- 1School of Life Sciences, Huaibei Normal University, Huaibei, China
- 2College of Horticulture, Nanjing Agricultural University, Nanjing, China
The red color is an attractive trait of fruit and determines its market acceptance. 5-Aminolevulinic acid (ALA), an eco-friendly plant growth regulator, has played a universal role in plant secondary metabolism regulation, particularly in flavonoid biosynthesis. It has been widely reported that ALA can up-regulate expression levels of several structural genes related to flavonoid metabolism and anthocyanin accumulation. However, the molecular mechanisms behind ALA-induced expression of these genes are complicated and still far from being completely understood. In this study, transcriptome analysis identified the differentially expressed genes (DEGs) associated with ALA-induced anthocyanin accumulation. Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis showed that the flavonoid biosynthesis (ko00941) pathway was significantly enhanced in the ALA-treated apple calli at 24, 48, and 72 h after the treatment. Expression pattern revealed that ALA up-regulated the expression of the structural genes related to not only anthocyanin biosynthesis (MdCHS, MdCHI, MdF3’H, MdDFR, MdANS, and MdUFGT) but also anthocyanin transport (MdGST and MdMATE). Two R2R3-MYB transcription factors (MdMYB10 and MdMYB9), which are the known positive regulators of anthocyanin biosynthesis, were significantly induced by ALA. Gene overexpression and RNA interference assays demonstrated that MdMYB10 and MdMYB9 were involved in ALA-induced anthocyanin biosynthesis. Moreover, MdMYB10 and MdMYB9 might positively regulate the transcription of MdMATE8 by binding to the promoter region. These results indicate that MdMYB10 and MdMYB9 modulated structural gene expression of anthocyanin biosynthesis and transport in response to ALA-mediated apple calli coloration at the transcript level. We herein provide new details regarding transcriptional regulation of ALA-induced color development.
Introduction
Fruit coloration acts as one of the most important factors determining commodity value of red cultivars. Finding eco-friendly, efficient, and easy-to-operate methods to improve fruit color has long been a major concern of fruit researchers. 5-Aminolevulinic acid (ALA), a key precursor in tetrapyrrole biosynthesis, has drawn increasing attention due to its important regulation roles in multiple physiological processes, including plant development, secondary metabolism, and fruit ripening (Akram and Ashraf, 2013). Promoting red fruit coloration is one of the most outstanding roles of ALA, which has been widely demonstrated in apple (Wang et al., 2006; Xie et al., 2013; Zheng et al., 2017), grape (Watanabe et al., 2006), pear (Xiao et al., 2012), peach (Guo et al., 2013; Ye et al., 2017), litchi (Feng et al., 2015), and strawberry (Li et al., 2016), indicating great application potential of ALA in modern fruit production. However, the molecular mechanism underlying ALA-induced fruit coloration is largely unknown.
The red pigment of fruit is mainly caused by anthocyanin, which is the major component of flavonoids. Anthocyanin is synthesized from the cytoplasmic face of endoplasmic reticulum in the cytosol and then transported into the vacuoles (Bae et al., 2006). In plant, anthocyanin biosynthesis starts from phenylalanine and is genetically catalyzed by the key enzymes encoded by structural genes, including PAL, CHS, CHI, F3’H, DFR, LDOX/ANS, and UFGT. The common biosynthetic pathways of anthocyanin have been well characterized in Zea mays (Wienand et al., 1986), petunia (Britsch and Grisebach, 1986), snapdragon (Martin et al., 1991), Arabidopsis thaliana (Shirley et al., 1995), and other plant species. Subsequently, a number of flavonoid biosynthesis pathways have been characterized in fruit crops, including apple (Takos et al., 2006), pear (Fischer et al., 2007), grape (Sparvoli et al., 1994), and strawberry (Pillet et al., 2015). Pigment also associated with anthocyanin transport. It was described that several members of glutathione S-transferase (GST) and the multidrug and toxic compound extrusion (MATE) family were involved in anthocyanin transport, such as Zea mays Bronze2 (bz2) (Marrs et al., 1995), petunia AN9 (Mueller et al., 2000), Arabidopsis TT12 and TT19 (Debeaujon et al., 2001), and grape AM1 and AM3 (Gomez et al., 2009). With regard to the preliminary mechanism, several studies suggest that ALA-promoted anthocyanin accumulation is closely related to higher expression levels of flavonoids biosynthetic genes, including CHS, DFR, ANS, and UFGT (Guo et al., 2013; Xie et al., 2013; Feng et al., 2016; Ye et al., 2017).
In general, the structural genes of anthocyanin biosynthesis are coordinately regulated at transcriptional level by a MBW ternary complex of MYB, basic helix-loop-helix (bHLH) domains, and WD40 proteins (Xu et al., 2015). In higher plants, MYB transcription factors (TFs) are one of the largest TF families characterized by the conserved MYB DNA-binding domain. R2R3 subfamily with two adjacent MYB domains are the principal member of MYB, which plays a crucial role in secondary metabolism, stress response, meristem formation, and the cell cycle (Martin and Paz-Ares, 1997). Based on the conservation of amino acid motifs in C terminal domains and of the DNA-binding domain, R2R3-MYB proteins have been divided into 25 subgroups (SG) in Arabidopsis (Dubos et al., 2010). In apple, 229 MYB protein families, containing 222 typical R2R3 MYB proteins, were subdivided into 45 subgroups (Cao et al., 2013). Among them several SGs were involved in the regulation of flavonoid biosynthesis. The SG6 mainly associated with anthocyanin biosynthesis and accumulation by activating structural genes, such as AtMYB75/PAP1, AtMYB90/PAP2, AtMYB113, and AtMYB114 in Arabidopsis (Gonzalez et al., 2008), PhAN2 in petunia (Quattrocchio et al., 1999), PpMYB10.3 in peach (Zhou et al., 2015, 2016), PyMYB10 in pear (Feng et al., 2010), and MdMYB10/MdMYB1/MdMYBA in apple (Takos et al., 2006; Ban et al., 2007; Espley et al., 2007). Members of SG5 were suggested responsible for proanthocyanidins (PAs) synthesis, including AtMYB123/TT2 (Nesi et al., 2001), FaMYB9 and FaMYB11 (Schaart et al., 2013), VvMYBPA2 (Terrier et al., 2009), MdMYB11 (An et al., 2015), and MdMYB12 (Wang et al., 2017). Moreover, MdMYB9, belonged to the SG5 subfamily, bound to the promoters of ANS, ANR, and LAR and promoted the accumulation of anthocyanins and PAs (An et al., 2015). SG7 typically regulating flavonol synthesis were described in Arabidopsis (Stracke et al., 2007), grape (Czemmel et al., 2009), and apple (Wang et al., 2017). In addition, some R2R3-MYB proteins of SG4, which encode transcriptional repressors, also exhibited different effects on flavonoids synthesis. In transgenic tobacco, overexpression of MdMYB3 resulted in anthocyanin accumulation and pigmentation (Vimolmangkang et al., 2013). In contrast, FaMYB1 acted as the transcriptional repressors to reduce expression of late flavonoid biosynthesis genes in tobacco (Aharoni et al., 2001). To date, ALA-promoted expression of anthocyanin biosynthetic genes has also been linked to several regulatory genes. MdMYB10 and MdbHLH33 appeared to play positive roles in ALA-induced anthocyanin accumulation in apple (Zheng et al., 2018), while in peach skin, ALA significantly activated the expression of PpMYB10 and PpWD40 but not PpbHLH3 (Ye et al., 2017). It seems that R2R3-MYB TFs of SG6 respond to ALA-stimulated coloration, but the directly conclusive evidences are limited. Thus, transcriptional network is necessary for us to better understand the molecular mechanism of ALA-induced anthocyanin accumulation.
Under artificial condition, calli can be continuously and uniformly produced to efficiently occupy the available space without seasonal restrictions and also provide a homogeneous system observation and analysis of different treatment effects. Lately, calli was widely used as the model system to identify key candidate genes’ functions and elucidate the mechanism of PGRs on plant growth regulation, especially in flavonoid metabolism (Ji et al., 2015; Sun et al., 2017; Premathilake et al., 2020). Therefore, in the present study, apple fruit calli undergo a transcriptome profiling analysis to identify the differentially expressed genes (DEGs) between the ALA-treated and untreated control apple calli at three illuminating time points. Two key candidate TFs identified by RNA-seq were then proved to regulate ALA-induced coloration in different transgenic cell lines. Our findings enrich the knowledge regarding the molecular mechanism behind ALA-improved fruit coloration, which can substantially accelerate the study of ALA functions and its application in modern fruit production.
Materials and Methods
Plant Materials and Sample Treatment
The apple calli used in this study were induced from the flesh of “Fuji” apple (Malus domestica Borkh.), which were collected from commercial apple orchards of eastern China, Fengxian in Jiangsu Province. Calli were grown on MS medium (1 mg L–1 6-BA and 1 mg L–1 2, 4-D) at 22°C in dark, and were subcultured at 20-day intervals (An et al., 2015). For sample treatment, calli cultured in separate flasks were gathered and randomly transferred to liquid medium with (ALA) or without (Control) 50 mg L–1 ALA for 3 h in a shaking incubator in dark. For anthocyanin induction, all samples were re-transferred to MS solid medium and cultured at 22°C under light of 100 μmol m–2 s–1 photon flux density. Calli were harvested after being illuminating for 24, 48, and 72 h, respectively. At each time point, approximate 3 g apple calli were sampled after light irradiation and prepared three replicates for each treatment. A total of 18 independent libraries were subjected to transcriptome sequencing. Control calli were named as Control-24 (including C1, C2, and C3; C, Control), Control-48 (including C7, C8, and C9), and Control-72 (including C13, C14, and C15). ALA-treated calli were named ALA-24 (including T4, T5, and T6; T, treatment), ALA-48 (including T10, T11, and T12), and ALA-72 (including T16, T17, and T18).
Extraction and Determination of Anthocyanin Content
Anthocyanin content in calli was measured according to Xie et al. (2013) with slight modifications. Calli were incubated in 1% (v/v) HCl-methanol for 24 h at room temperature in dark. After centrifugation at 8000 × g for 15 min, the absorbance of upper aqueous phase was measured at 530, 620, and 650 nm with a spectrophotometer. The content of anthocyanin was expressed as nmol of cyanidin-3-galactoside in 1 g of fresh sample using a molar extinction coefficient of 3.43 × 104 (Ubi et al., 2006). Mean values were obtained from three independent replicates.
RNA Extraction, Library Preparation, and RNA-Seq Analysis
Total RNA was isolated from calli using the RNAprep pure Plant Kit (Tiangen, Beijing, China) following the manufacturer’s instructions. DNase I was used for treating total RNA samples, and then mRNA was purified from total RNA using poly-T oligo-attached magnetic beads. The quality of RNA was examined by an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, United States). About 3 μg of high-quality RNA per sample was used to construct the RNA-seq libraries, and a total of 18 libraries (six for per time point) were generated using NEBNext® UltraTM RNA Library Prep Kit. The library preparations were sequenced for paired-end reads with the HiSeq X system (Illumina, San Diego, CA, United States) by Novogene (Beijing, China). The Illumina raw data have been deposited in the NCBI sequence read archive (SRA) database under accession number PRJNA525304.
To obtain the high-quality reads (clean reads), raw RNA-seq reads were filtered by removing the adaptor, reads with unknown sequences “N” > 10%, and low-quality reads (Qphred ≤ 20 bases). Clean reads were then mapped to apple genome (Malus domestica v3.0.a1) using TopHat v2.0.12 (Trapnell et al., 2009; Velasco et al., 2010). HTSeq v0.6.1 was used to count the reads numbers mapped to each gene (Anders et al., 2014). Gene expression levels were estimated with FPKM (Fragments Per Kilobase of transcript per Million mapped reads) (Trapnell et al., 2010).
To identify DEGs between the control and ALA treatment, read counts as imputing data were normalized by DESeq (Anders and Huber, 2010). False discovery rate (FDR) was controlled by p values adjusted according to Benjamini and Hochberg’s approach. Genes with an absolute value of the log2 (Fold Change) ≥ 1 and FDR < 0.05 were identified as DEGs. The principal component analysis (PCA) was conducted with the internal steps of the R package version 3.5.31. GOseq R package was used to analyze Gene Ontology (GO) enrichment of DEGs (Young et al., 2010). GO terms with a corrected p < 0.05 were considered significantly enriched. The KOBAS software was applied to identify the significantly enriched KEGG pathways among the DEGs (Mao et al., 2005).
Phylogenetic Analysis
All the phylogenetic trees were generated using the maximum-likelihood (ML) method with 1000 bootstraps in MEGA X2. The accession numbers and sequences of MYB transcription factors in multiple plant species that were included in the phylogenetic analysis are listed in Supplementary Table 1. The sequences of 21 MdMATE members of clade I and 24 different known MATEs are listed in Supplementary Table 2.
Construct Expression Vectors and Agrobacterium-Mediated Transformation System of Apple Calli
For overexpression of MdMYB10 and MdMYB9, their CDSs were amplified by PCR from cDNA of “Fuji” apple fruit flesh calli. The PCR product was cloned into the pBI121 plant transformation vector downstream of the CaMV 35S promoter. To silence MdMYB9, MdMYB10, and MdMATE8 expression, a partial CDS of MdMYB10 (421 bp), MdMYB9 (507 bp), and MdMATE8 (429 bp) was transferred to RNAi vector (pHELLSGATE2, pHG2) through Gateway BP reaction (Invitrogen). The recombinant and empty plasmids were transformed into competent A. tumefaciens strain EHA105 using the freeze/thaw method. One single transformed A. tumefaciens colony was inoculated in 30 ml of YEB medium supplemented with corresponding antibiotics and grown at 28°C with shaking at 150 r.p.m. When OD600 of culture liquid reached approximately 0.5, A. tumefaciens cells were centrifuged at 4000 × g for 5 min, and resuspended in 10 ml of infiltration buffer (10 mM MES, pH 5.6, and 100 μM acetosyringone). The fresh calli were dipped into A. tumefaciens suspension for 15 min at room temperature. The calli were then co-cultured on solid MS medium containing 2,4-D and 6-BA for 3 days at 25°C in dark. Subsequently, the calli were retransferred to MS medium (250 mg L–1 carbenicillin and 30 mg L–1 kanamycin) for positive transgenic selection after being washed five to eight times with sterile water. The anthocyanin contents of calli were measured according to the method described above. The expression levels in apple transgenic calli lines were confirmed by qRT-PCR amplification. All assays were replicated at least three times.
