
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Plant Sci. , 09 February 2021
Sec. Plant Cell Biology
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.627501
The pentatricopeptide repeat (PPR) proteins constitute one of the largest nuclear-encoded protein families in higher plants, with over 400 members in most sequenced plant species. The molecular functions of these proteins and their physiological roles during plant growth and development have been widely studied. Generally, there is mounting evidence that PPR proteins are involved in the post-transcriptional regulation of chloroplast and/or mitochondrial genes, including RNA maturation, editing, intron splicing, transcripts’ stabilization, and translation initiation. The cooperative action of RNA metabolism has profound effects on the biogenesis and functioning of both chloroplasts and mitochondria and, consequently, on the photosynthesis, respiration, and development of plants and their environmental responses. In this review, we summarize the latest research on PPR proteins, specifically how they might function in the chloroplast, by documenting their mechanism of molecular function, their corresponding RNA targets, and their specific effects upon chloroplast biogenesis and host organisms.
The family of pentatricopeptide repeat (PPR) proteins is well known because of its abundant members and essential functions in angiosperm species, having been discovered just 20 years ago in a genome sequencing analysis of Arabidopsis thaliana (Small and Peeters, 2000). The amino acid composition and structure of PPR proteins are similar to those of tetratricopeptide repeat (TPR) proteins, which usually mediate the interaction among proteins (Rovira and Smith, 2019); both families are encoded by nuclear genes and characterized by a tandem of multiple repeating units. PPR proteins belong to the α-solenoid RNA-binding proteins (RBPs) superfamily, and these RBPs are reported to regulate all steps of the life cycle of messenger RNA (mRNA) (Kramer et al., 2018; Müller-McNicoll et al., 2019). Nonetheless, PPR proteins are also closely related to the family of tandem repeat (TR) proteins, which includes armadillo (ARM), leucine-rich repeats (LRRs), tetratricopeptide, ankyrin (ANK), and WD40 proteins, whose functions have been extensively studied in plants, suggesting a potential role of PPR proteins during stress and development processes (Sharma and Pandey, 2016).
Mutations in PPR-encoding genes are always accompanied by defects in chloroplast biogenesis, pigmentation, and embryo and seed development (Sun et al., 2018; Tadini et al., 2018; Wang X. W. et al., 2020). The corresponding phenotypes affiliated with PPR genes disruption probably arise from the loss of one to several mitochondrial or chloroplast gene products that are necessary for organelle development (Barkan and Small, 2014); of course, there are a few exceptions, such as gun1 and defectively organized tributaries 4, which are involved in integrating multiple developmental and stress-related signals in both young seedlings and the leaves of adults (Petricka et al., 2008; Ruckle and Larkin, 2009; Cottage et al., 2010). Nevertheless, dozens of photosynthesis defective mutants can survive during the seedling stage until their seed reserves are depleted (Barkan and Small, 2014). Therefore, the embryo-lethal phenotypes caused by plastid defects may generally be due to a dysfunctional plastid translation system that subsequently prevents the biogenesis of several key factors required for normal chloroplast development and photosynthesis (Zoschke et al., 2016; Tadini et al., 2018). Although there is more evidence to suggest that PPR proteins participate specifically in organellar RNA processing, only a few PPR proteins have been functionally characterized in contrast to the vast majority of PPR members. This discrepancy is mainly attributed to the phenotypes of PPR genes disruption being very similar among individual PPR proteins, which makes gene functional studies have more challenges.
Based on its members’ motif structure, the PPR family can be further classified into two subfamilies, PLS and P. The P-class PPR proteins always contain from 2 to over 30 loosely conserved 35 amino acid PPR (P) motifs (Cheng et al., 2016). In addition, a few P-class subfamily members contain an additional small MutS-related (SMR) domain following an array of P-class PPR motifs, so these are classified as PPR-SMR subgroup (Liu et al., 2013). Actually, similar to SMR, there have been defined several subgroups according to their catalytic C-terminal domains, for example, TGM, TGM CCCH-zinc finger, LAGLIDADG, Mitochondrial RNA polymerase, and PRORP (Manna, 2015). In contrast, the PLS-class subfamily members usually consist of an array of triplets, namely, the canonical P motif, L motif, and S motif; both S and L motifs are related to the PPR motif but with a variable length of 31 amino acids (aa) and 35 or 36 aa, respectively (Barkan and Small, 2014). The PLS subfamily PPR proteins can be further categorized based on the attached domains found downstream of the PPR motifs: E, E+, and DYW, which occur in combination or alone (Cheng et al., 2016; Dedow and Bailey-Serres, 2019).
The classification of the PPR proteins family has undergone detailed revision as more species are sequenced and new PPR proteins are discovered. Based on a genomic analysis and comparison of 41 terrestrial plant species with evolutionary differences, the PPR motifs have been redefined as follows (Cheng et al., 2016): the P motif is further divided into P1 and P2, according to a difference in the first helix. Then, in the PLS motif, the L1 and L2 motifs, respectively, consisting of 35 aa and 36 aa can be distinguished, by a difference in the second helix. Similarly, the S motif is also divided into the regular 31-aa S1 motif and the 32-aa S2 motif. Besides the S1 and S2 motifs, a novel S-like motif (31 aa) named SS has been identified that is usually juxtaposed to other S motifs. Meanwhile, the E, E+, and DYW motifs were redefined, and two new 34-aa motifs, E1 and E2, were proposed. In this way, the PLS subfamily was re-divided into six subgroups and a combined P subfamily; here, we summarized the structure and classification of their PPR motifs based on this newly revised definition (Figure 1).
