- 1Department of Plant Sciences, University of Oxford, Oxford, United Kingdom
- 2The James Hutton Institute, Dundee, United Kingdom
- 3ICAR-Indian Institute of Soil Science, Bhopal, India
- 4ICAR-National Bureau of Agriculturally Important Microorganisms, Mau, India
- 5Department of Plant Sciences, School of Life Science, University of Hyderabad, Hyderabad, India
Pigeon pea (Cajanus cajan L. Millsp. ) is a legume crop resilient to climate change due to its tolerance to drought. It is grown by millions of resource-poor farmers in semiarid and tropical subregions of Asia and Africa and is a major contributor to their nutritional food security. Pigeon pea is the sixth most important legume in the world, with India contributing more than 70% of the total production and harbouring a wide variety of cultivars. Nevertheless, the low yield of pigeon pea grown under dry land conditions and its yield instability need to be improved. This may be done by enhancing crop nodulation and, hence, biological nitrogen fixation (BNF) by supplying effective symbiotic rhizobia through the application of “elite” inoculants. Therefore, the main aim in this study was the isolation and genomic analysis of effective rhizobial strains potentially adapted to drought conditions. Accordingly, pigeon pea endosymbionts were isolated from different soil types in Southern, Central, and Northern India. After functional characterisation of the isolated strains in terms of their ability to nodulate and promote the growth of pigeon pea, 19 were selected for full genome sequencing, along with eight commercial inoculant strains obtained from the ICRISAT culture collection. The phylogenomic analysis [Average nucleotide identity MUMmer (ANIm)] revealed that the pigeon pea endosymbionts were members of the genera Bradyrhizobium and Ensifer. Based on nodC phylogeny and nod cluster synteny, Bradyrhizobium yuanmingense was revealed as the most common endosymbiont, harbouring nod genes similar to those of Bradyrhizobium cajani and Bradyrhizobium zhanjiangense. This symbiont type (e.g., strain BRP05 from Madhya Pradesh) also outperformed all other strains tested on pigeon pea, with the notable exception of an Ensifer alkalisoli strain from North India (NBAIM29). The results provide the basis for the development of pigeon pea inoculants to increase the yield of this legume through the use of effective nitrogen-fixing rhizobia, tailored for the different agroclimatic regions of India.
Introduction
Pigeon pea (Cajanus cajan L. Millsp.) is grown by millions of resource-poor farmers in semiarid and tropical subregions of Asia and Africa as a major contributor to their food security (Mula and Saxena, 2010; Varshney et al., 2010). The initial domestication of pigeon pea was started in central India over 3,500 years ago, from its wild progenitor Cajanus cajanifolius (Vavilov, 1951; Saxena et al., 2014). Pigeon pea is the sixth most important legume in the world, representing 5% of the total pulse production (4.92 M ha), with India contributing more than 70% of the total (3.6 M ha) and harbouring a wide variety of cultivars (218 making up 73% of the total) (Saxena, 2006). It was estimated by the Food and Agriculture Organisation (FAO) that the worldwide annual production of pigeon pea in 2019 was 5.6 Mt, of which ~59% was produced by India alone [FAO statistics (www.fao.org/faostat)].
Pigeon pea is a perennial shrub normally cultivated as an annual crop and, in India, can be used in rotation and intercrop systems with different cereal crops. Moreover, pigeon pea develops a deep root system, making it drought tolerant. These traits encourage cultivation in rain-fed drylands, although the poor growth conditions (e.g., aridity, nutrient-poor soils) mean that yields remain low. Effective symbiosis may improve nitrogen (N) content in this pulse legume and, hence, seed quality and quantity. However, legume-rhizobium symbioses are sensitive to drought, and, therefore, N fixation can be inefficient (Serraj et al., 1999; Mula and Saxena, 2010; Varshney et al., 2012). Selecting from among the diversity of pigeon pea cultivars sown in India may lead to improved symbiotic partners, as in the case for other legumes like soybean (Yang et al., 2010). To increase pigeon pea yields, it is important to select superior rhizobial strains that perform well under a wide variety of various stresses. Such bacteria can be developed into pigeon pea inoculants, tailored to perform well under different agroclimatic conditions.
However, until now, genomic diversity studies have only been performed in countries in the American and African continents, showing that the preferred endosymbionts are Bradyrhizobium spp. In Trinidad and Tobago, the main symbiont was Bradyrhizobium elkanii (Ramsubhag et al., 2002), whereas, in the Dominican Republic, Bradyrhizobium yuanmingense dominated. In the Ivory Coast, two different clades can nodulate pigeon pea, one associated with the B. elkanii group and a second one later assigned as the new species B. ivorense (Fossou et al., 2016, 2020). Additionally, another new species isolated from pigeon pea in the Dominican Republic has been defined as Bradyrhizobium cajani (Araújo et al., 2017), illustrating the great diversity present within pigeon pea endosymbionts across the world. Ensifer (syn. Sinorhizobium) has been reported as a symbiont only rarely, but strains were isolated using pigeon pea as a trap plant in soybean fields in Brazil (Coutinho et al., 1999; Stepkowski et al., 2003). Diverse pigeon pea rhizobia have been reported in Indian soils and have a long history of usage as inoculants; nevertheless, rigorous diversity studies have not been performed on these endosymbionts. In this study, we applied a mechanistic-holistic approach to study the diversity of pigeon pea native endosymbionts across India.
The aim in this study was to characterise the pigeon pea endosymbiont population isolated from a diversity of soil types in South (Alfisols), Central (Vertisols), and North India (Inceptisols). To achieve this, we isolated representative Indian pigeon pea rhizobia, sequenced representative strains, assessed their ability to nodulate pigeon pea and promote its growth, and analysed their genetic and genomic features. We uncovered the diversity of this population and the relationship between pigeon pea and members of the genera Bradyrhizobium and Ensifer. Comparisons of symbiotic-related features and the putative proteomes of these strains reveal the preferred pigeon pea endosymbionts in India.
Materials and Methods
Strain Isolation From Nodules
Pigeon pea nodules were collected from three different regions in India: South India representing Alfisols (Telangana/Andhra Pradesh, Hyderabad University, HU strains), Central India representing Vertisols (Madhya Pradesh, Bhopal Rhizobia Pigeon pea, BRP strains), and North India, representing Inceptisols (Uttar Pradesh/Haryana/Punjab, National Bureau of Agricultural Important Microorganisms, NBAIM strains) (Supplementary Table S1). Nodules were surface sterilised by washing with ethanol (70%) for 1 min, followed by 2% sodium hypochlorite for 5 min, and finally washing with sterile-distilled water. The nodules were homogenised in 0.9% NaCl and directly streaked on Yeast Mannitol media (YM), supplemented with Congo red (0.0025%) (CRYEMA) for visual screening (Vincent, 1970; Somasegaran and Hoben, 1994). Plates were incubated at 28°C for up to 3–5 days. Selected colonies were streaked onto fresh CRYEMA plates to obtain pure cultures.
Eight pigeon pea inoculant strains were obtained from the Microbial Germplasm collection of the International Crops Research Institute for the Semiarid Tropics (ICRISAT), Hyderabad, India, were also included as reference strains (Supplementary Table S1).
Assessment of Bacterial Diversity by BOX-PCR
DNA extraction was achieved by alkaline lysis (0.05-M NaOH, 0.25% SDS) (Rivas et al., 2001). Isolated DNA was used as a template to generate BOX-PCR fingerprints, using the specific BOXA1R primer (CTACGGCAAGGCGACGCTGACG) (Versalovic et al., 1994). Amplification was carried out in a 25-μl PCR reaction containing 5–10 ng of isolated DNA and 1 U of OneTaq polymerase (NEB). BOX-PCR products were visualised on 2% agarose gels at 100 V until clear band separation. Gel images of the BOX-PCR fingerprint of each strain in the IU population were compared to find those that were the same and those that were different from each other.
Nodulation Test
Seeds of C. cajan cv. Asha (ICPL 87119) were surface sterilised with sodium hypochlorite (3% active chlorine) and 0.1% (v/v) Tween 20 for 6 min and rinsed three times with sterile-distilled water. Surface-sterilised seeds were germinated on 0.5% distilled water agar in petri plates at 28°C in the dark. Germinated seedlings were transferred to sterile test tubes containing 30 ml of vermiculite: perlite mixture (1:1) and 30 ml of B&D nutrient solution. The tubes were transferred to a growth chamber at a temperature of 28°C, 16/8-h day/night light regime, a 70% moisture level, and 100 μmol m−2 s−1 irradiances. Each tube was inoculated with 1 ml of bacterial liquid culture (108 CFU). Negative control tubes were left uninoculated. Five test tubes for each isolate were completely randomised in the growth chamber. Plants were harvested and scored for nodulation after 8 weeks of growth.
Assessment of Plant Growth Promotion
A representative strain from each BOX-PCR pattern was used as an inoculant with pigeon pea to assess its plant growth-promoting potential, using a temperature of 28°C, 16/8-h day/night light regime, a 70% moisture level, and 100 μmol m−2 s−1 irradiances. The experiment was run as a completely randomised design with five replications. Sterile 1 L pots were filled with a 1:1 mixture of vermiculite: perlite and 400 ml B&D nutrient solution (Broughton and Dilworth, 1971). Seeds were surface sterilised and germinated as described above. Seedlings were transferred to 1-L pots and inoculated with 1 ml of bacterial liquid culture (108 CFU). To prevent cross-contamination during watering, the pots were covered with plastic film with a hole for the shoot. Plants were fed weekly with a B&D nutrient solution and watered daily, or as required. The plants were harvested 8 weeks after inoculation, and shoot and root biomass obtained from five replicates was quantified after drying in an oven at 70°C for 5 days. The pigeon pea endosymbiont reference strains, IC3195, IC3342, IC4059, IC4060, and IC4062 were included as positive controls.
Genome Sequencing, Annotation and Analysis
Culture samples were outsourced to Microbes NG, Birmingham, United Kingdom for Illumina sequencing (MiSeq v2, PE 2 × 250 bp). The closest available reference genome for each sample was identified with Kraken v2 (Wood and Salzberg, 2014), and reads were mapped to the reference genome using bwa-mem v0.7.17 (Li and Durbin, 2009) to assess the quality of the data. De novo assembly was performed with SPAdes v3.14.1 (Bankevich et al., 2012). Automated annotation was made using Prokka v1.12 (Seemann, 2014). Geneious R10 (v10.2.6) was used to investigate genome annotation. The rRNA copy number was estimated by calculating the relative coverage of 16S rRNA vs. that of rpoB, a single-copy gene. All genomes were uploaded to GenBank (BioProject PRJNA679722). BioSample IDs are given in Supplementary Table S2.
Phylogenetic and Phylogenomic Analysis
nodC sequences from strains were extracted from annotated genomes or obtained from GenBank (Supplementary Table S3). Alignment was performed using MUSCLE software (Edgar, 2004). Distances were calculated according to the two-parameter model of Kimura (1980). Phylogenies of nodC were inferred using the neighbour-joining (NJ) method. All analyses were performed using MEGA X software (Kumar et al., 2018). All nodes with a bootstrap value lower than 70% were removed. The similarity of draught genome sequences of India-UK (IU) strains and ICRISAT (IC) strains (Table 1), together with genome sequences from closely related species considered as references, was analysed by calculating pairwise average nucleotide identity (ANI) values (Konstantinidis and Tiedje, 2005; Goris et al., 2007). ANI was performed using the Nucmer algorithm [Average nucleotide identity MUMmer (ANIm)] (Kurtz et al., 2004) as implemented in the JSpecies software v.1.2.1. Pairwise similarity percentage was transformed into a dissimilarity distance matrix and plotted as an NJ cladogram (Saitou and Nei, 1987) on MEGA X (Kumar et al., 2018). BioSample codes for each genome used can be found in Supplementary Table S3.
Cluster Synteny
nod cluster regions were extracted from GenBank files using Geneious R10 (v10.2.6). Synteny analysis was performed in CloVR-Comparative (Angiuoli et al., 2011; Agrawal et al., 2017) and visualised with Sybyl in this platform (Riley et al., 2011). Sybyl defines an orthologue when a protein sequence has an identity >70%, a coverage cutoff of 80%, and an e-value >1e−5.
Nodulation Outer Protein (Nop) Analysis
A local blast database was constructed with IU and IC draught proteomes. Well-characterised genes associated with Type 3 Secretion System (T3SS) machinery (rhcQ, rhcU, ttsI, nolU, and nolV) and its putative effectors (Nop: nopA, nopB, nopC, nopD, nopE, nopF, nopJ, nopL, nopM, nopP, nopT, nopX, nopAA, nopAC, and nopAR) were obtained from UniProt and NCBI databases (as shown in Supplementary Table S4), and blastp was performed. A blastp hit of at least 50% identity, 50% coverage, and e-value >1e−5 in protein sequence was considered an orthologue (as shown in Supplementary Table S4 for locus tags). Clustered heatmaps were generated using the pheatmap R package (Kolde, 2019).
Genetic Features Analysis
CMG-Biotools were used to infer core genomes and pangenomes of IU and IC strains using for orthologue analysis protein files (Vesth et al., 2013). Protein files were uploaded to OrthoVenn2 running locally. Orthovenn2 uses a cutoff 1e−5 to define paralogues (within genomes) and orthologues (between genomes).
