
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 21 July 2021
Sec. Plant Cell Biology
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.674031
This article is part of the Research TopicPlasmodesmata: Recent Progress and New InsightsView all 12 articles
Post-embryonic cells contain minute lipid bodies (LBs) that are transient, mobile, engage in organellar interactions, and target plasmodesmata (PD). While LBs can deliver γ-clade 1,3-β-glucanases to PD, the nature of other cargo is elusive. To gain insight into the poorly understood role of LBs in meristems, we investigated their dynamics by microscopy, gene expression analyzes, and proteomics. In developing buds, meristems accumulated LBs, upregulated several LB-specific OLEOSIN genes and produced OLEOSINs. During bud maturation, the major gene OLE6 was strongly downregulated, OLEOSINs disappeared from bud extracts, whereas lipid biosynthesis genes were upregulated, and LBs were enlarged. Proteomic analyses of the LB fraction of dormant buds confirmed that OLEOSINs were no longer present. Instead, we identified the LB-associated proteins CALEOSIN (CLO1), Oil Body Lipase 1 (OBL1), Lipid Droplet Interacting Protein (LDIP), Lipid Droplet Associated Protein1a/b (LDAP1a/b) and LDAP3a/b, and crucial components of the OLEOSIN-deubiquitinating and degradation machinery, such as PUX10 and CDC48A. All mRFP-tagged LDAPs localized to LBs when transiently expressed in Nicotiana benthamiana. Together with gene expression analyzes, this suggests that during bud maturation, OLEOSINs were replaced by LDIP/LDAPs at enlarging LBs. The LB fraction contained the meristem-related actin7 (ACT7), “myosin XI tail-binding” RAB GTPase C2A, an LB/PD-associated γ-clade 1,3-β-glucanase, and various organelle- and/or PD-localized proteins. The results are congruent with a model in which LBs, motorized by myosin XI-k/1/2, traffic on F-actin, transiently interact with other organelles, and deliver a diverse cargo to PD.
Plasmodesmata (PD) are notoriously difficult to investigate, and understanding PD functioning from the composition is fraught with difficulties. Both composition and architecture of PD are subject to regulation by cells that share them. Rather than functioning autonomously, they are subject to control by cells that construct, maintain, and operate them. Consequently, PD composition and function are context-dependent, differing between tissues in dependence on developmental and metabolic cellular states. Filtering out commonalities while recognizing the unique aspects of PD at specific locations and conditions, therefore, remains a challenge.
Information on PD composition and function has been gathered using a variety of plant and tissue systems and approaches. Frequently, investigations are focused on PD (ultra)structure and/or localization of suspected PD proteins by immunochemistry or transgenic expression of fluorescently tagged proteins, microinjection studies, and proteomic and lipidomic studies (reviewed in Faulkner and Maule, 2011; Sager and Lee, 2014; Heinlein, 2015a; Brault et al., 2019; Han et al., 2019; Reagan and Burch-Smith, 2020).
Despite the importance of determining PD composition and architecture, the question remains unanswered of how relevant PD components are delivered to PD and integrated into the functional fabric of the PD channel. The cellular mechanisms that deliver structural PD components also modulate gating events. While the exterior and interior of PD are targeted by distinct mechanisms, PD conductance is also subject to control by physiological processes on both sides of the channels. For example, nuclear-organellar signaling contributes to ROS-mediated plasmodesmal regulation, involving mitochondria and chloroplasts as sensors for cellular homeostasis (Burch-Smith and Zambryski, 2011).
Regarding supply routes, the most frequently studied route is the Brefeldin-sensitive excretion pathway, which delivers proteins to the cell wall (Sager and Lee, 2014; Han et al., 2019; Reagan and Burch-Smith, 2020). Cargos delivered via this pathway may include integral membrane proteins and GPI-anchored proteins. The latter can attach to exoplasmic membrane rafts, which move them laterally to the exterior of the PD (Mongrand et al., 2010). With regard to the PD interior, important leads emerged from studies that analyzed how viruses take advantage of existing cellular mechanisms, such as the cortical endoplasmic reticulum (ER) and the cytoskeleton, and how they interact with molecular complexes that control PD gating (reviewed in e.g., Heinlein, 2015b; Reagan and Burch-Smith, 2020). Little explored is the mechanism by which cytoplasmically produced lipid bodies (LBs) target PD to deliver a largely unknown cargo to the channel (Rinne et al., 2011; Veerabagu et al., 2020).
In seeds, LB production requires OLEOSINs (Huang, 1992; Tzen and Huang, 1992; Abell et al., 2002). OLEOSIN proteins are small 15–26 kD proteins that are co-translationally inserted into the bilayer of the ER, guided by an ER-resident signal recognition particle (Abell et al., 2004). The 5–6 nm long hydrophobic hairpin of OLEOSIN is embedded under strain in the bilayer, which facilitates its diffusion into the stable hydrophobic environment of a nascent LB, promoting its eventual release into the cytosol (Abell et al., 2002; Huang and Huang, 2017; Huang, 2018). This budding process is facilitated by a critical imbalance in leaflet surface tensions and involves SEIPIN proteins and interacting lipid biosynthesis genes (Cai et al., 2015; Barbosa and Siniossoglou, 2017). Once formed, OLEOSIN secures the integrity and small size of the LBs by stabilizing the monolayer and preventing coalescence and fusion (Siloto et al., 2006; Shimada et al., 2008; Hsiao and Tzen, 2011). In addition, CALEOSIN and STEROLEOSIN can bind competitively with an expanding monolayer, mediated by short ca. 2 nm hydrophobic hairpins (Huang, 2018).
Like seeds, bud meristems contain LBs, and eight of the nine Populus OLEOSIN genes are expressed in apices (Veerabagu et al., 2020). The capacity of LBs to deliver cargos to PD could be important in the shoot apical meristem (SAM), where PD are continuously produced within and between cell lineages (van der Schoot and Rinne, 1999). In the active SAM, individual cells are continuously displaced toward the periphery to be integrated into differentiating tissues. To secure the functional integrity of the SAM, all cells need to continuously update their relative position by exchanging signals, among others, through existing and newly formed PD (Rinne and van der Schoot, 1998). LBs potentially contribute to PD formation, maintenance, and cell-cell signaling by shuttling lipids, enzymes, and signaling molecules to the PD entrance (van der Schoot et al., 2011; Paul et al., 2014a). An LB shuttle function was demonstrated in N. benthamiana, where LBs delivered eGFP-tagged 1,3-β-glucanases to PD, identified by TMV MP-mRFP (Rinne et al., 2011). Similarly, transgenic Arabidopsis LBs, tagged with PtOLE6-eGFP, targeted primary and secondary PD in various cell types (Veerabagu et al., 2020). The LBs do not arrive at the PD by bulk cytoplasmic streaming but by processive trafficking on F-actin, mediated by myosin XI-k/1/2 (Veerabagu et al., 2020).
The PD in the SAM of woody perennials are unique in the sense that they are modified during the seasonal cycle. Under short days, the PD are shut down by Dormancy Sphincter Complexes (DSCs). DSCs act as circuit breakers that interrupt the symplasmic circuitry of the SAM, preventing electrical and metabolic coupling and exchange of transcription factors and other regulatory molecules, arresting the SAM in a dormant state (Paul et al., 2014a,b). Unlike classical sphincters where callose is present extracellularly, DSCs contain additional internal deposits that can be targeted by LBs (Rinne et al., 2001; Rinne and van der Schoot, 2003). When recruited to the PD, the LB-associated enzyme 1,3-β-glucanase aligns with its substrate, resulting in callose hydrolysis, restoration of the PD channel, and dormancy release (Rinne et al., 2001; Rinne and van der Schoot, 2003). It is unknown what other cargos LBs can deliver to PD during dormancy release, and to what degree it differs from what is present in active meristems. A consensus view is that cytoplasmic LB motility enriches LB cargos by facilitating organellar interactions and exchange of proteins and lipids (Bartz et al., 2007; Hodges and Wu, 2010; Murphy, 2012; Krahmer et al., 2013; Gao and Goodman, 2015; Zhi et al., 2017). Proteins might also be recruited directly from the cytoplasm, especially when molecular crowding at the monolayer is reduced. These include monolayer-embedded proteins, lipophilic signals, lipid-anchored proteins, electrostatically associated proteins, and molecules that opportunistically hitch a ride on moving LBs (reviewed in van der Schoot et al., 2011; Paul et al., 2014a; Huang, 2018).
As in buds, accumulated LBs constitute a unique proactive dormancy-release and signaling mechanism, LBs may be expected to store proteins related to these functions. LB cargos are likely to include proteins that assist docking at the plasma membrane (PM), possibly proteins that become integrated into the fabric of PD, and non-cell-autonomous signals (van der Schoot et al., 2011; Paul et al., 2014a,b). How the actomyosin system that guides LBs to PD (Veerabagu et al., 2020) connects to the PD is unknown. Notably, F-actin and myosin VIII have been localized at PD (White et al., 1994; Baluska et al., 2001; Golomb et al., 2008; White and Barton, 2011). In addition, the actin-binding and nucleation complex Arp2/3 was localized at PD (Van Gestel et al., 2003; Deeks and Hussey, 2005; Fiserova et al., 2006), and recently it has been shown that the class I formin FH2 acts as an actin nucleation factor that caps and stabilizes F-actin at PD (Diao et al., 2018). Some plant viruses that move through PD have usurped and hijacked the cytoplasmic actomyosin system to facilitate their cell-to-cell transport (Amari et al., 2011, 2014; Sager and Lee, 2014; Heinlein, 2015b). Although actin can facilitate delivery of viral complexes to the PD entrance, PD-associated actin might restrict the size of the PD channel as virus passage requires severing of this actin (Ding et al., 1996; Su et al., 2010).
While it is unknown how the F-actin on which LBs traffic is anchored to the PD, it is also unknown what enables LBs to dock at PD. Given the difference between LB and PD diameters in dormant meristems, respectively, ca. 1 μm and 60–220 nm (channel and external ring; Rinne and van der Schoot, 2004), it might involve proteins that interact with PD orifices or their immediate surroundings at the PM. Targeting of eGFP-tagged LBs to the PD/PM area yields deflated LBs that appear as juxtaposed fluorescent patches, sandwiching primary and secondary PD (Rinne et al., 2011; Veerabagu et al., 2020). We hypothesized earlier that LBs could dock at remorin-decorated membrane rafts that act as sorting devices (van der Schoot et al., 2011; Paul et al., 2014a) and involve hemifusion between the LB monolayer and the cytoplasmic leaflet of the PM. If so, this might be mediated by SNARE protein complexes, which can localize to LBs (Boström et al., 2007; Sollner, 2007; Murphy et al., 2009; Paul et al., 2014a).
The relation of LBs with PD is virtually unexplored. To gauge how LBs might contribute to cellular homeostasis, organellar interaction, and cargo delivery to PD, we investigated their accumulation and putative composition by microscopy, gene expression analyzes, and proteomics. An important goal was to create an inventory of candidate proteins, such as known LB proteins and proteins that may hitch a ride to the PD. The results indicate that OLEOSIN is responsible for LB accumulation but that at a later stage it is removed and replaced by LDIP/LDAP proteins to allow recruitment of cytoplasmic proteins through reduced molecular crowding at the monolayer. Removal and degradation of OLEOSIN probably involve the action of PUX10, the segregase CDC48A, and the 26S proteasome, all of which were present in the LB fraction. Other identified proteins included CLO1, OBL1, GPAT8, a PD/LB-localized 1,3-β-glucanase, and proteins that likely reflect LB motility, organellar interactions, storage, and docking to PD/PM sites. Confirming LB/PD localization and the role of individual proteins in PD functioning will require future investigations. The current data lay the groundwork for such investigations and expand a model (Veerabagu et al., 2020) in which cargo-enriched LBs are anchored by GTPase RAB-C2A to myosin XI-k/1/2 for actin-guided transport to PD.
Lipid bodies have been detected in meristems previously but their accumulation patterns have not been characterized. In this study, LB production, size, and number were analyzed from transmission electron microscope (TEM) sections of meristems of growing plants (APs), developing terminal buds (DEBs), and dormant terminal buds (DOBs) (Figures 1A–C). In all three, LBs were present in the SAM and the subjacent rib meristem/rib zone (RM/RZ) area but their numbers were low in actively growing apices, especially in the RM/RZ area (Figure 1D). The low number in this area reflects the high rate of metabolism and cell division related to stem elongation. During bud development, apical stem elongation ceases and LB numbers in the RM/RZ area and the SAM increased significantly, while further increase during dormancy development was minor (Figure 1D). Based on LB numbers detected in TEM sections, we approximated their total number for an average cell volume in both the SAM and RM/RZ. The calculation accounted for section thickness (80 nm), LB diameter, and the volume of the nucleus and organelles (Figure 1E, Supplementary Figure 1). These data showed that all cells contained multiple LBs, even the RM/RZ of actively growing long-day plants. In developing buds, the number of LBs in SAM cells had increased by ca. 25%, while in the RM/RZ it had increased by ca. 50%, corresponding to the cessation of cell division (Figure 1D), but beyond that, the increase in number was minor, and only in the SAM by ca. 10%. The increase in LB sizes showed a similar trend. In growing apices, LBs were small (Figures 1A,E), but their size had significantly increased in developing buds, both in the SAM and the RM/RZ (Figures 1B,E). During dormancy establishment, LB size only increased further in the RM/RZ (Figures 1C,E).
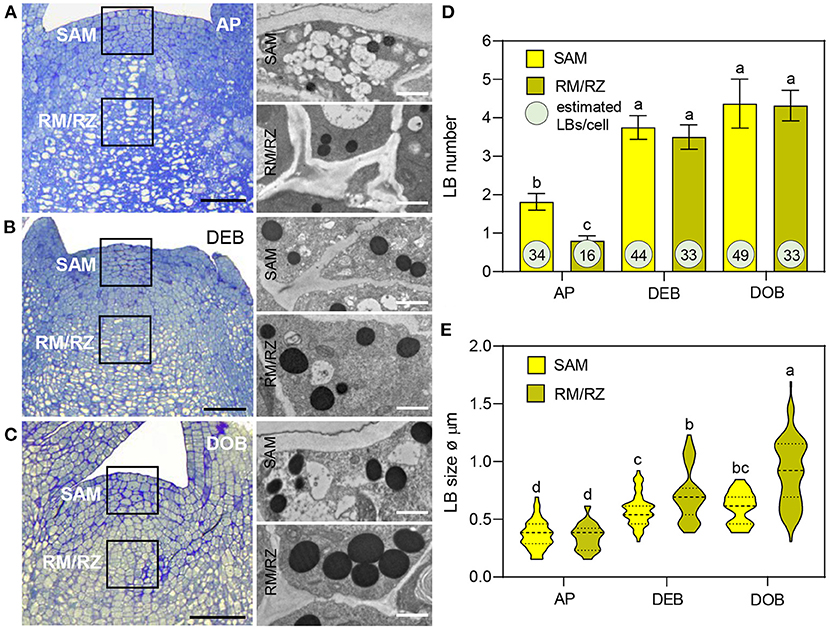
Figure 1. Lipid body (LB) production in the shoot apex of Populus. (A–C) show apices of (A) actively growing plants in long days (APs), (B) in short day-induced developing buds (DEBs) and (C) dormant buds (DOBs). TEM images (A–C) show LBs in the shoot apical meristem (SAM) and the rib meristem/rib zone (RM/RZ) (boxed meristem areas). (D) LB numbers were counted and (E) LB diameters were measured in successive TEM images. Bar diagram (D) shows means ± SD of LB numbers per cell per section. The encircled numbers are calculated LB numbers per cell volume based on average cell sizes, as explained in Supplementary Figure 1. (E) Violin plots show diameters of LBs including mean-line and quartiles. Different letters indicate statistically significant differences between treatments (one-way ANOVA, Fisher's post-hoc analysis P < 0.05). Bars are 50 (apices) and 1 μm (LBs).
