- 1Department of Biotechnology, Collage of Life Science and Technology, Huazhong University of Science and Technology, Wuhan, China
- 2School of Agricultural Sciences, Zhengzhou University, Zhengzhou, China
- 3Faculty of Biology and Food Engineering, Guangdong University of Petrochemical Technology, Maoming, China
Drought stress is one of the most environmental abiotic stresses affecting seed germination and crop growth. In the present study, the genetic characteristics of seed germination under drought stress in a Brassica napus double haploid population were analyzed. Five germination-related indexes, including germination percentage (GP), root length (RL), shoot length (SL), fresh weight (FW), and root-to-shoot length ratio (R/S) under control and drought stress, were calculated, and the drought stress index (DSI), including DSI-GP, DSI-RL, DSI-SL, DSI-FW, and DSI-R/S, was determined using the quantitative trait loci (QTLs) analysis based on high-density genetic linkage map. The phenotypic analysis indicated that the R/S is an effective morphological trait in the determination of drought tolerance in the seedling stage. Thirty-nine identified QTLs were observed for these traits and then integrated into 36 consensus QTLs, in which 18 QTLs were found to affect the DSI of four traits (GP, RL, SL, and R/S). Based on the co-linearity between genetic and physical maps of B. napus, 256 candidate genes were detected, and 128 genes have single-nucleotidepolymorphisms/insertion–deletion (SNP/InDel) variations between two parents, some of which were associated with the drought stress tolerance (for example, BnaC03g32780D, BnaC03g37030D, and BnaC09g27300D). The present results laid insights into drought tolerance and its genetic bases in B. napus.
Introduction
Worldwide, plants live in permanently changing environments that are often undesirable or stressful for growth and development. These unfavorable conditions include abiotic stresses, such as drought, heat, nutrient deficiency, cold, and salinity, and biotic stresses, such as herbivore attack and pathogen infections. One of the most important abiotic stresses impairing seed germination and yield in global agriculture is drought, which affects 40% of the world’s land area (Zhang et al., 2014).1 Drought significantly affects seed germination, leading to decrease of plant density and yield. In the future, it is expected that drought will probably increase in certain areas of the world. Unfortunately, global climate change could lead to increased drought in many parts of the world and thus could have a major impact on crops (Trenberth et al., 2013). Therefore, there is an urgent need to produce crop varieties capable of adapting to such conditions and maintaining high levels of productivity. These challenges have prompted scientists to do more to improve the ability of crops to resist such harsh conditions (Zhang et al., 2014).
Rapeseed (Brassica napus L., AACC, 2n = 4x = 38), which belongs to Brassicaceae, is one of the important crops, with soybean and oil palm considered as the largest oil crops in the world (Zhang D. et al., 2015). It is very sensitive to drought stress, which leads to severe yield reduction (Wan et al., 2009). The development of varieties with high tolerance to drought and drought-related stress could maintain the high yield production in some cases (Wan et al., 2009).
The genetic dissection of the quantitative traits that control the adaptation of crops to adverse conditions is a prerequisite for the development of many effective approaches, which aimed to improve the ability of accommodating itself to adversity and enhance yield production under abiotic stress (Li et al., 2014). It is worth mentioning that the performance of crops represents the final result of interactions between thousands of genes and environmental conditions as well as culture practices (Collins et al., 2008), and it is clear that analyzing quantitative trait loci (QTLs) is an effective method for dissecting the complex quantitative traits, which has been widely used in many crops, such as in wheat (Wang et al., 2018), rapeseed (Zhou et al., 2014), common bean (Sandhu et al., 2018), and peanut (Zhao et al., 2016). As QTLs were correlated with phenotype variation, the corresponding loci could be amplified and therefore used for phenotypic improvement (Raboanatahiry et al., 2017). QTL map studies for drought tolerance were conducted in many crops including Arabidopsis, rice, wheat, and soybean. However, the use of low marker density genetic linkage map was not able to reveal the comprehensive genetic diversity of drought tolerance in rapeseed (Li et al., 2014). Despite the progress made in the plant’s response to drought stress, many genetic bases of drought tolerance in rapeseed has yet to be clarified (Li et al., 2014; Zhang, 2015; Zhao et al., 2016; Liang et al., 2019). There has been significant progress in understanding the signaling process that controls plant resistance to drought from perception of signals to cellular mechanisms (Seki et al., 2007), and it was revealed that the signal pathways including many common factors, such as abscisic acid, photosynthesis, and reducing ROS contents under drought, were involved in drought resistance (Jackson et al., 2003; Li et al., 2006; Dudziak et al., 2019; Liang et al., 2019).
Recently, Chao et al. (2017) have developed a high-density genetic linkage map with 3207 markers based on the KN double haploid (DH) population. In the present study, the germination-related traits including germination percentage (GP), root length (RL), shoot length (SL), fresh weight (FW), and root-to-shoot length ratio (R/S), as well as the drought stress index (DSI) of these traits, were used to dissect the genetic basis of germination response under drought stress based on QTL mapping in B. napus, and candidate genes within the QTLs associated with drought tolerance during seed germination were identified.
Materials and Methods
Plant Material and Genetic Linkage Map
The segregation DH population, named KN, was used in this experiment. The KN DH population was derived from a cross between the two parental lines KenC-8 and N53-2 first constructed by Wang et al. (2013). The high-density single-nucleotide polymorphisms (SNP)-based linkage map, including 3106 SNP bins (including 17,978 SNPs) and 101 non-SNP markers (SSR and STS) with an average genetic distance of 0.96 cM between adjacent loci constructed by Chao et al. (2017), was used for QTL mapping in the current study.