Statistical Analyses
For PcoA plots construction, data were analysed in PRIMER 6 (PRIMER-E). Data were normalised and a similarity matrix was calculated using Euclidian distance. Strain samples that lacked a value in any tested variable were removed from the analysis. Permutational multivariate analysis of variance (PERMANOVA) was run in PRIMER 6 (PRIMER-E) using 9,999 unrestricted permutations of raw data. PERMANOVA produces pseudo-F values as a proxy for the difference between beta-diversity and alpha-diversity using a given factor. Statistical analyses were performed on PRISM 9 v9.0.2.
Results
Bacterial Diversity Revealed by BOX-PCR
A total of 111 strains [termed collectively India-UK (IU) strains] were isolated from C. cajan root nodules in three different regions of India with different soils: 32 from South India (Alfisols; Telangana/Andhra Pradesh, HU strains), 47 from Central India (Vertisols; Madhya Pradesh, BRP strains), and 32 from North India (Inceptisols; Uttar Pradesh/Haryana/Punjab, NBAIM strains) (Supplementary Table S1, Figure 1A). BOX-PCR showed a total of 59 different profiles (a to bg) (Supplementary Table S5). A single representative strain from each region was selected from each BOX-PCR profile group, resulting in a total of 65: 20 HU strains, 27 BRP strains, and 18 NBAIM strains (Supplementary Table S5).
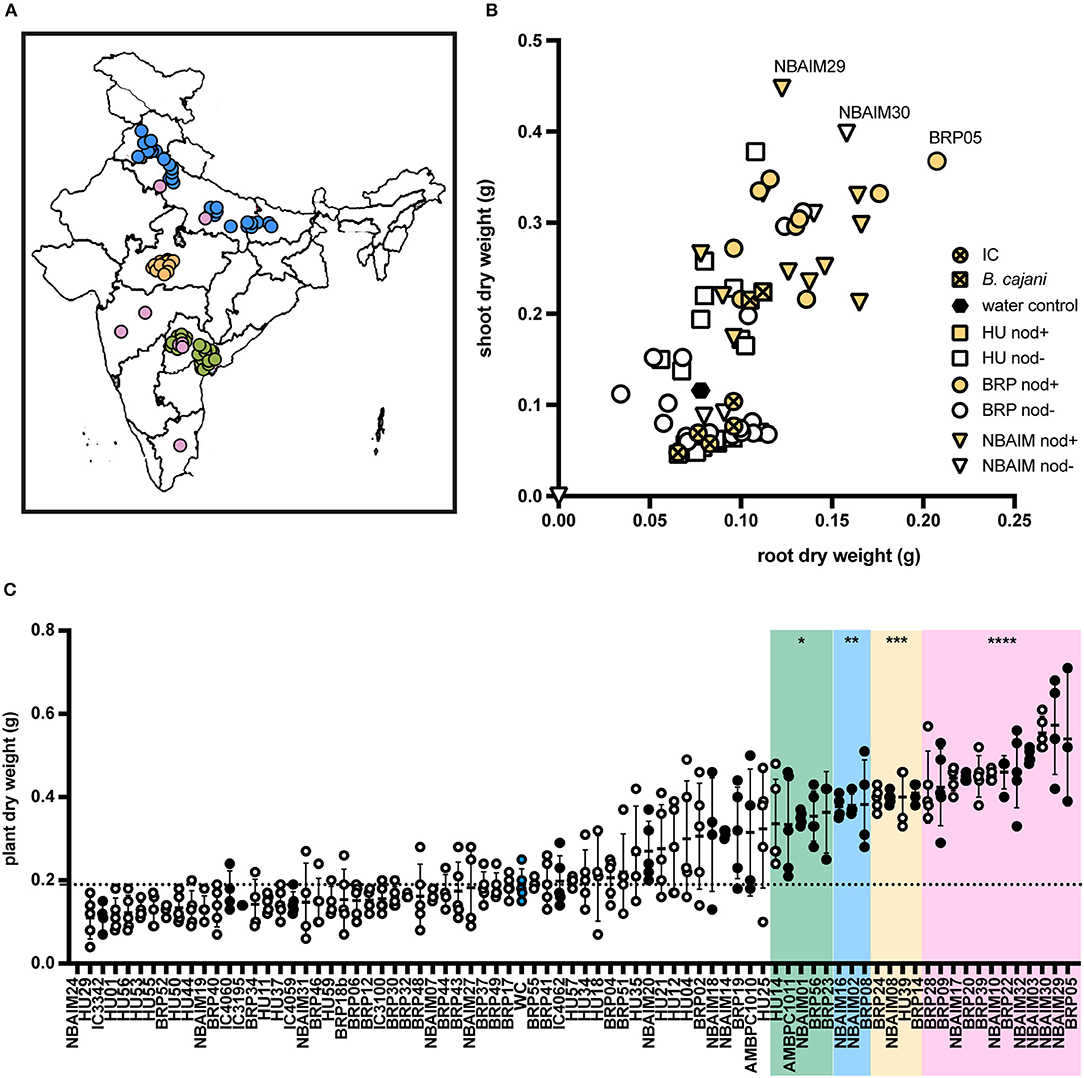
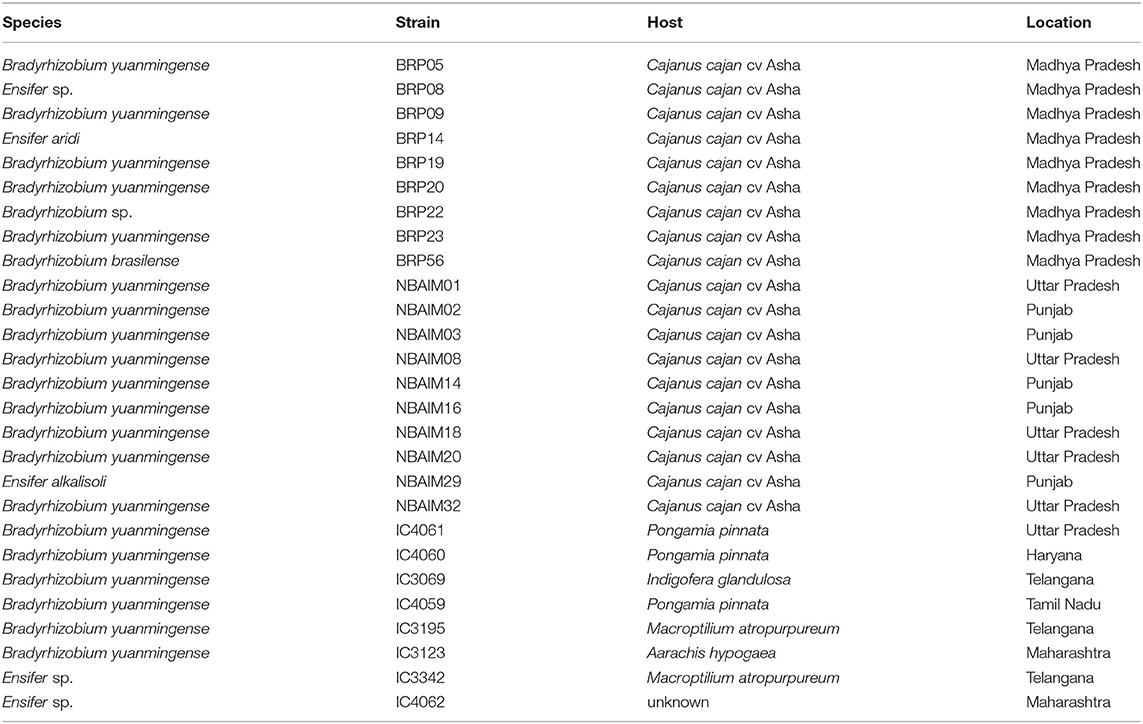
Figure 1. Isolation location and effect of bacterial inoculation on pigeon pea (Cajanus cajan L. Millsp.) of all India-UK (IU) strains and ICRISAT (IC) strains. (A) Map of India with isolation location for all IU and IC strains. In green, strains isolated by the University of Hyderabad (HU), yellow by ICAR-Indian Institute of Soil Science (BRP), blue by ICAR-National Bureau for Agriculturally Important Microorganisms (NBAIM), and pink ICRISAT pigeon pea reference strains. (B) Mean of dry weights of root (X-axis) and shoot (Y-axis) of C. cajan plants, following inoculation with bacterial strains. B. cajani reference strains AMBPC1010T and AMBPC1011 are represented by crossed squares, IC strains by crossed circles, HU isolates by squares, BRP isolates as circles, and NBAIM isolates as triangles. White symbols represent non-nodulating strains (nod-), and yellow symbols show nodulating strains (nod+) under controlled conditions. (C) Mean plant dry weight for each strain. White circles represent nod- strains, black circles nod+, and blue circles water control. A horizontal dashed line represents the water control dry weigh average. One-way ANOVA analysis (F = 15.43, R2 = 0.8076, p < 0.0001) was performed followed by Sidak's post-hoc test. Asterisks represent a significance level vs. water control (WC) (*p < 0.05, **p < 0.01, ***p < 0.005, and ****p < 0.0001).
Ability of Isolates to Nodulate and Influence Plant Growth
The 65 IU strains selected were used to investigate their effect on the growth of pigeon pea plants under controlled conditions (Supplementary Table S6). Some of these IC strains were included as positive controls since some of these are used as pigeon pea inoculants in India. In addition, B. cajani AMBPC1010T and B. cajani AMBPC1011, isolated from C. cajan in the Dominican Republic (Araújo et al., 2017), were included as controls. Nineteen of the tested strains produced nodules on pigeon pea: 9 (BRP) and 10 (NBAIM) (Supplementary Table S6). None of the strains selected from the Telangana/Andhra Pradesh region (HU) formed nodules on C. cajan under these test conditions. Results varied from the increased dry weight of both root and shoot to a detrimental effect when inoculated with NBAIM24 where plants were dead (Figures 1B,C). It is clear that IC strains have a less beneficial effect on the growth of these plants than any of the IU-nodulating strains isolated in this study. Even though IC strains are used as pigeon pea inoculants across India, none were originally isolated from pigeon pea plants (Table 1) (Rupela et al., 1991). The strains that show the most significant increases in plant dry weight compared to uninoculated water controls are NBAIM29 (nod+) and BRP05 (nod+) (Figure 1C). It is important to mention that there are non-nodulating strains, which under these conditions promote the growth of pigeon pea by an unknown mechanism. The nod− strain NBAIM30 has the best performance of a strain that does not form any nodules, with a positive effect on both shoot and root, outcompeting many nodulating strains (Figure 1C).
Phylogenomic Diversity
We sequenced 27 genomes, 19 IU strains, which formed nodules under the test conditions, together with 8 IC strains (Table 1), for comparison with reference strains and to decipher the taxonomic diversity among them. Based on ANIm phylogeny, IU and IC strains were associated either with Bradyrhizobium (22 strains) or Ensifer (5 strains) (Figure 2). Twenty strains are related to B. yuanmingense CCBAU 10071T, showing ANIm similarity values 96.8–98.3%. They all show lower ANIm values (82.3–90.7%) with the next most similar type strains: Bradyrhizobium forestalis INPA54BT, Bradyrhizobium liaoningense CCBAU 83689T, B. cajani AMBPC1010T, and Bradyrhizobium japonicum USDA 6T (Supplementary Table S7). We can consider that these twenty strains (BRP05, BRP09, BRP19, BRP20, BRP23, NBAIM01, NBAIM02, NBAIM03, NBAIM08, NBAIM14, NBAIM16, NBAIM18, NBAIM20, NBAIM32, IC3069, IC3965, IC3123, IC4059, IC4060, and IC4061) belong to B. yuanmingense; henceforth, they are defined as such in subsequent figures. Two strains cluster within Bradyrhizobium superclade II (Ormeño-Orrillo and Martínez-Romero, 2019). BRP56 has an ANIm of 96.5% with Bradyrhizobium brasilense UFLA03-321T, 95.5% with B. elkanii USDA 76T, and 95.4% with Bradyrhizobium pachyrhizi PAC 48T; henceforth, it is referred to as B. brasilense BRP56 in subsequent figures. Within the same superclade, BRP22 shows an ANIm similarity lower than 85.9% to all closely related type strains: Bradyrhizobium macuxiense BR 10303T, B. ivorense CI-1BT, Bradyrhizobium tropiciagri SEMIA 6148T, B. elkanii USDA 76T, B. brasilense UFLA 03-321T, and B. pachyrhizi PAC 48T. Strain BRP22 could represent a new species due to its ANIm similarity value lower than 96%, although new species descriptions based on a single strain are discouraged, given the requirement to demonstrate intraspecific diversity (De Lajudie et al., 2019). Therefore, we cannot, as yet, assign BRP22 to any given species, so it will subsequently be referred to as Bradyrhizobium sp. BRP22. For the five strains in the Ensifer group, NBAIM29 showed 98.9% similarity with Ensifer alkalisoli YIC4027T (E. alkalisoli NBAIM29), BRP14 showed 95.8% similarity with Ensifer aridi LMR002T (E. aridi BRP14), and BRP08, IC3342, and IC4062 showed just 91.2, 91.1, and 90.9% similarity, respectively, with the closest type strain, Ensifer terangae USDA 4894T, meaning that we cannot assign these latter three strains to any known species, i.e., Ensifer sp.BRP08, Ensifer sp.IC3342, and Ensifer sp.IC4062, respectively.