Cis-Acting Element Analysis
Cis-acting elements in the 1800-bp promoter region of MdMATE8 were identified using the PlantCARE database3.
Yeast One-Hybrid (Y1H) Assay
The promoter sequence (1800 bp) of MdMATE8 (MDP0000175055) was inserted into the pHIS2.1 vector (Clontech, Palo Alto, CA, United States), which contained the reporter gene HIS3. The coding sequences (CDS) of MdMYB10 and MdMYB9 were cloned into the pGADT7 vector (Clontech) to form pGADT7-MdMYB10 and pGADT7-MdMYB9, respectively. To detect the suitable concentration of 3-AT, the yeast strain Y187 transformed with the recombinant pHIS2 vectors were grown on SD/-Trp/-His medium containing different concentrations of 3-amino-1,2,4-triazole (3-AT). The constructs p53HIS2.1 and pGADT7-53 were used as the positive control. Negative controls were transformed with p53HIS2.1 + pGADT7, p53HIS2.1 + pGADT7-MdMYB10, p53HIS2.1 + pGADT7-MdMYB9, or pHIS2.1-MdMATE8pro + pGADT7, respectively. The transformants were selected on double dropout medium (DDO, SD/-Trp-Leu) media. The transformed yeast cells were plated onto triple dropout medium (TDO, SD/-Trp-Leu-His) containing 80 mM 3-AT, and cell growth was examined.
Total RNA Isolation and Quantitative PCR Expression Analysis
As for qPCR assay, total RNA was extracted using the cetyltrimethylammonium bromide (CTAB) method. The first strand of cDNA synthesis was performed using the TransScript® One-Step gDNA Removal and cDNA Synthesis Supermix (Transgen Biotech, China). The qRT-PCR reaction system was performed utilizing ChamQ SYBR qPCR master mix (Vazyme, Nanjing, China). The MdUBQ served as the housekeeping gene, and relative expressions of genes were calculated using the 2−ΔΔCT method (Livak and Schmitten, 2010). Each sample was quantified in triplicate. All qRT-PCR primers are listed in Supplementary Table 3.
Statistics Analysis
All of the results were collected from at least three independent parallel experiments. Statistical analysis was carried out using SPSS 20.0 statistical program. Significant differences (p < 0.05) of data were compared with control or among treatments by analysis of variance (ANOVA) and Duncan’s multiple range test. Graphs were generated in Excel 2016 and Origin Pro 7.5.
Results
Rapid Anthocyanin Accumulation in Apple Flesh Calli After ALA Treatment
To verify the effect of ALA on coloration development, the apple flesh calli were transferred to liquid Murashige and Skoog (MS) medium with (ALA) or without (Control) 50 mg L–1 ALA for 3 h in a shaking incubator in dark, and calli anthocyanin content was measured after being illuminated for 24, 48, and 72 h, respectively. Results showed that red color cumulatively increased in calli with time, and ALA exhibited an attractively promotive effect on coloration after illuminating for 48 h (Figure 1A). The anthocyanin content of ALA-treated calli did not significantly increase at 24 h, but reached 2.78- and 2.70-fold, respectively, of the control at 48 and 72 h (Figure 1B). These results suggest that ALA significantly induces anthocyanin accumulation in apple flesh calli. Therefore, this artificial calli system can be used to identify the candidate genes that are related to ALA-induced anthocyanin accumulation.
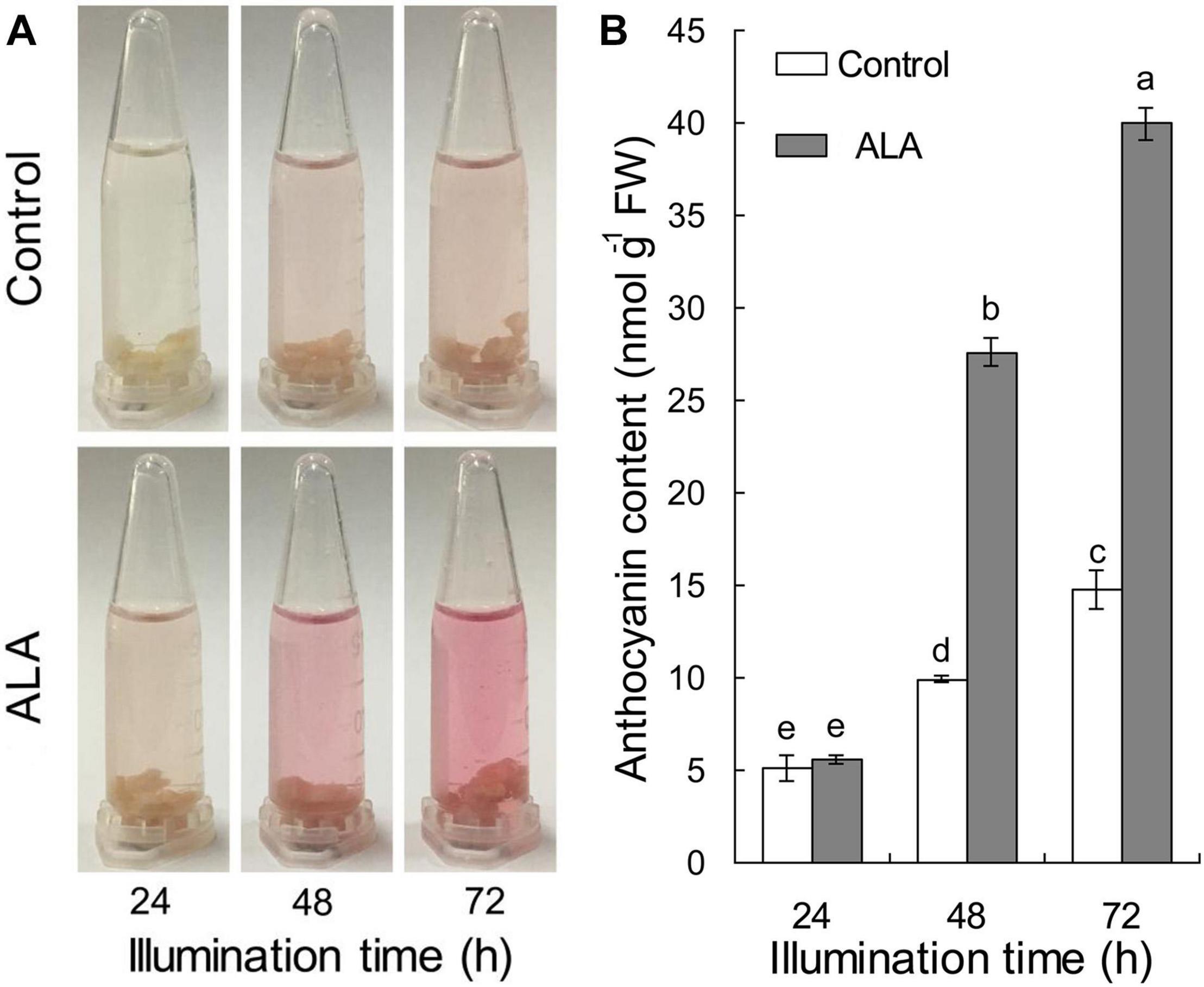
Figure 1. Time courses of anthocyanin accumulation after ALA treatment in fruit calli. The calli were incubated with 50 mg L–1 ALA or deionized water (Control) for 3 h under dark and then cultured in solid MS medium under light of 100 μmol m–2 s–1 photon flux density at 22°C for 72 h. (A) The process of color development in calli under light. (B) Anthocyanin content in calli. The different small letters represent significant differences (p < 0.05).
Overview of RNA Sequencing
A total of 514,483,442 raw reads were obtained from ALA-treated and control calli and were sampled at 24, 48, and 72 h. After quality filtering process, the number of clean reads for each library ranged from 22.7 to 32.0 million and the rate of clean reads/raw reads ranged from 93.82 to 96.26%. The Q30 values for all libraries were above 89.2% (Supplementary Table 4). An average of 90.62% reads were mapped to apple reference genome sequence, and 83.0% to 84.9% reads were mapped to exon region. Additionally, the correlation coefficient analysis showed that biological replicated libraries for two treatments per illumination time had highly consistent transcriptome profiles (Supplementary Table 4). These results reveal that the data we obtained are reliable and qualified for the following analysis.
Analysis of DEGs
To investigate differential gene expression between ALA-treated and control apple calli, gene expression levels were calculated based on the FPKM values (Supplementary Table 5). The DEGs were identified and filtered by a threshold of FDR ≤ 0.05 and an absolute Log2 (fold-change) value ≥ 1. A total of 1692 DEGs were identified as DEGs between ALA-treated and control calli (Supplementary Table 6). Except for the control calli illumination at 72 h, the PCA of the transcriptomic data revealed a high similarity among the three biological replicates within each treatment (Supplementary Figure 1). Among treatments, a clear separation of the ALA treatments from the control calli was observed. At 24 h after the treatment, the expression level of 487 and 354 genes were up- and down-regulated, respectively, in ALA-treated apple calli compared with control levels (Figure 2A). The expression level of 314 and 254 genes was up- and down-regulated, respectively, in ALA-treated apple calli relative to the corresponding control levels at 48 h after the treatment (Figure 2B). Furthermore, the expression level of 324 and 187 genes was up- and down-regulated, respectively, in ALA-treated apple calli versus control levels at 72 h (Figure 2C). More DEGs were found at 24 h between control and ALA-treated calli. DEGs among different time points were also identified. A total of 667, 411, and 415 DEGs were exclusively detected at 24, 48, and 72 h, respectively, suggesting that these genes might be involved in ALA-regulated physiology at a specific stage (Figures 2D–F). There were 29 common DEGs in all three illumination stages. Among these DEGs, 16 structural genes involved in flavonoid metabolism were identified (Figures 2D–F). These results imply that ALA stimulates significant changes of transcriptional level.
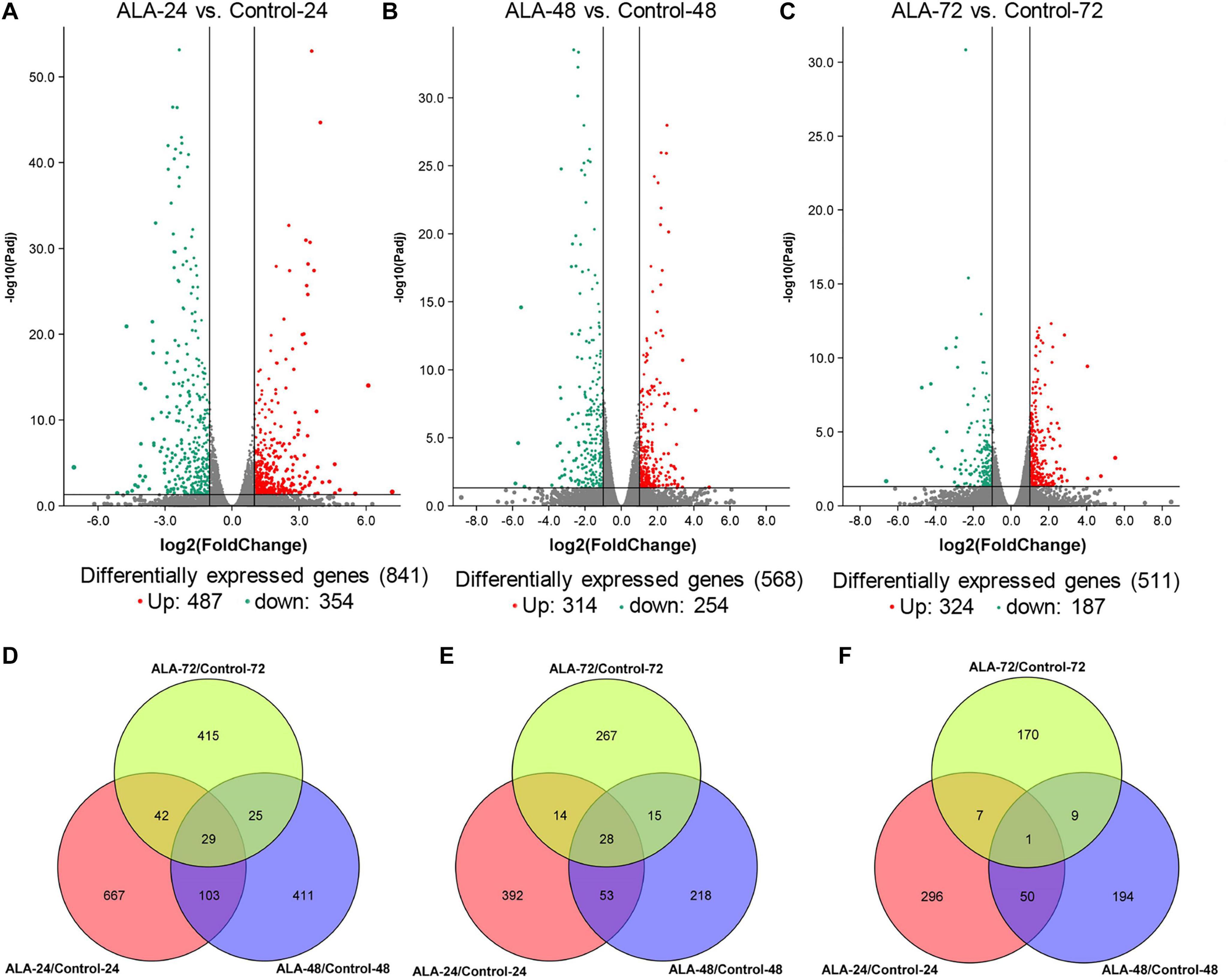
Figure 2. Differentially expressed genes (DEGs) identified by RNA-seq analysis in ALA-treated and control calli after illuminating for 24, 48, and 72 h. (A–C) Volcano plot of the RNA-seq showing DEGs in red and green. The X-axis represents the fold change in ALA-24 versus Control-24, ALA-48 versus Control-48, and ALA-72 versus Control-72, respectively. The Y-axis represents the negative –log10-transformed p values (p-adj < 0.05) for differences between the samples. (D) Quantity of total DEGs. (E) Quantity of up-regulated DEGs. (F) Quantity of down-regulated DEGs. Overlapping areas shows the shared DEGs at different time points.
