
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 15 March 2021
Sec. Plant Physiology
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.637352
This article is part of the Research TopicPlant Glycobiology - A Sweet World of Glycans, Glycoproteins, Glycolipids, and Carbohydrate-Binding ProteinsView all 33 articles
 Anna Konkina
Anna Konkina Mariola Klepadlo
Mariola Klepadlo Abdellah Lakehal
Abdellah Lakehal Zein El Zein
Zein El Zein Afroditi Krokida
Afroditi Krokida Mina Botros
Mina Botros Michail Iakovidis
Michail Iakovidis Pavel Chernobavskiy
Pavel Chernobavskiy Mohamed Elfatih Zerroumda
Mohamed Elfatih Zerroumda George Tsanakas
George Tsanakas Nikos Petrakis
Nikos Petrakis Athanasia-Maria Dourou
Athanasia-Maria Dourou Panagiotis Kalaitzis*
Panagiotis Kalaitzis*Plant responses to flooding, submergence and waterlogging are important for adaptation to climate change environments. Therefore, the characterization of the molecular mechanisms activated under hypoxic and anoxic conditions might lead to low oxygen resilient crops. Although in mammalian systems prolyl 4 hydroxylases (P4Hs) are involved in the oxygen sensing pathway, their role in plants under low oxygen has not been extensively investigated. In this report, an Arabidopsis AtP4H3 T-DNA knock out mutant line showed higher sensitivity to anoxic treatment possibly due to lower induction of the fermentation pathway genes, ADH and PDC1, and of sucrose synthases, SUS1 and SUS4. This sensitivity to anoxia was accompanied by lower protein levels of AGPs-bound epitopes such as LM14 in the mutant line and induction of extensins-bound epitopes, while the expression levels of the majority of the AGPs genes were stable throughout a low oxygen time course. The lower AGPs content might be related to altered frequency of proline hydroxylation occurrence in the p4h3 line. These results indicate active involvement of proline hydroxylation, a post-translational modification, to low oxygen response in Arabidopsis.
Oxygen is important for cellular respiration in all eukaryotes, including plants, which often encounter oxygen deficiency conditions during their life cycle such as root waterlogging and submergence and as a result have evolved various mechanisms of sensing and response to oxygen availability (Gibbs and Greenway, 2003; Loreti and Perata, 2020). Oxygen deprivation triggers various changes at different levels, such as metabolic shifts (Roberts et al., 1984; Drew, 1997; Planchet et al., 2017), alterations in gene expression (Branco-Price et al., 2005; Loreti et al., 2005; Vlad et al., 2007b), post-translational modifications (Iacopino and Licausi, 2020), and morphological changes (Perata and Alpi, 1993; Eysholdt-Derzsó and Sauter, 2017).
The activation of glycolysis for ATP production (Perata and Alpi, 1993; Gibbs and Greenway, 2003) and fermentative metabolism due to the upregulation of pyruvate decarboxylase (PDC) and alcohol dehydrogenase (ADH), leads to increased plant tolerance (Knox et al., 1995; Ismond et al., 2003; Kürsteiner et al., 2003). Moreover, knock-out mutants of Arabidopsis PDC1 and PDC2 resulted in higher susceptibility to submergence (Mithran et al., 2014), and mutants of rice and Arabidopsis ADH suppressed seed germination under anoxia (Jacobs et al., 1988; Matsumura et al., 1995; Banti et al., 2008). The glycolytic flow is maintained by the induction of energy efficient sucrose synthase (SUSs) genes such as SUS1 and SUS4 in Arabidopsis which were shown to be up-regulated under hypoxia (Santaniello et al., 2014).
In plants, oxygen sensing is mediated by the regulation of protein stability of group VII ERF (Ethylene Response Factor) cluster of transcription factors via the N-end rule pathway (Gibbs et al., 2011; Weits et al., 2014). A major component of this pathway is the Plant Cysteine Oxidase genes (PCO) which oxidize a Cys at the N-terminus of the VII ERFs polypeptide in an oxygen dependent manner, thus targeting VII ERFs to proteasomal degradation (Weits et al., 2014). The VII ERFs control the expression of anaerobic genes by binding to Hypoxia Response Promoter Element motifs present in anaerobic genes such as ADH, PDC, LBD41 (LOB Domain-Containing Protein 41), HRE1 as well as HRE2 (Hypoxia Responsive ERF 1 and 2), HRA1 (Hypoxia Response Attenuator) (Gasch et al., 2016; Giuntoli and Perata, 2018).
In the metazoans, the prolyl-4-hydroxylases (PHD, prolyl-4-hydroxylase domain) act as oxygen sensors by hydroxylation of HIF1α (Hypoxia Inducible Factor 1A) transcription factor (Wang and Semenza, 1993; Kaelin and Ratcliffe, 2008). Under normoxia PHDs mediate hydroxylation of proline in HIF1α, thus targeting it to proteasomal degradation (Pugh and Ratcliffe, 2003). Under hypoxia, HIFα accumulates, dimerizes with an HIFβ family member, translocates to the nucleus, and transcriptionally activates more than 100 genes (Mcneill et al., 2002; Kaelin and Ratcliffe, 2008; Myllyharju and Koivunen, 2013).
Plant prolyl-4-hydroxylases (P4H) are non-heme proteins belonging to the 2-oxoglutarate-dependent dioxygenase family (2-ODD) requiring molecular oxygen to catalyze proline hydroxylation of Hydroxyproline Rich Glycoproteins (HRGPs) and additional proteins (Clifton et al., 2006; Farrow and Facchini, 2014). This important post-translational modification (PTM) was detected in extensins (Velasquez et al., 2011, 2012), arabinogalactan proteins (AGP) (Schultz et al., 2004), as well as secreted peptide hormones (Kondo et al., 2006; Stührwohldt et al., 2020) and tyrosine-sulfated glycopeptides (Amano et al., 2007). In tomato (Solanum lycopersicum) only the SlAGP4 transcript abundance increased in response to an anoxic time course in mature green fruit pericarp tissue, while AGPs-bound epitopes were either constitutively expressed or upregulated (Fragkostefanakis et al., 2012). In addition to their involvement in various abiotic stresses in plants (Lamport et al., 2006; Mareri et al., 2019), fasciclin-like arabinogalactans (FLAs) and extensins (EXT) showed alterations in transcript levels in a deep-water rice cultivar of high tolerance to anaerobiosis, while three FLAs and two EXTs were upregulated (Minami et al., 2018). Several Arabidopsis AGPs were differentially expressed in a tissue-specific manner in roots under hypoxia and anoxia (Mustroph et al., 2009), while 22 Arabidopsis HRGPs, including five AGPs and five extensins, were identified in silico to exhibit alterations in expression patterns under anoxia (Schultz et al., 2002; Vlad et al., 2007a). Interestingly, five out of 13 Arabidopsis P4Hs were co-expressed with various AGPs suggesting putative interaction (Showalter et al., 2010). These data suggest possible involvement of AGPs in hypoxic and anoxic adaptation. Moreover, overexpression of Arabidopsis AtP4H1 resulted in higher expression of hypoxia-induced genes such as ADH1, PDC1, PDC2, and SUS1 (Asif et al., 2009).
In this study, the physiological role of AtP4H3 under hypoxic and anoxic conditions was investigated. An AtP4H3 T-DNA knock out mutant line exhibited reduced survival percentages under anoxia probably due to lower induction of the fermentation pathway genes, while the AGPs transcript abundance and bound-epitopes remained stable under hypoxic conditions. Moreover, analysis of Arabidopsis lines transformed with a construct of AtP4H3 promoter directing GUS expression indicated tissue specific expression mainly in root stele and columella cells, leaf tips and stipules under hypoxia.
The A. thaliana knock-out mutant line of AtP4H3 (At1g20270) N576446 (SALK_076446) was obtained from the European Arabidopsis Stock Centre (NASC)1. Seeds of Arabidopsis thaliana p4h3-2 mutants were surface-sterilized (30 s 100% ethanol followed by 5 min in 50% bleach), rinsed and allowed to imbibe at 4°C. After 4 days, seeds were transferred to plates with solid MS media (Appendix), and placed in a growth chamber (Snijders Scientific) in vertical orientation under long-day conditions [16 h light (100% intensity) and 8 h dark] with 70% humidity at 22°C during the light cycle and 16°C during the dark cycle. After 10 days, surviving seedlings were transferred to Murashige and Skoog (MS) medium without antibiotic selection. Two weeks later, plants were transferred into jars, and then to soil. Each plant was individually labeled, bagged, and placed separately to avoid cross-pollination.
To select for mutant plants following kanamycin selection on MS plates, 200 mg of leaf tissue from 6-week-old plants was frozen with liquid nitrogen and ground by pestle and mortar. 500 μl of DNA extraction buffer (200 mM Tris–HCl pH = 8.0; 250 mM NaCl; 25 mM EDTA pH = 8.0; 0.5% SDS) was added and samples were mixed by inversion several times. Then, an equal volume of phenol: chloroform 1:1 was added to the samples, vortexed for 20 s, centrifuged at 13000 rpm for 10 min and supernatants were transferred to new tubes. Cold isopropanol in 1:1 volume was added, samples were mixed by inversion and placed in −20°C for 1 h. Samples were centrifuged, washed with 70% ethanol, air-dried and resuspended in 150 μl of sterile distilled H2O. 20 μl of RNase A (10 mg/ml) was added and samples were then incubated for 1 h at 37°C. An equal volume of chloroform was added again, vortexed and centrifuged for 10 min. The supernatants were transferred to new tubes and washed with 2.5:1 volumes of 100% and 70% ethanol. Dried pellets were resuspended in 50 μl of sterile distilled H2O and stored at −20°C. DNA was quantified by an IMPLEN NanoPhotometerTM Pearl.
