- 1Cell and Molecular Sciences, The James Hutton Institute, Dundee, United Kingdom
- 2School of Biology, University of St Andrews, St Andrews, United Kingdom
- 3School of Life Sciences, University of Dundee, Dundee, United Kingdom
- 4INRAE UMR Institut Sophia Agrobiotech, Sophia Antipolis, France
- 5Entomology, Plant Pathology and Nematology Department, University of Idaho, Moscow, ID, United States
- 6Department of Plant Sciences, University of Idaho, Moscow, ID, United States
Potato cyst nematodes (PCN) are economically important pests with a worldwide distribution in all temperate regions where potatoes are grown. Because above ground symptoms are non-specific, and detection of cysts in the soil is determined by the intensity of sampling, infestations are frequently spread before they are recognised. PCN cysts are resilient and persistent; their cargo of eggs can remain viable for over two decades, and thus once introduced PCN are very difficult to eradicate. Various control methods have been proposed, with resistant varieties being a key environmentally friendly and effective component of an integrated management programme. Wild and landrace relatives of cultivated potato have provided a source of PCN resistance genes that have been used in breeding programmes with varying levels of success. Producing a PCN resistant variety requires concerted effort over many years before it reaches what can be the biggest hurdle—commercial acceptance. Recent advances in potato genomics have provided tools to rapidly map resistance genes and to develop molecular markers to aid selection during breeding. This review will focus on the translation of these opportunities into durably PCN resistant varieties.
Introduction
Potato is now the third most important food crop after rice and wheat for human consumption (CIP, 2020). Being an important source of carbohydrates, potato provides more protein and minerals than any other staple crop (Birch et al., 2012). With the increasing demand from a growing world population, optimised pest and disease management are of increasing importance for the sustainability of this crop. Potato cyst nematodes (PCN) are the most economically important parasitic nematodes of potato. They are soil inhabiting biotrophic sedentary endoparasites (Wyss, 1997) and are specialised pathogens of solanaceous species (Sabeh et al., 2019). The global economic impact of PCN has not been fully quantified, however, even a small infestation can incur considerable costs, including loss of income to growers, closure of local and international markets as well as expenses for regulatory activities such as surveys and monitoring systems (Hodda and Cook, 2009). There are estimates that the two species, Globodera rostochiensis and G. pallida, are responsible for the loss of 9% of the crop worldwide (Turner and Subbotin, 2013). A potential third PCN species was reported by Skantar et al. (2011) and has subsequently been described as G. ellingtonae (Handoo et al., 2012). It has been detected in Oregon and Idaho in the United States, and in the Andean region of South America (Lax et al., 2014). However, its pathogenicity to potato is inconsistent (Zasada et al., 2019).
PCN co-evolved with their hosts from the genus Solanum in South America (Canto Saenz and Mayer de Scurrah, 1977; Stone, 1985). For G. pallida, this co-evolution is estimated to have taken place over 20 million years, with a northward expansion from the south of Peru around Lake Titicaca (Picard et al., 2004, 2007). Both nematode species were probably introduced and became established in Europe in the 1850s as a consequence of the Irish potato famine (Evans et al., 1975), when tubers, with contaminated soil attached, were collected and brought to Europe as breeding material for late blight resistant potatoes. In the 1880s, damage to potato crops from nematodes was noted in Germany, and from the early 1900s in the United Kingdom (Evans et al., 1975). PCN have subsequently been spread, mainly via contaminated seed potatoes, to almost all countries where potato is grown. To date, they have been detected in 126 countries (79 for G. rostochiensis and 55 for G. pallida) (CABI, 2020a,b). PCN are quarantine pathogens, which means that phytosanitary regulations are applied to prevent the introduction or spread within a country, and once detected, eradication or containment measures are applied (reviewed in Pickup et al., 2018). Protection of seed land from infestations is vital to prevent spread into non-infested land within the EU, and testing of seed land prior to planting is required by the EU Council Directive 2007/33/EC. The regulations that apply to PCN provide a framework for the potato industry, but despite these regulations PCN continues to spread. The long delay between introduction and detection, which may require several crop cycles, confounds interpretation of how effective these regulations may be. The widespread presence of these nematodes in East Africa provides a contemporary example of the failure to prevent spread and the devastating consequences incurred by subsistence farmers who have become dependent on potato for their livelihoods (Mburu et al., 2020). Both species of PCN are also widespread in Algeria, the largest potato producer in Africa (Djebroune et al., 2021).
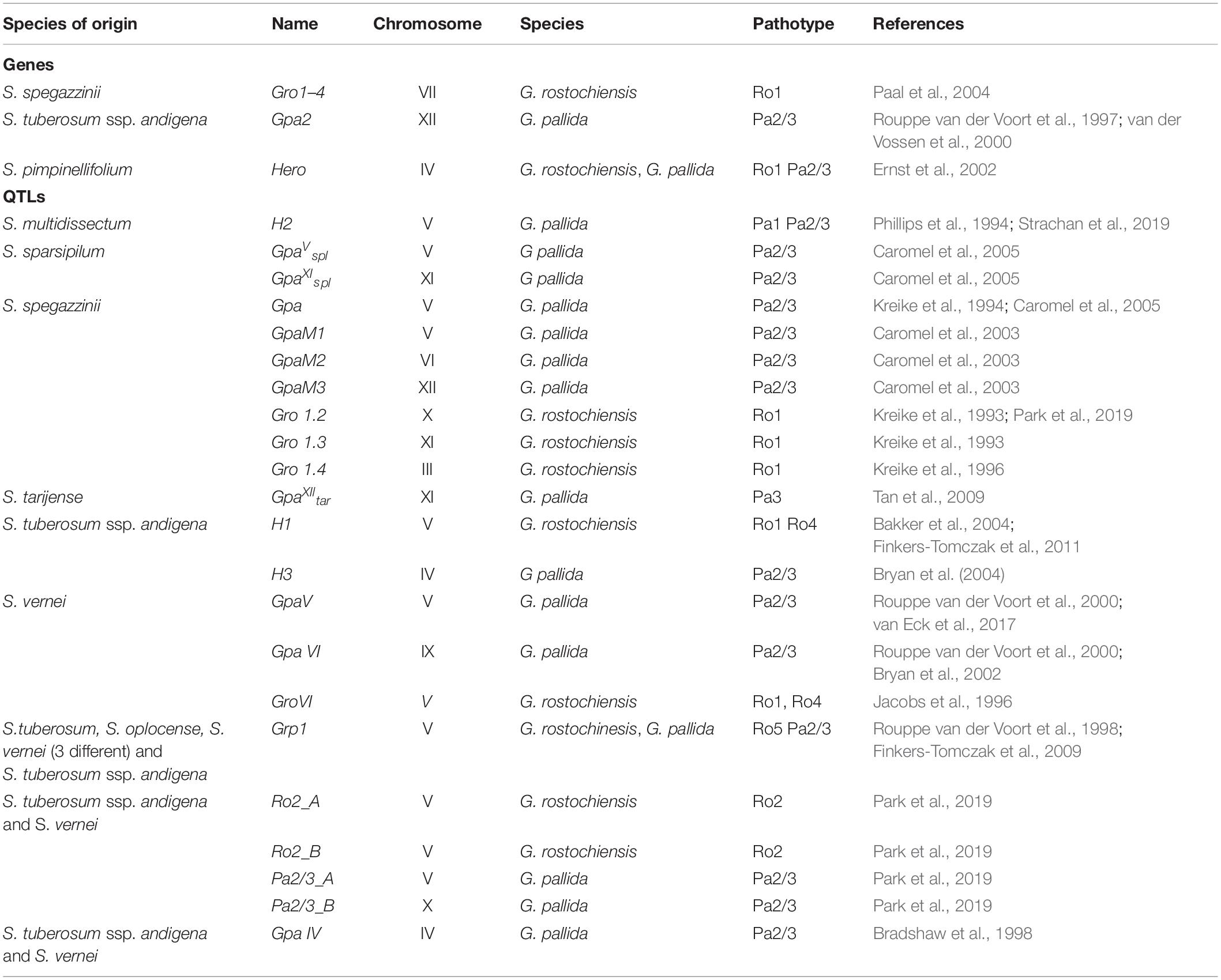
Disease symptoms caused by PCN are initially minimal, due to low infection levels in localised patches. Such symptoms are frequently interpreted as the effects of abiotic stress, and infestations frequently remain unidentified for many years until reduced yields or premature plant senescence are observed. PCN population dynamics has been studied since the 1950s, in order to predict yield losses by modelling different parameters such as level of field infestation at the start of the season, use of nematicides, soil type, rotation length and use of resistant/susceptible potato cultivars (e.g., Seinhorst, 1982; Been and Schomaker, 1998; Trudgill et al., 2014). Online tools such as NemaDecide1 and PCN Calculator2 were developed in an attempt to allow potato growers to simulate expected tuber yields and PCN population development when different conditions are applied. Figure 1A shows the time that passes between introduction of PCN to a field and reaching the detection/damage thresholds under different rotation schedules of susceptible crops in a very simplified model, only considering a constant multiplication rate, set at 15-fold per annum, and a fixed rate of natural decline, set at 25% per annum. In Figure 1B, an example of the predictions for population levels and yield trends is shown. The parameters used such as initial population level, decline rate, soil type, nematicide treatment, rotation length, cultivar resistance and tolerance can be adjusted with the PCN Calculator for G. pallida2.
Figure 1. (A) Schematic illustration of the increase of potato cyst nematodes (PCN) cysts with different rotation intervals, and the time until detection and damage thresholds are reached. This simplified sketch shows a natural decrease of viable cysts of 25% per year, and a 15-fold multiplication rate per year, no other factors are considered. (B) This illustrates the predictions of a model in which different factors including initial population level (in this example 80 eggs/g soil) soil type, nematicide treatment and rotation length, and cultivar characters including tolerance and resistance, can be altered to predict population and yield trends using a specified decline rate of 20% (https://pcncalculator.ahdb.org.uk).
The fact that PCN cysts are highly durable, and viable eggs can survive for decades in the absence of a host (Turner, 1996), together with difficulty in detection of new infestations, makes preventing the spread of PCN very challenging. An example of a successful eradication is from Western Australia, where in the late 1980s a total area of 35 ha was found to contain G. rostochiensis. This area was declared PCN free in 2010. However, at least part of this land has been converted to housing estates, precluding further sampling (Collins et al., 2010; IPPC, 2010). In Idaho, a containment and eradication programme is in progress following the detection in 2006 of two potato fields infested with G. pallida (Skantar et al., 2007; USDA, 2007). Preventing spread and eradication with soil fumigants is the focus of the regulatory programme in Idaho3. When PCN is well established but not widespread, preventing spread to uninfected fields becomes paramount. With widespread infestations, management involves various strategies (Back et al., 2018), including crop rotation, chemical and biological control, bio-fumigation and trap cropping. This review focuses on the use of host resistance to manage PCN.
Host Nematode Interactions
Potato cyst nematodes have long lasting biotrophic interactions with their hosts. Cysts, located in the soil after detaching from host roots, contain several hundred eggs, each of which contains a juvenile nematode (J). The moult from J1 to J2 takes place within the egg once embryogenesis is complete. The unhatched J2 is protected by a chitinous eggshell which is an impermeable barrier and provides physical protection against potential bacterial or fungal pathogens. Furthermore, the unhatched J2 is dehydrated and suspended in a perivitelline fluid that contains a high concentration of trehalose, which provides protection against low winter temperatures (Perry, 2002). Hatching of PCN eggs is triggered by exposure to chemicals released from the host roots, termed host root hatching factors. The requirement for hatching factors in root exudates to stimulate hatching of the J2s provides a means for synchronisation of the PCN life cycle with the presence of a host plant (Masler and Perry, 2018). The presence of hatching factors triggers a calcium dependent change in eggshell permeability, allowing water to enter the egg, reactivating the metabolic activity of the unhatched J2 (Clarke and Perry, 1985). The J2 nematode subsequently cuts its way out of the eggshell using its stylet (Doncaster and Seymour, 1973). Host resistance is not associated with the ability to reduce hatch of PCN—root exudates of resistant plants induce similar levels of hatch as those from susceptible plants (Turner and Stone, 1981). After hatching, the J2 moves through the soil, locating, and invading the host root. Physiologically active roots exude an array of molecules that form chemical gradients in soil that are perceived by the nematodes, allowing their orientation and migration toward the host. Despite the importance of the chemical signalling that governs the nematode chemotaxis, advances in nematode chemical ecology are relatively recent (Torto et al., 2018). To date, there is no evidence that host resistance affects PCN chemotaxis.
After penetrating the host, the nematodes migrate destructively through the root cells until reaching the inner cortical layer. Migration is facilitated by the production of a cocktail of plant cell wall degrading enzymes and modifying proteins including cellulases (Smant et al., 1998), pectate lyases (Popeijus et al., 2000), and expansins (Qin et al., 2004). The genes encoding these proteins are likely to have been acquired as a result of horizontal gene transfer from bacteria (Kikuchi et al., 2017). After reaching the inner cortical layer, the behaviour of the J2 changes and the nematode probes cells with its stylet to select an initial syncytial cell (ISC) that does not respond adversely by producing callose (Sobczak and Golinowski, 2011). Effectors are introduced into this ISC which then undergoes a transformation into a syncytium. Within the ISC the central vacuole degrades, the cytoplasm becomes enriched in subcellular organelles, and the nucleus becomes enlarged and ameboid in shape (Golinowski et al., 1996). The syncytium develops into a multinucleate structure through controlled breakdown of the cell walls separating the ISC from its neighbours, initially at the sites of plasmodesmata, with the subsequent fusion of protoplasts of adjoining cells (Grundler et al., 1998; Figure 2). Up to 300 cells can be included in the final structure. The syncytium provides a rich food source for the developing nematode but needs to be kept alive for the duration of the nematode life cycle as the nematode is not able to induce more than one such structure. The nematode produces a feeding tube during each cycle of feeding (Eves-van den Akker et al., 2015a). The precise nature of this structure remains unknown, but it likely acts as a molecular sieve and prevents damage to the syncytium when feeding.
