
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 17 May 2021
Sec. Plant Development and EvoDevo
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.674010
This article is part of the Research Topic Root Development: Towards Understanding Regulatory Networks and Complex Interactions Between Cell Populations View all 14 articles
Responsiveness to environmental conditions and developmental plasticity of root systems are crucial determinants of plant fitness. These processes are interconnected at a cellular level with cell wall properties and cell surface signaling, which involve arabinogalactan proteins (AGPs) as essential components. AGPs are cell-wall localized glycoproteins, often GPI-anchored, which participate in root functions at many levels. They are involved in cell expansion and differentiation, regulation of root growth, interactions with other organisms, and environmental response. Due to the complexity of cell wall functional and regulatory networks, and despite the large amount of experimental data, the exact molecular mechanisms of AGP-action are still largely unknown. This dynamically evolving field of root biology is summarized in the present review.
Plants are sessile organisms with cells surrounded by cell walls which mediate interactions with surrounding environment. Communication across the cell wall and related cell surface signaling is an essential, complex, and largely unexplored aspect of plant biology (Seifert and Blaukopf, 2010; Duman et al., 2020; Rui and Dinneny, 2020). The deposition and remodeling of the cell wall enables growth and development of plant organs, and cell-wall derived signals mediate responses to internal and external factors (Voxeur and Hofte, 2016; Gigli-Bisceglia et al., 2020).
Arabinogalactan proteins (AGPs) are ubiquitous in the cell wall and in extracellular exudates (Showalter, 2001). They take part in the regulatory and functional continuum of the plasmalemma, cell wall, and environment (Ellis et al., 2010). AGPs occur in all plant organs (Clarke et al., 1979; Fincher et al., 1983; Nguema-Ona et al., 2012; He et al., 2019) but molecular mechanisms of their function remain rather puzzling. They are involved in the regulation of plant growth and development, affect cell wall properties, structure, and architecture (Seifert, 2018, 2021; Tucker et al., 2018), play a role in stem development and differentiation (Ito et al., 2005; MacMillan et al., 2010; Liu et al., 2020), root growth and differentiation (Dolan et al., 1995; Bossy et al., 2009; Nguema-Ona et al., 2012), sexual reproduction (Cheung et al., 1995; Cheung and Wu, 1999; Nguema-Ona et al., 2012; Pereira et al., 2015; Su and Higashiyama, 2018), embryogenesis (Kreuger and van Holst, 1993; Yu and Zhao, 2012; Perez-Perez et al., 2018), fruit ripening (Leszczuk et al., 2020a, b), response to abiotic and biotic stress factors (Mareri et al., 2018; Seifert, 2021), and interactions with microorganisms (Nguema-Ona et al., 2013; Rashid, 2016).
The root system, not covered by a protective cuticle, is constantly interacting with the rhizosphere. It secretes protective mucilage and other compounds, interchanges signaling molecules with soil organisms, and adjusts root development according to the heterogeneous distribution of soil resources with an amazing degree of plasticity. Roots thus present a unique system to evaluate different aspects of AGP functions in the cell wall and extracellular spaces (Figure 1). In roots, AGPs are important regulators of elongation and differentiation of cells (Shi et al., 2003), including root hairs (Kirchner et al., 2018; Borassi et al., 2020). They represent important components of root exudates, aid in the formation of a rhizosheath (Galloway et al., 2020), modulate response to root pathogens and parasites (Gaspar et al., 2004; Bozbuga et al., 2018), and are involved in the establishment of root symbioses with beneficial microorganisms (Brewin, 2004). AGPs even form major components of the glue-like adhesive nanoparticles secreted by the roots of climbing plants (Huang et al., 2016). In this review we summarize selected aspects of AGP action related to root development and function (Figure 1), updating previous excellent reviews (Nguema-Ona et al., 2012, 2013) and covering recent advances in this field of root biology. Available AGP mutants with phenotypic manifestations in roots are summarized (Table 1).
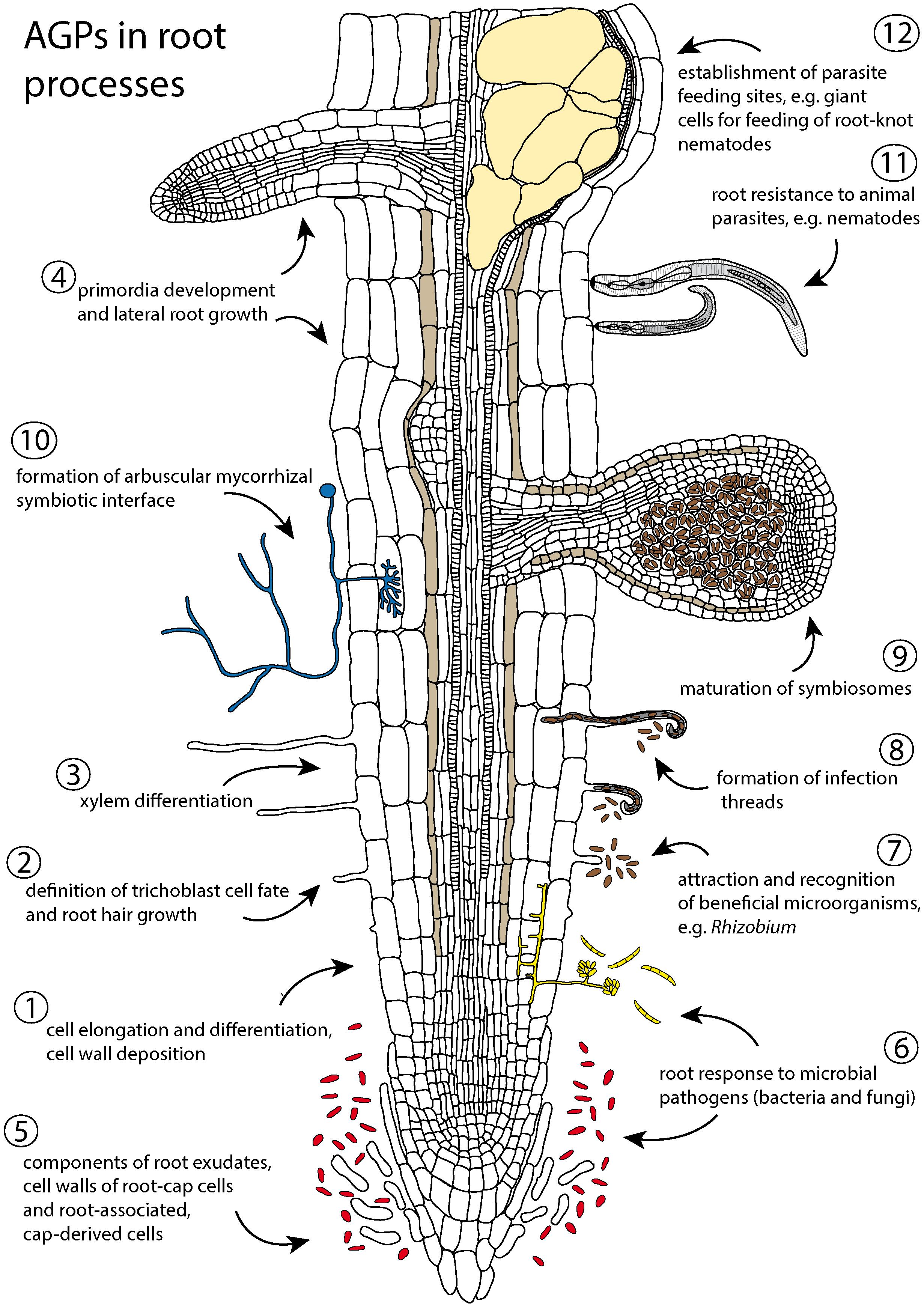
Figure 1. Schematic summary of the involvement of arabinogalactan proteins (AGPs) in root processes. (1–4) AGPs modulate cell wall properties and regulate developmental events in roots: (1) cell division, cell expansion and cell wall deposition (Shi et al., 2003; Yang et al., 2007; Zhang et al., 2011; Seifert, 2018, 2021; Tucker et al., 2018), (2) trichoblast definition and root hair growth (Šamaj et al., 1999; Lin et al., 2011; Marzec et al., 2015; Kirchner et al., 2018; Borassi et al., 2020), (3) xylem differentiation (Dolan et al., 1995; Bossy et al., 2009), and (4) early events of lateral root development (Yang et al., 2007; Johnson et al., 2011; Zhang et al., 2011). (5–12) AGPs are components of root exudates and cell walls of root cap cells and root-associated, cap-derived cells (border cells and border-like cells) and participate in responses to biotic and abiotic environmental factors: (5–6) help to protect roots against abiotic stress (e.g., drought, toxicity) and microbial pathogens (Cannesan et al., 2012; Koroney et al., 2016; Marquez et al., 2018; Driouich et al., 2019; Galloway et al., 2020), (7–10) participate in establishment of mutualistic interaction with N-fixing microorganisms (Berry et al., 2002; Brewin, 2004; Brewin et al., 2008; Tsyganova et al., 2009, 2019; Nguema-Ona et al., 2013), arbuscular fungi (Gollotte et al., 1995; Balestrini and Lanfranco, 2006; Schultz and Harrison, 2008) and beneficial endophytes (Basińska-Barczak et al., 2020; Nivedita et al., 2020), and (11–12) affect root susceptibility to parasites (Beneventi et al., 2013; Bozbuga et al., 2018).
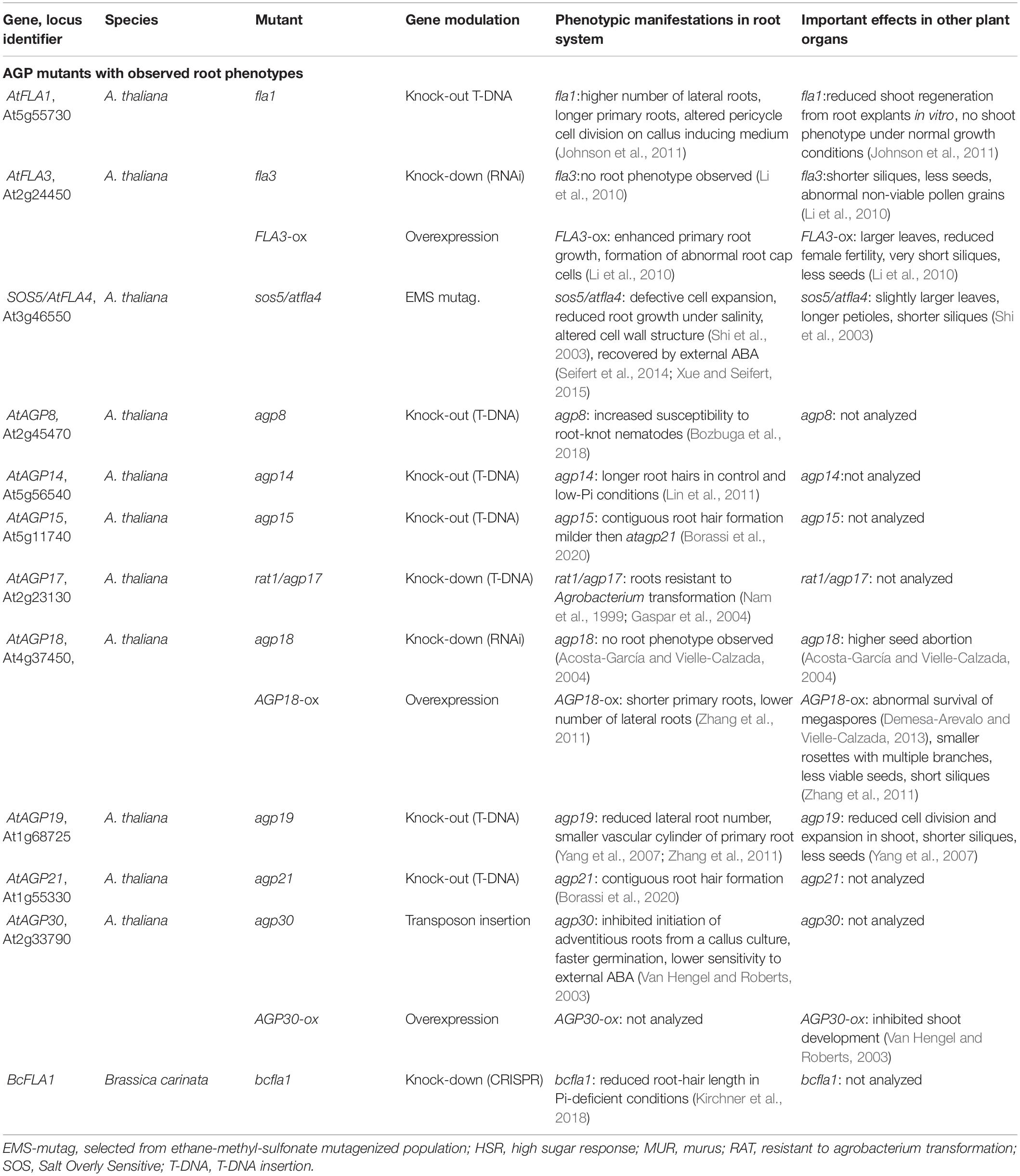
Table 1. Mutants with modulated expression of AGP genes showing phenotypic manifestations in root system.
