- 1Zhengzhou Research Base, State Key Laboratory of Cotton Biology, Zhengzhou University, Zhengzhou, China
- 2School of Life Sciences, Zhengzhou University, Zhengzhou, China
- 3Henan International Joint Laboratory of Crop Gene Resources and Improvements, School of Agricultural Sciences, Zhengzhou University, Zhengzhou, China
As a promising high-throughput reverse genetic tool in plants, virus-induced gene silencing (VIGS) has already begun to fulfill some of this promise in diverse aspects. However, review of the technological advancements about widely used VIGS system, tobacco rattle virus (TRV)-mediated gene silencing, needs timely updates. Hence, this article mainly reviews viral vector construction, inoculation method advances, important influential factors, and summarizes the recent applications in diverse plant species, thus providing a better understanding and advice for functional gene analysis related to crop improvements.
Introduction
Virus-induced gene silencing (VIGS) is a high-throughput reverse genetics technique that exploits an RNA-mediated antiviral defense mechanism [post-transcriptional gene silencing (PTGS)] for functional gene analysis (Ratcliff et al., 1997; Sunilkumar et al., 2006). VIGS was originally used to describe the recovery of viral symptoms on plants after virus infection (Kammen, 1997). Subsequently, researchers demonstrated that this is a manifestation of the plant’s natural defense mechanism induced by virus infection, and some endogenous genes that are homologous to viral genomes could also be silenced at the same time (Ratcliff et al., 1999). Thus, scientists modified the viral genome [complementary DNA (cDNA)] into a recombinant virus vector containing sequences that were homologous to host genes, which could trigger homologous endogenous gene silencing in plants.
Phytoene desaturase (PDS) and Agrobacterium-mediated tobacco rattle virus (TRV)-VIGS by leaf injection can be used as an example (Figure 1). Recombinant vectors containing 300–500 bp (base pair) cDNA fragments and devoid of homopolymeric regions of the PDS gene were introduced into plant cells during agroinfiltration with Agrobacterium cultures. After agroinfiltration, the T-DNA, including the viral genome, was transcribed into sing-strand RNA (ssRNA) in the host plant cells. Then, large amounts of double-stranded RNAs (dsRNAs) generated by RNA-dependent RNA polymerase (RdRp) encoded by the viral genome were detected by the host plant genome as aberrant sequences, and thus cleaved into short interfering RNA (siRNA) duplexes of 21–24 nucleotides by the action of Dicer-like proteins (Ding and Voinnet, 2007). SiRNAs were incorporated as single-stranded RNAs into RISC (RNA-induced silencing complex), which specifically screens and destroy the mRNAs (PDS transcripts) complementary to the siRNAs, resulting in degradation of mRNAs and a photo-bleaching plant phenotype (Waterhouse et al., 2001; Baulcombe, 2002; Zhang et al., 2014; Figure 1).
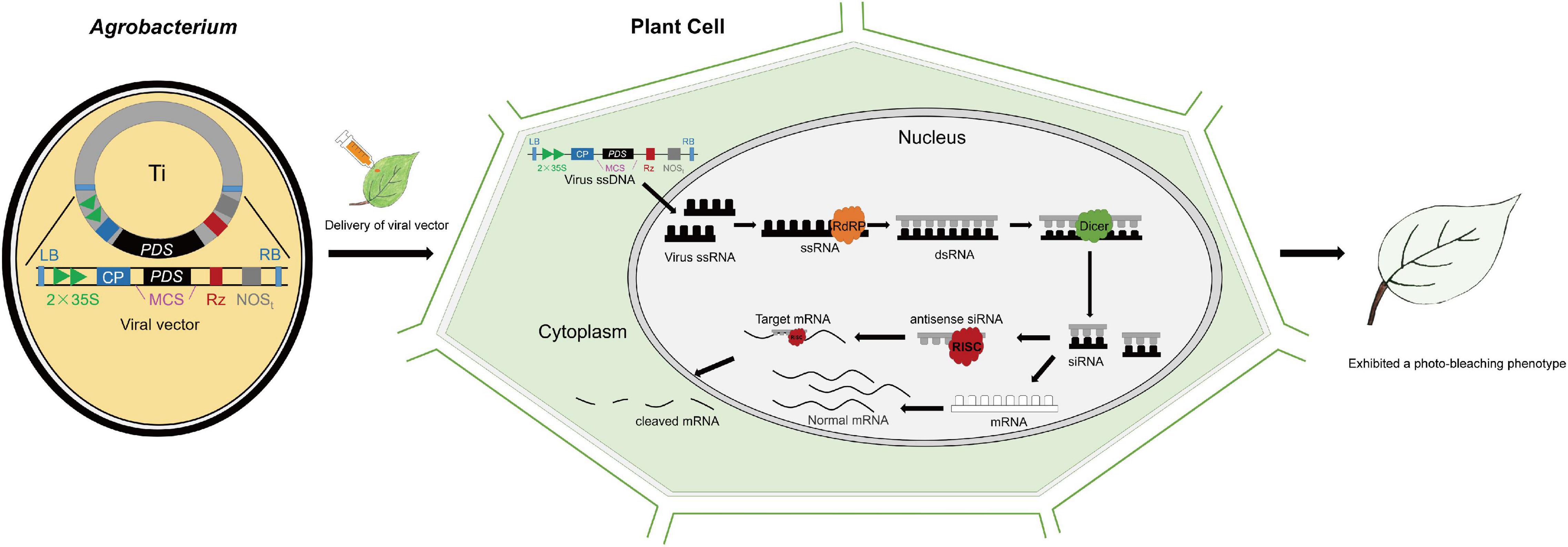
Figure 1. Virus-induced gene silencing (VIGS) mechanism. Phytoene desaturase (PDS) serves as an example. Upon infection, the T-DNA carrying the viral genome is transformed into the plant by Agrobacterium and then transcribed by the host’s RNA polymerase. RNA-dependent RNA Polymerase (RdRP) (yellow) produces double-stranded RNA (dsRNA) from the single-stranded RNA (ssRNA) viral transcript. The dsRNA is then recognized by DICER-like enzyme, Dicer (green) and cleaved into short interfering RNAs (siRNAs). Antisense siRNAs are recognized by RNA-induced silencing complex, RISC (red) and melted into ssRNAs, which then serve as templates for target gene degradation. The single-stranded siRNAs are amplified and spread as mobile silencing signals throughout the plant, thus resulting in target gene silencing in plant organs distant from the site of infection, symbolized by the photo-bleaching phenotype of the entire leaf. LB: left border; RB: right border; 2 × 35S: duplicated cauliflower mosaic virus 35S promoter; CP: coat protein; MCS: multiple cloning site; PDS: PDS cDNA fragment; Rz: self-cleaving ribozyme; NOSt: nopaline synthase terminator.
Compared with other traditional genetic tools, VIGS can rapidly (in 3–4 weeks) silence endogenous genes and display an easily observed silenced phenotype in contemporary plants, with no need for stable transformants. Partial sequence information is sufficient to silence the target gene, and there is a simple operation procedure. Therefore, VIGS technology has been widely used as a high-throughput genetic tool for genetic screening and functional genomics in many species (Lu et al., 2003; Burch-Smith et al., 2004), such as tomato (Liu et al., 2002a), tobacco (Senthil-Kumar and Mysore, 2011b), soybean (Liu et al., 2015), wheat, and corn (Zhang et al., 2017). Methodological advances of VIGS are very important in the post-genomic era. In this review, we firstly summarize the development history of the vector construction and inoculation methodology, and then focus on the significant efficiency influential factors in VIGS application, as well as the adoptable range of plant species of TRV-VIGS is updated here. Therefore, this review would provide a better reference for the methodological study of TRV-VIGS and functional analysis related to genetic improvements in crop breeding.
TRV-VIGS Vector Construction
Kumagai et al. (1995) constructed the first VIGS vector based on tobacco mosaic virus (TMV); they successfully knocked down NbPDS gene expression and obtained NbPDS silenced plants with an albino phenotype by inoculating in vitro RNA transcripts into Nicotiana benthamiana. Subsequently, many viral vectors were successfully modified and used for VIGS studies (Supplementary Table 1). RNA viruses were the earliest and most widely used viral carrier for the establishment of the VIGS system because of their small molecular weight and high infection efficiency (Bruun-Rasmussen et al., 2007). DNA viruses account for a small number of plant viruses with a large genome structure and limited movement in plants. Satellite viruses do not cause any diseases in plants themselves; they are generally not associated with any illness or interference with the true gene silenced phenotype. However, they are only suitable for use in a small number of host plants (Zhou et al., 2012; Supplementary Table 1). Most viruses used for VIGS cannot infect the plant growing points or meristems (Ratcliff et al., 2001; Harper et al., 2002; Supplementary Table 1), but the TRV-VIGS system successfully overcomes the host limitations of meristem transmission (Ratcliff et al., 2001; Liu et al., 2002b) by (1) effectively spreading to all plant tissues, including the meristems; (2) a wide host range [50 or more families (Solanaceae (Purkayastha and Dasgupta, 2009), Cruciferae (Zhang et al., 2004), and Gramineae (Scofield et al., 2005)), among others, in dicots and monocots]; and (3) developing mild viral symptoms after infection. Therefore, TRV vectors have been widely used for VIGS studies, and the viral vector modification of TRV that is important for silencing efficiency of the VIGS system is reviewed (Figure 2).
