
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 05 May 2021
Sec. Crop and Product Physiology
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.650068
Photoperiod is a crucial inducer of plant flowering. Cycling DOF factors (CDFs) play pivotal roles in the flowering of long-day (LD) and short-day (SD) plants. However, the functions of CDFs in the photoperiod regulated flowering remain unclear in day-neutral plants. In the present study, tomato (Solanum lycopersicum L. cv. “Ailsa Craig”) seedlings of the wild-type and transgenic lines of overexpressing CDFs were treated with different photoperiods. The flowering time and the expression pattern of SlCDFs and other FT-like genes were investigated. The results showed that tomato SlCDF1, SlCDF2, SlCDF3, SlCDF4, and SlCDF5 are homologs to Arabidopsis cycling DOF factor 1 (AtCDF1). SlCDF1–5 expression levels were influenced by the developmental stage and the tissue location, and notably, the expression patterns throughout light environments showed two opposite trends. Among the SlCDF1–5 overexpression transgenic lines, overexpressing SlCDF3 delayed flowering time in both LD (16 h light/8 h dark) and SD (8 h light/16 h dark) conditions. Furthermore, SlCDF3 led to an increase in the mRNA level of SlSP5G, a tomato FT-like gene, in LD conditions, while the transcription level of the other two FT-like genes, SlSP5G2 and SlSP5G3, were up-regulated in SD conditions. Taken together, at the transcription level, our results demonstrated that SlCDF3 played a significant role in controlling tomato flowering under LD and SD conditions, possibly through directly or indirectly regulating FT-like genes.
Light is one of the most important factors in plant growth and development, since light is not only the driving force of photosynthesis but acts as an important transduction signal to regulate photomorphogenesis and endogenous substance metabolism via triggering or repressing related gene expressions (Ma et al., 2001). Flowering is a crucial period in plants representing the shift from their vegetative to reproductive phase. For instance, long-day (LD) conditions induced Arabidopsis flowering (Andrés and Coupland, 2012), while short-day (SD) conditions promoted early flowering of the Sorghum (Wolabu et al., 2016). Although tomato is a typical day-neutral plant (DNP), the flowering time of tomato is delayed under LD conditions and promoted under short day (SD) conditions (Sawhney and Greyson, 1972).
Previous studies reveal that the responses of plant flowering to photoperiod is a very complex process and many key genes are involved in photoperiod-mediated flowering (Kinet, 1977; Song et al., 2015). In Arabidopsis, flowering locus T (FT) encodes florigen, a key protein in triggering flowering, and its transcription is regulated by photoperiods (Kobayashi et al., 1999). Furthermore, FT expression is directly and indirectly controlled by a series of upstream transcription factors. Among them, Flavin Kelch Box1 (FKF1), Gigantea (GI), and constans (CO) function as transcriptional activators, whereas short vegetative phase (SVP), flowering loucs C (FLC), tempranillo (TEM), and cycling DOF factor 1 (CDF1) function as transcriptional repressors (Andrés and Coupland, 2012). Phylogenetic tree analysis indicates that CDF proteins belong to a distinct group of DOF transcription factors, the D subfamily. CDF proteins contain a C2–C2 zinc finger at their N-termini, composed of a conserved 51-residue domain, which is a common feature for the DOF family. In addition, CDF proteins have three conserved motifs at their C-termini, which is the basis for their distinctive function (Corrales et al., 2014). There are five CDF genes (AtCDF1–5) in Arabidopsis and overexpression of AtCDF1 results in flowering delay (Song et al., 2012). Since AtCDF1, the first CDF protein was discovered in plants (Imaizumi et al., 2005), dozens of CDF related genes have been found in other plants, including Jatropha curcas JcDof1 and JcDof3 (Yang et al., 2010, 2011) and Brassica napus BnCDF1 (Xu and Dai, 2016). In the rice, overexpression of OsDof12 induces high expression of Heading date 3a (Hd3a) and early flowering under LD conditions. Conversely, osdof12 mutants have smaller petals and delay flowering time relative to wild-type plants (Iwamoto et al., 2009; Li et al., 2009). Subsequent studies have revealed that OsDof4 and OsDof12 are in the same subfamily, with similar functions in photoperiod-regulated flowering (Wu et al., 2017).
The in vivo and in vitro experiments have indicated that AtCDF1 specifically binds with DOF binding elements (DBEs) in the CO promoter to inhibit its expression (Imaizumi et al., 2005). And AtCDF1 could also directly bind to the promoter of FT to regulate flowering (Song et al., 2012). However, AtCDF1 is degraded when integrated with GI-FKF1 protein complexes (Ding et al., 2018). Moreover, Fornara et al. (2009) found that AtCDF2, AtCDF3, and AtCDF5 also participated in the regulation of flowering through modulation of the expression of flowering-related genes. Up to date, five CDF genes are identified in tomato: SlCDF1, SlCDF2, SlCDF3, SlCDF4, and SlCDF5. Overexpressed SlCDF3 but not SlCDF1 in Arabidopsis leads to late flowering through modulation of the expression of flowering-related genes including CO and FT (Corrales et al., 2014). However, the functions of SlCDF1–5 in the regulation of tomato flowering are still poorly understood. Our previous studies found three FT-like genes (SlSP5G, SlSP5G2, and SlSP5G3) played important roles in the photoperiod-modulated flowering in tomato (Cao et al., 2016, 2018). We hypothesize that there might be a cross-talk between SlCDF1–5 and FT-like in the regulation of tomato flowering. In this study, we generated SlCDF1–5 overexpression plants and analyzed the transcription levels of downstream FT-like genes in LD and SD conditions. Our results showed SlCDF3 delayed flowering time by regulating different FT-like genes, providing a molecular basis for elucidating the regulatory mechanism of flowering in the day-neutral plant.
