- 1Key Laboratory of Forest Genetics and Biotechnology, Ministry of Education of China, Co-Innovation Center for the Sustainable Forestry in Southern China, Nanjing Forestry University, Nanjing, China
- 2College of Biology and the Environment, Nanjing Forestry University, Nanjing, China
Liriodendron chinense (Lchi) is a Magnoliaceae plant, which is a basic angiosperm left behind by the Pleistocene and mainly distributed in the south of the Yangtze River. Liriodendron hybrids has good wood properties and is widely used in furniture and in other fields. It is not clear if they can adapt to different environmental conditions, such as drought and high and low temperatures, and the molecular mechanisms for this adaptation are unknown. Among plant transcription factors (TFs), the MYB gene family is one of the largest and is often involved in stress or adversity response signaling, growth, and development. Therefore, studying the role of MYBTFs in regulating abiotic stress signaling, growth, and development in Lchi is helpful to promote afforestation in different environments. In our research, a genome-wide analysis of the LchiMYB gene family was performed, including the phylogenetic relationship tree, gene exon-intron structure, collinearity, and chromosomal position. According to the evolutionary tree, 190 LchiMYBs were divided into three main branches. LchiMYBs were evenly distributed across 19 chromosomes, with their collinearity, suggesting that segment duplication events may have contributed to LchiMYB gene expansion. Transcriptomes from eight tissues, 11 stages of somatic embryogenesis, and leaves after cold, heat, and drought stress were used to analyze the function of the MYB gene family. The results of tissue expression analysis showed that most LchiMYB genes regulated bark, leaf, bud, sepal, stigma, and stamen development, as well as the four important stages (ES3, ES4, ES9, and PL) of somatic embryogenesis. More than 60 LchiMYBs responded to heat, cold, and drought stress; some of which underwent gene duplication during evolution. LchiMYB3 was highly expressed under all three forms of stress, while LchiMYB121 was strongly induced by both cold and heat stress. Eight genes with different expression patterns were selected and verified by quantitative real-time PCR (qRT-PCR) experiments. The results suggested that these LchiMYBs may regulate Lchi growth development and resistance to abiotic stress. This study shows the cross-regulatory function of LchiMYBs in the growth and development, asexual reproduction, and abiotic resistance of Lchi. This information will prove pivotal to directing further studies on the biological function of Lchi MYBTFs in genetic improvement and abiotic stress response.
Introduction
Plants respond to different environmental conditions through a series of physiological and metabolic processes (Chen et al., 2017; Li J. et al., 2019). At every stage of plant growth and development, there are many genes involved in regulating its biological processes (Dubos et al., 2010). Transcription factors (TFs), which bind to upstream sequences of genes, regulate gene expression and play a vital role in regulating biological processes (Ambawat et al., 2013; Li Y. et al., 2020). Generally, TFs are composed of at least four domains, including a transactivation domain, a DNA-binding domain, a nuclear localization signal, and oligomerization sites (Stracke et al., 2001; Dubos et al., 2010). According to the types of conserved domains, TFs can be divided into many families. The MYB gene family exists in all eukaryotes, especially in the plant kingdom. It is one of the largest families and has functional diversity (Dubos et al., 2010; Ambawat et al., 2013).
The highly conservative MYB domain of MYB family members comprises 1–4 neighboring incomplete tandem repeats. Each repeat unit consists of an average of 50–53 amino acid (aa) residues and forms three α-helices. The second and third repeats have a helix-turn-helix (HTH) secondary structure; these repeats bind in the upstream regions of the target genes (Stracke et al., 2001; Chen et al., 2017; Wei et al., 2020). However, MYB TFs possess many regulatory roles, as the more variable C-terminus of MYB proteins (Zhang et al., 2020). According to the number of neighboring MYB repeats, MYBs are divided into four capital subgroups: MYB-related (1R-MYB), R2R3-MYB (2R-MYB), R1R2R3-MYB (3R-MYB), and 4R-like MYB (4R-MYB). In the plant kingdom, 2R-MYBs are one of the maximal and most common MYBs and are considered to have two main evolutionary modes: the R1R2R3-MYB domain through the lack of an R1 repeat and the duplication of an R1 repeat in the R1-MYB domain (Dubos et al., 2010; Ambawat et al., 2013).
Since the discovery of the first plant MYB gene C1, the identification and functional characteristics of the plant MYB gene family have been widely reported (Dubos et al., 2010). A total of 519 TFs expressed in Arabidopsis are involved in regulating somatic embryogenesis (Gliwicka et al., 2013). Many TFs, such as LEAFY COTYLEDON (LEC), WUSCHEL (WUS), BABY BOOM (BBM), Arabinogalactan-proteins (AGPs), CLAVATA (CLV), and APETALA 2 (AP2), are involved in regulating the somatic embryogenesis process (Méndez-Hernández et al., 2019). WRKY, MYB, AIL, and bHLH regulate the later phase of somatic embryogenesis in rubber (Wang et al., 2021). In different tissues, AtMYB113 and AtMYB114 are expressed in vegetative tissues and regulate anthocyanin biosynthesis (Dubos et al., 2010). Eight BdMYB genes were expressed specifically in leaves of Brachypodium distachyon (Chen et al., 2017). Some R2R3-MYB genes had the highest expression in the pericarp of capsicum annuum L. (Wang J. et al., 2020).
Recently, numerous studies have focused on the response of the MYB family members to plant stress. In Arabidopsis, AtMYB15 responds to cold stress and has a negative regulatory role in the CBF signaling process (Shen et al., 2019). In rice, OsMYB2 responds to salt, cold, and drought stresses (Jiang et al., 2004; Katiyar et al., 2012). The TaMYB sdu1 gene in wheat (Triticum aestivum L.) has an important role in responding to salt and drought stresses (Rahaie et al., 2010). Overexpression of TaMYB34 in transgenic tobacco can enhance tolerance to drought, high temperature and high salt stress (Wei et al., 2020). In poplar, transgenic tobacco overexpressing PsnMYB108 gene has stronger salt tolerance than wild-type tobacco (Zhao et al., 2020).
The Liriodendron genus belongs to the Magnolia family. Other than the Pleistocene, there exist only two species in this genus, i.e., the East Asian [L. chinense (Hemsley) Sargent] species and the North-East American species (L. tulipifera Linn) (Chen et al., 2019). Two separate lineages of Liriodendron chinense (Lchi) are found on the Chinese mainland, comprising the eastern and western groups (Yang et al., 2016; Chen et al., 2019). Liriodendron hybrids is intraspecific hybrids crossed by L. chinense and L. tulipfera, which was a valuable ornamental tree species in both private and public spaces because of its fast growth, good wood properties, and wide use in furniture making and other fields (Hu et al., 2017; Li et al., 2021).
Complete genome sequencing of Lchi showed that magnolia is an ancient angiosperm, which is present before monocotyledon and dicotyledon differentiation (Chen et al., 2019). Therefore, it could provide valuable information on how plants from such an ancient phylogenetic branch cope with abiotic stress. Lchi is mainly distributed in the south of the Yangtze River (Chen et al., 2019); there are few studies on its ability to adapt to different environments, such as colder northern China, warmer southern China, or arid northwestern China. The MYB gene family has important functions in the growth, development, and response to abiotic stress, but studies on the whole genome of Lchi have rarely been reported.
In this research, the MYB family was systematically analyzed in the whole genome of Lchi, and 190 LchiMYBs were divided into three main groups (1R-MYB, 2R-MYB, and 3R-MYB). An overall analysis, including the determination of exon-intron structure, a gene duplication event, chromosome location, phylogenetic analysis, and synteny analysis, was performed. Furthermore, the specifically expressed MYB gene family members were identified through transcriptome analysis of vegetative and reproductive organs, somatic embryogenesis, and abiotic stressors of high and low temperatures and drought in Lchi. The results suggest that LchiMYBs have a vital role in the growth and development of Lchi and its adaptability to adversity.
This study provides insights into the regulatory function of LchiMYB in the growth and development, asexual reproduction, and abiotic resistance of Lchi, which provide a theoretical basis for the genetic improvement of Lchi, thereby increasing forest yield and promoting afforestation.
Materials and Methods
Datasets and Sequence Retrieval
The complete genome, transcript/protein sequences, and the genome feature file of Lchi were downloaded from https://www.ncbi.nlm.nih.gov/assembly/GCA_003013855.2. All MYB proteins of Arabidopsis thaliana were obtained from the TAIR database1. An MYB Hidden Markov profile (PF00249) was retrieved from the Pfam website2, and the MYB-domain-containing protein sequences in the Lchi genome were identified, using HMMER (v.3.0.1b, Cambridge University Press, England, United Kingdom) (with an E-value < 1E-5 and using this profile as a query. As a result, all candidate LchiMYBs were confirmed, using Pfam and the conservative Domains Database (CDD3) to validate the core domain sequences.
Sequence Analysis
The gene exon-intron structural feature of each LchiMYB was acquired from the genomic feature file (GFF/GTF) and displayed, using the GSDS website the R2R3 domain “seqlogos” map was made through the WebLogo (http://weblogo.berkeley.edu/)4, while the chromosome location and microsynteny of LchiMYBs were visualized, using the R software “Circlize”5 (Gu et al., 2014)5. The presumptive orthologous gene pairs (E-value cutoff < 1E-20, identity > 70%) were verified by the Python software “MCScanx”6 and then used for comparative synteny analysis among two model plants (Wang et al., 2012). The cis-acting elements of LchiMYB upstream regions were predicted by PlantCARE7. Ka/Ks values were assigned to LchiMYBs, using the Ka/Ks_calculator (Nekrutenko et al., 2002). The basic properties of LchiMYB proteins, including protein length, isoelectric point (pI), and molecular weight (MW), were analyzed, using the ExPasy website8.
Phylogenetic Analysis
Multiple sequence alignment (MSA) of Lchi and A. thaliana MYB-domain-containing proteins was done, using MUSCLE (v3.8.31), which was set to default parameters. The trimmed MSA file was used to construct the MYB phylogenetic tree, which was generated with trimAl (v1.4), set to the “automated1” mode (Capella-Gutierrez et al., 2009). The BEAST (v2.6.2) was used to perform MYB phylogenic analysis under the following settings: the maximum likelihood method, the substitution model was Dayhoff, priors set to the Yule Model, the birth rate was 1.0, the mcmc chain length was 10,000,000, bootstrapping set to 1,000, and the TreeAnnotator program set the posterior probability limit to 1.0 with a burn in percentage of 90 (Drummond et al., 2012). Then, the R software “ggtree” was used to visualize the phylogenic tree9 (Yu et al., 2018; Yu, 2020).
Somatic Embryogenesis Transcriptome of Liriodendron hybrids
Embryogenic callus was induced from the embryo of seeds of a Liriodendron hybrids, and successive stages of somatic embryogenesis were used for transcriptome sequencing. The data have not yet been published. The 11 stages are as follows: proembryogenic mass cultured for 20 days for embryogenic callus (PEMs), then liquid suspension cultured for 10 days (ES1), single cell cultured for 2 days (ES2), induced by ABA for 1 day (ES3) and 3 days (ES4), a 7-day globular embryo (ES5), a 13-day heart-shaped embryo (ES6), a 19-day torpedo embryo (ES7), a 25-day immature cotyledon embryo (ES8), a 31-day mature cotyledon embryo (ES9), and a 37-day plantlet (PL). The Fragments Per Kilobase of exon model per million mapped fragments (FPKM) of transcriptome data is shown in the Supplementary Table 1.
Various Tissue Transcriptomes of Lchi
The transcriptomes of various tissue types, namely, the phloem, stigma, xylem, bud, stamen, leaf, bark, and sepal, were used. The FPKM data of LchiMYBs are shown in the Supplementary Table 2. These transcriptome data have not yet been published.
Plant Materials, RNA Extraction, qRT-PCR Analysis, and Transcriptome Sequencing Analysis of Abiotic Stress in Liriodendron hybrids
Liriodendron hybrids seedlings generated through somatic embryogenesis were used as the starting material throughout this study. Before any experiments were performed, plantlets were taken out of the culture medium vessel and acclimatized in a greenhouse for 2 weeks (22°C, long-time photoperiod of 16-h light and 8-h darkness, and 75% relative humidity). For three abiotic adversity treatments, plants were transferred to a growth incubator (long-time photoperiod of 16-h light and 8-h darkness, and 75% relative humidity). To simulate cold, heat, and drought stress, plantlets were subjected to 4°C, 40°C, or 15% PEG 6000 treatment, respectively, for 1 h, 3 h, 6 h, 12 h, 1 day, and 3 days. Each treatment consisted of five replicates for each sampling time. All experimental leaf tissue samples were frozen in liquid nitrogen immediately and then conservatively at −80°C for RNA extraction by RNA-seq analysis and quantitative real-time PCR (qRT-PCR) experiments.
We used the FastPure Total RNA Isolation Kit (Vazyme, Nanjing, China) (RC401) to isolate total RNA in leaves. Then, agarose gel electrophoresis and a Nanodrop ND-1000 spectrophotometer were used to validate the quality of the extracted RNA. A HiScript® III 1st Strand cDNA Synthesis Kit (Vazyme, Nanjing, China) was used to synthesize cDNA with the extracted RNA as a template.
Lchi leaf tissue transcriptome after the three stressors was determined with RNA-seq on biological triplicates by using the extracted RNA samples and then mapping the clean reads, using our genome sequence as a reference. An RNA-seq analysis software pipeline was employed, consisting of hisat2 (v2.0.5) to map clean reads to the genomic sequence and Kallisto (v0.46.1) to count transcripts per million (TPM) values of individual genes. The TPM values of all mapped LchiMYBs (35269) are shown in Supplementary Table 3. DESeq2 (v 1.16.1) used analysis of variance (ANOVA; P.adjust < 0.05, | Log2FC| > 1) to determine the genes that were differentially expressed. TPM was used to calculate the transcript abundance of LchiMYB genes. Expression heatmaps were created, using the R package “pheatmap” (v1.0.12), with the log 2 (TPM + 1) values. The transcriptome data of the present study were archived and can also be obtained on NCBI (cold and heat: PRJNA679089; drought: PRJNA679101).
