- 1School of Pharmacy, Second Military Medical University, Shanghai, China
- 2Department of Pharmacy, Changzheng Hospital, Second Military Medical University, Shanghai, China
- 3Biomedical Innovation R&D Center, School of Medicine, Shanghai University, Shanghai, China
- 4Research and Development Center of Chinese Medicine Resources and Biotechnology, Shanghai University of Traditional Chinese Medicine, Shanghai, China
- 5State Key Laboratory of Subtropical Silviculture, Zhejiang A & F University, Hangzhou, China
Laccases are multicopper-containing glycoproteins related to monolignol oxidation and polymerization. These properties indicate that laccases may be involved in the formation of important medicinal phenolic acid compounds in Salvia miltiorrhiza such as salvianolic acid B (SAB), which is used for cardiovascular disease treatment. To date, 29 laccases have been found in S. miltiorrhiza (SmLACs), and some of which (SmLAC7 and SmLAC20) have been reported to influence the synthesis of phenolic acids. Because of the functional redundancy of laccase genes, their roles in S. miltiorrhiza are poorly understood. In this study, the CRISPR/Cas9 system was used for targeting conserved domains to knockout multiple genes of laccase family in S. miltiorrhiza. The expressions of target laccase genes as well as the phenolic acid biosynthesis key genes decrease dramatically in editing lines. Additionally, the growth and development of hairy roots was significantly retarded in the gene-edited lines. The cross-sections examination of laccase mutant hairy roots showed that the root development was abnormal and the xylem cells in the edited lines became larger and looser than those in the wild type. Additionally, the accumulation of RA as well as SAB was decreased, and the lignin content was nearly undetectable. It suggested that SmLACs play key roles in development and lignin formation in the root of S. miltiorrhiza and they are necessary for phenolic acids biosynthesis.
Introduction
Salvia miltiorrhiza, also known as Danshen, is a popular and effective traditional Chinese medicine used to treat cardio-cerebral vascular diseases (Guo et al., 2014). Its pharmacological activity relies on the existence of the lipid-soluble compounds such as tanshinones, along with the water-soluble phenolic acids, including rosmarinic acid (RA), salvianic acid (Danshensu), and salvianolic acid B (SAB). Previous studies have shown that SAB, which accumulates at higher levels in roots than in other organs, may relieve ischemic cardiovascular diseases by promoting angiogenesis to improve microcirculation (Yang et al., 2016). Currently, the biosynthesis pathway of phenolic acids in S. miltiorrhiza has only been elucidated from phenylalanine and tyrosine to RA, and the biosynthesis of compounds with complicated structures, such as SAB, has yet to be revealed (Figure 1A; Di et al., 2013). To determine the synthetic metabolism of these phenolic acid compounds, the analysis and comparison of structural analogs to mine key enzymes is a good method.
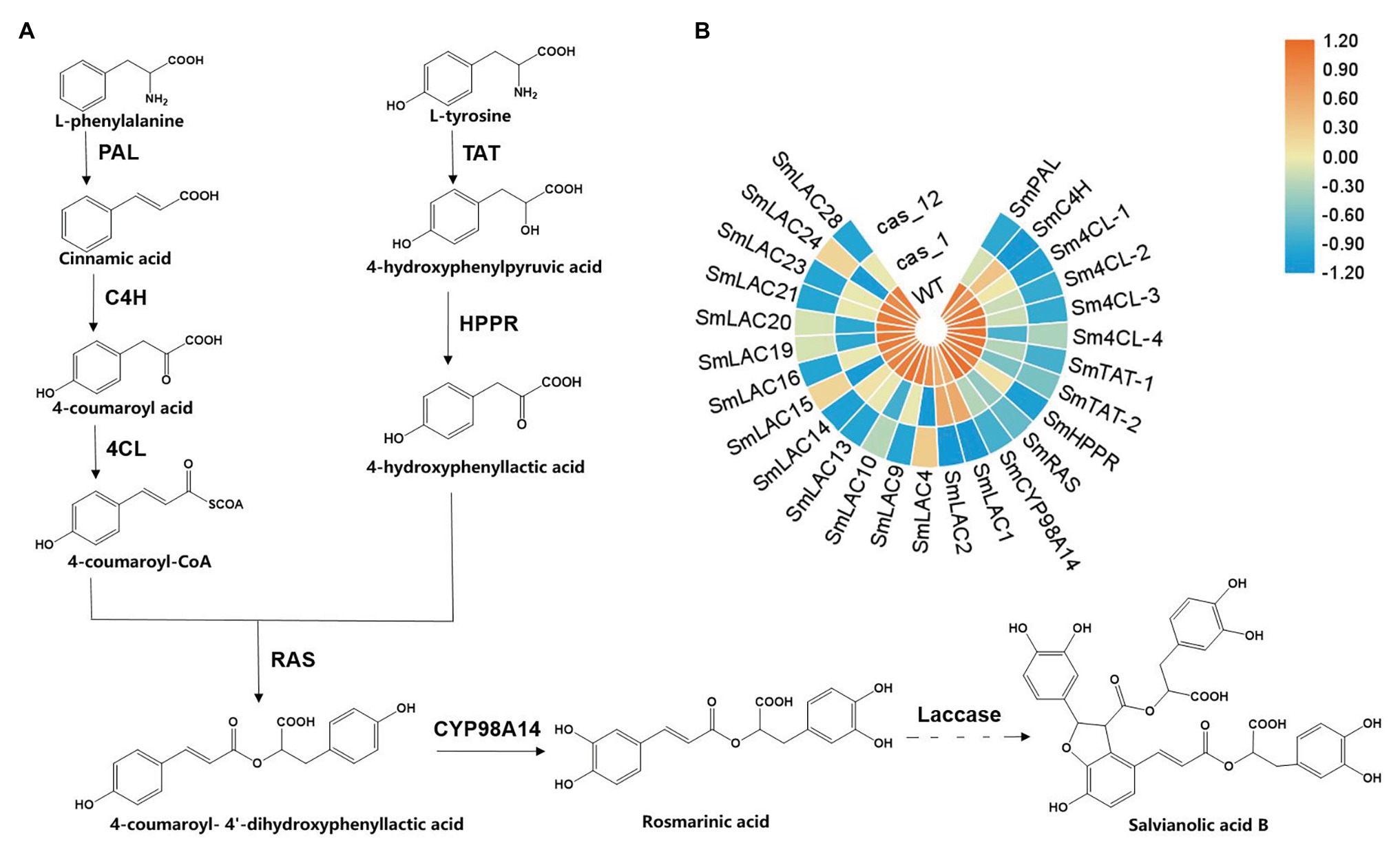
Figure 1. Scheme of the biosynthesis pathway of salvianolic acid B (SAB) in Salvia miltiorrhiza and expression profiles of key genes in transgenic hairy root lines. (A) Proposed phenolic acid biosynthesis pathway in S. miltiorrhiza. The solid line represents the verified biosynthesis process, whereas the dotted line represents the proposed biosynthetic process. (B) Heat maps of key genes of phenolic acid biosynthesis in the wild type (WT), Cas1-3, and Cas12-5 lines. Rows represent differentially expressed genes, and columns represent group comparisons. Blue and orange boxes represent lower and higher gene expression levels, respectively. The depth of color demonstrates the absolute value of the magnitude of the base-2 logarithmic fold change expression ratio. PAL, phenylalanine ammonia-lyase; C4H, cinnamic acid 4-hydroxylase; 4CL, 4-coenzyme A ligase; TAT, tyrosine aminotransferase; HPPR, 4-hydroxyphenylpylpyruvate reductase; RAS, rosmarinic acid synthase; and CYP98A14, cytochrome P450 monooxygenase.