Total RNA was isolated from roots of 7 day-old Arabidopsis Col-0 plants using PureLink® RNA Mini Kit by Ambion. 200–300 mg of frozen tissue was homogenized in liquid nitrogen and following steps were done according to the PureLink® RNA Mini Kit instructions. Quality control for RNA was performed with GelRed® Nucleic Acid Gel Stain (Biotium) on a 1.5% denaturing agarose gel electrophoresis with 2 μg RNA and a NanoPhotometer (NanoPhotometerTM Pearl, Implen). cDNA was synthesized using 5 μg of RNA with oligo (dT)15 primers and the SuperScript® Synthesis VILO cDNA kit (Invitrogen) according to the manufacturer’s guidelines.
A modified protocol based on Woodson and Handa (1987) was used to extract total protein from wild type and p4h3 plants (Woodson and Handa, 1987). Plant tissue was ground in liquid nitrogen and lysed afterward (Woodson and Handa, 1987). Extracted proteins were separated by sodium dodecyl sulfate-polyacrylamide 12% (pH 8.8) gel electrophoresis (SDS-PAGE) (Sambrook and Russell, 2006). The gel was transferred onto polyvinyl fluoride membrane (MILLIPORE Immobilon-P, 0.45 mm pore size, IPVH00010) at a constant current of 35 V for 2 h. Blots were soaked in blocking solution containing 5% non-fat dry milk (NFDM) in Tris Buffered Saline (TBS) solution [10 mM Tris HCl, 150 mM NaCl, 0.05% (v/v) Tween-20, 0.01% (w/v) Na Azide, pH 8.0] with shaking at 4°C for 3 h. Then membranes were incubated with LM14 (Moller et al., 2008), JIM20 (Knox et al., 1995), or MAC207 (Pennell et al., 1989) (Paul Knox Cell Wall Lab, University of Leeds, United Kingdom) rat monoclonal antibodies in 2.5% NFDM containing TBS-Tween solution at 4°C overnight. The Actin (Agrisera AB, Vännäs, Sweden) was used as a loading control. The bound antibodies were labeled with the Goat anti-Rat IgG Horse radish peroxidase (HRP conjugate) (MILLIPORE), washed and incubated with a chemiluminescent solution (SuperSignal West Pico Chemiluminescent Substrate, Thermo Scientific, 34077). After 1 min incubation, the membranes were exposed to X-Ray film.
Primers for sequencing and genotyping mutant lines were designed based on using http://signal.salk.edu/tdnaprimers.2.html and http://primer3.ut.ee/, respectively (Supplementary Table 1). To generate 35S:P4H3 transgenic plants, we delimited the open reading frame of AtP4H3 using the ORF Finder2. The DNA fragment was subcloned using the Gateway recombination technology (TOPO Cloning© system) into pCR©II-TOPO© vector according to the manufacturer’s instructions (INVITROGEN)3. Plasmids were verified by digestion and transferred to the pK7WG2D Gateway-compatible destination binary vector4 by recombination reactions. The generation of P4H3pro:GFP:GUS transgenic lines comprised of the amplification of a 1991 bp AtP4H3 promoter region upstream of the translation initiation codon of AtP4H3 by using Col-0 genomic DNA as template. PCR products were subcloned using the Gateway recombination technology (TOPO Cloning© system) into pCR©II-TOPO© vector according to the manufacturer’s instructions (INVITROGEN, see text footnote 3). Plasmids were verified by digestion and transferred to the pKGWFS7.0 Gateway-compatible binary vector (see text footnote 4) by recombination reactions. The AtP4H3 gene construct comprising the entire genomic sequence of 3497 bp for generating complementation lines was amplified from genomic DNA (wild type Col-0) and subcloned into pCR©II-TOPO© vector according to the manufacturer’s instructions (INVITROGEN, see text footnote 3) using the Gateway recombination technology (TOPO Cloning© system). Plasmids were verified by digestion and transferred to the pBGWFS7.0 Gateway-compatible binary vector (see text footnote 4) by recombination reactions. Agrobacterium tumefaciens strain GV3101 (pMP90) was used for transforming Arabidopsis plants (Zhang et al., 2006). Independent transformants on kanamycin selection were confirmed by PCR (Supplementary Table 1).
Localization of AtP4H3 promoter activity was monitored in 8-day old seedlings growing on MS medium containing P4H3pro:GUS:GFP. The staining procedure was performed according to Li (2011). For histochemical detection of GUS activity, 7-day old seedlings were collected in Eppendorf tubes and placed on ice along with cold 90% acetone. Following 20 min incubation at 37°C, acetone was removed and plants were washed with rinsing buffer. Then, a staining buffer with X-Gluc was added to each tube and seedlings were placed under vacuum for 5 min. Then the samples were infiltrated under vacuum, on ice, for 30 min and incubated at 37°C under darkness overnight. After an overnight incubation staining buffer was removed and seedlings were washed for 30 min consecutively in 20%, 35%, and 50% ethanol. FAA fixative was then added and seedlings were incubated for another 30 min. FAA was removed, 70% ethanol was added and seedlings were stored at 4°C. Images were collected under an optical microscope Leica MZ FL III at 10x objective with a DC 300F digital camera.
Hypoxia and anoxia conditions were replicated as outlined before (Vlad et al., 2007a). Seven day old seedlings were treated through an open system in Petri dishes with 1.5% O2 plus N2 (hypoxia) or 100% N2 (anoxia) in the dark at room temperature. The Petri dishes were placed in 1 L airtight jars connected in parallel and a 40–45 ml per min gas stream was passed through the system. Concentrations for O2 and CO2 in the outlet stream were monitored using a CO2/O2 analyzer (PCO2; Gas Data Ltd., Coventry, United Kingdom). Air-treated control plants were left under aerobic conditions. Time-course measurements began when the jars were fully purged with gas. For each time point (0-untreated/control, 1, 2, 4, and 6 h), samples were taken for roots and shoots separately, flash frozen in liquid nitrogen and stored at −80°C.
An anoxia survival assay was performed as outlined previously (Vlad et al., 2007a). Agar plates containing 7 day old seedlings were transferred in plates with anoxia (100% N2) for 7.5 h in the dark. Plates were then transferred to a growth chamber for post-anoxic recovery (16 h/8 h, 22°C/16°C, and light/dark photoperiod). Survival rates were measured at 3–4 days post-treatment and averages were taken from three biological replicates. Each replicate comprised of three technical replicates, with each replicate consisting of at least 50 plants for each line. Data were subjected to statistical analysis based on the regression analysis generalized linear model, using GenStat 12.1 software.
Real time PCR for Arabidopsis gene expression in response to hypoxia 1.5% O2 was conducted on a CFX Connect machine (BIORAD®) using SYBR® Select Master Mix (Applied Biosystems®). Specificity of primers was determined by dissociation kinetics for PCR products by the end of each run and subsequent analysis of these products using 2% agarose gel electrophoresis stained with GelRed (Biotium). Standard curves were established for each gene using serially diluted cDNA (8, 4, 2, and 1 μg). A linear relationship between the threshold cycle and the log of the starting cDNA concentration was determined for each gene in order to check the efficiency of the PCR. The Arabidopsis Actin gene (ACTIN2 – At3g18780) was used as an internal control.
For progeny test of seed stocks, one hundred seedlings were tested for kanamycin resistance to observe deviations from Mendelian segregation ratios using Chi-square (χ2) statistical tests. Gene expression data were analyzed using the 2–ΔΔCt method (Livak and Schmittgen, 2001) and presented as relative levels of gene expression. To determine relative fold differences for each sample, the Ct value for each gene was normalized to the Ct value for ACTIN2.
All measurements were conducted in triplicate (three technical replicates per biological replicate and three biological replicates) and results are expressed as mean ± standard errors of the relative expression (SEs). Then, all the data from the pairwise ΔCt of each technical replicate, were subjected to statistical analysis of variance (ANOVA) and a post hoc multicomparison test (Tukey’s honest significant difference criterion, 95% confidence interval) by using the statistics toolbox of MATLAB (The MathWorks Inc., Natick, MA, United States). The comparisons were contacted to identify the statistically significant differences between wild type (WT) and the p4h3-2 line, at each timestep of the time-course experiment, respectively. Differences at P < 0.05 were considered to be significant.
The reason why in some cases the expression between the control and the mutant line is not statistically significant, even though the expression levels with the error bars do not overlap, is that the statistical analysis was performed on the pairwise ΔCt values for each technical replicate of all three biological replicates. This methodology was selected in order to include all the data and not only the mean values of the three biological replicates (Goni et al., 2009; Pabingera et al., 2014).
The Arabidopsis genome comprises 13 putative P4Hs with various patterns of expression under anoxia, hypoxia and mechanical wounding (Vlad et al., 2007a). Only AtP4H3 was induced under anoxia and two hypoxic concentrations of 1.5% and 5% O2, while AtP4H4 was expressed under both hypoxic conditions (Vlad et al., 2007a).
The tissue-specific expression of AtP4H3 was determined in various organs and developmental stages by RT-PCR (Figure 1A). The AtP4H3 was expressed at the seedling stages (1–3 weeks) in both roots and leaves, in young inflorescences (6 weeks) and siliques (8 weeks), while there was no expression at the seed stage and only minimal at 8 week-old inflorescences (Figure 1A).
