- 1State Key Laboratory of Pharmaceutical Biotechnology, NJU Advanced Institute for Life Sciences (NAILS), School of Life Sciences, Nanjing University, Nanjing, China
- 2Institute for Advanced Studies, Wuhan University, Wuhan, China
- 3Research Center for Proteome Analysis, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, Shanghai, China
- 4State Key Laboratory of Soil and Sustainable Agriculture, Institute of Soil Science, Chinese Academy of Sciences (CAS), Nanjing, China
Many pieces of evidence show that the adaptive response of plants to salt stress requires the maturation of N-glycan on associated proteins. However, it is still little known about the salt-responsive glycoproteins that function in this process. In the present study, we identified salt-responsive glycoproteins in wild-type (WT) Arabidopsis and two mutants defective in N-glycan maturation, mns1 mns2 and cgl1. A total of 97 proteins with abundance changes of >1.5‐ or <0.67-fold were identified against salt stress by label-free liquid chromatography coupled mass spectrometry (LC-MS/MS) quantitative analyses. A comparison of differentially abundant glycoproteins (DAGs) indicated the substrate preferences regulated by MNS1/MNS2 and CGL1. In addition, the DAGs in mns1 mns2 hardly form functional regulatory networks in STRING analysis. Comparably, the regulatory network in cgl1 was visible and shared overlapping with that in WT. Such difference may supply the evidence to partially explain the lower salt sensitivity of mutant cgl1 than mns1 mns2. We further confirmed that two N-glycosylation clients, peroxidases PRX32 and PRX34, were involved in the salt stress response since the double mutants showed enhanced salt sensitivity. Together, our study provided proteomic evidence that N-glycans are crucial for modulating stress-responsive protein levels, and several novel glycoproteins responsible for salt stress tolerance in Arabidopsis were listed. Data are available via ProteomeXchange with identifier PXD006893.
Introduction
As sessile organisms, land plants often suffer from various stresses and adverse environments, such as extreme temperatures, drought, and high salinity. In agriculture, high salinity affects the geographical distribution of crops in nature and causes a large amount of yield loss (Qin et al., 2011; Zhu, 2016). Extensive studies have revealed that a variety of genes and/or pathways play synergistic roles in abiotic stress response in plants, and it is difficult to significantly improve plant resistance to external stress by modulating a single effective gene or protein.
Protein asparagine (Asn or N)-linked glycosylation is one of the most conserved co‐ and post-translational modification (PTM) in eukaryotic cells (Khoury et al., 2011; Moremen et al., 2012). After the tetradecyl glycan precursor, GlcNAc2Man9Glc3 (Glc for glucose, Man for mannose, and GlcNAc for N-acetylglucosamine) is transferred to selected Asn residue in the Asn-X-Ser/Thr motif (X can be any amino acid residue except Pro) on nascent polypeptide chains (Pless and Lennarz, 1977; Yan and Lennarz, 2002) by oligosaccharyltransferase complex (OST; Kelleher and Gilmore, 2006), the N-linked oligomannosidic glycans are further trimmed and modified in Golgi apparatus to synthesize mature N-glycans with the secretion of associated polypeptides or proteins (Lerouge et al., 1998; Nagashima et al., 2018). In Arabidopsis, the initial N-glycan processing events in the Golgi are catalyzed by two functionally redundant class I α-mannosidases (MNS1 and MNS2), which cleave three α-1,2-mannosyl residues to generate the substrate for CGL1/GnT1 (Liebminger et al., 2009; Kajiura et al., 2010; Supplementary Figure S1A). The GnT1/CGL1 catalyzes a GlcNAc addition required to remove two additional Man residues to add another GlcNAc, xylose, and fucose residues to form a complex N-glycan structure (Vonschaewen et al., 1993; Strasser et al., 1999).
In plants, N-glycosylation is involved in many biological processes, including ER-quality control of steroid hormone receptor (Jin et al., 2007; Hong et al., 2008), gametophyte recognition (Lindner et al., 2015; Muller et al., 2016), subcellular transport (Rips et al., 2014; Shen et al., 2014), plant innate immunity (Li et al., 2009; Nekrasov et al., 2009; Haweker et al., 2010; Xia et al., 2020), and stomatal development (Jiao et al., 2020) by regulating the stability, function, or sub-localization of the substrate protein. Moreover, several studies also establish the association of N-glycosylation with salt tolerance in Arabidopsis (Koiwa et al., 2003; Kang et al., 2008; Nagashima et al., 2018). Of note, the failure of complex N-glycan biosynthesis leads to the salt sensitivity of Arabidopsis seedlings (Frank et al., 2008; Kang et al., 2008; Strasser, 2016; Liu et al., 2018). Intriguingly, when focusing on two Arabidopsis mutants, mns1 mns2 (disrupted in Golgi MNSI) and cgl1-3 (defected in GnTI/CGL1), in both of which glycoproteins harbor high mannose type N-glycans, we find that two mutants show a different degree of salt sensitivity (Liu et al., 2018). Although an increasing number of N-glycosylation-related mutants have been identified and show changes in response to salt stress (Strasser, 2016), yet it is still less known on the underlying molecular mechanism, mainly due to the lack of understanding of salt stress-responsive glycoproteins. Large-scale screening and identification of N-glycosylated sites and N-glycopeptides are now available (Zielinska et al., 2010; Stadlmann et al., 2017). A quantitative N-glycoproteomic pipeline has been established, and N-glycosylation signatures of different cell populations, including cancer cells, were recently reported for better stratification of cancer patients (Boyaval et al., 2020; Fang et al., 2020). N-glycoproteomes of insects are attracting scientists for pest control (Scheys et al., 2018). Inflorescence tissue-specific N-glycopeptides (Xu et al., 2016) and cold stress-regulated glycoproteins (Ma et al., 2016) are also resolved by mass spectrum (MS).
To explore the molecular mechanism of protein N-glycosylation regulating the salt stress response, we performed a label-free proteomic analysis utilizing nanoflow liquid chromatography coupled MS (nanoLC-MS/MS) and comparatively analyzed the proteome profiles between wild-type (WT) and two mutants against salt stress. We also confirmed that two differentially abundant peroxidases, PRX32 and PRX34, were N-glycosylation substrates and function redundantly in salt tolerance by root growth assay and hydrogen peroxide (H2O2) measurement.
Materials and Methods
Plant Materials and Growth Conditions
The Arabidopsis thaliana WT was the Col-0 ecotype. The mutant lines mns1 (SALK_076002), mns2 (SALK_023251), prx32 (SALK_072340), prx34 (SALK_051769), and cgl1-3 (CS16367) mutant were purchased from ABRC (Arabidopsis Biological Resource Center at The Ohio State University). Double mutant mns1 mns2 was described previously (Liu et al., 2018), and prx32 prx34 was generated by cross prx32 with prx34 single mutant. Seed sterilization and plant growth conditions were described previously (Li et al., 2001).
Salt Treatment
The Arabidopsis seeds were surface-sterilized and sown on half-strength Murashige and Skoog (MS) solid medium [2.2 g dm−3 MS salts (Duchefa, BH Haarlem, The Netherlands), 10 g dm−3 sucrose, and 3 g dm−3 Gelrite (Duchefa, BH Haarlem, The Netherlands), pH 5.7–5.9]. After cold treatment at 4°C for 2 days, the plates were placed in a 22°C growth chamber with 16-h light/8-h dark photoperiod for 12 days, and then cultured in 1/2 MS liquid medium or 1/2 MS medium containing 200 mM NaCl for 6 h, respectively. For MS analysis, the seedlings were collected and weighed to ensure that each sample was more than 2 g.
Protein Extraction, Endo H Digestion, and Immunoblot Analysis
The plant materials including Arabidopsis seedlings and tobacco leaves were ground into fine powder in liquid N2 and added same volume of SDS loading buffer [125 mM Tris-HCl (pH 6.8), 4% (w/v) SDS, 20% (v/v) glycerol, 100 mM DTT, and 0.002% (w/v) bromophenol blue]. The extracts were thoroughly mixed by vortex and maintained on ice for 10 min. After heated at 95°C for 5 min, the mixture was centrifuged for 5 min at 16,000 g. The supernatant was treated with or without Endo Hf (New England Biolabs) treatment for 1.5 h at 37°C. Samples were then separated by 10 or 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). The total proteins from the supernatants were separated by 10% SDS-PAGE. The immunoblot analyses were performed with primary antibodies, including anti-BRI1 (Mora-Garcia et al., 2004), anti-RSW2 (Liu et al., 2018), and HRP-conjugated goat anti-rabbit IgG secondary antibody (Promega). For PRX32-HA or PRX34-HA detection, immunoblot analyses were performed with peroxidase-conjugated anti-HA antibodies (Sigma). The protein signals were detected with enhanced chemiluminescence Immobilon Western HRP Substrate (Millipore) or SuperSignal West Pico PLUS substrate kit (Thermo) by a CCD imager (Tanon).
Protein Preparation for MS Analysis
Total proteins were extracted with the modified trichloroethanoic acid (TCA) method described previously (Isaacson et al., 2006). Briefly, plant materials were ground into fine powder in liquid nitrogen and then lysed in 40 ml of cold TCA/acetone (v/v = 1/9, containing 65 mM DTT) solution, incubated at −20°C overnight. After centrifugation at 4°C at 10,000 g for 45 min, the supernatant was discarded, and the pellet was resuspended in 25 ml cold acetone. After centrifuged at 10,000 g for 45 min at 4°C again, the pellet was washed twice with cold acetone and dried in air. A total of 400 μl of UA buffer (8 M Urea, 150 mM Tris-HCl, pH 8.0) was used to dissolve the pellet before sonication. Finally, the supernatant was collected by centrifugation at 14,000 g for 30 min. Protein concentrations were determined by Bradford assay using bovine serum albumin as a standard (Bradford, 1976). The quality of the protein preparation was further evaluated by SDS-PAGE and visualized by Coomassie Brilliant Blue R-250 staining.
Protein Digestion and LC-MS/MS Analysis
An aliquot of 400 μg of protein was digested according to the filter aided sample preparation (FASP) procedure (Zielinska et al., 2010). After adding 30 μl of SDT buffer (4% SDS, 100 mM Tris-HCl, 100 mM DTT, pH 7.6), the clear supernatants obtained from the above procedure were boiled for 10 min to 30 kDa ultrafiltration units (Wisniewski et al., 2011). Filters were rinsed three times with 200 μl of UA buffer (8 M Urea, 150 mM Tris-HCl, pH 8.0) and then incubated in UA buffer with the addition of 100 μl 50 mM iodoacetamide for 30 min in the dark. Subsequently, the filters were washed three times with 100 μl UA buffer and 100 μl 40 mM ammonium bicarbonate, respectively. Next, the proteins were digested with 8 μg trypsin at 37°C. After 16 h incubation, peptides were eluted and transferred to a new 30 kDa filtration unit. A total of 50 μl lectin mixture containing ConA (Sigma, United States), WGA (Sigma, United States), and RCA120 (Sigma, United States; 125 μg each) were added to the top of the filters. After incubation for 1 h, 25 mM NH4HCO3 in H218O was used to wash unspecific peptides (Cambridge Isotope Laboratories, United States) twice. Deglycosylation with PNGase F (Roche, Switzerland) was performed in H218O at 37°C for 3 h. Within H218O, the deamidation conversion from Asn to Asp has a 2.9890 Da increase in molecular weight, which is the signature that distinguishes N-glycosylated peptides from O-glycosylated ones and those peptides that are not modified endogenously but survive the enrich anyway.
