- 1Research Institute for Dok-do and Ulleung-do Island, Kyungpook National University, Daegu, South Korea
- 2Institute of Natural Science, Yeungnam University, Gyeongsan, South Korea
- 3DMZ Botanic Garden, Korea National Arboretum, Yanggu, South Korea
- 4Department of Biology, School of Life Science, BK21 FOUR KNU Creative BioResearch Group, Kyungpook National University, Daegu, South Korea
- 5Department of Integrative Natural Sciences for the East Sea Rim, Kyungpook National University, Daegu, South Korea
- 6Department of Biological Sciences, Sungkyunkwan University, Suwon, South Korea
Functional gene transfer from organelles to the nucleus, known as intracellular gene transfer (IGT), is an ongoing process in flowering plants. The complete plastid genomes (plastomes) of two Ulleung island endemic violets, Viola ulleungdoensis and V. woosanensis, were characterized, revealing a lack of the plastid-encoded infA, rpl32, and rps16 genes. In addition, functional replacement of the three plastid-encoded genes in the nucleus was confirmed within the genus Viola and the order Malpighiales. Three strategies for the acquisition of a novel transit peptide for successful IGT were identified in the genus Viola. Nuclear INFA acquired a novel transit peptide with very low identity between these proteins, whereas the nuclear RPL32 gene acquired an existing transit peptide via fusion with the nuclear-encoded plastid-targeted SOD gene (Cu-Zn superoxide dismutase superfamily) as one exon, and translated both proteins in the cytosol using alternative mRNA splicing. Nuclear RPS16 contains an internal transit peptide without an N-terminal extension. Gene loss or pseudogenization of the plastid-borne rpl32 and rps16 loci was inferred to occur in the common ancestor of the genus Viola based on an infrageneric phylogenetic framework in Korea. Although infA was lost in the common ancestor of the order Malpighiales, the rpl32 and rps16 genes were lost multiple times independently within the order. Our current study sheds additional light on plastid genome composition and IGT mechanisms in the violet genus and in the order Malpighiales.
Introduction
Plastids are important cellular organelles that contain their own genomes and originate from cyanobacteria-like prokaryotes by endosymbiosis (Keeling, 2010). The plastid genome (plastome) undergoes reduction as a consequence of gene loss or endosymbiotic gene transfer (EGT) to the host cell nucleus (Timmis et al., 2004). Many transferred gene products need to be targeted back to the plastids, which involves repair of double-strand breaks by non-homologous end-joining of DNA released from organelles (Kleine et al., 2009). After EGT to the host nucleus, most angiosperm plastomes retain genes encoding 79 proteins, 30 tRNAs, and four rRNAs (Ruhlman and Jansen, 2014). Although the gene content is conserved in angiosperms, plastid-encoded gene loss (e.g., accD, clpP, infA, ndhA-K, rpl20, rpl22, rpl23, rpl32, rpoA, rps7, rps11, rps12, rps15, rps16, and rps18) has been documented across several lineages (Jansen et al., 2007; Park et al., 2018). These gene losses are often the result of functional gene transfer from plastids to the nucleus and are known to occur frequently and represent ongoing processes (Timmis et al., 2004). To conduct this unique function, the transferred plastid genes should acquire nuclear expression elements with transit peptides for the import of products into plastids (Bock and Timmis, 2008). However, the majority of functional transfers in angiosperms are restricted to nine of the 25 protein-encoding genes including acetyl-CoA carboxylase subunit β (accD), translation initiation factor A (infA), and four large and three small subunit ribosomal proteins (rpl20, rpl22, rpl23, rpl32, rps7, rps15, and rps16, respectively).
A number of possible mechanisms involved in functional gene replacement have been proposed, including direct intracellular gene transfer (IGT) to the nucleus, or gene substitution by a nuclear homolog. For example, IGT from plastids to the nucleus in angiosperms has been documented for five plastid genes: accD in Trifolium (Magee et al., 2010; Sabir et al., 2014), infA in Ranunculaceae (Park et al., 2015; Park and Park, 2020), rpl22 in Fabaceae and Fagaceae (Gantt et al., 1991; Jansen et al., 2011), Passiflora (Shrestha et al., 2020), rpl32 in Ranunculaceae (Park et al., 2015, 2020; Park and Park, 2020), Rhizophoraceae, and Salicaceae (Cusack and Wolfe, 2007; Ueda et al., 2007), rps7 in Passiflora (Shrestha et al., 2020), and rps15 in Papaver (Park et al., 2018). Among these, the transferred plastid accD, rpl32 (Rhizophoraceae and Salicaceae), and rps7 genes have adopted a transit peptide from an existing nuclear gene, whereas the infA, rpl22, rpl32 (Ranunculaceae), and rps15 genes acquired a novel transit peptide. Gene substitution for plastids in the nucleus has been documented for three genes: accD in Brassicaceae (Schulte et al., 1997; Babiychuk et al., 2011), Geranium (Park et al., 2017), and Poaceae (Konishi et al., 1996; Gornicki et al., 1997), rpl20 in Passiflora (Shrestha et al., 2020), rpl23 in spinach (Bubunenko et al., 1994), Geranium (Weng et al., 2016), rps16 in Lupinus (Keller et al., 2017), Medicago and Populus (Ueda et al., 2008), Passiflora (Shrestha et al., 2020), and Ranunculaceae (Park et al., 2020; Park and Park, 2020). The accD and rpl23 loci were substituted by a cytosolic homolog of eukaryotic origin, whereas rps16 and rpl20 have been substituted by cytosolic homologs of mitochondrial origin (i.e., the nuclear genome had already acquired mitochondrial rps16 and rpl20 via IGT).
The cosmopolitan genus Viola L. (Violaceae, Malpighiales) consists of approximately 600 species and is distributed mainly in temperate regions and high mountains in tropical regions (Yoo and Jang, 2010; Marcussen et al., 2014). Viola presumably originated in South America and subsequently dispersed into the Northern Hemisphere approximately 18 million years ago (Early Eocene), and allopolyploidy accounts for a significant portion of the speciation events within the genus (67–88%; Ballard et al., 1998; Marcussen et al., 2012, 2014). Several important characteristics, such as stigma morphology, pistil shape, petal color, chromosome number, and geographical distribution, in conjunction with several conventional molecular markers, have provided certain baseline classification systems and revealed important evolutionary processes. Nevertheless, we are still far from fully understanding the complex interspecific relationships involving Viola (Gingins de la Sarraz, 1823; Becker, 1925; Clausen, 1964; Ballard, 1996; Ballard et al., 1998; Marcussen et al., 2014). In particular, a high proportion of allopolyploidy within the genus, with numerous hybrid and polyploid complexes, in conjunction with highly variable morphological traits, has hampered violet systematists sufficiently to fully reveal complex reticulating events among the species. Korean Viola represents one of the most important angiosperm lineages, with 43–64 recognized taxa (Yoo and Jang, 2010), but little is known regarding the contribution of allopolyploidy in the speciation of phylogenetically and taxonomically complex groups. Of the numerous hybrid and polyploidy complexes, Viola woosanensis, endemic to Ulleung Island in Korea, has been suggested to be of hybrid origin involving other Ulleung Island endemic V. ulleungdoensis and East Asia-restricted V. chaerophylloides (Gil and Kim, 2016). As we assembled the plastomes of these Viola species to gain insights into the origin and evolution of Ulleung Island endemics, we discovered the absence of infA, rpl32, and rps16. Although plastomes of other congeneric species of Viola (i.e., V. mirabilis, V. radeana, V. phalacrocarpa, V. japonica, V. seoulensis, and V. websteri) have been characterized in Korea, the presence/absence of these genes has not been explored (Cheon et al., 2017, 2019, 2020). However, Menezes et al. (2018), in comparative plastid genome analysis among species in the order Malpighiales, reported the absence of rps16 and rpl32 in a single species of Violaceae (V. seoulensis) and in Salicaceae (Populus alba and Salix purpurea). This raises the question of how frequent the absence of rps16 and rpl32 genes is in the Viola genus and whether the absence of these genes in plastid genomes was transferred to another genome compartment or lost completely.
In this study, we assembled two plastome sequences of the violet species V. ulleungdoensis and V. woosanensis endemic to Ulleung Island, Korea, as genomic resources of insular endemics. We also documented the functional transfer of three genes, infA, rpl32, and rps16, from the plastid to the nucleus via IGT or gene substitution. Finally, we determined gene loss and functional transfer to the nucleus within the order Malpighiales and the loss of two genes (rps16 and rpl32) within the phylogenetic framework of the genus Viola in Korea.
