- 1Nottingham BBSRC Wheat Research Centre, School of Biosciences, University of Nottingham, Loughborough, United Kingdom
- 2Limagrain UK Limited, Bury St Edmunds, United Kingdom
- 3School of Biological Sciences, University of Bristol, United Kingdom
Wheat is one of the most important food and protein sources in the world and although, in recent years wheat breeders have achieved yield gains, they are not sufficient to meet the demands of an ever-growing population. Development of high yielding wheat varieties, resilient to abiotic and biotic stress resulting from climate change, has been limited by wheat’s narrow genetic base. In contrast to wheat, the wild relatives of wheat provide a vast reservoir of genetic variation for most, if not all, agronomic traits. Previous studies by the authors have shown the transfer of genetic variation from T. urartu into bread wheat. However, before the introgression lines can be exploited for trait analysis, they are required to have stable transmission of the introgressions to the next generation. In this work, we describe the generation of 86 doubled haploid (DH) wheat-T. urartu introgression lines that carry homozygous introgressions which are stably inherited. The DH lines were characterised using the Axiom® Wheat Relative Genotyping Array and 151 KASP markers to identify 65 unique T. urartu introgressions in a bread wheat background. DH production has helped accelerate the breeding process and facilitated the early release of homozygous wheat-T. urartu introgression lines. Together with the KASP markers, this valuable resource could greatly advance identification of beneficial alleles that can be used in wheat improvement.
Introduction
Triticum aestivum (2n = 6x = 42; AABBDD) was formed via two sequential hybridisations. The first, between the A-genome donor Triticum urartu Thum ex. Gandil (2n = 2x = 14; AuAu) (Chapman et al., 1976; Dvořák et al., 1993) and the B-genome donor, a species similar to Aegilops speltoides 2n = 2x = 14; SS) (Sarkar and Stebbins, 1956; Riley and Chapman, 1958; Feldman et al., 1995) occurred approximately 0.36 to 0.5 million years ago (Huang et al., 2002; Dvořák and Akhunov, 2005; Marcussen et al., 2014) to form tetraploid wheat Triticum turgidum ssp. dicoccoides (2n = 4x = 28; AABB). The second hybridisation occurred either only once or twice about 8,000–10,000 years ago between allotetraploid wheat and Aegilops tauschii, the D-genome donor (2n = 2x = 14; DD) (Kihara, 1944; McFadden and Sears, 1944; Matsuoka, 2011). Thus, wheat is a relatively new species and in addition, due to its evolutionary pathway, has gone through a significant genetic bottleneck which has been further compounded by selection for traits of importance for its domestication (Charmet, 2011). It is therefore expected that hexaploid wheat carries significantly less genetic variation for exploitation in breeding programmes and research than its ancestral species and distant wild relatives, some of which evolved millions of years ago (Dempewolf et al., 2017). As a result, new sources of genetic variation for T. aestivum are invaluable for a wide range of traits.
The presence of the Pairing Homoeologous 1 locus (Ph1) on the long arm of chromosome 5B of wheat (Okamoto, 1957; Riley and Chapman, 1958; Sears and Okamoto, 1958) restricts recombination at meiosis to homologous chromosomes (Sears, 1976). As a result, recombination cannot occur between the three related homoeologous genomes of wheat, i.e., the A, B and D genomes. The Ph1 locus also prevents recombination between the chromosomes in wheat/wild relative interspecific F1 hybrids where the genome(s) of the wild relative is/are homoeologous to that/those of wheat, i.e., wild relatives that belong to the tertiary gene pool of wheat. In contrast, recombination can occur more freely between wild relatives and ancestral species that share a homologous genome in common with wheat, i.e., species such as T. urartu that carries the A-genome of wheat (Chapman et al., 1976).
Triticum urartu has not been extensively used to introduce new genetic variation into wheat although accessions have been identified with resistance to diseases such as stem rust (Rouse and Jin, 2011), powdery mildew (Qiu et al., 2005) and root lesion nematode (Sheedy et al., 2012) and other traits such as high net photosynthetic rate (Austin et al., 1982; Austin, 1986) and those that affect bread making quality (Martín et al., 2008; Cuesta et al., 2015).
The transfer of chromosome segments or introgressions from wild relatives and ancestral species into wheat has been hindered in the past through the inability to know exactly what had been transferred. Much of the work was centred on the transfer of genetic variation for a trait of interest and relied very heavily on phenotyping for that trait in order to track the transfer. The molecular tools that are now available, however, enable the identification and characterisation of introgressions and the tracking of them through the different generations in a crossing programme.
The use of the new technologies at the Nottingham BBSRC Wheat Research Centre (WRC) has allowed the transfer of hundreds of introgressions from a variety of wild relatives into wheat, e.g., Amblyopyrum muticum (King et al., 2017), Thinopyrum bessarabicum (Grewal et al., 2018b), Ae. speltoides (King et al., 2018), T. urartu (Grewal et al., 2018a), Triticum timopheevii (Devi et al., 2019), Thinopyrum elongatum (Baker et al., 2020), and Aegilops caudata (Grewal et al., 2020b). However, without identifying what genetic variation is carried by the introgressions, the lines produced will remain quite simply as ‘seeds in a packet’ of unknown agronomic potential. It is therefore essential that these introgressions are phenotyped as extensively as possible for as many different traits as possible.
In order that these lines can be analysed in a range of environments throughout the world they need first be multiplied and then distributed to collaborators. In order to multiply the introgression lines, however, they must first be homozygous to ensure that they are stably inherited at each generation during multiplication. This paper describes the exploitation of the doubled haploid (DH) procedure to generate homozygous introgressions of T. urartu from heterozygous introgressions and their characterisation via molecular markers and genomic in situ hybridisation (GISH).
Materials and Methods
Plant Materials
A selection of the introgression lines generated and described by Grewal et al. (2018a) were used for DH production. In summary, T. aestivum cv. Chinese Spring ph1 mutant was pollinated with T. urartu [accessions 1010001, 1010002, and 1010006 obtained from the Germplasm Resources Unit (GRU) at John Innes Centre, United Kingdom]. The F1 interspecific hybrids produced were backcrossed to T. aestivum cv. Paragon, carrying the wild-type Ph1 locus, to obtain BC1 progeny which were then recurrently backcrossed with Paragon to produce a BC3 population. Selected BC3 plants were crossed to maize to initiate DH production. When this work was initially undertaken, no suitable SNP genotyping platform was available to enable marker-assisted selection and backcrossing. Thus, there was no indication of which BC3 lines carried T. urartu introgressions and in the absence of GISH capability to detect T. urartu derived A-genome segments, the BC3 plants were chosen at random for DH production.
Doubled Haploid Production
The DH production procedure used was as described by Laurie and Reymondie (1991) and used previously to generate wheat-Am. muticum DH lines (King et al., 2019). In summary, wheat spikes were emasculated and then pollinated with maize (cultivars Northern Extra Sweet, Prelude and Sundance). One day after pollination, internodes below pollinated spikes were filled with 10 mg l–1 of 2,4-dichlorophenoxyacetic acid (2,4-D solution) with a syringe and the holes sealed with petroleum jelly. The 2,4-D solution was also injected into each floret. After 14–21 days immature seeds were harvested, and haploid embryos were excised and cultured under sterile conditions. Subsequent colchicine treatment to obtain DH seedlings was carried out as described in Nemeth et al. (2015). Plants were grown to maturity in soil and self-fertilised seed was harvested.
Development of KASP Markers
Grewal et al. (2020a) generated 2374 SNPs between hexaploid wheat and ten wild relative species and converted 1000 of these SNPs into KASP assays. Of the remaining SNPs, 79 were selected (as set 1), that were polymorphic between wheat and T. urartu and Am. muticum for conversion to chromosome-specific KASP assays as described by Grewal et al. (2020a) and designated codes between WRC1001-WRC1079 (Supplementary Table 2).
