- 1Key Laboratory of Plant Biotechnology in Universities of Shandong Province, College of Life Sciences, Qingdao Agricultural University, Qingdao, China
- 2College of Agronomy, Qingdao Agricultural University, Qingdao, China
CONSTANS-LIKE (COL) genes play important roles in the regulation of plant growth and development, and they have been analyzed in many plant species. However, few studies have examined COL genes in mungbean (Vigna radiata). In this study, we identified and characterized 31 mungbean genes whose proteins contained B-Box domains. Fourteen were designated as VrCOL genes and were distributed on 7 of the 11 mungbean chromosomes. Based on their phylogenetic relationships, VrCOLs were clustered into three groups (I, II, and III), which contained 4, 6, and 4 members, respectively. The gene structures and conserved motifs of the VrCOL genes were analyzed, and two duplicated gene pairs, VrCOL1/VrCOL2 and VrCOL8/VrCOL9, were identified. A total of 82 cis-acting elements were found in the VrCOL promoter regions, and the numbers and types of cis-acting elements in each VrCOL promoter region differed. As a result, the expression patterns of VrCOLs varied in different tissues and throughout the day under long-day and short-day conditions. Among these VrCOL genes, VrCOL2 showed a close phylogenetic relationship with Arabidopsis thaliana CO and displayed daily oscillations in expression under short-day conditions but not long-day conditions. In addition, overexpression of VrCOL2 accelerated flowering in Arabidopsis under short-day conditions by affecting the expression of the flowering time genes AtFT and AtTSF. Our study lays the foundation for further investigation of VrCOL gene functions.
Introduction
Flowering time is a key factor that influences crop growth and development, and crops achieve higher yields when they flower at the correct time. To regulate flowering time, crops sense the interactions between endogenous and environmental factors to determine the transition from vegetative to reproductive growth (Wickland and Hanzawa, 2015; Beinecke et al., 2018; Eom et al., 2018; Xu and Chong, 2018). Several functional pathways have been identified that regulate the switch from vegetative to reproductive development. These include the photoperiodic, vernalization, ambient temperature, plant hormone, and autonomous flowering pathways (Boss et al., 2004; Jack, 2004; Baurle and Dean, 2006; Wickland and Hanzawa, 2015; Xu and Chong, 2018; Ronald and Davis, 2019; Taylor et al., 2019; Zhang et al., 2019). A number of genes in these pathways are known to be involved in flowering time regulation, including CONSTANS-LIKE (COL) genes, phosphatidyl ethanolamine-binding protein (PEBP) genes, and several members of the MADS-box gene family (Gangappa and Botto, 2014; Wickland and Hanzawa, 2015; Beinecke et al., 2018; del-Olmo et al., 2019; Jin et al., 2019, 2020; Jing et al., 2019; Lee et al., 2019; Nam et al., 2019; Ning et al., 2019; Parenicova et al., 2019).
COL genes belong to the zinc-finger transcription factor family and play central roles in plant growth and development (Khanna et al., 2009; Gangappa and Botto, 2014). COL proteins are identified based on their conserved structure, which includes one or two BBX (B-Box) domains and one CCT (CONSTANS, CO-like, and TIMING of CAB1) domain (Khanna et al., 2009; Gangappa and Botto, 2014). The BBX domain can be further divided into two types, B-Box1 and B-Box2, which are recognized by their consensus sequences and the distances between their zinc-binding residues, which are considered to be involved in protein-protein interactions (Khanna et al., 2009). The CCT domain has important functions in transcriptional regulation and nuclear protein transport (Robson et al., 2001; Khanna et al., 2009; Yan et al., 2011; Gangappa and Botto, 2014). The COL proteins are grouped into three classes based on the number and type of their conserved domains. Classes I and II have two distinct BBX domains and one CCT domain, whereas class III has only one BBX and one CCT domain. Classes I, II, and III contain 6, 7, and 4 members in Arabidopsis, respectively. In addition, several COL proteins contain valine-proline (VP) motifs in their C termini (Khanna et al., 2009; Gangappa and Botto, 2014).
Among these COL members, AtCO (AtBBX1) and its homologs are well studied in many plant species (Khanna et al., 2009; Gangappa and Botto, 2014; Luo et al., 2018; Luccioni et al., 2019; Serrano-Bueno et al., 2020). AtCO is expressed in a rhythmic manner and coordinates light pathway and circadian clock signal inputs in Arabidopsis (Putterill et al., 1993, 1995; Andres and Coupland, 2012; Song et al., 2013). Thus, AtCO plays an important role in the regulation of flowering time by the photoperiod-dependent pathway. Atco mutants exhibit delayed flowering time under long-day conditions (LD), but under short-day conditions (SD), their flowering times are similar to those of wild-type plants. By contrast, AtCO overexpression plants show early flowering time under both LD and SD conditions (Khanna et al., 2009; Gangappa and Botto, 2014). The AtCO protein binds to cis-acting elements in the promoter region of the flowering activator FLOWERING LOCUS T (AtFT) to active AtFT expression. Moreover, AtCO is regulated by many flowering factors, such as AtGI (GIGANTEA), AtCDF1 (CYCLING DOF FACTOR 1) and AtFKF1 (FLAVIN BINDING, KELCH REPEAT, F-BOX1) (Imaizumi et al., 2005; Sawa et al., 2007). OsHd1 (Heading date 1), the AtCO ortholog in rice, accelerates flowering under SD conditions but delays flowering under LD conditions through the regulation of the AtFT orthologs OsHd3a (Heading date 3a) and OsRFT1 (RICE FLOWERING LOCUS T1) (Yano et al., 2000; Komiya et al., 2008, 2009). The soybean AtCO orthologs GmCOL1, GmCOL2, GmCOL3, and GmCOL4 can complement the late flowering phenotype of Atco mutants (Wu et al., 2014). In addition to their functions in flowering time and circadian clock regulation, some COL proteins are also involved in abiotic or biotic stress responses, root development and stomatal opening (Khanna et al., 2009; Gangappa and Botto, 2014).
Mungbean is a diploid legume crop, and its seeds contain proteins and nutrients that are essential for human nutrition (Keatinge et al., 2011). The cultivated mungbean is thought to have been domesticated in India, from which it then spread to other areas (Fuller, 2007). Mungbean is considered to be an SD crop, and flowering time is a critical factor influencing its production (Vas Aggarwal and Poehlman, 1977; Imrie, 1996; Kim et al., 2015). Mungbean plants produce a large number of flowers, but only a few set pods. Approximately 70–90% of the flowers are shed, mainly the later-formed flowers of the racemes (Kumari and Verma, 1983; Mondal et al., 2011). Thus, it has been suggested that the prevention of late flowering is an important way to increase mungbean yield (Isobe et al., 1995; Kuroda et al., 1998; Mondal et al., 2011). The sequencing of the mungbean genome provides genetic resources for the investigation of gene functions (Kang et al., 2014), and the study of mungbean flowering time genes can therefore provide essential information for further modification of mungbean cultivars to increase yield. Until now, there has been limited information on the functions of genes involved in mungbean flowering time regulation. In this study, we identified mungbean COL genes and investigated their characteristics, including chromosomal distributions, gene structures, cis-acting elements and gene expression patterns. We also analyzed the functions of VrCOL2 in the regulation of flowering time. Our findings will provide useful information for further characterization of mungbean COL gene functions.
Materials and Methods
Plant Materials and Growth Conditions
The mungbean reference genome variety VC1973A was provided by Suk-Ha Lee at Seoul National University, Seoul, South Korea (Kang et al., 2014) and used for all experiments in this study. Mungbean seeds were geminated in tap water for 1 day and then planted in soil-filled pots. Seedlings were grown in growth chambers with 16 h 25°C light/8 h 25°C dark cycles for LD conditions and 10 h 25°C light/14 h 25°C dark cycles for SD conditions. Leaves of 5-week-old mungbean plants were sampled every 4 h after lights-on and used to analyze the diurnal rhythm of gene expression. Multiple tissues were collected from field-grown mungbean plants sown at the end of May in Qingdao, China, including roots, nodule roots, shoot apices, stems, leaves, flowers, pods and seeds (Shi et al., 2021). Tissues were collected in the afternoon (ZT 10–12) in early July for gene expression analysis, and all samples were stored at −80°C before RNA extraction. Arabidopsis plants were grown in growth chambers with 16 h 23°C light/8 h 21°C dark cycles for LD conditions and 10 h 23°C light/14 h 21°C dark cycles for SD conditions. Leaves of 2-week-old Arabidopsis were collected every 4 h after lights-on for gene expression analysis.
Identification of Mungbean VrCOL Members
The amino acid sequences of Arabidopsis BBXs were used as blast queries against the National Center for Biotechnology Information (NCBI) and mungbean genome databases1 to search for mungbean VrBBX proteins (Kang et al., 2014). The presence of conserved BBX and CCT domains in candidate genes was confirmed using the Pfam database and InterPro program with default parameters (Finn et al., 2017; El-Gebali et al., 2019).
Phylogenetic Analysis
The amino acid sequences of CO and COL proteins from Arabidopsis, soybean, Medicago, mungbean, rice, and maize were aligned using ClustalW2 (Oliver et al., 2005), and the resulting alignment was used to construct a phylogenetic tree in MEGA 7.0 using the neighbor-joining method with default parameters (Kumar et al., 2016). In addition, VrBBX proteins were aligned separately in ClustalW2 and used to construct a phylogenetic tree in MEGA 7.0 with the neighbor-joining method.
Chromosomal Distribution and Duplication Analyses
The physical positions of VrCOL genes were obtained from NCBI, and a chromosomal location map was constructed using MapInspect software (Mike Lischke, Berlin, Germany). Duplicated gene pairs were identified using OrthoMCL software as described by Fischer et al. (2011) and Jin et al. (2020). The duplicated gene pairs were defined as having greater than 60% amino acid sequence similarity and were visualized using Circos software (Krzywinski et al., 2009).
Analyses of Exon-Intron Organization, Conserved Domains, Sequence Logos, Protein Motifs, and Cis-Acting Elements
The genomic and CDS sequences of mungbean VrCOL genes were obtained from NCBI and used as inputs to the Gene Structure Display Server (GSDS) to analyze their gene structures (Hu et al., 2015). The full-length amino acid sequences of VrCOL proteins were used to analyze the positions of the conserved BBX and CCT domains using the InterPro program (Finn et al., 2017). The sequence logos of the conserved BBX1, BBX2, and CCT domains were analyzed using the WebLogo platform (Crooks et al., 2004). The conserved motifs present in the VrCOL proteins were identified using MEME tools, with an optimum motif width of 11–50 amino acid residues (Bailey et al., 2009). The cis-acting elements in each VrCOL promoter, 2 kb upstream of the initiation codon, were predicted by PlantCARE (Lescot et al., 2002).
Plasmid Construction and Plant Transformation
To investigate the functions of VrCOL2, a 35S: CDS-VrCOL2 plasmid was constructed. The VrCOL2 CDS was amplified from the cDNA of the sequenced mungbean variety VC1973A using primers with XhoI and XbaI digestion site sequences. The resulting PCR fragment was digested by the restriction endonucleases XhoI and XbaI to generate sticky ends. The pPTN1171 vector was digested with XhoI and XbaI to generate a linearized plasmid (Ping et al., 2014). Then the VrCOL2 and pPTN1171 fragments were ligated using T4 DNA ligase (Promega). The constructed plasmid was verified by sequencing. It was then introduced into Arabidopsis using the floral dip method (Bent, 2006), and successful transformation was confirmed by PCR. All primers are listed in Supplementary Table 1.