Materials and Methods
Plastid Genome Sequencing, Assembly, and Annotation
Fresh V. ulleungdoensis and V. woosanensis leaves from single individuals were collected from natural populations on Ulleung Island, off the east coast of the Korean Peninsula. Total DNA was isolated using the DNeasy Plant Mini Kit (Qiagen, Carlsbad, CA, United States), and the paired-end (PE) reads were generated using the Illumina HiSeq 4000 platform (Illumina, Inc., San Diego, CA, United States) at Macrogen Corporation (Seoul, South Korea). The PE reads were assembled de novo using Velvet v.1.2.10 (Zerbino and Birney, 2008) with multiple k-mers (99–143). From each assembly, the full length of the plastome contig, including only one copy of inverted repeat (IR), was generated. Gene annotation was conducted using Geneious R10 v.10.2.61 using the model plant tobacco plastome (NC_001879) as a reference, and tRNAs were confirmed using tRNAscan-SE v.2.0 (Chan and Lowe, 2019). The annotated plastome sequences have been deposited in GenBank with accession numbers MK228834 (V. ulleungdoensis) and MK228835 (V. woosanensis). Plastome maps were drawn using OGDRAW v1.3.1 (Greiner et al., 2019).2
Identification of Functional Gene Transfer to the Nucleus in Viola
To identify IGT events, the transcriptome from V. acuminata (SRR5320546) was assembled de novo using Trinity v.2.5.1 (Grabherr et al., 2011). Candidates for nuclear-encoded plastid genes were identified by performing “BlastN” searches using BLAST+ v.2.6.0 (Camacho et al., 2009) against the transcriptomes of rpl32 and rps16 gene sequences from Licania sprucei plastome (NC_024065), and infA gene sequences from Amborella trichopoda plastome (NC_005086). In the case of rps16, the “BlastN” search was additionally performed using the Medicago truncatula (AB365526) nuclear-encoded plastid rps16 sequences. The NCBI Conserved Domain Database (CDD; Marchler-Bauer et al., 2010) and TargetP v.1.1 (Emanuelsson et al., 2007) were used for functional domain annotation and prediction of transit peptides, respectively.
To confirm the nuclear-encoded plastid gene sequences, polymerase chain reaction (PCR) was performed using total genomic DNA from V. ulleungdoensis. Primer pairs were designed based on sequences of the nuclear-encoded INFA, SODcp-RPL32, and RPS16 transcripts from V. acuminata using Primer3 in Geneious R10 (Supplementary Table 1). The PCR products were purified using InClone™ gels and PCR purification kits (InClone Biotech Co., Seoul, Korea). Sequencing reactions were performed using purified PCR products using BigDye Terminator v.3.1 Cycle Sequencing Kit reagents (Applied Biosystems, Foster City, CA, United States) at GenoTech Corp. (Daejeon, Korea). Genomic sequences and transcripts were aligned using MUSCLE (Edgar, 2004) in Geneious R10.
In addition, seven Viola taxa were included: V. betonicifolia (SRR13316929), V. canadensis (ERR2040403), V. mandshurica (SRR5320533), V. orientalis (SRR5322130), V. pubescens (SRR5353310), V. tricolor (ERR2040402), and V. uliginosa (SRR1576988). Transcriptome assemblies, BlastN searches, identification of transit peptide and conserved domain, and alignments were performed as described above. Maximum likelihood (ML) analysis was performed using IQ-TREE v.1.6.8 (Nguyen et al., 2014), employing 1,000 bootstrap replications.
Identification of Functional Gene Transfer to the Nucleus in Order Malpighiales
To better understand the evolutionary fate of the plastid-encoded infA, rpl32, and rps16 genes, available Malpighiales transcriptomes from the 1KP project database3 and the genomics portal Phytozome v.12.1.64 were searched. Nuclear transcripts were identified by “BlastN” searches (e-value cutoff of 1e-6), employing the nuclear-encoded INFA, SODcp-RPL32, and RPS16 sequences from V. acuminata as query sequences. Phylogenetic analysis was performed to detect gene loss or apparent pseudogenes within the order Malpighiales using 19 plastomes from nine major families (Salicaceae, Passifloraceae, Violaceae, Euphorbiaceae, Malpighaceae, Erythroxylaceae, Clusiaceae, Linaceae, and Chrysobalanaceae; Supplementary Table 2). Euonymus japonicus (NC_028067) was selected as an outgroup based on a previous study (Choi and Park, 2016). The datasets were aligned with MAFFT (Katoh and Standley, 2013) in Geneious R10. ML analysis was performed using IQ-TREE v.1.6.8, employing 1,000 bootstrap replications.
Survey for Loss of Two Plastid-Encoded rpl32 and rps16 Genes in Viola
To document the presence/absence of two ribosomal protein genes, 19 species of Viola were sampled from major lineages of the genus in Korea including sections of Dischidium, Chamaemelanium, and Nomimium (Yoo and Jang, 2010; Supplementary Table 3). Total genomic DNA was isolated from silica gel-dried leaves using a DNeasy Plant Mini Kit (Qiagen, Carlsbad, CA, United States). To detect the rpl32 gene, the intergenic spacer (IGS) region between ndhF and trnL-UAG genes was amplified using newly designed PCR primers (Supplementary Table 1). In addition, to detect the rps16 gene, the IGS region between the trnQ-UUG and trnK-UUU genes was amplified using newly designed primers (Supplementary Table 1). PCR product purification and DNA sequencing were performed as described previously. Phylogenetic analysis of 19 species of Viola based on four chloroplast noncoding regions (three IGS sequences, atpF-H, psbA-trnH, and trnL-F, and one intronic sequence, rpl16) was performed to detect gene loss or apparent pseudogenes of rpl32 and rps16. Hybanthus concolor was used as an outgroup (Yoo and Jang, 2010; Supplementary Table 3). Sequences were aligned using MUSCLE in Geneious R10. ML analysis was performed with IQ-TREE v.1.6.8 with 1,000 bootstrap replications.
Results
General Features of Two Insular Viola Plastome Sequences
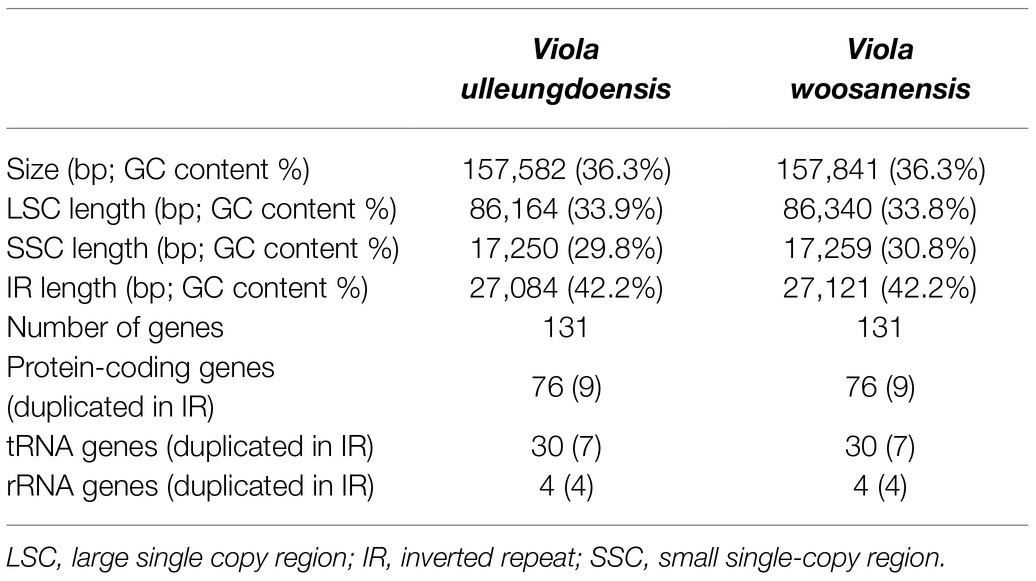
DNA sequencing generated approximately 4.2 and 3.9 Gb of paired-end reads for V. ulleungdoensis and V. woosanensis, respectively. The data were used to assemble plastomes with a deep coverage of 1,323× for V. ulleungdoensis and 511× for V. woosanensis. The V. ulleungdoensis and V. woosanensis plastomes were 157,582 and 157,841 bp, respectively, with a pair of inverted repeats (IRA and IRB) of 27,084 and 27,121 bp separated by small and large single copy (SSC and LSC) regions of 17,250 and 86,164, 17,259, and 86,340 bp, respectively (Figure 1 and Table 1). The two Viola plastomes encoded 110 genes including 76 protein-coding genes, 30 tRNA genes, and four rRNA genes. The plastid-encoded infA, rpl32, and rps16 genes appeared to have been lost in from their original plastomes. Mauve alignment (Darling et al., 2004) of seven Violaceae plastomes showed high structural and sequence similarities (Supplementary Figure 1). In addition, comparative analysis of seven Viola plastomes confirmed the loss of three plastid-encoded infA, rpl32, and rps16 genes shared by the Viola species examined in Korea (Supplementary Figure 1).
