- 1The Key Laboratory of Plant Development and Environmental Adaptation Biology, Ministry of Education, School of Life Sciences, Shandong University, Qingdao, China
- 2State Key Laboratory of Microbial Technology, Institute of Microbial Technology, Helmholtz International Lab for Anti-infectives, Shandong University-Helmholtz Institute of Biotechnology, Shandong University, Qingdao, China
Formation of nodules on legume roots results from symbiosis with rhizobial bacteria. Here, we identified two GATA transcription factors, MtHAN1 and MtHAN2, in Medicago truncatula, which are the homologs of HANABA TARANU (HAN) and HANABA TARANU LIKE in Arabidopsis thaliana. Our analysis revealed that MtHAN1 and MtHAN2 are expressed in roots and shoots including the root tip and nodule apex. We further show that MtHAN1 and MtHAN2 localize to the nucleus where they interact and that single and double loss-of-function mutants of MtHAN1 and MtHAN2 did not show any obvious phenotype in flower development, suggesting their role is different than their closest Arabidopsis homologues. Investigation of their symbiotic phenotypes revealed that the mthan1 mthan2 double mutant develop twice as many nodules as wild type, revealing a novel biological role for GATA transcription factors. We found that HAN1/2 transcript levels respond to nitrate treatment like their Arabidopsis counterparts. Global gene transcriptional analysis by RNA sequencing revealed different expression genes enriched for several pathways important for nodule development including flavonoid biosynthesis and phytohormones. In addition, further studies suggest that MtHAN1 and MtHAN2 are required for the expression of several nodule-specific cysteine-rich genes, which they may activate directly, and many peptidase and peptidase inhibitor genes. This work expands our knowledge of the functions of MtHANs in plants by revealing an unexpected role in legume nodulation.
Introduction
Legumes have the ability to survive in nitrogen-limited soils by forming root nodules that are colonized by nitrogen-fixing symbiotic bacteria. The formation of nodules requires a complex communication between bacteria and the host plants (Ferguson et al., 2010). The symbiosis begins with exudation secondary metabolites called flavonoids from legume roots which triggers Nod factor production by the rhizobia (Liu and Murray, 2016). The bacterial Nod factors are then perceived by the plant to initiate nodule formation, which forms from differentiated root cortex or pericycle (Via et al., 2016; Buhian and Bensmihen, 2018). The nodules in Medicago truncatula consist of several domains, including a persistent apical meristem (zone I), a 10- to 15-cell layer wide infection zone (zone II), a two- to three-cell layer wide interzone (zone II–III), and a growing nitrogen-fixing zone (zone III) (Farkas et al., 2014). Legume nodules initiate from cortical cells which form the nodule primordium. The nodule meristem is formed at the apex of the primordia (Timmers et al., 1999; Stougaard, 2001; Limpens and Bisseling, 2003). Many genes involved in nodulation and symbiotic nitrogen fixation have been isolated in M. truncatula (Roy et al., 2020), however, many regulators remain to be discovered due to functional redundancy.
Previous studies demonstrate the expression of several root meristem regulators in the nodule meristem, revealing a relationship between nodulation and lateral root development (Hirsch et al., 1997; Mathesius et al., 2000; de Billy et al., 2001; Bright et al., 2005; Desbrosses and Stougaard, 2011). MtPLETHORA3 (MtPLT3) and MtPLT4 are expressed not only in the central part of nodule meristem, but also in the root meristem (Schiessl et al., 2019). Knock-down of MtPLT genes results in a decrease of nodule number in M. truncatula (Franssen et al., 2015). Moreover, ASL18/LBD16a, a lateral root developmental element, functions in the downstream of NIN to drive nodule symbiosis in Lotus japonicus (Soyano et al., 2019). These studies indicate that root developmental programs are involved in nodule formation.
Nodule-specific cysteine-rich (NCR) peptides are small legume-specific peptides produced in rhizobium-infected cells, which have multiple functions in nodule development. In M. truncatula, about 600 genes code for NCR peptides (Guefrachi et al., 2014). Most NCR genes are expressed specifically in nodules, and none of them are induced by Nod factors during symbiosis (Guefrachi et al., 2014). The cationic NCR peptides have antibacterial activity, the application of which in vitro increases permeability of bacterial membranes leading to cell death (Tiricz et al., 2013).
Phytohormone signaling pathways are also important for the root nodule symbiosis (Liu et al., 2018). Activation of cytokinin signaling and the level of cytokinin is important for nodule formation and development. A loss-of-function mutation in the cytokinin receptor cre1 exhibited abundant infection-thread formation, but failed to initiate cortical cell division in M. truncatula (Murray et al., 2007). Auxin is also required for both infection thread formation and nodule development (Breakspear et al., 2014) which is regulated by the auxin influx carrier MtLAX2 (de Billy et al., 2001; Roy et al., 2017). Furthermore, ethylene, gibberellic acid (GAs) and abscisic acid (ABA) play negative roles in nodule formation. The ethylene-insensitive mutant mtskl (ortholog of Arabidopsis ein2) shows an increased number of infection threads and nodules (Penmetsa and Cook, 1997; Penmetsa et al., 2008). Exogenous application of GA3 inhibits NF-induced root hair deformation, infection thread formation and nodule development in L. japonicus and M. truncatula (Maekawa et al., 2009; Fonouni-Farde et al., 2016; Jin et al., 2016). Recent work shows that della triple mutant exhibits a strong impairment in infection thread formation and nodule development in M. truncatula (Jin et al., 2016). Treatment with abamine, a specific ABA biosynthesis inhibitor, can increase nodule number in L. japonicus (Suzuki et al., 2004), and exogenous application of ABA inhibits rhizobial infection and nodulation in many legume species (Suzuki et al., 2004; Ding et al., 2008).
GATA factors are transcription regulators that bind to the consensus sequence W-GATA-R [W, thymidine (T) or an adenosine (A); R, guanidine (G) or adenosine (A)] in DNA (Lowry and Atchley, 2000). All GATA transcription factors of Arabidopsis have a type IV zinc finger with the consensus C–X2–C–X17–20–C–X2–C (C, cysteine; X, any residue) followed by a highly basic amino acid stretch (Reyes et al., 2004). There are 29 GATA factors in the Arabidopsis genome, that are divided into four groups depending on sequence conservation, protein domains and gene structure (Reyes et al., 2004). Functional studies have identified the effects of GATA factors in a range of processes including cell elongation (Nishii et al., 2000; Shikata et al., 2004), floral meristem and shoot apical meristem development (Zhao et al., 2004), and seed germination (Liu et al., 2005). One that has been well studied is HANABA TARANU (HAN). HAN participates in many aspects of plant development. In the plant embryo, HAN is needed to position the inductive proembryo boundary. Mutation of HAN results in the apical redistribution of auxin as early as the eighth cell stage. Developmental defects of han are obvious by the 16th cell stage, when the tangential division often fails, and misaligned oblique division is usually observed in the upper tier (Nawy et al., 2010). In the reproductive stage, HAN plays roles in establishing boundaries and controlling WUS-expressing cells in shoots and flowers. The han mutants in Arabidopsis have flower developmental defects in all four whorls, including fused sepals, reduced numbers of petals and stamens and unfused carpels. The flowers of weak han mutant have two to four sepals, one to two petals, four or five stamens, and two asymmetric carpels (Zhao et al., 2004). The genetic combination han and clavata (clv) mutations results in highly fasciated SAMs (Zhao et al., 2004). As a boundary-expressing gene, HAN regulates flower development through communicating with the PINHEAD (PNH), JAGGED (JAG), BLADE-ON-PETIOLE 2 (BOP2), and CYTOKININ OXIDASE 3 (CKX3) (Ding et al., 2015).
In this study, we identified two members of GATA family in M. truncatula, MtHAN1 and MtHAN2, which are homologs of HAN and HANABA TARANU LIKE (HANL). We found that the expression of MtHAN1 and MtHAN2 are expressed in nodules and were nitrate regulated. Further genetic study showed that the mthan1 mthan2 double mutant developed more nodules than wild type. RNA-seq data showed that the DEGs in the mthan mutant included NCRs, peptidases and peptidase inhibitors and gene involved in multiple pathways including flavonoid biosynthesis and hormone signal transduction.
Materials and Methods
Plant Materials and Growth Conditions
Medicago truncatula ecotype R108 was used in this study. The mthan1-1 (NF13128), mthan1-2 (NF4035), mthan2-1 (NF4633), and mthan2-2 (NF5115) mutant lines were identified from a M. truncatula Tnt1 retrotransposon-tagged mutant collection (Tadege et al., 2008). The plants were cultivated in a growth chamber at 22°C day/20°C night, with a photoperiod of 16-h-day/8-h-night, and relative humidity was 70–80%. Nicotiana benthamiana used for BiFC experiments was grown under long-day conditions at 22°C day/20°C night.
Phylogenetic Analysis
Alignment of multiple protein sequences was performed using online CLUSTALW1. The neighbor-joining phylogenetic tree was constructed using the MEGA 4.1 software suite2. The most parsimonious trees with bootstrap values from 1,000 trials were shown.
Plasmids Construction and Plant Transformation
For constructing overexpression vectors, the full CDS of MtHAN1 and MtHAN2 were PCR amplified from wild type M. truncatula and cloned into the pENTR/D-TOPO cloning vector (Invitrogen), then transferred into the pEarleyGate103 vector by attL × attR recombination reactions (Invitrogen) (Earley et al., 2006). For analysis of gene expression patterns, the pBGWFS7 vector containing 2595bp and 2191bp promotors fragments of MtHAN1 and MtHAN2, respectively, were constructed (Karimi et al., 2002). For stable transformation, binary vector constructs were transformed into ecotype R108 using the disarmed Agrobacterium tumefaciens strain EHA105.
Subcellular Localization of MtHANs
For subcellular localization of MtHANs, the A. tumefaciens EHA105 strain harboring the plasmids pEarleyGate 103-MtHAN1 and pEarleyGate 103-MtHAN2 were infiltrated into epidermal cells of tobacco. 48 h later, fluorescence signals were examined using an LSM 880 (Zeiss) confocal laser scanning microscope.
Yeast Two-Hybrid Assays and BiFC Assay
To test physical interaction between MtHAN1 and MtHAN2, LR interactions were made for pENTR-MtHAN1 with pDEST22 and pENTR-MtHAN2 with pDEST32 (Invitrogen) to generate bait and prey plasmids construct. The yeast strain MAV203 was used for transformation of bait and prey plasmids. Yeast transformants were selected on synthetic minimal double dropout medium deficient in Trp and Leu (Clontech). For protein interaction tests, medium supplemented with SD-Leu-Trp-His (Clontech) and 1 mM 3-amino-1, 2, 4 triazole (Sigma) was used.
BiFC assays were conducted as described (Ou et al., 2011), and some modifications were made. LR reactions were used to transfer MtHAN1 and MtHAN2 into pEARLEY201-YN, pEARLEY202-YC, respectively, and then the plasmids were transformed into Agrobacterium EHA105. To test interactions, pEARLEY201-YN-MtHAN1 and pEARLEY202-YC-MtHAN2 were coinfiltrated into 4-weeks-old N. benthamiana leaves. After injection, N. benthamiana was incubated in darkness for 24 h and then 36 h in light. After that, the leaves were dissected for observation using a LSM 880 confocal laser scanning microscope (Zeiss) for capture of fluorescent images. The 488-nm line of an argon laser was chosen for excitation yellow fluorescent protein (YFP).