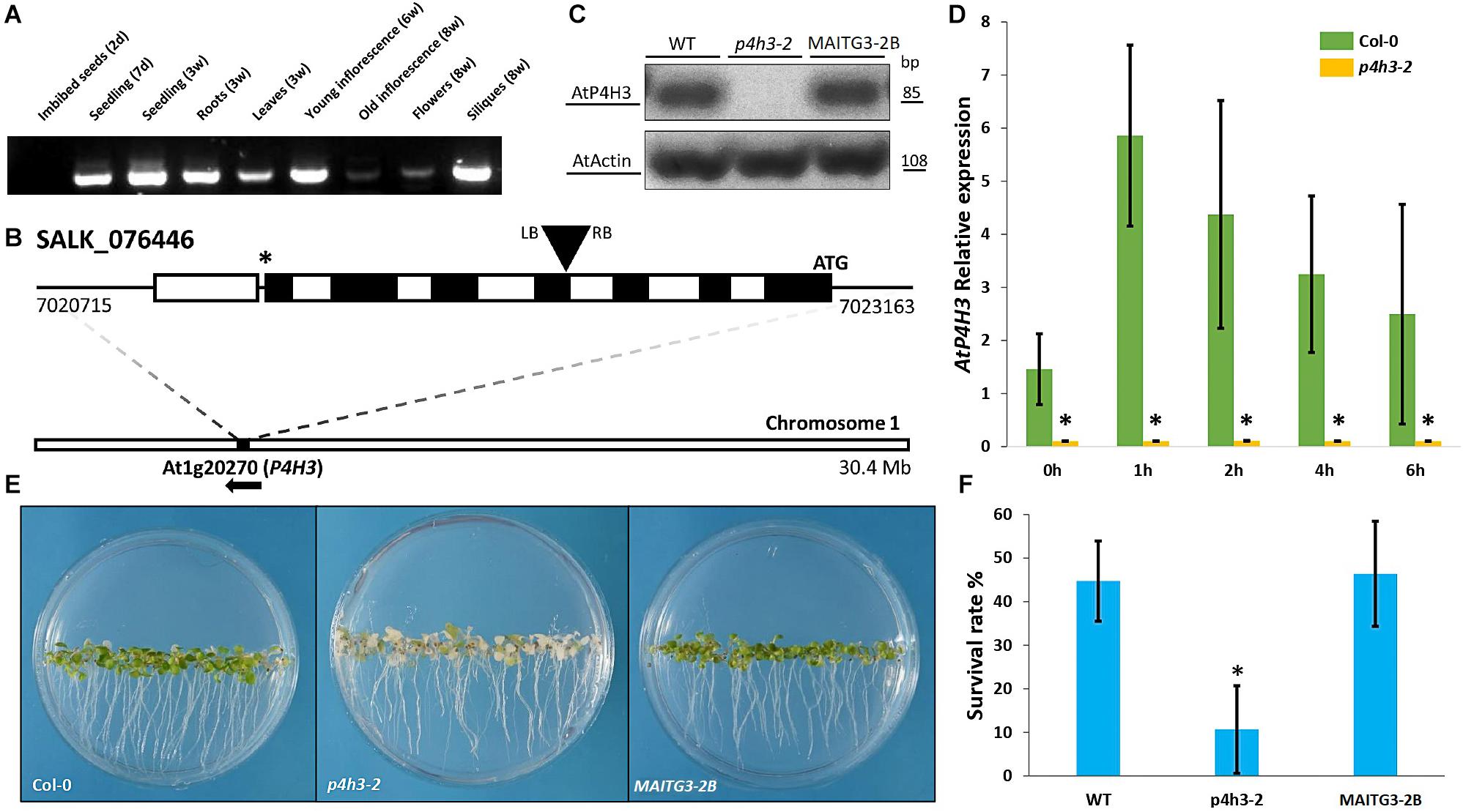
Figure 1. Effects of AtP4H3 on oxygen deprivation tolerance. (A) Tissues-specific expression of AtP4H3 gene in Arabidopsis thaliana (Col-0) by using reverse transcription-PCR. (B) T-DNA mutant line insertion schematic of the genomic region corresponding to P4H3. The positions of the T-DNA insertion (black triangle), the initiating codon (ATG), and the stop codon (*) are indicated, along with the location and direction of the gene (At1g20270) on chromosome 1. Genomic AtP4H3 sequences are represented by exons (black boxes), introns (white boxes), and untranslated regions (black line). The T-DNA orientation is indicated by left (LB) and right (RB) borders. Coordinates are in bp and Mb at the gene and chromosome level, respectively. Features are drawn to scale. (C) AtP4H3 transcript levels in wild type, T-DNA mutant line (p4h3-2) and complementation (compl) line #2 in 7 day-old seedlings. The RT-PCR was performed with qPCR primers. Actin was used as internal control. (D) Relative expression of AtP4H3 in Col-0 (green bars) and p4h3-2 mutant (gold bars) plants in response to a hypoxic time course by using qPCR. Relative expression levels are shown as fold change values and the error bars represent standard errors. (E) Phenotypes of the wild type (Col-0), p4h3-2, and complementation line 2 exposed to 100% N2 for 7.5 h after a 3-day post-anoxic recovery. (F) Survival rates of 7 day-old Arabidopsis plants after 100% N2 treatment for 7.5 h. Fifty seeds were placed per 10 mm Petri dishes. Survival rates were scored as a percentage following 3 days of post-anoxic recovery under normal conditions (21% O2, air). Data are shown as mean survival rates, error bars show Standard error. The experiment was repeated at least three times.
The physiological significance of AtP4H3 under oxygen deficiency conditions was investigated by characterizing the anoxic response of a p4h3-2 T-DNA line (Figure 1B). This mutant line was carrying an insertion at the 4th exon of AtP4H3 (SALK_076446) which was verified by genotyping (Figure 1B). The transcript abundance of AtP4H3 was determined in 7-day old seedlings of wild type, p4h3-2 T-DNA line and AtP4H3 complementation line #2 (MAITG3-2B) by using a RT-PCR approach (Figure 1C). No expression was observed in the p4h3-2 line while mRNA levels were detected in wild type and MAITG3-2B (Figure 1C). Homozygous p4h3-2 plants developed aerial lateral inflorescences inconsistently and further investigation will be required to characterize the phenotype (Supplementary Figure 1).
Seven day-old seedlings of wild type Col-0 and p4h3-2 were exposed to hypoxia and the AtP4H3 transcript levels were determined (Figure 1D). Low oxygen induced the AtP4H3 expression in wild type throughout the 6-h time course, as expected, peaking after 1 h of hypoxia, while no detectable mRNA levels were observed for the p4h3-2 line (Figure 1D).
Hypoxia tolerance assays indicated that Arabidopsis can easily survive at 1% O2 in the dark for several days (Licausi et al., 2010). The 7 day-old seedlings of p4h3-2 line, MAITG3-2B and wild type were exposed to anoxia for 7.5 h in order to determine their survival percentage (Figures 1E,F). After a 3-day post-anoxia period, most of the p4h3-2 seedlings turned white and died while most of the Col-0 and MAITG3-2B seedlings were green and continued to grow (Figures 1E,F). The p4h3-2 survival percentage was very low at the levels of 10.66% while in Col-0 and MAITG3-2B it was much higher at the levels of 44.74% and 46.46%, respectively (Figures 1E,F).
The spatial and temporal expression of AtP4H3 in young seedlings subjected to hypoxia and anoxia was investigated by analyzing stable Arabidopsis transgenic lines transformed with an P4H3pro:GUS:GFP construct. Several independent transgenic lines were generated and three of them were selected for further characterization. GUS staining indicated tissue specific expression of AtP4H3 consistently in the root stele (Figures 2A,c,f,h), root tips (Figures 2A,g), stipules (Figures 2A,d,i), and leaf tips (Figures 2A,e,j) of 5-day old seedlings. Hypoxia treatment for 3 h significantly increased GUS expression in columella cells, root stele and stipules while strong induction was observed in the stele of the root tip differentiation zone (Figure 2A). Moreover, a 1.5% O2 hypoxic and an anoxic time course experiment was performed to determine GUS expression patterns in the root tip (Figure 2B). The highest upregulation was observed after 4 h of hypoxia and anoxia with detection of GUS staining also in lateral root cup in addition to columella cells (Figure 2B). These results indicate that AtP4H3 is induced under oxygen deficiency conditions within 4 h of exposure and this induction is sustained up to 6 h (Figure 2).