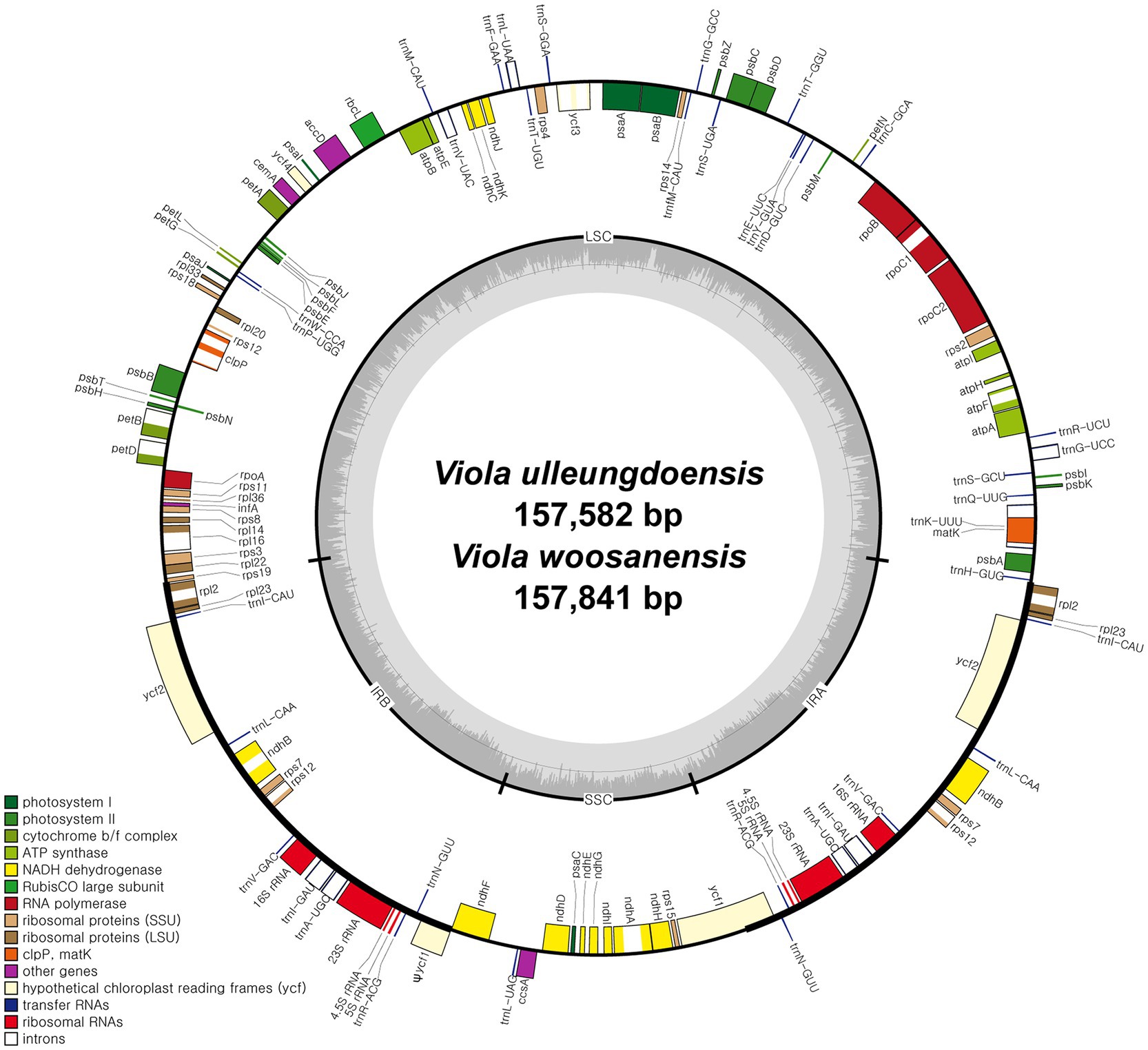
Figure 1. Complete plastome maps of Viola ulleungdoensis and Viola woosanensis. Genes located outside the circle are transcribed clockwise, whereas those located inside are transcribed counter clockwise. The gray bar area in the inner circle denotes the guanine-cytosine (GC) content of the genome, whereas the lighter gray area indicates the adenosine-thymine (AT) content of the genome. Large single copy, small single copy, and inverted repeat are indicated by LSC, SSC, and IR, respectively. Ψ indicates pseudogenes.
Functional Replacement of Three Protein-Coding Genes From Plastids to the Nucleus in Viola
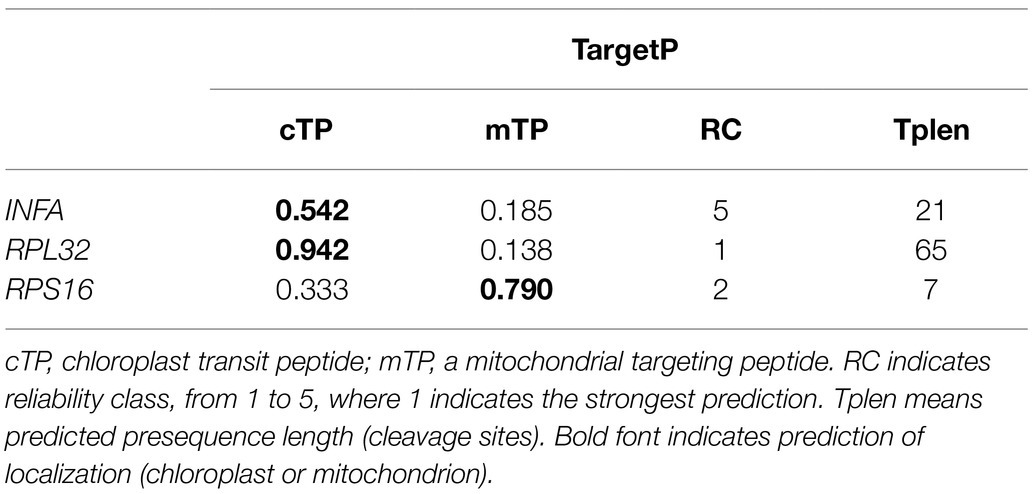
Our results revealed that the plastid-encoded infA, rpl32, and rps16 loci were lost in the assembled Viola plastomes. To investigate potential functional transfer to the nucleus, we performed BLAST searches against the V. acuminata transcriptome data using plastid gene sequences. A transcript with high sequence identity to infA was present, which has a transit peptide (TargetP: chloroplast = 0.542) and a conserved infA domain (Table 2 and Supplementary Figure 2). In addition, we also found an open reading frame (ORF) containing the conserved ribosomal protein L32 domain, which included extended sequences of 645 bp downstream from the conserved domain. The first 65 amino acids of the ORF were predicted by TargetP to be a transit peptide (chloroplast = 0.942), which is targeted to the plastid (Table 2). CDD analysis predicted the conserved domain of “Cu-Zn Superoxide Dismutase superfamily” involving the transit peptide and the conserved domain of “ribosomal protein L32” (Supplementary Figure 3A). To further investigate potential alternative mRNA splicing of the SODcp-RPL32 gene, we queried the V. acuminata transcriptome with the ORF. The complete sequence of plastid-targeted SOD was identified in the transcriptome (Supplementary Figure 3B). Although the rps16-like transcripts were not detected by a “BlastN” search against the plastid-encoded rps16 gene of L. sprucei, two different transcripts were detected using the query sequence of the nuclear-encoded RPS16 (M. truncatula, AB365526). Both transcripts also contained a transit peptide (TargetP: mitochondria = 0.790) and the conserved ribosomal protein S16 domain (Table 2 and Supplementary Figure 4).
Viola ulleungdoensis was surveyed to identify the nuclear-encoded INFA, SODcp-RPL32, and RPS16 loci (for plastids) using PCR and Sanger DNA sequencing. The gene sequence sizes for the nuclear-encoded INFA, RPS16 and SODcp-RPL32 genes were 459, 1,629, and 3,020 bp, respectively (Figure 2 and Supplementary Table 4). Alignment of the DNA and transcript nucleotide sequences confirmed that the nuclear-encoded INFA was an intronless gene, whereas the nuclear-encoded SODcp-RPL32 and RPS16 contained eight and two introns, respectively (Figure 2). The conserved domain of nuclear-encoded RPS16 was included in three exons (Figure 2). In the case of the nuclear-encoded SODcp-RPL32, the conserved domain of “Cu-Zn Superoxide Dismutase superfamily” was composed of the first to seventh exon, and the conserved domain of “ribosomal protein L32” was included in the eighth exon (Figure 2).
