
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 08 March 2021
Sec. Plant Systematics and Evolution
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.646622
 Li Yuan1
Li Yuan1 Jingwen Pan1
Jingwen Pan1 Shouhong Zhu1
Shouhong Zhu1 Yan Li1
Yan Li1 Jinbo Yao1
Jinbo Yao1 Qiulin Li1
Qiulin Li1 Shengtao Fang1
Shengtao Fang1 Chunyan Liu2
Chunyan Liu2 Xinyu Wang3
Xinyu Wang3 Bei Li1
Bei Li1 Wei Chen1*
Wei Chen1* Yongshan Zhang1*
Yongshan Zhang1*SUN-domain containing proteins are crucial nuclear membrane proteins involved in a plethora of biological functions, including meiosis, nuclear morphology, and embryonic development, but their evolutionary history and functional divergence are obscure. In all, 216 SUN proteins from protists, fungi, and plants were divided into two monophyletic clades (Cter-SUN and Mid-SUN). We performed comprehensive evolutionary analyses, investigating the characteristics of different subfamilies in plants. Mid-SUNs further evolved into two subgroups, SUN3 and SUN5, before the emergence of the ancestor of angiosperms, while Cter-SUNs retained one subfamily of SUN1. The two clades were distinct from each other in the conserved residues of the SUN domain, the TM motif, and exon/intron structures. The gene losses occurred with equal frequency between these two clades, but duplication events of Mid-SUNs were more frequent. In cotton, SUN3 proteins are primarily expressed in petals and stamens and are moderately expressed in other tissues, whereas SUN5 proteins are specifically expressed in mature pollen. Virus-induced knock-down and the CRISPR/Cas9-mediated knockout of GbSUN5 both showed higher ratios of aborted seeds, although pollen viability remained normal. Our results indicated divergence of biological function between SUN3 and SUN5, and that SUN5 plays an important role in reproductive development.
The nuclear envelope (NE) provides physical rigidity to the nucleus, protects the genome, organizes chromatin, functions in meiotic chromosome pairing, and positions the nucleus within the cell (Starr and Fridolfsson, 2010). The NE, consisting of an outer nuclear membrane (ONM) that is closely associated with perinuclear endoplasmic reticulum (ER) and an inner nuclear membrane (INM) that is connected to the ONM via the nuclear pores, plays a vital role in regulating transport into and out of the nucleus. The NE is also involved in the physical positioning of the nucleus and in the processes of cell division and nucleo-cytoplasmic signaling (Graumann et al., 2010). Significant progress has now been made in the study of novel plant NE proteins. These proteins include a Linker of Nucleoskeleton and Cytoskeleton (LINC) complex based on INM Sad1/Unc-84 (SUN)-domain proteins, ONM Klarsicht/ANC-1/Syne-1 Homology (KASH) proteins, and nuclear lamina associated proteins (CRWNs, KAKU4, and NEAPs) (Poulet et al., 2017). Evolution of KASH domain proteins has resulted in increasing complexity; some are highly conserved and appear in all species, but others are restricted in distribution (Zhou and Meier, 2014; Poulet et al., 2017). Nuclear lamina associated proteins present in plants but is absent in unicellular species, which may attribute to plants evolved a lamina-like structure (Poulet et al., 2017). However, SUN proteins appear throughout and may be one of the earliest evolving components of the plant NE (Murphy et al., 2010; Graumann et al., 2014; Poulet et al., 2017). This suggesting that SUN domain proteins are essential for most organisms. SUN-domain proteins are INM proteins that are part of the linker cytoskeletal elements with the nucleoskeleton (LINC) complexes (Crisp et al., 2006; Starr and Fridolfsson, 2010). SUN proteins are conserved in non-plant and plant systems and have evolved into Cter-SUN and Mid-SUN subfamilies differentiated by the position of the SUN domain within the protein (Field et al., 2012; Graumann et al., 2014). Furthermore, A Phylogenetic analysis of SUN-domain proteins exhibited an ancient divergence of CCSD (Cter-SUN) and PM3-type (Mid-SUN) protein, and the functional divergence of four orthologous groups (SUN1/2/3/5) within grass species (Murphy et al., 2010). Cter-SUN proteins have been described in Arabidopsis (AtSUN1, AtSUN2), maize (ZmSUN1, ZmSUN2), rice (OsSUN1, OsSUN2), yeast (Mps3), Sordaria macrospora (SmSUN1), and other organisms (Graumann et al., 2010; Murphy et al., 2010; Friederichs et al., 2012; Vasnier et al., 2014; Varas et al., 2015; Zhang et al., 2020). In yeast, the protein MPS3, critical for vegetative growth and sporulation, is involved in spindle polar body (SPB) replication, spindle formation during mitosis, and fusion of the nucleus (Jaspersen et al., 2002; Nishikawa et al., 2003; Antoniacci et al., 2004). In plants, SUN1 and SUN2 can interact with the KASH-domain of the WIP protein and SINE protein to anchor WIT, forming a NE bridge (also termed LINC complex) (Zhou et al., 2012; Tamura et al., 2013; Graumann et al., 2014; Groves et al., 2020). The LINC complex has multiple functions with structural roles in positioning of nuclei, maintaining the shape of nuclei, movement of the pollen nucleus, stomatal development, and in plant male fertility (Graumann et al., 2014; Tatout et al., 2014; Evans et al., 2020; Groves et al., 2020). Recently, the structure and dynamics research revealed that this bridging complex have a role in seed maturation and germination, Organ development, response to stress, and the regulation of gene activity by organizing chromatin in the 3D nuclear space (Evans et al., 2020). In addition, LINC complexs (SUN1) and lamin-like proteins (CRWN1/4) physically and functionally interact with chromatin-regulatory proteins (PWO1) that play roles in gene expression, nuclear size, and nuclear shape (Mikulski et al., 2019; Groves et al., 2020). A new NE protein, OPENER, was recently identified and binds SUN1/2 and is involved in embryonic development and nucleolar size (Wang et al., 2019). AtSUN1 and AtSUN2 play crucial roles in meiosis (Zhou et al., 2012, 2014). The double mutant of Atsun1-1/Atsun2-2 displayed greatly reduced fertility and severe meiotic defects, such as a delay in the progression of meiosis, an absence of full synapsis, the presence of unresolved interlock-like structures, and a reduction in the mean cell chiasma frequency (Varas et al., 2015). The double mutant of Ossun1/Ossun2 displayed similar severe defects in meiosis as Atsun1-1/Atsun2-2, but OsSUN2 has a more important role than OsSUN1 in rice meiosis (Zhang et al., 2020). The maize SUN2 (ZmSUN2) formed a distinct belt-like structure at the nuclear periphery that are converted to a half-belt in zygotene and then back to a belt in pachytene. The half belt structure of ZmSUN2 is disrupted in the chromosome segregation mutants, desynaptic (dy1), asynaptic1 (as1), and divergent spindle1 (dv1) (Murphy et al., 2014). This result suggests that the SUN belt is associated with meiotic telomere dynamics, chromosome synapsis. Mid-SUN proteins, different from Cter-SUN proteins, contain three TM domains (one at the N-terminus and two at the C-terminus), coiled-coil domains, and a Sun domain located in the central area. In yeast, SLP1 protein as the sole Mid-SUN protein is part of the complex with the YERP65 protein recruiting MPS3 localized in the NE (Friederichs et al., 2012). In S. macrospora, the deletion mutant of the SLP1 gene shows defects in both vegetative growth and sporulation (Vasnier et al., 2014). The Mid-SUN proteins have been described in Arabidopsis (AtSUN3, AtSUN4, and AtSUN5) and maize (ZmSUN3, ZmSUN4, and ZmSUN5) (Murphy et al., 2010; Graumann et al., 2014). AtSUN3 and AtSUN4 are located in the NE and ER, while ZmSUN3 and ZmSUN4 are located only in the NE (Murphy et al., 2010; Graumann et al., 2014). AtSUN3 and AtSUN4 are expressed in many tissues at moderate levels, while AtSUN5 is mainly expressed in pollen and various embryonic tissues (Graumann et al., 2014). In maize, ZmSUN3 and ZmSUN4 share similar expression patterns with those of AtSUN3 and AtSUN4 (Murphy et al., 2010). ZmSUN5 is specifically expressed in pollen, suggesting the function of nuclear migration down the pollen tube and possibly double fertilization (Murphy et al., 2010). In Arabidopsis, the single mutants in Atsun3-1, Atsun4-1, and Atsun5-1 do not show obvious growth or fertility defects, although changes in nuclear morphology can be detected (Graumann et al., 2014). The mutants of AtSUN3/Atsun3-1, Atsun4-1, and Atsun5-1 produced approximately 17.4% of aborted seeds/siliques, and the homozygous triple mutant was lethal (Graumann et al., 2014). In addition, Membrane yeast two-hybrid (MYTH) assay provides evidence for a complex which is formed by interaction of Mid-SUN proteins with Cter-SUN proteins through their coiled-coil domains on the NE, but the biological functions have not been confirmed in previous studies (Graumann et al., 2014). However, while SUN proteins are highly conserved, and exist in most organisms, the evolutionary history of Cter-SUNs and Mid-SUNs has not been systematically studied, and the function of mid-SUN proteins are poorly studied, especially SUN5. In this study, a combination of bioinformatics and molecular experiments were conducted to illuminate the evolution and the divergence of the Cter-SUN proteins and Mid-SUN proteins. Our results showed that the Cter-SUN and Mid-SUN proteins were monophyletic and have undergone different evolutionary histories from protists to plant species. Different expression patterns of the SUN members in cotton indicated the functional divergence among the subfamilies. Decreasing the expression of GbSUN5 caused the abortion of cotton seeds, indicating a probable function during fertilization, different from GbSUN3.
To obtain as many as SUN genes in sequenced eukaryote genomes as possible, several datasets and multiple steps were used to search for the sequences. Protein sequences of protists and fungi were downloaded from the Ensembl database1,2. Plant proteomics were downloaded from the Phytozome database3. The sequences of cotton and Arabidopsis were retrieved from CottonFGD4 and Tair datasets5. We also obtained the prokaryotic protein sequences from the Ensembl Bacteria database6. The amino acid sequences of AtSUN genes (Graumann et al., 2014) were used as queries for gene searches using BLASTP for SUN genes in the several datasets mentioned above within representative species with a cut-off E-value set at 1e–2. These sequences were further verified via Pfam (El-Gebali et al., 2019) batch searches with default settings for the threshold option, a Conserved Domain Database (CDD) batch search (Marchler-Bauer et al., 2017) and SMART database batch search (Letunic et al., 2015). Sequences with obvious errors and/or lengths less than 150aa were removed manually. Confirmed sequences were used for further analysis.
The amino acid sequences of SUN proteins were aligned using MUSCLE 3.8.31 (Edgar, 2004) with default parameters. MEGA-X (Kumar et al., 2018) was used to find the best model and to construct the maximum likelihood (ML) tree with bootstrap tests of 1000 replicates, the Gamma Distribution option, the partial deletion option, and the JTT + G model.
All SUN protein sequences were used to search against the Pfam (El-Gebali et al., 2019) HMMER (Potter et al., 2018), and CDD (Marchler-Bauer et al., 2017) databases to find other known domains/motifs apart from the SUN domains. To discover novel conserved motifs, the software Multiple Em for Motif Elicitation (MEME) v5.1.1 (Bailey et al., 2009) was employed online7 using the following parameters: Zero or One Occurrence Per Sequence (zoops), and the number of motifs was no greater than 20. The Gene Structure Display Server8 (Hu et al., 2015) was used for gene structure analysis. TMHMM v2.09 (Krogh et al., 2001) and Tmpred10 (Hofmann and Tmbase, 1993) were used to predict transmembrane helices (TMH). Coiled-coil (CC) domains were predicted by COILS-Server11 (Lupas et al., 1991).
