- Department of Life Science (BK21 program), Chung-Ang University, Seoul, South Korea
Protein phosphorylation by kinase is an important mechanism for adapting to drought stress conditions. Here, we isolated the CaDIMK1 (Capsicum annuum drought-induced MAP kinase 1) from dehydrated pepper leaf tissue and functionally characterized it. Subcellular localization analysis revealed that the CaDIMK1 protein was localized in the cytoplasm and nucleus. CaDIMK1-silenced pepper plants exhibited drought-susceptible phenotypes that were characterized by increased transpiration rates, low leaf temperatures, and decreased stomatal closure. In contrast, CaDIMK1-overexpressing (OX) transgenic Arabidopsis plants were hypersensitive to abscisic acid (ABA) from germination to adult growth stages. Furthermore, the CaDIMK1-OX plants were tolerant to drought stress. The transcript levels of several stress-related genes were high in CaDIMK1-OX plants than in wild-type plants. Taken together, our data demonstrate that CaDIMK1 acts as a positive modulator of drought tolerance and ABA signal transduction in pepper plants.
Introduction
Sessile plants are exposed to various environmental stresses that can lead to decreased crop yields. Water deficits caused by cold temperature, high salinity, and drought stresses can impact plant survival. Water is a key factor for plant growth, development, and survival. Plants have adapted to water-deficit conditions by altering many survival processes such as stomatal closure, stress-related gene expression, and abscisic acid (ABA) accumulation (Golldack et al., 2014; Basu et al., 2016; Ullah et al., 2018).
The phytohormone ABA plays a critical role in response to abiotic stress as well as plant growth and development (Finkelstein et al., 2002; Tuteja, 2007). When plants encounter a water-deficit condition, ABA is synthesized in several plant tissues, in particular in the leaves, leading to the initiation of signal transduction associated with a plant-adaptive response (Schroeder et al., 2001; Vahisalu et al., 2008; Lee et al., 2009, 2013). When the guard cells recognize ABA, the turgor and volume of the guard cells decrease, resulting in stomatal closure (Kim et al., 2010; Dong et al., 2018). The core ABA signal transduction pathway is composed of ABA receptors (PYR/PYL/RCAR) that directly bind to ABA and perceive ABA signals (Gonzalez-Guzman et al., 2012; Dittrich et al., 2019). This complex recognizes clade A protein phosphatase 2Cs (PP2Cs), including AHG1, PP2CA, HAB1, HAB2, ABI1, ABI2, AIP1, AIP2, and AIP3, and in turn inhibits phosphatase activity. The interactions between PP2Cs and PYR1/PYLs/RCARs lead to the release of the SnRK2 type kinases from PP2Cs, which activate downstream signaling, including transcription factors and ion channels (Brandt et al., 2012; Komatsu et al., 2013; Lim et al., 2015; Shinozawa et al., 2019). The biological functions of ABA have been widely studied; however, the precise mechanisms for ABA signaling and ABA-mediated drought stress remain largely unexplored.
All eukaryotes evolutionarily have the mitogen-activated protein kinase (MAPK) signaling module, which is associated with the regulation of plant growth, development, and stress response (Rodriguez et al., 2010; Xu and Zhang, 2015; Bigeard and Hirt, 2018). Three protein kinases commonly constitute the MAPK cascade: MAPK, MAPK kinase (MAP2K), and MAPK kinase (MAP3K), which are linked in a variety of ways to specific upstream activators and downstream substrates (Jonak et al., 2002; Mishra et al., 2006). Many MAPK cascades play a role in response to abiotic stress (Rodriguez et al., 2010), and its activation has been reported to be associated with ABA in various plant species (Danquah et al., 2014; de Zelicourt et al., 2016). Previous studies have revealed the components of MAPK cascades: 60 MAP3Ks, 20 MAPKs, and 10 MAP2Ks in Arabidopsis (Ichimura et al., 2002). The MAP3Ks constitute the largest group of kinases in the MAPK cascade and are classified into three groups: Raf-like kinase, ZR1-interacting kinase (ZIK), and MEKK (Jonak et al., 2002). In abiotic stress responses, several MAP3Ks are involved with the ABA core signaling pathway. Recently, MAP3Ks were found to be part of the activation of some SnRK2-type kinases through ABA-dependent and ABA-independent manner (Lin et al., 2020; Soma et al., 2020; Takahashi et al., 2020). Group A PP2C ABI1 interacts with MAP3K18 and inhibits its kinase activity (Mitula et al., 2015). Additionally, AIK1/MKKK20 modulates ABA sensitivity in terms of guard cell signaling, primary root growth, and development in Arabidopsis (Li Y. et al., 2017), and MAP3K YDA/YODA plays an essential role in stomatal patterning and inflorescence development (Bergmann et al., 2004; Wang et al., 2007). Loss-of-function mutants of AIK1/MKKK20 exhibit an increased number of stomata, consistent with clustered stomata in loss-of-function mutant of YDA/YODA.
In this study, the pepper MAP3K/MEKK gene, CaDIMK1 (Capsicum annuum drought-induced MAP kinase 1) was identified, which was highly induced by drought stress. ABA also increases the transcript level of CaDIMK1. CaDIMK1-silenced peppers and CaDIMK1-overexpressing (OX) transgenic Arabidopsis plants showed altered phenotypes to drought stress and ABA treatments, accompanied by different transpiration rates and stomatal apertures. CaDIMK1-OX plants also displayed an ABA hypersensitivity in germination and seedling growth stages. These data demonstrate that CaDIMK1 acts as a positive modulator in response to drought stress and ABA.
Materials and Methods
Plant Materials and Growing Conditions
In this study, Arabidopsis thaliana ecotype Columbia-0 was used for the OX transgenic plants. Seeds were disinfected with 70% ethanol and planted on MS plates with 0.5% sucrose. After cold stratification (4°C) for 2 days, seeds of each line were germinated at 24°C and 40% humidity for 7 days. The seedlings were then transplanted into plastic pots containing vermiculite, perlite, and peat moss (9:1:1 ratio). Pepper (C. annuum cv. Nockwang) and tobacco (Nicotiana benthamiana) plants were grown in pots containing a 1:1:1 ratio of a compost soil mix (vermiculite, perlite, and peat moss, 2:3:5, v/v/v), loam soil, and sand. All seedlings were grown under the following conditions: 24 ± 1°C, 60% humidity, and long-day condition (light/dark: 16 h/8 h).
Generation of Overexpression Transgenic Plants in Arabidopsis
The CaDIMK1 coding region was amplified using primer pairs (Supplementary Table 1). The PCR products of CaDIMK1 were cloned to the entry vector (pENTR/D-TOPO; Invitrogen) for gateway cloning and then subcloned to the destination vector for making fusion protein with green fluorescent protein (GFP) through LR reaction. The 35S:CaDIMK1-GFP plasmid was introduced to the strain GV3101 of Agrobacterium tumefaciens by electroporation. For plant transformation, we used the floral dip method (Clough and Bent, 1998). Homozygous T3 transgenic seeds were grown in selective media containing 50 μg ml–1 of phosphinothricin for further studies.
Virus-Induced Gene Silencing in Pepper Plants
Virus-induced gene silencing (VIGS) assay was performed to generate CaDIMK1-silenced pepper plants using the tobacco rattle virus (TRV) as previously described (Liu et al., 2002). Briefly, a 411–710-bp region of CaDIMK1 was amplified using the specific primers (Supplementary Table 1), which were ligated into a pTRV2 vector. The strain GV3101 of A. tumefaciens harboring constructs was infiltrated by syringe in both cotyledons of the pepper plant (each construct: OD600 = 0.2).
Protein Localization Assay
For the protein localization assay, GFP-tagged CaDIMK1 was transiently expressed in N. benthamiana through agroinfiltration. The GFP fluorescence signals were detected using LSM700 confocal microscope (Carl Zeiss) 2 days after infiltration.
Abscisic Acid, Drought, NaCl, and H2O2 Treatments
We treated with either a 100-μM ABA or 100-μM H2O2 solution and irrigated with a 200-mM NaCl solution in 4-week-old pepper leaves to analyze the induction of CaDIMK1 transcripts in pepper plants. Two-week-old pepper plants were treated with drought stress by withholding watering, and the dehydrated leaves were harvested at 0, 8, 10, and 12 days after treatment. For Arabidopsis, 3-week-old plants were applied with drought stress by removing them from the soil, followed by the leaves being harvested at the indicated time.
Germination Test and Seedling Growth Assay
For a germination test, seeds of each genotype were sown on 1/2 MS agar plates with 0.5, 0.75, and 1.0 μM ABA. The germinated seeds (radicle emergence) were measured daily for 6 days. Five days after plating, the numbers of seedlings with fully expanded green cotyledons were counted, and the root lengths were measured.
Stomata Aperture Assay and Thermal Imaging Analysis
To measure the stomatal aperture, we collected leaf peels from 4-week-old pepper (TRV2:CaDIMK1 and TRV2:00) plants and 3-week-old Arabidopsis (CaDIMK1-OX line #1, line #2, and wild-type) plants cultivated under well-watered condition. The collected leaf peels were incubated on stomata open buffer (SOB) for 3 h under a light intensity of 100 μmol m–2 s–1 to open the stomata fully. After being transferred into a new SOB containing various concentrations of ABA to induce stomata closing, stomata were observed using a Nikon Eclipse 80i microscope. The ratio of stomatal aperture width to length was calculated from at least 100 stomata of each plant line using ImageJ.
Pepper and Arabidopsis plants at the same development stage were applied with 50 μM ABA for 4 h to analyze leaf temperature changes in response to ABA. Thermal images of each plant line were taken by a T420 thermal imaging camera (FLIR systems).
Water Loss Measurement
The leaf tissues from 4-week-old pepper (TRV2:CaDIMK1 and TRV2:00) plants and rosette leaves from 3-week-old Arabidopsis (CaDIMK1-OX line #1, line #2, and wild-type) were harvested and placed in a growth chamber. Transpirational water losses of pepper and Arabidopsis were examined by measuring the fresh weight of leaf samples during 10 and 7 h, respectively, after detachment.
RNA Isolation and Semiquantitative Reverse Transcription-Polymerase Chain Reaction (RT-PCR) and Quantitative RT-PCR Assay
RT-PCR analysis was performed using plant total RNA samples isolated from 3-week-old pepper or 3-week-old Arabidopsis plants using TRI reagent (Invitrogen). For the removal of contamination of genomic DNA, RNA samples were applied with DNase. cDNA was synthesized by using iScript cDNA synthesis kit with 1 μg RNA and oligo-dT primers (Bio-Rad). CaDIMK1 and its homologous genes were amplified using primer pairs (Supplementary Table 1).
