
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 17 June 2021
Sec. Plant Physiology
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.665883
This article is part of the Research TopicAmino Acids in Plants: Regulation and Functions in Development and Stress DefenseView all 27 articles
Although it is known that brassinosteroids (BRs) play pleiotropic roles in plant growth and development, their roles in plant nutrient uptake remain unknown. Here, we hypothesized that BRs directly regulate ammonium uptake by activating the expression of rice AMT1-type genes. Exogenous BR treatment upregulated both AMT1;1 and AMT1;2 expression, while this induction was impaired in the BR-receptor gene BRI1 mutant d61-1. We then focused on brassinazole-resistant 1 (BZR1), a central hub of the BR signaling pathway, demonstrating the important role of this signaling pathway in regulating AMT1 expression and rice roots NH4+ uptake. The results showed that BR-induced expression of AMT1;2 was suppressed in BZR1 RNAi plants but was increased in bzr1-D, a gain-of-function BZR1 mutant. Further EMSA and ChIP analyses showed that BZR1 bound directly to the BRRE motif located in the promoter region of AMT1;2. Moreover, cellular ammonium contents, 15NH4+ uptake, and the regulatory effect of methyl-ammonium on root growth are strongly dependent on the levels of BZR1. Overexpression lines of BRI1 and BZR1 and Genetic combination of them mutants showed that BZR1 activates AMT1;2 expression downstream of BRI1. In conclusion, the findings suggest that BRs regulation of NH4+ uptake in rice involves transcription regulation of ammonium transporters.
Inorganic nitrogen (N) is an important plant nutrient and is absorbed from the rhizosphere in two forms, nitrate and ammonium. In the paddy field, high levels of NH4+ are of particular importance to the rice yield. Further understanding of the molecular basis and regulation of ammonium transport and its translocation to buds is needed to promote efficient nitrogen absorption and to improve crop yields. Ammonium transporter (AMT) proteins can induce high-affinity NH4+ uptake from the rhizosphere to root cells, and the transporter AMT2;1 has also been shown to play a crucial role in ammonium root-to-stem translocation (Giehl et al., 2017). Sequence analysis has identified 10 AMT members in the rice genome (Suenaga et al., 2003; Loque and von Wiren, 2004). Of these, AMT1;1, AMT1;2, and AMT1;3 are the main three AMTs. The expression patterns of these three proteins differ between different tissues with AMT1;2 and AMT1;3 mainly expressed in plant roots, while AMT1;1 is constitutively expressed in different tissues (Sonoda et al., 2003a). In rice, overexpression of AMT1;1 increases NH4+ uptake, improves plant growth and promotes yield production in limited N-fertilization conditions (Ranathunge et al., 2014). However, the overexpression of AMT1;3 has an opposite effect in regulating rice growth and NH4+ uptake to AMT1;1 (Bao et al., 2015). Studies have shown that transcriptional regulation of AMT genes strongly influences the plant’s N content and uptake of different forms of externally applied N. NH4+ can upregulate the expression of both AMT1;1 and AMT1;2, and inhibit the expression of AMT1;3. Under N starvation conditions, AMT1;3 is upregulated in response to NH4+ (Kumar et al., 2003; Sonoda et al., 2003a). Similar regulation at the posttranscriptional and posttranslational levels have also been observed in Arabidopsis. For example, AMT1;1 has been shown to be phosphorylated in C-terminal threonine residue to inhibit transporter activity in an NH4+-dependent manner (Yuan et al., 2007; Lanquar et al., 2009), and further, CBL-interacting serine/threonine protein kinase 23 (CIPK23) was reported to phosphorylate AMT1 to inhibit ammonium uptake (Straub et al., 2017). Indeterminate domain 10 (IDD10), a transcription factor, has recently been shown to directly activate AMT1;2 in rice (Xuan et al., 2013), furthermore, the ABI3/VP1 transcription factor RAVL1 activates AMT1;2 to directly modulate NH4+ uptake in rice (Xuan et al., 2016).
Transcriptome studies using gain-of-function mutants of the BES1 transcription factor and wild-type plants have identified the Arabidopsis genes regulated by brassinosteroids (BRs). These genes included AtAMT1;1 which was found to be upregulated by BR signaling activation (Goda et al., 2004; Yu et al., 2011). BRs are important phytohormones that bind to the cell surface receptor Brassinosteroid Insensitive 1 (BRI1), initiating a signaling cascade in which BRI1 binds to BRI1-Associated Receptor Kinase 1 (BAK1) leading to the downstream inactivation of the kinase Brassinosteroid Insensitive 2 (BIN2). The protein phosphatase PP2A dephosphorylates two master transcription factors Brassinozole-Resistant 1 (BZR1) and BRI1-EMS-Supperssor 1 (BES1), and the non-phosphorylated BZR1 and BES1 translocate to the nucleus to regulate the expression of BR responsive genes (Li and Chory, 1997; Li et al., 2002; Nam and Li, 2002; Kim and Wang, 2010; Yang et al., 2011; Guo et al., 2013; Tong and Chu, 2018). In addition, the stunting and BR-insensitive phenotype of bri1 BR receptor mutants can be rescued by the enhanced stability of BES1 and BZR1 in bes1-D and bzr1-D gain-of-function mutants (Wang et al., 2002; Yin et al., 2002). In rice, RAVL1, an upstream component of BR signaling, regulates BR homeostasis through binding to an E-box motif in the promoter regions of the BR receptor and biosynthesis genes (Je et al., 2010). BRs treatment enhances the expression of AMT1;1 and AMT1;2 in rice (Xuan et al., 2016); however, the detailed mechanism remains obscure.
These findings raise the questions of whether BRs play a role in nutrient uptake, and whether there is a direct regulatory link between BZR1 and AMTs in rice. In this study, we first analyzed the expression of AMT1 genes affected by BRs in the key BR signal transcription factor BZR1 and the BR receptor gene BRI1 mutants. We then investigated the expression patterns of AMT1 family members in the roots of the BRI1 mutant d61-1, bzr1-D siblings, as well as the BZR1 knockdown BZR1 RNAi. Genetic combinations between BRI1 and BZR1 were generated to examine activation of BRI1 and BZR1 in BR-mediated induction of AMT1;2. In addition, the cellular ammonium contents and 15N abundance were tested to investigate BZR1 function in ammonium uptake. Taken together, our results showed that BR-dependent ammonium uptake is partially controlled by BZR1.
The coding sequence of BZR1 was cloned into the pCambia1302 vector to construct the BZR1-GFP-expressing plasmid. The pCambia1302-BZR1 vector was subsequently transformed into Nipponbare rice calli to generate BZR1-GFP transgenic lines. The bri1-D in the Dongjin background as well as d61-1, bzr1-D, and BZR1 RNAi in the Nipponbare background were described previously (Yamamuro et al., 2000; Jeong et al., 2002; Bai et al., 2007; Qiao et al., 2017). BZR1 RNAi, d61-1, bzr1-D, bri1-D, bri1-D/BZR1 RNAi, BZR1-GFP, and d61-1/bzr1-D plants were grown in the greenhouse. Plants were first grown in distilled water (dH2O) for 1 week and subsequently transferred to brassinolide (BL) solution for analyzing BR effects on AMT1 expressions. Whole roots were harvested after 3 h of BL treatment. To examine the effects of NH4+ on the expression of BZR1, plants were grown in dH2O for 2 weeks before transfer to N-free nutrient solution for a further 3 days of growth (Abiko et al., 2005). The plants were then grown in the nutrient solution containing 0.5 mM (NH4)2SO4 at pH 5.5. The roots were sampled at 0 and 3 h after the transfer. To test the effects of methyl-ammonium (MeA) on root growth, we added 1 mM KNO3 as the only source of N and different concentrations of MeA to 0.5 × MS medium, and cultivated wild-type, BZR1 RNAi, and bzr1-D plants in the modified medium. The primary root length was measured and recorded on the sixth day.
Cellular total RNA was isolated by using the TRIzol reagent (Takara, Dalian, LN, China), and the RNA was treated with RQ-RNase-free DNase (Promega, Madison, WI, United States) to eliminate genomic DNA contamination. The GoScript Reverse Transcription kit (Promega, Madison, WI, United States) was used to synthesize cDNA. Quantitative RT-PCR was performed using the Illumina Research Quantity software Illumina Eco 3.0 (Illumina, San Diego, CA, United States), and each gene expression was normalized against that of the Ubiquitin level. The primers used for qRT-PCR are listed in Table 1.
Rice calli (8 g) expressing 35S: BZR1:GFP, and 35S:GFP were used for the ChIP assay. A pre-immune serum was used for pre-absorption before immunoprecipitation, and an anti-GFP monoclonal antibody (Clontech, Takara Bio, Japan) was used for immunoprecipitation. The immunoprecipitated DNAs were analyzed by ChIP-PCR for identification of the BZR1 binding region. The immunoprecipitated DNA was normalized by each input DNA in ChIP-PCR (Je et al., 2010). The primers used for the ChIP-PCR are shown in Table 1.
BZR1 ORF sequences were sub-cloned into the pET28a (+) expression vector to produce His:BZR1 recombinant protein in Escherichia coli strain BL21 DE3 after 4 h of 0.5 mM IPTG treatment at 28°C. To perform EMSA, 1 μg of His:BZR1 protein and 40k cpm of the 32P-labeled DNA probes were used. The protocol was followed as previously described (Je et al., 2010). Primers used in the EMSA are listed in Table 1.
