
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 30 April 2021
Sec. Plant Development and EvoDevo
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.631489
This article is part of the Research TopicMolecular Mechanisms of Flowering Plant ReproductionView all 19 articles
 Saurabh Singh1†
Saurabh Singh1† Reeta Bhatia2†
Reeta Bhatia2† Raj Kumar1
Raj Kumar1 Tusar K. Behera1
Tusar K. Behera1 Khushboo Kumari1
Khushboo Kumari1 Achintya Pramanik3
Achintya Pramanik3 Hemant Ghemeray1
Hemant Ghemeray1 Kanika Sharma3
Kanika Sharma3 R. C. Bhattacharya4
R. C. Bhattacharya4 Shyam S. Dey1*†
Shyam S. Dey1*†Mitochondrial markers can be used to differentiate diverse mitotypes as well as cytoplasms in angiosperms. In cauliflower, cultivation of hybrids is pivotal in remunerative agriculture and cytoplasmic male sterile lines constitute an important component of the hybrid breeding. In diversifying the source of male sterility, it is essential to appropriately differentiate among the available male sterile cytoplasms in cauliflower. PCR polymorphism at the key mitochondrial genes associated with male sterility will be instrumental in analyzing, molecular characterization, and development of mitotype-specific markers for differentiation of different cytoplasmic sources. Presence of auto- and alloplasmic cytonuclear combinations result in complex floral abnormalities. In this context, the present investigation highlighted the utility of organelle genome-based markers in distinguishing cytoplasm types in Indian cauliflowers and unveils the epistatic effects of the cytonuclear interactions influencing floral phenotypes. In PCR-based analysis using a set of primers targeted to orf-138, 76 Indian cauliflower lines depicted the presence of Ogura cytoplasm albeit the amplicons generated exhibited polymorphism within the ofr-138 sequence. The polymorphic fragments were found to be spanning over 200–280 bp and 410–470 bp genomic regions of BnTR4 and orf125, respectively. Sequence analysis revealed that such cytoplasmic genetic variations could be attributed to single nucleotide polymorphisms and insertion or deletions of 31/51 nucleotides. The cytoplasmic effects on varying nuclear-genetic backgrounds rendered an array of floral abnormalities like reduction in flower size, fused flowers, splitted style with the exposed ovule, absence of nonfunctional stamens, and petaloid stamens. These floral malformations caused dysplasia of flower structure affecting female fertility with inefficient nectar production. The finding provides an important reference to ameliorate understanding of mechanism of cytonuclear interactions in floral organ development in Brassicas. The study paves the way for unraveling developmental biology of CMS phenotypes in eukaryotic organisms and intergenomic conflict in plant speciation.
Mitochondria play a central role in energy production by oxidative phosphorylation, apoptosis, and other cellular processes (Chen et al., 2017; Shu et al., 2018). The migration of essential genes from mitochondrial genome to nuclear genome has been a consistent feature of genome evolution in eukaryotic organisms (Christensen, 2013; Horn et al., 2014). The integrated action of mitochondrial and nuclear genomes is pivotal for many of the cellular functions. The substantial structural complexity and variation in genome size of plant mitochondrial genome is resulted from the accumulation of diverse repeat sequences, frequent homologous recombination of large repeat sequences, and inclusion of foreign DNA fragments (Sloan, 2013; Gualberto and Kuhn, 2014; Gualberto et al., 2014; Chen et al., 2017). The plant mitogenomes also have the remarkable features of presence of highly diverse intergenetic regions, frequent mt-DNA rearrangements, slow evolution in mt-DNA sequences, and rapid structural evolution (Sloan, 2013; Chen et al., 2017). Attributed to these characteristics, the extensive genome reorganization and shuffling in gene order may occur in plant mitogenomes and unusual open reading frames (ORFs) are generated, some of which causes extreme phenotypes such as cytoplasmic male sterility (CMS) (Tanaka et al., 2012; Chen et al., 2017; Singh et al., 2019a). The CMS is a maternally inherited trait which is characterized by inability to produce viable pollen or male reproductive organs for sexual fertilization (Singh et al., 2019a). The mitochondrial genes determining CMS phenotype can be masked by fertility restorer genes (Rf) in nuclear genome. Presence of male sterile cytoplasm without the Rf gene in the nuclear genome results in CMS phenotype (Chen et al., 2017; Singh et al., 2019a). CMS is an important trait to provide new insights into plant nucleo-mitochondrial communication.
Cytoplasm-induced male sterility is a widespread phenomenon reported in over 150 plant species (Chase, 2007; Bohra et al., 2016; Chen et al., 2017). The CMS system is instrumental for utilization of heterosis in vegetable Brassicas for higher yield and quality traits (Dey et al., 2011a, 2018; Thakur et al., 2015; Singh et al., 2018a, 2019b,c). Cauliflower (Brassica oleracea var. botrytis L.) is an important member of vegetable Brassicas grown worldwide. It has been an important contributor in human diet owing to high content of dietary minerals, vitamins, glucosinolates, and other phytochemical compounds with antioxidant and anticancer properties (Singh et al., 2018a,b, 2019b). Availability of genetic mechanisms like sporophytic self-incompatibility (SSI) and CMS make the hybrid breeding in cauliflower remunerative (Dey et al., 2014; Sehgal and Singh, 2018; Singh et al., 2019b,c). However, nonavailability of strong S-alleles for reliable SSI system and instability of the S-alleles under varied climatic conditions made this system less attractive. Therefore, presently CMS is most widely used in hybrid breeding in cauliflower (Dey et al., 2013, 2014; Singh et al., 2021). The wild allies and other closely related Brassica coenospecies serve as the reservoir of different cytoplasms which are being used to develop CMS through intraspecific/interspecific/intergeneric hybridization mediated by sexual crossing or protoplast fusion (Cardi and Earle, 1997; Yamagishi and Bhat, 2014; Shu et al., 2016; Kang et al., 2017; Singh et al., 2019a; Bhatia et al., 2021a,b). Among the different CMS types, ogu (Ogura, 1968) and pol (Handa et al., 1995) CMS systems are widely used in hybrid breeding of vegetable Brassicas. Numerous CMS-related genes (orfs) have been identified and well characterized in different plant species. The key mitochondrial genes (orfs) associated with different CMS types in Brassicaceae are orf138 correlated with Ogura CMS (Ogura, 1968; Singh et al., 2019a), orf125 associated with kosena CMS of Radish (Iwabuchi et al., 1999), orf72 correlated with mur CMS of Diplotaxis muralis (Shinada et al., 2006), orf222 associated with “nap CMS” of Brassica napus (L’Homme et al., 1997), orf224/atp6 correlated with pol CMS (Wang et al., 1995), orf288 linked with hau CMS of B. juncea (Heng et al., 2018), orf263 correlated with “tour CMS” of Brassica tournefortii (Landgren et al., 1996), and orf220 associated with “tuber mustard CMS” of Brassica juncea (Yang et al., 2010). The gene-specific primers for these mitochondrial genomic regions can be efficiently utilized to differentiate different CMS types in Brassica crops. Several mitochondrial DNA-specific microsatellites (mt-SSR) markers have been developed to distinguish different CMS types and assessment of cytoplasmic diversity in different Brassica species like B. oleracea var. italic (Shu et al., 2015, 2016), B. oleracea var. capitata (Wang et al., 2012), B. rapa (Zamani-Nour et al., 2013; Zhang et al., 2013; Heng et al., 2015), B. napus, and B. juncea (Yu et al., 2014).
The CMS cauliflowers and other Brassica crops with Ogura or other alien cytoplasts display complex variations in reproductive features. These reproductive phenotypes are expressed as floral abnormalities such as homeotic-like transformation of stamens to pistil-like structures (pistillody), homeotic modification of stamens to petal-like structures (petaloidy), carpelloid stamens, adherence of functional stamens to style, partially opened flowers, splitted style with exposed ovule, splitted style, absence of nectaries, rudimentary nectaries, and fused flowers (Meur et al., 2006; Saha et al., 2016; Dey et al., 2017b; Kang et al., 2017; Singh et al., 2019a). These floral malformations are a result of incongruity between nuclear and mitochondrial genomes (Shu et al., 2015; Singh et al., 2019a). It is evident that loss of function, mutation, or insertion/deletion in MADS-box genes APETALA2, APETALA3, or PISTILLATA, which are class B genes of classic ABC model for flower development, renders homeotic transformation of stamens to carpels or petals to sepals (Zhang et al., 2011, 2018; Shu et al., 2015). The homeotic floral deformities have also been explained by the dosage imbalance of class B and class C genes of classic ABC model and aberrant mitochondrial gene expression (Meur et al., 2006; Liu et al., 2018). These floral malformations which are linked to genetic background and cytoplasm types, results in inefficient nectar production, impaired pollination, reduced female fertility, and consequently a drastic reduction in seed yield. The other alterations in flower phenotype and reproductive structures, such as aborted pollen, degenerative anthers, crooked or bent stigma, reduction in flower size, variation in filament size, etc., are universal in autoplasmic and alloplasmic CMS systems of Brassica crops (Shu et al., 2016; Dey et al., 2017a; Kang et al., 2017). In India, during the last three decades, a large number of CMS-based breeding lines/material have been generated in Indian cauliflowers by repeated backcrossing or somatic hybridization by exploiting various CMS sources. However, systematic characterization and identification of these floral malformations in CMS lines of the Indian cauliflowers have not been done so far.
Furthermore, mitochondrial markers can potentially be used to determine the CMS mitotypes and analyze genetic diversity in Indian cauliflower CMS lines. Moreover, mitotype-specific SSRs can be reliably used for distinguishing normal CMS lines from the one having varying degree of floral deformities. Identification of mt-SSRs and other mitochondria-specific markers associated with floral deformities will enable to screen out cauliflower CMS lines with different types of floral abnormalities at an early stage. Analysis of sequence-based variation and its association with floral reproductive development will be instrumental in elucidating developmental biology of floral organ development in CMS phenotypes of eukaryotic organisms.
The basic plant material consisted of 76 CMS accessions of cauliflower with different nuclear backgrounds, developed after more than nine generations of backcrossing (Table 1). Among the 76 genotypes, 63 CMS lines were developed at ICAR-Indian Agricultural Research Institute (IARI), Regional Station, Katrain, Kullu Valley, Himachal Pradesh, India through backcross introgression. Thirteen CMS based F1 hybrids were obtained from different private companies (Table 1). In addition, one fertile inbred line of cauliflower with normal cytoplasm, Sel-27 was used as control. The CMS lines along with control Sel-27 were grown in pro-trays under glasshouse conditions of Baragram Experimental Farm of ICAR-Indian Agricultural Research Institute (IARI), Regional Station, Katrain, Kullu Valley, Himachal Pradesh, India for extraction of nucleic acid. The deoxyribonucleic acid was isolated from fully expanding leaves (100 mg) from 25 to 30 days old seedlings using cetyltrimethyl ammonium bromide (CTAB) method with minor modifications (Murray and Thompson, 1980). The deoxyribonucleic acid samples were adjusted to 25–50 ng DNA/μl and were stored at −80°C until use.
Nineteen pairs of mitochondrial markers were used to detect nucleotide diversity among male sterile mitotypes of Indian cauliflowers (Supplementary Table 1) (Shu et al., 2016). One pair of primers (P1) was specific to orf138 determining Ogura CMS, two pairs of primers were specific to orf222 (P2 and P3), and one pair specific to orf222-orf224 (P4), orf224 (P5), atp6-orf224 (P6), and orf263 (P7). The six pairs of primers (P8–P13) were based on orf138-related genomic sequences, and other six pairs (P14–P19) were mitochondrial simple sequence repeat markers (mt-SSRs). The PCR amplifications were carried out in a reaction mixture of 25 μl consisted of 2 μl of genomic DNA template (50 ng), 1 μl of each forward and reverse primers, 12.50 μl of 2× PCR Green master mix (GoTaq DNA polymerase; Promega, United States), and 8.50 μl nuclease free water. The Eppendorf Mastercycler Nexus GSX1 was used for PCR amplification. Amplification was done following PCR cycling program: an initial denaturation of 95°C for 5 min, then 35 cycles of denaturation at 95°C for 30 s, annealing of primers at suitable temperature for 30 s and extension at 72°C for 1 min, and a final extension of 72°C for 7 min. The amplification by each polymorphic locus was repeated three times for the confirmation, and the PCR products were separated on 3% agarose gel electrophoresis in 1× TBE buffer (pH 8.0) at 100 mA voltage for 120 min. Ethidium bromide (EtBr) of 0.5 mg/ml was used for gel staining, and gel pictures were captured using digital gel documentation unit (BioSpectrum® Imaging SystemTM, United Kingdom). The determination of fragment sizes was done using PromegaTM 100 bp DNA step ladder.
For the cluster analysis based on mitochondrial markers, we studied the gel pictures and a binary matrix was used by converting the polymorphic loci data into 1 and 0. The amplified bands were scored visually as 1 for presence and 0 for absence. The qualitative differences in the band intensities were not taken into consideration for analysis. To categorize the CMS lines into distinct groups, the molecular data generated by combining the two types of analytical methods (mitochondrial DNA-specific markers and B. napus-based mt-SSRs) and subjected to cluster analysis via neighbor-joining (NJ) unweighted pair group method with arithmetic mean (UPGMA) using DARwin v6.0.017 (Perrier and Jacquemoud-Collet, 2006). The simple matching (SM) coefficient was computed to calculated genetic distance.