Phenolic acids are lignin-derived products. Lignins are aromacic biopolymers derived from p-coumaryl, coniferyl, and sinapyl alcohol (monolignol) oxidative coupling (Tang et al., 2019). These compounds deposit secondarily thickened cell walls, in which they play the essential roles in structural mechanical support, water transport, and pathogen attack resistance (Ferrer et al., 2008; Burton et al., 2010). The polymerization of monolignols is facilitated by laccase proteins (Falade et al., 2017). Laccases are members of a multicopper oxidase family belonging to the benzenediol oxygen reductases (EC 1.10.3.2) and are also known as p-diphenol oxidases or urushiol oxidase (Festa et al., 2008; Wang et al., 2015). Laccases exist widely in nature and can be extracted from various species, such as bacteria, fungi, plants, and mosses (Arregui et al., 2019). Laccases are usually constructed of three Cu-oxidase domains, and they are connected as follows: Cu-oxidase_3→Cu-oxidase→Cu-oxidase_2 domain. Domains one and three house the copper sites, and the second domain often helps form a substrate-binding cleft. Highly conserved features of laccase include a copper ion bound in a Type 1 site and three more copper ions bound in atrinuclear cluster that is sometimes described as a sum of Type 2 and Type 3 sites (Solomon et al., 1996). In oxidization reactions, the substrate is first binded and oxidized at the Type 1 site in Cu-oxidase_3 domain and then transferred to a Type 2/Type 3 trinuclear copper cluster, with the release of an electron. An oxygen molecule accepts the electron, ultimately generating water (Wang et al., 2019). Because of the low substrate specificity of this gene family, most phenolic and nonphenolic molecules can be oxidized by laccases, and they are extensively applied in sewage treatment, lignin degradation, and electricity-catalyzing reactions (Viswanath et al., 2014; Chandra and Chowdhary, 2015).
In planta, laccase was first identified from the Japanese lacquer tree, Rhus vernicifera, and its function was revealed in a series of studies. In Arabidopsis thaliana, 17 putative laccase genes have been identified from the genome, and AtLAC 4 (accession number AT2G38080), AtLAC 11 (accession number AT5G03260), and AtLAC 17 (accession number AT5G58910) have been verified to be related to monolignol polymerization (Berthet et al., 2011; Zhao et al., 2013). Oryza sativa laccase may be involved in pesticide catabolism or detoxification (Huang et al., 2016). Regarding the influence of phenolic compounds, the overexpression of GaLAC1 (accession number AY423714.1) from Gossypium arboretum in A. thaliana resulted in an increase in sinapic acid conversion into a monolactone-type dimer (Wang et al., 2004). In S. miltiorrhiza, after silencing a single SmLAC gene, the content of RA and SAB in transgenic hairy roots was reduced (Li Q. et al., 2019).
Because of the functional redundancy of laccase genes, single suppressed gene has little influence on the plant phenotypes (Simoes et al., 2020). Therefore, it is necessary to silence or knockout multiple genes in the laccase gene family to comprehensively evaluate their roles. Due to the high homology of conserved domains in 29 laccase genes, we used the CRISPR/Cas9 system to edit conserved domains to target several laccase genes simultaneously. The editing targets were selected among 29 SmLACs based on bioinformatics analysis and nucleotide and amino acid sequence alignments. Then, the targets were introduced into a binary vector to construct a single- or double-locus editing vector, which was transformed into hairy roots via the Agrobacterium rhizogenes method. The results showed that more than 20 SmLACs were knocked out with this system. Quantitative real-time PCR (qRT-PCR) and RNA-Seq experiments were performed to better understand the impact of SmLACs suppression, and the results demonstrated that the expression of target genes was decreased. Interestingly, the expression level of phenolic acid biosynthesis key genes was also reduced. The development of hairy roots was significantly suppressed in the gene-edited lines, and the RA and SAB contents were decreased dramatically. The examination of cross-sections of laccase mutant hairy roots showed that root development was abnormal and the xylem cells in the edited lines appeared larger and looser than those in the wild type (WT). Furthermore, lignin was nearly undetectable. In conclusion, this study showed that SmLACs play important roles in development and lignin formation in the root of S. miltiorrhiza and they are necessary for phenolic acids biosynthesis.
Materials and Methods
Conserved Domain Alignment and sgRNA Design
Conserved motifs in the complete amino acid sequences of SmLACs were identified using Multiple EM software for Motif Elicitation with the setting of a maximum of 10 motifs (Bailey et al., 2006). Alignments of SmLAC gene amino acid and nucleotide sequences were performed with MEGA 5.05 software with p-distance settings. The nucleotide sequences of highly conserved domains were selected and used in the online CRISPR/Cas9 design tool CHOPCHOP1 to search for potential gene editing sites. Every possible editing target followed by 5'-NGG was evaluated and ranked considering the off-target possibilities and editing location. Sequences with high marks are ideal targets for editing.
Vector Construction
Pairs of complementary oligos from selected sgRNAs were synthesized and annealed to generate dimers (Supplementary Table S1). For single-target-editing vector construction, the sgRNA1 dimer was directly cloned into the CRISPR/Cas9 system by BbsI digestion. For dual-targets-editing vector construction, the sgRNA2 dimer was first cloned into the CRISPR/Cas9 system, and the cassette including AtU6 and sgRNA2 was then amplified with KpnI and EcoRI restriction sites and subcloned into the single-target-editing vector. Finally, two CRISPR/Cas9 vectors were subcloned into the linearized pCAMBIA1300 (Cambia, Canberra, and Australia) plant expression vector by double restriction enzyme digestion and recombination.
Plant Materials
The S. miltiorrhiza cultivar (RA content, 3.5–3.8% DW and SAB content, 2.8–3.0% DW) used in this study was selected and developed over several years at the Second Military Medical University. The procedures for obtaining aseptic seedlings, Agrobacterium-mediated transformation, and hairy root cultures were performed as previously described (Zhou et al., 2018).
Transgenic Hairy Root Identification and Target Gene Mutation Detection
DNA was extracted from the obtained hairy roots via the CTAB method, and two pairs of primers were used to detect the rolB and cas9 genes from the C58C1 strain, as well as binary vector by PCR amplification. The fragments surrounding LAC10, LAC11, LAC13, LAC16, LAC21, LAC27, and LAC28 were amplified by PCR using the primers listed in Supplementary Table S1. Gene editing induced mutations was detected by aligning the sequencing chromatogram files of these PCR products with those of the WT controls. To identify the genotypes of the mutations, PCR products were cloned into a blunt zero vector (Transgen Biotech, Beijing) for sequencing.
Relative Expression Analysis via Quantitative Real-Time PCR
Total RNA was extracted from samples using the EasyPure Plant RNA Kit (Transgen Biotech, China) and then reverse transcribed to produce cDNA using a TransScript First-Strand cDNA Synthesis SuperMix Kit (TransGen Biotech). qRT-PCR assays were performed as reported previously (Zhou et al., 2018). All of the primers used for qRT-PCR are listed in Supplementary Table S1.
