
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 15 December 2020
Sec. Plant Metabolism and Chemodiversity
Volume 11 - 2020 | https://doi.org/10.3389/fpls.2020.608845
This article is part of the Research Topic Multiple Herbicide-Resistant Weeds and Non-target Site Resistance Mechanisms: A Global Challenge for Food Production View all 14 articles
 Laura Scarabel1*
Laura Scarabel1* Silvia Panozzo1
Silvia Panozzo1 Donato Loddo1
Donato Loddo1 Solvejg K. Mathiassen2
Solvejg K. Mathiassen2 Michael Kristensen2
Michael Kristensen2 Per Kudsk2
Per Kudsk2 Thomas Gitsopoulos3
Thomas Gitsopoulos3 Ilias Travlos4
Ilias Travlos4 Eleni Tani4
Eleni Tani4 Dimosthenis Chachalis5
Dimosthenis Chachalis5 Maurizio Sattin1
Maurizio Sattin1Annual ryegrass species (Lolium spp.) infest cereal crops worldwide. Ryegrass populations with multiple resistance to the acetyl coenzyme A carboxylase (ACCase) and acetolactate synthase (ALS) inhibitors are an increasing problem in several European countries. We investigated the resistance pattern and level of resistance in ryegrass populations collected in Denmark, Greece and Italy and studied the diversity of mechanisms endowing resistance, both target-site and metabolism based. All populations showed high resistance indexes (RI) to the ALS inhibitors, iodosufuron-methyl-sodium + mesosulfuron-methyl (RI from 8 to 70), whereas the responses to the two ACCase inhibitors, clodinafop-propargyl and pinoxaden, differed. The Greek and Italian populations were moderately to highly resistant to clodinafop (RI > 8) and showed low to moderate resistance to pinoxaden (RI ranged from 3 to 13) except for one Italian population. In contrast, the Danish Lolium populations showed low to moderate resistance to clodinafop (RI ranged from 2 to 7) and only one population was resistant to pinoxaden. Different mutant ACCase alleles (Leu1781, Cys2027, Asn2041, Val2041, Gly2078, Arg2088, Ala2096) and ALS alleles (Gly122, Ala197, Gln197, Leu197, Ser197, Thr197, Val205, Asn376, Glu376, Leu574) endowing resistance were detected in the Greek and Italian populations. In several plants, no mutated ALS and ACCase alleles were found showing a great heterogeneity within and among the Greek and Italian populations. Conversely, no mutant ACCase alleles were identified in the four Danish populations and only one mutant ALS allele (Leu574) was detected in two Danish populations. The expression level of nitronate monooxygenase (NMO), glutathione S-transferase (GST) and cytochrome P450s (CYP72A1 and CYP72A2) varied broadly among populations and individual plants within the populations. Constitutive up-regulation of GST, CYP72A1 and CYP72A2 was detected in resistant plants respect to susceptible plants in one Danish and one Italian population. It appears that the mechanisms underlying resistance are rather complex and diversified among Lolium spp. populations from the three countries, coevolution of both target-site resistance and metabolic based herbicide resistance appears to be a common feature in Denmark and Italy. This must be considered and carefully evaluated in adopting resistance management strategies to control Lolium spp. in cereal crops.
Ryegrass species (Lolium spp.) are obligate out-crossers with high genetic variability and fecundity (Pedersen et al., 2007; Holt et al., 2013). They are common weeds in many European countries and infest numerous cropping systems, including cereal crops, where they are considered a threat for the sustainability of cereal production. Historically, the control of Lolium spp. has been carried out with herbicides inhibiting acetyl coenzyme-A carboxylase (ACCase) but several populations have evolved resistance to this herbicide group. It is recognized that the diversification of the selection pressure by using herbicides with different sites of action is a key point for resistance management. Hence, the subsequent registration of acetolactate synthase (ALS) inhibitors introduced another herbicide site of action (SoA) to overcome this problem. These herbicides, able to control broad-leaved weeds as well as some grass species including Lolium spp., have been widely used by cereal growers. The recurrent treatments with herbicides having the same SoA have selected resistant Lolium spp. populations and this significantly reduces the number of herbicides available to control these weed species.
Several resistance mechanisms have been reported for Lolium spp. Among them, gene mutations reducing or blocking herbicide binding by conferring amino-acid changes in a target protein (Target Site Resistance, TSR) and enhanced metabolism causing accelerated herbicide degradation (one of the non-target-site resistance, NTSR mechanisms) are the main mechanisms in grass weeds such as Lolium spp. (Powles and Yu, 2010; Délye et al., 2013). TSR has been extensively studied in the last 20 years and many of the known mutations endowing herbicide resistance in the ALS (Yu and Powles, 2014) and ACCase (Kaundun, 2014) genes have been found in Lolium spp. populations (Tan et al., 2007; Scarabel et al., 2011; Han et al., 2016). Early works have established the presence of metabolic resistance to diverse herbicides such as chlorsulfuron, chlorotoluron in Lolium rigidum populations from Australia (Cotterman and Saari, 1992; Preston et al., 1996) and also the presence of both TSR and enhanced metabolism-based resistance (hereinafter referred as EMR) mechanisms in the same plant (Christopher et al., 1992; Tardif and Powles, 1994). From then on, EMR has been understudied and only recently, it was recognized as the predominant type of resistance to ACCase- and ALS inhibitors in grasses (Gaines et al., 2020). EMR, differently to TSR, can confer cross-resistance to herbicides with different SoA, including herbicides to which weeds have not been previously exposed (Petit et al., 2010). EMR is considered to be polygenically inherited, involving multiple genes encoding for metabolic enzymes such as cytochrome P450 monooxygenase (P450), glucosyl transferases (GT), glutathione S-transferases (GST), esterases and ABC transporters (Yuan et al., 2007; Duhoux and Délye, 2013; Duhoux et al., 2017). Four consistently over-expressed genes were identified in resistant individuals of Lolium rigidum, a close relative of Lolium multiflorum. These included two P450s, one nitronate monooxygenase (NMO) and one GT (Gaines et al., 2014, 2020). In L. multiflorum higher expressions of these four metabolism-related genes were reported in individuals of resistant populations from Denmark (Mahmood et al., 2016).
In Greece, the first case of L. rigidum resistant to both diclofop-methyl (ACCase inhibitor) and chlorsulfuron (ALS inhibitor) was reported in 2000 (Kotoula-Syka et al., 2000). Subsequently, other populations resistant only to chlorsulfuron were found and the resistance mechanism was attributed to enhanced activity of P450 in some populations and to TSR in others (Kaloumenos et al., 2012). In Italy, the first ACCase-resistant Lolium spp. population was recorded in the mid-1990s in central Italy (Bravin et al., 2001). Since then, ACCase-resistant cases have also spread to northern Italy and a few years ago Lolium spp. populations resistant to both ALS and ACCase inhibitors were recorded (Collavo et al., 2013) and they are now increasing. In Denmark, herbicide resistance in Lolium spp. appeared later than in Italy and Greece with the first case of Lolium multiflorum resistant to an ALS-inhibitor (iodosulfuron) registered in 2010 (Mathiassen, 2014). Since then, cases have been increasing and the first case of multiple-resistance to ACCase and ALS inhibitors was reported in 2010 (Heap, 2020).
Lolium spp. is very prone to evolve resistance and multi-resistant cases are increasing in the three countries. It was also shown that metabolic resistance evolves rapidly in L. rigidum when herbicides are used at low or suboptimal doses and this is an important point to consider for weed management (Neve and Powles, 2005; Manalil et al., 2011). Efforts should therefore be made to limit the evolution of resistance and to ensure the sustainability of cereal crops production. The aims of this work were (1) to determine the level of resistance to ALS and ACCase inhibitors through bioassays in twelve Lolium spp. populations collected in Denmark, Greece, and Italy; (2) to investigate the resistance mechanisms involved, both the detection of ALS and ACCase alleles endowing TSR and the presence of EMR mechanism. For the latter purpose, the gene expression of four herbicide metabolism related genes (NMO, GST, P450s CYP72A1 and CYP72A2) was investigated through qPCR.
