- 1International Crops Research Institute for the Semi-Arid Tropics (ICRISAT), Hyderabad, India
- 2The UWA Institute of Agriculture, The University of Western Australia, Perth, WA, Australia
- 3Plant and Environmental Sciences, Poole Agricultural Center, Clemson University, Clemson, SC, United States
- 4ICMR-National Institute of Nutrition (NIN), Hyderabad, India
- 5State Agricultural Biotechnology Centre, Centre for Crop and Food Innovation, Murdoch University, Murdoch, WA, Australia
The world faces a grave situation of nutrient deficiency as a consequence of increased uptake of calorie-rich food that threaten nutritional security. More than half the world’s population is affected by different forms of malnutrition. Unhealthy diets associated with poor nutrition carry a significant risk of developing non-communicable diseases, leading to a high mortality rate. Although considerable efforts have been made in agriculture to increase nutrient content in cereals, the successes are insufficient. The number of people affected by different forms of malnutrition has not decreased much in the recent past. While legumes are an integral part of the food system and widely grown in sub-Saharan Africa and South Asia, only limited efforts have been made to increase their nutrient content in these regions. Genetic variation for a majority of nutritional traits that ensure nutritional security in adverse conditions exists in the germplasm pool of legume crops. This diversity can be utilized by selective breeding for increased nutrients in seeds. The targeted identification of precise factors related to nutritional traits and their utilization in a breeding program can help mitigate malnutrition. The principal objective of this review is to present the molecular mechanisms of nutrient acquisition, transport and metabolism to support a biofortification strategy in legume crops to contribute to addressing malnutrition.
Introduction
Combating malnutrition in all its forms is one of the most significant global health challenges of the 21st century as it affects mostly women, infants, children, and adolescents. The World Health Organization (WHO) estimates that globally, more than 2 billion people suffer from micronutrient malnutrition, also known as “hidden hunger” (Ritchie and Roser, 2020). Modern breeding approaches, together with best agronomic practices during the Green Revolution, were instrumental in significantly increasing the production of major cereal crops. This increase led to a reduction in global hunger. However, several developing countries still face challenges related to malnutrition due to the consumption of cereal-based diets. Poor diets devoid of nutrient concentrations and bioavailability are among the principal risk factors for non-communicable diseases (NCDs), responsible for about 70% of deaths in 2015 (Forouzanfar et al., 2015).
Malnutrition Across the World
Malnutrition refers to inadequacies, excesses, or imbalances in an individual’s consumption of nutrients that adversely affect health and ultimately impair growth and fitness. It can be classified into three broad forms: undernutrition (wasting, stunting, and underweight), micronutrient-linked malnutrition (lack or excess of vitamins or and minerals) and overweight (obesity). It affects humans through increased morbidity, disability, stunted mental growth, and reduced National Socio-economic Development Plan (NSEDP) (FAO et al., 2017). Disturbingly, every second pregnant woman and about 40% of pre-school children in developing countries are estimated to be anemic; this leads to 20% of all maternal deaths. WHO estimated 5.3 million child deaths under the age of 5 during 2018, of which around 45% were linked to undernutrition.
Similarly, the share of neonatal deaths is projected to increase from roughly 46% in 2016 to 52% in 2030 (WHO, 2017). To make matters worse, around 2 billion people worldwide are anemic, mainly due to iron (Fe) deficiency (WHO, 2017). About 32.8% of women of reproductive stage and 32.5% of non-pregnant women, and 41.7% of children under the age of 5 are suffering from anemia globally (World Bank Data, 2016). Additionally, apart from zinc, iodine, and vitamin A deficiency, calcium, magnesium, and copper deficiencies are also prevalent in many developed and developing countries (Kumssa et al., 2015). Malnutrition has multifaceted consequences as it increases medical expenses and reduces productivity and economic growth. Malnutrition accounts for 11% of GDP losses in Asia and Africa, which is higher than the GDP loss experienced during the 2008–2010 financial crisis (Von Grebmer et al., 2016). Focusing only on delivering the energy needs of resource-poor people without considering their nutrient requirements will exacerbate the current state of malnutrition (Zarocostas, 2009). The Green Revolution was successful in increasing the productivity of major cereal crops multifold, preventing widespread famines and increasing the profitability of farmers in many developing countries (Bouis and Welch, 2010). However, a rise in micronutrient malnutrition in many nations suggests that agriculture needs to reevaluate its strategy to provide a healthy mix of sufficient calories along with essential nutrients. A sustainable solution to malnutrition would lie in linking agriculture to nutrition and health (Jones and Ejeta, 2016).
Human Nutrient Requirements
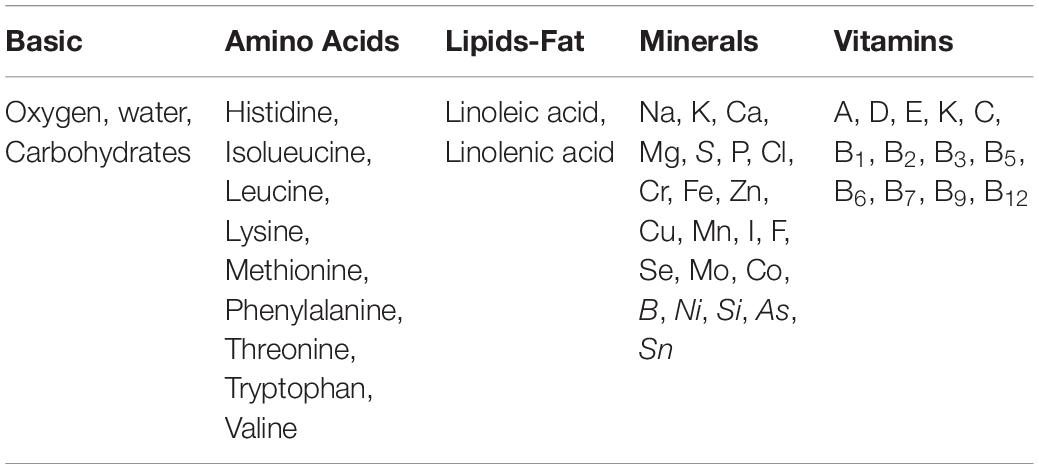
A human body needs more than 50 macronutrients (>0.1 % of dry mass) and micronutrients (<0.01 % of dry weight) from five groups of essential nutrients for proper growth and development (Table 1). Though these nutrients are needed in meager quantities, they enable the body to produce enzymes, hormones, and other essential substances that aid growth and development. The recommended dietary allowance (RDA) for various nutrients varies with gender and age (Supplementary File 1). Along with oxygen, water and carbohydrates, vitamins and minerals are vital substances for our bodies to develop and function properly. According to the National Center for Complementary and Integrative Health (NCCIH, 2018) thirteen known vitamins, namely A, C, D, E, and K, and B vitamins (B1, B2, B3, B5, B7, B6, B12, and B9) and fifteen minerals, namely Calcium (Ca), Phosphorus (P), Potassium (K), Sodium (Na), Chlorine (Cl), Magnesium (Mg), Iron (Fe), Zinc (Zn), Iodine (I), Chromium (Cr), Copper (Cu), Fluorine (F), Molybdenum (Mo), Manganese (Mn), and Sulfur (S) are essential for health. Researchers have highlighted the need for 22 minerals for human well-being (White and Broadley, 2009), the lack of which present a grave threat to the health and development of populations around the globe, especially children and pregnant women in low-income countries.
Role of Legumes in Eradicating Malnutrition
Malnutrition and poverty are closely related, which is also evident from FAO’s data on per capita income and level of malnutrition1. Around 82% of the extremely poor live in South Asia (SA) and sub-Saharan Africa (SSA), regions that host countries severely affected by one or other form of malnutrition. Considering the substantial socio-economic impact of legumes in these regions, their importance for food and nutritional security has been realized (Figure 1). In general, legume seeds have higher concentrations of essential minerals, vitamins, and protein than those of cereals (White and Broadley, 2009).
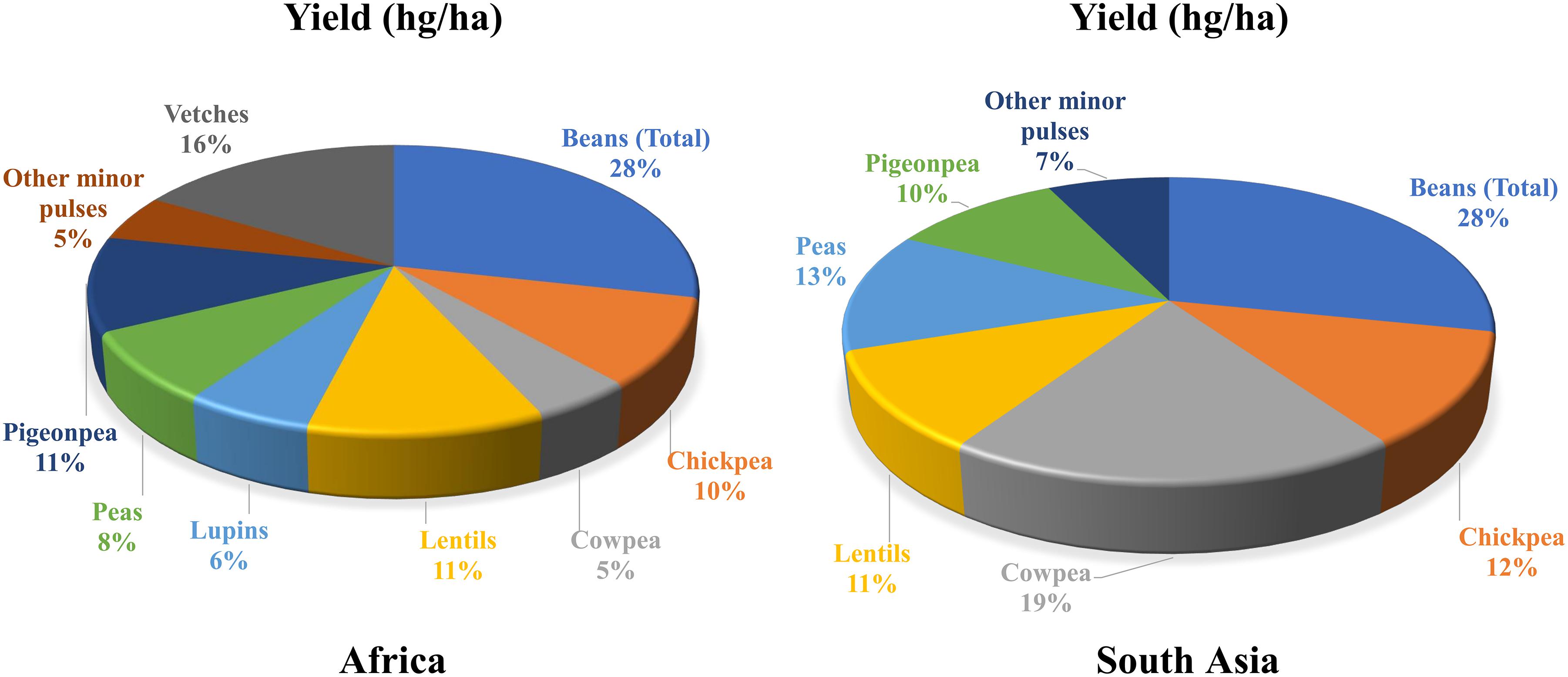
Figure 1. Share (%) of legumes’ yields in South Asia and Africa. Source: http://www.fao.org/faostat/en/#data/QC; accessed on April 16, 2018 Data Year: 2016. South Asia: Afghanistan, Bangladesh, Bhutan, India, Maldives, Nepal, Pakistan, and Sri Lanka. Africa: Algeria, Benin, Burkina Faso, Burundi, Cabo Verde, Cameroon, Central African Republic, Chad, Comoros, Congo, Côte d’Ivoire, Democratic Republic of the Congo, Djibouti, Egypt, Eritrea, Ethiopia, Gabon, Gambia, Guinea, Guinea-Bissau, Kenya, Lesotho, Liberia, Libya, Madagascar, Malawi, Mali, Mauritania, Morocco, Mozambique, Namibia, Niger, Nigeria, Réunion, Rwanda, Senegal, Sierra Leone, Somalia, South Africa, Togo, Sudan, Swaziland, Tunisia, Uganda, United Republic of Tanzania, Zambia, and Zimbabwe.
Legumes are an inexpensive source of protein (20–25%), minerals (Fe, Mg, K, P, and Zn) and vitamins (B1, B2, B3, B6, and B9) available to hundreds of millions of resource-poor people in SA and SSA. They are exceptionally notable because they complement the starches derived from cereals and root crops and help in efficient nutrient absorption. Legumes have a low-glycemic index and are rich in dietary fiber (8–27.5%), of which 3.3–13.8% correspond to soluble fiber (Sánchez-Chino et al., 2015). Storage protein is the major fraction of proteins in legumes. Almost 70% of the total protein comprises globulin, 10–20% each albumin and glutelins and low levels of prolamins (Sharif et al., 2018). Based on their lipid content, legumes can be classified into two main groups: those with low-fat content (1–6%), such as chickpea, lentil, bean, broad bean, etc., (Sánchez-Chino et al., 2015), and those with a high concentration of fat, which includes peanut and soybean (50 and 18%, respectively). The nutritional properties of legumes and their importance have been extensively reviewed in many articles (Jukanti et al., 2012; Mudryj et al., 2014; Sánchez-Chino et al., 2015; Foyer et al., 2016).
Considering the challenges related to hidden hunger and non-availability of nutritious food to a major portion of the global population, it is high time to initiate crop biofortification efforts. More than half of the world’s population faces impaired growth and fitness due to imbalances in an individual’s consumption of nutrients. Micronutrient malnutrition exists in several Asian and SSA countries due to the consumption of mainly cereals-based diets deprived of nutrient concentrations and bioavailability. Agriculture efforts focus on increasing nutrient content in cereals, but these are not enough to meet the global nutrition standards (Finkelstein et al., 2017). Legume crops have good genetic variation for nutritional traits, and therefore, there is a high potential for enhancing nutrients in seeds to contribute toward nutritional security. In this context, the discovery of genes and pathways accountable for nutrient acquisition and transport is pre-requisite. The targeted identification of precise factors related to nutritional traits and their utilization in a breeding program can help mitigate the challenge of malnutrition. A biofortification strategy by deploying breeding approaches for the nutritional improvement and microbiome for agronomic trait improvement in legumes has been presented (Rehman et al., 2019). However, it does not discuss molecular mechanisms and biochemical pathways for micronutrient acquisition and storage in legumes. This MS, therefore, provides up-to-date information on nutrient uptake mechanism and metabolism process that will serve as a foundation to initiate legume biofortification programs by integrating modern breeding approaches.
