
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 03 September 2021
Sec. Plant Metabolism and Chemodiversity
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.671487
This article is part of the Research Topic Exploring and Engineering Plant Specialized Metabolism: Latest Advances and New Horizons View all 10 articles
 Konstantinos Koudounas1†‡§
Konstantinos Koudounas1†‡§ Margarita Thomopoulou1†‡
Margarita Thomopoulou1†‡ Aimilia Rigakou2†
Aimilia Rigakou2† Elisavet Angeli1†
Elisavet Angeli1† Eleni Melliou2†
Eleni Melliou2† Prokopios Magiatis2†
Prokopios Magiatis2† Polydefkis Hatzopoulos1*†
Polydefkis Hatzopoulos1*†Specialized metabolism is an evolutionary answer that fortifies plants against a wide spectrum of (a) biotic challenges. A plethora of diversified compounds can be found in the plant kingdom and often constitute the basis of human pharmacopeia. Olive trees (Olea europaea) produce an unusual type of secoiridoids known as oleosides with promising pharmaceutical activities. Here, we transiently silenced oleuropein β-glucosidase (OeGLU), an enzyme engaged in the biosynthetic pathway of secoiridoids in the olive trees. Reduction of OeGLU transcripts resulted in the absence of both upstream and downstream secoiridoids in planta, revealing a regulatory loop mechanism that bypasses the flux of precursor compounds toward the branch of secoiridoid biosynthesis. Our findings highlight that OeGLU could serve as a molecular target to regulate the bioactive secoiridoids in olive oils.
Olive (Olea europaea L., Oleaceae) is an emblematic crop of the Mediterranean Basin and over the last decades, the cultivation has expanded to the Americas, Asia, and Oceania mostly due to the high nutritional value of olive oil. An essential trait that evolution has gifted to olives is the production of a certain type of secondary metabolites, known as oleosides, with an extremely narrow taxonomic distribution. Oleosides are terpene-derived secoiridoids, conjugates of glycosylated elenolic acid with a characteristic exocyclic 8,9-olefinic bond (Soler-Rivas et al., 2000), and are only present in the Oleaceae family and the genus Caiophora (Loasaceae) (Obied et al., 2008). Oleoside derivatives synergistically contribute to the beneficial aspects of olive oil in human health (Tripoli et al., 2005; Papanikolaou et al., 2019) and determine the flavor and quality of olive oil (Vitaglione et al., 2015). Recently, the European Medicinal Agency published a risk-benefit report on olives, and the European Food Safety Authority has already approved a health claim related to polyphenols in olive oil (EFSA, 2012; HMPC, 2017).
The dominant secoiridoid in olives accumulating up to 14% in young drupes is oleuropein (Amiot et al., 1986), an ester of elenolic acid with 2′-(3′,4′-dihydroxyphenyl)ethanol (hydroxytyrosol) that exhibits antioxidant, anti-inflammatory, antiproliferative, antimicrobial, and antiviral activities, hence being a metabolite of high interest for humans (Bulotta et al., 2013). The first identified enzyme that is engaged in the metabolism of oleuropein is the oleuropein-specific β-glucosidase (OeGLU; E.C. 3.2.1.206) (Koudounas et al., 2015, 2017). OeGLU is a homomultimeric enzyme, member of the Glycoside Hydrolase 1 family (GH1), and localized in the nucleus. Oleuropein is localized in the vacuoles or cytosol (Konno et al., 1999), thus physically separated from the OeGLU enzyme, and upon cell disruption, the dual-partner defensive system comes in contact. Deglycosylation of oleuropein by OeGLU produces an unstable aglycone form that is rapidly converted to a highly reactive molecule with a glutaraldehyde-like structure. This compound covalently binds to amino acids and exhibits strong protein denaturing/cross-linking activities, providing a mighty arsenal against herbivores (Konno et al., 1999).
In addition to a pivotal role in the plant chemical defense of Oleaceae species, the OeGLU-mediated enzymatic detonation of oleuropein is also crucial during olive oil extraction. Crushing of olive drupes followed by malaxation (coalescence of oil droplets through the mixing of olive paste) causes cell disruption. Oleuropein is exposed to OeGLU, and the aglycone products are massively produced, thus shaping the organoleptic properties of olive oil (Obied et al., 2008; Romero-Segura et al., 2012; Hachicha Hbaieb et al., 2015; Vitaglione et al., 2015).
Apart from OeGLU, only three enzymatic hubs are known to be engaged in the biosynthesis and biotransformations of secoiridoids in olive. The first one is an iridoid synthase (OeISY) which converts 8-oxogeranial into the iridoid scaffold in a two-step reduction-cyclization sequence (Alagna et al., 2016). Recently, two bi-functional cytochrome P450s (oleoside methyl ester synthase, OeOMES and secoxyloganin synthase, OeSXS) which convert 7-epi-loganin into the secoiridoid scaffold in two sequential oxidation steps were characterized (Rodríguez-López et al., 2021). Additionally, two methylesterase enzymes (elenolic acid methylesterase 1, OeEAME1 and elenolic acid methylesterase 2, OeEAME2) that generate oleacein and oleocanthal after the concerted activity of β-glucosidase on oleuropein and ligstroside, respectively, have been identified (Volk et al., 2019). Finally, a bona fide geraniol synthase (OeGES1) has been reported to generate geraniol – monoterpene alcohol that is the precursor of iridoids (Vezzaro et al., 2012). Despite the recent advances toward the elucidation of this pathway, the proposed biosynthetic pathway of oleuropein in Oleaceae involves at least 18 enzymatic steps, therefore, remains largely uncharacterized (Figure 1).
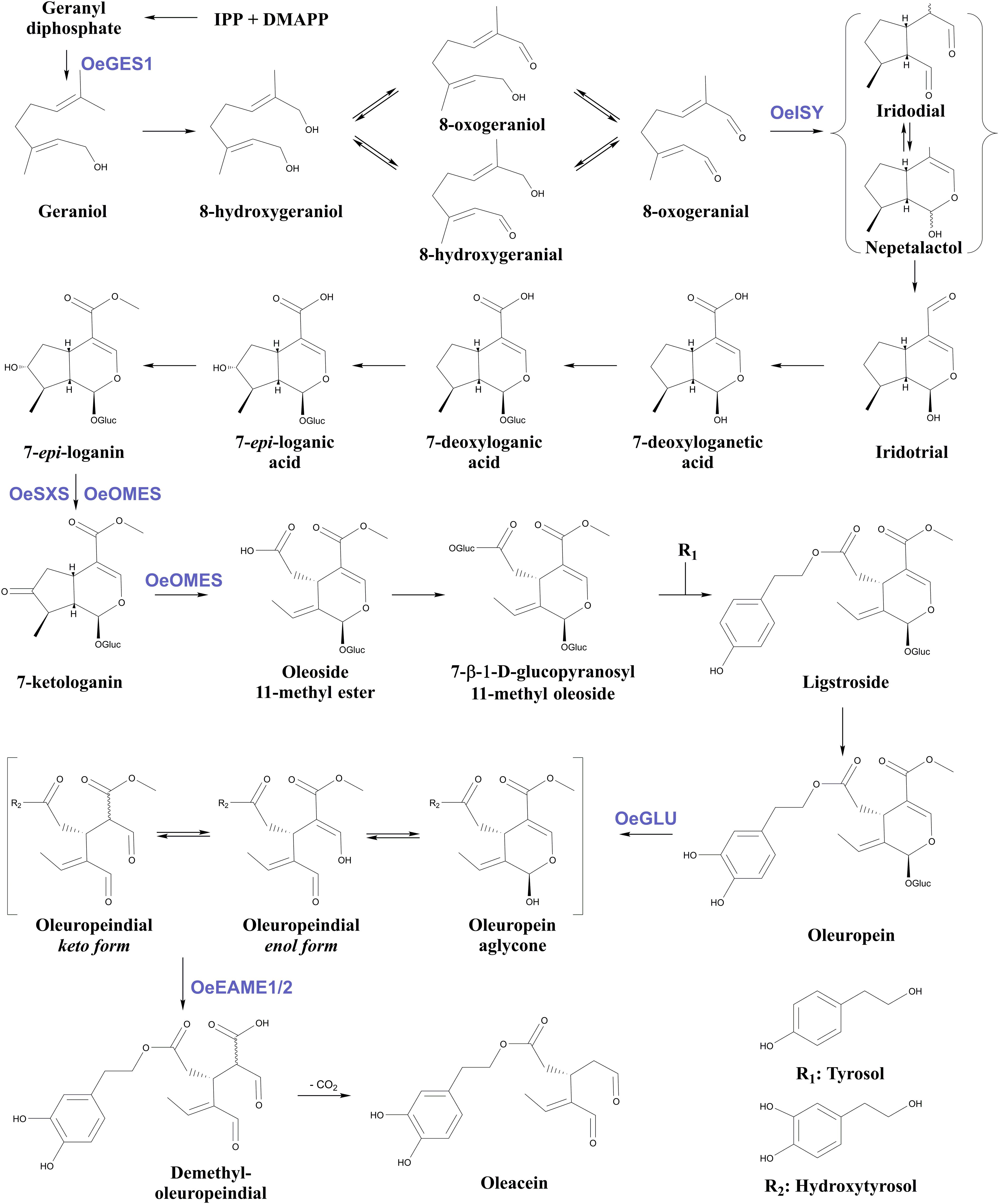
Figure 1. Proposed biosynthetic pathway of oleuropein and oleacein in Oleaceae. Characterized enzymes of the pathway as referred in the literature (Obied et al., 2008; Vezzaro et al., 2012; Koudounas et al., 2015; Alagna et al., 2016; Volk et al., 2019; Rodríguez-López et al., 2021). IPP, isopentenyl diphosphate; DMAPP, Dimethylallyl diphosphate; Glu, glucose; OeGES1, geraniol synthase; OeISY, iridoid synthase; OeOMES, oleoside methyl ester synthase; OeSXS, secoxyloganin synthase; OeGLU, oleuropein β-glucosidase; OeEAME1/2, elenolic acid methylesterase 1 and 2.