Initially, PCR amplification detected clear polymorphism for the primers P15 and P16 in terms of the length of amplified products. Based on the gel picture analysis, polymorphic fragments were selected for subsequent cloning and sequencing. Using polymorphic variants in the gel image, four representative polymorphic amplicons of both these groups of respective primers P15 and P16 were purified for sequencing. Two variants of each polymorphic fragment with different length were used for cloning and sequencing. In total, eight representative polymorphic amplicons (four from primer P15 and four from primer P16) were selected. Sequencing revealed that fragments amplified by primer P15 were of 251 bp in 46 CMS lines under study and 220 bp for the remaining CMS lines. The amplicons generated by primer P16 were of the size 420 bp in 42 CMS accessions under study and 471 bp in the remaining. The representative amplified PCR products were purified using Wizard® SV Gel and PCR Clean-Up System (Promega). For each of the polymorphic amplified fragment, the representative purified PCR products were cloned into pGEM®-T Easy vector system (Promega Corporation, United States) following the supplier’s guidelines. The products were transformed into Escherichia coli (DH5α)-competent cells. The positive clones were confirmed by colony PCR and restriction digestion. The five positive clones for each of the selected polymorphic fragments were subjected to Sanger Dideoxy sequencing (Eurofins Genomics India Pvt., Ltd.). The percent sequence identity and similarity analysis was carried out with NCBI (National Center for Biotechnology Information) web-based BLAST service1 (Altschul et al., 1990) for the comparative analysis of obtained sequences with available relevant mitochondrial genome sequences in Brassicaceae. The Sanger sequences obtained were assembled and subjected to multiple sequence alignment with reference mitochondrial genomes to determine any variation at nucleotide level using SeqMan Pro tool of DNASTAR version 15.32 (Lasergene Inc., Madison, WI, United States). The phylogenetic tree was constructed with MEGA X software (Kumar et al., 2018) based on different datasets of polymorphic primers. NCBI ORF finder3 was used to determine ORF numbers in the representative DNA sequence of polymorphic loci and to screen hypothetical protein translations. To verify predicted proteins, NCBI BLASTP or SMART BLAST (Altschul et al., 1990) was used. The amino acid multiple sequence alignment was performed using CLUSTAL Omega program (Sievers et al., 2011).
The sequences of selected polymorphic amplicons of Ogu307-33A, Ogu33-1A, Ogu16A, Ogu12A, Ogu12A-orf125, Ogu121-2A, Ogu-HL-3A, Ogu17A, and Ogu50A were submitted to the GenBank nucleotide sequence database of NCBI, and respective accession numbers were received after online publication as MN549523, MN549524, MN549525, MN549526, MN549527, MN549528, MN549529, MN549530, and MN549531, respectively. These accession numbers will be used hereafter.
The field study was carried out at Baragram Experimental Farm of ICAR-IARI, Regional Station, Katrain, Himachal Pradesh, India, situated along the river Beas. The recommended package of practices, suggested for growing a healthy crop at the Baragram Experimental Farm, were followed for better agronomic and phenotypic expression of crop (Singh et al., 2019c). To determine the effect of cytonuclear interactions on floral reproductive traits, a comparative phenotypic analysis of a set of 60 CMS lines of snowball group of Indian cauliflower along with their respective male fertile counterparts (maintainer lines) was carried out based on important floral morpho-physiological traits viz. (i) petal color, (ii) shape of style, (iii) presence of floral nectaries, (iv) presence of viable pollen, (v) type of ovary, (vi) petal size: petal length (mm) and petal width (mm), (vii) ratio of petal length to petal width, (viii) sepal size: sepal length (mm) and sepal width (mm), (ix) ratio of sepal length to sepal width, (x) short filament length (mm), (xi) long filament length (mm), (xii) short stamen length (mm), (xiii) long stamen length (mm), (xiv) style length (mm), (xv) ratio of stamen to style, (xvi) ratio of long stamen to short stamen, and (xvii) nectar quantity (μl) (Singh and Srivastava, 2006; Chang et al., 2011; Gautam et al., 2011; Dey et al., 2017b; Kang et al., 2017). The petal length to petal width ratio was estimated to determine changes in petal size. The long stamen to style ratio was determined to estimate relative position of stigma to stamen. The long to short stamen ratio was estimated to determine the effect of cytoplasm on stamen length (Singh and Srivastava, 2006). The presence or absence of pollen grains was determined on the basis of visual observation, and pollen viability was tested by staining with 2% acetocarmine (Singh and Srivastava, 2006). All the CMS lines and their respective maintainers were evaluated for different floral traits in randomized block design (RBD) with three replications to quantify the modifications in flower reproductive features due to cytonuclear conflict. The data set was subjected to statistical analysis using paired t-test. Observations were recorded from 12 flowers per genotype (four flowers from three plants) in each replication. The CMS lines were compared with their respective maintainers for detection of floral abnormalities (e.g., pistillody, carpelloid stamen, petaloid stamens, splitted style, partially opened flowers, fused flowers, absence of nectaries, etc.). Floral nectar is an important trait for comprehending the plant-pollinator mutualisms. For the analysis of effect of nuclear-genomic conflict on nectar production, the quantification of nectar production was done from the CMS lines and their respective maintainers. The data was taken from 10 flowers of each genotype during early morning hours with graduated capillary of 10 μl. The average data was used for further statistical analysis.
All the CMS lines were clustered into different groups based on cluster analysis and dendrogram construction based on Euclidean distance was done with the PCA and NJ UPGMA method using DARwin software version 6.0.017 (Perrier and Jacquemoud-Collet, 2006). For testing the reliability of NJ dendrogram, a bootstrap value of 1,000 replicates was used. To compare the CMS lines with their respective male fertile counterparts, an average of CMS lines and their male fertile counterparts (maintainers) for different floral traits was analyzed using paired t-test by subjecting the data to “R” statistical software.
The 76 CMS accessions (63 indigenously developed CMS lines and 13 CMS based hybrids) of Indian cauliflowers (Table 1) were screened using 19 pairs of mitochondrial markers. These markers consist of 13 pairs of mitochondrial gene-specific primers: P1–P13 and six pairs of mt-SSRs, P14–P19, designed based on the B. napus mitochondrial genome (Supplementary Table 1) (Honma et al., 2011; Wang et al., 2012; Shu et al., 2016). Among the 13 pairs of mitochondrial gene-specific primers, seven pairs specific to orf138 were amplified. They did not show any amplification in the control inbred line, Sel-27 devoid of Ogura cytoplasm (Figure 1A). However, none of the primers specific to pol, nap, and tour cytoplasms exhibited any amplification in the CMS lines. The six pairs of mitochondrial SSRs were amplified across the 76 Indian cauliflower CMS lines. The amplification pattern of the gene-based primers indicated that the cytoplasm type of all the CMS lines in Indian cauliflower was derived from Ogura cytoplasm; however, there was a variation at nucleotide level among the large genetic stock of Indian cauliflower. These results indicated presence of only Ogura cytoplasm in all the cauliflower lines and F1 hybrids under study, and none of the CMS lines and F1 hybrids were based on other types of cytoplasmic systems. Thus, these set of primers can be utilized effectively across the B. oleracea CMS germplasm to distinguish the diverse CMS systems harboring different cytoplasm types. However, different cytoplasm type-based CMS system is widely utilized across the Brassica crops and different cytoplasms are associated with different phenotypes. Thus, these mitochondrial markers would be helpful to distinguish CMS systems for their effective use in hybrid breeding program. Two pairs of mt-SSRs, P15 (BnTR4) and P16 (orf125) exhibited striking polymorphism among the CMS lines (Figures 1B,C). The amplicon size for the primers P15 and P16 varied between 200 and 280 bp and 410 and 470 bp, respectively (Figures 1B,C).
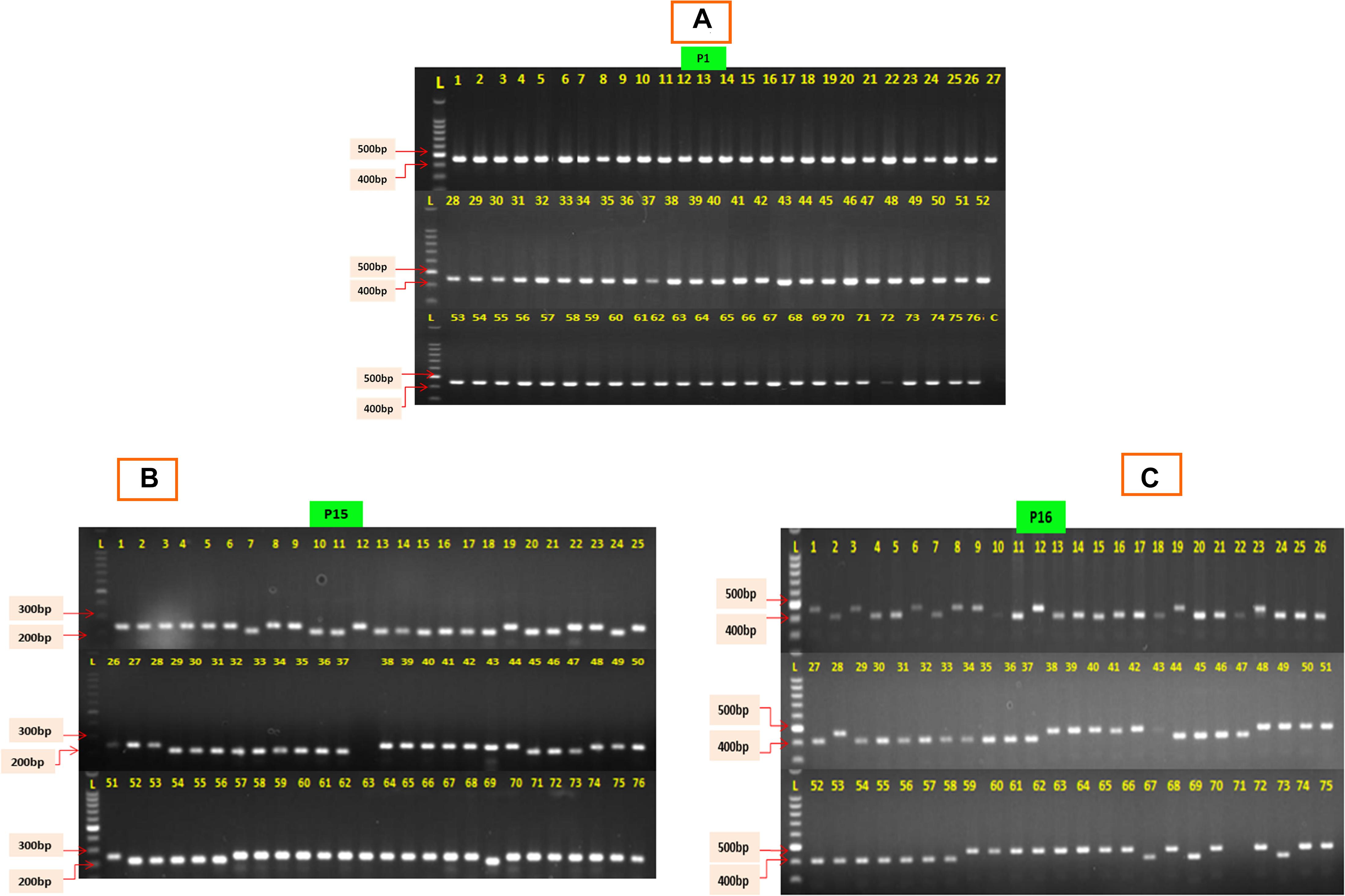
Figure 1. PCR amplification profile of 76 CMS lines of cauliflower. (A) The P1 depicts the PCR amplification with primer P1 specific to orf-138; L, ladder; C, control (Sel-27). (B) Amplification profile of 76 CMS lines exhibiting polymorphism with primer P15 (BnTR4). (C) Amplification profile of 76 CMS lines exhibiting polymorphism with primer P16 (orf125).
The principal component analysis (PCA) and NJ cluster analysis based on mitochondrial markers were used to estimate the diversity in origin of male sterile cytoplasm in cauliflower lines. The cluster analysis of the cauliflower CMS lines categorized them into three major distinct groups each with two subgroups and further subclusters in each subgroup (Supplementary Figures 1a,b). Broadly, six distinct subgroups were determined based on the origin of their cytoplasm source (Supplementary Figure 1b). The six CMS lines (A38 to A43) including four CMS-based hybrids CFH1522, Kimaya, Pahuja, and Snowpearl remained in one single group; the other eight CMS lines (A44 to A51) belonged to a different group. This group comprised four CMS-based hybrid from private seed companies; it might be that same original source of Ogura cytoplasm was used to develop female parent of these hybrids. Ten CMS lines (A52 to A61) belonged to the third group and other 15 CMS lines (A62 to A76) including six CMS-based hybrids (SM, Indam, KTCF-10A, Casper, Ponder, Brahma) formed a distinct fourth group. The nine CMS accessions (A29 to A37) represented in fifth group and the rest of the 28 CMS lines (A1 to A28) including three CMS-based hybrids of Acsen HyVeg private limited (HVCF-29, HVCF-18, and HVCF-16) formed the largest group (Supplementary Figure 1b). These results indicated that all the CMS lines are based on Ogura cytoplasm, but their source of origin might be different. Besides, cytonuclear genomic incompatibilities played a significant role in genetic variation during the course of speciation of these lines.
