
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 22 December 2020
Sec. Plant Development and EvoDevo
Volume 11 - 2020 | https://doi.org/10.3389/fpls.2020.617340
This article is part of the Research Topic Vernalization and Flowering Time: Celebrating 20 Years of FLC View all 15 articles
FLOWERING LOCUS C (FLC) is one of the best characterized genes in plant research and is integral to vernalization-dependent flowering time regulation. Yet, despite the abundance of information on this gene and its relatives in Arabidopsis thaliana, the role FLC genes play in other species, in particular cereal crops and temperate grasses, remains elusive. This has been due in part to the comparative reduced availability of bioinformatic and mutant resources in cereals but also on the dominant effect in cereals of the VERNALIZATION (VRN) genes on the developmental process most associated with FLC in Arabidopsis. The strong effect of the VRN genes has led researchers to believe that the entire process of vernalization must have evolved separately in Arabidopsis and cereals. Yet, since the confirmation of the existence of FLC-like genes in monocots, new light has been shed on the roles these genes play in both vernalization and other mechanisms to fine tune development in response to specific environmental conditions. Comparisons of FLC gene function and their genetic and epigenetic regulation can now be made between Arabidopsis and cereals and how they overlap and diversify is coming into focus. With the advancement of genome editing techniques, further study on these genes is becoming increasingly easier, enabling us to investigate just how essential FLC-like genes are to modulating flowering time behavior in cereals.
FLOWERING LOCUS C (FLC) genes are a clade of MADS-box transcription factors in plants and are major regulators in many aspects of plant development. They are mostly associated with vernalization-regulated flowering but also have important roles in seed dormancy (Chen et al., 2014; Chen and Penfield, 2018), ambient temperature regulated development (Balasubramanian et al., 2006; Lee et al., 2013), germination (Chiang et al., 2009), as well as being associated with other processes like bud dormancy, circadian rhythm, water use efficiency, and indirect defense against herbivory (McKay et al., 2003; Edwards et al., 2006; Kumar et al., 2016; Mohammadin et al., 2017). In fact, there are over 500 FLC binding sites in the Arabidopsis thaliana (henceforth Arabidopsis) genome indicating that FLC is involved in much more than vernalization (Deng et al., 2011). In flowering time regulation, FLC acts as a repressor protein and acts mainly by repressing the activation of key floral promoting genes such as FLOWERING LOCUS T (FT) and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1; Searle et al., 2006; Deng et al., 2011).
The existence of FLC-like genes in cereals remained elusive for many years while the wealth of information on Arabidopsis FLC continued to accumulate. Many believed that FLC was restricted to eudicots and that monocot plants evolved separate mechanisms to regulate development and flowering time. However, a turning point came when it was concretely established through genome synteny analysis and phylogenetic reconstructions that FLC relatives did indeed exist in cereals (Ruelens et al., 2013). In this pivotal publication, it was shown that a clade of genes within monocots were phylogenetically related to the FLC genes of Arabidopsis. Within this monocot FLC clade, there are two subclades: the OsMADS51 and OsMADS37 subclades, so called after the representation of these rice genes within each clade. The OsMADS51 subclade is subsequently divided into two groups: the ODDSOC1-like and ODDSOC2-like groups. The name “ODDSOC” came from their weak similarity to the flowering time gene SOC1 (Greenup et al., 2010). Members of the ODDSOC2 clade of genes are the most characterized out of all FLC-like genes in monocots thus far. The details of these relationships and their relationship to the Arabidopsis FLC genes can be seen in Figure 1. It must be noted that although the Arabidopsis and monocot FLC clades are related, it is likely that the ancestral gene function was partitioned differently within the groups. Therefore, direct comparisons of individual members across groups are not completely accurate.
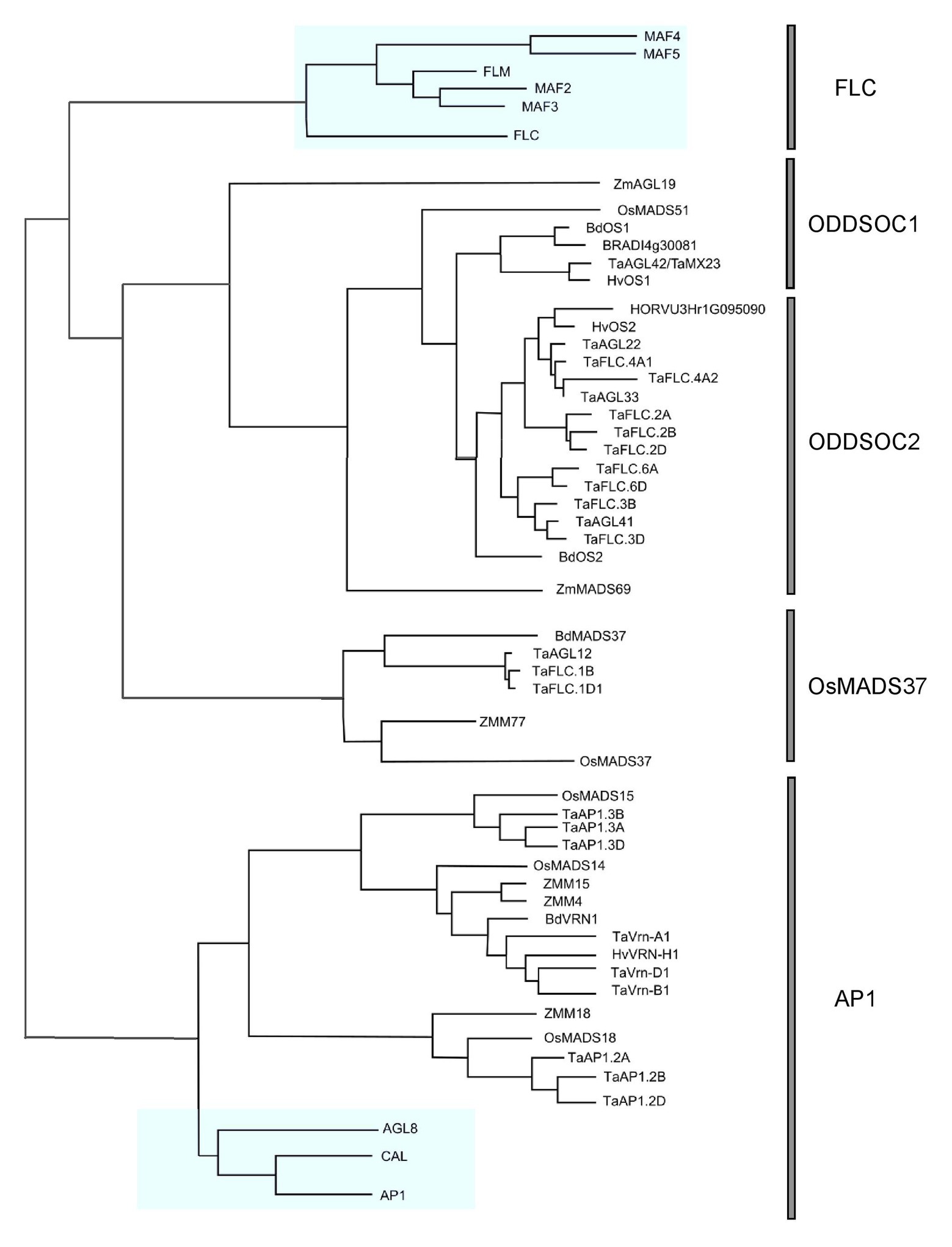
Figure 1. Phylogenetic relationships of all FLOWERING LOCUS C (FLC) genes annotated in cereals. A maximum likelihood phylogeny was generated using Geneious Pro v5.5.4 using MADS-box genes from eudicots and monocots. A reconstructed phylogeny containing only FLC and AP1 genes was drawn using InkScape. AP1 genes were used as an outgroup. Arabidopsis genes are highlighted in blue. A full version of the tree including all accession numbers can be found in Supplementary Figure S1.
Furthermore, FLC-like genes in cereals are Type II MADS-domain proteins despite having previously been annotated as Type I MADS-domain proteins, and have the typical MIKC protein structure (Zhao et al., 2006; Schilling et al., 2020).