Molecular Mechanisms of Mineral Acquisition and Transport
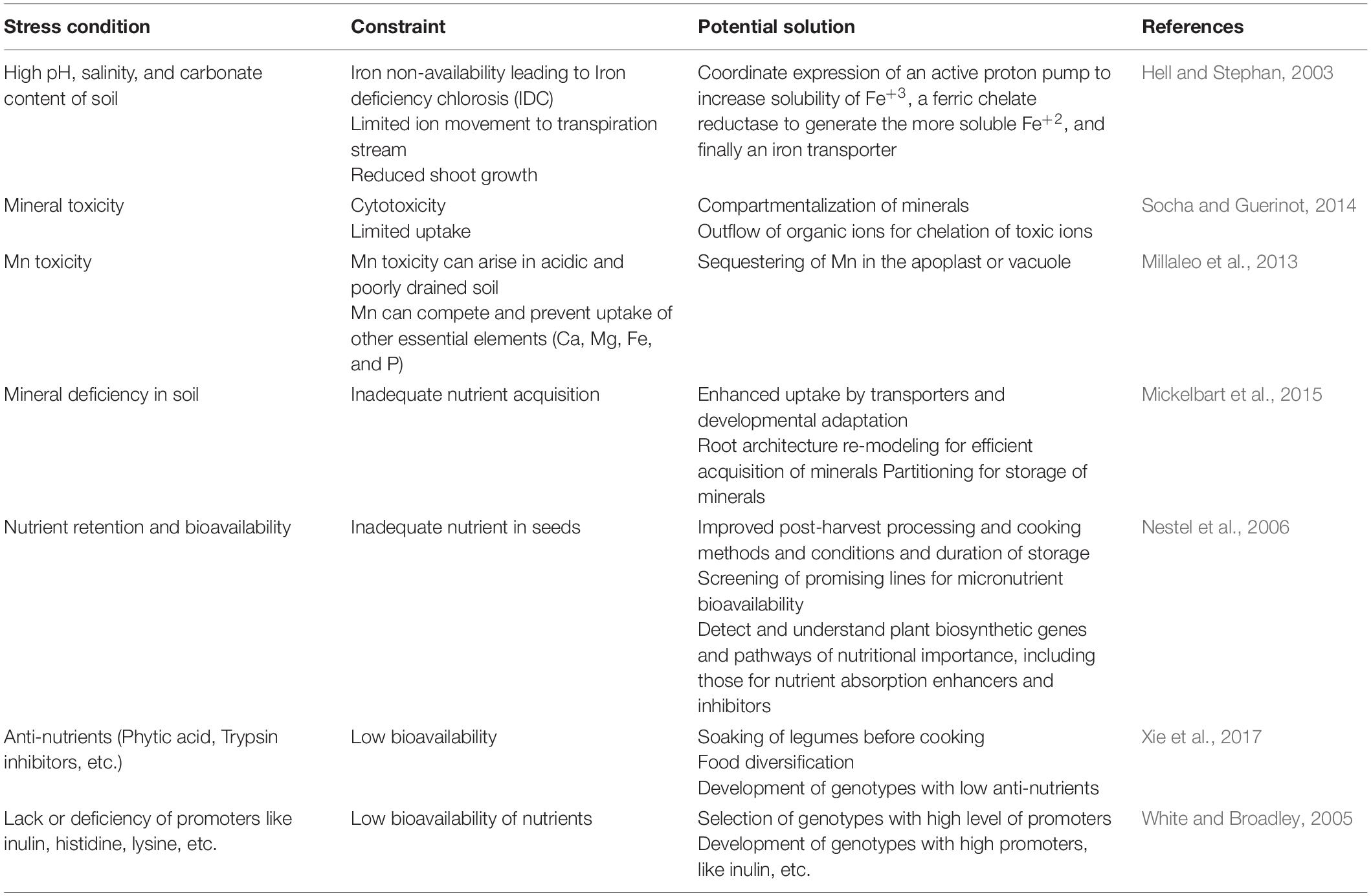
In order to maximize the bioavailability of nutrients, it is essential to understand the process of mineral acquisition, transport, and accumulation in legume seeds. Each of these processes is probably controlled by some genes, many of which are yet to be identified. Several studies have identified genes involved in translocation to different vegetative tissues and ultimately to seeds (Sperotto et al., 2014; Jeong et al., 2017). However, there is very limited knowledge of phloem-expressed genes involved in mineral loading and mobilization to different sink tissues (Braun et al., 2014). Therefore, while studies on specific transporters help us understand their function, whole-plant studies are required to ascertain transporters most relevant to seed mineral delivery. The acquisition and the mobilization of minerals in plants have been broadly studied (Walker and Waters, 2011; González-Guerrero et al., 2016; Xue et al., 2016). Several stresses can lead to the non-availability of key nutrition factors and result in improper crop growth. Details about these different stresses and their effect and a potential solution are provided in Table 2.
Iron (Fe) Transport
Legumes are “strategy I” plants that acidify the rhizosphere through an H+-ATPase (the enzyme of HA2, H+-ATPase family) to increase Fe3+ solubility (Santi and Schmidt, 2009). Then they reduce Fe3+ to Fe2+ with the help of chelate reductase, ferric reduction oxidase (FRO2)and finally Fe2+ taken up by root’s plasma membrane through a Fe2+ iron-regulated transporter (IRT1) or its homologues such as natural resistance-associated macrophage protein 1 (NRAMP1) or divalent metal-ion transporter 1 (DMT1) (Figure 2). Rhizosphere acidification is mainly associated with the release of protons followed by surplus uptake of cations (Fe+) over anions during nitrogen fixation (Sinclair and Krämer, 2012). Membrane recycling of IRT1 is controlled by ubiquitination in strategy I plants (Barberon et al., 2011). In legumes, Fe uptake and transportation to roots are mainly carried out by protein HA2, FRO2, and IRT1 (Walker and Connolly, 2008; Santi and Schmidt, 2009). Putative homologs for the transport of Fe from the leaf to the root through nutrient transporting genes such as FIT1, IRT1, OPT3, and bZIP23 have been identified in many legumes including peanut (AhIRT1; Xiong et al., 2012), Medicago truncatula (MtNRAMP1; Tejada-Jiménez et al., 2015), soybean (NRAMP genes; Qin et al., 2017), lentil (Ferritin-1, BHLH-1, and FER-like transcription factor protein and IRT1), and chickpea (CaFer1; Parveen et al., 2016).

Figure 2. (A) The different transporters in the uptake of nutrients from the soil and their translocation to aerial parts. (B) A schematic representation of mineral transport to roots through different pathways.
Relatively very little is known about Fe uptake, and regulation in legumes shoots (Thomine and Vert, 2013). Fe uptake in shoots is mediated by IRT-like transporters, and its movement in the xylem as ferric-citrate complexes has been observed in soybean (Palmer and Guerinot, 2009). Xylem unloading is a crucial step in the distribution and transportation of Fe to different tissues and sinks cell (Figure 2). Expression patterns show that ZIP transporters and YSL transporters are involved in metal unloading from xylem (Küpper and Kochian, 2010). Oligopeptide transporter (OPT) has been suggested to play a significant role in accurate long-distance Fe signaling from shoots to roots and in importing Fe into phloem companion cells in Arabidopsis (Kumar et al., 2017). Due to the abundance of nicotianamine (NA) in shoot tissues and its affinity to various ions, it can be assumed that YSL transporters are essential for metal transfer from the xylem to the leaves and the seeds, as evident from the expression of Arabidopsis genes AtYSL1 and AtYSL3 that increased during leaf senescence (Waters et al., 2006). NRAMP family genes are known to play a significant role in Fe homeostasis whereas YSL and OPTs play a major role in loading and unloading of Fe2+ NA complexes into and out of phloem (Palmer and Guerinot, 2009). Fe uptake and transportation in plants have been reviewed in several articles (Kobayashi and Nishizawa, 2012; Curie and Mari, 2017).
Zinc (Zn) Transport
Efficient uptake, transport, and accumulation of Zn in seeds are equally crucial for developing nutrient-rich crops (Astudillo et al., 2013). In legumes, Zn is mostly taken up across the plasma membrane of root cells as Zn2+. ZIP transporters have been involved in Zn uptake and transport from root to seeds (Colangelo and Guerinot, 2006; Palmgren et al., 2008). ZRT, IRT-like protein (ZIP), HMA heavy metal ATPase (HMA), Zinc-induced facilitator (ZIF), and metal tolerance protein (MTP) have been involved in Zn transport (Hussain et al., 2004). MTPs play a role in the mobilization of many metal ions such as Zn, Mn, Fe, Ni, Cd, and Co in the cytoplasm. In the case of M. truncatula, MtZIP1, MtZIP3, MtZIP4, MtZIP5, MtZIP6, and MtZIP7 genes were found to be upregulated under Zn deficiency in both roots and leaves, suggesting their active role in Zn transport (Hussain et al., 2004). The bZIP family is another important gene family involved in Zn transport in legumes. Studies in many dicots such as Arabidopsis, soybean (GmZIP1), common bean (PvZIP12, PvZIP13, PvZIP16, and PvbZIP1), Medicago (seven ZIP transporters), and Lotus japonicus have identified ZIP genes in different tissues like roots, leaves, and seeds (Lin et al., 2009; Astudillo et al., 2013). Mostly, Zn is transported through the symplastic pathway, but a considerable fraction may follow the apoplastic pathway through roots to reach the xylem (White et al., 2002; Figure 2). The cation diffusion facilitator (CDF) family members such as MTP1 and ZIF1 transporter play a role in Zn transport to the vacuole while NRAMPs have been identified in Zn mobilization from the vacuole (Haydon and Cobbett, 2007). Zn loading to the xylem is mediated through HMA, while within the xylem, is transverse as Zn2+ or in complex with histidines or Nicotianamine (Palmgren et al., 2008). While ZIP family members are actively involved in mediating Zn2+ influx to leaf tissue and also to the phloem, YSL is involved in loading Zn to the phloem and unloading to the seeds as Zn-NA complex (Haydon and Cobbett, 2007; Waters and Grusak, 2008).
Manganese (Mn) Transport
Manganese is an essential trace element in plants as it serves as a cofactor in many vital processes such as photosynthesis and lipid biosynthesis. Mn is available in the soil as Mn2+ for plant uptake (Figure 2). Very few transporters have been identified exclusively for Mn transport in plants. However, there are many transporters such as NRAMP, YSL, IRT1, CDF/MTP, P-Type-ATPase and VIT (vacuolar iron transporter) (Xia et al., 2010; Socha and Guerinot, 2014) that help in Mn transport. Transporters in Mn have broad specificity for other divalent cations such as Cd, Ca, Co, Zn, Fe, Cu, and Ni. In Arabidopsis, AtNRAMP1 was reported to be a high-affinity transporter for Mn transport in roots, and knockout lines for AtNRAMP1 showed susceptibility toward Mn deficiency (Cailliatte et al., 2010). ZIP1 remobilizes Mn from vacuoles to allow Mn translocation to the shoot through root vasculature (Milner et al., 2013). However, ZIP2 transporters do not seem to be the primary transporters of Mn in roots of many species, including M. truncatula. In the case of field pea and M. truncatula, PsIRT1, MtZIP4, and MtZIP7 genes can reestablish growth to the Mn uptake defective smf1 mutant in Mn-limited media indicating IRT/ZIP as a direct transporter of Mn in strategy I plants (Milner et al., 2013). A subset of cation channels such as Ca2+-permeable channels transport Mn2+ in the apical plasma membrane of Arabidopsis root hairs (Véry and Davies, 2000; Socha and Guerinot, 2014). Involvement of other routes in Mn transport can be plausible because of the presence of many transporters associated with Mn transport even in the absence of vacuolar iron transporter 1 (VIT1).
Phosphorus (P) Transport
Phosphorus uptake of plants from the soil is in the form of phosphate (Pi) either via root epidermal cells impelled through a proton gradient produced by plasma membrane H+-ATPases or with the help of arbuscular mycorrhizal fungi (AMF) found in legumes (Bucher, 2007; Figure 2). Several Pht1 genes are expressed in roots, aerial parts, and seeds, implying their potential involvement in internal Pi translocation. In the case of M. truncatula, Pi-transporters genes (MtPT1 and MtPT2) from the Pht1 family were found to be highly expressed in Pi-deprived roots (Liu et al., 2008). However, only MtPT5 showed high affinity for Pi uptake among the reported five (MtPT1, MtPT2, MtPT3, MtPT4, and MtPT5) Pht1 family genes in M. truncatula (Liu et al., 2008). In L. japonicus, three Pi transporter genes of the Pht1 family have been isolated (Maeda et al., 2006). In the case of soybean, 14 Pht1 genes (GmPT1-GmPT14) were identified in response to Pi availability in various tissues associated with its uptake and translocation (Qin et al., 2012). A high-affinity Pi transporter, GmPT5 helps in maintaining Pi homeostasis by regulating movement from roots to the region of aerial plant tissues in nodules of soybean (Qin et al., 2012). In chickpea, CaPHO1, CaPHO2, CaPHT1;4, CaPAP17, CaPPase4, and CaDGD1 were involved in Pi uptake, transport, allocation, and the mobilization/remobilization from roots and leaves to nodules (Esfahani et al., 2016). Pht1 transporters are mostly involved in transferring Pi into cells while other members of the Pht2, Pht3, and Pht4 families are associated with the transfer of Pi in the intercellular membrane.
Copper (Cu) Transport
Copper uptake from the soil follows similar strategies like Fe, entering the root cell through copper transporters (COPT) family transporter (Gayomba et al., 2013; Ryan et al., 2013). Cu is mostly available in the soil as Cu2+, which is transported to the root cell in its reduced form “Cu+” (Figure 2). Ferric reductase, FRO2, helps in reduction activity and also in Cu+ uptake by roots (Bernal et al., 2012). In Arabidopsis, Cu stress induces high Cu2+ chelate reductase activity regulated by SPL7, and this reductase was encoded by FRO4/5 at the root tips (Bernal et al., 2012; Ryan et al., 2013). After reduction, Cu+ is transported through the roots by copper transporter (COPT) proteins. COPT proteins have not been studied in detail in legumes. However, in Arabidopsis, COPT1 (in roots) and COPT2 (in shoots) are the core uptake transporters whereas COPT3 and COPT5 might be involved in intracellular Cu mobilization (Gayomba et al., 2013). Besides, COPT transporters ZIP2 and ZIP4 are also believed to support Cu uptake in plant cells in Arabidopsis. In Arabidopsis, the cysteine-rich metallothionein proteins (MT proteins) were upregulated during Cu stress, whereas in field pea, MT mRNA levels were mildly upregulated in Cu stress conditions.
Metabolic Pathways for Vitamins (β-Carotene, Folate, and Vitamin E) in Legumes
Understanding the pathways to and rate-limiting steps in the accumulation of various seed nutrients is a major challenge. Initial efforts in developing nutrient-rich crops have focused on overexpression of single genes that affect nutrient biosynthesis/uptake, transport or storage. Various studies have suggested that overexpression of a single gene is not sufficient to increase the accumulation of nutrients in seeds (Ishimaru et al., 2010). Considering the complex nature of nutrient accumulation in plants, multiple genes at different steps of translocation or biosynthetic pathways need to be manipulated simultaneously to increase seed nutrient concentrations. To enhance vitamins’ content in legumes, a cohesive understanding of the genetics of nutritional traits along with a knowledge of regulatory biochemical and molecular processes in the accumulation of nutrients are required (Asensi-Fabado and Munné-Bosch, 2010; Bhullar and Gruissem, 2013). A brief description of vitamins such as β-carotene, folate, tocopherol and anti-nutritional components such as phytic acid and raffinose biosynthesis are discussed below.
