- Key Laboratory of Application and Safety Control of Genetically Modified Crops, Rice Research Institute, Southwest University, Academy of Agricultural Sciences, Chongqing, China
Moderate curling generally causes upright leaf blades, which favors the establishment of ideal plant architecture and increases the photosynthetic efficiency of the population, both of which are desirable traits for super hybrid rice (Oryza sativa L.). In this study, we identified a novel curled-leaf mutant, curled flag leaf 2 (cfl2), which shows specific curling at the base of the flag leaf owing to abnormal epidermal development, caused by enlarged bulliform cells and increased number of papillae with the disordered distribution. Map-based cloning reveals that CFL2 encodes a cytochrome P450 protein and corresponds to the previously reported OsCYP96B4. CFL2 was expressed in all analyzed tissues with differential abundance and was downregulated in the clf1 mutant [a mutant harbors a mutation in the homeodomain leucine zipper IV (HD-ZIP IV) transcription factor Roc5]. Yeast one-hybrid and transient expression assays confirm that Roc5 could directly bind to the cis-element L1 box in the promoter of CFL2 before activating CFL2 expression. RNA sequencing reveals that genes associated with cellulose biosynthesis and cell wall-related processes were significantly upregulated in the cfl2 mutant. The components of cell wall, such as lignin, cellulose, and some kinds of monosaccharide, were altered dramatically in the cfl2 mutant when compared with wild-type “Jinhui10” (WT). Taken together, CFL2, as a target gene of Roc5, plays an important role in the regulation of flag leaf shape by influencing epidermis and cell wall development.
Introduction
The leaf is the primary photosynthesis organ in plants. The morphological characteristics of leaves, such as shape, size, and thickness, directly affect light utilization and thus influence the yield of crops (Zhang et al., 2015). Moderate curling is beneficial for the development of an erect leaf, which improves the population structure and increases the light utilization, and has important outcomes for breeding high-yield rice (Richards et al., 2002; Wang et al., 2020). Therefore, elucidation of the genetic mechanism of leaf curling is important both in understanding leaf development and in the improvement of plant architecture in rice.
Leaf curling is a complex agronomic character, which is regulated by both genotype and environment (Li et al., 2017). The rolling rice leaf is affected by bulliform cell development, adaxial–abaxial axis polarity, sclerenchyma formation, and epidermal structure (Xu et al., 2013). The morphology, number, size, and distribution of the bulliform cells in the epidermis are the predominant factors that affect leaf rolling. The transferred DNA (T-DNA) mutant BY240, in which the T-DNA was inserted in the promoter of ABAXIALLY CURLED LEAF1 (ACL1), shows increased number, and size of bulliform cells, which causes uncoordinated development of the abaxial and adaxial epidermis of the leaf, and results in abaxially rolled leaves. Overexpression of ACL2, the paralog of ACL1, can also induce abaxial leaf rolling (Li et al., 2010). Many additional genes are involved in leaf rolling by affecting the bulliform cell development, such as SEMI-ROLLED LEAF1 (SRL1/CLD1) (Xiang et al., 2012; Li et al., 2017), ZINC-FINGER HOMEODOMAIN1 (OsZHD1) (Xu et al., 2014), RICE OUTERMOST CELL-SPECIFIC GENE5 (Roc5) (Zou et al., 2011), ROLLED AND ERECT LEAF1 (REL1) (Chen et al., 2015), REL2 (Yang et al., 2016), LATERAL ORGAN BOUNDARIES DOMAIN (OsLBD3-7) (Li et al., 2016), and SHALLOT-LIKE2 (SLL2) (Zhang et al., 2015), but the mechanisms remain unclear. In addition, genes influence the bulliform cells through regulation of phytohormone, such as YABBY1 (OsYAB1) in gibberellin biosynthesis (Dai et al., 2007), BRASSINOSTEROID INSENSITIVE1-ASSOCIATED KINASE1 (OsI-BAK1) in brassinosteroid (BR) signaling (Khew et al., 2015), and CONSTITUTIVELY WILTED1 (OsCOW1/NAL7) and AUXIN RESPONSE FACTOR18 (OsARF18) in indole-3-acetic acid (IAA) synthetic pathway (Woo et al., 2007; Fujino et al., 2008; Huang et al., 2016). And, some genes related with secondary cell wall or cellulose formation, such as CELLULOSE SYNTHASE-LIKE D4 (OsCSLD4/NRL1) (Li et al., 2009; Hu et al., 2010; Luan et al., 2011), ROLLING LEAF14 (RL14) (Fang et al., 2012), and OsMYB103L (Yang et al., 2014), also influence the bulliform cell development.
Curling of the leaf usually correlates with polarity changes in the abaxial–adaxial axis. Homeodomain leucine zipper class III (HD-ZIPIII) family members are involved in the establishment of polarity during leaf development (McConnell et al., 2001; Otsuga et al., 2001; Juarez et al., 2004; Nagasaki et al., 2007). Five HD-ZIP III genes have been identified in rice. OSHB3 contributes to leaf polarity more than OSHB1 by involvement in vascular patterning and differentiation, whereas transgenic plant harboring mutated OSHB5 does not exhibit defective leaf polarity (Itoh et al., 2008). ADXIALIZED LEAF1 (ADL1) encodes a calpain-like cysteine proteinase, and loss-of-function of the maize ortholog DEFECTIVE KERNEL1 (DEK1) alters the adaxial–abaxial axis of the leaves, which shows ectopic bulliform-like cells in the abaxial epidermis (Hibara et al., 2009). OsAGO7 is a direct homolog of the Arabidopsis thaliana ZIP/Ago7 gene, which is involved in polar patterning, and controls the upward curling of rice leaves (Shi et al., 2007).
Sclerenchyma cells are lignified dead cells with thickened secondary cell walls that surround the vascular bundle and are involved in leaf rolling in rice (Zhang et al., 2009). SLL1 encodes a MYB transcription factor belonging to the KANADI family, and SRL2 encodes a novel plant-specific protein with unknown function. Mutants of both genes develop rolled leaves caused by the abnormal development of sclerenchyma cells on the abaxial side of the leaf (Zhang et al., 2009; Liu et al., 2016). Analysis of the srl2 sll1 double mutant shows that SLL1 and SRL2 participate in distinct metabolic pathways to regulate the development of sclerenchyma cells in the rice leaf (Zhang et al., 2009).
Epidermal structure also affects the leaf rolling. CURLY FLAG LEAF1 (CFL1), which encodes a protein containing a WW domain, interacts with HOMEODOMAIN GLABROUS1 (HDG1) to regulate negatively cuticle development, and the impaired cuticle formation is responsible for the curly leaf in cfl1 mutant (Wu et al., 2011). Obvious defects in the leaf epidermis and cuticle structures in the cld1 mutant, such as the disordered distribution of papillae and linear cork–silica cell pairs, indicate the importance of epidermal integrity for the maintenance of leaf shape (Li et al., 2017).
Although several of genes associated with leaf rolling have been identified and cloned, the mechanisms of abnormal leaf morphology are still poorly understood. In this study, we identified a novel curled-leaf mutant cfl2, generated by ethyl methane sulfonate (EMS) mutagenesis, which showed specific curling at the base of the flag leaf. Map-based cloning revealed that CFL2 encodes the OsCYP96B4 protein, which is a member of the cytochrome P450 monooxygenase family. Histological analysis and observation using scanning electron microscopy (SEM) revealed abnormal epidermis development in the cfl2 mutant. Biochemical and transcriptome analyses suggest that CFL2 is a downstream target gene of the HD-ZIP IV transcription factor Roc5, and upregulated genes are involved in cell wall-related processes in cfl2 mutant, which led to the abnormal development of the epidermis and thereby modulate flag leaf shape.
Materials and Methods
Plant Materials and Growth Conditions
The rice curled flag leaf mutant cfl2 and its two allelic mutants cfl2-1 and cfl2-2 were isolated from the progeny of indica restorer line “Jinhui10” seeds treated with EMS. The F2 mapping population was raised from the cross between the cfl2 and “Xinong1A,” a sterile line, with normal plant height and leaf morphology, selected by the Rice Research Institute of Southwest University. All plants were grown in paddy fields under natural conditions at the Rice Research Institute of Southwest University in Chongqing, China.
Microscopic Observations
For paraffin sectioning, the base of flag leaf of the wild-type “Jinhui10” (WT) and the cfl2 mutant were fixed with FAA solution (45% water, 45% ethanol, 5% formaldehyde, and 5% acetic acid) for 2 days at 4°C. The samples were embedded in paraffin after dehydrating with a graded ethanol series and infiltrating with ethanol–xylene in different proportions. Thin sections (approximately 8 μm thick) were cut using a rotary microtome (RM2245, Leica Microsystems, Hamburg, Germany). The sections were stained with fast green and safranin, then observed and photographed using a light microscope (Eclipse Ci-L, Nikon, Tokyo, Japan), as described previously (Ma et al., 2017). For frozen sections, the fresh flag leaf base was embedded in NEG-50 Frozen Section Medium and cut into 8 μm-thick sections at −20°C using a cryostat microtome (CryoStar NX50 OP, Thermo Fisher Scientific, United States), then observed and photographed through a light microscope (Eclipse Ci-L, Nikon, Tokyo, Japan). The area of bulliform cells was measured with AxioVision release 4.6 software. For SEM observations, fresh leaf samples were prepared and examined using a Hitachi SU3500 scanning electron microscope (Zhang et al., 2017).
Map-Based Cloning
A total of 837 individuals with curled flag leaves, which were selected from the 2,582 individuals of the F2 population raising from the cross between cfl2 mutant and “Xinong1A,” were used for CFL2 mapping. Simple sequence repeat (SSR) markers obtained from the publicly available rice databases1 were utilized for initial mapping, and insertion/deletion markers developed by our laboratory were used for fine mapping. For functional complementation, the CFL2 genomic fragment, consisting of 2,877 bp upstream of the start codon, the open reading frame, and 899 bp downstream of the stop codon, was amplified from the WT genomic DNA, digested using EcoRI and KpnI, and inserted into the binary vector pCAMBIA1301. The CFL2 complementation plasmid was introduced into the cfl2 mutant by Agrobacterium-mediated transformation as described previously (Sang et al., 2012). The primers used in this study are listed in Supplementary Table S4.
Subcellular Localization
The full-length coding sequence of CFL2 without the stop codon was amplified from WT and digested using SpeI and BamHI and then fused to the N-terminus of the GREEN FLUORESCENT PROTEIN (GFP) gene under the control of the enhanced Cauliflower mosaic virus (CaMV) 35S promoter in the expression vector pAN580 to generate CFL2–GFP construct. The CFL2–GFP plasmids were transformed into rice protoplasts by polyethylene glycol-mediated method. After overnight incubation at 28°C, fluorescence was observed using a LSM800 confocal laser microscope (Zeiss, Jena, Germany).
Protein Sequence Alignment and Phylogenetic Analysis
The full-length amino acid sequences of CYP96 clan members in rice and Arabidopsis were downloaded from the National Center for Biotechnology Information (NCBI) databases2. The protein sequence of ATHDG1 was downloaded from TAIR3, and the homologous genes in rice were identified from Phytozome4. Multiple sequence alignment was generated using ClustalX2.1. Phylogenetic trees were reconstructed with MEGA 5 using the maximum likelihood method and bootstrapping (with 1,000 replicates).