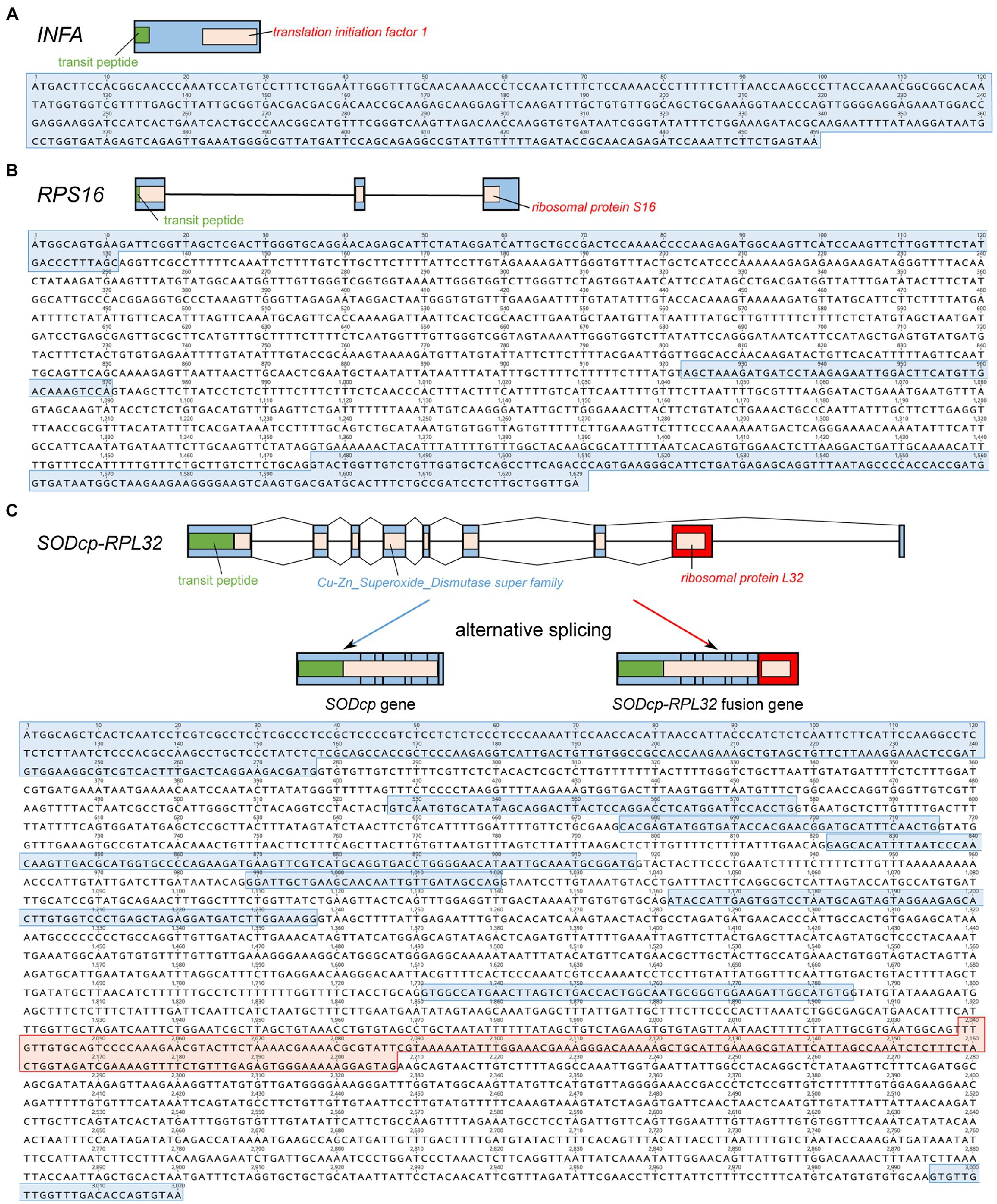
Figure 2. Organization of the nuclear-encoded INFA, SODcp-RPL32, and RPS16 genes from Viola ulleungdoensis. (A) Schematic diagram and nucleotide sequence of the nuclear-encoded INFA gene. (B) Schematic diagram and nucleotide sequence of the nuclear-encoded RPS16. (C) Schematic diagram and nucleotide sequence of nuclear-encoded SODcp-RPL32 as a chimeric gene with alternative splicing. Blue and red boxes denote exons and RPL32 gene regions, respectively. Green boxes indicate transit peptides.
Seven additional Viola transcriptomes were surveyed for the identification of three nuclear-encoded plastid-targeted genes and provided clear evidence for potential functional replacement of the plastid-encoded infA, rpl32, and rps16 to the nucleus. Interestingly, some Viola transcriptomes contain two or multiple copies. For example, V. acuminate transcriptomes contain two divergent INFA, RPS16 (for mitochondrial), and SODcp-RPL32 copies (Figure 3). Viola betonicifolia transcriptome contains three and two divergent copies of INFA and RPS16 (for mitochondrial), respectively. Viola tricolor transcriptome contains four and two divergent copies of INFA and RPS16 (for mitochondrial), respectively. Viola uliginosa transcriptome contains two divergent INFA and RPS16 (for mitochondrial) copies. Viola mandshurica transcriptome contains two divergent RPS16 (mitochondrial).
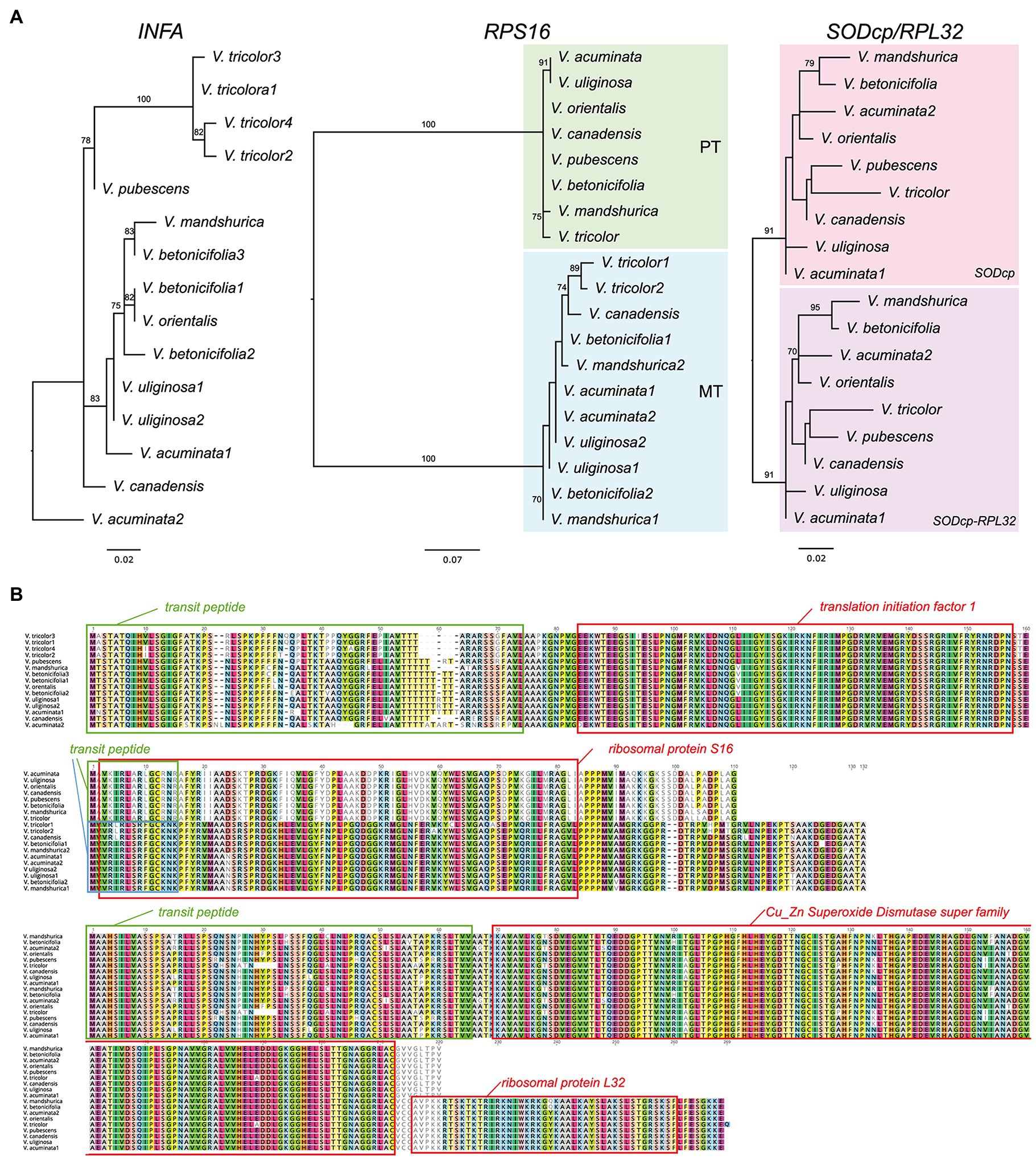
Figure 3. Nuclear-encoded INFA, RPS16, and SODcp-RPL32 genes in Viola. (A) Phylogenetic analysis of the nuclear-encoded genes from Viola. The numbers after each species indicate the paralogs of each nuclear gene. (B) Amino acid sequence alignment of the nuclear encoded INFA, RPS16, and SODcp-RPL32 copies from Viola. Boxes indicate transit peptide (green and blue) and conserved domains.
