- 1School of Biological Sciences, The University of Hong Kong, Pokfulam, China
- 2Centre for Soybean Research of the State Key Laboratory of Agrobiotechnology and School of Life Sciences, The Chinese University of Hong Kong, Shatin, China
Salinity is a major environmental factor that constrains soybean yield and grain quality. Given our past observations using the salt-sensitive soybean (Glycine max [L.] Merr.) accession C08 on its early responses to salinity and salt-induced transcriptomic modifications, the aim of this study was to assess the lipid profile changes in this cultivar before and after short-term salt stress, and to explore the adaptive mechanisms underpinning lipid homeostasis. To this end, lipid profiling and proteomic analyses were performed on the leaves of soybean seedlings subjected to salt treatment for 0, 0.5, 1, and 2 h. Our results revealed that short-term salt stress caused dynamic lipid alterations resulting in recycling for both galactolipids and phospholipids. A comprehensive understanding of membrane lipid adaption following salt treatment was achieved by combining time-dependent lipidomic and proteomic data. Proteins involved in phosphoinositide synthesis and turnover were upregulated at the onset of salt treatment. Salinity-induced lipid recycling was shown to enhance jasmonic acid and phosphatidylinositol biosyntheses. Our study demonstrated that salt stress resulted in a remodeling of membrane lipid composition and an alteration in membrane lipids associated with lipid signaling and metabolism in C08 leaves.
Introduction
Soybean (Glycine max [L.] Merr.) is one of the most important legume crops used as food and feed (Hasanuzzaman et al., 2016). As a major environmental constraint on crop yield, salinity triggers osmotic stress in roots followed by whole-plant ionic toxicity arising from Na+ and Cl– ion accumulation (Munns and Tester, 2008). Plants have evolved to adapt to high salt by salt-responsive signaling, balancing osmotic pressure in organic osmolyte formation, Na+ compartmentalization, enhanced scavenging of reactive oxygen species (ROS), phytohormone regulation, and cell structure modulation (Parvaiz and Satyawati, 2008; Phang et al., 2008). As modifying in membrane lipid composition provides an effective strategy in maintaining cell membrane integrity to confront stress (Elkahoui et al., 2004), the characterization of membrane lipids following salt treatment is crucial in formulating strategies to overcome salinity. Qualitative and quantitative lipid profiling following abiotic stresses such as drought (Gigon et al., 2004), cold (Barrero-Sicilia et al., 2017), or heat (Higashi et al., 2015) have been studied in Arabidopsis. Furthermore, salt-induced membrane lipid alterations documented in salt-sensitive varieties of Arabidopsis thaliana and Triticum sativum, and salt-tolerant varieties of Brassica napus and Zea mays indicated that adaptation to salinity stress relied on total membrane lipid composition (Guo et al., 2019). Other reports revealed a reduction in membrane lipids in the salt-sensitive barley after salinity stress (Hordeum vulgare L. cv. Manel) while no changes were evident in the salt-tolerant Hordeum maritimum and the halophyte Suaeda altissima (Chalbi et al., 2013; Tsydendambaev et al., 2013). Greater lipid accumulation and mobilization during seedling development occurred in the salt-sensitive variety of sunflower (Helianthus annuus L.) DRSH 1 than the salt-tolerant variety PSH 1962 (Gogna and Bhatla, 2020). However, lipid profiling to address membrane lipid compositional changes in salt-sensitive soybean leaves following salt treatment has not been reported.
The majority of membrane lipids in leaves are structural lipids including galactolipids (GLs) and phospholipids (PLs) (Guschina et al., 2014). Chloroplast membranes consist mainly of GLs including monogalactosyldiacylglycerol (MGDG) and digalactosyldiacylglycerol (DGDG) followed by anionic lipids comprising sulfoquinovosyldiacylglycerol (SQDG) and phosphatidylglycerol (PG) (Douce, 1974; Block et al., 1983). As the major components of photosystem II on the thylakoid membranes, MGDG and DGDG form an indispensable matrix for photosynthesis (Mizusawa and Wada, 2012), and their ratio is important in maintaining chloroplast membrane stability (Dörmann and Benning, 2002). The amounts of chloroplast lipids (MGDG, DGDG, SQDG, and PG) drastically decreased in B. napus leaves after cadmium stress (Nouairi et al., 2006). During drought stress, the ratio of DGDG/MGDG was elevated, mainly attributed by a fall in MGDG (Gigon et al., 2004; Torres-Franklin et al., 2007). Sodium chloride (NaCl)-induced decline in MGDG and DGDG was reported in leaves of Thellungiella halophila, Sulla carnosa, and Sulla coronaria (Sui and Han, 2014; Bejaoui et al., 2016).
Among the main PL classes, phosphatidic acid (PA) plays an important role in signal transduction and serves as a key intermediate in lipid metabolism (Athenstaedt and Daum, 1999). PA accumulation can be induced by biotic and abiotic stresses caused by salt, drought, cold, plant pathogens, and wounding (Tuteja and Sopory, 2008). Under duress, PA is produced through hydrolysis of the main structural PLs such as phosphatidylcholine (PC) and phosphatidylethanolamine (PE) by phospholipase D (PLD) (Pappan et al., 1998). It can also be generated by phosphorylation of diacylglycerol (DAG), which is the product of phospholipase C (PLC) hydrolysis on phosphatidylinositol (PI) (Arisz et al., 2009). PC also acts as a resource for lipid second messengers such as lysophosphatidylcholine (lysoPC), PA, and DAG (Exton, 1990). High salt and osmotic stress caused a rapid rise in PC turnover in Arabidopsis suspension−cultured cells along with an increase in phosphatidylinositol 4, 5-bisphosphate [PI(4,5)P2] and DAG pyrophosphate (Pical et al., 1999). Moreover, PC turnover facilitates substrates for production of chloroplast GLs (Karki et al., 2019). The chloroplast is the site of de novo fatty acid synthesis (FAS) with the conversion of pyruvate to acetyl-CoA and generation of carbon (C) chains 16:0, 18:0 and 18:1-acyl-carrier protein (ACP) for the assembly of PA (Hölzl and Dörmann, 2019), which can be further utilized for building chloroplast structural membranes (PG, MGDG, and DGDG) via the prokaryotic pathway (Boudière et al., 2014). This pathway is distinguished from the eukaryotic (Kennedy) pathway by the presence of a C chain 16 at the sn-2 position of the glycerol backbone (Boudière et al., 2014). For the eukaryotic pathway, 18:1-CoA esters bound to acyl-CoA binding proteins are imported into the endoplasmic reticulum (ER) and integrated to PC (Karki et al., 2019), which then undergoes desaturation to 18:2 and 18:3-CoA by acyl chain editing, dependent on the dynamic interconversion between PC and lysoPC (Bates et al., 2012). After PC desaturation, a proportion could be incorporated in the chloroplasts as DAG during GL biosynthesis (Hölzl and Dörmann, 2019; He et al., 2020).
Given the complexities in protein function upon stress treatment, proteomics represents a powerful approach to evaluate the proteome. Such analysis related to stresses such as drought, salinity, and extreme temperatures has been reported in several major crops (Ahmad et al., 2016). Furthermore, the integration of proteomics data with others from genomics, transcriptomics, or metabolomics provides wider scope in understanding biological processes (Zhang and Kuster, 2019). In soybean, proteome studies on abiotic stress have been reported on flood, drought, heat, and salinity (Das et al., 2016; Ji et al., 2016; Yin and Komatsu, 2017; Katam et al., 2020). A case in point is the integration of metabolome and proteome studies of the maize atg12 mutant which revealed a role for ATG12 in autophagic recycling by proteome remodeling and lipid turnover (McLoughlin et al., 2018).
To explore the adaptive mechanisms of soybean in lipid signaling and remodeling of metabolism following salt stress, lipid profiling was performed on leaves of seedlings from cultivar C08 (Lam et al., 2010; Qi et al., 2014) which is a salt-sensitive variety. In our previous study, C08 seedlings showed leaf drooping at 0.5 h under high salt stress followed by recovery (Liu et al., 2019). To study if this phenomenon could be closely linked to the dynamic changes in membrane lipid composition, integrated proteomic and lipidomic analyses were conducted. To address protein function in response to salt, label-free quantitative (LFQ) proteomic analysis on salt-treated soybean seedling leaves was conducted. Our study identified putative enzymes or protein components involved in lipid regulation and provide a comprehensive understanding of lipid remodeling following salt treatment in soybean.
Materials and Methods
Plant Materials and Growth Conditions
Soybean G. max [L.] Merr., accession C08 (Lam et al., 2010) seeds were germinated in vermiculite in a greenhouse. Seedlings were transferred to 1/2 strength Hoagland nutrient solution (Hoagland and Arnon, 1950) when the primary leaves were fully expanded. Detailed growth conditions have been previously described (Liu et al., 2019). For salt treatment, 14-day-old seedlings were transferred to 1/2 strength Hoagland solution supplemented with 0.9% (w/v) NaCl. NaCl was omitted in the control. Primary leaves were harvested at 0, 0.5, 1, and 2 h after treatment.
Lipid Extraction and Profiling
Total lipids were extracted following the protocol of Shiva et al. (2018) with minor modifications. Around six leaf punches from the primary leaf were sampled using a liquid nitrogen (N2) pre-cooled leaf puncher. The leaf punches were immediately immersed in 2 ml of isopropanol containing 0.01% butylated hydroxytoluene in a 20-ml vial at 75°C. The samples were heated at 75°C for 30 min to deactivate phospholipid-hydrolyzing enzymes. After cooling the samples to room temperature, 6 ml chloroform/methanol/water (30:41.5:3.5) were added, and the mixture was shaken at 100 rpm on an orbital shaker at room temperature for 24 h. To determine the dry weight after lipid extraction, the leaf punches were dried in an oven at 80°C for 72 h, cooled in an airtight desiccator, and weighed with a precision balance. The lipid content was estimated and normalized to the dry weight of the leaf punches. Five independent replicates of leaves from soybean seedlings were analyzed. Individual lipid species were denoted by the lipid class, followed by the total number of acyl Cs and the total number of C-C double bonds in the acyl chains, e.g., MGDG (34:6). The profile of membrane lipids was measured using an automated electrospray ionization-tandem mass spectrometry (Devaiah et al., 2006) at the Kansas Lipidomics Research Center.
Protein Extraction, Digestion and Peptide Preparation
Total leaf protein was extracted following the method of Lv et al. (2013) with minor modifications. Harvested primary leaves were ground in liquid N2 with a mortar and pestle. The powdered sample was suspended in 2 ml of sodium dodecyl sulfate (SDS) buffer (2% [w/v] SDS, 100 mM Tris–HCl, pH 8.0, 50 mM ethylenediaminetetraacetic acid disodium salt [EDTA-Na2], 20 mM dithiothreitol [DTT]) with 1 mM phenylmethanesulfonyl fluoride (Sigma) and 1× protease inhibitor cocktail (Sigma). The samples were heated at 95°C for 30 min. After clarification by centrifugation at 14,000 × g, the protein was precipitated with three volumes of acetone at −80°C for 1 h. The protein was pelleted by centrifugation at 14,000 × g and then resuspended in 8 M urea 50 mM Tris–HCl, pH 8.0. The precipitation procedure was repeated once. The air-dried protein pellet was further dissolved in 8 M urea 50 mM Tris–HCl, pH 8.0 at 37°C for over 3 h. Ten microgram protein per sample was reduced with 5 mM DTT at 37°C for 1 h and trypsin-digested at 37°C overnight using Trypsin Gold, Mass Spectrometry Grade (Promega) in a trypsin:protein ratio of 1:20 (w/w). The peptide mixture was desalted using a PierceTM C-18 spin column (Thermo Fisher ScientificTM) and then dissolved in 0.1% (v/v) formic acid. Three biological replicates were performed.