β-Glucuronidase Staining, X-Gal Staining and Microscopy
For β-Glucuronidase (GUS) staining analysis, nodules in different developmental stages were used. GUS activity was histochemically detected as previously described (Wang et al., 2019). The roots and nodules stained with GUS solution were fixed in 3% glutaraldehyde in a phosphate buffer and then dehydrated and embedded in wax. Then the samples were sectioned using a RM 2255 microtome (Leica). For X-gal staining, LacZ activity of nodules was assayed as previously described (Wang et al., 2018), and the sample were embedded in 5% (w/v) agarose, and 50-μm sections were made using a VT1200S vibratome (Leica).
Phenotypic Analysis
For root morphology analysis, mutant and wild type plants were transferred to MS plates after germination. The length of primary roots was measured on the seventh day.
Nitrate Treatment
Two-weeks-old seedlings growing in plastic tray were supplied with a solution containing 20 mM KNO3 for induction or 20 mM KCl as a control. Leaves and petioles were collected at 0, 2, 4, and 6 h after treatment and total RNA was extracted.
Nodule Induction
For rhizobial inoculation, seedlings of wild type, mthan1, mthan2, mthan1 mthan2, 35S:MtHAN1, and 35S:MtHAN2 were transferred into a plastic tray filled with perlite/sand (in a 3:1 ratio). Plants growing in the chamber were watered with nutrient solution without nitrate every 3 days. The Rhizobium Sinorhizobium meliloti 1021 strain was used for inoculation. Rhizobium was cultured with TY (tryptone, yeast extract, and sodium chloride) medium (supplemented with 10 mg mL–1 tetracycline, 200 mg mL–1 streptomycin, and 6 mmol L–1 calcium chloride) in 28°C shaker till it reaches OD600 value of 1.0 (Wang et al., 2018). After that, 5 mL of rhizobial suspension diluted to OD600 = 0.1 was inoculated to the roots of 5 days seedlings. Nodule numbers were identified 21 days post inoculation (dpi). For time course assays, nodules of wild type were harvested at 7, 14, and 21 dpi.
Nitrogenase Activity
Nitrogenase activity was measured by acetylene reduction assay (ARA) (Hardy et al., 1968). The roots with nodules of wild type, mthan1-1 mthan2-1, 35S:MtHAN1, and 35S:MtHAN2 were introduced into 120 mL bottles sealed with rubber stoppers, and each bottle contained three plants. 12 mL of air was replaced with 12 mL of acetylene and the bottles were incubated at room temperature for 2 h. 1 mL gas in each bottle was used to measure the production of ethylene in a Shimadzu GC 2014C gas chromatograph using a porapak N column (Shimadzu, Kyoto, Japan).
RNA Extraction, RT-PCR and Real-Time PCR Analysis
Total RNA of different tissues was extracted from 6-weeks-old plants, and RNA of nodules was extracted 3 weeks post inoculation with rhizobia. Total RNA was isolated using Trizol-RT Reagent (Molecular Research Center, INC). RT-PCR analysis was performed as described previously (Zhou et al., 2011). Real-time PCR was performed using the Fast Start Essential DNA Green Master mix (Roche) and the Bio-Rad CFX Connect TM sequence detection system was used for data acquisition and analysis. MtUBIQUITIN (Medtr3g110110) was used for normalization. The primers used for Real-Time PCR are listed in Supplementary Table 1 online.
Protoplast Transient Assay
To construct the effector plasmids, the CDS of MtHAN1 was cloned into pBI221 by LR interaction. For the reporter plasmids, the promoter fragments of NCR genes Medtr1g042940, Medtr5g072205 and Medtr5g072275 were cloned into the pGreenII-0800 vector, which contains the LUC reporter gene at C-terminus, using restriction sites Pst I and Bam H I. The reporter (2 μg) and effector plasmids (8 μg) were co-transfected into protoplasts isolated from rosette leaves of 3-weeks-old Col-0 plants by the PEG-mediated transfection method (Yoo et al., 2007). The LUC activity in each cell lysate was measured by the Luciferase Assay System Kit (Promega).
Results
Identification of the HAN Homologs in M. truncatula
To identify the homologs of HAN in M. truncatula, a BLASTP search using the Arabidopsis HAN against M. truncatula genome database was performed and two candidate genes, Medtr5g020230 and Medtr8g077510, were obtained. Phylogenetic analysis and protein alignment showed that these two genes were close homologs of HAN, HANL, and HANL2, thus, were named MtHAN1 and MtHAN2 (Figure 1A and Supplementary Figure 1). Subcellular localization of MtHAN1 and MtHAN2 (MtHANs) revealed that they localized to the nucleus and cytoplasm. The At-Hook Motif Containing Nuclear Localized (AHL) protein was used as a control (Figure 1B). A similar localization of MtHANs was also observed in protoplasts of M. truncatula (Figure 1C). A yeast two-hybrid (Y2H) assay was performed to test whether MtHAN1 and MtHAN2 can physically interact (Figure 1D). The results indicate that MtHAN1 and MtHAN2 do interact, and so presumably form a heterodimer as has been reported for HAN and its HANL homologs in Arabidopsis (Zhang et al., 2013). Moreover, in bimolecular fluorescence complementation (BiFC) assays MtHAN1 and MtHAN2 were found to only interact in the nucleus (Figure 1E), consistent with their predicted roles as transcription factors.
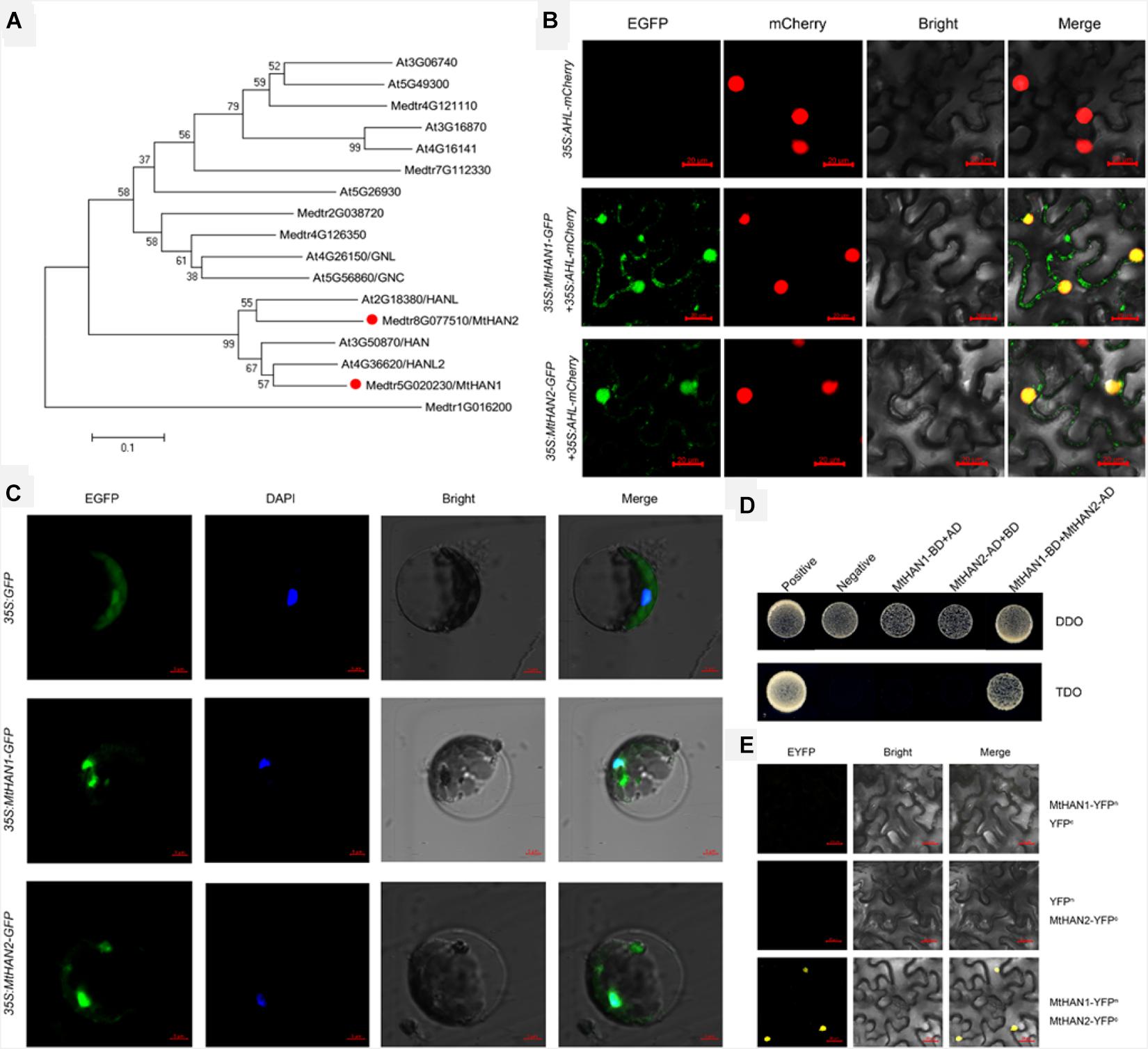
Figure 1. Medicago truncatula HANABA TARANU (HAN) homologues interact in nucleus. (A) Phylogenetic analysis of HAN genes among different species. (B) The subcellular localization of MtHAN1/2-GFP in Nicotiana benthamiana leaves. At-Hook Motif Containing Nuclear Localized (AHL)-mCherry was used as a marker for the nucleus. Bars = 20 μm. (C) The subcellular localization of MtHAN1/2-GFP in M. truncatula protoplasts. Free GFP was used as a control. Bars = 5 μm. (D) Physical interaction between MtHAN1 and MtHAN2 by yeast two-hybrid assay. Interaction was examined by yeast grown on TDO (SD-Leu-Trp-His) medium. (E) BiFC showing the interaction between MtHAN1 and MtHAN2 in nucleus. Bars = 20 μm.
Identification the Loss-of-Function Mutants of MtHAN1 and MtHAN2 in M. truncatula
To explore the functions of MtHAN1 and MtHAN2, a reverse genetic screening was performed in a Tnt1 retrotransposon-tagged mutant collection of M. truncatula (Cheng et al., 2014). Two independent Tnt1 insertion mutants, all with insertions in exon 2, for both MtHAN1 (mthan1-1 and mthan1-2) and MtHAN2 (mthan2-1 and mthan2-2), were isolated (Figure 2A). Amplification of full-length cDNA showed that the transcripts of MtHAN1 and MtHAN2 were abolished in their respective mutants (Figure 2B). These data indicate that all four alleles are knockout mutants. Phenotypic observation showed that there are no obvious developmental defects in the leaves or flowers of single mutants (Supplementary Figure 2). To assess functional redundancy between MtHAN1 and MtHAN2, double mutants were generated. However, the plants from two mutant combinations (mthan1-1 mthan2-1 and mthan1-2 mthan2-2) grew normally, and did not show any visible defects in leaf and flower development at the vegetative and reproductive stages (Supplementary Figure 2). These results imply that MtHANs may play different roles in M. truncatula, compared with those of HAN in Arabidopsis.