The olive tree is a non-model plant, member of a plant family that comprises perennial woody species (Green, 2004) recalcitrant to genetic transformation. Therefore, studies on the Oleaceae-specific biosynthetic pathway of secoiridoids and characterization of the aforementioned enzymatic hubs have been limited to heterologous in vivo systems (i.e., Nicotiana benthamiana, Saccharomyces cerevisiae, and Escherichia coli) and in vitro enzymatic assays, thus raising the question of the actual contribution of these enzymes in olives. Plant genomes are known to typically encode more than 30 members of the GH1 family (Ketudat Cairns and Esen, 2010) therefore olive possess other β-glucosidases that could deglycosylate oleuropein. However, we have previously determined that OeGLU is a single copy gene in the olive genome (Koudounas et al., 2015), and defensive GH1 β-glucosidases are known to be highly diversified and exhibit exceptional specificity against their respective substrates (Verdoucq et al., 2004; Xia et al., 2012).
In this study, we recruited the recently described Agrobacterium-mediated virus-induced gene silencing (VIGS) methodology (Koudounas et al., 2020) to transiently silence OeGLU in olive seedlings and validate whether OeGLU is the major oleuropein β-glucosidase in planta. Besides, downregulation of the OeGLU enzyme, transient silencing of OeGLU unexpectedly affected the biosynthesis of upstream and downstream secoiridoids suggesting the existence of a feedback regulatory loop in this pathway.
Ripe fruits were harvested from Olea europaea L. cv. “Koroneiki” grown in a natural environment at the Agricultural University of Athens. The black mesocarps were removed, and the woody endocarps were subjected to 10% sodium hydroxide for 10 min and then thoroughly washed to remove any fleshy remnants. The endocarps were dried naturally and stored until use. Seeds were extracted after gently cracking the woody endocarp with a bench vice. After surface sterilization followed by stratification for 7 days at 4°C, seeds were transferred to soil for germination and grown at 22°C in a Fitotron growth chamber (Weiss Gallenkamp, Loughborough, United Kingdom) with a 16/8 h light/darkness cycle and 100 μmol m–2 s–1 light intensity.
The pTRV1 (stock no.: CD3-1039) and pTRV2-MCS (stock no.: CD3-1040) plasmids encoding the bipartite RNA genome of tobacco rattle virus (TRV) were obtained from the Arabidopsis Biological Resource Center (ABRC1). RNA was extracted from olive young leaves using a phenol/chloroform procedure, and fragments of OeGLU (211 bp) and OeChlH (297 bp) were amplified by real-time polymerase chain reaction (RT-PCR) using the OeGLUi-F and OeGLUi-R, or OeChlH-F and OeChlH-R primers, respectively (Supplementary Table 1). To produce the pTRV2-OeGLU and pTRV2-OeChlH constructs, the PCR products were blunt-end ligated into the SmaI site of pUC19, and after verification by sequencing, the PCR products were directly sub-cloned into pTRV2-MCS (empty vector, EV) in antisense orientation relative to the TRV coat protein, utilizing the KpnI and XbaI restriction sites of pUC19.
The pTRV1 and pTRV2 constructs with either fragment of the targeted genes for silencing or an EV were electroporated in the Agrobacterium tumefaciens strain C58C1 RifR (GV3101) containing the T-DNA-deficient Ti plasmid pMP90. Positive transformants were harvested, and cells harboring the pTRV1 plasmid were mixed 1:1 with cells harboring the pTRV2 constructs in inoculation medium (10 mM 2-N-morpholino-ethanesulfonic acid pH 5.6, 10 mM MgCl2, and 150 μM acetosyringone). Olive plantlets having at least one pair of fully expanded leaves were Agroinoculated by gently pricking the leaves at the abaxial side as described (Koudounas et al., 2020). This procedure was repeated every 2 weeks until the observation of a phenotype. Leaves from the first two leaf pairs that emerged after Agroinoculation were harvested and used for molecular and biochemical studies. The presence of TRV in non-Agroinoculated (i.e., newly emerged) leaves was validated by detecting the transcripts of TRV coat protein (Supplementary Table 1; Senthil-Kumar and Mysore, 2014).
Reverse transcription was performed with SuperScript II RT (Invitrogen, Life Technologies, Carlsbad, CA, United States) using 200 ng of DNA-free RNA and the oligo(dT)17 primer. Quantitative gene expression analysis was performed in a PikoReal 96 Real-Time PCR system (Thermo Fisher Scientific, Waltham, MA, United States) using the SYBR Select Master Mix (Applied Biosystems, Life Technologies, Carlsbad, CA, United States) and calculated by the ΔΔCt method. OeGLU, OeGES1, OeISY, OeOMES, OeSXS, and OeEAME1/2 transcripts were amplified with the OeGLUq-F and OeGLUq-R, OeGES1q-F and OeGES1q-R, OeISYq-F and OeISYq-R, OeOMESq-F and OeOMESq-R, OeSXSq-F and OeSXSq-R, OeEAME1q-F and OeEAME1q-R, or OeEAME2q-F and OeEAME2q-R primers, respectively (Supplementary Table 1). Normalization of gene expression data was performed by using the OeActin housekeeping gene as a reference with primers OeActin_q-F and OeActin_q-R (Supplementary Table 1). Standard curves for both the target and the reference genes were generated to determine the amplification efficiency of each gene.
Harvested leaves were frozen in liquid nitrogen and ground into powder using a mortar and pestle. Crude proteins were extracted in ice-cold extraction buffer (100 mM Tris–HCl, pH 8.8, 100 mM EDTA, 1 mM PMSF, 100 mM KCL, 10 mM Na2SO3, and 100 mM glycine), samples were centrifuged, and the soluble fractions were quantified by the Bradford assay. The enzymatic reactions with oleuropein were performed by incubating 10 μg of the soluble protein extracts in 100-μl hydrolysis buffer (5 mM oleuropein, 150 mM sodium acetate, pH 5.5, and 0.05% bovine serum albumin [BSA]) at 37°C for 6 min. The deglycosylation degree of oleuropein was measured by high-performance liquid chromatography (HPLC) as described previously (Koudounas et al., 2015). The enzymatic reactions with p-nitrophenyl β-D-glucopyranoside (pNPGlu) were performed by incubating 10 μg of the soluble protein extracts in 500 μl hydrolysis buffer (10 mM pNPGlu, 150 mM sodium acetate, pH 5.5, and 0.05% BSA) at 37°C for 30 min, and reactions were stopped by mixing with an equal volume of 0.2 M sodium carbonate. The deglycosylated p-nitrophenyl was determined by measuring the absorbance at 405 nm. Relative activity was calculated by arbitrarily setting the oleuropein or pNPGlu β-glucosidase activity of soluble extracts from plants Agroinoculated with pTRV2-EV to 100%.
All solvents were of analytical grade and purchased from Merck. Syringaldehyde (98% purity) used as internal standard (IS) was purchased from Sigma-Aldrich (Steinheim, Germany). IS solution was prepared in acetonitrile at a concentration of 0.5 mg/mL and kept in a refrigerator. Prior to use, the IS solution was left to come to room temperature. Oleuropein was purchased from Extrasynthese (Genay, France), stock solution (15 mM) was prepared in ddH2O, and aliquots were stored in −20°C until usage.
Olive leaves were dried at room temperature in a dark place for a week. Then, the leaves were pulverized at room temperature, and 20 ml of MeOH were added to 100 mg of powdered dried olive leaves. The mixture was placed for 45 min in an ultrasonic bath followed by centrifugation at 4,000 rpm for 3 min. A portion of the methanolic extract was collected (10 mL), and 0.5 ml syringaldehyde in acetonitrile solution was added. The mixture was evaporated to dryness under vacuum in a rotary evaporator. The residue of the above procedure was dissolved in MeOD (600 μl), transferred to a 5 mm NMR tube, and the 1H NMR was recorded at 400 MHz. Oleuropein was identified and quantitated by integrating the peak of proton at 5.9 ppm, and based on the equation y = 0.512x + 0.0904, the results were expressed per 100 mg olive leaf sample (Mousouri et al., 2014). NMR spectra were recorded on a DRX 400 MHz and analyzed with MestreNova. A total of 32 scans were collected into 32K data points over a spectral width of 0–16 ppm with a relaxation delay of 1 s and an acquisition time of 1.7 s. Prior to Fourier transformation (FT), an exponential weighting factor corresponding to a line broadening of 0.3 Hz was applied. The spectra were phase corrected, and accurate integration was performed manually for the peaks of interest.
