- 1Department of Plant Biotechnology, College of Life Sciences and Biotechnology, Korea University, Seoul, South Korea
- 2Department of Molecular Biotechnology, College of Agriculture and Life Sciences, Bioenergy Research Center, Chonnam National University, Gwangju, South Korea
Salt stress constitutes a major form of abiotic stress in plants. Histone modification plays an important role in stress tolerance, with particular reference to salt stress resistance. In the current study, we found that HDA15 overexpression confers salt stress resistance to young seedling stages of transgenic plants. Furthermore, salt stress induces HDA15 overexpression. Transcription levels of stress-responsive genes were increased in transgenic plants overexpressing HDA15 (HDA15 OE). NCED3, an abscisic acid (ABA) biosynthetic gene, which is highly upregulated in HDA15 transgenic plants, enhanced the accumulation of ABA, which promotes adaptation to salt stress. ABA homeostasis in HDA15 OE plants is maintained by the induction of CYP707As, which optimize endogenous ABA levels. Lastly, we found that the double-mutant HDA15 OE/hy5 ko plants are sensitive to salt stress, indicating that interaction between HDA15 and ELONGATED HYPOCOTYL 5 (HY5) is crucial to salt stress tolerance shown by HDA15 OE plants. Thus, our findings indicate that HDA15 is crucial to salt stress tolerance in Arabidopsis.
Introduction
Abiotic stresses damage more than 50% of average crop yields (Lisar et al., 2012; Eun et al., 2019). Unpredicted changes in the global climate expose plants to various environmental stresses at different times in their life cycle. Overpopulation leads to serious shortages of agricultural water. Therefore, plant water stress may pose a serious issue in the future (De Oliveira et al., 2013; Kim et al., 2019). Water stress may be associated with drought, salt stress, or temperature stress, and water shortages lead to various physiological phenomena (Pardo, 2010). Salt stress, which is considered a major form of abiotic stress in plants, inhibits plant growth due to excessive absorption of Na+ and Cl−, causing ion imbalances and physiological disorders (Gupta and Huang, 2014). Salt stress exerts several effects on plant growth. It induces osmotic stress, causing plant growth to decrease by reducing water influx, which decreases turgor pressure, enhances stomatal closure, and reduces photosynthetic activity. Since excess Na+ causes specific ionic toxicities, regulating the exclusion of Na+ as well as transportation of Na+ is paramount. The higher the concentration of Na+ and Cl−, the lower the absorption of other nutrients such as K+ and Ca2+, a process that leads to nutritional imbalance (Abassi et al., 2016). Moreover, reactive oxygen species (ROS) may be generated as the result of plant exposure to salt stress. Lipid peroxidation, protein degradation, and DNA mutations increase with increasing ROS (Abassi et al., 2016). Because plants are sessile, they cannot avoid salt stress and must continually adapt and evolve a variety of mechanisms to reduce the adverse effects of salt stress. Some of these mechanisms include the production of antioxidant enzymes to detoxify ROS, biosynthesis of compatible osmolytes, such as proline or glycine betaine, and accumulation of abscisic acid (ABA) (Gupta and Huang, 2014).
ABA is a crucial plant hormone that regulates the expression of stress-responsive genes under osmotic stress by functioning as an endogenous messenger that transmits stress signals. Osmotic stress activates ABA biosynthesis and inhibits ABA degradation (Fernando and Schroeder, 2016; Nam et al., 2019; Seo et al., 2019). Furthermore, it is known that the Salt Overly Sensitive (SOS) signaling pathway plays an important role in salt stress tolerance and ion homeostasis (Zhu, 2016). Histone modification, especially histone acetylation and deacetylation, plays a crucial role in the response to salt stress (Chen et al., 2010; Ueda et al., 2017; Zheng et al., 2019). ELONGATED HYPOCOTYL 5 (HY5), a master transcription factor (TF), belongs to the basic leucine zipper (bZIP) family (Gangappa and Botto, 2016). It inhibits hypocotyl elongation and lateral root growth but mainly regulates the light signaling pathway in Arabidopsis (Gangappa and Botto, 2016; Yang et al., 2018). Additionally, HY5 also plays a role in abiotic stress and the ABA signaling pathway. Germination of hy5 mutant seeds is insensitive to the adverse effects of ABA, salt stress, and glucose, whereas hy5 plants are less tolerant to salt stress and osmotic stress (Chen et al., 2008). Moreover, interaction between HY5 and RSM1 facilitates the binding of HY5 to the ABI5 promoter, resulting in the upregulation of ABI5 in the presence of salt stress or ABA (Chen et al., 2008). HY5 also induces the expression of cold-related genes and anthocyanin biosynthesis genes (Catalá et al., 2011). Thus, besides its light-dependent functions, HY5 broadly promotes the adaptation of Arabidopsis plants to abiotic stresses.
Histone acetylation and deacetylation in plants are two histone modifications that have been studied extensively. Histone acetylation involves the addition of acetyl groups to lysine residues within the histone tail at the N-terminus, a process that is catalyzed by histone acetyltransferases (HATs). However, during histone deacetylation, histone deacetyl transferases (HDACs) catalyze the removal of acetyl groups from lysine residues in the histone tail (Konsoula and Barile, 2012; Liu et al., 2016). Reportedly, histone acetylation loosens the DNA structure by neutralizing the positive charges on lysine residues, making the structure easily accessible to TF complexes, which bind the gene promoter and regulate gene expression (Henikoff, 2005; Shahbazian and Grunstein, 2007). There are 12 HATs that belong to four families: the GNAT family, MYST family, CBP family, and TAFII250 family (Pandey et al., 2002). Eighteen HDACs have been identified in Arabidopsis, and these are divided into three superfamilies as follows: (i) The Reduced Potassium Dependence 3/Histone Deacetylase 1 (RPD3/HDA1) superfamily contains 12 HDACs, which are further subdivided into three classes: Class I (HDA6, HDA7, HDA9, and HDA19), Class II (HDA5, HDA14, HDA15, and HDA18), and Class III (HDA2 and its two isoforms). Another RPD3/HDA1 superfamily contains HDA8, HDA10, and HDA17, which are still unclassified. (ii) Silent Information Regulator 2 superfamily (SIR2) contains SRT1 and SRT2. (iii) Histone Deacetylase 2 (HD2)-related protein family contains plant-specific HDACs and comprise four members (HD2A/HDT1, HD2B/HDT2, HD2C/HDT3, and HD2D/HDT4) (Liu X. et al., 2014). Both HATs and HDACs are associated with salt stress in Arabidopsis. GCN5, a member of the GNAT family, plays a role in salt stress tolerance by mediating cell wall-related genes in response to salt stress (Zheng et al., 2019). An HDA6 mutant, axe1-5, and an HDA6 RNAi line showed sensitivity to salt stress during seed germination (Chen et al., 2010). An hda19 mutant in Col-0 background, hda9, as well as AtHD2C and AtHD2D overexpression lines are reportedly tolerant to salt stress (Sridha and Wu, 2006; Han et al., 2016; Zheng et al., 2016; Ueda et al., 2017). Conversely, quadruple mutants (hda5/14/15/18), an hda19 mutant in Ws background, and an hd2c mutant were also reported to be sensitive to salt stress (Chen et al., 2010; Luo et al., 2012; Ueda et al., 2017).
A previous study found that HDA15 forms a complex with PIF1 and PIF3 to regulate the expression of light-responsive genes (Liu et al., 2013; Gu et al., 2017). Moreover, four Nuclear Factor-YC homologs in Arabidopsis redundantly interact with HDA15 to target hypocotyl elongation-related genes (Tang et al., 2017). Recently, HDA15 was found to positively regulate the suppression of ROP genes and ABA negative regulators by forming a complex with Myb96. Moreover, loss of function in HDA15 was found to induce sensitivity to drought stress (Lee and Seo, 2019). Although HDA15 is reportedly involved in salt stress in plants, the detailed mechanism underlying this process is not entirely known. In order to study the mechanisms underlying salt stress, plants overexpressing HDA15 were generated and investigated to determine whether these plants were resistant than Col-0 plants under salt stress, anticipating that that this would lead to molecular mechanisms underlying HDA15 functions being revealed.
Materials and Methods
Plant Material, Growth Conditions, and Stress Treatments
Col-0 Arabidopsis thaliana (Col-0), HDA15 OE (transgenic, overexpression lines) were used in this study. The hda15 mutant (SALK_004027), hy5 mutant (hy5-215) was obtained from the Arabidopsis Biological Resource Center (ABRC). The seeds were surface sterilized and then incubated at 4°C in the dark for 3 days before germination. Half-strength MS media with 2% sucrose (pH = 5.7) were used as controls for plant growth. All seeds were grown at 23°C (16-h light/8-h dark) and 60% humidity for 3 days before being transferred to stressed media. For stress treatments, 4-day-old plants were subjected to 0, 175, or 200 mM NaCl stress treatment for 3 days. Plant phenotypes were captured photographically, survival rates were determined, and samples were collected for the chlorophyll assay. Forty plants were used per treatment (three replicates for each treatment), and the experiments were repeated three times independently. For vegetative salt stress treatment, after germination of seeds in 1/2 MS medium, they were raised for 10 days, and then seedlings of similar size were transferred to soil and allowed to grow for 3 more weeks before stress treatment. The plants were watered with 0 or 300 mM NaCl solutions for 8 days. Photographic images were obtained after 8 days from the initiation of stress, and the results were quantified by measuring the chlorophyll contents. Proline and MDA contents were also measured with the vegetative plant samples that were treated in the same manner.
Germination Test
The seeds were sterilized and maintained at 4°C in the dark for 3 days. The sterilized seeds were then directly germinated on media containing 0, 50, 100 mM NaCl or 0, 0.1, 0.3, 0.5, 1 μM ABA for 8 days. Each condition contained three replicated plates. The results were recorded as percentages (%) of green cotyledons.
DNA Construction and Transgenic Plant Generation
To generate HDA15 overexpression lines, the cDNA of HDA15 was amplified using RT-PCR with specific primers (Supplementary Table 1). The amplified fragments were then eluted and cloned into a TOPO vector (pCR8/GW/TOPO® TA Cloning Kit, Invitrogen). After sequence checking, HDA15 was transferred to a pMDC32 vector under the control of a 35S CAMV promoter using gateway recombination. For plant transformation, Col-0 plants were used to implement a floral dipping method using Agrobacterium tumefaciens (GV3101). T4 homozygous plants overexpressing HDA15 were utilized for all subsequent experiments.
To construct HDA15pro::GUS, we obtained ~1 kb of the HDA15 promoter via PCR using a specific primer (Supplementary Table 1). The promoter was first cloned to a TOPO vector (pCR™8/GW/TOPO® TA Cloning Kit, Invitrogen) and then subcloned into a pMDC162 vector. HDA15pro::GUS was subsequently transformed into Agrobacterium tumefaciens (GV3101) by electroporation and then Col-0 plants via floral dipping.
Lipid Peroxidation Assay
Three-week-old plants were challenged with 0 or 300 mM NaCl for 8 days, and the rosette leaves were collected and used. The assay was performed according to the previous study (Truong et al., 2017). Samples were homogenized using 0.1% (w/v) trichloroacetic acid (TCA) and centrifuged at 12,000 rpm, 4°C for 15 min. Thiobarbituric acid (0.5%) was diluted in 20% TCA and added to the supernatant, and the supernatant solution was incubated at 95°C for 25 min. The mixture was then placed on ice to terminate the reaction. Intensity of the developed color complex was measured at A532 and A600.