Orbitrap Liquid Chromatography-Tandem Mass Spectrometry (LC-MS/MS) and Quantitative Analysis
Eight hundred nanogram digested peptides from each sample was injected and analyzed using an Orbitrap FusionTM LumosTM TribridTM Mass Spectrometer (Thermo Fisher ScientificTM). Nano-liquid chromatography (LC) separation of digested peptides was carried out using a LC Ultimate 3000 RSLCnano system equipped with a C-18 5 μm-pre-column (300 μm i.d. × 5 mm) and an Acclaim Pepmap RSLC nanoViper C-18 column (75 μm i.d. × 25 cm). Mobile phase A (1.9% acetonitrile and 0.1% formic acid) and mobile phase B (98% acetonitrile and 0.1% formic acid) were used in LC. The LC mobile phase gradient profile was set as follows: 50°C chamber with 3 μL min–1 flow rate, beginning with 100% mobile phase A for 5 min, increasing mobile phase B from 0 to 6% in 3 min, to 18% in 40 min, to 30% in 10 min, then to 80% in 7 min, and finally re-equilibrating with 100% mobile phase A for 10 min. Raw data was acquired with Xcalibur software and analyzed using the Proteome Discoverer v2.3 software (Thermo Fisher ScientificTM). The resulted MS/MS spectra were searched against the G. max reference protein database (Williams 82 a2v1) downloaded from the Phytozome portal1 with Proteome Discoverer internal engine SEQUEST HT. The false discovery rate (FDR) was calculated and assigned to matched peptides by Percolator (Käll et al., 2007). Peptides with an FDR-value ≤ 0.01 were selected for subsequent analysis. LFQ was performed using Proteome Discoverer comprehensive LFQ consensus workflow. Briefly, unique peptides were used for quantification, MS1 precursor abundance was estimated based on its intensity. Peptide abundance was normalized by total peptide amount and summed into protein abundances. Each time point has three biological replicates. Protein fold changes were calculated based on protein abundance using salt-treated against untreated samples, and imputation was performed with replicate based resampling. An ANOVA test was employed to test differentially expressed proteins (DEPs), p-value was corrected by BH method (Benjamini and Hochberg, 1995). Proteins detected in all replicates of any one of the two samples with an adjusted p-value ≤ 0.05 were selected as DEPs. Common and specific sets of identified proteins as well as DEPs were visualized by Venny 2.1 (Oliveros, 2007). The protein abundance identified in all three replicates was used to conduct principal component analysis (PCA), and batch effects were removed by the removeBatchEffect function of limma package (Smyth and Speed, 2003).
Gene Ontology (GO) enrichment was performed on AgriGO v2.02 using GO slim annotation (Tian et al., 2017). The reference William 82 a2v1 was set as background. The Fisher exact test was employed and the p-value was corrected by the Yekutieli method (Yekutieli and Benjamini, 2001). GO terms were selected using a cut-off of the adjusted p-value ≤ 0.05 and the minimum number of mapping entries ≥5. Annotation from the Kyoto encyclopedia genes and genomes (KEGG) database was used for pathway enrichment analysis with KOBAS v2.0 (Xie et al., 2011). Protein sequences of input genes were searched against KEGG G. max terms by BLASTP software (Camacho et al., 2009). The hypergeometric test was further conducted to test enrichment. The matched hits were filtered by coverage ≥80% and identity ≥99%. The prediction of protein subcellular localisation was performed on ProtComp9.03.
Results
Salt-Induced GL and PL Changes in Soybean Leaves
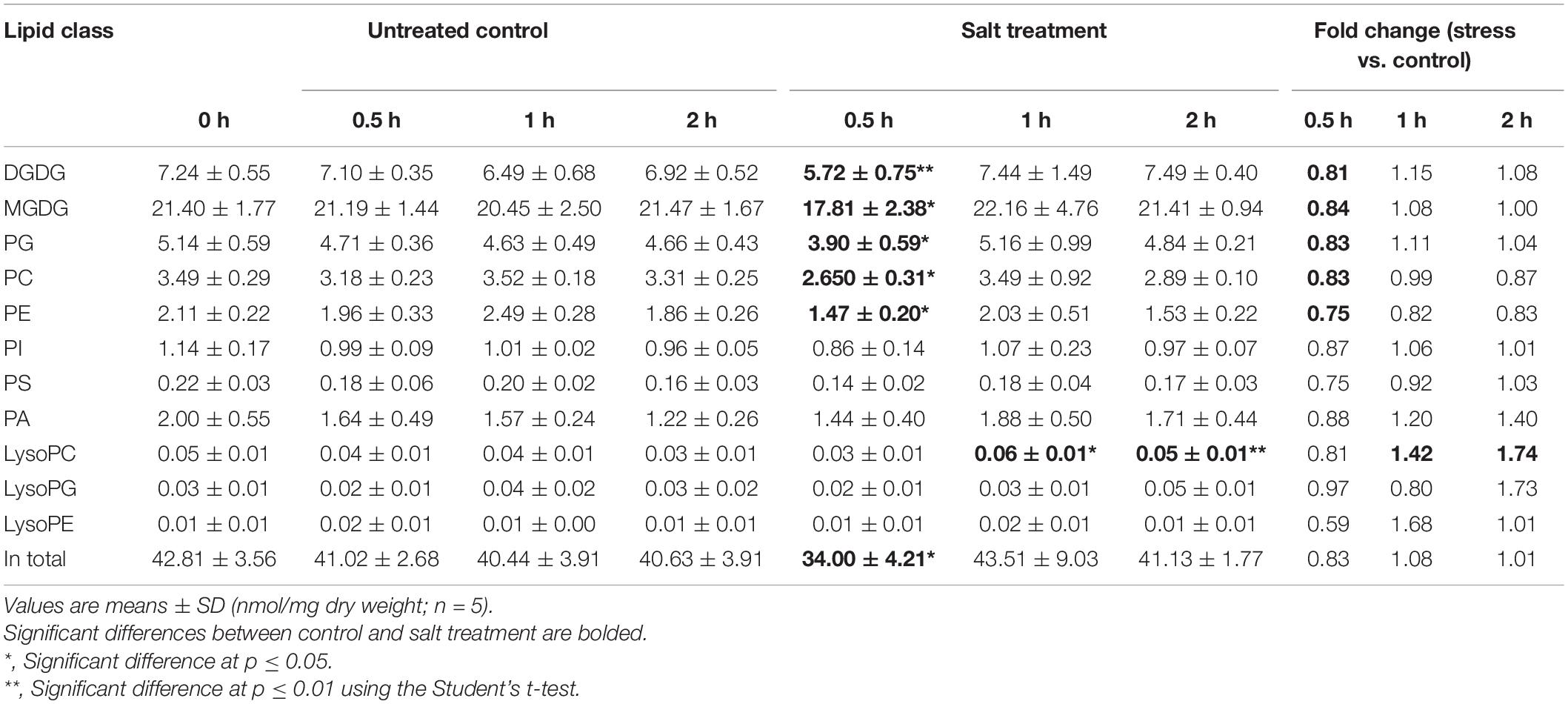
On tandem mass spectrometry (Table 1), lipid species including GLs (MGDG and DGDG), PLs (PC, PE, PG, PI, PA, and PS), and lysoPLs (lysoPC, lysoPE, and lysoPG) were identified in soybean leaf samples, and the five species most abundant in soybean leaves includes MGDG (50.0–52.8%), DGDG (16.0–18.2%), PG (11.5–12.0%), PE (7.8–8.2%), and PC (3.7–6.2%) (Supplementary Figure 1). The total content as well as several membrane lipid classes (DGDG, MGDG, PG, PC, and PE) in soybean leaves showed a rapid reduction (0.8-fold) at 0.5 h of salt treatment, which reverted to normal at 1 h (Table 1). LysoPC increased by 1.4-fold at 1 h and 1.7-fold at 2 h of salt treatment (Table 1). For PA, a critical intermediate in lipid biosynthesis in both the chloroplasts and the ER, PA (34:1) accumulated at 1 and 2 h, and PA (36:3) and PA (36:6) were elevated at 2 h (Figure 1 and Supplementary Table 1). For MGDG, DGDG, and PG, forming major components of chloroplast membranes primarily derived from PA in the plastids, abundant acyl species were MGDG (36:6), DGDG (34:3), DGDG (36:6), PG (32:1), and PG (34:2) (Supplementary Table 1). After salt treatment, MGDG (36:6), DGDG (36:6), and DGDG (34:3) significantly dipped at 0.5 h, but reverted to normal at 1 h (Figure 1 and Supplementary Table 1). Of the three, only DGDG (34:3) rose markedly at 2 h after salt treatment (Figure 1 and Supplementary Table 1). Moreover, the ratio of DGDG/MGDG, important in maintaining chloroplast membrane stability, increased at 1 and 2 h (Supplementary Figure 2). Amongst the PG acyl species, only PG (34:1) showed significant decrease at 0.5 h of salt treatment (Figure 1 and Supplementary Table 1).
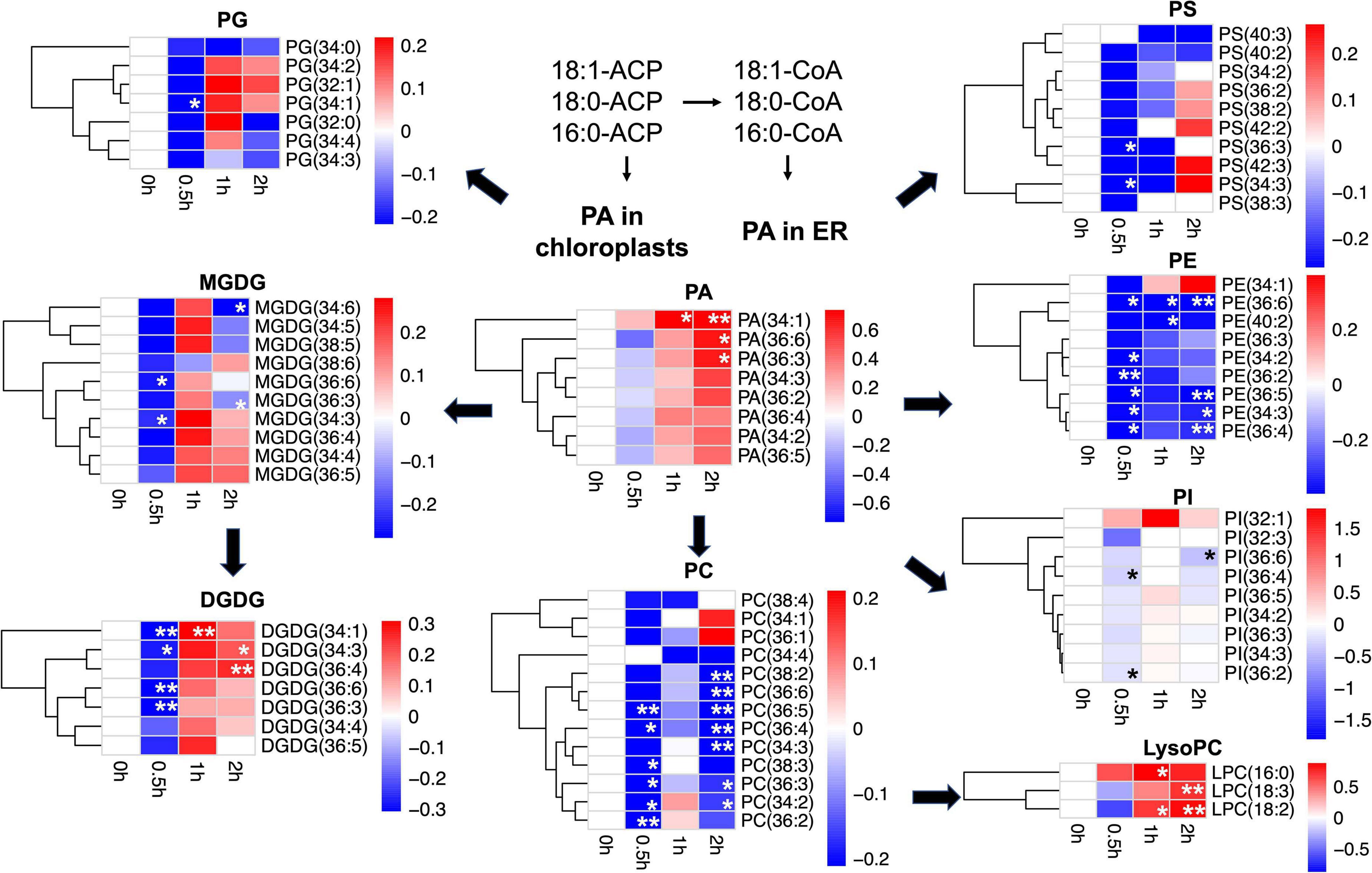
Figure 1. Changes in lipid acyl species under salt stress in leaves of soybean (Glycine max [L.] Merr.) accession C08. Heatmaps showing the log2-fold changes in each affected lipid acyl species at 0.5, 1, and 2 h of salt treatment. PA as lipid metabolic intermediates in the endoplasmic reticulum (ER) are derived from 16:0, 18:0-ACP, and 18:1-ACP plastidial de novo fatty acid synthesis. Total lipids were extracted from leaves of 14-day-old C08 seedlings grown in 1/2 strength Hoagland solution and treated with or without 0.9% (w/v) NaCl. The lipid molecular species were quantified by tandem mass spectrometry. Scale bar represents log2-fold changes. *, significant difference at p ≤ 0.05; **, significant difference at p ≤ 0.01 using the Student’s t-test. The raw data is provided in Supplementary Table 1. ACP, acyl carrier protein; DGDG, digalactosyldiacylglycerol; LysoPC, lysophosphatidylcholine; MGDG, monogalactosyldiacylglycerol; PA, phosphatidic acid; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PG, phosphatidylglycerol; PI, phosphatidylinositol; PS, phosphatidylserine.