Beta (β)-Carotene Biosynthesis
Plant carotenoids are the generic name for C40 tetraterpenoids with a conserved biosynthetic pathway that play a significant role in different processes including photosynthesis (DellaPenna and Pogson, 2006). There are two major groups of carotenoids; the first is oxygenated or xanthophyll that consists of lutein, violaxanthin, and neoxanthin, and the second is non-oxygenated or carotenes that include β-carotene and lycopene (DellaPenna and Pogson, 2006). Seeds of legumes are rich in carotenoids such as β-carotene, cryptoxanthin, lutein, and zeaxanthin (Abbo et al., 2005). For instance, β-carotene concentration in chickpea was higher than in genetically engineered “golden rice” endosperm but lower than in Golden Rice2, where β-carotene concentration was increased up to 23-fold (Abbo et al., 2005).
In legumes, plastid-confined MEP (2-C-methyl-D-erythritol 4-phosphate) pathway produces carbon flux, which is used for carotenoid biosynthesis (Giuliano, 2014). Carotenoid concentration is a highly heritable trait which is least affected by the environment (Owens et al., 2014). Identifying the metabolic bottlenecks associated with the carotenoid pathway can help in modifying strategies to develop carotenoid-rich crops. The key regulator gene of the carotenoid pathway is PSY; the overexpression of this gene or phytoene desaturase gene individually or a in combination has been practiced in several crops including soybean (Schmidt et al., 2015). In soybean, a 1500-fold increase in β-carotene content in dry seeds was observed compared to wild-type by introducing a chimeric gene from pea and a crtB gene from bacterium Pantoea using a biolistic method (Schmidt et al., 2015). In chickpea, four members of the PSY family that might have a positive effect on carotenoid concentration for various cotyledon colors were reported. A total of 32 genes for isoprenoid and carotenoid pathways in chickpea distributed across all eight chromosomes were also identified (Rezaei et al., 2016). Phytoene synthase and desaturase were found to have a major impact on pro-vitamin A and total carotenoid concentration through genetic transformation or overexpression of these genes. Xanthophylls are produced by converting pro-vitamin A compound with the help of β-carotene hydroxylation and can help in developing cultivars with higher pro-vitamin A as seen in potato, where silencing of β-carotene hydroxylase increased β-carotene concentration (da Silva Messias et al., 2014). Lutein, one of the main carotenoid types in chickpea, showed higher concentration in desi compared to kabuli type and was found to be adversely associated with seed weight (Abbo et al., 2005; Ashokkumar et al., 2014). Carotenoid concentration was higher in genotypes with green cotyledons in both pea and chickpea; a similar trend for lutein was observed in pea. Similarly, in transgenic soybean, increased concentration of β-carotene and seed protein content, with a decreased level of abscisic acid in cotyledons by overexpressing a seed-specific bacterial phytoene synthase gene was observed (Schmidt et al., 2015).
Folate Biosynthesis
Folates (Tetrahydrofolate and derivatives) are water-soluble B vitamins that act as cofactors in many vital metabolic functions, including the metabolism of amino acids, biosynthesis of nucleic acids in the human body. Legumes are a rich source of folates. A high concentration has been estimated in chickpea (351–589 μg/100 g), common bean (165–232 μg/100 g), and lentil (136–182 μg/100 g), (Blancquaert et al., 2014; Jha et al., 2015). Plants are the only source of folate for humans as the human body cannot synthesize it. Folate biosynthesis takes place in three subcellular compartments. Firstly, the Pterin and pABA moieties are synthesized in cytosol and plastids, respectively, while the rest of the reactions take place in the mitochondria. Pterin moiety synthesizes by converting GTP into dihydroneopterin triphosphate and formate with the help of GTP cyclohydrolase-I (Hossain et al., 2004). In legumes, pABA is synthesized from chorismate through two reactions in plastids. In mitochondria, after pyrophosphorylation of 6-hydroxymethyldihydropterin (HMDHP), it combines with pABA to form dihydropteroate with the help of enzymes HMDHP pyrophosphokinase and dihydropteroate synthase. After this reaction, glutamate residue is combined with the carboxy part of the pABA moiety of dihydropteroate to produce dihydrofolate with the help of enzyme dihydrofolate synthetase. Finally, folate is formed by the attachment of a glutamate tail to THF molecule catalyzed by dihydrofolate reductase.
Considering the complex nature of folate biosynthesis, metabolic engineering has emerged as a better approach to increase folate concentration in plants, such as by the overexpression of genes involved in pterin biosynthesis, a folate biosynthesis precursor (Hossain et al., 2004; Storozhenko et al., 2007; Blancquaert et al., 2014). Around a 150-fold increase in biosynthetic pteridines was reported in transformed lines of the common bean by introducing GTP cyclohydrolase I from Arabidopsis in three cultivars by particle bombardment (Rivera et al., 2016).
Vitamin E Biosynthesis
Tocopherol and tocotrienol derivatives are collectively called vitamin E. Improvement for vitamin E mostly focuses on enhancing vitamin E content in edible parts by regulating the activity of various enzymes involved in different steps of the synthesis, such as p-hydroxyphenylpyruvate dioxygenase, homogentisate phytyltransferase, homogentisate geranylgeranyl transferase, homogentisate solanesyltransferase2-methyl-6-phytyl-benzoquinol methyltransferase, tocopherol cyclase, and γ-tocopherol methyltransferase (Tang et al., 2016). Overexpression of γ-TMT resulted in an increased proportion of α-tocopherol in soybean (Sattler et al., 2004; Tavva et al., 2007) while overexpression of both MT and γ-TMT increased α-tocopherol 5-folds in soybean (Tavva et al., 2007). Overexpression for the combination of tyrA, HPPD, GGPP reductase and HPT resulted in an 11-fold increase in vitamin E content in soybean (Karunanandaa et al., 2005).
Metabolic Pathways of Anti-nutrients (Phytic Acid and Raffinose)
Phytic acid binds to mineral cations to form a mixed salt called phytate and sequesters inorganic phosphate in legumes. Myo-inositol is the precursor for many metabolites, including phytate, which plays an important role in plant stress adaptation. In addition to stress response, phytate plays a major role during seed germination to develop embryos and defense against oxidative stress. Considering its anti-nutritional role, breeding and transgenic approaches were used to reduce phytic acid in legumes (see Panzeri et al., 2011; Joshi-Saha and Reddy, 2015). In common bean, genes PvMIPSs and PvMIPSv (coding for myo-inositol 1phosphate), PvIMP (inositol monophosphatase), PvMIK (myo-inositol kinase), PvIPK2 (inositol 1,4,5-tris-phosphate kinase), PvITPKa and PvITPKb (inositol 1,3,4-triphosphate 5/6-kinase), and PvIPK1 (inositol 1,3,4,5,6 pentakisphosphate 2-kinase) have been identified and mapped on a reference genetic map through virtual mapping strategy (Fileppi et al., 2010). In common bean, a low phytic acid line (lpa1) 280-10 was selected and used for the identification of Mrp1 gene that down-regulates the phytic acid pathway at the transcriptional level (Panzeri et al., 2011). lpa mutants have also been identified in other legumes such as field pea and soybean using EMS-based mutagenesis (Warkentin et al., 2012). In chickpea, CaMIPS2 gene was found to be regulating the phytic acid biosynthesis pathway (Kaur et al., 2008). In soybean, identification of consistent metabolic changes in lpa mutants showed decreased content of myo-inositol and raffinose compared to the wild type and reported a significant role in reducing phytic acid (Frank et al., 2009). Silencing expression of multidrug resistance-associated protein (MRP) ATP-binding cassette (ABC) transporters in an embryo-specific manner resulted in low phytic acid and high inorganic phosphate in transgenic maize and soybean (using homologous soybean MRP gene) (Shi et al., 2007).
Raffinose is another major anti-nutrient affecting plant nutrition potential. In chickpea, raffinose content varied from 0.38 g/100 g to 0.99 g/100 g, while stachyose content ranged from 0.79 g/100 g to 1.87 g/100 g. Synthesis of galactinol is a key requirement for entering into the pathway of the raffinose family of oligosaccharides (RFO) biosynthesis. The key enzyme galactinol synthase synthesizes galactinol using UDP Galactose. Raffinose synthase helps to synthesize raffinose, and stachyose synthase helps to produce tetrasaccharide stachyose by utilizing galactinol, and both these reactions are reversible.
Understanding interactions between micronutrients, such as the synergic effect of Fe and pro-vitamin A carotenoids or the competitive effect of Fe and Zn and bioconversion factors, are essential for the development of nutrient-rich crops. Bioavailability of nutrients depends on endogenous (phytic acid, fiber, amino acids, and proteins) and exogenous factors in seeds. Legumes contain some promoters that enhance the bioavailability of minerals, even in the presence of anti-nutrients. Some promoter compounds are natural plant metabolites, and only minor changes in its accumulation in seeds may be necessary to impact the bioavailability of micronutrients. Inulin is a fructooligosaccharide found in small amounts in raw samples of lentil, chickpea, red kidney bean, common white bean, white bean and faba bean (Rastall and Gibson, 2015). It has a significant positive effect on improving the bioavailability of mineral nutrients in legumes.
Further studies are required to understand the types and amounts of prebiotics concerning in relation to increased bioavailability of minerals. Nicotianamine levels in plants have also shown a positive effect on enhancing Fe concentrations in seeds. Breeders should focus on enhancing the level of promoters such as inulin, β-carotene, histidine, lysine, riboflavin, and selenium, which can increase the bioavailability of Ca, Fe, Zn, Mg, and I (White and Broadley, 2005).
Agricultural Interventions Through Biofortification
Biofortification is the most sustainable approach to increase nutrient concentration and bioavailability in staple food crops. It refers to the procedure of improving the concentration of essential minerals, vitamins, essential amino acids, and fatty acids and reduces anti-nutritional factors enabling nutrient bioavailability in crop plants (Garcia-Casal et al., 2017). Biofortification approaches include the application of fertilizer to the soil or leaves, plant breeding, and genetic engineering (genetic modification and transgenesis) (Figure 3). It is the most economical and cost-effective way to provide nutrient-rich food to most vulnerable people and gives better yield and profit to farmers (Garcia-Casal et al., 2017).
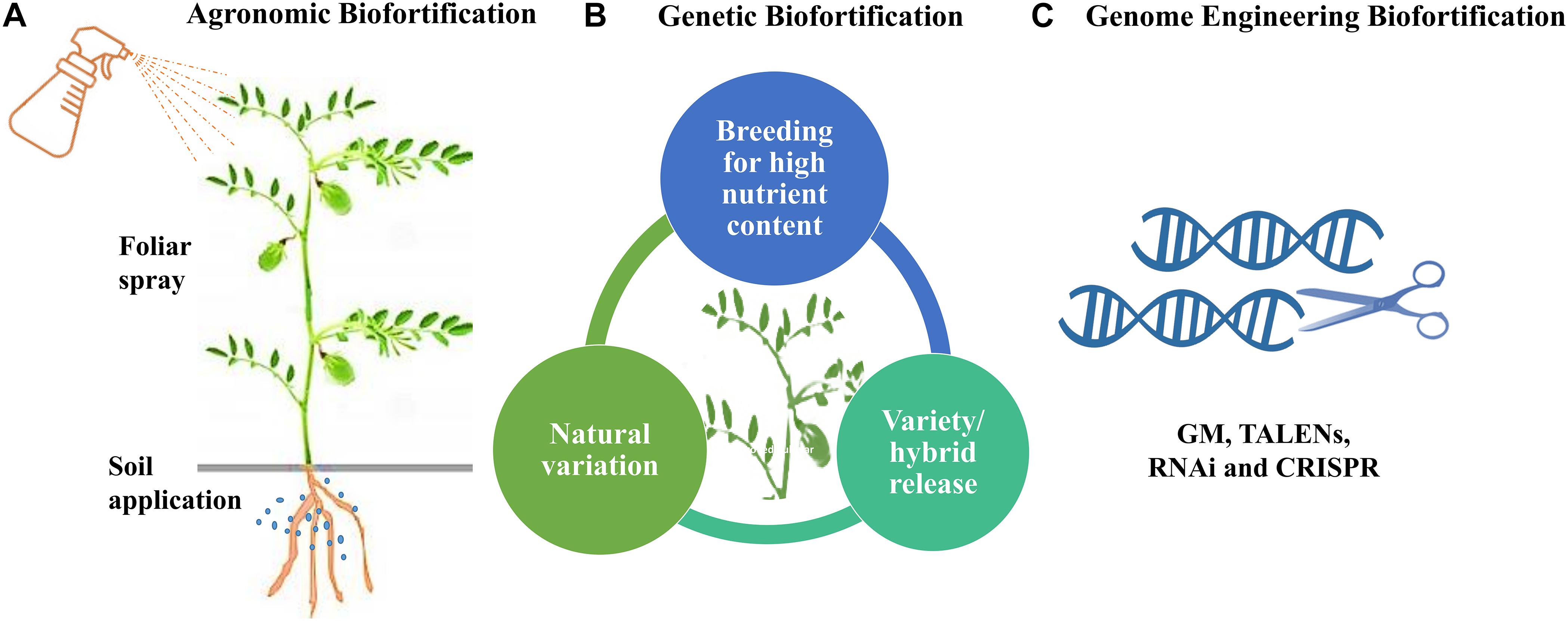
Figure 3. The three approaches for biofortification. (A) Agronomic Biofortification using soil and foliar spray. (B) Genetic Biofortification through breeding using conventional and genomics-assisted breeding. (C) Genome Engineering Biofortification including GM and DNA alteration technologies such as TALENs, RNAi and CRISPR.
Agronomic Biofortification
Fertilizer application for macronutrients (N, P, K, and S) and micronutrients (Zn, Ni, I, Co, Mo, and Se) have a significant impact on the accumulation of nutrients in seeds compared to other micronutrient fertilizers such as Fe that have limited phloem sap mobility. The concentration of minerals in the seed and cotyledon can be increased by optimizing the rate and timing of foliar application and using an approach that combines the application of soil and foliar spray to achieve a higher concentration of grain minerals. During foliar application, the minerals get absorbed by the leaf epidermis and then transported to sink via the xylem and phloem. Increasing the available soil concentration of Zn, Ni, I, and Se can significantly increase their concentrations in seeds, as confirmed by a study on pea and navy beans. Accumulation of Zn in the seed of field pea was positively influenced by the combined foliar application of Se and Zn. Se and I concentrations were improved in edible parts through the combined foliar application with increased Se and I (Poblaciones and Rengel, 2017). However, in the case of Fe, fertilization could not affect the mineral content of the seed. Application of fertilizer either in the soil or through foliar spray is a temporary solution compared to breeding approaches. Therefore, fertilizer application can be one of the effective ways to improve the concentration of nutrients in edible parts in combination with breeding or transgenic approaches.
Genetic Biofortification Through Breeding
Genetic biofortification includes the application of plant breeding techniques to produce crops with higher micronutrient content, moderate to low levels of anti-nutrients, and increased levels of substances that promote nutrient absorption (Bouis, 2003). Breeding approaches have great potential to increase micronutrient density by exploring the existing genetic variation to develop nutrient-rich crop varieties. Considering its sustainability and no regulatory and political restrictions, biofortification through breeding seems to be the most suitable approach for biofortification (Saltzman et al., 2017).