Experimental Design and Evaluation of the Phenotypic Indexes
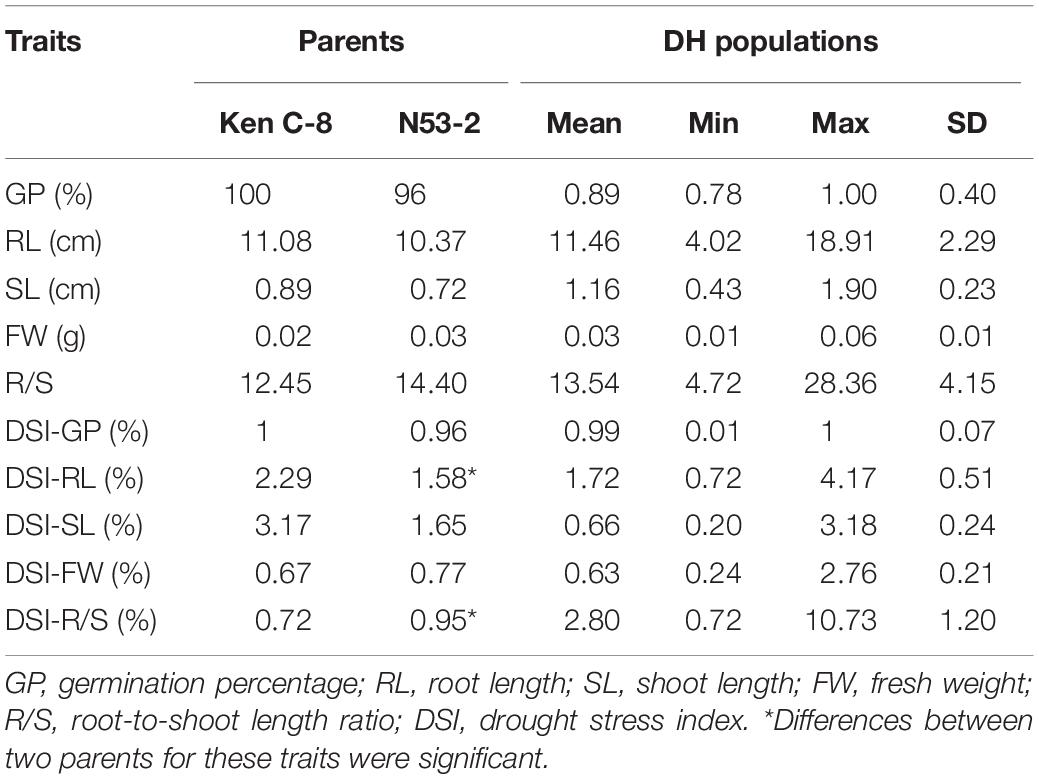
The seeds of two parents (KenC-8 and N53-2) and KNDH lines were disinfected firstly with 70% ethanol and prepared for seed germination test. Germination test was carried out in a seed germination box (L × W × H = 12 cm × 12 cm × 5 cm). Firstly, two layers of filter paper were placed on the bottom of the dish and then 15 ml of ddH2O (control) or 15% BEG6000 solution (drought stress treatment) was added (Michel and Kaufmann, 1973). Fifty seeds were sowed in each dish and covered and then the dishes were placed in the growing room. The germination rate was calculated after 7 days of cultivation, and three representative single seedlings from each line were selected to measure biological indicators including GP, RL, SL, FW, and R/S, and three biological repeats were implemented for the survey. Drought resistance was measured by the drought resistance coefficient (ratio of treatment to control) and DSI = traits under drought/traits under control × 100% (Zhang J. et al., 2015).
QTL Analysis
Quantitative trait locus analysis was performed as described by Wang et al. (2013). Windows QTL Cartographer 2.5 software was employed to identify the QTLs using the composite interval mapping (CIM) method (Zeng, 1994). The window size was 10 cM and the scan walking speed was 2 cM for the CIM method. The logarithm of odds (LOD) thresholds of QTL were determined by 1,000 permutation tests at the 95% confidence level (Doerge and Churchill, 1996). The BioMercator V4.2 program (Arcade et al., 2004) was used to integrate the QTL detected under control and drought stress with overlapping CIs into consensus QTL. The integration and nomenclature of QTLs were determined according to the method described by Wang et al. (2016). “K” was added at the end of the QTL name for QTLs detected under control to distinguish them from those detected under drought stress; for example, two QTLs, qRLK-6-1, and qSLK-11-1, were associated with root and SL under control, while two QTLs, qRL-9-1, and qSL-2-1, were associated with RL and SL under drought stress, respectively.
Identification of Candidate Genes
Candidate genes were detected according to the description of Cai et al. (2014). The physical and genetic map alignment relationship was confirmed using all SNPs in the genetic map, and 50 per probe sequence were obtained by Illumina Inc., as a query in the detection of homolog loci by the NBCI-Blstn local program against the “Darmor-bzh” reference B. napus genome (Chao et al., 2017; Rotmistrovsky et al., 2004). The detected regions on the genome that align with the confidence interval of QTLs were considered as QTL regions, and the genes located within the QTL were defined as candidate genes of the QTL, as described by the method of Chao et al. (2017). SNP/insertion–deletion (InDel) variations of candidate genes were analyzed according to the re-sequencing data of the two parents (SRA accession is SRP156346).2
Results
Phenotypic Variation of the Parents and the KN DH Population
The distribution of germination-related traits under drought in the two parents and 300 DH lines in KH populations is shown in Figure 1. Table 1 shows that significant differences were observed between the mean for DSI-RL and DSI-R/S traits of the two parents, while non-significant differences were observed for other traits under drought stress. SL, RL, GP, DSI-RL, and DSI-SL of the parent KenC-8 were higher than those of the parent N53-2, whereas the other traits (FW, R/S, DSI-FW, and DSI-R/S) were low compared to those of N53-2. On the other hand, there was transgressed segregation as well as continuous frequency distribution in all traits under drought stress (Figure 1). Compared to normal conditions, the variation’s average in RL and R/S increased and decreased in SL, while no change in variation’s average was detected in FW between control and drought conditions (Figure 1).
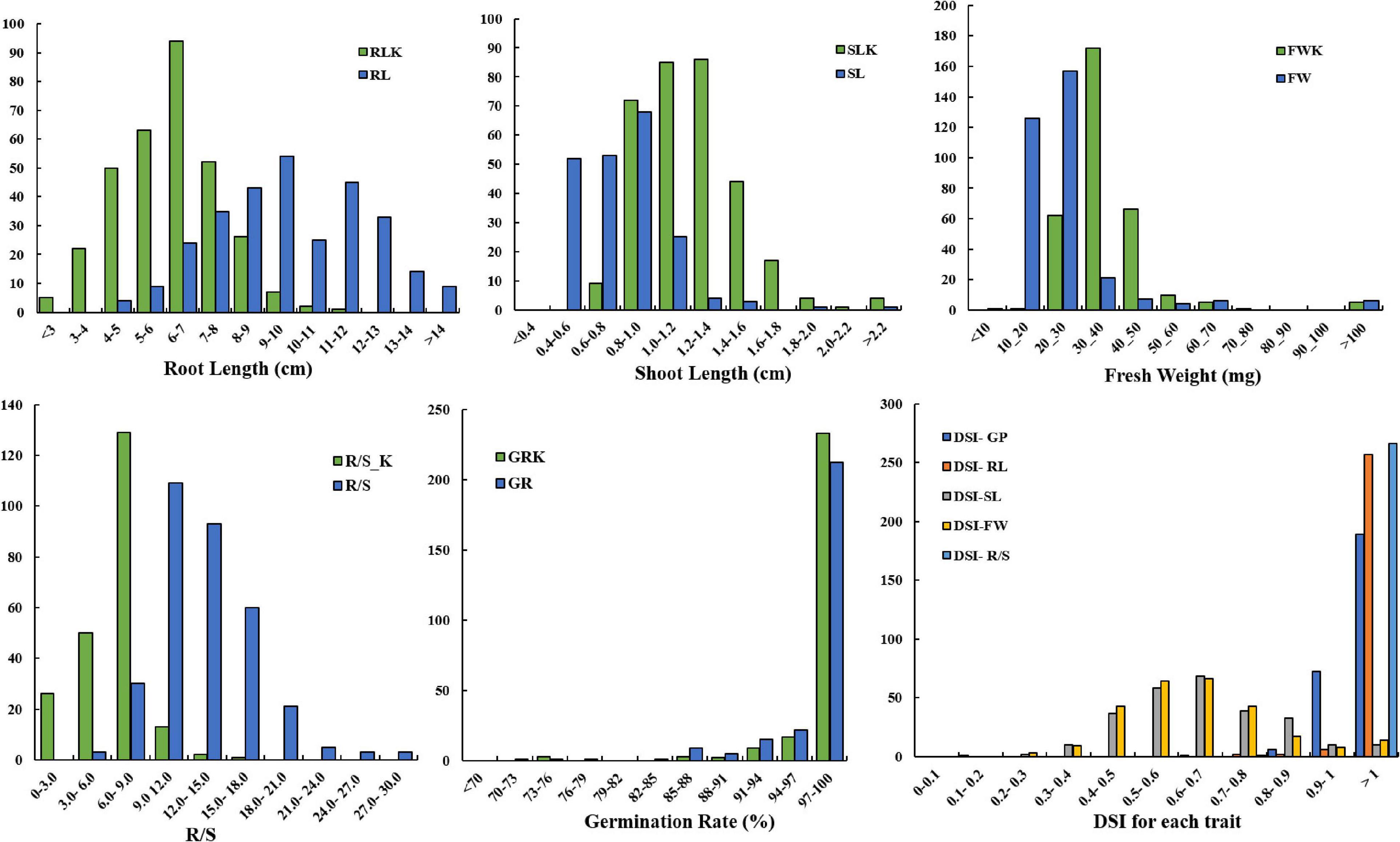
Figure 1. Distribution of germination traits under normal conditions and drought stress conditions. The units on the x-axis are the phenotypic values, and the units on the y-axis are the number of lines.