Proline Content
Three-week-old plants were challenged with 0 or 300 mM NaCl for 10 days. Rosette leaves were sampled and used for this assay. The assay was performed according to a previous study (Truong et al., 2017). Samples were homogenized in 3% aqueous sulfosalicylic acid and the supernatant was collected after centrifugation at 12,000 rpm for 10 min. The mixture of supernatant, acidic ninhydrin, and glacial acetic acid was cultured at 100°C for 1 h, then transferred to ice to terminate the reaction. Toluene was used to extract the reaction mixture, and absorbance was measured at A520.
qRT-PCR Analysis
For qRT-PCR analysis, 7-day-old plants were exposed to 150 mM NaCl or 1 μM ABA for 6 h. After that, samples were collected for RNA extraction and cDNA synthesis. Collected samples were used to isolate RNA, which was converted to cDNA following a previously described protocol (Jeong et al., 2018). Changes in the transcription levels of abiotic marker genes were detected via a qRT-PCR cycler (BioRad). Plant actin, Actin2, was used as the internal control. The experiment was repeated three times. The specific primers used in qRT-PCR analysis are listed (Supplementary Table 1).
β-Glucuronidase Assay
T4 homozygous seedlings were used for the β-glucuronidase (GUS) assay. Seven-day-old seedlings were subjected to 150 mM NaCl for 6 and 24 h. The seedlings were then collected and used to either extract RNA for GUS quantification or observe intensive staining under the microscope. The experiments were repeated three times independently to maintain consistency.
Abscisic Acid Content Measurement
For ABA content, 7-day-old seedlings were exposed to 150 mM NaCl for 24 h. Subsequently, 100-mg samples of seedlings were taken and ground in liquid nitrogen. Measurement of ABA content was performed according to a previous report (Liu N. et al., 2014) using a Phytodetek Elisa kit.
Chlorophyll Assay
For the chlorophyll assay, samples from the abovementioned phenotype stress treatment were used to carry out chlorophyll determination as previously described (Truong et al., 2017). Briefly, ground samples were extracted using 80% acetone in the dark at room temperature (RT) for 45 min. The supernatant was collected following centrifugation at 3,000 rpm at 4°C for 15 min. Absorbance of chlorophyll was measured at A663 and A645. The experiments were repeated thrice independently to maintain consistency.
Chromatin Immunoprecipitation Assay
Two-week-old plants were challenged using salt stress media containing 150 mM NaCl for 6 h. The samples were then subjected to fixation, sonication, and immunoprecipitation based on a previous protocol (Saleh et al., 2008). H3K14 and H4K16 antibodies were used in this assay. Purified DNA was analyzed via qRT-PCR with specifically designed primers at the 5′UTR and the exon.
Data Analysis
Each experiment was repeated thrice to obtain reproducible results. SD value was calculated from three replicates of each treatment. Statistical Analysis System (SAS) was utilized to perform one-way ANOVA and Tukey's test (α = 0.05). Different letters (a, b, or c) within a treatment group indicate significant differences based on one-way ANOVA (P < 0.05).
Results
HDA15 and Its Promoter Activity Is in Response to Salt Stress
In order to better understand the role of HDA15, we examined HDA15 expression in Col-0 plants exposed to salt stress for 6 and 24 h. The transcript level of HDA15 was increased at both 6 and 24 h, suggesting that HDA15 rapidly reacted to salt stress (Figure 1A). HDA15 expression was higher in flower tissues than in tissues such as roots, stems, cauline leaves, and rosette leaves (Figure 1B). Next, we questioned whether the increase seen in the transcription level of this gene was due to its promoter activity. Hence, homozygous T4 plants of the HDA15pro::GUS line were generated to visualize the promoter activities of HDA15 in response to salt stress. Upon examining GUS expression in HDA15pro::GUS plants grown under normal conditions, we determined that the GUS reporter gene was evenly expressed in all tissues of young plants (Figure 2A). However, following the exposure of HDA15pro::GUS plants to salt stress at 150 mM NaCl for 6 and 24 h, GUS expression becomes much stronger throughout the plants (Figure 2). As evidenced by the quantitation results of GUS expression (Figure 2B), it appears that the activity of the HDA15 promoter was significantly induced by salt stress.
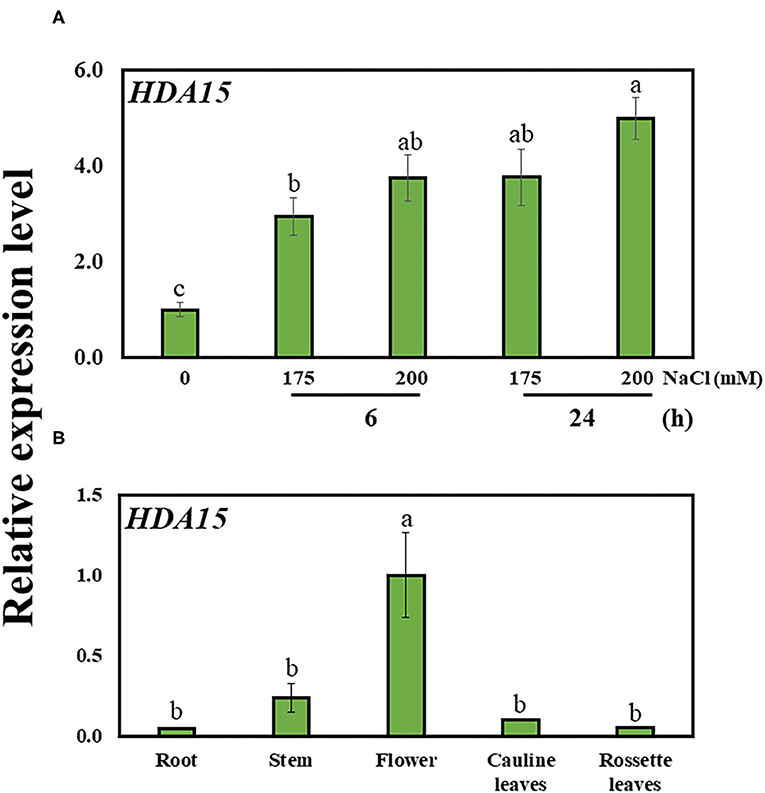
Figure 1. The expression of HDA15 in wild-type plants. (A) HDA15 expression in Col-0 plants in response to salt stress at 6 and 24 h. Seven-day-old Col-0 plants were exposed to 175 and 200 mM NaCl for 6 and 24 h followed by RNA extraction and cDNA synthesis for qRT-PCR. (B) The expression of HDA15 in different tissues in Col-0 plants. Five-week-old Col-0 plants in soil pots were used to collect stems, flowers, cauline leaves, and rosette leaves. For root samples, 2-week-old Col-0 plants germinated in MS media were utilized. Error bars represent the standard deviation of three replicates. Different letters (a, b, or c) within a treatment group indicate significant differences based on one-way ANOVA (P < 0.05).
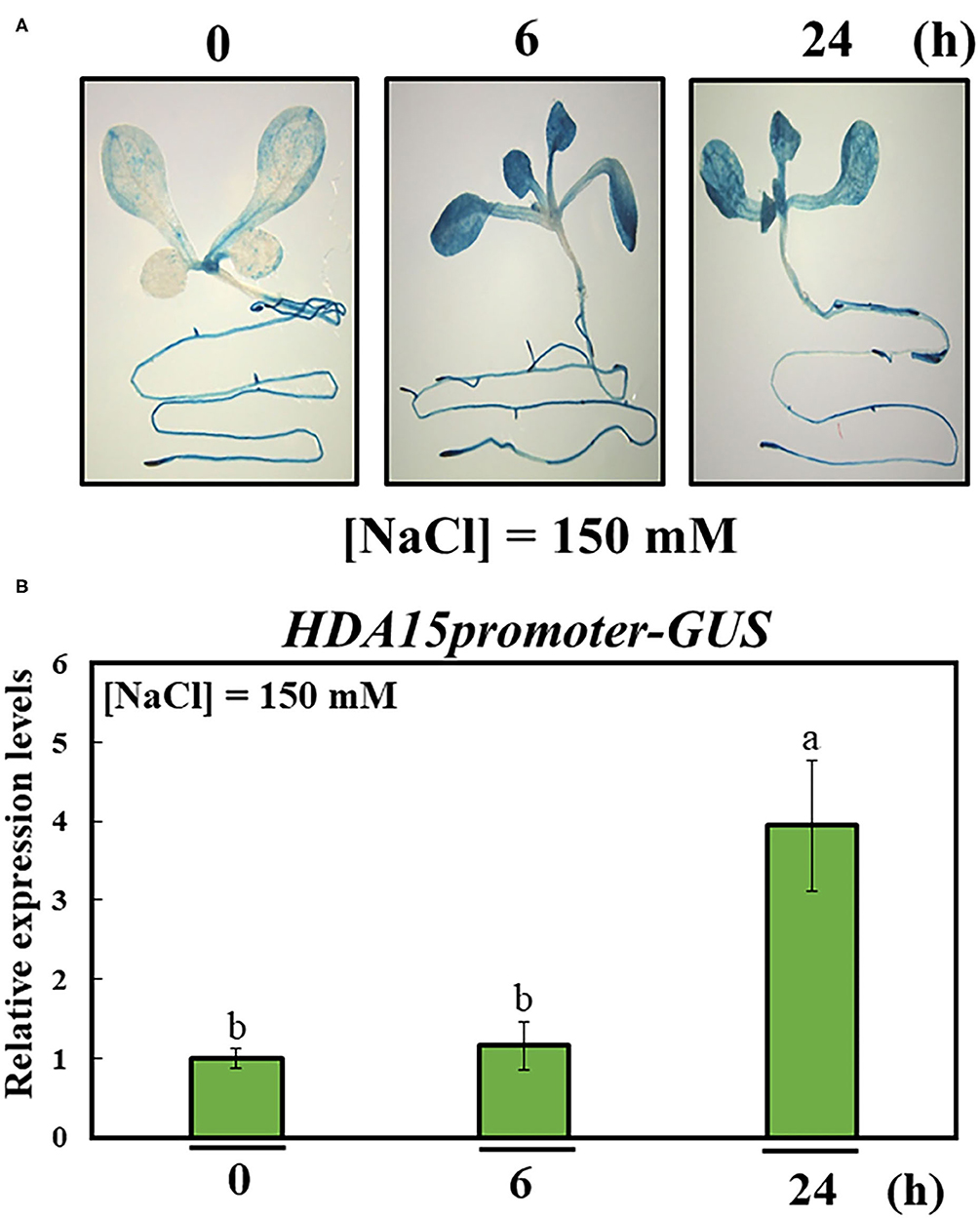
Figure 2. Activity of HDA15 promoter in response to salt stress. The promoter of HDA15 was cloned into a pMDC162 vector containing the GUS reporter gene via the Gateway system. Homozygous T4 seeds were used to perform β-glucuronidase (GUS) staining and quantification. (A) Visualization of the response of the HDA15 promoter to salt stress. Seven-day-old plants were stained with GUS solution for 2 h to visualize promoter activity in response to 150 mM NaCl at 6 and 24 h. (B) GUS quantification of the HDA15 promoter under salt stress. RNA was extracted from 7-day-old HDA15pro::GUS plants challenged by salt stress under the same conditions indicated above for GUS staining. The GUS primer was used to perform qRT-PCR. Actin2 was used as an internal control. Error bars represent the standard deviation of three replicates. Different letters (a, b) within a treatment group indicate significant differences based on one-way ANOVA (P < 0.05).
