
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci., 16 February 2021
Sec. Plant Development and EvoDevo
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.635500
This article is part of the Research TopicMolecular Mechanisms of Flowering Plant ReproductionView all 19 articles
Carpel is the ovule-bearing female reproductive organ of flowering plants and is required to ensure its protection, an efficient fertilization, and the development of diversified types of fruits, thereby it is a vital element of most food crops. The origin and morphological changes of the carpel are key to the evolution and adaption of angiosperms. Progresses have been made in elucidating the developmental mechanisms of carpel establishment in the model eudicot plant Arabidopsis thaliana, while little and fragmentary information is known in grasses, a family that includes many important crops such as rice (Oryza sativa), maize (Zea mays), barley (Hordeum vulgare), and wheat (Triticum aestivum). Here, we highlight recent advances in understanding the mechanisms underlying potential pathways of carpel development in grasses, including carpel identity determination, morphogenesis, and floral meristem determinacy. The known role of transcription factors, hormones, and miRNAs during grass carpel formation is summarized and compared with the extensively studied eudicot model plant Arabidopsis. The genetic and molecular aspects of carpel development that are conserved or diverged between grasses and eudicots are therefore discussed.
Carpels are a major defining feature of angiosperms. This distinctive female reproductive structure occupies the center of the flower (Figure 1), encloses ovules, and greatly improves reproductive efficiency compared with gymnosperms, involving a more complex and diversified process of pollination. Among species, the female reproductive organ, gynoecium or pistil, may occur as single carpel, multiple independent carpels, or a fused syncarpic structure (Gomariz-Fernández et al., 2017). A basic organization plan composed of three distinct regions can be found across angiosperms carpels despite their huge morphological diversity: the basal ovary, a style, and an apical stigma (Figure 1) (Becker, 2020). Poaceae, the grass family, is one of the largest families of angiosperms; it includes many agriculturally important crops, such as domesticated rice (Oryza sativa), barley (Hordeum vulgare), wheat (Triticum aestivum), and maize (Zea mays). The spikelet is a basic structure unit of the inflorescence in all grass family, which in rice contains a fertile floret and two depressed sterile lemmas (also called empty glumes). Within each rice floret, there are two bract-like organs called lemma and palea that surround the inner floral organs, the lodicule, stamen, and carpel (Figure 1B). Maize has a similar flower structure with rice, although maize flowers undergo sex determination after floral organ initiation, with only stamens and carpels developing in tassel and ear spikelet, respectively (Figure 1C). As the innermost floral organ, rice, barley, and wheat pistils display a similar structure, which is derived from the early fusion of three carpel primordia, with two partially fused styles and two feathery stigmas covered with papillae cells where pollen is deposited (Figure 1E) (Kellogg, 2001; Rudall et al., 2005; Dreni et al., 2013). In maize, the single pistil is formed by three connate carpels; however, only the two lateral-abaxial carpels will develop to an elongated silk, whereas the growth of medial-adaxial carpel is inhibited before enveloping the single ovule (Figure 1F) (Strable and Vollbrecht, 2019). In comparison, the pistil of model eudicot Arabidopsis thaliana consists of two fused lateral carpels that bear submarginal ovules in the two locules (Figure 1D) (Becker, 2020).
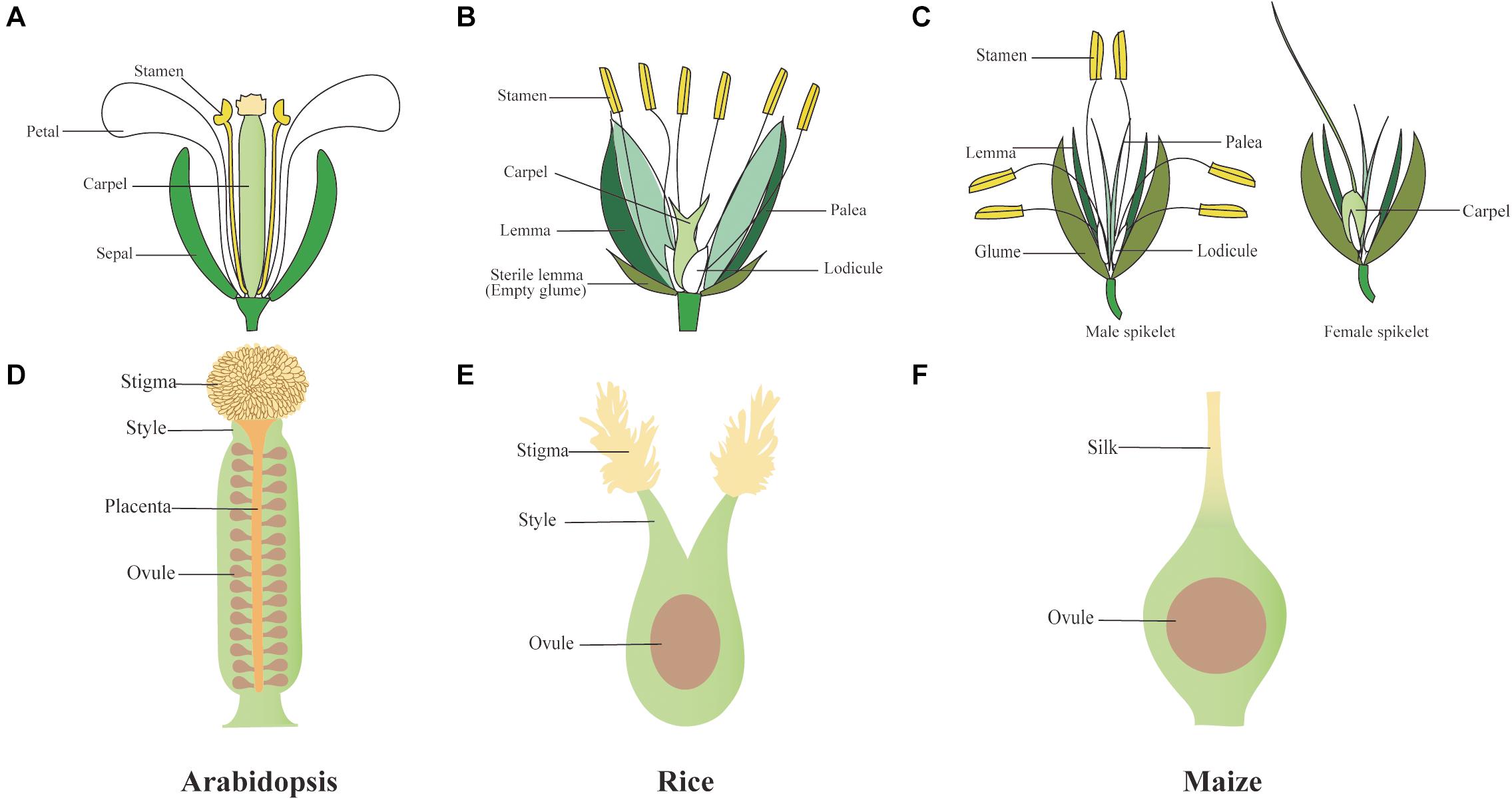
Figure 1. Schematic draws of Arabidopsis, rice, and maize florets and gynoecia. (A–C) Flower and spikelet structure of Arabidopsis (A), rice (B), and maize (C). (D) The pistil of Arabidopsis can be divided into ovary, style, stigma, and the ovules derive from a placenta (E) Rice syncarpic ovary harbors two styles and feathery stigmas, (F) while maize has a higher extent of fusion into one silk. Like in all grasses, only one ovule is developed in the pistil of rice and maize.
During the past two decades, advances in ancestral state reconstruction, phylogenetics and developmental genetics have shed some light on carpel evolution. Depending on whether carpel is considered to be modified from male or female structures in their presumed gymnosperm-like ancestor, several potential hypotheses for the origin of the flower and its carpel exist. The Mostly Male Theory (Frohlich, 2003) postulates the first carpels are formed after the ectopic ovules generated on male microsporophylls. The Out-of-Male (Theissen et al., 2002) and its sister theory Out-of-Female (Theissen et al., 2002) hypothesis propose that the bisexuality of the flower is arisen by expression movements of the male (B-class genes) and female-promoting gene (B-sister genes), thus leaving female structures at the apex and male structures in basal region. Baum and Hileman (2007) proposed a central role of gene LEAFY (LFY) in the origin of the flower, which regulates C-function MADS-box genes in the flowering plants ancestor; these C-class genes then would have specified apical female reproductive structure, which later became carpels (Vialette-Guiraud and Scutt, 2009; Ferrándiz et al., 2010). Recently, the carpel of Anaxagorea, the most basal genus of primitive angiosperms Annonaceae (Magnoliales), is studied (Li et al., 2020). Supports are provided in this study for another carpel origin theory called the stachyosporous theory, which suggests that carpel originated from a compound shoot of the ovular axis and foliar appendage (Hickey and Taylor, 1996; Li et al., 2020). Besides these theories, a basic ancestral angiosperm carpel developmental networks has been proposed (Becker, 2020), involving STYLISH (STY)-, SPATULA (SPT)-, ETTIN (ETT)-, and CRABS CLAW (CRC)-like genes in basal/apical and adaxial/abaxial polarity specification and growth, and HECATE (HEC) homologs regulating only longitudinal growth. Homologs of NGATHA (NGA) and STY/SHORT INTERNODES (SHI)/SHI RELATED SEQUENCE (SRS) may have already contributed in regulating stigmatic-like tissue development in the carpel of the hypothetic aMRCA (angiosperm most recent common ancestor), while it probably lacked a style (Fourquin et al., 2005; Endress and Doyle, 2009, 2015; Soltis and Soltis, 2016; Becker, 2020).
To date, most data on the gene regulatory networks involved in tissue differentiation and patterning during gynoecium morphogenesis processes are still derived from A. thaliana. The major roles of phytohormones, many transcription factors (TFs), and coregulators have been identified in specifying tissue orientations and gynoecium formation, which have been extensively reviewed in Arabidopsis (Reyes-Olalde et al., 2013; Pfannebecker et al., 2017a, b; Reyes-Olalde and de Folter, 2019; Simonini and Østergaard, 2019; Zúñiga-Mayo et al., 2019). Molecular genetic studies have uncovered developmental mechanisms of carpel establishment in two grass model species, rice and maize. In view of a similar carpel development pattern in grasses, these studies will facilitate related research in the future, including barley, wheat, and other cereals. Here, we update focused studies mainly in rice and maize, on the regulatory mechanisms, networks, and pathways underlying the establishment and regulation of carpel identity, morphogenesis and development, and their intrinsic connection with floral meristem determinacy (FMD).
Like any floral organ, carpels initiate from meristem primordia, which are small domes of undifferentiated cells, and then develop their specific properties and structure through a process called morphogenesis. There are some key regulators identified in grass carpel identity determination whose functions are either conserved or diverged versus Arabidopsis.
Genetic studies based on Arabidopsis and Antirrhinum majus led to the formulation of the ABC model of flower development and floral organ identity (Coen and Meyerowitz, 1991; Weigel and Meyerowitz, 1994) that predicts the combinatorial activity of three classes of genes as follows: A-class genes specify sepal identity in the first whorl, A- and B-class genes specify petal identity in the second whorl, and B- and C-class genes are necessary for stamen identity in the third whorl, whereas C-class genes alone are required for carpel identity and FMD in the fourth and last whorl in the center of the flower. Later studies in Petunia hybrida and Arabidopsis, proposed that a D class of genes is specifically required for ovule development and an E class of genes is essential for the function of all the other four classes, which led to an updated ABCDE model (Colombo et al., 1995; Pelaz et al., 2000, 2001; Honma and Goto, 2001; Favaro et al., 2003; Pinyopich et al., 2003). Except for the Arabidopsis A class APETALA2 (AP2) gene, all the ABCDE genes encode for MIKC-type MADS-box TFs working in tetrameric complexes (Riechmann et al., 1996; Theißen, 2001; Paøenicová et al., 2003; Theißen et al., 2016). While the conservation of the model for the outer whorls is amply debated (Litt, 2007; Causier et al., 2010), the C and D functions are widely conserved in carpel and ovule identity among angiosperms, which largely depends on MADS-box genes from the AGAMOUS subfamily (Figure 2A) (Dreni and Kater, 2014). In addition, complexes formed by their encoded proteins with E class proteins that are encoded by SEPALLATA subfamily MADS-box genes seem also conserved (Figure 2A) (Dreni and Kater, 2014).