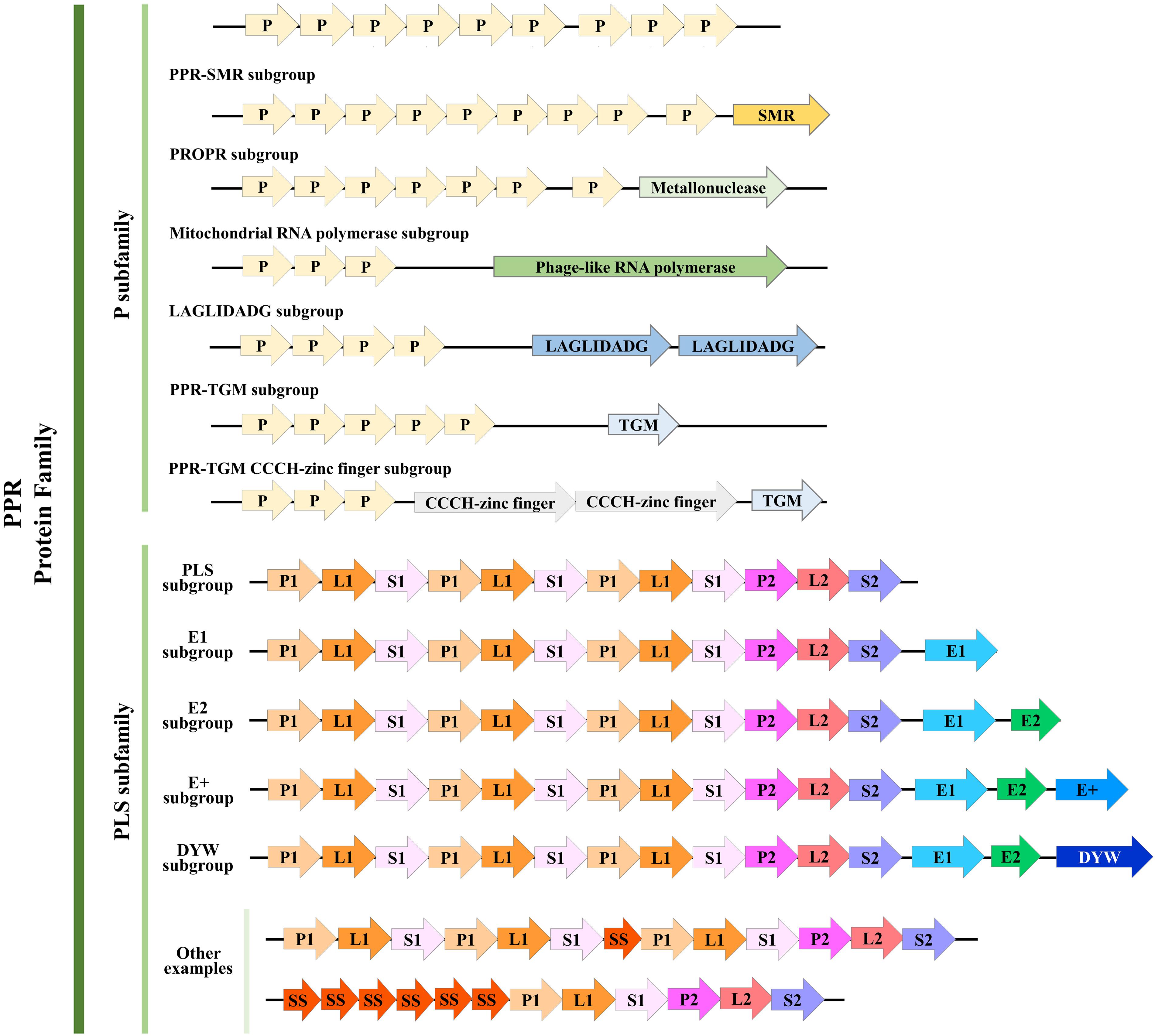
Figure 1. Structure of pentatricopeptide repeat (PPR) proteins. Refined classification and motif structure of PPR proteins (modified from Manna, 2015 and Cheng et al., 2016). The PPR family can be further classified into two subfamilies, P and PLS. P subfamily proteins consist of one or more tandem arrays of P motifs; whereas PLS subfamily proteins consist of triplets of P, L, and S motifs, occasionally interspersed with additional S motifs (P, L, and S can be further divided as we described above). A few P-class proteins contain additional motifs, such as the SMR, TGM, TGM CCCH-zinc finger, LAGLIDADG, Mitochondrial RNA polymerase, and PRORP. Almost all PLS-class proteins contain C-terminal E motifs (E1, E2, and E+; E+ motif was defined by Cheng et al., 2016), and many contain, in addition, a DYW motif. Some other examples that belong to the PLS subfamily were shown here. Arrows with different colors represent different motifs. Numbers of PPR motifs shown in each subgroup do not represent all members, and the motifs are not to scale.
Genomes have been sequenced in various plant species to date. To further reveal the extent of PPR proteins in plants at broader taxonomic scale, we analyzed about 30 species whose entire genomes were sequenced in the last several years. On average, we find about 500 PPR genes (filtered pseudo-genes and those alternatively spliced) in each of the various species, but up to 900 members in Papaver somniferum. With these data, we then constructed a phylogenetic tree of PPR proteins family in these plants (Figure 2). An outstanding question is this: why do plants have so many PPR proteins yet their bacterial ancestors do not? There are several theories whose adaptive hypotheses are not mutually exclusive, with some support for each of them (Barkan and Small, 2014). To narrow down the possibilities may require more extensive functional studies in PPR proteins and large-scale phylogenetic analysis across a vast number of plant species. However, for now, it is reasonable to conclude that PPR proteins clearly play a critical role in facilitating the chloroplast adaptation to the eukaryotic context (Stern et al., 2010).
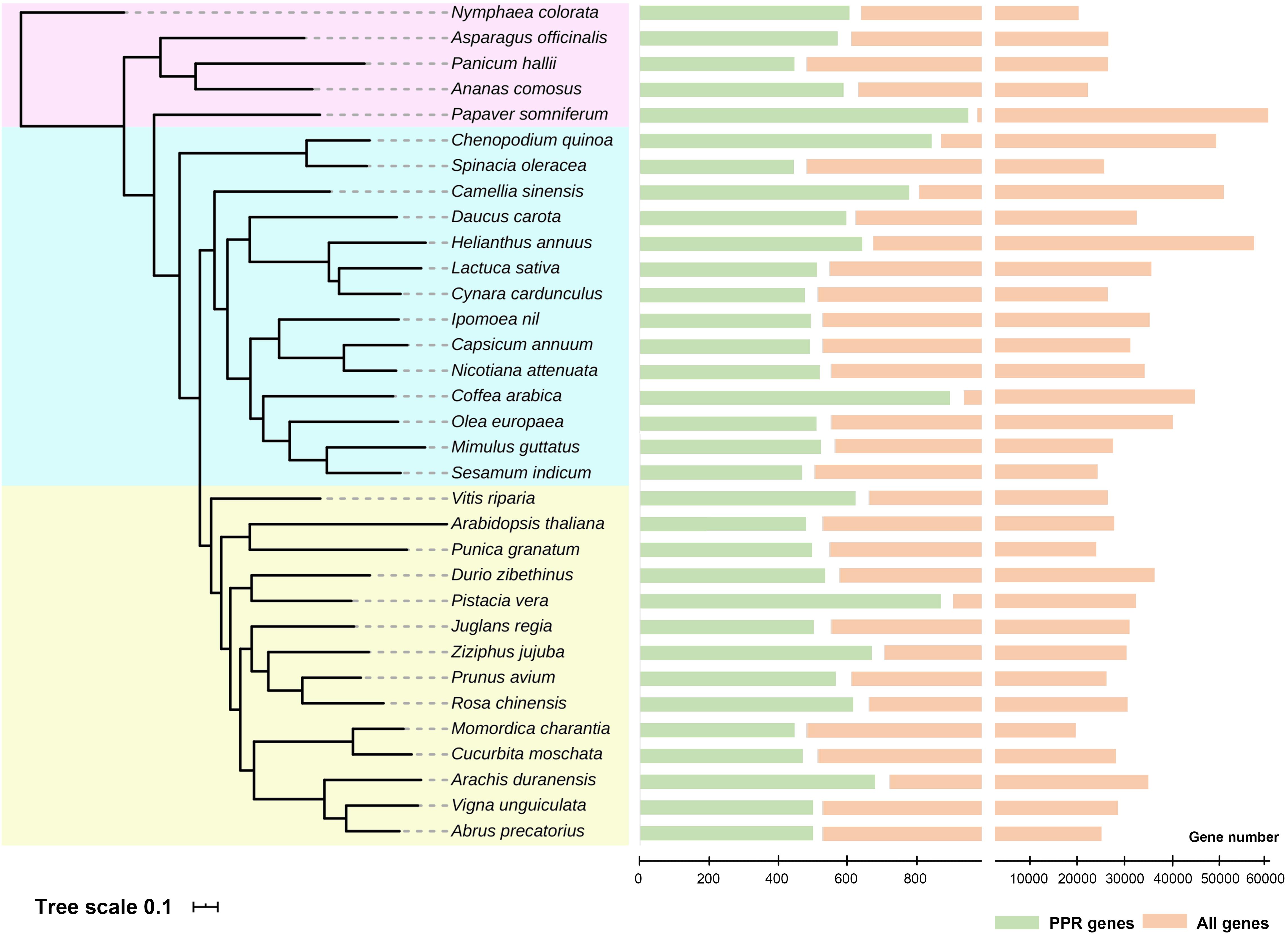
Figure 2. Phylogenetic tree and numbers of pentatricopeptide repeat (PPR) proteins in the entire genomes of recently sequenced plant species. All 33 species were divided into three groups (shown in different background colors), and the numbers of PPR genes were shown on the right. Detailed analysis methods were according to Yan et al. (2020). Genome data were obtained from NCBI, and these species were cited as follows: Lurin et al. (2004); Hellsten et al. (2013), Liu et al. (2014); Kim et al. (2014), Wang et al. (2014); Ming et al. (2015), Bertioli et al. (2016); Cruz et al. (2016), Hoshino et al. (2016); Iorizzo et al. (2016), Martínez-García et al. (2016); Scaglione et al. (2016), Badouin et al. (2017); Cui et al. (2017), Harkess et al. (2017); Jarvis et al. (2017), Qin et al. (2017); Reyes-Chin-Wo et al. (2017), Shirasawa et al. (2017); Sun et al. (2017), Teh et al. (2017); Xia et al. (2017), Xu C. et al. (2017), Xu S. et al. (2017), Guo et al. (2018); Lovell et al. (2018), Raymond et al. (2018); Tran et al. (2018), Girollet et al. (2019); Hovde et al. (2019), Lonardi et al. (2019); Zeng et al. (2019), and Zhang L. et al. (2020).
Functional chloroplasts are the main sites of photosynthesis, providing both relatively independent space environment and the key proteins for its successful operation, thus constructing the plant’s photoautotrophic condition. Plastid gene expression is indispensable for the development of chloroplasts and their maintenance of normal functions, and PPR proteins can affect it considerably. Thus, it is not surprising that defects in PPR proteins’ functions can yield similar phenotypes associated with chloroplast dysfunction (Manna, 2015). However, there are pronounced differences between the various PPR proteins.