Functional Transfer of Plastid Genes to the Nucleus in the Order Malpighiales
We determined the phylogenetic distribution of the plastid-encoded infA, rpl32, and rps16 genes in 19 representative species from nine families of the order Malpighiales with Euonymus japonicus (Celastraceae, Order Celastrales) as an outgroup (Figure 4 and Supplementary Figure 5A). Our analysis showed that loss of the infA gene occurred in the common ancestor of Malpighiales, whereas loss of the rpl32 and rps16 genes was lineage-specific within Malpighiales (Figure 4 and Supplementary Figure 5A).
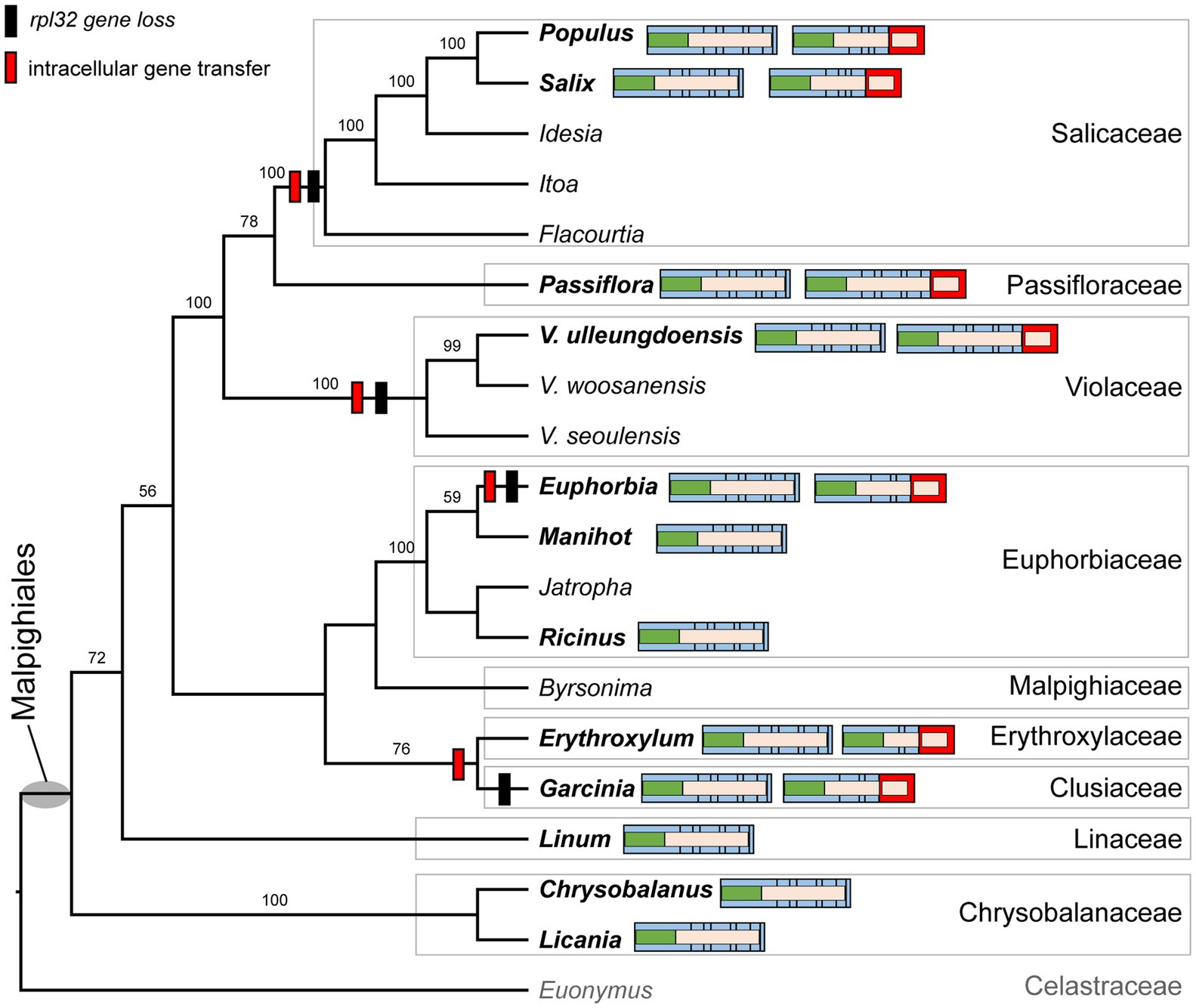
Figure 4. Phylogenetic distribution of nuclear-encoded SODcp-RPL32 in Malpighiales. Phylogenetic relationships were inferred from 19 representative species in nine families of Malpighiales using an outgroup, Euonymus (Celastraceae). Bootstrap values based on 1,000 replicates and >50% are shown for each node. Schematic diagrams of the variable structure of the nuclear-encoded SODcp and SODcp-RPL32 genes among selected Malpighiales are shown for 12 taxa in bold. Colors correspond to transit peptides, exons, and conserved domains (see Figure 2).
To investigate functional transfer to the nucleus, we queried the available transcriptome data from 11 Malpighiales species with the Viola INFA, SODcp-RPL32, and RPS16 sequences (Supplementary Table 4). Except for Garcinia and Ricinus, transcriptomes containing ORFs with the conserved domain of “translation initiation factor A” domain were identified at the C-termini (Supplementary Figure 5B). The extended region of Viola was highly divergent with very low sequence identity (13.7–53.2%) to other Malpighiales (Supplementary Figure 5B). Two genera, Euphorbia and Manihot, contained two versions of the nuclear-encoded INFA gene (Supplementary Figure 5B). With the exception of Manihot and Ricinus, two transcripts with the conserved domain of “ribosomal protein S16” identified at the N-terminus were detected in the transcriptome (Supplementary Figure 5C). Phylogenetic analyses of the RPS16 sequences from 11 Malpighiales species and both nuclear-encoded RPS16 (for plastids and mitochondria) of Viola indicated two different origins for the transcripts (Supplementary Figure 5C). In the case of rpl32, nuclear SODcp-RPL32 gene fusion was also identified in Populus, Salix, Euphorbia, Erythroxylum, Passiflora, and Garcinia transcriptomes (Figure 4). Amino acid sequence alignments of the chimeric gene products revealed structural degeneration or possible alternative mRNA splicing (Figure 3 and Supplementary Figure 6). Although the Erythroxylum plastome contained an intact rpl32 gene, we found evidence for nuclear SODcp-RPL32 gene fusion, suggesting the occurrence of an IGT event involving a common ancestor of the two families Erythroxylaceae and Clusiaceae (Figure 4).
Distribution of rpl32 and rps16 Gene Loss Among Species of Viola in Korea
As the presence of the plastid-encoded rpl32 and rps16 genes in the plastomes varied among representative lineages of the order Malpighiales, we further surveyed to confirm whether gene loss events are species- or genus-specific among representative species of Viola in Korea. Thus, we surveyed 19 species of Viola, including sections of Dischidium, Chamaemelanium, and Nomimium, and amplified two IGS regions of ndhF-trnL and trnK-trnQ via PCR, given the locations of rpl32 and rps16, respectively. The amplicon sizes of ndhF-trnL IGS ranged from 862 bp in V. lactiflora to 1,803 bp in V. veracunda. A BLAST search showed that the 19 examined species contained partial sequences of the rpl32 gene with premature stop codons (Supplementary Figure 7). The amplicon sizes of the trnK-trnQ IGS ranged from 659 bp in V. biflora to 761 bp in V. rossii. BLAST searches revealed that the examined 19 species lacked detectable rps16-like sequences (Supplementary Figure 7). The phylogenetic distribution of losses involving the two genes suggested that the IGT event occurred in the most recent common ancestor of the Viola lineage in Korea (Figure 5).