The expression patterns of stress-induced genes were analyzed using quantitative RT-PCR (qRT-PCR) (CFX96 TouchTMRT PCR detection system; Bio-Rad) with the iQTM SYBR Green Supermix (Bio-Rad). qRT-PCR was conducted following the manufacturer’s instructions. C. annuum Actin1 (CaACT1) and A. thaliana Actin8 (AtACT8) were used as an internal control for the normalization.
Protein Expression, Purification, and in vitro Kinase Assay
The expression and purification of GST-tagged CaDIMK1, CaDIMK1K32N, and OST1 recombinant proteins in bacterial cells were conducted as previously described (Lim et al., 2017). Briefly, the coding regions of each gene were inserted into a GST tagging Escherichia coli expression vector (pGEX4T-3), which were transferred into strain BL21 of E. coli cells. The GST-tagged proteins were expressed and purified by the glutathione S-transferase (GST) gene fusion system following the manufacturer’s instructions (GE Healthcare Bio-Sciences).
An in vitro kinase analysis used recombinant proteins that were reacted in a phosphorylation buffer (1 mM CaCl2, 1 mM dithiothreitol, 2.5 mM MgCl2, 2.5 mM MnCl2, and 20 mM Tris–HCl) with [γ-32P] ATP (7.5 μCi). After incubation at 30°C for 2 h, the reaction was terminated by boiling in a 5 × SDS-sample buffer with 25% β-mercaptoethanol, bromophenol blue (0.005%, G-250), glycerol (50%), SDS (10%), and Tris–HCl (250 mM, pH 6.8). The reacted kinases were separated using SDS-PAGE (10%). The SDS-PAGE gel was dried, and the phosphorylation signal was observed via autoradiography by Personal Molecular Imager (Bio-Rad).
Results
Isolation of Drought-Induced CaDIMK1
To isolate drought-induced MAP3 kinase, we used RNA-seq analysis and isolated eight MAP3 kinase genes from pepper plant leaves under drought stress: CA07g11510, CA07g11520, CA07g11530, CA07g11540, CA07g11550 (Jeong et al., 2020), CA07g11570, CA02g14340, CA02g14350, and CA02g14360 (Figure 1A). Domain analyses using a web-based tool (SMART)1 showed that MAP3Ks have a kinase domain, which phosphorylates tyrosine or serine-threonine amino acid residues of a target protein. We conducted qRT-PCR analysis to analyze the expression patterns of the MAP3Ks in the leaf tissues from pepper plants treated with drought stress. As shown in Figure 1B, all genes, except CA07g11550, were significantly induced by drought stress. From these genes, we selected CA07g11520, which had the highest differential expression level after drought treatment, for further investigation and was designated CaDIMK1.
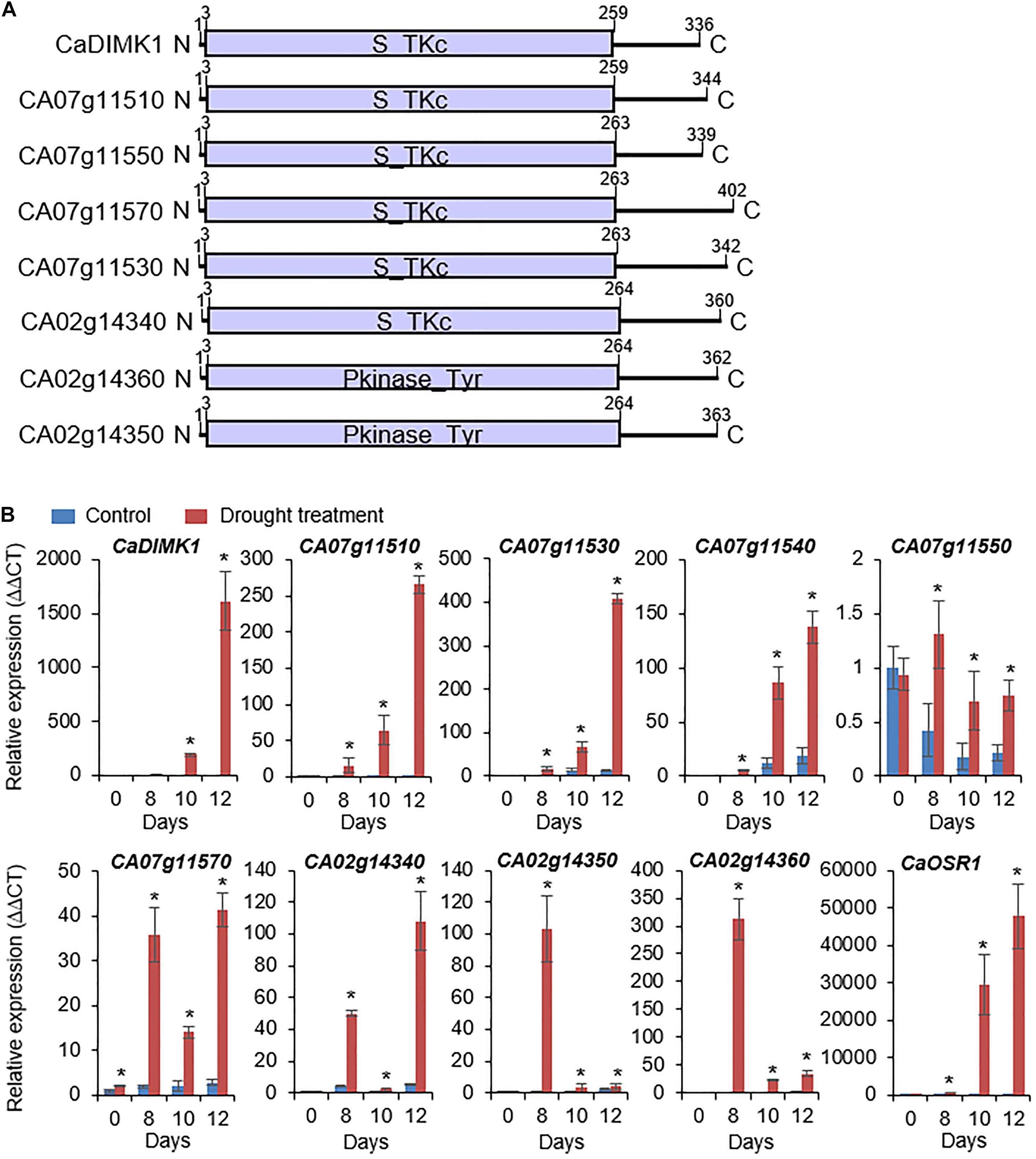
Figure 1. Drought-induced pepper MAP3 kinase genes. (A) Protein domain analysis of pepper MAP3Ks according to deduced amino acids. The kinase domain was marked via released data (web address: http://smart.embl-heidelberg.de). (B) Induction levels of MAP3K genes in pepper leaf tissue after drought stress treatment. The expression values are normalized by pepper Actin1 (CaACT1) gene as an internal control, and the induction level of each gene at 0 day was 1.0. As a positive control for drought treatment, CaOSR1 gene was amplified in parallel. Values are mean ± standard deviation, n = 3; asterisks indicate significant differences compared with nontreated control (Student’s t-test; *P < 0.05).
Capsicum annuum drought-induced MAP kinase 1 is composed of 336 amino acids (1,011-bp open reading frame) with an isoelectric point of 5.74 and a molecular weight of 37.49 kDa. When performing the BLASTP search on NCBI2, we found that CaDIMK1 (accession no. KAF3666587.1) shares relatively high identities/similarities (46.74–81.85%/63.73–86.61%) with the MAP3K proteins, in particular belonging to MEKK subfamily, from several higher plant species. A phylogenetic tree analysis was conducted using MAPKKK-MEKK proteins of the model plant Arabidopsis, together with drought- and/or ABA-responsive MEKK kinases from rice, cotton, tobacco, and tomato, characterized in previous studies (Ichimura et al., 2002; Shou et al., 2004; Rao et al., 2010; Wu et al., 2014; Mao et al., 2019; Na et al., 2019; Zhang et al., 2020). As shown in Figure 2, CaDIMK1, CA07g11520, CA07g11530, CA07g11540, CA07g11550, and CA07g11570 were clustered with AtMAPKKK15/16/17/18, SlMAPKKK51/53/55, OsMAPKKK62/63, and GhMEKK12. In contrast, CA02g14340, CA02g14350, and CA02g14360 were sorted into the same clade with AtMAPKKK19, 20, and 21. Previous studies have revealed an ABA-induced regulation of those Arabidopsis MAPKKK gene expression (Wang et al., 2011; Danquah et al., 2015) and functional involvement of some genes in ABA signaling (Mitula et al., 2015; Li Y. et al., 2017). Also, AtMAPKKK18 and GhMEKK12 play a positive role in drought tolerance of Arabidopsis and cotton, respectively (Li K. et al., 2017; Zhang et al., 2020). Based on these results, we proposed that CaDIMK1 could be involved in plant responses to ABA and drought stress.