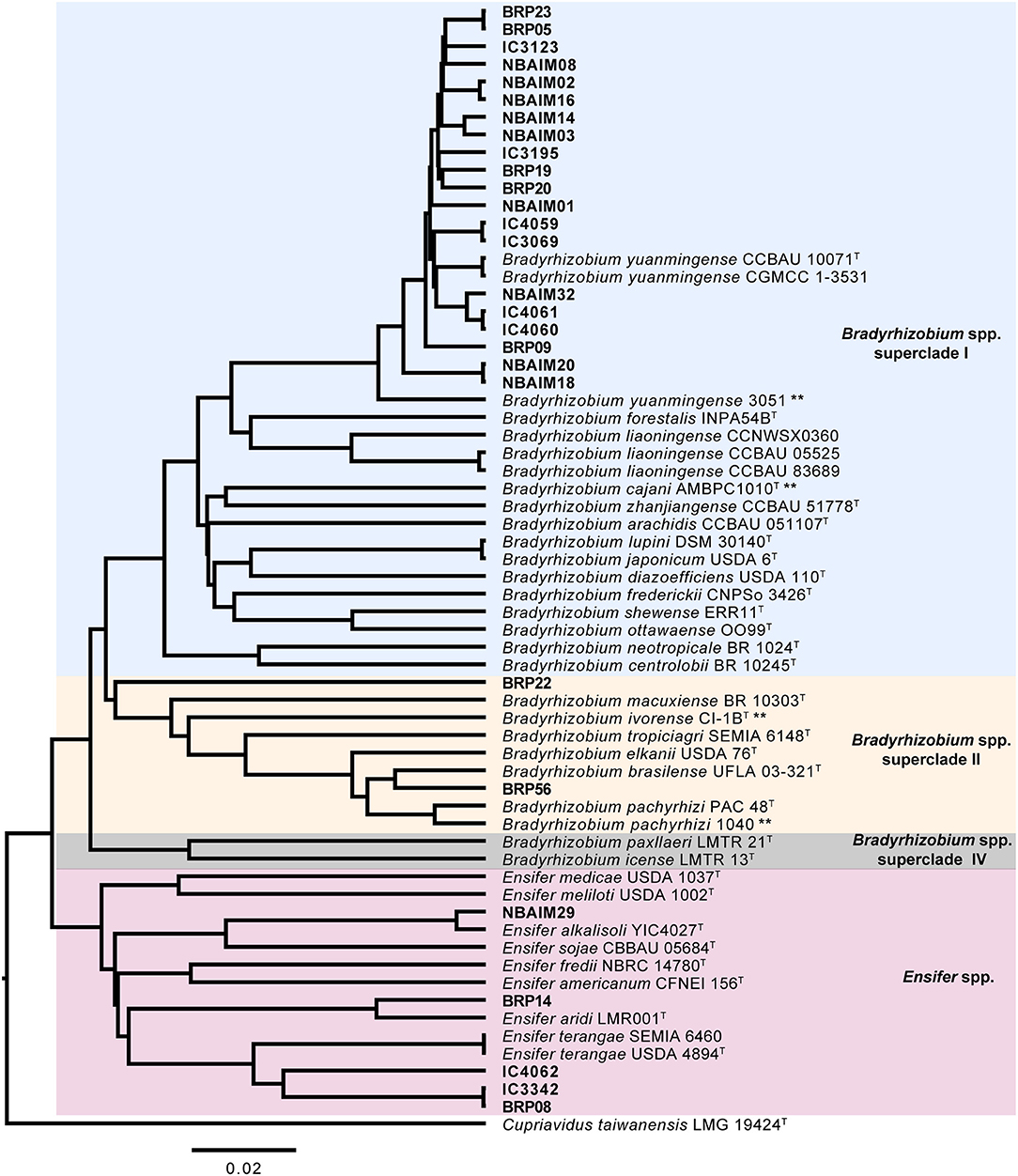
Figure 2. Average nucleotide identity MUMmer (ANIm) dendrogram. ANI-based UPMGA (unweighted pair group method with arithmetic mean) tree of IU, IC, and closely related reference strains. Bar, 2 nt substitutions per 100 nt. **Strains isolated from C. cajan.
Genome Characteristics
The IU and IC Bradyrhizobium strains have a genome of 7.5–9.2 Mb, a GC-content >62.7%, absence of replication plasmid genes (repABC), and 20 out of 22 have a single estimated copy of 16S rRNA (Supplementary Table S2). Most Bradyrhizobium genomes range between 7 and 10 Mb with an average of 8.6 Mb (Ormeño-Orrillo and Martínez-Romero, 2019). However, B. brasilense BRP56 presents a remarkably larger genome among the IU and IC strains at 9.2 Mb (Supplementary Table S2). This strain is phylogenetically related to B. elkanii, which characteristically contains genomes larger than 9 Mb (Reeve et al., 2017). Since there is an inherent difficulty in resolving repetitive regions with short reads by assemblers (Waters et al., 2018), we estimate the rRNA copy number as the coverage ratio between 16S rRNA and the single-copy housekeeping gene rpoB. Most Bradyrhizobium IU and IC strains showed a single predicted copy, except for B. yuanmingense IC3069 and B. yuanmingense NBAIM32. Even though it is uncommon within the genus Bradyrhizobium, strains with closed genomes like B. japonicum USDA 6T or Bradyrhizobium sp. BTAi1 have two copies of the rRNA cluster (Cytryn et al., 2008; Kaneko et al., 2011). There is a direct correlation between rRNA copy number and the time taken for a soil bacterium to respond to nutrient availability (Klappenbach et al., 2000), which could be translated into an adaptive advantage in a rhizosphere environment. In fact, B. yuanmingense NBAIM32 showed a significantly improved performance in plant growth experiments compared with other members of the By group (Figure 3, Supplementary Table S6). None of the IU and IC strains revealed the presence of plasmid-like replication genes (Supplementary Table S2). Although infrequent, plasmid presence was confirmed in Bradyrhizobium sp. BTAi1 and in Bradyrhizobium sp. DOA9 (Cytryn et al., 2008; Okazaki et al., 2015).
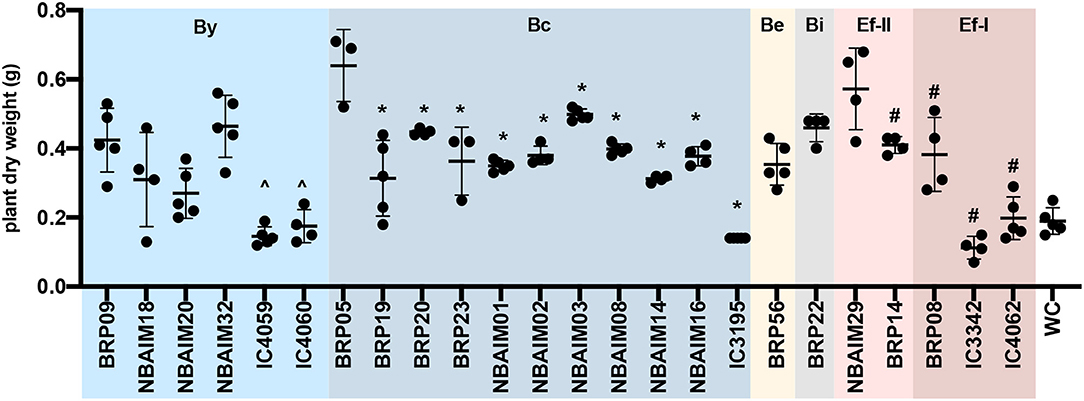
Figure 3. The dry weight of plants inoculated with IU and IC strains. Each dot represents a biological replicate. ∧Represents significant differences for each strain vs. BRP09 within the By group. *Represents significant differences for each strain vs. BRP05 within the Bc group. #Represents significant differences for each strain vs. NBAIM29. Analysis was performed using one-way ANOVA (F = 17.51, R2 = 0.8318, p < 0.0001), with Sidak's post-hoc test. WC, water control.
The IU and IC Ensifer strains showed a genome size of 6.5-7.4 Mb, 61-62 GC%, 3-6 estimated rRNA copies, and 2-3 plasmids (Supplementary Table S2). Most Ensifer spp. genomes have three copies of rRNA, as in Ensifer fredii NGR234 (Viprey et al., 2000), with the exception of Ensifer sp. IC4062, which shows six copies of rRNA. Ensifer IC strains have two copies of repABC, whereas IU strains have three, reflecting a different genomic organisation (Supplementary Table S2). These replicon numbers are in agreement with the work of Sugawara et al. (2013) with 48 different Ensifer spp., which showed 2–5 plasmids in Eckhart gels.
Phylogeny-Based on nodC
The nodC phylogenetic tree (Figure 4) shows that the twenty-seven sequenced strains fall into five main groups: B. yuanmingense (By), 8 strains; B. cajani (Bc), 12 strains; B. elkanii (Be), 1 strain; B. icense (Bi), 1 strain; and E. fredii (Ef), 5 strains. The IU and IC strains assigned as B. yuanmingense belong to either the By or Bc group. In the By group, B. yuanmingense NBAIM32, NBAIM18, NBAIM20, BRP09, IC4060, IC4061, IC3069, and IC4059 show 95% nodC nucleotide identity with B. yuanmingense reference strains, which were isolated in China from Glycine max (soybean) or Lespedeza cuneata, among which is B. yuanmingense CCBAU 10071T. These IU and IC By strains share 89–90% nodC identity with the closed group formed by B. diazoefficiens-related strains (Supplementary Table S8). The second main group, Bc, comprises B. yuanmingense strains IC3195, IC3123, BRP19, BRP20, NBAIM14, NBAIM03, BRP05, BRP23, NBAIM02, NBAIM08, and NBAIM16, which showed more than 96.8% nodC nucleotide identity with Bradyrhizobium zhanjiangense CCBAU 51778T and more than 92% with B. cajani AMBPC1010T (Figure 4). B. yuanmingense NBAIM01 is more distant and shares 88.8–89.3% identity with other Bc, IU, and IC strains, and 87–87.7% with the aforementioned reference strains (Supplementary Table S8). The closest nodC sequence to that of B. yuanmingense NBAIM01 is from Bradyrhizobium sp. LCT2 (91.23%). It is within this Bc group that 44% of the sequenced strains clade together, showing that this is the most common nodC type found in Indian C. cajan endosymbionts. B. brasilense BRP56 (Be group) has a nodC very similar to that of B. elkanii strains (99.7% identity) and 91.3% with that of B. ivorense CI-1BT, a pigeon pea endosymbiont isolated in the Ivory Coast (Fossou et al., 2020). Bradyrhizobium sp. BRP22 is found in group Bi, with its nodC sequence, showing 83.2% and 81.9% similarity, respectively, to B. icense LMTR 13T and Bradyrhizobium paxllaeri LMTR 21T (Figure 4).
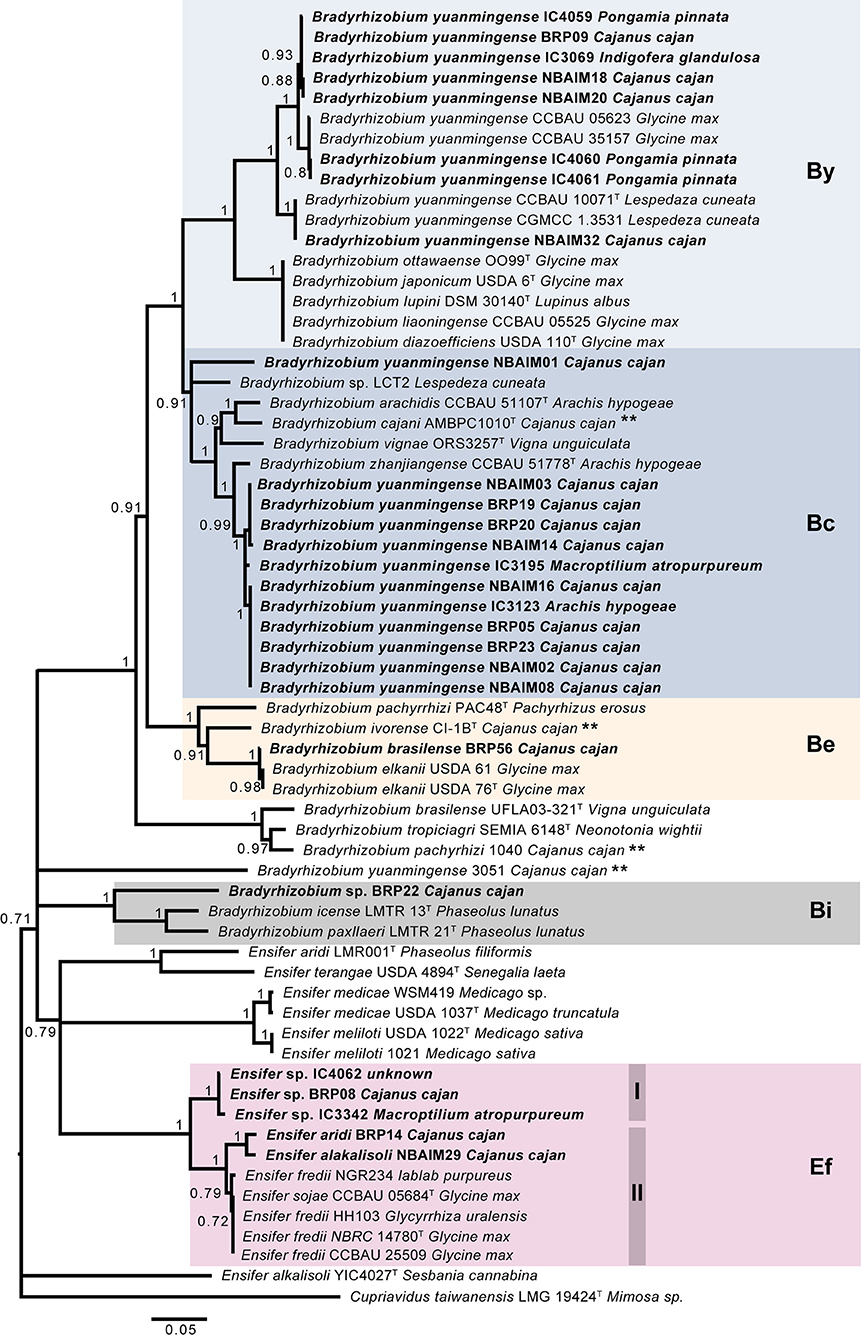
Figure 4. Tree-based on nodC phylogeny. Neighbour-joining (NJ) phylogenetic tree based on nodC sequence (1,458 nt) of IU and IC strains with closely related species. Each is shown together with the plant from which it was isolated. Bootstrap values (only values > 70%, expressed as a percentage of 1,000 replications) are shown at the branching points. Bar, 5 nt substitutions per 100 nt. **Strains isolated from C. cajan. By (B. yuanmingense), Bc (B. cajani), Be (B. elkanii), Bi (B. icense), and Ef (E. fredii).
