- Institute for Plant Sciences, Cluster of Excellence on Plant Sciences (CEPLAS), Biocenter, University of Cologne, Cologne, Germany
CONSTITUTIVE PHOTOMORPHOGENIC 1 functions as an E3 ubiquitin ligase in plants and animals. Discovered originally in Arabidopsis thaliana, COP1 acts in a complex with SPA proteins as a central repressor of light-mediated responses in plants. By ubiquitinating and promoting the degradation of several substrates, COP1/SPA regulates many aspects of plant growth, development and metabolism. In contrast to plants, human COP1 acts as a crucial regulator of tumorigenesis. In this review, we discuss the recent important findings in COP1/SPA research including a brief comparison between COP1 activity in plants and humans.
Introduction
Plants are versatile organisms that coordinate growth and development by constantly sensing and responding to various internal and external signals. By integrating the information from multiple signals at the molecular level, plants orchestrate complex downstream functions that maximize their evolutionary fitness. Among the external signals that influence plant development, light plays a pivotal role. Besides supplying energy for photosynthesis, light functions as an important developmental cue that shapes the life of plants starting from seed germination to senescence. Plants perceive different wavelengths of light via specialized photoreceptors and appropriately change the gene expression patterns that result in photomorphogenesis or light-driven plant growth.
Photoreceptors accomplish a major part of their signaling functions via regulating the activities of CONSTITUTIVE PHOTOMORPHOGENIC 1 (COP1), a master regulator of light signaling. Discovered more than two decades ago in the model plant Arabidopsis thaliana (Arabidopsis), COP1 is among the first-known repressors of photomorphogenesis (Deng et al., 1991). COP1, a Really Interesting New Gene (RING)-finger E3 ubiquitin ligase, exists widely in eukaryotes including mammals. It polyubiquitinates and facilitates the proteasome-mediated degradation of numerous substrates (Wang et al., 1999; Yi and Deng, 2005; Lau and Deng, 2012; Han et al., 2020; Table 1). E3 ubiquitin ligases identify and associate with protein substrates, recruit the ubiquitin conjugating enzyme E2, and assist or directly involve in the transfer of ubiquitin molecules from E2 to the substrates (Mazzucotelli et al., 2006). In plants, the E3 ligase activity of COP1 depends on its interaction with SUPPRESSOR OF PHYA-105 (SPA) proteins (Hoecker and Quail, 2001; Seo et al., 2003; Laubinger et al., 2004; Zhu et al., 2008; Ordoñez-Herrera et al., 2015).
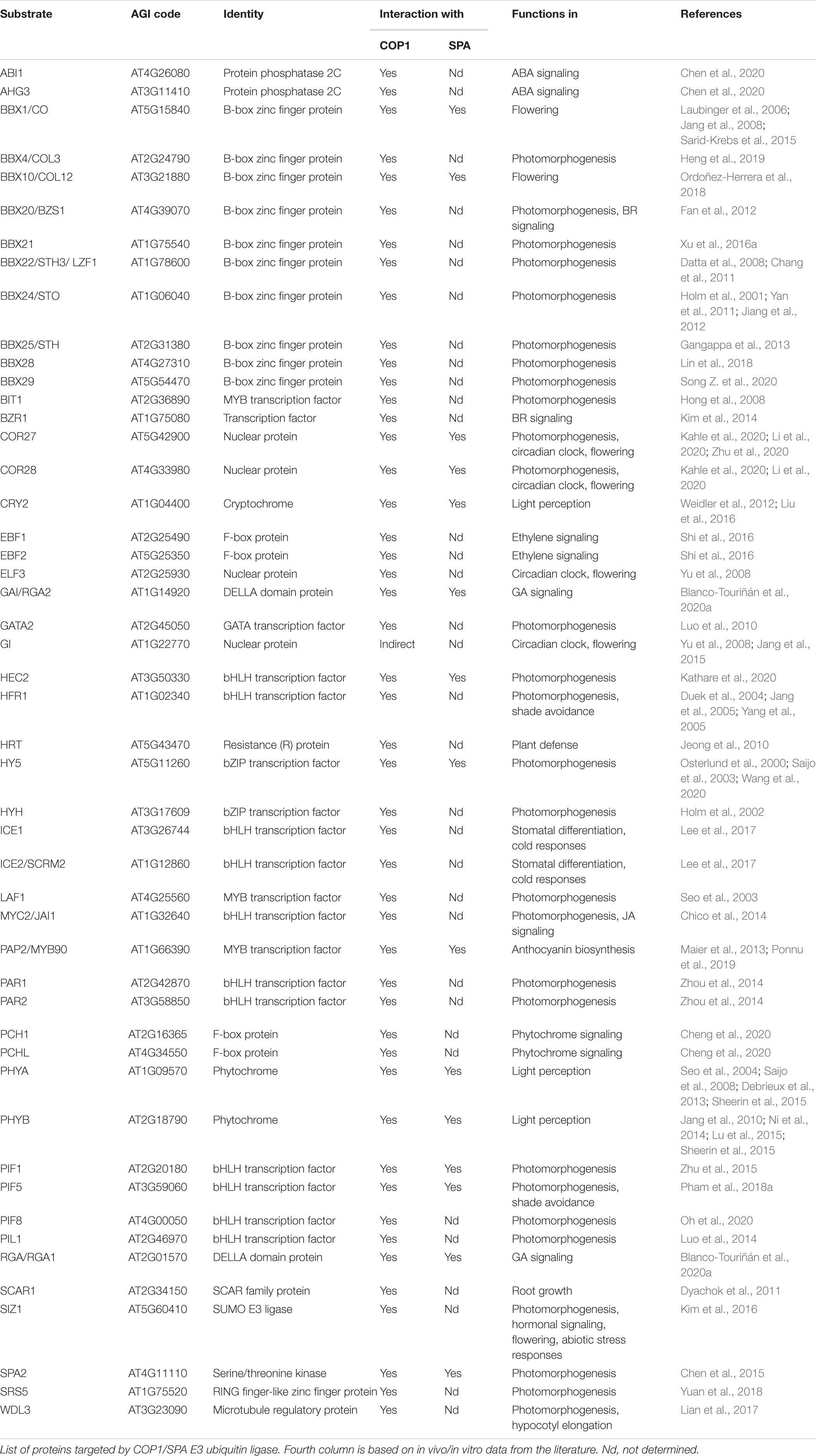
The COP1/SPA complex acts as a central repressor of light signaling in darkness, chiefly by ubiquitinating and thereby promoting the degradation of positive regulators of photomorphogenesis, which mostly are transcription factors. When plants are exposed to light, the photoreceptors suppress COP1/SPA activity, resulting in the stabilization of the COP1/SPA substrates which subsequently promote photomorphogenesis (Hoecker, 2017; Podolec and Ulm, 2018; Ponnu, 2020). COP1/SPA proteins function as part of a CULLIN4-DAMAGED DNA BINDING PROTEIN 1 (CUL4-DDB1)-based multi-subunit, higher-order E3 ligase complex in Arabidopsis (Chen et al., 2010). Depending on the proteins that are targeted for degradation, COP1/SPA regulates various light-promoted developmental processes in plants such as hypocotyl growth, anthocyanin biosynthesis, shade avoidance, flowering time, hormone signaling, and stomata development (Duek et al., 2004; Jang et al., 2005, 2008; Yang et al., 2005; Laubinger et al., 2006; Liu et al., 2008; Lau and Deng, 2012; Maier and Hoecker, 2015; Lee et al., 2017; Wang et al., 2019; Figure 1; Table 1). Besides facilitating the degradation of proteins that promote photomorphogenesis, the COP1/SPA complex stabilizes PHYTOCHROME INTERACTING FACTORs (PIFs) in darkness, which are negative regulators of light signaling (Bauer et al., 2004; Ling et al., 2017; Pham et al., 2018b).
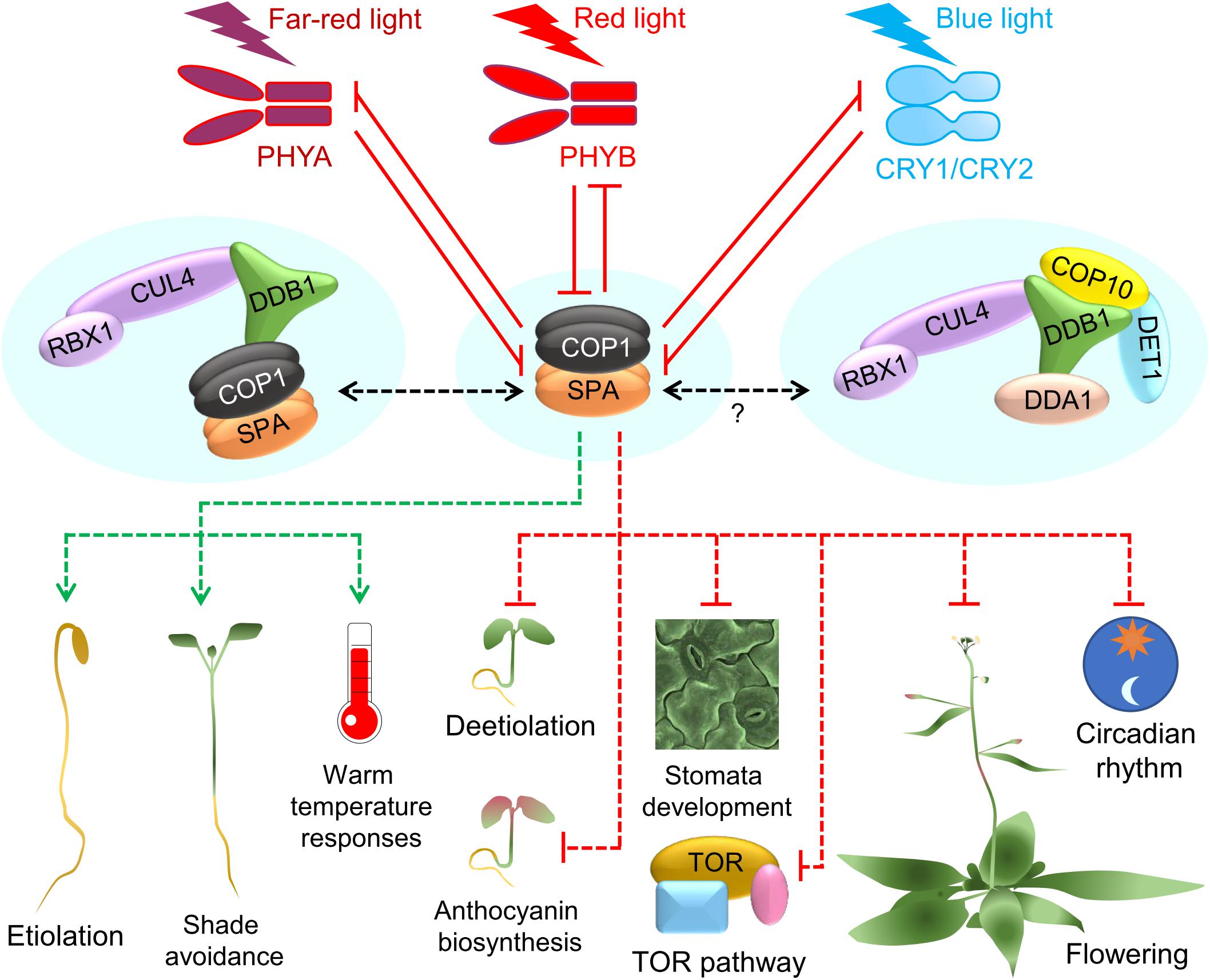
Figure 1. COP1/SPA acts as a central regulator of plant growth and development. A simplified schematic showing the major functions of the COP1/SPA complex as a repressor of light signaling in darkness. Phytochrome and cryptochrome photoreceptors inhibit COP1/SPA activity in the light. When photoreceptors are inactive, COP1/SPA polyubiquitinates a number of substrates and thereby promotes hypocotyl elongation, skotomorphogenesis, warm temperature and shade avoidance responses, but suppresses deetiolation, anthocyanin biosynthesis, stomata development, TOR kinase activity, circadian rhythm and flowering. The Arabidopsis COP1/SPA E3 ligase functions as part of a CUL4-DDB1-RBX1 complex and co-acts with the C3D complex. Green and red arrows represent promotion and suppression, respectively. Solid lines show direct regulation, and the dotted lines show indirect regulation. Question mark (?) shows the mechanisms that are not yet well understood.
Generally, COP1/SPA is an inhibitor of light signaling that is active primarily in darkness. However, in UVB light, COP1 promotes photomorphogenesis by interacting with the UVB receptor UV RESISTANCE LOCUS 8 (UVR8) which leads to the stabilization of HY5 (Oravecz et al., 2006; Liang et al., 2019). Moreover, COP1/SPA facilitates the light-induced degradation of PIFs by associating with phytochromes (PHYs) (Zhu et al., 2015; Pham et al., 2018b). Furthermore, COP1/SPA targets light-activated photoreceptors for degradation, such as PHYA, PHYB and CRYPTOCHROME2 (CRY2) under far red, red and blue light, respectively (Figure 1; Seo et al., 2004; Jang et al., 2010; Weidler et al., 2012; Debrieux et al., 2013; Liu et al., 2016).
Recent years have witnessed many breakthroughs in COP1 research. In this review, we provide an update on the structure and functions of COP1 and discuss the latest developments. Importance is given to the recent works performed in Arabidopsis COP1, with a small section dedicated to mammalian COP1. This review is intended to give a bird’s-eye view of COP1 research and the readers are requested to refer to the in-depth reviews written on specific topics.
Domains and Structure of the COP1/SPA E3 Ligase
Arabidopsis COP1 is a 76.2 kDa protein encoded by a single gene. It contains three distinct domains – the N-terminal zinc-binding RING domain, the central helical coiled-coil (CC) domain, and the C-terminal region (COP1-WD) containing tryptophan-aspartic acid repeats (WD40). The RING domain may interact with an ubiquitin conjugating enzyme E2, the CC domain confers the homodimerization of COP1 and heterodimerization with SPA proteins, and the COP1-WD binds the ubiquitination targets and photoreceptors (Deng and Quail, 1992; von Arnim and Deng, 1993; Torii et al., 1998; Uljon et al., 2016; Hoecker, 2017). The N-terminus of COP1 comprising the RING and CC domains (amino acids 1-282) is indispensable for COP1 function and can suppress the lethality of the cop1-5 null mutant when overexpressed (McNellis et al., 1994; Stoop-Myer et al., 1999; Stacey et al., 2000). A number of proteins were shown to interact with the N-terminus of COP1 including COP10, an E2 variant that also is part of a CUL4-based E3 complex (Suzuki et al., 2002; Yanagawa et al., 2004). While COP10, COP1-INTERACTING PROTEIN 8 (CIP8), and MIDGET (MID) bind specifically the RING domain of COP1, other CIP proteins (CIP1, CIP4 and CIP7) and the COP1-SUPPRESSOR 2 (CSU2) associate with the COP1-CC (Matsui et al., 1995; Yamamoto et al., 1998, 2001; Torii et al., 1999; Schrader et al., 2013; Xu et al., 2015). A single bipartite nuclear localization signal (NLS) between CC and WD, and a long cytoplasmic localization signal situated in the N-terminus facilitate light-mediated nuclear import and export of COP1 (von Arnim and Deng, 1994; Stacey et al., 1999; Subramanian et al., 2004). Nuclear-localized GFP-COP1 exhibits characteristic punctate structures called speckles or nuclear bodies in which interacting proteins also colocalize (Stacey et al., 1999).
A break-through crystallization study showed that the WDs of plant and mammalian COP1 have a similar seven-bladed β-propeller configuration (Uljon et al., 2016). An intact COP1-WD is essential for COP1 function in vivo which is consistent with its role in substrate recognition (McNellis et al., 1994; Uljon et al., 2016). Many substrates of COP1 share a short, conserved valine-proline (VP) motif as the COP1-WD binding site (Holm et al., 2001; Laubinger et al., 2006; Uljon et al., 2016; Lau et al., 2019; Ponnu et al., 2019; Yadav et al., 2020). This mechanism was also adopted by the photoreceptors, UVR8 and CRYs, which interact with COP1 via their conserved VP motifs (Lau et al., 2019; Ponnu et al., 2019). To accommodate diverse VP motifs of a wide range of interaction partners, the COP1-WD evolved an array of conserved amino acids that form a flexible VP-binding pocket (Uljon et al., 2016; Lau et al., 2019). Interestingly, the interaction partners of mammalian COP1, such as the human TRIB1 (homolog of Drosophila tribbles) can also bind Arabidopsis COP1-WD via its VP-motifs, pointing to the coevolution of the COP1-VP-binding pocket along with the VP-containing proteins (Uljon et al., 2016). In addition to substrates and photoreceptors, COP1-WD binds DDB1 in the CUL4 complex (Chen et al., 2010).