Structural proteins are a minor but essential component of the primary cell wall (Rui and Dinneny, 2020) and include proline-rich proteins (PRPs), glycine-rich proteins (GRPs), extensins (EXTs), and AGPs. AGPs are present in vascular plants, bryophytes (Bartels et al., 2017; Johnson et al., 2017; Ma et al., 2017; Classen et al., 2019) and green algae (Palacio-Lopez et al., 2019; Přerovská et al., 2021). AGPs or AGP-like proteins have also been detected in brown algae (Herve et al., 2016) and cyanobacteria (Jackson et al., 2012) opening discussion on their evolutionary origin (Knox, 2016).
Arabinogalactan proteins have the most extensive glycosylation of Pro/Hyp-rich glycoproteins. Their carbohydrate moiety forms 90 to 99% of their molecular mass, combining galactose and arabinose as major sugars with fucose, rhamnose, and glucuronic acid as minor sugars (Fincher et al., 1983; Ellis et al., 2010; Showalter and Basu, 2016; Silva J. et al., 2020). AGPs form a complex family (Showalter, 2001). Their classification has been modified several times over the last decades. Most recently they have been divided into several groups according to their molecular structure: classical AGP, AG peptides, Lys-rich AGPs, chimeric AGPs including FLAs (FASCICLIN-LIKE AGPs), ENODLs (EARLY NODULIN-LIKE AGPs), XYLPs (XYLOGEN-LIKE AGPs), other chimeric AGPs, and HAEs (AGP-EXT hybrids) (Showalter, 2001; Pereira et al., 2015; Mareri et al., 2018; He et al., 2019; Silva J. et al., 2020). Classical AGPs are characterized by the presence of an N-terminal signal sequence, which targets the protein to the endoplasmic reticulum (ER) and secretory pathway, a middle PAST-rich domain (rich in Pro, Ala, Ser, and Thr), and a C-terminal sequence, which is cleaved during the establishment of the GPI (glycosylphosphatidylinositol) anchor in the ER (Schultz et al., 1998). AG-peptides are short classical AGPs with only 10–15 amino acids. Fasciclin-like (FLA) AGPs are also similar to classical AGPs, but possess one or two fasciclin-like domains (FAS) in their protein core (He et al., 2019). Lys-rich AGPs contain a Lys-rich domain between PAST domain and C-terminus, ENODLs contain plastocyanin-like domains, XYLPs contain non-specific lipid transfer protein domains, and HAEs combine modules characteristic for AGPs and EXTs. For further details of classification see recent reviews (Ma et al., 2017; Silva J. et al., 2020).
Proposed mechanisms of AGP functions vary among groups or may be combined within a single protein. Crosslinking of glycoproteins, such as EXTs and AGPs, by cell wall peroxidases might reinforce the cell wall (Bradley et al., 1992; Kjellbom et al., 1997). AGPs are covalently linked to pectins or hemicelluloses (Immerzeel et al., 2006; Tan et al., 2013) and their action as “pectin plasticizers” was hypothesized (Lamport et al., 2006; Corral-Martinez et al., 2019). Another putative mechanism is an enzymatic release of mobile oligosaccharides from AGP glycan side chains that may act as signaling molecules possibly recognized by plasma membrane receptors (Showalter, 2001; Van Hengel and Roberts, 2002; Zagorchev et al., 2014; Silva J. et al., 2020). In spite of studies linking activity of plant chitinases with AGPs action in developmental processes (van Hengel et al., 2001; Dos Santos et al., 2006; Zielinski et al., 2021), this mechanism needs to be proven and membrane receptors recognizing AGP-borne oligosaccharide fragments are not yet characterized. AGPs crosslinked with other cell-wall polysaccharides, especially pectins, can also modulate the plasma membrane-cell wall continuum and cell to cell adhesion (Schultz et al., 1998; Showalter, 2001). FLAs can be involved in crosslinking and cell wall adhesion through the interactions of their FLA domains in the protein core – a mechanism proposed based on their similarity with animal fasciclins and their homophilic interactions, which influence developmental processes (Snow et al., 1989; Elkins et al., 1990). The crosslinking with pectins through PAC (Proline-rich Arabinogalactan protein and Conserved Cysteines) domain is another putative mechanism. This type of interaction was documented for AtAGP31 (Hijazi et al., 2014). The protein even interacted with itself through PAC domain in vitro (Hijazi et al., 2014).
Arabinogalactan proteins are often attached to the outer side of the plasma membrane by a GPI-anchor. GPI-anchored proteins act as signal transductors that may enable the targeting of partner receptor-like kinases or modulate ligand recognition specificity as co-receptors (Yeats et al., 2018; Zhou, 2019). A proposed function of AGPs may be related to the cleavage of GPI-anchors, which may generate intracellular messengers or extracellular signals to neighboring cells (Schultz et al., 1998; Showalter, 2001). However, this remains to be conclusively proven. The cleavage of the anchor may also release the plasma membrane from the cell wall matrix, influencing membrane dynamics, including the trafficking of membrane receptors between the plasmalemma and inner compartments (Seifert, 2020). AGPs might act as a cargo linkage/receptor during the endocytosis of extracellular material (Wang et al., 2019). The function of AGPs is likely related to the general adhesive properties of their peripheral carbohydrate moieties, which are Ca2+ and pH-dependent (Tan et al., 2018). AGPs are putative calcium capacitors (Lamport and Varnai, 2013; Lopez-Hernandez et al., 2020), which bind Ca2+ in a reversible and pH-dependent manner and thus enable Ca2+ oscillations and signal transduction (Lamport and Varnai, 2013; Lamport et al., 2018). Arabidopsis thaliana mutants with compromised glucuronidation of arabinogalactans and AGPs have reduced Ca2+-binding capacity, disrupted calcium wave propagation in roots, and show serious growth defects (Lopez-Hernandez et al., 2020). The complexity of putative functions and available study tools still did not provide consistent insight into physiological aspects of this protein family.
Arabinogalactan proteins are abundant throughout the plant body, including the roots. Su and Higashiyama (2018) summarized expression data for 130 of 151 AtAGP genes (including all subgroups; classical AGPs, AG peptides, FLA, XYPL, PAG, etc.) and many of them were expressed in roots. In Populus trichocarpa, 18 of 35 identified PtrFLA genes were analyzed by qRT-PCR and all of them were expressed in roots (Zang et al., 2015). In Triticum aestivum, all 34 identified TaFLA genes were expressed mostly in seeds and roots (Faik et al., 2006). In Oryza sativa, 10 of the 69 identified OsAGPs were abundantly expressed in roots (Ma and Zhao, 2010). AGP epitopes, localized via an immuno-histochemical approach, appeared differentially in various root tissues: pericycle sectors according to vascular tissue context (Knox et al., 1989; Casero et al., 1998), developing vascular tissues, trichoblasts, atrichoblasts, growing root hairs, root caps, and border cells, for review see Showalter (2001) and Nguema-Ona et al. (2012).
Early experiments with β-Glc-Yariv reagent, which interacts with AGPs, precipitates them from solution and disrupts their activity (Yariv et al., 1967; McCartney et al., 2003), indicated a significant role for AGPs in root growth. The β-Glc-Y-enriched medium strongly reduced growth of both the root and the shoot, but the compound itself accumulated only in root. Shoot growth inhibition thus seems to be a secondary effect of the affected root system (Willats and Knox, 1996). The impaired cell elongation of the cortical cells and the bulging of the rhizodermal cells within the elongation and differentiation zones are the primary effects of the treatment (Willats and Knox, 1996; Ding and Zhu, 1997). The ability of β-Glc-Yariv to trigger cell bulging and disorganization of cortical microtubules in roots of A. thaliana was later confirmed by Nguema-Ona et al. (2007). Although not specific for a particular AGP, β-Glc-Yariv highlighted the importance of AGPs in root growth and cell differentiation.
A more focused classification of functional mechanisms comes from the study of particular mutants. Disturbances of polysaccharide metabolism and AGP carbohydrate moieties were associated with reduced primary root growth in reb1/rhd1 (root epidermal bulger 1/root hair defective 1), a galactose biosynthesis mutant of A. thaliana (see below) (Baskin et al., 1992; Nguema-Ona et al., 2006). Its phenotype can be suppressed by supplementing growth media with 10 mM galactose, which recovered root cell expansion and anisotropic growth of control (Nguema-Ona et al., 2006). Other evidence supporting the role of AGPs and their sugar moieties in root elongation came from the mur1 (murus 1) mutant of A. thaliana with reduced fucosylation (see below), which induces a significant reduction of root elongation, and more interestingly, earlier and more frequent lateral root development (Van Hengel and Roberts, 2002). Developing primordia of mur1 do not label for fucose-containing epitopes (Freshour et al., 2003). Unfortunately, neither of those experiment identified affected phase of lateral root development.
The protein SOS5/AtFLA4 (SALT OVERLY SENSITIVE 5/FLA ARABINOGALACTAN PROTEIN 4) is one of the best characterized AGP members. A. thaliana sos5/fla4 mutant, with point mutation in the FAS domain of AtFLA4, displays reduced root growth under high salinity. This phenotype is caused by defected cell expansion (for more details see below) (Shi et al., 2003) and can be suppressed by external ABA application (Seifert et al., 2014). Another non-classical AGP influencing root growth and development is AtAGP30, which is not anchored by GPI into plasma membrane. The atagp30 mutant of A. thaliana fails to initiate adventitious roots from a callus culture, but growth of already established roots, lateral roots and root hairs are apparently unaffected (Van Hengel and Roberts, 2003; Van Hengel et al., 2004). AtAGP30 transcription starts in the primary root with germination, occurs mostly in the root tip and decreases as tissue differentiate (Van Hengel and Roberts, 2003; Van Hengel et al., 2004). Interestingly, its ectopic overexpression is detrimental for shoot development and stable overexpression transformants are not viable (Van Hengel and Roberts, 2003). A recent study linked AtAGP30 with restriction of cadmium (Cd) entrance and root tip tolerance to this stressor (Jing et al., 2019). It seems that the ability to maintain AtAGP30 expression under Cd stress is proportional to the level of Cd tolerance (Jing et al., 2019). Unfortunately, it is a pure speculation whether for example Cd retention in the cell wall or membrane protection due to AtAGP30 presence is involved.
AGP presence during lateral root development was indicated by positive antibody labeling in e.g., Musa spp. (Wu et al., 2017) or Solanum lycopersicum (Sala et al., 2017). However, there are not many reports connecting AGPs with lateral root development. Mutant atfla1 of A. thaliana produces a higher number of lateral roots compared to the wild type, which suggests the role of AtFLA1 in early events of lateral root development (Johnson et al., 2011). The phase of lateral root primordia development (initiation, development and outgrowth) which is affected in atfla1 and can cause the observed phenotype has not been defined. However, peculiar differences in pericycle division of atfla1 on callus inducing medium hint at initiation and/or starting divisions. AtFLA1 expression is not root-specific but was detected in the elongation zone of primary roots, and in the meristem and vasculature of lateral roots (Johnson et al., 2011). Cell division as well as cell expansion were affected also in atagp19 mutant (Yang et al., 2007; Zhang et al., 2011) resulting in plants with fewer lateral roots, and a smaller vascular cylinder of the primary root due to the lower number of procambial cells. Unfortunately, this is mentioned without any details (Yang et al., 2007), only later commented by Zhang et al. (2011). AtAGP19 along with AtAGP17 and AtAGP18 are members of a subfamily of lysine-rich classical AGPs. AGP19 is abundant in the central cylinder of roots (Yang et al., 2007, 2011). Interestingly, decreasing the arabinogalactosylation of AGPs reduces primary root growth (Gille et al., 2013), but induces longer lateral roots in A. thaliana (Ogawa-Ohnishi and Matsubayashi, 2015). It is possible that altered carbohydrate side chains of AGPs modify their ability to crosslink in muro resulting in changes to cell wall mechanical properties that manifests during cell expansion and organ growth.
Several other AGPs are linked with root growth. AtFLA3 is barely expressed in the mature roots of wild-type A. thaliana, but its ectopic overexpression stimulates primary root growth and triggers the formation of abnormal root cap cells (Li et al., 2010). In contrast, ectopic overexpression of AtAGP18 significantly inhibits root growth (Zhang et al., 2011). AtAGP18 regulates megaspore development (Demesa-Arevalo and Vielle-Calzada, 2013) but it is expressed also in roots, mostly in vascular tissues (Yang and Showalter, 2007), and its expression is under the control of ABA (Zhang et al., 2011). The AtAGP18-RNAi silenced lines have a high rate of seed abortion. Root growth phenotype was not observed in the same study but it was not analyzed in details (Acosta-García and Vielle-Calzada, 2004). AtAGP18 would therefore be an interesting candidate for future root-focused studies.
Several pieces of evidence implicate some AGPs in the regulation of root hair initiation and growth. Aberrant root-hair development in atagp21 is connected with contiguous root hair formation and high root hair density (Borassi et al., 2020). AtAGP21 is a part of the brassinosteroid regulatory circuits upstream of GL2 (GLABRA2), RHD6 (ROOT HAIR DEFECTIVE 6) and other downstream transcription factors determining the development of epidermal cells into root hairs. AtAGP21 itself is positively regulated by the BZR1 transcription factor and acts as a suppressor of GL2 (Borassi et al., 2020). A root-hair phenotype similar to atagp21 also occurs in other A. thaliana mutants with altered AGP content, such as O-glycosylation, fucosylation, or arabinosylation of AGPs, e.g., atagp15, hpgt (Ogawa-Ohnishi and Matsubayashi, 2015; Borassi et al., 2020). The hpgt1-1 hpgt2-1 hpgt3-1 triple-mutant is defective in O-glycosylation of AGPs due to the disruption of hydroxyproline galactosyltransferase 1–3 and forms longer and more dense root hairs compared to wild-type plants (Ogawa-Ohnishi and Matsubayashi, 2015). O-glycosylation of AtAGP21 is essential for its function, particularly secretion and cellular targeting (Borassi et al., 2020). Contiguous root hair development can also be triggered by β-Glc-Y (α-Man-Y has no effect) crosslinking AGPs and limiting their action in the cell wall, providing additional evidence for the role of AGPs in determining rhizodermal-cell fate in A. thaliana (Borassi et al., 2020). Another piece of evidence linking AGPs and root hair growth is a long-hair phenotype of agp14 mutant of A. thaliana (Lin et al., 2011) and a short-hair phenotype of higher-order glcat14 (β-glucuronosyl-transferases 14A-C) mutants of A. thaliana with increased AGP contents (Zhang et al., 2020).