The IU and IC Ensifer strains have a nodC similar to that of E. fredii (Ef group), which clade in two subgroups, Ef-I (Ensifer sp. BRP08, IC3342, and IC4062) and Ef-II (E. aridi BRP14 and E. alkalisoli NBAIM29). Within Ef-I, the nodC similarity is 99.3–99.9% and <94% with nodC from Ef-II. The Ef-II IU strains share 98.4% nodC identity, and circa, 96%, with E. fredii and E. sojae reference strains in the same Ef-II group (Figure 4).
Nod Cluster Synteny
Nod gene cluster synteny analysis was performed for strains in each nodC group: By, Bc, Be, Bi, and Ef (Figure 4). All IU and IC strains have nodABCIJ as a core cluster, which is present in all symbiotic nod factor (NF)-dependent rhizobia.
Bradyrhizobium
All newly sequenced strains in the By group show the presence of the same nodulation-related genes, nolY-[]-nolA-[]-D2-[]-D1YABCSUIJ-nolN-nodZ (Supplementary Figure S1). Representative strains B. yuanmingense BRP09, B. yuanmingense IC4060, and B. yuanmingense NBAIM32 were selected to further investigate their synteny with B. yuanmingense CCBAU 10071T and B. diazoefficiens USDA 110T (Figure 5A). B. diazoefficiens USDA 110T has three extra nodulation-related genes, nolZ, nolM, and nolO, not present in any of the newly sequenced strains. The By nod cluster is highly conserved, albeit with evidence of different insertion events. We can conclude that the By strains B. yuanmingense NBAIM18, NBAIM20, NBAIM32, BRP09, IC3069, IC4059, IC4060, and IC4061 have the same nod cluster as B. yuanmingense CCBAU 10071T. A highly conserved nod cluster, nolA-[]-nodD2D1YABCSUIJ-nolO-nodZ, is present in the newly sequenced strains belonging to the Bc group (Supplementary Figure S2). B. yuanmingense NBAIM08 and IC3195 were selected as representative and aligned with B. cajani AMBPC1010T and B. zhanjiangense CCBAU 51778T (Figure 5B). Notwithstanding transposase-related genes in B. yuanmingense IC3195 (which are not present in either of the type strains), we can conclude that the nod cluster and its genomic context are the same as that of B. cajani AMBPC1010T and B. zhanjiangense CCBAU 51778T. The nodC phylogeny shows that B. brasilense BRP56 belongs to the Be group, together with B. ivorense CI-1BT and B. elkanii USDA76T (Figure 4). The observed nod cluster is nolY-[]-nolA-[]-nodD2D1-[]-ABCSUIJ-nolO-nodZ (Figure 5C). B. brasilense BRP56 has a nodK gene among nodD1 and nodA, which is not annotated in CI-1BT, and neither is it in B. elkanii USDA 76T. Both reference strains have an open reading frame (ORF) in this region, which, in USDA 76T, has an amino acid (aa) identity of 97.7% to nodK of B. brasilense BRP56, whereas, in CI-1BT, it is just 59.4%. Furthermore, both B. brasilense BRP56 and B. elkanii USDA 76T have nopM downstream of nifA (Figure 5C). Overall, we suggest that B. brasilense BRP56 has a typical B. elkanii-type nod cluster. Bradyrhizobium sp. BRP22, together with B. icense LMTR 13T and B. paxllaeri LMTR 21T, belongs to the Bi nodC group (Figure 4). Analysis of their nod cluster synteny shows that all strains have nolA-[]-nodD2D1-[]-ABCSUIJ-nolO-nodZ-[]-noeE (Figure 5D). In addition, Bradyrhizobium sp. BRP22 and B. icense LMTR 13T have nopM downstream of noeE, whereas B. paxllaeri LMTR 21T shows a pseudogene (pg) with 50–54% aa identity to the N-terminal part of nopM. We conclude that Bradyrhizobium sp. BRP22 has a B. icense-type nod cluster.
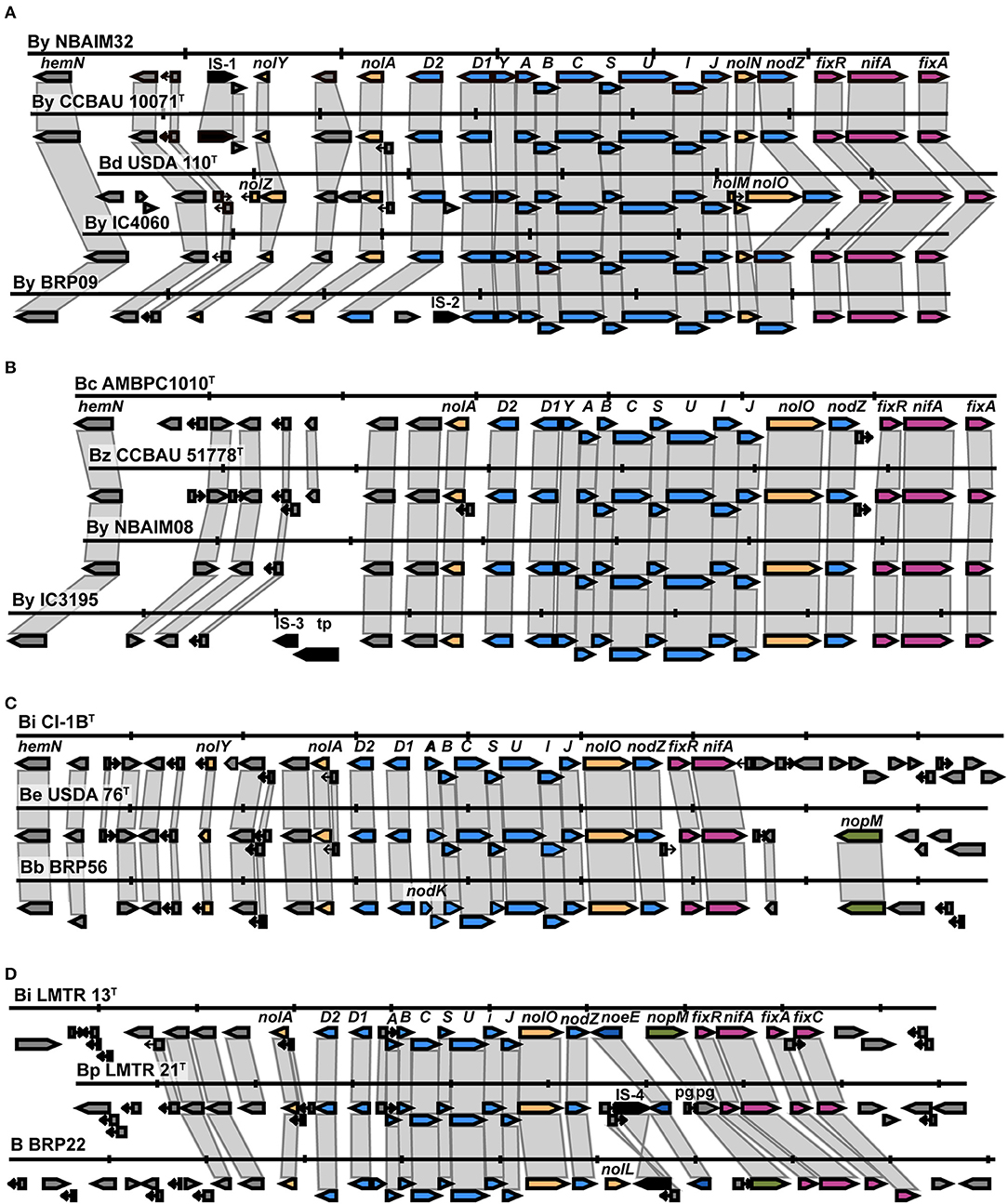
Figure 5. The nod cluster synteny of IU and IC strains in the By, Bc, Be, and Bi groups. (A) By group, (B) Bc group, (C) Be group, and (D) Bi group. Each row represents a single strain and shows the nod cluster organisation and its genomic context. The colour of the arrow reflects the genes: blue for nod, yellow for nol, pink for fix-nif, green for nop, dark blue for noe, and black for transposase/insertion-related genes. Vertical markers indicate 5 Kb in each genome. B (Bradyrhizobium sp.), By (B. yuanmingense), Bd (B. diazoefficiens), Bc (B. cajani), Bz (B. zhanjiangense), Bi CI-1BT (B. ivorense CI-1BT), Be (B. elkanii), Bb (B. brasilense), Bi LMTR13T (B. icense LMT13T) and Bp (B. paxllaeri).
In selected representative strains from each Bradyrhizobium nodC group (Figures 4, 5), this region of DNA was aligned (Supplementary Figure S3). As all strains have nolA-[]-nodD2-[]-nodD1-[]-nodABCSUIJ-[]-nodZ, this could be considered the minimum nod cluster necessary to nodulate pigeon pea. Other nodulation-related genes (nolY, nodK, nodY, nolO, nolN, nolL, or noeE) may be associated with host specificity and, therefore, play a part in the symbiotic performance.
Ensifer
The nod cluster regions for the five newly sequenced Ensifer strains were aligned and, together with reference strains in the same nodC phylogenetic clade, reveal a high degree of synteny within the group (Figure 6). The observed canonical nod cluster is nodABCIJ-nolO-noeI-[]-noeE. There are two genomic contexts for each nodC group (Ef-I and Ef-II), which suggests that they could have a different origin (Figure 6). Despite this, the IU and IC strains show great conservation of the nod cluster, except for the absence of nolL from E. alkalisoli NBAIM29. Instead, it has cysNC (adenylyl-sulphate kinase, A0A4S5J185). The lack of nolL could give E. alkalisoli NBAIM29 an advantage in plant recognition, which would explain its enhanced growth promotion phenotype observed in planta (Figure 3), although, as it is based only on a single strain, this is highly speculative.
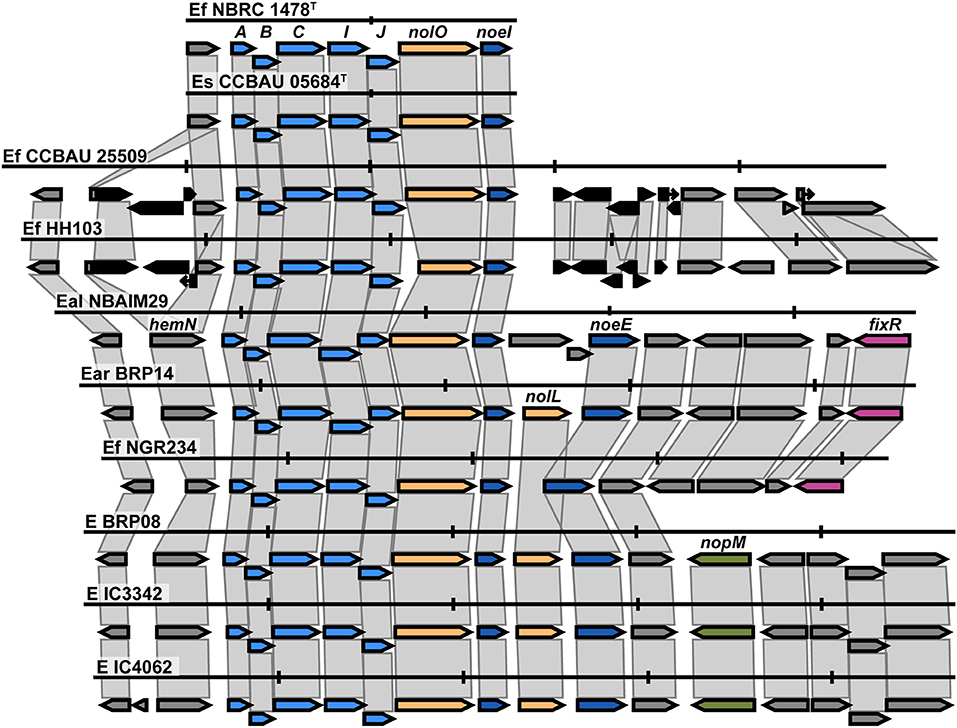
Figure 6. The nod cluster synteny of IU and IC strains in the Ensifer group together with type strains. Each row represents a single strain and shows the nod cluster organisation and its genomic context. The colour of the arrow reflects the genes: blue for the nod, yellow for nol, pink for fix-nif, and black for transposase/insertion-related genes. Vertical markers indicate 5 Kb in each genome. E (Ensifer sp.), Ef (E. fredii), Es (E. sojae), Eal (E. alkalisoli) and Ear (E. aridi).