GO Annotation and KEGG Pathway Analyses
Gene ontology (GO)-based term classification was performed to provide insights into DEG function. Among the total of 1692 DEGs, 1293 genes were annotated according to the GO database and were classified as “biological process,” “cellular component,” and “molecular function” (Figures 3A–C and Supplementary Table 7). However, the enriched items did not cover cellular components in the calli samples of ALA-72 versus Control-72 (Figure 3C). The two largest subcategories in the “biological process” category were “response to stimulus” including 523 DEGs and “biological regulation” including 466 DEGs. In addition, the subcategory of “flavonoid biosynthetic process” were notably enriched in the “biological process” at all three time points (Figures 3A–C). For the “cellular component” category, “membrane” including 506 DEGs and “intrinsic component of membrane” including 361 DEGs were the most abundant classes. Among the “molecular function” category, the two largest subcategories were “small molecule binding” and “anion binding” and included 346 and 340 DEGs, respectively (Supplementary Table 7).
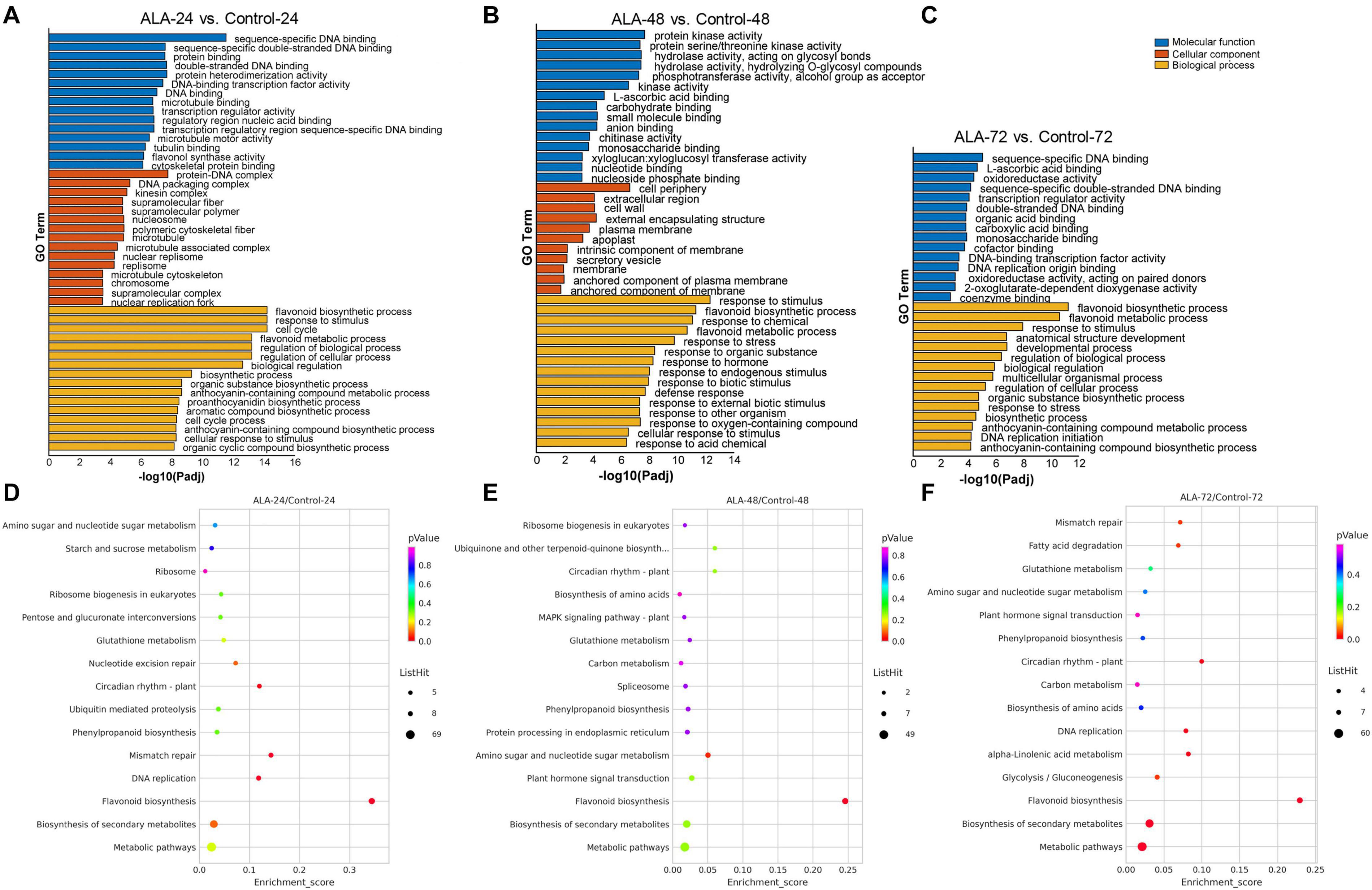
Figure 3. Gene ontology (GO) classifications and KEGG pathways enrichment of DEGs. (A–C) GO categories assigned to DEGs in ALA-24 versus Control-24 (A), ALA-48 versus Control-48 (B), and ALA-72 versus Control-72 (C), respectively. The X-axis represents the negative –log10-transformed p values (p-adj < 0.05) for differences between the samples. The left Y-axis shows categories according to the annotation of GO. (D–F) KEGG pathways of DEGs in ALA-24 versus Control-24 (D), ALA-48 versus Control-48 (E), and ALA-72 versus Control-72 (F), respectively. The Y-axis and X-axis present the KEGG pathways and the enrichment scores, respectively. Dot size corresponds to the number of distinct genes, whereas dot color reflects the p-adj value.
To further understand the biological pathways activated by ALA at each time point, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways enrichment were conducted. Corresponding to GO enrichment, the KEGG pathway analysis showed that the flavonoid biosynthesis (mdm00941) pathway was significantly enhanced in ALA-treated calli at all three time points after the treatment (Figures 3D–F). A total of 21, 15, and 14 DEGs were enriched in flavonoid biosynthesis at 24, 48, and 72 h, respectively (Supplementary Table 8). Among them, 11 DEGs were significantly enriched at all three time points. These results imply that flavonoid biosynthesis pathway significantly responds to ALA stimulation.
DEGs Related to Anthocyanin Biosynthesis and Transport
Anthocyanin biosynthesis is a dynamic and complex process catalyzed by multiple enzymes in the phenylpropanoid pathway. Here, 17 DEGs known to be involved in anthocyanin biosynthesis were identified according to GO and KEGG enrichment analysis (Figure 4). For example, MdCHS (MDP0000126567, MDP0000575740, MDP0000686661, and MDP0000686666), MdCHI (MDP0000134791, MDP0000252589, MDP0000759336, and MDP0000205890), and MdF3’H (MDP0000286933) were identified as the early biosynthetic genes (EBGs) in the early steps of the flavonoid biosynthesis pathway. Additionally, MdDFR (MDP0000494976), MdANS (MDP0000788934, MDP0000240641, MDP0000240643, and MDP0000360447), and MdUFGT (MDP0000543445, MDP0000933711, and MDP0000170162) were annotated as the late biosynthetic genes (LBGs) that were specifically involved in anthocyanin biosynthesis. Except for MdUFGT (MDP0000170162), which was down-regulated, 16 DEGs were significantly up-regulated in the ALA-treated apple calli, especially at 24 h after the treatment (Figure 4A). These results indicate that high expression levels of these structural genes in ALA-treated calli might be the main reason leading to anthocyanin accumulation.
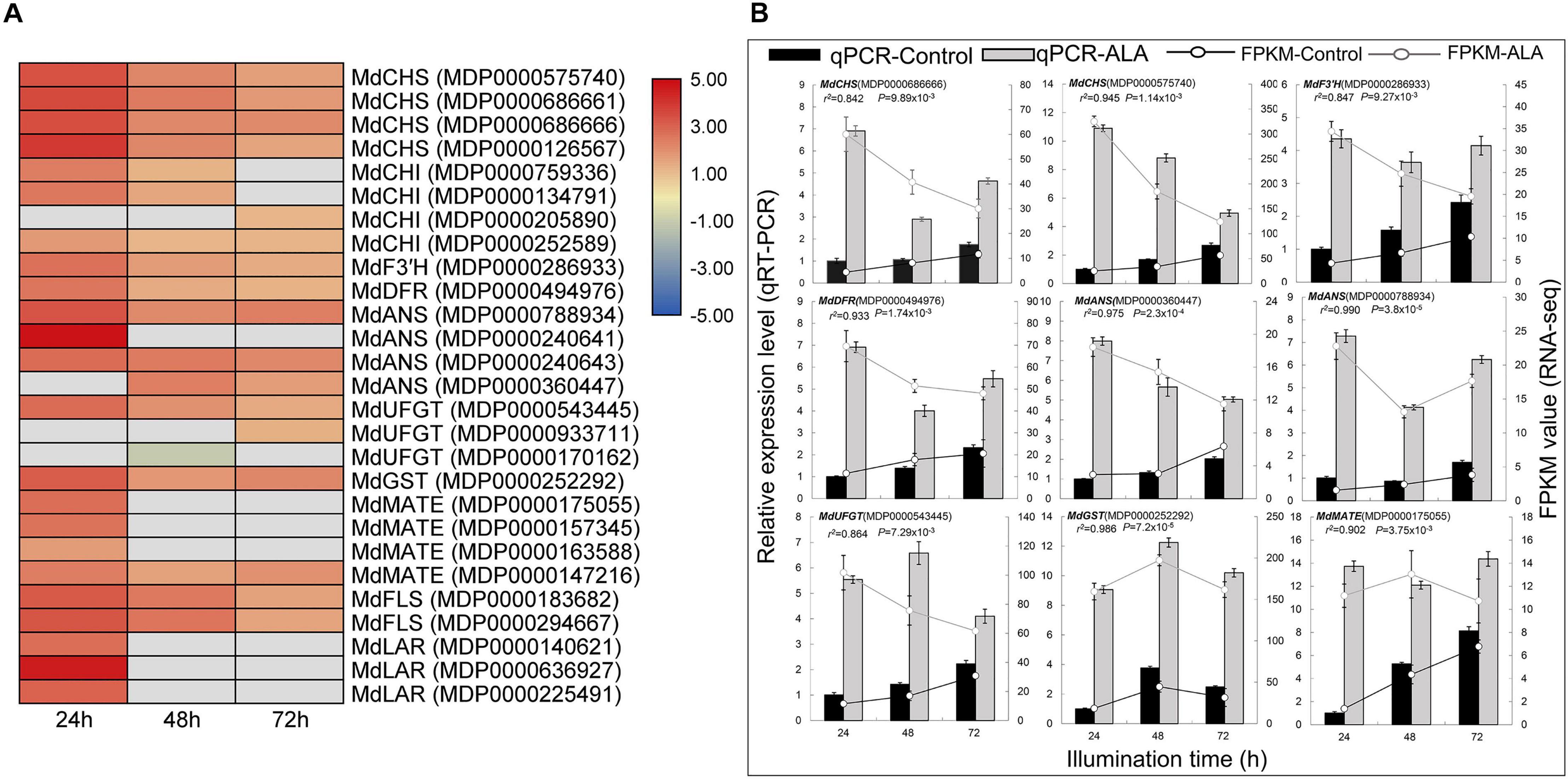
Figure 4. Expression pattern of genes involved in flavonoid biosynthesis and transport. (A) Heat map representation of transcriptional profiles of DEGs in the flavonoid biosynthesis pathway. The depth of color represents the changes of the value of log2 [fold change (FC)], and red represents up-regulation and blue represents down-regulation. The gray represents the relative differential expression level of those genes that did not reach screening threshold of DEGs in one or two time points after ALA treatment. (B) Verification of the expression of DEGs in the flavonoid biosynthesis pathway by qRT-PCR.
Flavonoid transporter also plays a vital role in flavonoid accumulation. The MDP0000252292, annotated as MdGSTF6, was up-regulated in ALA-treated calli, which has been proven to play a role in anthocyanin transporter of apple peel (Jiang et al., 2019). We also identified four MdMATE genes that were more highly expressed in ALA-treated calli, indicating that these genes might be involved in ALA-induced flavonoid accumulation (Figure 4A).
Moreover, two MdFLS (MDP0000183682, and MDP0000294667) and three MdLAR (MDP0000140621, MDP0000636927, and MDP0000225491) were identified, suggesting that ALA may also affect flavonol and proanthocyanidin (PA) biosynthesis (Figure 4A).
In addition, we noticed that several structural genes could not respond to ALA-induced flavonoid metabolism at all three illumination time points, such as MdCHI (MDP0000759336 and MDP0000134791), MdANS (MDP0000240641 and MDP0000360447), MdUFGT (MDP0000933711 and MDP0000170162), MdMATE (MDP0000175055, MDP0000157345, and MDP0000163588), and MdLAR (MDP0000140621, MDP0000636927, and MDP0000225491) (Figure 4A). Therefore, the results reveal that some genes of flavonoid metabolism respond to ALA only in specific time points.