Three technical replicates from three biological replicates per treatment were used in real-time quantitative polymer chain reaction (RT-qPCR) analyses and in vitro enzymatic assays. Three or six biological replicates of plants Agroinoculated with pTRV2-EV or pTRV2-OeGLU, respectively, were used for NMR spectra analysis. Data were analyzed using the GraphPad Prism program (GraphPad Software). The statistical significance of differences between control and OeGLU-silenced samples was tested by unpaired Student’s t-test.
Sequence data discussed in this article are available in GenBank under accession numbers: AY083162 (OeGLU), GABQ01080755 (OeChlH), GABQ01079399 (OeActin), AF406990 (pTRV1), AF406991 (pTRV2-MCS), JN408072 (OeGES1), KT954038 (OeISY), MT909123 (OeOMES), MT909125 (OeSXS), MK234850 (OeEAME1), and MK160486 (OeEAME2).
To silence OeGLU, we first performed bioinformatic analysis to identify specific regions that would not trigger any off-target silencing (Supplementary Figure 1). cDNA fragments of either OeGLU or the H subunit of Mg-protoporphyrin chelatase (OeChlH) were cloned in plasmids encoding the bipartite RNA genome of TRV. ChlH is involved in the biosynthesis of chlorophyll and serves as a positive control of the successful VIGS process (Senthil-Kumar and Mysore, 2014). Since the fragment used to trigger silencing of OeChlH had significant homology (Supplementary Figure 2A) with the corresponding cDNA from Nicotiana benthamiana (NbChlH), the efficiency of the construct to trigger silencing of ChlH was firstly validated in Agroinfiltrated tobacco plants. As expected, a yellowish leaf phenotype due to chlorophyll reduction was observed (Supplementary Figure 2B) indicating that NbChlH was successfully silenced. We next validated the efficiency of the TRV-based VIGS constructs by Agroinoculating olive seedlings. An intense yellowing phenotype indicative of successful silencing of the targeted gene in olive plants Agroinoculated with the pTRV2-OeChlH construct confirmed the successful application of the methodology (Supplementary Figure 3). Olive plants Agroinoculated with either the pTRV2-EV or the pTRV2-OeGLU constructs had no detectable phenotype (Supplementary Figure 3). Detection of the TRV coat protein transcripts in non-Agroinoculated leaves confirmed that TRV successfully infected the olive plants and moved systemically toward newly emerged leaves (Supplementary Figure 4).
To analyze the degree of silencing of OeGLU, we performed RT-qPCR analysis in olive young leaves, which clearly demonstrated that OeGLU was successfully silenced and the expression level was reduced by 80.82% compared to control conditions (Figure 2A). This result was further validated by semi-quantitative RT-PCR (Figure 2B). Additionally, we performed RT-qPCR analysis to monitor the expression levels of other characterized enzymes from the secoiridoid pathway in these plants. Silencing of OeGLU did not affect the transcript levels of either upstream (i.e., OeGES1, OeISY, and OeOMES) or downstream (i.e., OeEAME2) enzymes (Supplementary Figure 5).
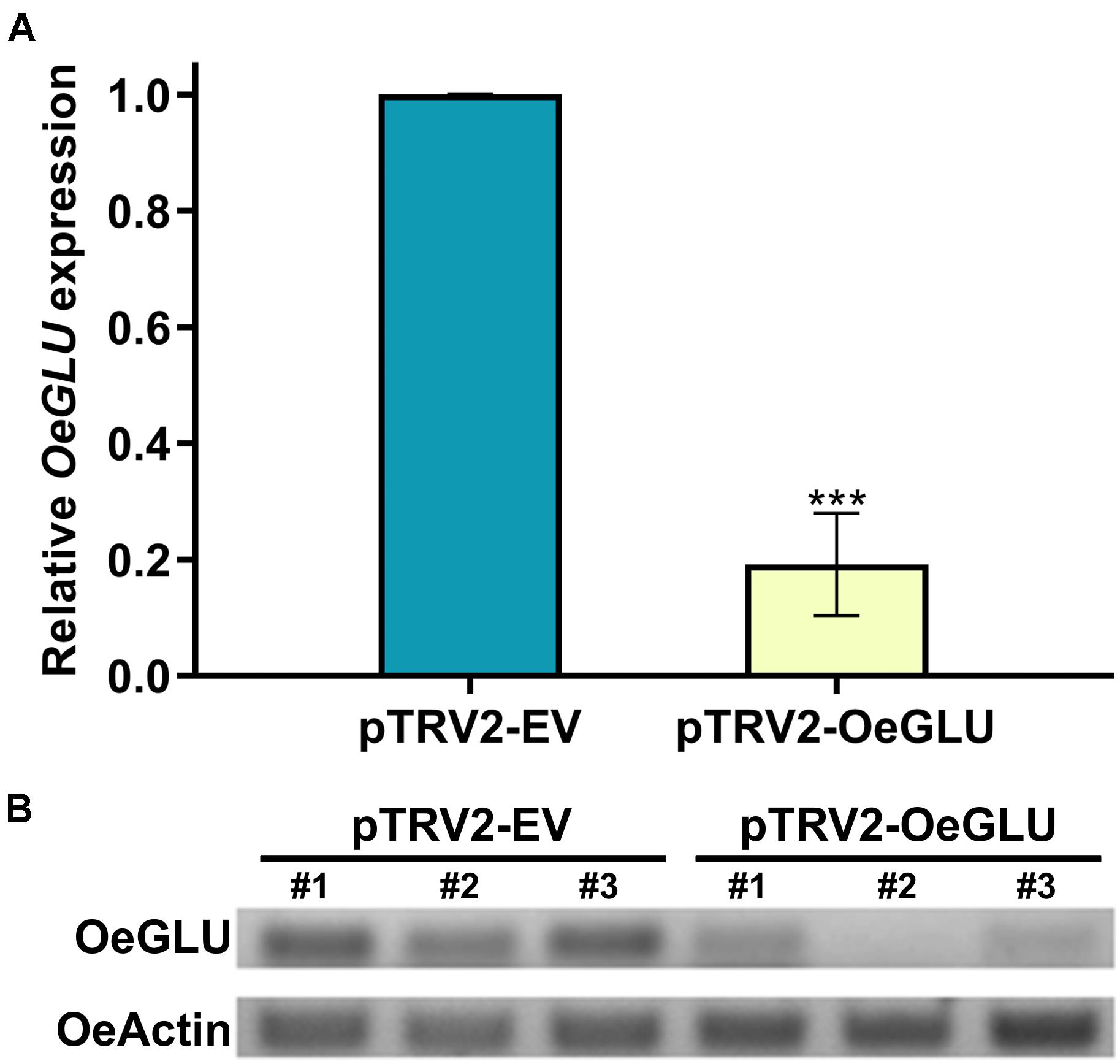
Figure 2. VIGS-mediated silencing of OeGLU in olive plants. Real Time quantitative PCR (RT-PCR) (A) and semi-quantitative RT-PCR (B) analysis of OeGLU expression level of plants Agroinoculated with either pTRV2-EV or pTRV2-OeGLU constructs. OeActin was used as an internal control to normalize the samples. Three technical replicates of three biological replicates per treatment (A) or three biological replicates per treatment (B) were analyzed. Mean ± SE and asterisks denote statistical significance (***P ≤ 0.001; Student’s t-test). Numbers indicate biological replicates. EV, empty vector; OeGLU, oleuropein β-glucosidase.
We next investigated whether this biochemical phenotype would have an impact on the deglycosylation of oleuropein. HPLC analysis of the in vitro enzymatic assays using protein extracts from plants that OeGLU was silenced with oleuropein demonstrated that the relative OeGLU enzymatic activity was drastically reduced (Figure 3A). The degree of deglycosylation of oleuropein was reduced by 65.96% compared to control conditions after 6 min of incubation. As expected, this result coincides with the RT-qPCR (Figure 2A) and is in accordance with the abundance of OeGLU transcripts in the RNAi-silenced plants. The progress of the enzymatic deglycosylation of oleuropein revealed that the relative activity of OeGLU during the first 3 min of incubation was null, at 6 min was 30.09%, and even after 15 min of incubation, it remained at 33.93% compared to control (Figure 3B).