Overexpression of HDA15 Enhanced the Tolerance of Transgenic Plants in Response to Salt Stress
To further investigate the function of HDA15 under salt stress, we generated overexpression lines of HDA15 (HDA15 OE) and tested the resulting phenotypes with 0, 175, and 200 mM NaCl treatment. No differences were observed between the growth performances of Col-0 and HDA15 OE plants under control conditions. However, HDA15 OE plants appeared greener under salt stress compared to Col-0 plants, most of which died after 3 days of being challenged by salt stress using 200 mM NaCl (Figure 3A). The results were also quantified as survival rates, which were equivalent to the proportion of plants with green cotyledons (Figure 3B). Exposure to 200 mM NaCl significantly decreased the survival rates of Col-0 plants (by ~95%) compared to those under normal conditions. However, exposure to 200 mM NaCl reduced the survival rates of HDA15 OE plants up to 35%, and this survival rate is higher than that of Col-0 under stress conditions (Figure 3B). In addition, chlorophyll contents were consistent with growth performance (Figure 3C). Although chlorophyll contents tended to decrease with salt stress in all types of plants, the decrease seen in HDA15 OE plants was less than that of Col-0.
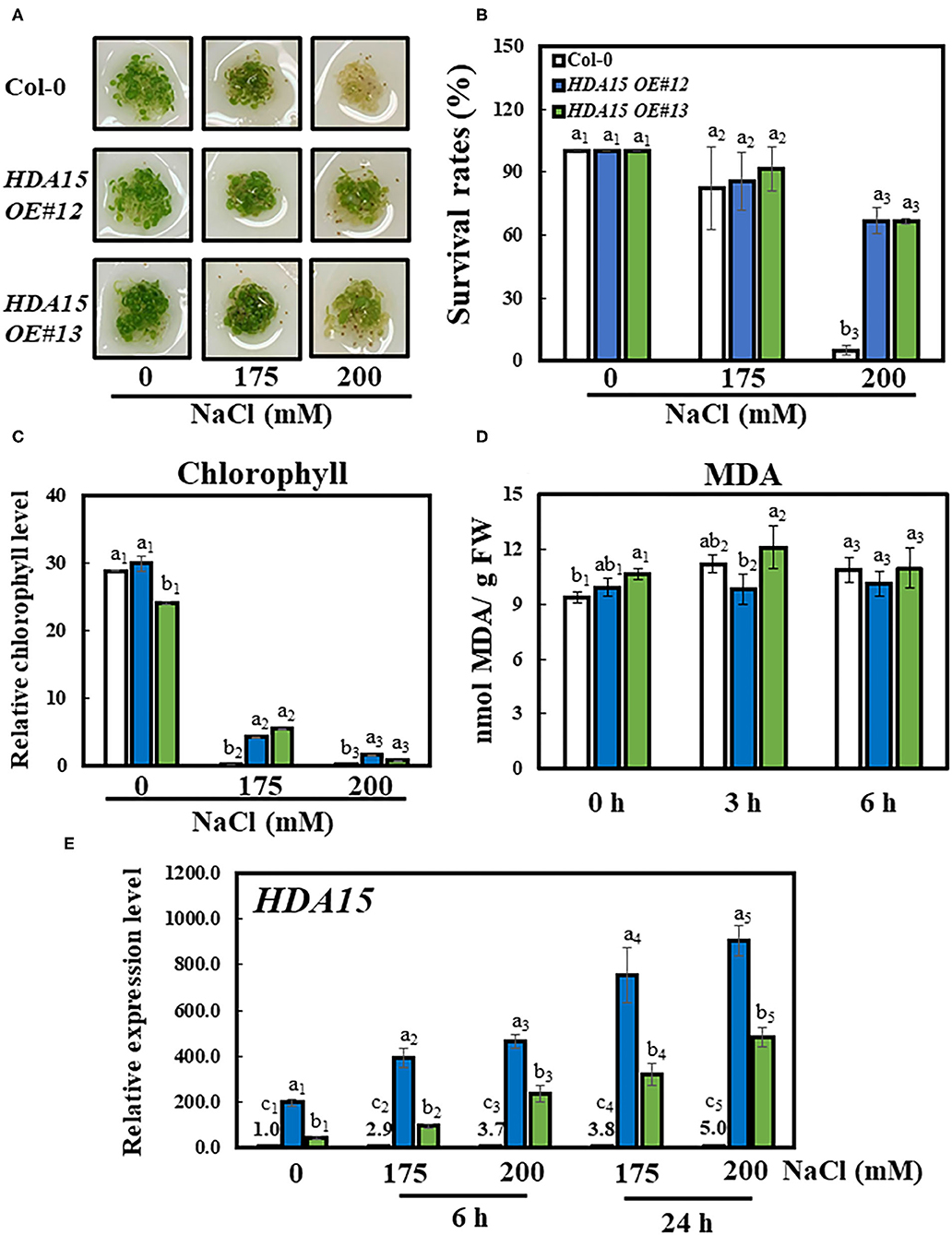
Figure 3. Growth performance of HDA15 transgenic plants, survival rates, and chlorophyll content in response to salt stress. Four-day-old plants, which were germinated in normal MS media, were transferred to different NaCl concentrations as indicated. The experiment was repeated three times independently to obtain a consistent result. (A) Phenotypes of Col-0 and HDA15 OE transgenic plants following a 3-day exposure to salt stress. (B) The survival rates of Col-0 and HDA15 OE transgenic plants following a 3-day exposure to salt stress. (C) Chlorophyll levels of Col-0 and HDA15 OE transgenic plants following a 3-day exposure to salt stress. (D) The lipid peroxidation levels of Col-0 and HDA15 OE plants in response to salt stress. Seven-day-old plants, which were germinated in normal MS media, were transferred to 150 mM NaCl for 0, 3, and 6 h. (E) HDA15 expression in Col-0 and HDA15 OE plants. Seven-day-old plants were exposed to 175 and 200 mM NaCl for 6 and 24 h, followed by RNA extraction and cDNA synthesis for qRT-PCR. Actin2 was used as an internal control. Error bars represent the standard deviation of three replicates. Different letters (a, b, or c) within a treatment group indicate significant differences based on one-way ANOVA (P < 0.05).
Under control conditions, the transcript levels of HDA15 OE#12 and HDA15 OE#13 plants were increased more than 200 and 43 times, respectively, compared to those of the Col-0 plants. Moreover, salt stress increased the transcript levels of HDA15 in transgenic plants by at least twice compared to those under normal conditions (Figure 3E). However, the hda15 knock out (ko) mutant did not show significant phenotypic alterations under salt stress when compared to Col-0 plants (Supplementary Figures 1A,B). In addition to investigating salt stress, we also examined the germination response of Col-0 and HDA15 OE plants in ABA media to determine whether HDA15 OE responds differently to ABA when compared with Col-0 (Supplementary Figures 1C,D). Under salt conditions, the percentage of germinated HDA15 OE plants was slightly less than that of Col-0 plants. The ratios of HDA15 OE plants with green cotyledons in response to exogenous ABA levels were slightly higher than those in Col-0 plants. Next, we measured lipid peroxidation, an indicator of oxidative stress. There was no significant difference between lipid peroxidation of Col-0 and HDA15 OE plants under salt stress (Figure 3D). Additionally, we tested the tolerance to salt stress of HDA15 OE plants in the vegetative stage (Supplementary Figure 2). Under salt stress, the leaves of Col-0 and HDA15 OE plants showed yellowing phenotype, indicating that salt stress is harmful to plant growth. As shown in Supplementary Figure 2A, HDA15 OE plants were less damaged than Col-0, which showed the same result in chlorophyll content measurement. According to these results, it is necessary to conduct more stress test experiments with various conditions to make a clear conclusion, but the HDA15 effect can be considered to have more influence on young seedlings.
Enhanced tolerance of HDA15 OE plants in response to salt prompted us to examine the transcription level of HDA15 and homologous genes including HDA5/14/18 in response to salt stress (Figure 4). The transcript levels of all three homologs were increased in response to high salt in Col-0 plants, confirming that Class II HDACs are responsive to salt stress. However, the transcript levels of the three homologs in HDA15 OE plants were not different from those of Col-0 plants under stress, indicating that HDA15 overexpression does not interfere with the expression of its homologs. To ensure our salt stress studies were properly conducted, we investigated the transcript level of an abiotic-stress marker gene, RD29B, a well-known gene that is induced by salt stress (Msanne et al., 2011). HDA15 transgenic plants showed increased transcript level of RD29B under salt stress (Figure 4), indicating that HDA15 OE plants regulate stress signaling more effectively than Col-0. Additionally, we detected a profile of downregulated genes in the hda15 ko mutant under normal conditions (RNA-Seq data; Zhao et al., 2019). These included some salt stress-responsive genes that downregulated transcript levels in the hda15 ko mutant, indicating that HDA15 serves as a positive regulator in the induction of some stress-responsive genes.
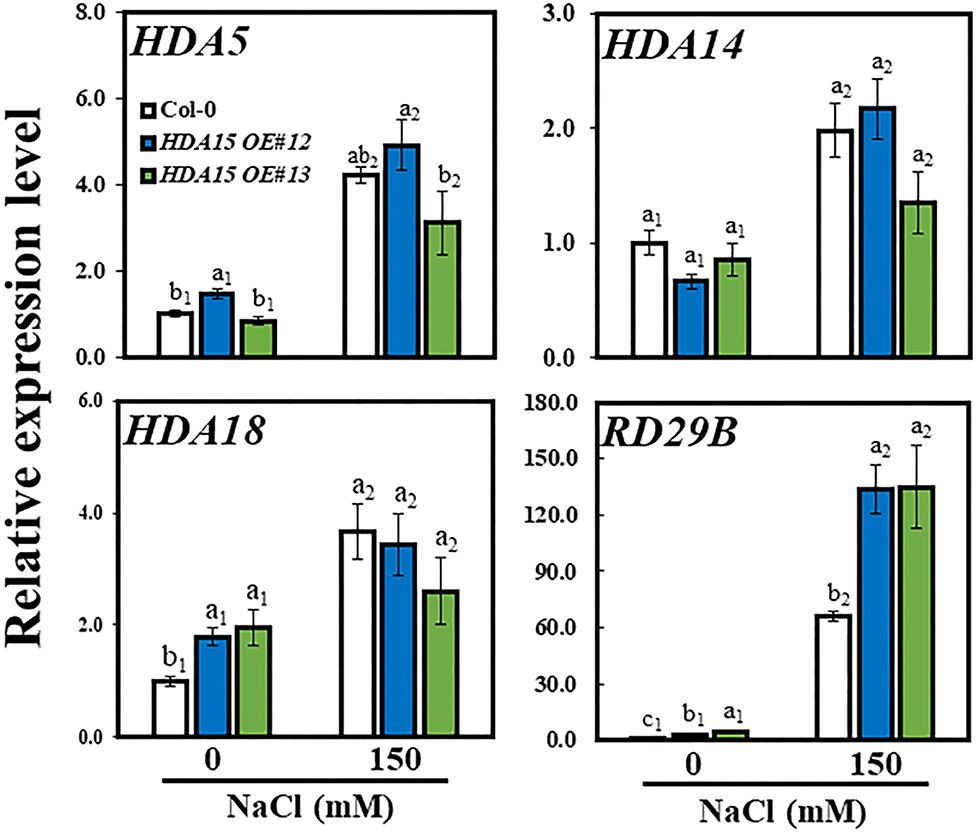
Figure 4. Expression levels of HDA15 homologs and a stress-related gene under salt stress. Seven-day-old plants were subjected to 150 mM NaCl for 6 h. RNA was then extracted, and cDNA was synthesized. Next, qRT-PCR was performed with primers for HDA5, HDA14, HDA18, and RD29B genes. Actin2 was used as an internal control. The experiment was repeated three times independently. Error bars represent the standard deviation of three replicates. Different letters (a, b, or c) indicate significant differences based on one-way ANOVA (P < 0.05).
