
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 07 April 2021
Sec. Plant Biotechnology
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.650820
The increase in the world population, the advent of new infections and health issues, and the scarcity of natural biological products have spotlighted the importance of recombinant protein technology and its large-scale production in a cost-effective manner. Microalgae have become a significant promising platform with the potential to meet the increasing demand for recombinant proteins and other biologicals. Microalgae are safe organisms that can grow rapidly and are easily cultivated with basic nutrient requirements. Although continuous efforts have led to considerable progress in the algae genetic engineering field, there are still many hurdles to overcome before these microorganisms emerge as a mature expression system. Hence, there is a need to develop efficient expression approaches to exploit microalgae for the production of recombinant proteins at convenient yields. This study aimed to test the ability of the DNA geminiviral vector with Rep-mediated replication to transiently express recombinant proteins in the freshwater microalgal species Chlamydomonas reinhardtii and Chlorella vulgaris using Agrobacterium-mediated transformation. The SARS-CoV-2 receptor binding domain (RBD) and basic fibroblast growth factor (bFGF) are representative antigen proteins and growth factor proteins, respectively, that were subcloned in a geminiviral vector and were used for nuclear transformation to transiently express these proteins in C. reinhardtii and C. vulgaris. The results showed that the geminiviral vector allowed the expression of both recombinant proteins in both algal species, with yields at 48 h posttransformation of up to 1.14 μg/g RBD and 1.61 ng/g FGF in C. vulgaris and 1.61 μg/g RBD and 1.025 ng/g FGF in C. reinhardtii. Thus, this study provides a proof of concept for the use of DNA viral vectors for the simple, rapid, and efficient production of recombinant proteins that repress the difficulties faced in the genetic transformation of these unicellular green microalgae. This concept opens an avenue to explore and optimize green microalgae as an ideal economically valuable platform for the production of therapeutic and industrially relevant recombinant proteins in shorter time periods with significant yields.
- A simple transformation method for Chlorella and Chlamydomonas is provided and feasible for scaling up recombinant protein production.
- The use of a geminiviral vector, which is normally used for transient plant transformation, has been successfully used for the expression of therapeutic proteins in algal systems.
- Microalgae are excellent, safe, and nutraceutically important organisms that are emerging as platforms for the development of various biological products.
Globally, there is a significant demand for the production of recombinant proteins due to their diversified properties in various applications, such as diagnosis, therapeutics, and industrial uses. Recombinant proteins are expressed in various hosts, such as bacteria (Swartz, 2001; Demain and Vaishnav, 2009; Ferrer-Miralles et al., 2009), yeast (Mattanovich et al., 2012; Kim et al., 2015), mammalian cells (Hacker and Balasubramanian, 2016; O’Flaherty et al., 2020), and plants (Mason and Arntzen, 1995; Cramer et al., 2000; Shanmugaraj et al., 2020a; Shanmugaraj and Phoolcharoen, 2021). Each expression system has its own advantages, disadvantages, and certain limitations, such as the availability of resources, inability to undergo molecular modifications, and requirement of sophisticated tools and techniques, resulting in toxic by-products and contamination risks (Barrera and Mayfield, 2013; Corchero et al., 2013). In the last decade, plants have emerged as an alternate platform for the expression of various functional recombinant proteins that have high therapeutic uses, with decreased manufacturing costs that are still in the pipeline for commercialization due to certain concerns related to environmental contamination, allergic reactions, and slow growth cycles (Salazar-González et al., 2015). Recombinant protein scale-ups in a safer way have drawn major attention to meet the current demand, with a focus on achieving higher quality and yields at mitigated production costs, thereby ensuring global coverage (Shanmugaraj and Phoolcharoen, 2021).
Microalgae comprise a diverse group of organisms with both prokaryotic and eukaryotic nature, making them an archetypal platform for recombinant technology. Microalgae are the source of a repertoire of natural biomolecules, with many of them having high protein, vitamin, and lipid contents, which have significant nutraceutical importance (Potvin and Zhang, 2010; Specht et al., 2010; Barrera and Mayfield, 2013; Yan et al., 2016; Bañuelos-Hernández et al., 2017; Ortega-Berlanga et al., 2018; Fayyaz et al., 2020; Sproles et al., 2021). Due to their ability to produce many compounds, algae have gained commercial and biotechnological interest, and some species are “generally regarded as safe” (GRAS) for their use as dietary supplements in humans and animals (Rosenberg et al., 2008). Microalgae represent the “best of both worlds” due to reasonable simple growth requirements with rapid growth and higher biomass and their ability to perform posttranslational modifications (Walker et al., 2005); thus, these are currently considered a promising platform for recombinant protein production with some particular advantages, including rapid transformation, no need for growth regulators, and the ability to properly synthesize complex proteins (Specht and Mayfield, 2014). Antigens from various pathogens, including the hepatitis B virus, Plasmodium falciparum, foot and mouth disease virus, Staphylococcus aureus, classical swine fever virus (CSFV), and influenza virus, have been produced in microalgae species, such as Chlamydomonas reinhardtii (freshwater), Dunaliella salina (marine), Schizochytrium spp. (marine), and Phaeodactylum tricornutum (Geng et al., 2003; Surzycki et al., 2009; Hempel et al., 2011; Gregory et al., 2012; Bayne et al., 2013). Photosynthetic unicellular algae, such as C. reinhardtii and Chlorella vulgaris, are most commonly used due to their high proteinaceous content with minimal nutrient requirements, such as salt-based media, carbon dioxide, and light, paving the way for much less difficult recombinant protein expression for many targeted applications (Potvin and Zhang, 2010; Tan et al., 2020). Microalgae are more advantageous for recombinant protein production, as they can be easily grown in contained bioreactors, minimizing the risk of airborne contaminants and protecting the environment from any potential flow of transgenes into the surroundings (Specht et al., 2010; Ahmad et al., 2020).
Successful transformation was achieved in microalgal species, such as Chlamydomonas, Chlorella, Volvox, Haematococcus, and Dunaliella, with very low levels of expression (Rosenberg et al., 2008). This may be due to barriers, such as thick cell walls, additional cellular membranes, target organelles, and the species being transformed (Barrera and Mayfield, 2013). Many transformation techniques, such as using cell wall-deficient strains (Hoffmann and Beck, 2005) and glass beads (Feng et al., 2008), electroporation (Feng et al., 2008), laboriously expensive mild cell disruption methods (Phong et al., 2018) such as bead milling (Günerken et al., 2015), ultrasonication (Piasecka et al., 2014), ionic liquids (Orr and Rehmann, 2016), microfluidization (Cha et al., 2011), pulsed arc technology (Goettel et al., 2013), cationic polymer-coated membranes (Yoo et al., 2014), and Agrobacterium-mediated transformation (Mayfield and Kindle, 1990; Kumar et al., 2004), have been developed for the stable nuclear expression of transgenes in a few microalgal species. Apart from these techniques, the transgene to be expressed requires codon optimization with construction in specific expression vectors using specialized promoters and selectable markers for higher expression levels in microalgae (Potvin and Zhang, 2010; Specht et al., 2010; Barrera and Mayfield, 2013). Although there has been a recent surge in the development of genetic engineering tools for microalgal transformation, one challenge for the field is to increase protein yields (Sproles et al., 2021). Hence, the present study aimed to develop an efficient transient expression approach for recombinant proteins using DNA viral vectors that target the nuclear genome in freshwater microalgal species.
The development and use of viral vectors have gained considerable attention in the recent decade for the heterologous expression of proteins in various hosts, most particularly in plant systems, and have emerged as a simple, easy, and effective mode of transformation with higher yields and good quality biological products (Chen et al., 2011; Dugdale et al., 2013; Salazar-González et al., 2015; Shanmugaraj et al., 2020a). One such system is based on geminiviral vectors that contain the replication elements Rep and Rep A from bean yellow dwarf virus (BeYDV), which are responsible for enhancing the expression levels of recombinant proteins. The vector backbone also has a cis-acting long intergenic region (LIR) and short intergenic region (SIR) elements with a duplicate enhancer and 35S CaMV constitutive promoter in the expression cassette. It has the 5′ untranslated region (UTR) from tobacco mosaic virus (TMV) and the P19 suppressor gene that is capable enough to target transgene expression in certain cases without the need for another vector. The use of geminiviral vectors has been proven efficient for transient expression (Chen et al., 2011) of various antigens, such as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2; Rattanapisit et al., 2020b); Ebola immune complex (Phoolcharoen et al., 2011); cholera toxin-B subunit; immunogen targeting the cytotoxic T-lymphocyte associated antigen-4 (Yiemchavee et al., 2021); growth factors, such as recombinant osteopontin (Rattanapisit et al., 2017, 2019), vascular endothelial growth factor (Bulaon et al., 2020), human epidermal growth factor (Hanittinan et al., 2020), and fibroblast growth factor (Rattanapisit et al., 2020a); and monoclonal antibodies (Shanmugaraj et al., 2020b), in Nicotiana benthamiana. Hence, these viral vectors, which have not been explored much in microalgae, can be used as an innovative approach in algal biotechnology to remove various barriers to nuclear transformation.