Due to the advancement of genome sequencing technology, genetic mapping and genome editing methods, the nature of the function of FLC-like genes in cereals is coming into focus. Furthermore, avenues are now opening to further advance our knowledge on these genes which may reveal diversification of their function from their Arabidopsis homologs. This review aims to highlight the key findings over the last two decades of the role FLC relatives play in cereals and how progress made in biotechnology will further our understanding of the molecular control of plant development, perhaps leading us to utilize these genes as biotechnological tools for crop improvement.
In cereals, the main determinants of vernalization-regulated flowering are the VERNALIZATION (VRN) genes VRN1, VRN2, and VRN3 (Yan et al., 2003, 2004, 2006). Generally, vernalization results in the upregulation of the floral promoter VRN1 which downregulates the floral repressor VRN2, alleviating its repressive effect on the flowering promoter VRN3, an orthologue of FT (reviewed in Trevaskis et al., 2007; Distelfeld et al., 2009; Ream et al., 2012). VRN3 then positively regulates VRN1 expression resulting in a positive feedback loop which induces flowering. This feedback loop in general determines the flowering habit of cereals with mutations in any of these proteins leading to altered spring or winter growth habit (Yan et al., 2004, 2006; Fu et al., 2005). Yet, variation in vernalization response can still remain in cultivars which have shared alleles of these genes (Rizza et al., 2016), opening up the potential for other genes to have functional significance in this process. The identification of FLC-like genes in cereal research remained elusive for many years leading to the conclusion that the vernalization pathway of Arabidopsis and cereals evolved completely separately (Yan et al., 2004; Winfield et al., 2009; Greenup et al., 2010). Incorporating the finding that an FLC clade exists in monocots, it appears more likely that the ancestral species of dicots and monocots contained both FLC and AP1/VRN1-like genes, and each group was differentially recruited during the evolution of vernalization responsiveness. As AP1 retains a role in regulating flowering in Arabidopsis, so too do FLC homologs play a role in similar processes in cereals. This section aims to highlight these roles FLC-like genes play in the vernalization process and flowering time regulation in crop species.
FLOWERING LOCUS C homologs were described as being involved in vernalization in cereals almost 20 years ago where Trevaskis et al. (2003) described TaMX23, a MADS-box gene repressed by vernalization in winter wheats. TaMX23 increases in abundance in early vegetative development and the effect of vernalization on TaMX23 expression depended on whether the cultivar was a spring or winter variety (Trevaskis et al., 2003). TaMX23 shares homology with both TaODDSOC2 (TaOS2; also known as TaAGL33) and TaAGL42 (Winfield et al., 2009). Although it is more similar in sequence to TaAGL42, its reported expression pattern reflects that of TaOS2.
TaOS2 is the most described FLC-like gene in wheat so far. Like FLC, all three homeologs of TaOS2 are downregulated by vernalization and repression is maintained 2 weeks post-vernalization (Winfield et al., 2009; Sharma et al., 2017; Appels et al., 2018). In winter varieties, TaOS2 expression is initially high in leaf tissue and gradually declines throughout development as temperature decreases, yet its expression is constitutively low in spring lines, indicating that the function is cultivar-dependent and relevant to the flowering habit of these lines (Winfield et al., 2009; Sharma et al., 2017). Creating premature stop codons using CRISPR/Cas9 gene editing in the D-homeolog revealed an effect of this gene on flowering time, as mutants flower 3 days earlier than wild type (Appels et al., 2018). It is encouraging that a knockout of a single homeolog in hexaploid wheat reveals a phenotype. A 3-day alteration in flowering time is no mean feat in wheat breeding and can have great implications on yield in a region-specific manner. Validating this phenotype in the field will be enlightening to discover whether the phenotype is maintained and in which environments are the greatest effects found. It is also possible that functional redundancy is at play and multiple mutations in all homeologs of TaOS2 might reveal more striking phenotypes to uncover the roles of these genes in flowering time regulation in wheat.
A second FLC-like gene, TaAGL42 (or TaODDSOC1), has also been described as being regulated by vernalization in wheat (Winfield et al., 2009; Sharma et al., 2017); however, TaAGL42 is upregulated in winter cultivars and is downregulated or stably expressed in spring varieties. Additionally, TaAGL42 expression was shown to increase rapidly in response to a sudden drop in temperature in two winter varieties, suggesting that this gene could be involved in cold acclimation and tolerance in these lines (Winfield et al., 2009). In conclusion, although the gene is cold-regulated in a variety-specific manner, the function of TaAGL42 remains unclear in wheat.
Relatives of these genes have also been described in barley, where their identification came about as a result of a desire to characterize new genes responsive to vernalization. Through the analysis of homologs of TaMX23 (Trevaskis et al., 2003), two genes were identified: one sharing homology to TaMX23 and another sharing homology with TaOS2 (Trevaskis et al., 2003; Winfield et al., 2009; Greenup et al., 2010). The two homologs were named HvODDSOC1 (HvOS1) and HvODDSOC2 (HvOS2), respectively, due to their weak sequence similarity to SOC1 in Arabidopsis. HvOS1 expression increased in response to vernalization, consistent with its homolog in wheat (Winfield et al., 2009). HvOS2 expression was repressed in response to vernalization in both the leaves and apices, and this repression was maintained post-vernalization (Greenup et al., 2010). The expression of HvOS2 was also strongest in winter barley varieties pre-vernalization and was dramatically reduced upon exposure to prolonged cold, while expression remained low and constant in spring varieties. This highlights the importance of choice of cultivar when studying FLC homologs in cereals. Overexpressing HvOS2 in the spring barley resulted in delayed flowering in these lines, strongly suggesting that HvOS2 acts as a repressor of the floral transition. In contrast, no phenotype was observed for HvOS2 knockdown lines created using RNA interference (RNAi); however, this is to be expected in a spring line where HvOS2 is low naturally and vernalization is not required. In a separate study, differences in the rate of reproductive development under insufficient vernalization conditions was also explained by a difference in HvOS2 expression levels between two winter varieties (Monteagudo et al., 2019), further supporting the idea of HvOS2 as a vernalization-dependent regulator of the floral transition.
Aside from the crops themselves, research has been conducted on FLC-like genes in the model temperate grass Brachypodium distachyon (henceforth Brachypodium). In fact, Brachypodium was the organism chosen to first analyze the response of monocot FLC homologs to vernalization after they were first reported by Ruelens et al. (2013). Three homologs were reported in Brachypodium: BdODDSOC1 (BdOS1), BdODDSOC2 (BdOS2), and BdMADS37. BdOS1 was shown to be upregulated by vernalization, like its homologs TaAGL42 and HvOS1 in wheat and barley, respectively. BdOS2 expression is also consistent with its homologs in these species, as it is downregulated by vernalization (Ruelens et al., 2013; Sharma et al., 2017). There is also evidence to suggest that BdOS2 pre-vernalization expression levels determine the vernalization requirement of individual Brachypodium accessions, with winter accessions having higher pre-vernalization expression levels of BdOS2 (Sharma et al., 2017). Overexpression of BdOS2 led to a delay in flowering time under vernalized conditions in the facultative accession Bd21–3, with the delay comparative to wild type plants which were not vernalized. This suggests that overexpression of BdOS2 keeps Bd21–3 in a non-vernalized state. It was also reported that BdOS2 knockdown via RNAi influenced the flowering time of Bd21–3; however, we and others have been unable to replicate these findings, calling these results into question. Attempts are currently being made to vigorously test the effect of low BdOS2 expression on flowering time regulation in Brachypodium, with most striking phenotypes expected in winter accessions, and not facultative lines like Bd21–3.
The third FLC homolog, BdMADS37, is also downregulated by vernalization and exists in a separate clade to the ODDSOC genes (Figure 1). No other reports about members of this gene group have been published since first described by Ruelens et al. (2013); however, BdMADS37 appeared as a potential candidate for a QTL explaining the differences in flowering time and vernalization requirement between spring and winter accessions under specific environmental conditions (Bettgenhaeuser et al., 2017). We have identified a fourth FLC homolog in Brachypodium, BRADI4g30081 (Figure 1), a paralog of BdOS1 which appears to be a truncated duplication of BdOS1, and expression has been detected in response to cold in the microarray dataset of Priest et al. (2014).
Much of what we have learned about FLC-like genes so far comes from basic research on flowering time regulation and vernalization in cereals. Therefore, many of these findings have been related to or are based on descriptions of the activities of VRN1. So far, a relationship between VRN1 and ODDSOC2 activity has been reported in wheat and its diploid relative Triticum monococcum, barley, and Brachypodium. In general, evidence exists to suggest that VRN1 is required to repress ODDSOC2 post-vernalization to enable rapid flowering.