Increased Transcript Levels of NCED3, an ABA Biosynthetic Gene, Enhance Salt Stress Tolerance of Plants via Enhanced Abscisic Acid Accumulation
A previous study suggested that HDA15 plays an important role in ABA signaling (Lee and Seo, 2019). Therefore, we checked the response of ABA biosynthetic genes NCED2, NCED3, NCED5, NCED6, and NCED9 (Figure 5). The results indicated that the transcript levels of three NCED genes, NCED2, NCED3, and NCED9, were enhanced under salt stress. In detail, the transcript levels of NCED2 and NCED9 were marginally increased under salt stress in HDA15 OE mutants compared to that of Col-0 plants. However, NCED3 transcript levels were ~1.5 times higher in two transgenic HDA15 plants than in Col-0 plants under salt stress treatment. The expression levels of both NCED5 and NCED6 in HDA15 OE plants were significantly downregulated in response to salt stress compared to those of Col-0 plants. NCED3 is upregulated by salt and drought (Barrero et al., 2006). It appears that suppression of NCED5 and NCED6 leads to the upregulation of NCED3. Therefore, we postulated that NCED3 may act as an important gene that affects ABA biosynthesis in response to salt stress in HDA15 OE plants. This result also indicated that ABA enhances salt stress tolerance of HDA15 OE plants, which led us to determine the ABA content in Col-0 and HDA15 OE plants under salt stress (Figure 6A). HDA15 OE plants showed a higher ABA content than Col-0 plants when challenged by salt stress. Consistent with these results, we observed that NCED3 expression was induced in OE plants exposed to ABA (Figure 6B). In addition, the transcript levels of HDA15 were increased in response to ABA (Supplementary Figure 4).
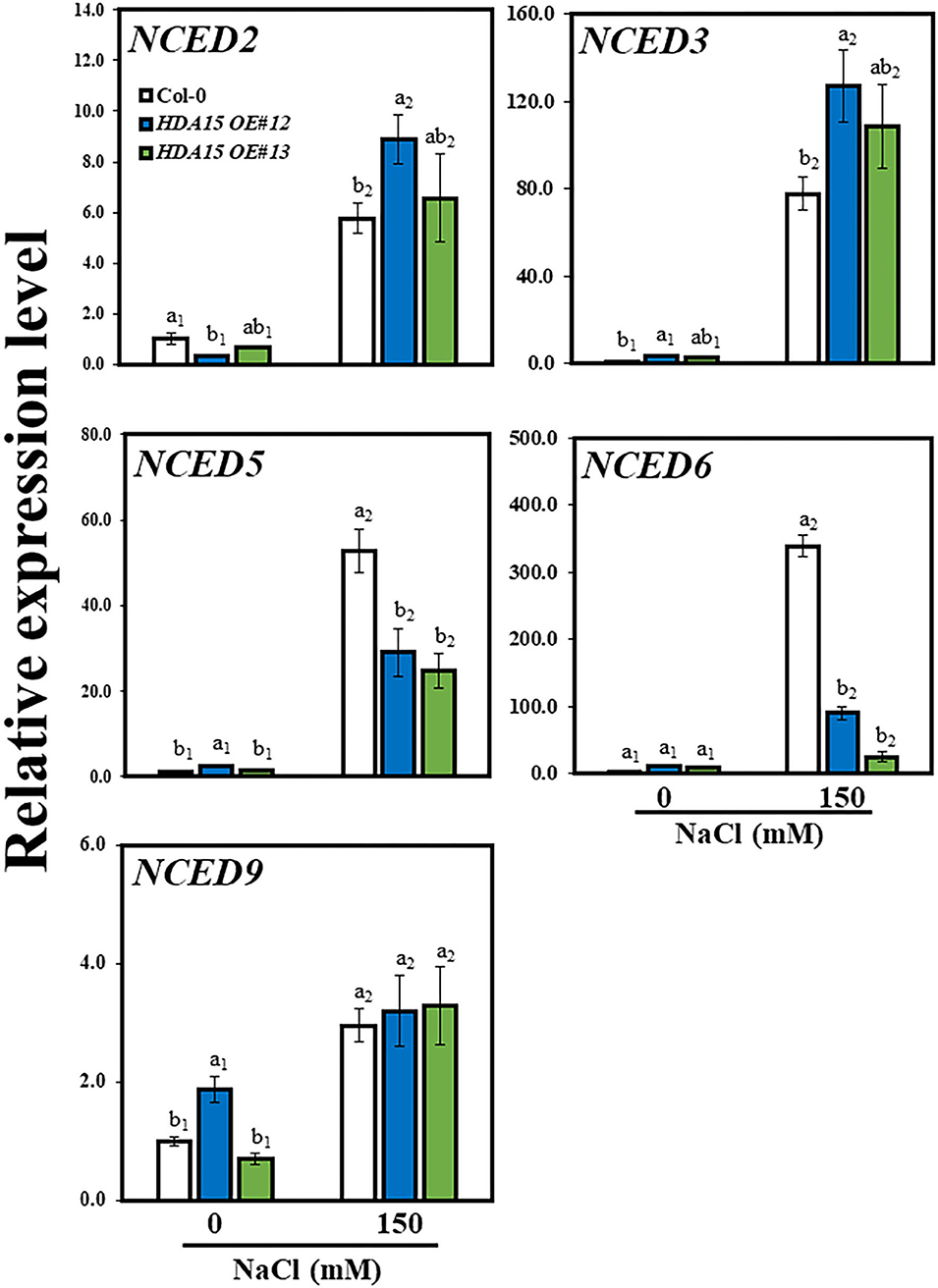
Figure 5. The expression levels NCED genes, which are abscisic acid (ABA) biosynthetic genes, under salt stress. Seven-day-old plants were subjected to 150 mM NaCl for 6 h. RNA was then extracted, and cDNA was synthesized. Next, qRT-PCR was performed with primers for NCED2, NCED3, NCED5, NCED6, and NCED9 genes. Actin2 was used as an internal control. The experiment was repeated three times independently. Error bars represent the standard deviation of three replicates. Different letters (a, b) indicate significant differences based on one-way ANOVA (P < 0.05).
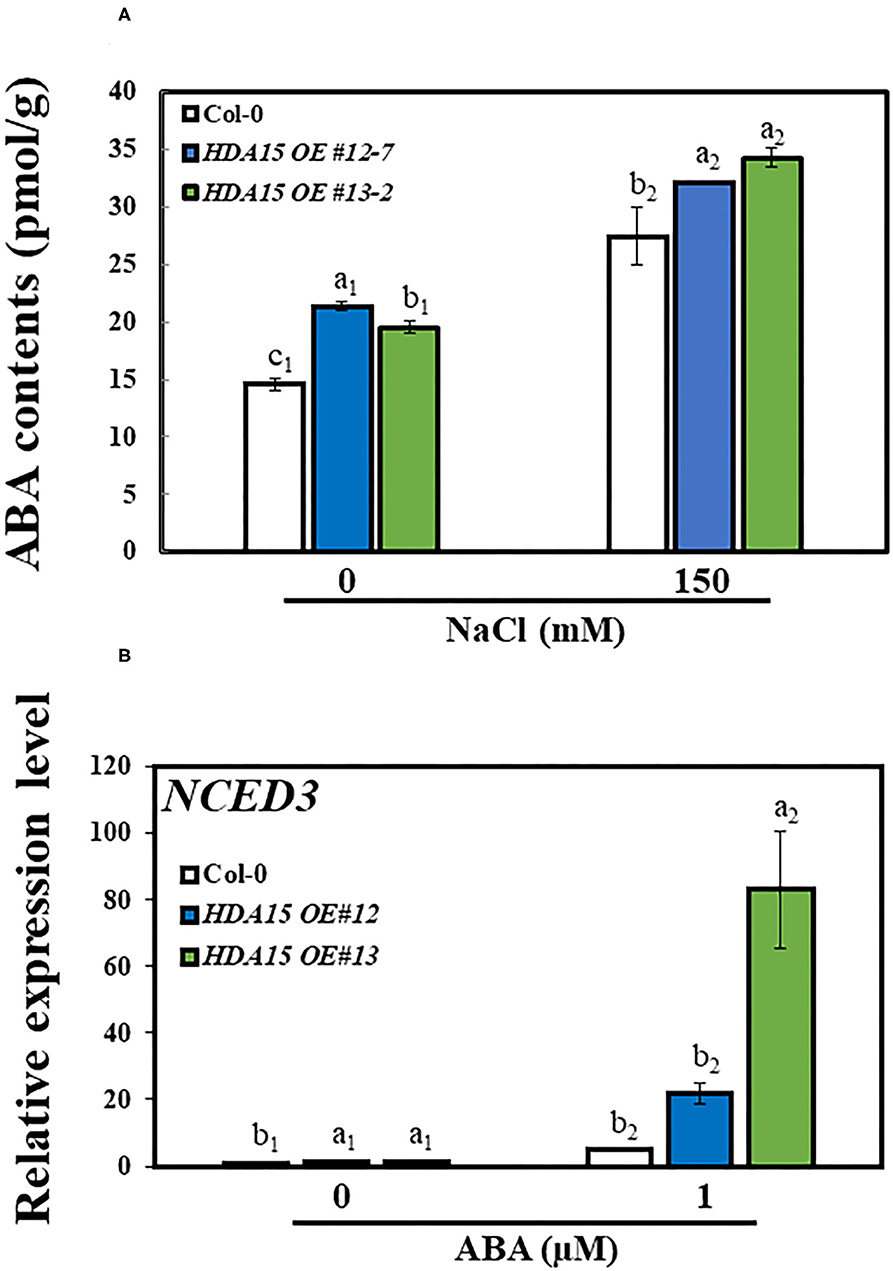
Figure 6. The accumulation of ABA in response to salt stress. (A) ABA contents. Seven-day-old plants were exposed to salt stress for 24 h, following which the contents of ABA were measured in Col-0 and HDA15 OE plants. (B) The expression level of NCED3 in response to ABA treatment in Col-0 and HDA15 OE plants. Seven-day-old plants were challenged with 1 μM ABA for the measurement of transcript levels in Col-0 and HDA15 OE plants. Error bars represent the standard deviation of three replicates. Different letters (a, b, or c) within a treatment group indicate significant differences based on one-way ANOVA (P < 0.05).