When PC and PE, the major PLs in the cell membranes formed from PA in the ER were analyzed, it was observed that PC acyl species including PC (36:3), PC (36:4), PC (36:5), and PC (34:2) were lower at 0.5 h after salt treatment, recovered at 1 h, and dropped again at 2 h (Figure 1 and Supplementary Table 1). In contrast, all detected molecular species of lysoPC, including lysoPC (16:0), lysoPC (18:2), and lysoPC (18:3) were elevated only at 1 or 2 h (Figure 1 and Supplementary Table 1). Similar to PC, most molecular species of PE declined after 0.5 h and again at 2 h, with PE (36:6) reduced at all the time points (Figure 1 and Supplementary Table 1). The ratio of PC/PE for 34:3, 34:2, 36:6, 36:5, and 36:2 increased at 1 h (Supplementary Table 2). Also, the PI species 36:2 and 36:4 were lower at 0.5 h and 36:6 declined after 2 h of salt treatment (Figure 1 and Supplementary Table 1). Moreover, the unsaturation levels of certain lipid species decreased, primarily in MGDG, PC, PE, PS, and PA at 1 and 2 h after salt treatment (Supplementary Table 3). These changes in various membrane lipid molecular species indicate that salt stress altered membrane lipids dynamically in C08 leaves.
Proteomic Analysis of C08 Leaves Under Salt Stress
Given that lipid turnover in soybean leaves is facilitated by a series of metabolic enzymes, regulatory proteins and their related transporters, to identify the molecular mechanisms in lipid modulation, proteome profiling was performed in parallel to lipid profiling. In total, 2,399, 2,563, 2,499, and 2,049 master proteins were detected at 0, 0.5, 1, and 2 h, respectively, after salt treatment (Table 2). When the entire proteome was analyzed by PCA, clear differences among time points were observed with three biological replicates of each time point tending to cluster together (Figure 2A). Venn diagram analysis of identified proteins from all time points showed that 1,640 master proteins were common, and 148, 117, 95, and 81 unique proteins exist at 0, 0.5, 1, and 2 h, respectively (Figure 2B).
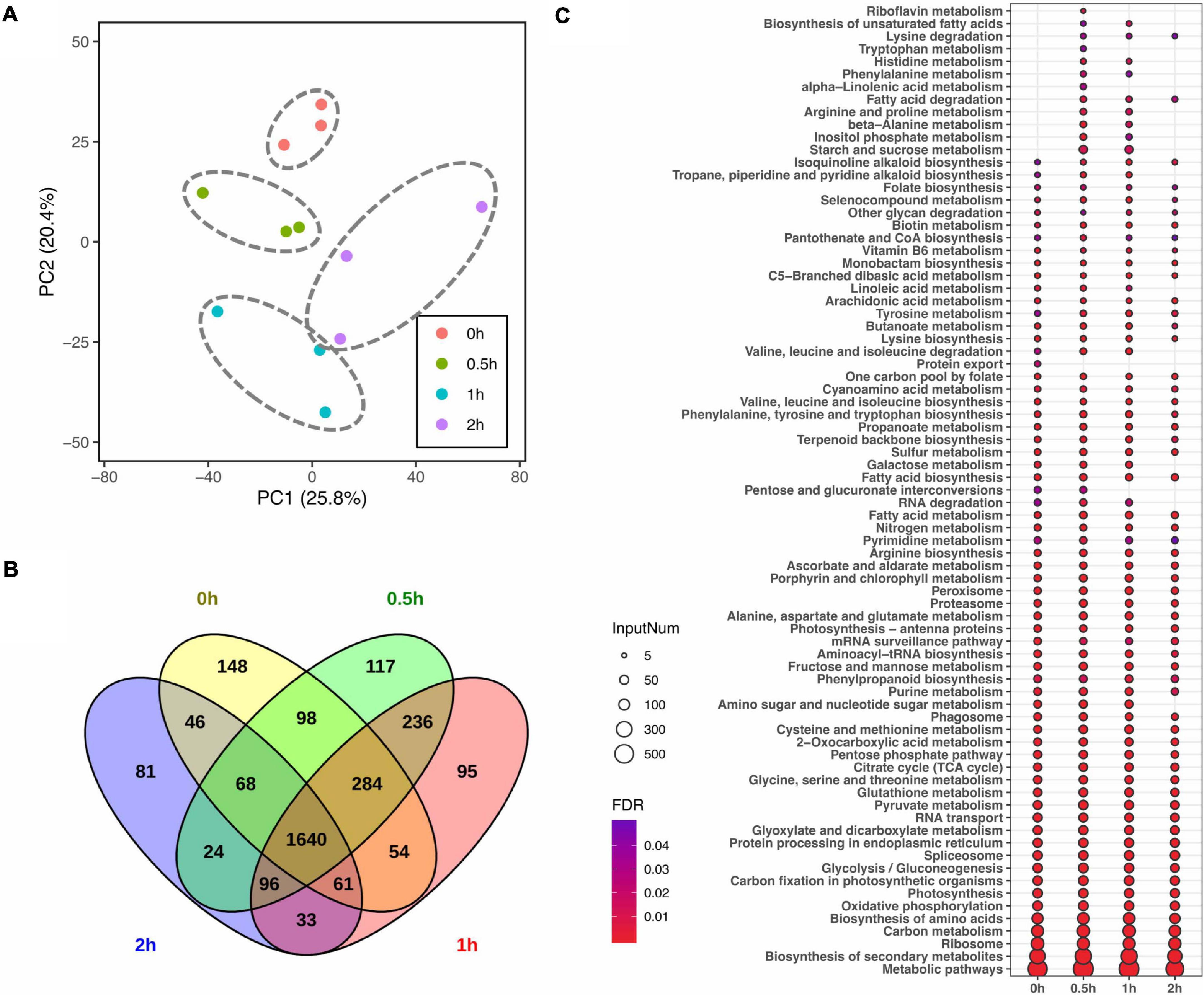
Figure 2. Impact of salt treatment for 0, 0.5, 1, and 2 h on leaf proteome of soybean (Glycine max [L.] Merr.) accession C08. Total leaf protein was extracted and analyzed by tandem mass spectrometry from 14-day-old C08 seedlings grown in 1/2 strength Hoagland solution and treated with or without 0.9% (w/v) NaCl. (A) PCA of proteome profiles. Normalized protein abundance was Log2 transformed and used as input data, centring and scaling was performed prior than calculating principle components. The first and second principal components were visualized in dot plot, and the variation was labeled in the brackets. (B) Venn diagram of identified proteins in each time point. (C) Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis of identified proteins from each time point. The color gradient represents the value of a false discovery rate, with a cut-off ≤0.05. The size of each circle represents the input number of proteins in the enriched pathway. The protein lists of enriched pathways are provided in Supplementary Table 4.
Pathway enrichment analysis was conducted by identifying KEGG pathways with all identified proteins with or without salt treatment. In comparison to 0 h, 11 and 7 metabolic or signaling pathways were enriched at 0.5 and 1 h, including pathways associated with amino acid (aa) biosynthesis and degradation, starch and sucrose metabolism, fatty acid (FA) metabolism and secondary metabolite biosynthesis pathways (Figure 2C and Supplementary Table 4). Furthermore, protein export was specially enriched at 0 h. The number of proteins enriched in biotin metabolism and FA biosynthesis increased after salt stress, while those associated with nucleotide excision repair and RNA degradation pathway declined (Figure 2C and Supplementary Table 4).
To identify differential protein abundance at each time point in C08 leaves under salt stress, LFQ analysis was carried out and the fold changes of protein abundance under stress vs. the control (i.e., 0.5/0, 1/0, and 2/0 h) calculated. The results revealed that 120 (81 upregulated), 181 (78 upregulated), and 206 (91 upregulated) DEPs were identified after 0.5, 1, and 2 h, respectively, of salt treatment in comparison to the untreated sample (Figure 3A and Supplementary Table 5). GO analysis of DEPs from 0.5, 1, and 2 h is shown in Figure 3B and Supplementary Table 6. In the biological process category, for enrichment at 1 h DEPs were associated mainly with amino acid metabolism, nitrogen compound biosynthesis, while DEPs at 2 h clustered more in steroid metabolism, lipid biosynthesis, oxidation reduction, and translation process (Figure 3B). Some stress responsive proteins including dehydrin and ROS scavenging enzyme glutathione S-transferase (GST) and heat shock proteins (HSPs) which accumulated after salt treatment corresponded with their induced transcription (Supplementary Table 7). Moreover, a major proportion DEPs was predicted to localize in the chloroplasts, mitochondria, nucleus, and cytoplasm after salt treatment (Supplementary Figure 3).
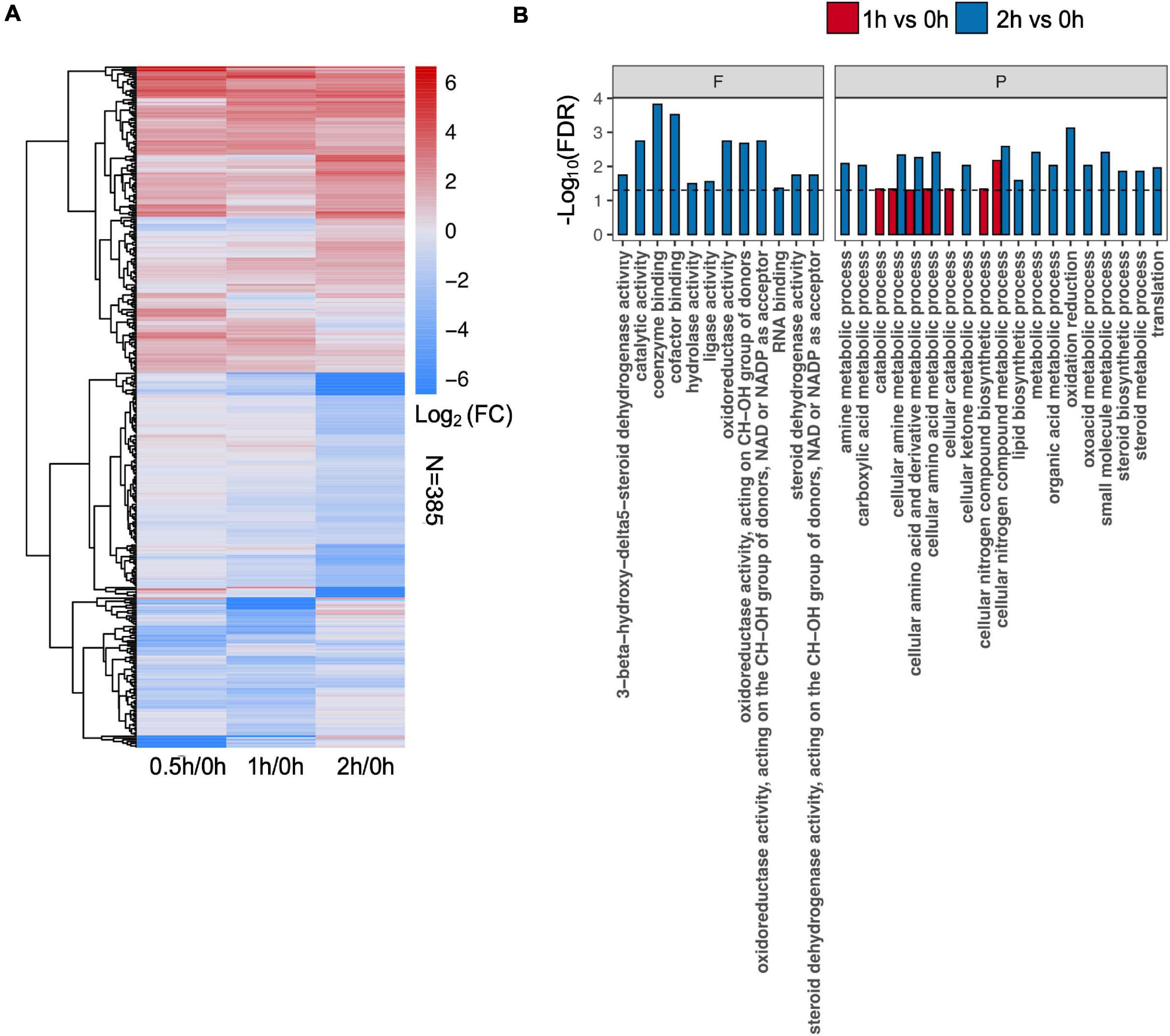
Figure 3. Effect of salt treatment on the soybean (Glycine max [L.] Merr.) accession C08 leaf proteome. (A) Heatmap displays the log2-fold changes in abundance of significant differentially expressed proteins (DEPs) (stress vs. normal control), adjusted p-value ≤ 0.05. The union set was used to generate the plot. (B) Gene ontology (GO) enrichment of DEPs at each time point compared to 0 h sample. Three categories of GO terms including “cellular component,” “biological process” (P), and “molecular function” (F) were used, only items in the last two were found enriched. Dashed line indicates FDR = 0.05. The protein lists of enriched items are provided in Supplementary Table 6.
