- 1RIKEN Center for Sustainable Resource Science, Yokohama, Japan
- 2Graduate School of Bioagricultural Sciences, Nagoya University, Nagoya, Japan
- 3Department of Molecular and Functional Genomics, Interdisciplinary Center for Science Research, Shimane University, Matsue, Japan
Oryza longistaminata, a wild rice, can propagate vegetatively via rhizome formation and, thereby, expand its territory through horizontal growth of branched rhizomes. The structural features of rhizomes are similar to those of aerial stems; however, the physiological roles of the two organs are different. Nitrogen nutrition is presumed to be linked to the vegetative propagation activity of rhizomes, but the regulation of rhizome growth in response to nitrogen nutrition and the underlying biological processes have not been well characterized. In this study, we analyzed rhizome axillary bud growth in response to nitrogen nutrition and examined the involvement of cytokinin-mediated regulation in the promotion of bud outgrowth in O. longistaminata. Our results showed that nitrogen nutrition sufficiency promoted rhizome bud outgrowth to form secondary rhizomes. In early stages of the response to nitrogen application, glutamine accumulated rapidly, two cytokinin biosynthesis genes, isopentenyltransferase, and CYP735A, were up-regulated with accompanying cytokinin accumulation, and expression of an ortholog of FINE CULM1, a negative regulator of axillary bud outgrowth, was severely repressed in rhizomes. These results suggest that, despite differences in physiological roles of these organs, the nitrogen-dependent outgrowth of rhizome axillary buds in O. longistaminata is regulated by a mechanism similar to that of shoot axillary buds in O. sativa. Our findings provide a clue for understanding how branched rhizome growth is regulated to enhance nutrient acquisition strategies.
Introduction
Seed reproduction and vegetative reproduction are the two major modes of propagation in plants. Some perennial plant species that vegetatively reproduce expand their territories through stolon or rhizome growth (Guo et al., 2020). Rhizomes grow horizontally underground and form branches as secondary and tertiary rhizomes by the outgrowth of axillary buds that develop at rhizome nodes. Some rhizome tips developmentally transform into aerial green shoots and appear aboveground, becoming new ramets.
Wild rice is known for its substantial ecological diversity. Oryza longistaminata, an African wild rice species, is mainly propagated vegetatively via rhizome formation and proliferates vigorously (Vaughan, 1994). Both O. longistaminata and Oryza sativa contain the AA genome and have a high degree of genome synteny as revealed by genome sequencing (Reuscher et al., 2018; Li et al., 2020). O. longistaminata has also been used to study the genetic basis of rhizome development (Hu et al., 2011; Yoshida et al., 2016; Fan et al., 2017; Kyozuka, 2017; Bessho-Uehara et al., 2018; Toriba et al., 2020). The basic structural features of rhizomes are similar to those of aerial stems; rhizomes are composed of phytomer units consisting of a node, an internode, and an axillary bud (Yoshida et al., 2016). Crown roots develop at most of the rhizome nodes, suggesting that the branching growth of rhizomes is a strategy for efficient nutrition acquisition from the soil. Whereas rhizomes and aerial stems have similar anatomical features, the physiological features are different. Rhizomes are photosynthetic sink organs in which the internodes can elongate even during the vegetative stage of the parental ramet, whereas aerial stems are photosynthetic source organs in which internode elongation is tightly linked with the reproductive transition.
Studies of the molecular basis for shoot branching (tillering) in O. sativa show that three phytohormones, auxin, cytokinin, and strigolactone play key roles in regulating this developmental process (Beveridge and Kyozuka, 2010; Wang and Li, 2011; Gallavotti, 2013). Apically-derived auxin negatively regulates cytokinin biosynthesis by repressing IPT4 and positively regulating strigolactone biosynthesis by D10 (Arite et al., 2007; Minakuchi et al., 2010). Strigolactone induces the expression of FC1 that negatively regulates axillary bud outgrowth, whereas expression of FC1 is repressed by cytokinin (Minakuchi et al., 2010). As for nutritional factors that influence shoot branching in O. sativa, phosphate-starvation induces de novo synthesis of strigolactones that suppress shoot branching (Umehara et al., 2010). In contrast, nitrogen sufficiency up-regulates glutamine-mediated IPT4 gene expression and plays a role in the promotion of shoot axillary bud outgrowth (Kamada-Nobusada et al., 2013; Ohashi et al., 2017; Sakakibara, 2021). A loss-of-function mutant of glutamine synthetase1;2 (GS1;2) reduced the nitrogen-dependent induction of IPT4 and the length of axillary buds, a phenotype that was rescued by the application of cytokinins (Ohashi et al., 2017). Nevertheless, how rhizome branching in O. longistaminata is regulated by nitrogen nutrition and the biological processes underlying rhizome axillary bud outgrowth remain to be elucidated.
In this study, we analyzed the growth of rhizome axillary buds in response to nitrogen nutrition and examined the involvement of cytokinin-mediated regulation in the promotion of bud outgrowth in O. longistaminata. Our results suggest that the nitrogen-dependent outgrowth of rhizome axillary buds in O. longistaminata is regulated by a mechanism similar to that for shoot axillary buds in O. sativa. This finding provides important insights for understanding the regulation of rhizome branching and growth in nutrient acquisition strategies.
Results
Effect of Nitrogen Nutrition on Rhizome Bud Outgrowth in O. longistaminata
To examine the growth response of rhizome axillary buds to an inorganic nitrogen source, young ramets of comparable growth stages (Figure 1A) were hydroponically grown in a low-nitrogen (Low-N, 200 μM NH4NO3) medium for 2 weeks. The young ramets were further grown under a Low-N or a high-nitrogen (High-N, 2 mM NH4NO3) medium for another 2 weeks, and growth of the axillary buds was measured. The resulting growth status of the axillary buds was categorized into one of four patterns: secondary rhizomes whose internodes elongated greater than 1 cm (Rhizome), a bud that grew slightly but the length was less than 1 cm (Bud), a degenerated bud undergoing necrosis (Degenerated), and a developmentally transformed green shoot (Shoot; Figure 1A). The outgrowth of rhizome axillary buds was promoted under the High-N treatment with the percentage of secondary rhizomes significantly higher than in the Low-N treatment (Figure 1B). By contrast, the percentage of degenerated buds in the Low-N treatment was significantly higher than for the High-N treatment. In both nitrogen treatments, about 30 to 40% of the axillary buds became green shoots (Figure 1B). These results suggest that an inorganic nitrogen source promotes rhizome bud outgrowth to form secondary rhizomes.