Presence of Gene-Encoding Nops
There are multiple pieces of evidence supporting a T3SS and Nops and their key roles in the establishment of symbiosis and host specificity in certain rhizobia-legume interactions (Pueppke and Broughton, 1999; López-Baena et al., 2016). We have confirmed the presence of T3SS machinery by finding orthologues (>50% aa identity and coverage) for rhcQ, rhcU, ttsI, nolV, and nolU from the well-characterised T3SS of B. vignae ORS3257 (Teulet et al., 2019) and E. fredii NGR234 (Freiberg et al., 1997) in all the newly sequenced strains presented in this study (Supplementary Table S4). To determine the putative range of T3SS effectors, we have based analysis on the Nops and used Nop sequences from well-characterised Ensifer and Bradyrhizobium spp. to find homologues in the IU and IC genomes (as shown in Supplementary Table S4).
Within the Bradyrhizobium strains, all IU and IC strains have orthologues for nopT, nopP2, nopM2, and nopM3. These T3SS effectors could be needed for establishing symbiosis between Bradyrhizobium spp. and pigeon pea (Figure 7A). The groups formed based on the presence and absence of Nop orthologues in Bradyrhizobium spp. (Figure 7A) are highly correlated with those observed in the nodC phylogeny and nod cluster synteny (Figures 4, 5). Cluster I (By) and cluster II (Bc) are distinguished from each other by the presence or absence of two groups of nop genes: group A (nopC, nopAA, nopM1, and nopX), group B (nopD, nopAR, nopL, and nopE), where cluster I (By) has group A genes but not those of group B and cluster II (Bc), vice versa (Figure 7A). There are a few orphan strains that present a different presence/absence pattern, e.g., B. yuanmingense BRP09 has all the Nop genes present in the other By strains (cluster I), plus nopAA (Figure 7A). However, we have not observed differences in plant dry weight between B. yuanmingense BRP09 and other IU strains of the By group (Figure 3), suggesting that the presence of nopAA is uncorrelated with plant performance. Bradyrhizobium yuanmingense NBAIM01 belongs to the Bc nod group (cluster II), although it shows a very different set of Nop homologues. Its lack of nopX and nopC (nolJ) could be counterbalanced by the presence of nopB, nopL, and/or nopE effectors since the plant performance of B. yuanmingense NBAIM01 is similar to that of other Bc members (Figure 3). Finally, B. yuanmingense BRP05 displays the common Nop genes for Bc strains (group II), plus nopA (Figure 7A). This strain promotes plant growth significantly more than any other Bc member or, indeed, any other Bradyrhizobium strains tested (Figure 3), which could be a result in part of the presence of this T3SS effector, although, without further strains showing similar characteristics, it is impossible to draw firm conclusions at this stage.
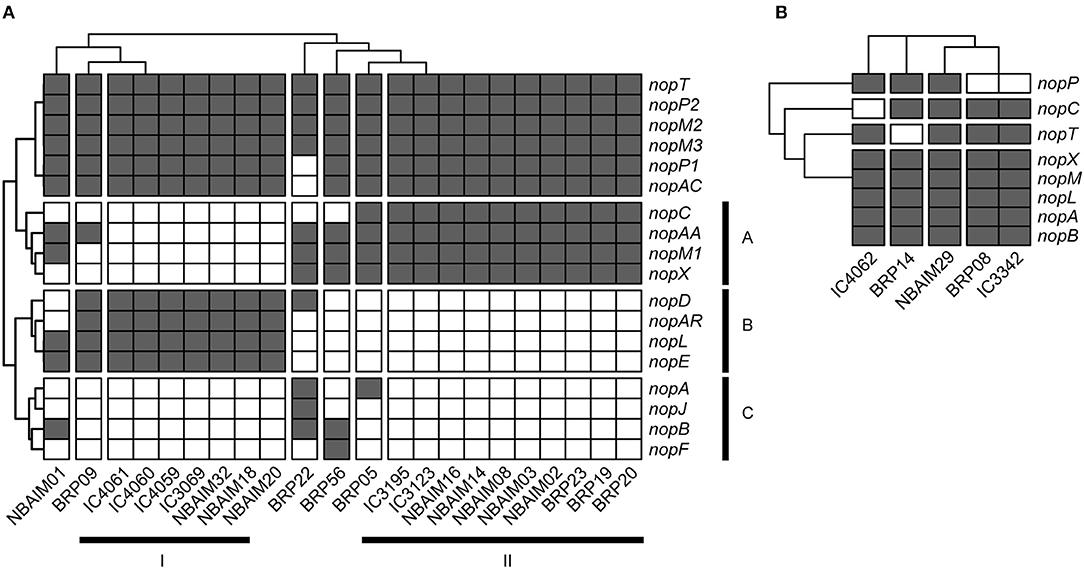
Figure 7. The presence and absence of nop genes. (A) Bradyrhizobium IU and IC strains are shown as a heat map. (B) Ensifer IU and IC strains are shown as a heat map.
In comparison to Bradyrhizobium, a total of only seven nop genes are present in Ensifer (Figure 7B). All Ensifer IU and IC strains show orthologues for nopA, nopB, nopL, nopM, and nopX. Nevertheless, only E. alkalisoli NBAIM29 has nopC, nopP, and nopT, and, together with the lack of nolL, could equip it for improved performance on pigeon pea (Figure 3). However, further strains showing the same characteristics are required to test this speculation.
Genetic Features
Putative proteomes of Bradyrhizobium and Ensifer IU and IC strains were analysed to infer their core genome and pangenomes (Figures 8A,D). Both groups of strains showed an open pangenome of 17,596 and 10,458, respectively, and a core genome of circa 3,500 genes for both. Non-core genes present in each group of strains could play a role in soil endurance, competition for root colonisation, host specificity, or symbiosis establishment and, therefore, may help explain the differences observed in plant growth promotion (Figure 3).
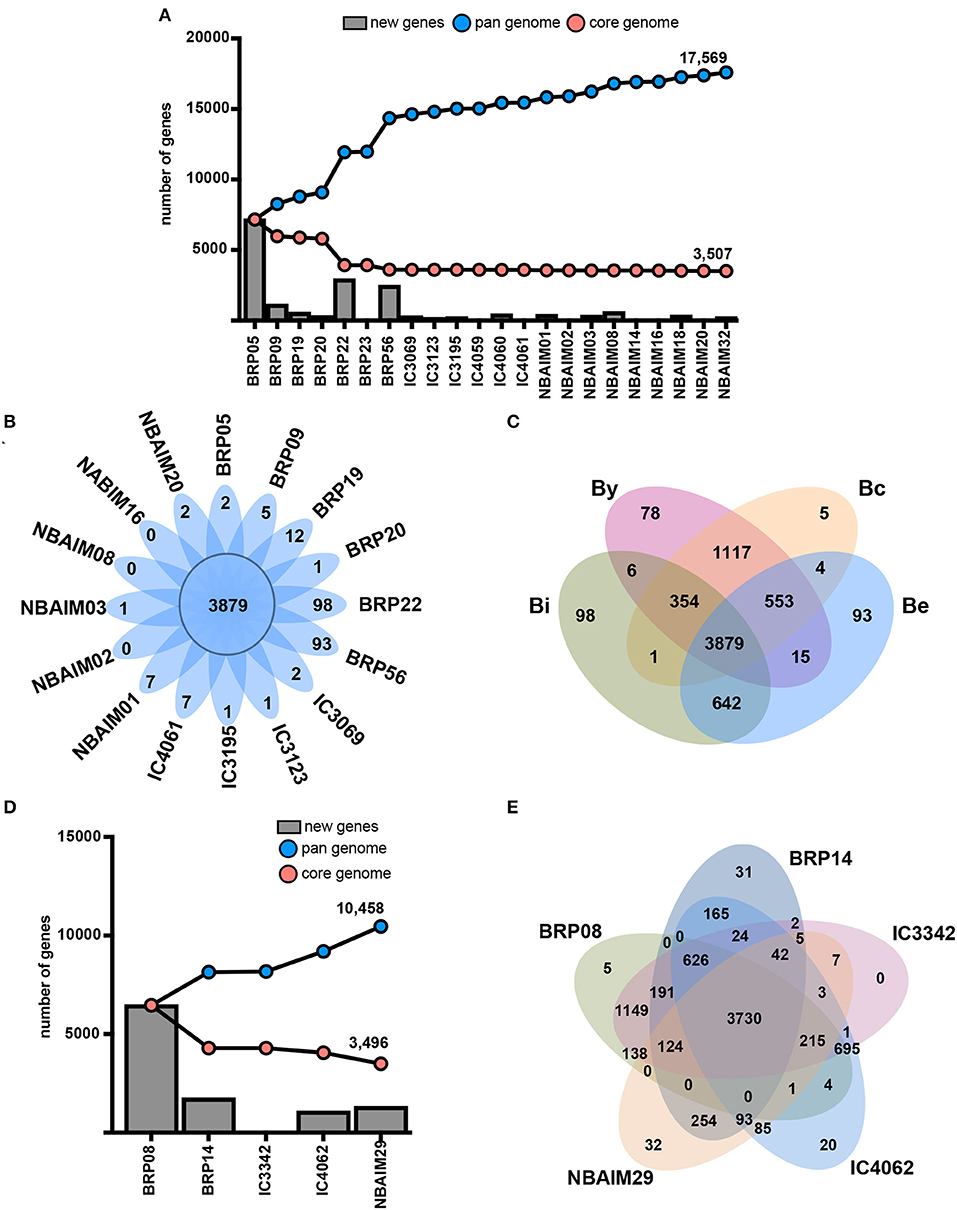
Figure 8. Genetic features. (A) Core genome and pangenome of Bradyrhizobium where X-axis shows strains and the Y-axis, the number of genes. Blue line: pangenome. Red line: core genome. New genes: bars. (B) Core and exclusive orthologue clusters for each Bradyrhizobium strain. (C) Venn diagram showing shared and exclusive orthologue families among nodulation-type groups. By: the B. yuanmingensenod-type group. Bc: the B. cajaninod-type group. Be: the B. elkaniinod-type group. Bi: the B. icensenod-type group. (D) Coregenome and pangenome of Ensifer where X-axis shows strains and the Y-axis, the number of genes. Blue line: pangenome. Red line: core genome. New genes: bars. (E) Core and exclusive orthologue clusters for each Ensifer strain.
Bradyrhizobium Orthologues
To reduce the complexity and computational time, we chose representative strains (shown in bold) between those sharing ANIm values greater than 99% similarity: B. yuanmingense BRP05-BRP23 (99.91%), IC3069-IC4069 (99.86%), NBAIM03-NBAIM14 (99.18%), NBAIM32-IC4060-IC4061 (99.27-99.28%), and NBAIM18-NBAIM20 (99.89%). All IU and IC strains share a total of 3,879 orthologue clusters with enrichment of DNA-related functions (GO:0006412, GO:0006313, and GO:0003700), transmembrane transport (GO0055085 and GO:0008643), and cell shape regulation (GO:0008360) (Figure 8B). Thirteen out of 16 analysed strains show exclusive orthologue clusters, which are not present in any other strain. Bradyrhizobium sp. BRP22 and B. brasilense BRP56 show the greatest number of exclusive groups of orthologues, 98 and 93, respectively (Figure 8B), which could be the result of the phylogenomic differences with other B. yuanmingense strains (Figures 2, 4, 5).
Since the most distinct feature observed among IU and IC strains is the nodulation-related gene groups (Figure 5, Supplementary Figure S3), we have compared the orthologue clusters shared among By, Bc, Be, and Bi (Figure 8C). The exclusive genes for each group represent orthologue clusters that are present in all strains within that specific group. By is the only group that exclusively shows enrichment in GO functions for carbohydrate transport (GO: 0008643 and GO: 0015407). Among the 1,117 orthologue groups shared between By and Bc strains there is enrichment in clusters associated with chemoreceptors (GO: 0007165), permeases (GO: 005585), and flagellum-dependent cell motility (GO: 0071973), which are not present in Bradyrhizobium sp. BRP22 and B. brasilense BRP56. Nevertheless, these strains could partially compensate for this absence through the catabolism of aromatic compounds (GO: 0019439), which are a component of pigeon pea root exudates (Ae et al., 1990).
Ensifer Orthologues
We compared the orthologues groups of IU and IC Ensifer strains, which all share 3,730 orthologue clusters (Figure 8E). In this core set, there is enrichment in different DNA-related biological processes (GO: 0006412, GO: 0006313, and GO: 0035556) and transmembrane transport (GO: 0055085). The comparison between IU and IC strains shows enrichment for IC strains in an orthologue cluster annotated as putative adenylate cyclase 3 (cya3, GO: 0035556), which could be the reason for the significantly different plant performance between IC and IU strains (Figure 3). Ensifer sp. IC3342, Ensifer sp. IC4062, Ensifer sp. BRP08, and E. aridi BRP14 strains shared 626 orthologues families that are not present in E. alkalisoli NBAIM29 (Figure 8E). Within this group of orthologues, there is enrichment in the biosynthesis pathway of rhizobactin 1021 (GO: 0019289). Since E. alkalisoli NBAIM29 has a significantly better plant performance (Figure 3), we hypothesise that synthesis of rhizobactin 1021 might be a cost, which E. alkalisoli NBAIM29 would not sustain.
