- School of Environmental and Municipal Engineering, Lanzhou Jiaotong University, Lanzhou, China
The formation of adventitious roots (ARs) is an ecologically and economically important developmental process in plants. The evolution of AR systems is an important way for plants to cope with various environmental stresses. This review focuses on identified genes that have known to regulate the induction and initiation of ARs and offers an analysis of this process at the molecular level. The critical genes involved in adventitious rooting are the auxin signaling-responsive genes, including the AUXIN RESPONSE FACTOR (ARF) and the LATERAL ORGAN BOUNDARIES-DOMAIN (LOB) gene families, and genes associated with auxin transport and homeostasis, the quiescent center (QC) maintenance, and the root apical meristem (RAM) initiation. Several genes involved in cell wall modulation are also known to be involved in the regulation of adventitious rooting. Furthermore, the molecular processes that play roles in the ethylene, cytokinin, and jasmonic acid signaling pathways and their crosstalk modulate the generation of ARs. The crosstalk and interaction among many molecular processes generates complex networks that regulate AR generation.
Introduction
Root systems are crucial for plant anchor and for the absorption of water and nutrients from the soil. Plant root system consists of primary, lateral, and adventitious roots (ARs) (Osmont et al., 2007). Although the capacity of AR formation is a genetic trait of many plant species (Sorin et al., 2006), in some cases, it can be induced by many environmental stresses, such as flooding, nutrient deficiency, heavy metals, and wounding (Steffens and Rasmussen, 2016). In ecosystems in which soil disturbance is frequent, adventitious rooting can be an alternative or supplement to seed propagation. Thus, AR formation is an important strategy for plant to cope with some environmental stresses (Ramirez-Carvajal and Davis, 2010). In addition, AR formation is an effective method for vegetative propagation of plants and has been successfully used in the production of horticultural species (Li et al., 2009; Steffens and Rasmussen, 2016). The process of AR formation is composed of three successively phases, that is induction, initiation, and expression (De Klerk et al., 1999). The key molecular events occur during root induction and initiation phases. The fate of root competent cells shifts from normal cells to root primordium cells during induction phase, which further develop to form root primordia during initiation phase. A great diverse of molecular and metabolic changes are involved in the induction and initiation of root primordia (Batish et al., 2008). Moreover, the formation of root primordia is regulated by a number of internal and external factors, such as hormones, wounding, and certain environmental stresses (Li et al., 2009; Steffens and Rasmussen, 2016). The physiological and biochemical aspects of AR formation are reasonably well known, however, the molecular processes underlying AR generation, particularly during the early phases, are still largely unknown (Ahkami et al., 2009; Ramirez-Carvajal et al., 2009; Gutierrez et al., 2012; Druege et al., 2016, 2019).
In recent years, there have been increasing interests focused on the molecular bases of adventitious rooting. Many studies have identified a large number of genes directly implicated in the induction and initiation of AR in many plant species (Table 1). These species are the herbaceous dicots Arabidopsis thaliana (Sorin et al., 2005, 2006; Gutierrez et al., 2009, 2012), Solanum lycopersicon (Negi et al., 2010; Vidoz et al., 2010), Medicago sativa (Delphine et al., 2008), Medicago truncatula (Nole-Wilson et al., 2005; Imin et al., 2007; Holmes et al., 2008, 2010), Cucumis sativus (Xuan et al., 2008; Li et al., 2011), and Petunia hybrid (Ahkami et al., 2009) and monocots Oryza sativa (Inukai et al., 2005; Liu et al., 2005, 2009, 2011) and Zea mays (Taramino et al., 2007), as well as the woody species Pinus radiata (Solé et al., 2008; Sozzani et al., 2010; Lucas et al., 2011), Pinus contorta (Hemerly et al., 1993; Mews et al., 1996; Lindrotha et al., 2001; Brinker et al., 2004), Populus trichocarpa (Rigal et al., 2012), Populus tremula × Populus alba (Ramirez-Carvajal et al., 2009), and Malus domestica (Smolka et al., 2009). However, different types of ARs are formed in these plant species as a normal developmental process or in response to various environmental stresses. Steffens and Rasmussen (2016) proposed a classification of AR subgroups and their definitions. Giving that different AR types are deferentially induced and regulated (Bellini et al., 2014; Pacurar et al., 2014; Druege et al., 2016, 2019; Steffens and Rasmussen, 2016), this review focused on the main four types of ARs, i.e., normal developed-stem roots and etiolation-induced hypocotyl roots in Arabidopsis seedlings; nodal and/or crown roots in rice and maize; de novo roots in leaf cultures in alfalfa and leaf explants in Arabidopsis and tobacco, and wounding-induced stem roots in woody cuttings such as Populus, Pinus, Malus, Eucalyptus, and Vitis plants.
All the genes that function in auxin perception, transport, and homeostasis, as well as in cell division, cell wall synthesis, cell wall weakening, root meristem formation, and quiescent center (QC) maintenance have been demonstrated to modulate AR formation (Brinker et al., 2004; Sorin et al., 2006; Holmes et al., 2010). Transcription factors (TFs), such as the APETALA2/ETHYLENE RESPONSIVE ELEMENT BINDING (AP2/ERF), MYB, NAC, WRKY, and bHLH families, have been found to mediate AR formation (Rigal et al., 2012; Druege et al., 2016, 2019). The previous studies have shown that, at the molecular level, complex networks are implicated in the regulation of AR generation in plants (Table 1).
Molecular Bases Associated With Auxin Signaling During Adventitious Rooting
Auxin has been well known to act as a central regulator controlling adventitious rooting in plants. Increasing evidences have shown that auxin signaling-related genes are the most important molecular bases for the initiation of AR in plants. Three kinds of auxin signaling genes are involved in the AR process, i.e., auxin synthesis- and homeostasis-related genes, auxin transport-related genes, and auxin-responsive genes.
Auxin Synthesis-, Homeostasis-, and Receptor-Related Genes Mediate AR Formation
The genes involved in IAA synthesis and homeostasis have been demonstrated to directly influence AR formation. YUCCA encodes flavin monooxygenase that is involved in tryptophan-dependent indole-3-acetic acid (IAA) biosynthesis (Cheng et al., 2006; Mashiguchi et al., 2011). The overexpression of YUCCA6 in Arabidopsis causes typical auxin overproduction phenotypes and increases free IAA to maintain local IAA levels in the root apical meristem (Rovere et al., 2013). Studies in Arabidopsis and rice showed that YUCCA6 directly mediate AR formation by increasing IAA levels not only via direct biosynthesis but also by affecting the expression of GRETCHEN HAGEN3 (GH3) family genes (Kim et al., 2007; Yamamoto et al., 2007). The GH3 family genes encode IAA-amido synthetases, which catalyze the conjugation of various amino acids to auxin and jasmonate, and consequently control the free IAA level (Ljung et al., 2002; Staswick et al., 2005; Sorin et al., 2006; Ludwig-Muller et al., 2009). The member GH3-3 was shown to be involved in the activation of AR initiation in both the hypocotyls and stems of Arabidopsis (Welander et al., 2014). WEAK ETHYLENE INSENSITIVE2/ANTHRANILATE SYNTHASE alpha 1 (WEI2/ASA1), WEAK ETHYLENE INSENSITIVE7/ANTHRANILATE SYNTHASE beta 1 (WEI7/ASB1), and TRYPTOPHAN SYNTHASE BETA 1 (TSB1) encode α- and β-subunits of anthranilate synthase, which is a rate-limiting enzyme of Trp-dependent IAA biosynthesis; disruption of their expression reduces auxin biosynthesis and results in a reduction in AR number in Arabidopsis (Stepanova et al., 2005). SUPERROOT1 (SUR1) encodes a C-S lyase that converts S-alkyl-thiohydroximate into thiohydroximate, which is the first step of indole glucosinolate biosynthesis. SUR2 encodes cytochrome P450-dependent monooxygenase CYP83B1, which blocks the synthesis of indole glucosinolates. The expression of these two genes increases endogenous IAA levels and thereby promoting adventitious rooting (Boerjan et al., 1995; Delarue et al., 1998; Mikkelsen et al., 2004). ARGONAUTE1 (AGO1) encodes a miRNA-binding protein that regulates post-transcriptional gene silencing (Morel et al., 2002; Vaucheret et al., 2004). De Almeida et al. (2010) demonstrated that AGO1 controls the genes related to auxin homeostasis and AR development in Eucalyptus globules. Studies in the ago1 mutants of Arabidopsis reveal that AGO1 alter auxin homeostasis by upregulating the expression of the auxin response factor ARF17 and GH3-like genes, leading to the defect in adventitious rooting (Sorin et al., 2005, 2006; Figure 1). IAA ALANINE RESISTANT4 (IAR4) encodes a putative mitochondrial pyruvate dehydrogenase E1α subunit that is involved in IAA metabolism and homeostasis. In Arabidopsis, this enzyme catalyzes the conversion of indole-3-pyruvate to IAA-CoA (Quint et al., 2009), resulting in low free IAA and AR formation. In Malus domestica, ADVENTITIOUS ROOTING RELATED OXYGENASE (ARRO-1) encodes 2-oxoacid-dependent dioxygenase, which can oxidize IAA leading to the reduction of AR initiation (Smolka et al., 2009). Calmodulin is involved in AR formation by modulating IAA content. A recent study showed that the poplar transgenic lines with overexpressed the calmodulin-like protein (CML) family gene, PdeCML23-1, exhibited more ARs and higher IAA accumulation in cuttings (Xiao et al., 2020).
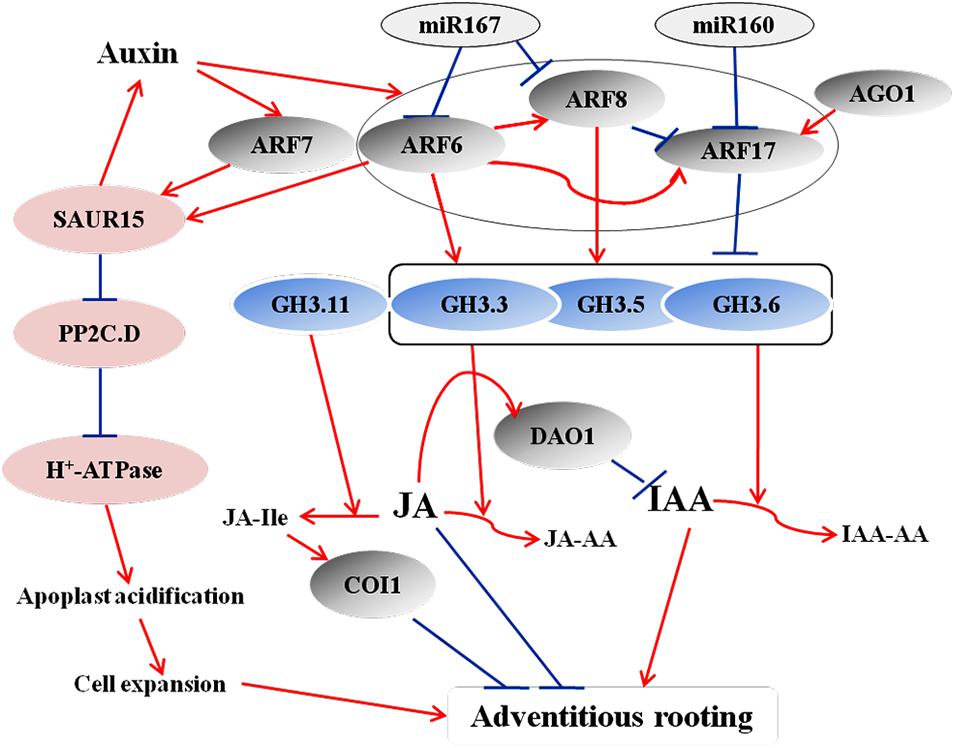
Figure 1. A model illustrating how ARFs, miRNAs, and GH3s proteins regulate adventitious rooting by controlling IAA and JA homeostasis during AR formation in Arabidopsis seedlings. Auxin controls adventitious root initiation by activating ARF6 and ARF8, leading to the downregulation of inhibitory COI1 signaling (Gutierrez et al., 2009, 2012). ARF6, ARF8, and ARF17 and their regulatory miRNAs interact in a complex network and act upstream of GH3.3, GH3.5, and GH3.6. ARF6 and ARF8 are positive regulators of these three GH3 genes, whereas ARF17 is a negative regulator. The three GH3 proteins control free IAA levels and JA homeostasis. The JA level determines the JA-Ile level. JA-Ile negatively regulates adventitious rooting by activating the COI1 signaling pathway. AGO1 upregulates GH3 genes by downregulating ARF17, thereby regulating adventitious rooting through this pathway, as well. Another feedback circuit consist of DAO1, IAA, and COI1-dependent JA signaling. COI1-dependent JA signaling actives the expression of DAO1, which in turn regulates IAA homeostasis by irreversible degradation (Lakehal et al., 2019b). Copeland (2020) and Yin et al. (2020) proposed a model of the involvement of SAUR15 in auxin signaling-mediated AR formation in Arabidopsis seedlings. ARFs directly bind to the promoter of SAUR15 to activate its expression. SAUR15 promote AR formation via two pathways. SAUR15 inhibit PP2C.D activity, leading to the release of membrane H+-ATPase activity, which then causes apoplastic acidification and cell expansion, thereby facilitating AR emergence. SAUR15 also activates auxin biosynthesis to promote AR formation. Red arrows represent positive regulation, and blue bars represent negative regulation.
Auxin receptor genes have also been demonstrated to be involved in AR formation. A recent study identified a homolog gene of Arabidopsis auxin receptor TIR1, PagFBL1, in poplar, which highly expressed in the stem cambium and secondary phloem during early phase of AR formation in cuttings. The transgenic cuttings with overexpression of PagFBL1 displayed higher ARs than the wild type, while knock-down of PagFBL1 reduced ARs. Furthermore, PagFBL1 interacts with PagIAA28.1 and PagIAA28 to release ARFs (Shu et al., 2019).
