
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 21 July 2021
Sec. Plant Breeding
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.659381
 Abhijit K. Das1†
Abhijit K. Das1† Munegowda M. Gowda1
Munegowda M. Gowda1 Vignesh Muthusamy1
Vignesh Muthusamy1 Rajkumar U. Zunjare1
Rajkumar U. Zunjare1 Hema S. Chauhan1
Hema S. Chauhan1 Aanchal Baveja1
Aanchal Baveja1 Vinay Bhatt1
Vinay Bhatt1 Gulab Chand1
Gulab Chand1 Jayant S. Bhat2
Jayant S. Bhat2 Satish K. Guleria3
Satish K. Guleria3 Supradip Saha1
Supradip Saha1 Hari S. Gupta1
Hari S. Gupta1 Firoz Hossain1*
Firoz Hossain1*Malnutrition is a widespread problem that affects human health, society, and the economy. Traditional maize that serves as an important source of human nutrition is deficient in vitamin-E, vitamin-A, lysine, and tryptophan. Here, favorable alleles of vte4 (α-tocopherol methyl transferase), crtRB1 (β-carotene hydroxylase), lcyE (lycopene ε-cyclase), and o2 (opaque2) genes were combined in parental lines of four popular hybrids using marker-assisted selection (MAS). BC1F1, BC2F1, and BC2F2 populations were genotyped using gene-based markers of vte4, crtRB1, lcyE, and o2. Background selection using 81–103 simple sequence repeats (SSRs) markers led to the recovery of recurrent parent genome (RPG) up to 95.45%. Alpha (α)-tocopherol was significantly enhanced among introgressed progenies (16.13 μg/g) as compared to original inbreds (7.90 μg/g). Provitamin-A (proA) (10.42 μg/g), lysine (0.352%), and tryptophan (0.086%) were also high in the introgressed progenies. The reconstituted hybrids showed a 2-fold enhancement in α-tocopherol (16.83 μg/g) over original hybrids (8.06 μg/g). Improved hybrids also possessed high proA (11.48 μg/g), lysine (0.367%), and tryptophan (0.084%) when compared with traditional hybrids. The reconstituted hybrids recorded the mean grain yield of 8,066 kg/ha, which was at par with original hybrids (mean: 7,846 kg/ha). The MAS-derived genotypes resembled their corresponding original hybrids for the majority of agronomic and yield-related traits, besides characteristics related to distinctness, uniformity, and stability (DUS). This is the first report for the development of maize with enhanced vitamin-E, vitamin-A, lysine, and tryptophan.
Malnutrition has become one of the alarming health problems leading to lower work efficiency and socio-economic losses worldwide (Allard, 1999). It affects two billion people especially in the developing countries (Arun et al., 2014). Malnutrition has been accounted for nearly 45% of deaths among children under the age of five (Azmach et al., 2013). Malnutrition contributes to loss in 11% gross domestic products (GDPs) in Asia and Africa, and in total, it could cost society up to US$3.5 trillion per year (International Food Policy Research Institute, 2018). Mild to moderate forms of micronutrient (vitamins and minerals) deficiency can severely affect human health and lead to mental impairment, thereby resulting in lower productivity in humans (Bouis, 2018). Global leaders have set the “Sustainable Development Goals” (SDGs) at United Nations in 2015 to alleviate poverty and hunger by 2030. It is estimated that with $1 investment in a proven nutrition programme, a benefit worth of $16 is achieved (International Food Policy Research Institute, 2017). Though various avenues like “food fortification,” “medical supplementation,” and “dietary diversification” are available as strategies to alleviate malnutrition, the development of biofortified crops with enhanced micronutrients still remains the most cost-effective way to accomplish these SDGs (Andersson et al., 2017).
Maize is a primary source of energy and food for hundreds of millions of people globally (Shiferaw et al., 2011). However, traditional maize possesses low vitamin-E (6–8 μg/g), vitamin-A (1–2 μg/g), lysine (0.15–0.20%), and tryptophan (0.03–0.04%), which play a crucial role in human metabolism (Zunjare et al., 2017, 2018a; Hossain et al., 2018; Das et al., 2019b). Vitamin-E quenches free radicals in the cell membrane, thereby protecting the polyunsaturated fatty acids (PUFA) from damage; besides, it also serves as an essential micronutrient for the proper functioning of the reproduction system. It further protects from cardiovascular disease, Alzheimer's disease, neurological disorder, and many age-related degenerations (Chander et al., 2008). Vitamin-E deficiency (VED) has been observed more in premature infants and elderly people (Das et al., 2018). It is estimated that over 20% of the examined people both in developed and in developing countries have suboptimal plasma alpha (α)-tocopherol, the most active form of vitamin-E (Li et al., 2012). Vitamin-A deficiency (VAD) is also a major health problem worldwide, and it causes visual impairment and results in low resistance to infectious diseases (Black et al., 2008). Vitamin-A deficiency affects about 20 million pregnant women, and one-third of them are clinically night-blind. It also affects 250 million children and accounts for increased childhood mortality and disease (WHO, 2009). Besides vitamins, lysine and tryptophan serve as essential amino acids for protein synthesis inside the human body, and deficiency of which leads to the most common symptoms such as loss of appetite, depression, delayed growth, and anxiety in children (Nuss and Tanumihardjo, 2010). Among nutritional disorders, the highest number of death occurs due to protein-related deficiencies worldwide (Bain et al., 2013; Hossain et al., 2019). Health benefits of quality protein maize (QPM) in human and the growth of poultry birds and pigs have also been reported (Gunaratna et al., 2010; Panda et al., 2014). Provitamin-A (proA)-rich biofortified maize was effective in reducing VAD in children (Gannon et al., 2014). Further, chickens fed with proA-rich maize kernel showed more redness in meat color and produced eggs with a higher amount of proA (Heying et al., 2014; Moreno et al., 2016; Odunitan-Wayas et al., 2016; Sowa et al., 2017). Over the last decade, efforts have been made for vitamin-E biofortification in different crops (Mene-Saffrane and Pellaud, 2017). In Arabidopsis, the introduction of γ-tocopherol methyltransferase (γ-TMT) resulted in an increased accumulation of α-tocopherol over γ-tocopherol in seeds (Shintani and DellaPenna, 1998; Mene-Saffrane and Pellaud, 2017). Similar results were also witnessed in other crops, including soybean (Arun et al., 2014) and lettuce (Cho et al., 2005).
Thus, the development of maize hybrids rich in these nutrients holds an immense promise to alleviate malnutrition in a holistic way. Maize mutants with a favorable allele of vte4 gene (α-tocopherol methyl transferase) accumulating 2–3 fold more vitamin-E have been identified (Li et al., 2012; Das et al., 2018). The natural variants of crtRB1 (β-carotene hydroxylase) and lcyE (lycopene ε-cyclase) that enhance vitamin-A by 2–10 fold are also available in maize germplasm (Harjes et al., 2008; Yan et al., 2010). The recessive opaque2 (o2) mutant allele increases lysine and tryptophan by 2-fold (Mertz et al., 1964; Hossain et al., 2018). Several QPM hybrids with o2 gene and proA-rich maize hybrids with crtRB1 and lcyE genes have been released worldwide (Cabrera-Soto et al., 2018; Hossain et al., 2019; Prasanna et al., 2020). In India, ICAR-Indian Agricultural Research Institute (ICAR-IARI), New Delhi, have developed (first of its kind) three proA-rich QPM hybrids for commercial cultivation (Muthusamy et al., 2014; Zunjare et al., 2018a). However, these hybrids still lack vitamin-E, which is essential for proper metabolism in humans (Das et al., 2019b). Thus, the development of maize hybrids rich in vitamin-E, vitamin-A, and essential amino acids holds immense significance in alleviating malnutrition in a sustainable and cost-effective way. Marker-assisted selection (MAS) is the ideal approach to combine desirable allele of multiple genes in maize breeding programme for the release of commercial hybrids (Yadava et al., 2018; Prasanna et al., 2020). Hence, the current study was undertaken to (i) combine favorable alleles of vte4, crtRB1, lcyE, and o2 genes into the parental lines of four released hybrids using MAS, (ii) evaluate the introgressed inbred lines and reconstituted hybrids for nutritional quality traits, and (iii) assess the genotypes for agronomic and yield-related traits. This is the first report of combining vitamin-E with vitamin-A, lysine, and tryptophan in a maize hybrid.
Four QPM inbreds improved for proA (HKI161-VA, HKI163-VA, HKI193-1-VA, and HKI193-2-VA) by Zunjare et al. (2018a) were selected for the enrichment of α-tocopherol (vitamin-E). These four inbreds are the parents of four proA-rich QPM hybrids (HQPM1-VA, HQPM4-VA, HQPM5-VA, and HQPM7-VA). The details of these hybrids are presented in Supplementary Table 1. The parental lines possess favorable alleles of o2, crtRB1, and lcyE genes, and are higher in lysine, tryptophan, and proA. However, all the four inbreds had a lower level of α-tocopherol. The donor inbred (HP465-41) developed at International Maize and Wheat Improvement Center (CIMMYT), Mexico, under the HarvestPlus programme was used for introgression of the favorable allele of vte4 into these parental lines. The donor line also possessed favorable alleles of crtRB1 and lcyE genes. Pedigree of donor and recipient parents is given in Supplementary Table 2.