The plant species tree was adapted from TimeTree12 (Kumar et al., 2017). The gene trees obtained from MEGA-X for each of two SUN clades were reconciled with the plant species tree individually by Notung-DM (Chen et al., 2000; Darby et al., 2017) with default parameters.
To analyze the expression profiles of GhSUN and GbSUN genes in different tissues and developmental stages, expression data for mRNA levels were retrieved from the genome-wide RNA-seq dataset in CottonFGD13 (Zhu et al., 2017) and the Cotton Omics Database14 (Hu et al., 2019), respectively. The heatmap charts were drawn according to gene expression values (FPKM).
Arabidopsis Columbia (Col-0) was used for the GbSUN5_At promoter transfer experiments and as wild-type controls. Plants were grown on soil in a growth chamber under long-day conditions (16 h light/8 h dark) at approximately 22–24°C. Gossypium hirsutum acc. TM-1 was used for the VIGS assay. TM-1 was grown in pots at 25°C in a growth chamber under a 16 h light–8 h dark cycle with 60% humidity.
Total RNA was extracted from different tissues using the RNAprep Pure plant Kit (Tiangen, Beijing, China). RNA was reverse transcribed to cDNA using a PrimeScript® RT reagent kit (Takara, Dalian, China). PCR amplifications were performed using SYBR® Premix Ex TaqTM (Tli RnaseH Plus) on an Applied Biosystems 7500 Fast Real-Time PCR System. The PCR conditions were as follows: primary denaturation at 95°C for 30 s followed by 40 amplification cycles of 5 s at 95°C and 30 s at 60°C. Cotton Actin7 (CottonFGD Gene ID: Gbar_A11G005750) was used as an internal control. Melting curve analysis was performed to ensure there was no primer-dimer formation. Information of the qRT-PCR primers are presented in Supplementary Table 1. The qRT-PCR was carried out with three biological replicates, each comprising three technical replicates. Relative gene expression levels were calculated using the 2–ΔΔCt method (Livak and Schmittgen, 2001).
To further analyze the expression pattern of GbSUN5_At, two GbSUN5_At upstream fragments of Hai 7124 (containing 1793 and 3149 bp at upstream locations of the ATG start codon) were amplified with the primer pair Ps1/Ps2 and Ps3/Ps4 (Supplementary Table 1) and recombined into PBI121 (GUS reporter gene) and pcambia2300 [green fluorescent protein (GFP) reporter gene], respectively, by In-Fusion® Cloning. The two constructs were both introduced into the host cells of Agrobacterium tumefaciens LBA3101. Agrobacterium-mediated transformation of Arabidopsis thaliana was transformed using the floral-dip method (Clough and Bent, 1998) and a modified procedure for Agrobacterium preparation (Logemann et al., 2006).
GUS (beta-glucuronidase) histochemical staining was performed using the GUS Staining Kit (Coolaber, Beijing, China). Tissues were stained with GUS solution overnight at 35°C after vacuum infiltration. Pollen was stained with DAPI (4′, 6-diamidino-2-phenylindole, dihydrochloride) as described in Coleman and Goff (1985).
The mature pollen from VIGS plants, knockout mutants, and the WT were fixed overnight in Carnoy’s fluid (60% ethanol, 30% chloroform, 10%, glacial acetic acid) and washed in a graded ethanol series (95% [×3], 75% [×3]). Fixed pollen was stained by Alexander’s staining solution for testing viability (Alexander, 1969).
We isolated a 450-bp fragment of GhSUN5_At from TM-1. The fragment was amplified using primers Psv5/Psv6 and subcloned into SpeI and SacI digested pCLCrVA, generating pCLCrVA-GhSUN5_At constructs (Tuttle et al., 2008). The vectors pCLCrVA-GhSUN5_At and pCLCrVB were introduced into A. tumefaciens strain GV3101 (Idris and Brown, 2004). More than 50 individual plants were infiltrated with a mixture of A. tumefaciens carrying pCLCrVA-GhSUN5_At and pCLCrVB (Idris and Brown, 2004). Untreated (CK) and empty vector (CLCrv: 00) transformed plants (n > 50) were used as experimental controls. The transcript levels of GhSUN5 in mature pollen of silenced plants were detected using primers Psv7Psv8 (primers indistinguishable between homologs). Those primers were listed in Supplementary Table 1.
For targeted editing of GhSUN5, a pair of sgRNAs was designed in the coding region of the SUN domain, and the tRNA-sgRNA fragment was ligated to the Prgeb32-GhU6.9-NPT II expression vector. Primers are listed in Supplementary Table 1. The construct was introduced into the host cells of A. tumefaciens LBA4404. Cotton cultivar Gh cv. HM-1, which exhibits a normal growth habit, was used as the transformation receptor. Agrobacterium-mediated transformation was conducted following a previous report (Jin et al., 2006).
In plants, members of the SUN gene family are characterized by three important units, transmembrane (TM), coiled-coil (CC), and SUN domains (Graumann et al., 2010; Murphy et al., 2010; Oda and Fukuda, 2011). To identify the SUN genes in major lineages, we performed searches against plants, protists, and fungi using the SUN proteins from A. thaliana (Graumann et al., 2014). In all, 216 sequences were retrieved from the genomes of 42 plants, 6 fungi, and 4 protists (Figure 1 and Supplementary Tables 2, 3). To explore the evolutionary history of eukaryotic SUN genes, we conducted phylogenetic analyses with full-length sequences from 26 representative species (Figure 1) using MEGA-X. Phylogenetic analysis of the retrieved proteins suggested that SUN proteins in eukaryotes evolved into two monophyletic clades (Cter-SUNs and Mid-SUNs) (Figure 2). In the selected species, the copy number of SUN genes ranged from 2 in Saccharomyces cerevisiae to 8 in soybeans, with the highest number being 13 in Selaginella moellendorffii. Two kinds of SUN genes were found in almost all selected species except for Chlamydomonas reinhardtii and Coccomyxa subellipsoidea C-169 (Supplementary Table 2). Two major clades of SUN genes in Mesangiospermae went through different evolutionary processes. Mid-SUN genes further evolved into two distinct subgroups (termed as SUN3 and SUN5) before the divergence of the ancestor of angiosperms, while Cter-SUNs retained one subfamily of SUN1 (Figure 2).
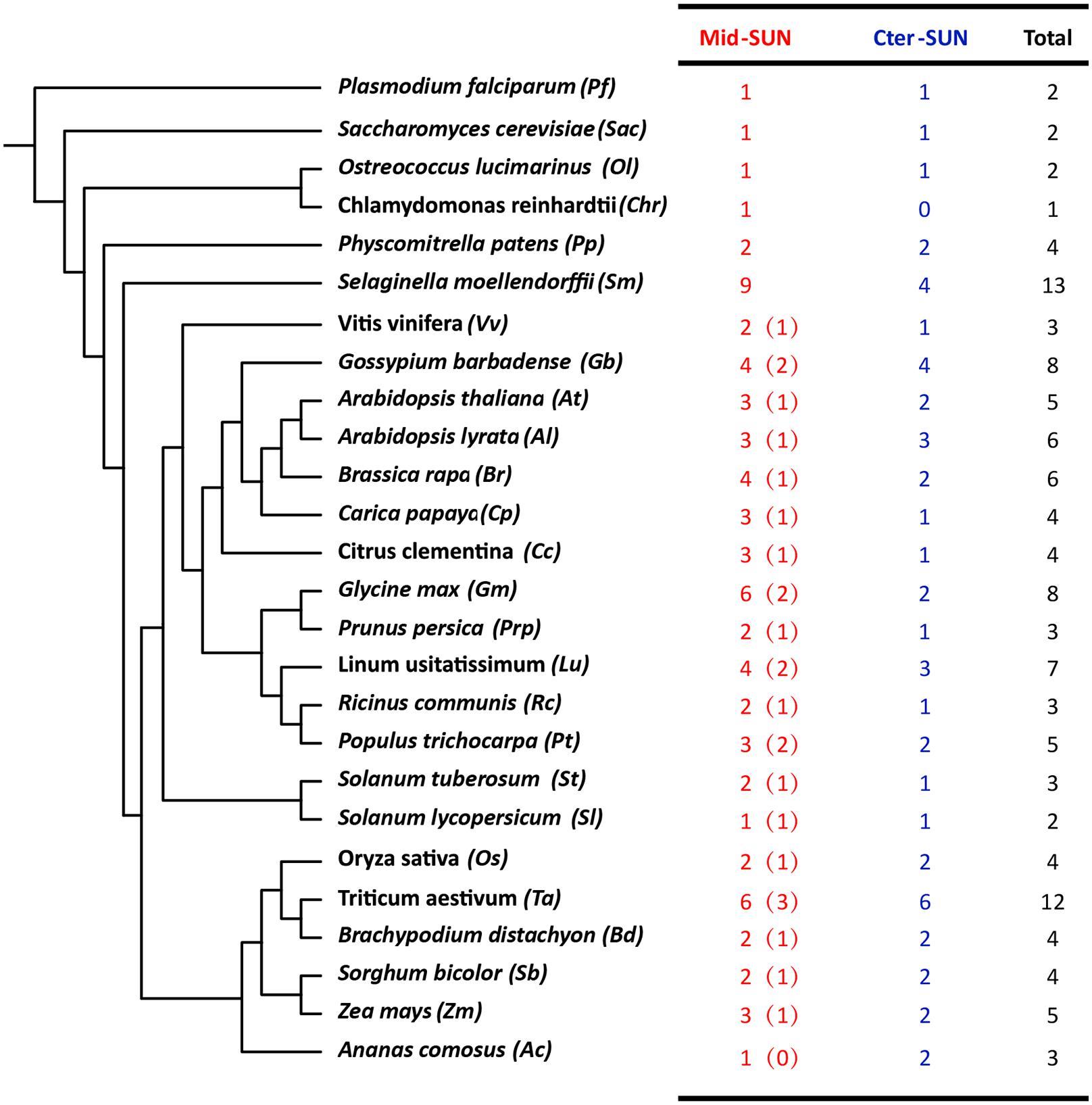
Figure 1. The number and classification of SUN genes in representative species. The numbers in the brackets of Mid-SUN denote the number of SUN5 genes in each Angiosperm species.