The fragments amplified by primer P15 were of 251 bp for 46 CMS lines under study and 220 bp for the remaining CMS lines. The representative polymorphic amplicons of both these groups depicting 251 and 220 bp sizes were selected randomly based on the size of the amplicons. They were purified and cloned in pGEM-T vector and sequenced. The obtained sequences were subjected to multiple sequence alignment with each other and reference mitochondrial genome sequences of Brassicaceae crops exhibiting high degree of sequence similarity (Table 2). The accession numbers for the sequenced fragments were obtained from GenBank NCBI as described in the Section “Materials and Methods.” The sequence analysis revealed that MN549523 shared highly conserved region with MN549524 and high degree of similarity with other reference genomic regions of mitochondrial genome sequences of KU831325.1, KJ820683.1, AB627043.1, Sequence 3 P15 from Shu et al. (2016), AP012988.1, and JF920286.1 (Figure 2 and Table 2). Whereas, the MN549525 exhibited high degree of similarity with MN549526 and reference mitogenome sequences used in sequence alignment analysis such as Sequence 4 P15 from Shu et al. (2016), AP018472.1, and AP012989.1 (Figure 2). The sequence alignment revealed a single nucleotide polymorphism (SNP) at position 78 (C/T) and in addition, the deletion of 31 nucleotide between positions 78 and 110 for MN549525 and MN549526 (Figure 2).
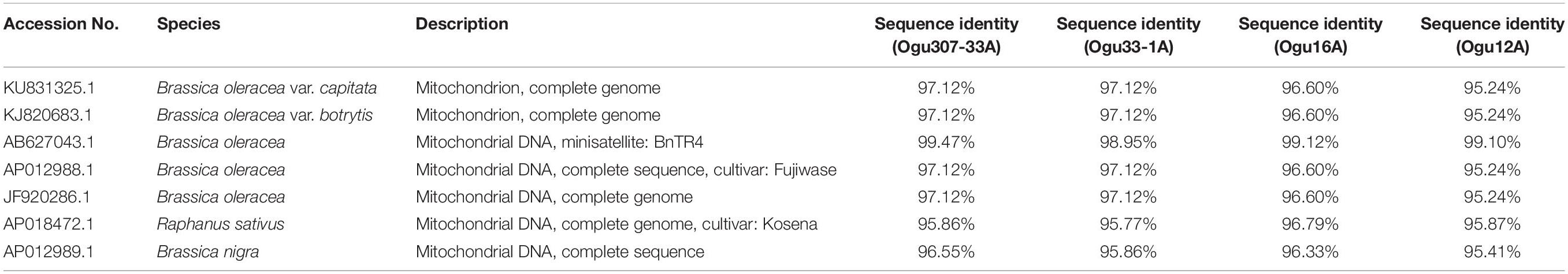
Table 2. Sequence similarity percentage of the selected polymorphic amplicons of cauliflower CMS accessions generated by mt-SSR (BnTR4) with corresponding mitogenome sequences.
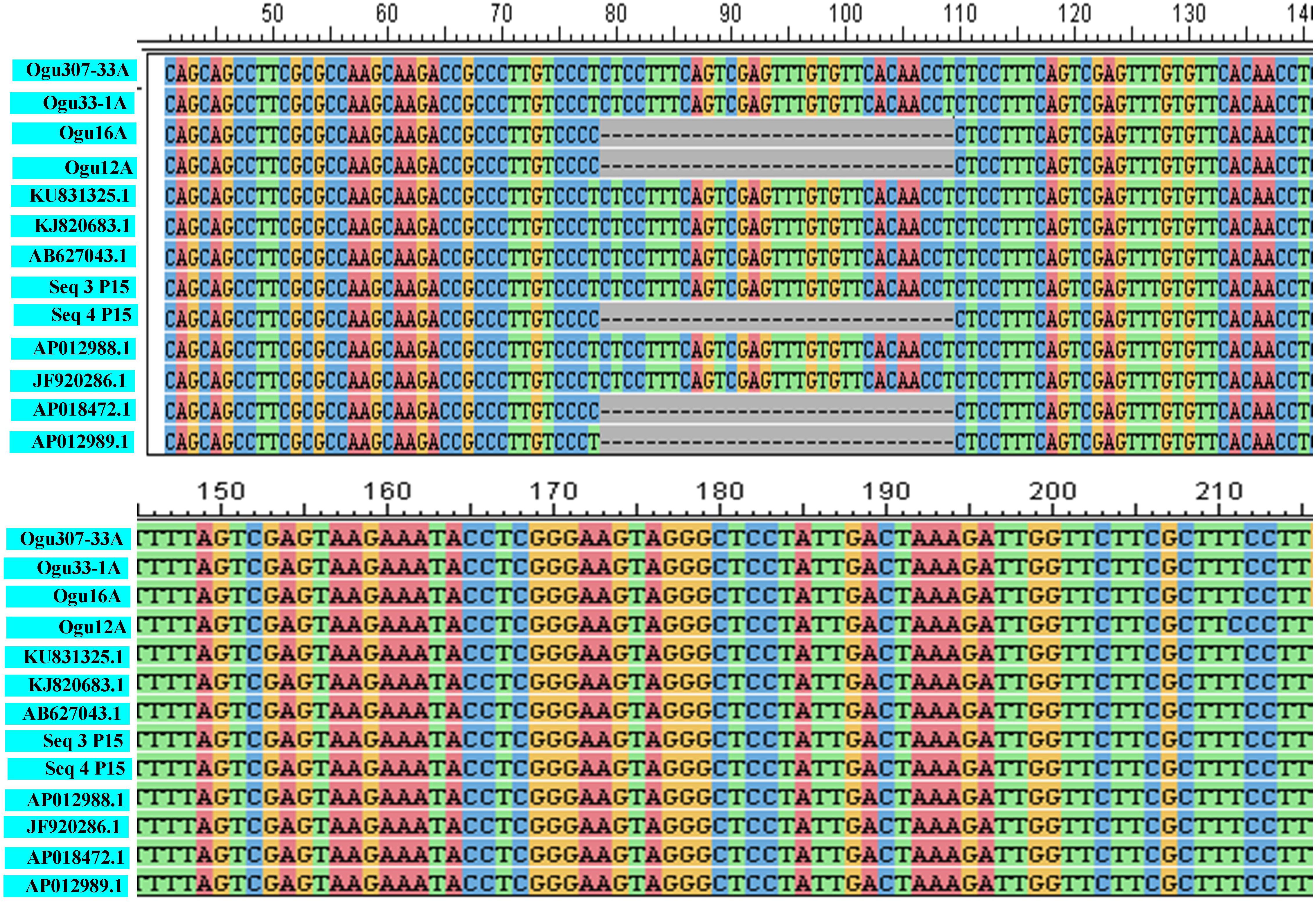
Figure 2. Sequence analysis and alignment of polymorphic amplicons generated by primer P15.Ogu307-33A (MN549523), Ogu33-1A (MN549524), Ogu16A (MN549525), and Ogu12A (MN549526) are selected polymorphic amplicons of CMS lines. Numbers in parenthesis are accession numbers obtained for the respective fragment sequence submitted to GenBank. KU831325.1 (Brassica oleracea var. capitata), KJ820683.1 (Brassica oleracea var. botrytis), AB627043.1 (Brassica oleracea), AP012988.1 (Brassica oleracea), JF920286.1 (Brassica oleracea), AP018472.1 (Raphanus sativus), and AP012989.1 (Brassica nigra) are related reference mitochondrial genome sequences of Brassicaceae crops available in NCBI gene bank database. Seq3 P15 and Seq4 P15 are reference sequences of broccoli from Shu et al. (2016).
The amplicons generated by primer P16 were of size 420 bp in 42 CMS accessions under study and 471 bp in the remaining. The obtained sequences were subjected to multiple sequence alignment with other mitogenome sequences of Brassicaceae crops available in NCBI database (Table 3). The sequence similarity analysis revealed that amplicons of MN549527 to MN549530 shares a high degree of sequence similarity with each other and CMS-related proteins in other Brassicaceae accessions AP018472.1, MG872827.1, JF920287.1, Seq6 P16 from Shu et al. (2016), AP012990.1, AP012989.1, and AB694744.1 (Figure 3). Likewise, MN549531 exhibited highly conserved region with Seq5 P16 from Shu et al. (2016), KU831325.1, and KJ820683.1 (Figure 3). The sequence alignment analysis revealed a deletion of size 51 bp between positions 371 and 423 in the exonic regions of ORFs for MN549527 to MN549530. While normal sequence without deletion was observed for MN549531.
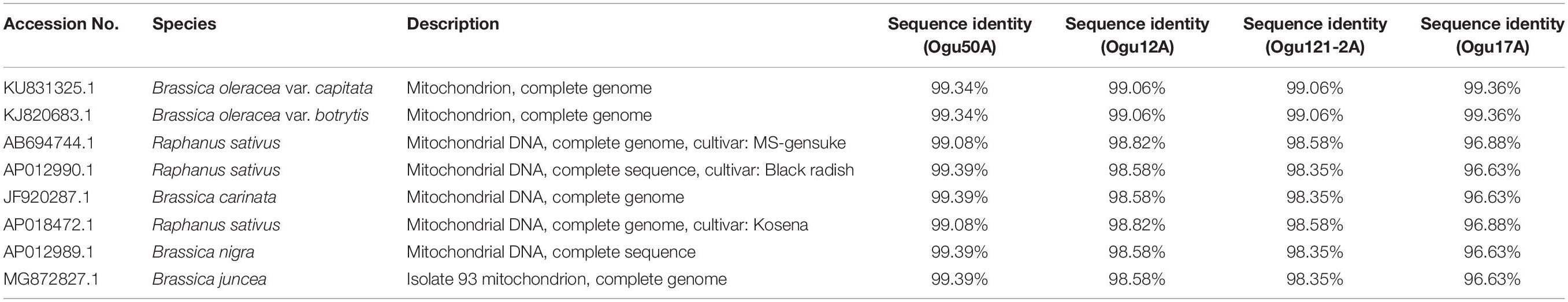
Table 3. Sequence similarity percentage of the selected polymorphic amplicons of cauliflower CMS accessions generated by mt-SSR (orf125) with corresponding mitogenome sequences.
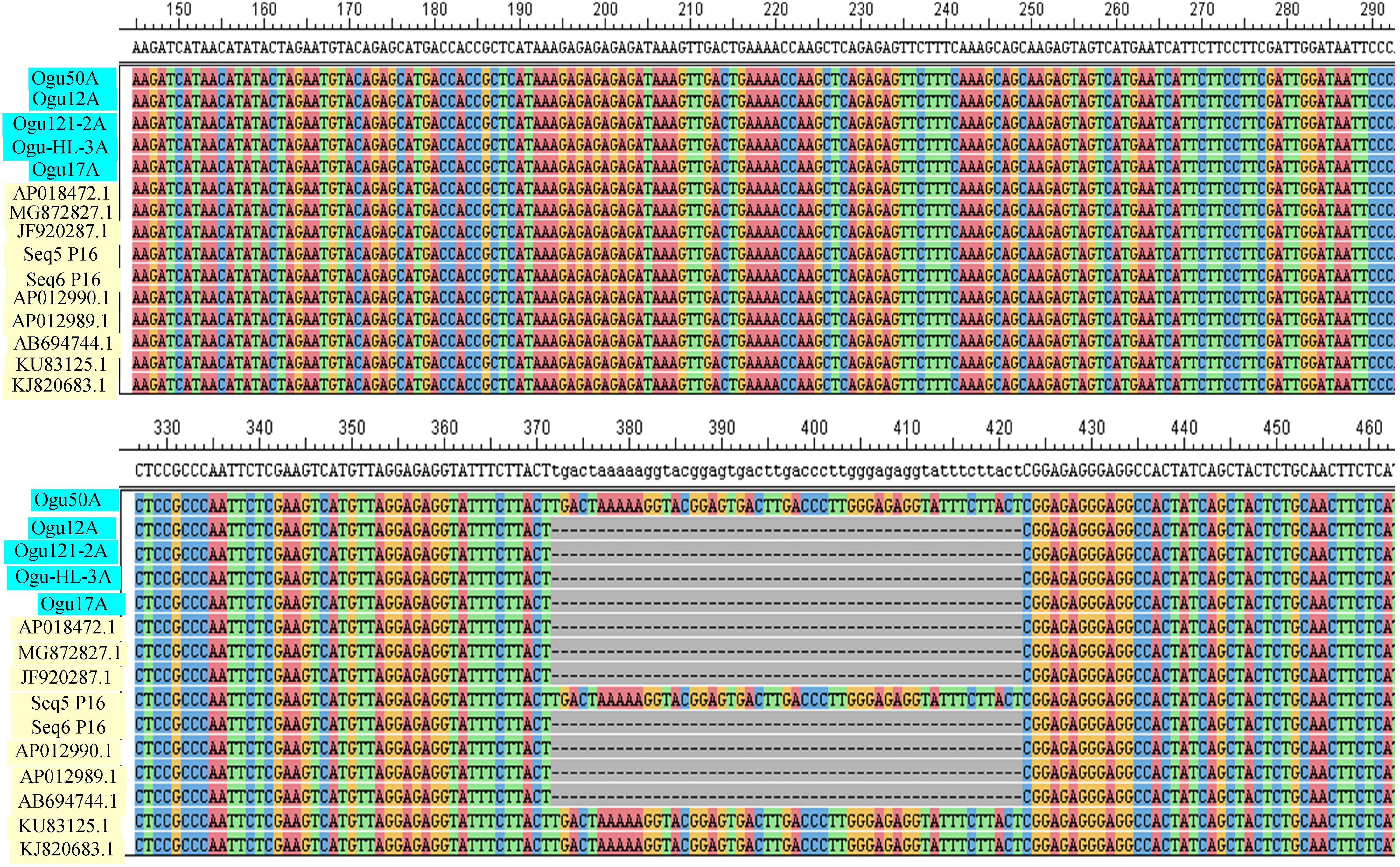
Figure 3. Sequence analysis and alignment of polymorphic amplicons generated by primer P16. Ogu50A (MN549531), Ogu12A-orf125 (MN549527), Ogu17A (MN549530), OguHL-3A (MN549529), and Ogu121-2A (MN549528) are selected polymorphic amplicons of cauliflower CMS mitotypes. Numbers in parenthesis are accession numbers obtained for the respective fragment sequence submitted to GenBank.KU831325.1 (Brassica oleracea var. capitata), KJ820683.1 (Brassica oleracea var. botrytis), AB694744.1 (Raphanus sativus), AP012990.1 (Raphanus sativus), JF920287.1 (Brassica carinata), AP018472.1 (Raphanus sativus), AP012989.1 (Brassica nigra), and MG872827.1 (Brassica juncea) are related reference mitochondrial genome sequences of Brassicaceae crops available in NCBI gene bank database. Seq5 and Seq6 P16 are reference sequences of broccoli from Shu et al. (2016).