Seeds from Lolium spp. plants were collected in winter cereal fields from three European countries (Denmark, Greece, and Italy) where the control of these grass species by ALS and ACCase inhibitors was poor. After a preliminary screening of the populations, conducted in each country, 12 populations, four from each country, with a high frequency of plants resistant to both ALS and ACCase inhibitors were selected (Table 1). Additionally, susceptible reference populations from each of the three countries were included. All seed samples were cleaned and stored in paper bags at 4°C until use.
Two whole-plant dose-response experiments were carried out at Legnaro (PD), Italy (45° 21′ N, 11° 58′ E). The experiments were performed as outdoor pot experiments during spring 2018 and autumn 2018 using commercial formulations of ALS and ACCase inhibitor herbicides.
To break dormancy, seeds were placed in Petri dishes on wet filter paper and vernalized at 4°C under dark conditions for 3 days. Then, seeds were placed in a germination cabinet and kept for 5 days at 25/15°C (day/night) with a 12 h photoperiod. Germinated seedlings at similar growth stages were transplanted into 16 cm diameter pots filled with a standard potting mixture (60% silty loam soil, 15% sand, 15% perlite, and 10% peat). The pots were placed outside in a semi-controlled environment and watered regularly to maintain the substrate at field capacity.
The experimental layout was a completely randomized design with three biological replicates (pots), each one with 9 seedlings. Herbicide application was carried out at BBCH 21-22 (Hess et al., 1997). All populations were treated with two ACCase inhibitors (pinoxaden and clodinafop-propargyl) and one of ALS inhibitor (mesosulfuron-methyl + iodosulfuron methyl-sodium) (Table 2). Herbicides were applied using a precision bench sprayer delivering 300 L ha–1 at a pressure of 215 kPa and speed of about 0.75 ms–1 with a boom equipped with three flat-fan (extended range) hydraulic nozzles (Teejet®, 11002). The herbicide doses applied ranging from 1/16 N to 4 N for the susceptible populations and from 1/8 N to 8 N for the resistant populations with N being the recommended field dose in Italy.
Four weeks after treatment, plant survival and shoot fresh weight were recorded for each pot and expressed as percentage of the mean of the non-treated control.
The dose-response data were analyzed using a non-linear regression analysis based on the log-logistic equation: Y = C + [(D - C)/[1 + (x/I50)b] where Y is the fresh weight or survival, C and D are the lower and upper asymptotes at very high and infinitely low doses, respectively, b is the slope of the curve around its inflection point, I50 is the dose giving a response equivalent to midway between the D and C parameters and x is the herbicide dose (g a.i.ha–1) (Seefeldt et al., 1995).
The ED50 (herbicide dose causing 50% plant mortality), GR50 (herbicide dose causing 50% reduction in fresh weight) and relative standard errors were estimated using GraphPad Prism 8 (GraphPad Software Inc., San Diego, CA, United States). For biological reasons and to improve the best-fit values of the parameters, the lower and upper asymptotes of plant survival and fresh weight data were constraints to 0 and 100%, respectively (Onofri, 2005). Data of each population and for each herbicide were analyzed together and an extra sum-of-squares F test, available in GraphPad Prism 8, was performed to determine if the data of the two experiments could be pooled, i.e., if one curve adequately fitted both data set.
Resistance indexes (RIs) were calculated as the ratio between the ED50 (or GR50) of the resistant and the susceptible population separately for each country. When the ED50 (or GR50) could not be determined because plant survival (or fresh weight) was higher than 50% even at the highest herbicide doses, the maximum applied dose was used as a proxy for ED50/GR50.
Five to ten ALS-resistant plants from the 12 Lolium populations were analyzed to detect the presence of mutant ALS and ACCase alleles. When no ACCase mutant alleles were detected another five ACCase-resistant plants were genotyped to confirm or reject the absence of mutant alleles indicating a putative non-target site resistance.
Total genomic DNA (gDNA) was extracted from 0.1 g leaf tissue using the CTAB method (Doyle and Doyle, 1987). A 1600 bp region of the CT domain of the plastidic ACCase gene was amplified by PCR on gDNA using the primers acclr9 and acclr6 (Table 3). The amplified region encompassed all the amino acid substitutions so far identified as conferring resistance. PCR amplifications were performed using GoTaq DNA Polymerase kit (Promega, United States) in a 25 μL mixture including 5 μL of 5 × Colorless GoTaq Flexi Buffer, dNTPs mix (0.2 mM each), MgCl2 (3 mM), forward and reverse primers (0.4 μM each), 0.125 μL GoTaq DNA Polymerase, and 25 ng of gDNA. The thermocycler program was as follows: 95°C for 2 min; 45 cycles of 95°C for 30 s, 57°C for 30 s, 72°C for 2 min; 72°C for 5 min. PCR products were purified with NucleoSpin Gel and PCR Clean-up kit (Macherey-Nagel GmbH & Co., Germany) following the manufacturer’s instructions. Once purified, PCR products obtained from each plant were sequenced by BMR Genomics (Padova, Italy) using primers LOL_FOR and LOL_FOR_SEQ (Table 3).
Similarly, a 1719 bp fragment of the ALS gene was amplified from each DNA extracted with primers LOL_ALS_F and ALS_LOL_R reported in Table 3. PCR amplifications were performed using GoTaq DNA Polymerase kit (Promega, United States) in a 25 μL total volume containing 5 μL of 5× Colorless GoTaq Flexi Buffer, 5% of DMSO (1.25 μL), dNTPs mix (0.2 mM each), MgCl2 (4 mM), forward and reverse primers (0.4 μM each), 0.125 μL GoTaq DNA Polymerase, and 25 ng of gDNA. The thermocycler program was as follows: 95°C for 2 min; 45 cycles of 95°C for 30 s, 60°C for 30 s, 72°C for 2 min; 72°C for 5 min. PCR products were purified as described for ACCase gene and sequenced by BMR Genomics using primers LOL_ALS_F and ALS_LOL_FS (Table 3).
Ten populations of Lolium spp. were chosen for this study, four susceptible populations and six resistant populations (DK29, DK90, GR24, GR30, IT533, and IT609) that had no (or sporadic) ALS and ACCase mutated plants. Resistant plants without mutant ALS and ACCase alleles were determined as described in the previous paragraph. All populations were sown in trays placed in a glasshouse at Flakkebjerg, Denmark. At BBCH 21, twenty plants from each population were separated into two individual plants (i.e., two clones) and transplanted into pots. A week later, one clone from each plant was sprayed with mesosulfuron-methyl + iodosulfuron-methyl sodium at 30 + 6 g ha–1 while the other clone remained non-treated. Plant responses to herbicide treatment was visually assessed 4 weeks after application. The treated clones were rated as susceptible or resistant while the non-treated clones were cut at the soil surface and frozen in liquid nitrogen immediately after harvest for subsequent gene expression analysis.
Total RNA was extracted from 50 mg of leaf material of three individual plants of the ten populations using RNeasy Plant Mini Kit (Qiagen, Stanford, CA, United States). The quality and concentration of RNA samples were determined as reported by Mahmood et al. (2016).
The qPCR reactions were performed with the GoTaq 1-step RT-qPCR System (Promega, Madison, WI, United States) using an Applied Bioscience ViiATM7 real-time PCR system with 384 wells (Thermo Fisher Scientific, Waltham, MA, United States). Reactions were performed in triplicates and a negative control consisting of reaction mix without template was also included for each primer. Briefly, 20 μL reaction mix included 10 μL GoTaq qPCR MasterMix 2×, 4 μL RNA template, 4 μL 0.5 pmol primers (1:1 mix of forward and reverse primers), 0.4 μL GoScript RT Mix 1-Step RT-qPCR 50x, 1.6 μL nuclease-free distilled water. Reaction conditions included 50°C for 5 min followed by 10 min incubation at 95°C, then 40 cycles of 95°C for 15 s and 60°C for 1 min. One internal control gene Rab GTPase (RGTP) and four herbicide metabolism genes NMO, GST, CYP72A1, and CYP72A2 were chosen. Primer sequences are identical to those described by Gaines et al. (2014) and available in Mahmood et al. (2016).