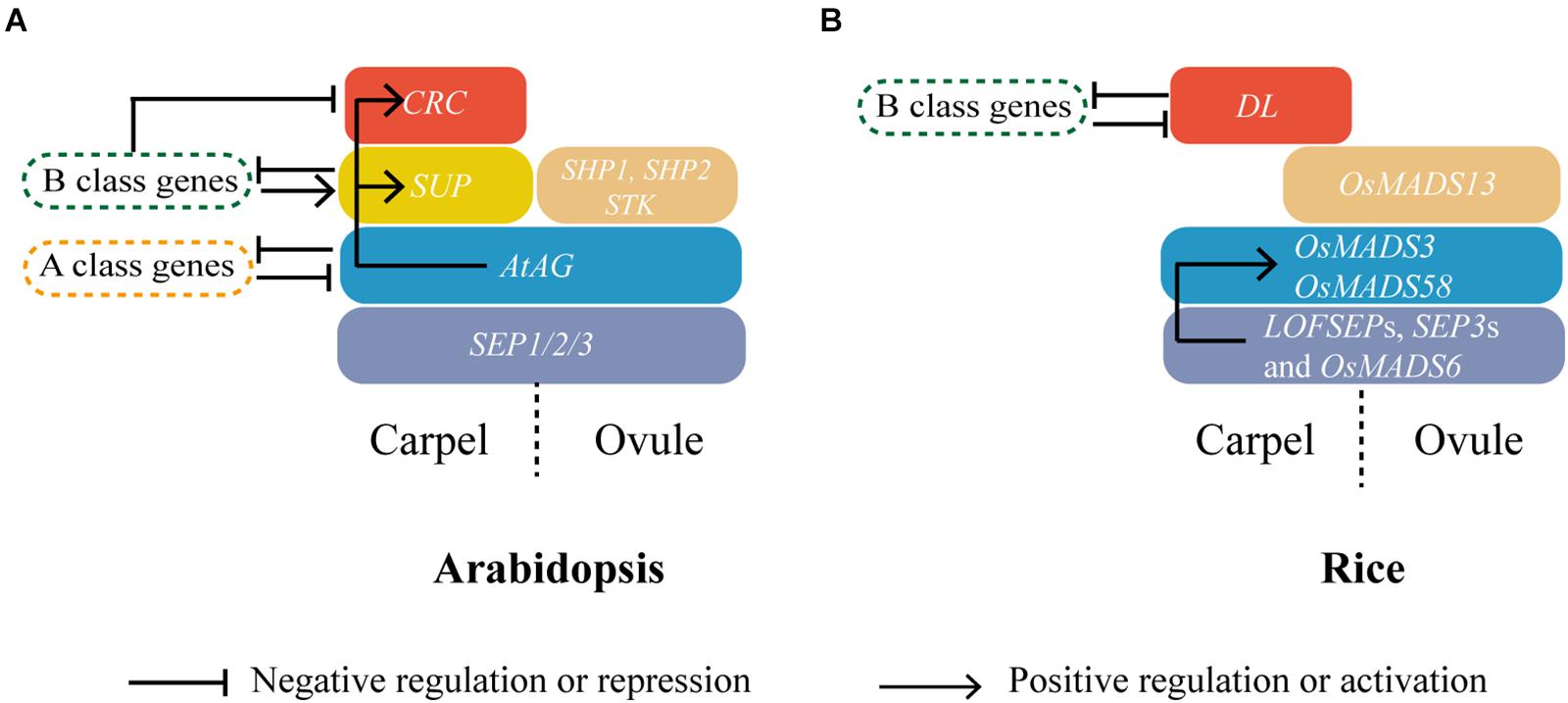
Figure 2. Carpel specification in dicot and monocot model plants Arabidopsis and rice. The expression domains of C-, D-, and E-class genes; SUP; and CRC, DL in carpel and ovule of Arabidopsis (A) and rice (B) are shown with blue, brown, purple, yellow, and red colors, respectively. Genetic interactions within these carpel identity regulators and other homeotic genes are also indicated. Direct regulations have been shown in Arabidopsis, whereas in rice a direct binding of OsMADS6 has only been shown to the second intron of OsMADS58 (Li et al., 2011a).
The AGAMOUS subfamily is named after its founding member gene, Arabidopsis AGAMOUS (AtAG). AtAG, as a C-class gene, is the master regulator of stamen and carpel formation (Bowman et al., 1989, 1991; Yanofsky et al., 1990; Lohmann and Weigel, 2002). FMD and reproductive organ identity are completely lost in the ag mutant, leading to a continuous repetition of sepal–petal–petal whorls forming indeterminate flowers (Bowman et al., 1989, 1991).
Rice has two AtAG orthologs, OsMADS3 and OsMADS58, which share a very similar expression profile; both express exclusively in whorl 3 (stamen) and whorl 4 (carpel), although OsMADS58 is expressed more uniformly throughout the floral meristem (FM) (Yamaguchi et al., 2006; Dreni et al., 2011). In rice osmads3-4 (Hu et al., 2011) and osmads3-fe1 (Yasui et al., 2017) single mutants, most flowers display a normal single pistil, whereas in the reported null allele osmads3-3 (Yamaguchi et al., 2006), 84% of the flowers showed increased carpel numbers and multiple stigmas, and enlarged or elongated ovules are also formed. Interestingly, this loss-of-function allele is generated by a T-DNA insertion in the large second intron (Yamaguchi et al., 2006), suggesting evolutionary conserved importance of this region in promoting the correct expression of AG-like genes, in agreement with studies conducted in Arabidopsis and Antirrhinum (Sieburth and Meyerowitz, 1997; Causier et al., 2009). Despite the similar expression patterns between OsMADS3 and OsMADS58, osmads58 single mutants display normal flowers; only 5% of those have bifurcated stigmas (Dreni et al., 2011). In both osmads3-3 osmads58 and osmads3-4 osmads58 double mutants, ectopic lodicules and green palea/carpel-like organs repeatedly replace stamen and carpel, respectively. With similar abaxial surface, Dreni et al. (2011) interpreted the organs replacing carpels in osmads3-3 osmads58 as palea-like, having a similar structure, and a body and two margin regions, which are characteristics of palea (Prasad et al., 2001; Prasad and Vijayraghavan, 2003). Surprisingly, this clear morphological transformation does not match with an equally obvious change in the expression of the only known putative marker genes: although the glume marker G1/ELE (long sterile lemma/elongated empty glume) is activated in the indeterminate FM of osmads3-3 osmads58, it does not mark clearly the emerging palea-like primordium that, in addition, continues to express DROOPING LEAF (DL), a gene required for carpel identity specification, like in the wild-type ovary wall (Dreni et al., 2011).
A residual carpel identity was discovered in the adaxial epidermis of the osmads3-3 osmads58 aberrant gynoecium, conferred by another AGAMOUS subfamily member, the D class ovule identity gene OsMADS13 (Dreni et al., 2007). OsMADS13 is expressed in the adaxial epidermis of the ovary wall of wild-type plants, and this pattern is maintained also in the adaxial epidermis of the palea/carpel-like organ of osmads3-3 osmads58 flower. Indeed, the residual carpel identity is completely eliminated by generating osmads3-3 osmads13 osmads58 triple mutants, whose palea-like structures grow even more and present an adaxial surface completely equivalent to a palea. Yet, DL expression remains strong in these organs (Dreni et al., 2011).
Recently, the identity of green organs replacing the carpel in the osmads3 osmads58 double mutants has been questioned by Sugiyama et al. (2019), who generated a new double-mutant osmads3-fe1 osmads58-7 for their analysis. Different from osmads3-3 osmads58 in which OsMADS58 is mutated by a dSpm transposon insertion in the second intron (Dreni et al., 2011), in osmads3-fe1 osmads58-7, the ∼600-bp promoter region and the first three exons of OsMADS3 are deleted, and there is a frame-shift mutation in exon 3 of OsMADS58 (Yasui et al., 2017; Sugiyama et al., 2019). Despite revealing that the green ectopic organs in the fourth whorl express the A-class and palea identity gene OsMADS15, which is also in agreement with the ABC model, Sugiyama et al. (2019) claim that the abaxial surface still retains features of the ovary cells. Based on this, they reinterpreted the fourth whorl organs of osmads3-fe1 osmads58-7 as carpel-like and proposed that the persisting expression of DL and OsMADS13 is supportive of carpel-like identity, while the palea-like structure is conferred by ectopic expression of the A class gene OsMADS15. In conclusion, Sugiyama et al. (2019) suggest that rice C-class genes are not essential for carpel identity. Future works will be needed to clarify this essential question.
Another recent study has suggested an important functional role for the conserved amino acidic residue 109 of AG proteins in grasses (Dreni et al., 2020). It is shown that an alternative splicing occurs on the OsMADS3 transcript of O. sativa, producing two variants of OsMADS3 that differ just for the presence or absence of a conserved serine residue, S109. Interestingly, this splicing variant is conserved and specific between grass families. Only the eudicot-like OsMADS3 isoform, lacking S109, is able to specify stamens and carpels in Arabidopsis, as revealed by functional complementation assay expressing the two rice isoforms in the Arabidopsis ag mutant (Dreni et al., 2020). It is further proposed that the different ability between OsMADS3+S109 and OsMADS3 in complementing the ag phenotype might be partially explained by their different interactions with diverse SEPALLATA-like proteins (Dreni et al., 2020). Therefore, the two isoforms of OsMADS3-like proteins might have partially distinct function in grasses.
In maize, there are three AG lineage genes, with ZAG1 ortholog to OsMADS58 and ZMM2 and ZMM23 orthologs to OsMADS3 (Schmidt et al., 1993; Theißen et al., 1995; Münster et al., 2002). Unlike rice, maize AG orthologs appear to express in spatially distinct domains, as ZAG1 and ZMM2 transcript levels are higher in carpel and stamen, respectively. A mutation in ZAG1 results in unfused carpels in female flowers, but the carpel identity is not affected (Mena et al., 1996). Functional analysis of ZMM2 and ZMM23 has not been reported yet.
The putative C class genes of wheat, WAG1 (wheat AGAMOUS1) and WAG2, have been identified as OsMADS58 and OsMADS3 orthologs, respectively (Meguro et al., 2003; Zhao et al., 2006; Wei et al., 2011). In early floral organ initiation stages, the transcription level of the two WAG genes is low, but transcripts are detected from booting to heading stages, and in situ hybridization analysis shows WAG expression in stamen, carpel, and ovule (Meguro et al., 2003; Yamada et al., 2009). Besides, ectopic expression of WAG1 and WAG2 can induce pistilloid ectopic stamens in an alloplasmic wheat line (Yamada et al., 2009). Collectively, these data suggest a partially conserved function of wheat AG orthologs in floral organ development, while their lower expressions detected in early stages may also indicate a minor function in carpel differentiation (Callens et al., 2018).
