- 1Department of Applied Biosciences, Kyungpook National University, Daegu, South Korea
- 2Department of Botany, Abdul Wali Khan University, Mardan, Pakistan
- 3Natural and Medical Science Research Center, University of Nizwa, Nizwa, Oman
Salinity has drastically reduced crop yields and harmed the global agricultural industry. We isolated 55 bacterial strains from plants inhabiting the coastal sand dunes of Pohang, Korea. A screening bioassay showed that 14 of the bacterial isolates secreted indole-3-acetic acid (IAA), 12 isolates were capable of exopolysaccharide (EPS) production and phosphate solubilization, and 10 isolates secreted siderophores. Based on our preliminary screening, 11 bacterial isolates were tested for salinity tolerance on Luria–Bertani (LB) media supplemented with 0, 50, 100, and 150 mM of NaCl. Three bacterial isolates, ALT11, ALT12, and ALT30, had the best tolerance against elevated NaCl levels and were selected for further study. Inoculation of the selected bacterial isolates significantly enhanced rice growth attributes, viz., shoot length (22.8–42.2%), root length (28.18–59%), fresh biomass (44.7–66.41%), dry biomass (85–90%), chlorophyll content (18.30–36.15%), Chl a (29.02–60.87%), Chl b (30.86–64.51%), and carotenoid content (26.86–70%), under elevated salt stress of 70 and 140 mM. Furthermore, a decrease in the endogenous abscisic acid (ABA) content (27.9–23%) and endogenous salicylic acid (SA) levels (11.70–69.19%) was observed in inoculated plants. Antioxidant analysis revealed an increase in total protein (TP) levels (42.57–68.26%), whereas it revealed a decrease in polyphenol peroxidase (PPO) (24.63–34.57%), glutathione (GSH) (25.53–24.91%), SOA (13.88–18.67%), and LPO levels (15.96–26.06%) of bacterial-inoculated plants. Moreover, an increase in catalase (CAT) (26–33.04%), peroxidase (POD) (59.55–78%), superoxide dismutase (SOD) (13.58–27.77%), and ascorbic peroxidase (APX) (5.76–22.74%) activity was observed. Additionally, inductively coupled plasma mass spectrometry (ICP-MS) analysis showed a decline in Na+ content (24.11 and 30.60%) and an increase in K+ (23.14 and 15.45%) and Mg+ (2.82 and 18.74%) under elevated salt stress. OsNHX1 gene expression was downregulated (0.3 and 4.1-folds), whereas the gene expression of OsPIN1A, OsCATA, and OsAPX1 was upregulated by a 7–17-fold in bacterial-inoculated rice plants. It was concluded that the selected bacterial isolates, ALT11, ALT12, and ALT30, mitigated the adverse effects of salt stress on rice growth and can be used as climate smart agricultural tools in ecofriendly agricultural practices.
Introduction
Soil salinity is abiotic stress that significantly limits agricultural productivity and food security (Shrivastava and Kumar, 2015; Kumar et al., 2020). According to estimates, there is more than 830 million ha of salt-affected agricultural land globally. Of the 230 million ha of irrigated land worldwide, 45 million ha (20%) has been affected by high salt concentrations (Munns, 2005; Hoang et al., 2016). It is predicted that 50% of arable land will be threatened by 2050 due to soil salinization (Khan et al., 2019b,e; Shultana et al., 2020a). The salinization of agricultural lands occurs because of the accumulation of salts in the soil, particularly sodium and chloride ions, which leads to hypertonic stress (Kumar et al., 2020). High sodium (Na+) accumulation limits water conductance, disturbs the nutrient balance, and negatively impacts intercellular potassium (K+) influx, with K being an essential element required for plant growth (Kumar et al., 2020). Plants under salinity stress undergo several morphological changes, such as reduced seed germination, seedling growth and yield, and related physiological and molecular changes, which impede their growth and development. Additionally, high salinity stress causes oxidative stress and enhances the production of reactive oxygen species (ROS), such as singlet oxygen (1O2), superoxide anion (O−2), and hydrogen peroxide (H2O2). High ROS generation damages cell membranes, lipids, and nucleic acid, and leads to programmed cell death (Jha and Subramanian, 2013; Habib et al., 2016; Khan et al., 2018; Santos et al., 2018). To reduce the toxic effect of salinity, the plant antioxidant system, including enzymatic [superoxide dismutase (SOD), peroxidase (POD), and catalase (CAT)] and non-enzymatic [glutathione (GSH), total protein (TP)] antioxidants, has to be activated to control the biosynthesis of ROS and maintain them at a low level (Jha and Subramanian, 2013; Habib et al., 2016; Santos et al., 2018; Khan et al., 2020b). SOD is a metalloenzyme that protects cells from oxidative damage, and it catalyzes the conversion of superoxide radicals to H2O2, whereas ascorbic peroxidase (APX) and CAT break down H2O2 to H2O and O2 produced by SOD (Santos et al., 2000; Jaleel et al., 2009; Habib et al., 2016).
Plants respond to salinity stress in a complex manner involving hormonal regulation, gene expression, and signal pathways (Jamil et al., 2011). Phytohormones are small chemicals that play a crucial role in plant growth and development (Yu et al., 2020). During stress conditions, plant endogenous abscisic acid (ABA), and salicylic acid (SA) are produced as a part of a stress response (Khan et al., 2020c; Yu et al., 2020). ABA plays an irreplaceable role in plant tolerance and adaptation to a variety of stresses, and it accumulates in plants under stress, stimulating stomatal closure, adaptive physiological responses, and change in gene expression (Xiong et al., 2002; Kim et al., 2010; Li et al., 2010; Sah et al., 2016; Khan et al., 2020c; Yu et al., 2020). SA is an endogenous growth regulator phytohormone that induces stress tolerance in plants, including salinity stress (Sakhabutdinova et al., 2003; Ali et al., 2015; Yu et al., 2020). Besides participating in plant biotic stresses, SA plays an important role in plant salt tolerance by enhancing antioxidant systems, synthesizing osmolytes, and promoting photosynthesis and plant growth under salt stress (Filgueiras et al., 2019; Ahanger et al., 2020; Khan et al., 2020b; Yu et al., 2020). The regulation of genes is one of the key phenomena in plants by which they respond and try to adapt to salt stress (Jamil et al., 2011). In rice, OsNHX1 is the most abundant Na+/H+ antiporter, and its gene expression is induced by different abiotic stresses, namely, high salinity and drought (Fukuda et al., 2004; Almeida et al., 2017). The phytohormone auxin regulates a variety of developmental processes in plant growth and root system architecture. A number of auxin influx genes, such as PIN and YUCCA, play a vital role in auxin biosynthesis, and they have been isolated, characterized, and can be exploited to help the plant adapt to various adverse environments, including high salinity environments (Khan et al., 2020a).
In the past three decades, diverse strategies, such as plant genetic engineering and molecular marker-assisted breeding approaches, have been used to develop saline-tolerant crops (Hamayun et al., 2017; Khan et al., 2019a, 2020a). However, these approaches are insufficient, labor-intensive, and time-consuming (Kumar et al., 2020). Currently, the use of plant growth-promoting bacteria (PGPB) to ameliorate abiotic stress is gaining importance in agricultural biotechnology and momentum for consideration (Nautiyal et al., 2013; Santos et al., 2018) by various direct and indirect mechanisms, such as the production of indole-3-acetic acid (IAA), exopolysaccharide (EPS) and organic acid siderophore, and phosphate solubilization (Nautiyal et al., 2013). These beneficial plant–microbe interactions are very frequent in nature and act as elicitors for tolerance to abiotic stress, including salinity stress (Nautiyal et al., 2013; Shultana et al., 2020a). Previously published reports showed that PGPB has been used for the mitigation of salinity stress in cucumber (Kang et al., 2014; Kartik et al., 2020), tomato (Kang et al., 2019), soybean (Khan et al., 2019b,c), rice (Nautiyal et al., 2013; Mukherjee et al., 2019) lettuce, and maize (Rojas-Tapias et al., 2012; Rafiq et al., 2020) plants.
Rice is a semiaquatic crop and the third most commonly valuable cereal crop after wheat and maize, feeding more than one-half of the world's population (Hoang et al., 2016). Rice is susceptible to salt stress when it is a seedling and during the early vegetative stage, and its productivity declines at low salt concentrations (Hoang et al., 2016; Tisarum et al., 2020). Hoang et al. (2016) reported that 10–50% yield loss was recorded at ECe 3.5–7.2 dSm−1. Several studies have emphasized the use of beneficial microorganisms, such as Azospirillum, Bacillus, Rhizobium, Serratia, and Pseudomonas, which can tolerate salinity stress and enhance the growth and productivity of crop plants (Nautiyal et al., 2013; Kang et al., 2014, 2019; Santos et al., 2018; Khan et al., 2019a,b). Therefore, we hypothesized that halotolerant microbes with multiple plant growth-promoting traits could be used in climate smart agriculture to enhance plant tolerance against salinity stress. Thus, we isolated bacteria and screened them for different plant growth promotion (PGP) traits, including IAA, siderophore production, EPS production, tolerance to NaCl stress, and identified isolates ALT11, ALT12, and ALT30 based on halotolerance and multiple PGP traits. The objective of this study was to understand the effect of selected isolates upon inoculation on the growth and chlorophyll content of rice under normal conditions and salinity stress of 70 and 140 mM. Furthermore, to elucidate the translocation of Na+, K+, and magnesium (Mg+) uptake, the production of various antioxidant (SOD, CAT, POD, APX) and non-enzymatic antioxidants [GSH, LPO, TP, polyphenol peroxidase (PPO)] was determined. Moreover, the expression pattern of salt-related genes under salinity stress in rice plants inoculated with or without selected bacterial isolates was investigated.
Materials and Methods
General Procedure
Plant roots with adhered soil were collected from Artemisia princeps, Chenopodium ficifolium, Oenothera biennis, and Echinochloa crus-galli growing in the sand dunes at Pohang beach. For bacterial isolation, root samples were preserved inside an icebox, and subsequent analyses were undertaken at the Crop Physiology Laboratory, Department of Applied Biosciences, Kyungpook National University, Korea. Plant roots with adhered soil were transferred into a conical flask (99 mL sterilized distilled water and shaken for 3 min). The samples were diluted through a series of 1-fold dilutions, and 0.1 mL of solution was spread on LB agar plates and incubated at 28°C. Bacterial colonies were collected based on their colony morphology, and they were preserved in 75% glycerol stock until further use.
Screening of Bacterial Isolates for Their Plant Growth-Promoting Capacity
Bacterial isolates were screened for different PGP characteristics, including IAA, siderophore production, phosphate solubilization, and organic acid and EPS production. For IAA production, a Salkowski reagent was used by adding 1 mL of supernatant and 1 mL of Salkowski reagent for 30 min in the dark. The development of a pink color indicated IAA production (Kang et al., 2020). Trypticase soy agar medium supplemented with Ca3(PO4)2 was used for phosphate solubilization, and the plates were incubated at 30°C for 7 days, and the formation of transparent halos around each colony was observed (Kubi et al., 2021). For siderophore production, chromeazurol “S” agar plates were incubated at 30°C until the appearance of orange halos, which contrasted with the blue background (Khan et al., 2020a). However, for determining bacterial EPS production, a Congo red assay [LB broth (25 g/L, sucrose (5%), Congo red (0.8 g/L)] was incubated for 4 days at 30°C (Kim et al., 2020).
Molecular Identification of Selected Bacterial Isolates
The selected bacterial strains were identified using the 16S rRNA gene through PCR using universal primer [27F primer (5′-AGAGTTTGATC (AC) TGGCT CAG-3′) and 1492R primer (5′-CGG(CT)TACCTTGTTA CGACTT-3′)]. BLASTn searching of EzTaxon and NCBI based on PCR amplification and sequencing of the 16S rRNA gene region was conducted to identify these bacterial strains at the molecular level. The BLASTn search identified isolates with 100% homology to their respective species. Additionally, the maximum-likelihood (ML) method was used to construct a phylogenetic tree for 16S with MEGA 10 after sequence alignment using Clustal W (version 7.222). To obtain a consensus tree and confirm the molecular identification of these strains, the respective sequences of the 16S rRNA gene region showing similarity to our endophytic isolates were aligned at 1,000 bootstrap replications.