The P subfamily of PPR proteins is generally believed to take part in RNA stabilization and translational activation and also promote the splicing of group II introns (Barkan and Small, 2014). Firstly, the function of P-class PPR proteins aims at stabilizing specific RNAs in chloroplast. In general, PPR proteins can bind and stabilize the 3′ or 5′ termini of transcription units, also including those termini at the end of a transcript arising from processing between open reading frames (ORFs) in polycistronic units. Moreover, the molecular mechanism depends on diversified PPR motifs bound to a high-affinity RNA target, which can act as a barrier to block exoribonucleases activity. The bound protein determines the actual site of the processed ends, while also preventing the adjacent RNA from undergoing degradation (Barkan and Small, 2014). A classic example, AtBFA2, is necessary for the stabilization of atpH/F transcripts in chloroplasts by binding to the atpF–atpA intergenic region, where it serves as a barrier to protect the atpH/F transcripts from destruction by exonucleases (Zhang et al., 2019). Compared with RNA stabilization, subsequent research has shown that translation activation plays a more crucial role in the enhancement of target genes’ expression (Zoschke et al., 2013). It was found that ZmPPR10 can bind to the atpI–atpH intergenic region in the chloroplast, thereby augmenting the efficiency of atpH translation. The examples of this function are still obscure, and the underlying mechanism may depend on substrate binding that results in the failed formation of an inhibitory structure by occupying the 5′ end of the hairpin. Meanwhile, blocking this site also reduces the activity of 5′→3′ exoribonucleases, which accordingly stabilized the ORF structure. Thus, RNA stabilization effects and translational enhancement are complementary, as proven repeatedly by studying the function of the LPE1 protein (Prikryl et al., 2011; Williams-Carrier et al., 2019).
P-type PPR proteins feature prominently among a plethora of nucleus-encoded proteins in the splicing of group II introns. Usually, the group II introns in plant organelles have lost the ability of self-splicing and so require other auxiliary factors for their splicing to occur (de Longevialle et al., 2010). It is likely that most of the P-type PPR proteins promote splicing by binding to RNA segments that would otherwise interfere with effective intron folding. Many progresses have been achieved in elucidating such functions in recent years. For example, EMB-7L strongly affects the splicing of multiple plastid or chloroplast transcripts in maize, namely, trans-rps12-intron1, rpl2, atpF, and ndhA (Yuan et al., 2019). In our research group, we have characterized two novel PPR factors in Arabidopsis: PDM3 and PDM4. In the pdm3 mutant, intron splicing of trnA, ndhB, and clpP-1 was significantly inhibited, and the ensuing phenotypes showed an abnormal chloroplast, inability to engage in photosynthetic autotrophy, and seedling death (Zhang et al., 2017). Concerning the pdm4 mutant, several group II introns, such as ndhA, petB, ycf3-int1, and petD, were also seriously affected, having a similar phenotype to the pdm3 mutant (Wang X. W. et al., 2020). Unlike multiple targets affected by PDM3 and PDM4, some P-type proteins have been found to act upon a specific target (Ito et al., 2018; Lee et al., 2019; Wang X. M. et al., 2020). Given that plastid intron splicing is a very important process during the gene expression, here we drew a diagram of proteins that facilitate plastid intron splicing (Supplementary Figure 1) based on previous studies (Kroeger et al., 2009; Stern et al., 2010) and the PPR proteins mentioned in this review. Nevertheless, some P subfamily members, for example, ATP4, PDM2, NUWA, and WSL5, are also required for RNA editing of plastid transcripts (Zoschke et al., 2012; Du et al., 2017; He et al., 2017; Liu et al., 2018).