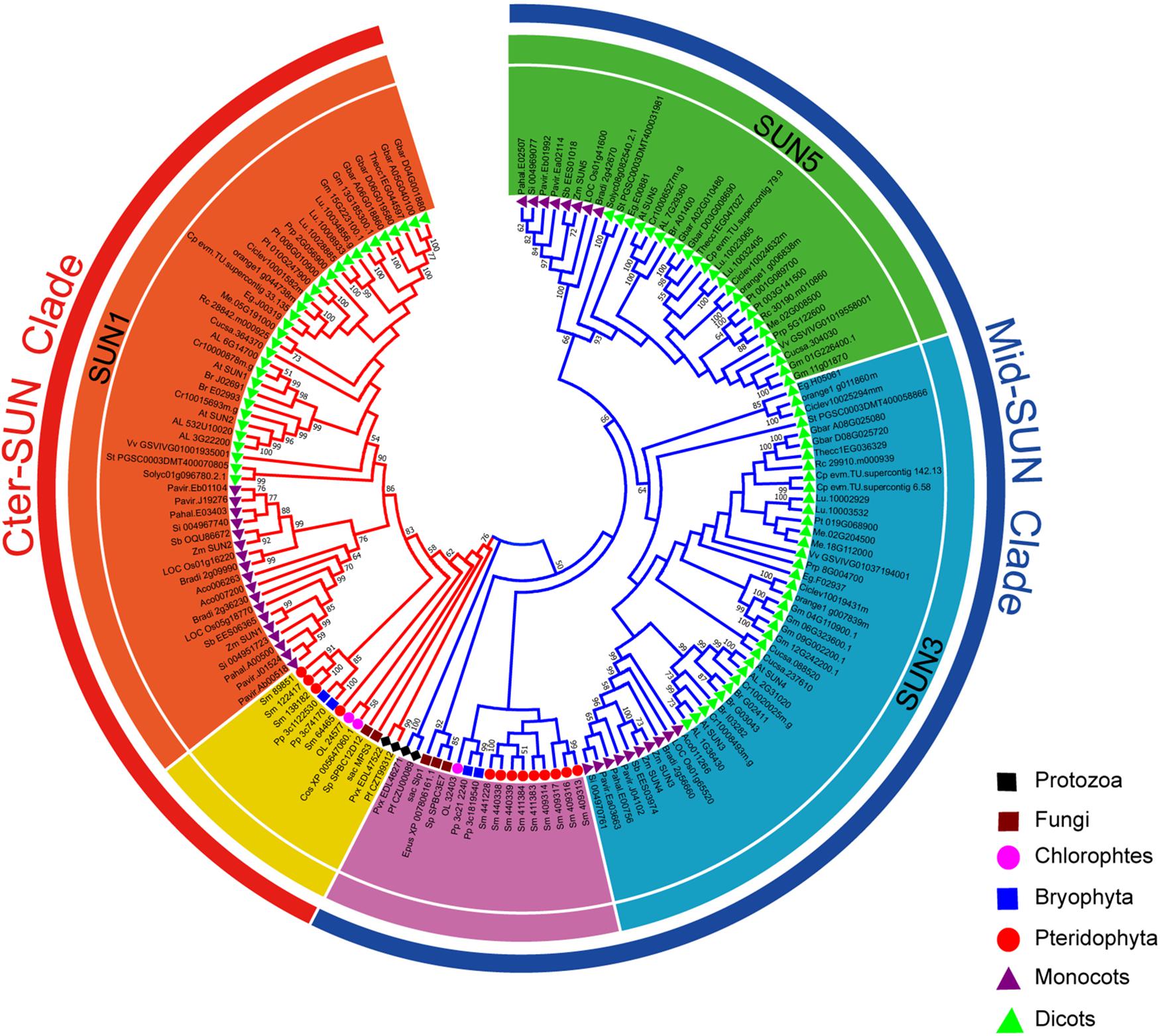
Figure 2. Phylogeny of representative SUN genes from protists, plants, and fungi. The tree topology generated via MEGA-X is shown here. Numbers on branches indicate the bootstrap percentage values calculated from 1000 replicates, and only values higher than 50% are shown. The Mid-SUN clade is indicated by the blue branch lines, and the Cter-SUN clade by the red branch lines. The Mid-SUN clade is separated into SUN3 and SUN5. The Cter-SUN clade retained one subfamily of SUN1. The peripheral groups of Cter-SUN and Mid-SUN are shown in yellow and purple, respectively.
To explore the origin of the SUN gene family, CLIME15 (Li et al., 2014) was performed for predicting the evolutionarily conserved modules (ECM) using At-SUN proteins. The results showed two distinct evolutionary histories between Cter-SUNs and Mid-SUNs across 138 eukaryotes. Mid-SUN proteins existed in more protists and fungi, with first appearance in Entamoeba histolytica, while Cter-SUN was lost in many fungi, with the first appearance in Plasmodium vivax (Additional File 1). Analysis of the SUN genes identified in the 10 species of protists and fungi revealed a similar distribution into two clades (Supplementary Figure 1). This further suggested that the division between these two branches dated from before the divergence of the protists.
According to the Pfam database at http://pfam.sanger.ac.uk, the SUN-domain protein family comprises over 30 different architectures that can be grouped into proteins with a central SUN domain (Mid-SUNs) and proteins containing a SUN domain at their C-terminus. Based on a combination of type and number of the TMH motif(s), coiled-coil (CC) (s), intrinsically disordered protein regions (IDPs), and the Sad1/UNC-84 domain, all of the SUN proteins from angiosperms can be further classified into two types (Cter-SUNs and Mid-SUNs). The Cter-SUN proteins usually contain a SUN domain at the C-terminus (C-sun) and a TMH motif at the N-terminus with CC and IDPs. The Mid-SUN proteins contain three TMH motifs (one TMH motif at the N-terminus and two TMH motifs at the C-terminus) and one other type of SUN domain (M-sun) with an internal CC and IDPs (Figure 3A and Supplementary Figure 2). The TMH in Cter-SUN showed moderately conserved amino acid residues. AtSUN1 and AtSUN2 were located in the INM of the NE, showing the transmembrane domain from nucleoplasm to the NE lumen (Zhou et al., 2012). AtSUN3 and AtSUN4 proteins expressed as fluorescent fusion proteins were membrane-associated and localized to the NE and ER (Graumann et al., 2014). The sequence logos of TMHs from two clades of proteins showed two types of TMH units. TMH1 of Cter-SUN and TMH2 of Mid-SUN shared similar amino acid residues enriched with Val, Ser, Phe, and Leu (Figure 3C), suggesting the similar directions from nucleoplasm to cytoplasm. TMH1 and TMH3 of Mid-SUN contained conserved Trp residues and moderately conserved Ser and Leu (Figure 3C), showing the same direction from cytoplasm to the lumen of the NE. Thus, the model of topological arrangements for generalized Cter-SUN and Mid-SUN proteins in the plants NE are presented in Figure 3B. Examination of the functional units of two subfamily members (SUN3 and SUN5) of Mid-SUN revealed little difference in angiosperms, except for the CC units. In S. moellendorffii, Physcomitrella patens, and Ostreococcus lucimarinus, none or partial TMHs existed in the proteins of Mid-SUN (Supplementary Figure 2).
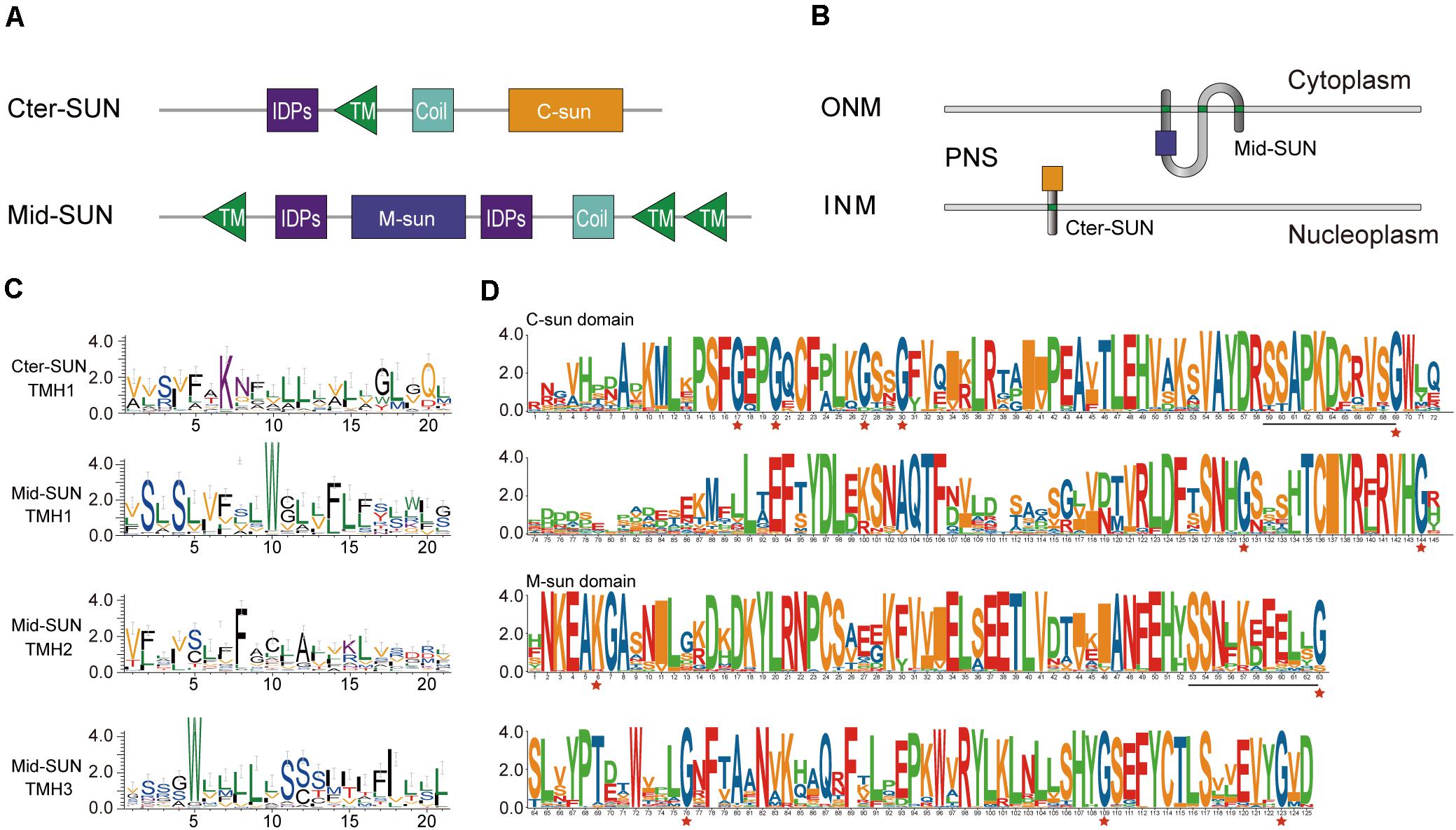
Figure 3. Protein organization and sequence features of SUN proteins. (A) Comparative protein motifs of the Cter-SUN and Mid-SUN protein subfamilies. Protein motifs are drawn based on a search using CDD, Pfam, HMMER, and MEME programs. (B) Possible protein arrangement models with the SUN domain in the perinuclear space are shown for the Cter-SUN and Mid-SUN. Models do not attempt to depict other domain organizations (such as IDPS and coiled-coil) and multimer interactions that may occur with the SUN or coiled-coil (not shown) domains. PNS, perinuclear space at the nuclear envelope. (C) Sequence features shown in the form of web logos representing TMH of two clades of Cter-SUNs and Mid-SUNs from 26 selected species. Logos were generated using the Weblogo3 application (http://weblogo.threeplusone.com/). (D) SUN domain-logo analysis of Cter-SUN and Mid-SUN proteins in 20 angiosperm species. The red stars indicate conserved Gly motifs in two types of SUN proteins. Common conserved motifs of C-SUN domain and M-SUN domain are underlined.
To better understand the difference between the two types of SUN proteins, exon/intron organization of different SUN genes was also examined. Each monophyletic clade shared similar exon/intron organization. Most members of Cter-SUNs usually contained one intron, while those in Mid-SUNs had multiple exons. It was intriguing to note that the SUN domain was separated by an intron in Cter-SUN, but the domain in Mid-SUN was maintained complete without an intron insertion (Supplementary Figure 3).
To further examine the divergence between the two types of Sad1/UNC-84 domains in SUN proteins, the conserved protein domains from 20 selected angiosperm species were filtered out. We performed sequence logo analysis for 68 SUN domain sequences from Mid-SUNs and 40 from Cter-SUNs. Examination of the domains revealed that their protein sequences shared unique characteristics within each of the two SUN phylogenetic clades (Figure 3D). The Cter-SUN proteins (SUN1) share the same type of Sad1/UNC-84 domain (hereby named the “C-sun” domain), which was also displayed by conserved motifs (motifs 4, 7, 11, and 13) (Supplementary Figure 2). Mid-SUN proteins (SUN3 and SUN5) shared another type of Sad1/UNC-84 domain (hereby named the “M-sun” domain), which was also displayed by conserved motifs (motifs 1, 2, 5, and 6) (Supplementary Figure 2). The C-sun domain contains a consensus conserved seven-Gly motif, while the M-sun domain contains highly conserved five-Gly motifs (Figure 3D). The C-sun domain starts with conserved Pro-Ser-Phe-Gly-Glu-Pro-Gly, ends with Thr-Cys-Iie-Tyr-Arg-X-Arg-Val-His-Gly, and has stretches of about 20 amino acid residues with lower conservation in the middle. The M-sun domain started with conserved Asn-Lys-Glu-Ala-Lys-Gly-Ala, ended with Cys-Thr-Leu-Ser-X-X-Glu-Val-Tyr-Gly and was more conserved than the C-sun domain. The Sad1/UNC-84 domains between SUN3 and SUN5 from selected angiosperm species shared consensus conserved amino acid residues. However, examination of both types of Sad1/UNC-84 domains in the identified SUN proteins revealed that they shared no consensus conserved motif except for the common SSxxKxxxxxG motif (Figure 3D), suggesting a significant difference of biological function in the two monophyletic clades, but they might have a common ancestor.