A list of 1742 SNPs was also combined from a core set of 960 SNPs already used to develop a KASP wheat panel by University of Bristol for LGC, Biosearch Technologies1 (last accessed on 11 June 2020) and a set of 782 SNPs that were identified as reliable for use in wheat via various genotyping platforms including as KASP markers (Burridge et al., 2018). After removing duplicates, 1572 SNPs were obtained which were all available as probes on the 820K Axiom® Array (Winfield et al., 2016). Through analysis of the KASP primers dataset of all the 820K Axiom® SNPs, generated using PolyMarker (Ramirez-Gonzalez et al., 2015) and available at CerealsDB2 (last accessed 11 June 2020), information on significant BLAST hits for each of these SNPs was obtained. The 229 SNPs that were observed to be present on a single contig in the wheat genome (Refseqv1.0) (IWGSC et al., 2018) were selected (as set 2). Pre-validated KASP assay primers for these SNPs were obtained via CerealsDB and designated codes between WRC1080-WRC1308 (Supplementary Table 2). A further 77 chromosome-specific KASP assays were also selected (as set 3) that were simultaneously developed from the 820K Axiom® Array to be polymorphic between wheat and Th. bessarabicum. These were designated codes WRC1317-WRC1393 (Supplementary Table 2).
Genotyping of Wheat-T. urartu DH Lines
All the DH lines were initially genotyped with the Axiom® 36K Wheat-Relative Genotyping Array (Thermo Fisher Scientific, United Kingdom) (Winfield et al., 2016; King et al., 2017) and subsequently genotyped with KASP markers (LGC, Biosearch Technologies, United Kingdom). The latter included the three sets of KASP assays as described above in addition to chromosome-specific KASP markers reported by Grewal et al. (2020a).
Genomic DNA was isolated from leaf tissue of 10-day-old seedlings in a 96-well plate as described by Thomson and Henry (1995). All DH lines were genotyped alongside four wheat genotypes (Chinese Spring, Paragon, Pavon, and Highbury) and the T. urartu accessions as controls.
Genotyping with the Axiom® 36K Wheat-Relative Array was performed by including all the DH lines on the array with other wheat-T. urartu backcrossed populations. Genotypes were called, using the methodology described by Grewal et al. (2018a), for a subset of probes present on the genetic map of T. urartu as reported by the same study.
All DH lines shown to have an introgression from T. urartu in the array genotyping were subsequently genotyped with KASP markers after a round(s) of self-fertilisation. The genotyping procedure was as described by Grewal et al. (2020b). In summary, the genotyping reactions were set up using the automated PIPETMAX® 268 (Gilson, United Kingdom) and performed in a ProFlex PCR system (Applied Biosystems by Life Technology) in a final volume of 5 μl with 1 ng genomic DNA, 2.5 μl KASP reaction mix (ROX), 0.068 μl primer mix and 2.43 μl nuclease free water. PCR conditions were set as 15 min at 94°C; 10 touchdown cycles of 10 s at 94°C, 1 min at 65–57°C (dropping 0.8°C per cycle); and 35 cycles of 10 s at 94°C, 1 min at 57°C. Fluorescence detection of the reactions was performed using a QuantStudio 5 (Applied Biosystems) and the data analysed using the QuantStudioTM Design and Analysis Software V1.5.0 (Applied Biosystems).
All KASP markers included in the final list of working assays were used in a BLAST analysis against the wheat genome sequence Refseqv1 (IWGSC et al., 2018) and the T. urartu genome sequence (Ling et al., 2018). The markers within a linkage group were ordered according to their physical positions on the T. urartu genome (Supplementary Table 3) and used to represent the approximate sizes of the chromosome segments introgressed from T. urartu into wheat (Figure 1).
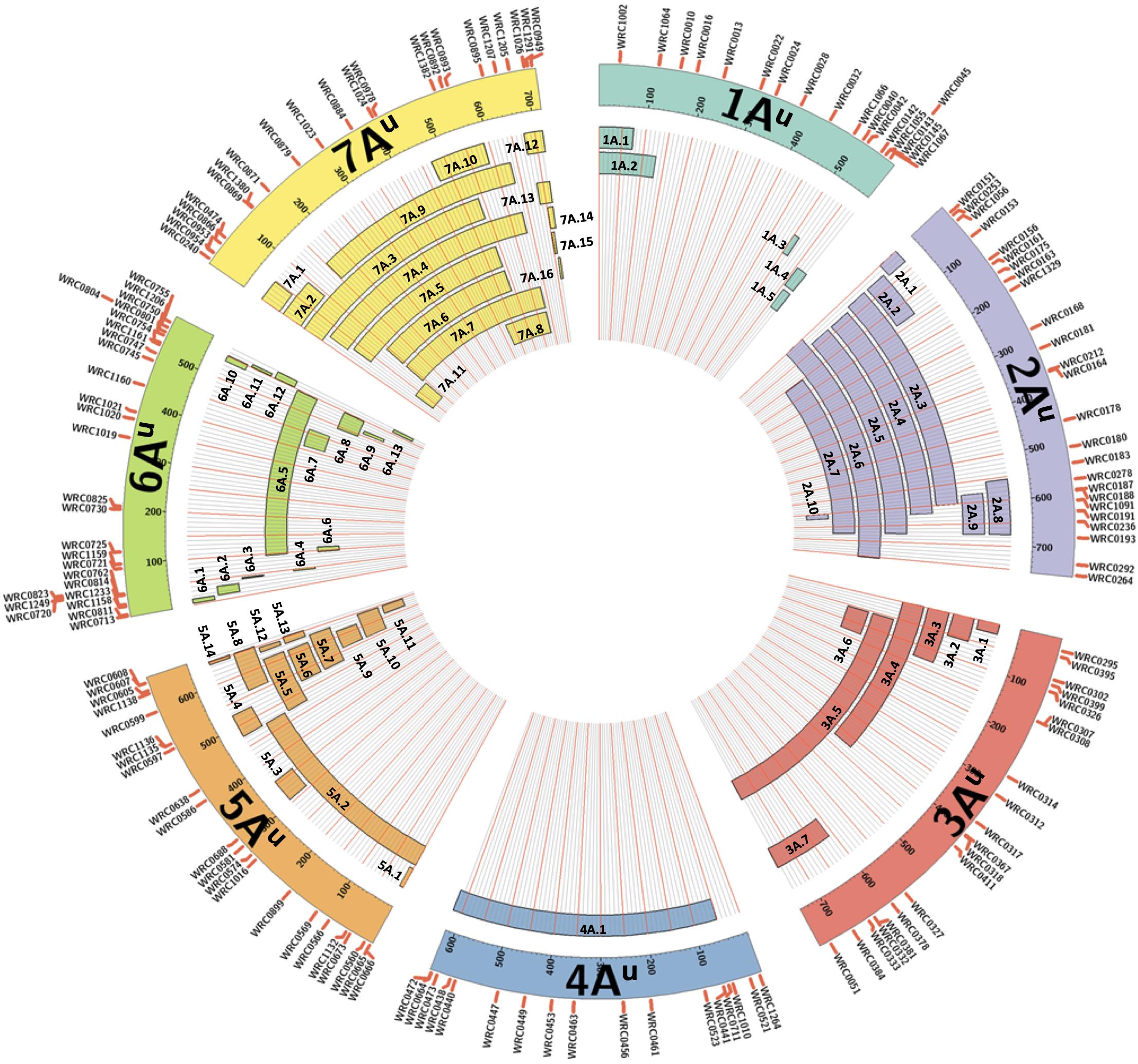
Figure 1. A graphical representation of the sizes of the 65 unique introgressions of T. urartu, along each T. urartu chromosome, found in the DH lines. In each of the seven chromosomes, the KASP markers are shown ordered according to their physical position (Ling et al., 2018). Each segment is given a unique name which is used to characterise the DH lines in Table 2.
Genomic in situ Hybridisation (GISH)
The protocol for genomic in situ hybridization (GISH) and preparation of chromosome spreads was as described in Kato et al. (2004), King et al. (2017), and Grewal et al. (2020b). In summary, genomic DNA was isolated from the three putative diploid progenitors of bread wheat, i.e., T. urartu (A genome), Ae. speltoides (B genome), and Ae. tauschii (D genome), using extraction buffer [0.1 M Tris-HCl (pH 7.5), 0.05 M EDTA (pH 8.0), 1.25% SDS]. Samples were incubated at 65°C for 1 h before being placed on ice and mixed with ice cold 6 M NH4C2H3O for 15 min. The samples were then spun down, the supernatant mixed with isopropanol to pellet the DNA and the isolated DNA further purified with phenol/chloroform and resuspended in 2 × SSC and 1 × TE buffer, pH 7.0. The genomic DNA of (1) T. urartu was labelled by nick translation with ChromaTideTM Alexa FluorTM 488-5-dUTP (Invitrogen; C11397; coloured green), (2) Ae. speltoides was labelled by nick translation with DEAC-dUTP (Jena Bioscience; NU-803-DEAC; coloured bluish purple), and (3) Ae. tauschii was labelled with ChromaTideTM Alexa FluorTM 594-5-dUTP (Invitrogen; C11400; coloured red).