The Interactions Among ARFs, GH3s, miRNAs, and SPLs During AR Formation
The ARF TFs control the expression of auxin-responsive genes at the transcriptional level and regulate plant development through auxin signaling (Guilfoyle and Hagen, 2007; Paponov et al., 2008; Bargmann et al., 2013; Chaiwanon and Wang, 2015). In Arabidopsis, ARF6, ARF8, and ARF17 control the expression of GH3.3, GH3.5, and GH3.6, and the upregulation of GH3s promote AR formation (Sorin et al., 2006; Gutierrez et al., 2012; Figure 1). ARF17 negatively regulates adventitious rooting by repressing the expression of GH3 genes, thereby altering auxin homeostasis; in contrast, ARF6 and ARF8 positively regulate adventitious rooting by inducing the expression of GH3 genes (Sorin et al., 2005, 2006; Gutierrez et al., 2009, 2012; Lakehal et al., 2019a). Another study proposed that the activation of GH3 genes might be due to the auxin-induced degradation of ARF6 and ARF8 (Pacurar et al., 2014). Furthermore, ARF6, ARF8, and ARF17 interact at the transcriptional level (Gutierrez et al., 2009). At the post-transcriptional level, miR160 and miR167 regulate the expression of ARF6, ARF8, and ARF17. ARF6, ARF8, and ARF17, in turn, affect the expression of miR160 and miR167 (Gutierrez et al., 2012). These results suggest that a subtle balance between the repressor ARF17 and the activators ARF6 and ARF8 control the initiation of ARs via a complex regulatory network (Sorin et al., 2005; Gutierrez et al., 2009; Figure 1). Recently, Lee et al. (2019) identified a novel auxin signaling module controlling AR formation in Arabidopsis hypocotyls, in which ARF7 and ARF19 act as positive regulators via activating the downstream members LBD16 and LBD18 (Figure 2). Cai et al. (2019) showed that, in poplar, the cuttings with overexpression of PeARF8.1 gene displayed more ARs than the wild type, and miR167a targets PeARF6s and PeARF8s to regulate AR formation. Similarly, Liu et al. (2020) showed that the poplar peu-miR160a negatively regulated the genes PeARF10.1, PeARF16.1, PeARF16.2, PeARF16.3, PeARF17.1, and PeARF17.2. Overexpressing PeARF17.1 or mPeARF17.2 significantly increased AR number in the cuttings. In this network, miRNAs function as modulators to fine-tune adventitious rooting (Gutierrez et al., 2009). A recent study by Lakehal et al. (2019a) showed that AUX/IAA family genes are also involved in AR formation in Arabidopsis hypocotyls, of which IAA6, IAA9, and IAA17 repress ARF6 and ARF8 by interacting with them. Furthermore, IAA6, IAA9, and IAA17 repress the expression of GH3.3, GH3.5, and GH3.6 during AR formation (Figure 3).
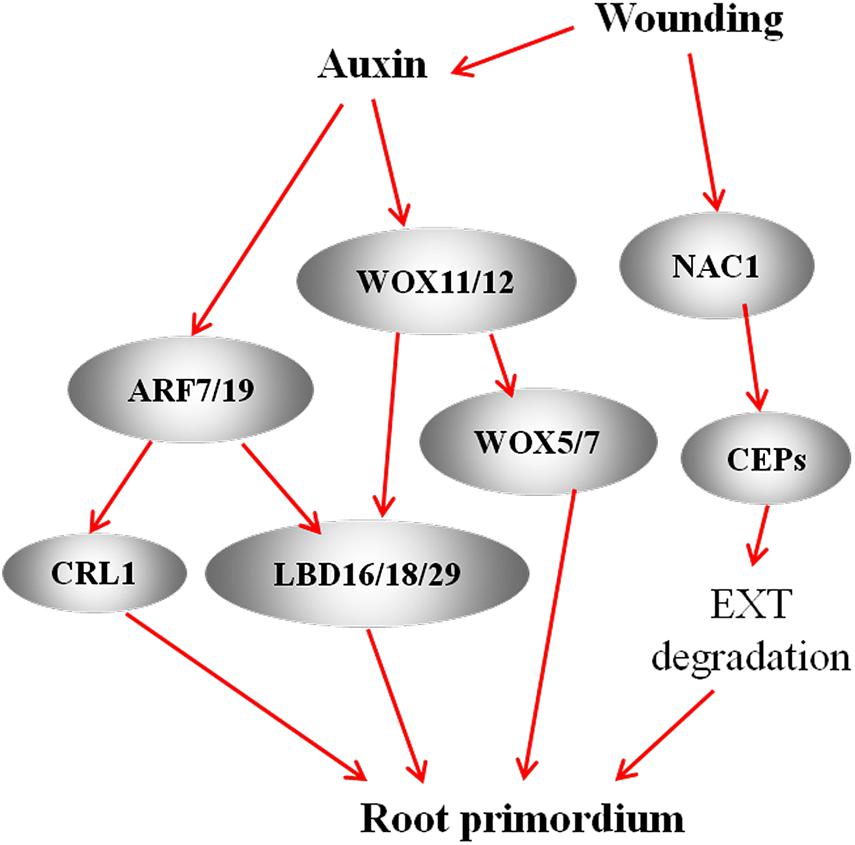
Figure 2. A model of interaction among WOXs, LBDs, and ARFs regulates the formation of AR primordia during AR formation. In the AR model of Arabidopsis seedlings, ARF7/19 activate LBD16/18/29 expression to promote AR formation (Lee et al., 2019). In the rice model, ARFs target CRL1 to regulate AR (Inukai et al., 2005). During de novo AR formation in Arabidopsis leaf explants, wounding-induced auxin activate the expression of WOX11 and WOX12, which upregulate LBD16 and LBD29 (Liu et al., 2014) and directly activate WOX5 and WOX7 to promote AR (Hu and Xu, 2016). In de novo AR model in leaf explant of Arabidopsis, wounding-induced the expression of NAC1 highly upregulated the expression of Cys endopeptidase-coding gene CEP1 and CEP2. CEP plays a role in degradation of extensin (EXT) in the cell wall and thus conduces to the emergence of AR primordia (Chen et al., 2016). Red arrows represent positive regulation.
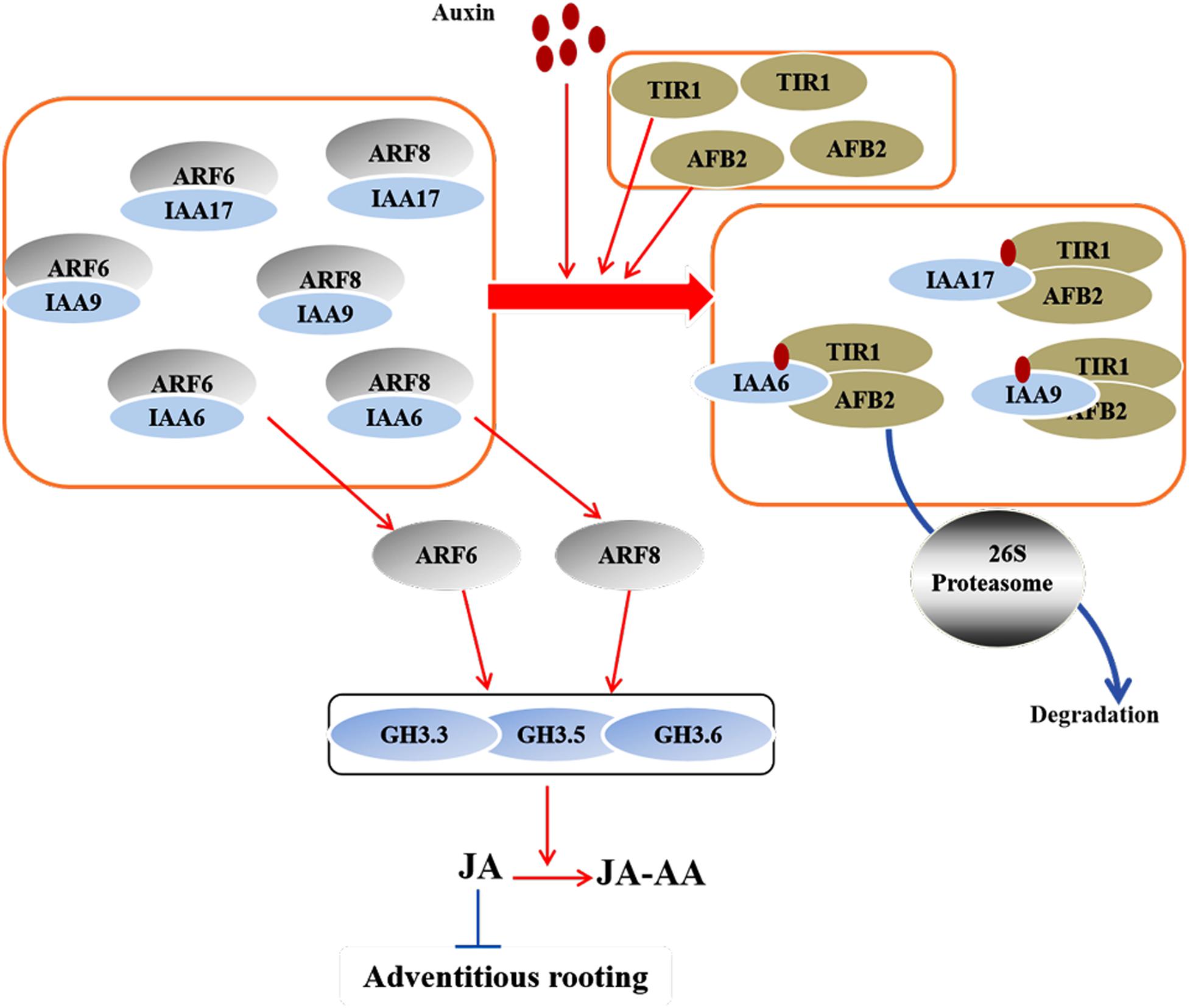
Figure 3. A model that was recently proposed by Lakehal et al. (2019a) illustrates the interaction between IAAs and ARFs during AR formation in Arabidopsis hypocotyls. The AUX/IAA family proteins IAA6, IAA9, and IAA17 interact with ARF6 and ARF8 to repress their activity at low IAA level. At high IAA level, the Aux/IAA proteins form an auxin coreceptor complex with TIR1 and AFB2 and are degraded through the 26S proteasome. The released ARF6 and ARF8 further initiate the expression of auxin-responsive genes for AR formation. In this model, IAA6, IAA9, and IAA17 repress the expression of GH3.3, GH3.5, and GH3.6, while ARF6 and ARF8 active their expression. Red arrows represent positive regulation, and blue bars represent negative regulation.
The stimulative role of miR156 in de novo root formation has been proved in Arabidopsis (Aung et al., 2017; Ye et al., 2020b), Malus xiaojinensis (Xu et al., 2017), and poplar (Cai et al., 2019). In Arabidopsis, miR156 repress its targets SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) TFs to maintain juvenile traits. The member SPL10 inhibits AR regeneration by reducing wounding-induced auxin burst in the juvenile leaf explants. In the adult leaf explants, the members SPL2/10/11 bind to the promoters of AP2/ERFs resulting in attenuation of AR induction by reducing auxin level (Gandikota et al., 2007; Ye et al., 2020a). In the M. xiaojinensis leaf explants, the high expression of miR156 is required for auxin-induced AR formation, and MxSPL26 is involved in this process (Xu et al., 2017). In the cuttings of E. grandis, EgSPL2 and EgSPL5 negatively affect AR formation (Abu-Abied et al., 2012). These results suggest that miR156-SPLs network control AR formation.
Recently, Yin et al. (2020) revealed the interaction between SMALL AUXIN-UP RNA15 (SAUR15) and ARFs in controlling AR formation. ARF6 and ARF7 directly bind to the promoter of SAUR15 to promote its expression. SAUR15 promote AR formation via enhancing auxin accumulation and inhibiting the activity of PP2C-D type 2C protein phosphatase, which activates the activity of membrane H+-ATPases leading to apoplastic acidification, thereby facilitating cell expansion and AR emergence (Copeland, 2020; Figure 1).
Changes in Expression of Auxin Transporter-Related Genes During AR Formation
Polar auxin transport (PAT) results in the differential distribution of auxin in certain tissues, which directly triggers the initiation of ARs (Ludwig-Muller et al., 2005). Studies in Arabidopsis hypocotyls showed that auxin moves out of the shoot apex and accumulates in the vascular parenchyma adjacent to pericycle founder cells, causing an IAA gradient that is essential for adventitious rooting (Rovere et al., 2013; Sukumar et al., 2013). Three main classes of auxin transporters determine the differential distribution of auxin in the tissues, including the influx carriers AUXIN1/LIKE AUX1 (AUX1/LAX) and two kinds of efflux carriers, i.e., the ATP-BINDING CASSETTE subfamily B (ABCB) and the PIN- FORMED (PIN) proteins. Arabidopsis has four highly conserved AUX1 family multigenes (i.e., AUX1 and LIKE AUX1 [LAX] genes LAX1, LAX2, and LAX3), eight PIN family members, and 29 ABCB members, which provide roust functional redundancy in regulating auxin transport and homeostasis (Guan et al., 2019).
PIN1 mediates AR induction by facilitating the lateral auxin efflux from the vascular system to the pericycle founder cells (Péret et al., 2012). The auxin induced PIN1 expression is essential for AR formation in cotyledon cuttings of Mangifera indica (Li et al., 2012) and auxin-dependent AR emergence in rice (Xu et al., 2005). PIN expression reinforces auxin gradients, further regulating the expression of meristem cell-specific genes PLETHORA1 (PLT1) and PLT2 (Benkova et al., 2003; Aida et al., 2004; Blilou et al., 2005). The PIN-LIKES (PILS) family proteins are novel putative auxin transport facilitators localized to the endoplasmic reticulum and participate in the regulation of intracellular IAA transport and auxin availability (Barbez et al., 2012). Recent studies examined the expression profile of PIN family members in olive and tomato cuttings. During IBA-induced AR formation in olive cuttings, OePIN2b and OePIN5b were highly upregulated, whereas, OePIN1a-c, OePIN3a-c, OePIN6, and OePIN8 were downregulated (Velada et al., 2020). In tomato cuttings, SlPIN2, SlPIN 3, SlPIN 4, and SlPIN 7 were involved in AR induction and initiation (Guan et al., 2019).
Auxin regulates PIN expression through the ARF family proteins (Vieten et al., 2005) and increases PIN levels and activity at the plasma membrane (Paciorek et al., 2005). Several proteins are involved in controlling the asymmetrical transport of PINs and polar auxin transport in Arabidopsis and rice (Geldner et al., 2003, 2004; Morita and Kyozuka, 2007), such as a Ser/Thr kinase encoded by PINOID (PID) (Benjamins et al., 2001) and a large guanine nucleotide exchange factor for ADP-ribosylation factor (GNOM), which is a small GTPase. The rice OsGNOM1 encoded by CRL2, can affect polar auxin transport by disturbing the vesicle trafficking of PIN1, thereby affecting the formation of AR primordia (Kitomi et al., 2008; Liu et al., 2009, 2011).