Alteration of de novo FA and JA Biosynthetic Enzymes in the Chloroplasts During Salt Stress
To further explore the possible mechanisms underlining redistribution of membrane lipids at the early stages of salt treatment, various proteins from the chloroplasts associated with FA and jasmonic acid (JA) biosyntheses were analyzed. Major enzymes related to de novo FAS were detected with no obvious changes, including the rate-limiting enzyme acetyl-CoA carboxylase (ACC), 3-oxoacyl-ACP synthase I, II, III, 3-hydroxyacyl-ACP dehydratase, enoyl-ACP reductase, oleoyl-ACP thioesterase, and stearoyl-ACP desaturase (Figure 4 and Supplementary Table 8).
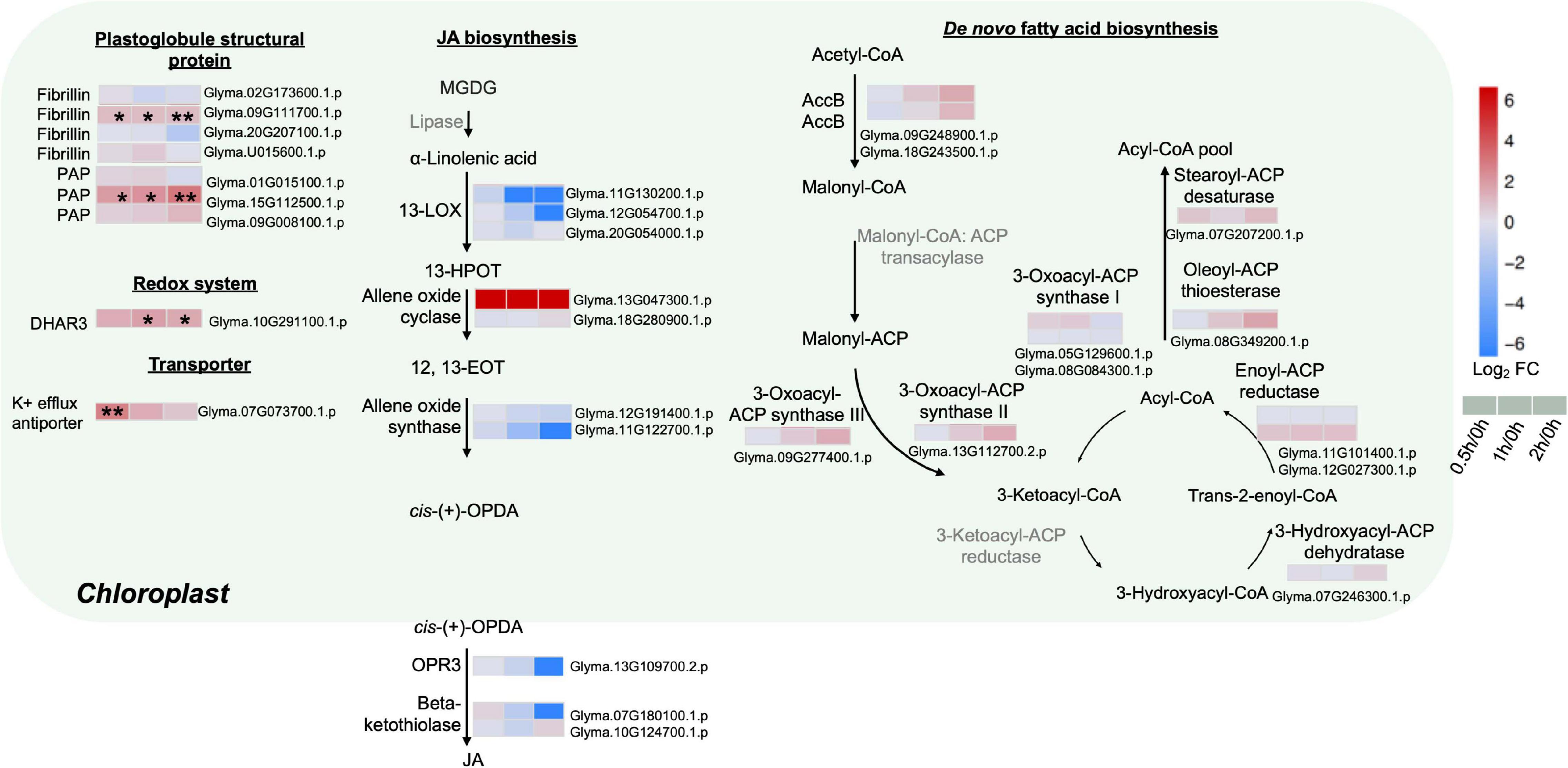
Figure 4. Proteins with differential abundance related to various metabolic pathways in the chloroplasts and peroxisomes during salt stress in leaves of soybean (Glycine max [L.] Merr.) accession C08. Each significant differentially expressed protein is presented as heatmaps with shades of red or blue according to the scale bar. Scale bar indicates log2-fold changes of protein abundance (stress vs. control) with an adjusted p-value ≤ 0.05. *, significant difference at p ≤ 0.05; **, significant difference at p ≤ 0.01 using ANOVA test. For tandem mass spectrometry, the total leaf protein was extracted from leaves of 14-day-old C08 seedlings grown in 1/2 strength Hoagland solution and treated with or without 0.9% (w/v) NaCl. Differential protein abundances were calculated by label-free quantification of MS peptide signals. AccB, biotin carboxyl carrier protein of acetyl-CoA carboxylase; ACP, acyl carrier protein; DHAR3, dehydroascorbate reductase 3; 12, 13-EOT, 12, 13-epoxyoctadeca-9, 11, 15-trienoic acid; 13-HPOT, 13 hydroperoxyoctadeca-9, 11, 15-trienoic acid; JA, jasmonic acid; 13-LOX, 13-lipoxygenases; OPR3, 12-oxophytodienoate reductase 3; PAP, plastid lipid-associated protein.
The enzymes related to the biosynthesis of JA, a fatty acid-derived phytohormone regulating abiotic stress responses in plants, were investigated (Figure 4 and Supplementary Table 8). Most key enzymes chloroplast-localized 13S-lipoxygenase (13-LOX), allene oxide cyclase (AOC), allene oxide synthase (AOS), 12-oxophytodienoate reductase 3 (OPR3) as well as the peroxisome-localized β-ketothiolase were identified with no obvious changes in our results (Figure 4 and Supplementary Table 8). Only one AOC (Glyma.13G047300.1.p) was detected only after salt treatment (Figure 4 and Supplementary Table 8).
As stress triggers membrane lipid degradation and the resultant polyunsaturated fatty acids (PUFAs) can be stored as triacylglycerol (TAG) in the plastoglobules (lipid bodies in the chloroplasts), it was not surprising that plastoglobule structural proteins, fibrillin and plastid-lipid-associated protein (PAP) (homologs of the fibrillin family) increased at 0.5, 1 and 2 h of salt treatment (Figure 4 and Supplementary Table 8). Fibrillin (Glyma.09G111700.1.p) and PAP (Glyma.15G112500.1.p) were induced continuously from 0.5 to 2 h of salt treatment (Figure 4 and Supplementary Table 8). Moreover, other plastidial proteins were also identified. The enzymatic antioxidant dehydroascorbate reductase 3 (DHR3) was elevated by three-fold at 2 h. The K+ efflux antiporter, which facilitates endosomal pH homeostasis and salt tolerance, increased at 0.5 h (Figure 4 and Supplementary Table 8).
Mitochondrial TCA Cycle and Cytosolic Glycolysis Enhanced During Salt Stress
Given that energy production processes such as glycolysis and TCA cycle are known to respond to early salt exposure, analysis on proteins related to these two pathways were conducted. The major enzymes of the TCA cycle including citrate synthase, malate dehydrogenase, succinate dehydrogenase, succinate synthetase, α-ketoglutarate, and isocitrate dehydrogenase were detected (Figure 5 and Supplementary Table 8). An isocitrate dehydrogenase (Glyma.10G058100.1.p) was upregulated at least 7.8-fold during salt treatment (Figure 5 and Supplementary Table 8). Some enzymes of the cytoplasmic glycolysis pathway also increased. A hexokinase (Glyma.07G124500.1.p) rose five-fold at 0.5 h, while an enolase (Glyma.16G204600.1.p) increased four-fold at 0.5 and 2 h after salt treatment (Figure 5 and Supplementary Table 8). Proteome data indicated a three-fold increase in pyruvate dehydrogenase (PDH) complex E1 (Glyma.14G186900.2.p) at 0.5 h of salt treatment (Figure 5 and Supplementary Table 8). PDH is a vital metabolic enzyme that links glycolysis to the TCA cycle by converting pyruvate to acetyl-CoA, and increasing the influx of acetyl-CoA from glycolysis into the TCA cycle. These results indicate that in C08 leaves, the TCA cycle and the glycolysis pathway were enhanced by salt treatment.