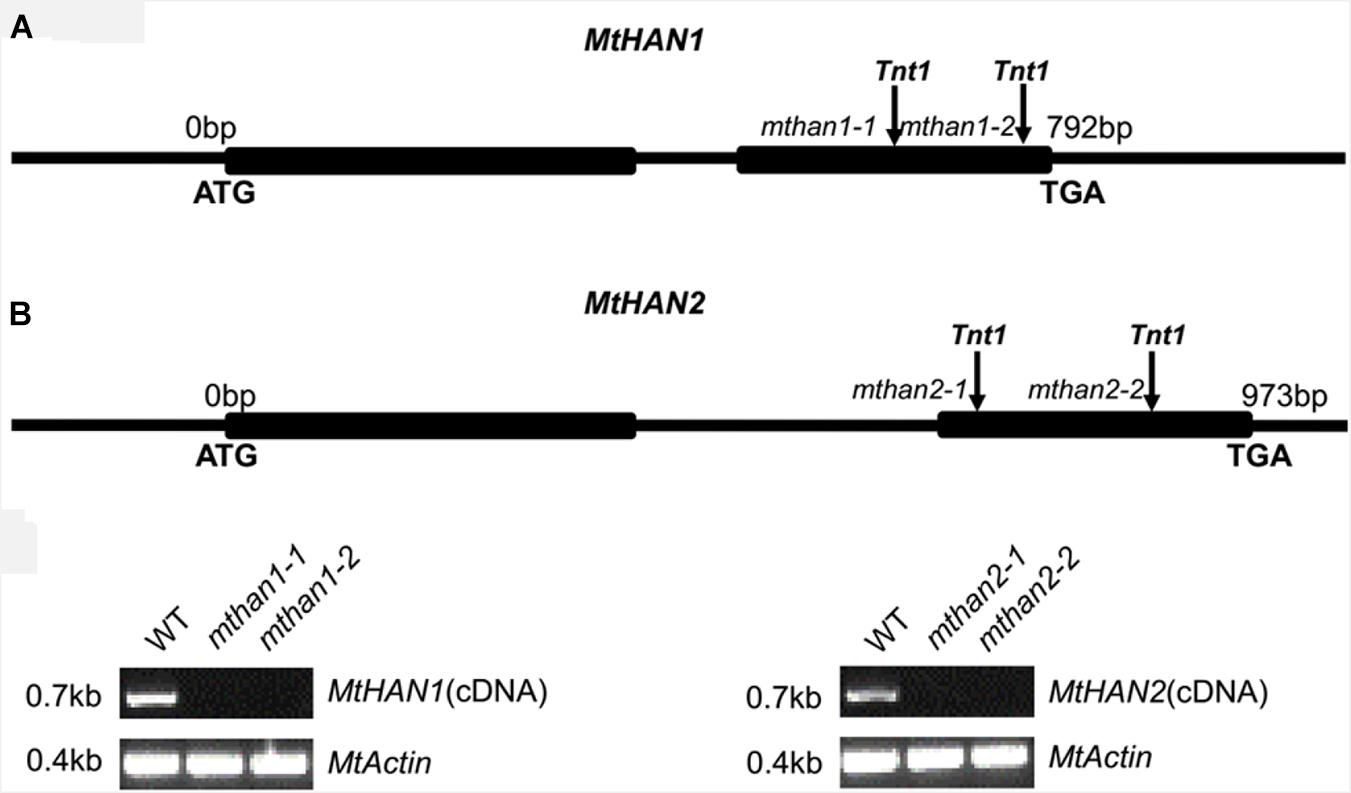
Figure 2. Molecular characterization of MtHANs in M. truncatula. (A) Schematic diagram of the gene structure of MtHAN1 and MtHAN2. Vertical arrows mark the positions of the Tnt1 insertions. Boxes represent exons and lines represent introns and UTRs. (B) RT-PCR analysis of MtHAN1 and MtHAN2 transcripts in the wild type (WT) and mutant alleles. MtActin was used as a loading control. 28 cycles were used for the RT-PCR.
MtHAN1 and MtHAN2 Play a Redundant Role in Root Development
To investigate the potential function of MtHAN1 and MtHAN2 in M. truncatula development, their expression patterns were analyzed. Tissue-expression analysis by qRT-PCR showed that both MtHAN1 and MtHAN2 were relatively highly expressed in the shoot bud, stem and petiole, with MtHAN2 also showing high expression in the root (Figure 3A). To more comprehensively determine the expression patterns of MtHAN1 and MtHAN2, promoter-GUS constructs were stably introduced into wild type, and GUS activity was examined in transgenic plants. The results showed that MtHAN1 and MtHAN2 displayed similar expression patterns in independent transgenic lines. GUS signals were detected in root tip of germinating seeds, in root vascular tissues, lateral root primordia, and primary root tips. No GUS staining was observed in the control (Figures 3B–E and Supplementary Figure 3). To assess if MtHAN1 and MtHAN2 function in root development, the root length and lateral root number were measured in wild type and mutants. The results showed that the lateral root number in the double mutants was similar to wild type, but the root length was increased in double mutants (Figure 4). These observations suggest that MtHAN1 and MtHAN2 are redundantly involved in the root elongation.
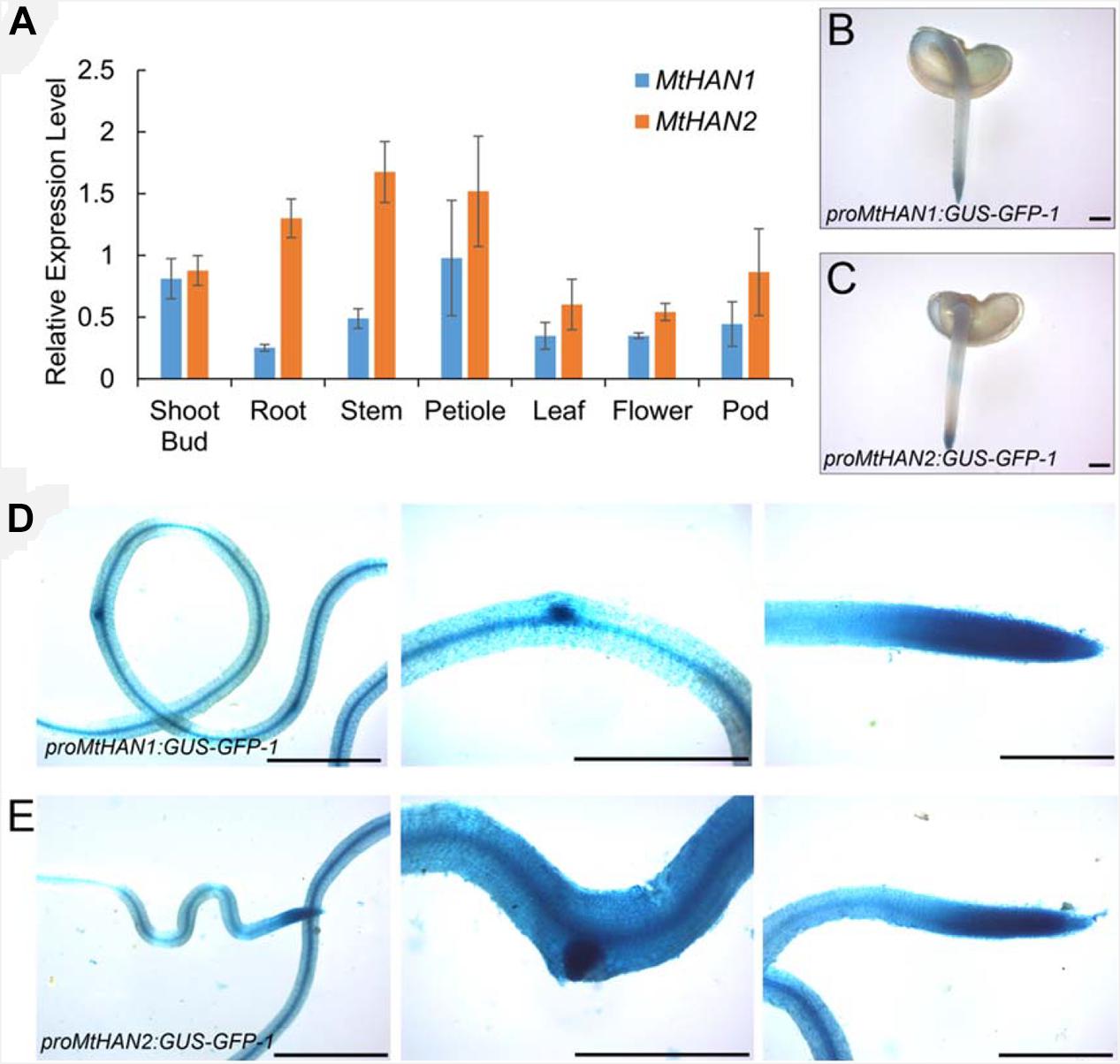
Figure 3. Expression patterns of MtHAN1 and MtHAN2. (A) The expression patterns of MtHAN1 and MtHAN2 in tissues of shoot bud, root, stem, petiole, leaf, flower, and pod. (B–E) GUS expression in roots that were transformed with either proMtHAN1:GUS–GFP in panel (B,D) or proMtHAN2:GUS–GFP in panel (C,E). (B,C) GUS staining in 5-days-old germinating seeds. (D,E) GUS staining in root vascular, lateral root primordial, and main root tip of 7-days-old root. Bars = 1 mm.