Primers of eight MYB genes selected by RNA-seq results with different expression patterns were used for quantifying the expression profile and were designed through the primer 3 website10. The RNA samples used for quantitative RT-PCR (qRT-PCR) were the same as those for RNA-seq. qRT-PCR was implemented with a Roche Lightcyler® 480II instrument, using 2x AceQ® qPCR SYBR® Green Master Mix (Without ROX) (Vazyme, Nanjing, China). The composition of the PCR mix was as follows: 10 μl of 2x AceQ® qPCR SYBR® Green Master Mix (without ROX), 0.4 μl of each primer, 1 μl of a cDNA template (10 ng/μl), and 8.2 μl of ddH2O for a final volume of 20 μl. The internal reference gene chosen for housekeeping was the Lchi 18S gene. The reaction was implemented as follows: 95°C for 30 s and then 45 cycles of 95°C for 10 s and 60°C for 30 s. All experiments were run in 96-well plates. Each experiment was carried out in biological triplicates, as well as three technical replicates. All data generated from qRT-PCR were calculated by the 2–ΔΔCT formula. The melt curve of primers is shown in the Supplementary Figure 1, and sequences are also shown in Supplementary Table 4.
Sequence analysis was used to obtain the number of LchiMYB family members, physical and chemical properties of the protein, replication events, and promoter functions of regulating growth, development, and abiotic stress response. Phylogenetic analysis further analyzed the grouping specificity of MYB genes and explored their expression patterns in the growth, development, and response to abiotic stress according to the grouping specific functions of these genes. Therefore, abiotic stress experiments and RNA-seq of tissue and somatic embryogenesis were carried out. Finally, eight genes with different expression patterns for abiotic stress were selected for qRT-PCR verification.
Results
Identification of LchiMYBs and Corresponding Protein Sequence Features
To investigate the MYB genes within the Liriodendron chinense (Lchi) genome, the MYB hidden Markov model (PF00249) was used as a query in hmmsearch searches against the Lchi genome-wide protein sequences. Many MYB protein sequences (>250 candidates) that had MYB or MYB-like repeats were obtained. In Pfam, these sequences were further used to verify the completeness of the MYB domain in the identified sequences, discarding those with or without incomplete MYB domains. Finally, a total of 190 non-redundant LchiMYB proteins were obtained, including 76 (40%) R2R3-MYBs (2R-MYBs), 109 (57.37%) MYB-related (1R-MYBs) proteins, and 5 (2.63%) R1R2R3-MYBs (3R-MYBs). Together, they accounted for 0.5387% of all annotated Lchi genes (35,269), which is a larger fraction than that in rice (0.3934%), yet smaller than that in Arabidopsis (0.5964%), which is of the same family. No 4R-MYB proteins were identified, possibly due to the incompleteness of the Lchi genome. The 1R-MYB proteins accounted for more than half of the total number of MYB proteins, thus constituting the largest group of MYB genes. According to the results of this study, most MYB genes resided within the R2R3-MYB subgroup, yet Lchi 2R-MYBs represent less than half of the total number of MYB proteins, which contrasts with previous studies on rice and Arabidopsis. Perhaps, this is one aspect that differentiates basic angiosperms, such as Lchi, from higher angiosperms.
The predicted LchiMYB genes were numbered from LchiMYB1 to LchiMYB165 based on their coordinate order on Lchi chromosomes, and LchiMYB166–LchiMYB190 were numbered based on their order on sequenced scaffolds. The characteristics of LchiMYB genes, including their chromosome position, protein MW, amino acid length, and pI, were further analyzed (Supplementary Table 5). The smallest protein was LchiMYB180 at 75 amino acids (aa) in length, and the largest was LchiMYB51 at 1,374 aa, while the MW ranged from 8.67 (LchiMYB100) to 149.2 (LchiMYB158) kDa, and the pI ranged from 4.34 (LchiMYB150) to 10.61 (LchiMYB48).
To detect the sequence characteristics of MYB and the base frequency of each amino acid site, 76 homologous R2R3-MYB sequences were analyzed by MSA, and the seqlog map of sequence frequency was obtained (Figure 1).
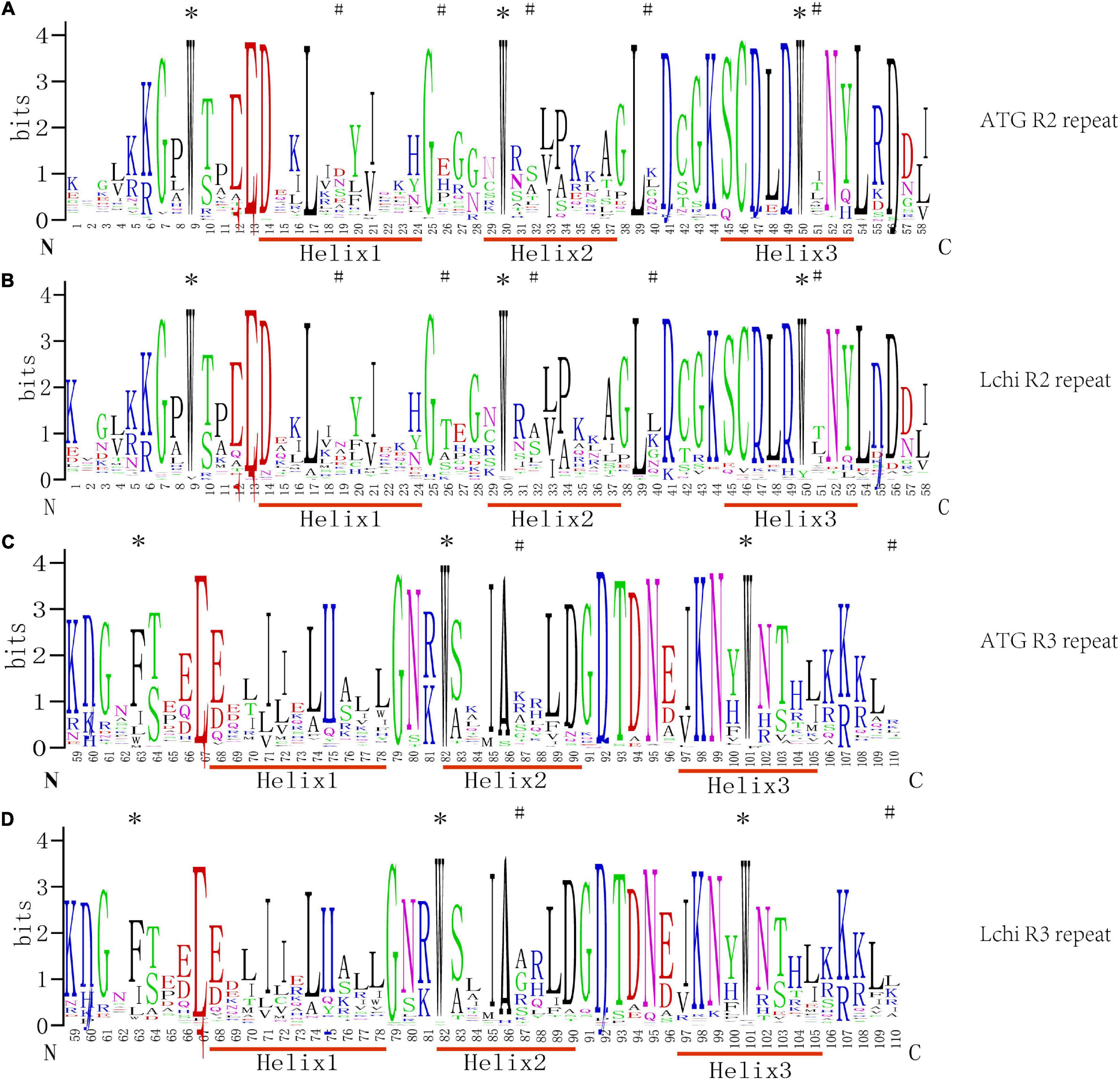
Figure 1. Sequence logos of the R2 and R3MYB repeats from Liriodendron chinense (Lchi) and Arabidopsis. Multiple sequence alignment analysis of 76 Lchi and 126 Arabidopsis R2R3-MYB domains was used in ClustalX2.1. Panels (A,B) were the R2 repeats, and panels (C,D) were the R3 repeats in Arabidopsis and Lchi. N and C represent the N-terminal and C-terminal, respectively, and the bit score shows the conservative information for all positions in the sequence; the higher the value, the higher the conservation, and the different colors were generated by the website randomly. The asterisks represent the highly conserved tryptophan (W) and phenylalanine (F) residues, and the # represents the different patterns of positions between Lchi and Arabidopsis.
In general, R2R3-MYB domains have, on average, approximately 108 amino acids between each MYB protein (Liu et al., 2020; Pucker et al., 2020). Outside the DNA-binding domain, however, the length and the composition of amino acids vary greatly. The LchiMYB proteins show a similar pattern, confirming the conserved nature of the MYB domain (Sun et al., 2019; Wang et al., 2019).
As shown in Figure 1, the amino acid distribution frequencies within the LchiMYB domain repeat closely and resemble those of many previously reported plant species; for example, grapes, Arabidopsis, poplars, and pears. However, compared with other plants, the R2 and R3 MYB repeats of LchiMYB proteins contain some unique amino acids, including several highly conserved and evenly distributed tryptophan (W) residues, which are usually used as a label of MYB proteins in plants. According to previous reports, the first conserved amino acid of the R3 repeat is often replaced by Phe (F) or Ile (I). However, Trp at position 63 is often replaced by the amino acid Leu (L) in LchiMYB proteins. In addition to the highly conserved tryptophan residues, the amino acids in other regions of the LchiMYB protein are also conserved, such as Gly-25 (G), Cys-46 (C), and Arg-49 (R) in the R2 repeat, Leu-54 (L) in the linker region, and Glu-66 (E), Thr-87 (T), and Arg-94 (R) in the R3 repeat. In other species, such conserved amino acids are also found, which are mainly related to the third helix and the second turn motif (Millard et al., 2019; Zhou et al., 2019). Additionally, some residues in Lchi were distinct from Arabidopsis, such as the patterns at positions 19, 26, 32, 40, and 51 of R2 and 87 and 110 of R3, which show the divergence of the MYB domain.
These results suggest that the diverse physical and chemical properties of LchiMYB, fewer R2R3 members, and the R2R3-MYB domain were very conservative in plants. Therefore, it is necessary to further explore the characteristics of the LchiMYB gene family.
Chromosome Distribution, Duplication Event, and Collinearity Analysis of LchiMYBs
We continued characterizing the LchiMYB genes by charting their chromosomal positions. This analysis showed that the 190 LchiMYB genes are unevenly distributed across the 19 Lchi chromosomes, with 25 LchiMYB genes anchored to 21 unassembled scaffolds (Supplementary Tables 6, 7 and Figure 2).
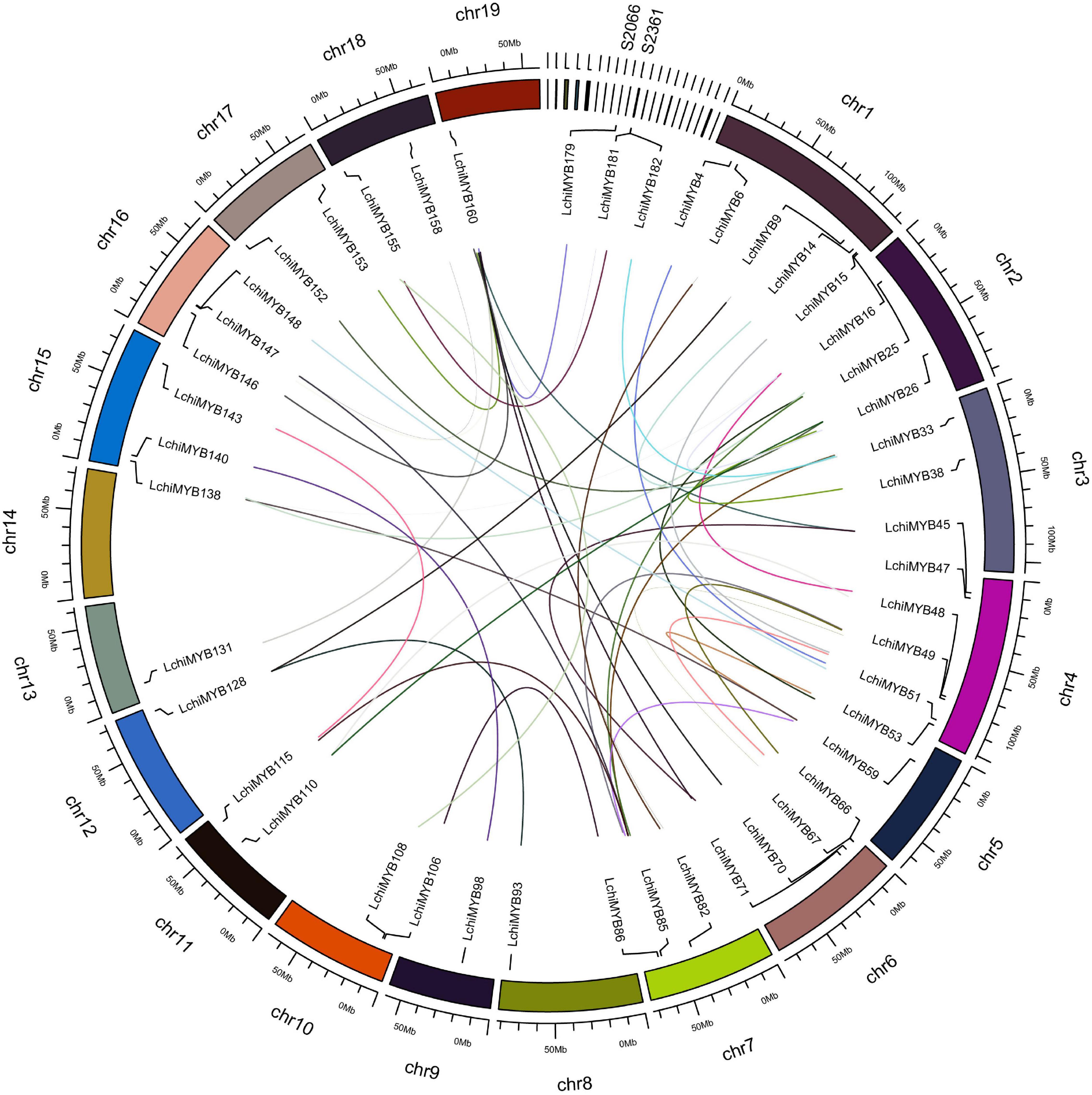
Figure 2. Chromosomal distribution and syntenic relationships of LchiMYBs. The colored curve lines represent duplicated MYB gene pairs. MYB family genes are located on all chromosomes and several scaffolds. Only duplicated gene pairs are shown; all 19 chromosomes are labeled, and only two scaffolds are labeled.