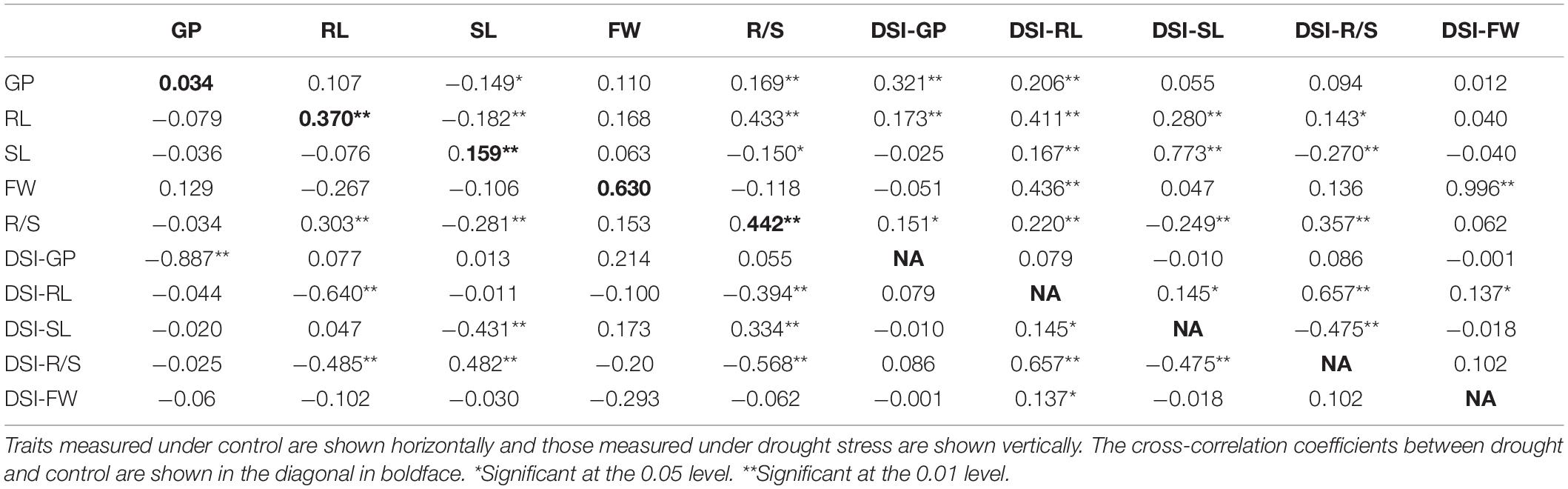
Correlation Among Seedling Traits Under Control and Drought Conditions
As shown in Table 2, a significantly positive correlation was observed for a pair of traits between control and drought stress. R/S was significantly and positively correlated with GP and RL; further analysis revealed that it was significantly and negatively correlated with SL and FW under drought stress. RL was significantly and positively correlated with R/S and slightly positively correlated with GP, while it was significantly and negatively correlated with DSI-RL and DSI-R/S under drought conditions. SL was significantly and negatively correlated with GP, RL, D/S, and DSI-SL, while it was significantly and positively correlated with DSI-R/S under drought stress. DSI-GP was significantly and positively correlated with RL and R/S under drought conditions. DSI-RL was significantly and positively correlated with all measured traits under drought stress. DSI-SL was significantly and positively correlated with RL and SL, while it was negatively correlated with R/S under drought. In addition, DSI-R/S was significantly and positively correlated with RL and R/S, while it was significantly and negatively correlated with R/S. Finally, DSI-FW was significantly and positively correlated with FW, while it was slightly positively correlated with GP and RL under drought stress.
