- 1Department of Field Crops, Faculty of Agriculture, Akdeniz University, Antalya, Turkey
- 2Department of Medicinal and Aromatic Plants, Akev University, Antalya, Turkey
- 3Sabanci University Nanotechnology Research and Application Center (SUNUM), Istanbul, Turkey
The seed-bearing capsule of sesame shatters at harvest. This wildish trait makes the crop unsuitable for mechanized harvesting and also restricts its commercial potential by limiting the cultivation for countries that have no access to low-cost labor. Therefore, the underlying genetic basis of the capsule shattering trait is highly important in order to develop mechanization-ready varieties for sustainable sesame farming. In the present study, we generated a sesame F2 population derived from a cross between a capsule shattering cultivar (Muganli-57) and a non-shattering mutant (PI 599446), which was used to construct a genetic map based on double-digest restriction-site-associated DNA sequencing. The resulting high-density genetic map contained 782 single-nucleotide polymorphisms (SNPs) and spanned a length of 697.3 cM, with an average marker interval of 0.89 cM. Based on the reference genome, the capsule shattering trait was mapped onto SNP marker S8_5062843 (78.9 cM) near the distal end of LG8 (chromosome 8). In order to reveal genes potentially controlling the shattering trait, the marker region (S8_5062843) was examined, and a candidate gene including six CDSs was identified. Annotation showed that the gene encodes a protein with 440 amino acids, sharing ∼99% homology with transcription repressor KAN1. Compared with the capsule shattering allele, the SNP change and altered splicing in the flanking region of S8_5062843 caused a frameshift mutation in the mRNA, resulting in the loss of function of this gene in the mutant parent and thus in non-shattering capsules and leaf curling. With the use of genomic data, InDel and CAPS markers were developed to differentiate shattering and non-shattering capsule genotypes in marker-assisted selection studies. The obtained results in the study can be beneficial in breeding programs to improve the shattering trait and enhance sesame productivity.
Introduction
Sesame (Sesamum indicum L.) is an important oilseed crop which belongs to the Sesamum genus of the Pedaliaceae family. It is cultivated in more than 60 countries in an area of 11.74 m ha, with a total production of 6.0 m tons (FAO, 2018). Although the production quantity is much lower than major oilseeds such as sunflower, rapeseed, and groundnut, the total production of sesame seed has increased by about 40% in the last decade (FAO, 2018). The high demand for this crop is related to its high-quality vegetable oil (Arslan et al., 2007; Uzun et al., 2008) and antioxidant lignans, including sesamin and sesamolin (Yoshida and Takagi, 1997; Moazzami and Kamal-Eldin, 2006). Its seeds have long been used in human nutrition, cosmetics, perfumery, soaps, insecticides, anti-aging treatments, and medicine. The health benefits of sesame have also been reported to reduce the rate of occurrence of certain cancers (Miyahara et al., 2001), constrain the growth of leukemia cells (Ryu et al., 2003), and decrease the susceptibility of low-density lipoprotein (LDL) to oxidation, which is recognized as a risk factor for atherosclerosis (Nakamura et al., 2020). In addition, sesame has agricultural advantages; it can survive in extreme conditions, can grow on only soil moisture without irrigation and fertilizer, and can be grown as a second crop (Ashri, 2007). These attributes make the crop an important income source especially for small-scale farmers in developing countries (Dossa et al., 2017).
Despite its agricultural, nutritional, and economic importance, the yield of sesame is lower than those of other major oilseeds (Hwang, 2005). It has some wildish traits, namely, capsule shattering (Uzun et al., 2004), indeterminate growth habit (Uzun and Cagirgan, 2006), and non-synchronous maturity, which are the main causes of low seed yield. Capsule shattering is the most serious problem in sesame production, as it causes seed losses at harvest that can be very high, up to 50% (Weiss, 1971). This wildish trait also restricts sesame’s commercial potential and limits its economic viability in countries that have no access to cheap labor because it makes the crop unsuitable for mechanized harvesting (Langham and Wiemers, 2002). Therefore, non-shattering and/or semi-shattering capsule genotypes should be useful for adapting sesame to mechanized harvesting.
A closed (non-shattered) capsule mutant trait was discovered by Langham (1946); however, over 99% of the sesame used for production in the world still has the shattering trait (Langham, 2001). Unwanted side effects such as cupped leaves, short seed pods, twisted stems, and low seed yield restrict the direct use of this non-shattering mutant type (Wongyai et al., 2001). Out-crosses between closed capsule mutants and advanced cultivars have been conducted in order to eliminate these side effects and also improve traits of agricultural importance (Uzun et al., 2004; Yol and Uzun, 2019). Although heterotic improvements in performance were obtained for seed yield and yield-related traits in these studies, there was no progress with respect to suitability for mechanized harvesting. Besides conventional breeding techniques, molecular markers have also been developed for the closed capsule mutant trait for use in marker-assisted selection (Uzun et al., 2003; Phumichai et al., 2017; Zhang H.Y. et al., 2018). However, there is a need to understand the mechanism of the shattering trait at the genomic level in order to increase the efficiency and efficacy of breeding studies.
High-throughput, “next-generation” sequencing technologies provide opportunity for the development of high-density maps to identify genes and quantitative trait loci (QTLs) for agronomically important traits (Nguyen et al., 2019). Especially reduced representation sequencing (RRS) approaches make low-cost and flexible genotyping in the target genomes possible (Scheben et al., 2017). Double-digest RAD sequencing (ddRAD-Seq) (Peterson et al., 2012) is one of the RRS methods for SNP discovery at a genome-wide scale. It includes digesting the DNA with selected restriction enzymes and then sequencing a specific size-selected range of generated fragments (Aguirre et al., 2019). It eliminates the shearing step of the original RAD protocol (Baird et al., 2008) and also provides more flexibility with respect to the targeted marker density and the number of fragments (Robledo et al., 2018) compared to RAD-Seq (Baird et al., 2008) and/or 2b-RAD (Wang et al., 2012). The ddRAD-Seq protocol has several advantages: the degree of genome coverage can be adjusted by the choice of different restriction enzymes, high efficiency and accuracy are provided in SNP discovery, and no reference genome is required in the bioinformatic process (Peterson et al., 2012). It has been used for high-density genetic map construction in strawberry (Davik et al., 2015), northern red oak (Konar et al., 2017), and platycladus (Jin et al., 2019). QTL mapping has been used for different traits in rapeseed (Chen et al., 2017), lettuce (Seki et al., 2020), and peanut (Liang et al., 2021) and in genetic diversity studies in orchid (Roy et al., 2017), onion (Lee et al., 2018), and sesame (Basak et al., 2019). However, to the best of our knowledge, there is no report about the construction of a genetic map in sesame with the use of SNPs discovered by ddRAD-Seq technology.
In this study, the F2 population that was developed by a cross between a capsule shattering sesame cultivar × non-shattering mutant line was sequenced by ddRAD-Seq. This genotypic data was used with the following aims: (i) to construct a genetic linkage map with SNPs, (ii) to reveal the genomic basis of the capsule shattering trait, and (iii) to develop markers for this trait. These findings will contribute to new breeding strategies in sesame in order to develop high-yielding genotypes that are also suitable for mechanized harvesting.
Materials and Methods
Plant Material and F2 Population Development
The F2 mapping population comprised of 120 individuals derived from a cross between a sesame cultivar (Muganli-57) and a sesame line. The male parent, Muganli-57, was registered by the West Mediterranean Agricultural Research Institute, Turkey. It has high seed yield, yellow seeds, about 50% oil content, and shattered capsules. The female parent (mutant line) was introduced from USDA GenBank with the accession number PI 599446, named Paloma. It is distinct from the cultivar in its agro-morphological traits. This line had low seed yield, brown seed color, curly leaves, short, round shape, and non-shattered capsules. Figure 1 shows the morphological differences between the leaf and capsules of these genetic materials.

Figure 1. The morphological differences for capsule shattering and non-shattering genotypes. (a,d,f) Leaf, capsule-forming stage, and harvested capsule of the non-shattered capsule phenotype, respectively. (b,c,e) Leaf, capsule-forming stage, and harvested capsule of the capsule shattering phenotype, respectively.
The parents were crossed in the greenhouse of Akdeniz University (36°53′ N, 38°30′ E and altitude of 15 m) during the 2015 growing season in Antalya, Turkey. A total of 1,496 F2 seeds were harvested from self-pollinated F1 plants and grown in four rows with a row-to-row distance of 70 cm and plant-to-plant distance within a row of 10 cm at the same location in 2017. These lines were phenotypically scored and labeled according to leaf and capsule shapes as capsule shattering and non-shattering genotypes in the maturation stage. A chi-square goodness of fit test was performed on the F2 population to confirm the segregation ratio for the capsule shattering trait. A total of 120 F2 individuals, which included 90 shattering and 30 non-shattering types as well as both parents, were selected for sequencing and the construction of the genetic map.
DNA Extraction and ddRAD-Seq
Healthy young leaves from 120 F2 individuals and the two parents were collected and stored at −80°C. Extraction of genomic DNA was carried out with a sodium dodecyl sulfate-based method (Ahmed et al., 2009) with minor modifications. DNA concentration and quality were measured with a Qubit 2.0 Fluorometer using a Qubit DNA Assay Kit (Life Technologies, Thermo Fisher Scientific).
A modified version of the ddRAD-Seq method (Peterson et al., 2012) was used for reduced representation genomic library construction. Briefly, the restriction enzymes VspI and MspI were employed to digest the genomic DNA of each individual. Ampure XP beads (Beckman Coulter Genomics) were used to clean the digested products followed by P1 and P2 adapter ligation with T4 ligase buffer. The 3′ end of the P1 adapter was modified because six-base cutter VspI restriction enzyme was selected instead of EcoRI, which is the enzyme used in the original ddRAD protocol. Following ligation, genotype-specific indexed PCR primers were used in PCR amplification for each reaction. The amplified products were then checked on an agarose gel and pooled in equal concentration. Size selection (400–500 bp) was carried out on the pooled products in a final step. The reduced representation genomic libraries were sequenced using an Illumina HiSeq 2,500 instrument at Macrogen (Seoul, Republic of Korea) for 150-bp paired-end sequencing. The ddRAD sequence data for studied samples have been deposited with links to BioProject accession number PRJNA633276 in the NCBI.