We sought to investigate whether cytoplasmic genetic diversity associated with SNPs or InDels at loci BnTR4 and orf125 (Figures 2, 3) has specifically impacted floral phenotypes of CMS lines. It was evident that the CMS lines exhibiting SNPs and deletions of 31 or 51 nucleotides in the ORFs exonic region was associated with varying degrees of floral abnormalities across the whole set of the CMS lines (Figure 4 and Table 4). The CMS lines with the absence of these deletions of nucleotides identified by polymorphic markers had normal flower structure with no deformities. More than 80% of the plants among CMS lines with deletions had one or another floral malformations indicating deformed flower structure. However, extent and type of deformities varied among the genotypes. The major deformities recorded in the CMS lines were (i) adherence of functional stamens with style, (ii) homeotic-like floral transformation petaloidy condition of stamens, (iii) partial petaloidy of functional stamens, (iv) splitted style along with adherence of stamens, (v) stigma hidden inside the petals, (vi) splitted style along with exposed ovules, (vii) unopened flower, (viii) stamens adherence with style and crooked stigma, (ix) partially opened flowers, (x) absence of nonfunctional stamens, (xi) fused flower, (xii) curved functional stamens with crooked stigma, and (xiii) absence of nectaries. Most of the CMS lines with none of the above floral deformities had normal female fertility and seed set (Figure 4 and Table 4).

Figure 4. Floral deformities associated with cyto-nuclear conflict. (a) Adherence of functional stamens with style, (b) petaloidy condition of stamens, (c) partial petaloidy of functional stamens, (d) splitted style along with adherence of stamens, (e) stigma hidden inside the petals and petaloid stamens, (f) splitted style with exposed ovules, (g) unopened flower, and (h,i) stamens adherence with style and bent stigma.
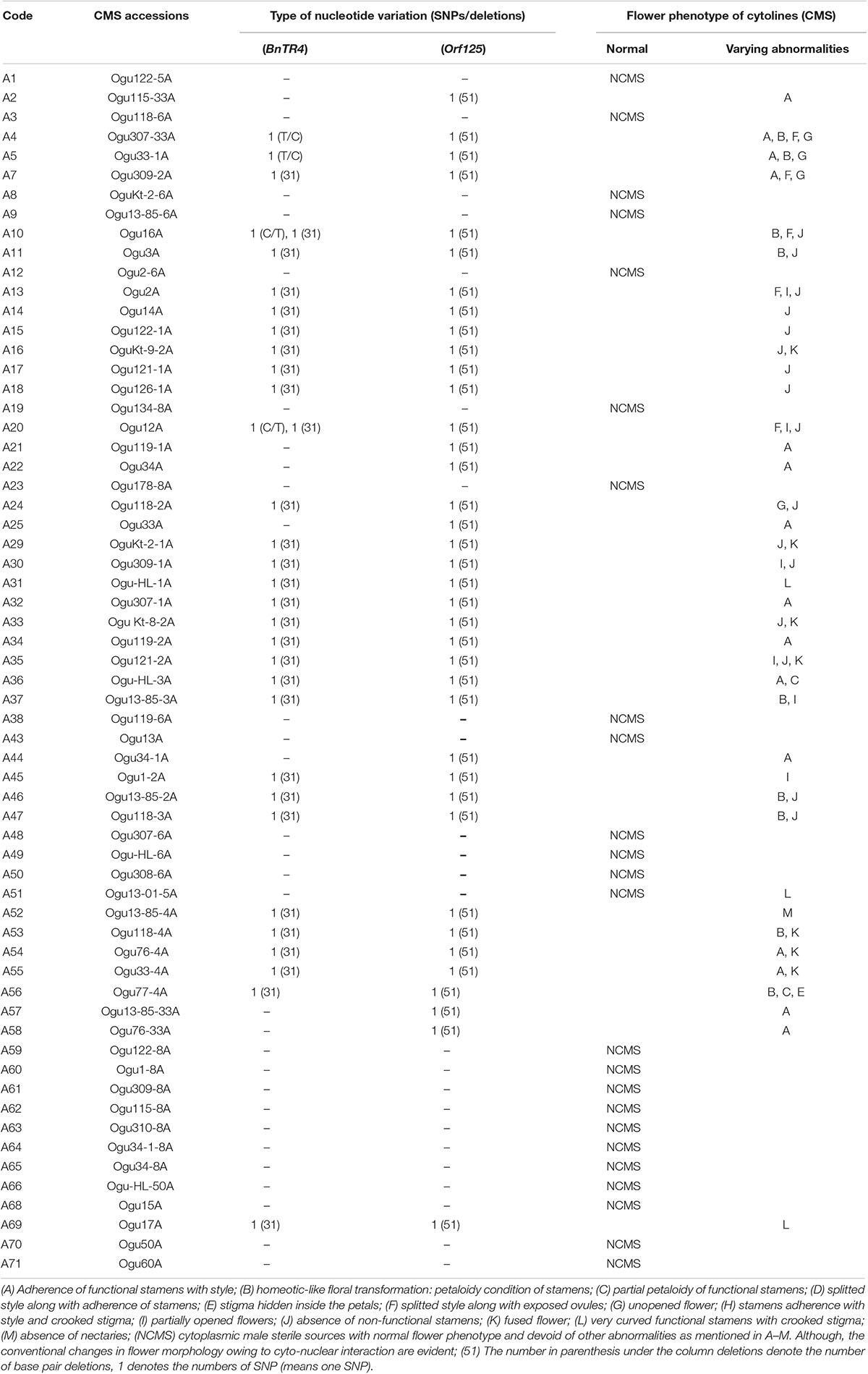
Table 4. Impact of cytoplasmic genetic variations based on InDels on floral phenotypes in varying nuclear backgrounds.
The ORF analysis revealed nucleotide deletion in the orf125 coding region of the male sterile cytoplasm. The protein sequence analysis of polymorphic amplicons and reference mitotypes showed deleted nucleotide-encoded 17 amino acids (Figure 5). Therefore, polymorphism of P15 (BnTR4) and P16 (orf125) could be ascribed to SNPs or fragment insertion/deletion near the mt-SSR loci. The amino acid sequence analysis in the region of ORF depicted the polymorphic region amplified by P16 mt-SSR which is located in the coding region of orf125 protein of B. oleracea (wild cabbage), B. juncea, B. rapa ssp. oleifera, Eruca vesicaria subsp. sativa, B. oleracea var. capitata, and B. oleracea var. botrytis (Supplementary Table 2) besides its location in B. carinata exonic region of orf108c. The protein sequence analysis revealed deletion of amino acids related to array of floral deformities with high degree of similarity with orf108c in B. carinata (Supplementary Table 2). The protein sequences devoid of deletion exhibited high similarity with orf125 in B. oleracea.

Figure 5. Amino acid sequence analysis of ORFs of P16 polymorphic amplicons. MN549531, MN549527, MN549530, MN549529, and MN549528 are selected protein sequences of ORFs of cauliflower CMS mitotypes. KU831325.1 (Brassica oleracea var. capitata), AB694744.1 (Raphanus sativus), and AP012990.1 (Raphanus sativus) are reference amino acid sequences of Brassicaceae obtained by NCBI ORF finder.
Polymorphism exhibited with P15 mt-SSR and mt-DNA sequences of other nine Brassicaceae mitotypes were broadly grouped into two major groups (Figure 6A). The phylogenetic analysis revealed a close affinity of cauliflower CMS lines, Ogu16A with Ogu12A and Ogu307-33A with Ogu33-1A. These cytoplasmic sources were clustered in one major group with one diploid accession of B. oleracea. This clustering in one group concurs with rapa/oleracea lineage propounded by Warwick and Black (1991) based on chloroplast genome. Another group represented diploid B. oleracea var. botrytis, B. oleracea var. capitata, B. oleracea var. italica, Raphanus sativus var. kosena, and B. nigra. This grouping also corresponded to rapa/oleracea lineage as proposed by Warwick and Black (1991) except one species of nigra lineage. Another group also confined to rapa/oleracea lineage with the exception of one species of nigra lineage. Similarly, the polymorphic amplicons based on P16 mitochondrial marker and corresponding mt-DNA sequences of 10 Brassicaceae mitotypes were clustered in two major groups (Figure 6B). The selected CMS sources, Ogu50A and Ogu17A were distantly placed from other CMS sources. OguHL-3A, Ogu12A, and Ogu121-2A exhibited a close affinity with each other and B. oleracea var. capitata based on phylogenetic analysis. The CMS line Ogu50A depicted high affinity with KJ820683.1 (B. oleracea var. botrytis L.) and Ogu17A showed high affinity with mitogenome of B. nigra and R. sativus var. kosena, and clustered in one major group along with Raphanus sativus cv. Ms-gensuke, Ogu50A, and B. oleracea var. botrytis (KJ820683.1). The black radish distantly placed itself from other members of its group.
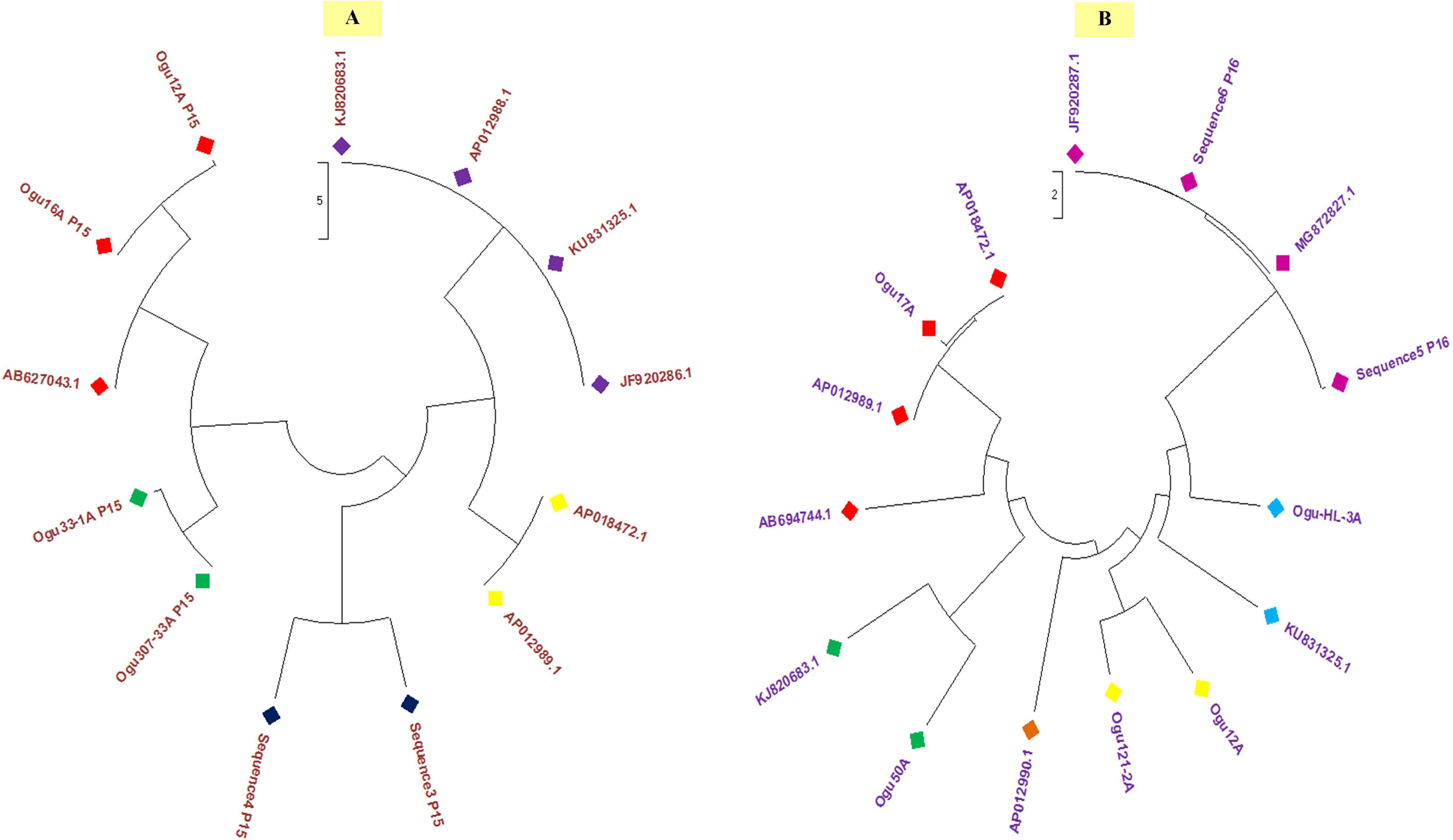
Figure 6. Phylogenetic analysis. (A) Phylogenetic tree based on sequence analysis of selected polymorphic fragments generated with primer P15 and related reference mitogenome sequences from NCBI gene bank database. (B) Phylogenetic tree based on sequence analysis of selected polymorphic amplicons generated with primer P16 and related reference mitogenome sequences from NCBI gene bank database.