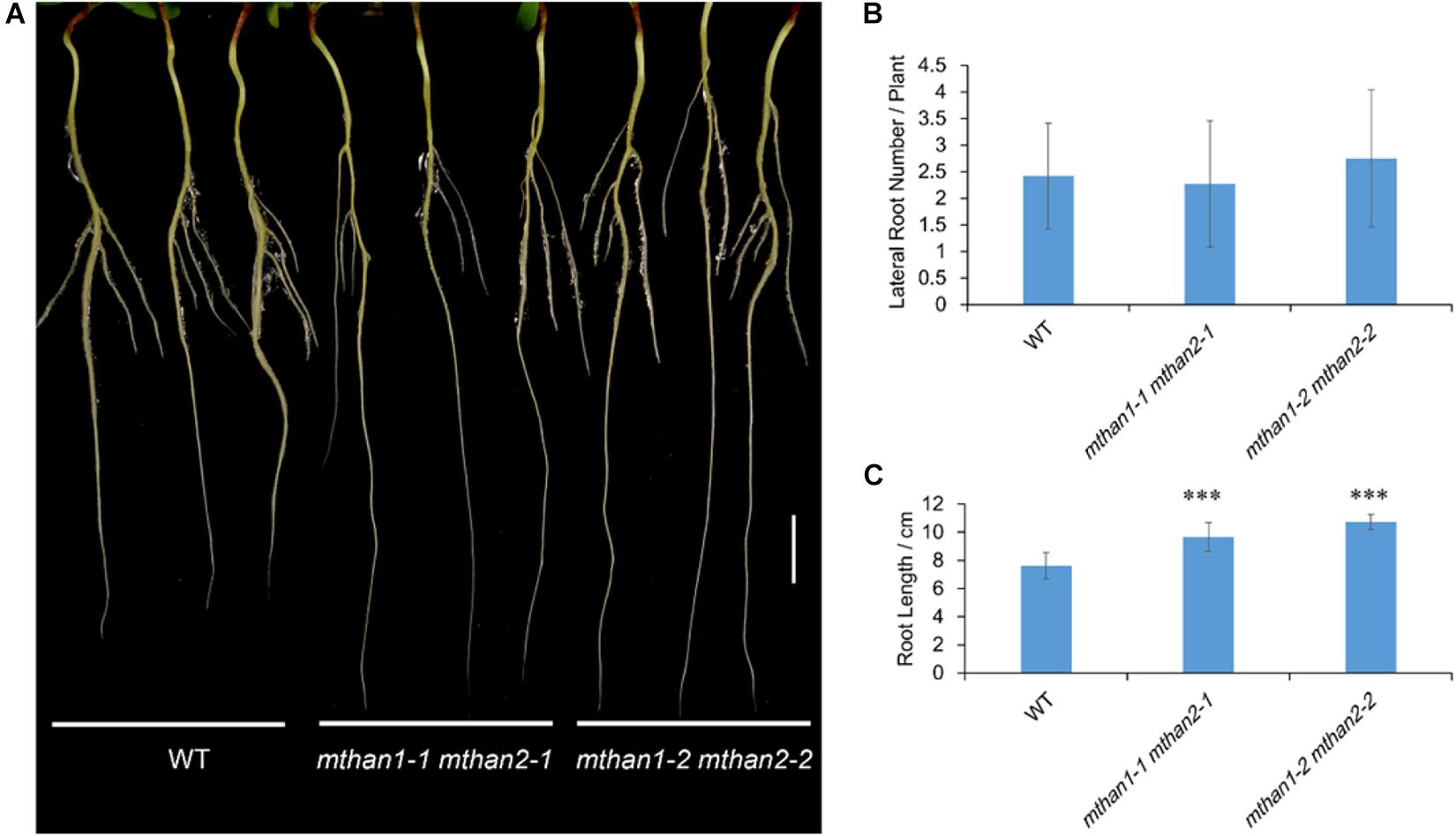
Figure 4. MtHAN1 and MtHAN2 are redundantly involved in the root elongation. (A) Seedling images of WT and mthan1 mthan2 double mutant plants on the seventh day after transferring to MS plates. Three representative seedlings of each plant line were photographed. Bar = 1 cm. (B) Measurement of lateral root number. (C) Measurement of primary root length. Values are the means ± SD (n = 12). ∗∗∗P < 0.001.
MtHAN1 and MtHAN2 Are Involved in Nodule Formation
To investigate whether MtHANs play roles in nodule development, the expression pattern of MtHANs in different organs during nodulation was analyzed. qRT-PCR data showed that both MtHAN1 and MtHAN2 were mainly expressed in the shoot bud, stem and root, but were expressed at lower levels in the nodule (Supplementary Figure 4A). These results are consistent with the data of MtHAN2 (the probeset of MtHAN1 is not available) derived from Medicago truncatula Gene Expression Atlas (MtGEA) (Supplementary Figure 4B). In developing nodules, the expression level of MtHAN1 was slightly increased, while the expression of MtHAN2 was decreased (Figure 5A). Moreover, we also detected the expression of MtHANs in young nodules and mature nodules at 21 dpi. The results showed that the transcriptional level of MtHAN1 was higher than that of MtHAN2 in both kinds of nodules (Supplementary Figure 4C). A previous report showed that expression of the GATA factor GATA, nitrate-inducible, carbon metabolism-involved (GNC) is nitrate inducible in Arabidopsis (Bi et al., 2005). Nitrate concentration is critical for nodule formation. High levels of nitrate are able to inhibit multiple aspects of nodule development, including flavonoid production, rhizobial infection, nodule initiation, nodule growth, and nitrogen fixation activity. In addition, high nitrate can accelerate nodule senescence or disintegration (Nishida and Suzaki, 2018). To test whether MtHANs are able to respond to nitrogen, their expression levels were measured after treatment with KNO3, using KCl as a control treatment. The results showed that the expressions levels of MtHAN1 and MtHAN2 were reduced after nitrate application for 2 h, and then, increased after 4 and 6 h treatment (Figure 5B).
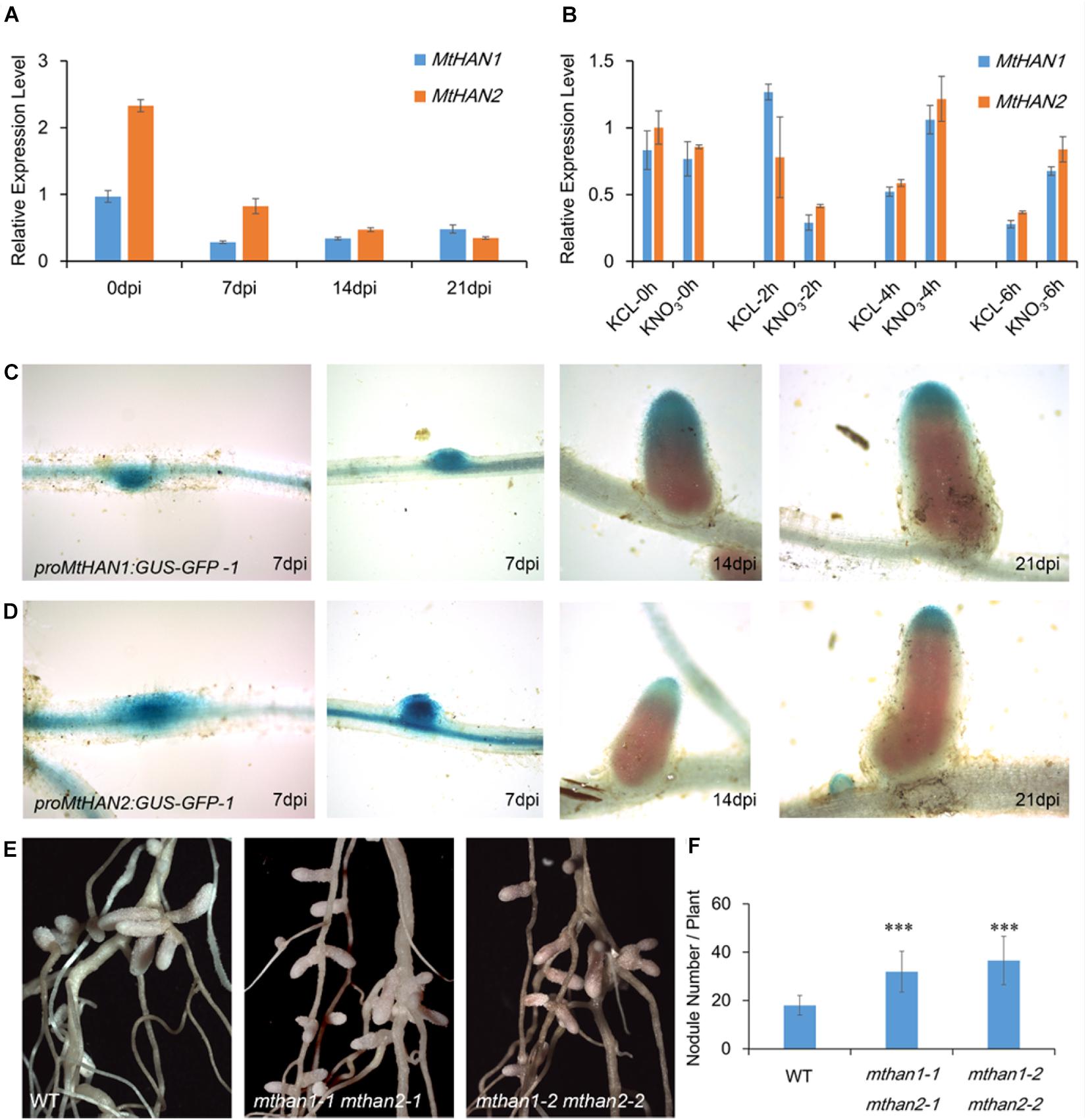
Figure 5. MtHAN1 and MtHAN2 express in nodules and the mthan1 mthan2 double mutant has an increased nodule number. (A) Expression pattern of MtHANs in different developmental stages of nodule. The root (0 dpi) was used as control. Values are the means ± SD of three biological replicates. (B) Expression pattern of MtHANs in leaves and petioles after nitrate treatment, KCl treatment as control. Values are the means ± SD of three biological replicates. (C,D) ProMtHAN1: GUS–GFP and proMtHAN2:GUS–GFP reporter activity associated with the progressive stages in nodule development. (E) Photos of nodules from WT and mthan1 mthan2 at 21 dpi. (F) The average number of nodules in WT and mthan1 mthan2 plants. Values are the means ± SD (n = 16). ∗∗∗P < 0.001.
To further investigate their roles in nodulation, the expression patterns of MtHANs was analyzed in nodules. The proMtHAN1:GUS and proMtHAN2:GUS transgenic plants were inoculated with S. meliloti strain 1021. At 7 dpi, GUS signals were observed in the developing nodule primordia and young nodule. At 14 and 21 dpi, MtHAN1 and MtHAN2 expression was associated predominantly with the meristem and infection zone of nodules. No GUS staining was observed in the control (Figures 5C,D and Supplementary Figure 3). We also confirmed this result by the cross-section of roots and nodules after staining (Supplementary Figure 5). These data indicate that MtHAN1 and MtHAN2 are correlated with nodule development. It is noted that MtHAN2 was expressed in the meristem as early as 14 dpi, therefore, the decreased expression observed for MtHAN2 at later stages of nodule development was possible due to the growing fixation zone.
To further assess the roles of MtHANs during nodulation, wild type and mutants plants were inoculated with S. meliloti 1021 (Supplementary Figure 6). We then measured the nodule number in mthan1-1 and mthan2-1. The result showed that the number of nodules in mthan1-1 was similar with that of wild type, while the number of nodules in mthan2-1 was slightly higher (Supplementary Figure 7A). Considering potential functional redundancy of the MtHANs, nodulation of the double mutants was evaluated. The nodule development was normal in mthan1 mthan2 double mutants (Figure 5E). However, the number of nodules in the mthan1 mthan2 double mutant was increased relative to wild type (Figure 5F). To assess whether there were any developmental defects in the mthan1 mthan2 nodules, we made a longitudinal section of histochemically stained mature nodules at 21 dpi with S. meliloti 1021 carrying hemA/lacZ fusion. The result showed that the nodules of mthan1 mthan2 had no obvious developmental defects (Supplementary Figure 8A,B). The measurement of nitrogenase activity also showed that nitrogenase activity of mthan1-1 mthan2-1 was not altered compared with that of wild type (Supplementary Figure 8C). In addition, we measured the length of shoot and roots, and the biomass of nodulated wild type and double mutant plants. The results showed that the shoot length of double mutant was reduced compared with that of wild type (Supplementary Figure 9A). However, there was no difference in root length and biomass between wild type and mutants (Supplementary Figures 9B,C). Then, MtHAN1 and MtHAN2 were overexpressed in wild type plants. qRT-PCR data showed that the expression levels of MtHANs were significantly increased in the transgenic plants, and overexpression of MtHANs didn’t influence leaf, flower and root development (Supplementary Figure 10). Furthermore, we analyzed the number of nodules in wild type, 35S:MtHAN1 and 35S:MtHAN2 plants at 21 dpi. The results showed no difference in the number of nodules between wild type and 35S:MtHANs plants (Supplementary Figure 11). Further analysis showed that overexpression of MtHAN1 or MtHAN2 didn’t affect nodule nitrogenase activity (Supplementary Figure 8C). These results indicate that MtHANs function in the repression of nodule formation.