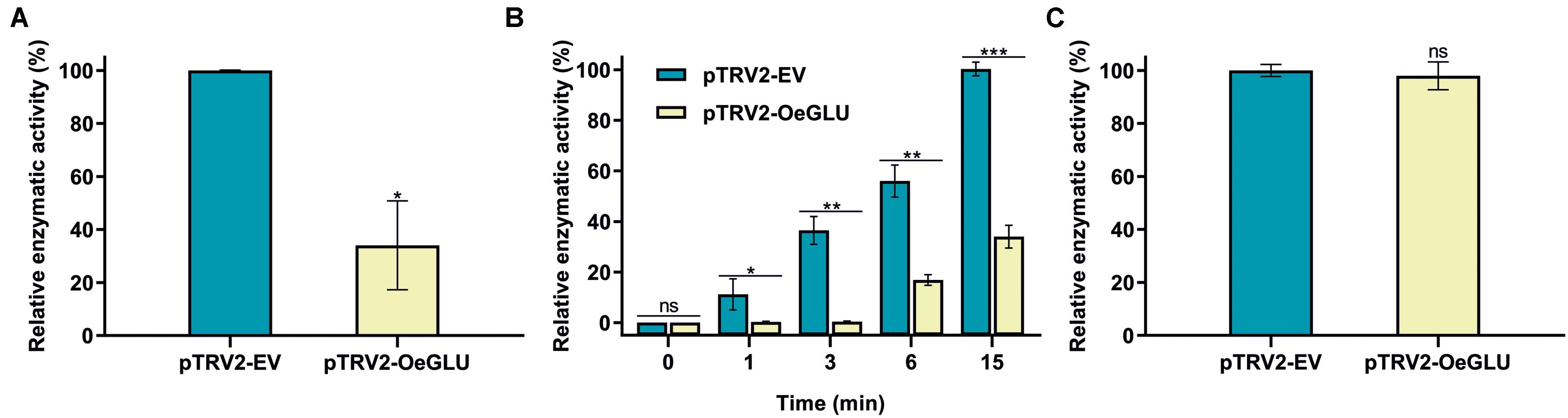
Figure 3. In vitro enzymatic assays of crude leaf protein extracts of olives Agroinoculated with either pTRV2-EV or pTRV2-OeGLU constructs. (A) Relative enzymatic activity against oleuropein at 6 min. (B) Relative enzymatic activity against oleuropein at regular time intervals. (C) Relative enzymatic activity against pNPGlu. Three technical replicates of three biological replicates per treatment were analyzed. Mean ± SE and asterisks denote statistical significance (***P ≤ 0.001, **P ≤ 0.01, *P ≤ 0.05, ns: not significant; Student’s t-test). EV, empty vector; OeGLU, oleuropein β-glucosidase; pNPGlu, p-nitrophenyl β-D-glucopyranoside.
This result can be attributed to the fact that the VIGS-based approach of silencing genes varies and rarely reaches 100% effectiveness, as would be in a knockout mutant. Therefore, a number of remaining “escaping” transcripts were translated to the reduced quantity of OeGLU, nevertheless adequate to catalyze the enzymatic reaction to less extend. In addition, other non-specific β-glucosidases present in the olive tree that exhibit a lower affinity for oleuropein could potentially deglycosylate this substrate but within a worth noting delayed time lapse.
The specificity of silencing of OeGLU was validated by screening for the relative enzymatic activity of pNPGlu deglycosylation – a general synthetic substrate often used with β-glucosidases (Figure 3C). Since the relative non-specific β-glucosidase activity of the crude extracts remained at similar levels, the silencing of OeGLU was highly selective.
We next questioned whether the reduction of OeGLU would have an impact in planta. Instead of substrate accumulation, the content of the oleuropein secoiridoid in OeGLU silenced plants was lower compared to control non-silenced plants (Figure 4 and Supplementary Table 2). In certain plants, oleuropein was even non-detectable as observed in the NMR spectra of the leaf extracts (Figures 4, 5A,B; Supplementary Figure 6). In our experimental setup, the lowest concentration of oleuropein that can be quantified is 70 μg of oleuropein per 100 mg of dry weight (Mousouri et al., 2014), therefore in these plants the amount of oleuropein was reduced by at least 1,000 times compared to control plants. Additionally, the content of oleuropein aglycones and any related downstream secoiridoid were almost non-detectable. This result is in agreement with the low amount of the precursor oleuropein since the concerted activity of OeGLU on oleuropein followed by methylesterase activity results in a pool of aglycone derivatives (Supplementary Figure 7). It is worth noting that the existence of the oleuropein aglycones in planta (Figure 5A) has not been highlighted in the literature potentially pointing out a continuous, nevertheless at a low level, catabolic pathway of oleuropein. How is oleuropein transported from the vacuoles to the nucleus to be deglycosylated by OeGLU and especially how the cells cope with the highly reactive aglycones remain to be addressed.
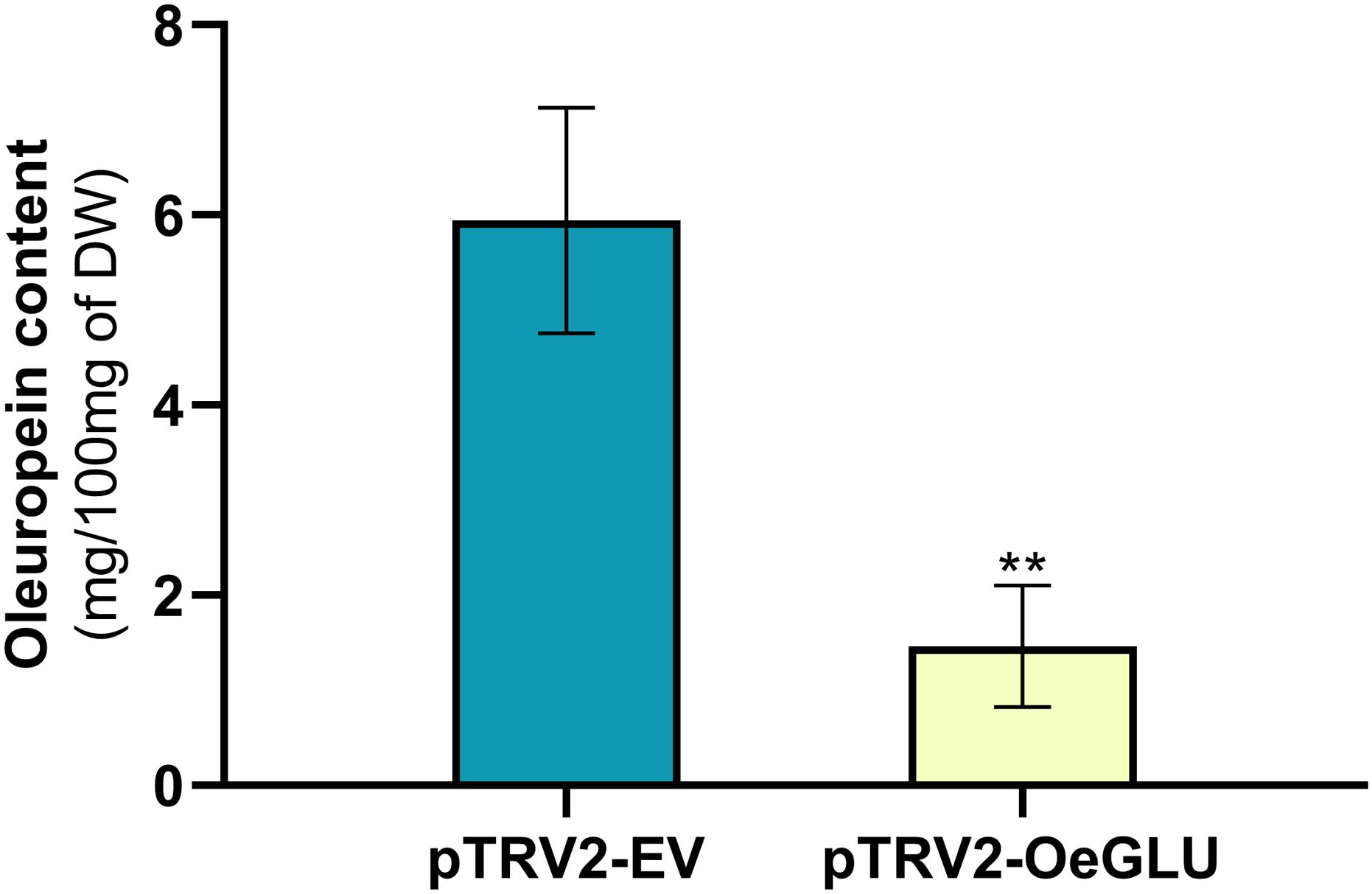
Figure 4. Oleuropein content (mg) per 100 mg (dry weight) of olive leaves. Olive plantlets Agroinoculated either with pTRV2-EV (n = 3) or with pTRV2-OeGLU (n = 6) constructs were screened (Supplementary Table 2). Mean ± SE and asterisks denote statistical significance (**P ≤ 0.01; Student’s t-test). EV, empty vector; OeGLU, oleuropein β-glucosidase.