Slides were probed using a probe mixture that contained the labelled genomic DNAs of T. urartu, Ae. speltoides and Ae. tauschii in the ratio 3:3:4, respectively, in 2 × SSC and 1 × TE buffer, pH 7.0, in a final volume of 10 μl per slide. Slides were counterstained with Vectashield mounting medium with 4′-6-diamidino-2-phenylindole dihydrochloride (DAPI) and analysed using a Zeiss Axio ImagerZ2 upright epifluorescence microscope (Carl Zeiss Ltd., Oberkochen, Germany) with filters for DAPI (blue), Alexa Fluor 488 (green), Alexa Fluor 594 (red), Alexa Fluor 546 and DEAC (aqua). Photographs were taken using a MetaSystems Coolcube 1 m CCD camera together with the Metafer v.4 software (MetaSystems GmbH, Altlussheim, Germany) and further image analysis was carried out using ISIS v.5.8.5 (MetaSystems GmbH, Altlussheim, Germany).
Results
Generation of Wheat-T. urartu DH Lines
Forty-nine BC3 plants were pollinated with maize in order to generate DH lines. In total, 183 DH seedlings were obtained from 26 BC3 plants (Table 1). Of these, 127 (69%) DH plants grew to maturity. A further 13 DH plants were found to be sterile resulting in the generation of 114 fertile DH plants (Table 1) that set F1 seed. Out of the 49 BC3 plants used for DH production, 20 (41%) eventually produced fertile DH plants.
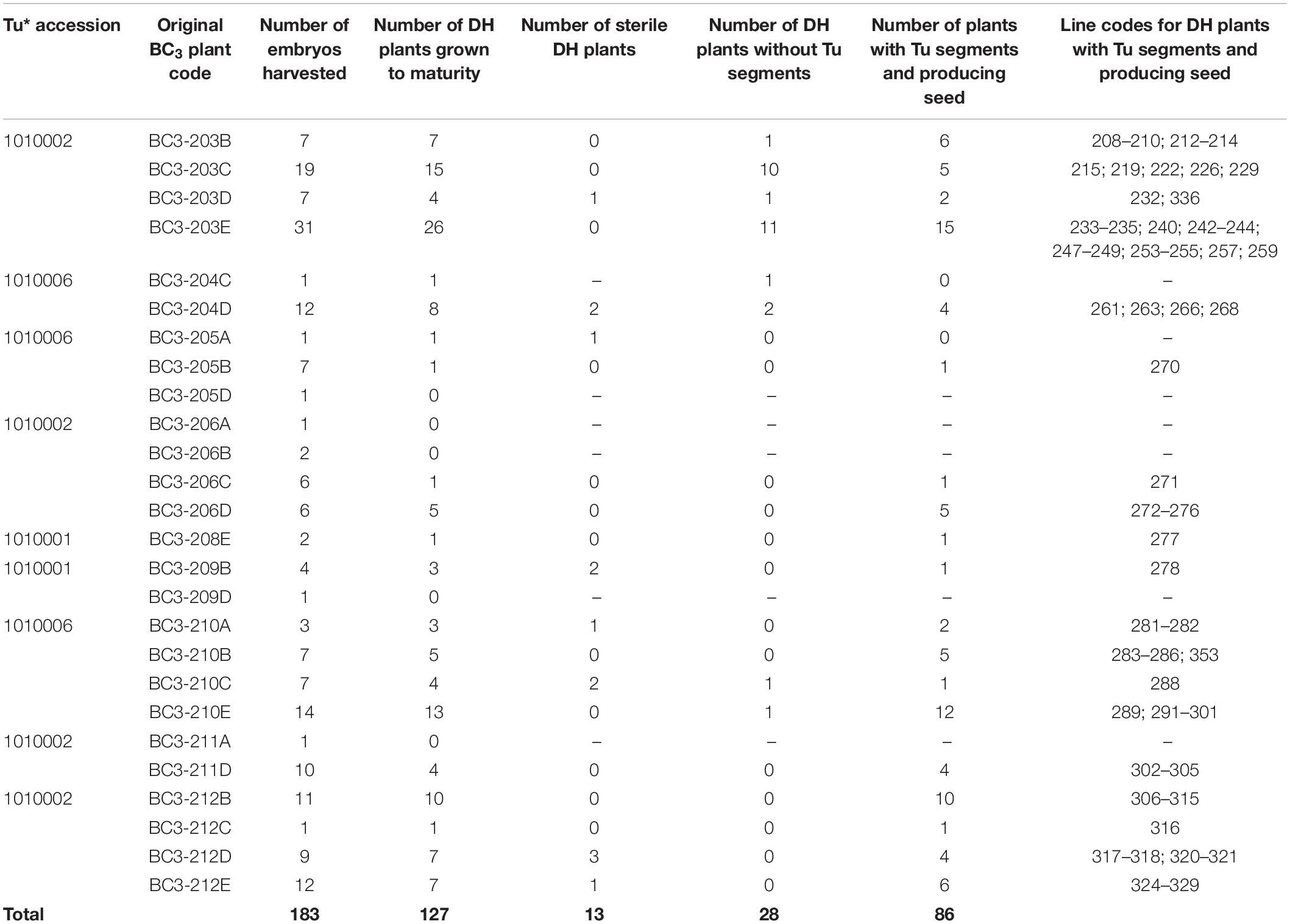
Table 1. Number of BC3 plants (and their original code) involved in the various stages of DH production including the number of DH plants that were fertile and carried T. urartu segments, their DH line codes and details of which T. urartu accession was present in each DH line.
Molecular Characterisation With the Axiom® Wheat-Relative Genotyping Array
The 114 fertile DH plants were genotyped using the Axiom® Wheat Relative Array. However, genotypes were called for a subset of the probes, 368 of those that were present on the genetic map of T. urartu (Grewal et al., 2018a). The resulting marker analysis indicated that 28 (25%) DH plants did not carry any T. urartu chromosomes and/or wheat-T. urartu segments. Thus, in total, 86 wheat-T. urartu fertile DH lines carrying T. urartu introgressions were obtained in this study (Table 1). The linkage group(s) of the T. urartu introgression(s) present in each of these 86 DH lines, as obtained through array genotyping, is shown in Table 2.
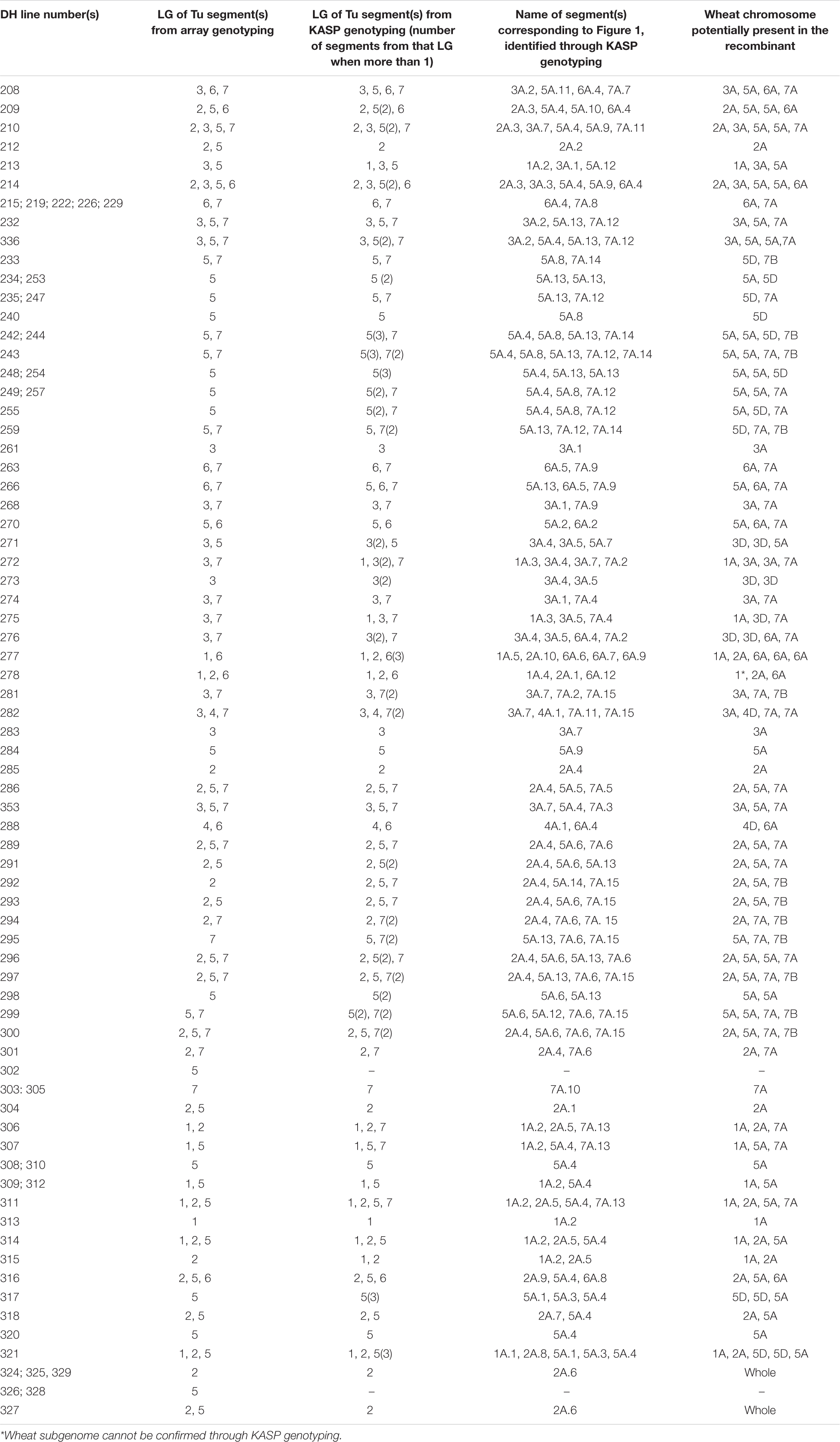
Table 2. List of all 86 wheat-T. urartu DH lines showing the linkage groups detected in each line via the Axiom array and KASP markers and the name of the unique introgression (corresponding to Figure 1) and its potential location in the wheat genome.
