
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 15 July 2021
Sec. Plant Abiotic Stress
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.629089
This article is part of the Research Topic Coping with Pollution - the Effects of Environmental Contaminants on Plant Growth and Physiology View all 8 articles
Cadmium is an environmental pollutant with high toxicity that negatively affects plant growth and development. To understand the molecular mechanisms of plant response to cadmium stress, we have performed a genome-wide transcriptome analysis on barley plants treated with an increased concentration of cadmium. Differential gene expression analysis revealed 10,282 deregulated transcripts present in the roots and 7,104 in the shoots. Among them, we identified genes related to reactive oxygen species metabolism, cell wall formation and maintenance, ion membrane transport and stress response. One of the most upregulated genes was PLANT CADMIUM RESISTACE 2 (HvPCR2) known to be responsible for heavy metal detoxification in plants. Surprisingly, in the transcriptomic data we identified four different copies of the HvPCR2 gene with a specific pattern of upregulation in individual tissues. Heterologous expression of all five barley copies in a Cd-sensitive yeast mutant restored cadmium resistance. In addition, four HvPCR2 were located in tandem arrangement in a single genomic region of the barley 5H chromosome. To our knowledge, this is the first example showing multiplication of the PCR2 gene in plants.
Cadmium (Cd) is a particularly dangerous environmental pollutant with a high toxicity to plants and other living organisms, including humans. Its concentration increases rapidly in the environment as an effect of anthropogenic factors such as mining, metal-work industries, urban traffic or the application of phosphate fertilizers (Nriagu and Pacyna, 1988). Cadmium contamination of soils can cause losses in agricultural yield and present a potential health risk for people from Cd transfer through the food chain.
Cadmium has no known function as a nutrient and its uptake by plants is dependent on soil concentration, bioavailability, redox potential, temperature, quantity of organic matter, and concentrations of other elements. Cd accumulation in plants affects various processes such as water and mineral uptake, photosynthesis and respiration, resulting in inhibition of growth and even death (Sanità Di Toppi and Gabbrielli, 1999). Cd ions compete with other nutrients for plant uptake and enter the plant via Fe2+/Fe3+, Zn2+, and Mn2+ transporters because of their limited specificity (Pedas et al., 2008; Sasaki et al., 2012). Thus, the plant damage caused by cadmium and other heavy metals comes from competition with essential mineral nutrients leading to deficient nutrition (Clarkson and Lüttge, 1989). Once Cd enters the root, it can be sequestrated in root vacuoles or translocated in the xylem through the apoplastic and/or symplastic pathway and transported into the aboveground parts of the plant (Salt and Rauser, 1995; Yang et al., 1998).
In addition, heavy metals may accumulate in cell compartments that impede the general metabolism of the plant (Thurman and Collins, 1983). As a response to biotic or abiotic stress, plants produce common stress-related transcripts like transcriptional factors, transporter proteins and other genes involved in signal transduction and secondary metabolite pathways (Atkinson and Urwin, 2012). Plants have developed a complex network of mechanisms to minimize the toxic effect of cadmium import. These active and passive strategies based on exclusion of the Cd2+ from the cellular environment are applied at several levels. As a first response, malate or citrate is secreted and binds to metal ions thereby restricting Cd root absorption (Delhaize and Ryan, 1995). If Cd is absorbed, it can be immobilized by cell wall or extracellular carbohydrates in the root (Sanità Di Toppi and Gabbrielli, 1999). Once Cd enters the cytosol, metal ions activate the synthesis of sulfur-containing ligands such as phytochelatins or metallothioneins (DalCorso et al., 2008) and these ligand-metal complexes are stored in special cellular compartments.
A previous study in Arabidopsis detected important genes encoding enzymes and proteins involved in sulfur assimilation, GSH metabolism, indole-3-acedic acid (IAA) and jasmonic acid (JA) biosynthesis activated by lead treatment (Liu et al., 2009). In rice, Lin et al. (2013) revealed that small heat shock proteins, sulfate assimilation and glutathione-S-transferase are specifically upregulated with Cd stress. Fusco et al. (2005) identified transcription factors putatively responsive to heavy metal stress indicating the complexity of the response of plants to Cd stress. It was showed that the TaHsfA4a gene plays a direct role in a Cd-induced transcriptional response in wheat and rice. Besides, metallothioneins are regulated by this heat-shock transcription factor under Cd exposure (Shim et al., 2009).
In this study, we analyzed genes and regulatory pathways related to the response of Cd stress in barley. Firstly, we measured the Cd concentration in both shoot and root tissue. Second, we monitored the cell cycle to examine the effect of Cd on cell progression through individual cell phases. Third, barley root and shoot tissues were subjected to transcriptome profiling. We identified several Cd responsive genes based on differential gene expression analysis between Cd-treated material and the control. Finally, we characterized the upregulated genes encoding Cd2+ transmembrane transporters PLANT CADMIUM RESITANCE 2 (HvPCR2) by heterologous overexpression in yeast.
Seeds used in the experiments were kindly provided by Genebank Gatersleben of the Leibniz Institute of Plant Genetics and Crop Plant Research. Barley seeds (Hordeum vulgare cv. Morex) were germinated at 20°C in the dark on filter paper moistened with deionized water. After 48 h the primary root length was measured for seeds that were then transferred to polypropylene pots containing 1L of test medium (changed every 24 h to maintain composition). Test media were aerated throughout the exposure time. Relative humidity was 50%, and day/night temperatures were 20/16°C during 16/8 h photoperiod.
The basal nutrient solution composition was KH2PO4, 0.4 mM; K2SO4, 0.4 mM; MgSO4 × 7H2O, 0.6 mM; NH4NO3, 1 mM; Ca(NO3)2 × 4H2O, 2 mM; FeNaEDTA, 75 μM; MnCl2 × 4H2O, 7 μM; ZnCl2, 3 μM; CuSO4 × 5H2O, 0.8 μM; H3BO3, 1.6 μM, and Na2MoO4 × 2H2O, 0.83 μM. The pH was adjusted with 1 M KOH to 5.5–6.0 (Podar, 2013). Test media, except control, were prepared by adding different volumes of stock solutions of CdCl2 to adjust the concentration.
To determine Cd accumulation in barley cv. Morex, plants were cultivated in control medium and medium supplemented with 80 μM cadmium, a previously determined effective concentration (EC50) causing a 50% inhibitory effect for root growth (Kintlová et al., 2017). Samples were collected and root length measured before and after 5 days of treatment. Both root and shoot tissue samples were washed first in 0.5 M EDTA to remove unabsorbed cadmium, then washed in distilled water. They were further dried in a hot air oven (50°C, 72 h). The cadmium concentration was then measured by atomic absorption spectrometry (AAS).
The experimental design consisted of three types of treatment to determine whether the damage of root tip cells caused by cadmium treatment is reversible or irreversible at set conditions. We examined control plants, cadmium-treated plants and washed cadmium-treated plants (designated in this study as Treatment 1–3). The experiment was repeated twice.
For all types of treatment, barley seeds were germinated (Hordeum vulgare cv. Morex) at 20°C in the dark on filter paper moistened with deionized water. After 48 h, 20y seeds were transferred to polypropylene pots containing 1L of test medium (changed every 24 h). Test media were aerated throughout the exposure time. Relative humidity was 50%, and day/night temperatures were 20/16°C during 16/8 h photoperiod. After 5 days, the plants in Treatment 3 were rinsed in deionized water 3 times and transferred to basal nutrition solution for 3 days, whereas Treatment 1 and 2 plants were handled immediately.
The plant nuclei extraction protocol and EdU labeling were performed on the same day for all types of treatment. Click-iT EdU Alexa Fluor 488 Flow Cytometry Assay Kit (Invitrogen, Carlsbad, CA, USA) was used for the cell proliferation assay. Twenty barley plants of each treatment were rinsed 3 times in deionized water and placed into Petri dish filled with 50 ml of fresh deionized water. 5-ethynyl-2′-deoxyuridine (EdU) was added to a final concentration of 9 μM and incubated for 2 h.
Roots with a length of about 1 cm were trimmed and incubated 20 min in 2% (v/v) formaldehyde fixative on ice. The roots were then washed in Tris buffer three times for 5 min on ice. About 70 meristem root tips per one treatment were excised and then hmogenized in 1 ml LB01 lysis buffer using a Polytron PT1200 homogenizer at 15,000 rpm for 13 s. The crude homogenate was filtered through a nylon mesh filter (50 μm pore size) into a new tube. The tubes were spun down at 400 g and 4°C for 10 min and the supernatant removed. The Click-iT reaction cocktail was prepared using 438 μl PBS, 10 μl CuSO4, 2.5 μl Fluorescent dye azide and 50 μl Reaction Buffer Additive for each sample. 500 μl of reaction cocktail was applied to the pelet, mixed well and incubated for 30 min in the dark at room temperature. The solution was spun down again and the pelet was resuspended in 500 μl LB01 buffer. The suspension was filtered through a nylon mesh filter (25 μm pore size) and stained with DAPI (2 μg/ml final concentration).