Pentatricopeptide repeat proteins are widely considered to be involved in RNA regulation and metabolism in plant organelles. Not surprisingly, the basis for sequence-specific RNA recognition by PPR tracts has been focused upon and studied for a long time. Fortunately, progress on this front has made great strides in understanding of how PPR proteins are able to recognize an RNA target (Barkan et al., 2012; Yin et al., 2013). The theory of the tandem array of PPR motifs is believed to facilitate the recognition and interaction with RNA, and it has been widely accepted in recent years (Yin et al., 2013). By using computational methods, Barkan et al. (2012) inferred a code for nucleotide recognition involving two amino acids in each repeat and then validated their model by designing and recoding a PPR protein to bind novel RNA sequences in vitro. These results proved that a PPR protein is capable of binding to an RNA sequence in a parallel orientation via a code-recognition mechanism, with nucleotide specificity depending chiefly on the amino acid identities at positions of 6 and 19 in each PPR repeat. In addition, using structural biology analysis, Yin et al. (2013) revealed the molecular mechanism for the specific and modular recognition of RNA bases A, G, and U by the ZmPPR10 protein; generally, crystal structures of ZmPPR10 were respectively elucidated in either RNA-bound or RNA-free states. In the absence of RNA binding, the 19 PPR repeats of ZmPPR10 are folded into a right-handed super-helical spiral. However, in the presence of its targeting ssRNA, PPR10 can assemble into an antiparallel, intertwined homodimer and exhibits substantial conformation changes. Finally, six corresponding PPR10 repeats were found to specifically recognize six nucleotides of psaJ, again testifying to the utility of that predicted code (Yin et al., 2013).
Except for the classical P motif, a PLS-type protein always harbors L or S variant PPR motifs and additional C-terminal domains (E, E+, and DYW), which is considered responsible for RNA editing at specific sites (Barkan and Small, 2014). In land plants, RNA editing is a post-transcriptional process that can be fulfilled by deaminating the specific cytidines to uridines, and most of the discovered trans-factors involved in this process belong to the PLS subfamily (Shikanai, 2015). It has been proven that the multiple organellar RNA editing factors (MORFs) can interact with the PLS-type PPR proteins and participate in RNA editing. Detailed, the PPR proteins recognize cytidine targets, whereas the MORF proteins modulate the RNA-binding activity of PPR proteins (Huang et al., 2019; Zhao et al., 2019). For instance, MORF9 binding induced significant compressed conformational changes of PPR protein, resulting in that the RNA-binding activity of PPR proteins was drastically increased (Yan et al., 2017). RNA editing can modify the genetic information on RNA molecules and generate translational start or stop codons, which are essential for timely and accurate gene expression (Kotera et al., 2005). In Arabidopsis chloroplast, more than 40 of such edited sites have been found (Ruwe et al., 2013), and the machinery and biological function of RNA editing has still not been fully clarified.
The E and DYW motifs are thought to be associated with RNA editing. QED1 and ECD1 in Arabidopsis (Wagoner et al., 2015; Jiang et al., 2018) and PPR6 and PPR16 in rice (Tang et al., 2017; Huang et al., 2020), which belong to the PLS-DYW subclass, all affect the RNA editing of different sites, and they can show varied levels of mutational phenotypes. Focusing here on PPR6, its corresponding ppr6 mutant showed early defective chloroplast development that led to albino leaves and eventual seedling death. In further comparing the sequences of 19 edited sites in the rice chloroplast, only the ndhB transcript differs between osppr6 mutants and wild-type plants. Compared with the wild-type, this cytidine was only edited to uridine about 30%, indicating that the osppr6 mutant still harbored 30% of its RNA editing activity. Therefore, OsPPR6 specifically edits the transcript of chloroplast ndhB, and the reduced editing efficiency of this site in osppr6 probably caused the disrupted function of the NADH dehydrogenase subunit 2 (Tang et al., 2017). A PSL-E member, PPR756 in rice, was proven to participate in editing events among three mitochondrial genes, rather than in the chloroplast per se (Zhang Q. et al., 2020). This finding further confirms that PLS-type PPR proteins typically act as site-specific editing factors during RNA editing in plant organelles. Interestingly, similar to exceptions of P-type PPR proteins, there are also some special PLS PPR proteins: OsWSL, OsSLA4, and PGL12, which are responsible for RNA splicing (Tan et al., 2014; Wang et al., 2018; Chen et al., 2019).
Chloroplast biogenesis is highly complex, and the intricate molecular mechanisms have not been fully characterized yet. However, the complexity of this process is understandable because of its ancestry, having originated via endosymbiosis with species of cyanobacteria (Cortleven and Schmülling, 2015). With its own separate genome, signaling connections are heavily relied upon to relay information between the nucleus and chloroplast genomes, and the development of functional and photosynthetically active chloroplasts depends on the proper formation and assembly of molecules (Pogson et al., 2015). Corroborating this, it has been demonstrated that PPR proteins participate in chloroplast gene expression and function (Barkan and Small, 2014).
Based on the above, we may summarize that various PPR proteins can foster a similar phenotype with respect to chloroplast, such as pale-yellowish pigmentation, altered PSII biogenesis and formation of grana thylakoid, hindered chlorophyll synthesis, and severe defects in photosynthesis. Upon further analysis, PPR proteins can directly or indirectly affect chloroplast biogenesis or development processes through a variety of action modes and functional types, which can lead to abnormal chloroplast development, or almost an entire loss of functioning chloroplast, leading to the death of embryos or seedlings. According to our reviewed material, we summarized the known PPR proteins’ respective activity in recent years in Supplementary Table 1 and drew a simple working model to further summarize the function of PPR proteins (Supplementary Figure 2).
In general, PPR proteins are divided into the PLS and P subfamilies, and they are usually involved in RNA editing or RNA stabilization, cleavage, and splicing. However, with the deepening and accumulating research, not all PPR proteins conform to this law. Maybe plenty of evolutionary analyses and crystal structures of PPR–RNA complexes will provide deeper insight into subtle classification and functional mechanism. Besides, more attention should be paid to PPR interacting proteins that could severely affect the PPR protein effects, such as MORFs and/or RNA editing factor interaction proteins (RIPs) (Zhao et al., 2019). Excitingly, some of new research studies suggest that artificial PPR proteins could be customized to bind specific endogenous RNA in vivo, thus providing infusive research prospects for the development of designer RBPs for applications in biotechnology and synthetic biology (McDermott et al., 2019).
JX designed the outlines in this review and edited the manuscript. XW and YA wrote the original manuscript. PX analyzed the genomic data. All authors read and approved the final manuscript.
This work was supported by the National Natural Science Foundation of China (Grant Number 31970653).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank the Charlesworth Group (www.cwauthors.com) for their linguistic assistance during the preparation of this manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.627501/full#supplementary-material
Supplementary Figure 1 | Proteins that facilitate plastid intron splicing. Introns are designated as subgroup I, IIA, and IIB, according to Michel et al. (1989). The oval represents the PPR proteins; quadrilateral refers to those proteins belonging to other families. Solid lines indicate the recent research progress mentioned in our present review paper; dashed lines indicate previous results. The arrows point to their targets.