The deglycosylated peptides were desalted and isolated using nanoliter flow rate capillary high-performance liquid chromatography (HPLC) Easy-nLC. The following gradient was used for the mobile phases [A, 0.1% formic acid acetonitrile aqueous solution (2% acetonitrile); B, 0.1% formic acid acetonitrile aqueous solution (84% acetonitrile)]: 0–45% of B over 0–100 min; 45–100% of B over 100–108 min; and 100% of B over 108–120 min. Purified peptide mixtures were analyzed by mass spectrometry using a Q-Exactive mass spectrometer (Thermo Fisher Scientific, United States). Precursor MS scans were acquired with a resolution of 70,000 at m/z 200. Ten most intense ions were fragmented by higher-energy collisional dissociation (HCD) in the quadrupole collision cell. The HCD fragment ion spectra were acquired in the orbitrap with a resolution of 17,500 at m/z 200. The following conditions were used: automatic gain control (AGC) target: 3e6, maximum ion accumulation times of 20 ms for full scans, and 60 ms for HCD.
Data Analysis
MS RAW data were searched against the TAIR10 protein database (Lamesch et al., 2012) in MaxQuant_1.3.0.5. The search followed an enzymatic cleavage rule of Trypsin/P and allowed a maximum of two missed cleavage sites and a peptide mass tolerance of 10 ppm. Carbamidomethylation of cysteine was defined as fixed modification, whereas protein N-terminal deamidation and methionine oxidation were defined as variable modifications for the database searches. The site false discovery rate (FDR) for peptide ≤0.01 (Zielinska et al., 2012). Label-free quantification was carried out using the MaxQuant software as described previously, and the intensity-based absolute quantification (iBAQ) of the identified peptides was selected to quantify protein abundance (Cox and Mann, 2008).
Experimental Design and Statistical Rationale
For label-free quantitative analyses, all six biological samples were analyzed with three replicates. N-glycopeptides identified in two or more independent analyses were accepted to quantify the abundance of the N-glycopeptide. The abundance changes of unique peptides were set to represent the corresponding glycoprotein profile in response to salt stress. In the case of more than one unique peptide were detected within a glycoprotein, the abundance was presented as an average value.
Bioinformatic Analysis
Candidate genes were classified with the online servers Agrigo (Du et al., 2010).1 Protein pathway analysis was performed using KEGG.2 The protein–protein interaction was analyzed with STRING.3
Root Growth Assay
Experiment procedures were as described previously (Kang et al., 2008). Briefly, surface-sterilized seeds were sown on cellophane membrane and placed on MS agar medium (4.4 g dm−3 MS salts, 30 g dm−3 sucrose, and 16 g dm−3 agar, pH 5.7–5.9). After cold treatment at 4°C for 2 days, the plates were incubated at 22°C for 3 days, and then seedlings together with the cellophane membrane were transferred to MS medium or MS medium supplemented with 160 mM NaCl for another 9 days. The seedlings were photographed with a Canon G12 digital camera, and root lengths were measured by ImageJ software.4
Reverse Transcription-PCR and Quantitative Real-Time PCR
Two-week-old WT, prx32, and prx34 mutant seedlings were collected for genotyping by reverse transcription-PCR (RT-PCR). Salt-treated samples were harvested for PRX32 and PRX34 gene expression analysis by quantitative real-time PCR (qPCR). Plant materials were extracted by Trizol (Invitrogen, United States), 1 μg total RNA from each sample was used to synthesize cDNA by PrimeScript 1st Strand cDNA Synthesis kit (Takara, Japan). qPCR was performed in triplicate using the LightCycler® 96 System (Roche Life Science) with SYBR green mix (TaKaRa). Gene expression of Actin2 was selected as an internal reference. The primers used for transcript amplification were listed as follows, ACT2-F (5'-GCCATCCAAGCTGTTCTCTC-3') and ACT2-R (5'-GCTCGTAGTCAACAGCAACAA-3') for Actin2, PRX32-F1 (5'-CTTCACTCTCCCACAACT-3') and PRX32-R1 (5'-TCGACCACATCATGTAGCA-3') for PRX32, and PRX34-F2 (5'-GTTGGTCTCGATCGTCCT-3') and PRX34-R2 (5'-ATGGAGCAGAGAGTTGGA-3') for PRX34.
Transient Expression of PRX32 and PRX34 Proteins and Tunicamycin Treatment
To generate PRX32 and PRX34 protein expression cassettes, the full-length coding sequences of PRX32 and PRX34 with 1× HA tag at C-terminus were obtained by PCR using cDNA from WT plant as a template with following primers, PRX32-F (5'-TCCCCCGGGCTGCAGGAATTCATGAATTTCTCTTATTCTTCCTTGT-3') and PRX32-R (5'-GATAAGCTTGATATCGAATTCAGCGTAATCTGGAACATCGTATGGGTACATAGAGCTGACAAAGTCAACGA-3'), PRX34-F (5'-TCCCCCGGGCTGCAGGAATTCATGCATTTCTCTTCGTCTTCAACA-3') and PRX34-R(5'-GATAAGCTTGATATCGAATTCAGCGTAATCTGGAACATCGTATGGGTACATAGAGCTAACAAAGTCAACGA-3'). The PCR products were cloned into binary vector pQG110 via Gibson Assembly (NEB). The resulted constructs were transformed to Agrobacterium tumefaciens GV3101. The bacteria were cultured, collected, and suspended in IFB buffer [0.5% (w/v) Glucose, 10 mM MgCl2, 10 mM MES, and 150 mM acetosyringone, pH 5.7] at 0.5 OD600. The bacterial mixture was then infiltrated into tobacco Nicotiana benthamiana leaves using a syringe. The injected plants were cultured for 30–48 h before harvesting for protein extraction. For tunicamycin treatment, 5 μM tunicamycin (Sigma) solution was injected into the same infiltration areas 12 h before harvest.
H2O2 Detection by DAB Staining
3,3'-Diaminobenzidine (DAB) staining was conducted as described previously (Daudi and O’Brien, 2012) with modifications. In brief, 2-week-old seedlings were cultured in 1/2 MS liquid medium containing 0 or 200 mM NaCl for 6 h, respectively. Seedlings were then washed with ddH2O three times before adding 5 ml of fresh 0.1% (w/v) DAB in 10 mM Na2HPO4 solution. After gently vacuum for 5 min to infiltrate the leaves, samples were incubated on a laboratory shaker for 4–5 h at 80–100 rpm in the dark. The chlorophyll was carefully bleached by boiling for 15 min before recording the staining results. H2O2 contents were quantified by grayscale analysis in ImageJ software.
Results
Identification of Glycosylated Peptides and Proteins Using LC-MS/MS
Two mutants mns1 mns2 and cgl1-3, together with the WT, were employed for comparative analyses. In both mns1 mns2 and cgl1-3 mutants, complex N-glycan modification was blocked. Therefore, glycoproteins were decorated with oligomannosidic N-glycans. This feature facilitates the enrichment of glycopeptides by lectins more efficiently (Zielinska et al., 2010; Song et al., 2011). The workflow for label-free quantitative N-glycoproteomic analysis and the principle for N-glycosites identification was shown (Figures 1A,B). The NaCl concentration and treatment duration were optimized by phenotype observation and protein abundance measurements (Figure 1C). Eventually, 200 mM NaCl for 6 h was used as the salt treatment condition. We also detected the band shifts of two glycoproteins BRI1 and RSW2 against Endo H treatment to assess if the salt treatment would block the N-glycan structure under such conditions. The results showed that the digested band patterns of two proteins did not change either in the WT or in the mutant background before and after salt treatment (Figure 1D), indicating that salt treatment had no significant effect on the structure of N-glycan, thus excluding the possible effect of N-glycan structural changes on protein enrichment in our study. After proteins were extracted, denatured, and digested with trypsin, the N-glycopeptides were enriched using FASP-based multiple lectins approach (Zielinska et al., 2010). To capture all three classes of high-mannose, hybrid, and complex types of N-glycosylated peptides (Supplementary Figure S1A), multi-lectin enrichment was employed. Multiple lectins were composed of concanavalin A (ConA), wheat germ agglutinin (WGA), and castor lectin RCA120, which bound mannose, N-acetylglucosamine, and galactose residues, respectively, on N-glycans (Supplementary Figure S1B). We first examined LC-MS/MS performance and the quality of the dataset. Pearson correlation analysis showed that the intensities of N-glycopeptides in three replicates for each Arabidopsis line, either with or without salt treatment, were highly correlated (r = 0.646–0.931; Supplementary Figure S2). We also assessed the number of identified N-glycopeptides and N-glycoproteins and found that more than half of N-glycopeptides were identified from three (40.45–57.27%) or two (57.30–80.77%) independent experiments (Supplementary Figure S3). These results showed that the N-glycoproteomic data were suitable for further analysis.