The role of AGPs in the determination of rhizodermal-cell fate is further supported by studies on other plant species. In Zea mays and Hordeum vulgare, specific AGP epitopes were detected on the surface of trichoblasts and root hairs, which differed from those of atrichoblasts (Šamaj et al., 1999; Marzec et al., 2015). Moreover, epitopes detected by LM2, LM14, and MAC207 antibodies, which are normally present at the surface of trichoblasts in H. vulgare, were absent in the rhizodermis of barley root-hairless mutant 1 (Marzec et al., 2015). In Brassica carinata, downregulation of BcFLA1, encoding a FLA AGP, via CRISPR/Cas9 significantly reduced root-hair length in phosphate-deficient conditions (Kirchner et al., 2018). BcFLA1 expression was enhanced by Pi deficiency, specifically in the low-P efficient cultivar of B. carinata. This cultivar is efficient in Pi uptake and increases the length of root hairs in Pi-deficient conditions considerably (Kirchner et al., 2018).
Interestingly, extensin related modifications of O-glycosylation did affect the root hair growth but not cell fate (Velasquez et al., 2015). Proline-rich extensin-like receptor kinase 13 (PERK13) was shown to provide negative control of root hair growth. A. thaliana mutant rhs10/perk13 (root hair specific 10/proline-rich extensin-like receptor kinase 13) has longer root hairs. PERK13 has AGP motifs in its extracellular domain, which may be important for its regulatory function (Hwang et al., 2016). It is proposed that AGP motifs sense the cell-wall integrity, triggering down-stream signal transduction (Cho, 2016). These results taken together indicate that AGPs might affect root hair formation via sensing or modification of cell wall properties, and can participate in signaling pathways controlling root-hair cell fate by an interaction with other proteins or cell wall components, e.g., receptor-like kinases or pectins.
As for other plant organs, the growth of roots is determined by cell division, elongation, and differentiation, which are tightly connected with cell wall characteristics. Cell wall composition and mechanical properties are developmentally regulated and respond to environmental factors (Cosgrove, 2005; Caffall and Mohnen, 2009; Somssich et al., 2016; Rui and Dinneny, 2020). Localization of GPI-anchored AGPs on the outer surface of the plasma membrane and their linkage to other cell wall components make them putative linkers of protoplast and the cell wall. β-D-glucosyl units of “active Yariv” reagent (Yariv et al., 1967) bind and precipitate AGPs, disrupting their action. Such treatment, similar to anti-AGP antibodies, induces rearrangement of microtubule cortical arrays in rhizodermal cells within minutes (Nguema-Ona et al., 2007) and stimulates an intense swelling of epidermal cells in the elongation zone in the longer term in A. thaliana (Ding and Zhu, 1997; Nguema-Ona et al., 2007). The impaired cell elongation was also observed in cell suspension cultures of Daucus carota (Willats and Knox, 1996).
A similar effect of AGPs on cell volume expansion is induced if the AGP glycosylation machinery is affected. Mutations in AGP-specific O-galactosyltransferases lead to defects in cell expansion. The galt2 galt5 (hydroxyproline-O-galactosyltransferase 2,5) mutant of A. thaliana has two disrupted AGP-specific galactosyltransferases, which are important for binding the galactose to the protein backbone and initializing O-glycosylation (Basu et al., 2013, 2015). Together with the lower glycosylation status of AGPs, the mutant displays reduced seed-coat cellulose content, swollen root-tip cells, and other root growth defects, e.g., inhibition of root growth, reduction of root hair length and density (Basu et al., 2015). Shorter roots were observed also in the quintuple mutant galt2 galt3 galt4 galt5 galt6, but surprisingly this mutant formed longer root hairs compared to wild type (Zhang et al., 2021). All these observations highlight the importance of O-glycosylation in cell growth and cell wall deposition (Basu et al., 2015; Showalter and Basu, 2016). In addition, the disruption of two Golgi-localized exo-β-1,3-galactosidases of glycoside hydroxylase family 43 (GH3) in the gh43 mutant of A. thaliana increases the content of cell-wall bound AGPs and triggers serious defects in root cell expansion and adhesion, e.g., root epidermal cell swelling and loss of anisotropic growth (Nibbering et al., 2020). These exo-β-1,3-galactosidases are putatively involved in the processing of AGPs during their maturation in the Golgi, regulating the length of the β-1,3-galactan backbone of AGPs, and altering the affinity of mature AGPs to other cell wall components (Nibbering et al., 2020).
The connection between AGP glycosylation and regulation of root cell expansion and cell wall properties is highlighted in other studies, where galactosylation and fucosylation are modified, affecting AGPs along with pectins and hemicelluloses. The A. thaliana mutant mur1 with a disrupted GDP-D-mannose-4,6-dehydratase enzyme of the GDP-L-fucose biosynthetic pathway contains less L-fucose in cell walls (Reiter et al., 1993; Bonin et al., 1997). L-fucose is a minor component of AGPs (Silva J. et al., 2020) as well as xyloglucans (Somssich et al., 2016). The L-fucose deficient mutant shows reduced root elongation by more than half compared to the wild type, and swollen root tips. Root growth inhibition is caused by a significant reduction in cell elongation, while the activity of root apical meristem is normal (Bonin et al., 1997; Van Hengel and Roberts, 2002). Alteration of root cell anisotropic growth occurs also in the reb1/rhd1 mutant (Baskin et al., 1992). Reduced root elongation and bulging trichoblasts observed in this mutant (Baskin et al., 1992; Andème-Onzighi et al., 2002) seem related to altered galactosylation of cell-wall xyloglucans (Nguema-Ona et al., 2006). The mutant has defective UDP D-galactose 4-epimerase enzyme (Seifert et al., 2002) and makes structurally different cell wall xyloglucans, which are devoid of galactose and fucose residues (Nguema-Ona et al., 2006). There is also an obvious link to AGPs and cytoskeletal structures, as the trichoblasts of reb1/rhd1 have disorganized microtubules and lack AGPs detectable by JIM14 and LM2 antibodies (Andème-Onzighi et al., 2002). However, the functional link is currently not known.
Mutant dim/dwf1 (diminuto/dwarf1) of A. thaliana in the brassinosteroid biosynthesis gene DIM/DWF1 (Klahre et al., 1998) is strongly affected in cell elongation and has reduced cellulose and lignin content (Hossain et al., 2012). The dim/dwf1 phenotype correlates with the amount of AGPs in the tissue, highlighting the role of AGPs in cell expansion (Takahashi et al., 1995) and implicating them in an executive part of the brassinosteroid signaling circuit (Jia et al., 2020).
Arabinogalactan protein-related growth defects often manifest strongly in the presence of high salinity or other stress factors, and are linked to cell wall integrity, maintenance, and adjustment of mechanical properties (Rui and Dinneny, 2020). Synthesis of L-arabinose, which is incorporated into AGPs, EXTs and some cell wall polysaccharides, depend on the MUR4/HSR8 (MURUS4/HIGH SUGAR RESPONSE 8) Golgi-localized UDP-D-xylose 4-epimerase. Plants of mur4/hsr8 show a significant reduction of L-arabinose (Reiter et al., 1997; Burget and Reiter, 1999; Burget et al., 2003) and a strong short-root phenotype under salinity, but not in either standard or osmotic stress (mannitol treatment) growth conditions (Zhao et al., 2019). Analysis of mur4/hsr8 mutant indicates defective cell wall structure but not signaling. This phenotype results in decreased root elongation and also cell-cell adhesion, resulting in epidermal discontinuity and bursting of cells (Zhao et al., 2019). Described defects were rescued by exogenous arabinose, but not glucose or xylose, confirming UDP-Ara biosynthesis consequence and affecting the level of AGP staining in roots (Zhao et al., 2019). Other enzymes affecting cell-wall AGPs are FUT4 and FUT6 (α-1,2-fucosyltransferases 4, 6), which are responsible for their fucosylation. Double mutant of A. thaliana fut4 fut6 has lower content of fucose and xylose in AGP extracts and short-root phenotype under conditions of salt stress (Tryfona et al., 2014).
The role of AGPs as pectin plasticizers and regulators of cell-wall extensibility under salt stress was proposed rather early (Zhu et al., 1993; Lamport et al., 2006; Olmos et al., 2017). Interestingly, AGPs isolated from roots (and other organs) of the seagrass Zostera marina reportedly had specific characteristics, distinguishing them from the AGPs of land plants (high degree of branching, high content of terminal α-L-arabinose), which might enhance the salt tolerance of this marine species (Pfeifer et al., 2020). In Urochloa decumbens, AGP epitopes accumulated in root cell walls of after aluminum treatment to maintain cell wall flexibility and increase the high-aluminum tolerance of this tropical grass (Silva T.F. et al., 2020). A recently proposed alternative model of AGP action under salinity stress is their function as carriers, binding Na+ ions and transferring them into the vacuole via vesicle trafficking (Olmos et al., 2017).
One of the best characterized AGPs in the context of salinity is SOS5/FLA4. The salt-sensitive mutant sos5/fla4 of A. thaliana has swollen root-tip cells due to abnormal cell expansion occurring under salt stress (Shi et al., 2003). Cell walls of sos5/fla4 have an altered structure. The pectin-rich middle lamella, essential for intercellular adhesion, is reduced and primary cell walls are thinner and less organized compared to the wild type (Shi et al., 2003). Interestingly, the hypertensive sos5/fla4 root phenotype under salt stress is milder in ABA-oversensitive mutants and suppressible by exogenous application of ABA (Seifert et al., 2014; Acet and Kadioglu, 2020). The protein might act synergistically with ABA as a putative modulator of ABA signaling upstream of cell wall biosynthesis (Seifert et al., 2014) and independent of the RBOHD and RBOHF (RESPIRATORY BURST OXIDASE HOMOLOG D,F) NADH oxidases (Xue and Seifert, 2015) of the ABA-signaling pathway controlling root growth (Jiao et al., 2013).
Interaction between ABA and SOS5/FLA4 modulates the content of H2O2 under salt stress (Acet and Kadioglu, 2020), indicating a more significant signaling rather than structural role for SOS5/FLA4. This is consistent with an identical phenotype reported previously for two AGP-specific galactosyltransferases (GALT2 and GALT5), fasciclin-like AGP (SOS5/FLA4) and two leucine rich repeat receptor kinases (FEI1 and FEI2) (Shi et al., 2003; Xu et al., 2008; Basu et al., 2015), which placed those components into a single regulatory pathway (Basu et al., 2016) and derived speculation that SOS5/FLA4 might act as a sensor of conditions in the apoplast via FEI kinases (Turupcu et al., 2018; Seifert, 2021). SOS5/FLA4 tagged with GFP was detected on the plasma membrane, soluble in the apoplast, and in endosomes (Xue et al., 2017). Its C-terminal fasciclin 1 domain (Fas1-2) is essential for its function, possibly involved in molecular interactions. The N-terminal Fas1 domain (Fas1-1) stabilizes proteins in the plasma-membrane (Xue et al., 2017), and it is a putative negative regulator of Fas1-2 binding to FEI1 kinase, which might augment the regulation of root growth according to environmental conditions (Turupcu et al., 2018; Seifert, 2021).
The roles of other individual AGPs and their subtypes still remain to be elucidated, but there is extensive experimental evidence (often coming from organs other than roots) which supports their role in cell wall biochemistry, deposition, and signaling. Modulation of EgrFLA1,2,3 expression levels in Eucalyptus grandis (MacMillan et al., 2010, 2015), PtFLA6 in Populus (Wang et al., 2015), AtFLA11, AtFLA12, and AtFLA16 in A. thaliana (MacMillan et al., 2010; Liu et al., 2020) altered stem cell-wall polysaccharide composition, cell-wall thickness, and stem mechanical properties. GhAGP3 and GhAGP4 are specifically expressed during the transition between cell elongation to the secondary cell wall deposition in developing cotton (Gossypium hirsutum) fibers, highlighting their roles during secondary cell wall formation (Liu et al., 2008). In Physcomitrella patens, application of AGP binding β-Glc-Yariv or the downregulation of AGP1 reduced the expansion of the protonema apical cell (Lee et al., 2005).
Roots provide an interface for interaction with rhizosphere biota. AGPs, putative environment-cell-wall-protoplast signal transductors (Seifert and Roberts, 2007), are important components of root exudates and root cell walls, especially in the root-cap and root-associated, cap-derived cells (Vicré et al., 2005; Cannesan et al., 2012; Koroney et al., 2016; Swamy et al., 2016; Driouich et al., 2019) and aid in the formation of the rhizosheath (Galloway et al., 2020). As such, they are likely mediators of root-microorganism interactions, participating in the attraction, recognition, and colonization of roots by beneficial microorganisms as well as in root responses to microbial pathogens (Nguema-Ona et al., 2012, 2013, 2014; Mareri et al., 2018) and parasites (Bozbuga et al., 2018).