Each of the recipient parents was crossed as female with donor parent as male during the winter season of 2014–2015 at Winter Nursery Center (WNC) of ICAR-Indian Institute of Maize Research (ICAR-IIMR), Hyderabad (17°19′N, 78°24′E, 542.6 MSL) (N, north; E, east; MSL, mean sea level). F1 plants were raised during the rainy season (2015) at ICAR-IARI, New Delhi (28°08′N, 77°12′E, 229 MSL), and tested for hybridity using vte4 and o2 specific markers. True heterozygotes were backcrossed as male with the respective recurrent parents. BC1F1 progenies were grown at WNC, Hyderabad, during the winter season of 2015–2016, and foreground positive (heterozygous for vte4 and o2; homozygous for crtRB1 and lcyE favorable allele) individuals were tagged before flowering. Individuals with more similarity to recurrent parents and high recovery of recurrent parent genome (RPG) were further backcrossed with the respective recurrent parents. BC2F1 progenies were grown during the rainy season (2016) at ICAR-IARI, New Delhi, and foreground positive plants (heterozygous: vte4 and o2; homozygous: crtRB1 and lcyE) were selected. Genotypes with high RPG and phenotypic similarity were advanced to raise BC2F2 progenies at WNC, Hyderabad, during the winter season of 2016–2017. The segregants homozygous for all the four genes (vte4, crtRB1, lcyE, and o2) were identified, and plants with high RPG and phenotypic similarity were selected. The BC2F3 progenies were raised during the rainy season (2017) at ICAR-IARI, New Delhi, and advanced to BC2F4 progenies for the generation of crosses during the winter season at WNC, Hyderabad. The reconstituted hybrids were evaluated during the rainy season (2018) at multilocations.
Genomic DNA (deoxy ribonucleic acid) was isolated from the leaf of young maize seedlings using the standard CTAB (cetyl trimethyl ammonium bromide) protocol (Murray and Thompson 1980). polymerase chain reaction (PCR) was performed using (i) InDel7 (InDel: insertion–deletion) and InDel118 markers for vte4, (ii) 3′TE-InDel marker for crtRB1 (TE: transposable element), (iii) 5′TE-InDel marker for lcyE, and (iv) phi057 (simple sequence repeat: SSR) for o2 (Supplementary Table 3). The product size of favorable alleles for InDel118 and InDel7 was 373 and 160 bp, whereas that of unfavorable alleles was 491 and 167 bp, respectively. For crtRB1-3′TE and lcyE-5′TE, the amplified product for favorable alleles was 543 and 650 bp, respectively. The favorable allele of phi057 in recurrent parents was of 165 bp, and the unfavorable donor allele was of 159 bp. For vte4, PCR was performed following the protocol of Li et al. (2012). Amplified product for InDel118 was resolved using 2% agarose gel, and that for InDel7 was resolved using 4% super fine resolution (SFR) agarose. For 3′TE-InDel of crtRB1, PCR was performed as suggested by Yan et al. (2010), and 1.5% agarose gel was used to resolve the amplified products. For 5′TE-InDel of lcyE, PCR was performed as suggested by Harjes et al. (2008), and products were resolved using 2% agarose gel. PCR amplified products of SSRs for o2 and background markers was resolved using 4% agarose as per PCR protocol as suggested by Hossain et al. (2018).
Two insertion/deletions (InDel7 and InDel118) at the promoter and 5′-UTR region of vte4 gene were identified by Li et al. (2012). Donor parent had favorable haplotype (0/0: deletion at both locations), whereas all the four recipient parents possessed unfavorable allele (7/118: insertion at both locations) of vte4. Since InDel7 and InDel118 are in close proximity within the vte4 gene, InDel118 was sufficient for genotyping of vte4 locus in backcross generations. Markers specific to 3′TE InDel (in exon 6 and 3′-UTR) of crtRB1 gene and 5′TE InDel (in 5′-UTR) of lcyE gene were used to select for high proA. Gene-based SSR (phi057) was used as a foreground marker for o2. The detail of the markers along with location in the genome used for the foreground selection is presented in Supplementary Table 3. Foreground selection was accomplished for the identification of plants heterozygous for vte4 and o2 and homozygous for the favorable allele of crtRB1 and lcyE in BC1F1 and BC2F1. In BC2F2 generation, plants homozygous for favorable allele all four genes were selected.
A set of 285 SSRs distributed throughout the maize genome covering all the 10 chromosomes was selected from the maize genome database (www.maizegdb.org) (Supplementary Table 4). These selected SSRs were used to identify polymorphic markers between the respective recurrent and donor parents. Respective polymorphic SSRs set between recurrent and donor parents were used for the identification of genotypes with high RPG in BC1F1, BC2F1, and BC2F2.
Fourteen BC2F4 progenies—four each from HKI161-VA and HKI193-1-VA; three each from HKI163-VA and HKI193-2-VA, along with the four recurrent parents—were grown in a randomized complete block design (RCBD) with two replications during the rainy season (2018) at ICAR-IARI Experimental Farm, New Delhi. Each entry was grown in a row of 3 m length, and plant-to-plant distance of 20 cm and row-to-row distance of 75 cm were maintained, and standard agronomic practices were followed to raise a good crop. In each row, 2–3 plants were self-pollinated to avoid xenia effects, and selfed seeds were used for the quality analysis. Grain yield, male and female flowering, plant and ear height, ear length, ear diameter, number of rows per ear, number of kernels per row, and 100 kernel weight were recorded from open pollinated plants. Further, all the 31 characteristics related to distinctness, uniformity, and stability (DUS) were also recorded in each of the introgressed lines and recurrent parent.
The introgressed lines were used to reconstitute hybrid combinations representing three versions (I, II, III) in each of the four original hybrids during the winter season (2017–2018) (Supplementary Table 5). The reconstituted 12 hybrid combinations and their corresponding original four hybrids along with a commercial check, Pusa Vivek QPM9 Improved (PVQ9I), were evaluated at three locations, viz. Bajaura (32°2′N, 77°9′E, 1,090 MSL), Delhi (28°08′N, 77°12′E, 229 MSL), and Dharwad (15°21′N, 75°05′E, MSL: 750 MSL) during the rainy season of 2018. PVQ9I is a proA-rich QPM hybrid released and notified for cultivation during 2017 in India. These three locations belong to three different maize-growing zones of the country. While Bajaura is situated in Northern Hills Zone, Delhi and Dharwad belong to North Western Plains Zone and Peninsular Zone, respectively. Hybrids were evaluated in RCBD with two replications. In each entry, 2–3 plants were self-pollinated to avoid contamination by foreign pollens, and selfed seeds were used for nutritional quality analysis. Grain yield, male and female flowering, plant height, ear height, ear length, ear girth, number of rows per ear, number of kernels per row, and 100 kernel weight were recorded from the open-pollinated plants. Further, the hybrids were also characterized for all the 31 DUS traits.
Seeds were stored at 4°C until extraction to avoid any degradation of quality traits. Extraction of tocopherol was performed following the protocol of Saha et al. (2013), but absolute ethanol was used instead of methanol. Twenty microliters of each sample were injected into the Dionex Ultimate 3000 UHPLC System (Ultra High Performance Liquid Chromatography; Thermo Scientific, Massachusetts, United States), and the fluorescence detector was used with an excitation wavelength of 290 nm and an emission wavelength of 325 nm to detect the peak. Reverse-phase column YMC-C30 (5 μm, 4.6 × 250mm; Waters Chromatography) was used. Methanol and TBME (tert-butyl methyl ether) in a ratio of 95:5 (v/v) were used as a mobile phase with a flow rate of 1 ml min−1. Different dilutions (50, 100, 500, and 1,000 ppm) of each of the tocopherol standards, viz. alpha (α)-, beta (β-), gamma (γ)-, and delta (δ)-tocopherol (SIGMA chemicals, United States), were used to prepare the standard curve, which was further used to determine the concentration of respective tocopherols in samples by the standard regression equation.
The protocol described by Kurilich and Juvik (1999) and Vignesh et al. (2012) was used for the extraction of β-carotene and β-cryptoxanthin, and quantification was performed using Dionex Ultimate 3000 UHPLC System. YMC carotenoid C30 column (5 μm, 4.6 × 250mm; YMC) was used to elute the sample that was detected with a diode array detector-3000 (RS) at 450nm. The mobile phase consisted of methanol:TBME (80:20, v/v) with a flow rate of 1 ml min−1. Concentration of β-carotene and β-cryptoxanthin was calculated from the standard regression curve prepared from different dilutions of corresponding standards (Sigma Aldrich, United States). Total β-carotene content and half of the β-cryptoxanthin content of each sample were used to calculate the proA concentration (Babu et al., 2013).
Lysine and tryptophan contents were extracted following the protocol standardized by Sarika et al. (2018), and the Dionex Ultimate 3000 UHPLC system was used for estimation. The AcclaimTM 120 C18 column (5 μm, 120 A°, and 4.6 × 150mm, Thermo Scientific) was used to elute the samples, and detection was done with a RS photodiode array detector (PDA) at 265 and 280 nm wavelength for lysine and tryptophan, respectively. Amino acid concentration of each sample was estimated from the standard regression curve prepared using external standards (AAS 18-5ML, Sigma Aldrich).
χ2-test for goodness of fit was performed using SPSS16 (SPSS Inc. Released 2007) to check the Mendelian segregation of genes in BC1F1, BC2F1, and BC2F2 generations. Simple sequence repeats-amplified fragments of foreground positive plants were scored as “A” to represent the recipient allele, “B” to represent the donor allele, and “H” for the heterozygote. Calculation of RPG recovery and graphical representation of background genome of selected individuals in backcross generations were done using Graphical Geno Types (GGT) version 3.0 (Van-Berloo, 1999). Windostat 8.5 software package was used for the calculation of analysis of variance (ANOVA) and standard error (SE) values for agronomic and biochemical data of improved inbreds and hybrid.