In order to develop a legume genetic biofortification program, the first step involves setting a target micronutrient level for each crop. Among the factors that affect genetic biofortification are available genetic variability and information about genes that control the absorption of the element by roots, translocation to shoots, mobilization in different vegetative parts, and deposition of the element in the edible parts in utilizable forms (Bouis and Welch, 2010). Besides, there are various environmental factors and cultural practices that can affect element accumulation in ds, and dietary factors that affect the absorption and utilization of minerals by the consumer (Bouis and Welch, 2010). Most biofortified products in Asia, Africa, and Latin America have been produced using breeding, while other technologies to develop biofortified products are under development (Garcia-Casal et al., 2017). HarvestPlus2 works with several CGIAR and National Agricultural Research centers from Africa, Asia and Latin America to develop and promote high nutrition content biofortified food crops. According to the HarvestPlus Annual Report (2015), several biofortified crops yellow cassava, orange flesh sweet potato with high levels of β-carotene (over 200 mg/g), iron beans (50–70% more iron content), orange maize, iron pearl millet, zinc rice and zinc wheat] developed through breeding have been released officially in more than 30 countries and are in the testing stage in more than 50 countries. Several studies have reported the efficacy of these released biofortified varieties in improving micronutrient deficiency among target populations (De Moura et al., 2014; Finkelstein et al., 2017).
Micronutrients constitute a very small portion of the total weight of a grain legume; therefore, precision estimation is a pre-requisite for effectively assessing genetic variation for breeding with stable and high element concentration. However, quick, accurate, and inexpensive methods for identifying nutrient-dense genotypes are yet to be identified. If wild relatives are found to be the source of micronutrients, pre-breeding approaches can be used to develop the parent for genetic biofortification. Transgressive segregation of heterosis can be exploited to create a genetic variation for the target nutrient trait in case it is difficult to achieve this through selection (Bouis and Welch, 2010). Any breeding program requires an understanding of the genetics of the target trait in order to select the parental line and breeding method. In addition, clarity on the correlation between nutritional traits and yield and yield-related traits will aid the selection of nutrient-rich lines with higher yields and desired traits (Bouis and Welch, 2010). For instance, a positive correlation between Fe and Zn content in seeds of common bean, peanut, mung bean, wheat, pearl millet, maize, and sesame indicates the scope to simultaneous improve these two traits (Cichy et al., 2009; Pixley et al., 2011; Velu et al., 2012). In the case of chickpea, negative correlation between Zn and grain yield across locations was reported, while a significant negative correlation of Fe with grain yield was observed at one location (Diapari et al., 2014).
Genome Engineering Biofortification
Transgenic approaches are necessary and even advantageous in comparison to breeding in the absence of natural variation in the gene pool of the target crop (Al-Babili and Beyer, 2005). The best example of genome engineering for the nutritional trait is “Golden Rice,” where an advanced transgenic line having 37 mg/g carotenoid was developed (Al-Babili and Beyer, 2005). Despite the positive side of transgenics, researchers have raised concerns related to allergies or intolerance associated with bioengineered or genetically modified crops along with the environmental side effects, and reduced biodiversity (Maghari and Ardekani, 2011; Raman, 2017).
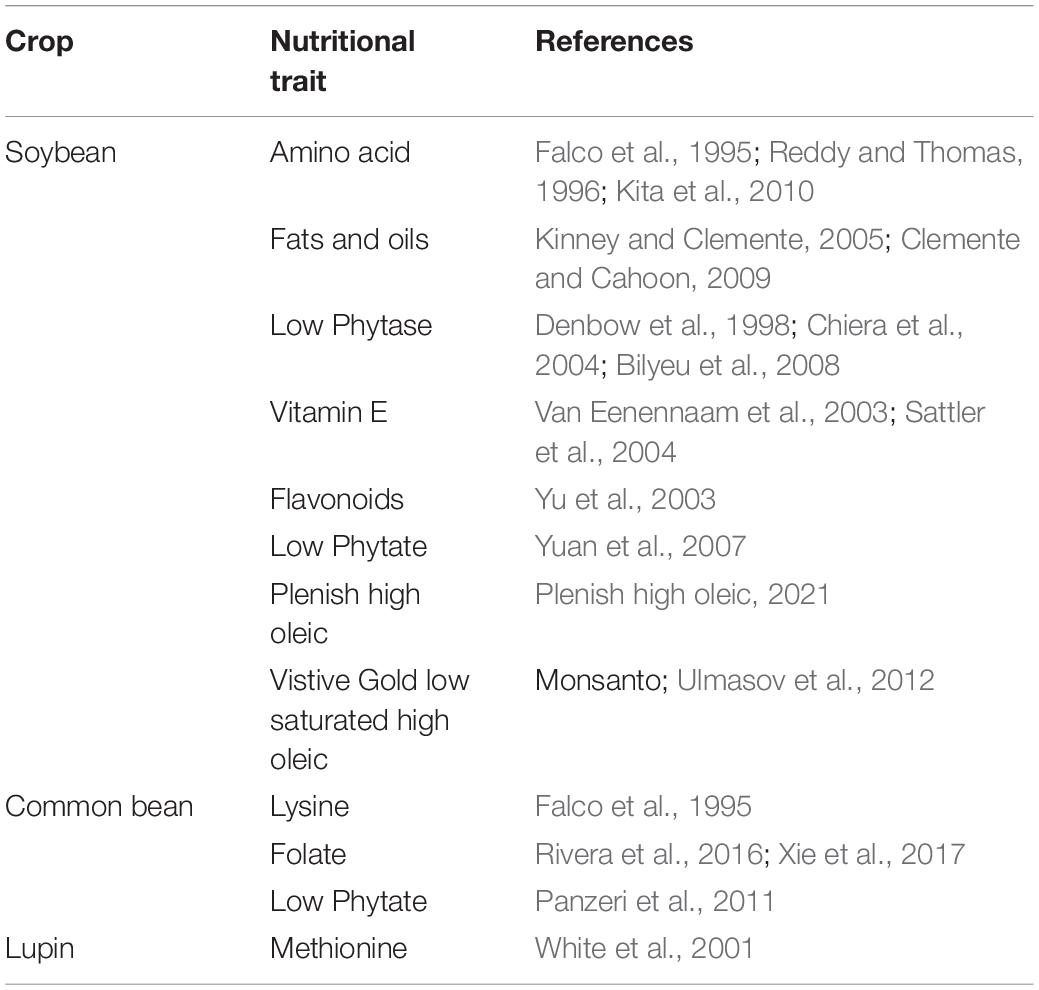
Recent advances in molecular biology have significantly changed the mutagenesis platforms for more targeted and accurate DNA alterations through transcription activator-like effector nucleases (TALENs), zinc-finger nucleases (ZFNs), and short palindromic repeat (CRISPR)–associated protein (Cas9) in legumes and other crops (Curtin et al., 2011; Haun et al., 2014; Michno et al., 2015; Sun et al., 2015; Tang et al., 2016). New technologies like TALENs, ZFNs, RNA interference (RNAi), and CRISPR/Cas9 need to be utilized for the improvement of nutritional traits in legumes. Of the different genetic engineering technologies, RNAi seems to have the edge over others as it is an advanced specific gene silencing technology and a very powerful innovation that can help to develop nutritionally rich and anti-nutrient low crops (Tang and Galili, 2004). RNAi technology has already been used to reduce the level of BOAA in grass pea, to reduce the content of Arah2, an allergen, by 25% in crude peanut extract (Dodo et al., 2008), and develop peanut oils having novel combinations of oleic acid content. RNAi has also been used to generate resistant common bean lines to Beans Golden Mosaic Virus (Bonfim et al., 2007). Suppression of SACPD gene through RNAi has increased resistance to several pathogens in soybean (Jiang et al., 2009). RNAi interventions targeting genes associated with lignin production resulted in enhanced resistance of soybean to Sclerotinia sclerotiorum because of reduced lignin concentration (Peltier et al., 2009). The RNAi approach has also helped in improving oleic acid in soybean. Apart from legumes, it has also been used to improve nutritional quality in maize, wheat, rice, cotton, jute, and tomato (Kusaba et al., 2003; Davuluri et al., 2005). Table 3 summarizes the list of genetic modifications that have taken place so far in legume biofortification.
Genomics Approaches to Nutritional Breeding
Genetic biofortification efforts through breeding methods have been partially effective in addressing the challenge of low nutrient content, though not to the extent desired. Therefore, it is essential to exploit the potential of genomics to accelerate the development of nutrition-rich improved cultivars. Details about genetic and genomic resources for important legumes have been extensively reviewed (Pandey et al., 2016; Bevan et al., 2017; Varshney et al., 2018; Roorkiwal et al., 2020). In the recent past, advancements in next-generation sequencing (NGS) technologies have led to a drastic reduction in cost and thereby resulted in making available genomic sequence for major legumes, enabling NGS-based methods for allele mining, candidate genes identification, and high-resolution genetic mapping. Though cost-effective genotyping platforms are available for deploying genomics-assisted breeding (GAB) in major legumes, the cost of high throughput and efficient estimation of nutrients poses a major challenge. The plant genome sequence offers an opportunity to dissect and understand the mechanism for functional characterization of genes involved in nutrient uptake and mobilization. Among legumes, the genome sequence of pigeon pea (Varshney et al., 2012), chickpea (Varshney et al., 2013), peanut (Bertioli et al., 2016; Chen et al., 2016), lupin (Hane et al., 2017), soybean (Schmutz et al., 2010), and common bean (Schmutz et al., 2014) have been completed and can provide the foundation for deploying genomics in legume breeding by detecting the genes responsible for nutritional traits.
Exploiting Genetic Variation of Micronutrients in Legumes
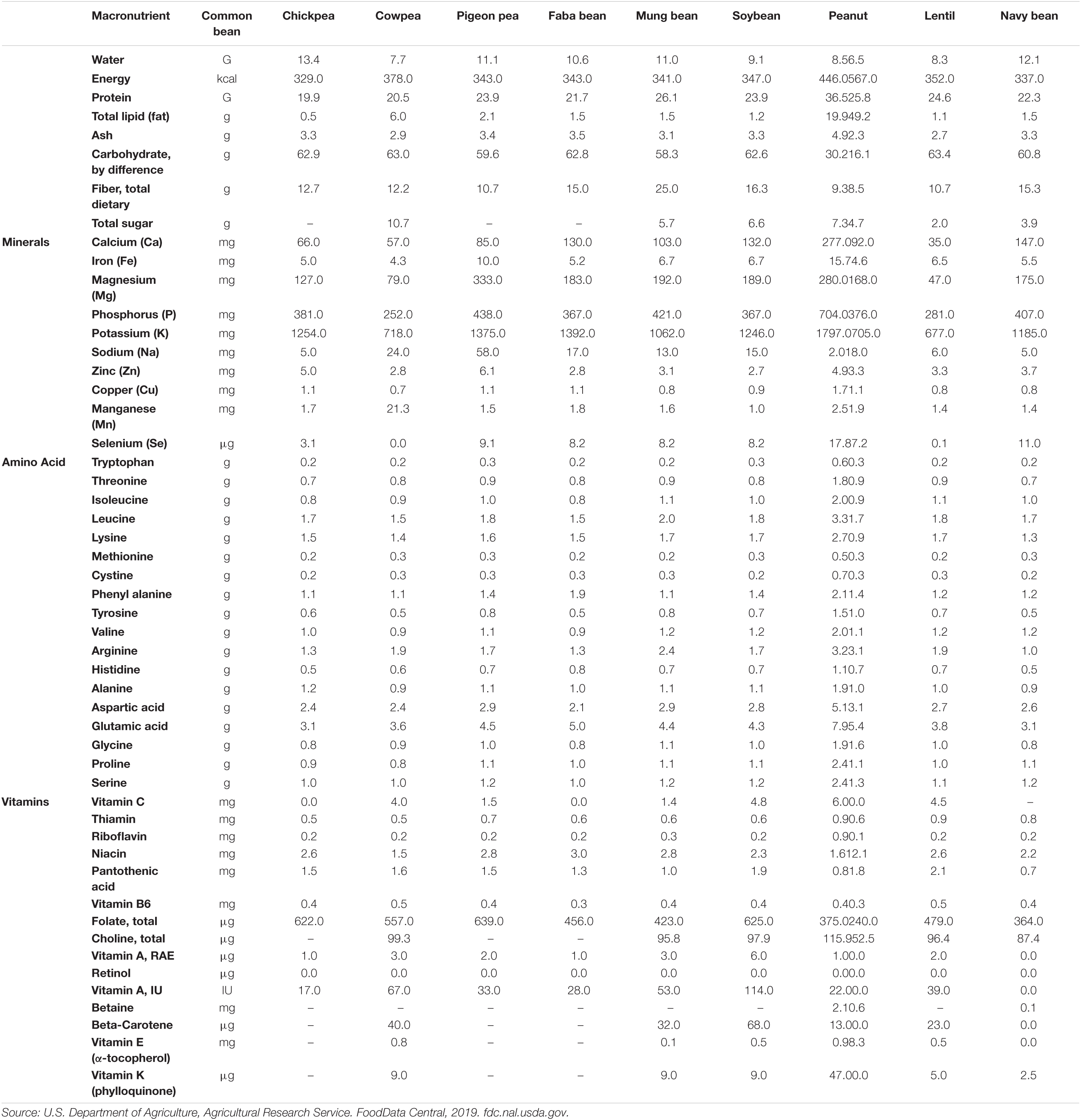
Screening of diverse germplasm is a pre-requisite to understanding the genetic variation for a trait of interest that can be used for breeding to increase the availability of that particular element (McCouch et al., 2013). Genetic variation enables a breeder to exploit heterosis, additive gene effects, and transgressive segregation to improve micronutrient concentration. When the required genetic variation is not available, transgenic approaches can provide additional sources of variation (Francis et al., 2017). Legumes are considered a rich source of nutrients and possess huge variation in the legume germplasm (Table 4). Generally, inductively coupled plasma-Mass spectrometry (ICP-MS) is utilized for mineral estimation; however, it requires expensive equipment, a skilled analyst, and extensive sample preparation. The colorimetric approach that has been used to measure minerals is semi-quantitative and laborious when applied for large-scale screening. In atomic absorption spectrometry (AAS), free atoms absorb light in the form of optical radiation for the quantitative detection of elements present in a sample. X-ray fluorescence spectroscopy (XRF) is also a consistent, high throughput, low-cost system to determine element concentrations in samples; it is classified as being either energy dispersive (EDXRF) or wavelength dispersive (WDXRF) (Singh et al., 2013).
Identification of QTLs/Genes to Interpret Genetic Architecture Concerning Nutrient Accumulation
Linking genetic data with data on nutrition content is an advanced and accurate approach to identify quantitative trait loci (QTLs) associated with a trait of interest. Nutritional profiling of genotypically characterized diverse set of germplasm (core collection, mini-core collection, a reference set, composite set) can link genetic data to global mineral nutrition (Ghandilyan et al., 2009; Norton et al., 2010). Recently, QTL mapping has been widely used to associate genetic variation with phenotypic variation and provide a reliable tool for gene discovery. The associated region or linked region (i.e., genetic markers) identified through QTL mapping can then be isolated or cloned for identification and analysis of the genes concerned. Molecular mapping of the genome segments that govern nutrient content/concentration has been done in many legumes (Table 5). Studies on understanding genes and processes to improve seed nutritional composition by identifying QTLs were limited to a few nutrients in legumes. For instance, QTLs for seed element concentration has been identified in L. japonicus (Klein and Grusak, 2009), M. truncatula (Sankaran et al., 2009), common bean (Blair et al., 2009, 2010, 2011; Cichy et al., 2009; Casañas et al., 2013), soybean (Zhang et al., 2009; Jegadeesan et al., 2010; Ramamurthy et al., 2014), chickpea (Sab et al., 2020), and lentil (Aldemir et al., 2017). Most of the studies conducted so far to map and tag the gene(s)/QTL(s) controlling micronutrient status in legumes were mostly found to have a quantitative mode of inheritance (Blair et al., 2010). For instance, in the case of Proteus vulgaris, two genes (PvIRT1 and PvIRT2) on chromosome-3 and two genes (PvbZIP2 and PvbZIP3) on chromosome-11 were aligned with QTLs for Fe and Zn (Jiang et al., 2008). The list of identified QTLs in several legumes for various nutritional traits has been presented in Table 5.