For our evolutionary analysis, 34 tomato DOF protein sequences reported by Corrales et al. (2014) were downloaded from http://solgenomics.net/ and 36 Arabidopsis DOF proteins reported by Lijavetzky et al. (2003) were downloaded from https://www.arabidopsis.org/. We used MegAlign and MEGA 7.0 software to perform amino acid sequence alignment and homology analysis of the target proteins, respectively, and CLUSTAL W to build neighbor-joining trees.
The structures of tomato CDF proteins were analyzed and predicted using the EzMol online tool1.
Tomato (Solanum lycopersicum L. var. Ailsa Craig) seeds were soaked in 50% bleach for 30 min. After sterilization, those seeds were rinsed thoroughly in running water and then incubated at 25°C under dark conditions. After germination, seedlings were sowed into the commercial substrate (PINDSTRUP, Beijing, China) and grown under white LED light (400–700 nm, peaked at 455 and 570 nm) in an environmentally controlled growth chamber with three different photoperiods: LD (16 h light/8 h dark), SD (8 h light/16 h dark), and DN (12 h light/12 h dark). The light intensity, relative humidity, and CO2 level in the growth chamber were set at 200 μmol m–2 s–1, 65%, and 400 μmol mol–1, respectively, while the day and night temperature was both set at 25°C.
To study the spatial expression patterns of SlCDF related genes, we extracted total RNA from the leaf, stem, and root samples of 5-weeks-old plants (three plants per sample and three samples per treatment) grown under DN conditions. Additionally, to study expression changes during development, seven leaves were collected from the top to the bottom of another three 7-weeks-old plants grown under DN conditions. To determine diurnal changes in the expression of SlCDFs, the third youngest and fully expanded leaves were collected every 4 h for 24 h (0, 4, 8, 12, 16, 20, and 24 h) from 5-weeks-old plants under SD, LD, and DN conditions. The plant samples were collected as the method described in our previous study (Cao et al., 2016). Total RNA was extracted using an RNeasy Plant mini kit (QIAGEN, Shanghai, China) following the manufacturer’s instructions. Plant genomic DNA was isolated using an Easy Pure Plant Genomic DNA kit (TransGen, Beijing, China) according to the manufacturer’s instructions.
The ORFs of SlCDF1 (Solyc03g115940), SlCDF2 (Solyc05g007880), SlCDF3 (Solyc06g069760), SlCDF4 (Solyc02g067230), and SlCDF5 (Solyc02g088070) were amplified by PCR from “Ailsa Craig” cultivar cDNAs that had been cloned into a pHELLSGATE8 vector (Invitrogen, Shanghai, China) by recombination using the ClonExpress II One Step Cloning Kit (Vazyme, Nanjing, China). PCR primers required for cloning genes and identifying positive transgenic plants are summarized in Supplementary Table 1. The pHELLSGATE8 vector carries a spectinomycin resistance gene for bacterial selection, a kanamycin resistance gene for the selection of transformed plants, and a CaMV35S promoter. The primers used are listed in Supplementary Table 1. The resulting plasmids were used to transform “Ailsa Craig” tomato plants following the method described by Ellul et al. (2003). The seeds from the transformed plants were harvested and plated onto a selective medium, and the kanamycin-resistant seedlings were transplanted to soil. Transgenic plants were further confirmed by PCR using genomic DNA as a template and amplified with CaMV35S forward and gene-specific reverse primers. Three transgenic lines with high expression were screened by qRT-PCR before the T1 generation; T1 generation seeds were screened by kanamycin and verified by qRT-PCR at the seedling stage before experimental observation.
After RNA extraction, cDNA synthesis was performed using the SuperscriptIII First-strand synthesis system (Invitrogen, Shanghai, China) as the manufacturer’s instructions. Real-time PCR was performed using SYBR Premix Ex Taq (Takara, Dalian, China) with a Bio-Rad CFX96 real-time PCR platform (Bio-Rad, Hercules, CA, United States). GAPDH transcripts were used as the internal control (Løvdal and Lillo, 2009). The qRT-PCR primers used in this study were listed in Supplementary Table 1. The efficiency of primers was calculated before carrying out the qRT-PCR reaction, all 10 gene primers of good quality with a PCR efficiency (in r-squared value) > 0.9 (Supplementary Table 1). The qRT-PCR was performed with three technical replicates.
All the data were subjected to one-way ANOVA analysis using SPSS 20.0 (SPSS, version 20.0, IBM Inc., Armonk, NY, United States). The significant differences between means were assessed by Duncan’s multiple range test (P < 0.05). Error bars in all figures represent standard deviations from the mean.
We constructed a phylogenetic tree based on the amino acid sequences of 36 Arabidopsis DOF and 34 tomato DOF proteins, finding that SlCDF1–5 in tomato were closely related to AtCDF1–5 (Supplementary Figure 1). Simultaneously, sequence alignment results also indicated that, other than AtCDF4, the remaining nine CDF proteins contained C2C2 zinc finger structures at their N-termini and three conserved sequences (21, 33, and 22 residues) at their C-termini (Figure 1A). Notably, AtCDF4 was observed to have only motif 2 at its C-terminus, indicating that it might have a distinct function. In addition to the amino acid sequences, the structures of introns and exons were also highly conserved. At the gene structure, except AtCDF4, all other nine CDF genes contained two exons and one intron (Figure 1B). The protein spatial conformation in Figure 1C showed that tomato SlCDF1–5 proteins mainly consist of a region of α-helices and β-strands as well as a considerable portion of random structures. Such spatial structures may facilitate CDF proteins binding to the promoters of target genes for transcriptional regulation.