In T. monococcum, it was observed that TmOS2 levels rose post-vernalization in mutant lines lacking functional TmVRN1 while levels remained low in wild type lines. Analysis of TmOS2 levels pre- and during vernalization showed that there was no difference in expression between wild type and mutant lines. It is only post-vernalization TmOS2 levels that are affected by TmVRN1 loss of function, suggesting that TmVRN1 is required to repress TmOS2 post-vernalization but not to reduce its activity initially (Greenup et al., 2010).
Similar to T. monococcum, HvOS2 expression is lowest in barley lines with dominant, active VRN1 alleles, consistent with the hypothesis that VRN1 represses OS2 in temperate cereals (Greenup et al., 2010). Supporting this hypothesis, it was subsequently reported that HvVRN1 binds to the HvOS2 promoter in the spring variety Golden Promise (Deng et al., 2015). Several HvVRN1 binding sites have also been identified throughout the HvOS2 locus (Monteagudo et al., 2019).
Likewise, there is an antagonistic relationship between BdOS2 and BdVRN1 expression patterns in Brachypodium. BdOS2 expression is elevated in BdVRN1 knockdown lines (Woods et al., 2016). As well as that, BdOS2 expression is elevated in lines overexpressing BdVRN2, associated with low BdVRN1 expression and delayed flowering (Woods et al., 2016). Interestingly, BdOS2 expression patterns are not significantly influenced by overexpression of BdVRN1 or knockdown of BdVRN2, indicating that the response of BdOS2 to BdVRN1 expression is qualitative and not dosage dependent.
This relationship between VRN1 and ODDSOC2 is conserved in hexaploid wheat, where TaOS2 expression post-vernalization is linked to the nature of VRN1 alleles found in a given cultivar (Dixon et al., 2019). This relationship has implications for flowering time behavior which will be discussed in more detail in the next section.
Temperature is a key environmental signal which regulates many facets of plant development. Flowering time in both Arabidopsis and cereals is regulated by ambient temperature, with increasing temperatures generally resulting in earlier flowering times (McMaster and Wilhelm, 2003; Balasubramanian et al., 2006; Ejaz and von Korff, 2017; Dixon et al., 2018). Underlying this trait in Arabidopsis are the activities of FLC and its relative FLOWERING LOCUS M (FLM). FLM, like FLC, negatively regulates the floral transition, however, it is mostly involved in ambient temperature-dependent flowering (Balasubramanian et al., 2006; Lee et al., 2013; Posé et al., 2013). FLM functions as part of a repressor complex with another MADS-domain transcription factor, SHORT VEGETATIVE PHASE (SVP). This complex represses the activities of flowering promoters under cold temperatures to delay flowering and the stability of the complex and of the proteins themselves are affected by increasing temperature, reducing their repressive effects in warm conditions (Lee et al., 2013; Posé et al., 2013; Capovilla et al., 2017). Temperature-dependent alternative splicing of FLM is integral to this response, where the relative abundance of certain transcripts compared to others determines the flowering phenotype in response to temperature (Capovilla et al., 2017; Lutz et al., 2017).
High levels of FLC itself also results in thermal unresponsiveness, therefore, suggesting that FLC suppresses thermal induction of flowering (Balasubramanian et al., 2006). These findings could suggest that vernalization is the dominant process that must be realized to allow Arabidopsis to be receptive to temperature, likely to prevent precocious flowering in winter.
In cereals, the activities of ODDSOC2 can also be linked to ambient-temperature regulated flowering. ODDSOC2 has been shown to be responsive to ambient temperature in both wheat and barley, which both show earlier flowering phenotypes in response to increasing temperature (McMaster and Wilhelm, 2003; Ejaz and von Korff, 2017; Dixon et al., 2018). In wheat, however, it was shown that certain cultivars exhibited delayed flowering in response to increasing ambient temperature (Dixon et al., 2019). It was revealed that this trait arose from the incomplete vernalization of this cultivar, leading to the re-activation of floral repressors including VRN2 and TaOS2. The increase in TaOS2 expression was linked to the VRN1 alleles found in this specific cultivar, which were unable to maintain repression of TaOS2 after incomplete vernalization, explaining in part the delayed flowering phenotype (Dixon et al., 2019).
HvOS2 was also shown to be responsive to ambient temperature in barley. HvOS2 expression increases under high temperature conditions, particularly under short day photoperiods – conditions which result in the slowest development of the shoot apex (Hemming et al., 2012; Ejaz and von Korff, 2017). Like TaOS2 in wheat, the response of HvOS2 is influenced by the VRN1 allele present in the variety analyzed, with lines with the winter Hvvrn1 allele having higher HvOS2 expression levels in response to high ambient temperature (Ejaz and von Korff, 2017). Hvvrn1 itself is downregulated under high temperature conditions, highlighting further the negative correlation between VRN1 and OS2 expression in cereals. Plants overexpressing HvOS2 also exhibited delayed reproductive development under both cool and high temperatures while lines with a RNAi-mediated knockdown of HvOS2 underwent more rapid reproductive growth at higher temperatures compared to wild type plants (Hemming et al., 2012). This is reflective of phenotypes obtained when FLM expression is modified in Arabidopsis (Posé et al., 2013).
Taken together, this evidence suggests that ODDSOC2 functions in winter cereal varieties to repress the reproductive transition under warm temperatures until the vernalization requirement is completely saturated. It can be speculated that this is an adaptation to prevent precocious flowering during the winter should a brief period of warmth occur. Interestingly, this ecologically significant process is linked to FLC gene activity across both Arabidopsis and cereals; however, unlike FLM, the mode of action of ODDSOC2 remains unknown. There is no evidence so far to suggest that ODDSOC2 is alternatively spliced to influence this process. It is also unknown whether this increase in ODDSOC2 expression occurs in varieties which can be induced to flower using warm temperatures and short-day conditions (Evans, 1987). Regardless, parallels can be drawn on the roles these FLC genes play in fine-tuning flowering time in a temperature-specific manner in both Arabidopsis and cereals.
A summary of the roles FLC genes play in vernalization, ambient temperature, and their relationship to VRN1 are outlined in Figure 2 and Table 1.
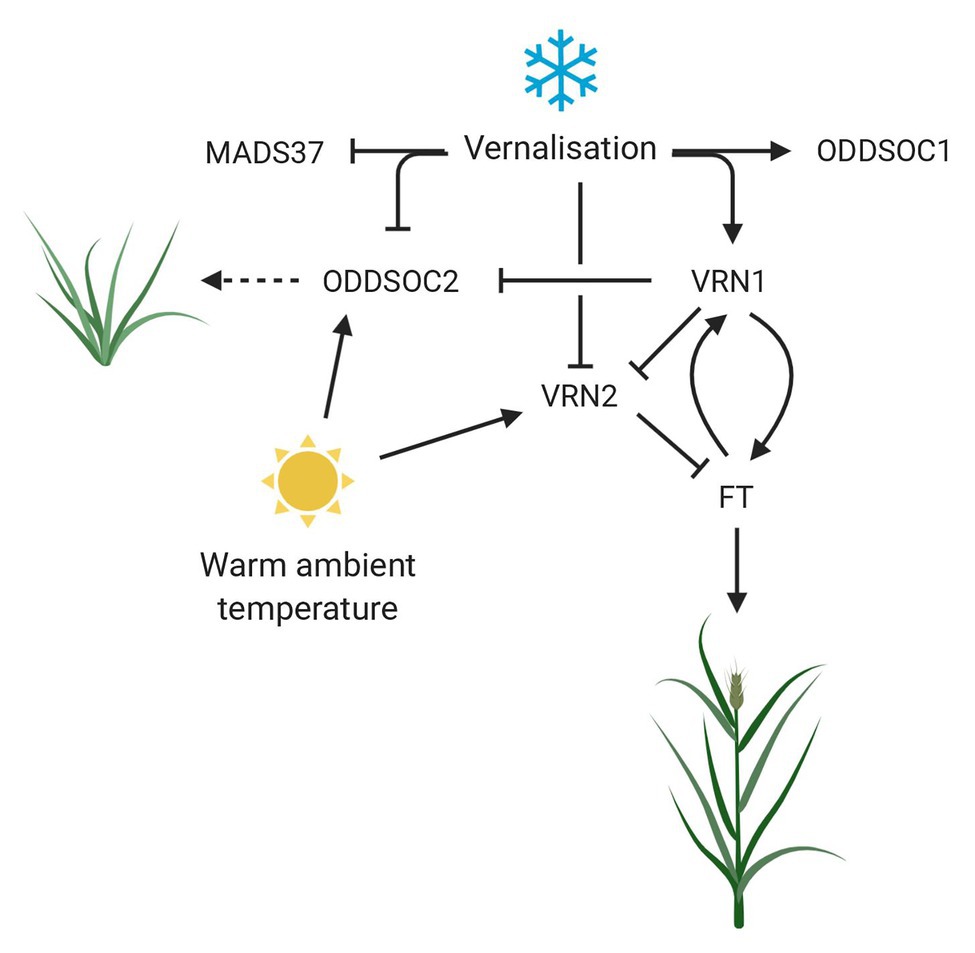
Figure 2. The role of FLC genes in the response to temperature in temperate cereals with a vernalization requirement. Vernalization represses the activities of floral repressors ODDSOC2 and VRN2, while promoting the upregulation of VRN1. VRN1 is required to maintain the repression of ODDSOC2 and VRN2 while promoting the flowering time gene FT, creating a positive feedback loop to lock in the floral transition. Warm ambient temperature during the vernalization process results in the upregulation of floral repressors to coordinate reproductive development with the most optimal environmental conditions. Dashed arrow implies function not confirmed. MADS37 and ODDSOC1 are included but their function is still unknown. Created with www.BioRender.com.