Since HDA15 mediates histone deacetylation, which usually inhibits the expression of its target genes, we conducted a histone chromatin immunoprecipitation (ChIP) assay to determine the acetylation levels of NCED3 in 5′UTR and exon regions. H3K14 and H4K16 levels at the 5′UTR were similar between control and salt stress conditions (Figures 7A,B). The NCED3 activator protein, NGATHA1, binds to the 5′UTR region of the NCED3 promoter under drought conditions (Sato et al., 2018). It appears that HDA15 does not interact with NCED3 activator to regulate the deacetylation levels at this 5′UTR region. However, a significant decrease in H3K14 and H4K16 levels was observed at the first exon region of the NCED3 (Figures 7C,D) in HDA15 OE plants under high salt stress. Since HDA15 acts as a negative regulator of certain genes, it is plausible that HDA15 increases deacetylation levels at the NCED3 exon region, leading to the inhibition of binding by unknown negative regulators.
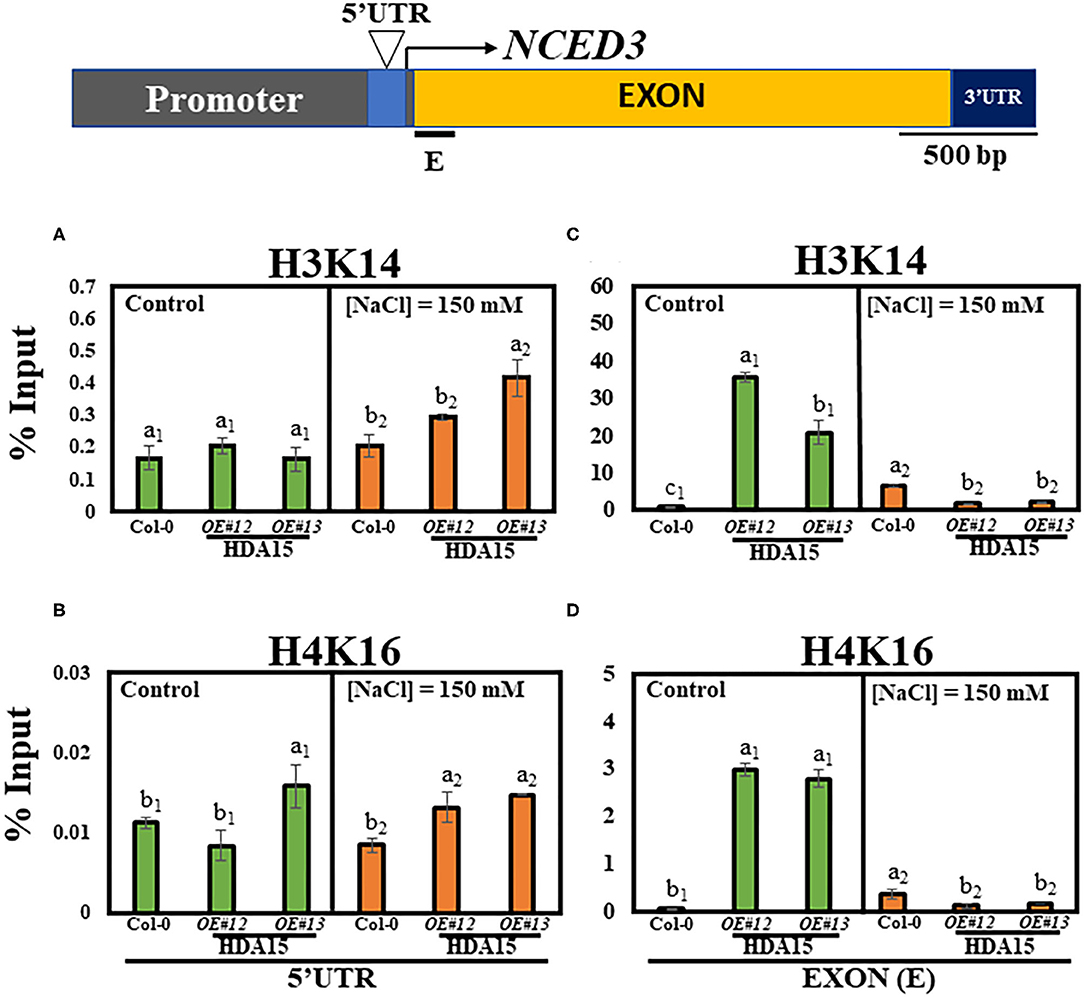
Figure 7. H3K14 and H4K16 levels in Col-0 and HDA15 OE transgenic plants in response to salt stress. Two-week-old plants were subjected to 150 mM salt stress for 6 h. The samples were then used for chromatin immunoprecipitation (ChIP) assay. Immunoprecipitated DNA was analyzed via qRT-PCR. (A) H3K14 levels at 5′UTR of NCED3 promoter under normal and salt stress conditions. (B) H4K16 levels at 5′UTR of NCED3 promoter under normal and salt stress conditions. (C) H3K14 levels at NCED3 exon under normal and salt stress conditions. (D) H4K16 levels at NCED3 exon under normal and salt stress conditions. Error bars represent the standard deviation of three replicates. Different letters (a, b, or c) within a treatment group indicate significant differences based on one-way ANOVA (P < 0.05).
Abscisic Acid Contents in HDA15 OE Plants Are Tightly Modulated via Abscisic Acid Homeostasis During Stress Response
Because NCED3 upregulation is partly responsible for ABA accumulation in HDA15 OE plants, we investigated the regulation of genes downstream of NCED3 under salt stress. We assessed the transcript levels of ABA2, ABA3, AAO3, and CYP707As (Figure 8). The transcript levels of ABA2, ABA3, and AAO3 in HDA15 OE plants were slightly higher than those of Col-0 under salt stress conditions. NCED3 reportedly serves a crucial role in ABA biosynthesis via its involvement in ABA biosynthetic pathways (Ma et al., 2018). In addition, two ABA catabolic genes, cytochrome P450 (CYP) 707A1 (CYP707A1) and CYP707A2, were upregulated in HDA15 OE plants in response to salt stress compared to those in Col-0 plants. However, there was no difference between the CYP707A3 transcript levels of Col-0 and HDA15 OE plants under stress conditions. Although ABA enables plants to adapt to abiotic stresses, excess internal ABA may negatively regulate plant growth (Belin et al., 2009; Yao and Finlayson, 2015). Thus, the increased expression of CYP707As may be a result of efforts to regulate ABA homeostasis in order to maintain cellular ABA levels that are optimal for plant growth. Furthermore, we determined that the expression of BG2, which hydrolyzes ABA-GE to ABA, induced by salt stress in HDA15 OE plants is higher compared to that in Col-0 plants. This also increases ABA accumulation in HDA15 OE plants under salt stress. We also examined the transcript levels of ABI genes. The transcript level of ABI5 was higher in HDA15 OE plants than that in Col-0 plants. During seedling development, ABI5 induces certain LEA genes, thereby enhancing adaptation of plants against abiotic stresses (Skubacz et al., 2016). Additionally, levels of ABI3 and ABI4 in HDA15 OE plants were upregulated in response to salt stress (Supplementary Figure 5). ABI3, ABI4, and ABI5 proteins reportedly act together during stress responses (Skubacz et al., 2016). Although ABI1 and ABI2 are negative regulators of the ABA signaling pathway, their transcript levels, especially that of ABI2, were also enhanced by salt stress in HDA15 OE plants. These results may be due to increased ABA content, which activates the negative feedback loop in the ABA signaling pathway (Merlot et al., 2001).
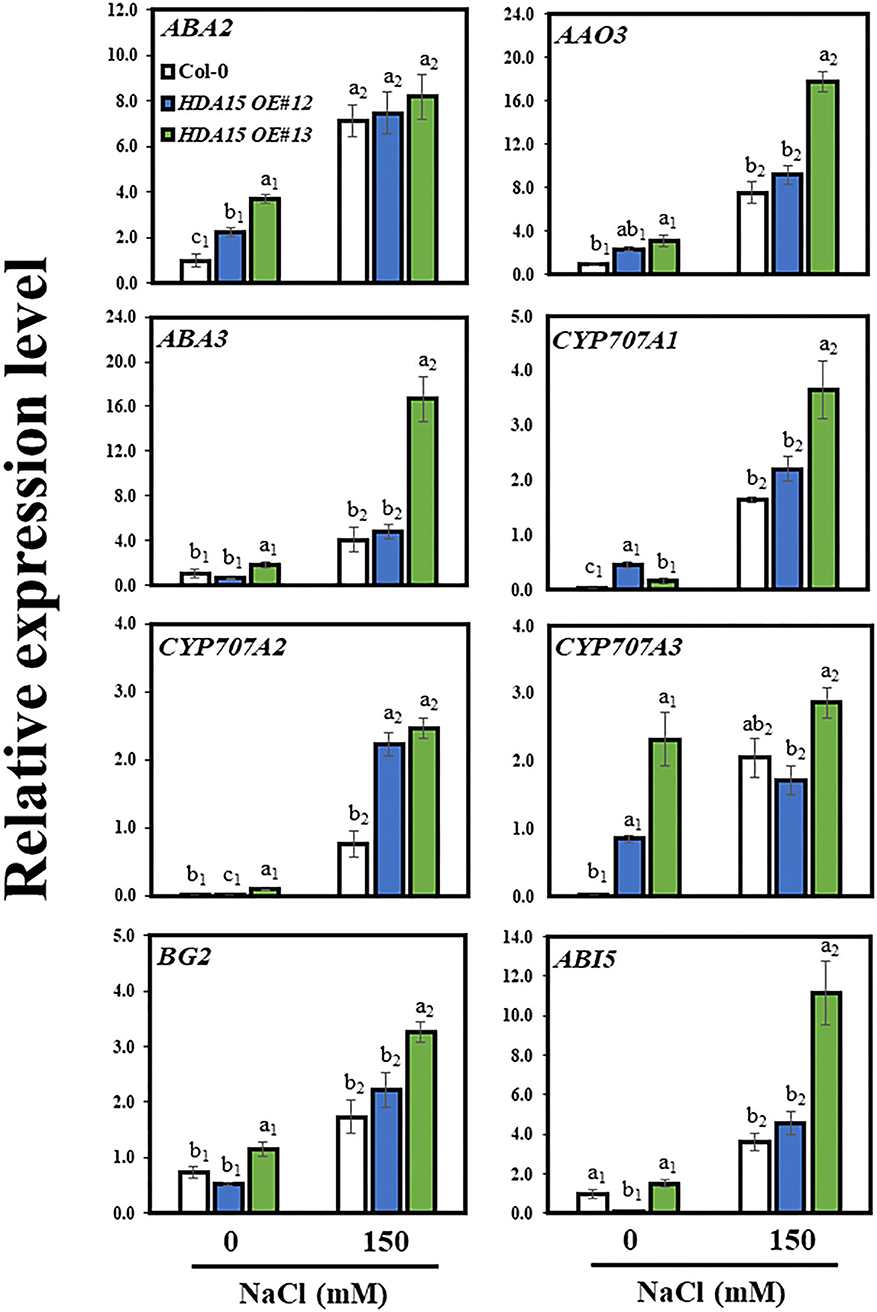
Figure 8. Expression of genes downstream of NCED3 in abscisic acid (ABA) biosynthetic and catabolic pathways. Seven-day-old plants were subjected to 150 mM NaCl for 6 h. RNA was then extracted, and cDNA was synthesized. Primers for ABA2, ABA3, AAO3, BG2, CYP707A1, CYP707A2, CYP707A3, CYP707A4, and ABI5 were used to perform qRT-PCR. Actin2 was used as an internal control. The experiment was repeated three times independently. Error bars represent the standard deviation of three replicates. Different letters (a, b, or c) were considered significant differences based on one-way ANOVA (P < 0.05).
