
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 26 August 2021
Sec. Plant Biotechnology
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.677553
This article is part of the Research Topic Chloroplast Biotechnology for Crop Improvement View all 12 articles
Carotenoids are pigments with important nutritional value in the human diet. As antioxidant molecules, they act as scavengers of free radicals enhancing immunity and preventing cancer and cardiovascular diseases. Moreover, α-carotene and β-carotene, the main carotenoids of carrots (Daucus carota) are precursors of vitamin A, whose deficiency in the diet can trigger night blindness and macular degeneration. With the aim of increasing the carotenoid content in fruit flesh, three key genes of the carotenoid pathway, phytoene synthase (DcPSY2) and lycopene cyclase (DcLCYB1) from carrots, and carotene desaturase (XdCrtI) from the yeast Xanthophyllomyces dendrorhous, were optimized for expression in apple and cloned under the Solanum chilense (tomatillo) polygalacturonase (PG) fruit specific promoter. A biotechnological platform was generated and functionally tested by subcellular localization, and single, double and triple combinations were both stably transformed in tomatoes (Solanum lycopersicum var. Microtom) and transiently transformed in Fuji apple fruit flesh (Malus domestica). We demonstrated the functionality of the S. chilense PG promoter by directing the expression of the transgenes specifically to fruits. Transgenic tomato fruits expressing DcPSY2, DcLCYB1, and DcPSY2-XdCRTI, produced 1.34, 2.0, and 1.99-fold more total carotenoids than wild-type fruits, respectively. Furthermore, transgenic tomatoes expressing DcLCYB1, DcPSY2-XdCRTI, and DcPSY2-XdCRTI-DcLCYB1 exhibited an increment in β-carotene levels of 2.5, 3.0, and 2.57-fold in comparison with wild-type fruits, respectively. Additionally, Fuji apple flesh agroinfiltrated with DcPSY2 and DcLCYB1 constructs showed a significant increase of 2.75 and 3.11-fold in total carotenoids and 5.11 and 5.84-fold in β-carotene, respectively whereas the expression of DcPSY2-XdCRTI and DcPSY2-XdCRTI-DcLCYB1 generated lower, but significant changes in the carotenoid profile of infiltrated apple flesh. The results in apple demonstrate that DcPSY2 and DcLCYB1 are suitable biotechnological genes to increase the carotenoid content in fruits of species with reduced amounts of these pigments.
Carotenoids are natural lipophilic pigments produced by plant plastids that play important roles in light harvesting during photosynthesis and in protecting the photosynthetic apparatus against excessive light radiation. These secondary metabolites also normally accumulate in fruits, flowers and seeds, providing them yellow, orange, and red colors to facilitate pollination and seed dispersal (Rosas-Saavedra and Stange, 2016). Carotenoids are substrates for the biosynthesis of hormones that are crucial for plant physiology such as abscisic acid (ABA) and strigolactones, involved in the stress and developmental signaling responses (Walter and Strack, 2011; Beltran and Stange, 2016). Several plastidial enzymes have been identified that are essential for carotenoid synthesis. Phytoene synthase (PSY) catalyzes the first step and is the key regulatory point controlling flux through the pathway (Figure 1). The carotenoid pathway has been thoroughly described (Alcaíno et al., 2016). Briefly, the condensation of three molecules of isopentyl pyrophosphate (IPP) with one molecule of di-methylallyl pyrophosphate (DMAPP) generates the geranylgeranyl diphosphate (GGPP) precursor. The condensation of two molecules of GGPP, carried out by PSY, yields phytoene (C40). This is the first committed step in carotenoid synthesis, which is also highly regulated. In plants, colorless phytoene undergoes several steps of desaturation and isomerization by phytoene desaturase (PDS), ζ–carotene desaturase (ZDS), carotenoid isomerase (CRTISO) and ζ–carotene isomerase (ζ-ISO) (Isaacson et al., 2002, 2004; Lu and Li, 2008). To produce lycopene in bacteria and yeast, the four enzymatic steps from phytoene to lycopene mentioned above are carried out by a single enzyme (CRTI) (Alcaíno et al., 2016). Subsequently, lycopene is cyclized by lycopene β-cyclase (LCYB) producing β-carotene, whereas for α-carotene production, both LCYB and lycopene ε-cyclase (LCYE) are required. Then, the hydroxylation of α-carotene produces lutein, a yellowish pigment that is abundant in leaves (Hornero-Méndez and Britton, 2002; Bang et al., 2007; Misawa, 2011; Figure 1). Two subsequent hydroxylations of β-carotene, catalyzed by carotenoid β-hydroxylase (CHYB) produce zeaxanthin, which can be epoxidized by zeaxanthin epoxidase (ZEP) to form violaxanthin, which can be used by violaxanthin de-epoxidase (VDE) to regenerate zeaxanthin (Cazzonelli and Pogson, 2010; Cazzonelli, 2011; Moise et al., 2014). Finally, violaxanthin is the precursor for ABA, which is produced in the cytoplasm (Figure 1). In mammals, carotenoids possess antioxidant activity by quenching reactive oxygen species (ROS) (Stahl and Sies, 2003) thus decreasing the risk of developing certain diseases caused by oxidative stress (Malone, 1991; Byers and Perry, 1992; Voorrips et al., 2000; Holick et al., 2002) including cancer, atheromas in vascular diseases, aging and macular degeneration (Marnett, 1987; Mordi, 1993; Bartley and Scolnik, 1995; Rao and Rao, 2007). Carotenoids are also precursors of vitamin A. Specifically, the most relevant provitamin A carotenoids, given their high antioxidant activity and wide distribution in food, are α- and β-carotene, some xanthophylls such as β-cryptoxanthin and some apo-carotenoids (Mínguez-Mosquera and Hornero-Méndez, 1997). Of these, β-carotene presents the highest provitamin A activity since each molecule produces two retinal molecules that are then reduced to vitamin A (retinol). Vitamin A is converted into the visual pigment rhodopsin (retinal), in the retina of the eye, and acts as a co-regulator of gene expression (retinoic acid) (Zhu et al., 2013). Vitamin A is also required for cell growth and for healthy immunity, among other physiological processes (Olson, 1996). Therefore, increasing the carotenoid content in crop plants to improve nutritional value and benefits for human health has been a major goal of many research programs worldwide. Over the last four decades, significant progress has been made in the manipulation of carotenoid content and composition in a large number of crops by either genetic engineering or conventional breeding (Fraser et al., 1999, 2009; Giuliano et al., 2008; Rosati et al., 2009; Farré et al., 2010, 2011; Ye and Bhatia, 2012; Giuliano, 2014). Conventional and assisted breeding have successfully enhanced β-carotene levels in maize, sweet potato and cassava, benefiting human health in several countries (Ceballos et al., 2013; Pixley et al., 2013). Genetic engineering also allows nutritional traits to be targeted to specific organs (e.g., cereal seeds) and multiple traits can be combined in the same plants without complex breeding programs (Naqvi et al., 2009, 2010). Other examples of efficient enhancement of carotenoids through metabolic engineering have been obtained in agronomical relevant crops (Alós et al., 2016), such as rice (Paine et al., 2005), tomato (D’Ambrosio et al., 2004), potato (Diretto et al., 2006), carrot (Jayaraj et al., 2008), canola (Shewmaker et al., 1999; Ravanello et al., 2003), cassava (Failla et al., 2012), sorghum (Lipkie et al., 2013), orange (Pons et al., 2014), and apple (Arcos et al., 2020).
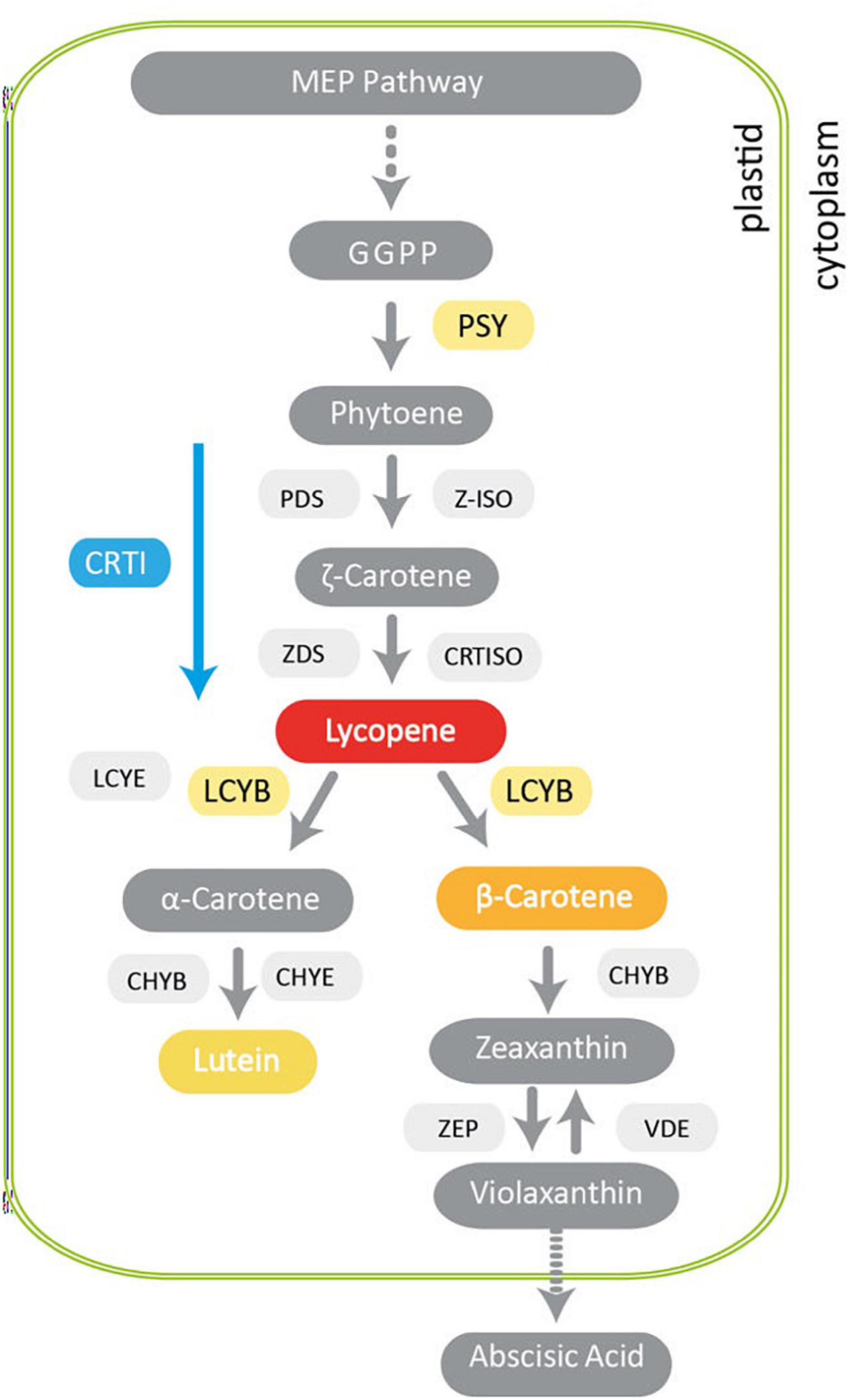
Figure 1. The main steps of carotenoid biosynthesis and related pathways in plants. The methylerythritol 4-phosphate (MEP) pathway produces isoprenoids in plastids leading to the production of geranylgeranyl pyrophosphate (GGPP). Phytoene synthase (PSY) is the first enzyme in the carotenogenic pathway and produces phytoene. In plants, phytoene is then desaturated and isomerized by phytoene desaturase (PDS), ζ–carotene desaturase (ZDS), ζ–carotene isomerase (Z-ISO) and prolycopene isomerase (CRTISO) forming lycopene. In bacteria, these reactions are carried out by a single multifunctional enzyme, phytoene desaturase (CRTI). β-carotene is produced if lycopene is cyclized only by lycopene β-cyclase (LCYB) whilst α-carotene is produced if both LCYB and lycopene ε-cyclase (LCYE) are involved. β-carotene hydroxylase (CHYB) and ε-carotene hydroxylase (CHYE) produce lutein. CHYB produces zeaxanthin, which can be converted into violaxanthin by zeaxanthin epoxidase (ZEP). The reverse step can be carried out by violaxanthin de-epoxidase (VDE). Violaxanthin is the precursor of abscisic acid. Dotted arrows indicate multiple enzymatic reactions. Solid gray arrows represent different steps of the carotenogenic pathway. The blue arrow indicates consecutive desaturations carried out by bacterial CRTI.