Here, the present study aimed to use the freshwater unicellular algal species C. vulgaris and C. reinhardtii, which are highly proteinaceous for genetically modifying their nuclear genome with viral vector transformation, using an antigen model SARS-CoV-2 receptor binding domain (SARS-CoV-2 RBD) and human basic fibroblast growth factor (bFGF) for biopharmaceutical expression. The SARS-CoV-2 RBD antigen and bFGF growth factor were particularly chosen because they have significant market demands and can be commercialized in the future by scaling up with this simple, rapid, and efficient nuclear transformation technique. We explored the potential of the geminiviral expression vector pBYR2eK2Md (pBYR2e) for nuclear transformation in microalgal species for recombinant protein production. In this context, both SARS-CoV-2 and bFGF were efficiently expressed by using the geminiviral vector in C. vulgaris and C. reinhardtii. The expression of recombinant SARS-CoV-2 and bFGF was determined by western blotting and quantified by enzyme-linked immunosorbent assay (ELISA) in both species. This study provides a proof of principle for the rapid production of biological compounds in microalgal species using DNA viral vectors that address the safety, cost, and easy scale-up in comparison with other production platforms and genetic engineering methods.
An axenic culture of C. reinhardtii was kindly provided by Professor Sergio Rosales Mendoza, UASLP, Mexico. C. vulgaris was obtained from the Thailand Institute of Scientific and Technological Research (TISTR), Pathum Thani, Thailand. C. reinhardtii and C. vulgaris were grown in Tris acetate phosphate (TAP) and f/2 media without silicates, respectively, from PhytoTech Labs. The two axenic microalgal cultures were incubated at 28 ± 1°C with cool fluorescent lamps at a photosynthetic photon flux density of 80–100 μmol/m2/s and a 16-h:8-h light:dark cycle on a rotary shaker (150 rpm) for 1 week. Every week thereafter, a subculture was started by routine transfer into fresh culture medium. For transformation, the C. reinhardtii and C. vulgaris species were inoculated at 5% (v/v) in fresh TAP and f/2 media and cultivated for 4–6 days at 150 rpm until growth approached the log phase (OD at 680 nm of 0.6–0.8).
The RBD region in the spike protein of SARS-CoV-2 (GenBank accession number: YP_009724390.1; F318-C617) was taken from NCBI and commercially synthesized (Genewiz, Suzhou, China). The bFGF nucleotide sequence (accession no: AAQ73204.1) was retrieved from GenBank and synthesized by GeneArt Gene Synthesis (Thermo Fisher Scientific). Furthermore, RBD and bFGF were fused with an 8X His tag at the C-terminus and cloned into a geminiviral vector (pBYR2e) by using XbaI and SacI restriction enzymes to create pBYR2e:SARS-CoV-2 RBD and pBYR2e:FGF, respectively. The expression vectors were transformed into Agrobacterium tumefaciens strain GV3101 by electroporation, and the resulting strains were confirmed by PCR (Rattanapisit et al., 2020a,b). Agrobacterium harboring the RBD and FGF genes was used for transient transformation in microalgal species.
The C. reinhardtii and C. vulgaris cultures were grown as per the conditions mentioned in section “Microalgae Growth Conditions.” Nuclear transformation was carried out in both C. reinhardtii and C. vulgaris species using 1.0 ml of A. tumefaciens culture harboring the SARS-CoV-2 RBD gene or bFGF gene with OD600 nm: 1.0 for 100 ml of each algal culture with OD680 nm: 0.6–0.8. Each culture medium was supplemented with 100 μM acetosyringone for efficient T-DNA transfer. Twenty-four hours postinfection, cefotaxime was added at a final concentration of 250 mg/l. The expression of the SARS-CoV-2 RBD and FGF in the transiently transformed microalgae was analyzed by collecting biomass samples at 24, 48, and 72 h postantibiotic addition. The cells were harvested by centrifugation at 5,000 rpm for 10 min at 4°C. The supernatant was discarded, and the pelleted biomass was used for protein extraction.
Recombinant protein expression was analyzed by western blotting. Cell samples collected at different time intervals were used for protein extraction. One hundred milligrams of fresh biomass was homogenized in 200 μl of extraction buffer containing 750 mM Tris–HCl at pH 8.0 with 15% sucrose, 100 mM β-mercaptoethanol, and 1 mM PMSF as described previously (Bañuelos-Hernández et al., 2017). The extracts were sonicated for four pulses for 10 s with an interval of 5 s. Samples were subsequently centrifuged at 8,000 rpm for 15 min, and the supernatants were collected for further analysis. Total soluble proteins (TSPs) in the extracts were determined by the Bradford method. TSP (15–20 μg, approximately 50 μl) was mixed with 4X reducing loading buffer, denatured by boiling for 10 min at 95°C, and subsequently subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis. Gels were blotted onto 0.45-μm PVDF nitrocellulose membranes. Protein transfer was performed using a Bio-Rad electroblotter for 1 h at 100 V in a methanol-based transfer buffer. The nitrocellulose blot was further processed for immune analysis. After blocking with 5% blocker (Bio-Rad) dissolved in 1X PBS for 1 h, the blots were incubated at 4°C overnight with a rabbit anti-His antibody tagged with HRP conjugate (ab1187, Abcam, United Kingdom) at a dilution of 1:5,000 in 3% fat-free milk dissolved in 1X PBS. The blot was washed three times with PBST (PBS containing 0.05% Tween 20). Antigen detection was revealed by incubating the blots with SuperSignal West Pico Plus Chemiluminescent Substrate solution following the instructions from the manufacturer (Thermo Fisher Scientific, http://www.thermoscientific.com) and exposing the film in the dark.
Severe acute respiratory syndrome coronavirus 2 RBD antigen (250 μg/ml) purified from recombinant N. benthamiana was used as the positive control. For quantitative ELISA, crude protein extracts from C. reinhardtii and C. vulgaris were diluted in 0.2 M carbonate buffer (pH 9.6). The diluted sample was added to an ELISA plate for protein adsorption overnight at 4°C. After blocking with 5% fat-free dry milk solution for 2 h at room temperature, the plates were incubated with anti-RBD serum (1:3,000) collected from sheep immunized with synthetic partial RBD peptide (CLKPFERDISTEIYQAGSTPCNGVEGFNCYFPLQ) overnight at 4°C. The plates were subsequently incubated with horseradish peroxidase-conjugated anti-sheep IgG at a dilution of 1:5,000 (A3415, Sigma-Aldrich) for 2 h at room temperature. A colorimetric reaction was induced by the addition of the TMB substrate solution (Promega, United States) with 30 min of incubation at 25°C. The absorbance values were measured at 450 nm. Standard curves were constructed using pure RBD protein from N. benthamiana to estimate the expression levels.
The bFGF expressed in transformed C. vulgaris and C. reinhardtii was quantified using the human FGF basic ELISA Kit (R&D Systems). The protocol was followed according to the manufacturer’s instructions.
The microalgae C. vulgaris and C. reinhardtii were grown in f/2 and TAP media, respectively, by adding 5% inoculum. The cultures were allowed to grow until the absorbance at 680 nm reached 0.8 and then transformed with Agrobacterium harboring SARS-CoV-2 RBD or bFGF in both microalgal species. The sequential flow representation and timeline for the production of recombinant proteins in microalgae are depicted in Figure 1. The geminiviral vector pBYR2e used for the current study drives the expression of transgenes under the control of the 35S promoter along with the suppressor P19 RNA silencing gene for higher protein expression (Diamos and Mason, 2019). The structural orientation of the vector pBYR2e used in this study is represented in Figure 2. Agrobacterium-mediated transformed cultures of C. vulgaris and C. reinhardtii with SARS-CoV-2 RBD and bFGF were collected at three time points (24, 48, and 72 h after antibiotic addition). The algal cultures were recovered by centrifugation, and the fresh biomass was weighed. The total soluble protein concentration was estimated by the Bradford assay using bovine serum albumin (Sigma) as the standard.