To better understand the evolutionary events that have occurred among these two subfamilies, we performed an analysis of gene duplication and loss using Notung software (Chen et al., 2000; Darby et al., 2017). We obtained the number of variations of SUN genes at different stages of evolution according to the constructed phylogenies and inferred whether the internal nodes within the each clade were associated with gene duplication, gene loss, or lineage divergence events (Figure 4).
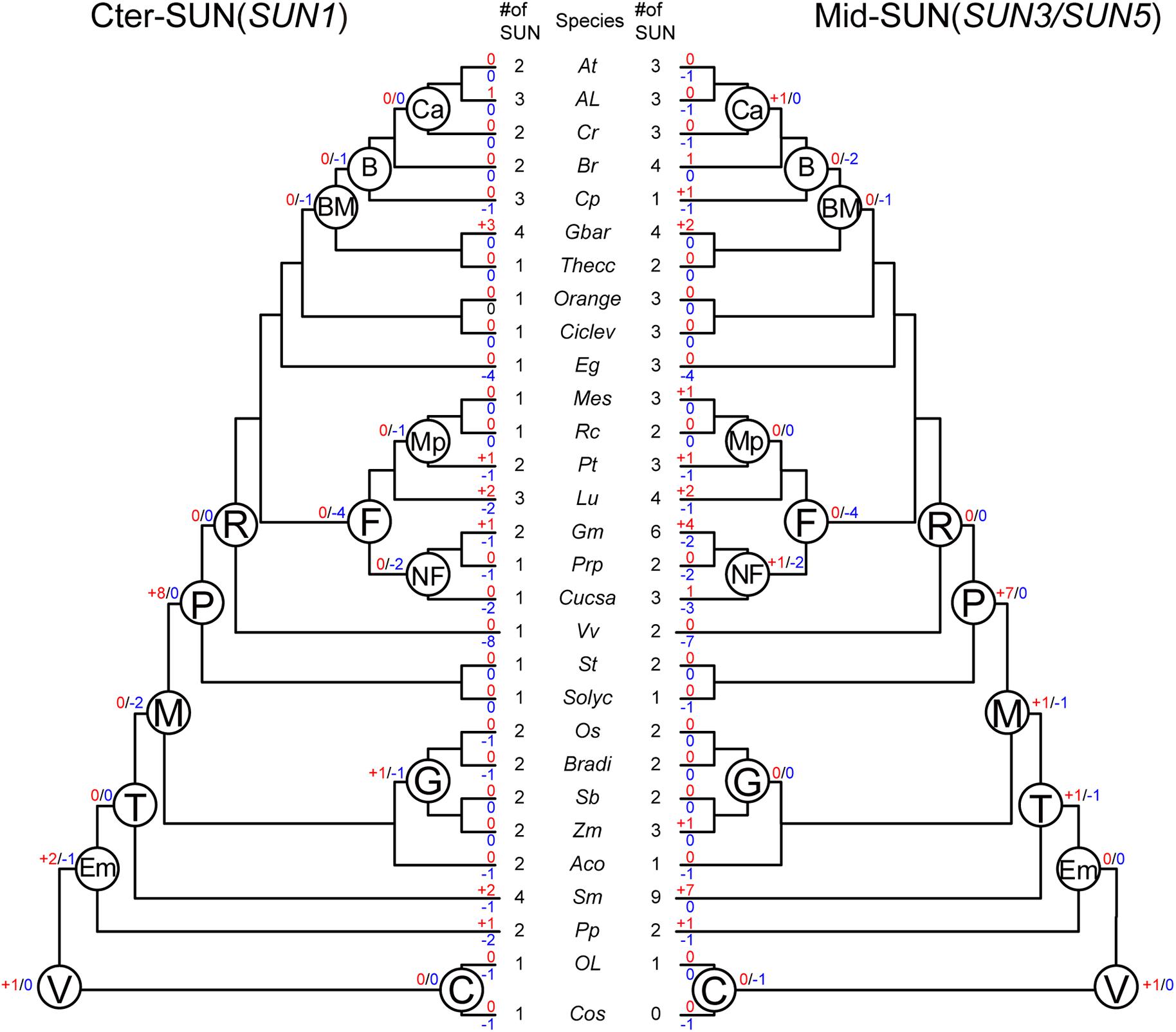
Figure 4. Evolutionary events in the SUN gene family in plants. Numbers of gene duplication (shown in blue after “+”) and loss (shown in red after “–”) events were inferred for each internal node as well as for current extant species. The numbers of SUN genes are also listed accordingly. Cos, Chlorella vulgaris. Ol, Ostreococcus lucimarinus. Pp, Physcomitrella patents. Sm, Selaginella moellendorffii. Aco, Ananas comosus. Zm, Zea mays. Sb, Sorghum bicolor. Bradi, Brachypodium distachyon. Os, Oryza sativa. Solyc, Solanum lycopersicum. St, Solanum tuberosum. Vv, Vitis vinifera. Cucsa, Cucumis sativus. Prp, Prunus persica. Gm, Glycine max. Lu, Linum usitatissimum. Pt, Populus trichocarpa. Rc, Ricinus communis. Me, Manihot esculenta. Eg, Eucalyptus grandis. Ciclev, Citrus clementina. Orange, Citrus sinensis. Thecc, Theobroma cacao. Gbar, Gossypium barbadense. Cp, Carica papaya. Br, Brassicales. Cr, Capsella rubella. Al, Arabidopsis lyrata. At, Arabidopsis thaliana. C, Chlorophyta. V, Viridiplantae. Em, Embryophyta. T, Tracheophyta. M, Mesangiospermae. P, Pentapetalae. R, Rosis. F, Fabid. NF, Nitrogen fixing. Mp, Malpighiales. BM, Brassicales Malvales. B, Brassicales. Ca, Camelineae. G, Grass.
In Vridiplantae, gene duplication was detected, and no gene loss was found in either SUN clade. Cter-SUNs did not experience gene duplication/loss events; Mid-SUN experienced one gene loss event in Chlorophyta. During the emergence of the embryophytes, no gene duplication/loss events occurred in Mid-SUNs, but two SUN genes were duplicated and one gene was lost in Cter-SUNs. In tracheophytes, Mid-SUNs experienced one gene loss and one gene duplication. However, during the emergence of Mesangiospermae, two genes were lost and none were duplicated in Cter-SUNs, while one gene was duplicated and lost in Mid-SUNs. These results suggest that Mid-SUN experienced rapid birth-and-death events (Karlin and Mcgregor, 1957; Nam et al., 2004) that may have resulted in the divergence between SUN3 and SUN5. In the grass lineage, Cter-SUNs exhibited one gene loss and one gene duplication; there were no gene duplication/loss events occurring in Mid-SUNs. With the emergence of Rosids, eight and seven genes were duplicated in Cter-SUNs and Mid-SUNs, respectively, and no gene was lost in either clade. In the Fabids, four genes were lost, while no gene was duplicated in either clade. With the emergence of the nitrogen-fixing plants, two genes were lost in Cter-SUNs and Mid-SUNs, but one gene was duplicated in the Mid-SUNs. The Brassicales, Malvales, and Brassicales lost several genes, and no duplicated genes were detected. Only Camelineae had no losses in either Cter-SUNs or Mid-SUNs, but only one duplicated gene was detected in Mid-SUNs (Figure 4).
Furthermore, we also examined gene-duplication/loss events in extant plant species. The gene duplication events in several extant plant species, such as Glycine max, were probably associated with their recent whole-genome duplication events. In contrast, in several other plant species including Vitis vinifera and Eucalyptus grandis, the phylogenies of SUN proteins showed drastic gene loss events. As for the general results, Mid-SUN experienced approximately equal numbers of gene loss events as Cter-SUN, while there were more gene duplications in Mid-SUN than in Cter-SUN (Supplementary Table 4). This observation suggests that more gene duplications in Mid-SUNs may be also a reason for the divergences between SUN3 and SUN5.
To test the hypothesis that Cter-SUN and Mid-SUN evolved independently and to examine the divergence of SUN3 and SUN5, we used RNA-seq data of island cotton (Hai-7124) and upland cotton (TM-1) to analyze the expression patterns of the SUN genes in different tissues. Members of SUN genes showed a similar expression pattern across various tissues in island cotton and upland cotton, suggesting similar function in regulating plant growth and development. SUN1s and SUN2s in cotton were expressed at medium levels in detected tissues. SUN3s are mainly expressed in petals and stamens and are moderately expressed in other tissues. Interestingly, SUN5s are specifically expressed in pollen (Figure 5). These results agree with the similar expression of those in maize and Arabidopsis (Murphy et al., 2010; Graumann et al., 2014).
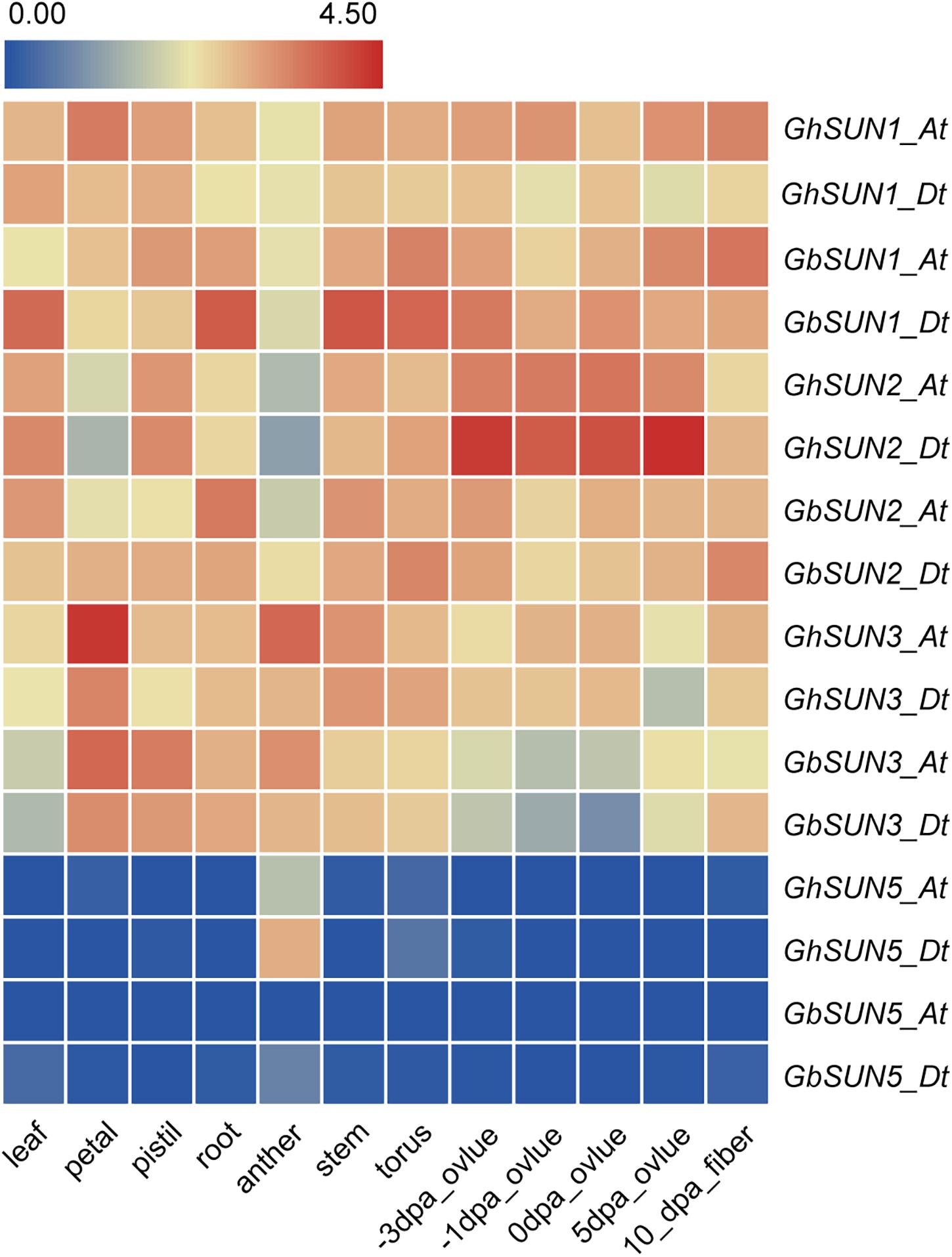
Figure 5. The expression profiles of SUN genes in a variety tissues of sea island cotton and upland cotton. The raw data for RNA-Seq of upland cotton and sea island cotton were downloaded from CottonFGD (https://cottonfgd.org/) and Cotton Omics (http://cotton.zju.edu.cn/), respectively. Gene expression levels are depicted with different colors on the scale. Red and blue represent high and low expression levels, respectively. dpa represents day post-anthesis.