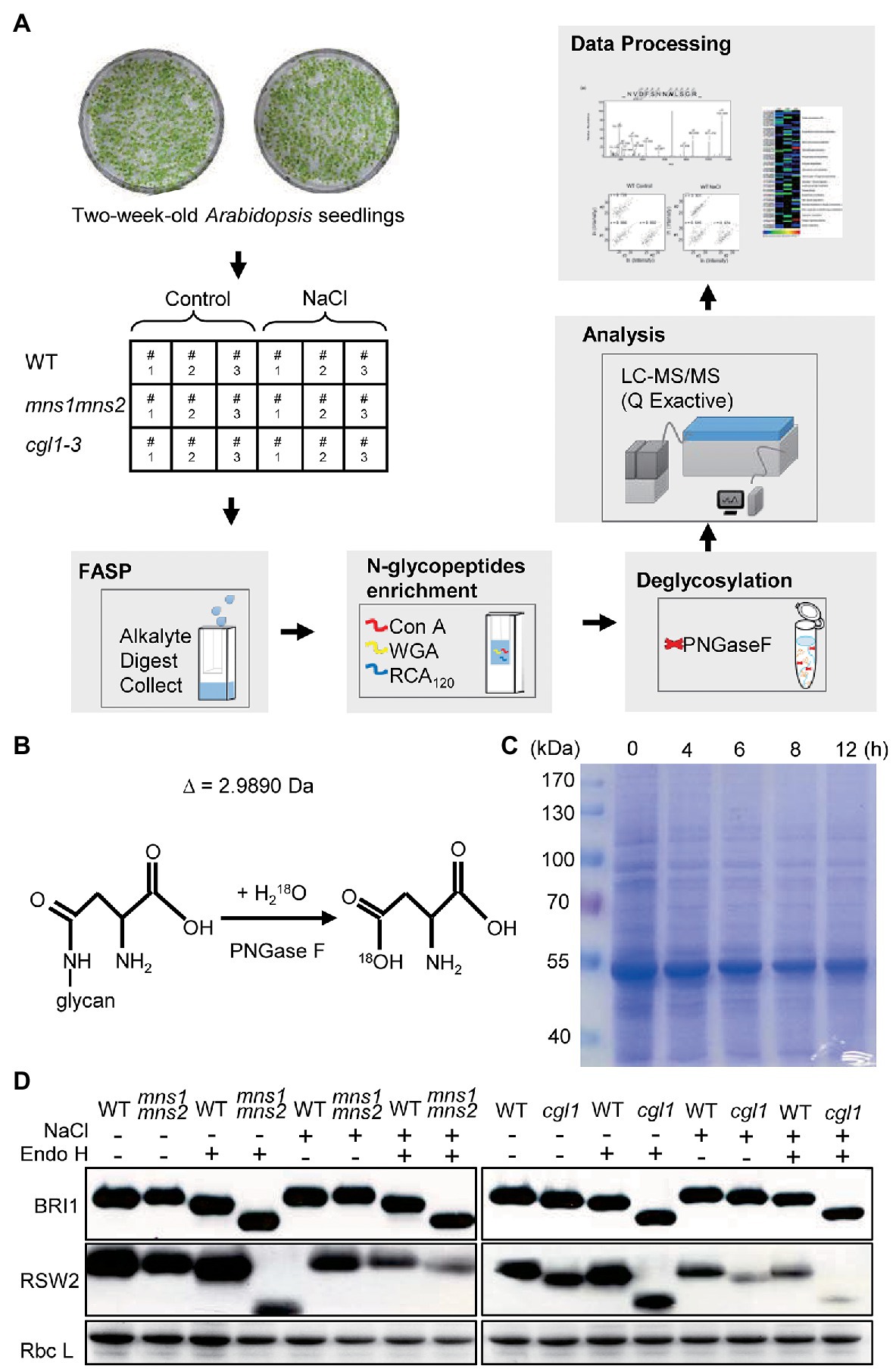
Figure 1. Schematic workflow of the experimental procedure. (A) A summary of the experimental design. Two-week-old seedlings were treated without (control) or 200 mM NaCl treatment (NaCl) for 6 h. Then, the total protein was extracted and digested into peptides using the filter aided sample preparation (FASP) method. N-glycopeptides were enriched by multiple mixed lectins and resolved by liquid chromatography coupled mass spectrometry (LC-MS/MS) after PNGase F digestion. The raw MS files were processed by MaxQuant. Three biological repeats were conducted. (B) Illustration of de-glycosylation of N-glycopeptides in H218O by PNGase F. In the presence of H218O, the conversion from Asn to Asp had a 2.9890 Da increase in molecular weight. (C) Detection of total protein abundance after salt treatment for different lengths of time. Two-week-old wild-type (WT) seedlings were treated in 200 mM NaCl for 0, 4, 6, 8, or 12 h. Total proteins were extracted and separated by 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis. Gels were stained by Coomassie Brilliant Blue R-250 (CBB). (D) N-glycan structure analyses by Endo H digestion. Total proteins were extracted from control and salt-treated seedlings. After Endo H digestion, the proteins were separated on 10% SDS-PAGE and used anti-BRI1 and anti-RSW2 antibodies to analyze protein-associated N-glycan. CBB staining was shown as a loading control.
A total of 727 glycosylation sites from 632 glycosylated polypeptides were identified, corresponding to 371 glycoproteins. Most N-glycosylation sites exhibited canonical N-X-S/T motif sequences (Figures 2A,B). The number of identified N-glycoproteins varied among samples. In control groups, 311 glycoproteins were identified in mns1 mns2, and 300 glycoproteins were identified in the cgl1-3 mutant, while only 203 were identified in the WT (Figure 2C). Different glycoproteins would be enriched despite the use of saturated multiple lectins, e.g., ConA bound more proteins from mns1 mns2 and cgl1 than the WT. Similarly, PNGase F was less effective on N-glycans from the WT. Therefore, in the WT, the reduced N-glycoproteins might be because the glycoproteins carrying complex N-glycans were less enriched than those with high mannose type N-glycans in the mutants. However, in the presence of high salinity, the number of glycoproteins detected in the WT slightly increased (from 203 to 244) but decreased in both mns1 mns2 (from 311 to 297) and cgl1-3 (from 300 to 263; Figure 2C). One possibility is that the abundance of glycoproteins in response to salt stress was modulated by the attached N-glycan structure and maturation failure of N-glycans destabilized the associated glycoprotein under salt stress, which corresponded to previous studies in which the maturation of N-glycans was essential for plants to tolerate salt stress.
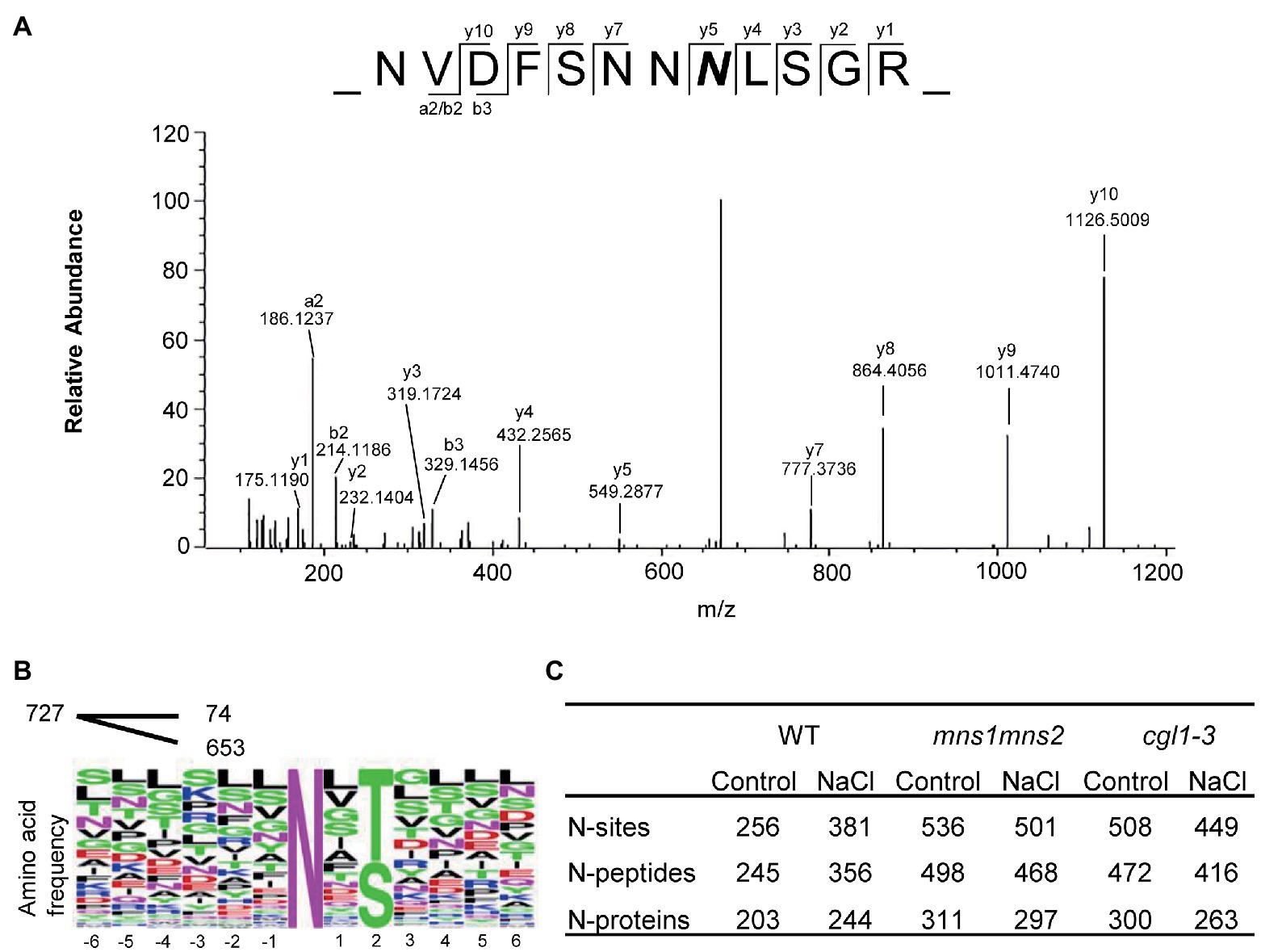
Figure 2. An overview of identified N-glycosylated motifs, peptides, and proteins. (A) Representative MS/MS spectrum of the peptide NVDFSNNNLSGR of the EF-Tu receptor (EFR). The 2.9890 Da mass increment of deamidation is indicated by a bold italic N. (B) Consensus sequence of identified N-glycosylation sites. Multiple glycosylated sequence motifs were analyzed, and relative frequency plots are shown. Of the total 727 sequence motifs, 74 are noncanonical motifs with an occurrence of <20 each. (C) Statistics of identified N-glycosites, N-glycopeptides, and N-glycoproteins by three replicates in six sample groups.
Characterization of Identified Glycoproteomes by GO Annotation
The function of identified N-glycoproteins was analyzed by gene ontology (GO) annotation from the following aspects: the participation of biological processes (BPs), molecular function (MF), and cell components (CCs; Ashburner et al., 2000). In general, the identified N-glycoproteins were involved in various BPs, including protein and carbohydrate metabolic processes, anatomical structure morphogenesis, photosynthesis, cellular development process, regulation of cell size, and stress response. For CC, the subcellular locations of identified proteins were in line with the expectation; the proteins were mainly secretory or membrane proteins and were distributed in the extracellular and endomembrane, including the endoplasmic reticulum (ER), Golgi apparatus, and vacuole, most of which undergo N-glycosylation. MF analysis showed that the identified N-glycosylated proteins were concentrated in catalytic, hydrolase, transferase, and kinase activities (Supplementary Figure S4). We further compared the glycoproteome profiles of the WT, mns1 mns2, and cgl1-3 between the salt treatment and corresponding control group. The results showed no significant difference in enriched GO items under salt stress (Supplementary Table S1). However, we found that more glycoproteins were detected in mns1 mns2 and cgl1-3 mutants than in the WT, which were mainly enriched in kinase and transferase activities (Supplementary Table S1), inferring that some kinases and transferases are significantly affected by the associated N-glycan structures under salt stress.
Global Changes of Protein Abundance in Response to Salt Stress
To further evaluate the effects of N-glycans on salt stress-responsive glycoproteins, we quantitatively compared the alterations of the N-glycoproteome. We set a threshold of protein abundance change to 1.5-fold, meaning that a protein with an abundance ≤0.67‐ or ≥1.5-fold after salt treatment was regarded as a salt responsive protein. In the WT, 37 of 141 proteins were quantified as salt responsive, of which 32 proteins increased by more than 1.5-fold and five proteins decreased to less than 0.67-fold. In mutants, the abundance of 45 of 236 glycoproteins in mns1 mns2 changed, of which 33 proteins accumulated and 12 proteins were decreased. Similarly, the abundance of 42 of 207 glycoproteins was changed in cgl1-3, of which 13 members were increased, and most members were decreased (Table 1). These analyses showed that the abundance of most salt-responsive proteins was increased in the WT compared to the mutants, particularly in cgl1-3, in which most glycoproteins were decreased. We also made a qualitative comparison between control and NaCl-treated samples. We defined a peptide/protein with quantitative values as presence, and otherwise, it was regarded as an absence. Pairwise analyses showed that three proteins (Glycosyl hydrolase family protein, BGLU34, EBP1) were exclusively detected in the WT control group, while eight proteins of the 11 exhibiting catalytic activity were only detected in the salt-treated WT. Similarly, eight proteins were only detected in the mns1 mns2 control group, and nine unique proteins, including BGLU24, FUC1, LYM1, IAR3, VSP2, sks3, PAP17, and two unknown proteins, were detected in salt-treated mns1 mns2; meanwhile, 21 proteins only presented in the cgl1-3 control group and, most of which were involved in the carbohydrate metabolic process (Supplementary Table S2). Eight proteins were affected in the same way in both mutants: the abundance of HBP1, GLP10, FLA9, and PRX34 increased, while STT3A, XXT5, AT1G59970, and RHS19 decreased in both cgl1-3 and mns1 mns2. In addition, three proteins, BGLU44, SCPL46, and PAP2, increased in mns1 mns2 but decreased in cgl1-3. NAI2 was decreased in mns1 mns2 but increased in cgl1-3 (Table 2). In addition, there were more differentially abundant glycoproteins (DAGs) exclusively detected in mns1 mns2 or cgl1-3 (Table 2). In conclusion, although the variation trends of DAGs in mns1 mns2 and cgl1-3 were similar, they were also different, which may provide clues to explain the different salt responses between the two mutants.