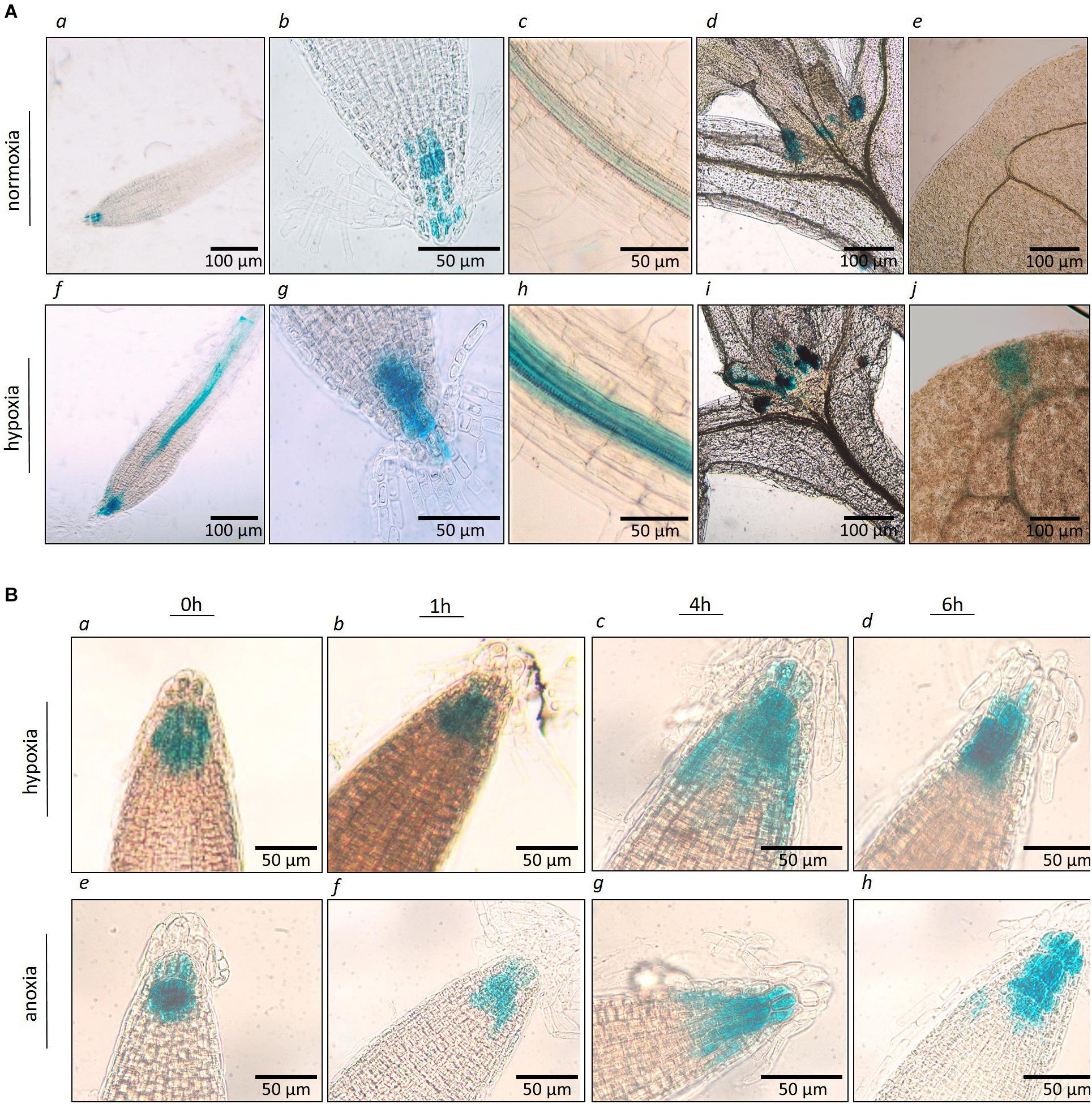
Figure 2. (A) Histochemical localization of AtP4H3 promoter-GUS expression in 5 day-old transgenic Arabidopsis seedlings treated with 21% (a–e) and 1.5% O2 (f–j) for 3 h. (a,f) AtP4H3 promoter activity in columella cells and vasculature in differentiation and elongation root zones. (b,g) AtP4H3 promoter activity in columella cells in root tips. (c,h) AtP4H3 promoter activity in the stele. (d,i) AtP4H3 promoter activity in shoot apex region. (e,j) AtP4H3 promoter activity in leaf tips. The assay consisted of three biological replicates, with each biological replicate including 3–5 technical replicates. (B) Histochemical localization of AtP4H3 promoter-GUS expression in 7 day-old transgenic Arabidopsis seedlings treated with hypoxia (1.5% O2) and anoxia. (a–d) AtP4H3 promoter activity in root tips under hypoxia (1.5% O2) time-course. (e–h) AtP4H3 promoter activity in root tips under anoxia time-course.
Considering the significantly lower survival percentage of p4h3-2 line, the possible effect of AtP4H3 on anaerobic response marker genes was investigated, despite the fact that AtP4H3 catalyzes a post-translational modification. The ADH1, PDC1, SUS1, SUS4 and the TFs, HRE1, and HRE2, were selected for expression analysis during a 1.5% O2 hypoxic time course by using qPCR analysis (Figure 3). The transcript levels of ADH1, PDC1, SUS1, and SUS4 were down-regulated in the mutant compared to Col-0 control for most of the 1-, 2-, 4-, and 6-h time points (Figure 3). The ADH1 expression, for most time points, was decreased by 50–60 fold in the p4h3-2 line, while lower fold change induction was observed for PDC1, SUS1, and SUS4 compared to Col-0 (Figure 3). Strong induction by 14- and 100-fold was detected in HRE1 and HRE2 transcript abundance in response to hypoxia, respectively (Figure 3). However, similar HRE1 and HRE2 mRNA levels were detected in both p4h3-2 and Col-0 seedlings, indicating that these hypoxia inducible TFs were not involved in the differential expression patterns of the anaerobic marker genes in p4h3-2 line under 1.5% O2 conditions (Figure 3).
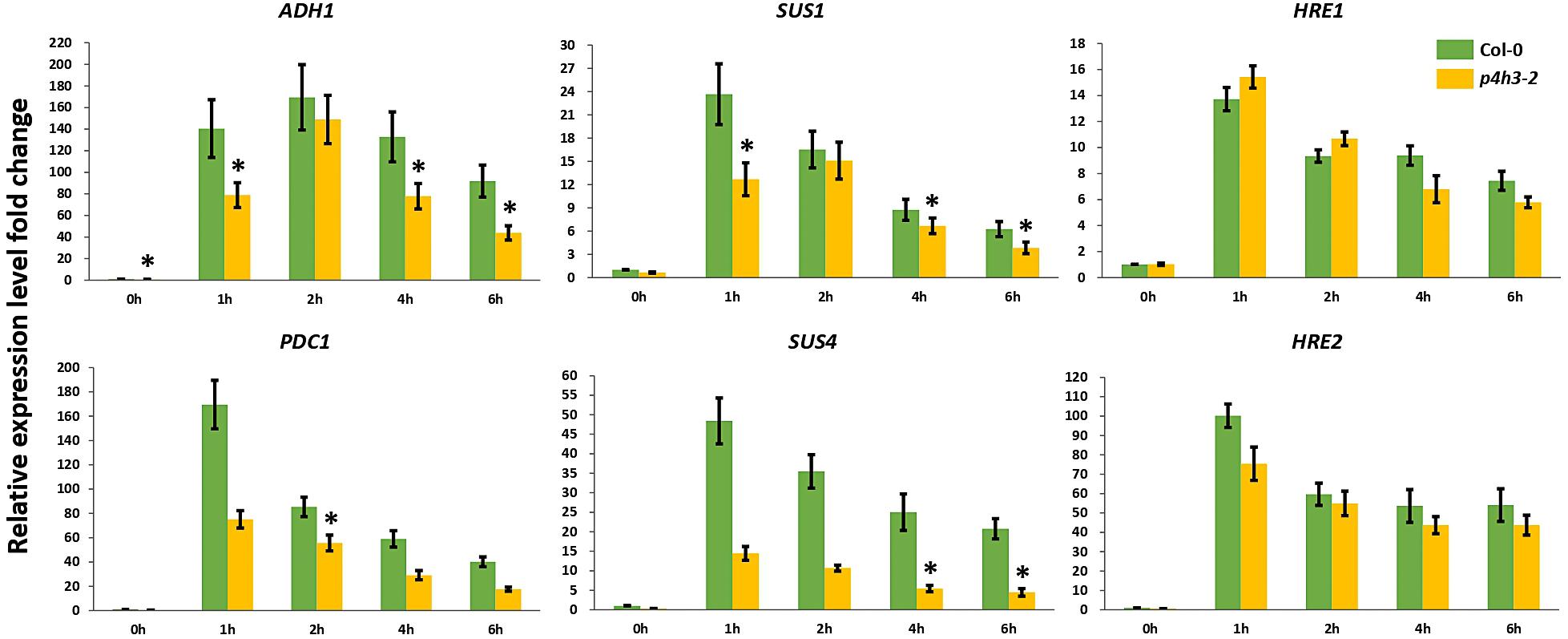
Figure 3. Expression of anaerobic marker genes under hypoxia. The expression of six genes (ADH1, PDC1, SUS1, SUS4, HRE1, and HRE2) was analyzed under hypoxia (1.5% O2) in 7 days-old Arabidopsis seedlings: Col-0 (green bars) and p4h3-2 (gold bars). Relative expression levels are shown as fold change values. Asterisks represents statistical significance between Col-0 and p4h3-2 within a treatment and the error bars represent the standard error in expression values.
Several Arabidopsis and tomato AGPs were shown to be differentially expressed under low oxygen and among them nine Arabidopsis AGPs were selected for further characterization considering microarray analysis data of low oxygen transcriptome in Arabidopsis (Liu et al., 2005; Showalter et al., 2010; Fragkostefanakis et al., 2012). This cluster of hypoxic AGPs is comprised of five classical AGPs and four (Fasciclin-like AGPs) FLAs (Supplementary Figure 2). All of them comprise signal peptides while the five classical AGPs and two FLAs, FLA8 and FLA12, contain a GPI anchor (Supplementary Figure 2).
Stable overall transcript levels of AGPs and FLAs were observed in the wild type Col-0 seedlings throughout the 6-h hypoxic time course by qPCR analysis (Figure 4).