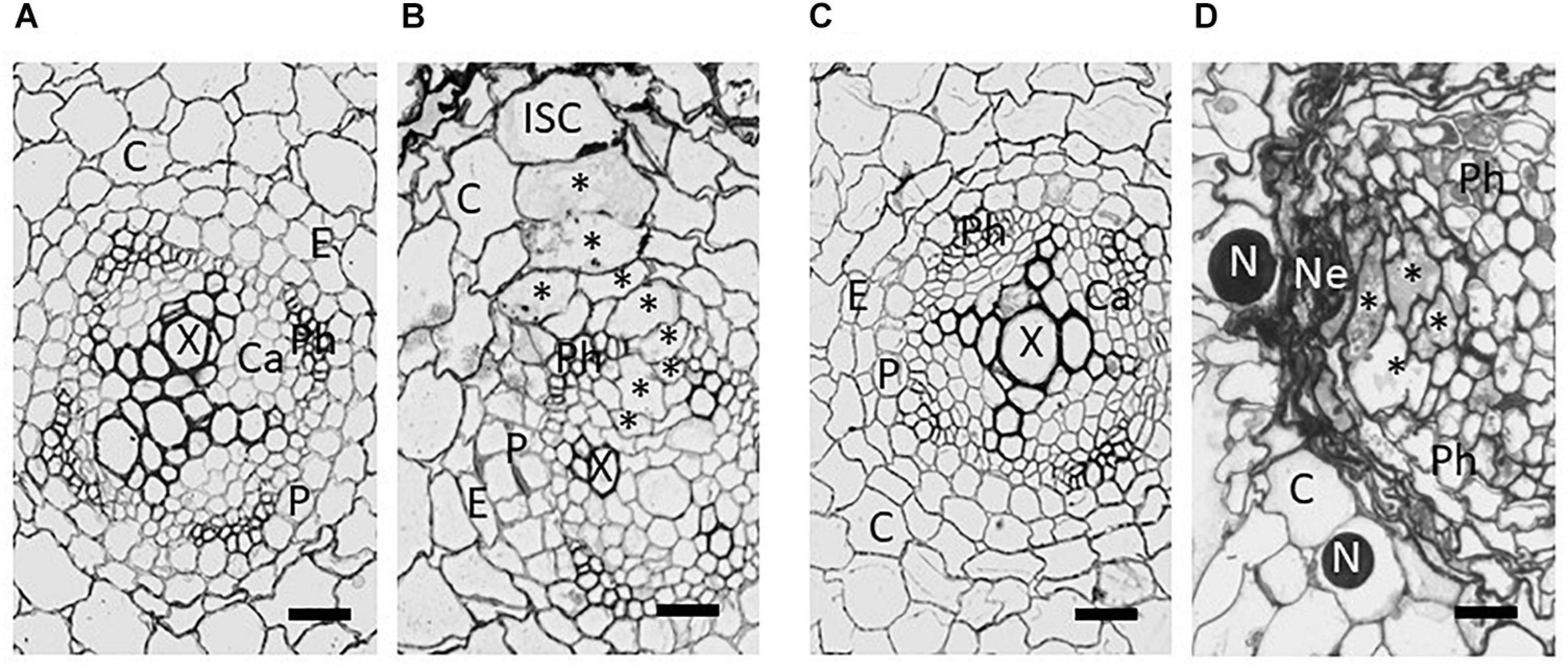
Figure 2. Brightfield images from tangential sections of potato roots showing development of syncytia induced by Globodera pallida in the roots of susceptible Solanum tuberosum cv. Pentland Ivory (A,B) and resistant reaction of Sa_12601 [S. tuberosum ssp. andigena CPC 2802 (H3)] (C,D). (A) Uninfected root and (B) syncytium at 4 dpi of susceptible cv Pentland Ivory, (C) uninfected and (D) necrosis and collapse of syncytium at 7 dpi of Sa_12601. C, cortex; Ca, pro-/cambium; E, endodermis; ISC, initial syncytial cell; N, nematode; Ne, necrosis; P, pericycle; Ph, phloem; X, xylem. Asterisks (*) indicate selected syncytial elements. Scale bars: 20 μm (from Varypatakis et al., 2020).
The interactions between PCN and its host are mediated by effectors. These proteins are secreted from the nematode into the host to manipulate its metabolism to the benefit of the nematode. PCN effectors, like those of other plant-parasitic nematodes, are produced in two subventral and one dorsal gland cell. Ultrastructural studies (Hussey and Mims, 1990), as well as transcriptome analysis (Thorpe et al., 2014) indicate that effectors important in the early stages of the host-parasite interaction (including migration) tend to be produced in the subventral gland cells, while those that are important at later stages are produced in the dorsal gland cell. In terms of the interaction between PCN and its host, effectors have been characterised that assist in the migration of the nematode through the root (e.g., Smant et al., 1998), in the induction of the syncytium and suppression of host defence responses (e.g., Mei et al., 2015).
The precise details of how the syncytium is formed remain unclear. However, it is known that nematode control of several plant hormone pathways is key to this process (Gheysen and Mitchum, 2019). Manipulation of these pathways can be achieved through effectors and plant peptide hormone mimics produced by the nematode (Ali et al., 2017). In particular, the role of auxin, which is a central regulator of plant organogenesis, has been investigated during the feeding site formation. Studies of auxin synthesis and perception mutants showed that manipulation of the auxin signalling pathway plays a critical role in the development of syncytia. For example, auxin insensitive mutants are unable to support normal syncytium development, while disruption of polar auxin transport gives rise to abnormally large syncytia (Goverse et al., 2000). In addition, the rearrangement of PIN and AUX/LAX auxin transporters in developing syncytial cells, where auxin responsive promoters are also activated, indicates that changes in local auxin concentration are important during syncytium formation (Grunewald et al., 2009).
As in other pathosystems, resistance against PCN is triggered by recognition of its effectors by R-genes, either directly or indirectly (Sacco et al., 2009). The effector gene RBP-1 was identified as the Gpa2 resistance cognate avirulence (Avr) factor (Sacco et al., 2009). It belongs to the SPRYSEC effector gene family (secreted SP1a and ryanodine receptor domain-containing effector), which is unique to cyst nematodes. Recently, a list of G. pallida candidate Avr genes, implicated in the interaction with plants containing the H3 or GpaV resistance sources, has been proposed by Varypatakis et al. (2020). The most common candidates belong to the SPRYSEC family. Other putative Avr candidate genes included cellulases, glutathione synthase and an orthologue of Heterodera glycines GLAND3, with unknown function.
Ultrastructural studies comparing the development of the syncytium in susceptible and resistant hosts showed structural differences evident within 2 days post infection (dpi) (Rice et al., 1985; Varypatakis et al., 2020). By 4 dpi susceptible plants showed incorporation of many additional cells into the syncytium and expansion into the vascular cylinder. However, no expansion was found in the resistant host apart from some degradation of cytoplasm. By 7 dpi most syncytia were degraded and compressed in a host with the H3 resistance derived from S. tuberosum ssp. andigena CPC 2802, and extensive necrosis of neighbouring cells was also observed (Figure 2; Varypatakis et al., 2020). Failure to incorporate cells within the vascular cylinder prevents development of a viable syncytium, which is needed by the feeding nematode to progress through moults to the adult stages. Some variation in the degree of syncytial degradation has been observed and this may account for the increased number of males observed in some hosts that are “partially resistant” and have syncytia which are compromised in their nutritional capabilities. Males do not require a sustained viable syncytium to reach maturity as they do not produce eggs, and hence a shift in the sex ratio toward males has been observed with some sources of resistance (Rice et al., 1985; Sobczak et al., 2005). Modification of the host root’s functions, caused by the initiation of the feeding sites and the establishment of the resistance response with extensive necrotic area can greatly affect plant growth and lead to crop failure at high nematode densities. The destructive root penetration during PCN invasion that occurs irrespective of the resistance, can also lead to colonisation by other root pathogens, exacerbating damage to the crop.
Virulence, Pathotypes, and Durability of Resistance
Nematode virulence is defined as the ability to reproduce on a host plant that contains specific genes, otherwise conferring resistance. Nematodes which cannot reproduce on the same resistant plant are called avirulent (Vanderplank, 1978; Turner et al., 1983; Blok et al., 2018). Virulence traits represent a characteristic of individual nematodes. Nematode populations can consist of a mixture of virulent and avirulent individuals. This proportion can change over time, depending on environmental conditions, such as use of resistant or susceptible hosts. The reproduction rate of virulent nematodes on resistant plants is typically lower than on a fully susceptible plant. Nematodes, like other pathogens, usually conform to the so-called trade-off concept, stating that resistance-breaking populations of pathogens pay a price by losing fitness on susceptible hosts compared to avirulent populations (Stearns, 1989; Laine and Barres, 2013). Nevertheless, a recent experimental evolution study (Fournet et al., 2016) found that this might not always be the case for G. pallida, for which virulent populations became fitter on resistant hosts, producing bigger cysts with more juveniles that hatched earlier. However, these experiments were conducted in petri-dishes with nutrient enriched agar, and thus might not represent the behaviour in the field.
In Europe, over 40 years ago, a pathotype scheme was developed (Kort et al., 1977), in order to identify which sources of resistance were most effective against European populations of G. rostochiensis and G. pallida. Globodera rostochiensis was subdivided into five pathotypes, Ro1–Ro5. Ro1 and Ro4 were differentiated by their low multiplication rate on S. tuberosum ssp. andigena CPC 1673, compared to Ro2, Ro3, and Ro5. Solanum kurtzianum hybrid 60.21.19 differentiated Ro2 from Ro3 and Ro5, and Ro1 from Ro4. Ro3 multiplied poorly on S. vernei 58.16/12/4, compared to Ro5. All G. rostochiensis pathotypes were unable to multiply on S. vernei hybrid 65.346/19. Globodera pallida was categorised into three European pathotypes, Pa1, Pa2, and Pa3; S. multidissectum hybrid P55/7 differentiated Pa1 from Pa2 and Pa3, and S. vernei (VTn)2 62.33.3 differentiated Pa2 from Pa3.
The Kort pathotype scheme provided a panel of PCN populations representing the phenotypic spectrum in Europe and identified several sources of resistance and their specificities for use by potato breeders. For G. rostochiensis, Ro1 is the dominant pathotype outside South America, and breeding efforts have focused on this pathotype. The occurrence of G. rostochiensis in the United Kingdom is probably the result of a single introduction (Evans et al., 1975; Bendezu et al., 1998), and resistance to G. rostochiensis Ro1 has proven to be remarkably durable. For G. pallida, the situation has been more complicated and the progress from breeding has been much slower. The differentiation of the pathotypes Pa2 and Pa3 on S. vernei (VTn)2 62.33.3 was not found to be robust, and this has led to questions about the validity of the pathotype scheme. Trudgill (1985) demonstrated that the reproduction factor of <1 used in the Kort scheme to indicate resistance was environmentally sensitive and proposed that the Pa2 and Pa3 pathotypes were a continuum (Pa2/3) that could not be distinguished reliably. Most European populations of G. pallida belong to the Pa2/3 pathotype, though genetic evidence supports a distinct introduction which includes the populations Chavornay and Luffness, classified as Pa3 in the Kort scheme (Hockland et al., 2012). At least three independent introductions of G. pallida from South America have occurred in Europe (Plantard et al., 2008; Hockland et al., 2012). In Scotland, mixtures of these three genotypes can be present in the same field (Eves-van den Akker et al., 2015b). Since this scheme was introduced, other sources of resistance to G. pallida have been found that are being used in breeding programmes and resistance-breaking populations of G. pallida have been detected. Taken together, these facts have confounded the issue of pathotype designation.
The PCN populations outside South America represent a small portion of the total genetic diversity of these species (Plantard et al., 2008; Grenier et al., 2010). This restricted genetic variation makes it possible to consider a potato breeding approach using host resistance for their control. While the H1 resistance to G. rostochiensis has shown to be highly durable, the resistance sources available for G. pallida are not expected to be as long-lasting due the greater genetic diversity of G. pallida populations. Also, in general, the resistances that have been identified for this species have not been as effective. It is possible that the combination of the more heterogenous nature of G. pallida populations with the more genetically complex nature of the quantitative resistances involved, will require the action of two genes for very high levels of durable resistance.
The use of resistant hosts exerts a selection pressure on nematode populations that can lead to an increase in the proportion of virulent vs. avirulent individuals (Whitehead, 1991; Hockland et al., 2012). Varypatakis et al. (2019) observed this effect when using different potato cultivars challenged with G. pallida populations previously grown repeatedly on potato cultivars with partial resistance to G. pallida. Beniers et al. (2019) also used partially resistant potato cultivars to show an increase of virulence in a population when the same source of resistance was applied over generations as have others (Turner, 1990; Phillips and Blok, 2008). Ultimately, this shift could lead to resistance-breaking PCN populations. This effect has already been reported for the resistant potato cultivar Innovator in some regions in Germany (Niere et al., 2014). Pyramiding different sources of resistance to G. pallida is likely to be needed to produce potato cultivars with durable, broad spectrum resistance. Indeed, Dalton et al. (2013) demonstrated a synergistic effect of increased resistance when combining two different PCN resistance loci in potato and Rigney et al. (2017) showed that this effect could be replicated with several G. pallida populations, though the durability of this material has not yet been assessed.
Resistance and Tolerance to Pcn and Their Assessment
Resistance and tolerance to pests and pathogens are two different strategies that plants and animals use to cope with biotic threats. Resistance to PCN is defined as the host plant’s ability to inhibit or limit reproduction, relative to a susceptible plant (Evans and Haydock, 1990; Blok et al., 2018; Pagán and García-Arenal, 2020). In contrast, tolerance toward PCN is defined as the ability of the potato to tolerate PCN infection without a reduction in yield. Unlike resistance, tolerance does not significantly affect the nematodes’ ability to infect the host and reproduce (Starr et al., 2002; Pagán and García-Arenal, 2020), and the two traits are independent of each other (Evans and Haydock, 1990; Trudgill, 1991). When confronted with high PCN population densities, resistant cultivars that are intolerant do not perform well (in terms of yield) although they will restrict reproduction of PCN, whereas susceptible tolerant cultivars allow PCN populations levels to rise and produce adequate yields (Trudgill, 1991). Combining resistance and tolerance is now regarded as essential by potato breeders in areas where PCN is widespread.