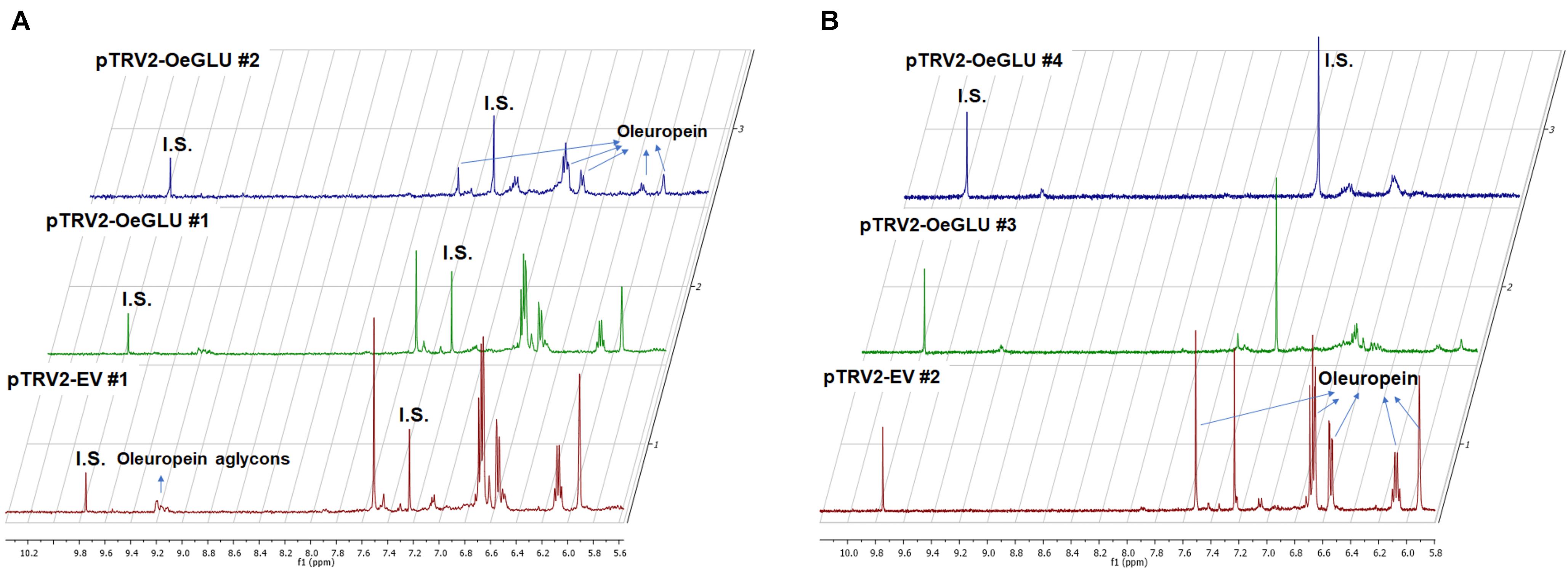
Figure 5. Representative 1D-qNMR spectra of the in planta oleuropein content. Comparison between the methanolic extracts of olive plants Agroinoculated with pTRV2-EV or pTRV2-OeGLU constructs (A,B). Numbers indicate biological replicates. EV, empty vector; IS, internal standard; OeGLU, oleuropein β-glucosidase; 1D qNMR, one-dimensional quantitative nuclear magnetic resonance.
In contrast, oleanolic acid or maslinic acid (triterpenic acids with characteristic peaks observed between 0.8 and 1.2 ppm), secondary compounds not related to the biosynthetic pathway of secoiridoids, were unaffected by the silencing of OeGLU (Figure 6A). The methyl group of oleuropein observed at 1.7 ppm and the characteristic H-1 at 5.90 ppm presented significant quantitative differences among the silenced and the control samples confirming that the effect in planta was apparently secoiridoid specific. No other peaks of related secoiridoids of any precursor molecule could be observed in the plants that OeGLU was silenced (Figures 5A,B, 6B). Control Agroinoculated plants contained high amount of oleuropein, and a detectable amount of decarboxymethylated oleuropein aglycone products, which are observed in the aldehyde region (Figures 4, 5A,B).
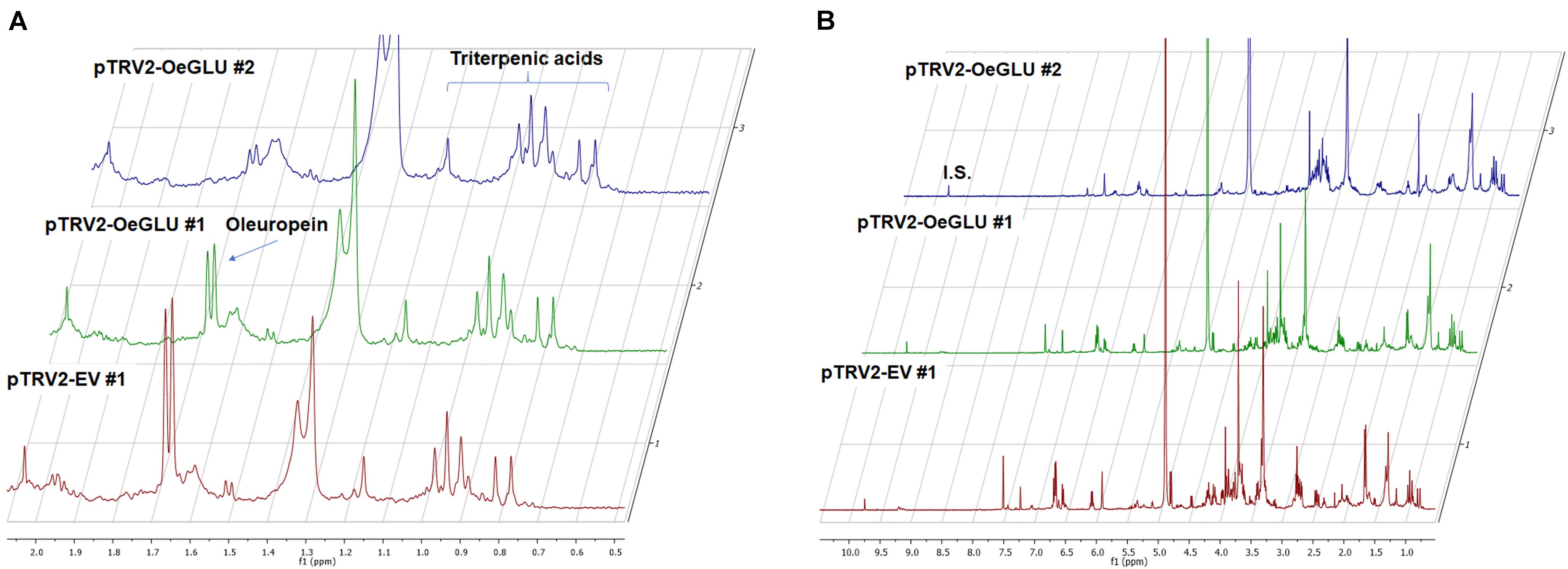
Figure 6. Representative 1D-qNMR spectra. Comparison between the methanolic extracts of plants Agroinoculated with pTRV2-EV and pTRV2-OeGLU constructs for their triterpenic acids content (A) or their full spectrum (B). Numbers indicate biological replicates. EV, empty vector; IS, internal standard; OeGLU, oleuropein β-glucosidase; 1D-qNMR, one-dimensional quantitative nuclear magnetic resonance.
Plant chemical defense is often formed as a dual-partner system composed of glycosylated secondary compounds and dedicated detonating β-glucosidases ensuring that a reactive defensive aglycone is released only after deglycosylation (Morant et al., 2008; Piasecka et al., 2015). In Oleaceae species, the oleuropein/OeGLU system serves as a mighty chemical arsenal against herbivores (Konno et al., 1999) and this enzymatic reaction also determines the quality and flavor of olive oil (Romero-Segura et al., 2012; Hachicha Hbaieb et al., 2015; Vitaglione et al., 2015). Previous in vitro determination of enzymatic properties of OeGLU (Koudounas et al., 2015, 2017) urged us to functionally characterize this enzyme in the olive tree.
Molecular analyses of the Agroinoculated olive plants confirmed the successful VIGS-mediated silencing of OeGLU, and transcript abundance was reduced by more than 80%. VIGS processes, as all the RNA interference (RNAi) approaches, are known to result in a broad range of silencing (Meins et al., 2005), and comparable transcript reduction was observed in the woody tree Jatropha curcas using TRV-based constructs (Ye et al., 2009). In vitro enzymatic assays with oleuropein revealed that the relative deglycosylation activity of leaf protein extracts from OeGLU-silenced plants was drastically reduced compared to control plants and was arrested to the ratio of almost one-third independently of the incubation time of the enzymatic reactions. In contrast, enzymatic assays with pNPGlu confirmed that the relative non-specific β-glucosidase activity of the crude extracts remained at similar levels. In agreement with previously determined biochemical characteristics and kinetics of heterologously expressed OeGLU (Koudounas et al., 2017), these results further confirm that OeGLU is the major oleuropein-specific β-glucosidase in olives since the deglycosylation activity of oleuropein was directly related to the abundance of OeGLU transcripts in planta. VIGS-mediated silencing is a powerful tool to study the biosynthetic pathway of secoiridoids in the olive tree, and this approach is expected to complement the functional characterization of other enzymatic hubs.