Rice and maize diverged from the Pooideae roughly 64 million years ago and their flowering times are regulated in different ways to the temperate cereals (Wang et al., 2015). Reflective of the tropical regions in which they evolved, these species have no vernalization requirement and flowering is promoted by short day photoperiods. Nonetheless, both rice and maize contain FLC homologs which have been shown to regulate flowering time. OsMADS51, a homolog of ODDSOC1, acts as a flowering promoter under short days in rice (subsp. Japonica; Kim et al., 2007). Knockout of this protein correlates with the downregulation of AP1 and FT homologs, which may explain the mode of action of this protein. In maize, ZmMADS69 acts as a flowering promoter under both long and short days, and is thought to have been a target of selection to expand the cultivation zone of maize (Liang et al., 2019). Therefore, despite paucity of information on FLC-like genes in cereals, it is clear that flowering time regulation is a fundamental feature of these genes across the cereals.
The epigenetic regulation of FLC in Arabidopsis is well described and FLC can be considered a model gene for the study of epigenetics in general (Whittaker and Dean, 2017). FLC activity is regulated through the chromatin environment at the FLC locus and via RNA-mediated silencing mechanisms. Chromatin modification to promote FLC activity is regulated mainly through the actions of the FRIGIDA (FRI) complex (FRI-C). The FRI-C increases levels of active chromatin markers, such as H3K36me3 and H3K4me3, through the recruitment of chromatin modification proteins (Choi et al., 2011; Li et al., 2018). These markers are targeted during vernalization, where they are removed, and replaced with H3K9me3 and H3K27me3, resulting in a silenced chromatin state (Bastow et al., 2004; Finnegan and Dennis, 2007; Angel et al., 2011; Whittaker and Dean, 2017). The accumulation of chromatin silencing markers is mediated by the PHD-PRC2 complex (Plant Homeodomain-Polycomb Repression Complex 2; Wood et al., 2006; De Lucia et al., 2008).
Silencing of FLC is additionally associated with the action of long non-coding RNAs (lncRNAs) and components of the autonomous pathway (Sheldon et al., 2000; Ietswaart et al., 2012; Whittaker and Dean, 2017). RNA binding proteins of the autonomous pathway function to process COOLAIR, a set of lncRNAs transcribed antisense of FLC (Swiezewski et al., 2009; Hornyik et al., 2010; Whittaker and Dean, 2017). The significance of these lncRNAs in the regulation of vernalization remains controversial (Helliwell et al., 2011; Luo et al., 2019); however, much data have been gathered to indicate a functional if not essential role. The physical association of COOLAIR with FLC chromatin is associated with the reduction of H3K36me3 and H3K4me3, rendering the chromatin inactive (Csorba et al., 2014; Fang et al., 2020). COOLAIR is also induced by vernalization to assist with the inactivation of FLC (Swiezewski et al., 2009; Kim and Sung, 2017). The silencing of FLC is associated with two other lncRNAs, COLDWRAP and COLDAIR, which recruit the PHD-PRC2 complex to specific chromatin regions (Heo and Sung, 2011; Kim and Sung, 2017).
Due to the relatively recent discovery of FLC-like genes in cereals, no research has been done to test whether FRI or the FRI-C functions in cereal plants. Studies have shown that homologs of the various FRI-C components can be detected in monocots (Choi et al., 2011) and rice FRI-like genes form distinct clades with Arabidopsis FRI-like genes (Michaels et al., 2004). According to the plant genome database EnsemblPlants, 25 and 13 proteins have been annotated as FRI-like for Triticum aestivum (cv. Chinese Spring) and Hordeum vulgare (cv. Morex), respectively. Homologs can also be identified for Brachypodium distachyon (9), Oryza sativa subsp. japonica (12), Sorghum bicolor (10), Triticum dicoccoides (16), Triticum turgidum (15), and Zea mays (13). It is possible that these uncharacterized proteins may act as scaffold proteins similar to FRI but for other pathways and functions.
In Arabidopsis, FLC is silenced during vernalization via a series of histone modifications by the PHD-PRC2 complex (De Lucia et al., 2008). The utilization of this complex in plants as a method to regulate vernalization-dependent flowering is conserved across Arabidopsis and cereals. The major regulator of vernalization in cereals, VRN1, acts as a promoter of flowering, rather than a repressor like FLC. Before vernalization, H3K27me3 repressive marks are deposited at the VRN1 locus in wheat and barley (Oliver et al., 2009; Diallo et al., 2012). During vernalization, H3K27me3 decreases while the active markers H3K4me3 and H3K36me3 increase (Oliver et al., 2009; Diallo et al., 2012). The same mode of epigenetic regulation of VRN1 is conserved in Brachypodium, and is regulated by ENHANCER OF ZESTE-LIKE 1 (EZL1), a homolog of CURLY LEAF (CLF), and a methyltransferase in the PRC2 complex in Arabidopsis (Lomax et al., 2018). VRN3/FT is also regulated in the same way in wheat and Brachypodium (Oliver et al., 2009; Huan et al., 2018).
Recruitment of the PRC2 to epigenetically regulate the vernalization response evolved in both Arabidopsis and cereals. However, the nature of the chromatin modifiers deposited at FLC and VRN1 is different due to their nature as a repressor and promoter, respectively. Therefore, this recruitment likely evolved after the independent evolution of the vernalization response pathway in monocots and dicots. Yet, some evidence exists to suggest that FLC-like genes are also regulated by the PRC2 in a similar manner to FLC in Arabidopsis. To analyze the effect of vernalization on histone modifications at HvOS2 in barley, H3K27me3 marks were analyzed at the presumed transcriptional start site of HvOS2 in plants with or without 7 weeks of vernalization (Greenup et al., 2010). There was no significant difference in H3K27 trimethylation at this region indicating that perhaps repression of HvOS2 post-vernalization is regulated in a different way to FLC. The region tested by Greenup et al. begins ~100 bp upstream of the transcriptional start site (TSS). Although H3K27me3 deposits increase during vernalization at the TSS of FLC in Arabidopsis, the greatest increase is at the exon 1/intron 1 junction termed the “nucleation region” (Bastow et al., 2004; Finnegan and Dennis, 2007; Yuan et al., 2016). On return to warmth, H3K27me3 spreads from the nucleation region across the FLC locus. Future experiments targeting other regions within the HvOS2 locus could reveal more similarities in the epigenetic regulation of both FLC and HvOS2. Regulation via other markers such as H3K9me3 or H3 acetylation levels could be investigated, as these markers are also involved in FLC regulation.