Transcriptomic Profiles of Nodules in mthan1 mthan2
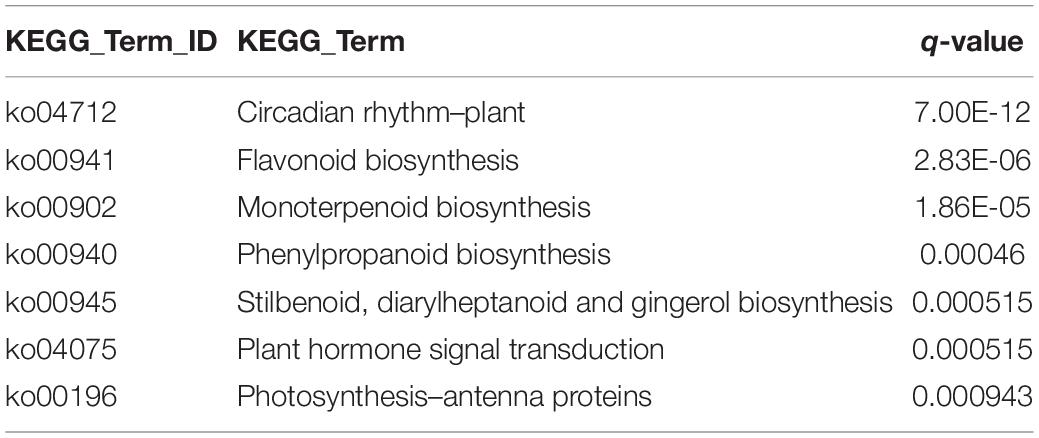
To understand the involvement of MtHANs in nodulation, we performed RNA-seq transcriptomic analysis using nodules from wild type and mthan1-1 mthan2-1 3 weeks after inoculation with S. meliloti 1021. Genes with more than twofold expression changes and the P-values less than 0.001 were identified as different expression genes (DEGs). In total, 1,485 DEGs were found significantly changed between wild type and mthan1 mthan2. Compared to wild type, 662 and 823 were up- and down-regulated, respectively, in mthan1-1 mthan2-1 double mutant (Figures 6A,B and Supplementary Table 2). Multiple DEGs are involved in processes that are important for nodulation. Nod factor perception leads to root hair swelling and branching, which requires cell wall relaxation followed by a redirection of cell wall material secretion (Xie et al., 2012). Gene ontology (GO) terms enrichment analysis showed that, among which, the most enriched GO terms were extracellular region (GO:0005576), carbohydrate metabolic process (GO:0005975), DNA binding transcription factor activity (GO:0003700), hydrolase activity, hydrolyzing O-glycosyl compounds (GO:0004553), cell wall (GO:0005618), external encapsulating structure (GO:0030312) and O-methyltransferase activity (GO:0008171) (Figure 6C). Notably, that among DEGs having nodulation (GO:0009877) as a GO term. Nodule morphogenesis (GO:0009878) and development involved in symbiotic interaction (GO:0044111) was also enriched (Figure 6D). Flavonoid biosynthesis and phytohormones play important roles in legume-rhizobia recognition or nodule initiation and development (Murray et al., 2007; Suzaki et al., 2012; Roy et al., 2017; Gifford et al., 2018). Based on KEGG analysis, several secondary metabolite biosynthesis pathways were identified, including flavonoid biosynthesis (ko00941), monoterpenoid biosynthesis (ko00902), phenylpropanoid biosynthesis (ko00940), stilbenoid, diarylheptanoid, and gingerol biosynthesis (ko000945) and plant hormone signal transduction (ko04075) (Table 1 and Supplementary Figures 12–16). Examination of several of the major nodulation genes in the double mutant showed that their expression was essentially unchanged (Supplementary Table 3). These results implied that MtHANs affect nodulation through multiple pathways.
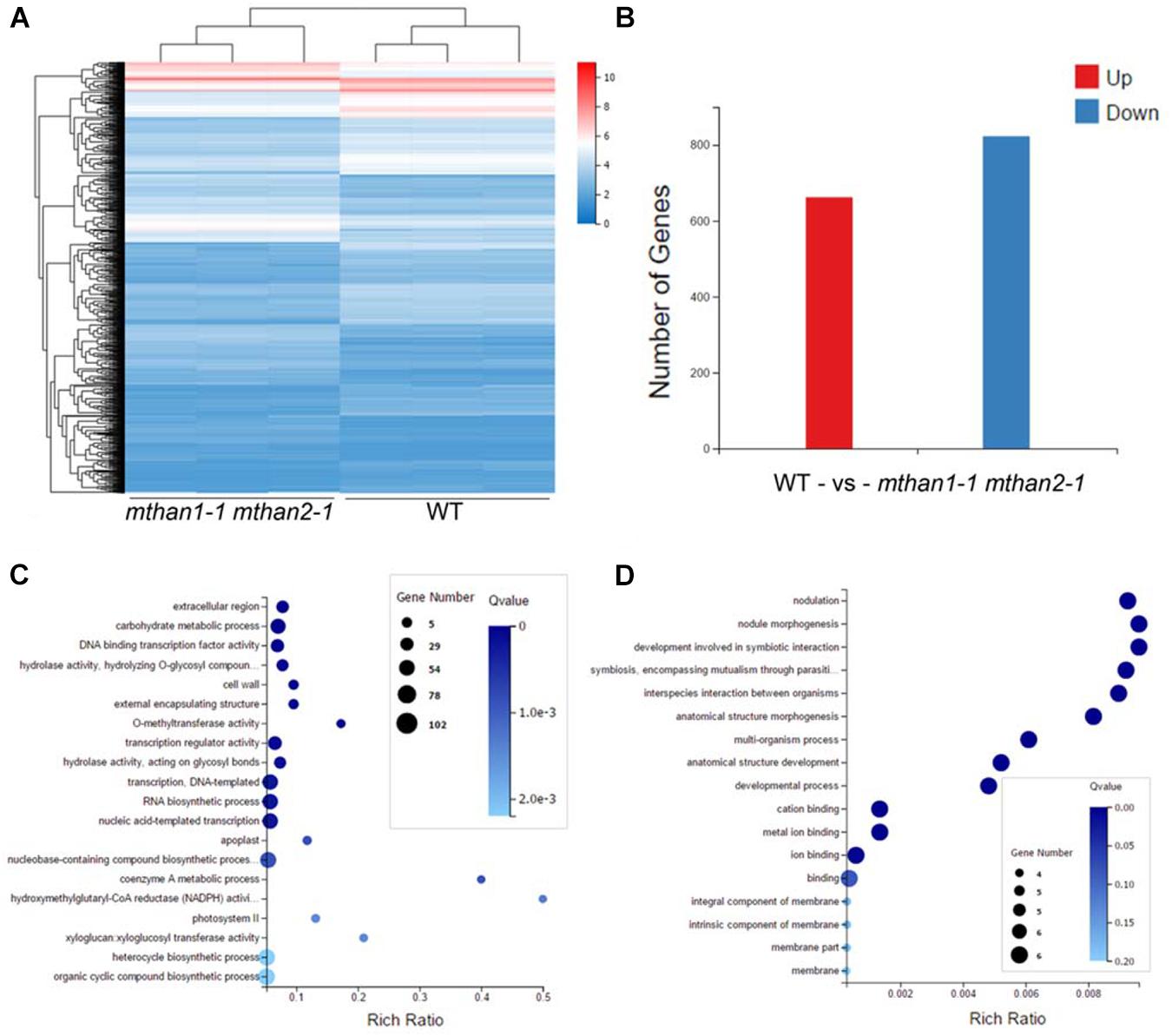
Figure 6. Different expression genes (DEGs) in mthan1-1 mthan2-1 nodules and gene ontology (GO) enrichment of DEGs. (A) The heatmap of fragments per kilobase of transcript per million reads mapped (FPKM) values of 1,485 differential expressed genes in three biological replicates of nodules from WT and mthan1-1 mthan2-1 plants. (B) The number of up-regulated and down-regulated genes in the mthan1-1 mthan2-1 nodules at 21 dpi. (C) The top 20 significantly enriched GO term of DEGs. (D) GO enrichment associated to nodulation and symbiosis.
The NCR Genes Were Under the Regulation of MtHANs
We noticed that amongst the mthan1 mthan2 DEGs were three NCR genes, Medtr1g042940, Medtr5g072205, and Medtr5g072275, that had significantly decreased expression in the mutant. This was confirmed using qRT-PCR (Figure 7A). Previous studies showed that chemically synthesized cationic NCR peptides have antibacterial activity in vitro, and can suppress bacterial cell division in nodules (Tiricz et al., 2013). In the expression pattern database3 (Roux et al., 2014), two of three NCRs could be identified. It showed that the gene corresponding to gene model Medtr1g042940 was expressed in FIId, FIIp, IZ, and ZIII and while another corresponding to Medtr5g072205 was expressed in FIId, which overlapped or more distal to the expression domains of MtHANs (Supplementary Figure 17). To investigate whether MtHANs are able to regulate the transcription of the three NCR genes, we first detected the expression level of the three NCR genes in 21 dpi nodules of mthan1-1 and mthan2-1. The result showed that there was a decrease in the expression of the gene corresponding to Medtr5g072205 and Medtr5g072275 in mthan1-1, while there was a decrease in the expression of the genes corresponding to Medtr1g042940 and Medtr5g072205 in mthan2-1 (Supplementary Figure 7B). These data suggest that MtHAN1 and MtHAN2 play the redundant roles in the regulation of expression of the NCR corresponding to Medtr5g072205, while MtHAN1 and MtHAN2, respectively, regulate the expression of the genes corresponding to Medtr5g072275 and Medtr1g042940. Then, the expression levels of the three NCR genes were measured in nodules of 35S:MtHAN1 and 35S:MtHAN2 transgenic plants. The result showed that transcripts related to Medtr1g042940 and Medtr5g072205 were up-regulated in nodules of MtHAN1-overexpressing plants, implying that MtHAN1 may activate the expression of these two NCR genes (Figure 7B). To test whether MtHAN1 is able to activate the expression of NCR genes, the promoter sequences of three NCR genes were analyzed. Multiple binding motif of GATA factors were found in these promoters (Figure 7C). Then, transient expression assays were performed. The luciferase reporter constructs driven by 2-kb promoters of the three NCR genes were co-transformed, respectively, with MtHAN1 effector proteins into Arabidopsis protoplasts (Figure 7D). The results showed that luminescence intensity driven by the promoter of Medtr1g042940 and Medtr5g072205 was increased significantly, compared with the GFP control effector protein (Figures 7E–G). These results imply that MtHAN1 is able to recognize the promoters of these two genes in protoplasts and activates their expression. Taken together, these finding indicates that the transcriptional regulation of MtHANs on NCR genes is very complicated. Although MtHAN1 is able to activate the expression of Medtr1g042940 and Medtr5g072205, the downregulation of these genes in the double mutant was partially dependent on the functional redundancy of MtHAN1 and MtHAN2.