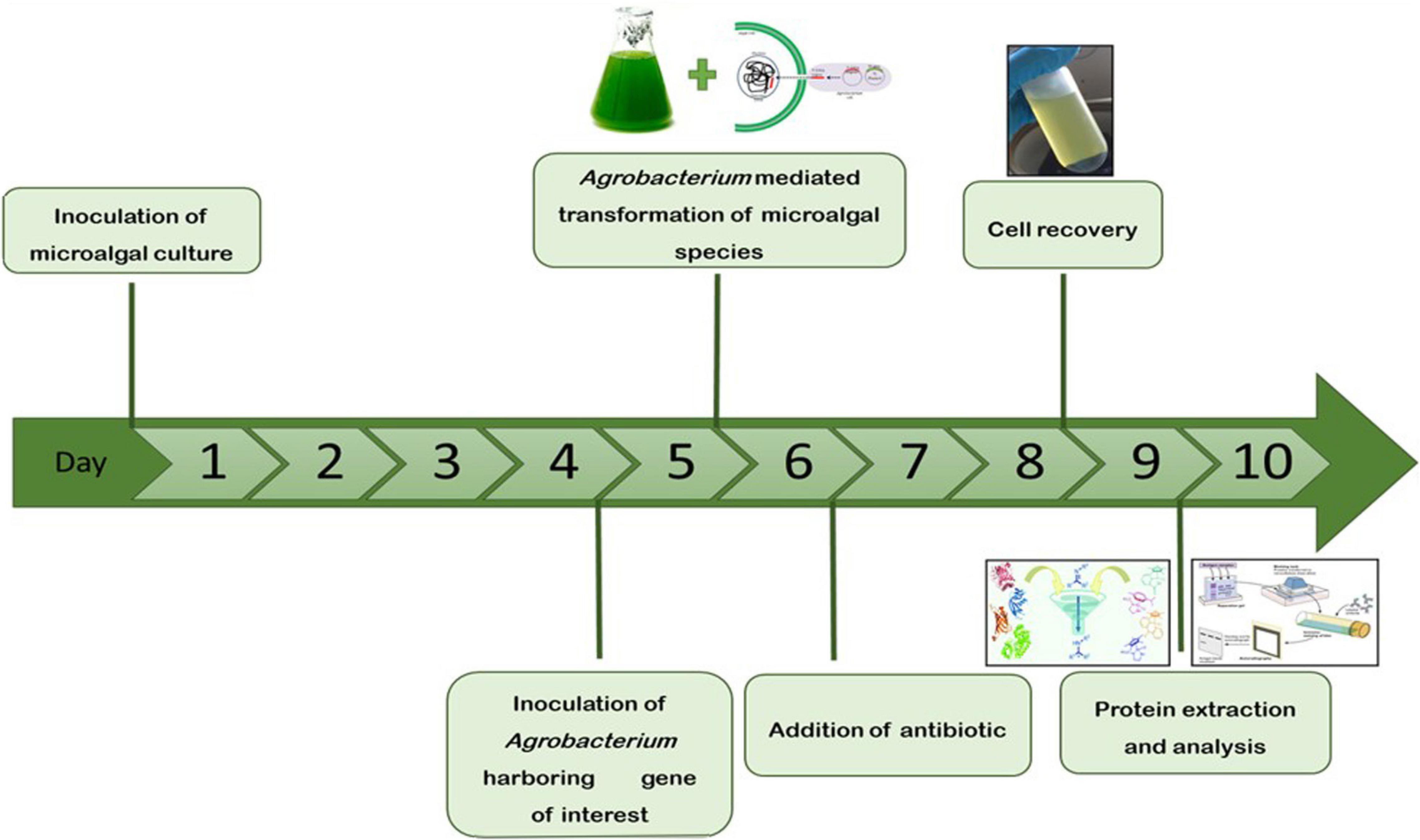
Figure 1. Schematic representation and timeline for the production of recombinant proteins in microalgae by transient gene expression.
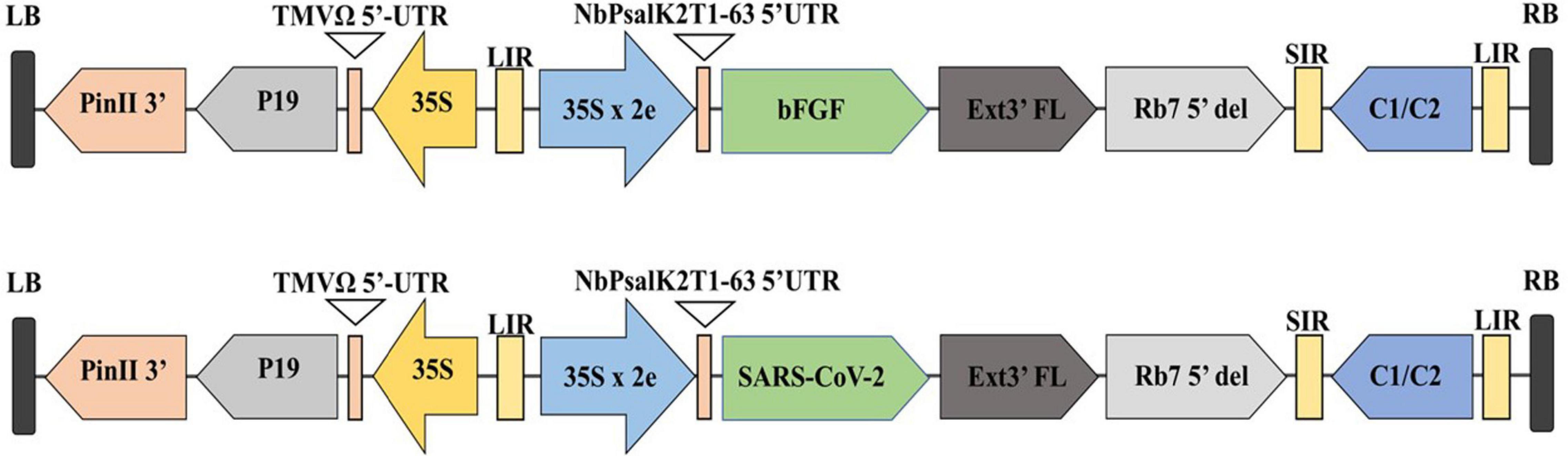
Figure 2. Diagrammatic representation of the T-DNA regions of the expression vector pBYR2eK2Md (pBYR2e). Modified from (Rattanapisit et al., 2020a,b). The bFGF and RBD genes were cloned into geminiviral vector using Xbal and Sad sites. RB and LB: The right and left borders of the T-DNA region in Agrobacterium; P35S: Cauliflower Mosaic Virus (CaMV) 35S promoter; P19: the RNA silencing suppressor from tomato bushy stunt virus (TBSV); NbPsalK2Tl-63 5′UTR: 5′ untranslated region; RBD: SARS-CoV-2 RBD; FGF: basic fibroblast growth factor gene; Ext3’FL: 3’ region of tobacco extension gene; Rb7 5′: 5′ of the Rb7 matrix attachment region/scaffold attachment region; SIR: short intergenic region of BeYDV: LIR: long intergenic region of BeYDV; C2/C1: Bean Yellow Dwarf Virus (BeYDV) ORFs Cl and C2 encoding for replication initiation protein (Rep) and Rep A: TMVfi 5′-UTR: 5′ untranslated region of tobacco mosaic virus Ω; Pinll 3’: the terminator from potato proteinase inhibitor II gene.
The fresh biomass collected from transformed cultures of C. vulgaris and C. reinhardtii with SARS-CoV-2 and bFGF was lysed and used for protein extraction. The expression of the recombinant RBD and FGF proteins was evaluated by SDS-PAGE and western blot analysis. The RBD protein was observed at the expected molecular weight of approximately 38 kDa in the western blot for both C. vulgaris and C. reinhardtii (Figure 3; lanes 2–4). The 3-day postinfiltrated leaf extract of N. benthamiana was used as the positive control (Figure 3; lane 5), and the wild-type extracts of both algae species did not show any protein expression (Figure 3; lane 1). RBD protein expression was observed in all the samples collected at 24, 48, and 72 h in both freshwater microalgal species and was further confirmed by immunoblotting assay.