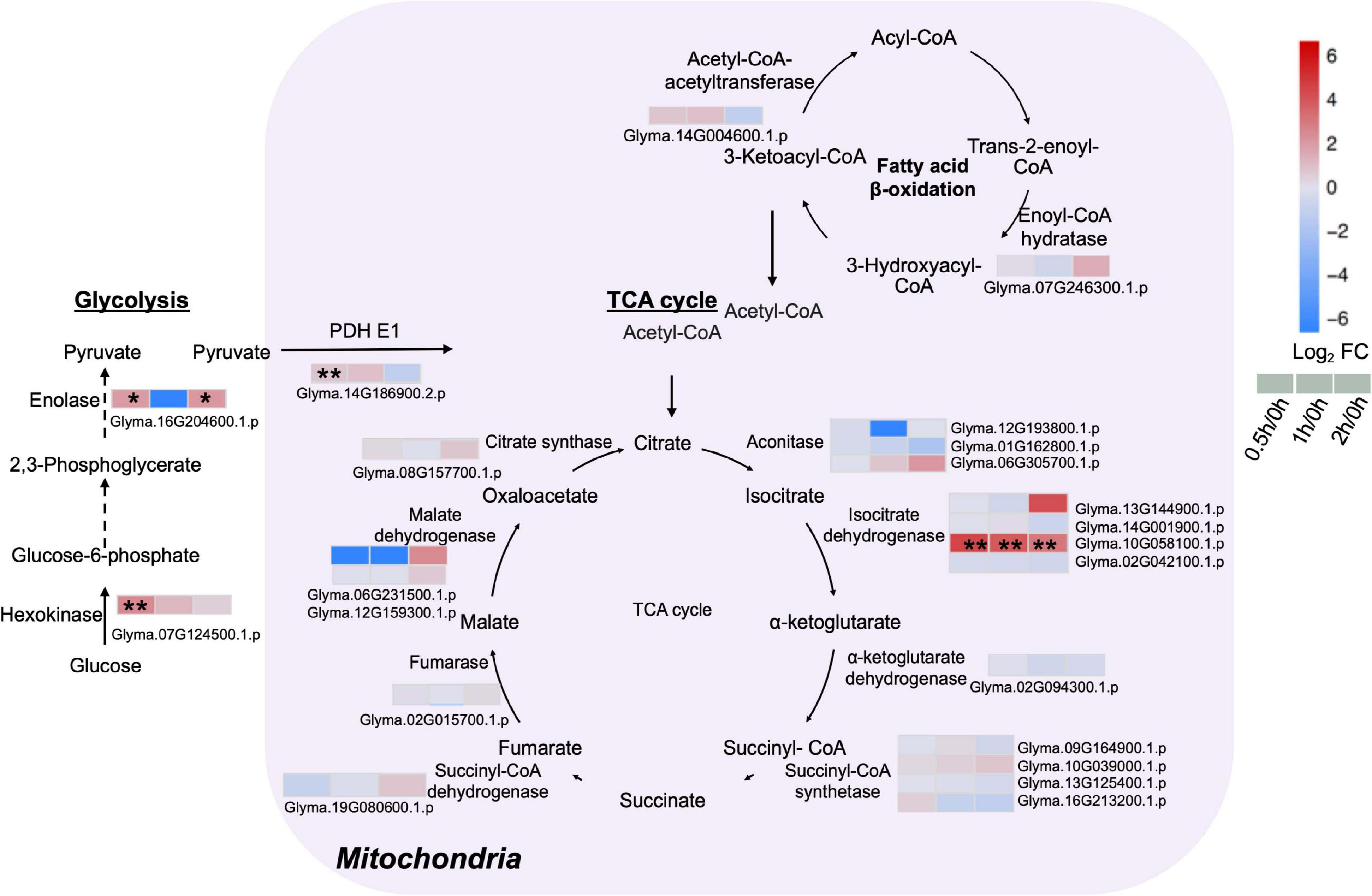
Figure 5. Proteins with differential abundance involved in the tricarboxylic acid (TCA) cycle, glycolysis and fatty acid β-oxidation pathway during salt stress in leaves of soybean (Glycine max [L.] Merr.) accession C08. Each significant differentially expressed protein is presented as heatmaps with shades of red or blue according to the scale bar. Scale bar indicates log2-fold changes of protein abundance (stress vs. control) with an adjusted p-value ≤ 0.05. *, significant difference at p ≤ 0.05; **, significant difference at p ≤ 0.01 using ANOVA test. Black arrow, reaction between two intermediates; dashed arrow, several steps in the reaction. For tandem mass spectrometry, the total leaf protein was extracted from leaves of 14-day-old C08 seedlings grown in 1/2 strength Hoagland solution and treated with or without 0.9% (w/v) NaCl. Differential protein abundances were calculated by label-free quantification of MS peptide signals. PDH E1, pyruvate dehydrogenase E1 component.
PI Signaling and PL Metabolism in the ER Are Affected by Salt Treatment
Our proteome data detected three phosphatidylinositol-4-phosphate 5-kinases (PI4P5Ks), of which one (Glyma.13G062700.1.p) was elevated continuously from 0.5 to 2 h (Figure 6 and Supplementary Table 8). Phosphatidylinositol phospholipase C (PI-PLC), responsible for converting PI (4,5)P2 to inositol 1,4,5-trisphosphate (IP3) and DAG, was detected but not altered in our dataset (Figure 6 and Supplementary Table 8).
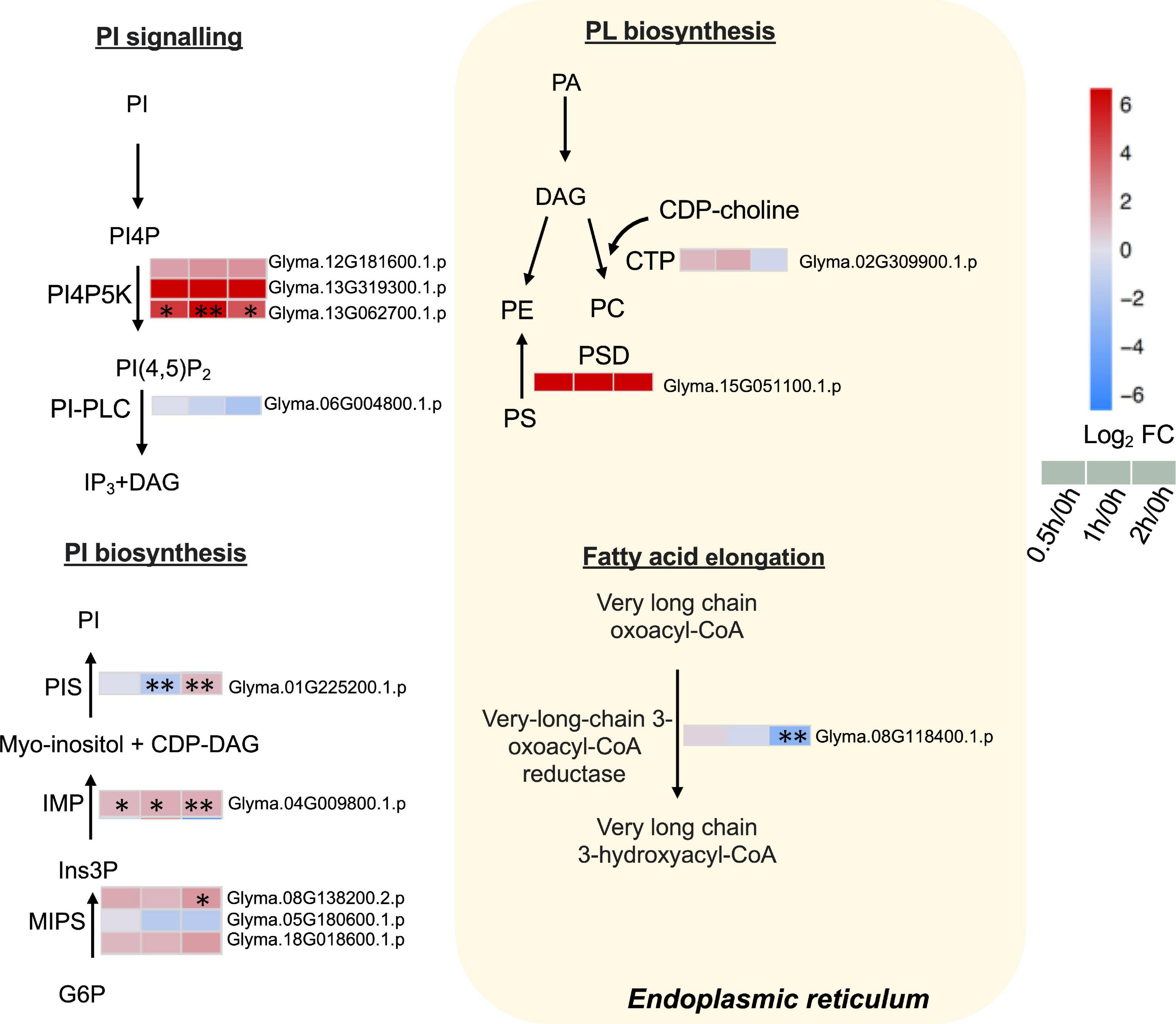
Figure 6. Proteins with differential abundance involved in phospholipid metabolic pathways during salt stress in leaves of soybean (Glycine max [L.] Merr.) accession C08. Each significant differentially expressed protein is presented as heatmaps with shades of red or blue according to the scale bar. Scale bar indicates log2-fold changes of protein abundance (stress vs. control) with an adjusted p-value ≤ 0.05. ∗, significant difference at p ≤ 0.05; ∗∗, significant difference at p ≤ 0.01 using ANOVA test. For tandem mass spectrometry, the total leaf protein was extracted from leaves of 14-day-old C08 seedlings grown in 1/2 strength Hoagland solution and treated with or without 0.9% (w/v) NaCl. Differential protein abundances were calculated by label-free quantification of MS peptide signals. CDP-choline, cytidine diphosphate choline; CDP-DAG, cytidine diphosphate diacylglycerol; PA, phosphatidic; CTP, choline-phosphate cytidylyltransferase; DAG, diacylglycerol; G6P, glucose 6-phosphate; Ins3P, myo–inositol–3–phosphate; IMP, inositol monophosphatase; MIPS, myo-inositol-1-phosphate synthase; PA, phosphatidic acid; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PI, phosphatidylinositol; PI(4,5)P2, phosphatidylinositol 4, 5-bisphosphate; PI4P, phosphatidylinositol 4-phosphate; PI4PK5, phosphatidylinositol-4-phosphate 5-kinase; PI-PLC, phosphoinositide phospholipase C; PIS, phosphatidylinositol synthase; PS, phosphatidylserine; PS decarboxylase, phosphatidylserine decarboxylase.
Furthermore, the key enzymes related to PI biosynthesis accumulated after salt treatment. The ER-localized PI synthase (PIS) decreased by 0.3-fold at 1 h and increased by 2.3-fold at 2 h (Figure 6 and Supplementary Table 8). The myo-inositol-3-phosphate synthase (MIPS, Glyma.08G138200.2.p) was induced 4.4-fold at 2 h and inositol monophosphatase (IMP) (Glyma.04G009800.1.p) was upregulated from 0.5 to 2 h salt treatment (Figure 6 and Supplementary Table 8). Moreover, when other enzymes related membrane PL biosynthesis in the ER were examined, the results revealed that choline-phosphate cytidylyltransferase (CTP) essential in PC formation was not altered after salt treatment (Figure 6 and Supplementary Table 8). PS decarboxylase (PSD) (Glyma.15G051100.1.p) that converts PS to PE, was only identified after salt treatment (Figure 6 and Supplementary Table 8). The very-long-chain 3-oxoacyl-CoA reductase (KCR), required for the elongation of fatty acids, had decreased at 2 h of salt treatment (Figure 6 and Supplementary Table 8).
Discussion
Salt Stress Affects Membrane Lipid Composition
Earlier studies have shown the repercussion of salt stress on metabolites (Li et al., 2017), carbohydrate pool rebalancing aa metabolism (Zhang et al., 2017) and protein turnover (Kosová et al., 2013) as well as the maintenance of primary N homeostasis in various crops (Wang et al., 2012). Here we report on how high salt impacted membrane lipid composition and reveal a correlation in lipid metabolism with the remodulation of C and N pools upon salt treatment. Integration of lipidome and proteome data in a time series better facilitated an understanding on lipid turnover and protein alteration under salt stress. Salinity causes cell membrane breakdown and electrolyte leakage in soybean (Phang et al., 2008) and affects lipid composition (Shoemaker and Vanderlick, 2002). Leaf drooping was observed upon 30 min exposure to 0.9% salt solution, and recovery was evident at 1 h (Liu et al., 2019). Consistent with this, our lipid profiling of salt-treated soybean leaves showed a sharp decrease in total lipid content at 0.5 h of salt treatment which returned to normal at 1 h. The reduction and recovery of total lipids indicate that lipid composition could be rebalanced within a short period of time upon salt treatment of C08 leaves.
