- 1Kenya Agricultural and Livestock Research Organization, Njoro, Kenya
- 2Department of Plant Pathology, University of Minnesota, Saint Paul, MN, United States
- 3Cereal Disease Laboratory, United States Department of Agriculture-Agricultural Research Service, Saint Paul, MN, United States
- 4Debre Zeit Agricultural Research Center, Ethiopian Institute of Agricultural Research, Bishoftu, Ethiopia
- 5International Maize and Wheat Improvement Center (CIMMYT), Texcoco, Mexico
- 6CIMMYT - World Agroforestry Centre (ICRAF), Nairobi, Kenya
Stem rust caused by Puccinia graminis f. sp. tritici (Pgt) remains a constraint to wheat production in East Africa. In this study, we characterized the genetics of stem rust resistance, identified QTLs, and described markers associated with stem rust resistance in the spring wheat line CI 14275. The 113 recombinant inbred lines, together with their parents, were evaluated at the seedling stage against Pgt races TTKSK, TRTTF, TPMKC, TTTTF, and RTQQC. Screening for resistance to Pgt races in the field was undertaken in Kenya, Ethiopia, and the United States in 2016, 2017, and 2018. One gene conferred seedling resistance to race TTTTF, likely Sr7a. Three QTL were identified that conferred field resistance. QTL QSr.cdl-2BS.2, that conferred resistance in Kenya and Ethiopia, was validated, and the marker Excalibur_c7963_1722 was shown to have potential to select for this QTL in marker-assisted selection. The QTL QSr.cdl-3B.2 is likely Sr12, and QSr.cdl-6A appears to be a new QTL. This is the first study to both detect and validate an adult plant stem rust resistance QTL on chromosome arm 2BS. The combination of field QTL QSr.cdl-2BS.2, QSr.cdl-3B.2, and QSr.cdl-6A has the potential to be used in wheat breeding to improve stem rust resistance of wheat varieties.
Introduction
Wheat accounts for about 20% of global human consumed calories and proteins with hexaploid wheat [Triticum aestivum (2n = 6x = 42; AABBDD)] being the most widely cultivated species (Gupta et al., 2008). The European Union, China, India, Russia, and the United States are the top five wheat producers (Food and Agriculture Organization [FAO], 2018). Production of wheat is challenged by both abiotic and biotic factors. Among biotic constraints, stem rust of wheat caused by Puccinia graminis f. sp. tritici (Pgt) is considered one of the most devastating fungal diseases with the potential to result in a 100% yield loss on susceptible varieties. The Pgt race Ug99, first isolated from Uganda in the year 1999 (Pretorius et al., 2000) and later characterized as race TTKSK (Jin et al., 2008), was the first Pgt race virulent to stem rust resistance gene Sr31 (Pretorius et al., 2000; Wanyera et al., 2006). The unique virulence combination in race TTKSK rendered more than 90% of global wheat varieties susceptible during testing in 2005 (Singh et al., 2006). Since then, multiple derivatives of TTKSK have been reported, including TTKST, virulent to Sr24 and identified in 2006 (Jin et al., 2008); TTTSK, virulent to Sr36 and identified in 2007 (Jin et al., 2009); and TTKTT, virulent to SrTmp and identified in 2014 (Newcomb et al., 2016; Patpour et al., 2016). Host resistance remains the most effective and economical approach to controlling stem rust, and this can be expressed at the seedling stage and at the adult plant stage (Priyamvada et al., 2011). Plants with major-effect resistance genes (also known as R-genes) may express intermediate to immune levels of resistance when challenged by an avirulent race or strain of the pathogen. Some of the major genes currently utilized in breeding programs to confer resistance to Pgt are Sr13, Sr22, Sr25, Sr26, Sr33, and Sr50 (Helguera et al., 2003; Mago et al., 2005, 2013, 2015; Simons et al., 2011; Periyanan et al., 2013; Singh et al., 2015). Genes with temporary designations, such as SrND643 and SrNing, are also being utilized to provide resistance against Pgt (Lopez-Vera et al., 2014; Basnet et al., 2015). The downfall of deploying major-effect genes individually is that they can be easily overcome by the pathogen, resulting in what is referred to as “boom-and-bust” cycles. The Ug99 race group clearly highlights the boom-and-bust cycle by the defeat of important R-genes, such as Sr24, Sr31, Sr36, and SrTmp that were widely deployed worldwide (Pretorius et al., 2000; Jin et al., 2008, 2009).
The adult plant resistance genes, such as Sr2 (likely pleiotropic with Yr30), Sr55 (pleiotropic with Lr67, Yr46, Pm46), Sr56, Sr57 (pleiotropic with Lr34, Yr18, Pm38), and Sr58 (pleiotropic with Lr46, Yr29, Pm39), confer slow rusting resistance at the adult plant stage with each having a minor resistance effect and, thus, possibly not providing adequate resistance when present alone under high disease pressure (Bansal et al., 2014; Herrera-Foessel et al., 2014; Singh et al., 2015). Combinations of effective major-effect race-specific resistance genes (R-genes) and/or minor-effect race non-specific resistance genes may result in durable resistance, or resistance that remains effective for a longer period of time (Parlevliet and van Ommeren, 1988; Priyamvada et al., 2011). Identification and deployment of new resistance genes into advanced wheat lines is necessary for new resistance gene combinations to be assembled.
CI 14275 is a Canadian breeding line (Q-2331-34) that was a part of the 1966 USDA-ARS International Spring Wheat Rust Nursery and that appeared effective to the Ug99 race group in Kenya when screened in 2005, 2006, and 2007 (Rouse et al., 2011). The USDA National Small Grains Collection lists the pedigree of CI 14275 as Thatcher∗6/Kenya Farmer//6∗Lee/Kenya Farmer. Thatcher has been reported to have Sr5, Sr9g, Sr12, and Sr16 genes (McIntosh et al., 1995). The effectiveness of resistance in Thatcher to stem rust at the adult plant stage is shown to involve Sr12 (Rouse et al., 2014b) and can be enhanced when combined with Sr57 (Lr34) (Kolmer et al., 2011). Kenya Farmer has Sr6, Sr7a, Sr9b, Sr10, Sr11, and Sr12 genes, whereas Lee has Sr5, Sr9g, Sr11, Sr12, and Sr16 genes (Knott, 1957; McIntosh et al., 1995). CI 14275 previously exhibited low infection types to multiple races with broad virulence at the seedling stage (Rouse et al., 2011). In the field, CI 14275 was highly resistant to a composite of races in St. Paul, Minnesota, in addition to being moderately resistant to the Ug99 race group in Kenya (Rouse et al., 2011). The objectives of this study were to characterize the genetics of stem rust resistance in the spring wheat line CI 14275, identify QTLs associated with the resistance, and develop markers linked to the identified resistance QTL.
Materials and Methods
Development of the Primary Population
One hundred thirteen recombinant inbred lines (RILs) (F7:9) were developed from a cross between LMPG-6 [a stem rust–susceptible spring wheat line (Knott, 1990)] and CI 14275 (LMPG-6/CI 14275) through single-seed descent at the USDA-ARS Cereal Disease Laboratory (CDL), Minnesota in the United States.
Development of the Validation Population
From the 113 RILs (F7:9) derived from LMPG-6/CI 14275, line #162 was selected because it expressed a consistent level of resistance to stem rust across the three environments: Ethiopia (0-20MSS), Kenya (0-15SMS), and St. Paul, Minnesota (5RMR-25RMR). Additionally, line #162 was among the early maturing lines. Line #162 was crossed with Kwale, a susceptible Kenyan cultivar, to generate 180 F3:4 families (Kwale/Line #162).
Phenotyping of the Primary Population in the Greenhouse
The LMPG-6/CI 14275 RIL population, together with the parents, were evaluated for their seedling infection types to Pgt races TTKSK (isolate 04KEN156/04; Ug99) and TRTTF (06YEM34-1) in a Biosafety level 3 (BSL-3) greenhouse and for infection types to Pgt US races TPMKC (74MN1409), TTTTF (01MN84A-1-2), and RTQQC (04MN74-1) at the USDA-ARS CDL greenhouse in Minnesota, following an inoculation method described by Rouse et al. (2011). Square plastic pots (6 cm × 6 cm × 6 cm) were filled with 100% vermiculite. Five seeds for each of four lines per pot were planted in two replicates and placed in a greenhouse with temperature and light conditions as described previously (Hundie et al., 2019). Pgt isolates were derived from single pustules, increased in isolation and stored at −80°C. When primary leaves were fully emerged, the inoculum was prepared by removing Pgt urediniospores from −80°C, heat shocking for 15 min in a 45°C water bath, rehydrating for approximately 2–4 h in a chamber maintained at 80% relative humidity by a KOH solution, and suspending in lightweight mineral oil (Soltrol 70; ConocoPhillips Inc., Houston). The plants were inoculated by spraying them with a suspension using an atomizer. After inoculation, the plants were then placed in a fume hood for 30 min to allow the oil to evaporate and, thereafter, transferred to a dew chamber at 100% relative humidity for 14–16 h. After the dew chamber incubation, plants were returned to the greenhouse bench and maintained at 18 ± 2°C with a photoperiod of 16 h.
Two weeks postinoculation, seedlings were assessed for seedling infection types (ITs) based on the 0–4 scale developed by Stakman et al. (1962). The description of infection types used to classify the reactions to Pgt are as follows: ‘0’ = no uredinia or any other sign of infection, ‘;’ = no uredinia but presence of hypersensitive necrotic flecks, ‘1’ = small uredinia surrounded by yellow chlorotic or necrotic areas, ‘2’ = small- to medium-sized uredinia in a dark green island surrounded by a chlorotic area, ‘3’ = medium-sized uredinia, surrounded by light green chlorosis, and ‘4’ = large uredinia with no or a limited amount of chlorosis. All observed infection types on the same leaf were recorded with the infection type(s) listed in order according to their prevalence. A comma (,) symbol was used to separate multiple ITs observed on the same plant with the most frequent IT recorded first. Whenever multiple infection types were observed on different plants of the same line, a forward slash (/) symbol was used to separate the infection types. A letter ‘C’ was recorded when extensive chlorosis was associated with the infection. The plus (+) and minus (−) symbols were used for the pustules that were relatively larger or smaller, respectively, than normally associated with a given IT. Plants with ITs ranging from 0 to 2 were categorized as resistant, and those ranging from 3 to 4 were categorized as susceptible. The seedling infection types assigned on a 0–4 scale were then converted to a 0–9 linear scale according to Gao et al. (2019) for use in analyses. We calculated the raw mean values of the linearized infection types for each pathogen race across the two replications for use in subsequent QTL analyses. RILs with mean linearized values of 6 or less were considered resistant, whereas those with values greater than 6 were categorized as susceptible.