Indole-3-Acetic Acid and Organic Acid Production by Bacterial Isolates
The selected bacterial isolates were grown in LB media (10 g tryptone, 5 g yeast extract, pH 7.2) and centrifuged (5,000 × g; 10 min). The culture filtrates were centrifuged after 3 days to separate cells into a free culture, which were further acidified to a pH of 2.8 and supplemented with 40 μL (D5)-IAA as an IAA internal standard by following the standard protocol (Jan et al., 2019). The acidified and standard supplemented free culture cells were extracted, methylated, and injected into a GC/MS-SIM (6890N network GC system and 5,973 network mass selective detector; Agilent Technologies, the United States) for the identification and quantification of IAA (Sahile et al., 2021). For organic acid analysis, the cultural broth was filtered [0.22 um SmartPor Syrange Filter (P/N SPU0213-1)] and 10 ul was used to conduct high-performance liquid chromatography (HPLC; Waters 600, Milford, MA, the United States). The retention time and peak area were compared using the standard from Sigma-Aldrich, the United States (Khan et al., 2020c).
Bioassay for Growth Promotion Under Salt Stress
Seeds of rice (Oryza sativa L. var. Jin-so-mi) were surface-sterilized and incubated, and uniformly germinated seedlings were transferred to sterilized pots filled with autoclaved rice paddy purchased from Korea (Adhikari et al., 2020). The entire experiment was conducted in a growth chamber: day/night cycle 14 h at 30°C/10 h at 25°C; relative humidity 60–70%; light intensity 250 μmol/m−2s−1. The experiments consisted of (A) control rice plants (Control; only distilled water); (B) rice plants inoculated with ALT11, ALT12, and ALT30; (C) 70 mM NaCl stress; (D) 70 mM NaCl stress with the inoculation of isolates ALT11, ALT12, and ALT30; (E) 140 mM NaCl stress; and (F) 140 mM NaCl stress with the inoculation of isolates ALT11, ALT12, and ALT30. Bacterial culture (grown in LB media) was kept in a 250-ml bottle and centrifuged at 6,000 × g (10 min; 4°C). The pellets obtained were diluted with sterilized distilled water. Each pot was inoculated with 50 ml of freshly diluted bacterial culture (108 CFU), and this inoculation was repeated another three times after a further 5 days. After 15 days, 80 ml of salt solution (70 and 140 mM NaCl) was given to each pot for 7 days. Two weeks after the salt stress treatment, the plants were harvested and morphological parameters, such as root length, shoot length, fresh and dry biomass, and chlorophyll content (SPAD-502, Konica Minolta, Japan), were measured. The leaf chlorophyll (Chl a & Chl b) and carotenoid contents were determined following the standard protocol (Khan et al., 2020c, 2021). The absorbance readings of the Chl a, Chl b, and carotenoid contents were taken at wavelengths of 663, 645, and 580 nm using a spectrophotometer.
Chlorophyll a (mg/g FW) = [{(12.7*A663) – (2.69*A645)} 1000*W]*V
Chlorophyll b (mg/g FW) = [{(22.9*A645) – (4.68*A663)} / 1000*W]*V
Carotenoids (μg/g FW) = A480+(0.638*A663) – (0.638*A645) Chlorophyll a, b and carotenoid contents were calculated using the following formulae:
where A+ Absorbance at respective wave length; W = fresh weight and V = extraction volume.
Each treatment was replicated three times, and each replica has five pots having one plant in each pot. The whole experiment was independently repeated three times.
Estimation of Enzymatic and Non-enzymatic Antioxidant Activities
For antioxidant analysis, leaves were homogenized in a 50 mM phosphate buffer (pH 7.5) containing 1.0% (w/v) polyvinylpyrrolidone (PVP), 0.1 mM EDTA, and 0.5% (w/v) Triton X-100. SOD activities were measured spectrophotometrically (Multiskan GO; Thermo Fisher Scientific, Waltham, MA, the United States) at 560 nm following the procedure described in the study by Marklund and Marklund (1974). GSH was measured following the procedure described in the study by Asaf et al. (2017a). Briefly, 0.2 g samples were ground and homogenized in 3 mL of 5% trichloroacetic acid (TAC). Then, they were centrifuged, the supernatant (0.1 mL) was mixed with 150 mM monosodium phosphate buffer (3 mL) and Ellman's reagent (0.5 mL), and this was measured spectrophotometrically at 412 nm. CAT activity was determined following the procedure described in the study by Radhakrishnan and Lee (2013), and the resulting absorbance was measured at a wavelength of 240 nm. Similarly, PPO was estimated using the same reaction mixture of POD excluding H2O2, and the resulting reaction was measured at 420 nm wavelength (Khan et al., 2019e). Additionally, superoxide anions (SOAs) were measured following the procedure described in the study by Khan et al. (2018) and Khan et al. (2019d) at 580 nm, lipid peroxidation was assayed following the procedure described in the study by Khan et al. (2020b) measured at 532 nm wavelength, and leaf APX activity was estimated at 290 nm following the procedure described in the study by Kang et al. (2019).
Quantification of Endogenous Phytohormones Under Salt Stress
The endogenous phytohormones ABA and SA were quantified. For SA analysis, 0.2 g of freeze-dried sample was used with HPLC, and SA was quantified using fluorescence detection (Jan et al., 2019). For ABA analysis, 3 mg of powdered sample was mixed with 30 ml of extraction solution and 10 ng of ABA standard ([±]-3,5,5,7,7,7-d6). Furthermore, the extraction was dried and methylated with diazomethane for GC-MS/SIM analysis. The monitor responses to ions at m/z of 190 and 162 for Me-ABA, and 194 and 166 for Me-(2H6)-ABA were obtained using Lab-Base (ThermoQuest, Manchester, the United Kingdom) data system software (Asaf et al., 2017b).
Inductively Coupled Plasma Mass Spectrometry Analysis
The Na+, K+, and Mg+ contents in rice plants inoculated with selected bacterial isolates were quantified using inductively coupled plasma mass spectrometry (ICP-MS). Lyophilized powder (0.2 g) samples were soaked in HCl (0.5 M) and oven-dried. The obtained digested samples were then analyzed using ICP-MS (Optima 7900DV; PerkinElmer, Waltham, MA, the United States) (Khan et al., 2020b).
Estimation of Gene Expression Through RT-PCR
Quantitative real-time PCR (qRT-PCR) was analyzed as described earlier by Shahid et al. (2019). Total RNA was extracted using a TRIzol reagent, while a DiaStar™ RT kit (SolGent, Korea) was used for extracting cDNA. RT-PCR was performed in the EcoTM real-time PCR machine using 2X Real-time PCR Master Mix (including SYBR® Green I BioFACT™ Korea), using synthesized cDNA and gene-specific primers (Supplementary Table 1). To normalize the level of relative expression of each gene, actin was used for each reaction and the expression level was calculated in control plants relatively with other NaCl-inoculated and non-inoculated rice-stressed plants.
Statistical Analysis
Data were collected in triplicate and subjected to the Duncan multiple range test using the SAS version 9.2 software. Furthermore, the data were graphically presented using the GraphPad Prism software (version 6.01, San Diego, CA, the United States).
Results
Isolation, Screening Bioassay, and Identification of Selected Bacterial Isolates
A total of 55 rhizospheric bacterial isolates were isolated and screened for different PGP traits, such as siderophore production, phosphate solubilization, EPS formation, and IAA production (Supplementary Table 2, Supplementary Figure 1). The screening results indicated that 20 isolates showed IAA activity. Fourteen isolates showed positive results for siderophore, whereas 12 isolates showed phosphate solubilization and EPS activity (Supplementary Figure 1). Based on multiple PGP traits, 11 isolates were screened for salt tolerance at three different concentrations of NaCl (50, 100, and 150 mM). Isolates ALT 11, ALT 12, and ALT 30 showed the highest tolerance of NaCl (Supplementary Figure 2). Therefore, these isolates were selected for further investigation using molecular identification. Our results revealed that rhizospheric bacteria ALT 11 showed sequence similarity with Bacillus. Additionally, the neighbor-joining method was employed to construct phylogenetic trees using 16S and MEGA 6 software after performing sequence alignment using Clustal W and a default parameter. The results revealed that isolates ALT11, ALT12, and ALT30 exhibited a high level of 16S rRNA sequence identity with Bacillus. Furthermore, these sequences were submitted to NCBI with accession number MW513461 for ALT11, MW513462 for ALT12, and MW513463 for ALT30 (Supplementary Figure 3).
In vitro Indole-3-acetic Acid and Organic Acid of Isolate
The identified rhizospheric bacterial strain was grown in LB medium for 5 days, and the culture filtrate was tested for IAA, which was determined using GC-MS/SIM, and organic acid using HPLC. The IAA results showed that ALT11 produced the highest amount (3.5 μg/mL) followed by ALT12 and ALT30 (Figure 1A). Furthermore, the organic acid results showed that isolates ALT11 and ALT12 produced, in addition to succinic and acetic acid, malic and lactic acids (Figure 1B). Although acetic and succinic acids were produced by all isolates, the highest amounts of succinic acid were observed in the CF of isolate ALT30, whereas acetic acid was highly produced by ALT11 and ALT12 (Figure 1B).
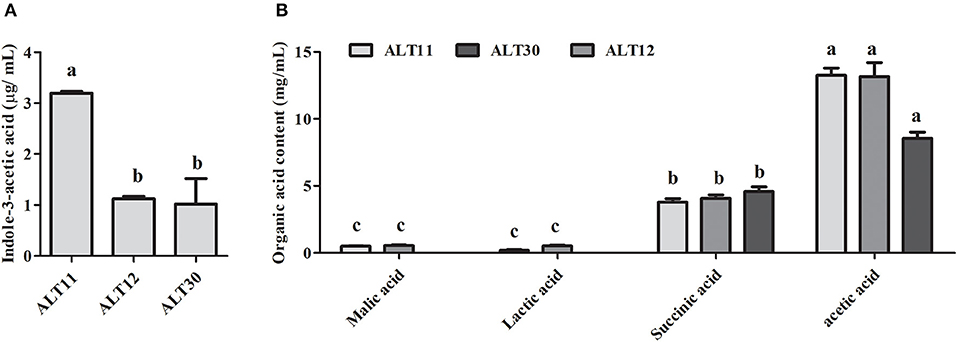
Figure 1. Quantification of indole-3-acetic acid (IAA) and organic acids produced by isolates ALT11, ALT12, and ALT30. (A) GC/MS-SIM analysis of the IAA content in the culture broth of isolate ALT1, (B) organic acid detection and quantification using high-performance liquid chromatography relative to their respective standard. Different letters indicate significant differences between the mean values of three replicates ± standard deviation.
Bacterial Isolates Regulate Rice Growth Under Salinity Stress
Our results showed that increasing salinity stress adversely affected the growth attributes of rice plants (Figure 2). Under salinity stress, significant decreases were observed in shoot length (22.8–42.2%), root length (28.18–59%), fresh weight (44.7–66.41%), and dry weight (85–90%) in the 70 and 140 mM NaCl treatments compared with the rice plants in the control treatment (Table 1). The first value between parentheses shows 70 mM, while the second value shows 140 mM of NaCl stress. However, the plant–microbe interaction reverted the effect of salinity stress and caused increases in shoot length (27.03–46.5%), root length (29.11–40.81%), fresh weight (45.94–53.33%), and dry weight (59.42–66.66%) in bacterial-inoculated salt-stressed plants compared with the plants treated with 70 and 140 mM NaCl (Table 1). Similarly, increases in salinity (70 and 140 mM) also reduced the chlorophyll content (18.30–36.15%), Chl a (29.02–60.87%), Chl b (30.86–64.51%), and total carotenoid content (21.63–62.28%) decreased compared with the control plants, whereas there were increases in chlorophyll content (35.87–47.76%), Chl a (22.41–59.93%), Chl b (27.28–52.89%), and total carotenoid content (26.86–70%) in the bacterial-inoculated salt-stressed plants compared with the plants only treated with NaCl (70 and 140 mM; Table 2). Under normal conditions, increases in growth characteristics, such as shoot/root length (17.99/27.28%), fresh/dry weight (29.10/93.54%), chlorophyll content (23.92%), Chl a (14.74%), Chl b (7.03%), and total carotenoid content (16.95%), were observed in bacterial-inoculated rice plants compared with the control rice plants (Tables 1, 2).

Figure 2. Effects of selected bacterial isolates, ALT11, ALT12, and ALT30, on the growth of rice plants (9 weeks old) under standard salinity and 70 and 140 mM NaCl stress.