The effector (35S:BZR1), reporters [pAMT1;2 and BRRE (BR Responsive Element)-mutated promoter mpAMT1;1-GUS fusions] and an internal control (35S:LUC) were co-transformed into protoplasts from Arabidopsis for testing transcriptional activation (Yamaguchi et al., 2010). 35S:BZR1 was cloned into the GAL4BD region of the p35S:GAL4BD vector and 2.5 kb of normal and BRRE-mutated (in which CGTGT/CG was replaced by TTTTTT) AMT1;2 promoters were cloned into the TATA region of the p35S:TATA:GUS vector (Xuan et al., 2013). PEG-mediated transformation and subsequent activity measurement were performed as previously described (Yoo et al., 2007).
Cellular ammonium contents in rice roots were calculated by using an F-kit (Roche, Basel, Switzerland) following the manufacturer’s instructions (Oliveira et al., 2002).
Wild-type, BZR1 RNAi, d61-1, bzr1-D, bri1-D, bri1-D/BZR1 RNAi, and d61-1/bzr1-D plants were cultivated for a 2-week nursery period in deionized water, following which the seedlings were transferred to N-free nutrient solution to continue culturing (Sonoda et al., 2003b). After culturing for 3 days, the protoplasmic absorption of 15NH4+ was analyzed. The detailed method for the calculation of 15NH4+ influx and the ratio of 15N to 14N in the total N pool was as previously described (Xuan et al., 2016).
Statistical analysis was performed with Prism 5 software (GraphPad, San Diego, CA, United States). All data were expressed as mean ± SE. Comparison between multiple groups was performed by using one-way ANOVA with values of P < 0.05 considered as significant, followed by Bonferroni’ s multiple comparison tests.
Brassinolide is the most active form of BR. To investigate whether BR affects the expression of rice AMT1, we used a series of BL concentration gradients of 0, 10, 100, and 200 nm to treat the wild-type, a weak allele of rice BRI1 mutant, d61-1, and the BRI1 overexpression line bri1-D (Jeong et al., 2002), and compared the changes in the AMT1 expression level under the different treatment conditions. Quantitative RT-PCR analysis showed different patterns in the response of the AMT gene to BR at the transcriptional level: AMT1;1 and AMT1;2 showed dose-dependent upregulation in response to BR treatment, while no obvious change was observed in AMT1; 3. Without BR treatment, the transcription levels of AMT1;2 and AMT1;3 in the d61-1 mutant were slightly lower than those of the control group, while in bri1-D, the transcription levels of these genes were higher than those of wild-type plants. Among the various genotypes treated with BR, the expression levels of AMT1;1 and AMT1;2 in wild-type plants were higher than the level of d61-1. The expression level of AMT1;1 and AMT1;2 in bri1-D was significantly higher than that of the wild-type plants. Also, expression of a BR biosynthetic gene D2 was analyzed. The result indicated that D2 expression was suppressed by BR treatment in a dose-dependent manner, and D2 expression level was higher in d61-1 while lower in bri1-D compared to wild-type plants (Figure 1). These results suggest that BR mediates the regulation of AMT1;1 and AMT1;2 transcription levels via the cell surface receptor BRI1.
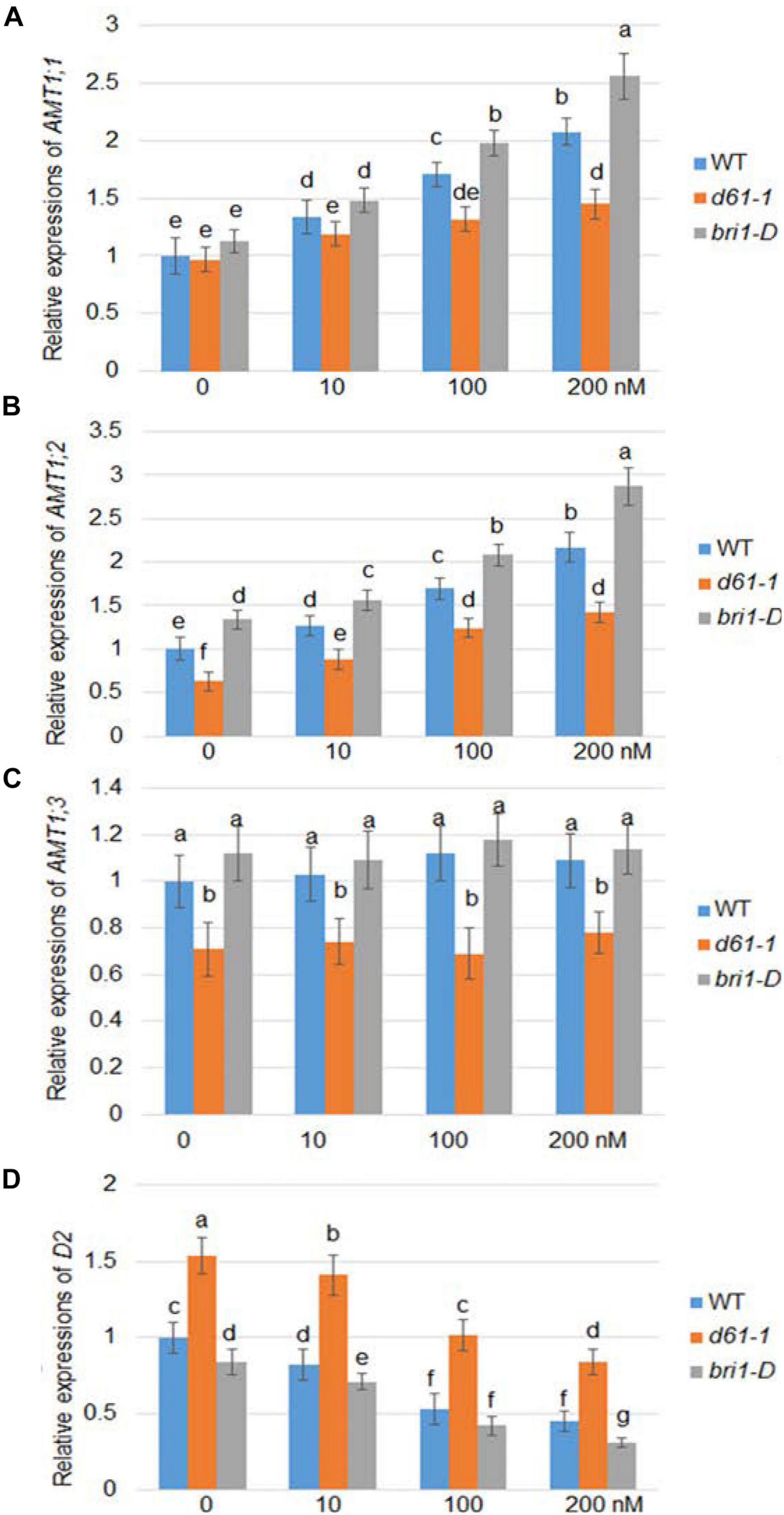
Figure 1. BR-dependent AMT1 expression in wild-type, BRI1 mutant d61-1, and BRI1 overexpression line bri1-D plant roots. Rice plants were grown for 7 days in distilled water, followed by 3 h with 0, 10, 100, or 200 nM brassinolide (BL). The expression levels of AMT1 1 (A), AMT1 2 (B), AMT1 3 (C), and D2 (D) in plants treated with BL was measured by qRT-PCR using Ubiquitin to normalize the expression of each gene. Error bars indicate means ± SE (n = 3). The experiments were repeated at least three times. Different letters represent statistically significant differences (P < 0.05).
As in Arabidopsis, rice BZR1 has been reported to be a key BR signaling transcription factor controlling the expression of downstream genes (Bai et al., 2007). To investigate the role of BZR1 in BR-mediated AMT1 induction, BZR1 RNAi (#1), the knockdown transgenic line, and bzr1-D dominant mutant line were constructed (Bai et al., 2007; Ren et al., 2020). The changes in AMT1;1 and AMT1;2 transcription levels in these lines after BL treatment were monitored. The transcriptional abundance of AMT1;1 was not affected by changes in BZR1 levels but could be increased in response to BL treatment in all plants (Figure 2A). Although the changes in the AMT1;2 transcription level in the BZR1 RNAi mutants and bzr1-D were not significantly associated with BL treatment, they showed a significant decrease and increase, respectively. Only the AMT1;2 expression levels correlated with the BZR1 level in a BR-dependent manner (Figure 2B). To verify BZR1 levels in the BZR1 RNAi plants, qRT-PCR was performed. The results indicated that BZR1 levels were reduced by about 60–70% in BZR1 RNAi lines (#1-#4). Also, AMT1;2 expression levels were significantly lower in the BZR1 RNAi lines (#1-#4) than in the wild-type plants (Figure 2C).