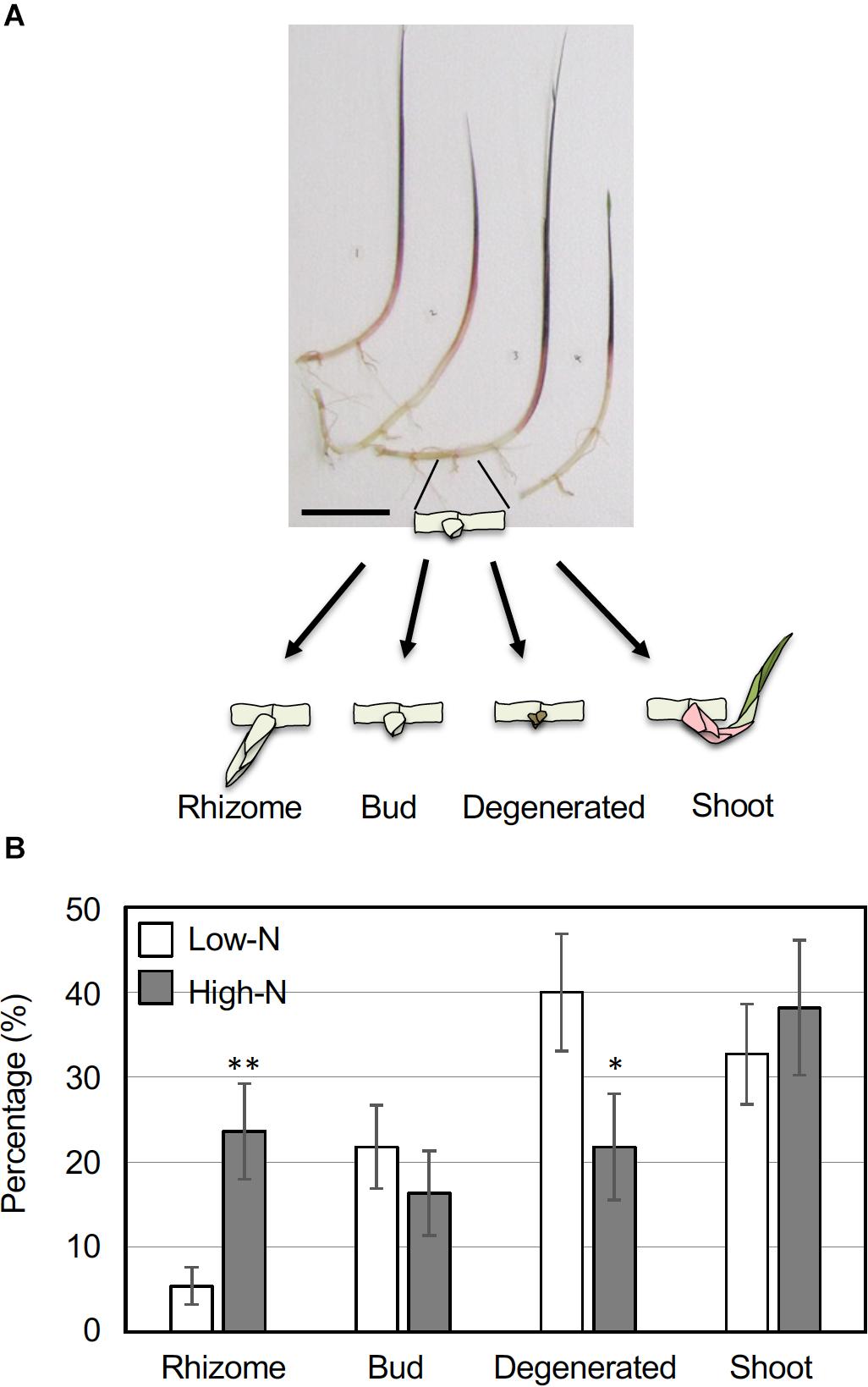
Figure 1. Growth response of O. longistaminata rhizome axillary buds to nitrogen nutrition. Young ramets having rhizomes at a similar growth stage were hydroponically grown in a low-N (200 μM NH4NO3) medium for 2 weeks. The ramets were subsequently transferred to either a low-N or high-N (2 mM NH4NO3) medium and incubated for an additional 2 weeks. Growth of the axillary buds on the rhizome nodes was monitored and classified into one of four categories: Rhizome (length > 10 mm), Buds (length < 10 mm), Degenerated, and Shoot. Detailed category definitions are explained in the text. (A) Typical ramets used in this experiment. Scale bar = 10 cm. (B) A comparison of rhizome growth in low-N and high-N media. Vertical bars represent the mean ± S.E. [n (ramet) = 13 for low-N, n = 14 for high-N]. Each ramet rhizome had 2 to 7 nodes, and the total number of nodes was 55 for low-N and 55 for high-N samples. *P < 0.05; **P < 0.01 (Student’s t test) compared to the low-N medium.
Phytohormone Profile in Rhizomes in Response to Nitrogen Application
In O. sativa, de novo cytokinin biosynthesis in roots and shoots is up-regulated by nitrogen sources, and cytokinins are involved in the promotion of axillary bud outgrowth of shoot stems (Kamada-Nobusada et al., 2013; Ohashi et al., 2017). To evaluate the effect of nitrogen application on the levels of phytohormones in rhizomes, we profiled the accumulation levels of the major phytohormones (cytokinins, ABA, and IAA) in crown roots, axillary buds, and nodes at 6 h after 2 mM NH4NO3 application. For the cytokinins, levels of trans-zeatin (tZ) and its precursors, such as tZ riboside and tZ riboside 5’-phosphates (tZRPs), and N6-(Δ2-isopentenyl)adenine riboside 5’-phosphates (iPRPs), precursors of iP, increased in crown roots, buds, and nodes of the NH4NO3 -treated rhizomes (Figure 2 and Supplementary Table 1). In contrast, the levels of other cytokinins and their precursors, such as cZ, cZR, and cZRPs, and most of the glucosides decreased after NH4NO3 application to the crown roots, but the levels of these compounds did not change in other plant organs (Figure 2 and Supplementary Table 1). Since tZRPs and iPRPs are initial products catalyzed by adenosine phosphate-isopentenyltransferase (IPT) and CYP735A (Sakakibara, 2006), these results suggest that de novo synthesis of cytokinins is activated by nitrogen application to the rhizomes. Interestingly, there was no significant difference in the levels of ABA or IAA resulting from supplemental NH4NO3 application to crown roots, axillary buds, or nodes (Supplementary Table 1).