Screening of donor and recipient parents using InDel118 and InDel7 markers of vte4 revealed the presence of favorable alleles in donor parent and unfavorable alleles in recurrent parents for both InDels. InDel118 produced easily distinguishable alleles between recipient and donor parents (allele difference of 118 bp), compared to InDel7 (allele size difference of 7 bp). Since InDel118 and InDel7 are closely linked and present within the same gene, the selection of InDel118 could invariably select InDel7 as well. Hence, only InDel118 was used for foreground selection to introgress the favorable allele of vte4. For o2 gene, the use of phi057 primer pair had amplified favorable allele in each of the four recurrent parents and the unfavorable allele in the donor parent. Recipient and donor parents both amplified the favorable alleles of crtRB1 and lcyE using 3′TE-InDel and 5′TE-InDel, respectively.
The donor parent was crossed as male with each of the recurrent parents. All the F1s were heterozygote for vte4 and o2 genes and homozygous for the favorable allele of crtRB1 and lcyE.
A total of 285 markers were used for the polymorphism survey between the donor and recurrent parents. Parental polymorphism survey identified 103, 81, 89 and 81 polymorphic SSRs amounting to 36.14, 28.42, 31.22, and 28.42% of polymorphism between the donor (HP465-41) and HKI161-VA, HKI163-VA, HKI193-1-VA, and HKI193-2-VA, respectively (Supplementary Table 4). Number of polymorphic markers ranged from 4 to 15 per linkage group. Respective polymorphic markers were used to recover the background genome of the recipient parent in each of the crosses.
Population size of BC1F1 ranged from 96 to 124 across four genetic backgrounds (Table 1). Genotyping of BC1F1 populations with InDel118 identified 62, 53, 51, and 66 heterozygous individuals in HKI161-VA, HKI163-VA, HKI193-1-VA, and HKI193-2-VA, respectively. Chi-square for the goodness of fit showed the Mendelian segregation pattern (1:1) for vte4 in each of the crosses (Table 1). Vte4 heterozygote genotypes from each of the populations were further screened for o2 homozygote (o2o2) using phi057. Thirty plants of HKI161-VA, 22 plants of HKI163-VA, 21 plants of HKI193-1-VA, and 24 plants of HKI193-2-VA were homozygous for favorable o2 allele. In the case of HKI161-VA-, HKI163-VA-, and HKI193-1-VA-based populations, Mendelian segregation of 1:1 was observed, while in HKI-193-2-VA-based population it significantly deviated from the Mendelian segregation. These selected plants were screened for crtRB1 and lcyE genes, and all were homozygous for the favorable alleles. Owing to cross-pollinated nature of maize, the genotyping for crtRB1 and lcyE was carried out to avoid any cross-pollination (by wild-type plants grown elsewhere in the field) that may lead to heterozygosity and later on fixation of an unfavorable allele in the progeny. Phenotypic selection for individuals that were heterozygous for vte4 and homozygous for o2, crtRB1, and lcyE reduced the number to 10 for each family. Recurrent parent genome among the selected segregants varied from 61.70 to 77.20% across the populations (Table 2). Based on the plant, ear, and grain characteristics, two selected ears each in HKI161-VA (HKI161-VA-19: 72.85% and HKI161-VA-57: 76.10%), HKI163-VA (HKI163-VA-8: 77.20% and HKI-VA-38: 75.65%), and HKI193-1 (HKI193-1-VA-10: 75.00% and HKI-VA-20: 75.30%) and one in HKI193-2 (HKI-193-2-VA: 76.75%) were advanced to raise BC2F1 populations (Table 2).
The population size in BC2F1 varied from 100 to 122. Genotyping with InDel118 revealed that 57, 63, 60, and 46 plants were heterozygotes in populations of HKI161-VA, HKI163-VA, HKI193-1-VA, and HKI193-2-VA, respectively (Table 1). The Chi-square (χ2) test for goodness of fit indicated the Mendelian segregation of 1:1. Each of the foreground positive plants (heterozygous for vte4) was screened for o2, crtRB1, and lcyE. As per expectation, all the plants heterozygous for vte4 were homozygous for favorable alleles of the three genes. Phenotypic selection further reduced them to 10 individuals in populations of HKI161-VA, HKI163-VA, and HKI193-1-VA, while it reduced them to nine individuals for HKI193-2-VA population. Recurrent parent genome in these selected segregants ranged from 79.95 to 90.60% across populations (Table 2). Nine segregants, HKI161-VA-19-4 (88.45%), HKI161-VA-57-5 (88.70%), HKI163-VA-8-52 (89.1%), HKI163-VA-8-82 (87.3%), HKI163-VA-38-5 (86.6%), HKI193-1-VA-10-5 (87.35%), HKI193-1-VA-20-37 (87.95%), HKI193-2-VA-1-93 (86.25%), and HKI193-2-VA-1-94 (86.25%), were finally selected based on plant, ear and grain characteristics. These selected plants were selfed to raise BC2F2 populations.
Four BC2F2 populations with a size of 96–120 individuals were raised. Foreground selection with InDel118 identified three genotypic classes in 1:2:1 without any segregation distortion in any of the populations (Supplementary Figure 1; Table 1). Individuals homozygous for favorable allele of vte4 (25, 28, 29 and 32) were identified in populations of HKI161-VA, HKI163-VA, HKI193-1-VA, and HKI193-2-VA, respectively (Table 1). All the foreground positive plants for InDel118 also possessed favorable allele of o2, crtRB1, and lcyE (Supplementary Figures 2–4; Table 1). Recurrent parent genome in phenotypically selected BC2F2 individuals ranged from 84.45 to 95.45% across the populations (Figure 1; Table 2). Based on high RPG and phenotype in relation to plant, ear and grain characteristics, three to four segregants in each of the crosses were selected. HKI161-VA-19-4-60 (95.45%) and HKI161-VA-57-5-4 (91.70%) of HKI161-VA families and HKI163-VA-8-52-26 (91.15%) and HKI163-VA-8-82-18 (90.20%) of HKI163-VA families were the best with high RPG. Among HKI193-1-VA- and HKI193-2-VA-based families, HKI193-1-VA-20-37-64 (93.35%), HKI193-1-VA-10-5-19 (91.45%), HKI193-2-VA-1-94-84 (88.75%), and HKI193-2-VA-1-94-70 (88.75%) were the most promising with higher RPG.
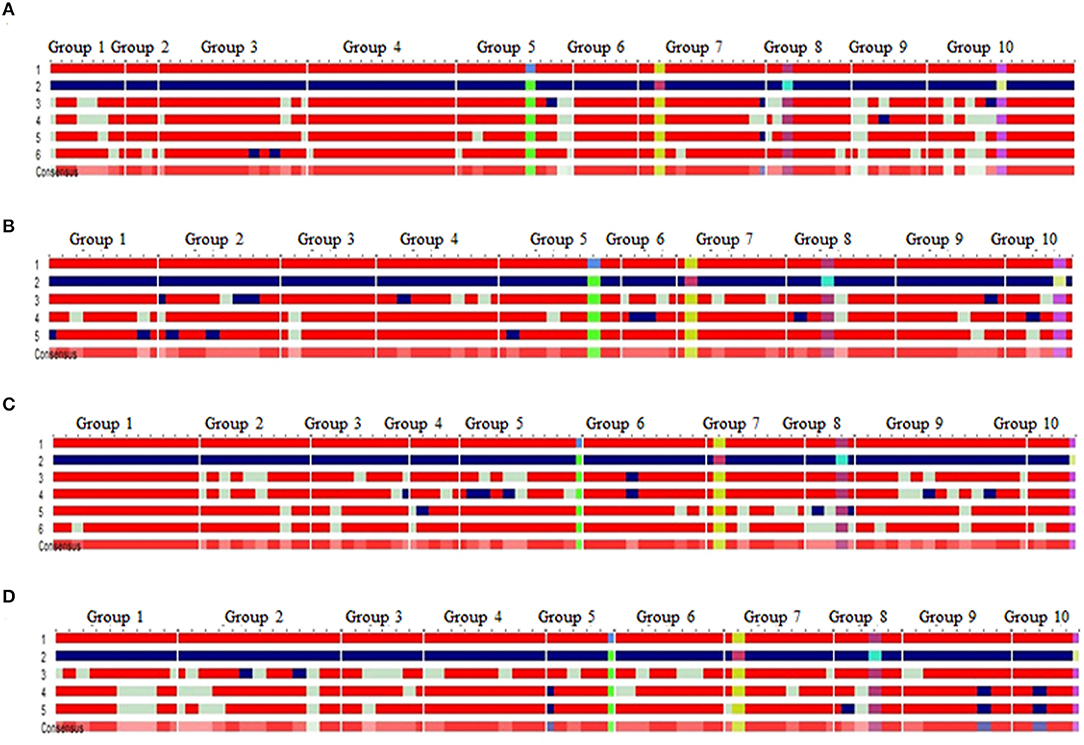
Figure 1. Background recovery of selected progenies in BC2F2 of the RP × DP [(A) HKI161-VA+VE, (B) HKI163-VA+VE, (C) HKI193-1-VA+VE, and (D) HKI193-2-VA+VE]; 1, recurrent parent (red); 2, donor parent (blue); light green color represent the locus in heterozygous condition; light red color in “consensus sequence” represents the locus having donor allele or heterozygosity, 3–5/6: introgressed progenies; favorable allele of vte4 (green box), favorable allele of phi057 (yellow box), favorable allele of lcyE (purple box), favorable allele of crtRB1 (pink box); group 1–10 represents the chromosome 1–10.