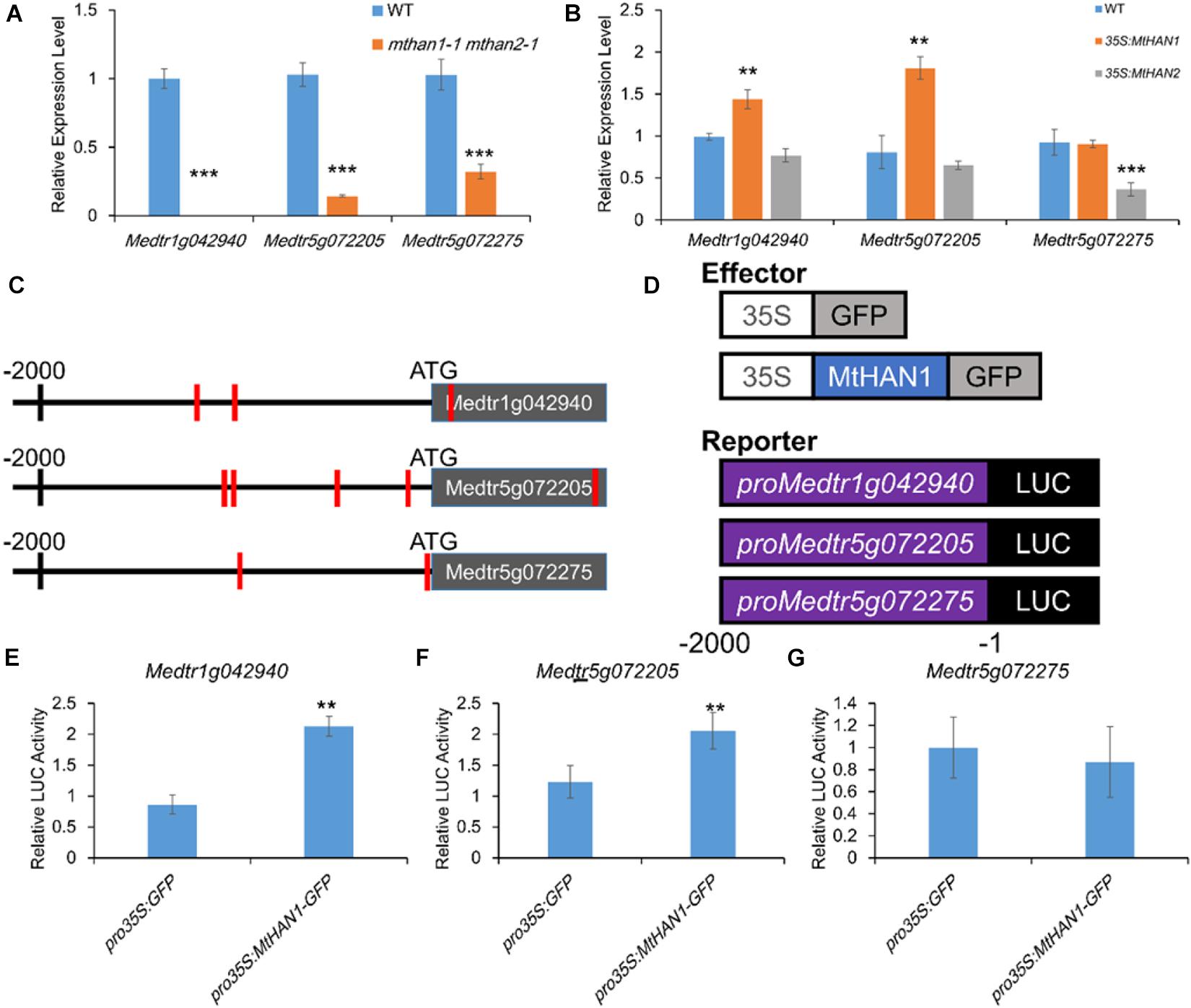
Figure 7. MtHAN1 and MtHAN2 regulate the expression of three NCR genes (gene models Medtr1g042940, Medtr5g072205, and Medtr5g072275) in nodules. (A) Relative expression of NCRs in nodules of mthan1-1 mthan2-1 at 21 dpi. Values are the means ± SD of three biological replicates. ***P < 0.001. (B) Relative expression of NCRs in nodules of 35S:MtHAN1 and 35S:MtHAN2 at 21 dpi. Values are the means ± SD of three biological replicates. **P < 0.01. (C) Schematic diagram of the genes corresponding to Medtr1g042940, Medtr5g072205, and Medtr5g072275. Red vertical line indicates the MtHANs-binding motif (W-GATA-R). (D) Schematic structures of the effector and reporter constructs for the transient expression assay, in which GFP and MtHAN1 were under the control of the Cauliflower mosaic virus (CaMV) 35S promoter, the firefly luciferase (LUC) reporter gene was driven by NCR promoters (Medtr1g042940, Medtr5g072205, and Medtr5g072275), and the Renilla luciferase (LUC; from Renilla reniformis) gene driven by the 35S promoter was used as an internal reference. (E–G) MtHAN1 promotes the transcription of two NCRs (gene models Medtr1g042940 and Medtr5g072205) in a protoplast transient assay. The GFP protein was used as negative control. Mean and SD values were obtained from three biological replicates. **P < 0.01, ***P < 0.001.
MtHAN1 and MtHAN2 Regulate the Expression of Peptidase and Peptidase Inhibitor Genes in Nodules
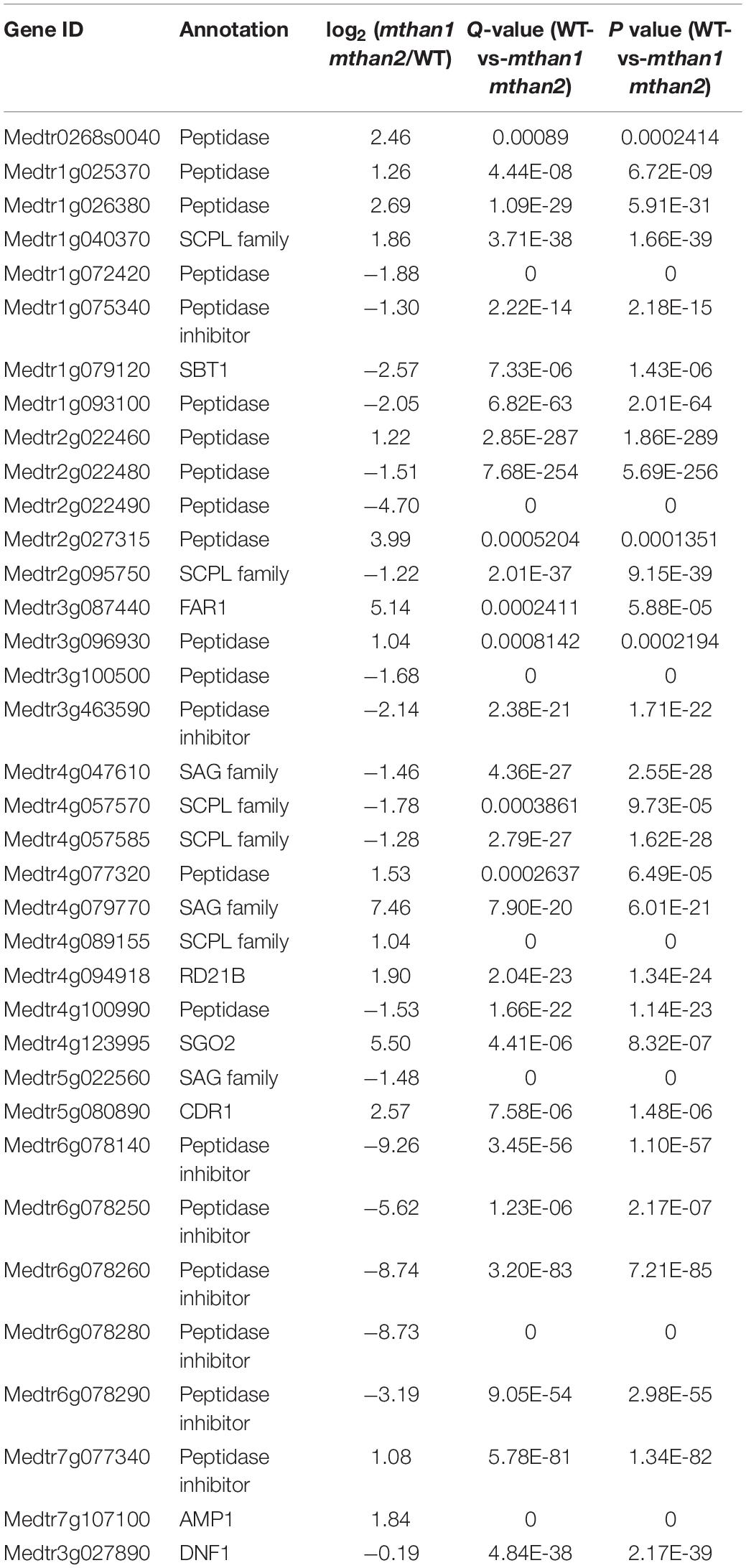
In plant, peptidases participate in many developmental processes, and biotic and abiotic stresses by regulating protein function. To ensure proper levels of proteolysis, the peptidase activity is precisely controlled by peptidase inhibitors at the protein level (Santamaria et al., 2014). All NCR peptides have a conserved signal peptide, which is cleaved by the signal peptidase DNF1 before secretion (Van de Velde et al., 2010). In this study, transcriptome profile analysis revealed that in addition to the three NCR genes, the expression of many peptidase or peptidase inhibitors also changed significantly (Table 2), even though the function of these genes has not been well studied in nodule formation. This includes a small decrease in the expression of DNF1, which has been directly implicated in nodule function through its role in processes NCR peptides. Taken together, these results suggest that MtHANs may regulate nodule formation by influencing the expression and processing of small peptides, including NCRs.
Discussion
GATA transcriptional factors were first identified because of their roles in the regulation of light responsive genes (Buzby et al., 1990; Gilmartin et al., 1990; Lam et al., 1990; Borello et al., 1993) and their ability to bind the GATA core sequence (Lowry and Atchley, 2000). Further studies showed that GATA factors play key roles in different organ development in Arabidopsis (Nishii et al., 2000; Shikata et al., 2004; Zhao et al., 2004; Liu et al., 2005; Nawy et al., 2010; Ding et al., 2015). In this study, the homologs of HAN and HANL, MtHAN1 and MtHAN2, were identified in M. truncatula. Surprisingly, subcellular localization shows that both MtHAN1 and MtHAN2 are localized not only in the nucleus, but also in the cytoplasm. However, MtHAN1 and MtHAN2 were only able interact with each other in nucleus, implying that like HAN and HANL in Arabidopsis, they can form the heterodimer in nucleus (Zhang et al., 2013).
Previous reports show that HAN plays a role in flower development in Arabidopsis. Loss-of-function of HAN leads to decrease in numbers of petals and stamens and fused sepals (Zhao et al., 2004). In addition, three other GATA family genes, HANL2, GNC, and GNL, play redundant roles with HANs in regulating flower development (Zhang et al., 2013). In M. truncatula, however, mthan1, mthan2 and their double mutant did not show any obvious defects in flowers. It is possible that MtHANs do not participate in flower development, or that the homologs of GNC and GNL may replace the functions of MtHANs in regulating flower development in M. truncatula. Either way, this finding suggests that the functional diversity of GATA factors between Arabidopsis and M. truncatula.