Threshold-cycle (Ct) values were calculated for each reaction. Gene-specific PCR efficiency was used to calculate the expression of target genes in relation to the expression of internal reference gene. Equivalent slopes for target and internal control gene were observed in amplification plots. The ΔCt value was calculated as follows: ΔCt (target genes) = Ct (target gene) – Ct (reference gene), where Ct is the cycle number at which PCR product exceeded a set threshold. Relative transcript level (RTL) was calculated through = 1 × 2–ΔCt (Pfaffl, 2006). The significance levels for each gene expression were calculated for all pairwise comparisons through a single factor ANOVA followed by Tukey HSD (Honestly Significant Difference) test.
The extra sum-of-squares F test conducted on the survival and fresh weight data of each population to compare the dose-response curves obtained in the two experiments indicated that most of the curves were significantly different at p < 0.05. Therefore, it was not possible to estimate a common curve except for the survival data obtained with mesosulfuron + iodosulfuron (Table 4).
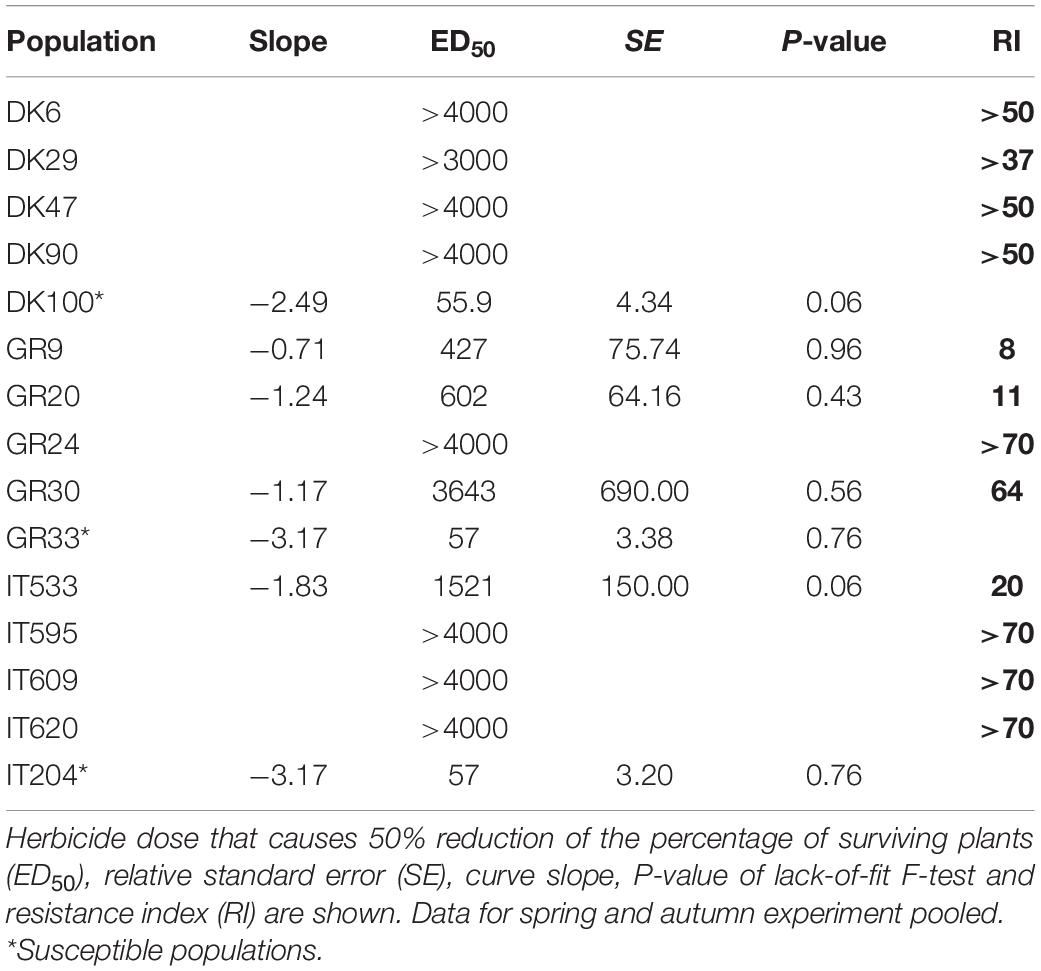
Table 4. Parameter estimates of the dose-response of Atlantis WG (field dose is 500 g ha–1 = 15 g mesosulfuron + 3 g iodosulfuron ha–1) on Lolium spp. populations.
For mesosulfuron + iodosulfuron the estimated ED50 values of the three susceptible reference populations were similar, 55.9 g ha–1 (=1.68 g mesosulfuron + 0.33 g iodosulfuron) for population DK100 and 57 g ha–1 (=1.71 g mesosulfuron + 0.34 g iodosulfuron) for populations GR33 and IT204 (Table 4). The four Danish populations exhibited high level of resistance and even at the highest herbicide dose tested (120 + 24 g a.i. ha–1 of mesosulfuron + iodosulfuron) plant survival was higher than 50%. Similarly, the four Italian populations were highly resistant with RIs > 70 for three populations IT595, IT609, IT620 and RI = 20 for population IT533, while the Greek populations showed RIs ranging from 8 to >70.
The reference populations showed higher ED50 values with clodinafop, ranging from 29 to 55 g a.i. ha–1, in the spring experiment than in the autumn experiment where the ED50 values ranged from 15 to 19 g a.i. ha–1. In general, the RIs of all populations were higher in the autumn experiment compared to the spring experiment, however, the ranking of populations was consistent. The four Danish populations showed low to moderate resistance to clodinafop. The RIs for plant survival were between 2.3 and 6.8 in the spring experiment and between 3.4 and 7.3 in the autumn experiment. The RIs based on fresh weight recorded slight differences between both experiments with RIs ranged between 0.9–4.8 and 0.6–8.5 in the spring and autumn experiments, respectively (Table 5).
All four Greek populations were highly resistant to clodinafop with ED50 and GR50 values that corresponded to the higher doses tested (i.e., 480 g a.i. ha–1) in both experiments. The resulted RIs were around 17 for both survival and fresh weight in the spring experiment and >32 or >135 in the autumn experiment. Similarly, all four Italian populations were highly resistant to clodinafop with higher values of RI recorded for the autumn experiment (Table 5).
Overall, the reference populations were completely controlled at half the recommended dose of pinoxaden (i.e., 22.5 g a.i. ha–1). The ED50 values of the three reference populations ranged between 9 and 12 g a.i ha–1 in both experiments. Three Danish populations (DK6, DK29, and DK47) showed a slight shift in the susceptibility to pinoxaden with RIs ranging from 1.1 to 2.6 based on plant survival and 0.9 to 3.1 based on fresh weight in both experiments while the fourth population DK90 had a higher RI. Three out of four Greek populations (GR9, GR24, GR30) were highly resistant to pinoxaden, with RIs ranging from 6 to 12.6 and from 19 to 39, based on survival in the spring and autumn experiment, respectively. The fourth population (GR20) had a lower resistance level respect to the other Greek populations (RI = 3.1 in the spring experiment and 11.4 in the autumn experiment). Finally, three of the four Italian populations (IT533, IT609, and IT620) were moderately resistant to pinoxaden, with RIs ranging between 6.3 and 7 for plant survival and between 2 and 9.2 for fresh weight. The RI values based on fresh weight were similar for the autumn experiment while the RIs based on survival were higher (Table 6).