Development of KASP Markers
The progeny of the 86 DH lines were further genotyped with KASP markers to validate the results of the array genotyping and identify the wheat chromosome(s) involved in the recombinant chromosomes.
Grewal et al. (2020a) produced a panel of 710 KASP markers validated across ten wild relative species of which 118 were polymorphic between bread wheat and T. urartu. This subset of markers, consisting of 115 chromosome-specific and 3 chromosome-nonspecific KASP assays (Table 3), were used to genotype the DH lines in the present study. All 118 markers were able to distinguish between wheat and T. urartu alleles and were thus able to detect a vast majority of the introgressions in the DH plants. However, there were still gaps in the marker set where the array genotyping had detected segments but no KASP markers were available.
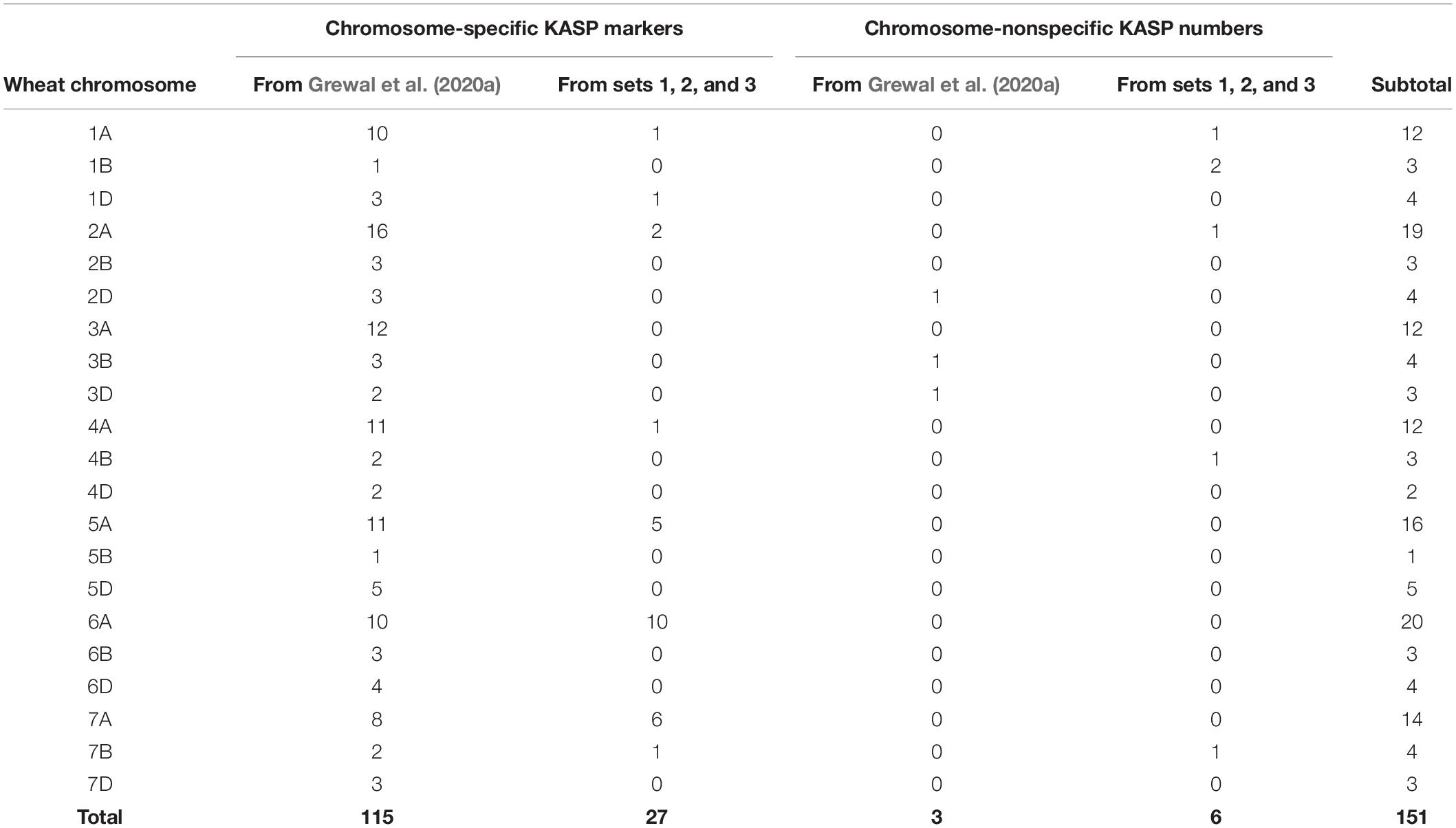
Table 3. Number of chromosome-specific and nonspecific KASP markers developed for each wheat chromosome in the current study (from sets 1, 2, and 3) versus previously developed KASP markers.
To achieve better detection of all T. urartu introgressions with KASP markers, a further three sets of KASP assays which were being developed in tandem with this work were used for genotyping the DH lines. From set 1, 13 (16%) out of the 79 assays failed to amplify the templates and produced no genotypes. Of the working assays (those that detected at least two wheat genotypes), 14 KASP markers were validated as being polymorphic between wheat and T. urartu. Ten of these KASP markers were chromosome-specific in wheat. From set two, 209 (90%) KASP assays amplified the control samples, of which 16 markers were found to be polymorphic for T. urartu with 14 of these being chromosome-specific in wheat. Eighteen (23%) out of the 77 assays failed from set 3 and 3 KASP markers were identified as being polymorphic for T. urartu from amongst the working assays. All three assays were chromosome specific in wheat. In summary, out of the 385 KASP assays designed to work across various wild relatives and wheat genotypes in the three sets, 334 (∼87%) were found to be polymorphic between wheat and various other wild relatives (data not shown) and of those 33 (∼10%) were found to work for T. urartu in a wheat background. Of these new KASP assays for T. urartu, 27 were chromosome-specific in wheat and 6 were chromosome-nonspecific (Table 3). Combining these with those previously developed by Grewal et al. (2020a) resulted in a total of 151 KASP markers that were used to characterise the wheat-T. urartu DH lines in this study (Table 3 and Figure 1).
Genotyping of DH Lines With Chromosome-Specific KASP Markers
Through KASP genotyping with chromosome-specific assays, we were able to detect 216 Triticum urartu introgressions across the 86 DH lines with 65 unique introgressions (segments or whole chromosomes) that were present in one or more DH lines (Table 2). The KASP markers were able to detect the presence of one (or more) additional T. urartu linkage group (LG) in 19 of the DH lines as compared to the Axiom array. Although, the reverse was true for six DH lines indicating that there were still significant gaps in the detection capacity of the KASP markers.