AGPs and chimeric arabinogalactan protein-extensins (AGPEs) take part in the mutual interactions between roots and microorganisms. AGP-epitopes were found at arbuscular mycorrhiza symbiotic interfaces (Gollotte et al., 1995; Balestrini and Lanfranco, 2006). The involvement of MtAMA1 (ARBUSCULAR MYCORRHIZA AGP 1) in arbuscular mycorrhiza is indicated by the specific expression of the MtAMA1 gene exclusively in arbuscule containing cortical cells of Medicago truncatula (Schultz and Harrison, 2008). Its mode of operation in the plant-fungi interface is still unknown, but signaling feedback from the cell wall might be anticipated. The authors speculate about a possible coreceptor on the plasma membrane or a mobile signaling molecule after its release from plasma membrane by the cleavage of the GPI anchor (Schultz and Harrison, 2008). Interestingly, two AGP-like (AGL) proteins were identified in the genome of Glomus intraradices, with a specific structure not found in plants or non-mycorrhizal fungi. These GiAGLs contain repeat domains that can form polyproline II helices with positively and negatively charged faces. The authors suggest their role in the interaction with host cell wall surface (Schultz and Harrison, 2008). Unfortunately, there are few recent references on this particular topic.
A symbiont as a source of AGPs at the host interface was recorded also from free-living cyanobacteria Nostoc, containing a putative AGP peptide genes (classical AGP, AG peptide, and FLA class) and cell surface epitopes responsive to AGP antibodies were detected at the Nostoc-Gunnera interface (Jackson et al., 2012). Their discovery suggests that the role of AGPs in the host-symbiont interface might develop from rather ancient cell surface interaction processes and AGP role might evolutionarily originate from very early symbioses (Jackson et al., 2012).
Interaction via AGPs during symbiotic infection by nitrogen-fixing rhizobia has been repeatedly proven, for review see Brewin (2004), Brewin et al. (2008), Nguema-Ona et al. (2013), Rashid (2016). Formation of new lateral root organs - nodules colonized by rhizobia, is a tightly orchestrated process, which is mainly initiated by microbial entry via an infection thread (Coba de la Pena et al., 2017; Ferguson et al., 2019). Rhizobia traveling through infection threads are embedded in a matrix containing AGPEs and other glycoproteins (Rathbun et al., 2002; Brewin, 2004; Reguera et al., 2010). Abnormal infection thread development in Pisum sativum mutants (sym33; sym 40) is associated with disrupted targeting of AGPEs (MAC265 antibody) exocytosis and authors speculate that this might be correlated with inefficient symbiosome formation in mutants (Tsyganova et al., 2009). Cell wall remodeling that takes place during onset of the symbiosome (Coba de la Pena et al., 2017; Tsyganova et al., 2019) is the potentially affected process. AGPs (localized with JIM1 antibody) are present in the nodule membranes during the maturation of symbiosomes in Pisum (Tsyganova et al., 2019). Their significance is still unclear but their presence was not observed in nodules of the sym31 mutant (Tsyganova et al., 2019) with undifferentiated bacterioids and symbiosome membranes staying in the juvenile state (Borisov et al., 1997). This indicates that AGPs play a role in symbiosome maturation and ontogeny (Tsyganova et al., 2019). AGPs are abundant also in the actinorhizal nodules of Alnus, especially during early nodulation stages (Berry et al., 2002).
In addition, AGP-encoding genes are upregulated in Oryza sativa roots upon colonization by Piriformospora indica (Nivedita et al., 2020), a beneficial growth-promoting fungal endophyte that improves salt-stress tolerance in many plant species (Waller et al., 2005; Trivedi et al., 2013). In Triticum aestivum, AGP-epitopes (detected by JIM14) occur abundantly in roots infected by Trichoderma ssp., a beneficial fungal antagonist of phytopathogens (Basińska-Barczak et al., 2020). These recent observations indicate that AGPs may also promote root interaction with beneficial endophytes.
Analyzing the role of AGPs in root response to pathogens, a suppressive role to early infection by microbial pathogens was demonstrated by AGPs extracted from border cells (BC) of Pisum sativum and border-like cells (BLC) of Brassica napus (Cannesan et al., 2012). AGPs from BL and BLC attracted zoospores of oomycete Aphanomyces euteiches and induced their encystment (loss of the motility due to loss of the flagella). The attraction was far more efficient for P. sativum extract in agreement with the fact that A. euteiches is the pathogen of P. sativum not B. napus. Root exudates, but not extracted AGPs, then strongly stimulated their germination (Cannesan et al., 2012). Root-associated, cap-derived cells (BC and BLC) thus act as a blind target, trapping the pathogen (extracellular root trap) and preventing its contact with the root proper (Hawes et al., 2000; Driouich et al., 2019; Ropitaux et al., 2020).
There is also substantial evidence that the composition of AGPs in roots or root exudates changes in response to pathogens or parasites. In Solanum tuberosum, AGPs (detected with LM2 and JIM15 antibodies) were upregulated in root exudates in response to elicitors derived from Pectobacterium atrosepticum, the pathogen causing soft rot disease in potato (Koroney et al., 2016). In Musa spp. roots, AGPs were upregulated by Fusarium oxysporum f. sp. cubense infection (Wu et al., 2017). Changes in AGP levels occurred in the roots of A. thaliana infected by Plasmodiophora brassiace, which caused clubroot disease. In this case, AGPs were mostly downregulated, but FLA5 was upregulated together with many cell-wall-modifying enzymes, alpha-expansins in particular (Irani et al., 2018). In the roots of Glycine max, repression of FLA encoding genes was induced by the fungal pathogen Macrophomina phaseolina (Marquez et al., 2018) trying to seize root tissues. Besides microbial pathogens, animal parasites induce changes in root AGP levels as well. In roots of a resistant cultivar of Glycine max, the upregulation of FLAs is triggered by the attack of root-knot nematodes (Beneventi et al., 2013).
Fluctuation of AGP levels occurs also during the attack of parasitic plant species of Cuscuta genus on the host-plant stems. Epidermal contact of Cuscuta reflexa stimulates the secretion of AGPs by the host plant, Lycopersicon esculentum, to enhance its adhesion to the host stem in the early phase of interaction (Albert et al., 2006). Downregulation of attAGP (attachment AGP) expression decreased the attachment capability of the parasite (Albert et al., 2006). The presence of AGPs in attachment “cement” was recorded on the surface (holdfast epidermal cells) of C. campestris and C. japonica stems (Hozumi et al., 2017) supporting the role of AGPs in parasite-host attachment. Accumulation of AGPs in the tip of developing haustoria appear after penetration of the host stem (Hozumi et al., 2017; Shimizu and Aoki, 2019) and expression analysis of Cuscuta developing haustoria identified them as FLAs. On the contrary, the later intrusive growth of Cuscuta haustorium triggers the depletion of AGPs in stem tissues facing the attack, which was shown for Pelargonium zonale penetrated by C. reflexa (Striberny and Krause, 2015). In Orobanchaceae root parasites, AGPs accumulate in the hyaline body, a specialized parenchymatous central core of the parasitic haustorium. The functional significance of this accumulation is, however, unclear (Pielach et al., 2014).
Mechanisms of AGP action in root biotic interaction are still unresolved and puzzling. Several mechanisms were proposed, including the recognition and attachment of microbes, formation of a protective biofilm against degradation of cell wall by pathogenic organisms, or antimicrobial action, for review see Nguema-Ona et al. (2013) and Mareri et al. (2018). In addition, the significance of AGPs in response to pathogens is frequently inconclusive. They may act together with EXTs to modify the cell wall cross-linking in response to pathogens, for review see Rashid (2016). In some studies, EXTs seemed more important. Among others, EXTs rather than AGPs correlated with the resistance to F. oxysporum f. sp. cubense, in spite of the pathogen-induced changes in AGP levels in Musa spp. cultivars (Wu et al., 2017). β-Glc-Y reagent failed to affect the interaction with Pectobacterium atrosepticum, although AGPs were upregulated in response to this pathogen in Solanum tuberosum roots. Root exudate pre-incubated with β-Glc-Y promoted the growth of the pathogen in a very similar way as non-incubated one (Koroney et al., 2016). Higher levels of AGPs and also EXTs were detected in roots of a Benincasa hispida cultivar resistant to F. oxysporum f. sp. Benincaseae (Xie et al., 2011). Various studies indicate that other cell-wall glycoproteins (EXTs or AGPEs), are at least equally important and change their levels in roots in response to pathogens or symbionts (Shailasree et al., 2004; Plancot et al., 2013; Wu et al., 2017; Castilleux et al., 2020).
Further and more conclusive functional characterization of AGPs roles in root-pathogen interactions thus requires direct evidence based e.g., on modulation of AGP-genes expression and analyses of induced phenotypes. There are only few studies revealing the role of individual AGPs in this process. A. thaliana rat1/agp17 (resistant to Agrobacterium transformation 1) mutant, defective in arabinogalactan protein AtAGP17, is resistant to Agrobacterium transformations of root segments (Nam et al., 1999; Gaspar et al., 2004). In spite of the difficulties with AtAGP17 transcript detection in roots (Gaspar et al., 2004; Yang et al., 2007, 2011), the protein seems highly abundant in root tissues (Yang et al., 2011). It affects the attachment of Agrobacterium to the root surface and modulates the systemic acquired resistance, which allows for successful infection (Gaspar et al., 2004). Two other AGPs, AtAGP12, and AtAGP24, enhanced their expression in the roots of A. thaliana after infection of Plectosphaerella cucumerina, a necrotrophic fungal pathogen. AtAGP24-GFP localized in close proximity to plasma membrane and the overexpression of AtAGP24 strongly increased the susceptibility to P. cucumerina, which is evidence for its involvement in the pathogen response (Dobon et al., 2015).
There is also direct evidence of the involvement of a particular AGP gene in root defense against animal parasites. The knock-out of AtAGP8 gene in A. thaliana leads to a significantly increased susceptibility toward root-knot nematode Meloidogyne incognita (Bozbuga et al., 2018). The susceptibility seems related to the cell wall composition and resistance of root tissue to form specific feeding sites, giant cells. These hypertrophied multinucleate cells re-differentiate from a small number of root cells being pierced by a nematode stylet. Their cells walls contain AGPs and are enriched with highly methyl-esterified homogalacturonans, xyloglucans and arabinans, allowing for plasticity and cell expansion (Bozbuga et al., 2018). Increased susceptibility to root cyst nematode was also observed in reb1/rhd1 mutant (Baum et al., 2000; Wubben et al., 2004) with lower AGP levels in roots (Ding and Zhu, 1997). Besides the atagp8 and reb1/rhd1 mutants, increased susceptibility to nematodes was found in two rhamnogalacturonan I pectin deficient mutants of A. thaliana (arabinan deficient 1,2), while mutants with suppressed mannan and galactan epitopes (mannan synthesis-related 1 and β-galactosidase 5) were more resistant (Gantulga et al., 2008; Harholt et al., 2012; Wang et al., 2013; Bozbuga et al., 2018).
Cell-wall localized AGPs work as modulators of cell expansion and differentiation, signal transductors on the cell surface, and effectors of responses to environmental conditions and other organisms. In roots, the multifaceted roles of AGPs are emphasized due to the requirement for high growth plasticity and constant exchange of signals with the environment. The data gained from observing plants with altered expression of AGPs or carbohydrate composition of cell wall, immunohistochemical studies, and structural analyses clearly link AGPs and their glycosylation status with cell wall properties, cell expansion and organ growth.
Despite the obvious significance of AGPs, we still have limited information about the roles of individual AGPs in roots and the whole plant. Abundance of AGPs, the complexity of their functions, and their obvious redundancy make this issue challenging. A detailed focus on loss-of-function mutants can move us ahead in understanding the mechanisms of AGP action in roots. Characterization of AGP mutants were summarized in this review alongside other studies on cell wall chemistry to provide an overview of the current state of this topic.
DH performed the literature survey, drafted and wrote the manuscript. ET performed the literature survey, drafted and wrote the manuscript, and made the figure and table. AS conceptualized and finalized the manuscript. All authors contributed to the article and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors thank Lena M. Hunt M.Sc. for English language correction.