RNA-Seq Analysis
Total RNA was extracted from the hairy roots of Cas1-3, Cas12-5, and WT using the TRIzol reagent (TransGen Biotech, China). RNA quality was examined on a Bioanalyzer 2,100 spectrophotometer (Agilent), and the RNA was stored at −80°C. RNA samples with an RIN (RNA Intergrity Number) >8.0 were considered suitable for subsequent experiments. cDNA libraries were prepared by using the NEBNext™ Ultra Directional RNA Library Prep Kit (NEB, United States), the NEBNext Poly(A) mRNA Magnetic Isolation Module, and NEBNext Multiplex Oligos according to the manufacturer’s instructions. The products were purified and enriched by PCR to produce the final cDNA libraries and were quantified on an Agilent 2,100 spectrophotometer. The tagged cDNA libraries were pooled in equal ratios and used for 150 bp paired-end sequencing in a single lane on the Illumina HiSeqXTen platform. The fragment counts of each gene were normalized via the fragments per kb per million (FPKM) method.
Histochemical Staining of Hairy Roots
Histochemical staining was performed on sections cut from the maturation zone. The hairy roots were embedded in 7% agarose before being transversely sectioned at a thickness of 50 μm using a vibratome (Leica VT1000S, Leica, Germany). The safranin O-green staining method was used to detect the deposition and composition of lignin (Bouvier et al., 2013). All sections were observed under an Olympus BX43 microscope (Japan).
Rosmarinic Acid, Salvianolic Acid B, and Lignin Content Determination
Hairy roots of different lines were collected and dried at 40°C, and 0.1 g samples were then used to measure RA and SAB contents by high-performance liquid chromatography-tandem mass spectrometry as reported previously (Agilent 1,200, United States; Zhou et al., 2018). Lignin content was estimated spectrophotometrically as thioglycolic acid (TGA) derivative. Lignin extraction and the determination processes were similar to those described previously (Sircar and Mitra, 2009).
Statistical Analysis
Eight independent transgenic lines from each suppression group, namely, Cas1-2, Cas1-3, Cas1-7, Cas1-10, Cas12-1, Cas12-5, Cas12-6, and Cas12-9 lines were selected for subsequent analysis. The WT lines were used as controls. All parameters were expressed as the mean ± SD of three replicates. All data were derived from three biological repeats with three technical replicates. One-way ANOVA was applied and significant differences between mean values of treatments were analyzed using the Student’s t-test.
Results
Conserved Domain Alignment of SmLACs and Editing Site Selection
The complete open reading frames (ORFs) of 29 SmLACs were obtained from a previous study, and each of them was analyzed for conserved domains with BLAST (Li Q. et al., 2019).
Two conserved domain sequences were selected as target loci for editing, and the online web tool CHOPCHOP was used to search for suitable sgRNA sequences for the CRISPR/Cas9 editing system. Potential editing sites were ranked according to software scores based on several factors, such as self-complementarity and possible editing efficiency. Finally, two 20 bp gene editing sites were identified: sgRNA1: GCCAACTATATATGCTCGAA, corresponding to the conserved Cu-oxidase_3 domain, and sgRNA2: CAATGCATAAACCACACCCC, corresponding to the conserved Cu-oxidase_2 domain (Figures 2A,B).
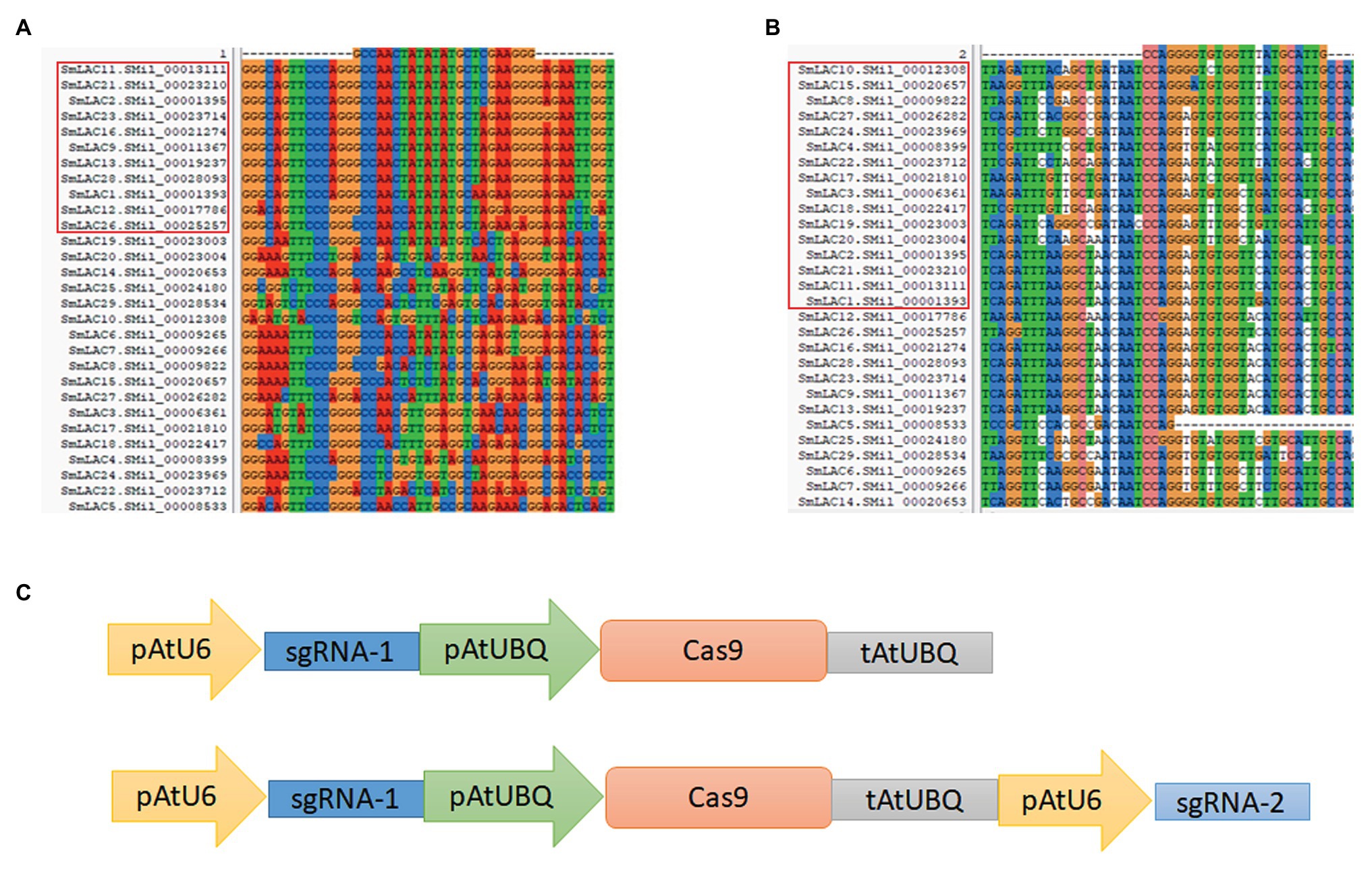
Figure 2. Overview of the CRISPR/Cas9 system for SmLAC disruption. (A,B) Partial sequence alignment of the first and second conserved copper domains among 29 SmLACs. The target sequence and PAM sequence (GGG and TGG) are listed above. The highly conserved regions from different SmLACs aligned with the editing targets are indicated (red frame). (C) Schematic view of two CRISPR/Cas9 knockout vectors used in this study. The cas9 protein was driven by the ubiquitin promoter, and the sgRNA was driven by the U6 promoter of Arabidopsis. The Cas1 vector carried only one AtU6-sgRNA cassette, and the Cas12 vector carried two AtU6-sgRNA cassettes to induce double mutations in SmLACs.