The introgressions were named after the T. urartu LG they were derived from and Table 2 provides details of which LG(s) and corresponding introgressions, as detected by KASP markers, are present in each DH line. In addition, Figure 1 shows the relative size of each of these introgressions as compared to the size of the T. urartu chromosomes (Ling et al., 2018). In total, we obtained 5 introgressed segments from LG 1, 9 segments and a pair of whole chromosomes from LG 2, 7 segments from LG3, only one large segment from LG4, 14 mostly small segments from LG 5, 13 segments from LG 6 and 15 segments of varying sizes from LG 7 of T. urartu in the wheat background. The maximum number of T. urartu introgressions in a DH line was five with DH lines 210, 214, 243, 277, and 321 all having five introgressions each (Table 2).
Genotyping of homozygous introgression lines with chromosome-specific KASP markers also allows the potential identification of the wheat chromosome recombined with the T. urartu segment (Grewal et al., 2020a,b) due to the absence of the wheat-specific allele in homozygous lines which will instead only show the presence of the wild-relative-specific allele for a marker that lies within the introgression. Since the lines characterised in this work are DH lines, they are expected to be homozygous for every introgression from T. urartu and thus, the chromosome-specific KASP markers should be able to identify the wheat chromosome recipient of the introgression.
Of the 65 introgressions, the A-genome specific KASP markers indicated that 54 were with an A-genome chromosome of wheat (Table 2) where the genotypes were called as homozygous for the wild relative alleles. During GISH analysis, the genomic probe used for detecting A-genome chromosomes in wheat is prepared from T. urartu genomic DNA and expectedly also detects Au chromosome segments of T. urartu in a wheat background. Thus, it is not possible to validate the presence and size of the T. urartu segments introgressed into the A genome of wheat using GISH.
Of the remaining 11 introgressions, the chromosome-specific KASP markers indicated that introgressions 5A.1 and 5A.3 were with Chr 5D of wheat and 7A.14 and 7A.15 were with Chr 7B of wheat (Table 2). However, all of these were potentially too small to be visualised using GISH. Similarly, 1A.4 was potentially too small to be detected by GISH and the markers were also unable to conclusively indicate which subgenome of wheat it had recombined with. Where the introgressions had not directly recombined with the A genome of wheat and were larger segments, GISH analysis was used to validate the KASP genotyping results.
GISH Analysis of Wheat-T. urartu DH Lines
Introgression 2A.6 was found to be a pair of whole 2Au chromosomes that replaced a pair of wheat 2D chromosomes in DH lines 324, 325, 327 and 329 making these 2Au(2D) substitution lines. Figure 2A shows the genotyping of DH 324 with KASP markers and validation of the result with GISH. The heterozygous calls for markers on Chr 2A and 2B of wheat indicate the presence of a very large segment from Chr 2Au of T. urartu but the lack of any homozygous calls indicates that the introgression has not recombined with a wheat chromosome. Instead, the null calls for markers on 2D indicate that the pair of 2D chromosomes is missing in this line. GISH analysis confirms the presence of 16 A genome chromosomes (the probe for the wheat A-genome chromosomes detects any Au genome chromosomes with the same colour) and 12 D-genome chromosomes thereby indicating that introgression 2A.6 is potentially a pair of Chr 2Au.
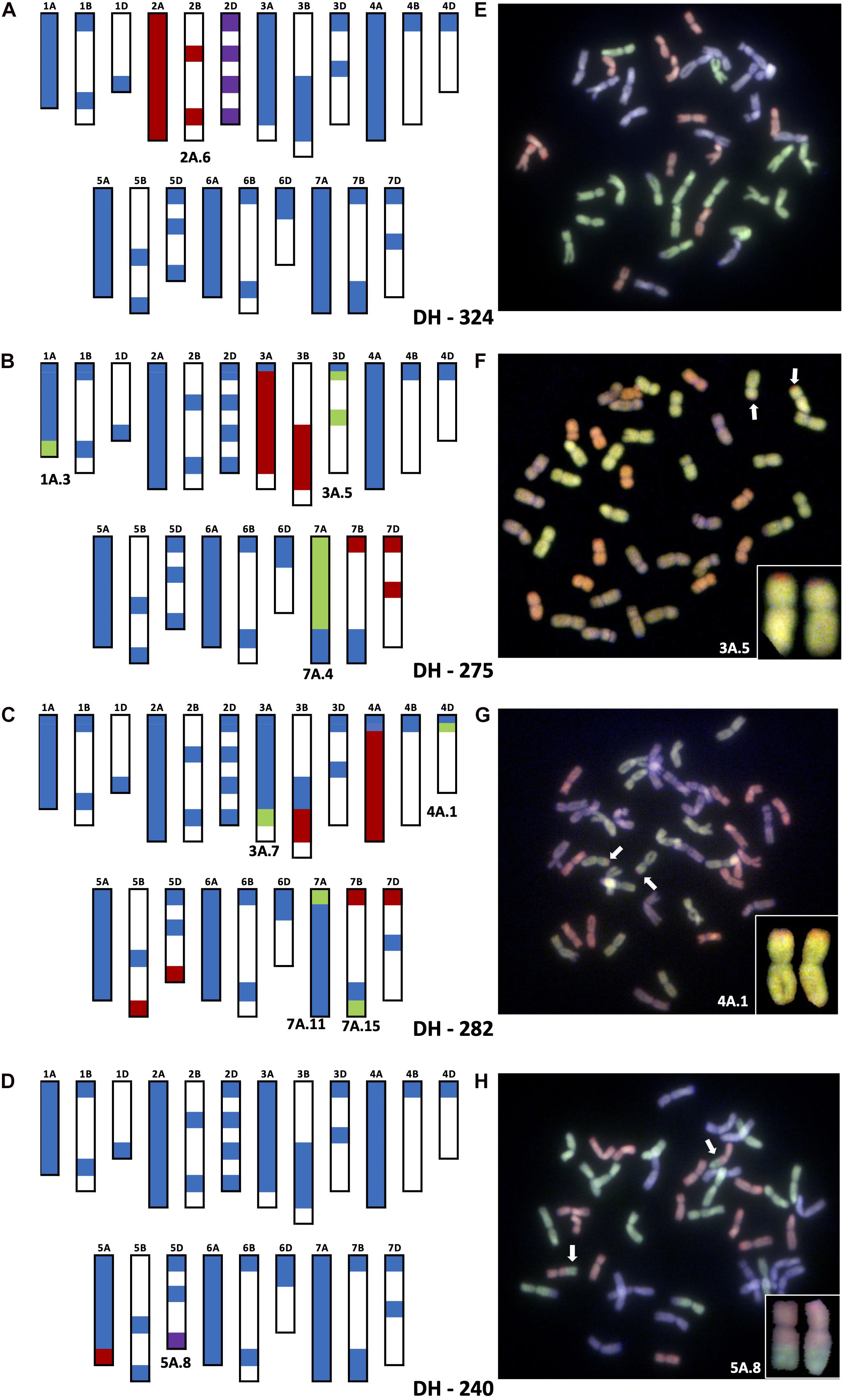
Figure 2. Molecular characterization of wheat–T. urartu DH lines showing genotyping analysis with chromosome–specific KASP assays (A–D) and Genomic in situ hybridization (GISH) images of root metaphase spreads (E–H). Panels (A,E) showing presence of whole chromosome 2Au (2A.6) substituting for Chr 2D in DH-324. Panels (B,F) showing the introgression of segment 3A.5 into Chr 3D of wheat in DH-275. Panels (C,G) showing the introgressions of segment 4A.1 into Chr 4D of wheat in DH-282. Panels (D,H) showing the introgression of segment 5A.8 into Chr 5D of wheat. In the genotyping data, all heterozygous calls are shown in red, homozygous wild relative calls in green, homozygous wheat calls in blue and null calls are shown in purple. White spaces indicate regions where KASP markers are not present. For the GISH, green represents the A-genome of wheat or Au-genome of T. urartu, greyish purple, the B-genome and red, the D-genome of wheat. Identifiable introgressions are indicated by white arrows.