Supplementary Figure 2 | Simple working models of PPR proteins. PPR proteins can interact with RNA to affect gene expression by several ways (modified and refined from Barkan and Small, 2014). (A) PPR protein could expose (or mask) cis-elements for RNA translation, RNA splicing, RNA cleavage, or RNA editing. (B) PPR protein can activate translation via preventing the formation of an RNA structure, which masks the ribosome-binding site. (C) PPR protein can stabilize RNAs by blocking 5′→3′ and 3′→5′ exoribonucleases. Different PPR proteins are shown in different colors.
Supplementary Table 1 | Phenotypes and functions of PPR proteins in plant growth and development according to this review were summarized in recent years (since 2015). Location: C, chloroplast; M, mitochondrion.
Badouin, H., Gouzy, J., Grassa, C. J., Murat, F., Staton, S. E., Cottret, L., et al. (2017). The sunflower genome provides insights into oil metabolism, flowering and Asterid evolution. Nature 546, 148–152. doi: 10.1038/nature22380
Barkan, A., Rojas, M., Fujii, S., Yap, A., Chong, Y. S., Bond, C. S., et al. (2012). A combinatorial amino acid code for RNA recognition by pentatricopeptide repeat proteins. PLoS Genet. 8:e1002910. doi: 10.1371/journal.pgen.1002910
Barkan, A., and Small, I. (2014). Pentatricopeptide repeat proteins in plants. Annu. Rev. Plant Biol. 65, 415–442. doi: 10.1146/annurev-arplant-050213-040159
Bertioli, D. J., Cannon, S. B., Froenicke, L., Huang, G., Farmer, A. D., Cannon, E. K., et al. (2016). The genome sequences of Arachis duranensis and Arachis ipaensis, the diploid ancestors of cultivated peanut. Nat. Genet. 48, 438–446. doi: 10.1038/ng.3517
Chen, L., Huang, L., Dai, L., Gao, Y., Zou, W., Lu, X., et al. (2019). PALE-GREEN LEAF12 encodes a novel pentatricopeptide repeat protein required for chloroplast development and 16S rRNA processing in Rice. Plant Cell Physiol. 60, 587–598. doi: 10.1093/pcp/pcy229
Cheng, S., Gutmann, B., Zhong, X., Ye, Y., Fisher, M. F., Bai, F., et al. (2016). Redefining the structural motifs that determine RNA binding and RNA editing by pentatricopeptide repeat proteins in land plants. Plant J. 85, 532–547. doi: 10.1111/tpj.13121
Cortleven, A., and Schmülling, T. (2015). Regulation of chloroplast development and function by cytokinin. J. Exp. Bot. 66, 4999–5013. doi: 10.1093/jxb/erv132
Cottage, A., Mott, E. K., Kempster, J. A., and Gray, J. C. (2010). The Arabidopsis plastid-signalling mutant gun1 (genomes uncoupled1) shows altered sensitivity to sucrose and abscisic acid and alterations in early seedling development. J. Exp. Bot. 61, 3773–3786. doi: 10.1093/jxb/erq186
Cruz, F., Julca, I., Gómez-Garrido, J., Loska, D., Marcet-Houben, M., Cano, E., et al. (2016). Genome sequence of the olive tree, Olea europaea. Gigascience 5:29. doi: 10.1186/s13742-016-0134-5
Cui, J., Cheng, J., Nong, D., Peng, J., Hu, Y., He, W., et al. (2017). Genome-wide analysis of simple sequence repeats in bitter gourd (Momordica charantia). Front. Plant Sci. 8:1103. doi: 10.3389/fpls.2017.01103
de Longevialle, A. F., Small, I. D., and Lurin, C. (2010). Nuclearly encoded splicing factors implicated in RNA splicing in higher plant organelles. Mol. Plant. 3, 691–705. doi: 10.1093/mp/ssq025
Dedow, L. K., and Bailey-Serres, J. (2019). Searching for a match: structure, function and application of sequence-specific RNA-Binding proteins. Plant Cell Physiol. 60, 1927–1938. doi: 10.1093/pcp/pcz072
Du, L., Zhang, J., Qu, S., Zhao, Y., Su, B., Lv, X., et al. (2017). The pentratricopeptide repeat protein Pigment-Defective Mutant2 is involved in the regulation of chloroplast development and chloroplast gene expression in Arabidopsis. Plant Cell Physiol. 58, 747–759. doi: 10.1093/pcp/pcx004
Girollet, N., Rubio, B., Lopez-Roques, C., Valière, S., Ollat, N., and Bert, P. F. (2019). De novo phased assembly of the Vitis riparia grape genome. Sci. Data 6:127. doi: 10.1038/s41597-019-0133-3
Guo, L., Winzer, T., Yang, X., Li, Y., Ning, Z., He, Z., et al. (2018). The opium poppy genome and morphinan production. Science 362, 343–347. doi: 10.1126/science.aat4096
Harkess, A., Zhou, J., Xu, C., Bowers, J. E., Van der Hulst, R., Ayyampalayam, S., et al. (2017). The asparagus genome sheds light on the origin and evolution of a young Y chromosome. Nat. Commun. 8:1279. doi: 10.1038/s41467-017-01064-8
He, S., Sun, Y., Yang, Q., Zhang, X., Huang, Q., Zhao, P., et al. (2017). A novel imprinted gene NUWA controls mitochondrial function in early seed development in Arabidopsis. PLoS Genet. 13:e1006553. doi: 10.1371/journal.pgen.1006553
Hellsten, U., Wright, K. M., Jenkins, J., Shu, S., Yuan, Y., Wessler, S. R., et al. (2013). Fine-scale variation in meiotic recombination in Mimulus inferred from population shotgun sequencing. Proc. Natl. Acad. Sci. U.S.A. 110, 19478–19482. doi: 10.1073/pnas.1319032110
Hoshino, A., Jayakumar, V., Nitasaka, E., Toyoda, A., Noguchi, H., Itoh, T., et al. (2016). Genome sequence and analysis of the Japanese morning glory Ipomoea nil. Nat. Commun. 7:13295. doi: 10.1038/ncomms13295
Hovde, B. T., Daligault, H. E., Hanschen, E. R., Kunde, Y. A., Johnson, M. B., Starkenburg, S. R., et al. (2019). Detection of Abrin-Like and Prepropulchellin-Like toxin genes and transcripts using whole genome sequencing and full-length transcript sequencing of Abrus precatorius. Toxins (Basel) 11:691. doi: 10.3390/toxins11120691