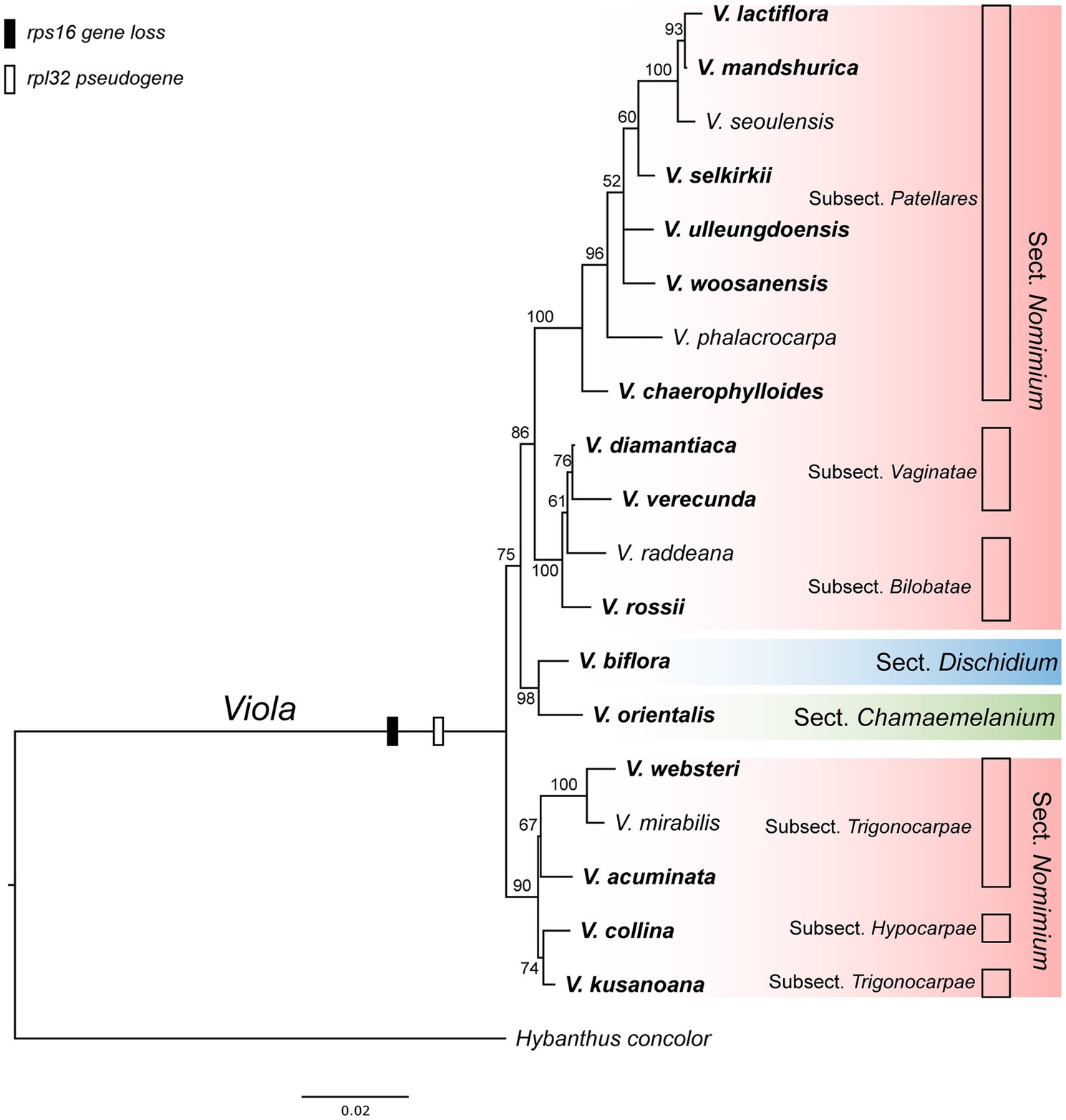
Figure 5. Phylogenetic distribution of plastid-encoded rpl32 and rps16 gene loss events in 19 representative species of the family Violaceae. The bootstrap value based on 1,000 replicates and >50% are shown for each node.
Discussion
In this study, we generated two complete plastomes of V. ulleungdoensis and V. woosanensis in the family Violaceae and explored the loss of three plastid-encoded genes, infA, rpl32, and rps16. Various analyses confirmed the transfer of these three plastid genes to the nuclear compartment in the genus Viola and in the order Malpighiales, further highlighting the dynamic process of IGT in angiosperms.
Sequenced plastomes of photosynthetic angiosperms generally contain 79 protein-coding genes, but the evolutionary timing of plastid gene loss events in multiple lineages has only been inferred (Jansen et al., 2007). These findings suggest that plastid gene transfer to the nucleus is still an ongoing process in angiosperms. However, successful functional gene transfer requires several steps because of the different nuclear machinery required for transcription and translation (Woodson and Chory, 2008). For example, a promoter, poly-A tail, and targeting sequence must be acquired after the integration of a plastid gene into the nuclear genome, and the subsequent pseudogenization and elimination of the original plastid gene sequence from its plastome. Transit peptides are critical for the transport of proteins into plastids in the cytosol. Three mechanisms for the acquisition of a targeting sequence in the nuclear genome have been reported (Ueda and Kadowaki, 2012); the acquisition of a novel transit peptide, acquisition of an existing transit peptide from a nuclear-encoded gene for plastids, and no acquisition due to the presence of an inherent transit peptide encoded in the gene. We determined that the Viola species examined in this study utilized all three strategies for the acquisition of transit peptides. For example, the transferred infA acquired a novel transit peptide based on the very low sequence identity between these proteins and no successful BLAST search results. In contrast, the transferred rpl32 locus acquired an existing transit peptide by inserting into the nuclear-encoded plastid-targeted SOD gene as one exon. This second mechanism is known to occur uniquely in Malpighiale lineages (Cusack and Wolfe, 2007), whereas transfer of the plastid rpl32 gene within another lineage (e.g., Ranunculaceae) followed a similar strategy to that involving infA. The generic structure of the nuclear SODcp-RPL32 gene and two types of transcripts from Viola species suggested that the nuclear gene mRNA transcript was alternatively spliced and translated in the cytosol for import of both protein products into plastids (Figure 2). Likewise, the mangrove (Bruguiera gymnorrhiza) contains a chimeric gene with an alternative mRNA splicing mechanism, whereas Popular undergoes double duplication and complete sub-functionalization, which contains the rpl32 protein with a SODcp-derived amino terminus and SODcp protein in plastids (Cusack and Wolfe, 2007; Ueda et al., 2007). The third strategy for the acquisition of transit peptides involves the plastid-encoded rps16 gene in Viola (Figure 2). The nuclear-encoded RPS16 for plastids lacks an N-terminal extension because of the presence of an internal targeting signal in the gene (Ueda et al., 2008). The transit peptide of RPS16 is derived from a duplicate copy of a nuclear homolog, which is targeted to the mitochondrion (Ueda et al., 2008).
The analyzed Viola species contained two or multiple versions of the nuclear-encoded plastid-targeted genes, showing patterns of asymmetrical gene duplication. These patterns suggested that the duplicated genes independently originated at the different time within the Viola or that stochastic loss after ancient gene duplication in the genus. Allopolyploidy has played an important role in the speciation of the genus Viola (Ballard, 1996; Marcussen et al., 2012, 2014). This evolutionary history of Viola suggested that the duplication of the nuclear-encoded plastid-targeted genes may occurs via whole genome duplication or polyploidy after their functional replacement to the nucleus. The study of IGT can help to the detection of ancient polyploidization (Park et al., 2020). To better understand the important role of polyploidy in the evolutionary history of Viola, it will be necessary to sequence additional transcriptomes from Viola species spanning all recognized sections and subsections.
As the presence/absence of the rpl32 and rps16 genes varied within the order Malpighiales, we further sequenced each IGS region that included genes from 19 congeneric species of Viola. Our results indicated that the rpl32 gene was pseudogenized, whereas the rps16 gene was completely lost from the plastome (Supplementary Figure 7). These results suggested that IGT involving the two genes must have occurred in the most-recent common ancestor of violet species in Korea, that is, most likely genus-specific; these became pseudogenes or were completely lost upon their transfer to the nuclear compartment.