Primers acclr9/acclr6 amplified a 1600 bp amplicon encompassing all codons of the ACCase gene known to confer resistance. Similarly, primers LOL_ALS_F/ALS_LOL_R amplified a 1719 bp amplicon encompassing all codons of the ALS gene conferring resistance. Both amplicons were sequenced for all the 83 plants (Table 7).
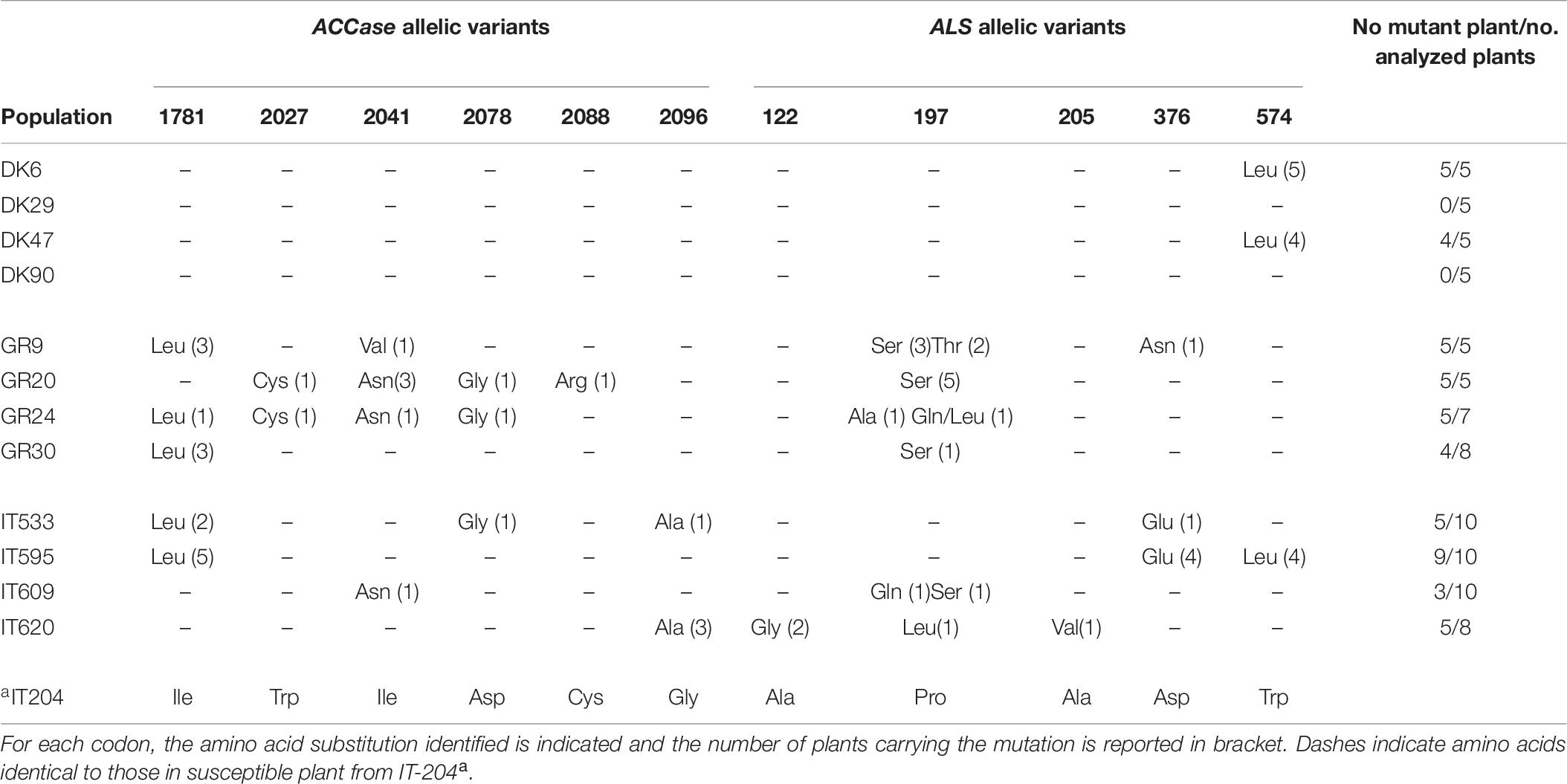
Table 7. ALS and ACCase allelic variants identified in resistant Lolium spp. plants from Danish, Greek, and Italian populations compared to the susceptible plant.
The sequencing of ACCase amplicons revealed that all the Greek and Italian populations had plants with a mutated ACCase allele (Table 7). Overall, in the Greek populations, six different ACCase mutant alleles were detected: Leu1781, Cys2027, Asn2041, Val2041, Gly2078, and Arg2088. In population GR30 only one type of mutant ACCase allele was detected (Leu1781), in population GR9 two types (Leu1781 and Val2041), in population GR20 four types (Cys2027, Asn2041, Gly2078, and Arg2088) as in population GR24 (Leu1781, Cys2027, Asn2041, and Gly2078). In addition, different mutant ACCase alleles were found in the same plant of population GR20.
Four different types of ACCase mutant alleles were found in plants from Italy. Populations IT595, IT609 and IT620 showed only one ACCase mutant allele each – Leu1781, Asn2041 and Ala2096, respectively. In population IT533, three mutant alleles were identified: Leu1781, Gly2078, and Ala2096. In contrast, no mutant ACCase alleles were identified in any of the 20 plants analyzed from the Danish populations. It is noteworthy that several plants with no mutated ACCase allele were also identified within two Greek populations (GR24 and GR30) and in all the Italian populations (Table 7).
Six types of mutant ALS alleles (Ala197, Gln197, Leu197, Ser197, Thr197, and Asn376) were identified in the plants of the Greek populations. In populations GR20 and GR30 only one type of ALS mutant allele was detected (Ser197), while in populations GR9 three types (Ser197, Thr197, and Asn376) and in GR24 three types (Ala197, Gln197, and Leu197) were identified. In three of the four Italian populations more than one mutant ALS allele was detected: Glu376 and Leu574 in IT595, Gln197 and Ser197 in IT609 and Gly122, Leu197 and Val205 in IT620. The fourth population, IT533, had only the Glu376 allelic variant. Conversely, the plants of the Danish populations showed only one type of mutant ALS allele (Leu574), and only in two populations (DK6 and DK47) (Table 7). The ALS and ACCase sequences presented can be found in a specific repository (Panozzo and Scarabel, 2020).
Six of the resistant populations (DK29, DK90, GR24, GR30, IT533, and IT609) that had plants with no ALS and ACCase mutant alleles were further studied to determine the expression patterns of four genes, NMO, GST, CYP72A1, CYP72A2 known to be involved in herbicide metabolism. Four susceptible Lolium spp. populations were also examined (DK39, DK100, DK22M and DK22P).
For all four genes, significant differences between plants of the same population were observed. For example, the expression of NMO in plant DK100-1 was around 5.5 fold higher than in plants 2 and 3 (Figure 1A) and the expression in plant IT609-3 was 9 and 3.7 fold higher respect to plant 1 and 2, respectively. These differences were observed within all populations, except population IT533, implying that in general Lolium populations are heterogeneous for the gene expression of one or more herbicide metabolism genes. No significant differences in the expression of NMO were observed between susceptible and resistant populations (Figure 1A).