Previously, we showed that very early under dormancy-inducing conditions three of the eight expressed OLEOSINs were upregulated (Veerabagu et al., 2020). In this study, we analyzed their expression in growing apices, in developing buds during the LB accumulation phase (Figure 1D), and in buds that were developing dormancy (Figure 2A). OLE6 appeared to be the most important of the three OLEOSINs, both quantitatively and in its responsiveness to bud development. It was strongly upregulated during the LB accumulation phase and almost completely downregulated during dormancy development. In contrast, the minor genes OLE3 and OLE5 were only slightly upregulated during LB production and somewhat downregulated during dormancy development (Figure 2A).
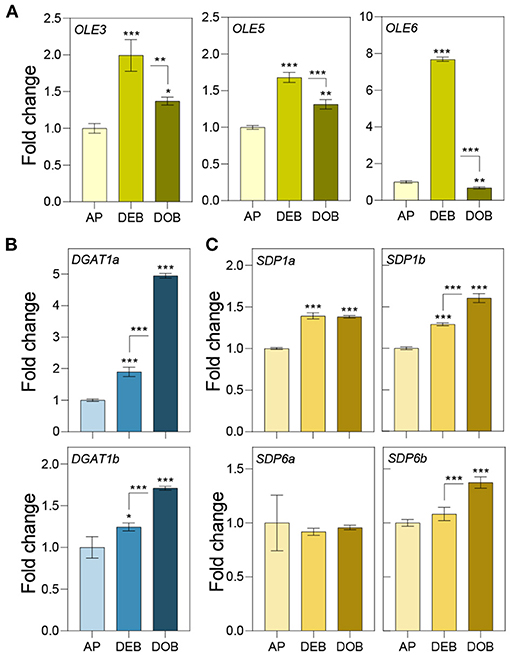
Figure 2. Expression of genes involved in lipid body production and lipid turnover. Relative expression of (A) OLEOSIN genes (OLE3, OLE5, and OLE6), (B) lipid biosynthesis genes (DGAT1a,b), and (C) lipase genes (SDP1a and b, and SDP6a and b) in apices under long day (APs), and in developing buds (DEBs), and dormant buds (DOBs) in short days. Values are calculated relative to the control (AP) and represent the means of three technical replicates ± SD of four plants. Asterisk(s) indicate statistical significance between treatments (one-way ANOVA, Tukey analysis; *P < 0.05; **P < 0.01; and ***P < 0.001).
To investigate if LB enlargement (Figures 1C,E) could be due to increased TAG biosynthesis, which is mediated by DGAT1 (Shockey et al., 2006), we identified two DGAT1 homologs, DGAT1a and DGAT1b, in the Populus trichocarpa genome (Supplementary Figure 2) and analyzed their expression during the same developmental stages. Both DGAT1a and DGAT1b, but especially DGAT1a, were upregulated during bud development and further during dormancy establishment (Figure 2B). This suggests that TAG biosynthesis may have increased concomitant with LB enlargement (Figure 1E). As the expression of lipases might counteract LB enlargement through TAG-hydrolysis, we also analyzed transcript levels of the lipase gene SUGAR DEPENDENT 1 (SDP1) and the mitochondrial gene SDP6, which is required for post-germinative seedling growth in Arabidopsis (Quettier and Eastmond, 2009). Two isoforms of both SDP1 and SDP6 were identified in the P. trichocarpa genome (Supplementary Figure 3). SDP1a and SDP1b were upregulated during bud development, whereas SDP1b was further upregulated during dormancy establishment. The SDP6a expression did not show any change, and SDP6b was only slightly upregulated during dormancy establishment (Figure 2C). Considering that LBs enlarged, the encoded enzymes might not have targeted the LBs, like in seeds where SPD1 transcript levels do not correlate with enzyme activity (Eastmond, 2006).
To identify integral LB proteins (referred to as class I) as well as proteins with amphipathic stretches (class II) and peripherally and transiently associated proteins (Bersuker and Olzmann, 2017; Pyc et al., 2017), we isolated and purified LBs from dormant buds using a protocol modified after Jolivet et al. (2004). Purified LBs were subjected to light and differential interference contrast (DIC) microscopy to confirm the absence of cellular fragments. Total fatty acid methyl ester (FAME) contents were quantified to verify the enrichment of fats in the final LB fraction (Supplementary Figure 4). LBs stained with the neutral lipid stain Nile red were further inspected under a confocal laser scanning microscope (CLSM) (Veerabagu et al., 2020). When visible contaminations were completely absent and all LBs were uniform and isodiametric in size, samples were prepared for protein precipitation, and extracted proteins were processed, trypsin-digested, and analyzed by liquid chromatography with tandem mass spectrometry (LC-MS/MS) as described in experimental procedures. MS/MS samples were analyzed using Mascot to search the P. trichocarpa database, whereas Scaffold was used to validate MS/MS-based peptide and protein identifications. We analyzed three independent pooled samples, obtained from different sets of plants that were grown at different times, representing biological replicates. To prevent loss of peripherally and transiently associated proteins that might be delivered to PD, such as, for example, the LB- and PD-localized γ-clade 1,3-β-glucanases, we deliberately omitted the commonly used salt step in the final wash of all three samples. However, the third sample received an extra wash, which diminished the number of identified proteins in the LB fraction. All proteins were identified with >99% probability, containing at least two unique peptides but often much more. In total, we identified 719 proteins in the LB fraction, of which many were still likely contaminants. To restrict the number of putative candidate proteins, we made a selection based on the previously reported presence of identified proteins at LBs or in LB fractions of plant and non-plant systems, localization at PD, or presence in PD-enriched fractions (Table 1).
A striking initial finding was that OLEOSIN proteins were not detected in the LB fraction of dormant buds while eight OLEOSIN genes were expressed in developing buds (Supplementary Table 1). This was unexpected as we anticipated that OLEOSINs would have remained on the LBs like in desiccated dormant seeds, where they stabilize the monolayer until germination commences (Siloto et al., 2006; Deruyffelaere et al., 2015; Shimada et al., 2018). As OLEOSINs were not detected, they were likely removed from LBs, possibly involving the ubiquitin-mediated 26S degradation system, which removes ubiquitinated OLEOSINs during Arabidopsis germination (Deruyffelaere et al., 2015, 2018; Kretzschmar et al., 2018).
To investigate the OLEOSIN disappearance further, we first validated that Populus OLEOSINs can be degraded by the ubiquitin-proteasome pathway. For this, we used PtOLE6, as OLE6 was highly expressed during the LB production phase (Figure 2A). Prediction of PtOLE6 ubiquitination sites with Bayesian Discriminant Algorithm Method (BDM-PUB; http://bdmpub.biocuckoo.org/index.php) showed the presence of at least six putative ubiquitination motifs (Supplementary Figure 5A). In Arabidopsis, OLE1-4 displayed one major and one or two minor ubiquitination sites (Deruyffelaere et al., 2015), and their alignment with PtOLE6 shows that the predicted K130 aligns with the major ubiquitination sites of AtOLE3-K159 and AtOLE4-K144 (Supplementary Figure 5B). We next investigated the involvement of the ubiquitin-proteasome-mediated degradation of PtOLE6 by overexpressing PtOLE6-eGFP in Arabidopsis. While PtOLE6-eGFP was degraded in the controls and the seedlings treated with the vacuolar cysteine protease inhibitor E64d (Deruyffelaere et al., 2015), seedlings treated with the proteasome inhibitor MG132 did not show degradation even after 72 h of imbibition (Figure 3A). Additionally, the MG132 treatment increased cytosolic accumulation of PtOLE6-eGFP (Figure 3B) in cytosolic aggregates, like in the case of AtOLE1 (Deruyffelaere et al., 2015).
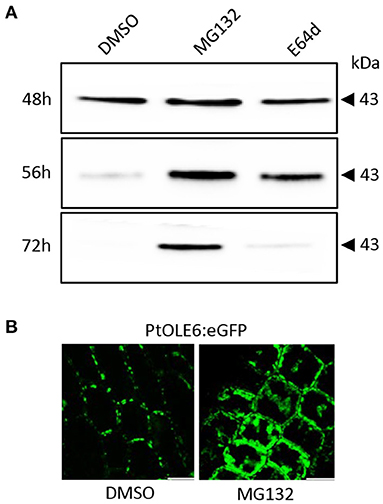
Figure 3. PtOLE6 degradation by the proteasome pathway. (A) MG132 inhibits proteasomal degradation of PtOLE6 in germinating Arabidopsis seeds. Arabidopsis PtOLE6-eGFP overexpressor seeds were germinated in DMSO control, MG132, and E64d for 48–72 h, and immunodetected using an anti-eGFP antibody. (B) Treatment with MG132 prevents degradation of PtOLE6-eGFP, resulting in cytosolic accumulation in epidermal cells of the hypocotyl adjacent to the radical. DMSO, control. Bar 50 μm.
Furthermore, we identified key components of the ubiquitin-mediated 26S degradation system in the LB fraction, such as Populus homologs of the Arabidopsis “plant UBX-domain-containing protein 10” (PUX10) and the segregase Cell Division Control Protein 48A (CDC48A) (Table 1) (Deruyffelaere et al., 2018; Kretzschmar et al., 2018). The bud LB fraction also contained the ubiquitin-activating enzyme UBA1, involved in conjugating ubiquitins to proteins, and several deubiquitinating proteases that prepare the unfolded proteins for degradation by the 20S core protease (Verma et al., 2002) (Table 1, Supplementary Table 1). In addition, the LB fraction contained multiple subunits of the 26S proteasome, such as the crucial 19S ATPase subunit RPT2A, which is important in meristem development (Lee et al., 2011), and 26S scaffolding components (Supplementary Table 1). Although these components are ubiquitous, they are likely to be involved in removing proteins from the LBs, namely, OLEOSINs, as they do so in germinating Arabidopsis seeds (Deruyffelaere et al., 2018).
The question remained how LBs retained their structural integrity during their enlargement (Figure 1) while OLEOSINs were absent from the LB fraction (Supplementary Table 1). A possible contributor to stability is the LB protein CALEOSIN1 (CLO1), which was the only CLO identified in the LB proteome (Table 1). However, we identified several other proteins that localize to LBs, such as the recently discovered class II protein Lipid Droplet Interacting Protein (LDIP) (Pyc et al., 2017; Coulon et al., 2020) and the Lipid Droplet Associated Proteins (LDAPs), both of which can contribute to LB stability (Gidda et al., 2016).
To assess the possibility that OLEOSINs were replaced by LDAPs/LDIP, we identified by phylogenetic analyses two P. trichocarpa homologs of PUX10 and CDC48A, six LDAP (LDAP1a, LDAP1b, LDPA2a, LDPA2b, LDAP3a, and LDAP3b), and one LDIP homolog (Supplementary Figures 6–9), and studied their expression in apices during bud development and dormancy establishment. Of the two PUX10 isoforms, PUX10b was upregulated during bud development and further during dormancy development (Figure 4A). Like in the case of PUX10a, the expression of one of the CDC48A isoforms, CDC48A2, was unaltered while CDC48A1 was upregulated (Figure 4B). The proteins encoded by the upregulated PUX10b and CDC48A1 genes remained present in the LB fraction of dormant buds (Table 1), suggesting they might also remove other ubiquitinated LB proteins. The expression patterns of the six LDAP genes were not identical, but all were upregulated during dormancy development, relative to expression levels in apices (Figures 5A–C). Notably, the major LDAP1b, which was upregulated during bud development, and excessively high (160-fold) during dormancy development, was little expressed in apices (Figures 5A,C). Also, the genes LDAP1a and LDAP3b were upregulated during bud development, while LDAP3a was not (Figure 5C). LDAP2a and LDAP2b were only slightly upregulated during bud development (Figure 5B). At this point, OLE6 was still very highly expressed (Figure 2A). The timeline shows that during dormancy development, when OLE6 was downregulated below the expression level in apices, all the five LDAP genes were strongly upregulated, especially LDAP1b (Figure 5A). In contrast, the upregulation of LDIP (ca. 5-fold; Figure 5D) was comparable to that of the other LDAPs.
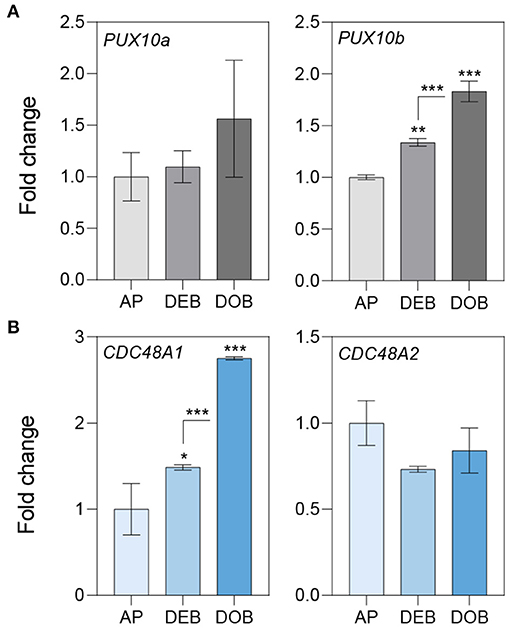
Figure 4. Expression of genes involved in OLEOSIN degradation. Relative expression of genes encoding (A) UBX Domain-Containing Proteins (PUX10a, b) and (B) Cell Division Control Proteins (CDC48A1,2) in apices (APs) under long day and in developing buds (DEBs) and dormant buds (DOBs) in short days. Values are calculated relative to the control (AP) and represent the means of three technical replicates ± SD of four plants. Asterisk(s) indicate statistical significance between treatments (one-way ANOVA, Tukey analysis; *P < 0.05; **P < 0.01; and ***P < 0.001).