The sgRNA1 sequence existed in 11 SmLACs (LAC1, LAC2, LAC9, LAC11, LAC12, LAC13, LAC16, LAC21, LAC23, LAC26, and LAC28), and the sgRNA2 sequence existed in 16 SmLACs (LAC1, LAC2, LAC3, LAC4, LAC8, LAC10, LAC11, LAC15, LAC17, LAC18, LAC19, LAC20, LAC21, LAC22, LAC24, and LAC27). Both the sgRNA1 and sgRNA2 sequences existed in four genes (LAC1, LAC2, LAC11, and LAC21).
CRISPR/Cas9 Expression Vector Construction
The CRISPR/Cas9 system was provided by Jian-Kang Zhu’s laboratory at the Shanghai Center for Plant Stress Biology, Chinese Academy of Sciences. In the vector, the sgRNA was driven by the U6 promoter, and the Cas9 protein was driven by the ubiquitin promoter from A. thaliana. To determine the editing efficiency for multiple genes in S. miltiorrhiza, we constructed two vectors. To construct the single-locus knockout vector, the 20 bp sgRNA1 sequence was inserted between the AtU6 and ubiquitin promoters. To generate the dual-site editing vector, a new cassette including the extra AtU6 promoter and sgRNA2 was introduced into the single-locus knockout vector (Figure 2C). The whole expression cassette was subcloned into the pCAMBIA1300 plant expression vector for A. tumefaciens-mediated transformation. The vector carrying sgRNA1 was referred to as Cas1, and the vector carrying both sgRNA1 and sgRNA2 was referred to as Cas12. The empty pCAMBIA1300 vector was used as a control.
Transgenic Hairy Root Identification and SmLACs’ Expression Analysis
After Agrobacterium rhizogenes-mediated transformation, hairy roots were regenerated from wounded young leaves and grew to maturity (Figures 3A–F). Sixteen strains of transgenic hairy roots were obtained in both the Cas1 and Cas12 lines, and PCR amplification was used to identify the positive transgenic hairy roots. The results showed that the CRISPR/Cas9 vector was successfully transformed into 15 transgenic hairy roots in Cas1 lines and 14 transgenic hairy roots in Cas12 lines (Figures 3G,H). Four transgenic hairy root lines from Cas1 and Cas12 lines were used in subsequent studies.
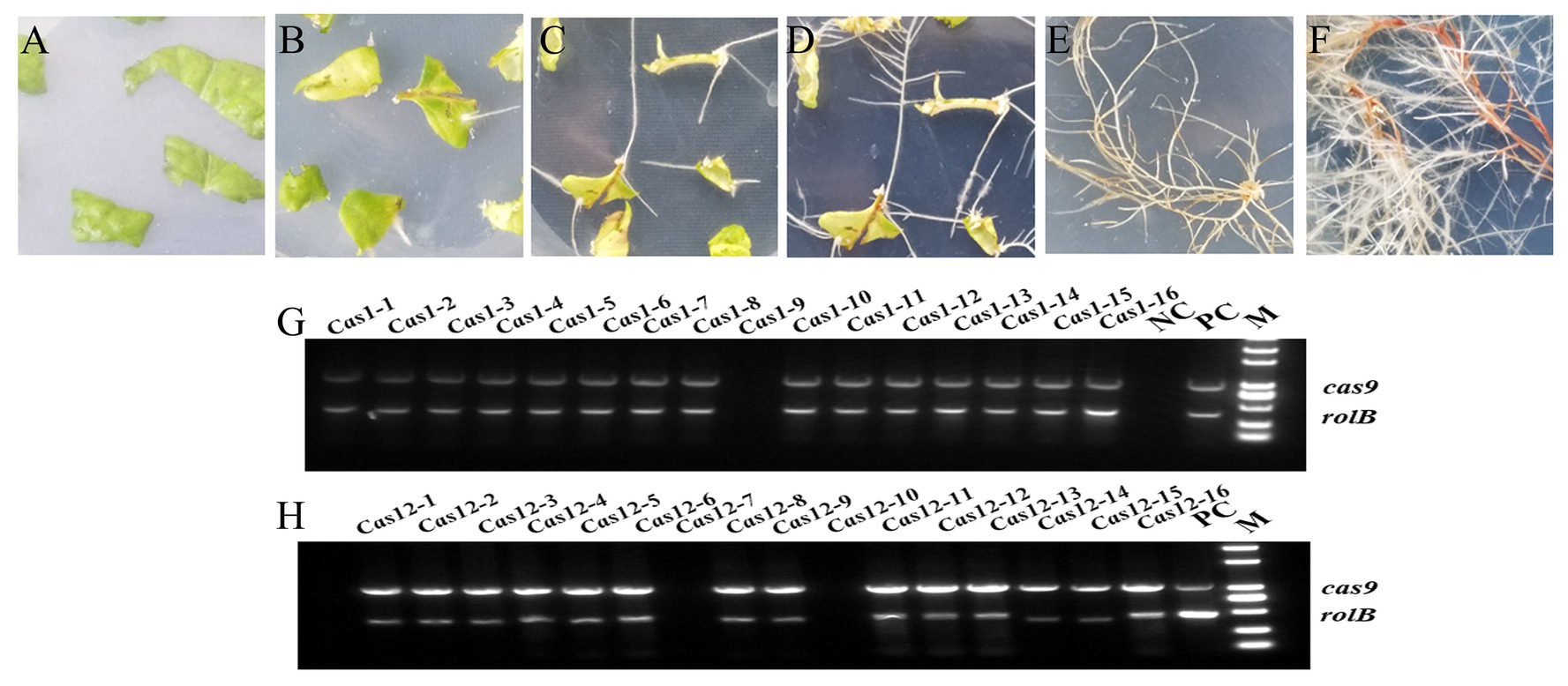
Figure 3. Cultivation transgenic hairy roots and identification of positive lines. (A) The leaf discs are spread on MS medium after Agrobacterium-mediated infection and undergo (B) callus formation, (C) hairy root growth, (D) bifurcation, (E) lines separation, and (F) growth cultivation to obtain mature roots for analysis. (G,H) Identification of the positive transgenic hairy roots using PCR amplification. The rolB gene from the Agrobacterium rhizogenes C58C1 strain and the cas9 gene from the constructed vector were amplified with two pairs of primers. The empty vector line was used as a positive control (PC), and WT hairy roots were used as a negative control (NC). M indicates marker 2000.
Five laccase genes from each editing domains were selected to preliminarily overview the effect of the CRISPR/Cas9 system by qRT-PCR. The results showed that the expression levels of most of the targeted genes were decreased dramatically after editing (Supplementary Figure S1). To better understand the impact of laccase editing, WT, Cas1-3 and Cas12-5 transgenic plants were evaluated via RNA-Seq experiments. A total of 57,609,184, 60,855,444, and 57,987,044 bp sequencing reads were successfully produced for WT, Cas1-3, and Cas12-5, respectively. Moreover, compared with the WT line, 3,108 and 2,666 differentially expressed genes were identified from the Cas1-3 and Cas12-5 lines with an FDR < 0.05 and log2|FC|>1.
The former identified 29 SmLAC genes as well as phenolic acid biosynthesis key genes were found on the basis of transcriptome statistics and a heat map was used to display the expression differences clearly (Figure 1B). The results showed that the expression levels of the edited laccase genes were decreased, and this result was in accordance with the qRT-PCR detection. In the Cas1-3 lines, the expression levels of six SmLACs (LAC4, LAC10, LAC15, LAC19, LAC20, and LAC24) were decreased after gene editing. Besides, nine SmLACs’ expression levels of Cas12-5 lines were also reduced (LAC1, LAC2, LAC9, LAC13, LAC14, LAC16, LAC21, LAC23, and LAC28). Compared with WT lines, the expression levels of genes encoding phenylalanine ammonia-lyase (PAL), cinnamic acid 4-hydroxylase (C4H): 4-coenzyme A ligase (4CL), tyrosine aminotransferase (TAT), 4-hydroxyphenylpylpyruvate reductase (HPPR), rosmarinic acid synthase (RAS), and cytochrome P450 monooxygenase (CYP98A14) in SmLAC editing lines were decreased dramatically, especially in Cas12-5 line. These results indicate that the expression levels of most SmLACs and phenolic acid biosynthesis key genes were markedly decreased in the transgenic edited lines.