Genotype-Metadata Correlation
We have analysed the pigeon pea population structure based on GC%, genome length, number of tRNAs, rRNA clusters, repABC, nod type, and presence/absence of nod and nop genes (Figure 9). The population separates based on the nod group each strain belongs to (Figure 9A). In addition, we have run PERMANOVA using bacterial species (B. yuanmingense, Bradyrhizobium sp., Ensifer sp., E. alkalisoli, and E. aridi), nod type (By, Bc, Be, Bi, Ef-I, and Ef-II), nop profile (B-nop-I, B-nop-II, B-nop-II, B-nop-IV, B-nop-V, B-nop-VI, B-nop-VII, E-nop-I, E-nop-II, E-nop-III, and E-nop-IV), location of isolation (Madhya Pradesh, Uttar Pradesh, Punjab, Haryana, Tamil Nadu, and Maharashtra), the plant host from which the strain was originally isolated (C. cajan cv. Asha, C. cajan cv. Bahar, Indigofera glandulosa, Arachis hypogaea, Macroptilium atropurpureum, and Pongamia pinnata), and the culture collection (or origin) (BRP, IC, and NBAIM) (Figure 9B). The main factor controlling the assembly of the pigeon pea endosymbiont population is the type of nod genes, followed by the species the strain belongs, to and finally, the nop gene set that each strain contains. The factors that had no significance were the location of isolation, the plant host from which the strain was originally isolated, and the culture collection from which the strain came (origin).
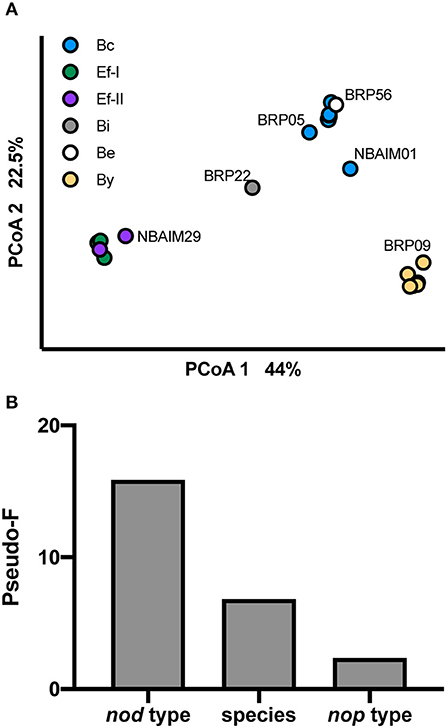
Figure 9. Population structure. (A) PCoA representing pigeon pea population and shown with visual separation by nod type. (B) Influence of different factors on pigeon pea endosymbiont population using PERMANOVA. Pseudo-F value as a proxy. N = 27.
Discussion
Among the BOX-PCR-reduced population of 65 strains, only 19 were able to establish symbiosis with pigeon pea under laboratory conditions. The isolation of non-symbiotic bacteria from nodule samples has previously been reported (Wu et al., 2011), as well as opportunistic infection (Zgadzaj et al., 2015). Moreover, Fossou et al. (2016) in their sampling of nodule-isolated strains from pigeon pea in Ivory Coast showed that 22% of the population did not display any rhizobia-like features. In addition, they were unable to amplify nitrogenase-encoding sequences (nifH) from 5% of the selected strains.
In our pigeon pea endosymbiont population of 27 IU and IC strains, the diversity is moderately driven by the species the strain belongs to (Figure 9). The main species-nodulating pigeon pea in India is B. yuanmingense (20 out of 27, Figure 2). In their study of pigeon pea endosymbionts in the Dominican Republic, Araújo et al. (2015) found that all strains investigated had a 99.8% identity to B. yuanmingense CCBAU 10071T. Moreover, one of the sequenced pigeon pea endosymbionts from this study, B. yuanmingense 3051 (ALSPC3051), shows a high ANIm similarity (95.7–96%) to B. yuanmingense IU and IC strains (Figure 2, Supplementary Table S7). It is interesting to note that B. yuanmingense may be a predominant symbiont in India since it has been isolated from several legumes throughout the country (Ojha et al., 2017; Rathi et al., 2018). B. brasilense BRP56 and Bradyrhizobium sp. BRP22 are rare strains in this population; however, within this superclade II exist other pigeon pea-nodulating species isolated in the Dominican Republic, Brazil, and Ethiopia, such as B. ivorense and B. elkanii (Stepkowski et al., 2003; Wolde-Meskel et al., 2005; Fossou et al., 2016, 2020).
Nodulation-related genes are the main diversity driver in the pigeon pea endosymbiont population (Figure 9). The nodC sequences defined clear groups, with Bc the most common (12 out of 27). This sequence is similar to the reference strains, including B. zhanjiangense CCBAU 51778T, a Chinese strain isolated from A. hypogaea (Li et al., 2019), and B. cajani AMBPC1010T, a C. cajan strain isolated in the Dominican Republic (Araújo et al., 2017). Despite differences observed in nodC, all Bradyrhizobium spp. share a common nod cluster nolA-[]-nodD2-[]-nodD1-[]-nodABCSUIJ-[]-nodZ (Supplementary Figure S3). However, different presence/absence patterns were observed for genes related to NF modifications, nolY, nodK, nodY, nolO, nolN, nolL, or noeE for each Bradyrhizobium nodC type group, By, Bc, Be, and Bi (Figures 4, 5). The IU and IC Bradyrhizobium strains either have nodY or nodK between nodD1 and nodA (Supplementary Figure S3), whose functions have not yet been elucidated (Menna and Hungria, 2011). The IU and IC strains from the Bc and Be groups have a nolY homologue upstream from nolA (Figures 5B,C). In B. diazoefficiens USDA 110T, a nolY mutant showed a significant disadvantage in nodule kinetics on Vigna radiata (mung bean), but this detrimental effect was not so strong in soybean (Dockendorff et al., 1994), suggesting that the presence or absence of nolY in Bradyrhizobium spp. could be related to their host range. Bradyrhizobium sp. BRP22 is the only strain with nolL and noeE homologues, which seem to modify the NF playing a role in host specificity (Figure 5D) (Corvera et al., 1999; Wei et al., 2008). Moreover, differences were observed in the presence and absence of the annotated carbamoyltransferases nolN and nolO (Supplementary Figure S3). Bc, Be, and Bi strains have the nolO homologue between nodJ and nodZ, whereas By strains have nolN. In B. diazoefficiens USDA 110T nolMNO is part of the nod operon, and nolNO, together with nodZ, acts in the NF 2-O-methylfucosylation. However, mutants in nolO or nolNO in this strain showed the same phenotype: delayed nodule formation and a reduced percentage of nodules per plant in legumes like soybean or mung bean (Luka et al., 1993). Since both genes encode carbamoyltransferases, which probably undertake the same function, it is possible that NF 2-O-methylfucosilation is essential to establish symbiosis in pigeon pea. Since differences in plant performance were not observed (Supplementary Figure S4B), we conclude that the common functional nod cluster for IU and IC Bradyrhizobium spp. is nolA-nodD2D1-nodY/K-nodABCSUIJ-nolO/nolN. Genes like nodY, nodK, noeE, and nolL only reflect phylogenetic diversity among these strains. The absence of nolY could have been positively selected, since the major group, Bc, does not show a nolY homologue. However, this selection has no impact on plant performance (Supplementary Figure S4A).
Based on the orthologue analysis, each Bradyrhizobium group could have developed a different strategy to endure in the pigeon pea rhizosphere (Figure 8C). Bc is the most common pigeon pea symbiont group in our population, and we hypothesise that it may be better adapted to the pigeon pea root environment. Together with By, both have homologues of chemoreceptors and flagella, which Be and Bi do not possess. These groups of genes are essential in rhizobium-legume symbioses (Jiang et al., 2016; Wheatley et al., 2020). However, Bradyrhizobium sp. BRP22 and B. brasilense BRP56 could partially compensate for this absence through the catabolism of aromatic compounds, which are present in pigeon pea root exudates (Ae et al., 1990). The By group shows enrichment in carbohydrate transporters. Since sugars are one of the main components of root exudates (Lynch and Whipps, 1990), it is possible that having a greater pool of carbohydrate transporters could give the By group an adaptative advantage in the pigeon pea rhizosphere, resulting in their increased prevalence in nodules.
Among IU and IC Bradyrhizobium spp., the Bc strain B. yuanmingense BRP05 promotes plant growth more significantly than any other strain tested (Figure 3), which could be, in part, a result of the presence of T3SS pili structures like nopA (Figure 7A). NopA is part of the external T3SS apparatus, and its deletion completely abolishes the secretion of other Nops, since it is a major component of the T3SS pili (Krishnan et al., 2003). However, in E. fredii USDA 257, the absence of nopA extends the host range to other soybean varieties, whereas, in cowpea, it has a slightly deleterious effect (Kim and Krishnan, 2014). Nevertheless, it is impossible to draw firmer conclusions without further strains showing similar characteristics to B. yuanmingense BRP05.
Ensifer spp. is an infrequent pigeon pea endosymbiont in the population since only five (of 22) IU and IC strains were assigned to this genus. There are a few records of Ensifer strains,-nodulating pigeon pea, including strains isolated in Cerrado soil in Brazil and India (Coutinho et al., 1999; Stepkowski et al., 2003). Their rarity is probably related to pigeon pea specificity rather than low Ensifer spp. numbers in soil, since, in India, Ensifer spp. are common endosymbionts of native legumes growing in alkaline soils (Gehlot et al., 2013; Tak et al., 2016; Sankhla et al., 2017; Rathi et al., 2018; Choudhary et al., 2020). Regarding nodulation genes, the most relevant feature is nolL, where its absence correlates with a significantly improved plant performance in E. alkalisoli NBAIM29 (Figures 3, 6). nolL determines 4-O-acetylation of the fucosyl residue in NF, and its deletion has been shown to have a negative effect on R. etli CE3 nodule kinetics in some Phaseolus vulgaris cultivars and in V. umbellata (Corvera et al., 1999). Furthermore, the heterologous expression of nolL in E. fredii USDA 257 extends its host range to other legumes like Leucaena leucocephala and L. halophilus (Berck et al., 1999). NolL plays a role in both host specificity and host range. Therefore, we hypothesise that the lack of the NF fucosyl acetylation might give E. alkalisoli NBAIM29 an advantage in plant recognition, explaining the phenotype observed in planta (Figure 3). However, with only one strain, this is highly speculative. Moreover, E. alkalisoli NBAIM29 is the only IU and IC Ensifer spp. strain that has nopC, nopP, and nopT; all of them are well-characterised T3SS effectors with functions related to host-range and interaction with the plant immune system. The deletion of any of these nop genes results in a reduction of nodules in the symbiosis between E. fredii and different legumes (soybean and P. vulgaris) (Boundy-Mills et al., 1994; Skorpil et al., 2005; Dai et al., 2008; López-Baena et al., 2009).
The comparison of orthologue groups between the IU and IC Ensifer spp. strains showed an exclusive group in IC annotated as an adenylate cyclase 3 (cya3), which modulate the extent of epidermal infection during nodulation (Tian et al., 2012). Indeed, a mutation in cya3 (cya5) in E. meliloti CXM1-105 significantly increased alfalfa shoot dry weight (Sharypova et al., 1999), which could be reflected in the different plant performances between IC and IU strains (Figure 3).
Remarkably, E. alkalisoli NBAIM29 lacks an orthologue family related to the synthesis of rhizobactin 1021 (rhbBCDEF), a siderophore that chelates iron (Fe) (Lynch et al., 2001). We hypothesise that the biosynthesis of a siderophore might be redundant in the pigeon pea rhizosphere since it exudes piscidic acid, an aromatic compound that solubilises phosphorous (P) by chelating Fe from P-Fe compounds (Ae et al., 1990). Siderophore biosynthesis would represent a metabolic cost to the other Ensifer strains, Ensifer sp. IC3342, Ensifer sp. IC4062, Ensifer sp. BRP08, and E. aridi BRP14, and could explain their significantly lower plant performance compared to that of E. alkalisoli NBAIM29 (Figure 3). It is possible that the presence of these Nop proteins, together with the lack of nolL and the rhizobactin 1021 biosynthesis pathways, endows E. alkalisoli NBAIM29 with improved plant recognition machinery that could translate into better performance with pigeon pea, but without further strains showing the same characteristics, it is impossible to tell at this stage.
Due to its intrinsic capacity to tolerate drought (grown on drylands), pigeon pea is a promising candidate for resilience to climate change; however, its yield remains low. The use of symbionts well-adapted to the growth conditions of pigeon pea could increase its productivity (Pellegrino et al., 2011; Pellegrino and Bedini, 2014; Pastor-Bueis et al., 2019). Our findings demonstrate that the most common pigeon pea endosymbiont in India is a B. yuanmingense strain with a B. cajani-B. zhanjiangens (Bc) nod type, defined mainly by the absence of nolY and the presence of nolO. Since we have not observed location to be a driving factor in population diversity, our findings may apply to much, if not all, of India. Due to its intrinsic capabilities for persisting and establishing symbiosis, in addition to its genetic and genomic features, we suggest that B. yuanmingense BRP05 could be a good candidate for inclusion in inoculum formulations for pigeon pea in India. However, testing a range of IU strains for symbiotic performance in field trials is essential to assess their real-world performance. Moreover, the less common Ensifer strains, like E. alkalisoli NBAIM29, may be better for alkaline conditions, where members of this genus often perform well (Gehlot et al., 2013; Tak et al., 2016; Sankhla et al., 2017; Rathi et al., 2018; Choudhary et al., 2020).
This study presents a first step in defining and collecting strains that can nodulate pigeon pea in Indian soils. Their ability to influence plant performance has been investigated in glasshouse experiments under sterile conditions. Therefore, extensive trialling in the field in India, using a range of different varieties of pigeon pea, is now suggested to evaluate their performance under these agronomic conditions. We are confident that such studies will lead to the selection of a group of highly effective strains for use in inoculant technology, improving the symbiotic performance of this essential legume in India.