Bioinformatic Analysis for SNP Discovery
The bioinformatic pipeline was operated using the Galaxy1 which is open, web-based computational platform. In the first step of analysis, raw reads were demultiplexed to genotype-specific fastq files with Je (v1.2.1) (Girardot et al., 2016). All sequencing files were then processed to filter out low-quality reads using fastp (v0.20.1) (Chen et al., 2018). In this step, reads having an average phred-scaled quality score less than 15 and containing restriction enzyme sequences were trimmed. Bowtie2 (v2.3.4.3) (Langmead and Salzberg, 2012) was used to align the cleaned reads to the sesame reference genome “Zhongzhi13 v2.0” (Wang et al., 2016) with the default parameters. The created genotype-specific individual BAM (binary sequence alignment file format) files were analyzed in a variant calling program, freebayes (bayesian genetic variant detector) (v13.1) (Garrison and Marth, 2012), with the calling model for simple diploids and filtering to include only sites with a total read coverage value of at least 20×. Bcftools merge (v1.10) (Li et al., 2009) was used to combine each genotype-specific variant file (.vcf) into a single variant file which was then converted to browser extensible data (BED) format to store the genomic regions as coordinates and associated annotations. Afterward, individual BAMs were re-analyzed on freebayes with the BED dataset, retaining sequence differences with at least 6× coverage to reveal reliable variants for each genotype, as before the created .vcf files were combined to a single variant file using the merge option from bcftools. This merged file was then further filtered with Tassel V5.2.52 (Glaubitz et al., 2014) to detect polymorphic sites using these parameters: minimum site count 80 and minimum–maximum heterozygous proportion 0.3–0.8. The filtered file was then converted to ABH genotype format for the mapping analysis.
Linkage Map Construction
Markers exhibiting missing data (> 20%) and segregation distortion (χ2-test, P < 0.001) were excluded. The remaining markers were used to construct a genetic linkage map with the JoinMap 4.1 program (Van Ooijen, 2011). Markers were grouped into linkage groups with a minimum logarithm of odds score of 10.0 and maximum recombination of 45%. The regression mapping algorithm was employed with the following parameters: goodness-of-fit jump threshold for removal of loci = 5.0, third round = yes and “ripple” was performed after adding each marker. The recombination rate was converted into map distance in cM (centimorgans) by the Kosambi mapping function (Kosambi, 1994). The capsule shattering trait was scored as a qualitative trait and analyzed with allelic data using JoinMap 4.1. The genetic linkage map was graphically visualized and modified in MapChart 2.32 (Voorrips, 2002).
Prediction of Gene Function
The flanking sequences of the SNP markers in the candidate gene were identified from the sesame reference genome. The potential gene coding sequence was predicted with FGENESH2 (Solovyev et al., 2006) based on sesame genome-specific parameters. The predicted gene regions from the shattering and non-shattering genotypes were amplified and sequenced with the Sanger method. These sequences were re-analyzed in FGENESH, and then functional annotation of the candidate gene was carried out manually by comparison with known proteins using the blastp algorithm in NCBI.
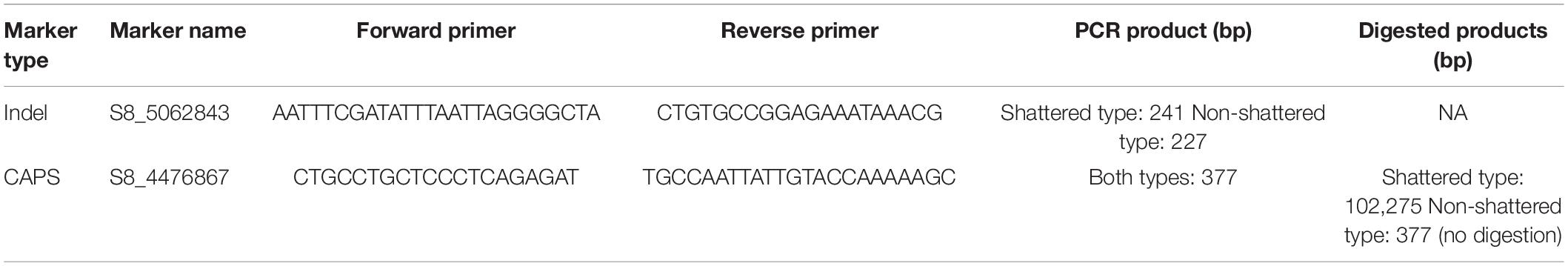
InDel and CAPS Marker Development
We designed cleaved amplified polymorphic sequence (CAPS) and insertion--deletion (InDel) markers associated with the capsule shattering trait. These were tested on two parents and F2 individuals. Primers were designed using Primer3 web, version 4.1.03, for both markers. PCR was performed in a 20-μl reaction volume with 1 μl of × 10 PCR buffer, 2.5 mM MgCl2, 0.5 μl of dNTP mix, 0.4 μl each of forward and reverse primers (10 pmol), 0.3 μl of Taq DNA polymerase (5 U/μl), 1.5 μl of genomic DNA, and Milli-Q water. Thermocycling started at 94°C for 5 min, followed by 45 cycles of 30 s at 92°C, 45 s at 57°C, 180 s at 72°C, and finally kept at 72°C for 10 min. For the CAPS marker, the appropriate restriction enzyme for the selected SNP region was identified in NEBcutter V2.04. After PCR amplification, the PCR product was restriction-digested in a total volume of 20 μl for 1 h according to the manufacturer’s protocol (New England Biolabs, MA, United States). Then, 10 μl of the products for CAPS and InDel markers was separated by electrophoresis on 2% agarose gel and visualized by UV light.
Results
Morphology and Inheritance of the Capsule Shattering Trait
The sesame cultivar, Muganli-57, had higher plant height and number of capsules per plant compared to the line, PI 599446 (Yol and Uzun, 2012). They also differ significantly from each other with respect to leaf and capsule phenotypes (Figure 1). Although the cultivar type had normal sesame plant architecture, the line showed curly and bigger leaves. Hairy and short seed pods (Figure 1) and high-density blooming were also observed in the line at maturity. Compared with Muganli-57, this type maintains a non-shattering capsule structure in harvest time (Figure 1). These visible phenotypes provide a clear difference between cultivar and PI 599446 in all growing period.
The seed pods of the F1 plants developed from the hybridization of the line (non-shattering capsule) with Muganli-57 were all completely shattered at maturity, indicating the dominance of the capsule shattering trait. To identify the genetic basis underlying the capsule shattering phenotype, a large population of 1,496 F2 individuals were planted in an experimental field. The segregation of shattered capsule to non-shattered capsule plants closely fitted the ratio of 3:1 (χ2 = 2.22, p > 0.05) (Table 1), suggesting that the capsule shattering trait is controlled by a single gene, in which the shattering allele is dominant over the non-shattering allele.
Genotyping
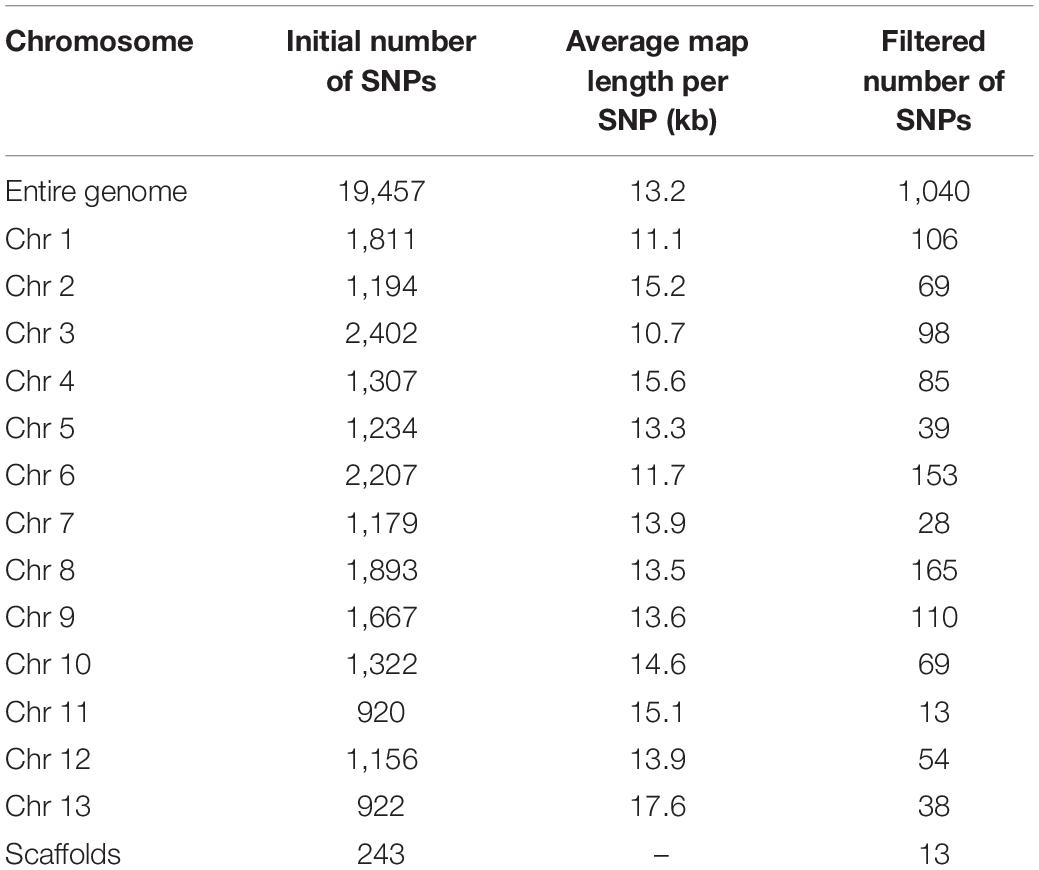
A total of 122 sampled individuals, that is 120 F2 progeny and two parents, were used in the construction of ddRAD-seq library. After sequencing, about 40.0 Gb of sequence reads was obtained. Two individuals, N6 and N12, were discarded from this data due to very low read numbers. The remaining raw data containing 488.32 M reads was filtered with quality parameters, and 468.17 M of clean data was obtained for further analysis (Supplementary Figure 1). The highest and lowest filtered read counts were 0.51 and 9.16 M for the lines N244 and N238, respectively, with the mean of 3.87 M reads per individual in the F2 population. The GC content ranged from 36.39 to 39.91% (Supplementary Figure 1). The mapping rates to the S. indicum L. reference genome for the shattered capsule cultivar (Muganli-57) and non-shattered capsule mutant line (PI 599446) were 78.6 and 80.41%, respectively, while that of the F2 population was 79.28% on average. A total of 19,457 SNPs were initially called from the F2 lines using the variant calling pipeline (see section “Materials and Methods”). The maximum and minimum average distance per SNP was identified as 10.7 and 17.6 kb on chromosomes 3 and 13, respectively, in the raw SNP data (Table 2). After filtering to select high-confidence polymorphic sites, a total of 1,040 polymorphic SNP markers were detected. Of these, 1,027 SNPs were distributed between all 13 sesame chromosomes, and the remaining 13 SNPs were unmapped. The highest number of SNPs was detected on chromosome 8 (165 SNPs), whereas the lowest number of SNPs was found on chromosome 11 (13 SNPs), with an overall mean of 79 SNPs per chromosome. Of the 1,040 polymorphic SNPs, more SNPs were of transition than transversion type, with the most frequent being 160 A/G and 158 C/T SNPs, accounting for 15.38 and 15.20%, respectively (Table 3).