The markers indicated that introgressions 3A.4 and 3A.5 had recombined with the D-genome of wheat and in particular chromosome 3D. Both introgressions are present in DH lines 271, 272 and 276, while DH 275 only contained segment 3A.5 (Table 2 and Figure 2B). The presence of 3A.5 in Chr 3D of DH 275 is indicated by the presence of heterozygous calls for markers on Chr 3A and Chr 3B but homozygous calls (for the T. urartu allele) for markers on Chr 3D (except those on the distal end of the short arm on Chr 3A and 3D which gave homozygous calls for the wheat allele) as shown in Figure 2B. This was validated by GISH analysis that showed a pair of recombinant chromosomes with a large A-genome or equivalent segment (likely Chr 3Au of T. urartu) and a very small segment of the D genome of wheat on the distal end of the short arm (Figure 2B). DH 275 also has introgressions 1A.3 and 7A.4 but since the markers indicate they have gone into Chr 1A and Chr 7A of wheat, respectively (through homozygous T. urartu calls for markers on these chromosomes), they cannot be observed through GISH.
In DH lines 282 and 288, introgression 4A.1 had recombined with Chr 4D of wheat (Table 2) as indicated by marker analysis in Figure 2C. The KASP markers on the very distal ends of the short arms of chromosomes 4A, 4B and 4D of wheat gave homozygous wheat calls but the majority of the markers on Chr 4A gave heterozygous calls while one marker on Chr 4D gave a homozygous call for the T. urartu allele indicating that except for a very small region at the distal end of the short arm, the majority of Chr 4D had been potentially replaced with Chr 4Au of wheat. The markers on the very distal ends of the long arms of Chr 5B and 5D also give heterozygous calls due to the 4AL-5AL translocation present in T. urartu and in wheat (Liu et al., 1992; King et al., 1994; Devos et al., 1995; Dvořák et al., 2018). Thus, since 4A.1 has a translocated segment from Chr 5AuL which is homoeologous to the corresponding regions in Chr 5BL and Chr 5DL, the markers on the latter two chromosomes also detect this segment (Figure 2C). This was validated by GISH analysis that showed a pair of recombinant chromosomes with a large A-genome or equivalent segment (likely Chr 4Au of T. urartu) and a very small segment of the D genome of wheat on the distal end of the short arm (Figure 2C). DH 282 also had introgressions 3A.7 and 7A.11 that recombined with Chr 3A and Chr 7A of wheat, respectively, as indicated by the markers in addition to a very small introgression 7A.15 which had potentially gone into Chr 7B of wheat (Table 2 and Figure 2C).
The markers also showed that introgressions 5A.8 and 5A.13 had recombined with both Chr 5A and Chr 5D of wheat in various DH lines. A number of combinations of these introgressions were found in DH lines 232–259 (Table 2). Figure 2D shows the genotype of DH 240 which was shown to have 5A.8 in Chr 5D. The heterozygous calls on the distal end of Chr 5AL indicated the presence of a T. urartu Chr 5AuL segment which is really translocation of Chr4AuL. The null call for the marker at the distal end of Chr 5D indicated that this segment had replaced the corresponding region of Chr 5D in this DH line (Figure 2D).
Root metaphase spreads were obtained for a total of 75 out of the 86 DH lines for GISH analysis. In addition to observing recombinant chromosomes, GISH also allowed the detection of all the wheat chromosomes. Even though no GISH images were obtained for DH lines 265 and 271, the markers suggested that these lines were missing a pair of 1D and 5D chromosomes, respectively. Supplementary Table 1 provides details of the number of chromosomes observed for each subgenome in wheat for all DH lines with GISH images.
Discussion
The primary aim of the work carried out at the Nottingham BBSRC Wheat Research Centre is to generate stable homozygous wheat-wild relative introgressions lines that can be distributed to the wider wheat community for trait discovery. In this work, we describe a strategy to generate large numbers of stable introgressions in wheat from T. urartu using the DH method on some of the lines from the back-crossed population generated by Grewal et al. (2018a) and molecular characterisation of these lines. This is similar to the work reported by King et al. (2019) where DHs were generated for wheat-Am. muticum introgression lines and genotyped using the Axiom® Wheat Relative Genotyping Array. However, the wheat-T. urartu DH introgression lines produced in this study have been characterised with chromosome-specific KASP markers in addition to the Axiom® Array. KASP markers were found to be more flexible and cost-effective as a genotyping platform and allowed, in most cases, to identification of wheat chromosomes into which the T. urartu segments had introgressed.
Production of Stable Homozygous Wheat-T. urartu DH Lines
From the 49 BC3 plants that were used to generate the DH population, a total of 114 fertile DH plants were generated from 20 of them (41%) with the array genotyping indicating that 86 of the DH lines carried a T. urartu segment or whole chromosomes (Tables 1, 2). This indicates that the DH technique was successful in the generation of homozygous introgression lines although at a low rate. A higher percentage (75%) of the DH lines produced carried a T. urartu segment in this work as compared to the wheat-Am. muticum DH lines where only 31% of the DH lines generated carried an Am. muticum introgression (King et al., 2019). However, it is difficult to assess the actual level of introgression transmission in these T. urartu lines as the original BC3 plants were not genotyped as the marker systems for the identification and characterization of the introgressions were still under development at the time of production and hence not available. Once homozygous, each introgression was stably inherited in every DH line. The generation of these DH lines accelerated the production of stable homozygous introgression lines which would have otherwise taken a number of rounds of self-fertilisation to remove heterozygosity. In addition, because the chromosome-specific KASP markers were not then available, progeny testing would have been needed in order to select lines homozygous for a segment, adding considerably to the logistics and costs of the development of the introgression lines. Due to the technical requirements of DH production and the availability of the chromosome-specific KASP markers, all lines currently in development at the WRC are being generated via a backcross and selfing programme.
Development of Chromosome-Specific KASP Markers for T. urartu
Thirty three new KASP markers were also developed in this work to add to the 118 reported previously (Grewal et al., 2020a) that are polymorphic between bread wheat and T. urartu. This set makes up only 9% of the markers tested in this work as the SNPs used for KASP assay conversion were identified from a variety of wild relatives and wheat accessions, as described in the methods. All KASP assays designed were tested on the DH lines generated in this work to identify as many KASP markers as possible for T. urartu. The other 301 KASP markers also developed in this work are polymorphic between wheat and a number of other wild relative species including (but not limited to) Am. muticum, Th. bessarabicum and T. timopheevii (data not shown). This is also a valuable set of KASP markers that can be used in other wheat wild relative breeding programmes.
It should be noted that a majority of the KASP markers (∼70%) in each linkage group, used for genotyping the DH lines in this work, were specific to the A-genome chromosomes (Table 3) and were well distributed across all seven A-genome chromosomes. This is due to the strategy of having subgenome-specific SNPs. T. urartu is the donor of the A-genome and therefore it is likely that sequences that would result in such SNPs with T. urartu would be present more frequently on the A-genome of wheat than the B and D genomes (on average there are between 5 and 7 markers on these two genomes in each linkage group). The current KASP marker set was able to detect most of the linkage groups that were detected in the DH lines by the Axiom array with the exception of a few lines where the Axiom array found the presence of a small segment from linkage group 5 of T. urartu but the KASP markers did not (Table 2). This indicates that more markers need to be developed to fill in any potential gaps in linkage group 5. However, the conversion of array SNPs into chromosome-specific KASP markers able to distinguish between heterozygous and homozygous individuals in polyploid species such as bread wheat, is complex and time-consuming (Makhoul et al., 2020). We are therefore currently developing strategies to identify single-copy regions in the wheat genome in order to generate more chromosome-specific SNPs between T. urartu and the B and D genome chromosomes of wheat. The use of single-copy regions to develop chromosome-specific markers could bias genotyping toward genome regions without strong homoeology, which might later make it difficult to find tightly linked markers for a trait of interest if the candidate genes were found to be in highly homoeologous regions. However, the objective of this work was firstly to introgress as many wild-relative segments into bread wheat as possible and secondly to develop a suitable genotyping platform that would allow effective marker-assisted breeding. When a wild relative introgression is identified using KASP markers, the distribution of any lines carrying that segment will be accompanied by information on the markers that can be used to track the segment in any crosses with locally adapted material.
Identifying the Size and the Recipient Wheat Genome of T. urartu Introgressions in DH Lines
The KASP markers allowed the identification of 65 unique T. urartu segments in the DH lines generated (Figure 1) and also gave an indication of the wheat chromosomes with which recombination had occurred. These 65 unique introgressions were contributed by 19 out of the 20 BC3 plants that were used for DH production (1 plant produced DH lines without any segments), averaging 3.4 introgressions from every donor BC3 plant. Some of the DH lines retained up to five introgressions potentially indicating a much higher number in the BC1 parent. These relatively large numbers of introgressions, even at the BC3 stage, are not surprising as T. urartu is the A-genome donor of bread wheat and homologous recombination, with the A-genome chromosomes of wheat was expected to occur at a high frequency in the F1 gametes as pairing between the chromosomes of T. urartu and the A-genome chromosomes of wheat has previously been shown to occur at a high frequency (Chapman et al., 1976; Dvořák, 1976). A majority of the introgressions (83%) were found to be with the A-genome of wheat (Table 2) as indicated by the homozygous wild relative calls for the A-genome specific KASP markers that were present within introgressions.