In situ Hybridization
The 387-bp CFL2 probe was amplified from the cDNA and labeled using the DIG RNA Labeling Kit (Roche, Basel, Switzerland) in accordance with the manufacturer’s protocols. The leaf and shoot apical meristem (SAM) of the WT were fixed in FAA overnight at 4°C. Pretreatment of sections, hybridization, and immunological detection were performed following previously described methods (Sang et al., 2012).
RNA Isolation and Quantitative Real-Time PCR Analysis
Total RNA of various plant tissues of the WT and mutants were isolated using the RNAprep Pure Plant Kit (Tiangen, Beijing, China). The concentration, purity, and integrity of extracted RNA were determined using a NanoDrop One spectrophotometer (Thermo Fisher Scientific) and agarose gel electrophoresis. RNA reverse transcription was carried out using the SuperScript III Reverse Transcriptase Kit (Invitrogen, Carlsbad, CA, United States) in accordance with the manufacturer’s instructions. Quantitative real-time PCR (qRT-PCR) was performed using NovoStart SYBR qPCR SuperMix Plus (Novoprotein, Shanghai, China) and a CFX ConnectTM Real-time System (Bio-Rad, Hercules, CA, United States) with three replicates.
Yeast One-Hybrid Assay
For the yeast one-hybrid assay (Y1H), the three tandems repeated L1 box AACATTTA and L1 mutant box AACCGTTA oligonucleotide sequences were annealed and inserted into the SacI and SalI sites in the pAbAi plasmid to generate pL1–AbAi and pL1mutant–AbAi. The two plasmids, linearized with BstBI, were transformed into Y1HGold cells on SD/-URA agar medium using the protocol for the Yeastmaker Yeast Transformation System to generate Y1HGold (L1/AbAi) and Y1HGold (L1 mutant/AbAi) strains. The full-length coding sequence (CDS) of Roc5 was amplified and cloned into the pGADT7 vector to generate Roc5–AD plasmids and then transformed to the two newly constructed Y1HGold bait strains after testing their background for AbAr expression.
Dual-Luciferase Assay
For the dual-luciferase assays, a 2.1 kb promoter segment of CFL2 was cloned and recombined into pGreenII 0800-LUC vector digested with KpnI and HindIII, thus generating the pCFL2:LUC reporter construct. The Renilla luciferase gene driven by CaMV35S promoter in the pGreenII 0800-LUC vector was used as an internal reference. The full-length CDS of Roc5 was amplified and inserted into the BamHI and EcoRI sites of the pGreenII 62-SK vector driven by the 35S promoter as the effector. The empty pGreenII 62-SK vector was used as a control. The dual-luciferase assays were performed in rice protoplasts, and the luciferase signal was detected using Dual-Luciferase Reporter Assay System and GloMax® 20/20 Luminometer (Promega, Madison, WI, United States) following the manufacturer’s instructions.
Transcriptome and Gene Ontology Enrichment Analyses
Total RNA was isolated from the base of the flag leaf of WT and cfl2 with three biological replicates at the early stage before the flag leaf emerged from the sheath. Library preparation and sequencing of six libraries were conducted by the Biomarker Technologies Corporation (Beijing, China) using a HiSeq 4000 platform (Illumina, San Diego, CA, United States) following the manufacturer’s protocol. The raw reads were filtered and then mapped to the rice reference genome using HISAT2 software with default parameters (Kim et al., 2015). Gene expression levels were quantified using StringTie and expressed as fragments per kilobase of transcript per million mapped reads (FPKM) (Florea et al., 2013). The differentially expressed genes (DEGs) were detected using the DESeq R package with the following criteria: false discovery rate < 0.05 and log2| fold change| > 1 (Love et al., 2014). The DEGs were used for gene ontology (GO) enrichment analysis with the agriGO online resource5. RNA sequencing data were deposited in the NCBI Sequence Read Archive (SRA) under accession PRJNA628038.
Analysis of Cell Wall Components
The base of flag leaves was collected from WT and cfl2 mutant for the cell wall component analysis. The lignin and cellulose content was measured using the correspondent kit (Solarbio, Catalog no. BC4205 and BC4285, respectively). The monosaccharide was extracted and analyzed by high-performance liquid chromatography (HPLC). To the above 0.1 g sample, 0.5 ml precooled trifluoroacetic acid (4 mol/l) was added and hydrolyzed for 4 h under 110°C, and the supernatant was collected after 8,000 g centrifugation for 10 min. Then, methanol (500 μl) was added, blow dried by nitrogen, and dissolved into 500 μl water. Solving liquid (50 μl) was taken and NaOH (50 μl) and PMP methanol solution (100 μl) were added and reacted for 100 min at 70°C avoiding light. HCl (100 μl) and water (400 μl) were added to the reaction mixture and then extracted by the same volume of chloroform three times. The 10 μl extractant was analyzed by HPLC (RIGOL L3000) with a column (RIGOL C18, 250 mm × 4.6 mm, 5 μm) at 30°C.
Results
Phenotypes of the cfl2 Mutant
The cfl2 mutant was identified from the progeny of EMS-treated seeds of the rice indica restorer line “Jinhui10.” In the paddy field, the cfl2 mutant was slightly and insignificantly shorter than WT at seedling stage, but the leaf morphology of the cfl2 mutant was identical to that of the WT, as well as the root and tiller (Figures 1A,B). At heading stage, no significant difference in plant height was observed between WT and the cfl2 mutant (Figure 1C and Supplementary Figure S1). The main agronomic traits of the cfl2 mutant were normal compared with those of WT (Supplementary Table S1). The flag leaf base of the cfl2 mutant was abnormal, in that the basal half of the leaf was curled inward, whereas the middle and distal portions were normally developed (Figure 1D). In addition, the blade of other leaves of the cfl2 mutant, including the second and third upper leaves, were normally unfolded as in the WT (Figures 1E,F). Two allelic mutants cfl2-1 and cfl2-2 were further identified from the mutant library. Although both of the allelic mutants exhibited a dwarfism phenotype, which is different from the cfl2, their flag leaves were also specifically curly (Supplementary Figure S2). These results indicate that the cfl2 was a specific curling mutant at the base of the flag leaf.

Figure 1. Phenotypic identification of the WT and cfl2 mutant. (A) 2 week-old seedlings of WT and cfl2. Scale bar = 12 cm. (B) WT and cfl2 plants in tillering stage. Scal bar = 20 cm. (C) Mature WT and cfl2 plants. Scale bar = 30 cm. (D) Panicle and flag leaf of WT and cfl2. Scale bar = 10 cm. (E,F) The top three leaves of WT and cfl2. Scale bars = 3 cm.
Flag Leaf Microstructure of the cfl2 Mutant
To further characterize the morphology of the flag leaf base, paraffin-embedded sections of the flag blade base at different stages were prepared. Both sides of the WT flag leaf blade were curled in a regular, circular manner, whereas the inner side of the cfl2 mutant was arranged irregularly at the early stage of blade development (Figures 2A,B). This irregular curling phenotype in the mutant continued after the flag leaf emergence (Figure 2C). When the flag leaf was fully expanded, both sides of the leaf blade showed outward expansion in the WT. In the cfl2 mutant, the outside half of the leaf blade showed outward expansion, whereas the inside half of the blade showed adaxial curling (Figures 2D,E), which ultimately formed a shallot-like structure (Figure 2F). Frozen cross sections of the mature flag leaf revealed that the number of bulliform cells on the sides of large and small vascular bundles was unchanged between the WT and cfl2 mutant. However, the average area of bulliform cells was significantly larger in the cfl2 mutant than in the WT (Figures 2G–L and Supplementary Table S5), which suggests that the curled leaf in the cfl2 mutant may be caused by the enlarged bulliform cells.
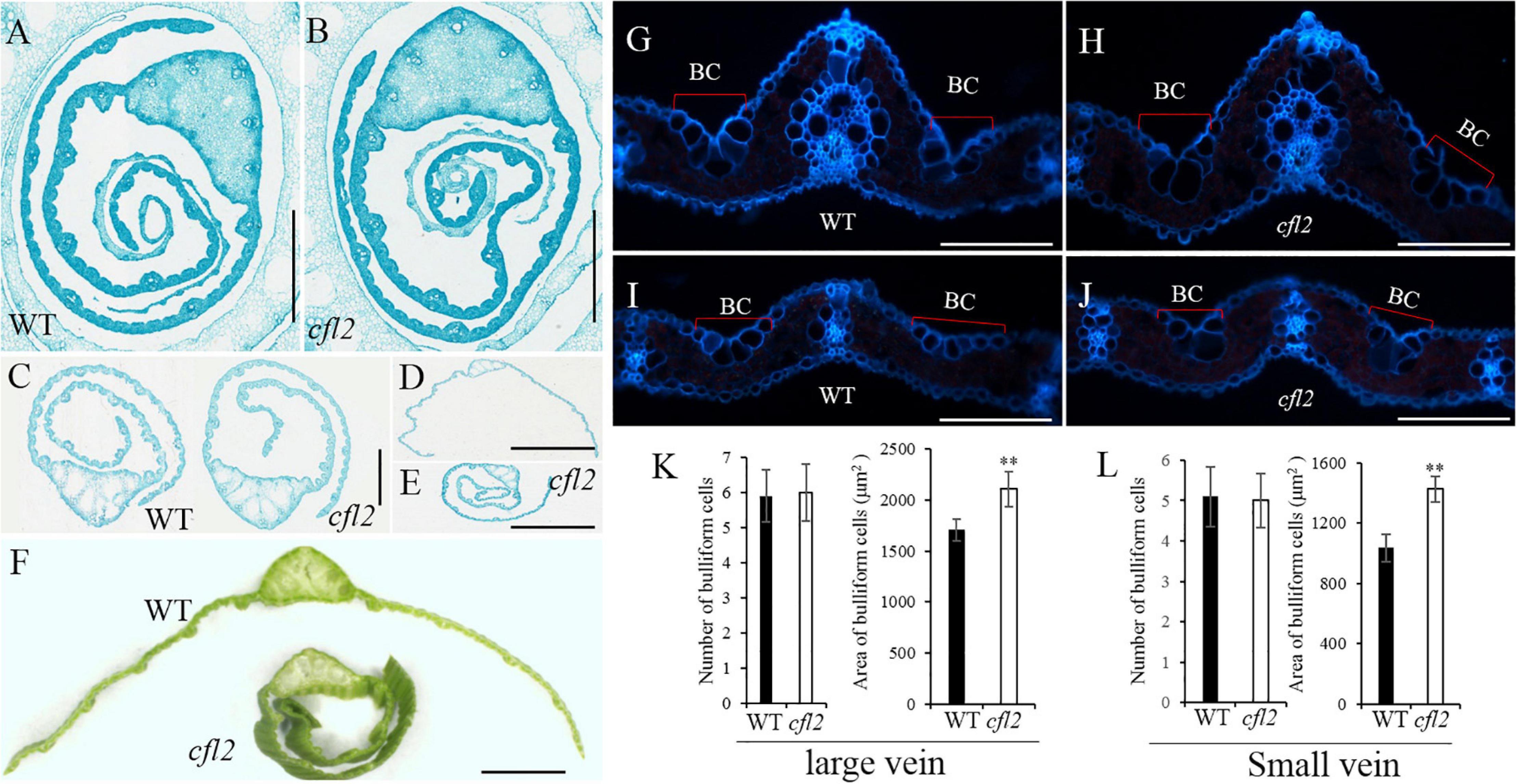
Figure 2. Flag leaf microstructure of WT and cfl2. (A,B) The WT and cfl2 flag leaf wrapped by sheath. Scale bars = 3 mm. (C) The emergence flag leaf of WT and cfl2. Scale bars = 1 mm. (D–F) The fully expanded flag leaf of WT and cfl2. Scale bar = 3 mm (D,E) and 1 mm (F). (G–J) Frozen cross sections of large vascular and small vascular bundles of WT and cfl2. BC, bulliform cells. Scale bars = 100 μm. (K,L) Statistical analysis of number and area of bulliform cells of large vascular and small vascular bundles in WT and cfl2. Values are means ± SD (n = 10). Student’s t-test was used for statistical analysis (∗∗P < 0.01).