Resistance
The cultivated tetraploid species S. tuberosum ssp. tuberosum does not show any resistance to PCN (Rousselle-Bourgeois and Mugniery, 1995). However, related wild potato species and landraces are a genetic resource for resistance to pests such as PCN, resilience to abiotic stresses and tuber quality characters. Many of them have been systematically tested for PCN resistance (e.g., Rousselle-Bourgeois and Mugniery, 1995; Castelli et al., 2003). To date, more than 50 potato species have been identified that show resistance to G. rostochiensis and/or G. pallida for at least one pathotype, in at least one accession. Wild potato species can be found in their natural habitats from the southwestern US to Argentina, with most species being native to Mexico and the Andean Highlands (Bethke et al., 2017). Initial approaches for identifying sources of resistance to PCN in potato involved the screening of over 1,200 accessions of wild species from the Commonwealth Potato Collection and led to the first identified PCN resistance locus (H1) from S. tuberosum ssp. andigena, which offers nearly complete resistance to G. rostochiensis pathotypes Ro1 and Ro4 (Ellenby, 1952). More recently, it was reported that G. ellingtonae does not reproduce on potato genotypes which carry the H1 resistance locus (Whitworth et al., 2018; Zasada et al., 2019).
Based on the “gene-for-gene” concept (Flor, 1971), quantitative resistance to PCN may be conferred by products of dominant resistance (R) genes. These genes recognise pathogen-produced effector proteins in infected cells, encoded by Avr genes, and trigger defence responses, which may include a hypersensitive reaction (Van der Biezen and Jones, 1998; Davies and Elling, 2015). The latter involves cells that are committed to cell death, depriving the nematodes of an adequate food source, and thus leading to resistance. While all known genomic loci conferring resistance to G. rostochiensis or G. pallida belong to the nucleotide-binding site and leucine-rich repeat (NB-LRR) family of R genes, it is notable that other gene classes may be important in providing resistance to PCN and other cyst nematodes. For example, the Cf-2 gene, which confers resistance against the fungal pathogen Cladosporium fulvum, encodes an extracellular receptor-like protein with a LRR domain, also provides resistance against G. rostochiensis (Lozano-Torres et al., 2012). For both C. fulvum and PCN, Cf-2 is activated by pathogen attempts to interfere with the apoplastic cysteine proteinase Rcr3. In addition, resistance against the soybean cyst nematode, H. glycines, can be underpinned by completely different molecular mechanisms that cause a toxic response specifically targeted at the syncytium. Resistance derived from the Rhg1 locus is based on a copy number variation of multiple genes, with three of these genes contributing to full resistance in soybean: a putative amino acid transporter, an α-SNAP [soluble NSF (N-ethylmaleimide sensitive fusion)-associated protein] and a protein containing a wound-inducible domain (Cook et al., 2012). Only the function of the α-SNAP has been elucidated to some extent. The defective resistance-type α-SNAP protein accumulates preferentially in the nematode feeding site relative to its wild type counterpart, causing cytotoxicity (Bayless et al., 2016). Similarly, the resistance conferred by Rhg4 is mediated through a defective variant form of a serine hydroxymethyl transferase expressed in the syncytium. This enzyme is otherwise essential for cellular one-carbon metabolism, causing a failure of nutrients reaching the nematode (Liu et al., 2012). No similar mechanisms against PCN have been identified to date but, interestingly, the proteins encoded by the soybean Rhg1 gene can function in different plant families and confers resistance to PCN in potato (Butler et al., 2019).
Tolerance
Factors that enhance tolerance of potato to PCN are not necessarily specific to PCN. Tolerance factors may involve enhanced root growth, the activation of general plant stress responses or combinations thereof, and can provide additional tolerance to e.g., drought or other pathogens (Trudgill, 1991; Blok et al., 2018). Environmental conditions have a major impact on tuber yield (Evans and Haydock, 1990), and tolerance is therefore environmentally sensitive and difficult to reliably assess.
The damage caused by PCN depends on the initial nematode density in the soil. Intolerant (sensitive) and susceptible cultivars show damage at even low rates of infestation (10 eggs/g soil). If damage is extensive, this can lead to low reproduction of the nematodes even though the plants are susceptible. Tolerant and susceptible cultivars, planted in infested land, can lead to a very high prevalence of PCN by increasing nematode population levels, while still giving acceptable yields, which thwarts PCN management programmes. But even a tolerant plant will suffer damage at high nematode densities, mostly due to reduced uptake of nutrients (Trudgill, 1987). When PCN is well-managed and populations have a low density, intolerant resistant potatoes may be grown, however, the best way to manage PCN would be the use of potato cultivars which are highly resistant and tolerant. Resistance ensures the reduction of nematode population levels and tolerance ensures reliable crop yields, even when the initial PCN levels are high (Blok et al., 2018). Tolerance to pathogens is usually not selected for in breeding programmes, therefore this trait is often lost, unless it is linked to another commercially useful trait (Trudgill, 1991). Very few quantitative trait loci (QTLs) for tolerance to cyst nematodes have been described so far (Williams et al., 2003, 2006; Ravelombola et al., 2020).
Assessment of Resistance
Any breeding programme that aims to include PCN resistance in a new cultivar requires a reliable, and ideally rapid, method for resistance screening. Screening methods to determine the level of resistance in potatoes are usually performed in a greenhouse in pots or rootrainer, or in a dark incubation chamber when using canisters (Figure 3). The choice of which test to use depends on the accuracy required, resources available and time constraints. PCN cysts, eggs or the J2 stage juveniles can be used as inoculum. For good practice, each assessment should include at least one susceptible and resistant control cultivar, be performed at least twice with a minimum of four biological replicates of each genotype grown, in a randomised planting design.

Figure 3. Methods for assessing PCN resistance in potatoes: (A) clay pots with potato plants growing in a sand/loam mixture which has been inoculated with cysts, (B) empty rootrainer cassette, 4 root systems in rootrainer with roots growing on surface of compost and female nematodes on root surface, (C) transparent canister with potato roots visible for counting female nematodes.
Canister tests can be used with relatively large numbers of progeny and therefore provide an efficient method to separate susceptible and resistant clones in a breeding programme. Potatoes are grown in the dark in transparent canisters with a lid, filled with moist soil and inoculated with PCN. After 8 weeks the number of female nematodes that are visible on the surface of the roots through the transparent canisters are counted (Phillips et al., 1980). The test in rootrainers is an alternative to the canister test. It is performed in a temperature-controlled greenhouse and is more labour intensive, as the plants require regular maintenance. Potato plants are grown in PCN infested soil in rootrainers for 7–8 weeks, then the number of female nematodes on the roots visible on the surface of the “root-ball” are counted after carefully removing them from the rootrainers (Strachan et al., 2019; Varypatakis et al., 2019). The most accurate assay for phenotyping potatoes is a pot test, where plants are grown for 12 weeks in a sand/loam 1:1 mixture that has been inoculated with PCN. When using cysts as inoculum, these need to be bagged to ensure that they are not counted as new cysts after the experiment is completed. The cysts are then washed out of the sand/loam mixture and counted (Fenwick, 1940; Reid et al., 2015).
The nematode reproduction rate Pf/Pi (multiplication factor with Pf and Pi being the final and initial female/cyst numbers, respectively) is a measurement used to assess the relative resistance of a potato genotype. If there are very few cysts, the eggs are extracted from the cysts and counted instead to assess reproduction. The current resistance evaluation system for PCN used in Europe was introduced in 2006 (EPPO, 2006). The reproduction rate of the G. pallida population Chavornay on the susceptible potato cultivar Desirée is assessed, and the susceptibility is divided into 9 susceptibility groups, with 1–3 being susceptible (>100–5%), 4–6 partially resistant (<25–3%) and 7–9 resistant (<3%). This system uses an agreed set of protocols and means that resistance ratings can be applied consistently to new cultivars.
All these bioassays require specialised materials and trained staff. They are time consuming and therefore expensive. Using genetically linked molecular markers to determine the presence or absence of resistance loci is faster and allows more progeny to be screened. However, bioassays ultimately need to be performed to confirm resistance. The accuracy of molecular markers is not always absolute, as homologous recombination can separate the marker and the resistance locus in each generation, the likelihood of recombination increasing with distance between marker and resistant locus on the genetic map. This problem may be overcome when the marker resides in the resistance gene itself, or if two markers flanking the resistance are used, as two recombination events in one generation are extremely rare.
Assessment of Tolerance
Tolerance to PCN is a complex trait and to date no molecular markers are available. Bioassays are therefore the only option to assess tolerance. To assess tolerance, the performance of different infected and non-infected potato genotypes is compared with that of genotypes with known tolerances (Cook and Evans, 1987; Trudgill, 1991). Field tests for the evaluation of tolerance are described in Trudgill (1991), whereas Arntzen and Wouters (1994) described a high correlation between pot and field evaluations of tolerance against PCN during early growth. The use of resistant and susceptible control potato cultivars with known levels of tolerance is advised in those assays, as the results are highly variable, both in replicates and in different experiments (Dale et al., 1988). If the assay is performed with and without nematicides, tolerant potatoes are expected to have the least increase of yield in conjunction with nematicide use (Trudgill and Cotes, 1983a). Tolerant cultivars often grow larger roots in heavily infested soil compared to nematicide treated or only lightly infested soil (Trudgill and Cotes, 1983b).
Breeding for Pcn Resistance and the Development of Molecular Markers for Marker Assisted Breeding
History of Breeding for PCN Resistant Potatoes
The modern cultivated potato derives mainly from a relatively small number of introductions into Europe from South America in the 16th, 17th, and 18th centuries centuries. By the end of the 18th century the crop had quite rapidly been adapted to tuberise in the long-day conditions found in Northern Europe through selection for early-tuberisation and higher tuber yields. However, early cultivars lacked sufficient genetic variation to provide resistance to major pests and diseases. This became a problem once the crop became widely grown as the predominant staple food crop in many countries in the 19th century. The most notable pathogens have been late blight (Phytophthora infestans) and latterly, PCN. This has led to the need to introgress genes for disease and pest resistance into potato varieties from the many wild and cultivated species of Central and South America, some of which can be crossed directly with cultivated potato.
Commercial potato breeding is still performed mainly at the tetraploid level, whereas many of the wild and primitive cultivated species containing disease resistance are diploid. The highly heterozygous nature of tetraploid potato breeding germplasm greatly compromises potato improvement. Potato breeding remains largely empirical and genetically unsophisticated, and most economically important traits are genetically complex, displaying continuous variation. Resistance to pest and disease threats are largely an exception, being generally controlled by the action of one or two dominant genes. This property has led to a focus on developing genetic markers for use in breeding for pest and disease resistance. Development of molecular genetic tools facilitating the mapping and cloning of genes that confer resistance has significantly accelerated the process of PCN-resistant cultivar development through marker-assisted selection (MAS) in recent years.
Efforts to breed resistance to PCN have been ongoing since the middle of the 20th century (Figure 4). The first cross to introgress the H1 resistance source, which confers resistance to Ro1 and Ro4 pathotypes of G. rostochiensis, was between S. tuberosum ssp. andigena CPC 1673 and the variety Kerr’s Pink in 1952. The resistance was found to be simply inherited, and after backcrossing and selection for commercially important traits as well as resistance, the varieties Maris Piper (England) and Pentland Javelin (Scotland) were released in 1966 and 1967, respectively. Maris Piper remains one of the dominant commercial cultivars in the United Kingdom. The H1 resistance has been widely deployed in breeding and is the most frequently used source of resistance to G. rostochiensis (Przetakiewicz and Milczarek, 2017) in current European and North American potato varieties. This resistance source has proven to be extremely durable to date.
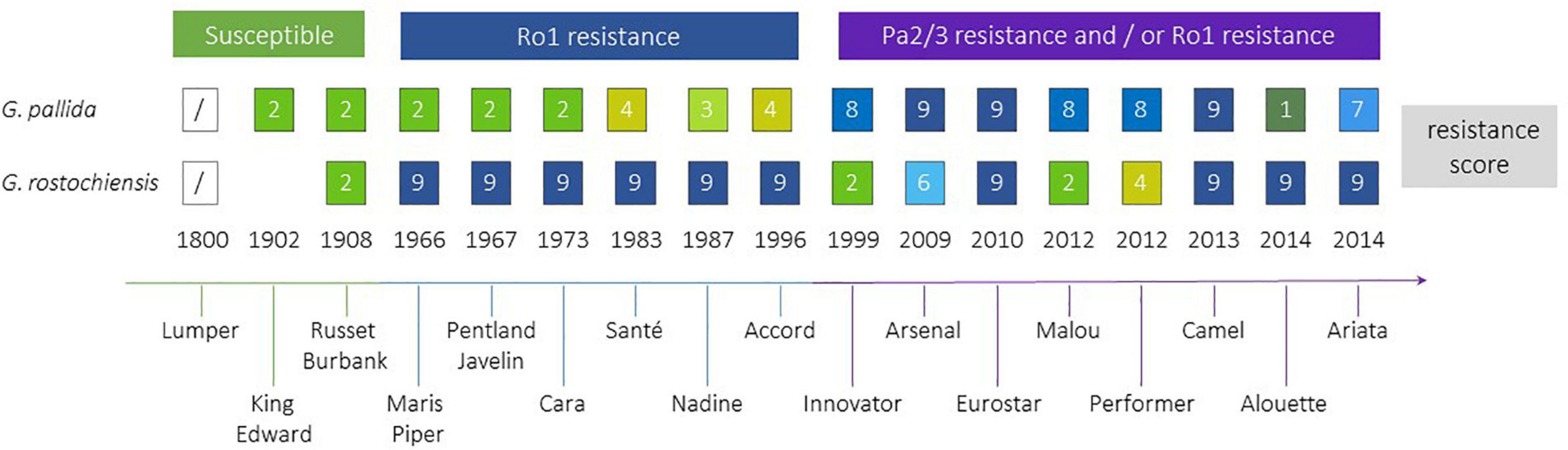
Figure 4. Selected potato cultivars, approximate time when they became commercially available, the introduction of resistances to Globodera rostochiensis and G. pallida. H1 resistance is used in all above cultivars against G. rostochiensis, whereas for G. pallida several sources of resistance have been introgressed. PCN resistance scores from 1 to 9 indicate increasing resistance levels from susceptible (1) to highly resistant (9) (EPPO, 2006).