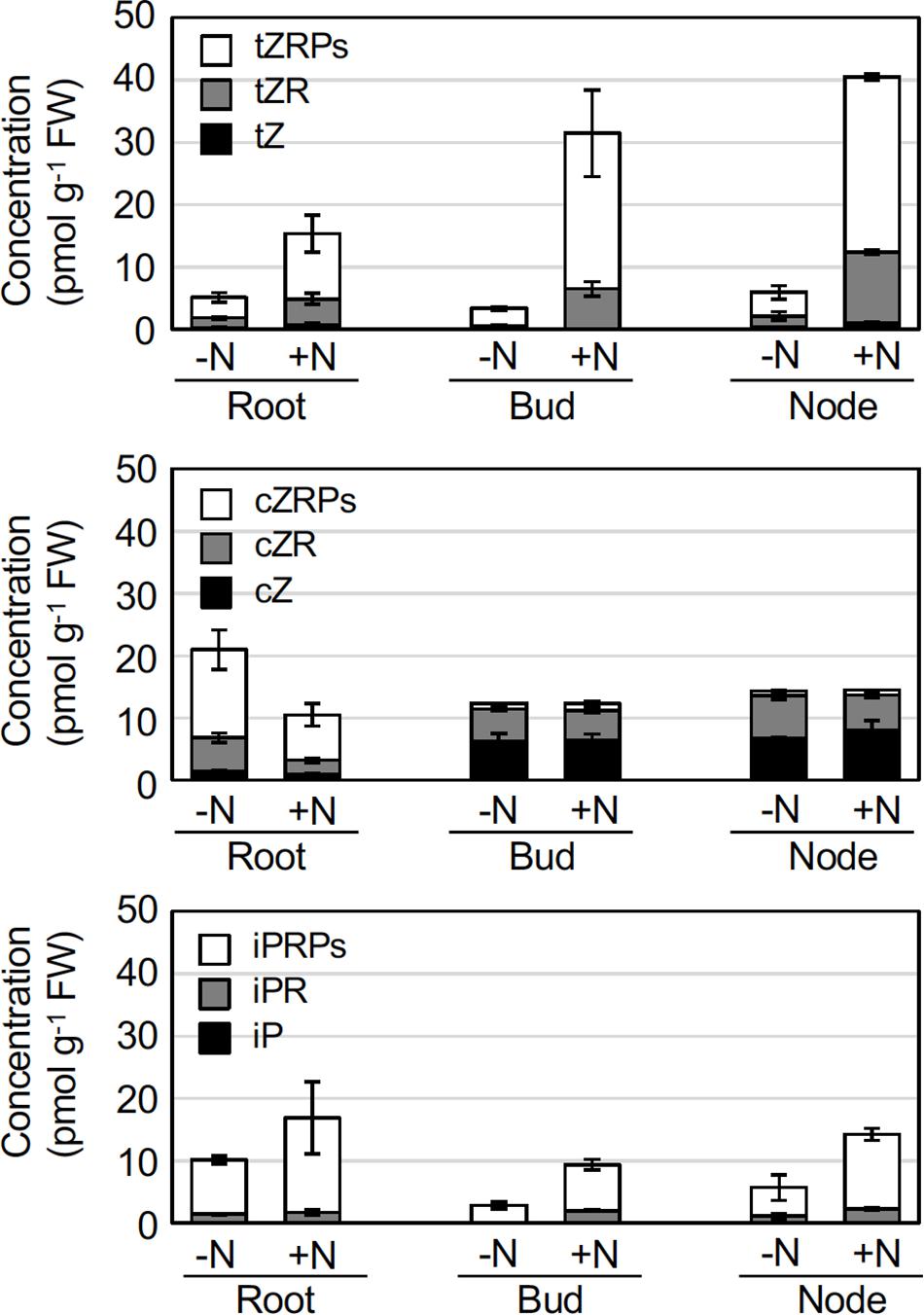
Figure 2. Effects of nitrogen applications on cytokinin concentrations in rhizome roots, buds, and nodes. Young ramets having rhizomes at a similar growth stage were hydroponically grown in a low-N (200 μM NH4NO3) medium for 2 weeks. The ramets were subsequently transferred to culture media containing no nitrogen (-N) or 2 mM NH4NO3 (+N). 6 h after transfer, rhizome roots, buds, and nodes were separately harvested, and the cytokinin hormone content was analyzed. Data are presented as the mean ± S.E. (n = 3). The complete data set is presented in Supplementary Table 1. tZ, trans-zeatin; tZR, tZ riboside; tZRPs, tZR 5’-phosphates; cZ, cis-zeatin; cZR, cZ riboside; cZRPs, cZR 5’-phosphates; iP, N6-(Δ2-isopentenyl)adenine; iPR, iP riboside; iPRPs, iPR 5’-phosphates; and FW, fresh weight.
To confirm a link between cytokinin accumulation and axillary bud outgrowth, we examined the effect of applying exogenous cytokinin on the rhizome. When we treated rhizomes with 1 μM tZ for 2 weeks, axillary bud growth was promoted (Supplementary Figure 1). These results support the hypothesis that cytokinin action is involved in nitrogen-dependent rhizome axillary bud outgrowth.
Expression of Cytokinin Biosynthesis Genes in Response to Nitrogen Supply
To understand the underlying events leading to the nitrogen-dependent accumulation of cytokinins, we analyzed the expression of genes involved in cytokinin biosynthesis by reverse transcription-quantitative PCR (RT-qPCR). To specify the responsive genes, we selected orthologs of three cytokinin biosynthesis genes, IPT, CYP735A, and LOG from the O. longistaminata genomic database1 (Supplementary Table 2), and first analyzed the accumulation of the corresponding transcripts in rhizome nodes that include axillary buds at 6 h after the 2 mM NH4NO3 application (Supplementary Figure 2). Among the analyzed genes, the transcripts for OlIPT4, OlIPT5, OlIPT8, and OlCYP735A4 accumulated in response to nitrogen application, suggesting that de novo cytokinin biosynthesis is up-regulated by nitrogen sources in the rhizome. NADH-GOGAT1 that encodes a glutamate synthase was used as an indicator for the nitrogen response (Hirose et al., 1997). We further analyzed the expression of OlIPT4, OlIPT8, and OlCYP735A4 in a time-course experiment. In the rhizome node that includes an axillary bud, the OlIPT4 transcript accumulated within 2 h after 2 mM NH4NO3 application (Figure 3A). Transcripts for OlIPT8 and OlCYP735A4 also accumulated in response to nitrogen application but more slowly. In roots, the OlIPT4 transcript accumulated gradually, but no clear induction of OlIPT8 or OlCYP735A4 was observed, although the expression of the nitrogen-responsive marker gene OlNADH-GOGAT1 was induced (Figure 3B).