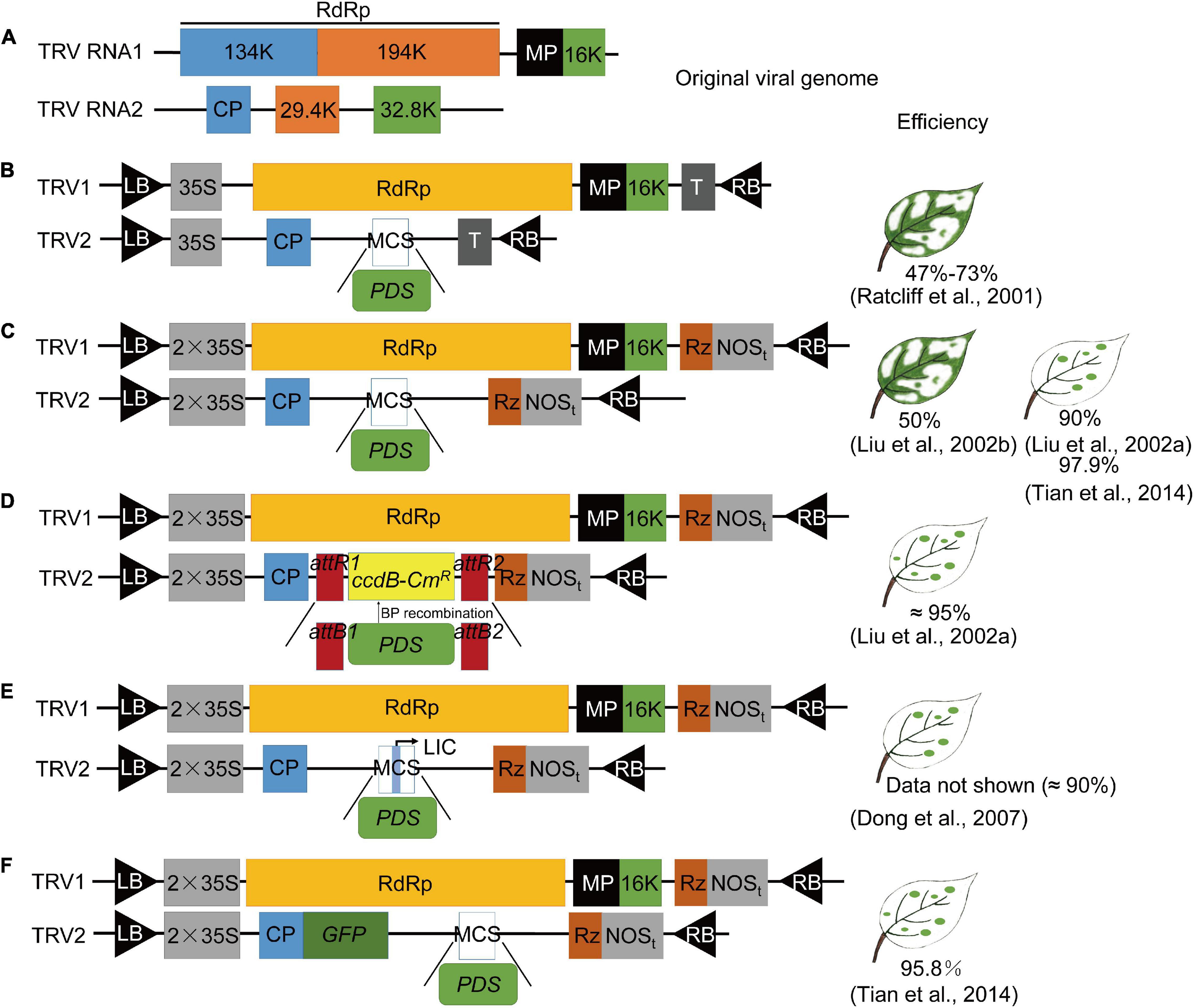
Figure 2. The development process of TRV vector construction. Phytoene desaturase, PDS was used as an example. (A) Genome organization of tobacco rattle virus (TRV). The TRV1 open reading frames (ORFs) correspond to 134 and 194 kDa replicases, a movement protein (MP), and a 16 kDa cysteine-rich protein (16K). The TRV2 ORFs correspond to the coat protein (CP) and 29.4 and 32.8 kDa proteins (29.4K and 32.8K). (B) TRV-based VIGS vector (Ratcliff et al., 2001). Ratcliff et al. constructed separate cDNA clones of TRV (strain PPK20) RNA1 and RNA2 under the control of cauliflower mosaic virus (CaMV) 35S promoters on the transferred (T) DNA of plant binary transformation vectors. They replaced the non-essential 29.4K and 32.8K genes with a multiple cloning site (MCS), leaving only the 5′ and 3′ untranslated regions and the viral coat protein. The cDNA clones were positioned between the left and right border (LB and RB) of the T-DNA and between CaMV 35S promoters (35S) and transcriptional terminators (T). The TRV open reading frames corresponded to the RdRp, MP, 16K, CP, and 29.4K and 32.8K proteins. (C) TRV-based VIGS vector (Liu et al., 2002a,b; Tian et al., 2014). TRV1: TRV cDNA clones were placed between the duplicated CaMV 35S promoter (2 × 35S) and the nopaline synthase terminator (NOSt) in a T-DNA vector. Rz, self-cleaving ribozyme. TRV2: TRV cDNA clones were placed between the duplicated CaMV 35S promoter (2 × 35S) and the nopaline synthase terminator (NOSt) in a T-DNA vector, and PDS was added to the MCS between CP and Rz. (D) Modified pTRV2 vector based on GATEWAY cloning technology containing attR1 and attR2 recombination sites (Liu et al., 2002a). The PCR products flanked by attB1 and attB2 sequences directionally recombined in vitro at the attR1 and attR2 sites contained in the plasmid when incubated with the BP CLONASE enzyme. Then, the PDS gene was cloned into the pTRV2-attR1-attR2 vector. (E) TRV-Ligation-independent cloning (LIC) vector (Dong et al., 2007). The TRV-LIC vector was created by inserting a cassette, containing adapters and two PstI sites, in two digestion and ligation reactions. Then, the CDS insely digested enzyme for digestion and ligation. (F) TRV-GFP vector (Tian et al., 2014). GFP CDS was fused with CP in the TRV2 vector to generate an easily traceable TRV vector in different parts of plants.
Tobacco rattle virus is a positive-sense RNA virus composed of the RNA1 and RNA2 genome. RNA1 encodes two replicases and one cysteine-enriched protein, which is sufficient for replication and movement within the plant even without RNA2. One coat protein (CP) and two non-structural proteins encoded by RNA2 allow virion formation and nematode-mediated transmission across plants (Ratcliff et al., 1999; Liu et al., 2002b; Figure 2A). Ratcliff et al. (1999) first used in vitro transcripts of the cDNA clones of TRV (strain PPK20) RNA1 and modified RNA2 [TRV-Green fluorescent protein (GFP) RNA2]. GFP was transcribed from the coat protein promoter (P) of the pea early browning virus and replaced the 29.4K (29.4 kDa) and 32.8K (32.8 kDa) open reading frames (ORF) to express GFP successfully in tobacco. Then, they constructed the original TRV-VIGS vector; the complete cDNA sequences of RNA1 and RNA2 were positioned between the left and right border (LB and RB) of the T-DNA (pBINTRA6) and between cauliflower mosaic virus (CaMV) 35S promoters (35S) and transcriptional terminators (T), and a multiple cloning site (MCS) was introduced during the cloning of RNA2 (TRV00, Ratcliff et al., 2001; Figure 2B). They inserted partial cDNA sequences of the NbPDS gene into MCS of TRV00, and successfully silenced NbPDS with mild viral symptoms and a silencing efficiency of 47–73%. In the following year, Liu et al. (2002a, b) developed the most commonly used TRV vector (TRV2-MCS, pYL156; Figure 2C) by using the duplicated CaMV 35S promoter (2 × 35S), instead of a single 35S, and adding a self-cleaving ribozyme (Rz, to increase virus infectivity) before the nopaline synthase terminator (NOSt) on the transferred Agrobacterium tumefaciens T-DNA of plant binary transformation vectors, which had a high silencing efficiency (90–97.9%). They successfully studied the function of tobacco RAR1 and other genes against TMV resistance (Liu et al., 2002a,b; Tian et al., 2014).
As TRV2-MCS is a labor-intensive and time-consuming cloning-dependent method, the same team modified the pTRV2 clone into a GATAWAY recombination system (TRV2-GATEWAY, pYL279), which allowed fast and easy cloning that was free of the restriction enzyme and ligation and could be used for large-scale functional genomics analysis (Liu et al., 2002a; Figure 2D). The construction of the recombinant TRV vector requires two or more cloning steps (Ratcliff et al., 2001). Thus far, compared with other VIGS systems, the gene silencing efficiency of TRV-MCS and TRV-GATEWAY in N. benthamiana has been widely adopted, and with these vectors, scientists successfully silenced the PDS gene in tomato and other species with a silencing efficiency around 90% (Liu et al., 2002a; Tian et al., 2014). These two improved TRV-VIGS vectors have been the most commonly used vectors for gene-functional studies (Chen et al., 2004; Senthil-Kumar and Mysore, 2011b; Liu et al., 2015; Zhang et al., 2017).
A new TRV2-Ligation-independent cloning (LIC) vector (pYY13) in which the inserts can be cloned independently of the connection (LIC), instead of using the expensive GATEWAY-based recombination system, has been developed (Dong et al., 2007; Figure 2E). The TRV2 vector, pYL170, which is identical to pYL156 (except for a plant selection marker), was used to generate the new TRV2-LIC vector by inserting a ccdB cassette containing adapters and two PstI sites using two digestion and ligation reactions (Burch-Smith et al., 2006). The insert requires the addition of adapter sequences, and together with pYY13, after treated with T4 DNA polymerase, both can be transformed into Escherichia coli for construction of the recombinant vector (Dong et al., 2007; Figure 2E). Compared with the commonly used TRV-MCS, the TRV-LIC vector is more efficient (about 90%). Therefore, the TRV-LIC vector allows high-throughput cloning of silenced fragments without the use of expensive recombinases (Dong et al., 2007).
With the deepening of TRV-VIGS application, researchers attempted to determine whether virus-infected and -affected sites could be monitored in more species. Tian et al. (2014) added the full cDNA sequence of GFP to the 3′ terminus of coat protein in the original TRV2 vector (pYL156) to form a fusion protein of CP and GFP and successfully tested the modified vector (Figure 2F) in many plants, including N. benthamiana, Arabidopsis thaliana, Rosa rugosa Thunb, and Fragaria ananassa. The proportion of TRV-GFP-PDS positive silenced plants (95.8%) was very close to that of TRV-PDS-VIGS positive plants (TRV2-MCS, 97.9%) in N. benthamiana, and the same phenotype could be replicated in other plants, indicating that the insertion of GFP did not change the gene silencing ability of TRV vectors (Tian et al., 2014). Transmission of the modified TRV-GFP virus can be easily detected using a fluorescence microscope and a handheld UV lamp. This improved TRV vector is a simple but visualizable and efficient genetic tool for functional genomics, especially in non-Solanaceae plants (Tian et al., 2014).
Apart from these vector construction modifications, there were also other studies about TRV-VIGS, like silencing multiple genes of Arabidopsis by inserting tandem gene sequences into TRV-MCS vector (Burch-Smith et al., 2006), silencing tandem constructs of the TRV-GATEWAY vector containing CHS (chalcone synthase) as a reporter gene to exam the function of floral-associated genes in petunia (Chen et al., 2004), or successfully elucidating the role of 24 highly homologous N. benthamiana ubiquitin E2 gene family members in plant immunity through extension PCR of different conserved gene fragments together with the TRV-GATEWAY system (Zhou and Zeng, 2017). TRV1 has also been modified for inserting plant gene fragments and invoking gene silencing without TRV2 (Deng et al., 2013). Taken together, these studies have contributed to the versatility of TRV-VIGS in plants, and more efficient TRV-based VIGS systems are expected to be developed in the future.
Methodology Progress of TRV-VIGS
The delivery of the constructed recombinant viral vector carrying partial cDNA sequences of target genes into plants for gene silencing is always very important for the efficacy of VIGS; therefore, scientists have developed many ways to introduce the viral vectors into plants. Initially, Ruiz et al. (1998) delivered the infectious potato virus X (PVX) RNA transcripts obtained by in vitro transcription into plants by mechanically rubbing them onto the leaves of 4- to 5-week-old wild-type N. benthamiana in the presence of a small amount of carborundum and successfully obtained the NbPDS gene silenced phenotype. However, the in vitro transcription of the viral cDNA was difficult and tedious to operate, and the stability of silencing efficiency was sometimes low in VIGS experiments. Later, researchers attempted to transform the recombinant viral vector into A. tumefaciens, which is better for viral cDNA transcription and is also sufficient for the exploration of the interaction between plants and viruses, and they successfully silenced target genes by syringe infiltration (Ratcliff et al., 2001). To make full use of TRV-VIGS in more species, many agroinfiltration methods were developed (Figure 3), such as leaf injection (syringe infiltration), Agrobacterium spray (airbrush infiltration), agrodrench, fruit agroinjection, vacuum infiltration, and even secondary inoculation by N. benthamiana leaf sap after inoculation with Agrobacterium cultures (Ratcliff et al., 2001; Liu et al., 2002a; Ekengren et al., 2003; Ryu et al., 2004; Hileman et al., 2005; Chung et al., 2006; Orzaez et al., 2006; Yuan et al., 2011; Figure 3). In this part, the inoculation method improvement of TRV-VIGS is reviewed to provide advice and new dimensions for future methodology studies of VIGS.