Chromosome 1 contains the most LchiMYB genes (17), followed by chromosome 4 (16). In contrast, chromosomes 14 and 17 only had three LchiMYB genes. There was no significant correlation between the number of LchiMYBs on a chromosome and the length of the chromosome.
The expansion of this gene family was mainly caused by gene replication events, including tandem and segment repeats. In this study, the collinearity of the MYB gene family was analyzed in the genome of Lchi, and gene replication events were explored. The results are shown in Figure 2. All chromosomes with an MYB gene are shown, and only duplicated genes are labeled on the chromosomes. A total of 40 segmental duplication events and 46 MYB genes were identified. Some LchiMYB genes had multiple replication events, such as LchiMYB48, LchiMYB110, and LchiMYB26. The LchiMYB genes are mainly distributed in synteny blocks on 18 chromosomes and two scaffolds. Generally, two or more genes arranged within 20 kb are defined as a gene cluster (Zhou et al., 2015). In this study, we identified two LchiMYB genes (LchiMYB20 and LchiMYB21) in a gene cluster. Ka/Ks (a non-synonymous/synonymous substitution ratio) values were calculated for all MYB gene pairs with segment duplication events (Supplementary Tables 8, 9). Twenty-two gene pairs had ka/ks values more than 1, and the ka/ks values of 18 gene pairs were less than 1, indicating that the LchiMYB gene was mainly subjected to positive selection pressure during evolution (Jia et al., 2003; Katiyar et al., 2012; Sun et al., 2019).
To further study the evolutionary status of the LchiMYB gene family, a genome-wide comparative syntenic analysis was conducted, using three species, namely, Lchi, Arabidopsis thaliana, and Oryza sativa (Katiyar et al., 2012; Ambawat et al., 2013), which belong to the basic angiosperms, dicotyledonous plants, and monocotyledonous plants, respectively (Figure 3).
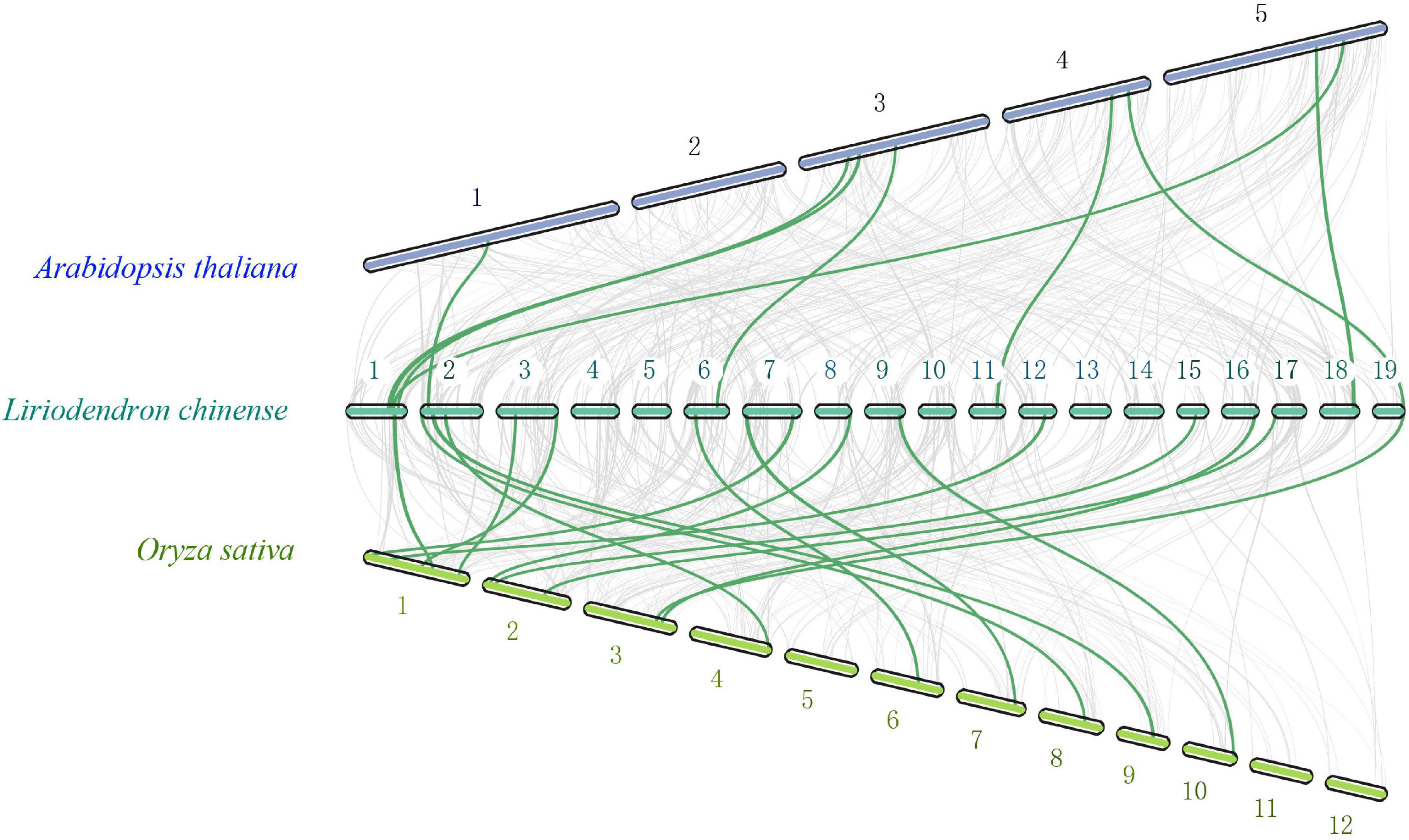
Figure 3. Collinearity analyses of MYB genes among Lchi, Oryza sativa, and Arabidopsis. The genomic linear blocks of Lchi, Oryza sativa, and Arabidopsis are represented by gray lines, while the syntenic MYB gene pairs are highlighted by the green lines. The number on the chromosome represents the chromosome ordinal number (Wang et al., 2012).
There were eight syntenic gene pairs between A. thaliana and Lchi, and 16 gene pairs between O. sativa and Lchi, which indicated that the evolutionary status of MYB in Lchi might be closer to that of O. sativa and consistent with that of its genome (Katiyar et al., 2012). The formation of these specific syntenic gene pairs may have resulted from the evolutionary status of magnolia species, in which basal angiosperms were formed before the separation of monocotyledons and dicotyledons (Chen et al., 2019). These results demonstrated that LchiMYBs are randomly distributed on chromosomes and mainly suffer from active selection. In the evolutionary process, it may have formed in the basal angiosperm stage. Thus, their gene structure, classification, and functional features need to be further explored.
Phylogeny Classification, Gene Exon–Intron Structure, and Cis Elements of LchiMYBs
To further explore the classification, gene exon-intron structure, and function of LchiMYBs, a Bayesian phylogenetic tree was constructed from the 190 LchiMYBs, which were classified into three groups (Supplementary Figure 2 and Supplementary Table 10). As R2R3-MYBs are very important in plants and have been reported in many kinds of research, 76 full-length R2R3-MYBs from Lchi were used to construct the Bayesian phylogenetic tree to classify LchiMYB genes (Liu et al., 2017). Based on the phylogenetic tree classification of Arabidopsis R2R3-MYB proteins, Lchi was divided into 20 subgroups (designated A1–A20) according to the similarity of protein sequences and the branches of the phylogenetic tree (Figure 4).
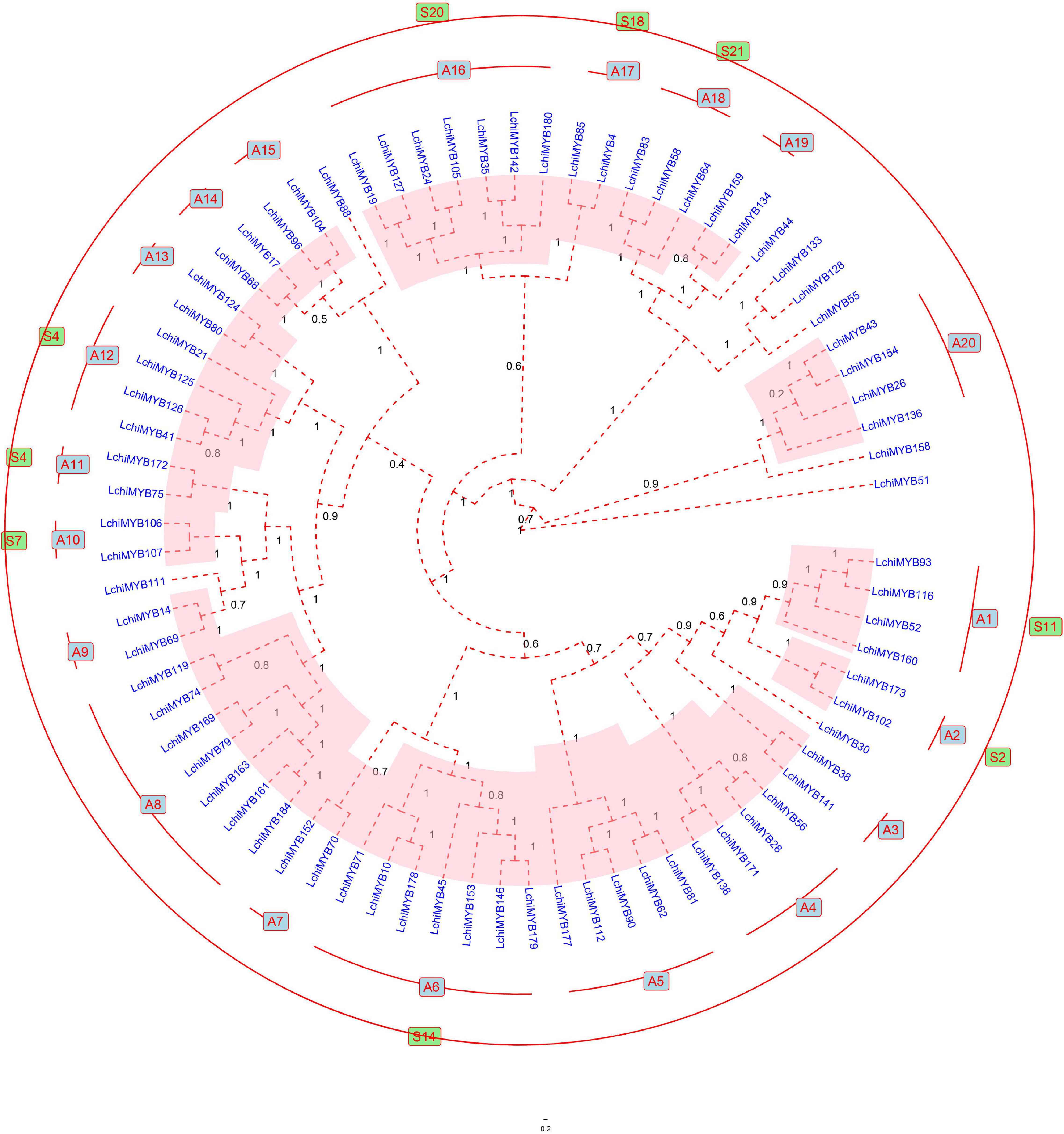
Figure 4. The Bayesian tree showing the phylogenetic relationships between 76 Lchi R2R3-MYB proteins. The protein sequences are divided into 20 main groups, each marked with a subgroup number (e.g., A1). The groups homologous to Arabidopsis are labeled outside the evolutionary tree and are indicated by the color green. The bootstrap values are supported by 1,000 replications and are shown beside the branches. Only 11 proteins did not cluster into any group.
The R2R3-MYB Bayesian phylogenetic tree of Lchi (76) and Arabidopsis (125) are presented in Supplementary Figure 3 and Supplementary Table 11. Most large subfamilies (A6, A8, and A15) have high bootstrap values, which are consistent with previous reports. However, certain smaller subfamilies (A3, A7, and A9) have no homologous groups with A. thaliana (Li and Lu, 2014; Zhao et al., 2014), and seven LchiMYB proteins were not grouped.
Following phylogenetic analysis, exon-intron structure analysis was performed for the 76 full-length Lchi R2R3-MYB genes. Most Lchi R2R3-MYB genes contained introns, except one gene (LchiMYB134) from the subgroup A19 (Supplementary Table 12 and Figure 5).