The auxin influx carrier AUX1 is also localized asymmetrically and takes up auxin in a pH-dependent, high-affinity manner in the root meristem cells (Blilou et al., 2005; Yang et al., 2006). Studies have shown that the genes AUX1 and LIKE-AUX1-3 (LAX3) in Arabidopsis (Péret et al., 2012; Rovere et al., 2016; Lee et al., 2019), AUX3 and AUX4 in Mangifera indica (Li et al., 2012), and SlLAX1 in tomato (Pattison and Catala, 2012) are essential for AR formation.
As auxin efflux carriers, ABCBs act in concert with the PIN proteins (Blakeslee et al., 2007; Mravec et al., 2008). Four Arabidopsis mutants, pin1, pin3, pin7, and abcb19, display significant reductions in AR formation, indicating that PIN and ABCB proteins mediate AR formation through affecting auxin efflux. Furthermore, the overexpression of ABCB19 increases auxin transport and IAA accumulation, driving the initiation of ARs in the hypocotyl of Arabidopsis, whereas suppression of ABCB19 led to a reduction in auxin level in competent cells, and resulted in less ARs (Christie et al., 2011; Sukumar et al., 2013). In Pinus taeda, 5NG4 encodes an auxin-induced transmembrane protein which probably acts as auxin transporter and is involved in AR formation in hypocotyl cuttings (Busov et al., 2004).
Besides, some proteins mediate AR formation by affecting the status of auxin transporters. For example, the Arabidopsis gene PP2A encodes a type 2A Ser/Thr protein phosphatase, which is shown to modulate AR formation by modifying the phosphorylation status of auxin transporters and affecting polar auxin transport (Ludwig-Muller et al., 2005).
Auxin-Responsive LOB-Domain TFs Are Involved in Adventitious Rooting
The plant-specific LATERAL ORGAN BOUNDARIES-DOMAIN (LOB-domain, LBD) TFs have been shown to modulate AR formation (Hochholdinger and Zimmermann, 2008) through regulating cell division and cell wall modification during AR generation. These LBD family members include the rice ADVENTITIOUS ROOTLESS1 (ARL1) (Liu et al., 2005) and CROWN ROOTLESS1 (CRL1), which encodes a member of AS2/LOB protein family (Inukai et al., 2005), the maize ROOTLESS CONCERNING CROWN AND SEMINAL ROOTS (RTCS) (Taramino et al., 2007), the Medicago MtLOB29 (Holmes et al., 2010), and the Arabidopsis LBD16, LBD18 and LBD29 (Shuai et al., 2002; Lee et al., 2019). ARL1 and CRL1 positively regulate the initiation of AR primordia in an auxin-dependent manner in rice (Inukai et al., 2005; Liu et al., 2005; Kitomi et al., 2008). RTCS and RTCL (RTCS-LIKE) regulate the initiation and maintenance of root primordia. The auxin-responsive RTCS inhibits AUXIN BINDING PROTEIN1 (ABP1), while activates calmodulin resulting in the activation of downstream calcium-dependent signaling (Taramino et al., 2007). Members of the LOB/ASYMMETRIC LEAVES2 (ASL2) TF family are expressed in response to auxin and are induced during AR generation in Medicago leaf cultures (Holmes et al., 2010). In Arabidopsis, auxin induces the expression of LBD16, LBD18, and LBD29 in hypocotyls and stems leading to AR formation; LBD16 plays a direct role in root initiation, while LBD29 acts an indirect regulator of root differentiation and development (Welander et al., 2014). Moreover, the rice CRL1 and Arabidopsis LBD16 and LBD18 are also directly promoted by ARF7 and ARF19 via binding to their promoters (Inukai et al., 2005; Lee et al., 2019; Figure 2).
Genes That Are Involved in AR Primordium Formation Play Regulative Roles in Quiescent Center (QC) Maintenance and Root Apical Meristem (RAM) Initiation
The initiation of RAM and maintenance of QC are key processes for AR induction. Two steps are required for cell fate transition to RAM formation. Firstly, regeneration-competent cells dedifferentiate to become root founder cells. Secondly, the root founder cells transform into root primordium cells and initiate mitosis to form RAM (Hu and Xu, 2016). The most prominent molecular processes occur during the QC and RAM formation. Many genes and several TF family genes have been identified to be involved in the regulation of QC and RAM initiation.
GRAS Family TFs
The SHORTROOT (SHR) and SCARECROW (SCR) genes encode closely related TFs belonging to the GRAS (GAI, RGA, and SCR-like) gene family, and their products play a role in establishing and maintaining the RAM (Aida et al., 2004; Xu et al., 2006; Sanchez et al., 2007; Solé et al., 2008). SCR controls cell division and differentiation leading to the formation of AR primordium in Arabidopsis (Heidstra et al., 2004; Wildwater et al., 2005; Legue et al., 2014). The SCARECROW-LIKE (SCL) family genes, such as PrSCL1 from Pinus radiata and CsSCL1 from Castanea sativa (Sanchez et al., 2007; Solé et al., 2008; Vielba et al., 2011), PrSHR from Populus radiata (Solé et al., 2008), and SCR from Populus trichocarpa (Rigal et al., 2012), are activated during the earliest stages of AR formation in the cuttings. In Arabidopsis, SCR interacts with SHR to activate downstream target genes, thereby regulating RAM and QC (Vernoux and Benfey, 2005) and the positioning of the stem cell niche (Lucas et al., 2011). SHR regulates the expression of direct target genes, including the D-type cyclin gene CYCD6;1 and the cyclin-dependent kinase genes CDKB2;1 and CDK2;2 (Sozzani et al., 2010), and induces endodermal cell identity and the expression of SCR. SCR controls asymmetrical cell divisions and limits the movement of SHR (Cui et al., 2007). These results indicate that SCR/SHR complex activates D-type CYCLIN genes and involves in initiation of cell dedifferentiation in Arabidopsis (Xu et al., 2006). Furthermore, a study found that the shr mutant exhibits a loss of PIN protein, indicating that SHR also affects PIN protein abundance (Lucas et al., 2011). Using a laser capture microdissection protocol for site-specific RNA isolation and analysis, Stevens et al. (2017) examined gene expression changes during AR formation in black walnut cuttings. The results indicated that, in rooting-competent cuttings, root primordium cells exhibited the greatest transcript abundance. In juvenile rooting-competent cells, the SCL expression displayed 23- to 24-fold increase, ARF6 and ARF8 as well as SHR expression displayed 2- to 4-fold increase, implying the importance of these genes for root primordium generation.
AP2/ERPB2 Family TFs
The five AINTEGUMENTA-like (AIL) family of AP2/ERPB2 domain TFs genes, including PLETHORA1 (PLT1), PLT2, AINTEGUMENTA (ANT), AINTEGUMENTA-Like (AIL), and BABY BOOM (BBM1), are expressed in all dividing tissues and are required for AR primordium formation (Xu et al., 2006; Galinha et al., 2007; Passarinho et al., 2008). In Arabidopsis, high expression of PLT1 and PLT2 was detected in the root meristem, which are required for QC identity and the patterning of the root stem cell niche within the RAM (Galinha et al., 2007; Imin et al., 2007; Holmes et al., 2008). PLT increases PIN expression leading to the flux of auxin into the RAM (Blilou et al., 2005). In Populus trichocarpa, the expression of PtPLT1.1 and PtBBM increase greatly during the organization and differentiation of AR primordia in the cuttings (Rigal et al., 2012). BBM encodes an embryo-expressed TF that is involved in the formation of meristematic cells during root primordium formation (Imin et al., 2007; Passarinho et al., 2008).
WUSCHEL-RELATED HOMEOBOX (WOX) Family TFs
The expression of WUSCHEL-RELATED HOMEOBOX (WOX) family TF genes characterizes the early derivatives of AR founder cells in Arabidopsis (Sarkar et al., 2007; Liu et al., 2014). The auxin-inducible WOX5 specifically expresses in the QC in Arabidopsis (Sarkar et al., 2007) and plays a pivotal role in the RAM formation in leaf cultures of Medicago truncatula (Chen et al., 2009). WOX5 can maintain the maximum auxin level at the root tip by affecting auxin distribution in Arabidopsis (Gonzali et al., 2005; Ditengou et al., 2008; Ding and Friml, 2010). In A. thaliana leaf explants, WOX11 and WOX12 were also shown to be involved in de novo root organogenesis (Liu et al., 2014). WOX11 acts redundantly with WOX12 to upregulate LBD16 and LBD29, resulting in the formation of root founder cells. Mutations in the WOX-binding elements caused reduced expression of LBD16 during AR generation, indicating that WOX11/12 directly activate LBD16 by binding to the elements of the LBD16 promoter (Xu, 2018Figure 2). Furthermore, WOX11/12 activates WOX5/7 transcription by directly binding to their promoters. Mutations in WOX5/7 result in the defect in primordium formation. These results indicate that the molecular process from WOX11/12 to WOX5/7 plays a critical role in root primordium initiation (Hu and Xu, 2016). In poplar, WOX family genes are also determined to be involved in AR formation. For example, Xu et al. (2015) demonstrated that overexpressing either PeWOX11a or PeWOX11b increased the AR number on the cuttings of poplar. A recent study cloned a Larix kaempferi gene LkWOX4 and overexpressed it in poplar, and the result showed that LkWOX4 overexpression significantly increased AR numbers and decreased AR lengths (Wang et al., 2020), imply that the common role of WOX members in modulating AR formation in different plant species.
Co-expression of AP2/ERPB2 and WOX TF Family Genes During RAM Formation
AP2/ERPB2 domain TFs genes BBM, PLT2, and WOX5 are highly expressed in the root formation cultures of Medicago truncatula, reflecting the auxin-induced enrichment of these genes within root stem cells (Holmes et al., 2010). In Arabidopsis, the WOX5, SCR, SHR, PLT1, and PLT2 genes are all involved in the control of the formation of RAM and QC (Aida et al., 2004; Xu et al., 2006). The Medicago truncatula MtWOX5, MtPLT1, MtPLT2, and MtBBM1 genes are also shown to regulate the formation of RAM (Imin et al., 2007). The Populus trichocarpa gene PtAIL1, which is an ortholog of Arabidopsis AtANT, was shown to promote the formation of root primordia. Rigal et al. (2012) demonstrated that, during AR formation in poplar cuttings, the ANT and AIL genes were activated by auxin and cytokinin.
Other TF Family Genes
The TF families zinc finger, WRKY, NAC, and bZIP have also been confirmed to be involved in RAM formation. For example, an auxin-inducible zinc finger TF gene LATERAL ROOT PRIMORDIA1 (LRP1) (Smith and Fedoroff, 1995; Holmes et al., 2010), a WRKY family member WRKY75 (Devaiah et al., 2007), a NAC family member NAC1 (petunia NAM and Arabidopsis ATAF1, ATAF2, and CUC2) (Chen et al., 2016), and a bZIP family member bZIP53 (Zhang et al., 2020) are involved in AR primordium formation and AR tip emergence. The auxin-inducible MYB-DOMAIN PROTEIN77 (MYB77) TF gene may be involved in AR initiation by interacting with ARFs and enhancing the expression of auxin-responsive genes (Shin et al., 2007).
Other Genes Involved in RAM Formation
In Arabidopsis, ROOT INITIATION DEFECTIVE2 (RID2) encodes a nuclear methyltransferase-like protein that likely contributes to the nucleolar activity of pre-rRNA processing via methylation, thereby promoting cell proliferation and consequently maintaining cell proliferation competence during dedifferentiation (Ohbayashi et al., 2011). ROOT PRIMORDIUM DEFECTIVE1 (RPD1), ROOT GROWTH DEFECTIVE1 (RGD1), RGD2, and RGD3 play roles in the maintenance of active cell proliferation in AR primordia (Konishi and Sugiyama, 2003, 2006). ROOT CLAVATA1-HOMOLOGUE1 (RCH1) and RCH2 encode the leucine-rich repeat receptor-like kinases, which are detected to specifically express in RAM cells (Dello Ioio et al., 2007). MICOTUBULE ORGANIZATION1 (MOR1) encodes a microtubule-associated protein, which is involved in auxin-responsive root meristem initiation (Konishi and Sugiyama, 2003). The gamma-glutamylcysteine synthetase-coding gene ROOT MERISTEMLESS1 (RML1) mediates meristem initiation and the maintenance of cell division during AR formation in Arabidopsis and Medicago (Vernoux et al., 2000b; Holmes et al., 2010). Gamma-glutamylcysteine synthetase acts as the first enzyme in glutathione biosynthesis, implying that glutathione pathway is involved in the regulation RAM. The CYCLIN-DEPENDENT KINASE2 (CDC2) acts as a developmental switch between mitotic cell division and post-mitotic cell differentiation and maintains the proliferation competence. In Pinus contorta, PcCDC2 is an auxin- and cytokinin-inducible gene, which functions as the role of CDC2 in cell division competence during auxin-induced AR formation in cuttings (Anders et al., 2001). The Olea europaea alternative oxidase gene OeAOX2 is highly expressed in AR primordia (Peixe et al., 2007) and its activity is highly induced during adventitious rooting in cuttings (Macedo et al., 2009). The heme oxygenase-1 gene HO-1 is involved in adventitious rooting in cucumber via the regulation of the HO-1-mediated target genes, such as DNAJ-like and the gene encoding calcium-dependent protein kinase (CDPK) (Lin et al., 2012).
Molecular Bases of Cell Wall Modification During the Emergence of AR Tips
The emergence of AR primordium through cortex and epidermis cells requires cell wall degradation and reconstruction (Steffens and Rasmussen, 2016). The EXTENSIN (EXT) functions as a wound healing protein and might limit the emergence of AR tips (Chen et al., 2016). The accumulation of EXT at the wounded site strengthens cell walls during wound healing in Arabidopsis (Merkouropoulos and Shirsat, 2003). Wounding induced the expression of EXT during rooting in Vitis vinifera stem cuttings (Thomas et al., 2003). In the leaf explants of Arabidopsis, wounding induces the expression of NAC1, which then enhances the expression of Cys-ENDOPEPTIDASE (CEP). The elevated CEP activity promotes the degradation of EXT. Thus, NAC1-CEP pathway promotes the emergence of AR tips via antagonizing EXT-mediated wound healing during AR emergence (Chen et al., 2016).