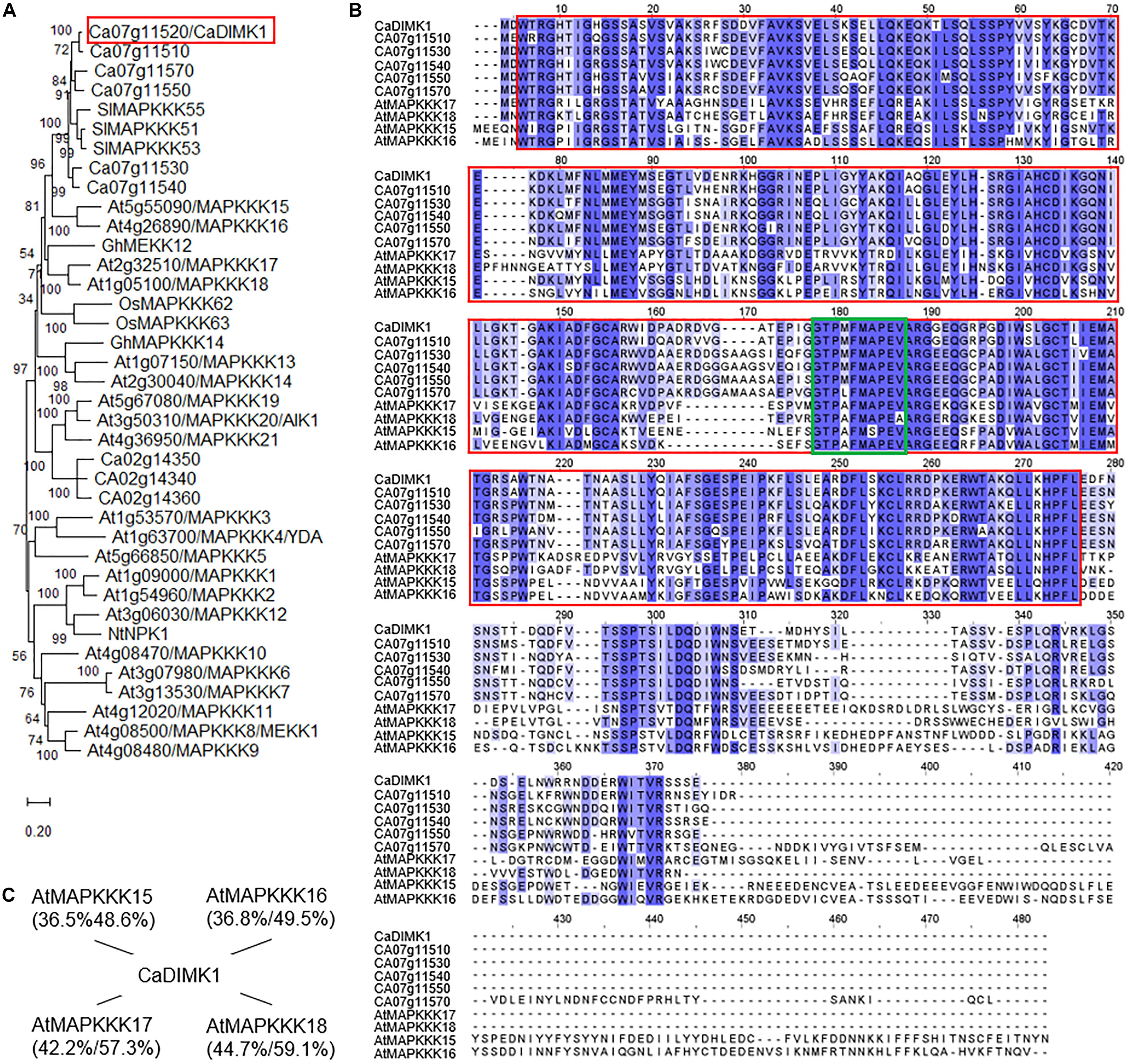
Figure 2. Amino acid sequence analysis of CaDIMK1. (A) Phylogenetic tree of pepper MAP3 kinases. The phylogenetic analysis was conducted via MEGA software (version 10.1) using the deduced amino acid sequences of MAP3 kinases from Arabidopsis, pepper, rice, cotton, tobacco, and tomato plants. The phylogenetic tree was built according to the neighbor-joining method, and bootstrap values were calculated from 1,000 bootstrap replications and are at each branch point. Scale bar indicates the evolutionary distance computed using the Poisson correction method. (B) Multiple alignment analysis of CaDIMK1 with its homologous Arabidopsis MAP3 kinases. Identical and similar amino acid residues are shaded according to the percentage identity in ClustalW2. Gaps introduced to maximize the alignment of homologous regions are marked by dashes. A red box indicates a serine/threonine protein kinase domain and a green box for a conserved kinase domain G(T/S)Px(W/Y/F)MAPEV in the MEKK-like group of the MAPKKK family. (C) Sequence homology of CaDIMK1 with its homologous Arabidopsis MAP3 kinases. In parenthesis, protein identity and similarity were calculated using EMBOSS needle (https://www.ebi.ac.uk/Tools/psa/emboss_needle/).
Molecular Characterization of CaDIMK1
We initially performed qRT-PCR analyses on pepper plants treated with ABA, NaCl, and H2O2 to determine whether CaDIMK1 is associated with abiotic stress responses (Figure 3A). Following exposure to ABA, high induction of CaDIMK1 expression started at 6 h. In contrast, NaCl treatment did not significantly affect the expression level of CaDIMK1 at all time points, except at 6 h. CaDIMK1 expression level showed a nine-fold decrease 2 h after treatment with H2O2. These results suggested that CaDIMK1 is presumably involved in both ABA signaling and drought stress response.
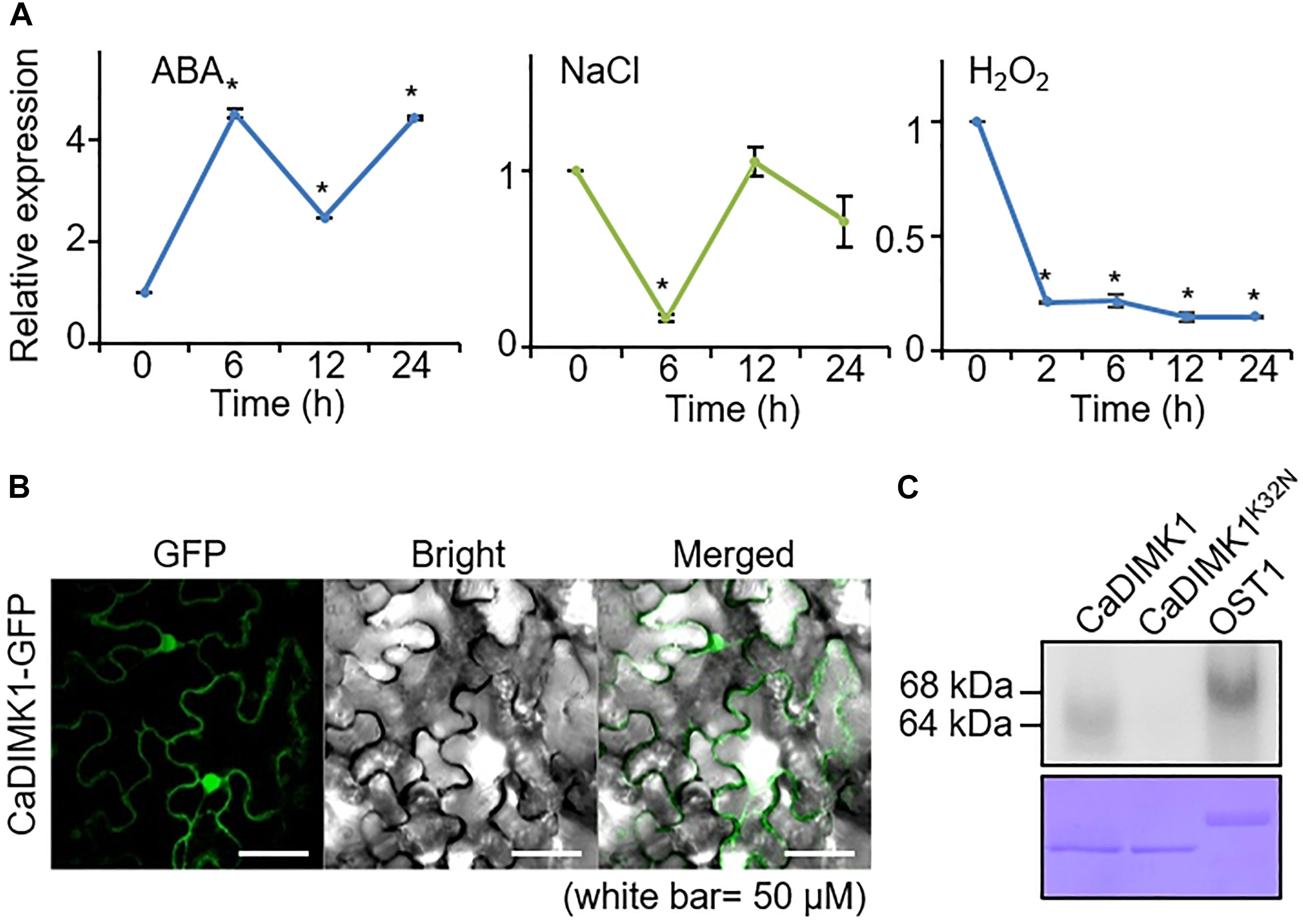
Figure 3. Expression of CaDIMK1 gene and subcellular localization of CaDIMK1 protein. (A) Induction levels of CaDIMK1 in pepper plant leaf tissue treated with abscisic acid (ABA; 100 μM), sodium chloride (NaCl; 200 mM), and hydrogen peroxide (H2O2; 100 μM). The expression values are normalized to pepper Actin1 (CaACT1) gene as a standard control. The induction level of CaDIMK1 at 0 h after treatment was 1.0. Values are mean ± SE, n = 3; asterisks indicate statistical differences compared with 0 h after treatment according to Student’s t-test (*P < 0.05). (B) Subcellular localization of CaDIMK1 protein in the epidermal cells of Nicotiana benthamiana. The transient expression of 35S:CaDIMK1-GFP construct was expressed in N. benthamiana leaves and detected using a confocal microscope. (C) In vitro auto kinase assay of GST-CaDIMK1 and GST-CaDIMK1K36N. [γ−−32P]-ATP was used for kinase assay. As a positive control, Arabidopsis OST1 was used. CBB, Coomassie brilliant blue staining.
Plant kinases are active in various areas within a cell; hence, we examined the localization of CaDIMK1 protein in the cell. When the fusion protein of CaDIMK1 with GFP was expressed in the epidermal cells of tobacco leaf tissues, the detected GFP fluorescence suggests that CaDIMK1 can function in the cytoplasm and nucleus (Figure 3B). CaDIMK1 has a serine-threonine kinase domain (Figure 1A); hence, we investigated the kinase activity of CaDIMK1 through in vitro kinase analysis (Figure 3C). We used CaDIMK1K32N with a substitution of lysine 32 for asparagine in the ATP-binding domain (Carrera et al., 1993) as a negative control and Arabidopsis OST1/SnRK2.6 as a positive control. As expected, auto kinase activity was shown in CaDIMK1, but not in CaDIMK1K32N.