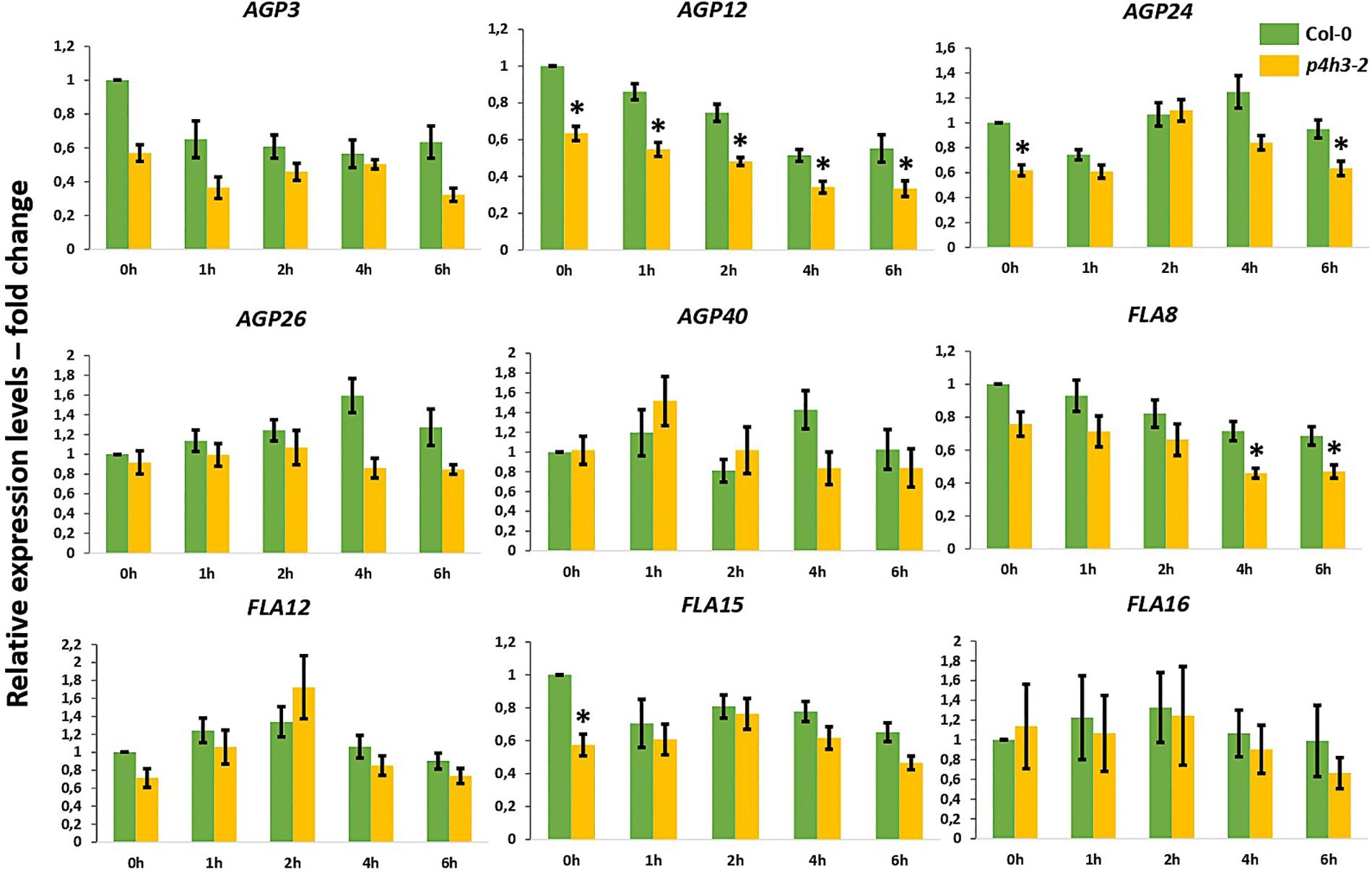
Figure 4. Expression of AGPs genes under hypoxia in the p4h3-2 line. The expression of nine AGPs – AGP3, AGP12, AGP24, AGP26, AGP40, FLA8, FLA12, FLA15, and FLA16 – was measured in hypoxic conditions (1.5% O2) in 7 day-old Arabidopsis seedlings. Col-0 (green bars) and p4h3-2 (gold bars) relative expression levels are shown as fold change values. Asterisks represent difference of significance between Col-0 and p4h3-2 within a treatment and error bars represent the standard error in expression values.
In the p4h3-2 line, the AGP12 showed lower levels of expression under normoxia (0 h) and this downregulation was continued compared to Col-0 throughout the time course (Figure 4). The FLA8 and AGP24 showed lower transcript levels after 4-, 6-h, and 6-h of hypoxia, respectively (Figure 4). These results might suggest indirect involvement of AtP4H3 also on the transcriptional regulation of AGPs despite catalyzing a post-translational modification. Comparison of the expression levels among the nine AGPs showed that AGP24 and AGP3 had the highest transcript abundance under normoxia (0 h) which was sustained also under hypoxia (Supplementary Figure 3). In the p4h3-2 mutant background, the mRNA levels of AGP24 increased by 2-fold in one time point (Supplementary Figure 3).
The soluble AGPs and extensins comprising the LM14-, MAC207-, and JIM20-epitopes were determined by western blot analysis (Figure 5). The LM14 antibody recognizes AGP-bound epitopes containing neutral sugars such as arabinose and galactose and are abundant in their side chains (Moller et al., 2008), while the MAC207 recognizes an epitope containing L-arabinose and o-glucuronic acid (Pennell et al., 1989). The JIM20 antibody recognizes extensin or HRGP glycoproteins (Knox et al., 1995).
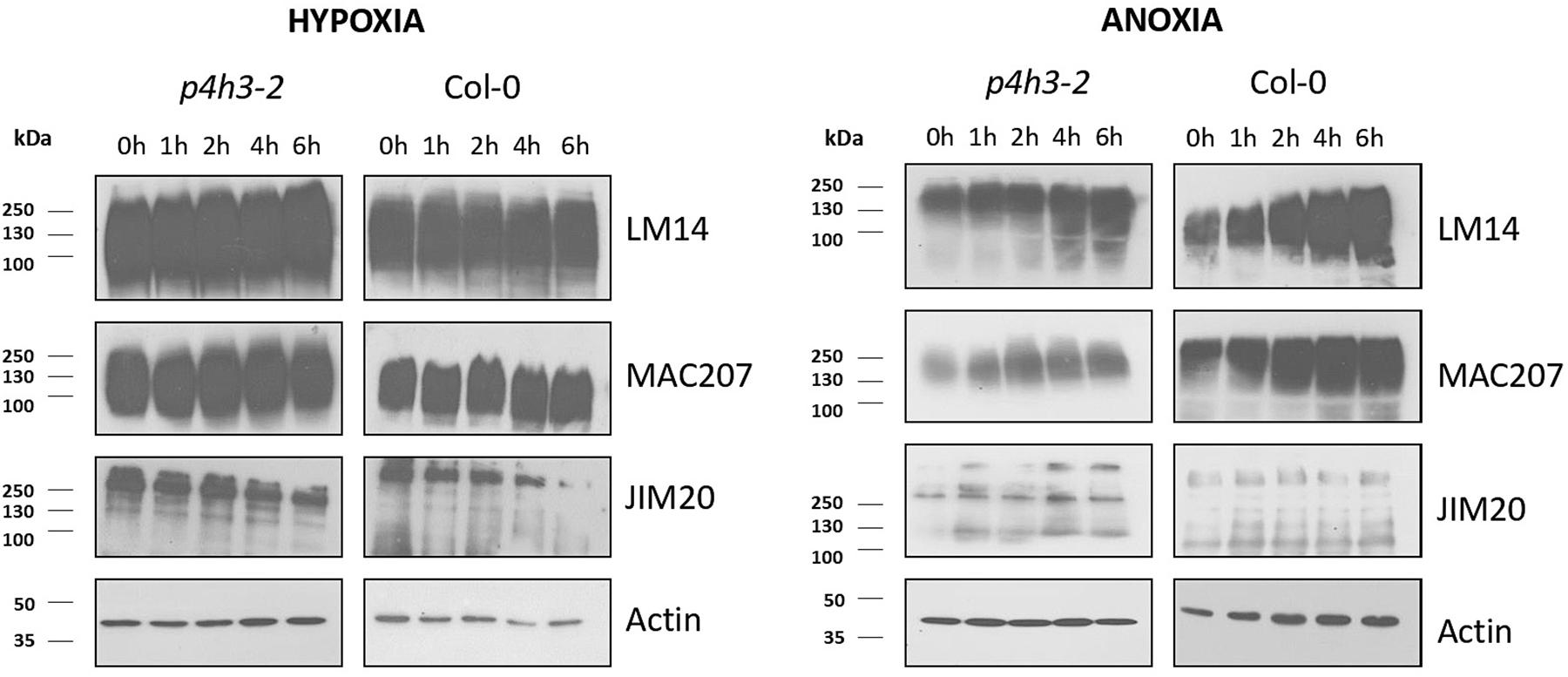
Figure 5. Western blot analysis of p4h3-2 and Col-0 seedlings using the LM14, MAC207, and JIM20 antibodies of AGPs. Total proteins (10 μl per lane) from Col-0 and p4h3-2 plants following hypoxia (left panel) and anoxia (right panel) treatments. Total proteins extracted from roots of 7 day-old seedlings subjected to hypoxia or anoxia for 0, 1, 2, 4, and 6 h were fractionated in SDS-PAGE in triplicate. Molecular masses are indicated on the left (kDa). At least three biological replicates were performed.