All flow cytometric analyses were performed on FACSAria SORP flow cytometer (BD Biosciences, Santa Clara, USA) equipped with blue (488 nm, 100 mW) and UV (355 nm, 100 mW) lasers and optical filters for FITC (530/30 band-pass) and DAPI (450/50 band-pass) fluorescence detection. At least 10,000 nuclei were recorded for each sample. Data were displayed on Alexa Fluor 488 vs. DAPI dot plots. To make statistical evaluation of the effect of cadmium treatment on cell proliferation, the region around cycling and non-cycling populations were drawn, respectively. The ratio of cycling/non-cycling was calculated for each sample and the mean and the standard deviation were calculated from three repetitions.
For transcriptome analysis we used the dataset available in NCBI SRA (sequence read archive) under accession numbers SRR5452097 to SRR5452108. These data were obtained from the barley plants grown in media supplemented with cadmium concentration that corresponds to established EC50 values as well as control plants for each treatment (Kintlová et al., 2017). Sequenced Read quantification of each dataset was performed using Salmon (Patro et al., 2017) and 63,658 H. vulgare CDS (Mascher, 2019) and 3,349,186 repeats (IBSC, 2016). Differential expression analysis was then conducted using R and the package DESeq2 (Love et al., 2014). Gene ontologies were retrieved from the available barley gene annotation (Mascher, 2019) and were further analyzed with the DE genes. Bioinformatic analysis of RNA-seq data was carried out in the computing and storage facilities MetaCentrum infrastructure.
The mRNA levels for selected differentially expressed transcripts was validated by qRT-PCR. As representatives, most up- and down-regulated transcripts from both root and shoot tissues were selected for analysis (Supplementary Table 3). Plant material was grown and treated as described above. The RNA from control and Cd-treated plants (80 μM) was isolated in triplicate by NucleoSpin RNA Plant kit (Macherey-Nagel, Germany) and transcribed to cDNA by Transcriptor high fidelity cDNA synthesis kit (Roche). The qRT-PCR reactions were performed on a Light Cycler 96 real-time PCR system (Roche, USA) using the 2x SYBR Master Mix (Top-Bio, Czech Republic). Relative expression of shoot and root transcripts was assessed using CFX Manager™ Software (Bio-Rad, Hercules, California, USA).
Amino acid identity for each HvPCR2 gene and previously described Arabidopsis PCR genes (Song et al., 2004, 2010) was determined using alignment algorithm MAFFT v7.450 (Katoh and Standley, 2013). The genomic sequence of Hordeum vulgare (IBSC_v2), Oryza sativa Indica group (ASM465v1) Zea mays (B73_RefGen_v4) and Musa acuminata (ASM31385v1) were used to analyze genomic loci containing PCR2 homologs, data were retrieved and visualized using EnsemblPlants website (Howe et al., 2020).
To test the function of HvPCR2 genes, all five identified copies were expressed in Saccharomyces cerevisiae cadmium hypersensitive mutant strain DTY168 (MATα, his6, leu2-3−112, ura3-52, ycf1::hisG) lacking the yeast cadmium factor 1 gene (Szczypka et al., 1994). HvPCR2.1-5 coding sequences were amplified from Morex cDNA (Supplementary Table 4) and cloned into p426 under the control of yeast GDP (glyceraldehyde-3-phosphate dehydrogenase) promoter (Mumberg et al., 1995). Vectors containing HvPCR2 fragments and empty vector p426 were introduced into the DTY168 strain by lithium acetate. Yeast transformants were grown overnight, OD600 was adjusted to 0.5, four 10-fold dilutions were made, and each culture was spotted on SD-Ura plates and SD-Ura plates supplemented with 50 μM CdCl2. Plates were incubated for 5–6 days at 28°C.
The effect of 80 μM Cd on barley after a 5-day treatment comprised of root shortening and overall growth retardation (Figure 1A). Root growth of Cd-treated plants was inhibited by ~50% in comparison with non-treated plants (Figure 1B; Supplementary Table 1). Further, we measured the Cd content in control and treated plants in order to obtain information about the distribution of Cd in barley plants in both root and shoot tissues. The Cd content in roots was in control conditions approximately double of the Cd concentration in shoots. Cd-treated plants accumulated roughly 6,600 times more Cd in roots than non-treated plants. This result shows a strong accumulation of Cd in the root and restriction of Cd translocation to the aerial tissues in high Cd stress (Figures 1C,D; Supplementary Table 2).
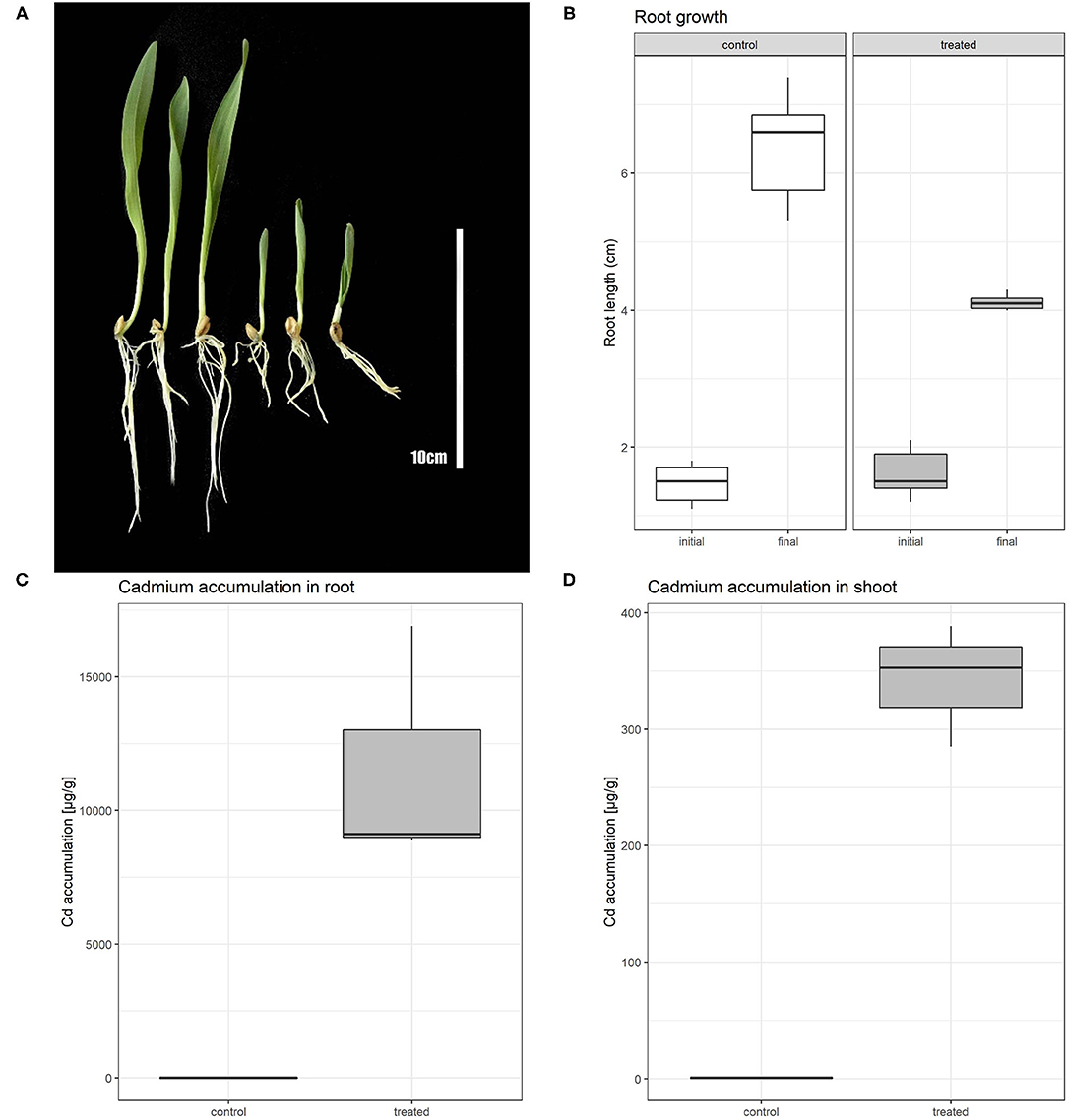
Figure 1. Phenotypic comparison of control and Cd treated plants (A); left section represents control plants, right section Cd treated plants. Boxplot showing root length of barley plants at the beginning of experiment (initial) and after 5 days of Cd or control treatment (final) (B). Boxplot of cadmium accumulation for both control and treated plants in root (C) and shoot (D).
The labeled thymidine analog (EdU) was used to monitor cell cycle abnormalities of Cd treated plants and to determine the Cd effect on root growth at the cellular level. In control plants, we observed an approximately equal number of cells before, during and after the S phase (Figure 2A). Cd-treated roots contained a significantly lower number of S phase cells (Figure 2B). Such a decrease may suggest that the cells entering the S phase are the most susceptible to Cd treatment. After removing the Cd by washing the roots, the number of cells in each state went back to the level similar to the non-treated control (Figure 2C). These findings indicate that the major reduction in root growth is caused by the suppression of the cell division process namely in G1/S cells.