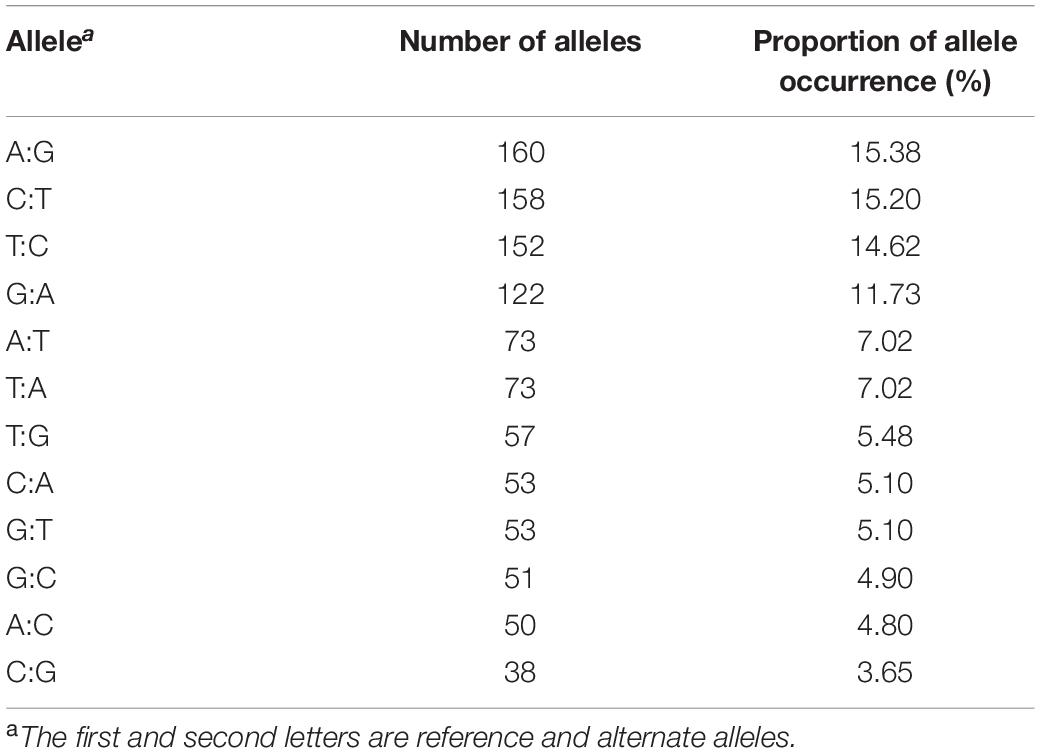
Table 3. Single-nucleotide polymorphism statistics in filtered data showing the number of alleles and the rate of allele occurrence.
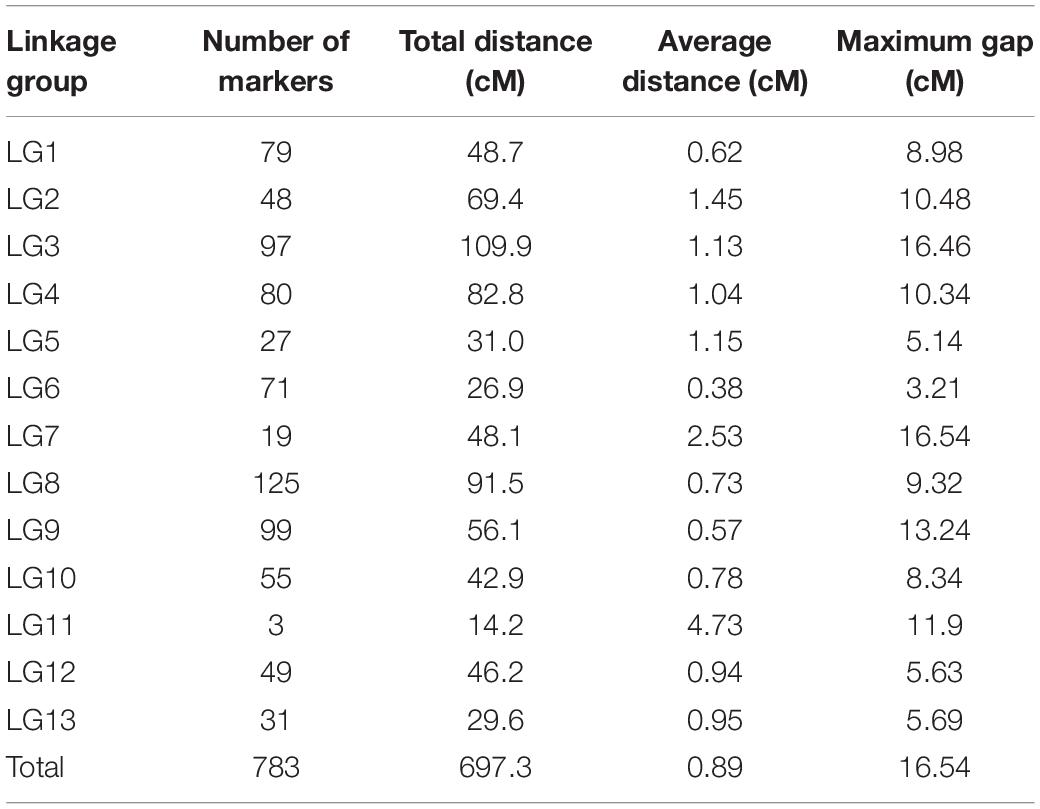
Linkage Map Construction
Before the construction of linkage maps, SNP markers that did not fit the expected 1:2:1 segregation ratio or included high missing data were excluded from further analysis. In the linkage map, a total of 782 SNPs were assigned to 13 linkage groups (LGs) (Table 4 and Figure 2). This map covered a total genetic distance of 697.3 cM, with the LGs ranging from 14.2 to 109.9 cM, with an average marker interval of 0.89 cM. The LG with the greatest number of markers was LG8, which included 125 markers, with an average distance of 0.73 cM. Conversely, LG11 had the fewest markers, had the lowest marker density (4.73 cM), and was also the shortest LG. LG13, the longest LG, had a genetic length of 109.90 cM and contained 97 markers, with an average distance between markers of 1.13 cM. The highest marker density was observed in LG6 (an average of 0.38 cM between markers), followed by LG1 (0.62 cM) (Table 4); the average distance between markers was under 1 cM in seven LGs (Table 4). The largest gap was of 16.54 cM, located in LG7, followed by 16.46- and 13.24-cM gaps in LG3 and LG9, respectively.

Figure 2. A genetic linkage map of sesame constructed using the F2 population of PI 599446 × Muganli-57.