Acet, T., and Kadioglu, A. (2020). SOS5 gene-abscisic acid crosstalk and their interaction with antioxidant system in Arabidopsis thaliana under salt stress. Physiol. Mol. Biol. Plants 26, 1831–1845. doi: 10.1007/s12298-020-00873-4
Acosta-García, G., and Vielle-Calzada, J.-P. (2004). A classical arabinogalactan protein is essential for the initiation of female gametogenesis in Arabidopsis. Plant Cell 16, 2614–2628. doi: 10.1105/tpc.104.024588
Albert, M., Belastegui-Macadam, X., and Kaldenhoff, R. (2006). An attack of the plant parasite Cuscuta reflexa induces the expression of attAGP, an attachment protein of the host tomato. Plant J. 48, 548–556. doi: 10.1111/j.1365-313X.2006.02897.x
Andème-Onzighi, C., Sivaguru, M., Judy-March, J., Baskin, T. I., and Driouich, A. (2002). The reb1-1 mutation of Arabidopsis alters the morphology of trichoblasts, the expression of arabinogalactan-proteins and the organization of cortical microtubules. Planta 215, 949–958. doi: 10.1007/s00425-002-0836-z
Balestrini, R., and Lanfranco, L. (2006). Fungal and plant gene expression in arbuscular mycorrhizal symbiosis. Mycorrhiza 16, 509–524. doi: 10.1007/s00572-006-0069-2
Bartels, D., Baumann, A., Maeder, M., Geske, T., Heise, E. M., von Schwartzenberg, K., et al. (2017). Evolution of plant cell wall: arabinogalactan-proteins from three moss genera show structural differences compared to seed plants. Carbohydr. Polym. 163, 227–235. doi: 10.1016/j.carbpol.2017.01.043
Basińska-Barczak, A., Błaszczyk, L., and Szentner, K. (2020). Plant cell wall changes in common wheat roots as a result of their interaction with beneficial fungi of Trichoderma. Cells 9:2319. doi: 10.3390/cells9102319
Baskin, T. I., Betzner, A. S., Hoggart, R., Cork, A., and Williamson, R. (1992). Root morphology mutants in Arabidopsis thaliana. Funct. Plant Biol. 19, 427–437. doi: 10.1071/PP9920427
Basu, D., Liang, Y., Liu, X., Himmeldirk, K., Faik, A., Kieliszewski, M., et al. (2013). Functional identification of a hydroxyproline-o-galactosyltransferase specific for arabinogalactan protein biosynthesis in Arabidopsis. J. Biol. Chem. 288, 10132–10143. doi: 10.1074/jbc.M112.432609
Basu, D., Tian, L., Debrosse, T., Poirier, E., Emch, K., Herock, H., et al. (2016). Glycosylation of a fasciclin-like arabinogalactan-protein (SOS5) mediates root growth and seed mucilage adherence via a cell wall receptor-like kinase (FEI1/FEI2) pathway in Arabidopsis. PLoS One 11:e0145092. doi: 10.1371/journal.pone.0145092
Basu, D., Wang, W., Ma, S., DeBrosse, T., Poirier, E., Emch, K., et al. (2015). Two hydroxyproline galactosyltransferases, GALT5 and GALT2, function in arabinogalactan-protein glycosylation, growth and development in Arabidopsis. PLoS One 10:e0125624. doi: 10.1371/journal.pone.0125624
Baum, T., Wubben, M. K. II, Su, H., and Rodermel, S. (2000). A screen for Arabidopsis thaliana mutants with altered susceptibility to Heterodera schachtii. J. Nematol. 32:166.
Beneventi, M. A., da Silva, O. B., de Sá, M. E. L., Firmino, A. A. P., de Amorim, R. M. S., Albuquerque, ÉV. S., et al. (2013). Transcription profile of soybean-root-knot nematode interaction reveals a key role of phythormones in the resistance reaction. BMC Genomics 14:322. doi: 10.1186/1471-2164-14-322
Berry, A. M., Rasmussen, U., Bateman, K., Huss-Danell, K., Lindwall, S., and Bergman, B. (2002). Arabinogalactan proteins are expressed at the symbiotic interface in root nodules of Alnus spp. New Phytol. 155, 469–479. doi: 10.1046/j.1469-8137.2002.00466.x
Bonin, C. P., Potter, I., Vanzin, G. F., and Reiter, W.-D. (1997). The MUR1 gene of Arabidopsis thaliana encodes an isoform of GDP-D-mannose-4, 6-dehydratase, catalyzing the first step in the de novo synthesis of GDP-L-fucose. Proc. Natl. Acad. Sci. 94, 2085–2090. doi: 10.1073/pnas.94.5.2085
Borassi, C., Gloazzo Dorosz, J., Ricardi, M. M., Carignani Sardoy, M., Pol Fachin, L., Marzol, E., et al. (2020). A cell surface arabinogalactan-peptide influences root hair cell fate. New Phytol. 227, 732–743. doi: 10.1111/nph.16487
Borisov, A. Y., Rozov, S., Tsyganov, V., Morzhina, E., Lebsky, V., and Tikhonovich, I. (1997). Sequential functioning of Sym-13 and Sym-31, two genes affecting symbiosome development in root nodules of pea (Pisum sativum L.). Mol. Gen. Genet. 254, 592–598. doi: 10.1007/s004380050456
Bossy, A., Blaschek, W., and Classen, B. (2009). Characterization and immunolocalization of arabinogalactan-proteins in roots of Echinacea purpurea. Planta Med. 75, 1526–1533. doi: 10.1055/s-0029-1185801
Bozbuga, R., Lilley, C. J., Knox, J. P., and Urwin, P. E. (2018). Host-specific signatures of the cell wall changes induced by the plant parasitic nematode, Meloidogyne incognita. Sci. Rep. 8:17302. doi: 10.1038/s41598-018-35529-7
Bradley, D. J., Kjellbom, P., and Lamb, C. J. (1992). Elicitor-and wound-induced oxidative cross-linking of a proline-rich plant cell wall protein: a novel, rapid defense response. Cell 70, 21–30. doi: 10.1016/0092-8674(92)90530-P
Brewin, N. J. (2004). Plant cell wall remodelling in the Rhizobium–legume symbiosis. Crit. Rev. Plant Sci. 23, 293–316. doi: 10.1080/07352680490480734
Brewin, N., Khodorenko, A., Tsyganov, V., Borisov, A., Tikhonovich, I., and Rathbun, E. (2008). “Legume AGP-extensins in Rhizobium infection,” in Biological Nitrogen Fixation: Towards Poverty Alleviation Through Sustainable Agriculture, eds F. Dakora, S. Chimphango, A. Valentine, C. Elmerich, and N. WE (Dordrecht: Springer), 185–187. doi: 10.1007/978-1-4020-8252-8_70
Burget, E. G., and Reiter, W.-D. (1999). The mur4 mutant of Arabidopsis is partially defective in the de novo synthesis of uridine diphosphol-arabinose. Plant Physiol. 121, 383–390. doi: 10.1104/pp.121.2.383
Burget, E. G., Verma, R., Mølhøj, M., and Reiter, W.-D. (2003). The biosynthesis of L-arabinose in plants: molecular cloning and characterization of a Golgi-localized UDP-D-xylose 4-epimerase encoded by the MUR4 gene of Arabidopsis. Plant Cell 15, 523–531. doi: 10.1105/tpc.008425
Caffall, K. H., and Mohnen, D. (2009). The structure, function, and biosynthesis of plant cell wall pectic polysaccharides. Carbohydr. Res. 344, 1879–1900. doi: 10.1016/j.carres.2009.05.021
Cannesan, M. A., Durand, C., Burel, C., Gangneux, C., Lerouge, P., Ishii, T., et al. (2012). Effect of arabinogalactan proteins from the root caps of pea and Brassica napus on Aphanomyces euteiches zoospore chemotaxis and germination. Plant Physiol. 159, 1658–1670. doi: 10.1104/pp.112.198507
Casero, P. J., Casimiro, I., and Knox, J. P. (1998). Occurrence of cell surface arabinogalactan-protein and extensin epitopes in relation to pericycle and vascular tissue development in the root apex of four species. Planta 204, 252–259. doi: 10.1007/s004250050254
Castilleux, R., Plancot, B., Gugi, B., Attard, A., Loutelier-Bourhis, C., Lefranc, B., et al. (2020). Extensin arabinosylation is involved in root response to elicitors and limits oomycete colonization. Ann. Bot. 125, 751–763. doi: 10.1093/aob/mcz068
Cheung, A. Y., Wang, H., and Wu, H.-M. (1995). A floral transmitting tissue-specific glycoprotein attracts pollen tubes and stimulates their growth. Cell 82, 383–393. doi: 10.1016/0092-8674(95)90427-1
Cheung, A., and Wu, H.-M. (1999). Arabinogalactan proteins in plant sexual reproduction. Protoplasma 208, 87–98. doi: 10.1007/BF01279078
Cho, H.-T. (2016). Arabinogalactan protein motif-containing receptor-like kinases are likely to play the negative feedback factor to maintain proper root hair length. Plant Signal. Behav. 11, 2007–2022. doi: 10.1080/15592324.2016.1226454
Clarke, A., Anderson, R., and Stone, B. (1979). Form and function of arabinogalactans and arabinogalactan-proteins. Phytochemistry 18, 521–540. doi: 10.1016/S0031-9422(00)84255-7
Classen, B., Baumann, A., and Utermoehlen, J. (2019). Arabinogalactan-proteins in spore-producing land plants. Carbohydr. Polym. 210, 215–224. doi: 10.1016/j.carbpol.2019.01.077
Coba de la Pena, T., Fedorova, E., Pueyo, J. J., and Lucas, M. M. (2017). The symbiosome: legume and Rhizobia co-evolution toward a nitrogen-fixing organelle? Front. Plant Sci. 8:2229. doi: 10.3389/fpls.2017.02229
Corral-Martinez, P., Driouich, A., and Segui-Simarro, J. M. (2019). Dynamic changes in arabinogalactan-protein, pectin, xyloglucan and xylan composition of the cell wall during microspore embryogenesis in Brassica napus. Front. Plant Sci. 10:332. doi: 10.3389/fpls.2019.00332
Cosgrove, D. J. (2005). Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol. 6, 850–861. doi: 10.1038/nrm1746
Demesa-Arevalo, E., and Vielle-Calzada, J. P. (2013). The classical arabinogalactan protein AGP18 mediates megaspore selection in Arabidopsis. Plant Cell 25, 1274–1287. doi: 10.1105/tpc.112.106237
Ding, L., and Zhu, J.-K. (1997). A role for arabinogalactan-proteins in root epidermal cell expansion. Planta 203, 289–294. doi: 10.1007/s004250050194
Dobon, A., Canet, J. V., Garcia-Andrade, J., Angulo, C., Neumetzler, L., Persson, S., et al. (2015). Novel disease susceptibility factors for fungal necrotrophic pathogens in Arabidopsis. PLoS Pathog. 11:e1004800. doi: 10.1371/journal.ppat.1004800
Dolan, L., Linstead, P., and Roberts, K. (1995). An AGP epitope distinguishes a central metaxylem initial from other vascular initials in the Arabidopsis root. Protoplasma 189, 149–155. doi: 10.1007/bf01280168
Dos Santos, A. L. W., Wiethölter, N., El Gueddari, N. E., and Moerschbacher, B. M. (2006). Protein expression during seed development in Araucaria angustifolia: transient accumulation of class IV chitinases and arabinogalactan proteins. Physiol. Plant. 127, 138–148. doi: 10.1111/j.1399-3054.2005.00637.x
Driouich, A., Smith, C., Ropitaux, M., Chambard, M., Boulogne, I., Bernard, S., et al. (2019). Root extracellular traps versus neutrophil extracellular traps in host defence, a case of functional convergence? Biol. Rev. Camb. Philos. Soc. 94, 1685–1700. doi: 10.1111/brv.12522
Duman, Z., Eliyahu, A., Abu-Abied, M., and Sadot, E. (2020). The contribution of cell wall remodeling and signaling to lateral organs formation. Isr. J. Plant Sci. 67, 110–127. doi: 10.1163/22238980-20191115
Elkins, T., Zinn, K., McAllister, L., HoffMann, F. M., and Goodman, C. S. (1990). Genetic analysis of a Drosophila neural cell adhesion molecule: interaction of fasciclin I and Abelson tyrosine kinase mutations. Cell 60, 565–575. doi: 10.1016/0092-8674(90)90660-7
Ellis, M., Egelund, J., Schultz, C. J., and Bacic, A. (2010). Arabinogalactan-proteins: key regulators at the cell surface? Plant Physiol. 153, 403–419. doi: 10.1104/pp.110.156000
Faik, A., Abouzouhair, J., and Sarhan, F. (2006). Putative fasciclin-like arabinogalactan-proteins (FLA) in wheat (Triticum aestivum) and rice (Oryza sativa): identification and bioinformatic analyses. Mol. Genet. Genomics 276, 478–494. doi: 10.1007/s00438-006-0159-z
Ferguson, B. J., Mens, C., Hastwell, A. H., Zhang, M., Su, H., Jones, C. H., et al. (2019). Legume nodulation: the host controls the party. Plant Cell Environ. 42, 41–51. doi: 10.1111/pce.13348
Fincher, G. B., Stone, B. A., and Clarke, A. E. (1983). Arabinogalactan-proteins: structure, biosynthesis, and function. Annu. Rev. Plant Physiol. 34, 47–70. doi: 10.1146/annurev.pp.34.060183.000403
Freshour, G., Bonin, C. P., Reiter, W.-D., Albersheim, P., Darvill, A. G., and Hahn, M. G. (2003). Distribution of fucose-containing xyloglucans in cell walls of the mur1 mutant of Arabidopsis. Plant Physiol. 131, 1602–1612. doi: 10.1104/pp.102.016444
Galloway, A. F., Akhtar, J., Marcus, S. E., Fletcher, N., Field, K., and Knox, P. (2020). Cereal root exudates contain highly structurally complex polysaccharides with soil-binding properties. Plant J. 103, 1666–1678. doi: 10.1111/tpj.14852
Gantulga, D., Turan, Y., Bevan, D. R., and Esen, A. (2008). The Arabidopsis At1g45130 and At3g52840 genes encode beta-galactosidases with activity toward cell wall polysaccharides. Phytochemistry 69, 1661–1670. doi: 10.1016/j.phytochem.2008.01.023
Gaspar, Y. M., Nam, J., Schultz, C. J., Lee, L.-Y., Gilson, P. R., Gelvin, S. B., et al. (2004). Characterization of the Arabidopsis lysine-rich arabinogalactan-protein AtAGP17 mutant (rat1) that results in a decreased efficiency of Agrobacterium transformation. Plant Physiol. 135, 2162–2171. doi: 10.1104/pp.104.045542
Gigli-Bisceglia, N., Engelsdorf, T., and Hamann, T. (2020). Plant cell wall integrity maintenance in model plants and crop species-relevant cell wall components and underlying guiding principles. Cell. Mol. Life Sci. 77, 2049–2077. doi: 10.1007/s00018-019-03388-8
Gille, S., Sharma, V., Baidoo, E. E., Keasling, J. D., Scheller, H. V., and Pauly, M. (2013). Arabinosylation of a Yariv-precipitable cell wall polymer impacts plant growth as exemplified by the Arabidopsis glycosyltransferase mutant ray1. Mol. Plant 6, 1369–1372. doi: 10.1093/mp/sst029
Gollotte, A., Gianinazzi-Pearson, V., and Gianinazzi, S. (1995). Immunodetection of infection thread glycoprotein and arabinogalactan protein in wild type Pisum sativum (L.) or an isogenic mycorrhiza-resistant mutant interacting with Glomus mosseae. Symbiosis 18, 69–85.