In contrast to barley, BdOS2 in Brachypodium showed high levels of H3K27me3 after vernalization in both spring and winter accessions (Sharma et al., 2017). H3K27me3 was enriched at the BdOS2 locus after vernalization for Bd21 and BdTR3C – spring and winter accessions, respectively, and the enrichment was maintained 1-week post-vernalization. For the winter allele of BdOS2 in BdTR3C, H3K27me3 can be found spanning the entire locus post-vernalization. The extensive methylation marks of H3K27 in the locus of BdTR3C compared to Bd21 may explain the mechanism as to how BdOS2 is stably repressed in the winter but not spring accession. It is possible that in strong winter varieties, FLC genes have evolved increasingly stringent or more complex methods of silencing to ensure flowering time is synchronized most optimally with the environment (Shindo et al., 2006; Hepworth et al., 2020). Analysis of winter varieties of FLC homologs in other cereal crops may reveal that the epigenetic regulation of FLC-like genes is more conserved than currently realized.
Brachypodium has also been shown to encode lncRNAs, similar to COOLAIR, which target FLC-like genes for downregulation during vernalization (Jiao et al., 2019). Two high confidence lncRNAs were detected for BdOS2, while one lncRNA could be detected for BdOS1. These lncRNAs were termed BdCOOLAIR1 and BdCOOLAIR2 for BdOS1 and BdOS2, respectively, as although they are not homologous to the AtCOOLAIR sequence, their position relative to their sense counterparts is similar. Expression of these lncRNAs is induced by vernalization, and their induction is significantly higher in a winter accession compared to a facultative accession, while their expression is absent in a spring accession. Knockdown of BdCOOLAIR2 via RNAi also affects the rate of silencing of BdOS2 in BdTR3C, though it is not essential for the complete silencing of BdOS2 (Jiao et al., 2019). This information suggests that lncRNAs complement the mechanisms which silence FLC-like genes in grasses, in a similar fashion to FLC regulation in Arabidopsis, although this mode of regulation is accession dependent. In addition, lncRNAs have been annotated for FLC genes in 6 other grass species, including wheat, although these still need to be experimentally verified.
Although thoroughly studied for its involvement in vernalization-dependent regulation of flowering time, FLC function is implicated in many other aspects of plant growth and development. Analysis of expression levels of FLC-like genes in cereals during development and under various experimental treatments also suggests that homologs of FLC play diverse roles in cereal physiology. For example, in the early gene expression experiments of Zhao et al. (2006), it was shown that at least one of the genes analyzed is expressed at, at least, one of the various life stages and in at least one of the various tissue types throughout wheat development. Expression can be detected from initial embryo imbibition to seed development post-anthesis while other genes are predominantly expressed in roots.
Curiously, in the dataset of both Zhao et al. (2006) and Schilling et al. (2020), TaAGL41 could not be detected at significant levels but could be detected by Sharma et al. (2017) for several cultivars. Further analysis using the Wheat Expression Browser (Ramírez-González et al., 2018) indicates that TaAGL41 is indeed expressed throughout development but the extent of its expression is cultivar-specific. This could suggest a role for this gene in fine tuning development in a cultivar-specific manner. The Wheat Expression Browser highlights that TaAGL41 is downregulated by cold in the spring cultivar Manitou (Li et al., 2015), yet the main stimulus which affected TaAGL41 across the dataset was infection by the wheat yellow rust pathogen Puccinia striiformis f. sp. Tritici (Dobon et al., 2016). Expression of two other high confidence FLC homologs in wheat, TaFLC.4A1 and TaFLC.4A2, could not be detected at significant levels in the developmental time course analyzed by Schilling et al. (2020), and their expression does not change considerably across the different varieties available on the Wheat Expression Browser. Rather, these genes appear to be mainly influenced by drought stress (Liu et al., 2015; Ramírez-González et al., 2018). The relationship between flowering time and stress adaptation is complex and the molecular mechanisms determining this relationship are still not fully understood; however, a link between flowering time regulators and stress is found in plants (reviewed in Kazan and Lyons, 2016). It is possible that FLC-like genes not only play roles regulating development but also that their function has diversified to fine tune other developmental and growth processes. This reflects findings in the Brassicaceae that although the core function of FLC across species is the regulation of flowering time, different members of the FLC clade have been recruited for species-specific roles typically within stress response pathways (Mateos et al., 2017). Further investigation into these expression patterns as well as generation of knockout mutants may reveal a novel role for these genes in stress response pathways in cereals.
Additionally, HvOS2 has been shown to negatively influence cell length and, therefore, leaf, internode, and spike length (Greenup et al., 2010). The data suggest that HvOS2 downregulation by vernalization allows the process of stem elongation and bolting as secondary regulation of the reproductive process.
As well as being expressed during seed development stages in wheat, FLC-like genes have been implicated in seed development in barley. HvOS1 and HvOS2 are differentially expressed in cultivars of different seed size and at different stages of seed development (Kapazoglou et al., 2012). Analysis of their expression patterns revealed that HvOS1 expression is induced more substantially in early seed development in cultivars with large seeds, while HvOS2 levels are significantly higher in later developmental stages in cultivars with small seeds. This pattern could suggest an association between the expression of FLC genes and seed size in barley and that each gene is important for different stages of development – either endosperm cellularization or seed maturation. Additionally, both genes contained the endosperm-specific element GCN4 in their promoters, along with elements for responses to abscisic acid, an important phytohormone for seed maturation as well as abiotic stress (Takaiwa et al., 1996; Finkelstein et al., 2002; Kapazoglou et al., 2012). Taken together, these data implicate a role of FLC homologs in seed development and suggest that perhaps there is an association between them and seed size. An association study including more cultivars with a variety of seed sizes could be undertaken to fully determine whether FLC-like genes regulate this important agronomic trait.
Outside of the temperate cereals, ZmAGL19, an FLC homolog in maize, is targeted by OPAQUE11, a central regulator of endosperm development and nutrient metabolism (Feng et al., 2018). OPAQUE11 is specifically expressed in the endosperm and positively regulates ZmAGL19 expression, suggesting that ZmAGL19 might be part of the seed development regulation process in maize.
Much has been learned about the roles FLC genes play in cereals, mostly indirectly through the study of flowering time in these species. The scientific community is now able to study FLC genes further due to the dramatically improved genetic resources available. Reference genome assemblies are now available for several hexaploid wheat cultivars, as well as tetraploid wheat, diploid progenitor species, and 2- and 6-row barley (Ling et al., 2013; Fox et al., 2014; Luo et al., 2017; Mascher et al., 2017; Appels et al., 2018; Maccaferri et al., 2019). Genes are also annotated to include SNP variations which can be easily identified using the online platform EnsemblPlants. Identifying homologs and SNP-variants across cultivars has never been easier for researchers without bioinformatics training. Access to tools such as these will increase the pace at which genes are identified and studied, increasing the potential to finally characterize the once enigmatic FLC gene family.
Additionally, populations of mutant plants have been created for widespread use in both hexaploid and tetraploid wheat and barley (Krasileva et al., 2017; Schreiber et al., 2019). TILLING lines containing homeolog-specific mutations in genes of interest can be ordered and crossed, creating specific combinations to study gene function and redundancy. In a more targeted approach, protocols for wheat transformation and mutation via virus-induced gene silencing and CRISPR/Cas9 are available (see wheat-training.com for resources). In combination with speed breeding, it is possible to fully characterize the effect of mutations in both model and crop plants in considerably less time (Watson et al., 2018). At this moment in time, comparable resources to the model plant Arabidopsis from which most of our information on FLC genes comes from are available. This review also highlights how relevant Brachypodium is as a model for basic and translational research for temperate cereals and that research using this small grass will continue to be a valuable option to study FLC genes. It is possible within the next few years that we will see a greater increase in FLC-related knowledge outside of Arabidopsis. The availability of these resources provides hope that much more knowledge can be gained on FLC function in cereals in years to come.
AK prepared the outline and wrote the manuscript. KG contributed to discussions and critical revision of the manuscript. All authors contributed to the article and approved the submitted version.