It was demonstrated that the expression of GNC is inducible by nitrate in Arabidopsis (Bi et al., 2005). In our experiments, the expression levels of MtHANs were initially suppressed and then later on increased after nitrate treatment in M. truncatula, further supporting a role for GATA factors in plant N-responses. Moreover, the expression patterns of MtHAN1 and MtHAN2 show that they are mainly expressed in nodule primordia and the infection zone of mature nodules, implying the involvement of MtHANs in nodulation. The nodule number was increased in the mthan1 mthan2 double mutant, indicating that MtHANs are negative regulators of nodule formation. NCR genes, which have different expression patterns, play important roles in the regulation of nodule number in M. truncatula. NCR247 is expressed in older cells of the proximal infection zone and in the interzone. It interacts with FtsZ in the rhizobia, which is required for septum formation, and is thought to inhibit bacterial cell division to promote differentiation of bacteroids in M. truncatula nodules (Farkas et al., 2014). Another NCR encoding gene, NFS1, is also expressed in the proximal infection zone and the interzone, and functions in provoking bacterial cell death and early nodule senescence (Yang et al., 2017). NFS2 is predominantly expressed the interzone of functional (Fix+) nodules, where it promotes bacterial lysis after differentiation (Wang et al., 2017). These studies indicate that some NCRs are expressed in different nodule zones, and play different roles in nodulation in M. truncatula. In this study, the expression levels of three NCR genes are significantly decreased in nodules of mthan1 mthan2. Given that NCRs are expressed and function in mature nodules, it seems unlikely they are involved in the increased nodule numbers observed in the mthan1 mthan2 double mutant. Nonetheless, the changed expression of DNF1, the signal peptidase that processes NCRs, and many other peptidase and peptidase inhibitors in mthan1 mthan2, suggests that this important nodule system is perturbed. On the other hand, global gene transcriptional analysis indicates that several other pathways are influenced, which are important for nodule development.
Nodule number in Medicago is regulated mainly by ethylene, which control infections, and AON, which systemically controls nodule number. AON operates through CLE peptides that are released in the root and travel to the shoot, where they are perceived by the SUNN receptor kinase resulting in inhibition of nodule formation (Mortier et al., 2010). In Arabidopsis HANs act synergistically with the SUNN homologue CLAVATA, also a CLE peptide receptor, to regulate the shoot meristem. One intriguing possibility is that the enhanced nodulation of MtHANs is due to interference with AON, an idea that can be investigated in the future.
In this study, we identify the homologs of HAN and HANL in M. truncatula and further characterize their mutants. We find that in contrast to the role of HAN in flower development in Arabidopsis, MtHANs negatively regulate nodule development. This work expands our knowledge of the functions of MtHANs in plants, and gives an intriguing lead for future studies of regulation of nodule number in legumes. Further molecular and genetic studies will shed light on the mechanisms how MtHANs regulate this important developmental process.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: SRA, PRJNA673405.
Author Contributions
YX, CZ, and LH designed research. YX and HFW performed most research and analyzed the data. ZL, LW, ZG, XZ, GY, and HLW contributed analytical tools and performed some nodule analysis experiments. YX, CZ, and LH wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by Grants from Ministry of Science and Technology of China (2016YFD0100500), The National Natural Science Foundation of China (31300209 and U1906201), and Shandong Province (ZR2019MC013), Agricultural Variety Improvement Project of Shandong Province (2019LZGC010), and project for innovation and entrepreneurship leader of Qingdao (19-3-2-3-zhc).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Prof. Kirankumar Mysore (Noble Research Institute), Dr. Jiangqi Wen (Noble Research Institute) for providing the Tnt1 mutants. We also would like to thank Haiyan Yu from State Key laboratory of Microbial Technology of Shandong University for help and guidance in microscope.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.616776/full#supplementary-material
Footnotes
- ^ http://www.genome.jp/tools/clustalw/
- ^ http://www.megasoftware.net/
- ^ https://iant.toulouse.inra.fr/symbimics
References
Bi, Y. M., Zhang, Y., Signorelli, T., Zhao, R., Zhu, T., and Rothstein, S. (2005). Genetic analysis of Arabidopsis GATA transcription factor gene family reveals a nitrate-inducible member important for chlorophyll synthesis and glucose sensitivity. Plant J. 44, 680–692. doi: 10.1111/j.1365-313X.2005.02568.x
Borello, U., Ceccarelli, E., and Giuliano, G. (1993). Constitutive, light-responsive and circadian clock-responsive factors compete for the different l box elements in plant light-regulated promoters. Plant J. 4, 611–619. doi: 10.1046/j.1365-313x.1993.04040611.x
Breakspear, A., Liu, C., Roy, S., Stacey, N., Rogers, C., Trick, M., et al. (2014). The root hair “infectome” of Medicago truncatula uncovers changes in cell cycle genes and reveals a requirement for Auxin signaling in rhizobial infection. Plant Cell 26, 4680–4701. doi: 10.1105/tpc.114.133496
Bright, L. J., Liang, Y., Mitchell, D. M., and Harris, J. M. (2005). The LATD gene of Medicago truncatula is required for both nodule and root development. Mol. Plant Microbe Interact. 18, 521–532. doi: 10.1094/MPMI-18-0521
Buhian, W. P., and Bensmihen, S. (2018). Mini-Review: nod factor regulation of phytohormone signaling and homeostasis during rhizobia-legume symbiosis. Front. Plant Sci. 9:1247. doi: 10.3389/fpls.2018.01247
Buzby, J. S., Yamada, T., and Tobin, E. M. (1990). A light-regulated DNA-binding activity interacts with a conserved region of a Lemna gibba rbcS promoter. Plant Cell 2, 805–814. doi: 10.1105/tpc.2.8.805
Cheng, X., Wang, M., Lee, H. K., Tadege, M., Ratet, P., Udvardi, M., et al. (2014). An efficient reverse genetics platform in the model legume Medicago truncatula. New Phytol. 201, 1065–1076. doi: 10.1111/nph.12575
de Billy, F., Grosjean, C., May, S., Bennett, M., and Cullimore, J. V. (2001). Expression studies on AUX1-like genes in Medicago truncatula suggest that auxin is required at two steps in early nodule development. Mol. Plant Microbe Interact 14, 267–277. doi: 10.1094/MPMI.2001.14.3.267
Desbrosses, G. J., and Stougaard, J. (2011). Root nodulation: a paradigm for how plant-microbe symbiosis influences host developmental pathways. Cell Host. Microbe 10, 348–358. doi: 10.1016/j.chom.2011.09.005
Ding, L., Yan, S., Jiang, L., Zhao, W., Ning, K., Zhao, J., et al. (2015). HANABA TARANU (HAN) bridges meristem and organ primordia boundaries through PINHEAD, JAGGED, BLADE-ON-PETIOLE2 and CYTOKININ OXIDASE 3 during flower development in Arabidopsis. PLoS Genet. 11:e1005479. doi: 10.1371/journal.pgen.1005479
Ding, Y., Kalo, P., Yendrek, C., Sun, J., Liang, Y., Marsh, J. F., et al. (2008). Abscisic acid coordinates nod factor and cytokinin signaling during the regulation of nodulation in Medicago truncatula. Plant Cell 20, 2681–2695. doi: 10.1105/tpc.108.061739
Earley, K. W., Haag, J. R., Pontes, O., Opper, K., Juehne, T., Song, K., et al. (2006). Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45, 616–629.
Farkas, A., Maroti, G., Durgo, H., Gyorgypal, Z., Lima, R. M., Medzihradszky, K. F., et al. (2014). Medicago truncatula symbiotic peptide NCR247 contributes to bacteroid differentiation through multiple mechanisms. Proc. Natl. Acad. Sci. U.S.A. 111, 5183–5188. doi: 10.1073/pnas.1404169111
Ferguson, B. J., Indrasumunar, A., Hayashi, S., Lin, M. H., Lin, Y. H., Reid, D. E., et al. (2010). Molecular analysis of legume nodule development and autoregulation. J. Integr. Plant Biol. 52, 61–76. doi: 10.1111/j.1744-7909.2010.00899.x
Fonouni-Farde, C., Tan, S., Baudin, M., Brault, M., Wen, J., Mysore, K. S., et al. (2016). DELLA-mediated gibberellin signalling regulates Nod factor signalling and rhizobial infection. Nat. Commun. 7:12636. doi: 10.1038/ncomms12636
Franssen, H. J., Xiao, T. T., Kulikova, O., Wan, X., Bisseling, T., Scheres, B., et al. (2015). Root developmental programs shape the Medicago truncatula nodule meristem. Development 142, 2941–2950. doi: 10.1242/dev.120774
Gifford, I., Battenberg, K., Vaniya, A., Wilson, A., Tian, L., Fiehn, O., et al. (2018). Distinctive patterns of flavonoid biosynthesis in roots and nodules of Datisca glomerata and Medicago spp. revealed by Metabolomic and gene expression profiles. Front. Plant Sci. 9:1463. doi: 10.3389/fpls.2018.01463
Gilmartin, P. M., Sarokin, L., Memelink, J., and Chua, N. H. (1990). Molecular light switches for plant genes. Plant Cell 2, 369–378. doi: 10.1105/tpc.2.5.369
Guefrachi, I., Nagymihaly, M., Pislariu, C. I., Van de Velde, W., Ratet, P., Mars, M., et al. (2014). Extreme specificity of NCR gene expression in Medicago truncatula. BMC Genomics 15:712. doi: 10.1186/1471-2164-15-712
Hardy, R. W., Holsten, R. D., Jackson, E. K., and Burns, R. C. (1968). The acetylene-ethylene assay for N2 fixation: laboratory and field evaluation. Plant Physiol. 43, 1185–1207. doi: 10.1104/pp.43.8.1185
Hirsch, A. M., Larue, T. A., and Doyle, J. (1997). Is the legume nodule a modified root or stem or an Organ sui generis? Crit. Rev. Plant Sci. 16, 361–392. doi: 10.1080/07352689709701954
Jin, Y., Liu, H., Luo, D., Yu, N., Dong, W., Wang, C., et al. (2016). DELLA proteins are common components of symbiotic rhizobial and mycorrhizal signalling pathways. Nat. Commun. 7:12433. doi: 10.1038/ncomms12433
Karimi, M., Inze, D., and Depicker, A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7, 193–195. doi: 10.1016/s1360-1385(02)02251-3
Lam, E., Kano-Murakami, Y., Gilmartin, P., Niner, B., and Chua, N. H. (1990). A metal-dependent DNA-binding protein interacts with a constitutive element of a light-responsive promoter. Plant Cell 2, 857–866. doi: 10.1105/tpc.2.9.857