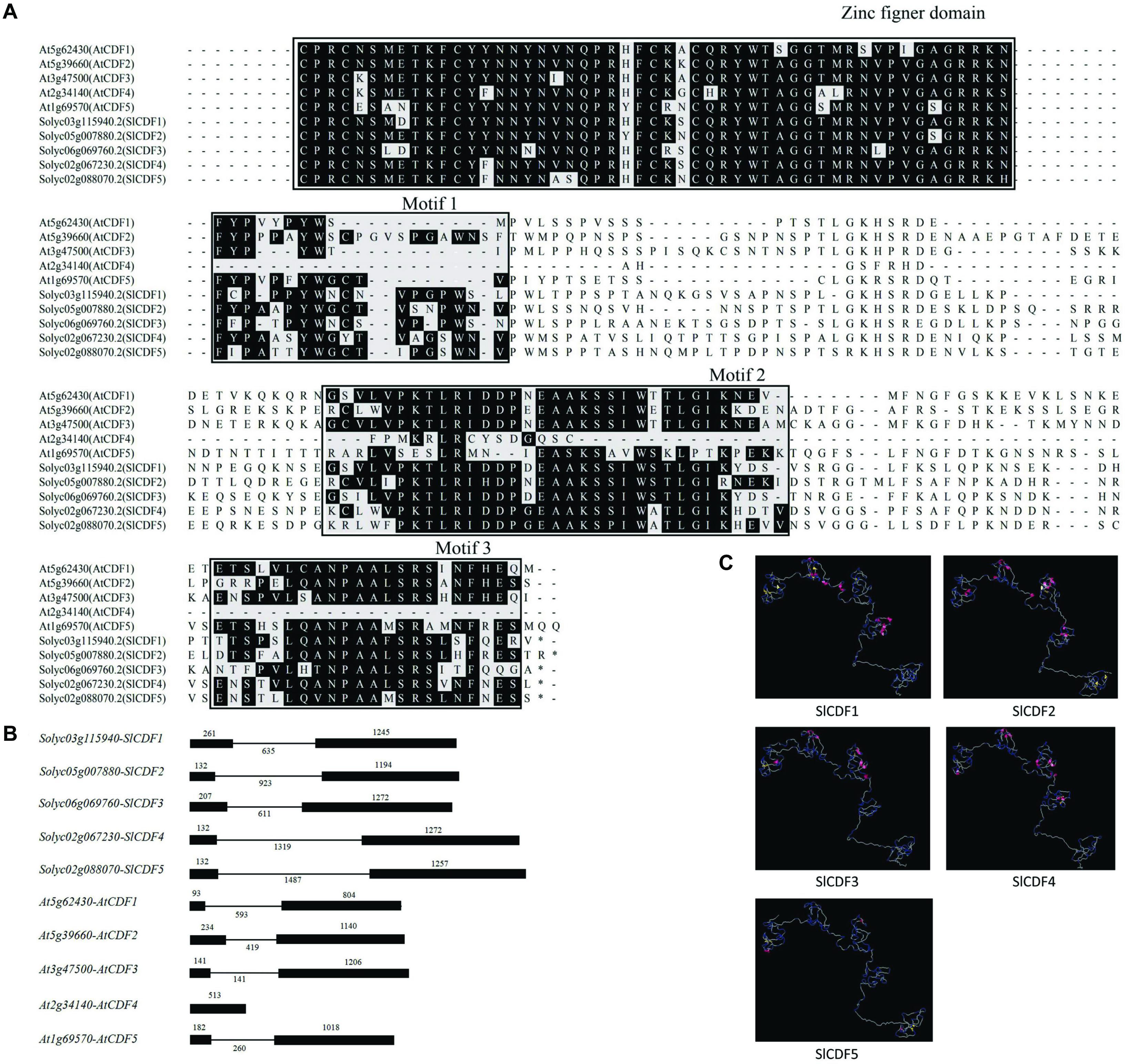
Figure 1. The corresponding amino acid sequences and spatial structures of tomato SlCDF1–5 genes were highly conserved. (A) Amino acid sequence alignment of Arabidopsis and tomato CDF proteins. The sequences in the black box were highly conserved. (B) Exon-intron structure analysis of CDF genes. The black solid boxes represent exons, while the horizontal lines represent introns. (C) The simulated three-dimensional structure of tomato SlCDF1–5. The pink and yellow areas represent α-helices and β-sheets, respectively.
The tissue-special expression patterns of related genes often indicate their functions. To investigate the roles of SlCDF1–5 genes in flowering, we first assayed their expression levels in different vegetative tissues. In total RNA extracted from leaf, stem, and root tissues of 5-weeks-old tomato plants grown under DN conditions, SlCDF1 and SlCDF2 were highly expressed in leaves, while SlCDF4 and SlCDF5 showed lower expression in roots. The SlCDF3 expression level was low in all detected tissues (Figure 2A). To further investigate the expression patterns of SlCDF1–5 across developmental stages, we assayed SlCDF1–5 expression in true leaves from top to bottom of 7-weeks-old tomato plants grown under DN conditions. SlCDF1 and SlCDF2 were highly expressed in all leaves and showed downward trends during development, while SlCDF3, SlCDF4, and SlCDF5 showed an opposite trend (Figure 2B).