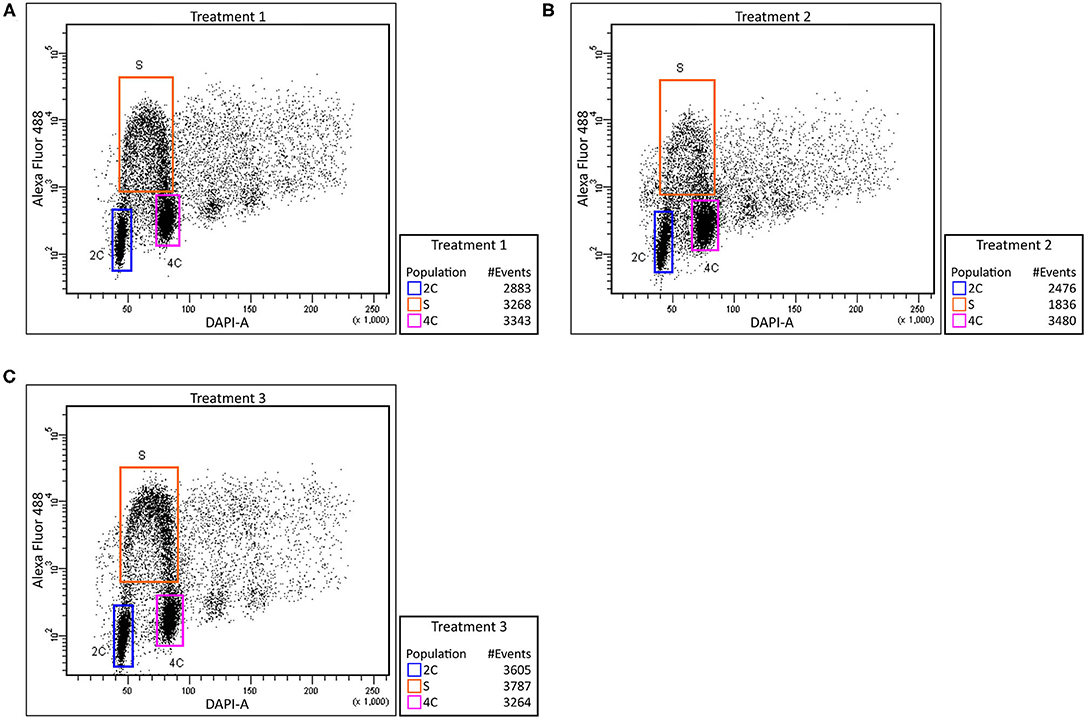
Figure 2. Effect of cadmium on root cell cycle. Barley root tip cells were labeled with EdU and analyzed using flow cytometry. Control plants (no added Cd)–Treatment 1 (A), plants treated with Cd–Treatment 2 (B), and plants washed and grown in control media after Cd exposure–Treatment 3 (C). Relative DNA content was measured using DAPI (X-axis) and extent of proliferating nuclei was measured by means of EdU incorporation based on Alexa Fluor 488 fluorescence (Y-axis). Regions of replicating (orange) and non-replicating (blue and purple) nuclei are indicated.
The entire dataset subjected to differential expression analysis contains 20.7 million of non-treated and 24.3 million of cadmium treated RNA reads (Kintlová et al., 2017). The analysis was performed using the DESeq2 package from Bioconductor (Love et al., 2014). We controlled the clustering of both control and cadmium replicates using multidimensional scaling for each tissue (Supplementary Figure 1). We identified 14,459 differentially expressed (DE) transcribed sequences in both root and shoot tissues using a p-value threshold of 0.05 (Figure 3). The overall count of DE after cadmium treatment is 14,459 (Figure 4; Supplementary Tables 6, 7) with 2,927 transcripts that have a significant change in their expression in both tissues while 7,355 were differentially expressed only in the root and 4,177 in the shoot. Separating the set of DE transcripts between up- and down-regulated reveals that most of the transcripts in the root and shoot are down-regulated (3,509 up vs. 3,846 down in root and 1,825 up vs. 2,352 down in shoot). Differentially expressed transcripts present in both tissues follow the same pattern of expression change with more down-regulated transcripts than up-regulated ones. From the 2,927 DE transcripts in both tissues, 903 are up-regulated, and 1,438 are down-regulated in roots and shoots. Nevertheless, 288 transcripts are down-regulated in roots while up-regulated in shoots, and 298 transcripts are up-regulated in roots while down-regulated in shoots (Figure 4).
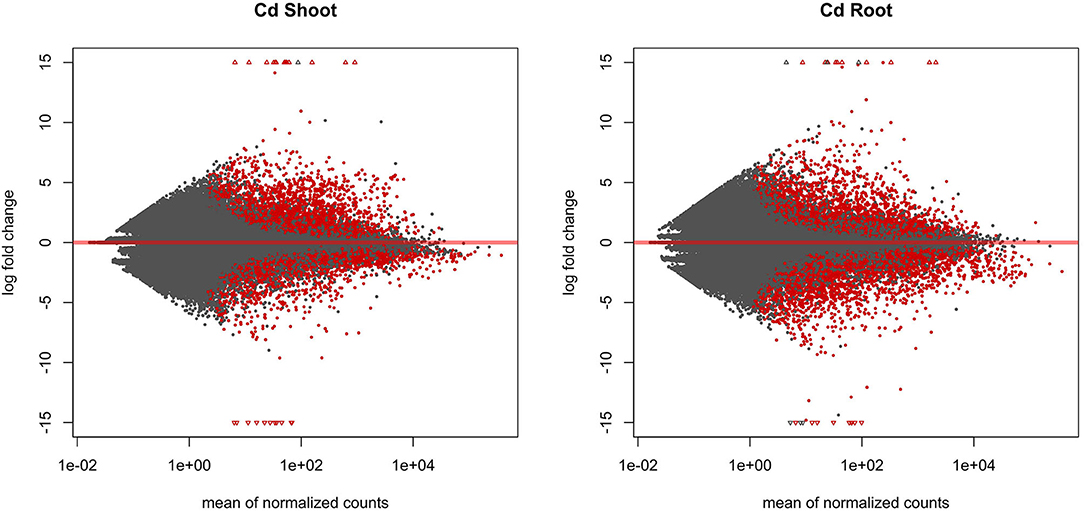
Figure 3. MA plot displaying the log-fold change (logFC) for each gene against the mean of normalized counts in shoot and root tissue. Differentially expressed genes with a p-value below 0.05 are highlighted in red.
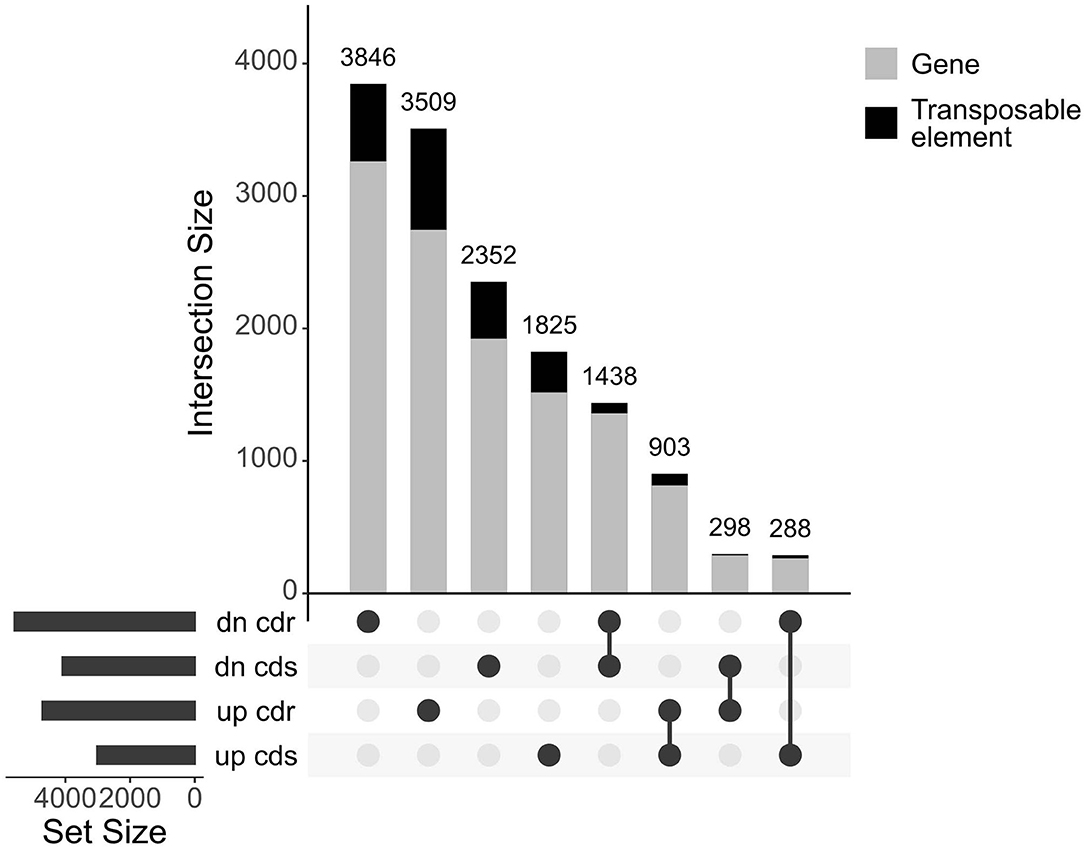
Figure 4. UpSet plot of the differentially expressed transcripts under cadmium treatment comparing response in root and shoot. Transcripts with a p-value below 0.05 were retained. Count of transcripts are displayed over the bars for each relationship of up and down (dn)-regulated transcripts from both root (cdr) and shoot (cds). The bars are colored according to the proportion of transcripts coming from both genes and transposable elements in gray and black, respectively.