In addition to conventional bi-parental mapping populations, efforts have also been made to exploit the available genetic variation for nutrient factors using genome-wide association studies. This approach has been used to identify markers associated with various key nutrition factors in common bean (Katuuramu et al., 2018; Caproni et al., 2020) and chickpea (unpublished). These identified genes/QTLs, after validation, may be deployed in to develop nutrient-rich legumes.
Prospects of the Role of Genomics in Nutritional Breeding
Next-generation sequencing-based genotyping technologies can be employed to understand the genetics of nutritional traits using precise marker-trait association (MTA), gene discovery, and functional marker development. Their potential has been proven for various agronomic traits in genetic mapping, marker-assisted selection (MAS), and genomic selection (GS) (Varshney et al., 2014, 2019). GAB approaches such as marker-assisted backcrossing (MABC) and marker-assisted recurrent selection (MARS) can be used for the improvement of single or multiple nutritional traits. Considering their higher cost, difficulty in estimation and the complex genetic mechanism controlling nutritional traits, deploying GS could be beneficial.
Next-generation sequencing-based high-density genotyping methods such as genotyping by sequencing (GBS) and whole-genome re-sequencing (WGRS) enable the identification of large-scale genome-wide SNPs for high resolution genetic and association mapping. For instance, in chickpea, kabuli reference genome and de novo-based GBS assays were used to identify high-quality SNPs for seed Fe and Zn content from 92 desi and kabuli chickpea accessions (Upadhyaya et al., 2016). Similarly, WGRS data on 300 lines from a chickpea reference set (Varshney et al., 2019) along with nutrient content estimation data is being used to identify markers associated with several key nutrient elements (unpublished data). Furthermore, 3000 lines from the global chickpea composite collection are being studied for micro- and macro-nutrient traits and re-sequenced in parallel to identify novel alleles associated with different nutrients (Varshney, 2016). In addition, recently popularized sequencing-based mapping approaches such as “QTL-Seq,” “MutMap,” “Seq-BSA,” “Indel-Seq,” and “Bulked segregant RNA-Seq (BSR-Seq)” can be adopted for mapping nutritional traits. Unique functional allelic variations selected from candidate genes were found to be linked with seed Fe and Zn concentrations in chickpea (Diapari et al., 2014). In the case of soybean, three candidate genes related to seed Fe and Zn storage in maturing seeds have been identified (Liu et al., 2011). In lentil, two SNP markers closely associated with seed Fe and Zn concentrations have been identified (Khazaei et al., 2017).
In addition to trait mapping, transcriptome sequencing has emerged as an alternative to genome sequencing for targeted expressed gene sequencing. Transcriptome sequencing provides an understanding of gene function and the molecular basis of various components related to nutrient mobilization in crops. Identification of candidate genes associated with nutritional traits is plausible from gene expression profiling data of transcriptome assemblies (Pandey et al., 2016). Expression pattern studies in several legume crops have identified genes involved in nutrient mobilization (Küpper and Kochian, 2010; Conte and Walker, 2011). The emerging and promising areas of proteomics that includes proteome mapping, comparative proteomics, post-translational modification, and protein-protein interaction could assist in future nutritional breeding programs (Pandey et al., 2016; Roorkiwal et al., 2020).
Furthermore, metabolomics-assisted breeding can greatly supplement the present breeding strategy for nutritional traits (Hossain et al., 2004; Storozhenko et al., 2007; Fernie and Schauer, 2009; Blancquaert et al., 2014). A complete study of metabolites is required to dissect the genetic basis of metabolic diversity in legumes. Several studies on plant metabolites have been carried out in crops like Arabidopsis, rice, and maize (Keurentjes et al., 2006; Schauer et al., 2006; Chan et al., 2010). The information on the genetic and molecular bases of natural variation in legume metabolomes is still limited. Metabolic profiling for phenylpropanoid and isoflavonoid biosynthesis in Medicago has been reported (Farag et al., 2008). The combination of metabolomics with transcriptomics, high-throughput phenotyping, and bioinformatics tools will enable the detection of candidate genes for nutritional traits.
Along with the study of metabolomics, “ionome” profiling is equally important to gain deeper insights into a physiological mechanism related to nutrient accumulation in seeds (Salt et al., 2008). To estimate mineral/micronutrients and their complex networks, ionomics has emerged as a potential area that enables genome-wide understanding of the dynamics of element accumulation in living systems (Baxter, 2010). It helps identify transporters, sensors, and other components that control the expression of metal transport proteins in legumes (Lahner et al., 2003). Ionome also assists in providing information about gene networks regulating various developmental and physiological processes related to the “ionome” of an individual and ultimately leading to the identification of potential candidate genes involved in element uptake, transport, and storage. Identified genes can be incorporated to develop nutrient-rich crops either through genetic modification or molecular breeding. Details about plant ionome have been extensively reviewed (Salt et al., 2008; Baxter, 2010; Huang and Salt, 2016). To sum up, an integrated approach that combines genomics with proteomics and metabolomics has the potential to identify the true candidate that can be directly deployed using GAB to develop nutrient-rich legume varieties.
Conclusion
The incredible advances in plant nutritional genomics provide effective and long-term solutions to the increasing problem of malnutrition. Efforts should be dedicated to identifying candidate genes using MTA and validation and understanding the genetic mechanism of nutrient uptake in crops. Modern breeding techniques like MAS and GS must be used to develop superior nutritionally rich genotypes. Many other modern technologies such as cisgenesis or intragenesis, RNAi, novel DNA editing technologies such as site-directed mutagenesis, and oligonucleotide-directed changes could be deployed to accelerate the process of varietal development. The focus should be not just on identifying nutrient-rich genotypes but also on the bioavailability of the target nutrient. Therefore, joint research efforts from breeders, biotechnologists, physiologists, and nutritionists are required to support and accelerate biofortification programs in legumes.
Author Contributions
RKV conceived the idea and provided critical inputs to the concept. MR and SP drafted sections of the manuscript and prepared figures. DT, RH, and RKV made a critical revision of the content of the manuscript. All authors contributed to the final reading and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank the Department of Biotechnology (DBT), Government of India and the Bill & Melinda Gates Foundation (OPP1114827), United States, for the partial financial support. MR, SP, and RKV acknowledge the funding support from the Department of Science and Technology, Government of India, through the INSPIRE Faculty Scheme and Early Career Research Award–SERB, National Post-Doctoral Fellowship and the JC Bose National Fellowship, respectively.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.682842/full#supplementary-material
Supplementary File 1 | Recommended Dietary Allowances (RDAs) for various nutrients.
Footnotes
References
Abbo, S., Molina, C., Jungmann, R., Grusak, M. A., Berkovitch, Z., Reifen, R., et al. (2005). Quantitative trait loci governing carotenoid concentration and weight in seeds of chickpea (Cicer arietinum L.). Theor. Appl. Genet. 111, 185–195. doi: 10.1007/s00122-005-1930-y
Al-Babili, S., and Beyer, P. (2005). Golden Rice – five yrs. on the road–five yrs. to go? Trends Plant Sci. 10, 565–573.
Aldemir, S., Ates, D., Temel, H. Y., Yagmur, B., Alsaleh, A., Kahriman, A., et al. (2017). QTLs for iron concentration in seeds of the cultivated lentil (Lens culinaris Medic.) via genotyping by sequencing. Turk J. Agric. 41, 243–255. doi: 10.3906/tar-1610-33
Asensi-Fabado, M. A., and Munné-Bosch, S. (2010). Vitamins in plants: occurrence, biosynthesis and antioxidant function. Trends Plant Sci. 15, 582–592. doi: 10.1016/j.tplants.2010.07.003
Ashokkumar, K., Tar’an, B., Diapari, M., Arganosa, G., and Warkentin, T. D. (2014). Effect of cultivar and environment on carotenoid profile of pea and chickpea. Crop Sci. 54, 2225–2235. doi: 10.2135/cropsci2013.12.0827
Astudillo, C., Fernandez, A. C., Blair, M. W., and Cichy, K. A. (2013). The Phaseolus vulgaris ZIP gene family: identification, characterization, mapping, and gene expression. Front. Plant Sci. 4:286.
Barberon, M., Zelazny, E., Robert, S., Conéjéro, G., Curie, C., Friml, J., et al. (2011). Monoubiquitin-dependent endocytosis of the iron-regulated transporter 1 (IRT1) transporter controls iron uptake in plants. Proc. Natl. Acad. Sci. U. S. A. 108, E450–E458.
Baxter, I. (2010). Ionomics: The functional genomics of elements. Brief Funct. Genomics. 9, 149–156. doi: 10.1093/bfgp/elp055
Beebe, S. E., Rojas-Pierce, M., Yan, X., Blair, M. W., Pedraza, F., Muñoz, F., et al. (2006). Quantitative trait loci for root architecture traits correlated with phosphorus acquisition in common bean. Crop Sci. 46, 413–423. doi: 10.2135/cropsci2005.0226
Bellaloui, N., Smith, J. R., and Mengistu, A. (2017). Seed nutrition and quality, seed coat boron and lignin are influenced by delayed harvest in exotically-derived soybean breeding lines under high heat. Front. Plant Sci. 8:1563.
Bernal, M., Casero, D., Singh, V., Wilson, G. T., Grande, A., Yang, H., et al. (2012). Transcriptome sequencing identifies SPL7-regulated copper acquisition genes FRO4/FRO5 and the copper dependence of iron homeostasis in Arabidopsis. Plant Cell. 24, 738–761. doi: 10.1105/tpc.111.090431
Bertioli, D. J., Cannon, S. B., Froenicke, L., Huang, G., Farmer, A. D., Cannon, E. K., et al. (2016). The genome sequences of Arachis duranensis and Arachis ipaensis, the diploid ancestors of cultivated peanut. Nat. Genet. 48, 438–446.
Bevan, M. W., Uauy, C., Wulff, B. B., Zhou, J., Krasileva, K., and Clark, M. D. (2017). Genomic innovation for crop improvement. Nature 543, 346–354.
Bhullar, N. K., and Gruissem, W. (2013). Nutritional enhancement of rice for human health: the contribution of biotechnology. Biotechnol. Adv. 31, 50–57. doi: 10.1016/j.biotechadv.2012.02.001
Bilyeu, K. D., Zeng, P., Coello, P., Zhang, Z. J., Krishnan, H. B., Bailey, A., et al. (2008). Quantitative conversion of phytate to inorganic phosphorus in soybean seeds expressing a bacterial phytase. Plant Physiol. 146, 468–477. doi: 10.1104/pp.107.113480
Blair, M. W., Astudillo, C., Grusak, M., Graham, R., and Beebe, S. (2009). Inheritance of seed iron and zinc content in common bean (Phaseolus vulgaris L.). Mol. Breed. 23, 197–207. doi: 10.1007/s11032-008-9225-z
Blair, M. W., Astudillo, C., Rengifo, J., Beebe, S. E., and Graham, R. (2011). QTL for seed iron and zinc concentrations in a recombinant inbred line population of Andean common beans (Phaseolus vulgaris L.). Theor. Appl. Genet. 122, 511–523. doi: 10.1007/s00122-010-1465-8
Blair, M. W., and Izquierdo, P. (2012). Use of the advanced backcross-QTL method to transfer seed mineral accumulation nutrition traits from wild to Andean cultivated common beans. Theor. Appl. Genet. 125, 1015–1031. doi: 10.1007/s00122-012-1891-x
Blair, M. W., Knewtson, S. J. B., Astudillo, C., Li, C. M., Fernandez, A. C., and Grusak, M. (2010). Variation and inheritance of iron reductase activity in the roots of common bean (Phaseolus vulgaris L.) and association with seed iron accumulation QTL. BMC Plant Biol. 10:215. doi: 10.1186/1471-2229-10-215
Blancquaert, D., De Steur, H., Gellynck, X., and Van Der Straeten, D. (2014). Present and future of folate biofortification of crop plants. J. Exp. Bot. 65, 895–906. doi: 10.1093/jxb/ert483
Bonfim, K., Faria, J. C., Nogueira, E. O., Mendes, E. A., and Aragão, F. J. (2007). RNAi-mediated resistance to Bean golden mosaic virus in genetically engineered common bean (Phaseolus vulgaris). Mol. Plant Microbe Interact. 20, 717–726. doi: 10.1094/mpmi-20-6-0717
Bouis, H. E., and Welch, R. M. (2010). Biofortification - A sustainable agricultural strategy for reducing micronutrient malnutrition in the global south. Crop Sci. 50, S20–S32.