The Arabidopsis CRABS CLAW (CRC), a YABBY family TF, is required for maintaining the proper growth of the gynoecium (Alvarez and Smyth, 1999). CRC is expressed throughout nectary development (Baum et al., 2001). In the gynoecium, CRC is first expressed in the pistil primordium, and subsequently, only in the abaxial epidermis of the carpel and in four internal domains adjacent to the cells that will form the placenta, then its expression disappears after the initiation of ovule primordia (Bowman and Smyth, 1999; Colombo et al., 2010). In the crc mutant, style growth is reduced, resulting in incomplete carpel fusion (Alvarez and Smyth, 1999). It is also reported that the Arabidopsis CRC, a direct target of AtAG (Gómez-Mena et al., 2005; Ó’Maoiléidigh et al., 2013), acts in mediating the transition from FM termination to gynoecium development (Figure 2A) (Yamaguchi et al., 2017). The rice CRC ortholog DL is proposed to be required for carpel identity, because in strong dl mutant alleles, carpels undergo homeotic transformation to stamens (Yamaguchi et al., 2004). In rice, DL expression can be first detected in a few cells at the lemma side of FM, and later during gynoecium development, DL expression starts in the carpel anlagen, shortly before carpel primordia begin to form. After carpel initiation, DL is expressed specifically and uniformly in carpel primordia, whereas no expression is detected in the FM or ovule primordium, which differs from both CRC and the typical AG-like C-function genes (Yamaguchi et al., 2004). DL seems to act mutually and antagonistically to the class B gene SUPERWOMAN1 (SPW1)/OsMADS16 to control carpel identity in rice (Figure 2B), as ectopic stamens replace the gynoecium in dl, whereas homeotic transformation of stamens into carpels occurs in spw1-1. Moreover, the SPW1 and DL transcripts can be detected in the ectopic stamens in the fourth whorl of dl and in the ectopic carpels of spw1-1 mutants, respectively (Nagasawa et al., 2003; Yamaguchi et al., 2004). In Arabidopsis, CRC expression is also found to be expanded in B class mutants ap3 and pi (Bowman and Smyth, 1999; Wuest et al., 2012). However, besides the homeotic transformation, unidentifiable floral organs are indeterminately produced in whorls 3 and 4 of the rice spw1-1 dl double mutants (Nagasawa et al., 2003; Sugiyama et al., 2019). These findings suggest that DL, besides regulating the boundary between whorls 3 and 4, may play an active role in carpel identity determination (Figure 2B). In comparison, Arabidopsis CRC seems to have a more specific function in regulating some aspects of carpel development and is not required to exclude the B class genes from the gynoecium, a function played instead by the Arabidopsis C2H2 zinc finger TF encoded by SUPERMAN (SUP), which is also positively regulated by B class genes themselves in the early stages (Figure 2A) (Schultz et al., 1991; Bowman et al., 1992; Bowman and Smyth, 1999; Sakai et al., 2000).
In wheat, the DL ortholog has been identified by homology screening (Ishikawa et al., 2009). TaDL is also expressed in the wheat pistil, as well as in the ectopic pistil-like primordia replacing stamens in a pistillody line (Hama et al., 2004; Ishikawa et al., 2009). Moreover, the wheat B class MADS-box genes are expressed as expected in the wild-type stamen primordia, but similar to the case in rice, transcripts are not detected in those ectopic carpels in the pistillody line (Hama et al., 2004). Taken together, these facts suggest that a mutual repression between DL and B class genes is conserved in grasses, although this is not yet clear whether it is direct or not.
In Arabidopsis, based on the ABCDE model, carpel identity is specified by the C function gene AtAG and E function genes SEPALLATA1/2/3 with the mutual repression between the A- and C-function genes in their respective domains (Figure 2A) (Pelaz et al., 2000, 2001; Causier et al., 2010). In contrast, the carpel identity regulation model in rice is more controversial, based on the different alleles studied, and/or diverse interpretations on osmads3, osmads58, and dl mutants, currently two different theories exist. Dreni et al. (2011) proposed that OsMADS3 and OsMADS58 still have conserved class C protein functions and redundantly regulate carpel identity, whereas Dreni et al. (2011) implicated DL as a fourth-whorl marker and negative regulator of class B genes, without a direct carpel identity specification function, as DL is unable to specify carpel identity in the absence of OsMADS3 and OsMADS58. In contrast, Sugiyama et al. (2019) claimed a dispensable role of OsMADS3 and OsMADS58 for carpel specification, which would be the first known case in angiosperms, and that DL is the key determinant of carpel identity. However, there is no functional carpel in either spw1-1 dl or osmads3 osmads58 double mutants, suggesting that AG-like genes or DL alone is not sufficient to induce carpel formation. Thus, it is more plausible to conclude that DL, OsMADS3, and OsMADS58 are all necessary for carpel identity specification and morphogenesis (Figure 2B). Besides the above three regulators, OsMADS13, the D function gene of rice (Dreni et al., 2007), also plays a role in carpel establishment, although limited to its adaxial side, as we described above (Figure 2B). This is in contrast with Arabidopsis, where class C and D genes redundantly confer ovule identity, but carpel identity is determined exclusively by C function genes (Figure 2A) (Pinyopich et al., 2003).
Grasses have diverse SEP-like genes that can be divided into two subclades, in rice the three genes OsMADS1, OsMADS5, and OsMADS34 belong to the LOFSEP clade, whereas the SEP3 clade consists of the other two members OsMADS7/45 and OsMADS8/24 (Malcomber and Kellogg, 2005; Zahn et al., 2005; Arora et al., 2007). Similar with Arabidopsis, higher-order knockdown and mutants of rice SEP genes cause the homeotic transformation of floral organs to leaf-like structures, and the rice SEP members also can form heterodimers with C- and D-class genes, suggesting a conserved pivotal role of SEP genes in carpel identity specification (Pelaz et al., 2001; Cui et al., 2010; Wu et al., 2017). However, functional diversification of rice SEP genes also exists compared with Arabidopsis, as inhibition of OsMADS1 or OsMADS34 alone, and OsMADS7 and OsMADS8 together are sufficient to cause severe phenotypes including leafy lemma and palea, homeotic transformation of lodicules, fewer stamens, increased carpels, and flowering time alterations (Cui et al., 2010; Gao et al., 2010). Carpels are either increased or completely missing in several combinations of lofsep mutants (Wu et al., 2017). The simultaneous knockdown of OsMADS7 and OsMADS8 causes different degrees of carpel fusion defects, also affecting the FMD with additional reproductive organ-like structures initiated inside the mutant carpels (Cui et al., 2010).
Recently, some potential regulations between rice SEP genes and AG genes have been discovered. As the expression of OsMADS3 and OsMADS58 significantly decreased in osmads1-z and lofsep double and triple mutants, it is proposed that rice LOFSEP members may act as upstream activators of C-class genes to regulate inner floral organs (Figure 2B) (Wu et al., 2017). Indeed, consistent with the delayed transition of the spikelet meristem to FM, the initiation of OsMADS3 expression is delayed, and OsMADS58 expression is missed from the FM in osmads1-z mutants (Hu et al., 2015). Moreover, another recent study (Osnato et al., 2020) has identified a substantial overlap with DEGs from the transcriptome data of osmads1 (Khanday et al., 2013; Hu et al., 2015) and osmads13 mutants, suggesting the two genes may regulate common downstream targets but in an antagonistic manner. The authors also proposed that OsMADS13 may act as a repressor in the carpel initiation pathway in the ovule domain (Osnato et al., 2020).
In short, these findings suggest that OsMADS3, OsMADS58, DL, OsMADS13, and SEP genes in rice have both conserved and diverse functions in carpel identity determination and morphogenesis, compared with Arabidopsis. These functional diversifications may play a pivotal role in establishing rice carpel morphology, and it is highly likely that similar cases also exist in other grass plants such as maize, barley, and wheat.
Unlike indeterminate meristems such as the shoot apical meristem and inflorescence meristem in Arabidopsis and some grass species, the FM is determinate, as the stem cells in FM will be consumed by the final floral organ initiated from it. The pattern of FM termination is diverse among angiosperm species. In wild-type rice flower, the carpel primordia bulge first arises at the lemma side of the FM, then at the flank of the meristem; the other carpel primordia develop toward the opposite side to enclose the meristem. The FM remains morphologically undifferentiated and later develops into the ovule (Itoh et al., 2005). But in Arabidopsis, the stem cell maintenance ceases when carpel primordium starts to be initiated, and ovules differentiate from the placenta after establishment of the carpel (Cucinotta et al., 2014). Thus, the central region of the FM is consumed by the ovule or carpel/placenta primordium in rice and in Arabidopsis, respectively.
Interactions between floral homeotic genes and the well-characterized CLAVATA (CLV)-WUSCHEL (WUS) negative feedback pathway, which coordinates stem cell proliferation with differentiation in the meristem, are part of FMD regulation (Brand et al., 2000; Schoof et al., 2000; Ikeda et al., 2009; Pautler et al., 2013). In Arabidopsis, the stem cell-promoting gene WUS is able to induce AtAG expression, whereas AtAG in turn can switch off WUS right after carpel primordia emergence, through several molecular mechanisms, to trigger FM termination in the center of the flower bud (reviewed by Shang et al., 2019). Mutations of the receptor kinase-encoding gene CLV1, another component of the CLV-WUS pathway, can trigger defects in AtAG expression (Clark et al., 1993). Grass class C genes also play an important role in FMD. Like the Arabidopsis ag mutant, osmads3 osmads58 double mutants show a dramatic loss of FMD, as an indefinite number of ectopic lodicules and green palea/carpel-like organs are repeatedly formed (Dreni et al., 2011; Sugiyama et al., 2019), and the FM marker gene OSH1 (O. sativa homeobox1) expression remains even in mature flowers (Sugiyama et al., 2019). Loss of function in maize ZAG1 also results in the partial loss of FMD in female flowers, showing a reiteration of carpels (Mena et al., 1996). The rice FON1 and FON2/FON4 genes are identified as orthologs of CLV1 and CLV3, respectively, an alteration in floral organ number and increased FM size have been observed in fon1-1 (Moon et al., 2006) and fon4-1 (Chu et al., 2006) mutants, suggesting that the function of the CLV3-CLV1 ligand–receptor system is conserved. The organ number is more severely affected in the inner whorls in those mutants, and nearly all fon4-1 and fon4-2 flowers have from 2 to 10 carpels (Chu et al., 2006). Both fon4-2 osmads3-4 and fon4-1 osmads58 double mutants display enhanced defects of FMD with more carpel-like structures than either single mutants. The fon4-2 osmads3-4 double mutant has a dramatically enlarged FM size and larger OSH1 expression region, compared with wild-type plants, whereas the expression of OsMADS3 and OsMADS58 in fon4-2 mirrored that in the wild type. In conclusion, FON4 synergistically interacts with C-class genes in rice FMD (Xu et al., 2017; Yasui et al., 2017).
As described above, ovule is the final organ differentiated from FM in grasses. Mutation in the ovule identity specification gene OsMADS13 also results in partial loss of FMD. Reiteration of carpels and prolonged expression of OSH1 have been observed in osmads13 mutants (Dreni et al., 2007; Yamaki et al., 2011). The number of carpelloid structures in the fon4-1 osmads13-3 double mutant is greatly increased in comparison with the single mutants, and the expression domain of OSH1 is wider. Besides, the expression level of OsMADS13 is increased and more widely detectable in the gynoecium of fon4-2 mutant (Xu et al., 2017). Thus, FON4 may function to repress OsMADS13 expression in the innermost whorl, or the increased expression of OsMADS13 may act as a molecular response to alleviate the expansion of the meristem in fon4-2 (Xu et al., 2017).
In Arabidopsis, LFY and UNUSUAL FLORAL ORGAN (UFO) are reported to be key factors in enhancing the floral fate of lateral meristems and regulating the ABC floral organ identity genes (Weigel et al., 1992; Lee et al., 1997; Parcy et al., 1998; Chae et al., 2008). In rice, ABERRANT PANICLE ORGANIZATION 2 (APO2, ortholog of LFY) and ABERRANT PANICLE ORGANIZATION 1 (APO1, ortholog of UFO) act cooperatively in controlling inflorescence and flower development. Both apo1 and apo2 mutants display partial loss of FMD in whorl 4 with indeterminate carpel formation, which is even more severe in the apo1-1apo2-1 double mutant (Ikeda et al., 2005; Ikeda-Kawakatsu et al., 2012). Furthermore, the expression of C class gene OsMADS3 is down-regulated in apo1. Considering that the osmads3 loss-of-function mutants form apo1-like flowers, it is reasonable to propose a positive regulation of the C class gene OsMADS3 by APO1, but not of B class genes as in Arabidopsis (Lohmann and Weigel, 2002; Ikeda et al., 2005).
Interactions within floral homeotic genes are also important in determining FM fate. Similar to Arabidopsis (Mizukami and Ma, 1995), FMD in rice seems to be sensitive to the amount of AG-like protein(s), as all the three double mutants (osmads3-3 osmads58, osmads3-3 osmads13, and osmads13 osmads58) result in an enhanced indeterminacy in the fourth whorl (Dreni et al., 2011). In addition, OsMADS13 and OsMADS3 do not regulate each other’s transcription, suggesting a synergistic role of these two genes (Li et al., 2011b).
During FM termination in Arabidopsis, AtAG represses WUS in part by directly activating three key target genes, the C2H2 zinc finger genes KNUCKLES (KNU) and SUP, and the YABBY gene CRC (Alvarez and Smyth, 1999; Payne et al., 2004; Gómez-Mena et al., 2005; Lee et al., 2005; Ó’Maoiléidigh et al., 2013). SUP and CRC contribute to FMD non-cell autonomously, at least in part by fine tuning auxin homeostasis (Yamaguchi et al., 2017; Lee et al., 2019).