The soluble AGPs comprising the LM14-, MAC207-, and JIM20-epitopes ranged from 250 up to 72 KDa (Figure 5). The LM14-bound AGPs showed stable protein levels in wild type Col-0 during the hypoxic time course and a marginal increase during the anoxic time course (Figure 5 and Supplementary Figure 4). Similar results were obtained in the p4h3-2 mutant background (Figure 5 and Supplementary Figure 4). The MAC207-bound epitopes exhibited stable levels of expression under hypoxic time course in wild type Col-0 and the p4h3-2 mutant line while a marginal upregulation was observed under anoxia (Figure 5 and Supplementary Figure 4). The JIM20-bound extensins exhibited stable levels with a slight decrease after 6 h of hypoxia in wild type Col-0 while under anoxia the JIM20 levels were stable (Figure 5 and Supplementary Figure 4). In the p4h3-2 mutant background, the JIM20 epitopes slightly decreased under hypoxia and increased under anoxia (Figure 5 and Supplementary Figure 4).
An anoxic time course of up to 12 h in wild type Col-0 and p4h3-2 mutant indicated slightly higher levels of LM14 and MAC207 epitopes suggesting possible contribution to cell homeostasis (Supplementary Figure 5). Marginally higher levels of AGPs-bound epitopes were detected in the wild type Col-0 compared to the p4h3-2 mutant line under anoxic conditions in two different anoxic time courses (Figure 5 and Supplementary Figures 4, 5) while no changes in AGPs content was observed under hypoxia (Figure 5 and Supplementary Figure 4).
Although several P4Hs are upregulated under oxygen deficiency, the role of proline hydroxylation on hypoxic response in plants has not been thoroughly investigated despite their role in metazoan oxygen sensing mechanism (Vlad et al., 2007a). Oxygen sensing mechanisms have been discovered in animal and plant kingdoms, while hypoxia sensing and response mechanisms utilize post-translational modifications to signal protein degradation in an oxygen dependent manner (Kaelin and Ratcliffe, 2008; Gibbs et al., 2011; Weits et al., 2014). The hydroxylation of proline residues generates a binding site for the von Hippel-Lindau (pVHL) tumor suppressor protein, which is a component of the ubiquitin ligase complex and thus, HIFa is polyubiquitylated and subjected to proteasomal degradation upon oxygen availability due to dependence of P4H activity on oxygen concentration (Kaelin and Ratcliffe, 2008; Zhang and Yang, 2012). Under hypoxia, HIFa accumulates, dimerizes with an HIFβ family member, translocates to the nucleus, and transcriptionally activates an array of genes (Mcneill et al., 2002; Kaelin and Ratcliffe, 2008). Plants have evolved another oxygen sensing system, which is comprised of different components. Instead of proline hydroxylation, the signaling modification is cysteine oxidation by PCO (Weits et al., 2014), while the proteolysis (PRT)6 N-degron pathway, equivalent to the non-plant Arg/N-degron pathway, is used for proteasomal degradation (Vicente et al., 2017; Dissmeyer, 2019).
The lower survival in the p4h3-2 line was associated with a significant downregulation trend of ADH and PDC1 expression (Figure 3). It is well established that increased activation of the fermentation pathway leads to better adaptation under oxygen deficiency (Ismond et al., 2003; Hwang et al., 2011; Mithran et al., 2014). Lower survival of Arabidopsis seedlings in hre1/hre2 mutant lines were associated with lower expression levels of ADH in two out of four time points in an 8-h hypoxic course (Licausi et al., 2010). In the p4h3-2 line, the decrease in survival of Arabidopsis seedlings was also associated with a suppression of ADH and PDC1 expression in three and one time point in a 6-h hypoxic time course, respectively (Figure 3). Moreover, the transcript levels of SUS1 and SUS4 were also downregulated in two time points in an 8-h hypoxic course in the hre1/hre2 mutant lines (Licausi et al., 2010). Similarly, in the p4h3-2 line, the expression of SUS1 and SUS4 was decreased in three and two time points, respectively (Figure 3). These results indicate similar responses to hypoxia, which might not be related to either HRE1 and/or HRE2, because their levels of expression were not altered in the p4h3-2 line (Figure 3).
One of the anaerobic genes, SUS1, was shown to be expressed in the phloem and contribute to hypoxia and anoxia tolerance, indicating partially similar to AtP4H3 spatial expression patterns (Martin et al., 1993; Yao et al., 2020). However, sus1/sus4 double mutants had significantly lower survival percentages under waterlogging, while similar to wild type survival was observed under anoxia, hypoxia and submergence (Bieniawska et al., 2007; Santaniello et al., 2014).
The spatial induction of AtP4H3 in root stele and columella cells, leaf tips and stipules of Arabidopsis seedlings subjected to hypoxia suggests tissue specific involvement in hypoxic adaptation. The strongest induction was observed in root stele and columella cells, while it has been reported that the respiration rates in stele of aerated roots were 6-fold higher in comparison to cortex tissue in banana plants (Aguilar et al., 2003), which indicates accelerated consumption of oxygen and therefore induction of oxygen deficiency in the stele under aerobic conditions. This suggest that the level of AtP4H3 induction might be regulated in an oxygen dependent manner.
The HRGP superfamily in Arabidopsis consists of 166 genes including 85 genes coding for various AGP subfamily members, from which 22 genes code for classical AGPs and 21 for FLAs (Showalter et al., 2010). Hypoxia and anoxia differentially regulate AGPs expression in tomato fruits, with transcript levels of some tomato AGPs significantly decreased under hypoxia and up- or down-regulated under anoxia (Fragkostefanakis et al., 2012). Among the five classical AGPs and the four FLAs, only AGP12 showed consistently lower expression levels in the p4h3-2 line throughout the 6-h hypoxic time course (Figure 4), indicating regulation at the transcriptional level despite the fact that AtP4H3 catalyzes a post-translational modification. This pattern of expression might be attributed to regulation by a transcription factor which is affected by the levels of expression of AtP4H3. In this case, the AtP4H3 might indirectly regulate the expression of AGP12. Overall, the other eight AGPs and FLAs exhibited stable levels of expression throughout the hypoxic time course Stable levels of gene expression (Figure 4) and protein content (Figure 5) were detected for the other eight AGPs and FLAs and the AGPs-bound epitopes during the hypoxic time course, respectively.
Previously, 25 TFs were identified to exhibit differentially expression patterns under hypoxia and were comprising proline hydroxylation motifs in their deduced amino acid sequence (Vlad et al., 2007b). However, there are no reports, up to now, on the hydroxylation of TFs in plants regulating protein stability. Alternatively, secreted peptide hormones such as PSY1, CEPs, CLV3, CLEs, CLE-PS2, and RGFs, which undergo posttranslational hydroxylation (Lee et al., 2020), might be involved in hypoxic response and regulate transcriptional responses through TFs. Among them, CLE and CEP peptides were shown to be associated with abiotic stress such as osmotic and drought tolerance in Arabidopsis (Smith et al., 2020; Takahashi et al., 2020).
The frequency of occurrence of proline hydroxylation determines the O-glycosylation of AGPs and extensins leading to either alterations of their structure and/or to their degradation. This might lead to marginal reduction in the content of AGP-bound epitopes in the p4h3-2 line in comparison to the wild type as observed in the anoxic time course. However, no changes in the AGPs content was detected between the p4h3-2 line and wild type in the hypoxic time course possibly due to redundant function of other P4Hs expressed under hypoxia.
A marginal, gradual decrease on the extensins-bound epitopes was detected throughout the hypoxic time course in both p4h3-2 line and wild type suggesting lower involvement in hypoxic response compared to AGPs. However, under anoxia an induction was observed only in the p4h3-2 line but not in the wild type indicating possible regulation either at the transcriptional level or alterations at their glycan structure. Further investigation of the transcriptome and proteome might provide additional information on the regulation of extensin levels under oxygen deficiency conditions.
Mainly anoxic but also hypoxic stress leads to a substantial decrease in ATP production and as a result to energy shortage (Gibbs and Greenway, 2003). In this context, the gradual increase in AGPs content throughout not only the 6-h but also the 12 h anoxic time course might suggest participation in the preservation of cell homeostasis under anoxia considering that protein synthesis is a high energy consumption cellular process (Branco-Price et al., 2005). The higher sensitivity of an AtP4H3 knock out mutant line to anoxic stress indicates the physiological significance of proline hydroxylation, a post-translation modification, in oxygen deficiency adaptation in Arabidopsis. Further investigation is required on the identification of proline hydroxylated proteins involved in this abiotic stress response.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
PK conceived and designed the work. PK, AKo, MK, AL, ZZ, AKr, MB, MI, ME, GT, PC, NP, and A-MD were involved in the experiments, analysis, and interpretation of the data. PK, AKo, MK, AL, ZZ, AKr, and MI were involved in drafting the work. PK, AKo, and MI revised and approved the final version. All authors contributed to the article and approved the submitted version.