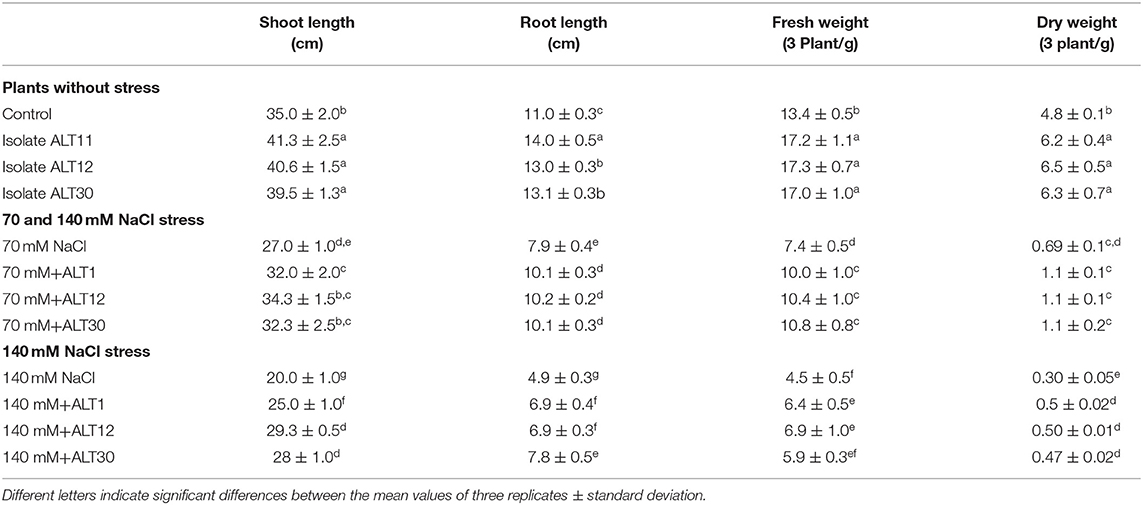
Table 1. Growth-promoting effect of isolates ALT11, ALT12, and ALT30 on rice growth under normal conditions and 70 and 140 mM NaCl stress.
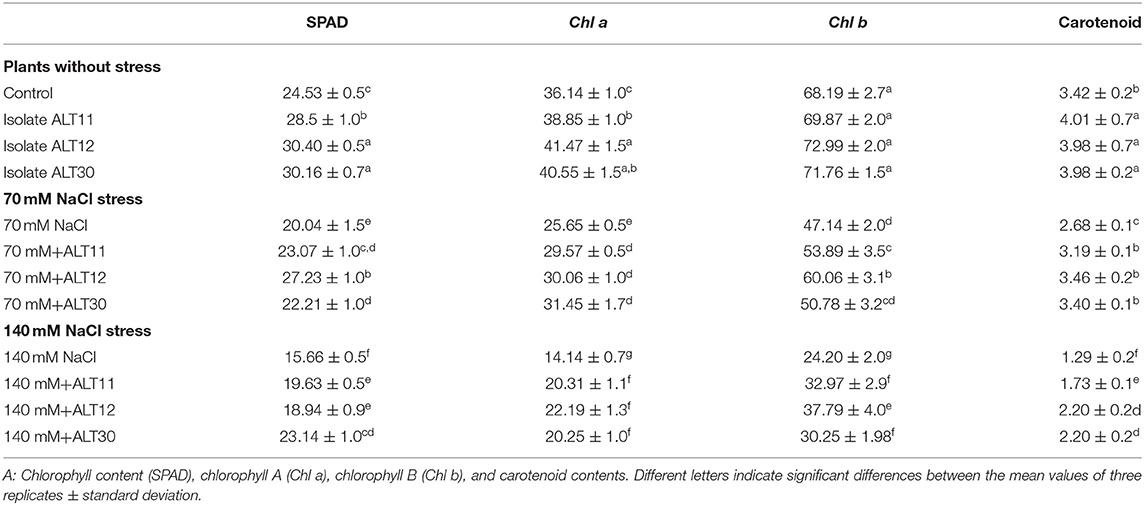
Table 2. The effect of NaCl stress on different chlorophyll contents of rice with and without the inoculation of isolates ALT11, ALT12, and ALT30.
Bacterial Isolates Regulate Rice Endogenous Phytohormones
The endogenous ABA and SA contents were investigated in both inoculated and non-inoculated rice plants under normal and NaCl stress (70 and 140 mM). The ABA results showed a significant increase in ABA content under salinity stress (125–165.43%). However, rice plants inoculated with all three strains showed a significant decrease in ABA content (27.9–23%) compared with rice plants only treated with NaCl (Figure 3A). The SA content showed an opposite response: A decrease in endogenous SA content was observed under 70 mM (18%) and 140 mM (58.32%) NaCl-stressed rice plants, whereas there was an increase in the SA content (11.70–69.19%) in bacterial-inoculated salt-stressed rice plants compared with plants only treated with NaCl (Figure 3B).
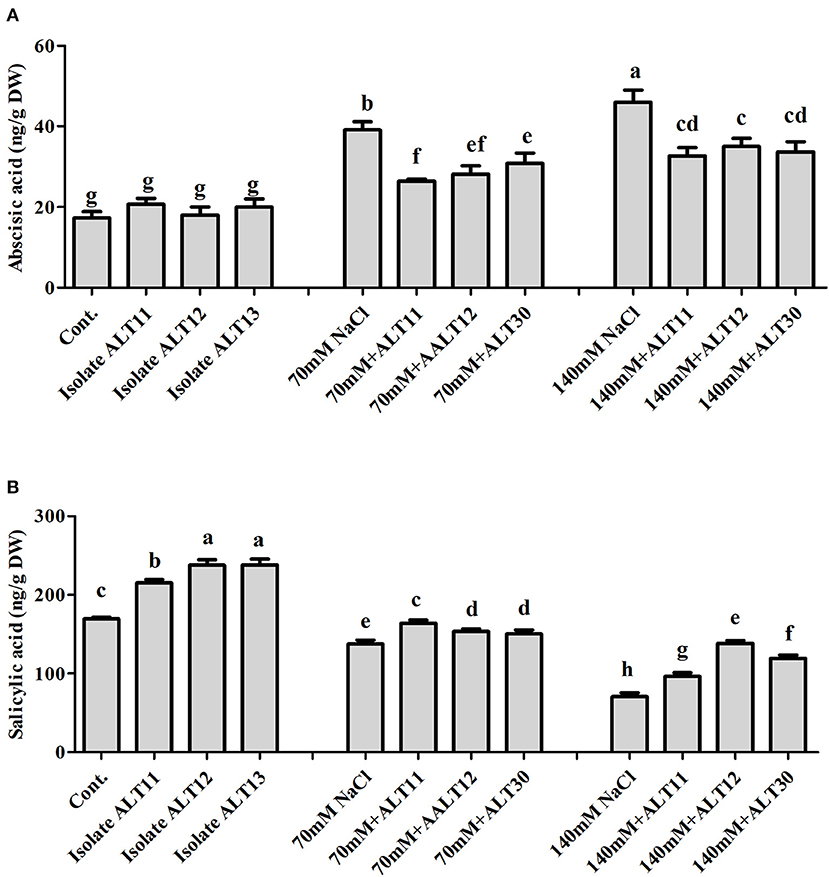
Figure 3. Endogenous abscisic acid (ABA) and salicylic acid (SA) quantification in rice plants inoculated with isolates ALT11, ALT12, and ALT30 under standard salinity and NaCl stress (70 and 140 mM). (A) Demonstrates ABA and (B) shows the amount of SA under normal conditions and NaCl stress. Different letters indicate significant differences between the mean values of three replicates ± standard deviation.
Regulation of Enzymatic and Non-enzymatic Antioxidants Under Salinity Stress
Different enzymatic and non-enzymatic antioxidant activities were determined in rice plants under NaCl stress and bacterial-inoculated plants. CAT, POD, PPO, GSH, SOA, and APX activities were increased with increasing salinity stress, whereas the opposite trend was observed for the TP content (Figures 4, 5). However, bacterial-inoculated rice plants showed an increase in TP content and a decrease in PPO, GSH, SOA, and LPO. Under salinity stress (70 and 140 mM), there were significant increases in PPO (3.7–5.4-fold), GSH (2.5–4-fold), SOA (0.8–1.7-fold), and LPO (1–4.3-fold; Figures 4A–D). On the other hand, rice plants inoculated with isolate ALT showed a significant decrease in PPO (24.63–34.57%), GSH (25.53–24.91%), SOA (13.88–18.67%), and LPO (15.96–26.06%) compared with plants treated with only 70 and 140 mM NaCl (Figures 4A–D). However, an increase in CAT (1.6–2.3-fold), POD (1–1.7-fold), SOD (41.89–60.89%), and APX (1.6–4.4-fold) was observed (Figures 5A–D). There were significant increases in CAT (26–33.04%), POD (59.55–78%), SOD (13.58–27.77%), and APX (5.76–22.74%) in bacterial-inoculated rice plants compared with salt-stressed plants (Figures 5A–D). The TP content showed the opposite response, and a decrease in TP content (45.23–70.31%) was observed in 70 and 140 mM NaCl-stressed rice plants, while there was an increase in the TP content (42.57–68.26%) in bacterial-inoculated salt-stressed plants compared with plants treated with NaCl (Figure 5E).
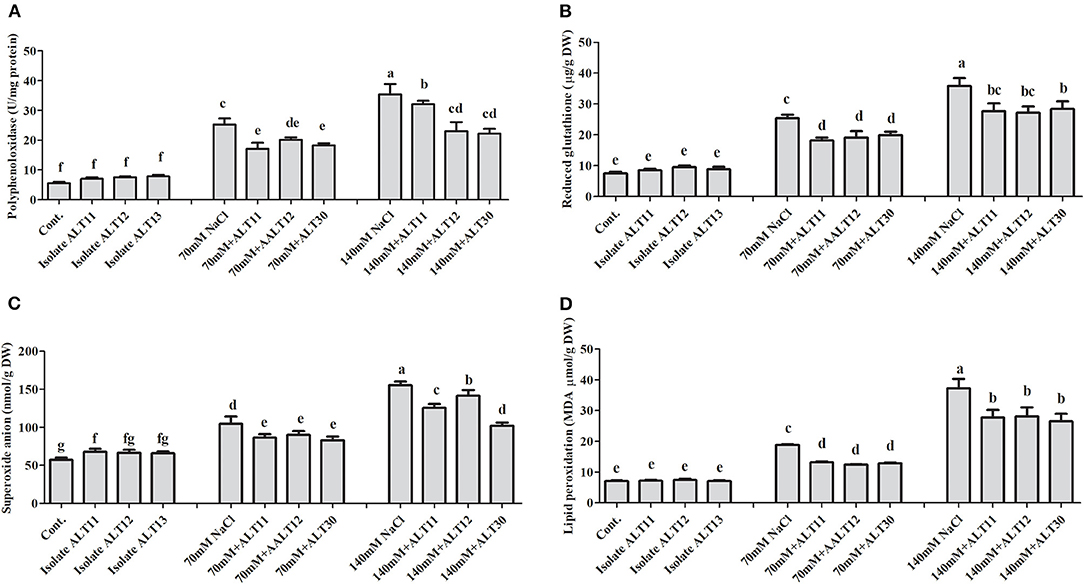
Figure 4. The effect of isolates ALT11, ALT12, and ALT30 on different antioxidants under normal conditions and NaCl stress (70 and 140 mM). (A) Polyphenol oxidase (PPO); (B) reduced glutathione (GSH); (C) superoxide anions (SOAs); (D) lipid peroxidation (MDA). Different letters indicate significant differences between the mean values of the three replicates ± standard deviation.