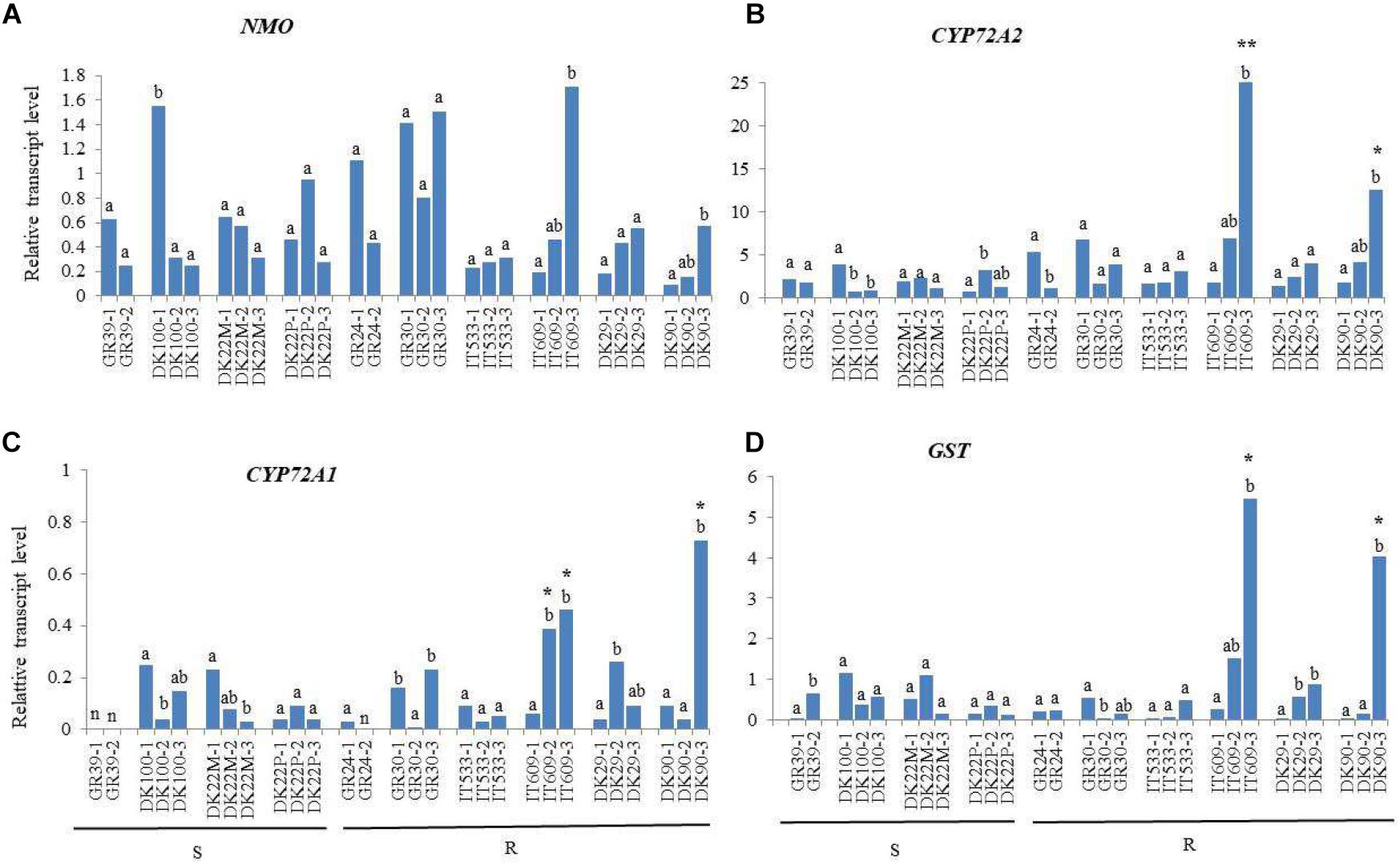
Figure 1. Relative transcript levels of NMO (A), CYP72A2 (B), CYP72A1 (C), and GST (D) in leaves of Lolium spp. plants resistant (R) or susceptible (S) to mesosulfuron-methyl + iodosulfuron-methyl relative to the internal reference gene RGTP. Significant differences between plants of the same population are indicated with different letters. Asterisks denote significant differences between R plant compared to all S plants tested (**) or compared to plants of only one S population (*). n = samples with Ct values below no template control.
Among the genes studied, CYP72A2 was expressed at the highest level in the resistant plant (IT609-3) with a relative expression value of 24.99 while the lowest expression was found in the susceptible plant DK22P-1 with RTL value of 0.72. The expression of CYP72A2 was significantly higher in plant IT609-3 compared to all the susceptible plants analyzed, and in the resistant plant DK90-3 compared to plants from the susceptible population DK22M, from 5 to 11-fold up-regulated. Other resistant plants, GR24-1, GR30-1 and IT609-2 exhibited higher expression respect to few susceptible plants. For example, CYP72A2 was 9.4 fold higher expressed in plant GR30-1 respect to the susceptible plant DK22P-1 but only 1.8 fold higher respect to plant DK100-1 (Figure 1B).
The CYP72A1 gene was expressed at a relatively low level as indicated by its generally low RTL values ranging from 0.03 to 0.73 However, a significant (p < 0.001) increased expression was found in plants IT609-2, IT609-3 and DK90-3. CYP72A1 was 8-fold up-regulated in the resistant plant IT609-3 compared on average to the susceptible plants of population DK22P and 7-fold up-regulated in the resistant plant IT609-2 compared to the same susceptible population. Plant DK90-3 exhibited the highest relative expression value for CYP72A1 (0.73) and the gene was 13-fold up-regulated compared on average to the susceptible plants of population DK22P and was also significantly up-regulated respect to plants DK100-2, DK100-3, DK22M-2 and DK22M-3 with 18, 5, 9 and 24-fold up-regulation, respectively (Figure 1C).
The GST gene showed significantly (p < 0.001) up-regulation in plant IT609-3 and DK90-3, whereas among the susceptible plants, population DK22P showed very low relative expression (on average RTL value of 0.19) as well as plant GR39-1 (RTL = 0.04) and DK22M-3 (RTL = 0.15). GST was 30-fold up-regulated in the resistant plant IT609-3 compared on average to the susceptible plants of population DK22P and was also significantly up-regulated compared to some plants of other susceptible populations, GR39-1, DK22M-1, DK22-M3 and DK100-2 with 136, 11, 36 and 14-fold up-regulation, respectively. Similarly, GST was 22-fold up-regulated in plant DK90-3 in comparison with the susceptible plants of population DK22P and was also significantly up-regulated compared to some plants of other susceptible populations, GR39-1, DK22-M3 and DK100-2 with 101, 27 and 11-fold up-regulation, respectively (Figure 1D). GST showed the highest differences in expression between susceptible and resistant populations respect to the other genes studied.
This study confirmed the occurrence of Lolium spp. populations multi-resistant to ALS and ACCase inhibitors in Denmark, Italy and Greece and highlighted differences in the pattern and level of resistance among countries and populations. TSR appears to be responsible for the resistance status of Greek populations, and for most of the Italians. Conversely, resistance in Danish populations is totally endowed by NTSR mechanism in case of ACCase inhibitors and by both NTSR and TSR in case of ALS inhibitors.
While all populations were highly resistant to mesosulfuron-methyl + iodosulfuron-methyl, the susceptibility to both ACCase inhibitors, clodinafop-propargyl and pinoxaden was generally much lower in the Danish populations compared to the Italian and Greek ones. A low resistance level has often been associated to the presence of non-target-site resistance mechanisms, however, in some cases the level of resistance can be higher due to the build-up over time of different NTSR mechanisms (Cocker et al., 2001; Kaundun, 2014). Among the investigated populations, the level of resistance to pinoxaden varied among populations in each country and among countries. These differences in the pattern and level of resistance could be related to different cropping practices and herbicides used in the three countries (Llewellyn and Powles, 2001; Owen et al., 2014). The herbicide pinoxaden is not authorized in Denmark while it is frequently used as a post-emergence application in Italian and Greek winter cereals fields. This supports the higher resistance indexes generally observed in the Italian and Greek populations. However, even if pinoxaden is not used, a low resistance level to pinoxaden has been detected in one Danish population. Instead, a low but clear resistance to clodinafop was found. This type of resistant phenotype as well as the absence of mutated ACCase alleles strongly suggests the presence of a NTSR mechanism.