QTL Mapping for Germination-Related Traits Under Control and Drought Conditions
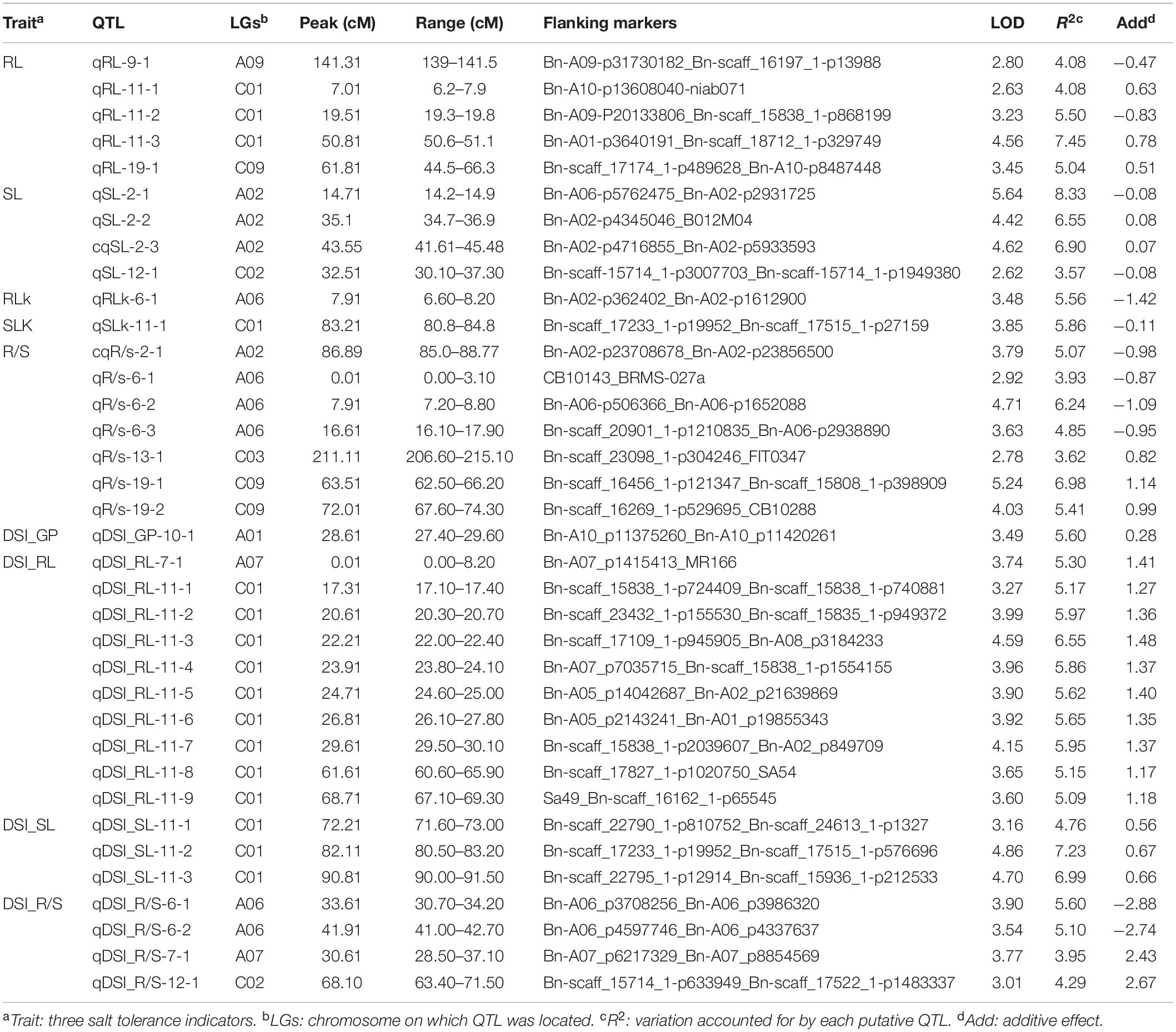
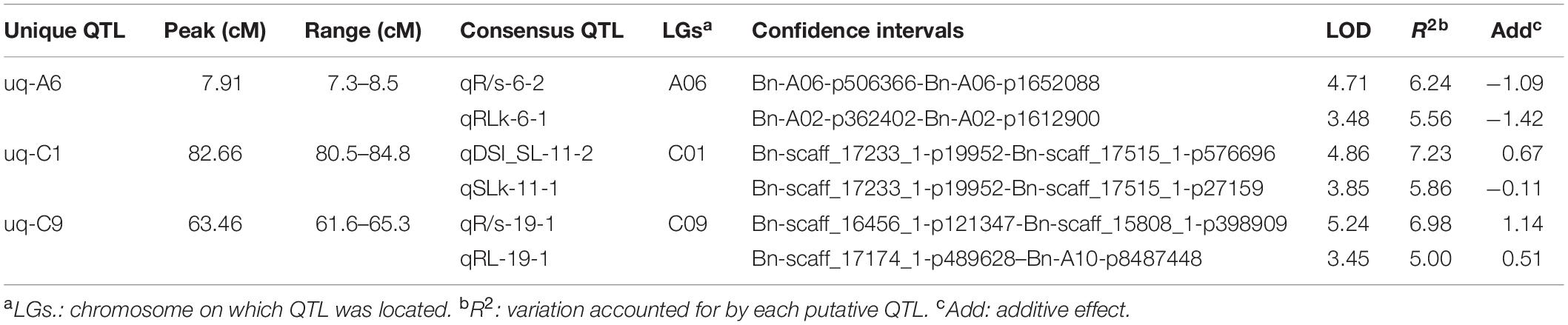
Using the high-density SNP linkage map and phenotypic data of seedling traits under control and drought conditions, a total of 39 QTLs were detected in KN DH populations (Supplementary Table 1). Thirty-nine identified QTLs were integrated into 36 consensus QTLs, of which 2 QTLs were detected under control, 16 QTLs were detected for individual traits under drought stress, and 18 QTLs controlled DSI for the five individual traits (Figure 2 and Table 3). 14 QTLs were located on the A sub-genome, while the remaining 22 QTLs were located on the C sub-genome. According to the integration results, three unique QTLs (uq-A6, uq-C1, and uq-C9) were detected on A06, C01, and C09, respectively (Table 4).

Figure 2. The locations of putative QTLs for drought tolerance. The numbers of linkage groups are shown at the top, and the genetic distance (cM) and markers are indicated on the left and right, respectively. The QTLs for germination traits are signed on the right of linkage groups.
For QTL analysis under control conditions, two QTLs (qRLK-6-1 and qSLK-11-1) were detected for RL and SL under control conditions on A06 and C01, respectively. The two QTLs have a negative effect on RL and SL, accounting for 5.56 and 5.86% of phenotypic variation (PV), respectively. No QTLs were detected for R/S, FW, and GP under control conditions.
For QTL analysis under drought conditions, a total of 16 QTLs were detected for three different traits (RL, SL, and R/S) under drought conditions (Supplementary Table 1). Compared to other chromosomes, A02 and C01 contained 19 QTLs associated with more than one trait. Seven QTLs associated with R/S trait under drought stress were located on A02, A06, C03, and C09. cqR/S-2-1 was detected on A02 and accounted for 6.39% of PV. Three QTLs (qR/S-6-1, qR/S-6-2, and qR/S-6-3) were detected on A06 and accounted for 3.13, 6.24, and 4.85% of PV, respectively. Three other QTLs for R/S, qR/S-13-1, qR/S-19-1, and qR/S-19-2 that accounted for 3.62, 6.98, and 5.41% of total PV were located on C03 and C09, which have a positive effect on the R/S ratio. For RL, five QTLs were mapped to three chromosomes. Three QTLs (qRL-11-1, qRL-11-2, and qRL-11-3) were located on C01 and showed 2.63, 5.50, and 7.45% of PV, respectively. In addition, two QTLs associated with RL (qRL-9-1 and qRL-19-1) were detected on A09 and C09, accounting for 2.80 and 3.45% of the PV, respectively. Four QTLs were detected for SL, of which three QTLs (qSL-2-1, qSL-2-2, and qSL-2-3) were located on A02, accounting for 5.64, 4.42, and 4.60% of PV, respectively. No QTLs were detected for FW and GP under drought conditions.