To verify the sequencing data, the relative expression levels of selected structural genes were further analyzed by qRT-PCR. The expression levels of these EBGs and LBGs were up-regulated to above 4.5-fold by ALA treatment (Figure 4B). These results suggest that these structural genes may play vital roles in ALA-induced anthocyanin accumulation.
Transcriptional Regulation of ALA-Induced Anthocyanin Metabolism
The changes of structural gene expression are controlled by transcriptional factors, which plays the key role in plant responding to external stimulates. In this study, a total of 171 DEGs predicted to encode transcription factors (TFs) were identified by iTAK software. The MYB family members were the predominant TF genes in response to the ALA treatment, followed by the AP2-EREBP, and WRKY. Moreover, bHLH family genes were also differentially expressed (Supplementary Table 9).
To further identify the key members of MYB family in the transcriptional regulation of ALA-induced anthocyanin accumulation in apple calli, a phylogenetic tree was constructed based on 39 selected R2R3-MYB proteins of phenylpropanoid pathway regulators and 27 MYB from DEGs (Figure 5A and Supplementary Table 1). Four candidate MYB transcription factors (MDP0000127691, MDP0000259614, MDP0000317257, and MDP0000573302) belonged to SG6, which phylogenetically related to positive regulators of anthocyanin biosynthesis such as VvMYBA1, VvMYBA2, and MrMYB1 (Niu et al., 2010; Rinaldo et al., 2015). Several candidate MYB TFs were revealed as the regulators of PA biosynthesis because they were grouped with SG5 MYB TFs including AtTT2, VvMYBPA2, FaMYB9, and OsMYB3. Additionally, two MYB TFs were identified as potential repressors, which branched with AtMYB4, AtMYB7, and AtMYB32 of SG4 (Fornalé et al., 2014).
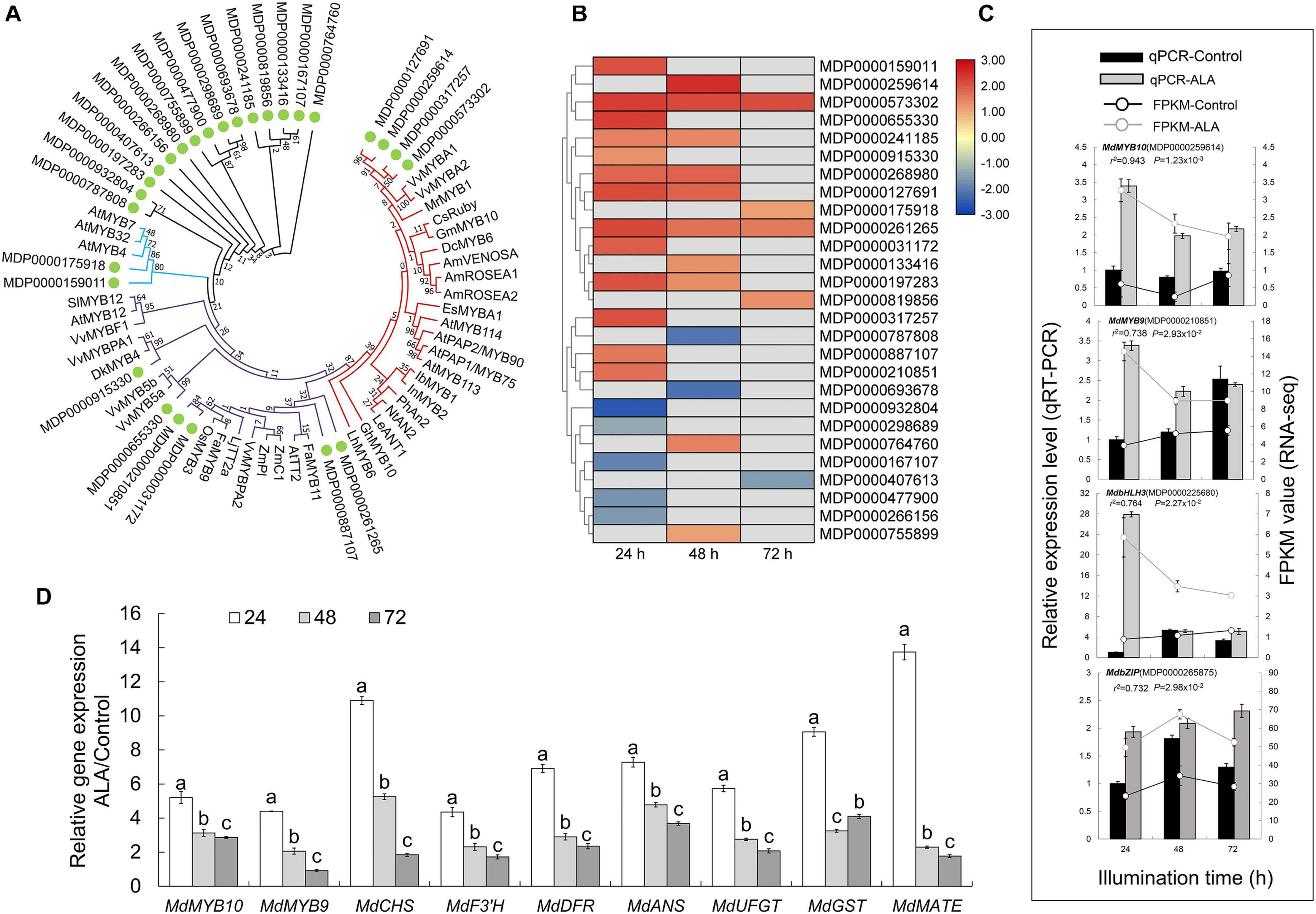
Figure 5. Analysis of differentially expressed MYB transcription factor genes related to putative flavonoid biosynthesis. (A) Phylogenetic tree with 39 phenylpropanoid pathway regulatory MYB TFs and 27 MYB from DEGs. (B) Heat map presenting the expression patterns of differentially expressed of 27 MYB. The depth of color represents the changes of the value of log2 [fold change (FC)], and red represents up-regulation and blue represents down-regulation. The gray represents genes that did not belong to DEGs at the corresponding time points. (C) Verification of the expression of four differentially expressed transcription factor genes by qRT-PCR analysis. (D) QRT-PCR analysis of the relative expression of MdMYB10 and MdMYBR9 and genes related to anthocyanin biosynthesis and transport in apple flesh calli at 24, 48, and 72 h after ALA treatment. The different letters in each gene represents significant differences (p < 0.05).
The expression profiles of the 27 MYBs showed that most of these genes were more highly expressed in the ALA-treated apple calli than in the control (Figure 5B). Moreover, most members of SG4-6 MYB TFs predicted as the candidates involved in ALA-regulated flavonoid metabolism were up-regulated and peaked at 24 or 48 h after the treatment, indicating that these TFs could rapidly respond to ALA stimulation. The relative expression levels of four selected transcription factor genes determined by qRT-PCR were consistent with the FPKM values based on the RNA-sequencing (Figure 5C). In Figure 5B, although MdMYB10 (MDP0000259614) and MdMYB9 (MDP0000210851) were not screened as DEGs at two time points in RNA-seq, MdMYB10 was up-regulated to 1.6-fold by ALA at 24 h (p < 0.05) and MdMYB9 was improved to 0.8-fold at 48 h (p = 0.01) after ALA-treatment. These results indicated that the expression levels of these two TFs were induced by ALA treatment, which was basically consistent with our qRT-PCR results in Figure 5C.
Among 12 MYB TFs of SG4-6, MdMYB10 (MDP0000259614 and MDP0000127691) and MdMYB9 (MDP0000210851) were characterized as known anthocyanin regulatory MYB transcription factors in apple (An et al., 2015; El-sharkawy et al., 2015). Their expression profiles were positively correlated with structural genes of anthocyanin accumulation under ALA treatment (Figure 5D and Supplementary Table 10). These results demonstrate that MdMYB10 and MdMYB9 may play key roles in ALA-regulated coloration.
Regulation Roles of MdMYB10 and MdMYB9 in ALA-Induced Anthocyanin Biosynthesis
To verify the roles of MdMYB10 and MdMYB9 in ALA-induced pigmentation, they were transiently overexpressed or silenced in “Fuji” apple fruit calli. The expression levels in their corresponding overexpression line were 12.4 and 6.7 times, respectively, higher than that in the control, which transformed the empty pBI121 plasmids (Supplementary Figures 2A,B). Moreover, exogenous ALA further up-regulated MdMYB10 or MdMYB9 expression level in their OE transgenic lines (Supplementary Figures 2A,B). In the RNA interference lines, the expression of MdMYB10 and MdMYB9 were approximately decreased to 61% and 55%, respectively, compared with the control, which was transformed with empty pHG2 plasmids (Supplementary Figures 2C,D). However, ALA did not significantly affect MdMYB10 and MdMYB9 expression in their RNAi calli. These results indicate that we obtain useable transformed apple calli of two key candidate genes.
Under light condition, the OE calli of MdMYB10 and MdMYB9 turned red more rapidly than the control(OE) (Figure 6A). Spectrophotometric analysis showed that MdMYB10(OE) and MdMYBR9(OE) accumulated significantly higher anthocyanin level, and the content was approximately 4.7 and 4.6 times, respectively, as high as that of the control(OE) calli (Figure 6B). Results confirmed their positive roles in anthocyanin biosynthesis. When exogenous ALA was applied, the color of MdMYB10 and MdMYB9 OE calli turned to deep red (Figure 6A), and the anthocyanin content further increased by 2.3 and 1.9 times, respectively, compared with each corresponding OE calli without ALA treatment (Figure 6B). Consistent with the color changes, overexpression of MdMYB10 or MdMYB9 up-regulated the expression level of structural genes of anthocyanin biosynthesis, and the effect was further promoted by ALA (Figure 6C). These results indicate that the promotive effect of ALA-induced anthocyanin accumulation is notably enhanced when MdMYB10 or MdMYB9 is overexpressed.
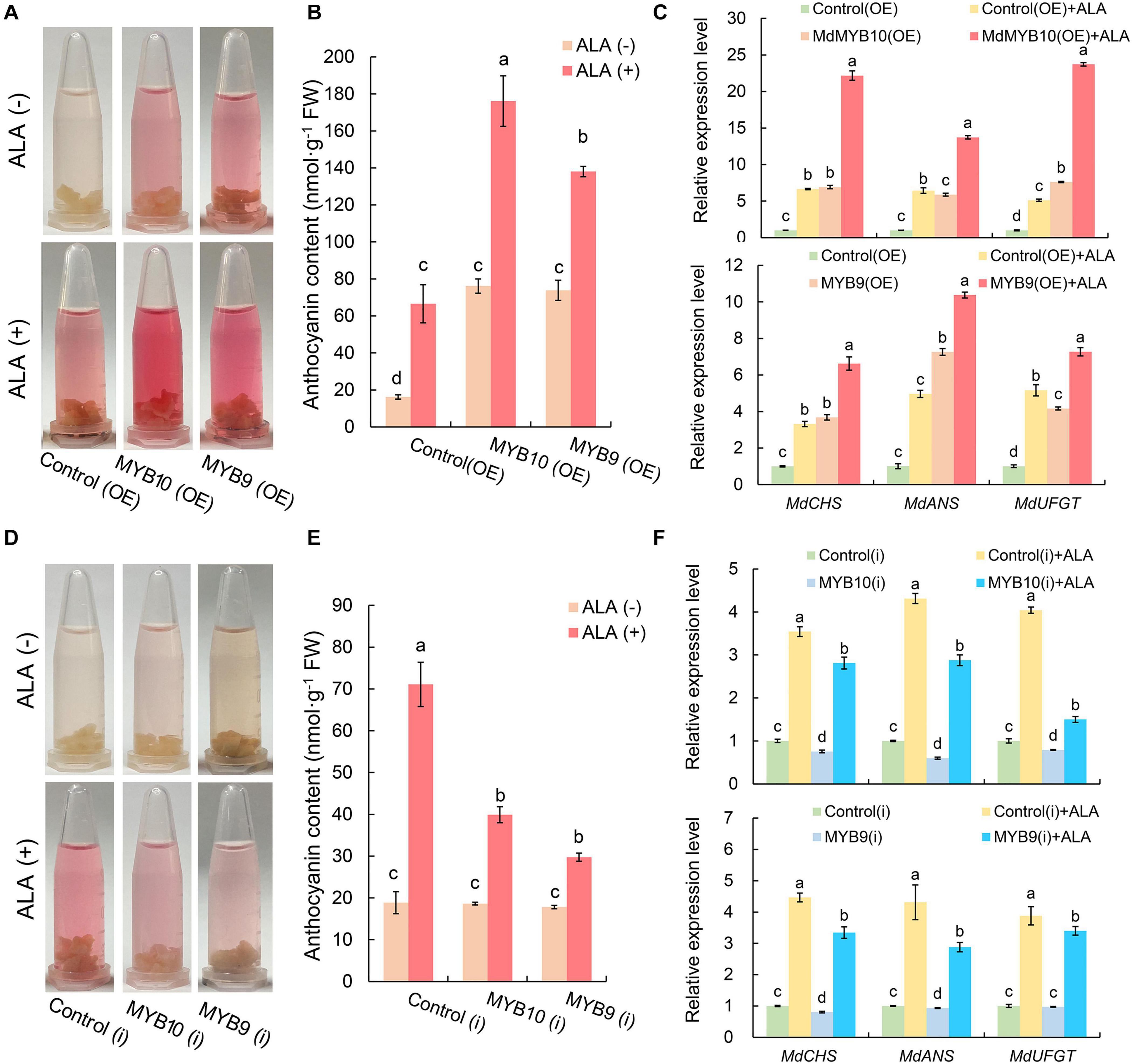
Figure 6. Effect of ALA on coloration level and anthocyanin content as well as the corresponding gene expression in MdMYB10 and MdMYB9 transgenic calli. Color (A) and anthocyanin content (B) in transgenic calli; OE, calli infiltrated with the plasmid for overexpressing of target genes; Control (OE), calli infiltrated with an empty pBI121 vector. (C) The expression of anthocyanin biosynthesis genes in MYB10(OE) and MYB9(OE). Color (D) and anthocyanin content (E) in transgenic calli; RNAi, calli infiltrated with the plasmid for silencing of target genes; Control(i), calli infiltrated with an empty pHG2 vector. (F) The expression of anthocyanin biosynthesis genes in MYB10(i) and MYB9(i). The different letters in each gene represent significant differences (p < 0.05).