Interaction Between HY5 and HDA15 Is Crucial for Salt Stress Tolerance
The TF, HY5, regulates the association between HDA15 protein and its target genes (Zhao et al., 2019). Thus, we speculated whether HY5 also plays a role in enhanced salt stress tolerance conferred by HDA15 overexpression. To address this issue, we generated a double-mutant by crossing hy5 mutant with HDA15 OE plants (Supplementary Table 1) and tested its tolerance in response to salt stress. All tested plants exhibited the normal phenotype under control conditions (Figures 9A–C). However, when grown in salt medium containing 175 or 200 mM NaCl, the HDA15 OE/hy5 ko plants were more sensitive to salt stress compared to other tested plants. HDA15 OE plants are the most tolerant to salt stress. The results of chlorophyll content determination also substantiated the phenotype test (Figure 9B). Thus, our observation indicated that HDA15 may also interact with HY5 to confer salt stress tolerance to plants. Moreover, our assessment of HY5 expression in HDA15 OE plants under salt stress indicated that HY5 transcript levels in HDA15 OE plants were upregulated (Supplementary Table 1). We also determined the transcript levels of RD29B and NCED3 in response to salt stress in HDA15 OE/hy5 ko plants and found that both these genes were downregulated (Figures 9D,E).
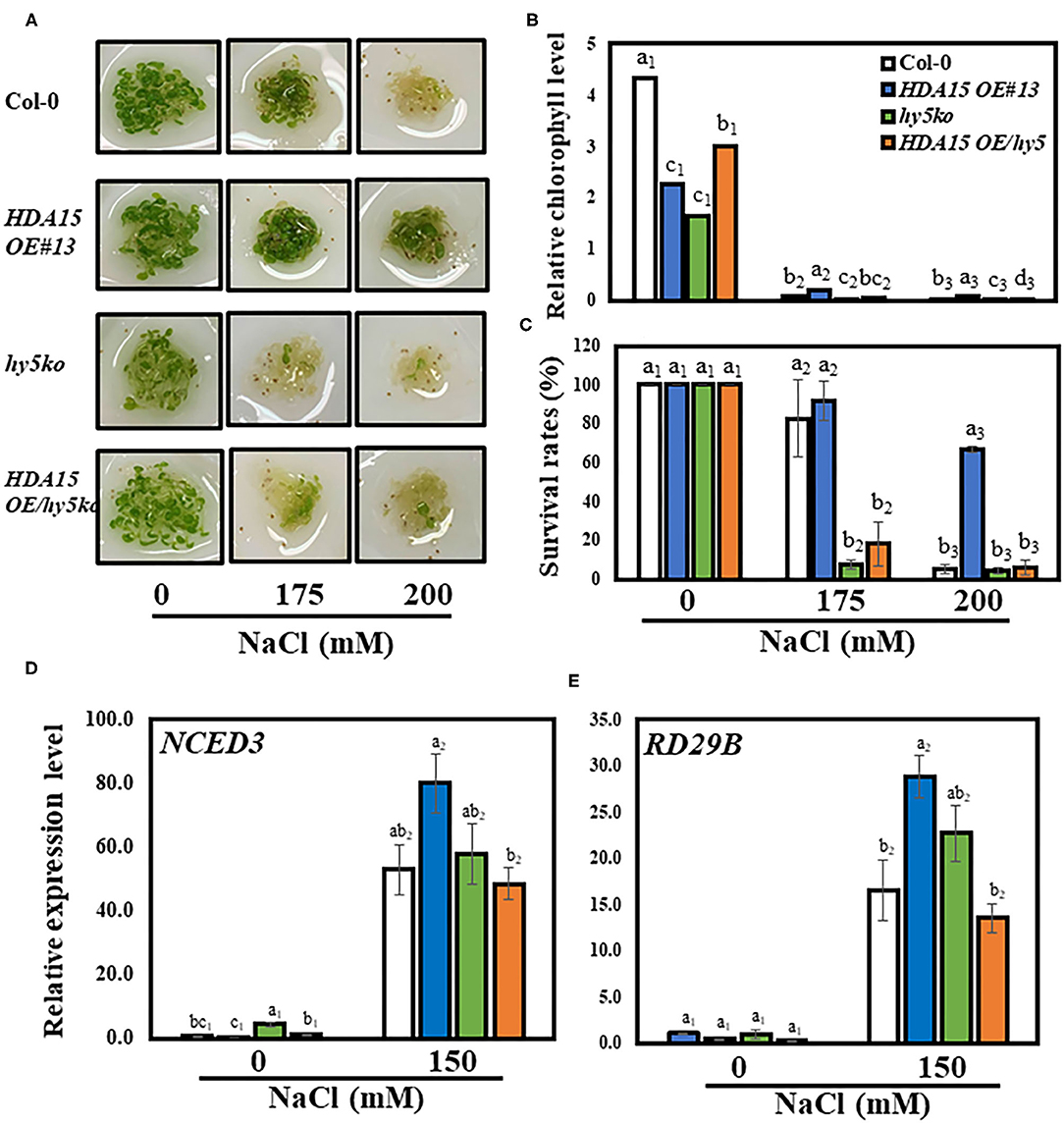
Figure 9. The co-action of HDA15 and HY5 in response to salt stress. Three-day-old plants germinated in normal MS media were transferred to different NaCl concentrations as indicated. The experiment was repeated three times independently to obtain a consistent result. (A) The phenotype of Col-0, HDA15 OE/hy5 ko, hy5 ko mutants, and HDA15 OE plants following a 4 d exposure to salt stress. (B) Chlorophyll levels of tested plants following a 4 d exposure to salt stress. (C) The expression levels of HY5 in Col-0 and HDA15 OE plants. (D,E) The expression levels of NCED3 and RD29B in Col-0, double mutants, HDA15 OE, and hy5 ko plants. Seven-day-old plants were exposed to 150 mM NaCl for 6 h followed by RNA extraction and cDNA synthesis for qRT-PCR. Actin2 was used as an internal control. Error bars represent the standard deviation of three replicates. Different letters (a, b, or c) within a treatment group indicate significant differences based on one-way ANOVA (P < 0.05).
Discussion
Presently, most studies have demonstrated the function of HDA15 in response to light (Liu et al., 2013; Gu et al., 2017). These indicate that both PIF1 and PIF3 interact with HDA15 in the dark to co-target and suppress a set of light-responsive chlorophyll biosynthetic and photosynthetic genes associated with hormone signaling (Liu et al., 2013; Gu et al., 2017). The role of HDA in response to salt stress has been revealed via studies on HDA6, HDA9, HDA19, AtHD2A, and AtHD2D (Sridha and Wu, 2006; Chen et al., 2010; Han et al., 2016; Zheng et al., 2016; Ueda et al., 2017). The most recent study indicated that HDA15 plays a positive role in ABA signaling by suppressing negative regulators of ABA, which enables plants to tolerate drought stress (Lee and Seo, 2019). Reportedly, a quadruple mutant showing loss of function of all four class II HDACs (HDA5/14/15/18) displayed sensitivity to salt stress (Ueda et al., 2017). However, no further detailed research has been conducted on the function of HDA15 alone under salt stress. Therefore, in this study, we aimed to characterize the function of the HDA15 in response to salt stress. We observed that the transcript level of HDA15 was increased in response to salt stress. Additionally, overexpression of HDA15 conferred salt stress tolerance to Arabidopsis, indicating the involvement of HDA15 in salt stress tolerance in these plants. We also examined the phenotype of hda15 ko mutants in response to salt stress, but these were similar to that of Col-0. The reason for this may be that HDA15 is one of several homologs belonging to class II of HDACs, which includes HDA5, HDA14, and HDA18. Only the quadruple mutant, hda5/14/15/18, showed a phenotype that was sensitive to salt stress (Ueda et al., 2017). Therefore, the loss of function of HDA15 alone may not exert a phenotypic effect in response to salt stress due to the compensatory role of other homologs. Furthermore, since HDA15 OE transgenic plants showed tolerant phenotypes against salt stress, we utilized Col-0 and HDA15 OE plants to characterize the function of HDA15. Previous studies also proposed that overexpression of AtHD2A and AtHD2D in Arabidopsis increased tolerance to salt stress (Sridha and Wu, 2006; Chen et al., 2010; Han et al., 2016; Zheng et al., 2016, 2019; Ueda et al., 2017).
As HDA15 is involved in ABA signaling and ABA accumulation enhances salt stress tolerance (Sah et al., 2016; Lee and Seo, 2019), we examined the effect of salt stress on the transcript levels of ABA biosynthetic genes. NCEDs are enzymes that mediate ABA biosynthesis. To date, five NCED genes are known to be present in Arabidopsis. These include AtNCED2, AtNCED3, AtNCED5, AtNCED6, and AtNCED9 (Ali et al., 2020). NCED3, which is induced by both ABA and NaCl, plays a vital role in osmotic stress-induced ABA biosynthesis (Iuchi et al., 2000; Tan et al., 2003; Barrero et al., 2006). Moreover, overexpression of OsNCED3 in rice conferred protection against osmotic stress (Huang et al., 2018). NCED3 induction increased ABA biosynthesis, resulting in enhanced ABA accumulation, which speeds up stomatal closure and upregulates the expression of stress-responsive genes, leading to increased stress tolerance in plants (Jakab et al., 2005). Drought-related stress causes the histone methyltransferase, ATX1, to modify H3K4me3, which then activates NCED3, resulting in drought- and ABA-related genes being upregulated (Kim et al., 2015). As shown in Arabidopsis, overexpression of GmWRKY16, a soybean WRKY TF, increases ABA accumulation, which is also observed when NCED3 is upregulated, enabling transgenic plants to resist drought and salt stresses (Ma et al., 2019). Thus, upregulation of NCED3 in HDA15 OE plants may be a key factor in osmotic stress tolerance, with particular reference to salt stress. The increased transcript levels of NCED3 led us to examine other downstream genes involved in the ABA biosynthetic pathway. NCED genes catalyze the synthesis of xanthoxin, which is converted to abscisic aldehyde by short-chain alcohol dehydrogenase/reductase (SDR/ABA2) and then to ABA by abscisic aldehyde oxidase (AAO3). The molybdenum cofactor sulfurase/ABA3 is required by aldehyde oxidase for its activity (Long et al., 2019). However, as ABA2, ABA3, and AAO3 play only minor roles in ABA biosynthetic pathway (Ma et al., 2018), their transcript levels were not significantly increased in HDA15 OE plants (Figure 8). In addition, the higher accumulation of ABA in HDA15 OE plants may be due to BG2 upregulation, which catalyzes inactive ABA-GE to ABA in one-step hydrolysis (Long et al., 2019). Induction of BG2 also helps plants to quickly enhance internal ABA levels for stress adaptation. On the other hand, cytochrome P450 monooxygenase 707A family members (CYP707As) are responsible for the degradation of ABA levels in plants. The mRNA levels of CYP707As are modulated by abiotic stresses (Yoon et al., 2020). Our results showed that the transcript levels of CYP707A1 and A2 in HDA15 OE plants were enhanced by salt stress. Such induction may help plants to prepare for ABA degradation when stress is removed. Although ABA is a key hormone in abiotic stress adaptation, where increased ABA content enables plants to tolerate unfavorable environments, excess ABA levels exert adverse effects on plant growth (Belin et al., 2009; Yao and Finlayson, 2015). Therefore, ABA homeostasis is important to balance plant growth and stress response in order to obtain the best growth performance under a given condition. In plants, the processes involved in the biosynthesis and catabolism of ABA must balance each other to maintain ABA contents at optimal levels during osmotic stress. Thus, CYP707A upregulation may be intended to maintain ABA levels within the optimal range in plants subjected to stress (Liu S. et al., 2014; Long et al., 2019). Therefore, while increasing internal ABA contents to cope with salt stress, HDA15 OE plants maintain ABA homeostasis by inducing the ABA catabolic compound, CYP707As, in order to balance and optimize ABA levels during the course of stress response and adaptation.