To detect the mutations in the Cas1-3 and Cas12-5 lines, selected SmLAC regions (approximately 500 bp) containing the target sequences were amplified using specific primers. The purified PCR products were sequenced directly, and the corresponding sequencing chromatogram files were decoded using the degenerate sequence decoding method. Salvia miltiorrhiza is a diploid plant, and the CRISPR/Cas9 system can lead to two mutation types in the target genes. A simplex peak suggests that a mutation appears on both strands and can be designated a homozygous mutation. An overlapping peak indicates that a transgenic hairy root is heterozygous and contains a mutation that appears on a single strand or that different mutations occur on the two DNA strands. All the PCR products from heterozygotes were cloned into a sequencing vector to identify mutations on different strands.
Among the successfully target-edited lines, most of the mutation events consisted of one or two nucleotide deletions or insertions. However, the loss of large fragments was also detected, as found in Cas12-5-LAC21 (Supplementary Figure S3). In most cases, the insertion or deletion of nucleotides influenced SmLAC translation, resulting in ORF shifts or early termination of target genes. The results indicated that it is feasible to knockout multiple targets with a single binary vector harboring several sgRNAs for gene editing in S. miltiorrhiza.
Phenotypic Analysis of CRISPR/Cas9-Generated Transgenic Hairy Roots
To evaluate the phenotype of the transgenic hairy roots, the growth status, microstructure, and metabolite content of the WT and edited lines were collected. Compared with the thriving growth of the control lines, hairy roots in SmLACs mutant lines showing retardation of root growth with the emergence of sparse root hairs (Figures 4A–F). The roots of the Cas1 lines were thin, and there is no or only one to two lateral roots distributed along the axial root (Figures 4G–I). Moreover, the root growth of the Cas12 lines was retarded and stopped at the elongation stage (Figures 4J–L). Additionally, the number of hairy roots extending from the callus was strikingly different among the control and gene-edited lines. Compared with the control lines, the SmLACs mutant lines developed fewer regenerated roots from callus.

Figure 4. Phenotype of SmLACs-edited hairy roots. Hairy root growing status at 1, 2, and 3 months after Agrobacterium-mediated infection of (A–C) WT lines, (D–F) empty vector lines, (G–I) Cas1 lines, and (J–L) Cas12 lines.
To investigate the root microstructure and changes in lignin after gene editing, transverse sections of the hairy roots of Cas1-3, Cas12-5, and WT plants were studied, and safranin O-green staining was performed. This staining results in the development of a red stain in the presence of lignified cell walls. It was found that lignin accumulation was rarely observable in the mutant lines compared with the WT lines. Moreover, there is no obvious lignin staining in xylem of SmLACs-edited lines. Higher magnification observations of root transverse sections suggested that very few vessels were lignified in the mutants, whereas lignification was observed in most of the xylem vessels in the WT lines. Additionally, the SmLACs-edited lines showed cellular structural abnormalities, reflected in differences in root cell shape and organization. The arrangement of the xylem and phloem of the SmLACs-edited lines was completely different from that in the controls. In the WT and empty vector lines, the xylem vessels of the roots were radially arranged, while the phloem was arranged in concentric rings (Figures 5A–F). However, as observed in the Cas1 lines, the xylem was arranged sparsely, and some cells in the phloem appeared to be collapsed (Figures 5G–I). According to the Cas12 lines, disorganized vascular patterning involving xylem cells developed in regions where phloem cells were distinctly enlarged and irregular (Figures 5J–L).
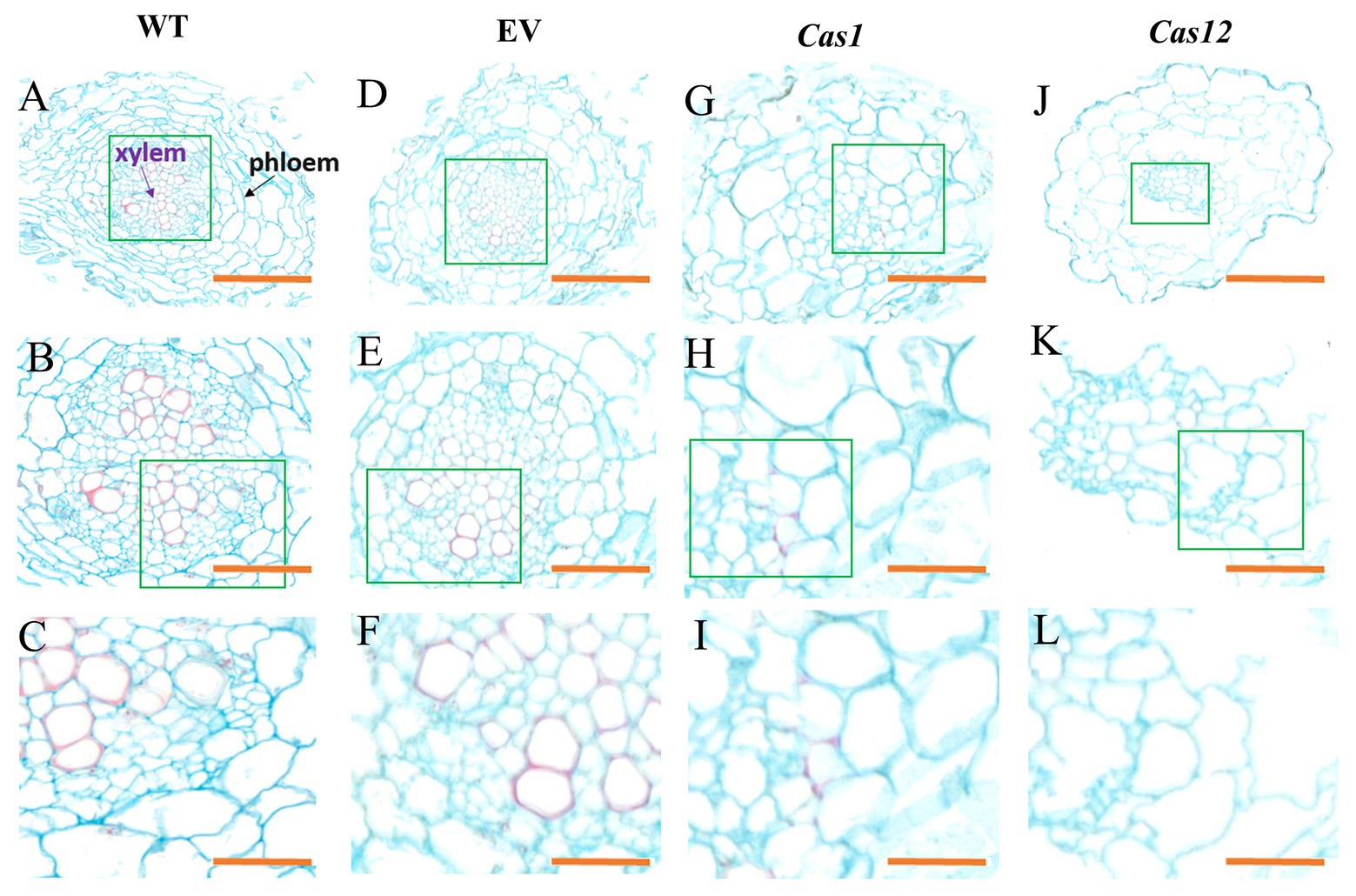
Figure 5. Phenotype of root microstructure in laccase mutant observed in cross-sections. The microstructure of (A–C) WT lines, (D–F) empty vector lines, (G–I) Cas1 lines, and (J–L) Cas12 lines. The purple arrow indicates the xylem, and the black arrow indicates the phloem. The green rectangles represent enlarged regions. Bars: (A,D,G,J) 100 μm; (B,E,H,K) 50 μm; and (C,F,I,L) 20 μm.