Breeding for resistance to G. pallida has been complicated by the more complex nature of most resistance sources to this species and the higher genetic variability of G. pallida populations outwith South America, compared to G. rostochiensis. A resistance source, denoted H2, found in the diploid wild species S. multidissectum was shown to confer resistance to some G. pallida populations. This resistance was transferred to cultivated potato by interploidy crosses (Bradshaw et al., 1996). However, this gene has not been widely deployed as it does not provide strong resistance to the Pa2/3 populations of G. pallida that are the most prevalent in Europe. Breeding efforts in the United Kingdom and mainland Europe focused on the South American diploid species S. vernei, which includes accessions with quantitative resistance to both PCN species. Resistance derived from S. vernei has been introgressed independently in United Kingdom and mainland European breeding programmes, but it took many years to develop cultivars showing high levels of resistance. Chromosome doubling of S. vernei was performed by chemical means. The resulting tetraploid plants were crossed for several generations to cultivars and breeding lines over 30 years, leading to the eventual release of varieties (e.g., Innovator, Arsenal, Eurostar), which contain high levels of resistance to G. pallida Pa1, 2/3 populations. The Grp1 resistance has also been used by breeders in Europe to produce cultivars such as Iledher, Seresta, and Aveka. However, G. pallida populations are already able to overcome this resistance (Grenier et al., 2020).
A third source of resistance for G. pallida to be successfully targeted in United Kingdom breeding programmes is the so called H3 source, originating from the landrace S. tuberosum spp. andigena (CPC 2775 and CPC 2802). It was originally thought to be determined by a single gene, but subsequent analysis has shown that H3 is genetically controlled by QTLs on chromosomes IV (locus subsequently known as GpaIV) and XI (Bryan et al., 2002, 2004). It took over 20 years (but only 2 breeding generations) to produce the partially resistant cultivar Eden and breeding line 12601ab1, which has a higher level of resistance than Eden to pathotypes Pa2/3. The latter also has excellent processing traits, leading to its heavy use in United Kingdom breeding programmes. Several cultivars carrying GpaIV have been released with moderate levels of resistance to Pa2/3, such as Vales Everest, Midas, Olympus and Rocket.
Ongoing efforts in potato breeding are aimed at “pyramiding” various sources of PCN resistance into single genotypes. Most of the known sources of resistance to G. pallida provide only partial resistance, and moreover are genetically complex, involving the action of more than one gene. The value of pyramiding different loci conferring G. pallida resistance has been shown in previous studies, notably that of Caromel et al. (2005) who showed that “stacking” two separate QTLs from the wild species S. sparsipilum into individual genotypes resulted in a stronger resistance phenotype than either QTL deployed individually. The ideal scenario would be to combine H1 with at least two of the major effect QTLs conferring G. pallida resistance (e.g., H2, GpaIV, GpaV). The successful accomplishment of this goal would be facilitated by the existence of breeding lines with high “dosages” of the relevant functional alleles, as well as genetic markers diagnostic for both the presence and dosage of the functional alleles of these genes.
Diagnostic Molecular Markers Linked to PCN Resistance and Their Applications
Progress in the genetic analysis of PCN resistance sources led to the mapping of these sources, and in some cases the functional alleles have been isolated (Table 1). This also allowed the development of molecular markers tightly linked to the resistance phenotypes that can be deployed in potato breeding. These markers enable more informed breeding approaches that do not rely solely on time-consuming and progress-limiting phenotypic assessments. The use of MAS in breeding for PCN resistance has been well adopted in most breeding programmes. A brief history of the main resistance mapping efforts and marker development is presented below.
The H1 gene was first localised on chromosome V using restriction fragment length polymorphism (RFLP) mapping technology (Gebhardt et al., 1993). Further attempts to clone H1 have led to the development of several tightly linked markers (Finkers-Tomczak et al., 2011). The RFLP marker CP113, linked in coupling to H1, was deployed to test for the presence of the marker in 53 tetraploid German potato varieties with unknown sources of resistance. This was perhaps the earliest attempt of using molecular markers to survey PCN resistance in potato. Unfortunately, this marker failed to detect the presence of H1 in resistant varieties, highlighting the need for validating the “transferability” of markers to populations other than those in which the marker was developed. This is a common problem in outbreeding crop species showing low levels of linkage disequilibrium. Two further markers, TG689 and 57R have been reported and deployed for H1 PCN resistance screening, showing more than 90% congruence between the marker assay and the PCN-resistance phenotype (Biryukova et al., 2008; Milczarek et al., 2011; Schultz et al., 2012). The marker TG689 has been used successfully in a breeding programme which led to the potato cultivar Missaukee, a processing cultivar with resistance to late blight, verticillium wilt and G. rostochiensis (Douches et al., 2010). The presence or absence of a 452 bp amplicon for the marker 57R proved to be the most effective diagnostic marker for the prediction of PCN resistance conferred by H1 (Park et al., 2018). This marker successfully identified H1 in 90 resistant potato cultivars and in 299 out of 300 from potato breeding programmes, and correctly categorised all susceptible potato cultivars (Schultz et al., 2012). Further development of single nucleotide polymorphisms (SNP) markers diagnostic for H1 has been reported by Meade et al. (2020) who deployed a highly effective pooled “whole-genome resequencing” approach using previously deployed markers (in this case 57R) to identify novel sequence polymorphisms that could be converted into a more generic competitive allele-specific PCR format than the gel-based system used for 57R.
Attempts to develop diagnostic molecular markers for GpaIV and GpaV major QTLs have been only partially successful, progress being hampered by the quantitative nature of these resistances. A PCR-based marker, SPUD 1636, was reported for the GpaV QTL which was developed by converting a closely linked amplified fragment length polymorphism (AFLP) marker (Bryan et al., 2002). This presence/absence marker generates a 226 bp amplicon from resistant sources, and the diagnostic value of the SPUD1636 marker to identify sources of resistance was demonstrated for some accessions of S. vernei as well as varieties and breeding lines (Bryan et al., 2002). The marker has also been adapted for MAS in a Spanish resistance breeding programme (Ortega and Lopez-Vizcon, 2012) and used for resistance screening by Sudha et al. (2016). However, it has turned out to be inefficient in selecting breeding lines (Milczarek et al., 2011). Subsequently, Sattarzadeh et al. (2006) reported the development of the PCR marker “HC” to tag the same resistance QTL from S. vernei. Rather than developing markers based on single polymorphic sites linked to the QTL, the authors performed “haplotype” analysis on a set of seven closely linked SNPs, all located at the QTL region spanning over several hundred kilobases (kb) and each showed significant associations with resistance phenotypes. Of the three haplotypes identified, haplotype “c” stood out as the one associated in coupling with the resistance. Two SNPs that were specific to the haplotype “c” were targeted to develop the PCR marker HC. Tests of the HC marker on 56 potato varieties beyond the experimental population indicated that HC was highly diagnostic for the presence of the S. vernei-derived resistance to G. pallida pathotype Pa2/3. The HC marker, since its development, has been used widely to screen for resistance (Sharma et al., 2014; Bhardwaj et al., 2019; Sudha et al., 2019). It generally shows good agreement with phenotypic tests although discrepancies occasionally occur. Schwarzfischer et al. (2010) showed that HC is an appropriate diagnostic tool to identify resistance in diverse German potato varieties. It has also been discovered that many European breeding lines contain the HC marker, indicating that this marker has been widely used in resistance breeding (Sattarzadeh et al., 2006).
For the “H3” source, the first marker to be used in breeding was the microsatellite STM3016, for which a particular allele (STM3016-122/177) was diagnostic for the GpaIV gene on chromosome IV (Moloney et al., 2010). Further work, based on the targeted resequencing of bacterial-artificial chromosome (BAC) clones from the region, led to the development of the PCR marker contig237, as well as three others (Moloney et al., 2010), which were highly predictive for GpaIV in a breeding population.
Other resistance sources to PCN targeted for marker development, are the gene Gro1–4, for which a sequence characterised amplified region (SCAR) marker was developed following efforts to isolate the gene (Paal et al., 2004). Additional primer pairs were developed (Asano et al., 2012) but neither has been reliable for screening purposes. A further source of resistance to G. rostochiensis is the gene GroVI, identified in S. vernei (Jacobs et al., 1996) and mapped to chromosome V at the same region as H1. Its allelic relation to H1 remains unclear. Two reported SCAR markers were developed by converting two RAPD markers, U14 and X02 that flank GroVI, which generate amplicons of 366 and 854 bp, respectively. The two markers mapped 19 cM distal and 4 cM proximal to GroVI (Jacobs et al., 1996). Milczarek et al. (2011) used the two markers to screen a collection of 72 cultivars for resistances and generated the expected bands but found them to be undiagnostic as they were observed in both resistant and susceptible cultivars. Like Gro1–4, the GroVI resistance has not been deployed as widely as H1, probably due to overlapping with H1 in its resistance specificity to the most common pathotype Ro1.
The majority of described resistance genes or QTLs for PCN are located within resistance “gene clusters” within the potato genome. Almost half of the described QTLs from different potato wild species map to the same region on chromosome V, where resistances for other potato pathogens are also described. This region, defined by RFLP markers GP21 and GP179 is called a “hotspot” for resistances (Gebhardt and Valkonen, 2001). Therefore, some of these resistances might share a common origin and possibly developed in ancestors and later evolved by diversification in wild potato species (Finkers-Tomczak et al., 2009). The resistances Grp1 and GpaV have been mapped to the same chromosomal region, raising the possibility that they might be conferred by the same genetic factors (Finkers-Tomczak et al., 2009). Figure 5 illustrates these findings, showing the alignment and clustering of known nematode R-gene loci to higher numbers of predicted NB-LRRs, or hotspots of resistance, in the doubled monoploid S. tuberosum group Phureja clone DM 1-3 516 R44 (DM), which was developed as the first potato reference genome.
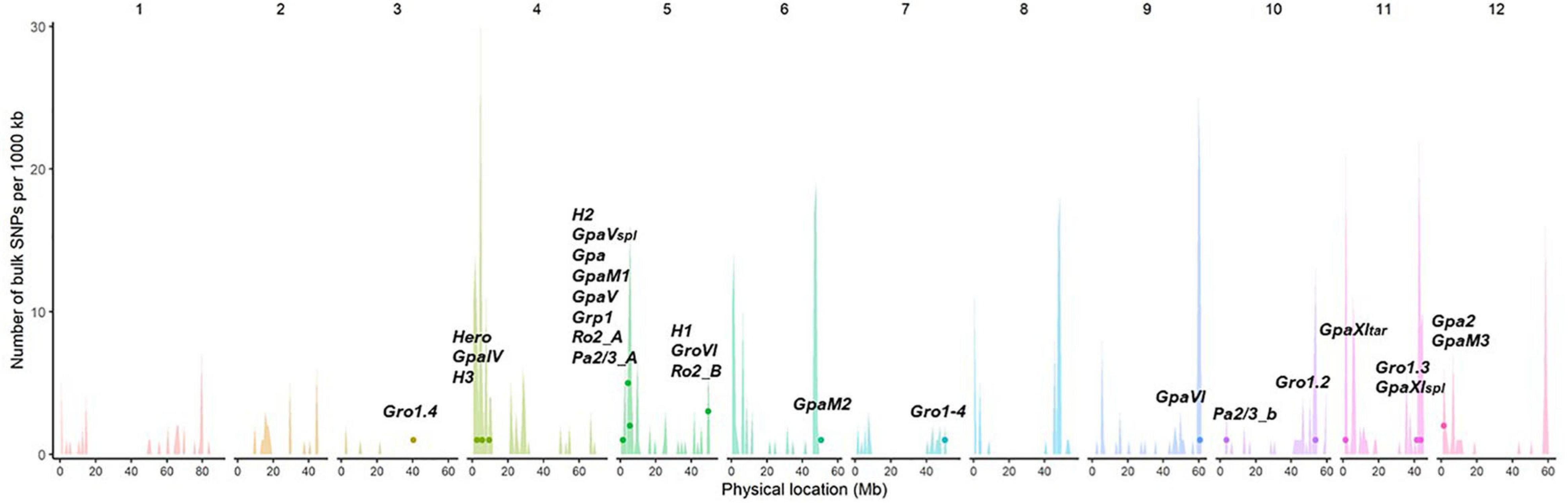
Figure 5. Potato cyst nematodes (PCN) resistance loci align with known R gene clusters. The unequal distribution and position of predicted NB-LRR genes in the genome of DM is illustrated in the histogram, with spikes indicating clusters of resistance. The approximate relative location in the reference genome DM (PGSC v4.03) of known nematode resistance genes and QTLs are displayed as dots. The histogram spikes indicate numbers of predicted NB-LRR genes per 1,000 kb across the potato genome.
Potato Genomics Tools Applied to the Analysis of PCN Resistance
The first nematode R gene, Hs1pro–1, was identified in sugar beet against the beet cyst nematode, Heterodera schachtii, in 1997 and relied on analysing chromosomal breakage points alongside a “satellite” marker (Cai et al., 1997). However, subsequent genetic analysis showed that resistance does not co-segregate with the presence of this gene (Sandal et al., 1997). Other plant resistances were studied with the aid of bulked segregant analysis (BSA) using molecular markers such as RFLPs; cleaved amplified polymorphic sequences (CAPS) and AFLPs (Michelmore et al., 1991). In 1998, the tomato gene Mi-1 was cloned, which provides resistance to both root knot nematodes (Meloidogyne incognita, M. javanica, M. arenaria) and several phloem-feeding insects, through the use of yeast-artificial chromosome libraries (Vos et al., 1998). Similarly, Gpa2 from potato was mapped to a 115kb region linked to a sequence-related NB-LRR gene, which provides resistance to potato virus X (Rouppe van der Voort et al., 2000; van der Vossen et al., 2000). Gro1–4 was isolated in a similar way (Paal et al., 2004). The identification of the Hero locus from tomato, although initially defined with RFLP markers in a segregating population, was more complex than Gpa2 or Gro1–4 (Ganal et al., 1995). A total of 14 potential candidate NB-LRRs were identified by sequencing 19 cosmids spanning the 180kb locus. Based on cosmid sequence information, these gene candidates were narrowed down by analysis of open reading frames, and the use of specific PCR markers. This allowed the functional analysis of just three potential candidate genes to identify the Hero gene conferring broad spectrum resistance to G. rostochiensis and some G. pallida pathotypes (Ernst et al., 2002). For other resistance genes, a map-based BSA approach has proven to be unsuitable. The H1 gene for example, mapped to the distal end of chromosome V that contains numerous NB-LRR homologues, has very limited recombination and a lack of co-linearity between different haplotypes (Bakker et al., 2004; Finkers-Tomczak et al., 2011). While this gene has not yet been isolated, despite great efforts to do so, newer approaches and technologies may allow this and other genes to be isolated from such complex loci.