RNA Extraction and Transcription Analysis
RNA isolation and quantitative real-time PCR (qRT–PCR) analysis were carried out as described in Li et al. (2019). Gene expression levels were normalized to an Actin gene from mungbean (Vradi03g00210) (Li et al., 2019). Each sample was analyzed using three biological replicates. All primers are listed in Supplementary Table 1.
Results
Identification of VrCOL Genes in Mungbean
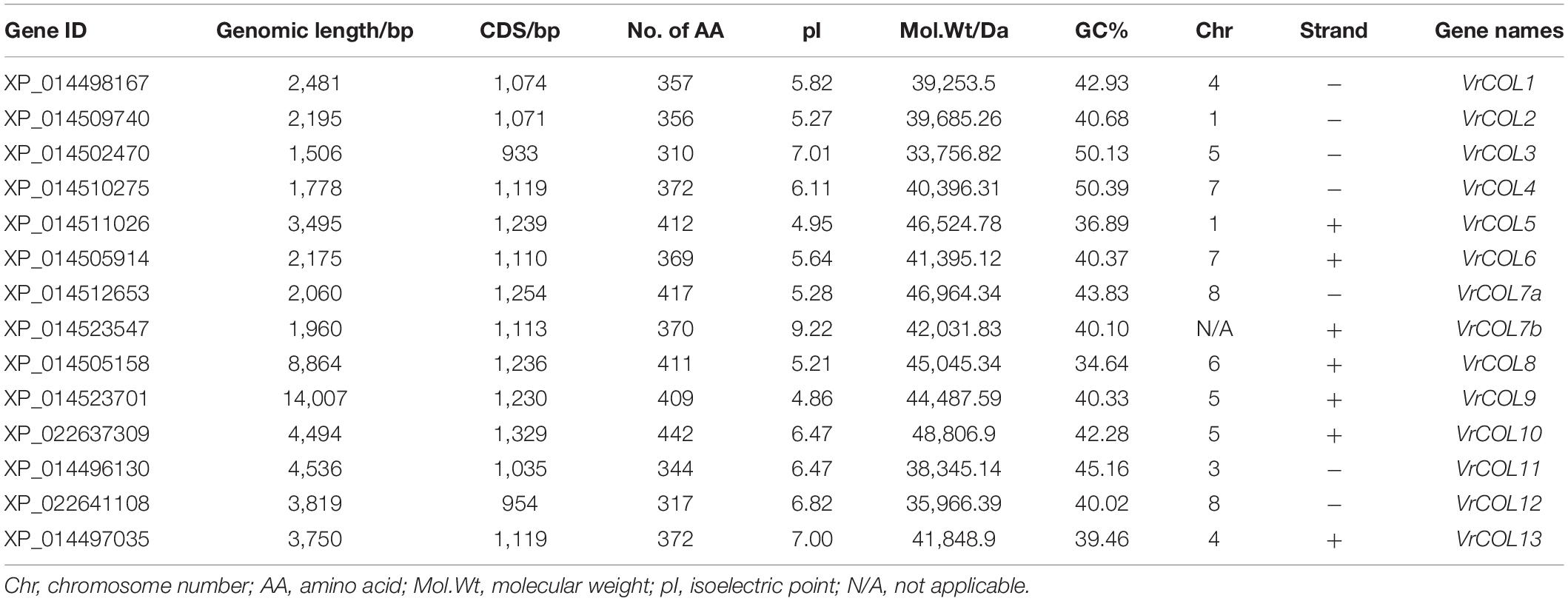
To search for mungbean VrCOL genes, we first identified mungbean proteins that contained BBX domains. The amino acid sequences of the conserved BBX domain (PF00643) and of Arabidopsis BBX proteins were used as blast queries against the mungbean genome database at NCBI. The presence of conserved BBX domains in each candidate mungbean gene was confirmed using Pfam and InterPro software, and a total of 31 VrBBX genes were identified in the mungbean genome (Figure 1). Among the VrBBX proteins, 17 contained only BBX domains, and 14 contained both BBX and CCT domains. The latter were designated VrCOL proteins (Figure 1 and Table 1). We then analyzed the numbers and types of BBX and CCT domains in the VrCOL proteins, and found two distinct BBX domains (BBX1 and BBX2) and one CCT domain (Supplementary Figure 1). Sequence logos of the BBX1 (CX2CX8CX4AXLCX2CDX3HX8HXR), BBX2 (CX2CX4AX3CX7CX2CDX3HX8H), and CCT (RYX2 KX3RX3KX2RYX2RKX2AX2RXR) domains were produced using WebLogo (Figure 2 and Supplementary Figure 1). Nine VrCOL proteins contained one BBX1, one BBX2, and one CCT domain, and five VrCOL proteins contained one BBX1 and one CCT domain (Figure 1 and Table 1).

Figure 1. Phylogenetic relationships and conserved domain analyses of the VrBBX proteins. (A) Phylogenetic relationship analysis of 31 VrBBX proteins. The amino acid sequences of mungbean proteins containing BBX domains were used to construct a phylogenetic tree with the neighbor-joining method. (B) The positions of conserved BBX1, BBX2, and CCT domains in the VrBBX proteins. The blue, purple and orange boxes in each VrBBX protein indicate the BBX1, BBX2, and CCT domains, respectively. (C) The positions of the conserved BBX1, BBX2, and CCT domains in each VrBBX protein.
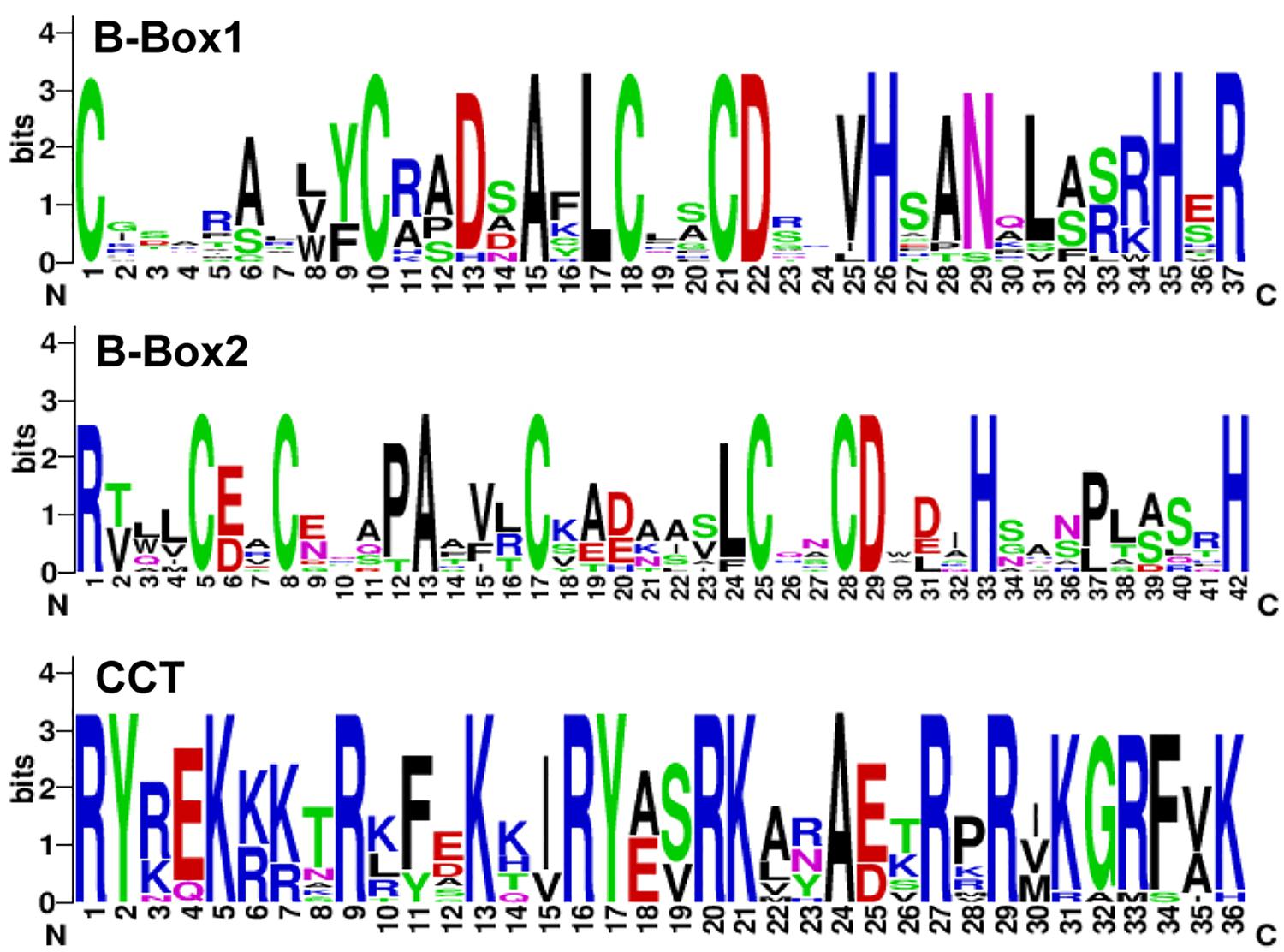
Figure 2. Sequence logos of the BBX1, BBX2, and CCT domains of VrCOL proteins. The conserved domains were analyzed using the WebLogo platform. The amino acid sequences of each conserved domain are presented on the x-axis, and the height of each letter corresponds to the conservation of each residue.
Multiple characteristics of the VrCOL members were analyzed based on their genomic and protein sequences (Table 1). The genomic lengths of VrCOL genes ranged from 1,506 (XP_014502470) to 14,007 bp (XP_014523701), the CDS lengths ranged from 933 (XP_014502470) to 1,329 bp (XP_022637309), and the amino acid numbers ranged from 310 to 442. The isoelectric points of VrCOL proteins varied from 4.86 (XP_014523701) to 9.22 (XP_014523547), and their molecular weights ranged from 33,756.82 Da (XP_014502470) to 48,806.9 Da (XP_022637309). The GC content, which influences gene stability to some degree, ranged from 34.64 to 50.39%, and 12 of the 14 VrCOL genes had lower than 50% GC content (Table 1).
Phylogenetic Analysis of the VrCOL Proteins
To analyze the evolutionary relationships among the VrCOL genes and obtain information from well-studied CO homologs in other species, a phylogenetic tree was constructed using 17 Arabidopsis, 26 soybean, 11 Medicago, 16 rice, 18 maize, and 14 mungbean CO and COL proteins (Gangappa and Botto, 2014; Wu et al., 2014, 2017; Hu et al., 2018). The VrCOL genes were named VrCOL1 to VrCOL13 based on their phylogenetic relationships with their soybean orthologs (Figure 3 and Table 1). The COL proteins were grouped into three classes based on their phylogenetic relationships (Khanna et al., 2009; Gangappa and Botto, 2014; Figure 3). Classes I, II, and III contained 4, 6, and 4 VrCOL members, respectively (Figure 3). The BBX1 and BBX2 domains were located close to one another in the class I and II proteins, with the exception of VrCOL10 (Figure 1), whereas class III proteins contained only one BBX domain (Figures 1, 3). Among these VrCOL members, VrCOL1 and VrCOL2 showed close relationships to Arabidopsis AtCO, soybean GmCOL1a, GmCOL1b, GmCOL2a, and GmCOL2b and rice OsHd1 (OsCOL-A), all of which have documented roles in the regulation of flowering time (Khanna et al., 2009; Gangappa and Botto, 2014; Wu et al., 2014). This result suggests that VrCOL1 and VrCOL2 may play critical roles in the flowering time regulation of mungbean.