The rice ortholog of CRC is DL, which also functions as carpel identity regulator and, non-cell autonomously, in FMD (Yamaguchi et al., 2004; Sugiyama et al., 2019). In rice dl mutant, ectopic stamens are variable in number (Yamaguchi et al., 2004). The osmads3-4 dl double mutants display a severe loss of FMD and produced extra whorls of lodicule-like organs in the floral center (Li et al., 2011b). However, the expression patterns of OsMADS3 and DL are not affected by each other; thus, OsMADS3 and DL may terminate the FM in a synergistic way. On the contrary, osmads13-3 dl flowers exhibited the similar defects to those of dl, and the transcripts of OsMADS13 is undetectable in dl flowers, suggesting that DL and OsMADS13 may function in the same pathway to specify the identity of carpel/ovule and FMD (Figure 3) (Li et al., 2011b). In Arabidopsis, CRC is a direct target of both B class genes and of the C class gene AtAG, which act as repressors and as activators, respectively (Bowman and Smyth, 1999; Gómez-Mena et al., 2005; Wuest et al., 2012; Ó’Maoiléidigh et al., 2013). In rice, DL expression also expands outward in the third whorl in B class mutants (Yamaguchi et al., 2004), but it does not disappear in the palea-like organs replacing the pistil in osmads3 osmads58 double and osmads3 osmads13 osmads58 triple mutants, suggesting that its expression is independent by C class genes (Dreni et al., 2011). In Arabidopsis, CRC genetically interacts in the fourth whorl with another FMD regulator, SUP, and common downstream genes such as cytokinin- and auxin-related genes are found between these two TFs (Prunet et al., 2017; Lee et al., 2019). As rice SUP counterparts have not been studied yet, it will be interesting to assess their function in rice FMD and carpel development.
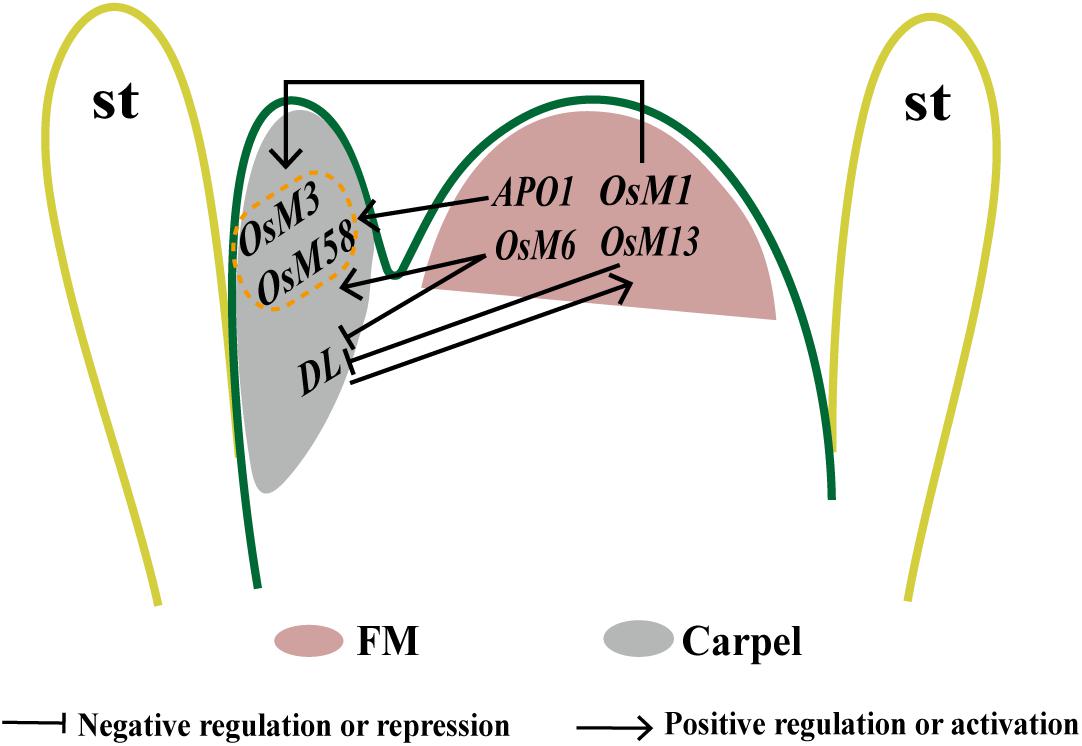
Figure 3. Proposed model to illustrate the genetic interaction between rice floral homeotic genes in FMD in whorl 4. OsMADS3 and OsMADS58 are possibly activated by E-function genes OsMADS1 and OsMADS6 and also APO1, the expression of DL may be repressed by OsMADS6 and OsMADS13. In turn, DL may indirectly regulate OsMADS13 in a positive way. The onset of OsMADS13 expression in the final stage of the FM leads its transition to ovule primordium.
Recently, the maize CRC and DL co-orthologs DRL1 and DRL2 have been also reported to regulate FM activity and impose FMD from lateral floral organs in a non-cell-autonomous way (Strable and Vollbrecht, 2019). The drl1 mutant ears are sterile, carpel walls remain unfused with underdeveloped silks, and an expanded nucellus-like structure is exposed in the center. The medial-adaxial carpel (determinate carpel) is greatly reduced in both drl2 and drl1 drl2 double mutants, and multiple lateral-abaxial carpels (indeterminate carpels) have been observed in the double mutants, indicating a prolonged FM activity (Strable and Vollbrecht, 2019). Genetic interaction and expression analyses using gene regulatory network inference suggest that DRL1 and ZAG1 target each other, and there is a common set of downstream genes involved in floral patterning and FMD (Strable and Vollbrecht, 2019).
As mentioned previously, physical and genetic interactions between OsMADS1 and the two rice C-class genes are essential for the regulation of carpel identity specification and FM activity (Hu et al., 2015); 74.67% of osmads1-z osmads3-4 flowers lack inner floral organs, but only have extra glume-like structures, and OSH1 expression is lost at a later stage in this double mutant, suggesting a function of OsMADS1 and OsMADS3 together in repressing FM activity (Figure 3) (Hu et al., 2015). As another MADS-box gene with E-class functions in rice, genetic analysis reveals a key role of the AGL6 subfamily member MOSAIC FLORAL ORGAN 1 (MFO1)/OsMADS6 in early flower development (Li et al., 2011a). In the flower of mfo1 mutants, the FMD is lost, and extra carpels or spikelets developed in mfo1 florets (Ohmori et al., 2009). A complex regulation network also exists within OsMADS6 and other carpel regulators. All flowers in osmads6 dl double mutant display an inflorescence-like structure in whorl 4, and ectopic expression of DL is present in the altered palea organ and ectopic carpels or abnormal ovules in osmads6-1. Taken together, it is proposed that OsMADS6 may repress the expression of DL, whereas DL does not affect OsMADS6 expression (Figure 3) (Li et al., 2011a).
Moreover, OsMADS6 also interacts genetically with OsMADS13, OsMADS3, and OsMADS58. The flowers of osmads6-1 osmads13-3 display more severe defects in carpel/ovule development and FMD than single mutants (Li et al., 2011a), its floral axis developing higher-order carpel-like organs expressing DL. There is no obvious regulation between OsMADS6 and OsMADS13 at the transcriptional level, although they have partial functional redundancy in carpel/ovule identity specification and FMD termination. Defects are intensified with even more carpels in the floral center of the osmads6-1 osmads3-4 and osmads6-1 osmads58 double mutants, and an inflorescence-like structure is produced in osmads6-1 osmads3-4 flowers (Li et al., 2011a). In osmads6-1, the expression of OsMADS3 seems much weaker and delayed, and expression of OsMADS58 is reduced at the early stages. Furthermore, OsMADS6 may regulate OsMADS58 expression directly through binding to a CArG element in its second intron (Li et al., 2011a), suggesting that OsMADS6 may promote C-class genes expression during early rice flower development (Figure 3).
Generally, FM activity is terminated after the formation of a fixed number and pattern of floral organs. In Arabidopsis, it has been shown that AtAG is a key regulator in abolishing FM by switching off WUS (Lohmann et al., 2001; Liu et al., 2011; Sun et al., 2019). In rice, interactions between FON4, APO1, and C- and D-function genes are shown to act in FMD. Also, the interactions between floral homeotic genes DL, OsMADS3, OsMADS58, OsMADS13, and E-function genes OsMADS1 and OsMADS6 suggest a regulatory module that fine-tunes floret patterning and FMD in rice (Figure 3) (Ohmori et al., 2009; Dreni et al., 2011; Li et al., 2011a,b; Hu et al., 2015; Strable and Vollbrecht, 2019).
Carpel is a relatively complex biological structure among the plant organs; it requires several tissues to acquire specific identities in the specific time and places. Studies in Arabidopsis have shown that gynoecium formation depends on precise hormonal and genetic interactions to produce this complex tissue organization (Marsch-Martínez et al., 2012; Marsch-Martínez and de Folter, 2016; Moubayidin and Østergaard, 2017). Several recent reviews well-summarized the role of plant hormones such as auxin, cytokinin, and their crosstalk and the genes that connect these hormonal pathways during gynoecium development processes such as apical-basal and mediolateral polarity establishment, identity specification, symmetry establishment, and patterning (Moubayidin and Østergaard, 2017; Deb et al., 2018; Zúñiga-Mayo et al., 2019). The developmental control by plant hormone in the carpel margin meristem, which is the inner tissues possessing meristematic characteristics in the Arabidopsis gynoecium, is also addressed in several other reviews (Reyes-Olalde et al., 2013; Reyes-Olalde and de Folter, 2019).
Auxin is an essential hormone for almost all developmental processes in plants; experimental and modeling approaches have demonstrated that organ initiation depends on a threshold concentration of auxin (Reinhardt et al., 2003). Disruption of the Arabidopsis and maize protein kinase PINOID (PID), which is involved in polar auxin transport, causes severe defects in flower initiation. Arabidopsis pid and maize ortholog bif2 (BARREN INFLORESCENCE2) mutants display defects in flower initiation that results in a pin-like inflorescence (Bennett et al., 1995; McSteen et al., 2007). Surprisingly, despite the fact that OsPID expresses throughout the panicle development as well as in pistil, none of the ospid mutants develops pin-like inflorescences. Instead, stigma development is completely eliminated in ospid mutants and normal ovules with eight-nucleate embryo sac failed to develop in most spikelets (He et al., 2019; Xu et al., 2019), whereas the overexpression of OsPID led to overproliferation of stigmas (Morita and Kyozuka, 2007). These observations suggest a key role of OsPID for stigma development and ovule initiation.
In ospid mutant, the expression of most auxin response factor genes is shown to be downregulated, and OsETTIN1, OsETTIN2, and OsMONOPTEROS (OsMP) lost their original spatiotemporal expression pattern during pistil development (Xu et al., 2019). As ETTIN and MP play pivotal roles in FM maintenance, OSH1 transcription signal is weaker, and FON4 transcription level is higher in ospid, it is proposed that OsPID may regulate stigma and ovule initiation by maintaining stem cell identity through auxin signaling (Xu et al., 2019). A recent study shows that OsPID also interacts physically with OsMADS16 (Wu et al., 2020); thus, OsPID may be involved in carpel development by interacting with floral homeotic genes. Indeed, another auxin-responsive gene OsMGH3 (OsMADS1 regulated GH3 domain-encoding gene) has been reported as a common downstream target of OsMADS1 and OsMADS6 (Prasad et al., 2005; Zhang et al., 2010). GH3 control cellular bioactive auxin by inactivating excess auxin as conjugates of amino acids and sugars to maintain auxin homeostasis (Woodward and Bartel, 2005). OsMGH3 knockdown plants display partial overlapped phenotypes with osmads1 and osmads6; carpel development and pollen viability are affected by reduced fertility in dsRNAiOsMGH3 mutant plants (Yadav et al., 2011). Besides, 10%–20% of OsMGH3 knockdown transgenic flowers show enlarged carpels, which protrudes out of the flower before anthesis (Yadav et al., 2011). Such enlarged carpel phenotypes are similar to Arabidopsis ettin and arf2 that are mutants in auxin response factors, suggesting the key role of auxin-responsive genes regulation in plant carpel development (Sessions et al., 1997; Okushima et al., 2005).