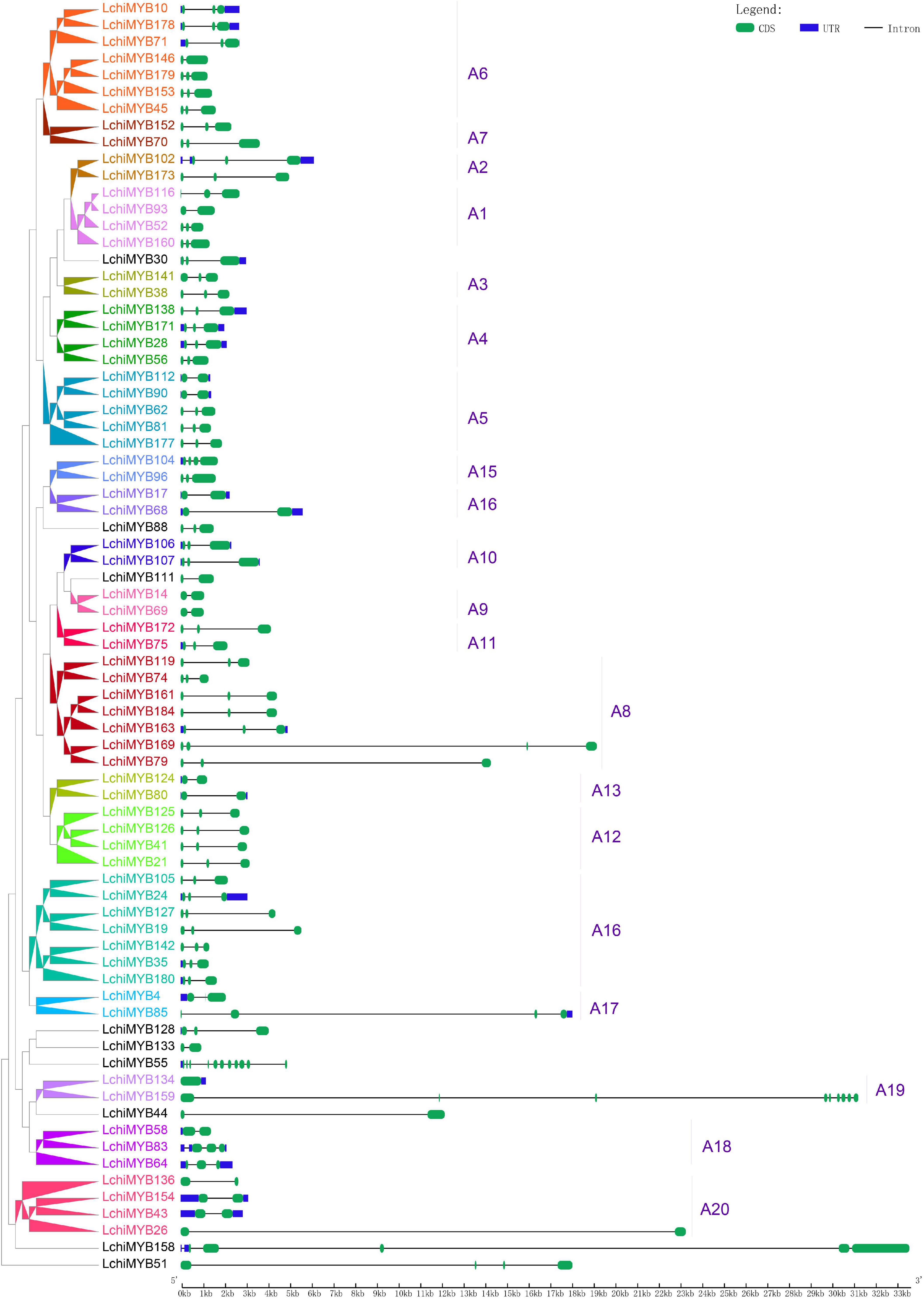
Figure 5. Lchi MYB gene exon-intron structures. Blue rectangles indicate untranslated 5′– and 3′– regions; green rectangles indicate exons; black straight lines represent introns; the tick marks represent the length of the gene. The corresponding groups were labeled (e.g., A1), with different colors on the branches and gene labels.
The number of introns in the MYB gene ranged from 0 to 3, except for three genes (LchiMYB158, LchiMYB159, and LchiMYB55), which had 4, 9, and 10, respectively. More than half (45/76) of the LchiMYB genes had three exons; the results of these gene structures are consistent with those of the same group with similar gene structures in the evolutionary tree, such as A2, A3, and A10. However, there are two exceptions: subgroups A17 and A19, which contained different members of introns. Overall, similar gene structures and phylogenetic tree groupings strongly support the reliability of our grouping classifications (Yanhui et al., 2006; Wilkins et al., 2009).
To further understand LchiMYB gene function, the PlantCARE website (see footnote 7) was used to identify known cis elements within the 1.5-kb upstream region of each gene (Figure 6).
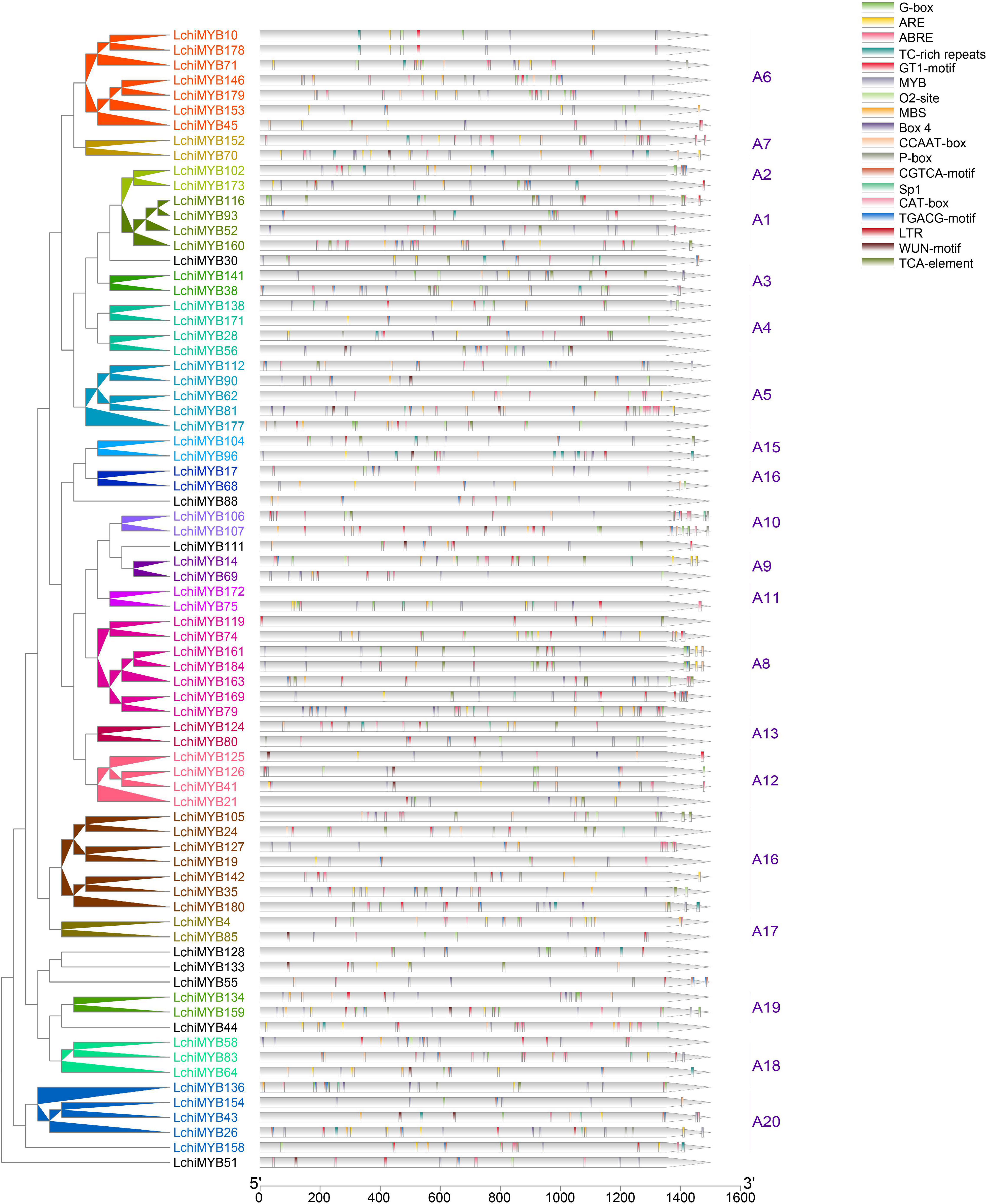
Figure 6. LchiMYB cis-acting elements. Displayed is an unrooted phylogenetic tree constructed, using Beast (v2.6.2) set to the Bayesian method. The legend contains 18 cis-acting elements identified within the 1.5-kb long LchiMYB promoter sequences; the tick marks represent the length of the promoter sequences. Different cis-acting elements are color coded. The corresponding groups were labeled (e.g., A1) with different colors on the branches and gene labels.
Two types of cis elements were detected: one related to plant growth and development, and the other related to stress responses (Supplementary Table 13) (Srideepthi et al., 2020). Cis elements related to growth and development include the MYB binding site (MYB), light responsive (Box-4, G-box, GT1-motif, Sp1), an MYBHv1 binding site (CCAAT-box), meristem-specific activation (O2-site), and meristem expression (CAT-box). Cis elements related to abiotic stress include anaerobic induction (ARE), methyl jasmonate (MeJA) response (TGACG-motif and CGTCA-motif), ABA-responsive element (ABRE), salicylic acid (SA) response (a TCA element), gibberellin responsive (P-box), defense and stress response (TC rich), drought response (MBS), low-temperature response (LTR), and wound-responsive (WUN-motif) (Chen et al., 2017). These results showed that LchiMYB genes regulate abiotic stress signaling, and that they potentially regulate each other during growth and development. Therefore, it is possible that, under abiotic stress conditions, LchiMYBs may change their expression patterns, and they also play a regulatory role in different growth and developmental stages.
Expression Profiling of LchiMYB Genes in Vegetative and Reproductive Organs of Lchi
To explore the spatial expression pattern of LchiMYBs, the expression profiles of different tissues were studied to determine the function of MYBs based on previous reports. LchiMYBs regulate development in different tissues and perform differential expression profiling in Lchi (Chen et al., 2017). As shown in Figure 7, a total of 150 (78.95%) genes were differentially expressed in eight tissues, namely bark, phloem, xylem, leaf, bud, sepal, stigma, and stamen.
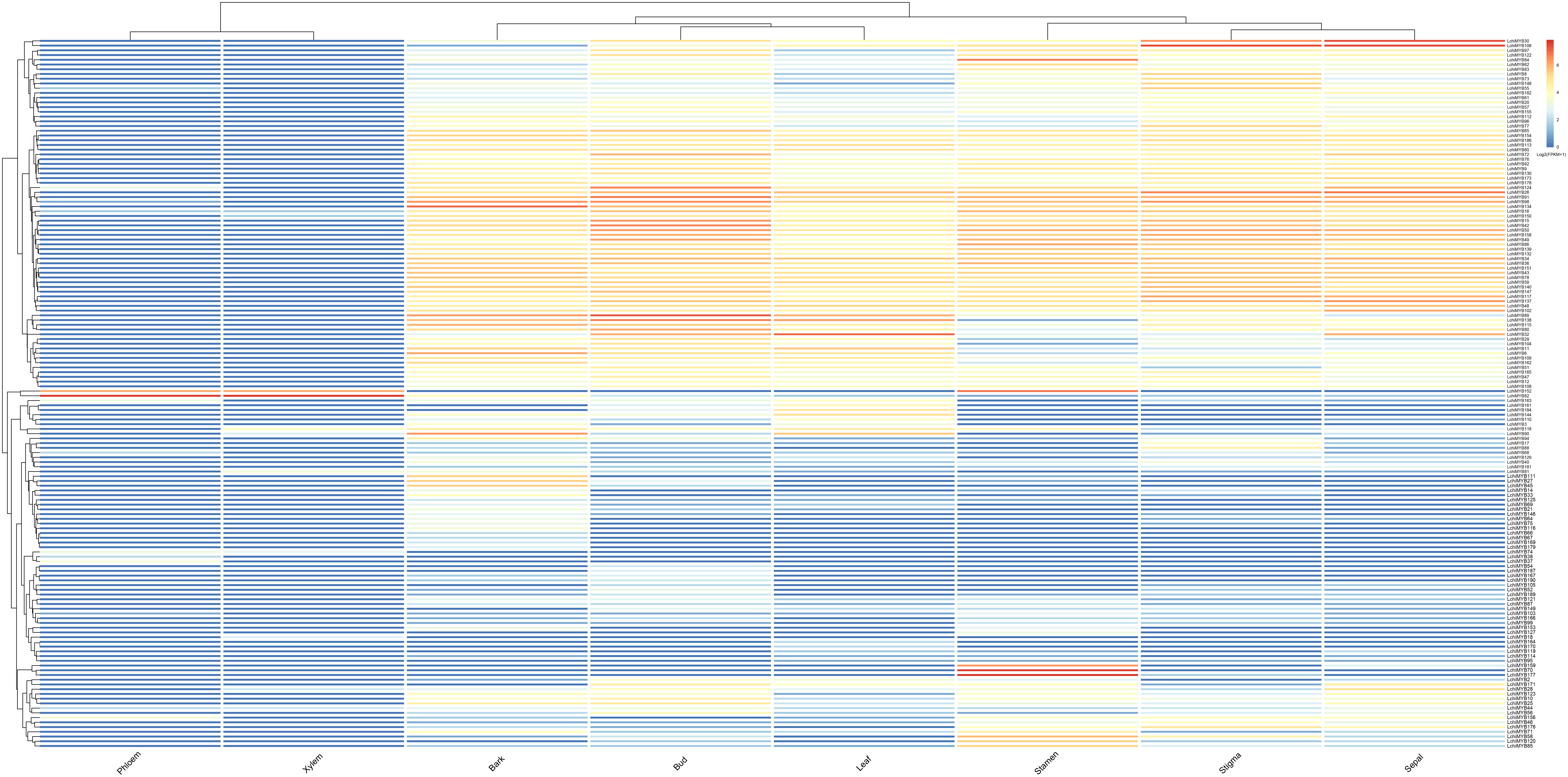
Figure 7. The heatmap of FPKM values in eight different tissues. The phloem, xylem, stigma, bark, bud, leaf, stamen, stigma, and sepal represent the eight different tissues. The transcript abundance level was normalized and hierarchically clustered by using the log 2 (FPKM + 1) comparison among genes per tissue and different tissues. Colored blocks show decreased (blue) or increased (red) transcript accumulation at different stages. The color legend indicates the fold change expression of LchiMYBs, changing from low (blue) to high (red) expression. The heatmap was generated, using the R package “pheatmap” (v 1.0.12).