ANOVA showed a significant variation among the recurrent and introgressed inbred lines for most of the quality traits except tryptophan (Supplementary Table 6). Introgressed progenies recorded higher α-tocopherol than their recurrent parents. The recurrent parents, viz. HKI161-VA (9.01 μg/g), HKI163-VA (7.91 μg/g), HKI193-1-VA (8.32 μg/g), and HKI193-2-VA (6.26 μg/g), recorded lower α-tocopherol. HP465-41 with higher α-tocopherol (18.38 μg/g) was used as donor parent. α-Tocopherol across introgressed progeny was significantly high (16.13 μg/g) as compared to 7.90 μg/g in the original lines. However, α-tocopherol varied from 17.51 to 19.73 μg/g (mean: 18.21 μg/g), 13.05 to 15.69 μg/g (mean: 14.4 μg/g), 16.19 to 18.14 μg/g (mean: 17.03 μg/g), and 12.87 to 15.35 μg/g (mean: 13.91 μg/g) among introgressed progenies of HKI161-VA, HKI163-VA, HKI193-1-VA, and HKI193-2-VA. HKI161-VA-57-5-4 was the best progeny with 19.73 μg/g of α-tocopherol, while other promising progenies were HKI163-VA-8-52-26 (15.69 μg/g), HKI193-1-VA-20-37-64 (18.14 μg/g), and HKI193-2-VA-1-94-84 (15.35 μg/g) (Table 3). The mean γ-tocopherol in HKI161-VA-derived progenies was 16.56 μg/g, while it was 2.59, 39.67, and 29.97 μg/g among progenies of HKI163-VA, HKI193-1-VA, and HKI193-2-VA, respectively (Table 3). The γ-tocopherol values among the original parents ranged from 7.37 to 40.62 μg/g. The mean δ-tocopherol and total tocopherol were 4.52 and 43.70 μg/g, respectively, across the introgressed progenies. These nutrients were low among the parents (4.00 and 32.60 μg/g, respectively). The concentration of β-tocopherol was quite low and could not be detected. A significant change in the proportion of different tocopherol components and total tocopherol was also observed in the improved lines. The mean α-tocopherol to total tocopherol increased from 30% among parental lines to 45% across introgressed progenies. Similarly, α-tocopherol to γ-tocopherol also recorded an enhancement to a level of 185% among the improved introgressed lines, while it was 60% among the parental lines. Zunjare et al. (2018a) reported high proA (10–12 μg/g) in the crtRB1 and lcyE introgressed maize genotypes compared to 1–2 μg/g in the traditional maize. Mean proA among introgressed progenies was high (10.42 μg/g) with a range of 7.43–12.42 μg/g. Similar levels of proA were observed in the parental lines, viz. HKI161-VA (10.44 μg/g), HKI163-VA (8.37 μg/g), HKI193-1-VA (10.78 μg/g), and HKI193-2-VA (12.42 μg/g) (Table 3). Quality protein maize maize cultivars possess higher tryptophan (>0.06%) and lysine (>0.30%) contents than normal maize (tryptophan: 0.03–0.04%, lysine: 0.15–0.20%) (Hossain et al., 2018). The lysine among the MAS-derived progenies was also high with a range from 0.309 to 0.408% with a mean of 0.352%. Mean tryptophan among the progenies was high (0.083%) with a range from 0.076 to 0.092%. Similarly, high level of lysine (0.364%) and tryptophan (0.086%) was recorded among the parents (Table 3).
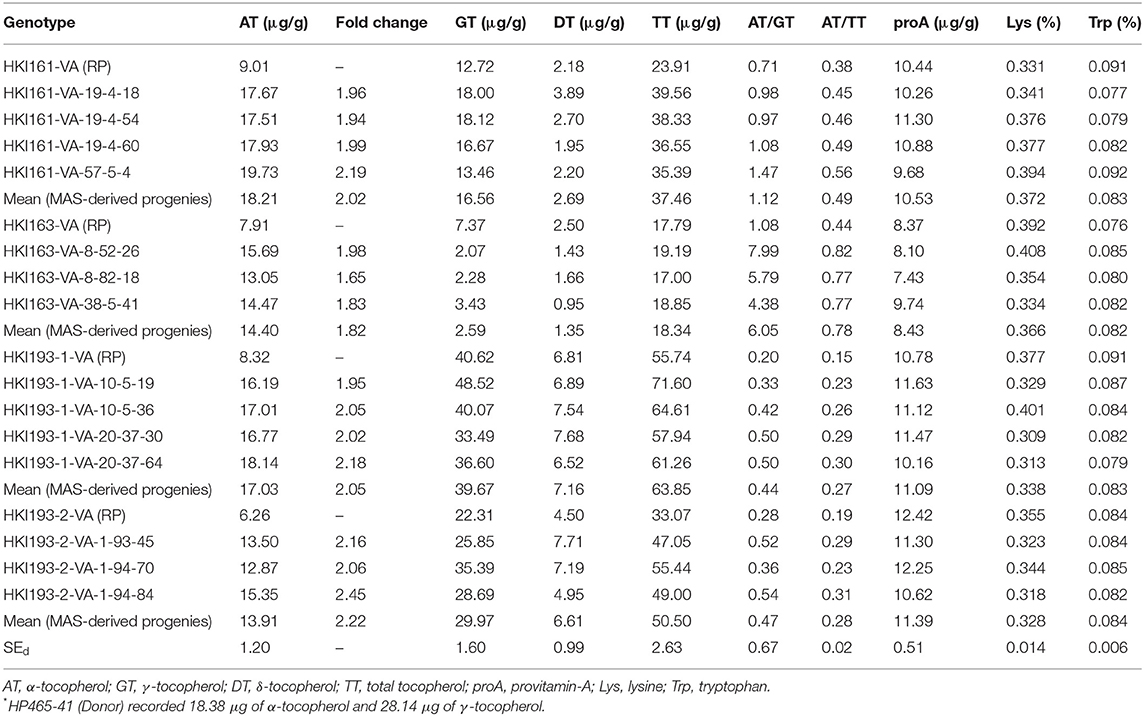
Table 3. Mean performance of vte4 introgressed progenies and their recurrent parents for different quality parameters.
Significant variation was observed among the recurrent and introgressed inbred lines for each of the phenotypic traits (Supplementary Table 7). However, the introgressed inbred lines were comparable to their original parents for the majority of agronomic and yield characteristics (Supplementary Table 8). Plant and ear height, flowering time, and yield-related traits were predominantly at par with the original inbreds. Improved lines were also similar to original parents for most of the DUS traits except for (i) anthocyanin coloration of brace root in progenies of HKI161-VA and (ii) anthocyanin coloration of glume of tassel excluding base in some of the progenies of HKI161-VA, HKI163-VA, HKI193-1-VA, and HKI193-2-VA. Attitude of the leaf blade was straight in HKI163-VA, while one of the progenies showed drooping type. Similarly, anthocyanin coloration in the leaf sheath was absent in HKI163-VA, while it was present in all the progenies of the line (Supplementary Table 9).
Significant variation was observed among the original and reconstituted hybrids for each of the quality traits (Supplementary Table 10). However, vte4-based reconstituted hybrids recorded higher mean α-tocopherol (16.83 μg/g) with a range of 14.77–19.31 μg/g. The original hybrids recorded low α-tocopherol (6.72–8.67 μg/g), while it was 8.74 μg/g in the check. HQPM1-VA+VE-III recorded the highest α-tocopherol (19.31 μg/g), while the other promising hybrids were HQPM4-VA+VE-III (17.11 μg/g), HQPM5-VA+VE-I (17.60 μg/g), and HQPM7-VA+VE-II (18.03 μg/g) (Table 4; Supplementary Figure 5). Overall, there was 2.11-fold increase in α-tocopherol. The mean γ-tocopherol and total tocopherol among the reconstituted hybrids were 22.35 μg/g (range: 13.73–32.91 μg/g) and 45.20μg/g (range: 37.66–53.72 μg/g) compared to 18.78 and 31.24 μg/g in the original hybrids. The same in the check hybrids were 19.88 and 32.73 μg/g, respectively. The value of δ-tocopherol in the reconstituted hybrids was higher (7.27 μg/g) than that of the original hybrids (5.43 μg/g) and check (4.20 μg/g) (Table 4). α-, γ-, and δ-tocopherol to the total tocopherols in the original hybrids were 25, 58, and 17%, whereas an increased proportion of α-tocopherol (36%) was recorded in improved hybrids with 48 and 16% of γ- and δ-tocopherol, respectively. The ratio of α- to γ-tocopherol and α- to total tocopherol increased to 0.82 and 0.38 from 0.46 and 0.26, respectively. Improved hybrids had high proA (mean: 11.48 μg/g, range: 9.61–13.38 μg/g) similar to original hybrids (mean: 11.43 μg/g, range: 9.62–12.38 μg/g) and check (10.84 μg/g) (Table 4; Supplementary Figure 6). The lysine (mean: 0.367%, range: 0.324–0.407%) and tryptophan (mean: 0.084%, range: 0.077–0.096%) in the reconstituted hybrids were also high and at par with the original hybrids (lysine, mean: 0.368%, range: 0.327–0.391%; tryptophan, mean: 0.087%, range: 0.078–0.091%) and check (lysine: 0.340%, tryptophan: 0.080%) (Table 4; Supplementary Figures 7, 8). PVQ9I recorded high proA (10.84 μg/g), lysine (0.340%), and tryptophan (0.080%).