In contrast to a single COP1 protein, Arabidopsis has four SPA proteins (SPA1 to SPA4) which have partially redundant, but also distinct functions (Hoecker et al., 1999; Laubinger et al., 2004, 2006; Menon et al., 2016). All four SPAs carry similar domains, with a CC and WD-repeat comparable to that of COP1 (Hoecker, 2017). But instead of a RING domain in COP1, SPA proteins have a weakly conserved N-terminal kinase-like domain. At least for SPA1, the kinase activity of the N-terminal domain was demonstrated recently (Paik et al., 2019; Wang et al., 2020). Accordingly, missense mutations in the kinase domain severely compromise SPA1 function in transgenic Arabidopsis plants (Holtkotte et al., 2016; Paik et al., 2019). The N-terminal domains of SPA1 and SPA2 are also responsible for the destabilization of these SPA proteins (Fittinghoff et al., 2006; Yang and Wang, 2006; Chen et al., 2016). Like the respective domains of COP1, the CC of SPA1 engages in homo- and heterodimerization (with other SPAs and with COP1), and the WD participates in substrate interactions. However, in contrast to COP1, detailed structural information is lacking for SPA-WD as no crystallization studies have been successful to date. Transgenic plants expressing individual point mutations in SPA1-WD revealed the indispensable functions of many residues in this region (Yang and Wang, 2006). In fact, the SPA1-WD contains the same conserved amino acids that make up the COP1-VP-binding pocket, suggesting that SPA proteins also engage in VP-mediated interactions (Ponnu et al., 2019). Consistent with this hypothesis, the C-termini of CRYs interact with SPA1-WD via their VP motifs (Ponnu et al., 2019).
One of the functional COP1/SPA complexes is likely a heterotetramer consisting of a COP1 homodimer with different combinations of two SPA proteins (SPA homo/heterodimers) (Zhu et al., 2008). Since photoreceptor-mediated suppression of COP1 activity involves direct interactions of light-activated photoreceptors with COP1 and SPA proteins (Podolec and Ulm, 2018), a COP1/SPA tetramer may interact with a tetramer of photoreceptors. In agreement with this idea, the Arabidopsis CRYs were recently demonstrated to form blue light-induced homo- and heterooligomers (Liu Q. et al., 2020; Shao et al., 2020; Palayam et al., 2021). A possible scenario of CRY-COP1/SPA interactions might be the binding of CRY-homo- or heterotetramers with a COP1/SPA heterotetramer, with each photoreceptor monomer in direct association with a COP1 or SPA monomer. In-depth structural and proteomic studies are required to confirm the presence of such large protein complexes in planta.
COP1/SPA as a Part of a CUL4-Based E3 Ligase
There is very good evidence that in both plants and animals, COP1 acts as a part of a CUL4-based E3 ligase consisting of the core proteins CUL4, DDB1 and a RING-BOX protein (RBX) (Wertz et al., 2004; Chen et al., 2006). CUL4 is a scaffold protein anchoring a DDB1-based adaptor module and the E2-recruiting protein RBX1 at its N and C termini, respectively. In the adaptor module, DDB1 forms the core linker that attaches DWD box-containing proteins as substrate receptors. A wide range of DWD proteins that interact with DDB1 provide functional specificities to the CUL4-DDB1 E3 ligases (Hua and Vierstra, 2011). In vitro biochemical experiments showed that COP1/SPA proteins function as DWD proteins and associate with DDB1 via their DWD boxes to form a CUL4-COP1/SPA complex (Chen et al., 2010). However, the crystal structure of COP1-WD revealed that the DWD box is buried within and cannot be accessed without destroying the WD (Uljon et al., 2016). Hence, further studies are necessary to decipher the details of specific interactions between COP1/SPA and DDB1.
In addition to the observed DDB1-COP1/SPA protein-protein interactions, genetic evidence also strongly supports that COP1/SPA acts as part of a CUL4-based E3 ubiquitin ligase. Viable, hypomorphic cul4 cosuppression lines exhibit constitutive photomorphogenesis in darkness and early flowering phenotypes similar to those of cop1 and spa mutants. Moreover, cosuppression of CUL4 synergistically interacts with a weak mutation in COP1 (Chen et al., 2010). Also, COP1/SPA action in PIF degradation or thermomorphogenesis involves CUL4 (Delker et al., 2014; Zhu et al., 2015; Gangappa and Kumar, 2017). Unclear, however, is the role of the RING finger in COP1, since the COP1 RING finger, like RBX1, can bind E2 (Schulman et al., 2000; Zheng et al., 2002; Choi et al., 2014), suggesting redundant activities in the CUL4-RBX1-DDB1-COP1/SPA complex. Indeed, in vitro, recombinant COP1 has ubiquitin ligase activity via its RING finger domain without the need for other CUL4 complex components (Saijo et al., 2003; Seo et al., 2003). In agreement with this finding, the RING finger of COP1 is essential for COP1 function in vivo (Torii et al., 1998; Stoop-Myer et al., 1999). It is therefore possible that COP1/SPA proteins may not always occur in association with the CUL4-RBX1-DDB1 complex but may also act as a CUL4-independent E3 ubiquitin ligase.
Not well understood is the co-action of COP1 with DET1, another essential protein in the suppression of photomorphogenesis in darkness (Pepper et al., 1994). DET1 associates with DDB1, COP10, and DDB1-ASSOCIATED 1 (DDA1) to form a COP10-DDB1-DET1-DDA1 (C3D) complex that binds CUL4 and functions as a CUL4-based E3 ligase (Chen et al., 2006; Irigoyen et al., 2014; Fonseca and Rubio, 2019). Since Arabidopsis COP1 interacted with DDB1 independently of DET1, it was proposed that the CUL4-DDB1-COP1/SPA complex may be structurally distinct from the CUL4-C3D complex (Chen et al., 2010; Lau and Deng, 2012; Irigoyen et al., 2014). However, a recent preprint demonstrated an in vivo association of COP1/SPA proteins with the C3D complex in dark-grown Arabidopsis cell cultures that constitutively express DET1 (Cañibano et al., 2020). This argues for the possibility that COP1/SPA proteins may form a part of the DET1-containing C3D complex in Arabidopsis. The direct interaction between COP1 and COP10, a component of the C3D complex (Suzuki et al., 2002), may support this idea. Also, multiple pieces of evidence show that DET1 and COP1/SPA act in concert in regulating photomorphogenesis and thermomorphogenesis (Chen et al., 2010; Delker et al., 2014; Gangappa and Kumar, 2017). det1 and cop1 mutants have similar pleiotropic phenotypes with highly comparable gene expression patterns, such as the constitutive and abnormally high expression of light-responsive genes (Mayer et al., 1996; Ma et al., 2003). A weak allele of det1, acting as an enhancer of a spa1 mutation, provided further evidence for DET1-COP1/SPA co-action throughout the developmental stages of plants (Nixdorf and Hoecker, 2010). In agreement with this, double mutant and biochemical analyses showed that CUL4 and DET1 act synergistically with COP1 in repressing photomorphogenesis (Chen et al., 2010). Further studies are required to decipher the molecular mechanism involved in the co-action between COP1/SPA proteins and the components of the C3D complex.
Functions of COP1/SPA in Seedling Etiolation
Dark-grown seedlings undergo skotomorphogenesis and show etiolation characterized by an elongated hypocotyl, closed and yellow cotyledons, and a tightly folded apical hook. The elongated hypocotyl increases the likelihood that a soil-covered seedling reaches the light. Hence, energy is diverted from cotyledon development to elongation growth. Once exposed to light, hypocotyl elongation is inhibited, hook and cotyledons open, and chlorophyll is synthesized, i.e., the seedling deetiolates. COP1/SPA prevents deetiolation in darkness by marking several transcription factors for degradation. This includes HY5, HY5 HOMOLOG (HYH) and LONG HYPOCOTYL IN FAR-RED 1 (HFR1) and a number of other positive regulators of the light responses. In spa quadruple and cop1 mutants, these transcription factors also accumulate in darkness, causing constitutive photomorphogenesis in complete darkness (Deng et al., 1991; Laubinger et al., 2004; Ordoñez-Herrera et al., 2015).
HY5 is a bZIP transcription factor that was recently shown to function mainly by activation of gene expression rather than by gene repression (Burko et al., 2020). Along with HYH and HFR1, HY5 suppresses the elongation of hypocotyls in light-grown seedlings (Lau and Deng, 2012; Hoecker, 2017; Yuan et al., 2018; Bhatnagar et al., 2020). HY5 is a non-canonical transcriptional regulator that carries a DNA-binding domain but no transcriptional activation domain. It was postulated earlier that HY5 may act in concert with other transcription factors that provide a domain for transcriptional activation. Indeed, members of a large family of B-box transcription factors (BBXs) were recently shown to interact with HY5 and to modulate HY5 activity (Datta et al., 2007, 2008; Gangappa et al., 2013; Wei et al., 2016; Lin et al., 2018; Burko et al., 2020; Bursch et al., 2020; Song Z. et al., 2020; Zhao et al., 2020). In particular, BBX20 to BBX22 were shown to be required for HY5 activity (Bursch et al., 2020). Interestingly, like HY5, many BBX proteins are also substrates of COP1 (Vaishak et al., 2019; Yadav et al., 2020). Hence, both types of transcription factors (bZIP and BBXs) that are mostly unrelated in sequence evolved to be targeted by COP1 in darkness. Many BBXs and HY5 do share COP1-WD-binding VP motifs mediating the interaction with COP1 (Gangappa and Botto, 2014; Yadav et al., 2020). BBX proteins also regulate the expression of other BBX proteins in a feedback regulatory loop involving COP1. For example, BBX4 promotes the expression of BBX11, a negative regulator of photomorphogenesis, but BBX11 interacts with COP1 to regulate the activity of BBX4 (Liu B. et al., 2020). A systematic approach is needed to identify the BBXs that interact with the COP1/SPA complex and to study the implications of these associations on light-mediated plant development.
In addition to ubiquitinating proteins that promote photomorphogenesis, COP1/SPA stabilizes PIFs in darkness to positively regulate skotomorphogenesis. PIFs are bHLH transcription factors that, like COP1/SPA, are required for seedling etiolation in darkness (Leivar et al., 2008; Pham et al., 2018b). They upregulate many elongation-related genes (Pham et al., 2018b). Interestingly, COP1 stabilizes PIFs via a non-canonical mechanism and apparently not via its E3 ligase activity: PIF3 and PIF4 abundance is regulated by BRASSINOSTEROID-INSENSITIVE 2 (BIN2), a GSK3-like kinase. BIN2 phosphorylates PIF3 and PIF4 which leads to their degradation under dark conditions (Bernardo-García et al., 2014; Ling et al., 2017; Figure 2). The COP1/SPA complex prevents PIF phosphorylation by BIN2, thereby inhibiting the degradation of PIF3 and PIF4. While COP1 interacts with BIN2 and thereby sequesters BIN2 from binding to PIFs, SPA1 occupies the BIN2-binding domain of PIF3 (Ling et al., 2017). By stabilizing PIF3 levels, the COP1/SPA complex promotes etiolation in darkness. While suppressing photomorphogenesis by stabilizing PIFs, the COP1/SPA complex also prevents the over-accumulation of PIFs, such as PIF1, via a co-degradation mechanism involving HFR1 in darkness (Xu et al., 2017).
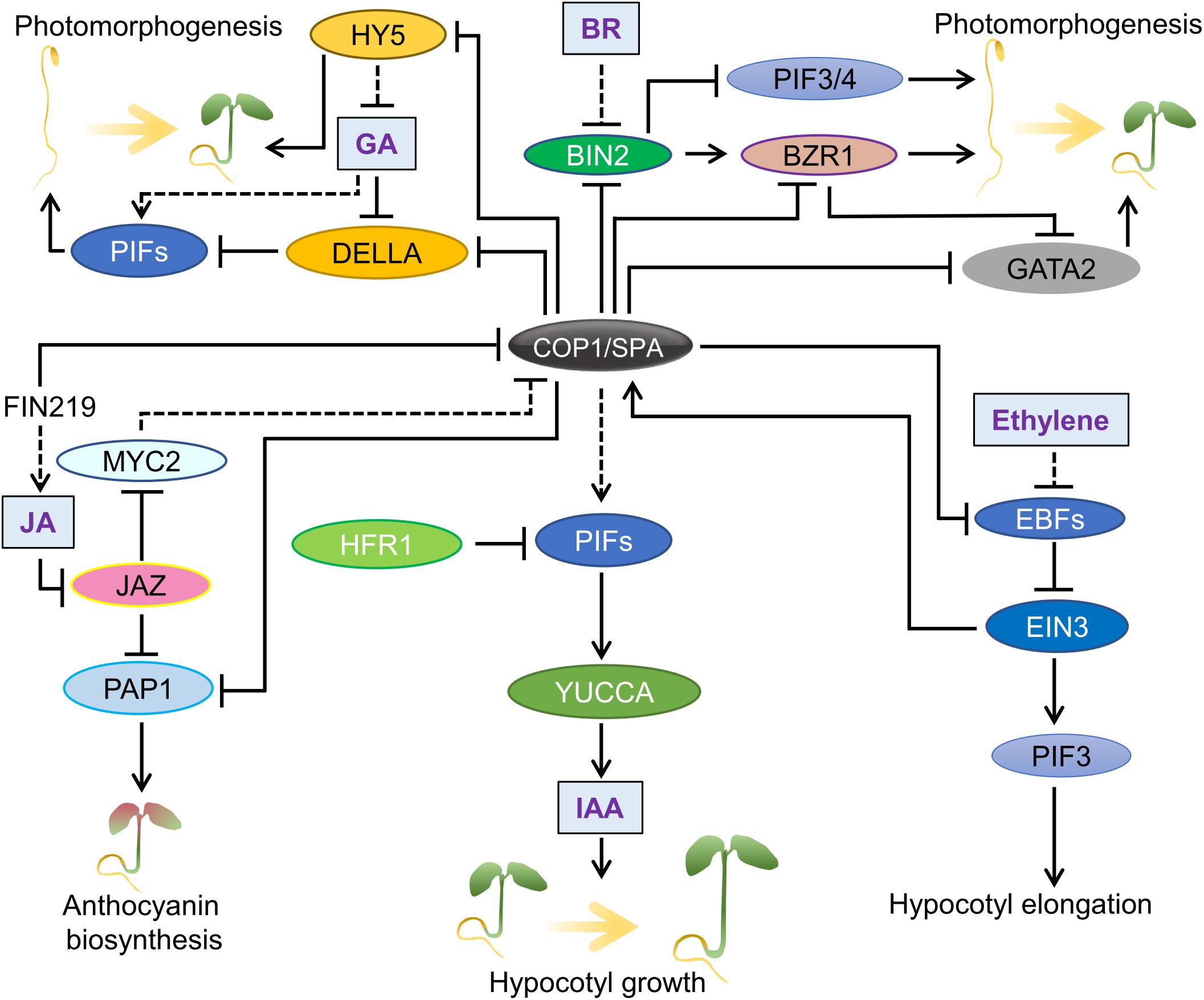
Figure 2. Function of the COP1/SPA complex in hormone responses. An overview of the major signaling pathways that involve hormones and the COP1/SPA complex. Effects of auxin (IAA), jasmonic acid (JA), gibberellic acid (GA), brassinosteroid (BR) and ethylene are shown. Solid lines show direct regulation and dotted lines represent indirect influence.
Under light conditions, photoreceptors such as CRYs, PHYs and UVR8 suppress the activity of the COP1/SPA complex to promote photomorphogenesis (Hoecker, 2017; Podolec and Ulm, 2018; Ponnu, 2020). As revealed by recent studies, the COP1/SPA complex also functions as a promoter of photomorphogenesis, via destabilizing PIFs under light conditions. The COP1/SPA complex mediates the light-induced degradation of PIF1 by functioning as a CUL4-COP1/SPA E3 ligase in a PHYB-dependent manner (Zhu et al., 2015). In this complex, SPA1 functions as a serine/threonine kinase and phosphorylates PIF1, which is necessary for light-induced PIF1 degradation (Paik et al., 2019). The PHOTOPERIODIC CONTROL OF HYPOCOTYL 1 (PCH1) and PCH1-LIKE (PCHL) that regulate the thermal reversion of PHYB, enhance the degradation of PIF1 in light, by promoting PHYB-PIF1 and COP1/SPA-PIF1 associations (Cheng et al., 2020).