Apple (Malus domestica) is one of the most widely produced fruits in the world and is consumed not only as a fresh fruit but also in processed forms, such as juice and jam1 (Harker et al., 2003). As well as improving nutritional value, fruit color is one of the major aims of apple breeding because it provides novel appearances and influences consumer preferences (Yuan et al., 2014). The accumulation of anthocyanins and carotenoids in the skin and pulp are responsible for color in mature fruits (Telias et al., 2011; Cerda et al., 2020). However, commercial varieties such as “Royal Gala” and “Fuji” present low levels of carotenoids in the flesh reaching about 2-6 μg/g fresh weight (FW; Ampomah-Dwamena et al., 2012; Cerda et al., 2020). Specifically, the flesh of Fuji fruits accumulates around 1.8 μg/g FW in total carotenoids (Arcos et al., 2020), 0.04–0.22 μg/g dry weight (DW) of lutein and 0.79–1.14 μg/g DW of β-carotene (Delgado-Pelayo et al., 2014). Low carotenoid levels can be ascribed to reduced biosynthesis, deficient accumulation, and/or efficient degradation. Previous studies determined that the PSY enzyme in apple is codified by a family of genes with different levels of expression in fruit flesh (Ampomah-Dwamena et al., 2012; Cerda et al., 2020) and that the most expressed PSY genes are functional (Ampomah-Dwamena et al., 2015; Cerda et al., 2020) suggesting that the synthesis pathway is not the limiting step for carotenoid accumulation. In addition, the ectopic expression of the Arabidopsis 1-deoxy-D-xylulose 5-phosphate reductoisomerase (AtDXR) in Fuji apple, a gene participating in the synthesis of carotenoid precursors, produces a threefold rise in total and individual carotenoids in leaves (Arcos et al., 2020). This information led to the proposal that apple is a potential target to become a functional food with high pro-vitamin A and antioxidant components, when genes that codify for biosynthetic enzymes are expressed. Therefore, this work focused on the generation of a biotechnological platform for the rapid functional assessment of whether specific biosynthetic genes can be used to increase the content of provitamin A in fruits. This platform initially uses tomato fruits as a model, before final testing in apples.
Fruits and vegetables are the main sources of carotenoids, and among these, the orange carrot stands out. To generate the biotechnological platform, we chose PSY2 and LCYB1 from Daucus carota and CrtI from Xanthophyllomyces dendrorhous. Carrot accumulates high levels of α and β-carotene in its taproot, similar to X. dendrorhous, a yeast that is able to accumulate as much as 0.5% DW in astaxanthin, a ketocarotenoid produced from β-carotene (Niklitschek et al., 2008). Carrot presents two paralogs for PSY and LCYB genes (Just et al., 2007). DcPSY2 and DcLCYB1 are highly expressed in carrot leaves and tap roots during plant development (Fuentes et al., 2012; Simpson et al., 2016), suggesting functional roles for carotenoid synthesis. However, no functional characterization for DcPSY2 has been published yet. Nevertheless, in the case of DcLCYB1, its overexpression results in 2–10 fold more carotenoids in transgenic tobacco (Moreno et al., 2016, 2020), and a threefold increase in carrots (Moreno et al., 2013). Indeed, DcLCYB1 also presents a plastidial localization (Moreno et al., 2013). Regarding XdCrtI, it codifies the enzyme that catalyzes the conversion of phytoene into lycopene in X. dendrorhous (Niklitschek et al., 2008). Although several studies have used CrtI from Erwinia uredevora to modify the synthesis of carotenoids in plants, as well as the use of other bacterial carotenogenic genes orthologous to PSY (CrtB) and LCYB (CrtY) (Römer et al., 2000; Ye et al., 2000; Fraser et al., 2002; Paine et al., 2005; Diretto et al., 2007), XdCrtI has yet to be used to increase carotenoids for this purpose. Here, we describe the design and development of carotenogenic gene expression vectors (pCP-CG) that utilize single or combinations of DcPSY2 and DcLCYB1 carrot carotenogenic genes and XdCrtI from yeast to increase the carotenoid level in apples. To guide transgenic gene expression, the selection of a suitable promoter is essential. In the case of fruits, one of the enzymes that participates in fruit ripening is polygalacturonase (PG), a hydrolase responsible for the degradation of polyuronides or pectins in the cell wall (DellaPenna et al., 1986). PG accumulates during fruit ripening, due to transcriptional activation of the PG gene (Sheehy et al., 1987; DellaPenna et al., 1989). The tomato PG promoter (pPG) was characterized by Montgomery et al. (1993), who showed that the first 806 pb upstream of the transcription start site are sufficient to confer fruit flesh (pulp) specific expression. Therefore, in this study, the carotenoid biosynthetic genes were cloned under the pPG from Solanum chilense (tomatillo). Once the biotechnological platform was built, the functionality of the vectors was evaluated through subcellular localization, by stable transformation of Solanum lycopersicum cv. Microtom and by transient expression in M. domestica. The results demonstrate that the pCP-CG vectors, specially pPSY2 and pLCYB1, increment the total carotenoid and β-carotene levels in tomatoes and/or apple fruits, proving their suitability in genetic engineering programs for the improvement of nutritional quality and color traits.
pCP vector construction was based on a backbone from the pB7FWG2-AtDXR vector, which includes the p35S:DXR:GFP:T35S cassette (Perello et al., 2016, kindly provided by Dr. Manuel Rodriguez-Concepción, CRAG-CSIC, Spain). This cassette was obtained after a double digestion with XbaI and SacI. The backbone fragment of 7,854 bp contains the RB and LB of the T-DNA and the BAR gene for BASTA or glufosinate (herbicide) resistance in plants. A specifically designed Multicloning Site (MCS) of 141 bp including EcoRI, ApaI, PstI, AvrII, EcoRV, BspEI, NcoI, and BglII restriction sites flanked by XbaI and SacI was synthesized by Genescript (United States2). The MCS was cloned in the XbaI and SacI sites of the backbone to yield a new and royalty-free binary vector, named pCP that confers resistance to BASTA herbicide (Supplementary Figure 1, 201403013, Inapi, Chile).
DNA sequences of carrot PSY2 (DQ192187) and LCYB1 (DQ192190), and CrtI (gi.68250374) of X. dendrorhous, were optimized based on M. domestica codon usage by means of the OPTIMIZER software3. The M. domestica table obtained from4 was used as a reference set. Vector NTI software was used to verify the correct reading frame of the constructs as well as the amino acid homology with the native sequences encoded by PSY2, CrtI, and LCYB1. The FGENESH 2.6 gene prediction program was also employed, selecting the item ‘‘organism dicotyledonous plants5”. A plastidial signal peptide (tp) (171 bp) from the small subunit of pea Ribulose Bisphosphate Carboxylase Oxygenase (RUBISCO; EC 4.1.1.39 Seq ID X00806) was included at the 5′ NTR of XdCrtI to direct the encoded protein to these organelles (Figure 2). A 806 bp fragment of the tomato (S. chilense, tomatillo) PG promoter (pPG) was selected to direct the expression of each gene. A 277 bp fragment of the Agrobacterium tumefaciens nopaline synthase terminator (NosT) was included at the 3′ end of each coding sequence (Accession number V000876). The sequences of interest, including pPG:DcPSY2:NosT, pPG:DcLCYB1:NosT, and pPG:tp:XdCrtI:NosT were synthesized and cloned in the pUC57 vector by GeneScript (United States). Afterward, pUC57:pPG:DcPSY2:NosT was digested with SpeI, pUC57:pPG:DcLCYB1:NosT with AvrII and pPG:tp:XdCrtI:NosT with XhoI, and each cassette was cloned into the MCS of pCP producing the single (pPSY2 and pLCYB1), double (pPSY2-CRTI) and triple vectors (pPSY2-CRTI-LCYB), collectively called pCP-CG vectors (Figure 2). Each vector was verified through sequencing (Macrogen Co.) and transformed into Agrobacterium tumefaciens (GV3101 strain). Positive clones were selected and used for stable transformation of S. lycopersicum var. Microtom and for transient transformation of M. domestica var. Fuji.
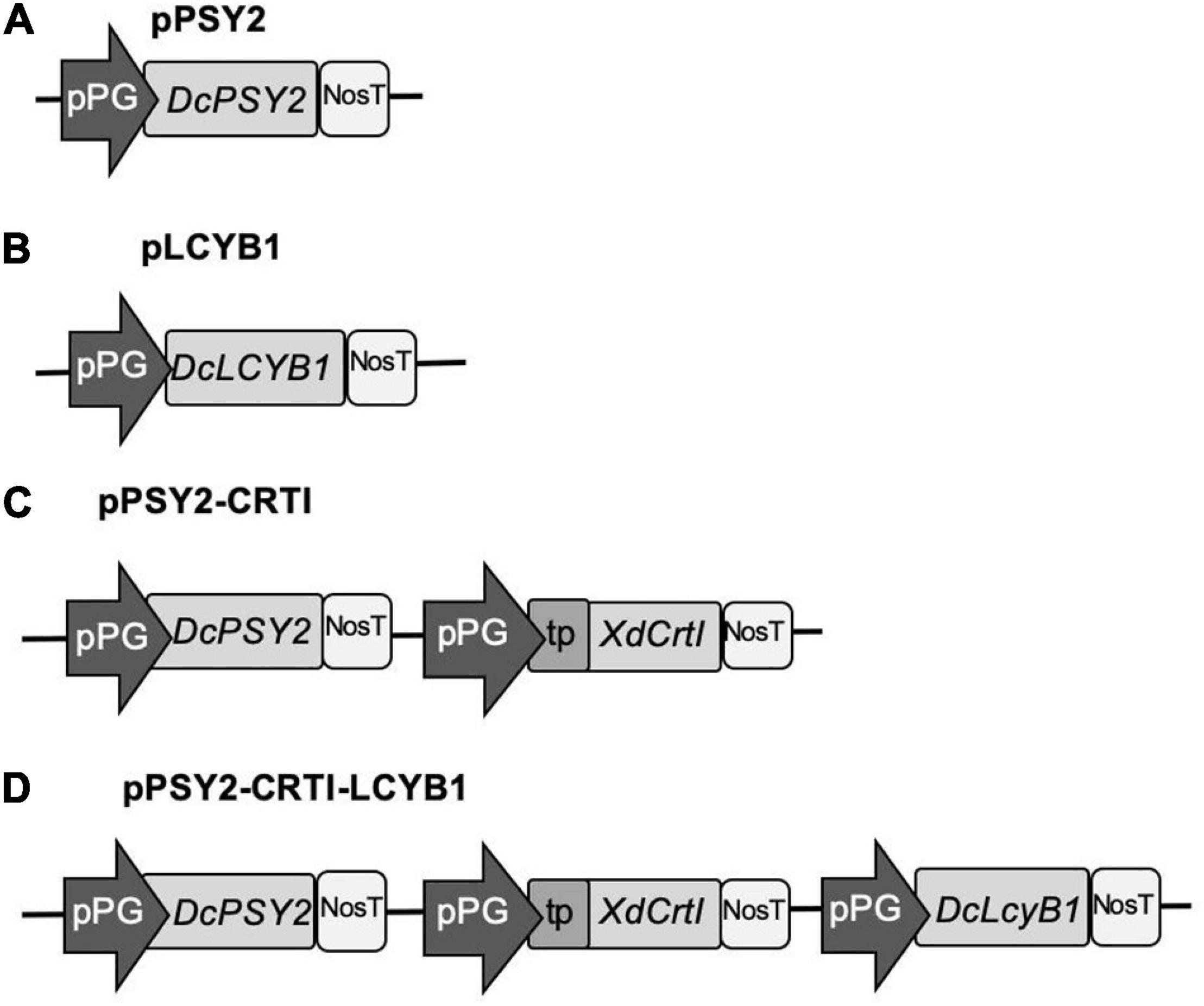
Figure 2. pCP-CG constructs used for the functional assessment of DcPSY2, DcLCYB1, and XdCRTI. The pCP backbone is derived from pB7FWG2.0, with the incorporation of a specifically designed Multiple Cloning Site (MCS; see Supplementary Figure 1). Different carotenogenic genes were cloned into the MCS, under the control of the tomato polygalacturonase promoter (pPG) and the Nos terminator (NosT). (A) pPSY2. The single gene and fruit-specific expression cassette PG:DcPSY2 was chemically synthesized and cloned into pCP. (B) pLCYB1. The single gene and fruit-specific expression cassette PG:DcLCYB2 was chemically synthesized and cloned into pCP. (C) pPSY2-CRTI. A double gene and fruit-specific expression cassette including the PG promoter driving the XdCRTI gene fused to a transit peptide (PG:tpXdCRTI) was chemically synthesized and cloned into pPSY2. (D) pPSY2-CRTI-LCYB1. The single gene and fruit-specific expression cassette PG:DcLCYB2 was cloned into pPSY2-CRTI.
For the subcellular localization analysis, the full-length and optimized for M. domestica DcPSY2 and XdCrtI sequences were amplified, including the PG promoter but excluding the stop codon from the previously sequenced pPSY2 and pPSY2-CRTI clones using primers listed in Supplementary Table 1. Amplicons were subcloned into pCR8/GW/TOPO (Invitrogen) and thereafter recombined into the binary vector pMDC107 (Curtis and Grossniklaus, 2003) in order to express the chimeric proteins DcPSY2:GFP and tp:XdCRTI:GFP, both under the control of pPG. As controls, the pPG:GFP (cloned in pCAMBIA 1302, Abcam) and the empty pMDC107 vector were included (Supplementary Figure 2). The constructs were transiently expressed by agroinfiltration in Nicotiana tabacum (tobacco) leaves and tomato fruits at breaker stage. A. tumefaciens suspensions containing the pPG:DcPSY2:GFP, pPG:tp:XdCRTI:GFP and both control constructs were infiltrated into 2 months old tobacco leaves and into the tomato fruit flesh following previously described protocols (Moreno et al., 2013; Arcos et al., 2020). Plants and fruits were maintained at 20–22°C, 30–50% humidity and while tobacco plants were kept under a 16 h white light photoperiod, tomato fruits were kept in the darkness for 4 days. Then, the infiltrated areas of the leaves and fruits were analyzed for GFP and chlorophyll fluorescence using a confocal LSM510 microscope. Images were processed with LSM5 Image Browser.