Mapping of Shattering Trait and Candidate Gene Prediction
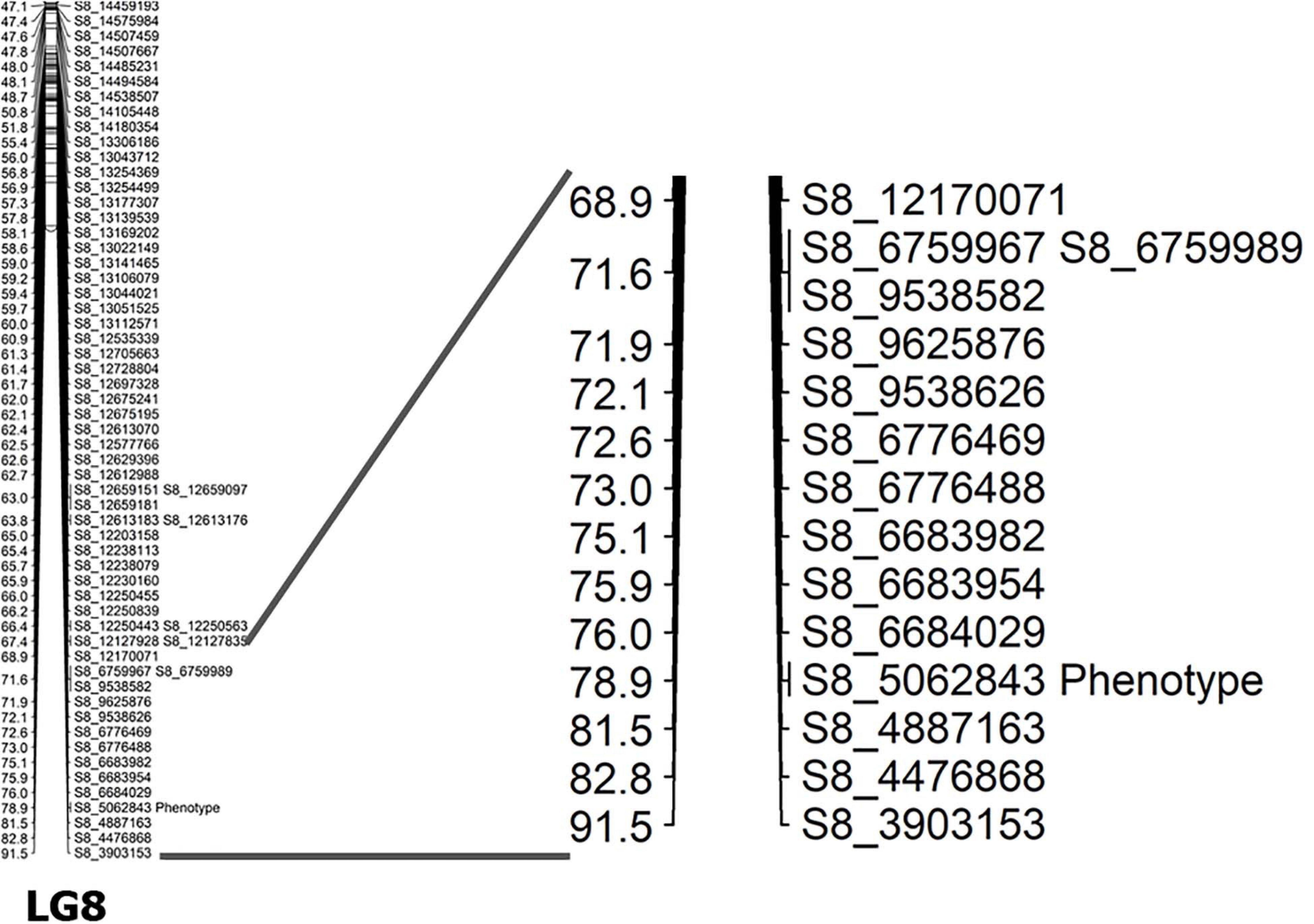
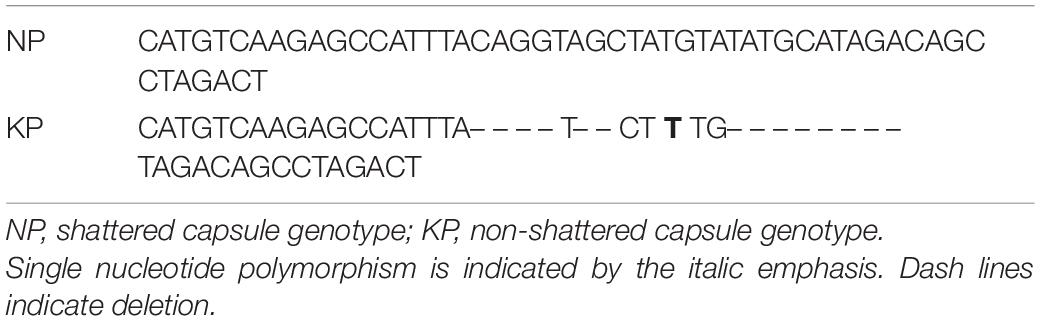
Capsule shattering is predicted to be controlled by a single gene, and this phenotype was scored as a qualitative trait in the mapping analysis. Based on the Zhongzhi13, v2.0, sesame reference genome, it was mapped to coincide with SNP marker S8_5062843 (78.9 cM) near the end of LG8 (chromosome 8) (Figure 3). The locus containing the SNP marker was visualized for capsule shattering and non-shattering parents and selected F2 lines (Figure 4). On chromosome 8, the nucleotide “A” was found in the reference genome at this SNP position, while the “T” nucleotide was observed in the non-shattering parent line (KP). Accordingly, the parental line (NP) with the shattered capsule trait had a sequence identical to the reference genome in the specified region. Heterozygosity was observed in some F2 lines such as N101 and N110 for this SNP, with individual reads matching both parental alleles; these lines had the shattered capsule phenotype. Other F2 lines such as K40, K86, N79, and N91 were homozygotes with the same alleles at corresponding chromosomal loci as their parents (Figure 4). When all F2 lines were examined, a 1:2:1 segregation ratio was obtained for this SNP (27:48:29, non-shattered capsule/heterozygous/shattered capsule) in F2 population. In addition, short deletions on either side of the SNP (S8_5062843) with a total length of 14 nts were detected (Figure 4) and showed the same segregation as the SNP for the sequence of this region on chromosome 8 in both parental lines as given in Table 5.
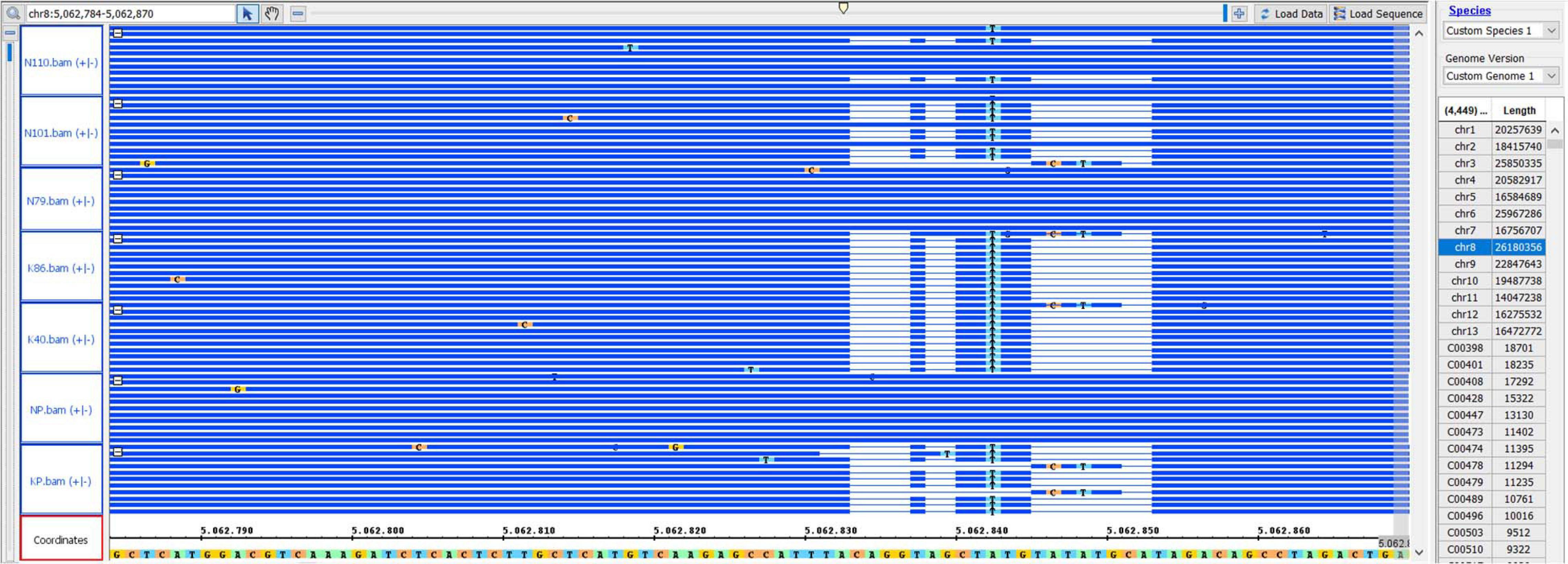
Figure 4. IGB shows the marker position of S8_5062843 for the selected genotypes. NP, shattered capsule parent; KP, non-shattering capsule parent. N110, N101, and N79 are F2 lines that had the shattered capsule trait. K40 and K86, non-shattering lines in F2. The coordinates show the reference genome sequence.
In order to reveal genes potentially determining the shattering trait, the SNP region (S8_5062843) was examined with FGENESH software. About 40 kb flanking sequence of the SNP was extracted from the sesame reference genome and analyzed. The results showed that a region covering the SNP marker (S8_5062843) encoded an mRNA including six exons, which was identified as a potential gene related to the shattering trait (data not shown). This sequence region was later amplified with PCR primers, and the sequence of the polymorphic region was confirmed by Sanger sequencing for shattering and non-shattering parents; this produced predicted gene sequences of 6,836 and 6,822 bp in length, respectively. Using tools at the Softberry website to predict gene structure, these sequences were re-analyzed, and six CDSs were predicted in both types (Figure 5). Compared with the reference (capsule shattering) mRNA sequence, the deletions flanking the SNP remove a splice site from the flanking region of S8_5062843, resulting in a shortened exon 2 (Table 5). This altered mRNA is predicted to cause a frameshift mutation starting with the 315th amino acid changing from valine to methionine (Val to Met), which is proposed to result in the loss of function of this gene for the non-shattering parent (Table 6). The potential gene sequence (capsule shattering type) was compared to the non-redundant protein database, and the candidate gene with 440 amino acids shared 99% sequence homology with the predicted transcription repressor KAN1 isoform X1 (S. indicum) [XP_011085885.1]. Other KAN1/KAN1-like proteins from different plants (XP_022869422.1, Olea europaea var. sylvestris; GFP94880.1, Phtheirospermum japonicum) also indicated sequence homology > 60% with our sequence. The mRNA sequence of the capsule shattering gene encoding a KAN1/KAN1-like protein has been submitted to NCBI with accession no. MT991091.