Cytokinin is a key regulator of meristem size and activity by controlling cell division and differentiation and thus influences numerous developmental programs in plants. The rice LONELY GUY (LOG) gene encodes a cytokinin-activating enzyme functioning in the final step of bioactive cytokinin synthesis. The mRNA of LOG, which is required to maintain meristem activity, is specifically detected in shoot meristem tips, and its loss-of-function mutant displays premature termination of the shoot meristem (Kurakawa et al., 2007). In the log-3 weak allele, 65.3% of spikelets form a slender pistil lacking an ovule, whereas 15.8% form no pistil. The FM volume in log-3 mutant is normally maintained until carpel protrusion, but subsequently its size is decreased compared to the wild type (Yamaki et al., 2011). In the log-3 fon1-1 double mutant, the FM size is recovered, and the ovules phenotype is largely rescued compared to the log-3 single mutant. In addition to prevent premature termination of the shoot meristem, LOG is also required for ovule formation through maintaining a sufficient volume of the FM to allow normal development of carpels (Yamaki et al., 2011).
In conclusion, studies conducted to date have revealed a close link between plant hormone pathways and carpel establishment in grasses. Although advances have been made in this area, there are still many questions awaiting to be answered, including how the transition between meristem activity and ovule initiation is regulated and whether there are even closer interactions between plant hormone pathways and the C, D, and E class genes relevant to carpel development.
MiRNAs are small ribonucleic acid molecules (typically 21 nt in length) that negatively regulate gene expression mainly by triggering mRNA cleavage, translational inhibition, or DNA cytosine and/or histone methylation of miRNA target genes (Voinnet, 2009; Liu et al., 2017). Studies conducted so far suggest the role of miRNAs as an important regulatory mechanism in all phases of plant life including flower development (Rubio-Somoza and Weigel, 2011; Xie et al., 2015; Smoczynska and Szweykowska-Kulinska, 2016; Liu et al., 2017).
It seems that miRNA-mediated regulation of floral genes as was observed in Arabidopsis also takes place in monocots, such as the suppression of AP2-like genes mediated by miRNA172 (Chen, 2004; Chuck et al., 2007; Zhu et al., 2009; Nair et al., 2010). Overexpression of miRNA172 converts sepals and petals into carpels in Arabidopsis, a phenotype similar to ap2 mutants (Chen, 2004). Rice plants overexpressing miR172 phenocopied another AP2-like gene mutant named snb, displaying multiple changes in flower organ development; the carpel was occasionally replaced by a mosaic organ with a lodicule base and anther apices (Zhu et al., 2009). Defective unfused carpels are also shown in the female flowers of maize tassel seed4 (ts4), a loss-of-function mutant of miRNA172e (Chuck et al., 2007). Mutations in hormonal balance and TFs such as APETALA2 (AP2), CUP-SHAPED COTYLEDON2 (CUC2) and HECATE all lead to partially or completely unfused carpels in Arabidopsis gynoecium, and CUC2 has been shown as a target of miR164 (Ripoll et al., 2011; Nahar et al., 2012; Schuster et al., 2015). Using the combination of miRNA sequencing, degradome, and physiological analyses, in total 20 of 162 known miRNAs have been identified to be differentially expressed between incompletely and completely fused carpels in maize (Li et al., 2017). Moreover, 60% target genes of the differentially expressed known miRNAs are found to encode TFs, including those reported to play a role in carpel fusion and development such as auxin response factor (ARF), TB1-CYC-PCFs (TCP), AP2, growth regulating factor (GRF), MYB, and NAC (Li et al., 2017). In barley, the AP2 ortholog Cly1 (cleistogamy1) is also targeted by miR172; however, the downregulation of Cly1 seems only to affect lodicule development (Nair et al., 2010). In wheat, mutations to the two AP2-like miR172 targets AP2L5 (also known as gene Q) and AP2L2 resulting in homeotic transformation of lodicule and the adjacent stamen into carpelloid structures (Debernardi et al., 2020). But different from the petal-to-stamen conversion observed in Arabidopsis ap2 mutants, which is just caused by the expansion of AtAG expression in the second whorl, the conversion of lodicules in wheat ap2l2 ap2l5 mutant is a result of both reduced expression of B-class genes and increased expression of AG-like genes (Debernardi et al., 2020).
In Arabidopsis, miRNA396 has been characterized to mediate carpel development by suppressing its GRF target genes. Pistils with a single carpel are observed in miR396 overexpression plants, these carpel abnormalities can be rescued by miR396-resistant version of GRFs (Liang et al., 2014). GRF interact with its transcription coactivator GRF-INTERACTING FACTOR (GIF) to form the GRF/GIF complex in plant cell nucleus. It is also proposed that the amount of GRF/GIF complex is essential for carpel establishment. While the gif single mutant displays normal pistils, the triple mutant gif1/gif2/gif3 produces abnormal pistils similar to 35S:MIR396a/grf5 plants (Liang et al., 2014). Similarly, transgenic rice plants overexpressing miR396 display altered floral organ morphology including abnormal stigma numbers, and this effect is correlated with a significant down-regulation of GRF6 and other members of this family (Liu et al., 2014). OsGRFs, modulated by their interaction with OsGIF1, directly activate the expression of targets, including OsJMJ706 and OsCR4, which has been reported to participate in the regulation of floral organ development (Sun and Zhou, 2008; Pu et al., 2012; Liu et al., 2014). Overall, increasing evidence suggests a role of microRNAs such as miRNA172 and miRNA396 in carpel development, especially in carpel fusion.
As productivity depends on grain number and grain weight in many corps (Sreenivasulu and Schnurbusch, 2012; Sakuma and Schnurbusch, 2020), genes and genetic pathways involved in carpel establishment can potentially be manipulated to increase their yield. For example, the increase in carpel number in fon4 mutants also results in some flower with two seeds that have normal embryos (Chu et al., 2006). Reducing the preprogrammed abortion of floral organs such as pistil primordia abortion in maize tassel spikelets and lateral floret abortion in barley could also be a potential way in yield improvement, as reviewed recently (Gauley and Boden, 2019; Sakuma and Schnurbusch, 2020). However, how to manipulate those genetic components that can improve carpel traits without affecting negatively inflorescence and floral development might be a big challenge. Besides, the completely female-sterile ospid mutant lacking stigma and style also shows its potential for applications in hybrid rice production. In traditional hybrid rice production, the two parental lines need to be planted within close proximity, and grains from the two parental lines have to be harvested separately as only grains from the male-sterile maternal line will be hybrid rice, which makes the process labor intensive. On the other hand, in seedling and young adult stages, ospid mutants appear quite normal, whereas extra stamens and viable pollen grains are produced in ospid flowers (He et al., 2019). Therefore, if sown together with male-sterile lines, an ospid mutant might be a good resource for rice hybrid breeding, which will greatly reduce labor and production costs compared with the traditional method, as any grains produced will be hybrid using sterile lines from both parents (He et al., 2019), whereas in this case, a rescue system will be needed for both parents.
During the last decade, great progress has been made toward understanding developmental mechanisms of carpel establishment. As summarized above, grasses have evolved specific mechanism to regulate carpel identity and stigma formation. However, many conserved regulators are maintained during grass family evolution. TFs in the AG lineage still play essential roles in grass female organ regulation, with conserved functions and conserved interaction partners. The miRNA-mediated carpel regulation pathways are also largely conserved among monocot and dicot plants. Even though some genes and pathways have been discovered in grass carpel development especially in rice and maize, the underlying modes of action are still largely unknown, which should be the future focus. With the increasing availability of gene editing technologies and artificially selected cultivars and the genome-scale expression profiling data, a deeper understanding in cereal crop carpel regulation genes and pathways will be obtained, which will undoubtedly benefit crop breeding programs for grain yield increase.
CS, GL, and DZ designed the manuscript. CS, LD, GL, and DZ wrote the manuscript. LD, GL, and DZ critically evaluated the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported financially by the National Key Research and Development Program of China, Ministry of Science and Technology (Grant Nos. 2016YFD0100804 and 2016YFE0101000), the National Natural Science Foundation of China (Grant Nos. 31970803 and 31861163002), and the Australian Research Council (DP170103352).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Alvarez, J., and Smyth, D. R. (1999). CRABS CLAW and SPATULA, two Arabidopsis genes that control carpel development in parallel with AGAMOUS. Development 126, 2377–2386.
Arora, R., Agarwal, P., Ray, S., Singh, A. K., Singh, V. P., Tyagi, A. K., et al. (2007). MADS-box gene family in rice: genome-wide identification, organization and expression profiling during reproductive development and stress. BMC Genomics 8:242. doi: 10.1186/1471-2164-8-242
Baum, D. A., and Hileman, L. C. (2007). “A developmental genetic model for the origin of the flower,” in Flowering and its Manipulation, ed. C. Ainsworth (Hoboken, NJ: Wiley).
Baum, S. F., Eshed, Y., and Bowman, J. L. (2001). The Arabidopsis nectary is an ABC-independent floral structure. Development 128, 4657–4667. doi: 10.1111/j.1756-1051.1997.tb00355.x
Becker, A. (2020). A molecular update on the origin of the carpel. Curr. Opin. Plant Biol. 53, 15–22. doi: 10.1016/j.pbi.2019.08.009
Bennett, S. R. M., Alvarez, J., Bossinger, G., and Smyth, D. R. (1995). Morphogenesis in pinoid mutants of Arabidopsis thaliana. Plant J. 8, 505–520. doi: 10.1046/j.1365-313X.1995.8040505.x
Bowman, J. L., Sakai, H., Jack, T., Weigel, D., Mayer, U., and Meyerowitz, E. M. (1992). SUPERMAN, a regulator of floral homeotic genes in Arabidopsis. Development 114, 599–615.
Bowman, J. L., and Smyth, D. R. (1999). CRABS CLAW, a gene that regulates carpel and nectary development in Arabidopsis, encodes a novel protein with zinc finger and helix-loop-helix domains. Development 126, 2387–2396.
Bowman, J. L., Smyth, D. R., and Meyerowitz, E. M. (1989). Genes directing flower development in Arabidopsis. Plant Cell 1, 37–52. doi: 10.2307/3869060
Bowman, J. L., Smyth, D. R., and Meyerowitz, E. M. (1991). Genetic interactions among floral homeotic genes of Arabidopsis. Development 112, 1–20.
Brand, U., Fletcher, J. C., Hobe, M., Meyerowitz, E. M., and Simon, R. (2000). Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289, 617–619. doi: 10.1126/science.289.5479.617
Callens, C., Tucker, M. R., Zhang, D., and Wilson, Z. A. (2018). Dissecting the role of MADS-box genes in monocot floral development and diversity. J. Exp. Bot. 69, 2435–2459. doi: 10.1093/jxb/ery086
Causier, B., Bradley, D., Cook, H., and Davies, B. (2009). Conserved intragenic elements were critical for the evolution of the floral C-function. Plant J. 58, 41–52. doi: 10.1111/j.1365-313X.2008.03759.x
Causier, B., Schwarz-Sommer, Z., and Davies, B. (2010). Floral organ identity: 20 years of ABCs. Semin. Cell Dev. Biol. 21, 73–79. doi: 10.1016/j.semcdb.2009.10.005
Chae, E., Tan, Q. K. G., Hill, T. A., and Irish, V. F. (2008). An Arabidopsis F-box protein acts as a transcriptional co-factor to regulate floral development. Development 135, 1235–1245. doi: 10.1242/dev.015842
Chen, X. (2004). A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303, 2022–2025. doi: 10.1126/science.1088060
Chu, H., Qian, Q., Liang, W., Yin, C., Tan, H., Yao, X., et al. (2006). The FLORAL ORGAN NUMBER4 gene encoding a putative ortholog of Arabidopsis CLAVATA3 regulates apical meristem size in rice. Plant Physiol. 142, 1039–1052. doi: 10.1104/pp.106.086736
Chuck, G., Meeley, R., Irish, E., Sakai, H., and Hake, S. (2007). The maize tasselseed4 microRNA controls sex determination and meristem cell fate by targeting Tasselseed6/indeterminate spikelet1. Nat. Genet. 39, 1517–1521. doi: 10.1038/ng.2007.20
Clark, S. E., Running, M. P., and Meyerowitz, E. M. (1993). CLAVATA1, a regulator of meristem and flower development in Arabidopsis. Development 119, 397–418.