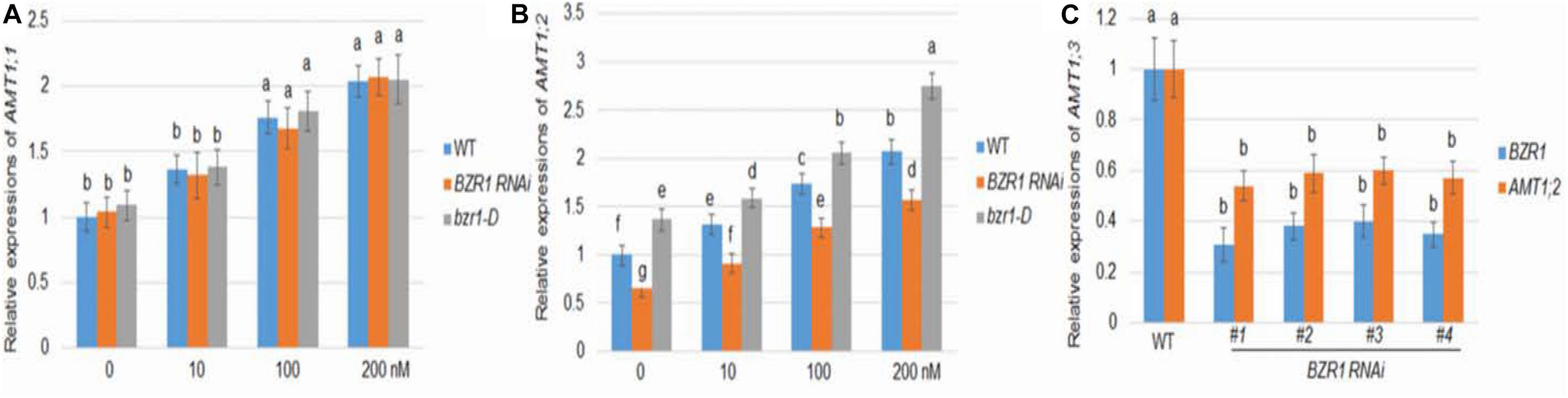
Figure 2. BR-mediated AMT1 expression in BZR1 RNAi and BZR1 constitutively active bzr1-D plant roots. Rice plants were grown in distilled water for 7 days and then treated with 0, 10, 100, or 200 nM BL for 3 h. (A) The expression levels of AMT1;1 was measured by qRT-PCR. (B) The expression levels of AMT1;2 was measured by qRT-PCR. Ubiquitin was used to normalize the expression of each gene. Data represent means ± SE (n = 3). (C) BZR1 and AMT1;2 expression levels in wild-type and BZR1 RNAi lines (#1-#4). Data indicate means ± SE (n = 3). The experiments were repeated at least three times. Different letters represent statistically significant differences (P < 0.05).
Since the changes in the transcription level of AMT1;2 are highly consistent with the changes in the expression level of BZR1, which indicated that BZR1 acts as a transcriptional activator upstream of AMT1;2. To verify whether the binding site of BZR1 includes the promoter of AMT1;2, we constructed transgenic plants of 35S:GFP and 35S:BZR1:GFP and performed ChIP assays. Promoter-sequence analysis showed that two BRRE (BR Responsive Element) motifs were located within a 2.5 kb stretch in front of the AMT1;2 start codon (Figure 3A). We designed four primer pairs to amplify the four fragments (P1−P4) of the AMT1;2 promoter and performed qPCR experiments to check the GFP-immunoprecipitates in 35S:GFP and 35S:BZR1:GFP transgenic siblings (Figure 3B). The ChIP results showed that BZR1 could bind directly to the AMT1;2 promoter, especially in the P4 region. Since the P4 fragment harbors two putative BRRE motifs (Figure 3A), we further performed EMSA experiments to determine which of the BRRE motifs was responsible for the BZR1 binding. We designed two specific probes each containing a BRRE motif. The B probe showed stronger binding to BZR1 (Figure 3A) while the binding of the A probe was slightly weaker. When the probes were mutated, their binding to BZR1 was lost. These results suggest that BZR1 can bind both BRRE motifs in the P4 region but the binding strength is different (Figure 3C).
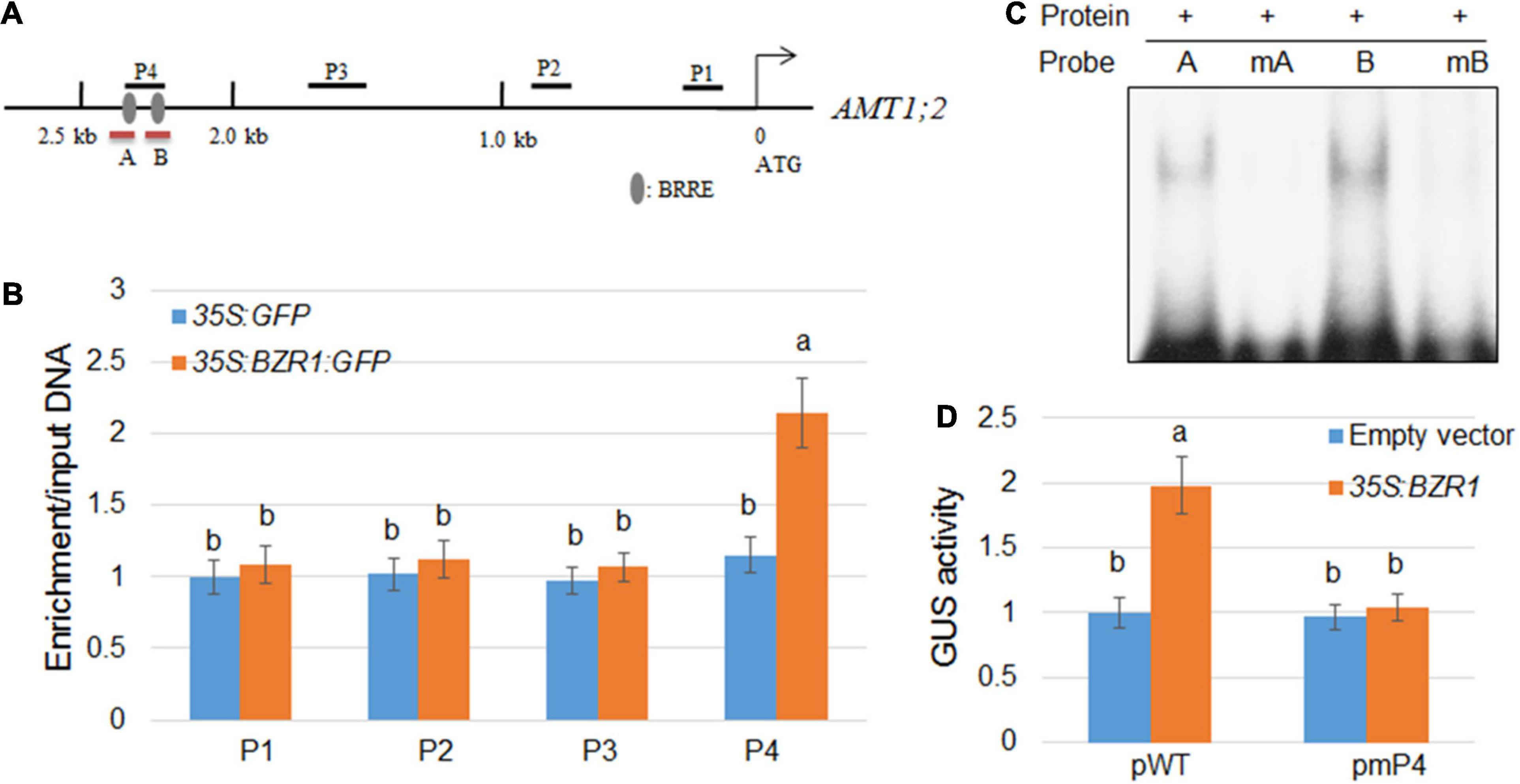
Figure 3. Activation of AMT1;2 by BZR1 via binding to BRRE elements in its promoter region. (A) Schematic diagram showing the position of BRRE in the AMT1;2 promoter and probes used for ChIP detection. The gray oval represents BRRE, the letters P (1, 2, 3, 4) indicate the positions of the probe. Red lines under the BRRE elements indicate the probe used in the EMSA analysis. (B) An anti-GFP antibody was used to amplify immunoprecipitated DNA in the 2.5 kb AMT1;2 promoter (P1–P4) for CHIP detection. The qPCR method was used to determine the relative ratios of immunoprecipitated DNA to input DNA. Input DNA was used for data normalization. Data represent means ± SE (n = 3). Transgenic plants expressing GFP and BZR1-GFP under control of 35S promoter were used. Different letters represent statistically significant differences (P < 0.05). (C) The affinity of BZR1 to each BRRE elements (A and B) located in P4 region was evaluated by EMSA. The BRRE motif sequence (CGTGT/CG) was replaced with the sequence TTTTTT in the mA and mB probes, and the mutant BRRE probes (mA and mB) were used as the negative controls. (D) 35S:BZR1 and each GUS-expressing vector were subjected to transient co-expression experiments under the control of the natural and BRRE-mutated AMT1;2 promoters. The activation of the native promoter (pWT) and BRRE element mutated promoter (pmP4) was measured in protoplasts that were co-transformed with 35S:BZR1. The GUS expression data was normalized by the luciferase gene driven by the 35S promoter. Error bars are ± SE of the means (n = 3). The experiments were repeated at least three times. Different letters represent statistically significant differences (P < 0.05).