Analysis of the transcripts related to repeated elements revealed that 2,285 elements (from total 14,459 DE transcribed sequences) are differentially expressed (Supplementary Figure 2). 59% are specifically differentially expressed in the root while 32% in the shoot. About 75% of the total of DE transposable elements are long terminal repeats (LTR). Transposable element related transcripts are more expressed in roots than shoots, similar to the genes-related ones, but unlike gene transcripts, they are more often up-regulated. Most of the expressed LTR belong to an unknown subfamily; otherwise, DE LTR Gypsy are more abundant than LTR Copia, followed by CACTA, nonLTR/LINE, and MITE elements (Supplementary Figure 3; Supplementary Table 6).
The 10 most abundant gene ontology (GO) annotations for both roots and shoots were assessed for DE transcripts. Comparison of the results reveals that regardless of the tissue, most of the genes influenced by cadmium are: involved in oxidation-reduction processes and phosphorylation, have mainly ATP binding or transferase activity functions and are part of the membrane. Moreover, the genes involved in protein phosphorylation and protein kinase activity are also influenced, but the majority of them are up-regulated while the genes involved in catalytic activity are mainly down-regulated (Figure 5).
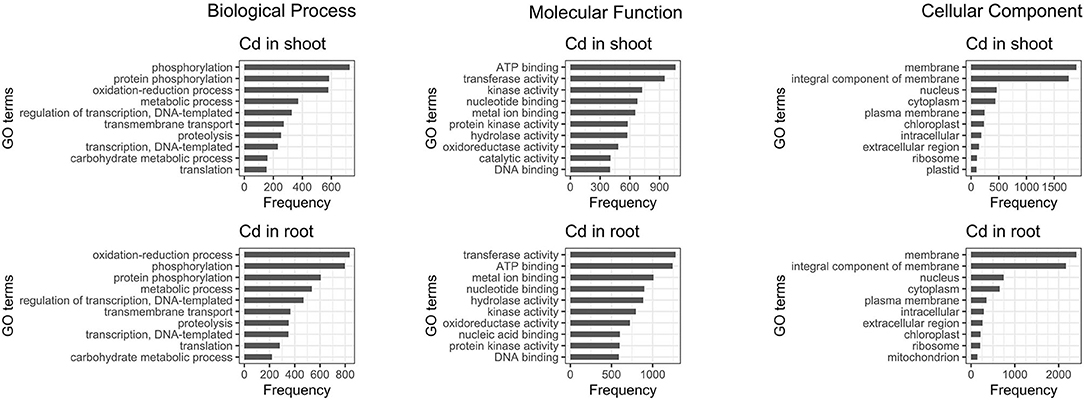
Figure 5. Ten most abundant gene ontology of differentially expressed genes present in roots and shoots. The 10 most abundant biological process, molecular function, and cellular component gene ontology (GO) are displayed for both root and shoot tissues.
In order to validate RNA-seq results, we selected a total of six transcripts for which we designed qPCR primers (Supplementary Table 3). HORVU.MOREX.r2.7HG0558200 is up-regulated in shoots, HORVU.MOREX.r2.6HG0456470 is down-regulated in shoots and HORVU.MOREX.r2.1HG0006620 is not differentially expressed in shoots. HORVU.MOREX.r2.2HG0162330 is up-regulated in roots, HORVU.MOREX.r2.7HG0538720 is down-regulated in roots and HORVU.MOREX.r2.7HG0561140 is not differentially expressed in roots (Supplementary Figure 4). The qPCR results correspond to the mRNA level identified by differential expression analysis for each of the six candidates (Supplementary Figure 5).
Among the most Cd upregulated transcripts, we identified homologs of PLANT CADMIUM RESISTANCE 2 (PCR2) gene. Predicted protein sequences show the highest similarity with AtPCR2 protein (Supplementary Table 5), therefore we have designated these genes HvPCR2.1 (HORVU.MOREX.r2.1HG0071220), HvPCR2.2 (HORVU.MOREX.r2.5HG0439660), HvPCR2.3 (HORVU.MOREX.r2.5HG0439680), and HvPCR2.4 (HORVU.MOREX.r2.5HG0439700). While the mRNA level of HvPCR2.1 is significantly upregulated by Cd in both root and shoot tissue, HvPCR2.2-4 are upregulated only in the shoot (Figure 6A). Surprisingly, HvPCR2.2-4 were found to be located in the same locus on barley chromosome 5H (Figure 6B). The fifth copy of HvPCR2 gene (HORVU.MOREX.r2.5HG0439690; denominated HvPCR2.5) is also physically present in this ~35 kb spanning region as well (Figure 6B). However, HvPCR2.5 does not exhibit significant upregulation upon Cd treatment. Similar tandem arrangement of PCR2 homologs have been found in rice (~30 kb locus on chromosome 3 containing six PCR2 homologs), corn (~90 kb locus on chromosome 5 containing three PCR2 homologs) and banana (~30 kb locus on chromosome 3 containing three PCR2 homologs) (Supplementary Figure 23).
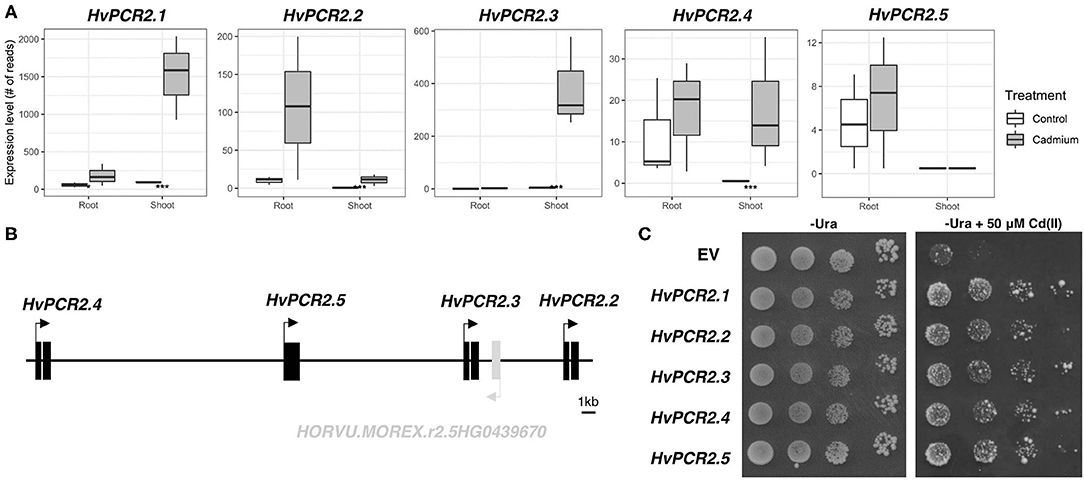
Figure 6. Expression, gene cluster structure, and functional analyses of HvPCR2. Transcription profiles of all HvPCR2 genes in control and after Cd exposure in root and shoot tissue (A). Box plot showing the expression level of a gene as count of reads for both root and shoot tissues in control and cadmium treated conditions. The difference between control and treatment was statistically tested and the adjusted P-value < 0.05 is displayed as an asterisk (*), similarly P-value < 0.005 is displayed as (***). Schematic diagram of the ~35 kb long genomic region of barley chromosome 5H containing HvPCR2.2-5 genes in tandem array (B). Expression of HvPCR2 genes restore the Cd tolerance in yeast mutant DTY168 (Δycf1) (C). Growth of DTY168 yeast cultures transformed with empty vector (EV) or constructs containing HvPCR2 genes on solid media with and without addition of Cd(II).
To elucidate the function of HvPCR2 genes in metal tolerance, we expressed them in the cadmium sensitive yeast strain DTY168 (Δycf1), which showed restricted growth at increased Cd concentrations. Growth of the DTY168 harboring empty vector p426 GDP was strongly inhibited by 50 μM CdCl2, whereas the same yeast mutant strain expressing either of the HvPCR2 genes was able to restore Cd tolerance (Figure 6C).
Cadmium is known to inhibit the growth of wheat (Sun et al., 2005) and other important crop relatives to barley (Rizwan et al., 2016; Ling et al., 2017). RNA-seq based comparative transcriptomics have been successfully used to uncover the mechanisms of plant stress response (Tombuloglu et al., 2015; Haak et al., 2017). In this study, we analyzed the transcriptome profiles of barley plants affected by elevated cadmium concentrations. We have selected Hordeum vulgare cv. Morex because of the availability of its genome and reference transcriptome sequence (Mascher, 2019).
To provide quantitative information about the presence of cadmium in barley plants, we measured the Cd concentration in roots and shoots separately (Figures 1C,D). We found that under cadmium treatment roots and shoots were able to accumulate a high concentration of this element. While Cd-treated shoots accumulate up to 300 times more Cd than in the non-treated condition, roots accumulated more than 6,600 times of this element. This difference indicates the limitations in the transport of cadmium in the aerial part of the plant.