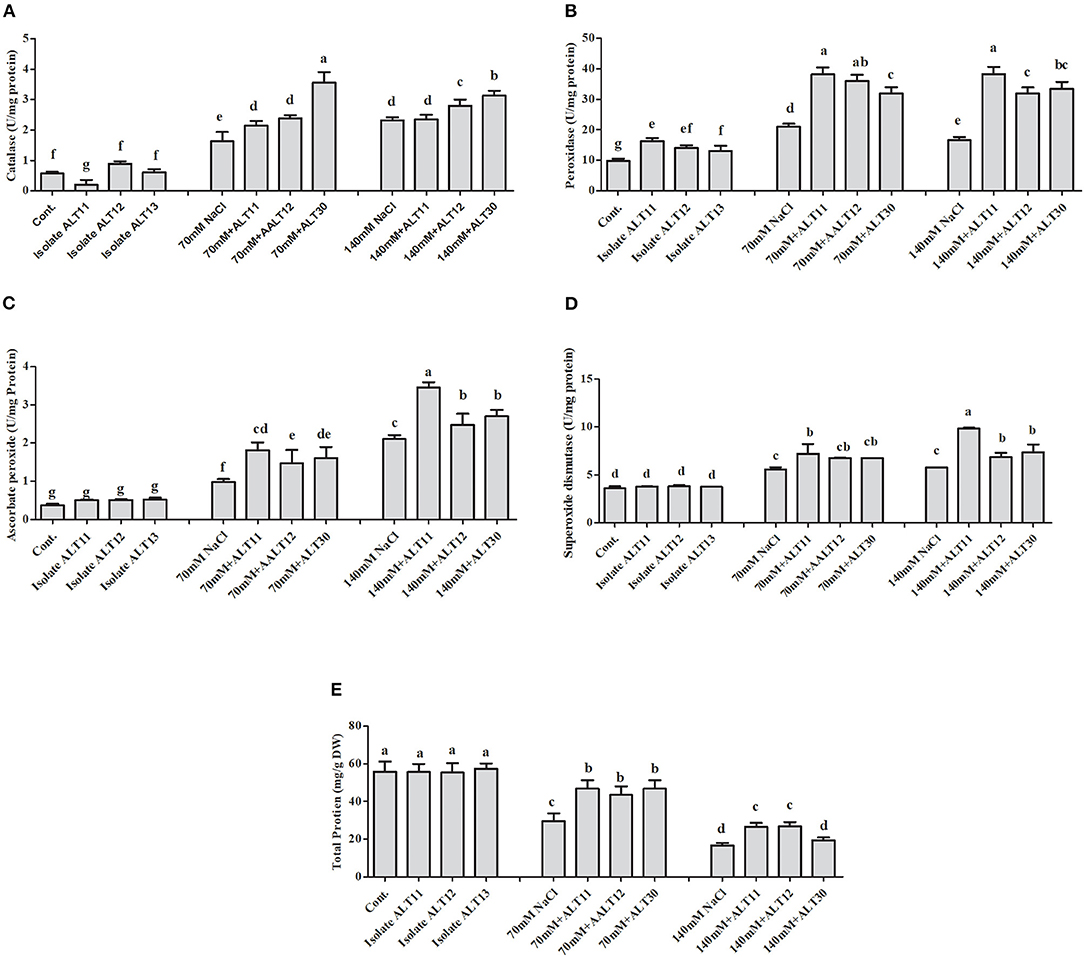
Figure 5. The effect of isolates ALT11, ALT12, and ALT30 on different antioxidants under normal conditions and NaCl stress (70 and 140 mM). (A) Catalase (CAT); (B) peroxidase (POD); (C) ascorbic peroxidase (APX); (D) superoxide dismutase (SOD); and (E) total protein (TP). Different letters indicate significant differences between the mean values of three replicates ± standard deviation.
Inductively Coupled Plasma Mass Spectrometric Analysis of Na+, K+, and Mg+ Ions Under Salt Stress
The ICP-MS results demonstrated that Na+, K+, and Mg+ accumulations were different in control, NaCl-stressed (70 and 140 mM), and bacterial-inoculated rice plants (Figure 6). The accumulation of Na increased at 70 mM (5.4-fold) and 140 mM (12.2-fold) in the NaCl-stressed rice plants compared with the control rice plants. However, there was a decrease in the Na content (24.11 and 30.60%) of rice plants inoculated with bacterial isolates compared with plants treated with only NaCl (Figure 6A). Additionally, as salinity increased (to 70 and 140 mM), the K (9.22 and 17.42%, respectively) and Mg (21.12 and 37.67%, respectively) content decreased (Figures 6B,C). However, increases in K (23.14 and 15.45%) and Mg (2.82 and 18.74%) contents were observed in bacterial-inoculated salt-stressed plants compared with plants only treated with 70 or 140 mM of NaCl (Figures 6B,C). Under normal conditions, in bacterial-inoculated rice plants, there is an increase in K (15.30%) and Mg (12.32%) contents, while no significant difference in Na (9.53%) content was observed compared with the control rice plants (Figures 6A–C).
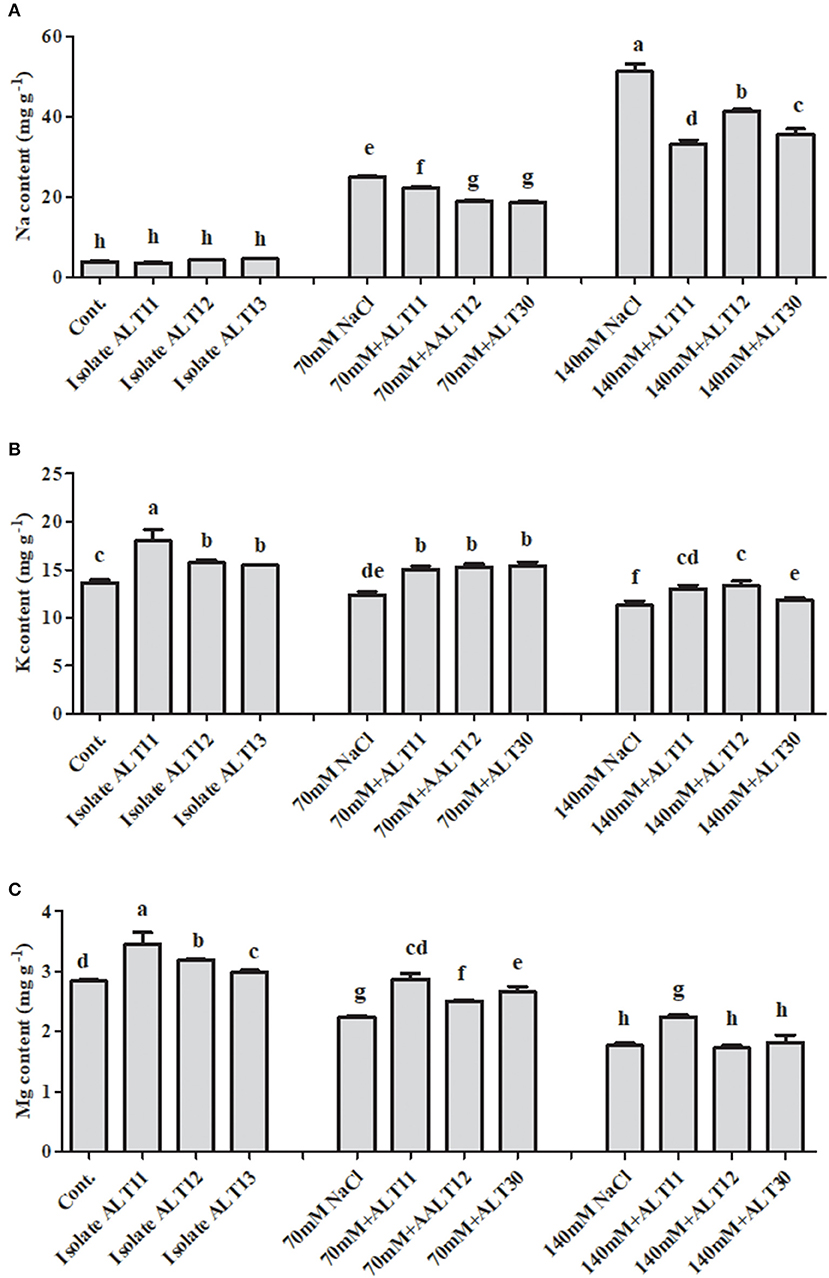
Figure 6. The effect of isolates ALT11, ALT12, and ALT30 on sodium (Na+), potassium (K+) and magnesium (Mg) contents under normal conditions and NaCl stress (70 and 140 mM). (A) Na+ content, (B) K+ content, and (C) Mg content in rice plants under normal conditions and NaCl stress. Different letters indicate significant differences between the mean values of three replicates ± standard deviation.
Estimation of Gene Expression
Gene expression was analyzed using an RT-PCR of rice under NaCl stress with and without inoculated bacterial isolates (Figure 7). The OsNHX1 gene expression results showed that increasing salinity stress to 70 and 140 mM caused a higher expression of OsNHX1 (0.9 and 8.5-fold, respectively) in rice plants (Figure 7A). However, decreases in the expression of OsNHX1 (0.3 and 4.1-fold) were observed in rice plants inoculated with bacterial isolates under NaCl stress (Figure 7A). Similarly, OsPIN1A gene expression revealed that OsPIN1A was downregulated in rice plants under 70 and 140 mm NaCl stress compared with the control rice plants, whereas there was a higher expression of genes in rice plants inoculated with bacterial isolates than in rice plants treated only with NaCl (Figure 7B). Under normal conditions, there were no significant differences in OsCATA. However, rice plants treated with 70 and 140 mM NaCl showed enhanced OsCATA expression (Figure 7C). The bacterial isolates in inoculated rice plants augmented salinity stress through an antioxidant defense system and enhanced OsCATA expression under NaCl stress (Figure 7C). The OsAPX1 gene was highly expressed (15–39-fold) under 70 and 140 mM NaCl stress. However, there was a higher expression of OsAPX1 at 70 mM (7-fold) and 140 mM (17-fold) in bacterial-inoculated rice plants than in rice plants treated only with NaCl (Figure 7D).
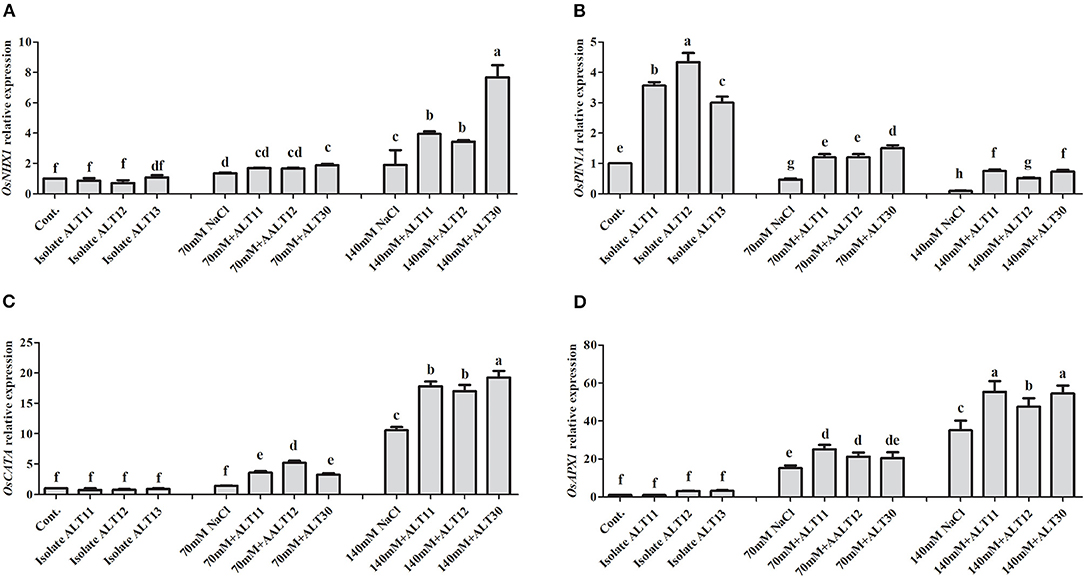
Figure 7. Gene expression in response to ALT11, ALT12, and ALT30 under normal conditions and NaCl stress. Relative expression of (A) OsNHX1, (B) OsPIN1A, (C) OsCATA, and (D) OsAPX1 genes under normal conditions and NaCl stress (70 and 140 mM) calculated using actin gene expression. Different letters indicate significant differences between the mean values of three replicates ± standard deviation.
