- 1College of Life Sciences, Qingdao Agricultural University, Qingdao, China
- 2Key Laboratory of Qingdao Major Crop Germplasm Resource Innovation and Application, Qingdao, China
- 3College of Agronomy, Qingdao Agricultural University, Qingdao, China
Aminoacylase-1 is a zinc-binding enzyme that is important in urea cycling, ammonia scavenging, and oxidative stress responses in animals. Aminoacylase-1 (ACY-1) has been reported to play a role in resistance to pathogen infection in the model plant Nicotiana benthamiana. However, little is known about its function in plant growth and abiotic stress responses. In this study, we cloned and analyzed expression patterns of ZmACY-1 in Zea mays under different conditions. We also functionally characterized ZmACY-1 in N. benthamiana. We found that ZmACY-1 is expressed specifically in mature shoots compared with other tissues. ZmACY-1 is repressed by salt, drought, jasmonic acid, and salicylic acid, but is induced by abscisic acid and ethylene, indicating a potential role in stress responses and plant growth. The overexpression of ZmACY-1 in N. benthamiana promoted growth rate by promoting growth-related genes, such as NbEXPA1 and NbEIN2. At the same time, the overexpression of ZmACY-1 in N. benthamiana reduced tolerance to drought and salt stress. With drought and salt stress, the activity of protective enzymes, such as peroxidase (POD), superoxide dismutase (SOD), and catalase (CAT) from micrococcus lysodeikticus was lower; while the content of malondialdehyde (MDA) and relative electrolytic leakage was higher in ZmACY-1 overexpression lines than that in wild-type lines. The results indicate that ZmACY-1 plays an important role in the balance of plant growth and defense and can be used to assist plant breeding under abiotic stress conditions.
Introduction
Water limitation, normally caused by drought and salt stress, is one of the most important factors affecting plant growth (Koevoets et al., 2016). Drought induces a series of changes at the morphological, physiological, and biochemical levels. Drought leads to stomatal closure; mechanical damage to protoplasts, cell walls, and biofilm systems; decline in photosynthetic efficiency and cell dehydration, which will eventually lead to metabolic disorder (Gupta et al., 2020). Salt stress leads to extracellular osmotic stress, ionic toxicity, production of reactive oxygen species (ROS), and accumulation of superoxide anions, thus disrupting ion balance and stability of biological macromolecules (Claeys and Inzé, 2013). Climate change threatens to exacerbate the problem of water limitation on plant growth and food production (Van Zelm et al., 2020).
To understand the physiological and molecular mechanisms of plant growth under stressed conditions, several signaling pathways have been elucidated (Van Wallendael et al., 2019). For example, DELLAS (Lantzouni et al., 2020) and AP2/ERF-type transcriptional factors (Xie et al., 2019) have been demonstrated as crucial regulators in plant growth and stress tolerance. In addition, amino acid metabolism, such as glutathione, arginine, and proline, have been reported to play important roles in stress responses. Glutathione is composed of glutamic acid, cysteine, and glycine; and glutathione reductase (GR) accumulates in salt-tolerant pea lines with long-term salt stress (Hernandez et al., 2000). Glutathione S-transferase regulates stress-activated signals by inhibiting the activity of apoptotic signal kinase 1 (Cho et al., 2001). High expression of glutathione S-transferase promotes the growth of tobacco seedlings under stress conditions (Roxas et al., 1997). Knocking out the arginine metabolism-related gene AtARG2 enhanced environmental tolerance (Shi et al., 2013). Proline protects macromolecules and cell membranes during dehydration and also acts as an osmotic agent and free radical scavenger. The accumulation of proline is involved in the scavenging of ROS (Schwörer et al., 2020).
Aminoacylase-1 is a homodimer zinc-binding enzyme in cell solutes, which catalyzes the hydrolysis of acylated L-amino acids to L-amino acids and acyl groups. It plays a role in the catabolism and recovery of acylated amino acids. N-acetyl-L-glutamic acid is an allosteric activator of carbamyl phosphate synthase, and carbamyl phosphate synthase is a key enzyme that converts NH4+ molecules in the urea cycle. When amino acid catabolism increases, N-acetylglutamate synthase is up-regulated to produce more N-acetyl-L-glutamate, which up-regulates carbamyl phosphate synthase and allows it to deal with excess NH4+. In addition, ACY-1 may play a role in regulating the cellular response to oxidative stress in animals by interacting with sphingosine kinase-1 (Maceyka et al., 2004).
Currently, aminoacylase-1 (DS2) has been found to play a role in plant defense systems and in the salicylic acid-dependent pathway. Silencing DS2 enhanced the resistance of Nicotiana benthamiana to Phytophthora infestans (Nakano et al., 2014). However, the functional importance of aminoacylase-1 in plant growth and abiotic stress responses is still unknown. In this study, we cloned ZmACY-1 and transformed it into the model plant N. benthamiana. We found an important role of ZmACY-1 in plant growth and stress resistance. This study highlights amino acid metabolism as a new way for improving plant performance in stressed conditions.
Results
Characterization of Aminoacylase-1 Homologs
First, in order to clone ZmACY-1, we analyzed its sequence in silico using the Bioxm software. The full-length open reading frame of ZmACY-1 is 1,317 base pairs, and it encodes 439 amino acids. The molecular formula of ZmACY-1 is C2173H3367N599O625S14, the relative molecular weight is 48.33 kDa, the theoretical isoelectric point is 6.02, the total number of positive charge residues (Arg + Lys) is 42, the total number of negative charge residues (Asp + Glu) is 50, and the coefficient of instability is 46.03. ZmACY-1 belongs to the zinc peptidase superfamily and M20 aminoacylase-1 subfamily, and it contains an M20-acylase domain and five zinc binding sites.
To investigate the conservation of ACY-1 between different plant species, we used NCBI Blast to search for homologous amino acid sequences and used the MEGA 5.1 software for phylogenetic analysis. We found that the ZmACY-1 amino acid sequence (NP_001150325.2) and SbACY-1 amino acid sequence (XP_002436680.1) in Sorghum bicolor showed up to 92.26% similarity. In addition, ZmACY-1 is closely related to SiACY-1 and PhACY-1 (Figure 1), indicating that ACY-1 is highly conserved in plants.