Hsieh, W. Y., Liao, J. C., Chang, C. Y., Harrison, T., Boucher, C., and Hsieh, M. H. (2015). The slow growth 3 pentatricopeptide repeat protein is required for the splicing of mitochondrial NADH dehydrogenase subunit7 intron 2 in Arabidopsis. Plant Physiol. 168, 490–501. doi: 10.1104/pp.15.00354
Huang, C., Li, Z. R., Yu, Q. B., Ye, L. S., Cui, Y. L., Molloy, D. P., et al. (2019). MORF2 tightly associates with MORF9 to regulate chloroplast RNA editing in Arabidopsis. Plant Sci. 278, 64–69. doi: 10.1016/j.plantsci.2018.10.020
Huang, W., Zhang, Y., Shen, L., Fang, Q., Liu, Q., Gong, C., et al. (2020). Accumulation of the RNA polymerase subunit RpoB depends on RNA editing by OsPPR16 and affects chloroplast development during early leaf development in rice. New Phytol. 228, 1401–1416. doi: 10.1111/nph.16769
Iorizzo, M., Ellison, S., Senalik, D., Zeng, P., Satapoomin, P., Huang, J., et al. (2016). A high-quality carrot genome assembly provides new insights into carotenoid accumulation and asterid genome evolution. Nat. Genet. 48, 657–666. doi: 10.1038/ng.3565
Ito, A., Sugita, C., Ichinose, M., Kato, Y., Yamamoto, H., Shikanai, T., et al. (2018). An evolutionarily conserved P-subfamily pentatricopeptide repeat protein is required to splice the plastid ndhA transcript in the moss Physcomitrella patens and Arabidopsis thaliana. Plant J. 94, 638–648. doi: 10.1111/tpj.13884
Jarvis, D. E., Ho, Y. S., Lightfoot, D. J., Schmöckel, S. M., Li, B., Borm, T. J., et al. (2017). The genome of Chenopodium quinoa. Nature 542, 307–312. doi: 10.1038/nature21370
Jiang, T., Zhang, J., Rong, L., Feng, Y., Wang, Q., Song, Q., et al. (2018). ECD1 functions as an RNA-editing trans-factor of rps14-149 in plastids and is required for early chloroplast development in seedlings. J. Exp. Bot. 69, 3037–3051. doi: 10.1093/jxb/ery139
Kim, S., Park, M., Yeom, S. I., Kim, Y. M., Lee, J. M., Lee, H. A., et al. (2014). Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species. Nat. Genet. 46, 270–278. doi: 10.1038/ng.2877
Kotera, E., Tasaka, M., and Shikanai, T. (2005). A pentatricopeptide repeat protein is essential for RNA editing in chloroplasts. Nature 43, 326–330. doi: 10.1038/nature03229
Kramer, M. C., Anderson, S. J., and Gregory, B. D. (2018). The nucleotides they are a-changin’: function of RNA binding proteins in post-transcriptional messenger RNA editing and modification in Arabidopsis. Curr. Opin. Plant Biol. 45, 88–95. doi: 10.1016/j.pbi.2018.05.010
Kroeger, T. S., Watkins, K. P., Friso, G., van Wijk, K. J., and Barkan, A. (2009). A plant-specific RNA-binding domain revealed through analysis of chloroplast group II intron splicing. Proc. Natl. Acad. Sci. U.S.A. 106, 4537–4542. doi: 10.1073/pnas.0812503106
Lee, K., Park, S. J., Colas des Francs-Small, C., Whitby, M., Small, I., and Kang, H. (2019). The coordinated action of PPR4 and EMB2654 on each intron half mediates trans-splicing of rps12 transcripts in plant chloroplasts. Plant J. 100, 1193–1207. doi: 10.1111/tpj.14509
Liu, M. J., Zhao, J., Cai, Q. L., Liu, G. C., Wang, J. R., Zhao, Z. H., et al. (2014). The complex jujube genome provides insights into fruit tree biology. Nat. Commun. 5:5315. doi: 10.1038/ncomms6315
Liu, S., Melonek, J., Boykin, L. M., Small, I., and Howell, K. A. (2013). PPR-SMRs: ancient proteins with enigmatic functions. RNA Biol. 10, 1501–1510. doi: 10.4161/rna.26172
Liu, X., Lan, J., Huang, Y., Cao, P., Zhou, C., Ren, Y., et al. (2018). WSL5, a pentatricopeptide repeat protein, is essential for chloroplast biogenesis in rice under cold stress. J. Exp. Bot. 69, 3949–3961. doi: 10.1093/jxb/ery214
Lonardi, S., Muñoz-Amatriaín, M., Liang, Q., Shu, S., Wanamaker, S. I., Lo, S., et al. (2019). The genome of cowpea (Vigna unguiculata [L.] Walp.). Plant J. 98, 767–782. doi: 10.1111/tpj.14349
Lovell, J. T., Jenkins, J., Lowry, D. B., Mamidi, S., Sreedasyam, A., Weng, X., et al. (2018). The genomic landscape of molecular responses to natural drought stress in Panicum hallii. Nat. Commun. 9:5213. doi: 10.1038/s41467-018-07669-x
Lurin, C., Andrés, C., Aubourg, S., Bellaoui, M., Bitton, F., Bruyère, C., et al. (2004). Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 16, 2089–2103. doi: 10.1105/tpc.104.022236
Lv, J., Shang, L., Chen, Y., Han, Y., Yang, X., Xie, S., et al. (2020). OsSLC1 encodes a pentatricopeptide repeat protein essential for early chloroplast development and seedling survival. Rice 13:25. doi: 10.1186/s12284-020-00385-5
Manna, S. (2015). An overview of pentatricopeptide repeat proteins and their applications. Biochimie 113, 93–99. doi: 10.1016/j.biochi.2015.04.004
Martínez-García, P. J., Crepeau, M. W., Puiu, D., Gonzalez-Ibeas, D., Whalen, J., Stevens, K. A., et al. (2016). The walnut (Juglans regia) genome sequence reveals diversity in genes coding for the biosynthesis of non-structural polyphenols. Plant J. 87, 507–532. doi: 10.1111/tpj.13207
McDermott, J. J., Watkins, K. P., Williams-Carrier, R., and Barkan, A. (2019). Ribonucleoprotein capture by in vivo expression of a designer pentatricopeptide repeat protein in Arabidopsis. Plant Cell 31, 1723–1733. doi: 10.1105/tpc.19.00177
Michel, F., Umesono, K., and Ozeki, H. (1989). Comparative and functional anatomy of group II catalytic introns–a review. Gene 82, 5–30. doi: 10.1016/0378-1119(89)90026-7
Ming, R., VanBuren, R., Wai, C. M., Tang, H., Schatz, M. C., Bowers, J. E., et al. (2015). The pineapple genome and the evolution of CAM photosynthesis. Nat. Genet. 47, 1435–1442. doi: 10.1038/ng.3435
Müller-McNicoll, M., Rossbach, O., Hui, J., and Medenbach, J. (2019). Auto-regulatory feedback by RNA-binding proteins. J. Mol. Cell Biol. 11, 930–939. doi: 10.1093/jmcb/mjz043