The three PCN resistance genes cloned so far belong to the NB-LRR family of R-genes. Gro1–4 encodes an NB-LRR protein with a Toll-interleukin receptor domain at the N-terminus (Paal et al., 2004), while Hero (Ernst et al., 2002) and Gpa2 (van der Vossen et al., 2000) encode CC-NB-LRR proteins, which have a coiled-coil domain at the N-terminus. Gro1–4 and Gpa2 originate from S. spegazzinii and S. tuberosum ssp. andigena, respectively, while the Hero resistance gene is derived from the wild tomato species S. pimpinellifolium. None of the other important PCN resistances have yet been isolated, despite significant efforts to do so. Nevertheless, mapping work has led to the development of several genetic markers for resistance that are used in potato breeding programmes, as described above.
With the availability of a reference potato genome (The Potato Genome Sequencing Consortium, 2011) followed by rapid advancements and cost reductions in next-generation sequencing (NGS), modern genotyping methods, for example genotyping-by-sequencing (GBS) (Elshire et al., 2011) with novel genetic approaches, such as genome-wide association studies (GWAS) (Visscher et al., 2012) for QTL discovery and marker-trait analyses, have emerged and have been applied to potato (e.g., Sharma et al., 2018). These approaches provide much higher genetic resolution than the previously used “low density” approaches. They also allow for high throughput analyses and larger populations to be rapidly screened (Deschamps et al., 2012). The release of the potato reference genome aimed to provide a resource for identifying important genes (Sharma et al., 2013) including NB-LRRs. Although this reference has been invaluable in genetic studies, it only offers an incomplete picture, particularly in highly repetitive regions or in the case of NB-LRR genes where the reference is from a largely susceptible background. Thus, more recently, potato genomes have been sequenced from the homozygous diploid inbred potato clone M6 of S. chacoense, which is used as a breeding line and in genetics analysis (Jansky et al., 2014). A heterozygous clone, RH, has also solved haplotype blocks with third generation sequencing technology (Leisner et al., 2018; Zhou et al., 2020). These additional genomes will allow a greater resolution and understanding of NB-LRR loci and aid in the isolation and cloning of target genes.
An alternative to conventional “genome-wide” mapping approaches that overcomes some of the limitations of the potato reference genomes is provided by targeted enrichment sequencing. This process reduces the genome complexity to the most likely candidates with the help of specific “baits” designed to a subset of all potato genes. For potato, the following enrichment strategies have been adopted to date and comprise whole exome capture (WEC), generic-mapping enrichment sequencing (GenSeq) which targets single copy, conserved genes dispersed throughout the potato genome, two defence gene related capture approaches, resistance gene enrichment sequencing (RenSeq), and receptor-like protein enrichment sequencing (Jupe et al., 2013; Giolai et al., 2016; Chen et al., 2018; Lin et al., 2020). RenSeq was developed to allow high resolution SNP markers to be obtained for the detection and mapping of NB-LRRs (Jupe et al., 2013). RenSeq has also proven important to identify full length candidate NB-LRRs in combination with long-read sequencing (Witek et al., 2016). Furthermore, when used as a diagnostic tool (dRenSeq), the application informs resistance breeders, providing a full and detailed picture of the known NB-LRRs present in wild species and parental material (Van Weymers et al., 2016; Armstrong et al., 2019).
The most recent approach to isolate and map resistance genes is independent of a reference genome and takes advantage of K-mers in conjunction with association studies. K-mers are subreads of a length = K and, unlike SNPs, can inform on small haplotypes. The genetic mapping processes of K-mers are similar to GWAS by analysing diverse accessions for association between sequence polymorphisms and traits, but this can be conducted independently of a reference genome. K-mer studies based on RenSeq derived reads are referred to as AgRenSeq and have been used, for example, to isolate four resistance genes in wheat (Arora et al., 2019). Other, non-RenSeq-based K-mer approaches have also recently been used to map resistance against wart disease in potatoes (Prodhomme et al., 2019).
As sequencing methods continue to improve, so will the outputs for the de novo assembly of genomes or loci of interest, particularly in highly repetitive regions and for the many potato species with high levels of heterozygosity. Indeed, newly emerging and rapidly improving long-read sequencing technologies such as Oxford Nanopore and PacBio, are already starting to provide potential solutions for potato genomics and new potato genomes are being released (van Lieshout et al., 2020; Zhou et al., 2020). With these technological advances and the computational tools that facilitate ever more complex analyses, mapping and cloning of resistance genes should become more routine in the future.
Discussion
While host resistance deployment remains a preferred strategy to control pests and pathogens of potato, introgression of resistance genes from wild and landrace species into commercially successful cultivars remains a considerable challenge. On the one hand, adaptation of pests and pathogens to overcome resistance can negate all of the efforts of breeders. On the other hand, meeting commercial requirements in which resistance is only one of many characters the breeder must deliver, can also negate their efforts. Education, incentives and interventions may all be required in order for PCN resistant cultivars to be more widely grown by the potato industry and for consumer acceptance. More options in cultivars are urgently needed that meet different requirements of the potato markets. Rapid advances in genomics improving transferability of markers for specific traits, such as PCN resistance, but also for a wide range of other agronomically important characters, should revolutionise potato breeding in the future.
One reason for the relative lack of success in breeding for G. pallida resistance is that efforts have tended to focus on the larger QTL and neglect the smaller effect loci (chromosomes IX and XI for S. vernei and H3, respectively). Caromel et al. (2005) demonstrated the potential of MAS for introgression of G. pallida resistance genes derived from S. sparsipilum in a potato breeding programme by “stacking” two separate QTLs into individual genotypes. This resulted in a more-robust resistance phenotype than was obtained by the presence of a single QTL. This study and those of Dalton et al. (2013) and Rigney et al. (2017) underpin the basis for current efforts to “pyramid” resistance genes and QTLs into the same genetic background. Combined with the implementation of genomic selection (GS), which takes advantage of the availability of genome-wide high density markers to estimate the breeding value and the analysis of more complex traits, molecular markers are gaining significant traction in the breeding of potatoes (Slater et al., 2016).
Resistance genes that act to protect potatoes from PCN are a precious commodity. There are limited numbers of resistance genes that are amenable to introgression that will likely make a significant impact in the foreseeable future for the management of G. pallida. While there is an urgent need for new cultivars with G. pallida resistance that meet commercial requirements, more effort will be needed to ensure that resistance remains effective and durable.
Author Contributions
UG and VB coordinated the manuscript. All authors listed have made direct intellectual contributions to the article and its underpinning research and have approved it for publication.
Funding
This work was funded by the Scottish Government Rural and Environment Science and Analytical Services Division and the USDA NIFA 2015-69004-23634 project GLOBAL.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
- ^ http://www.nemadecide.com
- ^ https://pcncalculator.ahdb.org.uk
- ^ https://www.aphis.usda.gov/aphis/ourfocus/planthealth/plant-pest-and-disease-programs/pests-and-diseases/nematode/pcn/pcn-home
References
Ali, M. A., Azeem, F., Li, H., and Bohlmann, H. (2017). Smart parasitic nematodes use multifaceted strategies to parasitize plants. Front. Plant Sci. 8:1699. doi: 10.3389/fpls.2017.01699
Armstrong, M. R., Vossen, J., Lim, T. Y., Hutten, R. C. B., Xu, J. F., Strachan, S. M., et al. (2019). Tracking disease resistance deployment in potato breeding by enrichment sequencing. Plant Biotechnol. J. 17, 540–549. doi: 10.1111/pbi.12997
Arntzen, F. K., and Wouters, T. C. A. E. (1994). Assessing the tolerance to Globodera pallida of resistant potato genotypes by means of field and pot tests. Potato Res. 37, 51–63. doi: 10.1007/BF02360432
Arora, S., Steuernagel, B., Gaurav, K., Chandramohan, S., Long, Y. M., Matny, O., et al. (2019). Resistance gene cloning from a wild crop relative by sequence capture and association genetics. Nat. Biotechnol. 37, 139–143. doi: 10.1038/s41587-018-0007-9
Asano, K., Kobayashi, A., Tsuda, S., Nishinaka, M., and Tamiya, S. (2012). DNA marker-assisted evaluation of potato genotypes for potential resistance to potato cyst nematode pathotypes not yet invading into Japan. Breed. Sci. 62, 142–150. doi: 10.1270/jsbbs.62.142
Back, M. A., Cortada, L., Grove, I. G., and Taylor, V. (2018). “Field management and control strategies,” in Cyst Nematodes, eds R. N. Perry, M. Moens, and J. T. Jones (Wallingford: CAB International), 305–336. doi: 10.1079/9781786390837.0305
Bakker, E., Achenbach, U., Bakker, J., van Vliet, J., Peleman, J., Segers, B., et al. (2004). A high-resolution map of the H1 locus harbouring resistance to the potato cyst nematode Globodera rostochiensis. Theor. Appl. Genet. 109, 146–152. doi: 10.1007/s00122-004-1606-z
Bayless, A. M., Smith, J. M., Song, J., McMinn, P. H., Teillet, A., August, B. K., et al. (2016). Disease resistance through impairment of alpha-SNAP-NSF interaction and vesicular trafficking by soybean Rhg1. Proc. Natl. Acad. Sci. U.S.A. 113, E7375–E7382. doi: 10.1073/pnas.1610150113
Been, T. H., and Schomaker, C. H. (1998). Quantitative Studies on the Management of Potato cyst Nematodes (Globodera spp.) in the Netherlands. Ph.D. Thesis, University Wageningen, Wageningen.
Bendezu, I. F., Russell, M. D., and Evans, K. (1998). Virulence of populations of potato cyst nematodes (Globodera spp.) from Europe and Bolivia towards differential potato clones frequently used for pathotype classification. Nematologica 44, 667–681. doi: 10.1163/005725998x00078
Beniers, J. E., Nollen, Y., van Eck, H. J., and Schouten, H. J. (2019). Increased virulence of Globodera pallida during repeated rearing on different resistant potato cultivars explained by a simple model. Plant Pathol. 68, 445–453. doi: 10.1111/ppa.12969
Bethke, P. C., Halterman, D. A., and Jansky, S. (2017). Are we getting better at using wild potato species in light of new tools? Crop Sci. 57, 1241–1258. doi: 10.2135/cropsci2016.10.0889
Bhardwaj, V., Salej, S., Ashwani, K., Vanishree, G., Sanjeev, S., Sundaresha, S., et al. (2019). Efficiency and reliability of marker assisted selection for resistance to major biotic stresses in potato. Potato J. 46, 56–66.
Birch, P. R. J., Bryan, G., Fenton, B., Gilroy, E. M., Hein, I., Jones, J. T., et al. (2012). Crops that feed the world 8: potato: are the trends of increased global production sustainable? Food Secur. 4, 477–508. doi: 10.1007/s12571-012-0220-1
Biryukova, V. A., Zhuravlev, A. A., Abrosimova, S. B., Kostina, L. I., Khromova, L. M., Shmyglya, I. V., et al. (2008). Use of molecular markers of potato golden nematode resistance genes H1 and GRO1. Russ. Agric. Sci. 34, 365–368. doi: 10.3103/s1068367408060013
Blok, V., Tylka, G., Smiley, R., deJong, W., and Taub, M. (2018). “Resistance breeding,” in Cyst Nematodes, eds R. N. Perry, M. Moens, and J. T. Jones (Wallingford: CAB International), 174–214.
Bradshaw, J., Dale, M., and Phillips, M. (1996). Breeding potatoes at SCRI for resistance to potato cyst nematodes. SCRI, Annu. Rep. 1995, 30–34.
Bradshaw, J. E., Meyer, R. C., Milbourne, D., McNicol, J. W., Phillips, M. S., and Waugh, R. (1998). Identification of AFLP and SSR markers associated with quantitative resistance to Globodera pallida (Stone) in tetraploid potato (Solanum tuberosum subsp. tuberosum) with a view to marker-assisted selection. Theor. Appl. Genet. 97, 202–210. doi: 10.1007/s001220050886
Bryan, G. J., McLean, K., Bradshaw, J. E., De Jong, W. S., Phillips, M., Castelli, L., et al. (2002). Mapping QTLs for resistance to the cyst nematode Globodera pallida derived from the wild potato species Solanum vernei. Theor. Appl. Genet. 105, 68–77. doi: 10.1007/s00122-002-0873-9
Bryan, G. J., McLean, K., Pande, B., Purvis, A., Hackett, C. A., Bradshaw, J. E., et al. (2004). Genetical dissection of H3-mediated polygenic PCN resistance in a heterozygous autotetraploid potato population. Mol. Breed. 14, 105–116. doi: 10.1023/b:molb.0000037999.13581.9c
Butler, K. J., Chen, S., Smith, J. M., Wang, X., and Bent, A. F. (2019). Soybean resistance locus Rhg1 confers resistance to multiple cyst nematodes in diverse plant species. Phytopathology 109, 2107–2115. doi: 10.1094/phyto-07-19-0225-r
CABI (2020a). Invasive Species Compendium Datasheet Globodera Pallida. Available Online at: https://www.cabi.org/isc/datasheet/27033 (accessed September 10, 2020).
CABI (2020b). Invasive species compendium datasheet Globodera rostochiensis. Available Online at: https://www.cabi.org/isc/datasheet/27034 (accessed September 10, 2020).
Cai, D. G., Kleine, M., Kifle, S., Harloff, H. J., Sandal, N. N., Marcker, K. A., et al. (1997). Positional cloning of a gene for nematode resistance in sugar beet. Science 275, 832–834. doi: 10.1126/science.275.5301.832
Canto Saenz, M., and Mayer de Scurrah, M. (1977). Races of the potato cyst nematode in the Andean region and a new system of classification. Nematologica 23, 340–349. doi: 10.1163/187529277x00066
Caromel, B., Mugniery, D., Kerlan, M. C., Andrzejewski, S., Palloix, A., Ellisseche, D., et al. (2005). Resistance quantitative trait loci originating from Solanum sparsipilum act independently on the sex ratio of Globodera pallida and together for developing a necrotic reaction. Mol. Plant Microbe Interact. 18, 1186–1194. doi: 10.1094/mpmi-18-1186
Caromel, B., Mugniery, D., Lefebvre, V., Andrzejewski, S., Ellisseche, D., Kerlan, M. C., et al. (2003). Mapping QTLs for resistance against Globodera pallida (Stone) Pa2/3 in a diploid potato progeny originating from Solanum spegazzinii. Theor. Appl. Genet. 106, 1517–1523. doi: 10.1007/s00122-003-1211-6
Castelli, L., Ramsay, G., Bryan, G., Neilson, S. J., and Phillips, M. S. (2003). New sources of resistance to the potato cyst nematodes Globodera pallida and G. rostochiensis in the Commonwealth Potato Collection. Euphytica 129, 377–386. doi: 10.1023/A:1022264111096
Chen, X. W., Lewandowska, D., Armstrong, M. R., Baker, K., Lim, T. Y., Bayer, M., et al. (2018). Identification and rapid mapping of a gene conferring broad-spectrum late blight resistance in the diploid potato species Solanum verrucosum through DNA capture technologies. Theor. Appl. Genet. 131, 1287–1297. doi: 10.1007/s00122-018-3078-6
CIP (2020). Potato facts and figures. Available online at: https://cipotato.org/potato/potato-facts-and-figures/ (accessed september 10, 2020).