Functions of COP1/SPA in Shade
Plants growing under dense canopies (low red to far-red ratio) undergo adaptive changes in response to shade. These responses include enhanced hypocotyl and internode elongation, hyponasty, reduced branching and accelerated flowering, and are collectively known as shade-avoidance syndrome (SAS) (Fiorucci and Fankhauser, 2017). Both COP1 and SPA proteins are essential for most facets of the SAS, except for earlier flowering (McNellis et al., 1994; Rolauffs et al., 2012). Shade enhances elongation growth by promoting the nuclear accumulation of COP1 (Pacín et al., 2013) which subsequently promotes the degradation of HFR1, a negative regulator of shade avoidance responses. Low levels of HFR1 lead to an activation of PIF-mediated gene expression which enhances hypocotyl elongation via auxin (Pacín et al., 2016; Iglesias et al., 2018). In addition, COP1 may act via HY5 and HYH which suppress stem growth in Arabidopsis (Nozue et al., 2015). Genetic interaction between COP1 and BBX transcription factors (BBX21, BBX22 and BBX25) demonstrated the involvement of BBXs in SAS responses (Crocco et al., 2010; Gangappa et al., 2013). Hence, BBX and HY5 may act in concert to regulate the SAS. The SAS responses are strongly inhibited by UVB via UVR8-COP1 interactions, resulting in stabilization of HY5 and suppression of PIFs (Favory et al., 2009; Hayes et al., 2017).
COP1/SPA Proteins in Temperature Responses
During their life cycle, plants are challenged with fluctuations in temperature. Extreme temperatures (frost, excessive heat) lead to stress responses, while smaller alterations lead to adaptive growth. Such adaptive growth responses to high ambient temperature (ca. 27–29°C for Arabidopsis) mimic growth responses in canopy shade: hypocotyls and internodes elongate and leaves show upward (hyponastic) growth, perhaps to cool the plant surface and to protect the meristem from soil heat (Crawford et al., 2012). Light and temperature signaling are strongly interconnected (Quint et al., 2016). Indeed, PHYB is a sensor of high ambient temperature because high temperature promotes dark reversion of PHYB from the active to the inactive conformation (Jung et al., 2016; Legris et al., 2016). Since active PHYB inhibits COP1/SPA activity, one can expect that COP1/SPA is also involved in temperature signaling. Experimental results confirm this idea: both COP1 and SPA are required for the response to high ambient temperature (Delker et al., 2014; Gangappa and Kumar, 2017; Lee et al., 2020).
At the cellular level, PIF4 coordinates light and warm temperature signals to promote hypocotyl elongation (Martínez et al., 2018; Vu et al., 2019). A DET1-COP1-HY5 hub controls PIF4 levels (Delker et al., 2014). While DET1 and COP1 stabilize PIF4 and promote hypocotyl elongation at elevated temperatures, HY5 negatively regulates thermosensory growth by competing with PIF4 targets (Gangappa and Kumar, 2017). Warm temperature-stabilized COP1/SPA complex reduces HY5 levels and stabilizes PIF4 (Legris et al., 2017; Martínez et al., 2018). Consistent with this observation, elevated temperatures were shown to promote nuclear accumulation of COP1 which leads to the suppression of HY5 activity and hypocotyl elongation (Park et al., 2017). In addition, elevated temperatures reduce anthocyanin biosynthesis via the COP1-HY5 module. Plants grown at warm temperatures accumulate less HY5 and also show reduced accumulation of anthocyanins (Kim et al., 2017). By directly binding and repressing MYB-LIK2 (MYBL2) transcription factor that negatively regulates anthocyanin biosynthesis (Matsui et al., 2008), HY5 acts as a positive regulator of anthocyanin accumulation (Wang et al., 2016). Lower HY5 at elevated temperatures leads to the enhanced expression of MYBL2, causing reduced anthocyanin biosynthesis (Kim et al., 2017). The enhancement of COP1 activity under elevated temperature seems to be tissue-specific, as COP1 was shown to be depleted at higher temperatures in the Arabidopsis rosettes which stabilizes GIGANTEA (GI) and accelerates flowering (Jang et al., 2015). In contrast, cold temperature depletes COP1 from the nuclei of root cells and causes HY5 accumulation which leads to the expression of cold-responsive genes (Catalá et al., 2011). Taken together, these observations point to the complexity of COP1-regulation by temperature signals. Future research should aim at elucidating the mechanisms that govern temperature-dependent nuclear import and export of COP1 in different tissues and at different developmental stages of plant growth.
In addition to COP1, SPA proteins are also required for PIF4-mediated thermomorphogenesis (Delker et al., 2014; Lee et al., 2020). SPAs were required for PIF4 activity, and SPA1-mediated phosphorylation of PIF4 was recently demonstrated in vitro. In contrast to BIN2-mediated phosphorylation that destabilizes PIF4 (Ling et al., 2017), SPA1-mediated phosphorylation appears to stabilize PIF4. Also, SPAs promote PHYB degradation in the SPA1-PHYB-PIF4 complex under warm temperatures, which may further stabilize PIF4 (Lee et al., 2020). The mechanistic details underlying the differential fates of PIF4 stability based on the phosphorylating kinases will be of interest in future research.
The COP1/SPA Complex in Hormonal Signaling
COP1/SPA activity intersects with multiple hormone signaling pathways in the control of growth, development, and stress responses (Wang et al., 2019). In particular, responses to shade and elevated ambient temperature involve the hormone auxin. Auxin is a universal promoter of hypocotyl and petiole growth. PIF transcription factors have been shown to enhance auxin levels in the shade by directly activating auxin biosynthesis genes such as YUCCAs, and COP1/SPA is required for this activation (Rolauffs et al., 2012; Pacín et al., 2016; Iglesias et al., 2018; Figure 2). Under warm temperatures and shade conditions, COP1 stabilizes PIFs leading to an enhancement in auxin levels (Gangappa and Kumar, 2017; Park et al., 2017). In contrast, under UVB light, UVR8 interacts with COP1 to suppress the E3 ubiquitin ligase activity, leading to destabilization of PIF4 and inhibition of hypocotyl growth via reduced auxin biosynthesis (Hayes et al., 2017; Tavridou et al., 2020). COP1 regulates polar auxin transport and hence root growth, perhaps by regulating PIN-FORMED (PIN) auxin transporters (Sassi et al., 2012).
Brassinosteroids (BRs) are plant hormones that are repressors of light-mediated de-etiolation and chlorophyll biosynthesis (Gupta and Nath, 2020). Consequently, BR-insensitive and deficient mutants show constitutive photomorphogenesis in darkness (Li et al., 1996; Nam and Li, 2002; Song et al., 2009). The signaling cross-talk mediated by COP1 and the BR pathway is an emerging field (Figure 2). One of the converging points of light and BR signaling may involve the GATA2 transcription factor that positively regulates light signaling. Both COP1 and BR signaling suppress the activities of GATA2 in darkness to promote skotomorphogenesis. While COP1 targets GATA2 for degradation, BRASSINAZOLE-RESISTANT 1 (BZR1) transcription factor inhibits GATA2 expression in darkness (Luo et al., 2010). It is possible that COP1 may affect BZR1 via the inhibition of BIN2 (Ling et al., 2017), a kinase that phosphorylates both BZR1 (He et al., 2002, 2) and PIF4 (Bernardo-García et al., 2014), and also act as a negative regulator of BR signaling. Consistent with this, elevated temperatures increase the nuclear accumulation of COP1, which sequesters BIN2 and enhances the function of PIF1 and BZR1 (Nieto et al., 2020). Alternatively, COP1 may reduce the BR sensitivity at cellular level by regulating MEMBRANE STEROID BINDING PROTEIN 1 (MSBP1) involved in suppressing the BR perception (Song et al., 2009). In darkness the activity of MSBP1 is suppressed in a COP1-dependent manner, which may increase BR perception and promote etiolation (Shi et al., 2011).
Gibberellins (GA) act similar to BR and suppress photomorphogenesis in darkness (Alabadí et al., 2004, 2008; Figure 2). GA and COP1 intersect at the DELLA proteins which are key negative regulators of GA signaling. In response to shade and elevated ambient temperatures, COP1 directly targets DELLAs for degradation and thereby promotes plant growth under these conditions (Blanco-Touriñán et al., 2020a). DELLAs are also degraded in response to GA via the SLEEPY1 (SLY1) ubiquitin ligase (Dill et al., 2004; Blanco-Touriñán et al., 2020b). SLY1 and COP1 interact with different domains in the DELLA proteins, suggesting independent mechanisms (Blanco-Touriñán et al., 2020a). DELLAs are negative regulators of PIFs, and by causing DELLA degradation, GA, promotes skotomorphogenesis by enhancing PIF activity. The COP1-HY5 pathway plays a crucial role in mediating GA-dependent skotomorphogenesis probably also via PIFs (Mazzella et al., 2014). Light promotes photomorphogenesis by suppressing GA signaling, mainly by reducing the GA content, and presumably also via regulating COP1-HY5-PIF module (Alabadí et al., 2008; Mazzella et al., 2014).
Ethylene is a gaseous hormone that acts contrastingly in dark and light. Under light conditions, ethylene promotes hypocotyl growth, but suppresses it in darkness (Yu and Huang, 2017; Figure 2). However, most of the ethylene-mediated processes in plants require light. COP1 co-operates with ethylene via stabilizing ETHYLENE INSENSITIVE 3 (EIN3), which promotes ethylene responses (Zhong et al., 2014). COP1 stabilizes EIN3 by degrading EIN3-BINDING F-BOX PROTEINs (EBFs) that target EIN3 for degradation (Shi et al., 2016). Interestingly, ethylene via EIN3 positively regulates the nuclear accumulation of COP1 in the light and thereby causes HY5 degradation to modulate hypocotyl growth (Yu et al., 2013). Taken together, the present knowledge suggests that under light conditions the COP1-HY5 module acts together with the ethylene-EIN3-PIF pathway to promote hypocotyl growth. The mechanisms of ethylene-COP1 coaction in darkness are not fully understood.
Jasmonic acid (JA), a plant hormone crucial in biotic stresses such as herbivory and pathogen attack, is also involved in the regulation of skotomorphogenesis (Huang et al., 2017; Figure 2). At lower levels of JA, JASMONATE-ZIM domain proteins (JAZs) suppress MYC transcription factors. JA degrades JAZ via recruiting an E3 ligase, leading to the activation of MYC2. In presence of light, MYC2 activates HY5. In addition, MYC2 can affect the function of both COP1 and SPA proteins (Gangappa et al., 2010; Zheng et al., 2017). Alternatively, JA can suppress the formation of the COP1/SPA complex and can attenuate the function of nuclear COP1 (Zheng et al., 2017). Whether JA-mediated COP1 suppression involves MYC2 is currently unclear. Another mode in which JA signaling affects COP1 activity and hypocotyl growth is through FAR-RED INSENSITIVE 219/JASMONATE RESISTANT 1 (FIN219/JAR1) that catalyzes the synthesis of the bioactive form of JA. FIN219 regulates hypocotyl elongation in shade conditions by promoting the cytoplasmic accumulation of COP1 (Swain et al., 2017). This action may antagonize shade-induced nuclear accumulation of COP1 (Pacín et al., 2013). Additionally, in blue light, FIN219-mediated export of COP1 from the nucleus act in concert with CRY-mediated suppression of COP1/SPA complex (Chen H.-J. et al., 2018). However, JA enhances the CRY-FIN219 association in blue light, thereby attenuating the CRY-mediated suppression of the COP1/SPA complex. This leads to enhanced COP1 activity and reduction of HY5 levels in the nucleus that promotes hypocotyl elongation (Chen H.-J. et al., 2018). JA-COP1 action is also implicated in the regulation of photomorphogenesis such as anthocyanin biosynthesis and protochlorophyllide formation (Wang et al., 2019). Similar to JA, cytokinins (CKs) promote photomorphogenesis and might act via other hormonal pathways. CKs may regulate plant growth via the COP1-HY5 module, but the underlying mechanisms remain unexplored (Vandenbussche et al., 2007).
Abscisic acid (ABA) functions opposite to light signals, antagonizing seed germination, seedling establishment, stomatal opening and root growth. ABA-INSENSITIVE 5 (ABI5), a bZIP transcription factor that plays a major role in ABA responses, colocalizes with COP1 (Lopez-Molina et al., 2003). Although direct interaction between COP1 and ABI5 is yet to be proven, genetic analyses suggest that COP1 acts downstream of ABI5 and promotes ABA signaling. COP1 positively regulates the ABA-mediated post-germination growth arrest of seedlings, by facilitating the binding of ABI5 to the promoters of ABA-responsive genes (Yadukrishnan and Datta, 2020). In addition, COP1 colocalizes with ABI5-binding protein AFP1, which promotes ABI5 degradation (Lopez-Molina et al., 2003). COP1-HY5 module also converges on ABA signaling via ABI4, another transcription factor that mediates ABA signaling. ABI4 acts antagonistically with HY5 in regulating COP1 and both ABI4 and HY5 are targeted by COP1 for degradation under dark and light conditions. Since the nuclear ABI4 is activated by a chloroplast-derived signal, COP1 integrates retrograde and light signaling pathways (Xu et al., 2016c). Besides acting at ABA-mediated seed germination and seedling establishment, COP1 participates in ABA-induced stomatal closure and functions as an important regulator of abiotic stress responses in Arabidopsis and pea (Moazzam-Jazi et al., 2018). Recently a mechanism of COP1-mediated stomatal closure via the regulation of ABA co-receptors has been described. COP1 directly interacts and facilitates the degradation of type 2C phosphatases (PP2Cs), which are ABA co-receptors that positively regulate stomatal opening in the absence of ABA. By degrading PP2Cs, COP1 promotes stomatal closure and acts together with ABA (Chen et al., 2020). The COP1-interacting protein CIP1 has also been reported to promote ABA signaling, but further details remain unknown (Ren et al., 2016).
The mechanisms underlying the activity of strigolactone (SL) in regulating plant growth and development are poorly understood. SL has been reported to stabilize HY5 via suppressing COP1 activity to promote photomorphogenesis. This is thought to occur via nuclear export of COP1 or COP1 degradation. Thus, SL may act in parallel with photoreceptors and repress COP1 (Tsuchiya et al., 2010; Jia et al., 2014). Also COP1 and HY5 associate with BBX20, a negative regulator of the SL pathway (Wei et al., 2016). The role of COP1/SPA complex in the SL pathway needs to be explored further in detail.
Other Functions of the COP1/SPA Complex
The numerous functions of the COP1/SPA complex are mediated by an increasing number of substrates that are being identified, with each substrate having specific functions in plant growth, development, and metabolism (see also Table 1). However, in contrast to mammalian COP1, the functions of plant COP1 in cell division, cell elongation, cytoskeletal network, etc., are relatively less explored. An earlier study suggests that in darkness, COP1 inhibits root elongation in Arabidopsis seedlings by targeting and degrading SCAR, a component of cytoskeletal dynamics (Dyachok et al., 2011). A direct link between COP1 and the cortical microtubule was found also in hypocotyl cells. In darkness, COP1 binds and promotes the degradation of WAVE-DAMPENED 2-LIKE 3 (WDL3), a microtubule-associated protein that promotes cell elongation and hypocotyl elongation (Lian et al., 2017). Surprisingly, WDL3 is a cytosolic protein (Lian et al., 2017) and therefore an unusual COP1 target since COP1 activity usually depends on its nuclear localization (von Arnim et al., 1997). Another example supporting a cytoplasmic activity of the COP1/SPA complex was reported in the microRNA (miRNA) biogenesis pathway of Arabidopsis (Cho et al., 2014). cop1 mutants are defective in miRNA biogenesis due to low levels of HYPONASTIC LEAVES 1 (HYL1), an RNA-binding protein involved in miRNA processing. HYL1 is cleaved by an unknown protein in the cytoplasm. Upon light exposure, COP1 translocates into the cytoplasm and may facilitate the degradation of this unknown protein. In this way, COP1 stabilizes HYL1 levels and thereby promotes microRNA biogenesis (Cho et al., 2014). However, the implication of possible COP1 activity in the cytosol requires further research. Interaction between COP1 and MIDGET (MID), a component of topoisomerase VI, via the very N-terminal domain of COP1 suggests that COP1 may have a direct effect on endoreduplication and genome integrity, the primary functions of MID (Schrader et al., 2013). As COP1 does not affect MID protein levels, the impact of the COP1-MID interaction in DNA replication and genome integrity is so far unclear (Schrader et al., 2013).