For verification of the data from RNA-seq, quantitative RT-PCR was used to profile the expression levels of GbSUN1, GbSUN3, and GbSUN5 in different tissues of Hai-7124. Cter-SUNs (GbSUN1 and GbSUN2) were found to be ubiquitously expressed, in agreement with published RNA-seq data. The expression levels of Mid-Sun genes in island cotton were clearly distinguished into two groups. Moderate expression of GbSUN3 was detected at different stages of anther development and in stigmas and roots, while there was lower expression in the other tissues. GbSUN5 showed specific expression during stamen development and pollen maturation. It was interesting that GbSUN3 showed expression patterns similar to GbSUN5 in anthers but expressed differently in the pollen on the flowering day (Figure 6).
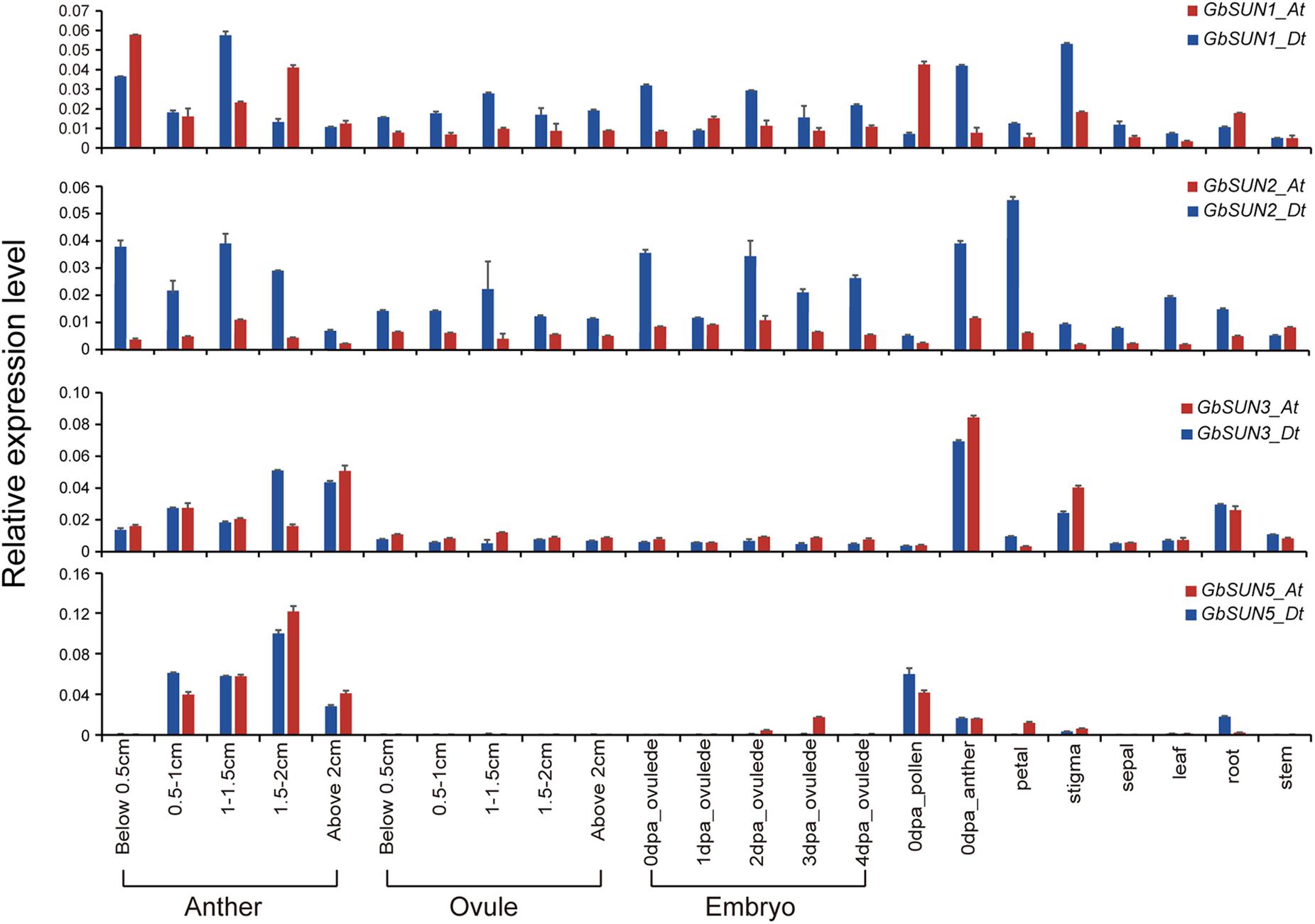
Figure 6. qRT-PCR confirmation of the expression levels of SUN genes in G. barbadense. Error bars represent the standard deviations of three independent experiments. Anthers and ovules at different stages were gathered with reference to the length of flower buds. dpa represents days post-anthesis. Values are means of at least three biological replicates.
The expression level of GbSUN3 gradually increased with anther development but had the lowest expression in the pollen on the flowering day. However, GbSUN5 had higher expression in the pollen on the flowering day than in post-dehiscence anthers, indicating a more important core role in pollen development, not in anthers (Figure 6). The different expression patterns among the three subgroups in island cotton suggested the functional divergence between SUN1, SUN3, and SUN5.
The expression profile of GbSUN5 restricted to the mature pollen was similar to that in maize (Murphy et al., 2010). To investigate whether GbSUN5 is specifically expressed in pollen, a 1.8-kb upstream fragment from GbSUN5_At and a 1.8-kb upstream fragment from AtSUN5 were transcriptionally fused to the GUS reporter gene and transformed into Arabidopsis Col-0. The results showed that no GUS staining was found in seedlings or early flowers (Figures 7A,B), while a weak GUS signal was detected in the stamens of immature flowers (Figure 7C). However, strong GUS staining was detected in mature pollen, whereas this was absent in anther tissues (Figure 7D). GUS staining was also detected in mature pollen and stigmas when the male flowers blossomed and the pollen dispersed (Figure 7E).

Figure 7. Analysis of GbSUN5_At promoter activity in different tissues. (A,B) No GUS staining was observed in the young rosettes or young flowers. (C) In immature flowers, a weak GUS signal was observed in stamens. (D) In mature anthers of the plant, strong GUS staining was observed in mature pollen grains, whereas no GUS staining was observed in the anthers. (E) GUS staining was also detected in mature pollen and stigmas several hours after anthesis.
To confirm the exact stage, a time course detection during pollen development was performed in GUS transgenic plants. The onset of SUN5 promoter activity coincided with the second pollen division that leads to tricellular pollen with sperm cells (Figures 8A,B). Similar results were observed in T1 lines with ProAtSUN5: GUS (Supplementary Figure 4). The results showed that the specific expression of SUN5 in mature pollen may be universal in plants, indicating a conservative biological function.

Figure 8. Temporal analysis of GbSUN5_At promoter activity in different stages of pollen development. (A) GUS staining in different stages of pollen development. (B) Fluorescence channels of the same pictures as in panel (A). Determination of pollen stages by visualization of nuclei by DAPI (4′,6-diamidino-2-phenylindole) fluorescence. (A) Bright field of the same pictures as in panel (B), showing GUS straining only in pollen grains at the tricellular stage when the sperm cells (white arrow) and vegetative cells (white arrowhead) are present. (C) The signals of ProGbSUN5_At-GFP in different stages of pollen germination. After 4–6 h of pollen germination, ProGbSUN5_At-GFP signals were observed in pollen grains and pollen tubes. After 8–10 h of pollen germination, ProGbSUN5_At-GFP signals were found in pollen tubes, whereas no GFP signal was observed in pollen grains. HAG, hours after germination.
To test whether GbSUN5_At was expressed in the grain germination, fusion of GFP with a 3-kb upstream fragment from GbSUN5_At transcriptionally was introduced into Arabidopsis. Expression of GbSUN5–GFP was investigated using fluorescence microscopy during pollen tube germination. The results showed that a strong GFP signal was observed in pollen grains, while a weak signal was detected in pollen tubes 4 h after pollen germination (HAP). At 6 HAP, the GFP signal weakened in pollen grains while being enhanced in pollen tubes. Interestingly, we could still observe a very bright fluorescent signal at 10 HAP (Figure 8C). These results suggested that the promoter of GbSUN5_At was active during the entire period of pollen development and germination.
To confirm the function of SUN5, we isolated a 450 bp fragment from upland cotton (TM1) and inserted it into the VIGS vector (CLCrv) to inhibit the endogenous expression of GhSUN5 by VIGS. GhCHLI (Mg-CHELATASE subunit I)-silenced plants showed a yellow bleaching phenotype as a control to judge whether the expression of GhSUN5 was silenced successfully in leaves. In Clcrv-GhSUN5 plants, qRT-PCR analysis revealed that the transcription levels of 22 of 50 VIGS plants were reduced to about 65% (Supplementary Figure 5A), while the pollen viability was similar to that of the CLCrv: 00 plants (Supplementary Figure 5B). At 10 days post-anthesis, abnormal cotton bolls were observed in GhSUN5-silenced plants, while the clcrv: 00 plants displayed the wild type (Supplementary Figure 5C). Moreover, some aborted seeds were found in the abnormal bolls (Supplementary Figure 5D). Compared with the Clcrv: 00 plants (approximate abortion rate 3.12%), Clcrv: GhSUN5 plants had 17.14% aborted seeds (Supplementary Figure 5E).
To further assess the function of GhSUN5, we used the CRISPR-Cas9 system to edit the sequence of the SUN domain against two target sites. Twelve independent transgenic lines were obtained. Three GhSUN5 knockout lines (two homozygous and one biallelic mutation) with either insertions or deletions at the SUN domain were identified (Figure 9A). Those knockout lines display no obvious growth and pollen abortion (Figures 9B,D). This is similar to the phenotype of VIGS Plants but with more extensive abortion in cotton bolls (Figures 9B,C,E), possibly due to more completely silence of the target gene expression by CRISPR-Cas9 system. These results further suggested that GhSUN5 genes did not have effects on pollen viability, and that they play a novel role in the karyogamy, sperm or/and early embryo development, similar to the SLP1 gene in S. macrospora and the SUN5 gene in mice and A. thaliana (Graumann et al., 2014; Vasnier et al., 2014; Shang et al., 2017).