Phenotyping of the Primary Population in the Field
The experimental lines were evaluated for stem rust severity in two African field locations: Njoro, Kenya, and Debre Zeit, Ethiopia, in 2016, 2017, and 2018. Evaluations in Africa took place during the off-season of approximately January through May. The lines were evaluated in St. Paul, Minnesota, in 2017 and 2018. Across the three locations, approximately 5 g seed was sown in each of two replicates planted in a randomized complete block design. In Njoro, Kenya, the field plots were two 0.7-m-long rows separated by a distance of 0.3 m. Disease spreader rows consisting of a mixture of susceptible cultivars Cacuke, Eagle 10, Robin, and six CIMMYT lines carrying Sr24 were planted to surround the entries 10–14 days before the experimental plots were planted. The disease spreader row mixture was also planted as hill plots on one side of the experimental plots to facilitate the buildup and spread of the disease. From the booting to heading growth stages (Zadok’s growth stages 37–60), freshly collected urediniospores of locally predominant Ug99 Pgt races TTKSK, TTKST, TTKTK, and TTKTT were bulked and suspended in distilled water, and approximately 1 mL of the suspension was injected into disease spreader row plants using a hypodermic syringe. Inoculations were conducted in the afternoons over 3 occasions with 7-day intervals. In Debre Zeit, Ethiopia, the experimental plots were planted as double 1-m-long rows, and disease spreader rows, including cultivars Arendeto, Digalu, Local Red, Morocco, and PBW343, were planted perpendicular to the plots and were artificially inoculated at Zadok’s growth stage 37–60 with bulked urediniospores from predominant Pgt races TTKSK, TKTTF, TRTTF, and JRCQC to initiate infection on the plots. In St. Paul, the experimental plots were single 1-m-long rows, separated by a distance of 0.3 m. The spreader rows that consisted of cultivars Baart, Morocco, and Thatcher were planted 1–2 weeks earlier than entries and sown perpendicular to the experimental plots. At the heading stage, the plots were spray-inoculated using an Ulva+ sprayer (Micron Sprayers Ltd., Bromyard, United Kingdom) with a light mineral oil suspension of bulked urediniospores of North American Pgt races QFCSC (isolate 06ND76C), TPMKC (isolate 74MN1409), RKRQC (isolate 99KS76A), RCRSC (isolate 77ND82A), QTHJC (isolate 75ND717C), and MCCFC (isolate 59KS19). Disease on the inoculated spreader rows initiated stem rust infection on the experimental plots.
When the spreaders had attained 50% severity in the three locations, stem rust severity was visually scored in the experimental plots based on the modified Cobb scale of 0–100, where 0 = immunity (no uredinia or any other sign of infection) and 100% = completely susceptible (Peterson et al., 1948). Infection response was rated as resistant (R), small uredinia surrounded by necrosis; moderately resistant (MR), medium-sized uredinia surrounded by necrosis or chlorosis; moderately susceptible (MS), medium-sized uredinia without necrosis; susceptible (S), large uredinia without necrosis; or MRMS, an infection response that included both the MR and MS categories (Roelfs et al., 1992). Coefficient of infection (COI) values were generated by multiplying the stem rust severity value for each line by a constant value for each infection response: 0 = 0, R = 0.2, RMR = 0.3, MR = 0.4, M = 0.6, MS = 0.8, S = 1.0 (Knott, 1989). Average coefficient of infection for the two replicates were determined and used for analyses. Raw mean coefficients of infection values across the two replicates were used for QTL analyses.
Phenotyping of the Validation Population in the Field
Twice-replicated plots of the 180 F3:4 validation lines were evaluated for field response to the Ug99 race group in Njoro, Kenya, in 2018. Planting; inoculation with Pgt races that included TTKSK, TTKST, TTKTK, and TTKTT; and evaluation was conducted as previously described.
DNA Extraction
Tissues were harvested from three-leaf-stage single plants of the RIL population (F7:9), from single F3 plants of the Kwale/line #162 population, and their parents into Eppendorf tubes (1.5 mL). The harvested leaves were dried and ground for 1–2 min using a Genogrinder (SPEX Sample Prep). Extraction buffer (300 μl: 200 mM Tris-HCL pH 8.0, 250 mM NaCl, 25 mM EDTA, 0.5% SDS, ddH20) was added to each well, the tubes were shaken gently before incubating at room temperature for 15 min, followed by centrifuging completed at 2500 rpm. Working under a fume hood, 300 μl of chloroform:isoamyl alcohol (24:1) was added to each well, the suspension was mixed, plates were centrifuged for 20 min at 2500 rpm, and the supernatant (200–300 μl) was transferred to separate tubes. Cold isopropanol (300 μl) was then added to each tube. Plates were shaken gently and placed in a refrigerator for 10–20 min. After decanting the supernatant, 300 μl of 70% ethanol was added to each tube, the tubes were centrifuged for 20 min at 2,500 rpm, the ethanol was poured out, and the DNA pellets were air-dried. When the pellets were completely dry, 100 μl of distilled H2O was added to each tube to resuspend the pellet and the tubes were left in a refrigerator overnight. DNA was quantified using a NanoDrop 1000 spectrophotometer (Thermo Fisher, Waltham, MA, United States), and concentrations were diluted to 50 ng/μl.
Genotyping of the Primary Population
Genotyping of the F7:9 RILs and their parents was conducted at the USDA-ARS small grains genotyping lab in Fargo, North Dakota, using the iSelect 90k SNP assay developed by Wang et al. (2014). The 90k SNP assay contains 81,587 SNPs and was developed for allohexaploid and allotetraploid wheat populations. The raw 90K SNP data together with the consensus map were uploaded onto Illumina GenomeStudio genotyping software version 2.0. The SNPs were manually called and the genotypes grouped as AA, AB, or BB. The missing genotypes were indicated as no call (NC). Following calculation of allele frequencies, a total of 12,243 polymorphic markers with both AA and BB allele frequency greater than 0.25 were selected.
QTL Mapping
The allele calls from GenomeStudio were used to create genetic linkage maps with the MSTMap algorithm (Wu et al., 2008) implemented in ASMap R package (Taylor and Butler, 2017) with the following parameters: distance function Kosambi, cut_off_p_value set at 1e-08, no_map_dist set at 15, no_map_size set at 0, missing_threshold set at 0.10, detect_bad_data set at “yes” and objective function set at “COUNT.” A total of 33 linkage groups, each with greater than five markers were generated. The 33 linkage groups included a total of 819 markers (Supplementary Table 1). The genotype calls were manually curated by coding single-marker, double-crossover events as NC. In addition, unusual heterozygous calls were manually curated by replacing these calls with NC. These 33 linkage groups were used for QTL analysis using the R/qtl software (Broman and Sen, 2009). The QTL that conferred resistance to races RTQQC, TTTTF, and TPMKC at the seedling stage and QTL that conferred resistance at the adult plant stage in the Kenya, Ethiopia, and United States environments were identified separately. Interval mapping was conducted using the scanone function, and single-QTL genome scan via Haley-Knott regression was performed. The statistical significance thresholds of the genome scan results were determined by running 1000 permutation tests with a 0.05 significance level. QTL were also reported when LOD values were greater than 3.0, and the corresponding markers were detected in multiple environments. Markers flanking the QTL were identified using the find.flanking function. Linkage groups were assigned to chromosomes based on the 90K consensus map. The physical positions of the QTL were identified by searching positions of the flanking markers in the T3/Wheat website: https://triticeaetoolbox.org/wheat/ (Blake et al., 2016). The probe sequence of the markers was determined by searching the GrainGenes website, https://wheat.pw.usda.gov/GG3/, when marker information was missing in the T3/Wheat website, and the sequences were blasted to the reference genome at the BLAST website: https://wheat-urgi.versailles.inra.fr/Seq-Repository/BLAST to find the physical position of the markers (Alaux et al., 2018).
Confirmation of Presence/Absence of Lr34/Sr57 in CI 14275
The marker csLV34 (Lagudah et al., 2006) linked to the pleiotropic rust resistance gene Lr34/Sr57/Yr18 was genotyped on the parents (LMPG-6 and CI 14275). Wheat cultivar Chinese Spring was used as the positive control for the gene.
Data Analyses
Chi-squared analyses were performed on the linearized seedling data to determine goodness-of-fit to the expected segregation ratios for different inheritance models. Using R software, analysis of variance (ANOVA) was performed using the average COI values with lines and environments both as fixed variables and replication at random. Correlation of stem rust severity between the environments was determined using Pearson correlation coefficients.
Results
Phenotyping in the Greenhouse
One hundred thirteen F7:9 RILs and their parents showed susceptible responses when tested against Pgt races TTKSK and TRTTF in two replicates each (Supplementary Table 2). A 33+ IT was observed on the resistant parent, CI 14275, and an IT of 3/33+ was observed on the susceptible parent, LMPG-6, when tested with race TTKSK (Table 1 and Supplementary Table 2). ITs of 33+ and 3+/33+ were observed on the parents CI 14275 and LMPG-6, respectively, when tested with race TRTTF (Table 1 and Supplementary Table 2). On the other hand, ITs of; 1/1+ 3C and 3+ were observed on the resistant and susceptible parents, respectively, when tested with race TTTTF, and the observed frequency of the population fit into the expected ratio of 1:1 (one gene) with a χ2 value of 0.704 (p = 0.40) (Table 1). Screening for race RTQQC resulted in 3/33+ and 3+/33+ ITs on CI 14275 and LMPG-6, respectively. Nearly all of the RILs were susceptible to RTQQC in at least one replication, and the rare low responses to RTQQC were usually not consistent across the two replications (Supplementary Table 2 and Table 1). For race TPMKC, ITs of 23−/23+ and 3+/33+ were observed on the resistant and susceptible parents, respectively, and the observed frequency of the population did not fit any simple expected ratio (Supplementary Table 2 and Table 1).