In the MdMYB10 and MdMYB9 RNAi lines, the coloration level and the anthocyanin content did not obviously change (Figures 6D,E), compared with control(i). ALA treatment also induced anthocyanin accumulation in MYB10(i) and MYB9(i); however, the promotive effects of ALA were markedly blocked and the anthocyanin content was 56% and 42%, respectively, of the control(i) treated with ALA (Figures 6D,E). The expression level of several structural genes decreased in the calli of MYB10(i) or MYB9(i) (Figure 6F). Compared with control(i) + ALA, silence of MdMYB10 or MdMYB9 significantly suppressed ALA-induced expression of structural genes (Figure 6F). These results demonstrate that ALA-induced anthocyanin biosynthesis is closely related to the normal transcriptional expression of MdMYB10 and MdMYB9.
MdMATE8 Involved in ALA-Regulated Anthocyanin Accumulation
In anthocyanin accumulation pathway, GSTs and MATEs are putatively associated with the vacuolar sequestration of anthocyanins. Recently, it was proved that ALA promoted anthocyanin accumulation through GST pathway (Fang et al., 2020), which was consistent with differentially expressed MdGSTF6 (MDP0000252292) in ALA-treated calli. Meanwhile, we also identified four MdMATE genes. To comprehensively analyze MATE transporter, the genome-wide analysis of the MATE genes in apple genome was conducted. At least 53 genes were initially isolated and named MdMATE1 to MdMATE53 according to their chromosomal information (Supplementary Figure 3A). A total of 53 MdMATE, 56 AtMATE, and 45 OsMATE were classified into four clades with ML (Maximum Likelihood) method (Supplementary Figure 3B). Some members of clade I were clustered with At3g59030 (AtTT12), and they may be involved in the transport of flavonoids (Debeaujon et al., 2001). Therefore, a phylogenetic analysis between 21 MdMATE members of clade I and 24 different known MATEs was conducted to further predict the functions of the MdMATE proteins (Figure 7A). One of the four MdMATE DEGs, named MdMATE8, was contained in seven MdMATE members, which were phylogenetically related to anthocyanin transporters such as MtMATE2 in barrel medic (Medicago truncatula) (Zhao et al., 2011), SlMTP77 in tomato (Solanum lycopersicum) (Mathews et al., 2003), and VvAM1 and VvAM3 in grapevine (Vitis vinifera) (Gomez et al., 2011), indicating that MdMATE8 might be involved in the anthocyanin accumulation in the apple calli. In addition, the other three DEGs (MdMATE1, MdMATE2, and MdMATE52) were clustered together with PA-transporting MATEs, such as FaTT12-1, VvMATE1, VvMATE2, AtTT12, and MtMATE1 (Figure 7A), implying that these three MdMATE might function in ALA-regulated PA accumulation. Thus, we chose MdMATE8 as the key candidate gene to analyze its molecular function in apple calli coloration and ALA-induced anthocyanin accumulation.
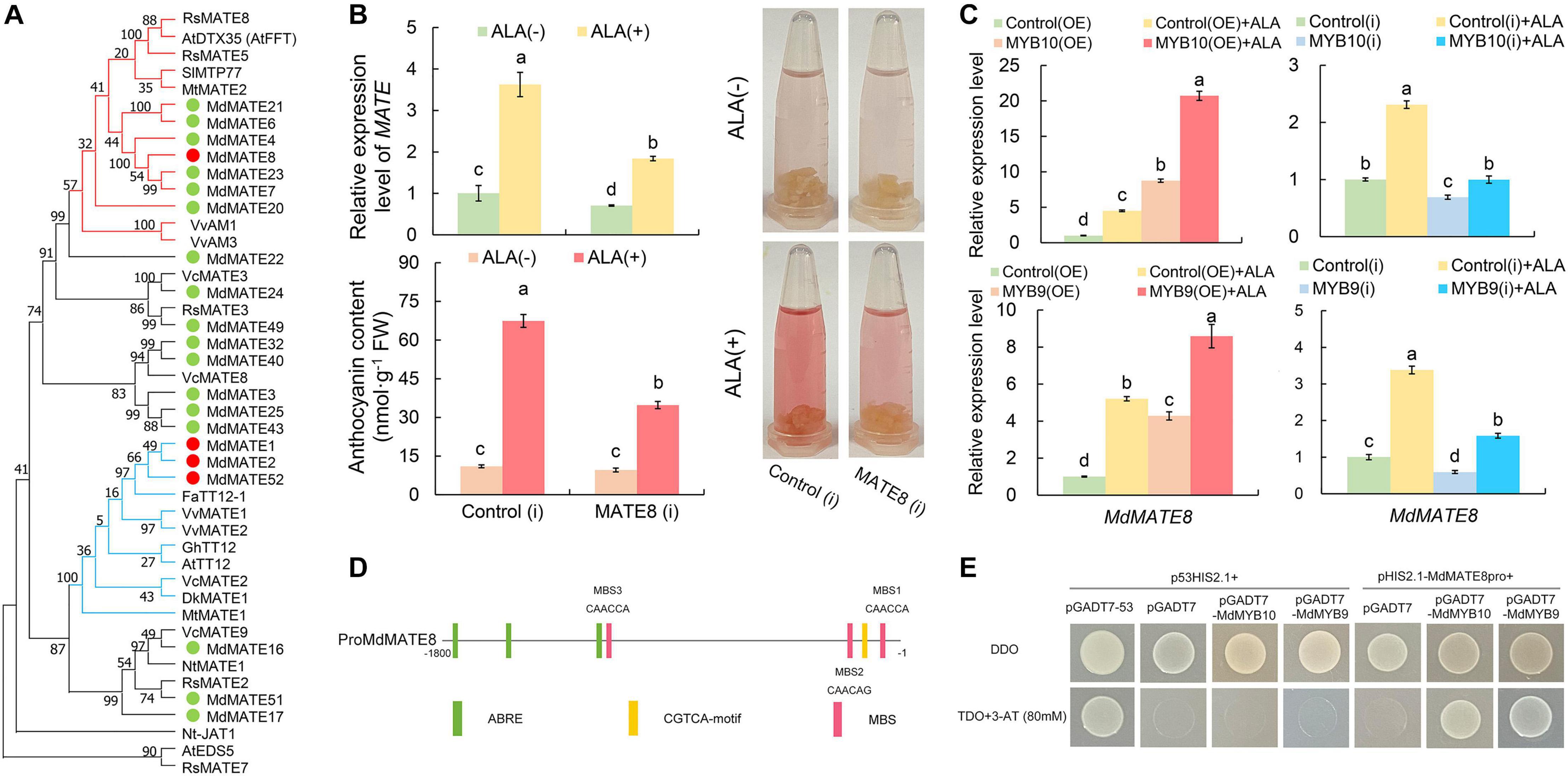
Figure 7. Functional characterization of MdMATE8. (A) Phylogenetic analysis of MdMATE proteins and selected MATE proteins from other species. (B) The expression levels, colors, and anthocyanin contents in the MdMATE8 transgenic calli with or without ALA treatment. (C) Transcript levels of MdMATE8 in MdMYB10 and MdMYB9 transgenic calli with or without ALA treatment. (D) The promoter region of MdMATE8. (E) Y1H assay showing the interaction between MdMYB10/MdMYB9 and the MdMATE8 promoter. The constructs p53HIS2.1 + pGADT7-53 were used as the positive control. The constructs p53HIS2.1 + pGADT7, p53HIS2.1 + pGADT7-MdMYB10, p53HIS2.1 + pGADT7-MdMYB9, or pHIS2.1-MdMATE8pro + pGADT7 were used as the negative controls, respectively. The different letters in each gene represent significant differences (p < 0.05).
Transient assays with apple calli were performed to test the function of MdMATE8. RNAi significantly decreased the expression of MdMATE8 to 58.5%, but the color levels and anthocyanin content did not change in comparison with control(i) calli (Figure 7B). However, the promotive effect of ALA on calli pigment was significantly impaired in the MdMATE8(i) transformed calli, and the anthocyanin content was 51.6% of the control(i) treated with ALA (Figure 7B). These results suggest that MdMATE8 may play important roles in ALA-induced anthocyanin accumulation.
Furthermore, the expression levels of MdMATE8 were detected in MdMYB10 and MdMYB9 transgenic calli lines (Figure 7C). Consistent with the anthocyanin content changes (Figures 7A,B), the expression level of MdMATE8 was significantly up-regulated in MdMYB10 or MdMYB9 overexpression calli, but down-regulated in their RNAi calli. The relative expression levels of MdMATE8 were more highly expressed in ALA-treated OE calli than in the control(OE) + ALA calli. However, the promotive capacity of ALA on expression of MdMATE8 was obviously inhibited in ALA-treated MdMYB10 or MdMYB9 RNAi calli, compared with the control(i) + ALA calli. These findings indicate that MdMYB10 and MdMYB9 were probably involved in transcriptional expression regulation of MdMATE8 in ALA-up-regulated anthocyanin accumulation.
To explore the region upstream of MdMATE8, the MdMATE8 promoter sequence was analyzed using the PlantCARE online tools. Some hormone response elements were detected in the promoter, including ABA (ABRE) and MeJA (CGTCA-motif) response elements. In addition, several putative MYB cis-elements were found in the promoter regions of MdMATE8, including CAACCA and CAACAG, as illustrated in Figure 7D. With the results above, we speculated that MdMYB10 or MdMYB9 might affect the transcriptional activity of the MdMATE8. To validate this possibility, we conducted the Y1H assay. The results showed that the transformants harboring the plasmid of pHIS2.1-MdMATE8 could not grow on SD/-Trp/-His media containing 80 mM of 3-AT (Supplementary Figure 4). Screening was carried out on TDO medium containing 80 mM 3-AT (Figure 7E). Like the positive control cells containing p53HIS2.1 + pGADT7-53, the yeast cells transformed with pHIS2.1-MdMATE8pro + pGADT7-MdMYB10, or pHIS2.1-MdMATE8pro + pGADT7-MdMYB9 were able to grow on TDO medium with 80 mM 3-AT, whereas the negative control yeast cells (p53HIS2.1 + pGADT7, p53HIS2.1 + pGADT7-MdMYB10, p53HIS2.1 + pGADT7-MdMYB9, and pHIS2.1-MdMATE8pro + pGADT7) were able to grow only on DDO medium (Figure 7E). These results showed that MdMYB10 and MdMYB9 can bind to the MdMATE8 promoter (Figure 7E). Therefore, all of these results suggest that MdMYB10 and MdMYB9 are involved in regulation of the expression of MdMATE8 in response to the ALA stimulation.
Discussion
ALA Effectively Induces Anthocyanin Accumulation in Calli Culture System
In red cultivars, the coloration of fruit largely determines fruit nutritional and commercial value. It has been well-documented that ALA plays an effective role in promoting fruit color formation (Wang et al., 2006; Xiao et al., 2012; Guo et al., 2013; Xie et al., 2013; Feng et al., 2016; Ye et al., 2017; Zheng et al., 2017). In orchards, exogenously sprayed high concentration ALA on fruit clusters of litchi at 45 days after full bloom significantly increased the anthocyanin content (Feng et al., 2015). Likewise, both exogenous 300 mg L–1 ALA and bagging remarkably promoted “Yunhongli 2” coloration at the early stage of fruit coloration (Xiao et al., 2012). Recently, direct root irrigation of ALA also improved fruit coloration and nutrition quality, and this approach was recommended when encountering fruit bag barriers (Zheng et al., 2018). Under lab conditions, researches showed that exogenous ALA elevated anthocyanin accumulation in fruit cubes placed into the growth chamber (Xie et al., 2013). The above researches suggest that ALA has a promising application prospect in fruit production. However, the mechanisms underlying ALA-induced anthocyanin accumulation remain largely unknown. Here, the calli induced from apple flesh also showed significantly higher anthocyanin content under ALA treatment than control after illuminating at 48 and 72 h (Figure 1), providing a more efficient way to study the regulation roles of ALA on anthocyanin accumulation.
ALA Rapidly Stimulates Transcriptome Changes
Secondary metabolites are catalytically synthesized by a series of enzymes in plant (Kim et al., 2004). Substantiality changes in the quantity and activity of these enzymes depend on regulation at the transcriptional or protein level (Xie et al., 2012; Yang et al., 2013; Jiang et al., 2014). Thus, regulation of transcription or protein levels is the fundamental reason of secondary metabolic rate changes. In litchi, the anthocyanin contents in pericarp remarkably increased after bags were removed 1–7 days; however, the greatest number of up-regulated DEGs was found between 0- and 1-day libraries (Zhang et al., 2016). Cyanidin-3-galactoside, the main cyanidin pigment in “Starkrimson” apple skin, rapidly accumulated at 4–8 days after bag was removed and peaked at 8 days, while the transcript levels of MdMYB1-1, MdbHLH3-2, and MdUFGT4 increased immediately when exposed to light at 0–2 days and then gradually decreased (Meng et al., 2015). Therefore, there appears a time course discrepancy between mRNA or protein expression level and secondary metabolite accumulation. Here, we found similar results. The DEGs were enriched at 24 h after ALA treatment (Figures 2A–C), but anthocyanin significantly accumulated at 48 h and 72 h (Figure 1), implying that ALA-induced changes of gene expression levels may be the main reason for anthocyanin accumulation in calli. Moreover, the KEGG analysis revealed that the flavonoid biosynthesis pathway was significantly enhanced in the ALA-treated apple calli compared with the untreated control (Figures 3D–F). Thus, the transcriptional data we obtained provides valuable information on elucidating the mechanism behind ALA-regulated color formation.