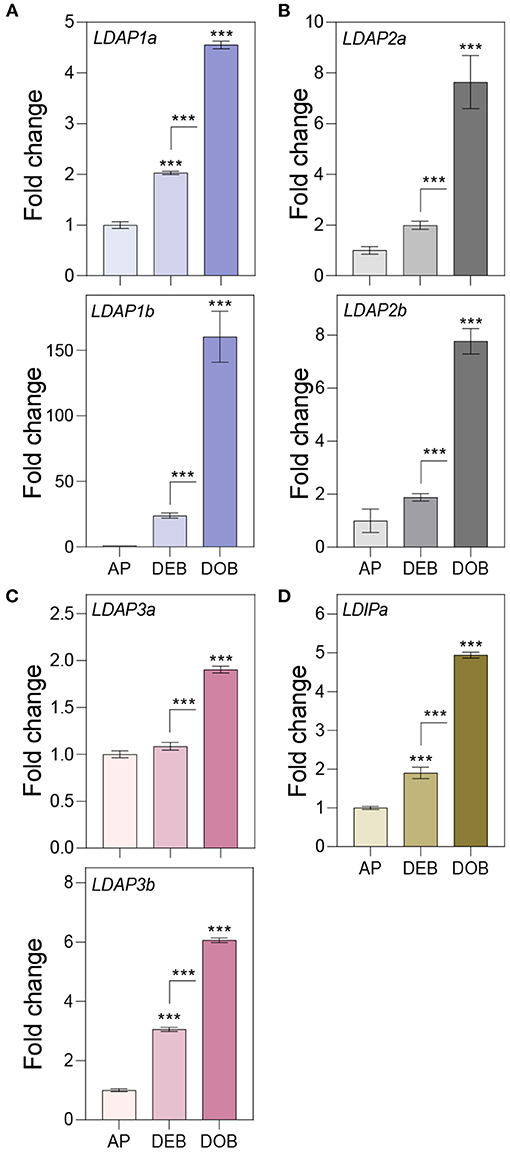
Figure 5. Expression of genes associating with the lipid body monolayer. Relative expression of genes encoding for (A–C) LDAPs and (D) LDIP in apices (APs) under a long day, and in developing buds (DEBs) and dormant buds (DOBs) in short days. Values are calculated relative to the control (AP) and represent the means of three technical replicates ± SD of four plants. Asterisk(s) indicate statistical significance between treatments (one-way ANOVA, Tukey analysis; ***P < 0.001).
In addition to the gene expression studies, we investigated if OLEOSIN and LDAP were present in extracts of buds at different developmental stages during growth and under dormancy-inducing conditions. In growing plants, antibodies detected OLEOSINs in younger developing buds above the bud maturation point (BMP), while LDAP1 was not detected (Figure 6A). In contrast, LDAP1 appeared to be abundant in full-grown buds below the BMP, which had ceased development and entered quiescence. Under dormancy-inducing conditions, OLEOSINs could not be detected in any bud, whereas LDAP1 was found in dormant buds and was even increasing over time in dormant buds (Figure 6B).
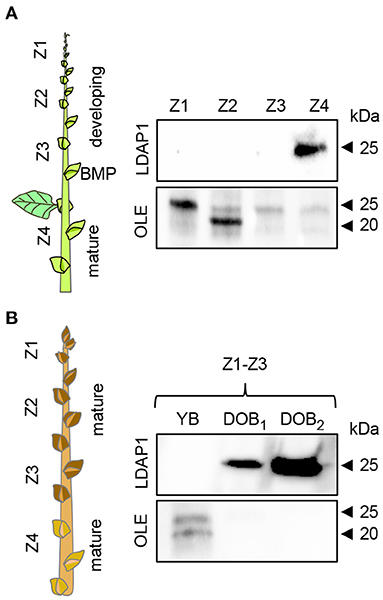
Figure 6. Immunodetection of LDAP1 and OLEOSIN proteins in axillary buds during their maturation under different daylength conditions. (A) Developing (zones Z1–Z3) and mature axillary buds (Z4) in reference to the bud maturation point (BMP) in long days. (B) Young axillary buds (YB) (Z1–Z3) in long days, and after 6 (DOB1, dormant buds 1) and 9 weeks (DOB2, dormant buds 2) in short days. Equal amounts of protein (10 mg) were loaded in wells.
That the Populus LDAPs can localize to LBs was demonstrated in leaf epidermal cells of N. benthamiana using a binary vector expressing the 35S promotor driven OLE6-eGFP fusion protein and binary vectors expressing LDAP1a-mRFP, LDAP1b-mRFP, LDAP2a-mRFP, LDAP3a-RFP, and LDAP3b-mRFP. The vectors were transformed into Agrobacterium tumefaciens and infiltrated into leaves for 2 days and investigated with CLSM. Captured images (Leica Application Suite X software) showed that all mRFP-tagged LDAPs were exclusively localized to OLE6:eGFP-tagged LBs (Figure 7). LDAP1b and LDAP3b did not localize to the smallest LBs, which only contained OLEOSIN. In brief, the upregulation of LDAPs, LDAPs presence in the LB fraction, and localization to LBs suggest that they were recruited to the monolayer after OLEOSIN removal. Together, these results suggest that OLEOSINs were degraded and replaced by LDAPs prior to bud completion and the establishment of a quiescent and dormant state.
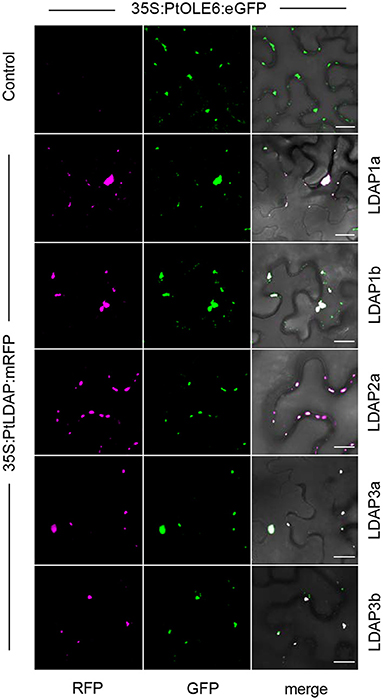
Figure 7. Localization of Lipid Droplet Associated Proteins (LDAPs) at lipid bodies in N. benthamiana. Representative confocal laser scanning microscopy images of leaf epidermal cells transiently co-expressing eGFP-tagged OLE6 and corresponding mRFP-tagged LDAPs (magenta). Single optical sections of RFP, GFP, and the merged images are shown for each set of experiments. Bars 50 μm.
The proteomic analysis of the bud LB fraction aimed to create an inventory of known and putative LB cargos that may contribute to cellular homeostasis, dormancy release and subsequent intercellular transport, and signaling in meristems. Their detection requires the omission of the salt wash (as shown above) that is commonly used to remove proteins that might associate with the monolayer during LB isolation, or are commonly considered contaminants. As argued above, this would also remove the peripherally associated proteins that hitch a ride to the PD. In buds, cytoplasmic proteins might associate with LBs when molecular crowding is reduced (Kory et al., 2015), and it seems possible that the enlarging LBs recruited a surplus of such proteins. These proteins may be stored at LBs and delivered to the PM and PD during dormancy release, serving membrane repair and renewed cell–cell communication. From the total fraction of 719 proteins, we selected 117 proteins, or ca. 16%, that potentially represent meaningful LB cargos (Table 1). As indicated, this selection was based on the reported presence of identical or similar proteins at LBs, in LB fractions, at PD, or in PD-enriched fractions (Fernandez-Calvino et al., 2011; Leijon et al., 2018). Since LBs frequently interact with other cell organelles and PD, we also included proteins that were reported or suggested to assist in LB-organelle-PM tethering, targeting, and hemifusion.
We detected in the LB fraction Caleosin 1 (CLO1), LDPI, LDAP3a, LDAP3b, LDAP1a, LDAP1b, Oil Body Lipase1 (OBL1a,b), GPAT8 (functionally redundant with GPAT4), and the 1,3-β-glucanase enzyme GH17_44 (Table 1). Like OLEOSIN, CLO1 has three structural domains, such as a pro-knot motif (Hsieh and Huang, 2004). Its major function is signaling, but at LBs, it contributes to monolayer stability (Chen et al., 1999). LDAPs associate with LB via amphipathic helices and similarly provide some structural integrity (Horn et al., 2013; Gidda et al., 2016; Kim et al., 2016). LDIP, which possesses a central hydrophobic sequence and few TMDs (Pyc et al., 2017), interacts with LDAP3 (Pyc et al., 2017) and may associate with nascent LBs (Coulon et al., 2020). OBL1 localizes to LBs in Arabidopsis seeds (Eastmond, 2006), and GH17_44 localizes to LBs in Populus meristems (Rinne et al., 2011). PUX10 and CDC48A are involved in the degradation of ubiquitinated proteins, such as OLEOSINs and might end up in the LB fraction attached to LB proteins. The LB fraction contained a dozen components of the 26S proteasome (Verma et al., 2002), such as the regulatory particle RPT2A and regulatory subunit RPN7, the ubiquitin-conjugating enzyme 13A (UBC13A), the ubiquitin-activating enzyme E11 (UAB1) (Table 1), and other 26S-associated proteins (Supplementary Table 1).
As LBs move in the actomyosin system motorized by myosin XI-k/1/2 (Veerabagu et al., 2020), it is of interest that the LB fraction contained two myosin XI tail binding GTPases. RAB-C2a, a homolog of the mammalian RAB18 that localizes to LBs, localizes in plants to peroxisomes that traffic on F-actin, like LBs (Hashimoto et al., 2008). RAB-D1 localizes to Golgi and endosomal vesicles (Pinheiro et al., 2009), which can interact with LBs (see discussion), while ARA6 localizes to LBs (Brasaemle et al., 2004; Fujimoto et al., 2004; Liu et al., 2004). The LB fraction contained only one actin, ACT7. This could be significant as ACT7 is associated with meristematic activity in germination and early plant development (McDowell et al., 1996; Kandasamy et al., 2001, 2009), is required in callus growth and present in PD fractions (Fernandez-Calvino et al., 2011). We also identified proteins that modulate actin dynamics, such as ADF4 and VAB2 (Table 1).
Proteins in the LB fraction that localize to PD components or the surrounding PM include, among others, (references in Table 1) Purple Acid Phosphatase (PAP1), two calreticulins (CRT1/CRT1A and CRT2/CRTB), two peroxidases (PRX36 and PRX37), the atypical 1,3-β-glucanase Zerzaust (ZET), 1,3-β-glucanase1 (BG1/GH17-37), Dehydrin HIRD11, Reticulon like protein B1 and B3 (RTN3), an inactive receptor kinase, Plasma Membrane Intrinsic Protein 1 and 3 (PIP1 and 3), Calnexin 1 (CNX1), and Bcl-2 Unfolding Protein BAG7 (Table 1; Supplementary Table 1). Of these, five were present in the Populus PD fraction (Leijon et al., 2018) and nine in that of Arabidopsis (Fernandez-Calvino et al., 2011). Several of these proteins were also previously localized to LBs, such as CNX1, RTN1, and RTN3 (Table 1).
Proteins previously found in LB fractions of plant and non-plant systems included LEA proteins, which protect cellular structure and PD during dehydration stress (Karlson et al., 2003). Among these were ERD7, LEA26, and LEA27 (Table 1). Furthermore, we identified alcohol dehydrogenases (ADH1-3), annexins (ANN1,2), transport family protein SecY, sterol methyltransferase (SMT2), embryo-specific protein ATS3, and Cytochrome P450 (Table 1).
The LB fraction also contained proteins involved in tethering, membrane trafficking, and membrane fusion, such as the CalB domain Synaptotagmin 2 (SYTB/SYT2), which is a C2 tethering protein, Syntaxins (SYP71) and other SYPs, the vesicle/protein transporter ADP-ribosylation factor 1 (ARF1), and the endosomal protein Guanine Nucleotide-Exchange (GEF), which recruits ARF1 to vesicles (Table 1, Supplementary Table 1). Moreover, a dozen Rab GTPases were present. These included ARA-3 (RAB8a), Golgi localized RAB-6A, and Suppressor of Variegation 11 (SVR11/RABE1B) (Table 1, Supplementary Table 1). The presence of CPOII subunits and COPI Coatomer subunits in conjunction with ARF1 is of interest, as they may mediate protein trafficking to and from LBs (Soni et al., 2009) (see discussion).
Other identified proteins potentially reflect organellar interaction, protein folding and unfolding at organelles and PD, lipid metabolism, storage, detoxification, and desiccation stress. Unfolding/folding proteins in the bud LB fraction included the chaperones/Heat Shock proteins Hsp-90-4, Hsp90-6, Hsp90-7, the Hsp70 proteins BIP2 and BIP3, and homologs of the cpn60 chaperonin TCP-1/CCT8. Of the TCP-1 chaperonin family proteins, 10 members were present (Table 1, Supplementary Table 1). Proteins related to lipid metabolism included Phospholipase C (PLC2), GDSL-motif esterase (acyltransferase), and the lipase Plat Domain Protein 2 (PLAT2). Also identified were three Histones, previously shown to be stored at LBs (Cermelli et al., 2006). The identified antioxidant enzymes may protect stem cells of the embryonic shoot from hypoxia-induced damage in the low-oxygen environment of the bud (Ophir et al., 2009; Meitha et al., 2015). They include Catalase 2 (CAT2), Thioredoxin-dependent peroxidase 1 (TPX1), the peroxidases PRX36 and PRX37, and Manganese Superoxide Dismutase (MSD1).
A direct comparison of the LB fraction of dormant buds and the PD fraction of cell suspension cultures of P. trichocarpa (Leijon et al., 2018) uncovered 19 identical proteins (Table 2). A part of these shared proteins was also identified in the PD fraction of Arabidopsis cell suspensions (Table 2). The presence of LDIP in the Populus PD fraction (Leijon et al., 2018) is of interest because LDIP is a LB protein (Pyc et al., 2017). Of interest is also the syntaxin SYP71, a plant-specific SNARE protein involved in vesicle docking and fusion (Suwastika et al., 2008) that is present in PD fractions of both Populus and Arabidopsis (Table 2). SNAREs can mediate LB tethering to distinct compartments, LB-LB fusion, and hemifusion with bilayer membranes (Boström et al., 2007; Murphy et al., 2009). The GTPase ARA-3 (RAB8a), which mediates vesicle docking, the PD-localized Calreticulin (CRT1/CRT1A) (Baluska et al., 2001), and the LEA protein ERD4 were present in both fractions. Likewise, the following proteins of the bud LB fraction were present in both PD fractions: ZET (Vaddepalli et al., 2017), an O-Glycosyl hydrolases family 17 protein, and the GPI-anchored protein SKU5 similar 1 (SKS1). Some proteins exclusively shared by the LB fraction and the Populus PD fraction were the Thioesterase superfamily protein, involved in fatty acid biosynthesis, the seed LB candidate protein Embryo-Specific Protein 3 (ATS3) (Vermachova et al., 2011; Kretzschmar et al., 2020), a leucine-rich receptor kinase (probably inactive), PAP1, the cytoplasmically localized Reversible Glycosylated Polypeptide 3 (RGP3), the aquaporin Plasma Membrane Intrinsic Protein 1 (PIP1), the PHB-domain protein PHB6, the antioxidant TPX1, and the hydrolase S-Formylglutathione Hydrolase (SFGH).