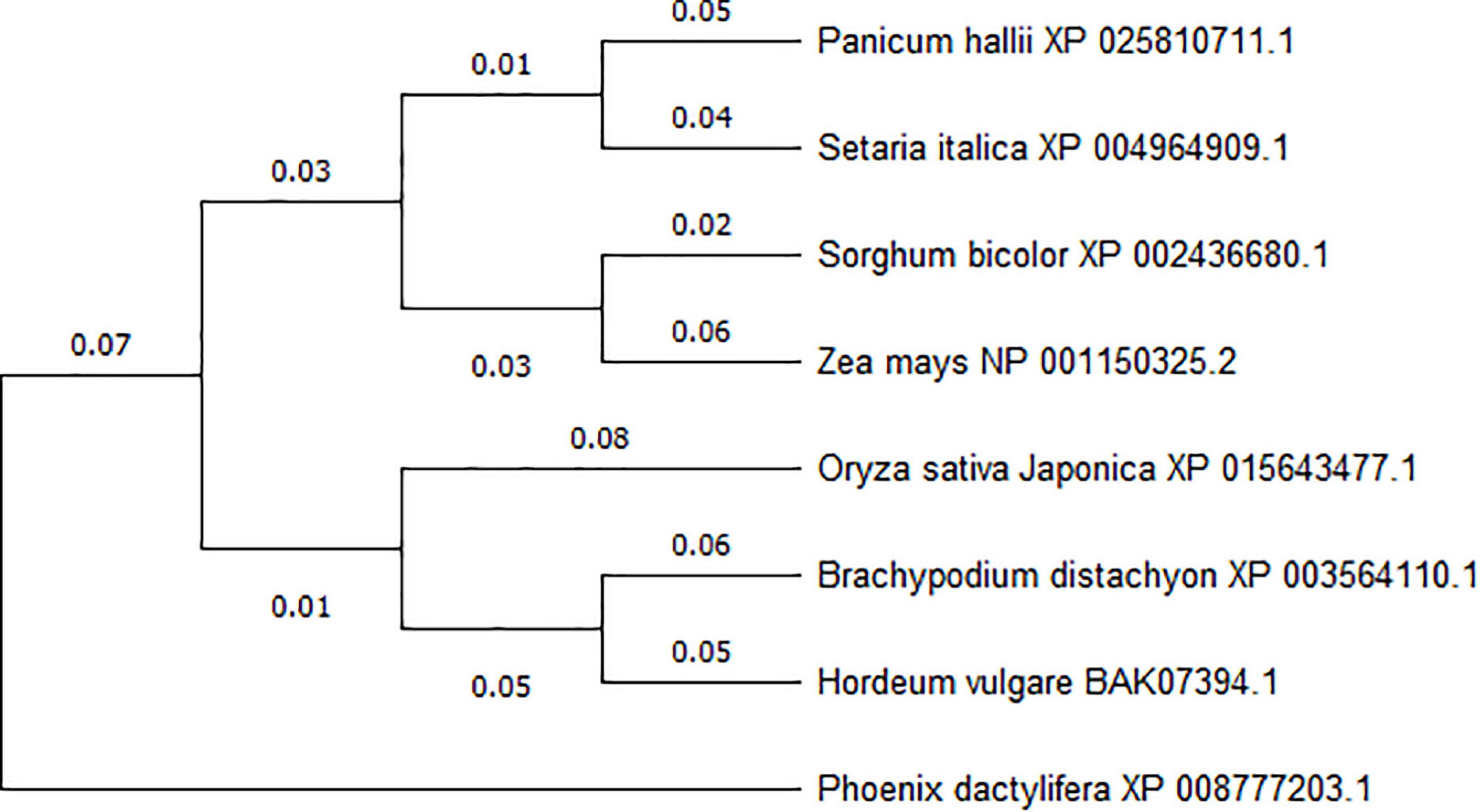
Figure 1. Phylogenetic analysis of ACY-1 based on amino acid sequences. A phylogenetic tree was generated from the deduced amino acid sequences of plant ACY-1 genes. Phylogenetic relationships were calculated using the maximum likelihood method based on DNAMAN and MEGA 5.1 software.
Expression Patterns of ZmACY-1
In order to detect the expression characteristics of ZmACY-1 in different stages and tissues of maize, we performed fluorescence quantitative PCR analysis on young roots, young stems, and young leaves of the trifoliate stage; mature roots, mature stems, mature leaves, aerial roots, tassel, ears of heading stage; and immature embryos 14 days after pollination (DAP). We found that the expression of ZmACY-1 was both tissue- and stage-specific (Figure 2A). ZmACY-1 is expressed highest in mature shoots where the expression of ZmACY-1 is about two times higher than that in young roots and young leaves, 2.75 times higher than that in mature roots, and 5.5 times higher than that in mature leaves. The fact that ZmACY-1 is most highly expressed in shoots indicates that ZmACY-1 is associated with plant growth rate. ZmACY-1 is also expressed in ears, indicating that it may also be involved in plant development.
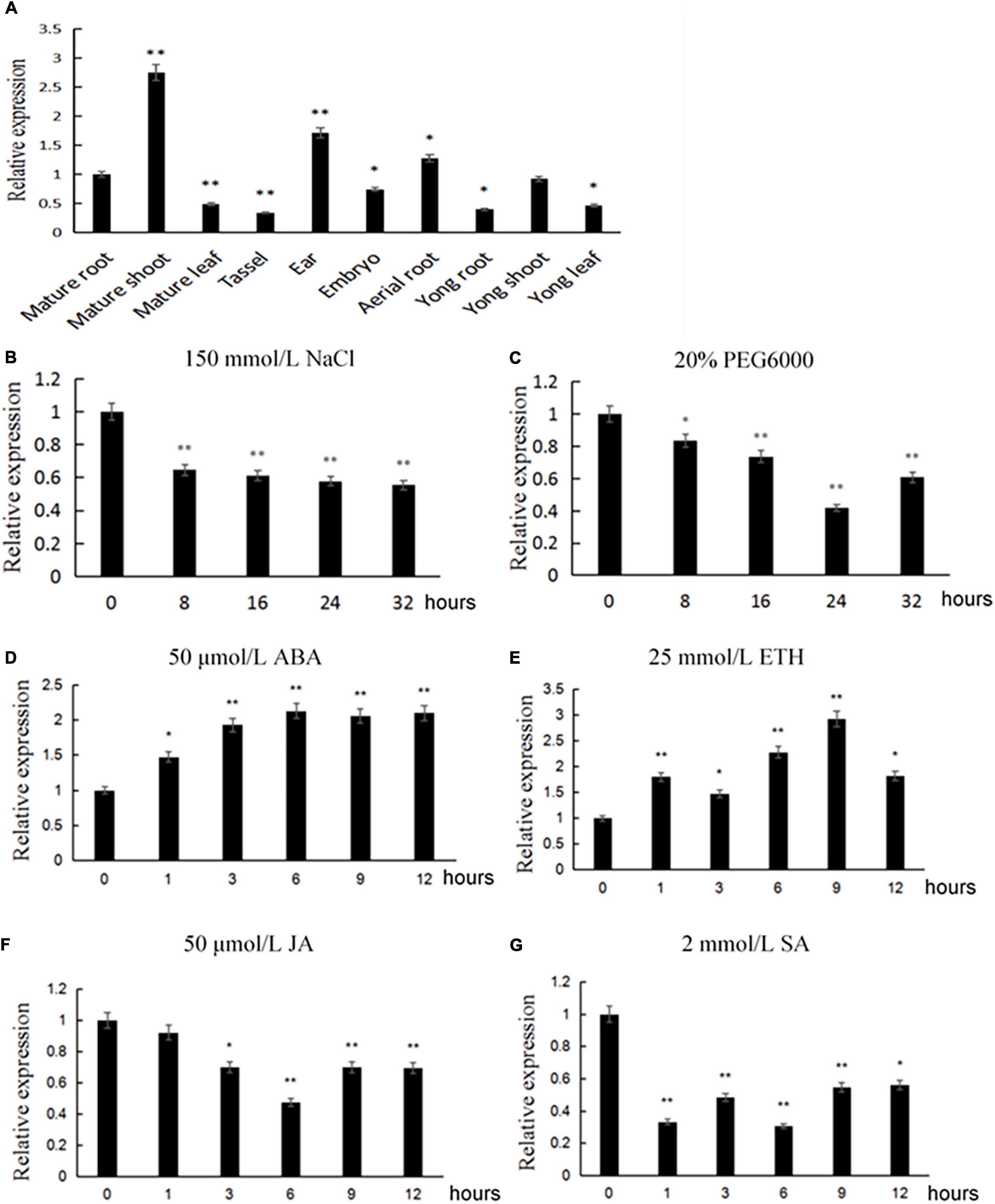
Figure 2. Expression pattern of ZmACY-1. The relative expression of ZmACY-1 under different conditions was measured. (A) Different tissues; (B) 150 mM NaCl; (C) 20% PEG 6000; (D) 50 μM abscisic acid; (G) 2 mM salicylic acid; (F) 50 μM jasmonic acid; (E) 25 mM ethylene. Three biological replicates were used. *P < 0.05, **P < 0.01.