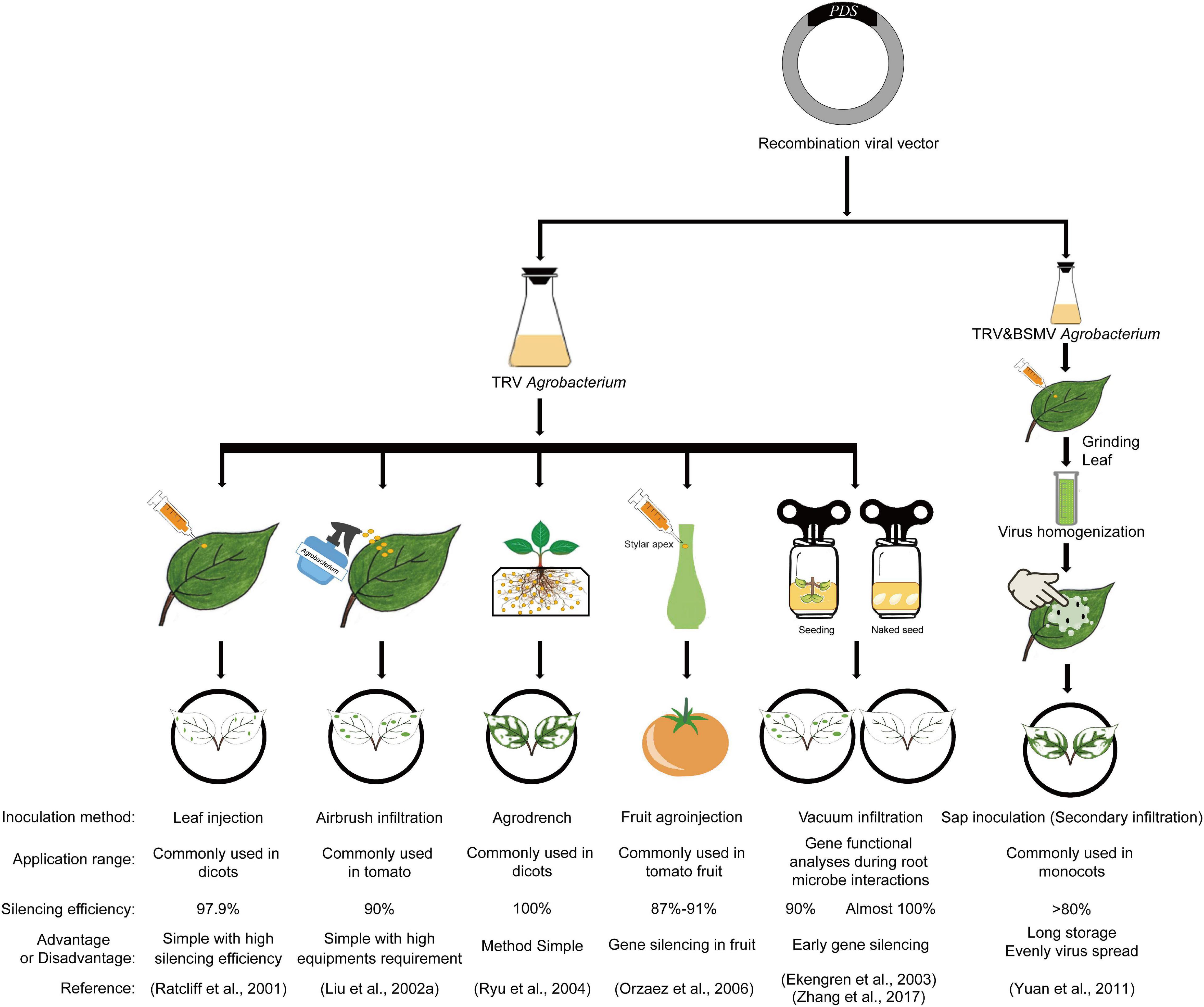
Figure 3. Schematic diagram of different VIGS inoculation methods. The phytoene desaturase, PDS gene was used as an example. Inoculation method, application range, silencing efficiency, advantages and disadvantages, and corresponding references are all included. TRV: Tobacco rattle virus; BSMV: Barley stripe mosaic virus.
Leaf Injection
Leaf injection (syringe infiltration) is the most common agroinfiltration method used for leaf or floral development-related gene studies in dicots with high silencing efficiency (Figure 3). Ratcliff et al. (2001) silenced the NbPDS gene in tobacco by infecting N. benthamiana leaves with a 2-ml needleless syringe carrying TRV Agrobacterium cultures, and then first proposed this method for VIGS. Liu et al. (2002b) successfully silenced the tomato PDS gene with a silencing efficiency of 50% by leaf injection (Liu et al., 2002b). Then, this method with little improvement and higher efficiency was widely used in Arabidopsis (Zhang et al., 2004), tomato (Ekengren et al., 2003), pepper (Choi et al., 2007), and cotton (Gao et al., 2011), among other dicotyledonous plants (Liu et al., 2015), for functional analysis of genes involved in plant (Lu et al., 2011) and floral development (Chen et al., 2005). However, it is laborious for large-scale screening, and monocotyledonous plants are difficult to infiltrate with this method; thus, new methods were developed for VIGS.
Spray Inoculation
Spray inoculation is mainly suitable for the VIGS of tomato genes with a success rate greater than 90% (Liu et al., 2002a; Dinesh-Kumar et al., 2003; Figure 3). In 2002, by transforming the recombinant TRV-VIGS plasmid into Agrobacterium and spraying the Agrobacterium cultures into tomato leaves with an art spray gun connected to a portable air compressor set at 75 psi, Liu et al. (2002a) successfully obtained the SlPDS silenced phenotype in tomato with a silencing efficiency of 90% (Liu et al., 2002a). Lu et al. (2011) successfully studied the function of the VTE1 (Tocopherol cyclase) gene involved in tomato vitamin E synthesis with this method (Lu et al., 2011). As the leaves of monocot plant are difficult to be injected with needleless syringe, spray inoculation and leaf friction methods are more adoptable for the VIGS studies of monocotyledonous genes (Dinesh-Kumar et al., 2003).
Agrodrench
Apart from leaf injection and spray inoculation, researchers also developed other methods to study early root development genes (Figure 3). By drenching the crown region (the soil adjacent to the roots) of young N. benthamiana seedlings with 3–5 ml of the TRV-containing Agrobacterium cultures (OD600 = 1.0), Ryu et al. (2004) successfully silenced the NbPDS gene in N. benthamiana and named the modified VIGS method agrodench (a simple, efficient, and fast inoculation method for Agrobacterium in VIGS experiments) (Ryu et al., 2004; Senthil-Kumar and Mysore, 2011b, 2014). This method is mainly used in diverse Solanaceous plants at the young seedlings stage and is more efficient than the leaf infiltration method for defining the roles of genes in roots (Ryu et al., 2004). The success rate is near 100% in N. benthamiana and about 60–70% in tomato, Nicotiana tabacum (tobacco), Petunia hybrida, Solanum tuberosum (potato), Capsicum annuum (pepper), and Solanaceae (eggplant) (Ryu et al., 2004). This indicates that the combination of different methods of agroinoculation at different stages may be a better choice for triggering heritable gene silencing in plants, which is very necessary and important for longer duration VIGS studies related to crop breeding or other fields.
Fruit Agroinjection
In addition to the previous infection methods carried out on leaves or roots, researchers also developed a method to study fruit genes. A transient methodology (fruit agroinjection) proposed by Orzaez et al. (2006) gently injected Agrobacterium cultures containing partial SlPDS cDNA into the fruit through the fruit stylar apex with a syringe, resulting in complete fruit infiltration with a silencing efficiency of 87–91% in tomato (Solanum lycopersicum) (Figure 3). This technology was not only good for gene silencing but also as a tool for fast transient expression in fruit (Orzaez et al., 2006), thus increasing the number of studies on fruit development and reproduction in different crops.
Vacuum Infiltration and Seed Imbibition
Vacuum infiltration is a timesaving agroinoculation method that can be carried out even at the plant seed stage, expanding the range of genes that can be studied by VIGS with a success rate of 90–100% (Ekengren et al., 2003; Hileman et al., 2005; Figure 3). Ekengren et al. (2003) poured tomato seedlings immersed in the Agrobacterium infection by turning it upside down into a vacuum device and successfully silenced the NPR1 and TGA genes. Then, Hileman et al. (2005) submerged the seedlings of different development stages into mixtures of Agrobacterium cultures and vacuum infiltrated poppy for 1 min, obtaining PapsPDS gene-silenced plants.
By vacuum infiltrating germinating wheat and corn seeds under a new Agrobacterium suspension containing a recombinant TRV vector and then co-cultivating with Agrobacterium cultures for a period of time, Zhang et al. (2017) successfully produced whole plant PDS gene-silenced wheat and corn. That was the first time that vacuum infiltration was implemented in a monocot at the seed imbibition stage. The next year, the same team developed a new agroinfiltration method (seed sock agroinoculation VIGS, SSA-VIGS) by soaking exposed cotton seeds in Agrobacterium culture during seed imbibition; they then performed functional analysis of GhBI-1 in response to salt stress. This was the first application of seed imbibition in dicotyledons (cotton) (Zhang et al., 2018). Seed imbibition avoids the extra use of expensive equipment, and the combination of vacuum infiltration and seed imbibition could be a timesaving method for VIGS in the future and might be applied for large-scale genomic studies in more plants.
Secondary Infiltration
Scientists have also developed a more complex inoculation procedure that is applicable for high-throughput applications of VIGS (Lu et al., 2003). By involving an intermediate step, researchers obtained a high-titer viral inoculum prepared from Agrobacterium infiltrated N. benthamiana leaves, and then mechanically inoculated it into plants for gene silencing. Sap inoculation (secondary inoculation) is usually conducted by gentle rubbing inoculated N. benthamiana leaf sap resuspended in 0.5 M phosphate buffer into carborundum-dusted Arabidopsis leaves. The delivery of leaf sap into Arabidopsis is usually implemented by leaf injection or sap spraying (airbrush infiltration) (Lu et al., 2003). According to the current researches, sap inoculation can also be used by seed imbibition for barley stripe mosaic virus (BSMV)-mediated VIGS and VOX (virus induced gene overexpression) in N. benthamiana and monocots (Yuan et al., 2011; Cheuk and Houde, 2017). These studies have improved researchers’ understanding of secondary inoculation in virus-related gene-functional studies and broadened the application range of genes by the sap inoculation method.
Important Factors of TRV-VIGS
As an experiment of TRV-VIGS carried out from viral vector selection, recombinant viral vector construction, Agrobacterium transformation (in vivo transcription of virus genome), the delivery of inoculum into plants, and gene silencing in plants, each step may strongly affect the silencing efficiency of the target gene. Therefore, this part discusses the influential factors of TRV-VIGS efficacy to provide reasonable suggestions for researchers.
Viral Vector Selection
Different viral vectors are suitable for VIGS research in diverse plants with different silencing efficiencies (for details, see Supplementary Table 1). TRV vectors have been widely used for VIGS across various species, both dicot and monocot, due to its advantages of mild viral symptoms and invasion into meristems (Gao et al., 2011; Senthil-Kumar and Mysore, 2011b; Yuan et al., 2011; Liu et al., 2015; Zhang et al., 2017). In Methods in Molecular Biology, edited by Courdavault and Besseau (2020), they report that most of the genes in plants, such as chili pepper, diploid potato, flax, and opium poppy, can be silenced with TRV-VIGS protocols, thus providing a better understanding of TRV-VIGS and more choices for gene function analysis in diverse plants. Therefore, TRV might be a really efficient gene silencing viral vector for VIGS, and more studies should be done on its mechanism, methodology, and application.