Many LchiMYBs were highly expressed in the bark, leaf, and stamen, followed by the bud, stigma, and sepal. Interestingly, only a few genes were expressed in the phloem and xylem tissues; perhaps, LchiMYBs have no obvious functions in these two tissues. For example, LchiMYB152 and –82 were only expressed highly in the phloem, xylem, and stamen, but not in other tissues. LchiMYB118 and –90 were highly expressed in the bark and leaf and nearly not expressed in other tissues. LchiMYB70, –177, and –2 were highly expressed in the stamen. This suggests that specific tissues are regulated by different genes.
Moreover, some LchiMYBs showed multi-tissue regulatory functions, like LchiMYB124, –26, –91, and –98, which were highly expressed in the stigma, bud, stamen, leaf, bark, and sepal but not in the phloem and xylem. Some genes were less expressed in all tissues, such as LchiMYB54, –187, –167, and –190, which were only less expressed in the bud and bark. These results showed that LchiMYBs have diversified regulation modes in different tissues, and there are tissue-specific genes.
Expression Profiling of LchiMYB Genes During 11 Successive Stages of Somatic Embryogenesis in Liriodendron hybrids
Embryogenic callus was induced by embryos of seeds gained from a Liriodendron hybrid, which was the offspring produced by artificial pollination (L. chinense × L. tulipifera), and the 11 successive stages of somatic embryogenesis were sequenced, namely PEMs, ES1, ES2, ES3, ES4, ES5, ES6, (ES7), (ES8), (ES9), and (PL). According to previous reports, the MYB gene has a regulatory function in somatic embryogenesis (Wang F. X. et al., 2020).
To further explore the time series expression pattern of LchiMYB genes, we analyzed their expression during successive stages of somatic embryogenesis (Wang F. X. et al., 2020). In total, 110 (57.89%) LchiMYB genes were differentially induced across various stages of somatic embryogenesis (Figure 8). Some MYB genes were highly expressed at all stages, while others were less expressed, with only few genes being specifically expressed at certain stages. For example, LchiMYB121, –134, –28, –56, –16, –26, and –9 were highly expressed at all stages, while LchiMYB114, –70, –152, –21, –82, –40, and –103 were less expressed at all stages.
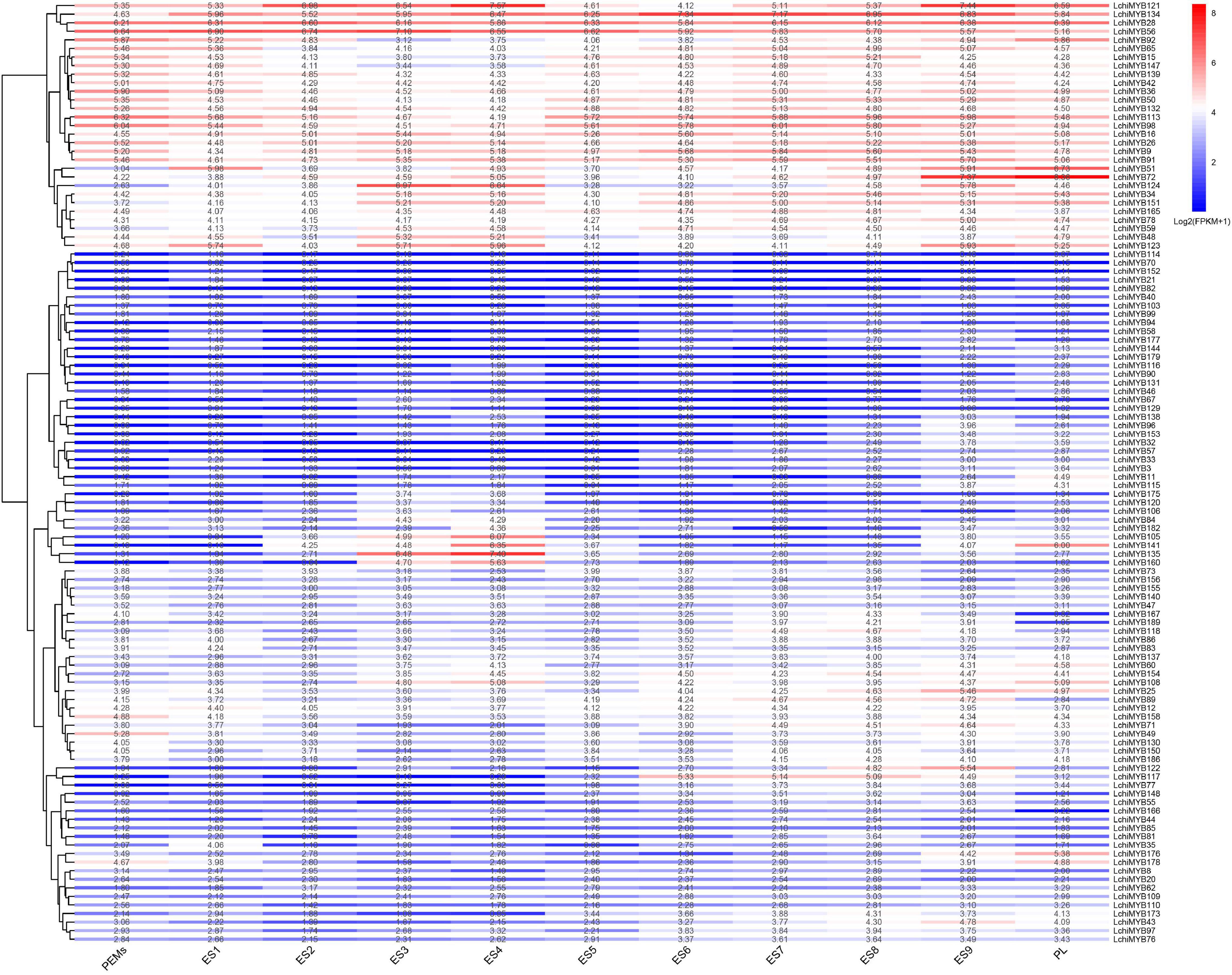
Figure 8. LchiMYB gene expression profiles were quantified from RNA-seq data at 11 stages of Liriodendron hybrid somatic embryogenesis: proembryogenic mass cultured for 20 days for embryogenic callus (PEMs), then liquid was suspension cultured for 10 days (ES1), single-cell cultured for 2 days (ES2), induced by ABA for 1 day (ES3) and 3 days (ES4), an 7-day globular embryo (ES5), a 13-day heart-shaped embryo (ES6), a 19-day torpedo embryo (ES7), a 25-day immature cotyledon embryo (ES8), a 31-day mature cotyledon embryo (ES9), and 37-day plantlet (PL). The transcript abundance level was normalized and hierarchically clustered, using the log 2 (FPKM + 1) comparison among genes per stage. Colored blocks show decreased (blue) or increased (red) transcript accumulation at different stages. The color legend indicates the fold change expression of LchiMYBs, running from low (blue) to high (red) expression. The heatmap was generated, using the R package “pheatmap” (v 1.0.12).
At the ES3 and ES4 stages, LchiMYB135, –160, and –135 were highly expressed, while showing low expression at other stages. LchiMYB141 showed high expression at the ES4 and PL stages but low expression at other stages. Notably, some genes had peak expression at a certain stage, such as LchiMYB51, –72, and –124 with a peak at the PL, ES9, and ES3 stages, respectively. These results showed that several LchiMYBs (R1-MYB and R2R3-MYB) potentially participate in somatic embryogenesis and were differentially expressed at different stages. The four stages imply that ES3, ES4, ES9, and PL may be the main stages regulated by LchiMYBs.
Diversified Expression Patterns of LchiMYBs Under Cold, Heat, and Drought Stress
Plants possess different abilities to perceive, respond, and adapt to various biotic and abiotic stressors, which is an important mechanism for plants to survive in adverse environmental conditions (Li J. et al., 2019; Zhou et al., 2019; Pucker et al., 2020). MYB TFs are essential for stress response. However, the mechanism by which LchiMYB genes respond to stress is still unknown. In this research, MYB gene expression patterns were explored under cold, heat, and drought stress at short intervals (0, 1, 3, 6, and 12 h) and long intervals (1 and 3 days). The TPM value of RNA-seq data from leaf tissues was used to visualize expression trends under different stressors. The TPM value of genes under the three abiotic stressors is shown in Supplementary Table 14.
As shown in Figures 9A–C, under cold stress (Figure 9A), the expression of LchiMYBs could be divided into three groups: high expression, low expression at all times, and high expression at long intervals. Under heat stress (Figure 9B), the expression of LchiMYBs could be divided into two groups: low expression at all times and high expression at several intervals. Interestingly, under drought stress (Figure 9C), the expression of LchiMYBs could also be divided into three groups: high expression, low expression at all times, and high expression at several intervals.
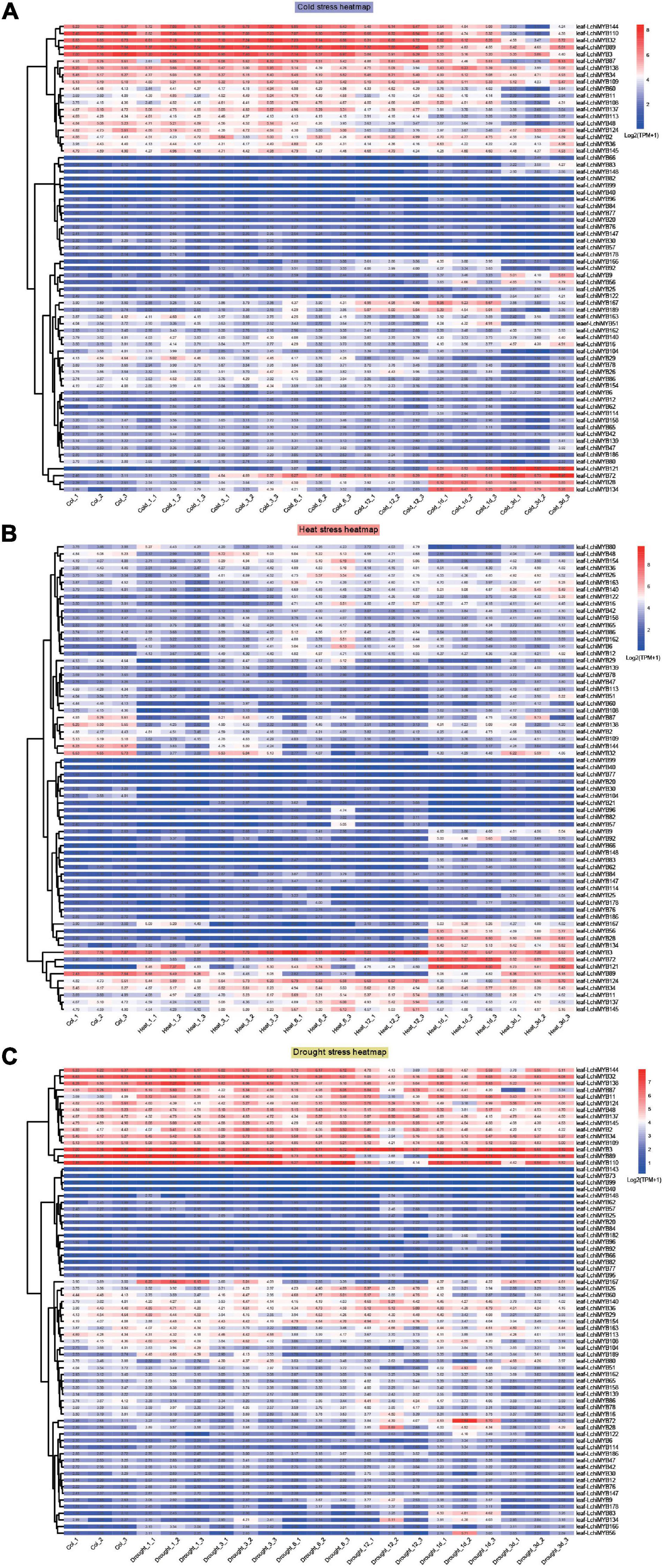
Figure 9. Expression patterns of LchiMYBs under cold (A), heat (B), and drought (C) stress at six successive time intervals. Ctrl_1, Ctrl_2, and Ctrl_3 represent three biological replicates of the control group; Cold_1, Cold_3, Cold_6, Cold_12, Cold_1d, Cold_3d, Heat_1, Heat_3, Heat_6, Heat_12, Heat_1d, Heat_3d, Drought_1, Drought_3, Drought_6, Drought_12, Drought_1d, and Drought_3d represent three repetitions of each time point (1 h, 3 h, 6 h, 12 h, 1 day, and 3 days); leaf-LchiMYBs represent the leaf tissues used for RNA-seq analysis. The standardization and hierarchical clustering of the transcriptional expression level were calculated by log 2 (TPM + 1) values. Colored blocks show decreased (blue) or increased (red) transcriptional expression among the different time points. The heatmap was generated by the R package “pheatmap” (v 1.0.12).