Clarke, A. J., and Perry, R. N. (1985). Eggshell calcium and the hatching of Globodera rostochiensis. Int. J. Parasitol. 15, 511–516. doi: 10.1016/0020-7519(85)90046-3
Collins, S., Vanstone, V., and Zhang, X. (2010). PCN “Area Freedom” for WA: Evaluation of the Current Status of Potato Cyst Nematode (Globodera rostochiensis) in Western Australia”. Final Report for Project PT04004 and MT04000. Sydney, NSW: Horticulture Australia Limited.
Cook, D. E., Lee, T. G., Guo, X., Melito, S., Wang, K., Bayless, A. M., et al. (2012). Copy number variation of multiple genes at Rhg1 mediates nematode resistance in soybean. Science 338, 1206–1209. doi: 10.1126/science.1228746
Cook, R., and Evans, K. (1987). “Resistance and tolerance,” in Principles and Practice of Nematode Control in Crops, eds R. H. Brown and B. R. Kerry (Marrickville, NSW: Academic Press), 179–231.
Dale, M. F. B., Phillips, M. S., Ayres, R. M., Hancock, M., Holliday, M., Mackay, G. R., et al. (1988). The assessment of the tolerance of partially resistant potato clones to damage by the potato cyst nematode Globodera pallida at different sites and in different years. Ann. Appl. Biol. 113, 79–88. doi: 10.1111/j.1744-7348.1988.tb03284.x
Dalton, E., Griffin, D., Gallagher, T. F., de Vetten, N., and Milbourne, D. (2013). The effect of pyramiding two potato cyst nematode resistance loci to Globodera pallida Pa2/3 in potato. Mol. Breed. 31, 921–930. doi: 10.1007/s11032-013-9845-9
Davies, L. J., and Elling, A. A. (2015). Resistance genes against plant-parasitic nematodes: a durable control strategy? Nematology 17, 249–263. doi: 10.1163/15685411-00002877
Deschamps, S., Llaca, V., and May, G. D. (2012). Genotyping-by-sequencing in plants. Biology 1, 460–483. doi: 10.3390/biology1030460
Djebroune, A., Chakali, G., de Andrade, E., Camacho, M. J., Rusinque, L., and Inácio, M. L. (2021). Integrative morphometric and molecular approach to update the impact and distribution of potato cyst nematodes Globodera rostochiensis and Globodera pallida (Tylenchida: Heteroderidae) in Algeria. Pathogens 10:216. doi: 10.3390/pathogens10020216
Doncaster, C. C., and Seymour, M. K. (1973). Exploration and selection of penetration sites by tylenchida. Nematologica 19, 137–145. doi: 10.1163/187529273x00277
Douches, D. S., Coombs, J., Felcher, K., Kirk, W. W., Long, C., and Bird, G. (2010). Missaukee: A round white potato variety combining chip-processing with resistance to late blight, verticillium wilt and golden cyst nematode. Am. J. Potato Res. 87, 10–18. doi: 10.1007/s12230-009-9111-1
Ellenby, C. (1952). Resistance to the potato root eelworm. Heterodera rostochiensis Wollenweber. Nature 170:1016. doi: 10.1038/1701016a0
Elshire, R. J., Glaubitz, J. C., Sun, Q., Poland, J. A., Kawamoto, K., Buckler, E. S., et al. (2011). A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS One 6:e19379. doi: 10.1371/journal.pone.0019379
EPPO (2006). Testing of potato varieties to assess resistance to Globodera rostochiensis and Globodera pallida. Bull. OEPP 36, 419–420. doi: 10.1111/j.1365-2338.2006.01032.x
Ernst, K., Kumar, A., Kriseleit, D., Kloos, D. U., Phillips, M. S., and Ganal, M. W. (2002). The broad-spectrum potato cyst nematode resistance gene (Hero) from tomato is the only member of a large gene family of NBS-LRR genes with an unusual amino acid repeat in the LRR region. Plant J. 31, 127–136. doi: 10.1046/j.1365-313X.2002.01341.x
Evans, K., Franco, J., and Descurrah, M. M. (1975). Distribution of species of potato cyst nematodes in South America. Nematologica 21, 365–369. doi: 10.1163/187529275x00103
Evans, K., and Haydock, P. P. J. (1990). A review of tolerance by potato plants of cyst nematode attack, with consideration of what factors may confer tolerancce and methods of assaying and improving it in crops. Ann. Appl. Biol. 117, 703–740. doi: 10.1111/j.1744-7348.1990.tb04836.x
Eves-van den Akker, S., Lilley, C. J., Jones, J. T., and Urwin, P. E. (2015a). Plant-parasitic nematode feeding tubes and plugs: new perspectives on function. Nematology 17, 1–9. doi: 10.1163/15685411-00002832
Eves-van den Akker, S., Lilley, C. J., Reid, A., Pickup, J., Anderson, E., Cock, P. J. A., et al. (2015b). A metagenetic approach to determine the diversity and distribution of cyst nematodes at the level of the country, the field and the individual. Mol. Ecol. 24, 5842–5851. doi: 10.1111/mec.13434
Fenwick, D. W. (1940). Methods for the recovery and counting of cysts of Heterodera schachtii from soil. J. Helminthol. 18, 155–172. doi: 10.1017/S0022149X00031485
Finkers-Tomczak, A., Bakker, E., de Boer, J., van der Vossen, E., Achenbach, U., Golas, T., et al. (2011). Comparative sequence analysis of the potato cyst nematode resistance locus H1 reveals a major lack of co-linearity between three haplotypes in potato (Solanum tuberosum ssp.). Theor. Appl. Genet. 122, 595–608. doi: 10.1007/s00122-010-1472-9
Finkers-Tomczak, A., Danan, S., van Dijk, T., Beyene, A., Bouwman, L., Overmars, H., et al. (2009). A high-resolution map of the Grp1 locus on chromosome V of potato harbouring broad-spectrum resistance to the cyst nematode species Globodera pallida and Globodera rostochiensis. Theor. Appl. Genet. 119, 165–173. doi: 10.1007/s00122-009-1026-1
Flor, H. H. (1971). Current status of gene-for-gene concept. Ann. Rev. Phytopathol. 9, 275–296. doi: 10.1146/annurev.py.09.090171.001423
Fournet, S., Eoche-Bosy, D., Renault, L., Hamelin, F. M., and Montarry, J. (2016). Adaptation to resistant hosts increases fitness on susceptible hosts in the plant parasitic nematode Globodera pallida. Ecol. Evol. 6, 2559–2568. doi: 10.1002/ece3.2079
Ganal, M. W., Simon, R., Brommonschenkel, S., Arndt, M., Phillips, M. S., Tanksley, S. D., et al. (1995). Genetic mapping of a wide spectrum nematode resistance gene (Hero) against Globodera rostochiensis in tomato. Mol. Plant Microbe Interact. 8, 886–891. doi: 10.1094/mpmi-8-0886
Gebhardt, C., Mugniery, D., Ritter, E., Salamini, F., and Bonnel, E. (1993). Identification of RFLP markers closely linked to the H1 gene conferring resistance to Globodera rostochiensis in potato. Theor. Appl. Genet. 85, 541–544. doi: 10.1007/bf00220911
Gebhardt, C., and Valkonen, J. P. T. (2001). Organization of genes controlling disease resistance in the potato genome. Annu. Rev. Phytopathol. 39, 79–102. doi: 10.1146/annurev.phyto.39.1.79
Gheysen, G., and Mitchum, M. G. (2019). Phytoparasitic nematode control of plant hormone pathways. Plant Physiol. 179, 1212–1226. doi: 10.1104/pp.18.01067
Giolai, M., Paajanen, P., Verweij, W., Percival-Alwyn, L., Baker, D., Witek, K., et al. (2016). Targeted capture and sequencing of gene-sized DNA molecules. Biotechniques 61, 315–322. doi: 10.2144/000114484
Golinowski, W., Grundler, F. M. W., and Sobczak, M. (1996). Changes in the structure of Arabidopsis thaliana during female development of the plant-parasitic nematode Heterodera schachtii. Protoplasma 194, 103–116. doi: 10.1007/bf01273172
Goverse, A., Overmars, H., Engelbertink, J., Schots, A., Bakker, J., and Helder, J. (2000). Both induction and morphogenesis of cyst nematode feeding cells are mediated by auxin. Mol. Plant Microbe Interact. 13, 1121–1129. doi: 10.1094/mpmi.2000.13.10.1121
Grenier, E., Fournet, S., Petit, E., and Anthoine, G. (2010). A cyst nematode ‘species factory’ called the andes. Nematology 12, 163–169. doi: 10.1163/138855409x12573393054942
Grenier, E., Kiewnick, S., Smant, G., Fournet, S., Montarry, J., Holterman, M., et al. (2020). Monitoring and tackling genetic selection in the potato cyst nematode Globodera pallida. EFSA Supp. Publ 17:1874E. doi: 10.2903/sp.efsa.2020.EN-1874
Grundler, F. M. W., Sobczak, M., and Golinowski, W. (1998). Formation of wall openings in root cells of Arabidopsis thaliana following infection by the plant-parasitic nematode Heterodera schachtii. Eur. J. Plant Pathol. 104, 545–551. doi: 10.1023/a:1008692022279
Grunewald, W., Cannoot, B., Friml, J., and Gheysen, G. (2009). Parasitic nematodes modulate pin-mediated auxin transport to facilitate infection. PLos Pathog. 5:e1000266. doi: 10.1371/journal.ppat.1000266
Handoo, Z. A., Carta, L. K., Skantar, A. M., and Chitwood, D. J. (2012). Description of Globodera ellingtonae n. sp. (Nematoda: Heteroderidae) from oregon. J. Nematol. 44, 40–57.