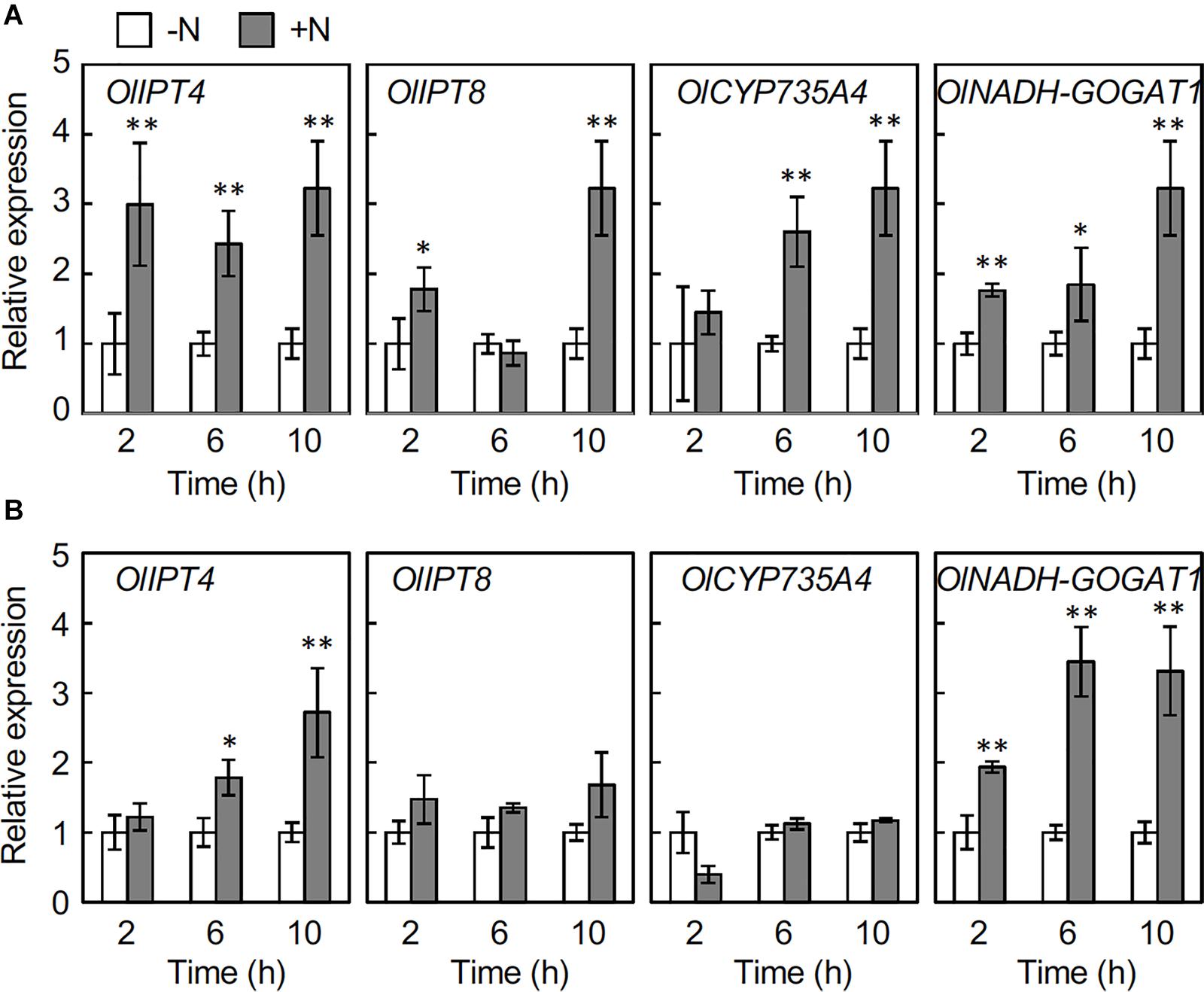
Figure 3. Changes in the expression of cytokinin biosynthesis genes in rhizomes responding to nitrogen application. Young ramets having rhizomes at a similar growth stage were hydroponically grown in a low-N (200 μM NH4NO3) medium for 2 weeks. The ramets were subsequently transferred to culture media containing no nitrogen (-N) or 2 mM NH4NO3 (+N). After incubation for the indicated times, rhizome roots and nodes with axillary buds were separately harvested. Total RNA prepared from the samples was subjected to RT-qPCR. Expression of the cytokinin biosynthesis genes in nodes including axillary buds (A) and roots (B). Transcript abundance was normalized with OlUBQ1 and expressed as relative values with respect to the value in the -N treatment. Values are means ± SE (n = 3). *P < 0.05; **P < 0.01 (Student’s t test) compared to the -N treatment.
Accumulation of Amino Acids in Response to the Nitrogen Supply
In O. sativa, a glutamine-related signal is a key indicator of IPT4 gene induction (Kamada-Nobusada et al., 2013; Ohashi et al., 2017). To obtain insight into the relationship between glutamine and IPT4 expression in O. longistaminata, we analyzed amino acid concentrations in rhizome nodes that include axillary buds and in roots in the presence or absence of supplemental nitrogen. Upon exposure to 2 mM NH4NO3, glutamine accumulated significantly in both roots and nodes within 2 h (Figure 4). Although the difference was rather smaller, threonine also significantly accumulated in both organs. Aspartate levels in nodes were higher than those in roots, but no substantial differences in the levels of other amino acids were found between organs.