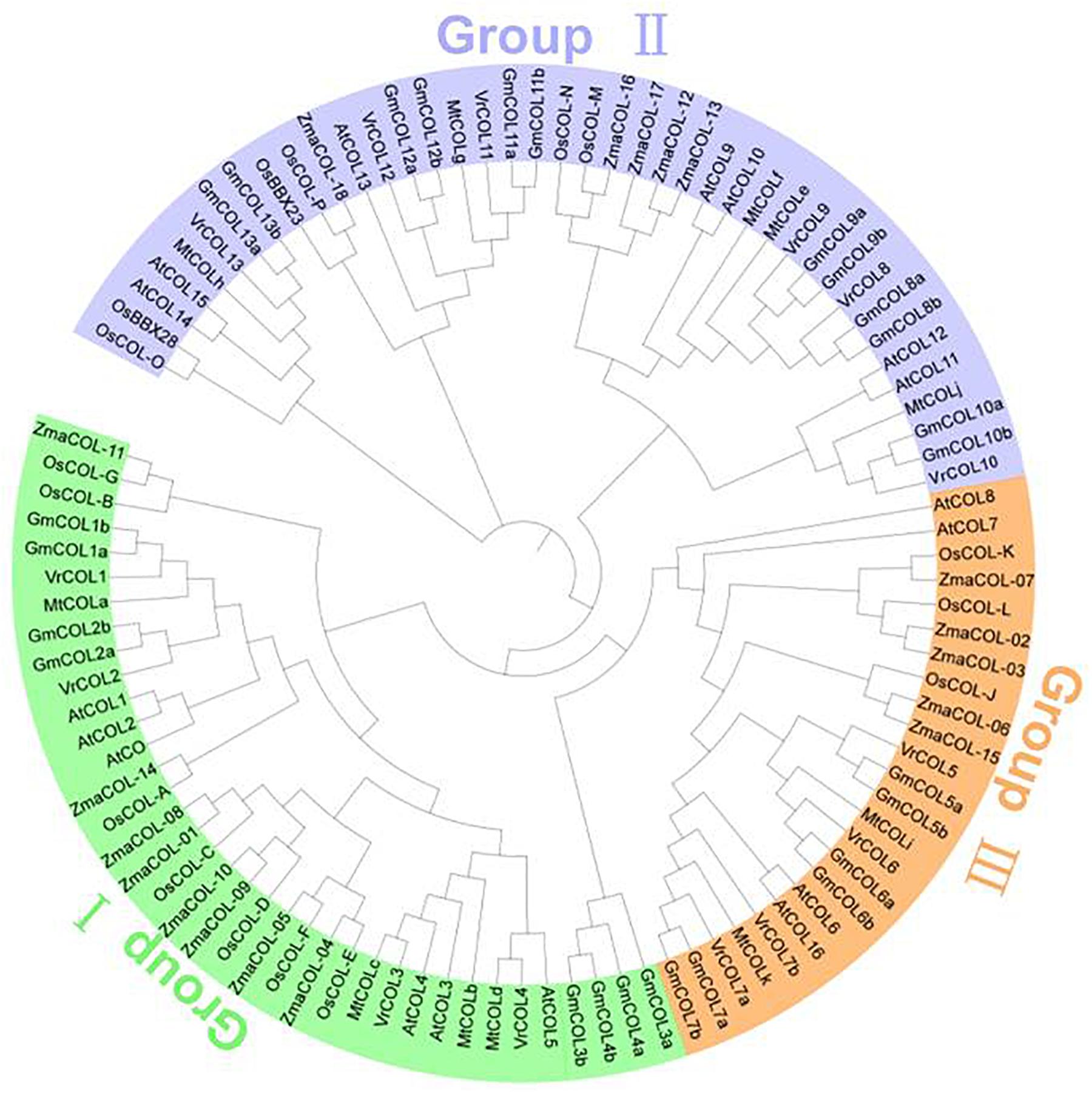
Figure 3. Evolutionary relationships among VrCOL proteins and COL proteins from other species. The amino acid sequences of COL proteins from Arabidopsis, soybean, Medicago, mungbean, rice and maize were used to construct a phylogenetic tree in MEGA 7.0 with the neighbor-joining method. VrCOL proteins are grouped into three classes and indicated with different colors.
Gene Structures and Conserved Motifs of the VrCOL Genes
To investigate the gene structures of the VrCOL genes, we downloaded their genomic and CDS sequences from NCBI and analyzed them using the GSDS program (Hu et al., 2015). All the VrCOL members contained 5′ UTR and 3′ UTR regions. Their exon numbers ranged from two to six, and their intron numbers ranged from one to six. All the group I and III VrCOL members contained two exons and one intron (Figure 4). By contrast, group II members contained various numbers of exons (3–6) and introns (2–6), suggesting potential functional diversity among these genes (Figure 4). To further investigate the conservation and diversity of VrCOL protein structures, we analyzed putative protein motifs in the VrCOLs. A total of 17 distinct motifs were identified, and all VrCOL proteins contained motifs 1 and 2, which appeared to represent the conserved BBX1 and CCT domains, respectively (Figure 4 and Supplementary Figure 2). Most members of the same class shared some conserved motifs. For example, class I proteins shared motifs 1, 2, 3, 9, and 16, class II members shared motifs 1, 2, 3, and 5, and class III members shared motifs 1, 2, 4, 8, 12, and 13 (Figure 4).
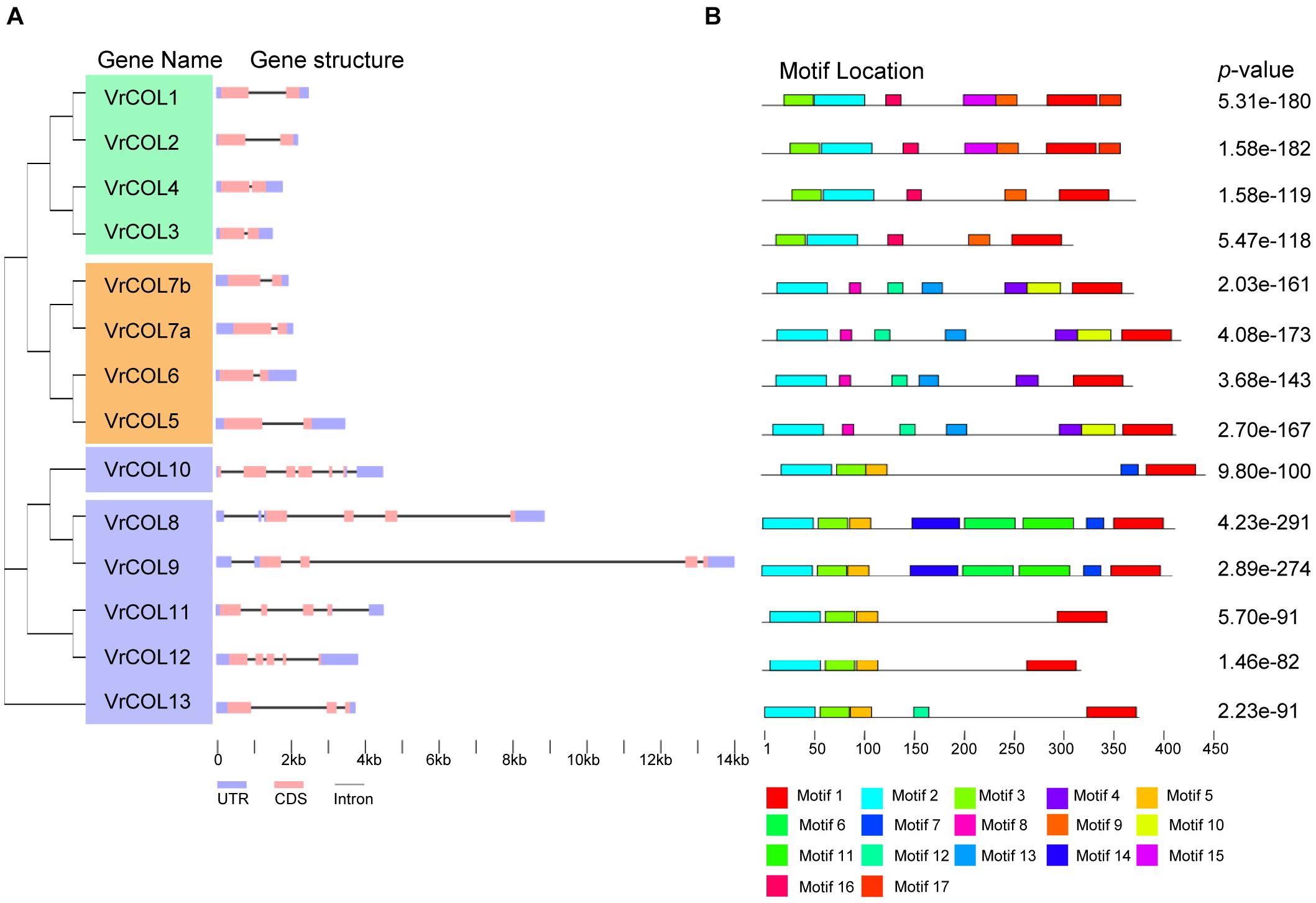
Figure 4. Gene structures and conserved motifs of the VrCOL proteins. (A) Exon-intron organization of the VrCOL genes. The length of each VrCOL gene is indicated, and the blue boxes, pink boxes and black lines indicate UTRs, exons and introns, respectively. (B) Conserved motifs of the VrCOL proteins. Conserved motifs were analyzed using MEME tools, and different motifs are indicated by different colored boxes.
Chromosomal Distribution and Duplication Analysis of the VrCOL Genes
Some genes have evolved from common ancestors, and the chromosomal locations of COL genes may provide insight into changes in gene distribution during evolution. To visualize the chromosomal locations of the VrCOL genes, we mapped them to their physical positions in the mungbean genome. VrCOL7b was discarded due to a lack of related positional information. Seven of the 14 VrCOL genes were located on the positive strand. Seven of the 11 mungbean chromosomes contained VrCOL genes, with the exception of chromosomes 2, 9, 10, and 11 (Figure 5 and Table 1). Chromosome 5 contained the greatest number of VrCOL genes (three), followed by chromosomes 1, 4, 7, and 8, with two genes on each. In addition, most of the VrCOL genes were located on the relatively long chromosomes (1, 5, 6, 7, and 8). Only three members (VrCOL1, VrCOL11, and VrCOL13) were located on the relatively short chromosomes 3 and 4 (Figure 5).
