinity stress at the seedling stage. Acta Physiol. Plantarum 42, 1–16. doi: 10.1007/s11738-020-03117-w
Wang, Y., Tang, H., Debarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40:e49. doi: 10.1093/nar/gkr1293
Wu, H., Zheng, L., Qanmber, G., Guo, M., Wang, Z., and Yang, Z. (2021). Response of phytohormone mediated plant homeodomain (PHD) family to abiotic stress in upland cotton (Gossypium hirsutum spp.). BMC Plant Biol. 21:13. doi: 10.1186/s12870-020-02787-5
Yamaguchi-Shinozaki, K., and Shinozaki, K. (1994). A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 6, 251–264. doi: 10.1105/tpc.6.2.251
Yan, W., Chen, D., Schumacher, J., Durantini, D., Engelhorn, J., Chen, M., et al. (2019). Dynamic control of enhancer activity drives stage-specific gene expression during flower morphogenesis. Nat. Commun. 10:1705. doi: 10.1038/s41467-019-09513-2
Yu, G., Smith, D. K., Zhu, H., Guan, Y., Lam, T. T. Y., and Mcinerny, G. (2016). GGtree : an r package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol. 8, 28–36. doi: 10.1111/2041-210X.12628
Yu, J., Jung, S., Cheng, C. H., Ficklin, S. P., Lee, T., Zheng, P., et al. (2014). CottonGen: a genomics, genetics and breeding database for cotton research. Nucleic Acids Res. 42, D1229–D1236. doi: 10.1093/nar/gkt1064
Yu, L., Liu, D., Chen, S., Dai, Y., Guo, W., Zhang, X., et al. (2020). Evolution and expression of the membrane attack complex and perforin gene family in the poaceae. Int. J. Mol. Sci. 21:5736. doi: 10.3390/ijms21165736
Zhang, B., and Horvath, S. (2005). A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. Biol. 4:17. doi: 10.2202/1544-6115.1128
Zhang, Q., Guan, P., Zhao, L., Ma, M., Xie, L., Li, Y., et al. (2020). Asymmetric epigenome maps of subgenomes reveal imbalanced transcription and distinct evolutionary trends in Brassica napus. Mol. Plant 14, 604–619. doi: 10.1016/j.molp.2020.12.020
Zhang, Z., Xiao, J., Wu, J., Zhang, H., Liu, G., Wang, X., et al. (2012). ParaAT: a parallel tool for constructing multiple protein-coding DNA alignments. Biochem. Biophys. Res. Commun. 419, 779–781. doi: 10.1016/j.bbrc.2012.02.101
Keywords: Gossypium, membrane attack complex/perforin (MACPF), cold stress, VIGS, WGCNA
Citation: Chen P, Jian H, Wei F, Gu L, Hu T, Lv X, Guo X, Lu J, Ma L, Wang H, Wu A, Mao G, Yu S and Wei H (2021) Phylogenetic Analysis of the Membrane Attack Complex/Perforin Domain-Containing Proteins in Gossypium and the Role of GhMACPF26 in Cotton Under Cold Stress. Front. Plant Sci. 12:684227. doi: 10.3389/fpls.2021.684227
Received: 23 March 2021; Accepted: 04 October 2021;
Published: 19 November 2021.
Edited by:
Jinfa Zhang, New Mexico State University, United StatesReviewed by:
Xianlong Zhang, Huazhong Agricultural University, ChinaLinghe Zeng, United States Department of Agriculture (USDA), United States
Copyright © 2021 Chen, Jian, Wei, Gu, Hu, Lv, Guo, Lu, Ma, Wang, Wu, Mao, Yu and Wei. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shuxun Yu, eXN4MTk1MzExQDE2My5jb20=; Hengling Wei, aGVuZ2xpbmd3ZWlAMTYzLmNvbQ==
†These authors have contributed equally to this work and share first authorship
 Pengyun Chen
Pengyun Chen Hongliang Jian1†
Hongliang Jian1† Lijiao Gu
Lijiao Gu Hantao Wang
Hantao Wang Aimin Wu
Aimin Wu Shuxun Yu
Shuxun Yu Hengling Wei
Hengling Wei