Hypersensitivity to Drought Stress in CaDIMK1-Silenced Pepper Plants
Capsicum annuum drought-induced MAP kinase 1 transcripts were considerably accumulated in the pepper leaves treated with drought stress and ABA (Figures 1B, 3A). Hence, we checked the functional role of CaDIMK1 in vivo. Since the pepper transformation has a technical limitation, we alternatively used the VIGS method in pepper plants and generated OX transgenic plants in Arabidopsis for the genetic studies of CaDIMK1. First, we produced CaDIMK1-silenced pepper plants (TRV2:CaDIMK1), showing a lower accumulation of CaDIMK1 transcripts than control pepper TRV2:00 (Supplementary Figure 1A). To analyze how silencing of CaDIMK1 affects pepper drought stress response, we subjected 2-week-old pepper plants of TRV2:00 and TRV2:CaDIMK1 to drought stress by withholding watering for 13 days (Figure 4A). Plants grown under well-watered conditions did not show any different phenotypes (Figure 4A, upper panel). However, relative to the control pepper plants, the CaDIMK1-silenced pepper plants showed wilted phenotypes under drought stress. After recovery by rewatering (as indicated by 14 days), the survival rate of TRV2:CaDIMK1 pepper plants (29.8 ± 5.7%) was dramatically lower than that of TRV2:00 pepper plants (65.3 ± 6.3%).
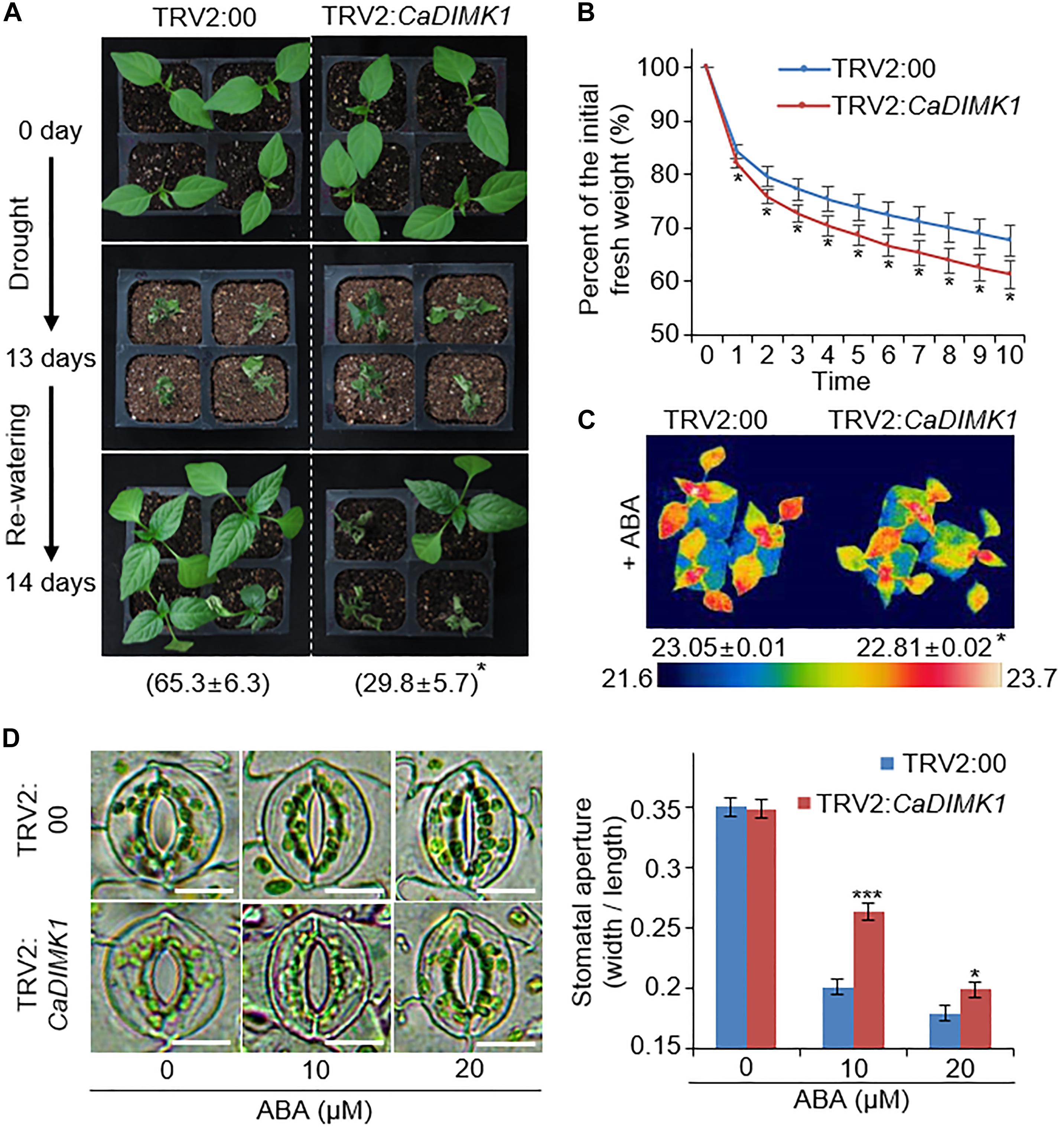
Figure 4. CaDIMK1 mediated drought tolerance via CaDIMK1-silenced pepper plants. (A) Drought sensitivity of TRV2:CaDIMK1 pepper plants. Three-week-old pepper plants expressed TRV2:00 and TRV2:CaDIMK1 constructs were grown in well-watered conditions (upper). The pepper was exposed to drought stress by withholding watering for 13 days (middle), followed by 1 day after rewatering (lower). The survival rates were counted at 1 day after rewatering. (B) Reduced fresh weight from the detached leaf tissues of TRV2:00 and TRV2:CaDIMK1 pepper plants during 10 h. (C) Reduced plant surface temperature of TRV2:CaDIMK1 pepper plants after 50 μM ABA treatment. Values are mean ± SE with three independent experiments (n = 10). (D) Reduced stomatal apertures of TRV2:00 and TRV2:CaDIMK1 pepper plants after various concentrations of ABA treatment. The stomata with guard cells were taken using a microscope when the stomatal pore size was measured. Leaves were incubated in stomata opening buffers with 0, 10, or 20 μM ABA. Values are mean ± SE with three independent experiments (n = 15). Asterisks indicate statistical differences between the TRV2:00 and TRV2:CaDIMK1 pepper plants according to Student’s t-test (*P < 0.05, ***P < 0.001). The scale bar represents 10 μm.
Preserving water by restricting transpirational water loss via stomata closure is critical for determining drought sensitivity. We measured the fresh weight of detached rosette leaves to investigate the rate of transpirational water loss. CaDIMK1-silenced pepper plants showed more significant water loss compared with the control plants (Figure 4B). In general, ABA treatment results in stomatal closing, causing enhanced leaf surface temperatures because of reduced evaporative cooling (Joo et al., 2019). We could not detect any differences in leaf surface temperatures between TRV2:CaDIMK1 and TRV2:00 plants under well-watered conditions. However, leaf surface temperatures in TRV2:CaDIMK1 were low compared with those in TRV2:00 plants following ABA treatment (Figure 4C). Consistently, there were no differences in the stomatal aperture between the TRV2:CaDIMK1 and TRV2:00 plants without ABA (Figure 4D). The application of ABA induced stomatal closing in both plants; however, the pore sizes in TRV2:CaDIMK1 plants were much larger than those in TRV2:00 plants. These results suggest that the downregulation of CaDIMK1 conferred reduced drought resistance via modulating the rate of water loss and ABA-mediated stomatal closing.
Increased ABA Sensitivity in CaDIMK1-OX Plants
We generated CaDIMK1- OX (CaDIMK1-OX) Arabidopsis transgenic plants to analyze the function of CaDIMK1 in response to drought stress and ABA (Supplementary Figure 1B). Two independent lines (#1 and #2) were selected for further genetic assays. Compared with wild-type Arabidopsis plants, we did not detect any statistical difference in phenotypes at all growth stages (Figures 5, 6). First, we investigated seed germination and seedling growth of CaDIMK1-OX plants in response to ABA. CaDIMK1-OX and wild-type seeds were normally germinated in the 0.5 × MS media in the absence of ABA. Application of ABA inhibited the seed germination of both plant lines, but the CaDIMK1-OX line had a lower germination rate than the wild-type plant at 7 days after plating (Figure 5A). The number of expanded cotyledons were significantly higher in CaDIMK1-OX lines than in the wild-type plants (Figures 5B,C). When seedlings of the two plant lines were vertically grown, the primary root growths of CaDIMK1-OX seedlings were significantly longer than those of wild-type plants (Figures 5D,E). These data indicated that enhanced expression of CaDIMK1 led to increased sensitivity to ABA in Arabidopsis seed germination and seedling stages.