In the present investigation, we sought to analyze whether cytonuclear interactions specifically affected floral qualitative traits in the CMS lines of Indian cauliflowers. Cauliflower CMS lines along with their respective maintainers were characterized for five traits namely style shape, petal color, presence of floral nectaries, presence of viable pollen, and type of ovary (Supplementary Table 3 and Supplementary Figures 2,3). All the CMS lines had normal ovary and varied nectaries. None of the CMS line showed the presence of viable pollen grains and few lines were devoid of anther and pollen grains. The CMS accessions had straight to slightly curved or fully curved stigma. Yellow-colored petals were predominant; however, white petals were recorded in Ogu33A, Ogu134-8A, Ogu13A, Ogu118-6A, OguKt-2-6A, and Ogu118-2A.
Comparative analysis for floral reproductive whorls and floral phenotypes of cauliflower CMS lines and their respective male fertile counterparts was conducted to determine the effects of mitonuclear coadaptation and disruption. The per se performance of several CMS lines and their respective maintainers recorded significant variation for different floral phenotypes (Supplementary Table 4 and Supplementary Figure 3). Petal length varied from 12.91 mm (Ogu76-4A) to 17.92 mm (Ogu2-6A). The longest petal was recorded in the CMS line Ogu2-6A followed by Ogu126-1A and Ogu310-8A. The petal width ranged from 4.18 mm (Ogu13-85-2A) to 7.83 mm (Ogu2-6A). The CMS line, Ogu2-6A followed by Ogu178-8A and Ogu33A-1301 had the widest petals. The ratio of petal length to petal width varied from 1.88 (Ogu33A-1301) to 3.82 (Ogu13-85-2A), and ratio was >2 except for Ogu-13-01-5A, Ogu307-1A, and Ogu-13-01-33A. The considerable differences were also observed for sepal size, and the sepal length and width varied from 6.23 mm (Ogu15A) to 10.28 mm (Ogu2-6A) and 2.21 mm (Ogu13-85-2A) to 3.53 mm (Ogu115-8A), respectively. The ratio of sepal length to sepal width ranged from 2.19 (Ogu15A) to 3.88 (OguKt-2-1A). Like petal size, sepal size too was significantly reduced in the male sterile lines. The length of short (nonfunctional) and long (functional) filament ranged from 2.30 mm (Ogu16A) to 6.38 mm (Ogu310-8A) and 3.40 mm (33A-1301) to 7.52 mm (Ogu310-8A), respectively. The introgression of sterile cytoplasm resulted in the elimination of nonfunctional filaments in nine CMS lines. Marked differences were also observed for stamen length, as the short stamen (nonfunctional) length varied from 3.41 mm (Ogu16A) to 8.13 mm (Ogu310-8A) and long stamen (functional) length from 4.79 mm (33A-1301) to 9.48 mm (Ogu2-6A). The ratio of functional (long stamen) to nonfunctional anthers (short stamen) was determined to detect any changes in CMS types, and it varied from 1.03 (OguKt-2-1A) to 2.07 (Ogu77-4A) and from 1.03 to 1.26 in maintainer lines. The nine CMS lines viz. Ogu121-1A, Ogu122-1A, Ogu126-1A, OguKt-9-2A, OguKt-8-2A, Ogu118-3A, Ogu2A, Ogu3A, and Ogu118-2A were completely devoid of nonfunctional stamens. The significant differences were also observed in all the CMS lines for style length and varied from 5.32 mm (Ogu14A) to 9.44 mm (Ogu16A). However, in the male fertile counterparts, the style length varied from 6.33 to 10.86 mm. These results clearly indicated the effect of cytonuclear genomic incompatibilities on reproductive whorls. Reduction in style length, stamen length, and even absence of short stamens was also recorded in the CMS lines in different nuclear backgrounds. Generally, the position of style was higher than the stamens in majority of the CMS lines except Ogu-Kt-2-1A, Ogu122-5A, Ogu118-4A, Ogu33-1A, Ogu33A, Ogu77-4A, OguKt-9-2A, Ogu3A, and Ogu14A (Supplementary Table 4). The cytonuclear conflict invariably reduced the stamen length in all the Indian cauliflower lines. The relative position of stamens and stigma was determined by estimating the ratio between functional stamens and style length. The ratio varied from 0.62 (33A-1301) to 1.30 (Ogu14A) in CMS lines and 0.98 to 1.46 in male fertile counterparts.
Comparative nectar quantity analysis in the CMS lines and their respective male fertile maintainers was conducted to determine the impact of cytonuclear interactions for this trait. Significant variation was observed in the CMS lines for nectar quantity (Table 5). The nectar quantity varied from 0.28 μl (Ogu15A) to 5.69 μl (Ogu1-8A) in the CMS lines, while in the male fertile counterpart, the nectar quantity varied from 0.60 to 13.94 μl. The highest quantity of nectar was found in the CMS line, Ogu1-8A followed by Ogu308-6A and Ogu2-6A. The results revealed a significant reduction in the nectar quantity of CMS lines as compared with their respective male fertile counterparts, indicating that cytonuclear interactions caused inefficient nectar production (Table 5).
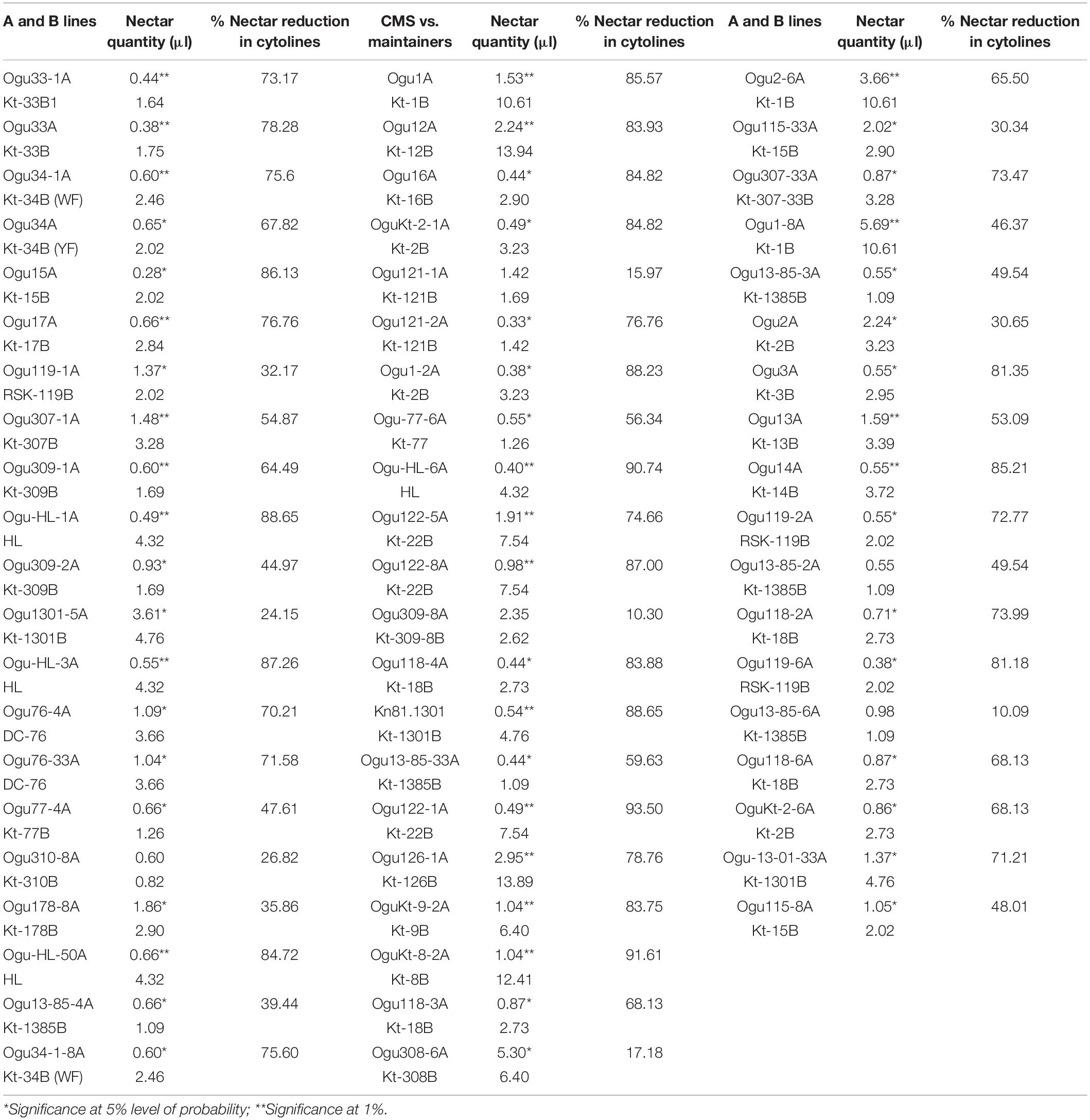
Table 5. Impact of cytonuclear interactions on nectar production of cytolines in varying nuclear backgrounds.
The CMS lines were grouped into different clusters based on floral reproductive whorls and phenotypic traits, and two major clusters (CI and CII) were formed with two subclusters in cluster II (Figure 7). The subclusters (C-IIA and C-IIB) in cluster II were further grouped into subclusters C-IIA-1, C-IIA-2, C-IIB-1, and C-IIB-2. Majority of CMS lines grouped into clusters II and I had only nine CMS lines. All the CMS accessions with the absence of nonfunctional stamens remained in cluster I. The six CMS lines with functional stamen lengths of 8.5 to 9.48 mm remained in the subcluster C-IIA-1. The nine CMS accessions grouped into subcluster C-IIA-2 with nonfunctional filament length of 3.1–3.8 mm, and majority of the CMS lines in this cluster had sepal width of <2.45 mm. The subcluster C-IIB-1a contained five CMS lines with petal length of <14.5 mm, style length of 6.28–6.6 mm, and functional stamen length of 6.49–6.61 mm. The subcluster C-IIB-1b had 10 CMS lines, and majority of them (60%) have petal length of 15–17 mm and sepal width of 2.9–3.4 mm. The subcluster C-IIB-2a comprised eight CMS lines, and majority of them had sepal length of >8.7 mm (i.e., 8.7–9.8 mm). The remaining 13 CMS lines were clustered into subcluster C-IIB-2b with petal length of >13 to <15 mm in majority of the lines.