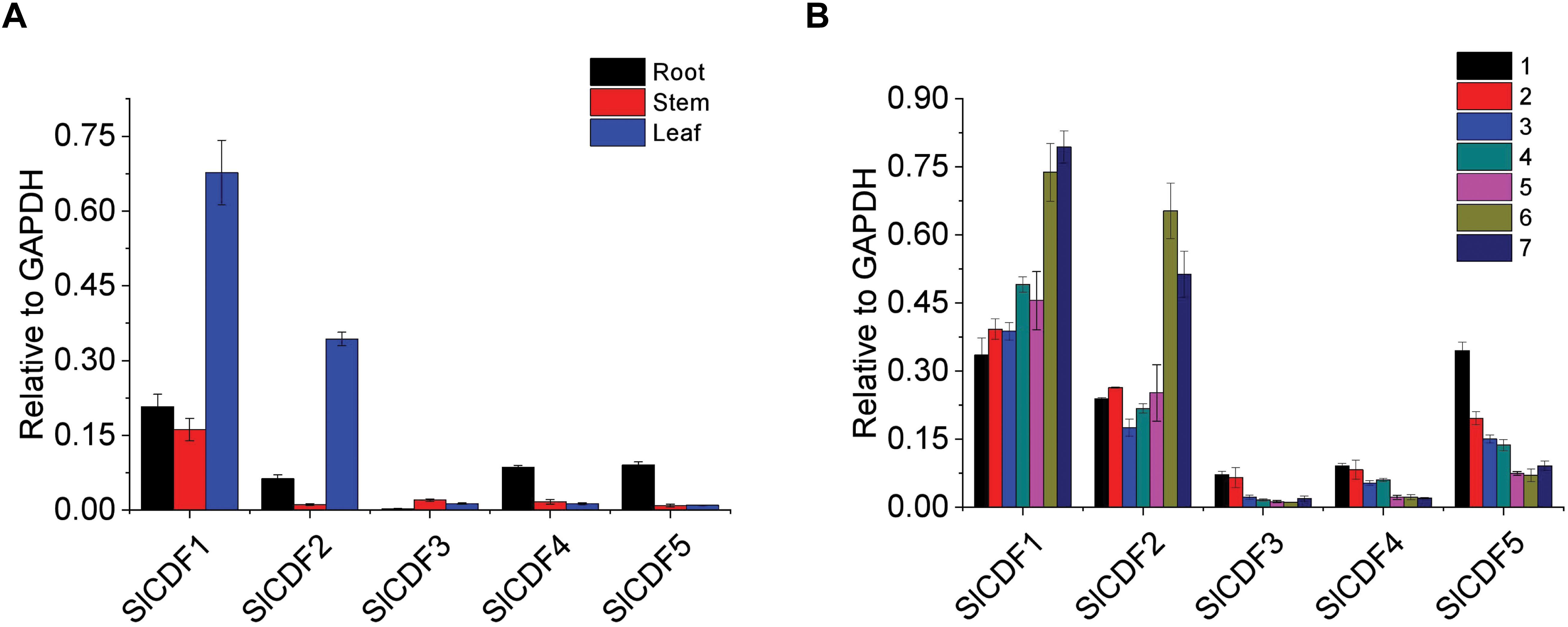
Figure 2. Transcription analyses of tomato SlCDF1–5 throughout development and among tissues. (A) SlCDF1–5 expression levels in tomato roots, stems, and leaves at 5-weeks-old tomato plants. (B) SlCDF1–5 expression levels in the first seven individual leaves from top to bottom in 7-weeks-old tomato plants. Three technical replicates were performed for each extract, vertical bars on the lines represent the SE (n = 3).
To reveal the relationships between SlCDF1–5 genes and photoperiod, we examined their diurnal expression patterns under different photoperiod conditions. Based on their expression patterns, SlCDF1–5 can be categorized into two groups. SlCDF1 and SlCDF3 expression peaked after being irradiated for 8 h, and then decreased in the following 8 h under LD conditions (Figure 3A). Under SD conditions, those gene expressions increased from 4 h before the onset of the light period and peaked at the end of the light period (Figure 3B). The other three SlCDF genes showed similar expression patterns, with SlCDF2, SlCDF4, and SlCDF5 expression peaking at the end of the dark period in LD conditions and at 4 h before the light period under SD conditions (Figures 3C,D). In addition, the same phenomenon was also observed under diurnal-neutral (DN) conditions (Supplementary Figure 2).
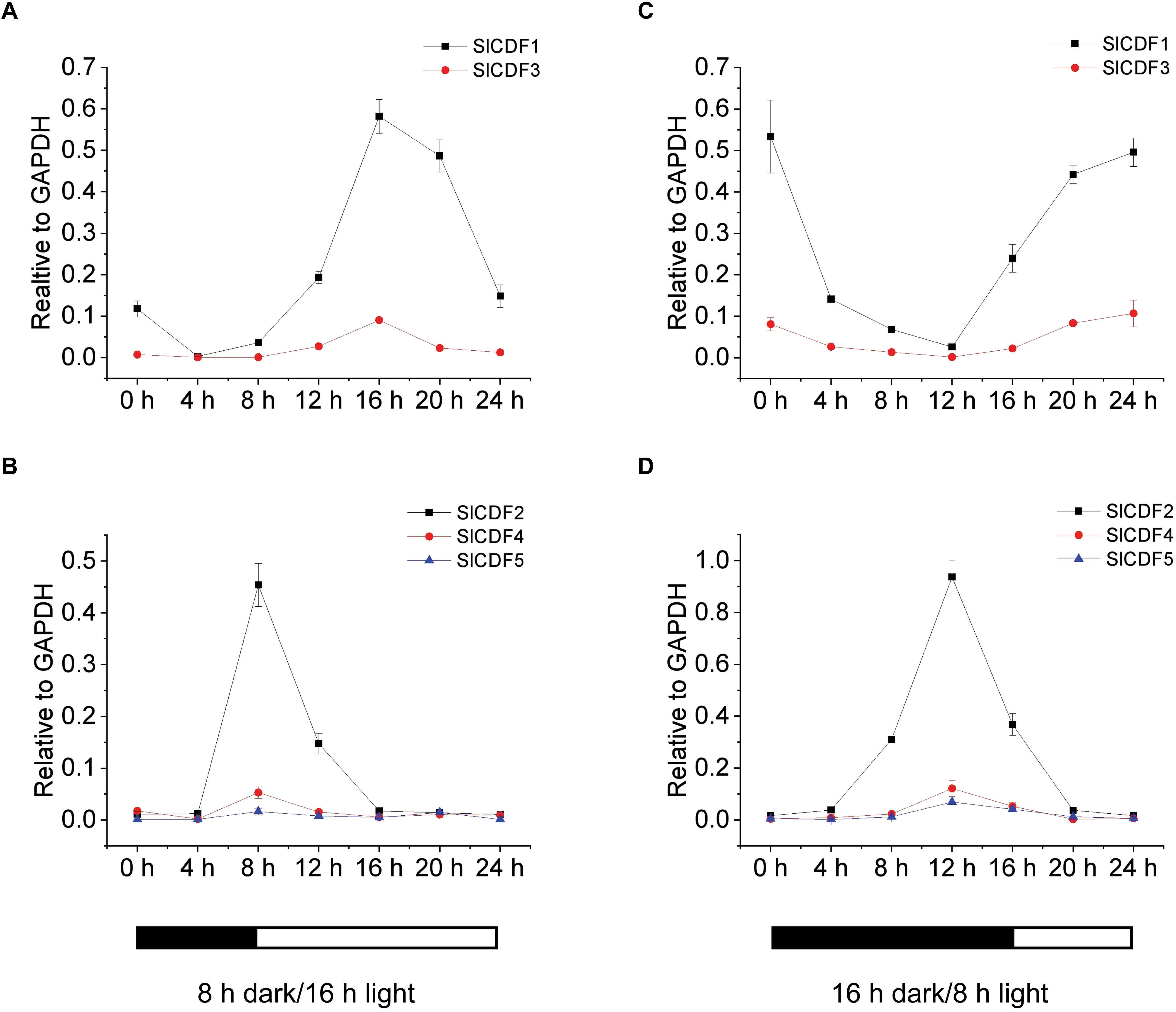
Figure 3. Transcription analyses of tomato SlCDF1–5 in response to different photoperiods. Expression of SlCDF1–5 was analyzed by qRT-PCR in 4-weeks-old tomato plants grown under a diurnal cycle of 16 h light/8 h dark (A,C) or 8 h light/16 h dark (B,D). White and black bars along the horizontal axis represent light and dark periods, respectively. Three technical replicates were performed for each extract, vertical bars on the lines represent the SE (n = 3).