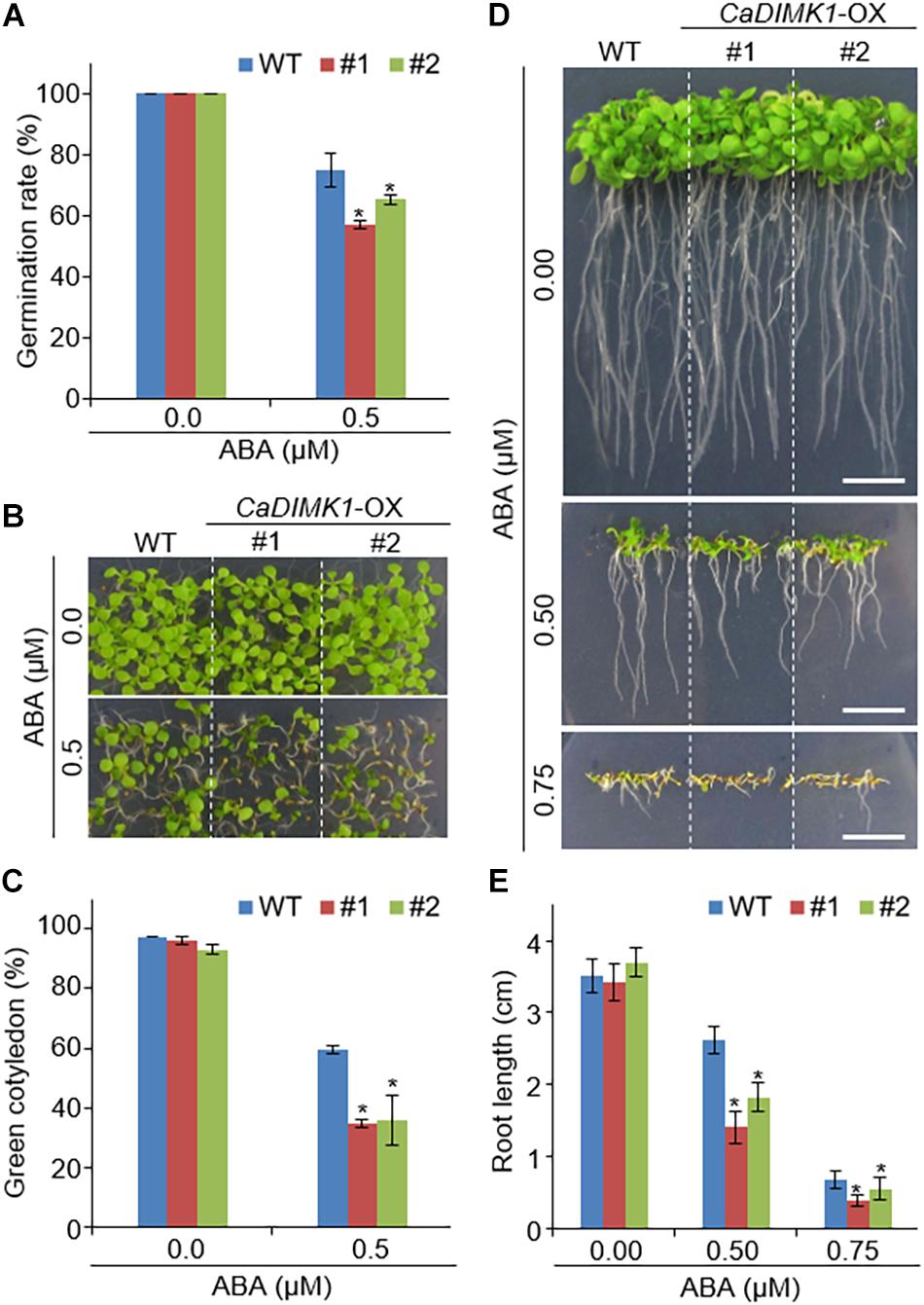
Figure 5. Increased sensitivity of CaDIMK1-OX plants to ABA. (A) Seed germination rates of transgenic lines and wild-type (WT) Col-0 plants on 0.5 × MS medium without ABA or supplemented with ABA and measured 7 days after sowing. (B,C) Cotyledon greening of CaDIMK1-OX and wild-type plants on 0.5× MS medium with or without ABA. Five days after plating, representative photographs were taken (B) and cotyledon greening in transgenic and wild-type plants was measured (C). Values are mean ± SE with six independent experiments (n = 36). (D,E) Root length of CaDIMK1-OX Arabidopsis and wild-type (WT) plants on 0.5 × MS medium containing 0.0, 0.5, and 0.75 μM ABA. Seven days after plating, representative photographs were taken (D) and primary root lengths in the transgenic and wild-type plants were measured (E). The scale bar represents 1 cm. Values are mean ± SE with three independent experiments (n = 25). Asterisks indicate statistical differences between wild-type and CaDIMK1-OX plants according to Student’s t-test (*P < 0.05).
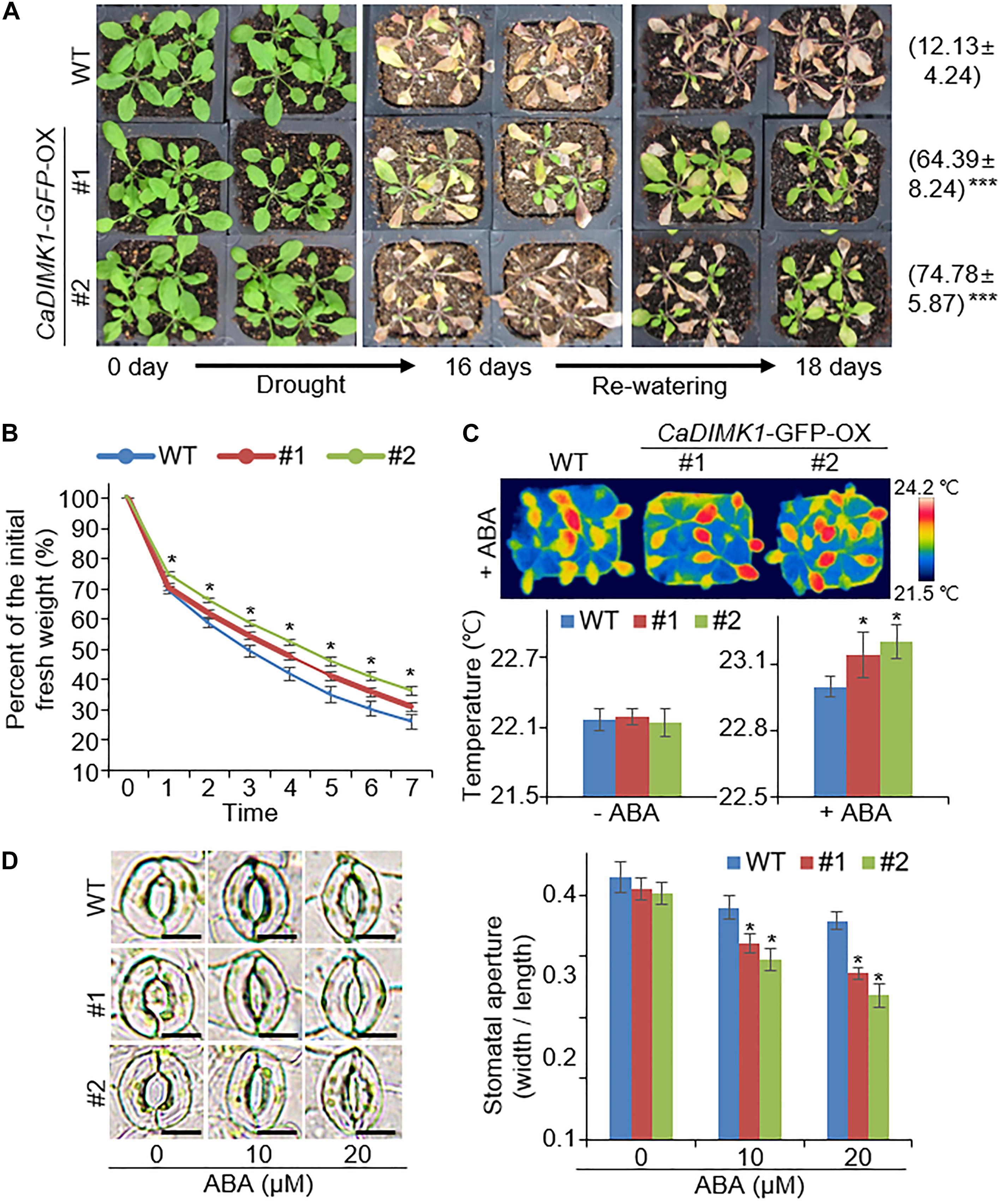
Figure 6. Increased drought stress tolerance of CaDIMK1-overexpressing (OX) plants. (A) Drought-tolerant phenotype of CaDIMK1-OX plants. Wild-type (WT) and CaDIMK1-OX Arabidopsis plants were cultivated for 3 weeks under well-watered conditions (left). Arabidopsis was exposed to drought stress by withholding watering for 16 days (middle), followed by 2 days after rewatering (right). The survival rates were counted 2 days after rewatering. Values are mean ± SE with three independent experiments (n = 42). (B) Transpiration water loss from wild-type and CaDIMK1-OX plant leaves during 7 h after leaf detachment. (C) Increased leaf surface temperatures of wild-type and CaDIMK1-OX Arabidopsis plants treated with 50μM ABA at 0 and 4 h. Values are mean ± SE with three independent experiments (n = 10). (D) Reduced stomatal opening in wild-type and CaDIMK1-OX transgenic plants treated with ABA. The stomata with guard cells were taken using a microscope when the stomatal pore size was measured. Leaves were incubated in stomata opening solutions with 0, 10, or 20 μM ABA. Values are mean ± SE with three independent experiments (n = 20). Asterisks indicate statistical differences between the wild-type and the transgenic plants according to Student’s t-test (*P < 0.05). The scale bar represents 10 μm.
Increased Drought Resistance in CaDIMK1-OX Plants
Based on the drought-induced expression of CaDIMK1 and the reduced drought resistance of CaDIMK1-silenced pepper (Figures 1, 4), we tested how overexpression of CaDIMK1 affects drought resistance in Arabidopsis plants (Figure 6). There were no differences between CaDIMK1-OX and wild-type plants under well-watered conditions (Figure 6A). Drought stress was applied by withholding watering for 16 days. Compared with wild-type plants, CaDIMK1-OX plants withered less, and more CaDIMK1-OX plants survived at 2 days after rewatering. The survival rate of CaDIMK1-OX was 64.39–74.78%, whereas that of wild-type plants was only 12.13% (Figure 6A). When measuring the water loss rate in rosette leaf tissues during 0–7 h after leaf detachment, we found that the fresh weight loss of CaDIMK1-OX leaves was significantly lower than that in the wild-type leaves (Figure 6B). To determine if this enhanced drought resistance is associated with ABA-mediated regulation of stomatal closure, we analyzed changes in leaf surface temperatures and stomatal apertures in response to ABA. Both CaDIMK1-OX and wild-type plants showed similar leaf temperatures under normal growth conditions; however, after ABA treatment, the leaf surface temperatures of the CaDIMK1-OX mutants were higher than those of the wild-type plants (Figure 6C). Consistently, the stomatal pore sizes of CaDIMK1-OX plants were smaller than those of wild-type plants after ABA treatment (Figure 6D). We also measured the transcript level of stress-responsive genes such as RAB18, RD29B, DREB2A, AHG1, PP2CA, and HAB1. qRT-PCR analyses revealed that these stress-responsive genes were highly induced in CaDIMK1-OX leaves than in wild-type leaves after treatment with drought stress (Figure 7). As shown by phenotypic analysis, CaDIMK1 plays an essential role in drought resistance by controlling ABA-dependent stomatal apertures and ABA-responsive gene expression.