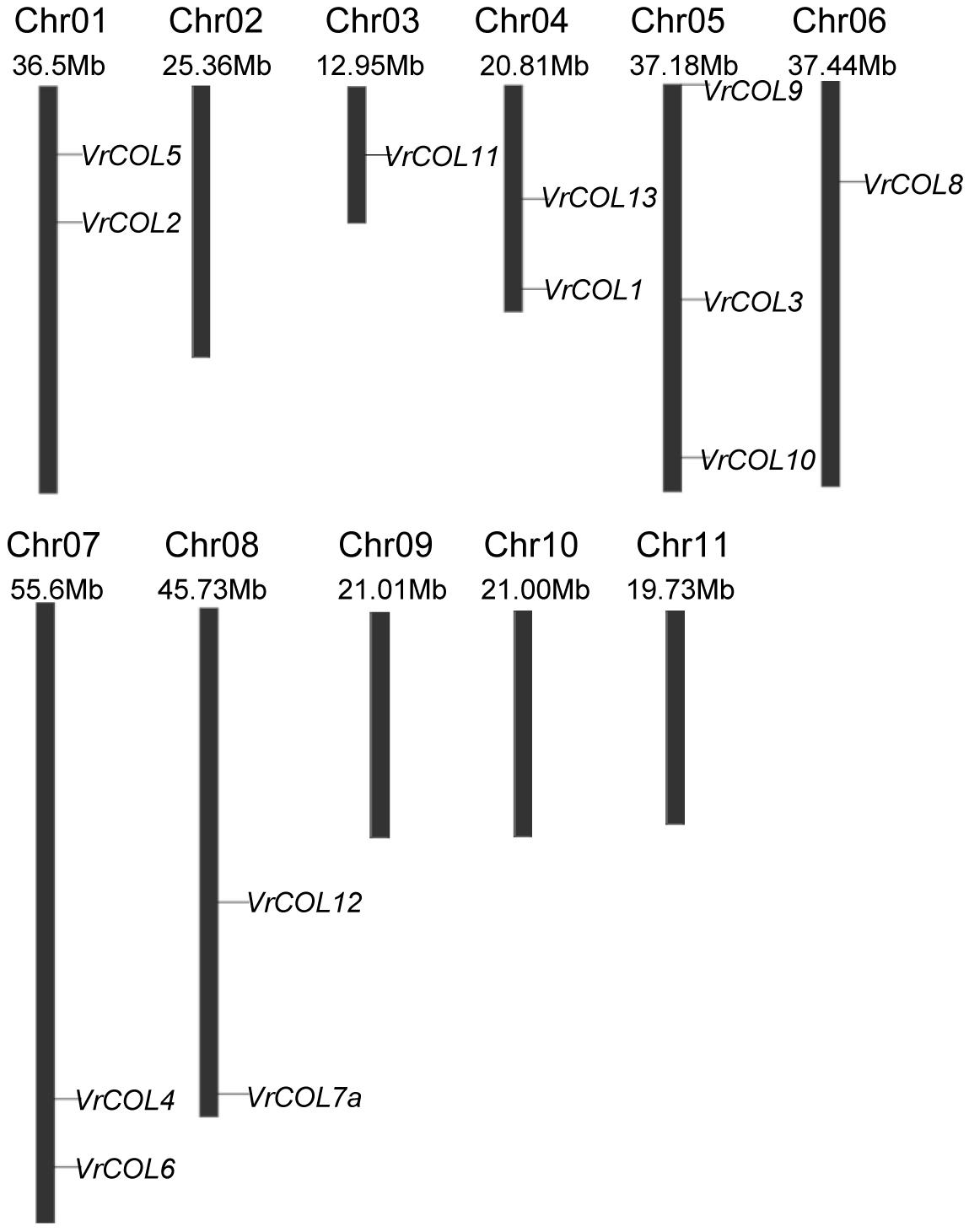
Figure 5. Chromosomal locations of the VrCOL genes. Chromosome number, chromosome length, and the positions of VrCOLs on the chromosome are indicated.
Mungbean has experienced one round of whole-genome duplication that produced many duplicated gene pairs (Kang et al., 2014; Li et al., 2019). To investigate the evolutionary relationships among the VrCOLs, we searched for duplicated gene pairs among them. Two interchromosomal duplication events were identified in chromosomes 1, 4, 5, and 6, producing the duplicated gene pairs VrCOL1/VrCOL2 and VrCOL8/VrCOL9 (Figure 6). The duplicated genes were clustered together in the phylogenetic tree (Figure 1). All the duplicated genes contained one BBX1, one BBX2 and one CCT domain and belonged to groups I and II; no duplicated gene pairs were found in group III. The duplicated genes VrCOL1 and VrCOL2 showed similar exon-intron organization and similar motifs, as did VrCOL8 and VrCOL9 (Figure 4), indicating that the duplicates may share similar functions.
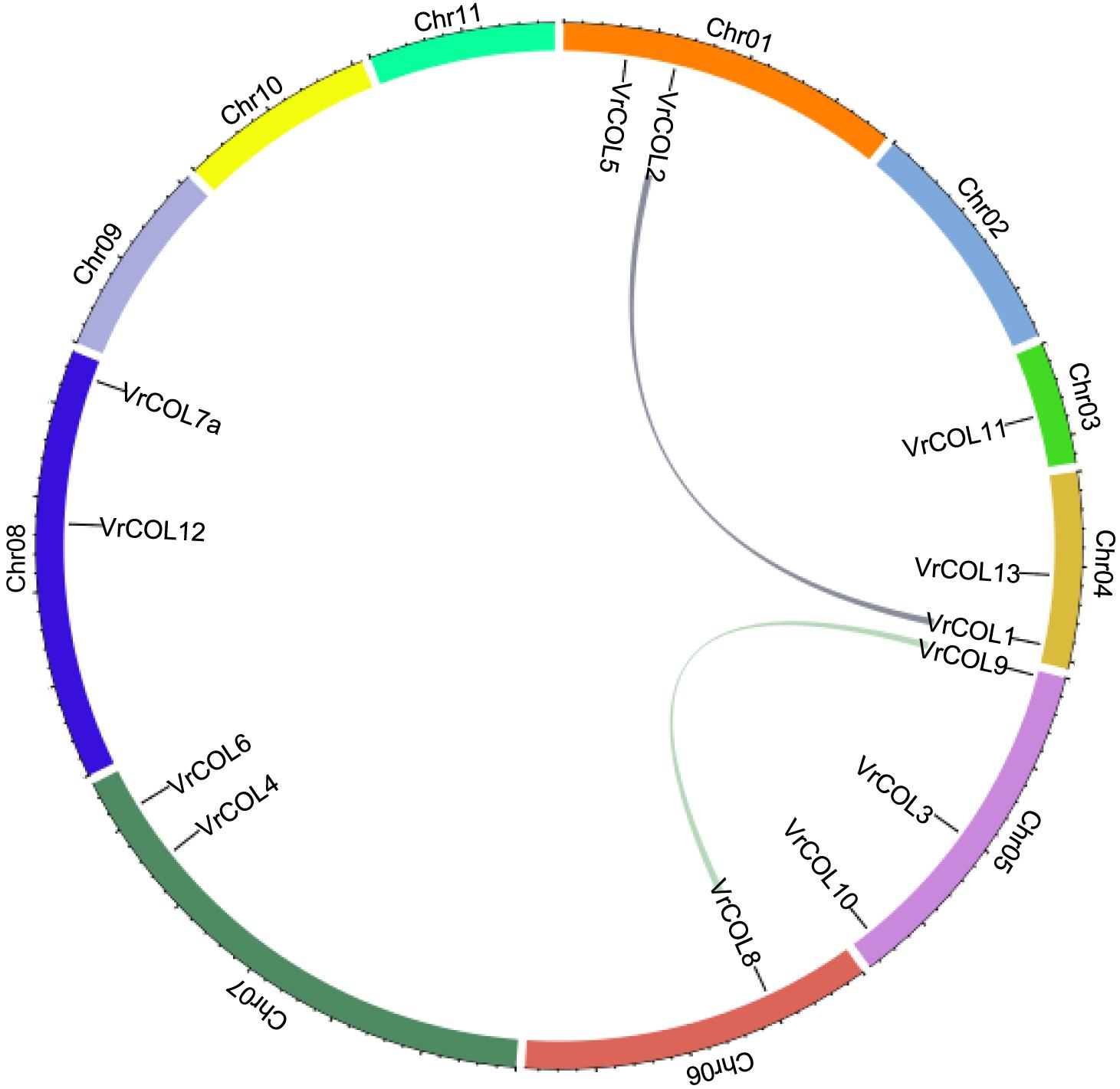
Figure 6. Duplication analyses of VrCOL proteins. The positions of VrCOL genes on the chromosomes are presented, and duplicated gene pairs are connected by colored lines.
Cis-Acting Element Analysis of the VrCOL Promoter Regions
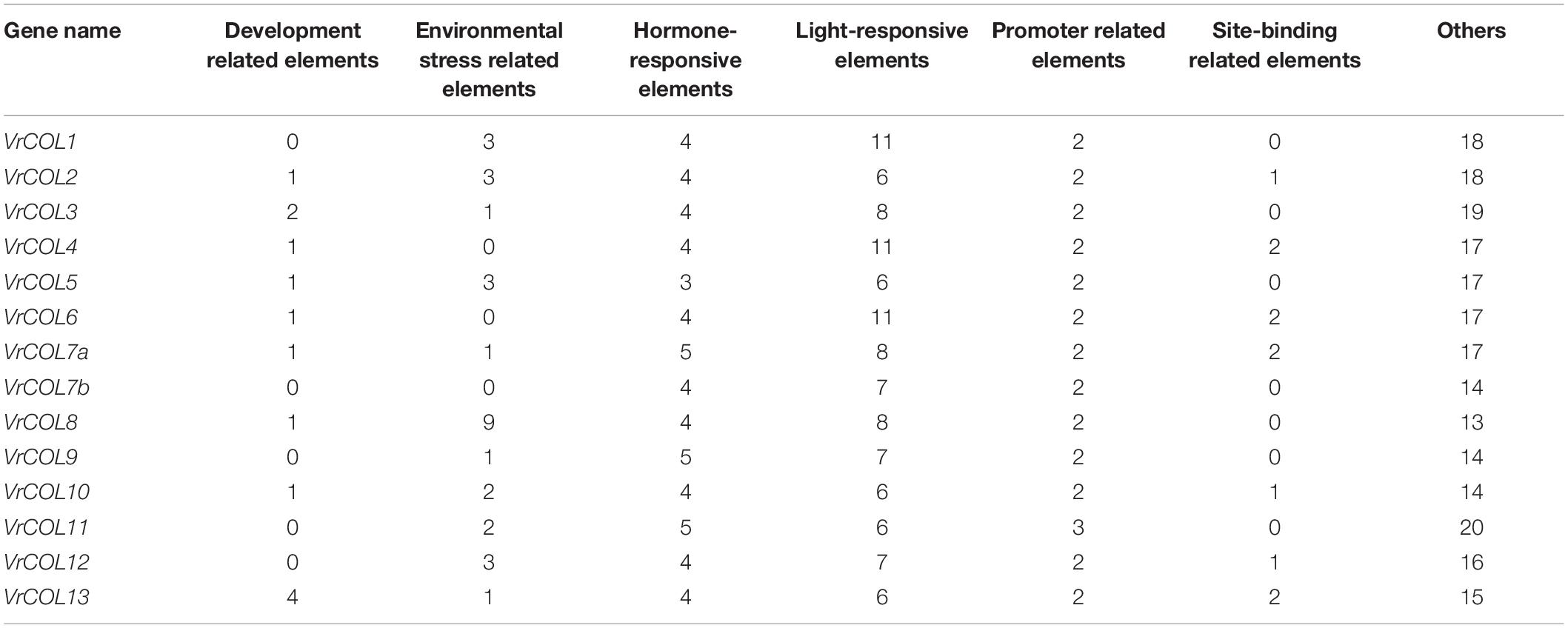
To predict the potential expression responses of VrCOL genes, we investigated the cis-acting elements in their promoters using PlantCARE (Lescot et al., 2002). A total of 82 cis-acting elements were found across the 14 VrCOL promoter regions (2 kb upstream of the initiation codon) (Supplementary Table 2). Forty-five of them had predicted functions, including six development-related elements, four environmental-stress-related elements, three site-binding-related elements, nine hormone-responsive elements, three promoter-related elements and twenty light-responsive elements (Table 2 and Supplementary Table 2). The various VrCOL promoter regions had different numbers and types of cis-acting elements, highlighting the functional diversity of these genes. All VrCOL promoters contained hormone-responsive elements, light-responsive elements and promoter-related elements. Light-responsive elements were the most abundant element in each VrCOL promoter, with the exception of VrCOL8 (Table 2), indicating that VrCOL genes may play critical roles in light-dependent signaling pathways. Environmental-stress-related elements were the most abundant element in the VrCOL8 promoter (nine elements), indicating that VrCOL8 may function in stress response (Table 2). All the VrCOL genes contained the promoter-related elements CAAT-Box and TATA-Box, which are basic promoter components. Thirteen of the 14 VrCOLs contained the hormone-responsive elements CGTCA-motif and TGACG-motif and the light-responsive element Box 4 (Supplementary Table 2), suggesting potential functions of these genes in related signaling pathways.