AK and The Geuten Lab are supported by KU Leuven grant C24/17/037 and FWO grant G065713N.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Emma Doyle (University College Dublin), Sam Balzarini (KU Leuven), Philip Ruelens (WUR), and Daniel Woods (UC Davis) for their helpful comments, suggestions, and insights. We also thank three reviewers of this article for their valuable comments.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.617340/full#supplementary-material
Angel, A., Song, J., Dean, C., and Howard, M. (2011). A Polycomb-based switch underlying quantitative epigenetic memory. Nature 476, 105–109. doi: 10.1038/nature10241
Appels, R., Eversole, K., Feuillet, C., Keller, B., Rogers, J., Stein, N., et al. (2018). Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361:eaar7191. doi: 10.1126/science.aar7191
Balasubramanian, S., Sureshkumar, S., Lempe, J., and Weigel, D. (2006). Potent induction of Arabidopsis thaliana flowering by elevated growth temperature. PLoS Genet. 2:e106. doi: 10.1371/journal.pgen.0020106
Bastow, R., Mylne, J. S., Lister, C., Lippman, Z., Martienssen, R. A., and Dean, C. (2004). Vernalization requires epigenetic silencing of FLC by histone methylation. Nature 427, 164–167. doi: 10.1038/nature02269
Bettgenhaeuser, J., Corke, F. M. K., Opanowicz, M., Green, P., Hernández-Pinzón, I., Doonan, J. H., et al. (2017). Natural variation in Brachypodium links vernalization and flowering time loci as major flowering determinants. Plant Physiol. 173, 256–268. doi: 10.1104/pp.16.00813
Capovilla, G., Symeonidi, E., Wu, R., and Schmid, M. (2017). Contribution of major FLM isoforms to temperature-dependent flowering in Arabidopsis thaliana. J. Exp. Bot. 68, 5117–5127. doi: 10.1093/jxb/erx328
Chen, M., Moore, K., Florance, H., MacGregor, D. R., Smirnoff, N., Paszkiewicz, K., et al. (2014). Maternal temperature history activates flowering locus T in fruits to control progeny dormancy according to time of year. Proc. Natl. Acad. Sci. 111, 18787–18792. doi: 10.1073/pnas.1412274111
Chen, M., and Penfield, S. (2018). Feedback regulation of COOLAIR expression controls seed dormancy and flowering time. Science 360, 1014–1017. doi: 10.1126/science.aar7361
Chiang, G. C. K., Barua, D., Kramer, E. M., Amasino, R. M., and Donohue, K. (2009). Major FLOWERING time gene, FLOWERING LOCUS C, regulates seed germination in Arabidopsis thaliana. Proc. Natl. Acad. Sci. 106, 11661–11666. doi: 10.1073/pnas.0901367106
Choi, K., Kim, J., Hwang, H. J., Kim, S., Park, C., Kim, S. Y., et al. (2011). The FRIGIDA complex activates transcription of FLC, a strong flowering repressor in Arabidopsis, by recruiting chromatin modification factors. Plant Cell 23, 289–303. doi: 10.1105/tpc.110.075911
Csorba, T., Questa, J. I., Sun, Q., and Dean, C. (2014). Antisense COOLAIR mediates the coordinated switching of chromatin states at FLC during vernalization. Proc. Natl. Acad. Sci. 111, 16160–16165. doi: 10.1073/pnas.1419030111
De Lucia, F., Crevillen, P., Jones, A. M. E., Greb, T., and Dean, C. (2008). A PHD-polycomb repressive complex 2 triggers the epigenetic silencing of FLC during vernalization. Proc. Natl. Acad. Sci. 105, 16831–16836. doi: 10.1073/pnas.0808687105
Deng, W., Casao, M. C., Wang, P., Sato, K., Hayes, P. M., Finnegan, E., et al. (2015). Direct links between the vernalization response and other key traits of cereal crops. Nat. Commun. 6:5882. doi: 10.1038/ncomms6882
Deng, W., Ying, H., Helliwell, C. A., Taylor, J. M., Peacock, W. J., and Dennis, E. S. (2011). FLOWERING LOCUS C (FLC) regulates development pathways throughout the life cycle of Arabidopsis. Proc. Natl. Acad. Sci. 108, 6680–6685. doi: 10.1073/pnas.1103175108
Diallo, A. O., Ali-Benali, M. A., Badawi, M., Houde, M., and Sarhan, F. (2012). Expression of vernalization responsive genes in wheat is associated with histone H3 trimethylation. Mol. Ecol. 287, 575–590. doi: 10.1007/s00438-012-0701-0
Distelfeld, A., Li, C., and Dubcovsky, J. (2009). Regulation of flowering in temperate cereals. Curr. Opin. Plant Biol. 12, 178–184. doi: 10.1016/j.pbi.2008.12.010
Dixon, L. E., Farré, A., Finnegan, E., Orford, S., Griffiths, S., and Boden, S. (2018). Developmental responses of bread wheat to changes in ambient temperature following deletion of a locus that includes FLOWERING LOCUS T1. Plant Cell Environ. 41, 1715–1725. doi: 10.1111/pce.13130
Dixon, L. E., Karsai, I., Kiss, T., Adamski, N. M., Liu, Z., Ding, Y., et al. (2019). VERNALIZATION1 controls developmental responses of winter wheat under high ambient temperatures. Development 146:dev172684. doi: 10.1242/DEV.172684
Dobon, A., Bunting, D. C. E., Cabrera-Quio, L. E., Uauy, C., and Saunders, D. G. O. (2016). The host-pathogen interaction between wheat and yellow rust induces temporally coordinated waves of gene expression. BMC Genomics 17:380. doi: 10.1186/s12864-016-2684-4
Edwards, K. D., Anderson, P. E., Hall, A., Salathia, N. S., Locke, J. C. W., Lynn, J. R., et al. (2006). FLOWERING LOCUS C mediates natural variation in the high-temperature response of the Arabidopsis circadian clock. Plant Cell 18, 639–650. doi: 10.1105/tpc.105.038315.1
Ejaz, M., and von Korff, M. (2017). The genetic control of reproductive development under high ambient temperature. Plant Physiol. 173, 294–306. doi: 10.1104/pp.16.01275
Evans, L. T. (1987). Short day induction of inflorescence initiation in some winter wheat varieties. Funct. Plant Biol. 14, 277–286.
Fang, X., Wu, Z., Raitskin, O., Webb, K., Voigt, P., Lu, T., et al. (2020). The 3' processing of antisense RNAs physically links to chromatin-based transcriptional control. Proc. Natl. Acad. Sci. 117, 15316–15321. doi: 10.1073/pnas.2007268117
Feng, F., Qi, W., Lv, Y., Yan, S., Xu, L., Yang, W., et al. (2018). OPAQUE11 is a central hub of the regulatory network for maize endosperm development and nutrient metabolism. Plant Cell 30, 375–396. doi: 10.1105/tpc.17.00616
Finkelstein, R. R., Gampala, S. S. L., and Rock, C. D. (2002). Abscisic acid signaling in seeds and seedlings. Plant Cell 14, 15–45. doi: 10.1105/tpc.010441
Finnegan, E., and Dennis, E. S. (2007). Vernalization-induced trimethylation of histone H3 lysine 27 at FLC is not maintained in mitotically quiescent cells. Curr. Biol. 17, 1978–1983. doi: 10.1016/j.cub.2007.10.026
Fox, S. E., Geniza, M., Hanumappa, M., Naithani, S., Sullivan, C., Preece, J., et al. (2014). De novo transcriptome assembly and analyses of gene expression during photomorphogenesis in diploid wheat Triticum monococcum. PLoS One 9:e96855. doi: 10.1371/journal.pone.0096855
Fu, D., Szűcs, P., Yan, L., Helguera, M., Skinner, J. S., Von Zitzewitz, J., et al. (2005). Large deletions within the first intron in VRN-1 are associated with spring growth habit in barley and wheat. Mol. Gen. Genomics. 273, 54–65. doi: 10.1007/s00438-004-1095-4
Greenup, A. G., Sasani, S., Oliver, S., Talbot, M. J., Dennis, E. S., Hemming, M. N., et al. (2010). ODDSOC2 is a MADS box floral repressor that is down-regulated by vernalization in temperate cereals. Plant Physiol. 153, 1062–1073. doi: 10.1104/pp.109.152488