The in planta amount of oleuropein was found significantly lower in OeGLU-silenced olive plants compared to controls. The content of upstream and downstream secoiridoids was also reduced or even non-detectable revealing that reduction of OeGLU transcripts affected unexpectedly this pathway. The characteristic peak at 5.95 ppm that corresponds to H-1 of oleoside 11-methyl ester (Park et al., 1999; Mousouri et al., 2014), the first secoiridoid of the biosynthetic pathway of oleuropein (Figure 1), was non-detectable. Additionally, no differences between control and OeGLU-silenced olive plants were observed in the characteristic peaks of precursor iridoids (Figure 1), such as 7-deoxyloganic acid (5.20, 7.41 ppm) (Teng et al., 2005), 7-epi-loganic acid (5.38, 7.37 ppm) (Damtoft et al., 1993), 7-epi-loganin (5.33, 7.41 ppm) (Itoh et al., 2005), and 7-ketologanin (5.51, 7.39 ppm) (Kamoldinov et al., 2011); therefore, none of these compounds was accumulated in the OeGLU-silenced plants.
In contrast, the amount of characteristic triterpenoids was unaffected by OeGLU silencing. Although a possible effect in other classes of terpenoids cannot be excluded, the effect was apparently secoiridoid specific. These results strongly suggest that a regulatory loop mechanism controls the flux toward the branch of secoiridoids in olives.
It is worth noting that 7-β-1-D-glucopyranosyl-11-methyl oleoside (Figure 1) has been proposed to be an intermediate in the biosynthetic pathway of olive secoiridoids (Obied et al., 2008) only as the precursor of ligstroside but it is unknown whether it is deglycosylated back to oleoside 11-methyl ester (Damtoft et al., 1993). This compound is found in trace amounts in olive leaves (De Nino et al., 1999) and has not been tested as a potential substrate of OeGLU since it is not commercially available. Even if OeGLU could potentially deglycosylate this diglucoside, the absence of the characteristic peak of its direct precursor – oleoside 11-methyl ester (Figure 1) – is an unexpected result.
In our experimental setup, olive leaves were dried at room temperature for a week prior to extraction and NMR spectra analysis. Although all secoiridoids and secoiridoid glucosides are not volatile compounds, this could potentially have an impact on volatile compounds found at the very early steps of the biosynthetic pathway before iridodial (Figure 1), like for example in the case of 8-hydroxygeraniol. On the other hand, no differences could be observed in the early iridoids among the samples, therefore most likely the biosynthesis of iridoids was supplied with comparable quantities of precursor molecules.
Among the characterized dual-partner defense systems, the absence of one partner typically does not affect the other. For example, cyanogenic glucosides are biosynthesized in barley (Hordeum vulgare) leaves despite the lack of co-localized cyanide releasing β-glucosidase and transient overexpression of a cyanogenic β-glucosidase from sorghum (Sorghum bicolor) reconstitutes cyanogenesis in barley leaves (Nielsen et al., 2006). In white clover (Trifolium repens) either both cyanogenic glucosides and the respective β-glucosidase or one or even none of the two partners are present (Olsen et al., 2007, 2008). In the glucosinolate-myrosinase defense system of Brassicaceae, knockout mutations of two functional myrosinases (TGG1 and TGG2) in Arabidopsis thaliana resulted in slightly higher glucosinolate content in certain developmental stages (Barth and Jander, 2006).
One example of unexpected phenotype was recently reported in the medicinal plant Catharanthus roseus after unbalancing the metabolic flux of strictosidine biosynthesis – a monoterpene indole alkaloid closely related to oleuropein with similar protein-crosslinking activity after deglycosylation (Guirimand et al., 2010; Sudžuković et al., 2016). VIGS-mediated silencing of either strictosidine β-glucosidase (Carqueijeiro et al., 2021) or the vacuolar exporter of strictosidine (Payne et al., 2017) resulted in necrotic symptoms. Possibly olives have evolved a regulatory framework within the secoiridoid pathway to avoid any substantial accumulation of cytotoxic intermediates that eventually could result in cell death.
A growing body of evidence highlights that the secondary metabolism may regulate plant defense responses which suggests the existence of synergies among distinct biosynthetic pathways (Erb and Kliebenstein, 2020). Mutants of enzymes involved in the biosynthesis of glucosinolates in Arabidopsis may affect the pathogen-triggered callose regulation (Clay et al., 2009), the biosynthesis of phenylpropanoids (Kim et al., 2020), hormonal signaling (Burow et al., 2015), or biosynthesis of other tryptophan-derived compounds (Frerigmann et al., 2016). Similar regulatory cross-talks exist in the biosynthesis of benzoxazinoids (Ahmad et al., 2011; Meihls et al., 2013; Li et al., 2018). Even though, feedback or feedforward regulation among specialized metabolism and distinct biosynthetic pathways was observed, in most cases, the underlying molecular etiology remains elusive.
Reports about regulatory loop mechanisms that self-govern a specialized biosynthetic pathway in plants are scarce. Identified examples include epistasis in carotenoid biosynthesis (Kachanovsky et al., 2012) and feedback regulation through intermediates in phenylpropanoids (Blount et al., 2000). To the best of our knowledge, the only known example partially resembling the phenotype observed in OeGLU-silenced olive plants has been reported in opium poppy (Papaver somniferum). Silencing of codeinone reductase (COR) resulted in accumulation of (S)-reticuline, an intermediate compound upstream of seven enzymatic steps proposing a feedback regulatory loop mechanism (Allen et al., 2004) and preventing intermediates of benzylisoquinoline synthesis to enter the morphine-specific branch. In agreement with the absence of any transcriptional effect in other enzymes engaged in the biosynthetic pathway of secoiridoids after silencing OeGLU in olives, silencing of COR did not affect the abundance of transcripts encoding for enzymes engaged upstream and downstream of (S)-reticuline (Allen et al., 2004). Although oleosides- and morphinan- type alkaloids are structurally and biosynthetically different, a fundamental regulatory mechanism could govern both secondary metabolic pathways. The subcellular compartmentalization of the secoiridoid pathway in Oleaceae engaging at least two physically separated compartments at the last enzymatic steps (i.e., vacuole and nucleus) (Koudounas et al., 2017) points out retrograde signaling and a sophisticated cross-talk within the cell.
Biosynthesis of specialized metabolites has energetic and metabolic costs for the plants (Gershenzon, 1994; Neilson et al., 2013; Cipollini et al., 2018). Possibly the absence of OeGLU in planta is perceived as a failure in the defensive pathway of secoiridoids and olives completely bypass the flux toward oleuropein to recoup the costs and avoid investing resources in the absence of defensive benefits. The molecular mechanism(s) by which olive cells sense the loss of a key enzymatic hub and bypass the flux toward the biosynthetic pathway of secoiridoids remain to be addressed. Although no difference was observed at the transcript levels of the selected genes engaged in this pathway, possibly the expression levels of other regulatory genes, such as transcription factors or specialized transporters, may have been altered after the silencing of OeGLU, and a comparative transcriptomic approach is expected to shed light on the etiology of the observed phenotype. Nonetheless, this study highlights an unexpected direct link between OeGLU and total content of secoiridoids, thus this enzyme could serve as a molecular target of high biotechnological interest in order to produce tailor-made olive oils with adjustable secoiridoid content having an optimized balance of flavor (Vitaglione et al., 2015) and high nutritional value with beneficial aspects in human health.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
KK and PH designed the research. KK, MT, and EA prepared the constructs, performed VIGS experiments, analyzed gene expressions, and performed in vitro enzymatic assays. EM and PM performed method development for qualitative and quantitative analyses of olive leaf ingredients by NMR. AR analyzed application of NMR methodology and performed data collection. KK, EM, PM, and PH wrote the manuscript. All authors contributed to the article and approved the submitted version.