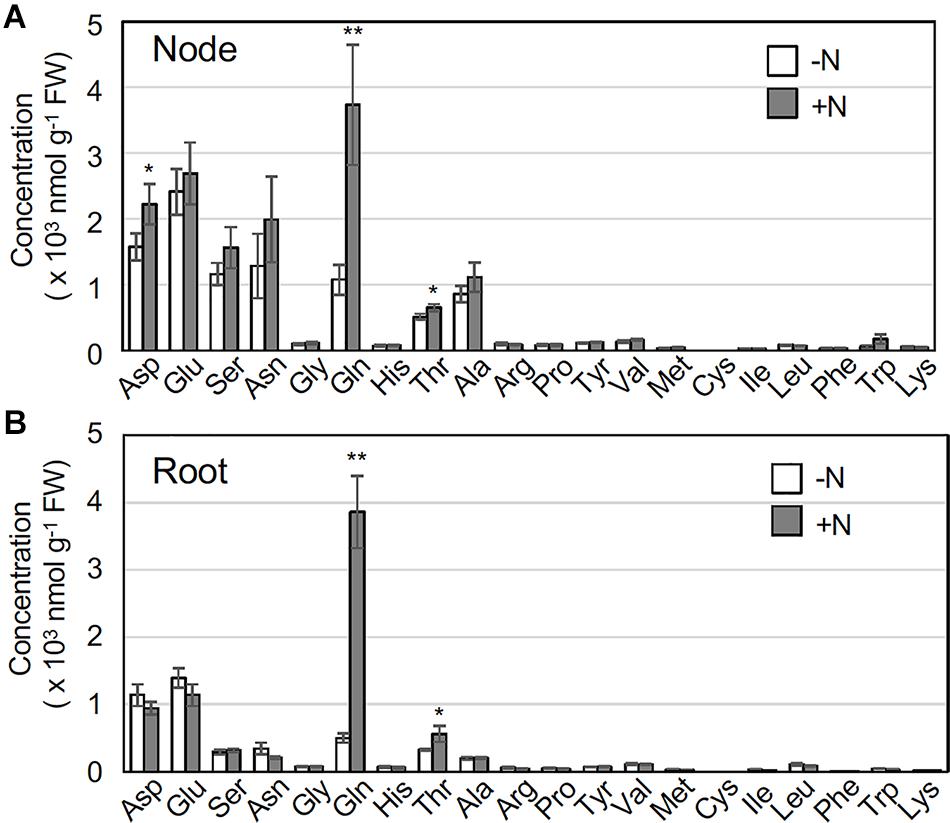
Figure 4. Effects of nitrogen application on amino acid concentrations in rhizome nodes and roots. Young ramets having rhizomes at a similar growth stage were hydroponically grown in a low-N (200 μM NH4NO3) medium for 2 weeks. The ramets were subsequently transferred to culture media containing no nitrogen (-N) or 2 mM NH4NO3 (+N) and incubated for 2 h. Rhizome nodes and roots were separately harvested and subjected to amino acid analysis. The amino acid concentrations in nodes including axillary buds (A) and roots (B). Data are presented as the mean ± S.E. (n = 3). *P < 0.05; **P < 0.01 (Student’s t test) compared to the -N treatment. FW, fresh weight.
To obtain more direct evidence, we examined the effect of an exogenous application of glutamine on the expression of OlIPT4. When we treated rhizomes with 50 mM glutamine, OlIPT4 expression in rhizome nodes was up-regulated in 6 h (Supplementary Figure 3), suggesting that glutamine-related signaling is involved in OlIPT4 induction.
Downregulation of OlFC1 Expression in Response to the Nitrogen Supply
To further delineate the underlying processes regulating rhizome axillary bud outgrowth, we analyzed the effect of nitrogen supply on the expression of OlFC1, a key regulator of shoot axillary bud growth. Transcript accumulation markedly decreased in rhizome axillary buds in response to nitrogen supply at 6 h, and the repression continued at 10 h after treatment (Figure 5). This result suggests that nitrogen sufficiency down-regulates OlFC1 expression.
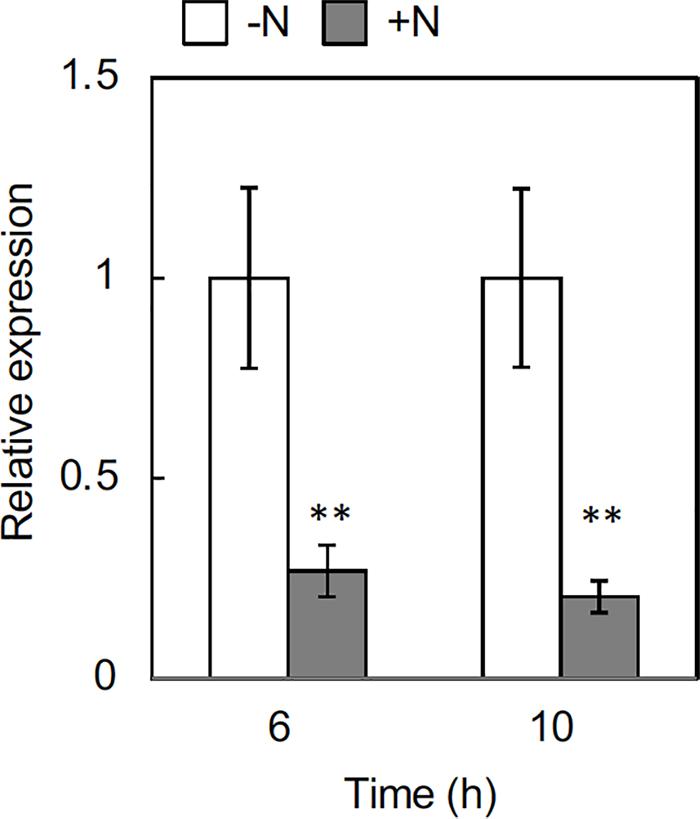
Figure 5. Effects of nitrogen application on the expression of OlFC1 in rhizome buds. Young ramets having rhizomes at a similar growth stage were hydroponically grown in a low-N (200 μM NH4NO3) medium for 2 weeks. The ramets were subsequently transferred to culture media containing no nitrogen (-N) or 2 mM NH4NO3 (+N) and incubated for 6 h and 10 h. Total RNA prepared from the samples was subjected to RT-qPCR analysis. Transcript levels were normalized by OlUBQ1 and expressed as relative values with respect to the value in the -N treatment. Values are the means ± SE (n = 3). **P < 0.01 (Student’s t test) compared to the -N treatment.
Discussion
In this study, we have shown that nitrogen nutrition promotes rhizome bud outgrowth to secondary rhizomes in O. longistaminata. The regulation of secondary rhizome formation in O. longistaminata relies on a mechanism similar to that for shoot axillary buds in O. sativa. In general, ample nitrogen nutrition promotes shoot growth but suppresses root system growth, with the opposite effect under nitrogen deficient conditions (Hermans et al., 2006; Gruber et al., 2013). Increasing the number of shoots (tillers) in response to nitrogen nutrition leads to larger amounts of aboveground biomass and more photosynthetic assimilation capacity. Also, prioritizing root system growth when nitrogen is deficient is a strategy for efficient nitrogen acquisition. In longer periods of O. longistaminata growth in the presence of different nitrogen treatments, growth of the secondary rhizomes was promoted and tertiary rhizomes branched under nitrogen-sufficient conditions (2 mM NH4NO3). Conversely, rhizome branching was suppressed and crown root growth was promoted under nitrogen-limiting conditions (200 μM NH4NO3; Supplementary Figure 4). Thus, the nitrogen-dependent promotion of rhizome elongation contributes to an expansion of the horizontal territory of vegetatively-propagated plants.