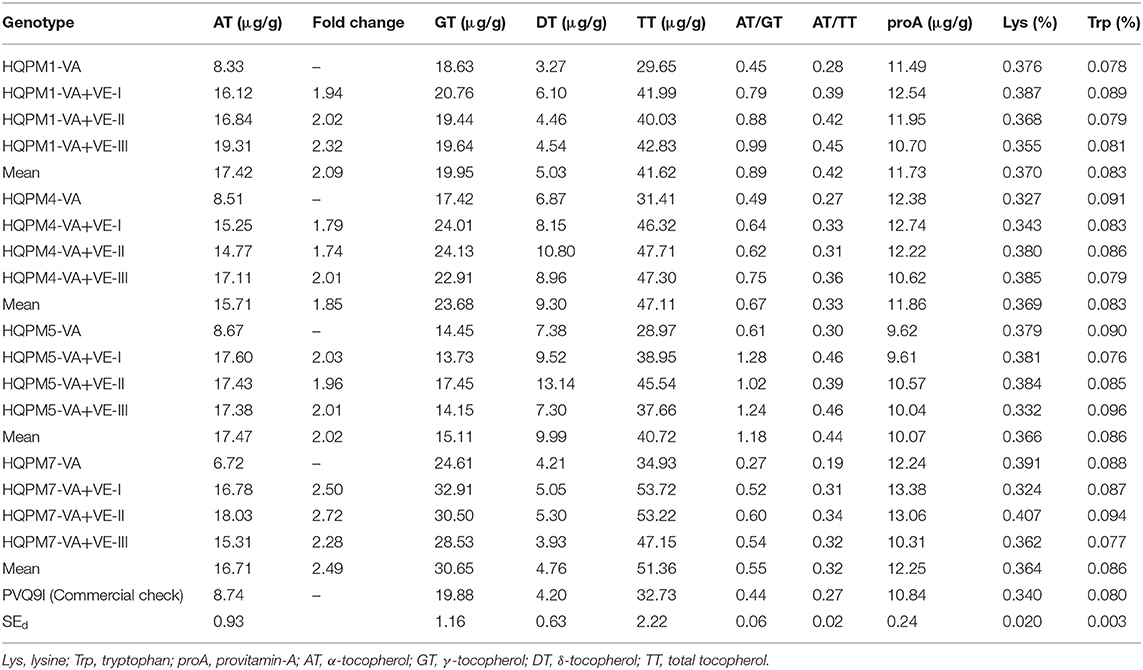
Table 4. Mean performance of vte4-derived reconstituted hybrids and their original hybrids for different quality parameters.
Significant variation was observed among the original and reconstituted hybrids for each of the phenotypic traits (Supplementary Table 11). However, original and reconstituted hybrids for different phenotypic traits, the grain yield and yield-attributing traits of most of the reconstituted hybrids were at par or higher with their respective original hybrids (Supplementary Table 12). The improved multinutrient-rich hybrids in the genetic background of HQPM1-VA, HQPM4-VA, HQPM5-VA, and HQPM7-VA recorded the mean grain yield of 8,530, 7,365, 8,332, and 7,896 kg/ha, while the same in the original hybrids were 8,207 kg/ha, 7,084 kg/ha, 8,546 kg/ha, and 7,548 kg/ha, respectively. Further, all the reconstituted hybrids recorded a higher yield than the commercial check, PVQ9I (6,337 kg/ha).
Each of the reconstituted hybrids was characterized for DUS traits. Original and improved hybrids were similar for most of the DUS traits except (i) HQPM4-VA+VE-I for anthocyanin coloration of glume excluding base and color of top of grains, (ii) HQPM5-VA+VE for anthocyanin coloration of brace root and color of grain, (iii) HQPM7-VA+VE-I, -II, and -III versions for anthocyanin coloration of brace root, (iv) HQPM7-VA+VE-III for color of top of grains and (v) HQPM5-VA+VE-II, HQPM5-VA+VE-III, HQPM7-VA+VE-I, and HQPM7-VA+VE-II for the width of leaf blade (Supplementary Table 13).
Traditional maize hybrids are deficient in essential nutrients including balanced protein enriched with lysine and tryptophan, and vitamins such as vitamin-A and vitamin-E (Prasanna et al., 2020). Maize mutants rich in these nutrients are available, viz. (i) o2 mutant with higher lysine and tryptophan (Mertz et al., 1964; Vasal, 2000), (ii) crtRB1 and lcyE mutants for high proA (Harjes et al., 2008; Yan et al., 2010), and (iii) vte4 mutant for higher vitamin-E or α-tocopherol (Li et al., 2012). The development of multinutrient maize cultivars with enriched lysine, tryptophan, vitamin-A, and vitamin-E holds immense significance to human health as multiple types of malnutrition can be addressed in a more sustainable way (Bouis, 2018). Stacking of genes in a single genetic background is challenging as it takes a long time and requires costly phenotyping in a large scale. However, MABB approach can achieve the goal with reduced time and cost (Collard et al., 2005; Muthusamy et al., 2014; Zunjare et al., 2018b). Hence, the study was undertaken to combine the mutant alleles of vte4, o2, crtRB1, and lcyE into one background to develop multinutrient-rich maize hybrids using the molecular breeding method. Four popular QPM hybrids already improved for proA (Zunjare et al., 2018a) were targeted for α-tocopherol enrichment.
Donor and recurrents parent carried the favorable allele of vte4 and o2, respectively, and both shared the favorable alleles of crtRB1 and lcyE. Gene-based markers, viz. InDel118, phi057, 3′TE-InDel, and 5′TE-InDel, were used for foreground selection of favorable alleles of vte4, o2, crtRB1, and lcyE, respectively. All of these markers are located within the target genes. Gene-based markers have a significant advantage over linked markers, which may lead to the selection of false-positive individuals due to genetic recombination between the target gene and the linked marker (Collard et al., 2005). The use of these gene-based markers ensures the accurate transfer of the target gene. Several researchers have successfully used these markers, viz. vte4 (Feng et al., 2015), o2 (Hossain et al., 2018), crtRB1 (Muthusamy et al., 2014; Zunjare et al., 2017, 2018a), and lcyE (Zunjare et al., 2018a,c) in MABB programme. vte4 gene followed the Mendelian segregation in BC1F1, BC2F1, and BC2F2 generations. o2 showed expected segregation in three of the BC1F1 populations, whereas one BC1F1 population (HKI193-2 based) showed a strong segregation distortion (SD). Hossain et al. (2018) also reported SD in some of the backcross populations segregating for o2. Distortion of a locus could be attributed to the existence of gametophytic factors, male sterility, mutants like defective kernel and embryo-specific mutation (Neuffer et al., 1997; Lu et al., 2002). The SD is possibly attributed to the presence of such type of loci in the genetic background of HKI193-2, which was quite different from the other three genetic backgrounds. Though in our populations, crtRB1 and lcyE did not segregate, strong SD for these two genes were reported in previous studies (Babu et al., 2013; Muthusamy et al., 2014; Liu et al., 2015; Zunjare et al., 2017, 2018a).
Recovery of background genome during conventional backcross breeding approach occurs at the rate of 1 – (1/2)n+1, where “n” denotes the number of backcrossing (Allard, 1999). Here, marker-assisted background selection along with phenotypic selection helped in a rapid recovery of background genome in selected progenies. Recovery of RPG of selected progenies was 84.45–90.0% with the highest being 95.45%. Zunjare et al. (2018a) could also recover 83.86–92.98% of the background genome by using 114–133 polymorphic markers in maize. Higher RPG could be recovered in just two backcrosses compared to conventional backcross breeding programme (Muthusamy et al., 2014; Hossain et al., 2018; Sarika et al., 2018; Zunjare et al., 2018a).