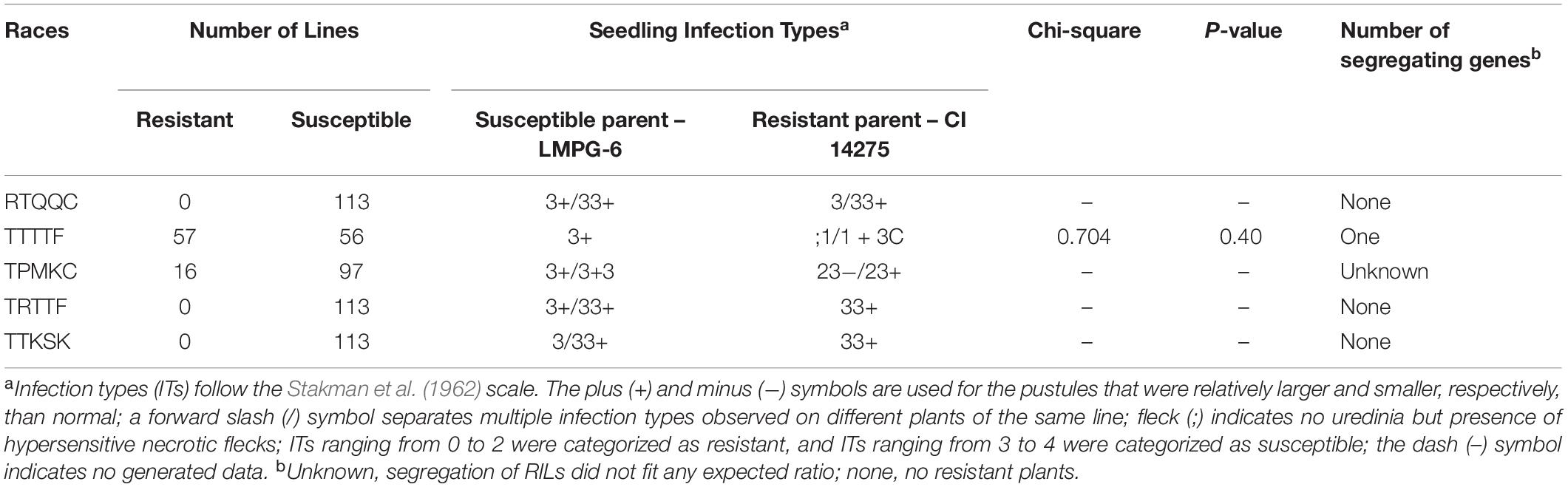
Table 1. Segregation of stem rust resistance in 113 recombinant inbred lines (RILs) along with response of parents of the cross LMPG-6/CI 14275 against five Puccinia graminis f. sp. tritici races.
Phenotyping in the Field
For the primary population, stem rust ratings from 0 to 90S were observed in Kenya 2016 (KEN16), 0 to 80S in Kenya 2017 (KEN17), and TR (trace R) to 60S in Kenya 2018 (KEN18) (Supplementary Table 3). Ratings from TR to 80S were observed in Ethiopia 2016 (ETH16), TMS (trace MS) to 50S in Ethiopia 2017 (ETH17), and TMR (trace MR) to 70S in Ethiopia 2018 (ETH18) (Supplementary Table 4). Ratings from 5RMR to 100S were observed in St. Paul 2017 (STP17) and TR to 80S in St. Paul 2018 (STP18) (Supplementary Table 5). Stem rust ratings from 0 to 70S were observed in the validation population. In the three Kenyan environments: KEN16, KEN17, and KEN18, the distribution of stem rust severity was somewhat skewed toward resistance (Figure 1). A symmetric distribution was observed in the ETH16, ETH17, and ETH18 environments (Figure 1). A somewhat symmetric distribution was observed in the STP17 and STP18 environments (Figure 1). ANOVA indicated significant effects for lines and environment, a significant interaction between the RILs and the environments, and a non-significant effect for replication (Table 2). Stem rust severity over the different environments had statistically significant correlation coefficients, ranging between 0.55 and 0.85 for correlations among all environments except with ETH17, with which the correlations were low but still statistically significant, ranging between 0.21 and 0.47 (Table 3).
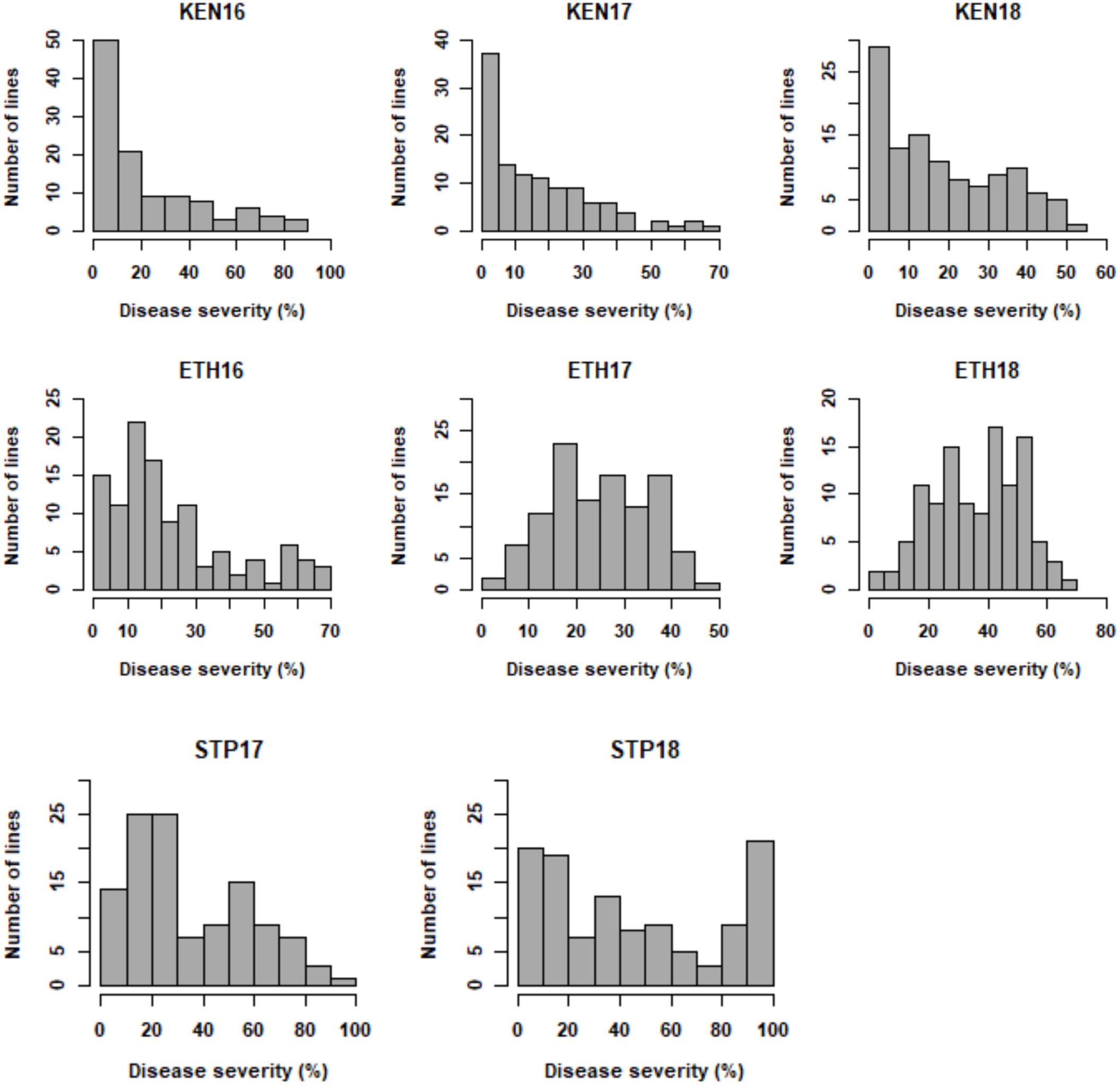
Figure 1. Frequency distribution for stem rust severity (%) in RILs developed from the cross LMPG-6/CI 14275 evaluated in Kenya (KEN), Ethiopia (ETH), and St. Paul (STP) in 2016, 2017, and 2018.

Table 2. Analysis of variance for stem rust severity of 113 recombinant inbred lines (RILs) and the parents of the cross LMPG-6/CI 14275 tested in eight environments (Kenya – 2016, 2017, and 2018; Ethiopia – 2016, 2017, and 2018; St. Paul – 2017 and 2018).
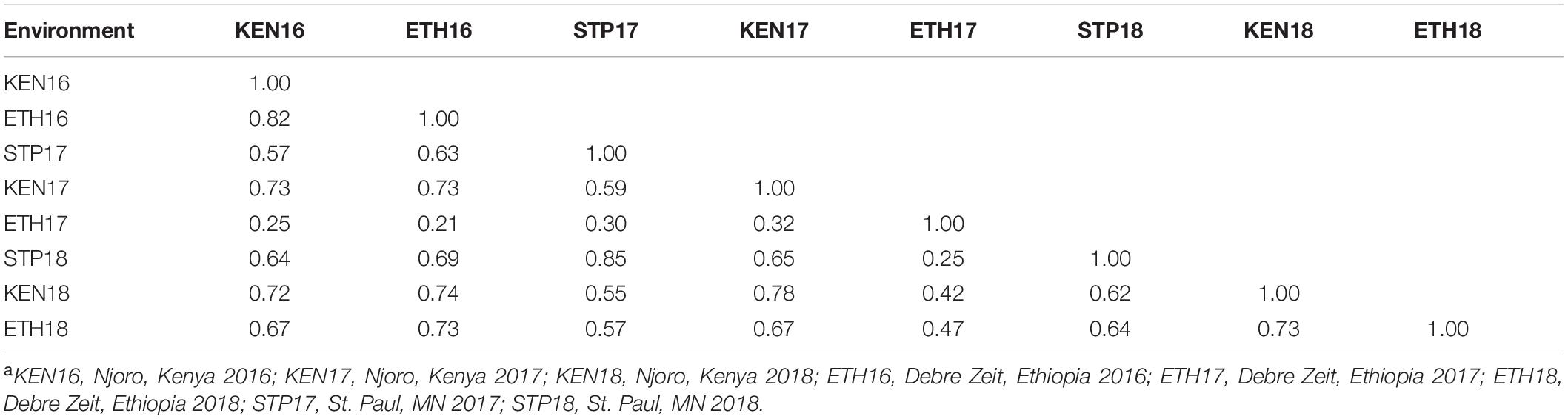
Table 3. Correlation coefficients among stem rust coefficient of infection (COI) values for 113 recombinant inbred lines (RILs) and the parents of the cross LMPG-6/CI 14275 in eight environments (Kenya – 2016, 2017, and 2018; Ethiopia – 2016, 2017, and 2018; St. Paul – 2017 and 2018)a.
Confirmation of Presence/Absence of Lr34/Sr57 in CI 14275
Both parents were negative for the positive allele of the Lr34 gene-based marker, indicating that the resistance in CI 14275 did not involve Lr34/Sr57. This result was confirmed by the lack of significant QTL on chromosome 7D, where Lr34 is located.