Due to the high similarity between the A-genome sequence of wheat and T. urartu, it was not possible to perform GISH analysis to observe T. urartu segments that had recombined with A-genome chromosomes in wheat. However, where homoeologous recombination had taken place between the T. urartu chromosomes and the wheat B or D genome chromosomes, it was possible to validate the genotyping using GISH analysis (Figure 2). The smallest segments that can currently be detected with GISH are about 18 Mbp and thus only the D-genome introgressions were large enough to visualise with GISH, where the markers had indicated small genomic regions of the wheat D genome chromosomes were present in recombinant chromosomes (Figure 2). This is due to the chromosome-specificity of the KASP markers which is able to firstly distinguish heterozygous from homozygous lines and secondly, in homozygous lines, to identify which wheat chromosomes had recombined with the T. urartu introgressions.
Only one segment from Chr 4Au of T. urartu was found in the DH lines in this work and furthermore, it had introgressed with the Chr 4D of wheat. This is probably due to the rearrangement of Chr 4A of bread wheat as compared to Chr 4Au. Even though both species have the 4AL/5AL translocation as this had occurred in the diploid ancestor before the formation of hexaploid wheat (King et al., 1994; Devos et al., 1995), Chr 4A of wheat underwent a further paracentric and pericentric inversion (Devos et al., 1995; Dvořák et al., 2018) which makes homoeologous recombination between Chr 4Au and Chr 4A difficult during meiosis as has been previously reported (Grewal et al., 2018a). Chr 4Au and Chr 4D show greater synteny in the short arm region than Chr 4Au and Chr 4A where introgression 4A.1 recombined with Chr 4D.
Multi-colour GISH analysis on 75 lines allowed the identification of the number of chromosomes in each line (Supplementary Table 1). While GISH can only indicate how many chromosomes are present per subgenome, the markers were able to identify where a pair of wheat chromosomes were missing. For example, in lines 324, 325, 327, and 329 all four chromosome 2D KASP markers gave a null call indicating that the pair of 2D chromosomes was missing in these lines. Similarly, genotyping with chromosome specific markers identified that DH lines 265 and 271 were missing a pair of 1D and 5D chromosomes, respectively. However, these markers are unable to indicate when only one of a pair of wheat chromosomes is missing.
All wheat-T. urartu DH lines have been deposited at the Germplasm Resource Unit (GRU) at John Innes Centre and are available to order via their website https://www.seedstor.ac.uk/ subject to the terms of the Material Transfer Agreement.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
SG, CY, SA, DS, SH-E, IK, and JK contributed to the backcross population creation. CN and AS contributed to the DH production. CY, DS, and SA contributed to the DNA extractions. AB contributed to the array genotyping. SA and DS contributed to the KASP genotyping. VG and CY contributed to the GISH analysis. SG contributed to the data analysis. IK, JK, and SG contributed to the conceptualization. SG, IK, and JK contributed to the manuscript writing. IK and JK contributed to the funding acquisition. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Biotechnology and Biological Sciences Research Council (Grant Numbers BB/J004596/1 and BB/P016855/1) as part of the Wheat Improvement Strategic Programme (WISP) and Designing Future Wheat (DFW) programme.
Conflict of Interest
CN and AS were employed by Limagrain UK Limited at the time this work was carried out.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.643636/full#supplementary-material
Footnotes
References
Austin, R. B. (1986). Dry matter yields and photosynthetic rates of diploid and hexaploid triticum species. Annal. Bot. 57, 847–857. doi: 10.1093/oxfordjournals.aob.a087169
Austin, R. B., Morgan, C. L., Ford, M. A., and Bhagwat, S. G. (1982). Flag leaf photosynthesis of Triticum aestivum and related diploid and tetraploid species. Annal. Bot. 49, 177–189. doi: 10.1093/oxfordjournals.aob.a086238
Baker, L., Grewal, S., Yang, C.-Y., Hubbart-Edwards, S., Scholefield, D., Ashling, S., et al. (2020). Exploiting the genome of thinopyrum elongatum to expand the gene pool of hexaploid wheat. Theor. Appl. Genet. 133, 2213–2226. doi: 10.1007/s00122-020-03591-3
Burridge, A. J., Wilkinson, P. A., Winfield, M. O., Barker, G. L. A., Allen, A. M., Coghill, J. A., et al. (2018). Conversion of array-based single nucleotide polymorphic markers for use in targeted genotyping by sequencing in hexaploid wheat (triticum aestivum). Plant Biotechnol. J. 16, 867–876. doi: 10.1111/pbi.12834
Chapman, V., Miller, T. E., and Riley, R. (1976). Equivalence of the a genome of bread wheat and that of Triticum urartu. Genet. Res. 27, 69–76.
Charmet, G. (2011). Wheat domestication: lessons for the future. C. R. Biol. 334, 212–220. doi: 10.1016/j.crvi.2010.12.013
Cuesta, S., Guzmán, C., and Alvarez, J. B. (2015). Molecular characterization of novel LMW-i glutenin subunit genes from triticum urartu thum. ex gandil. Theor. Appl. Genet. 128, 2155–2165. doi: 10.1007/s00122-015-2574-1
Dempewolf, H., Baute, G., Anderson, J., Kilian, B., Smith, C., and Guarino, L. (2017). Past and future use of wild relatives in crop breeding. Crop Sci. 57, 1070–1082. doi: 10.2135/cropsci2016.10.0885
Devi, U., Grewal, S., Yang, C. Y., Hubbart-Edwards, S., Scholefield, D., Ashling, S., et al. (2019). Development and characterisation of interspecific hybrid lines with genome-wide introgressions from triticum timopheevii in a hexaploid wheat background. BMC Plant Biol. 19:183. doi: 10.1186/s12870-019-1785-z
Devos, K. M., Dubcovsky, J., Dvorak, J., Chinoy, C. N., and Gale, M. D. (1995). Structural evolution of wheat chromosomes 4A, 5A, and 7B and its impact on recombination. Theor. Appl. Genet. 91, 282–288. doi: 10.1007/bf00220890
Dvořák, J. (1976). The Relationship between the genome of Triticum urartu and the A and B genomes of Triticum aestivum. Can. J. Genet. Cytol. 18, 371–377. doi: 10.1139/g76-045
Dvořák, J., and Akhunov, E. D. (2005). Tempos of gene locus deletions and duplications and their relationship to recombination rate during diploid and polyploid evolution in the aegilops-triticum alliance. Genetics 171, 323–332. doi: 10.1534/genetics.105.041632
Dvořák, J., Terlizzi, P. D., Zhang, H.-B., and Resta, P. (1993). The evolution of polyploid wheats: identification of the a genome donor species. Genome 36, 21–31. doi: 10.1139/g93-004
Dvořák, J., Wang, L., Zhu, T., Jorgensen, C. M., Luo, M. C., Deal, K. R., et al. (2018). Reassessment of the evolution of wheat chromosomes 4A, 5A, and 7B. Theor. Appl. Genet. 131, 2451–2462. doi: 10.1007/s00122-018-3165-8
Feldman, M., Lupton, F. G. H., and Miller, T. E. (1995). Wheats. Evolution of crop plants, 2nd Edn. London: Longman Scientific, 184–192.