Helliwell, C. A., Robertson, M., Finnegan, E., Buzas, D. M., and Dennis, E. S. (2011). Vernalization-repression of Arabidopsis FLC requires promoter sequences but not antisense transcripts. PLoS One 6:e21513. doi: 10.1371/journal.pone.0021513
Hemming, M. N., Walford, S. A., Fieg, S., Dennis, E. S., and Trevaskis, B. (2012). Identification of high-temperature-responsive genes in cereals. Plant Physiol. 158, 1439–1450. doi: 10.1104/pp.111.192013
Heo, J. B., and Sung, S. (2011). Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 331, 76–79. doi: 10.1126/science.1197349
Hepworth, J., Antoniou-kourounioti, R. L., Berggren, K., Selga, C., Tudor, E. H., Yates, B., et al. (2020). Natural variation in autumn expression is the major adaptive determinant distinguishing Arabidopsis FLC haplotypes. elife 9:e57671. doi: 10.7554/eLife.57671
Hornyik, C., Terzi, L. C., and Simpson, G. G. (2010). The Spen family protein FPA controls alternative cleavage and polyadenylation of RNA. Dev. Cell 18, 203–213. doi: 10.1016/j.devcel.2009.12.009
Huan, Q., Mao, Z., Chong, K., and Zhang, J. (2018). Global analysis of H3K4me3/H3K27me3 in Brachypodium distachyon reveals VRN3 as critical epigenetic regulation point in vernalization and provides insights into epigenetic memory. New Phytol. 219, 1373–1387. doi: 10.1111/nph.15288
Ietswaart, R., Wu, Z., and Dean, C. (2012). Flowering time control: another window to the connection between antisense RNA and chromatin. Trends Genet. 28, 445–453. doi: 10.1016/j.tig.2012.06.002
Jiao, F., Pahwa, K., Dochy, N., Manning, M., and Geuten, K. (2019). Cold induced antisense transcription of FLOWERING LOCUS C in distant grasses. Front. Plant Sci. 10:72. doi: 10.3389/fpls.2019.00072
Kapazoglou, A., Engineer, C., Drosou, V., Kalloniati, C., Tani, E., Tsaballa, A., et al. (2012). The study of two barley type I-like MADS-box genes as potential targets of epigenetic regulation during seed development. BMC Plant Biol. 12:166. doi: 10.1186/1471-2229-12-166
Kazan, K., and Lyons, R. (2016). The link between flowering time and stress tolerance. J. Exp. Bot. 67, 47–60. doi: 10.1093/jxb/erv441
Kim, S. L., Lee, S., Kim, H. J., Nam, H. G., and An, G. (2007). OsMADS51 is a short-day flowering promoter that functions upstream of Ehd1, OsMADS14, and Hd3a. Plant Physiol. 145, 1484–1494. doi: 10.1104/pp.107.103291
Kim, D. H., and Sung, S. (2017). Vernalization-triggered intragenic chromatin loop formation by long noncoding RNAs. Dev. Cell 40, 302.e4–312.e4. doi: 10.1016/j.devcel.2016.12.021
Krasileva, K. V., Vasquez-Gross, H. A., Howell, T., Bailey, P., Paraiso, F., Clissold, L., et al. (2017). Uncovering hidden variation in polyploid wheat. Proc. Natl. Acad. Sci. 114, E913–E921. doi: 10.1073/pnas.1619268114
Kumar, G., Arya, P., Gupta, K., Randhawa, V., Acharya, V., and Singh, A. K. (2016). Comparative phylogenetic analysis and transcriptional profiling of MADS-box gene family identified DAM and FLC-like genes in apple (Malusx domestica). Sci. Rep. 6:20695. doi: 10.1038/srep20695
Lee, J. H., Ryu, H. -S., Chung, K. S., Posé, D., Kim, S., Schmid, M., et al. (2013). Regulation of temperature-responsive flowering by MADS-box transcription factor repressors. Science 342, 628–632. doi: 10.1126/science.1241097
Li, Z., Jiang, D., and He, Y. (2018). FRIGIDA establishes a local chromosomal environment for FLOWERING LOCUS C mRNA production. Nat. Plants 4, 836–846. doi: 10.1038/s41477-018-0250-6
Li, Q., Zheng, Q., Shen, W., Cram, D., Brian Fowler, D., Wei, Y., et al. (2015). Understanding the biochemical basis of temperature-induced lipid pathway adjustments in plants. Plant Cell 27, 86–103. doi: 10.1105/tpc.114.134338
Liang, Y., Liu, Q., Wang, X., Huang, C., Xu, G., Hey, S., et al. (2019). ZmMADS69 functions as a flowering activator through the ZmRap2.7-ZCN8 regulatory module and contributes to maize flowering time adaptation. New Phytol. 221, 2335–2347. doi: 10.1111/nph.15512
Ling, H. Q., Wang, J., Zhao, S., Liu, D., Wang, J., Sun, H., et al. (2013). Draft genome of the wheat A-genome progenitor Triticum urartu. Nature 496, 87–90. doi: 10.1038/nature11997
Liu, Z., Xin, M., Qin, J., Peng, H., Ni, Z., Yao, Y., et al. (2015). Temporal transcriptome profiling reveals expression partitioning of homeologous genes contributing to heat and drought acclimation in wheat (Triticum aestivum L.). BMC Plant Biol. 15:152. doi: 10.1186/s12870-015-0511-8
Lomax, A., Woods, D. P., Dong, Y., Bouché, F., Rong, Y., Mayer, K. S., et al. (2018). An ortholog of CURLY LEAF/ENHANCER OF ZESTE like-1 is required for proper flowering in Brachypodium distachyon. Plant J. 93, 871–882. doi: 10.1111/tpj.13815
Luo, X., Chen, T., Zeng, X., He, D., and He, Y. (2019). Feedback regulation of FLC by FLOWERING LOCUS T (FT) and FD through a 5' FLC promoter region in Arabidopsis. Mol. Plant 12, 285–288. doi: 10.1016/j.molp.2019.01.013
Luo, M. C., Gu, Y. Q., Puiu, D., Wang, H., Twardziok, S. O., Deal, K. R., et al. (2017). Genome sequence of the progenitor of the wheat D genome Aegilops tauschii. Nature 551, 498–502. doi: 10.1038/nature24486
Lutz, U., Nussbaumer, T., Spannagl, M., Diener, J., Mayer, K. F. X., and Schwechheimer, C. (2017). Natural haplotypes of FLM non-coding sequences fine-tune flowering time in ambient spring temperatures in Arabidopsis. elife 6:e22114. doi: 10.7554/eLife.22114
Maccaferri, M., Harris, N. S., Twardziok, S. O., Pasam, R. K., Gundlach, H., Spannagl, M., et al. (2019). Durum wheat genome highlights past domestication signatures and future improvement targets. Nat. Genet. 51, 885–895. doi: 10.1038/s41588-019-0381-3
Mascher, M., Gundlach, H., Himmelbach, A., Beier, S., Twardziok, S. O., Wicker, T., et al. (2017). A chromosome conformation capture ordered sequence of the barley genome. Nature 544, 427–433. doi: 10.1038/nature22043
Mateos, J. L., Tilmes, V., Madrigal, P., Severing, E., Richter, R., Rijkenberg, C. W. M., et al. (2017). Divergence of regulatory networks governed by the orthologous transcription factors FLC and PEP1 in Brassicaceae species. Proc. Natl. Acad. Sci. 114, E11037–E11046. doi: 10.1073/pnas.1618075114
McKay, J. K., Richards, J. H., and Mitchell-Olds, T. (2003). Genetics of drought adaptation in Arabidopsis thaliana: I. Pleiotropy contributes to genetic correlations among ecological traits. Mol. Ecol. 12, 1137–1151. doi: 10.1046/j.1365-294X.2003.01833.x
McMaster, G. S., and Wilhelm, W. W. (2003). Phenological responses of wheat and barley to water and temperature: improving simulation models. J. Agric. Sci. 141, 129–147. doi: 10.1017/S0021859603003460
Michaels, S. D., Bezerra, I. C., and Amasino, R. M. (2004). FRIGIDA-related genes are required for the winter-annual habit in Arabidopsis. Proc. Natl. Acad. Sci. 101, 3281–3285. doi: 10.1073/pnas.0306778101
Mohammadin, S., Nguyen, T. P., Van Weij, M. S., Reichelt, M., and Schranz, M. E. (2017). Flowering locus C (FLC) is a potential major regulator of glucosinolate content across developmental stages of Aethionema arabicum (Brassicaceae). Front. Plant Sci. 8:876. doi: 10.3389/fpls.2017.00876
Monteagudo, A., Igartua, E., Contreras-Moreira, B., Gracia, M. P., Ramos, J., Karsai, I., et al. (2019). Fine-tuning of the flowering time control in winter barley: the importance of HvOS2 and HvVRN2 in non-inductive conditions. BMC Plant Biol. 19:113. doi: 10.1186/s12870-019-1727-9