PH acknowledges funding from the General Secretariat of Research and Technology, Greece, the National Emblematic Action “Olive Routes,” and the European Regional Development Fund of the European Union and Greek National Funds through the Operational Program Competitiveness, Entrepreneurship and Innovation, under the call RESEARCH–CREATE–INNOVATE (“ELIADA,” project code: T2EΔK -01315). MT acknowledges funding from Greece and the European Union (European Social Fund, ESF) through the Operational Program “Human Resources Development, Education and Lifelong Learning” in the context of the project “Strengthening Human Resources Research Potential via Doctorate Research” (MIS-5000432), implemented by the State Scholarships Foundation (IKY). AR acknowledges scholarship from World Olive Center for Health.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.671487/full#supplementary-material
Ahmad, S., Veyrat, N., Gordon-Weeks, R., Zhang, Y., Martin, J., Smart, L., et al. (2011). Benzoxazinoid Metabolites Regulate Innate Immunity against Aphids and Fungi in Maize. Plant Physiol. 157, 317–327. doi: 10.1104/pp.111.180224
Alagna, F., Geu-Flores, F., Kries, H., Panara, F., Baldoni, L., O’Connor, S. E., et al. (2016). Identification and characterization of the iridoid synthase involved in oleuropein biosynthesis in olive (Olea europaea) fruits. J. Biol. Chem. 291, 5542–5554. doi: 10.1074/jbc.M115.701276
Allen, R. S., Millgate, A. G., Chitty, J. A., Thisleton, J., Miller, J. A. C., Fist, A. J., et al. (2004). RNAi-mediated replacement of morphine with the nonnarcotic alkaloid reticuline in opium poppy. Nat. Biotechnol. 22, 1559–1566. doi: 10.1038/nbt1033
Amiot, M. J., Fleuriet, A., and Macheix, J. J. (1986). Importance and evolution of phenolic compounds in olive during growth and maturation. J. Agric. Food Chem. 34, 823–826. doi: 10.1021/jf00071a014
Barth, C., and Jander, G. (2006). Arabidopsis myrosinases TGG1 and TGG2 have redundant function in glucosinolate breakdown and insect defense. Plant J. 46, 549–562. doi: 10.1111/j.1365-313X.2006.02716.x
Blount, J. W., Korth, K. L., Masoud, S. A., Rasmussen, S., Lamb, C., and Dixon, R. A. (2000). Altering Expression of Cinnamic Acid 4-Hydroxylase in Transgenic Plants Provides Evidence for a Feedback Loop at the Entry Point into the Phenylpropanoid Pathway. Plant Physiol. 122, 107–116. doi: 10.1104/pp.122.1.107
Bulotta, S., Oliverio, M., Russo, D., and Procopio, A. (2013). “Biological activity of oleuropein and its derivatives,” in Natural Products: Phytochemistry, Botany and Metabolism of Alkaloids, Phenolics and Terpenes, eds K. G. Ramawat and J.-M. Mérillon (Berlin: Springer), 3605–3638.
Burow, M., Atwell, S., Francisco, M., Kerwin, Rachel, E., and Halkier, et al. (2015). The Glucosinolate Biosynthetic Gene AOP2 Mediates Feed-back Regulation of Jasmonic Acid Signaling in Arabidopsis. Mol. Plant 8, 1201–1212. doi: 10.1016/j.molp.2015.03.001
Carqueijeiro, I., Koudounas, K., Dugé de Bernonville, T., Sepúlveda, L. J., Mosquera, A., Bomzan, D. P., et al. (2021). Alternative splicing creates a pseudo-strictosidine β-D-glucosidase modulating alkaloid synthesis in Catharanthus roseus. Plant Physiol. 185, 836–856. doi: 10.1093/plphys/kiaa075
Cipollini, D., Walters, D., and Voelckel, C. (2018). “Costs of Resistance in Plants: from Theory to Evidence,” in Annual Plant Reviews Online, ed. J. A. Roberts (Hoboken: Wiley), 263–307.
Clay, N. K., Adio, A. M., Denoux, C., Jander, G., and Ausubel, F. M. (2009). Glucosinolate Metabolites Required for an Arabidopsis Innate Immune Response. Science 323, 95–101. doi: 10.1126/science.1164627
Damtoft, S., Franzyk, H., and Jensen, S. R. (1993). Biosynthesis of secoiridoid glucosides in Oleaceae. Phytochemistry 34, 1291–1299. doi: 10.1016/0031-9422(91)80018-V
De Nino, A., Mazzotti, F., Morrone, S. P., Perri, E., Raffaelli, A., and Sindona, G. (1999). Characterization of cassanese olive cultivar through the identification of new trace components by ionspray tandem mass spectrometry. J. Mass Spectrometry 34, 10–16. doi: 10.1002/(SICI)1096-9888(199901)34:1<10::AID-JMS744<3.0.CO;2-X
EFSA. (2012). Scientific Opinion on the substantiation of a health claim related to polyphenols in olive and maintenance of normal blood HDL cholesterol concentrations (ID 1639, further assessment) pursuant to Article 13(1) of Regulation (EC) No 1924/2006. EFSA J. 10:2848. doi: 10.2903/j.efsa.2012.2848
Erb, M., and Kliebenstein, D. J. (2020). Plant Secondary Metabolites as Defenses, Regulators, and Primary Metabolites: the Blurred Functional Trichotomy. Plant Physiol. 184, 39–52. doi: 10.1104/pp.20.00433
Frerigmann, H., Piślewska-Bednarek, M., Sánchez-Vallet, A., Molina, A., Glawischnig, E., Gigolashvili, T., et al. (2016). Regulation of Pathogen-Triggered Tryptophan Metabolism in Arabidopsis thaliana by MYB Transcription Factors and Indole Glucosinolate Conversion Products. Mol. Plant 9, 682–695. doi: 10.1016/j.molp.2016.01.006
Gershenzon, J. (1994). Metabolic costs of terpenoid accumulation in higher plants. J. Chem. Ecol. 20, 1281–1328. doi: 10.1007/BF02059810
Green, P. S. (2004). “Oleaceae,” in Flowering Plants ⋅ Dicotyledons: Lamiales (except Acanthaceae including Avicenniaceae), ed. J. W. Kadereit (Berlin: Springer), 296–306.
Guirimand, G., Courdavault, V., Lanoue, A., Mahroug, S., Guihur, A., Blanc, N., et al. (2010). Strictosidine activation in Apocynaceae: towards a “nuclear time bomb”? BMC Plant Biol. 10:182. doi: 10.1186/1471-2229-10-182
Hachicha Hbaieb, R., Kotti, F., García-Rodríguez, R., Gargouri, M., Sanz, C., and Pérez, A. G. (2015). Monitoring endogenous enzymes during olive fruit ripening and storage: correlation with virgin olive oil phenolic profiles. Food Chem. 174, 240–247. doi: 10.1016/j.foodchem.2014.11.033
HMPC. (2017). Final Assessment Report on Olea europaea L., Folium - First Version. Amsterdam: European Medicines Agency.
Itoh, A., Kumashiro, T., Yamaguchi, M., Nagakura, N., Mizushina, Y., Nishi, T., et al. (2005). Indole Alkaloids and Other Constituents of Rauwolfia serpentina. J. Nat. Products 68, 848–852. doi: 10.1021/np058007n
Kachanovsky, D. E., Filler, S., Isaacson, T., and Hirschberg, J. (2012). Epistasis in tomato color mutations involves regulation of phytoene synthase 1 expression by cis-carotenoids. Proc. Nat. Acad. Sci. U. S. A. 109, 19021–19026. doi: 10.1073/pnas.1214808109
Kamoldinov, K. S., Eshbakova, K. A., Bobakulov, K. M., and Abdullaev, N. D. (2011). Components of Fraxinus raibocarpa. Chem. Nat. Compounds 47, 448–449. doi: 10.1007/s10600-011-9958-5
Ketudat Cairns, J. R., and Esen, A. (2010). β-Glucosidases. Cell. Mol. Life Sci. 67, 3389–3405. doi: 10.1007/s00018-010-0399-2
Kim, J. I., Zhang, X., Pascuzzi, P. E., Liu, C.-J., and Chapple, C. (2020). Glucosinolate and phenylpropanoid biosynthesis are linked by proteasome-dependent degradation of PAL. New Phytologist 225, 154–168. doi: 10.1111/nph.16108
Konno, K., Hirayama, C., Yasui, H., and Nakamura, M. (1999). Enzymatic activation of oleuropein: a protein crosslinker used as a chemical defense in the privet tree. Proc. Nat. Acad. Sci. U. S. A. 96, 9159–9164. doi: 10.1073/pnas.96.16.9159
Koudounas, K., Banilas, G., Michaelidis, C., Demoliou, C., Rigas, S., and Hatzopoulos, P. (2015). A defence-related Olea europaea β-glucosidase hydrolyses and activates oleuropein into a potent protein cross-linking agent. J. Exp. Bot. 66, 2093–2106. doi: 10.1093/jxb/erv002
Koudounas, K., Thomopoulou, M., Angeli, E., Tsitsekian, D., Rigas, S., and Hatzopoulos, P. (2020). “Virus-Induced Gene Silencing in Olive Tree (Oleaceae),” in Virus-Induced Gene Silencing in Plants: Methods and Protocols, eds V. Courdavault and S. Besseau (New York: Springer), 165–182.