Marked differences between the WT and cfl2 mutant in the epidermis structure of the flag leaf were observed by SEM. The epidermal of bulliform cell pairs was smooth in the WT, but a distinct crack was observed in the cfl2 mutant (Figures 3A,B). The papillae were sparse and regularly arranged in both of the adaxial and abaxial epidermis of WT flag leaf, whereas the number of papillae was increased highly significant and their distribution disordered in the flag leaf of the cfl2 mutant (Figures 3A–D,I,J). No significant difference was observed between the WT and cfl2 mutant in the stomata and the crystal structure of cuticular wax on the adaxial and abaxial surfaces of the flag leaf (Figures 3E–H,K,L). These results demonstrated that the epidermal modifications were responsible for the curled flag leaf in the cfl2 mutant.
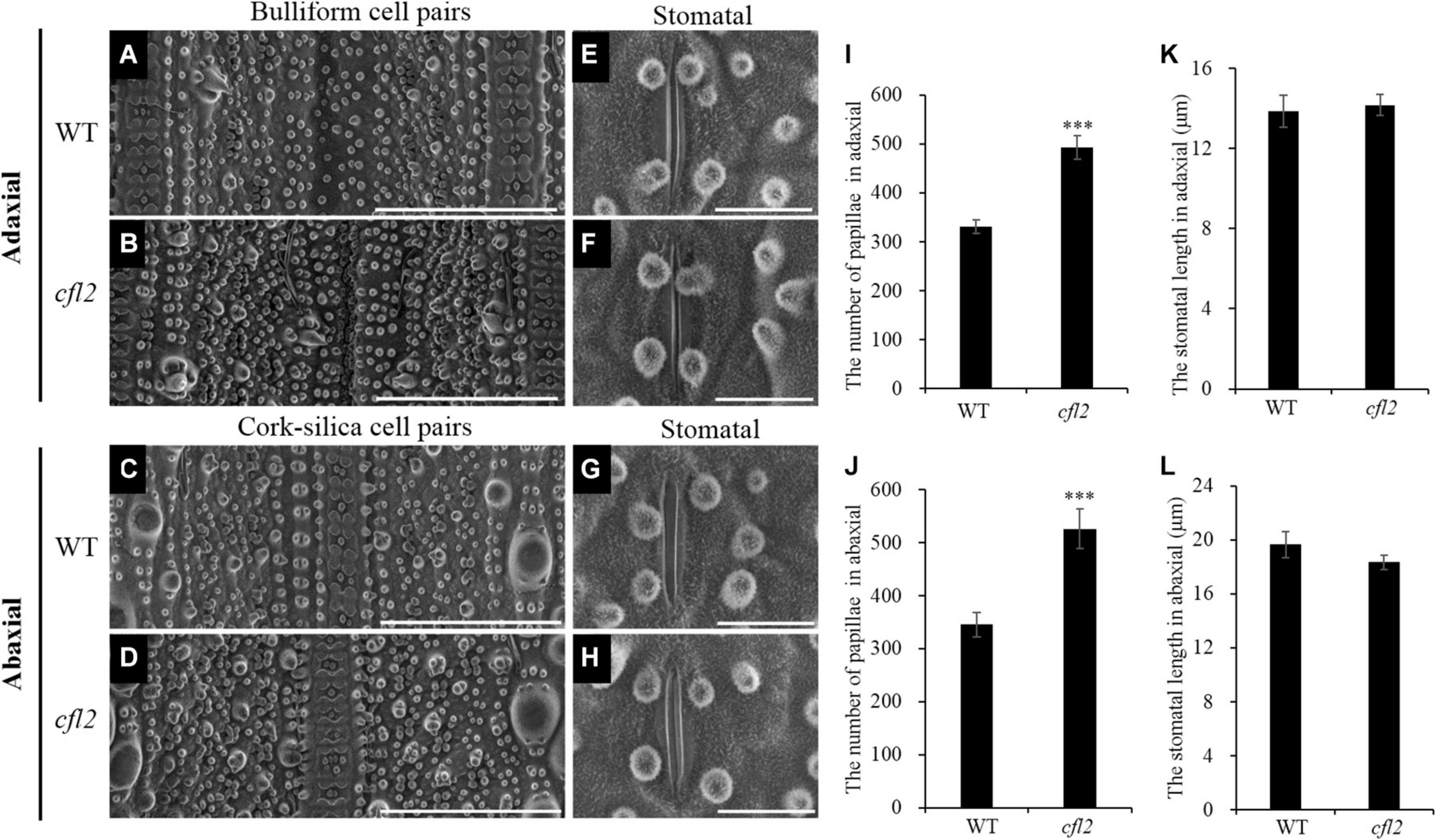
Figure 3. SEM observations of the flag leaf epidermis in WT and cfl2. (A,B) The bulliform cell pairs of WT and cfl2 in adaxial epidermis. Scale bars = 100 μm. (C,D) The cork–silica cell pair area of WT and cfl2 in abaxial epidermis. Scale bars = 100 μm. (E–H) The stomata of WT and cfl2 in adaxial and abaxial axis. Scale bars = 10 μm. (I,J) Statistical analysis of the number of papillae cell in adaxial and abaxial axis of WT and cfl2. (K,L) Statistical analysis of the stomatal length in adaxial and abaxial axis of WT and cfl2. Values are means ± SD (n = 6). Student’s t-test was used for statistical analysis (∗∗∗P < 0.001).
Map-Based Cloning of CFL2
To clone CFL2, the cfl2 mutant was crossed with “Xinong 1A,” an indica male-sterile line. The plant phenotypes, including the flag leaf morphology, were identical to those of the WT in all F1 individuals. Genetic analysis showed that segregation of the mutant phenotype in the F2 progeny conformed to a 3:1 ratio (837 of 2,582 individuals showed the mutant phenotype; χ2 = 0.49 < χ20.05 = 3.84), which indicated that the curled flag leaf trait of the cfl2 mutant was controlled by a single recessive nuclear gene. A total of 837 individuals consistent with the mutant phenotype were used for mapping analysis. CFL2 was fine mapped to the long arm of the chromosome 3 between InDel marker Ind03-11 and Ind03-6 with a physical interval of 78 kb (Figure 4A). Sequencing the 15 genes annotated in the Gramene database6 within the interval (Supplementary Table S2) revealed a single-nucleotide transition (C to T, causing a substitution of Ala to Val) within LOC_Os03g04680 in the cfl2 mutant. The two allelic mutants cfl2-1 and cfl2-2 were showed to be mutated in the coding sequence, leading to the substitution of the amino acid at different locations (Figure 4B).
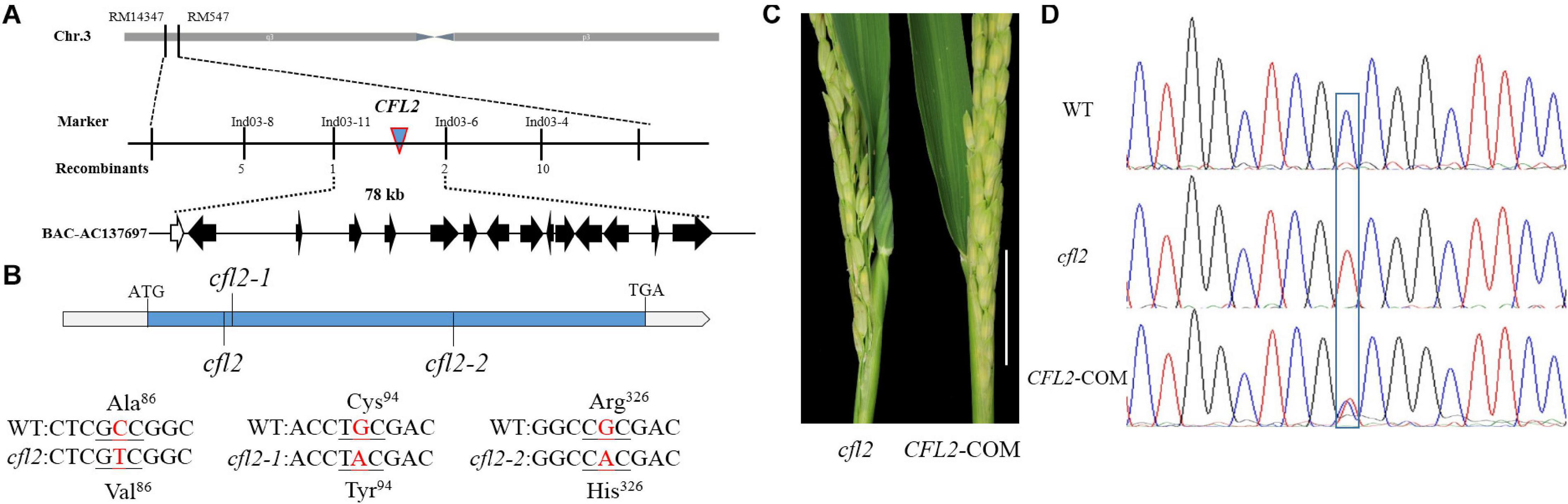
Figure 4. Map-based cloning of CFL2. (A) The CFL2 gene was fine mapped to the long arm of chromosome 3 between Ind03-11 and Ind03-6 with a physical interval 78 kb. (B) Gene structure of CFL2 and the mutation sites in cfl2 and its allelic mutant cfl2-1 and cfl2-2. (C) The base of the flag leaf morphology of the cfl2 mutant and complemented transgenic plant (CFL2-COM). Scale bar = 5 cm. (D) Sequence peak chromatograms of the mutation region in plants of the WT, cfl2, and CFL2-COM.
To verify that LOC_Os03g04680 is equivalent to the CFL2 gene, genome complementation was performed. The flag leaf base of the complemented line was recovered (Figure 4C), and the complemented lines were heterozygous (C/T) at the substitution site (Figure 4D). These results confirmed that the mutation of LOC_Os03g04680 was responsible for the curled flag leaf phenotype of the cfl2 mutant.
CFL2 Encodes a Cytochrome P450 Protein
LOC_Os03g04680 encodes a cytochrome P450 protein, belonging to the CYP96 subfamily, which was previously designated OsCYP96B4 (Nelson et al., 2004; Rengasamy et al., 2011). In total, 12 and 13 CYP96 members have been identified in the Arabidopsis and rice genome, respectively. Phylogenetic analysis showed that 25 proteins were categorized into two distinct evolutionary branches, which indicated that CYP96 subfamily proteins show strong functional conservations in monocotyledons and dicotyledons (Figure 5A). The CYP96 proteins contain a number of highly conserved domains, including six substrate-recognition sites (SRS), an I-helix, a K-helix (Glu-x-x-Arg domain), and a heme binding loop (Schuler and Werck-Reichhart, 2003). The amino acid sequence alignment showed that the cfl2 mutation site was not within a conserved domain. However, the mutation of the two allelic mutants cfl2-1 and cfl2-2 located the different conserved domain, especially the cfl2-2 mutation site, which was located in the I-helix domain (Figure 5B). The different mutation sites may be the cause of the different phenotypes in plant height of the three CFL2 allelic mutants.