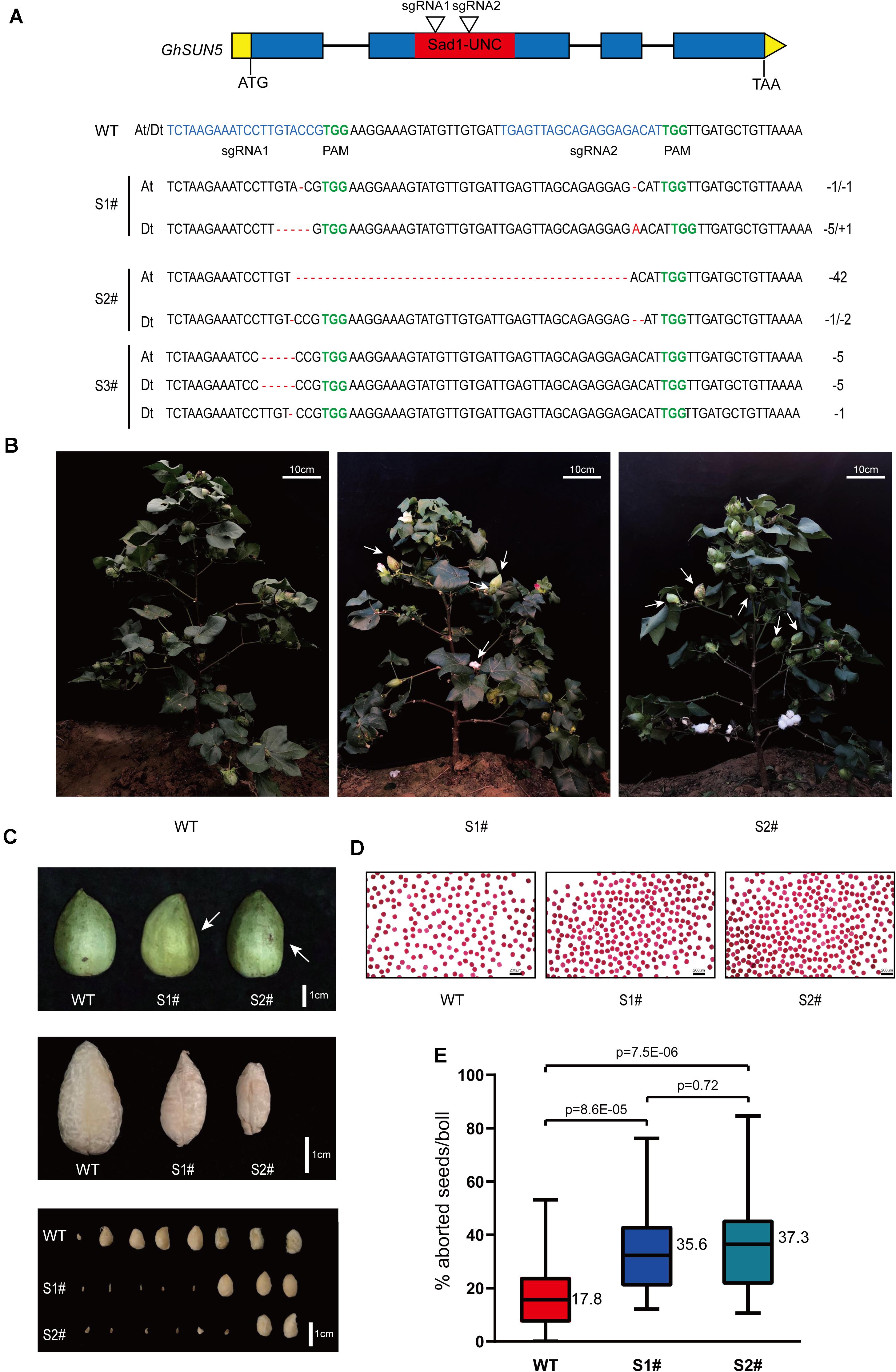
Figure 9. CRISPR-Cas9 targeted editing of the SUN domain in GhSUN5 genes. (A) Schematic map of two sgRNA target sites and three knockout cotton lines (two homozygous and one biallelic mutation). Exons, introns, SUN domain, and untranslated regions are shown as blue blocks, black lines, red blocks, and yellow boxes, respectively. Mutation sites in GhSUN5 are indicated in red. (B,C) Comparison of phenotypes of WT, S1#, and S2# mutants in the boll setting period. White arrows point to abortive cotton bolls (B) or aborted seeds (C). (D) Pollen vitality assays of S1# and S2# mutants. (E) Percentage of aborted seeds per boll in the wild type and the gene-edited plants. The data are shown as box-plot graphs; the horizontal lines across range bars represent the median (n > 30) and statistical analysis results from Student’s t-test.
SUN family proteins are highly conserved but partially differentiated throughout evolution in plant systems (Graumann et al., 2014; Tatout et al., 2014). In most organisms, the copy number of SUN genes varies slightly from ranged from 2 to 8 (Supplementary Table 2). Interestingly, we found that S. moellendorffii has the highest number of SUN genes (four Cter-SUN, nine Mid-SUN), which is significantly more than other species (Supplementary Table 2). A recent study showed that the Embryophyta possess at least two ONM KASH proteins (SINEs, WIPs, or TIK) except for S. moellendorffii, which interact with INM SUN proteins forming a NE bridge (LINC complexe) (Poulet et al., 2017). In addition, INM Cter-SUNs interacts with mid-SUN proteins forming LINC complexe by CC domain (Graumann et al., 2014). Thus, more Cter-SUNs and Mid-SUNs are required for LINC complexe in S. moellendorffii lacking ONM KASH proteins. In early eukaryotes such as fungi, protists, and Chlorophyta, SUN genes are present in single or low copy numbers and are essential for viability (Supplementary Table 2). For example, Cter-SUN proteins characterized in Schizosaccharomyces pombe and Caenorhabditis elegans are involved in duplication of the SPB, nuclear migration, and meiotic chromosome movements (Hagan and Yanagida, 1995; Malone et al., 1999). Following gene duplication, Cter-SUNs developed into SUN1 and SUN2, and these have been found to play vital roles in chromosome movements. In Arabidopsis and Oryza sativa, loss of SUN1 and SUN2 leads to a delay in the progression of meiosis, absence of full synapsis, defects in telomere clustering, and a reduction in the mean cell chiasma frequency. In addition, the expression pattern of Cter-SUN was similar between Arabidopsis and maize; SUN1 and SUN2 have been shown to be widely expressed in various tissues (Murphy et al., 2010; Graumann et al., 2014; Zhang et al., 2020). Consistent with this, the quantitative RT-PCR results showed that Sea island cotton SUN1 and SUN2 were expressed at low levels in most tissues examined in this study (Figure 6). This result indicates that SUN1 and SUN2 have maintained relatively conserved functions in meiosis from monocot to dicot plants. However, the functions of SUN1 and SUN2 proteins have diverged during evolution in some angiosperms. In Arabidopsis, AtSUN1 and AtSUN2 are thought to have completely redundant functions during meiosis. Homozygous single mutants exhibited normal vegetative growth, no obvious loss of fertility, and normal meiotic progression. The double mutant of sun1 and sun2 showed a significant reduction in fertility and severe meiotic defects (Varas et al., 2015). In contrast, OsSUN2 plays a more critical role than OsSUN1 in rice meiosis. The Ossun1 single mutant had a normal phenotype, but meiosis was disrupted in the Ossun2 mutant. These results are consistent with phylogenetic analyses of SUN1 and SUN2 in rice and Arabidopsis. AtSUN1 and AtSUN2 were closely related to each other, whereas OsSUN1 and OsSUN2 were assigned to two separated clades in phylogenetic analyses (Zhang et al., 2020). Interestingly, our phylogenetic analysis found that most monocotyledon SUN1 and SUN2 are divided into two separate clades, but almost all dicotyledon SUN1 and SUN2 were closely related to each other (Figure 2). This result suggested that Cter-SUNs (SUN1 and SUN2) show differential evolutionary patterns between dicotyledons and monocotyledons: Compared with dicotyledons, functional divergence may be more significant in monocotyledons.
Similarly, in the Cter-SUN clade, Mid-SUN also showed conservative and divergent evolution patterns in higher plants. Based on our phylogenetic study, Mid-SUN clade proteins were further evolved into SUN3 and SUN5 subfamilies. Previous studies have demonstrated that SUN3s are expressed at low to medium levels in most tissues, whereas SUN5s showed a very distinct and much more restricted pollen-related pattern of expression in Arabidopsis and Zea mays (Murphy et al., 2010; Graumann et al., 2014). This is consistent with expression profiles GbSUN3-At/Dt and GbSUN5-At/Dt (Figures 6–8). These results imply that SUN3 and SUN5 retained rather conserved functions in higher plants. However, some angiosperm SUN3 subfamily developed into SUN3 and SUN4 by duplication, but it is difficult to distinguish SUN3 from SUN4. In Arabidopsis, AtSUN3, AtSUN4, and AtSUN5 are thought to have redundant functions during growth and development; the single mutants display no obvious loss of growth or fertility defects, but the triple mutant sun3 sun4 sun5 was lethal, and in SUN3/sun3-1 sun4-1 sun5-1 plants, a slight reduction (about 17%) in fertility was observed (Graumann et al., 2014). Unlike Arabidopsis, when decreasing the expression of GbSUN5 by VIGS and disrupting GbSUN5 by CRISPR/Cas9 systems, aborted seeds were observed in silenced plants (Figure 9). These results suggest the functional divergence of GbSUN3 and GbSUN5. Consistent with this, GbSUN3 is mainly expressed in anthers instead of pollen, and GbSUN5 is specifically expressed in pollen (Figures 6–8). In addition, the cotton SUN3 subfamily did not develop SUN3 or SUN4, and Mid-SUN only contains SUN3 and SUN5. These findings led us to propose that there is functional divergence of GbSUN3 and GbSUN5 during double fertilization and embryo development, which is quite different from the lack of divergence of their Arabidopsis counterparts. At the same time, we noted that some of the SUN3 subfamilies in angiosperms developed into SUN3 and SUN4 by duplication, while the other SUN3 subfamilies did not replicate or lose SUN3 or SUN5, for example, Ananas comosus and Solanum lycopersicum. These data suggest that the functions of Mid-SUN proteins have had varied patterns of differentiation during the evolution of angiosperms.
Our results indicated that Cter-SUN had strikingly different patterns of evolution from Mid-SUN, but both types of SUN-domain proteins may have the same origin. All SUN amino acid sequences were used to search the Pfam databases, and we found that two types of SUN domain belonged to the same Pfam (Pf07738). In addition, although sequence features revealed a significant degree of divergence between c-sun and m-sun in plants through mass evolutionary events, we still found that they have a common SSxxKxxxxxG motif (Figure 3D). This indicates that Cter-SUN and Mid-SUN have a common ancestor and that they evolved independently. In prokaryotes, a few SUN proteins have been identified; these exist as single SUN domains. Individual strains of bacteria contain one gene encoding a SUN domain (Pf07738) (Supplementary Table 5). This observation suggests a role for the SUN domain in fundamental biological processes that have been conserved since prokaryotes throughout evolution. To explore the evolutionary history of prokaryote SUN genes, we conducted phylogenetic analyses with SUN domain sequences from prokaryotes and Arabidopsis. Based on our phylogenetic analyses, the prokaryote SUN genes also can be divided into two monophyletic clades (Cter-Sad1-UNC and Mid-Sad1-UNC) (Supplementary Figure 6). However, there is a single SUN type (Cter-SUN or Mid-SUN) in each prokaryote species. Thus, Cter-SUN and Mid-SUN have some overlapping functions, and absence of one of these types is not critical for survival.