Harholt, J., Jensen, J. K., Verhertbruggen, Y., Sogaard, C., Bernard, S., Nafisi, M., et al. (2012). ARAD proteins associated with pectic Arabinan biosynthesis form complexes when transiently overexpressed in planta. Planta 236, 115–128. doi: 10.1007/s00425-012-1592-3
Hawes, M. C., Gunawardena, U., Miyasaka, S., and Zhao, X. (2000). The role of root border cells in plant defense. Trends Plant Sci. 5, 128–133. doi: 10.1016/S1360-1385(00)01556-9
He, J., Zhao, H., Cheng, Z., Ke, Y., Liu, J., and Ma, H. (2019). Evolution analysis of the fasciclin-like arabinogalactan proteins in plants shows variable fasciclin-AGP domain constitutions. Int. J. Mol. Sci. 20:1945. doi: 10.3390/ijms20081945
Herve, C., Simeon, A., Jam, M., Cassin, A., Johnson, K. L., Salmean, A. A., et al. (2016). Arabinogalactan proteins have deep roots in eukaryotes: identification of genes and epitopes in brown algae and their role in Fucus serratus embryo development. New Phytol. 209, 1428–1441. doi: 10.1111/nph.13786
Hijazi, M., Roujol, D., Nguyen-Kim, H., Del Rocio Cisneros Castillo, L., Saland, E., Jamet, E., et al. (2014). Arabinogalactan protein 31 (AGP31), a putative network-forming protein in Arabidopsis thaliana cell walls? Ann. Bot. 114, 1087–1097. doi: 10.1093/aob/mcu038
Hossain, Z., McGarvey, B., Amyot, L., Gruber, M., Jung, J., and Hannoufa, A. (2012). DIMINUTO 1 affects the lignin profile and secondary cell wall formation in Arabidopsis. Planta 235, 485–498. doi: 10.1007/s00425-011-1519-4
Hozumi, A., Bera, S., Fujiwara, D., Obayashi, T., Yokoyama, R., Nishitani, K., et al. (2017). Arabinogalactan proteins accumulate in the cell walls of searching hyphae of the stem parasitic plants, Cuscuta campestris and Cuscuta japonica. Plant Cell Physiol. 58, 1868–1877. doi: 10.1093/pcp/pcx121
Huang, Y., Wang, Y., Tan, L., Sun, L., Petrosino, J., Cui, M.-Z., et al. (2016). Nanospherical arabinogalactan proteins are a key component of the high-strength adhesive secreted by English ivy. Proc. Natl. Acad. Sci. U.S.A. 113, E3193–E3202. doi: 10.1073/pnas.1600406113
Hwang, Y., Lee, H., Lee, Y. S., and Cho, H. T. (2016). Cell wall-associated ROOT HAIR SPECIFIC 10, a proline-rich receptor-like kinase, is a negative modulator of Arabidopsis root hair growth. J. Exp. Bot. 67, 2007–2022. doi: 10.1093/jxb/erw031
Immerzeel, P., Eppink, M. M., de Vries, S. C., Schols, H. A., and Voragen, A. G. J. (2006). Carrot arabinogalactan proteins are interlinked with pectins. Physiol. Plant. 128, 18–28. doi: 10.1111/j.1399-3054.2006.00712.x
Irani, S., Trost, B., Waldner, M., Nayidu, N., Tu, J., Kusalik, A. J., et al. (2018). Transcriptome analysis of response to Plasmodiophora brassicae infection in the Arabidopsis shoot and root. BMC Genomics 19:23. doi: 10.1186/s12864-017-4426-7
Ito, S., Suzuki, Y., Miyamoto, K., Ueda, J., and Yamaguchi, I. (2005). AtFLA11, a fasciclin-like arabinogalactan-protein, specifically localized in screlenchyma cells. Biosci. Biotech. Biochem. 69, 1963–1969. doi: 10.1271/bbb.69.1963
Jackson, O., Taylor, O., Adams, D. G., and Knox, J. P. (2012). Arabinogalactan proteins occur in the free-living cyanobacterium genus Nostoc and in plant–Nostoc symbioses. Mol. Plant Microbe Interact. 25, 1338–1349. doi: 10.1094/MPMI-04-12-0095-R
Jia, Z., Giehl, R. F. H., and von Wiren, N. (2020). The root foraging response under low nitrogen depends on DWARF1-mediated brassinosteroid biosynthesis. Plant Physiol. 183, 998–1010. doi: 10.1104/pp.20.00440
Jiao, Y., Sun, L., Song, Y., Wang, L., Liu, L., Zhang, L., et al. (2013). AtrbohD and AtrbohF positively regulate abscisic acid-inhibited primary root growth by affecting Ca2+ signalling and auxin response of roots in Arabidopsis. J. Exp. Bot. 64, 4183–4192. doi: 10.1093/jxb/ert228
Jing, Y., Shi, L., Li, X., Zheng, H., and He, L. (2019). AGP30: Cd tolerance related gene associate with mitochondrial pyruvate carrier 1. Plant Signal. Behav. 14:1629269. doi: 10.1080/15592324.2019.1629269
Johnson, K. L., Cassin, A. M., Lonsdale, A., Wong, G. K., Soltis, D. E., Miles, N. W., et al. (2017). Insights into the evolution of hydroxyproline-rich glycoproteins from 1000 plant transcriptomes. Plant Physiol. 174, 904–921. doi: 10.1104/pp.17.00295
Johnson, K. L., Kibble, N. A., Bacic, A., and Schultz, C. J. (2011). A fasciclin-like arabinogalactan-protein (FLA) mutant of Arabidopsis thaliana, fla1, shows defects in shoot regeneration. PLoS One 6:e25154. doi: 10.1371/journal.pone.0025154
Kirchner, T. W., Niehaus, M., Rossig, K. L., Lauterbach, T., Herde, M., Kuster, H., et al. (2018). Molecular background of Pi deficiency-induced root hair growth in Brassica carinata – a fasciclin-like arabinogalactan protein is involved. Front. Plant Sci. 9:1372. doi: 10.3389/fpls.2018.01372
Kjellbom, P., Snogerup, L., Stöhr, C., Reuzeau, C., McCabe, P. F., and Pennell, R. I. (1997). Oxidative cross-linking of plasma membrane arabinogalactan proteins. Plant J. 12, 1189–1196. doi: 10.1046/j.1365-313X.1997.12051189.x
Klahre, U., Noguchi, T., Fujioka, S., Takatsuto, S., Yokota, T., Nomura, T., et al. (1998). The Arabidopsis DIMINUTO/DWARF1 gene encodes a protein involved in steroid synthesis. Plant Cell 10, 1677–1690. doi: 10.1105/tpc.10.10.1677
Knox, J. P., Day, S., and Roberts, K. (1989). A set of cell surface glycoproteins forms an early marker of cell position, but not cell type, in the root apical meristem of Daucus carota L. Development 106, 47–56.
Knox, P. (2016). Delving in the deep for the origin of plant cell surface proteoglycans. New Phytol. 209, 1341–1343. doi: 10.1111/nph.13862
Koroney, A. S., Plasson, C., Pawlak, B., Sidikou, R., Driouich, A., Menu-Bouaouiche, L., et al. (2016). Root exudate of Solanum tuberosum is enriched in galactose-containing molecules and impacts the growth of Pectobacterium atrosepticum. Ann. Bot. 118, 797–808. doi: 10.1093/aob/mcw128
Kreuger, M., and van Holst, G.-J. (1993). Arabinogalactan proteins are essential in somatic embryogenesis of Daucus carota L. Planta 189, 243–248. doi: 10.1007/BF00195083
Lamport, D. T., and Varnai, P. (2013). Periplasmic arabinogalactan glycoproteins act as a calcium capacitor that regulates plant growth and development. New Phytol. 197, 58–64. doi: 10.1111/nph.12005
Lamport, D. T., Kieliszewski, M. J., and Showalter, A. M. (2006). Salt stress upregulates periplasmic arabinogalactan proteins: using salt stress to analyse AGP function. New Phytol. 169, 479–492. doi: 10.1111/j.1469-8137.2005.01591.x
Lamport, D. T., Tan, L., Held, M. A., and Kieliszewski, M. J. (2018). Pollen tube growth and guidance: Occam’s razor sharpened on a molecular arabinogalactan glycoprotein Rosetta stone. New Phytol. 217, 491–500. doi: 10.1111/nph.14845
Lee, K. J., Sakata, Y., Mau, S.-L., Pettolino, F., Bacic, A., Quatrano, R. S., et al. (2005). Arabinogalactan proteins are required for apical cell extension in the moss Physcomitrella patens. Plant Cell 17, 3051–3065. doi: 10.1105/tpc.105.034413
Leszczuk, A., Cybulska, J., Skrzypek, T., and Zdunek, A. (2020a). Properties of arabinogalactan proteins (AGPs) in apple (Malus x Domestica) fruit at different stages of ripening. Biology 9:225. doi: 10.3390/biology9080225
Leszczuk, A., Kalaitzis, P., Blazakis, K. N., and Zdunek, A. (2020b). The role of arabinogalactan proteins (AGPs) in fruit ripening-a review. Hortic. Res. 7:176. doi: 10.1038/s41438-020-00397-8
Li, J., Yu, M., Geng, L. L., and Zhao, J. (2010). The fasciclin-like arabinogalactan protein gene, FLA3, is involved in microspore development of Arabidopsis. Plant J. 64, 482–497. doi: 10.1111/j.1365-313X.2010.04344.x
Lin, W. D., Liao, Y. Y., Yang, T. J., Pan, C. Y., Buckhout, T. J., and Schmidt, W. (2011). Coexpression-based clustering of Arabidopsis root genes predicts functional modules in early phosphate deficiency signaling. Plant Physiol. 155, 1383–1402. doi: 10.1104/pp.110.166520
Liu, D., Tu, L., Li, Y., Wang, L., Zhu, L., and Zhang, X. (2008). Genes encoding fasciclin-like arabinogalactan proteins are specifically expressed during cotton fiber development. Plant Mol. Biol. Rep. 26, 98–113. doi: 10.1007/s11105-008-0026-7
Liu, E., MacMillan, C. P., Shafee, T., Ma, Y., Ratcliffe, J., van de Meene, A., et al. (2020). Fasciclin-like arabinogalactan-protein 16 (FLA16) is required for stem development in Arabidopsis. Front. Plant Sci. 11:615392. doi: 10.3389/fpls.2020.615392
Lopez-Hernandez, F., Tryfona, T., Rizza, A., Yu, X. L., Harris, M. O. B., Webb, A. A. R., et al. (2020). Calcium binding by arabinogalactan polysaccharides is important for normal plant development. Plant Cell 32, 3346–3369. doi: 10.1105/tpc.20.00027
Ma, H., and Zhao, J. (2010). Genome-wide identification, classification, and expression analysis of the arabinogalactan protein gene family in rice (Oryza sativa L.). J. Exp. Bot. 61, 2647–2668. doi: 10.1093/jxb/erq104
Ma, Y., Yan, C., Li, H., Wu, W., Liu, Y., Wang, Y., et al. (2017). Bioinformatics prediction and evolution analysis of arabinogalactan proteins in the plant kingdom. Front. Plant Sci. 8:66. doi: 10.3389/fpls.2017.00066
MacMillan, C. P., Mansfield, S. D., Stachurski, Z. H., Evans, R., and Southerton, S. G. (2010). Fasciclin-like arabinogalactan proteins: specialization for stem biomechanics and cell wall architecture in Arabidopsis and Eucalyptus. Plant J. 62, 689–703. doi: 10.1111/j.1365-313X.2010.04181.x
MacMillan, C. P., Taylor, L., Bi, Y., Southerton, S. G., Evans, R., and Spokevicius, A. (2015). The fasciclin-like arabinogalactan protein family of Eucalyptus grandis contains members that impact wood biology and biomechanics. New Phytol. 206, 1314–1327. doi: 10.1111/nph.13320
Mareri, L., Romi, M., and Cai, G. (2018). Arabinogalactan proteins: actors or spectators during abiotic and biotic stress in plants? Plant Biosyst. 153, 173–185. doi: 10.1080/11263504.2018.1473525
Marquez, N., Giachero, M. L., Gallou, A., Debat, H. J., Cranenbrouck, S., Di Rienzo, J. A., et al. (2018). Transcriptional changes in mycorrhizal and nonmycorrhizal soybean plants upon infection with the fungal pathogen Macrophomina phaseolina. Mol. Plant Microbe Interact. 31, 842–855. doi: 10.1094/MPMI-11-17-0282-R
Marzec, M., Szarejko, I., and Melzer, M. (2015). Arabinogalactan proteins are involved in root hair development in barley. J. Exp. Bot. 66, 1245–1257. doi: 10.1093/jxb/eru475
McCartney, L., Steele-King, C. G., Jordan, E., and Knox, J. P. (2003). Cell wall pectic (1→4)−β−d-galactan marks the acceleration of cell elongation in the Arabidopsis seedling root meristem. Plant J. 33, 447–454. doi: 10.1046/j.1365-313X.2003.01640.x
Nam, J., Mysore, K., Zheng, C., Knue, M., Matthysse, A., and Gelvin, S. (1999). Identification of T-DNA tagged Arabidopsis mutants that are resistant to transformation by Agrobacterium. Mol. Gen. Genet. 261, 429–438. doi: 10.1007/s004380050985