Many other genes have been shown to participate in the cell wall remodeling during AR emergence (Rigal et al., 2012). HYP-RICH GLYCOPROTEIN (HRGP), a cell wall protein family gene, is specifically activated during AR initiation in tobacco cuttings (Lund et al., 1997). ROOT HAIR DEFECTIVE3 (RHD3), a large GTP-binding protein-coding gene, is mediated in cell wall biosynthesis and actin organization, both of which are essential for cell expansion during AR formation in Arabidopsis and Populus (Hu et al., 2003; Xu et al., 2012). Expansins are responsible for the acid-induced loosening of cellulose and hemicellulose and are induced in hypocotyl bases during the early stages of AR induction in the cuttings of Pinus taeda (Hutchison et al., 1999). AtPME3 encodes a basic pectin methylesterase 3PME that plays a role in AR emergence in Arabidopsis. The absence of this protein leads to changes in the degree of methylesterification of galacturonic acids in cell wall components (Guenin et al., 2011). OeAOX promotes adventitious rooting in the cuttings of Olea europaea by linking phenylpropanoid metabolism and lignin metabolism (Macedo et al., 2012).
Molecular Bases for the Crosstalk of Plant Hormone Pathways During Adventitious Rooting
Ethylene-Auxin Crosstalk During AR Formation
Many studies have shown that AR formation also involves ethylene signaling pathway and the crosstalk with auxin (Negi et al., 2010; Vidoz et al., 2010; Figure 2). In Arabidopsis, auxin can enhance ethylene level by inducing the expression of several ACC synthase genes (Stepanova et al., 2005; Negi et al., 2008), thereby increasing ethylene levels and activating the expression of ARF19 via the two tissue-specific anthranilate synthases (Ruzicka et al., 2007; Stepanova et al., 2007; Swarup et al., 2007). Ethylene can promote IAA synthesis via inducing the expression of WEI2/ASA1 and WEI7/ASB1 genes (Ruzicka et al., 2007; Stepanova et al., 2007; Swarup et al., 2007) and promotes polar auxin transport via regulating the expression of the IAA-efflux carriers PIN1, PIN2, and PIN4, and the IAA-influx carriers AUX1 and LAX3 (Swarup et al., 2007; Negi et al., 2008, 2010). Both auxin and ethylene directly activate the expression ARF19 and ARF7 (Li et al., 2006). Therefore, ARF19 and ARF7 act as mediators of crosstalk between auxin and ethylene signaling pathways. The WEI2/ASA1 and WEI7/ASB1 proteins introduce into a feedback control between auxin and ethylene (Ivanchenko et al., 2008). For example, during flood-induced stem AR formation in tomato, WEI7 promotes the Trp-dependent auxin biosynthesis and triggers additional ethylene synthesis by enhancing the expression of two ACC synthase genes, LeACS3 and LeACS7, and DIAGEOTROPICA (DGT) is involved in this pathway (Vidoz et al., 2010). DGT has been shown to regulate auxin transport by affecting PIN localization (Ivanchenko et al., 2015). During etiolation-induced AR formation in Arabidopsis, ethylene initiates the ETHYLENE-INSENSITIVE2 (EIN2)-EIN3-like (EIL1) transcriptional cascade resulting in the inhibition of WEI2 (ASA1) and WEI7 (ASB) and YUCCA6 and consequently, reduces IAA level and AR formation (Veloccia et al., 2016).
Besides, many studies have shown that AP2/ERF family genes are also involved in AR formation. For example, the PtaERF003 gene, a member of the AP2/ERF family, promotes adventitious rooting in poplar cuttings through auxin signaling pathway (Trupiano et al., 2013). In a hybrid poplar clone, downregulated expression of PtaERF003 results in low auxin accumulation in competent cells and consequently, reduced AR formation (Trupiano et al., 2013). The S-adenosylmethionine synthase (SAMS) catalyzes the production of S-adenosylmethionine and is involved in ethylene synthesis. Brinker et al. (2004) found that the P. contorta SAMS-coding gene PcSAMS1 is involved in AR meristem formation.
Cytokinin-Auxin Crosstalk During AR Formation
Cytokinin represses the differentiation of primordia and AR formation in cucumber hypocotyls and in Populus tremula cuttings (Kuroha et al., 2002; Ramirez-Carvajal et al., 2009). Cytokinin negatively regulates auxin by inducing AUX/IAA proteins and downregulates PIN expression (Moubayidin et al., 2009; Su et al., 2011). Furthermore, in Arabidopsis, cytokinin inhibits PIN1 and LAX3 expression and further blocks auxin flow. It also limits the expression of WOX5 and YUCCA6 at the distal tip (Rovere et al., 2013). The type-B cytokinin response regulators (RRs) are cytokinin-responsive transcriptional activators, acting as histidine kinases (HKs) (Nishimura et al., 2004). Arabidopsis mutations with defect in type-B RRs are characterized by insensitivity to cytokinin and instinctive formation of ARs in hypocotyls (Argyros et al., 2008). In P. tremula cuttings, PtRR13 transcriptionally regulates downstream cytokinin signaling to repress adventitious rooting (Ramirez-Carvajal et al., 2009). Moreover, PtRR13 promotes the expression of CONTINUOUS VASCULAR RING1 (COV1), which encodes a negative regulator of vascularization, and PLEIOTROPIC DRUG RESISTANCE TRANSPORTER9 (PDR9), which encodes an auxin efflux pump, further affecting the vascular tissue formation during adventitious rooting. Thus PDR9 acts as a mediator of crosstalk between cytokinin and auxin signaling (Figure 4). In addition, PtRR13 represses adventitious rooting by inhibiting the expression of two ethylene-inducible TINY-like TFs belonging to the DEHYDRATION-RESPONSIVE ELEMENT-BINDING (DREB) protein subfamily of AP2/ERF TFs. TINY is a activator of ethylene signaling (Sun et al., 2008) and a possible intersection point between ethylene and cytokinin signaling (Ramirez-Carvajal et al., 2009).
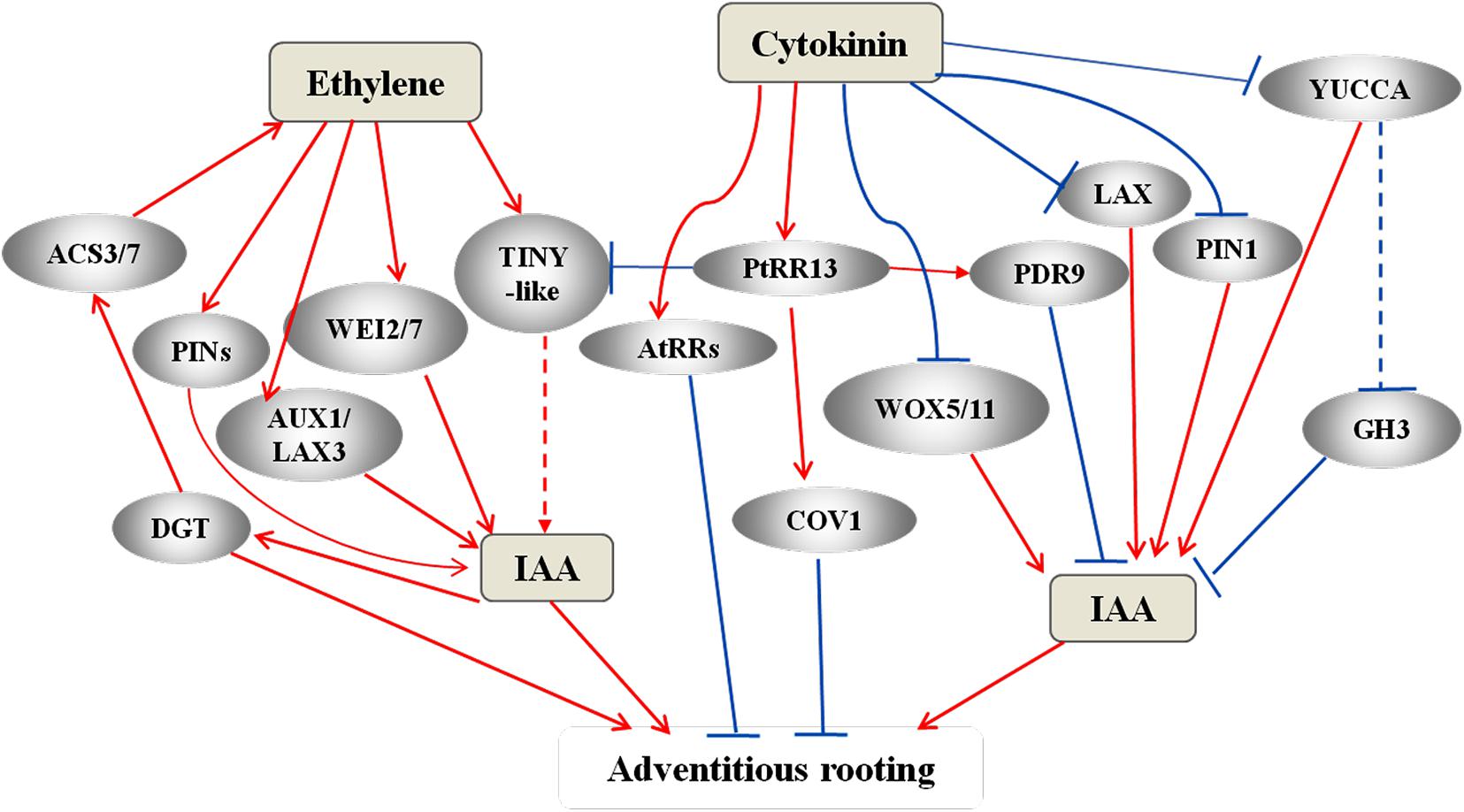
Figure 4. A complex network of IAA, ethylene, and cytokinin that has been proposed to regulate AR formation. In the AR models of Arabidopsis and flooded tomato, auxin triggers ethylene synthesis through the expression of DIAGEOTROPICA (DGT) and ACC synthesis genes ACS3 and ACS7 (Stepanova et al., 2005; Negi et al., 2008; Vidoz et al., 2010). Ethylene promote IAA synthesis via activating of WEI2 and WEI7 (Ruzicka et al., 2007; Stepanova et al., 2007; Swarup et al., 2007) and promotes polar auxin transport via activating PINs and AUX1 and LAX3 (Swarup et al., 2007; Negi et al., 2008, 2010). Cytokinin signaling activates RRs to repress AR in Arabidopsis (Argyros et al., 2008). In the AR model of poplar cuttings, the active PtRR13 repress AR formation by promoting the expression of COV1 and PDR9 and also by inhibiting the expression of TINY-like transcription factors (Ramirez-Carvajal et al., 2009). TINY is an activator of ethylene signaling (Sun et al., 2008) and a possible intersection point between ethylene and cytokinin signaling. Cytokinin also regulates auxin homeostasis and accumulation by negatively affecting the auxin carriers PIN1 and LAX3, and the WOX and YUCCA genes, thereby regulating adventitious rooting through these pathways, as well. Red arrows represent positive regulation, and blue bars represent negative regulation. The dotted lines or arrows represent the pathways need further identified.
During adventitious rooting in cultures of Medicago truncatula, cytokinin inhibits the PLT1, PLT2, and BBM1 expression and induces the expression of ANT, AIL1, and the TFs STM and WOX4 (Imin et al., 2007). WOX11 functions as an integrator of auxin and cytokinin signaling by directly repressing the expression of RR2 and is involved in the cytokinin-regulated development of ARs (Zhao et al., 2009; Kitomi et al., 2011).
Jasmonic Acid-Auxin Crosstalk During AR Formation
Jasmonic acid (JA), a stress-related hormone, inhibits adventitious rooting and works downstream of the IAA pathway (Gutierrez et al., 2012) through the GH3 proteins in Arabidopsis (Staswick et al., 2005; Staswick, 2009). GH3s catalyze the conjugation of IAA with amino acids and JA with amino acids, thereby reducing free IAA level and controlling JA homeostasis. Among them, GH3.11, also known as JASMONIC ACID RESISTANT1 (JAR1), is shown to suppress adventitious rooting in Arabidopsis hypocotyls. JAR1 conjugates JA to Ile to produce JA-Ile, an active form of JA, which further activates the JA receptor CORONATINE INSENSITIVE1 (COI1) (Fonseca et al., 2009; Figure 1) and promotes the interaction of COI1 with the co-receptor JAZ, which also is a transcriptional repressor of JA-responsive genes (Thines et al., 2007). GH3.3, GH3.5, and GH3.6 catalyze the production of JA-Asp, JA-Met, and JA-Trp, which are inactive forms of JA. Thus, these three GH3s have opposite effect to that of GH3.11 during adventitious rooting (Gutierrez et al., 2012). A study reveals a crosstalk between auxin and JA pathways that fine-tunes AR initiation in Arabidopsis hypocotyls (Gutierrez et al., 2012). Auxin affects JA homeostasis by upregulating the expression of GH3.3, GH3.5, and GH3.6, resulting in an increase in conjugational JA and a decline in the free JA. Auxin also induces the expression of JA biosynthetic genes in Arabidopsis (Paponov et al., 2008). Furthermore, during adventitious rooting in Arabidopsis, another linkage between IAA and JA involves the interaction between the auxin-responsive genes ARF6, ARF8, and ARF17 and their target genes GH3.3, GH3.5, and GH3.6. Recently, Lakehal et al. (2019a) proposed TIR1/AFB2-Aux/IAA-dependent auxin signaling, in which TRANSPORT INHIBITOR1 (TIR1) and AUXIN-SIGNALLING F-BOX (AFB2) proteins interact with IAA6, IAA9, and/or IAA17 to control JA homeostasis and AR initiation in Arabidopsis (Figure 4). Another feedback circuit between IAA and JA is mediated by DIOXYGENASE FOR AUXIN OXIDATION (DAO1) and COI1-dependent JA signaling. AtDAO1 catalyzes the production of 2-oxindole-3-acetic acid (oxIAA), thereby reducing free IAA level and AR initiation in Arabidopsis. The expression of DAO1 is induced by COI1-dependent JA signaling. DAO1 controls IAA level by catalyzing the conversion of IAA to oxIAA (Lakehal et al., 2019b).