Though all forms (α-, β-, γ-, and δ-) of tocopherols possess vitamin-E activity, α-tocopherol fractions possess six times more vitamin-E activity than γ-tocopherol (Das et al., 2020). Human liver preferentially absorbs α-tocopherol due to efficient hepatic α-tocopherol transfer protein (HTP) resulting in 10 times more accumulation than γ-tocopherol (Frank et al., 2012). Increased level of α-tocopherol with an increased proportion of α-/γ-tocopherol and α-/total tocopherol was recorded in each of the introgressed families. Maize genotypes carrying favorable haplotype of vte4 was reported to have a higher proportion of α-/γ-tocopherol and α-/total tocopherol than genotypes with unfavorable allele (Das et al., 2019a). Kernel α-tocopherol was enhanced by 1.82- to 2.22-fold in the MAS-derived inbreds and 1.85- to 2.49-folds among the reconstituted hybrids. Feng et al. (2015) reported 0.95- to 2.64-fold increase in α-tocopherol in the vte4 introgressed sweet corn inbreds, whereas Das et al. (2018) and Li et al. (2012) reported much higher concentration of α-tocopherol (4.3- and 3.2-fold, respectively) in selected maize genotypes with a favorable allele of vte4. InDel118 affects the level of transcription, and InDel7 regulates the translational efficiency of the vte4, thereby ensuring more kernel α-tocopherol (Li et al., 2012). In general, γ-tocopherol, δ-tocopherol, and total tocopherol were also increased in the vte4-based genotypes. This could be due to the higher flux of initial precursors of the pathway due to more conversion of γ-tocopherol to α-tocopherol in the last step of the tocopherol pathway. It is important to mention here that γ-tocopherol and δ-tocopherol also possess important functions as antioxidants (Evans et al., 2002). Thus, simultaneous enhancement of γ-tocopherol, δ-tocopherol, and total tocopherol along with α-tocopherol is beneficial for human health (Das et al., 2020). Kernel lysine and tryptophan in the introgressed progenies as well as in reconstituted hybrids were comparable to their respective recipient parents. Several mechanisms contribute to the enhancement of lysine and tryptophan in o2 genotypes, viz. (i) lysine-deficient zein fraction of protein is reduced with a consequent increase in lysine-rich non-zein proteins (Habben et al., 1994); (ii) reduction of transcription of lysine keto-reductase, which is a lysine-catabolizing enzyme (Kemper et al., 1999); and (iii) greater accumulation of several lysine-rich enzymes and proteins (Jia et al., 2013). The presence of favorable allele of crtRB1 and lcyE also led to the comparable but high concentration of proA in the introgressed progenies and hybrids. lcyE and crtRB1enhance kernel proA by regulating the pathway branching and hydroxylation, respectively (Harjes et al., 2008; Yan et al., 2010). Mutant crtRB1 and lcyE alleles produce lesser amount β-hydroxylase and β-cyclase enzymes, respectively, than the wild type, and result in a reduced conversion of β-carotene to downstream components and shifting more lycopene flux from α-branch to β-branch of the pathway (Harjes et al., 2008; Vallabhaneni et al., 2009; Yan et al., 2010). Though the frequency of co-occurrence of favorable alleles of crtRB1 and lcyE is very low in the maize germplasm (Azmach et al., 2013; Babu et al., 2013; Muthusamy et al., 2014; Gebremeskel et al., 2018), the greater combined effect of both over individual allele has been reported in a number of studies (Azmach et al., 2013; Zunjare et al., 2017; Gebremeskel et al., 2018). Further, the analysis revealed that a greater proportion of variations in nutritional quality traits were genetic in nature, and a minor variation was due to G × E interaction (α-T: 3.14%, γ-T: 3.40%, δ-T: 4.13%, TT: 3.32%, proA: 12.98%, lysine: 22.89%, and tryptophan: 10.77%). This suggested that these nutritional traits would not vary drastically when grown in a different environment (Hossain et al., 2018; Zunjare et al., 2018a). Recommended dietary allowance (RDA) for vitamin-E and vitamin-A is 10 mg/day and 4,800 μg/day for adults in humans, respectively (ICMR, 2010). While the RDA for lysine is 2,100 mg/day, the same for tryptophan is 288 mg/day for an adult of 60 kg weight (Gupta et al., 2015). Thus, the consumption of these biofortified maize hybrids (200 gm/day) can meet approximately the 32, 50, 38, and 55% of RDA for vitamin-E, vitamin-A, lysine, and tryptophan, respectively, as compared to 16, 8, 20, and 28% of normal maize (Dube et al., 2018).
Increased α-tocopherol content in each of these introgressed families provides direct evidence of the major effect of vte4 on the accumulation of α-tocopherol. However, a variation for kernel α-tocopherol within families even in the presence of the same favorable allele of vte4 could be due to background effect and differential interaction with genes in the pathway (Babu et al., 2013; Muthusamy et al., 2014). In contrast to the report of Li et al. (2012), introgressed progenies also revealed an increased quantity of γ-tocopherol and total tocopherol. Similar results were also reported by Feng et al. (2015) in the MAS-derived sweet corn inbreds with vte4. Most of the introgressed progenies except HKI161-VA-57-5-4 recorded kernel α-tocopherol concentration less than donor parent, which suggested the involvement of other minor quantitative trait loci (QTL)/genes, which possibly were not transferred from the donor line due to stringent background selection (Wong et al., 2003; Chander et al., 2008; Zhou et al., 2012). Rocheford et al. (2002), Wong et al. (2003), and Feng et al. (2013) detected minor QTL, which affects tocopherol composition in maize kernel. Though the selection for yield as such was not applied during the introgression process, the resemblance of reconstituted hybrids with original hybrids for grain yield was due to the high recovery of loci responsible for yield in recurrent parent through background selection (Hossain et al., 2018). However, interactions of a small fraction of the introgressed donor genome with the RPG resulted in a minor but significant difference in the grain yield of improved hybrids and respective original hybrids (Choudhary et al., 2014). Moreover, as grain yield is a quantitative trait, these variations could also be the influence of environment and genotype × environment. The α-tocopherol content of most of the reconstituted hybrids deviated from the mid-parent value of the introgressed inbred lines, suggesting interactions of various loci affecting tocopherol accumulation and non-additive gene action in the hybrids. Non-additive gene action for various tocopherol components in maize kernel has been reported by Das et al. (2019b). These newly derived hybrids rich in vitamin-E, vitamin-A, lysine, and tryptophan were high-yielding, and superior to the commercial heck, PVQ9I. These multinutrient-rich hybrids assume great significance in alleviating malnutrition through a holistic approach.
The benefit of biofortified maize hybrids for human health has been well-documented in several countries. However, studies on biofortification for kernel vitamin-E are limited in the case of maize. So far, maize hybrids rich in proA, lysine, and tryptophan have been developed and commercialized. However, this study is the first in reporting vitamin-E enrichment along with proA, lysine, and tryptophan in maize. Here, we demonstrated for the first time the development of multinutrient-rich maize with high vitamin-E, vitamin-A, lysine, and tryptophan by stacking of favorable alleles of vte4, o2, crtRB1, and lcyE in maize inbreds using MABB. These newly developed high-yielding maize hybrids with higher vitamin-E, vitamin-A, lysine, and tryptophan will help in addressing malnutrition in a more holistic and sustainable way.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Foreground and background selection in backcross populations was done by AD and HC. Phenotyping of introgressed inbreds and reconstituted hybrids was done by MG. Generation of backcross populations was done by VM and AD. Data analysis and interpretation were made by RZ. Biochemical analysis of tocopherols was performed by AD and HC. Biochemical analysis of carotenoids and amino acids was done by AB. Data recording of field experiments was done by VB and GC. Evaluation of multilocation trials was done by JB and SG. Protocol standardization was made by SS. Drafting of manuscript was made by AD, MG, VM, and FH. Design of experiments was made by FH, VM, and HG. Generation of funds was done by FH and HG. All authors contributed to the article and approved the submitted version.
Funding received from IARI, New Delhi through In-House project (CRSCIARISIL2014003) and Department of Biotechnology (DBT) through network project (BT/PR10922/AGII/106/944/2014) is thankfully acknowledged. MG acknowledges the receipt of Junior Research Fellowship of Indian Council of Agricultural Research (ICAR) during Master's programme.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
AD is thankful to Indian Council of Agricultural Research (ICAR), New Delhi, and ICAR-Indian Institute of Maize Research, Ludhiana, for providing the study leave for the doctoral programme. We thank CIMMYT-Mexico and HarvestPlus for sharing the donor inbred. Thanks are due to IIMR, Ludhiana, for providing the off-season nursery at Hyderabad. The help of Mr. Manish Kapasia, Technical Assistant, for the management of field activities and pollination programme is thankfully acknowledged.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.659381/full#supplementary-material
Andersson, M. S., Saltzman, A., Virk, P. S., et al. (2017). Progress update: crop development of biofortified staple food crops under Harvestplus. Afr. J. Food Agric. Nutr. Dev. 17, 11905–11935. doi: 10.18697/ajfand.78.HarvestPlus05
Arun, M., Subramanyam, K., Theboral, J., Sivanandhan, G., Rajesh, M., Dev, G. K., et al. (2014). Transfer and targeted overexpression of γ-tocopherol methyltransferase (γ-TMT) gene using seed-specific promoter improves tocopherol composition in Indian soybean cultivars. Appl. Biochem. Biotechnol. 172, 1763–1776. doi: 10.1007/s12010-013-0645-9
Azmach, G., Gedil, M., Menkir, A., and Spillane, C. (2013). Marker-trait association analysis of functional gene markers for provitamin A levels across diverse tropical yellow maize inbred lines. BMC Plant Biol. 13:227. doi: 10.1186/1471-2229-13-227
Babu, R., Rojas, N. P., Gao, S., Yan, J., and Pixley, K. (2013). Validation of the effects of molecular marker polymorphisms in LcyE and CrtRB1 on provitamin A concentrations for 26 tropical maize populations. Theor. Appl. Genet. 126, 389–399. doi: 10.1007/s00122-012-1987-3
Bain, L. E., Awah, P. K., Geraldine, N., Kindong, N. P., Siga, Y., Bernard, N., et al. (2013). Malnutrition in Sub–Saharan Africa: burden, causes and prospects. Pan Afr. Med. J. 15:120. doi: 10.11604/pamj.2013.15.120.2535
Black, R. E., Allen, L. H., Bhutta, Z. A., Caulfield, L. E., De Onis, M., Ezzati, M., et al. (2008). Maternal and child undernutrition: global and regional exposures and health consequences. Lancet 371, 243–260. doi: 10.1016/S0140-6736(07)61690-0
Bouis, H. E. (2018). “Reducing mineral and vitamin deficiencies through biofortification: progress under HarvestPlus”, in Hidden Hunger: Strategies to Improve Nutrition Quality, World Review of Nutrition and Dietetics, Vol. 118, eds H. K. Biesalski and R. Birner (Basel: Karger), 112–122. doi: 10.1159/000484342
Cabrera-Soto, L., Pixley, K. V., Rosales-Nolasco, A., Galicia-Flores, L. A., and Palacios-Rojas, N. (2018). Carotenoid and tocochromanol profiles during kernel development make consumption of biofortified “Fresh” maize an option to improve micronutrient nutrition. J. Agric. Food Chem.. 66, 9391–9398. doi: 10.1021/acs.jafc.8b01886
Chander, S., Guo, Y. Q., Yang, X. H., Yan, J. B., Zhang, Y. R., Song, T. M., et al. (2008). A Genetic dissection of tocopherol content and composition in maize grain using quantitative trait loci analysis and the candidate gene approach. Mol. Breed. 22, 353–365. doi: 10.1007/s11032-008-9180-8
Cho, E. A., Lee, C. A., Kim, Y. S., Baek, S. H., de los Reyes, B. G., and Yun, S. J. (2005). Expression of γ-tocopherol methyltransferase transgene improves tocopherol composition in lettuce (Latuca sativa L.). Mol. Cells 19, 16–22.