For QTL mapping for DSI, 18 QTLs affecting DSI of four traits (GP, RL, SL, and R/S) were identified on different chromosomes, including A06, A07, A10, C01, and C02 (Supplementary Table 2), and no QTL was detected for DSI-FW. The distribution of these QTLs was as follows: one QTL for GP was detected on A10 and accounted for 5.6% of PV. Ten QTLs for RL (one QTL on A07 and nine QTLs on C01) and PV ranged from 5.09 to 6.55%. Three QTLs for SL on C01 with PV ranged from 4.76 to 7.23%. In addition, four QTLs for R/S on A06, A07, and C02 with PV ranged from 3.95 to 5.60%.
Candidate Genes in Response to Drought Stress
According to the co-linearity between genetic and physical maps of B. napus, 256 candidate gene homologs with 234 genes involved in drought tolerance in Arabidopsis thaliana were detected (Supplementary Table 3). According to the data from re-sequencing of the two parents, 128 candidate genes have SNP/InDel variations between the two parents (Supplementary Table 4). The candidate genes were located within 36 QTLs on both the A and C genome and associated with three individual traits (RL, SL, and R/S) and DSI for all individual traits except FW.
Based on gene function annotation and genomic variation analysis, many important candidate genes involved in drought-related stress were identified. BnaC01g35030D that was homologous to the A. thaliana gene AT1G18080 was located on chromosome C01 within the two QTLs (qDSI_RL-11-1 and qDSI_SL-11-3) associated with both RL and SL under drought stress, which encoded a protein involved in cellular responses to abscisic acid. BnaC01g32600D was located on chromosome C01 within the two DSI-QTLs (qDSI_RL-11-2 and qDSI_SL-11-3) associated with both RL and SL under drought stress and encoded for an ortholog of the A. thaliana gene AT3G20340 involved in oxidative stress. BnaA06g02020D was located within QTLs qR/S-6-2 and qRLK-6-2, associated with R/S under drought and RL under control conditions, respectively, and orthologous to the A. thaliana gene “LysM-containing receptor-like kinase 3 (LYK3),” which functions in the abscisic acid-activated signaling pathway. BnaC01g34530D in QTL qSLK-11-1 was associated with SL under control, and it was orthologous to the A. thaliana gene “HALOTOLERANCE DETERMINANT 3 (HAL3A).” BnaC03g32780D within QTL qR/S-13-1 on chromosome C03 associated with R/S under drought was orthologous to the A. thaliana gene “ABI BINDING PROTEIN4 (AFP4).” BnaC03g37030D in QTL qR/S-13-1 was orthologous to the A. thaliana gene “DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2 (DREB2).” BnaC03g44440D was located in QTL qR/S-13-1 associated with R/S under drought conditions and orthologous to the A. thaliana gene “RING/U-box superfamily protein (XERICO).” BnaC03g45915D within QTL qR/S-13-1 was orthologous to the A. thaliana gene “LOW-TEMPERATURE-INDUCED 65, (RD29B).” BnaC03g45990D in QTL qR/S-13-1 associated with R/S under drought stress was an ortholog of the A. thaliana gene “COLD REGULATED 413 PLASMA MEMBRANE1 (COR413-PM1).” BnaC03g46570D was detected in QTL qR/S-13-1, orthologous to the A. thaliana gene “ASCORBATE PEROXIDSE 3 (APX3).” BnaC03g49530D in QTL qR/S-13-1 associated with R/S under drought conditions and orthologous to the A. thaliana gene “ABA REPRESSORE1, (ABR1).” BnaC03g64160D was detected within QTL qR/S-13-1 and was an ortholog of the A. thaliana gene “A. THALIANA MYB 4 (ATM4).” BnaC09g24550D in QTLs qR/S-19-1 and qRL-19-1 was associated with R/S and RL under drought stress, respectively, and orthologous to the A. thaliana gene “A. THALIANA INDUCAR OF CBP EXPRESSION 1 (ATICE1).” Two genes, BnaC09g27300D and BnaC09g27320D, were also located within QTL qR/S-19-1 and RL-19-1, and orthologous to the A. thaliana genes “RUBISCO ASSEMPLY FACTOR 2 (RAF2)” and “ATYY1”, respectively. BnaC09g36060D was detected in QTL qR/S-19-2 associated with R/S and orthologous to the A. thaliana gene “METHYLTHIOALKYLMALATE SYNTHASE 2 (MAM2)” (Supplementary Table 3). These important candidate genes provided insights into drought tolerance in B. napus, and their function could be verified further though a trans-gene method in the future.
Discussion
PV Under Drought Stress
Drought is one of the crises we are facing in the present time; therefore, it is necessary to develop new drought-tolerant crops with high seed germination vigor. Drought tolerance of crops is a very complex phenomenon because it is controlled by interaction between several factors, and some factors are only activated under drought stress. So, it is very difficult to evaluate these factors under non-stressed conditions. One of the effective methods to evaluate drought tolerance in crops is through the using of morphological traits under drought stress. In the present study, we used a number of morphological indexes to determine drought tolerance of B. napus and QTLs related to drought tolerance. It was revealed that the R/S was sensitive to drought stress, and it was consistent with the results of Dhanda et al. (2004) in wheat. On the other hand, higher correlation was also detected between R/S and DSI and other morphological traits (Table 2). In this study, data in Table 1 and Figure 1 illustrated that the parent N53-2 showed higher FW, R/S, and DSI than that of the parent KenC-8 under drought stress, which suggested that N53-2 was more tolerant to drought. We assumed that the use of DSI could help us in breeding and population screening under drought stress conditions. Moreover, all phenotypic traits showed transgressive segregation in the DH population, which suggested that the two parents contain all +Ve or -Ve alleles, which coincided with the results obtained in B. napus by Li et al. (2014). Here, we want to point out that R/S might be considered as an important phenotypic trait due to its sensitivity to drought stress; this finding in consistent with the results in wheat (Dhanda et al., 2004; Li et al., 2014). R/S was positively correlated with GP, RL, and DSI-SL under drought stress, while in the case of control, it showed a negative correlation with GP and SL; the results suggested that B. napus is likely to resist the effect of drought through increasing water uptake and reducing its loss. The increase in RL was essential to enhance water uptake, and the decrease in SL resulted in water loss (Dhanda et al., 2004; Ruta et al., 2010). In addition, Poorter and Remkes (1990) reported that, under stress conditions, the shoot biomass of plants decreased compared to roots, which relatively increased. As mentioned above, the R/S can be used as an important basis for assessing the ability of plants to withstand drought stress in the early growth stages.