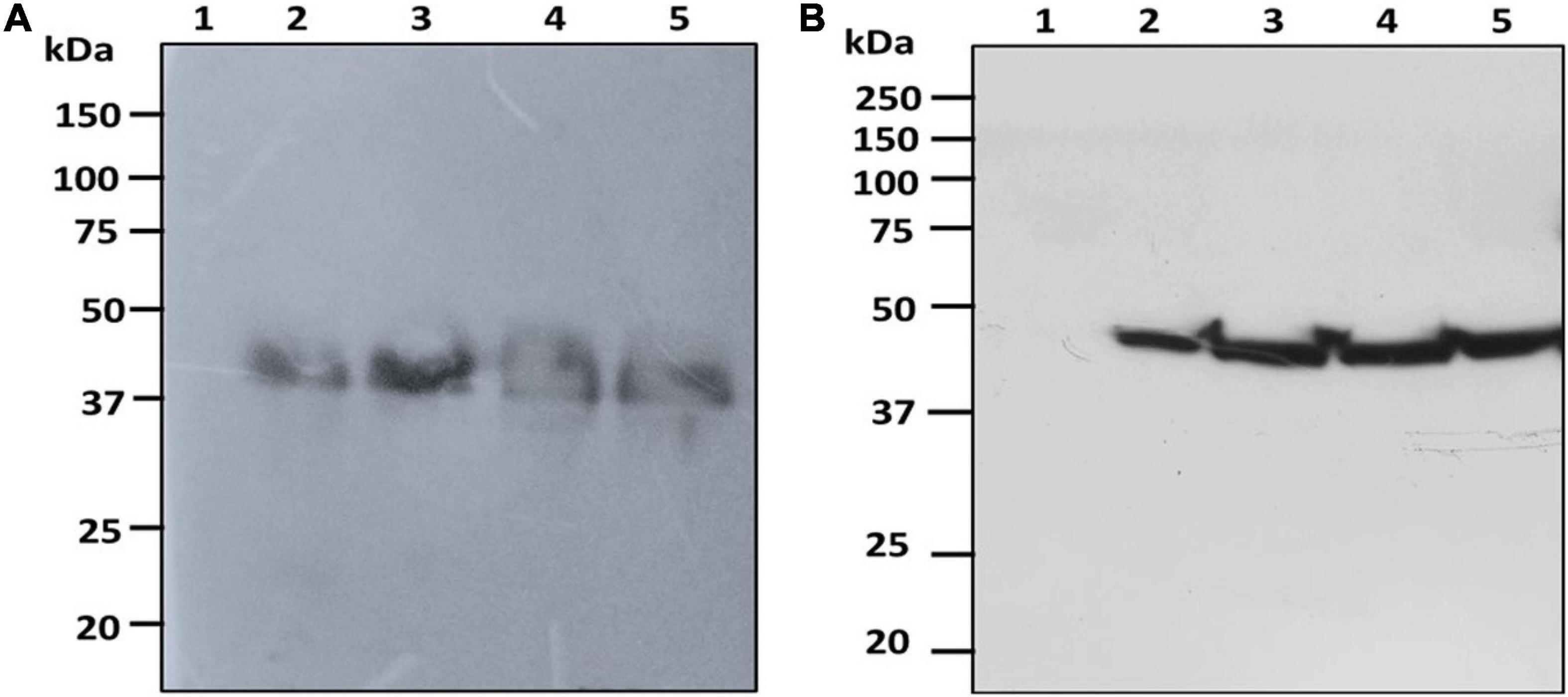
Figure 3. Western blot analysis of RBD protein of SARS-CoV-2 produced in microalgal species. The crude proteins were extracted from Chlorella (A) and Chlamydomonas (B) and the expression of RBD protein was analyzed on SDS-PAGE followed by Western blot probed with a rabbit anti-his antibody conjugated with HRP. Lane 1: Wild Type; Lane 2: Sample collected after 24 h of Agrobacterium transformation and cefotaxime addition; Lane 3: Sample collected after 48 h; Lane 4: Sample collected after 72 h; Lane 5: Positive control (Agro-infiltrated crude extract of N. benthamiana).
The pBYR2e:bFGF-transformed culture extracts of both C. vulgaris and C. reinhardtii were also analyzed by PAGE and western blotting using the rabbit anti-His antibody as an HRP conjugate for recombinant protein expression. The bFGF protein was observed at a molecular weight of approximately 24 kDa by western blotting for both C. vulgaris and C. reinhardtii (Figure 4; lanes 2–4). The extracts from both of the wild-type test algae did not show any protein expression (Figure 4; lane 1). FGF protein expression was low and was observed in all the samples collected at 24, 48, and 72 h in both microalgal species and was further quantified by ELISA. These results confirmed that geminiviral vectors can be used for the genetic transformation of green microalgal species. The expressed recombinant proteins were stable for a period of 1 month, and an in-depth molecular analysis needs to be performed to establish their strength, efficiency, and safety.
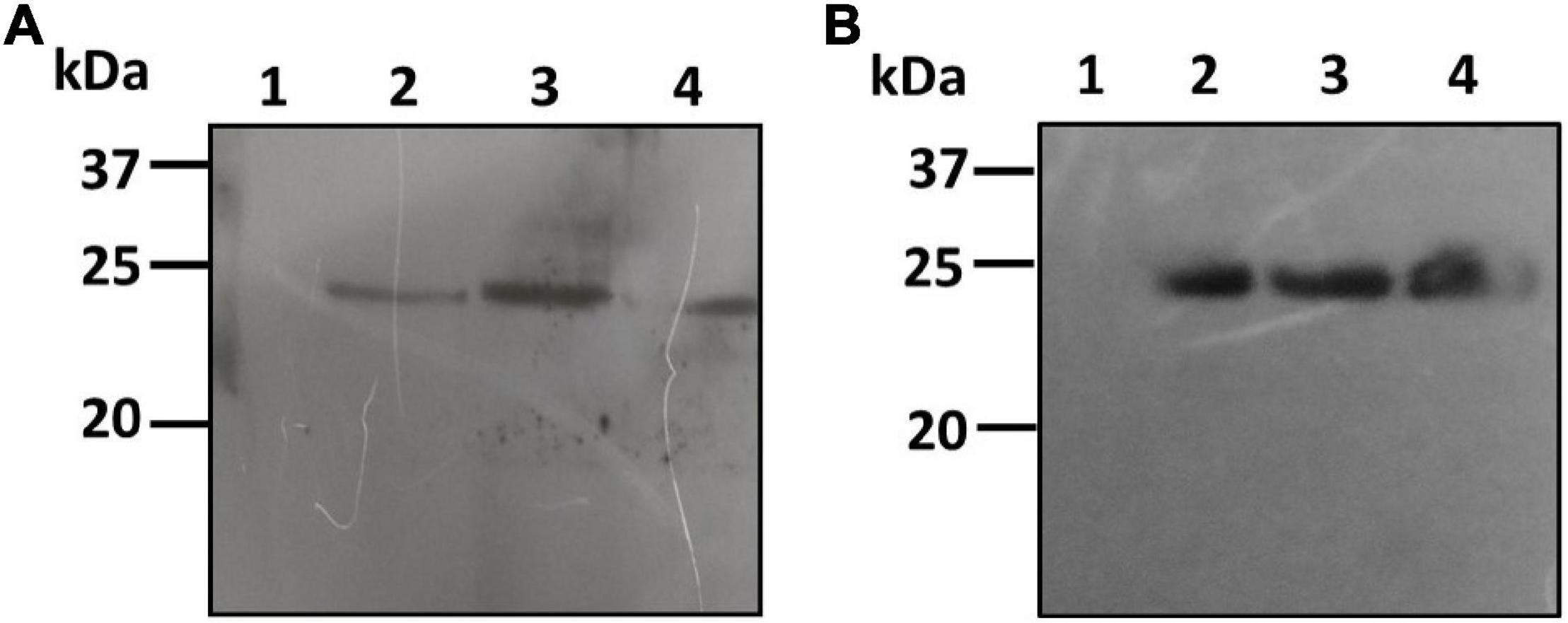
Figure 4. Western blot analysis of hFGF protein expressed microalgal species. The crude proteins were extracted from Chlorella (A) and Chlamydomonas (B) and the expression of hFGF protein was analyzed on SDS-PAGE followed by Western blot probed with a rabbit anti-his antibody conjugated with HRP. Lane 1: Wild Type; Lane 2: Sample collected after 24 h of Agrobacterium transformation and cefotaxime addition; Lane 3: Sample collected after 48 h; Lane 4: Sample collected after 72 h.
Basic fibroblast growth factor yields were determined by capture ELISA using a commercial kit. The bFGF protein yields at 24, 48, and 72 h reached 1.26, 1.61, and 0.365 ng/g fresh weight (FW) in C. vulgaris and 0.62, 1.025, and 0.895 ng/g FW in C. reinhardtii, respectively (Figure 5A). Although FGF expression was low, it accounted for 0.02–1% of TSP in both tested green algae, thereby confirming the authenticity and proper molecular channeling of the expressed protein.