LMPG-6/CI 14275 Genetic Map
The genetic map of LMPG-6/CI 14275 is listed in Supplementary Table 1. A total of 819 markers mapped to 33 linkage groups. The 33 linkage groups corresponded to 15 wheat chromosomes. Six wheat chromosomes were missing from the genetic map: 2D, 3A, 4B, 4D, 6D, and 7D. The remaining wheat chromosomes were represented in the map from anywhere between one and four linkage groups.
Identified QTLs at Seedling and Adult Plant Stages
The 0.05 LOD significance thresholds for each trait ranged from 3.63 to 3.81. We did not detect any significant QTL for response of the primary population to RTQQC, TPMKC, or ETH17. All significant QTL are reported in Table 4 in addition to QTL with an LOD of 3.0 or greater that were detected in multiple environments.
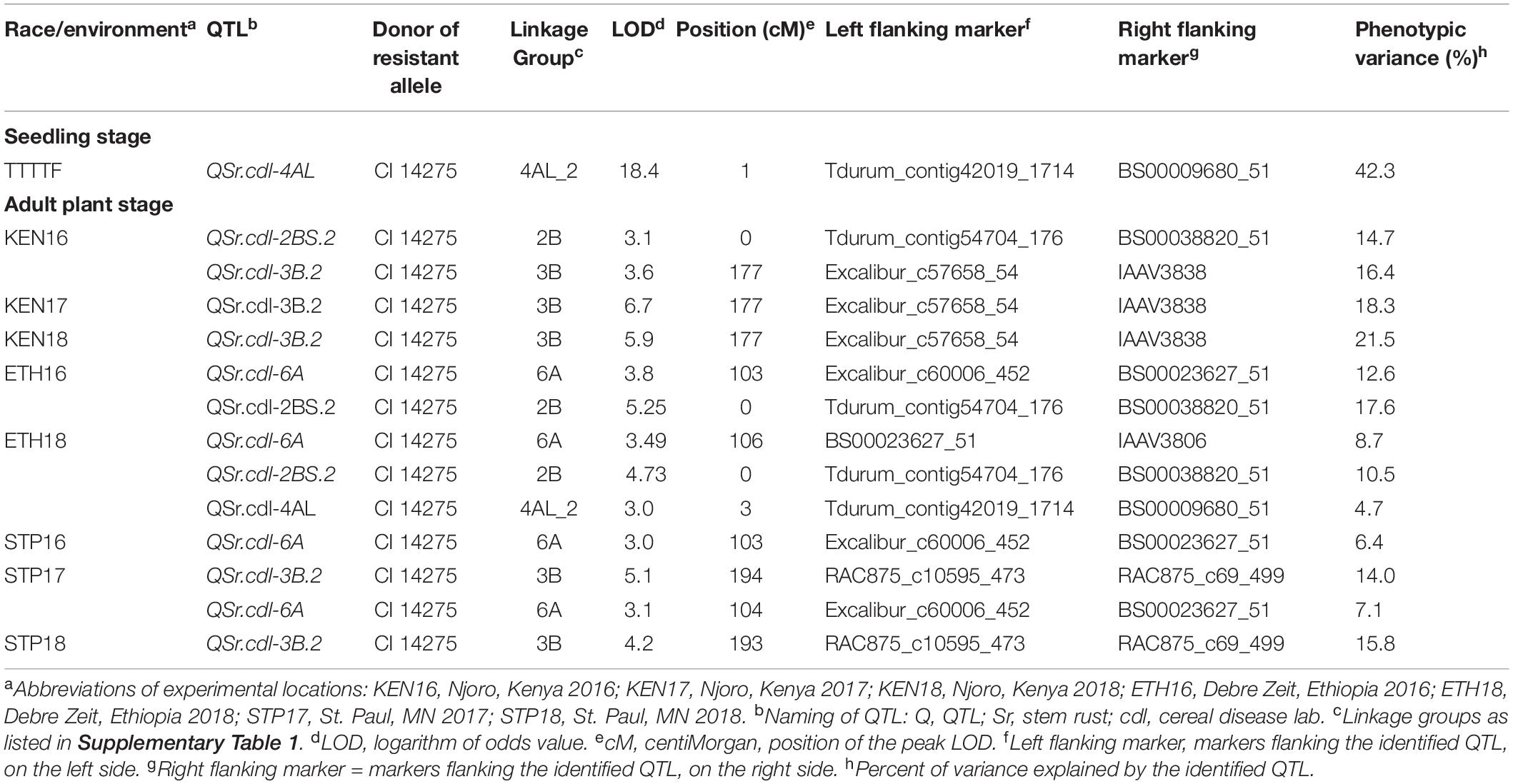
Table 4. Quantitative trait loci (QTL) identified in the recombinant inbred lines (RILs) population derived from the cross LMPG-6/CI 14275 for seedling resistance to Puccinia graminis f. sp. tritici races RTQQC, TPMKC, and TTTTF and for field resistance in Njoro, Kenya (2016, 2017, and 2018); Debre-Zeit, Ethiopia (2016, 2017, and 2018); and St. Paul, Minnesota (2017 and 2018).
QTL on Chromosome Arm 2BS
One QTL mapped to chromosome arm 2BS in the ETH16 and ETH18 environments and was designated QSr.cdl-2BS.2 (Table 4). This QTL also reached an LOD of 3.1 in KEN16. The QTL is flanked by the markers Tdurum_contig54704_176 and BS00038820_51 (Table 4). We designated this QTL with the suffix 2BS.2 because QSr.cdl-2BS was already designated (Rouse et al., 2014b). The phenotypic variance explained by the QTL ranged from 10.5 to 17.6% across the three environments (Table 4).
QTL on Chromosome 3B
One QTL mapped to chromosome 3B in the KEN17, KEN18, STP17, and STP18 environments and was designated QSr.cdl-3B.2 (Table 4). This QTL also reached an LOD of 3.6 in KEN16. We designated this QTL with the suffix 3B.2 because QSr.cdl-3B was already designated (Rouse et al., 2014b). The markers Excalibur_c57658_54, IAAV3838, RAC875_c10595_473, and RAC875_c69_499 were linked to QSr.cdl-3B.2 (Table 4). The phenotypic variance explained by the QTL in the five environments ranged from 14.0 to 21.5% (Table 4).
QTL on Chromosome Arm 4AL
One QTL mapped to chromosome arm 4AL for race TTTTF and was designated QSr.cdl-4AL (Table 4). The QTL QSr.cdl-4AL was flanked by markers Tdurum_contig42019_1714 and BS00009680_51 (Table 4). This QTL explained a large proportion (42.3%) of the phenotypic variance observed (Table 4). Interestingly, this QTL was observed in the field in ETH18 with an LOD of 3.0 (Table 4).
QTL on Chromosome Arm 6A
One QTL mapped to chromosome arm 6A in the ETH16, ETH18, STP16, and STP17 environments with LOD values of 3.8, 3.5, 3.0, and 3.1, respectively. This QTL did not pass the 0.05 LOD threshold in any of these environments, but it was detected above LOD 3.0 in four environments, which adds confidence to the validity of this QTL. We designated the QTL as QSr.cdl-6A (Table 4). The markers BS00023627_51, IAAV3806, and Excalibur_c60006_452 were linked to QSr.cdl-6A (Table 4). The phenotypic variance explained by the QTL in the four environments tested ranged from 6.4 to 12.6% (Table 4).
Combined 2BS, 3B, and 6A QTLs
Three QTLs, QSr.cdl-2BS.2, QSr.cdl-3B.2, and QSr.cdl-6A, were identified only at the adult plant stage in seven of the eight field environments. No QTL were reported in ETH17 because disease pressure was low although the line responses in ETH17 were significantly correlated with those in other environments. Based on the genotype data of the QTL peak markers, seven lines were found to have all three QTLs combined, and 11 lines lacked the three QTLs. The t-test results from the two groups (with and without the three QTLs) showed statistically significant differences (p < 0.01; 0.001) in seven environments (all environments except ETH17) (Table 5). The combined QTL provided large reductions in stem rust severity in the observed environments and appeared to be a highly effective combination even when the disease pressure was high (Table 5).
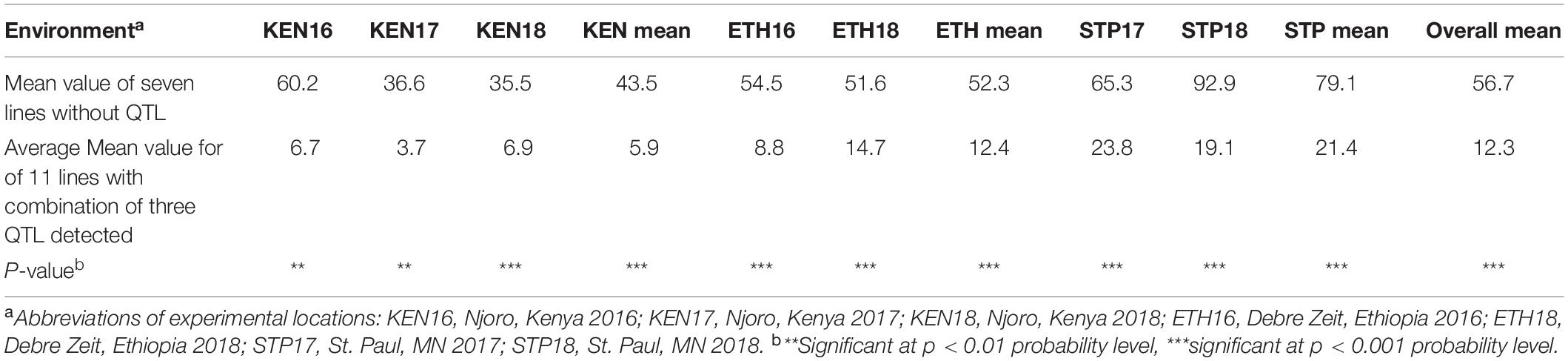
Table 5. Coefficient of infection (COI) (%) for LMPG-6/CI 14275 lines with and without three adult plant resistance (APR) quantitative trait loci (QTL): QSr.cdl-2BS.2, QSr.cdl-3B.2, and QSr.cdl-6A detected in six environments tested in this study, excluding ETH17.