HDA15 displays deacetylase activity, which mediates the modulation of gene repression (Liu et al., 2013; Perrella et al., 2013). Many transcriptional repressors recruit HDA proteins to form a complex to regulate their target genes. Interaction between HDACs and other proteins creates a structural link between DNA, histones, and core deacetylase enzymes (Perrella et al., 2013). For instance, PIF3, a negative regulator of light signaling, reportedly interacts with HDA15 to suppress chlorophyll biosynthesis and photosynthesis in the dark (Liu et al., 2013). HDA15 and HY5 reportedly co-modulate hypocotyl elongation, cell wall, and auxin-related genes (Zhao et al., 2019). HY5 is a master transcription regulator not only of light response but also of abiotic stress (Chen et al., 2008; Gangappa and Botto, 2016; Yang et al., 2018). We found that the salt stress-tolerant phenotype of HDA15 OE was absent in HDA15 OE/hy5 ko plants, indicating that HY5 and HDA15 may form a complex that regulates salt stress response in Arabidopsis. In addition to HDA15, HDA19 can also be recruited by the AP2/EREBP TF, AtERF7, in response to ABA and drought stress to form a suppressor complex with AtSin3 to suppress the expression of stress-responsive genes (Song et al., 2005). HDA19 also regulates ABA synthesis by forming a repressive complex with SNL2 when interacting with SNL1 (Wang et al., 2013). Moreover, the transcript levels of ABI3 are modulated by the BES1-TPL-HDA19 suppressor complex via histone deacetylation (Ryu et al., 2014). HDA19 RNAi plants were found to be insensitive to salt stress, and the MSI1-HDA19-SIN3 complex modulated ABA receptor genes involved in ABA signaling (Mehdi et al., 2016). Considered together, our results demonstrated that although HDA15 protein is expected to inhibit the expression of specific genes by promoting histone deacetylation, it may actually exert the opposite effect (Figure 7). Therefore, further studies, such as those utilizing clustered regularly interspaced short palindromic repeats (CRISPR) technology, are required to identify regions within target DNA that HDA affects in order to regulate gene expression. Such studies may contribute to the development of salt-resistant crop varieties.
Conclusions
In this study, we found that overexpression of HDA15 in Arabidopsis increased the transcription levels of NCED3, which further enhanced ABA synthesis, resulting in resistance to salt stress. We also found that HDA15 requires HY5 to increase the resistance of plants to salt stress. Investigation of the mechanism by which HDA15 increases the transcript levels of NCED3 in response to salt stress indicated that it possibly involves the inhibition of a negative regulator that binds this site via increased histone deacetylation of a specific region in the DNA of NCED3. However, further studies are needed to clearly identify negative regulators that are inhibited by HDA15 in order to fully understand its function under salt stress.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author Contributions
HT conducted the phenotype experiment, ChIP assay, qRT-PCR analysis, statistical analysis, and wrote the manuscript. SL performed the GUS experiment and ABA content assays. CT carried out the chlorophyll assay. WL conducted the MDA assay. E-HC did the proline assay. S-WH and HL designed the experiments and wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by a grant from the National Research Foundation of Korea (to HL, 2017; grant NRF-2017R1A2B4008706).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.640443/full#supplementary-material
References
Abassi, H., Jamil, M., Hag, A., Ali, S., Ahmad, R., Malik, Z., et al. (2016). Salt stress manifestation on plants, mechanism of salt tolerance, and potassium role in alleviating it: a review. Zemdirbyste Agric. 103, 229–238. doi: 10.13080/z-a.2016.103.030
Ali, S., Hayat, K., Iqbal, A., and Xie, L. (2020). Implications of abscisic acid in the drought stress tolerance of plants. Agronomy 10:1323. doi: 10.3390/agronomy10091323
Barrero, J. M., Rodríguez, P. L., Quesada, V., Piqueras, P., Ponce, M. R., and Micol, J. L. (2006). Both abscisic acid (ABA)-dependent and ABA-independent pathways govern the induction of NCED3, AAO3, and ABA1 in response to salt stress. Plant Cell Environ. 29, 2000–2008. doi: 10.1111/j.1365-3040.2006.01576.x
Belin, C., Megies, C., Hauserová, E., and Lopez-Molina, L. (2009). Abscisic acid represses growth of the Arabidopsis embryonic axis after germination by enhancing auxin signaling. Plant Cell 21, 2253–2268. doi: 10.1105/tpc.109.067702
Catalá, R., Medina, J., and Salinas, J. (2011). Integration of low temperature and light signaling during cold acclimation response in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 108, 16475–16480. doi: 10.1073/pnas.1107161108
Chen, H., Zhang, J., Neff, M. M., Hong, S.-W., Zhang, H., Deng, X.-W., et al. (2008). Integration of light and abscisic acid signaling during seed germination and early seedling development. PNAS 105, 4495–4500. doi: 10.1073/pnas.0710778105
Chen, L.-T., Luo, M., Wang, Y.-Y., and Wu, K. (2010). Involvement of Arabidopsis histone deacetylase HDA6 in ABA and salt stress response. J. Exp. Bot. 61, 3345–3353. doi: 10.1093/jxb/erq154
De Oliveira, A. B., Mendes-Alencar, N. L., and Gomes-Filho, E. (2013). “Comparison between the water and salt stress effects on plant growth and development,” in Responses of Organisms to Water Stress, ed S. Akinci (London: IntechOpen), 67–94.
Eun, H.-D., Ali, S., Jung, H., Kim, K., and Kim, W.-C. (2019). Profiling of ACC synthase gene (ACS11) expression in Arabidopsis induced by abiotic stresses. Appl. Biol. Chem. 62:42. doi: 10.1186/s13765-019-0450-4
Fernando, V. C. D., and Schroeder, D. F. (2016). “Role of ABA in Arabidopsis salt, drought, and desiccation tolerance,” in Abiotic and Biotic Stress in Plants–Recent Advances and Future Perspectives, Chapter 22, eds A. K. Shanker and C. Shanker (London: IntechOpen). 508–524.
Gangappa, S. N., and Botto, J. F. (2016). The multifaceted roles of HY5 in plant growth and development. Mol. Plant 9, 1353–1365. doi: 10.1016/j.molp.2016.07.002
Gu, D., Chen, C.-Y., Zhao, M., Zhao, L., Duan, X., Duan, J., et al. (2017). Identification of HDA15-PIF1 as a key repression module directing the transcriptional network of seed germination in the dark. Nucleic Acids Res. 45, 7137–7150. doi: 10.1093/nar/gkx283
Gupta, B., and Huang, B. (2014). Mechanism of salinity tolerance in plants: physiological, biochemical, and molecular characterization. Int. J. Genomics 2014:701596. doi: 10.1155/2014/701596
Han, Z., Yu, H., Zhao, Z., Hunter, D., Luo, X., Duan, J., et al. (2016). AtHD2D gene plays a role in plant growth, development, and response to abiotic stresses in Arabidopsis thaliana. Front. Plant Sci. 7:310. doi: 10.3389/fpls.2016.00310
Henikoff, S. (2005). Histone modifications: combinatorial complexity or cumulative simplicity? Proc. Natl. Acad. Sci. U.S.A. 102, 5308–5309. doi: 10.1073/pnas.0501853102
Huang, Y., Guo, Y., Liu, Y., Zhang, F., Wang, Z., Wang, H., et al. (2018). 9-cis-epoxycarotenoid dioxygenase 3 regulates plant growth and enhances multi-abiotic stress tolerance in rice. Front. Plant Sci. 9:162. doi: 10.3389/fpls.2018.00162
Iuchi, S., Kobayashi, M., Yamaguchi-Shinozaki, K., and Shinozaki, K. (2000). A stress-inducible gene for 9-cis-epoxycarotenoid dioxygenase involved in abscisic acid biosynthesis under water stress in drought-tolerant cowpea. Plant Physiol. 123, 553–562. doi: 10.1104/pp.123.2.553
Jakab, G., Ton, J., Flors, V., Zimmerli, L., Métraux, J.-P., and Mauch-Mani, B. (2005). Enhancing arabidopsis salt and drought stress tolerance by chemical priming for its abscisic acid responses. Plant Physiol. 139, 267–274. doi: 10.1104/pp.105.065698
Jeong, C. Y., Lee, W. J., Truong, H. A., Trịnh, C. S., Hong, S.-W., and Lee, H. (2018). AtMybL-O modulates abscisic acid biosynthesis to optimize plant growth and ABA signaling in response to drought stress. Appl. Biol. Chem. 61, 473–477. doi: 10.1007/s13765-018-0376-2
Kim, J.-M., Sasaki, T., Ueda, M., Sako, K., and Seki, M. (2015). Chromatin changes in response to drought, salinity, heat, and cold stresses in plants. Front. Plant Sci. 6:114. doi: 10.3389/fpls.2015.00114
Kim, S. G., Lee, J.-S., Bae, H. H., Kim, J.-T., Son, B.-Y., Kim, S.-L., et al. (2019). Physiological and proteomic analyses of Korean F1 maize (Zea mays L.) hybrids under water-deficit stress during flowering. Appl. Biol. Chem. 62:32. doi: 10.1186/s13765-019-0438-0
Konsoula, Z., and Barile, F. A. (2012). Epigenetic histone acetylation and deacetylation mechanisms in experimental models of neurodegenerative disorders. J. Pharmacol. Toxicol. Methods 66, 215–220. doi: 10.1016/j.vascn.2012.08.001
Lee, H. G., and Seo, P. J. (2019). MYB96 recruits the HDA15 protein to suppress negative regulators of ABA signaling in Arabidopsis. Nat. Commun. 10:1713. doi: 10.1038/s41467-019-09417-1
Lisar, S. Y. S., Motafakkerazad, R., Hossain, M. M., and Rahman, I. M. M. (2012). “Water stress in plants: causes, effects, and responses,” in Water Stress, eds I. Md, M. Rahaman, and H. Hasegawa (London: IntechOpen), 1–14.