Bouis, H. E. (2003). Micronutrient fortification of plants through plant breeding: can it improve nutrition in man at low cost? Proc. Nutr. Soc. 62, 403–411. doi: 10.1079/pns2003262
Braun, D. M., Wang, L., and Ruan, Y. L. (2014). Understanding and manipulating sucrose phloem loading, unloading, metabolism, and signalling to enhance crop yield and food security. J. Exp. Bot. 65, 1713–1735. doi: 10.1093/jxb/ert416
Bucher, M. (2007). Functional biology of plant phosphate uptake at root and mycorrhiza interfaces. New Phytol. 173, 11–26. doi: 10.1111/j.1469-8137.2006.01935.x
Cailliatte, R., Schikora, A., Briat, J. F., Mari, S., and Curie, C. (2010). High-affinity manganese uptake by the metal transporter NRAMP1 is essential for Arabidopsis growth in low manganese conditions. Plant Cell. 22, 904–917. doi: 10.1105/tpc.109.073023
Caldas, G. V., and Blair, M. W. (2009). Inheritance of seed condensed tannins and their relationship with seed-coat color and pattern genes in common bean (Phaseolus vulgaris L.). Theor. Appl. Genet. 119, 131–142. doi: 10.1007/s00122-009-1023-4
Caproni, L., Raggi, L., Talsma, E. F., Wenzl, P., and Negri, V. (2020). European landrace diversity for common bean biofortification: a genome-wide association study. Sci. Rep. 10:19775. doi: 10.1038/s41598-020-76417-3
Casañas, F., Pérez-Vega, E., Almirall, A., Plans, M., Sabaté, J., and Ferreira, J. J. (2013). Mapping of QTL associated with seed chemical content in a RIL population of common bean (Phaseolus vulgaris L.). Euphytica 192, 279–288. doi: 10.1007/s10681-013-0880-8
Chan, E. K. F., Rowe, H. C., and Kliebenstein, D. J. (2010). Understanding the evolution of defense metabolites in Arabidopsis thaliana using genome-wide association mapping. Genetics 185, 991–1007. doi: 10.1534/genetics.109.108522
Chen, X., Li, H., Pandey, M. K., Yang, Q., Wang, X., Garg, V., et al. (2016). Draft genome of the peanut A-genome progenitor (Arachis duranensis) provides insights into geocarpy, oil biosynthesis, and allergens. Proc. Natl. Acad. Sci. U. S. A. 113, 6785–6790. doi: 10.1073/pnas.1600899113
Chiera, J., Finer, J., and Grabau, E. (2004). Ectopic expression of a soybean phytase in developing seeds of Glycine max to improve phosphorus availability. Plant Mol. Biol. 56, 895–904. doi: 10.1007/s11103-004-5293-6
Cichy, K. A., Caldas, G. V., Snapp, S. S., and Blair, M. W. (2009). QTL analysis of seed iron, zinc, and phosphorus levels in an Andean bean population. Crop Sci. 49, 1742–1750. doi: 10.2135/cropsci2008.10.0605
Clemente, T. E., and Cahoon, E. B. (2009). Soybean oil: genetic approaches for modification of functionality and total content. Plant Physiol. 151, 1030–1040. doi: 10.1104/pp.109.146282
Colangelo, E. P., and Guerinot, M. L. (2006). Put the metal to the petal: metal uptake and transport throughout plants. Curr. Opin. Plant Biol. 9, 322–330. doi: 10.1016/j.pbi.2006.03.015
Conte, S. S., and Walker, E. L. (2011). Transporters contributing to iron trafficking in plants. Mol. Plant. 4, 464–476. doi: 10.1093/mp/ssr015
Curie, C., and Mari, S. (2017). New routes for plant iron mining. New Phytol. 214, 521–525. doi: 10.1111/nph.14364
Curtin, S. J., Zhang, F., Sander, J. D., Haun, W. J., Starker, C., Baltes, N. J., et al. (2011). Targeted mutagenesis of duplicated genes in soybean with zinc-finger nucleases. Plant Physiol. 156, 466–473. doi: 10.1104/pp.111.172981
da Silva Messias, R., Galli, V., Dos Anjos, E., Silva, S. D., and Rombaldi, C. V. (2014). Carotenoid biosynthetic and catabolic pathways: gene expression and carotenoid content in grains of maize landraces. Nutrients 6, 546–563. doi: 10.3390/nu6020546
Davuluri, G. R., van Tuinen, A., Fraser, P. D., Manfredonia, A., Newman, R., Burgess, D., et al. (2005). Fruit-specific RNAi-mediated suppression of DET1 enhances carotenoid and flavonoid content in tomatoes. Nat. Biotechnol. 23, 890–895. doi: 10.1038/nbt1108
DellaPenna, D., and Pogson, B. J. (2006). Vitamin synthesis in plants: tocopherols and carotenoids. Annu. Rev. Plant Biol. 57, 711–738. doi: 10.1146/annurev.arplant.56.032604.144301
Denbow, D. M., Grabau, E. A., Lacy, G. H., Kornegay, E. T., Russell, D. R., and Umbeck, P. F. (1998). Soybeans transformed with a fungal phytase gene improve phosphorus availability for broilers. Poult. Sci. 77, 878–881. doi: 10.1093/ps/77.6.878
Diapari, M., Sindhu, A., Bett, K., Deokar, A., Warkentin, T. D., and Tar’an, B. (2014). Genetic diversity and association mapping of iron and zinc concentrations in chickpea (Cicer arietinum L.). Genome 57, 459–468. doi: 10.1139/gen-2014-0108
Dodo, H. W., Konan, K. N., Chen, F. C., Egnin, M., and Viquez, O. M. (2008). Alleviating peanut allergy using genetic engineering: the silencing of the immunodominant allergen Ara h 2 leads to its significant reduction and a decrease in peanut allergenicity. Plant Biotechnol. J. 6, 135–145. doi: 10.1111/j.1467-7652.2007.00292.x
Dumont, E., Fontaine, V., Vuylsteker, C., Sellier, H., Bodèle, S., Voedts, N., et al. (2009). Association of sugar content QTL and PQL with physiological traits relevant to frost damage resistance in pea under field and controlled conditions. Theor. Appl. Genet. 118, 1561–1571. doi: 10.1007/s00122-009-1004-7
Esfahani, M. N., Kusano, M., Nguyen, K. H., Watanabe, Y., Ha, C. V., Saito, K., et al. (2016). Adaptation of the symbiotic Mesorhizobium–chickpea relationship to phosphate deficiency relies on reprogramming of whole-plant metabolism. Proc. Natl. Acad. Sci. U. S. A. 113, E4610–E4619.
FAO, IFAD, UNICEF, WFP, and WHO. (2017). The state of food security and nutrition in the world 2017. Building resilience for peace and food security. Rome: FAO.
Falco, S. C., Guida, T., Locke, M., Mauvais, J., Sanders, C., Ward, R. T., et al. (1995). Transgenic canola and soybean seeds with increased lysine. Biotechnology 13, 577–582. doi: 10.1038/nbt0695-577
Farag, M. A., Huhman, D. V., Dixon, R. A., and Sumner, L. W. (2008). Metabolomics reveals novel pathways and differential mechanistic and elicitor-specific responses in phenylpropanoid and isoflavonoid biosynthesis in Medicago truncatula cell cultures. Plant Physiol. 146, 387–402.
Fernie, A. R., and Schauer, N. (2009). Metabolomics-assisted breeding: a viable option for crop improvement? Trends Genet. 25, 39–48. doi: 10.1016/j.tig.2008.10.010
Fileppi, M., Galasso, I., Tagliabue, G., Daminati, M. G., Campion, B., Doria, E., et al. (2010). Characterisation of structural genes involved in phytic acid biosynthesis in common bean (Phaseolus vulgaris L.). Mol. Breed 25, 453–470. doi: 10.1007/s11032-009-9344-1
Finkelstein, J. L., Haas, J. D., and Mehta, S. (2017). Iron-biofortified staple food crops for improving iron status: a review of the current evidence. Curr. Opin. Biotechnol. 44, 138–145. doi: 10.1016/j.copbio.2017.01.003
Forouzanfar, M. H., Alexander, L., Anderson, H. R., Bachman, V. F., Biryukov, S., Brauer, M., et al. (2015). Global, regional, and national comparative risk assessment of 79 behavioural, environmental and occupational, and metabolic risks or clusters of risks in 188 countries, 1990-2013: a systematic analysis for the Global Burden of Disease Study (2013). Lancet 386, 2287–2323.
Foyer, C. H., Lam, H. M., Nguyen, H. T., Siddique, K. H., Varshney, R. K., Colmer, T. D., et al. (2016). Neglecting legumes has compromised human health and sustainable food production. Nat. Plants 2, 1–10. doi: 10.1007/978-981-13-0253-4_1
Francis, D., Finer, J. J., and Grotewold, E. (2017). Challenges and opportunities for improving food quality and nutrition through plant biotechnology. Curr. Opin. Biotechnol. 44, 124–129. doi: 10.1016/j.copbio.2016.11.009
Frank, T., Habernegg, R., Yuan, F. J., Shu, Q. Y., and Engel, K. H. (2009). Assessment of the contents of phytic acid and divalent cations in low phytic acid (lpa) mutants of rice and soybean. J. Food. Compost. Anal. 22, 278–284. doi: 10.1016/j.jfca.2008.11.022
Garcia-Casal, M. N., Peña-Rosas, J. P., Giyose, B., De Steur, H., and Van Der Straeten, D. (2017). Staple crops biofortified with increased vitamins and minerals: considerations for a public health strategy. Ann. N.Y. Acad. Sci. 1379, 38–47.
Gayomba, S. R., Jung, H. I., Yan, J., Danku, J., Rutzke, M. A., Bernal, M., et al. (2013). The CTR/COPT-dependent copper uptake and SPL7-dependent copper deficiency responses are required for basal cadmium tolerance in A. thaliana. Metallomics 5, 1262–1275. doi: 10.1039/c3mt00111c
Gelin, J. R., Forster, S., Grafton, S. K., McClean, P. E., and Rojas-Cifuentes, G. A. (2007). Analysis of seed zinc and other minerals in a recombinant inbred population of navy bean (Phaseolus vulgaris L.). Crop Sci. 47, 1361–1366. doi: 10.2135/cropsci2006.08.0510
Ghandilyan, A., Barboza, L., Tisné, S., Granier, C., Reymond, M., Koornneef, M., et al. (2009). Genetic analysis identifies quantitative trait loci controlling rosette mineral concentrations in Arabidopsis thaliana under drought. New Phytol. 184, 180–192. doi: 10.1111/j.1469-8137.2009.02953.x
Giuliano, G. (2014). Plant carotenoids: genomics meets multi-gene engineering. Curr. Opin. Plant Biol. 19, 111–117. doi: 10.1016/j.pbi.2014.05.006
González-Guerrero, M., Escudero, V., and Saéz, Á, and Tejada-Jiménez, M. (2016). Transition metal transport in plants and associated endosymbionts: arbuscular mycorrhizal fungi and rhizobia. Front. Plant Sci. 7:1088.
Guzmán-Maldonado, S. H., Martínez, O., Acosta-Gallegos, J. A., Guevara-Lara, F., and Paredes-López, O. (2003). Putative quantitative trait loci for physical and chemical components of common bean. Crop Sci. 43, 1029–1035. doi: 10.2135/cropsci2003.1029
Hane, J. K., Ming, Y., Kamphuis, L. G., Nelson, M. N., Garg, G., Atkins, C. A., et al. (2017). A comprehensive draft genome sequence for lupin (Lupinus angustifolius), an emerging health food: insights into plant–microbe interactions and legume evolution. Plant Biotechnol. J. 15, 318–330. doi: 10.1111/pbi.12615
Ritchie, H., and Roser, M. (2020). Micronutrient Deficiency. Germany: Published online at OurWorldInData.org.
HarvestPlus Annual Report (2015). HarvestPlus Annual Report. URL: https://www.harvestplus.org/knowledge-market/publications?f%5B0%5D=field_resource_type%3A11
Haun, W., Coffman, A., Clasen, B. M., Demorest, Z. L., Lowy, A., Ray, E., et al. (2014). Improved soybean oil quality by targeted mutagenesis of the fatty acid desaturase 2 gene family. Plant Biotechnol. J. 12, 934–940. doi: 10.1111/pbi.12201
Haydon, M. J., and Cobbett, C. S. (2007). Transporters of ligands for essential metal ions in plants. New Phytol. 174, 499–506. doi: 10.1111/j.1469-8137.2007.02051.x
Hell, R., and Stephan, U. W. (2003). Iron uptake, trafficking and homeostasis in plants. Planta 216, 541–551. doi: 10.1007/s00425-002-0920-4
Hossain, T., Rosenberg, I., Selhub, J., Kishore, G., Beachy, R., and Schubert, K. (2004). Enhancement of folates in plants through metabolic engineering. Proc. Natl. Acad. Sci. U. S. A. 101, 5158–5163. doi: 10.1073/pnas.0401342101
Huang, X. Y., and Salt, D. E. (2016). Plant ionomics: From elemental profiling to environmental adaptation. Mol. Plant. 9, 787–797. doi: 10.1016/j.molp.2016.05.003
Hussain, D., Haydon, M. J., Wang, Y., Wong, E., Sherson, S. M., Young, J., et al. (2004). P-type ATPase heavy metal transporters with roles in essential zinc homeostasis in Arabidopsis. Plant Cell. 16, 1327–1339. doi: 10.1105/tpc.020487
Von Grebmer, K., Bernstein, J., Nabarro, D., Prasai, N., Amin, S., Yohannes, Y., et al. (2016). 2016 Global hunger index: Getting to zero hunger. Washington: Intl Food Policy Res Inst.
Irzykowska, L., and Wolko, B. (2004). Interval mapping of QTLs controlling yield-related traits and seed protein content in Pisum sativum. J. Appl. Genet 45, 297–306.
Ishimaru, Y., Masuda, H., Bashir, K., Inoue, H., Tsukamoto, T., Takahashi, M., et al. (2010). Rice metal−nicotianamine transporter OsYSL2, is required for the long distance transport of iron and manganese. Plant J. 62, 379–390. doi: 10.1111/j.1365-313x.2010.04158.x
Jadhav, A. A., Rayate, S. J., Mhase, L. B., Thudi, M., Chitikineni, A., Harer, P. N., et al. (2015). Marker-trait association study for protein content in chickpea (Cicer arietinum L.). J. Genet. 94, 279–286. doi: 10.1007/s12041-015-0529-6
Jegadeesan, S., Yu, K., Poysa, V., Gawalko, E., Morrison, M. J., Shi, C., et al. (2010). Mapping and validation of simple sequence repeat markers linked to a major gene controlling seed cadmium accumulation in soybean [Glycine max (L.) Merr]. Theor. Appl. Genet. 121, 283–294. doi: 10.1007/s00122-010-1309-6
Jeong, J., Merkovich, A., Clyne, M., and Connolly, E. L. (2017). Directing iron transport in dicots: regulation of iron acquisition and translocation. Curr. Opin. Plant Biol. 39, 106–113. doi: 10.1016/j.pbi.2017.06.014
Jha, A. B., Ashokkumar, K., Diapari, M., Ambrose, S. J., Zhang, H., Tar’an, B., et al. (2015). Genetic diversity of folate profiles in seeds of common bean, lentil, chickpea and pea. J. Food Compost Anal. 42, 134–140. doi: 10.1016/j.jfca.2015.03.006
Jiang, W., Struik, P. C., van Keulen, H., Zhao, M., Jin, L. N., and Stomph, T. J. (2008). Does increased zinc uptake enhance grain zinc mass concentration in rice? Ann. Appl. Biol. 153, 135–147. doi: 10.1111/j.1744-7348.2008.00243.x
Jiang, D., Zhao, L., and Clapham, D. E. (2009). Genome-wide RNAi screen identifies Letm1 as a mitochondrial Ca2+/H+ antiporter. Science 326, 144–147. doi: 10.1126/science.1175145
Jones, A. D., and Ejeta, G. (2016). A new global agenda for nutrition and health: the importance of agriculture and food systems. Bull. World Health Organ. 94, 228–229. doi: 10.2471/blt.15.164509
Joshi-Saha, A., and Reddy, K. S. (2015). Repeat length variation in the 5’UTR of myo-inositol monophosphatase gene is related to phytic acid content and contributes to drought tolerance in chickpea (Cicer arietinum L.). J. Exp. Bot. 66, 5683–5690. doi: 10.1093/jxb/erv156
Jukanti, A. K., Gaur, P. M., Gowda, C. L., and Chibbar, R. N. (2012). Nutritional quality and health benefits of chickpea (Cicer arietinum L.): a review. Br. J. Nutr. 108, 11–26.
Katuuramu, D., Hart, J., Porch, T., Grusak, M., Glahn, R., and Cichy, K. (2018). Genome-wide association analysis of nutritional composition-related traits and iron bioavailability in cooked dry beans (Phaseolus vulgaris L.). Mol. Breed. 38:44. doi: 10.1007/s11032-018-0798-x
Karunanandaa, B., Qi, Q., Hao, M., Baszis, S. R., Jensen, P. K., Wong, Y. H., et al. (2005). Metabolically engineered oilseed crops with enhanced seed tocopherol. Metab. Eng. 7, 384–400. doi: 10.1016/j.ymben.2005.05.005
Kaur, H., Shukla, R. K., Yadav, G., Chattopadhyay, D., and Majee, M. (2008). Two divergent genes encoding L-myo-inositol 1-phosphate synthase1 (CaMIPS1) and 2 (CaMIPS2) are differentially expressed in chickpea. Plant Cell Environ. 31, 1701–1716. doi: 10.1111/j.1365-3040.2008.01877.x
Keurentjes, J. J., Fu, J., de Vos, C. H., Lommen, A., Hall, R. D., and Bino, R. J. (2006). The genetics of plant metabolism. Nat. Genet. 38, 842–849.