To explore the mechanism of SlCDF1–5 in the regulation of flowering under different photoperiods, corresponding overexpression lines were generated. The T0 generation plants were identified by PCR and qRT-PCR, and the three lines with the highest expression levels were selected and transplanted to the greenhouse for cultivation (Supplementary Figure 3). After kanamycin screening, genotyping PCR, and qRT-PCR verifying, T1 generation materials were subjected to different photoperiod conditions. The flowering time of the tomato is calculated as the number of true leaves needed before flowering. Under SD conditions, the number of true leaves of wild-type plants before flowering was eight on average, while the numbers of true leaves before flowering increased to nine under LD conditions (Figure 4A). Overexpression of SlCDF3 delayed flowering compared to wild-type plants in both LD and SD conditions. The number of leaves before flowering in SlCDF3-overexpressing plants (OE-5) increased to 10.7 under SD conditions and to 11.3 under LD conditions (Figure 4B). However, there were no significant differences between the flowering time of other SlCDFs overexpression lines and wild-type plants (Supplementary Figure 4).
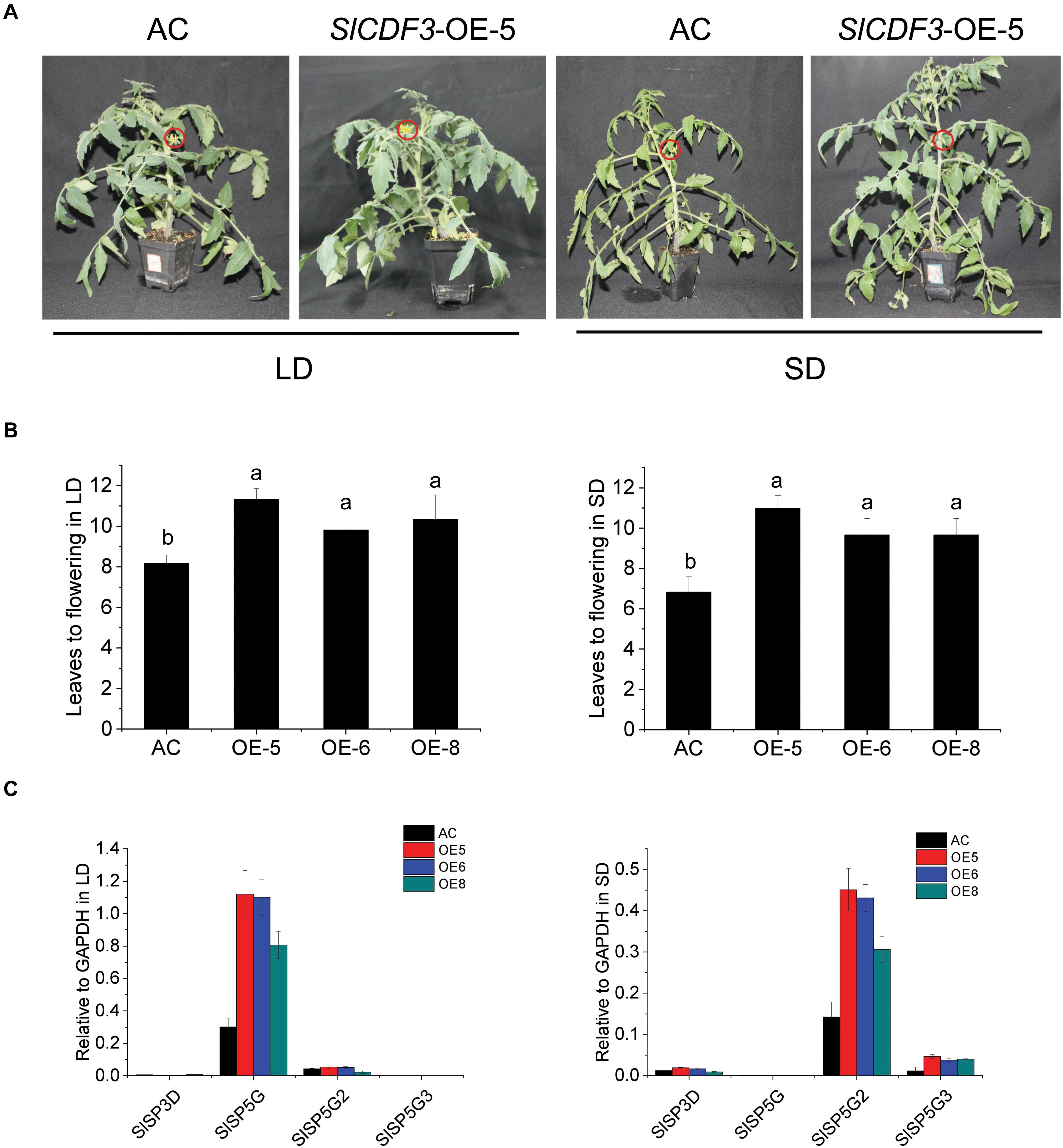
Figure 4. SlCDF3 delayed flowering by regulating different FT-like genes in long-day (LD) and short-day (SD) conditions. (A) Flowering phenotypes of SlCDF3-overexpressing tomato plants under LD or SD conditions. The red circles indicate flowers. (B) Flowering time of SlCDF3-overexpressing tomato plants represented by leave number under LD or SD conditions. Vertical bars on the lines represent the SE (n = 6). (C) The transcription levels of tomato FT-like genes in SlCDF3-overexpressing tomato plants under LD and SD conditions. Three technical replicates were performed for each extract, vertical bars on the lines represent the SE (n = 3).