Our results showed a reduction in the major MGDG acyl species and most DGDG acyl species at 0.5 h of salt stress (Figure 1 and Supplementary Table 1), consistent with salt-triggering GL reduction in leaves of rice, Cucumis sativus and Vigna unguiculata (De Paula et al., 1990; Yamane et al., 2004; Shu et al., 2012). Decreases in DGDG and MGDG are known to adversely affect photosynthetic membranes and membrane protein activity in the thylakoids (Simidjiev et al., 2000), and reduction of MGDG is a typical response to osmotic stress arising from salinity, drought or freezing in Arabidopsis (Welti et al., 2002; Gigon et al., 2004). Besides, an increased ratio in DGDG/MGDG at 1 and 2 h of salt treatment (Supplementary Figure 1) is consistent with a higher ratio in DGDG/MGDG in cowpea leaves after drought stress enhanced thylakoid stability and maintained the bilayer structure through the synthesis of stabilizing DGDG lipid species (Torres-Franklin et al., 2007). Therefore, a reduction in the major species of DGDG and MGDG and an increase in the DGDG/MGDG ratio of salt treatment may be a consequence of the disruption in chloroplast membrane stability and photosystem function at the onset of salt treatment.
Most PC and PE species declined at 0.5 h and again at 2 h after salt treatment (Figure 1 and Supplementary Table 1), consistent with a degradation in major acyl species of PC and PE under drought stress in Arabidopsis leaves and winter wheat seedlings (Gigon et al., 2004; Wang et al., 2020). PC and PE are the major PLs in the structural membranes, and PC is known to be involved in the adaption to abiotic stresses (Tasseva et al., 2004). Increase in PC species following salt stress has been reported in salt-tolerant plants including Catharanthus roseus and Mesembryanthemum crystallinum (Elkahoui et al., 2004; Barkla et al., 2018) while a decrease in PC species occurs in the salt-sensitive plants such as oats and wheat (Norberg and Liljenberg, 1991; Magdy et al., 1994).
Salt Stress Elevated Plastidial Proteins Related to Plastoglobules and JA Biosynthesis
Our results showed that AOC, a key enzyme in JA production, accumulated at 0.5, 1, and 2 h after salt treatment (Figure 4 and Supplementary Table 8). JA, a major phytohormone regulating abiotic stress responses, is important in early responses to osmotic and salt stress (Fujita et al., 2006; Chen et al., 2017) and its biosynthesis was triggered by salt in Arabidopsis (Ishiguro et al., 2001; Wang et al., 2015, 2018). AOC affects the rate of flux through the JA biosynthetic pathway (Yoeun et al., 2018). Overexpression of TaAOC1 conferred salt tolerance in wheat (Zhao et al., 2014). Using 2-DE and MALDI-TOF-MS/MS, higher AOC was reported in Medicago sativa following early salt stress (Xiong et al., 2017) while upregulated AtAOC2 protein abundance was reported in Arabidopsis under salt stress using 2-DE and iTRAQ LC-MS/MS (Pang et al., 2010). Our results lead support to lead support to an enhancement of JA production following salt treatment.
The upregulation of plastoglobule structural proteins was evident at the early stages of salt treatment (Figure 4 and Supplementary Table 8). Plastoglobules are known to play an essential role in lipid remodeling of the thylakoids (Rottet et al., 2015) and increase in size and number following FA accumulation under stress conditions (Bréhélin and Kessler, 2008; Rottet et al., 2015). They have been reported to be the site for the initiation of chloroplast stress-related JA biosynthesis (Hölzl and Dörmann, 2019). The accumulation of fibrillin FBN1-2 was found to affect JA biosynthesis under photosynthetic stress (Youssef et al., 2010). In Arabidopsis, FIBRILLIN2 protects photosystem II against abiotic stress by interacting with AOS and LOX of JA biosynthesis (Torres-Romero et al., 2020). Our results suggest that the breakdown of chloroplast membrane lipids (MGDG and DGDG) would lead to the production of PUFAs (Boudière et al., 2014), which act as substrates for JA biosynthesis at the onset of salt stress (Figure 4 and Supplementary Table 8).
Enhanced Mitochondrial TCA Cycle and Cytosolic Glycolysis at the Onset of Salt Treatment
Some enzymes involved in the TCA cycle and glycolysis pathway increased after salt treatment (Figure 5 and Supplementary Table 8), consistent with other proteome studies reported that major metabolic pathways in plants involved in energy generation (glycolysis, pyruvate decarboxylation and TCA cycle) induced by salt (Hossain et al., 2013). The TCA cycle is a crucial component of respiratory metabolism and an increased respiration after salinity exposure represents a short-term adjustment in demand for energy consumption (Bloom and Epstein, 1984). When salt-sensitive and salt-tolerant barley cultivars were subjected to short-term salinity exposure, both accumulated TCA cycle intermediates in the root elongation zone due to increased energy demand for cell division (Shelden et al., 2016). Pyruvate accumulated in both salt-sensitive and salt-tolerant soybean varieties under salt stress (Zhang et al., 2016). Pyruvate, the final product of the glycolysis pathway, can be converted to acetyl-CoA by the PDH complex and enter into the TCA cycle (Schertl and Braun, 2014). A higher level of certain enzymes in the glycolysis pathway and the TCA cycle during salt treatment (Figure 5 and Supplementary Table 8) enables accelerated respiration (Bloom and Epstein, 1984), to accommodate increased FAS and salt stress adaption in C08 leaves.
PI Signaling and Biosynthesis Triggered at the Onset of Salt Stress
Phosphoinositides, vital lipid signaling molecules in response to salt, are derived from PI by the action of lipid kinases and phosphatases (Heilmann and Heilmann, 2015). PI4P5K catalyzes phosphatidylinositol 4-phosphate (PI4P) and generate PI(4,5)P2 (Delage et al., 2013). Our data revealed an accumulation of PI4P5K at the initiation of salt treatment (Figure 6 and Supplementary Table 8). PI(4,5)P2 is a secondary messenger that functions in recruitment of signaling complexes to specific membrane locations (Martin, 1998). PI(4,5)P2 is synthesized by PI4P5K which is the flux-limiting step in plant phosphoinositide metabolism (Yang et al., 2007). PI4P5K could interact with PI(4,5)P2 and channel them toward different targets (Heilmann and Heilmann, 2015). While in response to salt stress, PI(4,5)P2 levels rapidly (<30 min) rose in rice leaves (Darwish et al., 2009). In Arabidopsis, the expression of PI4P5K was induced by salt stress within 1 h (Mikami et al., 1998). Our results also support the early response of PI4P5K following salt stress. Increase in PI4P5K will supply PI(4,5)P2 as substrates for phospholipases (Stenzel et al., 2008).
Enzymes involved in PI biosynthesis including IMP, MIPS, and PIS accumulated during salt treatment (Figure 6 and Supplementary Table 8). MIPS, the first step of myo-inositol generation, coverts glucose 6-phosphate (G6P) to myo−inositol−3−phosphate (Ins3P), which further forms free myo-inositol by IMP (Loewus and Murthy, 2000). MIPS−produced myo−inositol plays a pivotal role in protective mechanisms in salt−tolerant plant species in M. crystallinum (Nelson et al., 1998). PcINO1 encoding MIPS has been reported as a salt response protein in halophytic wild rice Porteresia coarctata Tateoka (Dastidar et al., 2006).
The overexpression of maize ZmPIS enhanced drought tolerance by elevating DGDG and MGDG contents and ABA synthesis in maize (Liu et al., 2013). Our results suggest that the increase of PI biosynthesis may subsequently accelerate the formation of products such as PI(4,5)P2, DAG, InsP3, and PA, boosting PI signaling cascades at the onset of salt stress.
Conclusion
In conclusion, our study shows the impact of short-term salt stress on lipid metabolism in a salt-sensitive soybean germplasm. Lipid remodeling was found to occur as early as 0.5 h of salt treatment coincident with the triggering of the PI signaling pathway. Membrane lipid recycling was observed to be related to an acceleration of plastidial JA biosynthesis and PI production. The TCA cycle and glycolysis pathways were activated for energy metabolism. In summary, salt stress caused an alteration in lipid composition and rapidly activated related protein responses in C08 leaves.
Data Availability Statement
The raw values obtained from the lipidomic data sets are available in Supplementary Table 1 (associated with Table 1 and Figure 1). Gene expression in Supplementary Table 7 was based on data retrieved from our previous study (Liu et al., 2019; NCBI SRA database with accession number SRP132150). The raw data and msf files for mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (https://www.ebi.ac.uk/pride/) partner repository with the dataset identifier PXD023092. The proteomics raw data are available in Supplementary Table 9. If any data sets are unavailable through the links stated above, they can be obtained from the corresponding author on the request.
Author Contributions
M-LC and H-ML planned and designed the research. AL and ZW performed the experiments. ZX analyzed the data. AL, ZX and M-LC wrote the manuscript. All authors have read and approved the manuscript.
Funding
This work was supported by the Wilson and Amelia Wong Endowment Fund to M-LC, Hong Kong Research Grants Council Area of Excellence Scheme (AoE/M-403/16), and the Innovation and Technology Fund (funding support to State Key Laboratory of Agrobiotechnology) of the HKSAR, China to H-ML and M-LC, and Lo Kwee-Seong Biomedical Research Fund awarded to H-ML. Any opinions, findings, conclusions or recommendations expressed in this event/publication do not reflect the views of the Government of the Hong Kong Special Administrative Region or the Innovation and Technology Commission.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Mary Roth and Ruth Welti (Kansas Lipidomics Research Center) for lipid profiling and data analysis as well as Hafiz Mamoon Rehman and Chun-Ning Leung for advice on proteomics data generation and analysis.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.644408/full#supplementary-material
Footnotes
- ^ https://phytozome.jgi.doe.gov/pz/portal.html
- ^ http://systemsbiology.cau.edu.cn/agriGOv2/
- ^ http://www.softberry.com/berry.phtml?topic=protcomppl&group=programs&subgroup=proloc
References
Ahmad, P., Abdel Latef, A. A. H., Rasool, S., Akram, N. A., Ashraf, M., and Gucel, S. (2016). Role of proteomics in crop stress tolerance. Front. Plant Sci. 7:1336. doi: 10.3389/fpls.2016.01336
Arisz, S. A., Testerink, C., and Munnik, T. (2009). Plant PA signaling via diacylglycerol kinase. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1791, 869–875. doi: 10.1016/j.bbalip.2009.04.006
Athenstaedt, K., and Daum, G. (1999). Phosphatidic acid, a key intermediate in lipid metabolism. Eur. J. Biochem. 266, 1–16. doi: 10.1046/j.1432-1327.1999.00822.x
Barkla, B. J., Garibay-Hernández, A., Melzer, M., Rupasinghe, T. W. T., and Roessner, U. (2018). Single cell-type analysis of cellular lipid remodelling in response to salinity in the epidermal bladder cells of the model halophyte Mesembryanthemum crystallinum. Plant. Cell. Environ. 41, 2390–2403. doi: 10.1111/pce.13352
Barrero-Sicilia, C., Silvestre, S., Haslam, R. P., and Michaelson, L. V. (2017). Lipid remodelling: unravelling the response to cold stress in arabidopsis and its extremophile relative Eutrema salsugineum. Plant Sci. 263, 194–200. doi: 10.1016/j.plantsci.2017.07.017
Bates, P. D., Fatihi, A., Snapp, A. R., Carlsson, A. S., Browse, J., and Lu, C. (2012). Acyl editing and headgroup exchange are the major mechanisms that direct polyunsaturated fatty acid flux into triacylglycerols. Plant Physiol. 160, 1530–1539. doi: 10.1104/pp.112.204438
Bejaoui, F., Salas, J. J., Nouairi, I., Smaoui, A., Abdelly, C., Martínez-Force, E., et al. (2016). Changes in chloroplast lipid contents and chloroplast ultrastructure in Sulla carnosa and Sulla coronaria leaves under salt stress. J. Plant Physiol. 198, 32–38. doi: 10.1016/j.jplph.2016.03.018
Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B. 57, 289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x
Block, M. A., Dorne, A. J., Joyard, J., and Douce, R. (1983). Preparation and characterization of membrane fractions enriched in outer and inner envelope membranes from spinach chloroplasts. II. Biochemical characterization. J. Biol. Chem. 258, 13281–13286.