Kompetitive Allele-Specific PCR (KASP) Markers
KASP assay primers were designed corresponding to 15 90K SNP markers linked to QTL, but only six of these KASP assays produced clear polymorphism in the validation population (Table 6). Simple ANOVAs for each marker were independently run to determine whether any of the KASP markers predicted the phenotype in the validation population. One marker linked to QSr.cdl-2BS.2, Excalibur_c7963_1722_C1, was associated with reduced stem rust severity in Njoro, Kenya, in 2018 (p-value = 0.003). This validated QSr.cdl-2BS.2, which was identified in the LMPG-6/CI 14275 population in the KEN16, ETH16, and ETH18 environments. Two KASP assays derived from markers linked to QSr.cdl-6A were polymorphic but were not associated with stem rust severity. We then discovered that line #162 did not possess the resistant alleles for markers linked to QSr.cdl-6A after reviewing the original 90K allele calls. Line #162 did possess the resistant alleles of markers linked to QSr.cdl-2BS.2 and QSr.cdl-3B.2. Line #162 was selected only based on phenotype, which explains the absence of QSr.cdl-6A. The observed polymorphism in the two KASP assays derived from markers linked to QSr.cdl-6A may be explained by broken linkage between the resistant allele of these markers and QSr.cdl-6A in the susceptible parent of the validation population: Kwale. Only one of the KASP assays that we designed for markers on chromosome 3B was polymorphic: Tdurum_contig32277_121. This marker was not associated with stem rust severity in the validation population. This raised the possibility that both parents of the validation population were fixed for the presence of QSr.cdl-3B.2. Because we considered QSr.cdl-3B.2 as likely conferred by Sr12, we tested the validation population with the Sr12 marker from Hiebert et al. (2016), NB-LRR-3, and found that the parents and population were fixed for presence of the Sr12-associated allele. Two additional KASP assays were designed based on markers linked to QSr.cdl-4AL. These assays were not associated with stem rust severity in Kenya, where the pathogen population is virulent to Sr7a.
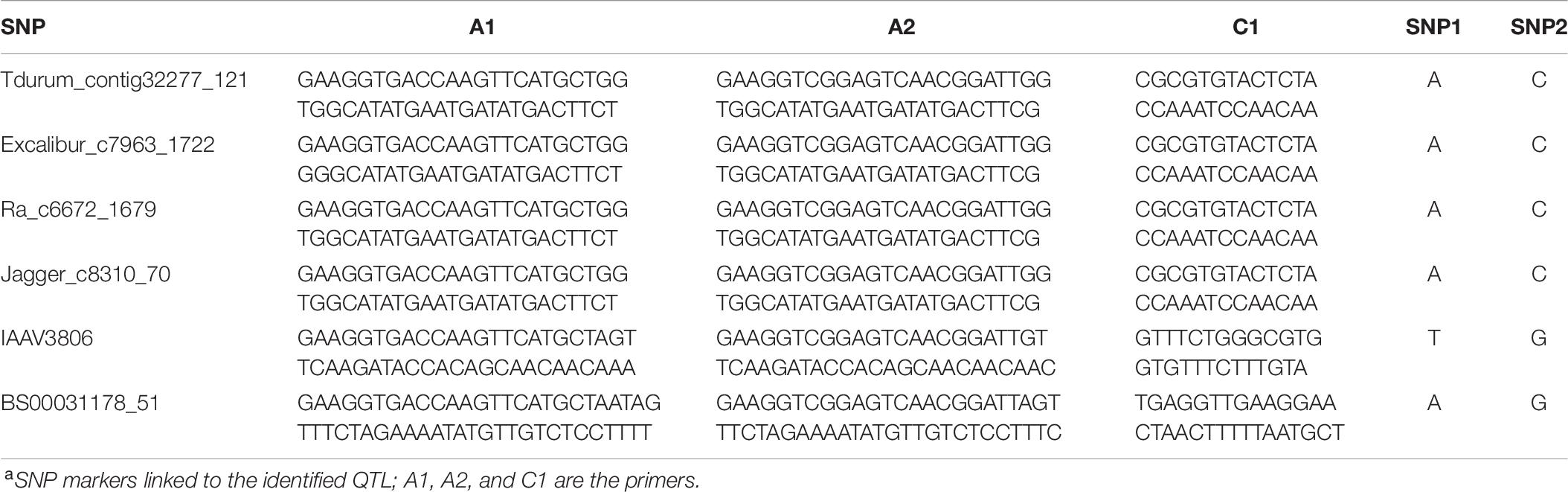
Table 6. Primer sequences of polymorphic Kompetitive allele-specific PCR (KASP) assays in the line #162/Kwale population derived from 90K single nucleotide polymorphism (SNP) markers linked to identified quantitative trait loci (QTL)a.
Discussion
The RIL population, together with the parents, was susceptible at the seedling stage when tested with Pgt races TTKSK, TRTTF, and RTQQC. This is an indication that the resistant parent, CI 14275, lacked an effective major gene conferring resistance to these races. This observation is different from that observed by Rouse et al. (2011) for the seedling response of CI 14275 to race TTKSK. Different environmental conditions between the two studies, particularly temperature (Zhang et al., 2017; Chen et al., 2018; Gao et al., 2019), may explain the variable responses although the differences in these findings may warrant further investigation. Similarly, the observed inconsistencies of infection types between replicates in some lines could have been due to the temperature effect on segregating stem rust resistance genes. The seedling resistance to race TTTTF was conferred by a single gene. Based on map location and race specificity, this gene is likely Sr7a (Turner et al., 2016; Saini et al., 2018). The distribution of disease severities for the RILs at the adult plant stage across all the environments was mostly continuous but somewhat skewed toward resistance in the Kenyan environments, suggesting a quantitative type of resistance. Lower correlation coefficients (0.21–0.32) were observed between ETH17 and the ETH16, KEN16, KEN17, STP17, and STP18 environments, suggesting genotype by environment interactions as indicated by the ANOVA results (Table 2). Stem rust severities of the entire population were relatively low in the ETH17 environment. The differences in environmental conditions, different races used for inoculation, and the amount of inoculum used in different environments could have also contributed to the observed significant genotype by environment interactions.
The identified QTL QSr.cdl-2BS.2 on chromosome arm 2BS with the explained phenotypic variance ranging from 10.5% to 17.6%, conferred adult plant resistance in the KEN16, ETH16, and ETH18 environments. The QTL was also identified in the validation population in Kenya in 2018. Several stem rust resistance genes have been mapped on chromosome arms 2BS and 2BL. The stem rust resistance genes effective to race TTKSK that mapped to chromosome arm 2BS are Sr36, Sr39, and Sr40 (Wu et al., 2009; Niu et al., 2011; Rouse et al., 2012). The gene Sr36 was identified in Triticum timopheevi, Sr39 was identified in Aegilops speltoides, and Sr40 was identified in Triticum araraticum. The resistance provided by the Sr36 gene was overcome by race TTTSK (virulent to Sr31 and Sr36) (Jin et al., 2009), whereas Sr39 and Sr40 genes appear effective against the Ug99 race group but have not been widely utilized in breeding programs due to their potential linkage drag. The alien species origin of genes Sr39 and Sr40 as well as susceptible ITs observed in the seedlings of the entire RIL population along with parents against race TTKSK ruled out the possibility of Sr39 or Sr40 conferring the observed resistance in this study.
The mapped genes on chromosome arm 2BL are Sr9h, Sr16, Sr28, and Sr47 (Tsilo et al., 2007; Hiebert et al., 2011; Rouse et al., 2014a). The genes Sr9h, Sr16, and Sr28 were identified in Triticum aestivum, whereas the gene Sr47 was identified in A. speltoides. The Sr16 gene was shown to be ineffective against Ug99 races (Jin et al., 2007). Because Sr16 and QSr.cdl-2BS.2 are on different chromosome arms, they cannot be the same. The Sr28 and Sr47 genes have been found effective against the Ug99 race group (Jin et al., 2007; Rouse et al., 2012), but the QTL QSr.cdl-2BS.2 identified in this study is unlikely either the Sr28 or Sr47 gene because of the observed susceptibility of CI 14275 at the seedling stage in response to race TTKSK. The Sr9 alleles Sr9a, Sr9b, Sr9d, Sr9e, Sr9f, and Sr9g are ineffective against Ug99 isolates, but the Sr9h allele has been found effective against race TTKSK and other races in the Ug99 race group with virulence to Sr31 (Rouse et al., 2014a). Based on the physical position of the peak marker, the QSr.cdl-2BS.2 found in this study is located around 58.3 Mb on the 90K consensus map (Wang et al., 2014). This map position excludes the possibility that QSr.cdl-2BS.2 could be conferred by stem rust resistance genes on the long arm of chromosome 2B. The QSr.umn-2B.2 reported by Bajgain et al. (2015) in a RB07/MN06113-8 population is between 22.7 and 40.7 Mb on 2B, and QSr.cdl-2BS reported by Rouse et al. (2014b) in the Thatcher/McNeal population is located between 12.0 and 31.7 Mb on 2B. The physical location of the reported QSr.cdl-2BS.2 is near these two previously reported QTL. Rouse et al. (2014b) found that the resistant allele of the reported QSr.cdl-2BS was contributed by Thatcher. Because Thatcher is a major component of the pedigree of CI 14275, it is likely that QSr.cdl-2BS from Rouse et al. (2014b) and QSr.cdl-2BS.2 from this study are conferred by the same gene(s) derived from Thatcher. Tdurum_contig54704_176, the peak marker associated with QSr.cdl-2BS.2, is located at 61 cM in the popseq consensus map, whereas the Excalibur_c7963_1722 marker that was picked up in the validation population is located at 60 cM in the popseq consensus map. A KASP marker was developed for Excalibur_c7963_1722 because it was found to be a flanking marker of QSr.cdl-2BS.2 in a preliminary QTL analysis. Several other studies have reported stem rust resistance QTLs on chromosome 2B (association mapping study QTL at 79.6 Mb on 2BL: Letta et al., 2013; GWAS study QTL located between 11.3 and 79.9 Mb: Prins et al., 2016; Cacuke/Huhwa and Cacuke/Yaye population between 65.8 and 73.9 Mb on 2BL: Randhawa et al., 2018). Our study is the first to both detect and validate an adult plant stem rust resistance QTL on chromosome arm 2BS.
QSr.cdl-3B.2 explains between 14.0 and 21.5% of the phenotypic variance and confers adult plant resistance in the KEN16, KEN17, KEN18, STP17, and STP18 environments. QSr.cdl-3B.2 is flanked by markers Excalibur_c57658_54, IAAV3838, RAC875_c10595_473, and RAC875_c69_499 in the various environments. The Sr2 gene is located on chromosome arm 3BS, whereas Sr12 is located near the centromere on chromosome 3B. Both were derived from Triticum turgidum. Sr2 is an adult plant resistance gene used in breeding as a source of durable resistance to stem rust although the gene may not provide adequate resistance under high disease pressure. Sr12 is present in all of the parents of CI 14275, cultivars Thatcher, Kenya Farmer, and Lee; therefore, Sr12 is almost certainly present in CI 14275 (unless there is a mistake in the pedigree or the gene postulations of the parents). The identified QSr.cdl-3B.2 in this study is located between 58.6 and 75.9 Mb on 3B and overlaps other reported QTLs on 3B (Thatcher/McNeal population located between 15.4 and 85.8 Mb, Rouse et al., 2014b; association mapping population located between 13.8 and 59.5 Mb, Bajgain et al., 2015; Spark/Rialto DH population located between 20.1 and 73.8 Mb, Getie et al., 2016; GWAS study, located between 68.9 and 71.0 Mb, Prins et al., 2016; LMPG-6/PI 362698-1 population located between 62.2 and 72.9 Mb, Zurn et al., 2018) and is likely the gene Sr12.