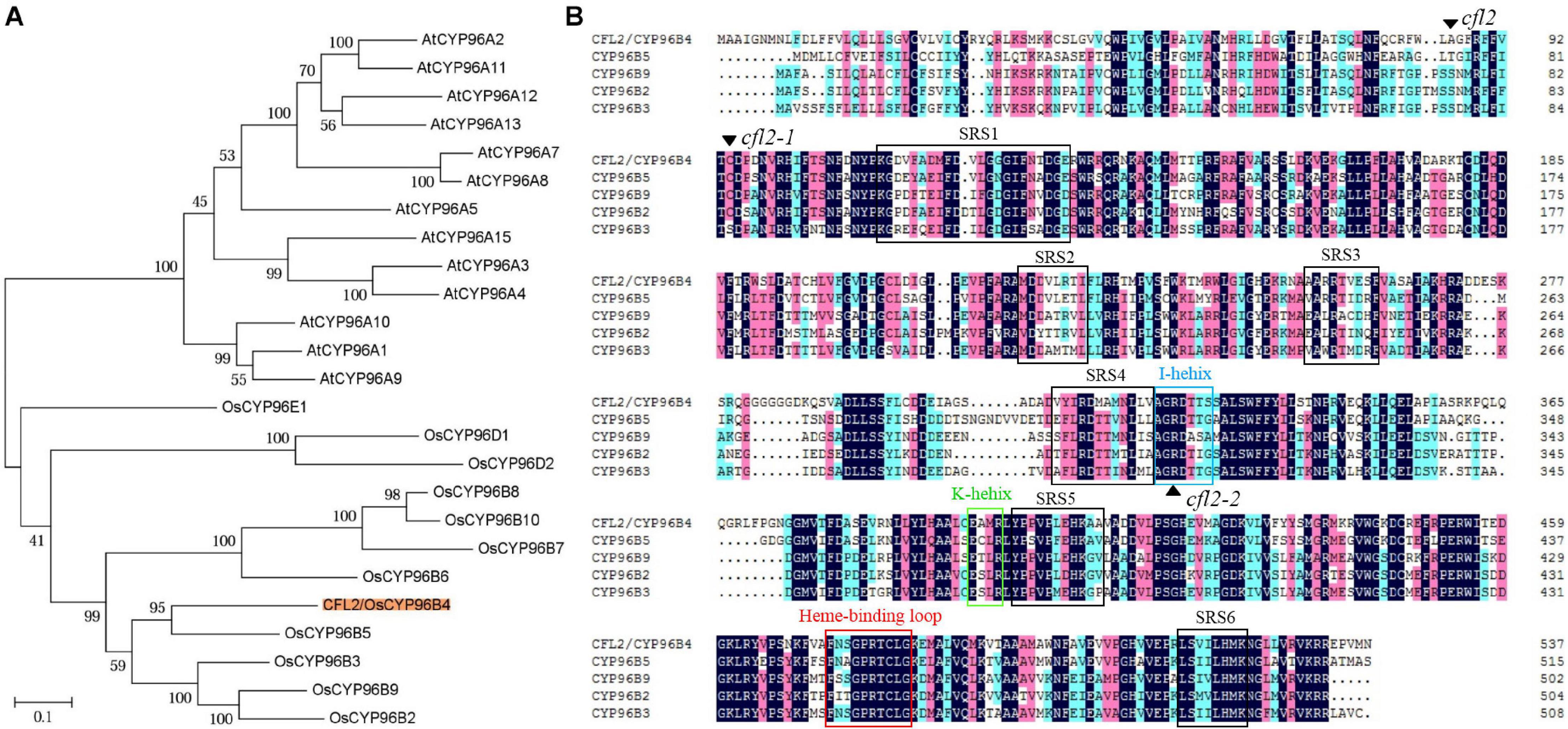
Figure 5. Protein bioinformatics analysis of CFL2. (A) Phylogenetic tree of CYP96 subfamily in rice and Arabidopsis. (B) Multiple sequence alignment of the CYP96 subfamily gene cluster on chromosome 3. The black arrowhead indicates the amino acid residue mutated in mutants. The open box with different color represent the conserved structures domains of CYP proteins, including SRS (substrate-recognition sites), I-helix (I-helix groove), K-helix (Glu-x-x-Arg domain), and a heme binding loop.
Expression Pattern of CFL2
To determine the subcellular localization of CFL2, the open reading frame of CFL2 was fused to GFP and driven by the CaMV35S promoter, and the plasmid was transformed into rice protoplast. The CFL2 fusion protein was co-localized with an endoplasmic reticulum (ER) marker, which suggests that CFL2 is located on the ER membrane (Figure 6A).
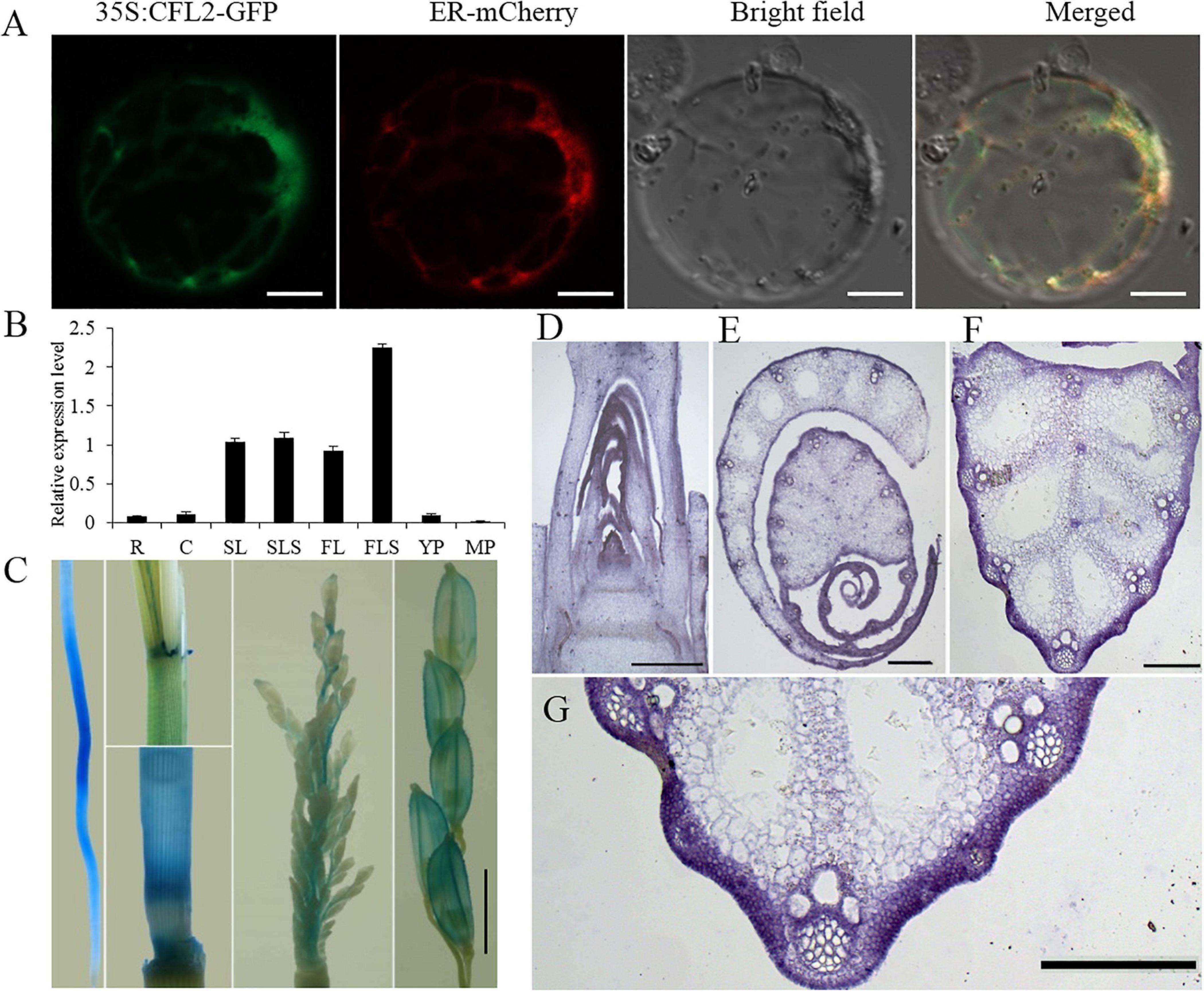
Figure 6. Protein subcellular localization and expression pattern of CFL2. (A) Subcellular localization of CFL2–GFP fusion protein in rice protoplasts. Scale bars = 10 μm. (B) qRT-PCR analysis of CFL2 in various organs, including the root (R), culm (C), second leaf (SL), second leaf sheath (SLS), flag leaf (FL), flag leaf sheath (FLS), young panicle (YP), and mature panicle (MP). Values are means ± SD (n = 3). (C) GUS stains in root, leaf sheath, culm, young panicle, and mature panicle. Scale bar = 2 mm. (D–H) In situ hybridization analysis of CFL2 expression in SAM (D), leaf (E), leaf vein (F), its partial enlarged drawing (G), and the control tissue using a sense probe (H). Scale bars = 1 cm (D–G) and 2 cm (H).
To investigate the expression pattern of CFL2, different tissues were analyzed by qRT-PCR. CFL2 was expressed in diverse tissues, including the root, culm, leaf and leaf sheath, youth panicle, and mature panicle. The highest level of expression was observed in the leaf blade and sheath, especially in flag leaf sheath (Figure 6B). GUS activity was ubiquitously in the root, culm sheath, and panicle of transgenic lines, which is consistent with the qRT-PCR results (Figure 6C). In situ RNA hybridization revealed that the CFL2 signal was detected in the outmost cells of SAM, leaf, and leaf vein (Figures 6D–H), which shows that CFL2 is an outermost cell layer-specific gene.
CFL2 May Be the Target Gene of Roc5
Given the defective leaf epidermis of the cfl2 mutant, and that CFL2 was specifically expressed in the outermost cell layer, we considered that CFL2 may be involved in the epidermis development. CFL1 encodes a WW-domain protein and negatively regulates epidermal cuticle development in rice. The homologous AtCFL1 in Arabidopsis can interact with HDG1, a transcription factor belonging to the HD-ZIP IV family, which is closely associated with the epidermis (Wu et al., 2011). Nine genes in the HD-ZIP IV family have been identified in rice (Ito et al., 2003), which form three distinct clades in phylogenetic trees, and the Roc4, Roc5, and Roc6 belong to the same clade with ATHDG1 (Figure 7A), which indicates that the three genes may show similar functions to ATHDG1 in the epidermis. The expression levels of Roc5 and CFL2 were significantly decreased in the clf1, an allelic mutant of Roc5 (Xing et al., 2017), compared with those of “Nipponbare” (Figure 7B). Gene sequence analysis revealed that a conservative L1 box-binding motif TAAATGTT (CFL2-P-L1) is present in the promoter region of CFL2 (Figure 7C). The Y1H assay result showed that Roc5 can bind to the CFL2-P-L1 box but cannot bind to the CFL2-P-L1 mutant box when selected with 200 ng/ml AbA in Y1H strains (Figure 7D). In addition, a dual-luciferase assay with rice protoplasts confirms that Roc5 can activate LUC transcription through binding to the CFL2 promoter (Figures 7E,F). These results suggested that Roc5 binds to the L1 box in the promoter of CFL2 and regulates its expression.