In the analysis of axillary bud growth patterns under Low-N and High-N treatments, there was no difference in the rate of differentiation to green shoots (Figure 1). Perhaps some of the axillary buds on rhizomes were already primed to become green shoots prior to the nitrogen treatment. Typically, the two basal nodes of ramets develop aerial shoot-type buds (Yoshida et al., 2016). The rate of axillary bud degeneration was higher in the low-N treatment. Deficiencies in the nitrogen supply could abort the growth of rhizome axillary buds in order to efficiently allocate nitrogen resources to a reduced number of rhizomes.
The expression of OlIPT4 was up-regulated by supplemental nitrogen or exogenously applied glutamine (Figure 3 and Supplementary Figure 3). Also, the expression of OlFC1 was repressed by an increased nitrogen supply (Figure 5). Studies on the regulation of shoot axillary bud outgrowth in O. sativa have shown that the expression of FC1 is repressed by cytokinin (Minakuchi et al., 2010). Our findings in this study suggest a model for the nitrogen-dependent promotion of rhizome axillary bud outgrowth (Figure 6) that is similar to how shoot axillary bud outgrowth is regulated in O. sativa. A supply of inorganic nitrogen rapidly increases the concentration of glutamine and triggers the induction of de novo cytokinin biosynthesis via OlIPT4 induction. The accumulated cytokinin suppresses the expression of OlFC1, thereby promoting outgrowth of the axillary bud.
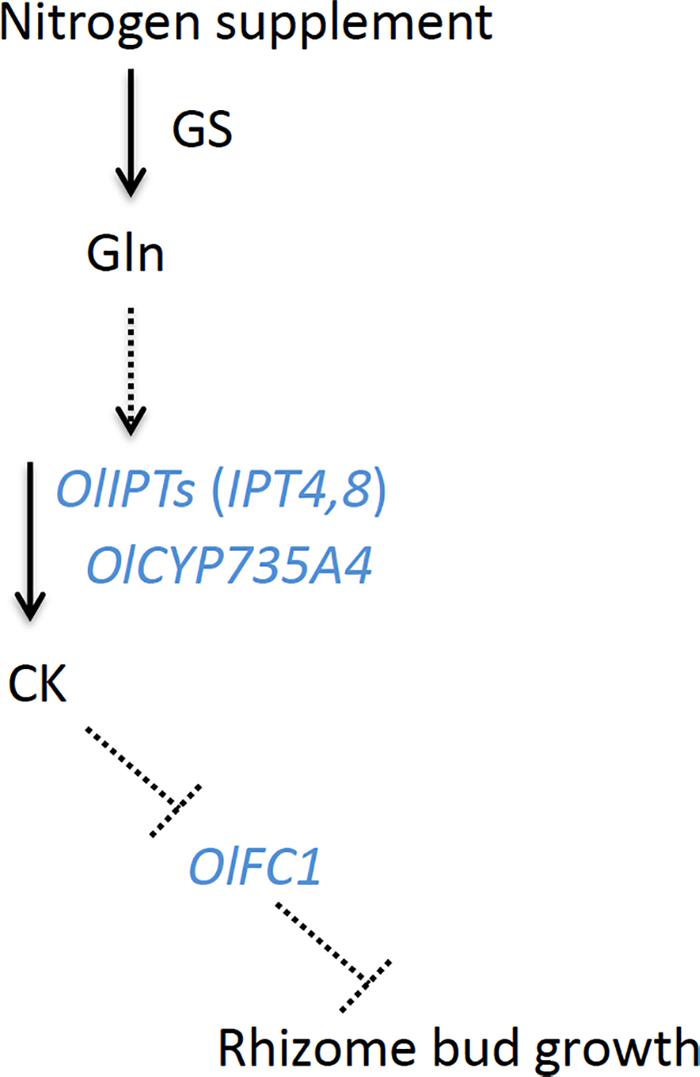
Figure 6. A model of hormonal control of rhizome axillary bud outgrowth in O. longistaminata. In this model, the nitrogen supply induces the accumulation of glutamine. Subsequently, a glutamine-related signal activates the expression of cytokinin biosynthesis genes, including OlIPT4, OlIPT8, and OlCYP735A4. The cytokinins negatively regulate the expression of OlFC1, the product of which functions to repress the outgrowth of the rhizome axillary bud. GS, glutamine synthetase; CK, cytokinin. Solid arrows represent the metabolic flow. Dashed arrows and T-end lines represent positive and negative regulation, respectively.
The degree of nitrogen-dependent induction of OlIPT4 and OlCYP735A4 expression in the node was greater than that in the root (Figure 3) and corresponded with the changes in tZ-type cytokinin accumulation (Figure 2). The regulation of cytokinin biosynthesis gene expression in response to nitrogen nutrition in O. sativa has been primarily analyzed in roots but not in nodes. IPT4 in O. sativa is mainly expressed in the phloem of vascular bundles (Kamada-Nobusada et al., 2013). O. longistaminata nodes might be a major organ for de novo cytokinin biosynthesis in response to nitrogen because nodes have a complex and dense vascular system (Yamaji and Ma, 2014).
FC1 expression is induced by strigolactones in O. sativa (Minakuchi et al., 2010; Xu et al., 2015), whereas strigolactone biosynthesis, important for tiller growth, is induced by phosphorus deficiency (Umehara et al., 2010). The biosynthetic pathways and molecular species of strigolactones are very diverse among plant species. In maize, nitrogen deficiency strongly induces the biosynthesis of zealactone, a strigolactone (Ravazzolo et al., 2019). The regulation strigolactone biosynthesis in O. longistaminata has not yet been investigated. The involvement of strigolactones in axillary bud elongation of underground stems in response to nitrogen nutrition and the interplay between cytokinin and strigolactone signalings will be the subjects of future research.