To explore the influence of the SmLACs mutations on phenolic acid synthesis, the determination of RA and SAB in regenerated hairy root extracts was performed via the HPLC/MS method; retention times were assessed in comparison with authentic standards as well as by searching data in the literature. The results showed that the accumulation of RA and SAB was decreased in all edited lines. The content of RA and SAB in Cas1-3 and Cas12-5 lines decreased considerably among the transgenic lines (Figures 6A,C). In addition, the lignin content was dramatically reduced in SmLACs suppressed lines (Figure 6B). These results clearly argued that SmLACs play a key role in lignin as well as phenolic acid biosynthesis.
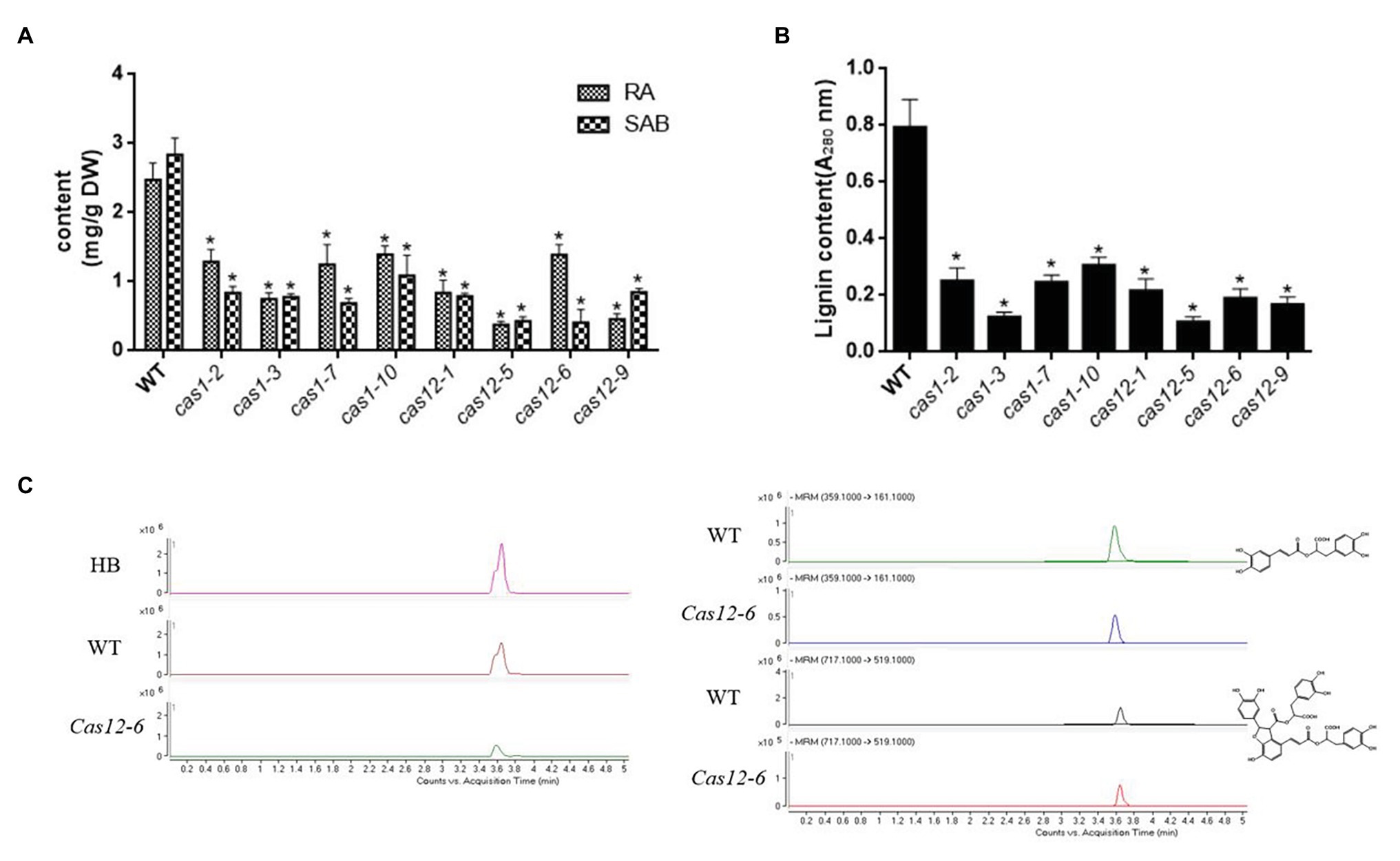
Figure 6. Accumulation of RA, SAB and lignin in transgenic hairy roots. (A) The contents of RA and SAB in transgenic hairy roots. (B) The contents of lignin in transgenic hairy roots. (C) Representative total ion chromatography (TIC) and MRM chromatography of a transgenic line (Cas12-6), the control line, and standard substance RA and SAB. MRM conditions for RA, SAB are as following. RA: precursor ion is 359.1, fragmentor energy is 120 V, collision energy is 14 eV, and product ion is 161.1. SAB: precursor ion is 717.1, fragmentor energy is 150 V, collision energy is 12 eV, and product ion is 519.1. All data presented here are means ± SD of three biological replicates, with error bars indicating SDs. Statistical significance was determined by the Student’s t-test (*p < 0.05).
Discussion
Phenolic acids can be derived from the processing of lignin, and in S. miltiorrhiza, the biosynthesis of phenolic acids starts with L-phenylalanine and L-tyrosine, which are separately transformed into the intermediates 4-coumaroyl CoA and 3,4-dihydroxyphenyllactic acid. Rosmarinic acid synthase and cytochrome P450-dependent monooxygenase catalyze two precursors to synthesize RA. Laccase has been proposed as a key enzyme involved in the synthetic process of RA to SAB conversion (Figure 1A). The role of laccase participated in RA conversion to SAB was speculated based on the key process of lignin polymerization. This reaction involves so-called “end-wise” polymerization in which oxidized monolignol radicals undergo cross-coupling reactions with radicals formed on the free-phenolic ends of growing lignin polymers. Permutations in the regiochemistry of the radical coupling step and variations in the mode of the post-coupling quinone methide rearomatization step allow numerous possible inter-mono-meric linkages in the final lignin polymer. The SAB could be regard as the dimer of RA and it is essential to understand laccase gene family in S. miltiorrhiza (Supplementary Figure S2).