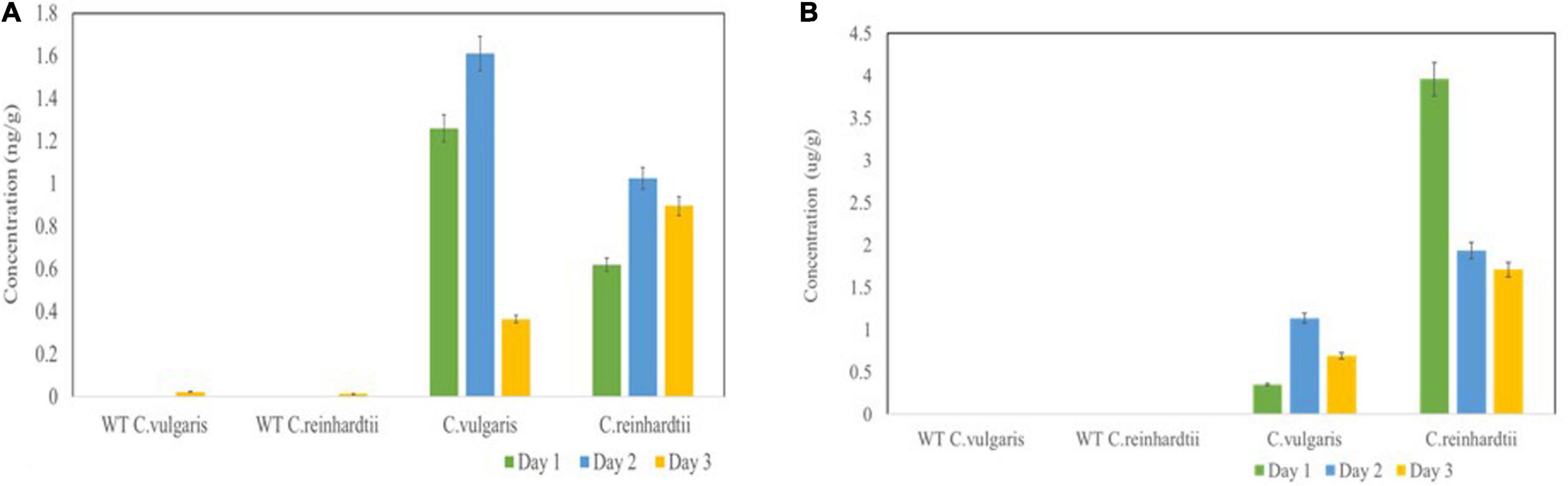
Figure 5. Expression levels of bFGF (A) and SARS-CoV-2 (B) in microalgal species was measured by ELISA. Microalgal species were transformed with Agrobacterium harboring the expression vector and the samples were collected on Day 1, 2, 3 and the expression was analyzed. The result was presented on the fresh weight (FW) basis. The data are expressed as mean ± standard deviation of two biological replicates with each sample taken in duplicates.
We also further quantified the recombinant RBD expressed in both tested algae species by ELISA using the purified RBD (125 μg/ml) from N. benthamiana as a standard. The RBD standard along with crude extracts from transformed C. vulgaris and C. reinhardtii was coated on 96-well plates and further incubated with anti-RBD serum from sheep immunized with a peptide derived from the RBD. The algae-made RBD showed specific binding with the anti-RBD antibody, whereas negative controls did not show any binding. These results confirmed that the use of DNA viral vectors can be used to produce recombinant antigens in green algae. The transformed C. vulgaris accumulated the RBD protein at concentrations of 0.35, 1.14, and 0.69 μg/g FW at 24, 48, and 72 h, respectively (Figure 5B), showing a high level of expression on day 2. C. reinhardtii showed a higher expression of RBD at concentrations of 3.96, 1.94, and 1.71 μg/g FW on days 1, 2, and 3, respectively (Figure 5B). Overall, the recombinant RBD protein expressed in the tested algae species ranged from 0.43 to 3.6% TSP. For the quantification studies, two biological replicates were performed, and each sample was subjected to two technical replicates. The data are presented as the means ± standard deviation.
The demand and use of recombinant proteins have become powerful tools globally due to their wide significance and important application in industry, diagnosis, and therapeutics (Rosales-Mendoza et al., 2020a). Hence, there is an immense need for the development and production of recombinant proteins in large quantities using various eukaryotic expression systems to provide several solutions, such as cost reduction and bioavailability of these biological substances in low- and middle-income countries in a safer and efficient way, by expanding the potential biosystems (Rosales-Mendoza et al., 2020b). Biopharmaceuticals are mainly produced in expression systems that include mammalian cells, bacteria, insect cells, yeast, and plants that are capable of meeting market demands with sufficient production levels but with some limitations (Shih and Doran, 2009; Dhara et al., 2018; Kis et al., 2019). The most commonly used expression system involves Chinese hamster ovary cells and plants for the production of various complex biomolecules that include vaccines, antibodies, hormones, clotting factors, enzymes, growth factors, and cytokines. The mammalian system requires complex nutritional requirements that involve high production costs with a high chance of contamination, requiring stringent sterility maintenance, whereas plant systems are not expensive, but there are certain technical and regulatory concerns for commercial scale production that remain to be addressed (Chen and Davis, 2016; Dhara et al., 2018; Burnett and Burnett, 2020; Shanmugaraj et al., 2020a).
Microalgae are diverse unicellular eukaryotic organisms with interesting properties that make them attractive hosts for recombinant protein production (Specht et al., 2010; Specht and Mayfield, 2014); these organisms are a rich source of high-value compounds, such as proteins, lipids, polyunsaturated fatty acids, and carotenoids, involved in the light-driven synthesis of therapeutic compounds at lower operating costs for upstream and downstream processes (Ahmad et al., 2020; Rosales-Mendoza et al., 2020b). Photosynthetic microalgae, such as C. reinhardtii and C. vulgaris, are considered safe for consumption as dietary supplements and form an attractive platform for the synthesis of high-value heterologous proteins due to their many beneficial attributes, such as ease of cultivation, lack of pathogens, and simple media components, allowing them to easily grow in containment photobioreactors under sterile conditions (Rosenberg et al., 2008; Potvin and Zhang, 2010; Specht et al., 2010; Yaakob et al., 2014; Rasala and Mayfield, 2015; Salazar-González et al., 2015). However, recombinant protein production levels are frequently very low, and thus, refinement of expression technologies is needed to enhance yields.
Hence, the present study was focused on the use of viral vectors for transient nuclear expression of recombinant proteins, which has not been explored previously in C. reinhardtii and C. vulgaris. A geminiviral expression system was selected based on replication elements from the BeYDV and the CaMV 35S promoter and enhancer (Chen et al., 2011; Diamos and Mason, 2019). The pBYR2e vector carrying the SARS-CoV-2 and bFGF proteins in this study was successfully transformed in N. benthamiana, a tobacco species with yields of 8 and 2.16 μg/g FW, respectively, 3 days postinfiltration (Rattanapisit et al., 2020a,b). The expression levels of recombinant proteins produced are high in plants, as the vector is particularly designed for plant systems, but the regulations, environmental concerns, and safety approval for oral consumption still need to be resolved (Streatfield, 2005; Laere et al., 2016).
The microalgal species C. reinhardtii and C. vulgaris were transiently transformed with Agrobacterium strains carrying the geminiviral vector called pBYR2e coding for the proteins of interest. To assess the potential of the expression system based on DNA replicons, we selected two biopharmaceuticals: SARS-CoV-2 RBD, which is an antigen derived from the spike protein of the novel coronavirus, and bFGF, which is a growth factor. These proteins were successfully expressed in both photosynthetic microalgal species, thereby proving the proof of principle to use these DNA viral vectors as a functional and efficient approach for transient expression of proteins without the involvement of any laborious processes. In a similar study by Bañuelos-Hernández et al. (2017), the Algevir system for transient expression of GP1 from the Zaire Ebola virus and LTB from Escherichia coli were assessed in Schizochytrium species, revealing the robustness of using viral vectors that employ viral replication elements for microalgal transformation (Bañuelos-Hernández et al., 2017). The maximum expression levels were attained for the SARS-CoV-2 RBD after 48 h of transformation, reaching maximum yields of 1.14 and 1.94 μg/g FW in C. vulgaris and C. reinhardtii, respectively. FGF expression was also found to be highest after 2 days of transformation with accumulation of 1.61 and 1.025 ng/g FW in C. vulgaris and C. reinhardtii, respectively. The Schizochytrium species that was transformed with the Algevir vector system showed maximum expression levels after 48 h postinduction with accumulation of 1.25 mg/g FW for GP1 and 0.12 mg/g FW for LTB (Bañuelos-Hernández et al., 2017). The recombinant human CL4mAb against the hepatitis B surface antigen (HBsAg) fused to green fluorescent protein and a tetrapeptide was expressed in the P. tricornutum nuclear genome, accumulating up to 8.7% TSP (Hempel et al., 2011). In the C. reinhardtii nuclear genome, the human erythropoietin (EPO) protein was expressed with the hsp70A/rbcS2 chimeric promoter (Eichler-Stahlberg et al., 2009). The expression of HIV-1 protein P24, which is a vaccine candidate for AIDS, was successfully achieved in Chlamydomonas using the PSAD promoter with 0.25% of the total cellular protein (Barahimipour et al., 2016). P. tricornutum was also successful in expressing recombinant mAb against a nucleoprotein part of the potent Marburg virus (Hempel et al., 2017). However, the expression of these recombinant proteins in algae is not comparable with those produced in tobacco plant systems. However, our study is the first report that has proven that the same plant vector can be used in microalgae transformation and that host-specific codon optimization and other genome editing techniques (Sproles et al., 2021) could enhance the yield of expression levels, which need to be further investigated.