However, MeJA (methyl jasmonate), another form of JA, is shown to promote adventitious rooting from thin cell layers of Nicotiana tabacum (Fattorini et al., 2009). MeJA increases the endogenous IAA and JA levels in Arabidopsis (Paponov et al., 2008). MeJA may maintain auxin homeostasis during adventitious rooting through regulating the coordination of PIN, IAA-amino acid hydrolase, and IAA-amido synthetase (Chen et al., 2007; Fattorini et al., 2009). For example, MeJA enhances the activity of PIN and IAA-amino acid hydrolase 6. Considering positive effects of MeJA also on IAA levels and the expression of ASA1 in Arabidopsis thin cell layers (Fattorini et al., 2017, 2018), further the early JA accumulation in the stem base of petunia cuttings (Ahkami et al., 2009) and furthermore the finding that reduced expression of the rate-limiting enzyme ALLENE OXIDE CYCLASE (PhAOC) in petunia inhibited wound-induced JA accumulation and AR formation (Lischweski et al., 2015), Druege et al. (2019) proposed that early wound-induced JA accumulation in cuttings stimulates AR induction, possibly via enhanced IAA accumulation. According to this theory, up-regulation of ASA1, IAA accumulation and intensity of AR formation in Arabidopsis leaf explants was related to early JA accumulation and dependent on JA signaling (Zhang et al., 2019).
Gibberellin (GA)-Auxin Crosstalk During AR Formation
The GA biosynthetic gene GA20ox1-OE plays an important role in GA homeostasis (Gallego-Giraldo et al., 2008). GA-INSENSITIVE DWARF1 (GID1) is a GA receptor gene (Ueguchi-Tanaka et al., 2005). Both hybrid aspens and Arabidopsis mutation lines with overexpression of AtGA20ox1, PttGID1.1, or PttGID1.3 genes display defects in AR formation, indicating that GA inhibits ARs by perturbing GA synthesis or signaling and polar auxin transport (Mauriat et al., 2014). A recent study cloned a histone deacetylase (HD2) gene PtHDT902 from Populus trichocarpa. Overexpressing PtHDT902 in Arabidopsis and poplar enhanced the expression of GA biosynthetic genes and inhibited AR formation in poplar (Ma et al., 2020), suggesting that HD2 promotes GA biosynthesis and thus suppresses AR formation.
Perspectives
In recent years, there have been significant advances in our understanding of the contributions of hormone signaling and gene networks to the regulation of the formation of ARs. An increasing number of genes involved in adventitious rooting and their interactions have been characterized and identified. The diversity of AR types and the complexity of the gene regulation networks pose a challenge to researchers. However, the molecular networks and mechanisms underlying the development of ARs need to be further clarified. At the transcriptional level, it is necessary to ascertain the key genes and gene expression profiles that regulate the different AR types and different stages of AR development. At the post-transcriptional level, the key proteins and protein profiles, as well as their regulation mechanisms during AR formation need to be explored. Another interesting issue is about the inherent differences between the easy-to-root and difficult-to-root plants at the molecular level. Such knowledge should ultimately lead to a better understanding the mechanisms underlying plant cell differentiation and AR development. In practice, we need to exploit the potential of ARs for improving the crop tolerances to various environmental stresses. Rapidly expanding knowledge of plant genomes and proteomes and new technologies will most likely shed more light on these questions in the future.
Author Contributions
The author confirms being the sole contributor of this work and has approved it for publication.
Funding
This work was funded by the National Natural Science Foundation of China (31760110 and 31560121).
Conflict of Interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Abu-Abied, M., Szwerdszarf, D., Mordehaev, I., Levy, A., Rogovoy, O., Belausov, E., et al. (2012). Microarray analysis revealed upregulation of nitrate reductase in juvenile cuttings of Eucalyptus grandis, which correlated with increased nitric oxide production and adventitious root formation. Plant J. 71, 787–799. doi: 10.1111/j.1365-313x.2012.05032.x
Ahkami, A. H., Lischewski, S., Haensch, K.-T., Porfirova, S., Hofmann, J., Rolletschek, H., et al. (2009). Molecular physiology of adventitious root formation in Petunia hybrida cuttings, involvement of wound response and primary metabolism. New Phytol. 181, 613–625. doi: 10.1111/j.1469-8137.2008.02704.x
Aida, M., Beis, D., Heidstra, R., Willemsen, V., Blilou, I., Galinha, C., et al. (2004). The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119, 109–120. doi: 10.1016/j.cell.2004.09.018
Anders, M. L., Anders, K., and von Arnold, S. (2001). Isolation of a PSTAIRE CDC2 cDNA from Pinus contorta and its expression during adventitious root development. Plant Physiol. Biochem. 39, 107–114. doi: 10.1016/s0981-9428(00)01229-8
Argyros, R. D., Mathews, D. E., Chiang, Y. H., Palmer, C. M., Thibault, D. M., Etheridge, N., et al. (2008). Type B response regulators of Arabidopsis play key roles in cytokinin signaling and plant development. Plant Cell 20, 2102–2116. doi: 10.1105/tpc.108.059584
Aung, B., Gao, R., Gruber, M. Y., Yuan, Z. C., Sumarah, M., and Hannoufa, A. (2017). MsmiR156 affects global gene expression and promotes root regenerative capacity and nitrogen fixation activity in alfalfa. Transgenic Res. 26, 541–557. doi: 10.1007/s11248-017-0024-3
Barbez, E., Kubeš, M., Rolčík, J., Béziat, C., Pěnčík, A., Wang, B., et al. (2012). A novel putative auxin carrier family regulates intracellular auxin homeostasis in plants. Nature 485, 119–122. doi: 10.1038/nature11001
Bargmann, B. O. R., Vanneste, S., Krouk, G., Nawy, T., Efroni, I., Shani, E., et al. (2013). A map of cell type-specific auxin responses. Mol. Syst. Biol. 9, 688. doi: 10.1038/msb.2013.40
Batish, D. R., Singh, H. P., Kaur, S., Kohli, R. K., and Yadav, S. S. (2008). Caffeic acid affects early growth and morphogenetic response of hypocotyl cuttings of mung bean (Phaseolus aureus). J. Plant Physio.l 165, 297–305. doi: 10.1016/j.jplph.2007.05.003
Bellini, C., Pacurar, D. I., and Perrone, I. (2014). Adventitious roots and lateral roots: similarities and differences. Annu. Rev. Plant Biol. 65, 639–666. doi: 10.1146/annurev-arplant-050213-035645
Benjamins, R., Quint, A., Weijers, D., Hooykaas, P., and Offringa, R. (2001). The PINOID protein kinase regulates organ development in Arabidopsis by enhancing polar auxin transport. Development 128, 4057–4067.
Benkova, E., Michniewicz, M., Sauer, M., Teichmann, T., Seifertova, D., Jurgens, G., et al. (2003). Local efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115, 591–602. doi: 10.1016/s0092-8674(03)00924-3
Blakeslee, J. J., Bandyopadhyay, A., Lee, O. R., Mravec, J., Tipapiwatanakun, B., Sauer, M., et al. (2007). Interactions among PIN-FORMED and P-glycoprotein auxin transporters in Arabidopsis. Plant Cell 19, 131–147. doi: 10.1105/tpc.106.040782
Blilou, I., Xu, J., Wildwater, M., Willemsen, V., Paponov, I., Friml, J., et al. (2005). The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433, 39–44. doi: 10.1038/nature03184
Boerjan, W., Cervera, M. T., Delarue, M., Beeckman, T., Dewitte, W., Bellini, C., et al. (1995). Superroot, a recessive mutation in Arabidopsis, confers auxin overproduction. Plant Cell 7, 1405–1419. doi: 10.2307/3870131
Brinker, M., van Zyl, L. L., Liu, W., Craig, D., Sederoff, R. R., Clapham, D. H., et al. (2004). Microarray analyses of gene expression during adventitious root development in Pinus contorta. Plant Physiol. 135, 1526–1539. doi: 10.1104/pp.103.032235
Busov, V. B., Johannes, E., Whetten, R. W., Sederoff, R. R., Spiker, S. L., Lanz-Garcia, C., et al. (2004). An auxin-inducible gene from loblolly pine (Pinus taeda L) is differentially expressed in mature and juvenile-phase shoots and encodes a putative transmembrane protein. Planta 218, 916–927. doi: 10.1007/s00425-003-1175-4
Cai, H., Yang, C., Liu, S., Qi, H., Wu, L., Xu, L. A., et al. (2019). MiRNA-target pairs regulate adventitious rooting in Populus: a functional role for miR167a and its target AUXIN RESPONSE FACTOR 8. Tree Physiol. 39, 1922–1936. doi: 10.1093/treephys/tpz085
Chaiwanon, J., and Wang, Z. Y. (2015). Spatiotemporal brassinosteroid signaling and antagonism with auxin pattern stem cell dynamics in Arabidopsis roots. Curr. Biol. 25, 1031–1042. doi: 10.1016/j.cub.2015.02.046
Chen, L. R., Chen, Y. J., Lee, C. Y., and Lin, T. Y. (2007). MeJA-induced transcriptional changes in adventitious roots of Bupleurum kaoi. Plant Sci. 173, 12–24. doi: 10.1016/j.plantsci.2007.03.013
Chen, S. K., Kurdyukov, S., Kereszt, A., Wang, X. D., Gresshoff, P. M., and Rose, R. J. (2009). The association of homeobox gene expression with stem cell formation and morphogenesis in cultured Medicago truncatula. Planta 230, 827–840. doi: 10.1007/s00425-009-0988-1
Chen, X., Cheng, J., Chen, L., Zhang, G., Huang, H., Zhang, Y., et al. (2016). Auxin-independent NAC pathway acts in response to explant-specific wounding and promotes root tip emergence during de novo root organogenesis in Arabidopsis. Plant Physiol. 170, 2136–2145. doi: 10.1104/pp.15.01733
Cheng, Y., Dai, X., and Zhao, Y. (2006). Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev. 20, 1790–1799. doi: 10.1101/gad.1415106
Christie, J. M., Yang, H., Richter, G. L., Sullivan, S., Thomson, C. E., Lin, J., et al. (2011). phot1 inhibition of ABCB19 primes lateral auxin fluxes in the shoot apex required for phototropism. PLoS Biol. 9:e1001076. doi: 10.1371/journal.pbio.1001076
Copeland, C. (2020). SAUR15 connects auxin perception to lateral and adventitious root formation. Plant Physiol. 184, 558–559. doi: 10.1104/pp.20.01089
Cui, H., Levesque, M. P., Vernoux, T., Jung, J. W., Paquete, A. J., Gallagher, K. L., et al. (2007). An evolutionarily conserved mechanism delimiting SHR movement defines a single layer of endodermis in plants. Science 316, 421–425. doi: 10.1126/science.1139531
De Almeida, M., Ruedell, C. M., Ricachenevsky, F. K., Sperotto, R. A., Pasquali, G., and Fett-Neto, A. G. (2010). Reference gene selection for quantitative reverse transcription-polymerase chain reaction normalization during in vitro adventitious rooting in Eucalyptus globulus Labill. BMC Mol. Biol. 11:73. doi: 10.1186/1471-2199-11-73
De Klerk, G. J., Krieken, W. V. D., and Jong, J. C. D. (1999). The formation of adventitious roots, new concepts, new possibilities. In Vitro Cell Dev. Biol. Plant 35, 189–199. doi: 10.1007/s11627-999-0076-z
Delarue, M., Prinsen, E., Vanonckelen, H., Caboche, M., and Bellini, C. (1998). sur2 mutations of Arabidopsis thaliana define a new locus involved in the control of auxin homeostasis. Plant J. 14, 603–611. doi: 10.1046/j.1365-313x.1998.00163.x
Dello Ioio, R., Linhares, F. S., Scacchi, E., Casamitjana-Martinez, E., Heidstra, R., Costantino, P., et al. (2007). Cytokinins determine Arabidopsis rootmeristem size by controlling cell differentiation. Curr. Biol. 17, 678–682. doi: 10.1016/j.cub.2007.02.047
Delphine, C., Frugier, F., Raices, M., Merchan, F., Giammaria, V., Gargantini, P., et al. (2008). A mutant ankyrin protein kinase from Medicago sativa affects Arabidopsis adventitious roots. Funct. Plant Biol. 35, 92–101. doi: 10.1071/fp07209
Devaiah, B. N., Karthikeyan, A. S., and Raghothama, K. G. (2007). WRKY75 transcription factor is a modulator of phosphate acquisition and root development in Arabidopsis. Plant Physiol. 143, 1789–1801. doi: 10.1104/pp.106.093971
Ding, Z., and Friml, J. (2010). Auxin regulates distal stem cell differentiation in Arabidopsis roots. Proc. Natl. Acad. Sci. U.S.A. 107, 12046–12051. doi: 10.1073/pnas.1000672107
Ditengou, F. A., Teale, W. D., Kocherspenger, P., Flittner, K. A., Kneuper, I., van der Graaff, E., et al. (2008). Mechanical induction of lateral root initiation in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 105, 18818–18823. doi: 10.1073/pnas.0807814105
Druege, U., Franken, P., and Hajirezaei, M. R. (2016). Plant hormone homeostasis, signaling, and function during adventitious root formation in cuttings. Front. Plant Sci. 7:381. doi: 10.3389/fpls.2016.00381
Druege, U., Hilo, A., Pérez-Pérez, J. M., Klopotek, Y., Acosta, M., Shahinnia, F., et al. (2019). Molecular and physiological control of adventitious rooting in cuttings: phytohormone action meets resource allocation. Ann. Bot. 123, 929–949. doi: 10.1093/aob/mcy234
Falasca, G., Altamura, M. M., D’Angeli, S., Zaghi, D., Costantino, P., and Mauro, M. L. (2010). The rolD oncogene promotes axillary bud and adventitious root meristems in Arabidopsis. Plant Physiol. Biochem. 48, 797–804. doi: 10.1016/j.plaphy.2010.06.002
Fattorini, L., Falasca, G., Kevers, C., Rocca, L. M., Zadra, C., and Altamura, M. M. (2009). Adventitious rooting is enhanced by methyl jasmonate in tobacco thin cell layers. Planta 231, 155–168. doi: 10.1007/s00425-009-1035-y
Fattorini, L., Hause, B., Gutierrez, L., Veloccia, A., Della Rovere, F., Piacentini, D., et al. (2018). Jasmonate promotes auxin induced adventitious rooting in dark-grown Arabidopsis thaliana seedlings and stem thin cell layers by a cross-talk with ethylene signalling and a modulation of xylogenesis. BMC Plant Biol. 18:182. doi: 10.1186/s12870-018-1392-4