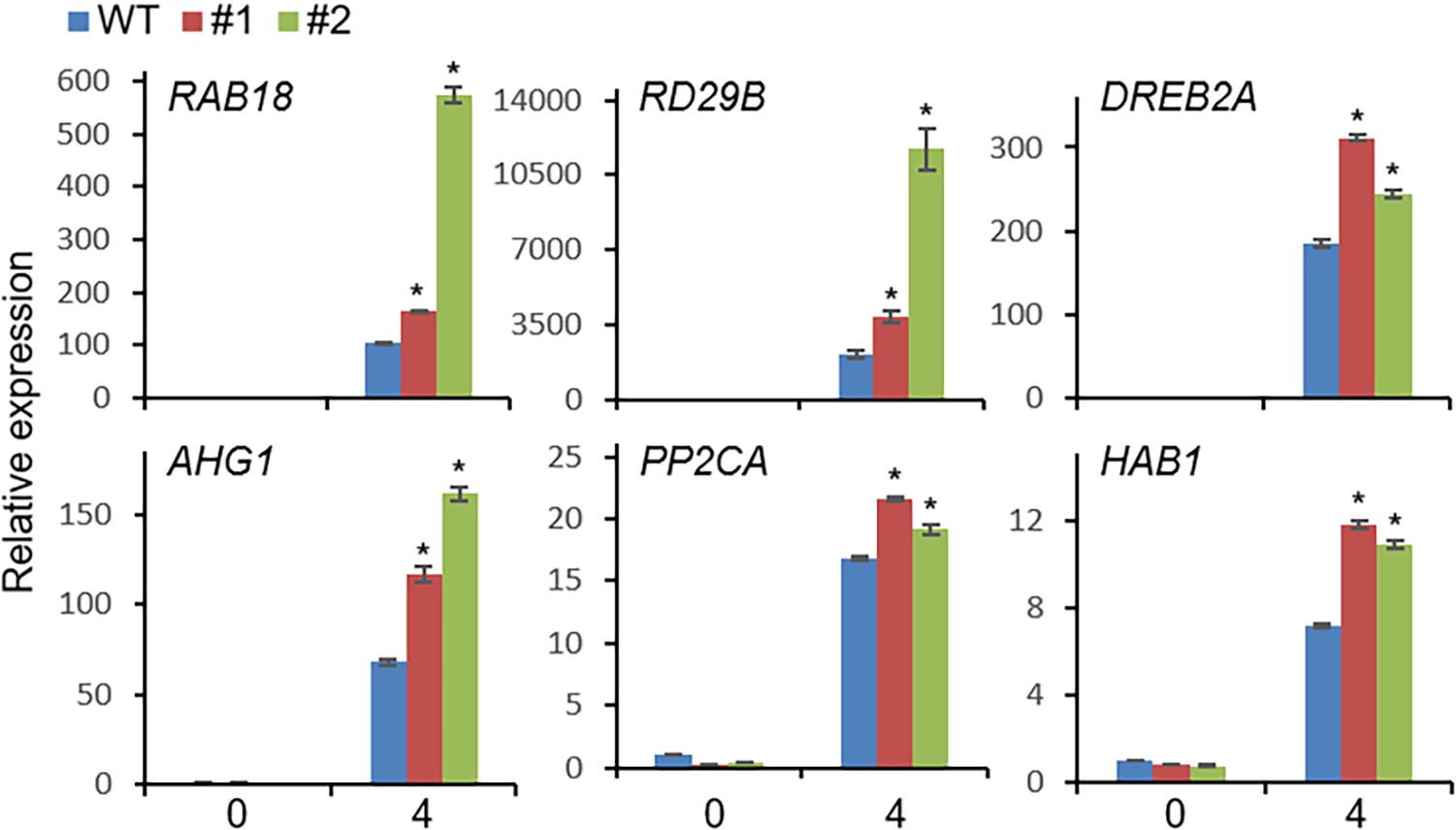
Figure 7. qRT-PCR assay of stress-responsive genes in CaDIMK1-overexpressing (OX) plants treated with drought stress at 0 and 4 h. The expression values are normalized to the Actin8 genes as an internal control. Values are mean ± SE, n = 3. Asterisks indicate statistical differences between wild-type and transgenic plants according to Student’s t-test (*P < 0.05).
Discussion
Plants modulate cellular activities using several processes such as transcription and posttranslational modifications to survive drought stress. Phosphorylation mediated by protein kinase is one of the posttranslational modifications, which plays a critical function in abiotic stress response and ABA signaling (Shi et al., 2018; Wang et al., 2018; Hong et al., 2020; Jeong et al., 2020). Previous studies reported that many protein kinases related to stress response were identified and functionally characterized; however, the exact process and function of these proteins remain elusive. In the ABA signal transduction pathway, SnRK2-type kinases are core components and act as positive regulators of drought stress response and ABA signal transduction pathway via modulation of stress-responsive gene transcription and channel activity (Geiger et al., 2009; Joshi-Saha et al., 2011; Brandt et al., 2012). Receptor-like kinases (RLKs) act as positive and negative regulators in the drought stress response (Ouyang et al., 2010; Hua et al., 2012; Marshall et al., 2012). RLKs perceive signals from intercellular spaces and transfer amplified signals to downstream substrate proteins (Shiu and Bleecker, 2003; Gish and Clark, 2011; Liang and Zhou, 2018). MAP kinase is also associated with plant responses to biotic and abiotic stresses (de Zelicourt et al., 2016; Zhang et al., 2016; Zhao et al., 2017); however, the exact functions in stress responses have been less studied than SnRK2-type kinases and RLKs. MAPK cascades are intracellular signaling pathways with sequential phosphorylation reactions to activate downstream partners in response to external signals (Rodriguez et al., 2010; Xu and Zhang, 2015). As revealed by physiological and molecular analysis, we have identified in this work the drought-induced pepper MEKK gene CaDIMK1, which plays an essential function in ABA signal transduction and drought response.
Plants initiate defense mechanisms under water-deficit conditions, such as accumulation of ABA and stress-responsive genes (Sato et al., 2018; Lu et al., 2019). Owing to low transformation efficiency in pepper plans, the VIGS analysis in pepper plants and overexpression assay in Arabidopsis were used for genetic investigation in this study. ABA accumulation in leaf tissue restricts transpirational water loss by closing the stomata, conferring drought tolerance. Downregulated CaDIMK1 by VIGS in pepper plants displayed a hypersensitive phenotype to drought stress, accompanied by large stomatal apertures that increase evaporation rates (Figure 4). Conversely, CaDIMK1-OX Arabidopsis displayed a drought-tolerant phenotype, which reduced transpirational water loss and stomatal pore size (Figure 6). These phenotype analyses suggest that different stomatal pore sizes in the silenced and overexpressed plants modulate water consumption, leading to altered drought phenotypes. Together with ABA signaling and drought stress, CaDIMK1 could be involved in different stress responses, including high salinity and osmotic stress, based on the altered expression of CaDIMK1 by treatment with NaCl and H2O2 (Figure 3A). Both drought stress and high salinity decrease the water availability to plant cells and also cause the accumulation of ROS such as hydrogen peroxide (Hasegawa et al., 2000). As homologs of CaDIMK1, AtMAPKKK15/16/17/18 are salt-inducible MEKK genes (Choi et al., 2017). In rice, OsMAPKKK63 is also induced by salt and its loss-of-function mutant shows decreased tolerance to salt stress (Na et al., 2019). Although CaDIMK1 gene expression in pepper leaves transiently decreased by salt stress compared with those genes, its functional involvement in response to salt stress may be supported by the data showing that CaDIMK1-OX plants were less sensitive to salt and mannitol during germination and seedling growth (Supplementary Figure 2).
The expression levels of stress- or ABA signal transduction-related genes are necessary to overcome drought stress, leading to plant survival (Zhu, 2016; Sato et al., 2018; Sharma et al., 2018; Lu et al., 2019; Joo et al., 2020). In this present study, the downstream substrate proteins of CaDIMK1 were not found; however, relative to wild-type plants, the transcript levels of stress- or ABA signal transduction-related genes were higher in CaDIMK1-OX plants. This indicates that CaDIMK1 may regulate and act upstream of these genes. In the ABA signaling pathway, clade A PP2Cs are core components that negatively control ABA signal transduction through dephosphorylation of SnRK2-type kinases (Robert et al., 2006; Nishimura et al., 2007; Umezawa et al., 2009; Vlad et al., 2009; Antoni et al., 2012). Arabidopsis clade A PP2C ABI1 also inhibits MAP3K protein MAPKKK18 and affects the stability of this kinase (Mitula et al., 2015). Interestingly, CaDIMK1-OX plants showed upregulation of clade A PP2Cs, including AHG1, PP2CA, and HAB1. Based on the relationship between MAP3Ks and clade A PP2Cs at the transcriptional and posttranslational levels, we proposed that CaDIMK1 may function upstream of clade A PP2Cs or CaDIMK1-mediated induction of PP2C genes may be part of the negative feedback regulation of the ABA signaling pathway. Under normal growth conditions, the phenotypes of the CaDIMK1-OX Arabidopsis plants and the expression levels of stress-related genes were not indistinguishable. Hence, the identification of processes that are upstream and downstream of CaDIMK1 will help comprehend the in vivo role of CaDIMK1 in plant cell to overcome drought stress.
In summary, altering the expression of CaDIMK1 affected seed germination, seedling growth, and drought stress response. This study suggests that CaDIMK1 is a positive regulator of ABA signal transduction and drought resistance. However, some uncertainty remains about how CaDIMK1 regulates stress-related genes and drought response via ABA signaling and which downstream target proteins are phosphorylated by CaDIMK1. Further studies are needed to determine downstream target proteins that physically interact with and are regulated by CaDIMK1.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author Contributions
MK, SJ, and CL performed the experiments and analyzed the results. CL and SL designed the experiments and wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by a grant from the Agriculture & Technology Development (Project No. PJ01589201) and the National Research Foundation of Korea (NRF) grants funded by the Korea Government (MSIT) (2021R1A2C2006338 and 2021R1A2C1007115), Rural Development Administration, South Korea.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.646707/full#supplementary-material
Supplementary Figure 1 | (A) Transcripts level of CaDIMK1 in the leaves of TRV2:00 and TRV2:CaDIMK1 plants. The CaACT1 gene was used as an internal control for normalization. (B) Transcripts level of CaDIMK1 in the CaDIMK1-OX Arabidopsis. The Actin8 gene was used as an internal control for normalization.