Solanum lycopersicum var. Microtom was cultivated in vitro in solidified Murashige and Skoog (MS: 4.4 g/L MS salts, 20 g/L sucrose, and 0.7% agar-agar) medium and transformed according to a previous report (Pino et al., 2010). Briefly, cotyledons of in vitro 8–10 days post-germination seedlings were cut and dipped into an A. tumefaciens culture containing the respective genetic construct for 15–20 min, and then transferred into a co-culture medium (4.4 g/L MS salts, 30 g/L sucrose, 6 g/L agar-agar, and 100 μM acetosyringone) for 2 days in darkness at 25°C (Supplementary Figure 3). The explants were then transferred to the Induction medium (4.4 g/L MS salts, 100 mg/L Myo-Inositol, 30 g/L sucrose, 6 g/L agar, 5 μM BAP, 0.3 mg/L BASTA, and 400 mg/L Timentin) for callus induction and initial shoot development which started between 4 and 6 weeks post-transformation. The new shoots were transferred to Elongation medium (4.4 g/L MS salts, 100 mg/L Myo-Inositol, 15 g/L sucrose, 6 g/L agar, 0.5 mg/L BASTA, and 400 mg/L Timentin). After 2 months, 4–6 cm high plants developed several branches and a minimum of 4–5 leaves. Then, the shoots were transferred to Rooting medium (4.4 g/L MS salts, 100 mg/L Myo-Inositol, 15 g/L sucrose, 6 g/L agar, 0.5 mg/L BASTA, 400 mg/L Timentin, and 5 mg/L IBA) for 2 weeks. Three months-old plants developed a root system including primary and secondary roots, following which they were acclimated to greenhouse conditions (photoperiod of 16 h light/8 h darkness and illuminated with white fluorescent light; 150 μmol/m2 s at 22–25°C) in plastic pots (20 × 10 cm) containing a mix of soil and vermiculite (2:1), until they finally reached maturity and started producing fruits approximately 1 month later. Mature T1 plants were used for further analysis (Supplementary Figure 3).
Tomato RNA extraction was performed as described in Meisel et al. (2005). Briefly, mesocarps from five transgenic tomatoes (500 mg) were ground in liquid nitrogen and divided to perform three independent RNA extractions. The powder was then mixed with extraction buffer (2% CTAB, 2% PVP40, 25 mM EDTA, 2 M NaCl, 100 mM Tris–HCl, 0.05% spermidine, and 2% B-mercaptoethanol) at 65°C for 15 min. The samples were centrifuged and the phase containing the RNA was precipitated with 10 M LiCl overnight at 4°C. After centrifugation at 12,000g during 20 min, samples were resuspended in 20 μL of DEPC water. Genomic DNA traces were eliminated by a 40 min DNase I (Fermentas) treatment. For cDNA synthesis, 2 μg of DNA-free RNA were incubated with 1.5 μL of 10 μM oligo AP (Supplementary Table 1) and 8.3 μL of RT mixture (0.5 μL of RNase Inhibitor Ribolock, Fermentas), 1 μL of 10 mM dNTPs (Fermentas), 2.8 μL of 25 mM MgCl2, and 4 μL of 5× RT Improm II buffer (Promega) for 5 min at 70°C. Then, the samples were incubated for 5 min on ice. Subsequently, 1.2 μL of RT Improm II was added to continue with cDNA synthesis according to RT Improm II manufacturer’s instructions. For conventional expression analysis (Supplementary Figure 4), PCR reactions (28–35 cycles) were performed using Taq DNA polymerase (New England BioLabs) according to the manufacturer’s recommendations and an annealing temperature adjusted to each set of primers (Supplementary Table 1) and using SlPSY to show the cDNA quality. Quantitative expression analysis (qRT-PCR) was performed in a LightCycler system (MX3000P, Stratagene), using SYBR Green double strand DNA binding dye, as described in Fuentes et al., 2012). Specific primers for optimized DcPSY2, DcLCYB1, and XdCRTI were used (Supplementary Table 1), based on the optimized sequences (Supplementary Material). Actin was selected as reference gene. Each qRT-PCR reaction was performed with three biological and two technical replicates. In all cases, the reaction specificities were tested with melting gradient dissociation curves and non-template controls (NTC).
Transient transformation of apples fruits was performed according to Arcos et al. (2020) with modifications. Fuji apple fruits at the mature green stage were infiltrated using the Agrobacterium-injection strategy containing the genetic construct of interest (pPSY2, pLCBY1, pPSY2-CRTI, or pPSY2-CRTI-LCYB1) and controls (WT). Each construct was infiltrated into a quarter of three fruits to have three biological replicas. Briefly, 30 mL of bacterial cultures in LB medium supplemented with Rifampicin (50 mg/L) and Spectinomycin (50 mg/L) were grown overnight at 28°C with agitation until an optical density (600 nm) of 0.2–0.3. The cultures were centrifuged, and the pellet resuspended in 30 mL of infiltration medium (10 mM MgSO4, 10 mM MES, 10 mM MgCl2). The culture was then infiltrated into the fruit flesh with a tuberculin syringe to a depth of 0.5 cm. For each apple, one half of the fruit was homogeneously agroinfiltrated. Agroinfiltrated fruits were maintained in an incubator with a photoperiod of 18 h light/6 h darkness, at 20–22°C and 30–50% of humidity. After 7 and 14 days post-infiltration, samples were taken for carotenoid quantification.
Mesocarps from five transgenic tomatoes (500 mg) of each line of approximately 40–45 days post-flowering were ground in liquid nitrogen and divided to carry out three independent carotenoid extractions. For transient analysis, three independent apple fruits were agroinfiltrated with each construct including both controls, and after 7 and 14 days, 300 mg of infiltrated flesh of each apple were ground in liquid nitrogen independently. In all cases, each sample was measured twice, by adding 4 mL of hexane:acetone:ethanol (2:1:1) to obtain an homogeneous mixture. This solution was transferred to a tube, shaken for 2 min, incubated on ice and darkness for a further 2 min, and then centrifuged at 10.000 rpm for 10 min at 4°C. Finally, the carotenoids were recovered from the upper phase and dried using gaseous nitrogen. Once dried, the samples were stored at −80°C or resuspended in 2 mL of acetone for carotenoid quantification. All extractions were carried out in on ice and in dim light conditions. Total carotenoids were quantified was carried out using a Shimadzu spectrophotometer at 474, 645/662 (chlorophyll a and b contribution) and at 520/750 nm (for turbidity). Quantification of individual carotenoids was undertaken in a Reverse Phase High Performance Liquid Chromatography (RP-HPLC) using a RP−18 Lichrocart 125−4 reverse phase column (Merck®). Pigment composition was determined according to Lichtenthaler and Buschmann (2001). The mobile phase was acetonitrile:methanol:isopropyl alcohol (85:10:5) with a 1.5 mL/min flow rate at room temperature in isocratic conditions. Carotenoid and chlorophyll concentrations were calculated according to the total pigments obtained at 474 nm by spectrophotometry, by retention times, absorption spectra and purity of the peaks, as described in Fuentes et al. (2012) and corroborated with the Carotenoids Handbook (Britton, 1995; Britton et al., 2004).
To generate the biotechnological platform to boost carotenoid content in apples, we chose PSY2 and LCYB1 from D. carota and CrtI from X. dendrorhous. The codon usage of each gene was optimized to maximize expression in apples (Supplementary Material).
The XdCrtI was destined to plastids using the transit peptide (tp) of the RUBISCO small subunit (Supplementary Material). This transit peptide has been used to direct Erwinia uredovora CrtI to tobacco leaf plastids, as determined by immunolocalization with gold particles (Misawa et al., 1993). We also amplified a 806 bp fragment upstream of the transcription start site of the PG promoter from S. chilense (tomatillo) (Supplementary Material) in order to direct expression of the carotenogenic genes to fruits. Using the different modules – the three carotenogenic genes, the tp and the PG promoter – four pCP-CG vectors were generated with different gene combinations, as described in section “Materials and Methods” (Figure 2).
Subsequently, we determined the correct plastid location of DcPSY2 and tp:XdCrtI; that of DcLCYB1:GFP has been reported previously (Moreno et al., 2013). Therefore, pPG:PSY2:GFP and pPG:tp:XdCrtI:GFP were transiently expressed in tomato fruits at breaker stage where both chloroplasts and chromoplasts are present in the flesh of the fruit. GFP fluorescence showed that DcPSY2:GFP had a speckled distribution (Figure 3a). This signal colocalized with the emission of chlorophyll (Figures 3b,c), strongly suggesting that DcPSY2:GFP has a chloroplastic localization. Similarly, GFP signal from tp:XdCrtI:GFP exhibited a distribution pattern of discrete foci (Figure 3e), which colocalized with the autofluorescence from chlorophyll (Figures 3f,g). Therefore, this result showed that the plastidial tp from the pea RUBISCO small subunit was functional in tomato. Considering that the GFP fluorescence distribution following infiltration with the control pPG:GFP construct was cytoplasmic in fruits, and much reduced in tobacco leaves (Supplementary Figure 2), we demonstrated the fruit specificity of the PG promoter from S. chilense. Overall, these results show that DcPSY2:GFP and tp:XdCrtI:GFP are chloroplastic.
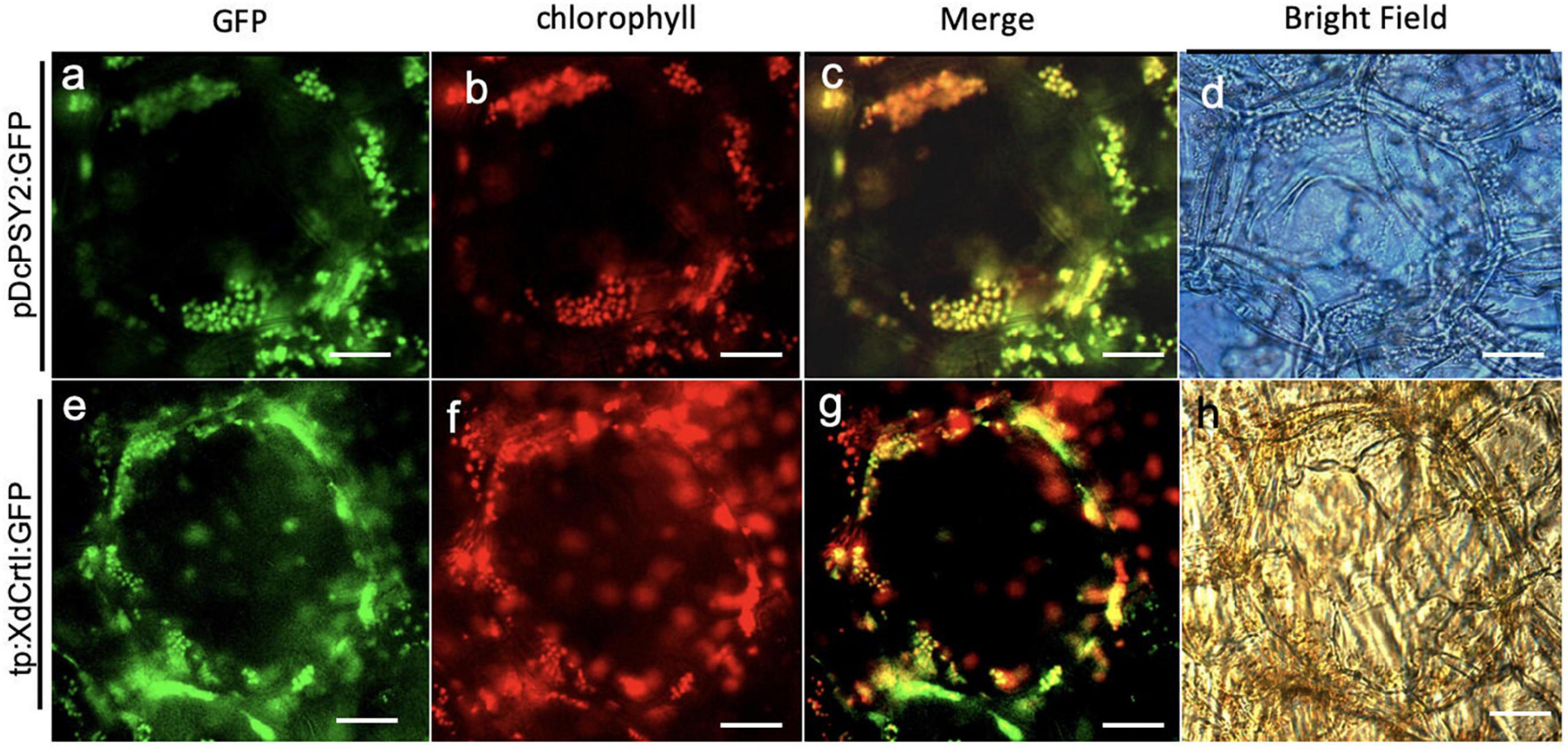
Figure 3. Subcellular localization of DcPSY2 and XdCRTI. Transient transformation of tomato fruits agroinfiltrated with DcPSY2:GFP or XdCrtI:GFP fused to a transit peptide (tp). The expression of both constructs was driven by the pPG promoter. Fluorescence was observed 4 days after transient transformation. (a) GFP fluorescence of DcPSY2:GFP. (b) Chlorophyll autofluorescence of (a). (c) Merge of panels (a,b), showing colocalization of chlorophyll and the GFP signal. (d) Bright field of (a–c). (e) GFP fluorescence of tp:XdCRTI:GFP. (f) Chlorophyll autofluorescence of (e). (g) Merge of panels (e,f), showing colocalization of chlorophyll and GFP signal. (h) Bright field of (e–g). The TRITC filter U-N31001 was used for visualizing GFP fluorescence, with an excitation wavelength between 465 and 495 nm. Emission was collected between 513 and 556 nm. Chlorophyll autofluorescence was assessed by using a U-MNG2 filter, with an excitation wavelength between 530 and 550 nm and emission collected at 590 nm. Scale bar = 50 μm.