Hockland, S., Niere, B., Grenier, E., Blok, V., Phillips, M., Den Nijs, L., et al. (2012). An evaluation of the implications of virulence in non-European populations of Globodera pallida and G. rostochiensis for potato cultivation in Europe. Nematology 14, 1–13. doi: 10.1163/138855411x587112
Hodda, M., and Cook, D. C. (2009). Economic impact from unrestricted spread of potato cyst nematodes in Australia. Phytopathology 99, 1387–1393. doi: 10.1094/phyto-99-12-1387
Hussey, R. S., and Mims, C. W. (1990). Ultrastructure of esophageal glands and their secretory granules in the root-knot nematode Meloidogyne incognita. Protoplasma 156, 9–18. doi: 10.1007/bf01666501
IPPC (2010). Eradication of Potato Cyst Nematode (PCN) from Western Australia. Available Online at: https://www.ippc.int/en/countries/Australia/pestreports/2010/09/eradication-of-potato-cyst-nematode-pcn-from-western-australia/ (accessed September 10, 2020)
Jacobs, J. M. E., van Eck, H. J., Horsman, K., Arens, P. F. P., Verkerk-Bakker, B., Jacobsen, E., et al. (1996). Mapping of resistance to the potato cyst nematode Globodera rostochiensis from the wild potato species Solanum vernei. Mol. Breed. 2, 51–60. doi: 10.1007/bf00171351
Jansky, S. H., Chung, Y. S., and Kittipadukal, P. (2014). M6: a diploid potato inbred line for use in breeding and genetics research. J. Plant Regist. 8, 195–199. doi: 10.3198/jpr2013.05.0024crg
Jupe, F., Witek, K., Verweij, W., Sliwka, J., Pritchard, L., Etherington, G. J., et al. (2013). Resistance gene enrichment sequencing (RenSeq) enables reannotation of the NB-LRR gene family from sequenced plant genomes and rapid mapping of resistance loci in segregating populations. Plant J. 76, 530–544. doi: 10.1111/tpj.12307
Kikuchi, T., Eves-van den Akker, S., and Jones, J. T. (2017). Genome evolution of plant-parasitic nematodes. Annu. Rev. Phytopathol. 55, 333–354. doi: 10.1146/annurev-phyto-080516-035434
Kort, J., Ross, H., Rumpenhorst, H. J., and Stone, A. R. (1977). International scheme for identifying and classifying pathotypes of potato cyst nematodes Globodera rostochiensis and G. pallida. Nematologica 23, 333–339. doi: 10.1163/187529277x00057
Kreike, C. M., Dekoning, J. R. A., Vinke, J. H., Vanooijen, J. W., Gebhardt, C., and Stiekema, W. J. (1993). Mapping of loci involved in quantitatively inherited resistance to the potato cyst nematode Globodera rostochiensis pathotype Ro1. Theor. Appl. Genet. 87, 464–470. doi: 10.1007/bf00215092
Kreike, C. M., Dekoning, J. R. A., Vinke, J. H., Vanooijen, J. W., and Stiekema, W. J. (1994). Quantitatively-inherited resistance to Globodera pallida is dominated by one major locus in Solanum spegazzinii. Theor. Appl. Genet. 88, 764–769. doi: 10.1007/bf01253983
Kreike, C. M., Kok-Westeneng, A. A., Vinke, J. H., and Stiekema, W. J. (1996). Mapping of QTLs involved in nematode resistance, tuber yield and root development in Solanum sp. Theor. Appl. Genet. 92, 463–470. doi: 10.1007/bf00223694
Laine, A. L., and Barres, B. (2013). Epidemiological and evolutionary consequences of life-history trade-offs in pathogens. Plant Pathol. 62, 96–105. doi: 10.1111/ppa.12129
Lax, P., Dueñas, J. C. R., Franco-Ponce, J., Gardenal, C. N., Doucet, M. E., and Arntzen, J. W. (2014). Morphology and DNA sequence data reveal the presence of Globodera ellingtonae in the andean region. Contrib. Zool. 83:227. doi: 10.1163/18759866-08304002
Leisner, C. P., Hamilton, J. P., Crisovan, E., Manrique-Carpintero, N. C., Marand, A. P., Newton, L., et al. (2018). Genome sequence of M6, a diploid inbred clone of the high-glycoalkaloid-producing tuber-bearing potato species Solanum chacoense, reveals residual heterozygosity. Plant J. 96, 482–482. doi: 10.1111/tpj.14075
Lin, X., Armstrong, M., Baker, K., Wouters, D., Visser, R. G. F., Wolters, P. J., et al. (2020). RLP/K enrichment sequencing; a novel method to identify receptor-like protein (RLP) and receptor-like kinase (RLK) genes. New Phytol. 227, 1264–1276. doi: 10.1111/nph.16608
Liu, S. M., Kandoth, P. K., Warren, S. D., Yeckel, G., Heinz, R., Alden, J., et al. (2012). A soybean cyst nematode resistance gene points to a new mechanism of plant resistance to pathogens. Nature 492, 256–260. doi: 10.1038/nature11651
Lozano-Torres, J. L., Wilbers, R. H. P., Gawronski, P., Boshoven, J. C., Finkers-Tomczak, A., Cordewener, J. H. G., et al. (2012). Dual disease resistance mediated by the immune receptor Cf-2 in tomato requires a common virulence target of a fungus and a nematode. Proc. Natl. Acad. Sci. U.S.A 109, 10119–10124. doi: 10.1073/pnas.1202867109
Masler, E. P., and Perry, R. N. (2018). “Hatch, survival and sensory perception,” in Cyst nematodes, eds R. N. Perry, M. Moens, and J. T. Jones (Wallingford: CAB International), 44–73. doi: 10.1079/9781786390837.0044
Mburu, H., Cortada, L., Haukeland, S., Ronno, W., Nyongesa, M., Kinyua, Z., et al. (2020). Potato cyst nematodes: a new threat to potato production in East Africa. Front. Plant Sci. 11:670. doi: 10.3389/fpls.2020.00670
Meade, F., Byrne, S., Griffin, D., Kennedy, C., Mesiti, F., and Milbourne, D. (2020). Rapid development of KASP markers for disease resistance genes using pooled whole-genome resequencing. Potato Res. 63, 57–73. doi: 10.1007/s11540-019-09428-x
Mei, Y., Thorpe, P., Guzha, A., Haegeman, A., Blok, V. C., MacKenzie, K., et al. (2015). Only a small subset of the SPRY domain gene family in Globodera pallida is likely to encode effectors, two of which suppress host defences induced by the potato resistance gene Gpa2. Nematology 17, 409–424. doi: 10.1163/15685411-00002875
Michelmore, R. W., Paran, I., and Kesseli, R. V. (1991). Identification of markers linked to disease-resistances genes by bulked segregant analysis - a rapid method to detect markers in specific genomic regions by using segregating populations. Proc. Natl. Acad. Sci. U.S.A. 88, 9828–9832. doi: 10.1073/pnas.88.21.9828
Milczarek, D., Flis, B., and Przetakiewicz, A. (2011). Suitability of molecular markers for selection of potatoes tesistant to Globodera spp. Am. J. Potato Res. 88, 245–255. doi: 10.1007/s12230-011-9189-0
Moloney, C., Griffin, D., Jones, P. W., Bryan, G. J., McLean, K., Bradshaw, J. E., et al. (2010). Development of diagnostic markers for use in breeding potatoes resistant to Globodera pallida pathotype Pa2/3 using germplasm derived from Solanum tuberosum ssp. andigena CPC 2802. Theor. Appl. Genet. 120, 679–689. doi: 10.1007/s00122-009-1185-0
Niere, B., Krüssel, S., and Osmers, K. (2014). Auftreten einer außergewöhnlich virulenten Population der Kartoffelzystennematoden. J. Kulturpflanzen 66, 426–427.
Ortega, F., and Lopez-Vizcon, C. (2012). Application of molecular marker-assisted selection (MAS) for disease resistance in a practical potato breeding programme. Potato Res. 55, 1–13. doi: 10.1007/s11540-011-9202-5
Paal, J., Henselewski, H., Muth, J., Meksem, K., Menendez, C. M., Salamini, F., et al. (2004). Molecular cloning of the potato Gro1-4 gene conferring resistance to pathotype Ro1 of the root cyst nematode Globodera rostochiensis, based on a candidate gene approach. Plant J. 38, 285–297. doi: 10.1111/j.1365-313X.2004.02047.x
Pagán, I., and García-Arenal, F. (2020). Tolerance of plants to pathogens: a unifying view. Annu. Rev. Phytopathol 58, 77–96. doi: 10.1146/annurev-phyto-010820-012749
Park, J., Hackett, C., Dandurand, L., Wang, X., and Jong, W. D. D. (2019). QTL for resistance to Globodera rostochiensis pathotype Ro2 and G. pallida pathotype Pa2/3 in autotetraploid potato. Am. J. Potato Res. 96, 552–563. doi: 10.1007/s12230-019-09745-4
Park, J., Yang, H., De Jong, W. S., and Wang, X. (2018). An evaluation of two H1-linked markers and their suitability for selecting Globodera rostochiensis resistant potatoes in the New York breeding program. Am. J. Potato Res. 95, 170–177. doi: 10.1007/s12230-017-9623-z
Perry, R. N. (2002). “Hatching,” in The Biology of Nematodes, ed. D. L. Lee (London: Taylor and Francis), 147–169.
Phillips, M. S., and Blok, V. C. (2008). Selection for reproductive ability in Globodera pallida populations in relation to quantitative resistance from Solanum vernei and S. tuberosum ssp. andigena CPC2802. Plant Pathol. 57, 573–580. doi: 10.1111/j.1365-3059.2007.01771.x
Phillips, M. S., Blok, V. C., Dale, M. F. B., and Trudgill, D. L. (1994). Virulence to plant resistance in potato cyst nematodes. Asp. Appl. Biol. 39, 139–143.
Phillips, M. S., Forrest, J. M. S., and Wilson, L. A. (1980). Screening for resistance to potato cyst nematode using closed containers. Ann. Appl. Biol. 96, 317–322. doi: 10.1111/j.1744-7348.1980.tb04782.x
Picard, D., Plantard, O., Scurrah, M., and Mugniery, D. (2004). Inbreeding and population structure of the potato cyst nematode (Globodera pallida) in its native area (Peru). Mol. Ecol. 13, 2899–2908. doi: 10.1111/j.1365-294X.2004.02275.x
Picard, D., Sempere, T., and Plantard, O. (2007). A northward colonisation of the andes by the potato cyst nematode during geological times suggests multiple host-shifts from wild to cultivated potatoes. Mol. Phylogenet. Evol. 42, 308–316. doi: 10.1016/j.ympev.2006.06.018
Pickup, J., Roberts, A. M. I, and Nijs, L. J. M. F. D. (2018). “Quarantine, distribution patterns and sampling,” in Cyst Nematodes, eds R. N. Perry, M. Moens, and J. T. Jones (Wallingford: CAB International), 128–153. doi: 10.1079/9781786390837.0128
Plantard, O., Picard, D., Valette, S., Scurrah, M., Grenier, E., and Mugniery, D. (2008). Origin and genetic diversity of Western European populations of the potato cyst nematode (Globodera pallida) inferred from mitochondrial sequences and microsatellite loci. Mol. Ecol. 17, 2208–2218. doi: 10.1111/j.1365-294X.2008.03718.x
Popeijus, H., Overmars, H., Jones, J., Blok, V., Goverse, A., Helder, J., et al. (2000). Degradation of plant cell walls by a nematode. Nature 406, 36–37. doi: 10.1038/35017641
Prodhomme, C., Esselink, D., Borm, T., Visser, R. G. F., van Eck, H. J., and Vossen, J. H. (2019). Comparative subsequence sets analysis (CoSSA) is a robust approach to identify haplotype specific SNPs; mapping and pedigree analysis of a potato wart disease resistance gene Sen3. Plant Methods 15:60. doi: 10.1186/s13007-019-0445-5
Przetakiewicz, A., and Milczarek, D. (2017). Evaluation of potato cultivars and breeding lines for resistance to Globodera rostochiensis and Globodera pallida. Plant Breed. Seed. Sci. 76, 3–8. doi: 10.1515/plass-2017-0014
Qin, L., Kudla, U., Roze, E. H. A., Goverse, A., Popeijus, H., Nieuwland, J., et al. (2004). A nematode expansin acting on plants. Nature 427, 30–30. doi: 10.1038/427030a
Ravelombola, W. S., Qin, J., Shi, A., Nice, L., Bao, Y., Lorenz, A., et al. (2020). Genome-wide association study and genomic selection for tolerance of soybean biomass to soybean cyst nematode infestation. PLoS One 15:e0235089. doi: 10.1371/journal.pone.0235089
Reid, A., Evans, F., Mulholland, V., Cole, Y., and Pickup, J. (2015). High-throughput diagnosis of potato cyst nematodes in soil samples. Methods Mol. Biol. 1302, 137–148. doi: 10.1007/978-1-4939-2620-6_11
Rice, S. L., Leadbeater, B. S. C., and Stone, A. R. (1985). Changes in cell structure in roots of resistant potatoes parasitized by potato cyst-nematodes. I. Potatoes with resistance gene H1 derived from Solanum tuberosum spp. andigena. Physiol. Plant Pathol. 27, 219–234. doi: 10.1016/0048-4059(85)90069-4
Rigney, B., Blok, V., Griffin, D., Dalton, E., and Milbourne, D. (2017). Consistent action of two partially effective loci conferring resistance to Globodera pallida Pa2/3 across multiple nematode field populations. Plant Pathol. 66, 1031–1040. doi: 10.1111/ppa.12640
Rouppe van der Voort, J., Lindeman, W., Folkertsma, R., Hutten, R., Overmars, H., van der Vossen, E., et al. (1998). A QTL for broad-spectrum resistance to cyst nematode species (Globodera spp.) maps to a resistance gene cluster in potato. Theor. Appl. Genet. 96, 654–661. doi: 10.1007/s001220050785
Rouppe van der Voort, J., van der Vossen, E., Bakker, E., Overmars, H., van Zandvoort, P., Hutten, R., et al. (2000). Two additive QTLs conferring broad-spectrum resistance in potato to Globodera pallida are localized on resistance gene clusters. Theor. Appl. Genet. 101, 1122–1130. doi: 10.1007/s001220051588
Rouppe van der Voort, J. R., Wolters, P., Folkertsma, R., Hutten, R., van Zandvoort, P., Vinke, H., et al. (1997). Mapping of the cyst nematode resistance locus Gpa2 in potato using a strategy based on comigrating AFLP markers. Theor. Appl. Genet. 95, 874–880. doi: 10.1007/s001220050638
Rousselle-Bourgeois, F., and Mugniery, D. (1995). Screening tuber-bearing Solanum spp. for resistance to Globodera rostochiensis Ro1 Woll. and G. pallida Pa2/3 stone. Potato Res. 38, 241–249. doi: 10.1007/BF02359906
Sabeh, M., Lord, E., Grenier, E., St-Arnaud, M., and Mimee, B. (2019). What determines host specificity in hyperspecialized plant parasitic nematodes? BMC Genom. 20:457. doi: 10.1186/s12864-019-5853-4
Sacco, M. A., Koropacka, K., Grenier, E., Jaubert, M. J., Blanchard, A., Goverse, A., et al. (2009). The cyst nematode SPRYSEC Protein RBP-1 elicits Gpa2-and RanGAP2-dependent plant cell death. PLoS Pathog. 5:14. doi: 10.1371/journal.ppat.1000564
Sandal, N. N., Salentijn, E. M. J., Kleine, M., Cai, D. G., Arens-de Reuver, M., Van Druten, M., et al. (1997). Backcrossing of nematode-resistant sugar beet: a second nematode resistance gene at the locus containing Hs1(pro-1)? Mol. Breed. 3, 471–480. doi: 10.1023/a:1009697119097
Sattarzadeh, A., Achenbach, U., Lubeck, J., Strahwald, J., Tacke, E., Hofferbert, H. R., et al. (2006). Single nucleotide polymorphism (SNP) genotyping as basis for developing a PCR-based marker highly diagnostic for potato varieties with high resistance to Globodera pallida pathotype Pa2/3. Mol. Breed. 18, 301–312. doi: 10.1007/s11032-006-9026-1
Schultz, L., Cogan, N. O. I., McLean, K., Dale, M. F. B., Bryan, G. J., Forster, J. W., et al. (2012). Evaluation and implementation of a potential diagnostic molecular marker for H1-conferred potato cyst nematode resistance in potato (Solanum tuberosum L.). Plant Breed. 131, 315–321. doi: 10.1111/j.1439-0523.2012.01949.x
Schwarzfischer, A., Behn, A., Groth, J., Reichmann, M., Kellermann, A., and Songe, Y. (2010). “Marker-assisted selection in practical potato breeding - experience and outlook,” in Proceedings of the 60. Jahrestagung der Vereinigung der Pflanzenzuchter Und Saatgutkaufleute Osterreichs, 24-26 November 2009, (Irdning: Raumberg-Gumpenstein), 81–85.
Seinhorst, J. W. (1982). The relationship in field experiments between population density of Globodera rostochiensis before planting potatoes and yield of potato tubers. Nematologica 28, 277–284. doi: 10.1163/187529282X00312
Sharma, R., Bhardwaj, V., Dalamu, D., Kaushik, S. K., Singh, B. P., Sharma, S., et al. (2014). Identification of elite potato genotypes possessing multiple disease resistance genes through molecular approaches. Sci. Hortic. 179, 204–211. doi: 10.1016/j.scienta.2014.09.018
Sharma, S. K., Bolser, D., de Boer, J., Sonderkaer, M., Amoros, W., Carboni, M. F., et al. (2013). Construction of reference chromosome-scale pseudomolecules for potato: integrating the potato genome with genetic and physical maps. G3 (Bethesda) 3, 2031–2047. doi: 10.1534/g3.113.007153
Sharma, S. K., MacKenzie, K., McLean, K., Dale, F., Daniels, S., and Bryan, G. J. (2018). Linkage disequilibrium and evaluation of genome-wide association mapping models in tetraploid potato. G3 (Bethesda) 8, 3185–3202. doi: 10.1534/g3.118.200377
Skantar, A., Handoo, Z., Carta, L., and Chitwood, D. (2007). Morphological and molecular identification of Globodera pallida associated with potato in Idaho. J. Nematol. 39, 133–144.