Khazaei, H., Podder, R., Caron, C. T., Kundu, S. S., Diapari, M., Vandenberg, A., et al. (2017). Marker–Trait Association Analysis of Iron and Zinc Concentration in Lentil (Medik.) Seeds. Plant Genome 7:10. doi: 10.3835/plantgenome2017.02.0007
Kinney, A. J., and Clemente, T. E. (2005). Modifying soybean oil for enhanced performance in biodiesel blends. Fuel Proc. Technol. 86, 1137–1147. doi: 10.1016/j.fuproc.2004.11.008
Kita, Y., Nakamoto, Y., Takahashi, M., Kitamura, K., Wakasa, K., and Ishimoto, M. (2010). Manipulation of amino acid composition in soybean seeds by the combination of deregulated tryptophan biosynthesis and storage protein deficiency. Plant Cell Rep. 29, 87–95. doi: 10.1007/s00299-009-0800-5
Klein, R. G., Mannuzza, S., Olazagasti, M. A., Roizen, E., Hutchison, J. A., Lashua, C., et al. (2012). Clinical and functional outcome of childhood attention-deficit/hyperactivity disorder 33 years later. Arch. Gen. Psych. 69, 1295–1303. doi: 10.1001/archgenpsychiatry.2012.271
Klein, M. A., and Grusak, M. A. (2009). Identification of nutrient and physical seed trait QTL in the model legume Lotus japonicus. Genome 52, 677–691. doi: 10.1139/g09-039
Kobayashi, T., and Nishizawa, N. K. (2012). Iron uptake, translocation, and regulation in higher plants. Annu. Rev. Plant Biol. 63, 131–152. doi: 10.1146/annurev-arplant-042811-105522
Kumar, R. K., Chu, H. H., Abundis, C., Vasques, K., Rodriguez, D. C., Chia, J. C., et al. (2017). Iron-nicotianamine transporters are required for proper long distance iron signaling. Plant Physiol. 175, 1254–1268. doi: 10.1104/pp.17.00821
Kumssa, D. B., Joy, E. J., Ander, E., Watts, M. J., Young, S. D., Walker, S., et al. (2015). Dietary calcium and zinc deficiency risks are decreasing but remain prevalent. Sci. Rep. 5:10974.
Küpper, H., and Kochian, L. V. (2010). Transcriptional regulation of metal transport genes and mineral nutrition during acclimatization to cadmium and zinc in the Cd/Zn hyperaccumulator Thlaspi caerulescens (Ganges population). New Phytol. 185, 114–129. doi: 10.1111/j.1469-8137.2009.03051.x
Kusaba, M., Miyahara, K., Iid, S., Fukuoka, H., Takano, T., Sassa, H., et al. (2003). Low glutelin content1: a dominant mutation that suppresses the glutelin multigene family via RNA silencing in rice. Plant Cell 15, 1455–1467. doi: 10.1105/tpc.011452
Lahner, B., Gong, J., Mahmoudian, M., Smith, E. L., Abid, K. B., Rogers, E. E., et al. (2003). Genomic scale profiling of nutrient and trace elements in Arabidopsis thaliana. Nat. Biotechnol. 21, 1215–1221. doi: 10.1038/nbt865
Li, H., Liu, H., Han, Y., Wu, X., Teng, W., Liu, G., et al. (2010). Identification of QTL underlying vitamin E contents in soybean seed among multiple environments. Theor. Appl. Genet. 120, 1405–1413. doi: 10.1007/s00122-010-1264-2
Lin, Y. F., Liang, H. M., Yan, S. Y., Boch, A., Clemens, S., Chen, C. C., et al. (2009). Arabidopsis IRT3 is a zinc-regulated and plasma membrane localized zinc/iron transporter. New Phytol. 182, 392–404. doi: 10.1111/j.1469-8137.2009.02766.x
Liu, G., Xu, H., Zhang, L., and Zheng, Y. (2011). Fe binding properties of two soybean (Glycine max L.) LEA4 proteins associated with antioxidant activity. Plant Cell Physiol. 52, 994–1002. doi: 10.1093/pcp/pcr052
Liu, J., Versaw, W. K., Pumplin, N., Gomez, S. K., Blaylock, L. A., and Harrison, M. J. (2008). Closely related members of the Medicago truncatula PHT1 phosphate transporter gene family encode phosphate transporters with distinct biochemical activities. J. Biol. Chem. 283, 24673–24681. doi: 10.1074/jbc.m802695200
Ma, Y., Coyne, C. J., Grusak, M. A., Mazourek, M., Cheng, P., Main, D., et al. (2017). Genome-wide SNP identification, linkage map construction and QTL mapping for seed mineral concentrations and contents in pea (Pisum sativum L.). BMC Plant Biol. 17:43.
Maeda, D., Ashida, K., Iguchi, K., Chechetka, S. A., Hijikata, A., Okusako, Y., et al. (2006). Knockdown of an arbuscular mycorrhiza-inducible phosphate transporter gene of Lotus japonicus suppresses mutualistic symbiosis. Plant Cell Physiol. 47, 807–817. doi: 10.1093/pcp/pcj069
Maghari, B. M., and Ardekani, A. M. (2011). Genetically modified foods and social concerns. Avicenna J. Med. Biotechnol. 3, 109–117.
McCouch, S., Baute, G. J., Bradeen, J., Bramel, P., Bretting, P. K., Buckler, E., et al. (2013). Agriculture: Feeding the future. Nature 499, 23–24.
Michno, J. M., Wang, X., Liu, J., Curtin, S. J., Kono, T. J., and Stupar, R. M. (2015). CRISPR/Cas mutagenesis of soybean and Medicago truncatula using a new web-tool and a modified Cas9 enzyme. GM Crops Food 6, 243–252. doi: 10.1080/21645698.2015.1106063
Mickelbart, M. V., Hasegawa, P. M., and Bailey-Serres, J. (2015). Genetic mechanisms of abiotic stress tolerance that translate to crop yield stability. Nat. Rev. Genet 16, 237–251. doi: 10.1038/nrg3901
Millaleo, R., Reyes-Díaz, M., Alberdi, M., Ivanov, A. G., Krol, M., and Hüner, N. P. (2013). Excess manganese differentially inhibits photosystem I versus II in Arabidopsis thaliana. J. Exp. Bot. 64, 343–354. doi: 10.1093/jxb/ers339
Milner, M. J., Seamon, J., Craft, E., and Kochian, L. V. (2013). Transport properties of members of the ZIP family in plants and their role in Zn and Mn homeostasis. J. Exp. Bot. 64, 369–381. doi: 10.1093/jxb/ers315
De Moura, F. F., Palmer, A. C., Finkelstein, J. L., Haas, J. D., Murray-Kolb, L. E., and Wenger, M. J. (2014). Are biofortified staple food crops improving vitamin a and iron status in women and children? New evidence from efficacy trials. Adv. Nutr. 5, 568–570. doi: 10.3945/an.114.006627
Mudryj, A. N., Yu, N., and Aukema, H. M. (2014). Nutritional and health benefits of pulses. Appl. Physiol. Nutr. Metab. 39, 1197–1204.
NCCIH (2018). Published online at www.nccih.nih.gov. URL: https://www.nccih.nih.gov/health/vitamins-and-minerals
Nestel, P., Bouis, H. E., Meenakshi, J. V., and Pfeiffer, W. (2006). Biofortification of staple food crops. J Nutr. 136, 1064–1067.
Norton, G. J., Deacon, C. M., Xion, L., Huang, S., Meharg, A. A., and Price, A. H. (2010). Genetic mapping of the rice ionome in leaves and grain: identification of QTLs for 17 elements including arsenic, cadmium, iron and selenium. Plant Soil 329, 139–153. doi: 10.1007/s11104-009-0141-8
Owens, B. F., Lipka, A. E., Magallanes-Lundback, M., Tiede, T., Diepenbrock, C. H., Kandianis, C. B., et al. (2014). A foundation for provitamin A biofortification of maize: genome-wide association and genomic prediction models of carotenoid levels. Genetics 198, 1699–1716. doi: 10.1534/genetics.114.169979
Palmer, C. M., and Guerinot, M. L. (2009). Facing the challenges of CuFe and Zn homeostasis in plants. Nat. Chem. Biol. 5, 333–340. doi: 10.1038/nchembio.166
Palmgren, M. G., Clemens, S., Williams, L. E., Krämer, U., Borg, S., Schjørring, J. K., et al. (2008). Zinc biofortification of cereals: problems and solutions. Trends Plant Sci. 13, 464–473. doi: 10.1016/j.tplants.2008.06.005
Pandey, M. K., Roorkiwal, M., Singh, V. K., Ramalingam, A., Kudapa, H., Thudi, M., et al. (2016). Emerging genomic tools for legume breeding: current status and future prospects. Front. Plant Sci. 7:455.
Panzeri, D., Cassani, E., Doria, E., Tagliabue, G., Forti, L., Campion, B., et al. (2011). A defective ABC transporter of the MRP family, responsible for the bean lpa1 mutation, affects the regulation of the phytic acid pathway, reduces seed myo−inositol and alters ABA sensitivity. New Phytol. 191, 70–83. doi: 10.1111/j.1469-8137.2011.03666.x
Parveen, S., Gupta, D. B., Dass, S., Kumar, A., Pandey, A., Chakraborty, S., et al. (2016). Chickpea ferritin cafer1 participates in oxidative stress response, and promotes growth and development. Sci. Rep. 6:31218.
Peltier, A. J., Hatfield, R. D., and Grau, C. R. (2009). Soybean stem lignin concentration relates to resistance to Sclerotinia sclerotiorum. Plant Dis. 93, 149–154. doi: 10.1094/pdis-93-2-0149
Pixley, K. V., Palacios-Rojas, N., and Glahn, R. P. (2011). The usefulness of iron bioavailability as a target trait for breeding maize (Zea mays L.) with enhanced nutritional value. Field Crops Res. 123, 153–160. doi: 10.1016/j.fcr.2011.05.011
Plenish high oleic (2021). Plenish high oleic. URL: https://www.pioneer.com/home/site/about/products/product-traits-technology/plenish/
Poblaciones, M. J., and Rengel, Z. (2017). Combined foliar selenium and zinc biofortification in field pea (Pisum sativum): accumulation and bioavailability in raw and cooked grains. Crop Pasture Sci. 68, 265–271. doi: 10.1071/cp17082
Qin, L., Han, P., Chen, L., Walk, T. C., Li, Y., Hu, X., et al. (2017). Genome-Wide Identification and Expression Analysis of NRAMP Family Genes in Soybean (Glycine Max L.). Front. Plant Sci. 8:1436.
Qin, L., Zhao, J., Tian, J., Chen, L., Sun, Z., Guo, Y., et al. (2012). The high-affinity phosphate transporter GmPT5 regulates phosphate transport to nodules and nodulation in soybean. Plant Physiol. 159, 1634–1643. doi: 10.1104/pp.112.199786
Ramamurthy, R. K., Jedlicka, J., Graef, G. L., and Waters, B. M. (2014). Identification of new QTLs for seed mineral, cysteine, and methionine concentrations in soybean [Glycine max (L.) Merr.]. Mol. Breed. 34, 431–445. doi: 10.1007/s11032-014-0045-z
Raman, R. (2017). The impact of Genetically Modified (GM) crops in modern agriculture: A review. GM Crops Food. 8, 195–208. doi: 10.1080/21645698.2017.1413522
Rivera, N. G. R., García-Salinas, C., Aragão, F. J. L., and de la Garza, R. I. D. (2016). Metabolic engineering of folate and its precursors in Mexican common bean (Phaseolus vulgaris L.). Plant Biotechnol. J. 14, 2021–2032. doi: 10.1111/pbi.12561
Rastall, R. A., and Gibson, G. R. (2015). Recent developments in prebiotics to selectively impact beneficial microbes and promote intestinal health. Curr. Opin. Biotechnol. 32, 42–46. doi: 10.1016/j.copbio.2014.11.002
Rehman, H. M., Cooper, J. W., Lam, H. M., and Yang, S. H. (2019). Legume biofortification is an underexploited strategy for combatting hidden hunger. Plant Cell Environ. 42, 52–70. doi: 10.1111/pce.13368
Reddy, A. S., and Thomas, T. L. (1996). Expression of a cyanobacterial delta 6-desaturase gene results in gamma-linolenic acid production in transgenic plants. Nat. Biotechnol. 14, 639–642. doi: 10.1038/nbt0596-639
Rezaei, M. K., Deokar, A., and Tar’an, B. (2016). Identification and Expression Analysis of Candidate Genes Involved in Carotenoid Biosynthesis in Chickpea Seeds. Front. Plant Sci. 7:1867.
Rezaei, M. K., Deokar, A., Arganosa, G., Roorkiwal, M., Pandey, S. K., Warkentin, T. D., et al. (2019). Mapping quantitative trait loci for carotenoid concentration in three F2 populations of chickpea. Plant Genome 12:190067. doi: 10.3835/plantgenome2019.07.0067
Roorkiwal, M., Bharadwaj, C., Barmukh, R., Dixit, G. P., Thudi, M., Gaur, P. M., et al. (2020). Integrating genomics for chickpea improvement: achievements and opportunities. Theor. Appl. Genet 133, 1703–1720. doi: 10.1007/s00122-020-03584-2
Ryan, B. M., Kirby, J. K., Degryse, F., Harris, H., McLaughlin, M. J., and Scheiderich, K. (2013). Copper speciation and isotopic fractionation in plants: uptake and translocation mechanisms. New Phytol. 199, 367–378. doi: 10.1111/nph.12276
Sab, S., Lokesha, R., Mannur, D. M., Somasekhar, Jadhav, K., Mallikarjuna, B. P., et al. (2020). Genome wide SNP discovery and mapping QTLs for seed iron and zinc concentrations in chickpea (Cicer arietinum L.). Front. Nutr. 7:559120. doi: 10.3389/fnut.2020.559120
Salt, D. E., Baxter, I., and Lahner, B. (2008). Ionomics and the study of the plant ionome. Annu. Rev. Plant Biol. 59, 709–733. doi: 10.1146/annurev.arplant.59.032607.092942
Saltzman, A., Birol, E., Oparinde, A., Andersson, M. S., Asare-Marfo, D., Diressie, M. T., et al. (2017). Availability, production, and consumption of crops biofortified by plant breeding: current evidence and future potential. Ann. N. Y. Acad. Sci. 1390, 104–114. doi: 10.1111/nyas.13314
Sánchez-Chino, X., Jiménez-Martínez, C., Dávila-Ortiz, G., Álvarez-González, I., and Madrigal-Bujaidar, E. (2015). Nutrient and nonnutrient components of legumes, and its chemopreventive activity: a review. Nutr Cancer 67, 401–410. doi: 10.1080/01635581.2015.1004729
Sankaran, R. P., Huguet, T., and Grusak, M. A. (2009). Identification of QTL affecting seed mineral concentration and content in the model legume Medicago truncatula. Theor. Appl. Genet. 119, 241–253. doi: 10.1007/s00122-009-1033-2
Santi, S., and Schmidt, W. (2009). Dissecting iron deficiency-induced proton extrusion in Arabidopsis roots. New Phytol. 183, 1072–1084. doi: 10.1111/j.1469-8137.2009.02908.x
Sarvamangala, C., Gowda, M. V. C., and Varshney, R. K. (2011). Identification of quantitative trait locifor protein content, oil content and oil quality in groundnut (Arachis hypogaea. L). Field Crops Res. 122, 49–59. doi: 10.1016/j.fcr.2011.02.010
Sattler, S. E., Cheng, Z., and DellaPenna, D. (2004). From Arabidopsis to agriculture: engineering improved vitamin E content in soybean. Trends Plant Sci. 9, 365–367. doi: 10.1016/j.tplants.2004.06.002
Schauer, N., Semel, Y., Roessner, U., Gur, A., Balbo, I., Carrari, F., et al. (2006). Comprehensive metabolic profiling and phenotyping of interspecific introgression lines for tomato improvement. Nat. Biotechnol. 24, 447–454. doi: 10.1038/nbt1192
Schmidt, M. A., Parrott, W. A., Hildebrand, D. F., Berg, R. H., Cooksey, A., Pendarvis, K., et al. (2015). Transgenic soya bean seeds accumulating β-carotene exhibit the collateral enhancements of oleate and protein content traits. Plant Biotechnol. J. 13, 590–600. doi: 10.1111/pbi.12286
Schmutz, J., Cannon, S. B., Schlueter, J., Ma, J., Mitros, T., Nelson, W., et al. (2010). Genome sequence of the palaeopolyploid soybean. Nature 463, 178–183.