Mutations in COP1/SPA genes cause pleiotropic phenotypes throughout plant development (McNellis et al., 1994; Laubinger et al., 2004). COP1/SPA aligns endogenous developmental pathways with the ambient light environment by controlling the protein stability of key developmental regulators. A pivotal function of COP1/SPA in the regulation of flowering time by photoperiod involves the transcription factor CONSTANS (CO). In darkness, CO is degraded via COP1/SPA which causes a delay in flowering under non-inductive short-day conditions, and therefore allows adjustment of flowering time to seasons (Laubinger et al., 2006; Jang et al., 2008; Liu et al., 2008; Sarid-Krebs et al., 2015; Xu et al., 2016b). Consistent with this finding, Arabidopsis cop1 mutants constitutively express floral integrator genes and undergo flowering in complete darkness when grown on sucrose-containing media (Nakagawa and Komeda, 2004). COP1 also suppresses flowering time by promoting the degradation of GI, a circadian clock-associated protein, in an EARLY FLOWERING 3 (ELF3)-dependent manner, and functions as an integrator of photoperiod and circadian signals (Yu et al., 2008). In darkness, COP1 represses TARGET OF RAPAMYCIN (TOR) kinase, the central regulator of energy signaling required for light-induced growth activation. By suppressing TOR in darkness, COP1 prevents light-induced stem cell activation at the shoot apex (Pfeiffer et al., 2016) and also the light-mediated enhancement in translation (Chen G.-H. et al., 2018).
A recent work demonstrated another mode of signal integration between light and the circadian clock via the regulation of COLD REGULATED 27 (COR27) and its homolog COR28 by the COP1/SPA complex. Both CORs are key regulators of the circadian clock and cold responses (Wang et al., 2017), and also function as negative regulators of light signaling. In darkness, CORs interact with the COP1/SPA complex via their VP motifs and thereby are degraded via the 26S proteasome (Kahle et al., 2020; Zhu et al., 2020). In light, CORs interact with HY5 and interfere with HY5 activity on the downstream genes to fine-tune hypocotyl growth in Arabidopsis (Kahle et al., 2020; Li et al., 2020; Zhu et al., 2020).
The differentiation of stomata from protodermal cells is also under the control of light and COP1/SPA. In darkness, stomata differentiation is suppressed by COP1/SPA-mediated degradation of INDUCER OF CAB EXPRESSION 1 (ICE1), a transcription factor required for stomata differentiation (Lee et al., 2017). Accordingly, cop1 mutants accumulate higher levels of ICE1 proteins in the nuclei of abaxial leaf epidermal cells, suggesting that the interaction partners of the COP1/SPA complex may vary according to the tissue types. In addition to stomatal differentiation, COP1 promotes ABA-mediated stomatal closure (Chen et al., 2020) and functions as an important regulator of abiotic stress in plants (Moazzam-Jazi et al., 2018). The role of COP1 in biotic stresses has also been documented. COP1 negatively regulates defense against Turnip Crinkle Mosaic Virus (TMV) in darkness by interacting and marking the resistance protein HYPERSENSITIVE RESPONSE TO TCV (HRT) for degradation (Jeong et al., 2010). Paradoxically, COP1 also contributes to the HRT-mediated defense against TMV by promoting the stability of RNA-binding proteins that are required for HRT-mediated TMV resistance (Lim et al., 2018). COP1/SPA also regulates specific metabolic pathways in darkness, such as anthocyanin biosynthesis, by targeting the PRODUCTION OF ANTHOCYANIN PIGMENT (PAP) transcription factors for degradation (Maier et al., 2013).
Regulation of COP1/SPA Activity
Upon illumination, plant photoreceptors suppress the E3 ligase activity of the COP1/SPA complex by directly interacting with COP1 and SPA proteins. This leads to a stabilization of COP1/SPA substrates and subsequent photomorphogenesis. Diverse mechanisms underlying the suppression of COP1/SPA by photoreceptors have been studied in detail, such as the nuclear export of COP1, dissociation of the COP1-SPA interaction, light-mediated degradation of SPA proteins, prevention of COP1 dimerization, and VP-mediated displacement of substrates by photoreceptors. Readers are encouraged to refer to recent reviews discussing these mechanisms in detail (Menon et al., 2016; Hoecker, 2017; Podolec and Ulm, 2018; Ponnu, 2020).
Genetic evidence indicates that both COP1 and SPA proteins are required for COP1/SPA activity in the suppression of photomorphogenesis in darkness. In vitro, COP1 alone has catalytic activity as a ubiquitin ligase due to the presence of an E2-binding zinc finger, implying that SPA proteins may have an essential regulatory function on the activity of COP1. SPA proteins are not necessary for nuclear accumulation of COP1 in darkness, as COP1 accumulates normally in the nucleus in dark-gown spa null mutants (Balcerowicz et al., 2017). On the other hand, SPA proteins via their DDB1-binding WD repeats contribute to the formation of a CUL4-DDB1-RBX1 ubiquitin ligase. In vitro experiments demonstrating catalytic activity of CUL4-DDB1-RBX1-COP1/SPA complexes in the presence and absence of SPAs are necessary to investigate the role of SPA proteins for such an E3 ligase. Since PIFs and COP1/SPA are both required to suppress photomorphogenesis in darkness, they might act together. Indeed, PIF1 was shown to interact with the WD-repeat of COP1 and SPA1. Moreover, pif and cop1/spa mutations have synergistic effects in dark-grown seedlings and the COP1/SPA target HY5 is not degraded in dark-grown pif mutants. Also, PIF1 enhances COP1 ubiquitination activity in vitro (Xu X. et al., 2014). The mechanistic nature of possible PIF-COP1/SPA co-action is so far unknown.
Since SPA proteins are specific to the green lineage, they may have evolved to place the activity of COP1 under the control of light, i.e., to allow inhibition of COP1 activity by light. Multiple pieces of evidence support this idea. Though both COP1 and SPA proteins can interact with photoreceptors, SPA proteins may regulate the affinity in vivo. Indeed, SPA proteins are required for the interaction of CRY1 with COP1 in vivo (Holtkotte et al., 2017). SPA proteins are also required for the light-induced nuclear exclusion of COP1 and thereby contribute to the light-induced inhibition of COP1 (Balcerowicz et al., 2017). Moreover, red and blue light leads to a dissociation of the COP1-SPA1 interaction, likely a very important mechanism of light-induced inhibition of COP1/SPA activity (Lian et al., 2011; Liu et al., 2011; Lu et al., 2015; Sheerin et al., 2015). Light leads to destabilization of SPA1 and very rapid degradation of SPA2 which will limit COP1 activity in the light (Balcerowicz et al., 2011; Chen et al., 2015, 2016). The kinase activity of SPA1 promotes photomorphogenesis by red light-induced phosphorylation, ubiquitination and subsequent degradation of PIF1 in a PHYB-dependent fashion (Paik et al., 2019).
Though COP1 protein levels do not change even after prolonged exposure to light (Zhu et al., 2008; Balcerowicz et al., 2011), COP1 homeostasis is controlled in darkness by CSU1, a RING-E3 ligase that was identified in a mutant screen for cop1 suppressors. CSU1 interacts with COP1, polyubiquitinates it, and thereby reduces COP1 levels in darkness. Similarly, CSU1 down-regulates SPA1 levels in darkness as well (Xu D. et al., 2014). CSU2, identified in the same suppressor screen, inhibits COP1 activity via a different mechanism. The interaction between CSU2 and COP1 via their respective CC domains suppresses COP1 E3 ligase activity in vitro. It is speculated that CSU2 may interfere with COP1 homodimerization or COP1/SPA heterodimerization (Xu et al., 2015). A mutation in CSU4 on the other hand genetically suppresses the cop1 mutation by interacting with CIRCADIAN CLOCK-ASSOCIATED 1 (CCA1) (Zhao et al., 2018). Another mechanism of COP1 regulation may be via the alternatively spliced COP1b, which potentially interferes with COP1 activity, at least in certain tissues (Zhou et al., 1998). Recently, the C3D component DET1 was also shown to regulate the levels of COP1 toward fine-tuning HY5 activity in Arabidopsis (Cañibano et al., 2020). However, the mechanisms underlying DET1-mediated destabilization of COP1 are not yet known.
Post-translational modifications also regulate COP1 activity. The attachment of small ubiquitin-like modifier (SUMO) peptides via the direct interaction of the SUMO E3 ligase SAP AND MIZ1 DOMAIN- CONTAINING LIGASE1 (SIZ1), enhances the COP1 E3 ligase activity toward HY5, but also leads to SIZ1 ubiquitination and proteasome-mediated degradation (Lin et al., 2016). csu3 suppressor of cop1 carried mutations in the serine/threonine kinase PINOID (PID), which regulates auxin homeostasis by phosphorylating auxin efflux carriers (Berkel et al., 2013). PID directly interacts and phosphorylates COP1 at serine 20, which suppresses COP1 activity (Lin et al., 2017).
Nuclear import and export of COP1 play an important role in regulating COP1 activity (von Arnim and Deng, 1994). Light, temperature, shade, and hormonal responses influence COP1 localization and thereby affect COP1 activity. While exposure to light promotes nuclear export of COP1 (Pacín et al., 2014, 1; Podolec and Ulm, 2018), warm temperature and shade promote nuclear accumulation of COP1 (Pacín et al., 2013; Park et al., 2017). Plant hormonal pathways involving auxin and JA regulate COP1 activity mainly via FIN219. Auxin induces FIN219 expression, which catalyzes JA biosynthesis (Hsieh and Okamoto, 2014). Under shade conditions, FIN219 directly interacts with COP1 and promotes its nuclear export (Swain et al., 2017).
Cop1 in Animals: a Comparison With Arabidopsis Cop1
Mammalian COP1, especially human COP1 (hCOP1) was identified as an ortholog of Arabidopsis COP1 (referred to as AtCOP1 in this section) that binds to the bZIP transcription factor c-Jun, an oncogenic protein (Bianchi et al., 2003). hCOP1 contains very similar domains as AtCOP1, i.e., a RING finger domain, a coiled-coil domain and a WD repeat. Like AtCOP1, hCOP1 is encoded by a single gene and functions as an E3 ubiquitin ligase, but in contrast to AtCOP1, it recognizes different targets which regulate cell division, DNA repair and apoptosis (Dornan et al., 2004; Song Y. et al., 2020). Various studies show that hCOP1 may be a potential oncogenic factor or a tumor suppressor, owing to its diverse roles in tumorigenesis (Lee et al., 2010). Several substrates and interacting proteins of hCOP1 were identified over the years (Song Y. et al., 2020). Hence, like AtCOP1, hCOP1 targets multiple substrates, and thereby, regulates multiple processes (Figure 3).
Figure 3. hCOP1 acts as part of multiple E3 ubiquitin ligases. A simplified representation of the hCOP1 function. hCOP1 itself acts as an E3 ligase and also cooperates with other E3 ligases in ubiquitinating and thereby promoting the degradation of different substrates.
Many functional parallels and differences can be drawn between AtCOP1 and hCOP1. Similar to AtCOP1, hCOP1 is nuclear-localized. Upon DNA damage, hCOP1 undergoes phosphorylation and autoubiquitination followed by degradation (Dornan et al., 2006). The phosphorylated residues of hCOP1 are not conserved in AtCOP1, suggesting that this mechanism of regulation is not conserved in AtCOP1. In vitro phosphorylation of AtCOP1 at Ser20, mediated by PID kinase, is so far the only evidence for COP1 phosphorylation (Lin et al., 2017). Also, though observed in vitro (e.g., Seo et al., 2003), there is no functional evidence for a biological role of AtCOP1 autoubiquitination. However, COP1-dependent degradation of SPA2 in light-grown seedlings was reported (Chen et al., 2015).
Like AtCOP1, hCOP1 is part of higher-order CUL4-RBX1-DDB1-based E3 ligase complex. However, unlike in Arabidopsis, the association between hCOP1 and DDB1 in the CUL4-E3 complex is not direct, but bridged by DET1 (Wertz et al., 2004). Work in a recent preprint demonstrated that AtCOP1 co-purifies with AtDET1 in vivo, raising the possibility that AtCOP1 acts together with AtDET1 as members of the same complex or of interacting complexes (Cañibano et al., 2020). Depending on the ubiquitination substrates, hCOP1 could act as an E3 ligase independently or in association with the CUL4-DDB1-RBX-DET1 (CDD) complex. hCOP1 ubiquitinates and promotes the degradation of the tumor suppressor protein p53 by interacting via the RING-domain, without involving any other E3 ligases (Dornan et al., 2004). By mediating the degradation of p53, hCOP1 acts as a suppressor of tumorigenesis. Similarly, hCOP1 interacts with another tumor suppressor protein p27 via the RING-domain that results in polyubiquitination and degradation of p27 (Choi et al., 2015). Unlike with p53 and p27, hCOP1 could directly or indirectly associate with CDD complex to degrade targets such as c-Jun. Even though the interaction between hCOP1 and c-Jun occurs via hCOP1-WD, the N-terminus of hCOP1 is essential for ubiquitination and degradation of c-Jun (Bianchi et al., 2003). Whether hCOP1 ubiquitinates c-Jun independently of CDD remains to be investigated. As the N-terminus is indispensable also for AtCOP1 function (Torii et al., 1998; Stoop-Myer et al., 1999), further research is required to determine whether AtCOP1 could act as an E3 ligase independently of the CUL4-DDB1 complex. Similarly, PEA3, an ETS family protein regulating cell proliferation, interacts with the CC and WD of hCOP1 and is subjected to ubiquitination and degradation via the CDD complex (Baert et al., 2010). ETS2, another member of the ETS family, is also targeted by hCOP1 to degradation, mediated by the CDD complex. However, similar to the interaction with c-Jun, an intact RING domain is essential for ETS2 degradation, raising the possibility that a part of ETS2 ubiquitination may occur via hCOP1 alone (Carrero et al., 2016). In addition, unlike c-Jun, DET1 of the CDD complex directly interacts with ETS2. hCOP1 also suppresses ETS1, but the involvement of the CDD complex is not reported so far. hCOP1 targets many other proteins for degradation such as MTA and FOXO; however, the mechanistic details are still unclear. Taken together, the N-terminus of both AtCOP1 and hCOP1 are essential for their functions.
hCOP1 also targets and promotes the degradation of C/EBPα, another tumor suppressor protein, via Tribbles (TRIB) as adapter proteins. Similarly, the interaction between AtCOP1 and GI is mediated by ELF3 as a bridge protein (Yu et al., 2008). Interestingly, ELF3 is also targeted for degradation by COP1 via another bridge protein BBX19 (Wang et al., 2015). hCOP1-WD and AtCOP1-WD both co-crystallized with TRIB peptides, indicating that the VP-binding pockets of AtCOP1 and hCOP1 are very similar and both are capable of associating with diverse VP domain-containing proteins (Uljon et al., 2016; Lau et al., 2019). TRIBs are pseudo serine/threonine protein kinases that do not usually phosphorylate the substrates on their own but instead function as scaffold proteins involved in protein binding (Eyers et al., 2017). TRIB1 also regulates hCOP1 activity via promoting nuclear accumulation of COP1. Interestingly, an intramolecular interaction within hCOP1 involving a pseudo-substrate latch and the VP-binding pocket of hCOP1 promotes the nuclear export of hCOP1. TRIB1 via its VP motif interacts with the VP-binding pocket of hCOP1, displacing the pseudo-substrate latch from hCOP1-WD, thereby masking the nuclear export signal (Kung and Jura, 2019). Future research could explore similar mechanisms that may operate in AtCOP1. CRYs are important regulators of AtCOP1 under blue light conditions (Holtkotte et al., 2017; Ponnu et al., 2019; Ponnu, 2020). However, hCRYs that function in circadian regulation do not interact directly with hCOP1. Recently, hCRYs are shown to disrupt the association between hCOP1 and C3D complex by directly interacting with DET1, suggesting the evolutionary conservation of CRY-mediated repression of COP1 E3 ligase activity (Rizzini et al., 2019).
Conclusion and Perspectives
Almost every facet of plant growth and development directly or indirectly associates with photomorphogenesis. The tremendous progress made in the last decades to understand the plant responses to light signals has unraveled many previously unknown mechanisms and established new signaling interconnections. Many important pathways that govern plant survival and establishment share the functions of key players that converge on the COP1/SPA complex. An increasing number of substrates and interaction partners that are being discovered connects COP1/SPA to almost every aspect of a plant’s lifecycle. Genetic, molecular, biochemical, and structural breakthroughs in recent times have provided new insights into the functions of the COP1/SPA E3 ligase.
However, many exciting and crucial questions remain to be answered, such as whether the COP1/SPA complex can function as an E3 ligase alone or only as a part of CUL4-based E3 complexes. Moreover, while both COP1 and SPA proteins are part of the CUL4-DDB1-RBX1 complex, they may also associate with the C3D complex. Notably, the mutants in the components of CUL4-based E3 complexes, including DET1, exhibit constitutive photomorphogenesis in darkness. As hCOP1 acts together with hDET1, it remains to be explored how AtDET1 and AtCOP1/AtSPA co-act in plants. Given that the COP1/SPA complex likely functions as a heterotetramer, large multimeric complexes may be formed in vivo when associated with CUL4-based E3 ligases. Therefore, it is likely that photoreceptors suppress the activity of these large complexes via additional mechanisms that are not known so far. In addition, such multimeric complexes may have functional specificities in tissue and developmental contexts. The pathways that affect these specificities and their implications are worth exploring.