Fattorini, L., Veloccia, A., Della Rovere, F., D’Angeli, S., Falasca, G., and Altamura, M. M. (2017). Indole-3-butyric acid promotes adventitious rooting in Arabidopsis thaliana thin cell layers by conversion into indole-3-acetic acid and stimulation of anthranilate synthase activity. BMC Plant Biol. 17:121. doi: 10.1186/s12870-017-1071-x
Feng, S. J., Xu, Y. M., Guo, C. K., Zheng, J. R., Zhou, B. Y., Zhang, Y. T., et al. (2016). Modulation of miR156 to identify traits associated with vegetative phase change in tobacco (Nicotiana tabacum). J. Exp. Bot. 67, 1493–1504. doi: 10.1093/jxb/erv551
Fonseca, S., Chico, J. M., and Solano, R. (2009). The jasmonate pathway: the ligand, the receptor and the core signalling module. Curr. Opin. Plant Biol. 12, 539–547. doi: 10.1016/j.pbi.2009.07.013
Friml, J., Wisniewska, J., Benkova, E., Mendgen, K., and Palme, K. (2002). Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature 415, 806–809. doi: 10.1038/415806a
Galinha, C., Hofhuis, H., Luijten, M., Willemsen, V., Blilou, I., Heidstra, R., et al. (2007). PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature 449, 1053–1057. doi: 10.1038/nature06206
Gallego-Giraldo, L., Ubeda-Tomás, S., Gisbert, C., García-Martínez, J. L., Moritz, T., and López-Díaz, I. (2008). Gibberellin homeostasis in tobacco is regulated by gibberellin metabolism genes with different gibberellin sensitivity. Plant Cell Physiol. 49, 679–690. doi: 10.1093/pcp/pcn042
Gandikota, M., Birkenbihl, R. P., Höhmann, S., Cardon, G. H., Saedler, H., and Huijser, P. (2007). The miRNA156/157 recognition element in the 39 UTR of the Arabidopsis SBP box gene SPL3 prevents early flowering by translational inhibition in seedlings. Plant J. 49, 683–693. doi: 10.1111/j.1365-313x.2006.02983.x
Geldner, N., Anders, N., Wolters, H., Keicher, J., Kornberger, W., Muller, P., et al. (2003). The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport, and auxin-dependent plant growth. Cell 112, 219–230. doi: 10.1016/s0092-8674(03)00003-5
Geldner, N., Richter, S., Vieten, A., Marquardt, S., Torres-Ruiz, R. A., Mayer, U., et al. (2004). Partial loss-of-function alleles reveal a role for GNOM in auxin transport-related, post-embryonic development of Arabidopsis. Development 131, 389–400. doi: 10.1242/dev.00926
Gonzali, S., Novi, G., Loreti, E., Paolicchi, F., Poggi, A., Alpi, A., et al. (2005). A turanose-insensitive mutant suggests a role for WOX5 in auxin homeostasis in Arabidopsis thaliana. Plant J. 44, 633–645. doi: 10.1111/j.1365-313x.2005.02555.x
Guan, L., Tayengwa, R., Cheng, Z., Peer, W. A., Murphy, A. S., and Zhao, M. (2019). Auxin regulates adventitious root formation in tomato cuttings. BMC Plant Biol. 19:435. doi: 10.1186/s12870-019-2002-9
Guenin, S., Mareck, A., Rayon, C., Lamour, R., Ndong, Y. A., Domon, J.-M., et al. (2011). Identification of pectin methylesterase 3 as a basic pectin methylesterase isoform involved in adventitious rooting in Arabidopsis thaliana. New Phytol. 192, 114–126. doi: 10.1111/j.1469-8137.2011.03797.x
Guilfoyle, T. J., and Hagen, G. (2007). Auxin response factors. Curr. Opin. Plant Biol. 10, 453–460. doi: 10.1016/j.pbi.2007.08.014
Gutierrez, L., Bussell, J. D., Pacurar, D. I., Schwambach, J., Pacurar, M., and Bellini, C. (2009). Phenotypic plasticity of adventitious rooting in Arabidopsis is controlled by complex regulation of auxin response factor transcripts and microRNA abundance. Plant Cell 21, 3119–3132. doi: 10.1105/tpc.108.064758
Gutierrez, L., Mongelard, G., Floková, K., Pǎcurar, D. I., Novák, O., Staswick, P., et al. (2012). Auxin controls Arabidopsis adventitious root initiation by regulating jasmonic acid homeostasis. Plant Cell 24, 2515–2527. doi: 10.1105/tpc.112.099119
Heidstra, R., Welch, D., and Scheres, B. (2004). Mosaic analyses using marked activation and deletion clones dissect Arabidopsis SCARECROW action in asymmetric cell division. Genes Dev. 18, 1964–1969. doi: 10.1101/gad.305504
Hemerly, A. S., Ferreira, P., de Almeida Engler, J., Van Montagu, M., Engler, G., and Inzé, D. (1993). cdc2a expression in Arabidopsis is linked with competence for cell division. Plant Cell 5, 1711–1723. doi: 10.2307/3869688
Hochholdinger, F., and Zimmermann, R. (2008). Conserved and diverse mechanisms in root development. Curr. Opin. Plant Biol. 11, 70–74. doi: 10.1016/j.pbi.2007.10.002
Holmes, P., Djordjevic, M. A., and Imin, N. (2010). Global gene expression analysis of in vitro root formation in Medicago truncatula. Funct. Plant Biol. 37, 1117–1131. doi: 10.1071/fp10159
Holmes, P., Goffard, N., Weiller, G. F., Rolfe, B. G., and Imin, N. (2008). Transcriptional profiling of Medicago truncatula meristematic root cells. BMC Plant Biol. 8:21. doi: 10.1186/1471-2229-8-21
Hu, X., and Xu, L. (2016). Transcription factors WOX11/12 directly activate WOX5/7 to promote root primordia initiation and organogenesis. Plant Physiol. 172, 2363–2373. doi: 10.1104/pp.16.01067
Hu, Y., Zhong, R., Morrison, W. H., and Ye, Z. H. (2003). The Arabidopsis RHD3 gene is required for cell wall biosynthesis and actin organization. Planta 217, 912–921. doi: 10.1007/s00425-003-1067-7
Hutchison, K. W., Singer, P. B., McInnis, S., Diaz-Sala, C., and Greenwood, M. S. (1999). Expansins are conserved in conifers and expressed in hypocotyls in response to exogenous auxin. Plant Physiol. 120, 827–831. doi: 10.1104/pp.120.3.827
Imin, N., Nizamidin, M., Wu, T., and Rolfe, B. G. (2007). Factors involved in root formation in Medicago truncatula. J. Exp. Bot. 58, 439–451. doi: 10.1093/jxb/erl224
Inukai, Y., Sakamoto, T., Ueguchi-Tanaka, M., and Shibata, Y. (2005). Crown rootless1, which is essential for crown root formation in rice, is a target of an AUXIN RESPONSE FACTOR in auxin signaling. Plant Cell 17, 1387–1396. doi: 10.1105/tpc.105.030981
Ivanchenko, M., Muday, G. K., and Dubrovsky, J. G. (2008). Ethylene–auxin interactions regulate lateral root initiation and emergence in Arabidopsis thaliana. Plant J. 55, 335–347. doi: 10.1111/j.1365-313x.2008.03528.x
Ivanchenko, M. G., Zhu, J., Wang, B., Medvecka, E., Du, Y., Azzarello, E., et al. (2015). The cyclophilin A DIAGEOTROPICA gene affects auxin transport in both root and shoot to control lateral root formation. Development 142, 712–721. doi: 10.1242/dev.113225
Kim, J. I., Sharkhuu, A., Jin, J. B., Li, P., Jeong, J. C., Baek, D., et al. (2007). yucca6, a dominant mutation in Arabidopsis, affects auxin accumulation and auxin-related phenotypes. Plant Physiol. 145, 722–735. doi: 10.1104/pp.107.104935
Kitomi, Y., Ito, H., Hobo, T., Aya, K., Kitano, H., and Inukai, Y. (2011). The auxin responsive AP2/ERF transcription factor CROWN ROOTLESS5 is involved in crown root initiation in rice through the induction of OsRR1, a type-A response regulator of cytokinin signaling. Plant J. 67, 472–484. doi: 10.1111/j.1365-313x.2011.04610.x
Kitomi, Y., Ogawa, A., Kitano, H., and Inukai, Y. (2008). CRL4 regulates crown root formation through auxin transport in rice. Plant Root 2, 19–28. doi: 10.3117/plantroot.2.19
Konishi, M., and Sugiyama, M. (2003). Genetic analysis of adventitious root formation with a novel series of temperature-sensitive mutants of Arabidopsis thaliana. Development 130, 5637–5647. doi: 10.1242/dev.00794
Konishi, M., and Sugiyama, M. (2006). A novel plant-specific family gene, ROOT PRIMORDIUM DEFECTIVE 1, is required for the maintenance of active cell proliferation. Plant Physiol. 140, 591–602. doi: 10.1104/pp.105.074724
Kuroha, T., Kato, H., Asami, T., Yoshida, S., Kamada, H., and Satoh, S. (2002). A transzeatin riboside in root xylem sap negatively regulates adventitious root formation on cucumber hypocotyls. J. Exp. Bot. 53, 2193–2200. doi: 10.1093/jxb/erf077
Lakehal, A., Chaabouni, S., Cavel, E., Le Hir, R., Ranjan, A., Raneshan, Z., et al. (2019a). A molecular framework for the control of adventitious rooting by TIR1/AFB2-Aux/IAA-dependent auxin signaling in Arabidopsis. Mol. Plant. 12, 1499–1514. doi: 10.1016/j.molp.2019.09.001
Lakehal, A., Dob, A., Novak, O., and Bellini, C. (2019b). A DAO1-mediated circuit controls auxin and jasmonate crosstalk robustness during adventitious root initiation in Arabidopsis. Int. J. Mol. Sci. 20, 4428. doi: 10.3390/ijms20184428
Lee, H. W., Cho, C., Pandey, S. K., Park, Y., Kim, M. J., and Kim, J. (2019). LBD16 and LBD18 acting downstream of ARF7 and ARF19 are involved in adventitious root formation in Arabidopsis. BMC Plant Biol. 19:46. doi: 10.1186/s12870-019-1659-4
Legue, V., Rigal, A., and Bhalerao, R. (2014). Adventitious root formation in tree species: involvement of transcription factors. Physiol. Plant 151, 192–198. doi: 10.1111/ppl.12197
Li, J., Dai, X., and Zhao, Y. (2006). A role for Auxin Response FACTOR 19 in auxin and ethylene signaling in Arabidopsis. Plant Physiol. 140, 899–908. doi: 10.1104/pp.105.070987
Li, J., Jia, H., Sun, P., Zhang, J., Xia, Y., Hu, J., et al. (2020). The WUSCHELa (PtoWUSa) is involved in developmental plasticity of adventitious root in poplar. Genes 11, 176. doi: 10.3390/genes11020176
Li, J., Zhang, J., Jia, H., Liu, B., Sun, P., Hu, J., et al. (2018). The WUSCHEL-related homeobox 5a (PtoWOX5a) is involved in adventitious root development in poplar. Tree Physiol. 38, 139–153. doi: 10.1093/treephys/tpx118
Li, K., Liu, Z., Xing, L., Wei, Y., Mao, J., Meng, Y., et al. (2019). miRNAs associated with auxin signaling, stress response, and cellular activities mediate adventitious root formation in apple rootstocks. Plant Physiol. Biochem. 139, 66–81. doi: 10.1016/j.plaphy.2019.03.006
Li, M. Y., Cao, Z. Y., Shen, W. B., and Cui, J. (2011). Molecular cloning and expression of a cucumber (Cucumis sativus L.) heme oxygenase-1 gene, CsHO1, which is involved in adventitious root formation. Gene 486, 47–55. doi: 10.1016/j.gene.2011.07.008
Li, S. W., Leng, Y., and Shi, R. F. (2017). Transcriptomic profiling provides molecular insights into hydrogen peroxide-induced adventitious rooting in mung bean seedlings. BMC Genomics 18:188. doi: 10.1186/s12864-017-3576-y
Li, S. W., Leng, Y., and Shi, R. F. (2020). Transcriptome characterization of gene profiling during early stage of nitric oxide-induced adventitious rooting in mung bean seedlings. J. Plant Growth Regul. 39, 430–455. doi: 10.1007/s00344-019-09993-y
Li, S. W., Shi, R. F., Leng, Y., and Zhou, Y. (2016). Transcriptomic analysis reveals the gene expression profile that specifically responds to IBA during adventitious rooting in mung bean seedlings. BMC Genomics 17:43. doi: 10.1186/s12864-016-2372-4
Li, S. W., Xue, L., Xu, S., Feng, H., and An, L. (2009). Mediators, genes and signaling in adventitious rooting. Bot. Rev. 75, 230–247. doi: 10.1007/s12229-009-9029-9
Li, Y. H., Zou, M. H., Feng, B.-H., Huang, X., Zhang, Z., and Sun, G. M. (2012). Molecular cloning and characterization of the genes encoding an auxin efflux carrier and the auxin influx carriers associated with the adventitious root formation in mango (Mangifera indica L.) cotyledon segments. Plant Physiol. Biochem. 55, 33–42. doi: 10.1016/j.plaphy.2012.03.012
Lin, Y., Li, M., Huang, L., Shen, W., and Ren, Y. (2012). Involvement of heme oxygenase-1 in b-cyclodextrin–hemin complex-induced cucumber adventitious rooting process. Plant Cell Rep. 31, 1563–1572. doi: 10.1007/s00299-012-1270-8
Lindrotha, A. M., Kvarnheden, A., and von Arnold, S. (2001). Isolation of a PSTAIRE CDC2 cDNA from Pinus contorta and its expression during adventitious root development. Plant Physiol. Biochem. 39, 107–114. doi: 10.1016/s0981-9428(00)01229-8
Lischweski, S., Muchow, A., Guthörl, D., and Hause, B. (2015). Jasmonates act positively in adventitious root formation in petunia cuttings. BMC Plant Biol. 15:229. doi: 10.1186/s12870-015-0615-1
Liu, H., Wang, S., Yu, X., Yu, J., He, X., Zhang, S., et al. (2005). ARL1, a LOB-domain protein required for adventitious root formation in rice. Plant J. 43, 47–56. doi: 10.1111/j.1365-313x.2005.02434.x
Liu, J., Sheng, L., Xu, Y., Li, J., Yang, Z., Huang, H., et al. (2014). WOX11 and 12 are involved in the first-step cell fate transition during de novo root organogenesis in Arabidopsis. Plant Cell 26, 1081–1093. doi: 10.1105/tpc.114.122887
Liu, S., Wang, J., Wang, L., Wang, X., Xue, Y., Wu, P., et al. (2009). Adventitious root formation in rice requires OsGNOM1 and is mediated by the OsPINs family. Cell Res. 9, 1110–1119. doi: 10.1038/cr.2009.70
Liu, S., Xue, Y., Wang, X., Zhang, B., Bi, Y., Qiu, M., et al. (2011). A dominant mutation in ARL2 causes impaired adventitious root development in rice. J. Plant Biol. 54, 227–236. doi: 10.1007/s12374-011-9160-0
Liu, S., Yang, C., Wu, L., Cai, H., Li, H., and Xu, M. (2020). The peu-miR160a-PeARF17.1/PeARF17.2 module participates in the adventitious root development of poplar. Plant Biotech. J. 18, 457–469. doi: 10.1111/pbi.13211
Ljung, K., Hull, A. K., Kowalczyk, M., Marchant, A., Celenza, J., and Cohen, J. D. (2002). Biosynthesis, conjugation, catabolism and homeostasis of indole-3-acetic acid in Arabidopsis thaliana. Plant Mol. Biol. 50, 309–332.