Since ACY-1 has an important role in response to Phytophthora infestans, we tested the expression of ZmACY-1 with different stress treatments in order to explore whether ZmACY-1 is affected by abiotic stress or hormone regulation. We treated trifoliate-stage maize seedlings with 150 mM NaCl, 20% PEG6000, and plant defense-related hormones (50 μM abscisic acid, 2 mM salicylic acid, 50 μM jasmonic acid, and 25 mM ethylene). We found that 150 mM NaCl (Figure 2B), 20% PEG6000 (Figure 2C), jasmonic acid (Figure 2F), and salicylic acid (Figure 2G) decreased the expression of ZmACY-1, while ABA (Figure 2D) and ethylene (Figure 2E) increased the expression of ZmACY-1. This suggested that ZmACY-1 may be involved in defense responses to drought stress, salt stress, and various plant hormones.
Functional Characterization of ZmACY-1 in Plant Growth
To further reveal the function of ZmACY-1 in plant growth, we constructed plasmids and generated transgenic lines in N. benthamiana. We obtained three independent transgenic lines, and the expression of ZmACY-1 in all the three transgenic lines was significantly higher than that in the wild type (Supplementary Figure 1). Then, these transgenic lines were phenotyped for yield-associated traits, such as germination, plant height, root length, stem diameter, leaf area, root area, shoot fresh weight, and fresh weight of roots and pods (Figure 3). We found that seed germination rate is higher in ZmACY-1 overexpression lines than in wild-type lines. The germination rate in transgenic lines reached 80–90% after 4 days, while wild-type controls only reached 60% germination (Figures 3A,B). In addition, the root length, leaf area, plant height, stem diameter, shoot fresh weight, and root fresh weight of the transgenic lines all overmatched those of the wild type (Figures 3E–L), which indicates that overexpression of ZmACY-1 promotes growth of N. benthamiana.
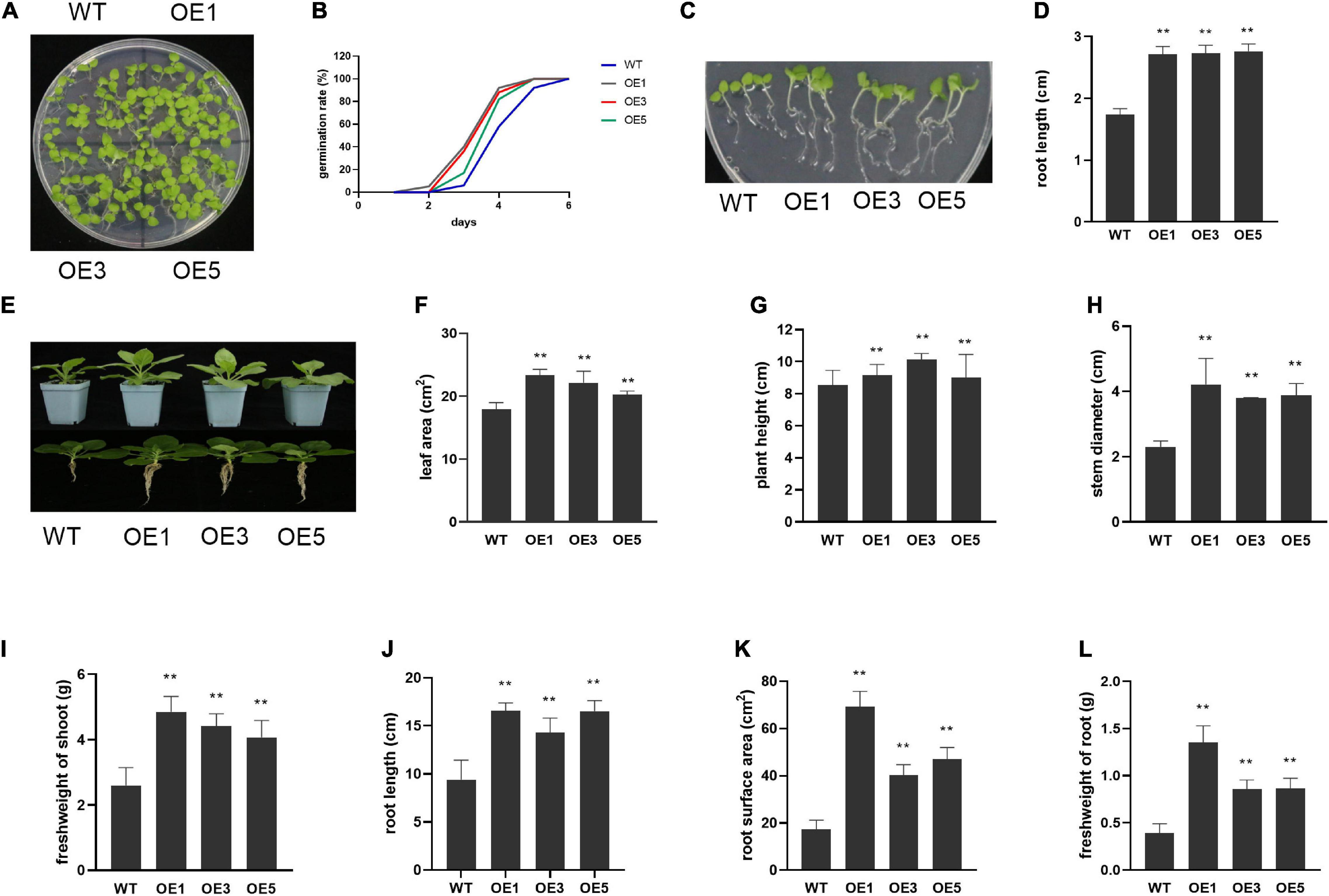
Figure 3. Function of ZmACY-1 in plant growth. (A) Ten-day-old N. benthamiana in MS-medium-supplemented plate; (B) Germination rate in MS-medium-supplemented plate; (C,D) Root length; (E–L) One-month-old N. benthamiana in pods. (F) Leaf area; (G) Plant height; (H) Stem diameter; (I) Shoot fresh weight; (J) Root length; (K) Root surface area; (L) Fresh weight of roots. Three biological replicates were used. *P < 0.05, **P < 0.01.