Oliver, S., Finnegan, E., Dennis, E. S., Peacock, W. J., and Trevaskis, B. (2009). Vernalization-induced flowering in cereals is associated with changes in histone methylation at the VERNALIZATION1 gene. Proc. Natl. Acad. Sci. 106, 8386–8391. doi: 10.1073/pnas.0903566106
Posé, D., Verhage, L., Ott, F., Yant, L., Mathieu, J., Angenent, G. C., et al. (2013). Temperature-dependent regulation of flowering by antagonistic FLM variants. Nature 503, 414–417. doi: 10.1038/nature12633
Priest, H. D., Fox, S. E., Rowley, E. R., Murray, J. R., Michael, T. P., and Mockler, T. C. (2014). Analysis of global gene expression in Brachypodium distachyon reveals extensive network plasticity in response to abiotic stress. PLoS One 9:e87499. doi: 10.1371/journal.pone.0087499
Ramírez-González, R. H., Borrill, P., Lang, D., Harrington, S. A., Brinton, J., Venturini, L., et al. (2018). The transcriptional landscape of polyploid wheat. Science 361:eaar6089. doi: 10.1126/science.aar6089
Ream, T. S., Woods, D. P., and Amasino, R. M. (2012). The molecular basis of vernalization in different plant groups. Cold Spring Harb. Symp. Quant. Biol. 77, 105–115. doi: 10.1101/sqb.2013.77.014449
Rizza, F., Karsai, I., Morcia, C., Badeck, F. W., Terzi, V., Pagani, D., et al. (2016). Association between the allele compositions of major plant developmental genes and frost tolerance in barley (Hordeum vulgare L.) germplasm of different origin. Mol. Breed. 36:156. doi: 10.1007/s11032-016-0571-y
Ruelens, P., De Maagd, R. A., Proost, S., Theißen, G., Geuten, K., and Kaufmann, K. (2013). FLOWERING LOCUS C in monocots and the tandem origin of angiosperm-specific MADS-box genes. Nat. Commun. 4:2280. doi: 10.1038/ncomms3280
Schilling, S., Kennedy, A., Pan, S., Jermiin, L. S., and Melzer, R. (2020). Genome-wide analysis of MIKC-type MADS-box genes in wheat: pervasive duplications, functional conservation and putative neofunctionalization. New Phytol. 225, 511–529. doi: 10.1111/nph.16122
Schreiber, M., Barakate, A., Uzrek, N., Macaulay, M., Sourdille, A., Morris, J., et al. (2019). A highly mutagenised barley (cv. Golden promise) TILLING population coupled with strategies for screening-by-sequencing. Plant Methods 15:99. doi: 10.1186/s13007-019-0486-9
Searle, I., He, Y., Turck, F., Vincent, C., Fornara, F., Kröber, S., et al. (2006). The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signalling in Arabidopsis. Genes Dev. 20, 898–912. doi: 10.1101/gad.373506
Sharma, N., Ruelens, P., D’hauw, M., Maggen, T., Dochy, N., Torfs, S., et al. (2017). A flowering locus C homolog is a vernalization-regulated repressor in Brachypodium and is cold regulated in wheat. Plant Physiol. 173, 1301–1315. doi: 10.1104/pp.16.01161
Sheldon, C. C., Rouse, D. T., Finnegan, E., Peacock, W. J., and Dennis, E. S. (2000). The molecular basis of vernalization: the central role of FLOWERING LOCUS C (FLC). Proc. Natl. Acad. Sci. 97, 3753–3758. doi: 10.1073/pnas.060023597
Shindo, C., Lister, C., Crevillen, P., Nordborg, M., and Dean, C. (2006). Variation in the epigenetic silencing of FLC contributes to natural variation in Arabidopsis vernalization response. Genes Dev. 20, 3079–3083. doi: 10.1101/gad.405306
Swiezewski, S., Liu, F., Magusin, A., and Dean, C. (2009). Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature 462, 799–802. doi: 10.1038/nature08618
Takaiwa, F., Yamanouchi, U., Yoshihara, T., Washida, H., Tanabe, F., Kato, A., et al. (1996). Characterization of common cis-regulatory elements responsible for the endosperm-specific expression of members of the rice glutelin multigene family. Plant Mol. Biol. 30, 1207–1221. doi: 10.1007/BF00019553
Trevaskis, B., Bagnall, D. J., Ellis, M. H., Peacock, W. J., and Dennis, E. S. (2003). MADS box genes control vernalization-induced flowering in cereals. Proc. Natl. Acad. Sci. 100, 13099–13104. doi: 10.1073/pnas.1635053100
Trevaskis, B., Hemming, M. N., Dennis, E. S., and Peacock, W. J. (2007). The molecular basis of vernalization-induced flowering in cereals. Trends Plant Sci. 12, 352–357. doi: 10.1016/j.tplants.2007.06.010
Wang, X., Wang, J., Jin, D., Guo, H., Lee, T. H., Liu, T., et al. (2015). Genome alignment spanning major poaceae lineages reveals heterogeneous evolutionary rates and alters inferred dates for key evolutionary events. Mol. Plant 8, 885–898. doi: 10.1016/j.molp.2015.04.004
Watson, A., Ghosh, S., Williams, M. J., Cuddy, W. S., Simmonds, J., Rey, M. D., et al. (2018). Speed breeding is a powerful tool to accelerate crop research and breeding. Nat. Plants 4, 23–29. doi: 10.1038/s41477-017-0083-8
Whittaker, C., and Dean, C. (2017). The FLC locus: a platform for discoveries in epigenetics and adaptation. Annu. Rev. Cell Dev. Biol. 33, 555–575. doi: 10.1146/annurev-cellbio-100616-060546
Winfield, M. O., Lu, C., Wilson, I. D., Coghill, J. A., and Edwards, K. (2009). Cold- and light-induced changes in the transcriptome of wheat leading to phase transition from vegetative to reproductive growth. BMC Plant Biol. 9:55. doi: 10.1186/1471-2229-9-55
Wood, C. C., Robertson, M., Tanner, G., Peacock, W. J., Dennis, E. S., and Helliwell, C. A. (2006). The Arabidopsis thaliana vernalization response requires a polycomb-like protein complex that also includes VERNALIZATION INSENSITIVE 3. Proc. Natl. Acad. Sci. 103, 14631–14636. doi: 10.1073/pnas.0606385103
Woods, D. P., McKeown, M., Dong, Y., Preston, J. C., and Amasino, R. M. (2016). Evolution of VRN2/Ghd7-like genes in vernalization-mediated repression of grass flowering. Plant Physiol. 170, 2124–2135. doi: 10.1104/pp.15.01279
Yan, L., Fu, D., Li, C., Blechl, A., Tranquilli, G., Bonafede, M., et al. (2006). The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proc. Natl. Acad. Sci. 103, 19581–19586. doi: 10.1073/pnas.0607142103
Yan, L., Loukoianov, A., Blechl, A., Tranquilli, G., Ramakrishna, W., SanMiguel, P., et al. (2004). The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science 303, 1640–1644. doi: 10.1126/science.1094305
Yan, L., Loukoianov, A., Tranquilli, G., Helguera, M., Fahima, T., and Dubcovsky, J. (2003). Positional cloning of the wheat vernalization gene VRN1. Proc. Natl. Acad. Sci. 100, 6263–6268. doi: 10.1073/pnas.0937399100
Yuan, W., Luo, X., Li, Z., Yang, W., Wang, Y., Liu, R., et al. (2016). A cis cold memory element and a trans epigenome reader mediate Polycomb silencing of FLC by vernalization in Arabidopsis. Nat. Genet. 48, 1527–1534. doi: 10.1038/ng.3712
Keywords: flowering time, cereals, FLOWERING LOCUS C, vernalisation, ambient temperature
Citation: Kennedy A and Geuten K (2020) The Role of FLOWERING LOCUS C Relatives in Cereals. Front. Plant Sci. 11:617340. doi: 10.3389/fpls.2020.617340
Received: 14 October 2020; Accepted: 02 December 2020;
Published: 22 December 2020.
Edited by:
Elizabeth Dennis, Commonwealth Scientific and Industrial Research Organisation (CSIRO), AustraliaReviewed by:
George Coupland, Max Planck Institute for Plant Breeding Research, GermanyCopyright © 2020 Kennedy and Geuten. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Koen Geuten, a29lbi5nZXV0ZW5Aa3VsZXV2ZW4uYmU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.