ACCase variant alleles endowing resistance were present in all the Greek and Italian populations studied. Overall, six different types of ACCase variant alleles were detected in the Greek populations: Leu1781, Cys2027, Asn2041, Val2041, Gly2078, and Arg2088 and four in the Italian populations: Leu1781, Asn2041, Gly2078, and Ala2096. Depending on the population considered, one to four different ACCase alleles were observed in the same population. Similar results were reported by Yu et al. (2007) who found different ACCase mutant alleles in Italian Lolium populations resistant to clethodim and showed that homozygous plants having Leu1781, Gly2078 or Arg2088 were also resistant to other ACCase inhibitors including clodinafop and pinoxaden. In a subsequent study conducted on Italian Lolium spp. populations resistant to pinoxaden, the same ACCase variant alleles as detected in our work were found, except for Cys2027 (Scarabel et al., 2011). In contrast, in all the four Danish populations investigated no ACCase variant alleles were detected indicating that target site resistance is not present.
The analyses of the ALS gene indicated that only one ALS variant allele (Leu574) endowing resistance to mesosulfuron-methyl + iodosulfuron-methyl was present in two Danish populations while in the other two, no ALS variants were detected. Conversely, in the Greek and Italian populations different ALS variant alleles were found, six (Ala197, Gln197, Leu197, Ser197, Thr197, Asn376) in the Greek populations and seven in the Italian ones (Gly122, Gln197, Leu197, Ser197, Val205, Glu376, Leu574) and diversity of ALS alleles was detected in some populations. The Italian populations showed amino acid substitutions at five different codons of the ALS gene. This is in accordance with the study of Yu et al. (2008), who, in a single Australian population, identified six different mutations in the ALS gene endowing resistance to chlorsulfuron. This is not surprising as Lolium spp. are obligate cross-pollinated species and therefore pollination among resistant plants from neighboring fields can occur within 3 km distance increasing the genetic heterogeneity of the Lolium plants (Busi et al., 2008). In the Greek populations, the majority of amino acid substitutions endowing resistance was observed at codon Pro-197 and this is in accordance with previous findings (Kaloumenos et al., 2012; Anthimidou et al., 2020). This substitution was frequently reported in numerous grass weeds and it usually confers resistance only to sulfonylureas (such as mesosulfuron-methyl + iodosulfuron-methyl) (Yu and Powles, 2014). Instead, the Leu574 ALS allele, present only in two Danish populations, endows high resistance to all chemical group of ALS inhibitors (Heap, 2020). The variability in the ALS mutations detected in the three countries confirms the differences observed in the cross-resistance pattern.
Some plants in two Greek populations (GR24 and GR30) and in all the Italian populations showed no amino acid substitutions endowing resistance to ALS and ACCase. This suggests that a different resistance mechanism (i.e., NTS) is likely present.
Enhanced metabolism-based resistance is considered the prevalent resistance mechanism in grass weeds and its complex genetic control (polygenic control) involves the regulation of specific genes (Délye, 2013). The cytochromes P450 belong to a supergene family and are involved in all the pathways of plant secondary metabolism (Werck-Reichhart and Feyereisen, 2000). They play a major role in the phase I of metabolic herbicide detoxification and in the coordination with the GST enzymes involved in phase II of herbicide detoxification (Cocker et al., 2001; Yuan et al., 2007). GSTs include a large, complex gene family in plants that catalyze the conjugation to various substrates and oxidatively produced compounds to reduced glutathione, which facilitates their metabolism and sequestration (Dalton et al., 2009). Nitronate monooxygenase is a flavin-dependent enzyme that oxidizes anionic alkyl nitronates. It is active on a broad range of substrates containing primary and secondary nitro groups and its involvement in detoxification of propionate-3-nitronate was reported in yeast and bacteria (Gadda and Francis, 2010). In A. thaliana, NMO was found to be associated with detoxification of the allelochemical benzoxazolin (Baerson et al., 2005).
The high expression of CYPs and GST is expected to enhance herbicide degradation in resistant plants (Délye, 2013). The expression level of both CYPs studied (CYP72A2 and CYP72A1) varied broadly from plant to plant and the same was observed for the gene GST. This variability of expression between plants implies that the populations are generally heterogeneous for the gene expression of one or more herbicide metabolism genes. Similar findings were reported by Duhoux and Délye (2013) who reported that the expression of five CYP genes, both constitutive and herbicide-induced, varied broadly from plant to plant in a French Lolium spp. population. Despite the variability among plants, our data showed that the expression levels of CYP72A1, CYP72A2, and GST were significantly higher in resistant plants in population IT609 and DK90 compared to the expression level of the susceptible plants. This proves that an enhanced herbicide degradation is present in these plants and highlights the evolution of metabolic based resistance in these Lolium populations. CYP72A gene was found to be involved in metabolic resistance to diclofop in L. rigidum (Gaines et al., 2014). Moreover, enhanced GST expression was shown to determine an acceleration in the herbicide degradation in a clodinafop-resistant L. rigidum population (Gaines et al., 2014) and in Danish L. multiflorum population (Mahmood et al., 2016).
No clear distinction in NMO expression was observed between resistant and susceptible plants. This gene was identified as candidate gene by RNAseq transcriptome analysis involved in metabolism-based diclofop resistance in L. rigidum (Gaines et al., 2014). NMO was found to be two-fold up-regulated in glufosinate-resistant Amaranthus palmeri respect to susceptible plants, both constitutively and herbicide-induced (Salas-Perez et al., 2018).
The no clear distinction of expression in the genes studied that are specific to the resistant plant may be related to the fact that additional genes not studied here are involved in the herbicide metabolization. Two cytochrome P450 genes (CYP81A12 and CYP81A21) were found to be overexpressed and associated with resistance to ALS inhibitors in Echinochloa phyllopogon (Iwakami et al., 2014). Recently, these genes were found to be involved in the detoxification of ACCase inhibitors in multiple herbicide resistant E. phyllopogon (Iwakami et al., 2019). Because of the high number and variability of CYPs and GSTs, plants have the potential to overcome the herbicide treatment trough a concerted action of several genes. It was demonstrated that the genetic control of P450 metabolism-based resistance mechanism in Lolium rigidum is governed by a set of genes that varied among plants, even in a given population (Busi et al., 2011).
In conclusion, in the present plant material, IT609-3 and DK90-3 exhibited high expression of GST, CYP72A1, and CYP72A2 genes constitutively, implying that herbicide resistance for these populations could be attributed to an elevated level of herbicide metabolism.
In the last decade there has been an increase in multi-resistant Lolium spp. populations in Europe (Heap, 2020). This work highlights the presence of a wide variety of multi-resistant Lolium spp. populations in Denmark, Greece, and Italy and provides strong evidence that two different resistance mechanisms (TSR and NTSR metabolism-based resistance) co-evolved.
A diversity of mutant ALS and ACCase alleles were detected among plants of the Greek and Italian populations indicating a high population heterogeneity. In the Danish populations, only one type of mutant ALS allele was found. However, in both situations, evolution of target-site resistance is suggested to be the result of a strong selection pressure imposed by the herbicides used. The frequent use of the ALS inhibitor, mesosulfuron + iodosulfuron and lower use of ACCase inhibitors in Danish cereals fields compared with the common herbicide usages in Italy and Greece may explain this difference.
The high variability among plants observed in the expression profile of the four genes involved in the metabolism of mesosulfuron + iodosulfuron indicates that even if plants are subjected to the same herbicide selective pressure, a different weed response to the chemical agent can occur. It is likely that since EMR is a polygenic trait, the evolution process over time allows accumulation of various favorable traits able to increase the survival of the plant and the transmission to the next generation.
The presence of different resistance mechanisms increases the complexity of resistant Lolium spp. management in cereal fields. From a practical point of view, TSR determines resistance to herbicides with the same SoA, while metabolism-based resistance can not only endow resistance to the selecting herbicides but also confers cross-resistance to herbicides having different SoA. Numerous genes can be involved in metabolism-based resistance and, according to their regulation, they can confer resistance to different chemicals (Gaines et al., 2014). Therefore, EMR mechanism observed in some Lolium populations investigated in this study is of concern because the simple rotation over the years of ALS and ACCase inhibiting herbicides will not be effective, as pointed out by Collavo et al. (2013).