Choudhary, M., Muthusamy, V., Hossain, F., Thirunavukkarasu, N., Pandey, N., Jha, S. K., et al. (2014). Characterization of β-carotene rich MAS-derived maize inbreds possessing rare genetic variation in β-carotene hydroxylase gene. Ind. J. Genet. Plant Breed. 74, 620–623. doi: 10.5958/0975-6906.2014.00900.6
Collard, B. C. Y., Jahufer, M. Z, Brouwer, J. B., and Pang, E.C.K. (2005). An introduction to markers, quantitative trait loci (QTL) mapping and marker-assisted selection for crop improvement: the basic concepts. Euphytica 142, 169–196 doi: 10.1007/s10681-005-1681-5
Das, A. K., Chhabra, R., Muthusamy, V., Chauhan, H. S., Zunjare, R. U., and Hossain, F. (2019b). Identification of SNP and InDel variations in the promoter and 5′ untranslated regions of γ-tocopherol methyl transferase (ZmVTE4) affecting higher accumulation of α-tocopherol in maize kernel. Crop J. 7, 469–479. doi: 10.1016/j.cj.2019.01.004
Das, A. K., Jaiswal, S. K., Muthusamy, V., Zunjare, R. U., Chauhan, H. S., Chand, G., et al. (2018). Molecular diversity and genetic variability of kernel tocopherols among maize inbreds possessing favourable haplotypes of γ-tocopherol methyl transferase (ZmVTE4). J. Plant Biochem. Biotechnol. 28, 253–262. doi: 10.1007/s13562-018-0470-x
Das, A. K., Muthusamy, V., Zunjare, R. U., Baveja, A., Chauhan, H. S., Bhat, J. S., et al. (2020). Genetic variability for kernel tocopherols and haplotype analysis of γ-tocopherol methyl transferase (vte4) gene among exotic-and indigenous-maize inbreds. J. Food Composit. Anal. 88:103446. doi: 10.1016/j.jfca.2020.103446
Das, A. K., Muthusamy, V., Zunjare, R. U., Chauhan, H. S., Sharma, P. K., Bhat, J. S., et al. (2019a). Genetic variability-, genotype × environment interactions-and combining ability-analyses of kernel tocopherols among maize genotypes possessing novel allele of γ-tocopherol methyl transferase (ZmVTE4). J. Cereal Sci. 86, 1–8. doi: 10.1016/j.jcs.2018.12.018
Dube, N., Chandra, M. P., Hossain, F., Pullakhandam, R., Thingnganing, L., and Bhardwaj, D. (2018). β-carotene bioaccessibility from biofortified maize (Zea mays) is related to its density and is negatively influenced by lutein and zeaxanthin. Food Funct. 9, 379–388. doi: 10.1039/C7FO01034F
Evans, J. C., Kodali, D. R., and Addis, P. B. (2002). Optimal tocopherol concentrations to inhibit soybean oil oxidation. J. Am. Oil Chem. Soc. 79, 47–51. doi: 10.1007/s11746-002-0433-6
Feng, F., Deng, F., Zhou, P., Yan, J., Wang, Q., Yang, R., et al. (2013). QTL mapping for the tocopherols at milk stage of kernel development in sweet corn. Euphytica 193, 409–417. doi: 10.1007/s10681-013-0948-5
Feng, F., Wang, Q., Liang, C., Yang, R., and Li, X. (2015). Enhancement of tocopherols in sweet corn by marker-assisted backcrossing of ZmVTE4. Euphytica 206, 513–521. doi: 10.1007/s10681-015-1519-8
Frank, J., Chin, X. W. D., Schrader, C., Eckert, G. P., and Rimbach, G. (2012). Do tocotrienols have potential as neuroprotective dietary factors? Ageing Res. Rev. 11, 163–180. doi: 10.1016/j.arr.2011.06.006
Gannon, B., Kaliwile, C., Arscott, S. A., Schmaelzle, S., Chileshe, J., Kalungwana, N., et al. (2014). Biofortified orange maize is as efficacious as a vitamin A supplement in Zambian children even in the presence of high liver reserves of vitamin A: a community-based, randomized placebo-controlled trial. Am. J. Clin. Nutr. 100, 1541–1550. doi: 10.3945/ajcn.114.087379
Gebremeskel, S., Garcia-Oliveira, A. L., Menkir, A., Adetimirin, V., and Gedil, M. (2018). Effectiveness of predictive markers for marker assisted selection of pro-vitamin A carotenoids in medium-late maturing maize (Zea mays L.) inbred lines. J. Cereal Sci. 79, 27–34. doi: 10.1016/j.jcs.2017.09.001
Gunaratna, N. S., De-Groote, H., Nestel, P., Pixley, K. V., and Mc-Cabe, G. P. (2010). A meta-analysis of community-based studies on quality protein maize. Food Policy 35, 202–210. doi: 10.1016/j.foodpol.2009.11.003
Gupta, H. S., Hossain, F., and Muthusamy, V. (2015). Biofortification of maize: an Indian perspective. Ind. J. Genet. Plant Breed. 75, 1–22. doi: 10.5958/0975-6906.2015.00001.2
Habben, J. E., Moro, G. L., Lopes, M. A., Or, E., Hamaker, B., and Larkins, B. A. (1994). “Altered patterns of protein synthesis in opaque-2 maize endosperm”, in Plant Molecular Biology, eds G. Coruzzi and P. Puigdomènech (Berlin: Heidelberg Springer), 301–307. doi: 10.1007/978-3-642-78852-9_29
Harjes, C. E., Rocheford, T. R., Bai, L., Brutnell, T. P., Kandianis, C. B., Sowinski, S. G., et al. (2008). Natural genetic variation in lycopene epsilon cyclase tapped for maize biofortification. Science 319, 330–333. doi: 10.1126/science.1150255
Heying, E. K., Tanumihardjo, J. P., Vasic, V., Cook, M., Palacios-Rojas, N., and Tanumihardjo, S. A. (2014). Biofortified orange maize enhances β-cryptoxanthin concentrations in egg yolks of laying hens better than tangerine peel fortificant. J. Agric. Food Chem. 62, 11892–11900. doi: 10.1021/jf5037195
Hossain, F., Muthusamy, V., Pandey, N., Vishwakarma, A. K., Baveja, A., Zunjare, R. U., et al. (2018). Marker-assisted introgression of opaque2 allele for rapid conversion of elite hybrids into quality protein maize. J. Genet. 97, 287–298. doi: 10.1007/s12041-018-0914-z
Hossain, F., Sarika, K., Muthusamy, V., Zunjare, R. U., and Gupta, H. S. (2019). “Quality protein maize for nutritional security,” in Quality Breeding in Field Crops, eds A. M. Iqbal Qureshi, Z. A. Dar, and S. H. Wani (Cham: Springer), 217–237. doi: 10.1007/978-3-030-04609-5_11
ICMR (2010). Nutrient Requirements and Recommended Dietary Allowance for Indians. A Report of the Expert Group of the Indian Council of Medical Research, New Delhi.