Discussion
Abiotic and biotic environmental stresses are among the most limiting factors to plant growth and productivity (Karlidag et al., 2010). Among these factors, salt stress is abiotic stress that negatively affects plant growth and crop productivity (Santos et al., 2018). High salinity induces numerous disorders in seed germination, growth, and yield (Yildirim et al., 2008). Salinity stress reduces plant growth due to an increase in Na uptake, increase in endogenous ABA content, excess ROS generation, and a reduction in photosynthetic rate and K uptake (Ansari et al., 2019). However, the interactions of PGPB reduce the extent of poor growth and help plants to survive in adverse saline conditions (Shukla et al., 2012). Our results suggest that rice plants inoculated with isolates ALT11, ALT12, and ALT30 showed better growth and higher tolerance to 70 and 140 mM NaCl stress (Figure 2, Table 1). Rice growth, biomass, and chlorophyll content were significantly decreased in plants only treated with NaCl, whereas there was an increase in the growth attributes and chlorophyll content of bacterial-inoculated rice plants under normal conditions and NaCl stress. This increase in the height and biomass of rice plants is probably due to the production of plant growth traits and tolerance to NaCl stress. Previous reports revealed that peanuts, radish, lettuce, strawberry, and wheat plants inoculated with bacterial isolates (Brachybacterium sp., Staphylococcus sp., Bacillus sp., and Kocuria sp.) showed higher growth rates and biomass (fresh/dry) than non-inoculated plants under salt stress (Yildirim et al., 2008, 2011; El-Tarabily and Youssef, 2010; Karlidag et al., 2010; Shukla et al., 2012; Desale et al., 2014; Liu et al., 2019). Amacher et al. (2000) revealed that PGPB reduces salinity stress in maize and wheat by ~50%. Similarly, leaf chlorophyll concentration is an indicator of salt tolerance and responds to increasing salinity stress (Habib et al., 2016; Shultana et al., 2020b). Salinity stress increases chlorophyllase activity, decreases chlorophyll synthesis, and destroys pigment proteins, decreasing chlorophyll pigment in alfalfa, lettuce, wheat, okra, and basil (Han and Lee, 2005; Bashan et al., 2006; Heidari and Golpayegani, 2012; Habib et al., 2016; Ansari et al., 2019). Our results showed that isolate inoculation of rice plants enhanced chlorophyll pigments and mitigated the harmful effects of salinity stress (Table 2). This increase in chlorophyll content in rice plants might be due to the beneficial effect of the inoculated isolates. Similarly, higher chlorophyll content was also reported for PGPB-inoculated salt-stressed rice, cucumber, and okra plants than for non-inoculated plants (Bal et al., 2013; Kang et al., 2014; Habib et al., 2016). Additionally, salinity stress affected carotenoid content, which plays an important role in the prevention of several degenerative stresses in plants (Babu et al., 2011). Other studies have shown that higher chlorophyll and photosynthetic contents in inoculated plants were related to higher absorption of iron and Mg (Hosseinzadah et al., 2011; Ansari et al., 2019). Mg is a macronutrient, and its absence adversely affects several functions in plants, including photosynthesis (Farhat et al., 2016). In plants, Mg is the most metabolically active mineral, with the highest concentration in chloroplasts (Waters, 2011). Mg shares a central atom in the tetrapyrrole ring of chlorophyll a and b molecules, which are considered essential pigments for photosynthetic light absorption (Waters, 2011; Tränkner et al., 2018). A decrease in Mg was found to adversely affect the photosynthesis rate (Hermans et al., 2005; Farhat et al., 2016). The ICP-MS analysis results for Mg showed that salinity stress (70 and 140 mM) decreased the level of Mg in rice plants compared with the control rice plants (Figure 6C). However, an increase in Mg content was observed in the bacterial-inoculated salt-stressed rice plant. Thus, a decrease in Mg content may contribute to a decrease in chlorophyll and photosynthetic pigments (Gomes et al., 2011). Previous studies also support our findings that plants subjected to salinity stress show a decrease in the Mg concentration (Hu and Schmidhalter, 2005; Jampeetong and Brix, 2009).
PGPB are a group of free-living microorganisms that colonize the roots of plants and play a beneficial role in various stages of plant growth, either directly, by increasing IAA, siderophore, and HCN, or indirectly, by inducing systemic resistance (Yildirim et al., 2008). EPS-producing PGPB play a significant role in alleviating salinity stress (Kumar et al., 2020), as EPS contains high molecular lipopolysaccharide proteins and polysaccharide lipids and binds with cations, such as Na, develop a soil sheath around the plant roots, and decrease the quantity of Na ions available for plant uptake (Shin et al., 2016; Kumar et al., 2020). EPS also helps in the establishment of plant–microbe interactions by providing a microenvironment in which microbes can survive under stress conditions, enhancing water retention, and regulating the diffusion of organic carbon sources (Kumar et al., 2020). Siderophores are a class of small-molecule compounds, which are highly specific for chelating Fe3+. Many PGPB acquire iron by producing siderophore and reduce the inhibitory effect of salinity stress by increasing photosynthesis and the chlorophyll content of plants (Ferreira et al., 2019; Liu et al., 2019; Kumar et al., 2020). Furthermore, several authors reported that siderophore-producing bacteria represent a promising alternative to chemical fertilizers due to tackling salt stress effects by enhancing the growth and biomass of soybean (El-Esawi et al., 2018), peanut (Paulucci et al., 2015), pepper (Wang et al., 2018), and wheat (Masalha et al., 2000). Moreover, laboratory assays conducted by Grobelak and Hiller (2017) and Shameer and Prasad (2018) reported an increase in plant growth as a result of inoculation with siderophore-producing bacteria, which constitutes direct evidence of the role of microbial siderophores in enhancing plant development. Additionally, phosphate is also important for plant growth. Phosphate-solubilizing halotolerant PGPB provides an opportunity to enhance P availability by solubilizing insoluble phosphate via various mechanisms, including secreting low molecular weight organic acids (Sharma et al., 2013; Etesami, 2018). The production of organic acids is the main mechanism used by microbes to mineralize inorganic P (Rodriguez et al., 2004). The organic acids produced by PGPR include malic, acetic, citric, oxalic, lactic, formic, gluconic, and 2-keto-gluconic acids (Vázquez et al., 2000; Vessey, 2003). Phosphate-solubilizing organic acid-producing microbes are considered important members of PGPR, and their application as biofertilizers has been shown to improve the growth of crops plants by increasing P availability to plants, reduce fertilizers inputs, and fulfill the metabolic demands of plant (Vessey, 2003; Khan et al., 2007; Panhwar et al., 2011). In salt-affected soil, the inoculation of phosphate-solubilizing halotolerant PGPB improves plant growth and suppresses the adverse effect of salt (Giri et al., 2004; Etesami and Beattie, 2018). Similarly, Bacillus aryabhattai and Bacillus megaterium are efficient halotolerant P-solubilizing microbes under saline conditions (Chookietwattana and Maneewan, 2012; Bhattacharyya et al., 2017). The production of IAA is a relatively common trait of most salt-tolerant PGPBs, and it increases the fitness of plant growth in salt-affected soils. IAA-producing, salt-tolerant Planococcus rifietoensis, Brachybacterium, and Haererohalobacter enhance the growth and yields of wheat and peanut plants under salinity stress (Shukla et al., 2012; Desale et al., 2014; Zhou et al., 2015). In this study, all selected isolates produced IAA, whereas isolate ALT11 produced the highest amount of IAA (Figure 1A). Furthermore, PIN protein plays an important role in facilitating auxin efflux, and salinity stress downregulates the expression of PIN genes. In this study, a decrease in OsPIN1A was observed in salt-stressed rice plants. However, in bacterial-inoculated rice plants, there was an increase in the expression of OsPIN1A genes (Figure 7B). Similar results were also reported by Fu et al. (2019) who found that salinity stress leads to a reduction in root meristem size through the downregulation of PIN genes, thereby reducing plant auxin levels. The increase in PIN gene expression in rice plants might be due to the production of IAA and the halotolerance of the selected isolates ALT11, ALT12, and ALT30.
The content of Na and K in rice was significantly affected under treatments of 70 and 140 mM NaCl. In this study, rice plants exposed to salinity stress had increased Na ion concentrations in their shoots (Figure 6A). This increase in Na content was negatively correlated with rice growth (root/shoot), biomass (fresh/dry), and chlorophyll content. Arora et al. (2010) reported that, under salinity stress, bacteria can bind Na ions through the secretion of EPS, which reduces Na toxicity in soil. A higher population of EPS-producing bacteria in the root rhizosphere reduces the concentration of Na availability for uptake and alleviates the NaCl effect on plants (Shultana et al., 2020b). In current observations, a decrease in Na content in bacterial-inoculated rice plants might be due to their EPS-producing activity (Supplementary Figure 1B). K is an inorganic solute, and it contributes to maintaining osmotic pressure, ionic strength, and osmotic adjustment in plants under normal and salt stress conditions (Santos et al., 2018). In general, the antagonism between Na and K ions promotes an increase in Na ions and a decrease in K content. Additionally, maize plants treated with an Azotobacter strain have enhanced K absorption and decreased Na content under salinity stress (Rojas-Tapias et al., 2012). According to Hauser and Horie (2010) and Shukla et al. (2012), salt-tolerant plants are able to maintain favorable K homeostasis during periods of salinity stress. The results of our study showed that plants inoculated with an isolate had enhanced K content and decreased Na content under salinity stress (Figure 6B). This increase in K content in rice plants was positively correlated with rice growth and biomass. Recent studies of plants inoculated with different PGPB demonstrate that microbes decrease Na content under salinity stress (Volkov, 2015; Ansari et al., 2019). Additionally, Etesami and Beattie (2018) and Shukla et al. (2012) showed that halotolerant bacteria could enhance K absorption in plants under salinity stress. Further, under higher salinity stress, the NHX family members were differentially expressed in bacterial-inoculated and non-inoculated rice plants. In rice, five OsNHX1 genes have been identified (Fukuda et al., 2011; Zhang et al., 2018) and play an important role in the compartmentalization of Na and K in the cytoplasm (Fukuda et al., 2011). Fukuda et al. (2011) and Fukuda et al. (2004) reported that the overexpression of OsNHX1 improves the salinity tolerance of transgenic rice. Similar results were observed in rice plants inoculated with all three bacterial strains (Figure 7A). Chen et al. (2007) reported that overexpression of OsNHX1 enhanced growth in maize plants in 200 mM of NaCl stress.
The protective role of ABA is pivotal in plant growth as it promotes stomatal closure to minimize water loss and mediate stress damage through the activation of stress response genes, which collectively increase plant stress tolerance (Kang et al., 2014). It has been widely described that the ABA content of plants increases under salinity stress. However, we found a significant decrease in ABA content in the presence of bacterial-inoculated plants compared with non-inoculated salt-stressed plants. There are several studies that narrate the same finding of low ABA content in PGPB-inoculated plants under abiotic stress, including salinity stress (Kang et al., 2014; Jan et al., 2019; Kim et al., 2020). Similarly, various bacteria have been reported with both increased (Salomon et al., 2014) and decreased ABA concentration (Zhang et al., 2008) in plants. In tomato plants, the ABA-utilizing bacteria was found to decrease ABA concentration (Belimov et al., 2014). However, some bacteria that cannot utilize ABA were also reported in maize, tomato, and pea plants to decrease ABA concentration by decreasing xylem and phloem ABA flows (Dodd et al., 2009; Jiang et al., 2012). These bacterial-mediated changes in ABA concentration may affect stomatal conductance to maintain photosynthesis (Salomon et al., 2014). Moreover, the mechanisms of bacterial moderation of plant ABA concentrations may differ depending on the traits of a particular strain. Identifying bacterial genes and enzymes responsible for ABA catabolism, as well as obtaining knockout mutants of the bacteria unable to utilize this compound, would further evaluate the physiological and ecological significance of this bacterial trait. However, SA induces systemic acquired resistance and improves plant growth under stress (Kang et al., 2014). In dicot plants, such as Nicotiana and Arabidopsis, SA exists at low levels, whereas in rice, the SA contents are higher under normal growth conditions (De Vleesschauwer et al., 2013), as various physiological and biochemical responses are regulated. ROS, such as SOAs and hydrogen peroxide, are normally produced at high rates and cause oxidative damages to the cell structure during salt stress conditions (Sharma et al., 2012; Rodrigues et al., 2013). However, a defensive system, called the antioxidant enzyme system, is also activated to remove the free radicals produced under stress conditions and maintain them at low levels (Habib et al., 2016). This system consists of several ROS-scavenging enzymes, such as POD, SOD, APX, CAT, which have the ability to mitigate abiotic stress, including salinity stress, remove free radicals, and avoid the toxicity of ROS produced in cells during stress (Santos et al., 2018). Improved control of the O2 level was observed in rice plants coinoculated with halotolerant isolate. However, O2 and H2O2 can attack and oxidize polyunsaturated fatty acids in the cell membrane, which results in an increase in lipid peroxidation (Pandey et al., 2017; Santos et al., 2018). In this study, rice plants coinoculated with all three bacterial isolates showed better control of lipid peroxidation under normal and salt stress conditions (Figure 4D). Similar results were also observed in cowpea plants coinoculated with Bradyrhizobium, Paenibacillus durus, and Paenibacillus graminis (Egamberdieva et al., 2013; Santos et al., 2018). GSH occurs in reduced GSH and oxidized forms and protects the cell membrane against ROS damage (Sytar et al., 2013; Santos et al., 2018). CAT and SOD are metalloproteins with an important role in controlling ROS levels (Santos et al., 2018). SOD catabolizes O2 to H2O2, which is further catabolized by CAT, and excess H2O2 is removed to produce H2O and O2 under salinity stress (Conklin and Barth, 2004; Jamil et al., 2011; Sytar et al., 2013; Pandey et al., 2017). In this study, rice plants inoculated with the isolates ALT11, ALT12, and ALT30 had increased SOD and CAT activities under salinity stress, whereas there was a decrease in H2O2 and lipid peroxidation. This result provides evidence of the beneficial effect of microbes on the positive balance of antioxidative enzymes that detoxify ROS metabolism (Santos et al., 2018). Similarly, high gene expressions of OsCATA and OsAPX1 were observed in bacterial-inoculated rice plants under salinity stress. CAT and APX enzymes are encoded by different gene families. In rice, enzymatic antioxidants such as CAT and APX help convert excess H2O2 to H2 and O2 (Rossatto et al., 2017; Peng et al., 2018). Our results also support the findings of studies by Rossatto et al. (2017) and Kim et al. (2007), who report a higher expression of OsAPX and OsCAT genes under salinity stress. The increase in the expression of these genes during stress conditions is correlated with an increase in antioxidant enzymatic activities.