Coen, E. S., and Meyerowitz, E. M. (1991). The war of the whorls: genetic interactions controlling flower development. Nature 353, 31–37. doi: 10.1038/353031a0
Colombo, L., Franken, J., Koetje, E., Van Went, J., Dons, H. J., Angenent, G. C., et al. (1995). The petunia MADS box gene FBP11 determines ovule identity. Plant Cell. 7, 1859–1868. doi: 10.1105/tpc.7.11.1859
Colombo, M., Brambilla, V., Marcheselli, R., Caporali, E., Kater, M. M., and Colombo, L. (2010). A new role for the SHATTERPROOF genes during Arabidopsis gynoecium development. Dev. Biol. 337, 294–302. doi: 10.1016/j.ydbio.2009.10.043
Cucinotta, M., Colombo, L., and Roig-Villanova, I. (2014). Ovule development, a new model for lateral organ formation. Front. Plant Sci. 5:117. doi: 10.3389/fpls.2014.00117
Cui, R., Han, J., Zhao, S., Su, K., Wu, F., Du, X., et al. (2010). Functional conservation and diversification of class e floral homeotic genes in rice (Oryza sativa). Plant J. 61, 767–781. doi: 10.1111/j.1365-313X.2009.04101.x
Deb, J., Bland, H. M., and Østergaard, L. (2018). Developmental cartography: coordination via hormonal and genetic interactions during gynoecium formation. Curr. Opin. Plant Biol. 41, 54–60. doi: 10.1016/j.pbi.2017.09.004
Debernardi, J. M., Greenwood, J. R., Jean Finnegan, E., Jernstedt, J., and Dubcovsky, J. (2020). APETALA 2-like genes AP2L2 and Q specify lemma identity and axillary floral meristem development in wheat. Plant J. 101, 171–187. doi: 10.1111/tpj.14528
Dreni, L., Jacchia, S., Fornara, F., Fornari, M., Ouwerkerk, P. B. F., An, G., et al. (2007). The D-lineage MADS-box gene OsMADS13 controls ovule identity in rice. Plant J. 52, 690–699. doi: 10.1111/j.1365-313X.2007.03272.x
Dreni, L., and Kater, M. M. (2014). MADS reloaded: evolution of the AGAMOUS subfamily genes. New Phytol. 201, 717–732. doi: 10.1111/nph.12555
Dreni, L., Osnato, M., and Kater, M. M. (2013). The ins and outs of the rice AGAMOUS subfamily. Mol. Plant 6, 650–664. doi: 10.1093/mp/sst019
Dreni, L., Pilatone, A., Yun, D., Erreni, S., Pajoro, A., Caporali, E., et al. (2011). Functional analysis of all AGAMOUS subfamily members in rice reveals their roles in reproductive organ identity determination and meristem determinacy. Plant Cell 23, 2850–2863. doi: 10.1105/tpc.111.087007
Dreni, L., Ravasio, A., Gonzalez-Schain, N., Jacchia, S., da Silva, G. J., Ricagno, S., et al. (2020). Functionally divergent splicing variants of the rice AGAMOUS ortholog OsMADS3 are evolutionary conserved in grasses. Front. Plant Sci. 11:637. doi: 10.3389/fpls.2020.00637
Endress, P. K., and Doyle, J. A. (2009). Reconstructing the ancestral angiosperm flower and its initial specializations. Am. J. Bot. 96, 22–66. doi: 10.3732/ajb.0800047
Endress, P. K., and Doyle, J. A. (2015). Ancestral traits and specializations in the flowers of the basal grade of living angiosperms. Taxon 64, 1093–1116. doi: 10.12705/646.1
Favaro, R., Pinyopich, A., Battaglia, R., Kooiker, M., Borghi, L., Ditta, G., et al. (2003). MADS-box protein complexes control carpel and ovule development in Arabidopsis. Plant Cell. 15, 2603–2611. doi: 10.1105/tpc.015123
Ferrándiz, C., Fourquin, C., Prunet, N., Scutt, C. P., Sundberg, E., Trehin, C., et al. (2010). Carpel development. Adv. Bot. Res. 55, 1–74. doi: 10.1016/S0065-2296(10)55001-8
Fourquin, C., Vinauger-Douard, M., Fogliani, B., Dumas, C., and Scutt, C. P. (2005). Evidence that CRABS CLAW and TOUSLED have conserved their roles in carpel development since the ancestor of the extant angiosperms. Proc. Natl. Acad. Sci. U.S.A. 102, 4649–4654. doi: 10.1073/pnas.0409577102
Frohlich, M. W. (2003). An evolutionary scenario for the origin of flowers. Nat. Rev. Genet. 4, 559–566. doi: 10.1038/nrg1114
Gao, X., Liang, W., Yin, C., Ji, S., Wang, H., Su, X., et al. (2010). The SEPALLATA-like gene OsMADS34 is required for rice inflorescence and spikelet development. Plant Physiol. 153, 728–740. doi: 10.1104/pp.110.156711
Gauley, A., and Boden, S. A. (2019). Genetic pathways controlling inflorescence architecture and development in wheat and barley. J. Integr. Plant Biol. 61, 296–309. doi: 10.1111/jipb.12732
Gomariz-Fernández, A., Sánchez-Gerschon, V., Fourquin, C., and Ferrándiz, C. (2017). The role of SHI/STY/SRS genes in organ growth and carpel development is conserved in the distant eudicot species Arabidopsis thaliana and Nicotiana benthamiana. Front. Plant Sci. 8:814. doi: 10.3389/fpls.2017.00814
Gómez-Mena, C., de Folter, S., Costa, M. M. R., Angenent, G. C., and Sablowski, R. (2005). Transcriptional program controlled by the floral homeotic gene AGAMOUS during early organogenesis. Development 132, 429–438. doi: 10.1242/dev.01600
Hama, E., Takumi, S., Ogihara, Y., and Murai, K. (2004). Pistillody is caused by alterations to the class-B MADS-box gene expression pattern in alloplasmic wheats. Planta 218, 712–720. doi: 10.1007/s00425-003-1157-6
He, Y., Yan, L., Ge, C., Yao, X. F., Han, X., Wang, R., et al. (2019). PINOID is required for formation of the stigma and style in rice. Plant Physiol. 180, 926–936. doi: 10.1104/pp.18.01389
Hickey, L. J., and Taylor, D. W. (1996). “Origin of the angiosperm flower,” in Flowering Plant Origin, Evolution & Phylogeny, eds D. W. Taylor and L. J. Hickey (Boston, MA: Springer), 176–231. doi: 10.1007/978-0-585-23095-5_8
Honma, T., and Goto, K. (2001). Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 409, 525–529. doi: 10.1038/35054083
Hu, L., Liang, W., Yin, C., Cui, X., Zong, J., Wang, X., et al. (2011). Rice MADS3 regulates ROS homeostasis during late anther development. Plant Cell. 23, 515–533. doi: 10.1105/tpc.110.074369
Hu, Y., Liang, W., Yin, C., Yang, X., Ping, B., Li, A., et al. (2015). Interactions of OsMADS1 with floral homeotic genes in rice flower development. Mol. Plant 8, 1366–1384. doi: 10.1016/j.molp.2015.04.009
Ikeda, K., Nagasawa, N., and Nagato, Y. (2005). Aberrant panicle organization 1 temporally regulates meristem identity in rice. Dev. Biol. 282, 349–360. doi: 10.1016/j.ydbio.2005.03.016
Ikeda, M., Mitsuda, N., and Ohme-Takagi, M. (2009). Arabidopsis WUSCHEL is a bifunctional transcription factor that acts as a repressor in stem cell regulation and as an activator in floral patterning. Plant Cell 21, 3493–3505. doi: 10.1105/tpc.109.069997
Ikeda-Kawakatsu, K., Maekawa, M., Izawa, T., Itoh, J. I., and Nagato, Y. (2012). ABERRANT PANICLE ORGANIZATION 2/RFL, the rice ortholog of Arabidopsis LEAFY, suppresses the transition from inflorescence meristem to floral meristem through interaction with APO1. Plant J. 69, 168–180. doi: 10.1111/j.1365-313X.2011.04781.x
Ishikawa, M., Ohmori, Y., Tanaka, W., Hirabayashi, C., Murai, K., Ogihara, Y., et al. (2009). The spatial expression patterns of DROOPING LEAF orthologs suggest a conserved function in grasses. Genes Genet. Syst. 84, 137–146. doi: 10.1266/ggs.84.137
Itoh, J. I., Nonomura, K. I., Ikeda, K., Yamaki, S., Inukai, Y., Yamagishi, H., et al. (2005). Rice plant development: from zygote to spikelet. Plant Cell Physiol. 46, 23-47. doi: 10.1093/pcp/pci501
Kellogg, E. A. (2001). Evolutionary history of the grasses. Plant Physiol. 125, 1198–1205. doi: 10.1104/pp.125.3.1198
Khanday, I., Ram Yadav, S., and Vijayraghavan, U. (2013). Rice LHS1/OsMADS1 controls floret meristem specification by coordinated regulation of transcription factors and hormone signaling pathways. Plant Physiol. 161, 1970–1983. doi: 10.1104/pp.112.212423
Kurakawa, T., Ueda, N., Maekawa, M., Kobayashi, K., Kojima, M., Nagato, Y., et al. (2007). Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature 445, 652–655. doi: 10.1038/nature05504
Lee, I., Wolfe, D. S., Nilsson, O., and Weigel, D. (1997). A leafy co-regulator encoded by unusual floral organs. Curr. Biol. 7, 95–104. doi: 10.1016/S0960-9822(06)00053-4
Lee, J. Y., Baum, S. F., Alvarez, J., Patel, A., Chitwood, D. H., and Bowman, J. L. (2005). Activation of CRABS CLAW in the nectaries and carpels of Arabidopsis. Plant Cell 17, 25–36. doi: 10.1105/tpc.104.026666
Lee, Z. H., Tatsumi, Y., Ichihashi, Y., Suzuki, T., Shibata, A., Shirasu, K., et al. (2019). CRABS CLAW and SUPERMAN coordinate hormone-, stress-, and metabolic-related gene expression during Arabidopsis stamen development. Front. Ecol. Evol. 7:437. doi: 10.3389/fevo.2019.00437
Li, H., Liang, W., Hu, Y., Zhu, L., Yin, C., Xu, J., et al. (2011a). Rice MADS6 interacts with the floral homeotic genes SUPERWOMAN1, MADS3, MADS58, MADS13, and DROOPING LEAF in specifying floral organ identities and meristem fate. Plant Cell 23, 2536–2552. doi: 10.1105/tpc.111.087262
Li, H., Liang, W., Yin, C., Zhu, L., and Zhang, D. (2011b). Genetic interaction of OsMADS3, DROOPING LEAF, and OsMADS13 in specifying rice floral organ identities and meristem determinacy. Plant Physiol. 156, 263–274. doi: 10.1104/pp.111.172080
Li, H., Peng, T., Wang, Q., Wu, Y., Chang, J., Zhang, M., et al. (2017). Development of incompletely fused carpels in maize ovary revealed by miRNA, target gene and phytohormone analysis. Front. Plant Sci. 8:463. doi: 10.3389/fpls.2017.00463
Li, Y., Du, W., Chen, Y., Wang, S., and Wang, X. F. (2020). 3D-Construction of vasculature of Anaxagorea and its implications in the integrated axial-foliar origin of the Angiosperm carpel. bioRxiv [Preprint]. doi: 10.1101/2020.05.22.111716
Liang, G., He, H., Li, Y., Wang, F., and Yu, D. (2014). Molecular mechanism of microRNA396 mediating pistil development in Arabidopsis. Plant Physiol. 164, 249–258. doi: 10.1104/pp.113.225144
Litt, A. (2007). An evaluation of A-function: evidence from the APETALA1 and APETALA2 gene lineages. Int. J. Plant Sci. 168, 73–91. doi: 10.1086/509662
Liu, H., Guo, S., Xu, Y., Li, C., Zhang, Z., Zhang, D., et al. (2014). OsmiR396d-regulated OsGRFs function in floral organogenesis in rice through binding to their targets OsJMJ706 and OsCR4. Plant Physiol. 165, 160–174. doi: 10.1104/pp.114.235564
Liu, W. W., Meng, J., Cui, J., and Luan, Y. S. (2017). Characterization and function of MicroRNA∗ s in plants. Front. Plant Sci. 8:2200. doi: 10.3389/fpls.2017.02200
Liu, X., Kim, Y. J., Müller, R., Yumul, R. E., Liu, C., Pan, Y., et al. (2011). AGAMOUS terminates floral stem cell maintenance inArabidopsis by directly repressing WUSCHEL throughrecruitment of Polycomb Group proteins. Plant Cell 23, 3654–3670. doi: 10.1105/tpc.111.091538
Lohmann, J. U., Hong, R. L., Hobe, M., Busch, M. A., Parcy, F., Simon, R., et al. (2001). A molecular link between stem cellregulation and floral patterning in Arabidopsis. Cell 105, 793–803. doi: 10.1016/S0092-8674(01)00384-1
Lohmann, J. U., and Weigel, D. (2002). Building beauty: the genetic control of floral patterning. Dev. Cell 2, 135–142. doi: 10.1016/S1534-5807(02)00122-3
Malcomber, S. T., and Kellogg, E. A. (2005). SEPALLATA gene diversification: brave new whorls. Trends Plant Sci. 10, 427–435. doi: 10.1016/j.tplants.2005.07.008
Marsch-Martínez, N., and de Folter, S. (2016). Hormonal control of the development of the gynoecium. Curr. Opin. Plant Biol. 29, 104–114. doi: 10.1016/j.pbi.2015.12.006
Marsch-Martínez, N., Reyes-Olalde, J. I., Ramos-Cruz, D., Lozano-Sotomayor, P., Zúñiga-Mayo, V. M., and de Folter, S. (2012). Hormones talking. Plant Signal. Behav. 7, 1698–1701. doi: 10.4161/psb.22422
McSteen, P., Malcomber, S., Skirpan, A., Lunde, C., Wu, X., Kellogg, E., et al. (2007). Barren Inflorescence2 encodes a co-ortholog of the Pinoid serine/threonine kinase and is required for organogenesis during inflorescence and vegetative development in maize. Plant Physiol. 144, 1000–1011. doi: 10.1104/pp.107.098558
Meguro, A., Takumi, S., Ogihara, Y., and Murai, K. (2003). WAG, a wheat AGAMOUS homolog, is associated with development of pistil-like stamens in alloplasmic wheats. Sex. Plant Reprod. 15, 221–230. doi: 10.1007/s00497-002-0158-0
Mena, M., Ambrose, B. A., Meeley, R. B., Briggs, S. P., Yanofsky, M. F., and Schmidt, R. J. (1996). Diversification of C-function activity in maize flower development. Science 274, 1537–1540. doi: 10.1126/science.274.5292.1537
Mizukami, Y., and Ma, H. (1995). Separation of AG function in floral meristem determinacy from that in reproductive organ identity by expressing antisense AG RNA. Plant Mol. Biol. 28, 767–784. doi: 10.1007/BF00042064
Moon, S., Jung, K. H., Lee, D. E., Lee, D. Y., Lee, J., An, K., et al. (2006). The rice FON1 gene controls vegetative and reproductive development by regulating shoot apical meristem size. Mol. Cells 21, 147–152.