Bloom, A., and Epstein, E. (1984). Varietal differences in salt-induced respiration in barley. Plant Sci. Lett. 35, 1–3. doi: 10.1016/0304-4211(84)90149-4
Boudière, L., Michaud, M., Petroutsos, D., Rébeillé, F., Falconet, D., Bastien, O., et al. (2014). Glycerolipids in photosynthesis: composition, synthesis and trafficking. Biochim. Biophys. Acta Bioenerg. 1837, 470–480. doi: 10.1016/j.bbabio.2013.09.007
Bréhélin, C., and Kessler, F. (2008). The plastoglobule: a bag full of lipid biochemistry tricks. Photochem. Photobiol. 84, 1388–1394. doi: 10.1111/j.1751-1097.2008.00459.x
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: architecture and applications. BMC Bioinformatics 10, 1–9. doi: 10.1186/1471-2105-10-421
Chalbi, N., Hessini, K., Gandour, M., Mohamed, S., Smaoui, A., Abdelly, C., et al. (2013). Are changes in membrane lipids and fatty acid composition related to salt-stress resistance in wild and cultivated barley? J. Plant Nutr. Soil Sci. 176, 138–147. doi: 10.1002/jpln.201100413
Chen, J., Zhang, J., Hu, J., Xiong, W., Du, C., and Lu, M. (2017). Integrated regulatory network reveals the early salt tolerance mechanism of Populus euphratica. Sci. Rep. 7:6769. doi: 10.1038/s41598-017-05240-0
Darwish, E., Testerink, C., Khalil, M., El-Shihy, O., and Munnik, T. (2009). Phospholipid signaling responses in salt-stressed rice leaves. Plant Cell. Physiol. 50, 986–997. doi: 10.1093/pcp/pcp051
Das, A., Eldakak, M., Paudel, B., Kim, D. W., Hemmati, H., Basu, C., et al. (2016). Leaf proteome analysis reveals prospective drought and heat stress response mechanisms in soybean. Biomed. Res. Int. 2016:6021047. doi: 10.1155/2016/6021047
Dastidar, K. G., Maitra, S., Goswami, L., Roy, D., Das, K. P., and Majumder, A. L. (2006). An insight into the molecular basis of salt tolerance of L-myo-inositol 1-P synthase (PcINO1) from Porteresia coarctata (Roxb.) Tateoka, a halophytic wild rice. Plant Physiol. 140, 1279–1296. doi: 10.1104/pp.105.075150
De Paula, F. M., Thi, A. T. P., De Silva, J. V., Justin, A. M., Demandre, C., and Mazliak, P. (1990). Effects of water stress on the molecular species composition of polar lipids from Vigna unguiculata L. leaves. Plant Sci. 66, 185–193. doi: 10.1016/0168-9452(90)90203-Z
Delage, E., Puyaubert, J., Zachowski, A., and Ruelland, E. (2013). Signal transduction pathways involving phosphatidylinositol 4-phosphate and phosphatidylinositol 4,5-bisphosphate: convergences and divergences among eukaryotic kingdoms. Prog. Lipid Res. 52, 1–14. doi: 10.1016/j.plipres.2012.08.003
Devaiah, S. P., Roth, M. R., Baughman, E., Li, M., Tamura, P., Jeannotte, R., et al. (2006). Quantitative profiling of polar glycerolipid species from organs of wild-type arabidopsis and a PHOSPHOLIPASE Dα1 knockout mutant. Phytochemistry 67, 1907–1924. doi: 10.1016/j.phytochem.2006.06.005
Dörmann, P., and Benning, C. (2002). Galactolipids rule in seed plants. Trends Plant Sci. 7, 112–118.
Douce, R. (1974). Site of biosynthesis of galactolipids in spinach chloroplasts. Science 183, 852–853. doi: 10.1126/science.183.4127.852
Elkahoui, S., Smaoui, A., Zarrouk, M., Ghrir, R., and Limam, F. (2004). Salt-induced lipid changes in Catharanthus roseus cultured cell suspensions. Phytochemistry 65, 1911–1917. doi: 10.1016/j.phytochem.2004.06.021
Fujita, M., Fujita, Y., Noutoshi, Y., Takahashi, F., Narusaka, Y., Yamaguchi-Shinozaki, K., et al. (2006). Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks. Curr. Opin. Plant Biol. 9, 436–442. doi: 10.1016/j.pbi.2006.05.014
Gigon, A., Matos, A.-R., Laffray, D., Zuily-Fodil, Y., and Pham-Thi, A.-T. (2004). Effect of drought stress on lipid metabolism in the leaves of Arabidopsis thaliana (Ecotype Columbia). Ann. Bot. 94, 345–351. doi: 10.1093/aob/mch150
Gogna, M., and Bhatla, S. C. (2020). Salt-tolerant and -sensitive seedlings exhibit noteworthy differences in lipolytic events in response to salt stress. Plant Signal. Behav. 15:1737451. doi: 10.1080/15592324.2020.1737451
Guo, Q., Liu, L., and Barkla, B. J. (2019). Membrane lipid remodeling in response to salinity. Int. J. Mol. Sci. 20:4264. doi: 10.3390/ijms20174264
Guschina, I. A., Everard, J. D., Kinney, A. J., Quant, P. A., and Harwood, J. L. (2014). Studies on the regulation of lipid biosynthesis in plants: application of control analysis to soybean. Biochim. Biophys. Acta Biomembr. 1838, 1488–1500. doi: 10.1016/j.bbamem.2014.02.008
Hasanuzzaman, M., Nahar, K., Rahman, A., Mahmud, J. A., Hossain, M. S., and Fujita, M. (2016). “Soybean production and environmental stresses,” in Environmental stresses in soybean production. Amsterdam, Netherlands: Elsevier, 61–102. doi: 10.1016/B978-0-12-801535-3.00004-8
He, M., Qin, C.-X., Wang, X., and Ding, N.-Z. (2020). Plant unsaturated fatty acids: biosynthesis and regulation. Front. Plant Sci. 11:390. doi: 10.3389/fpls.2020.00390
Heilmann, M., and Heilmann, I. (2015). Plant phosphoinositides complex networks controlling growth and adaptation. Biochim. Biophys. Acta 1851, 759–769. doi: 10.1016/j.bbalip.2014.09.018
Higashi, Y., Okazaki, Y., Myouga, F., Shinozaki, K., and Saito, K. (2015). Landscape of the lipidome and transcriptome under heat stress in Arabidopsis thaliana. Sci. Rep. 5:10533. doi: 10.1038/srep10533
Hoagland, D. R., and Arnon, D. I. (1950). The water-culture method for growing plants without soil. Circular. California Agric. Exp. Station 347:32.
Hölzl, G., and Dörmann, P. (2019). Chloroplast lipids and their biosynthesis. Annu. Rev. Plant Biol. 70, 51–81. doi: 10.1146/annurev-arplant-050718-100202
Hossain, Z., Khatoon, A., and Komatsu, S. (2013). Soybean proteomics for unraveling abiotic stress response mechanism. J. Proteome Res. 12, 4670–4684. doi: 10.1021/pr400604b
Ishiguro, S., Kawai-Oda, A., Ueda, J., Nishida, I., and Okada, K. (2001). The DEFECTIVE IN ANTHER DEHISCENCE1 gene encodes a novel phospholipase A1 catalyzing the initial step of jasmonic acid biosynthesis, which synchronizes pollen maturation, anther dehiscence, and flower opening in Arabidopsis. Plant Cell. 13, 2191–2209. doi: 10.1105/tpc.13.10.2191
Ji, W., Cong, R., Li, S., Li, R., Qin, Z., Li, Y., et al. (2016). Comparative proteomic analysis of soybean leaves and roots by iTRAQ provides insights into response mechanisms to short-term salt stress. Front. Plant Sci. 7:573. doi: 10.3389/fpls.2016.00573
Käll, L., Canterbury, J. D., Weston, J., Noble, W. S., and MacCoss, M. J. (2007). Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat. Methods 4, 923–925. doi: 10.1038/nmeth1113
Karki, N., Johnson, B. S., and Bates, P. D. (2019). Metabolically distinct pools of phosphatidylcholine are involved in trafficking of fatty acids out of and into the chloroplast for membrane production. Plant Cell. 31, 2768–2788. doi: 10.1105/tpc.19.00121
Katam, R., Shokri, S., Murthy, N., Singh, S. K., Suravajhala, P., Khan, M. N., et al. (2020). Proteomics, physiological, and biochemical analysis of cross tolerance mechanisms in response to heat and water stresses in soybean. PLoS One 15:e0233905. doi: 10.1371/journal.pone.0233905
Kosová, K., Prášil, I., and Vítámvás, P. (2013). Protein contribution to plant salinity response and tolerance acquisition. Int. J. Mol. Sci. 14, 6757–6789. doi: 10.3390/ijms14046757
Lam, H.-M., Xu, X., Liu, X., Chen, W., Yang, G., Wong, F.-L., et al. (2010). Resequencing of 31 wild and cultivated soybean genomes identifies patterns of genetic diversity and selection. Nat. Genet. 42, 1053–1059. doi: 10.1038/ng.715
Li, M., Guo, R., Jiao, Y., Jin, X., Zhang, H., and Shi, L. (2017). Comparison of salt tolerance in soja based on metabolomics of seedling roots. Front. Plant Sci. 8:1101. doi: 10.3389/fpls.2017.01101
Liu, A., Xiao, Z., Li, M.-W., Wong, F.-L., Yung, W.-S., Ku, Y.-S., et al. (2019). Transcriptomic reprogramming in soybean seedlings under salt stress. Plant. Cell. Environ. 42, 98–114. doi: 10.1111/pce.13186
Liu, X., Zhai, S., Zhao, Y., Sun, B., Liu, C., Yang, A., et al. (2013). Overexpression of the phosphatidylinositol synthase gene (ZmPIS) conferring drought stress tolerance by altering membrane lipid composition and increasing ABA synthesis in maize. Plant Cell. Environ. 36, 1037–1055. doi: 10.1111/pce.12040
Loewus, F. A., and Murthy, P. P. N. (2000). myo-Inositol metabolism in plants. Plant Sci. 150, 1–19. doi: 10.1016/S0168-9452(99)00150-8
Lv, D.-W., Subburaj, S., Cao, M., Yan, X., Li, X., Appels, R., et al. (2013). Proteome and phosphoproteome characterization reveals new response and defense mechanisms of Brachypodium distachyon leaves under salt stress. Mol. Cell. Proteomics 13, 632–652. doi: 10.1074/mcp.m113.030171
Magdy, M., Mansour, F., Hasselt, P. R., and Kuiper, P. J. C. (1994). Plasma membrane lipid alterations induced by NaCl in winter wheat roots. Physiol. Plant. 92, 473–478. doi: 10.1111/j.1399-3054.1994.tb08838.x
Martin, T. F. J. (1998). Phosphoinositide lipids as signaling molecules: common themes for signal transduction, cytoskeletal regulation, and membrane trafficking. Annu. Rev. Cell. Dev. Biol. 14, 231–264. doi: 10.1146/annurev.cellbio.14.1.231
McLoughlin, F., Augustine, R. C., Marshall, R. S., Li, F., Kirkpatrick, L. D., Otegui, M. S., et al. (2018). Maize multi-omics reveal roles for autophagic recycling in proteome remodelling and lipid turnover. Nat. Plants 4, 1056–1070. doi: 10.1038/s41477-018-0299-2
Mikami, K., Katagiri, T., Luchi, S., Yamaguchi-Shinozaki, K., and Shinozaki, K. (1998). A gene encoding phosphatidylinositol-4-phosphate 5-kinase is induced by water stress and abscisic acid in Arabidopsis thaliana. Plant J. 15, 563–568. doi: 10.1046/j.1365-313X.1998.00227.x
Mizusawa, N., and Wada, H. (2012). The role of lipids in photosystem II. Biochim. Biophys. Acta Bioenerg. 1817, 194–208. doi: 10.1016/j.bbabio.2011.04.008
Munns, R., and Tester, M. (2008). Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 59, 651–681. doi: 10.1146/annurev.arplant.59.032607.092911
Nelson, D. E., Rammesmayer, G., and Bohnert, H. J. (1998). Regulation of cell-specific inositol metabolism and transport in plant salinity tolerance. Plant Cell. 10, 753–764. doi: 10.2307/3870662