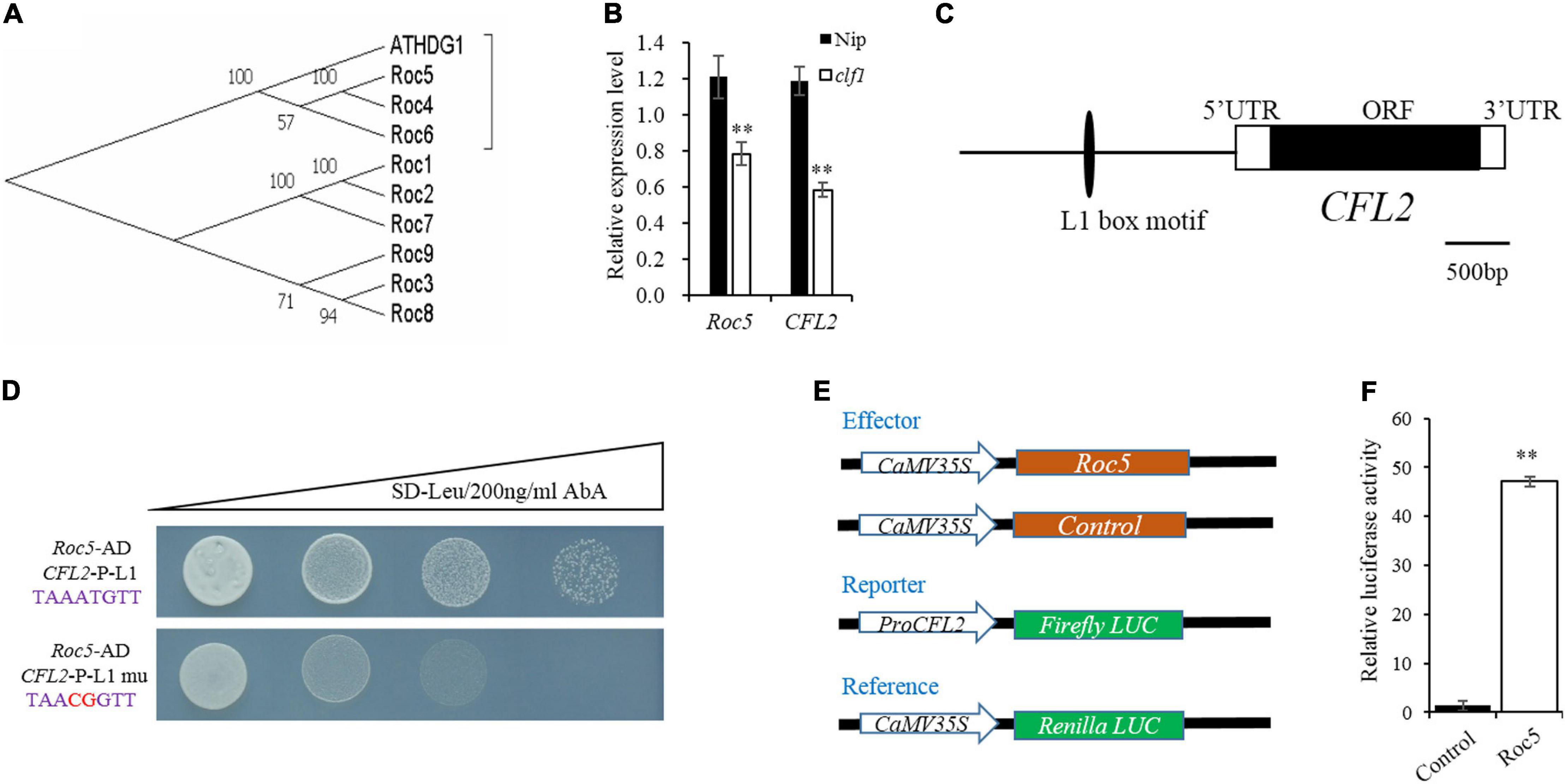
Figure 7. CFL2 may be a target gene of Roc5. (A) Phylogenetic tree of ATHDG1 and the nine numbers of HD-ZIP IV family gene in rice. (B) Expression of Roc5 and CFL2 in Nipponbare (Nip) and the clf1 mutant. Values are means ± SD (n = 3). Student’s t-test was used for statistical analysis (∗∗P < 0.01). (C) Distribution of the L1 box motif in the promoter regions of CFL2. (D) Yeast one-hybrid assay. (E) Schematic diagram show the constructs used in dual-luciferase assay. The empty pGreenII 62-SK vector was used as a control. (F) Roc5 activates the expression of CFL2 in rice protoplasts. Values are means ± SD (n = 3). Student’s t-test was used for statistical analysis (∗∗P < 0.01).
Identification and Functional Classification of DEGs Between WT and cfl2
To further explore the function of CFL2, the transcriptome sequencing of the flag leaf base from the cfl2 mutant and WT was performed. A significant positive correlation among three biological replicates (Pearson’s correlation > 0.867) was observed. After filtering, a total of 132,731,874 paired-end reads were obtained, and each biological replicate was uniquely mapped to the rice reference genome (Supplementary Table S3). A total of 1,033 DEGs between the cfl2 mutant and WT were identified, of which 789 genes were upregulated and 244 genes were downregulated. The results of GO enrichment analysis showed that genes were significantly over-represented in carbohydrate metabolic process, polysaccharide metabolic process, lipid transport, and cellulose biosynthetic and cell wall-related processes (Figure 8A). The cellulose synthase and cellulose synthase-like gene superfamily (CESA/CSL) is considered to be involved in cellulose and non-cellulosic matrix polysaccharide synthesis, which is an important component of plant cell walls (Wang et al., 2010). Among members of the superfamily, OsCESA1, OsCESA3, and OsCESA8 are required for cellulose synthesis in the primary cell walls, and CSLH1 and CSLF6 play important roles in the polysaccharide synthesis (Vega-Sanchez et al., 2012). Additionally, cell wall-related genes, such as expansins (OsEXP1), expansin precursor (LOC_Os03g06000, LOC_Os01g60770, and LOC_Os05g39990), and chitinase family genes (CHIT2, CHIT3, and CHIT8), were also upregulated in the cfl2 mutant (Figure 8B). The expression level of several cell wall-related genes was verified by qRT-PCR, which showed a similar expression pattern in cfl2 with the transcriptome data (Figure 8C). Meanwhile, we also found that the expression of cell wall-related genes showed the same trend in clf1 as in cfl2, such as BC1, CESA1, and CESA8 were increased significantly in clf1, when compared to “Nipponbare” (Supplementary Figure S4B). We further analyzed the cell wall composition of WT and cfl2 flag leaf. The main cell wall component, lignin, and cellulose were increased significantly, and the monosaccharide, such as mannose, glucose, and xylose were increased at an extremely significant level in cfl2 (Figure 8D and Supplementary Table S6). These results indicate the importance of CFL2 in the cell wall processes.
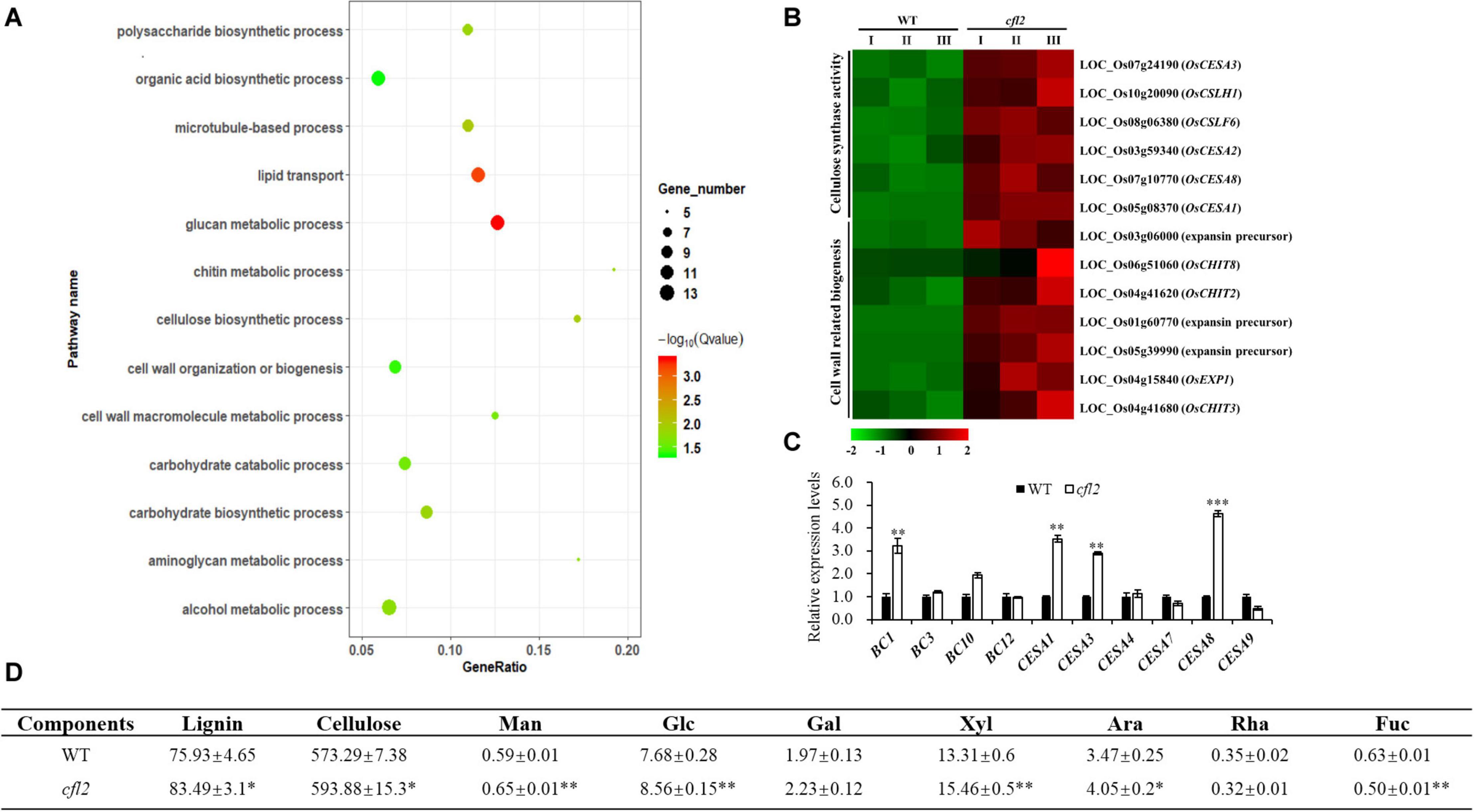
Figure 8. The analysis of cell wall-related process in cfl2. (A) GO enrichment of 789 upregulated genes in cfl2 transcriptome dates. The enriched pathways are listed on the left. (B) Heatmap showing the expression patterns of selected genes in the process of cellulose synthase and cell wall-related process. Data are from three biological replicates (I, II, and III). (C) Relative expression level of cell wall-related genes in WT and cfl2. Values are means ± SD (n = 3). Student’s t-test was used for statistical analysis (∗∗P < 0.01; ∗∗∗P < 0.001). (D) Compositional analysis of cell wall components among WT and cfl2. Each component was calculated as milligrams per gram. Man, mannose; Glc, glucose; Gal, galactose; Xyl, xylose; Ara, arabinose; Rha, rhamnose; Fuc, fucose. The results were mean ± SD of five or three independent experiments. Student’s t-test was used for statistical analysis (∗P < 0.05; ∗∗P < 0.01).