Supplementary Figure 2 | Increased tolerance of CaDIMK1-overexpressing (OX) plants to salt and osmotic stresses. (A) Root length of CaDIMK1-OX and wild-type (WT) plants on 0.5× MS medium containing 0, 400, and 500 mM Mannitol. (B) Root length of CaDIMK1-OX and wild-type plants on 0.5 MS medium containing 0, 100, 150 mM NaCl. After five days after plating, representative photographs were taken, and root lengths were measured. Values are mean ± SE with three independent experiments (n = 25). Asterisks indicate statistical differences between the wild-type and the transgenic plants according to the Student’s t-test (∗P < 0.05). The scale bar represents 1 cm.
Footnotes
References
Antoni, R., Gonzalez-Guzman, M., Rodriguez, L., Rodrigues, A., Pizzio, G. A., and Rodriguez, P. L. (2012). Selective inhibition of clade A phosphatases type 2C by PYR/PYL/RCAR abscisic acid receptors. Plant physiol. 158, 970–980. doi: 10.1104/pp.111.188623
Basu, S., Ramegowda, V., Kumar, A., and Pereira, A. (2016). Plant adaptation to drought stress. F1000Res 5:1554. doi: 10.12688/f1000research.7678.1
Bergmann, D. C., Lukowitz, W., and Somerville, C. R. (2004). Stomatal development and pattern controlled by a MAPKK kinase. Science 304, 1494–1497. doi: 10.1126/science.1096014
Bigeard, J., and Hirt, H. (2018). Nuclear signaling of plant MAPKs. Front. Plant Sci. 9:469. doi: 10.3389/fpls.2018.00469
Brandt, B., Brodsky, D. E., Xue, S., Negi, J., Iba, K., Kangasjarvi, J., et al. (2012). Reconstitution of abscisic acid activation of SLAC1 anion channel by CPK6 and OST1 kinases and branched ABI1 PP2C phosphatase action. Proc. Natl. Acad. Sci. U.S.A. 109, 10593–10598. doi: 10.1073/pnas.1116590109
Carrera, A. C., Alexandrov, K., and Roberts, T. M. (1993). The conserved lysine of the catalytic domain of protein kinases is actively involved in the phosphotransfer reaction and not required for anchoring ATP. Proc. Natl. Acad. Sci. U.S.A. 90, 442–446. doi: 10.1073/pnas.90.2.442
Choi, S., Lee, S., Na, Y., Jeung, S., and Kim, S. Y. (2017). Arabidopsis MAP3K16 and other salt-inducible MAP3Ks regulate ABA response redundantly. Mol. Cells 40, 230–242.
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Danquah, A., de Zelicourt, A., Boudsocq, M., Neubauer, J., Frei Dit Frey, N., Leonhardt, N., et al. (2015). Identification and characterization of an ABA-activated MAP kinase cascade in Arabidopsis thaliana. Plant J. 82, 232–244. doi: 10.1111/tpj.12808
Danquah, A., de Zelicourt, A., Colcombet, J., and Hirt, H. (2014). The role of ABA and MAPK signaling pathways in plant abiotic stress responses. Biotechnol. Adv. 32, 40–52. doi: 10.1016/j.biotechadv.2013.09.006
de Zelicourt, A., Colcombet, J., and Hirt, H. (2016). The Role of MAPK Modules and ABA during Abiotic Stress Signaling. Trends Plant Sci. 21, 677–685. doi: 10.1016/j.tplants.2016.04.004
Dittrich, M., Mueller, H. M., Bauer, H., Peirats-Llobet, M., Rodriguez, P. L., Geilfus, C. M., et al. (2019). The role of Arabidopsis ABA receptors from the PYR/PYL/RCAR family in stomatal acclimation and closure signal integration. Nat. Plants 5, 1002–1011. doi: 10.1038/s41477-019-0490-0
Dong, H., Bai, L., Zhang, Y., Zhang, G., Mao, Y., Min, L., et al. (2018). Modulation of guard cell turgor and drought tolerance by a peroxisomal acetate-malate shunt. Mol. Plant 11, 1278–1291. doi: 10.1016/j.molp.2018.07.008
Finkelstein, R. R., Gampala, S. S., and Rock, C. D. (2002). Abscisic acid signaling in seeds and seedlings. Plant cell 14(Suppl.), S15–S45.
Geiger, D., Scherzer, S., Mumm, P., Stange, A., Marten, I., Bauer, H., et al. (2009). Activity of guard cell anion channel SLAC1 is controlled by drought-stress signaling kinase-phosphatase pair. Proc. Natl. Acad. Sci. U.S.A. 106, 21425–21430. doi: 10.1073/pnas.0912021106
Gish, L. A., and Clark, S. E. (2011). The RLK/Pelle family of kinases. Plant J. 66, 117–127. doi: 10.1111/j.1365-313x.2011.04518.x
Golldack, D., Li, C., Mohan, H., and Probst, N. (2014). Tolerance to drought and salt stress in plants: unraveling the signaling networks. Front. Plant Sci. 5:151. doi: 10.3389/fpls.2014.00151
Gonzalez-Guzman, M., Pizzio, G. A., Antoni, R., Vera-Sirera, F., Merilo, E., Bassel, G. W., et al. (2012). Arabidopsis PYR/PYL/RCAR receptors play a major role in quantitative regulation of stomatal aperture and transcriptional response to abscisic acid. Plant Cell 24, 2483–2496. doi: 10.1105/tpc.112.098574
Hasegawa, P. M., Bressan, R. A., Zhu, J. K., and Bohnert, H. J. (2000). Plant cellular and molecular responses to high salinity. Annu. Rev. Plant Mol. Plant Physiol. 51, 463–499.
Hong, Y., Wang, Z., Liu, X., Yao, J., Kong, X., Shi, H., et al. (2020). Two chloroplast proteins negatively regulate plant drought resistance through separate pathways. Plant Physiol. 182, 1007–1021. doi: 10.1104/pp.19.01106
Hua, D., Wang, C., He, J., Liao, H., Duan, Y., Zhu, Z., et al. (2012). A plasma membrane receptor kinase, GHR1, mediates abscisic acid- and hydrogen peroxide-regulated stomatal movement in Arabidopsis. Plant Cell 24, 2546–2561. doi: 10.1105/tpc.112.100107
Ichimura, K., Shinozaki, K., Tena, G., Sheen, J., Henry, Y., Champion, A., et al. (2002). Mapk G: Mitogen-activated protein kinase cascades in plants: a new nomenclature. Trends plant sci. 7, 301–308.
Jeong, S., Lim, C. W., and Lee, S. C. (2020). The pepper MAP Kinase CaAIMK1 positively regulates ABA and drought stress responses. Front. Plant Sci. 11:720. doi: 10.3389/fpls.2020.00720
Jonak, C., Okrész, L., Bögre, L., and Hirt, H. (2002). Complexity, cross talk and integration of plant MAP kinase signalling. Curr. Opin. Plant Biol. 5, 415–424. doi: 10.1016/s1369-5266(02)00285-6
Joo, H., Lim, C. W., and Lee, S. C. (2019). A pepper RING-type E3 ligase, CaASRF1, plays a positive role in drought tolerance via modulation of CaAIBZ1 stability. Plant J. 98, 5–18. doi: 10.1111/tpj.14191
Joo, H., Lim, C. W., and Lee, S. C. (2020). The pepper RING-type E3 ligase, CaATIR1, positively regulates abscisic acid signalling and drought response by modulating the stability of CaATBZ1. Plant Cell Environ. 43, 1911–1924. doi: 10.1111/pce.13789
Joshi-Saha, A., Valon, C., and Leung, J. (2011). Abscisic acid signal off the STARting block. Mol. Plant 4, 562–580. doi: 10.1093/mp/ssr055
Kim, T. H., Bohmer, M., Hu, H., Nishimura, N., and Schroeder, J. I. (2010). Guard cell signal transduction network: advances in understanding abscisic acid, CO2, and Ca2+ signaling. Annu. Rev. Plant Biol. 61, 561–591. doi: 10.1146/annurev-arplant-042809-112226
Komatsu, K., Suzuki, N., Kuwamura, M., Nishikawa, Y., Nakatani, M., Ohtawa, H., et al. (2013). Group A PP2Cs evolved in land plants as key regulators of intrinsic desiccation tolerance. Nat. Commun. 4:2219.
Lee, S. C., Lan, W., Buchanan, B. B., and Luan, S. (2009). A protein kinase-phosphatase pair interacts with an ion channel to regulate ABA signaling in plant guard cells. Proc. Natl. Acad. Sci. U.S.A. 106, 21419–21424. doi: 10.1073/pnas.0910601106
Lee, S. C., Lim, C. W., Lan, W., He, K., and Luan, S. (2013). ABA signaling in guard cells entails a dynamic protein-protein interaction relay from the PYL-RCAR family receptors to ion channels. Mol. Plant 6, 528–538. doi: 10.1093/mp/sss078
Li, K., Yang, F., Zhang, G., Song, S., Li, Y., Ren, D., et al. (2017). AIK1, a mitogen-activated protein kinase, modulates abscisic acid responses through the MKK5-MPK6 kinase cascade. Plant Physiol. 173, 1391–1408. doi: 10.1104/pp.16.01386
Li, Y., Cai, H., Liu, P., Wang, C., Gao, H., Wu, C., et al. (2017). Arabidopsis MAPKKK18 positively regulates drought stress resistance via downstream MAPKK3. Biochem. Biophys. Res. Commun. 484, 292–297. doi: 10.1016/j.bbrc.2017.01.104
Liang, X., and Zhou, J. M. (2018). Receptor-like cytoplasmic kinases: central players in plant receptor kinase-mediated signaling. Annu. Rev. Plant Biol. 69, 267–299. doi: 10.1146/annurev-arplant-042817-040540
Lim, C. W., Baek, W., Jung, J., Kim, J. H., and Lee, S. C. (2015). Function of ABA in stomatal defense against biotic and drought stresses. Int. J. Mol. Sci. 16, 15251–15270. doi: 10.3390/ijms160715251
Lim, C. W., Baek, W., and Lee, S. C. (2017). The Pepper RING-Type E3 Ligase CaAIRF1 regulates ABA and drought signaling via CaADIP1 protein phosphatase degradation. Plant Physiol. 173, 2323–2339. doi: 10.1104/pp.16.01817
Lin, Z., Li, Y., Zhang, Z., Liu, X., Hsu, C. C., Du, Y., et al. (2020). A RAF-SnRK2 kinase cascade mediates early osmotic stress signaling in higher plants. Nat. Commun. 11:613.