In the current study, we identified two cis-elements targeted by the promoter of BZR1 which are closely related to transcriptional activation of AMT1;2. To further verify whether these cis-elements have similar functions in vivo, we used the Arabidopsis protoplast system to perform transient expression assays (Figure 3D). We also used the 35S:BZR1 plasmid and a vector expressing GUS, which is strictly controlled by one of four different types of 2.5 kb AMT1;2 promoters namely, the native (pWT) and three mutated (pmP4) promoters. In Arabidopsis protoplasts, the 35S:BZR1 plasmid and the GUS vector are co-transformed into a vector expressing GUS. In these mutated promoters, BRRE motif sequences (CGTGT/CG) were observed to be replaced by the sequence TTTTTT. To eliminate the error caused by conversion efficiency, 35S:LUC was used as an internal reference in each assay. By comparing the activity of the GUS genes under different promoter-driven conditions, we found that the GUS activity in Arabidopsis protoplasts was approximately twice that driven by the reporter gene promoter alone under the promoter pWT-driven conditions. Conversely, no detectable GUS activity was observed in the Arabidopsis protoplasts which were controlled by the mutant promoters (pmP4) (Figure 3D). These results indicated that direct combination with BZR1 is necessary for AMT1;2 promoter activation.
Root absorption of NH4+ is one of the main ways for plants to obtain N nutrients. To demonstrate whether BZR1 plays a role in this process, 15N-labeled ammonium was used to determine the efficiency of N absorption. The NH4+ concentrations in the root tissues of the wild-type, BZR1 RNAi, and bzr1-D lines were also determined. Seventeen day-old hydroponic seedlings were soaked in 200 μM of 15NH4+ solution for 6 min, after which the short-term import rate of the 15N-labeled ammonium in the plant roots was determined. Expressing the 15N influx in μmoles g–1 root dry weight h–1 (Yuan et al., 2007, 2013), the 15NH4+ influx in the BZR1 RNAi plants was only 68% of that of the wild-type, while the influx of 15NH4+ in the bzr1-D plants was greater than that of the wild-type plants (Figure 4A). We introduced the concept of “15N abundance,” representing the proportion of 15N to 14N in the total N pool, to further explore the role of BZR1 in the short-range transport of NH4+. Compared with the 15N internal flow results, the 15N abundance in the total 15N was lower in the BZR1 RNAi and higher in the bzr1-D plants compared to the wild-type (Figure 4B). The results of these short-term 15N absorption experiments indicate that BZR1 plays an important role in mediating NH4+ influx.
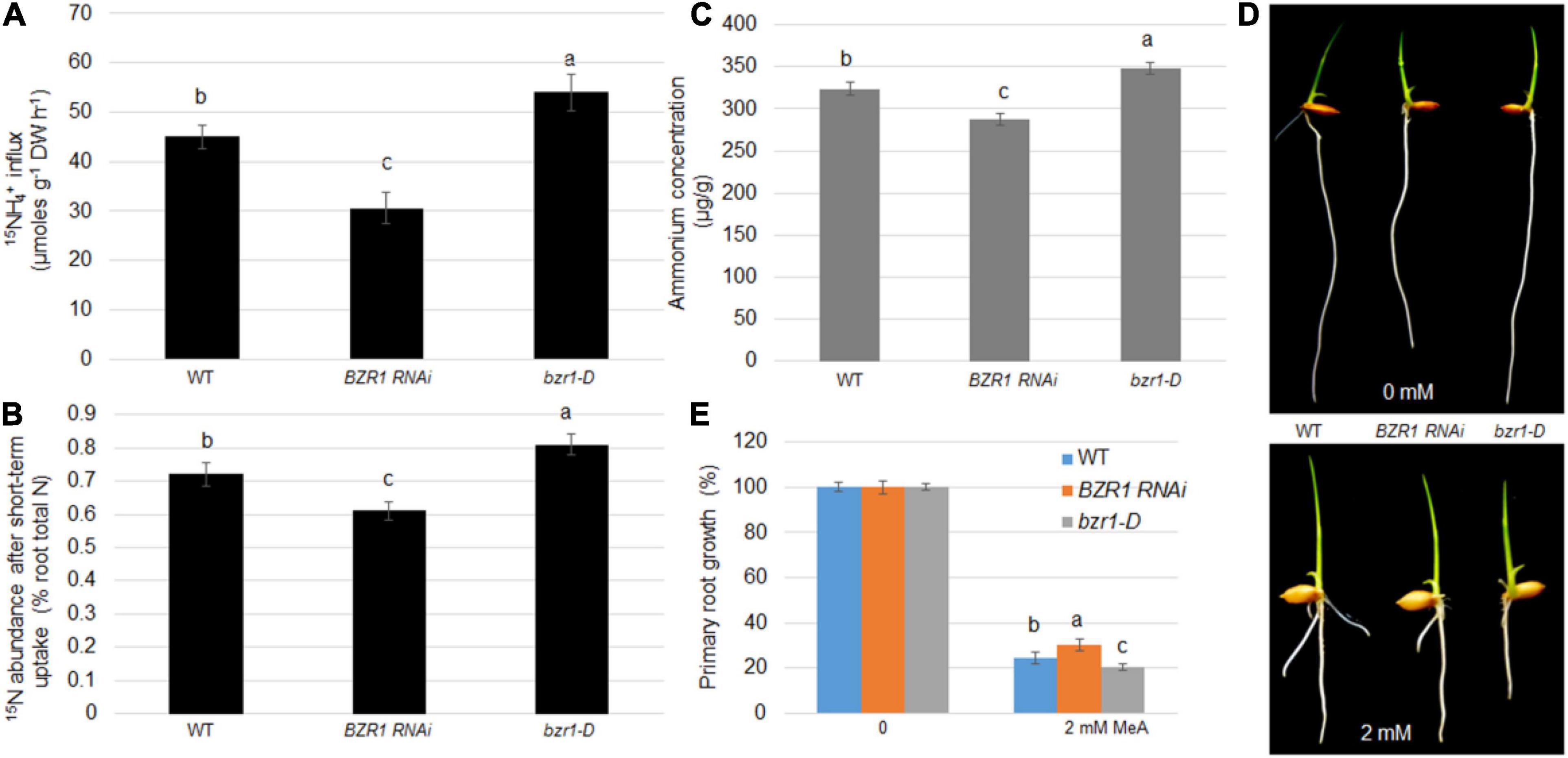
Figure 4. BZR1 effects on NH4+ uptake in plants. (A) The wild-type, BZR1 RNAi and BZR1 constitutively active bzr1-D plants were hydro-cultured in deionized water for 2 weeks, and then grown in a nitrogen-free nutrient solution for 3 days. After exposure to 200 μM 15N-labeled NH4+, the absorption of ammonium by rice roots was measured. Bars represent means ± SD (n = 6). (B) The wild-type, BZR1 RNAi, and bzr1-D plants were hydro-cultured in deionized water for 2 weeks, and then grown in a nitrogen-free nutrient solution for 3 days. After exposure to nutrient solution containing 200 μM 15N-labeled NH4+ for 6 min, the absorption of ammonium by rice roots and the 15NH4+ abundance in relation to the total 15N in roots was measured. Bars represent means ± SD (n = 6). (C) Intracellular NH4 levels of wild-type, BZR1, and bzr1-D grown for 3 days in 0.5 × MS were detected in plant roots. Data represent means ± SE (n = 3). (D) Under different conditions with 0 or 2 mM MeA, wild-type, BZR1 RNAi, and bzr1-D were grown in a modified 0.5 × MS medium containing 1 mM KNO3 for 6 days. MeA treatment significantly inhibited the growth of the primary root. In the absence of MeA treatment, the BZR1 RNAi primary root was shorter than that of the wild-type and bzr1-D. After 2 mM MeA treatment, bzr1-D showed a shorter primary root than BZR1 RNAi and wild-type. (E) The primary root growth was measured from wild-type, BZR1 RNAi, and bzr1-D with or without MeA supplementation as shown in (D). Data represent means ± SE (n > 15 plants). The experiments were repeated at least three times. Different letters represent statistically significant differences (P < 0.05).
BZR1 can regulate the expression of AMT1;2 genes in plant roots. We suspect that this regulation may be related to the long-term transport process of NH4+ in the roots. Therefore, the NH4+ content in the rice roots of 3-day-old wild-type, BZR1 RNAi, and bzr1-D seedlings grown on 0.5 × MS medium was determined. As expected, the content of NH4+ in the roots of BZR1 RNAi was lower than in the wild-type seedlings, while the roots of bzr1-D plants contained more NH4+ than the wild-type plants (Figure 4C). Furthermore, a toxic ammonium analog, MeA, was used as a replacement addition to the NH4+-free medium using a concentration gradient. Wild-type, BZR1 RNAi, and bzr1-D plants were grown in this medium and the length of the plant’s initial rooting was measured after 6 days of growth. In the absence of MeA, the primary roots of BZR1 RNAi were shorter than those of the wild-type and bzr1-D. With MeA treatment, the BZR1 RNAi root length was similar to that of the wild-type, while the bzr1-D roots were significantly shorter, the shortest in length among the three genetic lines (Figures 4D,E). These observations illustrated that the BZR1 RNAi response to MeA treatment is weaker than that of wild-type, while bzr1-D sensitivity to MeA treatment is relatively higher. Therefore, it can be inferred that BZR1 can participate in the process of rice root absorption of NH4+ in a long-term manner by regulating the expression of AMT1;2 genes.