Materials and Methods
Plant Materials and Growth Conditions
The perennial wild rice species O. longistaminata (IRGC10404) was used in this study. Plants were grown in soil in a temperature-controlled greenhouse with supplemental artificial light at 12 h light (30°C)/12 h dark (25°C) photoperiod. Young ramets of comparable growth stages (shoot lengths: 20 to 30 cm, rhizome lengths: 10 to 20 cm) were excised from parental ramets and grown for 2 weeks with aeration in a hydroponic culture solution (Kamachi et al., 1991) in which the NH4NO3 concentration was modified to either 200 μM NH4NO3 (low-N) or 2 mM NH4NO3 (high-N).
Quantification of Phytohormones
Extraction and semi-purification of phytohormones from about 100 mg fresh weight of plant samples were performed as described previously (Kojima et al., 2009; Kojima and Sakakibara, 2012). Cytokinins were quantified using an ultra-performance liquid chromatography (UPLC)-tandem quadrupole mass spectrometer (ACQUITY UPLCTM System/XEVO-TQS; Waters, Milford, MA, United States) with an octadecylsilyl (ODS) column (ACQUITY UPLC HSS T3, 1.8 μm, 2.1 mm × 100 mm, Waters; Kojima et al., 2009). ABA and IAA were quantified using an ultra-high performance liquid chromatography (UHPLC)-quadrupole-orbitrap mass spectrometer (UHPLC/Q-ExactiveTM; Thermo Fisher Scientific, Waltham, MA, United States) with an ODS column (ACQUITY UPLC HSS T3, 1.8 μm, 2.1 mm × 100 mm; Waters; Kojima and Sakakibara, 2012; Shinozaki et al., 2015).
Gene Cloning
Total RNA was prepared from plant samples using an RNeasy Plant Mini Kit with an RNase-free DNase Set (QIAGEN, Hilden, Germany). First-strand cDNA was synthesized by reverse transcription using a SuperScriptTM III First-Strand Synthesis System (Thermo Fisher Scientific) with oligo(dT)12–18 primers. cDNAs were amplified by PCR with specific primers and cloned into the pCRTM2.1-TOPO TA vector (Thermo Fisher Scientific).
Quantitative PCR Analysis
Quantitative PCR (qPCR) analysis was performed using a StepOnePlus Real-Time PCR system (Applied Biosystems, Waltham, MA, United States) with a KAPA SYBR® Fast qPCR Master Mix (2×) kit (KAPA Biosystems, London, United Kingdom). Transcript abundance was estimated using the relative quantification method (Livak and Schmittgen, 2001) with a UBQ1 homolog as the internal standard for normalization. Gene locus IDs and the specific primers are listed in Supplementary Table 3.
Determination of Free Amino Acids
Plant samples were powdered in liquid nitrogen and homogenized in 10 volumes of 10 mM HCl with 0.2 mM methionine sulfone as an internal control. The homogenate was centrifuged, and the supernatant was filtered through Ultrafree® -MC filters (Merck Millipore, Burlington, MA, United States). Derivatization of amino acids was carried out using the Pico-Tag® method (Waters), and the resulting derivatives were analyzed with an HPLC System (Alliance 2695 HPLC system/2475, Waters) using a Pico-Tag column as described in the manufacturer’s instruction manual.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
KS and HS designed the research project. KS, AT, NM, MKa, MKo, YT, and TH conducted the research. KS, MKo, YT, and HS analyzed the data. KS and HS wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by Advanced Low Carbon Technology Research and Development Program from the Core Research for Evolutional Science and Technology by JST (to HS; no. JPMJCR13B1) and Grants-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology, Japan (to HS; JP17H06473 and JP19H00931).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Drs. Motoyuki Ashikari, and Stephan Reuscher (Nagoya University) for their helpful discussion and providing the genome information of O. longistaminata.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.670101/full#supplementary-material
Footnotes
References
Arite, T., Iwata, H., Ohshima, K., Maekawa, M., Nakajima, M., Kojima, M., et al. (2007). DWARF10, an RMS1/MAX4/DAD1 ortholog, controls lateral bud outgrowth in rice. Plant J. 51, 1019–1029. doi: 10.1111/j.1365-313X.2007.03210.x
Bessho-Uehara, K., Nugroho, J. E., Kondo, H., Angeles-Shim, R. B., and Ashikari, M. (2018). Sucrose affects the developmental transition of rhizomes in Oryza longistaminata. J. Plant Res. 131, 693–707. doi: 10.1007/s10265-018-1033-x
Beveridge, C. A., and Kyozuka, J. (2010). New genes in the strigolactone-related shoot branching pathway. Curr. Opin. Plant Biol. 13, 34–39. doi: 10.1016/j.pbi.2009.10.003
Fan, Z., Cai, Z., Shan, J., and Yang, J. (2017). Bud position and carbohydrate play a more significant role than light condition in the developmental transition between rhizome buds and aerial shoot buds of Oryza longistaminata. Plant Cell Physiol. 58, 1281–1282. doi: 10.1093/pcp/pcx061
Gallavotti, A. (2013). The role of auxin in shaping shoot architecture. J. Exp. Bot. 64, 2593–2608. doi: 10.1093/jxb/ert141
Gruber, B. D., Giehl, R. F. H., Friedel, S., and von Wirén, N. (2013). Plasticity of the Arabidopsis root system under nutrient deficiencies. Plant Physiol. 163, 161–179. doi: 10.1104/pp.113.218453
Guo, L., Plunkert, M., Luo, X., and Liu, Z. (2020). Developmental regulation of stolon and rhizome. Curr. Opin. Plant Biol. 59:101970. doi: 10.1016/j.pbi.2020.10.003
Hermans, C., Hammond, J. P., White, P. J., and Verbruggen, N. (2006). How do plants respond to nutrient shortage by biomass allocation? Trends Plant Sci. 11, 610–617. doi: 10.1016/j.tplants.2006.10.007
Hirose, N., Hayakawa, T., and Yamaya, T. (1997). Inducible accumulation of mRNA for NADH-dependent glutamate synthase in rice roots in response to ammonium ions. Plant Cell Physiol. 38, 1295–1297. doi: 10.1093/oxfordjournals.pcp.a029120