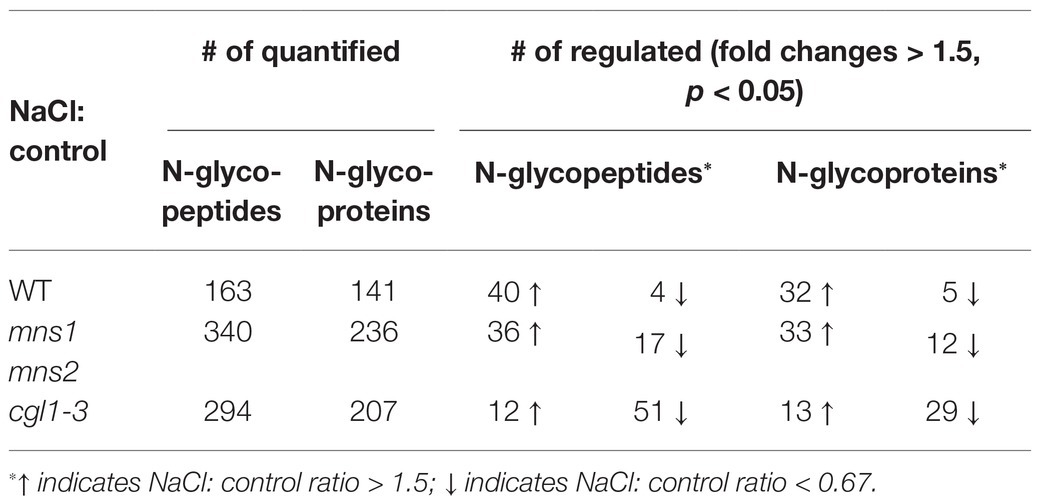
Table 1. Statistical analysis of differentially abundant N-glycopeptides and N-glycoproteins in response to salt stress.
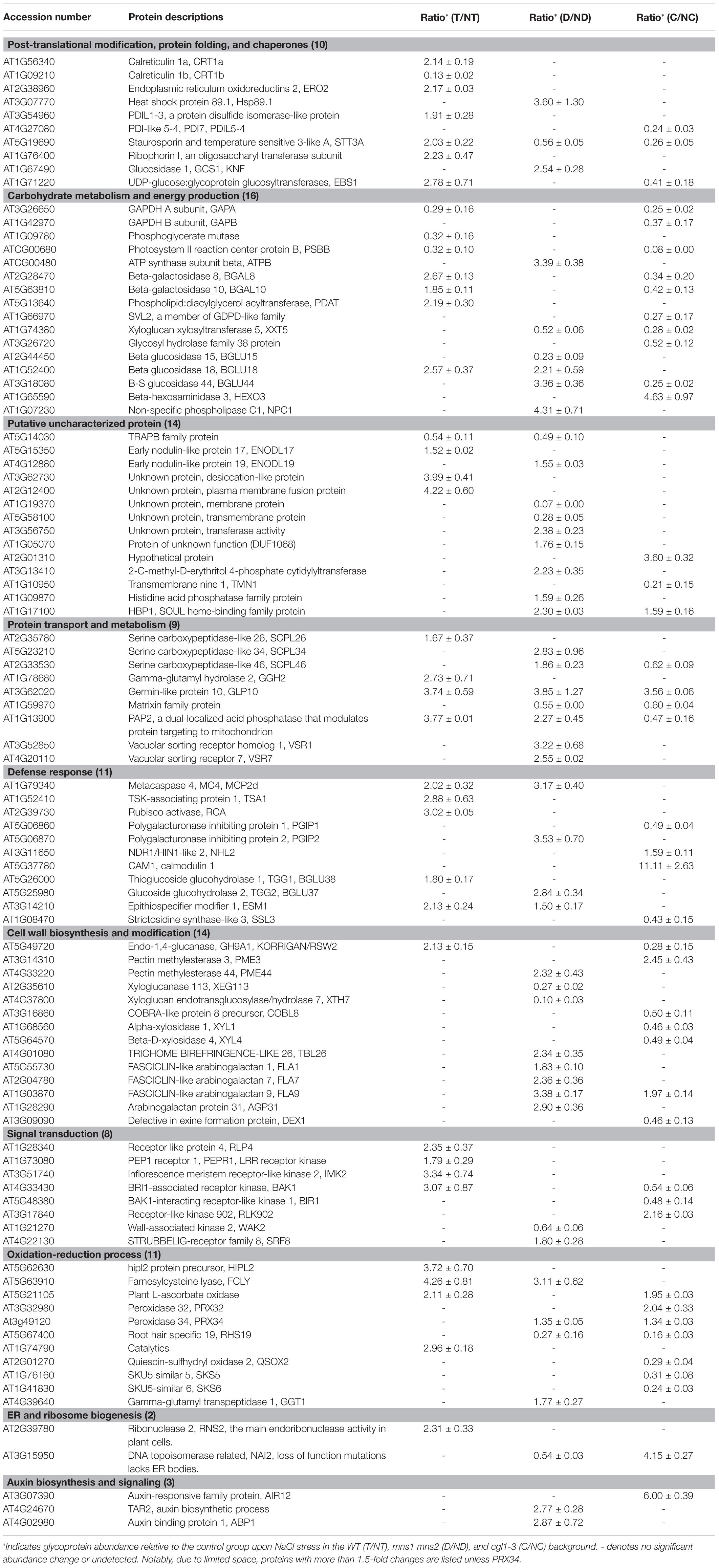
Table 2. Differentially abundant glycoproteins were identified in the WT, mns1 mns2, and cgl1-3 in response to salt stress.
Salt Responsive Glycoproteins Are Differentially Abundant in mns1 mns2 and cgl1 Mutants
The integral GO analysis of DAGs demonstrated that biological processes containing carbohydrate metabolic process, response to stress, and cellular component organization were modulated for salt tolerance (Supplementary Table S3). We further analyzed the biological functions of DAGs under salt stress in GO annotation and KEGG metabolic pathways. Consistent with the aforementioned results (Figure 2C), more decreased glycoproteins were observed in the two mutants, especially in cgl1-3 (143 vs. 67 in mns1 mns2; Figures 3A–C; Table 1). Notably, hydrolase activity was enriched in both mns1 mns2 and cgl1-3 mutants compared with the WT (Figures 3D–F). Next, we aligned the DAGs from the WT, mns1 mns2, and cgl1-3 with p < 0.05, and the abundance changes of the shared proteins were analyzed individually (Table 2). Under salt stress, the abundance of STT3A (OST Subunit), purple acid phosphatase 2 (PAP2), and germin-like protein 10 (GLP10) in the WT and two mutants changed, which are involved in the glycan chain transfer in the ER, root growth, and carbon metabolism, respectively (Figure 3D). STT3A increased in the WT but decreased in mns1 mns2 and cgl1-3; PAP2 increased in the WT and mns1mns2 but decreased in cgl1-3, and GLP10 increased in all three samples (Table 2). The commonly changed DAGs between the WT and cgl1-3 were stress response proteins (EBS1, GAPA, and KORRIGAN/RSW2) and active catalytic proteins (EBS1, BGAL8, PSBB, GAPA, KORRIGAN/RSW2, and BGAL10). Comparably, those DAGs related to the glycosyl compound catabolic process (BGLU18, FCLY, and ESM1) were changed in both WT and mns1 mns2. The DAGs shared by mns1 mns2 and cgl1-3 were mostly located in the membrane and cell wall (XXT5, HBP1, FLA9, BGLU44, and SCPL46; Table 2; Figure 3D). The further molecular functional analysis found that hydrolase activity was enriched in mns1 mns2 and cgl1-3 compared with the WT (Figures 3E,G), and most of them participated in cell wall biosynthesis and modification (Table 2), indicating that cell wall integrity and/or composition are crucial for salt tolerance. In addition, the DAGs in mutants had a broader distribution, including the Golgi apparatus, endosome, ER, vacuole, cell membrane, and extracellular regions. In the WT, the DAGs were most prevalent in the cytoplasm and endomembrane systems. The biological process of proteins identified in the WT was significantly enriched in response to stress and organic substances, while most DAGs from the mutants were related to the carbohydrate metabolic process (Figure 3G). These results suggested that salt-responsive glycoproteins in mns1 mns2 and cgl1-3 mutants cannot respond to stress correctly, which may be due to the lack of correct N-glycan modification, resulting in their abnormal subcellular localization.