Discussion
Curled Flag Leaf Phenotype in the cfl2 Mutant
Leaf rolling is a common mutant phenotype in rice. Normal curling of the leaf blade in the mature stage is beneficial for the spatial structure of the population and light utilization. To date, at least 31 rolled-leaf mutants have been characterized (Zhang et al., 2015), which showed the effect on all the leaves during the whole growth period and accompanied by multi-phenotypes, such as the reduced plant height (Hibara et al., 2009; Hu et al., 2010; Xu et al., 2014), the defective reproductive development (Khew et al., 2015; Huang et al., 2016; Zhao et al., 2016), and the abnormal root growth (Woo et al., 2007; Zhang et al., 2009). Few mutants in which only the flag leaves were affected have been reported. A rolled-leaf mutant was designated cfl1 because of the curly flag leaf phenotype, but all leaves of the mutant were affected after tillering stage (Wu et al., 2011). The flag leaf mutant sfl1 exhibits the screw flag leaf and panicle-at-bottom phenotypes, accompanied by additional changes in agronomic characters, such as the reduced plant height, flag leaf length and width, panicle length, and grain width (Alamin et al., 2017). The cfl2 mutant described in the current study was different from all other rolled-leaf mutants. No significant difference in other agronomic traits was observed in the cfl2 mutant (Supplementary Table S1). Leaves of the cfl2 mutant were normal during the entire growth period, except for the curled flag leaf base, and the distal portion of the flag leaf blade was normally expanded (Figures 1E,F). However, in addition to the specifically curled flag leaf, the dwarfism phenotypes were also observed in the two allelic mutants cfl2-1 and cfl2-2 (Supplementary Figure S2). The cfl2-1 mutation site located closed to the SRS1 domain, and cfl2-2 mutation site located in the I-helix domain, both of them showed to be more conserved than that of cfl2 (Figure 5B), which may explain the different phenotypes in plant height of the three allelic mutants. The unique phenotype of the curled flag leaf in cfl2, without defects in plant height or other agronomic traits, provides useful material for the study of the development mechanism of flag leaf.
Function of CFL2 in Leaf Development
As the outermost cell layer covering the plant body, the epidermis plays an important role as a protective barrier against biotic or abiotic agents and is an active interface that controls the vital exchange of gas, water, and nutrients with the environment (Javelle et al., 2011). To accommodate its multiple roles, the epidermis has developed a suite of characteristics, including cell types in L1, pavement cells, stomatal guard cells, trichomes, root hairs, papillate cells, and gland cells (Glover, 2000; Martin and Glover, 2007; Javelle et al., 2011). Some epidermis-defective mutants show abnormal leaf morphology, such as the cfl1 (Wu et al., 2011) and cld1 (Li et al., 2017) in rice, the cr4 in maize (Becraft et al., 1996), and the crinkly4 in Arabidopsis (Watanabe et al., 2004). These studies demonstrate that, as a dermal tissue system, the structural integrity of the epidermis is essential to maintain the normal development of leaves. The present SEM observations revealed an increased number and disordered distribution of papillae and an obvious crack on the epidermal of bulliform cell pairs and the linear cork–silica cell pairs in the flag leaf of the cfl2 mutant (Figures 3A–D,I,J). Other epidermal structures, such as the stomatal and crystal structure of cuticular wax, showed normal development compared with those of the WT (Figures 3E–H,K,L), which indicates that CFL2 affected the epidermal development only to a particular extent.
As the largest supergene families in plants, cytochrome P450 monooxygenases (CYP450s) play critical roles in the synthesis of lignin, ultraviolet protectants, pigments, defense compounds, fatty acids, hormones, and signaling molecules (Schuler and Werck-Reichhart, 2003). Phylogenetic analysis showed that CFL2 encoded a cytochrome P450 protein (OsCYP96B4) (Figure 5A), which belongs to the youngest family in CYP86 clan (Nelson et al., 2004). This family is an invention of angiosperms (Nelson and Werck-Reichhart, 2011), implying that the CYP96 family plays important roles in epidermal development. LCR, encoding cytochrome P450 CYP86A8, could be implicated in epidermis development and in preventing postgenital organ fusions (Wellesen et al., 2001); CYP96A15 involved in surface protection by participating in the synthesis of stem epidermis wax in Arabidopsis (Greer et al., 2007); CYP96B5 is involved in the formation of epidermal wax crystals affecting drought sensitivity in rice leaf (Zhang et al., 2020). Several mutants of OsCYP96B4 have been reported, which point the multi-functionality of CFL2/OsCYP96B4, in terms of an association with cell elongation and pollen germination (Rengasamy et al., 2011), a role in the fine-tuning of plant height (Zhang et al., 2014), mediation of growth and stress responses by fine-tuning the gibberellin-to-abscisic acid balance (Tamiru et al., 2015), a function in secondary cell wall formation (Wang et al., 2016), and an influence on a variety of metabolic pathways (Jiang et al., 2020). The positive transgenic plants with overexpression of CFL2 displayed developmental disorders including dwarf and growth retardation and were lethal ultimately (Supplementary Figure S3), which was consistent with previous results (Rengasamy et al., 2011). The dwarfism phenotypes of the two allelic mutants cfl2-1 and cfl2-2 also indicated that the CFL2/OsCYP96B4 play an important role in the plant height development (Supplementary Figures S2C,D). In contrast to the dwarfing or brittle sheath phenotypes reported in these allelic mutants of OsCYP96B4, the cfl2 mutant exhibits the specific curled phenotype at the base of the flag leaves without a significant change in plant height and other main agronomic traits (Figure 1, Supplementary Figure S1 and Supplementary Table S1). Besides, the allelic mutant cfl2-1 and cfl2-2 also showed the curly flag leaves accompanied by normal morphology of other leaves (Supplementary Figures S2E–H). Results of the GO enrichment analysis of the DEGs and the qRT-PCR showed that genes associated with cellulose biosynthesis and cell wall-related processes were significantly upregulated in the cfl2 mutant (Figures 8A–C), and the components of cell wall were altered dramatically in the cfl2 mutant compared with WT (Figure 8D), which suggest the importance of CFL2 in the cell wall-related processes in the flag leaf and further confirm the versatility of CFL2/OsCYP96B4. In addition, a previous report indicated that BSHT1/OsCYP96B4 was involved in the expression of the cell wall biosynthesis-related genes in rice plant, especially in sheath (Wang et al., 2016). Consistent with this research, the flag leaf sheath also showed the abnormal twist in the cfl2 mutant. The highest expression of CFL2 in the flag leaf sheath may also affect the cell wall-related processes in flag leaf sheath (Figure 6B), which is the possible reason for this phenotype.
The Relationship Between Roc5 and CFL2
In higher plants, L1 is the outermost cell layer of the SAM and differentiates into all above-ground organs, such as the leaves, stems, and flowers, which are essential for the normal development of the entire shoot (Javelle et al., 2011; Fang et al., 2015). Some genes specially expressed in L1 have been identified, such as ATML1, PDF2, ALE2, and ACR4 in Arabidopsis (Lu et al., 1996; Abe et al., 1999; Watanabe et al., 2004; Tanaka et al., 2007); ZmOCL1, ZmOCL3, ZmOCL4, and ZmOCL5 in maize (Ingram et al., 1999, 2000); and ONI1, ONI2, and ONI3 in rice (Ito et al., 2011; Tsuda et al., 2013; Akiba et al., 2014). The transcription factors of the HD-ZIP IV family are also associated with epidermis development in rice. Nine genes (Roc1–Roc9) in the HD-ZIP IV family have been identified in rice, of which five are specifically expressed in the epidermis (Ito et al., 2003). Interestingly, mutant of Roc5 showed similar phenotypes (no significant effect on plant height and enlarged bulliform cells) to the cfl2 mutant (Zou et al., 2011). The expression levels of Roc5 and CFL2 were significantly decreased in the clf1, an allelic mutant of Roc5, compared with those of “Nipponbare” (Figure 7B). The expression level of CFL2 was decreased, but the expression of Roc5 showed no significant difference in the cfl2 mutant when compared to the WT (Supplementary Figure S4A). Besides, both Roc5 and CFL2 showed specific expression in the L1 cell layer of SAM, leaf, and leaf vein (Figures 6D–G), and Roc5 could bind to the L1 box in the promoter of CFL2 (Figures 7C–F). These results indicated that transcription factor Roc5 directly regulated the expression of CFL2. The leaves of the oul1 mutant gradually curved in seedling stage and the curling phenotype became more evident during the growth period (Zou et al., 2011), whereas only the base of flag leaves was affected in the reproductive stage in the cfl2 mutant (Figures 1E,F). This apparent difference may reflect that, in addition to the regulation of Roc5, CFL2 is also affected by other proteins with currently unknown functions during leaf development, which will be our focus in future studies.
In summary, in contrast to other rolled-leaf mutants, cfl2 showed specific curling at the base of the flag leaf, whereas the distal portion of the flag leaf blade was normal and the other leaves of the cfl2 mutant maintained normal development throughout the growth period. Microscopic observations revealed that abnormal epidermis development caused the mutated curled leaf, as a result of enlarged bulliform cells and increased number of papillae with a disordered distribution in the cfl2 mutant. Biochemical experiments and transcriptome analysis indicate that CFL2 was controlled by Roc5, which bound to the L1 box in the promoter of CFL2, and affected epidermis development by influencing the expression of genes associated with cell wall-related processes, thereby regulating leaf morphology (Supplementary Figure S5).
Data Availability Statement
RNA sequencing data were deposited in the NCBI Sequence Read Archive (SRA) under accession PRJNA628038.
Author Contributions
XS, GH, and XZha planned and designed the research. YW, XZhu, and XW analyzed the data. XZha, YW, ZZ, YX, and JX performed the experiments. XZha, YW, ZZ, ZY, and XS conducted the fieldwork. XZha, YW, XZhu, XW, XS, and GH wrote, reviewed, and edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the grants from the Ministry of Science and Technology of China (Nos. 2016YFD0100501 and 2017YFD0100201). Fundamental Research Funds for the Central Universities (Grants No. XDJK2019C042) and China Postdoctoral Science Foundation (Grants No. 2018M640893).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Prof. Tiegang Lu and Zhiguo Zhang at the Chinese Academy of Agricultural Sciences for providing the seed of the clf1 mutant. We also thank Ph.D. student Yixin Cui at the College of Agronomy and Biotechnology of SWU for the transcriptome analyses.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.616977/full#supplementary-material
Supplementary Figure 1 | Internode comparison of WT and cfl2. (A,B) Panicle and internode length of WT and cfl2. Scale bars = 8 cm. (C) Statistical data of internodes in WT and cfl2. Mean of five independent experiments. I–VI: the 1st–6th internodes.
Supplementary Figure 2 | Phenotype of WT and the allelic mutants of CFL2. (A–D) Phenotype of WT, cfl2, and its two allelic mutants cfl2-1 and cfl2-2 in tillering stage. Scale bars = 20 cm. (E–H) The top three leaves of WT, cfl2, cfl2-1, and cfl2-2. Scale bars = 3 cm.