Norberg, P., and Liljenberg, C. (1991). Lipids of plasma membranes prepared from oat root cells. Plant Physiol. 96, 1136–1141. doi: 10.1104/pp.96.4.1136
Nouairi, I., Ammar, W. B., Youssef, N. B., Daoud, D. B., Ghorbal, M. H., Zarrouk, M., et al. (2006). Comparative study of cadmium effects on membrane lipid composition of Brassica juncea and Brassica napus leaves. Plant Sci. 170, 511–519. doi: 10.1016/j.plantsci.2005.10.003
Oliveros, J. C. (2007). VENNY. An interactive tool for comparing lists with Venn Diagrams. Available Online at: http://bioinfogp.cnb.csic.es/tools/venny/index.html.bioinfogp.cnb.csic.es/tools/venny/index.html
Pang, Q., Chen, S., Dai, S., Chen, Y., Wang, Y., and Yan, X. (2010). Comparative proteomics of salt tolerance in Arabidopsis thaliana and Thellungiella halophila. J. Proteome Res. 9, 2584–2599. doi: 10.1021/pr100034f
Pappan, K., Austin-Brown, S., Chapman, K. D., and Wang, X. (1998). Substrate selectivities and lipid modulation of plant phospholipase Dα, -β, and -γ. Arch. Biochem. Biophys. 353, 131–140. doi: 10.1006/abbi.1998.0640
Parvaiz, A., and Satyawati, S. (2008). Salt stress and phyto-biochemical responses of plants – a review. Plant Soil Environ. 54, 89–99. doi: 10.17221/2774-PSE
Phang, T.-H., Shao, G., and Lam, H.-M. (2008). Salt tolerance in soybean. J. Integr. Plant Biol. 50, 1196–1212. doi: 10.1111/j.1744-7909.2008.00760.x
Pical, C., Westergren, T., Dove, S. K., Larsson, C., and Sommarin, M. (1999). Salinity and hyperosmotic stress induce rapid increases in phosphatidylinositol 4, 5-bisphosphate, diacylglycerol pyrophosphate, and phosphatidylcholine in Arabidopsis thaliana cells. J. Biol. Chem. 274, 38232–38240. doi: 10.1074/jbc.274.53.38232
Qi, X., Li, M.-W., Xie, M., Liu, X., Ni, M., Shao, G., et al. (2014). Identification of a novel salt tolerance gene in wild soybean by whole-genome sequencing. Nat. Commun. 5:4340. doi: 10.1038/ncomms5340
Rottet, S., Besagni, C., and Kessler, F. (2015). The role of plastoglobules in thylakoid lipid remodeling during plant development. Biochim. Biophys. Acta Bioenerg. 1847, 889–899. doi: 10.1016/j.bbabio.2015.02.002
Schertl, P., and Braun, H.-P. (2014). Respiratory electron transfer pathways in plant mitochondria. Front. Plant Sci. 5:163. doi: 10.3389/fpls.2014.00163
Shelden, M. C., Dias, D. A., Jayasinghe, N. S., Bacic, A., and Roessner, U. (2016). Root spatial metabolite profiling of two genotypes of barley (Hordeum vulgare L.) reveals differences in response to short-term salt stress. J. Exp. Bot. 67, 3731–3745. doi: 10.1093/jxb/erw059
Shiva, S., Enninful, R., Roth, M. R., Tamura, P., Jagadish, K., and Welti, R. (2018). An efficient modified method for plant leaf lipid extraction results in improved recovery of phosphatidic acid. Plant Methods 14:14. doi: 10.1186/s13007-018-0282-y
Shoemaker, S. D., and Vanderlick, T. K. (2002). Stress-induced leakage from phospholipid vesicles: effect of membrane composition. Ind. Eng. Chem. Res. 41, 324–329. doi: 10.1021/ie010049t
Shu, S., Guo, S.-R., Sun, J., and Yuan, L.-Y. (2012). Effects of salt stress on the structure and function of the photosynthetic apparatus in Cucumis sativus and its protection by exogenous putrescine. Physiol. Plant. 146, 285–296. doi: 10.1111/j.1399-3054.2012.01623.x
Simidjiev, I., Stoylova, S., Amenitsch, H., Javorfi, T., Mustardy, L., Laggner, P., et al. (2000). Self-assembly of large, ordered lamellae from non-bilayer lipids and integral membrane proteins in vitro. Proc. Natl. Acad. Sci. 97, 1473–1476. doi: 10.1073/pnas.97.4.1473
Smyth, G. K., and Speed, T. (2003). Normalization of cDNA microarray data. Methods. 31, 265–273. doi: 10.1016/S1046-2023(03)00155-5
Stenzel, I., Ischebeck, T., König, S., Hołubowska, A., Sporysz, M., Hause, B., et al. (2008). The type B phosphatidylinositol-4-phosphate 5-kinase 3 is essential for root hair formation in Arabidopsis thaliana. Plant Cell 20, 124–141. doi: 10.1105/tpc.107.052852
Sui, N., and Han, G. (2014). Salt-induced photoinhibition of PSII is alleviated in halophyte Thellungiella halophila by increases of unsaturated fatty acids in membrane lipids. Acta Physiol. Plant. 36, 983–992. doi: 10.1007/s11738-013-1477-5
Tasseva, G., Richard, L., and Zachowski, A. (2004). Regulation of phosphatidylcholine biosynthesis under salt stress involves choline kinases in Arabidopsis thaliana. FEBS Lett. 566, 115–120. doi: 10.1016/j.febslet.2004.04.015
Tian, T., Liu, Y., Yan, H., You, Q., Yi, X., Du, Z., et al. (2017). AgriGO v2.0: A GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 45, W122–W129. doi: 10.1093/nar/gkx382
Torres-Franklin, M.-L., Gigon, A., de Melo, D. F., Zuily-Fodil, Y., and Pham-Thi, A.-T. (2007). Drought stress and rehydration affect the balance between MGDG and DGDG synthesis in cowpea leaves. Physiol. Plant 131, 201–210. doi: 10.1111/j.1399-3054.2007.00943.x
Torres-Romero, D., Gomez-Zambrano, A., Serrato, A. J., Sahrawy, M., and Merida, A. (2020). Fibrillin 2 interacts with other proteins to protect photosystem II against abiotic stress in Arabidopsis thaliana. bioRxiv. doi: 10.1101/2020.10.07.329979
Tsydendambaev, V. D., Ivanova, T. V., Khalilova, L. A., Kurkova, E. B., Myasoedov, N. A., and Balnokin, Y. V. (2013). Fatty acid composition of lipids in vegetative organs of the halophyte Suaeda altissima under different levels of salinity. Russ. J. Plant Physiol. 60, 661–671. doi: 10.1134/S1021443713050142
Tuteja, N., and Sopory, S. K. (2008). Chemical signaling under abiotic stress environment in plants. Plant Signal. Behav. 3, 525–536. doi: 10.4161/psb.3.8.6186
Wang, H., Zhang, M., Guo, R., Shi, D., Liu, B., Lin, X., et al. (2012). Effects of salt stress on ion balance and nitrogen metabolism of old and young leaves in rice (Oryza sativa L.). BMC Plant Biol. 12:194. doi: 10.1186/1471-2229-12-194
Wang, K., Guo, Q., Froehlich, J. E., Hersh, H. L., Zienkiewicz, A., Howe, G. A., et al. (2018). Two abscisic acid-responsive plastid lipase genes involved in jasmonic acid biosynthesis in Arabidopsis thaliana. Plant Cell. 30, 1006–1022. doi: 10.1105/tpc.18.00250
Wang, T., Tohge, T., Ivakov, A., Mueller-Roeber, B., Fernie, A. R., Mutwil, M., et al. (2015). Salt-related MYB1 coordinates abscisic acid biosynthesis and signaling during salt stress in arabidopsis. Plant Physiol. 169, 1027–1041. doi: 10.1104/pp.15.00962
Wang, Y., Zhang, X., Huang, G., Feng, F., Liu, X., Guo, R., et al. (2020). Dynamic changes in membrane lipid composition of leaves of winter wheat seedlings in response to PEG-induced water stress. BMC Plant Biol. 20:84. doi: 10.1186/s12870-020-2257-1
Welti, R., Li, W., Li, M., Sang, Y., Biesiada, H., Zhou, H.-E., et al. (2002). Profiling membrane lipids in plant stress responses. J. Biol. Chem. 277, 31994–32002. doi: 10.1074/jbc.M205375200
Xie, C., Mao, X., Huang, J., Ding, Y., Wu, J., Dong, S., et al. (2011). KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 39, W316–W322. doi: 10.1093/nar/gkr483
Xiong, J., Sun, Y., Yang, Q., Tian, H., Zhang, H., Liu, Y., et al. (2017). Proteomic analysis of early salt stress responsive proteins in alfalfa roots and shoots. Proteome Sci. 15, 1–19. doi: 10.1186/s12953-017-0127-z
Yamane, K., Rahman, S., Kawasaki, M., Taniguchi, M., and Miyake, H. (2004). Pretreatment with a low concentration of methyl viologen decreases the effects of salt stress on chloroplast ultrastructure in rice leaves (Oryza sativa L.). Plant Prod. Sci. 7, 435–441. doi: 10.1626/pps.7.435
Yang, J. I., Perera, I. Y., Brglez, I., Davis, A. J., Stevenson-Paulik, J., Phillippy, B. Q., et al. (2007). Increasing plasma membrane phosphatidylinositol(4,5)bisphosphate biosynthesis increases phosphoinositide metabolism in Nicotiana tabacum. Plant Cell. 19, 1603–1616. doi: 10.1105/tpc.107.051367
Yekutieli, D., and Benjamini, Y. (2001). The control of the false discovery rate in multiple testing under dependency. Ann. Stat. 29, 1165–1188. doi: 10.1214/aos/1013699998
Yin, X., and Komatsu, S. (2017). Comprehensive analysis of response and tolerant mechanisms in early-stage soybean at initial-flooding stress. J. Proteomics. 169, 225–232. doi: 10.1016/j.jprot.2017.01.014
Yoeun, S., Cho, K., and Han, O. (2018). Structural evidence for the substrate channeling of rice allene oxide cyclase in biologically analogous nazarov reaction. Front. Chem. 6:500. doi: 10.3389/fchem.2018.00500
Youssef, A., Laizet, Y., Block, M. A., Maréchal, E., Alcaraz, J. P., Larson, T. R., et al. (2010). Plant lipid-associated fibrillin proteins condition jasmonate production under photosynthetic stress. Plant J. 61, 436–445. doi: 10.1111/j.1365-313X.2009.04067.x
Zhang, B., and Kuster, B. (2019). Proteomics is not an island: multi-omics integration is the key to understanding biological systems. Mol. Cell. Proteomics 18, S1–S4. doi: 10.1074/mcp.E119.001693
Zhang, J., Yang, D., Li, M., and Shi, L. (2016). Metabolic profiles reveal changes in wild and cultivated soybean seedling leaves under salt stress. PLoS One 11:e0159622. doi: 10.1371/journal.pone.0159622
Zhang, Z., Mao, C., Shi, Z., and Kou, X. (2017). The amino acid metabolic and carbohydrate metabolic pathway play important roles during salt-stress response in tomato. Front. Plant Sci. 8:1231. doi: 10.3389/fpls.2017.01231
Keywords: Glycine max, lipid profiling, lipid signaling, membrane lipids, metabolism, salinity
Citation: Liu A, Xiao Z, Wang Z, Lam H-M and Chye M-L (2021) Galactolipid and Phospholipid Profile and Proteome Alterations in Soybean Leaves at the Onset of Salt Stress. Front. Plant Sci. 12:644408. doi: 10.3389/fpls.2021.644408
Received: 21 December 2020; Accepted: 18 February 2021;
Published: 17 March 2021.
Edited by:
Eric Marechal, UMR 5168 Laboratoire de Physiologie Cellulaire Vegetale (LPCV), FranceReviewed by:
Mie Shimojima, Tokyo Institute of Technology, JapanTill Peter Ischebeck, University of Göttingen, Germany
Copyright © 2021 Liu, Xiao, Wang, Lam and Chye. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hon-Ming Lam, aG9ubWluZ0BjdWhrLmVkdS5oaw==; Mee-Len Chye, bWxjaHllQGhrdS5oaw==
†These authors have contributed equally to this work
 Ailin Liu
Ailin Liu Zhixia Xiao2†
Zhixia Xiao2† Zhili Wang
Zhili Wang Hon-Ming Lam
Hon-Ming Lam Mee-Len Chye
Mee-Len Chye