Data Availability Statement
The data presented in the study are deposited in GenBank repository, BioProject PRJNA679722.
Author Contributions
BJ, PP, DR, AS, AP, and VR conceived the study and designed the manuscript. BJ, MM, AB, DC, SP, NA, SM, SK, PS, and MK performed the experiments. BJ, MM, and AT analysed the data. BJ prepared the manuscript. BJ, AT, AE, VR, EJ, PP, MM, DR, AP, AS, and AB critically reviewed the manuscript. All authors listed have made a substantial, direct and intellectual contribution to the study, and approved it for publication.
Funding
This study was supported by the Biotechnology and Biological Sciences Research Council [Grant No. BB/N013387/1], granted to PP; and by the Department of Biotechnology, Government of India [Grant No. BT/IN/UK-VNC/41/DLN/2015-16], granted to DR, AS, and AP.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank the Indian Council of Agricultural Research, New Delhi, and the Director IISS, Bhopal, India, for administrative support. We thank International Crops Research Institute for the Semiarid Tropics (ICRISAT), Hyderabad, India, for supplying pigeon pea seeds and rhizobia strains (IC). We would like to thank Prof. Fernando González-Andrés from Universidad de León, Spain, for sharing B. cajani AMBPC 1010T and B. cajani AMPBC 1011 strains. We would like to thank Lockdown for giving us the opportunity to put this study together for publishing.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.680981/full#supplementary-material
References
Ae, N., Arihara, J., Okada, K., Yoshihara, T., and Johansen, C. (1990). Phosphorus uptake by pigeon pea and its role in cropping systems of the Indian subcontinent. Science 248, 477–480. doi: 10.1126/science.248.4954.477
Agrawal, S., Arze, C., Adkins, R. S., Crabtree, J., Riley, D., Vangala, M., et al. (2017). CloVR-Comparative: automated, cloud-enabled comparative microbial genome sequence analysis pipeline. BMC Genomics 18:332. doi: 10.1186/s12864-017-3717-3
Angiuoli, S. V., Matalka, M., Gussman, A., Galens, K., Vangala, M., Riley, D. R., et al. (2011). CloVR: a virtual machine for automated and portable sequence analysis from the desktop using cloud computing. BMC Bioinformatics 12:356. doi: 10.1186/1471-2105-12-356
Araújo, J., Díaz-Alcántara, C.-A., Velázquez, E., Urbano, B., and González-Andrés, F. (2015). Bradyrhizobium yuanmingense related strains form nitrogen-fixing symbiosis with Cajanus cajan L. in dominican republic and are efficient biofertilizers to replace N fertilization. Sci. Hortic. 192, 421–428. doi: 10.1016/j.scienta.2015.06.009
Araújo, J., Flores-Félix, J. D., Igual, J. M., Peix, A., González-Andrés, F., Díaz-Alcántara, C. A., et al. (2017). Bradyrhizobium cajani sp. nov. isolated from nodules of Cajanus cajan. Int. J. Syst. Evol. Microbiol. 67, 2236–2241. doi: 10.1099/ijsem.0.001932
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Berck, S., Perret, X., Quesada-Vincens, D., Promé, J. C., Broughton, W. J., and Jabbouri, S. (1999). NolL of Rhizobium sp. strain NGR234 is required for O-acetyltransferase activity. J. Bacteriol. 181, 957–964. doi: 10.1128/JB.181.3.957-964.1999
Boundy-Mills, K., Kosslak, R., Tully, R., Pueppke, S., and Lohrke, S. (1994). Induction of the Rhizobium fredii nod box-independent nodulation gene nolJ requires a functional nodD1 gene. MPMI 7, 305–308. doi: 10.1094/MPMI-7-0305
Broughton, W. J., and Dilworth, M. J. (1971). Control of leghaemoglobin synthesis in snake beans. Biochem. J. 125, 1075–1080. doi: 10.1042/bj1251075
Choudhary, S., Tak, N., Bissa, G., Chouhan, B., Choudhary, P., Sprent, J. I., et al. (2020). The widely distributed legume tree Vachellia (Acacia) nilotica subsp. indica is nodulated by genetically diverse Ensifer strains in India. Symbiosis 80, 15–31. doi: 10.1007/s13199-019-00658-8
Corvera, A., Promé, D., Promé, J. C., Martínez-Romero, E., and Romero, D. (1999). The nolL gene from Rhizobium etli determines nodulation efficiency by mediating the acetylation of the fucosyl residue in the nodulation factor. MPMI 12, 236–246. doi: 10.1094/MPMI.1999.12.3.236
Coutinho, H. L. C., Oliveira, V. M., Lovato, A., Maia, A. H. N., and Manfio, G. P. (1999). Evaluation of the diversity of rhizobia in Brazilian agricultural soils cultivated with soybeans. Appl. Soil Ecol. 13, 159–167. doi: 10.1016/S0929-1393(99)00031-1
Cytryn, E. J., Jitacksorn, S., Giraud, E., and Sadowsky, M. J. (2008). Insights learned from pBTAi1, a 229-kb accessory plasmid from Bradyrhizobium sp. strain BTAi1 and prevalence of accessory plasmids in other Bradyrhizobium sp. strains. ISME J. 2, 158–170. doi: 10.1038/ismej.2007.105
Dai, W. J., Zeng, Y., Xie, Z. P., and Staehelin, C. (2008). Symbiosis-promoting and deleterious effects of NopT, a novel type 3 effector of Rhizobium sp. strain NGR234. J. Bacteriol. 190, 5101–5110. doi: 10.1128/JB.00306-08
De Lajudie, P. M., Andrews, M., Ardley, J., Eardly, B., Jumas-Bilak, E., Kuzmanović, N., et al. (2019). Minimal standards for the description of new genera and species of rhizobia and agrobacteria. Int. J. Syst. Evol. Microbiol. 69, 1852–1863. doi: 10.1099/ijsem.0.003426
Dockendorff, T. C., Sharma, A. J., and Stacey, G. (1994). Identification and characterization of the nolYZ genes of Bradyrhizobium japonicum. MPMI 7, 173–180. doi: 10.1094/MPMI-7-0173
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Fossou, R. K., Pothier, J. F., Zéz,é, A., and Perret, X. (2020). Bradyrhizobium ivorense sp. nov. as a potential local bioinoculant for Cajanus cajan cultures in Côte d'Ivoire. Int. J. Syst. Evol. Microbiol. 70, 1421–1430. doi: 10.1099/ijsem.0.003931
Fossou, R. K., Ziegler, D., Zézé, A., Barja, F., and Perret, X. (2016). Two major clades of Bradyrhizobia dominate symbiotic interactions with pigeonpea in fields of Côte d'Ivoire. Front. Microbiol. 7:1793. doi: 10.3389/fmicb.2016.01793
Freiberg, C., Fellay, R., Bairoch, A., Broughton, W. J., Rosenthal, A., and Perret, X. (1997). Molecular basis of symbiosis between Rhizobium and legumes. Nature 387, 394–401. doi: 10.1038/387394a0
Gehlot, H. S., Tak, N., Kaushik, M., Mitra, S., Chen, W. M., Poweleit, N., et al. (2013). An invasive Mimosa in India does not adopt the symbionts of its native relatives. Ann. Bot. 112, 179–196. doi: 10.1093/aob/mct112
Goris, J., Konstantinidis, K. T., Klappenbach, J. A., Coenye, T., Vandamme, P., and Tiedje, J. M. (2007). DNA–DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 57, 81–91. doi: 10.1099/ijs.0.64483-0
Jiang, N., Liu, W., Li, Y., Wu, H., Zhang, Z., Alexandre, G., et al. (2016). A chemotaxis receptor modulates nodulation during the Azorhizobium caulinodans-Sesbania rostrata symbiosis. Appl. Environ. Microbiol. 82, 3174–3184. doi: 10.1128/AEM.00230-16
Kaneko, T., Maita, H., Hirakawa, H., Uchiike, N., Minamisawa, K., Watanabe, A., et al. (2011). Complete genome sequence of the soybean symbiont Bradyrhizobium japonicum Strain USDA6T. Genes 2, 763–787. doi: 10.3390/genes2040763
Kim, W. S., and Krishnan, H. B. (2014). A nopA deletion mutant of Sinorhizobium fredii USDA257, a soybean symbiont, is impaired in nodulation. Curr. Microbiol. 68, 239–246. doi: 10.1007/s00284-013-0469-4
Kimura, M. (1980). A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120. doi: 10.1007/BF01731581
Klappenbach, J. A., Dunbar, J. M., and Schmidt, T. M. (2000). rRNA operon copy number reflects ecological strategies of bacteria. Appl. Environ. Microbiol. 66, 1328–1333. doi: 10.1128/AEM.66.4.1328-1333.2000
Kolde, R. (2019).Pheatmap: Pretty Heatmaps. R package version 1.0.12. Available online at: https://cran.r-project.org/web/packages/pheatmap/index.html
Konstantinidis, K. T., and Tiedje, J. M. (2005). Genomic insights that advance the species definition for prokaryotes. Proc. Natl. Acad. Sci. U.S.A. 102, 2567–2572. doi: 10.1073/pnas.0409727102
Krishnan, H. B., Lorio, J., Kim, W. S., Jiang, G., Kim, K. Y., Deboer, M., et al. (2003). Extracellular proteins involved in soybean cultivar-specific nodulation are associated with pilus-like surface appendages and exported by a type III protein secretion system in Sinorhizobium fredii USDA257. MPMI 16, 617–625. doi: 10.1094/MPMI.2003.16.7.617
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Kurtz, S., Phillippy, A., Delcher, A., Smoot, M., Shumway, M., Antonescu, C., et al. (2004). Versatile and open software for comparing large genomes. Genome Biol. 5:R12. doi: 10.1186/gb-2004-5-2-r12
Li, H., and Durbin, R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760. doi: 10.1093/bioinformatics/btp324
Li, Y. H., Wang, R., Sui, X. H., Wang, E. T., Zhang, X. X., Tian, C. F., et al. (2019). Bradyrhizobium nanningense sp. nov., Bradyrhizobium guangzhouense sp. nov. and Bradyrhizobium zhanjiangense sp. nov., isolated from effective nodules of peanut in Southeast China. Syst. Appl. Microbiol. 42:126002. doi: 10.1016/j.syapm.2019.126002
López-Baena, F. J., Monreal, J. A., Pérez-Montaño, F., Guasch-Vidal, B., Bellogín, R. A., Vinardell, J. M., et al. (2009). The absence of Nops secretion in Sinorhizobium fredii HH103 increases GmPR1 expression in Williams soybean. MPMI 22, 1445–1454. doi: 10.1094/MPMI-22-11-1445
López-Baena, F. J., Ruiz-Sainz, J. E., Rodríguez-Carvajal, M. A., and Vinardell, J. M. (2016). Bacterial molecular signals in the Sinorhizobium fredii-soybean symbiosis. Int. J. Mol. Sci. 17:755. doi: 10.3390/ijms17050755
Luka, S., Sanjuan, J., Carlson, R. W., and Stacey, G. (1993). nolMNO genes of Bradyrhizobium japonicum are co-transcribed with nodYABCSUIJ, and nolO is involved in the synthesis of the lipo-oligosaccharide nodulation signals. J. Biol. Chem. 268, 27053–27059. doi: 10.1016/S0021-9258(19)74217-3
Lynch, D., O'brien, J., Welch, T., Clarke, P., ÓcuiV, P, Crosa, J. H., et al. (2001). Genetic organization of the region encoding regulation, biosynthesis, and transport of rhizobactin 1021, a siderophore produced by Sinorhizobium meliloti. J. Bacteriol. 183, 2576–2585. doi: 10.1128/JB.183.8.2576-2585.2001
Lynch, J. M., and Whipps, J. M. (1990). Substrate flow in the rhizosphere. Plant Soil. 129, 1–10. doi: 10.1007/BF00011685
Menna, P., and Hungria, M. (2011). Phylogeny of nodulation and nitrogen-fixation genes in Bradyrhizobium: supporting evidence for the theory of monophyletic origin, and spread and maintenance by both horizontal and vertical transfer. Int. J. Syst. Evol. Microbiol. 61, 3052–3067. doi: 10.1099/ijs.0.028803-0
Mula, M. G., and Saxena, K. B. (2010). Lifting the Level of Awareness on Pigeonpea-a Global Perspective. Patancheru: International Crops Research Institute for the Semi-Arid Tropics.