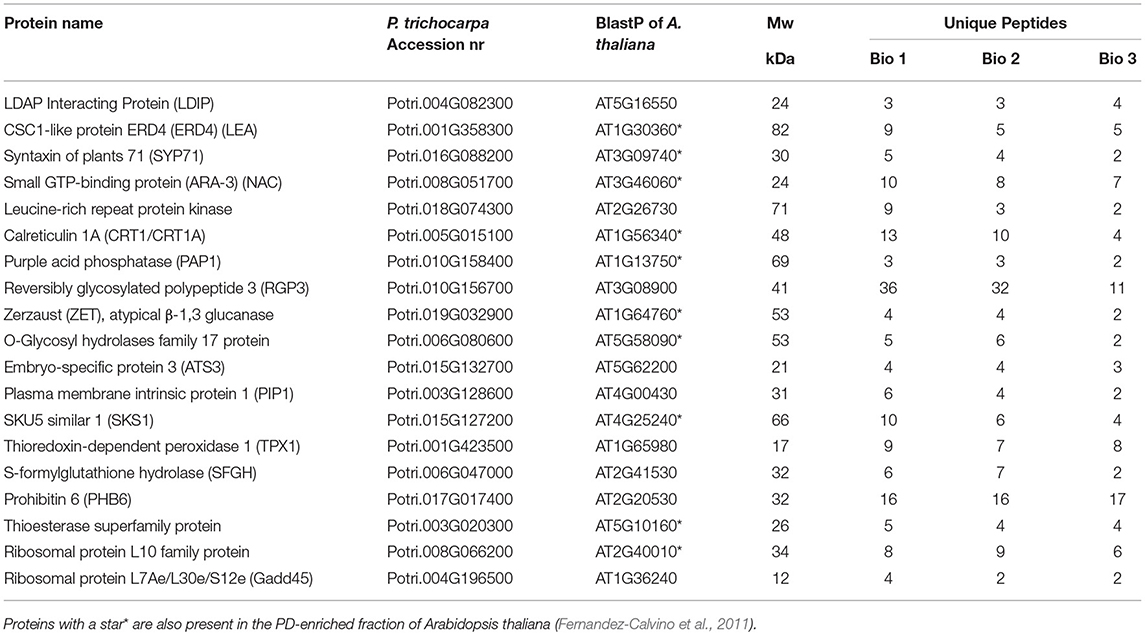
Table 2. Proteins that are shared by the bud LB fraction of Populus and the PD-enriched fraction of Populus trichocarpa Leijon et al. (2018).
OLEOSINs stabilize the LB monolayer, preventing coalescence and fusion during seed desiccation and dormancy (Siloto et al., 2006; Shimada et al., 2008; Hsiao and Tzen, 2011). Perennial buds also desiccate, albeit partially (Rinne et al., 2015), express OLEOSIN genes, produce OLEOSINs, accumulate LBs, and establish dormancy. The major gene OLE6 was strongly upregulated during bud formation (Figure 2A). Given these similarities, it came as a surprise that none of the eight Populus OLEOSINs were detected in the LB fraction of dormant buds (Table 1; Supplementary Table 1). OLE6 was completely downregulated prior to dormancy establishment (Figure 2A), while the minor genes OLE3 and OLE5 were expressed very little. The apparent absence of OLEOSINs in the LB fraction of dormant buds suggests they were degraded prior to dormancy establishment, likely by the machinery that also degrades OLEOSINs during seed germination (Deruyffelaere et al., 2018). Core components of this machinery were found in the Populus LB fraction, such as the adaptor protein PUX10, the segregase CDC48A (AAA ATPase Cell Division Cycle 48) and various 26S components, the ubiquitin-activating enzyme UBA1, the crucial 19S ATPase subunit RPT2A, and several deubiquitinating proteases, which are required for insertion of proteins into the 20S core (Table 1, Supplementary Table 1). PUX10 associates with LBs, and interacts with ubiquitin through its UBA domain and with CDC48A through its UBX domain, resulting in the degradation of ubiquitinated OLEOSINs by the 26S proteasome (Hsiao and Tzen, 2011; Deruyffelaere et al., 2015, 2018; Kretzschmar et al., 2018). As AtPUX10 is an integral class I LB protein that binds ubiquitins and recruits CDC48A (Deruyffelaere et al., 2018), it is perhaps not surprising that these components were present in the LB fraction of Populus buds. The predicted K130 ubiquitination site of PtOLE6 aligned with the major ubiquitination sites of AtOLE3-K159 and AtOLE4-K144 (Supplementary Figure 5B). Moreover, we found that in germinating seeds of an Arabidopsis, PtOLE6 overexpressor PtOLE6 is degraded like AtOLE1-4 (Deruyffelaere et al., 2015), and that degradation is partially inhibited by proteasome inhibitors E64d and blocked by MG132 (Figure 3). It seems likely, therefore, that this degradation mechanism is responsible for the early removal of OLEOSINs from LBs of buds, although direct localization of these proteins to the LBs remains to be demonstrated. During bud formation, the major gene OLE6 was strongly upregulated, producing OLEOSIN protein and many small LBs (Figures 1, 2). During dormancy establishment, OLE6 was downregulated below the level of growing apices (Figure 2A); and while PUX10b and CDC48A1were upregulated (Figures 4A,B), OLEOSINs were no longer detected (Figure 6B), and LBs had enlarged (Figure 1E). In correspondence with OLE6 downregulation, LDIPa, LDAP1, LDAP2, and LDAP3 were upregulated (Figure 5); and their encoded proteins, LDIP, LDAP1, and LDAP3, were detected in the bud LB fraction instead of OLEOSINs (Table 1). In brief, the data support the hypothesis that OLEOSIN is required for initiation of LB production in buds, but that prior to dormancy establishment OLEOSINs are removed by the PUX10/CDC48a degradation mechanism. PUX10 and CDC48A also likely degrade other LB proteins (Deruyffelaere et al., 2018), as they remain in LBs after OLEOSIN is removed.
That OLEOSINs are replaced by LDAP1 during LB enlargement is supported by the following facts. First, in long-day plants, OLEOSIN was detectable in extracts of developing buds and virtually absent in mature buds, while the reverse was true for LDAP1 (Figure 6A). Second, in short-day plants, OLEOSINs could not be detected in dormant buds, while LDAP1 was abundant (Figure 6B). Indeed, in general LDAPs might not interact with OLEOSIN-covered LBs, because LDAPs do not possess a hydrophobic hairpin and their relative hydrophilicity allows LB association only in detergent-sensitive manner (Gidda et al., 2016; Kim et al., 2016; Huang and Huang, 2017; Pyc et al., 2017). LDAPs and many of the identified LB fraction proteins might have been recruited to the expanding monolayer surface after OLEOSIN degradation, such as CLO1 (Table 1), which binds competitively with the monolayer (Huang, 2018). Noteworthy is that in N. benthamiana, the mRFP-tagged LDAP1b and LDAP3b, did not localize to the smallest OLE6-eGFP-tagged LBs (Figure 7), suggesting that these two proteins associate only with an expanded monolayer. LDIP anchors itself via an amphipathic helix in the monolayer and interacts with LDAP3 (Pyc et al., 2017) but is confined to a small area of the LB while LDAPs cover the entire monolayer (Coulon et al., 2020). LDIP was suggested to facilitate the neo-formation of small LBs (Coulon et al., 2020) in cooperation with SEIPINs (Cai et al., 2015; Barbosa and Siniossoglou, 2017). As LDIPa was upregulated during the LB accumulation phase in developing buds (Figure 5D), it seems possible that it did assist later LB emergence. LB expansion might be driven by increased TAG biosynthesis (Figures 1E, 2B), possibly involving the ER-localized enzyme GPAT8 (Table 1) (Gidda et al., 2009), which functions redundantly with GPAT4 (TAIR). Both localize to LBs in Arabidopsis (Fernández-Santaso et al., 2020) and in mammalian and insect cells (Wilfling et al., 2013).
Seed germination involves OLEOSIN degradation, LB enlargement, and recruitment of cytoplasmic proteins that mediate lipolysis and promote seedling growth (Deruyffelaere et al., 2015; Thazar-Poulot et al., 2015; D'Andrea, 2016). Germination cannot occur when the TAG lipase SDP1 is absent (Eastmond, 2006). While in buds SDP1a and SDP1b were slightly upregulated under short days (Figure 2C), the enzymes were absent from the LB fraction (Supplementary Table 1), and LB degradation was not observed. Either way, SPD1 transcript levels might not reflect enzyme activity because it might be post-transcriptionally regulated (Eastmond, 2006), or it is not delivered to LBs by the glyoxysomes that produce them (Thazar-Poulot et al., 2015) but kept in store for later bud activation. Notably, the retromer subunit VPS29 that mediates peroxisome tubulation to deliver SPD1 to the LBs was absent from the LB fraction (Supplementary Table 1).
LBs function in lipid homeostasis, protein sequestration, and membrane and cargo trafficking (Murphy, 2012). As a result, LB fractions can contain tens or even hundreds of proteins (Bartz et al., 2007; Hodges and Wu, 2010; Brocard et al., 2017; Zhi et al., 2017), most of which lack canonical LB functions. These “refugee” proteins (Hodges and Wu, 2010) are of interest, highlighting the dynamics of protein exchanges. Our proteomic analyzes were aimed at creating an inventory of such transiently associated proteins, as they could be involved in cellular homeostasis, dormancy release, and intercellular transport and signaling. The 719 identified proteins were present in the LB fractions of three independent experiments (Table 1, Supplementary Table 1), and among these were 117 proteins that included known LB proteins and proteins that were previously identified in LB fractions of various systems (Table 1). They relate to the ER, lipid metabolism, cytoskeleton, membrane trafficking, organellar interaction, membrane tethering and fusion, chaperone function, and signaling (Zehmer et al., 2009; Hodges and Wu, 2010; Gao and Goodman, 2015). Some of them might serve the unique needs of dormant bud meristems, which have reduced metabolism, face environmental and endogenous stresses, and maintain a dormancy-release mechanism (Rinne et al., 2001, 2011). In the absence of OLEOSINs, which mitigate desiccation and freezing stress (Siloto et al., 2006; Shimada et al., 2018), LDAPs, CLO1, and hydrophilins, such as LEA proteins (Table 1), might confer stability to LBs (Gidda et al., 2016; Kim et al., 2016). The bud, as a survival structure, must protect the enclosed embryonic shoot and its resident stem cells against environmental and endogenous onslaughts. Considering that the enclosed meristem resides in a hypoxic internal bud environment (Ophir et al., 2009; Meitha et al., 2015), the identified antioxidants might function to protect the stem cells. Notably, the LB fraction contains TPX1, the peroxidases PRX36, PRX37, CAT2, and MSD1 (Table 1), which, like in Drosophila (Bailey et al., 2015), can mitigate the effects of hypoxia-induced ROS. LBs may also detoxify stem cells by removing cytoplasmic free fatty acids, resulting from membrane degradation and remodeling during desiccation (Listenberger et al., 2003). Given the embryo-like nature of the embryonic shoot, the presence of histones H2A, H3, and H4 in the LB fraction is significant. Drosophila stores maternal H2A, H2B, H3, and H4 at LBs in its eggs as a repository for early embryogenesis (Cermelli et al., 2006; Li et al., 2012). Storage at LBs keeps histones available for immediate use while avoiding the cytotoxic effects of free histones (Gunjan and Verreault, 2003). In bud meristems, they could serve a similar purpose.
As shown previously, LBs do not arrive at PD by simple cytoplasmic streaming but by processive trafficking in the actomyosin system, which is severely impaired in an Arabidopsis 3KO myosinxi-k/1/2 mutant (Veerabagu et al., 2020). Myosin XI-K is the most important among the 13 class XI myosins of Arabidopsis and is responsible for the movement of mitochondria, peroxisomes, and endomembrane vesicles (Avisar et al., 2008, 2009; Sparkes et al., 2008; Peremyslov et al., 2015). The binding of LBs to myosin XIs, like that of other organelles, might require Rab GTPases. We identified in the LB fraction two “myosin XI tail-binding” proteins, the Rab GTPase homolog C2A (RAB-C2A) and the Rab GTPase homolog D1 (RAB-D1) (Table 1). In Arabidopsis, these factors interact with the C-terminal tail region of MYA2 (an isoform of myosin XI). While RAB-C2A specifically localizes to peroxisomes (Hashimoto et al., 2008), RAB-D1 associates with the Golgi apparatus (Pinheiro et al., 2009). A recent study on the Populus Rab family found that RAB-C2A did not localize at Golgi, TGN, or endosomes but instead at small unidentified vesicles (Zhang et al., 2018), which could have been peroxisomes but possibly also LBs. The presence of RAB-C2A in the LB fraction (Table 1) suggests that RAB-C2A is the receptor that anchors LBs to myosin XI-k/1/2, enabling motility, interaction with cell organelles, and targeting of PD. If so, the motility of LBs and peroxisomes on the same RAB-C2A/Myosin XI/actin assembly might explain their frequently observed interaction (Mathur et al., 2002; Hashimoto et al., 2008; Veerabagu et al., 2020). Notably, RAB18, a mammalian homolog of RAB-C2A, is present at LBs and in LB fractions (as shown in references in Table 1). The presence of Golgi-related RAB-D1 in the LB fraction (Table 1) suggests an interaction between LBs and Golgi- and endosomal vesicles (see below). LBs might move on ACT7, as it is the only actin identified in the bud LB (Table 1) and seed LB fractions of Chinese tallow (Zhi et al., 2017). ACT7 is required in seed germination, early plant development, and meristem proliferation (McDowell et al., 1996; Kandasamy et al., 2001, 2009), and it is found in the PD fraction of Arabidopsis callus cells (Fernandez-Calvino et al., 2011). The presence of actin-stabilizing VAB2 (Ma et al., 2012) and PD-localized actin-severing ADF4 in the LB fraction (Table 1) appears to reflect the dynamics of actin-mediated transport.
As argued by Gao and Goodman (2015), organelle markers, such as ER luminal chaperones and mitochondrial proteins, are so frequently associated with thoroughly purified LBs that it is unlikely that they all represent contaminants. At least some of them might reflect the frequently observed LB-organellar interactions (Murphy et al., 2009; Zehmer et al., 2009), facilitated by their movement on F-actin (Mathur et al., 2002; Van Gestel et al., 2003; Hashimoto et al., 2008; Veerabagu et al., 2020). We identified some mitochondrial proteins, vacuolar proteins, and lipid metabolism proteins, as well as Annexins, Calnexin, LEA proteins, Cytochrome P450s, transport proteins, Alcohol Dehydrogenases, and Embryo-Specific Protein3 (Table 1, Supplementary Table 1). Several of these are present in seed LB fractions and PD fractions (as shown in references in Table 1). The identified annexins putatively link LBs to the PM and PD, as they can bind F-actin, regulate membrane trafficking, bind phospholipids and Ca2+, inhibit callose synthase at PD, act as peroxidases, and induce membrane curvature (Delmer and Potikha, 1997; Clark et al., 2012; Boye et al., 2018).