The Genetic Bases and QTLs for Drought Tolerance
In the last decades, one of the most important approaches to clarifying quantitative trait crisis in plants was the use of molecular marker techniques for QTL analysis. Plant populations with different genetic structures have been developed to achieve these objectives. QTL analysis was performed as a response to biotic and abiotic stresses in many economically important crops, such as in peanut and wheat (Zhao et al., 2016; Wang et al., 2018). In the present study, the DH population named KN population was phenotyped and the QTL analysis was performed under both control and drought conditions, and some QTL/genes that controlled drought tolerance in B. napus were identified. In response to drought stress, many QTLs associated with drought tolerance were identified in tomato and wheat (Foolad et al., 2003; Wang et al., 2009) during seed germination and the early seedling stage, which indicated that a large number of genes are involved in drought tolerance mechanisms (Qie et al., 2014). In this study, a large number of QTLs related to drought resistance distributed on nine chromosomes in B. napus were detected. The locations of QTLs for some drought tolerance traits indicated that these traits might have smaller genetic bases. For example, many QTLs related to drought tolerance that contributed to different traits were detected in the same regions, such as qDSI_RL-11-1 and qDSI_SL-11-3, qDSI_RL-11-2 and qDSI_SL-11-3, and qDSI_RL-11-4 and qDSI_SL-11-3 on C01. In addition, qR/S-19-2 and qRL-19-1 were on C09, which suggests the close relationships between these traits and the probability of the presence of a single gene controlling more than one trait in this region; these results are consistent with results obtained under salinity stress in B. napus by Lang et al. (2017). Therefore, according to the present results, we considered that there are several key loci that control drought tolerance in B. napus (Table 3).
Several QTLs related to important drought-tolerant traits were detected on C01; therefore, this chromosome deserves specific importance in drought resistance in B. napus. For example, C01 carries QTL qDSI_SL-11-3 for SL and qDSI_RL-11-1 for RL, both of them adjoined with BnaC01g35030D (ortholog with A. thaliana AT1G18080) and were involved in abscisic acid stimulus. Two QTLs, qDSI_RL-11-2 and qDSI_SL-11-3, were located in the same position on C01 and associated with the BnaC01g32600D gene, which was orthologous to the gene for oxidative stress. Moreover, qDSI_RL-11-4 and qDSI_SL-11-3 were also located near each other and associated with B. napus gene BnaC01g30750D that is orthologous to ICE1, which is involved in ABA signaling and cold stress. However, QTLs qRL-11-3 for RL and qDSI_RL-11-2 affecting DSI were detected in the same region on C01. These results suggested the importance of these regions on chromosome C01 for drought tolerance.
Candidate Genes in Response to Drought Stress
The problem of selecting effective genes from hundreds of genes within QTLs is the biggest challenge, especially with the presence of many unfunctional genes (Gudys et al., 2018). For this reason, many techniques were used to help in the selection of target genes within QTLs, such as the ortholog of these genes to accessible model species and the upregulation/downregulation of the candidate genes when expressed in proteomic or transcriptional levels and based on the representation of the genes in specific physiological or biochemical pathways (Monclus et al., 2012; Bargsten et al., 2014; Kumar et al., 2017; Gudys et al., 2018). In this study, 256 genes were identified as candidate genes within 36 consensus QTLs. The transgressive segregation has been shown in the DH population due to the variation between the two parents detected in 128 genes. Based on the function annotation of the candidate genes, many genes orthologous to A. thaliana genes were associated with drought tolerance. BnaC03g44440D was located in QTL qR/S-13-1 that is associated with R/S under drought conditions and orthologous to the A. thaliana gene XERICO. Ko et al. (2006) reported that upregulation of XERICO improved drought tolerance in A. thaliana. BnaC03g37030D was located in QTL qR/S-13-1 and orthologous to the A. thaliana gene DREB2. Previous studies have reported that DREB overexpression enhanced drought tolerance in many plants such as A. thaliana, tobacco, and apple (Chen et al., 2016; Liao et al., 2017; Sharma et al., 2019). In this study, the gene BnaA02g09290D within QTL qSL-2-4 was orthologous to the A. thaliana gene MAPKKK15, which was reported to be involved in the drought tolerance mechanism in potato (Pieczynski et al., 2018). The two genes BnaC09g27300D and BnaC09g27320D were located within QTL qR/S-19-2 and qRL-19-1 and orthologous to the A. thaliana genes RAF2 and ATYY1, respectively. The AtYY1 gene was expressed under ABA, salt, and dehydration conditions, and overexpression of AtYY1 decreased ABA sensitivity in A. thaliana (Li et al., 2016). Furthermore, in the study conducted by Zhang H. et al. (2015), they reported that the RAF2 and SDIR1 complex had an essential role under stress by mediating the ABA signaling pathway. In addition, BnaC03g45915D within QTL qR/S-13-1 was orthologous to the A. thaliana gene RD29B, which was highly expressed under desiccation and played a vital role in drought tolerance (Prerostova et al., 2018). Our results illustrated that a lot of the genes related to drought tolerance in this study were located within QTLs associated with R/S, which supports the importance of this trait in QTL mapping. Also, several genes associated with drought tolerance were detected within the QTL for DSI on chromosome C01, such as BnaC01g35030D, BnaC01g32600D, and BnaC01g30750D, which indicated the role of these genes in drought tolerance in B. napus. In the end, we assumed that the above genes that were orthologous to drought stress tolerance genes in A. thaliana may be strongly associated with drought tolerance in B. napus. Further investigation and validation of these genes are underway to confirm their function in the drought tolerance in B. napus.
Conclusion
To illustrate the complexity of drought stress mechanisms and identification of genes underlying these mechanisms, we have used many genetic and genomic technologies. In this study, we used R/S as a phenotyping trait in QTL mapping and gene identification for drought tolerance at seedling stages in B. napus. Few studies used R/S as a phenotyping trait to QTLs in B. napus before. According to the QTL analysis, 39 QTLs associated with drought tolerance were identified and integrated to 36 consensus QTLs. Moreover, 256 drought tolerant-related candidate genes were identified, of which 128 genes have SNP/InDel variations between the two parents. In addition, our results indicated that specific regions on chromosome C01 were important for improved drought tolerance in B. napus due to the presence of many QTLs associated with drought tolerance. However, further studies are required to further confirm the candidate genes related to drought tolerance in B. napus.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Ethics Statement
The authors declare that the experiments comply with the current laws of the country in which they were performed.
Author Contributions
MG and HC conducted most of the experiments and data analysis for the overall study, and wrote and modified the manuscript. HL and WZ conducted parts of data analysis and provided a guide to the use of the related software. GL and ML designed and conceived the overall study and revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported financially by National Natural Science Foundation of China (32001583 and 32072131) and Key Research Plant Project of Shaanxi province (2020ZDLNY04-01).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.629970/full#supplementary-material
Supplementary Table 1 | Detailed information of identified QTLs for germination-related traits under control and drought stress.
Supplementary Table 2 | QTL for DSI for different agronomic traits identified under drought stress.
Supplementary Table 3 | The candidate genes homologues with 234 genes involved in drought tolerance in A. thaliana.
Supplementary Table 4 | 128 candidate genes have SNP/InDel variations between the two parents.
Abbreviations
QTL, quantitative trait locus; SNPs, single-nucleotide polymorphisms; InDel, insertion–deletion; DH, double haploid; GP, germination percentage; RL, root length; SL, shoot length; FW, fresh weight; R/S, root-to-shoot length ratio; DSI, drought stress index.