Insert Sequence
The optimization of cDNA libraries of the target gene is also very important to the silencing efficiency of TRV-VIGS. Experiments showed that the upper limit of the inserted sequence of viral vectors is about 1500 bp, and the lower limit was determined as 23 nucleotides (nt) identity (Thomas et al., 2001). Introns do not function in guiding VIGS in plants; thus, only exon sequences can be used for VIGS to successfully elicit gene silencing of plant endogenous genes (Ruiz et al., 1998). Liu and Page (2008) gave the following guidelines for constructs of TRV vectors: insert lengths should be in the range of 200–1500 bp and should be positioned in the middle of the cDNA without the homopolymeric regions.
However, the similarity of genes within the gene family also makes simultaneous silencing of multiple genes possible by targeting the conserved gene sequence (Zhou and Zeng, 2017). Multiple genes that are unrelated by nucleotide sequence can be silenced at the same time using VIGS by co-inoculating the respective VIGS constructs or inoculating the recombinant vector with tandem gene inserts (Burch-Smith et al., 2006). Therefore, to perform silencing of a single and specific gene, the insert must be selected according to the specific domain of the gene, or VIGS can be conducted by using the conserved sequence within a gene family to study the function of the whole gene family or in tandem gene inserts.
Agrobacterium Strain
The optimal A. tumefaciens strain used for VIGS varies with different plants and also affects the gene silencing efficiency. Studies have shown that Agrobacterium strain GV2260 works best in N. benthamiana, while strain GV3101 could also be used (Liu et al., 2002a). GV3101 works best for the silencing of TRV-VIGS in tomato, whereas LBA4404 and GV2260 could also be used, but the silencing efficiency is very low (Liu et al., 2002a; Dinesh-Kumar et al., 2003). Both Agrobacterium strain GV3101 and AGL-1 with TRV-MePDS-infiltrated distal leaves showed an albino phenotype at 20 days post-inoculation (dpi) (Zeng et al., 2019). A suitable temperature for the growth of Agrobacterium should be at or below 28°C for no longer than 2 days.
Inoculum Concentration
As the agroinoculation methods and characteristics of diverse infected plants are different, the concentration of the infection solution strongly affects the gene silencing efficiency of VIGS experiments. The solution used for the resuspension of concentrated Agrobacterium should be fresh for each use, and lower concentrations of the inoculum also work, but those higher than 1.0 OD600 may cause necrosis on the infiltrated N. benthamiana leaves (Dinesh-Kumar et al., 2003). Liu et al. (2002b) found the concentration (OD600 = 1.0) of Agrobacterium cultures that gave the best SlPDS gene silencing results in tomato, while Dinesh-Kumar et al. (2003) found that OD600 = 1.5 worked better for tomato. Therefore, for N. benthamiana, the OD600 should be adjusted to 1.0 and no more than 1.5 for tomato. Burch-Smith et al. (2006) proposed that the most suitable concentration of Agrobacterium infection solution is OD600 = 1.5 for TRV-VIGS experiment in Arabidopsis with an efficiency of almost 100%. Additionally, the naked cotton seeds soaked in Agrobacterium cultures with an OD600 of 1.5 for 90 min exhibited an optimal silencing efficiency of SSA-VIGS (Zhang et al., 2018). For vacuum infiltration, Zhang et al. (2017) found that the optimal condition for VIGS was vacuum treatment for 30–60 s with an Agrobacterium culture of OD600 = 0.3 and co-cultivation with the same concentration of Agrobacterium culture for 15 h. These experiments showed that different species require different concentrations of Agrobacterium inoculum; thus, the Agrobacterium inoculum concentration of each specific TRV-VIGS experiment should be optimized.
Environmental Factors
Studies have shown that environmental factors directly affect plant growth status and also virus accumulation and spread in plants, which are closely related to gene silencing efficiency and gene silencing duration (inheritance) of TRV-VIGS (Senthil-Kumar and Mysore, 2011b). Therefore, it is necessary to strictly control plant care when conducting VIGS experiments (Burch-Smith et al., 2004).
Age or Development Stages of Infected Plants
One study showed that Arabidopsis seedlings inoculated at the two- to three-leaf stage and grown under 16-h light displayed the photo-bleaching phenotype indicative of AtPDS silencing in almost 100% of the cases examined, while older seedlings inoculated at the same conditions exhibited reduced AtPDS transcript levels (95%) in the silenced plants (Burch-Smith et al., 2006). Studies also indicated that the four-leaf stage in N. benthamiana and two-leaf stage in tomato are the optimal ages for syringe infiltration of VIGS experiments. Another TRV-VIGS study in Streptocarpus rexii (Gesneriaceae) showed that the efficacy of TRV-VIGS showed a correlation with the age of S. rexii seedlings (Burch-Smith et al., 2006). The younger the plants were when infected with TRV, the more efficient the silencing of the SrPDS gene (Nishii et al., 2020).
Ambient Temperature
Previous reports showed that temperature is a key player in influencing the gene silencing phenotype development in plants with VIGS (Szittya et al., 2003; Fu et al., 2006; Tuttle et al., 2008). Generally, the gene silencing efficiency of TRV-VIGS was significantly reduced under high temperatures (temperatures higher than 27°C dramatically reduced virus titer levels in plants), while virus concentration and gene silencing efficiency were largely increased at low temperature (16–21°C for tomato) (Szittya et al., 2003; Caplan and Dinesh-Kumar, 2006). The most important reason for the success of the first long gene silencing report (duration of more than 2 years, and the acquisition of gene silencing T1 and T2 generation offspring) of TRV-VIGS is good plant care under low temperatures (18–20°C) (Senthil-Kumar and Mysore, 2011b). Therefore, efficient TRV-VIGS experiments should be conducted at relatively lower ambient temperatures (below 21°C for tomato) and then shifted to the optimum temperature for plant growth (24°C for tomato) after 1 or 2 days (Caplan and Dinesh-Kumar, 2006).
Humidity
An appropriate humidity is also beneficial for the silencing efficiency of TRV-VIGS. Fu et al. (2006) found that when cultured at a lower humidity (30–40%), more than 90% of tomatoes exhibited a SlPDS silenced phenotype. At the same time, they also found that TRV-RNA (RNA1 and RNA2) was more efficiently introduced into flowers and fruits at a lower humidity.
Therefore, good plant care (appropriate ambient temperature and humidity, adequate water and fertilizer, and free of pests and diseases) to maintain efficient silencing and healthy growth of gene-silenced plants is important for VIGS experiments and should be optimized in every single VIGS system.
Positive Controls
Tobacco rattle virus-VIGS can effectively downregulate the expression of PDS, H subunit of magnesium chelatase (ChlH), anthocyanidin synthase (ANS), and other genes in Solanaceae plants with distinct gene silencing phenotypes (Senthil-Kumar et al., 2008); thus, they are often utilized as positive controls to optimize and determine the efficacy of a VIGS system. For instance, Liu et al. (2002a) calculated the silencing efficiency of TRV-VIGS in tomato plants with SlPDS as a positive control (silencing plant with an albino phenotype) by leaf injection (50%) and Agrobacterium spray (90%). The transparent testa 2 (TT2) gene might be a positive control for genes that are expressed in seeds (seed coat). Seed bolls (containing seeds) used for TRV-mediated VIGS of TT2 gene showed a result of testa depigmentation with a silencing efficiency around 35% in flax seeds (Linum usitatissimum L.) (Hano et al., 2020). Only with an optimized positive control for each specific silenced region (organ) can a VIGS system provide the best silencing efficiency for application studies in that species. Thus more positive controls should be found and determined for some specific regions in VIGS system, such as root or other organs.
Inoculation Method
To realize the gene functions across different plant development stages, researchers have developed many VIGS inoculation methods (Figure 3). Leaf injection is the most commonly used inoculation method to silence genes related to leaf development, floral development, fruit development, metabolic processes, and plant-pathogen interactions (R genes) in different plants (Ratcliff et al., 2001; Liu et al., 2002a,b, 2004; Chen et al., 2004; Ryu et al., 2004). When studying genes related to seedling development, floral development, or other fields of monocot plants, Agrobacterium spray, agrodench, and sap inoculation can be used for VIGS of target genes (Scofield et al., 2005; Cloutier et al., 2007; Zhang et al., 2017). For tomato genes, using spray infiltration, the silencing success rate was about 90% (Dinesh-Kumar et al., 2003). For gene-functional analysis of root development, seed dormancy, and seed germination, the preferred methods are agrodench, vacuum infiltration, and seed imbibition (Zhang et al., 2017; unpublished data).
Tobacco rattle virus-VIGS studies showed that agrodench coupled with leaf inoculation provoked an effective gene silencing phenotype in tomato and tobacco plants that lasted for more than 2 years, and gene silencing was maintained in T1 and T2 generation offspring (Senthil-Kumar and Mysore, 2011b). Combined sap inoculation and seed imbibition, Cheuk and Houde (2017) also obtained better and more even gene expression levels of the BSMV homogenate than other VOX experiments conducted by leaf injection or leaf friction of leaf sap. Therefore, an integrated application of different inoculation methods might trigger better gene silencing results using VIGS.
These studies provide an important reference for the improvement and application of TRV-VIGS. By the comprehensive comparison and combination of different agroinoculation methods (multiple methods conducted together) under appropriate plant care, researchers can create a more suitable VIGS method for functional genomic analysis (Figure 4).
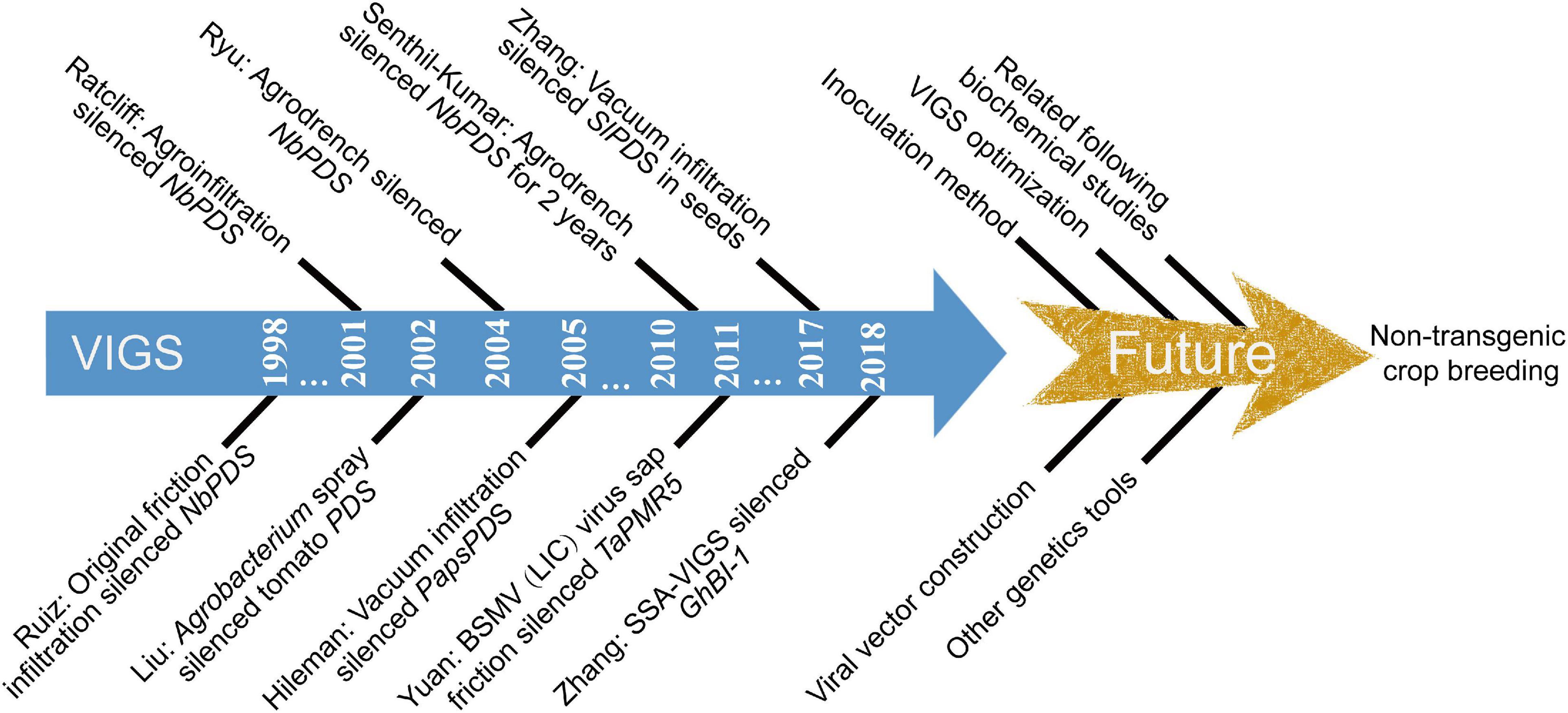
Figure 4. Important historical milestones of VIGS system. TRV: Tobacco rattle virus; BSMV: Barley stripe mosaic virus; LIC: Ligation-independent cloning; SSA-VIGS: Seed sock agroinoculation VIGS; NbPDS: Tobacco phytoene desaturase; TaPMR5: Wheat powdery mildew resistance 5 gene; SlPDS: Tomato phytoene desaturase; GhBI-1: Cotton Bax inhibitor-1.