Organellar interactions involve chaperone-like molecules that mediate membrane and vesicle transport, tethering, and membrane (hemi-)fusion. Chaperones transport, fold and unfold proteins during autophagy, and function at peroxisomes, mitochondria, chloroplasts, and ER (Boston et al., 1996; Kriechbaumer et al., 2012). We identified Synaptotagmin B, various Syntaxins, ARF1 and ARF-related proteins, and transport-related RAB GTPases (Table 1). The identified calnexins and several of the chaperones (Table 1) were also present in the LB fraction of the Chinese tallow (Zhi et al., 2017). In the SAM, LB-resident chaperones, delivered to PD, could assist in the documented unfolding and refolding of the non-cell-autonomous proteins that move cell-to-cell (Aoki et al., 2002). Essential in SAM function is the intercellular movement of the conserved transcription factor SHOOT MERISTEMLESS (STM) in Arabidopsis, and KNOTTED1 (KN1) in maize, which requires chaperon-assisted unfolding in the donor cell and chaperonin-assisted refolding in the destination cell (Kitagawa and Jackson, 2017). The LB fraction contained, among others, the CCT8 subunit of chaperonin CTP-1 (AT3G03960) that refolds STM/KN1 in the destination cell (Xu et al., 2011; Kitagawa and Jackson, 2017). Although CCT8/CTP-1 localization at LBs remains to be shown, LBs that dock at opposite sides of PD (Rinne et al., 2011; Veerabagu et al., 2020) could engage in unfolding proteins for export and refolding incoming proteins in the SAM.
Lipid bodies can target PD to deliver enzymes that enhance PD conductivity (Rinne et al., 2001, 2011; Veerabagu et al., 2020). Part of the identified proteins may similarly arrive at the PD interior and surrounding PM via the LB shuttle. This seems feasible, as many of the proteins in the LB fraction have been localized to PD, such as acid phosphatases (Esau and Charvat, 1975), calreticulins (Baluska et al., 2001), calnexins (Liu et al., 2017), and reticulon (Knox et al., 2015; Kriechbaumer et al., 2015).
Reticulon, present in the LB fraction of insect cells, adopts a hairpin-like topology in the outer ER leaflet (Beller et al., 2008). It is of interest, therefore, that reticulon3 (RTN3) is present in the bud LB fraction (Table 1), and that it is identified in the PD fraction (Fernandez-Calvino et al., 2011) and localized at PD (Knox et al., 2015; Kriechbaumer et al., 2015). This suggests the possibility that in meristems RTN3 could arrive at the desmotubule via LBs. The dehydrin HIRD11 (Table 1) may protect PM- and PD-localized proteins during winter, such as the PD-localized dehydrin in cold-acclimated tissue of dogwood (Karlson et al., 2003). Similarly, the γ-clade GH17-family protein (Table 1) localizes to PD (Rinne et al., 2011). RGP3 might also localize to PD, like RGP2 (Sagi et al., 2005). Proteins in the LB fraction, some of which localized to PD, were also identified in PD fractions of both Arabidopsis (Fernandez-Calvino et al., 2011) and P. trichocarpa (Leijon et al., 2018), such as ERD4, Syntaxin of Plants 71 (SYP71), small GTP-binding protein ARA-3 (NAC/RAB8a), Calreticulin 1A (CRT1A), PAP1, ZET, O-Glycosyl hydrolases family 17 protein, SKU5 similar 1 (SKS1), and Thioesterase superfamily protein (Table 2). The presence of the LB protein LDIP and the candidate LB protein ATS3 (Vermachova et al., 2011; Kretzschmar et al., 2020) in the Populus PD proteome (Leijon et al., 2018) and in the LB fraction (Table 1) is an independent confirmation that LBs target PD.
The number of proteins in the LB fraction that are destined for PD might be much larger than that which is found in enriched PD fractions of cell cultures because they contain only young primary PD (Fernandez-Calvino et al., 2011; Leijon et al., 2018). In complex three-dimensional tissues, such as bud meristems, LBs dock at both primary and secondary PD (Rinne et al., 2011; Veerabagu et al., 2020). The overlap with available PD proteomes might, therefore, underestimate the LB proteins that are delivered to PD in meristems.
The presence of some ER-luminal proteins and GPI-anchored proteins in the LB fraction appears puzzling. A possible explanation is that they might reside inside a subset of LBs. Most LBs are formed at the cytoplasmic leaflet of the ER, which then determines the composition of the monolayer and its associated proteins (Guo et al., 2009; Walther and Farese, 2009). Alternatively, an entire oil lens may be cut out from the ER, resulting in a bicelle LB, in which the monolayer is derived from both ER leaflets (Ploegh, 2007). Such LBs possess proteins that reside on both sites of the ER membrane and include luminal ER-anchored proteins. In addition, during “vesicular budding” (Walther and Farese, 2009), minute cytoplasmic bilayer vesicles are produced and tethered to ER, while neutral lipids are imported into the bilayer by a lipid shuttle. As a result, the growing LB can contain a minuscule aqueous inclusion inside the remnants of the luminal ER leaflet, as also observed in birch meristems (Rinne and van der Schoot, 2004). In the last two cases, LBs can contain luminal ER proteins, for example, calnexin and BIP (Table 1), which are indeed frequently found in LB fractions (Brasaemle et al., 2004; Liu et al., 2004; Umlauf et al., 2004; Ploegh, 2007). LBs might also recruit organellar proteins. For example, LBs interact with ER, the Golgi apparatus, and early and late endosomes (Beller et al., 2010), and the coatomer assemblies SAR1-COPII and GEF-ARF1-COPII are implicated in protein delivery to LBs (Soni et al., 2009). Moreover, Arf1/COP1 locates to LBs, probably directing ER proteins to bridges that connect the ER to LBs (Bartz et al., 2007; Beller et al., 2010; Wilfling et al., 2014; Gao and Goodman, 2015; Brocard et al., 2017; Huang et al., 2019). It is of interest, therefore, that COPII and SAR1/2, as well as COPI, GEF (BIG5), and ARF1, are present in the bud LB fraction (Table 1).
The current inventory of putative LB-associated proteins lays out a road map for future investigations into the shuttle function of LBs in plant meristems, serving cellular metabolism, organellar interaction, and uniquely for plants, transport to and through PD.
Hybrid aspen (Populus tremula × Populus tremuloides) clone T89 plants were micro propagated, moved to a glass house, and grown under an 18-h long day (Veerabagu et al., 2020). When the plants reached a height of 80–100 cm, half of them were subjected to 10-h short days to induce dormant buds. We investigated meristems in three distinct phases: actively proliferating apices of growing long-day plants (APs), apices inside developing buds (DEBs, 3–4 weeks old), and dormant apices in completed terminal buds (DOBs, 6–9 weeks old).
P. trichocarpa OLE6-eGFP was overexpressed in Arabidopsis thaliana (Col-0), and homozygous lines were obtained as previously described by Veerabagu et al. (2020). Transgenic Arabidopsis PtOLE6-eGFP seeds were germinated on Whatman paper, wetted with 2 ml of 0.2% DMSO (control), 200 μM MG132, or 40 μM E64d (Deruyffelaere et al., 2015). Seeds were first subjected to 72 h of stratification at 4°C in the dark and later germinated under continuous light for 48–72 h as indicated at 25°C.
Shoot apices of growing plants (APs), developing buds (DEBs, 4 weeks) and dormant buds (DOBs, 8 weeks) were fixed overnight in 2% (v/v) glutaraldehyde and 3% (v/v) paraformaldehyde at 4°C in 100 mM phosphate citrate buffer (Rinne et al., 2001). Because of the density of the bud tissues, the infiltration into LR white resin (LRW) was done gradually by increasing the LRW concentration stepwise from 30 to 70%, and, finally, incubating 4–7 days in 100% LRW. The resin was polymerized at 55°C for 24 h. Samples were trimmed longitudinally, and 1 μm thick median sections were stained with 1% (w/v) aqueous toluidine blue for light microscopical observation (Leica DM6B, Leica Microsystems, Wetzlar, Germany) and imaging with a digital camera (Leica DMC4500, Leica Microsystems, Wetzlar, Germany). Subsequently, ultra-thin 80 nm sections were cut from samples and observed with a TEM (FEI Morgagni 268 FEI Company, Eindhoven, The Netherlands), which is electronically set to produce high-contrast LBs.
The number and sizes of LBs were assessed manually in median longitudinal TEM sections, restricted to two areas in the SAM and in the subjacent rib meristem/rib zone region (RM/RZ) (Figures 1A–C). Per photoperiod 2–3 replicate apices were used, and LB number and sizes were assessed in two TEM sections. As the chance to section through a LB increases with LB size (diameter), the numbers were corrected relative to DOB. In addition, we calculated the total number of LBs per average cell volume, considering section thickness and LB diameter, as explained in Supplementary Figure 1.
Complementary DNA (cDNA) preparations derived from dormant buds were used as a template to amplify all the LDAPs. The LDAP entry clones were obtained by BP reaction in pDONR207 and verified by sequencing. The binary constructs for mRFP fusion protein expression under the control of the 35S promoter were obtained via LR reaction using the respective LDAP entry clones and the destination vector pB7WGR2 (Karimi et al., 2002). For qPCR analyses, we collected apices from growing long-day plants (APs), and from short-day plants with developing buds (DEBs, 3 weeks), and dormant buds (DOBs, 6 weeks). Three biological replicates, each containing pooled samples, were frozen in liquid nitrogen. RNA was extracted from 0.2–0.3 g of frozen samples and analyzed with quantitative qRT-PCR, as described before (Katyayini et al., 2019). All clones were constructed using the Gateway™ technology (Invitrogen, Waltham, MA, United States). The binary vector pK7FWG2 expressing PtOLE6-eGFP fusion protein was constructed as described previously by Veerabagu et al. (2020). cDNA preparations derived from dormant buds were used as a template to amplify all the LDAPs. The LDAP entry clones were obtained by BP reaction in pDONR207 and verified by sequencing. The binary constructs for mRFP fusion protein expression under the control of the 35S promoter were obtained via LR reaction using the respective LDAP entry clones and the destination vector pB7WGR2 (Karimi et al., 2002). Gene-specific primer sequences for qPCR analyses were designed using Primer3. The list of primers and genes used for qPCR and cloning are presented in Supplementary Tables 2, 3.
Dormant buds were collected from short-day (9 weeks) exposed plants and used for LB purification and repeated in three independent experiments (Bio 1–3). Freshly harvested buds weighing 5–8 g were kept on ice until all the hard bud scales were carefully removed. The material was rinsed thrice in cold distilled water and then processed for LB extraction and purification. For LB purification, we used a previously described method (Jolivet et al., 2004) with two modifications. First, we deliberately omitted the NaCl ionic elution step to avoid discarding the loosely attached LB-associated proteins, which we aimed to identify. Second, proteins were precipitated using ice-cold methanol instead of hexane. Bio 1–2 received the prescribed 1X Tween20 wash, but Bio 3 was washed once more with Tween20 to increase stringency. The purity of the LB fraction was checked under light and DIC microscope to confirm absence of cellular contaminations. Total FAME measurement was carried out to assess the enrichment of fats in the LB fractions (O'Fallon et al., 2007). A fraction of the floating “fat pads” containing purified LBs was collected and inspected under the confocal microscope to confirm its purity and integrity (Supplementary Figure 4C). The fat pad, which was enriched with LBs (Supplementary Figure 4D), was further processed for protein precipitation (Veerabagu et al., 2020).
To investigate the production of LDAP1 and OLEOSIN in Populus buds by immunodetection, axillary buds were obtained from different developmental zones of long and short-day plants, as indicated in Figure 6. Proteins were extracted using the 2x Laemmli buffer, and equal amounts of protein (10 mg) were loaded in each well. Western blot analyses were carried out using primary antibodies of A. thaliana anti-OLE1 polyclonal antibody (PhytoAB-PHY0954A) at a dilution ratio of 1:2000, Hevea brasiliensis anti-SRPP1 (LDAP1) monoclonal antibody (Abcam-ab138711, Abcam, Cambridge, United Kingdom) at a dilution ratio of 1:1000, and anti-eGFP monoclonal antibody (Thermo Fisher-MA1-952, Thermo-Fisher Scientific, Waltham, MA, United States) at a dilution ratio of 1:2000. Goat anti-rabbit IgG H&L (HRP) antibody phytoAB-PHY6000 at a dilution ratio of 1:5000 was used as a secondary antibody for OLE1. Goat anti-mouse IgG H&L (HRP) (Abcam-ab205719) at a dilution ratio of 1:5000 was used as a secondary antibody for both LDAP1 and eGFP antibodies. Immunoblot signals were detected with the Clarity Western ECL kit (Bio-Rad, Hercules, CA, United States) and imaged with a Chemidoc (Bio-Rad, Hercules, CA, United States) digital imager.
Degradation of OLEOSIN and inhibition of degradation were studied in imbibed seeds of transgenic PtOLE6-eGFP-expressing Arabidopsis seedlings (Figure 3). Proteins were extracted from imbibed PtOLE6-eGFP transgenic seeds and immunoblotted using an anti-eGFP monoclonal antibody (Thermo Fisher-MA1-952, Thermo-Fisher Scientific, Waltham, MA, United States) and detected in Western blots as described above.
In short, the LB pellets were rinsed with methanol and air-dried. Subsequently, proteins were extracted in appropriate volumes of 2x Laemmli buffer. The third biological replicate received an additional wash, which further purified the sample. The extracted proteins were loaded on a 10% Bio-Rad mini protein gel (Bio-Rad, Hercules, CA, United States) and allowed to run ~1.5 cm into the gel. The protein gel was stained with Coomassie Brilliant Blue (MilliporeSigma, Burligton, United States) for an hour and destained overnight. The region of the gel showing stained proteins was isolated and excised into six fractions. The gel pieces were reduced (DTT) and alkylated (iodoacetamide) before overnight digestion with trypsin. After digestion, the peptides were extracted from the gel by sonication in a 0.1% TFA solution and cleaned up by a reversed-phase (C18) spin-tip procedure. Eluted peptides were dried and re-dissolved in a loading solution (2% acetonitrile, 0.05% TFA in MilliQ water). Samples were analyzed by LC-MS/MS, with Ultimate™ 3000 RSLC nano system coupled with Q Exactive hybrid quadrupole-orbitrap (Thermo-Fisher Scientific, Waltham, MA, United States). Thermo raw files were converted using the msfileconvert module of the Proteowizard (v 3.0.7076) software suite. All MS/MS samples were analyzed using Mascot (Matrix Science, London, UK; version 2.6.1). The mascot was set up to search the P. trichocarpa database (txid_3694, 53583 entries). The mascot was searched with a fragment ion mass tolerance of 0.02 Da and a parent ion tolerance of 10.0 PPM. Carbamidomethylation of cysteine was specified in Mascot as a fixed modification. Deamidation of asparagine and glutamine, oxidation of methionine, and acetylation of the n-terminus were specified in the Mascot as variable modifications.