The recent discovery that SPA1 functions as a serine/threonine kinase will further advance our understanding of the role of SPA proteins in the COP1/SPA complex. Future research may explore the kinase activity of SPAs other than SPA1, processes that regulate SPA1 kinase activity and additional phosphorylation substrates of the SPA1 kinase. Since hCOP1 shares similarities with AtCOP1, interdisciplinary comparative studies employing advanced biochemical and structural tools may provide more insights into the functions of the COP1/SPA E3 ubiquitin ligase.
Author Contributions
Both authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Funding
Our research was funded by the Deutsche Forschungsgemeinschaft (DFG) HO2793/3-3 and under Germany’s Excellence Strategy EXC 2048/1, project ID: 390686111 to UH.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Alabadí, D., Gallego-Bartolomé, J., Orlando, L., García-Cárcel, L., Rubio, V., Martínez, C., et al. (2008). Gibberellins modulate light signaling pathways to prevent Arabidopsis seedling de-etiolation in darkness. Plant J. 53, 324–335. doi: 10.1111/j.1365-313X.2007.03346.x
Alabadí, D., Gil, J., Blázquez, M. A., and García-Martínez, J. L. (2004). Gibberellins repress photomorphogenesis in darkness. Plant Physiol. 134, 1050–1057. doi: 10.1104/pp.103.035451
Baert, J.-L., Monte, D., Verreman, K., Degerny, C., Coutte, L., and de Launoit, Y. (2010). The E3 ubiquitin ligase complex component COP1 regulates PEA3 group member stability and transcriptional activity. Oncogene 29, 1810–1820. doi: 10.1038/onc.2009.471
Balcerowicz, M., Fittinghoff, K., Wirthmueller, L., Maier, A., Fackendahl, P., Fiene, G., et al. (2011). Light exposure of Arabidopsis seedlings causes rapid de-stabilization as well as selective post-translational inactivation of the repressor of photomorphogenesis SPA2. Plant J. 65, 712–723. doi: 10.1111/j.1365-313X.2010.04456.x
Balcerowicz, M., Kerner, K., Schenkel, C., and Hoecker, U. (2017). SPA proteins affect the subcellular localization of COP1 in the COP1/SPA ubiquitin ligase complex during photomorphogenesis. Plant Physiol. 174, 1314–1321. doi: 10.1104/pp.17.00488
Bauer, D., Viczián, A., Kircher, S., Nobis, T., Nitschke, R., Kunkel, T., et al. (2004). Constitutive photomorphogenesis 1 and multiple photoreceptors control degradation of phytochrome interacting factor 3, a transcription factor required for light signaling in Arabidopsis. Plant Cell 16, 1433–1445. doi: 10.1105/tpc.021568
Berkel, K., van Boer, R. J., de Scheres, B., and ten Tusscher, K. (2013). Polar auxin transport: models and mechanisms. Development 140, 2253–2268. doi: 10.1242/dev.079111
Bernardo-García, S., de Lucas, M., Martínez, C., Espinosa-Ruiz, A., Davière, J.-M., and Prat, S. (2014). BR-dependent phosphorylation modulates PIF4 transcriptional activity and shapes diurnal hypocotyl growth. Genes Dev. 28, 1681–1694. doi: 10.1101/gad.243675.114
Bhatnagar, A., Singh, S., Khurana, J. P., and Burman, N. (2020). HY5-COP1: the central module of light signaling pathway. J. Plant Biochem. Biotechnol. 29, 590–610. doi: 10.1007/s13562-020-00623-3
Bianchi, E., Denti, S., Catena, R., Rossetti, G., Polo, S., Gasparian, S., et al. (2003). Characterization of human constitutive photomorphogenesis protein 1, a RING finger ubiquitin ligase that interacts with jun transcription factors and modulates their transcriptional activity. J. Biol. Chem. 278, 19682–19690. doi: 10.1074/jbc.M212681200
Blanco-Touriñán, N., Legris, M., Minguet, E. G., Costigliolo-Rojas, C., Nohales, M. A., Iniesto, E., et al. (2020a). COP1 destabilizes DELLA proteins in Arabidopsis. PNAS 117, 13792–13799. doi: 10.1073/pnas.1907969117
Blanco-Touriñán, N., Serrano-Mislata, A., and Alabadí, D. (2020b). Regulation of DELLA proteins by post-translational modifications. Plant Cell Physiol. 61, 1891–1901. doi: 10.1093/pcp/pcaa113
Burko, Y., Seluzicki, A., Zander, M., Pedmale, U. V., Ecker, J. R., and Chory, J. (2020). Chimeric activators and repressors define HY5 activity and reveal a light-regulated feedback mechanism[OPEN]. Plant Cell 32, 967–983. doi: 10.1105/tpc.19.00772
Bursch, K., Toledo-Ortiz, G., Pireyre, M., Lohr, M., Braatz, C., and Johansson, H. (2020). Identification of BBX proteins as rate-limiting cofactors of HY5. Nat. Plants 6, 921–928. doi: 10.1038/s41477-020-0725-0
Cañibano, E., Bourbousse, C., Garcia-Leon, M., Wolff, L., Garcia-Baudino, C., Barneche, F., et al. (2020). DET1-mediated COP1 regulation avoids HY5 activity over second-site targets to tune plant photomorphogenesis. bioRxiv [Preprint]. doi: 10.1101/2020.09.30.318253
Carrero, Z. I., Kollareddy, M., Chauhan, K. M., Ramakrishnan, G., and Martinez, L. A. (2016). Mutant p53 protects ETS2 from non-canonical COP1/DET1 dependent degradation. Oncotarget 7, 12554–12567. doi: 10.18632/oncotarget.7275
Catalá, R., Medina, J., and Salinas, J. (2011). Integration of low temperature and light signaling during cold acclimation response in Arabidopsis. PNAS 108, 16475–16480. doi: 10.1073/pnas.1107161108
Chang, C.-S. J., Maloof, J. N., and Wu, S.-H. (2011). COP1-mediated degradation of BBX22/LZF1 optimizes seedling development in Arabidopsis. Plant Physiol. 156, 228–239. doi: 10.1104/pp.111.175042
Chen, G.-H., Liu, M.-J., Xiong, Y., Sheen, J., and Wu, S.-H. (2018). TOR and RPS6 transmit light signals to enhance protein translation in deetiolating Arabidopsis seedlings. PNAS 115, 12823–12828. doi: 10.1073/pnas.1809526115
Chen, H.-J., Fu, T.-Y., Yang, S.-L., and Hsieh, H.-L. (2018). FIN219/JAR1 and cryptochrome1 antagonize each other to modulate photomorphogenesis under blue light in Arabidopsis. PLoS Genet. 14:e1007248. doi: 10.1371/journal.pgen.1007248
Chen, H., Huang, X., Gusmaroli, G., Terzaghi, W., Lau, O. S., Yanagawa, Y., et al. (2010). Arabidopsis CULLIN4-damaged DNA binding protein 1 interacts with constitutively photomorphogenic1-suppressor of phya complexes to regulate photomorphogenesis and flowering time. Plant Cell 22, 108–123. doi: 10.1105/tpc.109.065490
Chen, H., Shen, Y., Tang, X., Yu, L., Wang, J., Guo, L., et al. (2006). Arabidopsis CULLIN4 forms an E3 ubiquitin ligase with RBX1 and the CDD complex in mediating light control of development. Plant Cell 18, 1991–2004. doi: 10.1105/tpc.106.043224
Chen, Q., Bai, L., Wang, W., Shi, H., Botella, J. R., Zhan, Q., et al. (2020). COP1 promotes ABA-induced stomatal closure by modulating the abundance of ABI/HAB and AHG3 phosphatases. New Phytol. 229, 2035–2049. doi: 10.1111/nph.17001
Chen, S., Lory, N., Stauber, J., and Hoecker, U. (2015). Photoreceptor specificity in the light-induced and COP1-mediated rapid degradation of the repressor of photomorphogenesis SPA2 in Arabidopsis. PLoS Genet. 11:e1005516. doi: 10.1371/journal.pgen.1005516
Chen, S., Wirthmueller, L., Stauber, J., Lory, N., Holtkotte, X., Leson, L., et al. (2016). The functional divergence between SPA1 and SPA2 in Arabidopsis photomorphogenesis maps primarily to the respective N-terminal kinase-like domain. BMC Plant Biol. 16:165. doi: 10.1186/s12870-016-0854-9
Cheng, M.-C., Enderle, B., Kathare, P. K., Islam, R., Hiltbrunner, A., and Huq, E. (2020). PCH1 and PCHL directly interact with PIF1, promote its degradation, and inhibit its transcriptional function during photomorphogenesis. Mol. Plant 13, 499–514. doi: 10.1016/j.molp.2020.02.003
Chico, J.-M., Fernández-Barbero, G., Chini, A., Fernández-Calvo, P., Díez-Díaz, M., and Solano, R. (2014). Repression of jasmonate-dependent defenses by shade involves differential regulation of protein stability of MYC transcription factors and their JAZ repressors in Arabidopsis. Plant Cell 26, 1967–1980. doi: 10.1105/tpc.114.125047
Cho, S. K., Chaabane, S. B., Shah, P., Poulsen, C. P., and Yang, S. W. (2014). COP1 E3 ligase protects HYL1 to retain microRNA biogenesis. Nat. Commun. 5:5867. doi: 10.1038/ncomms6867
Choi, C. M., Gray, W. M., Mooney, S., and Hellmann, H. (2014). Composition, roles, and regulation of cullin-based ubiquitin E3 ligases. Arabidopsis Book 6:e0175. doi: 10.1199/tab.0175
Choi, H. H., Guma, S., Fang, L., Phan, L., Ivan, C., Baggerly, K., et al. (2015). Regulating the stability and localization of CDK inhibitor p27Kip1 via CSN6-COP1 axis. Cell Cycle 14, 2265–2273. doi: 10.1080/15384101.2015.1046655
Crawford, A. J., McLachlan, D. H., Hetherington, A. M., and Franklin, K. A. (2012). High temperature exposure increases plant cooling capacity. Curr. Biol. 22, R396–R397. doi: 10.1016/j.cub.2012.03.044
Crocco, C. D., Holm, M., Yanovsky, M. J., and Botto, J. F. (2010). AtBBX21 and COP1 genetically interact in the regulation of shade avoidance. Plant J. 64, 551–562. doi: 10.1111/j.1365-313X.2010.04360.x
Datta, S., Hettiarachchi, C., Johansson, H., and Holm, M. (2007). SALT TOLERANCE HOMOLOG2, a B-box protein in Arabidopsis that activates transcription and positively regulates light-mediated development. Plant Cell 19, 3242–3255. doi: 10.1105/tpc.107.054791
Datta, S., Johansson, H., Hettiarachchi, C., Irigoyen, M. L., Desai, M., Rubio, V., et al. (2008). LZF1/SALT TOLERANCE HOMOLOG3, an Arabidopsis B-box protein involved in light-dependent development and gene expression, undergoes COP1-mediated ubiquitination. Plant Cell 20, 2324–2338. doi: 10.1105/tpc.108.061747
Debrieux, D., Trevisan, M., and Fankhauser, C. (2013). Conditional involvement of constitutive photomorphogenic1 in the degradation of phytochrome A. Plant Physiol. 161, 2136–2145. doi: 10.1104/pp.112.213280
Delker, C., Sonntag, L., James, G. V., Janitza, P., Ibañez, C., Ziermann, H., et al. (2014). The DET1-COP1-HY5 pathway constitutes a multipurpose signaling module regulating plant photomorphogenesis and thermomorphogenesis. Cell Rep. 9, 1983–1989. doi: 10.1016/j.celrep.2014.11.043
Deng, X.-W., Caspar, T., and Quail, P. H. (1991). copl: a regulatory locus involved m hght-controlled development and gene expression in Arabidopsis. Genes Dev. 5, 1172–1182.