This article is based upon work from COST Action CA1821-ROXY-“Oxygen sensing a novel mean for biology and technology of fruit quality,” supported by COST (European Cooperation in Science and Technology). This work was also partially supported by the project “PlantUp: Upgrading Plant Capital” (MIS 5002803) which is implemented under the Action “Reinforcement of the Research and Innovation Infrastructure,” funded by the “Operational Programme Competitiveness, Entrepreneurship and Innovation” (NSRF 2014-2020) and co-financed by Greece and European Union (European Regional Development Fund).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would like to thank Mr. Theodoras Spano for excellent technical assistance.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.637352/full#supplementary-material
Aguilar, E. A., Turner, D. W., Gibb, D. J., Armstrong, W., and Sivasithamparam, K. (2003). Oxygen distribution and movement, respiration and nutrient loading in banana roots (Musa spp. L.) subjected to aerated and oxygen-depleted environments. Plant Soil 253, 91–102. doi: 10.1023/A:1024598319404
Amano, Y., Tsubouchi, H., Shinohara, H., Ogawa, M., and Matsubayashi, Y. (2007). Tyrosine-sulfated glycopeptide involved in cellular proliferation and expansion in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 104, 18333–18338. doi: 10.1073/pnas.0706403104
Asif, M. H., Trivedi, P. K., Misra, P., and Nath, P. (2009). Prolyl-4-hydroxylase (AtP4H1) mediates and mimics low oxygen response in Arabidopsis thaliana. Funct. Integr. Genomics 9, 525–535. doi: 10.1007/s10142-009-0118-y
Banti, V., Loreti, E., Novi, G., Santaniello, A., Alpi, A., and Perata, P. (2008). Heat acclimation and cross-tolerance against anoxia in Arabidopsis. Plant Cell Environ. 31, 1029–1037. doi: 10.1111/j.1365-3040.2008.01816.x
Bieniawska, Z., Paul Barratt, D. H., Garlick, A. P., Thole, V., Kruger, N. J., Martin, C., et al. (2007). Analysis of the sucrose synthase gene family in Arabidopsis. Plant J. 49, 810–828. doi: 10.1111/j.1365-313X.2006.03011.x
Branco-Price, C., Kawaguchi, R., Ferreira, R. B., and Bailey-Serres, J. (2005). Genome-wide analysis of transcript abundance and translation in arabidopsis seedlings subjected to oxygen deprivation. Ann. Bot. 96, 647–660. doi: 10.1093/aob/mci217
Clifton, I. J., McDonough, M. A., Ehrismann, D., Kershaw, N. J., Granatino, N., and Schofield, C. J. (2006). Structural studies on 2-oxoglutarate oxygenases and related double-stranded β-helix fold proteins. J. Inorg. Biochem. 100, 644–669. doi: 10.1016/j.jinorgbio.2006.01.024
Dissmeyer, N. (2019). Conditional protein function via N-Degron pathway-mediated proteostasis in stress physiology. Annu. Rev. Plant Biol. 70, 83–117. doi: 10.1146/annurev-arplant-050718-095937
Drew, M. C. (1997). Oxygen deficiency and root metabolism: injury and acclimation under hypoxia and anoxia. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 223–250. doi: 10.1146/annurev.arplant.48.1.223
Eysholdt-Derzsó, E., and Sauter, M. (2017). Root bending is antagonistically affected by hypoxia and ERF-mediated transcription via auxin signaling. Plant Physiol. 175, 412–423. doi: 10.1104/pp.17.00555
Farrow, S. C., and Facchini, P. J. (2014). Functional diversity of 2-oxoglutarate/Fe(II)-dependent dioxygenases in plant metabolism. Front. Plant Sci. 5:524. doi: 10.3389/fpls.2014.00524
Fragkostefanakis, S., Dandachi, F., and Kalaitzis, P. (2012). Expression of arabinogalactan proteins during tomato fruit ripening and in response to mechanical wounding, hypoxia and anoxia. Plant Physiol. Biochem. 52, 112–118. doi: 10.1016/j.plaphy.2011.12.001
Gasch, P., Fundinger, M., Müller, J. T., Lee, T., Bailey-Serres, J., and Mustropha, A. (2016). Redundant ERF-VII transcription factors bind to an evolutionarily conserved cis-motif to regulate hypoxia-responsive gene expression in arabidopsis. Plant Cell 28, 160–180. doi: 10.1105/tpc.15.00866
Gibbs, D., Lee, S. C., Md Isa, N., Gramuglia, S., Fukao, T., Bassel, G. W., et al. (2011). Homeostatic response to hypoxia is regulated by the N-end rule pathway in plants. Nature 479, 415–418. doi: 10.1038/nature10534
Gibbs, J., and Greenway, H. (2003). Mechanisms of anoxia tolerance in plants, growth, survival and anaerobic catabolism. Funct. Plant Biol. 30, 1–47. doi: 10.1071/PP98095
Giuntoli, B., and Perata, P. (2018). Group VII ethylene response factors in arabidopsis: regulation and physiological roles. Plant Physiol. 176, 1143–1155. doi: 10.1104/pp.17.01225
Goni, R., Garcia, P., and Foissac, S. (2009). The qPCR date statistical analysis. Integromics White Pap. 1–9.
Hwang, J. H., Lee, M. O., and Choy, Y. (2011). Expression profile analysis of hypoxia responses in arabidopsis roots and shoots. J. Plant Biol. 54, 373–383. doi: 10.1007/s12374-011-9172-9
Iacopino, S., and Licausi, F. (2020). The Contribution of Plant Dioxygenases to Hypoxia Signaling. Front. Plant Sci. 11:1008. doi: 10.3389/fpls.2020.01008
Ismond, K. P., Dolferus, R., Pauw, M., Dennis, E. S., and Good, A. G. (2003). Enhanced low oxygen survival in arabidopsis through increased metabolic flux in the fermentative pathway 1. Plant Physiol. 132, 1292–1302. doi: 10.1104/pp.103.022244
Jacobs, M., Dolferus, R., and Van Den Bossche, D. (1988). Isolation and biochemical analysis of ethyl methanesulfonate-induced alcohol dehydrogenase null mutants of Arabidopsis thaliana (L.) Heynh. Biochem. Genet. 26, 105–122. doi: 10.1007/BF00555492
Kaelin, W. G., and Ratcliffe, P. J. (2008). Oxygen sensing by metazoans: the central role of the hif hydroxylase pathway. Mol. Cell 30, 393–402. doi: 10.1016/j.molcel.2008.04.009
Kato-Noguchi, H., and Morokuma, M. (2007). Ethanolic fermentation and anoxia tolerance in four rice cultivars. J. Plant Physiol. 164, 168–173. doi: 10.1016/j.jplph.2005.09.017
Knox, J. P., Peart, J., and Neill, S. J. (1995). Identification of novel cell surface epitopes using a leaf epidermal-strip assay system. Planta An Int. J. Plant Biol. 196, 266–270. doi: 10.1007/BF00201383
Kondo, T., Sawa, S., Kinoshita, A., Mizuno, S., Kakimoto, T., Fukuda, H., et al. (2006). A plant peptide encoded by CLV3 identified by in situ MALDI-TOF MS analysis. Science 313, 845–848. doi: 10.1126/science.1128439
Kürsteiner, O., Dupuis, I., and Kuhlemeier, C. (2003). The Pyruvate decarboxylase1 Gene of arabidopsis is required during anoxia but not other environmental stresses [w]. Plant Physiol. 132, 968–978. doi: 10.1104/pp.102.016907
Lamport, D. T. A., Kieliszewski, M. J., and Showalter, A. M. (2006). Salt stress upregulates periplasmic arabinogalactan proteins: using salt stress to analyse AGP function. New Phytol. 169, 479–492. doi: 10.1111/j.1469-8137.2005.01591.x
Lee, K. H., Kwon, D. H., Song, J. T., and Seo, H. S. (2020). Production mechanisms, structural features and post-translational modifications of plant peptides. J. Plant Biol. 63, 259–265. doi: 10.1007/s12374-020-09255-5
Li, X. (2011). Histostaining for tissue expression pattern of promoter-driven GUS activity in Arabidopsis. Bio-101 e93. doi: 10.21769/BioProtoc.93
Licausi, F., Van Dongen, J. T., Giuntoli, B., Novi, G., Santaniello, A., Geigenberger, P., et al. (2010). HRE1 and HRE2, two hypoxia-inducible ethylene response factors, affect anaerobic responses in Arabidopsis thaliana. Plant J. 62, 302–315. doi: 10.1111/j.1365-313X.2010.04149.x
Liu, F., Vantoai, T., Moy, L. P., Bock, G., Linford, L. D., Quackenbush, J., et al. (2005). Global transcription profiling reveals comprehensive insights into hypoxic response in Arabidopsis. Plant Physiol. 137, 1115–1129. doi: 10.1104/pp.104.055475
Livak, K., and Schmittgen, T. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Loreti, E., and Perata, P. (2020). The many facets of hypoxia in plants. Plants 9:745. doi: 10.3390/plants9060745
Loreti, E., Poggi, A., Novi, G., Alpi, A., and Perata, P. (2005). A genome-wide analysis of the effects of sucrose on gene expression in arabidopsis seedlings under anoxia. Plant Physiol. 137, 1130–1138. doi: 10.1104/pp.104.057299
Mareri, L., Romi, M., and Cai, G. (2019). Arabinogalactan proteins: actors or spectators during abiotic and biotic stress in plants? Plant Biosyst. 153, 173–185. doi: 10.1080/11263504.2018.1473525
Martin, T., Frommer, W. B., Salanoubat, M., and Willmitzer, L. (1993). Expression of an Arabidopsis sucrose synthase gene indicates a role in metabolization of sucrose both during phloem loading and in sink organs. Plant J. 4, 367–377. doi: 10.1046/j.1365-313X.1993.04020367.x
Matsumura, H., Takano, T., Yoshida, K. T., and Takeda, G. (1995). A rice mutant lacking alcohol dehydrogenase. Breding Sci. 45, 365–367. doi: 10.1270/jsbbs1951.45.365
Mcneill, L. A., Hewitson, K. S., Gleadle, J. M., Horsfall, L. E., Oldham, N. J., Maxwell, P. H., et al. (2002). The use of dioxygen by HIF prolyl hydroxylase (PHD1). Bioorg. Med. Chem. Lett. 12, 1547–1550. doi: 10.1016/s0960-894x(02)00219-6
Minami, A., Yano, K., Gamuyao, R., Nagai, K., Kuroha, T., Ayano, M., et al. (2018). Time-course transcriptomics analysis reveals key responses of submerged deepwater rice to flooding. Plant Physiol. 176, 3081–3102. doi: 10.1104/pp.17.00858
Mithran, M., Paparelli, E., Novi, G., Perata, P., and Loreti, E. (2014). Analysis of the role of the pyruvate decarboxylase gene family in Arabidopsis thaliana under low-oxygen conditions. Plant Biol. 16, 28–34. doi: 10.1111/plb.12005
Moller, I., Marcus, S. E., Haeger, A., Verhertbruggen, Y., Verhoef, R., Schols, H., et al. (2008). High-throughput screening of monoclonal antibodies against plant cell wall glycans by hierarchical clustering of their carbohydrate microarray binding profiles. Glycoconj. J. 25, 37–48. doi: 10.1007/s10719-007-9059-7
Mustroph, A., Zanetti, M. E., Jang, C. J. H., Holtan, H. E., Repetti, P. P., Galbraith, D. W., et al. (2009). Profiling translatomes of discrete cell populations resolves altered cellular priorities during hypoxia in Arabidopsis. Plant Biol. 106, 18843–18848. doi: 10.1073/pnas.0906131106
Myllyharju, J., and Koivunen, P. (2013). Hypoxia-inducible factor prolyl 4-hydroxylases:common and specific roles. Biol. Chem. 394, 435–448. doi: 10.1515/hsz-2012-0328
Pabingera, S., Rödigerb, S., Kriegnera, A., Vierlingera, K., and Weinhäusela, A. (2014). A survey of tools for the analysis of quantitative PCR (qPCR) data. Biomol. Detect. Quantif. 1, 23–33. doi: 10.1016/j.bdq.2014.08.002
Pennell, R. I., Knox, J. P., Scofield, G. N., Selvendran, R. R., and Roberts, K. (1989). A family of abundant plasma membrane-associated glycoproteins related to the arabinogalactan proteins is unique to flowering plants. J. Cell Biol. 108, 1967–1977. doi: 10.1083/jcb.108.5.1967
Perata, P., and Alpi, A. (1993). Plant responses to anaerobiosis. Plant Sci. 93, 1–17. doi: 10.1016/0168-9452(93)90029-Y
Planchet, E., Lothier, J., and Limami, A. (2017). Hypoxic Respiratory Metabolism in Plants: Reorchestration of Nitrogen and Carbon Metabolisms Elisabeth. Berlin: Springer.