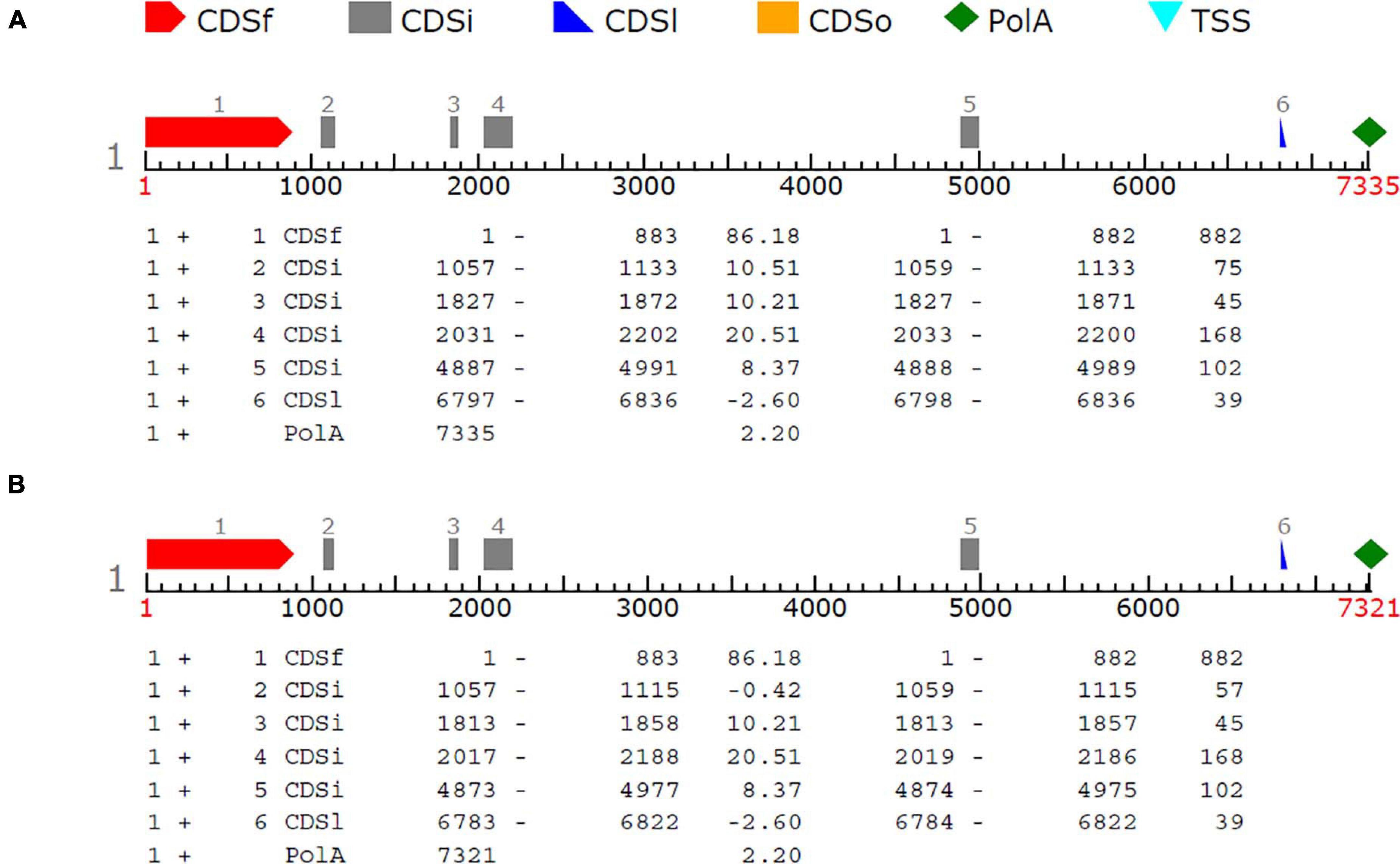
Figure 5. Gene structure of the candidate gene controlling the shattering trait. One gene and six CDSs were predicted in capsule shattering (A) and non-shattering (B) parents. CDSf, first exon (starting with start codon); CDSi, internal (internal exon); CDSl, last coding segment; PolyA, polyadenylation signal.
Marker Development and Validation
In order to differentiate between the capsule shattering and non-shattering genotypes, InDel (S8_5062843) and CAPS (S8_4476867) markers were designed (Table 7). We obtained a single PCR product using the InDel marker, with a size of about 241 bp in shattered genotypes, while the non-shattering genotype produced 227 bp (Supplementary Figure 2) because of deletions in the region of S8_5062843 (Table 5). CAPS markers require a SNP that falls within a restriction site, which is not the case for S8_5062843. However, a nearby SNP positioned at 4,476,867 bp in LG8/chr8 also segregated between the shattering and non-shattering lines, and this polymorphism eliminates a TaqI restriction site (Supplementary Figure 3). We designed a CAPS marker for the 4,476,867 C > A substitution that amplified a 377-bp PCR product. This product could not be digested in the non-shattering parent, but it yielded 275- and 102-bp restriction products with TaqI enzyme in the capsule shattering parent (Supplementary Figure 2). Heterozygous lines with respect to this SNP also showed both the original PCR product and the digested products (Supplementary Figure 2). These markers were also validated with sesame genotypes (Supplementary Figure 4).
Discussion
In the present study, we constructed a genetic map in sesame via the ddRAD-Seq strategy, which is a powerful SNP diagnostic technique for genotyping. This method produces a large number of sequence tags distributed throughout the genome at restriction sites, allowing a large number of functional polymorphic variants to be identified with low cost (Peterson et al., 2012). It also eliminated random shearing in the library preparation step, leading to high percentages of missing data, which is one of the major shortcomings of the genotyping-by-sequencing (GBS) method (Elshire et al., 2011). Recently, the ddRAD-Seq approach has been used successfully in different plants such as rapeseed (Chen et al., 2017), peanut (Liang et al., 2021), hazelnut (Helmstetter et al., 2020), and lettuce (Seki et al., 2020). However, to our knowledge, this study is the first time that it has been applied in sesame to construct a genetic map. With this technique, we identified 782 SNP markers distributed across 13 LGs; the number of LGs exactly matched the number of chromosomes in S. indicum (n = 13), showing the effectiveness of the ddRAD-Seq method to identify variants at a genome-wide level. However, large gaps were observed in LG1 and LG7 in our genetic map, and LG11 was only sparsely represented by three polymorphic SNPs (Figure 2). One of the disadvantages of this approach is that occasional genomic regions have very few restriction fragments of the right size to generate markers or may be lacking in polymorphisms. Our results were comparable to those by Shirasawa et al. (2016), who developed genetic maps for tomato with ddRAD-seq, which also left large gaps in some linkage groups. Before the construction of the linkage map, 258 SNP markers, which comprised 24.80% of the polymorphic SNPs, were excluded because of segregation distortion and missing data. Segregation distortion in plants results from gametophyte and/or zygotic selection or chromosomal rearrangements (Menz et al., 2002), and different genetic mechanisms of segregation distortion have been described (Li et al., 2011; Xu et al., 2013). Compared with other studies in sesame, higher values for segregation distortion were obtained by Zhang Y. et al. (2013) and Wu et al. (2014) in high-density genetic map construction. The population type and genetic relationships within the mapping population might also cause differences between gene ratios, leading to segregation distortion (Zhou et al., 2018). The genetic map in the current study covered 697.3 cM, with an average marker interval distance of 0.89 cM. Several genetic maps have been previously reported for sesame, mostly generated using SSR markers or SSRs in combination with other markers. Although these maps had higher length, lower marker densities were observed compared to our result. For example, a sesame genetic map was constructed with EST-SSR, AFLP, and RSAMPL markers mapped into 30 linkage groups covering a genetic length of 936.72 cM; the average distance between markers was 4.93 cM (Wei et al., 2009). A high-density map was constructed using 724 polymorphic markers which were SSR, AFLP, and RSAMPL; these were anchored in 14 linkage groups spanning a total of 1,216.00 cM, with a marker density of 1.86 cM per marker interval (Zhang H. et al., 2013). Another sesame genetic map that incorporated 424 SSR markers and comprised of 13 LGs was reported, with a total length of 1,869.8 cM and a mean density of 5.13 cM (Wang et al., 2017). With the advent of sequencing technologies, comparable high-density maps have been constructed with SNPs in sesame. An F2 population was genotyped with SLAF-seq approach, and the resulting map contained 1,233 markers and was 1,474.87 cM in length, with an average marker interval of 1.20 cM (Zhang Y. et al., 2013). This sequencing method was also used by Du et al. (2019) to construct a genetic map of 2,128.51cM in length, with an average distance of 0.99cM between adjacent markers. Wu et al. (2014) developed 224 F8:9 RILs which were sequenced with RAD-seq, the map, which was 844.46 cM in length, included 1,230 markers with an average of 0.69 cM between adjacent markers. The other reducing representation library sequencing approach, GBS, was employed for genotyping an F2 population and constructing a genetic map of 1,086.403 cM in length, with an average of 0.918 cM between adjacent bins (Zhang Y. et al., 2018). Compared to these studies, the total length of the map in our study is a little shorter, and this difference might be a result of the (i) type of mapping population, (ii) number of sequenced genotypes (Li et al., 2017), (iii) sequencing method, and/or (iv) genetic background of parents. Discarded linkage groups and fewer segregation distortion markers in mapping analysis might also be reasons for a shorter map (Wu et al., 2014). Changing the types of restriction enzyme in the library construction or increasing the size of the mapping population might be alternatives to improve this genetic map using the ddRAD-Seq approach (Yang et al., 2020).