Scaffold (version Scaffold 4.9., Proteome Software Inc., Portland, OR, United States) was used to validate MS/MS-based peptide and protein identifications. Peptide identifications were accepted if they could be established at >95% probability by the Scaffold Local FDR algorithm. Protein identifications were accepted if they could be established at >99% probability and contained at least two identified peptides. Protein probabilities were assigned by the Protein Prophet algorithm (Nesvizhskii et al., 2003). Proteins that contained similar peptides and could not be differentiated based on MS/MS analyses alone were grouped to satisfy the principles of parsimony. Proteins were considered identified only if in all three biological replicates they contained at least two unique peptides. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE* (Perez-Riverol et al., 2019) partner repository with the dataset identifier PXD020099 and 10.6019/PXD020099. Gene ontology information was retrieved using The Arabidopsis Information Resource (TAIR) (Berardini et al., 2015) and The Plant Genome Integrative Explorer Resource (PlantGenIE) (Sundell et al., 2015). The complete list of Populus lipid body proteins identified in all the three biological replicates and the proteins identified in individual biological replicates are presented in Supplementary Table 1. The P. trichocarpa LB proteins identified in this study were checked for their relative protein abundance of different cellular compartments using the Multiple Marker Abundance Profiling (MMAP) tool from the SUBA toolbox and presented in Supplementary Figure 10.
The corresponding binary constructs were transformed into the A. tumefaciens strain GV3101 pMP90 and further infiltrated into N. benthamiana leaves (Schutze et al., 2009) together with the p19 protein from tomato bushy stunt virus cloned in pBIN61 (Voinnet et al., 2000) to suppress gene silencing. After two days of infiltration, tobacco leaf epidermis was investigated with a Leica TCS SP5 CLSM (Leica Microsystems, Wetzlar, Germany), and eGFP (Veerabagu et al., 2020) and mRPF (Van Damme et al., 2004) fluorescence was recorded (eGFP: excitation/emission 488/500–540 nm; mRFP: excitation/emission 561/600–640 nm). The Leica Application Suite X software was used to arrange single optical sections of mRFP (in magenta), eGFP and the corresponding merged images. Degradation of PtOLE6-eGFP and inhibition of degradation were studied in seeds of transgenic PtOLE6-eGFP expressing Arabidopsis seedlings imbibed with 2 ml of.2% DMSO (control), 200 μM MG132, and 40μM E64d, and investigated with CLSM (Figure 3).
One-way ANOVA followed by Fisher's and Tukey's multiple comparisons tests were performed using GraphPad Prism Version 8.0.0 (www.graphpad.com). The MEGA6 program (http://www.megasoftware.net/) was used for ClustalW multiple sequence alignments and phylogenetic analysis, with maximum likelihood method and Poisson correction model. The Populus and Arabidopsis proteins of the phylogenetic analyses are presented in Supplementary Figures 2, 3, 6–9.
The original contributions presented in the study are publicly available. This data can be found here: https://www.ebi.ac.uk/pride/archive/projects/PXD020099.
MV, PR, LP, and CvdS designed the study. MV, PR, LP, and MS performed the experiments. MV and PR analyzed the data. MV, CvdS, and PR interpreted the data and wrote the manuscript. All authors contributed to the article and approved the submitted version.
The authors acknowledge the financial support of the Norwegian Research Council to CvdS and PR (FRIPRO Grant No: 263117).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors are grateful to Professor Klaus Harter, University of Tübingen, for kindly providing them the pB7WGR2 and pBIN61:p19 vectors. The authors thank Lene Cecile Hermansen and Hilde Raanaas Kolstad, Imaging centre, NMBU, for the assistance with the TEM and CLSM, and Ms. Marit Siira for greenhouse assistance.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.674031/full#supplementary-material
Abell, B. M., Hahn, M., Holbrook, L. A., and Moloney, M. M. (2004). Membrane topology and sequence requirements for oil body targeting of oleosin. Plant J. 37, 461–470. doi: 10.1111/j.1365-313X.2003.01988.x
Abell, B. M., High, S., and Moloney, M. M. (2002). Membrane protein topology of oleosin is constrained by its long hydrophobic domain. J. Biol. Chem. 277, 8602–8610. doi: 10.1074/jbc.M103712200
Amari, K., Di Donato, M., Dolja, V. V., and Heinlein, M. (2014). Myosins VIII and XI play distinct roles in reproduction and transport of Tobacco mosaic virus. PLoS Pathog. 10:e1004448. doi: 10.1371/journal.ppat.1004448
Amari, K., Lerich, A., Schmitt-Keichinger, C., Dolja, V. V., and Ritzenthaler, C. (2011). Tubule-guided cell-to-cell movement of a plant virus requires class XI myosin motors. PLoS Pathog. 7:e1002327. doi: 10.1371/journal.ppat.1002327
Aoki, K., Kragler, F., Xoconostle-Cazares, B., and Lucas, W. J. (2002). A subclass of plant heat shock cognate 70 chaperones carries a motif that facilitates trafficking through plasmodesmata. Proc. Natl. Acad. Sci. U.S.A. 99, 16342–16347. doi: 10.1073/pnas.252427999
Avisar, D., Abu-Abied, M., Belausov, E., Sadot, E., Hawes, C., and Sparkes, I. A. (2009). A comparative study of the involvement of 17 Arabidopsis myosin family members on the motility of Golgi and other organelles. Plant Physiol. 150, 700–709. doi: 10.1104/pp.109.136853
Avisar, D., Prokhnevsky, A. I., Makarova, K. S., Koonin, E. V., and Dolja, V. V. (2008). Myosin XI-K is required for rapid trafficking of Golgi stacks, peroxisomes, and mitochondria in leaf cells of Nicotiana benthamiana. Plant Physiol. 146, 1098–1108. doi: 10.1104/pp.107.113647
Bailey, A. P., Koster, G., Guillermier, C., Hirst, E. M. A., MacRae, J. I., Lechene, C. P., et al. (2015). Antioxidant role for lipid droplets in a stem cell niche of Drosophila. Cell 163, 340–353. doi: 10.1016/j.cell.2015.09.020
Baluska, F., Cvrckova, F., Kendrick-Jones, J., and Volkmann, D. (2001). Sink plasmodesmata as gateways for phloem unloading. Myosin VIII and calreticulin as molecular determinants of sink strength? Plant Physiol. 126, 39–46. doi: 10.1104/pp.126.1.39
Barbosa, A. D., and Siniossoglou, S. (2017). Function of lipid droplet-organelle interactions in lipid homeostasis. BBA-Mol. Cell Res. 1864, 1459–1468. doi: 10.1016/j.bbamcr.2017.04.001
Bartz, R., Zehmer, J. K., Zhu, M. F., Chen, Y., Serrero, G., Zhao, Y. M., et al. (2007). Dynamic activity of lipid droplets: protein phosphorylation and GTP-mediated protein translocation. J. Proteome Res. 6, 3256–3265. doi: 10.1021/pr070158j
Beller, M., Sztalryd, C., Southhall, N., Bell, M., Jackle, H., Auld, D. S., et al. (2008). COPI complex is a regulator of lipid homeostasis. PLoS 6:e292. doi: 10.1371/journal.pbio.0060292
Beller, M., Thiel, K., Thul, P. J., and Jackle, H. (2010). Lipid droplets: a dynamic organelle moves into focus. FEBS Lett. 584, 2176–2182. doi: 10.1016/j.febslet.2010.03.022
Berardini, T. Z., Reiser, L., Li, D. H., Mezheritsky, Y., Muller, R., Strait, E., et al. (2015). The Arabidopsis information resource: making and mining the “gold standard” annotated reference plant genome. Genesis 53, 474–485. doi: 10.1002/dvg.22877
Bersuker, K., and Olzmann, J. A. (2017). Establishing the lipid droplet proteome: mechanisms of lipid droplet protein targeting and degradation. BBA-Mol. Cell Biol. L 1862, 1166–1177. doi: 10.1016/j.bbalip.2017.06.006
Binns, D., Janusxewski, T., Chen, Y., Hill, J., Larkin, V. S., Zhao, Y., et al. (2006). An intimate collaboration between peroxisomes and lipid bodies. J. Cell Bio. 173, 719–731. doi: 10.1083/jcb.200511125
Boston, R. S., Viitanen, P. V., and Vierling, E. (1996). Molecular chaperones and protein folding in plants. Plant Mol. Biol. 32, 191–222. doi: 10.1007/BF00039383
Boström, P., Andersson, L., Rutberg, M., Perman, J., Lidberg, U., Johansson, B. R., et al. (2007). SNARE proteins mediate fusion between cytosolic lipid droplets and are implicated in insulin sensitivity. Nat. Cell Biol. 9, 1286–1293. doi: 10.1038/ncb1648
Boye, T. L., Jeppesen, J. C., Maeda, K., Pezeshkian, W., Solovyeva, V., Nylandsted, J., et al. (2018). Annexins induce curvature on free-edge membranes displaying distinct morphologies. Sci. Rep. 8:10309. doi: 10.1038/s41598-018-28481-z
Brasaemle, D. L., Dolios, G., Shapiro, L., and Wang, R. (2004). Proteomic analysis of proteins associated with lipid droplets of basal and lipolytically stimulated 3T3-L1 adipocytes. J. Biol. Chem. 279, 46835–46842. doi: 10.1074/jbc.M409340200
Brault, M. L, Petit, J. D., Immel, F., Nicolas, W. J., Glavier, M., Brocard, L., et al. (2019). Multiple C2 domains and transmembrane region proteins (MCTPs) tether membranes at plasmodesmata. EMBO Rep. 20:e47182. doi: 10.15252/embr.201847182
Brocard, L., Immel, F., Coulon, D., Esnay, N., Tuphile, K., Pascal, S., et al. (2017). Proteomic analysis of lipid droplets from Arabidopsis aging leaves brings new insight into their biogenesis and functions. Front. Plant Sci. 8:894. doi: 10.3389/fpls.2017.00894
Burch-Smith, T. M., and Zambryski, P. C. (2011). Plasmodesmata paradigm shift: regulation from without versus within. Annu. Rev. Plant Biol. 63, 239–260. doi: 10.1146/annurev-arplant-042811-105453
Cai, Y. Q., Goodman, J. M., Pyc, M., Mullen, R. T., Dyer, J. M., and Chapman, K. D. (2015). Arabidopsis SEIPIN proteins modulate triacylglycerol accumulation and influence lipid droplet proliferation. Plant Cell 27, 2616–2636. doi: 10.1105/tpc.15.00588
Cermelli, S., Guo, Y., Gross, S. P., and Welte, M. A. (2006). The lipid-droplet proteome reveals that droplets are a protein-storage depot. Curr. Biol. 16, 1783–1795. doi: 10.1016/j.cub.2006.07.062
Chen, J. C. F., Tsai, C. C. Y., and Tzen, J. T. C. (1999). Cloning and secondary structure analysis of caleosin, a unique calcium-binding protein in oil bodies of plant seeds. Plant Cell Physiol. 40, 1079–1086. doi: 10.1093/oxfordjournals.pcp.a029490
Clark, G. B., Morgan, R. O., Fernandez, M. P., and Roux, S. J. (2012). Evolutionary adaptation of plant annexins has diversified their molecular structures, interactions and functional roles. New Phytol. 196, 695–712. doi: 10.1111/j.1469-8137.2012.04308.x
Coulon, D., Brocard, L., Tuphile, K., and Brehelin, C. (2020). Arabidopsis LDIP protein locates at a confined area within the lipid droplet surface and favors lipid droplet formation. Biochimie 169, 29–40. doi: 10.1016/j.biochi.2019.09.018
D'Andrea, S. (2016). Lipid droplet mobilization: the different ways to loosen the purse strings. Biochimie 120, 17–27. doi: 10.1016/j.biochi.2015.07.010
Deeks, M. J., and Hussey, P. J. (2005). Arp2/3 and scar: plants move to the fore. Nat. Rev. Mol. Cell Biol. 6, 954–964. doi: 10.1038/nrm1765
Delmer, D. P., and Potikha, T. S. (1997). Structures and functions of annexins in plants. Cell. Mol. Life Sci. 53, 546–553. doi: 10.1007/s000180050070
Deruyffelaere, C., Bouchez, I., Morin, H., Guillot, A., Miquel, M., Froissard, M., et al. (2015). Ubiquitin-mediated proteasomal degradation of oleosins is involved in oil body mobilization during post-germinative seedling growth in Arabidopsis. Plant Cell Physiol. 56, 1374–1387. doi: 10.1093/pcp/pcv056
Deruyffelaere, C., Purkrtova, Z., Bouchez, I., Collet, B., Cacas, J. L., Chardot, T., et al. (2018). PUX10 is a CDC48A Adaptor protein that regulates the extraction of ubiquitinated oleosins from seed lipid droplets in Arabidopsis. Plant Cell 30, 2116–2136. doi: 10.1105/tpc.18.00275
Diao, M., Ren, S., Wang, Q., Qian, L., Shen, J., Liu, Y., et al. (2018). Arabidopsis formin 2 regulates cell-to-cell trafficking by capping and stabilizing actin filaments at plasmodesmata. eLife 7:e36316. doi: 10.7554/eLife.36316
Ding, B., Kwon, M. O., and Warnberg, L. (1996). Evidence that actin filaments are involved in controlling the permeability of plasmodesmata in tobacco mesophyll. Plant J. 10, 157–164. doi: 10.1046/j.1365-313X.1996.10010157.x
Eastmond, P. J. (2006). SUGAR-DEPENDENT1 encodes a patatin domain triacylglycerol lipase that initiates storage oil breakdown in germinating Arabidopsis seeds. Plant Cell 18, 665–675. doi: 10.1105/tpc.105.040543
Esau, K., and Charvat, I. D. (1975). An ultrastructural study of acid phosphatase localization in cells of Phaseolus vulgaris phloem by the use of the azo dye method. Tissue Cell, 7, 619–630. doi: 10.1016/0040-8166(75)90031-2
Faulkner, C., and Maule, A. (2011). Opportunities and successes in the search for plasmodesmal proteins. Protoplasma 248, 27–38. doi: 10.1007/s00709-010-0213-x
Fernandez-Calvino, L., Faulkner, C., Walshaw, J., Saalbach, G., Bayer, E., Benitez-Alfonso, Y., et al. (2011). Arabidopsis plasmodesmal proteome. PLoS ONE 6:e18880. doi: 10.1371/journal.pone.0018880