Petricka, J. J., Clay, N. K., and Nelson, T. M. (2008). Vein patterning screens and the defectively organized tributaries mutants in Arabidopsis thaliana. Plant J. 56, 251–263. doi: 10.1111/j.1365-313X.2008.03595.x
Pogson, B. J., Ganguly, D., and Albrecht-Borth, V. (2015). Insights into chloroplast biogenesis and development. Biochim. Biophys. Acta 1847, 1017–1024. doi: 10.1016/j.bbabio.2015.02.003
Prikryl, J., Rojas, M., Schuster, G., and Barkan, A. (2011). Mechanism of RNA stabilization and translational activation by a pentatricopeptide repeat protein. Proc. Natl. Acad. Sci. U.S.A. 108, 415–420. doi: 10.1073/pnas.1012076108
Qin, G., Xu, C., Ming, R., Tang, H., Guyot, R., Kramer, E. M., et al. (2017). The pomegranate (Punica granatum L.) genome and the genomics of punicalagin biosynthesis. Plant J. 91, 1108–1128. doi: 10.1111/tpj.13625
Raymond, O., Gouzy, J., Just, J., Badouin, H., Verdenaud, M., Lemainque, A., et al. (2018). The Rosa genome provides new insights into the domestication of modern roses. Nat. Genet. 50, 772–777. doi: 10.1038/s41588-018-0110-3
Reyes-Chin-Wo, S., Wang, Z., Yang, X., Kozik, A., Arikit, S., Song, C., et al. (2017). Genome assembly with in vitro proximity ligation data and whole-genome triplication in lettuce. Nat. Commun. 12:14953. doi: 10.1038/ncomms14953
Rovira, A. G., and Smith, A. G. (2019). PPR proteins - orchestrators of organelle RNA metabolism. Physiol. Plant 166, 451–459. doi: 10.1111/ppl.12950
Ruckle, M. E., and Larkin, R. M. (2009). Plastid signals that affect photomorphogenesis in Arabidopsis thaliana are dependent on GENOMES UNCOUPLED 1 and cryptochrome 1. New Phytol. 182, 367–379. doi: 10.1111/j.1469-8137.2008.02729.x
Ruwe, H., Castandet, B., Schmitz-Linneweber, C., and Stern, D. B. (2013). Arabidopsis chloroplast quantitative editotype. FEBS Lett. 587, 1429–1433. doi: 10.1016/j.febslet.2013.03.022
Scaglione, D., Reyes-Chin-Wo, S., Acquadro, A., Froenicke, L., Portis, E., Beitel, C., et al. (2016). The genome sequence of the outbreeding globe artichoke constructed de novo incorporating a phase-aware low-pass sequencing strategy of F1 progeny. Sci. Rep. 20:19427. doi: 10.1038/srep19427
Sharma, M., and Pandey, G. K. (2016). Expansion and function of repeat domain proteins during stress and development in plants. Front. Plant Sci. 6:1218. doi: 10.3389/fpls.2015.01218
Shikanai, T. (2015). RNA editing in plants: machinery and flexibility of site recognition. Biochim. Biophys. Acta 1847, 779–785. doi: 10.1016/j.bbabio.2014.12.010
Shirasawa, K., Isuzugawa, K., Ikenaga, M., Saito, Y., Yamamoto, T., Hirakawa, H., et al. (2017). The genome sequence of sweet cherry (Prunus avium) for use in genomics-assisted breeding. DNA Res. 24, 499–508. doi: 10.1093/dnares/dsx020
Small, I. D., and Peeters, N. (2000). The PPR motif - a TPR-related motif prevalent in plant organellar proteins. Trends Biochem. Sci. 25, 45–47. doi: 10.1016/s0968-0004(99)01520-0
Stern, D. B., Goldschmidt-Clermont, M., and Hanson, M. R. (2010). Chloroplast RNA metabolism. Annu. Rev. Plant Biol. 61, 125–155. doi: 10.1146/annurev-arplant-042809-112242
Sun, H., Wu, S., Zhang, G., Jiao, C., Guo, S., Ren, Y., et al. (2017). Karyotype stability and unbiased fractionation in the paleo-allotetraploid Cucurbita Genomes. Mol. Plant 10, 1293–1306. doi: 10.1016/j.molp.2017.09.003
Sun, Y., Huang, J., Zhong, S., Gu, H., He, S., and Qu, L. J. (2018). Novel DYW-type pentatricopeptide repeat (PPR) protein BLX controls mitochondrial RNA editing and splicing essential for early seed development of Arabidopsis. J. Genet. Genom. 45, 155–168. doi: 10.1016/j.jgg.2018.01.006
Tadini, L., Ferrari, R., Lehniger, M. K., Mizzotti, C., Moratti, F., Resentini, F., et al. (2018). Trans-splicing of plastid rps12 transcripts, mediated by AtPPR4, is essential for embryo patterning in Arabidopsis thaliana. Planta 248, 257–265. doi: 10.1007/s00425-018-2896-8
Tan, J., Tan, Z., Wu, F., Sheng, P., Heng, Y., Wang, X., et al. (2014). A novel chloroplast-localized pentatricopeptide repeat protein involved in splicing affects chloroplast development and abiotic stress response in rice. Mol. Plant 7, 1329–1349. doi: 10.1093/mp/ssu054
Tang, J., Zhang, W., Wen, K., Chen, G., Sun, J., Tian, Y., et al. (2017). OsPPR6, a pentatricopeptide repeat protein involved in editing and splicing chloroplast RNA, is required for chloroplast biogenesis in rice. Plant Mol. Biol. 95, 345–357. doi: 10.1007/s11103-017-0654-0
Teh, B. T., Lim, K., Yong, C. H., Ng, C. C. Y., Rao, S. R., Rajasegaran, V., et al. (2017). The draft genome of tropical fruit durian (Durio zibethinus). Nat. Genet. 49, 1633–1641. doi: 10.1038/ng.3972
Tran, H. T. M., Ramaraj, T., Furtado, A., Lee, L. S., and Henry, R. J. (2018). Use of a draft genome of coffee (Coffea arabica) to identify SNPs associated with caffeine content. Plant Biotechnol. J. 16, 1756–1766. doi: 10.1111/pbi.12912
Wagoner, J. A., Sun, T., Lin, L., and Hanson, M. R. (2015). Cytidine deaminase motifs within the DYW domain of two pentatricopeptide repeat-containing proteins are required for site-specific chloroplast RNA editing. J. Biol. Chem. 290, 2957–2968. doi: 10.1074/jbc.M114.622084
Wang, H. C., Chen, Z., Yang, Y. Z., Sun, F., Ding, S., Li, X. L., et al. (2020). PPR14 interacts with PPR-SMR1 and CRM protein Zm-mCSF1 to facilitate mitochondrial intron splicing in Maize. Front. Plant Sci. 11:814. doi: 10.3389/fpls.2020.00814