ALA Regulates Expressions of Structural Genes of Anthocyanin and Other Flavonoids Metabolism
Higher levels of anthocyanin are largely dependent on the higher transcript levels of a series of structural genes. Researches have reported that exogenous ALA up-regulated the expression levels of several key structural genes in the phenylpropanoid pathway (Guo et al., 2013; Xie et al., 2013; Feng et al., 2016; Ye et al., 2017; Zheng et al., 2018). Chalcone synthase (CHS), the key enzyme in the first committed step of the flavonoid biosynthetic pathway, catalyzes the stepwise condensation of 4-coumaroyl-CoA and malonyl-CoA into yellow chalcone (Martin, 1993). Exogenous ALA notably enhanced CHS gene expression in peach (Guo et al., 2013; Ye et al., 2017), apple (Xie et al., 2013; Feng et al., 2016; Zheng et al., 2017), and apple calli (Figure 4A). Leucoanthocyanidin dioxygenase (LDOX/ANS) and flavonoid 3-O-glucosyltransferase (UFGT) are the enzymes at the end of plant anthocyanin biosynthetic pathway, which catalyze colorless leucoanthocyanidins into colorful and stable anthocyanin (Kim et al., 2003). ALA strongly induced gene expression or enzyme activities of ANS and UFGT in peach (Guo et al., 2013), pear (Xiao et al., 2012), and apple (Xie et al., 2013). Similarly, here, both ANS and UFGT were increased by ALA at transcript levels (Figure 4A). Meanwhile, transcriptome analysis also showed that ALA significantly promoted expression levels of four CHI, one F3’H, and one DFR (Figure 4A). These results indicate that the biosynthetic genes of anthocyanin significantly respond to ALA induction.
For anthocyanin sequestration, several members of GST- and MATE-encoded enzymes are required for anthocyanin transport from cytoplasm to vacuole. Some members of GST have been identified to be responsible for anthocyanin transport in Arabidopsis (Kitamura et al., 2004), grapes (Gomez et al., 2011), litchi (Hu et al., 2016), peach (Zhao et al., 2020), and apple (El-sharkawy et al., 2015; Jiang et al., 2019). Some MATE family members also showed positive correlation with anthocyanin transport (Mathews et al., 2003; Marinova et al., 2007; Frank et al., 2011; Gomez et al., 2011; Zhao et al., 2011). In the present study, one GST and four MATE genes were up-regulated by ALA in calli (Figure 4A). Among them, MdGSTF6 has shown the positive role in anthocyanin accumulation in apple and ALA-treated apple calli (Jiang et al., 2019; Fang et al., 2020). MdMATE8, an ortholog of MtMATE2 (Zhao et al., 2011), SlMTP77 (Mathews et al., 2003), and VvAM1 and VvAM3 (Gomez et al., 2011), was predicted as the anthocyanin transporter (Figure 7A). Transient expression assays showed that the RNA interference of MdMATE8 notably inhibited anthocyanin accumulation in calli after ALA treatment (Figure 7B). These results suggest that MdGSTF6 and MdMATE8 may act as the key anthocyanin transporters for ALA-induced coloration.
In addition to anthocyanin, flavonol and proanthocyanidin are major subclasses of flavonoids. Anthocyanin and flavonol biosynthesis are produced from dihydroflavonol via two different branches, which are catalyzed by DFR or FLS, respectively. Overexpressing DFR or inactivation of FLS promoted anthocyanin accumulation (Davies et al., 2003; Stracke et al., 2009), while overexpression of FLS1 decreased seed coat pigmentation (Nguyen et al., 2016), suggesting existence of the substrate competition between FLS and DFR. Interestingly, root application of ALA not only promoted anthocyanin accumulation but also increased flavonol content (Zheng et al., 2017). ALA-improved flavonol accumulation has also been reported in guard cells of apple leaves and Arabidopsis cotyledons (An et al., 2016; Liu et al., 2016). Here, our results further showed that ALA simultaneously increased the transcription level of MdFLS and MdDFR (Figure 4A). Meanwhile, the expression of the key genes involved in PA biosynthesis (MdLARs) was up-regulated by ALA (Figure 4A). Additionally, three MATE DEGs were grouped with PA transporters (Figure 7A), and both MdMATE1 (MDP0000163588) and MdMATE2 (MDP0000157345) showed functional similarity to TT12 from A. thaliana and acted as vacuolar flavonoid/H+-antiporters active in PA accumulating cells (Debeaujon et al., 2001; Frank et al., 2011). These results demonstrate that ALA not only induces anthocyanin accumulation but also may participate in regulating other flavonoids’ metabolism. The knowledge of ALA affecting flavonol and proanthocyanidin accumulation pathways require further research.
MdMYB10 and MdMYB9 Are Involved in ALA-Induced Anthocyanin Accumulation in Apple Calli
Anthocyanin biosynthesis regulation particularly focuses on the MBW regulatory complex (Xu et al., 2015). R2R3-MYB TFs of SG6 predominantly play vital roles in modulation of anthocyanin metabolism in plant (Dubos et al., 2010). Among them, AtPAP1, AtPAP2, AtMYB113, and AtMYB114 have been demonstrated to control anthocyanin biosynthesis in Arabidopsis seedling vegetative tissues (Gonzalez et al., 2008). PhAN2 is predominantly correlated to anthocyanin biosynthesis in petunia flowers (Quattrocchio et al., 1999). Putative ortholog proteins of MYB10 of 20 different rosaceous fruits have been demonstrated to be involved in anthocyanin production (Lin-Wang et al., 2010). PyMYB10 has been characterized as being associated with anthocyanin biosynthesis in red-skinned pear cultivars (Feng et al., 2010). In strawberry, FaMYB10 activated the early and late steps of phenylpropanoid and flavonoid biosynthesis (Medina-Puche et al., 2014). In apple, it has been proposed that MdMYB1/A were responsible for the red coloration of the apple peel, while MdMYB10 mainly for the whole fruit as well as foliage (Peng and Moriguchi, 2013). Therefore, MdMYB10 is involved in anthocyanin biosynthesis, mainly in apple flesh. In the present study, we found that MdMYB10 expression was significantly increased and positively correlated with EBGs and LBGs of anthocyanin biosynthetic pathway in ALA-tread calli (Figures 5B,D). This result suggests that MdMYB10, a typical SG6 anthocyanin regulator, may be directly responsible for ALA-induced anthocyanin biosynthesis. Here, another typical R2R3-type MYB of SG5, MdMYB9, was screened out as a potential positive regulator mediated ALA-triggered color development (Figures 5A,B,D). In “Orin” apple transgenic calli, MdMYB9 has been functionally characterized as a positive regulator of MeJA-induced anthocyanin and PA production (An et al., 2015). Recent researches showed that MdMYB9 was involved in several TFs that regulated anthocyanin and PA biosynthesis in apple, such as MdNAC52 (Sun et al., 2019), MdBBX37 (An et al., 2020), and MdERF1B (Zhang et al., 2018). Therefore, MdMYB9 may also play a role in ALA-regulated anthocyanin accumulation.
On the basis of transient assays in apple calli, we confirmed that MdMYB10 and MdMYB9 are responsible for controlling anthocyanin biosynthesis in apple fruit (Figures 6A,B) (Espley et al., 2007; An et al., 2015). Previous studies observed that MdMYB10 and MdMYB9 could directly bind to the promoter region of MdDFR and MdANS, respectively (Espley et al., 2007; An et al., 2015). In the present study, overexpression of MdMYB10 and MdMYB9 significantly activated several key genes of the anthocyanin biosynthesis, and the effect was further enhanced in ALA-treated calli (Figures 6A–C), which may associate with ALA up-regulated MdMYB10 and MdMYB9 expression level in their OE transgenic lines (Supplementary Figures 2A,B). The results imply that several upstream TFs of MdMYB10 and MdMYB9 could respond to ALA-activated coloration to induce endogenous MdMYB10 and MdMYB9 expression in their OE calli. Thus, it seems that overexpression of MdMYB10 and MdMYB9 achieved by the 35S promoter could not have feedback to inhibit ALA-induced endogenous MdMYB10/9 gene expression. In contrast, the promotive roles of ALA on expression of MdCHS, MdANS, and MdUFGT were obviously suppressed in MdMYB10 or MdMYB9 silenced calli (Figure 6F). Therefore, MdMYB10 and MdMYB9 are essential for ALA-up-regulated expression of EBGs and LBGs of anthocyanin biosynthesis during calli coloration. Interestingly, ALA still up-regulated these structural gene expression levels in calli of two TFs RNAi lines (Figure 6F). These results suggest that ALA-induced structural gene expression partially depends on MdMYB10 and MdMYB9. In fact, MYB9 group is a rare type of anthocyanin-related MYB activator, which is lost in most dicot species (Zhou et al., 2016), whereas ALA is able to activate anthocyanin biosynthesis in various fruit species (Watanabe et al., 2006; Xiao et al., 2012; Guo et al., 2013; Feng et al., 2015; Li et al., 2016; Ye et al., 2017), suggesting that ALA-induced accumulation of anthocyanin is coordinately regulated by a set of R2R3-MYB genes. Moreover, among the DEGs of TFs, we also identified bHLH, AP2-EREBP, WRKY, bZIP, and NAC family members (Supplementary Table 9). These DEGs of TFs possibly function in ALA-activated coloration and will be clarified in further researches.
Additionally, our DEG data found that MdbHLH3 (MDP0000225680) was dramatically induced in 24 h under ALA treatment (Figure 5C and Supplementary Table 6). In apple calli, we also found that MdbHLH3 expression was up-regulated after ALA treatment (Zheng et al., 2018). An et al. (2015) found that MdbHLH3 could be recruited to the promoter regions of MdMYB9 to activate expression. Thus, it is reasonable to speculate that the MdMYB9 expression was initiated by MdbHLH3 in response to ALA. Importantly, MdbHLH3 was required for the regulatory activity of MdMYB10 and MdMYB9 (Xie et al., 2012; An et al., 2015). In MdMYB10 and MdMYB9 OE and RNAi transgenic calli, the anthocyanin content and selected structural gene expression level of ALA-treated calli were higher than that without ALA-treated calli (Figures 6A–F). We deem that MdbHLH3 up-regulated by ALA can active MdMYB9 expression or form more complexes of transcriptional regulation with MdMYB10 or MdMYB9 to bind to the promoters of structural genes and ultimately effectively promote anthocyanin accumulation in transgenic calli lines. The function of MdbHLH3 in ALA-induced coloration needs to be further conducted.
Compared with the intensive studies of the regulatory and structural genes involved in anthocyanin biosynthesis, the molecular mechanism associated with anthocyanin transport from cytoplasm to vacuole remains uncertain, especially in ALA-mediated anthocyanin accumulation. Recently, researches showed that anthocyanin transport is regulated by MBW complexes. In peach, transient overexpression of PpGST1 together with PpMYB10.1 led to much deeper coloration as compared with PpMYB10.1 alone (Zhao et al., 2020). In apple, the expression of MdGSTF6 was activated by MdMYB1 (Jiang et al., 2019). AtTT2 was also able to induce the AtTT12 promoter activity (Xu et al., 2014). Here, our results revealed that MdMYB10 and MdMYB9 showed high expression relation with MdMATE8 in ALA-stimulated color developing calli (Figure 5D). Overexpression of MdMYB10 and MdMYB9 up-regulated the expression of MdMATE8, but the expression was decreased in MdMYB10 or MdMYB9 RNAi calli (Figure 7C). Moreover, through Y1H screening, we found that MdMYB10 and MdMYB9 can bind to the MdMATE8 promoter (Figure 7E), indicating that MdMYB10 and MdMYB9 may activate MdMATE8 transcription by binding to its promoter. These findings provide a new understanding that ALA strongly up-regulates the expression of MdMYB10 and MdMYB9 to regulate anthocyanin biosynthesis as well as transport, thereby promoting anthocyanin accumulation.
Conclusion
In the current study, exogenous ALA-activated anthocyanin accumulation was analyzed on the transcriptome level in apple calli. The GO enrichment and KEGG analysis as well as anthocyanin metabolism pathway of DEGs demonstrated that ALA-induced expression changes of anthocyanin biosynthesis and transport structural genes may be the key reason for anthocyanin accumulation. The MYB family members were prominently involved in the transcriptional regulation of ALA-induced coloration. Two R2R3-MYB members, MdMYB10 and MdMYB9, are likely involved in the regulation of ALA-mediated anthocyanin biosynthesis via calli transient expression. Additionally, MATE gene family members’ identification, phylogenetic trees, expression pattern, expression level analysis, cis-acting elements’ prediction, and Y1H screening revealed that MdMYB10 and MdMYB9 may activate MdMATE8 transcription to regulate anthocyanin transport under ALA treatment. Overall, the study disclosed the key regulators and the putative mechanism behind the positive transcriptional regulation of ALA on anthocyanin accumulation in apple.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA525304.
Author Contributions
JZ, YA, and LW conceived and designed the experiments. JZ, LL, HT, and YA performed the experiments. JZ, LL, HT, and YA analyzed the data. JZ, YA, and LW drafted and modified the manuscript. All authors read and approved the manuscript.