Lucas, M., Swarup, R., Paponov, I. A., Swarup, K., Casimiro, I., Lake, D., et al. (2011). SHORT-ROOT regulates primary, lateral, and adventitious root development in Arabidopsis. Plant Physiol. 155, 384–398. doi: 10.1104/pp.110.165126
Ludwig-Muller, J., Julke, S., Bierfreund, N. M., Decker, E. L., and Reski, R. (2009). Moss (Physcomitrella patens) GH3 proteins act in auxin homeostasis. New Phytol. 181, 323–338. doi: 10.1111/j.1469-8137.2008.02677.x
Ludwig-Muller, J., Vertocnik, A., and Town, C. D. (2005). Analysis of indole-3-butyric acid-induced adventitious root formation on Arabidopsis stem segments. J. Exp. Bot. 56, 2095–2105. doi: 10.1093/jxb/eri208
Lund, S. T., Smith, A. C., and Hackett, W. P. (1997). Differential gene expression in response to auxin treatment in the wild type and rac, an adventitious rooting-1 competent mutant of tobacco. Plant Physiol. 114, 1197–1206. doi: 10.1104/pp.114.4.1197
Ma, X., Liang, X., Lv, S., Guan, T., Jiang, T., and Cheng, Y. (2020). Histone deacetylase gene PtHDT902 modifies adventitious root formation and negatively regulates salt stress tolerance in poplar. Plant Sci. 290, 110301. doi: 10.1016/j.plantsci.2019.110301
Macedo, E. S., Cardoso, H. G., Hernandez, A., Peixe, A. A., Polidoros, A., Ferreira, A., et al. (2009). Physiologic responses and gene diversity indicate olive alternative oxidase as a potential source for markers involved in efficient adventitious root induction. Physiol. Plant 137, 532–552. doi: 10.1111/j.1399-3054.2009.01302.x
Macedo, E. S., Sircar, D., Cardoso, H. G., Peixe, A., and Arnholdt-Schmitt, B. (2012). Involvement of alternative oxidase (AOX) in adventitious rooting of Olea europaea L. microshoots is linked to adaptive phenylpropanoid and lignin metabolism. Plant Cell Rep. 31, 1581–1590. doi: 10.1007/s00299-012-1272-6
Mashiguchi, K., Tanaka, K., Sakai, T., Sugawara, S., Kawaide, H., Natsume, M., et al. (2011). The main auxin biosynthesis pathway in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 108, 18512–18517. doi: 10.1073/pnas.1108434108
Mauriat, M., Petterle, A., Bellini, C., and Moritz, T. (2014). Gibberellins inhibit adventitious rooting in hybrid aspen and Arabidopsis by affecting auxin transport. Plant J. 78, 372–384. doi: 10.1111/tpj.12478
Merkouropoulos, G., and Shirsat, A. H. (2003). The unusual Arabidopsis extensin gene at Ext1 is expressed throughout plant development and is induced by a variety of biotic and abiotic stresses. Planta 217, 356–366. doi: 10.1007/s00425-003-1002-y
Mews, M., Baluska, F., and Volkmann, D. (1996). Tissue- and development-specific distribution of PSTAIR-proteins in cells of control and wounded maize root apices. J. Exp. Bot. 47, 819–829.
Mikkelsen, M. D., Naur, P., and Halkier, B. A. (2004). Arabidopsis mutants in the C-S lyase of glucosinolate biosynthesis establish a critical role for indole-3-acetaldoxime in auxin homeostasis. Plant J. 37, 770–777. doi: 10.1111/j.1365-313x.2004.02002.x
Morel, J. B., Godon, C., Mourrain, P., Beclin, C., Boutet, S., Feuerbach, F., et al. (2002). Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in post-transcriptional gene silencing and virus resistance. Plant Cell 14, 629–639. doi: 10.1105/tpc.010358
Morita, Y., and Kyozuka, J. (2007). Characterization of OsPID (the rice ortholog of PINOID) and its possible involvement in the control of polar auxin transport. Plant Cell Physiol. 48, 540–549. doi: 10.1093/pcp/pcm024
Moriuchi, H., Okamoto, C., Nishihama, R., Yamashita, I., Machida, Y., and Tanaka, N. (2004). Nuclear localization and interaction of RolB with plant 14-3-3 proteins correlates with induction of adventitious roots by the oncogene rolB. Plant J. 38, 260–275. doi: 10.1111/j.1365-313x.2004.02041.x
Moubayidin, L., Di Mambro, R., and Sabatini, S. (2009). Cytokinin–auxin crosstalk. Trends Plant Sci. 14, 557–562. doi: 10.1016/j.tplants.2009.06.010
Mravec, J., Kubeš, M., Bielach, A., Gaykova, V., Petrašek, J., Skůpa, P., et al. (2008). Interaction of PIN and PGP transport mechanisms in auxin distribution-dependent development. Development 135, 3345–3354. doi: 10.1242/dev.021071
Negi, S., Ivanchenko, M. G., and Muday, G. K. (2008). Ethylene regulates lateral root formation and auxin transport in Arabidopsis thaliana. Plant J. 55, 175–187. doi: 10.1111/j.1365-313x.2008.03495.x
Negi, S., Sukumar, P., Liu, X., Cohen, J. D., and Muday, G. K. (2010). Genetic dissection of the role of ethylene in regulating auxin-dependent lateral and adventitious root formation in tomato. Plant J. 61, 3–15. doi: 10.1111/j.1365-313x.2009.04027.x
Nishimura, C., Ohashi, Y., Sato, S., Kato, T., Tabata, S., and Ueguchi, C. (2004). Histidine kinase homologs that act as cytokinin receptors possess overlapping functions in the regulation of shoot and root growth in Arabidopsis. Plant Cell 16, 1365–1377. doi: 10.1105/tpc.021477
Nole-Wilson, S., Tranby, T. L., and Krizek, B. A. (2005). AINTEGUMENTA-like (AIL) genes are expressed in young tissues and may specify meristematic or division-competent states. Plant Mol. Biol. 57, 613–628. doi: 10.1007/s11103-005-0955-6
Ohbayashi, I., Konishi, M., Ebine, K., and Sugiyama, M. (2011). Genetic identification of Arabidopsis RID2 as an essential factor involved in pre-rRNA processing. Plant J. 67, 49–60. doi: 10.1111/j.1365-313x.2011.04574.x
Osmont, K. S., Sibout, R., and Hardtke, C. S. (2007). Hidden branches, developments in root system architecture. Annu. Rev. Plant Biol. 58, 93–113. doi: 10.1146/annurev.arplant.58.032806.104006
Paciorek, T., Zazimalova, E., Ruthardt, N., Petršek, J., Stierhof, Y. D., Kleine-Vehn, J., et al. (2005). Auxin inhibits endocytosis and promotes its own efflux from cells. Nature 435, 1251–1256. doi: 10.1038/nature03633
Pacurar, D. I., Pacurar, M. L., Bussell, J. D., Schwambach, J., Pop, T. I., Kowalczyk, M., et al. (2014). Identification of new adventitious rooting mutants amongst suppressors of the Arabidopsis thaliana superroot2 mutation. J. Exp. Bot. 65, 1605–1618. doi: 10.1093/jxb/eru026
Paponov, I. A., Paponov, M., Teale, W., Menges, M., Chakrabortee, S., Murray, J. A. H., et al. (2008). Comprehensive transcriptome analysis of auxin responses in Arabidopsis. Mol. Plant 1, 321–337. doi: 10.1093/mp/ssm021
Passarinho, P., Ketelaar, T., Xing, M., van Arkel, J., Maliepaard, C., Hendriks, M. W., et al. (2008). BABY BOOM target genes provide diverse entry points into cell proliferation and cell growth pathways. Plant Mol. Biol. 68, 225–237. doi: 10.1007/s11103-008-9364-y
Pattison, R. J., and Catala, C. (2012). Evaluating auxin distribution in tomato (Solanum lycopersicum) through an analysis of the PIN and AUX/LAX gene families. Plant J. 70, 585–598. doi: 10.1111/j.1365-313x.2011.04895.x
Peixe, A., Raposo, A., Lourenco, R., Cardoso, H., and Macedo, E. (2007). Coconut water and BAP successfully replaced zeatin in olive (Olea europaea L.) micropropagation. Sci. Hortic. 113, 1–7. doi: 10.1016/j.scienta.2007.01.011
Péret, B., Swarup, K., Ferguson, A., Seth, M., Yang, Y., Dhondt, S., et al. (2012). AUX/LAX genes encode a family of auxin influx transporters that perform distinct functions during Arabidopsis development. Plant Cell 24, 2874–2885. doi: 10.1105/tpc.112.097766
Pierre-Jerome, E., Moss, B. L., Lanctot, A., Hageman, A., and Nemhauser, J. L. (2016). Functional analysis of molecular interactions in synthetic auxin response circuits. Proc. Natl. Acad. Sci. U.S.A. 113, 11354–11359. doi: 10.1073/pnas.1604379113
Quint, M., Barkawi, L. S., Fan, K.-T., Cohen, J. D., and Gray, W. M. (2009). Arabidopsis IAR4 modulates auxin response by regulating auxin homeostasis. Plant Physiol. 150, 748–758. doi: 10.1104/pp.109.136671
Ramirez-Carvajal, G. A., and Davis, J. M. (2010). Cutting to the base identifying regulators of adventitious rooting. Plant Signal. Behav. 5, 281–283. doi: 10.4161/psb.5.3.10705
Ramirez-Carvajal, G. A., Morse, A. M., Dervinis, C., and Davis, J. M. (2009). The cytokinin type-B response regulator PtRR13 is a negative regulator of adventitious root development in Populus. Plant Physiol. 150, 759–771. doi: 10.1104/pp.109.137505
Rigal, A., Yordanov, Y. S., Perrone, I., Karlberg, A., Tisserant, E., Bellini, C., et al. (2012). The AINTEGUMENTA LIKE1 homeotic transcription factor PtAIL1 controls the formation of adventitious root primordia in poplar. Plant Physiol. 160, 1996–2006. doi: 10.1104/pp.112.204453
Rovere, D. F., Fattorini, L., D’Angeli, S., Veloccia, A., Falasca, G., and Altamura, M. M. (2013). Auxin and cytokinin control formation of the quiescent centre in the adventitious root apex of Arabidopsis. Ann. Bot. 112, 1395–1407. doi: 10.1093/aob/mct215
Rovere, D. F., Fattorini, L., Ronzan, M., Falasca, G., and Altamura, M. M. (2016). The quiescent centre and the stem cell niche in the adventitious roots of Arabidopsis thaliana. Plant Signal. Behav. 11, e1176660. doi: 10.1080/15592324.2016.1176660
Ruzicka, K., Ljung, K., Vanneste, S., Podhorska, R., Beeckman, T., Friml, J., et al. (2007). Ethylene regulates root growth through effects on auxin biosynthesis and transport-dependent auxin distribution. Plant Cell 19, 2197–2212. doi: 10.1105/tpc.107.052126
Sanchez, C., Vielba, J. M., Ferro, E., Covelo, G., Sole, A., Abarca, D., et al. (2007). Two SCARECROW-LIKE genes are induced in response to exogenous auxin in rooting-competent cuttings of distantly related forest species. Tree Physiol. 27, 1459–1470. doi: 10.1093/treephys/27.10.1459
Sarkar, A. K., Luijten, M., Miyashima, S., Lenhard, M., Hashimoto, T., Nakajima, K., et al. (2007). Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature 446, 811–814. doi: 10.1038/nature05703
Shin, R., Burch, A. Y., Huppert, K. A., Tiwari, S. B., Murphy, A. S., Guilfoyle, T. J., et al. (2007). The Arabidopsis transcription factor MYB77 modulates auxin signal transduction. Plant Cell 19, 2440–2453. doi: 10.1105/tpc.107.050963
Shu, W., Zhou, H., Jiang, C., Zhao, S., Wang, L., Li, Q., et al. (2019). The auxin receptor TIR1 homolog (PagFBL 1) regulates adventitious rooting through interactions with Aux/IAA28 in Populus. Plant Biotech. J. 17, 338–349. doi: 10.1111/pbi.12980
Shuai, B., Reynaga, C. G., and Springer, P. S. (2002). The LATERAL ORGAN BOUNDARIES gene defines a novel, plant-specific gene family. Plant Physiol. 129, 747–761. doi: 10.1104/pp.010926
Smith, D. L., and Fedoroff, N. V. (1995). LRP1, a gene expressed in lateral and adventitious root primordia of Arabidopsis. Plant Cell 7, 735–745. doi: 10.2307/3870175
Smolka, A., Welander, M., Olsson, P., Holefors, A., and Zhu, L. H. (2009). Involvement of the ARRO-1 gene in adventitious root formation in apple. Plant Sci. 177, 710–715. doi: 10.1016/j.plantsci.2009.09.009
Solé, A., Sánchez, C., Vielba, J. M., Valladares, S., Abarca, D., and Díaz-Sala, C. (2008). Characterization and expression of a Pinus radiata putative ortholog to the Arabidopsis SHORT-ROOT gene. Tree Physiol. 28, 1629–1639. doi: 10.1093/treephys/28.11.1629
Sorin, C., Bussell, J. D., Camus, I., Ljung, K., Kowalczyk, M., Geiss, G., et al. (2005). Auxin and light control of adventitious rooting in Arabidopsis require ARGONAUTE1. Plant Cell 17, 1343–1359. doi: 10.1105/tpc.105.031625
Sorin, C., Negroni, L., Balliau, T., Corti, H., Jacquemot, M. P., Davanture, M., et al. (2006). Proteomic analysis of different mutant genotypes of Arabidopsis led to the identification of 11 proteins correlating with adventitious root development. Plant Physiol. 140, 349–364. doi: 10.1104/pp.105.067868