Deng, X.-W., and Quail, P. H. (1992). Genetic and phenotypic characterization of cop1 mutants of Arabidopsis thaliana. Plant J. 2, 83–95. doi: 10.1111/j.1365-313X.1992.00083.x
Dill, A., Thomas, S. G., Hu, J., Steber, C. M., and Sun, T. (2004). The Arabidopsis F-Box protein SLEEPY1 targets gibberellin signaling repressors for gibberellin-induced degradation. Plant Cell 16, 1392–1405. doi: 10.1105/tpc.020958
Dornan, D., Shimizu, H., Mah, A., Dudhela, T., Eby, M., O’Rourke, K., et al. (2006). ATM engages autodegradation of the E3 ubiquitin ligase COP1 after DNA damage. Science 313, 1122–1126. doi: 10.1126/science.1127335
Dornan, D., Wertz, I., Shimizu, H., Arnott, D., Frantz, G. D., Dowd, P., et al. (2004). The ubiquitin ligase COP1 is a critical negative regulator of p53. Nature 429, 86–92. doi: 10.1038/nature02514
Duek, P. D., Elmer, M. V., van Oosten, V. R., and Fankhauser, C. (2004). The Degradation of HFR1, a Putative bHLH class transcription factor involved in light signaling, is regulated by phosphorylation and requires COP1. Curr. Biol. 14, 2296–2301. doi: 10.1016/j.cub.2004.12.026
Dyachok, J., Zhu, L., Liao, F., He, J., Huq, E., and Blancaflor, E. B. (2011). SCAR mediates light-induced root elongation in Arabidopsis through photoreceptors and proteasomes. Plant Cell 23, 3610–3626. doi: 10.1105/tpc.111.088823
Eyers, P. A., Keeshan, K., and Kannan, N. (2017). Tribbles in the 21st century: the evolving roles of tribbles pseudokinases in biology and disease. Trends Cell Biol. 27, 284–298. doi: 10.1016/j.tcb.2016.11.002
Fan, X.-Y., Sun, Y., Cao, D.-M., Bai, M.-Y., Luo, X.-M., Yang, H.-J., et al. (2012). BZS1, a B-box protein, promotes photomorphogenesis downstream of both brassinosteroid and light signaling pathways. Mol. Plant 5, 591–600. doi: 10.1093/mp/sss041
Favory, J.-J., Stec, A., Gruber, H., Rizzini, L., Oravecz, A., Funk, M., et al. (2009). Interaction of COP1 and UVR8 regulates UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis. EMBO J. 28, 591–601. doi: 10.1038/emboj.2009.4
Fiorucci, A.-S., and Fankhauser, C. (2017). Plant strategies for enhancing access to sunlight. Curr. Biol. 27, R931–R940. doi: 10.1016/j.cub.2017.05.085
Fittinghoff, K., Laubinger, S., Nixdorf, M., Fackendahl, P., Baumgardt, R.-L., Batschauer, A., et al. (2006). Functional and expression analysis of Arabidopsis SPA genes during seedling photomorphogenesis and adult growth. Plant J. 47, 577–590. doi: 10.1111/j.1365-313X.2006.02812.x
Fonseca, S., and Rubio, V. (2019). Arabidopsis CRL4 complexes: surveying chromatin states and gene expression. Front. Plant Sci. 10:1095. doi: 10.3389/fpls.2019.01095
Gangappa, S. N., and Botto, J. F. (2014). The BBX family of plant transcription factors. Trends Plant Sci. 19, 460–470. doi: 10.1016/j.tplants.2014.01.010
Gangappa, S. N., Crocco, C. D., Johansson, H., Datta, S., Hettiarachchi, C., Holm, M., et al. (2013). The Arabidopsis B-BOX protein BBX25 interacts with HY5, negatively regulating BBX22 expression to suppress seedling photomorphogenesis. Plant Cell 25, 1243–1257. doi: 10.1105/tpc.113.109751
Gangappa, S. N., and Kumar, S. V. (2017). DET1 and HY5 control PIF4-mediated thermosensory elongation growth through distinct mechanisms. Cell Rep. 18, 344–351. doi: 10.1016/j.celrep.2016.12.046
Gangappa, S. N., Prasad, V. B. R., and Chattopadhyay, S. (2010). Functional interconnection of MYC2 and SPA1 in the photomorphogenic seedling development of Arabidopsis. Plant Physiol. 154, 1210–1219. doi: 10.1104/pp.110.163717
Gupta, N., and Nath, U. (2020). Integration of light and hormone response during seedling establishment. J. Plant Biochem. Biotechnol. 29, 652–664. doi: 10.1007/s13562-020-00628-y
Han, X., Huang, X., and Deng, X. W. (2020). The photomorphogenic central repressor COP1: conservation and functional diversification during evolution. Plant Commun. 1:100044. doi: 10.1016/j.xplc.2020.100044
Hayes, S., Sharma, A., Fraser, D. P., Trevisan, M., Cragg-Barber, C. K., Tavridou, E., et al. (2017). UV-B perceived by the UVR8 photoreceptor inhibits plant thermomorphogenesis. Curr. Biol. 27, 120–127. doi: 10.1016/j.cub.2016.11.004
He, J.-X., Gendron, J. M., Yang, Y., Li, J., and Wang, Z.-Y. (2002). The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis. PNAS 99, 10185–10190. doi: 10.1073/pnas.152342599
Heng, Y., Jiang, Y., Zhao, X., Zhou, H., Wang, X., Deng, X. W., et al. (2019). BBX4, a phyB-interacting and modulated regulator, directly interacts with PIF3 to fine tune red light-mediated photomorphogenesis. PNAS 116, 26049–26056. doi: 10.1073/pnas.1915149116
Hoecker, U. (2017). The activities of the E3 ubiquitin ligase COP1/SPA, a key repressor in light signaling. Curr. Opin. Plant Biol. 37, 63–69. doi: 10.1016/j.pbi.2017.03.015
Hoecker, U., and Quail, P. H. (2001). The phytochrome A-specific signaling intermediate SPA1 interacts directly with COP1, a constitutive repressor of light signaling in Arabidopsis. J. Biol. Chem. 276, 38173–38178. doi: 10.1074/jbc.M103140200
Hoecker, U., Tepperman, J. M., and Quail, P. H. (1999). SPA1, a WD-repeat protein specific to phytochrome a signal transduction. Science 284, 496–499. doi: 10.1126/science.284.5413.496
Holm, M., Hardtke, C. S., Gaudet, R., and Deng, X.-W. (2001). Identification of a structural motif that confers specific interaction with the WD40 repeat domain of Arabidopsis COP1. EMBO J. 20, 118–127. doi: 10.1093/emboj/20.1.118
Holm, M., Ma, L.-G., Qu, L.-J., and Deng, X.-W. (2002). Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsis. Genes Dev. 16, 1247–1259. doi: 10.1101/gad.969702
Holtkotte, X., Dieterle, S., Kokkelink, L., Artz, O., Leson, L., Fittinghoff, K., et al. (2016). Mutations in the N-terminal kinase-like domain of the repressor of photomorphogenesis SPA1 severely impair SPA1 function but not light responsiveness in Arabidopsis. Plant J. 88, 205–218. doi: 10.1111/tpj.13241
Holtkotte, X., Ponnu, J., Ahmad, M., and Hoecker, U. (2017). The blue light-induced interaction of cryptochrome 1 with COP1 requires SPA proteins during Arabidopsis light signaling. PLoS Genet. 13:e1007044. doi: 10.1371/journal.pgen.1007044
Hong, S. H., Kim, H. J., Ryu, J. S., Choi, H., Jeong, S., Shin, J., et al. (2008). CRY1 inhibits COP1-mediated degradation of BIT1, a MYB transcription factor, to activate blue light-dependent gene expression in Arabidopsis. Plant J. 55, 361–371. doi: 10.1111/j.1365-313X.2008.03508.x
Hsieh, H.-L., and Okamoto, H. (2014). Molecular interaction of jasmonate and phytochrome A signalling. J. Exp. Bot. 65, 2847–2857. doi: 10.1093/jxb/eru230
Hua, Z., and Vierstra, R. D. (2011). The cullin-RING ubiquitin-protein ligases. Annu. Rev. Plant Biol. 62, 299–334. doi: 10.1146/annurev-arplant-042809-112256
Huang, H., Liu, B., Liu, L., and Song, S. (2017). Jasmonate action in plant growth and development. J. Exp. Bot. 68, 1349–1359. doi: 10.1093/jxb/erw495
Iglesias, M. J., Sellaro, R., Zurbriggen, M. D., and Casal, J. J. (2018). Multiple links between shade avoidance and auxin networks. J Exp Bot. 69, 213–228. doi: 10.1093/jxb/erx295
Irigoyen, M. L., Iniesto, E., Rodriguez, L., Puga, M. I., Yanagawa, Y., Pick, E., et al. (2014). Targeted degradation of abscisic acid receptors is mediated by the ubiquitin ligase substrate adaptor DDA1 in Arabidopsis. Plant Cell 26, 712–728. doi: 10.1105/tpc.113.122234
Jang, I.-C., Henriques, R., Seo, H. S., Nagatani, A., and Chua, N.-H. (2010). Arabidopsis phytochrome interacting factor proteins promote phytochrome B polyubiquitination by COP1 E3 ligase in the nucleus. Plant Cell 22, 2370–2383. doi: 10.1105/tpc.109.072520
Jang, I.-C., Yang, J.-Y., Seo, H. S., and Chua, N.-H. (2005). HFR1 is targeted by COP1 E3 ligase for post-translational proteolysis during phytochrome A signaling. Genes Dev. 19, 593–602. doi: 10.1101/gad.1247205
Jang, K., Gil Lee, H., Jung, S.-J., Paek, N.-C., and Joon Seo, P. (2015). The E3 Ubiquitin Ligase COP1 Regulates Thermosensory Flowering by Triggering GI Degradation in Arabidopsis. Sci. Rep. 5:12071. doi: 10.1038/srep12071
Jang, S., Marchal, V., Panigrahi, K. C. S., Wenkel, S., Soppe, W., Deng, X.-W., et al. (2008). Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. EMBO J. 27, 1277–1288. doi: 10.1038/emboj.2008.68
Jeong, R.-D., Chandra-Shekara, A. C., Barman, S. R., Navarre, D., Klessig, D. F., Kachroo, A., et al. (2010). Cryptochrome 2 and phototropin 2 regulate resistance protein-mediated viral defense by negatively regulating an E3 ubiquitin ligase. Proc. Natl. Acad. Sci. U.S.A. 107, 13538–13543. doi: 10.1073/pnas.1004529107
Jia, K.-P., Luo, Q., He, S.-B., Lu, X.-D., and Yang, H.-Q. (2014). Strigolactone-regulated hypocotyl elongation is dependent on cryptochrome and phytochrome signaling pathways in Arabidopsis. Mol. Plant 7, 528–540. doi: 10.1093/mp/sst093
Jiang, L., Wang, Y., Li, Q.-F., Björn, L. O., He, J.-X., and Li, S.-S. (2012). Arabidopsis STO/BBX24 negatively regulates UV-B signaling by interacting with COP1 and repressing HY5 transcriptional activity. Cell Res. 22:1046. doi: 10.1038/cr.2012.34
Jung, J.-H., Domijan, M., Klose, C., Biswas, S., Ezer, D., Gao, M., et al. (2016). Phytochromes function as thermosensors in Arabidopsis. Science 354, 886–889. doi: 10.1126/science.aaf6005
Kahle, N., Sheerin, D. J., Fischbach, P., Koch, L.-A., Schwenk, P., Lambert, D., et al. (2020). COLD REGULATED 27 and 28 are targets of CONSTITUTIVELY PHOTOMORPHOGENIC 1 and negatively affect phytochrome B signalling. Plant J. 104, 1038–1053. doi: 10.1111/tpj.14979
Kathare, P. K., Xu, X., Nguyen, A., and Huq, E. (2020). A COP1-PIF-HEC regulatory module fine-tunes photomorphogenesis in Arabidopsis. Plant J. 104, 113–123. doi: 10.1111/tpj.14908
Kim, B., Jeong, Y. J., Corvalán, C., Fujioka, S., Cho, S., Park, T., et al. (2014). Darkness and gulliver2/phyB mutation decrease the abundance of phosphorylated BZR1 to activate brassinosteroid signaling in Arabidopsis. Plant J. 77, 737–747. doi: 10.1111/tpj.12423
Kim, J. Y., Jang, I.-C., and Seo, H. S. (2016). COP1 controls abiotic stress responses by modulating AtSIZ1 function through its E3 ubiquitin ligase activity. Front. Plant Sci. 7:1182. doi: 10.3389/fpls.2016.01182
Kim, S., Hwang, G., Lee, S., Zhu, J.-Y., Paik, I., Nguyen, T. T., et al. (2017). High ambient temperature represses anthocyanin biosynthesis through degradation of HY5. Front. Plant Sci. 8:1787. doi: 10.3389/fpls.2017.01787
Kung, J. E., and Jura, N. (2019). The pseudokinase TRIB1 toggles an intramolecular switch to regulate COP1 nuclear export. EMBO J. 38:e99708. doi: 10.15252/embj.201899708
Lau, K., Podolec, R., Chappuis, R., Ulm, R., and Hothorn, M. (2019). Plant photoreceptors and their signaling components compete for COP1 binding via VP peptide motifs. EMBO J. 10:e102140. doi: 10.15252/embj.2019102140
Lau, O. S., and Deng, X. W. (2012). The photomorphogenic repressors COP1 and DET1: 20 years later. Trends Plant Sci. 17, 584–593. doi: 10.1016/j.tplants.2012.05.004
Laubinger, S., Fittinghoff, K., and Hoecker, U. (2004). The SPA quartet: a family of WD-repeat proteins with a central role in suppression of photomorphogenesis in Arabidopsis. Plant Cell 16, 2293–2306. doi: 10.1105/tpc.104.024216
Laubinger, S., Marchal, V., Gentilhomme, J., Wenkel, S., Adrian, J., Jang, S., et al. (2006). Arabidopsis SPA proteins regulate photoperiodic flowering and interact with the floral inducer CONSTANS to regulate its stability. Development 133, 3213–3222. doi: 10.1242/dev.02481
Lee, J.-H., Jung, J.-H., and Park, C.-M. (2017). Light inhibits COP1-mediated degradation of ICE transcription factors to induce stomatal development in Arabidopsis. Plant Cell 29, 2817-2830. doi: 10.1105/tpc.17.00371
Lee, S., Paik, I., and Huq, E. (2020). SPAs promote thermomorphogenesis by regulating the phyB-PIF4 module in Arabidopsis. Development 147:dev189233. doi: 10.1242/dev.189233
Lee, Y.-H., Andersen, J. B., Song, H.-T., Judge, A. D., Seo, D., Ishikawa, T., et al. (2010). Definition of ubiquitination modulator COP1 as a novel therapeutic target in human hepatocellular carcinoma. Cancer Res. 70, 8264–8269. doi: 10.1158/0008-5472.CAN-10-0749
Legris, M., Klose, C., Burgie, E. S., Rojas, C. C. R., Neme, M., Hiltbrunner, A., et al. (2016). Phytochrome B integrates light and temperature signals in Arabidopsis. Science 354, 897–900. doi: 10.1126/science.aaf5656
Legris, M., Nieto, C., Sellaro, R., Prat, S., and Casal, J. J. (2017). Perception and signalling of light and temperature cues in plants. Plant J. 90, 683–697. doi: 10.1111/tpj.13467
Leivar, P., Monte, E., Al-Sady, B., Carle, C., Storer, A., Alonso, J. M., et al. (2008). The Arabidopsis phytochrome-interacting factor PIF7, together with PIF3 and PIF4, regulates responses to prolonged red light by modulating phyB levels. Plant Cell 20, 337–352. doi: 10.1105/tpc.107.052142
Li, J., Nagpal, P., Vitart, V., McMorris, T. C., and Chory, J. (1996). A role for brassinosteroids in light-dependent development of Arabidopsis. Science 272, 398–401. doi: 10.1126/science.272.5260.398
Li, X., Liu, C., Zhao, Z., Ma, D., Zhang, J., Yang, Y., et al. (2020). COR27 and COR28 are novel regulators of the COP1–HY5 regulatory hub and photomorphogenesis in Arabidopsis. Plant Cell 32, 3139–3154. doi: 10.1105/tpc.20.00195
Lian, H.-L., He, S.-B., Zhang, Y.-C., Zhu, D.-M., Zhang, J.-Y., Jia, K.-P., et al. (2011). Blue-light-dependent interaction of cryptochrome 1 with SPA1 defines a dynamic signaling mechanism. Genes Dev. 25, 1023–1028. doi: 10.1101/gad.2025111
Lian, N., Liu, X., Wang, X., Zhou, Y., Li, H., Li, J., et al. (2017). COP1 mediates dark-specific degradation of microtubule-associated protein WDL3 in regulating Arabidopsis hypocotyl elongation. PNAS 114, 12321–12326. doi: 10.1073/pnas.1708087114
Liang, T., Yang, Y., and Liu, H. (2019). Signal transduction mediated by the plant UV-B photoreceptor UVR8. New Phytol. 221, 1247–1252. doi: 10.1111/nph.15469
Lim, G.-H., Hoey, T., Zhu, S., Clavel, M., Yu, K., Navarre, D., et al. (2018). COP1, a negative regulator of photomorphogenesis, positively regulates plant disease resistance via double-stranded RNA binding proteins. PLoS Pathog. 14:e1006894. doi: 10.1371/journal.ppat.1006894
Lin, F., Jiang, Y., Li, J., Yan, T., Fan, L., Liang, J., et al. (2018). B-Box Domain Protein28 negatively regulates photomorphogenesis by repressing the activity of transcription factor HY5 and undergoes COP1-mediated degradation. Plant Cell 30, 2006–2019. doi: 10.1105/tpc.18.00226