The EXPA gene family plays an important role in promoting leaf growth (Goh et al., 2012). Ein2 has been proved to be a positive regulator in the ethylene signaling pathway, which participates in many aspects of the plant life cycle (Li et al., 2015). Glutamine synthetase (GS) and asparagine synthetase (AS) have important roles in nitrogen metabolism (Németh et al., 2018; Rashmi et al., 2019). Thus, in order to study whether the overexpression of ZmACY-1 in N. benthamiana promoted the expression of plant growth-related and nitrogen assimilation-related genes, we detected the expression levels of NbEXPA1, NbEIN2, NbGS, and NbAS (Figure 4). We found that the expression levels of NbEXPA1, NbEIN2, NbGS, and NbAS in transgenic N. benthamiana were significantly higher than those in the wild type, indicating that ZmACY-1 functions by increasing the expression of growth-related and nitrogen assimilation-related genes.
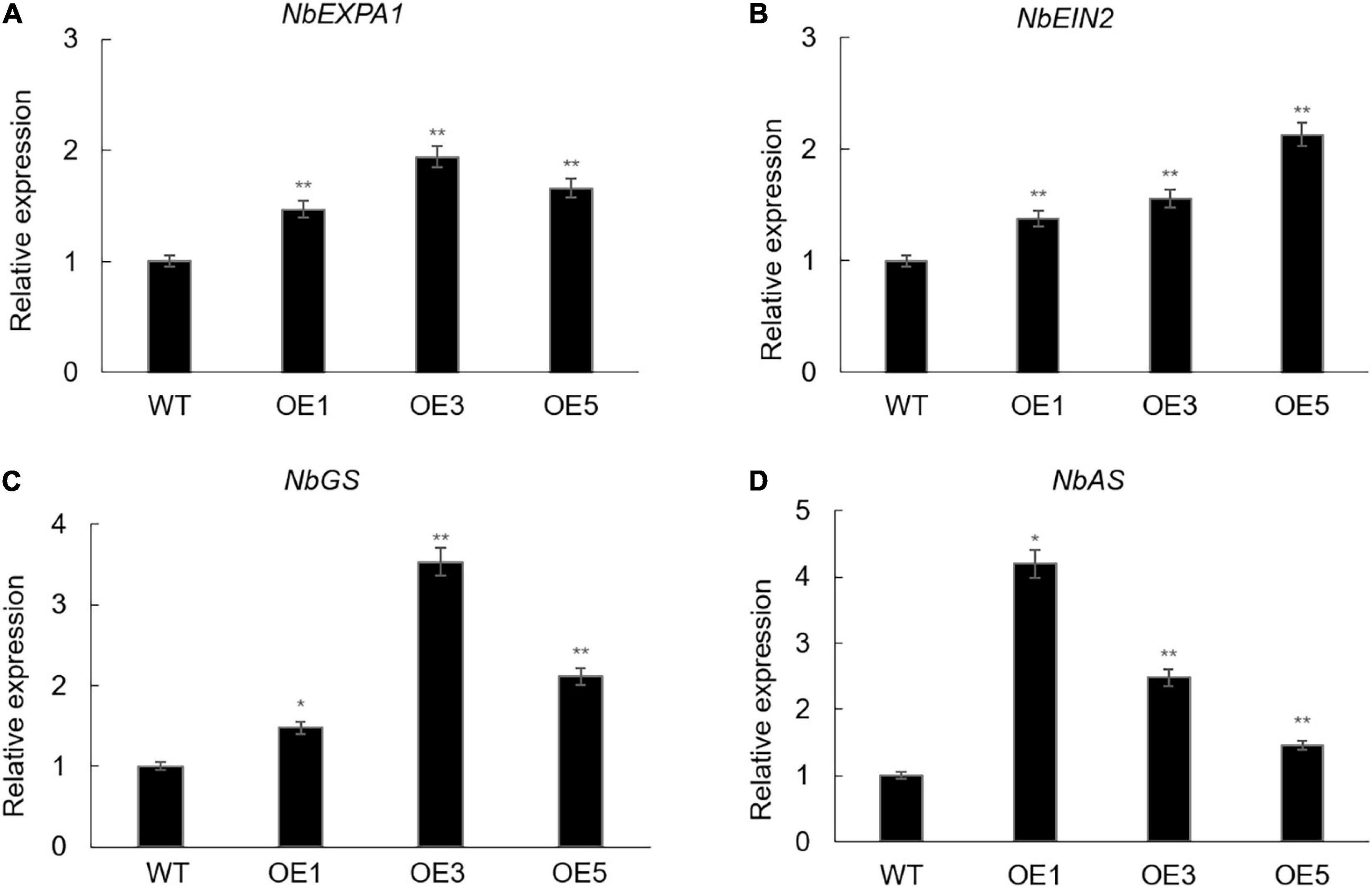
Figure 4. Downstream gene targets of ZmACY-1. (A) Relative expression of NbEXPA1; (B) Relative expression of NbEIN2; (C) Relative expression of NbGS; (D) Relative expression of NbAS. Three biological replicates were used. *P < 0.05, **P < 0.01.
It is predicted by pLoc-mPlant that the ZmACY-1 protein is mainly located in the chloroplast. Compared with the green fluorescence of the empty vector, the green fluorescence of the pCambia1300-ZmACY-1 fusion protein can be seen in both the cell membrane and the chloroplast, indicating that the ZmACY-1 protein plays a role in the cell membrane and chloroplast (Figure 5).
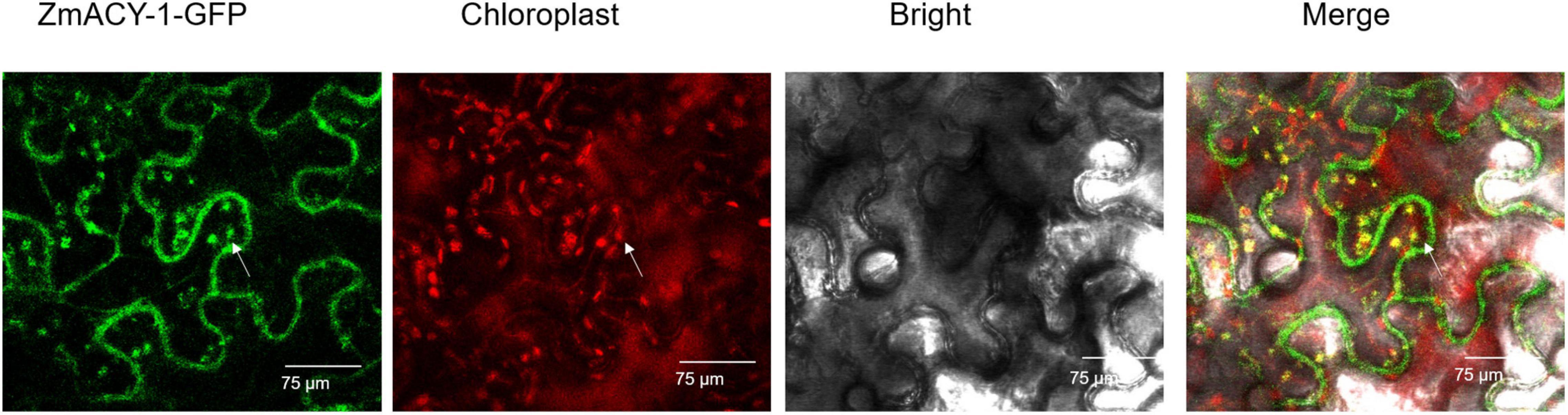
Figure 5. ZmACY-1 accumulates in the chloroplasts. Transient expression of ZmACY-1:GFP in N. benthamiana leaves. The full-length cDNA encoding ZmACY-1 was fused into the pCambia1300-GFP vector. The white arrow illustrates the overlap between ZmACY-1-GFP and the chloroplast signal.