To investigate the potential interaction between FT-like genes and the SlCDF3 gene, we measured the expression levels of FT-like genes in SlCDF3 overexpressing plants under LD and SD conditions (Figure 4C). Compared with wild-type plants, SlSP5G expression in SlCDF3-overexpressing plants was significantly increased under LD conditions, while SlSP5G2 and SlSP5G3 expression levels were increased under SD conditions. However, there were no distinct changes in the transcription levels of SlSP5G2 and SlSP5G3 in LD conditions and the expression level of SlSP5G in SD conditions. In addition, to further verify the special interaction between SlSP5G and SlCDF3, we also studied the expression levels of SlSP5G in T1 generation of other four SlCDFs-overexpressing tomato plants. These results showed that the transcription levels of SlSP5G in SlCDF1,4,5-overexpressing tomato plants were not significantly changed, with slightly decreased in SlCDF2-overexpressing transgenic lines (Supplementary Figure 5).
Photoperiod is one of the most important environmental factors in the regulation of plant flowering. Tomato is a typical day-neutral plant (DNP), but the flowering time of tomato is also regulated by photoperiod. At the transcription level, our present study demonstrated that SlCDF3 played an important role in the regulation of tomato flowering via inducing the expression of the downstream FT-like genes directly or indirectly.
The CDF proteins belong to the D subfamily of DOF transcription factors. The specific expression pattern of DOF family members has been assessed in many species. The expression levels of StDof29a, StDof32, and StDof34 from potato, a congener of tomato, are higher in leaves than other tissues (Li et al., 2009). However, OsDof11 is predominantly expressed in leaves and promotes flowering in rice by regulating Hd3a and OsMADS14 expression levels under LD conditions (Li et al., 2009). In our present study, the tissue-specific expression of SlCDF1–5 (Figures 2A,B) indicated SlCDF1–5 had potentially large differences in biological regulatory functions in tomato plants. According to the study of Corrales et al. (2014), SlCDF1–5 expression patterns could be divided into two groups under LD and continuous light conditions. In this study, we found that SlCDF1–5 showed two distinct expression patterns not only in LD conditions but also in DN and SD conditions (Figure 3 and Supplementary Figure 2). These results suggest that the transcription levels of SlCDF1–5 might be regulated by external light signals and circadian rhythm. Similar results were also reported in other flowering plants, including rice (Rdd1 and OsDof4) (Iwamoto et al., 2009; Li et al., 2009), Jatropha curcas (JcDof1 and JcDof3) (Yang et al., 2010, 2011), and potato (Kloosterman et al., 2013).
In most flowering plants, seasonal variation in flowering time largely depends on the expression levels of FT-like genes (Laurent and George, 2006; Abelenda et al., 2014). The day length-dependent FT expression is governed mainly by the transcriptional activator–CO (Sawa and Kay, 2011). In addition to CO, DOF family members also participate in the regulation of FT transcription. For example, AtCDF1, a repressor of CO transcription, controls photoperiodic flowering in Arabidopsis (Goralogia et al., 2017). Furthermore, overexpression of rice OsDof4 leads to earlier flowering under LD conditions, but late flowering under SD conditions (Wu et al., 2017). In our study, the increased transcription level of SlCDF3 induced delayed flowering in both LD and SD conditions (Figures 4A,B). This is in line with previous results in Arabidopsis (Imaizumi et al., 2005). However, except for SlCDF3, the increased expression of other SlCDFs did not delay the flowering under LD and SD conditions (Supplementary Figure 4). This could partly be explained by the different post-transcriptional regulation patterns or differences in protein stability of SlCDF1–5. Further detailed studies including the post-transcription regulation are still needed to reveal the mechanism of these genes in the regulation of tomato flowering.
Our previous studies found that there are three FT-like genes in tomatoes that respond to photoperiod: SlSP5G, SlSP5G2, and SlSP5G3 (Cao et al., 2018). Among them, SlSP5G is mainly expressed under LD conditions while SlSP5G2 and SlSP5G3 were expressed under SD conditions (Cao et al., 2016, 2018). To clarify the main reason for the delayed flowering in SlCDF3-overexpressing plants, we measured the expression of FT-like genes under LD and SD conditions (Figure 4C). Compared with wild-type plants, SlSP5G expression in SlCDF3-overexpressing plants was significantly increased under LD conditions, while SlSP5G2 and SlSP5G3 expression levels were increased under SD conditions (Figure 4C). Although the SlCDF3 protein level was not tested with SlCDF3-overexpressing plants in the present study, the up-regulation of the FT-like genes and the delayed flowering in SlCDF3-overexpressing plants suggested the possible regulation of SlCDF3 on plant flowering via interacting with FT-like genes at the transcription level. In LD conditions, SlCDF3 increases the transcription of SlSP5G and resulted late flowering; in SD conditions, SlCDF3 induces the expression levels of SlSP5G2 and SlSP5G3 might be one of the reason for inhibiting tomato flowering. Combined with the little effects of overexpressing SlCDF3 on the SlSP5G expression in SD conditions and the transcriptions of SlSP5G2 and SlSP5G3 in LD conditions, these results indicate that other transcription factors might be involved in the regulation of FT-like genes in different photoperiod conditions. Although we explained the variation of expression level in FT-like genes under LD and SD, whether SlCDF1–5 directly or indirectly regulates FT-like genes remains unknown, and further detailed studies on post-transcriptional modification and protein synthesis are still needed.