The mutant sesame line (PI 599466) had a non-shattering capsule and phenotypically differs from Muganli-57 (shattered capsule cultivar), with curling leaves and oval and smaller capsules at the vegetative stage (Figure 1). These agronomic differences facilitate the selection of genotypes with shattering/non-shattering capsules, especially during the seedling period, in hybridization studies. However, Liu et al. (2020) recently developed a mutant sesame line named JQA that has male infertility and also the curling leaf structure. This showed that the leaf curling observed in genotypes with non-shattering capsule should not be used as a direct phenotypic criterion in early growing stage for the selection of capsule shattering trait. The gene controlling capsule shattering and leaf curling was detected on the eighth chromosome in our study, whereas Liu et al. (2020) found the gene encoding male infertility and leaf shape (curling or rolling) on the 12th chromosome. Different gene pathways in the genome therefore control curling in sesame leaves. Complex developmental processes, including the control of polarity, regulate leaf transverse rolling in plants (Bowman et al., 2002; Micol and Hake, 2003). Studies from different researchers have found that three gene families, KANADI (Eshed et al., 2001), HD-ZIP III (Talbert et al., 1995), and YABBY (Eshed et al., 2004), regulate the establishment of the proximal/distal axis of leaves in Arabidopsis. Yan et al. (2008) cloned the SHALLOT-LIKE1 (SLL1)/RL9 gene by way of a map-based cloning strategy and identified that it encodes a GARP protein, an ortholog of the Arabidopsis KANADI family in rice. The sll1 mutant was defective in the regulation of leaf abaxial cell development, resulting in a leaf curling phenotype (Zhang et al., 2009). In our study, the candidate gene controlling the shattering trait was found on chromosome 8, encoding the KAN1/KAN1-like (KANADI) protein that regulates lateral organ polarity and is required for abaxial identity in both leaves and carpels in Arabidopsis (Kerstetter et al., 2001). The mutation in that gene is therefore hypothesized to cause loss of cell polarity in developing tissues, resulting in the development of mutants that have non-shattering capsules and curling leaves. Parallel with the current study, a loss of function of the KAN1-like protein was also detected in a curly leaf, non-shattering sesame mutant by Zhang H.Y. et al. (2018), who named this gene SiCl1. The fact that these mutations were discovered independently in the same gene in two independent studies and resulted in the same phenotype confirms that KAN1/SiCl1 determines the non-shattering trait. In Arabidopsis, KAN1 was demonstrated to be a transcriptional repressor, which shuts down the expression of several genes involved in tissue development and signaling through the interaction of its Myb-like domain with a 6-bp cis-acting target motif (Huang et al., 2014). Using ChIP-seq and a tiling array, over 200 genes were shown to be directly regulated by A. thaliana KAN1 (Merelo et al., 2013). The deletion reported here is at the C-terminal end of the Myb-like domain of sesame KAN1/SiCl1 and therefore likely to result in the loss of its target-binding function; this is also consistent with the recessive nature of the non-shattering trait. It can be hypothesized that, like its ortholog in A. thaliana, sesame KAN1 controls leaf patterning, capsule size, and capsule shattering through the regulation of numerous target genes. In future studies, generating mutations in the target genes of sesame KAN1/SiCl1 may enable a discrimination between the non-shattering trait and other undesirable phenotypic effects.
Capsule shattering is one of the most problematic issues in sesame farming because it causes seed losses at harvest of up to 50% (Weiss, 1971). Despite its importance, limited studies have been conducted to improve the understanding of the genomic basis of this trait. In the present study, the gene related to capsule shattering was determined, and new molecular marker technologies were developed. Previously, Uzun et al. (2003) crossed another non-shattering mutant line (cc3) with the Turkish variety Muganli-57, and a closely linked amplified fragment length polymorphism (AFLP) marker was identified in the resulting segregated population. Sequence-characterized amplified region markers associated with shattering resistance were later developed from F2 lines derived from a cross between Cplus1, a sesame line with shattering-resistant capsules, and KUAOX25, a line with shattering-susceptible capsules (Phumichai et al., 2017). In the present study, we developed InDel marker related to the shattering trait, and therefore the difference between shattered and non-shattered capsule genotypes can be visualized with basic PCR systems and simple agarose gel electrophoresis. The use of simple genotyping technologies is highly important because sesame breeding is conducted primarily in low-income Asian and African countries. This marker should be integrated into sesame breeding studies with the SSR marker ZMM2498, which determines the growth trait controlled by the dt gene on chromosome 9 (Zhang Y. et al., 2018). In sesame, plant growth is originally indeterminate, and this wildish character causes unwanted agricultural issues such as non-synchronous maturity and incompatibility to combine harvesting (Uzun et al., 2013). Determinate sesame makes a uniform date of maturity, which is one of the requirements for the mechanical harvesting (Uzun and Cagirgan, 2006) of non-shattering capsules. Thus, the markers S8_5062843 and ZMM2498 controlling the shattering and growth habit traits, respectively, can be used as important selection tools in the development of sesame genotypes suitable for mechanized harvesting.
Although non-shattering capsule mutants are theoretically suitable for mechanized harvesting, their low yields currently limit the use of these genotypes in sustainable sesame agriculture. Crossing between mutant and cultivars might be an alternative to obtain heterosis effects for increased yield. This breeding approach about crossing “local varieties × mutants,” “mutants × mutants,” and “mutants × introduced lines” was also suggested to obtain desired traits in sesame (Van Zanten, 2001). In this respect, Uzun et al. (2004) hybridized a non-shattering capsule mutant with local varieties and monitored the heterosis effect in terms of seed yield in all hybrids. In another study, the cultivar Muganli-57 and non-shattering capsule mutant (ACS 344) were introduced into a cross-breeding program, and higher values were acquired for the studied yield traits in capsule shattering types (Yol and Uzun, 2019). Although positive effects were observed on agronomic traits with these crosses, there was no improvement in the shattering trait concerning suitability for mechanized harvesting. Therefore, instead of agronomic approaches, the gene identified in this study should be studied further with gene editing technologies (CRISPR-Cas9, TALEN, and ZFN) to create new variations, which might confer non-shattering capsules without the other negative traits associated with the loss of function of this gene. This strategy was used by Zhai et al. (2019) to understand the functions of pod shattering resistance genes (IND and ALC) using CRISPR/Cas9 technology in rapeseed. A new indica rice line was also developed with the targeted CRISPR/Cas9-mediated editing of the qSH1 gene to obtain lower rates of seed shattering (Sheng et al., 2020). Therefore, the identified gene in the present study should also be assessed with gene editing technologies to make improvements on this trait.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA633276.
Author Contributions
EY conducted the data analysis, mapping, annotation and wrote the manuscript. MB and SK conducted the F2 populations and phenotyping. SL and BU supervised the project and reviewed and revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
Funding for this research was provided by the Scientific and Technological Research Council of Turkey (TUBITAK) through the project coded 217O225.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We appreciate the Scientific Research Projects Coordination Unit of Akdeniz University for continuous support. We are also grateful to USDA, ARS Plant Genetic Resources Conservation Unit, Griffin, GA, United States, for supplying the genetic material several times.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.679659/full#supplementary-material
Supplementary Figure 1 | Total number of reads and GC content (%) for F2 lines and two parents.
Supplementary Figure 2 | (a,b) Amplification of developed InDel (S8_5062843) and CAPS (S8_4476867) markers with selected genotypes, respectively. NP, shattered capsule parent, KP, non-shattered capsule parent. N91, N143, N204, N182, N231, and N258 had shattered capsule phenotypes in F2. K28, K93, and K1 had non-shattered capsule phenotypes in F2.
Supplementary Figure 3 | IGB shows the (a) single-nucleotide polymorphism (SNP) changes in the position of 4476867 on LG8. (b) Enzyme digestion with TaqI for this SNP. NP, shattered capsule parent; KP, non-shattered capsule parent.
Supplementary Figure 4 | (a,b) Validation of the developed markers: (a) for CAPS marker and (b) for InDel marker. NP, shattered capsule parent; KP, non-shattered capsule parent. ACS 169, ACS 195, and ACS 211 were shattered capsule sesame genotypes.
Footnotes
- ^ www.usegalaxy.eu
- ^ http://www.softberry.com/berry.phtml?topic=index&group=programs&subgroup=gfind
- ^ https://primer3.ut.ee/
- ^ http://nc2.neb.com/NEBcutter2/
References
Aguirre, N. C., Filippi, C. V., Zaina, G., Rivas, J. G., Acuña, C. V., Villalba, P. V., et al. (2019). Optimizing ddRADseq in non-model species: a case study in Eucalyptus dunnii Maiden. Agronomy 9:484. doi: 10.3390/agronomy9090484
Ahmed, I., Islam, M., Arshad, W., Mannan, A., Ahmad, W., and Mirza, B. (2009). High-quality plant DNA extraction for PCR: an easy approach. J. Appl. Genet. 50, 105–107. doi: 10.1007/bf03195661
Arslan, C., Uzun, B., Ulger, S., and Cagirgan, M. I. (2007). Determination of oil content and fatty acid composition of sesame mutants suited for intensive management conditions. J. Am. Oil Chem. Soc. 84, 917–920. doi: 10.1007/s11746-007-1125-6
Ashri, A. (2007). “Sesame (Sesamum indicum L.),” in Genetics Resources, Chromosome Engineering and Crop Improvement, Oilseed Crops, ed. R. J. Singh (Boca Raton, FL: CRC Press), 231–289.