The functional characterization of pPSY2, pLCYB1, pPSY2-CRTI, and pPSY2-CRTI-LCYB1 vectors was carried out by stable transformation of S. lycopersicum var. Microtom. The in vitro transformation and regeneration culture to obtain transgenic plants was adapted from the protocol described in Pino et al. (2010) and summarized in Suppplementary Figure 3. During plant development, no phenotypic differences were noticeable with respect to WT tomato plants, except in the case of those transformed with pPSY2-CRTI-LCYB1 that showed dramatic alterations at all stages during the regeneration process (Suppplementary Figure 3). These lines exhibited a slower induction of shoots and their vegetative development was also delayed in comparison with the other transgenic lines. Interestingly, these lines produced tomatoes when they were still in vitro, about 2 months after the regeneration process started. This was significantly earlier in comparison with the WT and other transformed lines, which only produced fruits once they were in soil in greenhouse conditions.
Transformed tomato plants were subjected to molecular analysis to select transgenic lines once they were acclimated to the greenhouse conditions, except for pPSY2-CRTI-LCYB1. Twelve pPSY2 lines, seven pLCYB1 lines, and eight pPSY2-CRTI lines were transgenic, based on the respective transgene amplification (data not shown). At fructification, tomato fruits from untransformed WT plants, as well as from lines transformed with pPSY2, pLCYB1, and pPSY2-CRTI were observed at different developmental and ripening stages ranging from incipient green fruits to red, fully mature fruits; no significant differences between them in terms of size and ripening time were noted, and all were similar to previous reports (Fraser et al., 2002; Apel and Bock, 2009). However, in the case of the pPSY2-CRTI-LCYB1 construct, it was not possible to regenerate fully developed plants; instead stem-like structures were obtained with a reduced number of incipiently developed leaves. Nonetheless, these plants showed an accelerated flower development with fully functional flowers that yielded fruits during in vitro culture (Suppplementary Figure 3). Since the tissue was limited, molecular analysis of these fruits was challenging. Thus, transgene expression was confirmed when some of these fruits reached maturity. We determined that fruits of seven pPSY2 lines, five pLCYB1 lines and seven pPSY2-CRTI lines expressed the transgene (Supplementary Figure 4), and three of each were selected for qRTPCR and phenotypic analysis (Figure 4) and carotenoid quantification (Figure 5). As explained above, it was not possible to regenerate fully developed lines transformed with pPSY2-CRTI-LCYB1. Therefore, the expression analysis was performed with a group of tomatoes obtained from in vitro culture and not from individual transgenic lines (Figure 4 and Supplementary Figure 4).
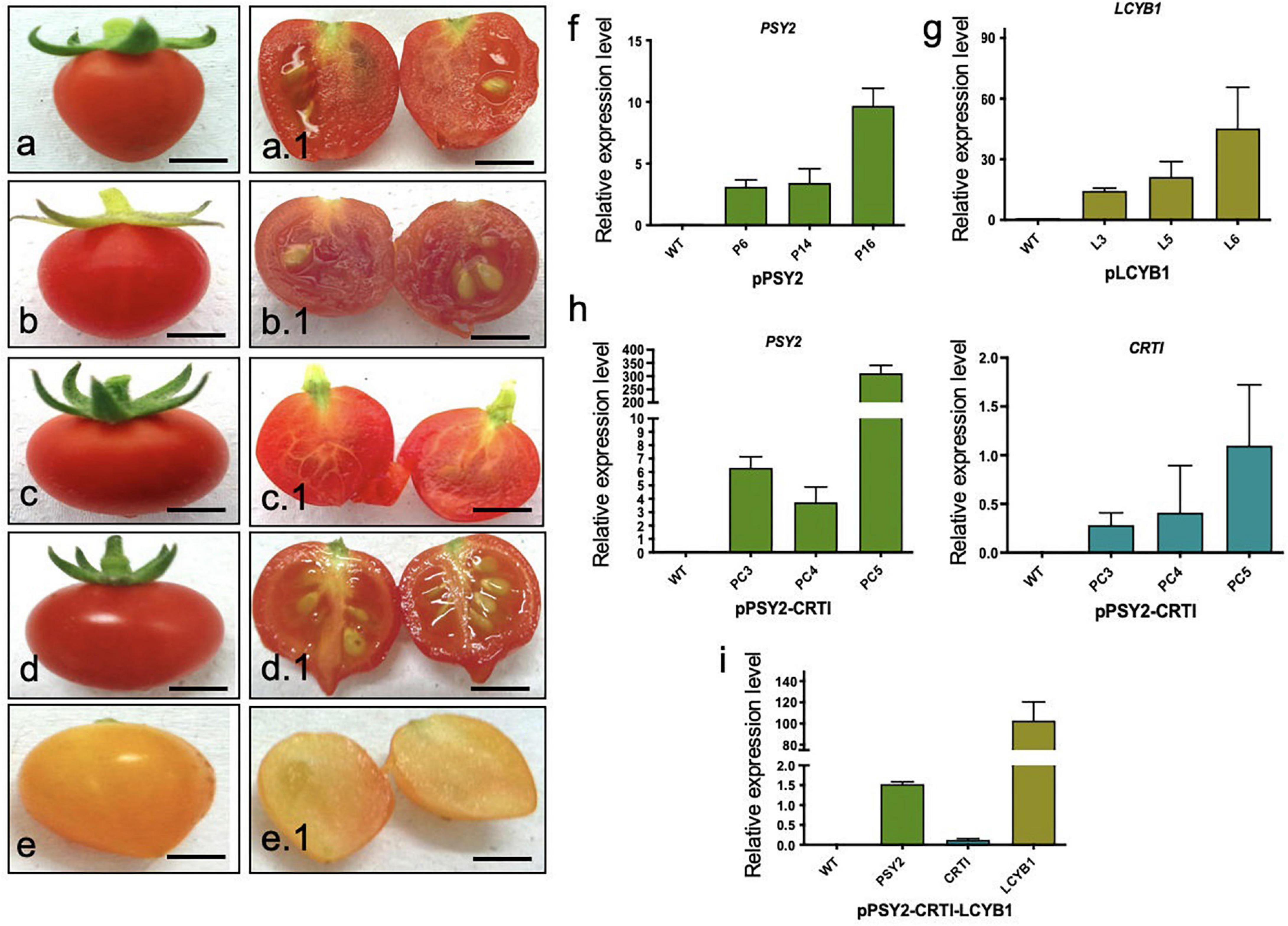
Figure 4. Fruit phenotypes and expression analysis in T1 transgenic fruits. (a) Representative WT tomato fruit. (a.1) Cross-section of (a). (b) Representative tomato fruit from pPSY2 line. (b.1) Cross-section of (b). (c) Representative tomato fruit from pLCYB1 line. (c.1) Cross-section of (c). (c) Representative tomato fruit from pPSY2-CRTI line. (d.1) Cross-section of (d). (e) Tomato fruit from pPSY2-CRTI-LCYB2. (e.1) cross-section of (e). Scale bar = 1 cm. (f) q-RTPCR in fruits of pPSY2, (g) pLCYB1, (h) pPSY2-CRTI, and (i) pPSY2-CRTI-LCYB1 transgenic lines. The relative expression was normalized to that of the Actin gene of S. lycopersicum. The expression analysis was performed with three biological replicas (cDNAs) from five fruits, each measured twice.
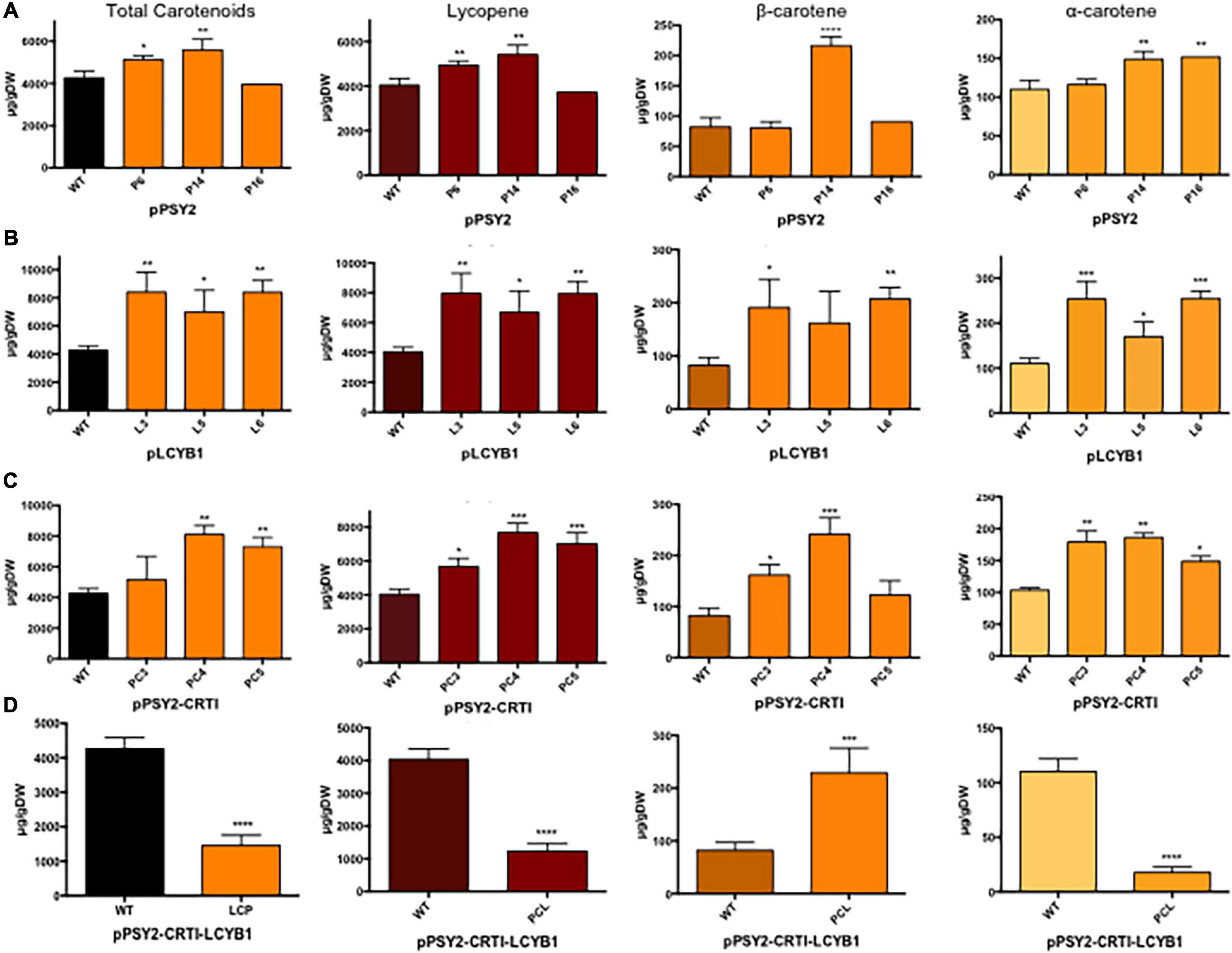
Figure 5. Total and individual carotenoid quantification in pCP-CG transgenic lines. (A) pPSY2, (B) pLCYB1, (C) pPSY2-CRTI, and (D) pPSY2-CRTI-LCYB1 transgenic lines. Total carotenoids were quantified in fruits by spectrophotometry at 474 nm and individual carotenoids by HPLC expressed in μg/gDW. Asterisks indicate significant differences respect to the control (WT) determined by one-way ANOVA and Dunnet post-test, *p < 0.05, **p < 0.01, ***p < 0.001. In (D) the results were analyzed with a two-way t-test, ****p < 0.0001.