This work displays the heterogeneity in the pattern and level of resistance to ALS and ACCase inhibitors in Danish, Greek, and Italian Lolium spp. populations and demonstrates the presence of target-site resistance and coexistence of metabolic based herbicide resistance mechanism in populations from Denmark and Italy. The potential of evolution of enhanced metabolism-based resistance is an important threat to consider for improving practices against resistance.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
All authors contributed to the design of this study, shared plant materials, contributed to give comments, revised the manuscript, and read and approved the final manuscript. DL, LS, MK, SM, and SP performed the experiments and analyzed the data. LS wrote the first draft of the manuscript.
This research (project RELIUM) was funded by the Italian Ministry of Agriculture and Forestry Politics (MIPAAF), the Danish Environmental Protection Agency (DEPA), and the Hellenic Agricultural Organisation (DEMETER) under the umbrella of the ERA-Net C-IPM (Coordinated Integrated Pest Management in Europe) funded by the European Union, Grant agreement No. 618110.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.608845/full#supplementary-material
Anthimidou, E., Ntoanidou, S., Madesis, P., and Eleftherohorinos, I. (2020). Mechanisms of Lolium rigidum multiple resistance to ALS and ACCase inhibiting herbicides and their impact on plant fitness. Pest. Biochem. Physiol. 164, 65–72. doi: 10.1016/j.pestbp.2019.12.010
Baerson, S. R., Sanchez-Moreiras, A., Pedrol-Bonjoch, N., Schulz, M., Kagan, I. A., Agarwal, A. K., et al. (2005). Detoxification and transcriptome response in Arabidopsis seedlings exposed to the allelochemical benzoxazolin-2(3H)-one. J. Biol. Chem. 280, 21867–21881. doi: 10.1074/jbc.M500694200
Bravin, F., Zanin, G., and Preston, C. (2001). Diclofop-methyl resistance in populations of Lolium spp. from central Italy. Weed Res. 41, 49–58. doi: 10.1046/j.1365-3180.2001.00217.x
Busi, R., Vila-Aiub, M. M., and Powles, S. (2011). Genetic control of a cytochrome P450 metabolism-based herbicide resistance mechanism in Lolium rigidum. Heredity 106, 817–824.
Busi, R., Yu, Q., Barrett-Lennard, R., and Powles, S. (2008). Long distance pollen-mediated flow of herbicide resistance genes in Lolium rigidum. Theor. Appl. Genet. 117, 1281–1290. doi: 10.1007/s00122-008-0862-8
Christopher, J. T., Powles, S. B., and Holtum, J. A. M. (1992). Resistance to acetolactate synthase-inhibiting herbicides in annual ryegrass (Lolium rigidum) involves at least two mechanisms. Plant Physiol. 100, 1909–1913. doi: 10.1104/pp.100.4.1909
Cocker, K. M., Northcroft, D. S., Coleman, J. O. D., and Moss, S. R. (2001). Resistance to ACCase-inhibiting herbicides and isoproturon in UK populations of Lolium multiflorum: mechanisms of resistance and implications for control. Pest Manag. Sci. 57, 587–597. doi: 10.1002/ps.330
Collavo, A., Strek, H., Beffa, R., and Sattin, M. (2013). Management of an ACCase inhibitor resistant Lolium rigidum population based on the use of ALS inhibitors: weed population evolution observed over a seven year field-scale investigation. Pest Manag. Sci. 69, 200–208. doi: 10.1002/ps.3449
Cotterman, J. C., and Saari, L. L. (1992). Rapid metabolic inactivation is the basis for cross-resistance to chlorsulfuron in diclofop-methyl-resistant rigid ryegrass (Lolium rigidum) biotype SR4/84. Pestic. Biochem. Physiol. 43, 182–192. doi: 10.1016/0048-3575(92)90032-u
Dalton, D. A., Boniface, C., Turner, Z., Lindahl, A., Kim, H. J., Jelinek, L., et al. (2009). Physiological roles of glutathione S-transferases in soybean root nodules. Plant Physiol. 150, 521–530. doi: 10.1104/pp.109.136630
Délye, C. (2013). Unravelling the genetic bases of non-target-site-based resistance (NTSR) to herbicides: a major challenge for weed science in the forthcoming decade. Pest Manag. Sci. 69, 176–187. doi: 10.1002/ps.3318
Délye, C., Jasieniuk, M., and Le Corre, V. (2013). Deciphering the evolution of herbicide resistance in weeds. Trends Genet. 29, 649–658. doi: 10.1016/j.tig.2013.06.001
Doyle, J. J., and Doyle, J. L. (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 19, 11–15.
Duhoux, A., Carrére, S., Duhoux, A., and Délye, C. (2017). Transcriptional markers enable identification of rye-grass (Lolium sp.) plants with non-target-site-based resistance to herbicide inhibiting acetolactate-synthase. Plant Sci. 257, 22–36. doi: 10.1016/j.plantsci.2017.01.009
Duhoux, A., and Délye, C. (2013). Reference genes to study herbicide stress response in Lolium sp.: upregulation of P450 genes in plants resistant to acetolactate-synthase inhibitors. PLoS One 8:e63576. doi: 10.1371/journal.pone.0063576
Gadda, G., and Francis, K. (2010). Nitronate monooxygenase, a model for anionic flavin semiquinone intermediates in oxidative catalysis. Arch. Biochem. Biophys. 493, 53–61. doi: 10.1016/j.abb.2009.06.018
Gaines, T. A., Duke, S. O., Morran, S., Rigon, C. A. G., Tranel, P. J., Küpper, A., et al. (2020). Mechanisms of evolved herbicide resistance. J. Biol. Chem. 295, 10307–10330. doi: 10.1074/jbc.REV120.013572
Gaines, T. A., Lorentz, L., Figge, A., Herrmann, J., Maiwald, F., Ott, M. C., et al. (2014). RNA−Seq transcriptome analysis to identify genes involved in metabolism−based diclofop resistance in Lolium rigidum. Plant J. 78, 865–876. doi: 10.1111/tpj.12514
Han, H., Yu, Q., Owen, M. J., Cawthray, G. R., and Powles, S. (2016). Widespread occurrence of both metabolic and target-site herbicide resistance mechanisms in Lolium rigidum populations. Pest Manag. Sci. 72, 255–263. doi: 10.1002/ps.3995
Heap, I. M. (2020). The International Survey of Herbicide Resistant Weeds. Available online at: http://weedscience.org/ (accessed August 24, 2020).