Supplementary Figure 3 | Phenotype of the CFL2 over-expressed transgenic lines. (A,B) Phenotype analysis of the five over-expressed CFL2 transgenic lines at the seeding stage (A) and the mature stage (B). Scale bar = 10 cm (A) and 20 cm (B). (C) The expression of CFL2 in the five over-expressed transgenic lines (OE). Values are means ± SD (n = 3). Student’s t-test was used for statistical analysis (∗∗∗P < 0.001).
Supplementary Figure 4 | Relative gene expression level analysis in wild type and mutants. (A) Relative expression level of Roc5 and CFL2 in WT and cfl2. (B) Relative expression level of cell wall-related genes in Nipponbare (Nip) and clf1. Values are means ± SD (n = 3). Student’s t-test was used for statistical analysis (∗∗P < 0.01).
Supplementary Figure 5 | A model of CFL2, controlled by Roc5, influences flag leaf development in rice by affecting cell wall-related process.
Supplementary Table 1 | Agronomic traits of WT and cfl2. Mean ± SD of seven biological repeats. Student’s t-test was used for statistical analysis.
Supplementary Table 2 | The list of annotated genes within the mapping interval.
Supplementary Table 3 | Summary of RNA sequencing data.
Supplementary Table 4 | Primers used in this study.
Supplementary Table 5 | Statistical results of bulliform cell number and areas.
Supplementary Table 6 | Original data of the cell wall components analysis.
Footnotes
- ^ http://rgp.dna.affrc.go.jp
- ^ http://www.ncbi.nlm.nih.gov
- ^ http://www.arabidopsis.org
- ^ https://phytozome.jgi.doe.gov
- ^ http://bioinfo.cau.edu.cn/agriGO/
- ^ http://www.gramene.org/
References
Abe, M., Takahashi, T., and Komeda, Y. (1999). Cloning and characterization of an L1 layer-specific gene in Arabidopsis thaliana. Plant Cell Physiol. 40, 571–580. doi: 10.1093/oxfordjournals.pcp.a029579
Akiba, T., Hibara, K., Kimura, F., Tsuda, K., Shibata, K., Ishibashi, M., et al. (2014). Organ fusion and defective shoot development in oni3 mutants of rice. Plant Cell Physiol. 55, 42–51. doi: 10.1093/pcp/pct154
Alamin, M., Zeng, D. D., Qin, R., Sultana, M. H., and Shi, C. H. (2017). Characterization and fine mapping of SFL1, a gene controlling screw flag leaf in rice. Plant Mol. Biol. Rep. 35, 491–503. doi: 10.1007/s11105-017-1039-x
Becraft, P. W., Stinard, P. S., and McCarty, D. R. (1996). CRINKLY4: a TNFR-Like receptor kinase involved in maize epidermal differentiation. Science 273, 1406–1409. doi: 10.1016/S0168-9525(97)90094-9
Chen, Q. L., Xie, Q. J., Gao, J., Wang, W. Y., Sun, B., Liu, B. H., et al. (2015). Characterization of rolled and erect leaf 1 in regulating leave morphology in rice. J. Exp. Bot. 66, 6047–6058. doi: 10.1093/jxb/erv319
Dai, M. Q., Zhao, Y., Ma, Q. F., Hu, Y., Hedden, P. F., Zhang, Q., et al. (2007). The rice YABBY1 gene is involved in the feedback regulation of gibberellin metabolism. Plant Physiol. 144, 121–133. doi: 10.1104/pp.107.096586
Fang, L. K., Zhao, F. M., Cong, Y. F., Sang, X. C., Du, Q., Wang, D. Z., et al. (2012). Rolling-leaf 14 is a 2OG-Fe (II) oxygenase family protein that modulates rice leaf rolling by affecting secondary cell wall formation in leaves. Plant Biotechnol. J. 10, 524–532. doi: 10.1111/j.1467-7652.2012.00679.x
Fang, Y. X., Hu, J., Xu, J., Yu, H. P., Shi, Z. Y., Xiong, G. S., et al. (2015). Identification and characterization of Mini1, a gene regulating rice shoot development. J. Integr. Plant Biol. 57, 151–161. doi: 10.1111/jipb.12230
Florea, L., Song, L., and Salzberg, S. L. (2013). Thousands of exon skipping events differentiate among splicing patterns in sixteen human tissues. F1000Research 2:188. doi: 10.12688/f1000research.2-188.v1
Fujino, K., Matsuda, Y., Ozawa, K., Nishimura, T., Koshiba, T., Fraaije, M. W., et al. (2008). NARROW LEAF 7 controls leaf shape mediated by auxin in rice. Mol. Genet. Genomics 279, 499–507. doi: 10.1007/s00438-008-0328-3
Glover, B. J. (2000). Differentiation in plant epidermal cells. J. Exp. Bot. 51, 497–505. doi: 10.1093/jexbot/51.344.497
Greer, S., Wen, M., Bird, D., Wu, X. M., Samuels, L., Kunst, L., et al. (2007). The cytochrome p450 enzyme CYP96A15 is the midchain alkane hydroxylase responsible for formation of secondary alcohols and ketones in stem cuticular wax of Arabidopsis. Plant Physiol. 145, 653–667. doi: 10.1104/pp.107.107300
Hibara, K. I., Obara, M., Hayashida, E., Abe, M., Ishimaru, T., Satoh, H., et al. (2009). The ADAXIALIZED LEAF1 gene functions in leaf and embryonic pattern formation in rice. Dev. Biol. 334, 345–354. doi: 10.1016/j.ydbio.2009.07.042
Hu, J., Zhu, L., Zeng, D. L., Gao, Z. Y., Guo, L. B., Fang, Y. X., et al. (2010). Identification and characterization of NARROW AND ROLLED LEAF 1, a novel gene regulating leaf morphology and plant architecture in rice. Plant Mol. Biol. 73, 283–292. doi: 10.1007/s11103-010-9614-7
Huang, J., Li, Z. Y., and Zhao, D. Z. (2016). Deregulation of the OsmiR160 target gene OsARF18 causes growth and developmental defects with an alteration of auxin signaling in rice. Sci. Rep. U.K. 6:29938. doi: 10.1038/srep29938
Ingram, G. C., Boisnard-Lorig, C., Dumas, C., and Rogowsky, P. M. (2000). Expression patterns of genes encoding HD-ZIP IV homeo domain proteins define specific domains in maize embryos and meristems. Plant J. 22, 401–414. doi: 10.1046/j.1365-313X.2000.00755.x
Ingram, G. C., Magnard, J.-L., Vergne, P., Dumas, C., and Rogowsky, P. M. (1999). ZmOCL1, an HDGL2 family homeobox gene, is expressed in the outer cell layer throughout maize development. Plant Mol. Biol. 40, 343–354. doi: 10.1023/A:1006271332400
Ito, M., Sentoku, N., Nishimura, A., Hong, S.-K., Sato, Y., and Matsueka, M. (2003). Roles of rice GL2-type homeobox genesin epidermis differentiation. Breed. Sci. 53, 245–253. doi: 10.1270/jsbbs.53.245
Ito, Y., Kimura, F., Hirakata, K., Tsuda, K., Takasugi, T., Eiguchi, M., et al. (2011). Fatty acid elongase is required for shoot development in rice. Plant J. 66, 680–688. doi: 10.1111/j.1365-313X.2011.04530.x
Itoh, J.-I., Hibara, K.-I., Sato, Y., and Nagato, Y. (2008). Developmental role and auxin responsiveness of class III homeodomain leucine zipper gene family members in rice. Plant Physiol. 147, 1960–1975. doi: 10.1104/pp.108.118679
Javelle, M., Vernoud, V., Rogowsky, P. M., and Ingram, G. C. (2011). Epidermis: the formation and functions of a fundamental plant tissue. New Phytol. 189, 17–39. doi: 10.1111/j.1469-8137.2010.03514.x
Jiang, L. M., Ramamoorthy, R., Ramachandran, S., and Kumar, P. P. (2020). Systems metabolic alteration in a semi-dwarf rice mutant induced by OsCYP96B4 gene mutation. Int. J. Mol. Sci. 21:1924. doi: 10.3390/ijms21061924
Juarez, M. T., Twigg, R. W., and Timmermans, M. C. (2004). Specification of adaxial cell fate during maize leaf development. Development 131, 4533–4544. doi: 10.1242/dev.01328
Khew, C. Y., Teo, C. J., Chan, W. S., Wong, H. L., Namasivayam, P., and Ho, C. L. (2015). Brassinosteroid insensitive 1-associated kinase 1 (OsI-BAK1) is associated with grain filling and leaf development in rice. J. Plant Physiol. 182, 23–32. doi: 10.1016/j.jplph.2015.05.003
Kim, D., Langmead, B., and Salzberg, S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi: 10.1038/nmeth.3317
Li, C., Zou, X. H., Zhang, C. Y., Shao, Q. H., Liu, J., Liu, B., et al. (2016). OsLBD3-7 overexpression induced adaxially rolled leaves in rice. PLoS One 11:e0156413. doi: 10.1371/journal.pone.0156413
Li, L., Shi, Z. Y., Li, L., Shen, G. Z., Wang, X. Q., An, L. S., et al. (2010). Overexpression of ACL1 (abaxially curled leaf 1) increased bulliform cells and induced abaxial curling of leaf blades in rice. Mol. Plant. 3, 807–817. doi: 10.1093/mp/ssq022
Li, M., Xiong, G. Y., Li, R., Cui, J. J., Tang, D., Zhang, B. C., et al. (2009). Rice cellulose synthase-like D4 is essential for normal cell-wall biosynthesis and plant growth. Plant J. 60, 1055–1069. doi: 10.1111/j.1365-313X.2009.04022.x
Li, W. Q., Zhang, M. J., Gan, P. F., Qiao, L., Yang, S. Q., Miao, H., et al. (2017). CLD1/SRL1 modulates leaf rolling by affecting cell wall formation, epidermis integrity and water homeostasis in rice. Plant J. 92, 904–923. doi: 10.1111/tpj.13728
Liu, X. F., Li, M., Liu, K., Tang, D., Sun, M. F., Li, Y. F., et al. (2016). Semi-Rolled Leaf 2 modulates rice leaf rolling by regulating abaxial side cell differentiation. J. Exp. Bot. 67, 2139–2150. doi: 10.1093/jxb/erw029
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8
Lu, P., Porat, R., Nadeau, J. A., and O’Neill, S. D. (1996). Identification of a meristem L1 layer-specific gene in Arabidopsis that is expressed during embryonic pattern formation and defines a new class of homeobox genes. Plant Cell 8, 2155–2168. doi: 10.1105/tpc.8.12.2155
Luan, W. J., Liu, Y. Q., Zhang, F. X., Song, Y. L., Wang, Z. Y., Peng, Y. K., et al. (2011). OsCD1 encodes a putative member of the cellulose synthase-like D sub-family and is essential for rice plant architecture and growth. Plant Biotechnol. J. 9, 513–524. doi: 10.1111/j.1467-7652.2010.00570.x
Ma, L., Sang, X. C., Zhang, T., Yu, Z. Y., Li, Y. F., Zhao, F. M., et al. (2017). ABNORMAL VASCULAR BUNDLES regulates cell proliferation and procambium cell establishment during aerial organ development in rice. New Phytol. 213, 275–286. doi: 10.1111/nph.14142