Monitoring the cell cycle by EdU labeling revealed a lower number of S phase cells in Cd-treated roots. Such a decrease may suggest that the cells before G1/S transition are the most susceptible to Cd treatment. The recovery of the number of cells after removing the Cd by washing the root indicates that the major reduction in root growth is caused by the suppression of the cell division process namely in G1 phase cells (Figure 2). While the negative influence of Cd toxicity on cell proliferation is well-known in barley and wheat (Zhang and Yang, 1994; Paradiso et al., 2008), this phenomenon is not yet fully understood. Presumably, a Cd-induced ROS accumulation may cause an oxidative posttranslational modification of cyclin D and A-type cyclin-dependent kinase, thereby reducing the functionality of essential G1/S transition regulators (Pena et al., 2012). Similar reductions in the number of S phase cells have been reported in Cd-treated Zea mays (Bertels et al., 2020), Sorghum bicolor (Zhan et al., 2017), and Vicia faba (Zabka et al., 2021) as well as in Al-treated barley (Jaskowiak et al., 2018). Although Cd is also known to cause abnormal mitosis and aberrant chromosomes in barley (Shi et al., 2016), our data suggest a substantial effect of Cd toxicity at the level of G1-to-S-phase transition.
In our study, we identified over 14,400 transcripts differentially expressed in Cd treated plants, with about half of them being down-regulated. The total number of DE transcripts were lower in shoots (7,104) than in roots (10,282), consistent with the Cd concentration level detected in both tissues (Figures 1C,D). Previous transcriptomic studies reporting the effect of cadmium on barley focused on Cd tolerance, rather than response to Cd excess (Cao et al., 2014; Derakhshani et al., 2020). As the general objective of our study was to describe the overall response to cadmium in barley, we identified higher number of differentially expressed transcripts.
First, we analyzed the repetitive fraction of the barley genome at the transcriptional level to quantify the impact of cadmium on the activity of individual elements. Abiotic stress is frequently accompanied by the transcriptional and transpositional activation of transposable elements (TEs) (Grandbastien, 1998). Moreover, cadmium related stress leads to the establishment of a new balance of expressed/repressed chromatin (Greco et al., 2012). We found that the vast majority of transcriptionally affected repeats represent LTR retrotransposons (Copia, Gypsy and unknown families) and CACTA, nonLTR/LINE and MITE elements (Supplementary Figure 3). Transcriptional response to cadmium treatment is more evident in root tissue (Supplementary Figures 2, 3) including both upregulation and downregulation of individual TEs (Supplementary Table 6).
DE transcripts are mostly connected with oxidation-reduction processes, protein phosphorylation, metabolic processes, regulation of transcription and transmembrane transport (Figure 5). Among them, the most upregulated transcripts match genes encoding proteins involved in ROS metabolism and oxidative stress such as tau class glutathione-S-transferase (GST, Supplementary Figures 6, 7), purple acid phosphatase (PAP, Supplementary Figure 8), multidrug resistance-associated protein 3 (MRP3, Supplementary Figure 9), and cytochrome P450 enzymes (P450, Supplementary Figures 10, 11). All of these are also known to play a role in the detoxification of xenobiotics or abiotic stress signaling (Bovet et al., 2003; Schenk et al., 2013; Kumar and Trivedi, 2018; Pandian et al., 2020). Cadmium is known to induce oxidative stress in plants (Grobelak et al., 2019); therefore, the enhancement of ROS scavenging mechanisms represents a straightforward reaction to elevated Cd concentration. In particular, increased expression of GST and P450 enzymes in response to Cd stress has been previously described in rice (Ogawa et al., 2009) and the simultaneous overexpression of both enzymes in Medicago truncatula led to improved tolerance to mercury contamination (Zhang et al., 2013). In addition, four genes encoding homologs to putative chromatin remodeler OXIDATIVE STRESS 3 (OX3) were upregulated (Supplementary Figure 12). Although the molecular mechanism of OX3 action is not completely understood, this enzyme provides tolerance to Cd and oxidative stress in Arabidopsis and fission yeast (Blanvillain et al., 2009).
Other coding genes showing significant upregulation after Cd treatment, include those involved in cell wall formation and modification. Transcripts coding for lignin biosynthesis components, namely phenylalanine ammonia lyase (PAL; Supplementary Figure 13), shikimate O-hydroxycinnamoyltransferase (HCT; Supplementary Figure 14), cinnamoyl-CoA reductase (CCR; Supplementary Figure 15), cinnamyl alcohol dehydrogenase (CAD; Supplementary Figure 16), laccase 7 (LAC7; Supplementary Figure 17), and peroxidase (POD; Supplementary Figure 18) have been highly upregulated in Cd response. PAL is the key enzyme of the lignin biosynthesis acting at the beginning of this metabolic pathway. Accordingly, the Arabidopsis pal1 pal2 pal3 pal4 quadruple mutant showed approximately four times decreased lignin content in its cell wall compared with the wild-type (Huang et al., 2010). PAL activity or expression is known to be induced by Cd stress in various plant species including pea (Głowacka et al., 2019), Medicago sativa (Gutsch et al., 2019), Salix matsudana (Yang et al., 2015), rice (Hsu and Kao, 2005), and wheat (Shakirova et al., 2016). HCT, CCR and CAD catalyze the synthesis of monolignols, which are then polymerized into lignin by LAC and POD enzymes (Liu et al., 2018). As the lignification is improving the cell wall impermeability and lignin itself binds cadmium (Parrotta et al., 2015), the enhanced expression of these enzymes is likely to result in a lower Cd uptake and a higher Cd sequestration capacity. Notably, altered lignin biosynthesis was proposed to contribute to cadmium tolerance in Zn/Cd hyperaccumulator Noccaea caerulescens (van de Mortel et al., 2006, 2008). Other highly Cd responsive transcripts encode homologs of wound induced protein 1 (HORVU.MOREX.r2.1HG0053620; Supplementary Figure 19) and Leucine-rich repeat receptor-like protein kinase (HORVU.MOREX.r2.2HG0164280, Supplementary Figure 19), both of which have been reported to function in cell wall integrity sensing and responding to biotic and abiotic stress (Logemann and Schell, 1989; Van der Does et al., 2017).
Metal ion transporters play significant role in heavy metal homeostasis and detoxification (Jain et al., 2018). In our dataset, five deregulated transcripts have been identified to encode the members of the ZRT/IRT-like protein (ZIP) family (Supplementary Figure 20). ZIP transmembrane proteins are known to mediate cellular uptake, intracellular trafficking and detoxification of heavy metal cations such as Zn, Fe, Cd, Cu, Mn, Co, and Ni (Ajeesh Krishna et al., 2020). The role of selected ZIP transporters has been studied extensively in monocots (Li et al., 2013; Evens et al., 2017; Zheng et al., 2018) including barley (Pedas et al., 2009; Tiong et al., 2015). Interestingly, QTL mapping in barley identified two ZIP transcripts among the candidate genes for Cd tolerance (Derakhshani et al., 2020). In our dataset, we identified three previously studied ZIP transporters–HvZIP1 (HORVU.MOREX.r2.3HG0273580), HvZIP3 (HORVU.MOREX.r2.2HG0158440), and HvZIP6 (HORVU.MOREX.r2.1HG0020460). HvZIP1 was found to be upregulated by Cd in both tissues, but the effect is much stronger in roots (Supplementary Figure 20). An identical expression pattern has been found for OsZIP1, a rice ortholog for HvZIP1, that functions as a heavy metal efflux transporter maintaining detoxification of Zn, Cu and Cd in rice (Liu et al., 2019). Upon cadmium exposure, the HvZIP3 shows upregulation in roots but downregulation in shoots (Supplementary Figure 20). The transgenic barley plants with reduced HvZIP3 expression showed a significant increase in grain Cd accumulation (Sun et al., 2015). Further, a co-expression of rice HvZIP3 homolog (OsZIP3) with rice heavy metal transporter genes HEAVY METAL ATPASE 2 (OsHMA2) and LOW-AFFINITY CATION TRANSPORTER1 (OsLCT1) resulted in a decrease in Cd accumulation and enhanced Cd tolerance (Tian et al., 2019). HvZIP6 shows strong upregulation in shoots only (Supplementary Figure 20). So far, HvZIP6 was studied only in relation to Zn homeostasis (Tiong et al., 2015) but OsZIP6 is known to transport Cd2+ cations (Kavitha et al., 2015), therefore HvZIP6 may take part in shoot Cd detoxification. Two previously unknown barley ZIP transporters encoded by HORVU.MOREX.r2.2HG0098160 and HORVU.MOREX.r2.7HG0603680, show downregulation after Cd treatment (Supplementary Figure 20), which may suggest these could mediate Cd uptake.
Another group of deregulated genes encoding cation transmembrane transporters are members of the NATURAL RESISTANCE ASSOCIATED MACROPHAGE PROTEIN (NRAMP) family. In Cd-treated roots, HvNRAMP5 (HORVU.MOREX.r2.4HG0337880) exhibited several-fold downregulation (Supplementary Figure 21), which is consistent with the role of HvNRAMP5 as a Cd uptake transporter (Astolfi et al., 2014; Wu et al., 2016). Conversely, the HvNRAMP5 mRNA level slightly increased after Cd excess in shoot, which may imply involvement of this gene in shoot Cd distribution in stress condition. The HvNRAMP2 (HORVU.MOREX.r2.4HG0331160) were downregulated in shoots (Supplementary Figure 21). As the genome-wide association mapping study has proposed OsNRAMP2 to be responsible for Cd accumulation (Zhao et al., 2018), the downregulation of this transcript in barley might serve as a defense against accumulation of high Cd concentrations in shoot. The two remainder transcriptionally deregulated transporters from the NRAMP family were HvNRAMP1 (HORVU.MOREX.r2.7HG0610240; upregulated in roots) and HvNRAMP6 (HORVU.MOREX.r2.3HG0215920; upregulated in shoots) (Supplementary Figure 21).