Nguema-Ona, E., Andème-Onzighi, C., Aboughe-Angone, S., Bardor, M., Ishii, T., Lerouge, P., et al. (2006). The reb1-1 mutation of Arabidopsis. Effect on the structure and localization of galactose-containing cell wall polysaccharides. Plant Physiol. 140, 1406–1417. doi: 10.1104/pp.105.074997
Nguema-Ona, E., Bannigan, A., Chevalier, L., Baskin, T. I, and Driouich, A. (2007). Disruption of arabinogalactan proteins disorganizes cortical microtubules in the root of Arabidopsis thaliana. Plant J. 52, 240–251. doi: 10.1111/j.1365-313X.2007.03224.x
Nguema-Ona, E., Coimbra, S., Vicré-Gibouin, M., Mollet, J.-C., and Driouich, A. (2012). Arabinogalactan proteins in root and pollen-tube cells: distribution and functional aspects. Ann. Bot. 110, 383–404. doi: 10.1093/aob/mcs143
Nguema-Ona, E., Vicre-Gibouin, M., Cannesan, M. A., and Driouich, A. (2013). Arabinogalactan proteins in root-microbe interactions. Trends Plant Sci. 18, 440–449. doi: 10.1016/j.tplants.2013.03.006
Nguema-Ona, E., Vicre-Gibouin, M., Gotte, M., Plancot, B., Lerouge, P., Bardor, M., et al. (2014). Cell wall O-glycoproteins and N-glycoproteins: aspects of biosynthesis and function. Front. Plant Sci. 5:499. doi: 10.3389/fpls.2014.00499
Nibbering, P., Petersen, B. L., Motawia, M. S., Jorgensen, B., Ulvskov, P., and Niittyla, T. (2020). Golgi-localized exo-beta1,3-galactosidases involved in cell expansion and root growth in Arabidopsis. J. Biol. Chem. 295, 10581–10592. doi: 10.1074/jbc.RA120.013878
Nivedita, Gazara, R. K., Khan, S., Iqrar, S., Ashrafi, K., and Abdin, M. Z. (2020). Comparative transcriptome profiling of rice colonized with beneficial endophyte, Piriformospora indica, under high salinity environment. Mol. Biol. Rep. 47, 7655–7673. doi: 10.1007/s11033-020-05839-z
Ogawa-Ohnishi, M., and Matsubayashi, Y. (2015). Identification of three potent hydroxyproline O-galactosyltransferases in Arabidopsis. Plant J. 81, 736–746. doi: 10.1111/tpj.12764
Olmos, E., Garcia De La Garma, J., Gomez-Jimenez, M. C., and Fernandez-Garcia, N. (2017). Arabinogalactan proteins are involved in salt-adaptation and vesicle trafficking in tobacco by-2 cell cultures. Front. Plant Sci. 8:1092. doi: 10.3389/fpls.2017.01092
Palacio-Lopez, K., Tinaz, B., Holzinger, A., and Domozych, D. S. (2019). Arabinogalactan proteins and the extracellular matrix of Charophytes: a sticky business. Front. Plant Sci. 10:447. doi: 10.3389/fpls.2019.00447
Pereira, A. M., Pereira, L. G., and Coimbra, S. (2015). Arabinogalactan proteins: rising attention from plant biologists. Plant Reprod. 28, 1–15. doi: 10.1007/s00497-015-0254-6
Perez-Perez, Y., Carneros, E., Berenguer, E., Solis, M. T., Barany, I., Pintos, B., et al. (2018). Pectin de-methylesterification and AGP increase promote cell wall remodeling and are required during somatic embryogenesis of Quercus suber. Front. Plant Sci. 9:1915. doi: 10.3389/fpls.2018.01915
Pfeifer, L., Shafee, T., Johnson, K. L., Bacic, A., and Classen, B. (2020). Arabinogalactan-proteins of Zostera marina L. contain unique glycan structures and provide insight into adaption processes to saline environments. Sci. Rep. 10:8232. doi: 10.1038/s41598-020-65135-5
Pielach, A., Leroux, O., Domozych, D. S., Knox, J. P., and Popper, Z. A. (2014). Arabinogalactan protein-rich cell walls, paramural deposits and ergastic globules define the hyaline bodies of rhinanthoid Orobanchaceae haustoria. Ann. Bot. 114, 1359–1373. doi: 10.1093/aob/mcu121
Plancot, B., Santaella, C., Jaber, R., Kiefer-Meyer, M. C., Follet-Gueye, M. L., Leprince, J., et al. (2013). Deciphering the responses of root border-like cells of Arabidopsis and flax to pathogen-derived elicitors. Plant Physiol. 163, 1584–1597. doi: 10.1104/pp.113.222356
Přerovská, T., Henke, S., Bleha, R., Spiwok, V., Gillarová, S., Yvin, J. C., et al. (2021). Arabinogalactan-like glycoproteins from Ulva lactuca (Chlorophyta) show unique features compared to land plants AGPs. J. Phycol. 57, 619–635. doi: 10.1111/jpy.13121
Rashid, A. (2016). Defense responses of plant cell wall non-catalytic proteins against pathogens. Physiol. Mol. Plant Pathol. 94, 38–46. doi: 10.1016/j.pmpp.2016.03.009
Rathbun, E. A., Naldrett, M. J., and Brewin, N. J. (2002). Identification of a family of extensin-like glycoproteins in the lumen of Rhizobium-induced infection threads in pea root nodules. Mol. Plant Microbe Interact. 15, 350–359. doi: 10.1094/MPMI.2002.15.4.350
Reguera, M., Abreu, I., Brewin, N. J., Bonilla, I., and Bolanos, L. (2010). Borate promotes the formation of a complex between legume AGP-extensin and Rhamnogalacturonan II and enhances production of Rhizobium capsular polysaccharide during infection thread development in Pisum sativum symbiotic root nodules. Plant Cell Environ. 33, 2112–2120. doi: 10.1111/j.1365-3040.2010.02209.x
Reiter, W. D., Chapple, C., and Somerville, C. R. (1997). Mutants of Arabidopsis thaliana with altered cell wall polysaccharide composition. Plant J. 12, 335–345. doi: 10.1046/j.1365-313X.1997.12020335.x
Reiter, W.-D., Chapple, C. C., and Somerville, C. R. (1993). Altered growth and cell walls in a fucose-deficient mutant of Arabidopsis. Science 261, 1032–1035. doi: 10.1126/science.261.5124.1032
Ropitaux, M., Bernard, S., Schapman, D., Follet-Gueye, M. L., Vicre, M., Boulogne, I., et al. (2020). Root border cells and mucilage secretions of soybean, Glycine max (Merr) L.: Characterization and role in interactions with the oomycete Phytophthora parasitica. Cells 9:2215. doi: 10.3390/cells9102215
Rui, Y., and Dinneny, J. R. (2020). A wall with integrity: surveillance and maintenance of the plant cell wall under stress. New Phytol. 225, 1428–1439. doi: 10.1111/nph.16166
Sala, K., Malarz, K., Barlow, P. W., and Kurczynska, E. U. (2017). Distribution of some pectic and arabinogalactan protein epitopes during Solanum lycopersicum (L.) adventitious root development. BMC Plant Biol. 17:25. doi: 10.1186/s12870-016-0949-3
Šamaj, J., Braun, M., Baluška, F., Ensikat, H.-J., Tsumuraya, Y., and Volkmann, D. (1999). Specific localization of arabinogalactan-protein epitopes at the surface of maize root hairs. Plant Cell Physiol. 40, 874–883. doi: 10.1093/oxfordjournals.pcp.a029617
Schultz, C. J., and Harrison, M. J. (2008). Novel plant and fungal AGP-like proteins in the Medicago truncatula-Glomus intraradices arbuscular mycorrhizal symbiosis. Mycorrhiza 18, 403–412. doi: 10.1007/s00572-008-0194-1
Schultz, C., Gilson, P., Oxley, D., Youl, J., and Bacic, A. (1998). GPI-anchors on arabinogalactan-proteins: implications for signalling in plants. Trends Plant Sci. 3, 426–431. doi: 10.1016/S1360-1385(98)01328-4
Seifert, G. J. (2018). Fascinating fasciclins: a surprisingly widespread family of proteins that mediate interactions between the cell exterior and the cell surface. Int. J. Mol. Sci. 19:1628. doi: 10.3390/ijms19061628
Seifert, G. J. (2020). On the potential function of type II arabinogalactan O-glycosylation in regulating the fate of plant secretory proteins. Front. Plant Sci. 11:563735. doi: 10.3389/fpls.2020.563735
Seifert, G. J. (2021). The FLA4-FEI pathway: a unique and mysterious signaling module related to cell wall structure and stress signaling. Genes 12:145. doi: 10.3390/genes12020145
Seifert, G. J., and Blaukopf, C. (2010). Irritable walls: the plant extracellular matrix and signaling. Plant Physiol. 153, 467–478. doi: 10.1104/pp.110.153940
Seifert, G. J., and Roberts, K. (2007). The biology of arabinogalactan proteins. Annu. Rev. Plant Biol. 58, 137–161. doi: 10.1146/annurev.arplant.58.032806.103801
Seifert, G. J., Barber, C., Wells, B., Dolan, L., and Roberts, K. (2002). Galactose biosynthesis in Arabidopsis: genetic evidence for substrate channeling from UDP-D-galactose into cell wall polymers. Curr. Biol. 12, 1840–1845. doi: 10.1016/S0960-9822(02)01260-5
Seifert, G. J., Xue, H., and Acet, T. (2014). The Arabidopsis thaliana FASCICLIN LIKE ARABINOGALACTAN PROTEIN 4 gene acts synergistically with abscisic acid signalling to control root growth. Ann. Bot. 114, 1125–1133. doi: 10.1093/aob/mcu010
Shailasree, S., Kini, K. R., Deepak, S., Kumudini, B. S., and Shetty, H. S. (2004). Accumulation of hydroxyproline-rich glycoproteins in pearl millet seedlings in response to Sclerospora graminicola infection. Plant Sci. 167, 1227–1234. doi: 10.1016/j.plantsci.2004.06.012
Shi, H., Kim, Y., Guo, Y., Stevenson, B., and Zhu, J.-K. (2003). The Arabidopsis SOS5 locus encodes a putative cell surface adhesion protein and is required for normal cell expansion. Plant Cell 15, 19–32. doi: 10.1105/tpc.007872
Shimizu, K., and Aoki, K. (2019). Development of parasitic organs of a stem holoparasitic plant in genus Cuscuta. Front. Plant Sci. 10:1435. doi: 10.3389/fpls.2019.01435
Showalter, A. (2001). Arabinogalactan-proteins: structure, expression and function. Cell. Mol. Life Sci. 58, 1399–1417. doi: 10.1007/PL00000784
Showalter, A. M., and Basu, D. (2016). Glycosylation of arabinogalactan-proteins essential for development in Arabidopsis. Commun. Integr. Biol. 9:e0125624. doi: 10.1080/19420889.2016.1177687
Silva, J., Ferraz, R., Dupree, P., Showalter, A. M., and Coimbra, S. (2020). Three decades of advances in arabinogalactan-protein biosynthesis. Front. Plant Sci. 11:610377. doi: 10.3389/fpls.2020.610377
Silva, T. F., Ferreira, B. G., Dos Santos Isaias, R. M., Alexandre, S. S., and Franca, M. G. C. (2020). Immunocytochemistry and density functional theory evidence the competition of aluminum and calcium for pectin binding in Urochloa decumbens roots. Plant Physiol. Biochem. 153, 64–71. doi: 10.1016/j.plaphy.2020.05.015
Snow, P. M., Bieber, A. J., and Goodman, C. S. (1989). Fasciclin III: a novel homophilic adhesion molecule in Drosophila. Cell 59, 313–323. doi: 10.1016/0092-8674(89)90293-6
Somssich, M., Khan, G. A., and Persson, S. (2016). Cell wall heterogeneity in root development of Arabidopsis. Front. Plant Sci. 7:1242. doi: 10.3389/fpls.2016.01242
Striberny, B., and Krause, K. (2015). Cell wall glycoproteins at interaction sites between parasitic giant dodder (Cuscuta reflexa) and its host Pelargonium zonale. Plant Signal. Behav. 10:e1086858. doi: 10.1080/15592324.2015.1086858
Su, S., and Higashiyama, T. (2018). Arabinogalactan proteins and their sugar chains: functions in plant reproduction, research methods, and biosynthesis. Plant Reprod. 31, 67–75. doi: 10.1007/s00497-018-0329-2
Swamy, M. K., Akhtar, M. S., and Sinniah, U. R. (2016). “Root exudates and their molecular interactions with rhizospheric microbes,” in Plant, Soil and Microbes, eds K. R. Hakeem and M. S. Akhtar (Cham: Springer), 59–77. doi: 10.1007/978-3-319-29573-2_4
Takahashi, T., Gasch, A., Nishizawa, N., and Chua, N.-H. (1995). The DIMINUTO gene of Arabidopsis is involved in regulating cell elongation. Genes Dev. 9, 97–107. doi: 10.1101/gad.9.1.97
Tan, L., Eberhard, S., Pattathil, S., Warder, C., Glushka, J., Yuan, C., et al. (2013). An Arabidopsis cell wall proteoglycan consists of pectin and arabinoxylan covalently linked to an arabinogalactan protein. Plant Cell 25, 270–287. doi: 10.1105/tpc.112.107334