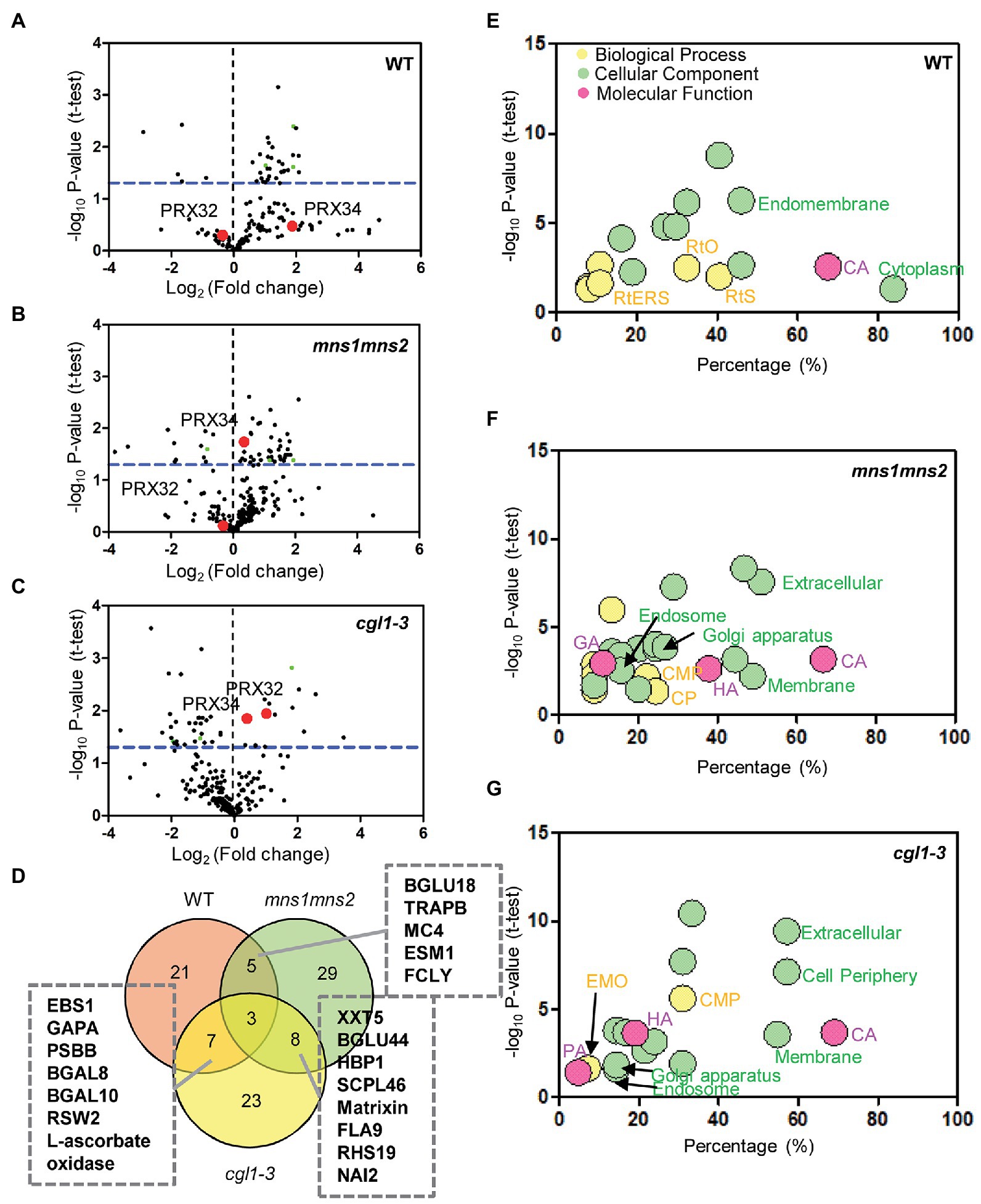
Figure 3. Gene ontology (GO) annotations of differentially abundant N-glycoproteins under salt stress. (A–C) Identification of differentially abundant N-glycoproteins in the WT (A), mns1 mns2 (B), and cgl1-3 (C) under salt stress. Each dot represents one protein. The blue horizontal bar represents the threshold value of p (≤0.05). The red dots represent PRX32 and PRX34. (D) Venn diagram demonstrating the overlap in the number of salt stress-responsive glycoproteins in the WT, mns1 mns2, and cgl1-3. Three common proteins quantified in the WT and two mutants are presented as green dots in (A–C). (E–G) GO analyses of salt stress-responsive proteins in the WT (E), mns1 mns2 (F), and cgl1-3 (G). Significantly enriched GO terms are marked on the diagram. CA, catalytic activity; HA, hydrolase activity; PA, phosphorylating activity; GA, glucosidase activity; RtO, response to organic substance; RtS, response to stress; RtERS, response to ER stress; CMP, carbohydrate metabolic process; CP, catabolic process; EMO, extracellular matrix organization.
Two members from the class III peroxidase family identified in the DAGs, PRX32 (At3g32980) and Peroxidase 34 (PRX34, At3g49120). The protein abundance of neither PRX32 nor PRX34 was significantly changed in the WT under salt stress (Supplementary Table S5). However, both PRX32 and PRX34 were significantly increased in the cgl1-1 mutant by more than 2-fold (p = 0.011429) and 1.3-fold (p = 0.014898), respectively. Interestingly, in the mns1mns2 mutant, only PRX34, the more abundant protein, displayed a 1.3-fold increase (p = 0.018209; red point in Figures 3A–C; Supplementary Table S5). We reasoned that the absence of N-glycan modification may affect the functions of PRX proteins in the mutants, and the increase of protein abundance may be due to the feedback regulation under salt stress. While the different changes of the two proteins probably contribute to the difference in salt tolerance between mns1 mns2 and cgl1-3.
KEGG pathway analysis of DAGs showed that proteins processing in the ER, biosynthesis of secondary metabolites, plant-pathogen interactions, N-glycan biosynthesis, and photosynthesis were influenced by salt stress in the WT and mutants (Supplementary Figure S5). However, mns1 mns2 mutations mainly affected glycolysis/gluconeogenesis, biosynthesis of antibiotics, tryptophan metabolism, terpenoid backbone biosynthesis, and carotenoid biosynthesis. In contrast, in the cgl1-3 mutant, plant hormone signal transduction, carbon metabolism, carbon fixation in photosynthetic organisms, ascorbate, aldarate metabolism, and microbial metabolism in diverse environments were predominantly enriched (Supplementary Table S4). Taken together, these results suggested that N-glycan modification mediated by MNS1/MNS2 and CGL1 may have different influences on salt-responsive glycoproteins to adapt to salt stress.
Protein–Protein Interaction Network Analysis by STRING
N-glycoproteins include many secretory and membrane-bound proteins involved in the intracellular networks and intercellular communications of plant development and adaptive responses to stress. After identifying salt-responsive N-glycoproteins, we evaluated the possible regulatory network among them using the STRING server.5 We classified the identified proteins into two groups, increased and decreased under salt stress. In the WT, the increased proteins contained multiple molecular chaperones (PDIL1-3, PDIL1-3, and EBS1) and oligosaccharyltransferase subunits [STT3A, STT3B, At2g01720 (OST1A), At1g76400 (OST1B), HAP6, DGL1, and At1g61790 (OST3/6)] and present a close interaction for N-glycosylation, protein folding, and ER quality control (Figure 4A). In the decreased group, only several proteins related to the Calvin cycle and light reaction were illustrated (Figure 4B). In comparison, the DAGs in mns1 mns2 hardly formed functional regulatory networks. This might be caused by decreased protein stability or abundance and made the detected proteins more dispersed (Figures 4C,D). In the cgl1 mutant, the networks were still visible in the phosphorylation pathway by increased proteins (Figure 4E) and N-glycosylation biogenesis, cell wall organization, and light reactions by decreased proteins (Figure 4F). Although the changing trend of DAGs was slightly different from those in the WT, the regulatory network in cgl1 was still detectable (Figure 4F). We also found several DAGs involved in oxidative stress response, including PRX32, PRX34, GGAT1, and RHS19 (Figures 4E,F), indicating that the peroxidase members responsible for cellular hydrogen peroxide clearance orchestrated with others to withstand salt stress.
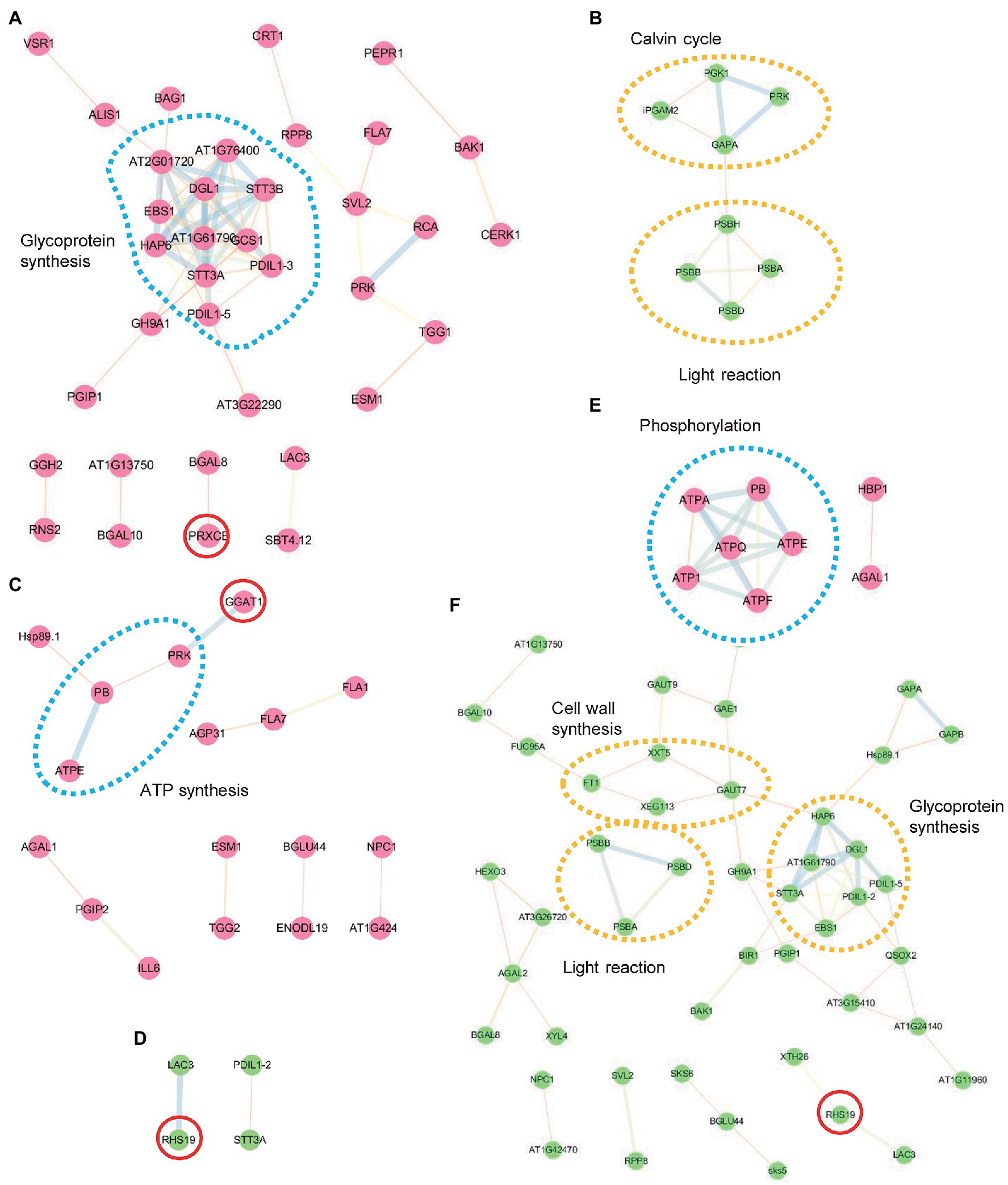
Figure 4. Protein interaction network analysis using the STRING database. Interaction network of differentially abundant glycoproteins (DAGs) was performed with the STRING system (https://string-db.org) based on known and predicted interactions. Protein–protein interactions among increased (red points) and decreased (green points) proteins in the WT (A,B), mns1 mns2 (C,D), and cgl1-3 (E,F) were analyzed separately. Lines of different font-weight represent different types of evidence for the associations. The dotted circles highlight the involved biological processes. Red circles are proteins associated with the oxidation-reduction process. Proteins without interactions are not shown.