Morita, Y., and Kyozuka, J. (2007). Characterization of OsPID, the rice ortholog of PINOID, and its possible involvement in the control of polar auxin transport. Plant Cell Physiol. 48, 540–549. doi: 10.1093/pcp/pcm024
Moubayidin, L., and Østergaard, L. (2017). Gynoecium formation: an intimate and complicated relationship. Curr. Opin. Genet. Dev. 45, 15–21. doi: 10.1016/j.gde.2017.02.005
Münster, T., Deleu, W., Wingen, L. U., Ouzunova, M., Cacharrón, J., Faigl, W., et al. (2002). Maize MADS-boxgenes galore. Maydica 47, 287–301.
Nagasawa, N., Miyoshi, M., Sano, Y., Satoh, H., Hirano, H., Sakai, H., et al. (2003). SUPERWOMAN1 and DROOPING LEAF genes control floral organ identity in rice. Development 130, 705–718. doi: 10.1242/dev.00294
Nahar, M. A. U., Ishida, T., Smyth, D. R., Tasaka, M., and Aida, M. (2012). Interactions of CUP-SHAPED COTYLEDON and SPATULA genes control carpel margin development in Arabidopsis thaliana. Plant Cell Physiol. 53, 1134–1143. doi: 10.1093/pcp/pcs057
Nair, S. K., Wang, N., Turuspekov, Y., Pourkheirandish, M., Sinsuwongwat, S., Chen, G., et al. (2010). Cleistogamous flowering in barley arises from the suppression of microRNA-guided HvAP2 mRNA cleavage. Proc. Natl. Acad. Sci. U.S.A. 107, 490–495. doi: 10.1073/pnas.0909097107
Ohmori, S., Kimizu, M., Sugita, M., Miyao, A., Hirochika, H., Uchida, E., et al. (2009). MOSAIC FLORAL ORGANS1, an AGL6-like MADS box gene, regulates floral organ identity and meristem fate in rice. Plant Cell 21, 3008–3025. doi: 10.1105/tpc.109.068742
Okushima, Y., Mitina, I., Quach, H. L., and Theologis, A. (2005). AUXIN RESPONSE FACTOR 2 (ARF2): a pleiotropic developmental regulator. Plant J. 43, 29–46. doi: 10.1111/j.1365-313X.2005.02426.x
Ó’Maoiléidigh, D. S., Wuest, S. E., Rae, L., Raganelli, A., Ryan, P. T., Kwaśniewska, K., et al. (2013). Control of reproductive floral organ identity specification in Arabidopsis by the C function regulator AGAMOUS. Plant Cell. 25, 2482–2503. doi: 10.1105/tpc.113.113209
Osnato, M., Lacchini, E., Pilatone, A., Dreni, L., Grioni, A., Chiara, M., et al. (2020). Transcriptome analysis reveals rice OsMADS13 as an important repressor of the carpel development pathway in ovules. J. Exp. Bot. 9:eraa460. doi: 10.1093/jxb/eraa460
Parcy, F., Nilsson, O., Busch, M. A., Lee, I., and Weigel, D. (1998). A genetic framework for floral patterning. Nature 395, 561–566. doi: 10.1038/26903
Paøenicová, L., De Folter, S., Kieffer, M., Horner, D. S., Favalli, C., Busscher, J., et al. (2003). Molecular and phylogenetic analyses of the complete MADS-Box transcription factor family in Arabidopsis: new openings to the MADS world. Plant Cell. 15, 1538–1551. doi: 10.1105/tpc.011544
Pautler, M., Tanaka, W., Hirano, H. Y., and Jackson, D. (2013). Grass meristems I: shoot apical meristem maintenance, axillary meristem determinacy and the floral transition. Plant Cell Physiol. 54, 302–312. doi: 10.1093/pcp/pct025
Payne, T., Johnson, S. D., and Koltunow, A. M. (2004). KNUCKLES (KNU) encodes a C2H2 zinc-finger protein that regulates development of basal pattern elements of the Arabidopsis gynoecium. Development 131, 3737–3749. doi: 10.1242/dev.01216
Pelaz, S., Ditta, G. S., Baumann, E., Wisman, E., and Yanofsky, M. F. (2000). B and C floral organ identity functions require SEPALLATTA MADS-box genes. Nature 405, 200–203. doi: 10.1038/35012103
Pelaz, S., Tapia-López, R., Alvarez-Buylla, E. R., and Yanofsky, M. F. (2001). Conversion of leaves into petals in Arabidopsis. Curr. Biol. 11, 182–184. doi: 10.1016/S0960-9822(01)00024-0
Pfannebecker, K. C., Lange, M., Rupp, O., and Becker, A. (2017a). An evolutionary framework for carpel developmental control genes. Mol. Biol. Evol. 34, 330–348. doi: 10.1093/molbev/msw229
Pfannebecker, K. C., Lange, M., Rupp, O., Becker, A., and Purugganan, M. (2017b). Seed plant-specific gene lineages involved in carpel development. Mol. Biol. Evol. 34, 925–942. doi: 10.1093/molbev/msw297
Pinyopich, A., Ditta, G. S., Savidge, B., Liljegren, S. J., Baumann, E., Wisman, E., et al. (2003). Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature 424, 85–88. doi: 10.1038/nature01741
Prasad, K., Parameswaran, S., and Vijayraghavan, U. (2005). OsMADS1, a rice MADS-box factor, controls differentiation of specific cell types in the lemma and palea and is an early-acting regulator of inner floral organs. Plant J. 43, 915–928. doi: 10.1111/j.1365-313X.2005.02504.x
Prasad, K., Sriram, P., Santhosh Kumar, C., Kushalappa, K., and Vijayraghavan, U. (2001). Ectopic expression of rice OsMADS1 reveals a role in specifying the lemma and palea, grass floral organs analogous to sepals. Dev. Genes Evol. 211, 281–290. doi: 10.1007/s004270100153
Prasad, K., and Vijayraghavan, U. (2003). Double-stranded RNA interference of a rice PI/GLO paralog, OsMADS2, uncovers its second-whorl-specific function in floral organ patterning. Genetics 165, 2301–2305. doi: 10.14746/ka.2014.2.06
Prunet, N., Yang, W., Das, P., Meyerowitz, E. M., and Jack, T. P. (2017). SUPERMAN prevents class B gene expression and promotes stem cell termination in the fourth whorl of Arabidopsis thaliana flowers. Proc. Natl. Acad. Sci. U.S.A. 114, 7166–7171. doi: 10.1073/pnas.1705977114
Pu, C. X., Ma, Y., Wang, J., Zhang, Y. C., Jiao, X. W., Hu, Y. H., et al. (2012). Crinkly4 receptor-like kinase is required to maintain the interlocking of the palea and lemma, and fertility in rice, by promoting epidermal cell differentiation. Plant J. 70, 940–953. doi: 10.1111/j.1365-313X.2012.04925.x
Reinhardt, D., Pesce, E. R., Stieger, P., Mandel, T., Baltensperger, K., Bennett, M., et al. (2003). Regulation of phyllotaxis by polar auxin transport. Nature 426, 255–260. doi: 10.1038/nature02081
Reyes-Olalde, J. I., and de Folter, S. (2019). Control of stem cell activity in the carpel margin meristem (CMM) in Arabidopsis. Plant Reprod. 32, 123–136. doi: 10.1007/s00497-018-00359-0
Reyes-Olalde, J. I., Zuñiga-Mayo, V. M., Chávez Montes, R. A., Marsch-Martínez, N., and de Folter, S. (2013). Inside the gynoecium: at the carpel margin. Trends Plant Sci. 18, 644–655. doi: 10.1016/j.tplants.2013.08.002
Riechmann, J. L., Krizek, B. A., and Meyerowitz, E. M. (1996). Dimerization specificity of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA, and AGAMOUS. Proc. Natl. Acad. Sci. U.S.A. 93, 4793–4798. doi: 10.1073/pnas.93.10.4793
Ripoll, J. J., Roeder, A. H. K., Ditta, G. S., and Yanofsky, M. F. (2011). A novel role for the floral homeotic gene APETALA2 during Arabidopsis fruit development. Development 138, 5167–5176. doi: 10.1242/dev.073031
Rubio-Somoza, I., and Weigel, D. (2011). MicroRNA networks and developmental plasticity in plants. Trends Plant Sci. 16, 258–264. doi: 10.1016/j.tplants.2011.03.001
Rudall, P. J., Stuppy, W., Cunniff, J., Kellogg, E. A., and Briggs, B. G. (2005). Evolution of reproductive structures in grasses (Poaceae) inferred by sister-group comparison with their putative closest living relatives, Ecdeiocoleaceae. Am. J. Bot. 92, 1432–1443. doi: 10.3732/ajb.92.9.1432
Sakai, H., Krizek, B. A., Jacobsen, S. E., and Meyerowitz, E. M. (2000). Regulation of SUP expression identifies multiple regulators involved in Arabidopsis floral meristem development. Plant Cell 12, 1607–1618. doi: 10.1105/tpc.12.9.1607
Sakuma, S., and Schnurbusch, T. (2020). Of floral fortune: tinkering with the grain yield potential of cereal crops. New Phytol. 225, 1873–1882. doi: 10.1111/nph.16189
Schmidt, R. J., Veit, B., Mandel, M. A., Mena, M., Hake, S., and Yanofsky, M. F. (1993). Identification and molecular characterization of ZAG1, the maize homolog of the Arabidopsis floral homeotic gene AGAMOUS. Plant Cell. 5, 729–737. doi: 10.2307/3869610
Schoof, H., Lenhard, M., Haecker, A., Mayer, K. F. X., Jürgens, G., and Laux, T. (2000). The stem cell population of Arabidopsis shoot meristems is maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100, 635–644. doi: 10.1016/S0092-8674(00)80700-X
Schultz, E. A., Pickett, F. B., and Haughn, G. W. (1991). The FLO10 gene product regulates the expression domain of homeotic genes AP3 and PI in Arabidopsis flowers. Plant Cell 3, 1221–1237. doi: 10.2307/3869229
Schuster, C., Gaillochet, C., and Lohmann, J. U. (2015). Arabidopsis HECATE genes function in phytohormone control during gynoecium development. Development 142, 3343–3350. doi: 10.1242/dev.120444
Sessions, A., Nemhauser, J. L., McColl, A., Roe, J. L., Feldmann, K. A., and Zambryski, P. C. (1997). ETTIN patterns the Arabidopsis floral meristem and reproductive organs. Development 124, 4481–4491.