For these evaluations, five mature fruits of each line were harvested and phenotypically analyzed. Fully ripe WT tomato fruits (2.2 ± 0.3 cm diameter) have a homogeneous red exocarp, are firm to the touch and have a central pedicel surrounded by several sepals (Figure 4a). Transverse sections showed a red fleshy mesocarp and endocarp with several yellowish seeds embedded in a locular cavity (Figure 4a.1). Transgenic tomato fruits from pPSY2 (Figures 4b,b.1), pLCYB1 (Figures 4c,c.1), and pPSY2-CRTI (Figures 4d,d.1) were phenotypically similar to fruits from WT plants (2.4 ± 0.3 cm diameter). On the other hand, pPSY2-CRTI-LCYB1 transgenic fruits exhibited a pale orange color at the ripening stage (Figures 4e,e.1), unlike the fruits of the WT and other transgenic lines, which turned red. In addition to the color difference, these transgenic fruits did not develop seeds and were therefore sterile lines (Figure 4e.1). Quantitative expression analysis showed that pPSY2 lines (P6, P14, and P16) present between 2.8 and 9.8-fold of PSY2 expression, pLCYB1 lines (L3, L5, and L6) present between 12 and 45.2-fold LCYB1 expression (Figures 4f,e, respectively), whereas pPSY-CRTI lines (PC3, PC4, and PC5) present between 3.6 and 300-fold expression of PSY2 and 0.2 and 1.2-fold CRTI expression (Figure 4h). In the case of the transgenic pPSY2-CRTI-LCYB1 fruits, CRTI expression was significantly lower than that of PSY2 and LCYB1 (Figure 4i).
Regarding the total content of carotenoids, and compared to WT plants, the pPSY2 lines present a 1.21–1.34-fold increase, pLCYB1 lines present a 1.7-2-fold increase, and pPSY2-CRTI lines showed a 1.21–1.99-fold increase whilst pPSY2-CRTI-LCYB1 lines had a 2.9-fold decrease (Figure 5). This suggests that the expression of LCYB1 is sufficient to enhance the total carotenoid content in fruits and that the simultaneous expression of these three transgenes negatively modulates the carotenoid pathway, reducing the total carotenoid content in tomato fruits (Figure 5).
Analysis of the individual carotenoid profiles revealed that transgenic tomatoes from the pPSY2 lines had up to 1.25-fold more lycopene, and only one line presented a 2.5-fold increase in β-carotene and two had a 1.27-fold increment in α-carotene with respect to WT (Figure 5A). Transgenic fruits from the pLCYB1 lines had a 1.75–2.0-fold increase in lycopene, 2.12–2.5-fold increase in β-carotene, and 1.36-2.27 fold increment in α-carotene with respect to the WT (Figure 5B). Fruits of pPSY2:CRTI lines presented a 1.87-1.95-fold increase in lycopene, two to threefold increase in β-carotene and 1.4-1.75-fold increment in α-carotene with respect to WT (Figure 5C), whereas pPSY2-CRTI-LCYB1 transgenic tomatoes present 3.6-fold less lycopene, a 2.57-fold increment in β-carotene, and 7.3-fold less α-carotene compared to untransformed controls (Figure 5D). The orange coloration phenotype of these tomatoes could be due to the combination of a reduced amount of red lycopene together with an increment in orange β-carotene (Figure 4e). These results suggest that the expression of carrot carotenogenic genes is capable of modulating carotenoid synthesis in a fruit that naturally accumulates large amounts of these pigments. Moreover, the increase in carotenoid content does not directly correlate with the expression level, as exemplified by comparing pPSY2 and pPSY2-CRTI lines. Interestingly, pLCYB1 was the genetic construct that triggered the highest increase in carotenoid content, constituting the most promising tool for future experiments.
In order to test if the pCP-CG constructs were able to modify the content of carotenoids in apple, transient assays were carried out in Fuji fruits at breaker stage (Suppplementary Figure 5). At 7 and 14 days post-agroinfiltration, tissue samples were taken for carotenoid analysis. After 7 days of incubation, apples agroinfiltrated with pPSY2 and pLCYB1 showed an increase of 1.65 to 1.50-fold in total carotenoid content (4.3 and 3.94 μg/gFW, respectively) in comparison to non-agroinfiltrated (WT) apples (2.61 μg/gFW) (Table 1). Carotenoid profile analysis revealed that for these constructs, the lutein levels remain stable with respect to the control (Table 1). On the contrary, the β-carotene content almost doubles (to 3.51 μg/gFW in pPSY2 and to 3.30 μg/gFW in pLCYB1), respect to the WT apple fruits (1.81 μg/gFW) (Table 1). Fruits agroinfiltrated with pPSY2-CRTI showed a significant increment in total carotenoids and β-carotene of 1.17- and 1.3-fold (3.05 and 2.35 μg/gFW, respectively) (Table 1). Similarly, apple fruits infiltrated with pPSY2-CRTI-LCYB1 exhibited an increment in their total carotenoid (3.07 μg/gFW) and β-carotene (2.23 μg/gFW) content in comparison to control WT fruits, but to a similar or lesser extent in comparison with fruits agroinfiltrated with other pCP-CG constructions (Table 1).
Samples taken 14-days post-infiltration revealed that the total carotenoid content doubled in fruits agroinfiltrated with each one of the four pCP-CG constructs in comparison to WT (Table 1). Similarly, a significant increase in β-carotene levels was found in the infiltrated fruits with the pCP-CG series (4.78 μg/gFW for pPSY2, 5.47 μg/gFW for pLCYB1, 3.40 μg/gFW for pPSY2-CRTI, and 3.43 μg/gFW for pPSY2-CRTI-LCYB1) in comparison to control WT fruits (0.94 μg/gFW; Table 1). On the other hand, the apple fruits transiently transformed with the pCP-CG constructs did not present changes in the lutein content (Table 1), maintaining the same correlation observed in the 7-day incubation experiment. Altogether, these results suggest that transient expression of the pCP-CG constructs are functional in apple fruits, and suggest that double or triple constructs with genes arranged in tandem do not trigger a synergistic effect on the carotenoid content.
Significant progress has been made in the generation of genetically modified crops with improved carotenoid content and nutritional properties (Alós et al., 2016). With the same goal, we designed and generated a biotechnological platform to evaluate key genes, with the aim of improving the carotenoid content, and thus the nutritional value of apples. The selection of the carotenogenic genes was based on our previous studies regarding carotenoid biosynthesis regulation in carrot, particularly in the taproot, which accumulates up to 1.5 mg/gDW in total carotenoids (Fuentes et al., 2012; Moreno et al., 2013, 2016; Rodriguez-Concepcion and Stange, 2013; Simpson et al., 2016, 2018). We have established that the increment in carotenoids correlates with a significant increase in the expression of almost all carotenogenic genes, especially DcPSY2, DcZDS2, DcPDS, DcLCYB1, DcLCYE, DcZEP, and DcNCED1 (Simpson et al., 2016). The two PSY paralogs in carrot, DcPSY1 and DcPSY2, present an induction of 3.2 and 316-fold during carrot tap root development, respectively, whereas LCYB1 rises by eightfold and LCYB2 by 100-fold at the mature stage of taproot development (Simpson et al., 2016).
Some of the carotenogenic genes from carrot have been previously characterized, demonstrating that they are functional in vivo. Both DcZDS1 and DcLCYB1 code for functional enzymes necessary for carrot development and carotenoid synthesis (Moreno et al., 2013; Flores-Ortiz et al., 2020). Total carotenoids rise threefold when DcLCYB1 is overexpressed in carrot and a 2–10-fold increment when expressed in tobacco, with concomitant increases in chlorophyll content and photosynthetic activity which together have a positive impact on plant fitness (Moreno et al., 2013, 2016, 2020). The ability of LCYB1 to confer these qualities by itself when expressed in plants was the reason for selecting this gene as a candidate to be assessed in the biotechnological platform developed in this work. On the other hand, DcPSY2 was selected as previous data suggested that it could play a relevant role during the stress response; we determined that its expression is induced by saline stress and ABA in carrot, and showed that AREB transcription factors bind to the DcPSY2 promoter (Simpson et al., 2018). Here we demonstrated that DcPSY2:GFP has the expected plastidial subcellular localization (Figure 3), and is therefore a possible candidate for boosting carotenoid content in both transgenic pPSY2 tomatoes (Figure 5A) and transiently infiltrated apple fruit flesh (Table 1). In the case of XdCrtI, its functionality was demonstrated by site directed mutagenesis using the double recombinant method in X. dendrorhous, whereby the deletion of XdCrtI in the homozygous strain causes a completely albino phenotype (Niklitschek et al., 2008). Here we determined that tp:XdCrtI:GFP with the pea small subunit RUBISCO transit peptide was correctly destined to plastids (Figure 3) and could be a suitable contender for enhancing total carotenoids and β-carotene when comparing pPSY2 and pPSY2-CRTI transgenic tomatoes (Figure 5C). The synthesis of carotenoids requires GGPP, a common precursor also needed for vitamin E (α-tocopherol), gibberellic acid and chlorophyll synthesis. Therefore, re-directing this precursor toward the synthesis of carotenoids by using constitutive promoters can have a negative impact on the metabolism of these other compounds. Consequently, a detrimental effect can be triggered both phenotypically and/or physiologically, with the generation of alterations such as dwarfism (Fray et al., 1995; Sandmann, 2001). A strategy to minimize this risk is to design genetic constructs with genes driven by tissue- or developmental-specific promoters. In this work, a 806 bp fragment of the fruit specific promoter of the tomato PG gene from S. chilense was chosen. This promoter has 99% identity to the 4,052–4,818 bp region of the S. lycopersicum PG promoter FJ465170.1 (Lau et al., 2009; Supplementary Figure 6), and targets specifically the transgenes to fruits (Figure 3 and Supplementary Figure 2). Although the expression of PG increases during apple ripening (Janssen et al., 2008), it has been established that the PG promoter does not depend on ethylene, and thus induces ripening in both climacteric fruits such as tomatoes (Lincoln et al., 1987), and non-climacteric fruit such as apples7. Importantly, the PG promoter from S. chilense is not subjected to intellectual property protection and is therefore available to be used without royalties. As a selection marker, we used bar that confers resistance to the BASTA (or phosphinothricin) herbicide. This gene is free of rights, and there are several currently commercialized transgenic plants8,9 harboring bar. The carotenogenic genes used here are also free of rights, even more so after optimizing their nucleotide sequence to make them more different from those native sequences present in carrot and yeast databases. For optimization, the sequence of interest was modified according to the preferential use of codons of the final organism, using the average of the codon usage of ribosomal genes or of a group of highly expressed genes (Puigbò et al., 2007). Here, the codon usage table of 567 mRNAs of M. domestica was available, and considered a sufficient population (163,086 codons) to successfully guide the codon usage preference, or at least to discriminate unusual codons (see text footnote 4). In the optimization of DcPSY2, XdCrtI, and DcLCYB1, most codons were conserved, since those used preferentially in carrots and yeast were the same as those in apples. However, there were codons that code for certain amino acids such as Tyr, Thr, Ser, Gln, Asn, Ile, Phe, and Glu where codon optimization was required.
Tomato represents a suitable model to evaluate our biotechnological platform designed for the functional assessment of candidate carotenogenic genes. The results obtained through this initial strategy allowed us, in a short period of time, to project possible results that were ultimately used to improve the carotenoid content in apples.
Mature tomato fruits accumulate significant amounts of the reddish lycopene, and very low levels of β-carotene and xanthophylls (Fraser and Bramley, 2004; Liu et al., 2015; D’Andrea and Rodriguez-Concepcion, 2019; Diretto et al., 2020). During ripening, carotenoid levels rise up to 400 times, of which lycopene represents 90% of the total, accumulating approximately 2,000 μg/g DW of this pigment (Fraser et al., 1994, 2002). This large accumulation of carotenoids is associated with several factors such as sink capacity and the activity of some plastidial proteins (Sun et al., 2018; D’Andrea and Rodriguez-Concepcion, 2019). Equally important is the regulatory role of the expression level of carotenogenic genes during ripening such as PSY1 and PDS that present an increase whilst LCYB and LCYE decrease their expression during ripening (Giuliano et al., 1993; Pecker et al., 1996; Ronen et al., 1999, 2000).
As mentioned above, transgenic tomatoes expressing apple-optimized DcPSY2, DcLCYB1, and XdCrtI showed a dissimilar increase in the levels of total carotenoids, lycopene, β-carotene and lutein (Figure 5) which correlated only partially with gene expression (Figures 4f–i). Specifically, pPSY2 lines present up to 1.34-fold rises in total carotenoids and 1.25-fold increases in lycopene in P6 and P14 lines, whereas only P14 has a 2.5-fold increase in β-carotene, and P14 and P16 have a 1.27-fold increment in α-carotene (Figure 5A); P6 and P14 have the lowest PSY2 expression level (Figure 4f). Nevertheless, this allows us to conclude that DcPSY2 codes for a functional enzyme capable of increasing carotenoids in tomato fruits but at a moderate level and that a reduced expression level is sufficient to increment β-carotene (P14). Fraser et al., 2002 obtained an increase of up to 2.1 times in the total carotenoid content and lycopene in fruits of the Alisa Craig tomato variety transformed with the CrtB gene. This gene codifies a PSY from E. uredovora, which was expressed in a fruit-specific manner. These results suggest that DcPSY2 might not be as efficient as CrtB, but this should be confirmed by comparing the same variety of tomato.