Applications of TRV-VIGS Technology
With the improvement of VIGS tools, TRV-VIGS has been utilized for the functional genomic analysis of more and more plant species, including most dicotyledonous species, some monocotyledonous plants, and even some trees. The gene function of many genes in different plant tissues and organs, from seeds, roots, stems, leaves, flowers, and fruits, have been revealed by TRV-VIGS in the past 20 years that relate to plant growth, development and reproduction, metabolic pathways, and response to biotic and abiotic stress, among others (Burch-Smith et al., 2004; Purkayastha and Dasgupta, 2009; Becker and Lange, 2010). A complete review of the literature on all aspects of VIGS application is out of the scope of this work; here, this article attempts to provide an update on the most recent applications of VIGS in plants.
Dicotyledon
Solanaceae
Since TRV-VIGS was first derived from tobacco, it is widely used for functional gene studies in other solanaceous plants (Purkayastha and Dasgupta, 2009; Senthil-Kumar and Mysore, 2011a, 2014). VIGS has been used to study the functional roles played by several kinases, such as (NPK1, WIPK, and SIPK, in defense against TMV-mediated by N-gene resistance in N. benthamiana (Jin et al., 2002, 2003; Sharma et al., 2003; Liu et al., 2004). The TRV-VIGS study of RanGAP2 showed that the interaction with Rx (an NB-LRR protein from potato) is required for conferring PVX resistance in N. benthamiana (Tameling and Baulcombe, 2007). VIGS silencing of two genes, an extracellular peroxidase and a cytochrome P450, revealed their defense-related roles in pepper (Kim et al., 2006; Choi et al., 2007). Another VIGS study demonstrated the role of a gene encoding arabinogalactan protein, attAGP in tomato during the attack by plant parasite Cuscuta reflexa (Albert et al., 2006). The TRV-mediated VIGS system has been applied for gene-functional analysis in eggplant (Liu et al., 2012). VIGS silencing of the VTE1 gene in tomato leaves with an Agrobacterium infection solution carrying the TRV vector demonstrated that VTE1 is involved in vitamin E synthesis in tomatoes (Lu et al., 2011). Organ aging issues in petunia flowers and functional analysis of peony genes have also been demonstrated with TRV-VIGS technology (Chen et al., 2005; Xie et al., 2019).
Virus-induced gene silencing has also been used for the functional analysis of root genes. Silencing of genes in roots is more effective by the agrodrench method than by syringe infiltration (Ryu et al., 2004). The functional roles of IRT1 (iron-regulated metal transporter), TTG1 (transparent testa glabra), RHL1 (root hairless1), β-tubulin, RML1 (root meristemless1), and nematode resistance (Mi) genes, which are involved in root development, have been demonstrated by this vector (Jablonska et al., 2007).
Recently, VIGS has unraveled the functional redundancy among group III members in their requirements for plant development and plant immunity-associated ROS production (Zhou and Zeng, 2017). Functional analysis of antiviral resistance signaling pathways has also been demonstrated by TRV-VIGS in N. benthamiana (Hashimoto et al., 2019). The TRV-mediated VIGS system was established in 4-week-old in vitro diploid potato plants by syringe infiltration (Zhao et al., 2020). Additionally, TRV- and PVX-based microRNA silencing (VbMS) approaches to silence endogenous miRNAs in N. benthamiana and tomato plants by Agrobacterium infiltration have also been developed, thus facilitating functional studies of miRNAs in plants (Zhao et al., 2020).
Cruciferae
With the development of VIGS technology, many gene-functional studies have been done in the model plant Arabidopsis of the Cruciferae family (Senthil-Kumar and Mysore, 2011a). Cai et al. (2006) reported some factors that potentially influence TRV-mediated VIGS of PDS and actin gene expression in Arabidopsis. In addition, the optimized procedure (by PDS gene silencing) for the TRV-based VIGS is a potentially powerful tool for deciphering the signal transduction pathways of disease resistance in Arabidopsis (Burch-Smith et al., 2006; Cai et al., 2006). VIGS experiments carrying the R gene rps4 were conducted in Arabidopsis and demonstrated that the cell death, upon infection with Pseudomonas syringae, is dependent on three plant signaling components, EDS1, SGT1, and HSP90 (Zhang et al., 2004).
Malvaceae
The most important commercial crop of the Malvaceae family, cotton has also adopted the TRV-VIGS system for functional genomics studies. VIGS silencing of GhNDR1 and GhMKK2 by cotyledon injection compromised cotton resistance to Verticillium wilt (Gao et al., 2011). The successfully functional characterization of KATANIN and WRINKLED1 in cotton provided evidence that the TRV-VIGS system can be used for rapid functional analysis of genes involved in cotton fiber development (Qu et al., 2012). SSA-VIGS is a powerful tool for elucidating the functions of genes (GhBI-1 gene, in response to salt stress) in cotton, especially for genes expressed in young seedlings and roots (Zhang et al., 2018). Knockdown of Gh_A05G1554 (GhDHN_03) and Gh_D05G1729 (GhDHN_04) dehydrin genes has revealed their potential role in enhancing osmotic and salt tolerance in cotton (Kirungu et al., 2020). Therefore, combining cotyledon injection together with the SSA-VIGS system (Zhang et al., 2018), almost all cotton genes can be characterized via VIGS assays in the future.
Leguminosae
Tobacco rattle virus-VIGS has also been used to study the functional roles of disease resistance genes in soybean. Through the use of VIGS assays in soybean, Liu et al. (2015) studied the relationship between TRV-VIGS technology and soybean mosaic virus (SMV) and demonstrated that resistance to SMV is not affected by early TRV inoculation in soybean.
Fruit Trees
The TRV-VIGS system has also been widely used for functional gene studies in fruit trees, including peach, cherry, strawberry, and litchi (Jia et al., 2010; Li et al., 2015, 2016; Xie et al., 2020). VIGS of the PpChlH gene proved that this gene plays a role in the synthesis of chlorophyll in peach leaves and fruits (Jia et al., 2010). VIGS silencing of the PacCYP707A1 (8′-hydroxylase) gene resulted in the silenced-cherries responding to dehydration during fruit development when adjusting ABA in cherries (Li et al., 2015). Similarly, UFGT1 (UDP-glucose: flavonoid 3-O-glycosyltransferase) plays a key role in the formation of litchi red peel, as documented by VIGS experiments (Li et al., 2016).
Ornamental Plants
A study demonstrated that the TRV-based VIGS technique could be adapted for high-throughput functional characterization of genes in the perennial tree peony (Xie et al., 2019). Nishii et al. (2020) utilized the broad host range TRV vector to target the SrPDS gene of S. rexii (Gesneriaceae) by agroinfiltration and sap inoculation and successfully obtained SrPDS silenced plants.
Other Commercial and Medicinal Plants
Moreover, several non-model plants, such as important commercial and medicinal plants, including cassava, flax, Papaver, Antirrhinum, mint, sweet basil, Ashwagandha, and olive tree, were also successfully explored with the TRV-VIGS system. Compared with the obvious cassava mosaic disease symptoms infiltrated by African cassava mosaic virus (ACMV)-based VIGS systems in previous studies, the TRV-mediated VIGS system in cassava plants showed mild disease symptoms, thus suggesting that the application of the TRV-VIGS system could promote functional genomics in cassava (Manihot esculenta Crantz) with a significant advantage (Zeng et al., 2019). A detailed protocol has been presented to perform TRV-VIGS assay by agroinoculation in cassava (Zaidi et al., 2020). Two detailed Agrobacterium-mediated infection protocols have also been described in flax (L. usitatissimum L.), based on whole plant vacuum and leaf syringe infiltration methods. The systemic impact on the gene transcript levels in the stem demonstrated that the VIGS system can be applied for the functional study of cell wall genes in flax (Chantreau and Neutelings, 2020).
Chen et al. (2020) investigated genes involved in alkaloid biosynthesis metabolism in opium poppy using syringe infiltration. Cruz et al. (2020) described the main steps of this biolistic (delivery of the transforming plasmids through a particle bombardment)-mediated TRV-VIGS in Catharanthus roseus and Rauvolfia tetraphylla of Apocynaceae. A recent report showed that TRV-VIGS can be used as a rapid gene function test for the complementation of a loss-of-function mutation in Antirrhinum majus L. (Tan et al., 2020). VIGS technology has also been established and adapted to target genes involved in the production of nepetalactone in Nepeta cataria (catnip) and Nepeta mussinii (catmint) of the Lamiaceae family (mint) (Palmer and O’Connor, 2020). A robust protocol for VIGS of sweet basil (Ocimum basilicum) ObChlH has recently been developed by vacuum infiltration with pTRV constructs (Misra et al., 2020). As a highly recalcitrant plant for genetic transformation, Ashwagandha (Withania somnifera), an important Indian medicinal plant, has an established procedure to carry out VIGS for gene function studies (Bomzan et al., 2020). TRV-mediated VIGS through agroinoculation of olive plantlets has been successfully performed for functional genomic analyses in the olive tree (Oleaceae) (Koudounas et al., 2020).
Monocotyledon
Not only can the TRV-VIGS system be used in most dicotyledonous species, it has also been adopted for gene-functional analysis of some monocotyledonous plants that are susceptible to TRV, such as wheat, corn, barley, rice, and Brachypodium sylvaticum in Gramineae and ornamental plants, such as orchids, a flowering plant of Orchidaceae.
Gramineae
Virus-induced gene silencing has been successfully applied for functional characterization of genes involved in leaf rust resistance of wheat (Scofield et al., 2005; Cloutier et al., 2007). Similarly, vacuum and co-cultivation agroinfiltration of (germinated) seeds resulted in TRV-mediated whole-plant TaPDS and ZmPDS gene silencing in wheat and maize (Zhang et al., 2017). These results proved that this system is suitable for functional analysis of genes involved in seed germination and early plant development stages (Zhang et al., 2017). Additionally, with the VIGS system, Li et al. (2019) demonstrated that the MAC3 (MOS4-associated complex) protein is a key regulatory factor necessary for corn endogenous immunity and disease resistance.