Wang, L., Yu, S., Tong, C., Zhao, Y., Liu, Y., Song, C., et al. (2014). Genome sequencing of the high oil crop sesame provides insight into oil biosynthesis. Genome Biol. 15:R39. doi: 10.1186/gb-2014-15-2-r39
Wang, X. M., Yang, Z., Zhang, Y., Zhou, W., Zhang, A., and Lu, C. (2020). Pentatricopeptide repeat protein PHOTOSYSTEM I BIOGENESIS FACTOR2 is required for splicing of ycf3. J. Integr. Plant Biol. 62, 1741–1761. doi: 10.1111/jipb.12936
Wang, X. W., Zhao, L., Man, Y., Li, X., Wang, L., and Xiao, J. (2020). PDM4, a pentatricopeptide repeat protein, affects chloroplast gene expression and chloroplast development in Arabidopsis thaliana. Front. Plant Sci. 11:1198. doi: 10.3389/fpls.2020.01198
Wang, Z. W., Lv, J., Xie, S. Z., Zhang, Y., and Ren, D. Y. (2018). OsSLA4 encodes a pentatricopeptide repeat protein essential for early chloroplast development and seedling growth in rice. Plant Growth Regul. 84, 249–260. doi: 10.1007/s10725-017-0336-6
Williams-Carrier, R., Brewster, C., Belcher, S. E., Rojas, M., Chotewutmontri, P., Ljungdahl, S., et al. (2019). The Arabidopsis pentatricopeptide repeat protein LPE1 and its maize ortholog are required for translation of the chloroplast psbJ RNA. Plant J. 99, 56–66. doi: 10.1111/tpj.14308
Xia, E. H., Zhang, H. B., Sheng, J., Li, K., Zhang, Q. J., Kim, C., et al. (2017). The tea tree genome provides insights into tea flavor and independent evolution of caffeine biosynthesis. Mol. Plant 10, 866–877. doi: 10.1016/j.molp.2017.04.002
Xu, C., Jiao, C., Sun, H., Cai, X., Wang, X., Ge, C., et al. (2017). Draft genome of spinach and transcriptome diversity of 120 Spinacia accessions. Nat. Commun. 8:15275. doi: 10.1038/ncomms15275
Xu, S., Brockmöller, T., Navarro-Quezada, A., Kuhl, H., Gase, K., Ling, Z., et al. (2017). Wild tobacco genomes reveal the evolution of nicotine biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 114, 6133–6138. doi: 10.1073/pnas.1700073114
Yan, J., Zhang, Q., Guan, Z., Wang, Q., Li, L., Ruan, F., et al. (2017). MORF9 increases the RNA-binding activity of PLS-type pentatricopeptide repeat protein in plastid RNA editing. Nat. Plants 3:17037. doi: 10.1038/nplants.2017.37
Yan, Q., Wu, F., Xu, P., Sun, Z., Li, J., Gao, L., et al. (2020). The elephant grass (Cenchrus purpureus) genome provides insights into anthocyanidin accumulation and fast growth. Mol. Ecol. Resour. 21, 526–542. doi: 10.1111/1755-0998.13271
Yin, P., Li, Q., Yan, C., Liu, Y., Liu, J., Yu, F., et al. (2013). Structural basis for the modular recognition of single-stranded RNA by PPR proteins. Nature 504, 168–171. doi: 10.1038/nature12651
Yuan, N., Wang, J., Zhou, Y., An, D., Xiao, Q., Wang, W., et al. (2019). EMB-7L is required for embryogenesis and plant development in maize involved in RNA splicing of multiple chloroplast genes. Plant Sci. 287, 110203. doi: 10.1016/j.plantsci.2019.110203
Zeng, L., Tu, X. L., Dai, H., Han, F. M., Lu, B. S., Wang, M. S., et al. (2019). Whole genomes and transcriptomes reveal adaptation and domestication of pistachio. Genome Biol. 20:79. doi: 10.1186/s13059-019-1686-3
Zhang, J., Xiao, J., Li, Y., Su, B., Xu, H., Shan, X., et al. (2017). PDM3, a pentatricopeptide repeat-containing protein, affects chloroplast development. J. Exp. Bot. 68, 5615–5627. doi: 10.1093/jxb/erx360
Zhang, L., Chen, F., Zhang, X., Li, Z., Zhao, Y., Lohaus, R., et al. (2020). The water lily genome and the early evolution of flowering plants. Nature 577, 79–84. doi: 10.1038/s41586-019-1852-5
Zhang, L., Zhou, W., Che, L., Rochaix, J. D., Lu, C., Li, W., et al. (2019). PPR protein BFA2 is essential for the accumulation of the atpH/F transcript in chloroplasts. Front. Plant Sci. 10:446. doi: 10.3389/fpls.2019.00446
Zhang, Q., Xu, Y., Huang, J., Zhang, K., Xiao, H., Qin, X., et al. (2020). The rice pentatricopeptide repeat protein PPR756 is involved in pollen development by affecting multiple RNA editing in mitochondria. Front. Plant Sci. 11:749. doi: 10.3389/fpls.2020.00749
Zhao, P., Wang, F., Li, N., Shi, D. Q., and Yang, W. C. (2020). Pentatricopeptide repeat protein MID1 modulates nad2 intron 1 splicing and Arabidopsis development. Sci. Rep. 10:2008. doi: 10.1038/s41598-020-58495-5
Zhao, X., Huang, J., and Chory, J. (2019). GUN1 interacts with MORF2 to regulate plastid RNA editing during retrograde signaling. Proc. Natl. Acad. Sci. U.S.A. 116, 10162–10167. doi: 10.1073/pnas.1820426116
Zoschke, R., Kroeger, T., Belcher, S., Schöttler, M. A., Barkan, A., and Schmitz-Linneweber, C. (2012). The pentatricopeptide repeat-SMR protein ATP4 promotes translation of the chloroplast atpB/E mRNA. Plant J. 72, 547–558. doi: 10.1111/j.1365-313X.2012.05081.x
Zoschke, R., Watkins, K. P., and Barkan, A. (2013). A rapid ribosome profiling method elucidates chloroplast ribosome behavior in vivo. Plant Cell 25, 2265–2275. doi: 10.1105/tpc.113.111567
Keywords: metabolism, gene expression, biogenesis, chloroplast, PPR protein
Citation: Wang X, An Y, Xu P and Xiao J (2021) Functioning of PPR Proteins in Organelle RNA Metabolism and Chloroplast Biogenesis. Front. Plant Sci. 12:627501. doi: 10.3389/fpls.2021.627501
Received: 09 November 2020; Accepted: 04 January 2021;
Published: 09 February 2021.
Edited by:
Peter Julian Nixon, Imperial College London, United KingdomReviewed by:
Shengjun Li, Qingdao Institute of Bioenergy and Bioprocess Technology (CAS), ChinaCopyright © 2021 Wang, An, Xu and Xiao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianwei Xiao, eGlhb2ppYW53ZWlAYmpmdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.