Regarding pLCYB1 transgenic lines, they have almost two times more total carotenoids and lycopene and up to 2.5 times more β-carotene and 2.27 times more α-carotene in all transgenic lines (Figure 5B). Although L3 presents the lowest expression level, it is one of the two transgenic lines with the highest carotenoid content together with L6, the line with the highest expression level (Figures 4g, 5B). Previous reports have shown that the fruit-specific expression of AtLCYB in tomato var. Moneymaker (Rosati et al., 2000) and the constitutive expression of the endogenous SlLCYB in tomato var. Red Setter (D’Ambrosio et al., 2004) resulted in an increase of total carotenoid content of 1.6 times and 2 times, respectively, which is similar to the increase we obtained in pLCYB1 tomatoes var. Microtom (Figure 5). Regarding individual carotenoids, AtLCYB transgenic tomato fruits yielded on average 3.1 times more β-carotene than the control whereas the lycopene content remained constant (Rosati et al., 2000). Similarly, SlLCYB transgenic lines presented an increase of over ten times in β-carotene (D’Ambrosio et al., 2004); unlike Rosati et al., 2000, this was accompanied by a 53-fold drop in the lycopene levels (D’Ambrosio et al., 2004). Interestingly, DcLCYB1 did not produce a detriment in lycopene content and actually produced an increase in α-carotene levels. The expression of DcLCYB1 alone produces a twofold increment in total carotenoids (Figure 5B), similar to transplastomic tomatoes that expressed LCYB from Narcissus pseudonarcissus that produced a 50% increment in total carotenoids (Apel and Bock, 2009). Expressing AtLCYB or EuCtrY enabled the enhancement of β-carotene without substantial reductions in lycopene (Rosati et al., 2000; Wurbs et al., 2007). This shows that DcLCYB1 is at least as equally efficient as LCYBs from other species. Indeed, tomatoes from pLCYB1 lines had a higher carotenoid content than tomatoes from the pPSY2 and pPSY2-CRTI lines (Figure 5), suggesting that in tomato the endogenous SlPSY, desaturases and isomerases are highly active. Our data revealed that the apple-optimized XdCrtI is functional in tomato. pPSY2-CRTI transgenic tomatoes exhibited a larger increase in total carotenoids and lycopene (twofold) in comparison to pPSY2 and also produced up to threefold more β-carotene and up to 1.75-fold more α-carotene compared to WT fruits (Figure 5C), especially in PC3 and PC4, the transgenic lines that have the lowest expression level of both PSY2 and CRTI. This suggests that a low expression level is enough to increment the carotenoid level whilst a very high level of expression may not produce a significant increment and sometimes causes a detriment in total or individual carotenoid levels (Figure 5C). In previous studies, EuCrtI was constitutively expressed in tomato var. Alisa Craig which resulted in a twofold increase in β-carotene levels with a significant reduction in both total carotenoid content and lycopene, producing orange-colored fruits (Römer et al., 2000). This metabolic effect was also due to the induction of endogenous PDS, ZDS, and LCYB and a reduction in PSY1 enzyme activity (Römer et al., 2000). These results suggest that the pPSY2-CRTI construct is more efficient in terms of total carotenoids, lycopene, and lutein production than pPSY2.
The simultaneous expression of three carotenogenic genes in tomatoes has not been previously reported, although it is a strategy that was previously used in rice and potato (Ye et al., 2000; Diretto et al., 2007). Both in rice that simultaneously express PSY, CRTI, and LCYB (Ye et al., 2000) and in potato, that express Erwinia CrtB (PSY), CrtI, and CrtY (LCYB) (Diretto et al., 2007), there were increases in lutein and β-carotene contents in transgenic lines. On the contrary, in our study, the expression of pPSY2-CRTI-LCYB1 in tomatoes produces a significant decrease in total carotenoid content. This decrease correlates with 3.6-fold falls in lycopene and 7.3-fold falls in α-carotene, but not in β-carotene levels, which increased significantly (by 2.57-fold, Figure 5D). We propose that these alterations are responsible for the orange phenotype of these transgenic fruits (Figure 4e). The decrease in total carotenoids observed in the pPSY1-CRTI-LCYB1 lines, although it is an unexpected result, is similar to that obtained by Römer et al. (2000) in tomatoes expressing 35SCaMV:CRTI. Even though there are several reasons that could explain this phenomenon, one hypothesis is that the constitutive and high expression of CRTI in transgenic lines, increases the expression levels of LCYB, thus causing a concomitant increment in β-carotene synthesis (Römer et al., 2000). A second hypothesis is that the CRTI enzyme is more efficient, delivering either a more abundant or a more suitable substrate to the LCYB enzyme which then transforms it into β-carotene. These two possibilities are not mutually exclusive and actually together could have a synergistic effect increasing the β-carotene content (Giuliano et al., 2000). In our case, the simultaneous expression of the three key genes may be redirecting the metabolic flux of the carotenoid pathway toward the production of β-carotene at the expense of the α-carotene branch and of the accumulation of those that are synthesized in previous stages, producing fruits with the pale orange appearance. In addition, the dwarf in vitro phenotype of these lines can also be explained by the redirection of the metabolic flow of GGPP precursors of the MEP pathway to carotenoid synthesis, with a detriment of the synthesis of gibberellic acid (GA), causing negative effects on the general development and physiology of the plant. GGPP is a precursor shared for the synthesis of carotenoids and for that of several growth regulators such as cytokines, ABA and GA (Cuttriss et al., 2007; Howitt et al., 2007; Rodríguez-Concepción, 2010). GA is an important phytohormone involved in plant development (Yamaguchi, 2008). Tomato plants (var. Alisa Craig) that overexpress PSY1 under the 35S promoter had fruits with higher levels of lycopene but presented a dwarf phenotype due to reduced levels of GA (Fray et al., 1995).
Taken together, our results allow us to suggest that pLCYB1 is the most suitable option for producing tomato fruits with a higher β-carotene and total carotenoid content.
To evaluate our biotechnological platform, we chose a transient expression assay because it provides a rapid and robust tool for assessing the activity of genetic constructs before undertaking a stable transformation (Sparkes et al., 2006). This is especially relevant in non-model fruits such as apples (Arcos et al., 2020). However, the infiltration of apple fruits has inherent challenges, given their anatomical and physiological characteristics such as large parenchymatous cells with substantial vacuoles (Spolaore et al., 2001). Together with the stage of maturity of the fruits during the infiltration, these factors contribute to making agroinfiltration a highly heterogeneous and complex process (Suppplementary Figure 5). Nevertheless, once key steps of the procedure have been standardized, it is a feasible approximation for functional assessment, providing a valuable tool. Our data showed that in Fuji apple fruits, all four pCP-GC constructs were capable of modifying the content of total and individual carotenoids after transient agroinfiltration (Table 1). Indeed, the 1.17–2.27-fold increment in total carotenoids obtained with pPSY2-CRTI was similar to those values previously reported by Arcos et al. (2020) with the same genetic construct (2GC) but using a different transient transformation strategy (vacuum infiltration). Interestingly, data at 14-days post-infiltration showed that the total carotenoid content and β-carotene levels were in several cases up to twice and triple, respectively, to those obtained at 7-days post agroinfiltration (Table 1). This could be associated with the stability of the enzymes after infiltration as well as the storage mechanism of the pigments once produced. In addition, it shows that apple fruits were capable of continuing the normal ripening process despite the exogenous bacterial application, at least for the periods evaluated. This also allows us to conclude that DcPSY2 is functional in apple fruits. Moreover, the increments seen in β-carotene also suggest that endogenous carotenogenic genes such as desaturases (PDS and ZDS), isomerases (Z-ISO and CRTISO) and LCYB are functional in apples.
The fact that the single constructs (pPSY2 and pLCYB1) duplicated the content of total carotenoids and β-carotene with respect to the double (pPSY2-CRTI) and triple constructs (pPSY2-CRTI-LCYB1) both at 7 and 14-days post infiltration, suggest that the transgenes in the double or triple constructs might not be properly expressed, or the stability of these transcripts/enzymes could be harmed. It is important to remember that each carotenogenic gene in the pCP constructs is driven by the same promoter, which could generate competition for the same transcription factors, limiting the recruitment of the transcription machinery, especially in the multiple constructs.
These results lead us to propose that apple fruits obtained from stable transformation will also accumulate at least two to three times more carotenoids. However, it is feasible to consider that the increment in the carotenoid levels would be even higher in the flesh considering our previous results, by expressing 35S:AtDXR in Fuji (Arcos et al., 2020). Specifically, leaves of AtDXR transgenic apples displayed up to a threefold rise in total carotenoids (508 mg g–1 FW) and in β-carotene (508 mg g–1 FW) when compared to wild-type plants, and twofold rises when transiently infiltrated in apple fruits (Arcos et al., 2020). In addition, in the flesh of banana fruits, a 2.3-fold increment in β-carotene was obtained by expressing PSY1 or PSY2 directed by a constitutive promoter when compared to the wild-type (Paul et al., 2017), and in kiwi fruit, a 1.3-fold increase in β-carotene was observed by expressing either GGPPS or PSY (Kim et al., 2010). Regarding the constructs generated, these results suggest that pPSY2 or pLCYB1 are the best candidates for the stable transformation of apples, and possibly for other fruits with pale flesh, in order to improve carotenoid content and thus nutritional properties.
1. The PG promoter from S. chilense var. tomatillo correctly directs gene expression to fruits in tomato and apple.
2. DcPSY2 codifies for a functional enzyme with a plastidial localization.
3. All four pCP-CG vectors are able to increase the total carotenoid content, particularly of β-carotene, in tomatoes and apples.
4. Of the pCP-CG vectors, those that possess the carrot carotenogenic genes DcPSY2 and DcLCYB1, would be the best candidates for increasing carotenoids in stably transformed apples.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
AA-M and CS conceived and designed the experiments. DA, AA-M, CF-O, and CP performed the experiments. DA, AA-M, CF-O, MH, and CS wrote the manuscript. All authors contributed to the article and approved the submitted version.
This study was funded by Programa de Investigación Asociativa (PIA) Anillo ACT192073 and Fondef D10I1022 (CS and MH) and Fondecyt 1180747 (CS) projects.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors acknowledge Manuel Rodriguez-Concepción (CRAG-CSIC, Spain) for providing the pB7FWG2-AtDXR vector and the Agencia Nacional de Investigación e Innovación (ANID) through the projects Anillo ACT192073, Fondef D10I1022, and Fondecyt 1180747 for financial support.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.677553/full#supplementary-material
Alcaíno, J., Baeza, M., and Cifuentes, V. (2016). Carotenoid distribution in nature. Subcell. Biochem. 79, 3–33. doi: 10.1007/978-3-319-39126-7_1
Alós, E., Rodrigo, M. J., and Zacarias, L. (2016). Manipulation of carotenoid content in plants to improve human health. Subcell. Biochem. 79, 311–343. doi: 10.1007/978-3-319-39126-7_12
Ampomah-Dwamena, C., Dejnoprat, S., Lewis, D., Sutherland, P., Volz, R. K., and Allan, A. C. (2012). Metabolic and gene expression analysis of apple (Malus × domestica) carotenogenesis. J. Exp. Bot. 63, 4497–4511. doi: 10.1093/jxb/ers134
Ampomah-Dwamena, C., Driedonks, N., Lewis, D., Shumskaya, M., Chen, X., Wurtzel, E. T., et al. (2015). The phytoene synthase gene family of apple (Malus x domestica) and its role in controlling fruit carotenoid content. BMC Plant Biol. 15:185.
Apel, W., and Bock, R. (2009). Enhancement of carotenoid biosynthesis in transplastomic tomatoes by induced lycopene-to-provitamin a conversion. Plant Physiol. 151, 59–66. doi: 10.1104/pp.109.140533
Arcos, Y., Godoy, F., Flores-Ortiz, C., Arenas-M, A., and Stange, C. (2020). Boosting carotenoid content in Malus domestica var. Fuji by expressing AtDXR through an Agrobacterium-mediated transformation method. Biotechnol. Bioeng. 117, 2209–2222. doi: 10.1002/bit.27358
Bang, H., Kim, S., Leskovar, D., and King, S. (2007). Development of a codominant CAPS marker for allelic selection between canary yellow and red watermelon based on SNP in lycopene β-cyclase (LCYB) gene. Mol. Breed. 20, 63–72. doi: 10.1007/s11032-006-9076-4
Bartley, G. E., and Scolnik, P. A. (1995). Plant carotenoids: pigments for photoprotection, visual attraction, and human health. Plant Cell 7, 1027–1038. doi: 10.2307/3870055
Beltran, J. C. M., and Stange, C. (2016). Apocarotenoids: a new carotenoid-derived pathway. Subcell. Biochem. 79, 239–272. doi: 10.1007/978-3-319-39126-7_9
Britton, G. (1995). Structure and properties of carotenoids in relation to function. FASEB J. 9, 1551–1558. doi: 10.1096/fasebj.9.15.8529834
Britton, G., Liaaen-Jensen, H., and Pfander, H. (2004). Carotenoids Handbook. Boston: Birkhäuser Basel.