Orchidaceae
Virus-induced gene silencing has also been adapted for functional validation of genes involved in floral growth and development of orchids (Hsieh et al., 2013). Chen et al. (2020) successfully silenced the PoFYF1/2 gene in Phalaenopsis orchids and revealed that PoFYF1/2 plays a role in suppressing senescence/abscission during early flower development.
Limitations
Virus-induced gene silencing is a promising genetic tool for functional genomic studies in plants. However, like any other technique, there are still some limitations of TRV-VIGS system. First, the timing of VIGS appearance, as well as gene silencing duration, is usually species-specific. The first long silencing duration report of the TRV-VIGS system in tobacco and tomato (Senthil-Kumar and Mysore, 2011b), together with other long silencing duration reports of BSMV-VIGS in wheat (Bruun-Rasmussen et al., 2007) and Apple latent spherical virus (ALSV)-VIGS in soybean (Igarashi et al., 2009; Yamagishi and Yoshikawa, 2009), will surely provide significant promise for the application of VIGS system in studies related to perennial crop breeding (Senthil-Kumar and Mysore, 2011a). So researchers should continue to prolong the gene silencing period of VIGS in the future.
Another challenge of this technique is its varying penetrance of the phenotype in vegetative and reproductive tissue that requires a larger number of plants to be screened for phenotypes. So far, studies have shown that silencing efficacy is often found regionally, dividing the whole plant or restricted to plant gene silenced regions without a few consecutive nodes (Wege et al., 2007; Becker and Lange, 2010). Thus, proper positive control selection and a combination of different inoculation methods are necessary for an efficient VIGS system in diverse tissues at different developmental stages of crops. However, most known positive controls are always visual phenotype of photo-bleaching on leaves, without any marker genes in other silenced regions. Hence, more positive control genes in different tissues should be screened in future VIGS system.
Additionally, the stability of the environmental control and operation techniques is also worth noting in future VIGS studies, because the environment can cause changes in the silencing effect. For example, compared with the BSMV-VIGS assay in wheat and TRV-VIGS in N. benthamiana, reports showed that the Chinese wheat mosaic virus (CWMV) vector is more effective in silencing endogenous genes and miRNAs at 17°C, thereby providing a powerful tool for gene function analysis in both N. benthamiana and wheat to fulfill the functions of the VIGS system at low temperatures (Yang et al., 2018). Since VIGS assay involves viruses that can be easily transmitted to other plants in a field environment, VIGS vector carrying inocula, seeds and other plant materials should be disposed under suitable biosafety regulation (Senthil-Kumar and Mysore, 2011a). What’s more, although TRV has a wide host range, it still could not infect all plants to some extent. Therefore, it is one of the important content for future researches to develop new VIGS vectors and also to modify the existing vectors to increase its host range and silencing efficiency (Senthil-Kumar et al., 2008). Note that many of these limitations are inherent to all VIGS vectors and are not specific to TRV.
Conclusion and Future Prospects
In the past two decades, many VIGS vectors have been developed (Supplementary Table 1), and TRV was preferentially used for VIGS assays in most dicots and some monocots, due to the high susceptibility of a wide range of hosts with mild viral symptoms after infection (Chen et al., 2005; Zhang et al., 2017; Xie et al., 2019). The increase in high-quality genome or transcriptome (EST) data has provided an excellent foundation and sufficient homologous sequence information for VIGS assays to target a specific gene, gene family, gene class, or miRNA mimic (Senthil-Kumar and Mysore, 2011a; Figure 1). As viral vector construction of TRV is an important factor of an efficient VIGS system, many modifications have been done based on the original TRV vector to obtain better silencing efficiency in different species (Figure 2). Multiple inoculation methods for introducing the viral constructs into plants have also been developed to extend the application range of genes that can be studied by this system (Figure 3). With the development of VIGS studies, scientists have found that the silencing efficiency of the VIGS system is strongly affected by the selection of Agrobacterium strain, inoculum concentration, environmental factors (plant care), and positive control; thus, many established VIGS protocols have been optimized. As VIGS protocols can be transposed to many other plants following minor adaptation, recently established TRV-VIGS protocols in many species will be a great help in deepening researchers’ understanding of gene functions in a variety of plant species (Courdavault and Besseau, 2020).
In total, all improvements of VIGS technology will speed up the application of this tool for identifying candidate genes involved in various aspects of plant biology (Figure 4), including plant-environment interactions, plant growth and development, metabolic processes, and other cellular processes in planta (Senthil-Kumar et al., 2008). A deeper understanding of the VIGS mechanism has provided researchers with the option of combining VIGS with other functional genomic approaches (conventional or molecular breeding and next-generation technology) for crop breeding studies (Cheng et al., 2010; Senthil-Kumar and Mysore, 2011a). VIGS can also be performed in a stable plant that overexpresses, downregulates (RNAi), or knocks out (mutant) an unrelated gene compared with the gene targeted by VIGS to characterize the function of a genetic pathway (Baena-Gonzalez et al., 2007).
In conclusion, a good VIGS system should be established with proper viral vector construction, inoculation method, optimized inoculum type and concentration, proper positive controls with good plant care and plant vigor, thus eliciting high gene silencing efficiency with a uniform phenotype and simple operation procedure with lower equipment requirements and largely extending the scope of genes that can be studied (Liu et al., 2002a; Benedito et al., 2004; Ryu et al., 2004; Yuan et al., 2011; Zhang et al., 2017; Cheuk and Houde, 2018). Finally, a proper VIGS assay together with other genetic tools and related biochemical studies (Lu et al., 2003) will surely provide a promising future for non-transgenic crop breeding with high production in diverse plants.
Author Contributions
GS, ZX, and MH researched data for the article, substantially contributed to discussion of the content, and wrote the manuscript. ZX, FW, BT, and GC substantially contributed to discussion of the content and reviewed the manuscript before submission. All authors have read and approved the manuscript to be published.
Funding
This work was supported by Joint Funds of the National Natural Science Foundation of China (U1904106), Program for Science & Technology Innovation Talents in Universities of Henan Province (19HASTIT014), and Henan Provincial Science and Technology Research Project (202102110009), and Youth Innovation Project of Key Discipline of Zhengzhou University (XKZDQN202002), and the State Key Laboratory of Cotton Biology Open Fund (CB2021A11).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.doi.org/10.6084/m9.figshare.14626272
References
Albert, M., Belastegui-Macadam, X., and Kaldenhoff, R. (2006). An attack of the plant parasite Cuscuta reflexa induces the expression of attAGP, an attachment protein of the host tomato. Plant J. 48, 54–56. doi: 10.1111/j.1365-313X.2006.02897.x
Baena-Gonzalez, E., Filip, R., and Johan, M. (2007). A central integrator of transcription networks in plant stress and energy signaling. Nature 448, 938–942. doi: 10.1038/nature06069
Becker, A., and Lange, M. (2010). VIGS-genomics goes functional. Trends Plant Sci. 15:1. doi: 10.1016/j.tplants.2009.09.002
Benedito, V. A., Visser, P. B., Angenent, G. C., and Krens, F. A. (2004). The potential of virus-induced gene silencing for speeding up functional characterization of plant genes. Genet. Mol. Res. 3, 323–341.
Bomzan, D. P., Shilpashree, H. B., Anjali, P., Kumar, S. R., and Nagegowda, D. A. (2020). Virus-induced gene silencing for functional genomics in Withania somnifera, an important indian medicinal plant. Methods Mol. Biol. 2172, 139–154. doi: 10.1007/978-1-0716-0751-0_11
Bruun-Rasmussen, M., Madsen, C. T., Jessing, S., and Albrechtsen, M. (2007). Stability of Barley stripe mosaic virus-Induced gene silencing in barley. Mol. Plant Microbe Interact. 20, 1323–1331. doi: 10.1094/MPMI-20-11-1323
Burch-Smith, T. M., Anderson, J. C., Martin, G. B., and Dinesh-Kumar, S. P. (2004). Applications and advantages of virus-induced gene silencing for gene function studies in plants. Plant J. 39, 734–746. doi: 10.1111/j.1365-313X.2004.02158
Burch-Smith, T. M., Schiff, M., Liu, Y. L., and Dinesh-Kumar, S. P. (2006). Efficient virus-induced gene silencing in Arabidopsis. Plant Physiol. 142, 21–27. doi: 10.1104/pp.106.084624
Cai, X. Z., Xu, Q. F., Wang, C. C., and Zheng, Z. (2006). Development of a virus-induced gene silencing system for functional analysis of the RPS2-dependent resistance signaling pathways in Arabidopsis. Plant Mol. Biol. 62, 223–232. doi: 10.1007/s11103-006-9016-z
Caplan, J., and Dinesh-Kumar, S. P. (2006). Using viral vectors to silence endogenous genes. Curr. Protoc. Microbiol. 16, 16I.6.1–16I.6.13. doi: 10.1002/9780471729259.mc16i06s01
Chantreau, M., and Neutelings, G. (2020). Virus-induced gene silencing of cell wall genes in flax (Linum usitatissimum). Methods Mol. Biol. 2172, 65–74. doi: 10.1007/978-1-0716-0751-0_6
Chen, J. C., Jiang, C. Z., Gookin, T. E., Hunter, D. A., Clark, D. G., and Reid, M. S. (2004). Chalcone synthase as a reporter in virus-induced gene silencing studies of flower senescence. Plant Mol. Biol. 55, 521–530. doi: 10.1007/s11103-004-0590-7
Chen, J. C., Jiang, C. Z., and Reid, M. S. (2005). Silencing a prohibitin alters plant development and senescence. Plant J. 44, 16–24. doi: 10.1111/j.1365-313X.2005.02505
Chen, R. J., Chen, X., Hagel, J. M., and Facchini, P. J. (2020). Virus-induced gene silencing to investigate alkaloid biosynthesis in Opium Poppy. Methods Mol. Biol. 2172, 75–92. doi: 10.1007/978-1-0716-0751-0_7
Cheng, S. F., Huang, Y. P., Wu, Z. R., Hu, C. C., Hsu, Y. H., and Tsai, C. H. (2010). Identification of differentially expressed genes induced by Bamboo mosaic virus infection in Nicotiana benthamiana by cDNA-amplified fragment length polymorphism. BMC Plant Biol. 10:286. doi: 10.1186/1471-2229-10-286
Cheuk, A., and Houde, M. (2017). A rapid and efficient method for uniform gene expression using the Barley stripe mosaic virus. Plant Methods 13:24. doi: 10.1186/s13007-017-0175-5
Cheuk, A., and Houde, M. (2018). A new barley stripe mosaic virus allows large protein overexpression for rapid function analysis. Plant Physiol. 176, 1919–1931. doi: 10.1104/pp.17.01412
Choi, H. W., Kim, Y. J., Lee, S. C., Hong, J. K., and Hwang, B. K. (2007). Hydrogen peroxide generation by the pepper extracellular peroxidase CaPO2 activates local and systemic cell death and defense response to bacterial pathogens. Plant Physiol. 145, 890–904. doi: 10.1104/pp.107.103325
Chung, S. M., Vaidya, M., and Tzfira, T. (2006). Agrobacterium is not alone: gene transfer to plants by viruses and other bacteria. Trends Plant Sci. 11, 1–4. doi: 10.1016/j.tplants.2005.11.001
Cloutier, S., McCallum, B. D., Loutre, C., Banks, T. W., Wicker, T., Feuillet, C., et al. (2007). Leaf rust resistance gene Lr1, isolated from bread wheat (Triticum aestivum L.) is a member of the large psr567 gene family. Plant Mol. Biol. 65, 93–106. doi: 10.1007/s11103-007-9201-8
Courdavault, V., and Besseau, S. (2020). [Methods in Molecular Biology] Virus-induced Gene Silencing in Plants. (Methods and Protocols). New York, NY: Humana Press, doi: 10.1007/978-1-0716-0751-0
Cruz, P. L., Restrepo, M. I., Bernonville, T. D. D., Oudin, A., Munsch, T., Lanoue, A., et al. (2020). A biolistic-mediated virus-induced gene silencing in apocynaceae to map biosynthetic pathways of Alkaloids. Methods Mol. Biol. 2172, 93–110. doi: 10.1007/978-1-0716-0751-0_8
Deng, X. B., Kelloniemi, J., Haikonen, T., Vuorinen, A. L., Elomaa, P., Teeri, T. H., et al. (2013). Modification of tobacco rattle virus RNA1 to serve as a VIGS vector reveals that the 29K movement protein is an RNA silencing suppressor of the virus. Mol. Plant Microbe Interact. 26, 503–514. doi: 10.1094/MPMI-12-12-0280-R
Dinesh-Kumar, S. P., Anandalakshmi, R., Marathe, R., Schiff, M., and Liu, Y. L. (2003). Virus-induced gene silencing. Methods Mol. Biol. 236, 287–293.