Martin, C., and Glover, B. J. (2007). Functional aspects of cell patterning in aerial epidermis. Curr. Opin. Plant Biol. 10, 70–82. doi: 10.1016/j.pbi.2006.11.004
McConnell, J. R., Emery, J., Eshed, Y., Bao, N., Bowman, J., and Barton, M. K. (2001). Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 411, 709–713. doi: 10.1038/35079635
Nagasaki, H., Ltoh, J.-I., Hayashi, K., Hibara, K. I, Satoh-Nagasawa, N., Nosaka, M., et al. (2007). The small interfering RNA production pathway is required for shoot meristem initiation in rice. Proc. Natl. Acad. Sci. U.S.A. 104, 14867–14871. doi: 10.1073/pnas.0704339104
Nelson, D., and Werck-Reichhart, D. (2011). A P450-centric view of plant evolution. Plant J. 66, 194–211. doi: 10.1111/j.1365-313x.2011.04529.x
Nelson, D. R., Schuler, M. A., Paquette, S. M., Werck-Reichhart, D., and Bak, S. (2004). Comparative genomics of rice and Arabidopsis. Analysis of 727 cytochrome P450 genes and pseudogenes from a monocot and a dicot. Plant Physiol. 135, 756–772. doi: 10.1104/pp.104.039826
Otsuga, D., Deguzman, B., Prigge, M. J., Drews, G. N., and Clark, S. E. (2001). REVOLUTA regulates meristem initiation at lateral positions. Plant J. 25, 223–236. doi: 10.1046/j.1365-313x.2001.00959.x
Rengasamy, R., Jiang, S. Y., Srinivasan, R., and Juergen, K. (2011). Oryza sativa cytochrome P450 family member OsCYP96B4 reduces plant height in a transcript dosage dependent manner. PLoS One 6:e28069. doi: 10.1371/journal.pone.0028069
Richards, R. A., Rebetzke, G. J., Condon, A. G., and van Herwaarden, A. F. (2002). Breeding opportunities for increasing the efficiency of water use and crop yield in temperate cereals. Crop Sci. 42, 111–121. doi: 10.2135/cropsci2002.1110
Sang, X. C., Li, Y. F., Luo, Z. K., Ren, D. Y., Fang, L. K., Wang, N., et al. (2012). CHIMERIC FLORAL ORGANS1, encoding a monocot-specific MADS box protein, regulates floral organ identity in rice. Plant Physiol. 160, 788–807. doi: 10.1104/pp.112.200980
Schuler, M. A., and Werck-Reichhart, D. (2003). Functional genomics of P450s. Annu. Rev. Plant Biol. 54, 629–667. doi: 10.1146/annurev.arplant.54.031902.134840
Shi, Z. Y., Wang, J., Wan, X. S., Shen, G. Z., Wang, X. Q., and Zhang, J. L. (2007). Over-expression of rice OsAGO7 gene induces upward curling of the leaf blade that enhanced erect-leaf habit. Planta 226, 99–108. doi: 10.1007/s00425-006-0472-0
Tamiru, M., Undan, J. R., Takagi, H., Abe, A., Yoshida, K., Undan, J. Q., et al. (2015). A cytochrome P450, OsDSS1, is involved in growth and drought stress responses in rice (Oryza sativa L.). Plant Mol. Biol. 88, 85–99. doi: 10.1007/s11103-015-0310-5
Tanaka, H., Watanabe, M., Sasabe, M., Hiroe, T., Tanaka, T., Tsukaya, H., et al. (2007). Novel receptor-like kinase ALE2 controls shoot development by specifying epidermis in Arabidopsis. Development 134, 1643–1652. doi: 10.1242/dev.003533
Tsuda, K., Akiba, T., Kimura, F., Ishibashi, M., Moriya, C., Nakagawa, K., et al. (2013). ONION2 fatty acid elongase is required for shoot development in rice. Plant Cell Physiol. 54, 209–217. doi: 10.1093/pcp/pcs169
Vega-Sanchez, M. E., Verhertbruggen, Y., Christensen, U., Chen, X., Sharma, V., Varanasi, P., et al. (2012). Loss of cellulose synthase-like F6 function affects mixed-linkage glucan deposition, cell wall mechanical properties, and defense responses in vegetative tissues of rice. Plant Physiol. 159, 56–69. doi: 10.1104/pp.112.195495
Wang, H. M., Shi, Y. F., Zhang, X. B., Xu, X., and Wu, J. L. (2020). Characterization of a novel rice dynamic narrow-rolled leaf mutant with feficiencies in aromatic amino acids. Int. J. Mol. Sci. 21:1521. doi: 10.3390/ijms21041521
Wang, L. P., Guo, K., Li, Y., Tu, Y. Y., Hu, H. Z., Wang, B. R., et al. (2010). Expression profiling and integrative analysis of the CESA/CSL superfamily in rice. BMC Plant Biol. 10:282. doi: 10.1186/1471-2229-10-282
Wang, X. L., Cheng, Z. J., Zhao, Z. C., Gan, L., Qin, R. Z., Zhou, K. M., et al. (2016). BRITTLE SHEATH1 encoding OsCYP96B4 is involved in secondary cell wall formation in rice. Plant Cell Rep. 35, 745–755. doi: 10.1007/s00299-015-1916-4
Watanabe, M., Tanaka, H., Watanabe, D., Machida, C., and Machida, Y. (2004). The ACR4 receptor-like kinase is required for surface formation of epidermis-related tissues in Arabidopsis thaliana. Plant J. 39, 298–308. doi: 10.1111/j.1365-313X.2004.02132.x
Wellesen, K., Durst, F., Pinot, F., Benveniste, I., Nettesheim, K., Wisman, E., et al. (2001). Functional analysis of the LACERATA gene of Arabidopsis provides evidence for different robes of fatty acid omega-hydroxylation in development. Proc. Natl. Acad. Sci. U.S.A. 98, 9694–9699. doi: 10.1073/pnas.171285998
Woo, Y. M., Park, H. J., Su’Udi, M., Yang, J. I, Park, J. J., Back, K., et al. (2007). Constitutively wilted 1, a member of the rice YUCCA gene family, is required for maintaining water homeostasis and an appropriate root to shoot ratio. Plant Mol. Biol. 65, 125–136. doi: 10.1007/s11103-007-9203-6
Wu, R. H., Li, S. B., He, S., Wassmann, F., Yu, C. H., Qin, G. J., et al. (2011). CFL1, a WW domain protein, regulates cuticle development by modulating the function of HDG1, a class IV homeodomain transcription factor, in rice and Arabidopsis. Plant Cell 23, 3392–3411. doi: 10.1105/tpc.111.088625
Xiang, J. J., Zhang, G. H., Qian, Q., and Xue, H. W. (2012). Semi-rolled leaf 1 encodes a putative glycosylphosphatidylinositol-anchored protein and modulates rice leaf rolling by regulating the formation of bulliform cells. Plant Physiol. 159, 1488–1500. doi: 10.1104/pp.112.199968
Xing, B., Chen, Z. H., and Zhang, Z. G. (2017). Map-based cloning of a curly leaf gene CLF1 in rice. Biotechnol. Bull. 33, 110–115. doi: 10.13560/j.cnki.biotech.bull.1985.2017-0694
Xu, J., Wang, L., Qian, Q., and Zhang, G. H. (2013). Research advance in molecule regulation mechanism of leaf morphogenesis in rice (Oryza sativa L.). Acta Agron. Sin. 39, 767–774. doi: 10.3724/SP.J.1006.2013.00767
Xu, Y., Wang, Y. H., Long, Q. Z., Huang, J. X., Wang, Y. L., Zhou, K. N., et al. (2014). Overexpression of OsZHD1, a zinc finger homeodomain class homeobox transcription factor, induces abaxially curled and drooping leaf in rice. Planta 239, 803–816. doi: 10.1007/s00425-013-2009-7
Yang, C. H., Li, D. Y., Liu, X., Ji, C. J., Hao, L. L., Zhao, X. F., et al. (2014). OsMYB103L, an R2R3-MYB transcription factor, influences leaf rolling and mechanical strength in rice (Oryza sativa L.). BMC Plant Biol. 14:158. doi: 10.1186/1471-2229-14-158
Yang, S. Q., Li, W. Q., Miao, H., Gan, P. F., Qiao, L., Chang, Y. L., et al. (2016). REL2, a gene encoding an unknown function protein which contains DUF630 and DUF632 domains controls leaf rolling in rice. Rice 9:37. doi: 10.1186/s12284-016-0105-6
Zhang, D., Yang, H. F., Wang, X. C., Qiu, Y. J., Tian, L. H., Qi, X. Q., et al. (2020). Cytochrome P450 family member CYP96B5 hydroxylates alkanes to primary alcohols and is involved in rice leaf cuticular wax synthesis. New Phytol. 225, 2094–2107. doi: 10.1111/nph.16267
Zhang, G. H., Xu, Q., Zhu, X. D., Qian, Q., and Xue, H. W. (2009). SHALLOT-LIKE1 is a KANADI transcription factor that modulates rice leaf rolling by regulating leaf abaxial cell development. Plant Cell 21, 719–735. doi: 10.1105/tpc.108.061457
Zhang, J., Liu, X. Q., Li, S. Y., Cheng, Z. K., and Li, C. Y. (2014). The rice semi-dwarf mutant sd37, caused by a mutation in CYP96B4, plays an important role in the fine-tuning of plant growth. PLoS One 9:e88068. doi: 10.1371/journal.pone.0088068
Zhang, J. J., Wu, S. Y., Jiang, L., Wang, J. L., Zhang, X., Guo, X. P., et al. (2015). A detailed analysis of the leaf rolling mutant sll2 reveals complex nature in regulation of bulliform cell development in rice (Oryza sativa L.). Plant Biol. 17, 437–448. doi: 10.1111/plb.12255
Zhang, T., Li, Y. F., Ma, L., Sang, X. C., Ling, Y. H., Wang, Y. T., et al. (2017). LATERAL FLORET 1 induced the three-florets spikelet in rice. Proc. Natl. Acad. Sci. U.S.A. 114, 9984–9989. doi: 10.1073/pnas.1700504114
Zhao, S. S., Zhao, L., Liu, F. X., Wu, Y. Z., Zhu, Z. F., Sun, C. Q., et al. (2016). NARROW AND ROLLED LEAF 2 regulates leaf shape, male fertility, and seed size in rice. J. Integr. Plant Biol. 58, 983–996. doi: 10.1111/jipb.12503
Keywords: cell wall, CFL2/OsCYP96B4, curled flag leaf, epidermis, L1 box, rice, Roc5
Citation: Zhang X, Wang Y, Zhu X, Wang X, Zhu Z, Li Y, Xie J, Xiong Y, Yang Z, He G and Sang X (2021) Curled Flag Leaf 2, Encoding a Cytochrome P450 Protein, Regulated by the Transcription Factor Roc5, Influences Flag Leaf Development in Rice. Front. Plant Sci. 11:616977. doi: 10.3389/fpls.2020.616977
Received: 13 October 2020; Accepted: 28 December 2020;
Published: 12 February 2021.
Edited by:
Cheng-Guo Duan, Chinese Academy of Sciences (CAS), ChinaReviewed by:
Hui Zhang, Shanghai Normal University, ChinaDasheng Zhang, Shanghai Chenshan Plant Science Research Center (CAS), China
Copyright © 2021 Zhang, Wang, Zhu, Wang, Zhu, Li, Xie, Xiong, Yang, He and Sang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guanghua He, aGVnaHN3dUAxNjMuY29t; Xianchun Sang, c2FuZ3hpYW5jaHVuQDE2My5jb20=
 Xiaobo Zhang
Xiaobo Zhang Xianchun Sang
Xianchun Sang