Koudounas, K., Thomopoulou, M., Michaelidis, C., Zevgiti, E., Papakostas, G., Tserou, P., et al. (2017). The C-Domain of Oleuropein β-Glucosidase Assists in Protein Folding and Sequesters the Enzyme in Nucleus. Plant Physiol. 174, 1371–1383. doi: 10.1104/pp.17.00512
Li, B., Förster, C., Robert, C. A. M., Züst, T., Hu, L., Machado, R. A. R., et al. (2018). Convergent evolution of a metabolic switch between aphid and caterpillar resistance in cereals. Sci. Adv. 4:eaat6797. doi: 10.1126/sciadv.aat6797
Meihls, L. N., Handrick, V., Glauser, G., Barbier, H., Kaur, H., Haribal, M. M., et al. (2013). Natural Variation in Maize Aphid Resistance Is Associated with 2,4-Dihydroxy-7-Methoxy-1,4-Benzoxazin-3-One Glucoside Methyltransferase Activity. Plant Cell 25, 2341–2355. doi: 10.1105/tpc.113.112409
Meins, F., Si-Ammour, A., and Blevins, T. (2005). RNA silencing systems and their relevance to plant development. Ann. Rev. Cell Dev. Biol. 21, 297–318. doi: 10.1146/annurev.cellbio.21.122303.114706
Morant, A. V., Jørgensen, K., Jørgensen, C., Paquette, S. M., Sánchez-Pérez, R., Møller, B. L., et al. (2008). β-Glucosidases as detonators of plant chemical defense. Phytochemistry 69, 1795–1813. doi: 10.1016/j.phytochem.2008.03.006
Mousouri, E., Melliou, E., and Magiatis, P. (2014). Isolation of Megaritolactones and Other Bioactive Metabolites from ‘Megaritiki’ Table Olives and Debittering Water. J. Agric. Food Chem. 62, 660–667. doi: 10.1021/jf404685h
Neilson, E. H., Goodger, J. Q. D., Woodrow, I. E., and Møller, B. L. (2013). Plant chemical defense: at what cost? Trends Plant Sci. 18, 250–258. doi: 10.1016/j.tplants.2013.01.001
Nielsen, K., Hrmova, M., Nielsen, J., Forslund, K., Ebert, S., Olsen, C., et al. (2006). Reconstitution of cyanogenesis in barley (Hordeum vulgare L.) and its implications for resistance against the barley powdery mildew fungus. Planta 223, 1010–1023. doi: 10.1007/s00425-005-0158-z
Obied, H. K., Prenzler, P. D., Ryan, D., Servili, M., Taticchi, A., Esposto, S., et al. (2008). Biosynthesis and biotransformations of phenol-conjugated oleosidic secoiridoids from Olea europaea L. Nat. Prod. Rep. 25, 1167–1179. doi: 10.1039/b719736e
Olsen, K. M., Hsu, S.-C., and Small, L. L. (2008). Evidence on the Molecular Basis of the Ac/ac Adaptive Cyanogenesis Polymorphism in White Clover (Trifolium repens L.). Genetics 179, 517–526. doi: 10.1534/genetics.107.080366
Olsen, K. M., Sutherland, B. L., and Small, L. L. (2007). Molecular evolution of the Li/li chemical defence polymorphism in white clover (Trifolium repens L.). Mol. Ecol. 16, 4180–4193. doi: 10.1111/j.1365-294X.2007.03506.x
Papanikolaou, C., Melliou, E., and Magiatis, P. (2019). “Olive Oil Phenols,” in Functional Foods, ed. V. Lagouri. (London: IntechOpen), 29–46.
Park, H. J., Lee, M. S., Lee, K. T., Sohn, I. C., Han, Y. N., and Miyamoto, K. I. (1999). Studies on constituents with cytotoxic activity from the stem bark of Syringa velutina. Chem. Pharmaceutical Bull. 47, 1029–1031. doi: 10.1248/cpb.47.1029
Payne, R. M. E., Xu, D., Foureau, E., Teto Carqueijeiro, M. I. S., Oudin, A., Bernonville, T. D. D., et al. (2017). An NPF transporter exports a central monoterpene indole alkaloid intermediate from the vacuole. Nat. Plants 3:16208. doi: 10.1038/nplants.2016.208
Piasecka, A., Jedrzejczak-Rey, N., and Bednarek, P. (2015). Secondary metabolites in plant innate immunity: conserved function of divergent chemicals. New Phytol. 206, 948–964. doi: 10.1111/nph.13325
Rodríguez-López, C. E., Hong, B., Paetz, C., Nakamura, Y., Koudounas, K., Passeri, V., et al. (2021). Two bi-functional cytochrome P450 CYP72 enzymes from olive (Olea europaea) catalyze the oxidative C-C bond cleavage in the biosynthesis of secoxy-iridoids – flavor and quality determinants in olive oil. New Phytol. 229, 2288–2301. doi: 10.1111/nph.16975
Romero-Segura, C., García-Rodríguez, R., Sánchez-Ortiz, A., Sanz, C., and Pérez, A. G. (2012). The role of olive β-glucosidase in shaping the phenolic profile of virgin olive oil. Food Res. Int. 45, 191–196. doi: 10.1016/j.foodres.2011.10.024
Senthil-Kumar, M., and Mysore, K. S. (2014). Tobacco rattle virus–based virus-induced gene silencing in Nicotiana benthamiana. Nat. Protoc. 9, 1549–1562. doi: 10.1038/nprot.2014.092
Soler-Rivas, C., Espín, J. C., and Wichers, H. J. (2000). Oleuropein and related compounds. J. Sci. Food Agric. 80, 1013–1023. doi: 10.1002/(SICI)1097-0010(20000515)80:7<1013::AID-JSFA571<3.0.CO;2-C
Sudžuković, N., Schinnerl, J., and Brecker, L. (2016). Phytochemical meanings of tetrahydro-β-carboline moiety in strictosidine derivatives. Bioorg. Med. Chem. 24, 588–595. doi: 10.1016/j.bmc.2015.12.028
Teng, R. W., Wang, D. Z., Wu, Y. S., Lu, Y., Zheng, Q. T., and Yang, C. R. (2005). NMR assignments and single-crystal X-ray diffraction analysis of deoxyloganic acid. Magn. Reson. Chem. 43, 92–96. doi: 10.1002/mrc.1502
Tripoli, E., Giammanco, M., Tabacchi, G., Di Majo, D., Giammanco, S., and La Guardia, M. (2005). The phenolic compounds of olive oil: structure, biological activity and beneficial effects on human health. Nutr. Res. Rev. 18, 98–112. doi: 10.1079/NRR200495
Verdoucq, L., Moriniere, J., Bevan, D. R., Esen, A., Vasella, A., Henrissat, B., et al. (2004). Structural determinants of substrate specificity in family 1 β-glucosidases: novel insights from the crystal structure of sorghum dhurrinase-1, a plant beta-glucosidase with strict specificity, in complex with its natural substrate. J. Biol. Chem. 279, 31796–31803. doi: 10.1074/jbc.M402918200
Vezzaro, A., Krause, S. T., Nonis, A., Ramina, A., Degenhardt, J., and Ruperti, B. (2012). Isolation and characterization of terpene synthases potentially involved in flavor development of ripening olive (Olea europaea) fruits. J. Plant Physiol. 169, 908–914. doi: 10.1016/j.jplph.2012.01.021
Vitaglione, P., Savarese, M., Paduano, A., Scalfi, L., Fogliano, V., and Sacchi, R. (2015). Healthy Virgin Olive Oil: a Matter of Bitterness. Crit. Rev. Food Sci. Nutr. 55, 1808–1818. doi: 10.1080/10408398.2012.708685
Volk, J., Sarafeddinov, A., Unver, T., Marx, S., Tretzel, J., Zotzel, J., et al. (2019). Two novel methylesterases from Olea europaea contribute to the catabolism of oleoside-type secoiridoid esters. Planta 250, 2083–2097. doi: 10.1007/s00425-019-03286-0
Xia, L., Ruppert, M., Wang, M., Panjikar, S., Lin, H., Rajendran, C., et al. (2012). Structures of alkaloid biosynthetic glucosidases decode substrate specificity. ACS Chem. Biol. 7, 226–234. doi: 10.1021/cb200267w
Keywords: Olea europaea, olive, secoiridoids, oleuropein, β-glucosidase, virus-induced gene silencing, tobacco rattle virus, one-dimensional quantitative nuclear magnetic resonance (1D-qNMR)
Citation: Koudounas K, Thomopoulou M, Rigakou A, Angeli E, Melliou E, Magiatis P and Hatzopoulos P (2021) Silencing of Oleuropein β-Glucosidase Abolishes the Biosynthetic Capacity of Secoiridoids in Olives. Front. Plant Sci. 12:671487. doi: 10.3389/fpls.2021.671487
Received: 23 February 2021; Accepted: 11 August 2021;
Published: 03 September 2021.
Edited by:
Thu Thuy Dang, University of British Columbia Okanagan, CanadaReviewed by:
Laura Perez Fons, Royal Holloway, University of London, United KingdomCopyright © 2021 Koudounas, Thomopoulou, Rigakou, Angeli, Melliou, Magiatis and Hatzopoulos. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Polydefkis Hatzopoulos, cGhhdEBhdWEuZ3I=
†ORCID: Konstantinos Koudounas, orcid.org/0000-0002-6000-6565; Margarita Thomopoulou, orcid.org/0000-0002-4999-7257; Elisavet Angeli, orcid.org/0000-0003-4104-909X; Aimilia Rigakou, orcid.org/0000-0002-1034-7484; Eleni Melliou, orcid.org/0000-0003-1419-1752; Prokopios Magiatis, orcid.org/0000-0002-0399-5344; Polydefkis Hatzopoulos, orcid.org/0000-0002-0074-2552
§Present address: Konstantinos Koudounas, EA2106 Biomolécules et Biotechnologies Végétales, Université de Tours, Tours, France
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.