The Sr12 gene (QSr.cdl-3B.2) alone did not provide strong adult plant resistance in three environments, suggesting the necessity of combining Sr12 and other QTL(s). The gene Lr34/Sr57 has been reported to enhance the effectiveness of Thatcher resistance in North America and Kenya (Kolmer et al., 2011), but the parents (LMPG-6 and CI 14275) were both negative for the Lr34 gene-based marker (CsLV34), indicating that the resistance in CI 14275 did not involve Lr34/Sr57. Lack of significant QTL on chromosome 7D in this study, where Lr34 is located, also is a strong indication that Lr34/Sr57 was not segregating in this population. The KASP markers linked to QSr.cdl-3B.2 were not associated with stem rust response in the validation population. To look into this further, we genotyped CI 14275, LMPG-6, line #162, and Kwale with the Sr12-associated marker from Hiebert et al. (2016): NB-LRR3. We found that LMPG-6 did not possess the Sr12 gene, but CI 14275 and both parents of the validation population (Line #162 and Kwale) did possess Sr12. Therefore, Sr12 is likely fixed in the validation population.
QSr.cdl-4AL, which explained a large proportion of the phenotypic variance (42.3%) for response to race TTTTF, is located at 73.3 Mb on chromosome arm 4AL. QSr.cdl-4AL is flanked by the markers Tdurum_contig42019_1714 and BS00009680_51. SrND643, a temporarily designated gene that is also located on chromosome arm 4AL, is reported to provide inadequate resistance under high disease pressure (Basnet et al., 2015). Even though SrND643 is located between 72.3 and 73.7 Mb at a similar location to identified QSr.cdl-4AL, SrND643 shows a low infection type of 2 to 22+ at the seedling stage against Ug99 races TTKSK and TTKST (Basnet et al., 2015). In contrast, line CI 14275 shows a high infection type of 3 to 33+ against Ug99 race TTKSK. These data suggest that QSr.cdl-4AL is not SrND643. Notably, CI 14275 and SrND643 were tested against the same isolate under the same temperature regime and in the same greenhouse but not in the same experiment. The stem rust resistance gene Sr7 with two characterized alleles Sr7a and Sr7b is located on chromosome arm 4AL, and these alleles confer resistance to some North American Pgt races (McIntosh et al., 1995; Turner et al., 2016). In this study, race TTTTF was virulent on Sr7b in the North American stem rust differential set with infection types ranging from 3− to 3+, ruling out the possibility of Sr7b being involved in the observed resistance. The QTL QSr.rwg-4A (believed to be Sr7a), is located between 59.1 and 73.9 Mb and expresses resistance to race TTTTF (Saini et al., 2018) in the durum wheat ‘Lebsock.’ The location of QSr.cdl-4AL and QSr.rwg-4A overlaps, and it is, therefore, likely that QSr.cdl-4AL identified in this study is Sr7a. Sr7a is demonstrated to be effective to race TTTTF by Turner et al. (2016). The detection of QSr.cdl-4AL in ETH18 may have been the result of an abundance of race TKTTF present in the disease nursery. Race TKTTF became predominant in Ethiopia starting in 2014 and was reported to be avirulent to Sr7a (Olivera et al., 2015).
QSr.cdl-6A, located between 51.4 and 52.0 Mb, conferred adult plant resistance in the ETH16, ETH18, STP16, and STP17 environments. QSr.cdl-6A explains phenotypic variance ranging from 6.4 to 12.6% and is flanked by the markers BS00023627_51, IAAV3806, and Excalibur_c60006_452 in various environments. The previously described stem rust resistance genes on chromosome arm 6AL are Sr13, Sr26, and Sr52. All three of these genes are derived from species other than bread wheat and confer seedling resistance to race TTKSK (Qi et al., 2011; Zhang et al., 2017). Because CI 14275 is not effective to race TTKSK at the seedling stage, these genes are not present in CI 14275. QSr.cdl-6A is close to both the QTL reported by Letta et al. (2013), which mapped between 60.6 and 61.8 Mb on 6A, and QTL reported in a genome-wide association study located between 10.0 and 59.6 Mb on 6A (Kankwatsa et al., 2017). The susceptibility of CI 14275 at the seedling stage to race TTKSK and pedigree information, however, rule out the possibilities of these genes (Sr13, Sr26, and Sr52) contributing to the observed resistance. The Sr8 locus alleles Sr8a and Sr8b are reported on chromosome arm 6AS (Singh and McIntosh, 1986). A QTL identified by Bajgain et al. (2015) in an association mapping study is located at 52.3 Mb, a similar position to QSr.cdl-6A. Prins et al. (2016) identified QTL on 6AS although the physical location of linked markers was not found. A race-specific gene on 6A, likely Sr8a, was identified at the seedling stage for race TRTTF (Dunckel et al., 2015). In Ethiopian nurseries, other than the predominance of race TTKSK, Pgt races, including TRTTF and JRCQC, have been reported (Olivera et al., 2012; Hailu et al., 2015). The races TRTTF and JRCQC are avirulent to Sr8a although race TTKSK, which is predominant in Kenyan and Ethiopian environments, is virulent to Sr8a (Olivera et al., 2012; Hailu et al., 2015). The susceptibility of the RIL population to TRTTF rules out the possibility of QSr.cdl-6A being Sr8a. Nirmala et al. (2017) mapped Sr8155B1, a gene conferring resistance to race TTKST on chromosome 6A. Sr8155B1 is located between 6.7 and 10.9 Mb, a different location from the QTL QSr.cdl-6A identified in this study. In addition, data from our laboratory indicate that TRTTF is avirulent to Sr8155B1, ruling out the possibility of CI 14275 possessing Sr8155B1 because of its susceptibility to TRTTF (Table 1). The KASP markers linked to QSr.cdl-6A are not associated with stem rust resistance in the validation population. Unfortunately, line #162 (the resistant parent in the validation population) possesses the susceptible alleles of the QSr.cdl-6A-linked markers. Selecting line #162 as a parent was undertaken based on the phenotypic data alone. Therefore, the absence of QSr.cdl-6A from line #162 precludes our ability to validate this QTL in the validation population. Additional experiments are needed to validate the effectiveness of QSr.cdl-6A.
Conclusion
This study reports QTL for wheat stem rust resistance on chromosomes 2BS, 3B, 4AL, and 6A. The identified QTL QSr.cdl-2BS.2, which conferred adult plant resistance in the KEN16, ETH16, and ETH18 environments, was validated in a second population in KEN18 and can be selected for by the validated linked marker Excalibur_c7963_1722. This QTL, therefore, has the potential of being used in marker-assisted selection. Our study is the first to both detect and validate an adult plant stem rust resistance QTL on chromosome arm 2BS. QSr.cdl-3B.2, which confers resistance in the KEN16, KEN17, KEN18, STP17, and STP18 environments is likely Sr12. QSr.cdl-4AL, which confers resistance to race TTTTF, is postulated to be Sr7a. QSr.cdl-6A, which confers resistance in the ETH16, ETH18, STP16, and STP17 environments, is potentially a new QTL, but the QTL requires validation in another population before it is recommended for use in breeding. The adult plant resistance of wheat line CI 14275 in Africa is characterized as the cumulative contribution of three QTL: QSr.cdl-2BS.2, QSr.cdl-3B.2, and QSr.cdl-6A.
Data Availability Statement
The datasets generated for this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
ZK, RD-M, and MNR contributed to writing of the initial drafts of the manuscript and provided insights on data analysis. YJ developed the mapping population and contributed to manuscript writing. EE helped with genotypic data analysis. WB, AG, GM, MSR, and SB facilitated the field studies in Africa. All authors contributed to the submitted manuscript.
Funding
We acknowledge the financial support provided by the United States Agency for International Development as part of the Feed the Future initiative under the CGIAR Fund, award number BFS-G-11-00002, and the predecessor fund the Food Security and Crisis Mitigation II grant, award number EEM-G-00-04-00013 and the USDA-ARS National Plant Disease Recovery System.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We acknowledge Sam Stoxen, Samantha Armintrout, and Greta Ciessau for their support in planting and maintaining field and greenhouse experiments, for technical help in the laboratory, and the assistance with manual SNP calling of 90K SNP data. We also extend our gratitude to the technical staff of the Kenya Agricultural and Livestock Research Organization (KALRO), Njoro, Kenya, and the Debre Zeit Agricultural Research Centre, Ethiopia, for their assistance in establishing and maintaining field experiments.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.609659/full#supplementary-material
Supplementary Table 1 | Genetic map of the LMPG-6/CI14275 population.
Supplementary Table 2 | Seedling infection types of 113 recombinant inbred lines (RILs) of the cross LMPG-6/CI 14275 and the parents against five Puccinia graminis f. sp. tritici races: RTQQC, TTTTF, TPMKC, TRTTF, and TTKSK.
Supplementary Table 3 | Stem rust severity and response of 113 recombinant inbred lines (RILs) of the cross LMPG-6/CI 14275 and the parents to predominant Ug99 races in Kenya in 2016 (KEN16), 2017 (KEN17), and 2018 (KEN18). Data for two replicates (Rep) are displayed.
Supplementary Table 4 | Stem rust severity and response of 113 recombinant inbred lines (RILs) of the cross LMPG-6/C I14275 and the parents to predominant Ug99 races in Ethiopia in 2016 (ETH16), 2017 (ETH17), and 2018 (ETH18). Data for two replicates (Rep) are displayed.
Supplementary Table 5 | Stem rust severity and response of 113 recombinant inbred lines (RILs) of the cross LMPG-6/C I14275 and the parents to US Pgt races in St. Paul, MN, in 2017 (STP17) and 2018 (STP18). Data for the two replicates are displayed.