Sozzani, R., Cui, H., Moreno-Risueno, M. A., Busch, W., Van Norman, J. M., Vernoux, T., et al. (2010). Spatiotemporal regulation of cell-cycle genes by SHORT-ROOT links patterning and growth. Nature 466, 128–132. doi: 10.1038/nature09143
Staswick, P. E. (2009). The tryptophan conjugates of jasmonic and indole-3-acetic acids are endogenous auxin inhibitors. Plant Physiol. 150, 1310–1321. doi: 10.1104/pp.109.138529
Staswick, P. E., Serban, B., Rowe, M., Tiryaki, I., Maldonado, M. T., Maldonado, M. C., et al. (2005). Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell 17, 616–627. doi: 10.1105/tpc.104.026690
Steffens, B., and Rasmussen, A. (2016). The physiology of adventitious roots. Plant Physiol. 170, 603–617. doi: 10.1104/pp.15.01360
Stepanova, A. N., Hoyt, J. M., Hamilton, A. A., and Alonso, J. M. (2005). A Link between ethylene and auxin uncovered by the characterization of two root-specific ethylene-insensitive mutants in Arabidopsis. Plant Cell 17, 2230–2242. doi: 10.1105/tpc.105.033365
Stepanova, A. N., Yun, J., Likhacheva, A. V., and Alonso, J. M. (2007). Multilevel interactions between ethylene and auxin in Arabidopsis roots. Plant Cell 19, 2169–2185. doi: 10.1105/tpc.107.052068
Stevens, M. E., Woeste, K. E., and Pijut, P. M. (2017). Localized gene expression changes during adventitious root formation in black walnut (Juglans nigra L.). Tree Physiol. 38, 877–894. doi: 10.1093/treephys/tpx175
Su, Y. H., Liu, Y. B., and Zhang, X. S. (2011). Auxin–cytokinin interaction regulates meristem development. Mol. Plant 4, 616–625. doi: 10.1093/mp/ssr007
Sugiyama, M. (2003). Isolation and initial characterization of temperature-sensitive mutants of Arabidopsis thaliana that are impaired in root redifferentiation. Plant Cell Physiol. 44, 588–596. doi: 10.1093/pcp/pcg077
Sukumar, P., Maloney, G. S., and Muday, G. K. (2013). Localized induction of the ATP-binding cassette B19 auxin transporter enhances adventitious root formation in Arabidopsis. Plant Physiol. 162, 1392–1405. doi: 10.1104/pp.113.217174
Sun, S., Yu, J. P., Chen, F., Zhao, T. J., Fang, X. H., Li, Y. Q., et al. (2008). TINY, a dehydration-responsive element (DRE)-binding protein-like transcription factor connecting the DRE- and ethylene-responsive element-mediated signaling pathways in Arabidopsis. J. Biol. Chem. 283, 6261–6271. doi: 10.1074/jbc.m706800200
Swarup, R., Perry, P., Hagenbeek, D., Van Der Straeten, D., Beemster, G. T., Sandberg, G., et al. (2007). Ethylene upregulates auxin biosynthesis in Arabidopsis seedlings to enhance inhibition of root cell elongation. Plant Cell 19, 2186–2196. doi: 10.1105/tpc.107.052100
Taramino, G., Sauer, M., Stauffer, J. L., Multani, D., Niu, X., Sakai, H., et al. (2007). The maize, Zea mays L. RTCS gene encodes a LOB domain protein that is a key regulator of embryonic seminal and post-embryonic shoot-borne root initiation. Plant J. 50, 649–659. doi: 10.1111/j.1365-313x.2007.03075.x
Thines, B., Katsir, L., Melotto, M., Niu, Y., Mandaokar, A., Liu, G. H., et al. (2007). JAZ repressor proteins are targets of the SCF (COI1) complex during jasmonate signalling. Nature 448, 661–665. doi: 10.1038/nature05960
Thomas, P., Lee, M. M., and Schiefelbein, J. (2003). Molecular identification of proline-rich protein genes induced during root formation in grape (Vitis vinifera L.) stem cuttings. Plant Cell Environ. 26, 1497–1504. doi: 10.1046/j.1365-3040.2003.01071.x
Trupiano, D., Yordanov, Y., Regan, S., Meilan, R., Tschaplinski, T., Scippa, G. S., et al. (2013). Identification, characterization of an AP2/ERF transcription factor that promotes adventitious, lateral root formation in Populus. Planta 238, 271–282. doi: 10.1007/s00425-013-1890-4
Ueguchi-Tanaka, M., Ashikari, M., Nakajima, M., Itoh, H., Katoh, E., Kobayashi, M., et al. (2005). GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437, 693–698. doi: 10.1038/nature04028
Vaucheret, H., Vazquez, F., Crete, P., and Bartel, D. P. (2004). The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev. 18, 1187–1197. doi: 10.1101/gad.1201404
Velada, I., Cardoso, H., Porfirio, S., and Peixe, A. (2020). Expression profile of PIN-formed auxin efflux carrier genes during IBA-induced in vitro adventitious rooting in Olea europaea L. Plants 9, 185. doi: 10.3390/plants9020185
Veloccia, A., Fattorini, L., Della Rovere, F., Sofo, A., D’Angeli, S., Betti, C., et al. (2016). Ethylene and auxin interaction in the control of adventitious rooting in Arabidopsis thaliana. J. Exp. Bot. 67, 6445–6458. doi: 10.1093/jxb/erw415
Vernoux, T., and Benfey, P. N. (2005). Signals that regulate stem cell activity during plant development. Curr. Opin. Genet. Dev. 15, 388–394. doi: 10.1016/j.gde.2005.06.008
Vernoux, T., Kronenberger, J., Grandjean, O., Laufs, P., and Traas, J. (2000a). PINFORMED 1 regulates cell fate at the periphery of the shoot apical meristem. Development 127, 5157–5165.
Vernoux, T., Wilson, R. C., Seeley, K. A., Reichheld, J.-P., Muroy, S., Brown, S., et al. (2000b). The ROOT MERISTEMLESS1/CADMIUM SENSITIVE2 gene defines a glutathione-dependent pathway involved in initiation and maintenance of cell division during postembryonic root development. Plant Cell 12, 97–109. doi: 10.2307/3871032
Vidoz, M. L., Loreti, E., Mensuali, A., Alpi, A., and Perata, P. (2010). Hormonal interplay during adventitious root formation in flooded tomato plants. Plant J. 63, 551–562. doi: 10.1111/j.1365-313x.2010.04262.x
Vielba, J. M., Díaz-Sala, C., Ferro, E., Rico, S., Lamprecht, M., Abarca, D., et al. (2011). CsSCL1 is differentially regulated upon maturation in chestnut microshoots and is specifically expressed in rooting-competent cells. Tree Physiol. 31, 1152–1160. doi: 10.1093/treephys/tpr086
Vieten, A., Vanneste, S., Wisniewska, J., Benkova, E., Benjamins, R., Beeckman, T., et al. (2005). Functional redundancy of PIN proteins is accompanied by auxin-dependent cross-regulation of PIN expression. Development 132, 4521–4531. doi: 10.1242/dev.02027
Wang, H., Xie, Y., Liu, W., Tao, G., Sun, C., Sun, X., et al. (2020). Transcription factor LkWOX4 is involved in adventitious root development in Larix kaempferi. Gene 758, 144942. doi: 10.1016/j.gene.2020.144942
Wang, X. F., He, F. F., Ma, X. X., Mao, C. Z., Hodgman, C., Lu, C. G., et al. (2011). OsCAND1 is required for crown root emergence in rice. Mol. Plant 4, 289–299. doi: 10.1093/mp/ssq068
Welander, M., Geier, T., Smolka, A., Ahlman, A., Fan, J., and Zhu, L. H. (2014). Origin, timing, and gene expression profile of adventitious rooting in Arabidopsis hypocotyles and stems. Am. J. Bot. 101, 255–266. doi: 10.3732/ajb.1300258
Wildwater, M., Campilho, A., Perez- Perez, J. M., Heidstra, R., Blilou, I., Korthout, H., et al. (2005). The RETINOBLASTOMA-RELATED gene regulates stem cell maintenance in Arabidopsis roots. Cell 123, 1337–1349. doi: 10.1016/j.cell.2005.09.042
Willemsen, V., Wolkenfelt, H., de Vrieze, G., Weisbeek, P., and Scheres, B. (1998). The HOBBIT gene is required for formation of the root meristem in the Arabidopsis embryo. Development 125, 521–531. doi: 10.1111/j.1365-2443.2008.01186.x
Xiao, Z., Zhang, Y., Liu, M., Zhan, C., Yang, X., Nvsvrot, T., et al. (2020). Coexpression analysis of a large-scale transcriptome identified a calmodulin-like protein regulating the development of adventitious roots in poplar. Tree Physiol. 40, 1405–1419. doi: 10.1093/treephys/tpaa078
Xu, J., Hofhuis, H., Heidstra, R., Sauer, M., Friml, J., and Scheres, B. (2006). A molecular framework for plant regeneration. Science 311, 385–388. doi: 10.1126/science.1121790
Xu, L. (2018). De novo root regeneration from leaf explants: wounding, auxin, and cell fate transition. Curr. Opin. Plant Biol. 41, 39–45. doi: 10.1016/j.pbi.2017.08.004
Xu, M., Liu, S., Xuan, L., Huang, M., and Wang, Z. (2016). Isolation and characterization of a poplar D-type cyclin gene associated with the SHORTROOT/SCARECROW network. Trees 30, 255–263. doi: 10.1007/s00468-015-1296-y
Xu, M., Xie, W., and Huang, M. (2012). Overexpression of PeRHD3 alters the root architecture in Populus. Biochem. Bioph. Res. Commun. 424, 239–244. doi: 10.1016/j.bbrc.2012.06.083
Xu, M., Xie, W., and Huang, M. (2015). Two WUSCHEL-related HOMEOBOX genes, PeWOX11a and PeWOX11b, are involved in adventitious root formation of poplar. Physiol. Plant 155, 446–456. doi: 10.1111/ppl.12349
Xu, M., Zhu, L., Shou, H., and Wu, P. (2005). A PIN1 Family Gene, OsPIN1, involved in auxin-dependent adventitious root emergence and tillering in rice. Plant Cell Physiol. 46, 1674–1681. doi: 10.1093/pcp/pci183
Xu, X., Li, X., Hu, X., Wu, T., Wang, Y., Xu, X., et al. (2017). High miR156 expression is required for auxin-induced adventitious root formation via MxSPL26 independent of PINs and ARFs in Malus xiaojinensis. Front. Plant Sci. 8:1059. doi: 10.3389/fpls.2017.01059
Xuan, W., Zhu, F. Y., Xu, S., Huang, B. K., Ling, T. F., Qi, J. Y., et al. (2008). The heme oxygenase/carbon monoxide system is involved in the auxin-induced cucumber adventitious rooting process. Plant Physiol. 148, 881–893. doi: 10.1104/pp.108.125567
Yamamoto, Y., Kamiya, N., Morinaka, Y., Matsuoka, M., and Sazuka, T. (2007). Auxin biosynthesis by the YUCCA genes in rice. Plant Physiol. 143, 1362–1371. doi: 10.1104/pp.106.091561
Yang, Y., Hammes, U. Z., Taylor, C. G., Schachtman, D. P., and Nielsen, E. (2006). High-affinity auxin transport by the AUX1 influx carrier protein. Curr. Biol. 16, 1–5.
Ye, B. B., Shang, G. D., Pan, Y., Xu, Z. G., Zhou, C. M., Mao, Y. B., et al. (2020a). AP2/ERF transcription factors integrate age and wound signals for root regeneration. Plant Cell 32, 226–241. doi: 10.1105/tpc.19.00378
Ye, B. B., Zhang, K., and Wang, J. W. (2020b). The role of miR156 in rejuvenation in Arabidopsis thaliana. J. Integr. Plant Biol. 62, 550–555. doi: 10.1111/jipb.12855
Yin, H., Li, M., Lv, M., Hepworth, S. R., Li, D., Ma, C., et al. (2020). SAUR15 promotes lateral and adventitious root development via activating H+-ATPases and auxin biosynthesis. Plant Physiol. 184, 837–851. doi: 10.1104/pp.19.01250
Zhang, G. F., Zhao, F., Chen, Y. Q., Pan, Y., Sun, L., Bao, N., et al. (2019). Jasmonate-mediated wound signalling promotes plant regeneration. Nature Plants 5, 491–497. doi: 10.1038/s41477-019-0408-x
Zhang, X., Zou, Z., Zhang, J., Zhang, Y., Han, Q., Hu, T., et al. (2011). Over-expression of sly-miR156a in tomato results in multiple vegetative and reproductive trait alterations and partial phenocopy of the sft mutant. FEBS Lett. 585, 435–439. doi: 10.1016/j.febslet.2010.12.036
Zhang, Y., Yang, X., Cao, P., Xiao, Z., Zhan, C., Liu, M., et al. (2020). The bZIP53–IAA4 module inhibits adventitious root development in Populus. J. Exp. Bot. 71, 3485–3498. doi: 10.1093/jxb/eraa096
Keywords: molecular bases, adventitious rooting, auxin, cytokinin, ethylene, hormone crosstalk
Citation: Li S-W (2021) Molecular Bases for the Regulation of Adventitious Root Generation in Plants. Front. Plant Sci. 12:614072. doi: 10.3389/fpls.2021.614072
Received: 05 October 2020; Accepted: 08 January 2021;
Published: 28 January 2021.
Edited by:
Uwe Druege, Research Centre for Horticultural Crops, University of Applied Sciences Erfurt, GermanyReviewed by:
Riccardo Di Mambro, University of Pisa, ItalyLin Xu, Center for Excellence in Molecular Plant Sciences, Chinese Academy of Sciences (CAS), China
Copyright © 2021 Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shi-Weng Li, bGlzaHdlbmdAbWFpbC5semp0dS5jbg==
 Shi-Weng Li
Shi-Weng Li