In previous study, although three laccase genes in S. miltiorrhiza (SmLAC7, SmLAC20, and SmLAC28) have been reported to influence phenolic acid synthesis, the roles of SmLACs in physiological processes remain less clear (Li Q. et al., 2019). Here, we studied laccase function from a comprehensive perspective, focusing not only on phenolic acid metabolism but also on the alteration of physiological processes in S. miltiorrhiza. In this study, we reported the roles of SmLACs in the alteration of lignin synthesis, which was essential for root development and phenolic acid ingredient metabolism. The retardation of hairy root growth and lateral root formation caused by CRISPR/Cas9 editing may result from the disturbance of diverse physiological and biochemical processes. The significant microstructural changes observed in transverse sections from SmLAC-edited lines also suggested that laccases play a key role in root development. In addition, the contents of RA and SAB as well as the expression of key genes in phenolic acid biosynthesis pathway were decreased dramatically, suggesting that SmLAC genes are involved in phenolic acid biosynthesis.
The laccase genes in S. miltiorrhiza can be clustered into seven groups according to previous study (Li Q. et al., 2019). In this research, the target of sgRNA1 belonged to group VII and the sgRNA2 belonged to I, II, III, IV, and V groups. The results showed that the accumulation of lignin and phenolic acids in most of cas12 lines was lower than that in cas1 lines. Additionally, the ratio of SAB to RA seemed to be lower than that of WT lines if laccase was involved in dimerization of RA to SAB. However, the contents of RA and SAB among the transgenic lines were not as expected. Furthermore, the growing status of cas12 lines seemed to be poorer than cas1 lines. These phenotypes indicated that the functions of SmLACs are redundant and their roles in different SmLACs groups are cross-functional. It is meaningful to figure out their function in following studies.
Despite the high degree of redundancy of laccase genes in S. miltiorrhiza, it is difficult to identify lignin-specific genes (Li C. et al., 2019). It is time consuming to generate multiple mutants to study the function of multiple genes. In gene family function studies in particular, the function of a single gene might be redundant, and multiple genes should be knocked out to explore their phenotypes and roles. Therefore, this study also provides new gene editing methods to study laccase gene family in S. miltiorrhiza. In previous studies, the CRISPR/Cas9 system as reported to target a single gene locus at a time, and this study performed dual-locus editing in S. miltiorrhiza (Li et al., 2017; Zhou et al., 2018; Deng et al., 2020). This work will facilitate the application to a new strategy for functional studies, targeting multiple genes and gene families in S. miltiorrhiza.
In conclusion, we revealed the function of laccase in S. miltiorrhiza. The laccase family may influence expression level of phenolic acid biosynthesis key genes, lignin biosynthesis, and hairy root growth and development resulting in the accumulation of phenolic acid content (Figure 7). The results provide a solid ground for further exploring laccases and uncover multiply functions of laccase in S. miltiorrhiza.
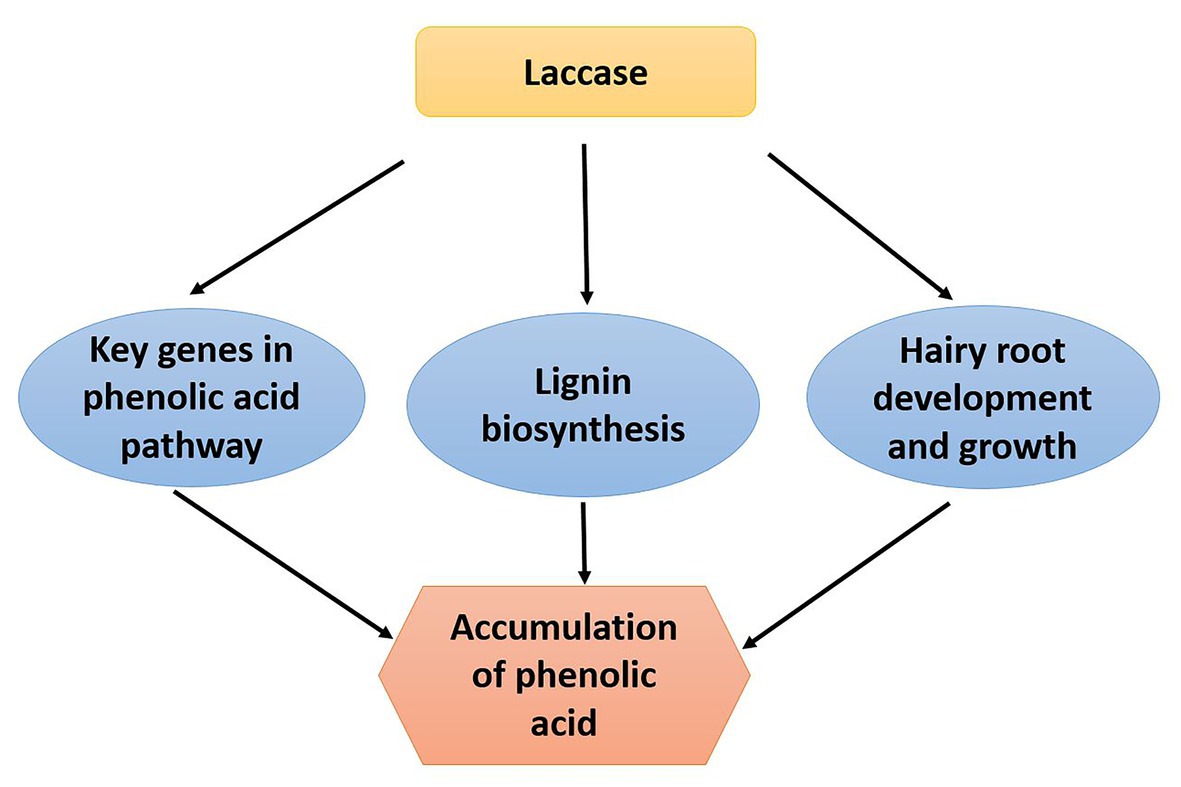
Figure 7. The mechanistic model depicting S. miltiorrhiza laccase gene family regulates phenolic acid accumulation. The laccase gene family of S. miltiorrhiza may regulate accumulation of phenolic acid by influencing expression level of phenolic acid biosynthesis key genes, lignin biosynthesis, and hairy root growth and development.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
ZZ, MG, WC, and LZ conceived and designed the entire research plans. ZZ, QL, JF, and LX performed most of the experiments. QL, YW, YX, QB, and KH provided technical assistance to ZZ. ZZ and LZ wrote the manuscript. MG and WC helped with the organization and editing. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by National Natural Science Foundation of China (grant nos. 31770329, 32070327, and 82003897), National Key R&D Program of China (2019YFC1711100), China Postdoctoral Science Foundation (2019M661602), Shanghai Rising-Star Program (18QB1402700), and Shanghai Local Science and Technology Development Fund Program guided by the Central Government (YDZX20203100002948).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.647768/full#supplementary-material
Footnotes
References
Arregui, L., Ayala, M., Gomez-Gil, X., Gutierrez-Soto, G., Hernandez-Luna, C. E., Herrera, D. L. S. M., et al. (2019). Laccases: structure, function, and potential application in water bioremediation. Microb. Cell Factories 18:200. doi: 10.1186/s12934-019-1248-0
Bailey, T. L., Williams, N., Misleh, C., and Li, W. W. (2006). Meme: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 34, W369–W373. doi: 10.1093/nar/gkl198
Berthet, S., Demont-Caulet, N., Pollet, B., Bidzinski, P., Cezard, L., Le Bris, P., et al. (2011). Disruption of laccase4 and 17 results in tissue-specific alterations to lignification of Arabidopsis thaliana stems. Plant Cell 23, 1124–1137. doi: 10.1105/tpc.110.082792
Bouvier, D. M., Bouchabke-Coussa, O., Voorend, W., Antelme, S., Cezard, L., Legee, F., et al. (2013). Disrupting the cinnamyl alcohol dehydrogenase 1 gene (bdcad1) leads to altered lignification and improved saccharification in Brachypodium distachyon. Plant J. 73, 496–508. doi: 10.1111/tpj.12053
Burton, R. A., Gidley, M. J., and Fincher, G. B. (2010). Heterogeneity in the chemistry, structure and function of plant cell walls. Nat. Chem. Biol. 6, 724–732. doi: 10.1038/nchembio.439