As the expression of AMT1;2 genes mediated by BR is inhibited in both the d61-1 and BZR1 RNAi mutants, it is reasonable to speculate that BZR1 and BRI1 may affect the expression of AMT1;2 genes. To investigate this, a series of genetic analyses were employed: d61-1 and bzr1-D, BZR1 RNAi, and bri1-D lines were individually hybridized and two genetic combinations were constructed. We then measured the effects of BR-dependent AMT1;2 genes expression in the d61-1, bzr1-D, d61-1/bzr1-D, and wild-type lines without or without BL treatment. High levels of AMT1;2 mRNA were observed in both the bzr1-D and d61-1/bzr1-D lines after 3 h of root treatment with BL. In d61-1, the mRNA level was somewhat lower than in d61-1/bzr1-D but higher than the wild-type in both lines (Figure 5A). The mRNA levels of AMT1;2 in three other genotypes, including bri1-D, BZR1 RNAi, and bri1-D/BZR1 RNAi plants, were subsequently determined. We observed a similar low-level expression of AMT1;2 mRNA in BZR1 RNAi and a high-level expression in bri1-D plants with or without BR treatment. However, the BZR1 RNAi blocked AMT1;2 induction in the bri1-D background (Figure 5B). These results demonstrated that BZR1 positively regulates AMT1;2 expression downstream of BRI1 irrespective of exogenous BR stimulation.
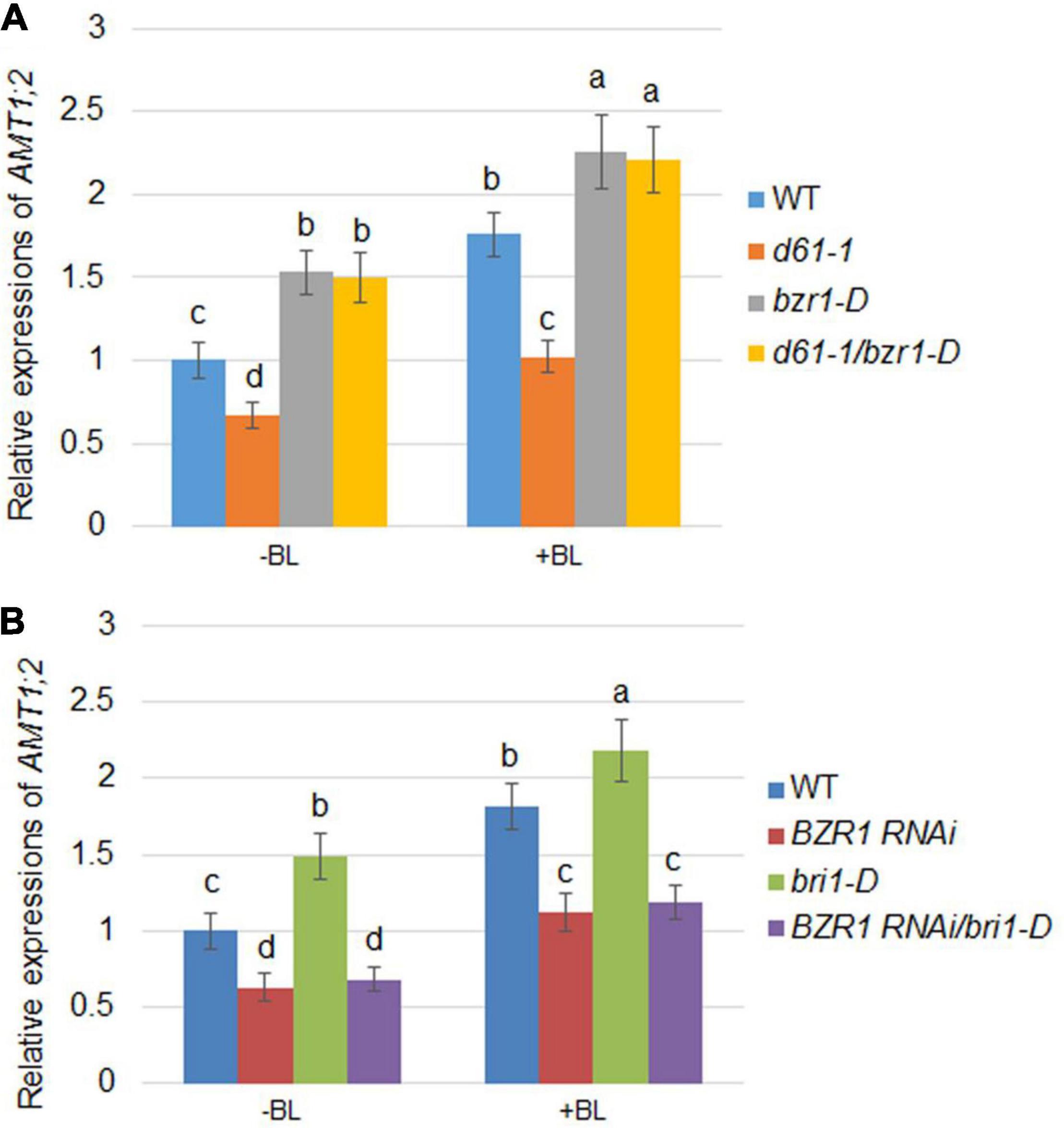
Figure 5. BR-induced AMT1;2 expression in the genetic combinations between BZR1 and BRI1. (A) The expression levels of AMT1;2 genes in wild-type, BRI1 mutant d61-1, gain-of-function mutant bzr1-D, and their cross offspring d61-1/bzr1-D before and 3 h after BL treatment. (B) The expression levels of AMT1;2 genes in the wild-type, BZR1 RNAi, BRI1 overexpression line bri1-D, and their cross offspring BZR1 RNAi/bri1-D before and 3 h after BL treatment. The above plants were grown in distilled water for 7 days with or without treatment of 100 nM BL for 3 h. The expression levels of AMT1;2 genes were measured by qRT-PCR. Sample mRNA levels were normalized to those of Ubiquitin. Data represent means ± SE (n = 3). The experiments were repeated at least three times. Different letters represent statistically significant differences (P < 0.05).
Since AMT1;2 is significantly regulated by NH4+, we assessed the effects of BRI1 and BZR1 on NH4+-induced AMT1;2 expression. To determine the NH4+-dependent AMT1;2 gene expression, we treated 17-day-old seedlings originally grown in dH2O and N-free nutrient medium with a 0.5 mM (NH4)2SO4 solution. The whole roots of the plant were then collected at 0 and 3 h after treatment. To examine whether BZR1 and BRI1 play the same role in NH4+-dependent expression of AMT1;2 as in BR-dependent induction, BZR1 and BRI1 mutants were used to examine the AMT1;2 expression levels. When compared with wild type, both bzr1-D and d61-1/bzr1-D contained higher mRNA levels of AMT1;2, while those in d61-1 were lower (Figure 6A). Furthermore, the bri1-D lines showed high levels of AMT1;2 mRNA, with reduced AMT1;2 mRNA expression in BZR1 RNAi, regardless of NH4+ treatment. Nevertheless, similar patterns of mRNA expression of AMT1;2 were observed in BZR1 RNAi and bri1-D/BZR1 RNAi (Figure 6B). Taken together, these results indicated that AMT1;2 expression was sensitive to the BZR1 level and that BZR1 acts downstream of BRI1 in this signaling pathway.
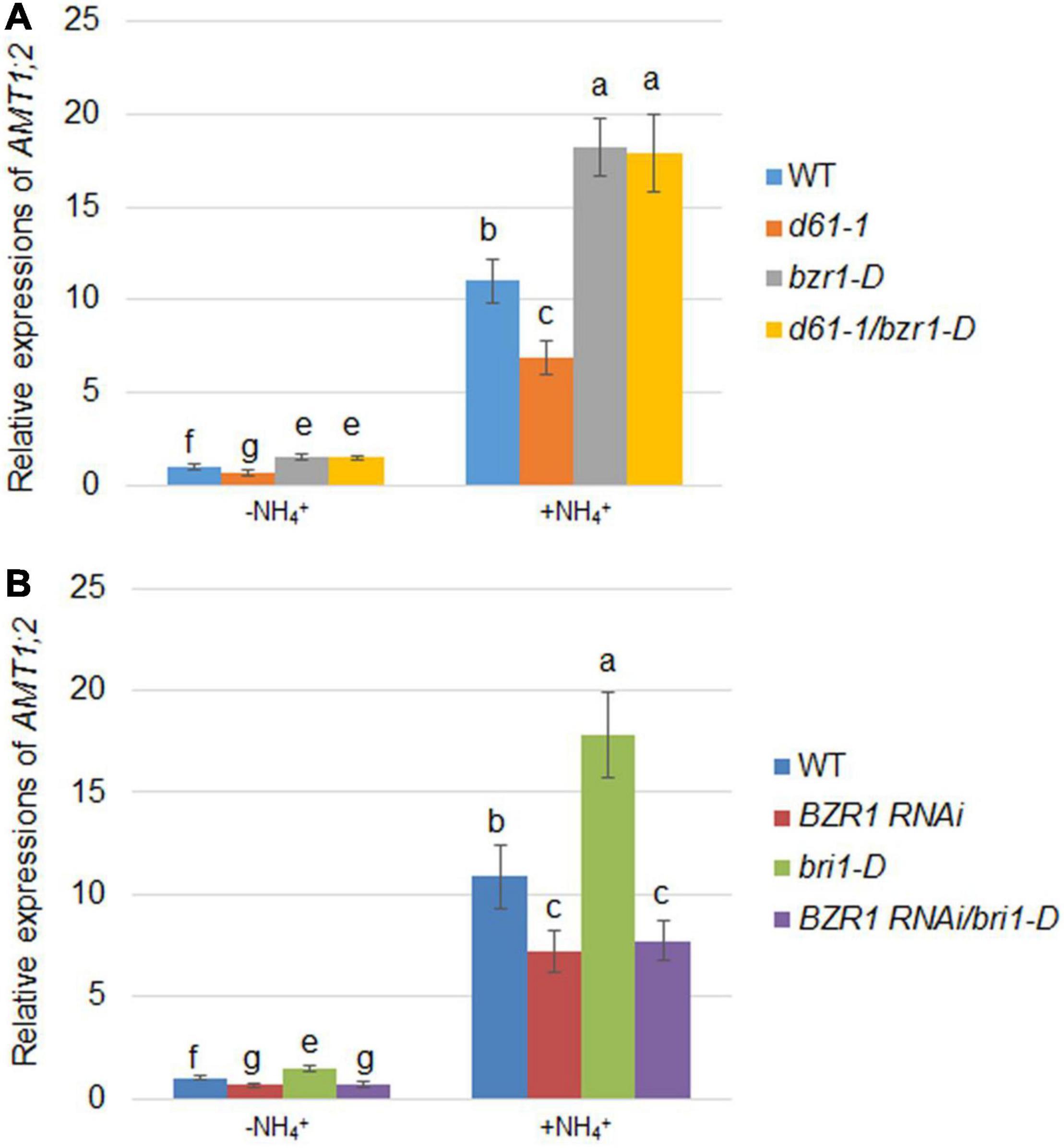
Figure 6. NH4+-induced AMT1;2 expression in BZR1, BRI1, and their genetic combinations. (A) NH4+-induced AMT1;2 expression was detected in the wild-type, BRI1 mutant d61-1, gain-of-function mutant bzr1-D, and their cross offspring d61-1/bzr1-D. (B) NH4+-induced AMT1;2 expression was detected in BZR1 RNAi, BRI1 overexpression line bri1-D, and their cross offspring BZR1 RNAi/bri1-D. Full root sampling was carried out on 17-day-old seedlings that were transferred to nutrient solution containing 0.5 mM (NH4)2SO4 for 0 and 3 h. Measurement of AMT1;2 expression levels using qRT-PCR. Sample mRNA levels were normalized with respect to those of Ubiquitin. Data represent means ± SE (n = 3). The experiments were repeated at least three times. Different letters represent statistically significant differences (P < 0.05).