Although nine plastid gene losses (infA, rpl20, rpl22, rpl32, rps7, rps16, rps19, ycf1, and ycf2) have been documented in Malpighiales (Menezes et al., 2018; Shrestha et al., 2020), molecular characterization of these events across the order has been limited. Of these gene loss events, we focused specifically on the fate of the plastid-encoded infA, rpl32, and rps16 loci in Malpighiales to better understand the dynamics of IGT. As plastid phylogenomic relationships of Malpighiales from nine families were reconstructed, we demonstrated that the infA gene loss occurred in the common ancestor of the order Malpighiales, whereas rpl32 and rps16 were lost independently multiple times within the order. Transcriptome data provided evidence for successful lineage- or species-specific IGT (Figure 4 and Supplementary Figures 5, 6). Plastid-encoded infA underwent multiple independent transfers to the nucleus (Millen et al., 2001). Our findings suggested a single transfer of the plastid infA to the nucleus in the most-recent common ancestor of the order Malpighiales although we did not find evidence for Garcinia and Ricinus. The transcriptomes of three genera, Manihot, Euphorbia, and Populus, contained two versions of the nuclear-encoded INFA, which may be associated with whole genome or segmental gene duplication. However, the phylogenetic placement of nuclear-encoded INFA suggests different origins for the duplicated copies. For example, Manihot and Euphorbia copies were duplicated by a species-specific event involving nuclear-encoded INFA (Supplementary Figure 6). In the case of Populus, INFA was duplicated in the common ancestor of Populus and Salix, and one copy loss occurred in Salix, resulting in the presence of two copies in Populus. The nuclear-encoded SODcp-RPL32 is a well-characterized example of gene transfer in Malpighiales (Cusack and Wolfe, 2007), in which the plastid rpl32 is integrated into an intron of the nuclear SOD gene in the Malpighiales ancestor. Comparative analyses of the nuclear-encoded SODcp-RPL32 in Malpighiales revealed that the transfer events were lineage-specific (Figure 4) and argued against the scenario of a single ancestral event. However, further studies, including more Malpighiales transcriptomes, are needed to confirm whether the formation of the SODcp-RPL32 chimeric gene is a general or specific phenomenon. Inactivation of plastid genes is favored after nuclear gene activation (Martin and Herrmann, 1998), and intermediate stages of the gene transfer process have been identified in Ranunculaceae (Park et al., 2020). We found co-existence of intact and transcribed gene copies in the Erythroxylum nucleus and plastids, suggesting that the IGT event occurred in the common ancestor of Erythroxylum and Garcinia. Likewise, Passiflora contains both copies of the nuclear-encoded SODcp-RPL32 and plastid rpl32 copies. A recent study also identified the SODcp-RPL32 chimeric gene in many Passiflora species that lack the rpl32 in their plastomes and in some Passiflora groups that contain plastid rpl32 (Shrestha et al., 2020). These results indicated that putative functional transfer of plastid rpl32 to the nucleus occurs in the common ancestor of the genus Passiflora. The nuclear-encoded plastid RPS16 is a good example of gene substitution for functional replacement involving the nucleus. It has been replaced by the nuclear-encoded mitochondrial RPS16, which had already been transferred to the nucleus via IGT (Keller et al., 2017). The transit peptides of the nuclear-encoded RPS16 contain a dual targeting signal for mitochondria and plastids, which is an internal signal without an N-terminal extension (Ueda et al., 2008). Among Malpighiales, plastid rps16 was lost independently at least seven times in our samples, and an expanded survey detected two versions of the nuclear-encoded RPS16 copies that were from two different origins (Supplementary Figure 5C). Although Ricinus and Garcinia plastomes contain the plastid rps16 gene, we found nuclear-encoded plastid RPS16 copies (Supplementary Figure 5C). These results suggested that gene substitution involving plastids occurred in the most-recent common ancestor of Malpighiales, and this may have facilitated the loss of the plastid-encoded rps16 gene from its plastomes. This finding also indicated that gene substitution of plastid-encoded rps16 was in the intermediate stage within Malpighiales.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/genbank/, MK228834 and https://www.ncbi.nlm.nih.gov/genbank/, MK228835.
Author Contributions
JY, SP, S-CK, and J-HP conceived and designed experiments. JY generated and analyzed the data and wrote a draft of the manuscript, which SP edited and S-CK revised. SP analyzed transcriptome results. H-YG collected samples and conducted PCR and sequencing procedures. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Basic Science Research Program through the National Research Foundation of Korea, which is funded by the Ministry of Education (2016R1A6A1A05011910 and 2019R1A2C1011221) to J-HP and JY.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.678580/full#supplementary-material
Footnotes
2. ^https://chlorobox.mpimp-golm.mpg.de/OGDraw.html
References
Babiychuk, E., Vandepoele, K., Wissing, J., Garcia-Diaz, M., De Rycke, R., Akbari, H., et al. (2011). Plastid gene expression and plant development require a plastidic protein of the mitochondrial transcription termination factor family. Proc. Natl. Acad. Sci. U. S. A. 108, 6674–6679. doi: 10.1073/pnas.1103442108
Ballard, H. E. (1996). Phylogenetic Relationships and Infrageneric Groups in Viola (Violaceae) Based on Morphology, Chromosome Numbers, Natural Hybridization and Internal Transcribed Spacer (ITS) Sequences. Madison, WI, USA: University of Wisconsin-Madison.
Ballard, H. E. Jr., Sytsma, K. J., and Kowal, R. R. (1998). Shrinking the violets: phylogenetic relationships of infrageneric groups in Viola (Violaceae) based on internal transcribed spacer DNA sequences. Syst. Bot. 23, 439–458. doi: 10.2307/2419376
Becker, W. (1925). “Viola L.” in Die natürlichen Pflanzenfamilien, vol. 21. Parietales und Opuntiales. ed. A. Engler (Leipzig: Wilhelm Engelmann), 363–376.
Bock, R., and Timmis, J. N. (2008). Reconstructing evolution: gene transfer from plastids to the nucleus. Bioessays 30, 556–566. doi: 10.1002/bies.20761
Bubunenko, M., Schmidt, J., and Subramanian, A. (1994). Protein substitution in chloroplast ribosome evolution: a eukaryotic cytosolic protein has replaced its organelle homologue (L23) in spinach. J. Mol. Biol. 240, 28–41. doi: 10.1006/jmbi.1994.1415
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421
Chan, P. P., and Lowe, T. M. (2019). “tRNAscan-SE: searching for tRNA genes in genomic sequences,” in Gene Prediction: Methods and Protocols. ed. M. Kollmar (New York, NY: Springer New York), 1–14.
Cheon, K.-S., Kim, D.-K., Kim, K.-A., and Yoo, K.-O. (2020). The complete chloroplast genome sequence of Viola japonica (Violaceae). Mitochondrial DNA B 5, 1297–1298. doi: 10.1080/23802359.2020.1731376
Cheon, K.-S., Kim, K.-A., Kwak, M., Lee, B., and Yoo, K.-O. (2019). The complete chloroplast genome sequences of four Viola species (Violaceae) and comparative analyses with its congeneric species. PLoS One 14:e0214162. doi: 10.1371/journal.pone.0214162
Cheon, K. S., Yang, J. C., Kim, K. A., Jang, S. K., and Yoo, K. O. (2017). The first complete chloroplast genome sequence from Violaceae (Viola seoulensis). Mitochondrial DNA A DNA Mapp. Seq. Anal. 28, 67–68. doi: 10.3109/19401736.2015.1110801
Choi, K. S., and Park, S.-J. (2016). The complete chloroplast genome sequence of Euonymus japonica (Celastraceae). Mitochondrial DNA A DNA Mapp. Seq. Anal. 27, 3577–3578. doi: 10.3109/19401736.2015.1075127
Clausen, J. (1964). Cytotaxonomy and distributional ecology of western North American violets. Madroño 17, 173–197.
Cusack, B. P., and Wolfe, K. H. (2007). When gene marriages don’t work out: divorce by subfunctionalization. Trends Genet. 23, 270–272. doi: 10.1016/j.tig.2007.03.010
Darling, A. C. E., Mau, B., Blattner, F. R., and Perna, N. T. (2004). Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14, 1394–1403. doi: 10.1101/gr.2289704
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Emanuelsson, O., Brunak, S., von Heijne, G., and Nielsen, H. (2007). Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2, 953–971. doi: 10.1038/nprot.2007.131
Gantt, J. S., Baldauf, S. L., Calie, P. J., Weeden, N. F., and Palmer, J. D. (1991). Transfer of rpl22 to the nucleus greatly preceded its loss from the chloroplast and involved the gain of an intron. EMBO J. 10, 3073–3078. doi: 10.1002/j.1460-2075.1991.tb07859.x
Gil, H.-Y., and Kim, S.-C. (2016). Viola woosanensis, a recurrent spontaneous hybrid between V. ulleungdoensis and V. chaerophylloides (Violaceae) endemic to Ulleung Island, Korea. J. Plant Res. 129, 807–822. doi: 10.1007/s10265-016-0830-3
Gingins de la Sarraz, F. C. J. (1823). Mémoire sur la famille des Violaces. Mém. Soc. Phys. His. Nat. Geneve 2, 1–28.