One strength of using green algae is the possibility of implementing the delivery of therapeutic proteins orally. Algal cells are known to be natural green factories that can emerge as excellent organisms with highly valuable and significant products through gene transformations (Georgianna and Mayfield, 2012; Dyo and Purton, 2018). These eukaryotic microorganisms, which are regarded as safe, unlike the bacterial system, are capable of undergoing complex protein folding and modifications to form active proteins that are specifically important for human use as antigens and antibodies (Ma et al., 2020). Each system needs to be evaluated for its attributes before developing an oral vaccine delivery platform (Rosales-Mendoza et al., 2016). From this study, we predict the use of DNA viral vectors to transiently transform essential edible microalgae for the expression of various therapeutically important proteins and use them as oral delivery vehicles. It is only a preliminary yet significant concept, and furthermore, detailed studies are needed to test its efficacy, safety, and limitations.
Although nuclear and chloroplast transformation methods are available for microalgal species, these technologies have been frequently associated with low yields and genetic instability of transformed clones associated with gene silencing and genetic rearrangements and require lengthy processes for the transformation and selection of candidate clones. Advances in technologies that target chloroplast transformation have led to improvements in protein yields (Mayfield and Kindle, 1990; Surzycki et al., 2009; Specht and Mayfield, 2014). Chloroplast-based expression offers high protein productivity due to the high expression derived from the high copy number of transformed genomes per chloroplast and the general absence of gene silencing with no position effects; the integration is site-directed and mediated by homologous recombination (Bock and Warzecha, 2010). A limitation to consider for chloroplast-expressed biopharmaceuticals is the lack of complex posttranslational modifications, such as glycosylation, that are relevant for some biopharmaceuticals. In addition, it should be considered that protein secretion into culture media is not possible under chloroplast-based expression. Currently, yields in chloroplast expression are typically in the 0.02–5% TSP range with notable exceptions (42% of TSP of VP28) (Surzycki et al., 2009). Few other algal species have shown yields up to 3 mg/l of bovine milk amyloid A protein by chloroplast expression and 15 mg/l of recombinant protein secretion in culture media by nuclear expression (Lauersen et al., 2013; Gimpel et al., 2015; Ramos-Vega et al., 2018). Recombinant hemagglutinin proteins from the influenza virus have been expressed in the microalga Schizochytrium sp. in a secretion modality with yields up to 20 mg/l (Bayne et al., 2013).
In comparison, the use of viral vectors for transient expression in microalgae is considered more attractive since its features could overcome some of the limitations of the stable nuclear and chloroplast approaches. Nuclear transformation allows the cellular machinery to undergo molecular modifications, whereas the intracellular accumulation of antigens allows the whole cell to serve as a vaccine delivery vehicle along with the host’s endogenous compounds (Bañuelos-Hernández et al., 2017). In addition, the present study involves the use of the geminiviral vector pBYR2e, which offers enhanced expression of target proteins in the presence of duplicate enhancers and offers a short production time period.
In summary, the use of DNA viral vectors for genetic engineering in algal species for transient expression of therapeutic proteins is a versatile method that has many advantages, such as a short production time with significant protein yields without the involvement of cumbersome procedures, such as stable nuclear and chloroplast transformation techniques. Another advantage is that there will be no genetic instability of the stable transformants, as is the case of nuclear and chloroplast stable transformation of microalgae (Bañuelos-Hernández et al., 2017). Therefore, the DNA viral vector system was initially tested with the photosynthetic unicellular algal species C. vulgaris and C. reinhardtii, which can be easily grown on a large scale and employ transient transformation (Qu et al., 2013; Song et al., 2015). In conclusion, this study provides a proof of concept for the use of DNA viral vectors with replication elements as a robust approach for the simple and efficient transient expression of recombinant proteins in microalgae, which will open new avenues in the genetic engineering of other microalgal species. The expression levels of SARS-CoV-2 RBD and bFGF in both green algal species were significant, paving the way for scale-up and detailed studies on potency, strength, and safety in a cost-effective manner to reach market targets. Furthermore, these edible green Chlamydomonas and Chlorella species can be employed for the production of other valuable metabolites using geminiviral vector transformation with advanced genome editing methods to enhance the quality and yields of the bioproducts, thus economically and environmentally broadening the feasibility of the algal platform.
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
SV conceived and designed the experiments and revised the manuscript. SV, SR-M, and WP supervised the study. AM carried out the transformation in microalgal species and performed the protein expression and quantification by ELISA, analyzed the data, and wrote the manuscript. All authors read and approved the final version of the manuscript.
This research project was supported by Rachadapisek Sompote Fund for Innovation No. CU_GI_62_08_33_01. AM is a recipient of the Second Century Fund (C2F), Chulalongkorn University.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
AM would like to thank Dr. Balamurugan Shanmugaraj for his valuable insight and Dr. Kaewta Rattanapisit for her kind support during the period of research.
Ahmad, N., Mehmood, M. A., and Malik, S. (2020). Recombinant protein production in microalgae: emerging trends. Protein Pept. Lett. 27, 105–110. doi: 10.2174/0929866526666191014124855
Bañuelos-Hernández, B., Monreal-Escalante, E., González-Ortega, O., Angulo, C., and Rosales-Mendoza, S. (2017). Algevir: an expression system for microalgae based on viral vectors. Front. Microbiol. 8:1100. doi: 10.3389/fmicb.2017.01100
Barahimipour, R., Neupert, J., and Bock, R. (2016). Efficient expression of nuclear transgenes in the green alga Chlamydomonas: synthesis of an HIV antigen and development of a new selectable marker. Plant Mol. Biol. 90, 403–418. doi: 10.1007/s11103-015-0425-8
Barrera, D. J., and Mayfield, S. P. (2013). “High-value recombinant protein production in microalgae,” in Handbook of Microalgal Culture :Applied Phycology and Biotechnology, 2nd Edn, eds A. Richmond and Q. Hu, (London: John Wiley & Sons), 532–544. doi: 10.1002/9781118567166.ch27
Bayne, A. C., Boltz, D., Owen, C., Betz, Y., Maia, G., Azadi, P., et al. (2013). Vaccination against influenza with recombinant hemagglutinin expressed by Schizochytrium sp. confers protective immunity. PLoS One 8:e61790. doi: 10.1371/journal.pone.0061790
Bock, R., and Warzecha, H. (2010). Solar-powered factories for new vaccines and antibiotics. Trends Biotechnol. 28, 246–252. doi: 10.1016/j.tibtech.2010.01.006
Bulaon, C. J. I., Shanmugaraj, B., Oo, Y., Rattanapisit, K., Chuanasa, T., Chaotham, C., et al. (2020). Rapid transient expression of functional human vascular endothelial growth factor in Nicotiana benthamiana and characterization of its biological activity. Biotechnol. Rep. 27:e00514. doi: 10.1016/j.btre.2020.e00514
Burnett, M. J. B., and Burnett, A. C. (2020). Therapeutic recombinant protein production in plants: challenges and opportunities. Plants People Planet 2, 121–132. doi: 10.1002/ppp3.10073
Cha, K. H., Lee, J. Y., Song, D. G., Kim, S. M., Lee, D. U., Jeon, J. Y., et al. (2011). Effect of microfluidization on in vitro micellization and intestinal cell uptake of lutein from Chlorella vulgaris. J. Agric. Food Chem. 59, 8670–8674. doi: 10.1021/jf2019243
Chen, Q., and Davis, K. R. (2016). The potential of plants as a system for the development and production of human biologics. F1000Res. 5:F1000 Faculty Rev-192.