More than 60 LchiMYB genes were differentially expressed in response to at least one stressor; most of them were induced by multiple stressors, implying that they play a cross-regulatory role in different signal transduction pathways in response to abiotic stress. For instance, LchiMYB144, –32, –89, and –3 were induced by all three stressors. LchiMYB3 was highly expressed under all stressors, while LchiMYB32 and LchiMYB144 were only less expressed under heat stress but had high expression under the other two stressors. Similarly, LchiMYB89 was highly expressed under heat and cold stress but only less expressed under drought stress, suggesting that they are pleiotropic regulators that regulate different environmental condition responses. For instance, LchiMYB121 responded to cold and heat stress but not to drought stress.
However, some LchiMYB genes responded to one stress simultaneously. For instance, three genes (LchiMYB72, –28, and –134) were significantly upregulated by cold treatment at 1– and 3-day intervals; four genes (LchiMYB145, –137, –11, and –34) were significantly induced by heat stress; and four genes (LchiMYB48, –137, –45, and –2) were significantly induced by drought stress. Additionally, some genes showed opposite expression patterns under different stressors. LchiMYB134 was significantly upregulated under cold and heat stress but downregulated by drought stress at the 3-d interval. Among these stressors, LchiMYBs was more upregulated by long intervals of heat and cold stress than by short intervals, indicating that long intervals were the main biological process of Lchi, and MYBs play an important role in it.
Combined with the above results, LchiMYBs play multiple roles in the response to adversity stress; some genes regulate cold, heat, or drought stress responses, while others regulate the response to two or three stressors. Some genes were not induced by abiotic stress and may be expressed under other types of stress. These results imply that the signaling pathways of plants in response to abiotic stress are complex; long intervals were an important response time interval for LchiMYBs. In conclusion, these findings provide valuable information for improving the environmental tolerance of Lchi, which can be achieved by genetic manipulation of LchiMYB genes.
qRT-PCR Verification of Lchi Under Three Abiotic Stressors
qRT-PCR was used to validate the transcriptome data acquired from abiotically stressed leaf tissues of seedlings. Eight genes that were expressed under three abiotic stressors with a different pattern were selected for qRT-PCR experiments (Chen et al., 2017; Liu et al., 2017): two were of the R2R3-MYB type (LchiMYB83 and –62), while the others were of the 1R-MYB type (Figures 10A–C). The qRT-PCR values are presented in Supplementary Table 15. Each LchiMYB gene showed a unique expression trend in response to the three abiotic stress conditions.
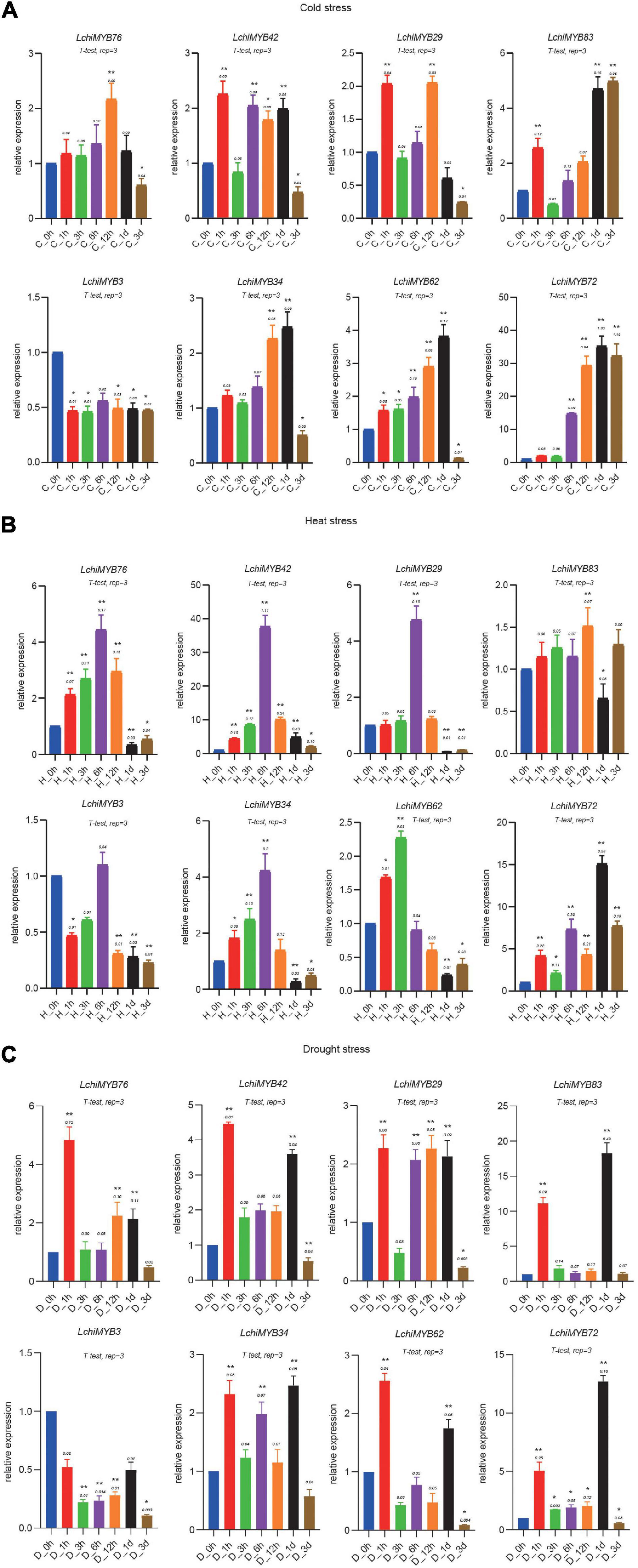
Figure 10. Expression analysis of LchiMYBs under cold (A), heat (B), and drought (C) stress treatments, analyzed by qRT-PCR. The Y-axis shows the relative expression level, and the X-axis indicates different time points of stress treatment; the mean ± standard error measurement value (SEM) (n = 3) is displayed. The t-test represents the Student’s test statistic method, and rep = 3 represents the three biological replicates. The group for each treatment time point compared with the control group; the significantly different (T-test) *p < 0.05 and **p < 0.01.
Under cold stress conditions, all but one LchiMYB gene (LchiMYB3) showed induced expression. Three genes (LchiMYB42, –29, and –83) showed a 2-fold expression response, as they were strongly upregulated after 1 h; after which, they showed a response again when 6 h to 1 day had passed (Figure 10A). The remaining four genes (LchiMYB76, –34, –62, and –72) showed a singular wave of upregulated expression at 12 h and 1 day, with LchiMYB72 responding very strongly with about 40-fold upregulation and LchiMYB76, showing only marginal upregulation (Figure 10A). This showed that 1 h, 12 h, 1 day, and 3 days were the key response intervals, and LchiMYBs were upregulated at these time points; perhaps, they have an important regulatory function during these periods.
Under heat stress, five LchiMYB genes showed an expression peak point at 6 h, while those of three genes were different, i.e., LchiMYB83, LchiMYB62, and LchiMYB72 showed an expression peak point at 12 h, 3 h, and 1 day, respectively (Figure 10B). These genes were induced by heat stress and compared with the control group; there was a peak in expression. Similarly, the LchiMYB72 showed 15-fold upregulation at the 1-d time point, and LchiMYB42 showed approximately 40-fold upregulation at the 6-h time point. These results indicate that the 3-h, 6-h, 12-h, and 1-d time points were the main response times for the MYB family under heat stress, and they might play an important role during this period.
Under drought stress, a different, more complex type of expression response was observed than that to temperature stress (Figure 10C). LchiMYB genes responded early at 1-h time point, except LchiMYB3, which was lower than at 0 h. LchiMYB42, –34, –83, –72, and –62 were highly expressed at the 1-d interval. Some genes, like LchiMYB29, were highly expressed at all time points, except at 3 h and 3 days. LchiMYB76 was highly expressed at the 1-h interval. In contrast to temperature stress, LchiMYB83 was upregulated by approximately 20-fold, and LchiMYB72 was upregulated by approximately 12-fold at the 1-d interval. These results suggested that the function of MYB is diverse under drought stress, and its signal pathway may be different from that under temperature stress; thus, the regulation mode of MYB genes was also different.
In particular, two MYB genes (LchiMYB72 and –42) were highly expressed in response to all three tested stressors, suggesting that they function at the intersection of separate biological signaling pathways. In contrast, LchiMYB3 showed low expression under all three stressors, indicating that it may have no prominent role in stress signaling.
Taken together, our RNA-seq and qPCR data indicated that LchiMYB genes were likely involved in the response of Lchi to abiotic stress at a particular time, making them important players in the complex signal regulation network and in the adaptation of the tree to various adverse environments.
Discussion
LchiMYBs Possess Less R2R3-MYBs Genome-Wide and in Highly Conserved Domain Sequences
MYB TFs are the largest family in plants and regulate many gene expressions, which are involved in many biological processes and metabolic pathways (Stracke et al., 2001; Dubos et al., 2010). In Lchi, there are no genome-wide reports of the MYB gene family and their biological functions, such as growth, development, and abiotic processes. In our study, for the first time, the MYB gene family was identified in a series of analyses, including tissue expression, somatic embryogenesis expression, and abiotic stress response.
A total of 190 MYB genes were identified, including 76 (40%) R2R3-MYB genes. This result was consistent with the potato MYB gene family, which has 111 R2R3-MYBs, accounting for 44% of all 251 MYB genes (Li Y. et al., 2020). In many plants, R2R3-MYBs are the largest subgroup in the MYB gene family. For instance, A. thaliana possesses 126 R2R3-MYBs, which account for 64.29% of all 196 MYBs (Dubos et al., 2010). The B. distachyon MYB gene family contains 85 (69.67%) MYB-R2R3s of the total 122 BdMYBs (Chen et al., 2017). Pepper (Capsicum annuum L.) has 108 R2R3-MYBs, accounting for 50% of all 216 MYBs (Wang J. et al., 2020), and O. sativa has 109 R2R3-MYBs (58.91%) of the total 185 MYBs (Dubos et al., 2010). These results indicated that R2R3-MYBs were the main subgroup in the plant MYB gene family, which may be associated with the evolutionary status. Lchi was considered a basal angiosperm with fewer R2R3-MYBs than higher angiosperms (Chen et al., 2019).
The sequences of the R2R3 domain were analyzed with A. thaliana, suggesting that they are highly conservative between different plants. The R2R3 repeats bind upstream of regulatory genes and induce them to express in different tissues or under various environments. Although Lchi was a basal angiosperm with an evolutionary status lower than higher angiosperms, such as monocotyledons and dicotyledons (Chen et al., 2019), R2R3-MYB sequences were still conserved in different plants, demonstrating that the function of the MYB gene family may be conserved in basal angiosperms (Stracke et al., 2001).
LchiMYB Family Expansion Originated From a Gene Segmental Replication Event
Gene duplication is an important mechanism by which new genes are generated. It contributes significantly to the expansion of the MYB gene family in the plant kingdom, leading to the diversification of gene function or driving gene evolution (Yanhui et al., 2006; Dubos et al., 2010). Our results showed that 46 (24.21%) LchiMYB genes had segmental repeat events in Lchi, which indicated that segmental duplication events were an important driving force for the expansion of LchiMYB genes.
Previous studies showed that the 3R-MYB gene is obtained by replicating one R1 repeat with a 2R-MYB domain, or 3R-MYB is formed by replicating two R2 repeats with a 1R-MYB domain (Millard et al., 2019; Liu et al., 2020; Pucker et al., 2020). For instance, the 1R-MYB gene LchiMYB73 may be produced by the loss of two R repeats of the 3R-MYB gene LchiMYB8. Most gene duplications resulted from segmental duplications, including the R2R3-MYB and 1R-MYB genes. As a result, there are more MYB genes in the Lchi genome.
Duplicated genes often lose their original function and/or obtain new functions, which increases the adaptability of plants to unfavorable environments. Previous studies showed that the retention of replication genes in the genome leads to diverse expression patterns and responses to various abiotic stressors (Gates et al., 2016; Roy, 2016; Liu et al., 2017; Mmadi et al., 2017). In this study, we found that some gene pairs showed different expression levels under different abiotic stressors. For instance, in one pair, LchiMYB86 was expressed, while LchiMYB106 was not expressed under the three tested stress conditions. Further analysis showed that LchiMYB86 contains certain cis elements, such as LTR and MBS elements, which respond to cold and drought stress in its promoter region. In contrast, these elements were not found in the upstream region of LchiMYB106.
LchiMYB Genes Regulate the Development of Vegetative and Reproductive Organs
According to previous reports, the potato StR2R3-MYB gene has functions in different tissues, such as the leaves, roots, shoots, sepals, stamens, flowers, petals, carpels, and fruit (Li Y. et al., 2020). In B. distachyon, BdMYB055, –011, –083, and –089 were highly expressed in different parts of floral organs (Chen et al., 2017). In Arabidopsis, AtMYB37, –38, and –84 regulated axillary meristem formation and vegetative processes (Dubos et al., 2010). In this research, eight tissue transcriptomes were evaluated to investigate the function of LchiMYBs. LchiMYBs are involved in vegetative and reproductive organs, including the bark, phloem, xylem, leaf, bud, sepal, stigma, and stamen. In particular, some LchiMYBs were specifically expressed in some tissues; for example, LchiMYB152 and –82 were mainly regulated in the phloem, xylem, and stamen but not in other tissues. In the bark and leaves, LchiMYB118 and –90 had a high expression pattern, while LchiMYB70, –177, and –2 were only expressed in the stamen. These expression patterns indicated that some LchiMYB genes were tissue specific and only regulated in their target organs and were not expressed in other tissues. Conversely, some genes were constitutively regulated and had a multi-tissue expression pattern. For example, LchiMYB124, –26, –91, and –98 were only expressed in the xylem and phloem but not in other tissues.