Byers, T., and Perry, G. (1992). Dietary carotenes, vitamin C, and vitamin E as protective antioxidants in human cancers. Annu. Rev. Nutr. 12, 139–159. doi: 10.1146/annurev.nu.12.070192.001035
Cazzonelli, C. I. (2011). Carotenoids in nature: insights from plants and beyond. Funct. Plant Biol. 38, 833–847. doi: 10.1071/fp11192
Cazzonelli, C. I., and Pogson, B. J. (2010). Source to sink: regulation of carotenoid biosynthesis in plants. Trends Plant Sci. 15, 266–274. doi: 10.1016/j.tplants.2010.02.003
Ceballos, H., Morante, N., Sánchez, T., Ortiz, D., Aragón, I., and Chávez, A. L. (2013). Rapid cycling recurrent selection for increased carotenoids content in cassava roots. Crop Sci. 53, 2342–2351. doi: 10.2135/cropsci2013.02.0123
Cerda, A., Moreno, J. C., Acosta, D., Godoy, F., Cáceres, J. C., and Cabrera, R. (2020). Functional characterisation and in silico modelling of MdPSY2 variants and MdPSY5 phytoene synthases from Malus domestica. J. Plant Physiol. 249, 153166. doi: 10.1016/j.jplph.2020.153166
Curtis, M. D., and Grossniklaus, U. (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133, 462–469. doi: 10.1104/pp.103.027979
Cuttriss, A. J., Chubb, A. C., Alawady, A., Grimm, B., and Pogson, B. J. (2007). Regulation of lutein biosynthesis and prolamellar body formation in Arabidopsis. Funct. Plant Biol. 34, 663–672. doi: 10.1071/fp07034
D’Ambrosio, C., Giorio, G., Marino, I., Merendino, A., Petrozza, A., Salfi, L., et al. (2004). Virtually complete conversion of lycopene into β-carotene in fruits of tomato plants transformed with the tomato lycopene β-cyclase (tlcy-b) cDNA. Plant Sci. 166, 207–214. doi: 10.1016/j.plantsci.2003.09.015
D’Andrea, L., and Rodriguez-Concepcion, M. (2019). Manipulation of plastidial protein quality control components as a new strategy to improve carotenoid contents in tomato fruit. Front. Plant Sci. 10:1071.
Delgado-Pelayo, R., Gallardo-Guerrero, L., and Hornero-Méndez, D. (2014). Chlorophyll and carotenoid pigments in the peel and flesh of commercial apple fruits varieties. Food Res. Int. 65, 272–281. doi: 10.1016/j.foodres.2014.03.025
DellaPenna, D., Alexander, D. C., and Bennett, A. B. (1986). Molecular cloning of tomato fruit polygalacturonase: analysis of polygalacturonase mRNA levels during ripening. Proc. Natl. Acad. Sci. 83, 6420–6424. doi: 10.1073/pnas.83.17.6420
DellaPenna, D., Lincoln, J. E., Fischer, R. L., and Bennett, A. B. (1989). Transcriptional analysis of polygalacturonase and other ripening associated genes in rutgers, rin, nor, and Nr tomato fruit. Plant Physiol. 90, 1372–1377. doi: 10.1104/pp.90.4.1372
Diretto, G., Al-Babili, S., Tavazza, R., Papacchioli, V., Beyer, P., and Giuliano, G. (2007). Metabolic engineering of potato carotenoid content through tuber-specific overexpression of a bacterial mini-pathway. PLoS One 2:e0000350.
Diretto, G., Frusciante, S., Fabbri, C., Schauer, N., Busta, L., Wang, Z., et al. (2020). Manipulation of β-carotene levels in tomato fruits results in increased ABA content and extended shelf life. Plant Biotechnol. J. 18, 1185–1199. doi: 10.1111/pbi.13283
Diretto, G., Tavazza, R., Welsch, R., Pizzichini, D., Mourgues, F., and Papacchioli, V. (2006). Metabolic engineering of potato tuber carotenoids through tuber-specific silencing of lycopene epsilon cyclase. BMC Plant Biol. 6:13.
Failla, M. L., Chitchumroonchokchai, C., Siritunga, D., De Moura, F. F., Fregene, M., and Manary, M. J. (2012). Retention during processing and bioaccessibility of β-carotene in high β-carotene transgenic cassava root. J. Agric. Food Chem. 60, 3861–3866. doi: 10.1021/jf204958w
Farré, G., Bai, C., Twyman, R. M., Capell, T., Christou, P., and Zhu, C. (2011). Nutritious crops producing multiple carotenoids - a metabolic balancing act. Trends Plant Sci. 16, 532–540. doi: 10.1016/j.tplants.2011.08.001
Farré, G., Sanahuja, G., Naqvi, S., Bai, C., Capell, T., and Zhu, C. (2010). Travel advice on the road to carotenoids in plants. Plant Sci. 179, 28–48. doi: 10.1016/j.plantsci.2010.03.009
Flores-Ortiz, C., Alvarez, L. M., Undurraga, A., Arias, D., Durán, F., Wegener, G., et al. (2020). Differential role of the two ζ-carotene desaturase paralogs in carrot (Daucus carota): ZDS1 is a functional gene essential for plant development and carotenoid synthesis. Plant Sci. 291:110327.
Fraser, P. D., and Bramley, P. M. (2004). The biosynthesis and nutritional uses of carotenoids. Prog. Lipid Res. 43, 228–265. doi: 10.1016/j.plipres.2003.10.002
Fraser, P. D., Enfissi, E. M. A., and Bramley, P. M. (2009). Genetic engineering of carotenoid formation in tomato fruit and the potential application of systems and synthetic biology approaches. Arch. Biochem. Biophys. 483, 196–204. doi: 10.1016/j.abb.2008.10.009
Fraser, P. D., Kiano, J. W., Truesdale, M. R., Schuch, W., and Bramley, P. M. (1999). Phytoene synthase-2 enzyme activity in tomato does not contribute to carotenoid synthesis in ripening fruit. Plant Mol. Biol. 40, 687–698.
Fraser, P. D., Romer, S., Shipton, C. A., Mills, P. B., Kiano, J. W., and Misawa, N. (2002). Evaluation of transgenic tomato plants expressing an additional phytoene synthase in a fruit-specific manner. Proc. Natl. Acad. Sci. U. S. A. 99, 1092–1097. doi: 10.1073/pnas.241374598
Fraser, P. D., Truesdale, M. R., Bird, C. R., Schuch, W., and Bramley, P. M. (1994). Carotenoid biosynthesis during tomato fruit development. evidence for tissue-specific gene expression. Plant Physiol. 105, 405–413. doi: 10.1104/pp.105.1.405
Fray, R. G., Wallace, A., Fraser, P. D., Valero, D., Hedden, P., and Bramley, P. M. (1995). Constitutive expression of a fruit phytoene synthase gene in transgenic tomatoes causes dwarfism by redirecting metabolites from the gibberellin pathway. Plant J. 8, 693–701. doi: 10.1046/j.1365-313x.1995.08050693.x
Fuentes, P., Pizarro, L., Moreno, J. C., Handford, M., Rodriguez-Concepcion, M., and Stange, C. (2012). Light-dependent changes in plastid differentiation influence carotenoid gene expression and accumulation in carrot roots. Plant Mol. Biol. 79, 47–59. doi: 10.1007/s11103-012-9893-2
Giuliano, G. (2014). Plant carotenoids: genomics meets multi-gene engineering. Curr. Opin. Plant Biol. 19, 111–117. doi: 10.1016/j.pbi.2014.05.006
Giuliano, G., Aquilani, R., and Dharmapuri, S. (2000). Metabolic engineering of plant carotenoids. Trends Plant Sci. 5, 406–409. doi: 10.1016/s1360-1385(00)01749-0
Giuliano, G., Bartley, G. E., and Scolnik, P. A. (1993). Regulation of carotenoid biosynthesis during tomato development. Plant Cell 5, 341–351.
Giuliano, G., Tavazza, R., Diretto, G., Beyer, P., and Taylor, M. A. (2008). Metabolic engineering of carotenoid biosynthesis in plants. Trends Biotechnol. 26, 139–145. doi: 10.1016/j.tibtech.2007.12.003
Harker, F. R., Gunson, F. A., and Jaeger, S. R. (2003). The case for fruit quality: an interpretive review of consumer attitudes, and preferences for apples. Postharvest Biol. Technol. 28, 333–347. doi: 10.1016/s0925-5214(02)00215-6
Holick, C. N., Michaud, D. S., Stolzenberg-Solomon, R., Mayne, S. T., Pietinen, P., and Taylor, P. R. (2002). Dietary carotenoids, serum β-carotene, and retinol and risk of lung cancer in the alpha-tocopherol, beta-carotene cohort study. Am. J. Epidemiol. 156, 536–547. doi: 10.1093/aje/kwf072
Hornero-Méndez, D., and Britton, G. (2002). Involvement of NADPH in the cyclization reaction of carotenoid biosynthesis. FEBS Lett. 515, 133–136. doi: 10.1016/s0014-5793(02)02453-5
Howitt, C. A., Pogson, B. J., Cuttriss, A. J., and Mimica, J. L. (2007). “Carotenoids,” in the Structure and Function of Plastids. Advances in Photosynthesis and Respiration, vol 23, eds R. R. Wise and J. K. Hoober (Dordrecht: Springer).
Isaacson, T., Ohad, I., Beyer, P., and Hirschberg, J. (2004). Analysis in vitro of the enzyme CRTISO establishes a poly-cis-carotenoid biosynthesis pathway in plants. Plant Physiol. 136, 4246–4255. doi: 10.1104/pp.104.052092
Isaacson, T., Ronen, G., Zamir, D., and Hirschberg, J. (2002). Cloning of tangerine from tomato reveals a Carotenoid isomerase essential for the production of β-carotene and xanthophylls in plants. Plant Cell 14, 333–342. doi: 10.1105/tpc.010303
Janssen, B. J., Thodey, K., Schaffer, R. J., Alba, R., Balakrishnan, L., and Bishop, R. (2008). Global gene expression analysis of apple fruit development from the floral bud to ripe fruit. BMC Plant Biol. 8:16. doi: 10.1186/1471-2229-8-16
Jayaraj, J., Devlin, R., and Punja, Z. (2008). Metabolic engineering of novel ketocarotenoid production in carrot plants. Transgenic Res. 17, 489–501. doi: 10.1007/s11248-007-9120-0
Just, B. J., Santos, C. A. F., Fonseca, M. E. N., Boiteux, L. S., Oloizia, B. B., and Simon, P. W. (2007). Carotenoid biosynthesis structural genes in carrot (Daucus carota): isolation, sequence-characterization, single nucleotide polymorphism (SNP) markers and genome mapping. Theor. Appl. Genet. 114, 693–704. doi: 10.1007/s00122-006-0469-x
Kim, M., Kim, S.-C., Song, K. J., Kim, H. B., Kim, I.-J., and Song, E.-Y. (2010). Transformation of carotenoid biosynthetic genes using a micro-cross section method in kiwifruit (Actinidia deliciosa cv. Hayward). Plant Cell Rep. 29, 1339–1349. doi: 10.1007/s00299-010-0920-y
Lau, J. M., Cooper, N. G., Robinson, D. L., and Korban, S. S. (2009). Sequence and in silico characterization of the tomato polygalacturonase (PG) promoter and terminator region. Plant Mol. Biol. Rep. 27, 250–256. doi: 10.1007/s11105-008-0081-0
Lichtenthaler, H. K., and Buschmann, C. (2001). Extraction of photosynthetic tissues: chlorophylls and carotenoids. Curr. Protoc. Food Anal. Chem. 1.
Lincoln, J. E., Cordes, S., Read, E., and Fischer, R. L. (1987). Regulation of gene expression by ethylene during Lycopersicon esculentum (tomato) fruit development. Proc. Natl. Acad. Sci. U. S. A. 84, 2793–2797. doi: 10.1073/pnas.84.9.2793
Lipkie, T. E., De Moura, F. F., Zhao, Z. Y., Albertsen, M. C., Che, P., and Glassman, K. (2013). Bioaccessibility of carotenoids from transgenic provitamin a biofortified sorghum. J. Agric. Food Chem. 61, 5764–5771. doi: 10.1021/jf305361s
Liu, L., Shao, Z., Zhang, M., and Wang, Q. (2015). Regulation of carotenoid metabolism in tomato. Mol. Plant 8, 28–39. doi: 10.1016/j.molp.2014.11.006
Lu, S., and Li, L. (2008). Carotenoid metabolism: biosynthesis, regulation, and beyond. J. Integr. Plant Biol. 50, 778–785. doi: 10.1111/j.1744-7909.2008.00708.x
Malone, W. F. (1991). Studies evaluating antioxidants and β-carotene as chemopreventives. Am. J. Clin. Nutr. 53, 305S–313S.
Marnett, L. J. (1987). Peroxyl free radicals: potential mediators of tumor initiation and promotion. Carcinogenesis 8, 1365–1373. doi: 10.1093/carcin/8.10.1365
Meisel, L., Fonseca, B., González, S., Baeza-Yates, R., Cambiazo, V., Campos, R., et al. (2005). A rapid and efficient method for purifying high quality total RNA from peaches (Prunus persica) for functional genomics analyses. Biol. Res. 38, 83–88.