References
Alaux, M., Rogers, J., Letellier, T., Flores, R., Alfama, F., Pommier, C., et al. (2018). Linking the international wheat genome sequencing consortium bread wheat reference genome sequence to wheat genetic and phenomic data. Genome Biol. 19:111. doi: 10.1186/s13059-018-1491-4
Bajgain, P., Rouse, M. N., Bulli, P., Bhavani, S., Gordon, T., Wanyera, R., et al. (2015). Association mapping of North American spring wheat breeding germplasm reveals loci conferring resistance to Ug99 and other African stem rust races. BMC Plant Biol. 15:249. doi: 10.1186/s12870-015-0628-9
Bansal, U., Bariana, H., Wong, D., Randhawa, M., Wicker, T., Hayden, M., et al. (2014). Molecular mapping of an adult plant stem rust resistance gene Sr56 in winter wheat cultivar Arina. Theor. Appl. Genet. 127, 1441–1448. doi: 10.1007/s00122-014-2311-1
Basnet, B. R., Singh, S., Lopez-Vera, E. E., Huerta-Espino, J., Bhavani, S., Jin, Y., et al. (2015). Molecular mapping and validation of SrND643: a new wheat gene for resistance to the stem rust pathogen Ug99 race group. Phytopathology 105, 470–476. doi: 10.1094/PHYTO-01-14-0016-R
Blake, V. C., Birkett, C., Matthews, D. E., Hane, D. L., Bradbury, P., and Jannink, J.-L. (2016). The Triticeae Toolbox: combining phenotype and genotype data to advance small-grains breeding. Plant Genome 9, 1–10. doi: 10.3835/plantgenome2014.12.0099
Broman, K. W., and Sen, S. (2009). A Guide to QTL Mapping with R/qtl (Statistics for Biology and Health). New York, NY: Springer Science+Business Media.
Chen, S., Zhang, W., Bolus, S., Rouse, M. N., and Dubcovsky, J. D. (2018). Identification and characterization of wheat stem rust resistance gene Sr21 effective against the Ug99 race group at high temperature. PLoS Genet. 14:e1007287. doi: 10.1371/journal.pgen.1007287
Dunckel, S. M., Olson, E. L., Rouse, M. N., Bowden, R. L., and Poland, J. A. (2015). Genetic mapping of race-specific stem rust resistance in the synthetic hexaploid W7984 X Opata mapping population. Crop Sci. 55, 2580–2588. doi: 10.2135/cropsci2014.11.0755
Food and Agriculture Organization [FAO] (2018). Food and Agricultural Organization of the United Nations; Food outlook biannual report on global food markets. Available online at: http://www.fao.org/3/CA0239EN/ca0239en.pdf (accessed April 23, 2019).
Gao, L., Babiker, E. M., Nava, I. C., Nirmala, J., Bedo, Z., Lang, L., et al. (2019). Temperature-sensitive wheat stem rust resistance gene Sr15 is effective against Puccinia graminis f. sp. tritici race TTKSK. Plant Pathol. 68, 143–151. doi: 10.1111/ppa.12928
Getie, B., Singh, D., Bansal, U., Simmonds, J., Uauy, C., and Park, R. F. (2016). Identification and mapping of resistance to stem rust in the European winter wheat cultivars Spark and Rialto. Mol. Breed. 36:114. doi: 10.1007/s11032-016-0537-0
Gupta, P. K., Mir, R. R., Mohan, A., and Kumar, J. (2008). Wheat genomics: present status and future prospects. Int. J. Plant Genom. 2008:896451. doi: 10.1155/2008/896451
Hailu, E., Woldaeb, G., Denbel, W., Alemu, W., and Abebe, T. (2015). Distribution of stem rust (Puccinia graminis f. sp. tritici) races in Ethiopia. Adv. Crop Sci. Tech. 3:173. doi: 10.4172/2329-8863.1000173
Helguera, M., Khan, I. A., Kolmer, J., Lijavetzky, D., Zhong-qi, L., and Dubcovsky, J. (2003). PCR assays for the Lr37-Yr17-Sr38 cluster of rust resistance genes and their use to develop isogenic hard red spring wheat lines. Crop Sci. 43, 1839–1847. doi: 10.2135/cropsci2003.1839
Herrera-Foessel, S. A., Singh, R. P., Lillemo, M., Huerta-Espino, J., Sridhar Bhavani, S., Singh, S., et al. (2014). Lr67/Yr46 confers adult plant resistance to stem rust and powdery mildew in wheat. Theor. Appl. Genet. 127, 781–789. doi: 10.1007/s00122-013-2256-9
Hiebert, C. W., Fetch, T. G., Zegeye, T., Thomas, J. B., Somers, D. J., Humphreys, D. G., et al. (2011). Genetics and mapping of seedling resistance to Ug99 stem rust in Canadian wheat cultivars ‘Peace’ and ‘AC Cadillac’. Theor. Appl. Genet. 122, 143–149. doi: 10.1007/s00122-010-1430-6
Hiebert, C. W., Kolmer, J. A., McCartney, C. A., Briggs, J., Fetch, T., Bariana, H., et al. (2016). Major gene for field stem rust resistance co-locates with resistance gene Sr12 in ‘Thatcher’ wheat. PLoS One 11:e0157029. doi: 10.1371/journal.pone.0157029
Hundie, B., Girma, B., Tadesse, Z., Edae, E., Olivera, P., Abera, E. H., et al. (2019). Characterization of Ethiopian wheat germplasm for resistance to four Puccinia graminis f. sp. tritici races facilitated by single-race nurseries. Plant Dis. 103, 2359–2366. doi: 10.1094/PDIS-07-18-1243-RE
Jin, Y., Singh, R. P., Ward, R. W., Wanyera, R., Kinyua, M., Njau, P., et al. (2007). Characterization of seedling infection types and adult plant infection responses of monogenic Sr gene lines to race TTKS of Puccinia graminis f. sp. tritici. Plant Dis. 91, 1096–1099. doi: 10.1094/PDIS-91-9-1096
Jin, Y., Szabo, L. J., Pretorius, Z. A., Singh, R. P., and Ward, R. (2008). Detection of virulence to resistance gene Sr24 within race TTKS of Puccinia graminis f. sp. tritici. Plant Dis. 92, 923–926. doi: 10.1094/PDIS-92-6-0923
Jin, Y., Szabo, L. J., Rouse, M. N., Fetch, T. Jr., Pretorius, Z. A., Wanyera, R., et al. (2009). Detection of virulence to resistance gene Sr36 within the TTKS race lineage of Puccinia graminis f. sp. tritici. Plant Dis. 93, 367–370. doi: 10.1094/PDIS-93-4-0367
Kankwatsa, P., Singh, D., Thomson, P. C., Babiker, E. M., Bonman, J. M., Newcomb, M., et al. (2017). Characterization and genome-wide association mapping of resistance to leaf rust, stem rust and stripe rust in a geographically diverse collection of spring wheat landraces. Mol. Breed. 37:113. doi: 10.1007/s11032-017-0707-8
Knott, D. (1989). The Wheat Rusts-Breeding for Resistance (Monographs on Theoretical and Applied Genetics, 12). Berlin: Springer.
Knott, D. R. (1957). The inheritance of rust resistance. III. The inheritance of stem rust resistance in nine Kenya varieties of common wheat. Can. J. Plant Sci. 37, 366–384. doi: 10.4141/cjps57-043
Knott, D. R. (1990). Near-isogenic lines of wheat carrying genes for stem rust resistance. Crop Sci. 30, 901–905. doi: 10.2135/cropsci1990.0011183X003000040029x
Kolmer, J. A., Garvin, D. F., and Jin, Y. (2011). Expression of a Thatcher wheat adult plant stem rust resistance QTL on chromosome arm 2BL is enhanced by Lr34. Crop Sci. 51, 526–533. doi: 10.2135/cropsci2010.06.0381
Lagudah, E. S., McFadden, H., Singh, R. P., Huerta-Espino, J., Bariana, H. S., and Spielmeyer, W. (2006). Molecular genetic characterization of the Lr34/Yr18 slow rusting resistance gene region in wheat. Theor. Appl. Genet. 114, 21–30. doi: 10.1007/s00122-006-0406-z
Letta, T., Maccaferri, M., Badebo, A., Ammar, K., Ricci, A., Crossa, J., et al. (2013). Searching for novel sources of field resistance to Ug99 and Ethiopian stem rust races in durum wheat via association mapping. Theor. Appl. Genet. 126, 1237–1256. doi: 10.1007/s00122-013-2050-8
Lopez-Vera, E. E., Nelson, S., Singh, R. P., Basnet, B. R., Haley, S. D., Bhavani, S., et al. (2014). Resistance to stem rust Ug99 in six bread wheat cultivars maps to chromosome 6DS. Theor. Appl. Genet. 127, 231–239. doi: 10.1007/s00122-013-2212-8
Mago, R., Bariana, H. S., Dundas, I. S., Spielmeyer, W., Lawrence, G. J., Pryor, A. J., et al. (2005). Development of PCR markers for the selection of wheat stem rust resistance genes Sr24 and Sr26 in diverse wheat germplasm. Theor. Appl. Genet. 111, 496–504. doi: 10.1007/s00122-005-2039-z
Mago, R., Verlin, D., Zhang, P., Bansal, U., Bariana, H., Jin, Y., et al. (2013). Development of wheat-Aegilops speltoides recombinants and simple PCR-based markers for Sr32 and a new stem rust resistance gene on the 2S#1 chromosome. Theor. Appl. Genet. 126, 2943–2955. doi: 10.1007/s00122-013-2184-8
Mago, R., Zhang, P., Vautrin, S., Šimková, H., Bansal, U., Luo, M. C., et al. (2015). The wheat Sr50 gene reveals rich diversity at a cereal disease resistance locus. Nat. Plants 1:15186. doi: 10.1038/nplants.2015.186
McIntosh, R. A., Wellings, C. R., and Park, R. F. (1995). Wheat Rusts: An Atlas of Resistance Genes. East Melbourne: CSIRO Publications, 93–99.