Chen, Q., He, J., Phoolcharoen, W., and Mason, H. S. (2011). Geminiviral vectors based on bean yellow dwarf virus for production of vaccine antigens and monoclonal antibodies in plants. Hum. Vaccin. 7, 331–338. doi: 10.4161/hv.7.3.14262
Corchero, J. L., Gasser, B., Resina, D., Smith, W., Parrilli, E., Vázquez, F., et al. (2013). Unconventional microbial systems for the cost-efficient production of high-quality protein therapeutics. Biotechnol. Adv. 31, 140–153. doi: 10.1016/j.biotechadv.2012.09.001
Cramer, C. L., Boothe, J. G., and Oishi, K. K. (2000). “Transgenic plants for therapeutic proteins: linking upstream and downstream strategies,” in Plant Biotechnology: New Products and Applications, eds J. Hammond, P. Mcgarvey, and V. Yusibov, (Berlin: Springer Berlin Heidelberg), 95–118. doi: 10.1007/978-3-642-60234-4_5
Demain, A. L., and Vaishnav, P. (2009). Production of recombinant proteins by microbes and higher organisms. Biotechnol. Adv. 27, 297–306. doi: 10.1016/j.biotechadv.2009.01.008
Dhara, V. G., Naik, H. M., Majewska, N. I., and Betenbaugh, M. J. (2018). Recombinant antibody production in CHO and NS0 cells: differences and similarities. BioDrugs 32, 571–584. doi: 10.1007/s40259-018-0319-9
Diamos, A. G., and Mason, H. S. (2019). Modifying the replication of geminiviral vectors reduces cell death and enhances expression of biopharmaceutical proteins in Nicotiana benthamiana Leaves. Front. Plant Sci. 9:1974. doi: 10.3389/fpls.2018.01974
Dugdale, B., Mortimer, C. L., Kato, M., James, T. A., Harding, R. M., and Dale, J. L. (2013). In plant activation: an inducible, hyperexpression platform for recombinant protein production in Plants. Plant Cell 25, 2429–2443. doi: 10.1105/tpc.113.113944
Dyo, Y. M., and Purton, S. (2018). The algal chloroplast as a synthetic biology platform for production of therapeutic proteins. Microbiology 164, 113–121. doi: 10.1099/mic.0.000599
Eichler-Stahlberg, A., Weisheit, W., Ruecker, O., and Heitzer, M. (2009). Strategies to facilitate transgene expression in Chlamydomonas reinhardtii. Planta 229, 873–883. doi: 10.1007/s00425-008-0879-x
Fayyaz, M., Chew, K. W., Show, P. L., Ling, T. C., Ng, I. S., and Chang, J.-S. (2020). Genetic engineering of microalgae for enhanced biorefinery capabilities. Biotechnol. Adv. 43:107554. doi: 10.1016/j.biotechadv.2020.107554
Feng, S., Xue, L., Liu, H., and Lu, P. (2008). Improvement of efficiency of genetic transformation for Dunaliella salina by glass beads method. Mol. Biol. Rep. 36, 1433–1439. doi: 10.1007/s11033-008-9333-1
Ferrer-Miralles, N., Domingo-Espín, J., Corchero, J. L., Vázquez, E., and Villaverde, A. (2009). Microbial factories for recombinant pharmaceuticals. Microb. Cell Fact. 8:17. doi: 10.1186/1475-2859-8-17
Geng, D., Wang, Y., Wang, P., Li, W., and Sun, Y. (2003). Stable expression of hepatitis B surface antigen gene in Dunaliella salina (Chlorophyta). J. Appl. Phycol. 15, 451–456. doi: 10.1023/b:japh.0000004298.89183.e5
Georgianna, D. R., and Mayfield, S. P. (2012). Exploiting diversity and synthetic biology for the production of algal biofuels. Nature 488, 329–335. doi: 10.1038/nature11479
Gimpel, J. A., Hyun, J. S., Schoepp, N. G., and Mayfield, S. P. (2015). Production of recombinant proteins in microalgae at pilot greenhouse scale. Biotechnol. Bioeng. 112, 339–345. doi: 10.1002/bit.25357
Goettel, M., Eing, C., Gusbeth, C., Straessner, R., and Frey, W. (2013). Pulsed electric field assisted extraction of intracellular valuables from microalgae. Algal Res. 2, 401–408. doi: 10.1016/j.algal.2013.07.004
Gregory, J. A., Li, F., Tomosada, L. M., Cox, C. J., Topol, A. B., Vinetz, J. M., et al. (2012). Algae-produced Pfs25 elicits antibodies that inhibit malaria transmission. PLoS One 7:e37179. doi: 10.1371/journal.pone.0037179
Günerken, E., D’hondt, E., Eppink, M. H., Garcia-Gonzalez, L., Elst, K., and Wijffels, R. H. (2015). Cell disruption for microalgae biorefineries. Biotechnol. Adv. 33, 243–260. doi: 10.1016/j.biotechadv.2015.01.008
Hacker, D. L., and Balasubramanian, S. (2016). Recombinant protein production from stable mammalian cell lines and pools. Curr. Opin. Struct. Biol. 38, 129–136. doi: 10.1016/j.sbi.2016.06.005
Hanittinan, O., Oo, Y., Chaotham, C., Rattanapisit, K., Shanmugaraj, B., and Phoolcharoen, W. (2020). Expression optimization, purification and in vitro characterization of human epidermal growth factor produced in Nicotiana benthamiana. Biotechnol. Rep. 28:e00524. doi: 10.1016/j.btre.2020.e00524
Hempel, F., Lau, J., Klingl, A., and Maier, U. G. (2011). Algae as protein factories: expression of a human antibody and the respective antigen in the diatom Phaeodactylum tricornutum. PLoS One 6:e28424. doi: 10.1371/journal.pone.0028424
Hempel, F., Maurer, M., Brockmann, B., Mayer, C., Biedenkopf, N., Kelterbaum, A., et al. (2017). From hybridomas to a robust microalgal-based production platform: molecular design of a diatom secreting monoclonal antibodies directed against the Marburg virus nucleoprotein. Microb. Cell Fact. 16:131.
Hoffmann, X. K., and Beck, C. F. (2005). Mating-induced shedding of cell walls, removal of walls from vegetative cells, and osmotic stress induce presumed cell wall genes in Chlamydomonas. Plant Physiol. 139, 999–1014. doi: 10.1104/pp.105.065037
Kim, H., Yoo, S. J., and Kang, H. A. (2015). Yeast synthetic biology for the production of recombinant therapeutic proteins. FEMS Yeast Res. 15, 1–16. doi: 10.1007/978-1-61779-770-5_1
Kis, Z., Shattock, R., Shah, N., and Kontoravdi, C. (2019). Emerging technologies for low-cost, rapid vaccine manufacture. Biotechnol. J. 14:1800376. doi: 10.1002/biot.201800376
Kumar, S. V., Misquitta, R. W., Reddy, V. S., Rao, B. J., and Rajam, M. V. (2004). Genetic transformation of the green alga–Chlamydononas reinhardtii by Agrobacterium tumefaciens. Plant Sci. Int. J. Exp. Plant Biol. 166, 731–738. doi: 10.1016/j.plantsci.2003.11.012
Laere, E., Ling, A. P. K., Wong, Y. P., Koh, R. Y., Mohd Lila, M. A., and Hussein, S. (2016). Plant-based vaccines: production and challenges. J. Bot. 2016:4928637.
Lauersen, K. J., Berger, H., Mussgnug, J. H., and Kruse, O. (2013). Efficient recombinant protein production and secretion from nuclear transgenes in Chlamydomonas reinhardtii. J. Biotechnol. 167, 101–110. doi: 10.1016/j.jbiotec.2012.10.010
Ma, K., Bao, Q., Wu, Y., Chen, S., Zhao, S., Wu, H., et al. (2020). Evaluation of microalgae as immunostimulants and recombinant vaccines for diseases prevention and control in aquaculture. Front. Bioeng. Biotechnol. 8:590431. doi: 10.3389/fbioe.2020.590431
Mason, H. S., and Arntzen, C. J. (1995). Transgenic plants as vaccine production systems. Trends Biotechnol. 13, 388–392. doi: 10.1016/s0167-7799(00)88986-6
Mattanovich, D., Branduardi, P., Dato, L., Gasser, B., Sauer, M., and Porro, D. (2012). Recombinant protein production in yeasts. Methods Mol. Biol. 824, 329–358. doi: 10.1007/978-1-61779-433-9_17
Mayfield, S. P., and Kindle, K. L. (1990). Stable nuclear transformation of Chlamydomonas reinhardtii by using a C. reinhardtii gene as the selectable marker. Proc. Natl. Acad. Sci. U.S.A. 87, 2087–2091. doi: 10.1073/pnas.87.6.2087
O’Flaherty, R., Bergin, A., Flampouri, E., Mota, L. M., Obaidi, I., Quigley, A., et al. (2020). Mammalian cell culture for production of recombinant proteins: a review of the critical steps in their biomanufacturing. Biotechnol. Adv. 43:107552. doi: 10.1016/j.biotechadv.2020.107552
Orr, V. C. A., and Rehmann, L. (2016). Ionic liquids for the fractionation of microalgae biomass. Curr. Opin. Green Sust. Chem. 2, 22–27. doi: 10.1016/j.cogsc.2016.09.006
Ortega-Berlanga, B., Bañuelos-Hernández, B., and Rosales-Mendoza, S. (2018). Efficient expression of an Alzheimer’s Disease vaccine candidate in the microalga Schizochytrium sp. using the algevir system. Mol. Biotechnol. 60, 362–368. doi: 10.1007/s12033-018-0077-4
Phong, W. N., Show, P. L., Ling, T. C., Juan, J. C., Ng, E.-P., and Chang, J.-S. (2018). Mild cell disruption methods for bio-functional proteins recovery from microalgae—Recent developments and future perspectives. Algal Res. 31, 506–516. doi: 10.1016/j.algal.2017.04.005
Phoolcharoen, W., Bhoo, S. H., Lai, H., Ma, J., Arntzen, C. J., Chen, Q., et al. (2011). Expression of an immunogenic Ebola immune complex in Nicotiana benthamiana. Plant Biotechnol. J. 9, 807–816. doi: 10.1111/j.1467-7652.2011.00593.x
Piasecka, A., Krzemińska, I., and Tys, J. (2014). Physical methods of microalgal biomass pretreatment. Int. Agrophys. 28, 341–348. doi: 10.2478/intag-2014-0024
Potvin, G., and Zhang, Z. (2010). Strategies for high-level recombinant protein expression in transgenic microalgae: a review. Biotechnol. Adv. 28, 910–918. doi: 10.1016/j.biotechadv.2010.08.006
Qu, L., Ren, L. J., Li, J., Sun, G. N., Sun, L. N., Ji, X. J., et al. (2013). Biomass composition, lipid characterization, and metabolic profile analysis of the fed-batch fermentation process of two different docosahexanoic acid producing Schizochytrium sp. strains. Appl. Biochem. Biotechnol. 171, 1865–1876. doi: 10.1007/s12010-013-0456-z
Ramos-Vega, A., Rosales-Mendoza, S., Bañuelos-Hernández, B., and Angulo, C. (2018). Prospects on the use of Schizochytrium sp. to develop oral vaccines. Front. Microbiol. 9:2506. doi: 10.3389/fmicb.2018.02506
Rasala, B. A., and Mayfield, S. P. (2015). Photosynthetic biomanufacturing in green algae; production of recombinant proteins for industrial, nutritional, and medical uses. Photosynth. Res. 123, 227–239. doi: 10.1007/s11120-014-9994-7
Rattanapisit, K., Abdulheem, S., Chaikeawkaew, D., Kubera, A., Mason, H. S., Ma, J. K., et al. (2017). Recombinant human osteopontin expressed in Nicotiana benthamiana stimulates osteogenesis related genes in human periodontal ligament cells. Sci. Rep. 7:17358.