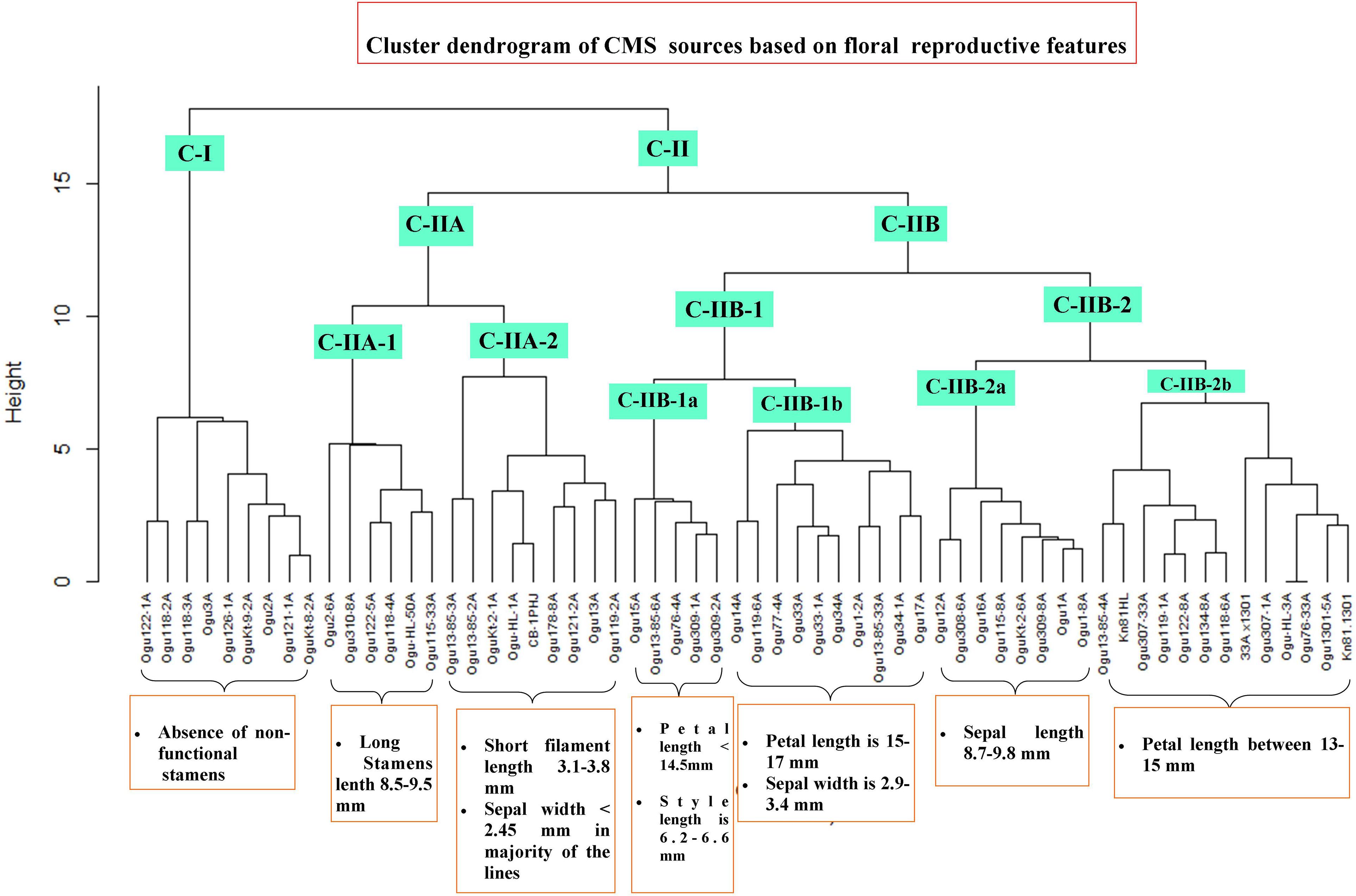
Figure 7. Cluster dendrogram of cauliflower cytoplasmic sources based on floral traits. Cluster C-I harbor all the CMS lines without nonfunctional stamens. Cluster C-II comprises different subclusters with varying degree of floral phenotypic variability and abnormalities.
The CMS phenotype is a common feature in higher plants controlled by mt-DNA and widely used to facilitate hybrid development in Brassica vegetables (Shu et al., 2016; Singh et al., 2018a, 2019a; Dey et al., 2019). The propensity to frequent recombination, mt-DNA rearrangements, and encoding of numerous genes makes plant mitogenome more complex relative to their metazoan counterpart. In this context, the role of mitochondrial markers in quick identification and differentiation of different CMS types has been reported earlier in other members of Brassicaceae (Wang et al., 2012; Shu et al., 2015, 2016). We are reporting the usefulness of mitochondria-specific markers in selecting CMS lines with normal flower structure and extent of mitogenome diversity for the first time in Indian cauliflowers. In this study, mt-DNA-specific primers associated with different CMS genes, orf138, orf222, orf224, orf222-224, orf263, atp6-orf224, and mt-SSRs were utilized. The results revealed that all the CMS lines of Indian cauliflower possessed orf138-related fragment specific to Ogura CMS establishing the wide use of Ogura cytoplasm in the development of CMS system (Chen et al., 2009; Han et al., 2018; Singh et al., 2019a). Therefore, it is urgently needed to diversify the source of the CMS in Indian cauliflowers to avoid any imminent threat of epidemic which may collapse the entire hybrid seed industry of Brassica oleracea vegetables (Dey et al., 2013). The polymorphic mt-SSR loci indicated the presence of cytoplasmic genetic variation among Ogura CMS-based male sterile genetic stock of Indian cauliflowers. The PCA and NJ cluster analysis based on combined analysis of mt-DNA-specific and mt-SSR markers classified the 76 cytolines into six different groups (Supplementary Figure 1B).
The polymorphic amplicons generated with primer P15 were of size between 220 and 280 bp, while for P16 of 410–470 bp size. The lengths of polymorphic products of A13 to A17, A24 to A27, A29 to A37, A69, A73, A2, A4–A5, and A52–A58 were identical as reported in B. oleracea var. capitata (Wang et al., 2012), which can differentiate OguCMSHY, OguCMSR1–2 (420 bp) from pol CMS and OguCMSR3 (471 bp). Similar findings were reported for five CMS lines of broccoli (Shu et al., 2016). Therefore, the original source of OguCMS in the studied cauliflower CMS lines could have come from OguCMSR1–2 and OguCMSHY. The sequence analysis revealed that SNPs and insertion/deletions could explain the cytoplasmic genetic variations among the CMS lines of Indian cauliflowers. Polymorphic amplicons targeting orf125 coding regions of mitochondrial genome demonstrated a deletion of 51 nucleotides in the CMS lines with varied floral deformities. Similarly, the deformed CMS lines had a deletion of 31 nucleotides in the coding region of BnTR4. Wang et al. (2012) successfully reported the use of chloroplast (cpSSRs) and mitochondrial (mt-SSRs) primers in demonstrating the polymorphism in alloplasmic CMS lines of cabbage. Shu et al. (2015; Shu et al., 2016) demonstrated the role of organelle SSRs and mt-DNA-specific and mt-SSR markers in distinguishing different CMS types, differentiating CMS lines with carpelloid stamens in broccoli. Cytoplasmic genetic variations associated with deletions of 31/51 nucleotides in the ORF exonic region demonstrated that insertions/deletions of nucleotides can explain the wide range of floral abnormalities in the CMS lines of Indian cauliflowers. Type and extent of deformities in different CMS lines varied even after the presence of deletions of 31/51 bp, indicating a possible role of nuclear genome, cytonuclear interaction, and environment in determining the type and prevalence of malformities. Therefore, further in-depth studies are needed to pinpoint the exact role of the promoter, coding regions, and epigenetics in determining the specific type and distribution of deformities in the CMS population. However, it was established in the present study that combined use of the primers P15 and P16 can identify the suitable source of male sterile cytoplasm which will provide normal floral structures after their introgression into B. oleracea nuclear backgrounds. Therefore, these two identified markers will be extremely useful to the B. oleracea geneticists and breeders in developing good CMS lines for use in commercial hybrid breeding and further genetic studies.
Cytonuclear genomic incompatibilities and introgression of alien cytoplasm have been reported to display complex floral structure variations in auto- and alloplasmic CMS lines of Brassica crops. The flowering regulation in higher plants is a highly complex process controlled by interactions of genes or genes × environment. MADS-box genes are crucial player in plants for vegetative and reproductive growth development (Meur et al., 2006; Zhang et al., 2011, 2018; Saha et al., 2016; Liu et al., 2018). The identity and development of each floral reproductive whorl is determined by varying combinations of genes of classical flowering ABCDE model (Sheng et al., 2019). It is well documented that loss of function, mutations, or insertions/deletions in MADS-box transcription factors such as APETALA2, APETALA3, or PISTILLATA, AGAMOUS, AGL11, SEP1, SEP2, SEP3, SEP4, SHP2, SHP2, etc. causes varying degree of floral malformations (Meur et al., 2006; Zhang et al., 2011, 2018; Saha et al., 2016; Liu et al., 2018; Sheng et al., 2019). Different types of floral abnormalities observed in the cauliflower cytolines (Figure 4) such as homeotic-like modification of stamens to petal-like structures (petaloidy), adherence of functional stamens to style, partially opened flowers, splitted style with exposed ovule, splitted style, absence of nectaries, rudimentary nectaries, and fused flowers could be explained by absence of amino acids causing dysfunction of one of MADS-box genes. Furthermore, the dosage imbalance of class B and class C genes of classical flowering model, aberrant mitochondrial gene expression results in homeotic-like floral deformities observed in the present investigation (Meur et al., 2006; Liu et al., 2018). Previously, the role of deletions of nucleotide in the ORF coding region of broccoli cytolines was suggested to be associated with carpelloid stamen phenotype (Shu et al., 2015, 2016); however, their role in causing an array of floral deformities is not reported. Identification of these specific deletions associated with several floral deformities would be instrumental in selection of desirable CMS types for hybrid breeding and will enhance the understanding about evolutionary relationships in Brassicas. Analysis of ORFs, similarity of genes, and protein sequence revealed that polymorphic loci were located at exonic regions of orf125 and orf108c of mitochondrial genomes of Brassicaceae species. It is quite likely that orf125 and orf108c mitochondrial genes are playing pivotal role in development of flower organs. The floral abnormalities which are associated with aberrant mitochondrial gene expression, mutation in mitochondrial genomes, cytoplasmic-nuclear conflict, cytoplasm types, and genetic backgrounds render scanty nectar production and impaired pollination and affect female fertility (Shu et al., 2015, 2019; Dey et al., 2017b; Kang et al., 2017). These characteristics consequently results in poor seed yield. Identification and roughing out undesirable cytolines in the initial stage of the breeding program will save huge time efforts in developing CMS lines for their successful use in hybrid development.
In the present investigation and our earlier reports (Dey et al., 2011b, 2017b), we had observed floral abnormalities in CMS lines in the successive nuclear backgrounds. Initially, the expression of floral deformities may be weak or incomplete because of partial substitution of nuclear genome. The exact expressions were observed only after complete nuclear substitution. The identification of specific insertion/deletion and SNPs associated with floral abnormalities in the initial stage of backcross introgression will be extremely helpful to the breeders to use only those CMS types which will later results in normal flower structures without any deformities. The proclivity of plant mitochondrial genome to frequent recombination, genomic rearrangements, and rapid structural evolution leading to heteroplasmy results in polymorphism in cauliflower cytoplasmic sources (McCauley, 2013; Yang et al., 2018). Predominantly, the uniparental, which is a maternal inheritance of chloroplast and mitochondrial genome, is reported in most of the angiosperms (Birky, 1995), including Brassica crops such as broccoli, cabbage, and cauliflower (Reboud and Zeyl, 1994; Zhang et al., 2012; Shu et al., 2015). Recently, the paternal (Erickson and Kemble, 1990; McCauley, 2013; Worth et al., 2014) and biparental (Hansen et al., 2007; Weihe et al., 2009) inheritance of mitogenome is also reported. Therefore, the findings of the present study pave the way for further analysis to detect any possible paternal inheritance of mitochondrial genome.
Pollinators play an important role in hybrid breeding of Brassicas, and pollinator visit is determined by flower phenotype like size, color, and form (Shu et al., 2019). Usually, the large flower size is positively linked with more numbers of pollinator visits, as large floral organ size is generally assumed to contain more nectar (Fenstter et al., 2006; Gomez et al., 2008; Parachnowitsch et al., 2019; Shu et al., 2019). Floral nectar is the primary reward besides pollen, floral oils, resins, and scents, offered to pollinators, and their proportionate ratio is vital in determining plant-pollinator interactions (Gomez et al., 2008). Floral organ size, morphology, proper development of nectaries, efficient nectar production, fruiting features, and pollinator attracting ability are considered potent indexes to evaluate cytolines (Shu et al., 2019). The cytoplasmic effects on floral phenotypes are well documented in Brassicas which are manifested through cytonuclear allelic interactions. The role of additive mitonuclear genetic effects in altering floral phenotypes is rare, while epistatic cytonuclear genetic effects are larger in magnitude (Dobler et al., 2014; Roux et al., 2016). In the present investigation, the effects of cytonuclear interactions on floral qualitative traits were insignificant in different nuclear backgrounds (Supplementary Table 3 and Supplementary Figures 2,3). However, varying degree of cytoplasmic effects on floral reproductive whorls, floral size, structure, and other floral-nectar phenotypes were observed over the successive backcross generations in the development of cauliflower CMS lines (Supplementary Table 4). The introgression of sterile cytoplasm significantly reduced the flower size in the form of reduction in petal length, petal width, and sepal length across the nuclear background series. The other predominant types of structural changes in reproductive whorls were flowers devoid of nonfunctional stamens, alteration in style length, reduction in functional stamens length, modification in position of style and anthers (style position was higher than anthers across the nuclear background series with a few exceptions), and inefficient nectar production. The genetic conflict of sterile cytoplasm-nuclear genome of B. oleracea var. botrytis could explain the floral deformities (Chen et al., 2017; Singh et al., 2019a). In Arabidopsis, the role of cytonuclear epistasis on various adaptive traits is suggested (Roux et al., 2016). Singh and Srivastava (2006) observed the narrowing of petals, reduction of filament length, and increase in style length in the B. juncea male sterile lines with the introgression of ogura, tournefortii, trachystoma, and siifolia wild cytoplasm. They also obtained the erratic results with the ogura cytoplasm in the different nuclear backgrounds because of cytonuclear epistasis. Although, it is also suggested that rigorous recurrent manual or honey bee-mediated selection during backcross breeding cycles could reduce floral deformities to a limited extent (Dey et al., 2017b). However, it is not possible to ameliorate these deformities through conventional backcrossing and different selection strategies.