Newcomb, M., Olivera, P. D., Rouse, M. N., Szabo, L. J., Johnson, J., Gale, S., et al. (2016). Kenyan isolates of Puccinia graminis f. sp. tritici from 2008 to 2014: virulence to SrTmp in the Ug99 race group and implications for breeding programs. Phytopathology 106, 729–736. doi: 10.1094/PHYTO-12-15-0337-R
Nirmala, J., Saini, J., Newcomb, M., Olivera, P., Gale, S., Klindworth, D., et al. (2017). Discovery of a novel stem rust resistance allele in durum wheat that exhibits differential reactions to Ug99 isolates. Genes Genomes Genet. 7, 3481–3490. doi: 10.1534/g3.117.300209
Niu, Z., Klindworth, D. L., Friesen, T. L., Chao, S., Jin, Y., Cai, X., et al. (2011). Targeted introgression of a wheat stem rust resistance gene by DNA marker-assisted chromosome engineering. Genetics 187, 1011–1021. doi: 10.1534/genetics.110.123588
Olivera, P., Newcomb, M., Szabo, L. J., Rouse, M., Johnson, J., Gale, S., et al. (2015). Phenotypic and genotypic characterization of race TKTTF of Puccinia graminis f. sp. tritici that caused a wheat stem rust epidemic in southern Ethiopia in 2013-14. Phytopathol 105, 917–928. doi: 10.1094/PHYTO-11-14-0302-FI
Olivera, P. D., Jin, Y., Rouse, M., Badebo, A., Fetch, T. Jr., Singh, R. P., et al. (2012). Races of Puccinia graminis f. sp. tritici with combined virulence to Sr13 and Sr9e in a field stem rust screening nursery in Ethiopia. Plant Dis. 96, 623–628. doi: 10.1094/PDIS-09-11-0793
Parlevliet, J. E., and van Ommeren, A. (1988). Accumulation of partial resistance in barley to barley leaf rust and powdery mildew through recurrent selection against susceptibility. Euphytica 37, 261–274. doi: 10.1007/BF00015122
Patpour, M., Hovmøller, M. S., Justesen, A. F., Newcomb, M., Olivera, P., Jin, Y., et al. (2016). Emergence of virulence to SrTmp in the Ug99 race group of wheat stem rust, Puccinia graminis f. sp. tritici, in Africa. Plant Dis. 100:522. doi: 10.1094/PDIS-06-15-0668-PDN
Periyanan, S., Moore, J., Ayliffe, M., Bansal, U., Wang, X., Huang, L., et al. (2013). The gene Sr33, an ortholog of barley Mla genes, encodes resistance to wheat stem rust race Ug99. Science 341, 786–788. doi: 10.1126/science.1239028
Peterson, R. F., Campbell, A. B., and Hannah, A. E. (1948). A diagrammatic scale for estimating rust intensity on leaves and stem of cereals. Can. J. Res. 26, 496–500. doi: 10.1139/cjr48c-033
Pretorius, Z. A., Singh, R. P., Wagoire, W. W., and Payne, T. S. (2000). Detection of virulence to wheat stem rust resistance gene Sr31 in Puccinia graminis f. sp. tritici in Uganda. Plant Dis. 84:203. doi: 10.1094/PDIS.2000.84.2.203B
Prins, R., Dreisigacker, S., Pretorius, Z., van Schalkwyk, H., Wessels, E., Smit, C., et al. (2016). Stem rust resistance in a geographically diverse collection of spring wheat lines collected from across Africa. Front. Plant Sci. 7:973. doi: 10.3389/fpls.2016.00973
Priyamvada, Saharan, M. S., and Tiwari, R. (2011). Durable resistance in wheat. Int. J. Genet. Mol. Biol. 3, 108–114. doi: 10.5897/IJGMB.9000018
Qi, L. L., Pumphrey, M. O., Friebe, B., Zhang, P., Qian, C., Bowden, R. L., et al. (2011). A novel Robertsonian translocation event leads to transfer of a stem rust resistance gene (Sr52) effective against race Ug99 from Dasypyrum villosum into bread wheat. Theor. Appl. Genet. 123, 159–167. doi: 10.1007/s00122-011-1574-z
Randhawa, M. S., Singh, R. P., Dreisigacker, S., Bhavani, S., Huerta-Espino, J., Rouse, M. N., et al. (2018). Identification and validation of a common stem rust resistance locus in two bi-parental populations. Front. Plant Sci. 9:1788. doi: 10.3389/fpls.2018.01788
Roelfs, A. P., Singh, R. P., and Saari, E. E. (1992). Rust Diseases of Wheat: Concepts and Methods of Disease Management. Mexico: CIMMYT.
Rouse, M. N., Nava, I. C., Chao, S., Anderson, J. A., and Jin, Y. (2012). Identification of markers linked to the race Ug99 effective stem rust resistance gene Sr28 in wheat (Triticum aestivum L.). Theor. Appl. Genet. 125, 877–885. doi: 10.1007/s00122-012-1879-6
Rouse, M. N., Nirmala, J., Jin, Y., Chao, S., Fetch, T. Jr., Pretorious, Z. A., et al. (2014a). Characterization of Sr9h, a wheat stem rust resistance allele effective to Ug99. Theor. Appl. Genet. 127, 1681–1688. doi: 10.1007/s00122-014-2330-y
Rouse, M. N., Talbert, L. E., Singh, D., and Sherman, J. D. (2014b). Complementary epistasis involving Sr12 explains adult plant resistance to stem rust in Thatcher wheat (Triticum aestivum L.). Theor. Appl. Genet. 127, 1549–1559. doi: 10.1007/s00122-014-2319-6
Rouse, M. N., Wanyera, R., Njau, P., and Jin, Y. (2011). Sources of resistance to stem rust race Ug99 in spring wheat germplasm. Plant Dis. 95, 762–766. doi: 10.1094/PDIS-12-10-0940
Saini, J., Faris, J. D., Zhang, Q., Rouse, M. N., Jin, Y., Long, Y., et al. (2018). Identification, mapping, and marker development of stem rust resistance genes in durum wheat ‘Lebsock’. Mol. Breed. 38:77. doi: 10.1007/s11032-018-0833-y
Simons, K., Abate, Z., Chao, S., Zhang, W., Rouse, M., Jin, Y., et al. (2011). Genetic mapping of stem rust resistance gene Sr13 in tetraploid wheat (Triticum turgidum ssp. durum L.). Theor. Appl. Genet. 122, 649–658. doi: 10.1007/s00122-010-1444-0
Singh, R. P., Hodson, D. P., Jin, Y., Huerta-Espino, J., Kinyua, M., Wanyera, R., et al. (2006). Current status, likely migration and strategies to mitigate the threat to wheat production from race Ug99 (TTKS) of stem rust pathogen. CAB Rev. Perspect. Agric. Vet. Sci. Nutr. Nat. Resour. 1, 1–13.
Singh, R. P., Hodson, D. P., Jin, Y., Lagudah, E. S., Ayliffe, M. A., Bhavani, S., et al. (2015). Emergence and spread of new races of wheat stem rust fungus: continued threat to food security and prospects of genetic control. Phytopathology 105, 872–884. doi: 10.1094/PHYTO-01-15-0030-FI
Singh, R. P., and McIntosh, R. A. (1986). Cytogenetical studies in wheat. XIV. Sr8b for resistance to Puccinia graminis f. sp. tritici. Can. J. Genet. Cytol. 28, 189–197. doi: 10.1139/g86-026
Stakman, E. C., Steward, D. M., and Loegering, W. Q. (1962). Identification of Physiologic Races of Puccinia graminis var. Tritici. Maryland: United States Department of Agriculture, Agricultural Research Service.
Taylor, J., and Butler, D. (2017). R package ASMap: efficient geneti linkage map construction and diagnosis. J. Stat. Softw. 79, 1–29. doi: 10.18637/jss.v079.i06
Tsilo, T. J., Jin, Y., and Anderson, J. A. (2007). Microsatellite markers linked to stem rust resistance allele Sr9a in wheat. Crop Sci. 47, 2013–2020. doi: 10.2135/cropsci2007.02.0087
Turner, M. K., Jin, Y., Rouse, M. N., and Anderson, J. A. (2016). Stem rust resistance in ‘Jagger’ winter wheat. Crop Sci. 56, 1719–1725. doi: 10.2135/cropsci2015.11.0683
Wang, S., Wong, D., Forrest, K., Allen, A., Chao, S., Huang, B. E., et al. (2014). Characterization of polyploid wheat genomic diversity using a high-density 90,000 single nucleotide polymorphism array. Plant Biotechnol. J. 12, 787–796. doi: 10.1111/pbi.12183
Wanyera, R., Kinyua, M. G., Jin, Y., and Singh, R. P. (2006). The spread of stem rust caused by Puccinia graminis f. sp. tritici, with virulence on Sr31 in wheat in Eastern Africa. Plant Dis. 90:113.
Wu, S., Pumphrey, M., and Bai, G. (2009). Molecular mapping of stem-rust resistance gene Sr40 in wheat. Crop Sci. 49, 1681–1686. doi: 10.2135/cropsci2008.11.0666
Wu, Y., Bhat, P. R., Close, T. J., and Lonardi, S. (2008). Efficient and accurate construction of genetic linkage maps from the minimum spanning tree of a graph. PLoS Genet. 4:e1000212. doi: 10.1371/journal.pgen.1000212
Zhang, W., Chen, S., Abate, Z., Nirmala, J., Rouse, M. N., and Dubcovsky, J. (2017). Identification and characterization of Sr13, a tetraploid wheat gene that confers resistance to the Ug99 stem rust race group. Proc. Natl. Acad. Sci. U.S.A. 114, E9483–E9492. doi: 10.1073/pnas.1706277114
Keywords: Triticum aestivum, Puccinia graminis f. sp. tritici, genetics, QTL, disease resistance, stem rust, Ug99
Citation: Kosgey ZC, Edae EA, Dill-Macky R, Jin Y, Bulbula WD, Gemechu A, Macharia G, Bhavani S, Randhawa MS and Rouse MN (2021) Mapping and Validation of Stem Rust Resistance Loci in Spring Wheat Line CI 14275. Front. Plant Sci. 11:609659. doi: 10.3389/fpls.2020.609659
Received: 23 September 2020; Accepted: 08 December 2020;
Published: 12 January 2021.
Edited by:
Jose Luis Gonzalez Hernandez, South Dakota State University, United StatesReviewed by:
Daryl LaVerne Klindworth, Edward T. Schafer Agricultural Research Center (USDA-ARS), United StatesCurt A. McCartney, University of Manitoba, Canada
Copyright © 2021 Kosgey, Edae, Dill-Macky, Jin, Bulbula, Gemechu, Macharia, Bhavani, Randhawa and Rouse. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zennah C. Kosgey, emVubmFoa0BnbWFpbC5jb20=; Matthew N. Rouse, bWF0dGhldy5yb3VzZUB1c2RhLmdvdg==
 Zennah C. Kosgey
Zennah C. Kosgey Erena A. Edae
Erena A. Edae Ruth Dill-Macky
Ruth Dill-Macky Yue Jin2,3
Yue Jin2,3 Worku Denbel Bulbula
Worku Denbel Bulbula Ashenafi Gemechu
Ashenafi Gemechu Godwin Macharia
Godwin Macharia Sridhar Bhavani
Sridhar Bhavani Mandeep S. Randhawa
Mandeep S. Randhawa Matthew N. Rouse
Matthew N. Rouse