In summary, LchiMYBs have an important function in the vegetative and reproductive organs of Lchi, thereby regulating tree growth and flower development. These key LchiMYB genes were useful for further exploration of their regulatory function through genetic manipulation and other molecular experiments. In the future, these data will be beneficial to improve the growth, trunk thickening, and reproduction of Lchi, thereby increasing the utilization value of wood and reforestation applications.
LchiMYB Genes Have Important Functions at the ABA-Induced Stage in Somatic Embryogenesis
According to a previous report, abscisic acid (ABA), ethylene, and cytokinins (CKs) have important functions during the induction of plant somatic embryogenesis (SE) (Méndez-Hernández et al., 2019). MYB1R1-, MYB98-, and MYB26-like were upregulated in later SE of Hevea brasiliensis (Wang et al., 2021). AtMYB115, AtMYB118, and EMK (EMBRYOMAKER) affected SE in Arabidopsis (Gliwicka et al., 2013). In contrast, ectopically, overexpressing the homeo domain TF WUSCHEL (WUS) and MYB TF MYB118 formed somatic embryos in Arabidopsis (Su Y. H. et al., 2014; Wang F. X. et al., 2020). In Lchi, however, MYB genes that regulate somatic embryogenesis directly were still not reported. The somatic embryogenesis of the Liriodendron hybrid is an important method of asexual propagation to obtain more seedlings. Thus, the LchiMYB expression pattern of somatic embryogenesis was explored. Eleven successive stages of somatic embryogenesis were used to obtain RNA-seq data and identify the function of LchiMYBs.
Some LchiMYBs have shown a powerful regulation function in these 11 stages, such as LchiMYB121, –134, –28, –56, –16, –26, and –9. Moreover, after a single cell was cultured for 2 days, it was induced by ABA for 1 day (ES3) and 3 days (ES4), and LchiMYBs were highly expressed, including LchiMYB135, –160, –141, and –135. At the ES9 and PL stages, LchiMYB51, –72, and –124 had peak expression. The expression pattern showed that somatic embryogenesis at specific stages may be regulated by some MYB genes, especially at ES3, ES4, ES9, and PL stages during these four periods. Presumably, some LchiMYBs were significantly induced by ABA treatment, such as at the ES3 and ES4 stages of somatic embryogenesis, suggesting that ABA-induced LchiMYBs regulate the gradual formation of somatic embryos.
Taken together, according to previous reports and the expression pattern, MYBs have some vital functions during somatic embryogenesis after being induced by ABA. These results shed a new light on the MYB genes involved in the somatic embryogenesis process and ABA-dependent expression patterns. Thus, these ABA-induced LchiMYBs were worth further investigation by molecular experiments and revealed their regulatory mechanism. In the future, the regulatory genes of these important stages will be helpful to improve the efficiency of asexual reproduction in Lchi, thereby improving the natural reproduction ability of Lchi and increasing the yield of trees.
The Cross-Regulatory Role of LchiMYB Families in Response to Cold, Heat, and Drought Abiotic Stress
Overexpression of the MYB gene enhances adaptability to abiotic stress. For example, in Arabidopsis, overexpression of two wheat genes (TaMYB33, TaMYB73) enhances salt tolerance (Li J. et al., 2019). Similarly, overexpression of AtMYB44 increased its tolerance to multiple abiotic stressors (Stracke et al., 2001; Katiyar et al., 2012). Overexpressing GmMYBJ1 enhances tolerance to drought and low-temperature stress, suggesting its potential function to improve abiotic stress tolerance (Su L.-T. et al., 2014). Some MYB TFs respond to abiotic stress by regulating the expression of the target genes. For instance, TaMYB19-B changes the expression of stress-related genes to enhance the ability of wheat to overcome adverse conditions. TaMYB73 participates in the salt stress response by regulating the expression of stress-related genes (Zhang et al., 2018; Li J. et al., 2019).
Many MYB genes may have a potential function in abiotic stress. For instance, in rice, cold stress significantly induced the expression of OsMYB2 and OsMYB511; osmotic stress and exogenous ABA significantly induced the expression of OsMYB511 and CMYB1 (Yanhui et al., 2006; Katiyar et al., 2012). In Arabidopsis, jasmonate treatment significantly induced the expression of MYB21/At3g27810, and MYB24/At5g40350 (Kranz et al., 1998). Additionally, 150 MYB genes were identified in soybeans, 43 of which were significantly upregulated under cold, salt, and drought stress (Baillo et al., 2019; Li J. et al., 2019). Under dehydration, salt, cold, and ABA treatments, PtsrMYB was significantly upregulated, which was highly consistent with AtMYB109 (Sun et al., 2014).
In the present study, we used RNA-seq of heat, cold, and drought stress to explore the functions of LchiMYBs, which helps to better understand the cross-regulatory function of MYB under multiple stressors. Among these stressors, different expression patterns were observed. LchiMYB expression patterns were divided into three groups under cold and drought stress but into two groups under heat stress. Some LchiMYBs were expressed under three stress conditions, while others were expressed only under one or two stress conditions. For instance, LchiMYB3 was highly expressed under all stresses, while LchiMYB144, –32, and –89 were induced under the three stressors with different expression patterns. LchiMYB32, –144, and –89 were highly expressed under two stressors but less expressed under one stressor. Additionally, LchiMYB121 was only induced under cold and heat stress. Therefore, these genes were the crucial regulatory factors of Lchi under abiotic stress. LchiMYB3 may be the main regulator, while other genes are secondary regulators, forming a complex abiotic stress signal network of cross-regulation, and the corresponding biological pathways need to be further studied.
Our qRT-PCR results showed that eight selected LchiMYB genes with different expression patterns were regulated by cold, heat, and drought stress. The expression of six LchiMYB genes was upregulated in response to all three stress conditions. A previous study demonstrated that MBS and LTR are major cis elements regulating drought and cold-responsive gene expression (Katiyar et al., 2012; Wang J. et al., 2020).
Most of the LchiMYB genes possess these elements. These results further imply that LchiMYB genes have a potential cross-regulatory function under abiotic stress.
There were few studies on the abiotic stress resistance of Lchi, especially in the whole genome. Because Lchi is mainly distributed to the south of the Yangtze River (Chen et al., 2019), these results provide a theoretical basis for the large-scale promotion of Lchi, which can be extended to the colder northern region, the higher temperature southern region, and the arid western region of China, and contribute to reducing the greenhouse effect and improving the ecological environment in the future.
Conclusion
Here, 190 MYB members were identified in the Lchi genome. These LchiMYBs were phylogenetically divided into three major groups, and the R2R3-MYB group (76) was classified into 20 subgroups. The protein sequence features, gene exon-intron structure characteristics, chromosomal locations, phylogeny, and the intra- and inter-species collinearity of the LchiMYBs were additionally analyzed. In the evolution process, the MYB family mainly experienced positive selection. The transcriptome of vegetative and reproductive organs showed that LchiMYB118 and –90 mainly regulated the development of vegetative organs, and LchiMYB70, –177, and –2 mainly regulated the development of reproductive organs. Global gene expression patterns of the 11 stages of somatic embryogenesis suggested that they probably have important functions in the regulation of different Lchi somatic embryogenesis stages, especially in the regulation of the ES3, ES4, ES9, and PL stages. Some MYB genes may cross-regulate Lchi under abiotic stress, such as LchiMYB3, –144, –32, and –89. Overall, this study offers further insight into the functional roles of LchiMYBs, indicating their regulatory function in growth, development, somatic embryogenesis, and abiotic stress. These results will be helpful to improve the growth and reproduction of Lchi by providing a strong theoretical basis for the functional verification of a single gene in the later stage, asexual propagation, and afforestation in different areas.
Data Availability Statement
The original contributions presented in the study are publicly available. This data can be found here: NCBI, accession numbers: PRJNA679089 and PRJNA679101.
Author Contributions
WW completed the analysis of LchiMYBs and qPCR experiments and wrote the manuscript. SZ performed the analysis. LZ, DW, SL, YLi, and YLu designed and performed the qPCR experiments. JZ provided the transcriptome data for somatic embryogenesis. ZH, TC, JS, and JC designed the project, performed the transcriptome data analysis, and wrote the manuscript. All authors agreed on the whole manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (32071784), the Key Research and Development Plan of Jiangsu Province (BE2017376), the Qinglan Project of Jiangsu Province, the Distinguished Professor Project of Jiangsu province, and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD). The funding bodies had no participation in this paper.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank LetPub (www.letpub.com) for its linguistic assistance during the preparation of this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.641280/full#supplementary-material
Supplementary Figure 1 | A quantitative real-time PCR (qRT-PCR) dissolution curve and a standard curve of eight genes. The primer specificity and amplification efficiency of eight genes were checked by melting curve analysis and standard curve analysis, respectively. LchiMYBs represent the genes, Lchi18S represents the internal reference gene, and red curves represent the melting and the standard.
Supplementary Figure 2 | The Bayesian tree indicating the phylogenetic relationships between 190 Lchi MYB proteins. The proteins are divided into three subgroups, each designated with a different color (e.g., green represents 1R-MYB). The bootstrap values were supported by 1,000 replications and are shown beside the branches.
Supplementary Figure 3 | The Bayesian tree indicating the phylogenetic relationships between 76 MYB proteins from Liriodendron chinense (Lchi) and 126 from Arabidopsis. The proteins are divided into 24 main groups, each labeled with a subgroup number (e.g., S1). The bootstrap values were supported by 1,000 replications and are shown beside the branches.
Supplementary Table 1 | Fragments per kilobase of the exon model per million mapped fragments (FPKM) value of somatic embryogenesis.
Supplementary Table 2 | FPKM value of various tissues for LchiMYBs.
Supplementary Table 3 | Transcripts per million (TPM) values of mapped LchiMYBs.
Supplementary Table 4 | Primer sequences of eight LchiMYBs.
Supplementary Table 5 | Protein size features of LchiMYBs.
Supplementary Table 6 | Chromosome length of LchiMYBs.
Supplementary Table 7 | Gene position of LchiMYBs.
Supplementary Table 8 | Synteny gene pairs of LchiMYBs.
Supplementary Table 9 | Selective pressure analysis of LchiMYB-duplicated gene pairs.
Supplementary Table 10 | Protein sequences of 190 LchiMYBs.
Supplementary Table 11 | Protein sequences of Lchi and ATG R2R3-MYBs.
Supplementary Table 12 | Number and length of the exon-intron structure of LchiR2R3-MYBs.
Supplementary Table 13 | Cis elements of LchiR2R3-MYBs.
Supplementary Table 14 | TPM value of LchiMYBs under cold, heat, and drought stress.
Supplementary Table 15 | Quantitative real-time PCR (qRT) value of LchiMYBs under cold, heat, and drought stress.
Abbreviations
Lchi, Liriodendron chinense; MYB, Myeloblastosis; AtMYB, A. thaliana MYB; LchiMYB, Liriodendron chinense MYB; qRT-PCR, quantitative real-time PCR; MW, molecular weight; aa, amino acid; TPM, transcripts per million; pI, isoelectric point; ATG, A. thaliana; MSA, multiple sequence alignment; TFs, transcription factors; HTH, helix-turn-helix; ABA, abscisic acid; FPKM, fragments per kilobase of the exon model per million mapped fragments.