To determine the effects of BRI1 and BZR1 on the absorption of NH4+ in plant roots, we examined the import of 15N-labeled ammonium in BZR1 and BRI1 genetic combinations. 15NH4+ influx was significantly lower in d61-1, while higher in bzr1-D or d61-1/bzr1-D than in the wild-type (Figure 7A). Also, the 15NH4+ influx was clearly lower in both BZR1 RNAi and BZR1 RNAi/bri1-D, while higher in bri1-D compared to wild-type plants (Figure 7B).
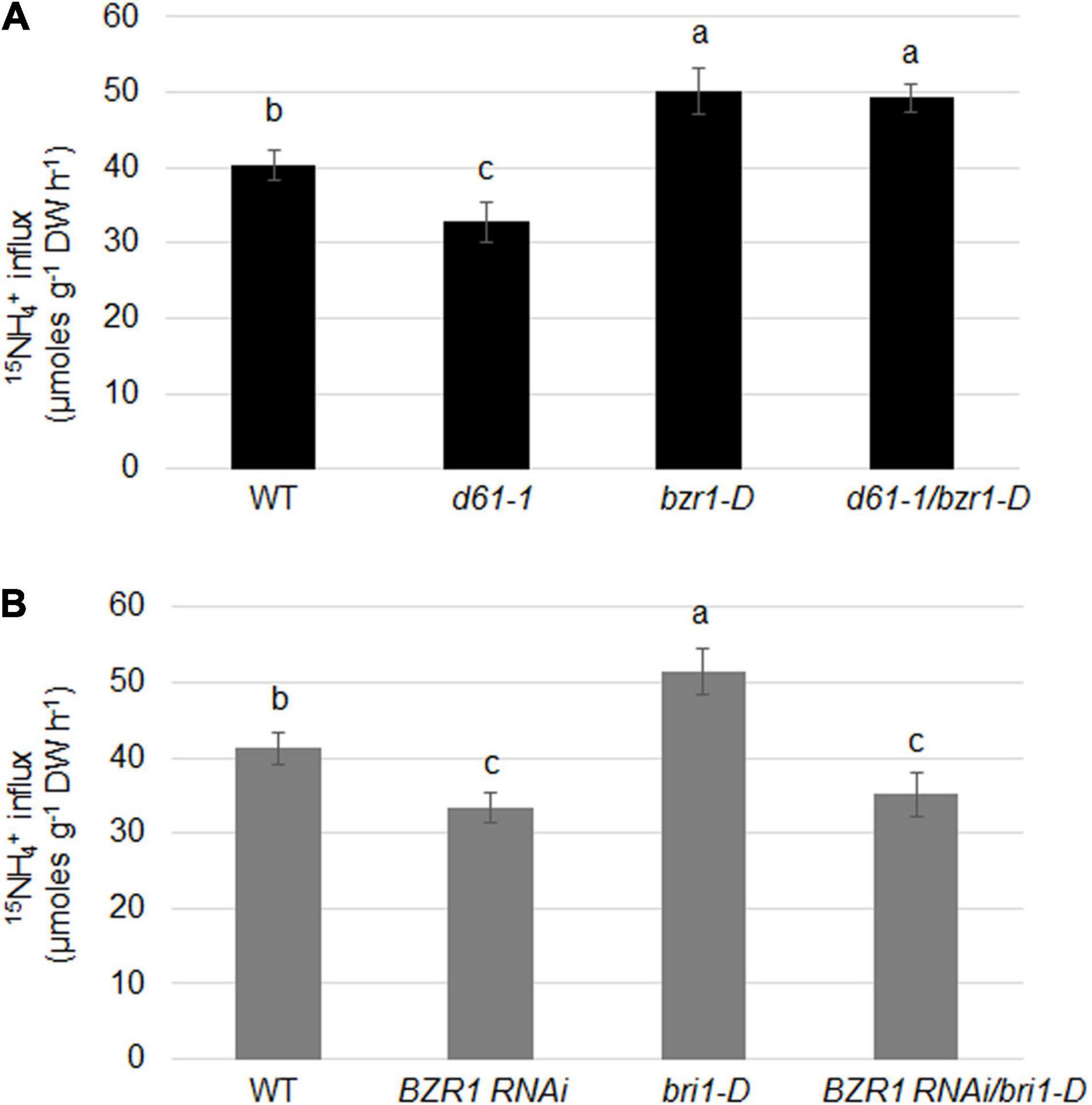
Figure 7. BRI1 and BZR1 effects on NH4+ uptake in plants. (A) 15NH4+ abundance in total 15N in roots of wild-type, BRI1 mutant d61-1, gain-of-function mutant bzr1-D, and their cross offspring d61-1/bzr1-D plants. (B) 15NH4+ abundance in total 15N in roots of wild-type, BZR1 RNAi, BRI1 overexpression line bri1-D, and their cross offspring BZR1 RNAi/bri1-D plants. The 2-week-old hydroponic seedlings were further transferred and cultured in nitrogen-free nutrient solution. Three days later, plant roots were immersed in 200 μM 15N-labeled NH4+ nutrient solution for 6 min. The rate of ammonium uptake by the roots was then determined. 15NH4+ abundance in total 15N in roots was measured after 6 min of 15NH4+ uptake. Bars represent means ± SD (n = 6). The experiments were repeated at least three times. Different letters represent statistically significant differences (P < 0.05).
Although BRs are known for their pleiotropic roles in the regulation of plant growth and development, their effect on nutrient uptake is unclear. The activation of BR signaling has been shown by transcriptome analysis of BR treated wild-type or bes1-D Arabidopsis plants to positively regulate the expression of the AMT gene AtAMT1;1 (Goda et al., 2004; Yu et al., 2011). Also, the BR signaling transcription factor RAVL1 activates AMT1;2 to enhance NH4+ uptake in rice (Xuan et al., 2016). The possibility of a link between BRs and AMT1 expression suggests that the process of root ammonium absorption may be coordinated with the physiological function of BRs in growth stimulation. In this study, we verified this issue by assessing the transcriptional levels of the AMT genes and the role they playing in ammonium uptake by the roots of rice lines, which genes involved in the regulation of BR homeostasis were modulated. We found high levels of BR susceptibility in all these genes which showed upregulated expression in response to BR treatment, especially the NH4+ transporter gene AMT1;2. Combining this result with the phenotype of higher NH4+ uptake, it can be inferred that this BR-dependent regulation is mediated by the direct binding between the transcription factor BZR1 and AMT1;2 promoters. This regulation occurs after sensing BR signals. Thus, AMT1;2-mediated uptake of NH4+ can be seen as a physiological response mediated by BR.