Chandra, R., and Chowdhary, P. (2015). Properties of bacterial laccases and their application in bioremediation of industrial wastes. Environ Sci Process Impacts 17, 326–342. doi: 10.1039/c4em00627e
Deng, C., Shi, M., Fu, R., Zhang, Y., Wang, Q., Zhou, Y., et al. (2020). Aba-responsive transcription factor bzip1 is involved in modulating biosynthesis of phenolic acids and tanshinones in Salvia miltiorrhiza. J. Exp. Bot. 71, 5948–5962. doi: 10.1093/jxb/eraa295
Di, P., Zhang, L., Chen, J., Tan, H., Xiao, Y., Dong, X., et al. (2013). 13C tracer reveals phenolic acids biosynthesis in hairy root cultures of Salvia miltiorrhiza. ACS Chem. Biol. 8, 1537–1548. doi: 10.1021/cb3006962
Falade, A. O., Nwodo, U. U., Iweriebor, B. C., Green, E., Mabinya, L. V., and Okoh, A. I. (2017). Lignin peroxidase functionalities and prospective applications. Microbiology 6:e00394. doi: 10.1002/mbo3.394
Ferrer, J. L., Austin, M. B., Stewart, C. J., and Noel, J. P. (2008). Structure and function of enzymes involved in the biosynthesis of phenylpropanoids. Plant Physiol. Biochem. 46, 356–370. doi: 10.1016/j.plaphy.2007.12.009
Festa, G., Autore, F., Fraternali, F., Giardina, P., and Sannia, G. (2008). Development of new laccases by directed evolution: functional and computational analyses. Proteins 72, 25–34. doi: 10.1002/prot.21889
Guo, H. D., Cui, G. H., Tian, J. X., Lu, P. P., Zhu, Q. C., Lv, R., et al. (2014). Transplantation of salvianolic acid B pretreated mesenchymal stem cells improves cardiac function in rats with myocardial infarction through angiogenesis and paracrine mechanisms. Int. J. Cardiol. 177, 538–542. doi: 10.1016/j.ijcard.2014.08.104
Huang, M. T., Lu, Y. C., Zhang, S., Luo, F., and Yang, H. (2016). Rice (Oryza sativa) laccases involved in modification and detoxification of herbicides atrazine and isoproturon residues in plants. J. Agric. Food Chem. 64, 6397–6406. doi: 10.1021/acs.jafc.6b02187
Li, B., Cui, G., Shen, G., Zhan, Z., Huang, L., Chen, J., et al. (2017). Targeted mutagenesis in the medicinal plant Salvia miltiorrhiza. Sci. Rep. 7:43320. doi: 10.1038/srep43320
Li, Q., Feng, J., Chen, L., Xu, Z., Zhu, Y., Wang, Y., et al. (2019). Genome-wide identification and characterization of Salvia miltiorrhiza laccases reveal potential targets for salvianolic acid B biosynthesis. Front. Plant Sci. 10:435. doi: 10.3389/fpls.2019.00435
Li, C., Li, D., Zhou, H., Li, J., and Lu, S. (2019). Analysis of the laccase gene family and mir397-/mir408-mediated posttranscriptional regulation in Salvia miltiorrhiza. PeerJ 7:e7605. doi: 10.7717/peerj.7605
Simoes, M. S., Carvalho, G. G., Ferreira, S. S., Hernandes-Lopes, J., de Setta, N., and Cesarino, I. (2020). Genome-wide characterization of the laccase gene family in Setaria viridis reveals members potentially involved in lignification. Planta 251:46. doi: 10.1007/s00425-020-03337-x
Sircar, D., and Mitra, A. (2009). Accumulation of p-hydroxybenzoic acid in hairy roots of Daucus carota 2: confirming biosynthetic steps through feeding of inhibitors and precursors. J. Plant Physiol. 166, 1370–1380. doi: 10.1016/j.jplph.2009.02.006
Solomon, E. I., Sundaram, U. M., and Machonkin, T. E. (1996). Multicopper oxidases and oxygenases. Chem. Rev. 96, 2563–2606. doi: 10.1021/cr950046o
Tang, Y., Liu, F., Xing, H., Mao, K., Chen, G., Guo, Q., et al. (2019). Correlation analysis of lignin accumulation and expression of key genes involved in lignin biosynthesis of ramie (Boehmeria nivea). Gene 10:389. doi: 10.3390/genes10050389
Viswanath, B., Rajesh, B., Janardhan, A., Kumar, A. P., and Narasimha, G. (2014). Fungal laccases and their applications in bioremediation. Enzyme Res. 2014:163242. doi: 10.1155/2014/163242
Wang, G. D., Li, Q. J., Luo, B., and Chen, X. Y. (2004). Ex planta phytoremediation of trichlorophenol and phenolic allelochemicals via an engineered secretory laccase. Nat. Biotechnol. 22, 893–897. doi: 10.1038/nbt982
Wang, W., Liu, F., Jiang, Y., Wu, G., Guo, L., Chen, R., et al. (2015). The multigene family of fungal laccases and their expression in the white rot basidiomycete flammulina velutipes. Gene 563, 142–149. doi: 10.1016/j.gene.2015.03.020
Wang, F., Xu, L., Zhao, L., Ding, Z., Ma, H., and Terry, N. (2019). Fungal laccase production from lignocellulosic agricultural wastes by solid-state fermentation: a review. Microorganisms 7:665. doi: 10.3390/microorganisms7120665
Yang, M. C., You, F. L., Wang, Z., Liu, X. N., and Wang, Y. F. (2016). Salvianolic acid B improves the disruption of high glucose-mediated brain microvascular endothelial cells via the ros/hif-1alpha/vegf and mir-200b/vegf signaling pathways. Neurosci. Lett. 630, 233–240. doi: 10.1016/j.neulet.2016.08.005
Zhao, Q., Nakashima, J., Chen, F., Yin, Y., Fu, C., Yun, J., et al. (2013). Laccase is necessary and nonredundant with peroxidase for lignin polymerization during vascular development in Arabidopsis. Plant Cell 25, 3976–3987. doi: 10.1105/tpc.113.117770
Keywords: Salvia miltiorrhiza, laccase, CRISPR/Cas9, hairy root, development, salvianolic acid B
Citation: Zhou Z, Li Q, Xiao L, Wang Y, Feng J, Bu Q, Xiao Y, Hao K, Guo M, Chen W and Zhang L (2021) Multiplexed CRISPR/Cas9-Mediated Knockout of Laccase Genes in Salvia miltiorrhiza Revealed Their Roles in Growth, Development, and Metabolism. Front. Plant Sci. 12:647768. doi: 10.3389/fpls.2021.647768
Edited by:
Guodong Wang, Institute of Genetics and Developmental Biology (CAS), ChinaCopyright © 2021 Zhou, Li, Xiao, Wang, Feng, Bu, Xiao, Hao, Guo, Chen and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Meili Guo, bWxndW9AMTI2LmNvbQ==; Wansheng Chen, Y2hlbndhbnNoZW5nQHNtbXUuZWR1LmNu; Lei Zhang, emhhbmdsZWlAc21tbXUuZWR1LmNu
†These authors have contributed equally to this work
 Zheng Zhou
Zheng Zhou Qing Li2†
Qing Li2† Meili Guo
Meili Guo