Rattanapisit, K., Jantimaporn, A., Kaewpungsup, P., Shanmugaraj, B., Pavasant, P., Namdee, K., et al. (2020a). Plant-produced basic fibroblast growth factor (bFGF) promotes cell proliferation and collagen production. Planta Med. Int. Open 07, e150–e157.
Rattanapisit, K., Shanmugaraj, B., Manopwisedjaroen, S., Purwono, P. B., Siriwattananon, K., Khorattanakulchai, N., et al. (2020b). Rapid production of SARS-CoV-2 receptor binding domain (RBD) and spike specific monoclonal antibody CR3022 in Nicotiana benthamiana. Sci. Rep. 10:17698.
Rattanapisit, K., Srifa, S., Kaewpungsup, P., Pavasant, P., and Phoolcharoen, W. (2019). Plant-produced recombinant Osteopontin-Fc fusion protein enhanced osteogenesis. Biotechnol. Rep. (Amst) 21:e00312. doi: 10.1016/j.btre.2019.e00312
Rosales-Mendoza, S., Angulo, C., and Meza, B. (2016). Food-grade organisms as vaccine biofactories and oral delivery vehicles. Trends Biotechnol. 34, 124–136. doi: 10.1016/j.tibtech.2015.11.007
Rosales-Mendoza, S., García-Silva, I., González-Ortega, O., Sandoval-Vargas, J. M., Malla, A., and Vimolmangkang, S. (2020a). The potential of algal biotechnology to produce antiviral compounds and biopharmaceuticals. Molecules 25:4049. doi: 10.3390/molecules25184049
Rosales-Mendoza, S., Solís-Andrade, K. I., Márquez-Escobar, V. A., González-Ortega, O., and Bañuelos-Hernandez, B. (2020b). Current advances in the algae-made biopharmaceuticals field. Expert Opin. Biol. Ther. 20, 751–766. doi: 10.1080/14712598.2020.1739643
Rosenberg, J. N., Oyler, G. A., Wilkinson, L., and Betenbaugh, M. J. (2008). A green light for engineered algae: redirecting metabolism to fuel a biotechnology revolution. Curr. Opin. Biotechnol. 19, 430–436. doi: 10.1016/j.copbio.2008.07.008
Salazar-González, J. A., Bañuelos-Hernández, B., and Rosales-Mendoza, S. (2015). Current status of viral expression systems in plants and perspectives for oral vaccines development. Plant Mol. Biol. 87, 203–217. doi: 10.1007/s11103-014-0279-5
Shanmugaraj, B., I Bulaon, C. J., and Phoolcharoen, W. (2020a). Plant molecular farming: a viable platform for recombinant biopharmaceutical production. Plants 9:842. doi: 10.3390/plants9070842
Shanmugaraj, B., Rattanapisit, K., Manopwisedjaroen, S., Thitithanyanont, A., and Phoolcharoen, W. (2020b). Monoclonal antibodies B38 and H4 produced in Nicotiana benthamiana neutralize SARS-CoV-2 in vitro. Front. Plant Sci. 11:589995. doi: 10.3389/fpls.2020.589995
Shanmugaraj, B., and Phoolcharoen, W. (2021). Addressing demand for recombinant biopharmaceuticals in the COVID-19 era. Asian Pacific J. Trop. Med. 14, 49–51. doi: 10.4103/1995-7645.306736
Shih, S. M., and Doran, P. M. (2009). Foreign protein production using plant cell and organ cultures: advantages and limitations. Biotechnol. Adv. 27, 1036–1042. doi: 10.1016/j.biotechadv.2009.05.009
Song, X., Zang, X., and Zhang, X. (2015). Production of high docosahexaenoic acid by Schizochytrium sp. using low-cost raw materials from food industry. J. Oleo Sci. 64, 197–204. doi: 10.5650/jos.ess14164
Specht, E., and Mayfield, S. (2014). Algae-based oral recombinant vaccines. Front. Microbiol. 5:60. doi: 10.3389/fmicb.2014.00060
Specht, E., Miyake-Stoner, S., and Mayfield, S. (2010). Micro-algae come of age as a platform for recombinant protein production. Biotechnol. Lett. 32, 1373–1383. doi: 10.1007/s10529-010-0326-5
Sproles, A. E., Fields, F. J., Smalley, T. N., Le, C. H., Badary, A., and Mayfield, S. P. (2021). Recent advancements in the genetic engineering of microalgae. Algal Res. 53:102158. doi: 10.1016/j.algal.2020.102158
Streatfield, S. J. (2005). Regulatory issues for plant-made pharmaceuticals and vaccines. Expert Rev. Vaccines 4, 591–601. doi: 10.1586/14760584.4.4.591
Surzycki, R., Greenham, K., Kitayama, K., Dibal, F., Wagner, R., Rochaix, J. D., et al. (2009). Factors effecting expression of vaccines in microalgae. Biologicals 37, 133–138. doi: 10.1016/j.biologicals.2009.02.005
Swartz, J. R. (2001). Advances in Escherichia coli production of therapeutic proteins. Curr. Opin. Biotechnol. 12, 195–201. doi: 10.1016/s0958-1669(00)00199-3
Tan, C. H., Show, P. L., Lam, M. K., Fu, X., Ling, T. C., Chen, C.-Y., et al. (2020). Examination of indigenous microalgal species for maximal protein synthesis. Biochem. Eng. J. 154:107425. doi: 10.1016/j.bej.2019.107425
Walker, T. L., Purton, S., Becker, D. K., and Collet, C. (2005). Microalgae as bioreactors. Plant Cell Rep. 24, 629–641. doi: 10.1007/978-1-4614-3348-4_26
Yaakob, Z., Ali, E., Zainal, A., Mohamad, M., and Takriff, M. S. (2014). An overview: biomolecules from microalgae for animal feed and aquaculture. J. Biol. Res. Thessaloniki 21:6. doi: 10.1186/2241-5793-21-6
Yan, N., Fan, C., Chen, Y., and Hu, Z. (2016). The potential for microalgae as bioreactors to produce pharmaceuticals. Int. J. Mol. Sci. 17:962. doi: 10.3390/ijms17060962
Yiemchavee, S., Wong-Arce, A., Romero-Maldonado, A., Shanmugaraj, B., Monsivais-Urenda, A. E., Phoolcharoen, W., et al. (2021). Expression and immunogenicity assessment of a plant-made immunogen targeting the cytotoxic T-lymphocyte associated antigen-4: a possible approach for cancer immunotherapy. J. Biotechnol. 329, 29–37. doi: 10.1016/j.jbiotec.2021.01.016
Keywords: Chlorella vulgaris, Chlamydomoans reinhardtii, nuclear transformation, geminiviral vector, green algae, therapeutic proteins, quantification, western blot
Citation: Malla A, Rosales-Mendoza S, Phoolcharoen W and Vimolmangkang S (2021) Efficient Transient Expression of Recombinant Proteins Using DNA Viral Vectors in Freshwater Microalgal Species. Front. Plant Sci. 12:650820. doi: 10.3389/fpls.2021.650820
Received: 08 January 2021; Accepted: 08 March 2021;
Published: 07 April 2021.
Edited by:
Goetz Hensel, Heinrich Heine University Düsseldorf, GermanyReviewed by:
Pau Loke Show, University of Nottingham Malaysia Campus, MalaysiaCopyright © 2021 Malla, Rosales-Mendoza, Phoolcharoen and Vimolmangkang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sornkanok Vimolmangkang, U29ybmthbm9rLnZAcGhhcm0uY2h1bGEuYWMudGg=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.