Our present study demonstrated the pivotal role of SlCDF3 in controlling tomato flowering time at the transcription level and identified the potential cross-talk between SlCDF3 and FT-like genes. SlCDF3-overexpressing led to late flowering by inducing transcription of SlSP5G under LD conditions, and by triggering the expression of SlSP5G2 and SlSP5G3 under SD conditions. These findings could enrich the current understanding of the tomato flowering signal transduction pathway and provide a theoretical basis for related studies on flowering in DNPs.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
ZB and KC conceived the original research plan and designed the experiment. DX, XL, and XW performed the experiments and analyzed the data. DX and KC wrote the manuscript, while ZB reviewed and edit the manuscript. All authors contributed to the article and approved the submitted version.
The authors would like to thank National Natural Science Foundation of China (Grant No. 31801903) and Science and Technology Innovation Program of the Chinese Academy of Agricultural Sciences, The Key Laboratory of Agricultural Engineering in the Middle and Lower Reaches of Yangze River, Ministry of Agriculture and Rural Affairs, P. R. China, for their support.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.650068/full#supplementary-material
Supplementary Figure 1 | Phylogenetic analysis of Arabidopsis and tomato DOF protein families. The Arabidopsis and tomato gene trees were inferred by the neighbor-joining method after alignment of the DOF domains of the 36 Arabidopsis and 34 tomato DOF proteins. The resulting Major Clusters of Orthologous Genes (MCOG) A, B, C, and D are indicated. White circles with tables represent Arabidopsis CDF proteins, and black circles represent tomato CDF proteins. The numbers nearby tree branches indicate their confidence. The scale bar represents 0.05 estimated amino acid substitutions per site.
Supplementary Figure 2 | Transcription analyses of tomato SlCDF1–5 in response to day-neutral (DN) conditions. SlCDF1–5 expression levels were assayed by qRT-PCR in 4-weeks-old tomato plants grown under a diurnal cycle of 12 h light/12 h dark. White and black bars along the horizontal axis represent light and dark periods, respectively. Three technical replicates were performed for each extract, vertical bars on the lines represent the SE (n = 3).
Supplementary Figure 3 | Transcription analyses of target genes in SlCDF1–5-overexpressing tomato plants. SlCDF1 (A), SlCDF2 (B), SlCDF3 (C), SlCDF4 (D), and SlCDF5 (E) expression assayed by qRT-PCR in T0 generation transgenic tomatoes grown under day-neutral (DN) conditions. Three technical replicates were performed for each extract, vertical bars on the lines represent the SE (n = 3).
Supplementary Figure 4 | Overexpression of SlCDF1, SlCDF2, SlCDF4, and SlCDF5 had no effect on flowering time of tomato under long day (LD) and short-day (SD) conditions. (A) Images of overexpression tomato plants flowering under LD and SD conditions. The red circles indicate flowers. (B) Leaf number at flowering in overexpression tomato plants under LD and SD conditions. Vertical bars on the lines represent the SE (n = 6).
Supplementary Figure 5 | Overexpression of SlCDF1, SlCDF2, SlCDF4, and SlCDF5 did not change the transcription of SlSP5G under long day (LD) conditions. Three technical replicates were performed for each extract, vertical bars on the lines represent the SE (n = 3).
Abelenda, J. A., Navarro, C., and Prat, S. (2014). Flowering and tuberization: a tale of two nightshades. Trends Plant Sci. 19, 115–122. doi: 10.1016/j.tplants.2013.09.010
Andrés, F., and Coupland, G. (2012). The genetic basis of flowering responses to seasonal cues. Nat. Rev. Genet. 13, 627–639. doi: 10.1038/nrg3291
Cao, K., Cui, L., Zhou, X., Ye, L., Zou, Z., and Deng, S. (2016). Four tomato FLOWERING LOCUS T-like proteins act antagonistically to regulate floral initiation. Front. Plant Sci. 6:1213. doi: 10.3389/fpls.2015.01213
Cao, K., Yan, F., Xu, D., Ai, K., Yu, J., Bao, E., et al. (2018). Phytochrome B1-dependent control of SP5G transcription is the basis of the night break and red to far-red light ratio effects in tomato flowering. BMC Plant Biol. 18:158. doi: 10.1186/s12870-018-1380-8
Corrales, A.-R., Nebauer, S. G., Carrillo, L., Fernández-Nohales, P., Marqués, J., Renau-Morata, B., et al. (2014). Characterization of tomato Cycling Dof Factors reveals conserved and new functions in the control of flowering time and abiotic stress responses. J. Exp. Bot. 65, 995–1012. doi: 10.1093/jxb/ert451
Ding, J., Böhlenius, H., Rühl, M. G., Chen, P., Sane, S., Zambrano, J. A., et al. (2018). GIGANTEA−like genes control seasonal growth cessation in Populus. New Phytol. 218, 1491–1503. doi: 10.1111/nph.15087
Ellul, P., Garcia-Sogo, B., Pineda, B., Rios, G., Roig, L., and Moreno, V. (2003). The ploidy level of transgenic plants in Agrobacterium-mediated transformation of tomato cotyledons (Lycopersicon esculentum L. Mill.) is genotype and procedure dependent. Theor. Appl. Genet. 106, 231–238. doi: 10.1007/s00122-003-1289-x