Footnotes
- ^ http://www.arabidopsis.org/
- ^ http://pfam.xfam.org
- ^ https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi
- ^ http://gsds.gao-lab.org/index.php
- ^ https://cran.r-project.org/web/packages/circlize/
- ^ https://pypi.org/project/jcvi/
- ^ http://bioinformatics.psb.ugent.be/webtools/plantcare/html/
- ^ https://www.expasy.org/resources/compute-pi-mw
- ^ http://www.bioconductor.org/packages/release/bioc/html/ggtree.html
- ^ https://www.yeastgenome.org/primer3
References
Ambawat, S., Sharma, P., Yadav, N. R., and Yadav, R. C. (2013). Myb transcription factor genes as regulators for plant responses: an overview. Physiol. Mol. Biol. Plants 19, 307–321. doi: 10.1007/s12298-013-0179-1
Baillo, E. H., Kimotho, R. N., Zhang, Z., and Xu, P. (2019). Transcription factors associated with abiotic and biotic stress tolerance and their potential for crops improvement. Genes 10:771. doi: 10.3390/genes10100771
Capella-Gutierrez, S., Silla-Martinez, J. M., and Gabaldon, T. (2009). Trimal: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973. doi: 10.1093/bioinformatics/btp348
Chen, J., Hao, Z., Guang, X., Zhao, C., Wang, P., et al. (2019). Liriodendron genome sheds light on angiosperm phylogeny and species–pair differentiation. Nat. Plants 5, 18–25. doi: 10.1038/s41477-018-0323-6
Chen, S., Niu, X., Guan, Y., and Li, H. (2017). Genome-wide analysis and expression profiles of the myb genes in brachypodium distachyon. Plant Cell Physiol. 58, 1777–1788. doi: 10.1093/pcp/pcx115
Drummond, A. J., Suchard, M. A., Xie, D., and Rambaut, A. (2012). Bayesian phylogenetics with beauti and the beast 1.7. Mol. Biol. Evol. 29, 1969–1973. doi: 10.1093/molbev/mss075
Dubos, C., Stracke, R., Grotewold, E., Weisshaar, B., Martin, C., et al. (2010). Myb transcription factors in arabidopsis. Trends Plant Sci. 15, 573–581. doi: 10.1016/j.tplants.2010.06.005
Gates, D. J., Strickler, S. R., Mueller, L. A., Olson, B. J. S. C., and Smith, S. D. (2016). Diversification of r2r3-myb transcription factors in the tomato family solanaceae. J. Mol. Evolut. 83, 26–37. doi: 10.1007/s00239-016-9750-z
Gliwicka, M., Nowak, K., Balazadeh, S., Mueller-Roeber, B., and Gaj, M. D. (2013). Extensive modulation of the transcription factor transcriptome during somatic embryogenesis in arabidopsis thaliana. PLoS One 8:e69261. doi: 10.1371/journal.pone.0069261
Gu, Z., Gu, L., Eils, R., Schlesner, M., and Brors, B. (2014). Circlize implements and enhances circular visualization in r. Bioinformatics 30, 2811–2812. doi: 10.1093/bioinformatics/btu393
Hu, A., Chen, Z., Wang, P., Yang, L., Wang, g., Wang, D., et al. (2017). Establishment of transient gene expression systems in protoplasts from liliodendron hybrid mesophyll cells. PLoS One. 12:e172475. doi: 10.1371/journal.pone.0172475
Jia, L., Clegg, M. T., and Jiang, T. (2003). Excess non-synonymous substitutions suggest that positive selection episodes occurred during the evolution of dna-binding domains in the arabidopsis r2r3-myb gene family. Plant Mol. Biol. 52, 627–642. doi: 10.1023/a:1024875232511
Jiang, C., Gu, X., and Peterson, T. (2004). Identification of conserved gene structures and carboxy-terminal motifs in the myb gene family of arabidopsis and oryza sativa l. SSP. Indica. Genome Biol. 5:R46. doi: 10.1186/gb-2004-5-7-r46
Katiyar, A., Smita, S., Lenka, S. K., Rajwanshi, R., Chinnusamy, V., et al. (2012). Genome-wide classification and expression analysis of myb transcription factor families in rice and arabidopsis. BMC Genomics 13:544. doi: 10.1186/1471-2164-13-544
Kranz, H. D., Denekamp, M., Greco, R., Jin, H., Leyva, A., et al. (1998). Towards functional characterisation of the members of the r2r3-myb gene family from arabidopsis thaliana. Plant J. 16, 263–276. doi: 10.1046/j.1365-313x.1998.00278.x
Li, C., and Lu, S. (2014). Genome-wide characterization and comparative analysis of r2r3-myb transcription factors shows the complexity of myb-associated regulatory networks in salvia miltiorrhiza. BMC Genomics 15:277. doi: 10.1186/1471-2164-15-277
Li, J., Han, G., Sun, C., and Sui, N. (2019). Research advances of myb transcription factors in plant stress resistance and breeding. Plant Signal. Behav. 14:1613131. doi: 10.1080/15592324.2019.1613131
Li, T., Yuan, W., Qiu, S., and Shi, J. (2021). Selection of reference genes for gene expression analysis in Liriodendron hybrids’ somatic embryogenesis and germinative tissues. Sci. Rep. 11:4957. doi: 10.1038/s41598-021-84518-w
Li, X., Tang, Y., Li, H., Luo, W., Zhou, C., et al. (2020). A wheat r2r3 myb gene tampc1-d4 negatively regulates drought tolerance in transgenic arabidopsis and wheat. Plant Sci. 299:110613. doi: 10.1016/j.plantsci.2020.110613
Li, Y., Lin-Wang, K., Liu, Z., Allan, A. C., Qin, S., et al. (2020). Genome-wide analysis and expression profiles of the str2r3-myb transcription factor superfamily in potato (solanum tuberosum l.). Int. J. Biol. Macromol. 148, 817–832. doi: 10.1016/j.ijbiomac.2020.01.167
Li, Y., Zhang, S., Zhang, N., Zhang, W., Li, M., et al. (2019). Myb-cc transcription factor, tamybsm3, cloned from wheat is involved in drought tolerance. BMC Plant Biol. 1:19. doi: 10.1186/s12870-019-1751-9
Liu, C., Hao, J., Qiu, M., Pan, J., and He, Y. (2020). Genome-wide identification and expression analysis of the myb transcription factor in japanese plum (prunus salicina). Genomics 112, 4875–4886. doi: 10.1016/j.ygeno.2020.08.018
Liu, C., Xie, T., Chen, C., Luan, A., Long, J., et al. (2017). Genome-wide organization and expression profiling of the r2r3-myb transcription factor family in pineapple (ananas comosus). BMC Genomics 18:896. doi: 10.1186/s12864-017-3896-y
Méndez-Hernández, H. A., Ledezma-Rodríguez, M., Avilez-Montalvo, R. N., Juárez-Gómez, Y. L., Skeete, A., et al. (2019). Signaling overview of plant somatic embryogenesis. Front. Plant Sci. 10:77. doi: 10.3389/fpls.2019.00077
Millard, P. S., Kragelund, B. B., and Burow, M. (2019). R2r3 myb transcription factors – functions outside the dna-binding domain. Trends Plant Sci. 24, 934–946. doi: 10.1016/j.tplants.2019.07.003
Mmadi, M. A., Dossa, K., Wang, L., Zhou, R., Wang, Y., et al. (2017). Functional characterization of the versatile myb gene family uncovered their important roles in plant development and responses to drought and waterlogging in sesame. Genes 8:362. doi: 10.3390/genes8120362
Nekrutenko, A., Makova, K. D., and Li, W. H. (2002). The k(a)/k(s) ratio test for assessing the protein-coding potential of genomic regions: an empirical and simulation study. Genome Res. 12, 198–202. doi: 10.1101/gr.200901
Pucker, B., Pandey, A., Weisshaar, B., and Stracke, R. (2020). The r2r3-myb gene family in banana (musa acuminata): genome-wide identification, classification and expression patterns. PLoS One 15:e239275. doi: 10.1371/journal.pone.0239275
Rahaie, M., Xue, G.-P., Naghavi, M. R., Alizadeh, H., and Schenk, P. M. (2010). A MYB gene from wheat (Triticum aestivum L.) is up-regulated during salt and drought stresses and differentially regulated between salt-tolerant and sensitive genotypes. Plant Cell Rep. 29, 835–844, doi: 10.1007/s00299-010-0868-y
Roy, S. (2016). Function of myb domain transcription factors in abiotic stress and epigenetic control of stress response in plant genome. Plant Signal. Behav. 11:e1117723. doi: 10.1080/15592324.2015.1117723
Shen, X., Hu, Z., Xiang, X., Xu, L., and Cao, J. (2019). Overexpression of a stamen-specific r2r3-myb gene bcmf28 causes aberrant stamen development in transgenic arabidopsis. Biochem. Biophys. Res. Commun. 518, 726–731. doi: 10.1016/j.bbrc.2019.08.119
Srideepthi, R., Krishna, M. S. R., Suneetha, P., Krishna, R. S., and Karthikeyan, S. (2020). Genome-wide identification, characterization and expression analysis of non-rd receptor like kinase gene family under colletotrichum truncatum stress conditions in hot pepper. Genetica 148, 283–296. doi: 10.1007/s10709-020-00104-4
Stracke, R., Werber, M., and Weisshaar, B. (2001). The r2r3-myb gene family in arabidopsis thaliana. Curr. Opin. Plant Biol. 4, 447–456. doi: 10.1016/s1369-5266(00)00199-0
Su, L.-T., Li, J.-W., Liu, D.-Q., Zhai, Y., Zhang, H.-J., Li, X.-W., et al. (2014). A novel MYB transcription factor, GmMYBJ1, from soybean confers drought and cold tolerance in Arabidopsis thaliana. Gene 538, 46–55. doi: 10.1016/j.gene.2014.01.024
Su, Y. H., Liu, Y. B., Bai, B., and Zhang, X. S. (2014). Establishment of embryonic shoot-root axis is involved in auxin and cytokinin response during arabidopsis somatic embryogenesis. Front. Plant Sci. 5:792. doi: 10.3389/fpls.2014.00792
Sun, P., Zhu, X., Huang, X., and Liu, J. H. (2014). Overexpression of a stress-responsive MYB transcription factor of Poncirus trifoliata confers enhanced dehydration tolerance and increases polyamine biosynthesis. Plant Physiol. Biochem. 78, 71–79. doi: 10.1016/j.plaphy.2014.02.022
Sun, W., Ma, Z., Chen, H., and Liu, M. (2019). Myb gene family in potato (solanum tuberosum l.): Genome-wide identification of hormone-responsive reveals their potential functions in growth and development. Int. J. Mol. Sci. 20:847. doi: 10.3390/ijms20194847
Wang, F. X., Shang, G. D., Wu, L. Y., Xu, Z. G., Zhao, X. Y., et al. (2020). Chromatin accessibility dynamics and a hierarchical transcriptional regulatory network structure for plant somatic embryogenesis. Dev. Cell 54, 742–757. doi: 10.1016/j.devcel.2020.07.003
Wang, J., Liu, Y., Tang, B., Dai, X., Xie, L., et al. (2020). Genome-wide identification and capsaicinoid biosynthesis-related expression analysis of the r2r3-myb gene family in capsicum annuum l. Front. Genet. 11:183. doi: 10.3389/fgene.2020.598183
Wang, N., Ma, Q., Ma, J., Pei, W., Liu, G., et al. (2019). A comparative genome-wide analysis of the r2r3-myb gene family among four gossypium species and their sequence variation and association with fiber quality traits in an interspecific g. Hirsutum x g. Barbadense population. Front. Genet 10:741. doi: 10.3389/fgene.2019.00741
Wang, Y., Li, H., Zhou, Y., Guo, D., Zhu, J., et al. (2021). Transcriptomes analysis reveals novel insight into the molecular mechanisms of somatic embryogenesis in hevea brasiliensis. BMC Genomics 22:501. doi: 10.1186/s12864-021-07501-9
Wang, Y., Tang, H., Debarry, J. D., Tan, X., Li, J., et al. (2012). Mcscanx: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40:e49. doi: 10.1093/nar/gkr1293
Wei, Q., Chen, R., Wei, X., Liu, Y., Zhao, S., Yin, X., et al. (2020). Genome-wide identification of R2R3-MYB family in wheat and functional characteristics of the abiotic stress responsive gene TaMYB344. BMC Genomics 21:792. doi: 10.1186/s12864-020-07175-9
Wilkins, O., Nahal, H., Foong, J., Provart, N. J., and Campbell, M. M. (2009). Expansion and diversification of the populus r2r3-myb family of transcription factors. Plant Physiol. 149, 981–993. doi: 10.1104/pp.108.132795
Yang, A., Dick, C. W., Yao, X., and Huang, H. (2016). Impacts of biogeographic history and marginal population genetics on species range limits: a case study of liriodendron chinense. Sci. Rep. 6:25632. doi: 10.1038/srep25632
Yanhui, C., Xiaoyuan, Y., Kun, H., Meihua, L., Jigang, L., et al. (2006). The myb transcription factor superfamily of arabidopsis: expression analysis and phylogenetic comparison with the rice myb family. Plant Mol. Biol. 60, 107–124. doi: 10.1007/s11103-005-2910-y
Yu, G. (2020). Using ggtree to visualize data on tree-like structures. Curr. Protoc. Bioinform. 69:e96. doi: 10.1002/cpbi.96
Yu, G., Lam, T. T., Zhu, H., and Guan, Y. (2018). Two methods for mapping and visualizing associated data on phylogeny using ggtree. Mol. Biol. Evol. 35, 3041–3043. doi: 10.1093/molbev/msy194
Zhang, T., Zhao, Y., Wang, Y., Liu, Z., and Gao, C. (2018). Comprehensive analysis of myb gene family and their expressions under abiotic stresses and hormone treatments in tamarix hispida. Front. Plant Sci. 9:1303. doi: 10.3389/fpls.2018.01303
Zhang, Y., Xu, S., Cheng, Y., Wang, J., Wang, X., Liu, R., et al. (2020). Functional identification of PsMYB57 involved in anthocyanin regulation of tree peony. BMC Genetics 21:124. doi: 10.1186/s12863-020-00930-7
Zhao, K., Cheng, Z., Guo, Q., Yao, W., Liu, H., Zhou, B., et al. (2020). Characterization of the poplar R2R3-MYB gene family and Over-Expression of PsnMYB108 confers salt tolerance in transgenic tobacco. Front. Plant Sci. 11:571881. doi: 10.3389/fpls.2020.571881
Zhao, P., Li, Q., Li, J., Wang, L., and Ren, Z. (2014). Genome-wide identification and characterization of r2r3myb family in solanum lycopersicum. Mol. Genet. Genom. 289, 1183–1207. doi: 10.1007/s00438-014-0879-4
Zhou, C., Chen, Y., Wu, Z., Lu, W., Han, J., et al. (2015). Genome-wide analysis of the myb gene family in physic nut (jatropha curcas l.). Gene 572, 63–71. doi: 10.1016/j.gene.2015.06.072
Keywords: Liriodendron chinense, genome-wide, transcription factor, MYB gene family, abiotic stress
Citation: Wu W, Zhu S, Zhu L, Wang D, Liu Y, Liu S, Zhang J, Hao Z, Lu Y, Cheng T, Shi J and Chen J (2021) Characterization of the Liriodendron Chinense MYB Gene Family and Its Role in Abiotic Stress Response. Front. Plant Sci. 12:641280. doi: 10.3389/fpls.2021.641280
Received: 14 December 2020; Accepted: 09 June 2021;
Published: 26 July 2021.
Edited by:
Xiaojuan Li, Beijing Forestry University, ChinaReviewed by:
Manuela Silva, University of Lisbon, PortugalXiao-Ren Chen, Yangzhou University, China
Copyright © 2021 Wu, Zhu, Zhu, Wang, Liu, Liu, Zhang, Hao, Lu, Cheng, Shi and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jinhui Chen, Y2hlbmpoQG5qZnUuZWR1LmNu
 Weihuang Wu
Weihuang Wu Sheng Zhu
Sheng Zhu Liming Zhu
Liming Zhu Dandan Wang
Dandan Wang Yang Liu
Yang Liu Siqin Liu
Siqin Liu Jiaji Zhang
Jiaji Zhang Zhaodong Hao
Zhaodong Hao Ye Lu
Ye Lu Tielong Cheng
Tielong Cheng Jisen Shi
Jisen Shi Jinhui Chen
Jinhui Chen