Jia, M., Wu, H., Clay, K. L., Jung, R., Larkins, B. A., and Gibbon, B. C. (2013). Identification and characterization of lysine-rich proteins and starch biosynthesis genes in the opaque2 mutant by transcriptional and proteomic analysis. BMC Plant Biol. 13:60. doi: 10.1186/1471-2229-13-60
Kemper, E. L., Neto, G. C., Papes, F., Moraes, K. C. M., Leite, A., and Arruda, P. (1999). The role of opaque2 in the control of lysine-degrading activities in developing maize endosperm. Plant Cell. 11, 1981–1993. doi: 10.1105/tpc.11.10.1981
Kurilich, A. C., and Juvik, J. A. (1999). Quantification of carotenoid and tocopherol antioxidants in Zea mays. J. Agric. Food Chem. 47, 1948–1955. doi: 10.1021/jf981029d
Li, Q., Yang, X., Xu, S., Cai, Y., Zhang, D., Han, Y., et al. (2012). Genome-wide association studies identified three independent polymorphisms associated with α-tocopherol content in maize kernels. PLoS ONE 7:e36807. doi: 10.1371/journal.pone.0036807
Liu, L., Jeffers, D., Zhang, Y., Ding, M., Chen, W., Kang, M. S., et al. (2015). Introgression of the crtRB1 gene into quality protein maize inbred lines using molecular markers. Mol. Breed. 35, 1–12. doi: 10.1007/s11032-015-0349-7
Lu, H., Romero-Severson, J., and Bernardo, R. (2002). Chromosomal regions associated with segregation distortion in maize. Theor. Appl. Genet. 105, 622–628. doi: 10.1007/s00122-002-0970-9
Mene-Saffrane, L., and Pellaud, S. (2017). Current strategies for vitamin E biofortification of crops. Curr. Opin. Biotechnol. 44, 189–197. doi: 10.1016/j.copbio.2017.01.007
Mertz, E. T., Bates, L. S., and Nelson, O. E. (1964). Mutant gene that changes protein composition and increases lysine content of maize endosperm. Science 145, 279–280. doi: 10.1126/science.145.3629.279
Moreno, J. A., Díaz-Gómez, J., Nogareda, C., Angulo, E., Sandmann, G., Portero-Otin, M., et al. (2016). The distribution of carotenoids in hens fed on biofortified maize is influenced by feed composition, absorption, resource allocation and storage. Sci. Rep. 6, 1–11. doi: 10.1038/srep35346
Muthusamy, V., Hossain, F., Thirunavukkarasu, N., Choudhary, M., Saha, S., Bhat, J. S., et al. (2014). Development of β-carotene rich maize hybrids through marker-assisted introgression of β-carotene hydroxylase allele. PLoS ONE 9:e113583. doi: 10.1371/journal.pone.0113583
Neuffer, M. G., Coe, E. H., and Wessler, S. R. (1997). Mutants of Maize. New York, NY: Cold Spring Harbor Laboratory Press.
Nuss, E. T., and Tanumihardjo, S. A. (2010). Maize: a paramount staple crop in the context of global nutrition. Comprehen. Rev. Food Sci. Food Saf. 9, 417–436. doi: 10.1111/j.1541-4337.2010.00117.x
Odunitan-Wayas, F. A., Kolanisi, U., Chimonyo, M., and Siwela, M. (2016). Effect of provitamin A biofortified maize inclusion on quality of meat from indigenous chickens. J. Appl. Poult. Res. 25, 581–590. doi: 10.3382/japr/pfw040
Panda, A. K., Zaidi, P. H., Rama, R. S. V., and Raju, M. V. L. N. (2014). Efficacy of quality protein maize in meeting energy and essential amino acid requirements in broiler chicken production. J. Appl. Anim. Res. 42, 133–139. doi: 10.1080/09712119.2013.822812
Prasanna, B. M., Palacios-Rojas, N., Hossain, F., Muthusamy, V., Menkir, A., Dhliwayo, T., et al. (2020). Molecular breeding for nutritionally enriched maize: status and prospects. Front. Genet. 10:1392. doi: 10.3389/fgene.2019.01392
Rocheford , T. R, Wong, J. C., Egesel, C. O., and Lambert, R.J. (2002). Enhancement of vitamin E levels in corn. J. Am. Coll. Nutr. 21, 191S−198S. doi: 10.1080/07315724.2002.10719265
Saha, S., Walia, S., Kundu, A., and Pathak, N. (2013). Effect of mobile phase on resolution of the isomers and homologues of tocopherols on a triacontyl stationary phase. Anal. Bioanal. Chem. 405, 9285–9295. doi: 10.1007/s00216-013-7336-9
Sarika, K., Hossain, F., Muthusamy, V., Zunjare, R. U., Baveja, A., Goswami, R., et al. (2018). Opaque16, a high lysine and tryptophan mutant, does not influence the key physico-biochemical characteristics in maize kernel. PLoS ONE 13:e0190945. doi: 10.1371/journal.pone.0190945
Shiferaw, B., Prasanna, B. M., Hellin, J., and Bänziger, M. (2011). Crops that feed the world 6. Past successes and future challenges to the role played by maize in global food security. Food Secur. 3, 307–327. doi: 10.1007/s12571-011-0140-5
Shintani, D., and DellaPenna, D. (1998). Elevating the vitamin E content of plants through metabolic engineering. Science 282, 2098–2100. doi: 10.1126/science.282.5396.2098
Sowa, M., Yu, J., Palacios-Rojas, N., Goltz, S. R., Howe, J. A., Davis, C. R., et al. (2017). Retention of carotenoids in biofortified maize flour and β-cryptoxanthin-enhanced eggs after household cooking. ACS Omega 2, 7320–7328. doi: 10.1021/acsomega.7b01202
Vallabhaneni, R., Gallagher, C. E., Licciardello, N., Cuttriss, A. J., Quinlan, R. F., and Wurtzel, E. T. (2009). Metabolite sorting of a germplasm collection reveals the hydroxylase3 locus as a new target for maize provitamin A biofortification. Plant Physiol. 151, 1635–1645. doi: 10.1104/pp.109.145177
Van-Berloo, R. (1999). Computer note. GGT: software for the display of graphical genotypes. J. Hered. 90, 328–329. doi: 10.1093/jhered/90.2.328
Vasal, S. K. (2000). The quality protein maize story. Food Nutr. Bull. 21, 445–450. doi: 10.1177/156482650002100420
Vignesh, M., Hossain, F., Nepolean, T., Saha, S., Agrawal, P. K., Guleria, S. K., et al. (2012). Genetic variability for kernel β-carotene and utilization of crtRB1 3'TE gene for biofortification in maize (Zea mays L.). Indian J. Genet. 72, 189–194.
WHO (2009). Global Health Risks: Mortality and Burden of Disease Attributable to Selected Major Risks. Geneva: World Health Organization.
Wong, J. C., Lambert, R. J., Tadmor, Y., and andRocheford, T. R. (2003). QTL associated with accumulation of tocopherols in maize. Crop Sci. 43, 2257–2266. doi: 10.2135/cropsci2003.2257
Yadava, D. K., Hossain, F., and Mohapatra, T. (2018). Nutritional security through crop biofortification in India: status and future prospects. Indian J. Med. Res. 148, 621–631. doi: 10.4103/ijmr.IJMR_1893_18
Yan, J., Kandianis, C. B., Harjes, C. E., Bai, L., Kim, E. H., Yang, X., et al. (2010). Rare genetic variation at Zea mays crtRB1 increases β-carotene in maize grain. Nat. Genet. 42, 322–327. doi: 10.1038/ng.551
Zhou, Y., Han, Y., Li, Z., Fu, Y., Fu, Z., Xu, S., et al. (2012). ZmcrtRB3 encodes a carotenoid hydroxylase that affects the accumulation of α-carotene in maize. J. Integr. Plant Biol. 54, 260–269. doi: 10.1111/j.1744-7909.2012.01106.x
Zunjare, R. U., Chhabra, R., Hossain, F., Muthusamy, V., Baveja, A., and Gupta, H. S. (2018b). Development and validation of multiplex-PCR assay for simultaneous detection of rare alleles of crtRB1 and lcyE governing higher accumulation of provitamin-A in maize. J. Plant Biochem. Biotechnol. 27, 208–214. doi: 10.1007/s13562-017-0432-8
Zunjare, R. U., Chhabra, R., Hossain, F., Muthusamy, V., and Gupta, H. S. (2018c). Molecular characterization of 5'UTR of lycopene epsilon cyclase (lcyE) gene among exotic and indigenous inbreds for its utilization in maize biofortification. 3Biotech 8:75. doi: 10.1007/s13205-018-1100-y
Zunjare, R. U., Hossain, F., Muthusamy, V., Baveja, A., Chauhan, H. S., Bhat, J. S., et al. (2018a). Development of biofortified maize hybrids through marker-assisted stacking of β-carotene hydroxylase, lycopene-ε-cyclase and opaque2 genes. Front. Plant Sci. 9:178. doi: 10.3389/fpls.2018.00178
Zunjare, R. U., Hossain, F., Muthusamy, V., Baveja, A., Chauhan, H. S., Thirunavukkarasu, N., et al. (2017). Influence of rare alleles of β-carotene hydroxylase (crtRB1) and lycopene epsilon cyclase (lcyE) genes on accumulation of provitamin A carotenoids in maize kernels. Plant Breed. doi: 10.1111/pbr.12548
Keywords: maize, biofortication, nutrition, marker-assist selection, hybrid, DUS traits
Citation: Das AK, Gowda MM, Muthusamy V, Zunjare RU, Chauhan HS, Baveja A, Bhatt V, Chand G, Bhat JS, Guleria SK, Saha S, Gupta HS and Hossain F (2021) Development of Maize Hybrids With Enhanced Vitamin-E, Vitamin-A, Lysine, and Tryptophan Through Molecular Breeding. Front. Plant Sci. 12:659381. doi: 10.3389/fpls.2021.659381
Received: 27 January 2021; Accepted: 21 June 2021;
Published: 21 July 2021.
Edited by:
Sean Mayes, University of Nottingham, United KingdomReviewed by:
Mulatu Geleta, Swedish University of Agricultural Sciences, SwedenCopyright © 2021 Das, Gowda, Muthusamy, Zunjare, Chauhan, Baveja, Bhatt, Chand, Bhat, Guleria, Saha, Gupta and Hossain. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Firoz Hossain, ZmhfZ3BiQHlhaG9vLmNvbQ==
†Present Address: Abhijit K. Das, ICAR-Indian Institute of Maize Research, Ludhiana, India
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.