Hess, M., Barralis, G., Bleiholder, H., Buhr, L., Eggers, T. H., Hack, H., et al. (1997). Use of the extended BBCH scale-general for the descriptions of the growth stages of mono and dicotyledonous weed species. Weed Res. 37, 433–441. doi: 10.1046/j.1365-3180.1997.d01-70.x
Holt, J. S., Welles, S. R., Silvera, K., Heap, I. M., Heredia, S. M., Martinez-Berdeja, A., et al. (2013). Taxonomic and life history bias in herbicide resistant weeds: implications for deployment of resistant crops. PLoS One 8:e71916. doi: 10.1371/journal.pone.0071916
Iwakami, S., Endo, M., Saika, H., Okuno, J., Nakamura, N., Yokoyama, M., et al. (2014). Cytochrome P450 CYP81A12 and CYP81A21 are associated with resistance to two acetolactate synthase inhibitors in Echinochloa phyllopogon. Plant Physiol. 165, 618–629. doi: 10.1104/pp.113.232843
Iwakami, S., Kamidate, Y., Yamaguchi, T., Ishizaka, M., Endo, M., Suda, H., et al. (2019). CYP81A P450s are involved in concomitant cross-resistance to acetolactate synthase and acetyl-CoA carboxylase herbicides in Echinochloa phyllopogon. New Phytol. 221, 2112–2122. doi: 10.1111/nph.15552
Kaloumenos, N. S., Tsioni, V. C., Daliani, E. G., Papavassileiou, S. E., Vassileiou, A. G., Laoutidou, P. N., et al. (2012). Multiple Pro-197 substitutions in the acetolactate synthase of rigid ryegrass (Lolium rigidum) and their impact on chlorsulfuron activity and plant growth. Crop Prot. 38, 35–43. doi: 10.1016/j.cropro.2012.03.002
Kaundun, S. S. (2014). Resistance to acetyl-CoA carboxylase-inhibiting herbicides. Pest Manag. Sci. 70, 1405–1417. doi: 10.1002/ps.3790
Kotoula-Syka, E., Tal, A., and Rubin, B. (2000). Diclofop-resistant Lolium rigidum from northern Greece with cross-resistance to ACCase inhibitors and multiple resistance to chlorsulfuron. Pest Manag. Sci. 56, 1054–1058. doi: 10.1002/1526-4998(200012)56:12<1054::AID-PS267<3.0.CO;2-M
Llewellyn, R. S., and Powles, S. B. (2001). High levels of herbicide resistance in rigid ryegrass (Lolium rigidum) in the wheat belt of Western Australia. Weed Technol. 15, 242–248. doi: 10.1614/0890-037X2001015[0242:HLOHRI]2.0.CO;2
Mahmood, K., Mathiassen, S. M., Kristensen, M., and Kudsk, P. (2016). Multiple herbicide resistance in Lolium multiflorum and identification of conserved regulatory elements of herbicide resistance genes. Front. Plant Sci. 7:1160. doi: 10.3389/fpls.2016.01160
Manalil, S., Busi, R., and Renton, M. (2011). Rapid evolution of herbicide resistance by low herbicide dosages. Weed Sci. 59, 210–217. doi: 10.1614/WS-D-10-00111.1
Mathiassen, S. K. (2014). “Herbicide resistance mapping in the EU Northern Zone,” in Herbicide Resistance in Europe: Challenges, Opportunities and Threats, Frankfurt.
Neve, P., and Powles, S. B. (2005). Recurrent selection with reduced herbicide rates results in the rapid evolution of herbicide resistance in Lolium rigidum. Theor. Appl. Genet. 110, 1154–1166. doi: 10.1007/s00122-005-1947-2
Onofri, A. (2005). Bioassay97: a new excel® VBA macro to perform statistical analyses on herbicide dose-response data. Riv. Ital. Agrometereol. 3, 40–45.
Owen, M. J., Martinez, N. J., and Powles, S. B. (2014). Multipe herbicide-resistant Lolium rigidum (annual ryegrass) now dominates across the Western Autralian grain belt. Weed Res. 54, 314–324. doi: 10.1111/wre.12068
Panozzo, S., and Scarabel, L. (2020). ACCase and ALS Gene Sequences of Multi-Resistant Lolium spp. from Three European Countries. Mendeley Data, V1. London: Mendeley. doi: 10.17632/53zzypkcvf.1
Pedersen, B. P., Neve, P., Andreasen, C., and Powles, S. B. (2007). Ecological fitness of a glyphosate –resistant Lolium rigidum population: growth and seed production along a competition gradient. Basic Appl. Ecol. 8, 258–268. doi: 10.1016/j.baae.2006.01.002
Petit, C., Duhieu, B., Boucansaud, K., and Délye, C. (2010). Complex genetic control of non-target-site based resistance to herbicides inhibiting acetyl-coenzyme A carboxylase and acetolactate synthase in Alopecurus myosuroides Huds. Plant Sci. 178, 501–509. doi: 10.1016/j.plantsci.2010.03.007
Pfaffl, M. W. (2006). “Relative quantification,” in Real-time PCR (BIOS Advanced Methods), ed. M. T. Dorak (Milton Park: Taylor & Francis Group), 63–82.
Powles, S. B., and Yu, Q. (2010). Evolution in action: plants resistant to herbicides. Annu. Rev. Plant Biol. 61, 317–347. doi: 10.1146/annurev-arplant-042809-112119
Preston, C., Tardif, F. J., Christopher, J. T., and Powles, S. B. (1996). Multiple resistance to dissimilar herbicide chemistries in a biotype of Lolium rigidum due to enhanced activity of several herbicide degrading enzymes. Pestic. Biochem. Phys. 54, 123–134. doi: 10.1006/pest.1996.0016
Salas-Perez, R. A., Saski, C. A., Noorai, R. E., Srivastava, S. K., Lawton-Rauh, A. L., Nichols, R. L., et al. (2018). RNA-Seq transcriptome analysis of Amaranthus palmeri with differential tolerance to glufosinate herbicide. PLoS One 13:e0195488. doi: 10.1371/journal.pone.0195488
Scarabel, L., Panozzo, S., Varotto, S., and Sattin, M. (2011). Allelic variation of the ACCase gene and response to ACCase-inhibiting herbicides in pinoxaden-resistant Lolium spp. Pest Manag. Sci. 67, 932–941. doi: 10.1002/ps.2133
Seefeldt, S. S., Jensen, J. E., and Fuerst, E. P. (1995). Log-logistic analysis of herbicide dose-response relationships. Weed Technol. 9, 218–227. doi: 10.1017/S0890037X00023253
Tan, M. K., Preston, C., and Wang, G. X. (2007). Molecular basis of multiple resistance to ACCase-inhibiting and ALS-inhibiting herbicides in Lolium rigidum. Weed Res. 47, 534–541. doi: 10.1111/j.1365-3180.2007.00591.x
Tardif, F. J., and Powles, S. B. (1994). Herbicide multiple-resistance in a Lolium rigidum biotype is endowed by multiple mechanisms- isolation of a subset with resistant acetyl-CoA carboxylase. Physiol. Plant. 91, 488–494. doi: 10.1111/j.1399-3054.1994.tb02978.x
Werck-Reichhart, D., and Feyereisen, R. (2000). Cytochromes P450: a success story. Genome Biol. 1:REVIEWS3003. doi: 10.1186/gb-2000-1-6-reviews3003
Yu, Q., Collavo, A., Zheng, M.-Q., Owen, M., Sattin, M., and Powles, S. B. (2007). Diversity of acetyl-coenzyme a carboxylase mutations in resistant Lolium populations: evaluation using clethodim. Plant Physiol. 145, 547–558. doi: 10.1104/pp.107.105262
Yu, Q., Han, H., and Powles, S. B. (2008). Mutations of the ALS gene endowing resistance to ALS-inhibiting herbicides in Lolium rigidum populations. Pest Manag. Sci. 64, 1229–1236. doi: 10.1002/ps.1624
Yu, Q., and Powles, S. B. (2014). Resistance to AHAS inhibitor herbicides: current understanding. Pest Manag. Sci. 70, 1340–1350. doi: 10.1002/ps.3710
Yuan, J. S., Tranel, P. J., and Stewart, C. N. (2007). Non-target-site herbicide resistance: a family business. Trends Plant Sci. 12, 6–13. doi: 10.1016/j.tplants.2006.11.001
Keywords: ryegrass, target-site resistance, enhanced gene expression, metabolism, multiple herbicide resistance
Citation: Scarabel L, Panozzo S, Loddo D, Mathiassen SK, Kristensen M, Kudsk P, Gitsopoulos T, Travlos I, Tani E, Chachalis D and Sattin M (2020) Diversified Resistance Mechanisms in Multi-Resistant Lolium spp. in Three European Countries. Front. Plant Sci. 11:608845. doi: 10.3389/fpls.2020.608845
Received: 21 September 2020; Accepted: 20 November 2020;
Published: 15 December 2020.
Edited by:
Joel Torra, Universitat de Lleida, SpainReviewed by:
Ricardo Alcántara-de la Cruz, Federal University of São Carlos, BrazilCopyright © 2020 Scarabel, Panozzo, Loddo, Mathiassen, Kristensen, Kudsk, Gitsopoulos, Travlos, Tani, Chachalis and Sattin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Laura Scarabel, bGF1cmEuc2NhcmFiZWxAY25yLml0
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.