Funding
This research was financially supported by the National Natural Science Foundation of China (31772253), the Anhui Provincial Natural Science Foundation, China (1908085QC108), and the University Natural Science Research Project of Anhui Province (KJ2020A0043).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.640606/full#supplementary-material
Footnotes
- ^ http://www.Bioconductor.org
- ^ http://www.megasoftware.net
- ^ http://bioinformatics.psb.ugent.be/webtools/plantcare/html/
References
Aharoni, A., De Vos, C. H., Wein, M., Sun, Z., Greco, R., Kroon, A., et al. (2001). The strawberry FaMYB1 transcription factor suppresses anthocyanin and flavonol accumulation in transgenic tobacco. Plant J. 28, 319–332. doi: 10.1046/j.1365-313X.2001.01154.x
Akram, N. A., and Ashraf, M. (2013). Regulation in plant stress tolerance by a potential plant growth regulator, 5-aminolevulinic acid. J. Plant Growth Regul. 32, 663–679. doi: 10.1007/s00344-013-9325-9
An, J. P., Wang, X. F., Espley, R. V., Lin-Wang, K., Bi, S. Q., You, C. X., et al. (2020). An Apple B-Box protein MdBBX37 modulates anthocyanin biosynthesis and hypocotyl elongation synergistically with MdMYBs and MdHY5. Plant Cell Physiol. 61, 130–143. doi: 10.1093/pcp/pcz185
An, X. H., Tian, Y., Chen, K. Q., Liu, X. J., Liu, D. D., Xie, X. B., et al. (2015). MdMYB9 and MdMYB11 are involved in the regulation of the JA-induced biosynthesis of anthocyanin and proanthocyanidin in apples. Plant Cell Physiol. 56, 650–662. doi: 10.1093/pcp/pcu205
An, Y. Y., Feng, X. X., Liu, L. B., Xiong, L. J., and Wang, L. J. (2016). ALA-induced flavonols accumulation in guard cells is involved in scavenging H2O2 and inhibiting stomatal closure in Arabidopsis cotyledons. Front. Plant Sci. 7:1713. doi: 10.3389/fpls.2016.01713
Anders, S., and Huber, W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11:R106. doi: 10.1186/gb-2010-11-10-r106
Anders, S., Pyl, P. T., and Huber, W. (2014). HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. doi: 10.1093/bioinformatics/btu638
Bae, R. N., Kim, K. W., Kim, T. C., and Lee, S. K. (2006). Anatomical observations of anthocyanin rich cells in apple skins. Hortscience 41, 733–736. doi: 10.21273/HORTSCI.41.3.733
Ban, Y., Honda, C., Hatsuyama, Y., Igarashi, M., Bessho, H., and Moriguchi, T. (2007). Isolation and functional analysis of a MYB transcription factor gene that is a key regulator for the development of red coloration in apple skin. Plant Cell Physiol. 48, 958–970. doi: 10.1093/pcp/pcm066
Britsch, L., and Grisebach, H. (1986). Purification and characterization of (2S)-flavanone 3-hydroxylase from Petunia hybrida. Eur. J. Biochem. 156, 569–577. doi: 10.1111/j.1432-1033.1986.tb09616.x
Cao, Z. H., Zhang, S. Z., Wang, R. K., Zhang, R. F., and Hao, Y. J. (2013). Genome wide analysis of the apple MYB transcription factor family allows the identification of MdoMYB121 gene confering abiotic stress tolerance in plants. PLoS One 8:e69955. doi: 10.1371/journal.pone.0069955
Czemmel, S., Stracke, R., Weisshaar, B., Cordon, N., Harris, N. N., Walker, A. R., et al. (2009). The grapevine R2R3-MYB transcription factor VvMYBF1 regulates flavonol synthesis in developing grape berries. Plant Physiol. 151, 1513–1530. doi: 10.1104/pp.109.142059
Davies, K. M., Schwinn, K. E., Deroles, S. C., Manson, D. G., Lewis, D. H., Bloor, S. J., et al. (2003). Enhancing anthocyanin production by altering competition for substrate between flavonol synthase and dihydroflavonol 4-reductase. Euphytica 131, 259–268. doi: 10.1023/a:1024018729349
Debeaujon, I., Peeters, A. J. M., Leon-Kloosterziel, K. M., and Koornneef, M. (2001). The TRANSPARENT TESTA12 gene of Arabidopsis encodes a multidrug secondary transporter-like protein required for flavonoid sequestration in vacuoles of the seed coat endothelium. Plant Cell 13, 853–871. doi: 10.2307/3871345
Dubos, C., Stracke, R., Grotewold, E., Weisshaar, B., Martin, C., and Lepiniec, L. (2010). MYB transcription factors in Arabidopsis. Trends Plant Sci. 15, 573–581. doi: 10.1016/j.tplants.2010.06.005
El-sharkawy, I., Dong, L., and Xu, K. N. (2015). Transcriptome analysis of an apple (Malus × domestica) yellow fruit somatic mutation identifies a gene network module highly associated with anthocyanin and epigenetic regulation. J. Exp. Bot. 66, 7359–7376. doi: 10.1093/jxb/erv433
Espley, R. V., Hellens, R. P., Putterill, J., Stevenson, D. E., Kutty-Amma, S., and Allan, A. C. (2007). Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J. 49, 414–427. doi: 10.1111/j.1365-313x.2006.02964.x
Fang, X., An, Y. Y., Zheng, J., Shangguan, L. F., and Wang, L. J. (2020). Genome-wide identification and comparative analysis of GST gene family in apple (Malus domestica) and their expressions under ALA treatment. 3 Biotech. 10:307. doi: 10.1007/s13205-020-02299-x
Feng, S., Li, M. F., Wu, F., Li, W. L., and Li, S. P. (2015). 5-Aminolevulinic acid affects fruit coloration, growth, and nutrition quality of Litchi chinensis Sonn. cv. Feizixiao in Hainan, tropical China. Sci. Hortic. 193, 188–194. doi: 10.1016/j.scienta.2015.07.010
Feng, S. Q., Wang, Y. L., Yang, S., Xu, Y. T., and Chen, X. S. (2010). Anthocyanin biosynthesis in pears is regulated by a R2R3-MYB transcription factor PyMYB10. Planta 232, 245–255. doi: 10.2307/23391635
Feng, X. X., An, Y. Y., Zheng, J., Sun, M., and Wang, L. J. (2016). Proteomics and SSH analyses of ALA-promoted fruit coloration and evidence for the involvement of a MADS-box gene, MdMADS1. Front. Plant Sci. 7:1615. doi: 10.3389/fpls.2016.01615
Fischer, T. C., Gosch, C., Pfeiffer, J., Halbwirth, H., Halle, C., Stich, K., et al. (2007). Flavonoid genes of pear (Pyrus communis). Trees 21, 521–529. doi: 10.1007/s00468-007-0145-z
Fornalé, S., Lopez, E., Salazar-Henao, J. E., Fernández-Nohales, P., Rigau, J., and Caparros-Ruiz, D. (2014). AtMYB7, a new player in the regulation of UV-sunscreens in Arabidopsis thaliana. Plant Cell Physiol. 55, 507–516. doi: 10.1093/pcp/pct187
Frank, S., Keck, M., Sagasser, M., Niehaus, K., Weisshaar, B., and Stracke, R. (2011). Two differentially expressed MATE factor genes from apple complement the Arabidopsis transparent testa 12 mutant. Plant Biol. 13, 42–50. doi: 10.1111/j.1438-8677.2010.00350.x
Gomez, C., Conejero, G., Torregrosa, L., Cheynier, V., Terrier, N., and Ageorges, A. (2011). In vivo grapevine anthocyanin transport involves vesicle-mediated trafficking and the contribution of anthoMATE transporters and GST. Plant J. 67, 960–970. doi: 10.1111/j.1365-313X.2011.04648.x
Gomez, C., Terrier, N., Torregrosa, L., Vialet, S., Fournierlevel, A., Verriès, C., et al. (2009). Grapevine MATE-type proteins act as vacuolar H+-dependent acylated anthocyanin transporters. Plant Physiol. 150, 402–415. doi: 10.2307/40537864
Gonzalez, A., Zhao, M., Leavitt, J. M., and Lloyd, A. M. (2008). Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 53, 814–827. doi: 10.1111/j.1365-313X.2007.03373.x
Guo, L., Cai, Z. X., Zhang, B. B., Xu, J. L., Song, H. F., and Ma, R. J. (2013). The mechanism analysis of anthocyanin accumulation in peach accelerated by ALA. Acta Hortic. Sin. 40, 1043–1050. doi: 10.16420/j.issn.0513-353x.2013.06.004
Hu, B., Zhao, J., Lai, B., Qin, Y., Wang, H., and Hu, G. (2016). LcGST4 is an anthocyanin-related glutathione S-transferase gene in Litchi chinensis sonn. Plant Cell Rep. 35, 831–843. doi: 10.1007/s00299-015-1924-4
Ji, X. H., Wang, Y. T., Zhang, R., Wu, S. J., An, M. M., Li, M., et al. (2015). Effect of auxin, cytokinin and nitrogen on anthocyanin biosynthesis in callus cultures of red-fleshed apple (Malus sieversii f. niedzwetzkyana). Plant Cell Tiss. Organ. Cult. 120, 325–337. doi: 10.1007/s11240-014-0609-y
Jiang, R., Tian, J., Song, T. T., Zhang, J., and Yao, Y. C. (2014). The Malus crabapple transcription factor McMYB10 regulates anthocyanin biosynthesis during petal coloration. Sci. Hortic. 166, 42–49. doi: 10.1016/j.scienta.2013.12.002
Jiang, S., Chen, M., He, N., Chen, X., Wang, N., Sun, Q., et al. (2019). MdGSTF6, activated by MdMYB1, plays an essential role in anthocyanin accumulation in apple. Hortic. Res. 6:40. doi: 10.1038/s41438-019-0118-6
Kim, S., Binzel, M. L., Park, S., Yoo, K. S., and Pike, L. M. (2004). Inactivation of DFR (Dihydroflavonol 4-reductase) gene transcription results in blockage of anthocyanin production in yellow onions (Allium cepa). Mol. Breed. 14, 253–263. doi: 10.1023/B:MOLB.0000047770.92977.04
Kim, S. H., Lee, J. R., Hong, S. T., Yoo, Y. K., An, G., and Kim, S. R. (2003). Molecular cloning and analysis of anthocyanin biosynthesis genes preferentially expressed in apple skin. Plant Sci. 165, 403–413. doi: 10.1016/s0168-9452(03)00201-2
Kitamura, S., Shikazono, N., and Tanaka, A. (2004). TRANSPARENT TESTA 19 is involved in the accumulation of both anthocyanins and proanthocyanidins in Arabidopsis. Plant J. 37, 104–114. doi: 10.1046/j.1365-313X.2003.01943.x
Li, Y., Zhang, Z. Q., and Wang, L. J. (2016). Applications of 5-aminolevulinic acid on the physiological and biochemical characteristics of strawberry fruit during postharvest cold storage. Ciência Rural. 46, 2103–2109. doi: 10.1590/0103-8478cr20151426
Lin-Wang, K., Bolitho, K., Grafton, K., Kortstee, A., Karunairetnam, S., McGhie, T. K., et al. (2010). An R2R3 MYB transcription factor associated with regulation of the anthocyanin biosynthetic pathway in Rosaceae. BMC Plant Biol. 10:50. doi: 10.1186/1471-2229-10-50
Liu, L. B., Xiong, L. J., An, Y. Y., Zheng, J., and Wang, L. J. (2016). Flavonols induced by 5-aminolevulinic acid are involved in regulation of stomatal opening in apple leaves. Hortic. Plant J. 2, 323–330. doi: 10.1016/j.hpj.2017.02.002
Livak, K. J., and Schmitten, T. D. (2010). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Mao, X., Cai, T., Olyarchuk, J. G., and Wei, L. (2005). Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 21, 3787–3793. doi: 10.2307/1592215
Marinova, K., Pourcel, L., Weder, B., Schwarz, M., Barron, D., Routaboul, J. M., et al. (2007). The Arabidopsis MATE transporter TT12 acts as a vacuolar flavonoid/H+-antiporter active in proanthocyanidin-accumulating cells of the seed coat. Plant Cell 19, 2023–2038. doi: 10.1105/tpc.106.046029
Marrs, K. A., Alfenito, M. R., Lloyd, A. M., and Walbot, V. (1995). A glutathione S-transferase involved in vacuolar transfer encoded by the maize gene Bronze-2. Nature 375, 397–400. doi: 10.1038/375397a0
Martin, C., and Paz-Ares, J. (1997). MYB transcription factors in plants. Trends Genet. 13, 67–73. doi: 10.1016/S0168-9525(96)10049-4
Martin, C., Prescott, A., Mackay, S., Bartlett, J., and Vrijlandt, E. (1991). Control of anthocyanin biosynthesis in flowers of Antirrhinum majus. Plant J. 1, 37–49. doi: 10.1111/j.1365-313X.1991.00037.x
Martin, C. R. (1993). Structure, function, and regulation of the chalcone synthase. Int. Rev. Cytol. 147, 233–284. doi: 10.1016/S0074-7696(08)60770-6
Mathews, H., Clendennen, S. K., Caldwell, C. G., Liu, X. L., Connors, K., Matheis, N., et al. (2003). Activation tagging in tomato identifies a transcriptional regulator of anthocyanin biosynthesis, modification, and transport. Plant Cell 15, 1689–1703. doi: 10.1105/tpc.012963
Medina-Puche, L., Cumplido-Laso, G., Amil-Ruiz, F., Hoffmann, T., Ring, L., Rodríguez-Franco, A., et al. (2014). MYB10 plays a major role in the regulation of flavonoid/phenylpropanoid metabolism during ripening of Fragaria x ananassa fruits. J. Exp. Bot. 65, 401–417. doi: 10.1093/jxb/ert377
Meng, R., Qu, D., Liu, Y. L., Gao, Z. Y., Yang, H. J., Shi, X. Y., et al. (2015). Anthocyanin accumulation and related gene family expression in the skin of dark-grown red and non-red apples (Malus domestica, Borkh.) in response to sunlight. Sci. Hortic. 189, 66–73. doi: 10.1016/j.scienta.2015.03.046
Mueller, L. A., Goodman, C. D., Silady, R. A., and Walbot, V. (2000). AN9, a petunia glutathione S-transferase required for anthocyanin sequestration, is a flavonoid-binding protein. Plant Physiol. 123, 1561–1570. doi: 10.1002/(SICI)1097-007X(199903/04)27:23.0.CO;2-I
Nesi, N., Jond, C., Debeaujon, I., Caboche, M., and Lepiniec, L. (2001). The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed. Plant Cell 13, 2099–2114. doi: 10.1105/tpc.13.9.2099
Nguyen, N. H., Kim, J. H., Kwon, J., Jeong, C. Y., Lee, W., Lee, D., et al. (2016). Characterization of Arabidopsis thaliana FLAVONOL SYNTHASE 1, (FLS1) -overexpression plants in response to abiotic stress. Plant Physiol. Biochem. 103, 133–142. doi: 10.1016/j.plaphy.2016.03.010
Niu, S. S., Xu, C. J., Zhang, W. S., Zhang, B., Li, X., Lin-Wang, K., et al. (2010). Coordinated regulation of anthocyanin biosynthesis in Chinese bayberry (Myrica rubra) fruit by a R2R3 MYB transcription factor. Planta 231, 887–899. doi: 10.1007/s00425-009-1095-z
Peng, T., and Moriguchi, T. (2013). The molecular network regulating the coloration in apple. Sci. Hortic. 163, 1–9. doi: 10.1016/j.scienta.2013.06.005
Pillet, J., Yu, H. W., Chambers, A. H., Whitaker, V. M., and Folta, K. M. (2015). Identification of candidate flavonoid pathway genes using transcriptome correlation network analysis in ripe strawberry (Fragaria × ananassa) fruits. J. Exp. Bot. 66, 4455–4467. doi: 10.1093/jxb/erv205
Premathilake, A. T., Ni, J., Shen, J., Bai, S., and Teng, Y. (2020). Transcriptome analysis provides new insights into the transcriptional regulation of methyl jasmonate-induced flavonoid biosynthesis in pear calli. BMC Plant Biol. 20:388. doi: 10.1186/s12870-020-02606-x
Quattrocchio, F., Wing, J., van der Woude, K., Souer, E., de Vetten, N., Mol, J., et al. (1999). Molecular analysis of the anthocyanin2 gene of Petunia and its role in the evolution of flower color. Plant Cell 11, 1433–1444. doi: 10.2134/agronj1997.00021962008900030003x
Rinaldo, A. R., Cavallini, E., Jia, Y., Moss, S. M., McDavid, D. A., Hooper, L. C., et al. (2015). A grapevine anthocyanin acyltransferase, transcriptionally regulated by VvMYBA, can produce most acylated anthocyanins present in grape skins. Plant Physiol. 169, 1897–1916.