Transcription Patterns of VrCOL Genes in Different Tissues
To shed light on the potential functions of VrCOL genes during plant development, we analyzed the expression of VrCOL genes in different tissues, including roots, nodule roots, shoot apices, stems, leaves, flowers, pods and seeds. VrCOL genes showed distinct expression patterns in different tissues (Figure 7). For example, VrCOL3 was highly expressed in all tissues examined, whereas VrCOL2 and VrCOL7a showed low expression in most tissues. Some genes were expressed at high levels in specific tissues, suggesting that they may have critical functions in those tissues. For example, VrCOL6 showed high expression in leaves but low expression in nodule roots and flowers.
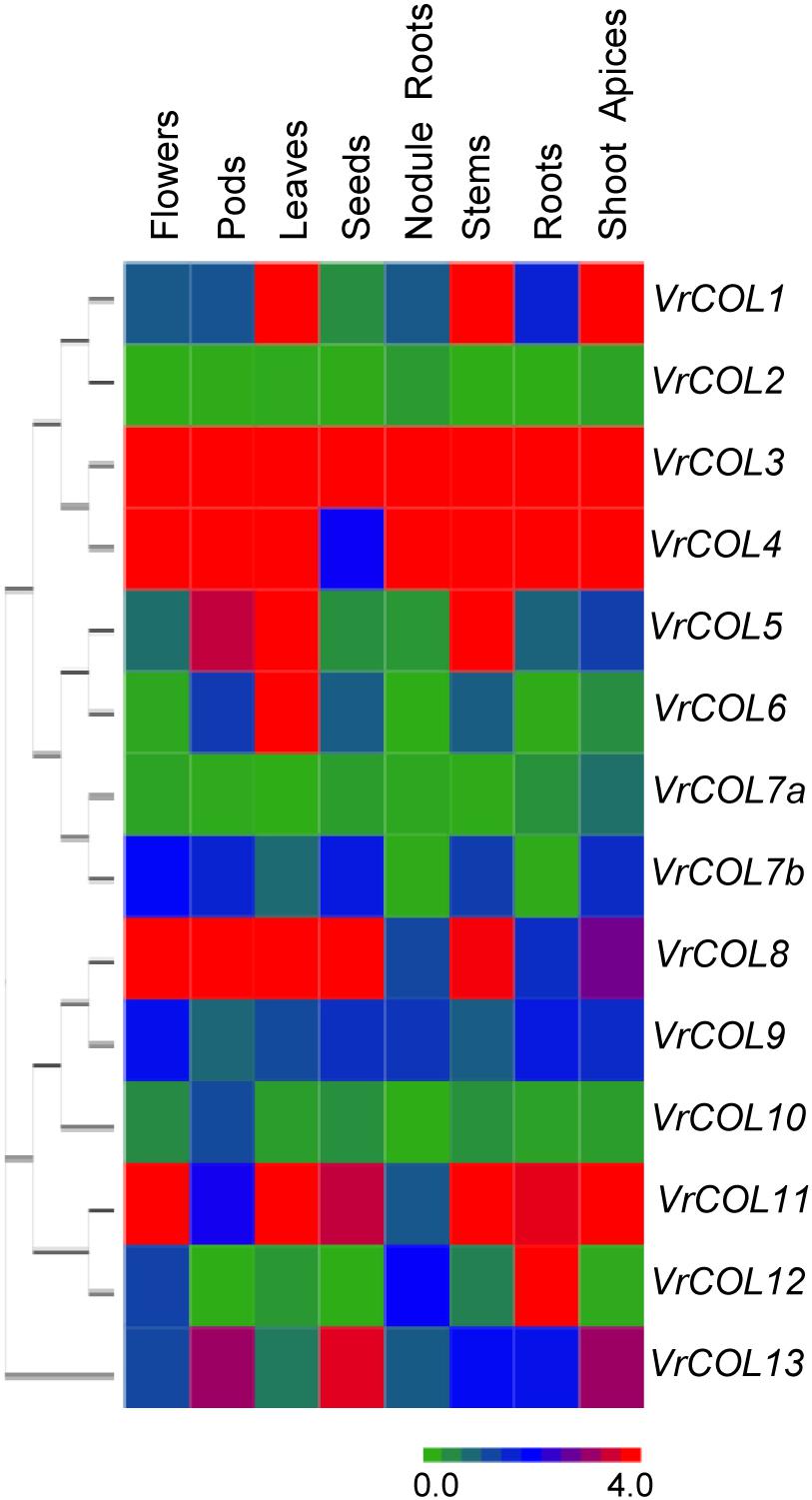
Figure 7. Relative expression levels of VrCOL genes in different tissues. The expression levels of VrCOL genes were analyzed by qRT–PCR. The expression level of VrCOL1 in flowers was set to 1, and other values were adjusted accordingly. The gene expression results were visualized using a heatmap generated with Multiple Experiment Viewer 4.9.0 (Saeed et al., 2003). Different colors in the heatmap indicate different expression levels.
Duplicated genes may retain some common functions and evolve some new functions (Kondrashov et al., 2002; Wang et al., 2015). To investigate the conservation and diversity of duplicated genes, we also analyzed their tissue-specific expression patterns. VrCOL1 and VrCOL2 differed in their expression levels across all tissues examined, indicating that they may have different responses to the environment in these tissues. VrCOL8 and VrCOL9 showed similar expression levels in roots and nodule roots but different expression levels in other tissues (Figure 7 and Supplementary Figure 3).
Diurnal Rhythm of VrCOL Gene Expression
In Arabidopsis, the expression levels of CO, COL1, and COL2 are regulated by the circadian clock and show diurnal oscillations (Suárez-López et al., 2001; Gangappa and Botto, 2014). We therefore investigated whether VrCOL genes exhibited diurnal expression rhythms in mungbean leaves under LD and SD conditions. Gene expression analysis revealed that VrCOL4, VrCOL6, VrCOL12, and VrCOL13 showed daily oscillations under both LD and SD conditions, whereas VrCOL1, VrCOL2, VrCOL5, VrCOL7a, VrCOL7b, VrCOL10, and VrCOL11 showed daily oscillations only under SD conditions (Figure 8). The duplicated genes VrCOL8 and VrCOL9 exhibited similar expression patterns under both LD and SD conditions, whereas VrCOL1 and VrCOL2 showed distinct expression patterns under both LD and SD throughout the day (Figure 8).
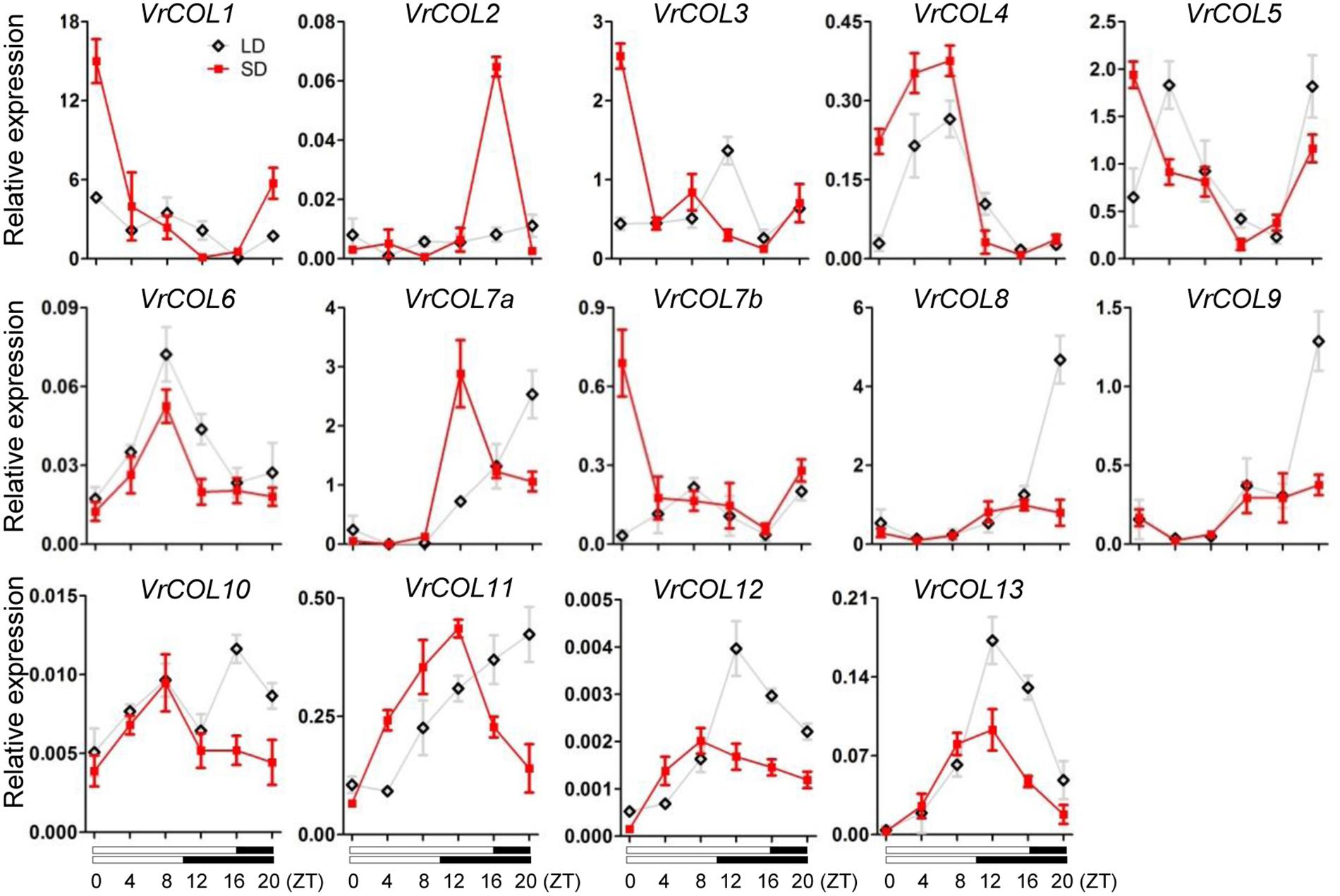
Figure 8. Relative expressions of VrCOLs in mungbean leaves throughout the day under SD and LD conditions. The SD condition was set as 8:00 am–6:00 pm light, 6:00 pm–8:00 am dark; the LD condition was set as 8:00 am–0:00 am light, 0:00 am–8:00 am dark. ZT, Zeitgeber Time. Expression level of VrCOLs was normalized to an Actin gene from mungbean. The gray and red lines indicate VrCOL expression levels under LD and SD conditions, respectively. Each sample was analyzed using three biological replicates.