Biological Validation of Quantitative Proteomics Results
To verify if the DAGs identified by the MS reflect the physiological responses of plants to salt stress, we select the candidate proteins for further validation. The GO functional analysis indicated that peroxidase directly or indirectly participated in the salt response (Table 2; Figure 4). The protein abundance of two peroxidases, PRX32 and PRX34, significantly increased in the salt stress-sensitive cgl1 mutant (Table 2; Supplementary Table S5). These two proteins are predicted harboring signal peptides and more than five potential N-sites with the NetNGlyc server (Figure 5A).6 We transiently expressed PRX32 and PRX34 in tobacco leaves in the absence or presence of tunicamycin, an N-glycosylation inhibitor. As shown in Figure 5B, the obvious accumulations of under-glycosylated forms of PRX32 and PRX34 were observed from recombinant PRX32-HA and PRX34-HA proteins. Similarly, after digestion with glycosidase Endo H, the band shifts and the accumulation of deglycosylated PRX32-HA proteins were detected (Figure 5C), implying that both proteins were N-glycosylated in vivo and their abundances were regulated by associated N-glycan. To further test if PRX32 and PRX34 were salt-responsive genes, qPCR analyses were performed. The basal expression level of PRX32 was low, and no obvious response to salt stress in WT and mutants was detected. By comparison, PRX34 was significantly upregulated against salt stress in mns1 mns2 and cgl1 mutants (Figures 5D,E). We also used Arabidopsis T-DNA insertional lines to test the response of prx32 and prx34 to high salinity. The T-DNA insertion of both lines laid in the 5'-untranslated region (UTR; Figure 6A), and semi-quantitative RT-PCR confirmed no detectable transcripts from prx32 and prx34 mutants either (Figure 6B). The single mutant of prx32 and prx34 displayed similar primary root length as WT under salt stress. However, the double mutations of prx32 prx34 were obviously sensitive to salt treatment, showing a further reduction in root length compared to the WT (p = 0.0009, n > 20; Figures 6C,D), suggesting that these two proteins might have overlapping effects on root growth under salt stress. In addition, DAB staining was conducted to explore whether the activity of PRX32 and PRX34 was relevant to the salt stress response. Agreed with a previous study that PRX34 functions in an apoplastic oxidative burst (O’Brien et al., 2012), H2O2 production induction by salt stress decreased prx34, prx32, and prx32 prx34 double mutants as well (Figures 6E,F). These results suggested the crucial roles of two functionally redundant PRX proteins in the salt response. PRX32 and PRX34 belong to the class III peroxidase (PRX) multigene superfamily, containing 73 encoding genes in Arabidopsis (Valerio et al., 2004); however, their biological functions remain largely unknown. It is possible that more PRX proteins were involved in the salt response. In addition, the results suggested that the identified DAGs are potential biologically functional salt-responsive glycoproteins.
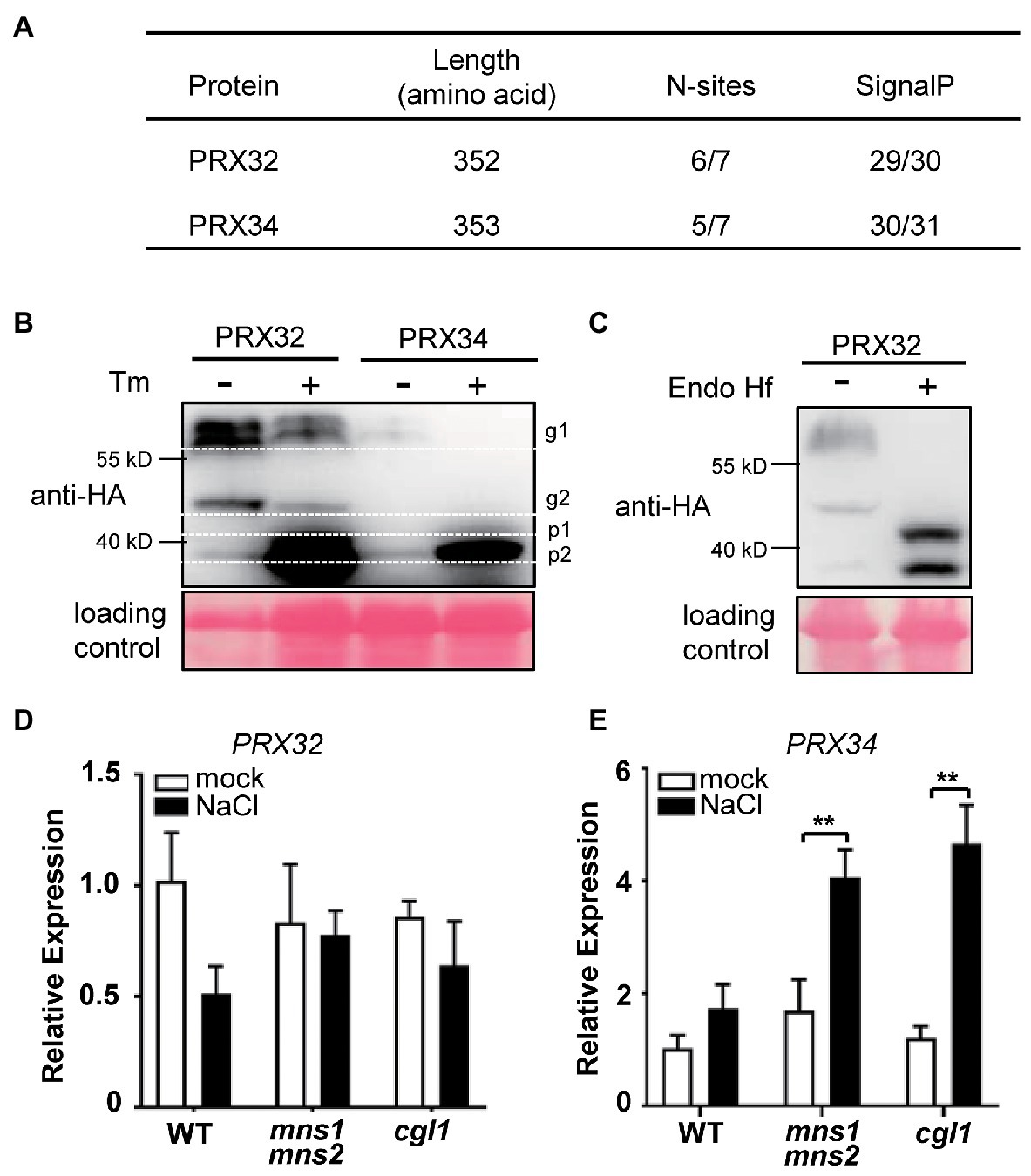
Figure 5. PRX32 and PRX34 are N-glycosylated, and their expressions are responsive to salt stress. (A) Prediction of N-glycosylation sites and signal peptides in PRX32 and PRX34. (B) N-glycosylation analysis of PRX32 and PRX34 proteins by tunicamycin treatment. PRX32-HA and PRX34-HA were transiently expressed in tobacco leaves in the absence or presence of tunicamycin and analyzed by immunoblots using anti-HA antibodies. g1 and g2 indicated two glycosylated bands; p1 and p2 represented two non-glycosylated bands. Ponceau S staining served as a loading control. The molecular weights of PRX32 and PRX34 are both 39 kD. (C) N-glycan analysis of PRX32 by Endo H digestion. The recombinant protein PRX32-HA was transiently expressed in tobacco leaves. After digested with glycosidase Endo Hf, the total proteins were separated on SDS-PAGE and detected with anti-HA antibodies. Ponceau S staining served as a loading control. (D,E) Quantitative real-time PCR (qPCR) analysis of gene expression in response to salt stress. Total RNAs were extracted from 2-week-old seedlings treated with or without salt stress described in Figure 1. The expression levels of PRX32 (D) and PRX34 (E) were analyzed by qPCR. **Indicates a significant difference at p < 0.001.
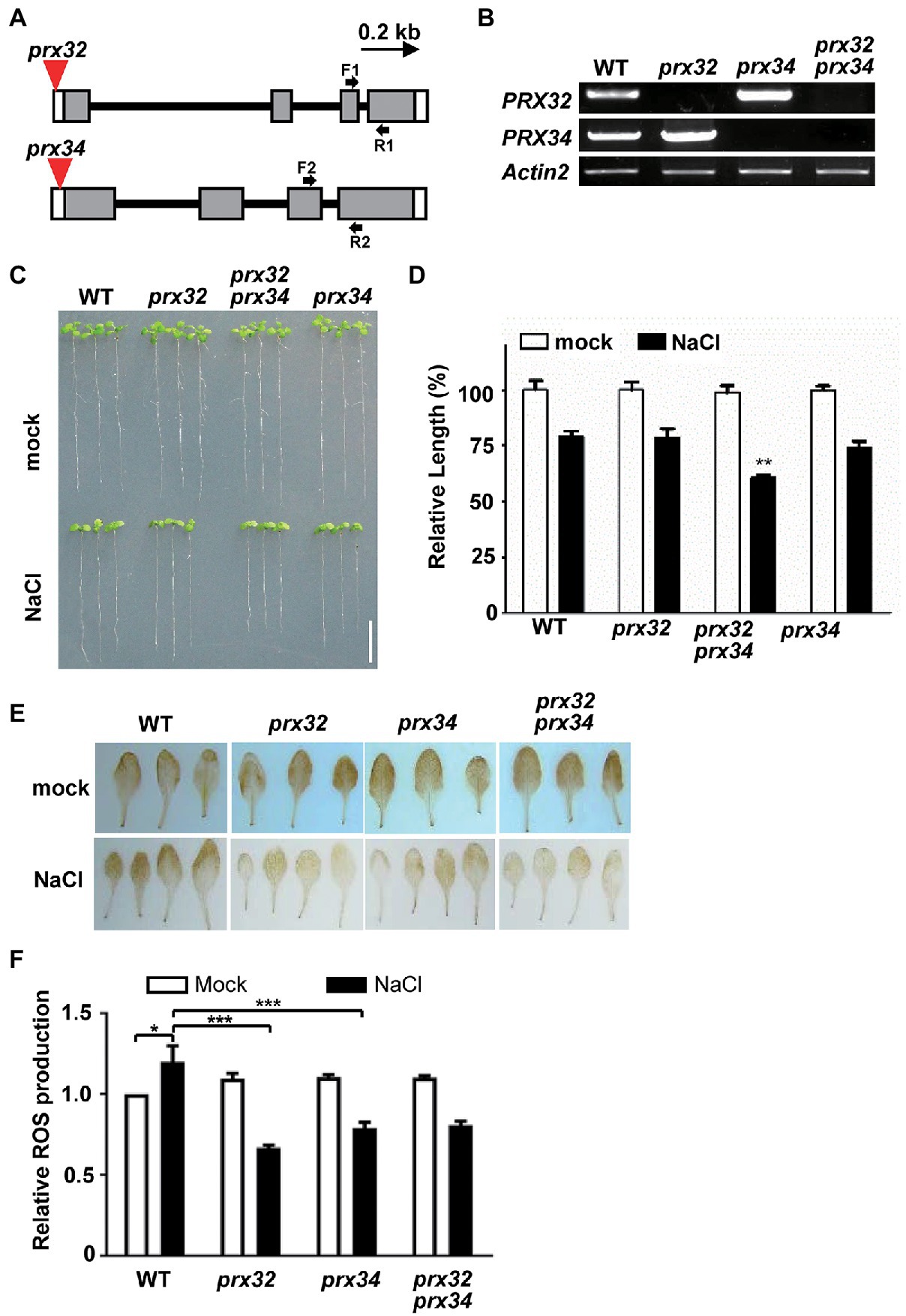
Figure 6. PRX32 and PRX34 are required for root growth under salt stress. (A) T-DNA insertion sites of prx alleles. Gray boxes represent exons and are linked with introns (lines). The open boxes denote untranslated regions (UTR). The location of the T-DNA insertion site in each mutant line is marked as a red triangle. A bar with an arrow from 5' to 3' representing 0.2 kb length is shown. (B) Reverse transcription-PCR (RT-PCR) analyses of PRX32 and PRX34 gene expression. Two-week-old Arabidopsis seedlings were used for RNA extraction, and the transcript of Actin2 was amplified as an internal control. (C) Root growth phenotypes against salt treatment. Five-day-old seedlings were transferred to MS medium supplemented without (mock) or NaCl to grow for 9 days before taking photos. Bar = 0.5 cm. (D) Statistics analysis of root length. The root length shown in (C) was statistically analyzed. The primary root length of the WT (mock) was set as 100%, and relative values were shown. **Indicated a significant difference from the value of the WT (NaCl) with p < 0.001, n > 20 (Student’s t-test). (E) Reactive oxygen species (ROS) production analysis by 3,3'-Diaminobenzidine (DAB) staining. Two-week-old seedlings were transferred to liquid Murashige and Skoog (MS) medium supplemented without (mock) or 200 mM NaCl to grow for 6 h before staining. (F) Statistics analysis of DAB staining. The measurement was performed in ImageJ software. The value of WT (mock) was set as 1.0, and relative values were shown. * and *** indicated significant differences at p < 0.01 and p < 0.0001, respectively (both n > 10, Student’s t-test).