Conclusions
This study suggests that the tested isolates (ALT11, ALT12, and ALT30) play a pivotal role in mitigating NaCl tolerance (70 and 140 mM) in rice. Inoculation with halotolerant bacterial isolates showed improved growth, biomass, and chlorophyll content by altering endogenous phytohormones, activating an antioxidant defense system, ion uptake, and regulating salt-related genes under normal and salinity stress in rice plants. Therefore, based on our findings, further studies should examine the genomics of our novel bacterial strains and conduct saline field trials to take this research to the farmer's fields for its practical application.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
MAK conducted the experiments. MH and SA helped in the writing of the manuscript. MAK and S-MK conducted hormonal and antioxidant analyses. MK and BW-Y conducted RT-PCR analysis. I-JL designed, supervised, and financed the research. All authors have read and agreed to its content and that the manuscript conforms to the journal's policies.
Funding
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2017R1D1A1B04035601).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.665590/full#supplementary-material
References
Adhikari, A., Lee, K.-E., Khan, M. A., Kang, S.-M., Adhikari, B., Imran, M., et al. (2020). Effect of silicate and phosphate solubilizing rhizobacterium enterobacter ludwigii GAK2 on Oryza sativa L. under cadmium stress. J. Microbiol. Biotechnol. 30, 118–126. doi: 10.4014/jmb.1906.06010
Ahanger, M. A., Aziz, U., Alsahli, A. A., Alyemeni, M. N., and Ahmad, P. (2020). influence of exogenous salicylic acid and nitric oxide on growth, photosynthesis, and ascorbate-glutathione cycle in salt stressed vigna angularis. Biomolecules 10:42. doi: 10.3390/biom10010042
Ali, A., Ahmad, W., Rashid, M., and Abbas, M. N. (2015). Salinity tolerance in rice as affected by application of abscisic acid and salicylic acid. Bangladesh J. Botany 45, 93–98. Available online at: https://www.cabdirect.org/cabdirect/abstract/20173187056
Almeida, D. M., Gregorio, G. B., Oliveira, M. M., and Saibo, N. J. M. (2017). Five novel transcription factors as potential regulators of OsNHX1 gene expression in a salt tolerant rice genotype. Plant Mol. Biol. 93, 61–77. doi: 10.1007/s11103-016-0547-7
Amacher, J. K., Koenig, R., and Kitchen, B. (2000). Salinity and plant tolerance. AG-SO, 3:1. Available online at: https://digitalcommons.usu.edu/cgi/viewcontent.cgi?article=1042&context=extension_histall
Ansari, M., Shekari, F., Mohammadi, M. H., Juhos, K., Végvári, G., and Biró, B. (2019). Salt-tolerant plant growth-promoting bacteria enhanced salinity tolerance of salt-tolerant alfalfa (Medicago sativa L.) cultivars at high salinity. Acta Physiol. Plantarum 41:195. doi: 10.1007/s11738-019-2988-5
Arora, M., Kaushik, A., Rani, N., and Kaushik, C. P. (2010). Effect of cyanobacterial exopolysaccharides on salt stress alleviation and seed germination. J. Environ. Biol. 31, 701–704.
Asaf, S., Khan, A. L., Khan, M. A., Imran, Q. M., Yun, B.-W., and Lee, I.-J. (2017a). Osmoprotective functions conferred to soybean plants via inoculation with Sphingomonas sp. LK11 and exogenous trehalose. Microbiol. Res. 205, 135–145. doi: 10.1016/j.micres.2017.08.009
Asaf, S., Khan, M. A., Khan, A. L., Waqas, M., Shahzad, R., Kim, A.-Y., et al. (2017b). Bacterial endophytes from arid land plants regulate endogenous hormone content and promote growth in crop plants: an example of Sphingomonas sp. and Serratia marcescens. J. Plant Interact. 12, 31–38. doi: 10.1080/17429145.2016.1274060
Babu, M., Singh, D., and Gothandam, K. M. (2011). Effect of salt stress on expression of carotenoid pathway genes in tomato. J. Stress Physiol. Biochem. 7, 87–94. Available online at: https://www.academia.edu/5771661/Effect_of_salt_stress_on_expression_of_carotenoid_pathway_genes_in_tomato
Bal, H. B., Nayak, L., Das, S., and Adhya, T. K. (2013). Isolation of ACC deaminase producing PGPR from rice rhizosphere and evaluating their plant growth promoting activity under salt stress. Plant Soil 366, 93–105. doi: 10.1007/s11104-012-1402-5
Bashan, Y., Bustillos, J., Leyva, L., Hernandez, J.-P., and Bacilio, M. (2006). Increase in auxiliary photoprotective photosynthetic pigments in wheat seedlings induced by Azospirillum brasilense. Biol. Fertil. Soils 42, 279–285. doi: 10.1007/s00374-005-0025-x
Belimov, A. A., Dodd, I. C., Safronova, V. I., Dumova, V. A., Shaposhnikov, A. I., Ladatko, A. G., et al. (2014). Abscisic acid metabolizing rhizobacteria decrease ABA concentrations in planta and alter plant growth. Plant Physiol. Biochem. 74, 84–91. doi: 10.1016/j.plaphy.2013.10.032
Bhattacharyya, C., Bakshi, U., Mallick, I., Mukherji, S., Bera, B., and Ghosh, A. (2017). Genome-guided insights into the plant growth promotion capabilities of the physiologically versatile bacillus aryabhattai strain AB211. Front. Microbiol. 8:411. doi: 10.3389/fmicb.2017.00411
Chen, M., Chen, Q., Niu, X., Zhang, R., Lin, H., Xu, C., et al. (2007). Expression of OsNHX1 gene in maize confers salt tolerance and promotes plant growth in the field. Plant Soil Environ. 53, 490–498. doi: 10.17221/2302-PSE
Chookietwattana, K., and Maneewan, K. (2012). Screening of efficient halotolerant phosphate solubilizing bacterium and its effect on promoting plant growth under saline conditions. World Appl. Sci. J. 16, 1110–1117. Available online at: https://www.idosi.org/wasj/wasj16(8)12/11.pdf
Conklin, P. L., and Barth, C. (2004). Ascorbic acid, a familiar small molecule intertwined in the response of plants to ozone, pathogens, and the onset of senescence. Plant Cell Environ. 27, 959–970. doi: 10.1111/j.1365-3040.2004.01203.x
De Vleesschauwer, D., Gheysen, G., and Höfte, M. (2013). Hormone defense networking in rice: tales from a different world. Trends Plant Sci. 18, 555–565. doi: 10.1016/j.tplants.2013.07.002
Desale, P., Patel, B., Singh, S., Malhotra, A., and Nawani, N. (2014). Plant growth promoting properties of Halobacillus sp. and Halomonas sp. in presence of salinity and heavy metals. J. Basic Microbiol. 54, 781–791. doi: 10.1002/jobm.201200778
Dodd, I. C., Theobald, J. C., Richer, S. K., and Davies, W. J. (2009). Partial phenotypic reversion of ABA-deficient flacca tomato (Solanum lycopersicum) scions by a wild-type rootstock: normalizing shoot ethylene relations promotes leaf area but does not diminish whole plant transpiration rate. J. Exp. Bot. 60, 4029–4039. doi: 10.1093/jxb/erp236
Egamberdieva, D., Berg, G., Lindström, K., and Räsänen, L. A. (2013). Alleviation of salt stress of symbiotic Galega officinalis L.(goat's rue) by co-inoculation of Rhizobium with root-colonizing Pseudomonas. Plant Soil 369, 453–465. doi: 10.1007/s11104-013-1586-3
El-Esawi, M. A., Alaraidh, I. A., Alsahli, A. A., Alamri, S. A., Ali, H. M., and Alayafi, A. A. (2018). Bacillus firmus (SW5) augments salt tolerance in soybean (Glycine max L.) by modulating root system architecture, antioxidant defense systems and stress-responsive genes expression. Plant Physiol. Biochem. 132, 375–384. doi: 10.1016/j.plaphy.2018.09.026
El-Tarabily, K. A., and Youssef, T. (2010). Enhancement of morphological, anatomical and physiological characteristics of seedlings of the mangrove Avicennia marina inoculated with a native phosphate-solubilizing isolate of Oceanobacillus picturae under greenhouse conditions. Plant Soil 332, 147–162. doi: 10.1007/s11104-010-0280-y
Etesami, H. (2018). Can interaction between silicon and plant growth promoting rhizobacteria benefit in alleviating abiotic and biotic stresses in crop plants? Agric. Ecosyst. Environ. 253, 98–112. doi: 10.1016/j.agee.2017.11.007
Etesami, H., and Beattie, G. (2018). Mining halophytes for plant growth-promoting halotolerant bacteria to enhance the salinity tolerance of non-halophytic crops. Front. Microbiol. 9:148. doi: 10.3389/fmicb.2018.00148
Farhat, N., Elkhouni, A., Zorrig, W., Smaoui, A., Abdelly, C., and Rabhi, M. (2016). Effects of magnesium deficiency on photosynthesis and carbohydrate partitioning. Acta Physiol. Plantarum 38:145. doi: 10.1007/s11738-016-2165-z
Ferreira, M. J., Silva, H., and Cunha, A. (2019). Siderophore-producing rhizobacteria as a promising tool for empowering plants to cope with iron limitation in saline soils: a review. Pedosphere 29, 409–420. doi: 10.1016/S1002-0160(19)60810-6
Filgueiras, C. C., Martins, A. D., Pereira, R. V., and Willett, D. S. (2019). The ecology of salicylic acid signaling: primary, secondary and tertiary effects with applications in agriculture. Int. J. Mol. Sci. 20:5851. doi: 10.3390/ijms20235851
Fu, Y., Yang, Y., Chen, S., Ning, N., and Hu, H. (2019). Arabidopsis IAR4 Modulates primary root growth under salt stress through ROS-mediated modulation of auxin distribution. Front. Plant Sci. 10:522. doi: 10.3389/fpls.2019.00522
Fukuda, A., Nakamura, A., Hara, N., Toki, S., and Tanaka, Y. (2011). Molecular and functional analyses of rice NHX-type Na+/H+ antiporter genes. Planta 233, 175–188. doi: 10.1007/s00425-010-1289-4
Fukuda, A., Nakamura, A., Tagiri, A., Tanaka, H., Miyao, A., Hirochika, H., et al. (2004). Function, intracellular localization and the importance in salt tolerance of a vacuolar Na+/H+ antiporter from rice. Plant Cell Physiol. 45, 146–159. doi: 10.1093/pcp/pch014
Giri, B., Kapoor, R., Agarwal, L., and Mukerji, K. (2004). Preinoculation with arbuscular mycorrhizae helps Acacia auriculiformis grow in degraded Indian wasteland soil. Commun. Soil Sci. Plant Anal. 35, 193–204. doi: 10.1081/CSS-120027643
Gomes, M. A. D. C., Suzuki, M. S., Cunha, M. D., and Tullii, C.F. (2011). Effect of salt stress on nutrient concentration, photosynthetic pigments, proline and foliar morphology of Salvinia auriculata Aubl. Acta Limnol. Brasiliensia 23, 164–176. doi: 10.1590/S2179-975X2011000200007
Grobelak, A., and Hiller, J. (2017). Bacterial siderophores promote plant growth: screening of catechol and hydroxamate siderophores. Int. J. Phytoremed. 19, 825–833. doi: 10.1080/15226514.2017.1290581
Habib, S. H., Kausar, H., and Saud, H. M. (2016). Plant growth-promoting rhizobacteria enhance salinity stress tolerance in okra through ROS-scavenging enzymes. Biomed. Res. Int. 2016:6284547. doi: 10.1155/2016/6284547
Hamayun, M., Hussain, A., Khan, S. A., Kim, H.-Y., Khan, A. L., Waqas, M., et al. (2017). Gibberellins producing endophytic fungus Porostereum spadiceum AGH786 rescues growth of salt affected soybean. Front. Microbiol. 8:686. doi: 10.3389/fmicb.2017.00686
Han, H. S., and Lee, K. D. (2005). Plant growth promoting rhizobacteria effect on antioxidant status, photosynthesis, mineral uptake and growth of lettuce under soil salinity. Res. J. Agric. Biol. Sci. 1, 210–215.