Fernández-Santaso, R., Izquierdo, Y., López, A., Muniz, L., Martinez, M., Cascóm, T., et al. (2020). Protein profiles of lipid droplets during the hypersensitive defense response of Arabidopsis against Pseudomonas infection. Plant Cell Physiol. 61, 1144–1157. doi: 10.1093/pcp/pcaa041
Fiserova, J., Schwarzerova, K., Petrasek, J., and Opatrny, Z. (2006). ARP2 and ARP3 are localized to sites of actin filament nucleation in tobacco BY-2 cells. Protoplasma 227, 119–128. doi: 10.1007/s00709-006-0146-6
Fujimoto, Y., Itabe, H., Sakai, J., Makita, M., Noda, J., Mori, M., et al. (2004). Identification of major proteins in the lipid droplet fraction isolated from the human hepathocyt cell line HuH7. Biochim. Biophys. Acta 1644, 47–59. doi: 10.1016/j.bbamcr.2003.10.018
Gao, Q., and Goodman, J. M. (2015). The lipid droplet-a well-connected organelle. Front. Cell Devel. Biol. 3:49. doi: 10.3389/fcell.2015.00049
Gidda, S. K., Park, S., Pyc, M., Yurchenko, O., Cai, Y. Q., Wu, P., et al. (2016). Lipid droplet-associated proteins (LDAPs) are required for the dynamic regulation of neutral lipid compartmentation in plant cells. Plant Physiol. 170, 2052–2071. doi: 10.1104/pp.15.01977
Gidda, S. K., Shockey, J. M., Rothstein, S. J., Dyer, J. M., and Mullen, R. T. (2009). Arabidopsis thalianaGPAT8 and GPAT9 are localized to the ER and possess distinct ER retrieval signals: functionaldivergence of the dilysine ER retrieval motif in plant cells. Plant Physiol. Biochem. 47, 867–879. doi: 10.1016/j.plaphy.2009.05.008
Golomb, L., Abu-Abied, M., Belausov, E., and Sadot, E. (2008). Different subcellular localizations and functions of Arabidopsis myosin VIII. BMC Plant Biol. 8:3. doi: 10.1186/1471-2229-8-3
Gunjan, A., and Verreault, A. (2003). A Rad53 kinase-dependent surveillsnce mechanism 3that regulates histone protein levels6 in S. cerevisiae. Cell 115, 537–549. doi: 10.1016/S0092-8674(03)00896-1
Guo, Y., Cordes, K. R., Farese, R. V., and Walther, T. C. (2009). Lipid droplets at a glance. J. Cell Sci. 122, 749–752. doi: 10.1242/jcs.037630
Han, X., Huang, L.-J., Feng, D., Jiang, W., Miu, W., and Li, N. (2019). Plasmodemata-related structural and functional proteins: the long-sought-after secrets of a cytoplasmic channel in plant cell walls. Int. J. Mol. Sci. 20:2946. doi: 10.3390/ijms20122946
Hashimoto, K., Igarashi, H., Mano, S., Takenaka, C., Shiina, T., Yamaguchi, M., et al. (2008). An isoform of Arabidopsis myosin XI interacts with small GTPases in its C-terminal tail region. J. Exp. Bot. 59, 3523–3531. doi: 10.1093/jxb/ern202
Heinlein, M. (2015a). “Plasmodesmata,” in Methods and Protocols. Methods in Molecular Biology, ed. M. Heinlein (New York, NY: Springer), 346. doi: 10.1007/978-1-4939-1523-1
Heinlein, M. (2015b). “Plasmodesmata: channnels for viruses on the move,” in Plasmodesmata. Methods and Protocols. Methods in Molecular Biology, ed. M. Heinlein (New York, NY: Springer) 25–52. doi: 10.1007/978-1-4939-1523-1_2
Hodges, B. D. M., and Wu, C. C. (2010). Proteomic insights into an expanded cellular role for cytoplasmic lipid droplets. J. Lipid Res. 51, 262–273. doi: 10.1194/jlr.R003582
Horn, P. J., James, C. N., Gidda, S. K., Kilaru, A., Dyer, J. M., Mullen, R. T., et al. (2013). Identification of a new class of lipid droplet-associated proteins in plants. Plant Physiol. 162, 1926–1936. doi: 10.1104/pp.113.222455
Hsiao, E. S. L., and Tzen, J. T. C. (2011). Ubiquitination of oleosin-H and caleosin in sesame oil bodies after seed germination. Plant Physiol. Biochem. 49, 77–81. doi: 10.1016/j.plaphy.2010.10.001
Hsieh, K., and Huang, A. H. C. (2004). Endoplasmic reticulum, oleosins, and oils in seeds and tapetum cells. Plant Physiol. 136, 3427–3434. doi: 10.1104/pp.104.051060
Huang, A. H. C. (1992). Oilbodies and oleosins in seeds. Annu. Rev. Plant Physiol. Plant Mol. Biol. 43, 177–200. doi: 10.1146/annurev.pp.43.060192.001141
Huang, A. H. C. (2018). Plant lipid droplets and their associated proteins: potential for rapid advances. Plant Physiol. 176, 1894–1918. doi: 10.1104/pp.17.01677
Huang, C. Y., and Huang, A. H. C. (2017). Unique motifs and length of hairpin in oleosin target the cytosolic side of endoplasmic reticulum and budding lipid droplet. Plant Physiol. 174, 2248–2260. doi: 10.1104/pp.17.00366
Huang, S. X., Jiang, L. W., and Zhuang, X. H. (2019). Possible roles of membrane trafficking components for lipid droplet dynamics in higher plants and green algae. Front. Plant Sci. 10:207. doi: 10.3389/fpls.2019.00207
Jolivet, P., Roux, E., d'Andrea, S., Davanture, M., Negroni, L., Zivy, M., et al. (2004). Protein composition of oil bodies in Arabidopsis thaliana ecotype WS. Plant Physiol. Biochem. 42, 501–509. doi: 10.1016/j.plaphy.2004.04.006
Kandasamy, M. K., Gilliland, L. U., McKinney, E. C., and Meagher, R. B. (2001). One plant actin isovariant, ACT7, is induced by auxin and required for normal callus formation. Plant Cell 13, 1541–1554. doi: 10.1105/TPC.010026
Kandasamy, M. K., McKinney, E. C., and Meagher, R. B. (2009). A Single vegetative actin isovariant overexpressed under the control of multiple regulatory sequences is sufficient for normal Arabidopsis development. Plant Cell 21, 701–718. doi: 10.1105/tpc.108.061960
Karimi, M., Inze, D., and Depicker, A. (2002). GATEWAY((TM)) vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7, 193–195. doi: 10.1016/S1360-1385(02)02251-3
Karlson, D. T., Fujino, T., Kimura, S., Baba, K., Itoh, T., and Ashworth, E. N. (2003). Novel plasmodesmata association of dehydrin-like proteins in cold-acclimated red-osier dogwood (Cornus sericea). Tree Physiol. 23, 759–767. doi: 10.1093/treephys/23.11.759
Katyayini, N. U., Rinne, P. L. H., and van der Schoot, C. (2019). Strigolactone-based node-to-bud signaling may restrain shoot branching in hybrid aspen. Plant Cell Physiol. 60, 2797–2811. doi: 10.1093/pcp/pcz170
Kim, E. Y., Park, K. Y., Seo, Y. S., and Kim, W. T. (2016). Arabidopsis small rubber particle protein homolog SRPs play dual roles as positive factors for tissue growth and development and in drought stress responses. Plant Physiol. 170, 2494–2510. doi: 10.1104/pp.16.00165
Kitagawa, M., and Jackson, D. (2017). Plasmodesmata-mediated cell-to-cell communication in the shoot apical meristem: how stem cells talk. Plants 6:12. doi: 10.3390/plants6010012
Knox, K., Wang, P. W., Kriechbaumer, V., Tilsner, J., Frigerio, L., Sparkes, I., et al. (2015). Putting the squeeze on plasmodesmata: arole for reticulons in primary plasmodesmata formation. Plant Physiol. 168, 1563–1572. doi: 10.1104/pp.15.00668
Kory, N., Thiam, A. R., Farese, R. V., and Walther, T. C. (2015). Protein crowding is a determinant of lipid droplet protein composition. Dev. Cell 34, 351–363. doi: 10.1016/j.devcel.2015.06.007
Krahmer, N., Hilger, M., Kory, N., Wilfling, F., Stoehr, G., Mann, M., et al. (2013). Protein correlation profiles identify lipid droplet proteins with high confidence. Mol. Cell. Proteomics 12, 1115–1126. doi: 10.1074/mcp.M112.020230
Kretzschmar, F. K., Doner, N. M., Krawczyk, H. E., Scholz, P., Schmitt, K., Valerius, O., et al. (2020). Identification of low-abundance lipid droplet proteins in seeds and seedlings. Plant Physiol. 182, 1326–1345. doi: 10.1104/pp.19.01255
Kretzschmar, F. K., Mengel, L. A., Muller, A. O., Schmitt, K., Blersch, K. F., Valerius, O., et al. (2018). PUX10 is a lipid droplet-localized scaffold protein that interacts with CELL DIVISION CYCLE48 and is involved in the degradation of lipid droplet proteins. Plant Cell 30, 2137–2160. doi: 10.1105/tpc.18.00276
Kriechbaumer, V., Botchway, S. W., Slade, S. E., Knox, K., Frigerio, L., Oparka, K., et al. (2015). Reticulomics: protein-protein interaction studies with two plasmodesmata-localized reticulon family proteins identify binding partners enriched at plasmodesmata, endoplasmic reticulum, and the plasma membrane. Plant Physiol. 169, 1933–1945. doi: 10.1104/pp.15.01153
Kriechbaumer, V., von Loffelholz, O., and Abell, B. M. (2012). Chaperone receptors: guiding proteins to intracellular compartments. Protoplasma 249, 21–30. doi: 10.1007/s00709-011-0270-9
Lee, K. H., Minami, A., Marshall, R. S., Book, A. J., Farmer, L. M., Walker, J. M., et al. (2011). The RPT2 Subunit of the 26S proteasome directs complex assembly, histone dynamics, and gametophyte and sporophyte development in Arabidopsis. Plant Cell 23, 4298–4317. doi: 10.1105/tpc.111.089482
Leijon, F., Melzer, M., Zhou, Q., Srivastava, V., and Bulone, V. (2018). Proteomic analysis of plasmodesmata from Populus cell suspension cultures in relation with callose biosynthesis. Front. Plant Sci. 9:1681. doi: 10.3389/fpls.2018.01681
Li, Z. H., Thiel, K., Thul, P. J., Beller, M., Kuhnlein, R. P., and Welte, M. A. (2012). Lipid droplets control the maternal histone supply of Drosophila embryos. Curr. Biol. 22, 2104–2113. doi: 10.1016/j.cub.2012.09.018
Listenberger, L. L., Han, X. L., Lewis, S. E., Cases, S., Farese, R. V., Ory, D. S., et al. (2003). Triglyceride accumulation protects against fatty acid-induced lipotoxicity. Proc. Natl. Acad. Sci. U.S.A. 100, 3077–3082. doi: 10.1073/pnas.0630588100
Liu, P. S., Ying, Y. S., Zhao, Y. M., Mundy, D. I., Zhu, M. F., and Anderson, R. G. W. (2004). Chinese hamster ovary K2 cell lipid droplets appear to be metabolic organelles involved in membrane traffic. J. Biol. Chem. 279, 3787–3792. doi: 10.1074/jbc.M311945200
Liu, Z., Jia, Y., Ding, Y., Shi, Y., Li, Z., Guo, Y., et al. (2017). Plasma membrane CRK1-mediated phosphorylation of 14-3-3 proteins induces their nuclear import to fine-tune CBF signalling during cold response. Mol. Cell 66, 117–128. doi: 10.1016/j.molcel.2017.02.016
Lizaso, A., Tan, K. T., and Lee, Y. H. (2013). Beta-adrenergic receptor-stimulated lipolysis requires the RAB7-mediated autolysosomal lipid degradation. Autophagy 9, 1228–1243. doi: 10.4161/auto.24893
Ma, B. Y., Qian, D., Nan, Q., Tan, C., An, L. Z., and Xiang, Y. (2012). Arabidopsis vacuolar H+-ATPase (V-ATPase) B subunits are involved in actin cytoskeleton remodeling via binding to, bundling, and stabilizing F-actin. J. Biol. Chem. 287, 19008–19017. doi: 10.1074/jbc.M111.281873
Martin, S., Driessen, K., Nixon, S. J., Zerial, M., and Parton, R. G. (2005). Regulated localization of Rab18 to lipid droplets: effects of lipolytic stimulation and inhibition of lipid droplet catabolism. J. Biol. Chem. 280, 42325–42335. doi: 10.1074/jbc.M506651200
Mathur, J., Mathur, N., and Hulskamp, M. (2002). Simultaneous visualization of peroxisomes and cytoskeletal elements reveals actin and not microtubule-based peroxisome motility in plants. Plant Physiol. 128, 1031–1045. doi: 10.1104/pp.011018
McDowell, J. M., An, Y. Q., Huang, S. R., McKinney, E. C., and Meagher, R. B. (1996). The Arabidopsis ACT7 actin gene is expressed in rapidly developing tissues and responds to several external stimuli. Plant Physiol. 111, 699–711. doi: 10.1104/pp.111.3.699
Meitha, K., Konnerup, D., Colmer, T. D., Considine, J. A., Foyer, C. H., and Considine, M. J. (2015). Spatio-temporal relief from hypoxia and production of reactive oxygen species during bud burst in grapevine (Vitis vinifera). Ann. Bot. 116, 703–711. doi: 10.1093/aob/mcv123
Mongrand, S., Stanislas, T., Bayer, E. M. F., Lherminier, J., and Simon-Plas, F. (2010). Membrane rafts in plant cells. Trends Plant. Sci. 15, 656–663. doi: 10.1016/j.tplants.2010.09.003
Müller, A. O., and Ischbeck, T. (2018). Characterization of the enzymatic activity and physiological function of the lipid droplet-associated triacylglycerol lipase AtOBL1. New Phyol. 217, 1062–1076. doi: 10.1111/nph.14902
Murphy, D. J. (2012). The dynamic roles of intracellular lipid droplets: from archaea to mammals. Protoplasma 249, 541–585. doi: 10.1007/s00709-011-0329-7
Murphy, S., Martin, S., and Parton, R. G. (2009). Lipid droplet-organelle interactions; sharing the fats. BBA-Mol. Cell Biol. L 1791, 441–447. doi: 10.1016/j.bbalip.2008.07.004
Nakamura, N., Banno, Y., and Tamiya-Koizumi, K. (2005). Arf1-dependent PLD1 is localized to oleic-acid-induced lipid droplets in NIH3T3 cells. Biochem. Biophys. Res. Commmun. 335, 117–123. doi: 10.1016/j.bbrc.2005.07.050
Nesvizhskii, A. I., Keller, A., Kolker, E., and Aebersold, R. (2003). A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75, 4646–4658. doi: 10.1021/ac0341261
O'Fallon, J. V. O., Busboom, J. R., Nelson, M. L., and Gaskin, C. T. (2007). A direct method for fatty acid methyl ester synthesis: application to wet meat tissues, oils, and feedstuffs. J. Anim. Sci. 85, 1511–1521. doi: 10.2527/jas.2006-491