Lin, F., Xu, D., Jiang, Y., Chen, H., Fan, L., Holm, M., et al. (2017). Phosphorylation and negative regulation of CONSTITUTIVELY PHOTOMORPHOGENIC 1 by PINOID in Arabidopsis. PNAS 2017:201702984. doi: 10.1073/pnas.1702984114
Lin, X.-L., Niu, D., Hu, Z.-L., Kim, D. H., Jin, Y. H., Cai, B., et al. (2016). An Arabidopsis SUMO E3 Ligase, SIZ1, negatively regulates photomorphogenesis by promoting COP1 activity. PLoS Genet. 12:e1006016. doi: 10.1371/journal.pgen.1006016
Ling, J.-J., Li, J., Zhu, D., and Deng, X. W. (2017). Noncanonical role of Arabidopsis COP1/SPA complex in repressing BIN2-mediated PIF3 phosphorylation and degradation in darkness. PNAS 2017:201700850. doi: 10.1073/pnas.1700850114
Liu, B., Long, H., Yan, J., Ye, L., Zhang, Q., Chen, H., et al. (2020). A HY5-COL3-COL13 regulatory chain for controlling hypocotyl elongation in Arabidopsis. Plant Cell Environ. 44, 130–142. doi: 10.1111/pce.13899
Liu, B., Zuo, Z., Liu, H., Liu, X., and Lin, C. (2011). Arabidopsis cryptochrome 1 interacts with SPA1 to suppress COP1 activity in response to blue light. Genes Dev. 25, 1029–1034. doi: 10.1101/gad.2025011
Liu, L.-J., Zhang, Y.-C., Li, Q.-H., Sang, Y., Mao, J., Lian, H.-L., et al. (2008). COP1-mediated ubiquitination of CONSTANS is implicated in cryptochrome regulation of flowering in Arabidopsis. Plant Cell 20, 292–306. doi: 10.1105/tpc.107.057281
Liu, Q., Su, T., He, W., Ren, H., Liu, S., Chen, Y., et al. (2020). Photooligomerization determines photosensitivity and photoreactivity of plant cryptochromes. Mol. Plant 13, 398–413. doi: 10.1016/j.molp.2020.01.002
Liu, Q., Wang, Q., Liu, B., Wang, W., Wang, X., Park, J., et al. (2016). The blue light-dependent polyubiquitination and degradation of Arabidopsis cryptochrome2 requires multiple E3 ubiquitin ligases. Plant Cell Physiol. 57, 2175–2186. doi: 10.1093/pcp/pcw134
Lopez-Molina, L., Mongrand, S., Kinoshita, N., and Chua, N.-H. (2003). AFP is a novel negative regulator of ABA signaling that promotes ABI5 protein degradation. Genes Dev. 17, 410–418. doi: 10.1101/gad.1055803
Lu, X.-D., Zhou, C.-M., Xu, P.-B., Luo, Q., Lian, H.-L., and Yang, H.-Q. (2015). Red-light-dependent interaction of phyB with SPA1 promotes COP1–SPA1 dissociation and photomorphogenic development in Arabidopsis. Mol. Plant 8, 467–478. doi: 10.1016/j.molp.2014.11.025
Luo, Q., Lian, H.-L., He, S.-B., Li, L., Jia, K.-P., and Yang, H.-Q. (2014). COP1 and phyB physically interact with PIL1 to regulate its stability and photomorphogenic development in Arabidopsis. Plant Cell 26, 2441–2456. doi: 10.1105/tpc.113.121657
Luo, X.-M., Lin, W.-H., Zhu, S., Zhu, J.-Y., Sun, Y., Fan, X.-Y., et al. (2010). Integration of light- and brassinosteroid-signaling pathways by a GATA transcription factor in Arabidopsis. Dev. Cell 19, 872–883. doi: 10.1016/j.devcel.2010.10.023
Ma, L., Zhao, H., and Deng, X. W. (2003). Analysis of the mutational effects of the COP/DET/FUS loci on genome expression profiles reveals their overlapping yet not identical roles in regulating Arabidopsis seedling development. Development 130, 969–981. doi: 10.1242/dev.00281
Maier, A., and Hoecker, U. (2015). COP1/SPA ubiquitin ligase complexes repress anthocyanin accumulation under low light and high light conditions. Plant Signal. Behav. 10:e970440. doi: 10.4161/15592316.2014.970440
Maier, A., Schrader, A., Kokkelink, L., Falke, C., Welter, B., Iniesto, E., et al. (2013). Light and the E3 ubiquitin ligase COP1/SPA control the protein stability of the MYB transcription factors PAP1 and PAP2 involved in anthocyanin accumulation in Arabidopsis. Plant J. 74, 638–651. doi: 10.1111/tpj.12153
Martínez, C., Nieto, C., and Prat, S. (2018). Convergent regulation of PIFs and the E3 ligase COP1/SPA1 mediates thermosensory hypocotyl elongation by plant phytochromes. Curr. Opin. Plant Biol. 45, 188–203. doi: 10.1016/j.pbi.2018.09.006
Matsui, K., Umemura, Y., and Ohme-Takagi, M. (2008). AtMYBL2, a protein with a single MYB domain, acts as a negative regulator of anthocyanin biosynthesis in Arabidopsis. Plant J. 55, 954–967. doi: 10.1111/j.1365-313X.2008.03565.x
Matsui, M., Stoop, C. D., von Arnim, A. G., Wei, N., and Deng, X. W. (1995). Arabidopsis COP1 protein specifically interacts in vitro with a cytoskeleton-associated protein. CIP1. PNAS 92, 4239–4243. doi: 10.1073/pnas.92.10.4239
Mayer, R., Raventos, D., and Chua, N. H. (1996). det1, cop1, and cop9 mutations cause inappropriate expression of several gene sets. Plant Cell 8, 1951–1959. doi: 10.1105/tpc.8.11.1951
Mazzella, M. A., Casal, J. J., Muschietti, J. P., and Fox, A. R. (2014). Hormonal networks involved in apical hook development in darkness and their response to light. Front. Plant Sci. 5:52. doi: 10.3389/fpls.2014.00052
Mazzucotelli, E., Belloni, S., Marone, D., De Leonardis, A., Guerra, D., Di Fonzo, N., et al. (2006). The E3 ubiquitin ligase gene family in plants: regulation by degradation. Curr. Genom. 7, 509–522. doi: 10.2174/138920206779315728
McNellis, T. W., von Arnim, A. G., Araki, T., Komeda, Y., Miséra, S., and Deng, X. W. (1994). Genetic and molecular analysis of an allelic series of cop1 mutants suggests functional roles for the multiple protein domains. Plant Cell 6, 487–500. doi: 10.1105/tpc.6.4.487
Menon, C., Sheerin, D. J., and Hiltbrunner, A. (2016). SPA proteins: SPAnning the gap between visible light and gene expression. Planta 244, 297–312. doi: 10.1007/s00425-016-2509-3
Moazzam-Jazi, M., Ghasemi, S., Seyedi, S. M., and Niknam, V. (2018). COP1 plays a prominent role in drought stress tolerance in Arabidopsis and Pea. Plant Physiol. Biochem. 130, 678–691. doi: 10.1016/j.plaphy.2018.08.015
Nakagawa, M., and Komeda, Y. (2004). Flowering of Arabidopsis cop1 mutants in darkness. Plant Cell Physiol. 45, 398–406. doi: 10.1093/pcp/pch047
Nam, K. H., and Li, J. (2002). BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 110, 203–212. doi: 10.1016/S0092-8674(02)00814-0
Ni, W., Xu, S.-L., Tepperman, J. M., Stanley, D. J., Maltby, D. A., Gross, J. D., et al. (2014). A mutually assured destruction mechanism attenuates light signaling in Arabidopsis. Science 344, 1160–1164. doi: 10.1126/science.1250778
Nieto, C., Luengo, L. M., and Prat, S. (2020). Regulation of COP1 function by brassinosteroid signaling. Front. Plant Sci. 11:1151. doi: 10.3389/fpls.2020.01151
Nixdorf, M., and Hoecker, U. (2010). SPA1 and DET1 act together to control photomorphogenesis throughout plant development. Planta 231, 825–833. doi: 10.1007/s00425-009-1088-y
Nozue, K., Tat, A. V., Devisetty, U. K., Robinson, M., Mumbach, M. R., Ichihashi, Y., et al. (2015). Shade avoidance components and pathways in adult plants revealed by phenotypic profiling. PLoS Genet. 11:e1004953. doi: 10.1371/journal.pgen.1004953
Oh, J., Park, E., Song, K., Bae, G., and Choi, G. (2020). PHYTOCHROME INTERACTING FACTOR8 inhibits phytochrome a-mediated far-red light responses in Arabidopsis. Plant Cell 32, 186–205. doi: 10.1105/tpc.19.00515
Oravecz, A., Baumann, A., Máté, Z., Brzezinska, A., Molinier, J., Oakeley, E. J., et al. (2006). CONSTITUTIVELY PHOTOMORPHOGENIC1 is required for the UV-B response in Arabidopsis. Plant Cell 18, 1975–1990. doi: 10.1105/tpc.105.040097
Ordoñez-Herrera, N., Fackendahl, P., Yu, X., Schaefer, S., Koncz, C., and Hoecker, U. (2015). A cop1 spa mutant deficient in COP1 and SPA proteins reveals partial co-action of COP1 and SPA during Arabidopsis post-embryonic development and photomorphogenesis. Mol. Plant 8, 479–481. doi: 10.1016/j.molp.2014.11.026
Ordoñez-Herrera, N., Trimborn, L., Menje, M., Henschel, M., Robers, L., Kaufholdt, D., et al. (2018). The transcription factor COL12 is a substrate of the COP1/SPA E3 ligase and regulates flowering time and plant architecture1. Plant Physiol. 176, 1327–1340. doi: 10.1104/pp.17.01207
Osterlund, M. T., Hardtke, C. S., Wei, N., and Deng, X. W. (2000). Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405:462. doi: 10.1038/35013076
Pacín, M., Legris, M., and Casal, J. J. (2013). COP1 re-accumulates in the nucleus under shade. Plant J. 75, 631–641. doi: 10.1111/tpj.12226
Pacín, M., Legris, M., and Casal, J. J. (2014). Rapid decline in nuclear COSTITUTIVE PHOTOMORPHOGENESIS1 abundance anticipates the stabilization of its target ELONGATED HYPOCOTYL5 in the light. Plant Physiol. 164, 1134–1138. doi: 10.1104/pp.113.234245
Pacín, M., Semmoloni, M., Legris, M., Finlayson, S. A., and Casal, J. J. (2016). Convergence of CONSTITUTIVE PHOTOMORPHOGENESIS 1 and PHYTOCHROME INTERACTING FACTOR signalling during shade avoidance. New Phytol. 211, 967–979. doi: 10.1111/nph.13965
Paik, I., Chen, F., Pham, V. N., Zhu, L., Kim, J.-I., and Huq, E. (2019). A phyB-PIF1-SPA1 kinase regulatory complex promotes photomorphogenesis in Arabidopsis. Nat. Commun. 10, 1–17. doi: 10.1038/s41467-019-12110-y
Palayam, M., Ganapathy, J., Guercio, A. M., Tal, L., Deck, S. L., and Shabek, N. (2021). Structural insights into photoactivation of plant Cryptochrome-2. Commun. Biol. 4, 1–11. doi: 10.1038/s42003-020-01531-x
Park, Y.-J., Lee, H.-J., Ha, J.-H., Kim, J. Y., and Park, C.-M. (2017). COP1 conveys warm temperature information to hypocotyl thermomorphogenesis. New Phytol. 215, 269–280. doi: 10.1111/nph.14581
Pepper, A., Delaney, T., Washburnt, T., Poole, D., and Chory, J. (1994). DET1, a negative regulator of light-mediated development and gene expression in Arabidopsis, encodes a novel nuclear-localized protein. Cell 78, 109–116. doi: 10.1016/0092-8674(94)90577-0
Pfeiffer, A., Janocha, D., Dong, Y., Medzihradszky, A., Schöne, S., Daum, G., et al. (2016). Integration of light and metabolic signals for stem cell activation at the shoot apical meristem. eLife Sci. 5:e17023. doi: 10.7554/eLife.17023
Pham, V. N., Kathare, P. K., and Huq, E. (2018a). Dynamic regulation of PIF5 by COP1-SPA complex to optimize photomorphogenesis in Arabidopsis. Plant J. 96, 260–273. doi: 10.1111/tpj.14074
Pham, V. N., Kathare, P. K., and Huq, E. (2018b). Phytochromes and phytochrome interacting factors. Plant Physiol. 176, 1025–1038. doi: 10.1104/pp.17.01384
Podolec, R., and Ulm, R. (2018). Photoreceptor-mediated regulation of the COP1/SPA E3 ubiquitin ligase. Curr. Opin. Plant Biol. 45, 18–25. doi: 10.1016/j.pbi.2018.04.018
Ponnu, J. (2020). Molecular mechanisms suppressing COP1/SPA E3 ubiquitin ligase activity in blue light. Physiol. Plant. 169, 418–429. doi: 10.1111/ppl.13103
Ponnu, J., Riedel, T., Penner, E., Schrader, A., and Hoecker, U. (2019). Cryptochrome 2 competes with COP1 substrates to repress COP1 ubiquitin ligase activity during Arabidopsis photomorphogenesis. PNAS 116, 27133–27141. doi: 10.1073/pnas.1909181116
Quint, M., Delker, C., Franklin, K. A., Wigge, P. A., Halliday, K. J., and van Zanten, M. (2016). Molecular and genetic control of plant thermomorphogenesis. Nat. Plants 2:15190. doi: 10.1038/nplants.2015.190
Ren, C., Zhu, X., Zhang, P., and Gong, Q. (2016). Arabidopsis COP1-interacting protein 1 is a positive regulator of ABA response. Biochem. Biophys. Res. Commun. 477, 847–853. doi: 10.1016/j.bbrc.2016.06.147
Rizzini, L., Levine, D. C., Perelis, M., Bass, J., Peek, C. B., and Pagano, M. (2019). Cryptochromes-mediated inhibition of the CRL4Cop1-complex assembly defines an evolutionary conserved signaling mechanism. Curr. Biol. 29, 1954.e4–1962.e4. doi: 10.1016/j.cub.2019.04.073
Rolauffs, S., Fackendahl, P., Sahm, J., Fiene, G., and Hoecker, U. (2012). Arabidopsis COP1 and SPA genes are essential for plant elongation but not for acceleration of flowering time in response to a low red light to far-red light ratio. Plant Physiol. 160, 2015–2027. doi: 10.1104/pp.112.207233
Saijo, Y., Sullivan, J. A., Wang, H., Yang, J., Shen, Y., Rubio, V., et al. (2003). The COP1–SPA1 interaction defines a critical step in phytochrome A-mediated regulation of HY5 activity. Genes Dev. 17, 2642–2647. doi: 10.1101/gad.1122903
Saijo, Y., Zhu, D., Li, J., Rubio, V., Zhou, Z., Shen, Y., et al. (2008). Arabidopsis COP1/SPA1 complex and FHY1/FHY3 associate with distinct phosphorylated forms of phytochrome A in balancing light signaling. Mol. Cell. 31, 607–613. doi: 10.1016/j.molcel.2008.08.003
Sarid-Krebs, L., Panigrahi, K. C. S., Fornara, F., Takahashi, Y., Hayama, R., Jang, S., et al. (2015). Phosphorylation of CONSTANS and its COP1-dependent degradation during photoperiodic flowering of Arabidopsis. Plant J. 84, 451–463. doi: 10.1111/tpj.13022
Sassi, M., Lu, Y., Zhang, Y., Wang, J., Dhonukshe, P., Blilou, I., et al. (2012). COP1 mediates the coordination of root and shoot growth by light through modulation of PIN1- and PIN2-dependent auxin transport in Arabidopsis. Development 139, 3402–3412. doi: 10.1242/dev.078212
Schrader, A., Welter, B., Hulskamp, M., Hoecker, U., and Uhrig, J. F. (2013). MIDGET connects COP1-dependent development with endoreduplication in Arabidopsis thaliana. Plant J. 75, 67–79. doi: 10.1111/tpj.12199
Schulman, B. A., Carrano, A. C., Jeffrey, P. D., Bowen, Z., Kinnucan, E. R., Finnin, M. S., et al. (2000). Insights into SCF ubiquitin ligases from the structure of the Skp1-Skp2 complex. Nature 408, 381–386. doi: 10.1038/35042620
Seo, H. S., Watanabe, E., Tokutomi, S., Nagatani, A., and Chua, N.-H. (2004). Photoreceptor ubiquitination by COP1 E3 ligase desensitizes phytochrome A signaling. Genes Dev. 18, 617–622. doi: 10.1101/gad.1187804
Seo, H. S., Yang, J.-Y., Ishikawa, M., Bolle, C., Ballesteros, M. L., and Chua, N.-H. (2003). LAF1 ubiquitination by COP1 controls photomorphogenesis and is stimulated by SPA1. Nature 423, 995–999. doi: 10.1038/nature01696
Shao, K., Zhang, X., Li, X., Hao, Y., Huang, X., Ma, M., et al. (2020). The oligomeric structures of plant cryptochromes. Nat. Struct. Mol. Biol. 27, 480–488. doi: 10.1038/s41594-020-0420-x
Sheerin, D. J., Menon, C., Oven-Krockhaus, S. Z., Enderle, B., Zhu, L., Johnen, P., et al. (2015). Light-activated phytochrome A and B interact with members of the SPA family to promote photomorphogenesis in Arabidopsis by reorganizing the COP1/SPA complex. Plant Cell 27, 189–201. doi: 10.1105/tpc.114.134775
Shi, H., Liu, R., Xue, C., Shen, X., Wei, N., Deng, X. W., et al. (2016). Seedlings transduce the depth and mechanical pressure of covering soil using COP1 and ethylene to regulate EBF1/EBF2 for soil emergence. Curr. Biol. 26, 139–149. doi: 10.1016/j.cub.2015.11.053
Shi, Q.-M., Yang, X., Song, L., and Xue, H.-W. (2011). Arabidopsis MSBP1 is activated by HY5 and HYH and is involved in photomorphogenesis and brassinosteroid sensitivity regulation. Mol. Plant 4, 1092–1104. doi: 10.1093/mp/ssr049