Hauser, F., and Horie, T. (2010). A conserved primary salt tolerance mechanism mediated by HKT transporters: a mechanism for sodium exclusion and maintenance of high K+/Na+ ratio in leaves during salinity stress. Plant Cell Environ. 33, 552–565. doi: 10.1111/j.1365-3040.2009.02056.x
Heidari, M., and Golpayegani, A. (2012). Effects of water stress and inoculation with plant growth promoting rhizobacteria (PGPR) on antioxidant status and photosynthetic pigments in basil (Ocimum basilicum L.). J. Saudi Soc. Agric. Sci. 11, 57–61. doi: 10.1016/j.jssas.2011.09.001
Hermans, C., Bourgis, F., Faucher, M., Strasser, R. J., Delrot, S., and Verbruggen, N. (2005). Magnesium deficiency in sugar beets alters sugar partitioning and phloem loading in young mature leaves. Planta 220, 541–549. doi: 10.1007/s00425-004-1376-5
Hoang, T. M. L., Tran, T. N., Nguyen, T. K. T., Williams, B., Wurm, P., Bellairs, S., et al. (2016). Improvement of salinity stress tolerance in rice: challenges and opportunities. Agronomy 6:54. doi: 10.3390/agronomy6040054
Hosseinzadah, F., Satei, A., and Ramezanpour, M. R. (2011). Effects of mycorrhiza and plant growth promoting rhizobacteria on growth, nutrients uptake and physiological characteristics in Calendula officinalis L. Middle East J. Sci. Res. 8, 947–953.
Hu, Y., and Schmidhalter, U. (2005). Drought and salinity: a comparison of their effects on mineral nutrition of plants. J. Plant Nutr. Soil Sci. 168, 541–549. doi: 10.1002/jpln.200420516
Jaleel, C. A., Riadh, K., Gopi, R., Manivannan, P., Inès, J., Al-Juburi, H. J., et al. (2009). Antioxidant defense responses: physiological plasticity in higher plants under abiotic constraints. Acta Physiol. Plantarum 31, 427–436. doi: 10.1007/s11738-009-0275-6
Jamil, A., Riaz, S., Ashraf, M., and Foolad, M. R. (2011). Gene expression profiling of plants under salt stress. CRC. Crit. Rev. Plant Sci. 30, 435–458. doi: 10.1080/07352689.2011.605739
Jampeetong, A., and Brix, H. (2009). Effects of NaCl salinity on growth, morphology, photosynthesis and proline accumulation of Salvinia natans. Aquat. Bot. 91, 181–186. doi: 10.1016/j.aquabot.2009.05.003
Jan, R., Khan, M. A., Asaf, S., Lubna, Lee, I.-J., and Kim, K. M. (2019). Metal resistant endophytic bacteria reduces cadmium, nickel toxicity, and enhances expression of metal stress related genes with improved growth of oryza sativa, via regulating its antioxidant machinery and endogenous hormones. Plants 8:363. doi: 10.3390/plants8100363
Jha, Y., and Subramanian, R. (2013). Paddy plants inoculated with PGPR show better growth physiology and nutrient content under saline condition. Chilean J. Agric. Res. 73, 213–219. doi: 10.4067/S0718-58392013000300002
Jiang, F., Chen, L., Belimov, A. A., Shaposhnikov, A. I., Gong, F., Meng, X., et al. (2012). Multiple impacts of the plant growth-promoting rhizobacterium Variovorax paradoxus 5C-2 on nutrient and ABA relations of Pisum sativum. J. Exp. Bot. 63, 6421–6430. doi: 10.1093/jxb/ers301
Kang, S.-M., Khan, A. L., Waqas, M., You, Y.-H., Kim, J.-H., Kim, J.-G., et al. (2014). Plant growth-promoting rhizobacteria reduce adverse effects of salinity and osmotic stress by regulating phytohormones and antioxidants in Cucumis sativus. J. Plant Interact. 9, 673–682. doi: 10.1080/17429145.2014.894587
Kang, S.-M., Shahzad, R., Bilal, S., Khan, A. L., Park, Y.-G., Lee, K.-E., et al. (2019). Indole-3-acetic-acid and ACC deaminase producing Leclercia adecarboxylata MO1 improves Solanum lycopersicum L. growth and salinity stress tolerance by endogenous secondary metabolites regulation. BMC Microbiol. 19:80. doi: 10.1186/s12866-019-1450-6
Kang, S. M., Adhikari, A., Lee, K. E., Khan, M. A., Khan, A. L., Shahzad, R., et al. (2020). Inoculation with Indole-3-Acetic acid-producing rhizospheric Rhodobacter sphaeroides KE149 Augments growth of adzuki bean plants under water stress. J. Microbiol. Biotechnol. 30, 717–725. doi: 10.4014/jmb.1911.11063
Karlidag, H., Esitken, A., Yildirim, E., Donmez, M. F., and Turan, M. (2010). Effects of plant growth promoting bacteria on yield, growth, leaf water content, membrane permeability, and ionic composition of strawberry under saline conditions. J. Plant Nutr. 34, 34–45. doi: 10.1080/01904167.2011.531357
Kartik, V. P., Jinal, H. N., and Amaresan, N. (2020). Inoculation of cucumber (Cucumis sativus L.) seedlings with salt-tolerant plant growth promoting bacteria improves nutrient uptake, plant attributes and physiological profiles. J. Plant Growth Regul. 1–13. doi: 10.1007/s00344-020-10226-w
Khan, M. A., Asaf, S., Khan, A. L., Adhikari, A., Jan, R., Ali, S., et al. (2019a). Halotolerant rhizobacterial strains mitigate the adverse effects of NaCl stress in soybean seedlings. BioMed. Res. Int. 2019:9530963. doi: 10.1155/2019/9530963
Khan, M. A., Asaf, S., Khan, A. L., Adhikari, A., Jan, R., Ali, S., et al. (2020a). Plant growth-promoting endophytic bacteria augment growth and salinity tolerance in rice plants. Plant Biol. 22, 850–862. doi: 10.1111/plb.13124
Khan, M. A., Asaf, S., Khan, A. L., Jan, R., Kang, S.-M., Kim, K.-M., et al. (2019b). Rhizobacteria AK1 remediates the toxic effects of salinity stress via regulation of endogenous phytohormones and gene expression in soybean. Biochem. J. 476, 2393–2409. doi: 10.1042/BCJ20190435
Khan, M. A., Asaf, S., Khan, A. L., Jan, R., Kang, S.-M., Kim, K.-M., et al. (2020c). Thermotolerance effect of plant growth-promoting Bacillus cereus SA1 on soybean during heat stress. BMC Microbiol. 20:175. doi: 10.1186/s12866-020-01822-7
Khan, M. A., Asaf, S., Khan, A. L., Jan, R., Kang, S. M., Kim, K. M., et al. (2020b). Extending thermotolerance to tomato seedlings by inoculation with SA1 isolate of Bacillus cereus and comparison with exogenous humic acid application. PLoS ONE 15:e0232228. doi: 10.1371/journal.pone.0232228
Khan, M. A., Asaf, S., Khan, A. L., Ullah, I., Ali, S., Kang, S.-M., et al. (2019c). Alleviation of salt stress response in soybean plants with the endophytic bacterial isolate Curtobacterium sp. SAK1. Ann. Microbiol. 69, 797–808. doi: 10.1007/s13213-019-01470-x
Khan, M. A., Hamayun, M., Iqbal, A., Khan, S. A., Hussain, A., Asaf, S., et al. (2018). Gibberellin application ameliorates the adverse impact of short-term flooding onGlycine max L. Biochem. J. 475, 2893–2905. doi: 10.1042/BCJ20180534
Khan, M. A., Khan, A. L., Imran, Q. M., Asaf, S., Lee, S.-U., Yun, B.-W., et al. (2019d). Exogenous application of nitric oxide donors regulates short-term flooding stress in soybean. PeerJ 7:e7741. doi: 10.7717/peerj.7741
Khan, M. A., Sahile, A. A., Jan, R., Asaf, S., Hamayun, M., Imran, M., et al. (2021). Halotolerant bacteria mitigate the effects of salinity stress on soybean growth by regulating secondary metabolites and molecular responses. BMC Plant Biol. 21:176. doi: 10.1186/s12870-021-02937-3
Khan, M. A., Ullah, I., Waqas, M., Hamayun, M., Khan, A. L., Asaf, S., et al. (2019e). Halo-tolerant rhizospheric Arthrobacter woluwensis AK1 mitigates salt stress and induces physio-hormonal changes and expression of GmST1 and GmLAX3 in soybean. Symbiosis 77, 9–21. doi: 10.1007/s13199-018-0562-3
Khan, M. S., Zaidi, A., and Wani, P. A. (2007). Role of phosphate-solubilizing microorganisms in sustainable agriculture — a review. Agron. Sustain. Dev. 27, 29–43. doi: 10.1051/agro:2006011
Kim, D. W., Shibato, J., Agrawal, G. K., Fujihara, S., Iwahashi, H., Kim Du, H., et al. (2007). Gene transcription in the leaves of rice undergoing salt-induced morphological changes (Oryza sativa L.). Mol Cells 24, 45–59.