Ojha, A., Tak, N., Rathi, S., Chouhan, B., Rao, S. R., Barik, S. K., et al. (2017). Molecular characterization of novel Bradyrhizobium strains nodulating Eriosema chinense and Flemingia vestita, important unexplored native legumes of the sub-Himalayan region (Meghalaya) of India. Syst. Appl. Microbiol. 40, 334–344. doi: 10.1016/j.syapm.2017.06.003
Okazaki, S., Noisangiam, R., Okubo, T., Kaneko, T., Oshima, K., Hattori, M., et al. (2015). Genome analysis of a novel Bradyrhizobium sp. DOA9 carrying a symbiotic plasmid. PLoS ONE 10:e0117392. doi: 10.1371/journal.pone.0117392
Ormeño-Orrillo, E., and Martínez-Romero, E. (2019). A genomotaxonomy view of the Bradyrhizobium genus. Front. Microbiol. 10:1334. doi: 10.3389/fmicb.2019.01334
Pastor-Bueis, R., Sánchez-Cañizares, C., James, E. K., and González-Andrés, F. (2019). Formulation of a highly effective inoculant for common bean based on an autochthonous elite strain of Rhizobium leguminosarum bv. phaseoli, and genomic-based insights into its agronomic performance. Front. Microbiol. 10:2724. doi: 10.3389/fmicb.2019.02724
Pellegrino, E., and Bedini, S. (2014). Enhancing ecosystem services in sustainable agriculture: biofertilization and biofortification of chickpea (Cicer arietinum L.) by arbuscular mycorrhizal fungi. Soil Biol. Biochem. 68, 429–439. doi: 10.1016/j.soilbio.2013.09.030
Pellegrino, E., Bedini, S., Avio, L., Bonari, E., and Giovannetti, M. (2011). Field inoculation effectiveness of native and exotic arbuscular mycorrhizal fungi in a Mediterranean agricultural soil. Soil Biol. Biochem. 43, 367–376. doi: 10.1016/j.soilbio.2010.11.002
Pueppke, S. G., and Broughton, W. J. (1999). Rhizobium sp. strain NGR234 and R. fredii USDA257 share exceptionally broad, nested host ranges. MPMI 12, 293–318. doi: 10.1094/MPMI.1999.12.4.293
Ramsubhag, A., Umaharan, P., and Donawa, A. (2002). Partial 16S rRNA gene sequence diversity and numerical taxonomy of slow growing pigeonpea (Cajanus cajan L. Millsp) nodulating rhizobia. FEMS Microbiol. Lett. 216, 139–144. doi: 10.1111/j.1574-6968.2002.tb11427.x
Rathi, S., Tak, N., Bissa, G., Chouhan, B., Ojha, A., Adhikari, D., et al. (2018). Selection of Bradyrhizobium or Ensifer symbionts by the native Indian caesalpinioid legume Chamaecrista pumila depends on soil pH and other edaphic and climatic factors. FEMS Microbiol. Ecol. 94:fiy180. doi: 10.1093/femsec/fiy180
Reeve, W., Van Berkum, P., Ardley, J., Tian, R., Gollagher, M., Marinova, D., et al. (2017). High-quality permanent draft genome sequence of the Bradyrhizobium elkanii type strain USDA 76T, isolated from Glycine max (L.) Merr. Stand. Genomic. Sci. 12, 26–26. doi: 10.1186/s40793-017-0238-2
Riley, D. R., Angiuoli, S. V., Crabtree, J., Dunning Hotopp, J. C., and Tettelin, H. (2011). Using Sybil for interactive comparative genomics of microbes on the web. Bioinformatics 28, 160–166. doi: 10.1093/bioinformatics/btr652
Rivas, R., Velázquez, E., Valverde, A., Mateos, P. F., and Martínez-Molina, E. (2001). A two primers random amplified polymorphic DNA procedure to obtain polymerase chain reaction fingerprints of bacterial species. Electrophoresis 22, 1086–1089. doi: 10.1002/1522-2683()22:6<1086::AID-ELPS1086>3.0.CO;2-6
Rupela, O. P., Kumar Rao, J. V. D. K., Sudarshana, M. R., Usha Kiran, M., and Anjaiah, V. (1991). Rhizobium germplasm Resources at ICRISAT Center. Patancheru: International Crops Research Institute for the Semi-Arid Tropics.
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425.
Sankhla, I. S., Tak, N., Meghwal, R. R., Choudhary, S., Tak, A., Rathi, S., et al. (2017). Molecular characterization of nitrogen fixing microsymbionts from root nodules of Vachellia (Acacia) jacquemontii, a native legume from the Thar Desert of India. Plant Soil. 410, 21–40. doi: 10.1007/s11104-016-2838-9
Saxena, K. B. (2006). Seed Production Systems in Pigeonpea ICRISAT. Patancheru: International Crops Research Institute for the Semi-Arid Tropics.
Saxena, R. K., Von Wettberg, E., Upadhyaya, H. D., Sanchez, V., Songok, S., Saxena, K., et al. (2014). Genetic diversity and demographic history of Cajanus spp. illustrated from genome-wide SNPs. PLoS ONE 9:e88568. doi: 10.1371/journal.pone.0088568
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Serraj, R., Sinclair, T. R., and Purcell, L. C. (1999). Symbiotic N2 fixation response to drought. J. Exp. Bot. 50, 143–155. doi: 10.1093/jxb/50.331.143
Sharypova, L. A., Yurgel, S. N., Keller, M., Simarov, B. V., Pühler, A., and Becker, A. (1999). The eff-482 locus of Sinorhizobium meliloti CXM1-105 that influences symbiotic effectiveness consists of three genes encoding an endoglycanase, a transcriptional regulator and an adenylate cyclase. Mol. Gen. Genet. 261, 1032–1044. doi: 10.1007/s004380051052
Skorpil, P., Saad, M. M., Boukli, N. M., Kobayashi, H., Ares-Orpel, F., Broughton, W. J., et al. (2005). NopP, a phosphorylated effector of Rhizobium sp. strain NGR234, is a major determinant of nodulation of the tropical legumes Flemingia congesta and Tephrosia vogelii. Mol. Microbiol. 57, 1304–1317. doi: 10.1111/j.1365-2958.2005.04768.x
Somasegaran, P., and Hoben, H. J. (1994). Handbook for Rhizobia : Methods in Legume-Rhizobium Technology. New York, NY: Springer-Verlag. doi: 10.1007/978-1-4613-8375-8
Stepkowski, T., Czaplińska, M., Miedzinska, K., and Moulin, L. (2003). The variable part of the dnaK gene as an alternative marker for phylogenetic studies of rhizobia and related alpha proteobacteria. Syst. Appl. Microbiol. 26, 483–494. doi: 10.1078/072320203770865765
Sugawara, M., Epstein, B., Badgley, B. D., Unno, T., Xu, L., Reese, J., et al. (2013). Comparative genomics of the core and accessory genomes of 48 Sinorhizobium strains comprising five genospecies. Genome Biol. 14:R17. doi: 10.1186/gb-2013-14-2-r17
Tak, N., Awasthi, E., Bissa, G., Meghwal, R. R., James, E. K., Sprent, J. S., et al. (2016). Multi locus sequence analysis and symbiotic characterization of novel Ensifer strains nodulating Tephrosia spp. in the Indian Thar Desert. Syst. Appl. Microbiol. 39, 534–545. doi: 10.1016/j.syapm.2016.08.002
Teulet, A., Busset, N., Fardoux, J., Gully, D., Chaintreuil, C., Cartieaux, F., et al. (2019). The rhizobial type III effector ErnA confers the ability to form nodules in legumes. Proc. Natl. Acad. Sci. U.S.A. 116, 21758–21768. doi: 10.1073/pnas.1904456116
Tian, C. F., Garnerone, A.-M., Mathieu-Demazière, C., Masson-Boivin, C., and Batut, J. (2012). Plant-activated bacterial receptor adenylate cyclases modulate epidermal infection in the Sinorhizobium meliloti–Medicago symbiosis. Proc. Natl. Acad. Sci. U.S.A. 109, 6751–6756. doi: 10.1073/pnas.1120260109
Varshney, R. K., Chen, W., Li, Y., Bharti, A. K., Saxena, R. K., Schlueter, J. A., et al. (2012). Draft genome sequence of pigeonpea (Cajanus cajan), an orphan legume crop of resource-poor farmers. Nat. Biotechnol. 30, 83–89. doi: 10.1038/nbt.2022
Varshney, R. K., Penmetsa, R. V., Dutta, S., Kulwal, P. L., Saxena, R. K., Datta, S., et al. (2010). Pigeonpea genomics initiative (PGI): an international effort to improve crop productivity of pigeonpea (Cajanus cajan L.). Mol. Breed. 26, 393–408. doi: 10.1007/s11032-009-9327-2
Vavilov, N. I. (1951). The Origin, Variation, Immunity and Breeding of Cultivated Plants: Selected Writings. Waltham: Chronica Botanica Company.
Versalovic, J., Schneider, M., De Bruijn, F., and Lupski, J. R. (1994). Genomic fingerprinting of bacteria using repetitive sequence-based polymerase chain reaction. Methods Mol. Biol. 5, 25–40.
Vesth, T., Lagesen, K., Acar, Ö., and Ussery, D. (2013). CMG-biotools, a free workbench for basic comparative microbial genomics. PLoS ONE 8:e60120. doi: 10.1371/journal.pone.0060120
Vincent, J. M. (1970). A Manual for the Practical Study of Root-Nodule Bacteria. Oxford: International Biological Programme, Blackwell Scientific.
Viprey, V., Rosenthal, A., Broughton, W. J., and Perret, X. (2000). Genetic snapshots of the Rhizobium species NGR234 genome. Genome Biol. 1:research0014.0011. doi: 10.1186/gb-2000-1-6-research0014
Waters, N. R., Abram, F., Brennan, F., Holmes, A., and Pritchard, L. (2018). riboSeed: leveraging prokaryotic genomic architecture to assemble across ribosomal regions. Nucleic Acids Res. 46, e68–e68. doi: 10.1093/nar/gky212
Wei, M., Yokoyama, T., Minamisawa, K., Mitsui, H., Itakura, M., Kaneko, T., et al. (2008). Soybean seed extracts preferentially express genomic loci of Bradyrhizobium japonicum in the initial interaction with soybean, Glycine max (L.) Merr. DNA Res. 15, 201–214. doi: 10.1093/dnares/dsn012
Wheatley, R. M., Ford, B. L., Li, L., Aroney, S. T. N., Knights, H. E., Ledermann, R., et al. (2020). Lifestyle adaptations of Rhizobium from rhizosphere to symbiosis. Proc. Natl. Acad. Sci. U.S.A. 117, 23823–23834. doi: 10.1073/pnas.2009094117
Wolde-Meskel, E., Terefework, Z., Frostegård, Å., and Lindström, K. (2005). Genetic diversity and phylogeny of rhizobia isolated from agroforestry legume species in southern Ethiopia. Int. J. Syst. Evol. Microbiol. 55, 1439–1452. doi: 10.1099/ijs.0.63534-0
Wood, D. E., and Salzberg, S. L. (2014). Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 15:R46. doi: 10.1186/gb-2014-15-3-r46
Wu, L. J., Wang, H. Q., Wang, E. T., Chen, W. X., and Tian, C. F. (2011). Genetic diversity of nodulating and non-nodulating rhizobia associated with wild soybean (Glycine soja Sieb. & Zucc.) in different ecoregions of China. FEMS Microbiol. Ecol. 76, 439–450. doi: 10.1111/j.1574-6941.2011.01064.x
Yang, S., Tang, F., Gao, M., Krishnan, H. B., and Zhu, H. (2010). R gene-controlled host specificity in the legume–rhizobia symbiosis. Proc. Natl. Acad. Sci. U.S.A. 107:18735. doi: 10.1073/pnas.1011957107
Keywords: Bradyrhizobium, Ensifer (Sinorhizobium), pigeon pea (Cajanus cajan), nod cluster, nodulation outer proteins (Nop), comparative genomics, India
Citation: Jorrin B, Maluk M, Atoliya N, Kumar SC, Chalasani D, Tkacz A, Singh P, Basu A, Pullabhotla S, Kumar M, Mohanty SR, East AK, Ramachandran VK, James EK, Podile AR, Saxena AK, Rao D and Poole PS (2021) Genomic Diversity of Pigeon Pea (Cajanus cajan L. Millsp.) Endosymbionts in India and Selection of Potential Strains for Use as Agricultural Inoculants. Front. Plant Sci. 12:680981. doi: 10.3389/fpls.2021.680981
Received: 15 March 2021; Accepted: 06 August 2021;
Published: 07 September 2021.
Edited by:
Julie Grossman, University of Minnesota Twin Cities, United StatesReviewed by:
Elisa Pellegrino, Sant'Anna School of Advanced Studies, ItalyAttila Kereszt, Eötvös Lóránd Research Network, Hungary
Copyright © 2021 Jorrin, Maluk, Atoliya, Kumar, Chalasani, Tkacz, Singh, Basu, Pullabhotla, Kumar, Mohanty, East, Ramachandran, James, Podile, Saxena, Rao and Poole. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Beatriz Jorrin, YmVhdHJpei5qb3JyaW5AcGxhbnRzLm94LmFjLnVr; Philip S. Poole, cGhpbGlwLnBvb2xlQHBsYW50cy5veC5hYy51aw==; DLN Rao, ZGxucmFvLmljYXJAZ21haWwuY29t; Anil Kumar Saxena, c2F4ZW5hNDYxQHlhaG9vLmNvbQ==; Appa Rao Podile, cG9kaWxlcmFvQGdtYWlsLmNvbQ==
 Beatriz Jorrin
Beatriz Jorrin Marta Maluk
Marta Maluk Nagvanti Atoliya
Nagvanti Atoliya Shiv Charan Kumar
Shiv Charan Kumar Danteswari Chalasani
Danteswari Chalasani Andrzej Tkacz
Andrzej Tkacz Prachi Singh4
Prachi Singh4 Anirban Basu
Anirban Basu Sarma VSRN Pullabhotla
Sarma VSRN Pullabhotla Murugan Kumar
Murugan Kumar Santosh Ranjan Mohanty
Santosh Ranjan Mohanty Vinoy K. Ramachandran
Vinoy K. Ramachandran Euan K. James
Euan K. James Appa Rao Podile
Appa Rao Podile Anil Kumar Saxena
Anil Kumar Saxena DLN Rao
DLN Rao Philip S. Poole
Philip S. Poole