Schmutz, J., McClea, P. E., Mamidi, S., Wu, G. A., Cannon, S. B., Grimwood, J., et al. (2014). A reference genome for common bean and genome-wide analysis of dual domestications. Nat. Genet. 46, 707–713.
Sharif, H. R., Williams, P. A., Sharif, M. K., Abbas, S., Majeed, H., Masamba, K. G., et al. (2018). Current progress in the utilization of native and modified legume proteins as emulsifiers and encapsulants–A review. Food Hydrocoll. 76, 2–16. doi: 10.1016/j.foodhyd.2017.01.002
Shi, J., Wang, H., Schellin, K., Li, B., Faller, M., Stoop, J. M., et al. (2007). Embryo-specific silencing of a transporter reduces phytic acid content of maize and soybean seeds. Nat. Biotechnol. 25, 930–937. doi: 10.1038/nbt1322
Sinclair, S. A., and Krämer, U. (2012). The zinc homeostasis network of land plants. Biochim. Biophys. Acta 1823, 1553–1567. doi: 10.1016/j.bbamcr.2012.05.016
Singh, U. M., Sareen, P., Sengar, R. S., and Kumar, A. (2013). Plant ionomics: a newer approach to study mineral transport and its regulation. Acta Physiol. Plant 35, 2641–2653. doi: 10.1007/s11738-013-1316-8
Socha, A. L., and Guerinot, M. L. (2014). Mn-euvering manganese: the role of transporter gene family members in manganese uptake and mobilization in plants. Front. Plant Sci. 5:106.
Sompong, U., Somta, P., Raboy, V., and Srinives, P. (2012). Mapping of quantitative trait loci for phytic acid and phosphorus contents in seed and seedling of mungbean (Vigna radiata (L.) Wilczek). Breed Sci. 62, 87–92. doi: 10.1270/jsbbs.62.87
Sperotto, R. A., Ricachenevsky, F. K., Williams, L. E., Vasconcelos, M. W., and Menguer, P. K. (2014). From soil to seed: micronutrient movement into and within the plant. Front. Plant Sci. 5:438.
Storozhenko, S., De Brouwer, V., Volckaert, M., Navarrete, O., Blancquaert, D., Zhang, G. F., et al. (2007). Folate fortification of rice by metabolic engineering. Nat. Biotechnol. 25, 1277–1279. doi: 10.1038/nbt1351
Sun, X., Hu, Z., Chen, R., Jiang, Q., Song, G., Zhang, H., et al. (2015). Targeted mutagenesis in soybean using the CRISPR–Cas9 system. Sci. Rep. 5:10342.
Tang, F., Yang, S., Liu, J., and Zhu, H. (2016). Rj4, a gene controlling nodulation specificity in soybeans, encodes a thaumatin-Like protein but not the one previously reported. Plant Physiol. 170, 26–32. doi: 10.1104/pp.15.01661
Tang, G., and Galili, G. (2004). Using RNAi to improve plant nutritional value: from mechanism to application. Trends Biotechnol. 22, 463–469. doi: 10.1016/j.tibtech.2004.07.009
Tar’an, B., Warkentin, T., Somers, D. J., Miranda, D., Vandenberg, A., Blade, S., et al. (2004). Identification of quantitative trait loci for grain yield, seed protein concentration and maturity in field pea (Pisum sativum L.). Euphytica 136, 297–306. doi: 10.1023/b:euph.0000032721.03075.a0
Tavva, V. S., Kim, Y. H., Kagan, I. A., Dinkins, R. D., Kim, K. H., and Collins, G. B. (2007). Increased alpha-tocopherol content in soybean seed overexpressing the Perilla frutescens gamma-tocopherol methyltransferase gene. Plant Cell Rep. 26, 61–70. doi: 10.1007/s00299-006-0218-2
Tejada-Jiménez, M., Castro-Rodríguez, R., Kryvoruchko, I., Lucas, M. M., Udvardi, M., Imperial, J., et al. (2015). Medicago truncatula natural resistance-associated macrophage protein1 is required for iron uptake by rhizobia-infected nodule cells. Plant Physiol. 168, 258–272. doi: 10.1104/pp.114.254672
Thomine, S., and Vert, G. (2013). Iron transport in plants: better be safe than sorry. Curr. Opin. Plant Biol. 16, 322–327. doi: 10.1016/j.pbi.2013.01.003
Ulmasov, T., Voelker, T., Wilkes, R., and Cornelius, J. (2012). “High-Oleic, Low-Saturate Soybeans Offer a Sustainable and Nutritionally Enhanced Solution for Food Applications Requiring High Oil Stability,” in ∗Designing Soybeans for 21st Century Markets, (USA: AOCS Press).
Upadhyaya, H. D., Bajaj, D., Das, S., Kumar, V., Gowda, C. L., Sharma, S., et al. (2016). Genetic dissection of seed-iron and zinc concentrations in chickpea. Sci. Rep. 6:24050.
Van Eenennaam, A. L., Lincoln, K., Durrett, T. P., Valentin, H. E., Shewmaker, C. K., and Thorne, G. (2003). Engineering vitamin E content: from Arabidopsis mutant to soy oil. Plant Cell. 15, 3007–3019. doi: 10.1105/tpc.015875
Varshney, R. K. (2016). Exciting journey of 10 years from genomes to fields and markets: Some success stories of genomics-assisted breeding in chickpea, pigeonpea and groundnut. Plant Sci. 242, 98–107. doi: 10.1016/j.plantsci.2015.09.009
Varshney, R. K., Chen, W., Li, Y., Bharti, A. K., Saxena, R. K., Schlueter, J. A., et al. (2012). Draft genome sequence of pigeonpea (Cajanus cajan), an orphan legume crop of resource-poor farmers. Nat. Biotechnol. 30, 83–89. doi: 10.1038/nbt.2022
Varshney, R. K., Song, C., Saxena, R. K., Azam, S., Yu, S., Sharpe, A. G., et al. (2013). Draft genome sequence of chickpea (Cicer arietinum) provides a resource for trait improvement. Nat. Biotechnol. 31, 240–246.
Varshney, R. K., Thudi, M., Pandey, M. K., Tardieu, F., Ojiewo, C., Vadez, V., et al. (2018). Accelerating genetic gains in legumes for the development of prosperous smallholder agriculture: integrating genomics, phenotyping, systems modelling and agronomy. J. Exp. Bot. 69, 3293–3312. doi: 10.1093/jxb/ery088
Varshney, R. K., Terauchi, R., and McCouch, S. R. (2014). Harvesting the promising fruits of genomics: applying genome sequencing technologies to crop breeding. PLoS Biol. 12:e1001883. doi: 10.1371/journal.pbio.1001883
Varshney, R. K., Pandey, M. K., Bohra, A., Singh, V. K., Thudi, M., and Saxena, R. K. (2019). Toward the sequence-based breeding in legumes in the post-genome sequencing era. Theor. Appl. Genet. 132, 797–816. doi: 10.1007/s00122-018-3252-x
Velu, G., Singh, R. P., Huerta-Espino, J., Peña, R. J., Arun, B., Mahendru-Singh, A., et al. (2012). Performance of biofortified spring wheat genotypes in target environments for grain zinc and iron concentrations. Field Crops Res. 137, 261–267. doi: 10.1016/j.fcr.2012.07.018
Véry, A. A., and Davies, J. M. (2000). Hyperpolarization-activated calcium channels at the tip of Arabidopsis root hairs. Proc. Natl. Acad. Sci. U. S. A. 97, 9801–9806. doi: 10.1073/pnas.160250397
Walker, E. L., and Connolly, E. L. (2008). Time to pump iron: iron-deficiency-signaling mechanisms of higher plants. Curr. Opin. Plant Biol. 11, 530–535. doi: 10.1016/j.pbi.2008.06.013
Walker, E. L., and Waters, B. M. (2011). The role of transition metal homeostasis in plant seed development. Curr. Opin. Plant Biol. 14, 318–324. doi: 10.1016/j.pbi.2011.03.025
Warkentin, T. D., Delgerjav, O., Arganosa, G., Rehman, A. U., Bett, K. E., Anbessa, Y., et al. (2012). Development and characterization of low-Phytate Pea. Crop Sci. 52, 74–78. doi: 10.2135/cropsci2011.05.0285
Waters, B. M., and Grusak, M. A. (2008). Whole-plant mineral partitioning throughout the life cycle in Arabidopsis thaliana ecotypes Columbia Landsberg erecta, Cape Verde Islands, and the mutant line ysl1ysl3. New Phytol. 177, 389–405. doi: 10.1111/j.1469-8137.2007.02288.x
Waters, B. M., Chu, H. H., Didonato, R. J., Roberts, L. A., Eisley, R. B., Lahner, B., et al. (2006). Mutations in Arabidopsis yellow stripe-like1 and yellow stripe-like3 reveal their roles in metal ion homeostasis and loading of metal ions in seeds. Plant Physiol. 141, 1446–1458. doi: 10.1104/pp.106.082586
White, C. L., Tabe, L. M., Dove, H., Hamblin, J., Young, P., Phillips, N., et al. (2001). Increased efficiency of wool growth and live weight gain in Merino sheep fed transgenic lupin seed containing sunflower albumin. J. Sci. Food Agric. 81, 147–154. doi: 10.1002/1097-0010(20010101)81:1<147::aid-jsfa751>3.0.co;2-e
White, P. J., Whiting, S. N., Baker, A. J. M., and Broadley, M. R. (2002). Does zinc move apoplastically to the xylem in roots of Thlaspi caerulescens? New Phytol. 153, 201–207. doi: 10.1046/j.0028-646x.2001.00325.x
White, P. J., and Broadley, M. R. (2005). Biofortifying crops with essential mineral elements. Trends Plant Sci. 10, 586–593. doi: 10.1016/j.tplants.2005.10.001
White, P. J., and Broadley, M. R. (2009). Biofortification of crops with seven mineral elements often lacking in human diets–iron, zinc, copper, calcium, magnesium, selenium and iodine. New Phytol. 182, 49–84. doi: 10.1111/j.1469-8137.2008.02738.x
WHO (2017). Factsheet: Children – reducing mortality. URL: : https://www.afro.who.int/health-topics/child-health
World Bank Data. (2016). World Bank Data. URL: https://data.worldbank.org/indicator/SH.ANM.CHLD.ZS on 21st May 2020.
Xia, J., Yamaji, N., Kasai, T., and Ma, J. F. (2010). Plasma membrane-localized transporter for aluminum in rice. Proc. Natl. Acad. Sci. U. S. A. 107, 18381–18385. doi: 10.1073/pnas.1004949107
Xie, W., Perry, G., Martin, C. J., Shim, Y. S., Navabi, A., and Pauls, K. P. (2017). Molecular characterization of dihydroneopterin aldolase and aminodeoxychorismate synthase in common bean-genes coding for enzymes in the folate synthesis pathway. Genome 60, 588–600. doi: 10.1139/gen-2016-0227
Xiong, H., Kobayashi, T., Kakei, Y., Senoura, T., Nakazono, M., Takahashi, H., et al. (2012). AhNRAMP1 iron transporter is involved in iron acquisition in peanut. J. Exp. Bot. 63, 4437–4446. doi: 10.1093/jxb/ers117
Xue, Y., Xia, H., Christie, P., Zhang, Z., Li, L., and Tang, C. (2016). Crop acquisition of phosphorus, iron and zinc from soil in cereal/legume intercropping systems: a critical review. Ann. Bot. 117, 363–377. doi: 10.1093/aob/mcv182
Yu, O., Shi, J., Hession, A. O., Maxwell, C. A., McGonigle, B., and Odell, J. T. (2003). Metabolic engineering to increase isoflavone biosynthesis in soybean seed. Phytochemistry 63, 753–763. doi: 10.1016/s0031-9422(03)00345-5
Yuan, F. J., Zhao, H. J., Ren, X. L., Zhu, S. L., Fu, X. J., and Shu, Q. Y. (2007). Generation and characterization of two novel low phytate mutations in soybean (Glycine max L. Merr.). Theor. Appl. Genet 115, 945–957. doi: 10.1007/s00122-007-0621-2
Zarocostas, J. (2009). Malnutrition and hunger rise as economic crisis hits world’s poorest people. BMJ 338:b1651. doi: 10.1136/bmj.b1651
Keywords: malnutrition, micronutrient, metabolic pathway, biofortification, trait-mapping, genomics
Citation: Roorkiwal M, Pandey S, Thavarajah D, Hemalatha R and Varshney RK (2021) Molecular Mechanisms and Biochemical Pathways for Micronutrient Acquisition and Storage in Legumes to Support Biofortification for Nutritional Security. Front. Plant Sci. 12:682842. doi: 10.3389/fpls.2021.682842
Received: 19 March 2021; Accepted: 06 May 2021;
Published: 07 June 2021.
Edited by:
Sergio J. Ochatt, INRA, UMR 1347 Agroécologie, FranceReviewed by:
Karl Kunert, University of Pretoria, South AfricaJun Fang, Key Laboratory of Soybean Molecular Design and Breeding, Northeast Institute of Geography and Agroecology, Chinese Academy of Sciences, China
Copyright © 2021 Roorkiwal, Pandey, Thavarajah, Hemalatha and Varshney. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rajeev K. Varshney, ci5rLnZhcnNobmV5QGNnaWFyLm9yZw==; cmFqZWV2LnZhcnNobmV5QG11cmRvY2guZWR1LmF1
†These authors have contributed equally to this work
 Manish Roorkiwal
Manish Roorkiwal Sarita Pandey
Sarita Pandey Dil Thavarajah
Dil Thavarajah R. Hemalatha4
R. Hemalatha4 Rajeev K. Varshney
Rajeev K. Varshney