Understanding the molecular mechanisms and pathways of CMS in cauliflower associated with floral abnormalities along with determining the paternal leakage (if any) need further in depth investigation. Identified molecular markers associated with different kinds of floral deformities will facilitate the selection of CMS lines with normal flower structure at the initial stage of breeding. The establishment of InDel-associated cytoplasmic genetic variations and their association with floral deformities provides reference for understanding developmental biology of floral organ development in CMS lines. The results will also pave the way for elaborating the role of cytonuclear interactions on CMS phenotypes in crop plants.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
SD and RB conceived and designed the study. SS and SD performed the experiments. SS did the data recording and lab experiments with input from KS, HG, and KK. SS analyzed the data. AP helped in recording of data related to the reproductive structure of the CMS lines. RK, SD, and RB monitored and contributed the materials. SS performed the sequencing and other molecular analysis. SS wrote the original draft of the manuscript. SD, TB, RK, and RCB edited the final manuscript. All the authors read and approved the final manuscript. All authors contributed to the article and approved the submitted version.
Financial support was provided by the NAHEP/CAAST and CRP on Hybrid Technology (Cauliflower) research grant by Indian Council of Agricultural Research (Grant No. NAHEP/CAAST/2018-19/07 and 12-144). Funding was provided to SS for his Ph.D. research program (NAHEP/CAAST) and SD (CRP on Hybrid Technology). ICAR-Indian Agricultural Research Institute provided Senior Research Fellowship (SRF) to SS during his Ph.D. program.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
SS is thankful to ICAR-IARI for providing senior scholarship during the Ph.D. program. The authors are also thankful to Dr. Amolkumar U. Solanki, ICAR-National Institute for Plant Biotechnology, New Delhi, India for timely help during analysis of sequence data and interpretation of sequencing results.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.631489/full#supplementary-material
Supplementary Figure 1 | Principal component analysis and neighbor-joining cluster analysis. The cluster analysis of cauliflower cytolines of varying nuclear genome background based on combined analysis of mt-Dna-specific and mt-Ssr primers is presented here. (a) Principal component analysis of 76 cytolines based on molecular data. (b) Nj dendrogram of cytolines depicting six distinct groups in different colors.
Supplementary Figure 2 | Floral structure of cauliflower cytolines in different nuclear backgrounds.
Supplementary Figure 3 | Comparative flower morphology of cytolines and their male fertile counterparts. The upper lane in each image represents male fertile maintainer lines and respective cytolines are in lower lane.
Supplementary Table 1 | List of mitochondrial primers used in the present investigation.
Supplementary Table 2 | Sequence identity and gene similarity analysis of polymorphic amplicons of cauliflower cytolines targeting orf125 location with corresponding sequences of Brassicaceae mitochondrial genomes.
Supplementary Table 3 | Impact of cytonuclear interactions on floral qualitative traits of cauliflower cytolines in different nuclear backgrounds.
Supplementary Table 4 | Impact of cytonuclear interactions on floral reproductive traits of cauliflower cytolines in different nuclear backgrounds.
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Bhatia, R., Dey, S. S., Sharma, K., Singh, S., Kumar, S., Pramanik, A., et al. (2021a). Back-cross introgression of ‘Tour’ cytoplasm from Brassica napus through in vitro embryo rescue reveals partial restoration of sterility in B. oleracea. Sci. Hortic. 282:110014. doi: 10.1016/j.scienta.2021.110014
Bhatia, R., Sharma, K., Parkash, C., Pramanik, A., Singh, D., Singh, S., et al. (2021b). Microspore derived population developed from an inter-specific hybrid (Brassica oleracea × B. carinata) through a modified protocol provides insight into B genome derived black rot resistance and inter-genomic interaction. Plant Cell Tiss. Organ Cult. doi: 10.1007/s11240-021-02018-1. [Epub ahead of print].
Birky, C. W. Jr. (1995). Uniparental inheritance of mitochondrial and chloroplast genes: mechanisms and evolution. Proc. Natl. Acad. Sci. U.S.A. 92, 11331–11338.
Bohra, A., Jha, U. C., Adhimoolam, P., Bisht, D., and Singh, N. P. (2016). Cytoplasmic male sterility (CMS) in hybrid breeding in field crops. Plant Cell Rep. 35, 967–993. doi: 10.1007/s00299-016-1949-3
Cardi, T., and Earle, E. D. (1997). Production of new CMS Brassica oleraacea by transfer of ‘Anand’ cytoplasm from B. rapa through protoplast fusion. Theor. Appl. Genet. 94, 204–212. doi: 10.1007/s001220050401
Chang, S., Yang, T., Du, T., Huang, Y., Chen, J., Yan, J., et al. (2011). Mitochondrial genome sequencing helps show the evolutionary mechanism of mitochondrial genome formation in Brassica. BMC Genomics. 12:497. doi: 10.1186/1471-2164-12-497
Chase, C. D. (2007). Cytoplasmic male sterility: a window to the world of plant mitochondrial-nuclear interactions. Trends Genet. 23, 81–90.
Chen, L., Liu, L., Jin, P., Gong, Y., Sun, X., and Ma, E. (2009). Cytological and molecular identification of cytoplasm in two male sterile lines in radish. Mol. Plant Breed. 7, 757–762.
Chen, Z., Zhao, N., Li, S., Grover, C. E., Nie, H., Wendel, J. F., et al. (2017). Plant mitochondrial genome evolution and Cytoplasmic male sterility. Crit. Rev. Plant. Sci. 36, 55–69. doi: 10.1080/07352689.2017.1327762
Christensen, A. (2013). Plant mitochondrial genome evolution can be explained by DNA repair mechanisms. Genome Biol. Evol. 5, 1079–1086.
Dey, S. S., Bhatia, R., Bhardwaj, I., Mishra, V., Sharma, K., Parkash, C., et al. (2017b). Molecular-agronomic characterization and genetic study reveals usefulness of refined Ogura cytoplasm based CMS lines in hybrid breeding of cauliflower (Brassica oleracea var. botrytis L.). Sci. Hortic. 224, 27–36.
Dey, S. S., Bhatia, R., Parkash, C., Kalia, P., and Barwal, R. N. (2014). Evaluation of cauliflower (Brassica oleracea var. botrytis) CMS (Ogura) lines for agronomic and floral traits. Ind. J. of Hortic 71, 424–427.
Dey, S. S., Bhatia, R., Parkash, C., Sharma, S., Dabral, M., Mishra, V., et al. (2017a). Alteration in important quality traits and antioxidant activities in Brassica oleracea with Ogura cybrid cytoplasm. Plant Breed. 136, 400–409.
Dey, S. S., Bhatia, R., Pramanik, A., Sharma, K., and Parkash, C. (2019). A unique strategy to improve the floral traits and seed yield of Brassica oleracea cytoplasmic male sterile lines through honeybee mediated selection. Euphytica 215:211. doi: 10.1007/s10681-019-2431-4
Dey, S. S., Bhatia, R., Sharma, S. R., Parkash, C., and Sureja, A. K. (2013). Effects of chloro1plast substituted Ogura male sterile cytoplasm on the performance of cauliflower (Brassica oleracea var. botrytis L.) F1 hybrids. Sci. Horic. 157, 45–51.
Dey, S. S., Bhatia, R., Sharma, S. R., Sharma, K., Parkash, C., and Kumar, R. (2018). Population dynamics in introgression of wild male sterile cytoplasm into Brassica oleracea: a small population based breeding model. Sci. Hortic. 232, 231–239.
Dey, S. S., Sharma, S. R., Bhatia, R., Kumar, P. R., and Parkash, C. (2011b). Development and characterization of “Ogura” based improved CMS lines of cauliflower. Indian J. Genet. 71, 37–42.
Dey, S. S., Sharma, S. R., Bhatia, R., Parkash, C., and Barwal, R. N. (2011a). Superior CMS (Ogura) lines with better combining ability improve yield and maturity in cauliflower (Brassica oleracea var. botrytis). Euphytica 182:187. doi: 10.1007/s10681-011-0425-y
Dobler, R., Rogell, B., Budar, F., and Dowling, D. K. (2014). A meta-analysis of the strength and nature of cytoplasmic genetic effects. J. Evol. Biol. 27, 2021–2034. doi: 10.1111/jeb.12468
Erickson, L., and Kemble, R. (1990). Paternal inheritance of mitochondria in rapeseed (Brassica napus). Mol. Gen. Genet. 222, 135–139.
Fenstter, C. B., Cheely, G., Dudash, M. R., and Reynolds, R. J. (2006). Nectar reward and advertisement in humming bird-pollinated Silene virginica (Caryophyllaceae). Am. J. Bot. 93, 1800–1807. doi: 10.3732/ajb.93.12.1800
Gautam, P. K., Rathore, R. K. S., Sutar, A. R., and Mangle, B. B. (2011). Characterization of different male sterile lines on morphological characters of India mustard [Brassica juncea (L.) Czern & Coss] along with their maintainer. Int. Multidiscip. Res. J. 1, 04–08.
Gomez, J. M., Bosch, J., Perfectti, F., Fernandez, J. D., Abdelaziz, M., and Camacho, J. P. M. (2008). Association between floral traits and rewards in Erysimummedio hispanicum (Brassicaceae). Ann. Bot. 101, 1413–1420. doi: 10.1093/aob/mcn053
Gualberto, J. M., and Kuhn, K. (2014). DNA-binding proteins in plant mitochondria: implications for transcription. Mitochondrion 19, 323–328. doi: 10.1016/j.mito.2014.02.004
Gualberto, J. M., Mileshina, D., Wallet, C., Niazi, A. K., Weber-Lotfi, F., and Dietrich, A. (2014). The plant mitochondrial genome: dynamics and maintenance. Biochimie 100, 107–120.
Han, F., Zhang, X., Yang, L., Zhuang, M., Zhang, Y., Li, Z., et al. (2018). iTRAQ-based proteomic analysis of Ogura-CMS cabbage and its maintainer line. Int. J. Mol. Sci. 9:3180. doi: 10.3390/ijms19103180
Handa, H., Gualberto, J. M., and Grienenberger, J. M. (1995). Characterization of mitochondrial orfB gene and its derivative, orf224, a chimeric open reading frame specific to one mitochondrial genome of the “Polima” malesterile cytoplasm in rapeseed (Brassica napus L.). Curr. Genet. 28, 546–552. doi: 10.1007/BF00518167
Hansen, A. K., Escobar, L. K., Gilbert, L. E., and Jansen, R. K. (2007). Paternal, maternal, and biparental inheritance of the chloroplast genome in Passiflora (Passifloraceae): implications for phylogenetic studies. Am. J. Bot. 94, 42–46. doi: 10.3732/ajb.94.1.42
Heng, S., Gao, J., Wei, C., Chen, F., Li, X., Wen, J., et al. (2018). Transcript levels of orf288 are associated with the hau cytoplasmic male sterility system and altered nuclear gene expression in Brassica juncea. J. Exp. Bot. 69, 455–466. doi: 10.1093/jxb/erx443
Heng, S., Shi, D., Hu, Z., Huang, T., Li, J., Liu, L., et al. (2015). Characterization and classification of one new cytoplasmic male sterility (CMS) line based on morphological, cytological and molecular markers in non-heading Chinese cabbage (Brassica rapa L.). Plant Cell Rep. 34, 1529–1537. doi: 10.1007/s00299-015-1804-y
Honma, Y., Yoshida, Y., Terachi, T., Toriyama, K., Mikami, T., and Kubo, T. (2011). Polymorphic minisatellites in the mictochondrial DNAs of Oryza and Brassica. Curr. Genet 57, 261–270.
Horn, R., Gupta, K. J., and Colombo, N. (2014). Mitochondrion role in molecular basis of cytoplasmic male sterility. Mitochondrion 19, 198–205.
Iwabuchi, M., Koizuka, N., Fujimoto, H., Sakai, T., and Imamura, J. (1999). Identification and expression of the kosena radish (Raphanus sativus cv. Kosena) homologue of the Ogura radish CMS-associated gene, orf138. Plant Mol. Biol. 39, 183–188.
Kang, L., Li, P., Wang, A., Ge, X., and Li, Z. (2017). A novel cytoplasmic male sterility in Brassica napus (inap CMS) with carpelloid stamens via protoplast fusion with Chinese woad. Front. Plant Sci. 8:529. doi: 10.3389/fpls.2017.00529
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549.