Functional Characterization of ZmACY-1 in Abiotic Stress
To further determine the function of ZmACY-1, we tested the performance of transgenic N. benthamiana under stress conditions. First, we performed a salt stress treatment by growing 1-month-old N. benthamiana in a 350 mM NaCl solution. We found that after 10 days of salt treatment, the transgenic plants exhibited a higher degree of chlorosis than the wild-type plants (Figure 6A). Under normal conditions, the chlorophyll content (Figure 6B) and shoot fresh weight (Figure 6C) in transgenic lines were significantly higher than those of the wild type; while after 10 days of salt stress, the chlorophyll content and shoot fresh weight of the transgenic lines were significantly lower than those of the wild type. This indicated that the overexpression of ZmACY-1 in N. benthamiana reduced salt stress tolerance.
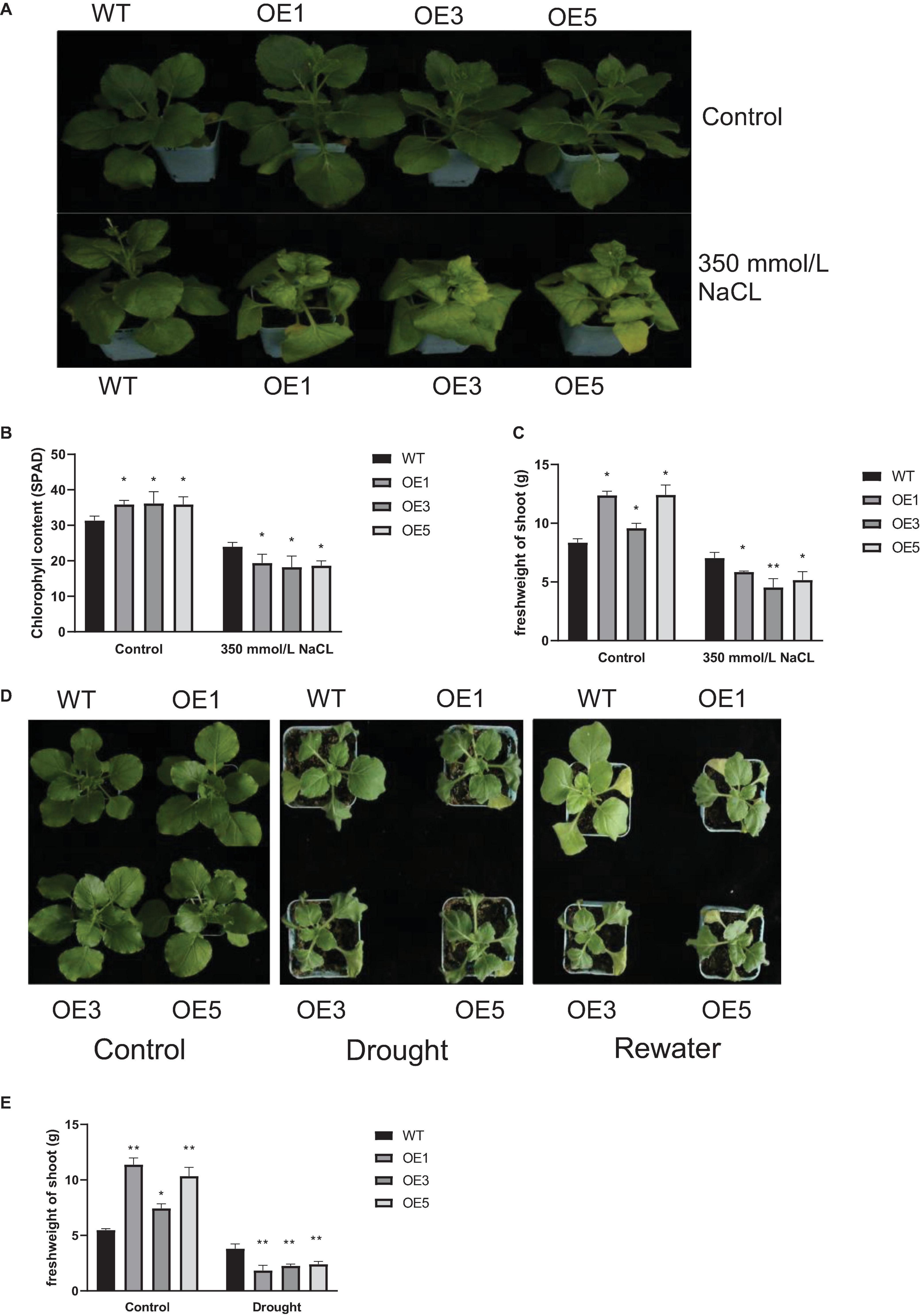
Figure 6. Function of ZmACY-1 in stress responses. (A) Phenotype of N. benthamiana under 350 mM NaCl stress treatment; (B) Chlorophyll content; (C) Shoot fresh weight; (D) Phenotype of N. benthamiana with drought treatment; (E) Shoot fresh weight. Three biological replicates were used. *P < 0.05, **P < 0.01.
Then, we performed drought treatment by withholding water on 1-month-old N. benthamiana. We found that after 7 days of drought treatment, the wilting degree of transgenic lines was more serious than that of the wild type, and that the recovery degree of the transgenic lines was significantly lower than that of the wild type (Figure 6D). Drought stress significantly reduced the shoot fresh weight of the wild-type and transgenic lines (Figure 6E). The shoot fresh weight of transgenic lines was significantly higher than that of the wild type under normal conditions. However, with drought stress, the shoot fresh weight was significantly lower than that of the wild type. These results showed that the overexpression of ZmACY-1 in N. benthamiana reduced drought stress resistance.
To determine the physiological mechanisms, we tested the contents of peroxidase (POD), superoxide dismutase (SOD), catalase (CAT) from micrococcus lysodeikticus, and malondialdehyde (MDA), and relative electrolytic leakage after 2 days of treatment with water, 20% PEG6000, and 350 mM NaCl. We found that drought and salt stress increased POD, SOD, and CAT activities, MDA content, and electrolytic leakage (Figure 7). However, with drought and salt stress, the activities of POD, SOD, and CAT in the transgenic lines were lower, and MDA content and electrolytic leakage was higher than those of the wild type. This further revealed that ZmACY-1 is involved in the process of stress responses.