Fornara, F., Panigrahi, K. C., Gissot, L., Sauerbrunn, N., Rühl, M., Jarillo, J. A., et al. (2009). Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev. Cell 17, 75–86. doi: 10.1016/j.devcel.2009.06.015
Goralogia, G. S., Liu, T. K., Zhao, L., and Panipinto, P. M. (2017). CYCLING DOF FACTOR 1 represses transcription through the TOPLESS co-repressor to control photoperiodic flowering in Arabidopsis. Plant J. 92, 244–262. doi: 10.1111/tpj.13649
Imaizumi, T., Schultz, T., Harmon, F., Ho, L., and Kay, S. (2005). FKF1F-BOX protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science 309, 293–297. doi: 10.1126/science.1110586
Iwamoto, M., Higo, K., and Takano, M. (2009). Circadian clock−and phytochrome−regulated Dof−like gene, Rdd1, is associated with grain size in rice. Plant Cell Environ. 32, 592–603. doi: 10.1111/j.1365-3040.2009.01954.x
Kinet, J. (1977). Effect of light conditions on the development of the inflorescence in tomato. Sci. Horticult. 6, 15–26. doi: 10.1016/0304-4238(77)90074-7
Kloosterman, B., Abelenda, J. A., Gomez, M. D. M. C., Oortwijn, M., De Boer, J. M., Kowitwanich, K., et al. (2013). Naturally occurring allele diversity allows potato cultivation in northern latitudes. Nature 495, 246–250. doi: 10.1038/nature11912
Kobayashi, Y., Kaya, H., Goto, K., Iwabuchi, M., and Araki, T. (1999). A pair of related genes with antagonistic roles in mediating flowering signals. Science 286, 1960–1962. doi: 10.1126/science.286.5446.1960
Laurent, C., and George, C. (2006). The quest for florigen: review of recent progress. J. Exp. Bot. 57, 3395–3403. doi: 10.1093/jxb/er1095
Li, D., Yang, C., Li, X., Gan, Q., Zhao, X., and Zhu, L. (2009). Functional characterization of rice OsDof12. Planta 229, 1159–1169. doi: 10.1007/s00425-009-0893-7
Lijavetzky, D., Carbonero, P., and Vicente-Carbajosa, J. (2003). Genome-wide comparative phylogenetic analysis of the rice and Arabidopsis Dof gene families. BMC Evol. Biol. 3:17. doi: 10.1186/1471-2148-3-17
Løvdal, T., and Lillo, C. (2009). Reference gene selection for quantitative real-time PCR normalization in tomato subjected to nitrogen, cold, and light stress. Anal. Biochem. 387, 238–242. doi: 10.1016/j.ab.2009.01.024
Ma, L., Li, J., Qu, L., Hager, J., Chen, Z., Zhao, H., et al. (2001). Light control of Arabidopsis development entails coordinated regulation of genome expression and cellular pathways. Plant Cell 13, 2589–2607. doi: 10.1105/tpc.13.12.2589
Sawa, M., and Kay, S. A. (2011). GIGANTEA directly activates flowering Locus T in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 108, 11698–11703. doi: 10.1073/pnas.1106771108
Sawhney, V. K., and Greyson, R. I. (1972). On the initiation of the inflorescence and floral organs in tomato (Lycopersicon esculentum). Can. J. Bot. 50, 1493–1495. doi: 10.1139/b72-183
Song, Y. H., Shim, J. S., Kinmonth-Schultz, H. A., and Imaizumi, T. (2015). Photoperiodic flowering: time measurement mechanisms in leaves. Annu. Rev. Plant Biol. 66, 441–464. doi: 10.1146/annurev-arplant-043014-115555
Song, Y. H., Smith, R. W., To, B. J., Millar, A. J., and Imaizumi, T. (2012). FKF1 conveys timing information for CONSTANS stabilization in photoperiodic flowering. Science 336, 1045–1049. doi: 10.1126/science.1219644
Wolabu, T. W., Zhang, F., Niu, L., Kalve, S., Bhatnagar−Mathur, P., Muszynski, M. G., et al. (2016). Three FLOWERING LOCUS T−like genes function as potential florigens and mediate photoperiod response in sorghum. New Phytol. 210, 946–959. doi: 10.1111/nph.13834
Wu, Q., Liu, X., Yin, D., Yuan, H., Xie, Q., Zhao, X., et al. (2017). Constitutive expression of OsDof4, encoding a C2-C2 zinc finger transcription factor, confesses its distinct flowering effects under long- and short-day photoperiods in rice (Oryza sativa L.). BMC Plant Biol. 17:166. doi: 10.1186/s12870-017-1109-0
Xu, J., and Dai, H. (2016). Brassica napus Cycling Dof Factor1 (BnCDF1) is involved in flowering time and freezing tolerance. Plant Growth Regul. 80, 315–322. doi: 10.1007/s10725-016-0168-9
Yang, J., Yang, M., Wang, D., Chen, F., and Shen, S. (2010). JcDof1, a Dof transcription factor gene, is associated with the light−mediated circadian clock in Jatropha curcas. Physiol. Plant. 139, 324–334. doi: 10.1111/j.1399-3054.2010.01363.x
Keywords: photoperiod, SlCDF3, FT-like gene, flowering time, tomato
Citation: Xu D, Li X, Wu X, Meng L, Zou Z, Bao E, Bian Z and Cao K (2021) Tomato SlCDF3 Delays Flowering Time by Regulating Different FT-Like Genes Under Long-Day and Short-Day Conditions. Front. Plant Sci. 12:650068. doi: 10.3389/fpls.2021.650068
Received: 06 January 2021; Accepted: 13 April 2021;
Published: 05 May 2021.
Edited by:
Antonio Ferrante, University of Milan, ItalyReviewed by:
Tomokazu Kawashima, University of Kentucky, United StatesCopyright © 2021 Xu, Li, Wu, Meng, Zou, Bao, Bian and Cao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhonghua Bian, Ymlhbnpob25naHVhQGNhYXMuY24=; Kai Cao, a2Nhb0BqYWFzLmFjLmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.