Mínguez-Mosquera, M. I., and Hornero-Méndez, D. (1997). Changes in provitamin a during paprika processing. J. Food Prot. 60, 853–857. doi: 10.4315/0362-028x-60.7.853
Misawa, N. (2011). Carotenoid β-ring hydroxylase and ketolase from marine bacteria - promiscuous enzymes for synthesizing functional xanthophylls. Mar. Drugs 9, 757–771. doi: 10.3390/md9050757
Misawa, N., Yamano, S., Linden, H., De Felipe, M. R., Lucas, M., and Ikenaga, H. (1993). Functional expression of the Erwinia uredovora carotenoid biosynthesis gene crtl in transgenic plants showing an increase of β-carotene biosynthesis activity and resistance to the bleaching herbicide norflurazon. Plant J. 4, 833–840. doi: 10.1046/j.1365-313x.1993.04050833.x
Moise, A. R., Al-Babili, S., and Wurtzel, E. T. (2014). Mechanistic aspects of carotenoid biosynthesis. Chem. Rev. 114, 164–193. doi: 10.1021/cr400106y
Montgomery, J., Pollard, V., Deikman, J., and Fischer, R. L. (1993). Positive and negative regulatory regions control the spatial distribution of polygalacturonase transcription in tomato fruit pericarp. Plant Cell 5, 1049–1062. doi: 10.2307/3869626
Mordi, R. C. (1993). Mechanism of β-carotene degradation. Biochem. J. 292, 310–312. doi: 10.1042/bj2920310
Moreno, J. C., Cerda, A., Simpson, K., Lopez-Diaz, I., Carrera, E., Handford, M., et al. (2016). Increased Nicotiana tabacum fitness through positive regulation of carotenoid, gibberellin and chlorophyll pathways promoted by Daucus carota lycopene β-cyclase (Dclcyb1) expression. J. Exp. Bot. 67, 2325–2338. doi: 10.1093/jxb/erw037
Moreno, J. C., Mi, J., Agrawal, S., Kössler, S., Turečková, V., and Tarkowská, D. (2020). Expression of a carotenogenic gene allows faster biomass production by redesigning plant architecture and improving photosynthetic efficiency in tobacco. Plant J. 103, 1967–1984. doi: 10.1111/tpj.14909
Moreno, J. C., Pizarro, L., Fuentes, P., Handford, M., Cifuentes, V., and Stange, C. (2013). Levels of Lycopene β-Cyclase 1 modulate carotenoid gene expression and accumulation in Daucus carota. PLoS One 8:e0058144.
Naqvi, S., Farré, G., Sanahuja, G., Capell, T., Zhu, C., and Christou, P. (2010). When more is better: multigene engineering in plants. Trends Plant Sci. 15, 48–56. doi: 10.1016/j.tplants.2009.09.010
Naqvi, S., Zhu, C., Farre, G., Ramessar, K., Bassie, L., and Breitenbach, J. (2009). Transgenic multivitamin corn through biofortification of endosperm with three vitamins representing three distinct metabolic pathways. Proc. Natl. Acad. Sci. U. S. A. 106, 7726–7727.
Niklitschek, M., Alcaíno, J., Barahona, S., Sepúlveda, D., Lozano, C., and Carmona, M. (2008). Genomic organization of the structural genes controlling the astaxanthin biosynthesis pathway of Xanthophyllomyces dendrorhous. Biol. Res. 41, 93–108.
Olson, J. A. (1996). Benefits and liabilities of vitamin a and carotenoids. J. Nutr. 126(4 Suppl.), 1208S–1212S.
Paine, J. A., Shipton, C. A., Chaggar, S., Howells, R. M., Kennedy, M. J., and Vernon, G. (2005). Improving the nutritional value of golden rice through increased pro-vitamin a content. Nat. Biotechnol. 23, 482–487. doi: 10.1038/nbt1082
Paul, J. Y., Khanna, H., Kleidon, J., Hoang, P., Geijskes, J., and Daniells, J. (2017). Golden bananas in the field: elevated fruit pro-vitamin a from the expression of a single banana transgene. Plant Biotechnol. J. 15, 520–532. doi: 10.1111/pbi.12650
Pecker, I., Gabbay, R., Cunningham, F. X., and Hirschberg, J. (1996). Cloning and characterization of the cDNA for lycopene beta-cyclase from tomato reveals decrease in its expression during fruit ripening. Plant Mol. Biol. 30, 807–819. doi: 10.1007/bf00019013
Perello, C., Llamas, E., Burlat, V., Ortiz-Alcaide, M., Phillips, M. A., and Pulido, P. (2016). Differential subplastidial localization and turnover of enzymes involved in isoprenoid biosynthesis in chloroplasts. PLoS One 11:e0150539. doi: 10.1371/journal.pone.0150539
Pino, L. E., Lombardi-Crestana, S., Azevedo, M. S., Scotton, D. C., Borgo, L., and Quecini, V. (2010). The Rg1 allele as a valuable tool for genetic transformation of the tomato “Micro-Tom” model system. Plant Methods 6:23. doi: 10.1186/1746-4811-6-23
Pixley, K., Rojas, N. P., Babu, R., Mutale, R., Surles, R., and Simpungwe, E. (2013). “Biofortification of maize with provitamin a carotenoids,” in Carotenoids and Human Health, ed. S. Tanumihardjo (Totowa, NJ: Humana Press).
Pons, E., Alquézar, B., Rodríguez, A., Martorell, P., Genovés, S., and Ramón, D. (2014). Metabolic engineering of β-carotene in orange fruit increases its in vivo antioxidant properties. Plant Biotechnol. J. 12, 17–27. doi: 10.1111/pbi.12112
Puigbò, P., Guzmán, E., Romeu, A., and Garcia-Vallvé, S. (2007). OPTIMIZER: a web server for optimizing the codon usage of DNA sequences. Nucleic Acids Res. 35, W126–W131.
Ravanello, M. P., Ke, D., Alvarez, J., Huang, B., and Shewmaker, C. K. (2003). Coordinate expression of multiple bacterial carotenoid genes in canola leading to altered carotenoid production. Metab. Eng. 5, 255–263. doi: 10.1016/j.ymben.2003.08.001
Rodríguez-Concepción, M. (2010). Supply of precursors for carotenoid biosynthesis in plants. Arch. Biochem. Biophys. 504, 118–122. doi: 10.1016/j.abb.2010.06.016
Rodriguez-Concepcion, M., and Stange, C. (2013). Biosynthesis of carotenoids in carrot: an underground story comes to light. Arch. Biochem. Biophys. 539, 110–116. doi: 10.1016/j.abb.2013.07.009
Römer, S., Fraser, P. D., Kiano, J. W., Shipton, C. A., Misawa, N., and Schuch, W. (2000). Elevation of the provitamin a content of transgenic tomato plants. Nat. Biotechnol. 18, 666–669. doi: 10.1038/76523
Ronen, G., Carmel-Goren, L., Zamir, D., and Hirschberg, J. (2000). An alternative pathway to β-carotene formation in plant chromoplasts discovered by map-based cloning of Beta and old-gold color mutations in tomato. Proc. Natl. Acad. Sci. U. S. A. 97, 11102–11107. doi: 10.1073/pnas.190177497
Ronen, G., Cohen, M., Zamir, D., and Hirschberg, J. (1999). Regulation of carotenoid biosynthesis during tomato fruit development: expression of the gene for lycopene epsilon-cyclase is down-regulated during ripening and is elevated in the mutant Delta. Plant J. 17, 341–351. doi: 10.1046/j.1365-313x.1999.00381.x
Rosas-Saavedra, C., and Stange, C. (2016). Biosynthesis of carotenoids in plants: enzymes and color. Subcell. Biochem. 79, 35–69. doi: 10.1007/978-3-319-39126-7_2
Rosati, C., Aquilani, R., Dharmapuri, S., Pallara, P., Marusic, C., and Tavazza, R. (2000). Metabolic engineering of beta-carotene and lycopene content in tomato fruit. Plant J. 24, 413–420. doi: 10.1046/j.1365-313x.2000.00880.x
Rosati, C., Diretto, G., and Giuliano, G. (2009). Biosynthesis and engineering of carotenoids and apocarotenoids in plants: state of the art and future prospects. Biotechnol. Genet. Eng. Rev. 26, 139–162. doi: 10.7313/upo9781907284489.006
Sandmann, G. (2001). Genetic manipulation of carotenoid biosynthesis: strategies, problems and achievements. Trends Plant Sci. 6, 14–17. doi: 10.1016/s1360-1385(00)01817-3
Sheehy, R. E., Pearson, J., Brady, C. J., and Hiatt, W. R. (1987). Molecular characterization of tomato fruit polygalacturonase. Mol. Gen. Genet. 208, 30–36. doi: 10.1007/bf00330418
Shewmaker, C. K., Sheehy, J. A., Daley, M., Colburn, S., and Ke, D. Y. (1999). Seed-specific overexpression of phytoene synthase: increase in carotenoids and other metabolic effects. Plant J. 20, 401X–412X.
Simpson, K., Cerda, A., and Stange, C. (2016). Carotenoid biosynthesis in Daucus carota. Subcell. Biochem. 79, 199–217. doi: 10.1007/978-3-319-39126-7_7
Simpson, K., Fuentes, P., Quiroz-Iturra, L. F., Flores-Ortiz, C., Contreras, R., and Handford, M. (2018). Unraveling the induction of phytoene synthase 2 expression by salt stress and abscisic acid in Daucus carota. J. Exp. Bot. 69, 4113–4126. doi: 10.1093/jxb/ery207
Sparkes, I. A., Runions, J., Kearns, A., and Hawes, C. (2006). Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 1, 2019–2025. doi: 10.1038/nprot.2006.286
Spolaore, S., Trainotti, L., and Casadoro, G. (2001). A simple protocol for transient gene expression in ripe fleshy fruit mediated by Agrobacterium. J. Exp. Bot. 52, 840–850.
Sun, T., Yuan, H., Cao, H., Yazdani, M., Tadmor, Y., and Li, L. (2018). Carotenoid metabolism in plants: the role of plastids. Mol. Plant 11, 58–74. doi: 10.1016/j.molp.2017.09.010
Telias, A., Bradeen, J. M., Luby, J. J., Hoover, E. E., and Allen, A. C. (2011). Regulation of anthocyanin accumulation in apple peel. Hortic. Rev. 38, 357–391.
Voorrips, L. E., Goldbohm, R. A., Van Poppel, G., Sturmans, F., Hermus, R. J. J., and Van Den Brandt, P. A. (2000). Vegetable and fruit consumption and risks of colon and rectal cancer in a prospective cohort study: the Netherlands cohort study on diet and cancer. Am. J. Epidemiol. 152, 1081–1092. doi: 10.1093/aje/152.11.1081
Walter, M. H., and Strack, D. (2011). Carotenoids and their cleavage products: biosynthesis and functions. Nat. Prod. Rep. 28, 663–692. doi: 10.1039/c0np00036a
Wurbs, D., Ruf, S., and Bock, R. (2007). Contained metabolic engineering in tomatoes by expression of carotenoid biosynthesis genes from the plastid genome. Plant J. 49, 276–288. doi: 10.1111/j.1365-313x.2006.02960.x
Yamaguchi, S. (2008). Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol. 59, 225–251. doi: 10.1146/annurev.arplant.59.032607.092804
Ye, V. M., and Bhatia, S. K. (2012). Pathway engineering strategies for production of beneficial carotenoids in microbial hosts. Biotechnol. Lett. 34, 1405–1414.
Ye, X., Al-Babili, S., Andreas, K., Jing, Z., Paola, L., Beyer, P., et al. (2000). Engineering the provitamin a (β-carotene) biosynthetic pathway into (carotenoid-free) rice endosperm. Science 287, 303–305. doi: 10.1126/science.287.5451.303
Yuan, K., Wang, C., Wang, J., Xin, L., Zhou, G., and Li, L. (2014). Analysis of the MdMYB1 gene sequence and development of new molecular markers related to apple skin color and fruit-bearing traits. Mol. Genet. Genom. 289, 1257–1265. doi: 10.1007/s00438-014-0886-5
Keywords: carotenoid, Malus domestica, Solanum lycopersicum, DcPSY2, DcLCYB1, Daucus carota, transgenic tomatoes, metabolic engineering
Citation: Arias D, Arenas-M A, Flores-Ortiz C, Peirano C, Handford M and Stange C (2021) Daucus carota DcPSY2 and DcLCYB1 as Tools for Carotenoid Metabolic Engineering to Improve the Nutritional Value of Fruits. Front. Plant Sci. 12:677553. doi: 10.3389/fpls.2021.677553
Received: 07 March 2021; Accepted: 26 May 2021;
Published: 26 August 2021.
Edited by:
Patricia León, National Autonomous University of Mexico, MexicoReviewed by:
Barbara Molesini, University of Verona, ItalyCopyright © 2021 Arias, Arenas-M, Flores-Ortiz, Peirano, Handford and Stange. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Claudia Stange, Y3N0YW5nZUB1Y2hpbGUuY2w=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.