Song, L. I, Zhou, X.-Y., Li, L. I, Xue, L.-J., Yang, X. I, and Xue, H.-W. (2009). Genome-wide analysis revealed the complex regulatory network of brassinosteroid effects in photomorphogenesis. Mol. Plant 2, 755–772. doi: 10.1093/mp/ssp039
Song, Y., Liu, Y., Pan, S., Xie, S., Wang, Z., and Zhu, X. (2020). Role of the COP1 protein in cancer development and therapy. Semin. Cancer Biol. 67, 43–52. doi: 10.1016/j.semcancer.2020.02.001
Song, Z., Yan, T., Liu, J., Bian, Y., Heng, Y., Lin, F., et al. (2020). BBX28/BBX29, HY5 and BBX30/31 form a feedback loop to fine-tune photomorphogenic development. Plant J. 104, 377–390. doi: 10.1111/tpj.14929
Stacey, M. G., Hicks, S. N., and von Arnim, A. G. (1999). Discrete domains mediate the light-responsive nuclear and cytoplasmic localization of Arabidopsis COP1. Plant Cell 11, 349–363. doi: 10.1105/tpc.11.3.349
Stacey, M. G., Kopp, O. R., Kim, T.-H., and von Arnim, A. G. (2000). Modular domain structure of Arabidopsis COP1. Reconstitution of activity by fragment complementation and mutational analysis of a nuclear localization signal in planta. Plant Physiol. 124, 979–990. doi: 10.1104/pp.124.3.979
Stoop-Myer, C., Torii, K. U., McNellis, T. W., Coleman, J. E., and Deng, X.-W. (1999). The N-terminal fragment of Arabidopsis photomorphogenic repressor COP1 maintains partial function and acts in a concentration-dependent manner. Plant J. 20, 713–717. doi: 10.1046/j.1365-313X.1999.00639.x
Subramanian, C., Kim, B.-H., Lyssenko, N. N., Xu, X., Johnson, C. H., and von Arnim, A. G. (2004). The Arabidopsis repressor of light signaling, COP1, is regulated by nuclear exclusion: mutational analysis by bioluminescence resonance energy transfer. PNAS 101, 6798–6802. doi: 10.1073/pnas.0307964101
Suzuki, G., Yanagawa, Y., Kwok, S. F., Matsui, M., and Deng, X.-W. (2002). Arabidopsis COP10 is a ubiquitin-conjugating enzyme variant that acts together with COP1 and the COP9 signalosome in repressing photomorphogenesis. Genes Dev. 16, 554–559. doi: 10.1101/gad.964602
Swain, S., Jiang, H.-W., and Hsieh, H.-L. (2017). FAR-RED INSENSITIVE 219/JAR1 contributes to shade avoidance responses of Arabidopsis seedlings by modulating key shade signaling components. Front. Plant Sci. 8:1901. doi: 10.3389/fpls.2017.01901
Tavridou, E., Pireyre, M., and Ulm, R. (2020). Degradation of the transcription factors PIF4 and PIF5 under UV-B promotes UVR8-mediated hypocotyl growth inhibition in Arabidopsis. Plant J. 101, 507–517. doi: 10.1111/tpj.14556
Torii, K. U., McNellis, T. W., and Deng, X. W. (1998). Functional dissection of Arabidopsis COP1 reveals specific roles of its three structural modules in light control of seedling development. EMBO J. 17, 5577–5587. doi: 10.1093/emboj/17.19.5577
Torii, K. U., Stoop-Myer, C. D., Okamoto, H., Coleman, J. E., Matsui, M., and Deng, X. W. (1999). The RING finger motif of photomorphogenic repressor COP1 specifically interacts with the RING-H2 motif of a novelArabidopsis protein. J. Biol. Chem. 274, 27674–27681. doi: 10.1074/jbc.274.39.27674
Tsuchiya, Y., Vidaurre, D., Toh, S., Hanada, A., Nambara, E., Kamiya, Y., et al. (2010). A small-molecule screen identifies new functions for the plant hormone strigolactone. Nat. Chem. Biol. 6, 741–749. doi: 10.1038/nchembio.435
Uljon, S., Xu, X., Durzynska, I., Stein, S., Adelmant, G., Marto, J. A., et al. (2016). Structural basis for substrate selectivity of the E3 Ligase COP1. Structure 24, 687–696. doi: 10.1016/j.str.2016.03.002
Vaishak, K. P., Yadukrishnan, P., Bakshi, S., Kushwaha, A. K., Ramachandran, H., Job, N., et al. (2019). The B-box bridge between light and hormones in plants. J. Photochem. Photobiol. B Biol. 191, 164–174. doi: 10.1016/j.jphotobiol.2018.12.021
Vandenbussche, F., Habricot, Y., Condiff, A. S., Maldiney, R., Straeten, D. V. D., and Ahmad, M. (2007). HY5 is a point of convergence between cryptochrome and cytokinin signalling pathways in Arabidopsis thaliana. Plant J. 49, 428–441. doi: 10.1111/j.1365-313X.2006.02973.x
von Arnim, A. G., and Deng, X. W. (1993). Ring finger motif of Arabidopsis thaliana COP1 defines a new class of zinc-binding domain. J. Biol. Chem. 268, 19626–19631. doi: 10.1016/S0021-9258(19)36562-7
von Arnim, A. G., and Deng, X.-W. (1994). Light inactivation of Arabidopsis photomorphogenic repressor COP1 involves a cell-specific regulation of its nucleocytoplasmic partitioning. Cell 79, 1035–1045. doi: 10.1016/0092-8674(94)90034-5
von Arnim, A. G., Osterlund, M. T., Kwok, S. F., and Deng, X. W. (1997). Genetic and developmental control of nuclear accumulation of COP1, a repressor of photomorphogenesis in Arabidopsis. Plant Physiol. 114, 779–788. doi: 10.1104/pp.114.3.779
Vu, L. D., Xu, X., Gevaert, K., and Smet, I. D. (2019). Developmental plasticity at high temperature. Plant Physiol. 181, 399–411. doi: 10.1104/pp.19.00652
Wang, C.-Q., Sarmast, M. K., Jiang, J., and Dehesh, K. (2015). The transcriptional regulator BBX19 promotes hypocotyl growth by facilitating COP1-mediated EARLY FLOWERING3 degradation in Arabidopsis. Plant Cell 27, 1128–1139. doi: 10.1105/tpc.15.00044
Wang, H., Kang, D., Deng, X.-W., and Wei, N. (1999). Evidence for functional conservation of a mammalian homologue of the light-responsive plant protein COP1. Curr. Biol. 9, 711–S2. doi: 10.1016/S0960-9822(99)80314-5
Wang, P., Cui, X., Zhao, C., Shi, L., Zhang, G., Sun, F., et al. (2017). COR27 and COR28 encode nighttime repressors integrating Arabidopsis circadian clock and cold response. J. Integr. Plant Biol. 59, 78–85. doi: 10.1111/jipb.12512
Wang, W., Chen, Q., Botella, J. R., and Guo, S. (2019). Beyond light: insights into the role of constitutively photomorphogenic1 in plant hormonal signaling. Front. Plant Sci. 10:557. doi: 10.3389/fpls.2019.00557
Wang, W., Paik, I., Kim, J., Hou, X., Sung, S., and Huq, E. (2020). Direct phosphorylation of HY5 by SPA1 kinase to regulate photomorphogenesis in Arabidopsis. bioRxiv [Preprint]. doi: 10.1101/2020.09.10.291773
Wang, Y., Wang, Y., Song, Z., and Zhang, H. (2016). Repression of MYBL2 by Both microRNA858a and HY5 leads to the activation of anthocyanin biosynthetic pathway in Arabidopsis. Mol. Plant 9, 1395–1405. doi: 10.1016/j.molp.2016.07.003
Wei, C.-Q., Chien, C.-W., Ai, L.-F., Zhao, J., Zhang, Z., Li, K. H., et al. (2016). The Arabidopsis B-box protein BZS1/BBX20 interacts with HY5 and mediates strigolactone regulation of photomorphogenesis. J. Genet. Genom. 43, 555–563. doi: 10.1016/j.jgg.2016.05.007
Weidler, G., Oven-Krockhaus, S. Z., Heunemann, M., Orth, C., Schleifenbaum, F., Harter, K., et al. (2012). Degradation of Arabidopsis CRY2 is regulated by SPA proteins and phytochrome A. Plant Cell 24, 2610–2623. doi: 10.1105/tpc.112.098210
Wertz, I. E., O’Rourke, K. M., Zhang, Z., Dornan, D., Arnott, D., Deshaies, R. J., et al. (2004). Human de-etiolated-1 regulates c-jun by assembling a CUL4A ubiquitin ligase. Science 303, 1371–1374. doi: 10.1126/science.1093549
Xu, D., Jiang, Y., Li, J., Lin, F., Holm, M., and Deng, X. W. (2016a). BBX21, an Arabidopsis B-box protein, directly activates HY5 and is targeted by COP1 for 26S proteasome-mediated degradation. PNAS 113, 7655–7660. doi: 10.1073/pnas.1607687113
Xu, D., Lin, F., Jiang, Y., Huang, X., Li, J., Ling, J., et al. (2014). The RING-Finger E3 ubiquitin ligase COP1 SUPPRESSOR1 negatively regulates COP1 abundance in maintaining COP1 homeostasis in dark-grown Arabidopsis seedlings. Plant Cell 26, 1981–1991. doi: 10.1105/tpc.114.124024
Xu, D., Lin, F., Jiang, Y., Ling, J., Hettiarachchi, C., Tellgren-Roth, C., et al. (2015). Arabidopsis COP1 SUPPRESSOR 2 represses COP1 E3 ubiquitin ligase activity through their coiled-coil domains association. PLoS Genet. 11:e1005747. doi: 10.1371/journal.pgen.1005747
Xu, D., Zhu, D., and Deng, X. W. (2016b). The role of COP1 in repression of photoperiodic flowering. F1000Res. 5:F1000 Faculty Rev-178. doi: 10.12688/f1000research.7346.1
Xu, X., Chi, W., Sun, X., Feng, P., Guo, H., Li, J., et al. (2016c). Convergence of light and chloroplast signals for de-etiolation through ABI4–HY5 and COP1. Nat. Plants 2:16066. doi: 10.1038/nplants.2016.66
Xu, X., Kathare, P. K., Pham, V. N., Bu, Q., Nguyen, A., and Huq, E. (2017). Reciprocal proteasome-mediated degradation of PIFs and HFR1 underlies photomorphogenic development in Arabidopsis. Development 144, 1831–1840. doi: 10.1242/dev.146936
Xu, X., Paik, I., Zhu, L., Bu, Q., Huang, X., Deng, X. W., et al. (2014). PHYTOCHROME INTERACTING FACTOR1 enhances the E3 ligase activity of CONSTITUTIVE PHOTOMORPHOGENIC1 to synergistically repress photomorphogenesis in Arabidopsis. Plant Cell 26, 1992–2006. doi: 10.1105/tpc.114.125591
Yadav, A., Ravindran, N., Singh, D., Rahul, P. V., and Datta, S. (2020). Role of Arabidopsis BBX proteins in light signaling. J. Plant Biochem. Biotechnol. 29, 623–635. doi: 10.1007/s13562-020-00597-2
Yadukrishnan, P., and Datta, S. (2020). Light and abscisic acid interplay in early seedling development. New Phytol. 229, 763–769. doi: 10.1111/nph.16963
Yamamoto, Y. Y., Deng, X.-W., and Matsui, M. (2001). CIP4, a New COP1 target, is a nucleus-localized positive regulator of Arabidopsis photomorphogenesis. Plant Cell 13, 399–411. doi: 10.1105/tpc.13.2.399
Yamamoto, Y. Y., Matsui, M., Ang, L.-H., and Deng, X.-W. (1998). Role of a COP1 interactive protein in mediating light-regulated gene expression in Arabidopsis. Plant Cell 10, 1083–1094. doi: 10.1105/tpc.10.7.1083
Yan, H., Marquardt, K., Indorf, M., Jutt, D., Kircher, S., Neuhaus, G., et al. (2011). Nuclear localization and interaction with COP1 are required for STO/BBX24 function during photomorphogenesis. Plant Physiol. 156, 1772–1782. doi: 10.1104/pp.111.180208
Yanagawa, Y., Sullivan, J. A., Komatsu, S., Gusmaroli, G., Suzuki, G., Yin, J., et al. (2004). Arabidopsis COP10 forms a complex with DDB1 and DET1 in vivo and enhances the activity of ubiquitin conjugating enzymes. Genes Dev. 18, 2172–2181. doi: 10.1101/gad.1229504
Yang, J., Lin, R., Sullivan, J., Hoecker, U., Liu, B., Xu, L., et al. (2005). Light regulates COP1-mediated degradation of HFR1, a transcription factor essential for light signaling in Arabidopsis. Plant Cell 17, 804–821. doi: 10.1105/tpc.104.030205
Yang, J., and Wang, H. (2006). The central coiled-coil domain and carboxyl-terminal WD-repeat domain of Arabidopsis SPA1 are responsible for mediating repression of light signaling. Plant J. 47, 564–576. doi: 10.1111/j.1365-313X.2006.02811.x
Yi, C., and Deng, X. W. (2005). COP1 – from plant photomorphogenesis to mammalian tumorigenesis. Trends Cell Biol. 15, 618–625. doi: 10.1016/j.tcb.2005.09.007
Yu, J.-W., Rubio, V., Lee, N.-Y., Bai, S., Lee, S.-Y., Kim, S.-S., et al. (2008). COP1 and ELF3 control circadian function and photoperiodic flowering by regulating GI stability. Mol. Cell. 32, 617–630. doi: 10.1016/j.molcel.2008.09.026
Yu, Y., and Huang, R. (2017). Integration of ethylene and light signaling affects hypocotyl growth in Arabidopsis. Front. Plant Sci. 8:57. doi: 10.3389/fpls.2017.00057
Yu, Y., Wang, J., Zhang, Z., Quan, R., Zhang, H., Deng, X. W., et al. (2013). Ethylene promotes hypocotyl growth and HY5 degradation by enhancing the movement of COP1 to the nucleus in the light. PLoS Genet. 9:e1004025. doi: 10.1371/journal.pgen.1004025
Yuan, T.-T., Xu, H., Zhang, Q., Zhang, L.-Y., and Lu, Y.-T. (2018). The COP1 target SHI-RELATED SEQUENCE 5 facilitates photomorphogenesis by directly activating photomorphogenesis-promoting genes in Arabidopsis. Plant Cell 30, 2368–2382. doi: 10.1105/tpc.18.00455
Zhao, X., Heng, Y., Wang, X., Deng, X. W., and Xu, D. (2020). A positive feedback loop of BBX11–BBX21–HY5 promotes photomorphogenic development in Arabidopsis. Plant Commun. 1:e100045. doi: 10.1016/j.xplc.2020.100045
Zhao, X., Jiang, Y., Li, J., Huq, E., Chen, Z. J., Xu, D., et al. (2018). COP1 SUPPRESSOR 4 promotes seedling photomorphogenesis by repressing CCA1 and PIF4 expression in Arabidopsis. PNAS 115, 11631–11636. doi: 10.1073/pnas.1813171115
Zheng, N., Schulman, B. A., Song, L., Miller, J. J., Jeffrey, P. D., Wang, P., et al. (2002). Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature 416, 703–709. doi: 10.1038/416703a
Zheng, Y., Cui, X., Su, L., Fang, S., Chu, J., Gong, Q., et al. (2017). Jasmonate inhibits COP1 activity to suppress hypocotyl elongation and promote cotyledon opening in etiolated Arabidopsis seedlings. Plant J. 90, 1144–1155. doi: 10.1111/tpj.13539
Zhong, S., Shi, H., Xue, C., Wei, N., Guo, H., and Deng, X. W. (2014). Ethylene-orchestrated circuitry coordinates a seedling’s response to soil cover and etiolated growth. PNAS 111, 3913–3920. doi: 10.1073/pnas.1402491111
Zhou, D.-X., Kim, Y.-J., Li, Y.-F., Carol, P., and Mache, R. (1998). COP1b, an isoform of COP1 generated by alternative splicing, has a negative effect on COP1 function in regulating light-dependent seedling development in Arabidopsis. Mol. Gen. Genet. 257, 387–391. doi: 10.1007/s004380050662
Zhou, P., Song, M., Yang, Q., Su, L., Hou, P., Guo, L., et al. (2014). Both PHYTOCHROME RAPIDLY REGULATED1 (PAR1) and PAR2 promote seedling photomorphogenesis in multiple light signaling pathways. Plant Physiol. 164, 841–852. doi: 10.1104/pp.113.227231
Zhu, D., Maier, A., Lee, J.-H., Laubinger, S., Saijo, Y., Wang, H., et al. (2008). Biochemical characterization of Arabidopsis complexes containing constitutively photomorphogenic1 and suppressor of phya proteins in light control of plant development. Plant Cell 20, 2307–2323. doi: 10.1105/tpc.107.056580
Zhu, L., Bu, Q., Xu, X., Paik, I., Huang, X., Hoecker, U., et al. (2015). CUL4 forms an E3 ligase with COP1 and SPA to promote light-induced degradation of PIF1. Nat. Commun. 6:7245. doi: 10.1038/ncomms8245
Keywords: COP1, SPA, CUL4, C3D, CDD, DDB1, E3 ligase, ubiquitination
Citation: Ponnu J and Hoecker U (2021) Illuminating the COP1/SPA Ubiquitin Ligase: Fresh Insights Into Its Structure and Functions During Plant Photomorphogenesis. Front. Plant Sci. 12:662793. doi: 10.3389/fpls.2021.662793
Received: 01 February 2021; Accepted: 04 March 2021;
Published: 24 March 2021.
Edited by:
Hanjo A. Hellmann, Washington State University, United StatesReviewed by:
Dongqing Xu, Nanjing Agricultural University, ChinaEnamul Huq, University of Texas at Austin, United States
Judy Callis, University of California, Davis, United States
Copyright © 2021 Ponnu and Hoecker. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ute Hoecker, aG9lY2tlcnVAdW5pLWtvZWxuLmRl; Jathish Ponnu, anBvbm51QHVuaS1rb2Vsbi5kZQ==
 Jathish Ponnu
Jathish Ponnu Ute Hoecker
Ute Hoecker