Two transcripts encoding HEAVY METAL ATPASE (HMA), a P-type ATPase cation transporter family, were identified as being responsive to Cd. HvHMA1 (HORVU.MOREX.r2.5HG0401460) is expressed only in shoot tissue and no transcripts are detected after Cd treatment (Supplementary Figure 22). This pattern is similar to results published previously (Mikkelsen et al., 2012) and suggests that Cd induced downregulation of protoplast- and aleurone layer-localized HvHMA1 helps to prevent the cadmium transport into grains and protoplasts. The mRNA level of HvHMA3 gene (HORVU.MOREX.r2.7HG0603650) was decreased by Cd treatment (Supplementary Figure 22). Unfortunately, the function of this HMA3 copy has not been studied yet, so it is hard to clarify the effect of its downregulation. On the other hand, different HvHMA3 gene (HORVU5Hr1G094430) has been proposed to play a crucial role in grain Cd accumulation (Wu et al., 2015; Lei et al., 2020).
Remarkably, four homologs of PLANT CADMIUM RESISTANCE 2 (PCR2), encoding a Cys-rich domain containing membrane Cd/Zn efflux transporter (Song et al., 2010), have been identified among the Cd upregulated transcripts in shoot (Figure 6A). PCR1 and PCR2 genes were previously studied in Arabidopsis and it was proposed that they play a role in Zn/Cd detoxification and distribution (Song et al., 2004, 2010). The Arabidopsis pcr2 loss-of-function mutant is sensitive to Cd (Song et al., 2010). In rice, overexpression of OsPCR1 and OsPCR3 mediated Cd tolerance and lowered Cd accumulation (Wang et al., 2019). As there are no data regarding the function of PCR2 genes in monocots, we expressed barley HvPCR2 genes in a Cd sensitive yeast mutant. All five HvPCR2 genes restored the Cd tolerance in yeast (Figure 6C), hence it is probable that they retain a similar function as Arabidopsis homologs. Moreover, we have found out that four HvPCR2 genes are located on barley chromosome 5H in tandem arrangement (Figure 6B), probably due to local gene duplication events. A similar arrangement of multiple PCR2 copies was found in genomic regions of rice, corn and banana (Supplementary Figure 23), suggesting that PCR2 multiplication might be common in monocots. Duplication of heavy metal transporter genes is known to contribute to Cd tolerance in metal hyperaccumulators Arabidopsis halleri and Noccaea caerulescens (Hanikenne et al., 2008; Ó Lochlainn et al., 2011). Beside increasing gene dosage, the duplication may also allow functional divergence. It is therefore possible that the increased copy number of PCR2 genes originated as an adaptive response to Cd toxicity and/or Zn homeostasis requirements.
In summary, we investigated the impact of cadmium excess at the transcriptional level in barley. Among Cd-induced transcripts, we have identified the HvPCR2 gene as one of the most cadmium responsive. Surprisingly, further analysis suggested local multiplication of the gene on barley chromosome 5H as well as in other monocot species. Using gene complementation in yeast mutants we confirmed the functionality of individual HvPCR2 copies. This study may provide a valuable resource for the potential exploitation of the role of the HvPCR2 gene complex in cadmium tolerance in plants.
The datasets generated for this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: Sequenced reads are available in NCBI SRA (sequence read archive) under accession SRR5452097 to SRR5452108 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA382490).
MK and VH conducted the research in the laboratory. Computational analysis done by NB. All authors were involved in planning the experiments and contributed to the manuscript writing.
The financial support of the Technology Agency of the Czech Republic (project no. TN01000048) was gratefully acknowledged.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Computational resources were supplied by the project e-Infrastruktura CZ (e-INFRA LM2018140) provided within the program Projects of Large Research, Development and Innovations Infrastructures. We would like to thank Václav Bačovský for critically evaluating our manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.629089/full#supplementary-material
Ajeesh Krishna, T. P., Maharajan, T., Victor Roch, G., Ignacimuthu, S., and Antony Ceasar, S. (2020). Structure, function, regulation and phylogenetic relationship of ZIP family transporters of plants. Front. Plant Sci. 11:662. doi: 10.3389/fpls.2020.00662
Astolfi, S., Ortolani, M. R., Catarcione, G., Paolacci, A. R., Cesco, S., Pinton, R., et al. (2014). Cadmium exposure affects iron acquisition in barley (Hordeum vulgare) seedlings. Physiol. Plant 152, 646–659. doi: 10.1111/ppl.12207
Atkinson, N. J., and Urwin, P. E. (2012). The interaction of plant biotic and abiotic stresses: from genes to the field. J. Exp. Bot. 63, 3523–3544. doi: 10.1093/jxb/ers100
Bertels, J., Huybrechts, M., Hendrix, S., Bervoets, L., Cuypers, A., and Beemster, G. T. S. (2020). Cadmium inhibits cell cycle progression and specifically accumulates in the maize leaf meristem. J. Exp. Bot. 71, 6418–6428. doi: 10.1093/jxb/eraa385
Blanvillain, R., Kim, J. H., Wu, S., Lima, A., and Ow, D. W. (2009). OXIDATIVE STRESS 3 is a chromatin-associated factor involved in tolerance to heavy metals and oxidative stress. Plant J. 57, 654–665. doi: 10.1111/j.1365-313X.2008.03717.x
Bovet, L., Eggmann, T., Meylan-Bettex, M., Polier, J., Kammer, P., Marin, E., et al. (2003). Transcript levels of AtMRPs after cadmium treatment: induction of AtMRP3. Plant. Cell Environ. 26, 371–381. doi: 10.1046/j.1365-3040.2003.00968.x
Cao, F., Chen, F., Sun, H., Zhang, G., Chen, Z.-H., and Wu, F. (2014). Genome-wide transcriptome and functional analysis of two contrasting genotypes reveals key genes for cadmium tolerance in barley. BMC Genomics 15:611. doi: 10.1186/1471-2164-15-611
Clarkson, D. T., and Lüttge, U. (1989). Mineral nutrition: divalent cations, transport, and compartmentalization. Prog. Bot. 51, 93–112. doi: 10.1007/978-3-642-75154-7_7
DalCorso, G., Farinati, S., Maistri, S., and Furini, A. (2008). How plants cope with cadmium: staking all on metabolism and gene expression. J. Integr. Plant Biol. 50, 1268–1280. doi: 10.1111/j.1744-7909.2008.00737.x
Delhaize, E., and Ryan, P. R. (1995). Aluminum toxicity and tolerance in plants. Plant Physiol. 107, 315–321. doi: 10.1104/pp.107.2.315
Derakhshani, B., Jafary, H., Zanjani, B. M., Hasanpur, K., Mishina, K., Tanaka, T., et al. (2020). Combined QTL mapping and RNA-Seq profiling reveals candidate genes associated with cadmium tolerance in barley. PLoS ONE 15, 1–19. doi: 10.1371/journal.pone.0230820
Evens, N. P., Buchner, P., Williams, L. E., and Hawkesford, M. J. (2017). The role of ZIP transporters and group F bZIP transcription factors in the Zn-deficiency response of wheat (Triticum aestivum). Plant J. 92, 291–304. doi: 10.1111/tpj.13655
Fusco, N., Micheletto, L., Dal Corso, G., Borgato, L., and Furini, A. (2005). Identification of cadmium-regulated genes by cDNA-AFLP in the heavy metal accumulator Brassica juncea L. J. Exp. Bot. 56, 3017–3027. doi: 10.1093/jxb/eri299
Głowacka, K., Zróbek-Sokolnik, A., Okorski, A., and Najdzion, J. (2019). The Effect of cadmium on the activity of stress-related enzymes and the ultrastructure of pea roots. Plants (Basel, Switzerland) 8:413. doi: 10.3390/plants8100413
Grandbastien, M.-A. (1998). Activation of plant retrotransposons under stress conditions. Trends Plant Sci. 3, 181–187. doi: 10.1016/S1360-1385(98)01232-1
Greco, M., Chiappetta, A., Bruno, L., and Bitonti, M. B. (2012). In Posidonia oceanica cadmium induces changes in DNA methylation and chromatin patterning. J. Exp. Bot. 63, 695–709. doi: 10.1093/jxb/err313
Grobelak, A., Swiatek, J., Murtaś, A., and Jaskulak, M. (2019). “Chapter 9–cadmium-induced oxidative stress in plants, cadmium toxicity, and tolerance in plants: from physiology to remediation,” in eds M. Hasanuzzaman, M. N. V. Prasad, and P. Fujita (Cambridge, MA: Academic Press), 213–231.