Shang, E., Ito, T., and Sun, B. (2019). Control of floral stem cell activity in Arabidopsis. Plant Signal. Behav. 14:1659706. doi: 10.1080/15592324.2019.1659706
Sieburth, L. E., and Meyerowitz, E. M. (1997). Molecular dissection of the AGAMOUS control region shows that cis elements for spatial regulation are located intragenically. Plant Cell. 9, 355–365. doi: 10.1105/tpc.9.3.355
Simonini, S., and Østergaard, L. (2019). Female Reproductive Organ Formation: A Multitasking Endeavor, 1st Edn. Amsterdam: Elsevier Inc.
Smoczynska, A., and Szweykowska-Kulinska, Z. (2016). MicroRNA-mediated regulation of flower development in grasses. Acta Biochim. Pol. 63, 687–692. doi: 10.18388/abp.2016_1358
Soltis, P. S., and Soltis, D. E. (2016). Ancient WGD events as drivers of key innovations in angiosperms. Curr. Opin. Plant Biol. 30, 159–165. doi: 10.1016/j.pbi.2016.03.015
Sreenivasulu, N., and Schnurbusch, T. (2012). A genetic playground for enhancing grain number in cereals. Trends Plant Sci. 17, 91–101. doi: 10.1016/j.tplants.2011.11.003
Strable, J., and Vollbrecht, E. (2019). Maize YABBY genes drooping leaf1 and drooping leaf2 regulate floret development and floral meristem determinacy. Development 146:dev171181. doi: 10.1242/dev.171181
Sugiyama, S. H., Yasui, Y., Ohmori, S., Tanaka, W., and Hirano, H. Y. (2019). Rice flower development revisited: regulation of carpel specification and flower meristem determinacy. Plant Cell Physiol. 60, 1284–1295. doi: 10.1093/pcp/pcz020
Sun, B., Zhou, Y., Cai, J., Shang, E., Yamaguchi, N., Xiao, J., et al. (2019). Integration of transcriptional repression and polycomb-mediated silencing of WUSCHEL in floral meristems. Plant Cell. 31, 1488–1505. doi: 10.1105/tpc.18.00450
Sun, Q., and Zhou, D. X. (2008). Rice jmjC domain-containing gene JMJ706 encodes H3K9 demethylase required for floral organ development. Proc. Natl. Acad. Sci. U.S.A. 105, 13679–13684. doi: 10.1073/pnas.0805901105
Theissen, G., Becker, A., Winter, K. U., Munster, T., Kirchner, C., and Saedler, H. (2002). How the land plants learned their floral ABCs: the role of MADS-box genes in the evolutionary origin of flowers. Dev. Genet. Plant Evol. 65, 173–205. doi: 10.1201/9781420024982.ch9
Theißen, G. (2001). Development of floral organ identity: stories from the MADS house. Curr. Opin. Plant Biol. 4, 75–85. doi: 10.1016/S1369-5266(00)00139-4
Theißen, G., Melzer, R., and Rümpler, F. (2016). MADS-domain transcription factors and the floral quartet model of flower development: linking plant development and evolution. Development 143, 3259–3271. doi: 10.1242/dev.134080
Theißen, G., Strater, T., Fischer, A., and Saedler, H. (1995). Structural characterization, chromosomal localization and phylogenetic evaluation of two pairs of AGAMOUS-like MADS-box genes from maize. Gene 156, 155–166. doi: 10.1016/0378-1119(95)00020-7
Vialette-Guiraud, A. C. M., and Scutt, C. P. (2009). “Carpel evolution,” in Annual Review of Plant Biology, eds G. Jander and C. Voelckel (Hoboken, NJ: John Wiley & Sons).
Voinnet, O. (2009). Origin, biogenesis, and activity of plant MicroRNAs. Cell 136, 669–687. doi: 10.1016/j.cell.2009.01.046
Wei, O., Peng, Z., Zhou, Y., Yang, Z., Wu, K., and Ouyang, Z. (2011). Nucleotide diversity and molecular evolution of the WAG-2 gene in common wheat (Triticum aestivum L.) and its relatives. Genet. Mol. Biol. 34, 606–615. doi: 10.1590/S1415-47572011000400013
Weigel, D., Alvarez, J., Smyth, D. R., Yanofsky, M. F., and Meyerowitz, E. M. (1992). LEAFY controls floral meristem identity in Arabidopsis. Cell 69, 843–859. doi: 10.1016/0092-8674(92)90295-N
Weigel, D., and Meyerowitz, E. M. (1994). The ABCs of floral homeotic genes. Cell 78, 203–209. doi: 10.1016/0092-8674(94)90291-7
Woodward, A. W., and Bartel, B. (2005). Auxin: regulation, action, and interaction. Ann. Bot. 95, 707–735. doi: 10.1093/aob/mci083
Wu, D., Liang, W., Zhu, W., Chen, M., Ferrandiz, C., Burton, R. A., et al. (2017). Loss of LOFSEP transcription factor function converts spikelet to leaf-like structures in rice. Plant Physiol. 176, 1646–1664. doi: 10.1104/pp.17.00704
Wu, H. M., Xie, D. J., Tang, Z. S., Shi, D. Q., and Yang, W. C. (2020). PINOID regulates floral organ development by modulating auxin transport and interacts with MADS16 in rice. Plant Biotechnol. J. 18, 1778–1795. doi: 10.1111/pbi.13340
Wuest, S. E., O’Maoileidigh, D. S., Rae, L., Kwasniewska, K., Raganelli, A., Hanczaryk, K., et al. (2012). Molecular basis for the specification of floral organs by APETALA3 and PISTILLATA. Proc. Natl. Acad. Sci. U.S.A. 109, 13452–13457. doi: 10.1073/pnas.1207075109
Xie, M., Zhang, S., and Yu, B. (2015). microRNA biogenesis, degradation and activity in plants. Cell. Mol. Life Sci. 72, 87–99. doi: 10.1007/s00018-014-1728-7
Xu, M., Tang, D., Cheng, X., Zhang, J., Tang, Y., Tao, Q., et al. (2019). OsPINOID regulates stigma and ovule initiation through maintenance of the floral meristem by auxin signaling. Plant Physiol. 180, 952–965. doi: 10.1104/pp.18.01385
Xu, W., Tao, J., Chen, M., Dreni, L., Luo, Z., Hu, Y., et al. (2017). Interactions between FLORAL ORGAN NUMBER4 and floral homeotic genes in regulating rice flower development. J. Exp. Bot. 68, 483–498. doi: 10.1093/jxb/erw459
Yadav, S. R., Khanday, I., Majhi, B. B., Veluthambi, K., and Vijayraghavan, U. (2011). Auxin-responsive OsMGH3, a common downstream target of OsMADS1 and OsMADS6, controls rice floret fertility. Plant Cell Physiol. 52, 2123–2135. doi: 10.1093/pcp/pcr142
Yamada, K., Saraike, T., Shitsukawa, N., Hirabayashi, C., Takumi, S., and Murai, K. (2009). Class D and B-sister MADS-box genes are associated with ectopic ovule formation in the pistil-like stamens of alloplasmic wheat (Triticum aestivum L.). Plant Mol. Biol. 71, 1–14. doi: 10.1007/s11103-009-9504-z
Yamaguchi, N., Huang, J., Xu, Y., Tanoi, K., and Ito, T. (2017). Fine-tuning of auxin homeostasis governs the transition from floral stem cell maintenance to gynoecium formation. Nat. Commun. 8:1125. doi: 10.1038/s41467-017-01252-6
Yamaguchi, T., Lee, D. Y., Miyao, A., Hirochika, H., An, G., and Hirano, H. (2006). Functional diversification of the two C-Class MADS Box genes OsMADS3 and OsMADS58 in Oryza sativa. Plant Cell. 18, 15–28. doi: 10.1105/tpc.105.037200.1
Yamaguchi, T., Nagasawa, N., Kawasaki, S., Matsuoka, M., Nagato, Y., and Hirano, H. Y. (2004). The YABBY gene DROOPING LEAF regulates carpel specification and midrib development in Oryza sativa. Plant Cell. 16, 500–509. doi: 10.1105/tpc.018044
Yamaki, S., Nagato, Y., Kurata, N., and Nonomura, K. I. (2011). Ovule is a lateral organ finally differentiated from the terminating floral meristem in rice. Dev. Biol. 351, 208–216. doi: 10.1016/j.ydbio.2010.12.006
Yanofsky, M. F., Ma, H., Bowman, J. L., Drews, G. N., Feldmann, K. A., and Meyerowitz, E. M. (1990). The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature 346, 35–39. doi: 10.1038/346035a0
Yasui, Y., Tanaka, W., Sakamoto, T., Kurata, T., and Hirano, H. Y. (2017). Genetic enhancer analysis reveals that FLORAL ORGAN NUMBER2 and OsMADS3 co-operatively regulate maintenance and determinacy of the flower meristem in rice. Plant Cell Physiol. 58, 893–903. doi: 10.1093/pcp/pcx038
Zahn, L. M., Kong, H., Leebens-Mack, J. H., Kim, S., Soltis, P. S., Landherr, L. L., et al. (2005). The evolution of the SEPALLATA subfamily of MADS-box genes: a preangiosperm origin with multiple duplications throughout angiosperm history. Genetics 169, 2209–2223. doi: 10.1534/genetics.104.037770
Zhang, J., Nallamilli, B. R., Mujahid, H., and Peng, Z. (2010). OsMADS6 plays an essential role in endosperm nutrient accumulation and is subject to epigenetic regulation in rice (Oryza sativa). Plant J. 64, 604–617. doi: 10.1111/j.1365-313X.2010.04354.x
Zhao, T., Ni, Z., Dai, Y., Yao, Y., Nie, X., and Sun, Q. (2006). Characterization and expression of 42 MADS-box genes in wheat (Triticum aestivum L.). Mol. Genet. Genomics 276, 334–350. doi: 10.1007/s00438-006-0147-3
Zhu, Q. H., Upadhyaya, N. M., Gubler, F., and Helliwell, C. A. (2009). Over-expression of miR172 causes loss of spikelet determinacy and floral organ abnormalities in rice (Oryza sativa). BMC Plant Biol. 9:149. doi: 10.1186/1471-2229-9-149
Keywords: carpel, carpel identity, meristem determinacy, plant hormones, miRNA, grass
Citation: Shen C, Li G, Dreni L and Zhang D (2021) Molecular Control of Carpel Development in the Grass Family. Front. Plant Sci. 12:635500. doi: 10.3389/fpls.2021.635500
Received: 30 November 2020; Accepted: 04 January 2021;
Published: 16 February 2021.
Edited by:
David Smyth, Monash University, AustraliaReviewed by:
Toshiro Ito, Nara Institute of Science and Technology (NAIST), JapanCopyright © 2021 Shen, Li, Dreni and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dabing Zhang, emhhbmdkYkBzanR1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.