Discussion
In the current study, we conducted an MS-based label-free proteomic analysis to study differentially abundant glycoproteins in mns1 mns2 and cgl1-3 mutants using lectin enrichment methods (Zielinska et al., 2010). The results showed that N-glycosylation biosynthesis was important for the homeostasis of the plant cell against high salinity. By analyzing DAGs, we found that the abundance of several subsets of glycoproteins responsible for glycoprotein biosynthesis, stress response, signal transduction, and oxidation-reduction process increased, while energy production and cell wall organization related glycoproteins were reduced under salt stress in the WT (Table 2; Figure 4F). In agreement with previous studies, our data suggested that plants primarily need to save energy for survival due to impaired photosynthesis system under stress conditions. However, the abundance modulation of salt-responsive glycoproteins was impaired in both mutants, presumably due to the lack of mature N-glycan. In addition, comprehensive studies between mns1 mns2 and cgl1 in response to high salinity revealed that two N-glycan modification steps mediated by MNS1/MNS2 and CGL1 might differentially modulate client glycoproteins, which is consistent with the difference of salt-sensitive phenotypes between mns1mns2 and cgl1-3 mutant (Liu et al., 2018). Thus, our results supply an integral overview of the requirements of highly coordinated N-glycosylation steps in plants to adapt to environmental stress.
Reactive oxygen species (ROS), including the superoxide anion, hydrogen peroxide, and hydroxyl radical, are normal byproducts of aerobic cell metabolism, but a large amount of ROS production and accumulation induced by salt stress in plant cells is dangerous for photosynthesis, cell wall biosynthesis, and many other biological processes (Munns and Tester, 2008; Yang and Guo, 2018). The active oxygen scavengers in plants include catalase (CAT), ascorbic acid peroxidase (APX), superoxide dismutase (SOD), glutathione reductase (GR), and some non-enzymatic molecules such as anti-ascorbic acid, glutathione, carotenoids, and anthocyanins (Apel and Hirt, 2004). Arabidopsis contains a complex ROS synthesis, regulation, and clearance-related network involving more than 150 genes (Mittler et al., 2004). Our results demonstrated that two functional peroxidases, PRX32 and PRX34, were potentially modulated by N-glycosylation and responsible for root development and salt sensitivity (Figures 5, 6). However, the exact roles of PRX32 and PRX34, together with other class III peroxidase members in maintaining root development against salt stress, still need to be investigated (Arnaud et al., 2017). Moreover, addressing the regulatory roles of their attached N-glycans is another challenge.
In the present study, we demonstrated that the protein abundance changes in mns1 mns2 and cgl1-3 differed by label-free quantitative analysis (Table 2). GO and KEGG pathway results also indicated that the modulated substrate proteins in mns1 mns2 and cgl1-3 were involved in different biological pathways under salt stress (Figure 3; Supplementary Figure S5). We suspected that each N-glycosylation step might influence the specific subsets of glycoproteins during stress responses. Several reports have shown that different N-glycoproteins may harbor different N-glycan structures. For instance, TGG1, one of two myrosinase enzymes in Arabidopsis, is decorated with oligomannosidic N-glycans, as seen in the MS (Liebminger et al., 2012). However, most attached N-glycans on the RSW2 protein, which is critical for cell wall synthesis, were modified with a complex glycan structure (Liebminger et al., 2013). The difference in N-glycan structures between TGG1 and RSW2 may reflect intrinsic protein structures for native cellular functions and may be the molecular basis for differential regulation by N-glycosylation steps.
Distinct from the case of responses to biotic stress, plant responses to abiotic stress often involve multiple genes in multiple metabolic pathways, and there is a complex association between different abiotic stress responses (Cabello et al., 2014). From the perspective of genomics and proteomics, the regulatory network of abiotic stress responses exhibits temporal and spatial complexity in the context of signal transduction and expression of regulatory genes and functional proteins such as transcription factors and enzymes responsible for stress-related metabolite synthesis or deposition (Zhu, 2016). In addition, the protein-quantification method using MS coupling label-free analysis may lead to different protein quantification results due to the differing abundance of N-glycopeptides identified from the same protein (Song et al., 2013). Therefore, it will be helpful to integrate valuable findings from different large-scale multi-omics experiments. Recently, several studies revealed the N-glycan microheterogeneity on glycoproteins in Arabidopsis (Xu et al., 2016; Zeng et al., 2017). It would be interesting to understand whether N-glycosyl regulation occurs under specific conditions. It is necessary to determine the N-glycan structures and quantitation on the glycopeptides to evaluate the precise roles of each N-glycan in the future.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: http://www.proteomexchange.org/, PXD006893.
Author Contributions
CL and ZH conceived and designed the experiments. CL, HZ, HC, DH, and XL performed the experiments. CL, GN, and PL conducted the bioinformatic analysis. CL and GN wrote the manuscript. PL and ZH revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was partially supported by the National Natural Science Foundation of China (31870263 and 31322008).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Arabidopsis Biological Resource Center at Ohio State University for supplying seeds of T-DNA insertional mutants, members of Lan laboratory for bioinformatic analysis supports, and members of Hong laboratory for helpful discussion.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.646425/full#supplementary-material
Footnotes
1. ^http://bioinfo.cau.edu.cn/agriGO/
2. ^http://www.genome.jp/kegg/
References
Apel, K., and Hirt, H. (2004). Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55, 373–399. doi: 10.1146/annurev.arplant.55.031903.141701
Arnaud, D., Lee, S., Takebayashi, Y., Choi, D., Choi, J., Sakakibara, H., et al. (2017). Cytokinin-mediated regulation of reactive oxygen species homeostasis modulates stomatal immunity in Arabidopsis. Plant Cell 29, 543–559. doi: 10.1105/tpc.16.00583
Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., et al. (2000). Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25–29. doi: 10.1038/75556
Boyaval, F., van Zeijl, R., Dalebout, H., Holst, S., van Pelt, G. W., Farina-Sarasqueta, A., et al. (2020). N-glycomic signature of stage II colorectal cancer and its association with the tumor microenvironment. Mol. Cell. Proteomics 20:100057. doi: 10.1074/mcp.RA120.002215
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. doi: 10.1016/0003-2697(76)90527-3
Cabello, J. V., Lodeyro, A. F., and Zurbriggen, M. D. (2014). Novel perspectives for the engineering of abiotic stress tolerance in plants. Curr. Opin. Biotechnol. 26, 62–70. doi: 10.1016/j.copbio.2013.09.011
Cox, J., and Mann, M. (2008). MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372. doi: 10.1038/nbt.1511
Daudi, A., and O’Brien, J. A. (2012). Detection of hydrogen peroxide by DAB staining in Arabidopsis leaves. Bio Protoc. 2:e263. doi: 10.21769/BioProtoc.263
Du, Z., Zhou, X., Ling, Y., Zhang, Z., and Su, Z. (2010). agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res. 38, W64–W70. doi: 10.1093/nar/gkq310
Fang, P., Ji, Y., Silbern, I., Doebele, C., Ninov, M., Lenz, C., et al. (2020). A streamlined pipeline for multiplexed quantitative site-specific N-glycoproteomics. Nat. Commun. 11:5268. doi: 10.1038/s41467-020-19052-w
Frank, J., Kaulfurst-Soboll, H., Rips, S., Koiwa, H., and von Schaewen, A. (2008). Comparative analyses of Arabidopsis complex glycan1 mutants and genetic interaction with staurosporin and temperature sensitive3a. Plant Physiol. 148, 1354–1367. doi: 10.1104/pp.108.127027
Haweker, H., Rips, S., Koiwa, H., Salomon, S., Saijo, Y., Chinchilla, D., et al. (2010). Pattern recognition receptors require N-glycosylation to mediate plant immunity. J. Biol. Chem. 285, 4629–4636. doi: 10.1074/jbc.M109.063073
Hong, Z., Jin, H., Tzfira, T., and Li, J. (2008). Multiple mechanism-mediated retention of a defective brassinosteroid receptor in the endoplasmic reticulum of Arabidopsis. Plant Cell 20, 3418–3429. doi: 10.1105/tpc.108.061879
Isaacson, T., Damasceno, C. M., Saravanan, R. S., He, Y., Catala, C., Saladie, M., et al. (2006). Sample extraction techniques for enhanced proteomic analysis of plant tissues. Nat. Protoc. 1, 769–774. doi: 10.1038/nprot.2006.102
Jiao, Q. S., Chen, T. S., Niu, G. T., Zhang, H. C., Zhou, C. F., and Hong, Z. (2020). N-glycosylation is involved in stomatal development by modulating the release of active abscisic acid and auxin in Arabidopsis. J. Exp. Bot. 71, 5865–5879. doi: 10.1093/jxb/eraa321
Jin, H., Yan, Z., Nam, K. H., and Li, J. (2007). Allele-specific suppression of a defective brassinosteroid receptor reveals a physiological role of UGGT in ER quality control. Mol. Cell 26, 821–830. doi: 10.1016/j.molcel.2007.05.015
Kajiura, H., Koiwa, H., Nakazawa, Y., Okazawa, A., Kobayashi, A., Seki, T., et al. (2010). Two Arabidopsis thaliana Golgi alpha-mannosidase I enzymes are responsible for plant N-glycan maturation. Glycobiol. 20, 235–247.