Ophir, R., Pang, X. Q., Halaly, T., Venkateswari, J., Lavee, S., Galbraith, D., et al. (2009). Gene-expression profiling of grape bud response to two alternative dormancy-release stimuli expose possible links between impaired mitochondrial activity, hypoxia, ethylene-ABA interplay and cell enlargement. Plant Mol. Biol. 71, 403–423. doi: 10.1007/s11103-009-9531-9
Ozeki, S., Xheng, J., Tauchi-Satu, K., Hatano, N., Taniguchi, H., and Fujimoto, T. (2005). Rab18 localizes to lipid droplets and induces their close apposition to the endoplasmic reticulum-derived membrane. J. Cell Sci. 118, 2601–2611. doi: 10.1242/jcs.02401
Paul, L. K., Rinne, P. L. H., and van der Schoot, C. (2014a). Refurbishing the plasmodesmal chamber: a role for lipid bodies? Front. Plant Sci. 5:40. doi: 10.3389/fpls.2014.00040
Paul, L. K., Rinne, P. L. H., and van der Schoot, C. (2014b). Shoot meristems of deciduous woody perennials: self-organization and morphogenetic transitions. Curr. Opin. Plant Biol. 17, 86–95. doi: 10.1016/j.pbi.2013.11.009
Peremyslov, V. V., Cole, R. A., Fowler, J. E., and Dolja, V. V. (2015). Myosin-powered membrane compartment drives cytoplasmic streaming, cell expansion and plant development. PLoS ONE 10:e0139331. doi: 10.1371/journal.pone.0139331
Perez-Riverol, Y., Csordas, A., Bai, J. W., Bernal-Llinares, M., Hewapathirana, S., Kundu, D. J., et al. (2019). The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 47, D442–D450. doi: 10.1093/nar/gky1106
Pinheiro, H., Samalova, M., Geldner, N., Chory, J., Martinez, A., and Moore, I. (2009). Genetic evidence that the higher plant Rab-D1 and Rab-D2 GTPases exhibit distinct but overlapping interactions in the early secretory pathway. J. Cell Sci. 122, 3749–3758. doi: 10.1242/jcs.050625
Ploegh, H. L. (2007). A lipid-based model for the creation of an escape hatch from the endoplasmic reticulum. Nature 448, 435–438. doi: 10.1038/nature06004
Pyc, M., Cai, Y. Q., Gidda, S. K., Yurchenko, O., Park, S., Kretzschmar, F. K., et al. (2017). Arabidopsis lipid droplet-associated protein (LDAP) interacting protein (LDIP) influences lipid droplet size and neutral lipid homeostasis in both leaves and seeds. Plant J. 92, 1182–1201. doi: 10.1111/tpj.13754
Quettier, A. L., and Eastmond, P. J. (2009). Storage oil hydrolysis during early seedling growth. Plant Physiol. Biochem. 47, 485–490. doi: 10.1016/j.plaphy.2008.12.005
Reagan, B. C., and Burch-Smith, T. M. (2020). Viruses reveal the secrets of plasmodesmal cell biology. Mol. Plant Microb. Int. 33, 26–39. doi: 10.1094/MPMI-07-19-0212-FI
Rinne, P. L. H., Kaikuranta, P. M., and van der Schoot, C. (2001). The shoot apical meristem restores its symplasmic organization during chilling-induced release from dormancy. Plant J. 26, 249–264. doi: 10.1046/j.1365-313X.2001.01022.x
Rinne, P. L. H., Paul, L. K., Vahala, J., Ruonala, R., Kangasjarvi, J., and van der Schoot, C. (2015). Long and short photoperiod buds in hybrid aspen share structural development and expression patterns of marker genes. J. Exp. Bot. 66, 6745–6760. doi: 10.1093/jxb/erv380
Rinne, P. L. H., and van der Schoot, C. (1998). Symplasmic fields in the tunica of the shoot apical meristem coordinate morphogenetic events. Development 125, 1477–1485. doi: 10.1242/dev.125.8.1477
Rinne, P. L. H., and van der Schoot, C. (2003). Plasmodesmata at the crossroads between development, dormancy, and defense. Can J. Bot. 81, 1182–1197. doi: 10.1139/b03-123
Rinne, P. L. H., and van der Schoot, C. (2004). Cell-cell communication as a key factor in dormancy cycling. J. Crop Improv. 10, 113–156. doi: 10.1300/J411v10n01_07
Rinne, P. L. H., Welling, A., Vahala, J., Ripel, L., Ruonala, R., Kangasjarvi, J., et al. (2011). Chilling of dormant buds hyperinduces FLOWERING LOCUS T and recruits GA-inducible 1,3-beta-glucanases to reopen signal conduits and release dormancy in Populus. Plant Cell 23, 130–146. doi: 10.1105/tpc.110.081307
Sager, R., and Lee, J. Y. (2014). Plasmodesmata in integrated cell signalling: insights from development and environmental signals and stresses. J. Exp. Bot. 65, 6337–6358. doi: 10.1093/jxb/eru365
Sagi, G., Katz, A., Guenoune-Gelbart, D., and Epel, B. L. (2005). Class 1 reversibly glycosylated polypeptides are plasmodesmal-associated proteins delivered to plasmodesmata via the Golgi apparatus. Plant Cell 17, 1788–1800 doi: 10.1105/tpc.105.031823
Sato, S., Fukasamwa, M., Yamakawa, Y., Natsume, T., Suzuki, T., Shoji, L., et al. (2006). Proteomic profiling of lipid droplet proteins in hepatoma cell lines expressing hepatits C virus core protein. J. Biochem. 139, 921–930. doi: 10.1093/jb/mvj104
Schroeder, B., Schulze, R. J., Weller, S., Sletten, A. C., Casey, C. A., and Mcniven, M. A. (2015). The small GTPase Rab7 as a central regulator of hepatocellular lipophagy. Hepatology 61, 1896–1907. doi: 10.1002/hep.27667
Schutze, K., Harter, K., and Chaban, C. (2009). Bimolecular fluorescence complementation (BiFC) to study protein-protein interactions in living plant cells. Methods Mol. Biol. 479, 189–202. doi: 10.1007/978-1-59745-289-2_12
Shimada, T. L., Hayashi, M., and Hara-Nishimura, I. (2018). Membrane dynamics and multiple functions of oil bodies in seeds and leaves. Plant Physiol. 176, 199–207. doi: 10.1104/pp.17.01522
Shimada, T. L., Shimada, T., Takahashi, H., Fukao, Y., and Hara-Nishimura, I. (2008). A novel role for oleosins in freezing tolerance of oilseeds in Arabidopsis thaliana. Plant J. 55, 798–809. doi: 10.1111/j.1365-313X.2008.03553.x
Shockey, J. M., Gidda, S. K., Chapital, D. C., Kuan, J. C., Dhanoa, P. K., Bland, J. M., et al. (2006). Tung tree DGAT1 and DGAT2 have nonredundant functions in triacylglycerol biosynthesis and are localized to different subdomains of the endoplasmic reticulum. Plant Cell 18, 2294–2313. doi: 10.1105/tpc.106.043695
Siloto, R. M. P., Findlay, K., Lopez-Villalobos, A., Yeung, E. C., Nykiforuk, C. L., and Moloney, M. M. (2006). The accumulation of oleosins determines the size of seed oilbodies in Arabidopsis. Plant Cell 18, 1961–1974. doi: 10.1105/tpc.106.041269
Sollner, T. H. (2007). Lipid droplets highjack SNAREs. Nat. Cell Biol. 9, 1219–1220. doi: 10.1038/ncb1107-1219
Soni, K. G., Mardones, G. A., Sougrat, R., Smirnova, E., Jackson, C. L., and Bonifacino, J. S. (2009). Coatomer-dependent protein delivery to lipid droplets. J. Cell Sci. 122, 1834–1841. doi: 10.1242/jcs.045849
Sparkes, I. A., Teanby, N. A., and Hawes, C. (2008). Truncated myosin XI tail fusions inhibit peroxisome, Golgi, and mitochondrial movement in tobacco leaf epidermal cells: a genetic tool for the next generation. J. Exp. Bot. 59, 2499–2512. doi: 10.1093/jxb/ern114
Su, S. Z., Liu, Z. H., Chen, C., Zhang, Y., Wang, X., Zhu, L., et al. (2010). Cucumber mosaic virus movement protein severs actin filaments to increase the plasmodesmal size exclusion limit in tobacco. Plant Cell 22, 1373–1387. doi: 10.1105/tpc.108.064212
Sundell, D., Mannapperuma, C., Netotea, S., Delhomme, N., Lin, Y. C., Sjodin, A., et al. (2015). The plant genome integrative explorer resource: PlantGenIE.org. New Phytol. 208, 1149–1156. doi: 10.1111/nph.13557
Suwastika, I. N., Uemura, T., Shiina, T., Sato, M. H., and Takeyasu, K. (2008). SYP71, a Plant-specific Qc-SNARE protein, reveals dual localization to the plasma membrane and the endoplasmic reticulum in Arabidopsis. Cell Struct. Funct. 33, 185–192. doi: 10.1247/csf.08024
Thazar-Poulot, N., Miquel, M., Fobis-Loisy, I., and Gaude, T. (2015). Peroxisome extensions deliver the Arabidopsis SDP1 lipase to oil bodies. Proc. Natl. Acad. Sci. U.S.A. 112, 4158–4163. doi: 10.1073/pnas.1403322112
Turro, S., Ingelmo-Torres, M., Estanyol, M., Tebar, F., Fernández, M. A., Albor, C. V., et al. (2006). Identification and characterization of associated with lipid droplet protein 1: a novel membrane assocaiated protein that resids on hepatic lipid droplets. Traffic 7, 1254–1269. doi: 10.1111/j.1600-0854.2006.00465.x
Tzen, J. T. C., and Huang, A. H. C. (1992). Surface-structure and properties of plant seed oil bodies. J. Cell Biol. 117, 327–335. doi: 10.1083/jcb.117.2.327
Umlauf, E., Csaszar, E., Moertelmaier, M., Schuetz, G. J., Parton, R. G., and Prohaska, R. (2004). Association of stomatin with lipid bodies. J. Biol. Chem. 279, 23699–23709. doi: 10.1074/jbc.M310546200
Vaddepalli, P., Fulton, L., Wieland, J., Wassmer, K., Schaeffer, M., Ranf, S., et al. (2017). The cell wall-localized atypical beta-1,3 glucanase ZERZAUST controls tissue morphogenesis in Arabidopsis thaliana. Development 144, 2259–2269. doi: 10.1242/dev.152231
Van Damme, D., van Poucke, K., Boutant, E., Ritzenthaler, C., Inz,é, D., and Geelen, D. (2004). In vivo dynamics and differential microtubule-binding activities of MAP65 proteins. Plant Physiol. 136, 3956–3967. doi: 10.1104/pp.104.051623
van der Schoot, C., Paul, L. K., Paul, S. B., and Rinne, P. L. H. (2011). Plant lipid bodies and cell-cell signaling: a new role for an old organelle? Plant Signal. Behav. 6, 1732–1738. doi: 10.4161/psb.6.11.17639
van der Schoot, C., and Rinne, P. (1999). Networks for shoot design. Trends Plant Sci. 4, 31–37. doi: 10.1016/S1360-1385(98)01362-4
Van Gestel, K., Slegers, H., von Witsch, M., Samaj, J., Baluska, F., and Verbelen, J. P. (2003). Immunological evidence for the presence of plant homologues of the actin-related protein Arp3 in tobacco and maize: subcellular localization to actin-enriched pit fields and emerging root hairs. Protoplasma 222, 45–52. doi: 10.1007/s00709-003-0004-8
Veerabagu, M., Paul, L. K., Rinne, P. L. H., and van der Schoot, C. (2020). Plant lipid bodies traffic on actin to plasmodesmata motorized by myosin XIs. Int. J. Mol. Sci. 21:1422. doi: 10.3390/ijms21041422
Verma, R., Aravind, L., Oania, R., McDonald, W. H., Yates, J. R., Koonin, E. V., et al. (2002). Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science 298, 611–615. doi: 10.1126/science.1075898
Vermachova, M., Purkrtova, Z., Santrucek, J., Jolivet, P., Chardot, T., and Kodicek, M. (2011). New protein isoforms identified within Arabidopsis thaliana seed oil bodies combining chymotrypsin/trypsin digestion and peptide fragmentation analysis. Proteomics 11, 3430–3434. doi: 10.1002/pmic.201000603
Voinnet, O., Lederer, C., and Baulcombe, D. C. (2000). A viral movement protein prevents spread of the gene silencing signal in Nicotiana benthamiana. Cell 103, 157–167. doi: 10.1016/S0092-8674(00)00095-7
Walther, T. C., and Farese, R. V. (2009). The life of lipid droplets. BBA-Mol. Cell Biol. L 1791, 459–466. doi: 10.1016/j.bbalip.2008.10.009
White, R. G., Badelt, K., Overall, R. L., and Vesk, M. (1994). Actin associated with plasmodesmata. Protoplasma 180, 169–184. doi: 10.1007/BF01507853
White, R. G., and Barton, D. A. (2011). The cytoskeleton in plasmodesmata: a role in intercellular transport? J. Exp. Bot. 62, 5249–5266. doi: 10.1093/jxb/err227
Wilfling, F., Thiam, A. R., Olarte, M. J., Wang, J., Beck, R., Gould, T. J., et al. (2014). Arf1/COPI machinery acts directly on lipid droplets and enables their connection to the ER for protein targeting. Elife 3:e01607. doi: 10.7554/eLife.01607
Wilfling, F., Wang, H., Haas, J. T., Krahmer, N., Gould, T. J., Uchida, A., et al. (2013). Triacylglycerol synthesis enzymes mediate lipid droplet growth by relocalizing from the ER to lipid droplets. Dev. Cell. 24, 384–399. doi: 10.1016/j.devcel.2013.01.013
Xu, X. M., Wang, J., Xuan, Z. Y., Goldshmidt, A., Borrill, P. G. M., Hariharan, N., et al. (2011). Chaperonins Facilitate KNOTTED1 Cell-to-cell trafficking and stem cell function. Science 333, 1141–e01144. doi: 10.1126/science.1205727
Zehmer, J. K., Huang, Y. G., Peng, G., Pu, J., Anderson, R. G. W., and Liu, P. S. (2009). A role for lipid droplets in inter-membrane lipid traffic. Proteomics 9, 914–921. doi: 10.1002/pmic.200800584
Zhang, J., Li, Y., Liu, B. B., Wang, L. J., Zhang, L., Hu, J. J., et al. (2018). Characterization of the Populus Rab family genes and the function of PtRabE1b in salt tolerance. BMC Plant Biol. 18:124. doi: 10.1186/s12870-018-1342-1
Keywords: LB/LD proteome, oleosin, LDIP, LDAP, Caleosin, ACT7, Myosin XI-binding Rab C2A, plasmodesmata
Citation: Veerabagu M, Rinne PLH, Skaugen M, Paul LK and van der Schoot C (2021) Lipid Body Dynamics in Shoot Meristems: Production, Enlargement, and Putative Organellar Interactions and Plasmodesmal Targeting. Front. Plant Sci. 12:674031. doi: 10.3389/fpls.2021.674031
Received: 28 February 2021; Accepted: 14 June 2021;
Published: 21 July 2021.
Edited by:
Jung-Youn Lee, University of Delaware, United StatesReviewed by:
Motoki Tominaga, Waseda University, JapanCopyright © 2021 Veerabagu, Rinne, Skaugen, Paul and van der Schoot. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Christiaan van der Schoot, Y2hyaXMudmFuZGVyc2Nob290QG5tYnUubm8=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.