BZR1 plays an important role in the coordination of ammonium uptake and BR signaling pathways. There are three lines of evidence that support this point. First, determination of the transcription level of AMT1 genes in plants treated with exogenous BL showed that the AMT1;2 in roots can respond to BL processing, increasing the level of transcription. BZR1 regulation plays a vital role in this process. Since the AMT1;2 transcription level in BZR1 RNAi seedlings was also lower than that of the wild-type while the transcription level in the bzr1-D plants was higher; this expression pattern was also observed without BL treatment. Second, there was also a correlation between higher AMT1;2 transcription levels and higher NH4+ uptake. We measured the rate of uptake of 15N-labeled NH4+ by rice roots expressing different BZR1 levels. NH4+ uptake rates were compared at 15N abundance and 15N influx levels, and it was found that the NH4+ uptake of BZR1 RNAi was significantly lower than that of the wild-type, which may be explained by down-regulation of AMT1;2. This concept is further supported by the findings in bzr1-D roots, where the enrichment of 15NH4+ was higher than that in the wild-type. Therefore, in the presence of ammonium, a non-significant increase in NH4+ influx may be related to the down regulation of AMT mRNA levels (Yuan et al., 2007) or AMT protein activity (Lanquar et al., 2009; Yuan et al., 2013) at posttranscriptional or posttranslational levels. Furthermore, increased BZR1 expression levels were also associated with increased ammonium abundance in roots. We observed that all these NH4+ uptake-related traits were affected in these lines with or without additional BR treatment. This observation suggests that BZR1 is a positive regulator of NH4+ uptake under any condition conducive to plant growth. To sum up, BZR1 is the coordination center between NH4+ absorption and general growth promotion mediated by BRs. AMT1;2 transcriptional level changes are significantly affected by BZR1 levels, suggesting a BZR1-dependent ammonium uptake pathway in roots. We have done further research on whether BZR1 directly regulates AMT1;2 expression. Nine motifs were identified in the 2.5 kb promoter region of AMT1;2 by promoter sequence analysis. EMSA, CHIP, and transient gene expression tests were used to further study the interaction between AMT1;2 and BZR1. The two BRRE motifs in the promoter have been shown to be sites that bind BZR1 and directly activate AMT1;2 transcription. In addition, although BZR1 can bind both BRRE motifs, the binding strength is different. It is likely that the positions of BRRE motifs in the promoter plays an important role in the regulation of AMT1;2. The comparison of BR- and NH4+-mediated expression of AMT1;2 in BZR1 RNAi, bzr1-D, and wild-type plants indicates that constitutive activation of AMT1;2 expression via BZR1 is independent of external signal stimulation. BRI1 performs a receptor function in BR signaling pathways and actively regulates BR-dependent AMT1;2 expression through its signal reception. In addition, the roots of d61-1, the BRI1 mutant, accumulate less NH4+ than the corresponding wild-type, suggesting a positive role of BRI1 in regulating AMT-mediated NH4+ uptake in roots. The key BR signaling regulators BZR1, the BR receptor BRI1 play the putative role in the regulation of AMT1;2 gene expression. Therefore, clarifying their inter-relationships enhances our understanding of the relationship between BR signaling and the mechanism of AMT regulation. We measured BR- or NH4+-mediated AMT1;2 gene expression levels and 15NH4+ absorption in three rice lines with different combinations of BZR1 and BRI1 gene expression levels, including the line bzr1-D in the d61-1 background and the line bri1-D in the BZR1 RNAi background. The activation of BRI1-induced AMT1;2 requires BZR1 involvement, while the process of activation by BZR1 does not require BRI1 activity. Therefore, BZR1 plays a key role in coordinating root uptake of ammonium and BR-dependent plant growth gene expression regulation. Our study further elucidated a relationship between the two key factors BZR1 and BRI1 in the BR signaling pathway associated with regulation of AMT1;2 gene expression and demonstrated that BZR1 was localized downstream of the BRI1 during regulation of AMT1;2 expression. It has been reported that BZR1 act as an integrator or master regulator to regulate plant growth, development, and immunity by directly interacting with key proteins from hormone signaling, stress signaling, cell elongation, flowering, immune signaling and so on (Eunkyoo et al., 2012; Lozano-Duran et al., 2013; Eunkyoo et al., 2014; Li and He, 2015; Nolan et al., 2017; Tian et al., 2018). However, whether BZR1 functions as heterodimer to regulate AMT1;2 needs to be further analyzed.
Ammonium strongly induces expression of AMT1;1 and AMT1;2, the two major AMTs in rice roots. However, the molecular mechanism of ammonium-mediated AMT1 gene expression is not clear. The present results on the effect of BRI1 on NH4+-induction of these genes suggest a complex mechanism for the regulation of AMT1 expression. Nonetheless, our results further support the existence of a common genetic component between plant nutrient uptake and the hormonal regulatory mechanisms involved in plant growth and development. In the current study, we found that although AMT1;1 responded to BR treatment and its expression was induced in roots, BZR1 does not appear to have the ability to regulate AMT1;1 expression. Different expression patterns were observed during the response of each member of the AMT1 gene family to BR processing. These findings suggest that different BR-mediated regulatory circuits may have different effects on the same AMT1 gene.
In addition, a recent study has shown that NH4+-induced miR444 positively regulates BR biosynthesis to regulate rice root growth. This microRNA targets five MADS-box transcription repressors directly upstream of BR-deficient dwarf 1 (OsBRD1), a key BR biosynthetic gene (Jiao et al., 2020), suggesting NH4+ signaling also activates BR biosynthesis. Together with our results, we propose that there is a complicated feedback regulatory mechanism between NH4+ and BR signaling. It would be useful to investigate the effects of NH4+ on BZR1 protein levels or modifications in the future. Our study also found a highly complex interaction between plant hormone signaling and nutrient absorption pathways. These complex regulatory networks involved in ammonium uptake in plants require further genetic and molecular studies to elucidate their detailed mechanisms.
Brassinosteroids play diverse functions in plant growth and development. In this study, we examined the role of BRs in ammonium uptake in rice. The data indicate that BR signaling activates AMT1;1 and AMT1;2 expression in the presence of the BR receptor BRI1, and the BR signaling transcription factor BZR1 directly activates AMT1;2. The further genetic study revealed that BZR1 activates AMT1;2 expression downstream of BRI1 to improve ammonium uptake in rice. These results indicate that BR signaling positively controls ammonium uptake partially via BZR1-mediated activation of AMT1;2, one of the key AMTs. Our analyses extend the knowledge of the BR-regulating module and its role in the regulation of nitrogen uptake in rice plants.
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
SY, YZ, and DY conceived this project. QS and YX provided the experimental design ideas and plant materials. YZ, SY, and DY carried out the experiments and generated the data. QS, YX, and DY contributed to the summary and analysis of the data. QS, YX, YZ, and SY wrote the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the Support Plan for Innovative Talents in Colleges and Universities of Liaoning Province (LR2017037) and Natural Science Foundation of Liaoning Province (2020-YQ-05).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abiko, T., Obara, M., Ushioda, A., Hayakawa, T., Hodges, M., and Yamaya, T. (2005). Localization of NAD-isocitrate dehydrogenase and glutamate dehydrogenase in rice roots: candidates for providing carbon skeletons to NADH-glutamate synthase. Plant Cell Physiol. 46, 1724–1734. doi: 10.1093/pcp/pci188
Bai, M. Y., Zhang, L. Y., Gampala, S. S., Zhu, S. W., Song, W. Y., Chong, K., et al. (2007). Functions of OsBZR1 and 14-3-3 proteins in brassinosteroid signaling in rice. Proc. Natl. Acad. Sci. U.S.A. 104, 13839–13844. doi: 10.1073/pnas.0706386104
Bao, A., Liang, Z., Zhao, Z., and Cai, H. (2015). Overexpressing of OsAMT1-3, a high affinity ammonium transporter gene, modifies rice growth and carbon-nitrogen metabolic status. Int. J. Mol. Sci. 16, 9037–9063. doi: 10.3390/ijms16059037
Eunkyoo, O., Zhu, J. Y., Bai, M. Y., Augusto, A. R., Yu, S., and Wang, Z. Y. (2014). Cell elongation is regulated through a central circuit of interacting transcription factors in the Arabidopsis hypocotyl. eLife 3:e03031. doi: 10.7554/eLife.03031
Eunkyoo, O., Zhu, J. Y., and Wang, Z. Y. (2012). Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat. Cell Biol. 14, 802–809. doi: 10.1038/ncb2545
Giehl, R. F. H., Laginha, A. M., Duan, F., Rentsch, D., Yuan, L., and von Wiren, N. (2017). A critical role of AMT2;1 in root-to-shoot translocation of ammonium in Arabidopsis. Mol. Plant 10, 1449–1460. doi: 10.1016/j.molp.2017.10.001
Goda, H., Sawa, S., Asami, T., Fujioka, S., Shimada, Y., and Yoshida, S. (2004). Comprehensive comparison of auxin-regulated and brassinosteroid-regulated genes in Arabidopsis. Plant Physiol. 134, 1555–1573. doi: 10.1104/pp.103.034736
Guo, H., Li, L., Aluru, M., Aluru, S., and Yin, Y. (2013). Mechanisms and networks for brassinosteroid regulated gene expression. Curr. Opin. Plant Biol. 16, 545–553. doi: 10.1016/j.pbi.2013.08.002
Je, B. I., Piao, H. L., Park, S. J., Park, S. H., Kim, C. M., Xuan, Y. H., et al. (2010). RAV-Like1 maintains brassinosteroid homeostasis via the coordinated activation of BRI1 and biosynthetic genes in rice. Plant Cell 22, 1777–1791. doi: 10.1105/tpc.109.069575
Jeong, D. H., An, S., Kang, H. G., Moon, S., Han, J. J., Park, S., et al. (2002). T-DNA insertional mutagenesis for activation tagging in rice. Plant Physiol. 130, 1636–1644. doi: 10.1104/pp.014357
Jiao, X., Wang, H., Yan, J., Kong, X., Liu, Y., Chu, J., et al. (2020). Promotion of BR biosynthesis by miR444 is required for ammonium-triggered inhibition of root growth. Plant Physiol. 182, 1454–1466. doi: 10.1104/pp.19.00190
Kim, T. W., and Wang, Z. Y. (2010). Brassinosteroid signal transduction from receptor kinases to transcription factors. Annu. Rev. Plant Biol. 61, 681–704. doi: 10.1146/annurev.arplant.043008.092057
Kumar, A., Silim, S. N., Okamoto, M., Siddiqi, M. Y., and Glass, A. D. (2003). Differential expression of three members of the AMT1 gene family encoding putative high-affinity NH4+ transporters in roots of Oryza sativa subspecies indica. Plant Cell Environ. 26, 907–914. doi: 10.1046/j.1365-3040.2003.01023.x
Lanquar, V., Loque, D., Hormann, F., Yuan, L., Bohner, A., Engelsberger, W. R., et al. (2009). Feedback inhibition of ammonium uptake by a phospho-dependent allosteric mechanism in Arabidopsis. Plant Cell 21, 3610–3622. doi: 10.1105/tpc.109.068593
Li, J., and Chory, J. (1997). A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90, 929–938. doi: 10.1016/S0092-8674(00)80357-8
Li, J., Wen, J., Lease, K. A., Doke, J. T., Tax, F. E., and Walker, J. C. (2002). BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110, 213–222. doi: 10.1016/S0092-8674(02)00812-7
Li, Q. F., and He, J. X. (2015). BZR1 interacts with HY5 to mediate brassinosteroid- and lightregulated cotyledon opening in Arabidopsis in darkness. Mol. Plant 9, 113–125.