Schaart, J. G., Dubos, C., Fuente, I. R. D. L., van Houwelingen, A. M. M. L., Vos, R. C. H. D., Jonker, H. H., et al. (2013). Identification and characterization of MYB-bHLHWD40 regulatory complexes controlling proanthocyanidin biosynthesis in strawberry (Fragaria × ananassa) fruits. Int. Rice Res. Notes 197, 454–467. doi: 10.1111/nph.12017
Shirley, B. W., Kubasek, W. L., Storz, G., Bruggemann, E., Koornneef, M., Ausubel, F. M., et al. (1995). Analysis of Arabidopsis mutants deficient in flavonoid biosynthesis. Plant J. 8, 659–671. doi: 10.1046/j.1365-313X.1995.08050659.x
Sparvoli, F., Martin, C., Scienza, A., Gavazzi, G., and Tonelli, C. (1994). Cloning and molecular analysis of structural genes involved in flavonoid and stilbene biosynthesis in grape (Vitis vinifera, L.). Plant Mol. Biol. 24, 743–755. doi: 10.1007/bf00029856
Stracke, R., De Vos, R. C. H., Bartelniewoehner, L., Ishihara, H., Sagasser, M., Martens, S., et al. (2009). Metabolomic and genetic analyses of flavonol synthesis in Arabidopsis thaliana support the in vivo involvement of leucoanthocyanidin dioxygenase. Planta 229, 427–445. doi: 10.2307/23390160
Stracke, R., Ishihara, H., Huep, G., Barsch, A., Mehrtens, F., Niehaus, K., et al. (2007). Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J. 50, 660–677. doi: 10.1111/j.1365-313X.2007.03078.x
Sun, J. J., Wang, Y. C., Chen, X. S., Gong, X. J., Wang, N., Ma, L., et al. (2017). Effects of methyl jasmonate and abscisic acid on anthocyanin biosynthesis in callus cultures of red-fleshed apple (Malus sieversii, f. niedzwetzkyana). Plant Cell Tiss. Organ. Cult. 130, 227–237. doi: 10.1007/s11240-017-1217-4
Sun, Q., Jiang, S., Zhang, T., Xu, H., Fang, H., Zhang, J., et al. (2019). Apple NAC transcription factor MdNAC52 regulates biosynthesis of anthocyanin and proanthocyanidin through MdMYB9 and MdMYB11. Plant Sci. 289:110286. doi: 10.1016/j.plantsci.2019.110286
Takos, A. M., Jaffé, W., Jacob, S. R., Bogs, J., Robinson, S. P., and Walker, A. R. (2006). Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol. 142, 1216–1232. doi: 10.2307/20206009
Terrier, N., Torregrosa, L., Ageorges, A., Vialet, S., Verries, C., Cheynier, V., et al. (2009). Ectopic expression of VvMybPA2 promotes proanthocyanidin biosynthesis in grapevine and suggests additional targets in the pathway. Plant Physiol. 149, 1028–1041. doi: 10.1104/pp.108.131862
Trapnell, C., Pachter, L., and Salzberg, S. L. (2009). TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111. doi: 10.1093/bioinformatics/btp120
Trapnell, C., Williams, B. A., Pertea, G., Mortazavi, A., Kwan, G., van Baren, M. J., et al. (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515. doi: 10.1038/nbt.1621
Ubi, B. E., Honda, C., Bessho, H., Kondo, S., Wada, M., Kobayashi, S., et al. (2006). Expression analysis of anthocyanin biosynthetic genes in apple skin: effect of UV-B and temperature. Plant Sci. 170, 571–578. doi: 10.1016/j.plantsci.2005.10.009
Velasco, R., Zharkikh, A., Affourtit, J., Dhingra, A., Cestaro, A., Kalyanaraman, A., et al. (2010). The genome of the domesticated apple (Malus × domestica Borkh). Nat. Genet. 42, 833–839. doi: 10.1038/ng.654
Vimolmangkang, S., Han, Y., Wei, G., and Korban, S. S. (2013). An apple MYB transcription factor, MdMYB3, is involved in regulation of anthocyanin biosynthesis and flower development. BMC Plant Biol. 13:176. doi: 10.1186/1471-2229-13-176
Wang, N., Xu, H., Jiang, S., Zhang, Z., Lu, N., Qiu, H., et al. (2017). MYB12 and MYB22 play essential roles in proanthocyanidin and flavonol synthesis in red-fleshed apple (Malus sieversii f. niedzwetzkyana). Plant J. 90, 276–292. doi: 10.1111/tpj.13487
Wang, Z. H., Tang, G. H., Li, Z. Q., and Wang, L. J. (2006). Promotion of 5-aminolevulinic acid and genistein on anthocyanin accumulation in apples. Acta Hortic. Sin. 33, 1055–1058. doi: 10.1360/yc-006-1280
Watanabe, K., Nishihara, E., Watanabe, S., Tanaka, T., Takahashi, K., and Takeuchi, Y. (2006). Enhancement of growth and fruit maturity in 2-year-old grapevines cv. Delaware by 5-aminolevulinic acid. Plant Growth Regul. 49, 35–42. doi: 10.1007/s10725-006-0024-4
Wienand, U., Weydemann, U., Niesbach-Klosgen, U., Peterson, P. A., and Saedler, H. (1986). Molecular cloning of the c2 locus of Zea mays, the gene coding for chalcone synthase. Mol. Gen. Genet. 203, 202–207. doi: 10.1007/bf00333955
Xiao, C. C., Zhang, S. L., Hu, H. J., Tian, R., Wu, J., Yang, Z. J., et al. (2012). Effects of bagging and exogenous 5-aminolevulinic acid treatment on coloration of ‘Yunhongli 2’. J. Nanjing Agric. Univ. 35, 25–29. doi: 10.7685/j.issn.1000-2030.2012.06.005
Xie, L., Wang, Z. H., Cheng, X. H., Gao, J. J., Zhang, Z. P., and Wang, L. J. (2013). 5-Aminolevulinic acid promotes anthocyanin accumulation in Fuji apples. Plant Growth Regul. 69, 295–303. doi: 10.1007/s10725-012-9772-5
Xie, X. B., Li, S., Zhang, R. F., Zhao, J., Chen, Y. C., Zhao, Q., et al. (2012). The bHLH transcription factor MdbHLH3 promotes anthocyanin accumulation and fruit colouration in response to low temperature in apples. Plant Cell Environ. 35, 1884–1897. doi: 10.1111/j.1365-3040.2012.02523.x
Xu, W., Grain, D., Bobet, S., Le Gourrierec, J., Thévenin, J., Kelemen, Z., et al. (2014). Complexity and robustness of the flavonoid transcriptional regulatory network revealed by comprehensive analyses of MYB-bHLH-WDR complexes and their targets in Arabidopsis seed. New Phytol. 202, 132–144. doi: 10.1111/nph.12620
Xu, W. J., Dubos, C., and Lepiniec, L. (2015). Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci. 20, 176–185. doi: 10.1016/j.tplants.2014.12.001
Yang, F., Cai, J., Yang, Y., and Liu, Z. (2013). Overexpression of microRNA828 reduces anthocyanin accumulation in Arabidopsis. Plant Cell Tiss. Organ. Cult. 115, 159–167. doi: 10.1007/s11240-013-0349-4
Ye, J. B., Yang, X. H., Chen, Q. W., Xu, F., and Wang, G. Y. (2017). Promotive effects of 5-aminolevulinic acid on fruit quality and coloration of Prunus persica (L.) Batsch. Sci. Hortic. 217, 266–275. doi: 10.1016/j.scienta.2017.02.009
Young, M. D., Wakefield, M. J., Smyth, G. K., and Oshlack, A. (2010). Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 11:R14. doi: 10.1186/gb-2010-11-2-r14
Zhang, H. N., Li, W. C., Wang, H. C., Shi, S. Y., Shu, B., Liu, L. Q., et al. (2016). Transcriptome profiling of light-regulated anthocyanin biosynthesis in the pericarp of Litchi. Front. Plant Sci. 7:963. doi: 10.3389/fpls.2016.00963
Zhang, J., Xu, H., Wang, N., Jiang, S. H., Fang, H. C., Zhang, Z. Y., et al. (2018). The ethylene response factor MdERF1B regulates anthocyanin and proanthocyanidin biosynthesis in apple. Plant Mol. Biol. 98, 205–218. doi: 10.1007/s11103-018-0770-5
Zhao, J., Huhman, D., Shadle, G., He, X. Z., Sumner, L. W., Tang, Y., et al. (2011). MATE2 mediates vacuolar sequestration of flavonoid glycosides and glycoside malonates in Medicago truncatula. Plant Cell 23, 1536–1555. doi: 10.1105/tpc.110.080804
Zhao, Y., Dong, W., Zhu, Y., Allan, A. C., Lin-Wang, K., and Xu, C. (2020). PpGST1, an anthocyanin-related glutathione S-transferase gene, is essential for fruit coloration in peach. Plant Biotechnol. J. 18, 1284–1295. doi: 10.1111/pbi.13291
Zheng, J., An, Y. Y., Feng, X. X., and Wang, L. J. (2017). Rhizospheric application with 5-aminolevulinic acid improves coloration and quality in ‘Fuji’ apples. Sci. Hortic. 224, 74–83. doi: 10.1016/j.scienta.2017.06.004
Zheng, J., An, Y. Y., and Wang, L. J. (2018). 24-Epibrassinolide enhances 5-ALA-induced anthocyanin and flavonol accumulation in calli of ‘Fuji’ apple flesh. Plant Cell Tiss. Organ. Cult. 134, 319–330. doi: 10.1007/s11240-018-1418-5
Zhou, H., Lin-Wang, K., Wang, H., Gu, C., Dare, A. P., Espley, R. V., et al. (2015). Molecular genetics of blood-fleshed peach reveals activation of anthocyanin biosynthesis by NAC transcription factors. Plant J. 82, 105–121. doi: 10.1111/tpj.12792
Keywords: ALA, anthocyanin accumulation, apple fruit calli, differentially expressed genes, MdMYB10, MdMYB9, MdMATE8
Citation: Zheng J, Liu L, Tao H, An Y and Wang L (2021) Transcriptomic Profiling of Apple Calli With a Focus on the Key Genes for ALA-Induced Anthocyanin Accumulation. Front. Plant Sci. 12:640606. doi: 10.3389/fpls.2021.640606
Received: 11 December 2020; Accepted: 25 February 2021;
Published: 26 March 2021.
Edited by:
Meng Xie, Brookhaven National Laboratory, United StatesReviewed by:
Yunjun Zhao, Brookhaven National Laboratory (DOE), United StatesAshutosh Pandey, National Institute of Plant Genome Research (NIPGR), India
Copyright © 2021 Zheng, Liu, Tao, An and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuyan An, YW55dXlhbjA0NDdAbmphdS5lZHUuY24=; Liangju Wang, d2xqQG5qYXUuZWR1LmNu
 Jie Zheng
Jie Zheng Longbo Liu
Longbo Liu Huihui Tao
Huihui Tao Yuyan An
Yuyan An Liangju Wang
Liangju Wang