Unlike prokaryote SUNs existing as a single SUN domain, most eukaryote SUN proteins also contain a TMH domain and a coiled-coil domain, and each type of SUN protein (Cter-SUN and Mid-SUN) is highly conserved, especially in the SUN domain (Supplementary Figures 1, 2). However, the homologs of Cter-SUNs MPS3 in S. cerevisiae have diverged significantly from the rest of the eukaryotes (Supplementary Figure 7). A blast search using most of Cter-SUN identified the S. pombe SUN domain protein with a significant E-value but not the Mps3 domain of S. cerevisiae. We discovered that Mps3 existed in a few yeast species and was well-conserved (Supplementary Figure 8). At the same time, ECM of yeast SLP1 (Mid-SUN) and MPS3 (Cter-SUN) were predicted by CLIME (Additional File 2). SLP1 homologs are widely distributed in 138 species, and this pattern coincides with AtSUN3/AtSUN4/AtSUN5. In contrast to AtSUN1/AtSUN2, MPS3 homologs exist only in a few yeasts, in agreement with the above results. It is noteworthy that AtSUN1/AtSUN2 homologs were absent in a few yeasts, while MPS3 homologs were present in those yeasts (Additional File 1). The results indicate that Cter-SUN likely experienced the divergence between yeasts and other eukaryotes. Also, SUN-KASH bridges span the nuclear periplasm and link the nucleoskeleton and cytoskeleton in most organisms, while MPS3 cannot bind to a KASH domain-containing protein due to the lack of many of the residues that are thought to be critical for SUN-KASH binding based on crystallographic studies (Friederichs et al., 2012). However, MPS3 directly or indirectly interacts with some SPB-related proteins such as MPS2, SPC42. NDJ1, SIR4, SPC29, and CSM4, proteins that are yeast-specific (Additional File 3) and are involved in telomere tethering, gene inactivation, formation of the chromosome bouquet, rapid telomere movement in meiotic prophase, SPB duplication, and tethering the half-bridge to the core SPB in S. cerevisiae (Jaspersen et al., 2002, 2006; Bupp et al., 2007; Conrad et al., 2008). The function of S. cerevisiae MPS3 is similar to other Cter-SUNs (Ding et al., 2007; Talamas and Hetzer, 2011; Friederichs et al., 2012). This finding suggests that a basic mechanism of Cter-SUN protein action is conserved in all eukaryotes but involves a different protein in yeast. In plants, Cter-SUN genes are highly conserved and have retained one subfamily of SUN1, but Mid-SUN genes further evolved into two distinct subgroups (SUN3 and SUN5) before the divergence of the ancestor of angiosperms (Figure 2). Based on the results, we propose a model to describe the evolutionary history of the SUN gene family (Figure 10). In this model, we suggest that a primitive SUN protein existed by itself as a single SUN domain in the common ancestor of all living things, and this protein experienced gene duplication, differentiation, and eventually developed independently into members of Cter-Sad1-UNC proteins and Mid-Sad1-UNC proteins in prokaryotes. These two types of SUNs acquired a coiled-coil domain and a TMH domain at the N-terminus (Cter-SUNs) or C-terminus (Mid-SUNs) of the SUN domain during the emergence of eukaryotes. Most Cter-SUNs were well conserved in eukaryotes and developed SUN1 except for a few yeasts in which this became MPS3. Prior to the emergence of angiosperms, Mid-SUNs were further duplicated and gradually evolved into SUN3 and SUN5. While we have a general idea of when these events occurred, more detailed biological functions of SUN5 and SUN3 still need to be determined. Further experiments need to be performed to reveal functional divergence of Mid-SUN in higher plants, and this will improve our understanding of the diversification and functional evolution of the whole SUN family.
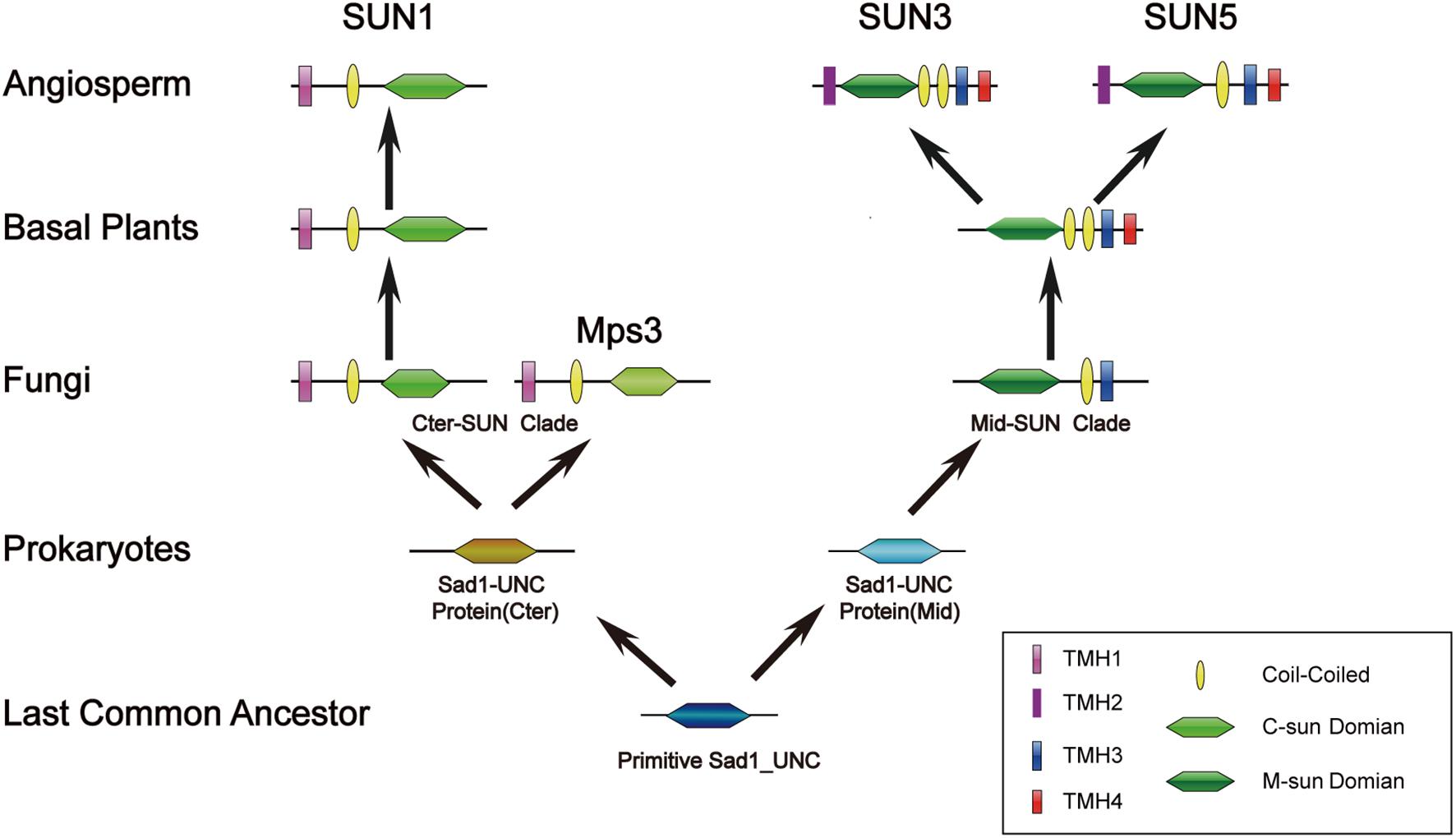
Figure 10. Evolutionary scenario of the SUN gene family in plants. The primitive Sad1_UNC (SUN) protein only exists as a SUN domain and may have emerged in the most recent common ancestor. The SUN genes were further duplicated and gradually evolved into two clades (Cter-SUN and Mid-SUN) in prokaryotes. Cter-SUN gained an N-terminal TMH and a CC domain, whereas Mid-SUN gained a C-terminal TMH and a CC domain in picoeukaryotes. In yeast, Cter-SUN evolved into Mps3, whereas Cter-SUN developed into SUN1 in other eukaryotes. Through the evolutionary divergence of Angiosperm plants, Mid-SUN proteins gained a C-terminal TMH and gradually evolved into SUN3 and SUN5.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
LY, WC, and YZ conceived the research and designed the experiments. LY performed the experiments. JP, SZ, QL, XW, BL, SF, and CL participated in the experiments. LY, WC, and YZ analyzed the data and experiment results. LY and YZ wrote the manuscript. All authors read and approved the final manuscript.
This research was supported by grants from the National Natural Science Foundation of China (31871680), and Agricultural Science and Technology Innovation Program of Chinese Academy of Agricultural Sciences.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We acknowledge Xiaoyang Wang and Weidong Zhu (Institute of Cotton Research of the Chinese Academy of Agricultural Sciences, Anyang, China) for technical assistance.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.646622/full#supplementary-material
Alexander, M. P. (1969). Differential staining of aborted and nonaborted pollen. Stain Technol. 44, 117–122. doi: 10.3109/10520296909063335
Antoniacci, L. M., Kenna, M. A., Uetz, P., Fields, S., and Skibbens, R. V. (2004). The spindle pole body assembly component mps3p/nep98p functions in sister chromatid cohesion. J. Biol. Chem. 279, 49542–49550. doi: 10.1074/jbc.M404324200
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. doi: 10.1093/nar/gkp335
Bupp, J. M., Martin, A. E., Stensrud, E. S., and Jaspersen, S. L. (2007). Telomere anchoring at the nuclear periphery requires the budding yeast Sad1-UNC-84 domain protein Mps3. J. Cell Biol. 179, 845–854. doi: 10.1083/jcb.200706040
Chen, K., Durand, D., and Farach-Colton, M. (2000). NOTUNG: a program for dating gene duplications and optimizing gene family trees. J. Comput. Biol. 7, 429–447. doi: 10.1089/106652700750050871
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Coleman, A. W., and Goff, L. J. (1985). Applications of fluorochromes to pollen biology. I. Mithramycin and 4’,6-diamidino-2-phenylindole (DAPI) as vital stains and for quantitation of nuclear DNA. Stain Technol. 60, 145–154. doi: 10.3109/10520298509113905
Conrad, M. N., Lee, C. Y., Chao, G., Shinohara, M., Kosaka, H., Shinohara, A., et al. (2008). Rapid telomere movement in meiotic prophase is promoted by NDJ1, MPS3, and CSM4 and is modulated by recombination. Cell 133, 1175–1187. doi: 10.1016/j.cell.2008.04.047
Crisp, M., Liu, Q., Roux, K., Rattner, J. B., Shanahan, C., Burke, B., et al. (2006). Coupling of the nucleus and cytoplasm: role of the LINC complex. J. Cell Biol. 172, 41–53. doi: 10.1083/jcb.200509124
Darby, C. A., Stolzer, M., Ropp, P. J., Barker, D., and Durand, D. (2017). Xenolog classification. Bioinformatics 33, 640–649. doi: 10.1093/bioinformatics/btw686
Ding, X., Xu, R., Yu, J., Xu, T., Zhuang, Y., and Han, M. (2007). SUN1 is required for telomere attachment to nuclear envelope and gametogenesis in mice. Dev. Cell 12, 863–872. doi: 10.1016/j.devcel.2007.03.018
Edgar, R. C. (2004). MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. doi: 10.1186/1471-2105-5-113
El-Gebali, S., Mistry, J., Bateman, A., Eddy, S. R., Luciani, A., Potter, S. C., et al. (2019). The Pfam protein families database in 2019. Nucleic Acids Res. 47, D427–D432. doi: 10.1093/nar/gky995
Evans, D. E., Mermet, S., and Tatout, C. (2020). Advancing knowledge of the plant nuclear periphery and its application for crop science. Nucleus 11, 347–363. doi: 10.1080/19491034.2020.1838697
Field, M. C., Horn, D., Alsford, S., Koreny, L., and Rout, M. P. (2012). Telomeres, tethers and trypanosomes. Nucleus 3, 478–486. doi: 10.4161/nucl.22167
Friederichs, J. M., Gardner, J. M., Smoyer, C. J., Whetstine, C. R., Gogol, M., Slaughter, B. D., et al. (2012). Genetic analysis of Mps3 SUN domain mutants in Saccharomyces cerevisiae reveals an interaction with the SUN-like protein Slp1. G3 (Bethesda) 2, 1703–1718. doi: 10.1534/g3.112.004614
Graumann, K., Runions, J., and Evans, D. E. (2010). Characterization of SUN-domain proteins at the higher plant nuclear envelope. Plant J. 61, 134–144. doi: 10.1111/j.1365-313X.2009.04038.x
Graumann, K., Vanrobays, E., Tutois, S., Probst, A. V., Evans, D. E., and Tatout, C. (2014). Characterization of two distinct subfamilies of SUN-domain proteins in Arabidopsis and their interactions with the novel KASH-domain protein AtTIK. J. Exp. Bot. 65, 6499–6512. doi: 10.1093/jxb/eru368
Groves, N. R., Biel, A., Moser, M., Mendes, T., Amstutz, K., and Meier, I. (2020). Recent advances in understanding the biological roles of the plant nuclear envelope. Nucleus 11, 330–346. doi: 10.1080/19491034.2020.1846836
Hagan, I., and Yanagida, M. (1995). The product of the spindle formation gene sad1+ associates with the fission yeast spindle pole body and is essential for viability. J. Cell Biol. 129, 1033–1047. doi: 10.1083/jcb.129.4.1033
Hofmann, K., and Tmbase, S. W. (1993). TMBASE-a database of membrane spanning protein segments. Biol. Chem. 374:166.