Pugh, C. W., and Ratcliffe, P. J. (2003). Regulation of angiogenesis by hypoxia: role of the HIF system. Nat. Med. 9, 677–684. doi: 10.1038/nm0603-677
Roberts, J. K., Callis, J., Jardetzky, O., Walbot, V., and Freeling, M. (1984). Cytoplasmic acidosis as a determinant of flooding intolerance in plants. Proc. Natl. Acad. Sci. U.S.A. 81, 6029–6033. doi: 10.1073/pnas.81.19.6029
Sambrook, J., and Russell, D. (2006). SDS-Polyacrylamide Gel Electrophoresis of Proteins. CSH Protoc. 2016:rot4540. doi: 10.1101/pdb.prot4540
Santaniello, A., Loreti, E., Gonzali, S., Novi, G., and Perata, P. (2014). A reassessment of the role of sucrose synthase in the hypoxic sucrose-ethanol transition in Arabidopsis. Plant Cell Environ. 37, 2294–2302. doi: 10.1111/pce.12363
Schultz, C. J., Ferguson, K. L., and Lahnstein, J. (2004). Post-translational modifications of arabinogalactan-peptides of Arabidopsis thaliana. Endoplasmic reticulum and glycosylphosphatidylinositol-anchor signal cleavage sites and hydroxylation of proline. J. Biol. Chem. 279, 45503–45511. doi: 10.1074/jbc.M407594200
Schultz, C. J., Rumsewicz, M. P., Johnson, K. L., Jones, B. J., Gaspar, Y. M., and Bacic, A. (2002). Using genomic resources to guide research directions. The arabinogalactan protein gene family as a test case. Plant Physiol. 129, 1448–1463. doi: 10.1104/pp.003459
Showalter, A. M., Keppler, B., Lichtenberg, J., Gu, D., and Welch, L. R. (2010). A bioinformatics approach to the identification, classification, and analysis of hydroxyproline-rich glycoproteins. Plant Physiol. 153, 485–513. doi: 10.1104/pp.110.156554
Smith, S., Zhu, S., Joos, L., Roberts, I., Nikonorova, N., Vu, L. D., et al. (2020). The CEP5 peptide promotes abiotic stress tolerance, as revealed by quantitative proteomics, and attenuates the AUX/IAA equilibrium in Arabidopsis. Mol. Cell. Proteomics 19, 1248–1262. doi: 10.1074/mcp.RA119.001826
Stührwohldt, N., Ehinger, A., Thellmann, K., and Schaller, A. (2020). Processing and formation of bioactive CLE40 peptide are controlled by post-translational proline hydroxylation. Plant Physiol 184, 1573–1584. doi: 10.1104/pp.20.00528
Takahashi, F., Kuromori, T., Urano, K., Yamaguchi-Shinozaki, K., and Shinozaki, K. (2020). Drought stress responses and resistance in plants: from cellular responses to long-distance intercellular communication. Front. Plant Sci. 11, 1–14. doi: 10.3389/fpls.2020.556972
Velasquez, M., Salter, J. S., Dorosz, J. G., Petersen, B. L., and Estevez, J. M. (2012). Recent advances on the posttranslational modifications of EXTs and their roles in plant cell walls. Front. Plant Sci. 3:93. doi: 10.3389/fpls.2012.00093
Velasquez, S. M., Velasquez, S. M., Ricardi, M. M., Dorosz, J. G., Fernandez, P. V., Nadra, A. D., et al. (2011). O-Glycosylated cell wall proteins are essential in root hair growth. Science 332, 1401–1403. doi: 10.1126/science.1206657
Vicente, J., Mendiondo, G. M., Movahedi, M., Peirats-Llobet, M., Juan, Y., Shen, Y., et al. (2017). The Cys-Arg/N-end rule pathway is a general sensor of abiotic stress in flowering plants. Curr. Biol. 27, 3183.e–3190.e. doi: 10.1016/j.cub.2017.09.006
Vlad, F., Spano, T., Vlad, D., Bou Daher, F., Ouelhadj, A., and Kalaitzis, P. (2007a). Arabidopsis prolyl 4-hydroxylases are differentially expressed in response to hypoxia, anoxia and mechanical wounding. Physiol. Plant. 130, 471–483. doi: 10.1111/j.1399-3054.2007.00915.x
Vlad, F., Spano, T., Vlad, D., Bou Daher, F., Ouelhadj, A., and Kalaitzis, P. (2007b). Involvement of arabidopsis Prolyl 4 hydroxylases in hypoxia, anoxia and mechanical wounding. Plant Signal. Behav. 2, 368–369. doi: 10.4161/psb.2.5.4462
Wang, G. L., and Semenza, G. L. (1993). General involvement of hypoxia-inducible factor 1 in transcriptional response to hypoxia. Proc. Natl. Acad. Sci. U.S.A. 90, 4304–4308. doi: 10.1073/pnas.90.9.4304
Weits, D. A., Giuntoli, B., Kosmacz, M., Parlanti, S., Hubberten, H., Riegler, H., et al. (2014). Plant cysteine oxidases control the oxygen-dependent branch of the N-end-rule pathway. Nat. Commun. 5, 1–10. doi: 10.1038/ncomms4425
Woodson, W. R., and Handa, A. K. (1987). Changes in protein patterns and in vivo protein synthesis during presenescence and senescence of hibiscus petals. J. Plant Physiol. 128, 67–75. doi: 10.1016/S0176-1617(87)80182-7
Yao, D., Gonzales-Vigil, E., and Mansfield, S. D. (2020). Arabidopsis sucrose synthase localization indicates a primary role in sucrose translocation in phloem. J. Exp. Bot. 71, 1858–1869. doi: 10.1093/jxb/erz539
Zhang, Q., and Yang, H. (2012). The roles of VHL-dependent ubiquitination in signaling and cancer. Front. Oncol. 2:35. doi: 10.3389/fonc.2012.00035
Keywords: Arabidopsis thaliana, prolyl-4-hydroxylases, anoxia, hypoxia, AGPs, FLAs, ADH, pDc
Citation: Konkina A, Klepadlo M, Lakehal A, Zein ZE, Krokida A, Botros M, Iakovidis M, Chernobavskiy P, Elfatih Zerroumda M, Tsanakas G, Petrakis N, Dourou A-M and Kalaitzis P (2021) An Arabidopsis Prolyl 4 Hydroxylase Is Involved in the Low Oxygen Response. Front. Plant Sci. 12:637352. doi: 10.3389/fpls.2021.637352
Received: 03 December 2020; Accepted: 09 February 2021;
Published: 15 March 2021.
Edited by:
Georg J. Seifert, University of Natural Resources and Life Sciences, Vienna, AustriaReviewed by:
Silvia Velasquez, University of Natural Resources and Life Sciences, Vienna, AustriaCopyright © 2021 Konkina, Klepadlo, Lakehal, Zein, Krokida, Botros, Iakovidis, Chernobavskiy, Elfatih Zerroumda, Tsanakas, Petrakis, Dourou and Kalaitzis. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Panagiotis Kalaitzis, cGFuYWdpb3RAbWFpY2guZ3I=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.