Landgren, M., Zetterstrand, M., Sundberg, E., and Glimelius, K. (1996). Alloplasmic male-sterile Brassica lines containing B. tournefortii mitochondria express an ORF 3’ of the atp6 gene and a 32 kDa protein. Plant Mol. Biol. 32, 879–890.
L’Homme, Y., Stahl, R. J., Li, X. Q., Hameed, A., and Brown, G. G. (1997). Brassica nap cytoplasmic male sterility is associated with expression of a mtDNA region containing achimeric gene similar to the pol CMS associated orf224 gene. Curr. Genet. 31, 325–335.
Liu, J., Li, C.-Q., Dong, Y., Yang, X., and Wang, Y.-Z. (2018). Dosage imbalance of B- and C-class genes causes petaloid-stamen relating to F1 hybrid variation. BMC Genomics 18:341. doi: 10.1186/s12870-018-1562-4
McCauley, D. E. (2013). Paternal leakage, heteroplasmy, and the evolution of plant mitochondrial genomes. New Phytol. 200, 966–977. doi: 10.1111/nph.12431
Meur, G., Gaikwad, K., Bhat, S. R., Prakash, S., and Kirti, P. B. (2006). Homeotic-like modification of stamens to petals is associated with aberrant mitochondrial gene expression in cytoplasmic male sterile Ogura Brassica juncea. J. Genet. 85, 133–139.
Murray, M. G., and Thompson, W. F. (1980). Rapid isolation of high molecular weight plant DNA. Nucleic Acid Res. 8, 4321–4325. doi: 10.1093/nar/8.19.4321
Ogura, H. (1968). Studies on the new male sterility in Japanese radish, with special references to the utilization of thissterility towards the practical raising of hybrid seeds. Mem. Fac. Agric. Kagoshima Univ. 6, 39–78.
Parachnowitsch, A. L., Manson, J. S., and Sletvold, N. (2019). Evolutionary ecology of nectar. Ann. Bot. 123, 247–261. doi: 10.1093/aob/mcy132
Perrier, X., and Jacquemoud-Collet, J. P. (2006). DARwin Software. Available online at: http://darwin.cirad.fr/ (accessed December 18, 2019).
Roux, F., Mary-Huard, T., Barillot, E., Wenes, E., Botran, L., Durand, S., et al. (2016). Cytonuclear interactions affect adaptive traits of the annual plant Arabidopsis thaliana in the field. Proc. Natl. Acad. Sci. U.S.A. 113, 3687–3692. doi: 10.1073/pnas.1520687113
Saha, G., Park, J.-I., Kim, H., Kang, K.-K., Cho, Y.-G., and Nou, I.-S. (2016). MADS-Box genes are associated with the petaloidy/sepaloidy of the stamens in cytoplasmic male sterile Brassica. Plant Breed. Biotech. 4, 40–50.
Sehgal, N., and Singh, S. (2018). Progress on deciphering the molecular aspects of cell-to-cell communication in Brassica self-incompatibility response. 3Biotech 8:347. doi: 10.1007/s13205-018-1372-2
Sheng, X.-G., Zhao, Z.-Q., Wang, J.-S., Yu, H.-F., Shen, Y.-S., Zeng, X.-Y., et al. (2019). Genome wide analysis of MADS-box gene family in Brassica oleracea reveals conservation and variation in flower development. BMC Plant Biol. 19:106. doi: 10.1186/s12870-019-1717-y
Shinada, T., Kikucho, Y., Fujimoto, R., and Kishitani, S. (2006). An alloplasmic male-sterile line of Brassica oleracea harboring the mitochondria from Diplotaxis muralis expresses a novel chimeric open reading frame, orf72. Plant Cell Physiol. 47, 549–555.
Shu, J., Liu, Y., Li, Z., Zhang, L., Fang, Z., Yang, L., et al. (2016). Detection of the diversity of cytoplasmic male sterility sources in broccoli (Brassica oleracea var. italica) using mitochondrial markers. Front. Plant Sci. 7:927. doi: 10.3389/fpls.2016.00927
Shu, J., Liu, Y., Li, Z., Zhang, L., Zhang, L., Fang, Z., et al. (2015). Organelle simple sequence repeat markers help to distinguish carpelloid stamen and normal cytoplasmic male sterile sources in broccoli. PLoS One 10:e0138750. doi: 10.1371/journal.pone.0138750
Shu, J., Liu, Y., Zhang, L., Li, Z., Fang, Z., Yang, L., et al. (2019). Evaluation and selection of sources of cytoplasmic male sterility in broccoli. Euphytica 215:125. doi: 10.1007/s10681-019-2453-y
Shu, J., Zhang, L., Liu, Y., Li, Z., Fang, Z., Yang, L., et al. (2018). Normal and abortive buds transcriptomic profiling of Broccoli ogu cytoplasmic male sterile line and its maintainer. Int. J. Mol. Sci. 19:2501. doi: 10.3390/ijms19092501
Sievers, F., Wilm, A., Dineen, D., Gibson, T. J., Karplus, K., Li, W., et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using CLUSTAL Omega. Mol. Sys. Biol. 7:539.
Singh, K. H., and Srivastava, K. K. (2006). Characterization of different cytoplasmic male sterility systems in Indian mustard (Brassica juncea L. Czern & Coss). Plant Breed. 125, 72–76.
Singh, S., Bhatia, R., Kumar, R., Das, A., Ghemeray, H., Behera, T. K., et al. (2021). Characterization and genetic analysis of OguCMS and doubled haploid based large genetic arsenal of Indian cauliflowers (Brassica oleracea var. botrytis L.) for morphological, reproductive and seed yield traits revealed their breeding potential. Genet. Resour. Crop Evol. 68, 1–21. doi: 10.1007/s10722-020-01090-4
Singh, S., Bhatia, R., Kumar, R., Sharma, K., Dash, S., and Dey, S. S. (2018a). Cytoplasmic male sterile and doubled haploid lines with desirable combining ability enhances the concentration of important antioxidant attributes in Brassica oleracea. Euphytica 214:207. doi: 10.1007/s10681-018-2291-3
Singh, S., Dey, S. S., Bhatia, R., Kumar, R., and Behera, T. K. (2019a). Current understanding of male sterility systems in vegetable Brassicas and their exploitation in hybrid breeding. Plant Reprod. 32, 231–256. doi: 10.1007/s00497-019-00371-y
Singh, S., Dey, S. S., Bhatia, R., Kumar, R., Sharma, K., and Behera, T. K. (2019c). Heterosis and combining ability in cytoplasmic male sterile and doubled haploid based Brassica oleracea progenies and prediction of heterosis using microsatellites. PLoS One 14:e0210772. doi: 10.1371/journal.pone.0210772
Singh, S., Dey, S. S., Kumar, R., Bhatia, R., Ghemeray, H., and Behera, T. K. (2019b). Genetic analysis and interaction among CUPRAC, FRAP, phytochemical and phenotypic traits in cauliflower (Brassica oleracea var. botrytis L.). Int. J. Chem. Stud. 7, 1484–1494.
Singh, S., Singh, R., Thakur, P., and Kumar, R. (2018b). “Phytochemicals, functionality and breeding for enrichment of cole vegetables (Brassica oleracea L.),” in Phytochemicals in Vegetables: A Valuable Sourcce of Bioactive Compounds, eds S. A. Petropoulos, I. C. F. R. Ferreira, and L. Barros (Sharjah: Bentham Science Publishers), 256–295.
Sloan, D. B. (2013). One ring to rile them all? Genome sequence provides new insights into the ‘master circle’ model of plant mitochondrial DNA structure. New Phytol. 200, 978–985. doi: 10.1111/nph.12395
Tanaka, Y., Tsuda, M., Yasumoto, K., Yamagishi, H., and Terachi, T. (2012). A complete mitochondrial genome sequence of Ogura-type male-sterile cytoplasm and its comparative analysis with that of normal cytoplasm in radish (Raphanus sativus L.). BMC Genomics 13:352. doi: 10.1186/1471-2164-13-352
Thakur, P., Vidyasagar, and Singh, S. (2015). Evaluation of cytoplasmic male sterile (CMS) progenies and maintainer lines for yield and horticultural traits in cabbage (Brassica oleracea var. capitata L.). SABRAO J. Breed. Genet. 47, 29–39.
Wang, H. M., Ketela, T., Keller, W. A., Gleddie, S. C., and Brown, G. G. (1995). Genetic correlation of the orf224/atp6 gene region with Polima CMS in Brassica somatic hybrids. Plant Mol. Biol. 27, 801–807.
Wang, Q., Zhang, Y., Fang, Z., Liu, Y., Yang, L., and Zhuang, M. (2012). Chloroplast and mitochondrial SSR help to distinguish allo-cytoplasmic male sterile types in cabbage (Brassica oleracea L. var. capitata). Mol. Breed. 30, 709–716. doi: 10.1007/s11032-011-9656-9
Warwick, S. I., and Black, L. D. (1991). Molecular systematics of Brassica and allied genera (Subtribe Brassicinae, Brassiceae) chloroplast genome and cytodeme congruence. Theor. Appl. Genet. 82, 8192. doi: 10.1007/BF00231281
Weihe, A., Apitz, J., Pohlheim, F., Salinas-Hartwig, A., and Borner, T. (2009). Biparental inheritance of plastidial and mitochondrial DNA and hybrid variegation in Pelargonium. Mol. Genet. Genomics 282, 587–593.
Worth, J. R. P., Yokogawa, M., and Isagi, Y. (2014). Outcrossing rates and organelle inheritance estimated from two natural populations of the Japanese endemic conifer Sciadopity sverticillata. J. Plant Res. 127, 617–626. doi: 10.1007/s10265-014-0646-y
Yamagishi, H., and Bhat, S. R. (2014). Cytoplasmic male sterility in Brassicaceae crops. Breed. Sci. 64, 38–47. doi: 10.1270/jsbbs.64.38
Yang, J. H., Liu, X. Y., Yang, X. D., and Zhang, M. F. (2010). Mitochondrially- targeted expression of a cytoplasmic male sterility associated orf220 gene causes male sterility in Brassica juncea. BMC Plant Biol 10:231. doi: 10.1186/1471-2229-10-231
Yang, K., Nath, U. K., Biswas, M. K., Kayum, M. A., Yi, G-e, Lee, J., et al. (2018). Whole-genome sequencing of Brassica oleracea var. capitata reveals new diversity of the mitogenome. PLoS One 13:e0194356. doi: 10.1371/journal.pone.0194356
Yu, X., Liu, Y., Lv, Y., Liu, Z., Chen, Z., Lu, G., et al. (2014). Development of molecular markers specific to petaloid-type cytoplasmic male sterility in tuber mustard (Brassica juncea var. tumida Tsen et Lee). Mol. Biol. Rep. 41, 769–778. doi: 10.1007/s11033-013-2916-5
Zamani-Nour, S., Clemens, R., and Mollers, C. (2013). Cytoplasmic diversity of Brassica napus L., Brassica oleracea L. and Brassica rapa L. as determined by chloroplast microsatellite markers. Genet. Resour. Crop Evol. 60, 953–965.
Zhang, R. J., Hu, S. W., Yan, J. Q., and Sun, G. L. (2013). Cytoplasmic diversity in Brassica rapa L. investigated by mitochondrial markers. Genet. Resour. Crop Evol. 60, 967–974.
Zhang, Y., Huang, S., Wang, X., Liu, J., Guo, X., Mu, J., et al. (2018). Defective APETALA2 genes lead to sepal modification in Brassica crops. Front. Plant Sci. 9:367. doi: 10.3389/fpls.2018.00367
Zhang, Y., Wang, X., Zhang, W., Yu, F., Tian, J., Li, D., et al. (2011). Functional analysis of the two Brassica AP3 genes involved in apetalous and stamen carpelloid phenotypes. PLoS One 6:e20930. doi: 10.1371/journal.pone.0020930
Keywords: Brassica oleracea, cytoplasmic male sterility, mitotype-specific markers, sequence analysis, cytonuclear interactions, floral malformations, floral reproductive traits
Citation: Singh S, Bhatia R, Kumar R, Behera TK, Kumari K, Pramanik A, Ghemeray H, Sharma K, Bhattacharya RC and Dey SS (2021) Elucidating Mitochondrial DNA Markers of Ogura-Based CMS Lines in Indian Cauliflowers (Brassica oleracea var. botrytis L.) and Their Floral Abnormalities Due to Diversity in Cytonuclear Interactions. Front. Plant Sci. 12:631489. doi: 10.3389/fpls.2021.631489
Received: 22 December 2020; Accepted: 22 March 2021;
Published: 30 April 2021.
Edited by:
Maria Manuela Ribeiro Costa, University of Minho, PortugalReviewed by:
Jingbo Zhang, St. John’s University, United StatesCopyright © 2021 Singh, Bhatia, Kumar, Behera, Kumari, Pramanik, Ghemeray, Sharma, Bhattacharya and Dey. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shyam S. Dey, c3NkZXlAaWFyaS5yZXMuaW4=; c2h5YW0uaWFyaUBnbWFpbC5jb20=
†ORCID: Saurabh Singh, orcid.org/0000-0001-8038-1808; Reeta Bhatia, orcid.org/0000-0002-0668-6272; Shyam S. Dey, orcid.org/0000-0001-9211-8820
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.