Kang, J. S., Frank, J., Kang, C. H., Kajiura, H., Vikram, M., Ueda, A., et al. (2008). Salt tolerance of Arabidopsis thaliana requires maturation of N-glycosylated proteins in the Golgi apparatus. Proc. Natl. Acad. Sci. U. S. A. 105, 5933–5938. doi: 10.1073/pnas.0800237105
Kelleher, D. J., and Gilmore, R. (2006). An evolving view of the eukaryotic oligosaccharyltransferase. Glycobiology 16, 47R–62R. doi: 10.1093/glycob/cwj066
Khoury, G. A., Baliban, R. C., and Floudas, C. A. (2011). Proteome-wide post-translational modification statistics: frequency analysis and curation of the swiss-prot database. Sci. Rep. 1:90. doi: 10.1038/srep00090
Koiwa, H., Li, F., McCully, M. G., Mendoza, I., Koizumi, N., Manabe, Y., et al. (2003). The STT3a subunit lsoform of the Arabidopsis oligosaccharyltransferase controls adaptive responses to salt/osmotic stress. Plant Cell 15, 2273–2284. doi: 10.1105/tpc.013862
Lamesch, P., Berardini, T. Z., Li, D., Swarbreck, D., Wilks, C., Sasidharan, R., et al. (2012). The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 40, D1202–D1210. doi: 10.1093/nar/gkr1090
Lerouge, P., Cabanes-Macheteau, M., Rayon, C., Fischette-Laine, A. C., Gomord, V., and Faye, L. (1998). N-glycoprotein biosynthesis in plants: recent developments and future trends. Plant Mol. Biol. 38, 31–48. doi: 10.1023/A:1006012005654
Li, J., Nam, K. H., Vafeados, D., and Chory, J. (2001). BIN2, a new brassinosteroid-insensitive locus in Arabidopsis. Plant Physiol. 127, 14–22. doi: 10.1104/pp.127.1.14
Li, J., Zhao-Hui, C., Batoux, M., Nekrasov, V., Roux, M., Chinchilla, D., et al. (2009). Specific ER quality control components required for biogenesis of the plant innate immune receptor EFR. Proc. Natl. Acad. Sci. U. S. A. 106, 15973–15978. doi: 10.1073/pnas.0905532106
Liebminger, E., Grass, J., Altmann, F., Mach, L., and Strasser, R. (2013). Characterizing the link between glycosylation state and enzymatic activity of the endo-beta1,4-glucanase KORRIGAN1 from Arabidopsis thaliana. J. Biol. Chem. 288, 22270–22280. doi: 10.1074/jbc.M113.475558
Liebminger, E., Grass, J., Jez, J., Neumann, L., Altmann, F., and Strasser, R. (2012). Myrosinases TGG1 and TGG2 from Arabidopsis thaliana contain exclusively oligomannosidic N-glycans. Phytochemistry 84, 24–30. doi: 10.1016/j.phytochem.2012.08.023
Liebminger, E., Huttner, S., Vavra, U., Fischl, R., Schoberer, J., Grass, J., et al. (2009). Class I alpha-mannosidases are required for N-glycan processing and root development in Arabidopsis thaliana. Plant Cell 21, 3850–3867. doi: 10.1105/tpc.109.072363
Lindner, H., Kessler, S. A., Muller, L. M., Shimosato-Asano, H., Boisson-Dernier, A., and Grossniklaus, U. (2015). TURAN and EVAN mediate pollen tube reception in Arabidopsis Synergids through protein glycosylation. PLoS Biol. 13:e1002139. doi: 10.1371/journal.pbio.1002139
Liu, C., Niu, G., Zhang, H., Sun, Y., Sun, S., Yu, F., et al. (2018). Trimming of N-glycans by the Golgi-localized alpha-1,2-mannosidases, MNS1 and MNS2, is crucial for maintaining RSW2 protein abundance during salt stress in Arabidopsis. Mol. Plant 11, 678–690. doi: 10.1016/j.molp.2018.01.006
Ma, J., Wang, D., She, J., Li, J., Zhu, J. K., and She, Y. M. (2016). Endoplasmic reticulum-associated N-glycan degradation of cold-upregulated glycoproteins in response to chilling stress in Arabidopsis. New Phytol. 212, 282–296. doi: 10.1111/nph.14014
Mittler, R., Vanderauwera, S., Gollery, M., and Van Breusegem, F. (2004). Reactive oxygen gene network of plants. Trends Plant Sci. 9, 490–498. doi: 10.1016/j.tplants.2004.08.009
Mora-Garcia, S., Vert, G., Yin, Y., Cano-Delgado, A., Cheong, H., and Chory, J. (2004). Nuclear protein phosphatases with Kelch-repeat domains modulate the response to brassinosteroids in Arabidopsis. Genes Dev. 18, 448–460. doi: 10.1101/gad.1174204
Moremen, K. W., Tiemeyer, M., and Nairn, A. V. (2012). Vertebrate protein glycosylation: diversity, synthesis and function. Nat. Rev. Mol. Cell Biol. 13, 448–462. doi: 10.1038/nrm3383
Muller, L. M., Lindner, H., Pires, N. D., Gagliardini, V., and Grossniklaus, U. (2016). A subunit of the oligosaccharyltransferase complex is required for interspecific gametophyte recognition in Arabidopsis. Nat. Commun. 7:10826. doi: 10.1038/ncomms10826
Munns, R., and Tester, M. (2008). Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 59, 651–681. doi: 10.1146/annurev.arplant.59.032607.092911
Nagashima, Y., von Schaewen, A., and Koiwa, H. (2018). Function of N-glycosylation in plants. Plant Sci. 274, 70–79. doi: 10.1016/j.plantsci.2018.05.007
Nekrasov, V., Li, J., Batoux, M., Roux, M., Chu, Z. H., Lacombe, S., et al. (2009). Control of the pattern-recognition receptor EFR by an ER protein complex in plant immunity. EMBO J. 28, 3428–3438. doi: 10.1038/emboj.2009.262
O’Brien, J. A., Daudi, A., Finch, P., Butt, V. S., Whitelegge, J. P., Souda, P., et al. (2012). A peroxidase-dependent apoplastic oxidative burst in cultured Arabidopsis cells functions in MAMP-elicited defense. Plant Physiol. 158, 2013–2027. doi: 10.1104/pp.111.190140
Pless, D. D., and Lennarz, W. J. (1977). Enzymatic conversion of proteins to glycoproteins. Proc. Natl. Acad. Sci. U. S. A. 74, 134–138. doi: 10.1073/pnas.74.1.134
Qin, F., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2011). Achievements and challenges in understanding plant abiotic stress responses and tolerance. Plant Cell Physiol. 52, 1569–1582. doi: 10.1093/pcp/pcr106
Rips, S., Bentley, N., Jeong, I. S., Welch, J. L., von Schaewen, A., and Koiwa, H. (2014). Multiple N-glycans cooperate in the subcellular targeting and functioning of Arabidopsis KORRIGAN1. Plant Cell 26, 3792–3808. doi: 10.1105/tpc.114.129718
Scheys, F., Van Damme, E. J. M., De Schutter, K., Staes, A., Gevaert, K., and Smagghe, G. (2018). Evolutionarily conserved and species-specific glycoproteins in the N-glycoproteomes of diverse insect species. Insect Biochem. Mol. Biol. 100, 22–29. doi: 10.1016/j.ibmb.2018.04.011
Shen, J., Ding, Y., Gao, C., Rojo, E., and Jiang, L. (2014). N-linked glycosylation of AtVSR1 is important for vacuolar protein sorting in Arabidopsis. Plant J. 80, 977–992. doi: 10.1111/tpj.12696
Song, W., Henquet, M. G., Mentink, R. A., van Dijk, A. J., Cordewener, J. H., Bosch, D., et al. (2011). N-glycoproteomics in plants: perspectives and challenges. J. Proteomics 74, 1463–1474. doi: 10.1016/j.jprot.2011.05.007
Song, W., Mentink, R. A., Henquet, M. G., Cordewener, J. H., van Dijk, A. D., Bosch, D., et al. (2013). N-glycan occupancy of Arabidopsis N-glycoproteins. J. Proteomics 93, 343–355. doi: 10.1016/j.jprot.2013.07.032
Stadlmann, J., Taubenschmid, J., Wenzel, D., Gattinger, A., Durnberger, G., Dusberger, F., et al. (2017). Comparative glycoproteomics of stem cells identifies new players in ricin toxicity. Nature 549, 538–542. doi: 10.1038/nature24015
Strasser, R. (2016). Plant protein glycosylation. Glycobiology 26, 926–939. doi: 10.1093/glycob/cww023
Strasser, R., Steinkellner, H., Boren, M., Altmann, F., Mach, L., Glossl, J., et al. (1999). Molecular cloning of cDNA encoding N-acetylglucosaminyltransferase II from Arabidopsis thaliana. Glycoconj. J. 16, 787–791. doi: 10.1023/A:1007127815012
Valerio, L., De Meyer, M., Penel, C., and Dunand, C. (2004). Expression analysis of the Arabidopsis peroxidase multigenic family. Phytochemistry 65, 1331–1342. doi: 10.1016/j.phytochem.2004.04.017
Vonschaewen, A., Sturm, A., Oneill, J., and Chrispeels, M. J. (1993). Isolation of a mutant Arabidopsis plant that lacks N-acetyl glucosaminyl transferase-I and is unable to synthesize Golgi-modified complex N-linked glycans. Plant Physiol. 102, 1109–1118. doi: 10.1104/pp.102.4.1109
Wisniewski, J. R., Zielinska, D. F., and Mann, M. (2011). Comparison of ultrafiltration units for proteomic and N-glycoproteomic analysis by the filter-aided sample preparation method. Anal. Biochem. 410, 307–309. doi: 10.1016/j.ab.2010.12.004
Xia, Y., Ma, Z., Qiu, M., Guo, B., Zhang, Q., Jiang, H., et al. (2020). N-glycosylation shields Phytophthora sojae apoplastic effector PsXEG1 from a specific host aspartic protease. Proc. Natl. Acad. Sci. U. S. A. 117, 27685–27693. doi: 10.1073/pnas.2012149117
Xu, S. L., Medzihradszky, K. F., Wang, Z. Y., Burlingame, A. L., and Chalkley, R. J. (2016). N-glycopeptide profiling in Arabidopsis inflorescence. Mol. Cell. Proteomics 15, 2048–2054. doi: 10.1074/mcp.M115.056101
Yan, Q., and Lennarz, W. J. (2002). Studies on the function of oligosaccharyl transferase subunits. Stt3p is directly involved in the glycosylation process. J. Biol. Chem. 277, 47692–47700. doi: 10.1074/jbc.M208136200
Yang, Y., and Guo, Y. (2018). Unraveling salt stress signaling in plants. J. Integr. Plant Biol. 60, 796–804. doi: 10.1111/jipb.12689
Zeng, W., Ford, K. L., Bacic, T., and Heazlewood, J. L. (2017). N-glycan micro-heterogeneity in glycoproteins of Arabidopsis. Mol. Cell. Proteomics 17, 413–421. doi: 10.1074/mcp.RA117.000165
Zhu, J. K. (2016). Abiotic stress signaling and responses in plants. Cell 167, 313–324. doi: 10.1016/j.cell.2016.08.029
Zielinska, D. F., Gnad, F., Schropp, K., Wisniewski, J. R., and Mann, M. (2012). Mapping N-glycosylation sites across seven evolutionarily distant species reveals a divergent substrate proteome despite a common core machinery. Mol. Cell 46, 542–548. doi: 10.1016/j.molcel.2012.04.031
Keywords: N-glycan, salt response, label-free mass spectrum, proteomics, Arabidopsis
Citation: Liu C, Niu G, Li X, Zhang H, Chen H, Hou D, Lan P and Hong Z (2021) Comparative Label-Free Quantitative Proteomics Analysis Reveals the Essential Roles of N-Glycans in Salt Tolerance by Modulating Protein Abundance in Arabidopsis. Front. Plant Sci. 12:646425. doi: 10.3389/fpls.2021.646425
Edited by:
Richard Strasser, University of Natural Resources and Life Sciences Vienna, AustriaReviewed by:
Klára Kosová, Crop Research Institute (CRI), CzechiaTong Zhang, Pacific Northwest National Laboratory (DOE), United States
Copyright © 2021 Liu, Niu, Li, Zhang, Chen, Hou, Lan and Hong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhi Hong, emhpaG9uZ0BuanUuZWR1LmNu; Ping Lan, cGxhbkBpc3Nhcy5hYy5jbg==
 Chuanfa Liu
Chuanfa Liu Guanting Niu
Guanting Niu Xiaowen Li1
Xiaowen Li1 Huchen Zhang
Huchen Zhang Ping Lan
Ping Lan Zhi Hong
Zhi Hong