Liu, Y., Schiff, M., and Dinesh-Kumar, S. P. (2002). Virus-induced gene silencing in tomato. Plant J. 31, 777–786. doi: 10.1046/j.1365-313x.2002.01394.x
Lu, S., Fadlalla, T., Tang, S., Li, L., Ali, U., Li, Q., et al. (2019). Genome-wide analysis of phospholipase D gene family and profiling of phospholipids under abiotic stresses in Brassica napus. Plant Cell Physiol. 60, 1556–1566. doi: 10.1093/pcp/pcz071
Mao, X., Zhang, J., Liu, W., Yan, S., Liu, Q., Fu, H., et al. (2019). The MKKK62-MKK3-MAPK7/14 module negatively regulates seed dormancy in rice. Rice 12:2.
Marshall, A., Aalen, R. B., Audenaert, D., Beeckman, T., Broadley, M. R., Butenko, M. A., et al. (2012). Tackling drought stress: receptor-like kinases present new approaches. Plant Cell 24, 2262–2278. doi: 10.1105/tpc.112.096677
Mishra, N. S., Tuteja, R., and Tuteja, N. (2006). Signaling through MAP kinase networks in plants. Arch. Biochem. Biophys. 452, 55–68. doi: 10.1016/j.abb.2006.05.001
Mitula, F., Tajdel, M., Ciesla, A., Kasprowicz-Maluski, A., Kulik, A., Babula-Skowronska, D., et al. (2015). Arabidopsis ABA-activated kinase MAPKKK18 is regulated by protein phosphatase 2C ABI1 and the ubiquitin-proteasome pathway. Plant Cell Physiol. 56, 2351–2367. doi: 10.1093/pcp/pcv146
Na, Y. J., Choi, H. K., Park, M. Y., Choi, S. W., Xuan Vo, K. T., Jeon, J. S., et al. (2019). OsMAPKKK63 is involved in salt stress response and seed dormancy control. Plant Signal. Behav. 14:e1578633. doi: 10.1080/15592324.2019.1578633
Nishimura, N., Yoshida, T., Kitahata, N., Asami, T., Shinozaki, K., and Hirayama, T. (2007). ABA-Hypersensitive Germination1 encodes a protein phosphatase 2C, an essential component of abscisic acid signaling in Arabidopsis seed. Plant J. 50, 935–949. doi: 10.1111/j.1365-313x.2007.03107.x
Ouyang, S. Q., Liu, Y. F., Liu, P., Lei, G., He, S. J., Ma, B., et al. (2010). Receptor-like kinase OsSIK1 improves drought and salt stress tolerance in rice (Oryza sativa) plants. Plant J. 62, 316–329. doi: 10.1111/j.1365-313x.2010.04146.x
Rao, K. P., Richa, T., Kumar, K., Raghuram, B., and Sinha, A. K. (2010). In silico analysis reveals 75 members of mitogen−activated protein kinase kinase kinase gene family in rice. DNA Res. 17, 139–153. doi: 10.1093/dnares/dsq011
Robert, N., Merlot, S., N’Guyen, V., Boisson-Dernier, A., and Schroeder, J. I. (2006). A hypermorphic mutation in the protein phosphatase 2C HAB1 strongly affects ABA signaling in Arabidopsis. FEBS Lett. 580, 4691–4696. doi: 10.1016/j.febslet.2006.07.047
Rodriguez, M. C., Petersen, M., and Mundy, J. (2010). Mitogen-activated protein kinase signaling in plants. Annu. Rev. Plant Biol. 61, 621–649.
Sato, H., Takasaki, H., Takahashi, F., Suzuki, T., Iuchi, S., Mitsuda, N., et al. (2018). Arabidopsis thaliana NGATHA1 transcription factor induces ABA biosynthesis by activating NCED3 gene during dehydration stress. Proc. Natl. Acad. Sci. U.S.A. 115, E11178–E11187.
Schroeder, J. I., Allen, G. J., Hugouvieux, V., Kwak, J. M., and Waner, D. (2001). Guard cell signal transduction. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 627–658.
Sharma, R., Singh, G., Bhattacharya, S., and Singh, A. (2018). Comparative transcriptome meta-analysis of Arabidopsis thaliana under drought and cold stress. PLoS One 13:e0203266. doi: 10.1371/journal.pone.0203266
Shi, S., Li, S., Asim, M., Mao, J., Xu, D., Ullah, Z., et al. (2018). The Arabidopsis calcium-dependent protein kinases (CDPKs) and their roles in plant growth regulation and abiotic stress responses. Int. J. Mol. Sci. 19:1900. doi: 10.3390/ijms19071900
Shinozawa, A., Otake, R., Takezawa, D., Umezawa, T., Komatsu, K., Tanaka, K., et al. (2019). SnRK2 protein kinases represent an ancient system in plants for adaptation to a terrestrial environment. Commun. Biol. 2:30.
Shiu, S. H., and Bleecker, A. B. (2003). Expansion of the receptor-like kinase/Pelle gene family and receptor-like proteins in Arabidopsis. Plant Physiol. 132, 530–543. doi: 10.1104/pp.103.021964
Shou, H., Bordallo, P., and Wang, K. (2004). Expression of the Nicotiana protein kinase (NPK1) enhanced drought tolerance in transgenic maize. J. Exp. Bot. 55, 1013–1019. doi: 10.1093/jxb/erh129
Soma, F., Takahashi, F., Suzuki, T., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2020). Plant Raf-like kinases regulate the mRNA population upstream of ABA-unresponsive SnRK2 kinases under drought stress. Nat. Commun. 11:1373.
Takahashi, Y., Zhang, J., Hsu, P. K., Ceciliato, P. H. O., Zhang, L., Dubeaux, G., et al. (2020). MAP3Kinase-dependent SnRK2-kinase activation is required for abscisic acid signal transduction and rapid osmotic stress response. Nat. Commun. 11:12.
Tuteja, N. (2007). Abscisic Acid and abiotic stress signaling. Plant Signal. Behav. 2, 135–138. doi: 10.4161/psb.2.3.4156
Ullah, A., Manghwar, H., Shaban, M., Khan, A. H., Akbar, A., Ali, U., et al. (2018). Phytohormones enhanced drought tolerance in plants: a coping strategy. Environ. Sci. Pollut. Res. Int. 25, 33103–33118. doi: 10.1007/s11356-018-3364-5
Umezawa, T., Sugiyama, N., Mizoguchi, M., Hayashi, S., Myouga, F., Yamaguchi-Shinozaki, K., et al. (2009). Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 106, 17588–17593. doi: 10.1073/pnas.0907095106
Vahisalu, T., Kollist, H., Wang, Y. F., Nishimura, N., Chan, W. Y., Valerio, G., et al. (2008). SLAC1 is required for plant guard cell S-type anion channel function in stomatal signalling. Nature 452, 487–491. doi: 10.1038/nature06608
Vlad, F., Rubio, S., Rodrigues, A., Sirichandra, C., Belin, C., Robert, N., et al. (2009). Protein phosphatases 2C regulate the activation of the Snf1-related kinase OST1 by abscisic acid in Arabidopsis. Plant Cell 21, 3170–3184. doi: 10.1105/tpc.109.069179
Wang, H., Ngwenyama, N., Liu, Y., Walker, J. C., and Zhang, S. (2007). Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis. Plant Cell 19, 63–73. doi: 10.1105/tpc.106.048298
Wang, P., Zhao, Y., Li, Z., Hsu, C. C., Liu, X., Fu, L., et al. (2018). Reciprocal regulation of the TOR kinase and ABA receptor balances plant growth and stress response. Mol. Cell. 69, 100–112.e6.
Wang, R. S., Pandey, S., Li, S., Gookin, T. E., Zhao, Z., Albert, R., et al. (2011). Common and unique elements of the ABA-regulated transcriptome of Arabidopsis guard cells. BMC Genomics 12:216. doi: 10.1186/1471-2164-12-216
Wu, J., Wang, J., Pan, C., Guan, X., Wang, Y., Liu, S., et al. (2014). Genome-wide identification of MAPKK and MAPKKK gene families in tomato and transcriptional profiling analysis during development and stress response. PLoS One 9:e103032. doi: 10.1371/journal.pone.0103032
Xu, J., and Zhang, S. (2015). Mitogen-activated protein kinase cascades in signaling plant growth and development. Trends Plant Sci. 20, 56–64. doi: 10.1016/j.tplants.2014.10.001
Zhang, J. B., Wang, X. P., Wang, Y. C., Chen, Y. H., Luo, J. W., Li, D. D., et al. (2020). Genome-wide identification and functional characterization of cotton (Gossypium hirsutum) MAPKKK gene family in response to drought stress. BMC Plant Biol. 20:217. doi: 10.1186/s12870-020-02431-2
Zhang, T., Chen, S., and Harmon, A. C. (2016). Protein-protein interactions in plant mitogen-activated protein kinase cascades. J. Exp. Bot. 67, 607–618. doi: 10.1093/jxb/erv508
Zhao, C., Wang, P., Si, T., Hsu, C. C., Wang, L., Zayed, O., et al. (2017). MAP kinase cascades regulate the cold response by modulating ICE1 protein stability. Dev. Cell 43, 618–629.e5.
Keywords: ABA, drought, kinase, phosphorylation, stomata
Citation: Kim M, Jeong S, Lim CW and Lee SC (2021) Mitogen-Activated Protein Kinase CaDIMK1 Functions as a Positive Regulator of Drought Stress Response and Abscisic Acid Signaling in Capsicum annuum. Front. Plant Sci. 12:646707. doi: 10.3389/fpls.2021.646707
Received: 28 December 2020; Accepted: 22 March 2021;
Published: 29 April 2021.
Edited by:
Kyung-Nam Kim, Sejong University, South KoreaReviewed by:
Jose M. Colmenero-Flores, Instituto de Recursos Naturales y Agrobiología, Spanish National Research Council (CSIC), SpainGirdhar Kumar Pandey, University of Delhi, India
Copyright © 2021 Kim, Jeong, Lim and Lee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chae Woo Lim, cHJvdGVhbkBjYXUuYWMua3I=; Sung Chul Lee, c2NsZWUxOTcyQGNhdS5hYy5rcg==
†These authors have contributed equally to this work
 Minchae Kim†
Minchae Kim† Soongon Jeong
Soongon Jeong Chae Woo Lim
Chae Woo Lim Sung Chul Lee
Sung Chul Lee