Overexpression of VrCOL2 Accelerates Flowering in Arabidopsis Under SD Conditions
VrCOL1 and VrCOL2 displayed close phylogenetic relationships with AtCO (Figure 1), and the amino acid sequences of VrCOL1 and VrCOL2 showed 49.35 and 50.93% similarities with AtCO, respectively. We speculated that VrCOL1 and VrCOL2 might influence flowering time in mungbean, and we therefore first analyzed the function of VrCOL2 in the regulation of flowering time in Arabidopsis in this study. To investigate the potential functions of VrCOL2 in flowering time regulation, VrCOL2 was transformed into Arabidopsis under the control of the 35S promoter. The empty vector was also transformed into Arabidopsis, and the transgenic plants showed no differences from wild type under both LD and SD conditions (Supplementary Figure 4). The VrCOL2 transgenic Arabidopsis lines showed high levels of VrCOL2 expression (Supplementary Figure 5). The wild-type Arabidopsis plants and three VrCOL2 overexpression lines exhibited approximately 14 rosette leaves after bolting under LD conditions, indicating that they had similar flowering times. By contrast, the wild-type Arabidopsis plants and three VrCOL2 overexpression lines showed approximately 47, 34, 34, and 31 rosette leaves after bolting under SD conditions, suggesting that VrCOL2 transgenic plants had earlier flowering times than wild-type plants (Figure 9). These results indicated that VrCOL2 regulated flowering time through a photoperiod-dependent pathway.
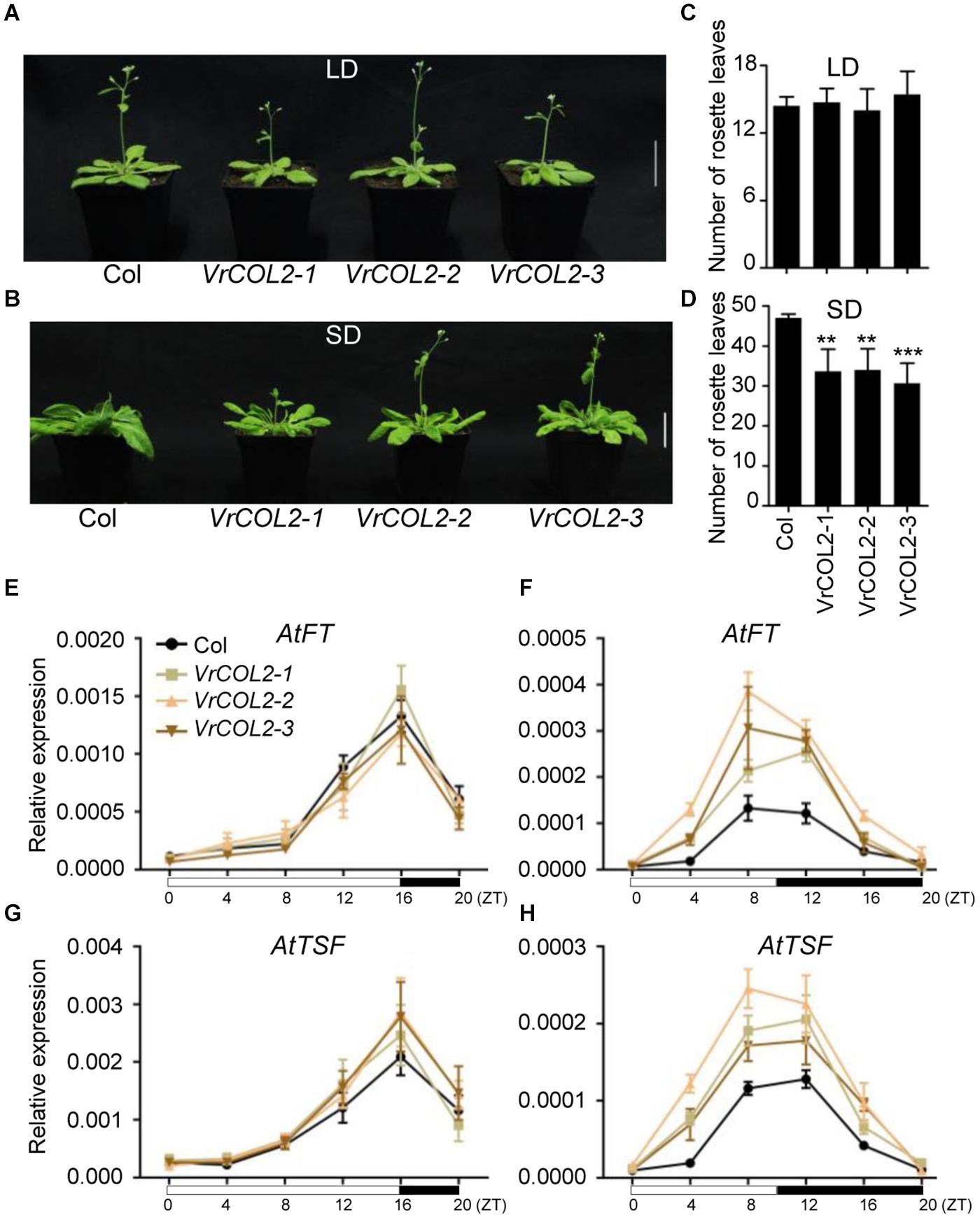
Figure 9. Overexpression of VrCOL2 accelerates flowering under SD conditions in Arabidopsis. Flowering time phenotypes of three VrCOL2 overexpression transgenic lines and wild-type Arabidopsis (Col) plants grown under LD (A) and SD conditions (B). Bar = 4 cm. The rosette leaf numbers of VrCOL2 overexpression transgenic lines and wild-type plants grown under LD (C) and SD conditions (D) as shown in panels (A,B). The expression levels of AtFT and AtTSF throughout the day in VrCOL2 transgenic lines and wild-type plants under LD (E,G) and SD (F,H) conditions. Leaves of 2-week-old VrCOL2 overexpression transgenic lines and wild-type plants were sampled every 4 h after lights-on. Expression levels of AtFT and AtTSF were normalized to an Actin gene from Arabidopsis. The SD condition was set as 8:00 am–6:00 pm light, 6:00 pm–8:00 am dark; the LD condition was set as 8:00 am–0:00 am light, 0:00 am–8:00 am dark.
AtFT and AtTSF accelerate flowering and are regulated by AtCO in Arabidopsis (Khanna et al., 2009; Gangappa and Botto, 2014), and we therefore investigated the expression of AtFT and AtTSF in wild-type and VrCOL2 transgenic plants under LD and SD conditions throughout the day. AtFT and AtTSF showed similar expression levels in VrCOL2 transgenic and wild-type plants in both light and dark conditions under LD treatment. By contrast, AtFT and AtTSF showed higher expression levels in VrCOL2 transgenic plants at several time points than in wild-type plants under SD conditions (Figure 9). These results further support the conclusion that VrCOL2 is involved in flowering time regulation under SD conditions.
Discussion
In recent decades, the investigation of CO and COL genes in many plant species has greatly increased our knowledge about the molecular mechanisms of flowering time regulation, stress response and root development (Khanna et al., 2009; Gangappa and Botto, 2014). Mungbean is a globally important legume crop, and the mechanisms of its flowering time regulation are still largely unknown. In this study, we identified and characterized 14 VrCOL genes from the mungbean genome and investigated the function of VrCOL2 in flowering time regulation.
The Arabidopsis, soybean, Medicago, and mungbean genomes contain 17, 26, 11, and 14 CO and COL members (Figure 1; Wong et al., 2014; Wu et al., 2014), and their genome sizes are 125 Mb (Initiative, 2000), 1100 Mb (Schmutz et al., 2010), 500 Mb (Young et al., 2011), and 579 Mb (Kang et al., 2014), respectively. Thus, genome size has no direct relationship with the number of COL genes in plants. Soybean has undergone two rounds of whole-genome duplication, whereas mungbean has experienced only one such duplication (Schmutz et al., 2010; Kang et al., 2014). As a result, the COL gene number in mungbean is approximately half that of soybean. Seven of the 11 (63.6%) mungbean chromosomes (Figure 5), seven of the eight (87.5%) Medicago chromosomes and 16 of the 20 (80.0%) soybean chromosomes contained COL genes (Wong et al., 2014; Wu et al., 2014), indicating that the distribution of COL genes has changed during evolution in legumes.
Plant genome evolution produces many duplicated gene pairs and provides resources for new gene functions (Kondrashov et al., 2002). Two duplicated gene pairs, VrCOL1/VrCOL2 and VrCOL8/VrCOL9 (Figure 6), were found among the mungbean VrCOLs. The duplicated genes showed close relationships in the phylogenetic tree and contained similar motifs (Figures 1, 4), indicating that they evolved from the same origin and may share similar functions. However, the duplicated gene pairs contained different numbers and types of cis-acting elements in their promoter regions and exhibited different expression levels in some tissues (Figure 7 and Table 2), suggesting that they might have evolved novel functions compared with their original gene. For example, VrCOL8 and VrCOL9 shared similar numbers of several cis-acting elements in their promoter regions, including promoter-related elements and site-binding related elements, but differed in the numbers of development-related elements, environmental-stress-related elements, hormone-responsive elements and light-responsive elements (Table 2 and Supplementary Table 2). VrCOL8 and VrCOL9 showed similar expression levels in roots and nodule roots, but their expression differed in flowers, pods, leaves, seeds, stems, and shoot apices (Figure 7 and Supplementary Figure 3). This result suggests that they may have retained some common functions from the original gene in roots and nodule roots but evolved novel functions in other tissues.
The expression of VrCOL genes in different tissues provides clues to their potential functions, and many VrCOL genes (such as VrCOL6 and VrCOL12) showed tissue-specific expression patterns (Figure 7). However, several VrCOL genes (including VrCOL2, VrCOL7a, and VrCOL10) showed low expression levels in all tissues tested, despite the fact that their promoter regions contained many cis-acting elements (Figure 7, Table 2, and Supplementary Table 2). Gene expression is influenced by many factors. For example, many circadian clock and flowering time regulation genes are controlled by photoperiod. Their expression changes under different photoperiods and during the day and night (Suárez-López et al., 2001; Jack, 2004; Wickland and Hanzawa, 2015; Xu and Chong, 2018). For example, VrCOL2 appeared to be a daily oscillation gene whose expression changed during the day under SD conditions but was low throughout the day under LD conditions (Figure 8). The different field-grown mungbean tissues were collected in the afternoon under relatively LD conditions in July, and that may explain why VrCOL2 showed low expression levels in the tissue expression analysis (Figure 7).