Baird, N. A., Etter, P. D., Atwood, T. S., Currey, M. C., Shiver, A. L., Lewis, Z. A., et al. (2008). Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS One 3:e3376. doi: 10.1371/journal.pone.0003376
Basak, M., Uzun, B., and Yol, E. (2019). Genetic diversity and population structure of the mediterranean sesame core collection with use of genome-wide SNPs developed by double digest RAD-Seq. PLoS one 14:e0223757. doi: 10.1371/journal.pone.0223757
Bowman, J. L., Eshed, Y., and Baum, S. F. (2002). Establishment of polarity in angiosperm lateral organs. Trends Genet. 18, 134–141. doi: 10.1016/s0168-9525(01)02601-4
Chen, J., Wang, B., Zhang, Y., Yue, X., Li, Z., and Liu, K. (2017). High-density ddRAD linkage and yield-related QTL mapping delimits a chromosomal region responsible for oil content in rapeseed (Brassica napus L.). Breed. Sci. 67, 296–306. doi: 10.1270/jsbbs.16116
Chen, S., Zhou, Y., Chen, Y., and Gu, J. (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, 884–890.
Davik, J., Sargent, D. J., Brurberg, M. B., Lien, S., Kent, M., et al. (2015). A ddRAD based linkage map of the cultivated strawberry, Fragaria xananassa. PLoS One 10:e0137746. doi: 10.1371/journal.pone.0137746
Dossa, K., Konteye, M., Niang, M., Doumbia, Y., and Cisse, N. (2017). Enhancing sesame production in West Africa’s Sahel: a comprehensive insight into the cultivation of this untapped crop in Senegal and Mali. Agric. Food Secur. 6:68.
Du, H., Zhang, H., Wei, L., Li, C., Duan, Y., and Wang, H. (2019). A high-density genetic map constructed using specific length amplified fragment (SLAF) sequencing and QTL mapping of seed-related traits in sesame (Sesamum indicum L.). BMC Plant Biol. 19:588.
Elshire, R. J., Glaubitz, J. C., Sun, Q., Poland, J. A., Kawamoto, K., Buckler, E. S., et al. (2011). A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS One 6:e19379. doi: 10.1371/journal.pone.0019379
Eshed, Y., Baum, S. F., Perea, J. V., and Bowman, J. L. (2001). Establishment of polarity in lateral organs of plants. Curr. Biol. 11, 1251–1260. doi: 10.1016/s0960-9822(01)00392-x
Eshed, Y., Izhaki, A., Baum, S. F., Floyd, S. K., and Bowman, J. L. (2004). Asymmetric leaf development and blade expansion in Arabidopsis are mediated by KANADI and YABBY activities. Development 131, 2997–3006. doi: 10.1242/dev.01186
FAO (2018). Available online at: http://www.fao.org/faostat. (accessed November 26, 2020)
Garrison, E., and Marth, G. (2012). Haplotype-based variant detection from short-read sequencing. arXiv.org [preprint] 1207.3907.
Girardot, C., Scholtalbers, J., Sauer, S., Su, S., and Furlong, E. E. M. (2016). Je, a versatile suite to handle multiplexed NGS libraries with unique molecular identifiers. BMC Bioinformatics 17:419.
Glaubitz, J. C., Casstevens, T. M., Lu, F., Harriman, J., and Elshire, R. J. (2014). TASSEL-GBS: a high capacity genotyping by sequencing analysis pipeline. PLoS One 9:e90346. doi: 10.1371/journal.pone.0090346
Helmstetter, A. J., Oztolan-Erol, N., Lucas, S. J., and Buggs, R. J. A. (2020). Genetic diversity and domestication of hazelnut (Corylus avellana L.) in Turkey. Plants People Planet 2, 326–339. doi: 10.1002/ppp3.10078
Huang, T., Harrar, Y., Lin, C., Reinhart, B., Newell, N. R., Talavera-Rauh, F., et al. (2014). Arabidopsis KANADI1 acts as a transcriptional repressor by interacting with a specific cis-element and regulates auxin biosynthesis, transport, and signaling in opposition to HD-ZIPIII factors. Plant Cell 26, 246–262. doi: 10.1105/tpc.113.111526
Hwang, L. S. (2005). “Sesame oil,” in Bailey’s Industrial Oil and Fat Products, ed. F. Shahidi (New Jersey: John Wiley & Sons), 537–576.
Jin, Y., Zhao, W., Nie, S., Liu, S. S., El-Kassaby, Y. A., Wang, X. R., et al. (2019). Genome-wide variant identification and high-density genetic map construction using RADseq for Platycladus orientalis (Cupressaceae). G3 5, 3663–3672. doi: 10.1534/g3.119.400684
Kerstetter, R., Bollman, K., Taylor, R., Bomblies, K., and Poethig, R. S. (2001). KANADI regulates organ polarity in Arabidopsis. Nature 411, 706–709. doi: 10.1038/35079629
Konar, A., Choudhury, O., Bullis, R., Fiedler, L., Kruser, J. M., and Stephens, M. T. (2017). High-quality genetic mapping with ddRADseq in the non-model tree Quercus rubra. BMC Genom. 18:417.
Kosambi, D. D. (1994). The estimation of map distance from recombination values. Ann. Eugen. 12, 12172–12175.
Langham, D. G. (1946). Genetics of sesame, III. “open sesame” and mottled leaf. J. Hered. 37, 149–152.
Langham, D. R. (2001). “Shatter resistance in sesame,” in Sesame Improvements by Induced Mutations, Proc Final FAO/IAEA Co-ord Res Meeting, ed. L. Van Zanten (Vienna: IAEA), 51–61.
Langham, D. R., and Wiemers, T. (2002). “Progress in mechanizing sesame in the US through breeding,” in Trends in New Crops and New Uses, eds J. Janickand and A. Whipkey (Virginia: American Society for Horticultural Science Press), 157–173.
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Lee, J. H., Natarajan, S., Biswas, M. K., Shirasawa, K., Isobe, S., Kim, H. T., et al. (2018). SNP discovery of Korean short day onion inbred lines using double digest restriction site-associated DNA sequencing. PLoS One 13:e0201229. doi: 10.1371/journal.pone.0201229
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Li, L., Zhao, S., Su, J., Fan, S., Pang, C., Wei, H., et al. (2017). High-density genetic linkage map construction by F2 populations and QTL analysis of early-maturity traits in upland cotton (Gossypium hirsutum L.). PLoS One 12:e0182918. doi: 10.1371/journal.pone.0182918
Li, X. H., Wang, X. J., Wei, Y. L., and Brummer, E. C. (2011). Prevalence of segregation distortion in diploid alfalfa and its implications for genetics and breeding applications. Theor. Appl. Genet. 123, 667–679. doi: 10.1007/s00122-011-1617-5
Liang, Y., Cason, J. M., Baring, M. R., and Septiningsih, E. M. (2021). Identification of QTLs associated with Sclerotinia blight resistance in peanut (Arachis hypogaea L.). Genet. Resour. Crop Evol. 68, 629–637. doi: 10.1007/s10722-020-01012-4
Liu, H., Zhou, F., Zhou, T., Yang, Y., and Zhao, Y. (2020). Fine mapping of a novel male-sterile mutant showing wrinkled-leaf in sesame by BSA-Seq technology. Ind. Crops Prod. 156:112862. doi: 10.1016/j.indcrop.2020.112862
Menz, M. A., Klein, R. R., Mullet, J. E., Obert, J. A., Unruh, N. C., and Klein, P. E. (2002). A high-density genetic map of Sorghum bicolor (L.) Moench based on 2926 AFLP, RFLP and SSR markers. Plant Mol. Biol. 48, 483–499.
Merelo, P., Xie, Y., Brand, L., Ott, F., Weigel, D., Bowman, J. L., et al. (2013). Genome-wide identification of KANADI1 target genes. PLoS One 8:e77341. doi: 10.1371/journal.pone.0077341
Miyahara, Y. H., Hibasami, H., Katsuzaki, H., Imai, K., and Komiya, T. (2001). Sesamolin from sesame seed inhibits proliferation by inducing apoptosis in human lymphoid leukemia Molt 4B cells. Int. J. Mol. Med. 7, 369–371.
Moazzami, A. A., and Kamal-Eldin, A. (2006). Sesame seed is a rich source of dietary lignans. J. Am. Oil Chem. Soc. 83, 719–723. doi: 10.1007/s11746-006-5029-7
Nakamura, Y., Okumura, H., Ono, Y., Kitagawa, Y., Rogi, T., and Shibata, H. (2020). Sesame lignans reduce LDL oxidative susceptibility by downregulating the platelet-activating factor acetylhydrolase. Eur. Rev. Med. Pharmacol. Sci. 24, 2151–2161.
Nguyen, K. L., Grondin, A., Courtois, B., and Gantet, P. (2019). Next-generation sequencing accelerates crop gene discovery. Trends Plant Sci. 24, 263–274. doi: 10.1016/j.tplants.2018.11.008
Peterson, B. K., Weber, J. N., Kay, E. H., Fisher, H. S., and Hoekstra, H. E. (2012). Double Digest RADseq: an inexpensive method for de novo SNP discovery and genotyping in model and non-model species. PLoS One 7:e37135. doi: 10.1371/journal.pone.0037135
Phumichai, C., Matthayatthaworn, W., Chuenpom, N., and Wongkaew, A. (2017). Identification of a SCAR marker linked to a shattering resistance trait in sesame. Turk. J. Field Crops 22, 258–265.
Robledo, D., Palaiokostas, C., Bargelloni, L., Martínez, P., and Houston, R. (2018). Applications of genotyping by sequencing in aquaculture breeding and genetics. Rev. Aquac. 10, 670–682. doi: 10.1111/raq.12193
Roy, S. C., Moitra, K., and De Sarker, D. (2017). Assessment of genetic diversity among four orchids based on ddRAD sequencing data for conservation purposes. Physiol. Mol. Biol. Plants 23, 169–183. doi: 10.1007/s12298-016-0401-z
Ryu, S. N., Kim, K. S., and Kang, S. S. (2003). Growth inhibitory effects of sesamolin from sesame seeds on human leukemia HL-60 cells. Korean J. Pharmacogn. 34, 237–241.