Skantar, A. M., Handoo, Z. A., Zasada, I. A., Ingham, R. E., Carta, L. K., and Chitwood, D. J. (2011). Morphological and molecular characterization of Globodera populations from Oregon and Idaho. Phytopathology 101, 480–491. doi: 10.1094/phyto-01-10-0010
Slater, A. T., Cogan, N. O. I., Forster, J. W., Hayes, B. J., and Daetwyler, H. D. (2016). Improving genetic gain with genomic selection in autotetraploid potato. Plant Genome 9:15. doi: 10.3835/plantgenome2016.02.0021
Smant, G., Stokkermans, J., Yan, Y. T., de Boer, J. M., Baum, T. J., Wang, X. H., et al. (1998). Endogenous cellulases in animals: isolation of beta-1,4-endoglucanase genes from two species of plant-parasitic cyst nematodes. Proc. Natl. Acad. Sci. U.S.A 95, 4906–4911. doi: 10.1073/pnas.95.9.4906
Sobczak, M., Avrova, A., Jupowicz, J., Phillips, M. S., Ernst, K., and Kumar, A. (2005). Characterization of susceptibility and resistance responses to potato cyst nematode (Globodera spp.) infection of tomato lines in the absence and presence of the broad-spectrum nematode resistance Hero gene. Mol. Plant Microbe Interact. 18, 158–168. doi: 10.1094/mpmi-18-0158
Sobczak, M., and Golinowski, W. (2011). “Cyst nematodes and syncytia,” in Genomics and Molecular Genetics of Plant-Nematode Interactions, eds J. Jones, G. Gheysen, and C. Fenoll (Dordrecht: Springer Academic Publisher), 61–82. doi: 10.1007/978-94-007-0434-3_4
Starr, J. L., Bridge, J., and Cook, R. (2002). “Resistance to plant-parasitic nematodes: history, current use and future potential,” in Plant Resistance to Parasitic Nematodes, eds J. L. Starr, R. Cook, and J. Bridge (Wallingford: CABI Publishing), 1–22. doi: 10.1079/9780851994666.0001
Stearns, S. C. (1989). Trade-offs in life-history evolution Funct. Ecol 3, 259–268. doi: 10.2307/2389364
Stone, A. R. (1985). Co-evolution of potato cyst nematodes and their host: implication for pathotypes and resistance. Bull. OEPP 15, 131–137. doi: 10.1111/j.1365-2338.1985.tb00212.x
Strachan, S. M., Armstrong, M. R., Kaur, A., Wright, K. M., Lim, T. Y., Baker, K., et al. (2019). Mapping the H2 resistance effective against Globodera pallida pathotype Pa1 in tetraploid potato. Theor. Appl. Genet. 132, 1283–1294. doi: 10.1007/s00122-019-03278-4
Sudha, R., Mhatre, P. H., Lekshmanan, D. K., Venkatasalam, E. P., Bairwa, A., Bhardwaj, V., et al. (2019). Phenotypic and molecular characterization of potato germplasm for potato cyst nematode resistance. Indian J. Genet. Plant. Breed. 79, 394–403. doi: 10.31742/ijgpb.79.2.2
Sudha, R., Venkatasalam, E. P., Bairwa, A., Bhardwaj, V., Dalamu, and Sharma, R. (2016). Identification of potato cyst nematode resistant genotypes using molecular markers. Sci. Hortic. 198, 21–26. doi: 10.1016/j.scienta.2015.11.029
Tan, A. M. Y., Park, T. H., Alles, R., Hutten, R. C., Visser, R. G., and van Eck, H. J. (2009). GpaXIltar originating from Solanum tarijense is a major resistance locus to Globodera pallida and is localised on chromosome 11 of potato. Theor. Appl. Genet. 119, 1477–1487. doi: 10.1007/s00122-009-1149-4
The Potato Genome Sequencing Consortium (2011). Genome sequence and analysis of the tuber crop potato. Nature 475, 189–195. doi: 10.1038/nature10158
Thorpe, P., Mantelin, S., Cock, P. J., Blok, V. C., Coke, M. C., Eves-van den Akker, S., et al. (2014). Genomic characterisation of the effector complement of the potato cyst nematode Globodera pallida. BMC Genom. 15:923. doi: 10.1186/1471-2164-15-923
Torto, B., Cortada, L., Murungi, L. K., Haukeland, S., and Coyne, D. L. (2018). Management of cyst and root knot nematodes: a chemical ecology perspective. J. Agric. Food Chem. 66, 8672–8678. doi: 10.1021/acs.jafc.8b01940
Trudgill, D. L. (1985). Potato cyst nematodes: a critical review of the current pathotyping scheme. Bull. OEPP 15, 273–279. doi: 10.1111/j.1365-2338.1985.tb00228.x
Trudgill, D. L. (1987). Effects of rates of a nematicide and of fertiliser on the growth and yield of cultivars of potato which differ in their tolerance of damage by potato cyst nematodes (Globodera rostochiensis and G. pallida). Plant Soil 104, 235–243. doi: 10.1007/BF02372537
Trudgill, D. L. (1991). Resistance to and tolerance of plant parasitic nematodes in plants. Annu. Rev. Phytopathol. 29, 167–192. doi: 10.1146/annurev.py.29.090191.001123
Trudgill, D. L., and Cotes, L. M. (1983a). Differences in the tolerance of potato cultivars to potato cyst nematodes (Globodera rostochiensis and Globodera pallida) in field trials with and without nemticides. Ann. Appl. Biol. 102, 373–384. doi: 10.1111/j.1744-7348.1983.tb02707.x
Trudgill, D. L., and Cotes, L. M. (1983b). Tolerance of potato to potato cyst nematodes (Globodera rostochiensis and G. pallida) in relation to the growth and efficiency of the root system. Ann. Appl. Biol. 102, 385–397. doi: 10.1111/j.1744-7348.1983.tb02708.x
Trudgill, D. L., Phillips, M. S., and Elliott, M. J. (2014). Dynamics and management of the white potato cyst nematode Globodera pallida in commercial potato crops. Ann. Appl. Biol. 164, 18–34. doi: 10.1111/aab.12085
Turner, S. J. (1990). The identification and fitness of virulent potato cyst nematode populations (Globodera pallida) selected on resistant Solarium vernei hybrids for up to eleven generations. Ann. Appl. Biol. 117, 385–397. doi: 10.1111/j.1744-7348.1990.tb04225.x
Turner, S. J. (1996). Population decline of potato cyst nematodes (Globodera rostochiensis, G. pallida) in field soils in Northern Ireland. Ann. Appl. Biol. 129, 315–322. doi: 10.1111/j.1744-7348.1996.tb05754.x
Turner, S. J., and Stone, A. R. (1981). Hatching of potato cyst nematodes (Globodera rostochiensis, G. pallida) in root exudates of Solanum vernei hybrids. Nematologica 27, 315–318. doi: 10.1163/187529281x00548
Turner, S. J., Stone, A. R., and Perry, J. N. (1983). Selection of potato cyst-nematodes on resistant Solanum vernei hybrids. Euphytica 32, 911–917. doi: 10.1007/BF00042173
Turner, S. J., and Subbotin, S. A. (2013). “Cyst nematodes,” in Plant Nematology, eds R. N. Perry and M. Moens (Wallingford: CAB International), 109–143.
USDA (2007). Potato Cyst Nematode Eradication in Idaho. Available Online at: https://www.aphis.usda.gov/plant_health/ea/downloads/pcn-ameded-ea.pdf (accessed 10.09.2020)
Van der Biezen, E. A., and Jones, J. D. (1998). Plant disease-resistance proteins and the gene-for-gene concept. Trends Biochem. Sci. 23, 454–456. doi: 10.1016/s0968-0004(98)01311-5
van der Vossen, E. A., van der Voort, J. N., Kanyuka, K., Bendahmane, A., Sandbrink, H., Baulcombe, D. C., et al. (2000). Homologues of a single resistance-gene cluster in potato confer resistance to distinct pathogens: a virus and a nematode. Plant J. 23, 567–576. doi: 10.1046/j.1365-313x.2000.00814.x
van Eck, H. J., Vos, P. G., Valkonen, J. P. T., Uitdewilligen, J. G. A. M. L., Lensing, H., de Vetten, N., et al. (2017). Graphical genotyping as a method to map Ny(o,n)sto and Gpa5 using a reference panel of tetraploid potato cultivars. Theor. Appl. Genet. 130, 515–528. doi: 10.1007/s00122-016-2831-y
van Lieshout, N., van der Burgt, A., de Vries, M. E., ter Maat, M., Eickholt, D., Esselink, D., et al. (2020). Solyntus, the new highly contiguous reference genome for potato (Solanum tuberosum). G3 (Bethesda) 10, 3489–3495. doi: 10.1534/g3.120.401550
Van Weymers, P. S. M., Baker, K., Chen, X. W., Harrower, B., Cooke, D. E. L., Gilroy, E. M., et al. (2016). Utilizing “Omic” technologies to identify and prioritize novel sources of resistance to the oomycete pathogen phytophthora infestans in potato germplasm collections. Front. Plant Sci. 7:11. doi: 10.3389/fpls.2016.00672
Vanderplank, J. E. (1978). “Genetic and molecular basis of plant pathogenesis,” in Part of Advanced Series in Agricultural Sciences, (Berlin: Springer-Verlag).
Varypatakis, K., Jones, J. T., and Blok, V. C. (2019). Susceptibility of potato varieties to populations of Globodera pallida selected for increased virulence. Nematology 21, 995–998. doi: 10.1163/15685411-00003283
Varypatakis, K., Veronneau, P.-Y., Thorpe, P., Cock, P. J. A., Lim, J. T.-Y., Armstrong, M. R., et al. (2020). The genomic impact of selection for virulence against resistance in the potato cyst nematode. Globodera pallida. Genes 11:1429. doi: 10.3390/genes11121429
Visscher, P. M., Brown, M. A., McCarthy, M. I., and Yang, J. (2012). Five Years of GWAS Discovery. Am. J. Hum. Genet. 90, 7–24. doi: 10.1016/j.ajhg.2011.11.029
Vos, P., Simons, G., Jesse, T., Wijbrandi, J., Heinen, L., Hogers, R., et al. (1998). The tomato Mi-1 gene confers resistance to both root-knot nematodes and potato aphids. Nat. Biotechnol. 16, 1365–1369. doi: 10.1038/4350
Whitehead, A. G. (1991). Selection for virulence in the potato cyst-nematode. Globodera pallida. Ann. Appl. Biol. 118, 395–402. doi: 10.1111/j.1744-7348.1991.tb05639.x
Whitworth, J. L., Novy, R. G., Zasada, I. A., Wang, X., Dandurand, L.-M., and Kuhl, J. C. (2018). Resistance of potato breeding clones and cultivars to three species of potato cyst nematode. Plant Dis. 102, 2120–2128. doi: 10.1094/pdis-12-17-1978-re
Williams, K. J., Lewis, J. G., Bogacki, P., Pallotta, M. A., Willsmore, K. L., Kuchel, H., et al. (2003). Mapping of a QTL contributing to cereal cyst nematode tolerance and resistance in wheat. Aust. J. Agric. Res. 54, 731–737. doi: 10.1071/AR02225
Williams, K. J., Willsmore, K. L., Olson, S., Matic, M., and Kuchel, H. (2006). Mapping of a novel QTL for resistance to cereal cyst nematode in wheat. Theor. Appl. Genet. 112, 1480–1486. doi: 10.1007/s00122-006-0251-0
Witek, K., Jupe, F., Witek, A. I., Baker, D., Clark, M. D., and Jones, J. D. G. (2016). Accelerated cloning of a potato late blight-resistance gene using RenSeq and SMRT sequencing. Nat. Biotechnol. 34, 656–660. doi: 10.1038/nbt.3540
Wyss, U. (1997). “Root parasitic nematodes: an overview,” in Cellular and Molecular Aspects of Plant-Nematode Interactions, eds C. Fenoll, F. M. W. Grundler, and S. A. Ohl (Netherlands: Springer), 5–22. doi: 10.1007/978-94-011-5596-0_2
Zasada, I. A., Ingham, R. E., Baker, H., and Phillips, W. S. (2019). Impact of Globodera ellingtonae on yield of potato (Solanum tuberosum). J. Nematol. 51, 1–10. doi: 10.21307/jofnem-2019-073
Keywords: nematodes, Globodera, resistance, virulence, molecular markers, genomics, potato breeding, integrated pest management
Citation: Gartner U, Hein I, Brown LH, Chen X, Mantelin S, Sharma SK, Dandurand L-M, Kuhl JC, Jones JT, Bryan GJ and Blok VC (2021) Resisting Potato Cyst Nematodes With Resistance. Front. Plant Sci. 12:661194. doi: 10.3389/fpls.2021.661194
Received: 30 January 2021; Accepted: 03 March 2021;
Published: 25 March 2021.
Edited by:
Juan Emilio Palomares-Rius, Consejo Superior de Investigaciones Científicas (CSIC), SpainReviewed by:
Maria Lurdes Inacio, Instituto Nacional Investigaciao Agraria e Veterinaria (INIAV), PortugalAurelio Ciancio, Istituto per la Protezione Sostenibile delle Piante, Italy
Isabel Luci Conceição, University of Coimbra, Portugal
Copyright © 2021 Gartner, Hein, Brown, Chen, Mantelin, Sharma, Dandurand, Kuhl, Jones, Bryan and Blok. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Vivian C. Blok, Vml2aWFuLkJsb2tAaHV0dG9uLmFjLnVr
 Ulrike Gartner
Ulrike Gartner Ingo Hein
Ingo Hein Lynn H. Brown1,3
Lynn H. Brown1,3 Xinwei Chen
Xinwei Chen Sophie Mantelin
Sophie Mantelin Sanjeev K. Sharma
Sanjeev K. Sharma Louise-Marie Dandurand
Louise-Marie Dandurand Joseph C. Kuhl
Joseph C. Kuhl John T. Jones
John T. Jones Glenn J. Bryan
Glenn J. Bryan Vivian C. Blok
Vivian C. Blok