CO and CO-homologous genes, such as OsHd1, play critical roles in flowering time regulation (Khanna et al., 2009; Gangappa and Botto, 2014). VrCOL2 showed close relationships with Arabidopsis CO, soybean GmCOL1a, GmCOL1b, GmCOL2a, and GmCOL2b and rice OsHd1 (OsCOL-A), and accelerated flowering under SD but not LD conditions in transgenic Arabidopsis lines (Figure 9). AtCO regulates AtFT and AtTSF to accelerate flowering (Putterill et al., 1993, 1995; Andres and Coupland, 2012; Song et al., 2013), and the expression of AtFT and AtTSF increased in VrCOL2 transgenic Arabidopsis lines at several time points under SD but not LD conditions (Figure 9). Moreover, VrCOL2 showed daily oscillations only under SD conditions, but not LD conditions (Figure 8), indicating that VrCOL2 might only have functions under SD conditions. VrCOL2 therefore affects the expression of downstream AtFT and AtTSF genes via photoperiod-dependent pathways. Moreover, AtCO protein accumulation is also regulated by the circadian clock. AtCO mRNA is highly abundant from late afternoon to dawn, but AtCO protein accumulates only in the late afternoon under LD conditions (Putterill et al., 1995; Shim and Imaizumi, 2015; Song et al., 2015; Shim et al., 2017). Although VrCOL2 was controlled by the 35S promoter and expressed under both LD and SD conditions, the accumulation of VrCOL2 proteins was unknown. Whether the accumulation of VrCOL2 protein depends on day length, in turn affecting flowering time by influencing AtFT and AtTSF expression requires further investigation. In addition, AtCO promotes flowering under LD conditions and suppresses flowering time under SD conditions (Luccioni et al., 2019), but rice OsHd1 accelerates flowering under SD conditions and delays flowering under LD conditions (Yano et al., 2000; Komiya et al., 2008, 2009). Mungbean (Imrie, 1996; Kim et al., 2015) and rice are SD plants, and Arabidopsis is an LD plant, and this may explain why CO homologs have different functions in different plant species. These results suggest that CO and its homologs are involved in flowering time regulation under photoperiod-dependent pathways and have distinct roles in different plant species. Thus, in summer LD conditions, the expression of VrCOL2 may be low and have little effect on the acceleration of flowering. In the autumn, as days become shorter, the expression of VrCOL2 may increase and accelerate mungbean flowering. In addition, VrCOL1 and VrCOL2 form a duplicated gene pair and show a close relationship with one another (Figures 1, 6), and VrCOL1 showed high expression levels in many tissues, indicating that VrCOL1 may share similar functions to VrCOL2 in flowering time regulation, a possibility that requires further investigation. Much more work is needed to fully elucidate the mechanisms by which VrCOL2 affects flowering time and circadian clock regulation in mungbean.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
SL conceived and designed the research. CL, QZ, HZ, and CC conducted the experiments and analyzed the data. SL and HZ wrote the manuscript. All authors read and approved the manuscript.
Funding
This research was funded by the National Key R&D Project (Grant 2016YFD0101005 and 2016YFD0100304), the National Natural Science Foundation of China (Grant 31971898), and the Qingdao Agricultural University Scientific Research Foundation (Grant 6631119010 and 6651118005).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Suk-Ha Lee at Seoul National University, Seoul, South Korea, for supplying mungbean VC1973A seeds. This manuscript has been released as a pre-print at Researchsquare, (Liu et al., 2020).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.608603/full#supplementary-material
Footnotes
References
Andres, F., and Coupland, G. (2012). The genetic basis of flowering responses to seasonal cues. Nat. Rev Genet. 13, 627–639. doi: 10.1038/nrg3291
Bailey, T., Boden, M., Buske, F., Frith, M., Grant, C., Clementi, L., et al. (2009). MEME suite: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. doi: 10.1093/nar/gkp335
Baurle, I., and Dean, C. (2006). The timing of developmental transitions in plants. Cell 125, 655–664. doi: 10.1016/j.cell.2006.05.005
Beinecke, F. A., Grundmann, L., Wiedmann, D. R., Schmidt, F. J., Caesar, A. S., Zimmermann, M., et al. (2018). The FT/FD-dependent initiation of flowering under long-day conditions in the day-neutral species Nicotiana tabacum originates from the facultative short-day ancestor Nicotiana tomentosiformis. Plant J. 96, 329–342. doi: 10.1111/tpj.14033
Bent, A. (2006). Arabidopsis thaliana floral dip transformation method. Methods Mol. Biol. 343, 87–103. doi: 10.1385/1-59745-130-4:87
Boss, P. K., Bastow, R. M., Mylne, J. S., and Dean, C. (2004). Multiple pathways in the decision to flower: enabling, promoting, and resetting. Plant Cell 16(Suppl.), S18–S31. doi: 10.1105/tpc.015958
Crooks, G. E., Hon, G., Chandonia, J. M., and Brenner, S. E. (2004). WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190. doi: 10.1101/gr.849004
del-Olmo, I., Poza-Viejo, L., Piñeiro, M., Jarillo, J. A., and Crevillén, P. (2019). High ambient temperature leads to reduced FT expression and delayed flowering in Brassica rapa via a mechanism associated with H2AZ dynamics. Plant J. 100, 343–356. doi: 10.1111/tpj.14446
El-Gebali, S., Mistry, J., Bateman, A., Eddy, S., Luciani, A., Potter, S., et al. (2019). The Pfam protein families database in 2019. Nucleic Acids Res. 47, 427–432. doi: 10.1093/nar/gky995
Eom, H., Park, S. J., Kim, M. K., Kim, H., Kang, H., and Lee, I. (2018). TAF15b, involved in the autonomous pathway for flowering, represses transcription of FLOWERING LOCUS C. Plant J. 93, 79–91. doi: 10.1111/tpj.13758
Finn, R., Attwood, T., Babbitt, P., Bateman, A., Bork, P., Bridge, A., et al. (2017). InterPro in 2017–beyond protein family and domain annotations. Nucleic Acids Res. 45, 190–199. doi: 10.1093/nar/gkw1107
Fischer, S., Brunk, B. P., Chen, F., Gao, X., Harb, O. S., Iodice, J. B., et al. (2011). Using OrthoMCL to assign proteins to OrthoMCL-DB groups or to cluster proteomes into new ortholog groups. Curr. Protoc. Bioinformatics 35, 1–19. doi: 10.1002/0471250953.bi0612s35
Fuller, D. Q. (2007). Contrasting patterns in crop domestication and domestication rates: recent archaeobotanical insights from the old world. Ann. Bot. 100, 903–924. doi: 10.1093/aob/mcm048
Gangappa, S. N., and Botto, J. F. (2014). The BBX family of plant transcription factors. Trends Plant Sci. 19, 460–470. doi: 10.1016/j.tplants.2014.01.010
Hu, B., Jin, J., Guo, A., Zhang, H., Luo, J., and Gao, G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297.
Hu, T., Wei, Q., Wang, W., Hu, H., Mao, W., Zhu, Q., et al. (2018). Genome-wide identification and characterization of CONSTANS-like gene family in radish (Raphanus sativus). PLoS One 13:e0204137. doi: 10.1371/journal.pone.0204137
Imaizumi, T., Schultz, T. F., Harmon, F. G., Ho, L. A., and Kay, S. A. (2005). FKF1 F-Box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science 309, 293–297. doi: 10.1126/science.1110586
Imrie, B. C. (1996). “Mung bean,” in The New Rural Industries: A Handbook for Farmers and Investors, ed. K. Hyde (Canberra, ACT: Rural Industries Research and Development Corporation), 355–360.