Footnotes
- ^ http://www.unep-wcmc.org/medialibrary/2011/10/19/3faf1880/Global-Drylands-FINAL-LR.pdf
- ^ https://www.ncbi.nlm.nih.gov/
References
Arcade, A., Labourdette, A., Falque, M., Mangin, B., Chardon, F., Charcosset, A., et al. (2004). BioMercator: integrating genetic maps and QTL towards discovery of candidate genes. Bioinformatics 20, 2324–2326. doi: 10.1093/bioinformatics/bth230
Bargsten, J., Nap, J., Sanchez-Perez, G., and van Dijk, A. (2014). Prioritization of candidate genes in QTL regions based on associations between traits and biological processes. BMC Plant Biol. 14:330. doi: 10.1186/s12870-014-0330-3
Cai, G., Yang, Q., Yi, B., Fan, C., Edwards, D., Batley, J., et al. (2014). A complex recombination pattern in the genome of allotetraploid Brassica napus as revealed by a high-density genetic map. PLoS One 9:e109910. doi: 10.1371/journal.pone.0109910
Chao, H., Wang, H., Wang, X., Guo, L., Gu, J., Zhao, W., et al. (2017). Genetic dissection of seed oil and protein content and identification of networks associated with oil content in Brassica napus. Sci. Rep. 7:46295.
Chen, H., Liu, L., Wang, L., Wang, S., and Cheng, X. (2016). VrDREB2A, a DREB-binding transcription factor from Vigna radiata, increased drought and high-salt tolerance in transgenic Arabidopsis thaliana. J. Plant Res. 129, 263–273. doi: 10.1007/s10265-015-0773-0
Collins, N. C., Tardieu, F., and Tuberosa, R. (2008). Quantitative trait loci and crop performance under abiotic stress: where do we stand? Plant Physiol. 147, 469–486. doi: 10.1104/pp.108.118117
Dhanda, S. S., Sethi, G. S., and Behl, R. K. (2004). Indices of drought tolerance in wheat genotypes at early stages of plant growth. J. Agron. Crop Sci. 190, 6–12. doi: 10.1111/j.1439-037x.2004.00592.x
Doerge, R. W., and Churchill, G. A. (1996). Permutation tests for multiple loci affecting a quantitative character. Genetics 142, 285–294. doi: 10.1093/genetics/142.1.285
Dudziak, K., Zapalska, M., Börner, A., Szczerba, H., Kowalczyk, K., and Nowak, M. (2019). Analysis of wheat gene expression related to the oxidative stress response and signal transduction under short-term osmotic stress. Sci. Rep. 9:2743.
Foolad, M. R., Zhang, L. P., and Subbiah, P. (2003). Genetics of drought tolerance during seed germination in tomato: inheritance and QTL mapping. Genome 46, 536–545. doi: 10.1139/g03-035
Gudys, K., Guzy-Wrobelska, J., Janiak, A., Dziurka, M. A., Ostrowska, A., Hura, K., et al. (2018). Prioritization of candidate genes in QTL Regions for physiological and biochemical traits underlying drought response in barley (Hordeum vulgare L.). Front. Plant Sci. 9:769. doi: 10.3389/fpls.2018.00769
Jackson, M. B., Saker, L. R., Crisp, C. M., Else, M. A., and Janowiak, F. (2003). Ionic and pH signalling from roots to shoots of flooded tomato plants in relation to stomatal closure. Plant Soil 253, 103–113. doi: 10.1023/a:1024588532535
Ko, J. H., Yang, S. H., and Han, K. H. (2006). Upregulation of an Arabidopsis RING-H2 gene, XERICO, confers drought tolerance through increased abscisic acid biosynthesis. Plant J. 47, 343–355. doi: 10.1111/j.1365-313x.2006.02782.x
Kumar, J., Gupta, D. S., Gupta, S., Dubey, S., Gupta, P., and Kumar, S. (2017). Quantitative trait loci from identification to exploitation for crop improvement. Plant Cell Rep. 36, 1187–1213. doi: 10.1007/s00299-017-2127-y
Lang, L., Xu, A., Ding, J., Zhang, Y., Zhao, N., Tian, Z., et al. (2017). Quantitative trait locus mapping of salt tolerance and identification of salt-tolerant genes in Brassica napus L. Front. Plant Sci. 8:1000. doi: 10.3389/fpls.2017.01000
Li, S., Assmann, S. M., and Albert, R. (2006). Predicting essential components of signal transduction networks: a dynamic model of guard cell abscisic acid signaling. PLoS Biol. 4:e312. doi: 10.1371/journal.pbio.0040312
Li, T., Wu, X., Li, H., Song, J., and Liu, J. (2016). A dual-function transcription factor, AtYY1, is a novel negative regulator of the Arabidopsis ABA response network. Mol. Plant 9, 650–661. doi: 10.1016/j.molp.2016.02.010
Li, Z., Mei, S., Mei, Z., Liu, X., Fu, T., Zhou, G., et al. (2014). Mapping of QTL associated with waterlogging tolerance and drought resistance during the seedling stage in oilseed rape (Brassica napus). Euphytica 197, 341–353. doi: 10.1007/s10681-014-1070-z
Liang, Y., Kang, K., Gan, L., Ning, S., Xiong, J., Song, S., et al. (2019). Drought-responsive genes, late embryogenesis abundant group3 (LEA3) and vicinal oxygen chelate, function in lipid accumulation in Brassica napus and Arabidopsis mainly via enhancing photosynthetic efficiency and reducing ROS. Plant Biotechnol. J. 17, 2123–2142. doi: 10.1111/pbi.13127
Liao, X., Guo, X., Wang, Q., Wang, Y., Zhao, D., Yao, L., et al. (2017). Overexpression of MsDREB6.2 results in cytokinin-deficient developmental phenotypes and enhances drought tolerance in transgenic apple plants. Plant J. 89, 510–526. doi: 10.1111/tpj.13401
Michel, B. E., and Kaufmann, M. R. (1973). The osmotic potential of polyethylene glycol 6000. Plant Physiol. 51, 914–916. doi: 10.1104/pp.51.5.914
Monclus, R., Leplé, J., Bastien, C., Bert, P., Villar, M., Marron, N., et al. (2012). Integrating genome annotation and QTL position to identify candidate genes for productivity, architecture and water-use efficiency in Populus spp. BMC Plant Biol. 12:173. doi: 10.1186/1471-2229-12-173
Pieczynski, M., Wyrzykowska, A., Milanowska, K., Boguszewska-Mankowska, D., Zagdanska, B., Karlowski, W., et al. (2018). Genome-wide identification of genes involved in the potato response to drought indicates functional evolutionary conservation with Arabidopsis plants. Plant Biotechnol. J. 16, 603–614. doi: 10.1111/pbi.12800
Poorter, H., and Remkes, C. (1990). Leaf area ratio and net assimilation rate of 24 wild species differing in relative growth rate. Oecologia 83, 553–559. doi: 10.1007/bf00317209
Prerostova, S., Dobrev, P. I., Gaudinova, A., Knirsch, V., Körber, N., Pieruschka, R., et al. (2018). Cytokinins: their impact on molecular and growth responses to drought stress and recovery in Arabidopsis. Front. Plant Sci. 9:655. doi: 10.3389/fpls.2018.00655
Qie, L., Jia, G., Zhang, W., Schnable, J., Shang, Z., Li, W., et al. (2014). Mapping of quantitative trait locus (QTLs) that contribute to germination and early seedling drought tolerance in the interspecific cross Setaria italica×Setaria viridis. PLoS One 9:e101868. doi: 10.1371/journal.pone.0101868
Raboanatahiry, N., Chao, H., Guo, L., Gan, J., Xiang, J., Yan, M., et al. (2017). Synteny analysis of genes and distribution of loci controlling oil content and fatty acid profile based on QTL alignment map in Brassica napus. BMC Genomics 18:776. doi: 10.1186/s12864-017-4176-6
Rotmistrovsky, K., Jang, W., and Schuler, G. D. (2004). A web server for performing electronic PCR. Nucleic Acids Res. 32, 108–112.