Gutsch, A., Sergeant, K., Keunen, E., Prinsen, E., Guerriero, G., Renaut, J., et al. (2019). Does long-term cadmium exposure influence the composition of pectic polysaccharides in the cell wall of Medicago sativa stems? BMC Plant Biol. 19:271. doi: 10.1186/s12870-019-1859-y
Haak, D. C., Fukao, T., Grene, R., Hua, Z., Ivanov, R., Perrella, G., et al. (2017). Multilevel regulation of abiotic stress responses in plants. Front. Plant Sci. 8:1564. doi: 10.3389/fpls.2017.01564
Hanikenne, M., Talke, I. N., Haydon, M. J., Lanz, C., Nolte, A., and Motte, P. (2008). Evolution of metal hyperaccumulation required cis-regulatory changes and triplication of HMA4. Nature 453, 391–395. doi: 10.1038/nature06877
Howe, K. L., Contreras-Moreira, B., De Silva, N., Maslen, G., Akanni, W., Allen, J., et al. (2020). Ensembl genomes 2020—enabling non-vertebrate genomic research. Nucleic Acids Res. 48, D689–D695. doi: 10.1093/nar/gkz890
Hsu, Y. T., and Kao, C. H. (2005). Abscisic acid accumulation and cadmium tolerance in rice seedlings. Physiol. Plant. 124, 71–80. doi: 10.1111/j.1399-3054.2005.00490.x
Huang, J., Gu, M., Lai, Z., Fan, B., Shi, K., Zhou, Y.-H., et al. (2010). Functional analysis of the arabidopsis andlt;emandgt;palandlt;/emandgt; gene family in plant growth, development, and response to environmental stress. Plant Physiol. 153, 1526 LP−1538 LP. doi: 10.1104/pp.110.157370
IBSC (2016). Positions and Classifications of Repetitive Elements in the Genome of Barley cv. Morex. Gatersleben: e!DAL–Plant Genomics and Phenomics Research Data Repository (PGP), IPK Gatersleben.
Jain, S., Muneer, S., Guerriero, G., Liu, S., Vishwakarma, K., Chauhan, D. K., et al. (2018). Tracing the role of plant proteins in the response to metal toxicity: a comprehensive review. Plant Signal. Behav. 13:e1507401. doi: 10.1080/15592324.2018.1507401
Jaskowiak, J., Tkaczyk, O., Slota, M., Kwasniewska, J., and Szarejko, I. (2018). Analysis of aluminum toxicity in Hordeum vulgare roots with an emphasis on DNA integrity and cell cycle. PLoS ONE 13:e0193156. doi: 10.1371/journal.pone.0193156
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kavitha, P. G., Kuruvilla, S., and Mathew, M. K. (2015). Functional characterization of a transition metal ion transporter, OsZIP6 from rice (Oryza sativa L.). Plant Physiol. Biochem. 97, 165–174. doi: 10.1016/j.plaphy.2015.10.005
Kintlová, M., Blavet, N., Cegan, R., and Hobza, R. (2017). Transcriptome of barley under three different heavy metal stress reaction. Genomics Data 13, 15–17. doi: 10.1016/j.gdata.2017.05.016
Kumar, S., and Trivedi, P. K. (2018). Glutathione S-transferases: role in combating abiotic stresses including arsenic detoxification in Plants. Front. Plant Sci. 9:751. doi: 10.3389/fpls.2018.00751
Lei, G. J., Fujii-Kashino, M., Wu, D. Z., Hisano, H., Saisho, D., Deng, F., et al. (2020). Breeding for low cadmium barley by introgression of a Sukkula-like transposable element. Nat. Food 1, 489–499. doi: 10.1038/s43016-020-0130-x
Li, S., Zhou, X., Huang, Y., Zhu, L., Zhang, S., Zhao, Y., et al. (2013). Identification and characterization of the zinc-regulated transporters, iron-regulated transporter-like protein (ZIP) gene family in maize. BMC Plant Biol. 13:114. doi: 10.1186/1471-2229-13-114
Lin, C. Y., Trinh, N. N., Fu, S. F., Hsiung, Y. C., Chia, L. C., Lin, C. W., et al. (2013). Comparison of early transcriptome responses to copper and cadmium in rice roots. Plant Mol. Biol. 81, 507–522. doi: 10.1007/s11103-013-0020-9
Ling, T., Gao, Q., Du, H., Zhao, Q., and Ren, J. (2017). Growing, physiological responses and Cd uptake of Corn (Zea mays L.) under different Cd supply. Chem. Speciat. Bioavailab. 29, 216–221. doi: 10.1080/09542299.2017.1400924
Liu, Q., Luo, L., and Zheng, L. (2018). Lignins: biosynthesis and biological functions in plants. Int. J. Mol. Sci. 19:335. doi: 10.3390/ijms19020335
Liu, T., Liu, S., Guan, H., Ma, L., Chen, Z., Gu, H., et al. (2009). Transcriptional profiling of Arabidopsis seedlings in response to heavy metal lead (Pb). Environ. Exp. Bot. 67, 377–386. doi: 10.1016/j.envexpbot.2009.03.016
Liu, X. S., Feng, S. J., Zhang, B. Q., Wang, M. Q., Cao, H. W., Rono, J. K., et al. (2019). OsZIP1 functions as a metal efflux transporter limiting excess zinc, copper, and cadmium accumulation in rice. BMC Plant Biol. 19:283. doi: 10.1186/s12870-019-1899-3
Logemann, J., and Schell, J. (1989). Nucleotide sequence and regulated expression of a wound-inducible potato gene (wun1). Mol. Gen. Genet. 219, 81–88. doi: 10.1007/BF00261161
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8
Mascher, M. (2019). Pseudomolecules and Annotation of the Second Version of the Reference Genome Sequence Assembly of Barley cv. Morex [Morex V2]. Gatersleben: e!DAL–Plant Genomics and Phenomics Research Data Repository (PGP).
Mikkelsen, M. D., Pedas, P., Schiller, M., Vincze, E., Mills, R. F., Borg, S., et al. (2012). Barley HvHMA1 Is a heavy metal pump involved in mobilizing organellar Zn and Cu and plays a role in metal loading into grains. PLoS ONE 7:e49027. doi: 10.1371/journal.pone.0049027
Mumberg, D., Müller, R., and Funk, M. (1995). Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156, 119–122. doi: 10.1016/0378-1119(95)00037-7
Nriagu, J. O., and Pacyna, J. M. (1988). Quantitative assessment of worldwide contamination of air, water, and soils by trace metals. Nature 333, 134–139. doi: 10.1038/333134a,0
Ó Lochlainn, S., Bowen, H. C., Fray, R. G., Hammond, J. P., King, G. J., White, P. J., et al. (2011). Tandem quadruplication of HMA4 in the zinc (Zn) and cadmium (Cd) hyperaccumulator Noccaea caerulescens. PLoS ONE 6:e17814. doi: 10.1371/journal.pone.0017814
Ogawa, I., Nakanishi, H., Mori, S., and Nishizawa, N. K. (2009). Time course analysis of gene regulation under cadmium stress in rice. Plant Soil 325:97. doi: 10.1007/s11104-009-0116-9
Pandian, B. A., Sathishraj, R., Djanaguiraman, M., Prasad, P. V. V., and Jugulam, M. (2020). Role of cytochrome P450 enzymes in plant stress response. Antioxidants 9, 1–15. doi: 10.3390/antiox9050454
Paradiso, A., Berardino, R., de Pinto, M. C., Sanità di Toppi, L., Storelli, M. M., Tommasi, F., et al. (2008). Increase in ascorbate–glutathione metabolism as local and precocious systemic responses induced by cadmium in durum wheat plants. Plant Cell Physiol. 49, 362–374. doi: 10.1093/pcp/pcn013
Parrotta, L., Guerriero, G., Sergeant, K., Cai, G., and Hausman, J.-F. (2015). Target or barrier? the cell wall of early- and later-diverging plants vs cadmium toxicity: differences in the response mechanisms. Front. Plant Sci. 6:133. doi: 10.3389/fpls.2015.00133
Patro, R., Duggal, G., Love, M. I., Irizarry, R. A., and Kingsford, C. (2017). Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419. doi: 10.1038/nmeth.4197
Pedas, P., Schjoerring, J. K., and Husted, S. (2009). Identification and characterization of zinc-starvation-induced ZIP transporters from barley roots. Plant Physiol. Biochem. 47, 377–383. doi: 10.1016/j.plaphy.2009.01.006
Pedas, P., Ytting, C. K., Fuglsang, A. T., Jahn, T. P., Schjoerring, J. K., and Husted, S. (2008). Manganese efficiency in barley: identification and characterization of the metal ion transporter HvIRT1. Plant Physiol. 148, 455–466. doi: 10.1104/pp.108.118851
Pena, L. B., Barcia, R. A., Azpilicueta, C. E., Méndez, A. A. E., and Gallego, S. M. (2012). Oxidative post translational modifications of proteins related to cell cycle are involved in cadmium toxicity in wheat seedlings. Plant Sci. 196, 1–7. doi: 10.1016/j.plantsci.2012.07.008
Podar, D. (2013). “Plant growth and cultivation,” in Plant Mineral Nutrients. Methods in Molecular Biology (Methods and Protocols), Vol. 953, ed F. J. M. Maathuis (Totowa, NJ: Humana Press), 23–45. doi: 10.1007/978-1-62703-152-3_2
Rizwan, M., Ali, S., Adrees, M., Rizvi, H., Zia-ur-Rehman, M., Hannan, F., et al. (2016). Cadmium stress in rice: toxic effects, tolerance mechanisms, and management: a critical review. Environ. Sci. Pollut. Res. 23, 17859–17879. doi: 10.1007/s11356-016-6436-4