Initiative, A. G. (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. doi: 10.1038/35048692
Isobe, K., Kokubun, M., and Tsuboki, Y. (1995). Effects of soybean raceme-order on pod set and seed growth in three cultivars. Jpn. J. Crop Sci. 64, 281–287. doi: 10.1626/jcs.64.281
Jack, T. (2004). Molecular and genetic mechanisms of floral control. Plant Cell 16(Suppl.), S1–S17. doi: 10.1105/tpc.017038
Jin, H., Tang, X., Xing, M., Zhu, H., Sui, J., Cai, C., et al. (2019). Molecular and transcriptional characterization of phosphatidyl ethanolamine-binding proteins in wild peanuts Arachis duranensis and Arachis ipaensis. BMC Plant Biol. 19:484. doi: 10.1186/s12870-019-2113-3
Jin, H., Xing, M., Cai, C., and Li, S. (2020). B-box proteins in Arachis duranensis: genome-wide characterization and expression profiles analysis. Agronomy 10:23. doi: 10.3390/agronomy10010023
Jing, Y., Guo, Q., Zha, P., and Lin, R. (2019). The chromatin−remodeling factor PICKLE interacts with CONSTANS to promote flowering in Arabidopsis. Plant Cell Environ. 42, 2495–2507. doi: 10.1111/pce.13557
Kang, Y. J., Kim, S. K., Kim, M. Y., Lestari, P., Kim, K. H., Ha, B. K., et al. (2014). Genome sequence of mungbean and insights into evolution within Vigna species. Nat. Commun. 5:5443. doi: 10.1038/ncomms6443
Keatinge, J. D. H., Easdown, W. J., Yang, R. Y., Chadha, M. L., and Shanmugasundaram, S. (2011). Overcoming chronic malnutrition in a future warming world: the key importance of mungbean and vegetable soybean. Euphytica 180, 129–141. doi: 10.1007/s10681-011-0401-6
Khanna, R., Kronmiller, B., Maszle, D. R., Coupland, G., Holm, M., Mizuno, T., et al. (2009). The Arabidopsis B-box zinc finger family. Plant Cell 21, 3416–3420. doi: 10.1105/tpc.109.069088
Kim, S. K., Nair, R. M., Lee, J., and Lee, S. H. (2015). Genomic resources in mungbean for future breeding programs. Front. Plant Sci. 6:626. doi: 10.3389/fpls.2015.00626
Komiya, R., Ikegami, A., Tamaki, S., Yokoi, S., and Shimamoto, K. (2008). Hd3a and RFT1 are essential for flowering in rice. Development 135, 767–774. doi: 10.1242/dev.008631
Komiya, R., Yokoi, S., and Shimamoto, K. (2009). A gene network for long-day flowering activates RFT1 encoding a mobile flowering signal in rice. Development 136, 3443–3450. doi: 10.1242/dev.040170
Kondrashov, F. A., Rogozin, I. B., Wolf, Y. I., and Koonin, E. V. (2002). Selection in the evolution of gene duplications. Genome Biol. 3:RESEARCH0008. doi: 10.1186/gb-2002-3-2-research0008
Krzywinski, M., Schein, J., Birol, I., Connors, J., Gascoyne, R., Horsman, D., et al. (2009). Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645. doi: 10.1101/gr.092759.109
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Kumari, P., and Verma, S. K. (1983). Genotypic differences in flower production, shedding and yield in mungbean. J. Agric. Sci. 99, 219–223. doi: 10.1016/0147-5975(83)90022-1
Kuroda, T., Saitoh, K., Mahmood, T., and Yanagawa, K. (1998). Differences in flowering habit between determinate and indeterminate types of soybean. Plant Prod. Sci. 1, 18–24. doi: 10.1017/S0007485320000280
Lee, C., Kim, S., Jin, S., Susila, H., Youn, G., Nasim, Z., et al. (2019). Genetic interactions reveal the antagonistic roles of FT/TSF and TFL1 in the determination of inflorescence meristem identity in Arabidopsis. Plant J. 99, 452–464. doi: 10.1111/tpj.14335
Lescot, M., Dehais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Li, S., Wang, R., Jin, H., Ding, Y., and Cai, C. (2019). Molecular characterization and expression profile analysis of heat shock transcription factors in mungbean. Front. Genet. 9:736. doi: 10.3389/fgene.2018.00736
Liu, C., Zhang, Q., Zhu, H., Cai, C., and Li, S. (2020). Functional characterization of mungbean CONSTANS-LIKE genes reveals a key role for CONSTANS-LIKE 2 in the control of flowering time in A. thaliana under short-day conditions. Researchsquare doi: 10.21203/rs.3.rs-24842/v2
Luccioni, L., Krzymuski, M., Sánchez−Lamas, M., Karayekov, E., Cerdán, P. D., and Casal, J. J. (2019). CONSTANS delays Arabidopsis flowering under short days. Plant J. 97, 923–932. doi: 10.1111/tpj.14171
Luo, X., Gao, Z., Wang, Y., Chen, Z., Zhang, W., Huang, J., et al. (2018). The NUCLEAR FACTOR−CONSTANS complex antagonizes polycomb repression to de−repress FLOWERING LOCUS T expression in response to inductive long days in Arabidopsis. Plant J. 95, 17–29. doi: 10.1111/tpj.13926
Mondal, M. M. A., Fakir, M. S. A., Juraimi, A. S., Hakim, M. A., Islam, M. M., and Shamsuddoha, A. T. M. (2011). Effects of flowering behavior and pod maturity synchrony on yield of mungbean [Vigna radiata (L.) Wilczek]. Aust J. Crop Sci. 5, 945–953. doi: 10.1111/j.1439-0523.2010.01842.x
Nam, J., dePamphilis, C. W., Ma, H., and Nei, M. (2019). Antiquity and evolution of the MADS-box gene family controlling flower development in plants. Mol. Biol. Evol. 20, 1435–1447. doi: 10.1093/molbev/msg152
Ning, Y., Chen, Q., Lin, R., Li, Y., Li, L., Chen, S., et al. (2019). The HDA19 histone deacetylase complex is involved in the regulation of flowering time in a photoperiod−dependent manner. Plant J. 98, 448–464. doi: 10.1111/tpj.14229
Oliver, T., Schmidt, B., Nathan, D., Clemens, R., and Maskell, D. (2005). Using reconfigurable hardware to accelerate multiple sequence alignment with clustalW. Bioinformatics 21, 3431–3432. doi: 10.1093/bioinformatics/bti508
Parenicova, L., de Folter, S., Kieffer, M., Horner, D. S., Favalli, C., Busscher, J., et al. (2019). Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: new openings to the MADS world. Plant Cell 15, 1538–1551. doi: 10.1105/tpc.011544
Ping, J., Liu, Y., Sun, L., Zhao, M., Li, Y., She, M., et al. (2014). Dt2 is a gain-of-function MADS-Domain factor gene that specifies semideterminacy in soybean. Plant Cell 26, 2831–2842. doi: 10.1105/tpc.114.126938
Putterill, J., Robson, F., Lee, K., and Coupland, G. (1993). Chromosome walking with YAC clones in Arabidopsis: isolation of 1700 kb of contiguous DNA on chromosome 5, including a 300 kb region containing the flowering-time gene CO. Mol. Gen. Genet. 239, 145–157. doi: 10.1007/BF00281613
Putterill, J., Robson, F., Lee, K., Simon, R., and Coupland, G. (1995). The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80, 847–857. doi: 10.1016/0092-8674(95)90288-0
Robson, F., Costa, M. M. R., Hepworth, S. R., Vizir, I., Pineiro, M., Reeves, P. H., et al. (2001). Functional importance of conserved domains in the flowering-time gene CONSTANS demonstrated by analysis of mutant alleles and transgenic plants. Plant J. 28, 619–631. doi: 10.1046/j.1365-313x.2001.01163.x
Ronald, J., and Davis, S. J. (2019). Focusing on the nuclear and subnuclear dynamics of light and circadian signaling. Plant Cell Environ. 42, 2871–2884. doi: 10.1111/pce.13634
Saeed, A. I., Sharov, V., White, J., Li, J., Liang, W., Bhagabati, N., et al. (2003). TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34, 374–378. doi: 10.2144/03342mt01
Sawa, M., Nusinow, D. A., Kay, S. A., and Imaizumi, T. (2007). FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science 318, 261–265. doi: 10.1126/science.1146994
Schmutz, J., Cannon, S., Schlueter, J., Ma, J., Mitros, T., Nelson, W., et al. (2010). Genome sequence of the palaeopolyploid soybean. Nature 463, 178–183. doi: 10.1038/nature08670
Serrano-Bueno, G., Said, F. E., de los Reyes, P., Lucas−Reina, E. I., Ortiz−Marchena, M. I., Romero, J. M., et al. (2020). CONSTANS−FKBP12 interaction contributes to modulate photoperiodic flowering in Arabidopsis. Plant J. 101, 1287–1302. doi: 10.1111/tpj.14590
Shi, R., Xu, W., Liu, T., Cai, C., and Li, S. (2021). VrLELP controls flowering time under short-day conditions in Arabidopsis. J. Plant Res. 134, 141–149. doi: 10.1007/s10265-020-01235-7
Shim, J. S., and Imaizumi, T. (2015). Circadian clock and photoperiodic response in Arabidopsis: from seasonal flowering to redox homeostasis. Biochemistry 54, 157–170. doi: 10.1021/bi500922q
Shim, J. S., Kubota, A., and Imaizumi, T. (2017). Circadian clock and photoperiodic flowering in Arabidopsis: CONSTANS is a hub for signal integration. Plant Physiol. 173, 5–15. doi: 10.1104/pp.16.01327
Song, Y. H., Ito, S., and Imaizumi, T. (2013). Flowering time regulation: photoperiod-and temperature-sensing in leaves. Trends Plant Sci. 18, 575–583. doi: 10.1016/j.tplants.2013.05.003
Song, Y. H., Shim, J. S., Kinmonth-Schultz, H. A., and Imaizumi, T. (2015). Photoperiodic flowering: time measurement mechanisms in leaves. Annu. Rev. Plant Biol. 66, 441–464. doi: 10.1146/annurev-arplant-043014-115555
Suárez-López, P., Wheatley, K., Robson, F., Onouchi, H., Valverde, F., and Coupland, G. (2001). CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410, 1116–1120. doi: 10.1038/35074138
Taylor, C. M., Kamphuis, L. G., Zhang, W., Garg, G., Berger, J. D., Mousavi-Derazmahalleh, M., et al. (2019). INDEL variation in the regulatory region of the major flowering time gene LanFTc1 is associated with vernalization response and flowering time in narrow−leafed lupin (Lupinus angustifolius L.). Plant Cell Environ. 42, 174–187. doi: 10.1111/pce.13320
Vas Aggarwal, D., and Poehlman, J. (1977). Effects of photoperiod and temperature on flowering in mungbean (Vigna radiata (L.) WILCZEK). Euphytica 26, 207–219. doi: 10.1007/BF00032086
Wang, Z., Zhou, Z., Liu, Y., Liu, T., Li, Q., Ji, Y., et al. (2015). Functional evolution of phosphatidyl ethanolamine binding proteins in soybean and Arabidopsis. Plant Cell 27, 323–336. doi: 10.1105/tpc.114.135103
Wickland, D. P., and Hanzawa, Y. (2015). The FLOWERING LOCUS T/TERMINAL FLOWER 1 gene family: functional evolution and molecular mechanisms. Mol. Plant 8, 983–997. doi: 10.1016/j.molp.2015.01.007
Wong, A. C. S., Hecht, V. F. G., Picard, K., Diwadkar, P., Laurie, R. E., Wen, J., et al. (2014). Isolation and functional analysis of CONSTANS-LIKE genes suggests that a central role for CONSTANS in flowering time control is not evolutionarily conserved in Medicago truncatula. Front. Plant Sci. 5:486. doi: 10.3389/fpls.2014.00486
Wu, F., Price, B. W., Haider, W., Seufferheld, G., Nelson, R., and Hanzawa, Y. (2014). Functional and evolutionary characterization of the CONSTANS gene family in short-day photoperiodic flowering in soybean. PLoS One 9:e85754. doi: 10.1371/journal.pone.0085754
Wu, W., Zheng, X. M., Chen, D., Zhang, Y., Ma, W., Zhang, H., et al. (2017). OsCOL16, encoding a CONSTANS-like protein, represses flowering by up-regulating Ghd7 expression in rice. Plant Sci. 260, 60–69. doi: 10.1016/j.plantsci.2017.04.004
Xu, S., and Chong, K. (2018). Remembering winter through vernalisation. Nat. Plants 4, 997–1009. doi: 10.1038/s41477-018-0301-z
Yan, H., Marquardt, K., Indorf, M., Jutt, D., Kircher, S., Neuhaus, G., et al. (2011). Nuclear localization and interaction with COP1 are required for STO/BBX24 function during photomorphogenesis. Plant Physiol. 156, 1772–1782. doi: 10.1104/pp.111.180208
Yano, M., Katayose, Y., Ashikari, M., Yamanouchi, U., Monna, L., Fuse, T., et al. (2000). Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 12, 2473–2483. doi: 10.1105/tpc.12.12.2473
Young, N., Debellé, F., Oldroyd, G., Geurts, R., Cannon, S. B., Udvardi, M. K., et al. (2011). The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature 480, 520–524. doi: 10.1038/nature10625
Keywords: mungbean, flowering time, CONSTANS, VrCOL2, genome wide
Citation: Liu C, Zhang Q, Zhu H, Cai C and Li S (2021) Characterization of Mungbean CONSTANS-LIKE Genes and Functional Analysis of CONSTANS-LIKE 2 in the Regulation of Flowering Time in Arabidopsis. Front. Plant Sci. 12:608603. doi: 10.3389/fpls.2021.608603
Received: 23 September 2020; Accepted: 11 January 2021;
Published: 04 February 2021.
Edited by:
Enamul Huq, University of Texas at Austin, United StatesReviewed by:
Dong-Hwan Kim, Chung-Ang University, South KoreaSourav Datta, Indian Institute of Science Education and Research, India
Copyright © 2021 Liu, Zhang, Zhu, Cai and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shuai Li, bGkyMDE0c2h1YWlAcWF1LmVkdS5jbg==
†These authors have contributed equally to this work
 Chenyang Liu
Chenyang Liu Qianqian Zhang
Qianqian Zhang Hong Zhu
Hong Zhu Chunmei Cai1
Chunmei Cai1 Shuai Li
Shuai Li