Scheben, A., Batley, J., and Edwards, D. (2017). Genotyping-by-sequencing approaches to characterize crop genomes: choosing the right tool for the right application. Plant Biotechnol. J. 15, 149–161. doi: 10.1111/pbi.12645
Seki, K., Komatsu, K., Hiraga, M., Tanaka, K., Uno, Y., and Mastumura, H. (2020). Identification of two QTLs for resistance to Fusarium wilt race 1 in lettuce (Lactuca sativa L.). Euphytica 216:174.
Sheng, X., Sun, Z., Wang, X., Tan, Y., Yu, D., and Yuan, G. (2020). Improvement of the rice “easy-to-shatter” trait via CRISPR/Cas9-mediated mutagenesis of the qSH1 gene. Front. Plant Sci. 11:619.
Shirasawa, K., Hirakawa, H., and Isobe, S. (2016). Analytical workflow of double-digest restriction site-associated DNA sequencing based on empirical and in silico optimization in tomato. DNA Res. 23, 145–153. doi: 10.1093/dnares/dsw004
Solovyev, V., Kosarev, P., Seledsov, I., and Vorobyev, D. (2006). Automatic annotation of eukaryotic genes, pseudogenes and promoters. Genome Biol. 7, 1–12.
Talbert, P. B., Adler, H. T., Parks, D. W., and Comai, L. (1995). The REVOLUTA gene is necessary for apical meristem development and for limiting cell divisions in the leaves and stems of Arabidopsis thaliana. Development 121, 2723–2735. doi: 10.1242/dev.121.9.2723
Uzun, B., and Cagirgan, M. I. (2006). Comparison of determinate and indeterminate lines of sesame for agronomic traits. Field Crops Res. 96, 13–18. doi: 10.1016/j.fcr.2005.04.017
Uzun, B., and Arslan, Ç, and Furat, S. (2008). Variation in fatty acid compositions, oil content and oil yield in a germplasm collection of sesame (Sesamum indicum L.). J. Am. Oil Chem. Soc. 85, 1135–1142. doi: 10.1007/s11746-008-1304-0
Uzun, B., Lee, D., Donini, P., and Cagirgan, M. I. (2003). Identification of a molecular marker linked to the closed capsule mutant trait in sesame using AFLP. Plant Breed. 122, 95–97. doi: 10.1046/j.1439-0523.2003.00787.x
Uzun, B., Ozbas, M. O., Canci, H., and Cagirgan, M. I. (2004). Heterosis for agronomic traits in sesame hybrids of cultivars x closed capsule mutants. Acta Agric. Scand. B Soil Plant Sci. 54, 108–112. doi: 10.1080/0909471030021998
Uzun, B., Yol, E., and Furat, S. (2013). Genetic advance, heritability and inheritance in determinate growth habit of sesame. Aust. J. Crop Sci. 7, 978–983.
Van Ooijen, J. W. (2011). Multipoint maximum likelihood mapping in a full-sib family of an outbreeding species. Genet. Res. 93, 343–349. doi: 10.1017/S0016672311000279
Van Zanten, L. (2001). “Sesame improvement by induced mutations: results of the co-ordinated research project and recommendation for future studies,” in Sesame Improvements by Induced Mutations, Proc Final FAO/IAEA Co-ord Res Meeting, ed. L. Van Zanten (Vienna: IAEA), 1–12.
Voorrips, R. E. (2002). MapChart: software for the graphical presentation of linkage maps and QTLs. J. Hered. 93, 77–78. doi: 10.1093/jhered/93.1.77
Wang, L., Xia, Q., Zhang, Y., Zhu, X., and Zhu, X. (2016). Updated sesame genome assembly and fine mapping of plant height and seed coat color QTLs using a new high-density genetic map. BMC Genom. 17:31.
Wang, L., Zhang, Y., Zhu, X., Zhu, X., Li, D., and Zhang, X. (2017). Development of an SSR-based genetic map in sesame and identification of quantitative trait loci associated with charcoal rot resistance. Sci. Rep. 7:8349.
Wang, S., Meyer, E., McKay, J. K., and Matz, M. V. (2012). 2b-RAD: a simple and flexible method for genome-wide genotyping. Nat. Methods 9, 808–810. doi: 10.1038/nmeth.2023
Wei, L. B., Zhang, H. Y., Zheng, Y. Z., Miao, H. M., Zhang, T. Z., and Guo, W.-Z. (2009). A genetic linkage map construction for sesame (Sesamum indicum L.). Genes Genom. 31, 199–208. doi: 10.1007/bf03191152
Wongyai, W., Saengkaewsook, W., and Veerawudh, J. (2001). “Sesame mutation induction: improvement of nonshattering capsule by using gamma rays and EMS,” in Sesame Improvements by Induced Mutations, Proc Final FAO/IAEA Co-ord Res Meeting, ed. L. Van Zanten (Vienna: IAEA), 71–78.
Wu, K., Liu, H., Yang, M., Tao, Y., Ma, H., and Wu, W. (2014). High-density genetic map construction and QTLs analysis of grain yield-related traits in sesame (Sesamum indicum L.) based on RAD-Seq technology. BMC Plant Biol. 14:274.
Xu, X., Li, L., Dong, X., Jin, W., Melchinger, A. E., and Chen, S. (2013). Gametophytic and zygotic selection leads to segregation distortion through in vivo induction of a maternal haploid in maize. J. Exp. Bot. 64, 1083–1096. doi: 10.1093/jxb/ers393
Yan, S., Yan, C. J., Zeng, X. H., Yang, Y. C., Fang, Y. W., Tian, C. Y., et al. (2008). ROLLED LEAF 9, encoding a GARP protein, regulates the leaf abaxial cell fate in rice. Plant Mol. Biol. 68, 239–250. doi: 10.1007/s11103-008-9365-x
Yang, W., Wang, Y., Jiang, D., Tian, C., Zhu, C., and Li, G. (2020). ddRADseq-assisted construction of a high-density SNP genetic map and QTL fine mapping for growth-related traits in the spotted scat (Scatophagus argus). BMC Genom. 21:278.
Yol, E., and Uzun, B. (2012). Geographical patterns of sesame accessions grown under mediterranean environmental conditions, and establishment of a core collection. Crop Sci. 52, 2206–2214. doi: 10.2135/cropsci2011.07.0355
Yol, E., and Uzun, B. (2019). Inheritance of indehiscent capsule character, heritability and genetic advance analyses in the segregation generations of dehiscent x indehiscent capsules in sesame. Tarim Bilim. Derg. 25, 79–85. doi: 10.15832/ankutbd.539005
Yoshida, H., and Takagi, S. (1997). Effects of seed roasting temperature and time on quality characteristics of sesame (Sesamum indicum L.) oil. J. Sci. Food Agric. 75, 19–26. doi: 10.1002/(sici)1097-0010(199709)75:1<19::aid-jsfa830>3.0.co;2-c
Zhai, Y., Cai, S., Hu, L., Yang, Y., Amoo, O., and Fan, C. (2019). CRISPR/Cas9-mediated genome editing reveals differences in the contribution of indehiscent homologues to pod shatter resistance in Brassica napus L. Theor. Appl. Genet. 132, 2111–2123. doi: 10.1007/s00122-019-03341-0
Zhang, G. H., Xu, Q., Zhu, X. D., Qian, Q., and Xue, H. W. (2009). SHALLOT-LIKE1 is a KANADI transcription factor that modulates rice leaf rolling by regulating leaf abaxial cell development. Plant Cell 21, 719–735. doi: 10.1105/tpc.108.061457
Zhang, H. Y., Miao, H. M., Wei, L. B., Li, C., Duan, Y., and Xu, F. (2018). Identification of a SiCL1 gene controlling leaf curling and capsule indehiscence in sesame via cross-population association mapping and genomic variants screening. BMC Plant Biol. 18:296.
Zhang, H., Miao, H., Wei, L., Li, C., Zhao, R., and Wang, C. (2013). Genetic analysis and QTL mapping of seed coat color in sesame (Sesamum indicum L.). PLoS One 8:e63898. doi: 10.1371/journal.pone.0063898
Zhang, Y., Wang, L., Gao, Y., Li, D., Yu, J., and Zhou, R. (2018). Genetic dissection and fine mapping of a novel dt gene associated with determinate growth habit in sesame. BMC Genet. 19:38.
Zhang, Y., Wang, L., Xin, H., Li, D., Ma, C., Ding, X., et al. (2013). Construction of a high-density genetic map for sesame based on large scale marker development by specific length amplified fragment (SLAF) sequencing. BMC Plant Biol. 13:141. doi: 10.1186/1471-2229-13-141
Zhou, F., Liu, Y., Liang, C., Wang, W., Li, C., Guo, Y., et al. (2018). Construction of a high-density genetic linkage map and QTL mapping of oleic acid content and three agronomic traits in sunflower (Helianthus annuus L.) using specific-locus amplified fragment sequencing (SLAF-seq). Breed. Sci. 68, 596–605. doi: 10.1270/jsbbs.18051
Keywords: neglected crop, annotation, breeding, high throughput sequencing, linkage map
Citation: Yol E, Basak M, Kızıl S, Lucas SJ and Uzun B (2021) A High-Density SNP Genetic Map Construction Using ddRAD-Seq and Mapping of Capsule Shattering Trait in Sesame. Front. Plant Sci. 12:679659. doi: 10.3389/fpls.2021.679659
Received: 12 March 2021; Accepted: 23 April 2021;
Published: 01 June 2021.
Edited by:
Zerihun Tadele, University of Bern, SwitzerlandReviewed by:
Mohammad Rashed Hossain, Bangladesh Agricultural University, BangladeshShouvik Das, Indian Agricultural Research Institute (ICAR), India
Copyright © 2021 Yol, Basak, Kızıl, Lucas and Uzun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Engin Yol, ZW5naW55b2xAYWtkZW5pei5lZHUudHI=
 Engin Yol
Engin Yol Merve Basak2
Merve Basak2 Stuart James Lucas
Stuart James Lucas Bulent Uzun
Bulent Uzun