Kim, T.-H., Böhmer, M., Hu, H., Nishimura, N., and Schroeder, J. I. (2010). Guard Cell signal transduction network: advances in understanding abscisic acid, CO2, and Ca2+ signaling. Annu. Rev. Plant Biol. 61, 561–591. doi: 10.1146/annurev-arplant-042809-112226
Kim, Y.-N., Khan, M. A., Kang, S.-M., Hamayun, M., and Lee, I.-J. (2020). Enhancement of drought-stress tolerance of Brassica oleracea var. italica L. by newly isolated Variovorax sp. YNA59. J. Microbiol. Biotechnol. 30, 1500–1509. doi: 10.4014/jmb.2006.06010
Kubi, H. A. A., Khan, M. A., Adhikari, A., Imran, M., Kang, S.-M., Hamayun, M., et al. (2021). Silicon and plant growth-promoting Rhizobacteria pseudomonas psychrotolerans CS51 mitigates salt stress in Zea mays L. Agriculture 11:272. doi: 10.3390/agriculture11030272
Kumar, A., Singh, S., Gaurav, A. K., Srivastava, S., and Verma, J. P. (2020). Plant growth-promoting bacteria: biological tools for the mitigation of salinity stress in plants. Front. Microbiol. 11:1216. doi: 10.3389/fmicb.2020.01216
Li, X.-J., Yang, M.-F., Chen, H., Qu, L.-Q., Chen, F., and Shen, S.-H. (2010). Abscisic acid pretreatment enhances salt tolerance of rice seedlings: proteomic evidence. Biochim. Biophys. Acta 1804, 929–940. doi: 10.1016/j.bbapap.2010.01.004
Liu, J., Tang, L., Gao, H., Zhang, M., and Guo, C. (2019). Enhancement of alfalfa yield and quality by plant growth-promoting rhizobacteria under saline-alkali conditions. J. Sci. Food Agric. 99, 281–289. doi: 10.1002/jsfa.9185
Marklund, S., and Marklund, G. (1974). Involvement of the superoxide anion radical in the autoxidation of pyrogallol and a convenient assay for superoxide dismutase. Eur. J. Biochem., 47, 469–474. doi: 10.1111/j.1432-1033.1974.tb03714.x
Masalha, J., Kosegarten, H., Elmaci, Ö., and Mengel, K. (2000). The central role of microbial activity for iron acquisition in maize and sunflower. Biol. Fertil. Soils 30, 433–439. doi: 10.1007/s003740050021
Mukherjee, P., Mitra, A., and Roy, M. (2019). Halomonas rhizobacteria of Avicennia marina of Indian sundarbans promote rice growth under saline and heavy metal stresses through exopolysaccharide production. Front. Microbiol. 10:1207. doi: 10.3389/fmicb.2019.01207
Munns, R. (2005). Genes and salt tolerance: bringing them together. New phytol. 167, 645–663. doi: 10.1111/j.1469-8137.2005.01487.x
Nautiyal, C. S., Srivastava, S., Chauhan, P. S., Seem, K., Mishra, A., and Sopory, S. K. (2013). Plant growth-promoting bacteria Bacillus amyloliquefaciens NBRISN13 modulates gene expression profile of leaf and rhizosphere community in rice during salt stress. Plant Physiol. Biochem. 66, 1–9. doi: 10.1016/j.plaphy.2013.01.020
Pandey, S., Fartyal, D., Agarwal, A., Shukla, T., James, D., Kaul, T., et al. (2017). Abiotic stress tolerance in plants: myriad roles of ascorbate peroxidase. Front. Plant Sci. 8:581. doi: 10.3389/fpls.2017.00581
Panhwar, Q., Othman, R. A., Rahman, Z., Meon, S., Razi, I., and Naher, U. (2011). Contribution of phosphate-solubilizing bacteria in phosphorus bioavailability and growth enhancement of aerobic rice. Spanish J. Agric. Res. 9, 810–820. doi: 10.5424/sjar/20110903-330-10
Paulucci, N. S., Gallarato, L. A., Reguera, Y. B., Vicario, J. C., Cesari, A. B., García De Lema, M. B., et al. (2015). Arachis hypogaea PGPR isolated from Argentine soil modifies its lipids components in response to temperature and salinity. Microbiol. Res. 173, 1–9. doi: 10.1016/j.micres.2014.12.012
Peng, Y., Hou, F., Bai, Q., Xu, P., Liao, Y., Zhang, H., et al. (2018). Rice calcineurin B-Like protein-interacting protein kinase 31 (OsCIPK31) is involved in the development of panicle apical spikelets. Front. Plant Sci. 9:1661. doi: 10.3389/fpls.2018.01661
Radhakrishnan, R., and Lee, I. J. (2013). Ameliorative effects of spermine against osmotic stress through antioxidants and abscisic acid changes in soybean pods and seeds. Acta Physiol. Plant. 35, 263–269. doi: 10.1007/s11738-012-1072-1
Rafiq, K., Akram, M. S., Shahid, M., Qaisar, U., and Rashid, N. (2020). Enhancement of salt tolerance in maize (Zea mays L.) using locally isolated Bacillus sp. SR-2-1/1. Biologia 75, 1425–1436. doi: 10.2478/s11756-020-00435-9
Rodrigues, A. C., Bonifacio, A., Antunes, J. E. L., Da Silveira, J. A. G., and Figueiredo, M. D. V. B. (2013). Minimization of oxidative stress in cowpea nodules by the interrelationship between Bradyrhizobium sp. and plant growth-promoting bacteria. Appl. Soil Ecol. 64, 245–251. doi: 10.1016/j.apsoil.2012.12.018
Rodriguez, H., Gonzalez, T., Goire, I., and Bashan, Y. (2004). Gluconic acid production and phosphate solubilization by the plant growth-promoting bacterium Azospirillum spp. Naturwissenschaften 91, 552–555. doi: 10.1007/s00114-004-0566-0
Rojas-Tapias, D., Moreno-Galván, A., Pardo-Díaz, S., Obando, M., Rivera, D., and Bonilla, R. (2012). Effect of inoculation with plant growth-promoting bacteria (PGPB) on amelioration of saline stress in maize (Zea mays). Applied Soil Ecol. 61, 264–272. doi: 10.1016/j.apsoil.2012.01.006
Rossatto, T., Do Amaral, M. N., Benitez, L. C., Vighi, I. L., Braga, E. J. B., De Magalhães Júnior, A. M., et al. (2017). Gene expression and activity of antioxidant enzymes in rice plants, cv. BRS AG, under saline stress. Physiol. Mol. Biol. Plants 23, 865–875. doi: 10.1007/s12298-017-0467-2
Sah, S. K., Reddy, K. R., and Li, J. (2016). Abscisic acid and abiotic stress tolerance in crop plants. Front. Plant Sci. 7:571. doi: 10.3389/fpls.2016.00571
Sahile, A. A., Khan, M. A., Hamayun, M., Imran, M., Kang, S.-M., and Lee, I.-J. (2021). Novel Bacillus cereus strain, ALT1, enhance growth and strengthens the antioxidant system of soybean under cadmium stress. Agronomy 11:404. doi: 10.3390/agronomy11020404
Sakhabutdinova, A. R. D., Fatkhutdinova, D., Bezrukova, M., and Shakirova, F. M. (2003). Salicylic acid prevents the damaging action of stress factors on wheat plants. Bulg. J. Plant Physiol. 29, 314–319.
Salomon, M. V., Bottini, R., De Souza Filho, G. A., Cohen, A. C., Moreno, D., Gil, M., et al. (2014). Bacteria isolated from roots and rhizosphere of Vitis vinifera retard water losses, induce abscisic acid accumulation and synthesis of defense-related terpenes in in vitro cultured grapevine. Physiol. Plant. 151, 359–374. doi: 10.1111/ppl.12117
Santos, A. D. A., Silveira, J. A. G. D., Bonifacio, A., Rodrigues, A. C., and Figueiredo, M. D. V. B. (2018). Antioxidant response of cowpea co-inoculated with plant growth-promoting bacteria under salt stress. Braz. J. Microbiol. 49, 513–521. doi: 10.1016/j.bjm.2017.12.003
Santos, R., Hérouart, D., Puppo, A., and Touati, D. (2000). Critical protective role of bacterial superoxide dismutase in rhizobium-legume symbiosis. Mol. Microbiol. 38, 750–759. doi: 10.1046/j.1365-2958.2000.02178.x
Shahid, M., Imran, Q. M., Hussain, A., Khan, M., Lee, S. U., Mun, B. G., et al. (2019). Comprehensive analyses of nitric oxide-induced plant stem cell-related genes in Arabidopsis thaliana. Genes. 10:190. doi: 10.3390/genes10030190
Shameer, S., and Prasad, T. N. V. K. V. (2018). Plant growth promoting rhizobacteria for sustainable agricultural practices with special reference to biotic and abiotic stresses. Plant Growth Regul. 84, 603–615. doi: 10.1007/s10725-017-0365-1
Sharma, P., Jha, A. B., Dubey, R. S., and Pessarakli, M. (2012). Reactive oxygen species, oxidative damage, and antioxidative defense mechanism in plants under stressful conditions. J. Botany 2012:217037. doi: 10.1155/2012/217037
Sharma, S. B., Sayyed, R. Z., Trivedi, M. H., and Gobi, T. A. (2013). Phosphate solubilizing microbes: sustainable approach for managing phosphorus deficiency in agricultural soils. SpringerPlus 2:587. doi: 10.1186/2193-1801-2-587
Shin, W., Siddikee, A., Joe, M. M., Benson, A., Kim, K., Selvakumar, G., et al. (2016). Halotolerant plant growth promoting bacteria mediated salinity stress amelioration in plants. Korean J. Soil Sci. Fertil. 49, 355–367. doi: 10.7745/KJSSF.2016.49.4.355
Shrivastava, P., and Kumar, R. (2015). Soil salinity: a serious environmental issue and plant growth promoting bacteria as one of the tools for its alleviation. Saudi J. Biol. Sci. 22, 123–131. doi: 10.1016/j.sjbs.2014.12.001
Shukla, P. S., Agarwal, P. K., and Jha, B. (2012). Improved salinity tolerance of Arachis hypogaea (L.) by the interaction of halotolerant plant-growth-promoting rhizobacteria. J. Plant Growth Regul. 31, 195–206. doi: 10.1007/s00344-011-9231-y
Shultana, R., Kee Zuan, A. T., Yusop, M. R., and Saud, H. M. (2020a). Characterization of salt-tolerant plant growth-promoting rhizobacteria and the effect on growth and yield of saline-affected rice. PLoS ONE 15:e0238537. doi: 10.1371/journal.pone.0238537
Shultana, R., Tan Kee Zuan, A., Yusop, M. R., Mohd Saud, H., and Ayanda, A. F. (2020b). Effect of salt-tolerant bacterial inoculations on rice seedlings differing in salt-tolerance under saline soil conditions. Agronomy 10:1030. doi: 10.3390/agronomy10071030
Sytar, O., Kumar, A., Latowski, D., Kuczynska, P., Strzałka, K., and Prasad, M. N. V. (2013). Heavy metal-induced oxidative damage, defense reactions, and detoxification mechanisms in plants. Acta Physiol. Plant. 35, 985–999. doi: 10.1007/s11738-012-1169-6
Tisarum, R., Theerawitaya, C., Samphumphuang, T., Polispitak, K., Thongpoem, P., Singh, H. P., et al. (2020). Alleviation of salt stress in upland rice (Oryza sativa L. ssp. indica cv. Leum Pua) using arbuscular mycorrhizal fungi inoculation. Front. Plant Sci. 11:348. doi: 10.3389/fpls.2020.00348
Tränkner, M., Tavakol, E., and Jákli, B. (2018). Functioning of potassium and magnesium in photosynthesis, photosynthate translocation and photoprotection. Physiol. Plant. 163, 414–431. doi: 10.1111/ppl.12747
Vázquez, P., Holguin, G., Puente, M., López-Cortés, A., and Bashan, Y. (2000). Phosphate-solubilizing microorganisms associated with the rhizosphere of mangroves in a semiarid coastal lagoon. Biol. Fertil. Soils 30, 460–468. doi: 10.1007/s003740050024
Vessey, J. K. (2003). Plant growth promoting rhizobacteria as biofertilizers. Plant Soil 255, 571–586. doi: 10.1023/A:1026037216893
Volkov, V. (2015). Salinity tolerance in plants. quantitative approach to ion transport starting from halophytes and stepping to genetic and protein engineering for manipulating ion fluxes. Front. Plant Sci. 6:873. doi: 10.3389/fpls.2015.00873
Wang, W., Wu, Z., He, Y., Huang, Y., Li, X., and Ye, B.-C. (2018). Plant growth promotion and alleviation of salinity stress in Capsicum annuum L. by Bacillus isolated from saline soil in Xinjiang. Ecotoxicol. Environ. Safety 164, 520–529. doi: 10.1016/j.ecoenv.2018.08.070
Waters, B. M. (2011). Moving magnesium in plant cells. New Phytol. 190, 510–513. doi: 10.1111/j.1469-8137.2011.03724.x
Xiong, L., Schumaker, K. S., and Zhu, J.-K. (2002). Cell signaling during cold, drought, and salt stress. Plant Cell 14, S165–S183. doi: 10.1105/tpc.000596
Yildirim, E., Turan, M., and Donmez, M. F. (2008). Mitigation of salt stress in radish (Raphanus sativus l.) by plant growth: promoting rhizobacteria. Romanian Biotechnol. Lett. 13, 3933–3943.
Yildirim, E., Turan, M., Ekinci, M., Dursun, A., and Çakmakçi, R. (2011). Plant growth promoting rhizobacteria ameliorate deleterious effect of salt stress on lettuce. Sci. Res. Essays 6, 4389–4396. doi: 10.5897/SRE11.219
Yu, Z., Duan, X., Luo, L., Dai, S., Ding, Z., and Xia, G. (2020). How plant hormones mediate salt stress responses. Trends Plant Sci. 25, 1117–1130. doi: 10.1016/j.tplants.2020.06.008
Zhang, H., Xie, X., Kim, M. S., Kornyeyev, D. A., Holaday, S., and Paré, P. W. (2008). Soil bacteria augment Arabidopsis photosynthesis by decreasing glucose sensing and abscisic acid levels in planta. Plant J. 56, 264–273. doi: 10.1111/j.1365-313X.2008.03593.x
Zhang, Y., Fang, J., Wu, X., and Dong, L. (2018). Na+/K+ balance and transport regulatory mechanisms in weedy and cultivated rice (Oryza sativa L.) under salt stress. BMC Plant Biol. 18:375. doi: 10.1186/s12870-018-1586-9
Keywords: salinity stress, growth attributes, hormones, antioxidants, genes expression, rice
Citation: Khan MA, Hamayun M, Asaf S, Khan M, Yun B-W, Kang S-M and Lee I-J (2021) Rhizospheric Bacillus spp. Rescues Plant Growth Under Salinity Stress via Regulating Gene Expression, Endogenous Hormones, and Antioxidant System of Oryza sativa L. Front. Plant Sci. 12:665590. doi: 10.3389/fpls.2021.665590
Received: 08 February 2021; Accepted: 19 May 2021;
Published: 11 June 2021.
Edited by:
Eric Ruelland, UMR7618 Institut d'écologie et des sciences de l'environnement de Paris (IEES), FranceReviewed by:
Alia Dellagi, AgroParisTech Institut des Sciences et Industries du Vivant et de L'environnement, FranceGyöngyvér Mara, Sapientia Hungarian University of Transylvania, Romania
Ma. del Carmen Orozco-Mosqueda, Michoacana University of San Nicolás de Hidalgo, Mexico
Copyright © 2021 Khan, Hamayun, Asaf, Khan, Yun, Kang and Lee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: In-Jung Lee, aWpsZWVAa251LmFjLmty
 Muhammad Aaqil Khan
Muhammad Aaqil Khan Muhammad Hamayun
Muhammad Hamayun Sajjad Asaf
Sajjad Asaf Murtaza Khan
Murtaza Khan Byung-Wook Yun
Byung-Wook Yun Sang-Mo Kang1
Sang-Mo Kang1 In-Jung Lee
In-Jung Lee