Liu, N., Ding, Y., Fromm, M., and Avramova, Z. (2014). Endogenous ABA extraction and measurement from Arabidopsis leaves. Bio Protoc. 4:e1257. doi: 10.21769/BioProtoc.1257
Liu, S., Lv, Y., Wan, X.-R., Li, L.-M., Hu, B., and Li, L. (2014). Cloning and expression analysis of cDNAs encoding ABA 8'-hydroxylase in peanut plants in response to osmotic stress. PLoS ONE 9:e97025. doi: 10.1371/journal.pone.0097025
Liu, X., Chen, C.-Y., Wang, K.-C., Luo, M., Tai, R., Yuan, L., et al. (2013). PHYTOCHROME INTERACTING FACTOR3 associates with the histone deacetylase HDA15 in repression of chlorophyll biosynthesis and photosynthesis in etiolated Arabidopsis seedlings. Plant Cell 25, 1258–1273. doi: 10.1105/tpc.113.109710
Liu, X., Yang, S., Yu, C.-W., Chen, C.-Y., and Wu, K. (2016). Histone acetylation and plant development. Enzymes 40, 173–199. doi: 10.1016/bs.enz.2016.08.001
Liu, X., Yang, S., Zhao, M., Luo, M., Yu, C.-W., Chen, C.-Y., et al. (2014). Transcriptional repression by histone deacetylases in plants. Mol. Plant 7, 764–772. doi: 10.1093/mp/ssu033
Long, H., Zheng, Z., Zhang, Y., Xing, P., Wan, X., Zheng, Y., et al. (2019). An abscisic acid (ABA) homeostasis regulated by its production, catabolism, and transport in peanut leaves in response to drought stress. PLoS ONE 14:e0213963. doi: 10.1371/journal.pone.0213963
Luo, M., Wang, Y.-Y., Liu, X., Yang, S., Lu, Q., Cui, Y., et al. (2012). HD2C interacts with HDA6 and is involved in ABA and salt stress response in Arabidopsis. J. Exp. Bot. 63, 3297–3306. doi: 10.1093/jxb/ers059
Ma, Q., Xia, Z., Cai, Z., Li, L., Cheng, Y., Liu, J., et al. (2019). GmWRKY16 enhances drought and salt tolerance through an ABA-mediated pathway in Arabidopsis thaliana. Front. Plant Sci. 9:1979. doi: 10.3389/fpls.2018.01979
Ma, Y., Cao, J., He, J., Chen, Q., Li, X., and Yang, Y. (2018). Molecular mechanism for the regulation of ABA homeostasis during plant development and stress responses. Int. J. Mol. Sci. 19:3643. doi: 10.3390/ijms19113643
Mehdi, S., Derkacheva, M., Ramström, M., Kralemann, L., Bergquist, J., and Hennig, L. (2016). The WD40 domain protein MSI1 functions in a histone deacetylase complex to fine-tune abscisic acid signaling. Plant Cell 28, 42–54. doi: 10.1105/tpc.15.00763
Merlot, S., Gosti, F., Guerrier, D., Vavasseur, A., and Giraudat, J. (2001). The ABI1 and ABI2 protein phosphatases 2C act in a negative feedback regulatory loop of the abscisic acid signalling pathway. Plant J. 25, 295–303. doi: 10.1046/j.1365-313x.2001.00965.x
Msanne, J., Lin, J., Stone, J. M., and Awada, T. (2011). Characterization of abiotic stress-responsive Arabidopsis thaliana RD29A and RD29B genes and evaluation of transgenes. Planta 234, 97–107. doi: 10.1007/s00425-011-1387-y
Nam, K.-H., Kim, D. Y., Kim, H. J., Pack, I.-S., Kim, H. J., Chung, Y. S., et al. (2019). Global metabolite profiling based on GC–MS and LC–MS/MS analyses in ABF3-overexpressing soybean with enhanced drought tolerance. Appl. Biol. Chem. 62:15. doi: 10.1186/s13765-019-0425-5
Pandey, R., Müller, A., Napoli, C. A., Selinger, D. A., Pikaard, C. S., Richards, E. J., et al. (2002). Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res. 30, 5036–5055. doi: 10.1093/nar/gkf660
Pardo, J. M. (2010). Biotechnology of water and salinity stress tolerance. Curr. Opin. Biotechnol. 21, 185–196. doi: 10.1016/j.copbio.2010.02.005
Perrella, G., Lopez-Vernaza, M. A., Carr, C., Sani, E., Gosselé, V., Verduyn, C., et al. (2013). Histone deacetylase complex1 expression level titrates plant growth and abscisic acid sensitivity in Arabidopsis. Plant Cell 25, 3491–3505. doi: 10.1105/tpc.113.114835
Ryu, H., Cho, H., Bae, W., and Hwang, I. (2014). Control of early seedling development by BES1/TPL/HDA19-mediated epigenetic regulation of ABI3. Nat. Commun. 5:4138. doi: 10.1038/ncomms5138
Sah, S. K., Reddy, K. R., and Li, J. (2016). Abscisic acid and abiotic stress tolerance in crop plants. Front. Plant Sci. 7:571. doi: 10.3389/fpls.2016.00571
Saleh, A., Alvarez-Venegas, R., and Avramova, Z. (2008). An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nat. Protoc. 3, 1018–1025. doi: 10.1038/nprot.2008.66
Sato, H., Takasaki, H., Takahashi, F., Takamasa, S., Satoshi, I., Nobutaka, M., et al. (2018). Arabidopsis thaliana NGATHA1 transcription factor induces ABA biosynthesis by activating NCED3 gene during dehydration stress. Proc. Natl. Acad. Sci. U.S.A. 115, E11178–E11187. doi: 10.1073/pnas.1811491115
Seo, J. S., Zhao, P., Jung, C., and Chua, N.-H. (2019). PLANT U-BOX PROTEIN 10 negatively regulates abscisic acid response in Arabidopsis. Appl. Biol. Chem. 62:39. doi: 10.1186/s13765-019-0446-0
Shahbazian, M. D., and Grunstein, M. (2007). Functions of site-specific histone acetylation and deacetylation. Annu. Rev. Biochem. 76, 75–100. doi: 10.1146/annurev.biochem.76.052705.162114
Skubacz, A., Daszkowska-Golec, A., and Szarejko, I. (2016). The role and regulation of ABI5 (ABA-Insensitive 5) in plant development, abiotic stress responses, and phytohormone crosstalk. Front. Plant Sci. 7:1884. doi: 10.3389/fpls.2016.01884
Song, C.-P., Agarwal, M., Ohta, M., Guo, Y., Halfter, U., Wang, P., et al. (2005). Role of an Arabidopsis AP2/EREBP-type transcriptional repressor in abscisic acid and drought stress responses. Plant Cell 17, 2384–2396. doi: 10.1105/tpc.105.033043
Sridha, S., and Wu, K. (2006). Identification of AtHD2C as a novel regulator of abscisic acid responses in Arabidopsis. Plant J. 46, 124–133. doi: 10.1111/j.1365-313X.2006.02678.x
Tan, B.-C., Joseph, L. M., Deng, W.-T., Liu, L., Li, Q.-B., Cline, K., et al. (2003). Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J. 35, 44–56. doi: 10.1046/j.1365-313X.2003.01786.x
Tang, Y., Liu, X., Liu, X., Li, Y., Wu, K., and Hou, X. (2017). Arabidopsis NF-YCs mediate the light-controlled hypocotyl elongation via modulating histone acetylation. Mol. Plant 10, 260–273. doi: 10.1016/j.molp.2016.11.007
Truong, H. A., Jeong, C. Y., Lee, W. J., Lee, B. C., Chung, N., Kang, C.-S., et al. (2017). Evaluation of a rapid method for screening heat stress tolerance using three Korean wheat (Triticum aestivum L.) cultivars. J. Agric. Food Chem. 65, 5589–5597. doi: 10.1021/acs.jafc.7b01752
Ueda, M., Matsui, A., Tanaka, M., Nakamura, T., Abe, T., Sako, K., et al. (2017). The distinct roles of class I and II RPD3-like histone deacetylases in salinity stress response. Plant Physiol. 175, 1760–1773. doi: 10.1104/pp.17.01332
Wang, Z., Cao, H., Sun, Y., Li, X., Chen, F., Carles, A., et al. (2013). Arabidopsis paired amphipathic helix proteins SNL1 and SNL2 redundantly regulate primary seed dormancy via abscisic acid-ethylene antagonism mediated by histone deacetylation. Plant Cell 25, 149–166. doi: 10.1105/tpc.112.108191
Yang, B., Song, Z., Li, C., Jiang, J., Zhou, Y., Wang, R., et al. (2018). RSM1, an Arabidopsis MYB protein, interacts with HY5/HYH to modulate seed germination and seedling development in response to abscisic acid and salinity. PLoS Genet. 14:e1007839. doi: 10.1371/journal.pgen.1007839
Yao, C., and Finlayson, S. A. (2015). Abscisic acid is a general negative regulator of Arabidopsis axillary bud growth. Plant Physiol. 169, 611–626. doi: 10.1104/pp.15.00682
Yoon, Y., Seo, D. H., Shin, H., Kim, H. J., Kim, C. M., and Jang, G. (2020). The role of stress-responsive transcription factors in modulating abiotic stress tolerance in plants. Agronomy 10:788. doi: 10.3390/agronomy10060788
Zhao, L., Peng, T., Chen, C.-Y., Ji, R., Gu, D., Li, T., et al. (2019). HY5 Interacts with the histone deacetylase HDA15 to repress hypocotyl cell elongation in photomorphogenesis. Plant Physiol. 180, 1450–1466. doi: 10.1104/pp.19.00055
Zheng, M., Liu, X., Lin, J., Liu, X., Wang, Z., Xin, M., et al. (2019). Histone acetyltransferase GCN5 contributes to cell wall integrity and salt stress tolerance by altering the expression of cellulose synthesis genes. Plant J. 97, 587–602. doi: 10.1111/tpj.14144
Zheng, Y., Ding, Y., Sun, X., Xie, S., Wang, D., Liu, X., et al. (2016). Histone deacetylase HDA9 negatively regulates salt and drought stress responsiveness in Arabidopsis. J. Exp. Bot. 67, 1703–1713. doi: 10.1093/jxb/erv562
Keywords: Arabidopsis, HDA15 gene, salinity, stress tolerance, histone modification, ABA accumulation
Citation: Truong HA, Lee S, Trịnh CS, Lee WJ, Chung E-H, Hong S-W and Lee H (2021) Overexpression of the HDA15 Gene Confers Resistance to Salt Stress by the Induction of NCED3, an ABA Biosynthesis Enzyme. Front. Plant Sci. 12:640443. doi: 10.3389/fpls.2021.640443
Received: 11 December 2020; Accepted: 22 March 2021;
Published: 30 April 2021.
Edited by:
Kyung-Nam Kim, Sejong University, South KoreaReviewed by:
Qibin M. A., South China Agricultural University, ChinaDaisuke Todaka, The University of Tokyo, Japan
Copyright © 2021 Truong, Lee, Trịnh, Lee, Chung, Hong and Lee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hojoung Lee, bGhvam91bmdAa29yZWEuYWMua3I=; Suk-Whan Hong, c3Vrd2hhbkBjaG9ubmFtLmFjLmty
 Hai An Truong
Hai An Truong Seokjin Lee1
Seokjin Lee1 Won Je Lee
Won Je Lee Hojoung Lee
Hojoung Lee