Gornicki, P., Faris, J., King, I., Podkowinski, J., Gill, B., and Haselkorn, R. (1997). Plastid-localized acetyl-CoA carboxylase of bread wheat is encoded by a single gene on each of the three ancestral chromosomesets. Proc. Natl. Acad. Sci. U. S. A. 94, 14179–14184. doi: 10.1073/pnas.94.25.14179
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., et al. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652. doi: 10.1038/nbt.1883
Greiner, S., Lehwark, P., and Bock, R. (2019). OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47, W59–W64. doi: 10.1093/nar/gkz238
Jansen, R. K., Cai, Z., Raubeson, L. A., Daniell, H., Depamphilis, C. W., Leebens-Mack, J., et al. (2007). Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc. Natl. Acad. Sci. U. S. A. 104, 19369–19374. doi: 10.1073/pnas.0709121104
Jansen, R. K., Saski, C., Lee, S.-B., Hansen, A. K., and Daniell, H. (2011). Complete plastid genome sequences of three rosids (Castanea, Prunus, Theobroma): evidence for at least two independent transfers of rpl22 to the nucleus. Mol. Biol. Evol. 28, 835–847. doi: 10.1093/molbev/msq261
Katoh, P. F., and Standley, D. M. (2013). MAFFT multiple sequence alignment software v7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Keeling, P. J. (2010). The endosymbiotic origin, diversification and fate of plastids. Philos. Trans. R. Soc. B Biol. Sci. 365, 729–748. doi: 10.1098/rstb.2009.0103
Keller, J., Rousseau-Gueutin, M., Martin, G. E., Morice, J., Boutte, J., Coissac, E., et al. (2017). The evolutionary fate of the chloroplast and nuclear rps16 genes as revealed through the sequencing and comparative analyses of four novel legume chloroplast genomes from Lupinus. DNA Res. 24, 343–358. doi: 10.1093/dnares/dsx0006
Kleine, T., Maier, U. G., and Leister, D. (2009). DNA transfer from organelles to the nucleus: the idiosyncratic genetics of endosymbiosis. Annu. Rev. Plant Biol. 60, 115–138. doi: 10.1146/annurev.arplant.043008.092119
Konishi, T., Shinohara, K., Yamada, K., and Sasaki, Y. (1996). Acetyl-CoA carboxylase in higher plants: most plants other than Gramineae have both the prokaryotic and the eukaryotic forms of this enzyme. Plant Cell Physiol. 37, 117–122. doi: 10.1093/oxfordjournals.pcp.a028920
Magee, A. M., Aspinall, S., Rice, D. W., Cusack, B. P., Sémon, M., Perry, A. S., et al. (2010). Localized hypermutation and associated gene losses in legume chloroplast genomes. Genome Res. 20, 1700–1710. doi: 10.1101/gr.111955.110
Marchler-Bauer, A., Lu, S., Anderson, J. B., Chitsaz, F., Derbyshire, M. K., DeWeese-Scott, C., et al. (2010). CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 39(suppl_1), D225–D229. doi: 10.1093/nar/gkq1189
Marcussen, T., Heier, L., Brysting, A. K., Oxelman, B., and Jakobsen, K. S. (2014). From gene trees to a dated allopolyploid network: insights from the angiosperm genus Viola (Violaceae). Syst. Biol. 64, 84–101. doi: 10.1093/sysbio/syu071
Marcussen, T., Jakobsen, K. S., Danihelka, J., Ballard, H. E., Blaxland, K., Brysting, A. K., et al. (2012). Inferring species networks from gene trees in high-polyploid North American and Hawaiian violets (Viola, Violaceae). Syst. Biol. 61, 107–126. doi: 10.1093/sysbio/syr096
Martin, W., and Herrmann, R. G. (1998). Gene transfer from organelles to the nucleus: How much, what happens, and why? Plant Physiol. 118, 9–17. doi: 10.1104/pp.118.1.9
Menezes, A. P. A., Resende-Moreira, L. C., Buzatti, R. S. O., Nazareno, A. G., Carlsen, M., Lobo, F. P., et al. (2018). Chloroplast genomes of Byrsonima species (Malpighiaceae): comparative analysis and screening of high divergence sequences. Sci. Rep. 8:2210. doi: 10.1038/s41598-018-20189-4
Millen, R. S., Olmstead, R. G., Adams, K. L., Palmer, J. D., Lao, N. T., Heggie, L., et al. (2001). Many parallel losses of infA from chloroplast DNA during angiosperm evolution with multiple independent transfers to the nucleus. Plant Cell 13, 645–658. doi: 10.1105/tpc.13.3.645
Nguyen, L.-T., Schmidt, H. A., von Haeseler, A., and Minh, B. Q. (2014). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Park, S., An, B., and Park, S. (2018). Reconfiguration of the plastid genome in Lamprocapnos spectabilis: IR boundary shifting, inversion, and intraspecific variation. Sci. Rep. 8:13568. doi: 10.1038/s41598-018-31938-w
Park, S., An, B., and Park, S. (2020). Recurrent gene duplication in the angiosperm tribe Delphinieae (Ranunculaceae) inferred from intracellular gene transfer events and heteroplasmic mutations in the plastid matK gene. Sci. Rep. 10:2720. doi: 10.1038/s41598-020-59547-6
Park, S., Jansen, R. K., and Park, S. (2015). Complete plastome sequence of Thalictrum coreanum (Ranunculaceae) and transfer of the rpl32 gene to the nucleus in the ancestor of the subfamily Thalictroideae. BMC Plant Biol. 15:40. doi: 10.1186/s12870-015-0432-6
Park, S., and Park, S. (2020). Large-scale phylogenomics reveals ancient introgression in Asian Hepatica and new insights into the origin of the insular endemic Hepatica maxima. Sci. Rep. 10:16288. doi: 10.1038/s41598-020-73397-2
Park, S., Ruhlman, T. A., Weng, M.-L., Hajrah, N. H., Sabir, J. S. M., and Jansen, R. K. (2017). Contrasting patterns of nucleotide substitution rates provide insight into dynamic evolution of plastid and mitochondrial genomes of geranium. Genome Biol. Evol. 9, 1766–1780. doi: 10.1093/gbe/evx124
Ruhlman, T. A., and Jansen, R. K. (2014). The plastid genomes of flowering plants. Methods Mol. Biol. 1132, 3–38. doi: 10.1007/978-1-62703-995-6_1
Sabir, J., Schwarz, E., Ellison, N., Zhang, J., Baeshen, N. A., Mutwakil, M., et al. (2014). Evolutionary and biotechnology implications of plastid genome variation in the inverted-repeat-lacking clade of legumes. Plant Biotechnol. J. 12, 743–754. doi: 10.1111/pbi.12179
Schulte, W., Töpfer, R., Stracke, R., Schell, J., and Martini, N. (1997). Multi-functional acetyl-CoA carboxylase from Brassica napus is encoded by a multi-gene family: indication for plastidic localization of at least oneisoform. Proc. Natl. Acad. Sci. U. S. A. 94, 3465–3470. doi: 10.1073/pnas.94.7.3465
Shrestha, B., Gilbert, L. E., Ruhlman, T. A., and Jansen, R. K. (2020). Rampant nuclear transfer and substitutions of plastid genes in Passiflora. Genome Biol. Evol. 12, 1313–1329. doi: 10.1093/gbe/evaa123
Timmis, J. N., Ayliffe, M. A., Huang, C. Y., and Martin, W. (2004). Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat. Rev. Genet. 5, 123–135. doi: 10.1038/nrg1271
Ueda, M., Fujimoto, M., Arimura, S.-I., Murata, J., Tsutsumi, N., and Kadowaki, K.-I. (2007). Loss of the rpl32 gene from the chloroplast genome and subsequent acquisition of a preexisting transit peptide within the nuclear gene in Populus. Gene 402, 51–56. doi: 10.1016/j.gene.2007.07.019
Ueda, M., and Kadowaki, K.-I. (2012). Chapter two - gene content and gene transfer from mitochondria to the nucleus during evolution. Adv. Bot. Res. 63, 21–40. doi: 10.1016/B978-0-12-394279-1.00002-8
Ueda, M., Nishikawa, T., Fujimoto, M., Takanashi, H., Arimura, S.-I., Tsutsumi, N., et al. (2008). Substitution of the gene for chloroplast RPS16 was assisted by generation of a dual targeting signal. Mol. Biol. Evol. 25, 1566–1575. doi: 10.1093/molbev/msn102
Weng, M.-L., Ruhlman, T. A., and Jansen, R. K. (2016). Plastid–nuclear interaction and accelerated coevolution in plastid ribosomal genes in Geraniaceae. Genome Biol. Evol. 8, 1824–1838. doi: 10.1093/gbe/evw115
Woodson, J. D., and Chory, J. (2008). Coordination of gene expression between organellar and nuclear genomes. Nat. Rev. Genet. 9, 383–395. doi: 10.1038/nrg2348
Yoo, K.-O., and Jang, S.-K. (2010). Infrageneric relationships of Korean Viola based on eight chloroplast markers. J. Syst. Evol. 48, 474–481. doi: 10.1111/j.1759-6831.2010.00102.x
Keywords: intracellular gene transfer, Viola, infA, rpl32, rps16, Malpighiales
Citation: Yang J, Park S, Gil H-Y, Pak J-H and Kim S-C (2021) Characterization and Dynamics of Intracellular Gene Transfer in Plastid Genomes of Viola (Violaceae) and Order Malpighiales. Front. Plant Sci. 12:678580. doi: 10.3389/fpls.2021.678580
Edited by:
Goetz Hensel, Heinrich Heine University Düsseldorf, GermanyReviewed by:
Maria D. Logacheva, Skolkovo Institute of Science and Technology, RussialKi Oug Yoo, Kangwon National University, South Korea
Copyright © 2021 Yang, Park, Gil, Pak and Kim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Seung-Chul Kim, c29uY2h1czk2QHNra3UuZWR1; c29uY2h1czIwMDlAZ21haWwuY29t; Jae-Hong Pak, amhwYWtAa251LmFjLmty
†These authors have contributed equally to this work and share first authorship
 JiYoung Yang1†
JiYoung Yang1† Seung-Chul Kim
Seung-Chul Kim