Hu, F., Wang, D., Zhao, X., Zhang, T., Sun, H., Zhu, L., et al. (2011). Identification of rhizome-specific genes by genome-wide differential expression analysis in Oryza longistaminata. BMC Plant Biol. 24:11. doi: 10.1186/1471-2229-11-18
Kamachi, K., Yamaya, T., Mae, T., and Ojima, K. (1991). A role for glutamine synthetase in the remobilization of leaf nitrogen during natural senescence in rice Leaves. Plant Physiol. 96, 411–417. doi: 10.1104/pp.96.2.411
Kamada-Nobusada, T., Makita, N., Kojima, M., and Sakakibara, H. (2013). Nitrogen-dependent regulation of de novo cytokinin biosynthesis in rice: the role of glutamine metabolism as an additional signal. Plant Cell Physiol. 54, 1881–1893. doi: 10.1093/pcp/pct127
Kojima, M., Kamada-Nobusada, T., Komatsu, H., Takei, K., Kuroha, T., Mizutani, M., et al. (2009). Highly sensitive and high-throughput analysis of plant hormones using ms-probe modification and liquid chromatographytandem mass spectrometry: an application for hormone profiling in Oryza sativa. Plant Cell Physiol. 50, 1201–1214. doi: 10.1093/pcp/pcp057
Kojima, M., and Sakakibara, H. (2012). Highly sensitive high-throughput profiling of six phytohormones using MS-probe modification and liquid chromatography-tandem mass spectrometry. Methods Mol. Biol. 918, 151–164. doi: 10.1007/978-1-61779-995-2_11
Kyozuka, J. (2017). Analysis of rhizome development in Oryza longistaminata, a wild rice species. Plant Cell Physiol. 58, 1283. doi: 10.1093/pcp/pcx064
Li, W., Li, K., Zhang, Q. J., Zhu, T., Zhang, Y., Shi, C., et al. (2020). Improved hybrid de novo genome assembly and annotation of African wild rice, Oryza longistaminata, from Illumina and PacBio sequencing reads. Plant Genome 13:e20001. doi: 10.1002/tpg2.20001
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-Δ Δ CT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Minakuchi, K., Kameoka, H., Yasuno, N., Umehara, M., Luo, L., Kobayashi, K., et al. (2010). FINE CULM1 (FC1) works downstream of strigolactones to inhibit the outgrowth of axillary buds in rice. Plant Cell Physiol. 51, 1127–1135. doi: 10.1093/pcp/pcq083
Ohashi, M., Ishiyama, K., Kojima, S., Kojima, M., Sakakibara, H., Yamaya, T., et al. (2017). Lack of cytosolic glutamine synthetase1;2 activity reduces nitrogen-dependent biosynthesis of cytokinin required for axillary bud outgrowth in rice seedlings. Plant Cell Physiol. 58, 679–690. doi: 10.1093/pcp/pcx022
Ravazzolo, L., Trevisan, S., Manoli, A., Boutet-Mercey, S., Perreau, F., and Quaggiotti, S. (2019). The control of zealactone biosynthesis and exudation is involved in the response to nitrogen in maize root. Plant Cell Physiol. 60, 2100–2112. doi: 10.1093/pcp/pcz108
Reuscher, S., Furuta, T., Bessho-Uehara, K., Cosi, M., Jena, K. K., Toyoda, A., et al. (2018). Assembling the genome of the African wild rice Oryza longistaminata by exploiting synteny in closely related Oryza species. Commun. Biol. 1:162. doi: 10.1038/s42003-018-0171-y
Sakakibara, H. (2006). Cytokinins: activity, biosynthesis, and translocation. Annu. Rev. Plant Biol. 57, 431–449. doi: 10.1146/annurev.arplant.57.032905.105231
Sakakibara, H. (2021). Cytokinin biosynthesis and transport for systemic nitrogen signaling. Plant J. 105, 421–430. doi: 10.1111/tpj.15011
Shinozaki, Y., Hao, S., Kojima, M., Sakakibara, H., Ozeki-Iida, Y., Zheng, Y., et al. (2015). Ethylene suppresses tomato (Solanum lycopersicum) fruit set through modification of gibberellin metabolism. Plant J. 83, 237–251. doi: 10.1111/tpj.12882
Toriba, T., Tokunaga, H., Nagasawa, K., Nie, F., Yoshida, A., and Kyozuka, J. (2020). Suppression of leaf blade development by BLADE-ON-PETIOLE orthologs is a common strategy for underground rhizome growth. Curr. Biol. 30, 509.e–516.e. doi: 10.1016/j.cub.2019.11.055
Umehara, M., Hanada, A., Magome, H., Takeda-Kamiya, N., and Yamaguchi, S. (2010). Contribution of strigolactones to the inhibition of tiller bud outgrowth under phosphate deficiency in rice. Plant Cell Physiol. 51, 1118–1126. doi: 10.1093/pcp/pcq084
Vaughan, D. A. (1994). The Wild Relatives of Rice: A Genetic Resources Handbook. Los Banos, CA: IRRI.
Wang, Y., and Li, J. (2011). Branching in rice. Curr. Opin. Plant Biol. 14, 94–99. doi: 10.1016/j.pbi.2010.11.002
Xu, J., Zha, M., Li, Y., Ding, Y., Chen, L., Ding, C., et al. (2015). The interaction between nitrogen availability and auxin, cytokinin, and strigolactone in the control of shoot branching in rice (Oryza sativa L.). Plant Cell Rep. 34, 1647–1662. doi: 10.1007/s00299-015-1815-8
Yamaji, N., and Ma, J. F. (2014). The node, a hub for mineral nutrient distribution in graminaceous plants. Trends Plant Sci. 19, 556–563. doi: 10.1016/j.tplants.2014.05.007
Keywords: axillary bud outgrowth, cytokinin, nitrogen, Oryza longistaminata, rhizome
Citation: Shibasaki K, Takebayashi A, Makita N, Kojima M, Takebayashi Y, Kawai M, Hachiya T and Sakakibara H (2021) Nitrogen Nutrition Promotes Rhizome Bud Outgrowth via Regulation of Cytokinin Biosynthesis Genes and an Oryza longistaminata Ortholog of FINE CULM 1. Front. Plant Sci. 12:670101. doi: 10.3389/fpls.2021.670101
Received: 20 February 2021; Accepted: 08 April 2021;
Published: 30 April 2021.
Edited by:
Jan Hejatko, Central European Institute of Technology (CEITEC), CzechiaReviewed by:
Helena G. Carvalho, Universidade do Porto, PortugalWeibing Yang, Institute of Plant Physiology and Ecology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, China
Copyright © 2021 Shibasaki, Takebayashi, Makita, Kojima, Takebayashi, Kawai, Hachiya and Sakakibara. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hitoshi Sakakibara, c2FrYWtpQGFnci5uYWdveWEtdS5hYy5qcA==
 Kyohei Shibasaki1
Kyohei Shibasaki1 Hitoshi Sakakibara
Hitoshi Sakakibara