Limpens, E., and Bisseling, T. (2003). Signaling in symbiosis. Curr. Opin. Plant Biol. 6, 343–350. doi: 10.1016/s1369-5266(03)00068-2
Liu, C. W., and Murray, J. D. (2016). The role of flavonoids in nodulation host-range specificity: an update. Plants (Basel) 5:33. doi: 10.3390/plants5030033
Liu, H., Zhang, C., Yang, J., Yu, N., and Wang, E. (2018). Hormone modulation of legume-rhizobial symbiosis. J. Integr. Plant Biol. 60, 632–648. doi: 10.1111/jipb.12653
Liu, P. P., Koizuka, N., Martin, R. C., and Nonogaki, H. (2005). The BME3 (Blue Micropylar End 3) GATA zinc finger transcription factor is a positive regulator of Arabidopsis seed germination. Plant J. 44, 960–971. doi: 10.1111/j.1365-313X.2005.02588.x
Lowry, J. A., and Atchley, W. R. (2000). Molecular evolution of the GATA family of transcription factors: conservation within the DNA-binding domain. J. Mol. Evol. 50, 103–115. doi: 10.1007/s002399910012
Maekawa, T., Maekawa-Yoshikawa, M., Takeda, N., Imaizumi-Anraku, H., Murooka, Y., and Hayashi, M. (2009). Gibberellin controls the nodulation signaling pathway in Lotus japonicus. Plant J. 58, 183–194. doi: 10.1111/j.1365-313X.2008.03774.x
Mathesius, U., Weinman, J. J., Rolfe, B. G., and Djordjevic, M. A. (2000). Rhizobia can induce nodules in white clover by “hijacking” mature cortical cells activated during lateral root development. Mol. Plant Microbe Interact 13, 170–182. doi: 10.1094/MPMI.2000.13.2.170
Mortier, V., Den Herder, G., Whitford, R., Van de Velde, W., Rombauts, S., D’Haeseleer, K., et al. (2010). CLE peptides control Medicago truncatula nodulation locally and systemically. Plant Physiol. 153, 222–237. doi: 10.1104/pp.110.153718
Murray, J. D., Karas, B. J., Sato, S., Tabata, S., Amyot, L., and Szczyglowski, K. (2007). A cytokinin perception mutant colonized by Rhizobium in the absence of nodule organogenesis. Science 315, 101–104. doi: 10.1126/science.1132514
Nawy, T., Bayer, M., Mravec, J., Friml, J., Birnbaum, K. D., and Lukowitz, W. (2010). The GATA factor HANABA TARANU is required to position the proembryo boundary in the early Arabidopsis embryo. Dev. Cell 19, 103–113. doi: 10.1016/j.devcel.2010.06.004
Nishida, H., and Suzaki, T. (2018). Nitrate-mediated control of root nodule symbiosis. Curr. Opin. Plant Biol. 44, 129–136. doi: 10.1016/j.pbi.2018.04.006
Nishii, A., Takemura, M., Fujita, H., Shikata, M., Yokota, A., and Kohchi, T. (2000). Characterization of a novel gene encoding a putative single zinc-finger protein, ZIM, expressed during the reproductive phase in Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 64, 1402–1409. doi: 10.1271/bbb.64.1402
Ou, B., Yin, K. Q., Liu, S. N., Yang, Y., Gu, T., Wing Hui, J. M., et al. (2011). A high-throughput screening system for Arabidopsis transcription factors and its application to Med25-dependent transcriptional regulation. Mol. Plant 4, 546–555. doi: 10.1093/mp/ssr002
Penmetsa, R. V., and Cook, D. R. (1997). A legume ethylene-insensitive mutant hyperinfected by its rhizobial symbiont. Science 275, 527–530. doi: 10.1126/science.275.5299.527
Penmetsa, R. V., Uribe, P., Anderson, J., Lichtenzveig, J., Gish, J. C., Nam, Y. W., et al. (2008). The Medicago truncatula ortholog of Arabidopsis EIN2, sickle, is a negative regulator of symbiotic and pathogenic microbial associations. Plant J. 55, 580–595. doi: 10.1111/j.1365-313X.2008.03531.x
Reyes, J. C., Muro-Pastor, M. I., and Florencio, F. J. (2004). The GATA family of transcription factors in Arabidopsis and rice. Plant Physiol. 134, 1718–1732. doi: 10.1104/pp.103.037788
Roux, B., Rodde, N., Jardinaud, M. F., Timmers, T., Sauviac, L., Cottret, L., et al. (2014). An integrated analysis of plant and bacterial gene expression in symbiotic root nodules using laser-capture microdissection coupled to RNA sequencing. Plant J. 77, 817–837. doi: 10.1111/tpj.12442
Roy, S., Liu, W., Nandety, R. S., Crook, A., Mysore, K. S., Pislariu, C. I., et al. (2020). Celebrating 20 years of genetic discoveries in legume nodulation and symbiotic nitrogen fixation. Plant Cell 32, 15–41. doi: 10.1105/tpc.19.00279
Roy, S., Robson, F., Lilley, J., Liu, C. W., Cheng, X., Wen, J., et al. (2017). MtLAX2, a functional homologue of the Arabidopsis Auxin influx transporter AUX1, is required for nodule organogenesis. Plant Physiol. 174, 326–338. doi: 10.1104/pp.16.01473
Santamaria, M. E., Diaz-Mendoza, M., Diaz, I., and Martinez, M. (2014). Plant protein peptidase inhibitors: an evolutionary overview based on comparative genomics. BMC Genomics 15:812. doi: 10.1186/1471-2164-15-812
Schiessl, K., Lilley, J. L. S., Lee, T., Tamvakis, I., Kohlen, W., Bailey, P. C., et al. (2019). NODULE INCEPTION recruits the lateral root developmental program for symbiotic nodule organogenesis in Medicago truncatula. Curr. Biol. 29, 3657–3668.e5. doi: 10.1016/j.cub.2019.09.005
Shikata, M., Matsuda, Y., Ando, K., Nishii, A., Takemura, M., Yokota, A., et al. (2004). Characterization of Arabidopsis ZIM, a member of a novel plant-specific GATA factor gene family. J. Exp. Bot. 55, 631–639. doi: 10.1093/jxb/erh078
Soyano, T., Shimoda, Y., Kawaguchi, M., and Hayashi, M. (2019). A shared gene drives lateral root development and root nodule symbiosis pathways in Lotus. Science 366, 1021–1023. doi: 10.1126/science.aax2153
Stougaard, J. (2001). Genetics and genomics of root symbiosis. Curr. Opin. Plant Biol. 4, 328–335. doi: 10.1016/s1369-5266(00)00181-3
Suzaki, T., Yano, K., Ito, M., Umehara, Y., Suganuma, N., and Kawaguchi, M. (2012). Positive and negative regulation of cortical cell division during root nodule development in Lotus japonicus is accompanied by auxin response. Development 139, 3997–4006. doi: 10.1242/dev.084079
Suzuki, A., Akune, M., Kogiso, M., Imagama, Y., Osuki, K., Uchiumi, T., et al. (2004). Control of nodule number by the phytohormone abscisic Acid in the roots of two leguminous species. Plant Cell Physiol. 45, 914–922. doi: 10.1093/pcp/pch107
Tadege, M., Wen, J., He, J., Tu, H., Kwak, Y., Eschstruth, A., et al. (2008). Large-scale insertional mutagenesis using the Tnt1 retrotransposon in the model legume Medicago truncatula. Plant J. 54, 335–347. doi: 10.1111/j.1365-313X.2008.03418.x
Timmers, A. C., Auriac, M. C., and Truchet, G. (1999). Refined analysis of early symbiotic steps of the Rhizobium-Medicago interaction in relationship with microtubular cytoskeleton rearrangements. Development 126, 3617–3628.
Tiricz, H., Szucs, A., Farkas, A., Pap, B., Lima, R. M., Maroti, G., et al. (2013). Antimicrobial nodule-specific cysteine-rich peptides induce membrane depolarization-associated changes in the transcriptome of Sinorhizobium meliloti. Appl. Environ. Microbiol. 79, 6737–6746. doi: 10.1128/AEM.01791-13
Van de Velde, W., Zehirov, G., Szatmari, A., Debreczeny, M., Ishihara, H., Kevei, Z., et al. (2010). Plant peptides govern terminal differentiation of bacteria in symbiosis. Science 327, 1122–1126. doi: 10.1126/science.1184057
Via, V. D., Zanetti, M. E., and Blanco, F. (2016). How legumes recognize rhizobia. Plant Signal. Behav. 11:e1120396. doi: 10.1080/15592324.2015.1120396
Wang, H., Wang, H., Liu, R., Xu, Y., Lu, Z., and Zhou, C. (2018). Genome-wide identification of TCP family transcription factors in medicago truncatula reveals significant roles of miR319-Targeted TCPs in nodule development. Front. Plant Sci. 9:774. doi: 10.3389/fpls.2018.00774
Wang, H., Xu, Y., Hong, L., Zhang, X., Wang, X., Zhang, J., et al. (2019). HEADLESS regulates auxin response and compound leaf morphogenesis in Medicago truncatula. Front. Plant Sci. 10:1024. doi: 10.3389/fpls.2019.01024
Wang, Q., Yang, S., Liu, J., Terecskei, K., Abraham, E., Gombar, A., et al. (2017). Host-secreted antimicrobial peptide enforces symbiotic selectivity in Medicago truncatula. Proc. Natl. Acad. Sci. U.S.A. 114, 6854–6859. doi: 10.1073/pnas.1700715114
Xie, F., Murray, J. D., Kim, J., Heckmann, A. B., Edwards, A., Oldroyd, G. E., et al. (2012). Legume pectate lyase required for root infection by rhizobia. Proc. Natl. Acad. Sci. U.S.A. 109, 633–638. doi: 10.1073/pnas.1113992109
Yang, S., Wang, Q., Fedorova, E., Liu, J., Qin, Q., Zheng, Q., et al. (2017). Microsymbiont discrimination mediated by a host-secreted peptide in Medicago truncatula. Proc. Natl. Acad. Sci. U.S.A. 114, 6848–6853. doi: 10.1073/pnas.1700460114
Yoo, S. D., Cho, Y. H., and Sheen, J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572. doi: 10.1038/nprot.2007.199
Zhang, X., Zhou, Y., Ding, L., Wu, Z., Liu, R., and Meyerowitz, E. M. (2013). Transcription repressor HANABA TARANU controls flower development by integrating the actions of multiple hormones, floral organ specification genes, and GATA3 family genes in Arabidopsis. Plant Cell 25, 83–101. doi: 10.1105/tpc.112.107854
Zhao, Y., Medrano, L., Ohashi, K., Fletcher, J. C., Yu, H., Sakai, H., et al. (2004). HANABA TARANU is a GATA transcription factor that regulates shoot apical meristem and flower development in Arabidopsis. Plant Cell 16, 2586–2600. doi: 10.1105/tpc.104.024869
Keywords: Medicago truncatula, HANABA TARANU, expression analysis, nodule development, NCR genes, Tnt1 mutants
Citation: Xu Y, Wang H, Lu Z, Wen L, Gu Z, Zhang X, Yu G, Wang H, Zhou C and Han L (2021) Developmental Analysis of the GATA Factor HANABA TARANU Mutants in Medicago truncatula Reveals Their Roles in Nodule Formation. Front. Plant Sci. 12:616776. doi: 10.3389/fpls.2021.616776
Received: 13 October 2020; Accepted: 18 March 2021;
Published: 29 April 2021.
Edited by:
Stephan Pollmann, Center for Plant Biotechnology and Genomics, National Institute of Agricultural and Food Research and Technology, SpainReviewed by:
Manuel González-Guerrero, Polytechnic University of Madrid, SpainJeremy Dale Murray, Institute of Plant Physiology and Ecology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences (CAS), China
Copyright © 2021 Xu, Wang, Lu, Wen, Gu, Zhang, Yu, Wang, Zhou and Han. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lu Han, bGhhbkBzZHUuZWR1LmNu
 Yiteng Xu
Yiteng Xu Hongfeng Wang
Hongfeng Wang Zhichao Lu
Zhichao Lu Lizhu Wen1
Lizhu Wen1 Chuanen Zhou
Chuanen Zhou Lu Han
Lu Han