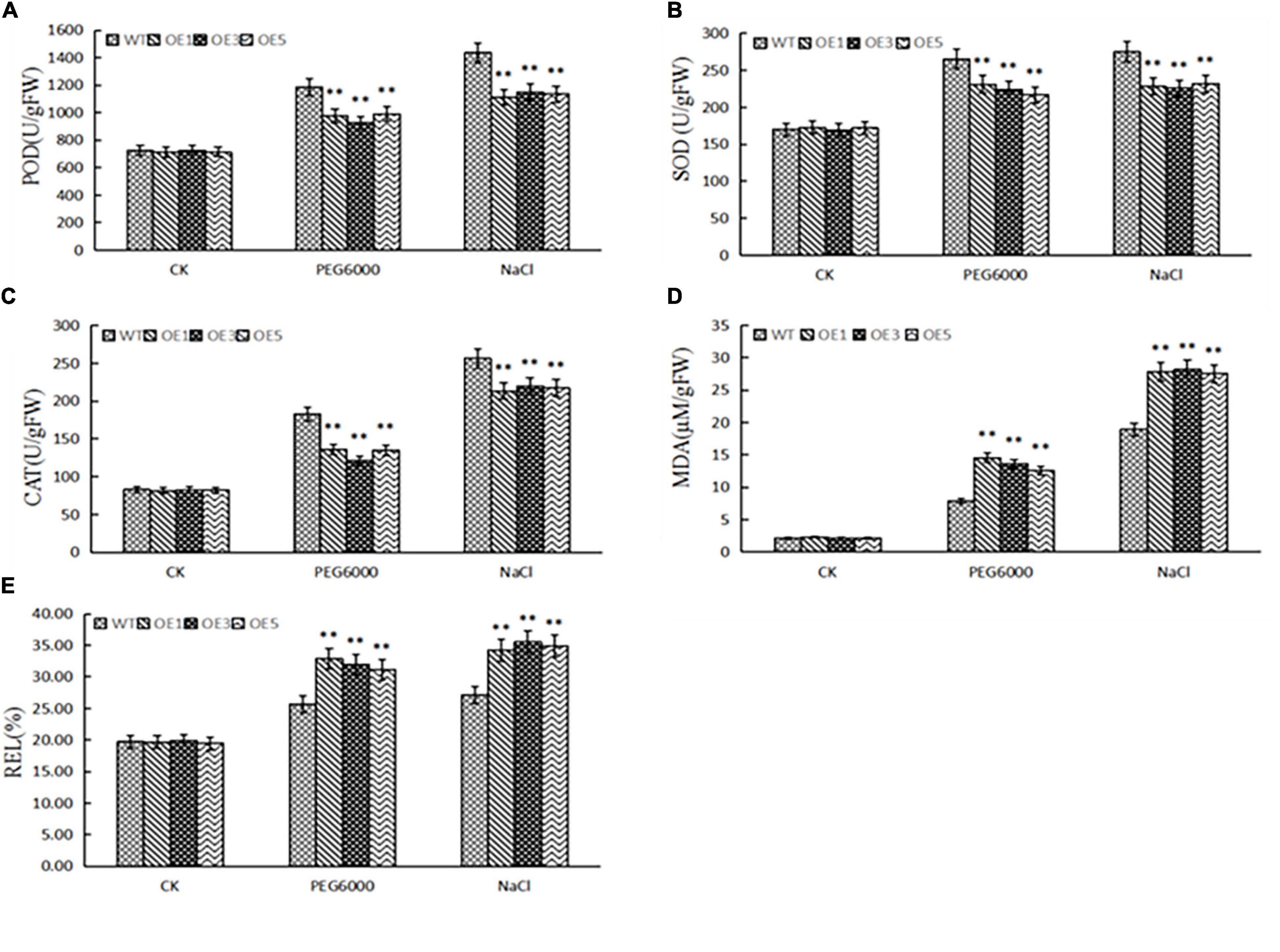
Figure 7. Physiological indices related to salt and drought stress. (A) POD activity; (B) SOD activity; (C) CAT activity; (D) MDA activity; (E) Electrolytic leakage. Three biological replicates were used. *P < 0.05, **P < 0.01.
Discussion
Aminoacylase-1 is a dimer zinc-binding enzyme that can catalyze the hydrolysis of acylated L-amino acids into L-amino acids and acyl groups, and is considered to play a role in the catabolism and recovery of acylated amino acids. In this study, we provided experimental evidence for the function of ZmACY-1 in plants. The finding that ZmACY-1 promotes plant growth while reducing stress tolerance may be a new signaling pathway for plant growth-defense trade-off.
The tissue and spatiotemporal specific expression of ZmACY-1, and regulation by stress and hormones, indicated that ZmACY-1 is involved in growth and development in response to environmental changes (Berens et al., 2019). In the trifoliate-stage maize seedlings, ZmACY-1 is induced by ABA and ethylene while repressed by jasmonic acid and salicylic acid after 3 h; while ZmACY-1 is repressed by both drought and salt stress after 8 h. This indicates that ZmACY-1 responds to plant hormones more quickly than salt and drought stress. The transcription levels of hormone genes NbEIN2, NbGS, and NbAS were higher in the ZmACY-1 transgenic lines than those in the wild type, which is consistent with the function of EIN2, GS, and AS (Li et al., 2015; Németh et al., 2018; Rashmi et al., 2019). ZmACY-1 might promote plant growth rate and enhance nitrogen assimilation ability by positively regulating the expression of plant growth-related genes. By further elucidating how exactly ZmACY-1 controls these genes, we can modify plant growth precisely.
Some genes have been identified in plant growth-defense trade-off, such as DELLA, ERF-type TF, and proline. The stability of DELLA promotes stress tolerance but inhibits growth (Verma et al., 2016). Similarly, ERF6 promotes stress tolerance but inhibits growth through the GA pathway (Dubois et al., 2013). In contrast with DELLA and ERF6, ZmACY-1 promotes plant growth under normal conditions. NEK6 is induced by salt stress stimulating growth (Zhang et al., 2011). Different with NEK6, ZmACY-1 is repressed by salt stresses. Therefore, it may be a new pathway in the growth defense trade-off.
Understanding the signaling pathways in growth-defense trade-off can avoid unnecessary yield loss (Todaka et al., 2015). Aminoacylases are involved in arginine metabolism and methionine (SAM) cycle, which are closely related to stress responses. The data are consistent with the previous study that abiotic stress affects nitrogen assimilation and amino acid metabolism in plants (Charlier and Bervoets, 2019). Under normal conditions, by overexpressing ZmACY-1, we can improve plant yield, while under stressed conditions, we have to carefully control the expression of ZmACY-1 in order to maintain stress tolerance. However, the physiological and molecular mechanisms of ZmACY-1 in maize growth and abiotic stress responses need to be further revealed (Wang et al., 2018). By constructing plasmids and generating maize transgenic lines, the effects of ZmACY-1 on plant growth and the mechanism of abiotic stress responses can be verified.
Experimental Procedures
Plant Materials and Growth Conditions
Maize inbred line Zheng 58 and N. benthamiana were provided by the Crop Breeding Laboratory of Qingdao Agricultural University. Zheng 58 was planted in the field at the Jiaozhou Experimental Station of Qingdao Agricultural University (36° N, 120° E) under normal management. Roots, stems, and leaves of three-leaf stage seedlings; and roots, stems, leaves, tassels, female ears, and aerial roots of mature plants at the heading stage were collected 7 days after pollination for quantitative RT-PCR. N. benthamiana was planted in a growth chamber at 23–26°C. All stress and plant hormones treatments were performed at the three-leaf stage. For salt and drought stresses, 150 mM NaCl and 20% PEG6000 were used. Samples were taken after 8, 16, 24, and 32 h of treatment. For the plant hormone treatment, 50 μM abscisic acid, 2 mM salicylic acid, 50 μM jasmonic acid, or 25 mM ethephon was used. Samples were taken after 1, 3, 6, 9, and 12 h of treatment. All the above experiments were repeated three times, with two plants as biological replicates each time.