Tan, L., Tees, D., Qian, J., Kareem, S., and Kieliszewski, M. J. (2018). Intermolecular interactions between glycomodules of plant cell wall arabinogalactan-proteins and extensins. Cell Surf. 1, 25–33. doi: 10.1016/j.tcsw.2018.03.001
Trivedi, D. K., Bhatt, H., Pal, R. K., Tuteja, R., Garg, B., Johri, A. K., et al. (2013). Structure of RNA-interacting cyclophilin A-like protein from Piriformospora indica that provides salinity-stress tolerance in plants. Sci. Rep. 3:3001. doi: 10.1038/srep03001
Tryfona, T., Theys, T. E., Wagner, T., Stott, K., Keegstra, K., and Dupree, P. (2014). Characterisation of FUT4 and FUT6 α-(1→2)-fucosyltransferases reveals that absence of root arabinogalactan fucosylation increases Arabidopsis root growth salt sensitivity. PLoS One 9:e93291. doi: 10.1371/journal.pone.0093291
Tsyganova, A. V., Seliverstova, E. V., Brewin, N. J., and Tsyganov, V. E. (2019). Comparative analysis of remodelling of the plant-microbe interface in Pisum sativum and Medicago truncatula symbiotic nodules. Protoplasma 256, 983–996. doi: 10.1007/s00709-019-01355-5
Tsyganova, A. V., Tsyganov, V. E., Findlay, K. C., Borisov, A. Y., Tikhonovich, I. A., and Brewin, N. J. (2009). Distribution of legume arabinogalactan protein-extensin (AGPE) glycoproteins in symbiotically defective pea mutants with abnormal infection threads. Cell Tissue Biol. 3, 93–102. doi: 10.1134/s1990519x09010131
Tucker, M. R., Lou, H., Aubert, M. K., Wilkinson, L. G., Little, A., Houston, K., et al. (2018). Exploring the role of cell wall-related genes and polysaccharides during plant development. Plants 7:42. doi: 10.3390/plants7020042
Turupcu, A., Almohamed, W., Oostenbrink, C., and Seifert, G. J. (2018). A speculation on the tandem fasciclin 1 repeat of FLA4 proteins in angiosperms. Plant Signal. Behav. 13:e1507403. doi: 10.1080/15592324.2018.1507403
Van Hengel, A. J., and Roberts, K. (2002). Fucosylated arabinogalactan-proteins are required for full root cell elongation in Arabidopsis. Plant J. 32, 105–113. doi: 10.1046/j.1365-313X.2002.01406.x
Van Hengel, A. J., and Roberts, K. (2003). AtAGP30, an arabinogalactan-protein in the cell walls of the primary root, plays a role in root regeneration and seed germination. Plant J. 36, 256–270. doi: 10.1046/j.1365-313X.2003.01874.x
Van Hengel, A. J., Barber, C., and Roberts, K. (2004). The expression patterns of arabinogalactan-protein AtAGP30 and GLABRA2 reveal a role for abscisic acid in the early stages of root epidermal patterning. Plant J. 39, 70–83. doi: 10.1111/j.1365-313X.2004.02104.x
van Hengel, A. J., Tadesse, Z., Immerzeel, P., Schols, H., Van Kammen, A., and de Vries, S. C. (2001). N-acetylglucosamine and glucosamine-containing arabinogalactan proteins control somatic embryogenesis. Plant Physiol. 125, 1880–1890. doi: 10.1104/pp.125.4.1880
Velasquez, S. M., Marzol, E., Borassi, C., Pol-Fachin, L., Ricardi, M. M., Mangano, S., et al. (2015). Low sugar is not always good: impact of specific O-glycan defects on tip growth in Arabidopsis. Plant Physiol. 168, 808–813. doi: 10.1104/pp.114.255521
Vicré, M., Santaella, C., Blanchet, S., Gateau, A., and Driouich, A. (2005). Root border-like cells of Arabidopsis. Microscopical characterization and role in the interaction with rhizobacteria. Plant Physiol. 138, 998–1008. doi: 10.1104/pp.104.051813
Voxeur, A., and Hofte, H. (2016). Cell wall integrity signaling in plants: “to grow or not to grow that’s the question”. Glycobiology 26, 950–960. doi: 10.1093/glycob/cww029
Waller, F., Achatz, B., Baltruschat, H., Fodor, J., Becker, K., Fischer, M., et al. (2005). The endophytic fungus Piriformospora indica reprograms barley to salt-stress tolerance, disease resistance, and higher yield. Proc. Natl. Acad. Sci. U.S.A. 102, 13386–13391. doi: 10.1073/pnas.0504423102
Wang, H., Jiang, C., Wang, C., Yang, Y., Yang, L., Gao, X., et al. (2015). Antisense expression of the fasciclin-like arabinogalactan protein FLA6 gene in Populus inhibits expression of its homologous genes and alters stem biomechanics and cell wall composition in transgenic trees. J. Exp. Bot. 66, 1291–1302. doi: 10.1093/jxb/eru479
Wang, L., Cheng, M., Yang, Q., Li, J., Wang, X., Zhou, Q., et al. (2019). Arabinogalactan protein-rare earth element complexes activate plant endocytosis. Proc. Natl. Acad. Sci. U.S.A. 116, 14349–14357. doi: 10.1073/pnas.1902532116
Wang, Y., Mortimer, J. C., Davis, J., Dupree, P., and Keegstra, K. (2013). Identification of an additional protein involved in mannan biosynthesis. Plant J. 73, 105–117. doi: 10.1111/tpj.12019
Willats, W. G., and Knox, J. P. (1996). A role for arabinogalactan-proteins in plant cell expansion: evidence from studies on the interaction of β−glucosyl Yariv reagent with seedlings of Arabidopsis thaliana. Plant J. 9, 919–925. doi: 10.1046/j.1365-313X.1996.9060919.x
Wu, Y., Fan, W., Li, X., Chen, H., Takac, T., Samajova, O., et al. (2017). Expression and distribution of extensins and AGPs in susceptible and resistant banana cultivars in response to wounding and Fusarium oxysporum. Sci. Rep. 7:42400. doi: 10.1038/srep42400
Wubben, M. J. II, Rodermel, S. R., and Baum, T. J. (2004). Mutation of a UDP-glucose-4-epimerase alters nematode susceptibility and ethylene responses in Arabidopsis roots. Plant J. 40, 712–724. doi: 10.1111/j.1365-313X.2004.02257.x
Xie, D., Ma, L., Samaj, J., and Xu, C. (2011). Immunohistochemical analysis of cell wall hydroxyproline-rich glycoproteins in the roots of resistant and susceptible wax gourd cultivars in response to Fusarium oxysporum f. sp. Benincasae infection and fusaric acid treatment. Plant Cell Rep. 30, 1555–1569. doi: 10.1007/s00299-011-1069-z
Xu, S. L., Rahman, A., Baskin, T. I., and Kieber, J. J. (2008). Two leucine-rich repeat receptor kinases mediate signaling, linking cell wall biosynthesis and ACC synthase in Arabidopsis. Plant Cell 20, 3065–3079. doi: 10.1105/tpc.108.063354
Xue, H., and Seifert, G. J. (2015). Fasciclin like arabinogalactan protein 4 and respiratory burst oxidase homolog D and F independently modulate abscisic acid signaling. Plant Signal. Behav. 10:e989064. doi: 10.4161/15592324.2014.989064
Xue, H., Veit, C., Abas, L., Tryfona, T., Maresch, D., Ricardi, M. M., et al. (2017). Arabidopsis thaliana FLA4 functions as a glycan-stabilized soluble factor via its carboxy-proximal Fasciclin 1 domain. Plant J. 91, 613–630. doi: 10.1111/tpj.13591
Yang, J., and Showalter, A. M. (2007). Expression and localization of AtAGP18, a lysine-rich arabinogalactan-protein in Arabidopsis. Planta 226, 169–179. doi: 10.1007/s00425-007-0478-2
Yang, J., Sardar, H. S., McGovern, K. R., Zhang, Y., and Showalter, A. M. (2007). A lysine-rich arabinogalactan protein in Arabidopsis is essential for plant growth and development, including cell division and expansion. Plant J. 49, 629–640. doi: 10.1111/j.1365-313X.2006.02985.x
Yang, J., Zhang, Y., Liang, Y., and Showalter, A. M. (2011). Expression analyses of AtAGP17 and AtAGP19, two lysine-rich arabinogalactan proteins, in Arabidopsis. Plant Biol. 13, 431–438. doi: 10.1111/j.1438-8677.2010.00407.x
Yariv, J., Lis, H., and Katchalski, E. (1967). Precipitation of Arabic acid and some seed polysaccharides by glycosylphenylazo dyes. Biochem. J. 105, 1C–2C. doi: 10.1042/bj1050001c
Yeats, T. H., Bacic, A., and Johnson, K. L. (2018). Plant glycosylphosphatidylinositol anchored proteins at the plasma membrane-cell wall nexus. J. Int. Plant Biol. 60, 649–669. doi: 10.1111/jipb.12659
Yu, M., and Zhao, J. (2012). The cytological changes of tobacco zygote and proembryo cells induced by beta-glucosyl Yariv reagent suggest the involvement of arabinogalactan proteins in cell division and cell plate formation. BMC Plant Biol. 12:126. doi: 10.1186/1471-2229-12-126
Zagorchev, L., Kamenova, P., and Odjakova, M. (2014). The role of plant cell wall proteins in response to salt stress. Sci. World J. 2014:764089. doi: 10.1155/2014/764089
Zang, L., Zheng, T., Chu, Y., Ding, C., Zhang, W., Huang, Q., et al. (2015). Genome-wide analysis of the fasciclin-like arabinogalactan protein gene family reveals differential expression patterns, localization, and salt stress response in Populus. Front. Plant Sci. 6:1140. doi: 10.3389/fpls.2015.01140
Zhang, Y., Held, M. A., and Showalter, A. M. (2020). Elucidating the roles of three beta-glucuronosyltransferases (GLCATs) acting on arabinogalactan-proteins using a CRISPR-Cas9 multiplexing approach in Arabidopsis. BMC Plant Biol. 20:221. doi: 10.1186/s12870-020-02420-5
Zhang, Y., Held, M. A., Kaur, D., and Showalter, A. M. (2021). CRISPR-Cas9 multiplex genome editing of the hydroxyproline-O-galactosyltransferase gene family alters arabinogalactan-protein glycosylation and function in Arabidopsis. BMC Plant Biol. 21:16. doi: 10.1186/s12870-020-02791-9
Zhang, Y., Yang, J., and Showalter, A. M. (2011). AtAGP18 is localized at the plasma membrane and functions in plant growth and development. Planta 233, 675–683. doi: 10.1007/s00425-010-1331-6
Zhao, C., Zayed, O., Zeng, F., Liu, C., Zhang, L., Zhu, P., et al. (2019). Arabinose biosynthesis is critical for salt stress tolerance in Arabidopsis. New Phytol. 224, 274–290. doi: 10.1111/nph.15867
Zhou, K. (2019). Glycosylphosphatidylinositol-anchored proteins in Arabidopsis and one of their common roles in signaling transduction. Front. Plant Sci. 10:1022. doi: 10.3389/fpls.2019.01022
Zhu, J. K., Shi, J., Singh, U., Wyatt, S. E., Bressan, R. A., Hasegawa, P. M., et al. (1993). Enrichment of vitronectin-and fibronectin-like proteins in NaCl-adapted plant cells and evidence for their involvement in plasma membrane–cell wall adhesion. Plant J. 3, 637–646. doi: 10.1111/j.1365-313X.1993.00637.x
Zielinski, K., Dubas, E., Gersi, Z., Krzewska, M., Janas, A., Nowicka, A., et al. (2021). Beta-1,3-Glucanases and chitinases participate in the stress-related defence mechanisms that are possibly connected with modulation of arabinogalactan proteins (AGP) required for the androgenesis initiation in rye (Secale cereale L.). Plant Sci. 302:110700.
Keywords: AGP, arabinogalactan proteins, root growth, root hairs, interactions, fasciclin-like, GPI anchor
Citation: Hromadová D, Soukup A and Tylová E (2021) Arabinogalactan Proteins in Plant Roots – An Update on Possible Functions. Front. Plant Sci. 12:674010. doi: 10.3389/fpls.2021.674010
Received: 28 February 2021; Accepted: 19 April 2021;
Published: 17 May 2021.
Edited by:
Svetlana Shishkova, National Autonomous University of Mexico, MexicoReviewed by:
Azeddine Driouich, Université de Rouen, FranceCopyright © 2021 Hromadová, Soukup and Tylová. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Edita Tylová, ZWRtdW56QG5hdHVyLmN1bmkuY3o=
†These authors share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.