Grewal, S., Hubbart-Edwards, S., Yang, C., Devi, U., Baker, L., Heath, J., et al. (2020a). Rapid identification of homozygosity and site of wild relative introgressions in wheat through chromosome-specific KASP genotyping assays. Plant Biotechnol. J. 18, 743–755. doi: 10.1111/pbi.13241
Grewal, S., Othmeni, M., Walker, J., Edwards, S. H., Yang, C.-Y., Scholefield, D., et al. (2020b). Development of wheat-Aegilops caudata introgression lines and their characterisation using genome-specific KASP markers. Front. Plant Sci. 11:606. doi: 10.3389/fpls.2020.00606
Grewal, S., Hubbart-Edwards, S., Yang, C., Scholefield, D., Ashling, S., Burridge, A., et al. (2018a). Detection of T. urartu Introgressions in wheat and development of a panel of interspecific introgression lines. Front. Plant Sci. 9:1565. doi: 10.3389/fpls.2018.01565
Grewal, S., Yang, C., Edwards, S. H., Scholefield, D., Ashling, S., and Burridge, A. J. I, et al. (2018b). Characterisation of Thinopyrum bessarabicum chromosomes through genome-wide introgressions into wheat. Theor. Appl. Genet. 131, 389–406. doi: 10.1007/s00122-017-3009-y
Huang, S., Sirikhachornkit, A., Su, X., Faris, J., Gill, B., Haselkorn, R., et al. (2002). Genes encoding plastid acetyl-CoA carboxylase and 3-phosphoglycerate kinase of the triticum/aegilops complex and the evolutionary history of polyploid wheat. Proc. Natl. Acad. Sci. USA 99, 8133–8138. doi: 10.1073/pnas.072223799
IWGSC, Appels, R., Eversole, K., Feuillet, C., Keller, B., Rogers, J., et al. (2018). Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361:eaar7191.
Kato, A., Lamb, J. C., and Birchler, J. A. (2004). Chromosome painting using repetitive DNA sequences as probes for somatic chromosome identification in maize. Proc. Natl. Acad. Sci. USA 101, 13554–13559. doi: 10.1073/pnas.0403659101
Kihara, H. (1944). Discovery of the DD-analyser, one of the ancestors of triticum vulgare (abstr) (in japanese). Agric. Hortic. 19, 889–890.
King, I. P., Purdie, K. A., Liu, C. J., Reader, S. M., Pittaway, T. S., Orford, S. E., et al. (1994). Detection of interchromosomal translocations within the triticeae by RFLP analysis. Genome 37, 882–887. doi: 10.1139/g94-125
King, J., Grewal, S., Yang, C. Y., Hubbart, S., Scholefield, D., Ashling, S., et al. (2017). A step change in the transfer of interspecific variation into wheat from Ambylopyrum muticum. Plant Biotechnol. J. 15, 217–226. doi: 10.1111/pbi.12606
King, J., Grewal, S., Yang, C.-Y., Hubbart Edwards, S., Scholefield, D., Ashling, S., et al. (2018). Introgression of aegilops speltoides segments in triticum aestivum and the effect of the gametocidal genes. Annal. Bot. 121, 229–240. doi: 10.1093/aob/mcx149
King, J., Newell, C., Grewal, S., Hubbart-Edwards, S., Yang, C.-Y., Scholefield, D., et al. (2019). Development of stable homozygous wheat/amblyopyrum muticum (aegilops mutica) introgression lines and their cytogenetic and molecular characterization. Front. Plant Sci. 10:34. doi: 10.3389/fpls.2019.00034
Laurie, D. A., and Reymondie, S. (1991). High frequencies of fertilization and haploid seedling production in crosses between commercial hexaploid wheat varieties and maize. Plant Breed. 106, 182–189. doi: 10.1111/j.1439-0523.1991.tb00499.x
Ling, H. Q., Ma, B., Shi, X., Liu, H., Dong, L., Sun, H., et al. (2018). Genome sequence of the progenitor of wheat a subgenome triticum urartu. Nature 557, 424–428. doi: 10.1038/s41586-018-0108-0
Liu, C. J., Atkinson, M. D., Chinoy, C. N., Devos, K. M., and Gale, M. D. (1992). Nonhomoeologous translocations between group 4, 5 and 7 chromosomes within wheat and rye. Theor. Appl. Genet. Theor. Appl. Genet. 83, 305–312. doi: 10.1007/BF00224276
Makhoul, M., Rambla, C., Voss-Fels, K. P., Hickey, L. T., Snowdon, R. J., and Obermeier, C. (2020). Overcoming polyploidy pitfalls: a user guide for effective SNP conversion into KASP markers in wheat. Theor. Appl. Genet. 133, 2413–2430. doi: 10.1007/s00122-020-03608-x
Marcussen, T., Sandve, S. R., Heier, L., Spannagl, M., Pfeifer, M., Jakobsen, K. S., et al. (2014). Ancient hybridizations among the ancestral genomes of bread wheat. Science 345:1250092.
Martín, M. A., Martín, L. M., and Alvarez, J. B. (2008). Polymorphisms at the Gli-Au1 and Gli-Au2 loci in wild diploid wheat (triticum urartu). Euphytica 163, 303–307. doi: 10.1007/s10681-008-9656-y
Matsuoka, Y. (2011). Evolution of polyploid triticum wheats under cultivation: the role of domestication, natural hybridization and allopolyploid speciation in their diversification. Plant Cell. Physiol. 52, 750–764. doi: 10.1093/pcp/pcr018
McFadden, E. S., and Sears, E. R. (1944). The artificial synthesis of Triticum spelta. Rec. Genet. Soc. 13, 26–27.
Nemeth, C., Yang, C. Y., Kasprzak, P., Hubbart, S., Scholefield, D., Mehra, S., et al. (2015). Generation of amphidiploids from hybrids of wheat and related species from the genera aegilops, secale, thinopyrum, and triticum as a source of genetic variation for wheat improvement. Genome 58, 71–79. doi: 10.1139/gen-2015-0002
Qiu, Y. C., Zhou, R. H., Kong, X. Y., Zhang, S. S., and Jia, J. Z. (2005). Microsatellite mapping of a triticum urartu tum. derived powdery mildew resistance gene transferred to common wheat (triticum aestivum L.). Theor. Appl. Genet. 111, 1524–1531. doi: 10.1007/s00122-005-0081-5
Ramirez-Gonzalez, R. H., Uauy, C., and Caccamo, M. (2015). Poly marker: a fast polyploid primer design pipeline. Bioinformatics 31, 2038–2039. doi: 10.1093/bioinformatics/btv069
Riley, R., and Chapman, V. (1958). Genetic control of the cytologically diploid behavious of hexaploid wheat. Nature 182, 259–260.
Rouse, M. N., and Jin, Y. (2011). Stem rust resistance in a-genome diploid relatives of wheat. Plant Dis. 95, 941–944. doi: 10.1094/pdis-04-10-0260
Sarkar, P., and Stebbins, G. L. (1956). Morphological evidence concerning the origin of the B genome in wheat. Am. J. Bot. 42, 297–304.
Sears, E. R. (1976). Genetic control of chromosome pairing in wheat. Annu. Rev. Genet. 10, 31–51. doi: 10.1146/annurev.ge.10.120176.000335
Sears, E. R., and Okamoto, M. (1958). Intergenomic chromosome relationships in hexaploid wheat. Proc. X Int. Congr. Genet. 2, 258–259.
Sheedy, J. G., Thompson, J. P., and Kelly, A. (2012). Diploid and tetraploid progenitors of wheat are valuable sources of resistance to the root lesion nematode pratylenchusthornei. Euphytica 186, 377–391. doi: 10.1007/s10681-011-0617-5
Thomson, D., and Henry, R. (1995). Single-step protocol for preparation of plant tissue for analysis by PCR. BioTechniques 19:400.
Keywords: wheat, Triticum urartu, doubled haploids, introgressions, KASP markers, genotyping
Citation: Grewal S, Guwela V, Newell C, Yang C-y, Ashling S, Scholefield D, Hubbart-Edwards S, Burridge A, Stride A, King IP and King J (2021) Generation of Doubled Haploid Wheat-Triticum urartu Introgression Lines and Their Characterisation Using Chromosome-Specific KASP Markers. Front. Plant Sci. 12:643636. doi: 10.3389/fpls.2021.643636
Received: 18 December 2020; Accepted: 19 April 2021;
Published: 13 May 2021.
Edited by:
Jacqueline Batley, University of Western Australia, AustraliaReviewed by:
Masahiro Kishii, International Maize and Wheat Improvement Center (Mexico), MexicoSurinder Banga, Punjab Agricultural University, India
Copyright © 2021 Grewal, Guwela, Newell, Yang, Ashling, Scholefield, Hubbart-Edwards, Burridge, Stride, King and King. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Julie King, anVsaWUua2luZ0Bub3R0aW5naGFtLmFjLnVr
 Surbhi Grewal
Surbhi Grewal Veronica Guwela
Veronica Guwela Claire Newell2
Claire Newell2 Cai-yun Yang
Cai-yun Yang Stephen Ashling
Stephen Ashling Duncan Scholefield
Duncan Scholefield Stella Hubbart-Edwards
Stella Hubbart-Edwards Amanda Burridge
Amanda Burridge Alex Stride
Alex Stride Ian P. King
Ian P. King Julie King
Julie King