Ding, S. W., and Voinnet, O. (2007). Antiviral immunity directed by small RNAs. Cell 130, 413–426. doi: 10.1016/j.cell.2007.07.039
Dong, Y. Y., Burch-Smith, T. M., Liu, Y. L., Mamillapalli, P., and Dinesh-Kumar, S. P. (2007). A ligation-independent cloning tobacco rattle virus vector for high-throughput virus-induced gene silencing identifies roles for NbMADS4-1 and -2 in floral development. Plant Physiol. 145, 1161–1170. doi: 10.1104/pp.107.107391
Ekengren, S. K., Liu, Y. L., Schiff, M., Dinesh-Kumar, S. P., and Martin, G. B. (2003). Two MAPK cascades, NPR1, and TGA transcription factors play a role in Pto-mediated disease resistance in tomato. Plant J. 36, 905–917. doi: 10.1046/j.1365-313x.2003.01944
Fu, D. Q., Zhu, B. Z., Zhu, H. L., Zhang, H. X., Xie, Y. H., Jiang, W. B., et al. (2006). Enhancement of virus-induced gene silencing in tomato by low temperature and low humidity. Mol. Cells 21, 153–160.
Gao, X. Q., Britt, R. C. Jr., Shan, L. B., and He, P. (2011). Agrobacterium-mediated virus-induced gene silencing assay in cotton. J. Vis. Exp. 54:e2938. doi: 10.3791/2938
Hano, C., Drouet, S., and Laine, E. (2020). Virus-induced gene silencing (vigs) in flax (Linum usitatissimum L.) seed coat: description of an effective procedure using the transparent testa 2 gene as a selectable marker. Methods Mol. Biol. 2172, 233–242. doi: 10.1007/978-1-0716-0751-0_17
Harper, G., Hull, R., Lockhart, B., and Olszewski, N. (2002). Viral sequences integrated into plant genomes. Annu. Rev. Phytopathol. 40, 119–136. doi: 10.1146/annurev.phyto.40.120301.105642
Hashimoto, M., Yamaji, Y., and Komatsu, K. (2019). Analysis of antiviral resistance signaling pathways by virus-induced gene silencing in Nicotiana benthamiana. Methods Mol. Biol. 2028, 85–95. doi: 10.1007/978-1-4939-9635-3_4
Hileman, L. C., Drea, S., Martino, G. D., Litt, A., and Irish, V. F. (2005). Virus-induced gene silencing is an effective tool for assaying gene function in the basal eudicot species Papaver somniferum (opium poppy). Plant J. 44, 334–341. doi: 10.1111/j.1365-313X.2005.02520
Hsieh, M. H., Pan, Z. J., Lai, P. H., Lu, H. C., Yeh, H. H., Hsu, C. C., et al. (2013). Virus-induced gene silencing unravels multiple transcription factors involved in floral growth and development in Phalaenopsis orchids. J. Exp. Bot. 64, 3869–3884. doi: 10.1093/jxb/ert218
Igarashi, A., Yamagata, K., Sugai, T., Takahashi, Y., Sugawara, E., Tamura, A., et al. (2009). Apple latent spherical virus vectors for reliable and effective virus-induced gene silencing among a broad range of plants including tobacco, tomato. Arabidopsis thaliana, cucurbits, and legumes. Virology 386, 407–416. doi: 10.1016/j.virol.2009.01.039
Jablonska, B., Ammiraju, J. S. S., Bhattarai, K. K., Mantelin, S., de Ilarduya, O. M., Roberts, P. A., et al. (2007). The Mi-9 gene from Solanum arcanum conferring heat-stable resistance to root-knot nematodes is a homolog of Mi-1. Plant Physiol. 143, 1044–1054. doi: 10.1104/pp.106.089615
Jia, H. F., Guo, J. X., Qin, L., and Shen, Y. Y. (2010). Virus-induced PpCHLH gene silencing in peach leaves (Prunus persica). J. Hortic. Sci. and Biotechnol. 85, 528–532. doi: 10.1111/pbi.13291
Jin, H., Axtell, M. J., Dahlbeck, D., Ekwenna, O., Zhang, S., Staskawicz, B., et al. (2002). NPK1, an MEKK1-like mitogen-activated protein kinase kinase kinase, regulates innate immunity and development in plants. Dev. Cell. 3, 291–297. doi: 10.1016/s1534-5807(02)00205-8
Jin, H., Liu, Y., Yang, K. Y., Kim, C. Y., Baker, B., and Zhang, S. (2003). Function of a mitogen activated protein kinase pathway in N gene-mediated resistance in tobacco. Plant J. 33, 719–731. doi: 10.1046/j.1365-313x.2003.01664.x
Kammen, A. V. (1997). Virus-induced gene silencing in infected and transgenic plants. Trends Plant Sci. 2, 409–411. doi: 10.1016/S1360-1385(97)01128-X
Kim, Y. C., Kim, S. Y., Paek, K. H., Choi, D., and Park, J. M. (2006). Suppression of CaCYP1, a novel cytochrome P450 gene, compromises the basal pathogen defense response of pepper plants. Biochem. Biophys. Res. Commun. 30, 638–645. doi: 10.1016/j.bbrc.2006.04.124
Kirungu, J. N., Magwanga, R. O., Pu, L., Cai, X. Y., Xu, Y. C., Hou, Y. Q., et al. (2020). Knockdown of Gh_A05G1554 (GhDHN_03) and Gh_D05G1729 (GhDHN_04) dehydrin genes, reveals their potential role in enhancing osmotic and salt tolerance in cotton. Genomics 112:2. doi: 10.1016/j.ygeno.2019.11.003
Koudounas, K., Thomopoulou, M., Angeli, E., Tsitsekian, D., Rigas, S., and Hatzopoulos, P. (2020). Virus-induced gene silencing in Olive tree (Oleaceae). Methods Mol. Biol. 2172, 165–182. doi: 10.1007/978-1-0716-0751-0_13
Kumagai, M. H., Donson, J., Della-Cioppa, G., Harvey, D., and Grill, L. K. (1995). Cytoplasmic inhibition of carotenoid biosynthesis with Virus-derived RNA. Proc. Natl. Acad. Sci. U S A. 92, 1679–1683. doi: 10.1073/pnas.92.5.1679
Li, C. C., An, X. H., Zhang, Z. Q., Liu, K., and Sun, J. (2019). Optimization of TRV-VIGS system and identification of top rot resistance related genes in Maize. J. Agric. Sci. 33, 2111–2118.
Li, Q., Chen, P., Dai, S. J., Sun, Y. F., Yuan, B., Kai, W. B., et al. (2015). PacCYP707A2 negatively regulates cherry fruit ripening while PacCYP707A1 mediates drought tolerance. J. Exp. Bot. 66, 3765–3774. doi: 10.1093/jxb/erv169
Li, X. J., Zhang, J. Q., Wu, Z. C., Lai, B., Huang, X. M., Qin, Y. H., et al. (2016). Functional characterization of a glucosyl?transferase gene, LcUFGT1, involved in the formation of cyaniding glucoside in the pericarp of Litchi chinensis. Physiol. Plant 156, 139–149. doi: 10.1111/ppl.12391
Liu, E., and Page, J. E. (2008). Optimized cDNA libraries for virus-induced gene silencing (VIGS) using tobacco rattle virus. Plant Methods 4:5. doi: 10.1186/1746-4811-4-5
Liu, H. P., Fu, D. Q., Zhu, B. Z., Yan, H. X., Shen, X. Y., Zuo, J. H., et al. (2012). Virus-induced gene silencing in Eggplant (Solanum melongena). J. Integr. Plant Biol. 54, 422–429. doi: 10.1111/j.1744-7909.2012.01102.x
Liu, X. B., Liu, N., Li, F. K., Wu, L. Z., Zhang, J., and Wang, D. M. (2015). Establishment of TRV-mediated transient gene-silencing system in soybean. Scientia Agricultura Sinica 48, 2479–2486. doi: 10.3864/j.issn.0578-1752.2015.12.021
Liu, Y., Schiff, M., and Dinesh-Kumar, S. P. (2002a). Virus-induced gene silencing in tomato. Plant J. 31, 777–786. doi: 10.1046/j.1365-313x.2002.01394
Liu, Y. L., Schiff, M., Marathe, R., and Dinesh-Kumar, S. P. (2002b). Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J. 30, 415–429. doi: 10.1046/j.1365-313x.2002.01297
Liu, Y., Schiff, M., and Dinesh-Kumar, S. P. (2004). Involvement of MEK1 MAPKK, NTF6 MAPK, WRKY/MYB transcription factors, COI1 and CTR1 in N-mediated resistance to tobacco mosaic virus. Plant J. 38, 800–809. doi: 10.1111/j.1365-313X.2004.02085.x
Lu, L. L., Fu, D. Q., Liu, H. P., Wang, X. H., Luo, Y. B., Zhu, B. Z., et al. (2011). Construction and identification VIGS vector about vitamin E biosynthesis gene of tomato. Northern Horticulture 16, 139–142.