Loque, D., and von Wiren, N. (2004). Regulatory levels for the transport of ammonium in plant roots. J. Exp. Bot. 55, 1293–1305. doi: 10.1093/jxb/erh147
Lozano-Duran, R., Macho, A. P., Boutrot, F., Segonzac, C., Somssich, I. E., and Zipfel, C. (2013). The transcriptional regulator BZR1 mediates trade-off between plant innate immunity and growth. elife 2:e00983. doi: 10.7554/eLife.00983
Nam, K. H., and Li, J. (2002). BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 110, 203–212. doi: 10.1016/S0092-8674(02)00814-0
Nolan, T. M., Brennan, B., Yang, M., Chen, J., Zhang, M., Li, Z., et al. (2017). Selective autophagy of BES1 mediated by DSK2 balances plant growth and survival. Dev. Cell 41, 33–46. doi: 10.1016/j.devcel.2017.03.013
Oliveira, I. C., Brears, T., Knight, T. J., Clark, A., and Coruzzi, G. M. (2002). Overexpression of cytosolic glutamine synthetase. Relation to nitrogen, light, and photorespiration. Plant Physiol. 129, 1170–1180. doi: 10.1104/pp.020013
Qiao, S., Sun, S., Wang, L., Wu, Z., Li, C., Li, X., et al. (2017). The RLA1/SMOS1 Transcription factor functions with OsBZR1 to regulate brassinosteroid signaling and rice architecture. Plant Cell 29, 292–309. doi: 10.1105/tpc.16.00611
Ranathunge, K., El-Kereamy, A., Gidda, S., Bi, Y. M., and Rothstein, S. J. (2014). AMT1;1 transgenic rice plants with enhanced NH4(+) permeability show superior growth and higher yield under optimal and suboptimal NH4(+) conditions. J. Exp. Bot. 65, 965–979. doi: 10.1093/jxb/ert458
Ren, Y., Tian, X., Li, S., Mei, E., He, M., Tang, J., et al. (2020). Oryza sativa mediator subunit OsMED25 interacts with OsBZR1 to regulate brassinosteroid signaling and plant architecture in rice. J. Integr. Plant Biol. 62, 793–811. doi: 10.1111/jipb.12914
Sonoda, Y., Ikeda, A., Saiki, S., von Wiren, N., Yamaya, T., and Yamaguchi, J. (2003a). Distinct expression and function of three ammonium transporter genes (OsAMT1;1-1;3) in rice. Plant Cell Physiol. 44, 726–734. doi: 10.1093/pcp/pcg083
Sonoda, Y., Ikeda, A., Saiki, S., Yamaya, T., and Yamaguchi, J. (2003b). Feedback regulation of the ammonium transporter gene family AMT1 by glutamine in rice. Plant Cell Physiol. 44, 1396–1402. doi: 10.1093/pcp/pcg169
Straub, T., Ludewig, U., and Neuhauser, B. (2017). The Kinase CIPK23 inhibits ammonium transport in Arabidopsis thaliana. Plant Cell 29, 409–422. doi: 10.1105/tpc.16.00806
Suenaga, A., Moriya, K., Sonoda, Y., Ikeda, A., Von Wiren, N., Hayakawa, T., et al. (2003). Constitutive expression of a novel-type ammonium transporter OsAMT2 in rice plants. Plant Cell Physiol. 44, 206–211. doi: 10.1093/pcp/pcg017
Tian, Y., Min, F., Qin, Z., Lv, H., and Bai, M. Y. (2018). Hydrogen peroxide positively regulates brassinosteroid signaling through oxidation of the BRASSINAZOLE-RESISTANT1 transcription factor. Nat. Commun. 9:1063. doi: 10.1038/s41467-018-03463-x
Tong, H., and Chu, C. (2018). Functional specificities of brassinosteroid and potential utilization for crop improvement. Trends Plant Sci. 23, 1016–1028. doi: 10.1016/j.tplants.2018.08.007
Wang, Z. Y., Nakano, T., Gendron, J., He, J., Chen, M., Vafeados, D., et al. (2002). Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev. Cell 2, 505–513. doi: 10.1016/S1534-5807(02)00153-3
Xuan, Y. H., Duan, F. Y., Je, B. I., Kim, C. M., Li, T. Y., Liu, J. M., et al. (2016). Related to ABI3/VP1-Like 1(RAVL1) regulates brassinosteroid-mediated activation of AMT1;2 in rice (Oryza sativa). J. Exp. Bot. 68, 727–737. doi: 10.1093/jxb/erw442
Xuan, Y. H., Priatama, R. A., Huang, J., Je, B. I., Liu, J. M., Park, S. J., et al. (2013). Indeterminate domain 10 regulates ammonium-mediated gene expression in rice roots. New Phytol. 197, 791–804. doi: 10.1111/nph.12075
Yamaguchi, M., Ohtani, M., Mitsuda, N., Kubo, M., Ohme-Takagi, M., Fukuda, H., et al. (2010). VND-INTERACTING2, a NAC domain transcription factor, negatively regulates xylem vessel formation in Arabidopsis. Plant Cell 22, 1249–1263. doi: 10.1105/tpc.108.064048
Yamamuro, C., Ihara, Y., Wu, X., Noguchi, T., Fujioka, S., Takatsuto, S., et al. (2000). Loss of function of a rice brassinosteroid insensitive1 homolog prevents internode elongation and bending of the lamina joint. Plant Cell 12, 1591–1605. doi: 10.2307/3871176
Yang, C. J., Zhang, C., Lu, Y. N., Jin, J. Q., and Wang, X. L. (2011). The mechanisms of brassinosteroids’ action: from signal transduction to plant development. Mol. Plant 4, 588–600. doi: 10.1093/mp/ssr020
Yin, Y., Wang, Z. Y., Mora-Garcia, S., Li, J., Yoshida, S., Asami, T., et al. (2002). BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 109, 181–191. doi: 10.1016/S0092-8674(02)00721-3
Yoo, S.-D., Cho, Y.-H., and Sheen, J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572. doi: 10.1038/nprot.2007.199
Yu, X., Li, L., Zola, J., Aluru, M., Ye, H., Foudree, A., et al. (2011). A brassinosteroid transcriptional network revealed by genome-wide identification of BESI target genes in Arabidopsis thaliana. Plant J. 65, 634–646. doi: 10.1111/j.1365-313X.2010.04449.x
Yuan, L., Gu, R., Xuan, Y., Smith-Valle, E., Loque, D., Frommer, W. B., et al. (2013). Allosteric regulation of transport activity by heterotrimerization of Arabidopsis ammonium transporter complexes in vivo. Plant Cell 25, 974–984. doi: 10.1105/tpc.112.108027
Keywords: brassinosteroids, ammonium uptake, BZR1, AMT1;2, rice
Citation: Yang S, Yuan DP, Zhang Y, Sun Q and Xuan YH (2021) BZR1 Regulates Brassinosteroid-Mediated Activation of AMT1;2 in Rice. Front. Plant Sci. 12:665883. doi: 10.3389/fpls.2021.665883
Received: 09 February 2021; Accepted: 10 May 2021;
Published: 17 June 2021.
Edited by:
Sakiko Okumoto, Texas A&M University, United StatesReviewed by:
Timothy O. Jobe, University of Cologne, GermanyCopyright © 2021 Yang, Yuan, Zhang, Sun and Xuan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qian Sun, c3VucWlhbjUzMjhAc3lhdS5lZHUuY24=; Yuan Hu Xuan, eHVhbnl1YW5odTExNUBzeWF1LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.