Phylogenetic Analysis of ACY-1 Genes
All sequences were downloaded from the NCBI database. The Bioxm software was used to compare the sequences using default parameters. The MEGA 5.1 software was used to analyze the sequences with phylogenetic tree analysis default parameters.
Generation of Overexpression Lines in N. benthamiana
ZmACY-1-XbaI-F and ZmACY-1-BamHI-R were used as primers for high-fidelity PCR amplification. The pCambia1300 expression vector and PMD19-ZmACY-1 PCR product were double digested with XbaI and BamHI at 37°C for 3 h. Then, the DNA Ligation (Takara, Dalian, China) was used for ligation. The recombinant plasmid was transformed into Escherichia coli DH5α and sent to Qingdao QingkeZixi Biotechnology Co., Ltd., (Qingdao, China), for sanger sequencing. The plasmid was transformed into N. benthamiana using LBA4404 competent cells. Transgenic N. benthamiana lines were incubated and propagated under light at 25°C.
RNA Isolation, cDNA Generation, and Quantitative RT-PCR
RNA isolation was performed following the Plant RNA Extraction Kit (Takara, Dalian, China) protocol. RNA quality was tested by agarose gel analysis. cDNA was synthesized by master mix kit (Takara, Dalian, China). Quantitative RT-PCR was performed using a TB Green Premix Ex Taq II (Tli RNaseH Plus) kit (Takara, Dalian, China). The endogenous gene NbEF1a was used as a reference.
Phenotyping
Transgenic and wild-type N. benthamiana seeds were sown in Petri dishes containing MS medium. Each plate contained 100 seeds. Seed germination rate was measured from the third day. To compare the growth rate of transgenic lines and wild type lines, seeds of each line were aseptically sown on the same plate. Twenty-five seeds were used for each line. Transgenic and wild-type N. benthamiana seedlings were cultured in an MS medium for 15 days and then transplanted into soil. After another 15 days, plant height, leaf area, stem thickness, root length, number of leaves, root area, and above-ground weight were measured. Fully developed pods in the middle of the mature stage of tobacco were collected for weighing and fruit length measurement.
Protein Subcellular Localization
The pCambia1300-GFP vector was digested with restriction endonucleases PstI (Takara). The CDS sequences of ZMACY-1 was amplified with gene-specific primer pairs, forward primer: gcttggatcctcgagctgcagATGCCGCCGCCTCTCCGC, reverse primer:acgggtcatgagctcctgcagGCCTTGGAACGAGCTTAGTGC. The PCR products were recovered using the FastPure Gel DNA Extraction Mini Kit (Vazyme Biotech Co., Ltd., Nanjing, China). The ZmACY-1 gene was connected to the pCambia1300-GFP vector using the ClonExpress II One Step Cloning Kit (Vazyme Biotech Co., Ltd., Nanjing, China) The recombinant plasmids were transferred into Agrobacterium tumefaciens strain GV3101. The transformed A. tumefaciens was cultured for 24 h at 28°C in L-broth supplemented with 50 μg/ml kanamycin, centrifuged at 5,000 × g for 10 min at room temperature and resuspended to a density (OD600) of 1. A. tumefaciens cells were infiltrated into the abaxial air spaces of the N. benthamiana plants. After 38 h of infiltration, the expression position of the ZMACY-1 proteins was observed with a Leica SP8 laser confocal microscope (Leica Microsystems, Inc., Buffalo Grove, IL, United States) using filter blocks to select for spectral emission at 488 nm.
Physiology Indexes
Physiology indexes were tested after 350 mM NaCl or 20% PEG6000 treatment.
For MDA, 0.2 g leaves were collected into 3 ml 5% trichloroacetic acid (TCA). After grinding, the mixture was centrifuged at 5,000 rpm for 10 min. The supernatant was transformed in a test tube. An equal volume of 0.5% thiobarbital acid (TBA) was added. After mixing, the samples were incubated in a boiling water bath for 30 min, and absorbance values at 450, 532, and 600 nm were tested. MAD content was calculated using Eq. 1:
For relative conductivity, 0.2 g leaves were collected in a test tube. Deionized water 20 ml was added until the leaves were immersed. Then, the samples were incubated at room temperature for 4 h. Conductivity R1 was first measured using a conductivity meter. After boiling for 25 min in a water bath, conductivity R2 was measured. Relative conductivity was calculated using Eq. 2:
For SOD activity, 0.2 g leaves were collected in a mortar. Pre-chilled 0.1 mol/L Tris–HCL buffer solution 20 ml was added for grinding. Then, the samples were transferred to a 2-ml centrifuge tube and centrifuged at 8,000 rpm for 30 min at 4°C. The supernatant of the crude enzyme solution was stored at −20°C. The SOD activity was measured using the nitrogen blue tetrazole (NBT) method, and the treated crude enzyme liquid sample was measured with a microplate reader at a wavelength of 560 nm. The SOD activity was calculated using Eq. 3:
A0 is the absorption value of the control tube at 560 nm, A is the absorbance value of the sample tube at 560 nm, VT is the total volume of enzyme extract (ml), V1 is the volume of enzyme solution added during measurement (ml), and W is the fresh weight of sample (g).
The POD activity was measured using the guaiacol colorimetric method. The crude enzyme solution sample was measured for absorbance at 470 nm using a microplate reader, and tested every 1 min for five times in total. The POD activity was calculated using Eq. 4:
ΔA470 is the change in light absorption within reaction time (t), C is the concentration of protein in enzyme solution (μg/μl), VS is the volume of enzyme solution used for measurement (ml), and t is the reaction time (min).
Accession Numbers
Sequence data can be found in the NCBI databases under the following accession numbers: ZmACY-1: NP_001150325.2, NbEXPA1: NM_001325646.1, NbEIN2: XM_016579720.1; NbGS: XM_016631331.1, and NbAS: XM_016658168.1.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
DC, XS, and XG designed the study. DC, JL, FJ, and QW performed the experiments. DC, JL, and FJ analyzed the data and wrote the draft manuscript. JL, YP, and MZ discussed and revised the draft manuscript. All authors read and approved the final manuscript.
Funding
This study was supported by the National Key Research and Development Program of China (2016YFD0102104), the National Natural Science Foundation of China (Grant No: 31201218), and the Taishan Scholars Program of Shandong Province (No. tsqn201909134).
Conflict of Interest
The Qingdao Agricultural University has filed a provisional patent application on Aminoacylase-1 in China (202010088971.1) and a provisional patent application on Aminoacylase-1 in Luxembourg (102487) arising from this work.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.593001/full#supplementary-material
Supplementary Figure 1 | Expression patterns of ZmACY in overexpressed lines. Wild-type and transgenic N. benthamiana were planted in a growth chamber at 23–26°C for 7 days. Three biological replicates were used. **P < 0.01.
Supplementary Table 1 | Primers used in this study.