Lu, R., Malcuit, I., Moffett, P., Ruiz, M. T., Peart, J., Wu, A. J., et al. (2003). High throughput virus-induced gene silencing implicates heat shock protein 90 in plant disease resistance. EMBO J. 22, 5690–5699. doi: 10.1093/emboj/cdg546
Misra, R. C., Sharma, S., Garg, A., and Ghosh, S. (2020). Virus-induced gene silencing in sweet basil (Ocimum basilicum). Methods Mol. Biol. 2172, 123–138. doi: 10.1007/978-1-0716-0751-0_10
Nishii, K., Fei, Y., Hudson, A., Möller, M., and Molnar, A. (2020). Virus-induced gene silencing in Streptocarpus rexii (Gesneriaceae). Mol. Biotechnol. 62, 317–325. doi: 10.1007/s12033-020-00248-w
Orzaez, D., Mirabel, S., Wieland, W. H., and Granell, A. (2006). Agroinjection of tomato fruits. a tool for rapid functional analysis of transgenes directly in fruits. Plant Physiol. 140, 3–11. doi: 10.1104/pp.105.068221
Palmer, L., and O’Connor, S. E. (2020). Virus-induced gene silencing in Nepeta. Methods Mol. Biol. 2172, 111–121. doi: 10.1007/978-1-0716-0751-0_9
Purkayastha, A., and Dasgupta, I. (2009). Virus-induced gene silencing: a versatile tool for discovery of gene functions in plants. Plant Physiol. Biochem. 47, 967–976. doi: 10.1016/j.plaphy.2009.09.001
Qu, J., Ye, J., Geng, Y. F., Sun, Y. W., Gao, S. Q., Zhang, B. P., et al. (2012). Dissecting functions of KATANIN and WRINKLED1 in cotton fiber development by virus-induced gene silencing. Plant Physiol. 160, 738–748. doi: 10.1104/pp.112.198564
Ratcliff, F., Harrison, B. D., and Baulcombe, D. C. (1997). A similarity between viral defense and gene silencing in plants. Science 276, 1558–1560. doi: 10.1126/science.276.5318.1558
Ratcliff, F., Martin-Hernandez, A. M., and Baulcombe, D. C. (2001). Tobacco rattle virus as a vector for analysis of gene function by silencing. Plant J. 25, 237–245. doi: 10.1046/j.0960-7412.2000.00942
Ratcliff, F. G., MacFarlane, S. A., and Baulcombe, D. C. (1999). Gene silencing without DNA: rna-mediated cross-protection between viruses. Plant Cell 11, 1207–1215. doi: 10.1105/tpc.11.7.1207
Ruiz, M. T., Voinnet, O., and Baulcombe, D. C. (1998). Initiation and maintenance of virus-induced gene silencing. Plant Cell 10, 937–946. doi: 10.1105/tpc.10.6.937
Ryu, C. M., Anand, A., Kang, L., and Mysore, K. S. (2004). Agrodrench: a novel and effective agroinoculation method for virus-induced gene silencing in roots and diverse Solanaceous species. Plant J. 40, 322–331. doi: 10.1111/j.1365-313X.2004.02211
Scofield, S. R., Huang, L., Brandt, A. S., and Gill, B. S. (2005). Development of a virus-induced gene silencing system for hexaploid wheat and its use in functional analysis of the Lr21-mediated leaf rust resistance pathway. Plant Physiol. 138, 2165–2173. doi: 10.1104/pp.105.061861
Senthil-Kumar, M., Ajith, A., Uppalapati, S. R., and Mysore, K. S. (2008). Virus-induced gene silencing and its applications. CAB Rev. Perspect. Agric. Vet. Sci. Nutr. Nat. Resour. 3, 1–18. doi: 10.1079/PAVSNNR20083011
Senthil-Kumar, M., and Mysore, K. S. (2011a). New dimensions for VIGS in plant functional genomics. Trends Plant Sci. 16, 656–665. doi: 10.1016/j.tplants.2011.08.006
Senthil-Kumar, M., and Mysore, K. S. (2011b). Virus-induced gene silencing can persist for more than 2 years and also be transmitted to progeny seedlings in Nicotiana benthamiana and tomato. Plant Biotechnol. J. 9, 797–806. doi: 10.1111/j.1467-7652.2011.00589
Senthil-Kumar, M., and Mysore, K. S. (2014). Tobacco rattle virus based virus-induced gene silencing in Nicotiana benthamiana. Nat. Protoc. 9, 1549–1562. doi: 10.1038/nprot.2014.092
Sharma, P. C., Ito, A., Shimizu, T., Terauchi, R., Kamoun, S., and Saitoh, H. (2003). Virus-induced silencing of WIPK and SIPK genes reduces resistance to a bacterial pathogen, but has no effect on the INF1-induced hypersensitive response (HR) in Nicotiana benthamiana. Mol. Genet. Genomics 269, 583–591. doi: 10.1007/s00438-003-0872-9
Sunilkumar, G., Campbell, L. M., Puckhaber, L., Stipanovic, R. D., and Rathore, K. S. (2006). Engineering cottonseed for use in human nutrition by tissue-specific reduction of toxic gossypol. Proc. Natl. Acad. Sci. U S A. 103, 18054–18059. doi: 10.1073/pnas.0605389103
Szittya, G., Silhavy, D., Molnár, A., Havelda, Z., Lovas, Á, Lakatos, L., et al. (2003). Low temperature inhibits RNA silencing-mediated defense by the control of siRNA generation. EMBO J. 22, 633–640. doi: 10.1093/emboj/cdg74
Tameling, W. I., and Baulcombe, D. C. (2007). Physical association of the NB-LRR resistance protein Rx with a Ran GTPase-activating protein is required for extreme resistance to Potato virus X. Plant Cell 19, 1682–1694. doi: 10.1105/tpc.107.050880
Tan, Y., Bukys, A., Molnár, A., and Hudson, A. (2020). Rapid, high efficiency virus-mediated mutant complementation and gene silencing in Antirrhinum. Plant Methods 16:145. doi: 10.1186/s13007-020-00683-5
Thomas, C. L., Jones, L., Baulcombe, D. C., and Maule, A. J. (2001). Size constraints for targeting post-transcriptional gene silencing and for RNA-directed methylation in Nicotiana benthamiana using a potato virus X vector. Plant J. 25, 417–425. doi: 10.1046/j.1365-313x.2001.00976
Tian, J., Pei, H. X., Zhang, S., Chen, J. W., Chen, W., Yang, R. Y., et al. (2014). TRV-GFP: a modified Tobacco rattle virus vector for efficient and visualizable analysis of gene function. J. Exp. Bot. 65, 311–322. doi: 10.1093/jxb/ert381
Tuttle, J. R., Idris, A. M., Brown, J. K., Haigler, C. H., and Robertson, D. (2008). Geminivirus-mediated gene silencing from Cotton leaf crumple virus is enhanced by low temperature in cotton. Plant Physiol. 148, 41–50. doi: 10.1104/pp.108.123869
Waterhouse, P. M., Wang, M.-B., and Lough, T. (2001). Gene silencing as an adaptive defence against viruses. Nature 411, 834–842. doi: 10.1038/35081168
Wege, S., Scholz, A., Gleissberg, S., and Becker, A. (2007). Highly efficient virus-induced gene silencing (VIGS) in California poppy (Eschscholzia californica): an evaluation of VIGS as a strategy to obtain functional data from non-model plants. Ann. Bot. 100, 641–649. doi: 10.1093/aob/mcm118
Xie, L. H., Zhang, Q. Y., Sun, D. Y., Yang, W. Z., Hu, J. Y., Niu, L. X., et al. (2019). Virus-induced gene silencing in the perennial woody Paeonia ostii. Peer J. 7:e7001. doi: 10.7717/peerj.7001
Xie, Y. G., Ma, Y. Y., Bi, P. P., Wei, W., Liu, J., Hu, Y., et al. (2020). Transcription factor FvTCP9 promotes strawberry fruit ripening by regulating the biosynthesis of abscisic acid and anthocyanins. Plant Physiol. Biochem. 146, 374–383. doi: 10.1016/j.plaphy.2019.11.004
Yamagishi, N., and Yoshikawa, N. (2009). Virus induced gene silencing in soybean seeds and the emergence stage of soybean plants with Apple latent spherical virus vectors. Plant Mol. Biol. 71, 15–24. doi: 10.1007/s11103-009-9505-y
Yang, J., Zhang, T. Y., Liao, Q. S., He, L., Li, J., Zhang, H. M., et al. (2018). Chinese Wheat Mosaic virus-induced gene silencing in monocots and dicots at low temperature. Front. Plant Sci. 9:1627. doi: 10.3389/fpls.2018.01627
Yuan, C., Li, C., Yan, L. J., Jackson, A. O., Liu, Z. Y., Han, C. G., et al. (2011). A high throughput barley stripe mosaic virus vector for virus induced gene silencing in monocots and dicots. PLoS one 6:e26468. doi: 10.1371/journal.pone.0026468
Zaidi, S. S. E. A., Kumar, V., Matias, L. E., and Hervé, V. (2020). Virus-induced gene silencing (vigs) in cassava using geminivirus agroclones. Methods Mol. Biol. 2172, 51–64. doi: 10.1007/978-1-0716-0751-0_5
Zeng, H. Q., Xie, Y. W., Liu, G. Y., Wei, Y. X., Hu, W., and Shi, H. T. (2019). Agrobacterium-mediated gene transient overexpression and Tobacco Rattle Virus (TRV)-based gene silencing in cassava. Int. J. Mol. Sci. 20:3976. doi: 10.3390/ijms20163976
Zhang, J., Yu, D. S., Zhang, Y., Liu, K., Xu, K. D., Zhang, F. L., et al. (2017). Vacuum and co-cultivation agroinfiltration of (germinated) seeds results in Tobacco Rattle Virus (TRV) mediated whole-plant virus-induced gene silencing (VIGS) in wheat and maize. Front. Plant Sci. 8:393. doi: 10.3389/fpls.2017.00393
Zhang, J. X., Wang, F. R., Zhang, C. Y., Zhang, J. H., Chen, Y., Liu, G. D., et al. (2018). A novel VIGS method by agroinoculation of cotton seeds and application for elucidating functions of GhBI-1 in salt-stress response. Plant Cell Rep. 37, 1091–1100. doi: 10.1007/s00299-018-2294-5
Zhang, X. L., Zhang, Z. W., Na, R., Yang, S. Q., Gao, J., and Zhao, J. (2014). The effects of different potato genotypes and different inoculation methods on VIGS silence efficiency. Huabei Nongxuebao (Acta Agriculturae Boreali-Sinica) 29, 36–40.
Zhang, Y., Dorey, S., Swiderski, M., and Jones, J. D. (2004). Expression of RPS4 in tobacco induces an AvrRps4-independent HR that requires EDS1. SGT1 and HSP90. Plant J. 40, 213–224. doi: 10.1111/j.1365-313X.2004.02201
Zhao, J. P., Jiang, H. L., Wang, J. Y., Wang, Z. H., Dong, J. G., and Song, J. Q. (2020). Virus-induced gene silencing in diploid and tetraploid potato species. Methods Mol. Biol. 2172, 39–50. doi: 10.1007/978-1-0716-0751-0_4
Zhou, B. J., and Zeng, L. R. (2017). Elucidating the role of highly homologous Nicotiana benthamiana ubiquitin E2 gene family members in plant immunity through an improved virus-induced gene silencing approach. Plant Methods 13:59. doi: 10.1186/s13007-017-0210-6
Keywords: TRV-VIGS, vector construction, agroinfiltration, secondary inoculation, methodology modification
Citation: Shi G, Hao M, Tian B, Cao G, Wei F and Xie Z (2021) A Methodological Advance of Tobacco Rattle Virus-Induced Gene Silencing for Functional Genomics in Plants. Front. Plant Sci. 12:671091. doi: 10.3389/fpls.2021.671091
Received: 23 February 2021; Accepted: 10 May 2021;
Published: 04 June 2021.
Edited by:
Guo-qing Song, Michigan State University, United StatesReviewed by:
Dongyan Zhao, Cornell University, United StatesAhmad A. Omar, Zagazig University, Egypt
Copyright © 2021 Shi, Hao, Tian, Cao, Wei and Xie. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fang Wei, ZmFuZ3dlaUB6enUuZWR1LmNu; Zhengqing Xie, enF4aWVAenp1LmVkdS5jbg==
†These authors have contributed equally to this work
 Gongyao Shi
Gongyao Shi Mengyuan Hao
Mengyuan Hao Baoming Tian1,3
Baoming Tian1,3 Gangqiang Cao
Gangqiang Cao Fang Wei
Fang Wei Zhengqing Xie
Zhengqing Xie