Hu, B., Jin, J., Guo, A. Y., Zhang, H., Luo, J., and Gao, G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. doi: 10.1093/bioinformatics/btu817
Hu, Y., Chen, J., Fang, L., Zhang, Z., Ma, W., Niu, Y., et al. (2019). Gossypium barbadense and Gossypium hirsutum genomes provide insights into the origin and evolution of allotetraploid cotton. Nat. Genet. 51, 739–748. doi: 10.1038/s41588-019-0371-5
Idris, A. M., and Brown, J. K. (2004). Cotton leaf crumple virus is a distinct western hemisphere begomovirus species with complex evolutionary relationships indicative of recombination and reassortment. Phytopathology 94, 1068–1074. doi: 10.1094/phyto.2004.94.10.1068
Jaspersen, S. L., Giddings, T. H. Jr., and Winey, M. (2002). Mps3p is a novel component of the yeast spindle pole body that interacts with the yeast centrin homologue Cdc31p. J. Cell Biol. 159, 945–956. doi: 10.1083/jcb.200208169
Jaspersen, S. L., Martin, A. E., Glazko, G., Giddings, T. H. Jr., Morgan, G., Mushegian, A., et al. (2006). The Sad1-UNC-84 homology domain in Mps3 interacts with Mps2 to connect the spindle pole body with the nuclear envelope. J. Cell Biol. 174, 665–675. doi: 10.1083/jcb.200601062
Jin, S., Zhang, X., Nie, Y., Guo, X., Liang, S., and Zhu, H. (2006). Identification of a novel elite genotype for in vitro culture and genetic transformation of cotton. Biol. Plant. 50, 519–524.
Karlin, S., and Mcgregor, J. (1957). The classification of birth and death processes. Trans. Am. Math. Soc. 86, 366–400.
Krogh, A., Larsson, B., von Heijne, G., and Sonnhammer, E. L. (2001). Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580. doi: 10.1006/jmbi.2000.4315
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Kumar, S., Stecher, G., Suleski, M., and Hedges, S. B. (2017). TimeTree: a resource for timelines, timetrees, and divergence times. Mol. Biol. Evol. 34, 1812–1819. doi: 10.1093/molbev/msx116
Letunic, I., Doerks, T., and Bork, P. (2015). SMART: recent updates, new developments and status in 2015. Nucleic Acids Res. 43, D257–D260. doi: 10.1093/nar/gku949
Li, Y., Calvo, S. E., Gutman, R., Liu, J. S., and Mootha, V. K. (2014). Expansion of biological pathways based on evolutionary inference. Cell 158, 213–225. doi: 10.1016/j.cell.2014.05.034
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Logemann, E., Birkenbihl, R. P., Ülker, B., and Somssich, I. E. (2006). An improved method for preparing Agrobacterium cells that simplifies the Arabidopsis transformation protocol. Plant Methods 2:16. doi: 10.1186/1746-4811-2-16
Lupas, A., Van Dyke, M., and Stock, J. (1991). Predicting coiled coils from protein sequences. Science 252, 1162–1164. doi: 10.1126/science.252.5009.1162
Malone, C. J., Fixsen, W. D., Horvitz, H. R., and Han, M. (1999). UNC-84 localizes to the nuclear envelope and is required for nuclear migration and anchoring during C. Elegans development. Development 126, 3171–3181.
Marchler-Bauer, A., Bo, Y., Han, L., He, J., Lanczycki, C. J., Lu, S., et al. (2017). CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 45, D200–D203. doi: 10.1093/nar/gkw1129
Mikulski, P., Hohenstatt, M. L., Farrona, S., Smaczniak, C., Stahl, Y., Kalyanikrishna, et al. (2019). The chromatin-associated protein PWO1 interacts with plant nuclear lamin-like components to regulate nuclear size. Plant Cell 31, 1141–1154. doi: 10.1105/tpc.18.00663
Murphy, S. P., Gumber, H. K., Mao, Y., and Bass, H. W. (2014). A dynamic meiotic SUN belt includes the zygotene-stage telomere bouquet and is disrupted in chromosome segregation mutants of maize (Zea mays L.). Front. Plant Sci. 5:314. doi: 10.3389/fpls.2014.00314
Murphy, S. P., Simmons, C. R., and Bass, H. W. (2010). Structure and expression of the maize (Zea mays L.) SUN-domain protein gene family: evidence for the existence of two divergent classes of SUN proteins in plants. BMC Plant Biol. 10:269. doi: 10.1186/1471-2229-10-269
Nam, J., Kim, J., Lee, S., An, G., Ma, H., and Nei, M. (2004). Type I MADS-box genes have experienced faster birth-and-death evolution than type II MADS-box genes in angiosperms. Proc. Natl. Acad. Sci. U.S.A. 101, 1910–1915. doi: 10.1073/pnas.0308430100
Nishikawa, S., Terazawa, Y., Nakayama, T., Hirata, A., Makio, T., and Endo, T. (2003). Nep98p is a component of the yeast spindle pole body and essential for nuclear division and fusion. J. Biol. Chem. 278, 9938–9943. doi: 10.1074/jbc.M210934200
Oda, Y., and Fukuda, H. (2011). Dynamics of Arabidopsis SUN proteins during mitosis and their involvement in nuclear shaping. Plant J. 66, 629–641. doi: 10.1111/j.1365-313X.2011.04523.x
Potter, S. C., Luciani, A., Eddy, S. R., Park, Y., Lopez, R., and Finn, R. D. (2018). HMMER web server: 2018 update. Nucleic Acids Res. 46, W200–W204. doi: 10.1093/nar/gky448
Poulet, A., Probst, A. V., Graumann, K., Tatout, C., and Evans, D. (2017). Exploring the evolution of the proteins of the plant nuclear envelope. Nucleus 8, 46–59. doi: 10.1080/19491034.2016.1236166
Shang, Y., Zhu, F., Wang, L., Ouyang, Y. C., Dong, M. Z., Liu, C., et al. (2017). Essential role for SUN5 in anchoring sperm head to the tail. Elife 6:e28199. doi: 10.7554/eLife.28199
Starr, D. A., and Fridolfsson, H. N. (2010). Interactions between nuclei and the cytoskeleton are mediated by SUN-KASH nuclear-envelope bridges. Annu. Rev. Cell Dev. Biol. 26, 421–444. doi: 10.1146/annurev-cellbio-100109-104037
Talamas, J. A., and Hetzer, M. W. (2011). POM121 and Sun1 play a role in early steps of interphase NPC assembly. J. Cell Biol. 194, 27–37. doi: 10.1083/jcb.201012154
Tamura, K., Iwabuchi, K., Fukao, Y., Kondo, M., Okamoto, K., Ueda, H., et al. (2013). Myosin XI-i links the nuclear membrane to the cytoskeleton to control nuclear movement and shape in Arabidopsis. Curr. Biol. 23, 1776–1781. doi: 10.1016/j.cub.2013.07.035
Tatout, C., Evans, D. E., Vanrobays, E., Probst, A. V., and Graumann, K. (2014). The plant LINC complex at the nuclear envelope. Chromosome Res. 22, 241–252. doi: 10.1007/s10577-014-9419-7
Tuttle, J. R., Idris, A. M., Brown, J. K., Haigler, C. H., and Robertson, D. (2008). Geminivirus-mediated gene silencing from Cotton leaf crumple virus is enhanced by low temperature in cotton. Plant Physiol. 148, 41–50. doi: 10.1104/pp.108.123869
Varas, J., Graumann, K., Osman, K., Pradillo, M., Evans, D. E., Santos, J. L., et al. (2015). Absence of SUN1 and SUN2 proteins in Arabidopsis thaliana leads to a delay in meiotic progression and defects in synapsis and recombination. Plant J. 81, 329–346. doi: 10.1111/tpj.12730
Vasnier, C., de Muyt, A., Zhang, L., Tesse, S., Kleckner, N. E., Zickler, D., et al. (2014). Absence of SUN-domain protein Slp1 blocks karyogamy and switches meiotic recombination and synapsis from homologs to sister chromatids. Proc. Natl. Acad. Sci. U.S.A. 111, E4015–E4023. doi: 10.1073/pnas.1415758111
Wang, W., Zhang, X., and Niittylä, T. (2019). OPENER is a nuclear envelope and mitochondria localized protein required for cell cycle progression in Arabidopsis. Plant Cell 31, 1446–1465. doi: 10.1105/tpc.19.00033
Zhang, F., Ma, L., Zhang, C., Du, G., Shen, Y., Tang, D., et al. (2020). The SUN domain proteins OsSUN1 and OsSUN2 play critical but partially redundant roles in meiosis. Plant Physiol. 183, 1517–1530. doi: 10.1104/pp.20.00140
Zhou, X., and Meier, I. (2014). Efficient plant male fertility depends on vegetative nuclear movement mediated by two families of plant outer nuclear membrane proteins. Proc. Natl. Acad. Sci. U.S.A. 111, 11900–11905. doi: 10.1073/pnas.1323104111
Zhou, X., Graumann, K., Evans, D. E., and Meier, I. (2012). Novel plant SUN-KASH bridges are involved in RanGAP anchoring and nuclear shape determination. J. Cell Biol. 196, 203–211. doi: 10.1083/jcb.201108098
Zhou, X., Graumann, K., Wirthmueller, L., Jones, J. D., and Meier, I. (2014). Identification of unique SUN-interacting nuclear envelope proteins with diverse functions in plants. J. Cell Biol. 205, 677–692. doi: 10.1083/jcb.201401138
Keywords: SUN proteins, evolution, divergence, cotton, reproductive development
Citation: Yuan L, Pan J, Zhu S, Li Y, Yao J, Li Q, Fang S, Liu C, Wang X, Li B, Chen W and Zhang Y (2021) Evolution and Functional Divergence of SUN Genes in Plants. Front. Plant Sci. 12:646622. doi: 10.3389/fpls.2021.646622
Received: 27 December 2020; Accepted: 18 February 2021;
Published: 08 March 2021.
Edited by:
Jeremy Coate, Reed College, United StatesReviewed by:
David Edgar Evans, Oxford Brookes University, United KingdomCopyright © 2021 Yuan, Pan, Zhu, Li, Yao, Li, Fang, Liu, Wang, Li, Chen and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Chen, MTUwOTM5MDY1NDdAMTYzLmNvbQ==; Yongshan Zhang, MTM5Mzg2OTgyOTlAMTYzLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.