Salt, D. E., and Rauser, W. E. (1995). MgATP-Dependent transport of phytochelatins across the tonoplast of oat roots. Plant Physiol. 107, 1293–1301. doi: 10.1104/pp.107.4.1293
Sanità Di Toppi, L., and Gabbrielli, R. (1999). Response to cadmium in higher plants. Environ. Exp. Bot. 41, 105–130. doi: 10.1016/S0098-8472(98)00058-6
Sasaki, A., Yamaji, N., Yokosho, K., and Ma, J. F. (2012). Nramp5 is a major transporter responsible for manganese and cadmium uptake in rice. Plant Cell 24, 2155–2167. doi: 10.1105/tpc.112.096925
Schenk, G., Miti,ć, N., Hanson, G. R., and Comba, P. (2013). Purple acid phosphatase: a journey into the function and mechanism of a colorful enzyme. Coord. Chem. Rev. 257, 473–482. doi: 10.1016/j.ccr.2012.03.020
Shakirova, F. M., Allagulova, C. R., Maslennikova, D. R., Klyuchnikova, E. O., Avalbaev, A. M., and Bezrukova, M. V. (2016). Salicylic acid-induced protection against cadmium toxicity in wheat plants. Environ. Exp. Bot. 122, 19–28. doi: 10.1016/j.envexpbot.2015.08.002
Shi, Q., Wang, J., Zou, J., Jiang, Z., Wu, H., Wang, J., et al. (2016). Cadmium localization and its toxic effects on root tips of barley. Zemdirbyste-Agriculture 103, 151–158. doi: 10.13080/z-a.2016.103.020
Shim, D., Hwang, J.-U., Lee, J., Lee, S., Choi, Y., An, G., et al. (2009). Orthologs of the class A4 heat shock transcription factor HsfA4a confer cadmium tolerance in wheat and rice. Plant Cell 21, 4031–4043. doi: 10.1105/tpc.109.066902
Song, W. Y., Choi, K. S., Kim, D. Y., Geisler, M., Park, J., Vincenzetti, V., et al. (2010). Arabidopsis PCR2 is a zinc exporter involved in both zinc extrusion and long-distance zinc transport. Plant Cell 22, 2237–2252. doi: 10.1105/tpc.109.070185
Song, W. Y., Martinoia, E., Lee, J., Kim, D., Kim, D. Y., Vogt, E., et al. (2004). A novel family of Cys-rich membrane proteins mediates cadmium resistance in Arabidopsis. Plant Physiol. 135, 1027–1039. doi: 10.1104/pp.103.037739
Sun, H., Chen, Z. H., Chen, F., Xie, L., Zhang, G., Vincze, E., et al. (2015). DNA microarray revealed and RNAi plants confirmed key genes conferring low Cd accumulation in barley grains. BMC Plant Biol. 15:259. doi: 10.1186/s12870-015-0648-5
Sun, Q., Wang, X. R., Ding, S. M., and Yuan, X. F. (2005). Effects of exogenous organic chelators on phytochelatins production and its relationship with cadmium toxicity in wheat (Triticum aestivum L.) under cadmium stress. Chemosphere 60, 22–31. doi: 10.1016/j.chemosphere.2004.10.068
Szczypka, M. S., Wemmie, J. A., Moye-Rowley, W. S., and Thiele, D. J. (1994). A yeast metal resistance protein similar to human cystic fibrosis transmembrane conductance regulator (CFTR) and multidrug resistance-associated protein. J. Biol. Chem. 269, 22853–22857. doi: 10.1016/S0021-9258(17)31723-4
Thurman, D. A., and Collins, J. C. L. (1983). “Metal tolerance mechanism in higher plants review,” in Proceedings of International Conference on Heavy Metals in the Environmental (Heidelberg), 298–300.
Tian, S., Liang, S., Qiao, K., Wang, F., Zhang, Y., and Chai, T. (2019). Co-expression of multiple heavy metal transporters changes the translocation, accumulation, and potential oxidative stress of Cd and Zn in rice (Oryza sativa). J. Hazard. Mater. 380:120853. doi: 10.1016/j.jhazmat.2019.120853
Tiong, J., Mcdonald, G., Genc, Y., Shirley, N., Langridge, P., and Huang, C. Y. (2015). Increased expression of six ZIP family genes by zinc (Zn) deficiency is associated with enhanced uptake and root-to-shoot translocation of Zn in barley (Hordeum vulgare). New Phytol. 207, 1097–1109. doi: 10.1111/nph.13413
Tombuloglu, G., Tombuloglu, H., Sakcali, M. S., and Unver, T. (2015). High-throughput transcriptome analysis of barley (Hordeum vulgare) exposed to excessive boron. Gene 557, 71–81. doi: 10.1016/j.gene.2014.12.012
van de Mortel, J. E., Almar Villanueva, L., Schat, H., Kwekkeboom, J., Coughlan, S., Moerland, P. D., et al. (2006). Large Expression differences in genes for iron and zinc homeostasis, stress response, and lignin biosynthesis distinguish roots of arabidopsis thaliana and the related metal hyperaccumulator thlaspi caerulescens. Plant Physiol. 142, 1127 LP−1147 LP. doi: 10.1104/pp.106.082073
van de Mortel, J. E., Schat, H., Moerland, P. D., van Themaat, E. V. E. R. L., van der Ent, S., Blankestijn, H.„, et al. (2008). Expression differences for genes involved in lignin, glutathione and sulphate metabolism in response to cadmium in Arabidopsis thaliana and the related Zn/Cd-hyperaccumulator Thlaspi caerulescens. Plant. Cell Environ. 31, 301–324. doi: 10.1111/j.1365-3040.2007.01764.x
Van der Does, D., Boutrot, F., Engelsdorf, T., Rhodes, J., McKenna, J. F., Vernhettes, S., et al. (2017). The Arabidopsis leucine-rich repeat receptor kinase MIK2/LRR-KISS connects cell wall integrity sensing, root growth and response to abiotic and biotic stresses. PLoS Genet. 13:e1006832. doi: 10.1371/journal.pgen.1006832
Wang, F., Tan, H., Han, J., Zhang, Y., He, X., Ding, Y., et al. (2019). A novel family of PLAC8 motif-containing/PCR genes mediates Cd tolerance and Cd accumulation in rice. Environ. Sci. Eur. 31:82. doi: 10.1186/s12302-019-0259-0
Wu, D., Sato, K., and Ma, J. F. (2015). Genome-wide association mapping of cadmium accumulation in different organs of barley. New Phytol. 208, 817–829. doi: 10.1111/nph.13512
Wu, D., Yamaji, N., Yamane, M., Kashino-Fujii, M., Sato, K., and Ma, J. F. (2016). The HvNramp5 transporter mediates uptake of cadmium and manganese, but not iron. Plant Physiol. 172, 1899–1910. doi: 10.1104/pp.16.01189
Yang, J., Li, K., Zheng, W., Zhang, H., Cao, X., Lan, Y., et al. (2015). Characterization of early transcriptional responses to cadmium in the root and leaf of Cd-resistant Salix matsudana Koidz. BMC Genomics 16:705. doi: 10.1186/s12864-015-1923-4
Yang, M., Lin, X., and Yang, X. (1998). Impact or Cd on growth and nutrient accumulation or different plant species. Chinese J. Appl. Ecol. 9, 89–94.
Zabka, A., Winnicki, K., Polit, J. T., Wróblewski, M., and Maszewski, J. (2021). Cadmium (II)-induced oxidative stress results in replication stress and epigenetic modifications in root meristem cell nuclei of vicia faba. Cells 10:640. doi: 10.3390/cells10030640
Zhan, Y., Zhang, C., Zheng, Q., Huang, Z., and Yu, C. (2017). Cadmium stress inhibits the growth of primary roots by interfering auxin homeostasis in Sorghum bicolor seedlings. J. Plant Biol. 60, 593–603. doi: 10.1007/s12374-017-0024-0
Zhang, Y., Liu, J., Zhou, Y., Gong, T., Wang, J., and Ge, Y. (2013). Enhanced phytoremediation of mixed heavy metal (mercury)-organic pollutants (trichloroethylene) with transgenic alfalfa co-expressing glutathione S-transferase and human P450 2E1. J. Hazard. Mater. 260, 1100–1107. doi: 10.1016/j.jhazmat.2013.06.065
Zhang, Y., and Yang, X. (1994). The toxic effects of cadmium on cell division and chromosomal morphology of Hordeum vulgare. Mutat. Res. 312, 121–126. doi: 10.1016/0165-1161(94)90016-7
Zhao, J., Yang, W., Zhang, S., Yang, T., Liu, Q., Dong, J., et al. (2018). Genome-wide association study and candidate gene analysis of rice cadmium accumulation in grain in a diverse rice collection. Rice (N. Y). 11:61. doi: 10.1186/s12284-018-0254-x
Keywords: barley, cadmium, transcriptomic analysis, HvPCR2, gene duplication
Citation: Kintlová M, Vrána J, Hobza R, Blavet N and Hudzieczek V (2021) Transcriptome Response to Cadmium Exposure in Barley (Hordeum vulgare L.). Front. Plant Sci. 12:629089. doi: 10.3389/fpls.2021.629089
Received: 13 November 2020; Accepted: 11 June 2021;
Published: 15 July 2021.
Edited by:
Eduardo Gusmão Pereira, Universidade Federal de Viçosa, BrazilReviewed by:
Huseyin Tombuloglu, Imam Abdulrahman Bin Faisal University, Saudi ArabiaCopyright © 2021 Kintlová, Vrána, Hobza, Blavet and Hudzieczek. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Vojtěch Hudzieczek, aHVkemllY3pla0BpYnAuY3o=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.