Ruta, N., Liedgens, M., Fracheboud, Y., Stamp, P., and Hund, A. (2010). QTLs for the elongation of axile and lateral roots of maize in response to low water potential. Theor. Appl. Genet. 120, 621–631. doi: 10.1007/s00122-009-1180-5
Sandhu, K. S., You, F. M., Conner, R. L., Balasubramanian, P. M., and Hou, A. (2018). Genetic analysis and QTL mapping of the seed hardness trait in a black common bean (Phaseolus vulgaris) recombinant inbred line (RIL) population. Mol. Breed. 38, 34–34.
Seki, M., Umezawa, T., Urano, K., and Shinozaki, K. (2007). Regulatory metabolic networks in drought stress responses. Curr. Opin. Plant Biol. 10, 296–302. doi: 10.1016/j.pbi.2007.04.014
Sharma, V., Goel, P., Kumar, S., and Singh, A. (2019). An apple transcription factor, MdDREB76, confers salt and drought tolerance in transgenic tobacco by activating the expression of stress-responsive genes. Plant Cell Rep. 38, 221–241. doi: 10.1007/s00299-018-2364-8
Trenberth, K. E., Dai, A., van der Schrier, G., Jones, P. D., Barichivich, J., Briffa, K. R., et al. (2013). Global warming and changes in drought. Nat. Clim. Change 4, 17–22.
Wan, J., Griffiths, R., Ying, J., McCourt, P., and Huang, Y. (2009). Development of drought-tolerant canola (Brassica napus L.) through genetic modulation of ABA-mediated stomatal responses. Crop Sci. 49, 1539–1554. doi: 10.2135/cropsci2008.09.0568
Wang, J., Chapman, S. C., Bonnett, D. G., and Rebetzke, G. J. (2009). Simultaneous selection of major and minor genes: use of QTL to increase selection efficiency of coleoptile length of wheat (Triticum aestivum L.). Theor. Appl. Genet. 119, 65–74. doi: 10.1007/s00122-009-1017-2
Wang, R., Liu, Y., Isham, K., Zhao, W., Wheeler, J., Klassen, N., et al. (2018). QTL identification and KASP marker development for productive tiller and fertile spikelet numbers in two high-yielding hard white spring wheat cultivars. Mol. Breed. 38, 135–135.
Wang, X., Chen, L., Wang, A., Wang, H., Tian, J., Zhao, X., et al. (2016). Quantitative trait loci analysis and genome-wide comparison for silique related traits in Brassica napus. BMC Plant Biol. 16:71. doi: 10.1186/s12870-016-0759-7
Wang, X., Wang, H., Long, Y., Li, D., Yin, Y., Tian, J., et al. (2013). Identification of QTLs associated with oil content in a high-oil Brassica napus cultivar and construction of a high-density consensus map for QTLs comparison in B. napus. PLoS One 8:e80569. doi: 10.1371/journal.pone.0080569
Zeng, Z. B. (1994). Precision mapping of quantitative trait loci. Genetics 136, 1457–1468. doi: 10.1093/genetics/136.4.1457
Zhang, B. (2015). MicroRNA: a new target for improving plant tolerance to abiotic stress. J. Exp. Bot. 66, 1749–1761. doi: 10.1093/jxb/erv013
Zhang, D., Pan, Q., Cui, C., Tan, C., Ge, X., Shao, Y., et al. (2015c). Genome-specific differential gene expressions in resynthesized Brassica allotetraploids from pair-wise crosses of three cultivated diploids revealed by RNA-seq. Front. Plant. Sci. 6:957. doi: 10.3389/fpls.2015.00957
Zhang, H., Cui, F., Wu, Y., Lou, L., Liu, L., Tian, M., et al. (2015). The RING finger ubiquitin E3 ligase SDIR1 targets SDIR1-INTERACTING PROTEIN1 for degradation to modulate the salt stress response and ABA signaling in Arabidopsis. Plant Cell 27, 214–227. doi: 10.1105/tpc.114.134163
Zhang, J., Mason, A., Wu, J., Liu, S., Zhang, X., Luo, T., et al. (2015). Identification of putative candidate genes for water stress tolerance in canola (Brassica napus). Front. Plant Sci. 6:1058. doi: 10.3389/fpls.2015.01058
Zhang, X., Lu, G., Long, W., Zou, X., Li, F., and Nishio, T. (2014). Recent progress in drought and salt tolerance studies in Brassica crops. Breed Sci 64, 60–73. doi: 10.1270/jsbbs.64.60
Zhao, Y., Zhang, C., Chen, H., Yuan, M., Nipper, R., Prakash, C. S., et al. (2016). QTL mapping for bacterial wilt resistance in peanut (Arachis hypogaea L.). Mol. Breed. 36, 13–13.
Keywords: Brassica napus, seed germination, drought tolerance, QTL analysis, candidate genes
Citation: Gad M, Chao H, Li H, Zhao W, Lu G and Li M (2021) QTL Mapping for Seed Germination Response to Drought Stress in Brassica napus. Front. Plant Sci. 11:629970. doi: 10.3389/fpls.2020.629970
Received: 16 November 2020; Accepted: 22 December 2020;
Published: 09 February 2021.
Edited by:
Chunyu Zhang, Huazhong Agricultural University, ChinaReviewed by:
Chuchuan Fan, Huazhong Agricultural University, ChinaWei Qian, Southwest University, China
Lei Shi, Huazhong Agricultural University, China
Copyright © 2021 Gad, Chao, Li, Zhao, Lu and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guangyuan Lu, bHV3aXpAMTYzLmNvbQ==; Maoteng Li, bGltYW90ZW5nNDI2QGh1c3QuZWR1LmNu
†These authors have contributed equally to this work
 Mahmoud Gad1†
Mahmoud Gad1† Weiguo Zhao
Weiguo Zhao Maoteng Li
Maoteng Li