References
Berens, M. L., Wolinska, K. W., Spaepen, S., Ziegler, J., Nobori, T., Nair, A., et al. (2019). Balancing trade-offs between biotic and abiotic stress responses through leaf age-dependent variation in stress hormone cross-talk. Proc. Natl. Acad. Sci. U.S.A. 116, 2364–2373. doi: 10.1073/pnas.1817233116
Charlier, D., and Bervoets, I. (2019). Regulation of arginine biosynthesis, catabolism and transport in Escherichia coli. Amino Acids 51, 1103–1127. doi: 10.1007/s00726-019-02757-8
Cho, S. G., Lee, Y. H., Park, H. S., Ryoo, K., Kang, K. W., Park, J., et al. (2001). Glutathione S-transferase mu modulates the stress-activated signals by suppressing apoptosis signal-regulating kinase 1. J. Biol. Chem. 276, 12749–12755. doi: 10.1074/jbc.m005561200
Claeys, H., and Inzé, D. (2013). The agony of choice: how plants balance growth and survival under water-limiting conditions. Plant Physiol. 162, 1768–1779. doi: 10.1104/pp.113.220921
Dubois, M., Skirycz, A., Claeys, H., Maleux, K., Dhondt, S., De Bodt, S., et al. (2013). Ethylene Response Factor6 acts as a central regulator of leaf growth under water-limiting conditions in Arabidopsis. Plant Physiol. 162, 319–332. doi: 10.1104/pp.113.216341
Goh, H. H., Sloan, J., Dorca-Fornell, C., and Fleming, A. (2012). Inducible repression of multiple expansin genes leads to growth suppression during leaf development. Plant Physiol. 159, 1759–1770. doi: 10.1104/pp.112.200881
Gupta, A., Rico-Medina, A., and Caño-Delgado, A. (2020). The physiology of plant responses to drought. Science 368, 266–269.
Hernandez, J. A., Jiménez, A., Mullineaux, P., and Sevilia, F. (2000). Tolerance of pea (Pisum sativum L.) to long-term salt stress is associated with induction of antioxidant defences. Plant Cell Environ. 23, 853–862.
Koevoets, I. T., Venema, J. H., Elzenga, J. T., and Testerink, C. (2016). Rootss withstanding their environment: exploiting root system architecture responses to abiotic stress to improve crop tolerance. Front. Plant Sci. 7:1335. doi: 10.3389/fpls.2016.01335
Lantzouni, O., Alkofer, A., Falter-Braun, P., and Schwechheimer, C. (2020). Growth-Regulating Factors interact with DELLAs and regulate growth in cold stress. Plant Cell 32, 1018–1034. doi: 10.1105/tpc.19.00784
Li, W., Ma, M., Feng, Y., Li, H., Wang, Y., Ma, Y., et al. (2015). EIN2-directed translational regulation of ethylene signaling in Arabidopsis. Cell 163, 670–683. doi: 10.1016/j.cell.2015.09.037
Maceyka, M., Nava, V. E., Milstien, S., and Spiegel, S. (2004). Aminoacylase 1 is a sphingosine kinase 1-interacting protein. FEBS Lett. 568, 30–34. doi: 10.1016/j.febslet.2004.04.093
Nakano, M., Nishihara, M., Yoshioka, H., Ohnishi, K., Hikichi, Y., and Kiba, A. (2014). Silencing of DS2 aminoacylase-like genes confirms basal resistance to Phytophthora infestans in Nicotiana benthamiana. Plant Signal. Behav. 9:e28004. doi: 10.4161/psb.28004
Németh, E., Nagy, Z., and Pécsváradi, A. (2018). Corrigendum: chloroplast glutamine synthetase, the key regulator of nitrogen metabolism in wheat, performs its role by fine regulation of enzyme activity via negative cooperativity of its subunits. Front. Plant Sci. 9:466. doi: 10.3389/fpls.2018.00466
Rashmi, D., Barvkar, V. T., Nadaf, A., Mundhe, S., and Kadoo, N. Y. (2019). Integrative omics analysis in Pandanus odorifer (Forssk.) Kuntze reveals the role of Asparagine synthetase in salinity tolerance. Sci. Rep. 9:932.
Roxas, V. P., Smith, R. K., Allen, E. R., and Allen, R. D. (1997). Overexpression of glutathione S-transferase/glutathioneperoxidase enhances the growth of transgenic tobacco seedlings during stress. Nat. Biotechnol. 15, 988–991. doi: 10.1038/nbt1097-988
Schwörer, S., Berisa, M., Violante, S., Qin, W., Zhu, J., Hendrickson, R. C., et al. (2020). Proline biosynthesis is a vent for TGFβ-induced mitochondrial redox stress. EMBO J. 39:e103334.
Shi, H., Ye, T., Chen, F., Cheng, Z., Wang, Y., Yang, P., et al. (2013). Manipulation of arginase expression modulates abiotic stress tolerance in Arabidopsis: effect on arginine metabolism and ROS accumulation. J. Exp. Botany 64, 1367–1379. doi: 10.1093/jxb/ers400
Todaka, D., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2015). Recent advances in the dissection of drought-stress regulatory networks and strategies for development of drought-tolerant transgenic rice plants. Front. Plant Sci. 6:84. doi: 10.3389/fpls.2015.00084
Van Zelm, E., Zhang, Y., and Testerink, C. (2020). Salt tolerance mechanisms of plants. Annu. Rev. Plant Biol. 71, 403–433.
Van Wallendael, A., Soltani, A., Emery, N. C., Peixoto, M. M., Olsen, J., and Lowry, D. B. (2019). A molecular view of plant local adaptation: incorporating stress-response networks. Annu. Rev. Plant Biol. 70, 559–583. doi: 10.1146/annurev-arplant-050718-100114
Verma, V., Ravindran, P., and Kumar, P. P. (2016). Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 16:86. doi: 10.1186/s12870-016-0771-y
Wang, J., Zhou, L., Shi, H., Chern, M., Yu, H., Yi, H., et al. (2018). A single transcription factor promotes both yield and immunity in rice. Science 361, 1026–1028. doi: 10.1126/science.aat7675
Xie, Z., Nolan, T. M., Jiang, H., and Yin, Y. (2019). AP2/ERF transcription factor regulatory networks in hormone and abiotic stress responses in Arabidopsis. Front. Plant Sci. 10:228. doi: 10.3389/fpls.2019.00228
Keywords: aminoacylase-1, salt stress, drought stress, plant growth, trade-off, Nicotiana benthamiana
Citation: Chen D, Li J, Jiao F, Wang Q, Li J, Pei Y, Zhao M, Song X and Guo X (2021) ZmACY-1 Antagonistically Regulates Growth and Stress Responses in Nicotiana benthamiana. Front. Plant Sci. 12:593001. doi: 10.3389/fpls.2021.593001
Received: 09 August 2020; Accepted: 21 June 2021;
Published: 23 July 2021.
Edited by:
Honghong Wu, Huazhong Agricultural University, ChinaReviewed by:
Xianchen Zhang, Anhui Agricultural University, ChinaJiangman He, University of California, Riverside, United States
Copyright © 2021 Chen, Li, Jiao, Wang, Li, Pei, Zhao, Song and Guo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiyun Song, c29uZ3h5QHFhdS5lZHUuY24=; Xinmei Guo, eG1ndW8yMDA5QDEyNi5jb20=
†These authors have contributed equally to this work
 Dongbin Chen
Dongbin Chen Junhua Li2,3†
Junhua Li2,3† Fuchao Jiao
Fuchao Jiao Qianqian Wang
Qianqian Wang Jun Li
Jun Li Meiai Zhao
Meiai Zhao Xinmei Guo
Xinmei Guo